Abstract
Murine adenovirus 2 (MAdV-2) infects cells of the mouse gastrointestinal tract. Like human adenoviruses, it is a member of the genus Mastadenovirus, family Adenoviridae. The MAdV-2 genome has a single fibre gene that expresses a 787 residue-long protein. Through analogy to other adenovirus fibre proteins, it is expected that the carboxy-terminal virus-distal head domain of the fibre is responsible for binding to the host cell, although the natural receptor is unknown. The putative head domain has little sequence identity to adenovirus fibres of known structure. In this report, we present high-resolution crystal structures of the carboxy-terminal part of the MAdV-2 fibre. The structures reveal a domain with the typical adenovirus fibre head topology and a domain containing two triple β-spiral repeats of the shaft domain. Through glycan microarray profiling, saturation transfer difference nuclear magnetic resonance spectroscopy, isothermal titration calorimetry and site-directed mutagenesis, we show that the fibre specifically binds to the monosaccharide N-acetylglucosamine (GlcNAc). The crystal structure of the complex reveals that GlcNAc binds between the AB and CD loops at the top of each of the three monomers of the MAdV-2 fibre head. However, infection competition assays show that soluble GlcNAc monosaccharide and natural GlcNAc-containing polymers do not inhibit infection by MAdV-2. Furthermore, site-directed mutation of the GlcNAc-binding residues does not prevent the inhibition of infection by soluble fibre protein. On the other hand, we show that the MAdV-2 fibre protein binds GlcNAc-containing mucin glycans, which suggests that the MAdV-2 fibre protein may play a role in viral mucin penetration in the mouse gut.
Keywords: adenovirus fibre, infection inhibition, ligand, crystal structure, affinity, gastrointestinal tract
Introduction
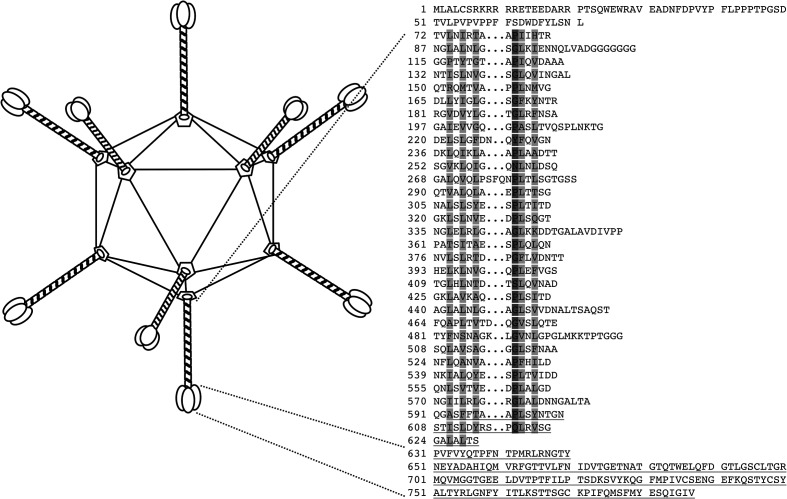
Adenoviruses are non-segmented, linear, double-stranded DNA viruses, taxonomically classified in the family Adenoviridae. First isolated from adenoid tissues [1], these non-enveloped icosahedral viruses are 70–90 nm in diameter and have trimeric fibre proteins protruding from their vertices (Fig. 1) [2, 3]. The vertices themselves are formed by pentameric penton base proteins, while trimeric hexons occupy the faces of the capsid. Viral infection begins when the fibre engages a host cell receptor. Several of these receptors have been identified and characterized for human adenoviruses [4], whereas the receptors for animal adenoviruses are largely undefined.
Fig. 1.
Adenovirus capsid and the MAdV-2 fibre protein. Schematic drawing of the icosahedral adenovirus capsid (left). The capsid surface is made up of hexon proteins, while trimeric fibre proteins emanate from the pentameric penton vertices. Sequence of the MAdV-2 fibre protein (right). The penton base-binding domain is predicted to contain residues 1–71, the fibre shaft contains up to 32 triple β-spiral repeats and the fibre head domain starts at residue 631.
Like human adenoviruses (HAdVs), murine adenovirus 2 (MAdV-2) is a member of the genus Mastadenovirus [5]. It belongs to the species Murine mastadenovirus B. MAdV-2 was isolated from the faeces of house mice, and in vitro studies showed its ability to cause cytopathic changes in mouse cell lines, particularly those of gastrointestinal origin, but not in monkey or human cell lines [6, 7]. Consistent with this tropism in cell culture, MAdV-2 infects the gastrointestinal tract upon the infection of mice, but causes no overt disease, and replicates in primary mouse intestinal cells [8, 9]. The genomic sequence of MAdV-2 has been determined and compared with that of MAdV-1 and MAdV-3, with which it is most closely related [10–12]. Analysis of the MAdV-2 fibre sequence suggested the presence of a virus-anchoring domain (up to residue 71), a central shaft domain containing 32 putative triple β-spiral repeats (residues 72–630; 13) and a C-terminal head domain (amino acids 631–787; Fig. 1). The head domain has little sequence identity (10–16 %) with known fibre head structures.
Here we report the crystal structure of the carboxy-terminal residues 594–787 of the MAdV-2 fibre. The structure revealed the expected homo-trimeric protein, containing two distal β-spiral repeats of the fibre shaft domain (residues 594–630) and the complete globular head domain consisting of amino acids 631–787. Glycan microarray profiling identified N-acetylglucosamine (GlcNAc) as a ligand for this protein, which was confirmed by saturation transfer difference (STD) nuclear magnetic resonance (NMR) spectroscopy (STD-NMR) and isothermal titration calorimetry (ITC). The crystal structure of GlcNAc in complex with the fibre trimer was also determined, revealing a GlcNAc-binding cleft involving the side-chains of amino acids Asn647, Tyr650 and Glu652 of the AB-loop and the backbone of residues 679–682 of the CD loop of the fibre head domain. GlcNAc binding of the MAdV-2 fibre may play a role in the gastrointestinal tropism of the virus and binding of the protein to mucin carbohydrates supports this.
Results and Discussion
In analogy with other adenoviruses [13–16], the carboxy-terminal head domain of the MAdV-2 fibre is likely to be important for primary receptor binding. However, the sequence identity of the C-terminal amino acids 583–787 with known adenovirus fibre head structures is rather low. Therefore, we set out to determine the crystallographic structure of the C-terminal domain of the MAdV-2 fibre and to obtain information about receptor binding.
Expression vector design, purification, crystallization and structure solution
Inspection of the 787 amino acid-long MAdV-2 fibre sequence revealed the presence of an N-terminal basic nuclear localization signal (up to residue 28), a putative penton base-binding motif (residues 35–41), an oligo-glycine sequence potentially forming a hinge region (residues 108–116) and the presence of putative triple β-spiral repeats starting at amino acid 72 and potentially ending at residue 630 [17–19]. The C-terminal to the triple β-spiral region is the fibre head domain. We expressed C-terminal fragments starting at residues 517 or 586 and ending at the natural C-terminus of the fibre protein (residue 787). The proteins were expressed in Escherichia coli and purified.
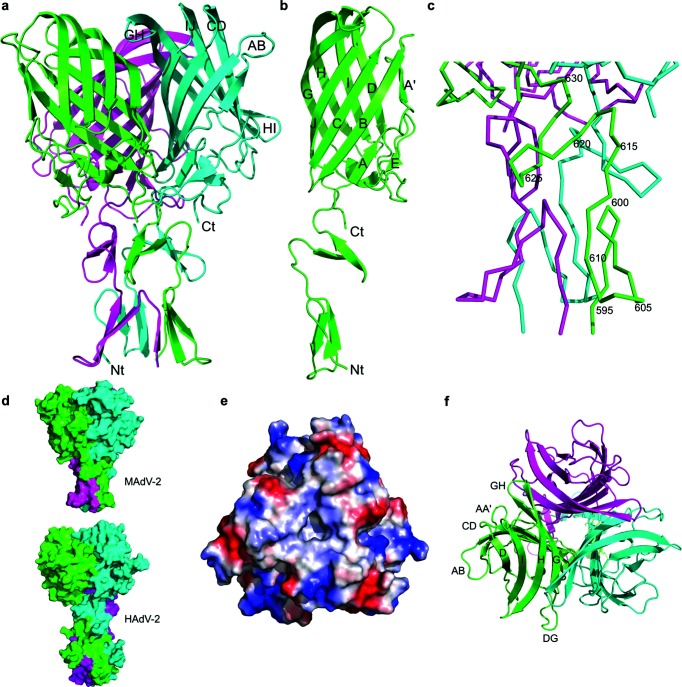
Crystals of the shorter construct (amino acids 586–787) were obtained at pH 7.5 in several conditions and space groups after 10 to 30 days. In contrast, the longer fragment (containing residues 517–787) failed to produce crystals. For the structure solution, a dataset from a methylmercury chloride-treated crystal was collected (Table S1, available in the online version of this article). The dataset was processed to 2.76 Å resolution and found to have considerable anomalous signal. Twelve mercury sites corresponding to four cysteine residues per monomer in a single trimer per asymmetric unit were identified. Refinement of these sites resulted in high-quality phases, allowing the construction of a model. Two high-resolution native structures were solved by molecular replacement (at 1.8 Å resolution and at 1.7 Å resolution). Automatic and manual building and refinement resulted in models with at least amino acids 594–787 for each of the protein chains. The structures revealed the presence of the fibre head domain along with two β-spiral repeats of the fibre shaft domain (Fig. 2a). The head domain starts at residue 631, while residues 592–630 form part of the shaft domain. The refined models have good geometry and most of the residues are in favoured regions of the Ramachandran plot (Table S1). The remaining N-terminal residues of the protein as well as the vector-supplied tags could not be modelled.
Fig. 2.
