Abstract
The transcription factor nuclear factor erythroid-2 (NF-E2)-related factor 2 (NRF2) is a central regulator of redox, metabolic, and protein homeostasis that intersects with many other signaling cascades. While the understanding of the complex nature of NRF2 signaling continues to grow, there is only one therapeutic targeting NRF2 for clinical use, dimethyl fumarate, used for the treatment of multiple sclerosis. The discovery of new therapies is confounded by the fact that NRF2 levels vary significantly depending on physiological and pathological context. Thus, properly timed and targeted manipulation of the NRF2 pathway is critical in creating effective therapeutic regimens. In this review, we summarize the regulation and downstream targets of NRF2. Furthermore, we discuss the role of NRF2 in cancer, neurodegeneration, and diabetes, as well as cardiovascular, kidney, and liver disease, with a special emphasis on NRF2-based therapeutics, including those that have made it into clinical trials.
Keywords: NRF2, KEAP1, therapeutics, cancer, disease, clinical trials
Introduction:
Originally characterized as a master regulator of redox homeostasis, the transcription factor nuclear factor erythroid-2 (NF-E2)-related factor 2 (NRF2) continues to emerge as a critical mediator of a diverse array of cellular functions. Through the ongoing identification of novel regulators, target genes, and disease contexts, a role for NRF2 has been indicated not only in redox homeostasis, but also drug/xenobiotic metabolism, DNA repair, mitochondrial function, iron, lipid and carbohydrate metabolism, proteostasis, and proliferation, all of which contribute to cell survival (1). While the beneficial role of NRF2 is well established, it has become increasingly clear that careful regulation of this pathway is critical for disease prevention. As the complex interplay between NRF2 physiology and pathology is revealed through careful mechanistic studies, a need for new compounds that allow for context-dependent modulation of the NRF2 signaling cascade continues to grow.
Due to new research revealing the previously unappreciated complexity of the NRF2 signaling network, an increasing number of NRF2-based therapies continue to be brought to light; however, the progression of therapeutics from bench to bedside continues to lag behind. Adding to this deficiency is the fact that the number of pathological contexts in which NRF2 plays a role, either directly or indirectly, is expanding rapidly. In this review, we summarize the currently identified regulators and downstream targets of NRF2, its crosstalk with other signaling pathways, as well as the identified role of NRF2 in cancer and other diseases. We also highlight current therapies that have progressed to clinical trials, and other pharmacological modulators of NRF2 that could be developed into therapeutics for the treatment and prevention of disease.
Overview of the NRF2 signaling pathway:
Key discoveries in the NRF2 field: An historical perspective
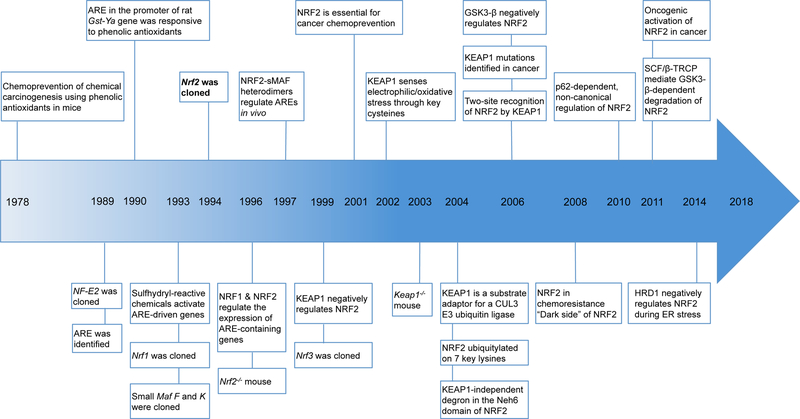
The NRF2 field is relatively young, as the majority of the key discoveries have occurred over the past four decades (Figure 1). Although NRF2 was first cloned in 1994, the original notion of transcriptional control of the antioxidant response originated in the 1970s, when a number of studies demonstrated that the anti-carcinogenic effects of phenolic antioxidants were due to their ability to increase activation of phase II detoxifying enzymes, such as glutathione-S-transferase (GST) (2; 3). While it was unknown at the time that GST is regulated by NRF2, this was some of the first evidence that activation of enzymes that detoxify reactive intermediates, many of which are now established NRF2 transcriptional targets, could be useful for preventing chemical-induced carcinogenesis. In 1989, a protein that binds to a consensus sequence containing an activator protein 1 (AP-1) core motif, nuclear factor erythroid 2 (NF-E2) was identified (4). NF-E2 was the first of six mammalian cap’n’collar (CNC), basic leucine zipper (bZIP) transcription factors found to bind this motif, with the others, NRF1 (5), NRF2 (6), NRF3 (7), BACH1 (8), and BACH2 (9) being identified shortly thereafter. This NF-E2/AP-1 sequence was later identified in the promoter of the rat GST-Ya gene, and was responsive to t-butylhydroquinone (t-BHQ), resulting in the coining of the term “antioxidant response element” (ARE) (10). It was then discovered that a wide array of sulfhydryl reactive chemicals induce ARE-driven gene expression, indicating that the transcription of phase II detoxifying enzymes was sensitive to electrophiles (11), a key functional feature of many currently identified activators of NRF2.
Figure 1. Key discoveries in the NRF2 field.
Timeline depicting important discoveries in the NRF2 field over the past four decades.
Many of the studies detailing the molecular interactions that drive NRF2 signaling were conducted in the mid to late 1990s. During this time, the domains of NRF2 were characterized (12), it was demonstrated that NRF2 dimerizes with small MAF (sMAF) proteins to activate transcription of ARE-containing genes (13), and it was determined that NRF2 is negatively regulated at the protein level by Kelch-like ECH associated protein 1 (KEAP1) (14). Also, the first Nrf2−/− mouse was generated, which developed normally (15), but exhibited a significant reduction in the levels of GST and NAD(P)H:quinone oxidoreductase (NQO1) in the intestines and liver compared to wild type mice following administration of phenolic antioxidants (13). Further, the Nrf2−/− mouse was critical in demonstrating that NRF2 plays a direct role in chemoprevention, as the chemoprotective effects of oltipraz, a dithiolethione, were lost in Nrf2−/− mice (16).
Once the chemoprotective role of NRF2 was introduced, the field shifted towards identifying ways to activate the NRF2 pathway. As many of the activators of key phase II detoxifying enzymes were electrophiles, the belief was that the redox sensitivity of NRF2 was conferred by a regulator capable of sensing oxidative/electrophilic stress. In 2002, it was shown in vitro that KEAP1 senses electrophilic stress via specific cysteine residues, with adduction of electrophilic inducers to the thiol group disrupting the KEAP1-NRF2 interaction (17). A year later, it was demonstrated in cells that different KEAP1 cysteines were sensitive to different inducers, a concept known as the “cysteine code” (18). Around the same time, studies showed that KEAP1 recruits the Cullin 3-Ring box 1 (CUL3-RBX1) E3 ubiquitin ligase complex that ubiquitylates seven key lysine residues that target NRF2 for degradation (19; 20). Not long after, it was discovered that KEAP1-independent degradation of NRF2 can occur as a result of glycogen synthase kinase β (GSK3-β) phosphorylation of key residues in NRF2, which triggers the recruitment of the S-phase kinase associated protein 1-Cullin1-Rbx1/β-transducin repeat containing protein (SCF/β-TrCP) E3 ligase complex (21; 22).
In 2006, the discovery that KEAP1 is mutated in non-small lung cell carcinomas, leading to chronically elevated levels of NRF2, presented the first evidence that NRF2 could contribute to cancer progression and chemoresistance, later termed the “dark side” of NRF2 (23; 24). This led to a shift in the field towards investigating the role of high NRF2 expression in cancer, and the premise that inhibition of NRF2 may be necessary to treat certain cancers. Another landmark discovery regarding the dark side was that autophagic dysfunction, which results in the p62-dependent sequestration of KEAP1, leads to prolonged activation of NRF2 in a non-canonical, cysteine-independent manner (25; 26). As the NRF2 field continues to expand, our understanding of the complexity of this pathway continues to advance.
Negative regulators and modes of activation of NRF2
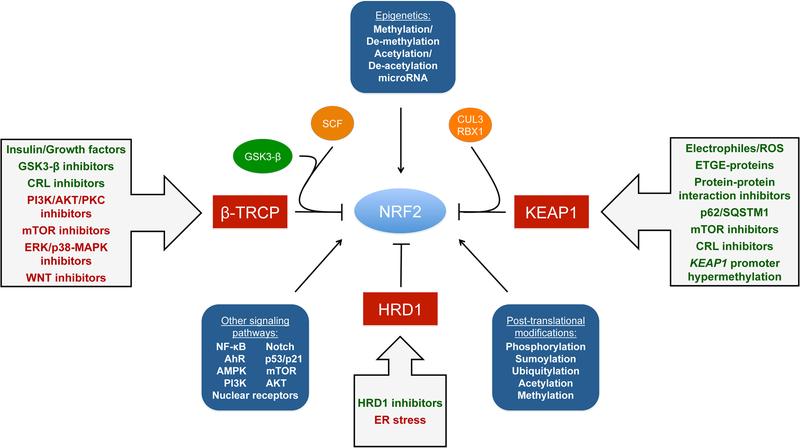
Regulation of NRF2 mainly occurs through the controlled maintenance of NRF2 protein levels. There are three E3 ubiquitin ligase complexes responsible for the ubiquitylation and degradation of NRF2, the CUL3-RBX1-KEAP1 complex, the SCF/β-TrCP complex, and HRD1. Each mediates the degradation of NRF2 upon different stimuli in specific subcellular compartments. The CUL3-RBX1-KEAP1 complex responds to electrophilic/oxidative stress in the cytosol. The SCF/β-TrCP complex, which can be nuclear or cytosolic, is more sensitive to metabolic changes, and is regulated by GSK3-β. HRD1 is localized to the ER and has only been demonstrated to ubquitylate NRF2 during ER stress (27). It is important to note that other signaling pathways, epigenetic factors, and post-translational modifications also regulate NRF2. Furthermore, activation or inhibition of the NRF2 pathway can be achieved by targeting the negative regulation of NRF2 (Figure 2).
