The prevalence of multidrug-resistant Pseudomonas aeruginosa has stimulated development of alternative therapeutics. Bacteriophage peptidoglycan hydrolases, termed lysins, represent an emerging antimicrobial option for targeting Gram-positive bacteria.
KEYWORDS: ESKAPE, Pseudomonas aeruginosa, antibiotic resistance, antimicrobial, endolysin, lysin, lysocin, peptidoglycan hydrolase, protein delivery
ABSTRACT
The prevalence of multidrug-resistant Pseudomonas aeruginosa has stimulated development of alternative therapeutics. Bacteriophage peptidoglycan hydrolases, termed lysins, represent an emerging antimicrobial option for targeting Gram-positive bacteria. However, lysins against Gram-negatives are generally deterred by the outer membrane and their inability to work in serum. One solution involves exploiting evolved delivery systems used by colicin-like bacteriocins (e.g., S-type pyocins of P. aeruginosa) to translocate through the outer membrane. Following surface receptor binding, colicin-like bacteriocins form Tol- or TonB-dependent translocons to actively import bactericidal domains through outer membrane protein channels. With this understanding, we developed lysocins, which are bioengineered lysin-bacteriocin fusion molecules capable of periplasmic import. In our proof-of-concept studies, components from the P. aeruginosa bacteriocin pyocin S2 (PyS2) responsible for surface receptor binding and outer membrane translocation were fused to the GN4 lysin to generate the PyS2-GN4 lysocin. PyS2-GN4 delivered the GN4 lysin to the periplasm to induce peptidoglycan cleavage and log-fold killing of P. aeruginosa with minimal endotoxin release. While displaying narrow-spectrum antipseudomonal activity in human serum, PyS2-GN4 also efficiently disrupted biofilms, outperformed standard-of-care antibiotics, exhibited no cytotoxicity toward eukaryotic cells, and protected mice from P. aeruginosa challenge in a bacteremia model. In addition to targeting P. aeruginosa, lysocins can be constructed to target other prominent Gram-negative bacterial pathogens.
INTRODUCTION
Antimicrobial resistance is a threat to global public health. One of the predominant antibiotic-resistant microorganisms responsible for high mortality rates is Pseudomonas aeruginosa. This Gram-negative pathogen is (i) the leading cause of mortality in cystic fibrosis patients, (ii) the main causative agent of burn wound infections, (iii) the most frequent Gram-negative bacterium associated with nosocomial and ventilator-acquired pneumonia, and (iv) the second most common cause of catheter-associated urinary tract infections (1). Additionally, P. aeruginosa is responsible for 3 to 7% of all bloodstream infections (BSIs) and 23 to 26% of Gram-negative bacteremias, translating to mortality rates ranging from 27 to 48% (2). With the antipseudomonal efficacy of standard-of-care (SOC) antibiotics progressively diminishing due to a combination of intrinsic, acquired, and adaptive resistance mechanisms utilized by the bacteria, the lack of therapeutic options has stimulated the World Health Organization to label P. aeruginosa as a critical priority for the research, discovery, and development of new antibiotics (3, 4).
Bacteriophage (phage)-encoded peptidoglycan (PG) hydrolases, termed lysins, represent an alternative class of antimicrobials to small-molecule antibiotics (5). During the phage replicative cycle, lysins degrade the PG of host bacteria to induce hypotonic lysis and phage progeny liberation. The extrinsic application of purified recombinant lysins has been validated for antibacterial efficacy toward several Gram-positive bacterial pathogens as a result of the PG constituting part of the exposed outer structural component of the cell (6). However, expanding the use of these enzymes to target Gram-negative bacteria has, in many cases, been impeded by the outer membrane (OM). To overcome this, lysins have been modified to permit OM translocation. For example, the peptide component of Artilysins, which contain an OM-permeabilizing peptide fused to a lysin, locally distorts the lipopolysaccharide layer to allow the lysin to penetrate through the OM (7). Unfortunately, like most Gram-negative lysins, Artilysins appear to be inactive in human serum (HuS), limiting their therapeutic applicability to superficial, nonsystemic bacterial infections (8–10). An alternative engineering strategy has been described using bacteriocin-lysin hybrid molecules to actively transport lysins across protein channels embedded in the OM of Gram-negative bacteria (11, 12). For instance, the construction of a lysin-colicin A chimeric molecule yielded a construct (Colicin-Lysep3) capable of traversing the OM of Escherichia coli. Nevertheless, as seen with colicin A, E. coli resistance to Colicin-Lysep3 can seemingly develop by mutating BtuB (the vitamin B12 receptor which also acts as the Colicin-Lysep3 receptor) or OmpF (the pore-forming channel used for Colicin-Lysep3 periplasmic import) (13, 14). Notably, E. coli with defective BtuB and OmpF mutations remains virulent (15, 16). There is also no experimental evidence to support Colicin-Lysep3 antibacterial activity in HuS.
Lysins can be engineered to kill P. aeruginosa in HuS by exploiting functional domains from S-type pyocins, which are chromosomally encoded colicin-like bacteriocins produced by P. aeruginosa for intraspecies competition (17). These SOS-inducible, high-molecular-weight proteinaceous toxins are evolutionarily conserved among many Gram-negative bacteria, including Enterobacter cloacae, E. coli, Klebsiella pneumoniae, and Yersinia pestis (18). In general, S-type pyocins bind a specific receptor on the surface of target Pseudomonas bacteria. By using the Tol or Ton import system, these bacteriocins deliver enzymatic (lipid II degradation, DNase, rRNase, or tRNase) or nonenzymatic (inner membrane [IM] pore formation) bactericidal domains to intracellular targets by active translocation across the OM through the channel created by the receptor or another adjacent OM protein.
One example of an S-type pyocin produced by P. aeruginosa is pyocin S2 (PyS2). PyS2 consists of (i) an N-terminal receptor-binding domain, (ii) an α-helical domain, (iii) a domain with homology to colicins of E. coli, and (iv) a C-terminal DNase domain (1, 19, 20) (Fig. 1A). Once secreted by the producing cell, the PyS2 domain I binds the ferripyoverdine receptor FpvAI, which is a gated TonB-dependent transporter naturally upregulated in iron-depleted environments to actively import the small siderophore ferripyoverdine. This interaction allows a short segment of disordered amino acids in FpvAI, known as the TonB box (TBB), to bind TonB1 in the periplasm; TonB1, ExbB, and ExbD are IM proteins that constitute the Ton import system (19). The labile portion of the FpvAI plug domain is unfolded by the proton motive force (PMF) associated with the Ton system. The unstructured region of PyS2 domain I (amino acids [aa] 1 to 45) passes through the newly created channel within FpvAI to present its own TBB (aa 11 to 15) to another nearby TonB1 protein in the periplasm. Once a PyS2-TonB1 translocon forms, the remainder of the bacteriocin undergoes PMF-dependent unfolding and OM translocation.
FIG 1.
GN4 and PyS2-GN4 purification and antipseudomonal activity. (A) To construct the PyS2-GN4 lysocin, PyS2 domain IV (aa 559 to 690) was deleted and replaced with the GN4 lysin (aa 1 to 144). (B) The GN4 lysin (16 kDa) and the wild-type PyS2-GN4 (76 kDa), PyS2-GN4ΔTBB (75 kDa), and PyS2-GN4KO (76 kDa) lysocins were purified to homogeneity according to sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis. (C) The muralytic activity of purified GN4 and PyS2-GN4 was determined by spotting 25 pmol of each on autoclaved Pseudomonas. CEWL and buffer were, respectively, used as positive and negative controls. Clearing zones signify PG degradation. (D to G) Using the plate lysis assay, the antipseudomonal activities of GN4, PyS2-GN4, PyS2-GN4ΔTBB, and PyS2-GN4KO, as indicated, were determined in growth medium by spotting 0.01 to 400 pmol of purified protein on P. aeruginosa strain 453. (H) The plate lysis assay was further used to analyze the antipseudomonal activity of 0.01 to 400 pmol of PyS2-GN4 against P. aeruginosa strain 453 in 50% HuS. Growth inhibition zones observed using the plate lysis assay indicate antipseudomonal activity.