Structure of the C-terminal domain of the MAdV-2 fibre containing two shaft repeats and the head domain. (a) Structure of the trimer with the three chains coloured differently. The fibre head domain is 6.1 nm wide and 4.5 nm high. The N- and C-termini and AB, CD, GH, HI and IJ loops of the cyan-coloured chain are indicated. (b) Structure of the monomer in which the β-strands of the fibre head are labelled. (c) Close-up of the shaft domain, with every fifth residue in one of the chains labelled. (d) Surface representations of the MAdV-2 (top) and HAdV-2 fibre (bottom) structures with the head domain in the same orientation. Note the relative rotation of the shaft of around 120° along its long axis. (e) Qualitative electrostatic surface of the trimer seen from the end of the fibre or ‘top’. Positively (blue) and negatively charged (red) regions are apparent. (f) Top view of the trimer with the three monomers coloured differently and in the same orientation as panel (e). Clearly visible β-strands and loops are labelled.
The fibre head domain
Each monomer of the MAdV-2 fibre head contains a β-sandwich, like other adenovirus fibre heads. Together, they form a 4.5 nm high β-propeller measuring 6.1 nm in diameter. Each monomer consists of 157 residues (631–787), i.e. fewer residues than other mastadenovirus fibre heads for which the structure is known. For comparison, the HAdV-5 fibre head contains 185 amino acids [20] and the canine adenovirus 2 (CAdV-2) fibre head 197 [21]. The difference is due to the loops on the top and the side of the fibre head being shorter in MAdV-2 when compared to the other two (Fig. S1). The β-sandwich of each MAdV-2 fibre head monomer consists of two anti-parallel β-sheets, ABCJ and GHID, as is the case for other adenovirus fibre heads. Like the CAdV-2 fibre head, strand A of the MAdV-2 fibre head is kinked and thus divided into strands A and A′ (Fig. 2b). Most of the loops connecting strands are short, except the DG loop, which contains residues 690–728. The DG loop contains an additional β-strand E and a helical stretch (residues 716–718). Strand E contains residues 702–704 and aligns with strand A.
Superposition of trimers from the two native models did not show any significant differences, and they aligned with root mean square deviation (RMSD) values of less than 0.5 Å. The closest structural homologue is the CAdV-2 fibre head domain [Protein Data Bank (PDB) code 2J1K; 21] with an alignment RMSD of 2.6 Å for C-α atoms and a Z-score of 15.8. The MAdV-2 fibre head domain shares the AA′BCJ-GHID topology with the CAdV-2 fibre head, even though fewer than 12 % of the amino acids are identical after structural alignment. However, when compared with the CAdV-2 fibre head, there are noticeable differences: the AB loop is in a different conformation, the CD and GH loops are shorter for the MAdV-2 fibre head, and the DG and HI loops contain short helical regions in the CAdV-2 fibre head that are not present in the MAdV-2 fibre head (Fig. S1). Other human and animal mastadenovirus fibre heads are also similar, with Z-scores ranging from 11.6 to 14.5. When the structure is compared to the fibre head domains of members of other genera, Z-scores of 10.5–12, 6.4–7 and 5–5.5 are obtained for aviadenovirus fibre heads, atadenovirus fibre heads and siadenovirus fibre heads, respectively. Similarity is also observed with reovirus fibre heads (Z-scores of 8.8–10.5).
The fibre shaft domain repeats
Beta-spiral repeats are present in adenovirus fibre shaft domains as well as the receptor-binding proteins of some other viral fibres, such as the mammalian and avian reovirus attachment proteins, sigma1 [22] and sigmaC [23], and the bacteriophage PRD1 fibre [24]. They are characterized by two small β-strands connected by a loop that usually contains a glycine or proline, which facilitates the formation of a type II β-turn [17]. In the MAdV-2 structure, two such β-spiral repeats are present (residues 594–623), and one partial repeat can also be observed between residues 624 and 630 (Figs 1, 2c). The first repeat is a canonical proline-type and contains a cis-peptide bond between residues Ala599 and Pro600 with a corresponding type II β-turn, as described for reovirus fibre proteins [22, 23]. The second repeat is not of the canonical glycine- or proline-type, with Gln618 in the position where normally a glycine or proline residue is observed and the insertion of an extra residue in the turn. The last half-repeat is followed by a three-amino acid linker consisting of hydrophobic amino acids, which connects the shaft to the C-terminal head domain (631-PVF-633).
Little flexibility can be anticipated in the MAdV-2 fibre shaft–head junction due to the nature of its constituent amino acids. In the case of HAdV-2, the six-residue linker (393-NKNDDK-398) is made up of hydrophilic residues and flexible. The flexibility of the fibre has been shown to influence receptor interaction, although not specifically at the shaft–head junction [25]. Interestingly, a relative rotation of around 120° is observed between the head and shaft domains when the MAdV-2 structure is compared with the HAdV-2 head–shaft structure (PDB code 1QIU; 13). This is illustrated in Fig. 2(d), where the head domains of the MAdV-2 and HAdV-2 fibres are shown in the same orientation and it can then be seen that the C-terminal shaft repeat that faces the reader is different (green for MAdV-2 and light blue for HAdV-2). Because these are the only two adenovirus fibre shaft structures known at high enough resolution, we cannot predict which conformation is present in other adenovirus fibres. The relative head–shaft orientations in the reovirus fibres sigma1 (PDB code 1KKE; 22) and sigmaC (PDB code 2BT8; 23), and in the bacteriophage PRD1 fibre (PDB code 1YQ8; 24), resemble the orientation in the HAdV-2 fibre more closely than that in the MAdV-2 fibre.
Stability of the MAdV-2 fibre
The inter-subunit contacts in the MAdV-2 fibre extend from the shaft repeats to almost the top of the assembly. These contacts are extensive and include several hydrophobic residues in the shaft as well as between head domain monomers. The fibre head and shaft repeats jointly have a monomeric surface area of 11×103 Å2, of which 17 % is buried upon trimer formation. Considering the fibre head only (residues 631–787), these values reduce to 8×103 Å2 and 12 %, respectively. The shaft domain, being intertwined, buries 20 % of its 3×103 Å2 monomeric surface area. There are 19 inter-monomer hydrogen bonds in total, 11 of which are located within the shaft repeats. This suggests that the C-terminal part of the shaft domain contributes significantly to maintaining the integrity of the trimer, which has also been observed for the HAdV-2 fibre [26]. In addition, two salt bridges are formed at each monomeric interface, one of which is formed by shaft residue Arg615 interacting with the C-terminal carboxyl group (Val787). The calculated energy decrease upon trimer formation is approximately −80 kcal mol−1, of which −38 kcal mol−1 is contributed by the shaft repeats alone.
Potential binding sites for known adenovirus protein receptors
We evaluated whether the structure of the MAdV-2 fibre head would support binding to coxsackievirus and adenovirus receptor (CAR) and CD46 [13, 27]. When the structure of the MAdV-2 fibre head is superposed on the structure of the CAdV-2 fibre head in complex with the D1 domain of CAR [21, 28], the DG loops are in a very different conformation (Fig. S2). Apart from Thr441 of CAdV-2, none of the residues that are important for CAR binding are conserved in the MAdV-2 fibre head. A basic residue involved in a salt bridge in CAR binding (Arg384 in CAdV-2) is structurally equivalent to Asp655 in the MAdV-2 fibre head, which has the opposite charge. Furthermore, Arg700 of the MAdV-2 fibre head would clash with CAR D1 if it were to bind in the same orientation. The structure of the MAdV-2 fibre head also does not appear to be compatible with binding to CD46. The HI loops of the HAdV-11 and HAdV-21 fibre head domains, which are important for CD46 binding [15, 27, 29], are much longer than the HI loop of the MAdV-2 fibre head (Fig. S3). Consequently, residues that are important for CD46 binding in the HAdV-11 fibre head (Arg279, Arg280, Asp284 and Glu285) are not structurally conserved. It thus appears that CAR and CD46 are unlikely to function as MAdV-2 fibre head ligands.
Carbohydrate-binding screen
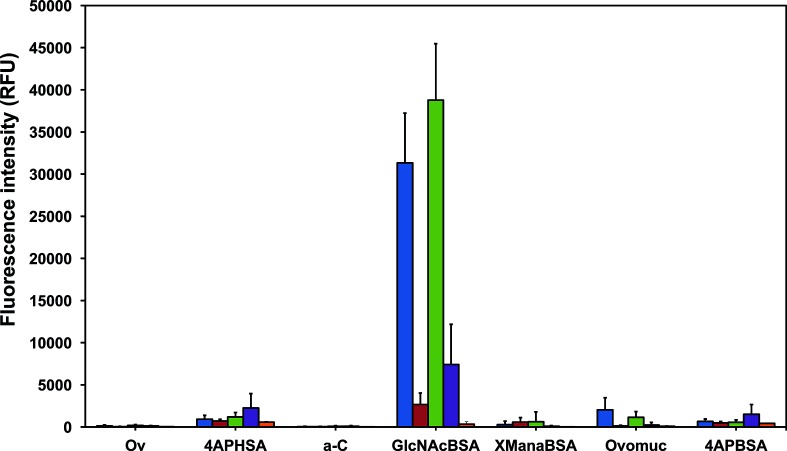
The surface of the MAdV-2 fibre head has a mixed distribution of predicted positively and negatively charged patches, especially on the peripheral areas and on top of the trimer (Fig. 2e, f). The CAdV-2 fibre head engages sialic acid using one of its peripherally located basic patches [21], and its structural similarity to the MAdV-2 fibre head indicated a potential carbohydrate-binding function. Binding of the HAdV-37 fibre head and the HAdV-52 short fibre head to cell surface sialic acid and of the TAdV-3 fibre head domain to 3′-sialyllactose has also been shown previously [30–32]. To explore this possibility, we screened both MAdV-2fib(517-787) and MAdV-2fib(586-787) with a glycan microarray consisting of a large variety of carbohydrate molecules alone and linked to proteins. We found that both MAdV-2fib(517-787) and MAdV-2fib(586-787) constructs bound with high intensity to GlcNAc-BSA and with lower binding intensity to ovomucoid (Fig. 3, Ovomuc), which contains complex N-linked oligosaccharides, including di- to penta-antennary structures with terminal and bisecting GlcNAc residues [33–36]. This binding was inhibited in the presence of 100 mM GlcNAc, demonstrating carbohydrate-mediated binding for both constructs [37]. Other trimeric His-tagged adenovirus capsid proteins did not bind to GlcNAc in the same glycan micro-array [32, 38], providing a further control.
