Abstract
Vagal Nerve Stimulation (VNS) Therapy® is a United States Food and Drug Administration approved neurotherapeutic for medically refractory partial epilepsy and treatment-resistant depression. The molecular mechanisms underlying its beneficial effects are unclear. We hypothesized that one mechanism involves neuronal activity-dependent modifications of central nervous system excitatory synapses. To begin to test this hypothesis, we asked whether VNS modifies the activity of neurons in amygdala and hippocampus. Neuronal recordings from adult, freely moving rats revealed that activity in both amygdala and hippocampus was modified by VNS immediately after its application, and changes were detected following 1 week of stimulation. To investigate whether VNS modifies the proteome of excitatory synapses, we established a label-free, quantitative liquid chromatography-tandem mass spectrometry workflow that enables global analysis of the constituents of the postsynaptic density (PSD) proteome. PSD proteins were biochemically purified from amygdala/piriform cortex of VNS- or dummy-treated rats following 1-week stimulation, and individual PSD protein levels were quantified by liquid chromatography-tandem mass spectrometry analysis. We identified 1899 unique peptides corresponding to 425 proteins in PSD fractions, of which expression levels of 22 proteins were differentially regulated by VNS with changes greater than 150%. Changes in a subset of these proteins, including significantly increased expression of neurexin-1α, cadherin 13 and voltage-dependent calcium channel α2δ1, the primary target of the antiepileptic drug gabapentin, and decreased expression of voltage-dependent calcium channel γ3, were confirmed by western blot analysis of PSD samples. These results demonstrate that VNS modulates excitatory synapses through regulating a subset of the PSD proteome. Our study reveals molecular targets of VNS and point to possible mechanisms underlying its beneficial effects, including activity-dependent formation of excitatory synapses.
Keywords: epilepsy, LC-MS/MS, neurophysiology, postsynaptic density, proteomics, vagal nerve stimulation
Vagal nerve stimulation (VNS) Therapy® is an Food and Drug Administration (FDA)-approved treatment for medically refractory partial epilepsy and treatment-resistant depression that is widely used clinically (Beekwilder and Beems 2010). However, the mechanisms by which VNS exerts these beneficial effects are not well understood. Recently, we demonstrated that brief (60-min) or chronic (1-week) VNS treatment is sufficient to elevate seizure threshold locally in the amygdala in the kindling model of limbic epilepsy (Alexander and McNamara 2012). One plausible mechanism by which VNS may exert this beneficial effect involves activity-dependent modification of central nervous system (CNS) excitatory synapses. Changes in neuronal activity levels have long-lasting consequences on several aspects of CNS neurons, including changes in protein expression levels at the postsynaptic density (PSD)(Trinidad et al. 2013). The PSD is a postsynaptic membrane specialization of excitatory synapses, which is composed of a diversity of proteins, including ligand-gated receptors, scaffold proteins, signaling molecules, and regulatory enzymes. Dynamic changes in the composition of PSD proteins can lead to modifications of signaling strength of excitatory synapses in health and disease (Sheng and Hoogenraad 2007; Bayés and Grant 2009).
To begin to address the possibility that VNS exerts its beneficial effect through activity-dependent regulation of excitatory synapse, we first conducted in vivo electrophysiology experiments to measure changes in neuronal activity in amygdala and hippocampus in response to VNS. We then established a workflow of unbiased proteomic analysis to ask whether and how activity evoked by VNS modifies the molecular composition of the PSD in the amygdala/piriform cortex, a brain area implicated in temporal lobe epilepsy and depression. We provide in vivo evidence that VNS modifies neuronal activity in amygdala and hippocampus at the individual neuron and network levels. We further report the development of a workflow utilizing label-free nano-scale capillary liquid chromatography-tandem mass spectrometry (LC-MS/MS) that permits identification and quantification of the content of proteins directly from biochemically purified PSD fractions. This comprehensive profile of the PSD proteome revealed significant changes in a subset of PSD proteins induced by VNS, which were confirmed by western blot analysis. The identities of PSD proteins modified by VNS provide clues to possible mechanisms by which VNS may exert its beneficial effects.
Materials and methods
Reagents and materials
All reagents were obtained from Sigma, St Louis, MO, USA unless specified otherwise. The VNS device, including a VNS stimulation electrode, a VNS stimulator (Demipulse model 103), a handheld computer and a programming wand, was provided by Cyberonics Inc. (Houston, TX, USA). Both the stimulation electrode and stimulator were implanted in the animals. The stimulation electrode was a helical wire with an internal diameter of 1.5 mm, approximately 12 inches long, encased in silicone and designed for use in small animals. One end of the electrode had two platinum wires encased in flexible silicon cuffs, which could be easily wrapped around the vagus nerve and carotid artery. The other end of the stimulation electrode had a male single-pin lead that inserted into the stimulator. The stimulator was 1.8 in × 1.3 in × 0.27 in size and 16 g in mass and was programmed using a handheld computer (Dell Axim X50; Dell Inc., Round Rock, TX, USA) and a programming wand. Software on the handheld computer allowed adjustment of stimulator output current, signal frequency, pulse width, signal ON time and signal OFF time.
Animals and surgery
All experiments were carried out in adult male Sprague–Dawley rats (300–400 g; Charles River Laboratories Inc., Wilmington, MA, USA) in accordance with the National Institutes of Health’s guidelines for the care and use of animals and with approved animal protocols from the Institutional Animal Care and Use Committee at Duke University. Rats were housed under a 12 : 12 light/dark cycle with access to food and water ad libitum. Rats undergoing surgical procedures were under ketamine (60 mg/kg) and xylazine (5 mg/kg) anesthesia. VNS implantation was performed as described previously (Alexander and McNamara 2012). Control, dummy-treated animals underwent identical surgical procedures as the VNS-implanted groups, although VNS leads were connected to dummy Demipulse stimulators that were incapable of delivering current. Henceforth, control animals shall be referred to as dummy-treated.
Microelectrode array implantation surgery was performed in a stereotaxic apparatus immediately following implantation of the VNS device. Twelve 35-μm S-isonel-coated tungsten microwires were lowered into each amygdala (2.8 mm posterior to bregma, 4.9 mm lateral to bregma, and 7.4 mm below dura), and 8 wires were lowered into CA3 (3.6 mm posterior to bregma, 3.0 mm lateral to bregma, and 3.3 mm below dura). Animals were treated with ketoprofen (5 mg/kg, s.c.) for analgesia and baytril antibiotic (10 mg/kg, s.c.) for 2 days following surgery and were allowed at least 1 week to recover from surgery before beginning experiments.
Electrophysiology
Microwire electrodes were connected to a printed circuit board, which was connected to a pre-amplifier by a miniature connector. Neural activity was recorded from the 32 implanted microwires and processed using a Multineuron Acquisition Processor (Plexon Inc., Dallas, TX, USA). Local field potentials (LFPs) were pre-amplified (500 ×), filtered (0.3–400 Hz), and digitized at 1000 Hz using a Digital Acquisition card (National Instruments, Austin, TX, USA) and a Multi-Neuron Acquisition Processor (Plexon Inc.). Simultaneous single unit recordings were made from the same wires that were used to collect LFP data (Nicolelis et al., 1997). Waveforms were categorized as single units online by means of a voltage threshold and principal component analysis. Waveforms were subsequently sorted offline using Offline Sorter (Plexon Inc.) to confirm the quality of the recorded spikes and separated into individual single units. Time stamps of the sorted waveforms were used to determine the firing rate of neurons over the course of the experiments.
VNS procedures
For the electrophysiology arm of the study, VNS devices were activated only after action potential waveforms from single neurons became apparent, which typically occurred approximately two to three weeks following electrode implantation. At that time, recordings were taken before and during delivery of VNS at 0.25, 0.5, 0.75, or 1.0 mA of current. The stimulation parameters used were 30 s ON and 5 min OFF, with each ON period consisting of 500 μs pulses delivered at 20 Hz. A typical stimulation schedule for an animal proceeded as follows (Fig. 1a):
Fig. 1.
