Using 894 phylogenetically diverse genomes of the Mycobacterium tuberculosis complex (MTBC), we simulated in silico the ability of the Hain Lifescience GenoType MTBC assay to differentiate the causative agents of tuberculosis. Here, we propose a revised interpretation of this assay to reflect its strengths (e.g., it can distinguish some strains of Mycobacterium canettii and variants of Mycobacterium bovis that are not intrinsically resistant to pyrazinamide) and limitations (e.g., Mycobacterium orygis cannot be differentiated from Mycobacterium africanum).
KEYWORDS: Mycobacterium tuberculosis, genotyping, intrinsic antibiotic resistance
ABSTRACT
Using 894 phylogenetically diverse genomes of the Mycobacterium tuberculosis complex (MTBC), we simulated in silico the ability of the Hain Lifescience GenoType MTBC assay to differentiate the causative agents of tuberculosis. Here, we propose a revised interpretation of this assay to reflect its strengths (e.g., it can distinguish some strains of Mycobacterium canettii and variants of Mycobacterium bovis that are not intrinsically resistant to pyrazinamide) and limitations (e.g., Mycobacterium orygis cannot be differentiated from Mycobacterium africanum).
INTRODUCTION
The in vitro diagnostic (IVD) CE-marked Hain Lifescience GenoType MTBC assay is the oldest, and likely the most widely used, commercial assay to differentiate the causative agents of tuberculosis (TB) (1). Strictly speaking, these agents comprise Mycobacterium canettii, which is almost exclusively limited to the Horn of Africa, on the one hand and several species/ecotypes of the Mycobacterium tuberculosis complex (MTBC) on the other, although most researchers and guidelines consider M. canettii to be part of the MTBC (2, 3). Clinically, the early identification of the precise causative agent of TB is important because it can serve as a marker for intrinsic resistance or may inform the attribution of the source of infection (e.g., in cases of Mycobacterium bovis, intrinsic resistance to pyrazinamide can usually be ruled in and a human source for the infection is unlikely [4]).
Throughout the past decade, the interpretation of the GenoType MTBC, but not its design, has been revised to reflect changes in our understanding of the causative agents of TB (1, 3, 5). More recently, several new animal species/ecotypes have been discovered, which prompted us to investigate to what extent these could be differentiated with the Hain assay using a collection of 894 diverse genomes representing M. canettii and major phylogenetic groups of MTBC (Fig. S1 and Table S1) (6). This was possible because Hain Lifescience has filed a European patent (7) for its assay, which relies on a 23 rRNA probe to identify M. canettii/MTBC as a whole, whereas mutations in gyrB and the RD1BCG deletion differentiate individual species/ecotypes (Figures S1 and S2 and Table S2) [8]. Specifically, we typed all 894 genomes in silico for the single-nucleotide polymorphism (SNP) and deletion markers from the patent (see Supplemental Methods S1).
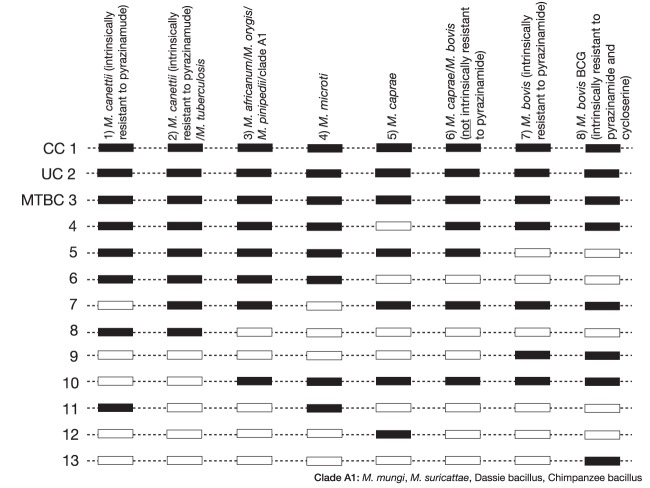
The current package insert of the GenoType MTBC lists seven binding patterns for M. canettii or MTBC isolates (patterns 2 to 8 in Fig. 1 and Table S1). In 2010, however, Fabre et al. demonstrated experimentally that a minority of M. canettii strains yield a novel pattern, which does not feature in the package insert (9). Our simulation confirmed these results. Specifically, two of the M. canettii strains with the unusual experimental pattern (i.e., Percy157 and Percy525) from Fabre et al., for which genomes were available and which could therefore be included in our study, also yielded the novel pattern in silico (pattern 1 in Fig. 1 and Table S1) (9). The remaining five M. canettii genomes from Fabre et al. (i.e., Percy22, Percy32, Percy50, Percy79, and Percy301) could not be differentiated from M. tuberculosis in silico, which was in agreement with the experimental findings (pattern 2 in Fig. 1 and Table S1) (9). Given the highly recombinogenic nature of M. canettii, it is not surprising that