Fig. 3.
Annotated binding profile of two isoforms of MAdV-2 fibre protein. Bar chart representing the binding intensity of the long [MAdV-2fib(517-787)] and short [MAdV-2fib(586-787)] forms of the MAdV-2 fibre to carbohydrates on a microarray surface. Binding was detected using a fluorescently labelled anti-His antibody. The data represent the average of four replicate experiments and the error bars depict one standard deviation of the mean calculated over four microarray slides. Blue, MAdV-2fib(517-787); red, MAdV-2fib(517-787) in the presence of 100 mM GlcNAc; green, MAdV-2fib(586-787); purple, MAdV-2fib(586-787) in the presence of 100 mM GlcNAc; orange, anti-His antibody control. Refer to the text and Table S2 for definitions of glycan abbreviations.
The viral proteins did not bind to the glucose neoglycoconjugate (XGlcBSA) presented on the microarray (Fig. S4), which indicated the possible importance of the N-acetyl group of GlcNAc for ligand binding. GlcNAc was also present on this microarray in the N,N′-diacetylchitobiose core (GlcNAc-β-(1→4)-GlcNAc) of N-linked oligosaccharides on glycoproteins such as fetuin and transferrin and in the N-acetyllactosamine (Gal-β-(1→4)-GlcNAc] neoglycoconjugate (LacNAcBSA) (Table S2), but no significant binding to these ligands was observed. These data suggested that terminal or non-reducing GlcNAc residues are the optimal ligands.
Ovalbumin is substituted with high mannose-, hybrid- and complex-type N-linked glycosylation with terminal GlcNAc, and bisecting GlcNAc residues [39]. However, no binding was observed with ovalbumin on the microarray (Ov, Fig. 3), which may be due to a relatively low proportion of the relevant binding structure in ovalbumin compared to ovomucoid or to a sterically hindered presentation of the non-reducing GlcNAc residues on ovalbumin. The viral proteins also did not bind to α-crystallin (Fig. 3, a-C), which contains O-linked GlcNAc. However, only 10 % of α-crystallin is glycosylated [39, 40], so it is not possible to determine whether the linkage or abundance of the GlcNAc residues is responsible for the lack of binding. Binding above the threshold was also observed with mannose α-linked to BSA, but the variability of binding was greater than the signal intensity itself (Fig. 3, XManαBSA), and the signal was not inhibited upon co-incubation with 50 mM mannose. In addition, all samples bound in a non-inhibitable manner to the exposed linker molecules 4AP-BSA and 4AP-HSA, which were placed on the slide as controls. It is possible that the high charge of the linker is responsible for the observed binding to the mannose neoglycoconjugate, as the linker molecules bind both the MAdV-2 fibre proteins and the anti-6XHis antibody.
Binding of GlcNAc to MAdV-2 fibre in solution
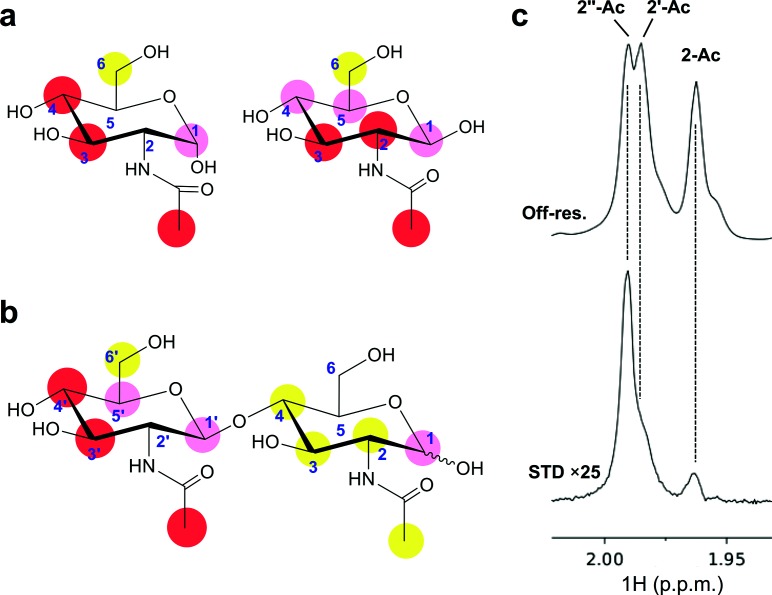
STD-NMR and ITC were employed to validate the glycan microarray screening results and to analyse GlcNAc binding to MAdV-2 fibre heads in solution. STD-NMR experiments performed on GlcNAc in the presence of the MAdV-2 fibre head gave rise to STD-positive peaks (Figs 4a, S5a), which confirmed that GlcNAc binding also occurs in aqueous solution. The highest degree of saturation was observed at the signals of protons H3 and H4 of the α-anomer and H2 and H3 of the β-anomer (the signals for H4 and H5 of the β-anomer overlapped and could not be quantified independently). The N-acetyl methyl peak also showed a high degree of saturation in both anomers. Comparatively, the H6 signals received the lowest relative STD intensity. Further STD-NMR experiments were conducted to analyse the recognition of the GlcNAc unit in more complex carbohydrates. Experiments performed on N,N′-diacetylchitobiose [GlcNAc-β-(1→4)-GlcNAc] showed greater STD intensities at the signals belonging to the non-reducing GlcNAc residue, suggesting that this sugar residue is recognized preferentially. Again, the highest STD responses were displayed by H3, H4 and the acetyl methyl signal of the non-reducing residue (Figs 4b, S5b). Experiments performed on chitobiose, the deacetylated version of N,N′-diacetylchitobiose, showed a much lower STD response than its diacetylated analogue (Fig. S2d), suggesting that the acetyl group could play a role in the binding, either establishing apolar interactions or neutralizing the positive charge of the sugar amine. STD experiments on N-acetyllactosamine [Gal-β-(1→4)-GlcNAc] showed no STD response for the disaccharide (although STD peaks were observed in the spectra, belonging to trace amounts of monomeric GlcNAc present in the sample) (Fig. S5e). This further supports the observation that non-reducing residues are preferentially recognized. Finally, experiments performed on the trisaccharide N,N′,N′′-triacetylchitotriose [GlcNAc-β-(1→4)-GlcNAc-β-(1→4)-GlcNAc] again showed a preferential recognition of the non-reducing portion, as was evident from the anomeric region of the spectrum (Fig. S5c), as well as the N-acetyl methyl signals (Fig. 4c). The N-acetyl methyl protons of the non-reducing terminus received the highest amount of magnetization, followed by that of the internal residue and the reducing unit, respectively.
Fig. 4.
Binding of N-acetylglucosamine to MAdV-2fib(586-787) studied by STD-NMR. (a) Epitope mapping obtained from an experiment performed on GlcNAc in the presence of protein. Chemical structures of α-GlcNAc (left) and β-GlcNAc (right). These anomers are in equilibrium in solution and resolved for most signals in the 1H-NMR spectrum. Labels indicate STD intensity for each signal, relative to the STD intensity of the H4 signal of the α-anomer. Complete spectra are in Fig. S5(a) (red circles, I>70 %; pink circles, 70 %>I>40 %; yellow circles, 40 %>I>20 %). (b) Binding of N,N′-diacetylchitobiose [GlcNAc-β-(1→4)-GlcNAc] to MAdV-2fib(586-787). Epitope mapping from an experiment performed with N,N′-diacetylchitobiose, with labels indicating the STD intensity for each signal relative to the STD intensity of the H4′ signal. See spectra in Fig. S5(b). (c) Binding of N,N′,N′′-triacetylchitotriose [GlcN′′Ac-β-(1→4)-GlcN′Ac-β-(1→4)-GlcNAc]. Detailed view of spectral region for the N-acetyl methyl signals (2′′Ac, non-reducing residue; 2′Ac, middle residue; Ac, reducing residue). Top spectrum: off-resonance (reference) spectra; bottom spectrum: STD spectrum (see full spectra in Fig. S5c).
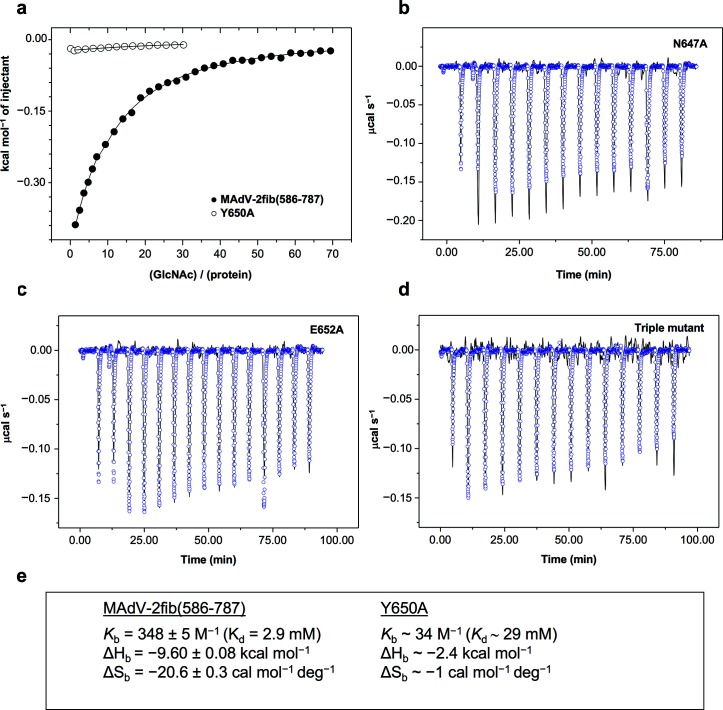
We then undertook an ITC study to determine the thermodynamic parameters of MAdV-2fib(586-787) binding to GlcNAc (Fig. 5a). Assuming one binding site per monomer of the trimer, a Kd value of 2.9 mM was determined (Fig. 5e). This affinity is comparable to that of turkey adenovirus 3 fibre head to sialyllactose [32], but low compared to some other adenovirus fibre–carbohydrate interactions, which are in the micromolar range [41, 42]. However, this corresponds to the binding of one GlcNAc unit to a single binding site in solution. The biological interacting partner may be a more complex carbohydrate or a glycosylated cell surface molecule with a more extensive interaction footprint on the fibre head, such as that of the GD1a glycan binding with several carbohydrate rings to the surface of the HAdV-37 fibre head [16]. Multivalency may also play a role [43]. The avidity effect of three binding sites per fibre and multiple fibres binding simultaneously to GlcNAc units could lead to significantly tighter binding of the virus to entities incorporating multiple terminal GlcNAc groups [44].