The molecular basis for PyS2 bactericidal activity has been extrapolated from an understanding of how enzymatic colicins of E. coli accomplish this feat (18). Following OM translocation, periplasmic chaperones refold PyS2. An unknown IM protease liberates domain IV from the remainder of the bacteriocin, allowing the DNase to translocate through an IM protein channel to access its cytosolic DNA substrate. Like all S-type pyocin-producing strains, P. aeruginosa that produces PyS2 also expresses an immunity protein, termed Imm2, that transiently binds to and neutralizes the bactericidal domain to prevent cellular suicide. As a product of being transcribed in the same operon, PyS2 and Imm2 are coexpressed to form an immediate complex (17). After secretion, it is believed that the PyS2-Imm2 complex dissociates in a PMF-dependent manner while translocating across the OM.
By exploiting the molecular characteristics that allow S-type pyocins to traverse the OM, we devised a strategy for delivering lysins to their PG substrate in P. aeruginosa. We describe and experimentally evaluate lysocins, which collectively consist of a lysin modified with S-type pyocin functional domains permitting periplasmic import. As proof of concept, we engineered the PyS2-GN4 lysocin by fusing PyS2 domains I to III to the P. aeruginosa phage PAJU2 lysin GN4. The antibacterial efficacy of the purified lysocin was assessed in vitro and in vivo to elucidate whether a functional lysin can be transported across the OM of Pseudomonas by means of a naturally evolved delivery system.
RESULTS
GN4 and PyS2-GN4 muralytic and antipseudomonal activity.
The amino acid coordinates for the four domains of PyS2 are as follows: domain I, aa 1 to 209; domain II, aa 210 to 312; domain III, aa 313 to 558; and domain IV, aa 559 to 690 (Fig. 1A). The GN4 lysin, a muramidase from phage PAJU2 of P. aeruginosa, consists of a single globular domain (aa 1 to 144). To construct the PyS2-GN4 lysocin, domain IV (the DNase domain) was deleted from PyS2 and replaced with the GN4 lysin (Fig. 1A).
To confirm that the native GN4 lysin is capable of cleaving pseudomonal PG, purified GN4 (Fig. 1B) was spotted on autoclaved (causing OM disruption) P. aeruginosa, along with chicken egg white lysozyme (CEWL) as a control. Clearing zones corresponding to muralytic activity of GN4 (and CEWL) confirmed that the lysin can cleave pseudomonal PG (Fig. 1C). Additionally, the purified PyS2-GN4 construct (Fig. 1B) degraded Pseudomonas PG, suggesting that GN4 retains its enzymatic activity as a fusion protein (Fig. 1C).
Next, the antipseudomonal activity of GN4 and PyS2-GN4 was investigated in vitro against viable P. aeruginosa. Spotting purified GN4 and PyS2-GN4 on P. aeruginosa lawns revealed that GN4 alone was ineffective (Fig. 1D), whereas PyS2-GN4 displayed antipseudomonal activity when ≥0.64 pmol was applied (Fig. 1E). Removing the TBB (aa 11 to 15) from PyS2-GN4 inhibited antipseudomonal activity by preventing OM translocation (Fig. 1F). This experimental evidence, that a functional lysin can be delivered to the PG of live P. aeruginosa through S-type pyocin fusion, validates the lysocin antimicrobial approach.
The GN4 lysin has a putative active site consisting of a Glu-(8 aa)-Asp-(5 aa)-Thr catalytic triad motif conserved in other lysins that function as glycosylases (21). For PyS2-GN4, these residues are E573, D582, and T588. The purified active-site knockout (KO) mutant PyS2-GN4 with the substitutions E573A, D582A, and T588A (PyS2-GN4E573A,D582A,T588A or PyS2-GN4KO) (Fig. 1B) was incapable of generating distinct growth inhibition zones when applied to P. aeruginosa, indicating that PyS2-GN4 antipseudomonal activity is predicated on the muralytic activity of the GN4 lysin (Fig. 1G).
As a first step in evaluating the in vivo therapeutic applicability of lysocins for P. aeruginosa BSIs, the antibacterial properties of PyS2-GN4 were analyzed in HuS. Activity could be observed when ≥0.13 pmol of lysocin was spotted on P. aeruginosa in 50% HuS (Fig. 1H). Compared to growth medium only (Fig. 1E), the increased clarity and diameter of the growth inhibition zones produced by PyS2-GN4 in HuS (Fig. 1H) indicates that the antibacterial effect was amplified. This finding could be due to lower free iron availability in serum, which upregulates the FpvAI receptor.
Lysocin killing kinetics and antibiofilm activity.
The bactericidal activity of PyS2-GN4 was initially assayed in iron-deficient Casamino Acids (CAA) medium as a function of antimicrobial concentration and time. The medium was supplemented with the iron chelator ethylenediamine hydroxyphenylacetic acid (EDDHA) to simulate free-iron deprivation in human blood. During the 12 h of incubation, 0.1 to 100 μg/ml lysocin killed P. aeruginosa bacteria at similar rates, resulting in nearly 2-, 3-, and 4-log reductions in bacterial viability at 2, 4, and 12 h, respectively (Fig. 2A). The killing kinetics of PyS2-GN4 were considerably reduced when it was diluted below 0.1 μg/ml. Lysocin concentrations of ≥10 μg/ml (132 nM) sterilized the bacterial culture when incubated in CAA medium and 50% HuS for 24 h (Fig. 2B). In terms of thermal stability, PyS2-GN4 fully retained bactericidal activity following short-term incubation at temperatures of ≤45°C (see Fig. S1 in the supplemental material). These collective experimental findings indicate that PyS2-GN4 is bactericidal at ≥0.1 μg/ml after 4 h, capable of sterilizing high concentrations of Pseudomonas at nanomolar concentrations in the absence and presence of HuS, and relatively thermostable.
FIG 2.
PyS2-GN4 killing kinetics and antibiofilm efficacy. The dose-response lysocin killing kinetics were determined by incubating P. aeruginosa strain 453 at 106 CFU/ml statically with 0.01 to 100 μg/ml PyS2-GN4 for 24 h at 37°C. Bacterial viability was assessed (A) in 2-h increments over the first 12 h in growth medium only and (B) at 24 h with or without HuS. (C) P. aeruginosa strain PAO1 biofilms were grown for 72 h at 37°C in CAAg medium and subsequently treated for 24 h with buffer or 0.03 to 500 μg/ml GN4, PyS2-GN4, or tobramycin. The residual biomass of the biofilms was qualitatively measured by means of crystal violet staining. SC, sterility control; GC, growth control. All error bars correspond to ±SEM, while dashed lines mark the limits of detection.
With antibacterial efficacy established in vitro against planktonic P. aeruginosa, the effect of PyS2-GN4 on biofilms was measured. Using a 24-well polystyrene plate, P. aeruginosa biofilms were established for 72 h in CAA medium with 0.2% (wt/vol) glucose (CAAg medium) and subsequently treated with GN4, PyS2-GN4, or tobramycin for a total of 24 h (Fig. 2C). Residual biofilm biomass was qualitatively assessed by staining with crystal violet. Similar to results with planktonic bacteria, the GN4 lysin alone was ineffective against the Pseudomonas biofilm. Alternatively, PyS2-GN4 and tobramycin disrupted biofilm biomass at concentrations of ≥0.16 μg/ml. More residual crystal violet was observed in tobramycin wells treated with 0.16 to 500 μg/ml than in those with PyS2-GN4 treatment, suggesting that the lysocin was more efficient at degrading biofilms (Fig. 2C).
Antibacterial activity range.
The lysocin antibacterial activity range was determined against a collection of P. aeruginosa strains and nonpseudomonal bacteria. Of the 11 P. aeruginosa strains tested, 4 were lysocin sensitive (Table 1). PyS2-GN4 displayed MIC values of ≤4 μg/ml toward the P. aeruginosa reference strain PAO1 and the 452, 453, and MDR-M-3 clinical isolates. As expected, multiplex PCR confirmed that these sensitive strains carry the fpvAI gene chromosomally while the remaining strains harbor fpvAII or fpvAIII. Natural resistance to PyS2-GN4 by strains lacking the FpvAI receptor is further evidence that lysocin activity is mediated through active transport to the periplasm of sensitive strains. None of the nonpseudomonal bacterial species were lysocin sensitive (Table S1).