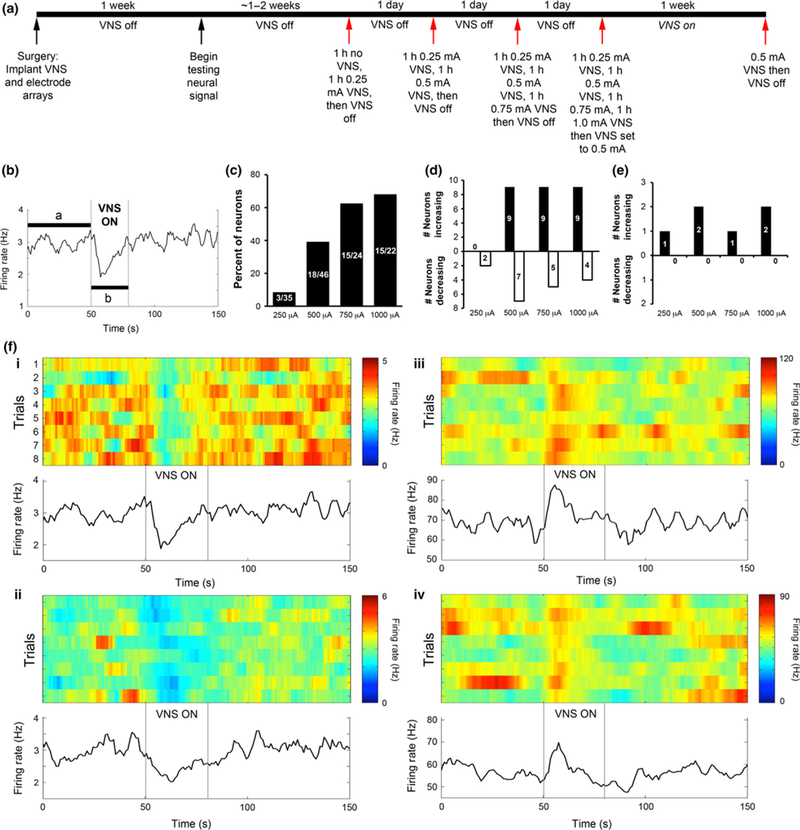
Vagal nerve stimulation (VNS) affects neuronal activity of amygdala neurons in a VNS intensity-dependent manner. (a) Timeline of procedures for recording neuronal activity from animals undergoing VNS stimulation. (b) Example data from one neuron demonstrating the method for determining whether a neuron shows a significant change in firing rate. Number of spikes per second of data were collected continuously while VNS cycled ON (30 s) and OFF (5 min). VNS ON times were aligned for each cycle, and the mean firing rate before, during, and following the VNS ON period was calculated across eight continuous cycles. The mean and 95% confidence interval of firing rate was measured during the 50 s before VNS ON (a). If firing rates during the VNS ON time (b) fell outside of the 95% CI of the pre-VNS mean at any time, then the neuron was classified as showing a significant change in firing rate. (c), Percent of amygdala neurons that showed a significant change in firing rate for each VNS intensity. Fractional numbers listed on the bars indicate the number of neurons showing a significant change in firing rate and the total number of neurons recorded from at each VNS intensity. (d and e), Number of neurons that showed a significant increase (black bars) or significant decrease (white bars) in firing rate during VNS ON for each VNS intensity. Neurons that showed a significant change in firing rate were classified according to their average baseline firing rate, and the conservative threshold of 10 Hz was used to differentiate a likely mixed population of projection neurons and interneurons (< 10 Hz firing rate, data shown in d) from a likely population of only interneurons (> 10 Hz firing rate, data shown in e). (f) Examples of neurons with high (i) and low (ii) average baseline firing rates that showed a significant change in firing rate during VNS ON. (i and ii) slow (<10 Hz) spiking neurons, which likely contains both interneurons and projection neurons, show variable effects on firing rate; some show significant increases in firing rates and others show significant decreases in firing rates. Shown are two examples of neurons that decrease firing rate. (iii–iv) Fast spiking interneurons increased firing rate with VNS.
Day 1: One hour with no VNS followed by 1 h of recording while VNS was programmed to 0.25 mA. VNS was then programmed to 0 mA.
Day 2: One hour of 0.25 mA VNS followed by 1 h of 0.5 mA VNS. VNS was then programmed to 0 mA.
Day 3: One hour of 0.25 mA VNS, 1 h of 0.5 mA VNS, then 1 h of 0.75 mA VNS. VNS was then programmed to 0 mA.
Day 4: One hour of 0.25 mA VNS, 1 h of 0.5 mA VNS, 1 h of 0.75 mA VNS, then 1 h of 1.0 mA VNS. VNS was then programmed to 0.5 mA.
Day 11 (1 week following Day 4 recordings): 1 h of 0.5 mA VNS. VNS was then programmed to 0 mA.
Recordings were made during each of the days listed above for periods of 1–4 h.
For the proteomics arm of the study, VNS devices were activated following a 1-week post-operative recovery period and remained active for 1 week (Fig. 5a). One week of VNS was chosen based on the finding that 1-week of VNS was sufficient to elevate seizure threshold in the amygdala kindling model (Alexander and McNamara 2012). The stimulation parameters used were 30 s ON and 5 min OFF, with each ON period consisting of 500 μs pulses delivered at 20 Hz, and a stimulation intensity of 0.5 mA. This cycle of stimulation was delivered for the entire 1-week period of stimulation. Control animals underwent all of the same procedures as VNS-treated animals. That is, all procedures for interrogating and programming stimulators were identical, except that stimulators in dummy-treated animals remained at the 0 mA current intensity setting. Following VNS activation, devices were not interrogated or programmed again.
Fig. 5.
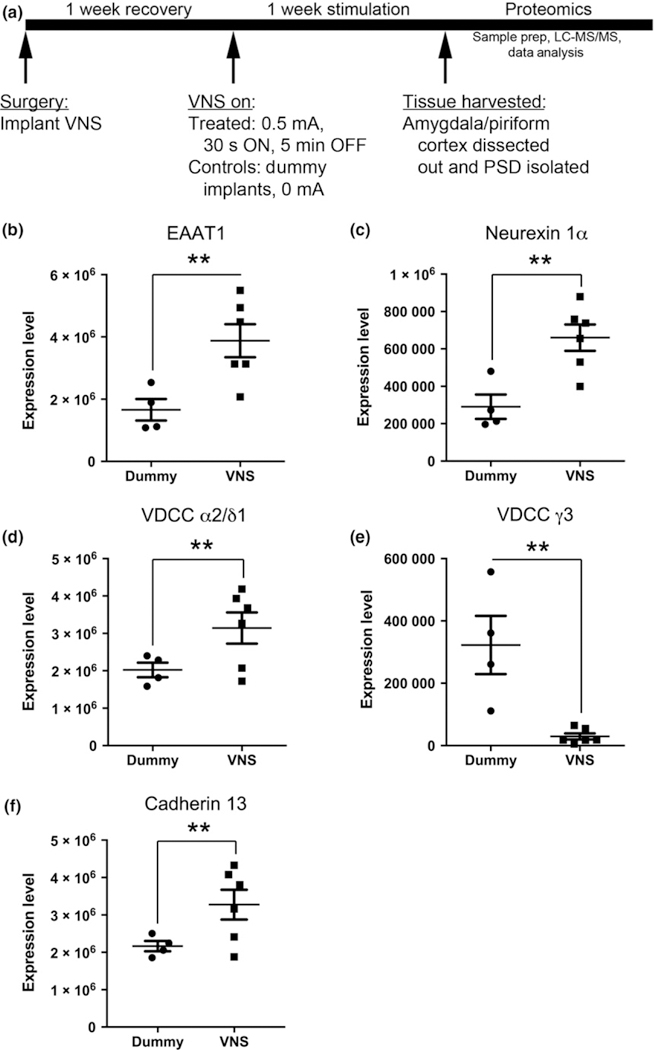
(a) Experimental timeline for assessing changes in the postsynaptic density (PSD) proteome induced by vagal nerve stimulation (VNS). (b–e) Bar graphs displaying expression levels of five proteins for individual control or VNS-treated animals. Proteins include excitatory amino acid transporter 1 (EAAT1) (b), neurexin 1a (c), VDCC α2/δ1 (d), VDCC γ3 (e) and cadherin 13 (f). **p < 0.01, one-way ANOVA.
Preparation of PSD samples
One week after VNS activation, rats were killed and the amygdala/piriform cortex structures were rapidly dissected on ice including the cortex below the rhinal fissure. The remaining cerebral cortices and hippocampi were also dissected and saved for western blot analyses. The tissues were then quickly frozen in liquid nitrogen. Subcellular fractionation was performed as described previously (Huang et al. 2008). The entire procedure was conducted at 4°C or on ice. In brief, amygdala/pyriform cortex samples dissected from each animal were pooled and homogenized in 10 volumes of homogenization buffer (0.32 M sucrose, 4 mM HEPES, 1 mM Na3OV4, 1 mM phenyl-methylsulfonyl fluoride, and proteinase mix inhibitor tablets (Roche Molecular Biochemicals, Indianapolis, IN, USA), pH 7.4) and centrifuged at 735 g for 10 min. After being spun at 325 g for 10 min, the supernatant (S1) was collected and centrifuged at 16 000 g for 15 min to provide a crude synaptosomal pellet (P2). Crude synaptosomes underwent osmotic shock by addition of ice-cold deionized H2O and rapidly returned to osmotic balance with 1 M HEPES pH 7.4. Following centrifugation at 16 000 g for 30 min, the pellet (P3) consisting of an enriched synaptosomal membrane fraction was collected. P3 was resuspended in homogenization buffer, placed on a discontinuous sucrose gradient of 0.32, 0.8 and 1.2 M sucrose, and spun at 150 000 g for 2 h in a swinging bucket rotor. The synaptosomal membrane fraction was recovered from the interface between 0.8 and 1.2 M sucrose. The membranes were diluted 1 : 4 with 4 mM HEPES, pH 7.4 and centrifuged at 150 000 g for 30 min. The pellet was resuspended in buffer containing 0.5% Triton X-100 (50 mM HEPES, 2 mM EDTA, 1 mM Na3OV4, 1 mM phenylmethylsulfonyl fluoride and 0.5% Triton X-100, pH 7.4) and rotated for 20 min at 4°C. The solution was centrifuged at 32 000 g for 20 min, resulting in a supernatant composed of presynaptic membranes and a Triton insoluble pellet composed of crude PSD (PSD-1T). After washed with homogenization buffer, the PSD-1T pellet was treated with 0.5% Triton X-100 for 20 min on ice and centrifuged at 32 000 g for 20 min and the pellet (PSD-2T) was recovered and washed three times with AmBis solution (50 mM, pH 7.4); the final pellet was designated ‘PSD’ and submitted for in situ digestion for LC-MS/MS analysis. Protein enrichment index was estimated by normalizing the immunoreactivity and protein amount of PSD-95 and synaptophysin in subcellular fractions over homogenate fraction. Similar to other proteome studies (Husi et al. 2000; Jordan et al. 2004; Li et al. 2004; Peng et al. 2004; Yoshimura et al. 2004; Cheng et al. 2006; Moron et al. 2007; Trinidad et al. 2008), some common contaminants, including keratins, myelin basic protein, glial fibrillary acidic protein, and mitochondrial proteins, were present in our PSD samples.