Fig. 5.
Isothermal titration calorimetry of MAdV-2fib(586-787) protein and its site-directed mutants with GlcNAc. (a) Variation of the heat evolved per mole of ligand on the ligand/protein molar ratio for the wild-type protein (black circles) and the Y650A mutant (white circles). Continuous lines are the fit of a one-site model per monomer to the experimental data. (b,c,d) Comparison of experimental traces registered upon GlcNAc injection on N647A, E652A or triple mutant (lines), or buffer (blue symbols). (e). Best fitting thermodynamic parameters of GlcNAc ITC-titration into the wild-type and the Y650A mutant.
Crystal structure of the MAdV-2 fibre head bound to GlcNAc
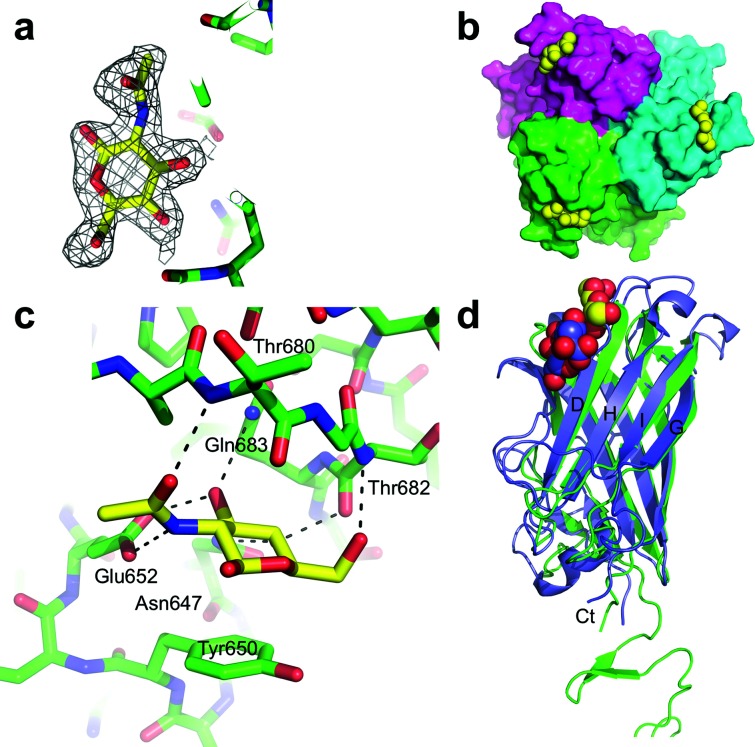
Crystals of MAdV-2fib(586-787) were soaked in 10 mM GlcNAc and a 2.0 Å resolution diffraction dataset was collected. Unbiased electron density maps showed the density for the ligand on each of the three protein chains (Fig. 6a). GlcNAc binds to a groove on the top of the trimer located towards the side of the molecule, between the AB and CD loops of the fibre head domain and involving residues of the D-strand (Fig. 6b). The sugar ring of GlcNAc makes CH–pi interactions with the side-chain of Tyr650 on the AB loop (Fig. 6c). In the complex structure, the O3 and O4 atoms point towards the protein, while O1 and O6 point towards the solvent, suggesting that GlcNAc linked to more complex molecules through O1 or O6 may also bind to the MAdV-2 fibre. Two other residues from the AB loop, Asn647 and Glu652, make hydrogen bond interactions with GlcNAc atoms. The N-acetyl group of the ligand is stabilized by a hydrogen bond involving its O7 atom and the main chain nitrogen of Thr680. Thr682 and Gln683 engage GlcNAc with two additional hydrogen bonds. All of these ligand atoms are situated on the opposite side of the acetyl group. The fact that the non-reducing residue appears to be selectively recognized in STD-NMR experiments agrees with the pose adopted by the monosaccharide in the crystallographic structure of the complex, in which substitution at position 4 would result in steric clashes with the protein surface, but position 1 points toward the solvent and is compatible with derivatization.
Fig. 6.
Binding site of GlcNAc on the MAdV-2 fibre head. (a) Omit map (2Fo-Fc) contoured at 1 sigma displayed around the GlcNAc molecule in monomer A. (b) Top or end-of-fibre view of three GlcNAc molecules (yellow) binding between the AB and CD loops of the MAdV-2 fibre head. (c) Close-up of one of the GlcNAc binding sites. Residues with atoms within hydrogen-bonding distance are labelled. Putative hydrogen bonds are marked with black interrupted lines. The sugar ring stacks onto the side-chain of Tyr650 (also labelled). (d) Superposition of the structure of the MAdV-2 fibre head (green) bound to GlcNAc (yellow and red) and the CAdV-2 fibre head bound to sialic acid (PDB 2WBV; slate blue and red).
To confirm the importance of the identified amino acids for GlcNAc interaction, we made three site-directed mutants and the corresponding triple mutant. Amino acid mutants were generated and proteins were expressed and tested for their binding to GlcNAc by ITC. When the stacking residue Tyr650 was mutated to an alanine, there was a 10-fold reduction (ΔΔG of about 1.4 kcal mol−1) in binding (Fig. 5a, e), which agrees with the values associated with other CH–pi interactions [45]. An even higher reduction in binding was observed when Asn647 was mutated to alanine (Fig. 5b), and no binding over control buffer (circles) was observed with the Glu652 to the alanine mutant (Fig. 5c). A triple mutant combining the three point mutants also failed to generate any binding response on ITC isotherms (Fig. 5d).
The GlcNAc site on the MAdV-2 fibre head is at a similar location to the binding site of sialic acid on the CAdV-2 fibre head (Fig. 6d), although the interacting residues and the nature of the ligand are different [28]. In both cases, residues of the D-strand are involved, but because this strand is shorter in the CAdV-2 fibre head, the ligand binds in a different position. This allows interaction with Arg515 of the I-strand, which makes a salt bridge with the carboxyl group of the sialic acid. In contrast, the GlcNAc molecules are stabilized by pi-stacking onto Tyr650 of the MAdV-2 fibre head. In both cases, hydrogen bonds and hydrophobic contacts complete the interactions at the binding sites. HAdV-19p, -37 and -52 also bind sialic acid containing compounds [14, 31, 42], but not in comparable positions (Fig. S6). In the case of the highly homologous HAdV-19p and HAdV-37 fibre heads, binding is at the top centre of the molecules, involving residues from the GH and IJ loops (PDB codes IUXB and 1UXA). In the HAdV-52 fibre head structure, the sialic acid analogue also binds to the top of the molecule, but somewhat shifted to the side and involving residues of the end of the DG loop from one monomer and of the GH loop of a neighbouring monomer (PDB code 4XL8).
GlcNAc competition binding assays
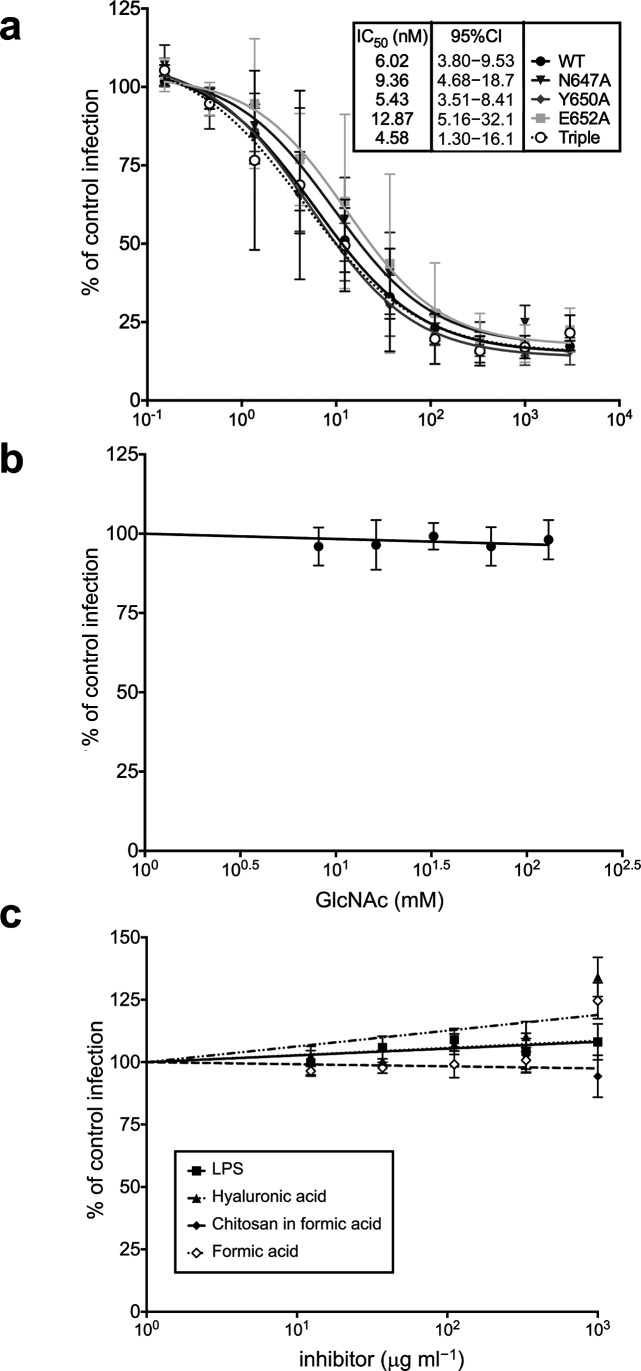
We undertook two lines of investigation to determine whether GlcNAc binding is important for MAdV-2 infection. Our first approach was to use a molar excess of recombinant MAdV-2 fibre head to compete MAdV-2 infection of CMT-93 epithelial cells of mouse rectal carcinoma origin and then to test the effects of mutations that reduce or preclude GlcNAc binding on fibre head activity in this assay. In the initial experiments, we found that wild-type MAdV-2fib(517-787) [IC50=16 nM, 95 % confidence interval (CI)=6–39 nM] was a much more potent competitor than MAdV-2fib(586-787) (IC50=60 µM, 95 % CI=22–346 µM), so the point mutations were only tested in the longer construct. However, none of the mutations altered the ability of MAdV-2fib(517-787) to compete with MAdV-2 infection (Fig. 7a). It is possible that the extra part of the shaft domain confers more stability to the protein (structural stability or stability against degradation by proteases), or that it contains additional receptor-binding epitopes. The fact that MAdV-2fib(517-787) inhibits MAdV-2 infection with an IC50 of 16 nM and that the Kd of GlcNAc binding to the fibre head is 2.9 mM suggests that a higher affinity binding interaction is provided by MAdV-2fib(517-787). The nature and location of this binding interaction are currently unknown.
