Abstract
A modified pan-PV consensus-degenerate hybrid oligonucleotide primer (CODEHOP) PCR was developed for generic and sensitive detection of a broad-spectrum of human papillomaviruses (HPVs) infecting the cutaneous epithelium. To test the analytical sensitivity of the assay we examined 149 eyebrow hair follicle specimens from immunocompetent male patients. HPV DNA was detected in 60 % (89/149) of analysed eyebrow samples with a total of 48 different HPV sequences, representing 21 previously described HPVs and 27 putative novel HPV types. Evidence for ten novel HPV subtypes and seven viral variants, clustering to three out of five genera containing cutaneous HPVs, was also obtained. Thus, we have shown that the modified pan-PV CODEHOP PCR assay is able to identify multiple HPV types, even from different genera, in the same clinical sample. Overall, these results demonstrate that the pan-PV CODEHOP PCR is an excellent tool for screening and identification of novel cutaneous HPVs, even in samples with low viral loads.
Keywords: CODEHOP, human papillomavirus, HPV, multiple infections
Papillomaviruses (PVs) are a large and diverse group of small, circular, double-stranded DNA viruses, classified into genera, species and types based on the comparison of L1 gene nucleotide sequences [1]. PVs infecting humans (HPVs) are grouped within five genera (Alpha-PV, Beta-PV, Gamma-PV, Mu-PV and Nu-PV), with mucosal/genital HPV types mostly clustering within the Alpha-PV genus. Cutaneous HPV types, representing approximately 75 % of all officially described HPVs to date, are highly divergent and are distributed into all five HPV genera [2].
HPVs are aetiologically linked with the development of various benign and malignant lesions of the skin and mucosa [3]. The mucosal high-risk HPVs (HPV16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58 and 59) are associated with the development of more than 99 % of cervical cancers, 70–90 % of anal and vaginal cancers, 47 % of penile cancers, 40 % of vulvar cancers and 25–30 % of oropharyngeal cancers. Mucosal low-risk HPV types, most commonly HPV6 and 11, are associated with more than 90 % of anogenital warts and laryngeal papillomas [4]. However, the role of cutaneous HPV types in skin cancer is still unclear. Cutaneous HPVs have not only been found in benign skin warts and non-melanoma skin cancer [5–7], but also on healthy skin worldwide [8–11]. Continued efforts to improve identification of novel cutaneous HPVs could lead to a better understanding of their phylogenetic diversity and clarification of their role in the development of skin cancer. Due to the extraordinary diversity of HPVs infecting the skin [12], the development of primer sets enabling amplification of a diverse range of HPV types is critical for this effort.
Over the past years, the sensitivity of molecular methods for detection of HPV infections has increased significantly. However, the majority of assays only detect infection with HPV types or species within a single genus. Only a few methods have demonstrated the ability to detect a broad-range of HPV types from different species and genera [13–15]. Traditionally, simple PCR assays using single or multiple pairs of degenerate primers or nested PCR assays using two pairs of degenerate primers were employed for identification and characterization of novel PVs [16]. Previously, we devised the consensus-degenerate hybrid oligonucleotide primer (CODEHOP) for broad-range detection of all members of a gene family [17], and have utilized this approach for novel virus identification [18]. We developed a pan-PV CODEHOP PCR-based assay to detect multiple HPV types distributed over all five PV genera in different types of warts as well as precancerous and cancerous lesions of the cervix [19, and personal communication]. However, the detection of HPVs was more challenging in eyebrow hair follicles because of frequent co-infections with multiple HPV types as well as low viral copy numbers. In this study, an improved version of the original pan-PV CODEHOP primers was used in a combination with a single-tube ‘hanging droplet’ PCR [20] to survey cutaneous HPV types in eyebrow samples.
The ‘hanging droplet’ pan-PV CODEHOP PCR was designed as a ‘re-amplification PCR’ in which the same primers were used in both rounds of amplifications, as reported previously [15, 21]. All reactions were set up on a Mastercycler proS thermal cycler (Eppendorf, Germany) and performed with a HotStarTaq Plus DNA Polymerase kit (Qiagen, Germany). The primary 20 µl reaction mixture was placed in a reaction tube and covered with one drop of mineral oil (Sigma-Aldrich, USA). After addition of 5 µl of the sample, the mixture had a final volume of 25 µl, containing 0.2 µM of each forward CODEHOP primer, DGDM_F1 (5′-GAG CTT ATA AAC ACA GAT ATT GAA GAT GGI GAY ATG-3′) (I, inosine) and DGDM_F2 (5′-GAG CTT ATA AAC ACA GTT ATT GAG GAY GGN GAY ATG-3′), 0.4 µM of the reverse CODEHOP primer PAP14_R (5′-GTA ACA AAC ACC TGG TTA CCC CAA CAG ATI CCA TTR TT-3′), 400 µM of each dNTP (PCR Nucleotide Mix; Roche Diagnostics, Germany), 1X CoralLoad PCR Buffer, 2 mM of MgCl2 and 0.625 U of HotStarTaq Plus DNA polymerase. In addition, 25 µl of the above-mentioned reaction mixture, including fresh DNA polymerase (1 U) and all three primers at the same concentrations, were loaded onto the inside of each reaction tube cap forming a ‘hanging droplet’ when the reaction cap was closed. Using the thermocycler without the heated lid, a first round of amplification was performed, in which the reaction mixture was first heated for 5 min at 95 °C, followed by 25 cycles of 1 min at 94 °C, 1 min at 50 °C and 1 min at 72 °C, followed by a final elongation step of 10 min at 72 °C. For the second round of amplification, the ‘hanging droplet’ was incorporated into the reaction mixture (final volume of 50 µl) using a quick centrifugation step (1 min at 2000 g), and 45 amplification cycles were performed using the same cycling conditions as for the first round of PCR.
The sensitivity and specificity of the optimized assay were evaluated on cloned DNA of several HPV types from Alpha-PV (HPV6, 11, 16, 18, 31, 33, 35, 39, 45, 52, 56, 58 and 59), Beta-PV (HPV150) and Gamma-PV (HPV4, 156 and 179) genera, at input levels of 5, 50, 500 and 1000 viral copies per reaction in a background of 50 ng of human placental DNA (Sigma) or human genomic DNA (Thermo Fisher Scientific). Non-specific amplification was tested using five replicates of 20, 50, 100 and 200 ng of human genomic DNA (Thermo Fisher Scientific) per reaction. The analytical sensitivity of the assay was determined to be five copies per reaction for HPV16 and 18, 50 copies per reaction for HPV6, 11, 33, 35, 45, 52, 56, 58, 66 and 179, and 500 copies per reaction for HPV4, 31, 39, 59, 150 and 156. These results indicated that the pan-PV CODEHOP PCR-based assay was a sensitive tool for broad-based HPV-screening in clinical samples with low viral copy numbers.