Figure 2. Regulation of NRF2 and possible modes of activation.
Schematic representation of NRF2 modes of regulation. NRF2 is regulated at the post-transcriptional and post-translational level, as well as via epigenetic factors and interaction with other signaling pathways. Modulation of NRF2 protein levels can be achieved through activation or inhibition of the KEAP1-CUL3-RBX1 complex, SCF-β-TrCP complex, or HRD1. Electrophilic/oxidative modification of key cysteines, competitive binding of ETGE containing proteins, protein-protein interaction inhibitors, increased levels of p62/SQSTM1, mTOR inhibitors, and CUL3-Ring E3 ligase (CRL) inhibitors (i.e. MLN4924), can all disrupt the KEAP1-NRF2 interaction. Hypermethylation of the KEAP1 promoter can also increase expression of NRF2. The SCF/β-TrCP-NRF2 interaction can be modulated by insulin/growth factors, or GSK3-β, CRL, PI3K-AKT-PKC, mTOR, ERK/p38-MAPK, and WNT inhibitors. Inhibitors of HRD1 (i.e. LS-102) could be utilized to prevent ER-stress associated degradation of NRF2.
NRF2 target genes
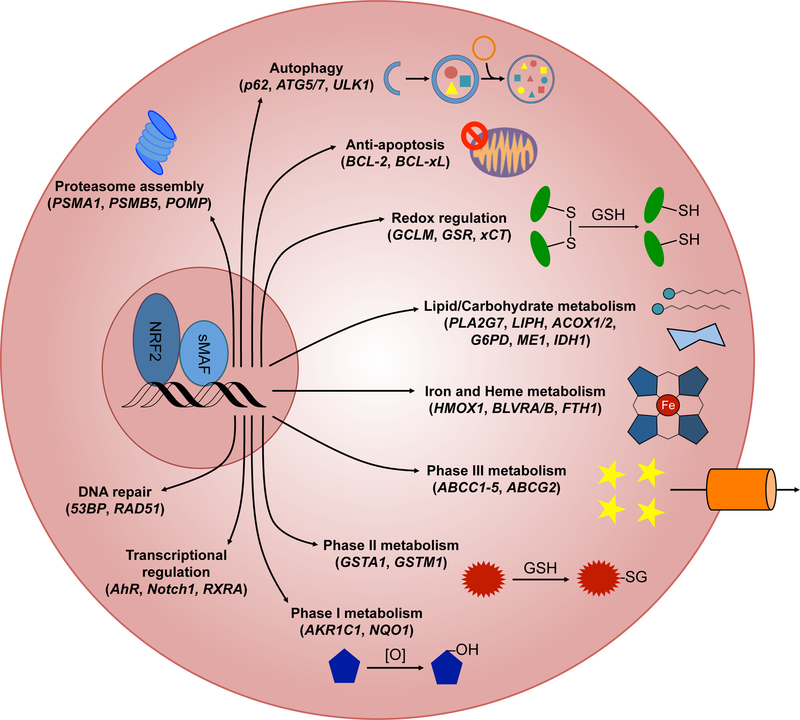
There are over 250 currently identified NRF2 target genes involved in a multitude of cellular processes, including: redox regulation; phase I, II, and III drug/xenobiotic metabolism; protein homoeostasis; DNA repair; carbohydrate and lipid metabolism; iron homeostasis; transcriptional regulation; and mitochondrial function (Figure 3).
Figure 3. Cellular pathways driven by NRF2 target genes.
NRF2 heterodimerizes with sMAF proteins to initiate the transcription of ARE-containing target genes. Verified NRF2 target genes are involved in proteasome assembly, autophagy, prevention of apoptosis, maintaining redox balance, lipid and carbohydrate metabolism, heme metabolism, iron homeostasis, all three phases of drug/xenobiotic metabolism, transcriptional regulation of other transcription factors, and DNA repair. Representative target genes are included in parentheses below each transcriptional response.
Redox regulation
Perhaps the best-known function of NRF2 is maintaining redox homoeostasis, mainly via the synthesis and redox cycling of GSH and thiol-based antioxidant enzymes. For example, the catalytic and modulatory subunits (GCLM and GCLC) of glutamate cysteine ligase (GCL) are NRF2 target genes, with GCL being responsible for the de novo synthesis of glutathione. Additionally, glutathione peroxidases (GPX2 and GPX4), which utilize GSH to reduce peroxides, glutathione reductase (GSR), that reduces oxidized glutathione, and peroxiredoxins (PRDX1 and PRDX6), which reduce peroxides directly, are all NRF2 target genes (28). The reduction of oxidized protein thiols by the coordinated action of thioredoxin 1 and thioredoxin reductase 1 (TXN1 and TXNRD1), as well as the import of cysteine via the xCT transporter (SLC7A11) which is critical for GSH production, are also transcriptionally regulated by NRF2 (28).
Drug/xenobiotic metabolism
NRF2 regulates all three phases of drug/xenobiotic metabolism. Examples of NRF2 targets involved in phase I metabolism are aldo-keto reductase family members (i.e. AKR1C1, AKR1B1, and ARK1B10) (29), aldehyde dehydrogenase family members (i.e. ALDH1A1, ALDH3A1, and ALDH7A1) (30), and NQO1 (13), which are involved in the reduction of toxicants/drugs to active metabolites. This allows phase II NRF2 transcriptional targets, including glutathione S-transferases (i.e. GSTA1–4 and GSTM1–4) (31) and UDP glucuronosyltransferases (i.e. UGT1A1 and UGT2B7) (32; 33) to conjugate these intermediates to glutathione (GSH) or glucuronic acid, respectively. Finally, NRF2 also regulates the transcription of phase III membrane transporters (the ATP-binding cassette family members/multidrug resistance proteins (i.e. ABCC1-5 and ABCG2)) (34; 35) that excrete xenobiotics/drugs from the cell.
Protein homeostasis
NRF2 downstream genes are involved in maintaining proteostasis via autophagy and the ubiquitin proteasome system (UPS). It was recently identified that a number of autophagy initiation proteins, including autophagy related (ATG) 5 and 7, as well as unc-51 like autophagy activating kinase (ULK) 1 and 2, contain putative AREs (36). NRF2 may directly regulate mTOR, a master regulator of both protein translation and autophagy (37). Furthermore, p62/SQSTM1, a protein that targets ubiquitylated proteins for autophagic degradation, is an NRF2 target. NRF2 also controls expression of proteasomal subunits (i.e. proteasome subunit alpha 1 (PSMA1) and proteasome subunit beta 5 (PSMB5)) (38), as well as the proteasome maturation protein (POMP) (39), which are critical for proteasome assembly.
Lipid, carbohydrate, and iron metabolism
NRF2 regulates a number of metabolic enzymes necessary for processing glucose and fatty acids for anabolic metabolism. Pentose phosphate pathway enzymes, including glucose-6-phosphate dehydrogenase (G6PD), 6-phosphogluconate dehydrogenase (PGD), and transaldolase (TALDO1), which are critical in regenerating NADPH, are all NRF2 target genes (40). NRF2 also modulates the expression of malic enzyme (ME1) and isocitrate dehydrogenase 1 (IDH1), two key TCA cycle enzymes (40; 41). Lipid catabolism is regulated by NRF2, as lipase H (LIPH), phospholipase A2 (PLA2G7), and acetyl-CoA oxidase 2 (ACOX2) (42) are NRF2 targets. Finally, NRF2 regulates heme metabolism and iron homeostasis through the transcription of heme oxygenase 1 (HMOX1), biliverdin A and B (BVRD½), and the light and heavy chains of ferritin (FTH1/FLH1) (43).
Transcriptional regulation, DNA repair, and prevention of apoptosis
Functional AREs have been identified in the promoters of a number of transcription factors, including aryl hydrocarbon receptor (AhR), neurogenic locus notch homolog protein 1 (NOTCH1), and retinoic receptor α (RXRA), indicating NRF2 can indirectly control the transcription of a host of non-ARE-containing genes, discussed in detail below. NRF2 also plays a role in DNA damage repair and preventing apoptosis, as p53-binding protein 1 (53BP1), DNA repair protein RAD51 homolog 1 (RAD51) (44), anti-apoptotic proteins B-cell lymphoma 2 (BCL2), and B-cell lymphoma-extra-large (BCLXL) (45; 46), are transcriptionally regulated by NRF2. The diversity of NRF2 targets indicates the central role of NRF2 in mediating cellular function.
Crosstalk with other signaling pathways
While NRF2 directly regulates a host of cellular responses via its transcriptional targets, it can also mediate cellular function via crosstalk with other major signaling cascades. These pathways, which include NOTCH1, AhR, nuclear factor-κB (NF-κB), cellular tumor antigen p53 (p53), AMP-activated protein kinase (AMPK), PI3K-AKT, and mTOR, are critical in responding to stress, highlighting a key role for NRF2 in maintaining cell survival.
The NOTCH signaling pathway mediates cell cycle and apoptosis during embryonic development and the determination of cell fate. The NOTCH1 gene has a functional ARE in the proximal region of its promoter, and NOTCH signaling is diminished in Nrf2−/− mouse embryonic fibroblasts (47). This NRF2-NOTCH signaling axis is critical during liver regeneration, as Nrf2−/− mice demonstrate delayed liver regrowth compared to wild type mice following partial hepatectomy; however, crossing these mice with a mouse strain that over-expresses the NOTCH-intracellular domain (NICD), completely rescues the NRF2 null phenotype (47). Conversely, NRF2 can be regulated by NOTCH, as the RBPjκ sequence, which is required for canonical NOTCH transcription, is highly conserved in the NRF2 promoter region across animals. Furthermore, NICD-overexpressing mice exhibit hyperactivation of NRF2, displaying a phenotype very similar to liver-specific Keap1−/− mice (48). Thus, crosstalk between NRF2 and NOTCH appears to play an integral role in mediating cytoprotection, particularly in the context of liver regeneration.
