Abstract
Background
Carriage of carbapenem-resistant Enterobacteriaceae (CRE) in humans may contribute to the dissemination of CRE and impact on communities and healthcare facilities. Carbapenem-resistant Escherichia coli (CREC) is one of the major type of CRE in the human gut. Here, we describe a cross-sectional study to investigate the prevalence of CREC, and in particular the mcr-1 carrying CREC, in health volunteers in China.
Methods
During September to December 2016, 3859 non-duplicated stool specimens were collected from healthy volunteers who received regular physical examinees in healthcare centers located in 19 provinces across China. Enrichment culture supplemented meropenem was used to isolate CREC. Carbapenemase producing determinants and the mcr-1 gene were determined by PCR amplification and sequencing. Isolates were further analyzed by antibiotic susceptibility test, genotyping, and whole genome analysis.
Findings
A total of 92 non-duplicated CREC were isolated from 3859 stool specimens, among which 43 CREC are carbapenemase positive. In addition, the co-existence of blaNDM and mcr-1 was found in 14 CREC, which also showed resistance to the majority of all antimicrobial agents analyzed. The genetic background of these CREC isolates are highly diversified based on molecular typing. Furthermore, whole genome sequence indicated that NDM-5 is the predominant determinant conferring carbapenem resistance in CREC, and that NDM-5 carrying plasmids (IncX3) are very similar.
Interpretation
The incidence of CREC carriage in healthy people in China was small; however, the co-existence of CREC with mcr-1 is disconcerting. Therefore, pre-screening prior to admission and monitoring of patients on high-dependency wards is highly recommended to control and prevent the dissemination of CRE in hospitals.
Outstanding Question
The high prevalence of CREC in the healthy people should not be underestimated, as it may increase the risk of infection. This knowledge could have impact on the pre-screening and monitoring of CRE before patient administration.
Keywords: CREC, NDM, MCR-1, Gut, Carriage, Healthy people
Research in context
Evidence before this study
Carbapenems Resistant Enterobacteriaceae (CRE) has become a global threat to public health, and numerous studies have indicated the wide prevalence of CRE in the hospital high-dependency units. However, the carriage of CRE, especially individuals admitted to and subsequently discharged from hospitals, has been underestimated and poorly studied.
Added value of this study
In this study, for the first time, a large scale observation study has been conducted to determine the carriage of CRE in healthy volunteers in China, and the results indicated a 2.38% (95% CI, 1.95%-2.91%) national-wide carriage of CRE in the healthy people. It is surprisingly that NDM-5 is the dominant determinants in the CRE isolates. This study provides the first insight of the carriage of CRE in a large cohort of healthy people.
Implications of all the available evidence
The outcome of this study suggest that the surveillance and monitoring of the carriage of CRE in the healthy people should be further evaluated, particularly for patients admitted to hospital high-dependency wards (e.g. oncology, ICU) to prevent nosocomial outbreaks.
Alt-text: Unlabelled Box
1. Introduction
Antimicrobial resistance has become a global concern that threatens our ability to treat common infections caused by bacterial organisms. Among the major multidrug-resistant organisms, the emergence and dissemination of carbapenem-resistant Enterobacteriaceae (CRE) is recognized as one of the most serious threats to public health worldwide [1], [2], [3], [4], [5], [6]. Infections caused by CRE are often associated with high mortality, primarily due to the limited availability of therapeutic options [7], [8]. The recent increase in CRE were mainly due to the acquisition of carbapenem hydrolyzing enzymes such as Klebsiella pneumoniae carbapenemase (KPC) and the New Delhi metallo-β-lactamase (NDM) [9], [10]. The carbapenem resistance in K. pneumoniae is commonly mediated by blaKPC, often observed in the hospitalized patients, while that in E. coli was predominantly cause by blaNDM, which identified in both hospitalized patients and healthy people.
To date, over 20 NDM variants have been identified since the discovery of NDM-1 in India [11]. The rapid plasticity of blaNDM in such a short period of time may enhance its growing importance not only in the human but also in the food animals. These NDM variants are highly similar with only several amino acids difference. Their hydrolysis activity toward carbapenem also varies. NDM-5 (Val88Leu, Met154Leu) and its closely related variants, NDM-17 (Val88Leu, Met154Leu, Glu170Lys) and NDM-20 (Val88Leu, Met154Leu, Arg270His), possess stronger carbapenemase activity comparing with NDM-1, and the plasmids carrying NDM-5, NDM-17, or NDM-20 share significant high identity over 99% [11]. In China, NDM-5 has been frequently observed during surveillance of both clinical and food animal sectors [12], [13], [14].
Colistin is considered as the last resort treatment for life-threatening infections caused by CRE. The recent emergence of the plasmid-mediated gene mcr-1 conferring resistance to colistin in CRE strains further complicates therapeutic options [15], leaving only a limited number of drugs for the treatment of infections. Recently, Li et al., reported the rapid increase of NDM and KPC as well as the emergence of mcr-1 in one of the largest hospitals in Henan province [16]. Usually, CRE infections occur in hospitalized patients, nursing homes, and other healthcare settings. Historically, CRE was usually thought to be acquired within hospital institutions; however, recently, several studies have suggested that CRE infections may be attributable to CRE colonized in the normal flora from healthy people [17]. Previous study also indicated that there is an increased risk of CRE infection and mortality in patients with CRE colonization at the time of intensive care unit (ICU) admission [18]. However, the prevalence of CRE in the normal flora of healthy people has been poorly studied, and the potential risk of carriage of CRE in the healthy population may have been underestimated.
Enterobacteriaceae, especially Escherichia coli, are the common and normal flora in healthy people, but are opportunistic pathogens and can cause a variety of nosocomial infections. Our recent study showed that high incidence of CRE was found in food animals, which may be transmitted to humans through the food chain, resulting in the colonization of CRE in healthy people [14]. The carriage of CRE might not only pose a potential risk for healthy people, but also contribute to the dissemination of these microorganisms in hospital settings. Therefore, to better understand the current epidemiological trends and characteristics of CRE in healthy people, we investigated the prevalence of carbapenem-resistant E. coli (CREC) isolates collected from healthy volunteers in 19 provinces across China. We also characterized the main mechanism(s) conferring carbapenem resistance as well as investigated the emergence of mcr-1 in CREC.
2. Materials and Methods
2.1. Study Design
The aim of this study is to investigate the carriage of CREC, especially the mcr-1 carrying CREC, in healthy people. We performed a cross-sectional multi-center study from September 2016 to December 2016. Stool specimens were collected from healthy people who received the regular physical examinees in 52 general hospitals located in 19 provinces including Beijing, Fujian, Guangdong, Guangxi, Guizhou, Hainan, Heilongjiang, Henan, Hubei, Hunan, Inner Mongolia, Jilin, Jiangxi, Ningxia, Shandong, Shanxi, Shaanxi, Tibet and Zhejiang, which accounted for a population of 844.5 million (~ 60%) in China. All the volunteers have been inquired about the medical history and antibiotic prescription. Only volunteers, who have not been hospitalized and taken any antibiotic in the last three months, will be selected for the screening in this study. At least 50 samples were collected from each province. In China, CRE screening is not a mandatory procedure in the clinical medicine, but it is required if the patient will be administrated into the ICU, and the screening guide is following the procedure issued by the United States CDC.