In order to analyse the ability of the pan-PV CODEHOP PCR to identify multiple HPV infections, a total of 149 pooled eyebrow hair follicle specimens, containing from 5 to 8 hairs each, were obtained from 149 immunocompetent male patients with clinically evident anogenital warts, as described previously [22]. The study was approved by the Institutional Review Board of the Ministry of Health of Republic of Slovenia (consent reference 97/11/09). All patients additionally provided a written informed consent and were sampled in compliance with the Helsinki Declaration. Total DNA was extracted from all specimens as described previously [22] and the quality of all DNA samples was verified by real-time PCR amplification of a 150 bp fragment of the human beta-globin gene [23].
The PCR products obtained using the pan-PV CODEHOP PCR-based assay (360–380 bp) were visualized by 2 % agarose gel electrophoresis, purified with a QIAquick PCR purification kit or in the case of multiple bands using the QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany). Both strands of each amplicon were sequenced using the same primers as for PCR at Microsynth AG (Balgach, Switzerland).
All HPV-specific amplicons were additionally cloned into pJET1.2/blunt cloning vectors using the CloneJET PCR Cloning Kit (Thermo Fisher Scientific). HPV plasmids were then transformed into One Shot TOP10 Chemically Competent E. coli cells (Invitrogen, USA), as per the manufacturer’s instructions. Clones containing targeted inserts were identified using colony PCR, performed with the FastStart PCR Master (Roche Diagnostics) and primers specific for the plasmid (pJET1.2 Forward Sequencing and pJET1.2 Reverse Sequencing Primers; Thermo Fisher Scientific). Positive clones were sequenced on both strands using pJET1.2 sequencing primers at Microsynth AG (Balgach, Switzerland). All obtained nucleotide sequences were compared to HPV sequences available in the GenBank database, using the blast server (www.ncbi.nlm.nih.gov/blast/). Novel putative HPV types, subtypes and viral variants were identified when their L1 nucleotide sequences showed less than 90 %, between 90 and 98 %, or between 98 and 99 % sequence identities with all previously known HPV types, respectively [1]. Results of the blast comparison were additionally verified by aligning the sequences of interest with L1 nucleotide sequences of the most closely related HPVs using the mega software, version 7 [24]. Nucleotide sequences indicating the presence of novel HPV types/subtypes/variants were subsequently assigned their in-house SIBX identifying numbers and submitted to the GenBank database.
Full-length L1 ORFs from 180 reference HPV types (www.hpvcenter.se) and 19 completely sequenced but officially unrecognized HPV genomes (obtained mostly by metagenomics) were included in the phylogenetic analysis of putatively novel HPV types/subtypes/variants obtained in the present study. Multiple alignments and pairwise nucleotide alignments were performed with mega v7 [24]. Phylogenetic trees were inferred by the Bayesian method using beast v1.8.4 [25] and Markov chain Monte Carlo (MCMC) simulations were performed during 2×107 generations, sampling one state every 1000 generations, with a burn-in of 10 %. Statistical convergence of MCMC was assessed by calculating the effective sample size using tracer v1.6 (http://beast.community/tracer). The maximum clade credibility tree across all of the plausible trees generated by beast was then computed using the TreeAnnotator program available in the beast package.
Using the modified pan-PV CODEHOP PCR-based assay, the overall HPV prevalence in eyebrow hair follicle specimens was estimated at 60 % (89/149), which is similar to other studies using different generic or genus-specific primer pairs [22, 26]. On the other hand, in other studies, in which a combination of PCR amplification of short amplicons followed by reverse hybridization or microarrays was used for the identification of HPV types, the HPV prevalence was estimated at >80 % [27–29]. These methods generally improve the rate of HPV detection in comparison to amplicon detection using agarose gel electrophoresis followed by HPV typing by sequencing, but reverse hybridization techniques and microarrays only allow the identification of a limited number of HPV types. In our study, a total of 48 different HPV isolates were identified, representing 21 previously described HPV types and 27 putative novel HPV types (SIBX isolates), clustering within three of the five genera containing cutaneous HPVs (Table 1). Moreover, the pan-PV CODEHOP PCR detected sequences representing ten novel putative HPV subtypes and seven novel viral variants.
Table 1. HPV sequences identified in eyebrow hair follicles using the pan-PV CODEHOP PCR.
| HPV/SIBX sequence | HPV species | Novel sequence | Closely related type (%) | No. of positive samples | ||
|---|---|---|---|---|---|---|
| (Accesion no.) | Type | Subtype | Variant | (Prevalence, %) | ||
| HPV6 (X00203) | Alpha-10 | x | 1 (0.7 %) | |||
| HPV11 (M14119) | Alpha-10 | x | 1 (0.7 %) | |||
| HPV5 (M17463) | Beta-1 | x | 1 (0.7 %) | |||
| HPV12 (X74466) | Beta-1 | x | 17 (11.4 %) | |||
| HPV21 (U31779) | Beta-1 | x | 2 (1.3 %) | |||
| HPV24 (U31782) | Beta-1 | x | 1 (0.7 %) | |||
