Key Points
Question
Can a deep-learning model classify hyperkalemia from the electrocardiogram (ECG) in patients with chronic kidney disease?
Findings
In this validation study, a deep neural network was trained using more than 1.5 million ECGs recorded from 1994 to 2017 from approximately 450 000 patients seen at the Mayo Clinic in Minnesota and validated on nearly 62 000 ECGs from the Mayo Clinic in Minnesota, Florida, and Arizona. Using 2 or 4 ECG leads, a deep-learning model detected hyperkalemia with high sensitivity and negative predictive value, with an area under the curve between 0.853 and 0.901.
Meaning
Deep learning may enable noninvasive screening for hyperkalemia in at-risk patients with chronic kidney disease.
Abstract
Importance
For patients with chronic kidney disease (CKD), hyperkalemia is common, associated with fatal arrhythmias, and often asymptomatic, while guideline-directed monitoring of serum potassium is underused. A deep-learning model that enables noninvasive hyperkalemia screening from the electrocardiogram (ECG) may improve detection of this life-threatening condition.
Objective
To evaluate the performance of a deep-learning model in detection of hyperkalemia from the ECG in patients with CKD.
Design, Setting, and Participants
A deep convolutional neural network (DNN) was trained using 1 576 581 ECGs from 449 380 patients seen at Mayo Clinic, Rochester, Minnesota, from 1994 to 2017. The DNN was trained using 2 (leads I and II) or 4 (leads I, II, V3, and V5) ECG leads to detect serum potassium levels of 5.5 mEq/L or less (to convert to millimoles per liter, multiply by 1) and was validated using retrospective data from the Mayo Clinic in Minnesota, Florida, and Arizona. The validation included 61 965 patients with stage 3 or greater CKD. Each patient had a serum potassium count drawn within 4 hours after their ECG was recorded. Data were analyzed between April 12, 2018, and June 25, 2018.
Exposures
Use of a deep-learning model.
Main Outcomes and Measures
Area under the receiver operating characteristic curve (AUC) and sensitivity and specificity, with serum potassium level as the reference standard. The model was evaluated at 2 operating points, 1 for equal specificity and sensitivity and another for high (90%) sensitivity.
Results
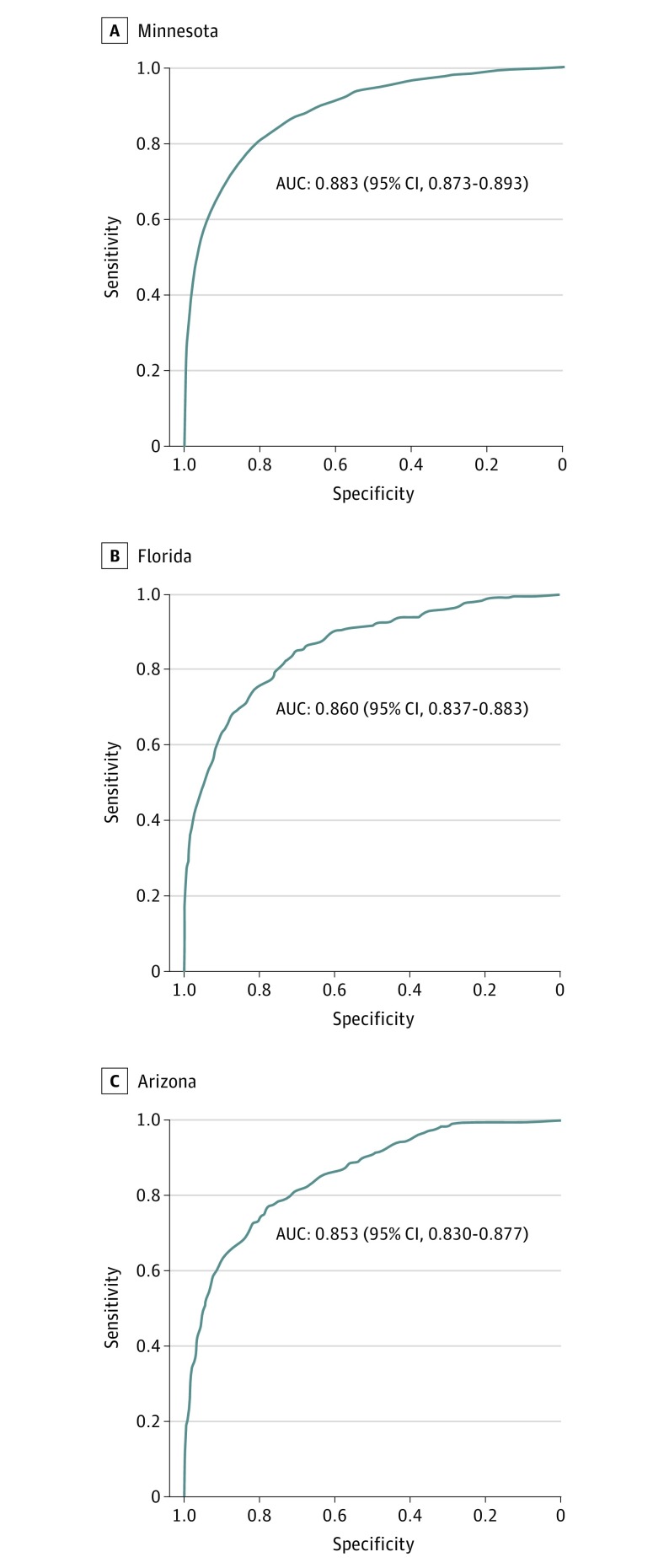
Of the total 1 638 546 ECGs, 908 000 (55%) were from men. The prevalence of hyperkalemia in the 3 validation data sets ranged from 2.6% (n = 1282 of 50 099; Minnesota) to 4.8% (n = 287 of 6011; Florida). Using ECG leads I and II, the AUC of the deep-learning model was 0.883 (95% CI, 0.873-0.893) for Minnesota, 0.860 (95% CI, 0.837-0.883) for Florida, and 0.853 (95% CI, 0.830-0.877) for Arizona. Using a 90% sensitivity operating point, the sensitivity was 90.2% (95% CI, 88.4%-91.7%) and specificity was 63.2% (95% CI, 62.7%-63.6%) for Minnesota; the sensitivity was 91.3% (95% CI, 87.4%-94.3%) and specificity was 54.7% (95% CI, 53.4%-56.0%) for Florida; and the sensitivity was 88.9% (95% CI, 84.5%-92.4%) and specificity was 55.0% (95% CI, 53.7%-56.3%) for Arizona.
