Ebola virus (EBOV) is the causative agent of Ebola virus disease (EVD), a deadly disease and major public health threat worldwide. A safe and highly efficacious vesicular stomatitis virus-based vaccine against EBOV is the only platform that has successfully completed phase III clinical trials and has been used in recent and ongoing outbreaks. Earlier studies showed that antibodies are the main mode of protection when this vaccine is administered 28 days before EBOV challenge. Recently, we showed this vaccine can provide protection when administered as early as 3 days before challenge and before antibodies are detected. This study seeks to identify the mechanisms of rapid protection, which in turn will pave the way for improved vaccines and therapeutics. Additionally, this study provides insight into host gene expression signatures that could provide early biomarkers to identify infected individuals who are at highest risk of poor outcomes.
KEYWORDS: VSV-EBOV, Ebola virus, filovirus, macaque, vaccine
ABSTRACT
Ebola virus (EBOV) is a single-stranded RNA virus that causes Ebola virus disease (EVD), characterized by excessive inflammation, lymphocyte apoptosis, hemorrhage, and coagulation defects leading to multiorgan failure and shock. Recombinant vesicular stomatitis virus expressing the EBOV glycoprotein (VSV-EBOV), which is highly efficacious against lethal challenge in nonhuman primates, is the only vaccine that successfully completed a phase III clinical trial. Additional studies showed VSV-EBOV provides complete and partial protection to macaques immunized 7 and 3 days before EBOV challenge, respectively. However, the mechanisms by which this live-attenuated vaccine elicits rapid protection are only partially understood. To address this, we carried out a longitudinal transcriptome analysis of host responses in whole-blood samples collected from cynomolgus macaques vaccinated with VSV-EBOV 28, 21, 14, 7, and 3 days before EBOV challenge. Our findings indicate the transcriptional response to the vaccine peaks 7 days following vaccination and contains signatures of both innate antiviral immunity as well as B-cell activation. EBOV challenge 1 week after vaccination resulted in large gene expression changes suggestive of a recall adaptive immune response 14 days postchallenge. Lastly, the timing and magnitude of innate immunity and interferon-stimulated gene expression correlated with viral burden and disease outcome in animals vaccinated 3 days before challenge.
INTRODUCTION
Infection with Ebola virus (EBOV), an enveloped virus containing a negative-sense single-stranded genome, results in Ebola virus disease (EVD), characterized by excessive inflammation, lymphocyte death, vascular impairment, and coagulation defects with high fatality rates. Currently there are 3 main variants of EBOV used for research purposes: Mayinga, Kikwit, and Makona. The latter is responsible for the 2014-to-2016 epidemic that resulted in ∼28,600 cases and ∼11,300 fatalities (1). This unprecedented epidemic accelerated the progression of several vaccines through clinical trials (2). One vaccine platform that has reliably demonstrated 100% efficacy in nonhuman primate (NHP) models and successfully completed a phase III clinical trial is the recombinant live-attenuated vesicular stomatitis virus (VSV) expressing the EBOV glycoprotein (GP) in place of the VSV G protein (VSV-EBOV) (3). A single dose of VSV-EBOV provides complete protection against lethal EBOV challenge in cynomolgus macaques vaccinated 28 days before challenge (4). Moreover, administration of VSV-EBOV 30 min or 24 h after challenge confers approximately 50% protection in rhesus macaques (5, 6). Additionally, several people, including those exposed during the 2014-to-2016 epidemic, received VSV-EBOV as a postexposure treatment and did not exhibit any signs of EVD (7, 8). Multiple phase I-II clinical trials have demonstrated VSV-EBOV to be safe and immunogenic (9). Importantly, the phase III ring vaccination trial in Guinea demonstrated VSV-EBOV is efficacious in humans (10).