this species yields two different patterns (10, 11). All representatives of this species, including the two strains that gave the new binding pattern experimentally and in silico, have been found to be resistant to pyrazinamide when tested with the Bactec MGIT 960 at 100 μg/ml, the only critical concentration recognized by the Clinical and Laboratory Standards Institute and the World Health Organization (WHO) (9, 12–17). Although it is unclear whether this phenotype is due to a single mechanism shared by all strains (e.g., rpsA T5A) or whether different mutations are responsible in different strains (e.g., panD M117T or a series of pncA mutations [Table S3]), we recommend that the package insert is updated to include this novel pattern as “M. canettii (intrinsically resistant to pyrazinamide)” (14, 18–20).
FIG 1.
Proposed interpretation of binding patterns of Hain Lifescience GenoType MTBC. Eight binding patterns are possible for samples that contain a single strain of MTBC or M. canettii. The first binding pattern is not currently included in the package insert of the GenoType MTBC (5, 9). With the exception of pattern 4 for Mycobacterium microti, the interpretations of the remaining patterns were updated to include information about intrinsic resistance to antibiotics and/or to reflect the improved understanding of the phylogenetic diversity among the causative agents of TB. More information about clade A1 can be found elsewhere (6). Additional binding patterns are possible for samples that are negative, contain other bacteria, or when the assay was not carried out correctly (in these cases, one or more of the conjugate control [CC], universal control [UC], or MTBC bands would be negative [5]).
Moreover, our findings suggest the following changes for the remaining seven binding patterns (Fig. 1, Fig. S1, and Table S1). First, pattern 3, currently used to differentiate Mycobacterium africanum from the rest of the MTBC and from M. canettii, has to be revised, since our analysis showed this pattern cannot distinguish M. africanum from Mycobacterium orygis, Mycobacterium pinnipedii, or the clade A1 ecotypes (i.e., Mycobacterium mungi, Mycobacterium suricattae, the chimpanzee bacillus, and the dassie bacillus) (6, 21, 22). Second, for the sake of clarity, we would separate M. bovis and Mycobacterium caprae, as they belong to two independent phylogenetic groups and are usually recognized as separate species/ecotypes (3). In contrast, the bacillus Calmette-Guérin (BCG) was derived from a M. bovis strain and is best described as M. bovis BCG to emphasize its intrinsic resistance to pyrazinamide (4). Finally, the current package insert features two binding patterns for “M. bovis subsp. caprae,” of which one is described to occur in only 5% of cases of M. caprae (5). Our collection featured seven genomes consistent with this rarer pattern. However, the seven genomes did not group together phylogenetically (Fig. S1). Three of the strains were isolated in 2009 from primates that were placed in quarantine upon entering the United States (23, 24). Their genomes grouped together with the M. caprae genomes on the phylogeny and shared the lepA V424V marker for this species (25). In contrast, the other four genomes were more closely related to that of M. bovis but lacked the pncA H57D mutation that is responsible for intrinsic pyrazinamide resistance in this species (8, 14). Three of these isolates were isolated from humans in Malawi and the fourth from a Nilgau antelope from a German zoo. For the latter sample, we knew the spoligotyping pattern, which we used to query the M. bovis spoligotype database (26). The spoligotype for the antelope isolate from 1996 (SB1898) appears to be very rare, as only one identical representative was found, which was submitted from Spain in 2009. Thus, it is unclear whether these four strains represent a novel ecotype or species, but because they are phylogenetically closer to M. bovis than to M. caprae, we recommend that pattern 6 should be reported as “M. caprae/M. bovis (not intrinsically resistant to pyrazinamide).”