Conclusions and Relevance
In this study, using only 2 ECG leads, a deep-learning model detected hyperkalemia in patients with renal disease with an AUC of 0.853 to 0.883. The application of artificial intelligence to the ECG may enable screening for hyperkalemia. Prospective studies are warranted.
This validation study evaluates the performance of a deep-learning model in detection of hyperkalemia from the electrocardiogram in patients with chronic kidney disease.
Introduction
Hyperkalemia is potentially life threatening for the 35 million American adults with chronic kidney disease (CKD).1 Owing to the effects of kidney dysfunction on potassium homeostasis, as well as the recommendation that these patients be treated with renin-angiotensin-aldosterone system (RAAS) inhibitors that preserve renal and cardiac function but also inhibit renal potassium secretion, the risk of hyperkalemia in CKD is substantially elevated.2
When present, hyperkalemia is often asymptomatic and associated with cardiac arrhythmias and death.3 Serum potassium monitoring can reduce the risk of hyperkalemia in patients with CKD by upwards of 71%.4 However, guideline-directed potassium monitoring5,6 is severely underused: up to one-third of patients taking RAAS inhibitors receive no monitoring, and only 10% receive potassium monitoring before and after RAAS inhibitors are started.7,8 We sought to improve detection of hyperkalemia in patients with CKD by developing and validating a noninvasive screening test using the electrocardiogram (ECG).
Hyperkalemia causes cardiotoxic effects and has been associated with a defined series of ECG abnormalities, including peaking of T waves, QRS prolongation, and PR shortening.9 However, in clinical practice, the sensitivity of physician readers in the ECG diagnosis of hyperkalemia has been estimated to be as low as 34% to 43%.10 Deep learning is a type of artificial intelligence that uses representation methods to identify meaningful patterns from complex digital files and has been used in medicine to identify lesions in mammograms or retinal images.11,12 We hypothesized that a deep-learning model (DLM) could effectively rule out, or screen for, hyperkalemia. To test this hypothesis, we trained and validated a DLM to classify hyperkalemia from ECGs in patients with CKD.
Methods
This study was approved by the institutional review board of the Mayo Clinic. Clinical data including digitally stored ECGs, serum potassium values, creatinine levels, race/ethnicity, age, body mass index (calculated as weight in kilograms divided by height in meters squared), and sex were obtained from Mayo Clinic. Patients’ informed consent was exempted by the institutional review board because of the retrospective nature of the study using fully anonymized ECG and health data. Race/ethnicity and sex information were self-reported by each patient, and used to assess algorithm robustness across populations.
Deep-Learning Model
Deep learning is a method premised on learning complex hierarchical representation from the data that constitute multiple levels of abstraction. Deep learning uses many hidden layers of neurons to produce increasingly abstracted, nonlinear representations of the underyling data. It is well suited for classification of complex graphic data including the ECG; early US Food and Drug Administration–cleared biomedical applications of deep learning have been in the domain of image processing for computed tomographic images of tumors or retinal camera images of diabetic retinopathy.
The model architecture was configured as a convolutional neural network with 11 layers, with the first 10 layers being convolutional and the last as a fully connected softmax layer (eMethods and eFigure 1 in the Supplement). The network function receives a 10-second ECG signal from any number of simultaneously acquired ECG leads and produces a parameter output between 0 and 1.
We trained 2 independent models on 4 and 2 leads of the ECG. Results using cross-validation on the development set indicated that the performance increase obtained by using additional leads beyond leads I, II, V3, and V5 was minimal. We trained a network on leads I and II because these leads can easily be recorded by dry electrode contact with the hands and left leg13; these ECG leads have been used to enable patient self-monitoring.14,15
Development and Validation Data sets
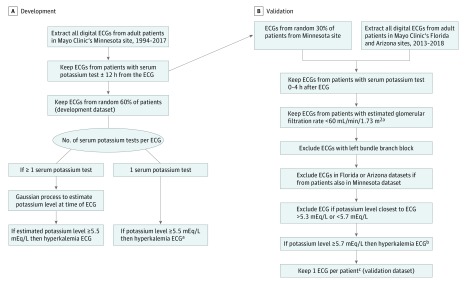
A data set was generated consisting of all 12-lead ECGs recorded on all adult patients at the Mayo Clinic in Rochester, Minnesota, between 1994 and 2017 who had also received at least 1 serum potassium test within 12 hours before or after the ECG. The data set included 2 835 059 twelve-lead ECGs and 4 277 183 potassium tests from 787 661 patients. A time stamp to the nearest minute was available for the blood draws and ECG recordings. The data set was partitioned randomly by patient, with 60% of patients used for DLM development (n = 449 380 of 787 661; development dataset) and 30% for the Minnesota validation data set (n = 228 421 of 787 661; Figure 1A). The remaining 10% of patients (n = 109 860 of 787 661) were reserved for future analysis. The DLM was trained and internally tested using the development data set.
Figure 1. Development and Validation Data Sets Generation and Electrocardiogram (ECG) Labeling of Hyperkalemia.
To convert potassium to millimoles per liter, multiply by 1.
aEstimated glomerular filtration rate calculated from the Chronic Kidney Disease Epidemiology Collaboration equation, using the mean serum creatinine values within 24 hours prior to time of ECG recording.
bGiven error in serum potassium, and to avoid potentially irrelevant misclassification rates around hyperkalemia threshold, ECGs with serum potassium greater than 5.3 mEq/L or less than 5.7 mEq/L were excluded.
cRandom ECG selected for Minnesota dataset. Most recent ECG selected for Arizona and Florida datasets.