Fig. 7.
Infection inhibition assays. (a) Inhibition of MAdV-2 infection by wild-type (WT) and the indicated mutants of MAdV-2fib(517-787). IC50 values with their corresponding 95 % confidence intervals (CIs) were determined by non-linear regression. MAdV-2 infection is not inhibited by (b) soluble GlcNAc or (c) GlcNAc-containing polymers. The data are expressed relative to control cells infected in the absence of inhibitor (100 %). The results are the means of at least three independent experiments.
Our second approach was to compete MAdV-2 infection with an excess of monomeric GlcNAc (up to 130 mM) and natural GlcNAc-containing polymers [chitosan, hyaluronan and lipopolysaccharide (LPS); up to 1 mg ml−1). However, none of the compounds inhibited MAdV-2 infection (Fig. 7b, c). Collectively, these results suggest that MAdV-2 infection is not dependent upon GlcNAc binding to the fibre head. However, the presentation of GlcNAc on the polymers used is only monomeric in effect, as there is only one terminal non-reducing GlcNAc residue available. Therefore, the lack of infection inhibition may not be surprising if the natural GlcNAc-containing ligand is multivalent in presentation. The CAdV-2 fibre head binds sialic acid, but this interaction is also thought not to be important for host cell recognition [28, 46]. Instead, CAdV-2 entry is dependent on CAR. We do not think that the MAdV-2 fibre head binds CAR, but it may bind another protein receptor.
Mucin binding of the MAdV-2 fibre
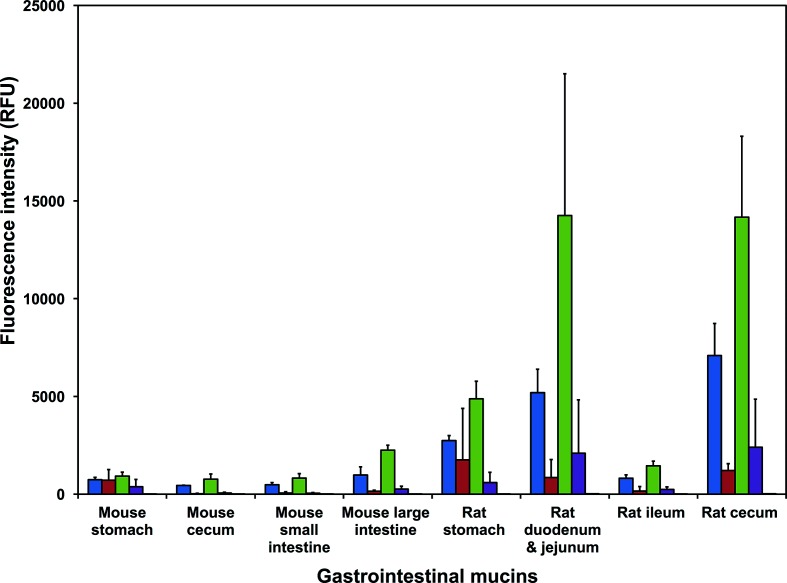
The epithelial cells of the gastrointestinal tract are coated with a thick layer of mucus, with mucins as the main components. Mucins are glycoproteins that are densely substituted with O-linked oligosaccharides and some N-linked oligosaccharides. The glycosylation of the gastrointestinal tract mucins varies depending on species, health status and location within the gastrointestinal tract [47]. We reasoned that mucin carbohydrates may be potential ligands of MAdV-2 fibres during infection, before reaching the underlying epithelial cells [48]. To test this, we incubated MAdV-2fib(517-787) and MAdV-2fib(586-787) on a gastrointestinal-tract mucin microarray consisting of purified natural mucins from eight animal species (cow, sheep, horse, pig, chicken, deer, mouse and rat), mucins from two human-derived cell lines and several reference glycoproteins (Table S3) [49]. All probes on the mucin microarray were recognized by MAdV-2fib(517-787) and MAdV-2fib(586-787) with varying intensity, with the exception of the deer abomasum mucin and the glycoprotein transferrin (Table S3 and Fig. S7). In general, MAdV-2fib(586-787) showed greater binding to the probes on the mucin microarray than MAdV-2fib(517-787), which reflects a slightly altered affinity or avidity of the shorter fibre protein compared to the longer one, or, alternatively, differences in the efficiency of recognition by the anti-His-tag antibody used for detection. Both constructs bound more intensely to several gastrointestinal tract post-stomach mucins from other species than to mouse gastrointestinal tract mucins. The greatest binding was to the mucins from ovine spiral colon, equine small intestine, equine right ventral colon, equine dorsal colon, human colon carcinoma cell-derived LS174T, porcine ceca, rat duodenum and jejunum and rat cecum (Fig. S7). Co-incubation with 100 mM GlcNAc reduced the binding of the viral proteins to most of the gastrointestinal tract mucins by 10 % or more (Table S4 and Fig. S3). Most intense binding events were inhibited by GlcNAc, except for the interactions with the equine right ventral colon and dorsal colon mucins (Table S4).
Both MAdV-2fib(517-787) and MAdV-2fib(586-787) bound to the mouse gastrointestinal tract mucins from stomach, small intestine, cecum and large intestine (or colon), albeit at a low intensity [<2500 relative fluorescent units (RFU)]. Binding to the post-stomach mucins was inhibited by co-incubation with 100 mM GlcNAc (>81 %; Fig. 8 and Table S3) for both proteins. However, only binding of MAdV-2fib(586-787) to mouse stomach-derived mucin was GlcNAc-inhibited. In contrast, both constructs bound to mucins from the rat gastrointestinal tract with greater intensity overall than was seen for binding to mouse gastrointestinal tract mucins, and this binding was uniformly GlcNAc-inhibited (Fig. 8 and Table S4). There were several instances of binding in a non-GlcNAc-dependent manner (Table S3 and Fig. 8). For example, the addition of 100 mM GlcNAc did not reduce the binding of MAdV-2fib(517-787) to fetuin, asialofetuin or human α1-acid glycoprotein. It is not clear whether this presumably non-GlcNAc-mediated binding is due to binding to other, or more complex, carbohydrate structures, or whether it is due to protein–protein interactions.
Fig. 8.
MAdV-2 fibre binding to mouse and rat gastrointestinal tract mucins. Bar chart representing the binding intensity of MAdV-2fib(586-787) and (MAdV-2fib(517-787) forms of the MAdV-2 fibre to mouse and rat gastrointestinal tract mucins on a mucin microarray in the presence and absence of 100 mM GlcNAc. Binding was detected using a fluorescently labelled anti-His antibody. The data for the uninhibited proteins represent the mean of three technical replicate microarray slides and the error bars represent one standard deviation of the mean (sd). The data for the inhibited proteins represent the mean of two technical replicates and the error bars represent one sd. Blue, MAdV-2fib(517-787); red, MAdV-2fib(517-787) in the presence of 100 mM GlcNAc; green, MAdV-2fib(586-787); purple, MAdV-2fib(586-787) in the presence of 100 mM GlcNAc; orange, anti-His antibody control.
Taken together, the data indicate that the carbohydrate motif recognized on the gastrointestinal tract mucins present in the biological milieu can be inhibited by GlcNAc and might contain this sugar. The MAdV-2 fibre protein may play a role in infection by aiding viral penetration through the mouse gastrointestinal tract mucus layer. Binding is not strong enough to retain the virus in the mucus layer and may facilitate penetration of the protective mucus layer to access the underlying epithelial cells. Strong MAdV-2 binding to gastrointestinal tract mucins would likely result in virus retention in the mucus layer and elimination from the gastrointestinal tract by peristalsis. The more intense binding of the MAdV-2 fibre proteins to rat gastrointestinal tract mucins (Fig. 8), which could make the virus vulnerable to this strategy, may help explain why rats are not susceptible to MAdV-2 [50].
Conclusion
Mouse adenoviruses are likely the first branch of the lineage of mastadenoviruses, the oldest representatives of the genus Mastadenovirus, and thus the most different from human mastadenoviruses. Our structures reveal a MAdV-2 fibre head domain with the same framework as human adenovirus fibres, but with a smaller size, due to smaller surface loops. We also show the second partial structure of an adenovirus fibre shaft domain. The relative orientation between the head and shaft domains varies by about 120° between the MAdV-2 and HAdV-2 fibre structures.
GlcNAc is a ligand for MAdV-2 fibre (when fibre is expressed in isolation in E. coli) and GlcNAc binds with mM affinity at a site between the AB and CD loops of the head domain. Solution experiments with di- and tri-saccharides revealed a preferred interaction with GlcNAc units at the non-reducing end and highlighted the importance of the acetyl group. The MAdV-2 fibre head interacting with monomeric GlcNAc does not appear to be important for host cell recognition by the virus, as soluble free GlcNAc and polymers presenting a single non-reducing GlcNAc residue did not inhibit infection. Therefore, if GlcNAc binding is indeed biologically important, it would likely require a multimeric presentation of non-reducing terminal GlcNAc residues, such as those found in complex N-linked glycans of mammalian glycoproteins and ovomucoid [36], or in clustered glycolipids on the cell surface. Interestingly, the short fibre of HAdV-52 has been shown to bind linear oligomeric forms of sialic acid much more strongly than monomeric sialic acid molecules [31]. Human mastadenovirus B adenoviruses have been found to bind to GlcNAc polyanionic polymers of the heparan sulphate proteoglycan type [51, 52]. However, the MAdV-2 fibre protein does not contain the basic sequence motifs BBXB or BBBXXB (where B is a basic amino acid and X any amino acid) associated with binding to such polyanionic polymers.