NRF2 signaling also overlaps with pathways that respond to exogenous stressors. For example, AhR, which forms a complex with aryl hydrocarbon receptor nuclear translocator (ARNT), can be bound by polycyclic aromatic hydrocarbons, such as dioxin, and translocate from the cytosol to the nucleus to regulate transcription of xenobiotic-responsive element (XRE) containing genes (49). NRF2 and a number of its target genes, such as GST (50) and NQO1 (51), also contain an XRE. Moreover, it was shown that NRF2 could directly regulate AhR transcription via an ARE (52), as well as the bardoxolone-imidazolide (synthetic, triterpenoid, CDDO-Im) inducible transcription of AhR target genes (53).
Another major pathway that has a bidirectional relationship with NRF2 is the NF-κB pathway. NF-κB has been shown to repress transcription of genes containing AREs, as both NRF2 and p65/RelA, a component of the NF-κB transcriptional complex, require CREB-binding protein (CBP) to transcribe their respective target genes (54). Thus, if p65 is bound to CBP it prevents NRF2-driven transcription of the ARE. Interestingly, a number of NRF2 activators, including sulforaphane (SF) (55), bardoxolone methyl (CDDO-Me) (56), and curcumin (57) have all been shown to suppress NF-κB signaling. In contrast, Nrf2−/− mice exhibit increased signs of NF-κB activation compared to wild type mice following stimulation with pro-inflammatory stimuli (58). It has also been demonstrated that inhibitor of NF-κB kinase β-subunit (IKKβ), a well-established negative regulator of NF-κB, is ubiquitylated and degraded by the KEAP1-CUL3-RBX1 complex (59). Lastly, a number of NRF2 target genes, such as NQO1, TRX1, and HMOX1, have all been shown to modify NF-κB-driven transcription, indicating these pathways crosstalk at both the upstream and downstream levels (60).
NRF2 directly interacts with the p53/p21 cascade, an important target for anti-cancer therapies due to its regulation of apoptosis. Specifically, co-expression of NRF2 and p53 results in the suppression of SLC7A11, NQO1, and GST in a variety of cell lines (61). It has been shown that NRF2 may control the expression of E3-ubiquitin protein ligase MDM2, a negative regulator of p53, implying that NRF2 can directly regulate the activity of p53 (62). Furthermore, p21 (CDKN1A), a p53 target gene involved in the regulation of cell cycle, apoptosis, and differentiation, directly binds to the DLG and ETGE motifs of NRF2, resulting in its stabilization and transcription of ARE-containing genes (63). The interaction between NRF2 and p53 is further confirmed by the fact that Nrf2−/−;p53+/− mice are more susceptible to nitrosamine-induced carcinogenesis than their Nrf2−/− or p53+/− counterparts (64), indicating that both are critical in mediating cell survival during stress.
NRF2 also interacts with key pathways that respond to changes in metabolism. For example, AMPK, a sensor of the overall energetic state of the cell via the AMP:ATP ratio, phosphorylates NRF2 at S550, enhancing its nuclear translocation (65). Similarly, GSK3-β, which as mentioned earlier is necessary for the degradation of NRF2 by β-TrCP, is phosphorylated by the PI3K-AKT signaling axis (66) or mTOR (67), indicating the stability of NRF2 can be determined by these two pathways. It has also been shown that mTOR phosphorylates p62/SQSTM1, enhancing its interaction with KEAP1, leading to autophagic degradation of the p62-KEAP1 complex and upregulation of NRF2 (68). Overall, NRF2 interacts with a number of key signaling cascades responsible for dictating cell growth and survival.
Other modes of NRF2 regulation
Epigenetic control of the NRF2 signaling pathway, as well as a number of single nucleotide polymorphisms in KEAP1 and NRF2/NFE2L2 that affect NRF2 expression, has been extensively studied in the context of cancer. Promoter methylation regulates the expression of NRF2 and KEAP1, as well as a number of NRF2 target genes including GST, NQO1, GPX, and UGT1A1 (69). Chemotherapy can decrease methylation of the NRF2 promoter, as 5-fluorouracil (5-FU)-induced ROS production results in hypomethylation of the NRF2 promoter and elevated NRF2 levels in drug-resistant colon cancer (70). NRF2 activity can be affected by hypermethylation of the KEAP1 promoter, which results in increased expression of NRF2. Not all epigenetic modifications enhance NRF2 activity, as hypermethylation of the NRF2 promoter suppresses NRF2 in late stage prostate cancer (71). The acetylation/deacetylation, methylation/demethylation of histones, as well as microRNAs have also been shown to affect NRF2 activation (69). Additional studies are needed to determine the potential epigenetic targets for modulating the NRF2 pathway in disease.
Aside from ubiquitylation and phosphorylation of key residues in the Neh2 and Neh6 domains to control its stability, NRF2 can also be modified by a number of other post-translational modifications (PTMs), including methylation, acetylation, and SUMOylation. Activity of NRF2 is increased by acetylation of key lysine residues in the Neh1 domain by p300/CBP, enhancing its DNA binding capability, which is reversed by deacetylation (72). Methylation of NRF2 on R437 also affects its transcriptional activity (73). As mentioned above, phosphorylation of NRF2 targets it for degradation; however, phosphorylation of other serine residues by a number of upstream kinases also occurs (S40, S215, S408, S550 and S577) (65; 74; 75). Phosphorylation of these serine residues has been proposed to increase NRF2 translocation to the nucleus, although the effect on the transcriptional activity of NRF2 may depend on the kinase/serine combination. Another identified NRF2 PTM is SUMOylation, which enhances the ubiquitylation and degradation of NRF2 in HepG2 cells (76), whereas in pancreatic β-islet cells, it enhances NRF2 activity and promotes cell survival (77), indicating the cell type-specific relevance of NRF2 PTMs.
Targeting NRF2 in disease:
Modulating NRF2 for the prevention and treatment of cancer and other chronic diseases
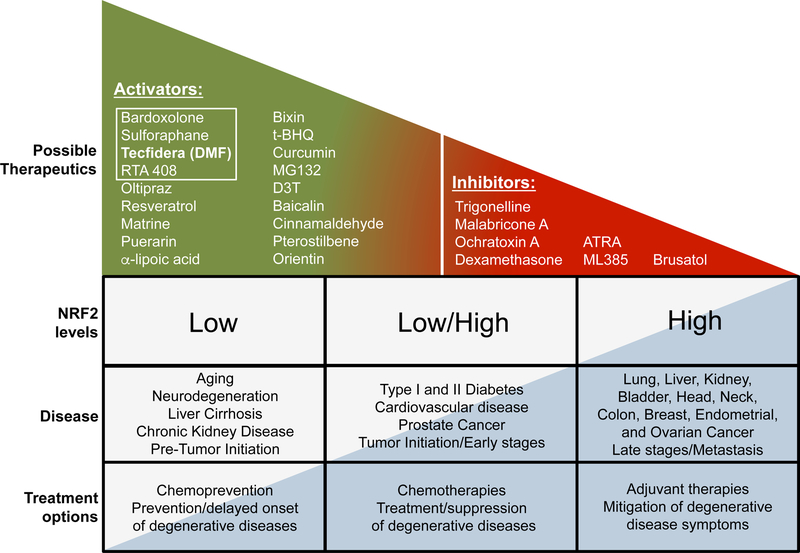
There is a developing theme in the NRF2 field that targeting NRF2 in disease is both context and time dependent. Harnessing the beneficial effects of pharmacological activation of NRF2 remains an important aspect of NRF2-based chemoprevention, and intervention in other chronic diseases, such as neurodegeneration, diabetes, cardiovascular disease, and chronic kidney and liver disease. However, a growing number of studies have revealed that NRF2 is already high in certain cancer and disease stages, indicating that pharmacological agents designed to mitigate the potentially harmful or transformative effects associated with prolonged activation of NRF2 should also be considered. Examples of current NRF2 activators and inhibitors, as well as NRF2 expression levels in disease are summarized in Figure 4.
Figure 4. Targeting NRF2 in disease is time and expression level dependent.
Graphical representation of NRF2 activators/inhibitors, and the current treatment options based on NRF2 levels in cancer and other chronic diseases. A number of NRF2 activators and inhibitors have been characterized that could be developed into translational therapeutics. The white box outlines therapies that have made it into clinical trials. DMF (Tecfidera), the only FDA approved drug, is indicated in bold. NRF2 levels and treatment possibilities vary depending on the timing and stage of disease. NRF2 activators are thought to provide the most therapeutic benefit prior to cancer initiation or onset of neurodegenerative or chronic inflammatory diseases. Some diseases, such as diabetes, cardiovascular disease, prostate cancer, inflammatory diseases, and post-initiation/early stage cancer have been reported to have either low or high levels of NRF2 depending on the context. Treatment options following onset of disease target NRF2 to intervene and prevent/delay progression. Many cancer types exhibit constitutively high levels of NRF2. For diseases where high levels of NRF2 are having a deleterious effect, an NRF2 inhibitor or adjuvant approach would be most effective. Cancers and late stage diseases with high levels of NRF2 are generally associated with poor prognosis, and treatments at this stage are designed to mitigate symptoms and enhance the efficacy of other therapies.
NRF2 in chemoprevention
To date, hundreds of studies have evaluated the in vitro and in vivo chemopreventive properties of NRF2-activating compounds (78; 79). Importantly, most of these chemopreventive compounds (or their bioactive metabolites) are electrophilic and activate NRF2 through the modification of cysteines, although peptides and small molecules that bind to the Kelch domain of KEAP1 have also been developed (80). To date, SF (natural, isothiocyanate), CDDO-Me (synthetic, triterpenoid), RTA 408 (synthetic, triterpenoid, Omaveloxolone), and DMF (synthetic, Tecfidera) are the only NRF2 activators that have entered clinical trials in the US, with only Tecfidera having been approved for the treatment of multiple sclerosis (MS). Clearly, the development of safe, potent, and specific NRF2-based therapies is still an area of exploration for cancer prevention. However, there is controversy regarding the use of NRF2 activators which may promote the dark side of NRF2 and support progression of pre-existing tumors. Thus, target populations and dosing schemes should be very carefully timed and planned depending on NRF2 level and the cancer stage.