To validate the model externally, we randomly selected a subset of the ECG-potassium pairs in patients with CKD from the Mayo Clinic in Rochester, Minnesota. In addition, additional unique data pairs were obtained from additional Mayo Clinic sites in Jacksonville, Florida, and Scottdale, Arizona. We kept the geographically distinct cohorts separate, in 3 validation data sets, to assess the performance of the model on data with different patient sociodemographics, extent of CKD, and treatment patterns. The validation data sets were created as per Figure 1B. For the Florida and Arizona data sets, any 12-lead ECG recorded from January 1, 2013, through March 31, 2018, was obtained. The Minnesota data set was composed of the previously partitioned original data set (30%). All ECGs within 4 hours before a serum potassium draw were identified.16 We included patients with stage 3 or greater CKD. A priori, we excluded ECGs with left bundle branch block because of the concern that the peaking of T waves and the widening of the QRS complex that occurs with hyperkalemia could be masked with a baseline left bundle branch block. If multiple ECGs were recorded within 4 hours of a potassium draw, the ECG closest in time to potassium was selected. In total, 61 965 ECG-potassium pairs were used for validation.
Hyperkalemia
We defined hyperkalemia as a serum potassium value of at least 5.5 mEq/L (to convert to millimoles per liter, multiply by 1) because this is a commonly used cutoff to prompt treatment for hyperkalemia.17,18 Several studies have shown a rapid increase in the risk of death as serum potassium levels exceed 5.5 mEq/L.3 The approach used to label hyperkalemia in the development data set is described in the eMethods in the Supplement. For validation, to avoid potentially irrelevant misclassification rates around the 5.5-mEq/L threshold, an analysis of ECGs with serum potassium levels either 5.3 mEq/L or less or at least 5.7 mEq/L was the focus of the analysis. The exclusion of ECGs with potassium levels near the threshold is similar to a phase II biomarker design.19 The 0.2-mEq/L potassium laboratory draw error rate was estimated based on our prior work.20
Statistical Analysis
The sample size for each validation data set was determined to be 6133 ECGs and was based on the maximal sample size needs of the area under the receiving operating characteristic curve (AUC) (eMethods in the Supplement).
The DLM was evaluated at 2 operating points selected from the development data set, one selected for equal sensitivity and specificity and the other for high (90%) sensitivity. These thresholds were applied to the validation data sets to characterize the sensitivity and specificity of the algorithm. Exact 95% confidence intervals were used for all measures of diagnostic performance except for AUC. The confidence interval for AUC was determined based on Sun and Su optimization of the Delong method using the pROC package in R (R Foundation).21 Statistical significance for differences in patient characteristics was defined as a 2-sided P value of less than .05. Measures of diagnostic performance were summarized using 2-sided 95% confidence intervals. Analyses were computed using software R, version 3.4.2 (R Foundation).
Results
The development data set included 1 576 581 ECGs, of which 30 184 (2.0%) were associated with a serum potassium value of at least 5.5 mEg/L (Table 1). About 3.8% of all ECGs were obtained from patients with end-stage renal disease (ESRD) (estimated glomerular filtration rate [eGFR], <15 mL/min/1.73 m2). Hyperkalemia ECGs were more likely to be recorded in patients who were older and men and who had stage 3 or greater CKD.
Table 1. Baseline Characteristics of Development and Validation Data sets.
| Characteristic | ECGs | |||
|---|---|---|---|---|
| All (n = 1 576 581) | Hyperkalemia (n = 30 184)a | Not Hyperkalemia (n = 1 546 397) | P Value | |
| Development data set | ||||
| Men, No. (%) | 875 002 (55.5) | 18 533 (61.4) | 856 469 (55.4) | <.001 |
| Age, mean (SD), y | 64.5 (15.8) | 66.4 (15.4) | 64.5 (15.8) | <.001 |
| BMI, mean (SD) | 29.2 (7.0) | 29.1 (7.9) | 29.2 (7.0) | .04 |
| eGFR, mean (SD), mL/min /1.73 m2c | 65.1 (27.3) | 38.0 (26.5) | 65.6 (27.0) | <.001 |
| Potassium, mean (SD), mEq/L | 4.26 (.53) | 5.85 (.43) | 4.23 (.47) | <.001 |
| ECGs with >1 serum potassium test, No. (%)d | 371 681 (23.6) | 16 395 (54.3) | 355 286 (23.0) | <.001 |
| Patients, No. | 449 380 | 19 243 | 430 137 | <.001 |
| ECGs per patient, mean (SD)b | 3.51 (5.52) | 1.65 (1.65) | 3.46 (5.39) | <.001 |
| Validation data sets | Minnesota (n = 50 099) | Florida (n = 6011) | Arizona (n = 5855) | |
| Men, No. (%) | 26 552 (53.0) | 3170 (52.7) | 3276 (56.0) | NA |
| Age, mean (SD), y | 69.0 (13.8) | 71.6 (13.9) | 72.2 (13.8) | NA |
| Race/ethnicity, No. (%) | ||||
| White | NA | 5077 (84.5) | 5271 (9.0) | NA |
| Black | NA | 650 (1.8) | 193 (3.3) | NA |
| Asian | NA | 103 (1.7) | 91 (1.6) | NA |
| Other | NA | 181 (3.0) | 300 (5.1) | NA |
| BMI, mean (SD)e | 29.2 (6.6) | 28.6 (6.3) | 28.4 (6.3) | NA |
| eGFR, mean (SD), mL/min/1.73 m2f | 42.5 (13.8) | 4.2 (16.4) | 41.1 (16.1) | NA |
| eGFR <15 mL/min/1.73 m2, No. (%) | 2908 (5.8) | 499 (8.3) | 406 (6.9) | NA |
| Potassium, mean (SD), mEq/L | 4.3 (.6) | 4.37 (.67) | 4.45 (.62) | NA |
| Potassium, range, mEq/L | 1.4-9.2 | 2.20-8.10 | 2.10-8.00 | NA |
| Potassium ≥5.7 mEq/L, No. (%)g | 1282 (2.6) | 287 (4.8) | 270 (4.6) | NA |
| Time to potassium test after ECG, mean (SD), h | 0.89 (0.98) | 0.84 (0.94) | 0.69 (0.75) | NA |
Abbreviations: BMI, body mass index (calculated as weight in kilograms divided by height in meters squared); ECG, electrocardiogram; eGFR, estimated glomerular filtration rate; NA, not applicable.