In situ digestion of PSD sample
PSD pellets were bath sonicated three times for 30 s in the presence of 100 μL 1% PPS Silent Surfactant (Protein Discovery; Knoxville, TN, USA)/50 mM ammonium bicarbonate, pH 8.0 and then centrifuged at 32 000 g for 5 min to form a secondary pellet. To this secondary pellet was added 100 μL of 1% PPS Silent Surfactant/50 mM ammonium bicarbonate, 5 mM dithiolthreitol, and samples were heated for 30 min at 70°C. Free sulfhydryls were alkylated in the presence of 10 mM iodoacetamide for 60 min at 20°C and an in situ proteolytic digestion of the pellet suspension was accomplished by the addition of 500 ng sequencing grade trypsin (Promega, Madison, WI, USA) for 18 h at 37°C. The samples were then acidified to pH 2.5 with 100% trifluoroacetic acid and incubated at 60°C for 1 h to hydrolyze the PPS Silent Surfactant. Insoluble hydrolyzed surfactant was cleared by centrifugation at 32 000 g for 5 min. To estimate total peptide quantity, a bicinchoninic acid assay (Pierce, Rockford, IL, USA) was performed against a calibration curve of trypsin-digested bovine serum albumin, and the final concentration of all samples was adjusted to 200 ng/μL with 2% acetonitrile and 0.1% trifluoroacetic acid.
Nano-flow liquid chromatography electrospray ionization tandem mass spectrometry
Chromatographic separation of peptide mixtures was performed on a Waters NanoAquity UPLC equipped with a 1.7 μm BEH130 C18 75 μm I.D. × 250 mm reversed-phase column. The mobile phase consisted of (A) 0.1% formic acid in water and (B) 0.1% formic acid in acetonitrile. Following a 5 μL injection (1 μg total), peptides were trapped for 5 min on a 5 μm Symmetry C18 180 μm I.D. × 20 mm column at 20 μL/min in 99.9% A. The analytical column was then switched in-line and the mobile phase was held for 5 min at 5% B before applying a linear elution gradient of 5% B to 40% B over 90 min at 300 nL/min. The analytical column was connected to a fused silica PicoTip emitter (New Objective, Cambridge, MA, USA) with a 10 μm tip orifice and coupled to the mass spectrometer through an electrospray interface.
MS were acquired on an LTQ-Orbitrap XL mass spectrometer (Thermo Fisher, Waltham, MA, USA) operating in positive-ion mode with an electrospray voltage of 2.0 kV. For the sample preparation reproducibility study, three independently prepared samples were analyzed once with one of the samples being analyzed in technical triplicate. For the 10 animal VNS study, each sample was acquired in technical duplicate. For all acquisitions, the instrument was set to acquire a precursor MS scan from m/z 400–2000 with r = 60 000 at m/z 400 and a target AGC (Automatic gain control) setting of 1e6 ions. MS/MS spectra of the top five most abundant precursor ions were acquired in the Orbitrap at r = 7500 at m/z 400. Max fill times were set to 1000 ms for full MS scans and 500 ms for MS/MS scans with minimum MS/MS triggering thresholds of 1000 counts. For all experiments, fragmentation occurred in the LTQ linear ion trap with a CID energy setting of 35% and a dynamic exclusion of 60 s for previously fragmented precursor ions.
Peptide identification, label-free quantitation, and ingenuity pathway analysis
Label-free quantitation and integration of qualitative peptide identifications was performed using Rosetta Elucidator (v 3.3; Rosetta Inpharmatics, Seattle, WA, USA). All raw LC-MS/MS data within an experiment were imported and subjected to chromatographic retention time and accurate mass alignment using the PeakTeller® algorithm within Rosetta Elucidator with a minimum peak time width set to 6 s, alignment search distance set to 4 min and the refine alignment option enabled. Quantitation of all signals in the precursor MS spectra was performed by Elucidator by measuring the peak height of each precursor’s selected ion chromatogram. Fold-change values between treatment groups (individual animals for reproducibility study or dummy-treated vs. stimulated for VNS study) were calculated on the protein level by averaging the sum of all features (an individual isotopomer within a precursor ion’s charge state envelope) associated with the precursor ion within a biological replicate. Only unique peptides were considered when summing to protein level expression values. Homologous Protein grouping was accomplished within Protein Teller algorithm executed within Rosetta Elucidator (Keller et al. 2002; Nesvizhskii et al. 2003). Here, the p-values were calculated based on a previously described ratio error model (Weng et al. 2006).
Qualitative peptide identifications were made by generating DTA files for all features that had associated MS/MS spectra. For features with multiple MS/MS spectra, the spectra were combined prior to DTA creation. All DTA files were submitted to Mascot (Matrix Science, Boston, MA, USA) searches against a SwissProt protein database containing entries only from the rodentia rodentia (including mus musculus and rattus norvegicus) taxonomy (Downloaded June 2010, 24 248 forward entries). The entire database was concatenated with the sequence-reversed version of each entry. Tolerances of 5 ppm precursor and 0.02 Da product ions were applied to all searches. All data were searched using trypsin specificity with up to two missed cleavages. Carbamidomethylation (+ 57.0214 Da on C) was set as a fixed modification, whereas oxidation (+ 15.9949 Da on M) was considered a variable modification. A peptide false discovery rate of approximately 1.0% was applied using the PeptideProphet algorithm within Elucidator using independent reverse decoy database validation (Keller et al. 2002; Nesvizhskii et al. 2003).
PSD proteins found to be differentially expressed as a function of VNS (p < 0.05) in the LC-MS/MS-based discovery experiment were analyzed by Ingenuity Pathway Analysis (build version: 400896M; content version: 28820210).
Immunoblotting analysis
Western blot analyses were employed to validate the PSD fractions and the change in expression level of a protein of interest that was discovered from the proteomics study. In brief, equivalent amounts of protein (1–2 μg for PSD and 10–20 μg for other subcellular fractions) prepared from individual animals were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis. After transferring onto nitrocellulose membrane, the blots were incubated overnight with primary antibodies followed by incubation with secondary antibodies (1 : 5000) for 1 h at 20°C. The antibodies and dilutions used in this study were as follow: neurexin 1α mouse monoclonal antibody (sc-136001, Lot# E2814, 1 : 250), excitatory amino acid transporter 1 (EAAT1) rabbit polyclonal antibody (sc-15316, Lot# J2513, 1 : 500), L-type Ca2+ CP-γ3 goat polyclonal antibody (sc-164818, Lot# D0212, 1 : 500), Cacna2d1 rabbit polyclonal antibody (sc-133438, Lot# E3041, 1 : 250), and Synaptophysin rabbit polyclonal antibody (sc-9116, Lot# A0708, 1 : 1000) were purchased from Santa Cruz Biotechnology; PSD-95 mouse monoclonal antibody (ab13552–50, Lot#726065, 1 : 1000) and actin rabbit polyclonal antibody (A2066, Lot# 095M4765V, 1 : 5000) were from Abcam (Cambridge, MA, USA) and Sigma, respectively. Cadherin 13 goat polyclonal antibody (AF3264, Lot#XEX041610A, 1:1000) was from R & D Systems, Minneapolis, MN, USA. All antibodies recognized the immunoreactivities that agreed with the expected molecular mass of proteins of interest. Immunoblots were developed with enhanced chemiluminescence (Amersham Biosciences, Piscataway, NJ, USA).
Statistical analyses
Electrophysiology: To determine whether amygdala neurons changed firing rate in response to VNS, firing rate distributions (1 s bins) were generated for each of at least six trials of VNS OFF then ON (5 min 30 s per trial corresponding to 5 min VNS OFF, 30 s VNS sec ON cycles). Binned firing rates were then averaged for all trials. Using the 50 s preceding VNS onset, the averages and 95% confidence intervals of firing rate per second were calculated based on a Poisson distribution (during time A shown in Fig. 1b). A neuron was considered to have changed firing rate if the firing frequency of at least one of the 1-s bins was beyond the 95% confidence interval limits. This process was repeated for each neuron at each VNS stimulation intensity (250, 500, 750, and 1000 μA).