M. orygis has been isolated from many different animals, and there is a growing recognition that it is a zoonotic source of human TB (27). Our in silico typing approach confirmed that M. orygis could be specifically identified by a mutation at codon 329 of gyrB (8). Since this marker is contained within the gyrB amplicon, we suggest that it could be added to the Hain assay, as this would avoid misclassifications such as that in Rahim et al., in which cattle from Bangladesh were erroneously reported to have been infected with M. africanum instead of with M. orygis (28).
The findings in this study are important for two reasons. First, most of our proposed changes can be implemented easily by updating the package insert of the Hain Lifescience GenoType MTBC (5). More broadly, given that whole-genome sequencing is now increasingly being used as a routine diagnostic tool, it would be possible to implement our in silico surveillance approach in real time to automatically flag unusual isolates for experimental follow-up. In fact, if clinical sequencing providers, such as Public Health England in the United Kingdom, were to offer this as a professional service, it could generate much-needed revenue to reduce the cost of sequencing to public health systems and, therefore, to the tax payer, while enabling commercial companies to conduct postmarketing surveillance for genotypic assays comprehensively and cost effectively—a win-win situation for all parties.
Supplementary Material
ACKNOWLEDGMENTS
Calculations were performed at the sciCORE (http://scicore.unibas.ch/) scientific computing core facility at University of Basel.
F.C. received personal fees from Next Gen Diagnostics LLC. S.J.P. is a consultant for Next Gen Diagnostics and Specific. C.U.K. is a consultant for the World Health Organization (WHO) Regional Office for Europe, QuantuMDx Group Ltd., and the Foundation for Innovative New Diagnostics, which involves work for the Cepheid Inc., Hain Lifescience, and the WHO. C.U.K. is an advisor to GenoScreen. The Bill & Melinda Gates Foundation, Janssen Pharmaceutica, and PerkinElmer covered C.U.K.’s travel and accommodation to present at meetings. The Global Alliance for TB Drug Development Inc. and Otsuka Novel Products GmbH have supplied C.U.K. with antibiotics for in vitro research. C.U.K. is collaborating with YD Diagnostics.
F.C. received support from the Wellcome Trust (201344/Z/16/Z). S.N. received support from the German Center for Infection Research, the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy (grant EXC 22167-390884018), and the Leibniz Science Campus EvoLUNG (Evolutionary Medicine of the Lung). S.G. was supported by the Swiss National Science Foundation (grants 310030_166687, IZRJZ3_164171, IZLSZ3_170834, and CRSII5_177163), the European Research Council (grant 309540-EVODRTB), and SystemsX.ch. The funders had no role in the study design, data collection, interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00159-19.
REFERENCES
- 1.Richter E, Weizenegger M, Rüsch-Gerdes S, Niemann S. 2003. Evaluation of genotype MTBC assay for differentiation of clinical Mycobacterium tuberculosis complex isolates. J Clin Microbiol 41:2672–2675. doi: 10.1128/JCM.41.6.2672-2675.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clinical and Laboratory Standards Institute. Laboratory detection and identification of mycobacteria, 2nd edition CLSI document M48 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 3.Gagneux S. 2018. Ecology and evolution of Mycobacterium tuberculosis. Nat Rev Microbiol 16:202–213. doi: 10.1038/nrmicro.2018.8. [DOI] [PubMed] [Google Scholar]
- 4.Köser CU, Feuerriegel S, Summers DK, Archer JA, Niemann S. 2012. Importance of the genetic diversity within the Mycobacterium tuberculosis complex for the development of novel antibiotics and diagnostic tests of drug resistance. Antimicrob Agents Chemother 56:6080–6087. doi: 10.1128/AAC.01641-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hain Lifescience GmbH. 2017. GenoType MTBC VER 1.X. Instructions for use IFU-301-11. Hain Lifescience GmbH, Nehren, Germany. [Google Scholar]
- 6.Brites D, Loiseau C, Menardo F, Borrell S, Boniotti MB, Warren R, Dippenaar A, Parsons SDC, Beisel C, Behr MA, Fyfe JA, Coscolla M, Gagneux S. 2018. A new phylogenetic framework for the animal-adapted Mycobacterium tuberculosis complex. Front Microbiol 9:2820. doi: 10.3389/fmicb.2018.02820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hain Life Science GmbH. December 2004. Method for detection and differentiation of Mycobacterium tuberculosis complex. European patent EP1490518B1.