SI conversion factor: To convert potassium to millimoles per liter, multiply by 1.
Hyperkalemia defined as serum potassium level associated with ECG ≥5.5 mEq/L.
For patients with more than 1 ECG in the data set, ECGs were recorded more than 24 hours apart such that potassium values were not used as labels for more than 1 ECG.
eGFR estimated by the Chronic Kidney Disease Epidemiology Collaboration formula: 175 × (Scr)-1.154 × (Age)-0.203 × (0.742 if female), with no accounting for race/ethnicity because it was not available.
When more than 1 serum potassium available within 12 hours prior and 12 hours after ECG, a Gaussian process was used to estimate potassium at time of ECG recording.
Missing values: BMI values were missing for 11 786 patients from Minnesota, 518 patients from Florida, and 771 patients from Arizona.
GFR estimated by Chronic Kidney Disease Epidemiology Collaboration for the Rochester data set using the following formula: 175 × (Scr)-1.154 × (Age)-0.203 × (0.742 if female), with no accounting for race for the Minnesota data set. Race/ethnicity was available and used for the Florida and Arizona data sets.
ECG excluded if potassium was more than 5.3 mEq/L or less than 5.7 mEq/L.
Table 2 summarizes patient demographics from the 3 sites of validation: Minnesota, Florida, and Arizona. Of the total number of patients, 53% to 56% were men. Mean eGFR was 40.2 to 42.5 mL/min/1.73 m2. Mean time to serum potassium draw after ECG was 41 to 53 minutes. The patients were diverse from a racial/ethnic and kidney disease perspective. Mean age was 69 years in Minnesota compared with 71 to 72 years in Florida and Arizona. More than 10% of patients in Florida were black (n = 650 of 6011) compared with 3.3% in Arizona (n = 193 of 5855). Only 5.8% in Minnesota (n = 2908 of 50 099) had ESRD compared with 6.9% (n = 406 of 5855) to 8.3% (n = 499 of 6011) at the other sites. Fewer than 3% in Minnesota had hyperkalemia (n = 1282 of 50 099) compared with 4.6% (n = 270 of 5855) to 4.8% (n = 287 of 6011) in Florida and Arizona, respectively.
Table 2. Validation Data Set Performance for Hyperkalemia From 2 and 4 Leads of the ECG.
| Validation Data Set | Value (95% CI) | |||
|---|---|---|---|---|
| 2-Lead ECGa | 4-Lead ECGb | |||
| Sensitivity = Specificity | High Sensitivityc | Sensitivity = Specificity | High Sensitivity | |
| Minnesota (n = 50 099) | ||||
| AUC | 0.883 (0.873-0.893) | 0.883 (0.873-0.893) | 0.901 (0.892-0.911) | 0.901 (0.892-0.911) |
| Sensitivity, % | 79.9 (77.6-82.0) | 90.2 (88.4-91.7) | 81.3 (79.0-83.4) | 89.3 (87.5-91.0) |
| Specificity, % | 81.3 (80.9-81.6) | 63.2 (62.7-63.6) | 84.2 (83.9-84.5) | 70.0 (69.6-70.4) |
| NPV, % | 99.4 (99.3-99.4) | 99.6 (99.5-99.7) | 99.4 (99.3-99.5) | 99.6 (99.5-99.7) |
| PPV, % | 10.1 (9.5-10.7) | 6.0 (5.7-6.4) | 11.9 (11.2-12.6) | 7.2 (6.8-7.7) |
| Florida (n = 6011) | ||||
| AUC | 0.860 (0.837-0.883) | 0.860 (0.837-0.883) | 0.885 (0.863-0.907) | 0.885 (0.863-0.907) |
| Sensitivity, % | 80.5 (75.4-84.9) | 91.3 (87.4-94.3) | 84.0 (79.2-88.0) | 92.3 (88.6-95.1) |
| Specificity, % | 75.2 (74.0-76.3) | 54.7 (53.4-56.0) | 77.1 (75.8-78.0) | 60.5 (59.2-61.7) |
| NPV, % | 98.7 (98.3-99.0) | 99.2 (98.8-99.5) | 99.0 (98.6-99.2) | 99.4 (99.0-99.6) |
| PPV, % | 14.0 (12.3-15.7) | 9.2 (8.1-10.3) | 15.4 (13.7-17.3) | 10.5 (9.3-11.7) |
| Arizona (n = 5855) | ||||
| AUC | 0.853 (0.830-0.877) | 0.853 (0.830-0.877) | 0.880 (0.860-0.901) | 0.880 (0.860-0.901) |
| Sensitivity, % | 78.1 (72.7-82.9) | 88.9 (84.5-92.4) | 82.6 (77.5-86.9) | 92.6 (88.8-95.4) |
| Specificity, % | 75.3 (74.1-76.4) | 55.0 (53.7-56.3) | 77.0 (75.9-78.1) | 60.3 (59.0-61.6) |
| NPV, % | 98.6 (98.2-98.9) | 99.0 (98.6-99.3) | 98.9 (98.6-99.2) | 99.4 (99.1-99.6) |
| PPV, % | 13.3 (11.6-15.0) | 8.7 (7.7-9.8) | 14.8 (13.0-16.7) | 10.1 (9.0-11.4) |
Abbreviations: AUC, area under the receiver operating characteristic curve; ECG, electrocardiogram; NPV, negative predictive value; PPV, positive predictive value.
2-Lead ECG using leads I and II.
4-Lead ECG using leads I, II, V3, and V5.
Operating point at sensitivity of 90%.
The algorithm performed well in identifying hyperkalemia in the validation data sets (Table 3). For detection of hyperkalemia using ECG leads I and II, the DLM achieved an AUC of 0.883 (95% CI, 0.873-0.893) in Minnesota; 0.860 (95% CI, 0.837-0.883) in Florida; and 0.853 (95% CI, 0.830-0.877) in Arizona (Figure 2). The AUC was 0.02 to 0.03 higher using ECG leads I, II, V3, and V5.