To determine whether hippocampal LFPs were affected by VNS, we first generated spectrograms of the LFPs to visualize changes in spectral power over time during VNS ON and OFF cycles. Upon identifying the theta band (4–10 Hz) as affected by VNS upon visual inspection of spectrograms, we generated power spectral density plots of the LFPs during the 30 s immediately before VNS ON and the 30 s of VNS ON. Only 30 s of time before VNS ON was used as the baseline period rather than the entire 50 s as shown in Fig. 1(b) because power spectral density values are positively related to the duration of data analyzed, so power values can only be compared if the data compared are of equivalent duration. Because the VNS ON period was 30 s, we limited the baseline time period to 30 s. Power values for frequencies between 4 and 10 Hz were measures for each VNS cycle during baseline (pre-VNS) periods and VNS ON periods and averaged over each trial for each animal, each VNS intensity, and each recording session. Power values were compared by Wilcoxon matched-pairs signed rank test (GraphPad Prism, La Jolla, CA, USA).
Proteomics: Relative protein quantities were calculated by summing each individually identified peptide to its respective protein following accurate-mass retention time alignment and then rationing the average protein quantity within the dummy-treated group against the average protein quantity within the VNS-treated group. A one-way ANOVA was applied to the means to determine whether a protein showed a significant change in expression level with VNS. The false discovery rate method, according to Storey (2002), was used to correct for multiple comparisons (Storey 2002). Only those proteins with a p-value < 0.05 and a fold-change higher than 1.5 were considered as being differentially expressed.
Immunoblotting: The immunoreactivities of individual bands on western blots were measured by ImageJ software (National Institutes of Health, Bethesda, MD, USA). Values obtained from dummy animals were averaged for each protein, and values obtained from VNS-treated animals were normalized to control values. Percent of control values are presented in graphs, and unpaired Student t-tests were applied to these values to determine whether a protein of interest showed a significant change in expression level with VNS.
Results
VNS modifies neuronal activity in basolateral amygdala and hippocampus
Our prior findings that VNS elevates the threshold for evoking seizures locally in amygdala (Alexander and McNamara 2012) led us to ask whether VNS modifies neuronal activity within the amygdala and a synaptically related structure, the hippocampus, of naive rats (Pikkarainen et al. 1999). Toward that end, rats were implanted with VNS devices and multielectrode arrays targeting the basolateral nucleus of the amygdala and area CA3 of hippocampus. Once spikes from individual amygdala neurons became apparent, recordings were made while VNS cycled ON and OFF using various stimulation intensities (Fig. 1a). We found that VNS modified the firing rate of amygdala neurons and that increasing VNS intensity resulted in an increasing number of neurons with firing rate significantly affected (Fig. 1b and c). We next asked whether VNS differentially affected neurons based on the neuron type. Individual neurons were divided into presumed interneurons and a mixed population of projection neurons and low-firing rate interneurons based on an average baseline firing rate threshold of 10 Hz (Pare and Gaudreau 1996; Likhtik et al 2006). For the neurons with firing rates less than 10 Hz, the responses to VNS were variable; some neurons exhibited increased firing rate during the VNS ON cycle while other neurons showed decreased firing rate (Fig. 1d and fi–ii). Each of the two neurons with average firing rates greater than 10 Hz exhibited an increased firing rate during VNS ON and with every VNS intensity tested (Fig. 1e and fiii–iv).
While recording activity of individual neurons within amygdala in response to acute VNS, simultaneous recordings of LFP from CA3 of hippocampus were performed. Spectrograms of hippocampal LFPs while VNS cycled ON and OFF revealed a modification of the LFP in the theta band during VNS ON periods (Fig. 2a). Power spectral density plots revealed that VNS-induced stimulation intensity-dependent increases in theta power in the 5.5–6.5 Hz range and simultaneous decreases in power in the 6.5–8.5 Hz range. Although the lowest intensity of VNS (250 μA) did not significantly affect LFP power (p > 0.05, Wilcoxon matched-pairs signed rank test), all other VNS intensities tested significantly affected theta power (500, 750, and 1000 μA, p < 0.01 for each, Wilcoxon matched-pairs signed rank test, Fig. 2bi).
Fig. 2.
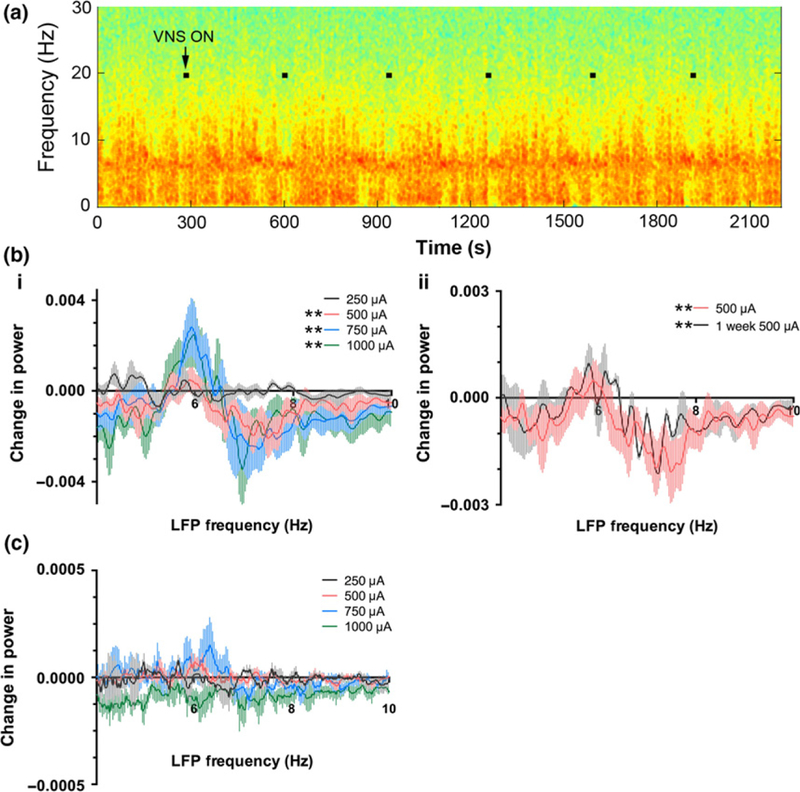
Vagal nerve stimulation (VNS) affects hippocampal field potential properties by increasing power in the low theta frequency range (5.5–6.5 Hz) and simultaneously decreasing power in the higher theta frequency range (6.5–8.5 Hz). This effect was dependent on the VNS intensity and persisted after 1 week of VNS treatment. (a) Example spectrogram of a local field potential recording from CA3 of hippocampus during VNS OFF and ON periods. Warmer colors represent greater spectral power, and cooler colors represent less spectral power. Black squares on the spectrogram show periods of VNS ON. (b) Quantification of the effect of VNS on theta (4–10 Hz) spectral power for the population of recordings and animals. Spectral power in the theta band was measured for 30 s before the onset of VNS and during the 30 s of VNS ON. The difference in power at each frequency point was plotted for each VNS intensity (i) and for 500 μA stimulation after 1 week on VNS (500 μA) treatment (ii), which is plotted with the identical 500 μA data as shown in i for comparison. All VNS intensities except 250 μA produced a significant shift in hippocampal theta power. (c) VNS did not affect amygdala theta power. Identical analysis as described for (b) was applied to local field potential (LFP) measured from amygdala. *p < 0.05, Wilcoxon matched-pairs signed rank test.
Upon completion of recording neuronal responses to acute VNS, VNS devices were programmed to continuously cycle ON and OFF for 1 week, a stimulation pattern conforming to a standard VNS protocol used in patients at a current intensity of 500 μA (Fig. 1a). After 1 week, neuronal responses to 500 μA VNS were again queried. Responses of hippocampal LFPs were similar to the response to acute VNS in that 500 μA VNS continued to produce a significant shift of hippocampal LFP theta power to lower frequencies (p < 0.01, Wilcoxon matched-pairs signed rank test, Fig. 2bii). Our ability to record responses of LFPs notwithstanding, we were unable to record a sufficient number of amygdala neurons to test the impact of chronic VNS on firing rate.
Identification of PSD proteins by quantitative LC-MS/MS analysis
The PSD is a postsynaptic membrane specialization that is essential for structure, function, and plasticity of excitatory synapses in the CNS. The protein composition of the PSD is regulated by neuronal activity (Sheng and Hoogenraad 2007). Our findings that VNS is able to modify neuronal activity led us to investigate whether VNS regulates the molecular composition of excitatory synapses. To globally quantify protein expression in excitatory synapses of amygdala/piriform cortex, the proteome of the PSD was examined with a label-free mass spectrometry based proteomic strategy. The amygdala/piriform cortex was dissected, and the PSD fraction was isolated with subcellular fractionation (Kennedy et al. 1983; Sheng and Hoogenraad 2007). Western blot analyses revealed the presence of PSD-95, a postsynaptic protein, and the absence of synaptophysin, a presynaptic protein, thereby supporting the integrity of the PSD fraction (Fig. 6a and h and Figure S3). The samples were further digested, submitted to LC-MS/MS and run in duplicate (Fig. 3).
Fig. 6.