We previously established antibodies as the primary mode of protection conferred by VSV-EBOV when administered 28 days before challenge (11). We also reported complete and partial (66%) protection when cynomolgus macaques were immunized with VSV-EBOV 7 and 3 days before challenge, respectively (12). Given the lack of detectable EBOV GP-specific antibodies at the time of challenge, these data suggest VSV-EBOV can initially confer rapid protection through mechanisms other than humoral immunity. However, these mechanisms remain incompletely understood. To address this, we carried out a longitudinal transcriptome analysis of blood samples collected from cynomolgus macaques that were vaccinated 28, 21, 14, 7, and 3 days before EBOV-Makona challenge from our recent study (12).
Data presented herein show that animals vaccinated 14 days or more before challenge exhibited small transcriptional changes postchallenge. In contrast, animals vaccinated 7 days prior to challenge had a strong transcriptional response characterized by high expression of interferon (IFN)-stimulated genes (ISGs), viral RNA sensors, inhibitors of viral RNA synthesis, and markers of B-cell activation at the time of challenge. Interestingly, these animals exhibited large transcriptional changes 14 to 28 days postchallenge, suggestive of the generation of a recall adaptive immune response. Animals vaccinated 3 days before EBOV-Makona challenge also exhibited increased expression of transcripts associated with innate immunity and antiviral defense at the time of challenge, albeit significantly reduced in magnitude and breadth compared to the animals vaccinated 7 days before challenge. The kinetics and magnitude of these early innate immune gene expression changes were predictive of disease outcome. These data reveal that innate immune responses engendered by VSV-EBOV contribute to protection against lethal challenge within 3 days and, more importantly, can pave the way for the development of a robust humoral immune response.
RESULTS
To elucidate the mechanisms by which VSV-EBOV provides rapid protection against EBOV challenge, we leveraged samples collected during our previous study investigating the time to protective immunity following VSV-EBOV vaccination (12). Fifteen cynomolgus macaques were divided into groups of 2 or 3 animals that were immunized with a single intramuscular injection of 5 × 107 PFU of VSV-EBOV at 28, 21, 14, 7, or 3 days before lethal EBOV-Makona challenge (see Fig. S1A in the supplemental material). Additionally, a negative-control group was immunized with the VSV-Marburg virus GP vaccine (VSV-MARV), which does not provide cross protection against EBOV, 28 days before EBOV challenge and succumbed to infection 5 to 6 days postchallenge (DPC). RNA was extracted from blood samples collected from each group on 0, 3, 6, 14, 28, and 42 DPC, and transcriptome sequencing (RNA-Seq) was used to measure host gene expression changes (Fig. S1A).
Study timeline and ImmGen and DCQ analysis of transcriptional changes detected 7 days post-VSV-EBOV vaccination. (A) Study time line. (B) Heat map showing expression profile of DEGs detected 7 DPV in the day −7 group across various immune cell populations as predicted by ImmGen’s MyGeneSet application. Red indicates high while blue indicates low likelihood of expression within indicated immune cell population. B, B cells; T, T cells; NK, natural killer cells; DC, dendritic cells; MF, macrophages; MO, monocytes. (C) Changes in immune cell subsets based on DEGs detected in the day −7 group on the day of challenge were predicted using Immquant software, which implements the DCQ deconvolution algorithm using the IRIS database of gene expression. * and ** denote P values of ≤0.05 and ≤0.01, respectively, compared to baseline (BL) by unpaired t test. Mean and SEM are shown. Download FIG S1, TIF file, 1.4 MB (1.5MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
VSV-EBOV vaccination elicits innate immune responses and activates B cells.
Our previous studies using peripheral blood mononuclear cells (PBMCs) demonstrated the host response to VSV-EBOV peaks 7 days postvaccination (DPV) and returns to baseline 14 DPV (13). However, we were unable to assess responses at earlier time points and only had access to PBMCs. Therefore, in this study, we analyzed gene expression changes in whole-blood (WB) samples collected 3, 7, 14, and 21 DPV in order to gain a broader understanding of the response to vaccination with VSV-EBOV (the same vaccine that was deployed in the West African epidemic and is currently used in the Democratic Republic of the Congo [DRC] outbreak) (12). Our previous studies using VSV-EBOV demonstrated the host transcriptional response to vaccination is resolved 28 DPV (13). Thus, for this analysis, samples collected on the day of challenge from the day −3, −7, −14, and −21 vaccination groups were compared to those obtained from the day −28 group (baseline [BL]) in order to determine gene expression changes induced by VSV-EBOV vaccination on 3, 7, 14, and 21 DPV.