Table 3. Confusion Matrix for Classification of Hyperkalemia, Deep-Learning Model vs Serum Potassium.
| Validation Data Set | No. (%)a | ||||
|---|---|---|---|---|---|
| True Positiveb | False Positive | False Negative | True Negative | Accuracyc | |
| Minnesota (n = 50 099) | |||||
| SN = SP, 2 ECG leads | 1032 (2.1) | 9578 (19.1) | 250 (0.5) | 39 239 (78.3) | 40 271 (80.4) |
| SN90, 2 ECG leads | 1153 (2.3) | 17 809 (35.5) | 129 (0.3) | 31 008 (61.9) | 32 161 (64.2) |
| SN = SP, 4 ECG leads | 1058 (2.1) | 8495 (17.0) | 224 (0.4) | 40 322 (80.5) | 41 380 (82.6) |
| SN90, 4 ECG leads | 1153 (2.3) | 15 372 (30.7) | 129 (0.3) | 33 445 (66.7) | 34 598 (69.0) |
| Florida (n = 6011) | |||||
| SN = SP, 2 ECG leads | 225 (3.7) | 1374 (22.9) | 62 (1.0) | 4350 (72.4) | 4575 (76.1) |
| SN90, 2 ECG leads | 262 (4.4) | 2499 (41.6) | 25 (0.4) | 3225 (53.6) | 3487 (58.0) |
| SN = SP, 4 ECG leads | 239 (4.0) | 1308 (21.8) | 48 (0.8) | 4416 (73.4) | 4655 (77.4) |
| SN90, 4 ECG leads | 263 (4.4) | 2149 (35.7) | 24 (0.4) | 3575 (59.5) | 3838 (63.9) |
| Arizona (n = 5855) | |||||
| SN = SP, 2 ECG leads | 209 (3.6) | 1322 (22.6) | 61 (1.0) | 4263 (72.8) | 4472 (76.4) |
| SN90, 2 ECG leads | 237 (4.0) | 2434 (41.6) | 33 (0.6) | 3151 (53.8) | 3388 (57.8) |
| SN = SP, 4 ECG leads | 223 (3.8) | 1275 (21.8) | 47 (0.8) | 4310 (73.6) | 4533 (77.4) |
| SN90, 4 ECG leads | 249 (4.3) | 2095 (35.8) | 21 (0.3) | 3490 (59.6) | 3739 (63.9) |
Abbreviations: SN, sensitivity, SP, specificity; SN90, sensitivity at 90%; ECG, electrocardiogram.
SI conversion factor: To convert potassium to millimoles per liter, multiply by 1.
Percentage value is value divided by the total sample size.
Hyperkalemia defined as serum potassium ≥5.5 mEq/L.
Accuracy calculated as number of true positives and true negative divided by total sample size.
Figure 2. Validation Data Set Performance for Hyperkalemia From Lead I and II of the Electrocardiogram.
AUC indicates area under the receiver operating characteristic curve.
Using the operating point with equal sensitivity and specificity, for ECG leads I and II, the DLM’s sensitivity and specificity were 79.9% (95% CI, 77.6%-82.0%) and 81.3% (95% CI, 80.9%-81.6%) in Minnesota; 80.5% (95% CI, 75.4%-84.9%) and 75.2% (95% CI, 74.0%-76.3%) in Florida; and 78.1% (95% CI, 72.7%-82.9%) and 75.3% (95% CI, 74.1%-76.4%) in Arizona. Given the less than 5% prevalence of hyperkalemia, these findings correspond to a negative predictive value of 98.6% to 99.4% across the validation data sets.
The operating point with high sensitivity reflects an output that would be used for a screening tool. The algorithm’s sensitivity and specificity for validation were 90.2% (95% CI, 88.4%-91.7%) and 63.2% (95% CI, 62.7%-63.6%) in Minnesota; 91.3% (95% CI, 87.4%-94.3%) and 54.7% (95% CI, 53.4%-56.0%) in Florida; and 88.9% (95% CI, 84.5%-92.4%) and 55.0% (95% CI, 53.7%-56.3%) in Arizona. These findings correspond to a negative predictive value of 99.0% to 99.6%.
We evaluated algorithm performance after adjusting for age, sex, race/ethnicity, eGFR, and body mass index to ensure consistency across a wide range of putative confounding variables (eTable 1 in the Supplement). The DLM had significantly better performance than use of patient demographics alone to detect hyperkalemia; the addition of patient demographics did not significantly improve the DLM’s performance (eFigure 2 in the Supplement).
The number of false-positive, false-negative, true-positive, and true-negative results for each model, as well as accuracy, is presented in eTable 3 in the Supplement. Between 50% and 70% of patients in the validation data sets did not have hyperkalemia predicted by the DLM, with less than 1% of all test results being false-negative; on the other hand, up to 42% of all test results were false-positive. For the DLM using 2 ECG leads and the high sensitivity operating point, we performed a medical record review of the 58 patients from the Florida (n = 25) and Arizona (n = 33) validation data sets with false-negative results. The median blood potassium was 5.9 mEq/L (range, 5.7-7.2, mEq/L). Of these patients, 34 (59%) were hospitalized, 18 (31%) had diabetes, 9 (16%) had ESRD, and in 8 (14%), the ECG demonstrated atrial fibrillation. A subsequent blood potassium measurement within 8 hours was less than 5.5 mEq/L in 30 patients (52%). Eight patients (14%) were treated for hyperkalemia. Two patients (3%) had platelet counts greater than 500 ×103/µL (to convert to ×109/L, multiply by 1) and in 1 patient (2%), the blood sample was hemolyzed.
We performed a sensitivity analysis with the exclusion of data from patients with ESRD (eTable 2 in the Supplement) because there are several case reports demonstrating that the usual electrocardiographic manifestations of hyperkalemia are less frequent in persons with ESRD.22 The exclusion of data from patients with ESRD, which decreased the validation sets’ size by 10%, did not substantially change the AUC.