Enrichment culture supplemented with 0.3 μg/ml meropenem was used to isolate the CREC. Briefly, about one-gram stool was inoculated into 5 ml of buffered peptone water in a 10 ml tube and incubated at 37 °C overnight. A 10 μl aliquot of the Enterobacteriaceae Enrichment broth was then spread onto a SS (Salmonella Shigella) agar plate supplemented with 0.3 μg/ml meropenem and incubated at 37 °C overnight. The pure colonies of E. coli were selected based on their color and morphology, and the isolates were identified using matrix-assisted laser desorption ionization/time of flight mass spectrometry (MALDI-TOF MS) (Bruker Daltonik GmbH, Bremen, Germany).
Ethical approval was given by The Second Affiliated Hospital of Zhejiang University. Individual consent forms were translated into Mandarin and consent was obtained for all healthy volunteers before sampling.
2.2. Gene Screening and Whole Genome Sequencing
First, the presence of carbapenem-resistant genes (blaNDM, blaKPC, blaIMP, blaVIM, blaOXA-48) and colistin-resistant gene (mcr-1) were screened using PCR, and then the positive products were validated with Sanger sequencing. All positive isolates were subjected to the whole genome sequencing. Genomic DNA of isolate was extracted using the Wizard Genomic DNA Purification Kit (Promega, Beijing, China), following the manufacturer's instructions. Indexed Illumina sequencing libraries were prepared using TruSeq DNA PCR-Free Sample Preparation Kit (Illumina Inc., San Diego, CA) following the standard protocol and sequenced on the Illumina HiSeq 2500 platform (Annoroad Biotec Co.) according to the manufacturer's protocols. The whole genome sequencing data was analyzed using multiple programs including SPAdes (whole genome sequence assembly), SRST2 (antibiotic resistance gene and MLST analysis), BRIG (BLAST ring comparison), Harvest and iTOL (phylogenetic tree analysis) as previously described [14]. After the whole genome sequencing, gap filing was conducted to complete the NDM-5 carrying plasmid using PCR and Sanger sequencing.
2.3. Antimicrobial Susceptibility Testing
The minimum inhibitory concentrations (MICs) for the CREC isolates against a panel of antibiotics were determined using broth micro-dilution method according to the Clinical and Laboratory Standards Institute (CLSI) guidelines. The results of antimicrobial susceptibility for carbapenems (imipenem, meropenem), cephems (cefepime, ceftazidime, cefotaxime, cefazolin, cefuroxime), piperacillin, β-lactam/β-lactamase combination (piperacillin-tazobactam, ampicillin-sulbactam, ticarcillin-clavulanate, cefoperazone-sulbactam (2:1), ceftazidime-clavulanate, cefotaxime-clavulanate), aminoglycosides (gentamicin, amikacin), fluoroquinolones (ciprofloxacin, levofloxacin), trimethoprim-sulfamethoxazole, minocycline, aztreonam, chloramphenicol, fosfomycin, and nitrofurantoin were interpreted according to CLSI guidelines, while the resistance for colistin and tigecycline was interpreted according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST).
2.4. Molecular Typing
All the blaNDM–positive E. coli were subjected to XbaI-digested pulsed-field gel electrophoresis (PFGE) typing. Salmonella enterica serotype Braenderup H9812 was used as a size marker. A dendrogram was generated from the homology matrix with a coefficient of 0.5% using the unweighted pair-group method using arithmetic averages (“UPGMA”) to describe the relationships among PFGE profiles. Isolates were considered to belong to the same PFGE group if their Dice similarity index was ≥ 85%.
2.5. Statistical Analysis
The prevalence of CREC in this study are reported as a pooled prevalence estimate with a 95% confidence interval to represent resistance from 2015 to 2016 and the analysis method was described as previous [19]. Briefly, random effects meta-analysis was conducted for individual data of province-specific prevalence to establish a properly combined prevalence estimate for the national estimate by considering between-province variation. All the statistical analysis was performed in R version 3.3.1. Pooled prevalence was estimated using metafor package.
3. Results
3.1. Overview of the CREC
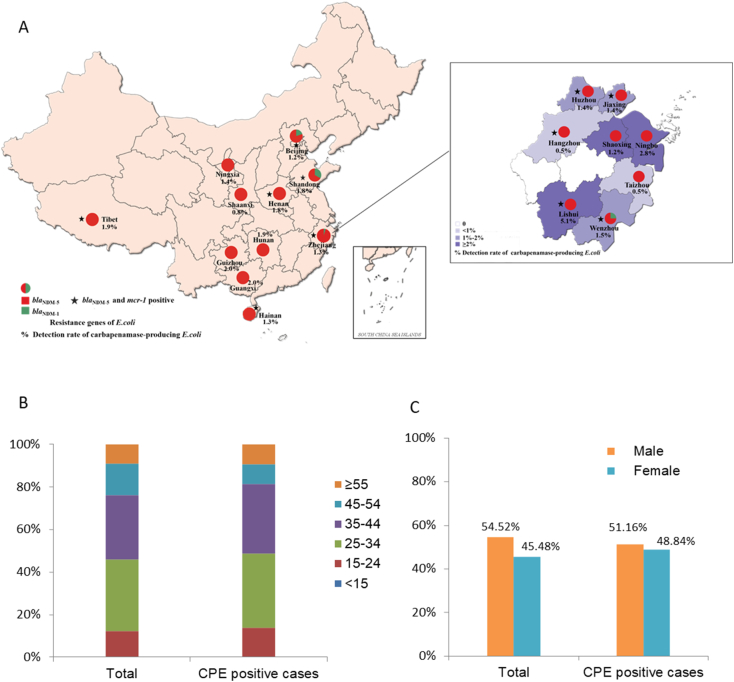
A total of 3859 non-duplicated stool specimens were collected from healthy volunteers in 19 provinces during September 2016 to December 2016. The prevalence of CREC recovered from different sampling locations, 15 out of the 19 Provinces and Municipalities yielded CREC isolates, except Guangdong, Hubei, Jiangxi and Shanxi (Table 1, Fig. 1A). The age and gender information of the volunteers was also showed in Fig. 1B and C, and the majority of the volunteers was between 25 and 44, accounting for about 60% of the population. The age and gender distribution was very similar in the CRE positive cases (age 38.16 ± 13.45, Female 48.84% vs Male 51.16%) and the study population (age 37.51 ± 11.98, Female 45.48% vs Male 54.52%). Of 3859 isolates, a total of 92 (2.38%, 95% CI, 1.95%–2.91%) non-duplicated CREC isolates were obtained (Table 1). We detected blaNDM, but not other carbapenem-resistant genes, in 43 out of 92 isolates. From the 43 blaNDM genes, 40 (93.0%; 95% CI, 81.4%–97.6%) were blaNDM-5, and 3 (7.0%; 95% CI, 2.4%–18.6%) blaNDM-1. In addition, the co-existence of mcr-1 and blaNDM was detected in 14 CREC. Interestingly, all blaNDM in the mcr-1 carrying E. coli belong to blaNDM-5. These E. coli isolates with both NDM-5 and MCR-1 were detected in six provinces (Table 1, Fig. 1). Approximately 40.2% of the total samples were from Zhejiang province, which were obtained from all 11 prefectural-level cities in Zhejiang province. The prevalence of CREC in Zhejiang is 2.26% (35 of 1551, 95% CI, 1.63–3.12%), which is similar to the average level nationwide (2.38%; 95% CI, 1.95–2.91%). The E. coli isolates carrying both NDM-5 and MCR-1 were detected in 5/11 cities (Table 2, Fig. 1).
