Helicobacter pylori is a major global pathogen, and its infection represents a key factor in the etiology of various gastric diseases, including gastritis, peptic ulcers, and gastric carcinoma. The efficacy of current standard treatment for H. pylori infection including two broad-spectrum antibiotics is compromised by toxicity toward the gut microbiota and the development of drug resistance, which will likely only be resolved through novel and selective antibacterial strategies.
KEYWORDS: Helicobacter pylori, zinc linolenate, antibacterial agents, drug resistance
ABSTRACT
Helicobacter pylori is a major global pathogen, and its infection represents a key factor in the etiology of various gastric diseases, including gastritis, peptic ulcers, and gastric carcinoma. The efficacy of current standard treatment for H. pylori infection including two broad-spectrum antibiotics is compromised by toxicity toward the gut microbiota and the development of drug resistance, which will likely only be resolved through novel and selective antibacterial strategies. Here, we synthesized a small molecule, zinc linolenate (ZnLla), and investigated its therapeutic potential for the treatment of H. pylori infection. ZnLla showed effective antibacterial activity against standard strains and drug-resistant clinical isolates of H. pylori in vitro with no development of resistance during continuous serial passaging. The mechanisms of ZnLla action against H. pylori involved the disruption of bacterial cell membranes and generation of reactive oxygen species. In mouse models of multidrug-resistant H. pylori infection, ZnLla showed in vivo killing efficacy comparable and superior to the triple therapy approach when use as a monotherapy and a combined therapy with omeprazole, respectively. Moreover, ZnLla treatment induces negligible toxicity against normal tissues and causes minimal effects on both the diversity and composition of the murine gut microbiota. Thus, the high degree of selectivity of ZnLla for H. pylori provides an attractive candidate for novel targeted anti-H. pylori treatment.
INTRODUCTION
Helicobacter pylori infects over 50% of the world’s population, and its infection is well recognized as a major cause of chronic gastritis and of the development of peptic ulcer and gastric cancer (1). In many studies, H. pylori eradication has been shown to have a prophylactic effect against gastric cancer (2–4). Triple therapy recommended by current treatment guidelines, including a proton pump inhibitor (PPI) and two broad-spectrum antibiotics (5), is frequently conducted to eradicate H. pylori. However, the emergence of antibiotic-resistant strains of H. pylori and various side effects compromises the effectiveness of these treatments (6, 7). In addition, some situations, such as the use of the antibiotics, could nonspecifically eliminate the commensal bacteria and disrupt the microbiota balance (8, 9). The resultant alterations in the gut microbiota may perturb host metabolism and immunity and thus contribute to an increased risk of disease development (10). Actually, gut microbiota dysbiosis has been shown to significantly influence the development of a variety of human diseases, such as obesity and metabolic syndrome (11), autism spectrum disorder (12), Parkinson’s disease and other neurologic diseases (13), allergy (14), atherosclerosis (15), inflammatory bowel disease (16), and colorectal cancer (17). To overcome these challenges, it is imperative to develop selective anti-H. pylori agents with enhanced efficacy and potentially diminished resistance.
A number of free fatty acids (FFAs), with potent antibacterial activities against a broad range of bacteria (18–21), including H. pylori, are of considerable research interests. In particular, FFAs seem to induce no acquired resistance to the antibacterial effect and are considered safe, which make them attractive as antibacterial agents for commercial exploitation. The antibacterial potency and spectrum of antibacterial action of each FFA are influenced by the length of the carbon chain and the presence, number, position, and orientation of double bonds (21). For instance, polyunsaturated fatty acids (PUFAs), including linolenic acid (Lla; C18:3) (22, 23), eicosapentaenoic acid (C20:5) (24), and docosahexaenoic acid (C22:6) (25), have been reported to show highly potent activity against H. pylori. In addition, the inhibitory effects of these PUFAs on H. pylori growth are comparable to each other (24). Although the efficiency is unsatisfying compared with that of conventional antibiotics, this could be solved by chemically generating derivatives of these PUFAs with other antibacterial agents for more desirable “drug-like” characteristics. As Lla is an essential fatty acid for humans and is more affordable than the two others, it is a good candidate for new formulations.
In recent years, the medicinal uses of metal complexes have received much attention, since metals can exhibit a wide variety of coordination properties and reactivities which can be used to form complex with drugs as ligands (26). Metal complexes contain a variety of structural and electronic features with the ability to form specific interactions with other biomolecules. This may increase the bioavailability of either the metal ion or the drug ligand or both (27). Among them, zinc is regarded as an essential trace metal element for human body, which plays a vital role in the maintenance of balance and the adjustment of an enormous number of biological processes (28). Aside from dietary zinc supplementation, Zn(II) compounds and zinc-based complexes have been studied for therapeutic use in the common cold, sickle cell anemia, skin diseases, acute diarrhea, gastric ulcers, and various other disorders (29–31). Furthermore, zinc is known to exhibit moderately antibacterial activity; especially, zinc complexes of some drugs, such as famotidine (32) or rifampin (33), showed higher antibacterial activities than did the free agents. Clearly, a zinc complex could be used for the development of novel drugs with promising pharmacological applications and offer unique therapeutic opportunities.
In the present study, we formulated the zinc complex of Lla (zinc linolenate [ZnLla]) and evaluated its potent and selective antibacterial activity against various standard and resistant strains of H. pylori. Furthermore, we report that the mechanisms of action of ZnLla involve membrane permeation and the generation of intracellular reactive oxygen species. In vivo efficacy studies were performed to demonstrate significant decolonization of H. pylori with inhibited toxicity against normal tissues and commensal bacteria. In summary, our data suggest that ZnLla could be suitable for use as a specific anti-H. pylori agent to eradicate multidrug-resistant H. pylori.
RESULTS
Preparation and characterization of zinc linolenate.
The compound was synthesized by reacting zinc chloride with the ligand linolenic acid (Lla) after activation. The purified Zinc linolenate (ZnLla) was characterized by a series of measurements. Fourier transform infrared (FTIR) spectra were collected to confirm that Lla was successfully linked to Zn2+ to prepare ZnLla. The spectra of the complex contained all the absorption bands due to the ligand molecule (Fig. 1A). Also, a new absorption band at 3,444.7 cm−1 indicative of coordination of the ligand with Zn2+ also appeared, which was absent in free Lla and may correspond to the stretching of the aliphatic OH group of Lla. The formation of ZnLla was also confirmed by measuring Zn2+ concentrations during the synthesis process. With the occurrence of complexation reaction, the Zn2+ concentration gradually decreased, and after 12 h, nearly 26% were left unreacted (see Fig. S1A in the supplemental material). We also attempted to analyze this complex using mass spectrometry. The matrix-assisted laser desorption ionization–time of flight (MALDI-TOF)/TOF mass spectra display an intense signal at m/z 637.511 ([M+H]+) that corresponds to the chemical formula Zn(Lla)2(H2O), a hydrated compound with a water molecule (Fig. 1B). In addition, UV-Vis spectrometry analysis showed similar spectra of Lla and ZnLla (Fig. S1B), indicating that there are no alterations in the π-π* and shoulder transitions between them. The 1H and 13C nuclear magnetic resonance (NMR) spectra of ZnLla had the same number of peaks, even the same spin-spin coupling and splitting in the 1H NMR, as in the spectrum of the free Lla (Fig. S2), confirming the presence of the full ligand skeleton and the proposed composition of the complex.
FIG 1.
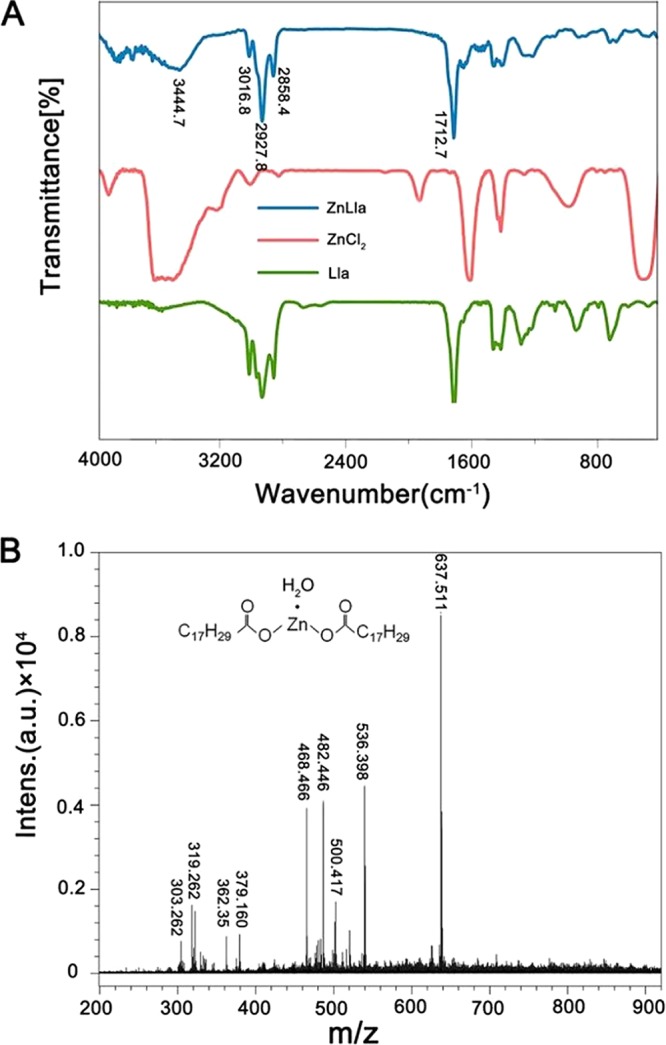
The FTIR and MALDI-TOF/TOF spectra of ZnLla. (A) FTIR of ZnCl2, Lla, and ZnLla. The unsaturated C-H vibration peak of double bond and the characteristic vibration peak of carboxyl group of Lla are 2,927.8 and 1,712.7, respectively. A new absorption band at 3,444.7 cm−1 was shown for ZnLla. (B) MALDI-TOF/TOF of ZnLla, m/z 637.511 corresponds to the chemical formula, Zn(Lla)2(H2O) ([M+H]+), a hydrated compound with a water molecule (MS: analysis calculated for C36H61O5Zn, m/z 637.381 [M+H]+). Intens., intensity; a.u., arbitrary units.
ZnLla has selective anti-H. pylori efficacy in vitro.