- 8.Huard RC, Fabre M, de Haas P, Lazzarini LC, van Soolingen D, Cousins D, Ho JL. 2006. Novel genetic polymorphisms that further delineate the phylogeny of the Mycobacterium tuberculosis complex. J Bacteriol 188:4271–4287. doi: 10.1128/JB.01783-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fabre M, Hauck Y, Soler C, Koeck JL, van Ingen J, van Soolingen D, Vergnaud G, Pourcel C. 2010. Molecular characteristics of “Mycobacterium canettii” the smooth Mycobacterium tuberculosis bacilli. Infect Genet Evol 10:1165–1173. doi: 10.1016/j.meegid.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 10.Supply P, Marceau M, Mangenot S, Roche D, Rouanet C, Khanna V, Majlessi L, Criscuolo A, Tap J, Pawlik A, Fiette L, Orgeur M, Fabre M, Parmentier C, Frigui W, Simeone R, Boritsch EC, Debrie AS, Willery E, Walker D, Quail MA, Ma L, Bouchier C, Salvignol G, Sayes F, Cascioferro A, Seemann T, Barbe V, Locht C, Gutierrez MC, Leclerc C, Bentley SD, Stinear TP, Brisse S, Medigue C, Parkhill J, Cruveiller S, Brosch R. 2013. Genomic analysis of smooth tubercle bacilli provides insights into ancestry and pathoadaptation of Mycobacterium tuberculosis. Nat Genet 45:172–179. doi: 10.1038/ng.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boritsch EC, Khanna V, Pawlik A, Honore N, Navas VH, Ma L, Bouchier C, Seemann T, Supply P, Stinear TP, Brosch R. 2016. Key experimental evidence of chromosomal DNA transfer among selected tuberculosis-causing mycobacteria. Proc Natl Acad Sci U S A 113:9876–9881. doi: 10.1073/pnas.1604921113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Somoskovi A, Dormandy J, Mayrer AR, Carter M, Hooper N, Salfinger M. 2009. “Mycobacterium canettii” isolated from a human immunodeficiency virus-positive patient: first case recognized in the United States. J Clin Microbiol 47:255–257. doi: 10.1128/JCM.01268-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koeck JL, Fabre M, Simon F, Daffé M, Garnotel E, Matan AB, Gérôme P, Bernatas JJ, Buisson Y, Pourcel C. 2011. Clinical characteristics of the smooth tubercle bacilli ‘Mycobacterium canettii’ infection suggest the existence of an environmental reservoir. Clin Microbiol Infect 17:1013–1019. doi: 10.1111/j.1469-0691.2010.03347.x. [DOI] [PubMed] [Google Scholar]
- 14.Feuerriegel S, Köser CU, Richter E, Niemann S. 2013. Mycobacterium canettii is intrinsically resistant to both pyrazinamide and pyrazinoic acid. J Antimicrob Chemother 68:1439–1440. doi: 10.1093/jac/dkt042. [DOI] [PubMed] [Google Scholar]
- 15.Blouin Y, Cazajous G, Dehan C, Soler C, Vong R, Hassan MO, Hauck Y, Boulais C, Andriamanantena D, Martinaud C, Martin E, Pourcel C, Vergnaud G. 2014. Progenitor “Mycobacterium canettii” clone responsible for lymph node tuberculosis epidemic, Djibouti. Emerg Infect Dis 20:21–28. doi: 10.3201/eid2001.130652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bouzid F, Astier H, Osman DA, Javelle E, Hassan MO, Simon F, Garnotel E, Drancourt M. 2018. Extended spectrum of antibiotic susceptibility for tuberculosis, Djibouti. Int J Antimicrob Agents 51:235–238. doi: 10.1016/j.ijantimicag.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 17.Briquet A, Vong R, Roseau JB, Javelle E, Cazes N, Riviere F, Aletti M, Otto MP, Ficko C, Duron S, Fabre M, Pourcel C, Simon F, Soler C. 2019. Clinical features of Mycobacterium canettii infection: a retrospective study of 20 cases among French soldiers and relatives. Clin Infect Dis. doi: 10.1093/cid/ciz107. [DOI] [PubMed] [Google Scholar]