| HPV118 (GQ246951) | Beta-1 | x | 1 (0.7 %) | |||
| HPV124 (GQ845446) | Beta-1 | x | 1 (0.7 %) | |||
| HPVRTRX7 (U85660) | Beta-1 | x | 1 (0.7 %) | |||
| SIBX22 (LK022298) | Beta-1 | x | HPV118 (88.0 %) | 3 (2.0 %) | ||
| SIBX25 (LK022301) | Beta-1 | x | HPV124 (91.3 %) | 3 (2.0 %) | ||
| SIBX62 (KP768220) | Beta-1 | x | HPV12 (93.2 %) | 1 (0.7 %) | ||
| SIBX65 (KP768223) | Beta-1 | x | HPV124 (88.9 %) | 1 (0.7 %) | ||
| HPV9 (X74464) | Beta-2 | x | 8 (5.4 %) | |||
| HPV15 (X74468) | Beta-2 | x | 9 (6.0 %) | |||
| HPV37 (U31786) | Beta-2 | x | 2 (1.3 %) | |||
| HPV38 (U31787) | Beta-2 | x | 1 (0.7 %) | |||
| HPV174 (HF930491) | Beta-2 | x | 4 (2.7 %) | |||
| HPV122 (GQ845444) | Beta-2 | x | 4 (2.7 %) | |||
| SIBX26 (LK022302) | Beta-2 | x | HPV113 (84.1 %) | 3 (2.0 %) | ||
| SIBX29 (LK022305) | Beta-2 | x | HPV209 (97.4 %) | 3 (2.0 %) | ||
| SIBX61 (KP768219) | Beta-2 | x | HPV174 (86.1 %) | 1 (0.7 %) | ||
| SIBX63 (KP768221) | Beta-2 | x | HPV174 (94.1 %) | 1 (0.7 %) | ||
| HPV75 (Y15173) | Beta-3 | x | 4 (2.7 %) | |||
| HPV115 (FJ947080) | Beta-3 | x | 1 (0.7 %) | |||
| SIBX59 (KP768217) | Beta-3 | x | HPV76 (96.6 %) | 2 (1.3 %) | ||
| SIBX64 (KP768222) | Beta-3 | x | SIBX59 (97.6 %) | 1 (0.7 %) | ||
| HPV150 (FN677755) | Beta-5 | x | 3 (2.0 %) | |||
| SIBX54 (LM653102) | Beta-5 | x | HPV96 (83.0 %) | 7 (4.7 %) | ||
| SIBX42 (LK022318) | Gamma-1 | x | HPV158 (83.6 %) | 1 (0.7 %) | ||
| SIBX44 (LK022320) | Gamma-1 | x | SIBX42 (99.7 %) | 1 (0.7 %) | ||
| SIBX45 (LK022321) | Gamma-1 | x | HPV158 (83.3 %) | 10 (6.7 %) | ||
| SIBX33 (LK022309) | Gamma-8 | x | mSE37 (75.2 %) | 1 (0.7 %) | ||
| SIBX37 (LK022313) | Gamma-8 | x | SIBX33 (97.9 %) | 1 (0.7 %) | ||
| SIBX35 (LK022311) | Gamma-9 | x | HPV129 (73.4 %) | 1 (0.7 %) | ||
| HPV142 (HM999994) | Gamma-10 | x | 1 (0.7 %) | |||
| SIBX39 (LK022315) | Gamma-10 | x | HPV142 (85.4 %) | 11 (7.4 %) | ||
| SIBX57 (KP768215) | Gamma-11 | x | HPV126 (89.5 %) | 1 (0.7 %) | ||
| SIBX60 (KP768218) | Gamma-11 | x | SIBX57 (98.4 %) | 2 (1.3 %) | ||
| HPV165 (JX444072) | Gamma-12 | x | 1 (0.7 %) | |||
| SIBX23 (LK022299) | Gamma-12 | x | mCG3 (78.6 %) | 2 (1.3 %) | ||
| SIBX27 (LK022303) | Gamma-12 | x | mCG3 (86.5 %) | 5 (3.4 %) | ||
| SIBX34 (LK022310) | Gamma-12 | x | HPV165 (80.2 %) | 1 (0.7 %) | ||
| SIBX40 (LK022316) | Gamma-12 | x | HPV157 (77.9 %) | 2 (1.3 %) | ||
| SIBX41 (LK022317) | Gamma-12 | x | SIBX40 (99.3 %) | 3 (2.0 %) | ||
| SIBX50 (LK022326) | Gamma-12 | x | HPV157 (99.7 %) | 1 (0.7 %) | ||
| SIBX43 (LK022319) | Gamma-15 | x | HPV179 (78.3 %) | 2 (1.3 %) | ||
| SIBX38 (LK022314) | Gamma-17 | x | HPV144 (75.8 %) | 1 (0.7 %) | ||
| SIBX24 (LK022300) | Gamma-18 | x | HPV156 (87.3 %) | 1 (0.7 %) | ||
| HPV161 (JX413109) | Gamma-19 | x | 1 (0.7 %) | |||
| SIBX47 (LK022323) | Gamma-19 | x | HPV166 (92.3 %) | 3 (2.0 %) | ||
| SIBX49 (LK022325) | Gamma-19 | x | HPV166 (86.1 %) | 1 (0.7 %) | ||
| SIBX51 (LM653099) | Gamma-19 | x | mZJ01 (74.9 %) | 2 (1.3 %) | ||
| SIBX55 (LM653103) | Gamma-19 | x | SIBX51 (99.7 %) | 7 (4.7 %) | ||
| SIBX58 (KP768216) | Gamma-19 | x | HPV166 (84.8 %) | 1 (0.7 %) | ||
| SIBX56 (LM653104) | Gamma-20 | x | HPV163 (80.8 %) | 5 (3.4 %) | ||
| SIBX28 (LK022304) | Gamma-24 | x | mFD1 (83.0 %) | 14 (9.4 %) | ||
| SIBX30 (LK022306) | Gamma-24 | x | mSE355 (79.3 %) | 4 (2.7 %) | ||
| SIBX31 (LK022307) | Gamma-24 | x | mFD1 (83.9 %) | 16 (10.7 %) | ||
| SIBX32 (LK022308) | Gamma-24 | x | SIBX31 (99.7 %) | 1 (0.7 %) | ||
| SIBX36 (LK022312) | Gamma-24 | x | mFD1 (86.3 %) | 1 (0.7 %) | ||
| SIBX46 (LK022322) | Gamma-24 | x | SIBX31 (97.9 %) | 8 (5.4 %) | ||
| SIBX48 (LK022324) | Gamma-24 | x | mKN1 (99.1 %) | 8 (5.4 %) | ||
| SIBX52 (LM653100) | Gamma-24 | x | SIBX30 (91.0 %) | 1 (0.7 %) | ||
| SIBX53 (LM653101) | Gamma-24 | x | HPV197 (80.2 %) | 6 (4.0 %) | ||
| TOTAL | 48 | 10 | 7 | |||
SIBX, novel putative HPV types/subtypes/variants identified in this study.
The phylogenetic relationships of all identified SIBX sequences and their type/subtype/variant-specific prevalences are provided in Table 1. The most frequently detected HPV sequences, which were identified in eight or more clinical samples, included HPV9 (Beta-2), HPV12 (Beta-1), HPV15 (Beta-2), SIBX28 (Gamma-24), SIBX31 (Gamma-24), SIBX39 (Gamma-10), SIBX45 (Gamma-1), SIBX46 (Gamma-24) and SIBX48 (Gamma-24). Even though previous studies similarly observed that Beta- and Gamma-PVs are highly prevalent in eyebrow samples [22, 26–29], a direct comparison of type-specific prevalence across studies is difficult due to methodological differences [15, 22, 26–32].
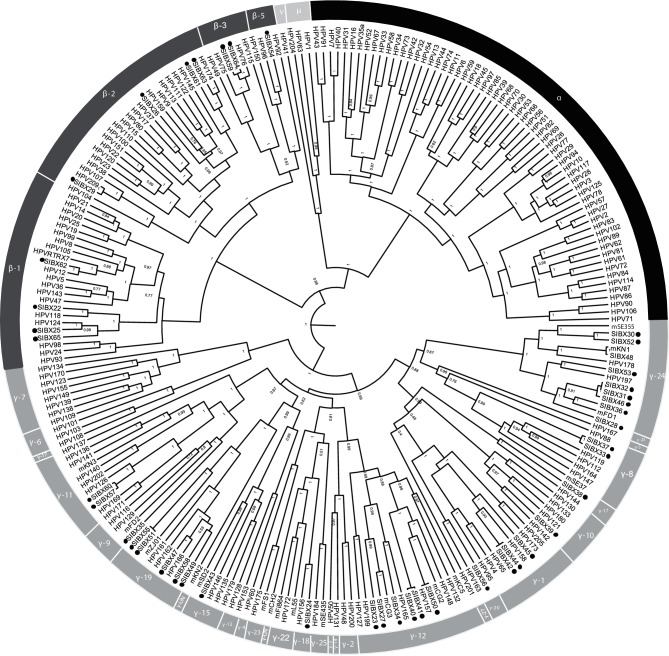
Phylogenetic clustering of the pan-PV CODEHOP PCR amplicons matched the clustering described previously by other authors [33]. The phylogenetic analysis and pairwise comparisons revealed that all SIBX sequences were clustered within defined species of Beta-PV and Gamma-PV genera. Eleven SIBX sequences grouped within the Beta-PV genus. Five represented novel putative HPV types and six represented novel putative HPV subtypes (Fig. 1, Table 1). The majority of SIBX sequences (33/44; 75 %) clustered within the Gamma-PV genus. Twenty-two represented novel putative HPV types, four represented novel putative HPV subtypes and seven represented novel putative HPV variants.