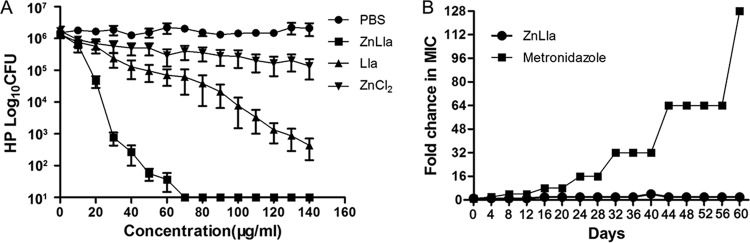
The antibacterial activities of ZnLla, ZnCl2, and Lla against four H. pylori strains were evaluated in vitro by determining their MICs and minimal bactericidal concentrations (MBCs). Growth inhibition of four H. pylori standard strains tested by serial broth dilution method showed MIC values of 4 to ∼8 μg/ml for ZnLla but ≥128 μg/ml for Lla, ZnCl2, or an antiulcer agent, zinc acexamate (Table 1). MBC for this study was defined as the minimal concentration that kills 99.9% (3 log) of targeted bacteria during 120 min of incubation. Accordingly, the MBC value of ZnLla was determined to be 30 μg/ml, whereas ZnCl2 had no bactericidal effect against H. pylori at all tested concentrations, and the MBC for Lla was 130 μg/ml (Fig. 2A). Thus, ZnLla showed the most potent bactericidal effect and completely killed H. pylori within 120 min. Despite its strong bactericidal activity, ZnLla showed little toxicity to human normal gastric epithelial GES-1 cells or gastric carcinoma AGS cells, as exposure to ZnLla at various concentrations (even 100 times the MIC, ∼400 μg/ml) showed unaffected cell viability based on a CCK-8 assay (Fig. S3A and B).
TABLE 1.
Broth microdilution MICs of ZnLla, Lla, ZnCl2, and zinc acexamate for H. pylori standard strains and drug-resistant clinical isolates
| Strain | Drug sensitivity (drug[s])a | MIC (μg/ml) for: |
||||
|---|---|---|---|---|---|---|
| ZnLla | Lla | ZnCl2 | Zinc acexamate | MTZ | ||
| HP26695 | S | 8 | 128 | >128 | >128 | 1 |
| NSH57 | S | 8 | 128 | >128 | >128 | 1 |
| MSD132 | S | 8 | 128 | >128 | >128 | 2 |
| G27 | S | 4 | 128 | >128 | >128 | 1 |
| HPYF-2 | S | 8 | 128 | >128 | >128 | 2 |
| HP159 | R (LVX, CLR, MTZ) | 8 | 128 | >128 | >128 | 64 |
| HP286 | R (LVX, CLR, MTZ) | 8 | 128 | >128 | >128 | 32 |
| HP289 | R (LVX, CLR, MTZ) | 8 | 128 | >128 | >128 | 64 |
| HP290 | R (LVX, CLR, MTZ) | 8 | 128 | >128 | >128 | 64 |
| HP285 | R (CLR, MTZ) | 4 | 128 | >128 | >128 | 64 |
| HP287 | R (CLR, MTZ) | 8 | 128 | >128 | >128 | 32 |
| HPGC-2 | R (CLR, MTZ) | 8 | 128 | >128 | >128 | 16 |
| HP163 | R (LVX, MTZ) | 8 | 128 | >128 | >128 | 32 |
| HP160 | R (MTZ) | 2 | 128 | >128 | >128 | 32 |
| HPYF-4 | R (MTZ) | 8 | 128 | >128 | >128 | 32 |
| HP161 | R (CLR) | 8 | 128 | >128 | >128 | 2 |
| HPYF-3 | R (CLR) | 4 | 128 | >128 | >128 | 2 |
| HPYF-5 | R (CLR) | 8 | 128 | >128 | >128 | 1 |
| HP162 | R (LVX) | 2 | 128 | >128 | >128 | 2 |
| HPGC1226 | R (AMX, MTZ) | 4 | 128 | >128 | >128 | 8 |
S, drug sensitive; R, drug resistant; LVX, levofloxacin; CLR, clarithromycin; MTZ, metronidazole; AMX, amoxicillin.
FIG 2.
ZnLla has bactericidal activity and a low risk for resistance. (A) In vitro bactericidal activities of ZnLla, Lla, and ZnCl2 at different concentrations against H. pylori G27 (HP). Data represent medians ± standard deviation (SD) of the results from three independent experiments. (B) Development of resistance to ZnLla and metronidazole in G27. The fold change was the normalized ratio of the MIC obtained for a given subculture to the MIC that obtained for first-time exposure. Serial passaging of H. pylori with subinhibitory concentrations of metronidazole leads to high resistance against metronidazole. However, the development of resistance is not observed with ZnLla. Representative results from two independent experiments are shown.
Resistance to antibiotics is of particular concern as a major cause of clinical failure to eradicate H. pylori (34, 35). Next, we further tested the antibacterial activity of ZnLla against clinically isolated drug-resistant H. pylori strains. ZnLla effectively inhibited the growth of all 16 drug-resistant strains with MICs of 2 to ∼8 μg/ml (Table 1), which further substantiated its potency in overcoming drug resistance toward effective anti-H. pylori therapy. As expected, ZnCl2 or Lla alone could not effectively kill drug-resistant strains. Note that other metal-Lla complexes or zinc complexes of other FFAs showed no antibacterial activity against H. pylori (Table S1). ZnLla was also examined for the possible development of insensitivity toward the bacteria upon subculture. Development of resistance was not observed in H. pylori during continuous serial passaging in the presence of various concentrations of ZnLla over 60 days (Fig. 2B). In contrast, H. pylori rapidly developed resistance to metronidazole within a few days of exposure (Fig. 2B).
A high degree of specificity is also a desirable trait for antibacterial agents targeting H. pylori. To assess specificity, a general antibacterial pattern of ZnLla was evaluated using 23 bacterial species. ZnLla exhibited no activity against most of non-Helicobacter strains in the panel even at 128 μg/ml, besides exerting a somewhat inhibitory effect on Neisseria gonorrhoeae and Bacillus cereus, with both MICs of 64 μg/ml at least 8-fold higher than that for H. pylori (Fig. 3). Therefore, ZnLla may be considered a specific anti-H. pylori molecule.
FIG 3.
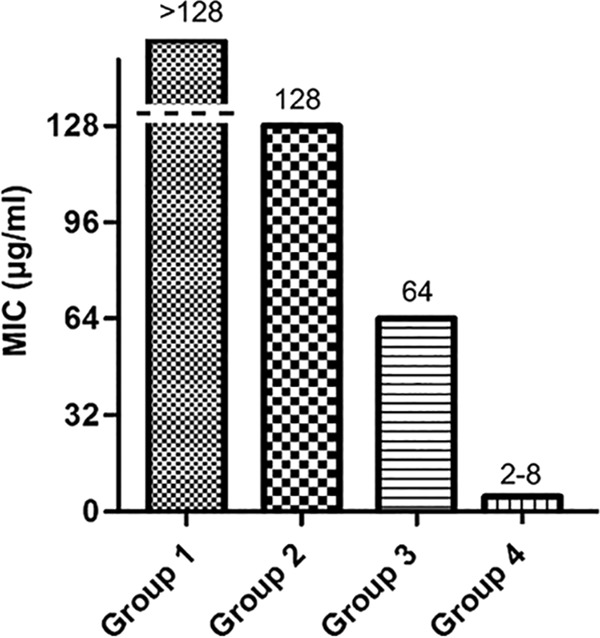
ZnLla has no antibacterial activities against non-Helicobacter strains. The MICs were determined by broth microdilution assays as described in Materials and Methods. The strains used were divided into four groups based on MIC values. The MIC for group 1 is >128 μg/ml, and the strains include Staphylococcus aureus ATCC 6538, Pseudomonas putida PAO1, Enterococcus faecalis FA2-2, Salmonella Typhimurium ATCC 14028, Moraxella catarrhalis ATCC 25238, Acinetobacter baumannii ATCC 19606, Mycobacterium smegmatis MC2 155, Bacillus subtilis 168, Enterococcus faecium ATCC 19434, Streptococcus pneumoniae ATCC 49619, Proteus mirabilis ATCC 29906, Haemophilus influenzae ATCC 49766, Aggregatibacter actinomycetemcomitans D7S-1, and Listeria monocytogenes EGD-e. The MIC for group 2 is 128 μg/ml, and the strains include Enterobacter cloacae ATCC 13047, Campylobacter jejuni NCTC11168, Escherichia coli MG1655, Klebsiella pneumoniae ATCC 35657, Lactococcus lactis MG1363, Stenotrophomonas maltophilia ATCC 51331, and Morganella morganii ATCC 25830. The MIC for group 3 is 64 μg/ml, and the strains include Neisseria gonorrhoeae ATCC 19424 and Bacillus cereus ATCC 14579. The MIC for group 4 is 2 to 8 μg/ml, and the strains include 20 H. pylori strains.
ZnLla disrupts H. pylori membranes and generates reactive oxygen species.
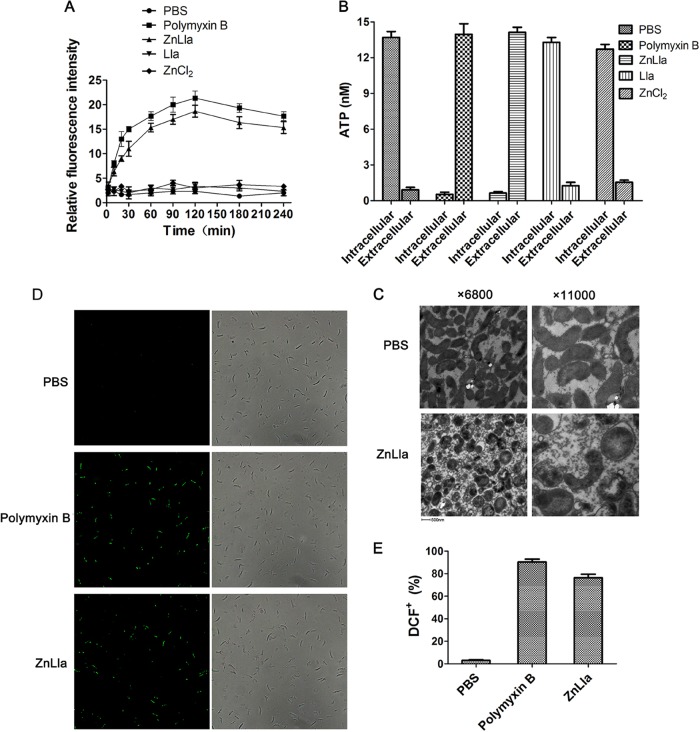
The bactericidal mechanism of ZnLla was explored by the vesicle leakage assessment as well as bacterial morphology observation. We first used the hydrophobic fluorescence probe N-phenyl-1-naphthylamine (NPN) to determine the outer membrane disruption of H. pylori in response to ZnLla treatment. There was no difference in NPN uptake in single-Lla- or ZnCl2-treated H. pylori compared to the phosphate-buffered saline (PBS) control (Fig. 4A). When treated with ZnLla, H. pylori showed a significant increase in the fluorescence signal of NPN, similar to that treated with polymyxin B, which disrupts the outer membrane permeability barrier (Fig. 4A and S4), indicating that ZnLla increased the outer membrane permeability of H. pylori. We also measured the release of ATP from the cytoplasm through the plasma membrane to examine the effect of ZnLla on the plasma membrane of H. pylori. ZnLla- or polymyxin B-treated cells released virtually all of their intracellular ATP into the medium, whereas Lla- or ZnCl2-treated cells did not (Fig. 4B and S5), demonstrating plasma membrane disruption caused by ZnLla treatment. Morphological alterations of H. pylori exposed to ZnLla for 120 min were assessed by transmission electron microscopy (TEM). Untreated bacteria appeared normal with intact cell wall and dense cytoplasm (Fig. 4C). The cells exposed to ZnLla lost their characteristic spiral shape and turned into transparent cocci. Moreover, ZnLla treatment induced vacuole-like swelling structures in the cell cytoplasm and membrane detachments between the outer and plasma membranes, accompanied by leakage of cytoplasmic contents (Fig. 4C). Taken together, these results indicate that ZnLla is able to selectively kill H. pylori via bacterial membrane disruption. These effects were also observed in free Lla- or liposomal Lla-treated H. pylori (36).