- 18.Alexander DC, Ma JH, Guthrie JL, Blair J, Chedore P, Jamieson FB. 2012. Gene sequencing for routine verification of pyrazinamide resistance in Mycobacterium tuberculosis: a role for pncA but not rpsA. J Clin Microbiol 50:3726–3728. doi: 10.1128/JCM.00620-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang S, Chen J, Shi W, Liu W, Zhang W, Zhang Y. 2013. Mutations in panD encoding aspartate decarboxylase are associated with pyrazinamide resistance in Mycobacterium tuberculosis. Emerg Micro Infect 2:e34. doi: 10.1038/emi.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi W, Cui P, Niu H, Zhang S, Tonjum T, Zhu B, Zhang Y. 2019. Introducing RpsA point mutations Δ438A and D123A into the chromosome of M. tuberculosis confirms their role in causing resistance to pyrazinamide. Antimicrob Agents Chemother. doi: 10.1128/AAC.02681-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kjeldsen MK, Bek D, Rasmussen EM, Priemé A, Thomsen V. 2009. Line probe assay for differentiation within Mycobacterium tuberculosis complex. Evaluation on clinical specimens and isolates including Mycobacterium pinnipedii. Scand J Infect Dis 41:635–641. doi: 10.1080/00365540903127425. [DOI] [PubMed] [Google Scholar]
- 22.van Ingen J, Rahim Z, Mulder A, Boeree MJ, Simeone R, Brosch R, van Soolingen D. 2012. Characterization of Mycobacterium orygis as M. tuberculosis complex subspecies. Emerg Infect Dis 18:653–655. doi: 10.3201/eid1804.110888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joshi D, Harris NB, Waters R, Thacker T, Mathema B, Krieswirth B, Sreevatsan S. 2012. Single nucleotide polymorphisms in the Mycobacterium bovis genome resolve phylogenetic relationships. J Clin Microbiol 50:3853–3861. doi: 10.1128/JCM.01499-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orloski K, Robbe-Austerman S, Stuber T, Hench B, Schoenbaum M. 2018. Whole genome sequencing of Mycobacterium bovis isolated from livestock in the United States, 1989–2018. Front Vet Sci 5:253. doi: 10.3389/fvets.2018.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Domogalla J, Prodinger WM, Blum H, Krebs S, Gellert S, Muller M, Neuendorf E, Sedlmaier F, Buttner M. 2013. Region of difference 4 in Alpine Mycobacterium caprae isolates indicates three variants. J Clin Microbiol 51:1381–1388. doi: 10.1128/JCM.02966-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith NH, Upton P. 2012. Naming spoligotype patterns for the RD9-deleted lineage of the Mycobacterium tuberculosis complex; https://www.Mbovis.org. Infect Genet Evol 12:873–876. doi: 10.1016/j.meegid.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 27.Lipworth S, Jajou R, de Neeling A, Bradley P, van der Hoek W, Maphalala G, Bonnet M, Sanchez-Padilla E, Diel R, Niemann S, Iqbal Z, Smith G, Peto T, Crook D, Walker T, van Soolingen D. 2019. SNP-IT tool for identifying subspecies and associated lineages of Mycobacterium tuberculosis complex. Emerg Infect Dis 25:482–488. doi: 10.3201/eid2503.180894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rahim Z, Thapa J, Fukushima Y, van der Zanden AGM, Gordon SV, Suzuki Y, Nakajima C. 2017. Tuberculosis caused by Mycobacterium orygis in dairy cattle and captured monkeys in Bangladesh: a new scenario of tuberculosis in South Asia. Transbound Emerg Dis 64:1965–1969. doi: 10.1111/tbed.12596. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.