TABLE 1.
Antimicrobial MIC and MBC values for numerous P. aeruginosa strains
| Antimicrobial | P. aeruginosa strain | FpvA type | MIC (μg/ml) | MBC (μg/ml) |
|---|---|---|---|---|
| PyS2-GN4 | PAO1 | I | 2 | |
| MDR-M-3 | I | 2 | ||
| 443 | III | >256 | ||
| 445 | II | >256 | ||
| 446 | II | >256 | ||
| 448 | II | >256 | ||
| 449 | II | >256 | ||
| 450 | III | >256 | ||
| 451 | II | >256 | ||
| 452 | I | 4 | ||
| 453 | I | 0.25 | 0.25 | |
| Colistin | 453 | I | 0.5 | 0.5 |
| Meropenem | 453 | I | 8 | 8 |
| Piperacillin-tazobactam | 453 | I | 16 | 128 |
| Tobramycin | 453 | I | 0.125 | 0.25 |
Benchmarking lysocin against SOC antibiotics.
PyS2-GN4 was benchmarked against four SOC antibiotics used clinically for P. aeruginosa BSIs. Using P. aeruginosa strain 453, the MIC and minimum bactericidal concentration (MBC) for PyS2-GN4 were obtained and compared to those of colistin, meropenem, piperacillin-tazobactam, and tobramycin (Table 1). The respective MIC values for PyS2-GN4, colistin, meropenem, piperacillin-tazobactam, and tobramycin were 0.25, 0.5, 8, 16, and 0.125 μg/ml. The MBC values for PyS2-GN4, colistin, meropenem, piperacillin-tazobactam, and tobramycin were, respectively, 0.25, 0.5, 8, 128, and 0.25 μg/ml.
Visualizing the mechanism of PyS2-GN4 antipseudomonal activity.
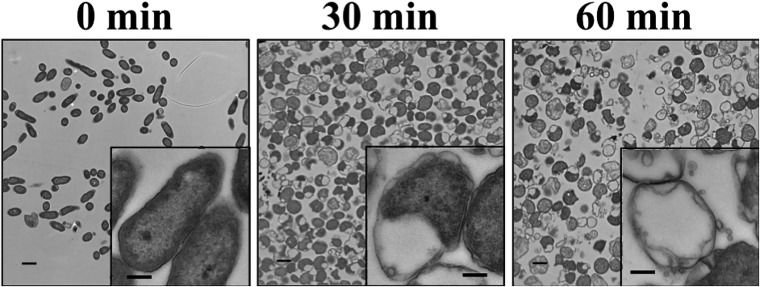
P. aeruginosa bacteria treated with lysocin were visualized by transmission electron microscopy (TEM) to better understand the mechanism of PyS2-GN4 antipseudomonal activity (Fig. 3). Untreated bacteria were rod shaped with uniform intracellular density. Conversely, P. aeruginosa bacteria at 30 min post-lysocin treatment transitioned from rod shaped to a more spherical morphology. This phenotype is indicative of bacteria with a defective cell wall, visual evidence of GN4 muralytic activity. Furthermore, as a result of the cleavage of the PG, the integrity of the OM and IM appears to be partially compromised through hypotonic pressure, resulting in cytoplasmic leakage and PMF disruption. At 60 min post-lysocin treatment, a significant portion of the bacterial population consisted of intact, nonviable cells either lacking or with noticeably reduced cytoplasmic content.
FIG 3.
Visualizing lysocin-treated P. aeruginosa by TEM. P. aeruginosa strain 453 was incubated with 50 μg/ml PyS2-GN4 in CAA medium with EDDHA for a total of 1 h at 37°C. The bacteria were then fixed and visualized by TEM at 0, 30, and 60 min posttreatment. Total magnifications of ×2,600 (scale bar, 1 μm) and ×13,000 (insets; scale bar, 200 nm) are shown.
Lysocin cytotoxicity.
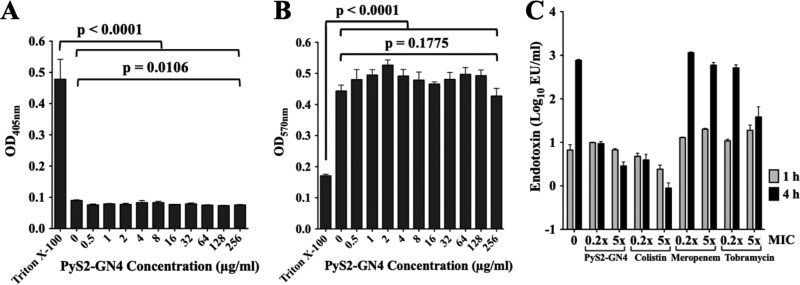
Lysocin cytotoxicity was initially measured using two different eukaryotic cell types. Human red blood cells (hRBCs) (Fig. 4A) and human promyeloblast HL-60 cells (Fig. 4B) were incubated with 0.5 to 256 μg/ml PyS2-GN4 for 8 h. In contrast to the Triton X-100 controls, no cytotoxicity was observed in the presence of lysocin. Next, endotoxin release was measured in growth medium after P. aeruginosa was treated with lysocin or SOC antibiotics (Fig. 4C). Compared to the level at the 1-h time point, the increase in endotoxin detected for the untreated control at 4 h may be attributed to cell division events (22). Endotoxin release stimulated by PyS2-GN4 and colistin, which has potent anti-endotoxin activity, was approximately 100- to 1,000-fold less than that with meropenem and tobramycin after 4 h of treatment (23). Colistin binds and neutralizes liberated endotoxin; in contrast, the small amount of endotoxin detected in the lysocin-treated samples relates to the ability of PyS2-GN4 to kill Pseudomonas with minimal disruption of the OM (Fig. 3), allowing endotoxin to remain anchored to the bacterial surface.
FIG 4.
Lysocin cytotoxicity toward eukaryotic cells and bacterial endotoxin release. (A) hRBCs were incubated in triplicate with buffer or 0.5 to 256 μg/ml PyS2-GN4 for 8 h at 37°C, and hemolysis, as a function of hemoglobin release, was assayed spectrophotometrically at 405 nm. Triton X-100 was used as a positive control for hemolysis. (B) Human promyeloblast HL-60 cells were incubated in triplicate with buffer or 0.5 to 256 μg/ml PyS2-GN4 for 8 h at 37°C, and viability, as a function of formazan product formation, was measured spectrophotometrically at 570 nm. Triton X-100 was used as a control for cytotoxicity. (C) Endotoxin release was measured in duplicate experiments after treating P. aeruginosa strain 453 at 106 CFU/ml for 1 or 4 h at 37°C in growth medium with 0.2× and 5× MIC of PyS2-GN4, colistin, meropenem, or tobramycin. An untreated control was included. All error bars correspond to ±SEM. P values were calculated using a one-way analysis of variance. EU, endotoxin units.
In vivo antipseudomonal efficacy using a murine model of bacteremia.
The in vivo antipseudomonal efficacy of PyS2-GN4 was examined using a murine model of bacteremia. Mice were injected intraperitoneally (i.p.) with P. aeruginosa strain 453 and then treated i.p. at 3 h postinfection with various doses of lysocin; survival was monitored for 10 days. At 3 h postinfection, mice were bacteremic, with bacterial concentrations in the heart, spleen, liver, and kidney ranging from ∼104 to 106 CFU/ml (Fig. 5A). In this model, only 37% of buffer-treated control animals survived the duration of the experiment (Fig. 5B). Alternatively, when mice were treated with 2.5, 5, 12.5, and 25 mg/kg lysocin, 73%, 80%, 93%, and 100%, respectively, were protected from death. Organs of surviving lysocin-treated mice did not contain detectable Pseudomonas bacteria at day 10 (data not shown).
FIG 5.