FIG 4.
ZnLla disrupts H. pylori cell membranes and generates reactive oxygen species. (A) Uptake of N-phenyl-1-naphthylamine (NPN) by H. pylori G27 cells after treatment of PBS (negative control) and 80 μg/ml polymyxin B (positive control), ZnLla, Lla, or ZnCl2. (B) The release of ATP from H. pylori cells after each treatment as shown in panel A. (C) Morphology alterations of H. pylori cells following exposure to PBS or ZnLla (80 μg/ml). (D) Fluorescence microscopic images (left) and bright-field images (right) of DCF+-labeled H. pylori stained with DCFDA after the incubation with PBS and 80 μg/ml polymyxin B or ZnLla. (E) The ratios of DCF+ cells incubated with each treatment calculated by manually counting under a microscope. Data represent medians ± SD of the results from three independent experiments.
In addition to primary lethal actions, some antibiotics have been shown to induce reactive oxygen species (ROS) generation that contributes to cell death. The antibacterial activity of the zinc ion has also been often attributed to ROS formation. We then performed 2′,7′-dichlorofluorescin-diacetate fluorometric assays to determine if ZnLla generated ROS in the bacteria. As shown in Fig. 4D, much higher levels of ROS were produced in polymyxin B-treated cells than the very low levels of ROS formed in the bacteria incubated with PBS, consistent with previous reports that polymyxin B induces ROS production in Acinetobacter baumannii (37) and Escherichia coli (38). As expected, H. pylori treated with ZnLla showed much higher fluorescent intensities, indicating larger amounts of ROS formation in the bacteria (Fig. 4D and E and S6). These data primarily suggested that the potent bactericidal effect of ZnLla on H. pylori also involved ROS generation.
ZnLla selectively kills H. pylori in vivo.
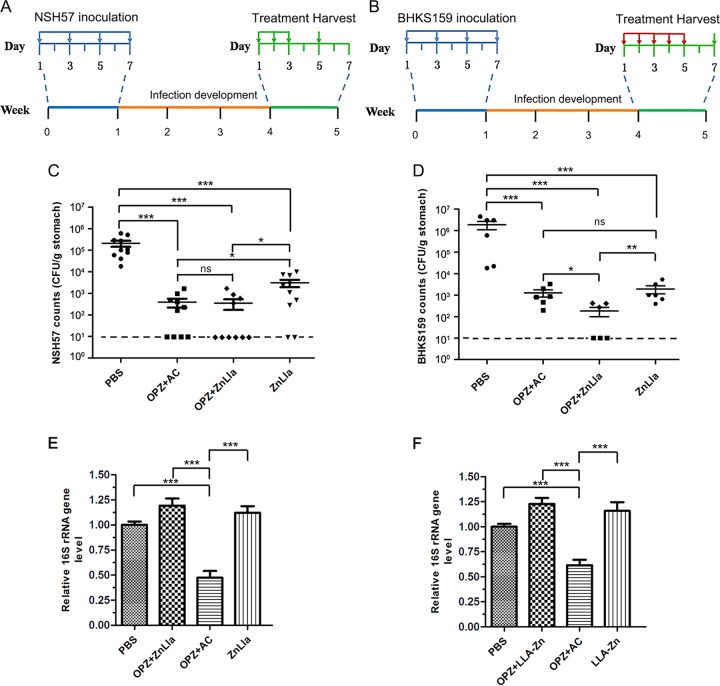
Next, we used H. pylori-infected mouse models to evaluate the therapeutic efficacy of ZnLla in vivo. We first infected mice with H. pylori NSH57 (39), a mouse-adapted derivative of the G27 strain, by oral gavage every other day for four times (Fig. 5A). Three weeks after inoculation, infected mice were divided into four groups and treated with PBS (control), triple therapy, ZnLla (monotherapy), or ZnLla plus omeprazole (dual therapy). In the study, therapeutic efficacy was evaluated by enumerating and comparing H. pylori counts in the mouse stomach. As shown in Fig. 5C, the dual therapy of ZnLla combined with omeprazole showed bacterial killing ability similar to that of the triple therapy, with a decrease in bacterial burden by ∼1,000× compared with the PBS group. Monotherapy with ZnLla gave a mean bacterial count 2 orders of magnitude lower than that in the PBS group, suggesting less efficacy than the dual therapy (Fig. 5C). As NSH57 is an antibiotic-sensitive strain, its mouse-infected model could not really mimic clinical drug resistance. We then constructed a second mouse model using a mouse-adapted BHKS159 strain domesticated from human clinical HP159 strain (with resistance to metronidazole, clarithromycin, and levofloxacin) to further determine if ZnLla could kill multidrug-resistant H. pylori strains in vivo. We found that after 5-day drug treatment, mice treated with ZnLla both as a monotherapy and dual therapy showed a >3-log reduction in bacterial burden compared with the PBS group (Fig. 5B and D). Importantly, the dual therapy caused a significant reduction in colony counts by ∼4.0 orders of magnitude, which was superior to the triple therapy reducing it by ∼3.1 orders of magnitude (Fig. 5D). These results suggested a high effectiveness of ZnLla in vivo against H. pylori infection.
FIG 5.
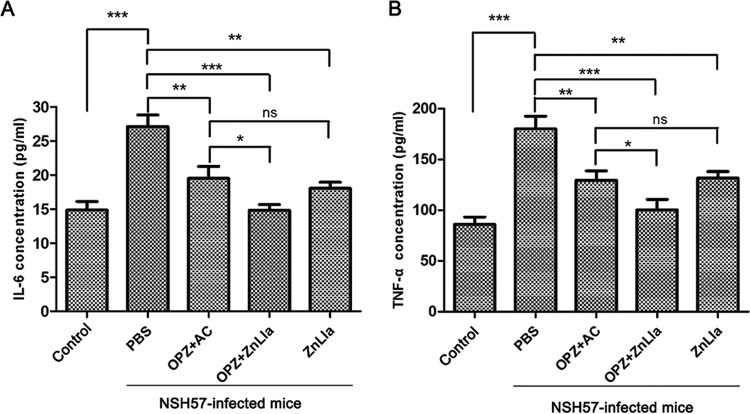
Anti-H. pylori efficacy in vivo. (A and B) The study protocols of H. pylori inoculation, infection development, and treatments in C57BL/6J mice. Strain NSH57 (drug sensitive) and BHKS159 (multidrug resistant) were used for two different mouse models, respectively. (C and D) Quantification of bacterial burden in the stomach of mice infected with H. pylori NSH57 (C) and BHKS159 (D) treated with PBS, triple therapy (omeprazole [OPZ]+AMX+CLR [AC]), omeprazole and ZnLla (OPZ+ZnLla), and ZnLla, respectively. Error bars represent the SD derived from 10 mice (NSH57 infected) or 6 mice (BHKS159 infected) per group. If no colonies were present, calculations were made using the limit of detection. (E and F) The killing effect of ZnLla against commensal bacteria was determined by measuring the bacterial load in the feces (E) and ileal contents (F) of mice after each treatment. The bacterial load was determined by quantitative real-time PCR. The 16S rRNA gene level was normalized to the tissue weight (n ≥ 6). All of the data are represented as average ± SD of the results from three independent experiments and analyzed by Student’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, no significant difference (P > 0.05).
In addition, the mono- or dual therapy of ZnLla had no significant effect on commensal bacterial load in the feces and ileal contents of mice (Fig. 5E and F), in good agreement with no growth inhibition of bacteria from 23 non-Helicobacter genera in vitro (Fig. 3). However, the triple treatment killed the commensal bacteria by half (Fig. 5E and F). These results collectively indicate that ZnLla could selective kill H. pylori both in vivo and in vitro.
ZnLla causes minor changes to gut microbiota.
