Abstract
Background
Androgen receptor splice variant 7 (AR-V7) has been implicated in resistance to abiraterone and enzalutamide treatment in men with metastatic castrationresistant prostate cancer (mCRPC). Tissue- or cell-based in situ detection of AR-V7, however, has been limited by lack of specificity.
Objective
To address current limitations in precision measurement of AR-V7 by developing a novel junction-specific AR-V7 RNA in situ hybridization (RISH) assay compatible with automated quantification.
Design, setting, and participants
We designed a RISH method to visualize single splice junctions in cells and tissue. Using the validated assay for junction-specific detection of the full-length AR (AR-FL) and AR-V7, we generated quantitative data, blinded to clinical data, for 63 prostate tumor biopsies.
Outcome measurements and statistical analysis
We evaluated clinical correlates of ARFL/AR-V7 measurements, including association with prostate-specific antigen progression-free survival (PSA-PFS) and clinical and radiographic progression-free survival (PFS), in a subset of patients starting treatment with abiraterone or enzalutamide following biopsy.
Results and limitations
Quantitative AR-FL/AR-V7 data were generated from 56 of the 63 (88.9%) biopsy specimens examined, of which 44 were mCRPC biopsies. Positive ARV7 signals were detected in 34.1% (15/44) mCRPC specimens, all of which also coexpressed AR-FL. The median AR-V7/AR-FL ratio was 11.9% (range 2.7–30.3%). Positive detection of AR-V7 was correlated with indicators of high disease burden at baseline. Among the 25 CRPC biopsies collected before treatment with abiraterone or enzalutamide, positive AR-V7 detection, but not higher AR-FL, was significantly associated with shorter PSA-PFS (hazard ratio 2.789, 95% confidence interval 1.12–6.95; p = 0.0081).
Conclusions
We report for the first time a RISH method for highly specific and quantifiable detection of splice junctions, allowing further characterization of AR-V7 and its clinical significance.
Patient summary
Higher AR-V7 levels detected and quantified using a novel method were associated with poorer response to abiraterone or enzalutamide in prostate cancer.
Keywords: Androgen receptor, RNA in situ hybridization, Splice variant, AR-V7
1. Introduction
Androgen receptor splice variant 7 (AR-V7) is one of the AR aberrations implicated in the development of castration-resistant prostate cancer (CRPC) [1,2]. AR-V7 originates from contiguous splicing of AR exons 1, 2, and 3 and the cryptic exon 3 (CE3) within the canonical intron 3 of the AR gene [1]. Specific detection of AR-V7 can be achieved by targeting the exon 3/CE3 splice junction via reverse transcription polymerase chain reaction (RT-PCR) [3]. A number of previous studies have demonstrated the prognostic value of AR-V7 detection by RT-PCR in men with metastatic CRPC (mCRPC) treated with abiraterone and/or enzalutamide. These studies used biological substrates such as prostate cancer tissues [4–8] and liquid biopsy samples, including circulating tumor cells (CTCs) [9–11], plasma exosomes [12], peripheral blood mononuclear cells (PBMCs) [13], and even whole blood samples [14,15]. While these approaches generally allow sensitive and specific detection of AR-V7, they are limited by a number of analytical and preanalytical challenges mainly attributable to low amounts of AR-V7 mRNA in liquid biopsy samples [16]. Critically, determination of AR-V7 status and its quantification were not possible in a significant proportion of mCRPC patients who were CTC-negative, even though the CTC-based AR-V7 test has been analytically validated and implemented in a clinical laboratory [17].
An alternative and potentially complementary approach to RT-PCR–based detection is RNA in situ hybridization (RISH). In contrast to the RT-PCR approach, RISH allows visualization of gene expression with spatial and morphological context [18]. Traditional RISH methods have been hampered by low sensitivity and a low signal-to-noise ratio, as well as the time-consuming effort required to develop experimental protocols for each detection target [19]. The RNAscope method is a recently developed RISH technique that uses an integrated probe design and signal amplification strategy to amplify target-specific signals by thousands fold without amplifying the background noise [20]. Importantly, this technique is compatible with routine formalinfixed paraffin-embedded (FFPE) tissues. Following an initial report on AR-V7 RISH by RNAscope [10], two recent reports showed that AR-V7 detected in FFPE tissue specimens by two different RISH methods was associated with CRPC and prognostic in those treated with AR-targeting therapies [21,22]. However, these RISH methods, while revolutionary in RNA detection, require multiple tiling probes covering a target sequence of ~1 kb, and therefore lack the resolution for detecting a variant-specific splice junction. For AR-V7 detection, the published methods [10,21,22] targeted the 1.3-kb CE3 sequence. Because the CE3 sequence is also present in AR genomic DNA and AR pre-mRNA that are retained in the nucleus before being spliced and exported to the cytoplasm, detection of the CE3 sequence described in these previous studies should not be equated to detection of AR-V7. Indeed, detection of pre-mRNA was reported in a previous study [21] and detection of AR genomic DNA cannot be ruled out, particularly in mCRPC specimens with AR amplification. In addition, specificity for AR-V7 detection that targets the CE3 sequence may be further compromised by simultaneous detection of AR-V9, another androgen receptor variant that shares the same 3’ CE3 sequence [23]. Therefore, accurate detection and quantification of ARV7 mRNA in intact cells would not be possible given the lack of resolution and detection specificity of existing RISH methods.