Lysocin antipseudomonal in vivo efficacy using a murine model of bacteremia. Mice (n =100) were i.p. infected with 108 CFU of P. aeruginosa strain 453. (A) The bacterial counts in organs were determined 3 h postinfection in order to confirm the animals were bacteremic. (B) Infected mice were i.p. treated at 3 h postinfection with either buffer (n = 35) or 2.5 (n = 15), 5 (n = 15), 12.5 (n = 15), or 25 mg/kg (n = 20) lysocin. Survival was monitored over 10 days. All error bars correspond to ±SEM. P values were calculated using a log rank (Mantel-Cox) test.
DISCUSSION
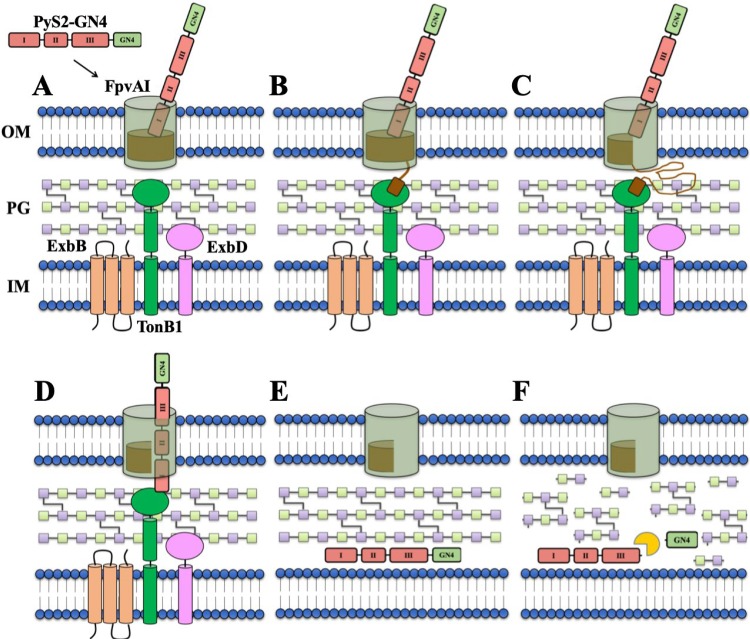
Here, we describe and experimentally validate in vitro and in vivo the use of lysocins, which represent a class of bioengineered antimicrobials that deliver phage lysins to their PG substrate in Gram-negative bacteria. More explicitly, the P. aeruginosa-specific PyS2-GN4 lysocin was designed by fusing PyS2 domains I to III to the GN4 lysin. With a general understanding of the molecular characteristics associated with colicin-like bacteriocins and lysins, a speculative model describing the mechanism of PyS2-GN4 antipseudomonal activity can be formulated from the collective experimental results presented in this study (Fig. 6). First, the lysocin targets P. aeruginosa due to domain I binding with high specificity to FpvAI (Fig. 6A). This interaction induces a conformational change in the receptor structure, allowing the FpvAI TBB to interact with TonB1 in the periplasm (Fig. 6B). The formation of this complex causes the PMF-driven unfolding (and opening) of the labile half of the FpvAI plug domain (Fig. 6C). The N-terminal unstructured region of the lysocin passes through the newly created opening to allow its own TBB to form a translocon with TonB1 (Fig. 6D). The PMF energizes the remainder of PyS2-GN4 to unfold and translocate into the periplasm, where the protein subsequently refolds (Fig. 6E). Finally, the GN4 lysin is putatively proteolytically liberated (experiments to prove this are ongoing) and cleaves the PG through hydrolysis of the β-1,4 glycosidic bond between N-acetylmuramic acid and N-acetylglucosamine (Fig. 6F). Due to cytoplasmic pressure, loss of PG structural integrity rapidly promotes membrane destabilization, cytoplasmic leakage, and PMF disruption, thereby killing the bacterial cell.
FIG 6.
Schematic overview of the proposed mechanism of PyS2-GN4 antipseudomonal activity. (A) When added extrinsically as a purified recombinant protein, domain I of PyS2-GN4 binds to the FpvAI receptor located on the surface of target P. aeruginosa bacteria. (B) This protein-protein interaction induces a conformational change in the receptor structure, resulting in the FpvAI TBB in the periplasm to recruit and bind TonB1. (C) The formation of this complex allows for the PMF-dependent unfolding of the labile half of the FpvAI plug domain. (D) Next, the unstructured region of lysocin domain I passes through the channel created in order to enable its own TBB to bind another nearby TonB1 protein in the periplasm. (E) The newly formed lysocin-TonB1 translocon stimulates the PMF-driven unfolding and import of the remainder of the lysocin into the periplasm. (F) Upon refolding, GN4 is proteolytically released and cleaves the PG to provoke partial membrane destabilization, cytoplasmic leakage, PMF disruption, and cell death.
The receptor bound by domain I of the pyocin is limited to certain bacterial species and strains. The lysocin approach is therefore narrow spectrum, with minimal effects on the normal microflora. This is supported in vitro by the inability of PyS2-GN4 to kill P. aeruginosa strains lacking the FpvAI receptor (Table 1) and nonpseudomonal bacterial species (see Table S1 in the supplemental material), significantly reducing the possibility of antibacterial activity on bystander commensal microorganisms in vivo.
Like PyS2-GN4, several potential pyocin-related lysocins appear to bind and translocate through receptors involved in ferrisiderophore import. Expression of these receptors is inversely regulated by free-iron availability. Considering that the free-iron concentration in HuS is ∼10−24 M, Pseudomonas in the bloodstream would be highly susceptible to lysocins due to receptor upregulation stimulated by free-iron depletion, as exemplified in Fig. 1H, 2B, and 5B (24). This understanding highlights the therapeutic potential of lysocins as narrow-spectrum antimicrobials for P. aeruginosa.
In addition to their potency toward planktonic bacteria, lysocins can potentially be used to break down pseudomonal biofilms. Mucoid P. aeruginosa biofilms are a major cause of morbidity and mortality in cystic fibrosis patients because of their ability to promote chronic lung infections (25). The physical barrier of biofilms permits constituent bacterial cells to resist immune cell opsonization and phagocytosis while also increasing tolerance to toxic oxygen radicals and antibiotics (26). Our proof-of-concept studies provide evidence that lysocins can possibly be used as effective antibiofilm agents (Fig. 2C).
Antibiotic-mediated endotoxin release during treatment of Pseudomonas bacteremia can have immediate adverse effects on patient morbidity. Once released, the endotoxin lipid A moiety stimulates immune cells to secrete proinflammatory cytokines, promoting endothelial damage and severe hemodynamic and metabolic disorders (27). As depicted in Fig. 4C, lysocin-treated P. aeruginosa released ∼100-fold less endotoxin after 4 h than the SOC antibiotics meropenem and tobramycin. In addition to translocating into the periplasm without perturbing the OM, lysocins employ an antibacterial mechanism that kills bacteria while simultaneously preventing destructive cell lysis, keeping endotoxin anchored to the intact OM of the nonviable cells (Fig. 3 and 6).
PyS2 functional domains were exploited for these initial proof-of-concept experiments as this bacteriocin was one of the first S-type pyocins discovered and, consequently, has been extensively characterized (19, 20, 28–34). These PyS2-related studies provided insights for designing an effective antipseudomonal lysocin. With the lysocin methodology now validated, explicit properties can be strategically engineered to improve downstream therapeutic applicability and potential. For example, PyS2-GN4 has strain specificity conferred by domain I, which binds FpvAI. The three P. aeruginosa FpvA receptor types are FpvAI, FpvAII, and FpvAIII, and each is equally distributed among clinical isolate populations, suggesting that one-third of clinically relevant P. aeruginosa strains will be sensitive to PyS2-GN4 (35). While this indicates that PyS2-GN4 alone may not be optimally effective, different strategies can be used to broaden strain coverage. Lysocins can be constructed with pyocin receptor-binding domains that recognize more conserved receptors. For instance, the receptor-binding domain of pyocin S5 binds the highly conserved ferripyochelin FptA receptor and demonstrates species-specific bactericidal activity, with the exception of strains naturally expressing this pyocin and its immunity protein (36). Strain coverage can also be expanded by formulating lysocin cocktails that bind all three FpvA receptors or, as exemplified previously, by fusing multiple unique receptor-binding domains together (37); such studies are currently in progress.