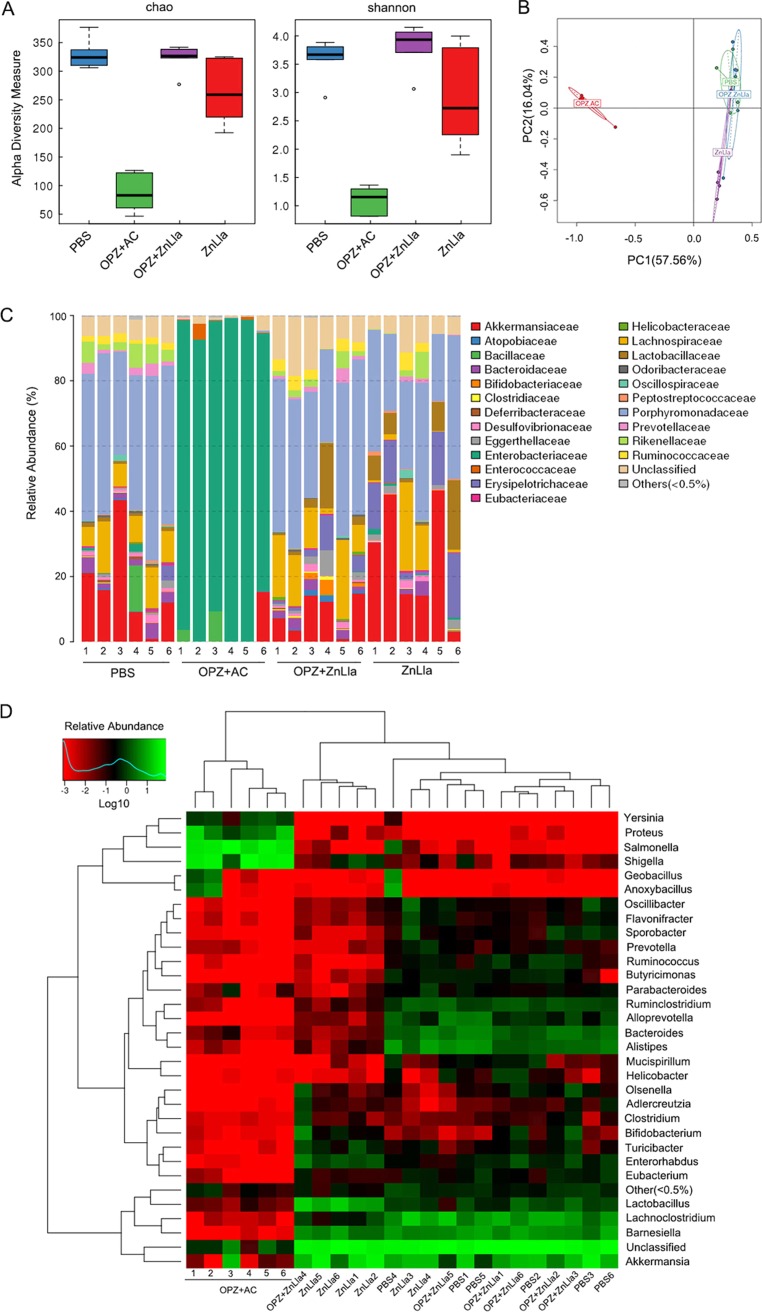
To characterize the changes in the gut microbiota after ZnLla or triple-therapy treatment, we collected stool samples from each group and employed 16S rRNA gene (V3–V4 region) sequencing to analyze the gut microbiota alterations. When data from all samples were pooled, we did not observe a statistically significant change in diversity between the microbiota of the PBS group and ZnLla mono- or dual-therapy group, as assessed by Chao1 and Shannon indices (Fig. 6A), which measure the total number of operational taxonomic units (OTUs) and, in case of the Shannon index, the richness, abundance, and evenness of the OTU distribution. However, a sharply significant decrease in diversity was evident after the triple therapy. We then used principal-component analysis (PCA) to compare relative distances of the microbial sequence data in each group. As shown in Fig. 6B, the triple-therapy group clustered away from other groups, while two ZnLla therapy groups were overall close to the PBS group, suggesting that these bacterial communities were more similar to each other than to that of the triple-therapy group. Minor differences in microbiota composition among the PBS and ZnLla mono- and dual-therapy groups were also demonstrated by the weighted and unweighted Unifrac analyses (Fig. S7). Thus, the α- and β-diversity analyses revealed that the compositions of microbiota remained similar after ZnLla treatment, whereas those after the triple-therapy treatment were distinctively changed.
FIG 6.
ZnLla causes minimal alterations in the diversity and composition of the gut microbiota. (A) Alpha-diversity estimates calculated from the sequenced data using Chao1 (left) and Shannon (right) indices for the four groups treated with PBS, triple therapy (OPZ+AC), omeprazole and ZnLla (OPZ+ZnLla), and ZnLla, respectively. The five lines, from bottom to top, are the minimum value, the first quartile, median, third quartile, and maximum values, and the abnormal value is shown as “o.” Statistical significance was calculated between the corresponding time points of the two groups. (B) PCA of the four groups based on OTU abundance. The number in brackets represents the contributions of principal components to differences among samples. A dot represents each sample. (C) Relative abundances of bacterial families in each sample receiving different treatments identified from the sequenced data. Data are clustered according to sample number of each group along the x axis. (D) Heat map comparing average abundances of species-level classification of the 16S rRNA gene sequences from each sample receiving different treatments.
We next compared the composition of the microbiota to identify bacterial taxa that differ among all groups. A bar plot analysis at the phylum level (Fig. S8) roughly showed that Bacteroidetes, Firmicutes, and Verrucomicrobia predominated in all samples from the PBS and ZnLla mono- or dual-therapy groups. Furthermore, the three groups were observed to display a similar microbiota composition from species bar plot and heat map analyses (Fig. 6C and D). However, in the triple-therapy groups, the microbiota exhibited a significant decline in the level of the above three phyla, and the Proteobacteria phylum (also family Enterobacteriaceae) underwent a large expansion in all mice, comprising nearly 90% of the total microbial abundance (Fig. S8). In particular, some bacterial families, including Porphyromonadaceae, Lachnospiraceae, Erysipelotrichaceae, Eggerthellaceae, and Lactobacillaceae, underwent apparent extinction in all mice after triple-therapy treatment (Fig. 6C), suggesting that the large-scale extinction of commensal species was accompanied by an overgrowth of Enterobacteriaceae. As shown in Fig. 6D, a few overgrown unique genera (Yersinia, Proteus, Salmonella, and Shigella) included in this bacterial family were found in the triple-therapy groups. Together, these findings indicated that compared with the triple-therapy treatment, ZnLla treatment causes only minor changes in the diversity and composition of the murine gut microbiota.
Immune response to ZnLla treatment.
To investigate whether ZnLla treatment can influence the immune response generated during H. pylori infection, the concentrations of selected proinflammatory cytokines (interleukin 1β, interleukin 6 [IL-6], IL-8, and tumor necrosis factor alpha [TNF-α]) in plasma obtained from each mouse were determined using commercial enzyme-linked immunosorbent assay (ELISA) kits. As shown in Fig. 7, H. pylori infection without treatment resulted in high secretion of IL-6 and TNF-α, in agreement with findings from previous studies (40). However, when infected mice were treated with the monotherapy or dual therapy of ZnLla, the levels of these two proinflammatory cytokines released to plasma were significantly reduced (P < 0.001) compared with those from the PBS control groups (Fig. 7A and B). In addition, dual therapy of ZnLla led to slightly lower levels of IL-6 and TNF-α than did the triple-therapy treatment (Fig. 7A and B), indicating that no acute toxicity was found in response to ZnLla treatment. Note that the concentrations of the other cytokines (IL-1β and IL-8) in plasma obtained from each mouse were not changed significantly (Fig. S9).
FIG 7.
Proinflammatory cytokine production of mice untreated/treated with ZnLla. (A and B) Concentrations of plasma IL-6 (A) and TNF-α (B) levels in uninfected mice (control) and infected mice receiving different treatments were determined by enzyme-linked immunosorbent assay (ELISA) kits. All of the data are represented as average ± SD of the results from three independent experiments and analyzed by Student’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, no significant difference (P > 0.05).
ZnLla toxicity evaluation in vivo.
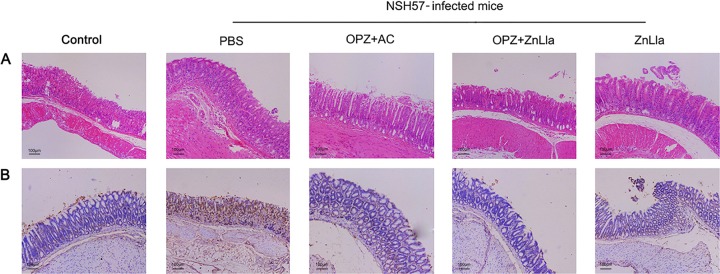
Last, the toxicity of ZnLla was further explored. No obvious change in animal body weight was observed following treatment with PBS, triple therapy, ZnLla, or omeprazole plus ZnLla (Fig. S3C), indicating the low toxicity of ZnLla in vivo. The longitudinal sections of gastric tissues obtained from the mice were collected and then stained with hematoxylin and eosin to evaluate the toxicity of ZnLla toward the stomach. Inflammation was significant in the gastric sections of H. pylori-infected mice treated with PBS, in which the ulcer crater and muscularis mucosae layer were heavily infiltrated with immune cells (Fig. 8A). However, the gastric tissues treated with ZnLla alone or combined with omeprazole maintained structural integrity with a clear layer of epithelial cells, similar to the gastric samples from the uninfected controls. Moreover, ZnLla administration did not increase the level of gastric epithelial apoptosis (Fig. 8B), as measured by terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay. The plasma levels of alanine aminotransferase, aspartate aminotransferase, creatinine, and urea nitrogen showed no significant change after ZnLla treatment (Table S2), verifying the lack of significant acute damage toward the liver and kidney. The low toxicity of ZnLla was further confirmed by ZnLla overdose (10 times the effective dosage) treatment. The absence of any detectable change in body weight (Fig. S3C), gastric histopathologic examination (Fig. S10), or blood biochemical index (Table S2) within a 5-day overdose treatment suggests that orally administered ZnLla is safe.
FIG 8.
Histological staining analysis of mice untreated/treated with ZnLla. (A and B) Representative images of H&E-stained (A) or TUNEL-stained (B) stomach from uninfected mice (control) and infected mice receiving different treatments. ZnLla treatment showed the same level of safety as the control (n ≥ 5, scale bar = 100 μm).
DISCUSSION
The development of drug resistance and detrimental side effects of the administered antibiotics are of serious concern in the treatment of H. pylori eradication. Clarithromycin-resistant H. pylori has been categorized by the World Health Organization as a “high priority” for research and development of new antibiotics (41). A novel anti-H. pylori agent should (i) show effective antibacterial action against both standard and clinical drug-resistant H. pylori strains, (ii) exert potent and selective anti-H. pylori activity and no effect on the gut microbiota, (iii) not induce the development of drug resistance, and (iv) possess wide safety. The present investigation was therefore designed to find and characterize potent anti-H. pylori molecules achieving these goals. In this study, we synthesized a novel anti-H. pylori agent, zinc linolenate, as a single bactericidal agent that can specifically target H. pylori in vivo and in vitro. Our results demonstrated a comparable antimicrobial efficacy of ZnLla monotherapy or significantly improved efficacy of dual therapy (ZnLla plus PPI) in reducing H. pylori bacterial loads in the stomach of drug-resistant H. pylori-infected mice compared with the current worldwide standard treatment, triple therapy. This in vivo anti-H. pylori activity of ZnLla was further evaluated with a new mouse model infected by a drug-resistant strain, BHKS159, which is more comprehensive than that infected by a drug-sensitive strain (Sydney strain 1 or NSH57) used in most reports. Thus, ZnLla may be suitable as a therapeutic agent for the treatment of drug-resistant H. pylori infection.
The use of two broad-spectrum antibiotics included in the triple therapy may disrupt the balance and diversity of gut microbiota, which is vital for bodily function. Also, accumulating evidence suggests that an unbalanced commensal microbiota is highly associated with dysregulated vaccine immune responses (42) and with the occurrence and progression of various diseases (43–45). Recent research has also shown that the efficacy of cancer immunotherapy with immune checkpoint antibodies can be diminished with the elimination of commensal bacteria by the administration of antibiotics (46–48). Therefore, the development of novel antibacterial agents that can selectively target and kill H. pylori with maintaining populations of commensal bacterial and microbiota stability is highly impending and important. In the current study, we demonstrated that ZnLla had selective antibacterial activities against H. pylori in vitro, exerting no antibacterial effects on other 23 bacteria examined (Fig. 3). In our H. pylori-infected mouse models, ZnLla had no effect on the population of commensal bacteria and the diversity and composition of microbiota, whereas the triple therapy killed nearly half of the commensal bacteria and caused severe microbiota alterations (Fig. 6). Therefore, ZnLla could be used as a selective and effective antibacterial agent against H. pylori in the stomach without harming the gut microbiota.