Fig. 1.
Phylogenetic comparison of putatively novel HPV sequences detected in this study compared to classified reference HPV sequences. Dots highlight the SIBX isolates. Node values represent Bayesian posterior probability values.
Since each CODEHOP includes a pool of extra-long primers with two distinct domains, degenerate core and consensus clamp, which perform differently during early and late PCR cycles due to dynamically changing and complex binding interactions, the differences in the sensitivity of the assay for different HPV types are hard to model using straightforward thermodynamic methods [17–19]. As shown in Table 2, the mentioned differences cannot be explained solely based on the number of nucleotide mismatches (not in the complete primer sequence nor the core part of the sequence) and/or the primer’s melting temperatures. The observed twofold differences in the analytical sensitivity of the assay are, therefore, most probably a consequence of reaction conditions, suggesting that even though the primer design might theoretically be perfect, their performance in the assay must be determined empirically.
Table 2. Alignment of pan-PV CODEHOP primers with HPV types used for the analysis of the assay’s analytical sensitivity and those identified in eyebrow hair follicle specimens, respectively, and type-specific melting temperatures of individual primers.
HPV types used for the analysis of the assay’s analytical sensitivity are shown in bold. The degenerated cores of pan-PV CODEHOP primers are highlighted in grey.
| No. of mismatches | ||||||
|---|---|---|---|---|---|---|
| HPV type | Species | Complete sequence (36 nt) |
Core sequence (12 nt) | Sensitivity (copies/reaction) | DGDM-F1 primer GAGCTTATAAACACAGATATTGAAGATGGIGAYATG | Tm (°C) |
| HPV18 | Alpha-7 | 5 | 0 | 5 | - - a - - - - a - - - - - - - - t - t - g - - - - - - - - - - - - - - - | 52.1 |
| HPV16 | Alpha-9 | 5 | 0 | 5 | - - - t - a - - - - - - - - - - t - - - - c - g - - - - - - - - - - - - | 51.2 |
| HPV56 | Alpha-6 | 10 | 0 | 50 | - c a t - a - - t - - t - - - c c - - - a - - g - - - - - - - - - - - - | 37.5 |
| HPV66 | Alpha-6 | 14 | 1 | 50 | - c a t - a g - t - - t - - c c c g - - a - - g - - c - - - - - - - - - | 10.3 |
| HPV45 | Alpha-7 | 6 | 0 | 50 | - - a - - - - a - - - - - - c a t - - - - - - g - - - - - - - - - - - - | 49.0 |
| HPV33 | Alpha-9 | 6 | 0 | 50 | - - a - - - - - - - - t - - t a t - - - - - - g - - - - - - - - - - - - | 46.5 |
| HPV35a | Alpha-9 | 10 | 1 | 50 | - - - t - a c - - - - - - - t - t a c - a c - - - - c - - - - - - - - - | 25.4 |
| HPV52 | Alpha-9 | 10 | 0 | 50 | c - - - - c - - t - - - - g t - t a - - a c - g - - - - - - - - - - - - | 33.5 |
| HPV58 | Alpha-9 | 9 | 0 | 50 | - - a - - - t - t - - t t - t a t - - - - - - g - - - - - - - - - - - - | 46.5 |
| HPV6* | Alpha-10 | 9 | 0 | 50 | - - a - - - - - t - c - - g t - t - - - a c - g - - - - - - - - - - - - | 33.5 |
| HPV11* | Alpha-10 | 9 | 0 | 50 | - - a - - - - - t - c - - g t - t - - - a c - g - - - - - - - - - - - - | 33.5 |
| HPV179 | Gamma-15 | 13 | 0 | 50 | c - a t - a g - - - - t t - t c c a - - - c - g - - - - - - - - - - - - | 33.5 |
| HPV4 | Gamma-1 | 6 | 0 | 500 | - - - - - - g - - - - t t - - t - c - - - c - - - - - - - - - - - - - - | 40.8 |
| HPV31 | Alpha-9 | 9 | 0 | 500 | - - a t - a - a - - - t t - - - t - - - a c - - - - - - - - - - - - - - | 40.8 |
| HPV39 | Alpha-7 | 7 | 0 | 500 | - - a - - a g - - - - - - - c c c - - - - - - g - - - - - - - - - - - - | 48.1 |
| HPV59 | Alpha-7 | 7 | 0 | 500 | - - a t - a - - - - - t - - - c c a - - - - - - - - - - - - - - - - - - | 54.0 |
| HPV150* | Alpha-5 | 9 | 0 | 500 | - - a t - a - a g - - t - - - a t c - - - - - - - - - - - - - - - - - - | 53.8 |
| HPV156 | Gamma-18 | 10 | 0 | 500 | - - a t - a t - t c - - - - t t - c - - - c - - - - - - - - - - - - - - | 40.8 |
| HPV5 | Beta-1 | 5 | 0 | - - a - - a - a - - - - - - - t - - - - a - - - - - - - - - - - - - - - | ||
| HPV12 | Beta-1 | 8 | 0 | - - - t - a - a g - - - - - t t t c - - - - - - - - - - - - - - - - - - | ||
| HPV21 | Beta-1 | 10 | 0 | - - a t - a - - t - - t t - - - c a - - a c - - - - - - - - - - - - - - | ||
| HPV24 | Beta-1 | 9 | 0 | a - - t - a g - - - - t t - - - t a - - - c - - - - - - - - - - - - - - | ||
| HPV118 | Beta-1 | 12 | 1 | - - a t - g g - g - - t - - t t t - - - - c - g - - - - a - - - - - - - | ||
| HPV124 | Beta-1 | 8 | 0 | - - - - - - g - - - - t t - - t - c - - a c - g - - - - - - - - - - - - | ||
| HPVRTRX7 | Beta-1 | 7 | 0 | - - a t - a - a - - - - - - t t t - - - - - - - - - - - - - - - - - - - | ||
| HPV9 | Beta-2 | 7 | 0 | - - a t - a - g - - - - - - - - t a - - - - - g - - - - - - - - - - - - | ||
| HPV15 | Beta-2 | 4 | 0 | - - a - - - - a - - - t - - - - t - - - - - - - - - - - - - - - - - - - | ||