Constitutive NRF2 activation in cancer
Considering the survival advantage conferred by NRF2, it is not surprising that cancer cells hijack the NRF2 pathway. Mutations in the Kelch or intervening region (IVR) domains of KEAP1 reduce its interaction with NRF2, resulting in constitutive activation in lung cancer cell lines and tissues (81). Additionally, several somatic mutations in KEAP1, as well as loss of heterozygocity (LOH) of the wild type allele, increase NRF2 and confer chemoresistance (23). Importantly, studies have corroborated that in lung adenocarcinoma, KEAP1 is as frequently mutated as TP53, whereas in lung squamous carcinoma both KEAP1 and NRF2 are among the most mutated genes (82; 83). The Cancer Genome Atlas (TCGA) project reported mutations and copy number alterations for KEAP1 and NRF2, as well as deletions or mutations of CUL3, in over 30% of squamous cell carcinoma cases (84). Studies have also identified reduced expression of RBX1 as a result of deletion or promoter hypermethylation (85; 86). Mechanisms that result in constitutive activation of NRF2, and the cancer types in which they are observed, are summarized in Table 1.
Table 1.
Genetic alterations in NRF2 pathway genes resulting in high expression of NRF2, as well as the tumor types in which they have been described.
| Gene | Alteration | Tumor types | References |
|---|---|---|---|
| KEAP1 | Somatic mutations (±LOH) | Lung (adenocarcinoma, squamous cell carcinoma, papillary carcinoma), breast, gallbladder, sporadic papillary renal cell carcinoma, clear cell renal cell carcinoma, ovarian, colorectal, prostate, liver, endometrial, head and neck | (23; 81–85; 87–93) |
| Promoter hypermethylation | Lung, prostate, malignant glioma, colorectal, breast, head and neck, papillary thyroid carcinoma, clear cell renal cell carcinoma | (85; 86; 94–99) | |
| Succination by fumarate | Hereditary type 2 papillary renal cell carcinoma | (100) | |
| CNV - deletion | Head and neck | (85) | |
| NRF2 | Somatic mutations | Clear cell renal cell carcinoma, lung squamous cell carcinoma, head and neck, skin squamous cell carcinoma, esophagus, larynx, endometrial, sporadic type 2 papillary renal cell carcinoma | (82; 84; 90; 91; 93; 101; 102) |
| CNV - amplification | Squamous cell lung cancer | (84) | |
| Splice variants | Lung, head and neck | (103) | |
| CUL3 | Somatic mutations | Sporadic type 2 papillary renal cell carcinoma, clear cell renal cell carcinoma, head and neck | (85; 90; 91) |
| CNV - deletion | Clear cell renal cell carcinoma, squamous cell lung cancer, head and neck | (84; 85; 91) | |
| Promoter hypermethylation | Papillary thyroid carcinoma | (86) | |
| RBX1 | CNV - deletion | Head and neck, ovarian, papillary thyroid carcinoma | (85; 86; 104) |
| Promoter hypermethylation | Ovarian | (104) |
NRF2 inhibitors in overcoming cancer resistance
Screening of natural product extracts resulted in the discovery of the first NRF2 inhibitor, brusatol (natural, quassinoid) (105). Brusatol sensitized several cancer cell lines to chemotherapeutic drugs by reducing the protein levels of NRF2 and its target genes (105). In combination with cisplatin, brusatol enhanced the chemotherapeutic sensitivity of lung cancer cells in a xenograft model, and overall tumor burden in a mouse lung cancer model (LSL-KrasG12D/+) (105). While two recent publications have determined that brusatol is a global translation inhibitor (106; 107), it continues to be a useful tool to study NRF2 inhibition in cancer chemosensitization, since short-lived proteins such as NRF2 are most susceptible to protein translation inhibition.
Other NRF2 inhibitors include ARE expression modulator 1 (AEM1), a small molecule inhibitor with an unknown mechanism of action (108), and ML385, a molecule that binds to the Neh1 domain of NRF2, blocking dimerization with MAFG and binding to the ARE (109). ML385 is the most intriguing of the current inhibitors, but it is unorthodox that it leads to a dramatic dose and time dependent decrease in NRF2 mRNA and protein levels. This is unlikely to result from direct inhibition of ARE-binding, and thus warrants further investigation. Other natural products that negatively regulate NRF2 activity without a precisely defined mode of action include trigonelline (110), malabaricone A (111), ochratoxin A (112), and wogonin (113). Additionally, nuclear receptors can bind directly to NRF2 and inhibit its transactivation activity, so some studies have proposed dexamethasone/budesonide (glucocorticoid receptor agonists) and all-trans retinoic acid (ATRA, retinoid acid receptor agonist) as NRF2 inhibitors (114). However, since the primary targets of these compounds are nuclear receptors and not NRF2 specifically, these are unlikely to be developed as NRF2-targeted therapeutics.
NRF2 in neurodegeneration
Similar to cancer, the role of NRF2 in neurodegenerative diseases is complex. Interestingly, reduced expression of NRF2 is associated with an age-related decline in neural stem cell function (115), and the NRF2 response to oxidative stress also diminishes with age (116). During neurodegeneration, NRF2 can be activated or suppressed depending on the affected cell type and stage of disease. For example, some studies have indicated that NQO1 and HMOX1 are decreased in Alzheimer’s disease (AD) brains; whereas others indicate that NRF2 remains confined to the cytosol, resulting in decreased target gene expression (117). NRF2 and NQO1 expression is elevated in infiltrating macrophages and astrocytes found in active, but not inactive, MS lesions; whereas NQO1, HMOX1 and PRDX levels, as well as nuclear localization of NRF2, are consistently elevated in the substantia nigra of Parkinson’s disease (PD) patients (117). In contrast, NRF2 protein levels are decreased in the primary motor cortex and spinal cord of patients with amyotrophic lateral sclerosis (ALS) (118). These discrepancies could be cell type and brain region specific but may also occur as a result of the stage of disease investigated. Since the responsiveness of the NRF2 pathway decreases with age, NRF2 activation could occur in the early stages but decline during later stages of disease.
A number of pharmacological activators of NRF2 improve neurodegenerative phenotypes. Bardoxolone-methylamide (synthetic, triterpenoid, CDDO-MA) has been shown to improve memory and decrease amyloid-β plaque formation in transgenic AD mice (119). Similarly, puerarin (natural, phytoestrogen), SF, orientin (natural, flavone), and baicalin (natural, flavone) all improved the AD phenotype (120–123). 3H-1,2-dithiole-3-thione (natural, dithiolethione, D3T) and bardoxolone-ethylamide/trifluoroethylamide (synthetic, triterpenoids, CDDO-EA/TFEA) protected wild type, but not Nrf2−/− mice, from 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-induced PD (124; 125). DMF is the only current NRF2 activator to make it through phase III clinical trials for the treatment of MS, however recent studies using SF, CDDO-TFEA, and matrine (natural, alkaloid) were shown to reduce MS phenotypes in mice (126–128). It is curious that DMF is the only FDA approved drug, despite not being the most potent activator of NRF2, which could be a result of NRF2-independent immunomodulation (129), synergistic effects with other pathways, or because the degree of NRF2 activation is critical depending on the disease context.
Dual role of NRF2 in diabetes
Dysregulation of NRF2 has also been demonstrated in type I and II diabetes. Increased oxidative stress is a prevalent feature of diabetes that leads to cellular dysfunction and metabolic changes in a number of tissues. However, much like NRF2 in cancer and neurodegeneration, the role of NRF2 in diabetes is complex and tissue/cell type dependent. SF and cinnamic aldehyde (CA, natural, flavonoid) were both shown to suppress oxidative damage and restore normal kidney function in a streptozotocin-induced mouse model of type I diabetes (130). Oral administration of CDDO-Im resulted in enhanced NRF2 activity and attenuation of the diabetic phenotype in db/db mice (131). CDDO-Im, CDDO-Me, oltipraz, and curcumin improved insulin sensitivity and glucose tolerance in both genetic and high fat-diet induced diabetic models (132–135).
Interestingly, Keap1flox/- mice, which have constitutively higher levels of NRF2, also exhibited delayed onset of diabetes when crossed with db/db mice (131); yet, other studies indicated that KEAP1 knockdown enhanced the diabetic phenotype in Lepob/ob mice and mice fed a high fat diet (136; 137), indicating dietary and genetic factors controlling NRF2 may affect the onset and progression of diabetes differently. CDDO-Me was shown to enhance kidney function and decrease body weight in patients with diabetic nephropathy (138); however, the study was terminated due to increased risk of cardiovascular events. Importantly, improved specificity and the proper clinical context could still yield a positive outcome for this and other NRF2-based drugs (139).
NRF2 in cardiovascular, kidney, and liver disease
Cardiovascular disease (CVD) covers a wide range of conditions from hypertension to coronary artery disease. Much like other chronic diseases, the progression of many CVDs can take decades. A common feature of CVD is increased oxidative damage to endothelial cells and cardiomyocytes, which can prevent proper cardiac function and result in chronic damage to the heart and vasculature leading to heart failure. Pharmacological activation of NRF2 via SF, t-BHQ, MG132, resveratrol, and α-lipoic acid have all been shown to rescue models of heart failure (140). Conversely, chronic activation of NRF2 in a mutant αB crystallin mouse model of cardiac hypertrophy results in reductive stress (141), indicating that controlled activation of NRF2 can prevent cardiac dysfunction, whereas prolonged activation may result in detrimental effects.
Chronic oxidative stress and inflammation are also prevalent features of chronic kidney disease (CKD) and chronic liver disease. CDDO-Me is still the most promising therapy that has reached the clinical trial phase for CKD, at least for patients without cardiac risk factors (142), and CDDO-Im has been shown to protect against drug-induced liver injury, as well as reduce hepatic lipid accumulation in models of non-alcoholic fatty liver disease (143). These studies indicate that activation of NRF2 confers protection against oxidative stress in diseases associated with chronic inflammation and ROS production; nevertheless, prolonged activation of the NRF2 pathway may result in metabolic changes that also contribute to disease progression via reductive stress.