Apart from the undesired toxicity, the triple therapy is also prone to the progressive emergence of drug resistance that weakens its therapeutic efficacy against H. pylori-induced gastric diseases (49, 50). The superior anti-H. pylori activity conferred by ZnLla can be explained by the unique capability of ZnLla to disrupt membrane integrity and produce intracellular ROS. The two highly destructive actions for bacterial killing dramatically reduced the opportunity for drug resistance to emerge (51, 52). As such, ZnLla did not show any tendency to develop resistance against H. pylori (Fig. 2B). The antimicrobial activity of ZnLla can result from properties of the metal and fatty acid components in the metal-organic framework. As H. pylori is sensitive to unsaturated free fatty acids through their incorporation into phospholipids (53), cell membrane disruption and leakage of cytoplasmic contents caused by ZnLla treatment could be attributed to the action of the Lla component. Moreover, after ZnLla solubilizes the bacterial membrane, the molecules will also be directly delivered to the bacterial cytoplasm with interference with bacterial intracellular pathways. As a reservoir of Zn2+, ZnLla provided their gradual release and also produced intracellular ROS, detected by the dichloro-dihydro-fluorescein diacetate (DCFH-DA) fluorometric assays, which also played a role in the inhibitory effect of ZnLla on H. pylori growth. Since the two components may affect H. pylori growth at multiple levels, other mechanisms of antibacterial action of ZnLla remain to be clarified.
In addition to the high efficacy and specificity of ZnLla in killing H. pylori in vivo and in vitro, the toxicity and safety evaluations also show that ZnLla is safe. Treatment with high doses of ZnLla had no effect on potential cytotoxic effect, mouse body weight, or gastric mucosal integrity. Further, treatment of mice with ZnLla did not induce the host immune response. Interestingly, ZnLla had significantly reduced the serum levels of H. pylori-induced proinflammatory cytokines IL-6 and TNF-α compared with the PBS control treatment. Especially, their serum levels in the dual-therapy groups including ZnLla and omeprazole treatment were slightly lower than those in the triple-therapy groups, suggesting that ZnLla is superior to the conventional antibiotics in the case of suppressing host immune responses. The anti-inflammatory effects of ZnLla result from actions of the components, zinc (54) and Lla (55), which were both reported to play roles in gastric mucosal protection through various pathways (56–58). It is generally believed that H. pylori stimulates cytokine production by epithelial cells that recruit and activate immune and inflammatory cells, and it contributes to the disease pathogenesis (59). As such, inhibition of the proinflammatory response by ZnLla, as observed here, is expected to exert cytoprotective effects on the gastric lining and halt the progression of gastric disease induced by H. pylori infection.
MATERIALS AND METHODS
Chemicals, bacterial strains, and culture media.
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA) or Sinopharm Chemical Reagent Co. Ltd. (Shanghai, China) and used as received, unless otherwise specified. H. pylori strains 26695 (60), G27, NSH57 (39), and MSD132 (61) and 16 clinical isolates were used in this study and were routinely cultured either in brain heart infusion (BHI) broth (Becton, Dickinson, Sparks, MD, USA) medium containing 10% fetal calf serum (FCS) or in Columbia blood agar (Sigma-Aldrich) plates containing 5% FCS under a microaerophilic environment (10% CO2, 85% N2, and 5% O2 and 90% relative humidity) at 37°C for 48 to 72 h in a double-gas CO2 incubator (model CB160; Binder, Germany). Standard strains G27, NSH57, and MSD132 were kindly provided by Nina R. Salama, Fred Hutchinson Cancer Research Center, USA. A total of 16 clinical strains of H. pylori were isolated from biopsy samples from 16 patients with gastric cancer or gastric ulcer, obtained from the First Affiliated Hospital of Nanchang University and the First Affiliated Hospital of Nanjing Medical University, China, using standard protocols. The strains were identified based on colony appearance, Gram staining, and positive reactions in the rapid urease test. Staphylococcus aureus ATCC 6538, Pseudomonas aeruginosa PAO1, Enterococcus faecalis FA2-2, Salmonella enterica subsp. enterica serovar Typhimurium ATCC 14028, Acinetobacter baumannii ATCC 19606, Bacillus subtilis 168, Enterococcus faecium ATCC 19434, Proteus mirabilis ATCC 29906, Listeria monocytogenes EGD-e, Enterobacter cloacae ATCC 13047, Escherichia coli MG1655, Klebsiella pneumoniae ATCC 35657, Lactococcus lactis MG1363, Stenotrophomonas maltophilia ATCC 51331, Morganella morganii ATCC 25830, and Bacillus cereus ATCC 14579 were cultured aerobically in BHI broth at 37°C. Moraxella catarrhalis ATCC 25238, Streptococcus pneumoniae ATCC 49619, and Aggregatibacter actinomycetemcomitans D7S-1 were cultured in BHI broth at 37°C with 5% CO2. Haemophilus influenzae ATCC 49766 was grown in BHI broth supplemented with 10 μg/ml hemin and 10 μg/ml NAD at 37°C with 5% CO2. Neisseria gonorrhoeae ATCC 19424 was grown in BHI broth supplemented with 5% FCS at 37°C with 5% CO2. Mycobacterium smegmatis MC2 155 was grown at 37°C in Middlebrook 7H9 broth (Becton, Dickinson) supplemented with 0.2% glycerol and 0.05% Tween 80. Campylobacter jejuni NCTC11168 was cultured as H. pylori.
Synthesis of zinc-linolenic acid complex.
Ten milliliters of ZnCl2 (0.163 g, 1.2 mmol) was heated at 165°C in the reactor kettle for 30 min and allowed to cool. A 10-ml solution of NaOH (0.08 g, 2 mmol) was slowly dripped into α-linolenic acid solution (0.557 g, 2 mmol in 10 ml water), and the entire solution was stirred at 60°C for 6 h in nitrogen. The mixture was then added into the activated ZnCl2 described above, reacted at 60°C for 12 h in nitrogen, and cooled overnight. Finally, pure Zn-linolenic acid complex (ZnLla) was obtained after dialysis (molecular weight cutoff [MWCO], 1,000), washing with water, and freeze-drying to remove unreacted free ions and Lla. The faint-yellow precipitate was collected, weighed (0.35g), and dissolved in ethanol for further experiments. ZnLla was characterized using a MALDI-TOF/TOF mass spectrometer (ultrafleXtreme; Bruker Daltonic, Germany), nuclear magnetic resonance (NMR) platform (Avance 500 MHz; Bruker, Switzerland), FTIR spectroscopy (Tensor27; Bruker, USA), UV-visible spectrophotometer (UH5300, Hitachi, Japan), and zinc ion tester (ZN-1A; Haiheng Electronic Technology, Shanghai, China).
Antibacterial activity.
Antibacterial activities were evaluated by examining the MIC, which was determined by a broth microdilution assay, as follows. Twofold serial dilutions of the test compounds were prepared in a 96-well microtiter plate containing 100 μl of BHI broth supplemented with 10% FCS. A 2-day-old H. pylori liquid culture was diluted 10 times in BHI broth and was inoculated into each well to give a final concentration of 5 × 105 to 1 × 106 CFU/ml. The plates were incubated for 3 days in a microaerophilic atmosphere at 37°C. After incubation, the plates were examined visually, and the MIC was determined to be the lowest concentration which resulted in no turbidity. For quality control and comparative analyses, the antibiotic metronidazole was also tested with each batch of ZnLla. MICs for aerobic bacteria were determined by the agar dilution streak method recommended in Clinical and Laboratory Standards Institute document M07-A7 (62). The broth was diluted with saline and applied to plates, delivering a final concentration of approximately 105 CFU/spot. The minimum bactericidal concentration (MBC) is determined as the lowest concentration of antibacterial agent that reduces the viability of the initial bacterial inoculum by 99.9%.
Drug resistance study.
Fresh BHI containing 10% FCS was inoculated with H. pylori G27 and incubated at 37°C under continuous shaking. Cells were grown to log phase and dispensed into 96-well microtiter plates with 100 μl fresh medium per well (adjusted to approximately 1 × 106 cells). Metronidazole and ZnLla were added at concentrations of 0.25×, 0.5×, 1×, 2×, and 4× the MIC. After a 2-day incubation at 37°C under continuous shaking, growth was determined with a microplate reader at an optical density at 600 nm (OD600), and cells from the second highest concentration showing visible growth were used to inoculate the subsequent culture. This procedure was repeated for up to 30 cycles (60 days), and alterations in MICs after every 2 cycles during the course of continued exposure were determined.
ZnLla in vitro cytotoxicity study.
Cell viability was evaluated using the Cell Counting Kit-8 assay (Sigma-Aldrich). The human gastric carcinoma cell line AGS (RRID:CVCL_0139) and the immortalized human gastric epithelial cell line GES-1 (RRID:CVCL_EQ22) were purchased from the Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences (SIBS), Chinese Academy of Sciences (CAS; Shanghai, China) and cultured in RPMI 1640 medium (Gibco, Grand Island, NY, USA) supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum (FBS) at 37°C in a humidified 5% CO2 atmosphere. For the study, 1 × 104 cells per well were grown in 96-well culture plates. After an overnight culture, cells were incubated with ZnLla, Lla, or ZnCl2 at predetermined concentrations for 24 h and were then treated with CCK-8 reagent for assessment of cytotoxicity. Untreated cells incubated with PBS served as a control.
Outer membrane permeability assay.
The outer membrane permeabilization was assessed by measuring the uptake of N-phenyl-1-naphthylamine (NPN) (63). An overnight culture of H. pylori was harvested, washed, and adjusted to an OD600 of 0.3. Approximately 1 × 107 bacterial cells were incubated with 100 μM NPN in 200 μl of PBS. Then, polymyxin B, ZnLla, Lla, or ZnCl2 (16, 80, and 400 μg/ml for each) was added and mixed with the treated bacteria. PBS was used as a negative control. Fluorescence measurements were taken after shaking at each time point, using a Synergy HTX multimode microplate reader (BioTek Instruments, Winooski, VT, USA) at an excitation wavelength of 350 nm and an emission wavelength of 420 nm. The NPN assays were performed at room temperature and repeated three times, and the results are expressed as relative fluorescence units.
Plasma membrane permeability assay.