Table 1.
Overview of sampling size and CREC in different provinces.
| Sampling provinces | NO. of samples | NO. of CREC | Positive rate of CREC (%; 95% CI) | NO. of NDM-5-producing CREC | NO. of NDM-1-producing CREC | NO. of mcr-1 and blaNDM positive CREC |
|---|---|---|---|---|---|---|
| Guizhou | 50 | 4 | 8.0 (3.15–18.84) | 1 | 0 | 0 |
| Heilongjiang | 50 | 3 | 6.0 (2.06–16.22) | 0 | 0 | 0 |
| Henan | 56 | 3 | 5.36 (1.84–14.61) | 1 | 0 | 1 |
| Hunan | 54 | 2 | 3.7 (1.02–12.54) | 1 | 0 | 0 |
| Beijing | 418 | 13 | 3.11 (1.83–5.25) | 4 | 1 | 1 |
| Shandong | 165 | 5 | 3.03 (1.30–6.90) | 2 | 1 | 1 |
| Fujian | 100 | 3 | 3.0 (1.02–8.45) | 0 | 0 | 0 |
| Inner Mongoria | 135 | 4 | 2.96 (1.16–7.37) | 0 | 0 | 0 |
| Tibet | 108 | 3 | 2.78 (0.95–7.85) | 2 | 0 | 1 |
| Hainan | 237 | 6 | 2.53 (1.17–5.41) | 3 | 0 | 2 |
| Shaanxi | 132 | 3 | 2.27 (0.78–6.47) | 1 | 0 | 0 |
| Zhejiang | 1551 | 35 | 2.26 (1.63–3.12) | 19 | 1 | 8 |
| Jilin | 100 | 2 | 2.0 (0.55–7.00) | 0 | 0 | 0 |
| Guangxi | 204 | 4 | 1.96 (0.77–4.93) | 4 | 0 | 0 |
| Ningxia | 140 | 2 | 1.43 (0.39–5.06) | 2 | 0 | 0 |
| Guangdong | 57 | 0 | 0.0 (0.0–0.0) | 0 | 0 | 0 |
| Hubei | 52 | 0 | 0.0 (0.0–0.0) | 0 | 0 | 0 |
| Jiangxi | 106 | 0 | 0.0 (0.0–0.0) | 0 | 0 | 0 |
| Shanxi | 144 | 0 | 0.0 (0.0–0.0) | 0 | 0 | 0 |
| Total | 3859 | 92 | 2.38 (1.95–2.91) | 40 | 3 | 14 |
Fig. 1.
Summary of geography information. A) Positive regions for NDM and/or MCR carrying E. coli in China. The green and red indicated the presence of NDM-1 and NDM-5 positive E. coli, while the star indicates the presence of MCR-1 positive E. coli. The numbers of isolates are indicated in the brackets. B) The age distribution of the total sample population and Carbapenemase Producing E. coli (CPE) positive case. C) The gender distribution of the total sample population and CPE positive.
Table 2.
Overview of sampling size and CREC in different cities in Zhejiang province.
| Sampling provinces | NO. of samples | NO. of CREC | Positive rate of CREC (%; 95% CI) | NO. of NDM-5-producing CREC | NO. of NDM-1-producing CREC | NO. of mcr-1 and blaNDM positive CREC |
|---|---|---|---|---|---|---|
| Lishui | 98 | 6 | 6.12 (2.84–12.72) | 5 | 0 | 4 |
| Ningbo | 144 | 5 | 3.47 (1.49–7.87) | 4 | 0 | 0 |
| Wenzhou | 259 | 9 | 3.47 (1.84–6.47) | 3 | 1 | 1 |
| Shaoxing | 83 | 2 | 2.41 (0.66–8.37) | 1 | 0 | 0 |
| Huzhou and Jiaxing | 218 | 4 | 1.83 (0.72–4.62) | 3 | 0 | 2 |
| Taizhou | 205 | 2 | 0.98 (0.27–3.49) | 1 | 0 | 0 |
| Hangzhou | 377 | 7 | 1.86 (0.90–3.78) | 2 | 0 | 1 |
| Quzhou | 66 | 0 | 0.0 | 0 | 0 | 0 |
| Jinhua | 101 | 0 | 0.0 | 0 | 0 | 0 |
| total | 1551 | 35 | 2.26 (1.63–3.12) | 19 | 1 | 8 |
3.2. Characterization of CREC
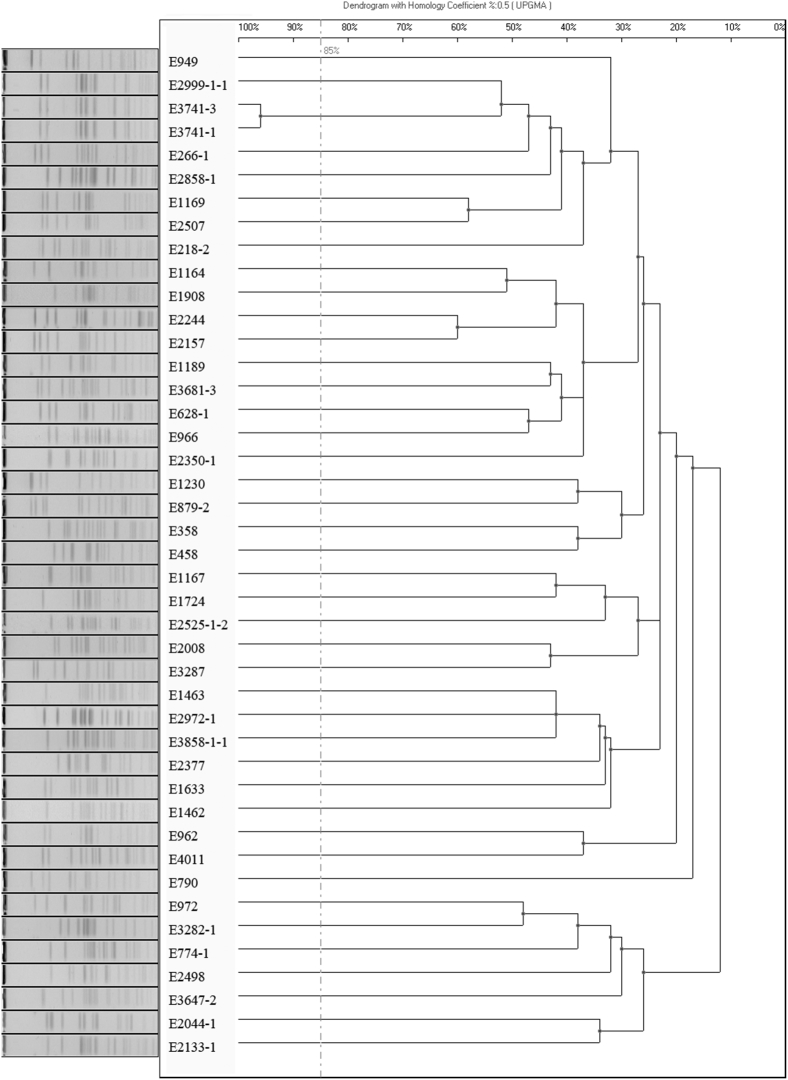
All NDM-producing E. coli were resistant to almost all β-lactams. The percentage of isolates susceptible to amikacin, colistin, tigecycline, and nitrofurantoin were 76.7%, 72.1%, 88.4%, and 83.7%, respectively (Table 3). All 14 MCR-1 and NDM-5 producing CREC exhibited resistance to all β-lactams, but remained susceptible to amikacin (92.9%), tigecycline (85.7%), and nitrofurantoin (92.9%) (Table 4). Moreover, PFGE analysis on the 43 blaNDM–positive E. coli isolates indicated that 42/43 PFGE patterns were distinctly different using 85% genetic similarity as cut-off, suggesting non-clonal dissemination is responsible for their transmission across China (Fig. 2).