| HPV37 | Beta-2 | 6 | 0 | - - a t - a - a - - - - - - - - t g - - - - - - - - - - - - - - - - - - | ||
| HPV38 | Beta-2 | 10 | 1 | - - a t - g - a - - - t - g t - t a - - - - - - - - c - - - - - - - - - | ||
| HPV122 | Beta-2 | 7 | 0 | - - - t - a - a - - - t t - - - t a - - - - - - - - - - - - - - - - - - | ||
| HPV174 | Beta-2 | 7 | 0 | - - a t - a - a - - - - - - t - t - - - - - - g - - - - - - - - - - - - | ||
| HPV75 | Beta-3 | 7 | 0 | - - a t - a g - - - - t - - - - t - - - - - - g - - - - - - - - - - - - | ||
| HPV115 | Beta-3 | 9 | 0 | - - a t - a - g - - - t - - t a t - - - - - - g - - - - - - - - - - - - | ||
| HPV142 | Gamma-10 | 10 | 0 | - - a - - g - a - c - t - g t - t c - - a - - - - - - - - - - - - - - - | ||
| HPV161 | Gamma-19 | 12 | 0 | - - a a g a - a - - c a - - t - t a - - - c - g - - - - - - - - - - - - | ||
| HPV165 | Gamma-12 | 12 | 0 | c - - t - a g - t g - t - a t - t - - - - c - g - - - - - - - - - - - - | ||
| No. of mismatches | ||||||
|---|---|---|---|---|---|---|
| HPV type | Species | Complete sequence (36 nt) | Core sequence (12 nt) | Sensitivity (copies/reaction) | DGDM-F2 primer GAGCTTATAAACACAGTTATTGAGGAYGGNGAYATG | Tm (°C) |
| HPV18 | Alpha-7 | 5 | 0 | 5 | - - a - - - - a - - - - - - - - - - t - g - - a - - - - - - - - - - - - | 47.6 |
| HPV16 | Alpha-9 | 3 | 0 | 5 | - - - t - a - - - - - - - - - - - - - - - c - - - - - - - - - - - - - - | 61.8 |
| HPV56 | Alpha-6 | 9 | 0 | 50 | - c a t - a - - t - - t - - - c c - - - a - - - - - - - - - - - - - - - | 43.6 |
| HPV66 | Alpha-6 | 12 | 0 | 50 | - c a t - a g - t - - t - - c c c g - - a - - - - - - - - - - - - - - - | 43.6 |
| HPV45 | Alpha-7 | 4 | 0 | 50 | - - a - - - - a - - - - - - c a - - - - - - - - - - - - - - - - - - - - | 55.9 |
| HPV33 | Alpha-9 | 4 | 0 | 50 | - - a - - - - - - - - t - - t a - - - - - - - - - - - - - - - - - - - - | 54.4 |
| HPV35a | Alpha-9 | 9 | 0 | 50 | - - - t - a c - - - - - - - t - - a c - a c - a - - - - - - - - - - - - | 31.8 |
| HPV52 | Alpha-9 | 8 | 0 | 50 | c - - - - c - - t - - - - g t - - a - - a c - - - - - - - - - - - - - - | 34.0 |
| HPV58 | Alpha-9 | 7 | 0 | 50 | - - a - - - t - t - - t t - t a - - - - - - - - - - - - - - - - - - - - | 52.6 |
| HPV6* | Alpha-10 | 7 | 0 | 50 | - - a - - - - - t - c - - g t - - - - - a c - - - - - - - - - - - - - - | 34.0 |
| HPV11* | Alpha-10 | 7 | 0 | 50 | - - a - - - - - t - c - - g t - - - - - a c - - - - - - - - - - - - - - | 34.0 |
| HPV179 | Gamma-15 | 12 | 0 | 50 | c - a t - a g - - - - t t - t c c a - - - c - - - - - - - - - - - - - - | 34.0 |
| HPV4 | Gamma-1 | 8 | 0 | 500 | - - - - - - g - - - - t t - - t a c - - - c - a - - - - - - - - - - - - | 12.2 |
| HPV31 | Alpha-9 | 9 | 0 | 500 | - - a t - a - a - - - t t - - - - - - - a c - a - - - - - - - - - - - - | 23.6 |
| HPV39 | Alpha-7 | 6 | 0 | 500 | - - a - - a g - - - - - - - c c c - - - - - - - - - - - - - - - - - - - | 51.7 |
| HPV59 | Alpha-7 | 8 | 0 | 500 | - - a t - a - - - - - t - - - c c a - - - - - a - - - - - - - - - - - - | 33.5 |
| HPV150* | Alpha-5 | 9 | 0 | 500 | - - a t - a - a g - - t - - - a - c - - - - - a - - - - - - - - - - - - | 33.1 |
| HPV156 | Gamma-18 | 12 | 0 | 500 | - - a t - a t - t c - - - - t t a c - - - c - a - - - - - - - - - - - - | 18.0 |
| HPV5 | Beta-1 | 7 | 0 | - - a - - a - a - - - - - - - t a - - - a - - a - - - - - - - - - - - - | ||
| HPV12 | Beta-1 | 8 | 0 | - - - t - a - a g - - - - - t t - c - - - - - a - - - - - - - - - - - - | ||
| HPV21 | Beta-1 | 11 | 0 | - - a t - a - - t - - t t - - - c a - - a c - a - - - - - - - - - - - - | ||
| HPV24 | Beta-1 | 9 | 0 | a - - t - a g - - - - t t - - - - a - - - c - a - - - - - - - - - - - - | ||
| HPV118 | Beta-1 | 10 | 1 | - - a t - g g - g - - t - - t t - - - - - c - - - - - - a - - - - - - - | ||
| HPV124 | Beta-1 | 8 | 0 | - - - - - - g - - - - t t - - t a c - - a c - - - - - - - - - - - - - - | ||
| HPVRTRX7 | Beta-1 | 7 | 0 | - - a t - a - a - - - - - - t t - - - - - - - a - - - - - - - - - - - - | ||
| HPV9 | Beta-2 | 5 | 0 | - - a t - a - g - - - - - - - - - a - - - - - - - - - - - - - - - - - - | ||
| HPV15 | Beta-2 | 4 | 0 | - - a - - - - a - - - t - - - - - - - - - - - a - - - - - - - - - - - - | ||
| HPV37 | Beta-2 | 6 | 0 | - - a t - a - a - - - - - - - - - g - - - - - a - - - - - - - - - - - - | ||
| HPV38 | Beta-2 | 9 | 0 | - - a t - g - a - - - t - g t - - a - - - - - a - - - - - - - - - - - - | ||
| HPV122 | Beta-2 | 7 | 0 | - - - t - a - a - - - t t - - - - a - - - - - a - - - - - - - - - - - - | ||
| HPV174 | Beta-2 | 5 | 0 | - - a t - a - a - - - - - - t - - - - - - - - - - - - - - - - - - - - - | ||
| HPV75 | Beta-3 | 5 | 0 | - - a t - a g - - - - t - - - - - - - - - - - - - - - - - - - - - - - - | ||
| HPV115 | Beta-3 | 7 | 0 | - - a t - a - g - - - t - - t a - - - - - - - - - - - - - - - - - - - - | ||