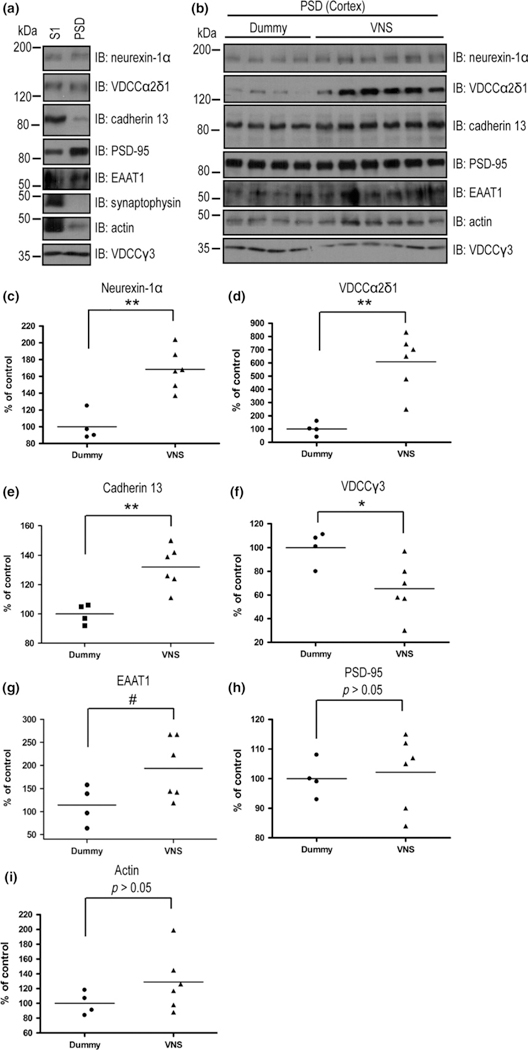
Vagal nerve stimulation (VNS) regulates the expression of postsynaptic density (PSD) proteins in rat cortex. (a) Expression of proteins of interest in the PSD fraction. Proteins of S1 (20 μg) or PSD (5 μg) fractions isolated from rat cortex were analyzed by immunoblotting analysis with the indicated antibodies. (b) VNS regulated the levels of PSD proteins. PSD fractions (5 μg) isolated from the cortices of dummy (N = 4) or VNS (N = 6) treated rats were analyzed by immunoblotting analysis with indicated antibodies. (c–i) Quantitative analyses of western blot immunoreactivities shown in (b). *p < 0.05, **p < 0.01, #p = 0.05, Student’s t-test.
Fig. 3.
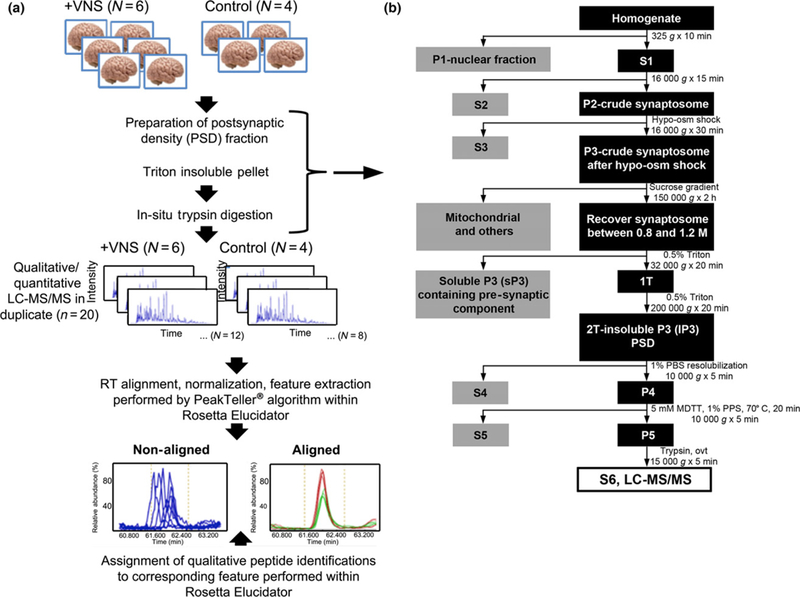
Protein measurements were made from animals that received 1 week of vagal nerve stimulation (VNS) treatment (500 μA). Work flow of sample preparation and data analysis. (a) Sample preparation and data acquisition strategy followed by data analysis workflow utilized in label-free quantitative proteomic analysis of rat postsynaptic density (PSD) fractions as a function of VNS. (b) Detailed description of brain preparation sample treatment from the time of isolation of brain homogenate from each animal through isolation of the S6 fraction, on which liquid chromatography-tandem mass spectrometry (LC-MS/MS) was performed.
LC-MS/MS of the purified PSD fraction revealed 1899 unique peptides, which corresponded to 436 unique proteins after searching a SwissProt-Rodentia protein database and annotated at a 1% peptide false discovery rate (Table S1). For 11 of these proteins, both the rat and mouse variants appeared in our dataset. For each of these 11 proteins, the mouse variant was removed from the data set, yielding 425 proteins (Table S1). The remaining 425 proteins were classified into 15 distinct functional categories based on literature reports of functions of various proteins (Fig. 4a, Table S1). Among the PSD proteins identified by our assay, approximately 60% (247 proteins) were identified by at least two unique peptides.
Fig. 4.
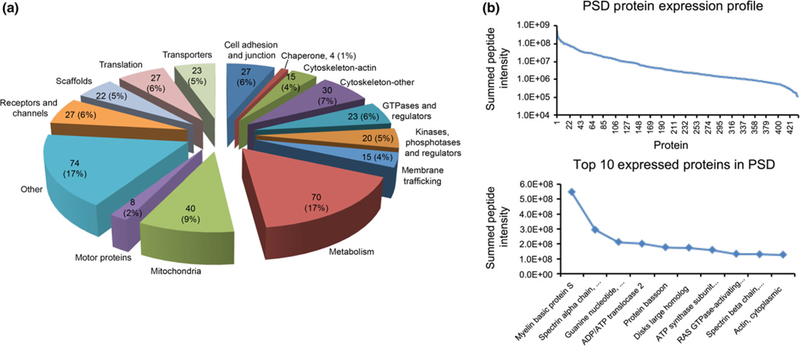
(a) Pie graph representing the relative distribution of proteins identified in this study grouped by functional classification. Each pie piece represents a functional classification of proteins identified. The number of proteins identified that fell into each functional classification and the relative proportion of all protein classes that each class represented in this study are listed on the respective pie pieces. (b) Individual peptide intensities, measured by their area-under-the-curve, belonging to the same protein were summed and the Log10 of this summed protein intensity for all 436 proteins are plotted on the top graph. The bottom plot expands the top 10 most heavily expressed proteins in the postsynaptic density (PSD) samples and lists them by name.
To globally assess the average expression range of identified proteins within the dataset, all of the individual peptide intensities (measured by their area-under-the-curve) belonging to the same protein were summed and the Log10 of this summed protein intensity was plotted (Fig. 4b). This expression profile exhibited a measured dynamic range of summed protein intensities over three orders of magnitude. The most abundant proteins based on summing all identified peptides specific to the respective proteins appeared to include both structural proteins, such as myelin basic protein, spectrin, and actin, as well as proteins with other cellular functions such as ADP/ATP translocase and PSD-95 (Disks large homolog 4).
Our findings confirm previous analyses of the PSD proteome and validate our methods. First, of the 425 proteins we identified, 299 (70%) have been previously identified in the PSD fraction using proteomic analyses (Husi et al. 2000; Jordan et al. 2004; Li et al. 2004; Peng et al. 2004; Yoshimura et al. 2004; Cheng et al. 2006; Moron et al. 2007; Trinidad et al. 2008). Second, the proteins identified here fell into similar functional protein categories as previous studies have shown (Peng et al. 2004; Cheng et al. 2006). Thus, our findings confirm previous PSD proteome findings and support the validity of our methods for isolating the PSD fraction from brain homogenate and characterizing its proteome using an LC-MS/MS-based strategy.
Although label-free quantitation strategies offer a high degree of experimental flexibility, particular care needs to be taken in sample preparation reproducibility, including generation of PSD fractions. To evaluate the technical variation of LC-MS/MS analysis, following in situ trypsindigestion, each digested PSD sample was subjected to duplicate LC-MS/MS acquisitions prior to accurate-mass retention time alignment within Rosetta Elucidator. Following qualitative assignment of peptide identification for each detected peak within the aligned dataset, all peptide intensities were normalized to the robust mean of the ion intensities across all samples, excluding the highest and lowest 10% of the measured signals. Protein expression levels were quantified by measuring and summing the area under the curve intensities of each detectable peptide within a protein across all individual samples. The results indicated that the average analytical variation in measured protein abundance (n = 379 unique proteins) across the three replicate injections was 11.9% (relative standard deviation).
To assess the reproducibility of the overall analytical approach, a dedicated reproducibility study on triplicate PSD fractions was performed on naive adult male Sprague–Dawley rats (Figure S1). The increase in variability resulting from additional sample manipulation steps incurred during the PSD preparation protocol appeared to be negligible, as the average % relative standard deviation in measured protein intensity across the same n = 379 proteins was 13.7%, a minimal increase from 11.9%.