A potential ligand-presenting molecule may be provided by mucin-type glycosylation, which is highly abundant in the mucus layer and on cell surfaces of the mammalian gastrointestinal tract. Indeed, multimerically presented, non-reducing terminal GlcNAc moieties are a common feature on mucins. Moreover, most mucin type O-linked glycans in the mouse gastrointestinal tract are core 2 type, which displays the non-reducing terminal GlcNAc residue Gal-β-(1→3)-[GlcNAc-β-(1→6)]-GalNAc multimerically [53]. Thus, GlcNAc binding may contribute to create multiple low-affinity interactions that are important for faecal–oral transmission and the intestinal tropism of MAdV-2 in vivo and are not apparent in cell culture. In addition, because adenoviruses find their utility in therapeutic applications and are actively pursued as gene delivery tools, the GlcNAc-binding MAdV-2 fibre head can potentially be used to direct adenovirus vectors to cells displaying this glycan on their surface. Future work will be required to determine the biological significance of these findings in the context of virus infection.
Methods
Expression and purification of MAdV-2fib(586-787) and MAdV-2fib(517-787)
Gene fragments coding for residues MAdV-2fib(517-787) and MAdV-2fib(586-787) were PCR-amplified from MAdV-2 genomic DNA and cloned into pET28a(+) (Merck, Darmstadt, Germany). Four different site-directed mutants (containing the mutations N647A, Y650A, or E652A, or all three together) were generated for both fibre constructs using the QuikChange procedure (Agilent Technologies, Santa Clara, CA, USA). The proteins were expressed and purified as for the TAdV-3 fibre head [54], except for the fact that 0.3 M sodium chloride was used in the cell resuspension buffer instead of 0.7 M. We used 10 mM Tris-HCl (pH 8.5) for the final storage buffer.
Crystallization, data collection and data processing
Sitting-drop vapour diffusion crystallization trials for MAdV-2fib(517-787) and MAdV-2fib(586-787) proteins were conducted at 21 °C. Plate-like crystals of MAdV-2fib(586-787) were obtained when 4.0 M ammonium formate, 0.1 M HEPES/NaOH (pH 7.5) was used as precipitant, using 1+1 µl drops and 0.15 ml reservoir solution. Crystals from this condition (native 1) were mounted in LithoLoops (Molecular Dimensions, Newmarket, Suffolk, UK) and vitrified directly in liquid nitrogen. For heavy-atom derivatization, methylmercury chloride was added to the reservoir in a final concentration of 5 mM and left overnight; 2 µl reservoir solution was then added to the drop and crystals were soaked for between 30 s and 1 min prior to vitrification. Crystals were also obtained when 10 % (w/v) PEG 4000, 0.1 M HEPES/NaOH (pH 7.5), 0.1 M magnesium chloride was used as the reservoir solution (native 2). These crystals were transferred to 10 % (w/v) PEG 4000, 0.1 M HEPES/NaOH (pH 7.5), 0.1 M magnesium chloride, 20 % (v/v) glycerol and soaked for 30 s prior to vitrification. The crystals used in 24 h soaking experiments with 10 mM GlcNAc (Sigma-Aldrich, St Louis, MO, USA) were of space group I213 and were obtained when 1.125 M lithium sulphate, 0.075 M HEPES/NaOH (pH 7.5), 25 % (v/v) glycerol was used as the reservoir solution. These crystals were vitrified directly. Diffraction datasets were collected at 100 K.
Crystallographic data integration was carried out using XIA2 [55] or directly with IMOSFLM [56], followed by symmetry determination, scaling and merging using the Collaborative Computational Project Number 4 (CCP4) software suite [57]. To solve the structure, the derivative dataset was input into AUTOSHARP pipeline version 3.10.2 [58]. The resulting model was used as a template for molecular replacement to solve the structures of the other crystal forms. Refinement was performed with REFMAC5 [59]. The models were validated with MOLPROBITY [60] and the interaction properties were determined by PISA [61]. Structural homologues were identified by the DALI server [62]. Figure preparation was carried out with PYMOL (Schrödinger LLC, Cambridge, MA, USA).
Carbohydrate and mucin microarray profiling
Neoglycoconjugate microarrays were constructed as previously described and contained 70 carbohydrate targets (Table S2) either on glycoproteins or attached to protein backbones through linkers [48, 63]. Screening was performed essentially as previously described [32]. The titration of several viral protein concentrations in Tris-buffered saline supplemented with calcium and magnesium ions [TBS; 20 mM Tris-HCl (pH 7.2), 100 mM sodium chloride, 1 mM calcium chloride, 1 mM magnesium chloride] containing 0.05 % Tween 20 (TBS-T) indicated that a 30 µg ml−1 concentration of MAdV-2 fibre protein and 2.7 µg ml−1 of anti-6XHis IgG-CF640R monoclonal antibody (Sigma-Aldrich) in TBS-T was optimal for the detection of viral protein binding. Two-step incubations were carried out in quadruplicate. After incubation, washing and drying, microarray slides were scanned immediately on an Agilent G2505 microarray scanner using the Cy5 channel (633 nm excitation, 90 % PMT, 5 µm resolution). Binding inhibition assays were performed in parallel in the presence of 100 mM GlcNAc to verify specific binding [37]. The experimental threshold for binding was taken as five times the background [32, 64].
Mucin microarray profiling was performed to identify potential mucin and glycoprotein ligands that would bind to MAdV-2fib(586-787) and MAdV-2fib(517-787) in a GlcNAc-dependent manner. The gastrointestinal tract mucin microarrays were constructed as previously described [48]. Screening was performed against 44 different gastrointestinal tract mucins and glycoproteins (Table S2). The titration of several viral protein concentrations in 0.05 % TBS-T indicated that a 20 µg ml−1 concentration of MAdV-2 fibre protein and 4 µg ml−1 of anti-6xHis IgG-CF640R monoclonal antibody (Sigma-Aldrich) in TBS-T was optimal for the detection of viral protein binding. Two-step incubations were carried out in triplicate and inhibition studies were carried out in duplicate. After incubation, microarray slides were washed and dried as described above and then scanned immediately on an Agilent G2505 microarray scanner using the Cy5 channel (633 nm excitation, 90 % PMT, 5 um resolution). Inhibition assays were performed in parallel in the presence of 100 mM GlcNAc to verify specific binding [32]. Data extraction and analysis were carried out as described previously [37].
Saturation transfer difference (STD) nuclear magnetic resonance (NMR) spectroscopy (STD-NMR)
All NMR spectra were acquired at 25 °C in a Bruker AVANCE 500 MHz spectrometer equipped with a 5 mm inverse probe head. STD-NMR experiments were performed in fully deuterated 10 mM potassium phosphate buffer (pD 7.7). The samples contained 4 mM of the tested ligands and 20 µM of the MAdV-2 fibre head protein. Two proton spectra were obtained in interleaved fashion, one by saturation of the spins of the protein on-resonance at 0 p.p.m. and another by saturation of the spins of the protein off-resonance at 100 p.p.m., with a train of Gaussian-shaped pulses of 50 ms each, for a total irradiation time of 2 s. A T2 relaxation filter consisting of a 15 ms 5 kHz spin-lock was used to reduce the protein background signals. STD spectra were obtained by subtracting the on-resonance from the off-resonance spectra. STD intensities were measured by comparing each STD spectrum with the correspondent off-resonance spectrum and normalizing to the ligand peak receiving the highest degree of saturation (in all cases, the H4 proton of terminal non-reducing GlcNAc residues). The resonances of all the ligands tested were assigned or confirmed by 1H-1H TOCSY, 1H-1H NOESY and phase-sensitive 1H-13C HSQC. The data were analysed using TopSpin 3.0 (Bruker, Rivas-Vaciamadrid, Spain) and the figures were prepared using MestReNova v8.0.2 (Mestrelab, Santiago de Compostela, Spain).
Isothermal titration calorimetry measurements
Isothermal titration calorimetry (ITC) was performed at 25 °C with a Microcal VP-ITC microcalorimeter (GE Healthcare). Proteins were exhaustively dialyzed against 10 mM Tris-HCl (pH 7.5) and GlcNAc solutions were prepared in the final dialysate. Titrations were performed by stepwise injections of 40 mM GlcNAc into the reaction cell loaded with the protein at concentrations of 0.13–0.28 mM. The heat of GlcNAc dilution was determined separately and subtracted from the total heat produced following each injection. Protein concentrations were determined spectrophotometrically at 280 nm using the theoretical monomeric molar extinction coefficients. The titration data were analysed using ITC–ORIGIN software (GE Healthcare).
MAdV-2 infection competition assays
The wild-type MAdV-2 was a gift from Susan Compton (Yale University School of Medicine) and was propagated in CMT-93 mouse rectal carcinoma cells (ATCC CCL-223), purified by CsCl density gradient centrifugation and quantified as described for MAdV-1 [65]. Serial dilutions of MAdV-2 were used to infect CMT-93 cell monolayers in black-wall, clear-bottom 96-well plates. Two days post-infection, the cells were fixed and stained with an anti-hexon antibody (8C4, Fitzgerald Industries International, Acton, MA, USA) and an Alexa Fluor 488-conjugated secondary antibody (Thermo Fisher Scientific, Waltham, MA, USA). Total monolayer fluorescence was measured using a Typhoon 9400 variable mode imager (GE Healthcare). Images were analysed using ImageJ software [66], and a virus concentration producing 50–80 % maximal signal was chosen for inhibition studies.