In the present study, we developed a novel RISH detection method targeting a single splice junction using probes straddling the targeted junction. We applied this novel method to detect and quantify AR-V7, by targeting the exon 3/CE3 junction, and full-length AR (AR-FL), by targeting the exon 7/exon 8 junction. Following validation of junction-specific detection of the AR transcripts in cell lines and in FFPE specimens from mCRPC patients, we applied the prototype technology and quantified AR-V7/ARFL levels in biopsies from mCRPC patients. We then conducted exploratory clinical correlative analysis for men treated with abiraterone or enzalutamide. We present the first example of visualization of splice junctions in morphologically intact cells, and demonstrate for the first time a highly specific and quantifiable AR-V7 RISH test for detection of clinically significant levels of AR-V7 mRNA in mCRPC patients.
2. Patients and methods
2.1. Patients
Two biopsy cohorts, one from the Johns Hopkins University School of Medicine (JHU cohort) and one from the Institute of Cancer Research and Royal Marsden NHS Foundation Trust (UK cohort), were used in this study. For the JHU cohort, 35 patients with metastatic prostate cancer gave informed consent to undergo the biopsy procedure under a study protocol approved by the institutional review board. Within this unselected and diverse cohort (Supplementary Table 1), nine patients with mCRPC underwent treatment with abiraterone or enzalutamide immediately following the biopsy procedure. For the UK cohort, 28 retrospective biopsies, including mainly bone marrow and prostate biopsies (Supplementary Table 1) were selected from patients treated with first-line abiraterone or enzalutamide (mainly abiraterone) following the biopsies. All study participants had given written informed consent and were enrolled in institutional protocols approved by a multicenter research ethics committee (Chelsea Research Ethics Committee, reference 04/Q0801/60). There were no other selection criteria; all samples tested are included in Supplementary Table 1. All experimental processes were performed while blinded to the sample type and related data.
2.2. RISH by BaseScope
The BaseScope (Advanced Cell Diagnostics, Inc., Hayward, CA) for AR-FL/AR-V7 were developed to achieve junction-specific detection of the AR transcripts. The BaseScope assay is based on the RNAscope technology [20] but uses an additional signal amplification step and requires only one “double Z” (1 ZZ) probe pair for single-molecule detection. The 1-ZZ probe for AR-V7 was designed to target the AR-V7–specific junction of exon 3 and CE3 (AR-E3/CE3) (ZZ probe target sequence GAC TCT GGG AGA AAA ATT CCG GGT TGG CAA TTG CAA GCA TCT C), and the 1-ZZ probe for AR-FL was designed to target the splice junction of exon 7 and exon 8 (AR-E7/E8) (ZZ probe target sequence GCT CAC CAA GCT CCT GGA CTC CGT GCA GCC TAT TGC GAG A),as illustrated schematically in Figure 1A. For each sample, four probes were used in four adjacent sections: AR-E7/E8, AR-E3/CE3, 1ZZ Hs-POLR2A as a positive control, and 1-ZZ DapB as a negative control. Slides with negative POLR2A staining (n = 4 in the JHU cohort and n = 3 in the UK cohort), indicative of poor tissue quality, were excluded from analysis. Automated quantification of AR transcripts was performed using RNAscope Spot Studio software (Supplementary material).