| HPV142 | Gamma-10 | 10 | 0 | - - a - - g - a - c - t - g t - - c - - a - - a - - - - - - - - - - - - | ||
| HPV161 | Gamma-19 | 10 | 0 | - - a a g a - a - - c a - - t - - a - - - c - - - - - - - - - - - - - - | ||
| HPV165 | Gamma-12 | 10 | 0 | c - - t - a g - t g - t - a t - - - - - - c - - - - - - - - - - - - - - | ||
| No. of mismatches | ||||||
|---|---|---|---|---|---|---|
| HPV type | Species | Complete sequence (38 nt) | Core sequence (12 nt) | Sensitivity (copies/reaction) | PAP14-R primer AAYAATGGIATCTGTTGGGGTAACCAGGTGTTTGTTAC | Tm (°C) |
| HPV18 | Alpha-7 | 9 | 2 | 5 | - - - - - - - - - g - t - - c - - - c a - - - t - - a t - a - - - - - - - - | 61.2 |
| HPV16 | Alpha-9 | 4 | 1 | 5 | - - - - - - - - - - - t - - - - - - - - - - - - - - a c - a - - - - - - - - | 59.0 |
| HPV56 | Alpha-6 | 6 | 1 | 50 | - - - - - - - - - - - t - - c - - - - - - - - t - - a t - a - - - - - - - - | 65.3 |
| HPV66 | Alpha-6 | 4 | 1 | 50 | - - - - - - - - - - - a - - c - - - - - - - - t - - - - - a - - - - - - - - | 47.6 |
| HPV45 | Alpha-7 | 5 | 1 | 50 | - - - - - - - - - - - t - - - - - - c a - - - t - - - t - - - - - - - - - - | 60.3 |
| HPV33 | Alpha-9 | 4 | 1 | 50 | - - - - - - - - - - - t - - - - - - - - c - - t - - - - - a - - - - - - - - | 55.1 |
| HPV35a | Alpha-9 | 4 | 1 | 50 | - - - - - - - - - - - t - - - - - - a - - - - - - - a t - - - - - - - - - - | 64.4 |
| HPV52 | Alpha-9 | 5 | 1 | 50 | - - - - - - - - - - - a - - - - - - - - c - - t - - - t - - - - - - - c - - | 55.8 |
| HPV58 | Alpha-9 | 6 | 1 | 50 | - - - - - - - - - - - t - - c - - - - - c - - t - - - t - a - - - - - - - - | 51.2 |
| HPV6* | Alpha-10 | 4 | 1 | 50 | - - - - - - - - - - - t - - - - - - - - - - - t - - a c - - - - - - - - - - | 77.7 |
| HPV11* | Alpha-10 | 5 | 1 | 50 | - - - - - - - - - - - t - - c - - - - - a - - - - - c t - - - - - - - - - - | 62.9 |
| HPV179 | Gamma-15 | 11 | 1 | 50 | - - - - - - - - - - - t - - c - - - t c c - - t g - c t - a - - - a - - - - | 55.7 |
| HPV4 | Gamma-1 | 4 | 1 | 500 | - - - - - - - - - - - t - - - - - - - a - - - t - - - t - - - - - - - - - - | 60.2 |
| HPV31 | Alpha-9 | 5 | 1 | 500 | - - - - - - - - - - - t - - - - - - - - c - - t - - - t - a - - - - - - - - | 47.6 |
| HPV39 | Alpha-7 | 8 | 1 | 500 | - - - - - - - - - - - a - - - - - - c a - - - t - - a t - a - - - c - - - - | 40.8 |
| HPV59 | Alpha-7 | 9 | 1 | 500 | - - - - - - - - - - - a - - - - - - c a c - - t - - a t - - - - - t - a - - | 48.9 |
| HPV150* | Alpha-5 | 7 | 1 | 500 | - - - - - - - - - - - t - - - - - - - - - - - t - - a a - c - - - t - a - - | 64.8 |
| HPV156 | Gamma-18 | 9 | 1 | 500 | - - - - - - - - - - - a c t - - - - - - - - - - - - a t g t - - - a - a - - | 61.5 |
| HPV5 | Beta-1 | 9 | 0 | - - - - - - - - - - - - c t g - - - - c - - - t - - a a - - - - - a - c - - | ||
| HPV12 | Beta-1 | 7 | 0 | - - - - - - - - - - - - c t g - - - - c - - - t - - - a - - - - - - - c - - | ||
| HPV21 | Beta-1 | 7 | 1 | - - - - - - - - - - - a - - - - - - t t - - - t - - a t - a - - - - - - - - | ||
| HPV24 | Beta-1 | 7 | 1 | - - - - - - - - - - - t - - - - - - - c - - - t - - a t - - - - c a - - - - | ||
| HPV118 | Beta-1 | 7 | 0 | - - - - - - - - - - - - - t a - - - - - - - - t - - a t - a - - - - - g - - | ||
| HPV124 | Beta-1 | 6 | 0 | - - - - - - - - - - - - - t a - - - - - - - - t - - a t - - - - - - - g - - | ||
| HPVRTRX7 | Beta-1 | 8 | 1 | - - - - - - - - - - - t c t g - - - - c - - - t - - - a - - - - - - - c - - | ||
| HPV9 | Beta-2 | 6 | 1 | - - - - - - - - - - - t - t a - - - - - - - - t - - - a - a - - - - - - - - | ||
| HPV15 | Beta-2 | 6 | 1 | - - - - - - - - - - - a c t - - - - - - - - - t - - - a - - - - - a - - - - | ||
| HPV37 | Beta-2 | 8 | 1 | - - - - - - - - - - - t - t a - - - - - - - - t - - a a - - - - - a - c - - | ||
| HPV38 | Beta-2 | 7 | 1 | - - - - - - - - - - - t - t a - - - - - c - - t - - a a - - - - - - - - - - | ||
| HPV122 | Beta-2 | 6 | 1 | - - - - - - - - - - - t - t g - - - - - - - - t - - - a - - - - - - - a - - | ||
| HPV174 | Beta-2 | 9 | 0 | - - - - - - - - - - - - c t g - - - - - - - - t - - a a - a - - - a - a - - | ||
| HPV75 | Beta-3 | 7 | 1 | - - - - - - - - - - - a - - - - - - - a a - - t - - a t - a - - - - - - - - | ||
| HPV115 | Beta-3 | 8 | 1 | - - - - - - - - - - - a - - c - - - - a a - - t - - - c - t - - - - - c - - | ||
| HPV142 | Gamma-10 | 6 | 1 | - - - - - - - - - g - - c t a - - - - - - - - - - - - c - - - - - - - a - - | ||
| HPV161 | Gamma-19 | 5 | 1 | - - - - - - - - - - - a - - - - - - - - a - - t - - - t - - - - - - - g - - | ||
| HPV165 | Gamma-12 | 6 | 2 | - - - - - c - - - - - t - - - - - - - - - - - - a - t t - a - - - - - - - - | ||
*HPV types also found in eyebrow samples. I, inosine; Tm, melting temperature.