Table 3.
Antimicrobial susceptibility profiles of 43 clinical NDM-producing E. coli strains.
| Drug class | Antimicrobial agents | MIC50 μg/ml |
MIC90 μg/ml |
Range μg/ml |
%R | %I | %S |
|---|---|---|---|---|---|---|---|
| Aminoglycoside | Gentamicin | 32 | > 32 | ≤ 2–>32 | 69.8 | 2.3 | 27.9 |
| Amikacin | ≤ 8 | > 128 | ≤ 8–>128 | 23.3 | 0.0 | 76.7 | |
| β-lactams and β-lactamase Inhibitor | Piperacillin-tazobactam | 256/4 | > 256/4 | ≤ 8/4–>256/4 | 67.4 | 27.9 | 4.7 |
| Ampicillin-sulbactam | > 64/32 | > 64/32 | ≤ 4/2–>64/32 | 97.7 | 0.0 | 2.3 | |
| Ticarcillin-clavulanate | 128/64 | 128/64 | ≤ 8/4–128/64 | 95.3 | 0.0 | 4.7 | |
| Cefoperazone-sulbactam (2:1) | > 128/64 | > 128/64 | 64/32–>128/64 | 100.0 | 0.0 | 0.0 | |
| Carbapenem | Imipenem | > 16 | > 16 | 8–>16 | 100.0 | 0.0 | 0.0 |
| Meropenem | > 16 | > 16 | 8–>16 | 100.0 | 0.0 | 0.0 | |
| Cephalosporin | Cefepime | > 32 | > 32 | 2–>32 | 90.7 | 7.0 | 2.3 |
| Ceftazidime | > 32 | > 32 | 32–>32 | 100.0 | 0.0 | 0.0 | |
| Cefotaxime | > 32 | > 32 | 32–>32 | 100.0 | 0.0 | 0.0 | |
| Cefazolin | > 32 | > 32 | 32–>32 | 100.0 | 0.0 | 0.0 | |
| Cefuroxim | > 64 | > 64 | 64–>64 | 100.0 | 0.0 | 0.0 | |
| Fluoroquinolone | Ciprofloxacin | 8 | > 8 | ≤ 0.25–>8 | 67.4 | 4.7 | 27.9 |
| Levofloxacin | 16 | > 16 | ≤ 0.5–>16 | 58.1 | 7.0 | 34.9 | |
| Sulfonamide and Trimethoprim Antibiotic Combinations | Trimethoprim-sulfamethoxazole | 8/152 | 8/152 | ≤ 1/19–8/152 | 95.3 | 0.0 | 4.7 |
| Fosfomycin | 16 | > 512 | ≤ 4–>512 | 39.5 | 2.3 | 58.1 | |
| Tetracycline | Minocycline | 8 | 16 | ≤ 2–16 | 46.5 | 20.9 | 32.6 |
| Tigecycline | 0.5 | 2 | ≤ 0.25–4 | 2.3 | 9.3 | 88.4 | |
| Others | Aztreonam | 4 | 64 | ≤ 2–64 | 41.9 | 4.7 | 53.5 |
| Chloramphenicol | 32 | 32 | ≤ 8–32 | 74.4 | 2.3 | 23.3 | |
| Colistin | ≤ 1 | 4 | ≤ 1–>16 | 27.9 | 0.0 | 72.1 | |
| Nitrofurantoin | ≤ 32 | 64 | ≤ 32–128 | 9.3 | 7.0 | 83.7 | |
| Piperacillin | > 256 | > 256 | ≤ 8–>256 | 97.7 | 0.0 | 2.3 |
S, susceptible; I, intermediate resistant; R, resistant; MIC90 and MIC50 values were defined as the lowest concentration of the antibiotic at which 90% and 50% of the isolates were inhibited, respectively.
Table 4.
MICs of 30 antimicrobial agents for 14 E. coli strains co-harboring mcr-1 and blaNDM-5.a
| NO. of samples | Province | MIC (μg/ml) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IPN | MPN | FEP | CAZ | CTX | CZ | CXM | PIP | PIT | AMS | TCA | SCF(2:1) | CAC | CTC | ||
| E218-2 | Zhejiang | 8 | > 16 | > 32 | > 32 | > 32 | > 32 | > 64 | 256 | 64/4 | > 64/32 | 128/64 | 128/64 | > 128/64 | 128/64 |
| E628-1 | Zhejiang | 16 | > 16 | > 32 | > 32 | > 32 | > 32 | > 64 | > 256 | 128/4 | > 64/32 | 128/64 | > 128/64 | > 128/64 | > 128/64 |
| E1164 | Zhejiang | > 16 | > 16 | > 32 | > 32 | > 32 | > 32 | > 64 | 256 | 32/4 | > 64/32 | 128/64 | > 128/64 | > 128/64 | > 128/64 |
| E1167 | Zhejiang | > 16 | > 16 | > 32 | > 32 | > 32 | > 32 | > 64 | > 256 | > 256/4 | > 64/32 | 128/64 | > 128/64 | 128/64 | > 128/64 |
| E1189 | Zhejiang | 8 | > 16 | > 32 | > 32 | > 32 | > 32 | > 64 | > 256 | 32/4 | > 64/32 | 128/64 | 128/64 | > 128/64 | > 128/64 |
| E1230 | Zhejiang | 16 | > 16 | > 32 | > 32 | > 32 | > 32 | > 64 | 256 | 32/4 | > 64/32 | 128/64 | > 128/64 | 128/64 | > 128/64 |
| E1462 | Zhejiang | 16 | 16 | 32 | 32 | 32 | 32 | 64 | 256 | 256/4 | 64/32 | 128/64 | 128/64 | 128/64 | 128/64 |
| E1463 | Zhejiang | 16 | > 16 | > 32 | > 32 | > 32 | > 32 | > 64 | > 256 | 256/4 | > 64/32 | 128/64 | > 128/64 | > 128/64 | > 128/64 |
| E774-1 | Tibet | > 16 | > 16 | > 32 | > 32 | > 32 | > 32 | > 64 | > 256 | 128/4 | > 64/32 | 128/64 | > 128/64 | > 128/64 | > 128/64 |
| E1908 | Henan | 16 | 16 | > 32 | > 32 | > 32 | > 32 | > 64 | > 256 | 16/4 | > 64/32 | 128/64 | 64/32 | ≤ 1/0.5 | ≤ 1/0.5 |
| E2244 | Beijing | 16 | > 16 | > 32 | > 32 | > 32 | > 32 | > 64 | > 256 | 64/4 | > 64/32 | ≤ 8/4 | > 128/64 | > 128/64 | > 128/64 |