To analyze the effect of biology on the reproducibility of our assay, we compared the difference in the PSD proteome among four dummy-treated or six VNS-stimulated adult male Sprague–Dawley rats. The average biological variation for a given protein across the four dummy-treated animals was 35.8% while the average variation across the six VNS-treated animals was 37.7%. As expected, these average variations are both higher than the average technical variation of the platform (13.7%). These results demonstrate that the quantitative LC/MS/MS approach employed for this study was reproducible and effective for global analysis of the proteome of PSD in response to a treatment paradigm.
Proteomic analysis of PSD protein content regulation by VNS
We next sought to determine whether 1 week of VNS modifies the PSD proteome in amygdala/piriform cortex. To that end, we implanted adult male Sprague–Dawley rats with active VNS devices or sham devices that deliver no current (dummy-treated animals). After animals were treated with VNS for 1 week, the PSD samples of amygdala/piriform cortex were isolated from all animals and then subjected to our proteomic analysis (Fig. 5a). Of the 425 proteins identified in our PSD samples, 1 week of VNS (N = 6 animals) induced a significant change in protein content in 56 proteins in comparison to dummy-treated animals (N = 4 animals) (n = 27 proteins up-regulated; n = 29 proteins down-regulated, p < 0.05). We found that there were 22 proteins with a significant (p < 0.01) change in protein content greater than 1.5 fold. Of the 22 proteins, the expression in the PSD fraction of VNS-treated animals was significantly increased for 10 proteins and significantly decreased for 12 proteins (Table 1). Analyses of five exemplary proteins, including neurexin-1α, α2δ1, γ3 subunit of voltage-gated calcium channel, cadherin 13, and EAAT1, revealed consistent and significant changes in protein content in VNS-treated animals compared to controls (Fig. 5b–f). To gain insight into how VNS affects signaling pathways in the CNS, the 56 proteins differentially expressed in PSD samples were subjected to Ingenuity pathway analysis. Consistent with the role of PSD, the top canonical pathway identified was glutamate receptor signaling, the top network identified was cell-to-cell signaling and interaction in the nervous system, the top disease associated with our identified proteins was neurological diseases, the top molecular and cellular function was cellular assembly and organization, and the top two hits for physiological system development and function were behavior and nervous system development and function (Table S2).
Table 1.
Proteins differentially expressed by > 1.5-fold in VNS-treated animals and control animals and reaching a p value < 0.01
| Proteins increased in VNS animals |
Proteins decreased in VNS animals |
||||
|---|---|---|---|---|---|
| Protein description | Fold change | p-value | Protein description | Fold | p-value |
| Band 4.1-like protein 3 | 4.63 | 1.2E-10 | Alpha-actinin-1 | 13.14 | 1.3E-05 |
| Excitatory amino acid transporter 1 | 2.34 | 1.3E-05 | Voltage-dependent calcium channel gamma-3 subunit | 10.91 | 5.0E-03 |
| Neurexin-1-alpha | 2.27 | 2.1E-05 | Ras-related protein Rab-5A – Mus musculus (Mouse) | 2.27 | 1.4E-03 |
| Casein kinase I isoform delta | 1.78 | 4.2E-07 | Cyclin-dependent kinase-like 5 | 2.05 | 1.1E-03 |
| Myosin-10 | 1.70 | 5.1E-04 | 60S ribosomal protein L19 | 2.03 | 9.6E-05 |
| Vacuolar proton pump subunit E 1 | 1.62 | 5.4E-03 | ProSAP-interacting protein 1 | 1.82 | 8.1E-03 |
| Thy-1 membrane glycoprotein | 1.59 | 1.4E-05 | Mitochondrial import receptor subunit TOM20 homolog | 1.75 | 1.1E-03 |
| Voltage-dependent calcium channel subunit alpha-2/delta-1 | 1.55 | 3.0E-03 | Keratin, type II cytoskeletal 6A | 1.74 | 1.0E-03 |
| Vacuolar ATP synthase catalytic subunit A | 1.55 | 1.8E-04 | Alcohol dehydrogenase 1 | 1.72 | 9.6E-05 |
| Cadherin-13 | 1.51 | 1.6E-03 | 40S ribosomal protein S14 | 1.58 | 7.9E-05 |
| Disks large-associated protein 3 | 1.57 | 9.9E-03 | |||
| Excitatory amino acid transporter 2 | 1.52 | 5.5E-03 | |||
To characterize the differences in global protein expression profiles between dummy-treated and VNS-treated animals, we subjected the z-score corrected intensity values from each of the 425 proteins to a three-dimensional principal component analysis (Figure S2a). Although this analysis takes into account expression changes of all proteins, the vast majority of which were not expected to change as a function of VNS, the analysis was able to globally differentiate the dummy and VNS-treated groups on PC2. Plotting only those 22 proteins found to be differentially regulated in the three-dimensional principal component analysis, a clear differentiation based on VNS treatment can be observed for the most significant principal component, PC1 (Figure S2b).
Immunoblotting analysis of PSD content regulation by VNS
To confirm the changes of PSD protein content discovered by our label-free mass spectrometry method in PSD fractions isolated from amygdala-pyriform cortex, we selected five proteins, neurexin-1α, VDCC α2δ1, VDCC γ3, EAAT1, and cadherin 13, each of which represented the pathways involved in the VNS effects. PSD fractions isolated from the cerebral cortices of dummy-treated and VNS-treated animals were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and western blot analysis was conducted using antibodies against selected proteins in the PSD. Immunoreactivities for neurexin-1α, α2δ1 and γ3 subunits of voltage-gated calcium channels, cadherin 13 as well as EAAT1 were found in the cortical PSD fractions (Fig. 6a). Immunoreactivity for PSD-95, a well-characterized PSD protein, was also found in PSD fractions (Fig. 6a). Expression levels of PSD-95 did not differ between VNS-treated and dummy-treated animals (Fig. 6b and h), consistent with no significant change in PSD-95 expression between groups identified by proteomic analysis. The constant expression of PSD-95 allowed us to use this protein to ensure protein and loading control in our western blot analyses. Similar to mass spectrometry studies of PSD fractions from amygdala-pyriform cortex, 1 week of VNS treatment resulted in significant increases in content of neurexin-1α, VDCCα2δ1, and cadherin 13 (p < 0.01, unpaired t-tests, Fig. 6c–e) and significant reductions of γ3 subunit of voltage-gated calcium channels (p < 0.05, unpaired t-test, Fig. 6f). Levels of EAAT1 exhibited a trend toward a significant increase (p = 0.05, unpaired t-test, Fig. 6g) and PSD-95 and actin levels did not differ between treatment groups (p > 0.05, unpaired t-test, Fig. 6h–i). These results confirm and extend our findings of differential expression of PSD proteins identified by the label-free mass spectrometry analyses.
Discussion
Vagal nerve stimulation is an FDA-approved method for treatment of both epileptic seizures and depression that are unresponsive to medications. The mechanisms underlying the beneficial effects of VNS for these conditions are incompletely understood. Toward that end, we conducted electrophysiological recordings of the effects of vagal nerve stimulation in adult rats, the results of which led us to ask whether VNS modified the proteome of excitatory synapse. The principal findings are: (i) VNS modified the activity of neurons within amygdala and hippocampus as assessed by recordings of both single neurons and populations performed in awake, freely moving animals; (ii) we established a highly reproducible label-free mass spectrometry-based proteomic strategy for analysis of the PSD proteome; (iii) among the 425 proteins identified in the PSD, treatment with VNS for 1 week modified the content of 22 proteins when compared with implanted but unstimulated animals, the changes confirmed by western blot analyses for a subset of proteins; (iv) three of the proteins whose expression was increased by VNS, namely neurexin-1α, cadherin 13, and α2δ1, have established roles in formation of excitatory synapses. We conclude that VNS modifies the activity of neurons within amygdala and hippocampus and also modifies the protein composition of excitatory synapses. The nature of the VNS-induced modifications of the proteome of excitatory synapses provides clues to the biological mechanisms of its beneficial effects.
VNS is an FDA-approved therapy for medically refractory epilepsy and treatment-resistant depression in humans. Our prior study demonstrated that VNS is able to elevate threshold for evoking seizures by electrical stimulation locally in the amygdala in the kindling model of temporal lobe epilepsy in rats (Alexander and McNamara 2012). These observations led us to ask whether VNS modifies the activity of neurons within amygdala as assessed by electro-physiological recordings in awake, behaving rats. We also assessed neuronal activity within the synaptically connected hippocampus using LFPs recorded from populations of neurons. Consistent with our hypothesis, VNS modified the activity of individual neurons within amygdala in a stimulation-dependent manner. VNS also modified neuronal activity within hippocampus, specifically producing an increase of theta power in the 5.5–6.5 Hz frequency.