Clinical trials targeting NRF2 and drug repositioning for NRF2-based therapies
As discussed above, a number of NRF2 activators have proceeded to clinical trials for the treatment of various pathologies; however, only DMF has been approved for the treatment of relapsing MS. One of the most popular current regimens is dietary supplementation with SF, which is being or has been tested in the treatment of chronic obstructive pulmonary disease (COPD) [ClinicalTrials.gov Identifier (CTI): NCT01335971, (144)], exposure to ozone and other airborne pollutants [CTI: NCT01625130, (145); CTI: NCT01437501, (146)], cystic fibrosis [CTI: NCT01315665], asthma [CTI: NCT01845493], sickle cell disease [CTI: NCT01715480, (147)], and head and neck cancer [CTI: NCT03182959, NCT03268993, and NCT03402230] with limited or yet to be reported efficacy. A number of recent clinical trials also tested SF in the treatment of symptoms associated with autism spectrum disorders [CTI: NCT0256148] and schizophrenia [CTI: NCT01716858, (148)], as well as an ongoing study in type II diabetic patients [CTI: NCT02801448]. Interestingly, topical SF is being tested in protecting skin from UV-associated damage [CTI: NCT03126539]. Other NRF2 activators are also being investigated, such as curcumin for later stage diabetic nephropathy [CTI: NCT03262363], and a recently completed trial of resveratrol for CKDs [CTI: NCT02433925]. RTA 408 is being tested for patients with mitochondrial myopathies [CTI: NCT02255422] and Friedrich’s ataxia [CTI: NCT02255435].
The idea of drug repositioning has also been gaining traction. Two recent examples are DMF for the treatment of PD (149), and bardoxolone in the treatment of sickle cell disease (150) and pulmonary hypertension (CTI: NCT02036970). Another intriguing possibility is utilizing a medicinal chemistry approach to derivatize pre-existing therapeutics to improve their specificity and efficacy. Moreover, there has been a recent shift towards non-electrophilic small molecule modifiers of the NRF2-KEAP1 interaction to help minimize off target effects (80). Studies designed to discover novel therapies, as well as optimize current approaches, should both be considered.
Summary points
The relationship between NRF2 and pathological states is complex and requires careful evaluation prior to the development of effective therapies
Activation of the NRF2 pathway can be achieved by targeting the negative regulators of NRF2
Inhibition of the NRF2 pathway has proven to be a challenge, with the most promising strategy being to interfere with NRF2-sMAF-ARE binding
NRF2 controls a host of transcriptional targets, either directly via AREs, or indirectly via regulation of other transcription factors, indicating its central role in cell survival
Activating NRF2 has had great success in experimental models of disease; however, many cancers/metabolic diseases have chronically elevated levels of NRF2, indicating a need for targeted NRF2 inhibitors in certain contexts
While a number of NRF2 activators have made it to clinical trials, only DMF is FDA approved for the treatment of MS
Repositioning of NRF2 therapeutics, through repurposing or derivatization of existing drugs may, help improve the number of NRF2-based therapies
Future issues:
How do we narrow the gap in advancing therapies from bench to bedside? While the past two decades of research have clearly indicated the dual role of NRF2 in disease, the number of clinically approved therapeutics is limited to DMF
What is the proper time course for targeting NRF2 in disease? The beneficial or detrimental activation of NRF2 is time and context dependent, presenting a significant challenge in developing NRF2-based therapeutic regimens moving forward
Why are NRF2 activators different? Many NRF2 activators have been reported, but all are not equal, showing differential activation of ARE-regulated genes and disease related effects, possibly due to synergistic effects with other modes of regulation
Can specific inhibitors of NRF2 be developed? Targeted inhibitors of NRF2 are needed, as the current options exhibit off target effects
Acknowledgements:
The authors are funded by the following grants from the National Institutes of Health: ES026845 (DDZ), DK109555 (DDZ), ES023758 (EC & DDZ), ES004940 (DDZ), and ES006694 (a center grant).
References:
- 1.Tebay LE, Robertson H, Durant ST, Vitale SR, Penning TM, et al. 2015. Mechanisms of activation of the transcription factor Nrf2 by redox stressors, nutrient cues, and energy status and the pathways through which it attenuates degenerative disease. Free Radic Biol Med 88:108–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wattenberg LW. 1978. Inhibitors of chemical carcinogenesis. Adv Cancer Res 26:197–226 [DOI] [PubMed] [Google Scholar]
- 3.Benson AM, Batzinger RP, Ou SY, Bueding E, Cha YN, Talalay P. 1978. Elevation of hepatic glutathione S-transferase activities and protection against mutagenic metabolites of benzo(a)pyrene by dietary antioxidants. Cancer Res 38:4486–95 [PubMed] [Google Scholar]
- 4.Mignotte V, Eleouet JF, Raich N, Romeo PH. 1989. Cis- and trans-acting elements involved in the regulation of the erythroid promoter of the human porphobilinogen deaminase gene. Proc Natl Acad Sci U S A 86:6548–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan JY, Han XL, Kan YW. 1993. Cloning of Nrf1, an NF-E2-related transcription factor, by genetic selection in yeast. Proc Natl Acad Sci U S A 90:11371–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moi P, Chan K, Asunis I, Cao A, Kan YW. 1994. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc Natl Acad Sci U S A 91:9926–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kobayashi A, Ito E, Toki T, Kogame K, Takahashi S, et al. 1999. Molecular cloning and functional characterization of a new Cap’n’ collar family transcription factor Nrf3. J Biol Chem 274:6443–52 [DOI] [PubMed] [Google Scholar]
- 8.Oyake T, Itoh K, Motohashi H, Hayashi N, Hoshino H, et al. 1996. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol Cell Biol 16:6083–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muto A, Hoshino H, Madisen L, Yanai N, Obinata M, et al. 1998. Identification of Bach2 as a B-cell-specific partner for small maf proteins that negatively regulate the immunoglobulin heavy chain gene 3’ enhancer. EMBO J 17:5734–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rushmore TH, Pickett CB. 1990. Transcriptional regulation of the rat glutathione S-transferase Ya subunit gene. Characterization of a xenobiotic-responsive element controlling inducible expression by phenolic antioxidants. J Biol Chem 265:14648–53 [PubMed] [Google Scholar]
- 11.Prestera T, Holtzclaw WD, Zhang Y, Talalay P. 1993. Chemical and molecular regulation of enzymes that detoxify carcinogens. Proc Natl Acad Sci U S A 90:2965–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Itoh K, Igarashi K, Hayashi N, Nishizawa M, Yamamoto M. 1995. Cloning and characterization of a novel erythroid cell-derived CNC family transcription factor heterodimerizing with the small Maf family proteins. Mol Cell Biol 15:4184–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, et al. 1997. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun 236:313–22 [DOI] [PubMed] [Google Scholar]
- 14.Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, et al. 1999. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev 13:76–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan K, Lu R, Chang JC, Kan YW. 1996. NRF2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development. Proc Natl Acad Sci U S A 93:13943–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramos-Gomez M, Kwak MK, Dolan PM, Itoh K, Yamamoto M, et al. 2001. Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc Natl Acad Sci U S A 98:3410–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, et al. 2002. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci U S A 99:11908–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang DD, Hannink M. 2003. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol 23:8137–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. 2004. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol 24:10941–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, et al. 2004. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol 24:7130–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McMahon M, Thomas N, Itoh K, Yamamoto M, Hayes JD. 2004. Redox-regulated turnover of Nrf2 is determined by at least two separate protein domains, the redox-sensitive Neh2 degron and the redox-insensitive Neh6 degron. J Biol Chem 279:31556–67 [DOI] [PubMed] [Google Scholar]
- 22.Rada P, Rojo AI, Chowdhry S, McMahon M, Hayes JD, Cuadrado A. 2011. SCF/{beta}-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol Cell Biol 31:1121–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, et al. 2006. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med 3:e420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X-J, Sun Z, Villeneuve NF, Zhang S, Zhao F, et al. 2008. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis 29:1235–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, et al. 2010. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol 12:213–23 [DOI] [PubMed] [Google Scholar]
- 26.Lau A, Wang XJ, Zhao F, Villeneuve NF, Wu T, et al. 2010. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol 30:3275–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harder B, Jiang T, Wu T, Tao S, Rojo de la Vega M, et al. 2015. Molecular mechanisms of Nrf2 regulation and how these influence chemical modulation for disease intervention. Biochem Soc Trans 43:680–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayes JD, Dinkova-Kostova AT. 2014. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem Sci 39:199–218 [DOI] [PubMed] [Google Scholar]