The release of ATP from cells was determined as an indicator of plasma membrane permeability after growing H. pylori G27 to an OD600 of 0.3. Approximately 1 × 107 bacterial cells were treated with polymyxin B, ZnLla, ZnCl2, or Lla (16, 80, and 400 μg/ml) for 2 h. PBS was used as a negative control. After treatment, each bacterial suspension was centrifuged, the supernatant was separated from the cell pellet, and the cell pellet was resuspended in 1 ml of PBS. A 100-μl aliquot of BacTiter-Glo reagent in the microbial cell viability assay kit (Promega) with 500 μg of lysostaphin was added to 100 μl of either the supernatant or resuspended cell pellet in an OptiPlate-96F solid-bottom black plate. The mixture was shaken for 2 min at 200 rpm, and luminescence was determined on a Packard Fusion plate reader. The concentration of ATP was determined by comparison to the luminescence obtained from the addition of the BacTiter-Glo reagent to an ATP standard curve.
ROS generation by ZnLla.
The ROS generated by ZnLla was analyzed using 2′,7′-dichlorofluorescin-diacetate (DCFDA) as a fluorescent probe. DCFDA upon enzymatic hydrolysis by intracellular esterases forms nonfluorescent 2′,7′-dichlorodihydrofluorescein (DCF-H), which is subsequently oxidized to highly fluorescent DCF in the presence of ROS. Therefore, the degree of DCF fluorescence intensity gives a quantitative evaluation of the amount of ROS formed in cells. Approximately 1 × 107 H. pylori G27 cells were incubated with 10 μM DCFDA for 1 h at 37°C. After incubation, the bacteria were pelleted by centrifugation and washed two times to remove the DCFDA outside the cells. Then, the cleaned bacterial cells were exposed to ZnLla or polymyxin B (16, 80, and 400 μg/ml) for 2 h. PBS was used as a negative control. The fluorescence intensity of the generated DCF was then determined at excitation and emission wavelengths of 488 and 535 nm, respectively. The percentage of bacteria exhibiting fluorescent signal was estimated by manual counting under a fluorescence microscope (MTB2004; Zeiss, Carl Zeiss, Germany).
Transmission electron microscopy.
The effects of ZnLla on the structure of H. pylori were examined with TEM. Sample processing was performed essentially according to a published procedure (64). Briefly, an overnight culture of H. pylori G27 was treated with ZnLla (80 μg/ml) and incubated for 2 h. PBS was used as a negative control. Samples were centrifuged at 3,000 × g for 5 min and bacterial pellets fixed by resuspending in 2% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4). Bacterial pellets were then embedded in 2% agarose and postfixed with 1% osmium tetroxide overnight at room temperature. After washing, samples were dehydrated multiple times in increasing concentrations of ethanol and embedded in Durcupan resin (Sigma-Aldrich). Fifty-five-nanometer sections were examined using a JEM-1200 transmission electron microscope (JEOL; Akishima, Tokyo, Japan) equipped with a 4-K Eagle digital camera (FEI, Hillsboro, OR, USA).
Mouse-adapted BHKS159 strain.
BHKS159 is a mouse-adapted H. pylori strain obtained after multiple passages through mice infected by human clinical isolate HP159 with resistance to metronidazole, clarithromycin, and levofloxacin. Six-week-old specific-pathogen-free female C57BL/6 mice were used for this study. Animal studies were approved by the Institutional Animal Care and Use Committee (IACUC) of Nanjing Medical University (NJMU) and conducted in accordance with the international standards for animal welfare and institutional guidelines. Mice were obtained from the Animal Core Facility at NJMU and housed at the same place. First, each of five C57BL/6 female mouse received 0.3 ml of 1 × 109 CFU/ml H. pylori HP159 in BHI broth administered intragastrically through oral gavage every 48 h and repeated four times, and the infection was allowed to develop for 3 weeks. Then, mice were sacrificed, and the output from one mouse was pooled (passage 1), isolated by Columbia blood agar plates, amplified by growth in liquid culture, and used to inoculate four new mice (passage 2). For the third passage, 10 mice were infected. These mice were sacrificed at 3 weeks, and 9 of the 10 inoculated mice were found to have detectable infection, with bacterial loads ranging from 3.0 to 5.5 log CFU/g. Two different strains from two mice, giving bacterial loads of >5.0 log CFU/g, were amplified and used to inoculate 10 new mice (passage 4). At 3 weeks, the mice were sacrificed. All mice were infected with bacterial loads ranging from 4.5 to 6.2 log CFU/g. Single-colony isolates were obtained from the mice with 6.2 log CFU/g, and one isolate, designated BHKS159, was used for further study. After each isolation, the H. pylori strains were identified based on Gram staining and positive reactions in the rapid urease test.
Anti-H. pylori efficacy in vivo.
Six-week-old specific-pathogen-free female C57BL/6 mice were used for this study. Each mouse received 0.3 ml of 1 × 109 CFU/ml H. pylori NSH57 in BHI broth administered intragastrically through oral gavage every 48 h, repeated four times (on days 1, 3, 5, and 7, respectively), and the infection was allowed to develop for 3 weeks. The mice were randomly assigned to four treatment groups (n = 10) to receive ZnLla alone, ZnLla with omeprazole (a proton pump inhibitor), triple therapy, or PBS. Mice were first administered omeprazole (a proton pump inhibitor) through oral gavage at a dose of 400 μmol/kg of body weight, followed by a lag time of 30 min before administration of the assigned treatments. ZnLla (24 mg/kg) and the triple-therapy formulation (28.5 mg/kg amoxicillin and 14.3 mg/kg clarithromycin) were administered once daily for a consecutive 3 days by oral gavage. The control group received an equivalent volume of PBS. Forty-eight hours after the last treatment, mice were killed, and the stomach, the liver, the spleen, the kidney, and the ileum were removed from the abdominal cavity. The stomach was cut along the greater curvature, and the gastric content was removed and rinsed with PBS. The stomachs were cut into two longitudinal sections, and each section was weighed. The sections were used for assessment of bacterial colonization and histology/epithelial apoptosis. For bacterial colonization, a gastric tissue section was suspended in 1 ml PBS and homogenized for H. pylori recovery. The homogenate was serially diluted and spotted onto a Columbia agar plate containing vancomycin (20 μg/ml), amphotericin (2 μg/ml), and bacitracin (30 μg/ml). The plates were then incubated at 37°C under microaerobic conditions for 5 days, and bacterial colonies were enumerated, and adjusted for dilutions, and expressed as CFU per gram of stomach. A new mouse model was also constructed using mouse-adapted multidrug-resistant BHKS159 strain to further assess anti-H. pylori efficacy in vivo. A five-day drug treatment (orally administration once daily for 5 consecutive days) was used in this model to determine if ZnLla has in vivo killing efficacy superior to that of the triple-therapy approach.
In vivo toxicity study.
Forty-eight hours after the last oral administration, H. pylori NSH57-infected mice receiving different treatments and uninfected mice (control) were killed, and the stomachs were removed for histological analysis. The longitudinal sections of gastric tissue were fixed in buffered paraffin and embedded in paraffin wax. A section of about 5 mm was stained with hematoxylin and eosin (H&E) to analyze tissue inflammation. Epithelial cell apoptosis was evaluated by a terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay (R&D Systems, Minneapolis, MN, USA). Sections were visualized by Hamamatsu NanoZoomer 2.0HT and the images processed using the NDP viewing software. Blood was collected from the orbital sinus for the analysis of proinflammatory cytokines (IL-1β, TNF-α, IL-6, and IL-8) levels determined by enzyme-linked immunosorbent assay (ELISA) kits (R&D Systems). ZnLla overdose (240 mg/kg, 10-fold effective dosage) treatment was also performed in uninfected mice to further evaluate ZnLla toxicity in vivo. Mouse body weight was monitored during the experiment period by weighing the mice daily. The blood was collected for the analysis of alanine aminotransferase (ALT), aspartate aminotransferase (AST), creatinine, and urea nitrogen levels, as described by Saldanha et al. (65). The stomach, small intestine, liver, and kidney were kept in formalin for H&E staining.
The killing effect of ZnLla against commensal bacteria was determined by measuring the bacterial load in the feces and ileal contents of H. pylori NSH57-infected mice receiving different treatments. The bacterial load was determined by quantitative real-time PCR using a protocol originally described by Stefka et al. (66). Total genomic DNA was extracted from the fecal samples (collected 24 hours after the last oral administration) and the contents of the cecum (removed from the killed mouse) using the FastDNA spin kit for soil (MP Biomedicals, Solon, OH, USA), according to the manufacturer’s instructions. The bacterial load was quantified against a standard curve derived from a pCR4TOPO-TA vector containing a nearly full-length copy of the 16S rRNA gene from a member of the Porphyromonadaceae. Bacterial DNA was amplified with universal primers 8F and 338R using the PowerUp SYBR green master mix (Applied Biosystems) and the StepOnePlus system (Applied Biosystems). Samples were denatured at 95°C for 2 minutes and cycled 40 times through 95°C for 20 s, 58°C for 20 s, and 72°C for 30 s, and then denaturation curves were determined from 58°C through 95°C. All quantitative PCR (qPCR) assays were conducted in an Applied Biosystems 7500 real-time PCR system. The results were normalized to ileal content/fecal weight.
Bacterial community composition and diversity.
The fecal bacterial community composition was determined by partial 16S rRNA gene sequencing of the extracted community. The V4 hypervariable regions of the 16S rRNA gene were amplified with the primers 515-F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806-R (5′-GGACTACHVGGGTWTCTAAT-3′), as previously described (67). These primers included Illumina adapters and error-correcting 12-bp barcodes. PCR was performed in triplicate for each sample in a total reaction volume of 20 μl. The PCR products were checked using 2% agarose gel electrophoresis, purified using the Axy-Prep DNA gel extraction kit, and quantified using the fluorescence quantitation (Qubit 2.0 fluorometer) and Qubit double-stranded DNA (dsDNA) high-sensitivity (HS) assay kit (Invitrogen, Carlsbad, CA, USA). The purified amplicons from a total of 24 samples of four groups (6 samples/group) were sequenced on an Illumina MiSeq PE250 sequencer and then processed and analyzed using the programs USEARCH (version 8.1.1831) (68) and QIIME (version 1.9.1) (69). Chimeric reads were filtered out from the sequenced data using the Gold ChimeraSlayer reference database (version microbiomutil-r20110519) (70), and the filtered sequences that shared at least 97% pairwise nucleotide identity were binned into operational taxonomic units (OTUs). To obtain species-level classification for the clustered OTUs, a custom reference database built from the NCBI 16S rRNA sequence and taxonomy database was used. Taxonomic assignments were performed using the blast method of QIIME’s assign_taxonomy.py script. Alpha-diversity estimates were calculated using Chao1 and Shannon indices. Phylogeny-based unweighted and weighted UniFrac distance matrices (71) were calculated using QIIME and visualized using the function “aheatmap” in the Nonnegative Matrix Factorization (NMF) package of the software R (72) to measure beta diversity. Principal-component analysis (PCA) was performed with the package “ade4” implemented in the statistical software R (version 3.1.1; http://www.r-project.org/) to display the differences in OTU compositions in different samples. Heatmaps were generated with the package “gplots” implemented in the software R using average species-level OTU counts that were normalized, log transformed, and offset by one. The data were filtered for species that were present in at least 90% of all samples with an overall relative variance as indicated in the figure legends to allow better visualization.