What neuronal circuits mediate these effects of VNS on neuronal activity in amygdala and hippocampus? The absence of known direct connections from the vagal nerve to neurons within amygdala and hippocampus implies that the effects are mediated by some intermediary(ies). We favor the candidacy of ascending neuromodulatory systems arising from the locus ceruleus and/or dorsal raphe. That is, afferent fibers of the vagus nerve project to the nucleus of the solitary tract, which in turn projects heavily to the locus ceruleus and dorsal raphe (Van Bockstaele et al. 1999; Nemeroff et al. 2006), nuclei from which noradrenergic and serotonergic neurons, respectively, arise. Several lines of evidence support the noradrenergic neurons in LC in particular. First, acute VNS of adult rats rapidly increases the firing of LC but not dorsal raphe neurons (Dorr 2006). Second, both acute and chronic VNS, with stimulation parameters identical to those used in our study, increase norepinephrine (NE) but not serotonin content in hippocampus (Roosevelt et al. 2006; Raedt et al. 2011; Manta et al. 2012). Third, direct electrical stimulation of NE-containing LC neurons increases theta power in the hippocampus (Walling et al. 2010), whereas stimulation of serotonin-containing raphe neurons either decreases or has no effect on theta power (Yamamoto et al. 1979). Importantly, local application of both NE and serotonin modify the activity of neurons within the amygdala, the direction of the effect based on the cell type, and receptors expressed (Cheng et al. 1998; Rainnie 1999; Buffalari and Grace 2007; Kaneko et al. 2008; Miyajima et al. 2010; McCool 2014).
The molecular mechanisms by which VNS produces its beneficial effects in medically refractory epilepsy are incompletely understood. One plausible mechanism involves modification of excitatory synapses, in part because plasticity of excitatory synapses is thought to contribute to the hyperexcitability of epilepsy (Sutula and Steward 1987; Goussakov et al. 2000). The PSD is a membrane specialization of the postsynaptic component of excitatory synapses in the CNS and is amenable to isolation and molecular analysis. The facts that the protein composition of the PSD is regulated by neuronal activity (Trinidad et al. 2013) and that VNS itself modifies neuronal activity strengthened the rationale for asking whether VNS regulates the molecular composition of excitatory synapses. To address this question, we conducted an unbiased, quantitative analysis of the proteins contained within excitatory synapses of amygdala/piriform cortex. Toward that end, we developed a label-free mass spectrometry-based proteomic strategy. Unlike approaches utilizing isobaric tags for quantifying proteins or stable isotope labeled amino acids, the label-free strategy utilized here allows for direct analysis of a tissue proteome across any number of unique treatment groups or biological replicates.
Our LC-MS/MS method identified the majority of proteins previously reported in the biochemically purified PSD fraction, and pathway analysis showed these proteins identified in our study are consistent with proteins expressed in PSD samples, confirming that our workflow is sufficiently robust to examine the comprehensive PSD proteome. Furthermore, our findings extend previous reports on the PSD proteome. Of the 425 proteins identified in our PSD samples, 126 proteins (~ 30%) have not previously been identified in the PSD proteome (bold proteins in Table S1); the majority fall into previously recognized categories, but some proteins were classified as other because of the lack of an appropriate annotation. Notably, previous proteomic analyses examined PSDs isolated from forebrain, whereas this study centered on the amygdala/pyriform cortex, a difference that may account for detection of previously unidentified proteins.
Treatment of naive animals with VNS for 1 week caused altered content of 56 proteins in the PSD. Might these provide any clues to biological consequences of VNS that underlie its beneficial effects? The pathway analysis revealed that cell-cell signaling and interaction as well as cellular assembly and organization were involved in the VNS effects. This in turn suggests an interesting possibility that VNS may increase formation of excitatory synapses. Consistent with this hypothesis, three proteins of interest that showed increased expression in VNS-treated animals and contribute to the formation of excitatory synapses are neurexin 1α, α2δ1, and cadherin 13 (Table 1 and Figs 5 and 6). Importantly, we found that each of these proteins was significantly, and consistently, increased among VNS-treated animals. Neurexin 1α functions as a trans-synaptic adhesion molecule through its binding neuroligins, and this binding mediates formation and expansion of excitatory synapses (Boucard et al. 2005; Singh et al. 2016). Likewise, cadherin 13 is a member of family of cell adhesion molecules and RNAi-mediated reduction of cadherin 13 impaired synapse formation in cultured neurons (Paradis et al. 2007). Finally, α2δ1 promotes formation of excitatory synapses in cultured retinal ganglion cells by serving as the receptor for thrombospondin derived from astrocytes (Eroglu et al. 2009). Although neurexin 1α has been localized to presynaptic terminals, we and others (Peng et al. 2004; Cheng et al. 2006; Bayes et al. 2012) detected its presence in a PSD fraction, perhaps because of its tightly binding neuroligin 1, which was also detected in our PSD prep. Interestingly, mutations of neurexin 1α and cadherin 13 have been associated with autism and other neuropsychiatric diseases, perhaps a consequence of disordered synaptogenesis. In sum, the VNS-induced increases in PSD content of these three proteins suggests that one mechanism by which VNS elevates seizure threshold would be to promote formation of excitatory synapses, presumably onto inhibitory neurons. We hypothesize that the effects of VNS on neuronal activity is a result of increased NE signaling. Whether NE itself may have promoted synaptogenesis of excitatory synapses in our preparation or acted through a secondary mechanism to enhance excitatory synapse formation requires further study.
Additional potential mechanisms of VNS action include reduced dendritic spine density and weakening α-amino-3-hydroxy-5-methylisoxazole-4-propionate receptors at excitatory synapse between principal neurons, mechanisms suggested by the consistently reduced content of voltage-dependent calcium channel γ3 subunit found among VNS-treated animals as well as the reduced content of cyclin-dependent kinase-like 5 (Kato et al. 2010; Ricciardi et al. 2012). Finally, pathway analysis suggested that glutamate receptor signaling is a primary target pathway for VNS. Consistent with this assertion, VNS increased expression of the glutamate transporter EAAT1 in VNS-treated animals, thereby likely contributing to increased synaptic glutamate clearance.
In summary, our study demonstrates that VNS modifies both neuronal activity in amygdala and hippocampus and the composition of excitatory synapses in the CNS. Results of our unbiased analyses of the PSD proteome and validation by immunoblotting suggest plausible biological mechanisms that may underlie the beneficial effects of VNS.
Supplementary Material
Acknowledgments
This work was supported by a grant from Cyberonics, Inc. Yang Zhong Huang is supported by the CURE Taking Flight Award. James O. McNamara served on the Cyberonics Mechanism of Action advisory board from 2003–2010.
Abbreviations used:
- AMRT
accurate-mass retention time
- CNS
central nervous system
- EAAT1
excitatory amino acid transporter 1
- FDA
Food and Drug Administration
- FDR
false discovery rate
- iTRAQ
isobaric tags for quantifying proteins
- LC
locus ceruleus
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- LFP
local field potential
- NE
norepinephrine
- PCA
principal component analysis
- PSD
postsynaptic density
- RSD
relative standard deviation
- SILAC
stable isotope labeled amino acids
- TFA
trifluoroacetic acid
- VDCCα2δ1
voltage-dependent calcium channel α2δ1
- VNS
vagal nerve stimulation
Footnotes
Supporting information
Additional Supporting Information may be found online in the supporting information tab for this article:
conflict of interest disclosure
The remaining authors have no conflicts of interest.