Fig. 1 –
Development of the BaseScope RNA in situ hybridization assay for detection of splice junctions specific to AR-FL and AR-V7. (A) Schematic illustration of the BaseScope assay and the splice junctions targeted for probe design. Top: Overview of the BaseScope assay workflow. Sections containing fixed tissues or cells were permeabilized, and exposed mRNA were hybridized with a single pair of BaseScope probes (ZZ pair) that straddle the exon/exon junction of interest. Following amplification by an advanced, next-generation signal amplification system, junction-specific signals can be visualized as punctate dots under a standard bright-field microscope. Bottom: AR splice junctions targeted for BaseScope probe design. The splice junction between AR exons 7 and 8 (E7/E8) was targeted for specific detection of the full-length AR (AR-FL), while the splice junction between exon 3 and cryptic exon 3 (CE3) (E3/CE3) was targeted for specific detection of AR-V7. (B) Specificity of the BaseScope AR probes as demonstrated by signals detected in prostate cancer cell lines with known AR-FL/AR-V7 profiles. The cell lines PC3 (AR-FL–negative, AR-V7–negative), LNCaP (AR-FL–positive, AR-V7–negative), and LNCaP95 (AR-FL–positive, AR-V7–positive) were stained using the following 1 ZZ BaseScope probes: AR-E7/E8 for AR-FL and ARE3/CE3 for AR-V7. (C) The BaseScope assay detects mature mRNA exclusively in cytoplasm. LNCaP95 cells (AR-FL–positive, AR-V7–positive) were stained with standard RNAscope (top) and BaseScope (bottom) assays. Both cytoplasmic and intranuclear (arrows) signals were detected with 18 ZZ AR-E1 and 20 ZZ AR-V7 probes used in the standard RNAscope (top) assay, while the 1 ZZ probes used in the BaseScope (bottom) assays detected punctate signals representing mature mRNA exclusively in the cytoplasm. (D) Comparison of AR-V7 signals detected by the standard RNAscope and BaseScope assays in a metastatic castration-resistant prostate cancer (mCRPC) biopsy specimen. The same mCRPC biopsy core was processed and stained for AR-V7 using 20 ZZ AR-V7 probes in the standard RNAscope assay (top) and the BaseScope assay (bottom) using the 1 ZZ AR-E3/CE3 probe. Note the intense intranuclear AR-V7 signal (arrow) with the RNAscope assay.
2.3. Statistical analysis
The baseline clinical characteristics in the JHU cohort (n = 28, excluding 4 disqualified samples and 3 samples diagnosed with small cell carcinoma/neuroendocrine [SC/NE]), and UK cohort (n = 16, including all those collected before abiraterone or enzalutamide treatment) were separately compared according to AR-V7 status (positive vs negative). Categorical and continuous variables were compared using Fisher’s exact test and a Mann-Whitney test, respectively.
Exploratory evaluations of an association between AR status and treatment outcome were conducted among the combined cohort of all patients treated with abiraterone or enzalutamide (n = 25) following the biopsy procedure. Outcome measures included prostate-specific antigen progression-free survival (PSA-PFS) and clinical/radiographic progression-free survival (PFS). Survival time differences were analyzed using a log-rank test. In all tests, p ≤ 0.05 was considered statistically significant. Statistical analyses were performed using GraphPad Prism version 7.0 (GraphPad Software, San Diego, CA, USA).
3. Results
3.1. Junction specific AR RISH assay development
The splice junction between AR exon 3 and CE3 (E3/CE3) is specific to AR-V7 mRNA. Detection of this junction (ie, specific detection of AR-V7) has not been possible in morphologically intact cells and the native tissue environment because of technical constraints of the existing RNAscope RISH assay requiring 20 ZZ probes targeting the 1.3-kb CE3 sequence [10,21,22]. We designed and optimized a novel AR-V7 RISH probe consisting of a 1-ZZ pair of oligonucleotide sequences straddling the AR E3/CE3 junction, in parallel with a novel 1-ZZ probe for the AR-FL that straddles the splice junction between AR exon 7 and exon 8 (E7/E8; Fig. 1A). To validate the specificity of these novel junction-specific AR probes, we first performed RISH in human prostate cancer cell lines with known AR-FL/ARV7 expression profiles. As shown in Figure 1B, probes each consisting of 1-ZZ pairs (termed BaseScope probes) detected punctate cytoplasmic signals consistent with the known AR-FL/AR-V7 status of the cell lines. The improvement in specificity of the BaseScope assay over the RNAscope assay was shown by comparison of two RISH assays in LNCaP95 cells (positive for both AR-FL and AR-V7). Consistent with previous findings [21], the RNAscope probes (20 ZZ over 1 kb) designed to target the entire CE3 sequence detected both cytoplasmic dots from mature AR-V7 mRNA and nonspecific intranuclear signals from ARV7 pre-mRNA (Fig. 1C), precluding accurate quantification. By contrast, the junction-specific AR-V7 probe (<50 bases) detected signals for mature AR-V7 mRNA exclusively in the cytoplasm (Fig. 1C). Parallel comparison of these two AR-V7 RISH assays in a metastatic CRPC biopsy specimen further confirmed this distinction (Fig. 1D).