- 29.Jung KA, Choi BH, Nam CW, Song M, Kim ST, et al. 2013. Identification of aldo-keto reductases as NRF2-target marker genes in human cells. Toxicol Lett 218:39–49 [DOI] [PubMed] [Google Scholar]
- 30.Alnouti Y, Klaassen CD. 2008. Tissue distribution, ontogeny, and regulation of aldehyde dehydrogenase (Aldh) enzymes mRNA by prototypical microsomal enzyme inducers in mice. Toxicol Sci 101:51–64 [DOI] [PubMed] [Google Scholar]
- 31.Chanas SA, Jiang Q, McMahon M, McWalter GK, McLellan LI, et al. 2002. Loss of the Nrf2 transcription factor causes a marked reduction in constitutive and inducible expression of the glutathione S-transferase Gsta1, Gsta2, Gstm1, Gstm2, Gstm3 and Gstm4 genes in the livers of male and female mice. Biochem J 365:405–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yueh MF, Tukey RH. 2007. Nrf2-Keap1 signaling pathway regulates human UGT1A1 expression in vitro and in transgenic UGT1 mice. J Biol Chem 282:8749–58 [DOI] [PubMed] [Google Scholar]
- 33.Nakamura A, Nakajima M, Higashi E, Yamanaka H, Yokoi T. 2008. Genetic polymorphisms in the 5’-flanking region of human UDP-glucuronosyltransferase 2B7 affect the Nrf2-dependent transcriptional regulation. Pharmacogenet Genomics 18:709–20 [DOI] [PubMed] [Google Scholar]
- 34.Maher JM, Dieter MZ, Aleksunes LM, Slitt AL, Guo G, et al. 2007. Oxidative and electrophilic stress induces multidrug resistance-associated protein transporters via the nuclear factor-E2-related factor-2 transcriptional pathway. Hepatology 46:1597–610 [DOI] [PubMed] [Google Scholar]
- 35.Singh A, Wu H, Zhang P, Happel C, Ma J, Biswal S. 2010. Expression of ABCG2 (BCRP) is regulated by Nrf2 in cancer cells that confers side population and chemoresistance phenotype. Mol Cancer Ther 9:2365–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pajares M, Jimenez-Moreno N, Garcia-Yague AJ, Escoll M, de Ceballos ML, et al. 2016. Transcription factor NFE2L2/NRF2 is a regulator of macroautophagy genes. Autophagy 12:1902–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bendavit G, Aboulkassim T, Hilmi K, Shah S, Batist G. 2016. Nrf2 Transcription Factor Can Directly Regulate mTOR: LINKING CYTOPROTECTIVE GENE EXPRESSION TO A MAJOR METABOLIC REGULATOR THAT GENERATES REDOX ACTIVITY. J Biol Chem 291:25476–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kwak MK, Wakabayashi N, Greenlaw JL, Yamamoto M, Kensler TW. 2003. Antioxidants enhance mammalian proteasome expression through the Keap1-Nrf2 signaling pathway. Mol Cell Biol 23:8786–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jang J, Wang Y, Kim HS, Lalli MA, Kosik KS. 2014. Nrf2, a regulator of the proteasome, controls self-renewal and pluripotency in human embryonic stem cells. Stem Cells 32:2616–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thimmulappa RK, Mai KH, Srisuma S, Kensler TW, Yamamoto M, Biswal S. 2002. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res 62:5196–203 [PubMed] [Google Scholar]
- 41.Mitsuishi Y, Taguchi K, Kawatani Y, Shibata T, Nukiwa T, et al. 2012. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell 22:66–79 [DOI] [PubMed] [Google Scholar]
- 42.Paek J, Lo JY, Narasimhan SD, Nguyen TN, Glover-Cutter K, et al. 2012. Mitochondrial SKN-1/Nrf mediates a conserved starvation response. Cell Metab 16:526–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Agyeman AS, Chaerkady R, Shaw PG, Davidson NE, Visvanathan K, et al. 2012. Transcriptomic and proteomic profiling of KEAP1 disrupted and sulforaphane-treated human breast epithelial cells reveals common expression profiles. Breast Cancer Res Treat 132:175–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jayakumar S, Pal D, Sandur SK. 2015. Nrf2 facilitates repair of radiation induced DNA damage through homologous recombination repair pathway in a ROS independent manner in cancer cells. Mutat Res 779:33–45 [DOI] [PubMed] [Google Scholar]
- 45.Niture SK, Jaiswal AK. 2012. Nrf2 protein up-regulates antiapoptotic protein Bcl-2 and prevents cellular apoptosis. J Biol Chem 287:9873–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Niture SK, Jaiswal AK. 2013. Nrf2-induced antiapoptotic Bcl-xL protein enhances cell survival and drug resistance. Free Radic Biol Med 57:119–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wakabayashi N, Shin S, Slocum SL, Agoston ES, Wakabayashi J, et al. 2010. Regulation of notch1 signaling by nrf2: implications for tissue regeneration. Sci Signal 3:ra52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wakabayashi N, Skoko JJ, Chartoumpekis DV, Kimura S, Slocum SL, et al. 2014. Notch-Nrf2 axis: regulation of Nrf2 gene expression and cytoprotection by notch signaling. Mol Cell Biol 34:653–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matsushita N, Sogawa K, Ema M, Yoshida A, Fujii-Kuriyama Y. 1993. A factor binding to the xenobiotic responsive element (XRE) of P-4501A1 gene consists of at least two helix-loop-helix proteins, Ah receptor and Arnt. J Biol Chem 268:21002–6 [PubMed] [Google Scholar]
- 50.Yeager RL, Reisman SA, Aleksunes LM, Klaassen CD. 2009. Introducing the “TCDD-inducible AhR-Nrf2 gene battery”. Toxicol Sci 111:238–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nioi P, Hayes JD. 2004. Contribution of NAD(P)H:quinone oxidoreductase 1 to protection against carcinogenesis, and regulation of its gene by the Nrf2 basic-region leucine zipper and the arylhydrocarbon receptor basic helix-loop-helix transcription factors. Mutat Res 555:149–71 [DOI] [PubMed] [Google Scholar]
- 52.Miao W, Hu L, Scrivens PJ, Batist G. 2005. Transcriptional regulation of NF-E2 p45-related factor (NRF2) expression by the aryl hydrocarbon receptor-xenobiotic response element signaling pathway: direct cross-talk between phase I and II drug-metabolizing enzymes. J Biol Chem 280:20340–8 [DOI] [PubMed] [Google Scholar]
- 53.Shin S, Wakabayashi N, Misra V, Biswal S, Lee GH, et al. 2007. NRF2 modulates aryl hydrocarbon receptor signaling: influence on adipogenesis. Mol Cell Biol 27:7188–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu GH, Qu J, Shen X. 2008. NF-kappaB/p65 antagonizes Nrf2-ARE pathway by depriving CBP from Nrf2 and facilitating recruitment of HDAC3 to MafK. Biochim Biophys Acta 1783:713–27 [DOI] [PubMed] [Google Scholar]
- 55.Heiss E, Herhaus C, Klimo K, Bartsch H, Gerhauser C. 2001. Nuclear factor kappa B is a molecular target for sulforaphane-mediated anti-inflammatory mechanisms. J Biol Chem 276:32008–15 [DOI] [PubMed] [Google Scholar]
- 56.Ahmad R, Raina D, Meyer C, Kharbanda S, Kufe D. 2006. Triterpenoid CDDO-Me blocks the NF-kappaB pathway by direct inhibition of IKKbeta on Cys-179. J Biol Chem 281:35764–9 [DOI] [PubMed] [Google Scholar]
- 57.Singh S, Aggarwal BB. 1995. Activation of transcription factor NF-kappa B is suppressed by curcumin (diferuloylmethane) [corrected]. J Biol Chem 270:24995–5000 [DOI] [PubMed] [Google Scholar]
- 58.Thimmulappa RK, Lee H, Rangasamy T, Reddy SP, Yamamoto M, et al. 2006. Nrf2 is a critical regulator of the innate immune response and survival during experimental sepsis. J Clin Invest 116:984–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee DF, Kuo HP, Liu M, Chou CK, Xia W, et al. 2009. KEAP1 E3 ligase-mediated downregulation of NF-kappaB signaling by targeting IKKbeta. Mol Cell 36:131–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wakabayashi N, Slocum SL, Skoko JJ, Shin S, Kensler TW. 2010. When NRF2 talks, who’s listening? Antioxid Redox Signal 13:1649–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Faraonio R, Vergara P, Di Marzo D, Pierantoni MG, Napolitano M, et al. 2006. p53 suppresses the Nrf2-dependent transcription of antioxidant response genes. J Biol Chem 281:39776–84 [DOI] [PubMed] [Google Scholar]
- 62.You A, Nam CW, Wakabayashi N, Yamamoto M, Kensler TW, Kwak MK. 2011. Transcription factor Nrf2 maintains the basal expression of Mdm2: An implication of the regulation of p53 signaling by Nrf2. Arch Biochem Biophys 507:356–64 [DOI] [PubMed] [Google Scholar]
- 63.Chen W, Sun Z, Wang XJ, Jiang T, Huang Z, et al. 2009. Direct interaction between Nrf2 and p21(Cip1/WAF1) upregulates the Nrf2-mediated antioxidant response. Mol Cell 34:663–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Iida K, Itoh K, Maher JM, Kumagai Y, Oyasu R, et al. 2007. Nrf2 and p53 cooperatively protect against BBN-induced urinary bladder carcinogenesis. Carcinogenesis 28:2398–403 [DOI] [PubMed] [Google Scholar]
- 65.Joo MS, Kim WD, Lee KY, Kim JH, Koo JH, Kim SG. 2016. AMPK Facilitates Nuclear Accumulation of Nrf2 by Phosphorylating at Serine 550. Mol Cell Biol 36:1931–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Salazar M, Rojo AI, Velasco D, de Sagarra RM, Cuadrado A. 2006. Glycogen synthase kinase-3beta inhibits the xenobiotic and antioxidant cell response by direct phosphorylation and nuclear exclusion of the transcription factor Nrf2. J Biol Chem 281:14841–51 [DOI] [PubMed] [Google Scholar]
- 67.Zhang HH, Lipovsky AI, Dibble CC, Sahin M, Manning BD. 2006. S6K1 regulates GSK3 under conditions of mTOR-dependent feedback inhibition of Akt. Mol Cell 24:185–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ichimura Y, Waguri S, Sou YS, Kageyama S, Hasegawa J, et al. 2013. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol Cell 51:618–31 [DOI] [PubMed] [Google Scholar]