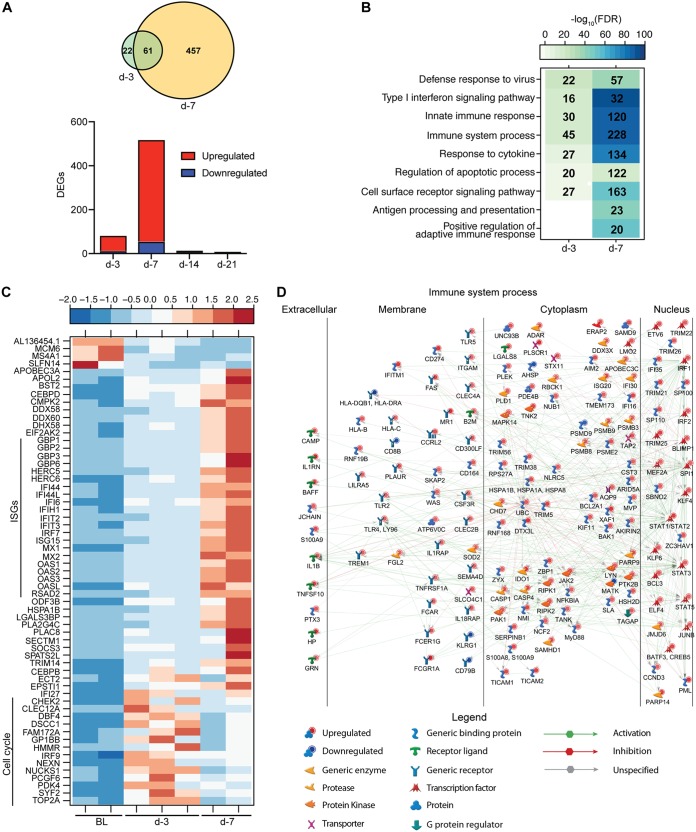
We detected 83 differentially expressed genes (DEGs) 3 DPV and 518 DEGs 7 DPV. In contrast, very few DEGs were detected 14 (12 DEGs) and 21 (6 DEGs) DPV (Fig. 1A). Functional enrichment revealed that DEGs detected 3 DPV enriched to Gene Ontology (GO) terms associated with defense response and innate immunity (Fig. 1B). Some of the DEGs detected 3 DPV were also detected 7 DPV, albeit with a higher magnitude of expression (Fig. 1A and C). These 61 DEGs consisted primarily of interferon-stimulated genes (ISGs) important for antiviral defense, such as OAS1, MX1, and IFIT2, as well as viral sensor genes DDX58 and DDX60 (Fig. 1C). Other DEGs that were most upregulated 3 DPV and, to a lesser extent, 7 DPV play a role in the cell cycle (DBF4, TOP2A, and CHEK2) (Fig. 1C).
FIG 1.
VSV-EBOV vaccination elicits transcriptional changes suggestive of antiviral innate immunity and B-cell activation. (A) The bar graph depicts the number of differentially expressed genes (DEGs; defined as those with a fold change [FC] of ≥2 and a false-discovery rate [FDR]-corrected P value of ≤0.05) that have human homologues detected in animals vaccinated 3, 7, 14, and 21 days before challenge relative to animals vaccinated 28 days before challenge on the day of challenge (DPC 0). Red indicates upregulated DEGs, while blue indicates downregulated DEGs. The Venn diagram displays overlap between DEGs detected in day −3- and day −7-vaccinated animals. (B) Functional enrichment of DEGs detected in the day −3- and day −7-vaccinated animals. Color intensity represents the statistical significance (shown as –log10 of the FDR-corrected P value); the range of colors is based on the lowest and highest –log10 (FDR) values for the entire set of Gene Ontology (GO) processes. The number of DEGs enriching to each GO term is listed within each box; blank boxes represent lack of significant enrichment to a given GO term. (C) Heat map representing expression (shown as normalized RPKM values) of DEGs detected in the day −3- and day −7-vaccinated animals on the day of challenge. The range of colors is based on scaled and centered RPKM values of the entire set of genes: red represents increased expression, while blue represents decreased expression. Each column represents 1 animal; BL represents the transcriptional profile of the day −28 animals at 0 DPC. (D) Network showing direct interactions between DEGs detected on the day of challenge in the day −7 group enriched to the GO term “Immune system process.”