Additional sensitivity analyses were conducted with all patients with CKD but with no potassium range exclusion (eTable 3 in the Supplement). The inclusion of all ECGs and serum potassium values decreased the AUC across all models by 0.02 to 0.04; with a higher prevalence of hyperkalemia, the positive predictive value increased by 3 to 5 percentage points.
Discussion
In patients with CKD, hyperkalemia is common and life threatening.23,24 On average, patients with CKD spend up to 9% of their time with serum potassium levels 5.5 mEq/L or greater.2 The 1-day odds of mortality are up to 13 times higher for patients with CKD with outpatient serum potassium levels of at least 6.0 mEq/L compared with patients without CKD with potassium levels of less than 5.5 mEq/L.3 Although treatments for hyperkalemia are effective and readily available,25 including the US Food and Drug Administration–approved potassium binders patiromer26 and zirconium cyclosilicate,27 the diagnosis of hyperkalemia, particularly outside of the hospital or clinic, is challenging because patients are often asymptomatic, and guideline-directed blood potassium monitoring is severely underperformed. The ability to noninvasively screen for hyperkalemia using ECG data would represent a major advance in patient care of this life-threatening condition.
In training and validating using a retrospective database of more than 1.6 million ECGs, the DLM had a high AUC of 0.853 to 0.901 for identifying hyperkalemia among patients with CKD from 2 or 4 ECG leads. The model was robust across diverse patients, geography, and year. At a high-sensitivity operating point, the DLM performed well as a potential screening tool to rule out hyperkalemia, with a negative predictive value greater than 99%. The model performance was better than other common screening tests, such as mammography for breast cancer (AUC, 0.78, positive predictive value, 3%-12%)28 and stool DNA testing for colorectal cancer and advanced precancerous lesions (AUC, 0.73).29 For patients with CKD with a clinical indication for serum potassium evaluation, such as with RAAS inhibitor medical management, the application of artificial intelligence to the ECG may enable noninvasive screening for hyperkalemia.
A prospective study is warranted to determine the association of the DLM in patients with the most to gain from improvements in the state-of-art hyperkalemia detection: those who take RAAS inhibitors (including ACE inhibitors, angiotensin receptor blockers, and mineralocorticoid receptor antagonists [MRAs]), of whom only 10% receive comprehensive guideline-recommended potassium monitoring.7 In patients with heart failure, MRAs reduce cardiovascular mortality, but only a small fraction (less than one-third of eligible patients) receive them at hospital discharge.30 A heightened fear of MRA-associated hyperkalemia and hyperkalemia-associated mortality31 and the complex potassium monitoring requirements for MRA initiation and titration6 are 2 potential reasons for underuse. A DLM that enables noninvasive potassium monitoring could assuage the fears of health care clinicians and enable more patients with CKD with heart failure to receive target doses of MRAs and other guideline-recommended medications, potentially improving care and outcomes.
This DLM used 2- or 4-lead ECG inputs from a traditional, supine 12-lead ECG in the clinic or hospital setting. It remains to be determined whether algorithm performance would be similar using other ECG inputs, such as from a 2-lead ambulatory ECG device, where the signal-to-noise ratio is lower, and in other settings such as the home; a DLM with this application would be convenient and patient friendly.15 For those patients with a clinical indication for blood potassium monitoring, such as patients with CKD taking RAAS inhibitors, a not-hyperkalemia result would reassure patient and physician of the absence of hyperkalemia, therefore avoiding a blood test. A hyperkalemia result would prompt a confirmatory and already clinically indicated serum potassium draw.
To our knowledge, this is the first deep-learning approach to evaluation of potassium levels from the ECG. Previous approaches have used standard regression models to estimate potassium and have highlighted the importance of T-wave width, T-wave amplitude, and descending T-wave slope as predictors of hyperkalemia.16,32 In contrast with traditional statistics methods, DLMs do not offer algorithmic transparency: we are unable to understand precisely how the algorithm’s heuristic arrived at its final destination. To try to understand how the DNN builds up its understanding of images for hyperkalemia over many layers, we performed feature visualization on the mean ECG beat33,34,35 (eFigure 3 in the Supplement); sometimes, expected changes in QRS and T-wave morphology were seen. In other high-probability visualizations, QRS and T-wave features were not present because there are features the DNN extracts that surpass human visualization.
Limitations
Our work is best understood in the context of its limitations. The data used were retrospective, and we did not have access to clinical data such as medication use and medical history. However, given that the network was trained using data from nearly 450 000 patients over 23 years, it is highly likely that most drugs and disease states were represented in the training set, ensuring robustness. This is supported by performance from 3 validation sets composed of geographically and racially/ethnically diverse populations. Furthermore, we evaluated algorithm performance after adjusting for age, sex, race/ethnicity, eGFR, and body mass index, and the association of the DLM output with serum hyperkalemia did not substantially or significantly change in the multivariable logistic regression analysis (eTable 1 in the Supplement).
Because the DLM was developed and validated using 12-lead ECG data in the clinic/hospital setting, additional, prospective testing is required to analyze the performance of the DLM using ECG data inputs in the home setting and, importantly, to determine whether the model improves hyperkalemia detection, care, and outcomes.
Hyperkalemia is often present in the setting of acute CKD. The DLM does not offer an analysis of renal function. However, the patients who most likely require an assessment of renal function are those patients with possible hyperkalemia, for whom the envisioned screening test would require a confirmatory blood test that typically includes renal function in addition to potassium level.
As a proposed screening test, of highest clinical concern for the DLM is false-negative results, in which the DNN predicts normokalemia, but blood tests show hyperkalemia; in this situation, the ECG test result may lead to false reassurance and undertreatment of life-threatening hyperkalemia. However, it is important to acknowledge that false-negative results may not necessarily be false because the ECG-based test, which depends on the response of cardiac tissue to blood potassium levels, is more physiologic than chemistry-based blood tests; an ECG-derived potassium level may be more germane to health and arrhythmia risk than the blood test. Another possibility is that there may have been errors in the blood tests in these patients, leading to falsely elevated potassium test results despite normokalemia. The ECG-based tests are not susceptible to mechanical, temperature, contamination, or the other potential errors associated with processing blood. One study found that 30.2% of patients with normokalemia had a pseudohyperkalemic blood test result.20 In false-negative patients, the medical record review suggested that 3 of 58 patients may have had spuriously elevated potassium blood test results owing to thrombocytosis or hemolysis. Among 52% of patients with a DLM false-negative result, a subsequent potassium measurement within 8 hours was less than our 5.5 mEq/L threshold, often without specific treatment.