Although the novel prototype AR-V7 RISH assay appeared to detect fewer transcripts than the RNAscope assay owing to significantly fewer ZZ pairs for detection (Fig. 1C,D), the junction-specific detection made it possible to conduct automated quantification of AR-V7–specific signals (Supplementary Fig. 1). As shown in Supplementary Figure 2, quantitative measurements of AR-V7, AR-FL, and AR-V7/AR-FL ratios from the novel assay were consistent with values derived from RT-PCR in a set of metastatic biopsies from CRPC patients (n = 13) with matching FFPE and frozen specimens. AR-V7 can also be detected in a tissue microarray containing autopsy specimens from CRPC patients (Supplementary Fig. 3), although no statistically significant correlation between RISH and RNA-Seq was found (n = 7; Supplementary Fig. 3). Therefore, we have demonstrated the validity and feasibility of AR-V7 quantification by the novel RISH assay.
3.2. AR-V7/AR-FL quantification in biopsy specimens and correlation with baseline clinical characteristics
Having established the novel junction-specific AR RISH method, we generated quantitative AR-V7 and AR-FL RISH data from two independent biopsy cohorts while blinded to the sample identity. The first cohort consisted of 35 biopsies from patients with metastatic prostate cancer collected at JHU (Supplementary Table 1). After excluding four samples that did not meet the quality control criteria (no signal with the POLR2A-positive control probe), samples were grouped into SC/NE (n = 3), castration-sensitive prostate cancer (CSPC; n = 3), and CRPC (n = 25) on the basis of pathology reports and clinical notes. Representative images showing AR-V7/AR-FL measurements were shown in Figure 2A, and quantitative values for all 31 samples were shown in Figure 2B. Notably, samples with AR-V7 signals were always concurrently positive for AR-FL and, without exception, ARFL measurement values were higher than those for AR-V7 (Fig. 2B and Supplementary Table 1).
Fig. 2 –
Detection and quantification of AR-FL and AR-V7 in two independent biopsy cohorts. (A) Representative images and quantified RNA in situ hybridization (RISH) scores for AR-FL (probe AR-E7/E8, top) and AR-V7 (probe AR-E3/CE3, bottom) mRNA detection in tissue biopsies from patients with metastatic prostate cancer. (B) AR quantification by junction-specific RISH in the JHU cohort. Left panel: Quantified AR-FL and AR-V7 mRNA expression in three small cell carcinoma/neuroendocrine (SC/NE) biopsies, three castration-sensitive prostate cancer (CSPC) biopsies, and 25 castration-resistant prostate cancer (CRPC) biopsies from the JHU cohort. The line indicates the value for the AR-V7 cutoff (0.4). Right panel: Relative AR-V7/AR-FL values and ratios (for those that were AR-V7–positive defined by the cutoff) in each of the 25 CRPC specimens. (C) AR quantification by junction-specific RISH in the UK cohort. Left panel: Quantified AR-FL and AR-V7 mRNA expression in six CSPC biopsies and 19 CRPC biopsies from the UK cohort. The line denoted the AR-V7 cutoff (0.4). Right panel: Relative AR-V7/AR-FL values and ratios (for those that were AR-V7–positive defined by the cutoff) in each of the 19 CRPC specimens.
Because AR-V7 values exhibited a continuous range (Supplementary Table 1), it was necessary to define AR-V7 “positivity” before clinical correlative analysis. We used a cutoff value of 0.4 to define AR-V7 “positivity” (Supplementary material). Using this cutoff, six of the 12 samples (50%) that had an AR-V7 RISH value above zero were AR-V7–positive (Fig. 2B and Supplementary Table 1). AR-V7 positivity was associated with prior treatment with ketoconazole, abiraterone, or enzalutamide, but not with any other baseline variable in this set of 28 biopsies (Supplementary Table 2). After defining the cutoff, a second cohort of 28 biopsies (UK cohort) was evaluated (Supplementary Table 1) using the same RISH method, among which nine biopsies were AR-V7–positive according to the predefined cutoff (Fig. 2C). In this cohort, 16 samples had baseline data available at the sampling time before treatment with abiraterone or enzalutamide (Supplementary Table 2). AR-V7 positivity was associated with serum PSA, but not with any other baseline variables in this cohort (Supplementary Table 2). Quantitative AR-V7/AR-FL RISH values from the combined 56 biopsy samples are presented in Supplementary Figure 4. Notably, all CSPC specimens (n = 9) and SC/NE samples (n = 3) were negative for AR-V7 according to this novel detection method (Supplementary Fig. 4). Among the CRPC specimens (n = 44), the AR-V7– positive rate was 34.1% (15/44), and the median AR-V7/ARFL ratio was ~11.9% among AR-V7–positive samples (Supplementary Fig. 4).