Natural resistance has hindered development of S-type pyocins clinically as antimicrobial agents. P. aeruginosa is genetically programmed to express an immunity protein that renders the bacterium insusceptible to the bactericidal effects of any chromosomally encoded S-type pyocins, whether produced inherently or by neighboring Pseudomonas bacteria. This is circumvented when lysocins are constructed since the C-terminal bactericidal domain of the pyocin targeted by the immunity protein is replaced with a lysin. The binding specificity of immunity proteins prevents recognition and neutralization of the lysin component of lysocins. Consequently, P. aeruginosa intrinsically resistant to the parental pyocin will be vulnerable to the lysocin.
An attempt was made to investigate the ability of P. aeruginosa to develop resistance to PyS2-GN4 using serial passage experiments under iron-depleted growth conditions. The experimental design consisted of initially determining the MIC (see Materials and Methods) of the lysocin in iron-chelated CAA medium. Pseudomonas growing at the highest lysocin concentration was to be used in a subsequent MIC assay; this process was to be repeated at least 15 times. Unfortunately, the bacteria were incapable of growing with or without lysocin under these iron-depleted conditions. Although acquired resistance to native pyocins is often attributed to chromosomal alterations that generate defective or downregulated FpvA receptors, thus inhibiting import of FpvA-dependent lysocins, obstructing the ferripyoverdine import system in Pseudomonas would result in avirulent strains (29, 38–40). Lysocin translocation can alternatively be impeded by modifying components of Tol or Ton import systems. However, inactivating TolA or TolQ of the Tol system was proposed to be lethal in P. aeruginosa, while TonB mutants are avirulent and incapable of growing in iron-depleted environments due to their inability to acquire iron mediated by pyoverdin, pyochelin, and heme uptake (41, 42). A third potential resistance mechanism involves mutating the chemical composition of PG to inhibit lysin muralytic activity. Lysin resistance has not been observed to date, which is attributable to phage coevolving with their bacterial hosts over millions of years. This has resulted in evolving lytic enzymes that cleave conserved and immutable targets in the PG, making resistance formation a very rare event.
In addition to Colicin-Lysep3 and PyS2-GN4, another bacteriocin-lysin hybrid molecule has been described. Pesticin, a colicin-like bacteriocin that targets Y. pestis and uropathogenic E. coli, contains a bactericidal domain that naturally functions as a lysozyme-like muramidase. By replacing this domain with the T4 lysozyme, the resulting hybrid molecule transported the T4 lysozyme to the periplasm of E. coli (12). This outcome was not unexpected as the T4 lysozyme is structurally superimposable and functionally identical to the pesticin muramidase domain. Importantly, a characteristic that differentiates PyS2-GN4 from Colicin-Lysep3 and from the pesticin hybrid molecule is lysocin’s ability to retain bactericidal activity in HuS; experimental evidence to support activity in serum for the other two hybrid molecules is lacking.
In conclusion, we devised and validated a strategy that allows extrinsically applied lysins to overcome the challenge of both bypassing the OM of P. aeruginosa and exhibiting antibacterial activity in serum. More specifically, we successfully bioengineered a highly specific delivery system that transports functional lysins to their PG substrate in HuS, resulting in PG cleavage and bacterial death. While antibacterial efficacy was confirmed against P. aeruginosa in these proof-of-concept studies, lysocins can be developed to target other antibiotic-resistant Gram-negative bacteria, including E. coli, Y. pestis, and the ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) pathogens K. pneumoniae and E. cloacae, thereby fulfilling an urgent global health care need.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains used in this study are outlined in Table S2 in the supplemental material. P. aeruginosa strains numbered 443 to 453 were clinical isolates from the clinical laboratory of Weill Cornell Medical Center (New York, NY). Further details relating to the site of isolation and clinical disease are unavailable. P. aeruginosa strain MDR-M-3, a multidrug resistant clinical isolate originating from a patient with cystic fibrosis, was obtained from Columbia University Medical Center (New York, NY). All Gram-negative strains were routinely grown in Luria-Bertani (LB) or CAA medium (5 g/liter Casamino Acids, 5.2 mM K2HPO4, 1 mM MgSO4). Gram-positive strains were grown in Trypticase soy broth (B. cereus and S. aureus), brain heart infusion broth (E. faecium), or Todd-Hewitt broth with 1% (wt/vol) yeast extract (Streptococcus pyogenes).
Molecular cloning and mutagenesis.
Genes encoding translated GN4 (GenBank accession number YP_002284361) and PyS2 (GenBank accession number NP_249841) were synthesized and codon optimized for protein expression in E. coli (GeneWiz, Inc.). The gn4 and pys2-gn4 genes were cloned into the E. coli expression vector pET28a using an NEBuilder HiFi DNA Assembly Cloning kit (New England Biolabs). pys2 nucleotides 1 to 1674 and gn4 were amplified using PCR. The 50-μl PCR mixture consisted of 1 ng of template DNA, 1× Q5 reaction buffer, 0.2 mM (each) deoxynucleoside triphosphate (dNTP), 0.5 μM each oligonucleotide primer, and 1 U of Q5 DNA polymerase (New England Biolabs). The primers used to amplify gn4 (GN4_F/GN4_R), the pys2 fragment of pys2-gn4 (PyS2_F/PyS2-GN4_R), and the gn4 fragment of pys2-gn4 (PyS2-GN4_F/GN4_R) are listed in Table S3. The thermocycler heating conditions were 98°C for 30 s, 35 cycles of 98°C for 10 s, 60°C for 30 s, and 72°C for 30 s/kb, followed by 72°C for 2 min. Next, a 20-μl reaction mixture consisting of 1× NEBuilder HiFi DNA Assembly Master Mix, gn4 or pys2-gn4 PCR product(s), and NcoI/BamHI-linearized pET28a was incubated at 50°C for 15 min and transformed into E. coli DH5α. Following sequence confirmation, pET28a::gn4 and pET28a::pys2-gn4 were transformed into E. coli BL21(DE3).
PyS2-GN4 with a deletion of the TBB (PyS2-GN4ΔTBB) was created by amplifying pET28a::pys2-gn4 with phosphorylated primers bordering pys2-gn4 nucleotides 33 to 45. PyS2-GN4E573A,D582A,T588A (PyS2-GN4KO) was generated using two sequential site-directed mutagenesis reactions. Each 50-μl PCR mixture consisted of 50 ng of template DNA, 1× Q5 reaction buffer, 0.2 mM each dNTP, 0.5 μM each oligonucleotide primer (Table S3), and 1 U of Q5 DNA polymerase. For the TBB deletion mutant, pET28a::pys2-gn4 was amplified with PyS2-GN4ΔTBB_F/PyS2-GN4ΔTBB_R to create pET28a::pys2-gn4ΔTBB. For the active-site mutant, pET28a::pys2-gn4 was initially amplified with PyS2-GN4_KO_1F/PyS2-GN4_KO_1R to generate pET28a::pys2-gn4E573A,D582A (where gn4E573A,D582A indicates the E573A and D582A substitutions encoded by gn4). Next, pET28a::pys2-gn4E573A,D582A was amplified with PyS2-GN4_KO_2F/PyS2-GN4_KO_2R to obtain pET28a::pys2-gn4KO. The thermocycler heating conditions were 98°C for 30 s, 25 cycles of 98°C for 10 s, 60°C for 30 s, and 72°C for 30 s/kb, followed by 72°C for 2 min. The PCR products were ligated using T4 DNA ligase (New England Biolabs) and transformed into E. coli DH5α. Following sequence confirmation, pET28a::pys2-gn4ΔTBB and pET28a::pys2-gn4KO were transformed into E. coli BL21(DE3).
Protein expression and purification.