- 69.Guo Y, Yu S, Zhang C, Kong AN. 2015. Epigenetic regulation of Keap1-Nrf2 signaling. Free Radic Biol Med 88:337–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kang KA, Piao MJ, Kim KC, Kang HK, Chang WY, et al. 2014. Epigenetic modification of Nrf2 in 5-fluorouracil-resistant colon cancer cells: involvement of TET-dependent DNA demethylation. Cell Death Dis 5:e1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Khor TO, Fuentes F, Shu L, Paredes-Gonzalez X, Yang AY, et al. 2014. Epigenetic DNA methylation of antioxidative stress regulator NRF2 in human prostate cancer. Cancer Prev Res (Phila) 7:1186–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kawai Y, Garduno L, Theodore M, Yang J, Arinze IJ. 2011. Acetylation-deacetylation of the transcription factor Nrf2 (nuclear factor erythroid 2-related factor 2) regulates its transcriptional activity and nucleocytoplasmic localization. J Biol Chem 286:7629–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu X, Li H, Liu L, Lu Y, Gao Y, et al. 2016. Methylation of arginine by PRMT1 regulates Nrf2 transcriptional activity during the antioxidative response. Biochim Biophys Acta 1863:2093–103 [DOI] [PubMed] [Google Scholar]
- 74.Huang HC, Nguyen T, Pickett CB. 2002. Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. J Biol Chem 277:42769–74 [DOI] [PubMed] [Google Scholar]
- 75.Sun Z, Huang Z, Zhang DD. 2009. Phosphorylation of Nrf2 at multiple sites by MAP kinases has a limited contribution in modulating the Nrf2-dependent antioxidant response. PLoS One 4:e6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Malloy MT, McIntosh DJ, Walters TS, Flores A, Goodwin JS, Arinze IJ. 2013. Trafficking of the transcription factor Nrf2 to promyelocytic leukemia-nuclear bodies: implications for degradation of NRF2 in the nucleus. J Biol Chem 288:14569–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.He X, Lai Q, Chen C, Li N, Sun F, et al. 2018. Both conditional ablation and overexpression of E2 SUMO-conjugating enzyme (UBC9) in mouse pancreatic beta cells result in impaired beta cell function. Diabetologia [DOI] [PubMed]
- 78.Kensler TW, Wakabayashi N, Biswal S. 2007. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annual review of pharmacology and toxicology 47:89–116 [DOI] [PubMed] [Google Scholar]
- 79.Jaramillo MC, Zhang DD. 2013. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev 27:2179–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rojo de la Vega MD M; Chapman E; Zhang DD 2016. NRF2-targeted therapeutics: New targets and modes of NRF2 regulation. Curr Opin Toxicol 1 [DOI] [PMC free article] [PubMed]
- 81.Padmanabhan B, Tong KI, Ohta T, Nakamura Y, Scharlock M, et al. 2006. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol Cell 21:689–700 [DOI] [PubMed] [Google Scholar]
- 82.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, et al. 2014. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505:495–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, et al. 2010. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature 466:869–73 [DOI] [PubMed] [Google Scholar]
- 84.Cancer Genome Atlas Research N. 2012. Comprehensive genomic characterization of squamous cell lung cancers. Nature 489:519–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Martinez VD, Vucic EA, Thu KL, Pikor LA, Lam S, Lam WL. 2015. Disruption of KEAP1/CUL3/RBX1 E3-ubiquitin ligase complex components by multiple genetic mechanisms: Association with poor prognosis in head and neck cancer. Head Neck 37:727–34 [DOI] [PubMed] [Google Scholar]
- 86.Martinez VD, Vucic EA, Pikor LA, Thu KL, Hubaux R, Lam WL. 2013. Frequent concerted genetic mechanisms disrupt multiple components of the NRF2 inhibitor KEAP1/CUL3/RBX1 E3-ubiquitin ligase complex in thyroid cancer. Mol Cancer 12:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li QK, Singh A, Biswal S, Askin F, Gabrielson E. 2011. KEAP1 gene mutations and NRF2 activation are common in pulmonary papillary adenocarcinoma. J Hum Genet 56:230–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nioi P, Nguyen T. 2007. A mutation of Keap1 found in breast cancer impairs its ability to repress Nrf2 activity. Biochem Biophys Res Commun 362:816–21 [DOI] [PubMed] [Google Scholar]
- 89.Shibata T, Kokubu A, Gotoh M, Ojima H, Ohta T, et al. 2008. Genetic alteration of Keap1 confers constitutive Nrf2 activation and resistance to chemotherapy in gallbladder cancer. Gastroenterology 135:1358–68, 68 e1–4 [DOI] [PubMed] [Google Scholar]
- 90.Ooi A, Dykema K, Ansari A, Petillo D, Snider J, et al. 2013. CUL3 and NRF2 mutations confer an NRF2 activation phenotype in a sporadic form of papillary renal cell carcinoma. Cancer Res 73:2044–51 [DOI] [PubMed] [Google Scholar]
- 91.Sato Y, Yoshizato T, Shiraishi Y, Maekawa S, Okuno Y, et al. 2013. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet 45:860–7 [DOI] [PubMed] [Google Scholar]
- 92.Konstantinopoulos PA, Spentzos D, Fountzilas E, Francoeur N, Sanisetty S, et al. 2011. Keap1 mutations and Nrf2 pathway activation in epithelial ovarian cancer. Cancer Res 71:5081–9 [DOI] [PubMed] [Google Scholar]
- 93.Wong TF, Yoshinaga K, Monma Y, Ito K, Niikura H, et al. 2011. Association of keap1 and nrf2 genetic mutations and polymorphisms with endometrioid endometrial adenocarcinoma survival. Int J Gynecol Cancer 21:1428–35 [DOI] [PubMed] [Google Scholar]
- 94.Wang R, An J, Ji F, Jiao H, Sun H, Zhou D. 2008. Hypermethylation of the Keap1 gene in human lung cancer cell lines and lung cancer tissues. Biochem Biophys Res Commun 373:151–4 [DOI] [PubMed] [Google Scholar]
- 95.Barbano R, Muscarella LA, Pasculli B, Valori VM, Fontana A, et al. 2013. Aberrant Keap1 methylation in breast cancer and association with clinicopathological features. Epigenetics 8:105–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Muscarella LA, Barbano R, D’Angelo V, Copetti M, Coco M, et al. 2011. Regulation of KEAP1 expression by promoter methylation in malignant gliomas and association with patient’s outcome. Epigenetics 6:317–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang P, Singh A, Yegnasubramanian S, Esopi D, Kombairaju P, et al. 2010. Loss of Kelch-like ECH-associated protein 1 function in prostate cancer cells causes chemoresistance and radioresistance and promotes tumor growth. Mol Cancer Ther 9:336–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hanada N, Takahata T, Zhou Q, Ye X, Sun R, et al. 2012. Methylation of the KEAP1 gene promoter region in human colorectal cancer. BMC Cancer 12:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fabrizio FP, Costantini M, Copetti M, la Torre A, Sparaneo A, et al. 2017. Keap1/Nrf2 pathway in kidney cancer: frequent methylation of KEAP1 gene promoter in clear renal cell carcinoma. Oncotarget 8:11187–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ooi A, Wong JC, Petillo D, Roossien D, Perrier-Trudova V, et al. 2011. An antioxidant response phenotype shared between hereditary and sporadic type 2 papillary renal cell carcinoma. Cancer Cell 20:511–23 [DOI] [PubMed] [Google Scholar]
- 101.Shibata T, Ohta T, Tong KI, Kokubu A, Odogawa R, et al. 2008. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc Natl Acad Sci U S A 105:13568–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kim YR, Oh JE, Kim MS, Kang MR, Park SW, et al. 2010. Oncogenic NRF2 mutations in squamous cell carcinomas of oesophagus and skin. The Journal of pathology 220:446–51 [DOI] [PubMed] [Google Scholar]
- 103.Goldstein LD, Lee J, Gnad F, Klijn C, Schaub A, et al. 2016. Recurrent Loss of NFE2L2 Exon 2 Is a Mechanism for Nrf2 Pathway Activation in Human Cancers. Cell Rep 16:2605–17 [DOI] [PubMed] [Google Scholar]
- 104.Martinez VD, Vucic EA, Thu KL, Pikor LA, Hubaux R, Lam WL. 2014. Unique Pattern of Component Gene Disruption in the NRF2 Inhibitor KEAP1/CUL3/RBX1 E3-Ubiquitin Ligase Complex in Serous Ovarian Cancer. BioMed Research International 2014:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ren D, Villeneuve NF, Jiang T, Wu T, Lau A, et al. 2011. Brusatol enhances the efficacy of chemotherapy by inhibiting the Nrf2-mediated defense mechanism. Proc Natl Acad Sci U S A 108:1433–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Vartanian S, Ma TP, Lee J, Haverty PM, Kirkpatrick DS, et al. 2016. Application of Mass Spectrometry Profiling to Establish Brusatol as an Inhibitor of Global Protein Synthesis. Mol Cell Proteomics 15:1220–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Harder B, Tian W, La Clair JJ, Tan AC, Ooi A, et al. 2017. Brusatol overcomes chemoresistance through inhibition of protein translation. Mol Carcinog 56:1493–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bollong MJ, Yun H, Sherwood L, Woods AK, Lairson LL, Schultz PG. 2015. A Small Molecule Inhibits Deregulated NRF2 Transcriptional Activity in Cancer. ACS Chemical Biology 10:2193–8 [DOI] [PubMed] [Google Scholar]
- 109.Singh A, Venkannagari S, Oh KH, Zhang Y-Q, Rohde JM, et al. 2016. Small Molecule Inhibitor of NRF2 Selectively Intervenes Therapeutic Resistance in KEAP1-Deficient NSCLC Tumors. ACS Chemical Biology 11:3214–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Arlt A, Sebens S, Krebs S, Geismann C, Grossmann M, et al. 2013. Inhibition of the Nrf2 transcription factor by the alkaloid trigonelline renders pancreatic cancer cells more susceptible to apoptosis through decreased proteasomal gene expression and proteasome activity. Oncogene 32:4825–35 [DOI] [PubMed] [Google Scholar]