DEGs detected 7 DPV mapped to GO terms associated with innate immune response, initiation of adaptive immunity, apoptosis, and signaling (Fig. 1B). Of the DEGs exclusively identified 7 DPV that enriched to “Immune system process,” 157 are predicted to directly interact as proteins (Fig. 1D). These genes play a role in antiviral innate immunity, including transcription factor genes STAT1 and IRF1, inflammatory mediator genes (i.e., MYD88, NFKBIA, and IL1B), and pathogen recognition receptor genes, such as TLR 2 and 4 (Fig. 1D). We also detected increased expression of genes involved in antigen processing and presentation, such as proteasome subunit genes PSMB9 and PSMB8 and major histocompatibility complex genes HLA-DQB1 and HLA-B, as well as TAP2, B2M, and MR1 (Fig. 1D). Moreover, we observed signatures of T-cell activation based on increased expression of HSH2D and TAGAP, as well as B-cell activation, as evidenced by increased expression of SPI1, JCHAIN, BAFF, LYN, PTK2B, and BLIMP1 (Fig. 1D). The 12 DEGs detected 14 DPV consisted of mitochondrial genes (MT-CO1 and MT-ND3) and ribosomal protein genes (RPL6 and RPS29), suggesting resolution of immune responses (data not shown).
We next used the Immunological Genome Project Consortium (ImmGen) database, which visualizes the distribution of gene expression across immune cell populations (14) in order to infer the source of DEGs detected 7 DPV. This analysis showed that vaccine-induced DEGs are highly expressed by antigen-presenting cells (monocytes and dendritic cells) and, to a lesser extent, lymphocytes (Fig. S1B). Additionally, we utilized Immquant, which implements a digital cell quantification algorithm (15), to predict changes in immune cell subsets based on DEGs detected 7 DPV. Using the Immune Response In Silico (IRIS) database (16), this analysis predicted the transcriptional changes to be associated with a significant increase in monocytes, activated dendritic cells, and CD4 Th2 T cells (Fig. S1C).
Innate immune responses generated in animals vaccinated 3 days before challenge correlate with disease outcome.
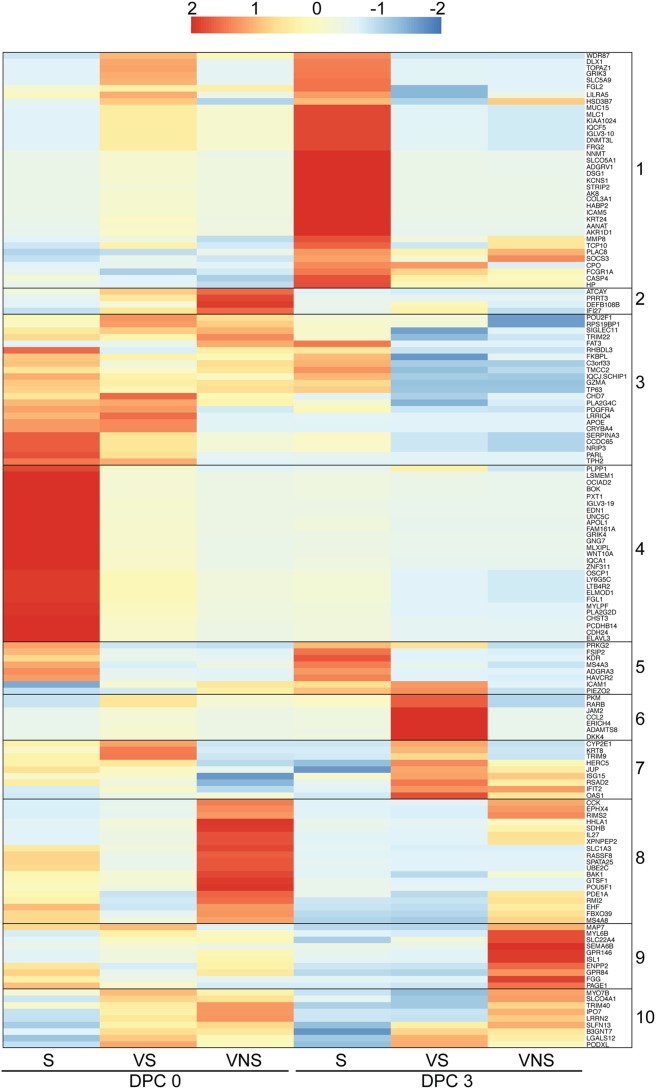
Each of the animals vaccinated 3 days before challenge displayed distinct outcomes following challenge (12). One animal (viremic nonsurvivor [VNS]) succumbed 8 DPC after exhibiting signs consistent with EVD (rash, thrombocytopenia, increased liver enzymes, and inflammatory mediators) and viremia on 6 and 8 DPC. The second animal (viremic survivor [VS]) developed mild signs of EVD and detectable viral titers 6 DPC but cleared the virus 9 DPC. The