Using E. coli BL21(DE3), GN4, PyS2-GN4, PyS2-GN4ΔTBB, and PyS2-GN4KO were expressed for 4 h at 37°C with shaking at 200 rpm in LB medium containing 50 μg/ml kanamycin. Protein expression was induced at mid-log phase (optical density at 600 nm [OD600] of 0.5) with 1 mM isopropyl β-d-1-thiogalactopyranoside. The cells were then harvested, washed, resuspended in 50 mM Tris-HCl, pH 7.5, 200 mM NaCl, and 1 mM phenylmethanesulfonyl fluoride, and lysed using an Emulsiflex-C5 homogenizer (Avestin). The lysate was cleared by centrifugation at 13,000 rpm for 1 h at 4°C. The soluble lysate fraction was dialyzed against 10 mM sodium phosphate, pH 7.0, followed by sterile filtration (0.2-μm pore size) to generate the crude lysate.
The crude lysate was applied to a HiTrap CM FF column (GE Healthcare Life Sciences) in 10 mM sodium phosphate, pH 7.0, using an AKTA fast protein liquid chromatography (FPLC) system (GE Healthcare Life Sciences). Protein was eluted from the column using a linear gradient from 0 to 250 mM NaCl. Elution fractions containing the protein of interest were dialyzed against 50 mM Tris-HCl, pH 7.5, and 200 mM NaCl and concentrated using an Amicon Ultra Ultracel-10K (GN4) or -50K (lysocin) filter (EMD Millipore). The protein samples were then applied to either a HiLoad 16/60 Superdex 75 (GN4) or 200 (lysocin) Prep Grade column (GE Healthcare Life Sciences) in 50 mM Tris-HCl, pH 7.5, and 200 mM NaCl. Highly pure GN4 and lysocin elution fractions were combined, concentrated, sterile filtered, and stored at −80°C until further needed.
Plate lysis assay using autoclaved or viable pseudomonas.
For determining muralytic activity, 25 pmol of each purified protein sample was spotted on 0.75% (wt/vol) agarose embedded with autoclaved P. aeruginosa in 50 mM Tris-HCl, pH 7.5. Clearing zones observed after 24 h of incubation at 37°C correspond to muralytic activity. For elucidating antipseudomonal activity toward viable bacteria, 0.01 to 400 pmol of each purified protein sample was spotted on 0.75% (wt/vol) agarose comprising P. aeruginosa strain 453 at an initial concentration of 5 × 106 CFU/ml in either CAA medium or CAA-HuS (1:1; HuS from pooled human male AB plasma; Sigma-Aldrich). Growth inhibition zones observed after 24 h of incubation at 37°C correspond to antipseudomonal activity. Buffer was spotted as a negative control.
Dose-response cell viability assay.
For the 12-h experiments, 0.01 to 100 μg/ml of PyS2-GN4 was incubated statically at 37°C with P. aeruginosa strain 453 at 106 CFU/ml in CAA medium with 0.5 mg/ml EDDHA. At 2-h increments, an aliquot was removed from each sample and plated on CAA agar to determine viable bacterial counts. For the 24-h experiments, lysocin at 0.1 to 100 μg/ml was incubated statically at 37°C with P. aeruginosa strain 453 at 106 CFU/ml in either CAA medium or CAA-HuS (1:1) with EDDHA. After 24 h, the samples were plated on CAA agar to assess bacterial viability. An untreated control was used for each data set. All samples were investigated in duplicate.
Biofilm disruption assay.
The biofilm disruption assay was modified from a previously described method (43). Wells of a 24-well flat-bottom polystyrene tissue culture plate were inoculated with P. aeruginosa strain PAO1 at 5 × 105 CFU/ml in 2 ml of CAAg medium. Sterility controls consisting of growth medium only were included. Biofilms were grown at 37°C for 72 h with humidity at 120 rpm. Biofilms were washed twice with phosphate-buffered saline (PBS) and treated statically for 24 h with buffer or antimicrobial at 0.03 to 500 μg/ml in 2.5 ml of CAAg supplemented with EDDHA. After treatment, each well was washed twice, stained for 10 min with 0.05% (wt/vol) crystal violet, and washed an additional three times. To qualitatively measure biofilm biomass, residual crystal violet stain in each well was solubilized with 2 ml of 33% (vol/vol) glacial acetic acid and imaged. Each sample was analyzed in duplicate.
Multiplex PCR to determine FpvA receptor type.
As previously described, six oligonucleotide primers were used for the simultaneous amplification of different fpvA gene types (44). fpvAI- (326 bp), fpvAII- (897 bp), and fpvAIII-specific (506 bp) gene fragments were, respectively, amplified with FpvAI_F/FpvAI_R, FpvAII_F/FpvAII_R, and FpvAIII_F/FpvAIII_R primers (Table S3). The 25-μl multiplex PCR consisted of 0.2 mM dNTPs, 0.5 μM of each oligonucleotide primer, 1× Taq reaction buffer, 1 U of Taq DNA polymerase (New England Biolabs), and 1 μl of an overnight P. aeruginosa culture. The thermocycler heating conditions were 95°C for 5 min, 35 cycles of 95°C for 30 s, 55°C for 30 s, and 68°C for 30 s/kb, followed by 68°C for 10 min.
Measuring MIC and MBC.
The MIC and MBC values were calculated using a modified version of the broth microdilution assay as previously described by the Clinical and Laboratory Standards Institute (45). The specific modifications were to the bacterial concentration and growth medium used. Briefly, using a 96-well flat-bottomed microtiter plate, bacteria at 104 CFU/ml were incubated statically in triplicate with 0.002 to 256 μg/ml antimicrobial in either Mueller-Hinton broth (MHB; Gram-positive bacteria) or CAA medium (Gram-negative bacteria) for 48 h at 37°C. CAA medium was used for Gram-negative bacteria to simulate low-iron conditions. Alternatively, MHB was used for Gram-positive bacteria due to their inability to grow in CAA medium. Bacterial growth was assessed by measuring the OD600 using a SpectraMax M5 microplate reader (Molecular Devices). The MIC was defined as the lowest antimicrobial concentration that inhibits bacterial growth. To determine the MBC, the contents from each well originating from the MIC microtiter plate was plated on CAA agar to quantitate bacterial viability. The MBC was defined as the lowest antimicrobial concentration required to kill ≥99.9% of the initial bacterial inoculum. Growth and sterility controls were included.
Transmission electron microscopy.
P. aeruginosa strain 453 at 108 CFU/ml was incubated statically with lysocin at 50 μg/ml in CAA medium with EDDHA for a total of 1 h at 37°C. At 0, 30, and 60 min, an aliquot was removed and fixed with 100 mM sodium cacodylate, pH 7.4, containing 4% (vol/vol) paraformaldehyde and 2% (vol/vol) glutaraldehyde. TEM images were obtained by The Rockefeller University Electron Microscopy Resource Center.
Cytotoxicity assays.
For the hemolytic assays, blood was obtained from healthy adult donors. This study was approved by our Institutional Review Board and all adult subjects provided written informed consent. In this assay, human blood initially collected in an EDTA-containing conical tube was obtained from The Rockefeller University Hospital. hRBCs were harvested by centrifugation at 800 × g for 10 min, washed three times with PBS, and resuspended in buffer to a 10% (vol/vol) concentration. Next, using a 96-well flat-bottomed microtiter plate, 100 μl of hRBC solution was mixed 1:1 in triplicate with a final concentration of 0.5 to 256 μg/ml PyS2-GN4 in buffer. PBS with or without 0.01% (vol/vol) Triton X-100 was used as a positive and negative control for hemolysis, respectively. The microtiter plate was incubated for 8 h at 37°C. Intact hRBCs were removed by centrifugation. To quantitate the relative concentration of hemoglobin release, 100 μl of the sample supernatant was transferred to a new 96-well microtiter plate, and the absorbance was measured at an OD405 using the microplate reader.
Cytotoxicity toward human promyeloblast HL-60 cells was determined using a CellTiter 96 Non-Radioactive Cell Proliferation Assay (Promega). Briefly, HL-60 cells (ATCC CCL-240) were harvested at 1,500 rpm for 5 min, washed twice with PBS, and resuspended to a concentration of 2 × 106 viable cells/ml based on trypan blue exclusion tests. Using a 96-well flat-bottomed microtiter plate, 1 × 105 HL-60 cells were mixed in triplicate with a final concentration of 0.5 to 256 μg/ml PyS2-GN4. As a positive and negative control for cytotoxicity, HL-60 cells were incubated in PBS with and without 0.01% Triton X-100, respectively. The samples were incubated for 8 h at 37°C with 5% CO2. Next, the dye solution was added to each sample. Viable cells convert the tetrazolium component of the dye solution into a formazan product. The microtiter plate was incubated for another 4 h. Solubilization/stop solution, which solubilizes the formazan product, was then added, and the plate was further incubated overnight at 37°C. The relative amount of formazan product was measured at an OD570 using a microplate reader.