Finally, the DLM is a screening test with low specificity, with upwards of 42% false-positive results, which may cause anxiety and inconvenience for patients. The risk of false-positive results will also increase over time with repeated testing. There are several potential strategies to reduce false-positive results. A personalized DLM, trained on multiple ECG-potassium pairs from a person, could improve specificity, but we did not have enough repeated measures data to develop such a model. Additional refinement of the ECG-based exclusion criteria for the DLM, such as left ventricular hypertrophy with severe repolarization abnormalities, may help reduce the likelihood of a false-positive result. Further investigation is needed to identify these and other risk factors for false-positive results. Finally, when a possible hyperkalemia result does occur, whether a true-positive or false-positive result, the education and communication of the results, as well as the messaging of the instructions for repeated ECG-based or subsequent lab testing, will be of the utmost importance.
Conclusions
Using 2 leads of an ECG acquired from patients with CKD, a DLM detected elevated potassium with an AUC of 0.853 to 0.883 and a sensitivity of 88.9% to 91.3%. The application of artificial intelligence to the ECG may enable screening for hyperkalemia. A prospectively validated screening test in the home setting is needed to improve care and outcomes in patients with renal and cardiac disease.
eFigure 1. Deep Neural Network Architecture
eMethods. Hyperkalemia Labeling in Development Dataset, Validation Dataset Creation, and Validation Dataset Sample Size Estimation
eFigure 2. Performance for Hyperkalemia from Lead I and II of Electrocardiogram, Deep Learning Model Versus Patient Demographics
eTable 1. Association of Deep Learning Model and Patient Demographics with Hyperkalemia
eTable 2. Sensitivity Analysis: Validation Dataset Performance for Hyperkalemia from the ECG, Excluding Patients with End-Stage Renal Disease
eTable 3. Sensitivity Analysis: Validation Dataset Performance for Hyperkalemia from the ECG, no ECG-Potassium Pairs Excluded
eFigure 3. Deep Neural Network Feature Visualization
References
- 1.Fitch K, Woolley JM, Engel T, Blumen H. The clinical and economic burden of hyperkalemia on medicare and commercial payers. Am Health Drug Benefits. 2017;10(4):202-210. [PMC free article] [PubMed] [Google Scholar]
- 2.Luo J, Brunelli SM, Jensen DE, Yang A. Association between serum potassium and outcomes in patients with reduced kidney function. Clin J Am Soc Nephrol. 2016;11(1):90-100. doi: 10.2215/CJN.01730215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Einhorn LM, Zhan M, Hsu VD, et al. The frequency of hyperkalemia and its significance in chronic kidney disease. Arch Intern Med. 2009;169(12):1156-1162. doi: 10.1001/archinternmed.2009.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raebel MA, Ross C, Xu S, et al. Diabetes and drug-associated hyperkalemia: effect of potassium monitoring. J Gen Intern Med. 2010;25(4):326-333. doi: 10.1007/s11606-009-1228-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kidney Disease Outcomes Quality Initiative (K/DOQI) K/DOQI clinical practice guidelines on hypertension and antihypertensive agents in chronic kidney disease. Am J Kidney Dis. 2004;43(5)(suppl 1):S1-S290. [PubMed] [Google Scholar]
- 6.Yancy CW, Jessup M, Bozkurt B, et al. ; American College of Cardiology Foundation; American Heart Association Task Force on Practice Guidelines . 2013 ACCF/AHA guideline for the management of heart failure: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol. 2013;62(16):e147-e239. doi: 10.1016/j.jacc.2013.05.019 [DOI] [PubMed] [Google Scholar]
- 7.Schmidt M, Mansfield KE, Bhaskaran K, et al. Adherence to guidelines for creatinine and potassium monitoring and discontinuation following renin-angiotensin system blockade: a UK general practice-based cohort study. BMJ Open. 2017;7(1):e012818. doi: 10.1136/bmjopen-2016-012818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooper LB, Hammill BG, Peterson ED, et al. Consistency of laboratory monitoring during initiation of mineralocorticoid receptor antagonist therapy in patients with heart failure. JAMA. 2015;314(18):1973-1975. doi: 10.1001/jama.2015.11904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Surawicz B. Relationship between electrocardiogram and electrolytes. Am Heart J. 1967;73(6):814-834. doi: 10.1016/0002-8703(67)90233-5 [DOI] [PubMed] [Google Scholar]
- 10.Wrenn KD, Slovis CM, Slovis BS. The ability of physicians to predict hyperkalemia from the ECG. Ann Emerg Med. 1991;20(11):1229-1232. doi: 10.1016/S0196-0644(05)81476-3 [DOI] [PubMed] [Google Scholar]
- 11.Salazar-Licea LA, Pedraza-Ortega JC, Pastrana-Palma A, Aceves-Fernandez MA. Location of mammograms ROI’s and reduction of false-positive. Comput Methods Programs Biomed. 2017;143:97-111. [DOI] [PubMed] [Google Scholar]
- 12.Ting DSW, Cheung CY, Lim G, et al. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA. 2017;318(22):2211-2223. doi: 10.1001/jama.2017.18152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lau JK, Lowres N, Neubeck L, et al. iPhone ECG application for community screening to detect silent atrial fibrillation: a novel technology to prevent stroke. Int J Cardiol. 2013;165(1):193-194. [DOI] [PubMed] [Google Scholar]
- 14.Halcox JPJ, Wareham K, Cardew A, et al. Assessment of remote heart rhythm sampling using the alivecor heart monitor to screen for atrial fibrillation: the REHEARSE-AF study. Circulation. 2017;136(19):1784-1794. [DOI] [PubMed] [Google Scholar]