Statistical analysis.
Statistical analyses were performed and graphs generated using Microsoft Excel and the GraphPad 5.2 software. The results are represented as means ± standard error of the mean (SEM). Statistical analyses were performed using Student’s t test. The values are significant at a P value of <0.05.
Data availability.
The raw sequencing data in this paper are available at the NCBI Sequence Read Archive (SRA) under BioProject accession number PRJNA505765.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by The National Key R&D Programs of China (grant 2018YFC0311003 to H.B. and grant 2016YFC1302201 to Y.X.), The National Science Foundation of the Jiangsu Higher Education Institutions of China (grant 18KJA310002 to H.B.), and the Jiangsu Specially-Appointed Professor Program of China (to H.B.).
We thank Nina Salama and Karen Ottemann for providing H. pylori strains. We also thank Zining Cui, Ming Zhao, and Kui Hong for helpful discussions.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00004-19.
REFERENCES
- 1.Fock KM, Graham DY, Malfertheiner P. 2013. Helicobacter pylori research: historical insights and future directions. Nat Rev Gastroenterol Hepatol 10:495–500. doi: 10.1038/nrgastro.2013.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choi IJ, Kook MC, Kim YI, Cho SJ, Lee JY, Kim CG, Park B, Nam BH. 2018. Helicobacter pylori therapy for the prevention of metachronous gastric cancer. N Engl J Med 378:1085–1095. doi: 10.1056/NEJMoa1708423. [DOI] [PubMed] [Google Scholar]
- 3.Oh S, Kim N, Kwon JW, Shin CM, Choi YJ, Lee DH, Jung HC. 2016. Effect of Helicobacter pylori eradication and ABO genotype on gastric cancer development. Helicobacter 21:596–605. doi: 10.1111/hel.12317. [DOI] [PubMed] [Google Scholar]
- 4.Leung WK, Wong IOL, Cheung KS, Yeung KF, Chan EW, Wong AYS, Chen L, Wong ICK, Graham DY. 2018. Effects of Helicobacter pylori treatment on incidence of gastric cancer in older individuals. Gastroenterology 155:67–75. doi: 10.1053/j.gastro.2018.03.028. [DOI] [PubMed] [Google Scholar]
- 5.Chey WD, Leontiadis GI, Howden CW, Moss SF. 2017. ACG clinical guideline: treatment of Helicobacter pylori infection. Am J Gastroenterol 112:212–239. doi: 10.1038/ajg.2016.563. [DOI] [PubMed] [Google Scholar]
- 6.Alfizah H, Norazah A, Hamizah R, Ramelah M. 2014. Resistotype of Helicobacter pylori isolates: the impact on eradication outcome. J Med Microbiol 63:703–709. doi: 10.1099/jmm.0.069781-0. [DOI] [PubMed] [Google Scholar]
- 7.Graham DY, Fischbach L. 2010. Helicobacter pylori treatment in the era of increasing antibiotic resistance. Gut 59:1143–1153. doi: 10.1136/gut.2009.192757. [DOI] [PubMed] [Google Scholar]
- 8.Wang ZJ, Chen XF, Zhang ZX, Li YC, Deng J, Tu J, Song ZQ, Zou QH. 2017. Effects of anti-Helicobacter pylori concomitant therapy and probiotic supplementation on the throat and gut microbiota in humans. Microb Pathog 109:156–161. doi: 10.1016/j.micpath.2017.05.035. [DOI] [PubMed] [Google Scholar]
- 9.Dethlefsen L, Huse S, Sogin ML, Relman DA. 2008. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol 6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ianiro G, Tilg H, Gasbarrini A. 2016. Antibiotics as deep modulators of gut microbiota: between good and evil. Gut 65:1906–1915. doi: 10.1136/gutjnl-2016-312297. [DOI] [PubMed] [Google Scholar]
- 11.Rajani C, Jia W. 2018. Disruptions in gut microbial-host co-metabolism and the development of metabolic disorders. Clin Sci (Lond) 132:791–811. doi: 10.1042/CS20171328. [DOI] [PubMed] [Google Scholar]
- 12.Vuong HE, Yano JM, Fung TC, Hsiao EY. 2017. The microbiome and host behavior. Annu Rev Neurosci 40:21–49. doi: 10.1146/annurev-neuro-072116-031347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de la Fuente-Nunez C, Meneguetti BT, Franco OL, Lu TK. 2018. Neuromicrobiology: how microbes influence the brain. ACS Chem Neurosci 9:141–150. doi: 10.1021/acschemneuro.7b00373. [DOI] [PubMed] [Google Scholar]
- 14.Plunkett CH, Nagler CR. 2017. The influence of the microbiome on allergic sensitization to food. J Immunol 198:581–589. doi: 10.4049/jimmunol.1601266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jie Z, Xia H, Zhong SL, Feng Q, Li S, Liang S, Zhong H, Liu Z, Gao Y, Zhao H, Zhang D, Su Z, Fang Z, Lan Z, Li J, Xiao L, Li J, Li R, Li X, Li F, Ren H, Huang Y, Peng Y, Li G, Wen B, Dong B, Chen JY, Geng QS, Zhang ZW, Yang H, Wang J, Wang J, Zhang X, Madsen L, Brix S, Ning G, Xu X, Liu X, Hou Y, Jia H, He K, Kristiansen K. 2017. The gut microbiome in atherosclerotic cardiovascular disease. Nat Commun 8:845. doi: 10.1038/s41467-017-00900-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuo T, Kamm MA, Colombel JF, Ng SC. 2018. Urbanization and the gut microbiota in health and inflammatory bowel disease. Nat Rev Gastroenterol Hepatol 15:440–452. doi: 10.1038/s41575-018-0003-z. [DOI] [PubMed] [Google Scholar]
- 17.Tilg H, Adolph TE, Gerner RR, Moschen AR. 2018. The intestinal microbiota in colorectal cancer. Cancer Cell 33:954–964. doi: 10.1016/j.ccell.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 18.Cartron ML, England SR, Chiriac AI, Josten M, Turner R, Rauter Y, Hurd A, Sahl HG, Jones S, Foster SJ. 2014. Bactericidal activity of the human skin fatty acid cis-6-hexadecanoic acid on Staphylococcus aureus. Antimicrob Agents Chemother 58:3599–3609. doi: 10.1128/AAC.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamashita S, Igarashi M, Hayashi C, Shitara T, Nomoto A, Mizote T, Shibasaki M. 2015. Identification of self-growth-inhibiting compounds lauric acid and 7-(Z)-tetradecenoic acid from Helicobacter pylori. Microbiology 161:1231–1239. doi: 10.1099/mic.0.000077. [DOI] [PubMed] [Google Scholar]
- 20.Matsui H, Takahashi T, Murayama SY, Kawaguchi M, Matsuo K, Nakamura M. 2017. Protective efficacy of a hydroxy fatty acid against gastric Helicobacter infections. Helicobacter 22:e12430. doi: 10.1111/hel.12430. [DOI] [PubMed] [Google Scholar]
- 21.Yoon BK, Jackman JA, Valle-Gonzalez ER, Cho NJ. 2018. Antibacterial free fatty acids and monoglycerides: biological activities, experimental testing, and therapeutic applications. Int J Mol Sci 19:E1114. doi: 10.3390/ijms19041114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun CQ, O’Connor CJ, Roberton AM. 2003. Antibacterial actions of fatty acids and monoglycerides against Helicobacter pylori. FEMS Immunol Med Microbiol 36:9–17. doi: 10.1016/S0928-8244(03)00008-7. [DOI] [PubMed] [Google Scholar]
- 23.Thamphiwatana S, Gao W, Obonyo M, Zhang L. 2014. In vivo treatment of Helicobacter pylori infection with liposomal linolenic acid reduces colonization and ameliorates inflammation. Proc Natl Acad Sci U S A 111:17600–17605. doi: 10.1073/pnas.1418230111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson L, Cockayne A, Spiller RC. 1994. Inhibitory effect of polyunsaturated fatty acids on the growth of Helicobacter pylori: a possible explanation of the effect of diet on peptic ulceration. Gut 35:1557–1561. doi: 10.1136/gut.35.11.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Correia M, Michel V, Matos AA, Carvalho P, Oliveira MJ, Ferreira RM, Dillies MA, Huerre M, Seruca R, Figueiredo C, Machado JC, Touati E. 2012. Docosahexaenoic acid inhibits Helicobacter pylori growth in vitro and mice gastric mucosa colonization. PLoS One 7:e35072. doi: 10.1371/journal.pone.0035072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chylewska A, Biedulska M, Sumczynski P, Makowski M. 2018. Metallopharmaceuticals in therapy–a new horizon for scientific research. Curr Med Chem 25:1729–1791. doi: 10.2174/0929867325666171206102501. [DOI] [PubMed] [Google Scholar]
- 27.Renfrew AK. 2014. Transition metal complexes with bioactive ligands: mechanisms for selective ligand release and applications for drug delivery. Metallomics 6:1324–1335. doi: 10.1039/c4mt00069b. [DOI] [PubMed] [Google Scholar]
- 28.Jansen J, Karges W, Rink L. 2009. Zinc and diabetes–clinical links and molecular mechanisms. J Nutr Biochem 20:399–417. doi: 10.1016/j.jnutbio.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 29.Hoque KM, Binder HJ. 2006. Zinc in the treatment of acute diarrhea: current status and assessment. Gastroenterology 130:2201–2205. doi: 10.1053/j.gastro.2006.02.062. [DOI] [PubMed] [Google Scholar]
- 30.Matsukura T, Tanaka H. 2000. Applicability of zinc complex of l-carnosine for medical use. Biochemistry (Mosc) 65:817–823. [PubMed] [Google Scholar]
- 31.Battistini F, Cordero C, Urcuyo FG, Rojas RF, Ollague W, Zaias N. 1983. The treatment of dermatophytoses of the glabrous skin: a comparison of undecylenic acid and its salt versus tolnaftate. Int J Dermatol 22:388–389. doi: 10.1111/j.1365-4362.1983.tb01215.x. [DOI] [PubMed] [Google Scholar]
- 32.Amin M, Iqbal MS, Hughes RW, Khan SA, Reynolds PA, Enne VI, Sajjad Ur R, Mirza AS. 2010. Mechanochemical synthesis and in vitro anti-Helicobacter pylori and uresase inhibitory activities of novel zinc(II)-famotidine complex. J Enzyme Inhib Med Chem 25:383–390. doi: 10.3109/14756360903179518. [DOI] [PubMed] [Google Scholar]
- 33.Pati R, Sahu R, Panda J, Sonawane A. 2016. Encapsulation of zinc-rifampicin complex into transferrin-conjugated silver quantum-dots improves its antimycobacterial activity and stability and facilitates drug delivery into macrophages. Sci Rep 6:24184. doi: 10.1038/srep24184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gerrits MM, van Vliet AH, Kuipers EJ, Kusters JG. 2006. Helicobacter pylori and antimicrobial resistance: molecular mechanisms and clinical implications. Lancet Infect Dis 6:699–709. doi: 10.1016/S1473-3099(06)70627-2. [DOI] [PubMed] [Google Scholar]