Endotoxin release.
P. aeruginosa strain 453 at 106 CFU/ml was treated with 0.2× and 5× MIC of PyS2-GN4, colistin, meropenem, or tobramycin in CAA medium for either 1 or 4 h at 37°C. The samples were subsequently centrifuged at 2,000 × g for 10 min. The supernatant was collected and passed through a 0.2-μm-pore-size syringe filter. Endotoxin concentration in the filtered supernatant was quantitated using a ToxinSensor Chromogenic LAL Endotoxin Assay kit (GenScript). All data are given as the means ± standard errors of the means (SEM) of duplicate experiments.
Murine model of bacteremia.
A murine model of bacteremia using P. aeruginosa was adapted from previous studies (46–50). Briefly, male 6-week-old C57BL/6 mice (Charles River Laboratories) were i.p. infected with 108 CFU of P. aeruginosa strain 453 and then i.p. treated 3 h postinfection with a single dose of either PBS or PyS2-GN4 at 2.5 to 25 mg/kg. Survival was monitored for 10 days. The collective results were obtained from four independent experiments and analyzed by Kaplan-Meier survival curves using GraphPad Prism. The Rockefeller University Institutional Animal Care and Use Committee approved all mouse experiments (protocol 17025).
Supplementary Material
ACKNOWLEDGMENTS
We thank Lars Westblade from Weill Cornell Medical College and Daniel Green from Columbia University Medical Center for supplying the P. aeruginosa clinical isolates used in this study.
This work was supported in part by funding from ContraFect Corporation.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00342-19.
REFERENCES
- 1.McCaughey LC, Josts I, Grinter R, White P, Byron O, Tucker NP, Matthews JM, Kleanthous C, Whitchurch CB, Walker D. 2016. Discovery, characterization and in vivo activity of pyocin SD2, a protein antibiotic from Pseudomonas aeruginosa. Biochem J 473:2345–2358. doi: 10.1042/BCJ20160470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hattemer A, Hauser A, Diaz M, Scheetz M, Shah N, Allen JP, Porhomayon J, El-Solh AA. 2013. Bacterial and clinical characteristics of health care- and community-acquired bloodstream infections due to Pseudomonas aeruginosa. Antimicrob Agents Chemother 57:3969–3975. doi: 10.1128/AAC.02467-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lister PD, Wolter DJ, Hanson ND. 2009. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 22:582–610. doi: 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO. 2017. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. World Health Organization, Geneva, Switzerland: http://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf?ua=1. [Google Scholar]
- 5.Fischetti VA. 2010. Bacteriophage endolysins: a novel anti-infective to control Gram-positive pathogens. Int J Med Microbiol 300:357–362. doi: 10.1016/j.ijmm.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heselepoth RD, Swift SM, Linden SB, Mitchell MS, Nelson DC. 2018. Enzybiotics: endolysins and bacteriocins, p 1–42. In Harpe DR, Abedon ST, Burrowes BH, McConville ML (ed), Bacteriophages. Springer Nature, Cham, Switzerland. [Google Scholar]
- 7.Briers Y, Walmagh M, Van Puyenbroeck V, Cornelissen A, Cenens W, Aertsen A, Oliveira H, Azeredo J, Verween G, Pirnay JP, Miller S, Volckaert G, Lavigne R. 2014. Engineered endolysin-based “Artilysins” to combat multidrug-resistant gram-negative pathogens. mBio 5:e01379. doi: 10.1128/mBio.01379-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larpin Y, Oechslin F, Moreillon P, Resch G, Entenza JM, Mancini S. 2018. In vitro characterization of PlyE146, a novel phage lysin that targets Gram-negative bacteria. PLoS One 13:e0192507. doi: 10.1371/journal.pone.0192507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thandar M, Lood R, Winer BY, Deutsch DR, Euler CW, Fischetti VA. 2016. Novel engineered peptides of a phage lysin as effective antimicrobials against multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother 60:2671–2679. doi: 10.1128/AAC.02972-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Briers Y, Lavigne R. 2015. Breaking barriers: expansion of the use of endolysins as novel antibacterials against Gram-negative bacteria. Future Microbiol 10:377–390. doi: 10.2217/fmb.15.8. [DOI] [PubMed] [Google Scholar]
- 11.Yan G, Liu J, Ma Q, Zhu R, Guo Z, Gao C, Wang S, Yu L, Gu J, Hu D, Han W, Du R, Yang J, Lei L. 2017. The N-terminal and central domain of colicin A enables phage lysin to lyse Escherichia coli extracellularly. Antonie Van Leeuwenhoek 110:1627–1635. doi: 10.1007/s10482-017-0912-9. [DOI] [PubMed] [Google Scholar]
- 12.Lukacik P, Barnard TJ, Keller PW, Chaturvedi KS, Seddiki N, Fairman JW, Noinaj N, Kirby TL, Henderson JP, Steven AC, Hinnebusch BJ, Buchanan SK. 2012. Structural engineering of a phage lysin that targets gram-negative pathogens. Proc Natl Acad Sci U S A 109:9857–9862. doi: 10.1073/pnas.1203472109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chai T, Wu V, Foulds J. 1982. Colicin A receptor: role of two Escherichia coli outer membrane proteins (OmpF protein and btuB gene product) and lipopolysaccharide. J Bacteriol 151:983–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cavard D, Lazdunski C. 1981. Involvement of Btub and Ompf proteins in binding and uptake of colicin-A. Fems Microbiology Lett 12:311–316. doi: 10.1111/j.1574-6968.1981.tb07664.x. [DOI] [Google Scholar]
- 15.Sampson BA, Gotschlich EC. 1992. Elimination of the vitamin B12 uptake or synthesis pathway does not diminish the virulence of Escherichia coli K1 or Salmonella typhimurium in three model systems. Infect Immun 60:3518–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hejair HMA, Zhu Y, Ma J, Zhang Y, Pan Z, Zhang W, Yao H. 2017. Functional role of ompF and ompC porins in pathogenesis of avian pathogenic Escherichia coli. Microb Pathog 107:29–37. doi: 10.1016/j.micpath.2017.02.033. [DOI] [PubMed] [Google Scholar]
- 17.Ghequire MG, De Mot R. 2014. Ribosomally encoded antibacterial proteins and peptides from Pseudomonas. FEMS Microbiol Rev 38:523–568. doi: 10.1111/1574-6976.12079. [DOI] [PubMed] [Google Scholar]
- 18.Cascales E, Buchanan SK, Duche D, Kleanthous C, Lloubes R, Postle K, Riley M, Slatin S, Cavard D. 2007. Colicin biology. Microbiol Mol Biol Rev 71:158–229. doi: 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White P, Joshi A, Rassam P, Housden NG, Kaminska R, Goult JD, Redfield C, McCaughey LC, Walker D, Mohammed S, Kleanthous C. 2017. Exploitation of an iron transporter for bacterial protein antibiotic import. Proc Natl Acad Sci U S A 114:12051–12056. doi: 10.1073/pnas.1713741114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joshi A, Grinter R, Josts I, Chen S, Wojdyla JA, Lowe ED, Kaminska R, Sharp C, McCaughey L, Roszak AW, Cogdell RJ, Byron O, Walker D, Kleanthous C. 2015. Structures of the ultra-high-affinity protein-protein complexes of pyocins S2 and AP41 and their cognate immunity proteins from Pseudomonas aeruginosa. J Mol Biol 427:2852–2866. doi: 10.1016/j.jmb.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun Q, Kuty GF, Arockiasamy A, Xu M, Young R, Sacchettini JC. 2009. Regulation of a muralytic enzyme by dynamic membrane topology. Nat Struct Mol Biol 16:1192–1194. doi: 10.1038/nsmb.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirikae T, Nakano M, Morrison DC. 1997. Antibiotic-induced endotoxin release from bacteria and its clinical significance. Microbiol Immunol 41:285–294. doi: 10.1111/j.1348-0421.1997.tb01203.x. [DOI] [PubMed] [Google Scholar]