3.3. Comparison of AR-V7 RISH and AR-V7 immunohistochemistry (IHC)
Detection of clinically significant AR-V7 can also be achieved by IHC using antibodies raised against the ARV7–specific peptide [8,24]. However, detection of nonspecific, unidentified protein targets in AR/AR-V7–negative cells has been reported [8]. To allow comparison of AR-V7 RISH and IHC results, we developed an optimized AR-V7 IHC method (Supplementary material) that uses a new AR-V7 antibody that specifically detected AR-V7 protein in cells with known AR-V7 status (Fig. 3A). In addition, areas of positive IHC staining corresponded to positive RISH staining in a sample with mixed SC/NE and adenocarcinoma histology (Fig. 3B). To further characterize the novel ARV7 RISH test, we compared AR-V7 measurements obtained with RISH and IHC methods (Supplementary material) in matched sections from 36 mCRPC biopsies (mainly from the UK cohort). The IHC results robustly correlated with the RISH results (Fig. 3C,D, Supplementary Table 3).
Fig. 3 –
Comparison of AR-FL/AR-V7 levels quantified by RNA in situ hybridization (RISH) and immunohistochemistry (IHC). (A) Western blot and IHC using the RevMab-RM7 AR-V7 antibody in prostate cancer cells with known AR profiles. Western blot showed the 80~kDa AR-V7 band consistent with known AR-V7 status in LNCaP (AR-V7–negative) and LNCaP95 (AR-V7–positive) cells. Different doses of LNCaP95 protein lysates were loaded. Nonspecific staining was shown at approximately 30 and 23 kDa. b-Actin was blotted as a loading control. In IHC experiments, PC3 cells showed negative AR-V7 IHC staining, LNCaP95 cells showed moderate AR-V7 staining, and HeLa cells transiently transfected with AR-V7 showed the highest level of ARV7 IHC staining (heterogeneity reflected the transfection efficiency). (B) AR-V7 IHC staining was compared with the AR-E3/CE3 BaseScope assay in a metastatic castration-resistant prostate cancer CRPC biopsy with mixed SC/NE and adenocarcinoma histology. (C) Representative images and quantified scores comparing IHC and RISH results in biopsies from the UK cohort. (D) Comparison of AR-V7 IHC values in AR-V7–positive (n = 10) and AR-V7– negative biopsies (n = 26) defined by junction-specific RISH. The p value was determined using an unpaired t test.
3.4. Association with treatment outcome
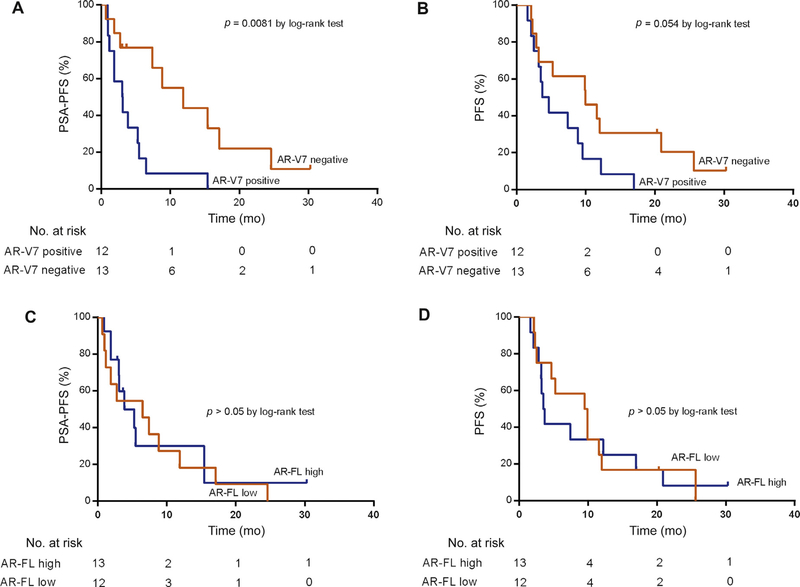
We conducted exploratory treatment outcome analyses after combining biopsies collected from patients treated with abiraterone or enzalutamide in the two cohorts. A total of 25 patients (n = 9 in the JHU cohort and n = 16 in the UK cohort) were biopsied before treatment with abiraterone or enzalutamide. PSA response rates were not significantly different by AR-V7 status, although a numerically better PSA response rate was observed in subjects with AR-V7 scores below the cutoff (Supplementary Fig. 5). AR-V7 status was significantly associated with shorter PSA-PFS (p = 0.0081; Fig. 4A) and showed a trend towards an association with PFS (p = 0.054; Fig. 4B). However, AR-FL status was not associated with either PSA-PFS or PFS in this combined cohort (Fig. 4C,D).