- 35.Savoldi A, Carrara E, Graham DY, Conti M, Tacconelli E. 2018. Prevalence of antibiotic resistance in Helicobacter pylori: a systematic review and meta-analysis in World Health Organization regions. Gastroenterology 155:1372–1382.e17. doi: 10.1053/j.gastro.2018.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Obonyo M, Zhang L, Thamphiwatana S, Pornpattananangkul D, Fu V, Zhang L. 2012. Antibacterial activities of liposomal linolenic acids against antibiotic-resistant Helicobacter pylori. Mol Pharm 9:2677–2685. doi: 10.1021/mp300243w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sampson TR, Liu X, Schroeder MR, Kraft CS, Burd EM, Weiss DS. 2012. Rapid killing of Acinetobacter baumannii by polymyxins is mediated by a hydroxyl radical death pathway. Antimicrob Agents Chemother 56:5642–5649. doi: 10.1128/AAC.00756-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dong TG, Dong S, Catalano C, Moore R, Liang X, Mekalanos JJ. 2015. Generation of reactive oxygen species by lethal attacks from competing microbes. Proc Natl Acad Sci U S A 112:2181–2186. doi: 10.1073/pnas.1425007112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baldwin DN, Shepherd B, Kraemer P, Hall MK, Sycuro LK, Pinto-Santini DM, Salama NR. 2007. Identification of Helicobacter pylori genes that contribute to stomach colonization. Infect Immun 75:1005–1016. doi: 10.1128/IAI.01176-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lindholm C, Quiding-Jarbrink M, Lonroth H, Hamlet A, Svennerholm AM. 1998. Local cytokine response in Helicobacter pylori-infected subjects. Infect Immun 66:5964–5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Govindaraj Vaithinathan A, Vanitha A. 2018. WHO global priority pathogens list on antibiotic resistance: an urgent need for action to integrate One Health data. Perspect Public Health 138:87–88. doi: 10.1177/1757913917743881. [DOI] [PubMed] [Google Scholar]
- 42.Lynn MA, Tumes DJ, Choo JM, Sribnaia A, Blake SJ, Leong LEX, Young GP, Marshall HS, Wesselingh SL, Rogers GB, Lynn DJ. 2018. Early-life antibiotic-driven dysbiosis leads to dysregulated vaccine immune responses in mice. Cell Host Microbe 23:653–660.e5. doi: 10.1016/j.chom.2018.04.009. [DOI] [PubMed] [Google Scholar]
- 43.Tang WH, Hazen SL. 2017. The gut microbiome and its role in cardiovascular diseases. Circulation 135:1008–1010. doi: 10.1161/CIRCULATIONAHA.116.024251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sommer F, Anderson JM, Bharti R, Raes J, Rosenstiel P. 2017. The resilience of the intestinal microbiota influences health and disease. Nat Rev Microbiol 15:630–638. doi: 10.1038/nrmicro.2017.58. [DOI] [PubMed] [Google Scholar]
- 45.Biragyn A, Ferrucci L. 2018. Gut dysbiosis: a potential link between increased cancer risk in ageing and inflammaging. Lancet Oncol 19:e295–e304. doi: 10.1016/S1470-2045(18)30095-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gopalakrishnan V, Helmink BA, Spencer CN, Reuben A, Wargo JA. 2018. The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell 33:570–580. doi: 10.1016/j.ccell.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Lei YM, Jabri B, Alegre ML, Chang EB, Gajewski TF. 2015. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 350:1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, Fidelle M, Flament C, Poirier-Colame V, Opolon P, Klein C, Iribarren K, Mondragon L, Jacquelot N, Qu B, Ferrere G, Clemenson C, Mezquita L, Masip JR, Naltet C, Brosseau S, Kaderbhai C, Richard C, Rizvi H, Levenez F, Galleron N, Quinquis B, Pons N, Ryffel B, Minard-Colin V, Gonin P, Soria JC, Deutsch E, Loriot Y, Ghiringhelli F, Zalcman G, Goldwasser F, Escudier B, Hellmann MD, Eggermont A, Raoult D, Albiges L, Kroemer G, Zitvogel L. 2018. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359:91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 49.Kanizaj TF, Kunac N. 2014. Helicobacter pylori: future perspectives in therapy reflecting three decades of experience. World J Gastroenterol 20:699–705. doi: 10.3748/wjg.v20.i3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O’Connor A, Lamarque D, Gisbert JP, O’Morain C. 2017. Treatment of Helicobacter pylori infection 2017. Helicobacter 22:e12410. doi: 10.1111/hel.12410. [DOI] [PubMed] [Google Scholar]
- 51.Desbois AP, Smith VJ. 2010. Antibacterial free fatty acids: activities, mechanisms of action and biotechnological potential. Appl Microbiol Biotechnol 85:1629–1642. doi: 10.1007/s00253-009-2355-3. [DOI] [PubMed] [Google Scholar]
- 52.Nederberg F, Zhang Y, Tan JP, Xu K, Wang H, Yang C, Gao S, Guo XD, Fukushima K, Li L, Hedrick JL, Yang YY. 2011. Biodegradable nanostructures with selective lysis of microbial membranes. Nat Chem 3:409–414. doi: 10.1038/nchem.1012. [DOI] [PubMed] [Google Scholar]
- 53.Khulusi S, Ahmed HA, Patel P, Mendall MA, Northfield TC. 1995. The effects of unsaturated fatty acids on Helicobacter pylori in vitro. J Med Microbiol 42:276–282. doi: 10.1099/00222615-42-4-276. [DOI] [PubMed] [Google Scholar]
- 54.Gammoh NZ, Rink L. 2017. Zinc in infection and inflammation. Nutrients 9:E624. doi: 10.3390/nu9060624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Das UN. 2011. Essential fatty acids and their metabolites as modulators of stem cell biology with reference to inflammation, cancer, and metastasis. Cancer Metastasis Rev 30:311–324. doi: 10.1007/s10555-011-9316-x. [DOI] [PubMed] [Google Scholar]
- 56.Salama SM, Gwaram NS, AlRashdi AS, Khalifa SA, Abdulla MA, Ali HM, El-Seedi HR. 2016. A zinc morpholine complex prevents HCl/ethanol-induced gastric ulcers in a rat model. Sci Rep 6:29646. doi: 10.1038/srep29646. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 57.Martins JL, Rodrigues OR, da Silva DM, Galdino PM, de Paula JR, Romao W, da Costa HB, Vaz BG, Ghedini PC, Costa EA. 2014. Mechanisms involved in the gastroprotective activity of Celtis iguanaea (Jacq.) Sargent on gastric lesions in mice. J Ethnopharmacol 155:1616–1624. doi: 10.1016/j.jep.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 58.Salga MS, Ali HM, Abdulla MA, Abdelwahab SI. 2012. Gastroprotective activity and mechanism of novel dichlorido-zinc(II)-4-(2-(5-methoxybenzylideneamino)ethyl)piperazin-1-iumphenolate complex on ethanol-induced gastric ulceration. Chem Biol Interact 195:144–153. doi: 10.1016/j.cbi.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 59.Salama NR, Hartung ML, Muller A. 2013. Life in the human stomach: persistence strategies of the bacterial pathogen Helicobacter pylori. Nat Rev Microbiol 11:385–399. doi: 10.1038/nrmicro3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, Nelson K, Quackenbush J, Zhou L, Kirkness EF, Peterson S, Loftus B, Richardson D, Dodson R, Khalak HG, Glodek A, McKenney K, Fitzegerald LM, Lee N, Adams MD, Hickey EK, Berg DE, Gocayne JD, Utterback TR, Peterson JD, Kelley JM, Cotton MD, Weidman JM, Fujii C, Bowman C, Watthey L, Wallin E, Hayes WS, Borodovsky M, Karp PD, Smith HO, Fraser CM, Venter JC. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 61.Dorer MS, Cohen IE, Sessler TH, Fero J, Salama NR. 2013. Natural competence promotes Helicobacter pylori chronic infection. Infect Immun 81:209–215. doi: 10.1128/IAI.01042-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Clinical and Laboratory Standards Institute. 2008. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 7th ed Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 63.Helander IM, Mattila-Sandholm T. 2000. Fluorometric assessment of Gram-negative bacterial permeabilization. J Appl Microbiol 88:213–219. doi: 10.1046/j.1365-2672.2000.00971.x. [DOI] [PubMed] [Google Scholar]
- 64.O’Rourke J, Bode G. 2001. Morphology and ultrastructure In Mobley HLT, Mendz GL, Hazell SL (ed), Helicobacter pylori: physiology and genetics. ASM Press, Washington, DC. [PubMed] [Google Scholar]
- 65.Saldanha CA, Garcia MP, Iocca DC, Rebelo LG, Souza ACO, Bocca AL, Almeida Santos MDFM, Morais PC, Azevedo RB. 2016. Antifungal activity of amphotericin B conjugated to nanosized magnetite in the treatment of paracoccidioidomycosis. PLoS Negl Trop Dis 10:e0004754. doi: 10.1371/journal.pntd.0004754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stefka AT, Feehley T, Tripathi P, Qiu J, McCoy K, Mazmanian SK, Tjota MY, Seo GY, Cao S, Theriault BR, Antonopoulos DA, Zhou L, Chang EB, Fu YX, Nagler CR. 2014. Commensal bacteria protect against food allergen sensitization. Proc Natl Acad Sci U S A 111:13145–13150. doi: 10.1073/pnas.1412008111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Edgar RC. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 69.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methe B, DeSantis TZ, Human Microbiome C, Petrosino JF, Knight R, Birren BW. 2011. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lozupone CA, Hamady M, Kelley ST, Knight R. 2007. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol 73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gaujoux R, Seoighe C. 2010. A flexible R package for nonnegative matrix factorization. BMC Bioinformatics 11:367. doi: 10.1186/1471-2105-11-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing data in this paper are available at the NCBI Sequence Read Archive (SRA) under BioProject accession number PRJNA505765.