- 23.Warren HS, Kania SA, Siber GR. 1985. Binding and neutralization of bacterial lipopolysaccharide by colistin nonapeptide. Antimicrob Agents Chemother 28:107–112. doi: 10.1128/AAC.28.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raymond KN, Dertz EA, Kim SS. 2003. Enterobactin: an archetype for microbial iron transport. Proc Natl Acad Sci U S A 100:3584–3588. doi: 10.1073/pnas.0630018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oliver A, Canton R, Campo P, Baquero F, Blazquez J. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251–1254. doi: 10.1126/science.288.5469.1251. [DOI] [PubMed] [Google Scholar]
- 26.Hoffmann N, Rasmussen TB, Jensen PO, Stub C, Hentzer M, Molin S, Ciofu O, Givskov M, Johansen HK, Hoiby N. 2005. Novel mouse model of chronic Pseudomonas aeruginosa lung infection mimicking cystic fibrosis. Infect Immun 73:2504–2514. doi: 10.1128/IAI.73.4.2504-2514.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bone RC. 1991. The pathogenesis of sepsis. Ann Intern Med 115:457–469. doi: 10.7326/0003-4819-115-6-457. [DOI] [PubMed] [Google Scholar]
- 28.Okawa I, Kageyama M, Egami F. 1973. Purification and properties of pyocin S2. J Biochem 73:281–289. [PubMed] [Google Scholar]
- 29.Ohkawa I, Shiga S, Kageyama M. 1980. Effect of iron concentration in the growth medium on the sensitivity of Pseudomonas aeruginosa to pyocin S2. J Biochem 87:323–331. doi: 10.1093/oxfordjournals.jbchem.a132740. [DOI] [PubMed] [Google Scholar]
- 30.Denayer S, Matthijs S, Cornelis P. 2007. Pyocin S2 (Sa) kills Pseudomonas aeruginosa strains via the FpvA type I ferripyoverdine receptor. J Bacteriol 189:7663–7668. doi: 10.1128/JB.00992-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith K, Martin L, Rinaldi A, Rajendran R, Ramage G, Walker D. 2012. Activity of pyocin S2 against Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 56:1599–1601. doi: 10.1128/AAC.05714-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sano Y, Matsui H, Kobayashi M, Kageyama M. 1993. Molecular structures and functions of pyocins S1 and S2 in Pseudomonas aeruginosa. J Bacteriol 175:2907–2916. doi: 10.1128/jb.175.10.2907-2916.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCaughey LC, Ritchie ND, Douce GR, Evans TJ, Walker D. 2016. Efficacy of species-specific protein antibiotics in a murine model of acute Pseudomonas aeruginosa lung infection. Sci Rep 6:30201. doi: 10.1038/srep30201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sano Y, Kobayashi M, Kageyama M. 1993. Functional domains of S-type pyocins deduced from chimeric molecules. J Bacteriol 175:6179–6185. doi: 10.1128/jb.175.19.6179-6185.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bodilis J, Ghysels B, Osayande J, Matthijs S, Pirnay JP, Denayer S, De Vos D, Cornelis P. 2009. Distribution and evolution of ferripyoverdine receptors in Pseudomonas aeruginosa. Environ Microbiol 11:2123–2135. doi: 10.1111/j.1462-2920.2009.01932.x. [DOI] [PubMed] [Google Scholar]
- 36.Elfarash A, Dingemans J, Ye L, Hassan AA, Craggs M, Reimmann C, Thomas MS, Cornelis P. 2014. Pore-forming pyocin S5 utilizes the FptA ferripyochelin receptor to kill Pseudomonas aeruginosa. Microbiology 160:261–269. doi: 10.1099/mic.0.070672-0. [DOI] [PubMed] [Google Scholar]
- 37.Kageyama M, Kobayashi M, Sano Y, Masaki H. 1996. Construction and characterization of pyocin-colicin chimeric proteins. J Bacteriol 178:103–110. doi: 10.1128/jb.178.1.103-110.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith AW, Hirst PH, Hughes K, Gensberg K, Govan JR. 1992. The pyocin Sa receptor of Pseudomonas aeruginosa is associated with ferripyoverdin uptake. J Bacteriol 174:4847–4849. doi: 10.1128/jb.174.14.4847-4849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baysse C, Meyer JM, Plesiat P, Geoffroy V, Michel-Briand Y, Cornelis P. 1999. Uptake of pyocin S3 occurs through the outer membrane ferripyoverdine type II receptor of Pseudomonas aeruginosa. J Bacteriol 181:3849–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyer JM, Neely A, Stintzi A, Georges C, Holder IA. 1996. Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infect Immun 64:518–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takase H, Nitanai H, Hoshino K, Otani T. 2000. Requirement of the Pseudomonas aeruginosa tonB gene for high-affinity iron acquisition and infection. Infect Immun 68:4498–4504. doi: 10.1128/IAI.68.8.4498-4504.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dennis JJ, Lafontaine ER, Sokol PA. 1996. Identification and characterization of the tolQRA genes of Pseudomonas aeruginosa. J Bacteriol 178:7059–7068. doi: 10.1128/jb.178.24.7059-7068.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Izano EA, Sadovskaya I, Wang H, Vinogradov E, Ragunath C, Ramasubbu N, Jabbouri S, Perry MB, Kaplan JB. 2008. Poly-N-acetylglucosamine mediates biofilm formation and detergent resistance in Aggregatibacter actinomycetemcomitans. Microb Pathog 44:52–60. doi: 10.1016/j.micpath.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Chial M, Ghysels B, Beatson SA, Geoffroy V, Meyer JM, Pattery T, Baysse C, Chablain P, Parsons YN, Winstanley C, Cordwell SJ, Cornelis P. 2003. Identification of type II and type III pyoverdine receptors from Pseudomonas aeruginosa. Microbiology 149:821–831. doi: 10.1099/mic.0.26136-0. [DOI] [PubMed] [Google Scholar]
- 45.Clinical and Laboratory Standards Institute. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 46.Kalle M, Papareddy P, Kasetty G, Morgelin M, van der Plas MJ, Rydengard V, Malmsten M, Albiger B, Schmidtchen A. 2012. Host defense peptides of thrombin modulate inflammation and coagulation in endotoxin-mediated shock and Pseudomonas aeruginosa sepsis. PLoS One 7:e51313. doi: 10.1371/journal.pone.0051313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Desmard M, Davidge KS, Bouvet O, Morin D, Roux D, Foresti R, Ricard JD, Denamur E, Poole RK, Montravers P, Motterlini R, Boczkowski J. 2009. A carbon monoxide-releasing molecule (CORM-3) exerts bactericidal activity against Pseudomonas aeruginosa and improves survival in an animal model of bacteraemia. FASEB J 23:1023–1031. doi: 10.1096/fj.08-122804. [DOI] [PubMed] [Google Scholar]
- 48.Felts AG, Giridhar G, Grainger DW, Slunt JB. 1999. Efficacy of locally delivered polyclonal immunoglobulin against Pseudomonas aeruginosa infection in a murine burn wound model. Burns 25:415–423. doi: 10.1016/S0305-4179(99)00017-0. [DOI] [PubMed] [Google Scholar]
- 49.Shivshetty N, Hosamani R, Ahmed L, Oli AK, Sannauallah S, Sharanbassappa S, Patil SA, Kelmani CR. 2014. Experimental protection of diabetic mice against lethal P. aeruginosa infection by bacteriophage. Biomed Res Int 2014:793242. doi: 10.1155/2014/793242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dings RP, Haseman JR, Leslie DB, Luong M, Dunn DL, Mayo KH. 2013. Bacterial membrane disrupting dodecapeptide SC4 improves survival of mice challenged with Pseudomonas aeruginosa. Biochim Biophys Acta 1830:3454–3457. doi: 10.1016/j.bbagen.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.