In order to determine the ability of the modified pan-PV CODEHOP PCR to amplify multiple HPV types within the same sample, multiple clones of the pan-PV amplicons were sequenced. Single HPV infections were detected in 33/89 (37 %) tested samples. The majority of eyebrow hair follicle specimens (56/89; 63 %) contained multiple HPV sequences, with HPV types from two different genera detected in 53 % (30/56) of samples with multiple HPV infections. In total, 25 clinical samples contained sequences from two different HPV species, 17 samples contained sequences from three different HPV species, six samples contained sequences from four different HPV species and three samples contained sequences from five different HPV species. Over two-thirds of HPV-positive eyebrow specimens (64/89; 72 %) contained sequences representing potentially novel HPV types/subtypes/variants, of which 81 % (52/64) were identified in multiple infections. Officially recognized HPV types were detected in 60 % (53/89) of HPV-positive samples, with 60 % (32/53) of those same samples containing a co-infection.
Even though as many as 8–12 cloned amplicons were sequenced for each HPV-positive clinical sample, the number of samples with multiple infections was probably underestimated, since amplicon cloning was not performed or failed for 10 samples. Deep sequencing techniques may be necessary to identify all HPV sequences amplified by the pan-PV CODEHOP PCR within a single sample [33].
Continued efforts to improve the identification of novel cutaneous HPVs are crucial for a better understanding of HPV phylogenetic diversity and a clarification of their role in the development of skin cancer. In the current study, we have shown that the modified pan-PV CODEHOP PCR assay is able to identify multiple HPV types, even from different genera, in the same clinical sample. Moreover, sequences representing 27 novel putative HPV types were identified. These results demonstrate that the modified pan-PV CODEHOP PCR is an excellent tool for HPV screening and identification of novel cutaneous HPVs, even in samples with low viral loads.
Funding information
The study was supported by the European Union’s Seventh Framework Program for research, technological development and demonstration under the CoheaHr project (grant agreement no. HEALTH-F3-2013-603019).
Acknowledgements
The authors would like to thank Pavle Košorok, MD, PhD, and Kristina Fujs Komloš, PhD, for the collection of eyebrow hair follicle specimens.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
The study was approved by the Institutional Review Board of the Ministry of Health of Republic of Slovenia (consent reference 97/11/09). All patients additionally provided a written informed consent and were sampled in compliance with the Helsinki Declaration.
Footnotes
Abbreviations: CODEHOP, consensus-degenerate hybrid oligonucleotide primer; HPV, human papillomavirus; I, inosine; MCMC, Markov chain Monte Carlo; PV, papillomavirus.
The GenBank/EMBL/DDBJ accession numbers for nucleotide sequences of novel/putative HPV types reported in this paper are: SIBX22 (LK022298), SIBX23 (LK022299), SIBX24 (LK022300), SIBX25 (LK022301), SIBX26 (LK022302), SIBX27 (LK022303), SIBX28 (LK022304), SIBX29 (LK022305), SIBX30 (LK022306), SIBX31 (LK022307), SIBX32 (LK022308), SIBX33 (LK022309), SIBX34 (LK022310), SIBX35 (LK022311), SIBX36 (LK022312), SIBX37 (LK022313), SIBX38 (LK022314), SIBX39 (LK022315), SIBX40 (LK022316), SIBX41 (LK022317), SIBX42 (LK022318), SIBX43 (LK022319), SIBX44 (LK022320), SIBX45 (LK022321), SIBX46 (LK022322), SIBX47 (LK022323), SIBX48 (LK022324), SIBX49 (LK022325), SIBX50 (LK022326), SIBX51 (LM653099), SIBX52 (LM653100), SIBX53 (LM653101), SIBX54 (LM653102), SIBX55 (LM653103), SIBX56 (LM653104), SIBX57 (KP768215), SIBX58 (KP768216), SIBX59 (KP768217), SIBX60 (KP768218), SIBX61 (KP768219), SIBX62 (KP768220), SIBX63 (KP768221), SIBX64 (KP768222), SIBX65 (KP768223).
References
- 1.de Villiers EM, Fauquet C, Broker TR, Bernard HU, Zur Hausen H. Classification of papillomaviruses. Virology. 2004;324:17–27. doi: 10.1016/j.virol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- 2.de Villiers EM. Cross-roads in the classification of papillomaviruses. Virology. 2013;445:2–10. doi: 10.1016/j.virol.2013.04.023. [DOI] [PubMed] [Google Scholar]
- 3.Cubie HA. Diseases associated with human papillomavirus infection. Virology. 2013;445:21–34. doi: 10.1016/j.virol.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 4.Forman D, de Martel C, Lacey CJ, Soerjomataram I, Lortet-Tieulent J, et al. Global burden of human papillomavirus and related diseases. Vaccine. 2012;30:F12–F23. doi: 10.1016/j.vaccine.2012.07.055. [DOI] [PubMed] [Google Scholar]
- 5.Forslund O, Iftner T, Andersson K, Lindelof B, Hradil E, et al. Cutaneous human papillomaviruses found in sun-exposed skin: Beta-papillomavirus species 2 predominates in squamous cell carcinoma. J Infect Dis. 2007;196:876–883. doi: 10.1086/521031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arroyo Mühr LS, Hultin E, Bzhalava D, Eklund C, Lagheden C, et al. Human papillomavirus type 197 is commonly present in skin tumors. Int J Cancer. 2015;136:2546–2555. doi: 10.1002/ijc.29325. [DOI] [PubMed] [Google Scholar]
- 7.Hošnjak L, Kocjan BJ, Pirš B, Seme K, Poljak M. Characterization of two novel gammapapillomaviruses, HPV179 and HPV184, isolated from common warts of a renal-transplant recipient. PLoS One. 2015;10:e0119154. doi: 10.1371/journal.pone.0119154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Antonsson A, Erfurt C, Hazard K, Holmgren V, Simon M, et al. Prevalence and type spectrum of human papillomaviruses in healthy skin samples collected in three continents. J Gen Virol. 2003;84:1881–1886. doi: 10.1099/vir.0.18836-0. [DOI] [PubMed] [Google Scholar]
- 9.Ekström J, Bzhalava D, Svenback D, Forslund O, Dillner J. High throughput sequencing reveals diversity of human papillomaviruses in cutaneous lesions. Int J Cancer. 2011;129:2643–2650. doi: 10.1002/ijc.26204. [DOI] [PubMed] [Google Scholar]