For competition studies, increasing concentrations of recombinant proteins diluted in Dulbecco’s modified Eagle’s medium (DMEM) with 10 % foetal bovine serum (FBS) were incubated with CMT-93 cells on ice for 45 min. Virus was added, and the samples were incubated at 37 °C for 2 h. The cells were then washed and cultured with DMEM containing 10 % FBS for 48 h prior to quantification. Alternatively, increasing concentrations of monomeric GlcNAc in water, chitosan in 1 % formic acid, hyaluronan in water, or LPS in water (all from Sigma-Aldrich) were incubated with MAdV-2 in serum-free medium (SFM) for 45 min at 4 °C in the presence or absence of CMT-93 cells. The cells were then washed with cold serum-free DMEM and cultured with DMEM containing 10 % FBS for 48 h prior to quantification. The IC50 values were calculated using Prism 7.0a and are indicated for the trimeric form of the fibre head.
Supplementary Data
Funding information
This research was sponsored by grant BFU2014-53425-P (to M. J. v. R.), coordinated grants CTQ2015-64597-P-C02-01 and CTQ2015-64597-P-C02-02 (to J. J. B. and F. J. C., respectively), grant BFU2015-70052-R (to M. M.) and the Spanish Adenovirus Network (AdenoNet, BIO2015-68990-REDT), all from the Spanish Agencia Estatal de Investigación. Financial support to M. M. from the CIBER of Respiratory Diseases (CIBERES) from the Spanish Institute of Health Carlos III is also acknowledged. These grants are co-financed by the European Regional Development Fund of the European Union. A. K. S. and T. H. N. were recipients of pre-doctoral fellowships from La Caixa and CSIC-VAST, respectively. The expression vectors were designed and created in Hungary, and this was financed by the Hungarian Scientific Research Fund (OTKA K100163). M. K. thanks Enterprise Ireland for a Commercialisation Fund grant (CF/2015/0089), A. K. acknowledges the National University of Ireland for a Cancer Care West Hardiman PhD scholarship and L. J. acknowledges the EU FP7 programme in support of the GlycoHIT consortium (grant no. 260600). This work was supported by R01 AI104920 (to J. G. S.) from the National Institute for Allergy and Infectious Diseases (www.niaid.nih.gov). S. S. W. was also supported by the Helen Riaboff Whiteley Endowment to the University of Washington and by Public Health Service, National Research Service Awards T32 AI083203 from the National Institute for Allergy and Infectious Diseases and T32 GM007270 from the National Institute of General Medical Sciences.
Acknowledgements
Crystallographic data were collected at beamline XALOC-BL13 (ALBA Synchrotron, Barcelona, Spain) with the collaboration of Fernando Gil, Jordi Benach and Jordi Juanhuix and at beamlines ID29 and ID23-2 (ESRF, Grenoble, France) with the help of Marta Sanz-Gaitero, Christoph Mueller-Dieckmann and Bárbara Machado Calisto.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
No experiments with humans or animals were performed.
Footnotes
The atomic models of the derivative structure, the two native structures and the GlcNAc-soaked structure are available in the Protein Data Bank under accession codes 5N83, 5N8D, 5NBH and 5NC1, respectively.
Abbreviations: BSA, bovine serum albumin; CAdV, canine adenovirus; CAR, coxsackievirus and adenovirus receptor; GlcNAc, N-acetylglucosamine; HAdV, human adenovirus; ITC, isothermal titration calorimetry; LPS, lipopolysaccharide; MAdV, murine adenovirus; PDB, Protein Data Bank; STD-NMR, saturation transfer difference nuclear magnetic resonance spectroscopy; TAdV, turkey adenovirus.
Four supplementary tables and seven supplementary figures are available with the online version of this article.
References
- 1.Rowe WP, Huebner RJ, Gilmore LK, Parrott RH, Ward TG. Isolation of a cytopathogenic agent from human adenoids undergoing spontaneous degeneration in tissue culture. Proc Soc Exp Biol Med. 1953;84:570–573. doi: 10.3181/00379727-84-20714. [DOI] [PubMed] [Google Scholar]
- 2.San Martín C. Latest insights on adenovirus structure and assembly. Viruses. 2012;4:847–877. doi: 10.3390/v4050847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu X, Veesler D, Campbell MG, Barry ME, Asturias FJ, et al. Cryo-EM structure of human adenovirus D26 reveals the conservation of structural organization among human adenoviruses. Sci Adv. 2017;3:e1602670. doi: 10.1126/sciadv.1602670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnberg N. Adenovirus receptors: implications for targeting of viral vectors. Trends Pharmacol Sci. 2012;33:442–448. doi: 10.1016/j.tips.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 5.Harrach B, Benkő M, Both GW, Brown M, Davison AJ, et al. Family Adenoviridae. In: King AM, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy: Classification and Nomenclature of Viruses. Ninth Report of the International Committee on Taxonomy of Viruses. San Diego, CA: Elsevier; 2011. pp. 125–141. (editors) [Google Scholar]
- 6.Hashimoto K, Sugiyama T, Sasaki S. An adenovirus isolated from the feces of mice I. Isolation and identification. Jpn J Microbiol. 1966;10:115–125. doi: 10.1111/j.1348-0421.1966.tb00298.x. [DOI] [PubMed] [Google Scholar]
- 7.Takeuchi A, Hashimoto K. Electron microscope study of experimental enteric adenovirus infection in mice. Infect Immun. 1976;13:569–580. doi: 10.1128/iai.13.2.569-580.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sugiyama T, Hashimoto K, Sasaki S. An adenovirus isolated from the feces of mice. II. Experimental infection. Jpn J Microbiol. 1967;11:33–42. doi: 10.1111/j.1348-0421.1967.tb00318.x. [DOI] [PubMed] [Google Scholar]
- 9.Wilson SS, Bromme BA, Holly MK, Wiens ME, Gounder AP, et al. Alpha-defensin-dependent enhancement of enteric viral infection. PLoS Pathog. 2017;13:e1006446. doi: 10.1371/journal.ppat.1006446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hemmi S, Vidovszky MZ, Ruminska J, Ramelli S, Decurtins W, et al. Genomic and phylogenetic analyses of murine adenovirus 2. Virus Res. 2011;160:128–135. doi: 10.1016/j.virusres.2011.05.023. [DOI] [PubMed] [Google Scholar]
- 11.Meissner JD, Hirsch GN, Larue EA, Fulcher RA, Spindler KR. Completion of the DNA sequence of mouse adenovirus type 1: sequence of E2B, L1, and L2 (18-51 map units) Virus Res. 1997;51:53–64. doi: 10.1016/S0168-1702(97)00079-8. [DOI] [PubMed] [Google Scholar]
- 12.Klempa B, Krüger DH, Auste B, Stanko M, Krawczyk A, et al. A novel cardiotropic murine adenovirus representing a distinct species of mastadenoviruses. J Virol. 2009;83:5749–5759. doi: 10.1128/JVI.02281-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bewley MC, Springer K, Zhang YB, Freimuth P, Flanagan JM. Structural analysis of the mechanism of adenovirus binding to its human cellular receptor, CAR. Science. 1999;286:1579–1583. doi: 10.1126/science.286.5444.1579. [DOI] [PubMed] [Google Scholar]
- 14.Burmeister WP, Guilligay D, Cusack S, Wadell G, Arnberg N. Crystal structure of species D adenovirus fiber knobs and their sialic acid binding sites. J Virol. 2004;78:7727–7736. doi: 10.1128/JVI.78.14.7727-7736.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Persson BD, Schmitz NB, Santiago C, Zocher G, Larvie M, et al. Structure of the extracellular portion of CD46 provides insights into its interactions with complement proteins and pathogens. PLoS Pathog. 2010;6:e1001122. doi: 10.1371/journal.ppat.1001122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nilsson EC, Storm RJ, Bauer J, Johansson SM, Lookene A, et al. The GD1a glycan is a cellular receptor for adenoviruses causing epidemic keratoconjunctivitis. Nat Med. 2011;17:105–109. doi: 10.1038/nm.2267. [DOI] [PubMed] [Google Scholar]
- 17.van Raaij MJ, Mitraki A, Lavigne G, Cusack S. A triple beta-spiral in the adenovirus fibre shaft reveals a new structural motif for a fibrous protein. Nature. 1999;401:935–938. doi: 10.1038/44880. [DOI] [PubMed] [Google Scholar]
- 18.Zubieta C, Schoehn G, Chroboczek J, Cusack S. The structure of the human adenovirus 2 penton. Mol Cell. 2005;17:121–135. doi: 10.1016/j.molcel.2004.11.041. [DOI] [PubMed] [Google Scholar]
- 19.Liu H, Wu L, Zhou ZH. Model of the trimeric fiber and its interactions with the pentameric penton base of human adenovirus by cryo-electron microscopy. J Mol Biol. 2011;406:764–774. doi: 10.1016/j.jmb.2010.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xia D, Henry LJ, Gerard RD, Deisenhofer J. Crystal structure of the receptor-binding domain of adenovirus type 5 fiber protein at 1.7 A resolution. Structure. 1994;2:1259–1270. doi: 10.1016/S0969-2126(94)00126-X. [DOI] [PubMed] [Google Scholar]
- 21.Seiradake E, Lortat-Jacob H, Billet O, Kremer EJ, Cusack S. Structural and mutational analysis of human Ad37 and canine adenovirus 2 fiber heads in complex with the D1 domain of coxsackie and adenovirus receptor. J Biol Chem. 2006;281:33704–33716. doi: 10.1074/jbc.M605316200. [DOI] [PubMed] [Google Scholar]
- 22.Chappell JD, Prota AE, Dermody TS, Stehle T. Crystal structure of reovirus attachment protein sigma1 reveals evolutionary relationship to adenovirus fiber. EMBO J. 2002;21:1–11. doi: 10.1093/emboj/21.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guardado Calvo P, Fox GC, Hermo Parrado XL, Llamas-Saiz AL, Costas C, et al. Structure of the carboxy-terminal receptor-binding domain of avian reovirus fibre sigmaC. J Mol Biol. 2005;354:137–149. doi: 10.1016/j.jmb.2005.09.034. [DOI] [PubMed] [Google Scholar]