| E2498 | Hainan | > 16 | > 16 | > 32 | > 32 | > 32 | > 32 | > 64 | > 256 | 128/4 | > 64/32 | 128/64 | > 128/64 | > 128/64 | > 128/64 |
| E2525-1-2 | Hainan | > 16 | > 16 | > 32 | > 32 | > 32 | > 32 | > 64 | > 256 | > 256/4 | > 64/32 | 128/64 | > 128/64 | > 128/64 | > 128/64 |
| E3647-2 | Shandong | > 16 | > 16 | > 32 | > 32 | > 32 | > 32 | > 64 | > 256 | 256/4 | > 64/32 | 128/64 | > 128/64 | 128/64 | > 128/64 |
| NO. of samples | Province | MIC (μg/ml) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GM | AN | CIP | LVF | SXT | MNO | AZT | C | FOS | FT | TGC | PE | ||
| E218-2 | Zhejiang | 32 | ≤ 8 | 8 | 8 | 8/152 | 4 | ≤ 2 | 32 | 512 | ≤ 32 | 0.5 | 4 |
| E628-1 | Zhejiang | > 32 | ≤ 8 | > 8 | 8 | 8/152 | 16 | ≤ 2 | 32 | 512 | ≤ 32 | 1 | 8 |
| E1164 | Zhejiang | > 32 | ≤ 8 | 8 | 8 | 8/152 | 4 | ≤ 2 | 32 | > 512 | ≤ 32 | ≤ 0.25 | 4 |
| E1167 | Zhejiang | > 32 | > 128 | > 8 | 16 | 8/152 | 16 | 16 | 32 | ≤ 4 | ≤ 32 | ≤ 0.25 | 4 |
| E1189 | Zhejiang | > 32 | ≤ 8 | ≤ 0.25 | ≤ 0.5 | 8/152 | 8 | 32 | 32 | > 512 | ≤ 32 | ≤ 0.25 | ≤ 1 |
| E1230 | Zhejiang | 16 | ≤ 8 | 2 | 2 | 8/152 | ≤ 2 | ≤ 2 | 32 | 512 | ≤ 32 | ≤ 0.25 | 4 |
| E1462 | Zhejiang | 4 | ≤ 8 | 8 | 16 | 8/152 | 16 | ≤ 2 | 32 | 16 | 128 | 1 | 4 |
| E1463 | Zhejiang | ≤ 2 | ≤ 8 | > 8 | > 16 | 8/152 | 16 | 4 | 32 | ≤ 4 | ≤ 32 | 4 | ≤ 1 |
| E774-1 | Tibet | > 32 | 16 | 8 | > 16 | 8/152 | 16 | ≤ 2 | ≤ 8 | 128 | ≤ 32 | 2 | 4 |
| E1908 | Henan | ≤ 2 | ≤ 8 | 4 | 4 | 8/152 | 4 | ≤ 2 | 32 | ≤ 4 | ≤ 32 | ≤ 0.25 | 2 |
| E2244 | Beijing | > 32 | ≤ 8 | > 8 | > 16 | 8/152 | 16 | 64 | 32 | > 512 | ≤ 32 | 0.5 | 2 |
| E2498 | Hainan | 32 | ≤ 8 | ≤ 0.25 | ≤ 0.5 | 8/152 | 4 | ≤ 2 | 32 | > 512 | ≤ 32 | ≤ 0.25 | 4 |
| E2525-1-2 | Hainan | ≤ 2 | ≤ 8 | ≤ 0.25 | ≤ 0.5 | 8/152 | 16 | 16 | 32 | ≤ 4 | ≤ 32 | 1 | 4 |
| E3647-2 | Shandong | > 32 | ≤ 8 | > 8 | 8 | 8/152 | 16 | 64 | 32 | > 512 | ≤ 32 | 0.5 | 2 |
IPN, Imipenem; MPN, Meropenem; FEP, Cefepime; CAZ, Ceftazidime; CTX, Cefotaxime; CZ, Cefazolin; CXM, Cefuroxim; PIP, Piperacillin; PIT, Piperacillin-tazobactam; AMS, Ampicillin-sulbactam; TCA, Ticarcillin-clavulanate; SCF (2:1), Cefoperazone-sulbactam (2:1); CAC, Ceftazidime-clavulanate; CTC, Cefotaxime-clavulanate; GM, Gentamicin; AN, Amikacin; CIP, Ciprofloxacin; LVF, Levofloxacin; SXT, Trimethoprim-sulfamethoxazole; MNO, Minocycline; AZT, Aztreonam; C, Chloramphenicol; FOS, Fosfomycin; FT, Nitrofurantoin; TGC, Tigecycline; PE, colistin.
The bold values indicated these strains were susceptible to the drugs according to EUCAST and CLSI standards.
Fig. 2.
PFGE analysis of carbapenem-resistant E. coli, XbaI was used for digestion of the genomic DNA.
3.3. The Genetic Background of CREC
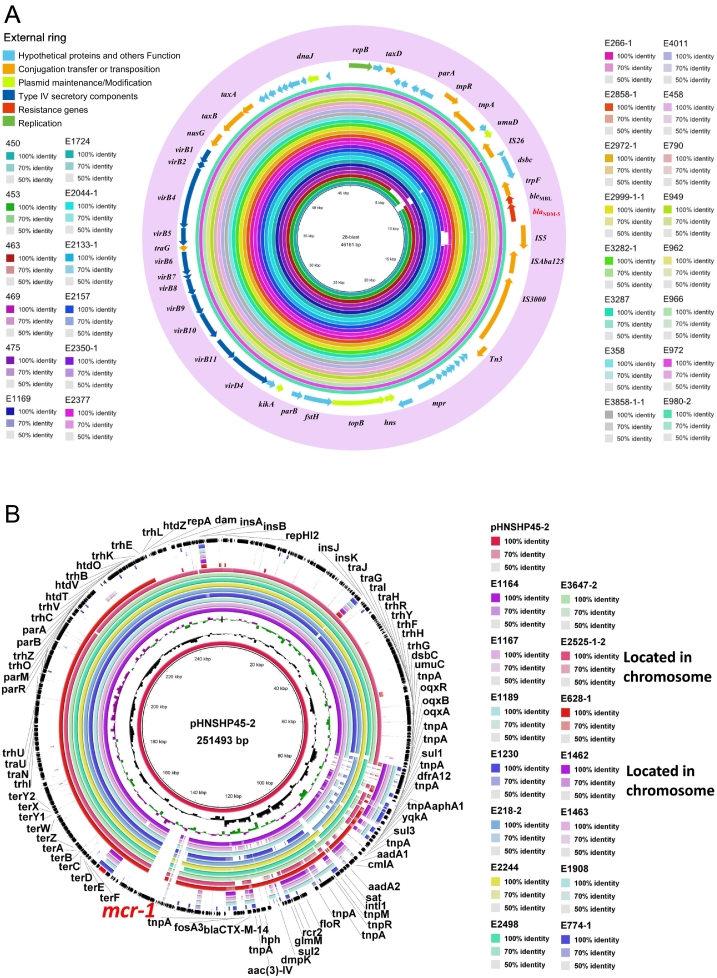
To further understand the genetic background of CREC and its role in the dissemination of blaNDM and mcr-1, whole genome sequencing was conducted to reveal plasmid and core genome information. The draft whole genome sequence of 14 mcr-1 positive E. coli (MCRPEC) isolated showed that NDM-5 was located on contigs with lengths ranging from 41 to 46 kb in all NDM-5 carrying CREC, which was further confirmed by S1-PFGE (~ 46 kb) (data not shown). The complete sequence of the blaNDM-5 carrying plasmid (designated pNDM5-ZJ628R,NCBI Accession No. MH500845) from isolate E628-1 was acquired through gap filling by sequencing PCR amplicons. pNDM5-ZJ628R belongs to the IncX3 family, which was 46,161-bp in length with a G/C content of 46.6%. This plasmid contains 60 putative open reading frames (ORFs) and the gene components of the plasmid are show in Fig. 3A. Of the 40 NDM-5 carrying CREC, 23 plasmid sequences were closed for the full length of the plasmid (~ 46 kb) and selected for comparison with five other IncX3 plasmids carrying blaNDM-5 identified from animal farms and showed over 99% identity (Fig. 3A). The draft sequences of the other NDM-5 carrying plasmids showed all were highly similar to pNDM5-ZJ628R.