- 10.Foulongne V, Sauvage V, Hebert C, Dereure O, Cheval J, et al. Human skin microbiota: high diversity of DNA viruses identified on the human skin by high throughput sequencing. PLoS One. 2012;7:e38499. doi: 10.1371/journal.pone.0038499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bolatti EM, Chouhy D, Hošnjak L, Casal PE, Kocjan BJ, et al. Corrigendum: natural history of human papillomavirus infection of sun-exposed healthy skin of immunocompetent individuals over three climatic seasons and identification of HPV209, a novel betapapillomavirus. J Gen Virol. 2017;98:2205–2206. doi: 10.1099/jgv.0.000900. [DOI] [PubMed] [Google Scholar]
- 12.Chouhy D, Bolatti EM, Pérez GR, Giri AA. Analysis of the genetic diversity and phylogenetic relationships of putative human papillomavirus types. J Gen Virol. 2013;94:2480–2488. doi: 10.1099/vir.0.055137-0. [DOI] [PubMed] [Google Scholar]
- 13.Boxman IL, Berkhout RJ, Mulder LH, Wolkers MC, Bouwes Bavinck JN, et al. Detection of human papillomavirus DNA in plucked hairs from renal transplant recipients and healthy volunteers. J Invest Dermatol. 1997;108:712–715. doi: 10.1111/1523-1747.ep12292090. [DOI] [PubMed] [Google Scholar]
- 14.Forslund O, Ly H, Higgins G. Improved detection of cutaneous human papillomavirus DNA by single tube nested 'hanging droplet' PCR. J Virol Methods. 2003;110:129–136. doi: 10.1016/S0166-0934(03)00109-5. [DOI] [PubMed] [Google Scholar]
- 15.Chouhy D, Gorosito M, Sánchez A, Serra EC, Bergero A, et al. New generic primer system targeting mucosal/genital and cutaneous human papillomaviruses leads to the characterization of HPV 115, a novel Beta-papillomavirus species 3. Virology. 2010;397:205–216. doi: 10.1016/j.virol.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kocjan BJ, Bzhalava D, Forslund O, Dillner J, Poljak M. Molecular methods for identification and characterization of novel papillomaviruses. Clin Microbiol Infect. 2015;21:808–816. doi: 10.1016/j.cmi.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 17.Rose TM, Schultz ER, Henikoff JG, Pietrokovski S, McCallum CM, et al. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res. 1998;26:1628–1635. doi: 10.1093/nar/26.7.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rose TM. CODEHOP-mediated PCR–a powerful technique for the identification and characterization of viral genomes. Virol J. 2005;2:20. doi: 10.1186/1743-422X-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Staheli JP, Ryan JT, Bruce AG, Boyce R, Rose TM. Consensus-degenerate hybrid oligonucleotide primers (CODEHOPs) for the detection of novel viruses in non-human primates. Methods. 2009;49:32–41. doi: 10.1016/j.ymeth.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walsh EE, Falsey AR, Swinburne IA, Formica MA. Reverse transcription polymerase chain reaction (RT-PCR) for diagnosis of respiratory syncytial virus infection in adults: use of a single-tube "hanging droplet" nested PCR. J Med Virol. 2001;63:259–263. doi: 10.1002/1096-9071(200103)63:3<259::AID-JMV1010>3.0.CO;2-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chouhy D, Bolatti EM, Piccirilli G, Sánchez A, Fernandez Bussy R, et al. Identification of human papillomavirus type 156, the prototype of a new human gammapapillomavirus species, by a generic and highly sensitive PCR strategy for long DNA fragments. J Gen Virol. 2013;94:524–533. doi: 10.1099/vir.0.048157-0. [DOI] [PubMed] [Google Scholar]
- 22.Kocjan BJ, Poljak M, Seme K, Potocnik M, Fujs K, et al. Distribution of human papillomavirus genotypes in plucked eyebrow hairs from Slovenian males with genital warts. Infect Genet Evol. 2005;5:255–259. doi: 10.1016/j.meegid.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 23.van Duin M, Snijders PJ, Schrijnemakers HF, Voorhorst FJ, Rozendaal L, et al. Human papillomavirus 16 load in normal and abnormal cervical scrapes: an indicator of CIN II/III and viral clearance. Int J Cancer. 2002;98:590–595. doi: 10.1002/ijc.10232. [DOI] [PubMed] [Google Scholar]
- 24.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hampras SS, Giuliano AR, Lin HY, Fisher KJ, Abrahamsen ME, et al. Natural history of cutaneous human papillomavirus (HPV) infection in men: the HIM study. PLoS One. 2014;9:e104843. doi: 10.1371/journal.pone.0104843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neale RE, Weissenborn S, Abeni D, Bavinck JN, Euvrard S, et al. Human papillomavirus load in eyebrow hair follicles and risk of cutaneous squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2013;22:719–727. doi: 10.1158/1055-9965.EPI-12-0917-T. [DOI] [PubMed] [Google Scholar]
- 28.Schneider I, Lehmann MD, Kogosov V, Stockfleth E, Nindl I. Eyebrow hairs from actinic keratosis patients harbor the highest number of cutaneous human papillomaviruses. BMC Infect Dis. 2013;13:186. doi: 10.1186/1471-2334-13-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iannacone MR, Gheit T, Pfister H, Giuliano AR, Messina JL, et al. Case-control study of genus-beta human papillomaviruses in plucked eyebrow hairs and cutaneous squamous cell carcinoma. Int J Cancer. 2014;134:2231–2244. doi: 10.1002/ijc.28552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asgari MM, Kiviat NB, Critchlow CW, Stern JE, Argenyi ZB, et al. Detection of human papillomavirus DNA in cutaneous squamous cell carcinoma among immunocompetent individuals. J Invest Dermatol. 2008;128:1409–1417. doi: 10.1038/sj.jid.5701227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Koning MN, Weissenborn SJ, Abeni D, Bouwes Bavinck JN, Euvrard S, et al. Prevalence and associated factors of betapapillomavirus infections in individuals without cutaneous squamous cell carcinoma. J Gen Virol. 2009;90:1611–1621. doi: 10.1099/vir.0.010017-0. [DOI] [PubMed] [Google Scholar]
- 32.Deng Q, Li J, Pan Y, Liu F, He Z, et al. Prevalence and associated risk factors of human papillomavirus in healthy skin specimens collected from rural Anyang, China, 2006–2008. J Invest Dermatol. 2016;136:1191–1198. doi: 10.1016/j.jid.2016.02.014. [DOI] [PubMed] [Google Scholar]
- 33.Bzhalava D, Eklund C, Dillner J. International standardization and classification of human papillomavirus types. Virology. 2015;476:341–344. doi: 10.1016/j.virol.2014.12.028. [DOI] [PubMed] [Google Scholar]