References
- Alexander GM and McNamara JO (2012) Vagus nerve stimulation elevates seizure threshold in the kindling model. Epilepsia 53, 2043–2052. [DOI] [PubMed] [Google Scholar]
- Bayés A and Grant SGN (2009) Neuroproteomics: understanding the molecular organization and complexity of the brain. Nat. Rev. Neurosci. 10, 635–646. [DOI] [PubMed] [Google Scholar]
- Bayés A, Collins MO, Croning MDR, van de Lagemaat LN, Choudhary JS and Grant SGN (2012) Comparative study of human and mouse postsynaptic proteomes finds high compositional conservation and abundance differences for key synaptic proteins. PLoS ONE 7, e46683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beekwilder JP and Beems T (2010) Overview of the clinical applications of vagus nerve stimulation. J. Clin. Neurophysiol. 27, 130–138. [DOI] [PubMed] [Google Scholar]
- Boucard AA, Chubykin AA, Comoletti D, Taylor P and Sudhof TC (2005) A splice code for trans-synaptic cell adhesion mediated by binding of neuroligin 1 to alpha- and beta-neurexins. Neuron 48, 229–236. [DOI] [PubMed] [Google Scholar]
- Buffalari DM and Grace AA (2007) Noradrenergic modulation of basolateral amygdala neuronal activity: opposing influences of alpha-2 and beta receptor activation. J. Neurosci. 27, 12358–12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng LL, Wang SJ and Gean PW (1998) Serotonin depresses excitatory synaptic transmission and depolarization-evoked Ca2+ influx in rat basolateral amygdala via -HT1A receptors. Eur. J. Neurosci. 10, 2163–2172. [DOI] [PubMed] [Google Scholar]
- Cheng D, Hoogenraad CC, Rush J et al. (2006) Relative and absolute quantification of postsynaptic density proteome isolated from rat forebrain and cerebellum. Mol. Cell Proteomics 5, 1158–1170. [DOI] [PubMed] [Google Scholar]
- Dorr AE (2006) Effect of vagus nerve stimulation on serotonergic and noradrenergic transmission. J. Pharmacol. Exp. Ther. 318, 890–898. [DOI] [PubMed] [Google Scholar]
- Eroglu C, Allen NJ, Susman MW et al. (2009) Gabapentin receptor alpha2delta-1 is a neuronal thrombospondin receptor responsible for excitatory CNS synaptogenesis. Cell 139, 380–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goussakov IV, Fink K, Elger CE and Beck H (2000) Metaplasticity of mossy fiber synaptic transmission involves altered release probability. J. Neurosci. 20, 3434–3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YZ, Pan E, Xiong Z-Q and McNamara JO (2008) Zinc-mediated transactivation of TrkB potentiates the hippocampal mossy fiber-CA3 pyramid synapse. Neuron 57, 546–558. [DOI] [PubMed] [Google Scholar]
- Husi H, Ward MA, Choudhary JS, Blackstock WP and Grant SG (2000) Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nat. Neurosci. 3, 661–669. [DOI] [PubMed] [Google Scholar]
- Jordan BA, Fernholz BD, Boussac M, Xu C, Grigorean G, Ziff EB and Neubert TA (2004) Identification and verification of novel rodent postsynaptic density proteins. Mot. Cell Proteomics 3, 857–871. [DOI] [PubMed] [Google Scholar]
- Kaneko K, Tamamaki N, Owada H et al. (2008) Noradrenergic excitation of a subpopulation of GABAergic cells in the basolateral amygdala via both activation of nonselective cationic conductance and suppression of resting K+ conductance: a study using glutamate decarboxylase 67-green fluorescent protein knock-in mice. Neuroscience 157, 781–797. [DOI] [PubMed] [Google Scholar]
- Kato AS, Gill MB, Yu H, Nisenbaum ES and Bredt DS (2010) TARPs differentially decorate AMPA receptors to specify neuropharmacology. Trends Neurosci. 33, 241–248. [DOI] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E and Aebersold R (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392. [DOI] [PubMed] [Google Scholar]
- Kennedy MB, Bennett MK and Erondu NE (1983) Biochemical and immunochemical evidence that the “major postsynaptic density protein” is a subunit of a calmodulin-dependent protein kinase. Proc. Natl Acad. Sci. USA 80, 7357–7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li KW, Hornshaw MP, Van Der Schors RC et al. (2004) Proteomics analysis of rat brain postsynaptic density. Implications of the diverse protein functional groups for the integration of synaptic physiology. J. Biol. Chem. 279, 987–1002. [DOI] [PubMed] [Google Scholar]
- Likhtik E, Pelletier JG, Popescu AT and Pare D (2006) Identification of basolateral amygdala projection cells and interneurons using extracellular recordings. J. Neurophysiol. 96, 3257–3265. [DOI] [PubMed] [Google Scholar]
- Manta S, El Mansari M, Debonnel G and Blier P (2012) Electrophysiological and neurochemical effects of long-term vagus nerve stimulation on the rat monoaminergic systems. Int. J. Neuropsychopharmacol. 16, 459–470. [DOI] [PubMed] [Google Scholar]
- McCool BA (2014) Lateral/basolateral amygdala serotonin type-2 receptors modulate operant self-administration of a sweetened ethanol solution via inhibition of principal neuron activity. Frontiers in Integrative Neuroscience. 2014;8:5. doi: 10.3389/fnint.2014.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyajima M, Ozaki M, Wada K and Sekiguchi M (2010) Noradrenaline-induced spontaneous inhibitory postsynaptic currents in mouse basolateral nucleus of amygdala pyramidal neurons: comparison with dopamine-induced currents. Neurosci. Lett. 480, 167–172. [DOI] [PubMed] [Google Scholar]
- Moron JA, Abul-Husn NS, Rozenfeld R, Dolios G, Wang R and Devi LA (2007) Morphine administration alters the profile of hippocampal postsynaptic density-associated proteins: a proteomics study focusing on endocytic proteins. Mol. Cell Proteomics 6, 29–42. [DOI] [PubMed] [Google Scholar]
- Nemeroff CB, Mayberg HS, Krahl SE, McNamara J, Frazer A, Henry TR, George MS, Charney DS and Brannan SK (2006) VNS therapy in treatment-resistant depression: clinical evidence and putative neurobiological mechanisms. Neuropsychopharmacology 31, 1345–1355. [DOI] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E and Aebersold R (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658. [DOI] [PubMed] [Google Scholar]
- Nicolelis MA, Ghazanfar AA, Faggin BM, Votaw S and Oliveira LM (1997) Reconstructing the engram: simultaneous, multisite, many single neuron recordings. Neuron. 18, 529–537. [DOI] [PubMed] [Google Scholar]
- Paradis S, Harrar DB, Lin Y, Koon AC, Hauser JL, Griffith EC, Zhu L, Brass LF, Chen C and Greenberg ME (2007) An RNAi-based approach identifies molecules required for glutamatergic and GABAergic synapse development. Neuron 53, 217–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pare D and Gaudreau H (1996) Projection cells and interneurons of the lateral and basolateral amygdala: distinct firing patterns and differential relation to theta and delta rhythms in conscious cats. J. Neurosci. 16, 3334–3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Kim MJ, Cheng D, Duong DM, Gygi SP and Sheng M (2004) Semiquantitative proteomic analysis of rat forebrain postsynaptic density fractions by mass spectrometry. J. Biol. Chem. 279, 21003–21011. [DOI] [PubMed] [Google Scholar]
- Pikkarainen M, Rönkkö S, Savander V, Insausti R and Pitkänen A (1999) Projections from the lateral, basal, and accessory basal nuclei of the amygdala to the hippocampal formation in rat. J. Comp. Neurol. 403, 229–260. [PubMed] [Google Scholar]
- Raedt R, Clinckers R, Mollet L et al. (2011) Increased hippocampal noradrenaline is a biomarker for efficacy of vagus nerve stimulation in a limbic seizure model. J. Neurochem. 117, 461–469. [DOI] [PubMed] [Google Scholar]
- Rainnie DG (1999) Serotonergic modulation of neurotransmission in the rat basolateral amygdala. J. Neurophysiol. 82, 69–85. [DOI] [PubMed] [Google Scholar]
- Ricciardi S, Ungaro F, Hambrock M et al. (2012) CDKL5 ensures excitatory synapse stability by reinforcing NGL-1-PSD95 interaction in the postsynaptic compartment and is impaired in patient iPSC-derived neurons. Nat. Cell Biol. 14, 911–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roosevelt RW, Smith DC, Clough RW, Jensen RA and Browning RA (2006) Increased extracellular concentrations of norepinephrine in cortex and hippocampus following vagus nerve stimulation in the rat. Brain Res. 1119, 124–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M and Hoogenraad CC (2007) The postsynaptic architecture of excitatory synapses: a more quantitative view. Annu. Rev. Biochem. 76, 823–847. [DOI] [PubMed] [Google Scholar]
- Singh SK, Stogsdill JA, Pulimood NS, Dingsdale H, Kim YH, Pilaz L-J, Kim IH et al. (2016) Astrocytes assemble thalamocortical synapses by bridging NRX1α and NL1 via hevin. Cell 164, 183–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD (2002) A direct approach to false discovery rates. J. R. Stat. Soc. Ser B 64, 479–498. [Google Scholar]
- Sutula T and Steward O (1987) Facilitation of kindling by prior induction of long-term potentiation in the perforant path. Brain Res. 420, 109–117. [DOI] [PubMed] [Google Scholar]
- Trinidad JC, Thalhammer A, Specht CG, Lynn AJ, Baker PR, Schoepfer R and Burlingame AL (2008) Quantitative analysis of synaptic phosphorylation and protein expression. Mol Cell Proteomics 7, 684–696. [DOI] [PubMed] [Google Scholar]
- Trinidad JC, Thalhammer A, Burlingame AL and Schoepfer R (2013) Activity-dependent protein dynamics define interconnected cores of co-regulated postsynaptic proteins. Mol. Cell Proteomics 12, 29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bockstaele EJ, Peoples J and Telegan P (1999) Efferent projections of the nucleus of the solitary tract to peri-locus coeruleus dendrites in rat brain: evidence for a monosynaptic pathway. J. Comp. Neurol. 412, 410–428. [DOI] [PubMed] [Google Scholar]
- Walling SG, Brown RAM, Milway JS, Earle AG and Harley CW (2010) Selective tuning of hippocampal oscillations by phasic locus coeruleus activation in awake male rats. Hippocampus 21, 1250–1262. [DOI] [PubMed] [Google Scholar]
- Weng L, Dai H, Zhan Y, He Y, Stepaniants SB and Bassett DE (2006) Rosetta error model for gene expression analysis. Bioinformatics 22, 1111–1121. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Watanabe S, Oishi R and Ueki S (1979) Effects of midbrain raphe stimulation and lesion on EEG activity in rats. Brain Res. Bull. 4, 491–495. [DOI] [PubMed] [Google Scholar]
- Yoshimura Y, Yamauchi Y, Shinkawa T, Taoka M, Donai H, Takahashi N, Isobe T and Yamauchi T (2004) Molecular constituents of the postsynaptic density fraction revealed by proteomic analysis using multidimensional liquid chromatography-tandem mass spectrometry. J. Neurochem. 88, 759–768. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