- 111.Manna A, De Sarkar S, De S, Bauri AK, Chattopadhyay S, Chatterjee M. 2015. The variable chemotherapeutic response of Malabaricone-A in leukemic and solid tumor cell lines depends on the degree of redox imbalance. Phytomedicine 22:713–23 [DOI] [PubMed] [Google Scholar]
- 112.Limonciel A, Jennings P. 2014. A review of the evidence that ochratoxin A is an Nrf2 inhibitor: implications for nephrotoxicity and renal carcinogenicity. Toxins (Basel) 6:371–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhong Y, Zhang F, Sun Z, Zhou W, Li ZY, et al. 2013. Drug resistance associates with activation of Nrf2 in MCF-7/DOX cells, and wogonin reverses it by down-regulating Nrf2-mediated cellular defense response. Mol Carcinog 52:824–34 [DOI] [PubMed] [Google Scholar]
- 114.Namani A, Li Y, Wang XJ, Tang X. 2014. Modulation of NRF2 signaling pathway by nuclear receptors: implications for cancer. Biochim Biophys Acta 1843:1875–85 [DOI] [PubMed] [Google Scholar]
- 115.Corenblum MJ, Ray S, Remley QW, Long M, Harder B, et al. 2016. Reduced Nrf2 expression mediates the decline in neural stem cell function during a critical middle-age period. Aging Cell 15:725–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhang H, Davies KJ, Forman HJ. 2015. Oxidative stress response and Nrf2 signaling in aging. Free Radic Biol Med 88:314–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Johnson DA, Johnson JA. 2015. Nrf2--a therapeutic target for the treatment of neurodegenerative diseases. Free Radic Biol Med 88:253–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sarlette A, Krampfl K, Grothe C, Neuhoff N, Dengler R, Petri S. 2008. Nuclear erythroid 2-related factor 2-antioxidative response element signaling pathway in motor cortex and spinal cord in amyotrophic lateral sclerosis. J Neuropathol Exp Neurol 67:1055–62 [DOI] [PubMed] [Google Scholar]
- 119.Dumont M, Wille E, Calingasan NY, Tampellini D, Williams C, et al. 2009. Triterpenoid CDDO-methylamide improves memory and decreases amyloid plaques in a transgenic mouse model of Alzheimer’s disease. J Neurochem 109:502–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhou Y, Xie N, Li L, Zou Y, Zhang X, Dong M. 2014. Puerarin alleviates cognitive impairment and oxidative stress in APP/PS1 transgenic mice. Int J Neuropsychopharmacol 17:635–44 [DOI] [PubMed] [Google Scholar]
- 121.Zhang R, Miao QW, Zhu CX, Zhao Y, Liu L, et al. 2015. Sulforaphane ameliorates neurobehavioral deficits and protects the brain from amyloid beta deposits and peroxidation in mice with Alzheimer-like lesions. Am J Alzheimers Dis Other Demen 30:183–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yu L, Wang S, Chen X, Yang H, Li X, et al. 2015. Orientin alleviates cognitive deficits and oxidative stress in Abeta1–42-induced mouse model of Alzheimer’s disease. Life Sci 121:104–9 [DOI] [PubMed] [Google Scholar]
- 123.Chen C, Li X, Gao P, Tu Y, Zhao M, et al. 2015. Baicalin attenuates alzheimer-like pathological changes and memory deficits induced by amyloid beta1–42 protein. Metab Brain Dis 30:537–44 [DOI] [PubMed] [Google Scholar]
- 124.Burton NC, Kensler TW, Guilarte TR. 2006. In vivo modulation of the Parkinsonian phenotype by Nrf2. Neurotoxicology 27:1094–100 [DOI] [PubMed] [Google Scholar]
- 125.Kaidery NA, Banerjee R, Yang L, Smirnova NA, Hushpulian DM, et al. 2013. Targeting Nrf2-mediated gene transcription by extremely potent synthetic triterpenoids attenuate dopaminergic neurotoxicity in the MPTP mouse model of Parkinson’s disease. Antioxid Redox Signal 18:139–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pareek TK, Belkadi A, Kesavapany S, Zaremba A, Loh SL, et al. 2011. Triterpenoid modulation of IL-17 and Nrf-2 expression ameliorates neuroinflammation and promotes remyelination in autoimmune encephalomyelitis. Sci Rep 1:201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li B, Cui W, Liu J, Li R, Liu Q, et al. 2013. Sulforaphane ameliorates the development of experimental autoimmune encephalomyelitis by antagonizing oxidative stress and Th17-related inflammation in mice. Exp Neurol 250:239–49 [DOI] [PubMed] [Google Scholar]
- 128.Liu N, Kan QC, Zhang XJ, Xv YM, Zhang S, et al. 2014. Upregulation of immunomodulatory molecules by matrine treatment in experimental autoimmune encephalomyelitis. Exp Mol Pathol 97:470–6 [DOI] [PubMed] [Google Scholar]
- 129.Schulze-Topphoff U, Varrin-Doyer M, Pekarek K, Spencer CM, Shetty A, et al. 2016. Dimethyl fumarate treatment induces adaptive and innate immune modulation independent of Nrf2. Proc Natl Acad Sci U S A 113:4777–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zheng H, Whitman SA, Wu W, Wondrak GT, Wong PK, et al. 2011. Therapeutic potential of Nrf2 activators in streptozotocin-induced diabetic nephropathy. Diabetes 60:3055–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Uruno A, Furusawa Y, Yagishita Y, Fukutomi T, Muramatsu H, et al. 2013. The Keap1-Nrf2 system prevents onset of diabetes mellitus. Mol Cell Biol 33:2996–3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Shin S, Wakabayashi J, Yates MS, Wakabayashi N, Dolan PM, et al. 2009. Role of Nrf2 in prevention of high-fat diet-induced obesity by synthetic triterpenoid CDDO-imidazolide. Eur J Pharmacol 620:138–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yu Z, Shao W, Chiang Y, Foltz W, Zhang Z, et al. 2011. Oltipraz upregulates the nuclear factor (erythroid-derived 2)-like 2 [corrected](NRF2) antioxidant system and prevents insulin resistance and obesity induced by a high-fat diet in C57BL/6J mice. Diabetologia 54:922–34 [DOI] [PubMed] [Google Scholar]
- 134.Saha PK, Reddy VT, Konopleva M, Andreeff M, Chan L. 2010. The triterpenoid 2-cyano-3,12-dioxooleana-1,9-dien-28-oic-acid methyl ester has potent anti-diabetic effects in diet-induced diabetic mice and Lepr(db/db) mice. J Biol Chem 285:40581–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.He HJ, Wang GY, Gao Y, Ling WH, Yu ZW, Jin TR. 2012. Curcumin attenuates Nrf2 signaling defect, oxidative stress in muscle and glucose intolerance in high fat diet-fed mice. World J Diabetes 3:94–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Xu J, Kulkarni SR, Donepudi AC, More VR, Slitt AL. 2012. Enhanced Nrf2 activity worsens insulin resistance, impairs lipid accumulation in adipose tissue, and increases hepatic steatosis in leptin-deficient mice. Diabetes 61:3208–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.More VR, Xu J, Shimpi PC, Belgrave C, Luyendyk JP, et al. 2013. Keap1 knockdown increases markers of metabolic syndrome after long-term high fat diet feeding. Free Radic Biol Med 61C:85–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.de Zeeuw D, Akizawa T, Audhya P, Bakris GL, Chin M, et al. 2013. Bardoxolone methyl in type 2 diabetes and stage 4 chronic kidney disease. N Engl J Med 369:2492–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhang DD. 2013. Bardoxolone brings Nrf2-based therapies to light. Antioxid Redox Signal 19:517–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhou S, Sun W, Zhang Z, Zheng Y. 2014. The role of Nrf2-mediated pathway in cardiac remodeling and heart failure. Oxid Med Cell Longev 2014:260429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rajasekaran NS, Varadharaj S, Khanderao GD, Davidson CJ, Kannan S, et al. 2011. Sustained activation of nuclear erythroid 2-related factor 2/antioxidant response element signaling promotes reductive stress in the human mutant protein aggregation cardiomyopathy in mice. Antioxid Redox Signal 14:957–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Nezu M, Suzuki N, Yamamoto M. 2017. Targeting the KEAP1-NRF2 System to Prevent Kidney Disease Progression. Am J Nephrol 45:473–83 [DOI] [PubMed] [Google Scholar]
- 143.Tang W, Jiang YF, Ponnusamy M, Diallo M. 2014. Role of Nrf2 in chronic liver disease. World J Gastroenterol 20:13079–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wise RA, Holbrook JT, Criner G, Sethi S, Rayapudi S, et al. 2017. Correction: Lack of Effect of Oral Sulforaphane Administration on Nrf2 Expression in COPD: A Randomized, Double-Blind, Placebo Controlled Trial. PLoS One 12:e0175077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Duran CG, Burbank AJ, Mills KH, Duckworth HR, Aleman MM, et al. 2016. A proof-of-concept clinical study examining the NRF2 activator sulforaphane against neutrophilic airway inflammation. Respir Res 17:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Egner PA, Chen JG, Zarth AT, Ng DK, Wang JB, et al. 2014. Rapid and sustainable detoxication of airborne pollutants by broccoli sprout beverage: results of a randomized clinical trial in China. Cancer Prev Res (Phila) 7:813–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Doss JF, Jonassaint JC, Garrett ME, Ashley-Koch AE, Telen MJ, Chi JT. 2016. Phase 1 Study of a Sulforaphane-Containing Broccoli Sprout Homogenate for Sickle Cell Disease. PLoS One 11:e0152895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Shiina A, Kanahara N, Sasaki T, Oda Y, Hashimoto T, et al. 2015. An Open Study of Sulforaphane-rich Broccoli Sprout Extract in Patients with Schizophrenia. Clin Psychopharmacol Neurosci 13:62–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lastres-Becker I, Garcia-Yague AJ, Scannevin RH, Casarejos MJ, Kugler S, et al. 2016. Repurposing the NRF2 Activator Dimethyl Fumarate as Therapy Against Synucleinopathy in Parkinson’s Disease. Antioxid Redox Signal 25:61–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Owusu-Ansah A, Ihunnah CA, Walker AL, Ofori-Acquah SF. 2016. Inflammatory targets of therapy in sickle cell disease. Transl Res 167:281–97 [DOI] [PMC free article] [PubMed] [Google Scholar]