- 15.Yasin OZ, Attia Z, Dillon JJ, et al. Noninvasive blood potassium measurement using signal-processed, single-lead ecg acquired from a handheld smartphone. J Electrocardiol. 2017;50(5):620-625. doi: 10.1016/j.jelectrocard.2017.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Velagapudi V, O’Horo JC, Vellanki A, et al. Computer-assisted image processing 12 lead ECG model to diagnose hyperkalemia. J Electrocardiol. 2017;50(1):131-138. [DOI] [PubMed] [Google Scholar]
- 17.Rossignol P, Dobre D, McMurray JJ, et al. Incidence, determinants, and prognostic significance of hyperkalemia and worsening renal function in patients with heart failure receiving the mineralocorticoid receptor antagonist eplerenone or placebo in addition to optimal medical therapy: results from the Eplerenone in Mild Patients Hospitalization and Survival Study in Heart Failure (EMPHASIS-HF). Circ Heart Fail. 2014;7(1):51-58. [DOI] [PubMed] [Google Scholar]
- 18.Yusuf AA, Hu Y, Singh B, Menoyo JA, Wetmore JB. Serum potassium levels and mortality in hemodialysis patients: a retrospective cohort study. Am J Nephrol. 2016;44(3):179-186. doi: 10.1159/000448341 [DOI] [PubMed] [Google Scholar]
- 19.Pepe MS, Etzioni R, Feng Z, et al. Phases of biomarker development for early detection of cancer. J Natl Cancer Inst. 2001;93(14):1054-1061. [DOI] [PubMed] [Google Scholar]
- 20.Friedman PA, Scott CG, Bailey K, et al. Errors of classification with potassium blood testing: the variability and repeatability of critical clinical tests. Mayo Clin Proc. 2018;93(5):566-572. doi: 10.1016/j.mayocp.2018.03.013 [DOI] [PubMed] [Google Scholar]
- 21.Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aslam S, Friedman EA, Ifudu O. Electrocardiography is unreliable in detecting potentially lethal hyperkalaemia in haemodialysis patients. Nephrol Dial Transplant. 2002;17(9):1639-1642. doi: 10.1093/ndt/17.9.1639 [DOI] [PubMed] [Google Scholar]
- 23.Kovesdy CP. Management of hyperkalaemia in chronic kidney disease. Nat Rev Nephrol. 2014;10(11):653-662. doi: 10.1038/nrneph.2014.168 [DOI] [PubMed] [Google Scholar]
- 24.Weiner ID, Wingo CS. Hyperkalemia: a potential silent killer. J Am Soc Nephrol. 1998;9(8):1535-1543. [DOI] [PubMed] [Google Scholar]
- 25.Sood MM, Sood AR, Richardson R. Emergency management and commonly encountered outpatient scenarios in patients with hyperkalemia. Mayo Clin Proc. 2007;82(12):1553-1561. doi: 10.1016/S0025-6196(11)61102-6 [DOI] [PubMed] [Google Scholar]
- 26.Weir MR, Bakris GL, Bushinsky DA, et al. ; OPAL-HK Investigators . Patiromer in patients with kidney disease and hyperkalemia receiving RAAS inhibitors. N Engl J Med. 2015;372(3):211-221. [DOI] [PubMed] [Google Scholar]
- 27.Packham DK, Rasmussen HS, Lavin PT, et al. Sodium zirconium cyclosilicate in hyperkalemia. N Engl J Med. 2015;372(3):222-231. [DOI] [PubMed] [Google Scholar]
- 28.Pisano ED, Gatsonis C, Hendrick E, et al. ; Digital Mammographic Imaging Screening Trial (DMIST) Investigators Group . Diagnostic performance of digital versus film mammography for breast-cancer screening. N Engl J Med. 2005;353(17):1773-1783. doi: 10.1056/NEJMoa052911 [DOI] [PubMed] [Google Scholar]
- 29.Imperiale TF, Ransohoff DF, Itzkowitz SH. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014;371(2):187-188. [DOI] [PubMed] [Google Scholar]
- 30.Rao KK, Enriquez JR, de Lemos JA, et al. Use of aldosterone antagonists at discharge after myocardial infarction: results from the National Cardiovascular Data Registry Acute Coronary Treatment and Intervention Outcomes Network (ACTION) Registry-Get with the Guidelines (GWTG). Am Heart J. 2013;166(4):709-715. [DOI] [PubMed] [Google Scholar]
- 31.Juurlink DN, Mamdani MM, Lee DS, et al. Rates of hyperkalemia after publication of the Randomized Aldactone Evaluation Study. N Engl J Med. 2004;351(6):543-551. doi: 10.1056/NEJMoa040135 [DOI] [PubMed] [Google Scholar]
- 32.Attia ZI, DeSimone CV, Dillon JJ, et al. Novel bloodless potassium determination using a signal-processed single-lead ECG. J Am Heart Assoc. 2016;5(1):e002746. doi: 10.1161/JAHA.115.002746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Erhan D, Courville A, Bengio Y. Visualizing Higher-Layer Features of a Deep Network: Technical Report 1341. Montreal, Quebec, Canada: University of Montreal; 2009. [Google Scholar]
- 34.Olah C, Mordvintsev A, Schubert L Feature visualization. https://distill.pub/2017/feature-visualization/. Published 2017. Accessed May 1, 2018.
- 35.Mordvintsev A, Olah C, Tyka M. Inceptionism: going deeper into neural networks. https://ai.googleblog.com/2015/06/inceptionism-going-deeper-into-neural.html. Accessed May 1, 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eFigure 1. Deep Neural Network Architecture
eMethods. Hyperkalemia Labeling in Development Dataset, Validation Dataset Creation, and Validation Dataset Sample Size Estimation
eFigure 2. Performance for Hyperkalemia from Lead I and II of Electrocardiogram, Deep Learning Model Versus Patient Demographics
eTable 1. Association of Deep Learning Model and Patient Demographics with Hyperkalemia
eTable 2. Sensitivity Analysis: Validation Dataset Performance for Hyperkalemia from the ECG, Excluding Patients with End-Stage Renal Disease
eTable 3. Sensitivity Analysis: Validation Dataset Performance for Hyperkalemia from the ECG, no ECG-Potassium Pairs Excluded
eFigure 3. Deep Neural Network Feature Visualization