Fig. 3.
BLAST ring comparison of NDM-5 and MCR-1 plasmids. A) The comparison of NDM-5 carrying plasmid sequences in CREC isolates. Each color represents a NDM-5 carrying plasmid. The internal ring is the reference sequence of NDM-5 carrying plasmid pNDM-ZJ628R, and the outside rings are other 23 plasmids from this study and 5 plasmids of animal origin, which are highly similar to pNDM-ZJ628R. B) The comparison of mcr-1 carrying plasmid sequences in mcr-1 positive CREC isolates. Each color represents a mcr-1 carrying plasmid. The internal ring is the reference sequence of mcr-1 carrying plasmid pHNSHP45-2, and the outside rings are other 14 plasmids from this study, which are similar to pNDM-ZJ628R.
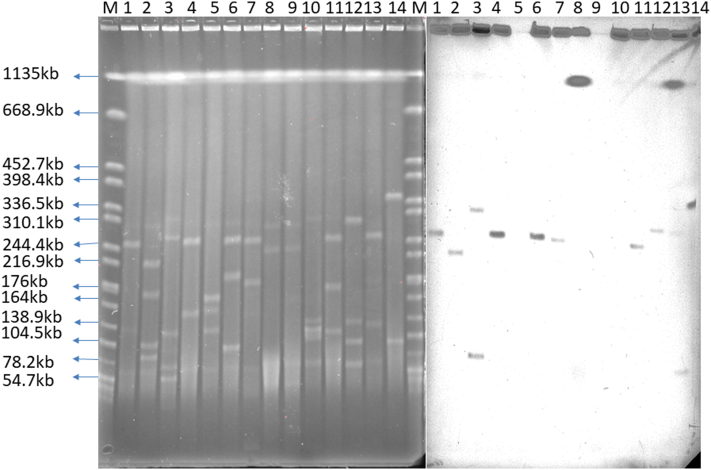
S1-PFGE analysis showed mcr-1 was mostly located on plasmids with lengths of 240–330 kb, and two were located on the chromosome (Fig. 5). Whole genome sequencing confirmed that the plasmid Inc. type is IncHI2, which was similar to plasmid pHNSHP45-2 [20]. Comparison of mcr-1 carrying plasmid sequences are shown in Fig. 3B. The sequencing of nine isolates carrying mcr-1 shares over 90% identity and 90% coverage with plasmid pHNSHP45-2.
Fig. 5.
S1-PFGE analysis of mcr-1 positive CREC. Bands M: H9812, 1:E218-2, 2:E628-1, 3:E774-1, 4:E1164, 5:E1167, 6:E1189, 7:E1230, 8:E1462, 9:E1463, 10:E1980, 11:E2244, 12:E2498, 13:E2525-1, 14:E3647-2, M:H9812.
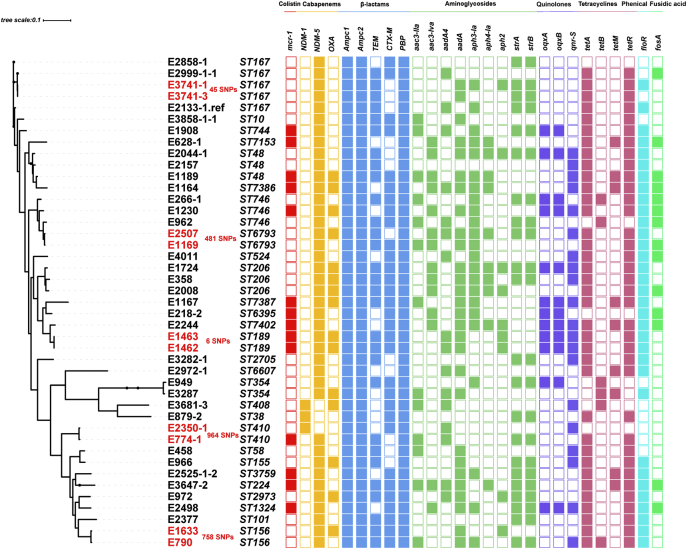
Core genome analysis of CREC and MCRPEC revealed 6 distinct lineages (Fig. 4). A total of 165,857 core SNPs were used to analyze the relationship among these isolates, and most isolates identified from the same region or adjacent region could be found in the same or closing lineage. We found these strains can be allocated across 32 ST clades, and none of these ST clades was dominant, affirming our PFGE analysis. Interestingly, the average number of antibiotic resistance genes are 18.7 ± 4.6 in the mcr-1-negative CREC, and 24.3 ± 3.7 in the mcr-1-positive CREC, indicating that mcr-1-carrying CREC are likely to carry more ARGs comparing with mcr-1-negative CREC in the normal flora of healthy people (P-value < 0.05).
Fig. 4.
The phylogenetic relationship of CREC and mcr-1 positive CREC, and their other antibiotic resistance genes. The left phylogenetic tree were constructed using the core SNPs with ST types. The color squares represent the antibiotic resistance genes sorted by the class of antibiotics.
4. Discussions
Since the first discovery of NDM-1 in 2009, over 20 NDM variants have now been reported. A nationwide Chinese survey demonstrated that the acquisition of two carbapenemases, KPC and NDM, was responsible for phenotypic resistance in 90% of the CRE strains tested (58% and 32%, respectively) [2]. In E. coli, the proportion of NDM was higher than that of KPC [2]. So far, majority of the epidemiology studies of CRE have focused on clinical data, as it was deemed that healthy individuals were usually not affected by the carriage of CRE. Hitherto, there have only few studies about the prevalence of CRE in the normal flora of a healthy population [17], [21]. Here, our study presents a comprehensive epidemiology study of the carriage of CRE in the healthy Chinese population.