Fig. 4 –
Exploratory clinical outcome analysis of AR-V7 and AR-FL status determined by BaseScope assay in patients treated with abiraterone or enzalutamide (n = 25). (A) Kaplan-Meier analysis of prostate-specific antigen progression-free survival (PSA-PFS) by AR-V7 status. The median PSA-PFS was 3.1 mo in AR-V7–positive patients and 11.8 mo in AR-V7–negative patients (AR-V7 positivity hazard ratio [HR] for PSA-PFS 2.789, 95% confidence interval [CI] 1.12–6.95; p = 0.0081 by log-rank test). (B) Kaplan-Meier analysis of clinical or radiographic progression-free survival (PFS) by AR-V7 status. The median clinical or radiographic PFS was 4.2 mo in AR-V7–positive patients and 9.9 mo in AR-V7–negative patients (AR-V7 positivity HR for PFS 2.118, 95% CI 0.89–5.02; p = 0.054 by log-rank test). (C) Kaplan-Meier analysis of PSA-PFS by AR-FL status. The median PSA-PFS was 3.9 mo in AR-FL high patients and 6.5 mo in AR-FL low patients (AR-FL high status HR for PSA-PFS 1.167, 95% CI 0.4957–2.745; p = 0.7145 by log-rank test). (D) Kaplan-Meier analysis of clinical or radiographic PFS by AR-FL status. The median clinical or radiographic PFS was 3.6 mo in AR-FL high patients and 9.8 mo in AR-FL low patients (AR-FL high status HR for PFS 1.073, 95% CI 0.464–2.475; p = 0.8675 by log-rank test).
4. Discussion
Here we present the first example of visualization of splice junctions in morphologically intact cells using a novel RISH assay, and quantitative analysis of AR-FL/AR-V7 mRNA levels in FFPE biopsies obtained from mCRPC patients. Although the study was limited by cohort size, AR-V7 status was correlated with clinical characteristics and clinical outcomes after treatment with abiraterone or enzalutamide. This novel AR-V7 RISH test may help to address some of the limitations of the RT-PCR–based test, for which clinical development may be limited by preanalytical and analytical challenges because of reliance on detection of CTCs and low levels of the analytes in liquid biopsy samples [16]. For example, the CTC-based test requires relatively fresh blood samples delivered and processed within 24 h of collection. In addition, reporting of AR-V7 status would not be possible for patients with no detectable CTCs, although they usually present with lower disease burden and favorable treatment outcome [25]. For AR-V7 tests using biological substrates other than CTCs (exosomes, PBMCs, and whole blood), full analytical performance data have not been reported [12–15]. Although tissue-based tests require an invasive sampling procedure and may be further compromised by tissue heterogeneity, the role of molecular aberrations detected in tissue biopsies remains important [26]. It may be possible to develop treatment or patient selection markers on the basis of a biopsy, as indicated in a recent article suggesting the feasibility of obtaining molecular information representative of the patient by sampling a single metastasis[27]. Therefore, the newly developed capability for detection and quantification of a critical AR aberration in biopsy specimens, upon further work, may address a significant hurdle in measurement science for treatment and patient selection.
In situ detection of AR-V7 can also be achieved by IHC. Two recent studies demonstrated the prognostic value of AR-V7 detection by IHC in tissue specimens or CTCs immobilized on glass slides [8,24]. However, nonspecific signals from this antibody were acknowledged [8]. While antibody-based tests have a number of advantages, development of an optimized antibody is technically challenging and time-consuming. In our comparison of RISH and IHC (Fig. 3), we used a new AR-V7 antibody that was determined to be more specific than those evaluated in previous studies [8,24]. Although the measurements were generally concordant (Fig. 3), discrepancies were found (Supplementary Table 3), potentially reflecting measurement variations that may be related to nonspecific detection by IHC or different regulation of translation from mRNA to protein, as well as protein degradation among cases. Nevertheless, there is merit in further developing IHC-based detection methods for AR-V7, particularly since AR splice variant protein may have a longer half-life than its parent mRNA transcript [28]. Importantly, however, the RISH method described here can also be adapted for application in the CTC platforms described earlier [24] to allow further comparison of RISH and IHC.
Owing to the small sample size limited by difficulty in obtaining an adequate number of pretreatment biopsies, our clinical correlative analysis is exploratory and we did not conduct multivariable analysis adjusting for other prognostic factors. The small sample size also limited our ability to further optimize and validate the cutoff used to define AR-V7 status. As a result of these limitations, the potential clinical utility of the tissue-based RISH test (eg, in CTC-negative patients) remains to be determined. The main goal of the present study was to develop and validate a novel in situ AR-V7 test for detection of clinically significant levels of AR-V7, using a novel prototype method that had recently undergone substantial improvement with respect to detection sensitivity (personal communication between J.L. and X.M.). The present study achieved this goal with the clinical resources currently available to the study investigators. Full clinical validation may be conducted in tissue or immobilized CTC specimens collected from ongoing clinical trials, and prospective studies can be designed to evaluate the potential utility of this novel test in drug development and patient management.