- 24.Merckel MC, Huiskonen JT, Bamford DH, Goldman A, Tuma R. The structure of the bacteriophage PRD1 spike sheds light on the evolution of viral capsid architecture. Mol Cell. 2005;18:161–170. doi: 10.1016/j.molcel.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 25.Wu E, Pache L, von Seggern DJ, Mullen TM, Mikyas Y, et al. Flexibility of the adenovirus fiber is required for efficient receptor interaction. J Virol. 2003;77:7225–7235. doi: 10.1128/JVI.77.13.7225-7235.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong JS, Engler JA. Domains required for assembly of adenovirus type 2 fiber trimers. J Virol. 1996;70:7071–7078. doi: 10.1128/jvi.70.10.7071-7078.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Persson BD, Reiter DM, Marttila M, Mei YF, Casasnovas JM, et al. Adenovirus type 11 binding alters the conformation of its receptor CD46. Nat Struct Mol Biol. 2007;14:164–166. doi: 10.1038/nsmb1190. [DOI] [PubMed] [Google Scholar]
- 28.Seiradake E, Henaff D, Wodrich H, Billet O, Perreau M, et al. The cell adhesion molecule "CAR" and sialic acid on human erythrocytes influence adenovirus in vivo biodistribution. PLoS Pathog. 2009;5:e1000277. doi: 10.1371/journal.ppat.1000277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cupelli K, Müller S, Persson BD, Jost M, Arnberg N, et al. Structure of adenovirus type 21 knob in complex with CD46 reveals key differences in receptor contacts among species B adenoviruses. J Virol. 2010;84:3189–3200. doi: 10.1128/JVI.01964-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arnberg N, Kidd AH, Edlund K, Nilsson J, Pring-Akerblom P, et al. Adenovirus type 37 binds to cell surface sialic acid through a charge-dependent interaction. Virology. 2002;302:33–43. doi: 10.1006/viro.2002.1503. [DOI] [PubMed] [Google Scholar]
- 31.Lenman A, Liaci AM, Liu Y, Frängsmyr L, Frank M, et al. Polysialic acid is a cellular receptor for human adenovirus 52. Proc Natl Acad Sci USA. 2018;115:E4264. doi: 10.1073/pnas.1716900115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh AK, Berbís MÁ, Ballmann MZ, Kilcoyne M, Menéndez M, et al. Structure and sialyllactose binding of the carboxy-terminal head domain of the fibre from a siadenovirus, Turkey adenovirus 3. PLoS One. 2015;10:e0139339. doi: 10.1371/journal.pone.0139339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parente JP, Wieruszeski JM, Strecker G, Montreuil J, Fournet B, et al. A novel type of carbohydrate structure present in hen ovomucoid. J Biol Chem. 1982;257:13173–13176. [PubMed] [Google Scholar]
- 34.Yamashita K, Kamerling JP, Kobata A. Structural study of the carbohydrate moiety of hen ovomucoid. Occurrence of a series of pentaantennary complex-type asparagine-linked sugar chains. J Biol Chem. 1982;257:12809–12814. [PubMed] [Google Scholar]
- 35.Yamashita K, Kamerling JP, Kobata A. Structural studies of the sugar chains of hen ovomucoid. Evidence indicating that they are formed mainly by the alternate biosynthetic pathway of asparagine-linked sugar chains. J Biol Chem. 1983;258:3099–3106. [PubMed] [Google Scholar]
- 36.Yet MG, Chin CC, Wold F. The covalent structure of individual N-linked glycopeptides from ovomucoid and asialofetuin. J Biol Chem. 1988;263:111–117. [PubMed] [Google Scholar]
- 37.Gerlach JQ, Kilcoyne M, Eaton S, Bhavanandan V, Joshi L. Non-carbohydrate-mediated interaction of lectins with plant proteins. Adv Exp Med Biol. 2011;705:257–269. doi: 10.1007/978-1-4419-7877-6_12. [DOI] [PubMed] [Google Scholar]
- 38.Menéndez-Conejero R, Nguyen TH, Singh AK, Condezo GN, Marschang RE, et al. Structure of a reptilian adenovirus reveals a phage tailspike fold stabilizing a vertebrate virus capsid. Structure. 2017;25:1562–1573. doi: 10.1016/j.str.2017.08.007. [DOI] [PubMed] [Google Scholar]
- 39.Harvey DJ, Wing DR, Küster B, Wilson IB. Composition of N-linked carbohydrates from ovalbumin and co-purified glycoproteins. J Am Soc Mass Spectrom. 2000;11:564–571. doi: 10.1016/S1044-0305(00)00122-7. [DOI] [PubMed] [Google Scholar]
- 40.Chalkley RJ, Burlingame AL. Identification of novel sites of O-N-acetylglucosamine modification of serum response factor using quadrupole time-of-flight mass spectrometry. Mol Cell Proteomics. 2003;2:182–190. doi: 10.1074/mcp.M300027-MCP200. [DOI] [PubMed] [Google Scholar]
- 41.Guardado-Calvo P, Muñoz EM, Llamas-Saiz AL, Fox GC, Kahn R, et al. Crystallographic structure of porcine adenovirus type 4 fiber head and galectin domains. J Virol. 2010;84:10558–10568. doi: 10.1128/JVI.00997-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lenman A, Liaci AM, Liu Y, Årdahl C, Rajan A, et al. Human adenovirus 52 uses sialic acid-containing glycoproteins and the coxsackie and adenovirus receptor for binding to target cells. PLoS Pathog. 2015;11:e1004657. doi: 10.1371/journal.ppat.1004657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lundquist JJ, Toone EJ. The Cluster Glycoside Effect. Chem Rev. 2002;102:555–578. doi: 10.1021/cr000418f. [DOI] [PubMed] [Google Scholar]
- 44.Lortat-Jacob H, Chouin E, Cusack S, van Raaij MJ. Kinetic analysis of adenovirus fiber binding to its receptor reveals an avidity mechanism for trimeric receptor-ligand interactions. J Biol Chem. 2001;276:9009–9015. doi: 10.1074/jbc.M009304200. [DOI] [PubMed] [Google Scholar]
- 45.Jiménez-Moreno E, Jiménez-Osés G, Gómez AM, Santana AG, Corzana F, et al. A thorough experimental study of CH/π interactions in water: quantitative structure-stability relationships for carbohydrate/aromatic complexes. Chem Sci. 2015;6:6076–6085. doi: 10.1039/C5SC02108A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soudais C, Boutin S, Hong SS, Chillon M, Danos O, et al. Canine adenovirus type 2 attachment and internalization: coxsackievirus-adenovirus receptor, alternative receptors, and an RGD-independent pathway. J Virol. 2000;74:10639–10649. doi: 10.1128/JVI.74.22.10639-10649.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johansson ME, Hansson GC. Immunological aspects of intestinal mucus and mucins. Nat Rev Immunol. 2016;16:639–649. doi: 10.1038/nri.2016.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Etzold S, Juge N. Structural insights into bacterial recognition of intestinal mucins. Curr Opin Struct Biol. 2014;28:23–31. doi: 10.1016/j.sbi.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 49.Kilcoyne M, Gerlach JQ, Gough R, Gallagher ME, Kane M, et al. Construction of a natural mucin microarray and interrogation for biologically relevant glyco-epitopes. Anal Chem. 2012;84:3330–3338. doi: 10.1021/ac203404n. [DOI] [PubMed] [Google Scholar]
- 50.Smith AL, Barthold SW. Factors influencing susceptibility of laboratory rodents to infection with mouse adenovirus strains K87 and FL. Arch Virol. 1987;95:143–148. doi: 10.1007/BF01311342. [DOI] [PubMed] [Google Scholar]
- 51.di Paolo NC, Kalyuzhniy O, Shayakhmetov DM. Fiber shaft-chimeric adenovirus vectors lacking the KKTK motif efficiently infect liver cells in vivo. J Virol. 2007;81:12249–12259. doi: 10.1128/JVI.01584-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tuve S, Wang H, Jacobs JD, Yumul RC, Smith DF, et al. Role of cellular heparan sulfate proteoglycans in infection of human adenovirus serotype 3 and 35. PLoS Pathog. 2008;4:e1000189. doi: 10.1371/journal.ppat.1000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Holmén Larsson JM, Thomsson KA, Rodríguez-Piñeiro AM, Karlsson H, Hansson GC. Studies of mucus in mouse stomach, small intestine, and colon. III. Gastrointestinal Muc5ac and Muc2 mucin O-glycan patterns reveal a regiospecific distribution. Am J Physiol Gastrointest Liver Physiol. 2013;305:G357–G363. doi: 10.1152/ajpgi.00048.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh AK, Ballmann MZ, Benkő M, Harrach B, van Raaij MJ. Crystallization of the C-terminal head domain of the fibre protein from a siadenovirus, turkey adenovirus 3. Acta Cryst F. 2013;69:1135–1139. doi: 10.1107/S174430911302397X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Winter G. xia2 : an expert system for macromolecular crystallography data reduction. J Appl Cryst. 2010;43:186–190. doi: 10.1107/S0021889809045701. [DOI] [Google Scholar]
- 56.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Cryst D. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, et al. Overview of the CCP4 suite and current developments. Acta Cryst D. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 59.Murshudov GN, Skubák P, Lebedev AA, Pannu NS, Steiner RA, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Cryst D. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Cryst D. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 62.Holm L, Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang L, Cummings RD, Smith DF, Huflejt M, Campbell CT, et al. Cross-platform comparison of glycan microarray formats. Glycobiology. 2014;24:507–517. doi: 10.1093/glycob/cwu019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Manimala JC, Roach TA, Li Z, Gildersleeve JC. High-throughput carbohydrate microarray profiling of 27 antibodies demonstrates widespread specificity problems. Glycobiology. 2007;17:17C–23. doi: 10.1093/glycob/cwm047. [DOI] [PubMed] [Google Scholar]
- 65.Gounder AP, Myers ND, Treuting PM, Bromme BA, Wilson SS, et al. Defensins potentiate a neutralizing antibody response to enteric viral infection. PLoS Pathog. 2016;12:e1005474. doi: 10.1371/journal.ppat.1005474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.