To date, over 20 NDM has been discovered, and blaNDM-1 appears to be overwhelmingly dominated in the clinical sector. Amit Ranjan et al. reported that the majority of MBL producers were NDM-1 (69%) followed subsequently by NDM-5 (19%) in extra-intestinal pathogenic E. coli strains [22]. Another study showed the similar finding in China from 2013 to 2015, 80% was NDM-1, and 17.8% was NDM-5 [23]. Surprisingly, in our study, the number of NDM-5 producing E. coli isolated in the gut flora (40 strains) was much higher than that of NDM-1 producing E. coli (3 strains), suggest the prevalence of NDM has been shifting from NDM-1 to NDM-5 in the community of China, but this shift has not been observed in India and other countries yet.
Since the first discovery of blaNDM-5 in China, blaNDM-5 has been identified in a variety of Enterobacteriaceae, and the Inc. type primarily carrying blaNDM-5 is IncX3. In this study, pNDM5-ZJ628R was identified as an IncX3 plasmid, with a typical backbone structure for this plasmid type, including regions involved in replication, partitioning, plasmid maintenance, transcriptional activation, and conjugation/type IV secretion [24], [25], which is very similar to the plasmids carrying blaNDM-5 in the CREC and MCRPEC (Fig. 3A and B).
The comparative analysis of 23 blaNDM-5 carrying plasmids (the contigs close to the full length of plasmid) revealed that these plasmids possessed very high similarity to the closed sequenced plasmid pNDM5-ZJ628R. BLAST homology analysis showed that pNDM5-ZJ628R had 100% identity and 100% query coverage with pP855-NDM5 (accession number: MF547508) and pNDM5_IncX3 (accession number: KU761328), a 46,161-bp IncX3 plasmid isolated from E. coli (accession number: P855MEM) of pig origin and Klebsiella pneumoniae (SZ204) of human origin [26]. Recently, a novel blaNDM-17 was discovered in E. coli from poultry in China. This blaNDM-17 carrying plasmid also belongs to IncX3 (GenBank accession no. KX833071), sharing 99% identity (46,142/46,161 bp) and 100% coverage with NDM5_IncX3 (KU761328), a 46,161-bp IncX3 plasmid isolated from Klebsiella pneumoniae (SZ204) [27]. In comparison with NDM-5, NDM-17 has a single E170K substitution and showed increased resistance to carbapenems, which may suggested blaNDM-5 IncX3 plasmid could be a pivotal branch for further evolution of NDM carbapenemases. The IncX3 plasmid carrying blaNDM-1 pNDM-HN380 (54,035 bp) [ 22] was reported in Hong Kong in 2012 was also very similar to pNDM5-ZJ628R (46,161 bp) apart from an 8 kb region, which may suggest this plasmid could be the origin of the IncX3 plasmid. Although IncX3 plasmids are considered low-prevalence, narrow-host-range plasmids of Enterobacteriaceae [12], our MLST analysis showed IncX3 plasmids have a wide host adaption capability in E. coli. These plasmids may have served as a common vehicle in disseminating blaNDM in E. coli among human, animal, and environment sectors, and might be responsible for the rapid spread of NDM-carrying isolates [13].
The overwhelming prevalence of blaNDM-5 in E. coli in the healthy population seem to be identical with the trend of blaNDM-5 in E. coli from the Chinese poultry production chain. A recent study reported blaNDM-5 accounted for 52.2% of all CRE isolates (84/161); not only in poultry farms, but also slaughterhouses, supermarkets, and poultry farm workers [14], which suggests a dynamic transition for blaNDM among human, animal and the environment.
It is worth noting that almost one third of the total carbapenemase producing E. coli isolates (14 out of 43) carry mcr-1, suggesting an emerging co-existence of blaNDM and mcr-1 in Enterobacteriaceae in human normal flora from healthy volunteers. The co-existence of blaNDM-5 and mcr-1 has been occasionally observed at a low prevalence in individual clinical samples [28], [29], [30], and to the best of our knowledge, this is the first report of the co-existence of blaNDM-5 and mcr-1 in the normal flora of humans. Interestingly, 23% of CRE isolates of animal origin from Shandong province showed a high co-existence of blaNDM and mcr-1, which is consistent with the findings in this study [14]. Although whole genome sequencing could not retrieve the complete sequence of mcr-1 carrying plasmids, the comparative analysis between whole genome draft sequences and two common mcr-1 carrying plasmids (pHNSHP45 and pHNSHP45-2) showed the majority of mcr-1 carrying contigs share high similarity to plasmid pHNSHP45-2. Moreover, MLST analysis also indicates the wide host adaptability of pHNSHP45-2 like plasmids in E. coli isolates from different regions in China.
The relative small number of samples is one of the limitations for this study, comparing with the population size in China, as this study is just an observation study for understanding the prevalence of CREC in the healthy people. Although the CRE positive case number is small, but it suggests the prevalence of CREC in the healthy people does exist, and the colonization of CREC in the healthy people may contribute to the later infection in the hospital. Now, according to the CHINET Surveillance Program, the prevalence of CREC in the hospital in about 2.0% based on 36,375 EREC isolates [31], which is similar to the prevalence of CREC in the healthy people. Another limitation is the limited information of geography from the volunteers, such as antibiotic consumption, poultry consumption, etc., which makes it difficult to identify the risks related to the colonization of CREC in the healthy people.
This is the first comprehensive study to reveal the emerging co-existence of blaNDM-5 and mcr-1 in the normal gut floral of healthy individuals. Our findings further suggest that the dissemination of blaNDM-5 may be augmented by the presence IncX3-type plasmids. Due to the high prevalence of CRE in the healthy human population, it is urgent to implement a screening procedure to monitor the presence of CRE for patients admitted onto hospital high dependency units e.g. oncology to prevent the dissemination of CRE in high-risk patients.
Contributions
ZS and YH contributed equally in this study. SW and RZ designed this study. YH, QS, FH, HZ, and RZ collected all the samples and conducted epidemiology study. ZS, YH, QS, SW, and RZ analyzed the data. ZS, SW, TRW, and RZ wrote the report. All authors reviewed, revised, and approved the final report.
Declaration of Interests
We declare no competing interests.
Acknowledgements
This work was supported in part by the National Natural Science Foundation of China (81661138002, 31761133004). TRW are also supported by a Medical Research Council grant DETER-XDR-CHINA (MR/P007295/1). We acknowledge the valuable contribution of Bing Wang (University of Neberaska-Lincoln, US) in providing statistical support and guidance as funded by NIH.
Contributor Information
Rong Zhang, Email: zhang-rong@zju.edu.cn.
Shaolin Wang, Email: shaolinwang@cau.edu.cn.