5. Conclusions
We demonstrated for the first time a highly specific and quantifiable AR-V7 RISH test for detection of clinically significant levels of AR-V7 mRNA in prostate tissue specimens. Our data lend further credence to the clinical importance of AR splice variants and describe a novel assay that merits further clinical qualification in both tissue and CTCs in future clinical trials.
Supplementary Material
Acknowledgments
The authors would like to thank Ms. Helen Fedor for her excellent technical assistance with the research biopsies, as well as coordinated efforts from GU Oncology, Interventional Radiology, and Pathology at the Johns Hopkins University School of Medicine for patient recruitment, specimen procurement, and specimen processing. We thank Celestia Higano, Bruce Montgomery, Evan Yu, Heather Cheng, Elahe Mostaghel, Paul Lange, Martine Roudier, Lawrence True, and Robert Vessella for their contributions to the University of Washington Medical Center Prostate Cancer Donor Rapid Autopsy Program. We also thank the patients who participated in this study and their families.
Funding/Support and role of the sponsor: Work by the Johns Hopkins University School of Medicine was supported by National Institutes of Health Grants R01 CA185297 and P30 CA006973, Department of Defense Prostate Cancer Research Program Grants W81XWH-15-2-0050, Johns Hopkins Prostate SPORE Grant P50 CA058236, and the Prostate Cancer Foundation. Work in the de Bono laboratory was supported by funding from the US Department of Defense, the Prostate Cancer Foundation, Movember, Prostate Cancer UK, Stand Up To Cancer, Cancer Research UK, and the UK Department of Health through an Experimental Cancer Medicine Centre grant. Work by the University of Washington and Fred Hutchinson Cancer Research Center was supported by the Department of Defense Prostate Cancer Research Program (W81XWH-14-2-0183,W81XWH-13-2-0093 and W81XWH-15-1-0430), Pacific Northwest Prostate Cancer SPORE (P50CA97186), a PO1 NIH grant(PO1CA163227), GRECC Veterans Affairs Research Service, the Institute for Prostate Cancer Research, and the Prostate Cancer Foundation. Adam Sharp is supported by the Medical Research Council, the Academy of Medical Sciences and Prostate Cancer UK. BaseScope assay-related materials were provided by Advanced Cell Diagnostics, Inc. (Newark, CA, USA). The sponsors played a role in manuscript approval.
Financial disclosures: Jun Luo certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: Jun Luo has served as consultant/ advisor for Sanofi, Sun Pharma, and Janssen Pharmaceuticals. Emmanuel S. Antonarakis has served as consultant/advisor for Sanofi, Dendreon, Medivation, Janssen Biotech, ESSA, and Astellas Pharma. Changxue Lu, Jun Luo, and Emmanuel S. Antonarakis are co-inventors of a technology related to AR splice variants that was licensed to A&G Pharmaceuticals, Tokai, and Qiagen. Johann S. de Bono, Adam Sharp, Daniel Nava Rodrigues, and Ines Figueiredo are employees of The Institute of Cancer Research, which has a commercial interest in abiraterone. Johann S. de Bono has served as a consultant/advisory member for Astellas Pharma, AstraZeneca, Bayer, Genmab, Genentech, GlaxoSmithKline, Janssen, Medivation, Orion Pharma, Pfizer, Tesaro, and Sanofi. Peter S. Nelson has served as a consultant/advisor for Janssen, Astellas, and Genentech. CA Courtney Anderson Mindy Wang, Xingyong Wu, Yuling Luo, Nan Su, Emily Park, and Xiao-Jun Ma are employees of Advanced Cell Diagnostics. The remaining authors have nothing to disclose.
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.eururo.2017.08.009.