References
- 1.Schwaber M.J., Carmeli Y. Carbapenem-resistant Enterobacteriaceae: a potential threat. JAMA. 2008;300(24):2911–2913. doi: 10.1001/jama.2008.896. [DOI] [PubMed] [Google Scholar]
- 2.Zhang R., Liu L., Zhou H. Nationwide surveillance of clinical carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBioMedicine. 2017;19:98–106. doi: 10.1016/j.ebiom.2017.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y., Wang Q., Yin Y. Epidemiology of carbapenem-resistant Enterobacteriaceae infections: report from China CRE Network. Antimicrob Agents Chemother. 2018 Jan 25;62(2) doi: 10.1128/AAC.01882-17. pii: e01882-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonomo R.A., Burd E.M., Conly J. Carbapenemase-producing organisms: A global scourge! Clin Infect Dis. 2018 Apr 3;66(8):1290–1297. doi: 10.1093/cid/cix893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamamoto N., Asada R., Kawahara R. Prevalence of, and risk factors for, carriage of carbapenem-resistant Enterobacteriaceae among hospitalized patients in Japan. J Hosp Infect. 2017;97(3):212–217. doi: 10.1016/j.jhin.2017.07.015. [DOI] [PubMed] [Google Scholar]
- 6.Castanheira M., Huband M.D., Mendes R.E., Flamm R.K. Meropenem-vaborbactam tested against contemporary Gram-negative isolates collected worldwide during 2014, including carbapenem-resistant, KPC-producing, multidrug-resistant, and extensively drug-resistant Enterobacteriaceae. Antimicrob Agents Chemother. 2017;61(9) doi: 10.1128/AAC.00567-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwaber M.J., Klarfeld-Lidji S., Navon-Venezia S., Schwartz D., Leavitt A., Carmeli Y. Predictors of carbapenem-resistant Klebsiella pneumoniae acquisition among hospitalized adults and effect of acquisition on mortality. Antimicrob Agents Chemother. 2008;52(3):1028–1033. doi: 10.1128/AAC.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciobotaro P., Flaks-Manov N., Oved M. Predictors of persistent carbapenem-resistant Enterobacteriaceae carriage upon readmission and score development. Infect Control Hosp Epidemiol. 2016;37(2):188–196. doi: 10.1017/ice.2015.278. [DOI] [PubMed] [Google Scholar]
- 9.Munoz-Price L.S., Poirel L., Bonomo R.A. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis. 2013;13(9):785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumarasamy K.K., Toleman M.A., Walsh T.R. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis. 2010;10(9):597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Z., Li J., Wang X. Novel variant of New Delhi metallo-beta-lactamase, NDM-20, in Escherichia coli. Front Microbiol. 2018;9:248. doi: 10.3389/fmicb.2018.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang P., Xie Y., Feng P., Zong Z. blaNDM-5 carried by an IncX3 plasmid in Escherichia coli sequence type 167. Antimicrob Agents Chemother. 2014;58(12):7548–7552. doi: 10.1128/AAC.03911-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang F., Xie L., Wang X. Further spread of blaNDM-5 in Enterobacteriaceae via IncX3 plasmids in Shanghai, China. Front Microbiol. 2016;7:424. doi: 10.3389/fmicb.2016.00424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y., Zhang R., Li J. Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat Microbiol. 2017;2:16260. doi: 10.1038/nmicrobiol.2016.260. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y.Y., Wang Y., Walsh T.R. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 16.Li Y., Sun Q.L., Shen Y. Rapid increase in prevalence of carbapenem-resistant Enterobacteriaceae (CRE) and emergence of colistin resistance gene mcr-1 in CRE in a hospital in Henan, China. J Clin Microbiol. 2018;56(4) doi: 10.1128/JCM.01932-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruppé E., Armand-Lefèvre L., Estellat C. Acquisition of carbapenemase-producing Enterobacteriaceae by healthy travellers to India, France, February 2012 to March 2013. Euro Surveill. 2014;19(14) doi: 10.2807/1560-7917.es2014.19.14.20768. (pii: 20768) [DOI] [PubMed] [Google Scholar]
- 18.McConville T.H., Sullivan S.B., Gomez-Simmonds A., Whittier S., Uhlemann A.C. Carbapenem-resistant Enterobacteriaceae colonization (CRE) and subsequent risk of infection and 90-day mortality in critically ill patients, an observational study. PLoS One. 2017;12(10) doi: 10.1371/journal.pone.0186195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang P., Shen Z., Zhang C. Surveillance of antimicrobial resistance among Escherichia coli from chicken and swine, China, 2008–2015. Vet Microbiol. 2017;203:49–55. doi: 10.1016/j.vetmic.2017.02.008. [DOI] [PubMed] [Google Scholar]
- 20.Zhi C., Lv L., Yu L.F., Doi Y., Liu J.H. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis. 2016;16(3):292–293. doi: 10.1016/S1473-3099(16)00063-3. [DOI] [PubMed] [Google Scholar]
- 21.Tischendorf J., de Avila R.A., Safdar N. Risk of infection following colonization with carbapenem-resistant Enterobactericeae: a systematic review. Am J Infect Control. 2016;44(5):539–543. doi: 10.1016/j.ajic.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ranjan A., Shaik S., Mondal A. Molecular epidemiology and genome dynamics of New Delhi metallo-beta-lactamase-producing extraintestinal pathogenic Escherichia coli strains from India. Antimicrob Agents Chemother. 2016;60(11):6795–6805. doi: 10.1128/AAC.01345-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu X., Xu X., Wang X. Diversity of New Delhi metallo-beta-lactamase-producing bacteria in China. International Journal of Infectious Diseases: IJID: Official Publication of the International Society for Infectious Diseases. 2017;55:92–95. doi: 10.1016/j.ijid.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 24.Ho P.L., Li Z., Lo W.U. Identification and characterization of a novel incompatibility group X3 plasmid carrying blaNDM-1 in Enterobacteriaceae isolates with epidemiological links to multiple geographical areas in China. Emerg Microbes Infect. 2012;1(11) doi: 10.1038/emi.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krishnaraju M., Kamatchi C., Jha A.K. Complete sequencing of an IncX3 plasmid carrying blaNDM-5 allele reveals an early stage in the dissemination of the blaNDM gene. Indian J Med Microbiol. 2015;33(1):30–38. doi: 10.4103/0255-0857.148373. [DOI] [PubMed] [Google Scholar]
- 26.Du H., Chen L., Tang Y.W., Kreiswirth B.N. Emergence of the mcr-1 colistin resistance gene in carbapenem-resistant Enterobacteriaceae. Lancet Infect Dis. 2016;16(3):287–288. doi: 10.1016/S1473-3099(16)00056-6. [DOI] [PubMed] [Google Scholar]
- 27.Liu Z., Wang Y., Walsh T.R. Plasmid-mediated novel blaNDM-17 gene encoding a carbapenemase with enhanced activity in a sequence type 48 Escherichia coli strain. Antimicrob Agents Chemother. 2017;61(5) doi: 10.1128/AAC.02233-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng B., Lv T., Xu H. Discovery and characterisation of an Escherichia coli ST206 strain producing NDM-5 and MCR-1 from a patient with acute diarrhoea in China. Int J Antimicrob Agents. 2018;51(2):273–275. doi: 10.1016/j.ijantimicag.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 29.Bulman Z.P., Chen L., Walsh T.J. Polymyxin combinations combat Escherichia coli harboring mcr-1 and blaNDM-5: preparation for a postantibiotic era. MBio. 2017;8(4) doi: 10.1128/mBio.00540-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mediavilla J.R., Patrawalla A., Chen L. Colistin- and carbapenem-resistant Escherichia coli harboring mcr-1 and blaNDM-5, causing a complicated urinary tract infection in a patient from the United States. MBio. 2016;7(4) doi: 10.1128/mBio.01191-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fupin Hu, Yan Guo, Demei Zhu. Antimicrobial resistance profile of clinical isolates in hospitals across China: report from the CHINET Surveillance Program, 2017. Chin J Infect Chemother. 2018;18(3) [Google Scholar]