References
- [1].Antonarakis ES, Armstrong AJ, Dehm SM, Luo J. Androgen receptor variant-driven prostate cancer: clinical implications and therapeutic targeting. Prostate Cancer Prostatic Dis 2016;19:231–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Sharp A, Welti J, Blagg J, de Bono JS. Targeting androgen receptor aberrations in castration-resistant prostate cancer. Clin Cancer Res 2016;22:4280–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hu R, Dunn TA, Wei S, et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormonerefractory prostate cancer. Cancer Res 2009;69:16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Qu Y, Dai B, Ye D, et al. Constitutively active AR-V7 plays an essential role in the development and progression of castration-resistant prostate cancer. Sci Rep 2015;5:7654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hörnberg E, Ylitalo EB, Crnalic S, et al. Expression of androgen receptor splice variants in prostate cancer bone metastases is associated with castration-resistance and short survival. PLoS One 2011;6:e19059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Efstathiou E, Titus M, Wen S, et al. Molecular characterization of enzalutamide-treated bone metastatic castration-resistant prostate cancer. Eur Urol 2015;67:53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Steinestel J, Luedeke M, Arndt T, et al. Detecting predictive androgen receptor modifications in circulating prostate cancer cells. Oncotarget 2015;23:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Welti J, Rodrigues DN, Sharp A, et al. Analytical validation and clinical qualification of a new immunohistochemical assay for androgen receptor splice variant-7 protein expression in metastatic castration-resistant prostate cancer. Eur Urol 2016;70:599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Antonarakis ES, Lu C, Luber B, et al. Androgen receptor splice variant 7 and efficacy of taxane chemotherapy in patients with metastatic castration-resistant prostate cancer. JAMA Oncol 2015;1:582–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Antonarakis ES, Lu C, Wang H, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med 2014;371:1028–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Onstenk W, Sieuwerts AM, Kraan J, et al. Efficacy of cabazitaxel in castration-resistant prostate cancer is independent of the presence of AR-V7 in circulating tumor cells. Eur Urol 2015;68:939–45. [DOI] [PubMed] [Google Scholar]
- [12].Del Re M, Biasco E, Crucitta S, et al. The detection of androgen receptor splice variant 7 in plasma-derived exosomal RNA strongly predicts resistance to hormonal therapy in metastatic prostate cancer patients. Eur Urol 2017;71:680–7. [DOI] [PubMed] [Google Scholar]
- [13].Qu F, Xie W, Nakabayashi M, et al. Association of AR-V7 and prostate-specific antigen RNA levels in blood with efficacy of abiraterone acetate and enzalutamide treatment in men with prostate cancer. Clin Cancer Res 2017;23:726–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Todenhofer T, Azad A, Stewart C, et al. AR-V7 transcripts in whole blood RNA of patients with metastatic castration resistant prostate cancer correlate with response to abiraterone acetate. J Urol 2017;197:135–42. [DOI] [PubMed] [Google Scholar]
- [15].Liu X, Ledet E, Li D, et al. A whole blood assay for AR-V7 and ARv567es in patients with prostate cancer. J Urol 2016;196:1758–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Luo J Development of AR-V7 as a putative treatment selection marker for metastatic castration-resistant prostate cancer. Asian J Androl 2016;18:580–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lokhandwala PM, Riel SL, Haley L, et al. Analytical validation of androgen receptor splice variant 7 detection in a Clinical Laboratory Improvement Amendments (CLIA) laboratory setting. J Mol Diagn 2017;19:115–25. [DOI] [PubMed] [Google Scholar]
- [18].Levsky JM, Singer RH. Fluorescence in situ hybridization: past, present and future. J Cell Sci 2003;116:2833–8. [DOI] [PubMed] [Google Scholar]
- [19].Speel EJ, Hopman AH, Komminoth P. Tyramide signal amplification for DNA and mRNA in situ hybridization. Methods Mol Biol 2006;326:33–60. [DOI] [PubMed] [Google Scholar]
- [20].Wang F, Flanagan J, Su N, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn 2012;14:22–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Guedes LB, Morais CL, Almutairi F, et al. Analytic validation of RNA in situ hybridization (RISH) for AR and AR-V7 expression in human prostate cancer. Clin Cancer Res 2016;22:4651–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Saylor PJ, Lee RJ, Arora KS, et al. Branched chain RNA in situ hybridization for androgen receptor splice variant AR-V7 as a prognostic biomarker for metastatic castration-sensitive prostate cancer. Clin Cancer Res 2017;23:363–9. [DOI] [PubMed] [Google Scholar]
- [23].Kohli M, Ho Y, Hillman DW, et al. Androgen receptor variant ARV9 is co-expressed with AR-V7 in prostate cancer metastases and predicts abiraterone resistance. Clin Cancer Res 2017;23: 4704–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Scher HI, Lu D, Scgreiber NA, et al. Association of AR-V7 on circulating tumor cells as a treatment-specific biomarker with outcomes and survival in castration-resistant prostate cancer. JAMA Oncol 2016;2:1441–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Antonarakis ES, Lu C, Luber B, et al. Clinical significance of androgen receptor splice variant-7 mRNA detection in circulating tumor cells of men with metastatic castration-resistant prostate cancer treated with first- and second-line abiraterone and enzalutamide. J Clin Oncol 2017;35:2149–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Robinson D, Van Allen EM, Wu YM, et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015;161:1215–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kumar A, Coleman I, Morrissey C, et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat Med 2016;22:369–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ferraldeschi R, Welti J, Powers MV, et al. Second-generation HSP90 inhibitor onalespib blocks mRNA splicing of androgen receptor variant 7 in prostate cancer cells. Cancer Res 2016;76:2731–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.