third animal exhibited no viremia or EVD symptoms (survivor [S]). To uncover gene signatures that determine disease outcome, we analyzed transcriptional changes 0, 3, and 6 DPC using maSigPro, which provides a set of statistically significant DEGs for the entire time course rather than at each time point (17). DPC 0 of the day −28 group served as a control, and each animal (VNS, VS, and S) was treated as a separate condition. This approach identified 689 protein-coding genes with temporal expression changes that were significantly different between the VNS, VS, and S animals. We next carried out unsupervised gene clustering using a Pearson correlation-based distance measure to identify groups of coregulated genes on 0 and 3 DPC (before the detection of viremia) and filtered genes that distinguished disease outcome, resulting in 152 genes (Fig. 2).
FIG 2.
Animals vaccinated 3 days before challenge display distinct transcriptional responses that correlate with disease outcome. Shown is a heat map representing hierarchical clustering of genes significantly associated with clinical outcome identified by maSigPro on 0 and 3 DPC. Clusters of genes that are coregulated are numbered 1 to 10. The range of colors is based on rlog transformation of normalized read counts: red represents increased expression, while blue represents decreased expression. Each column represents the nonviremic survivor (S), viremic survivor (VS), or viremic nonsurvivor (VNS).
Genes in cluster 4 were either exclusively or very highly expressed in the S animal 0 DPC compared to the VS and VNS animals. These genes play a role in regulation of wound healing (APOL1, FGL1, and EDN1), cell adhesion and migration (CDH24 and CHST3), innate immunity (LTB4R2, LY6G5C, and WNT10A), and humoral immunity (IGLV3 to -19) (Fig. 2). Additionally, genes highly expressed by the S animal at both 0 and 3 DPC (cluster 5) are involved in cell adhesion (KDR and ICAM1) and Th1-mediated immunity (HAVCR2). Expression of genes in cluster 4 was reduced in the survivor by 3 DPC, indicating a regulated innate immune response. Newly upregulated genes 3 DPC in the S animal (cluster 1) included ICAM5, a cell adhesion molecule gene, MMP8, which degrades extracellular matrix, IGLV3-10 and FCGR1A, which play a role in humoral immune response, and SOCS3, which negatively regulates cytokine signaling. Genes in cluster 3 were expressed by all animals on the day of challenge but remained upregulated only in the S animal on 3 DPC, such as TRIM22, an antiviral gene induced by interferon, GZMA, which is important in lysing target cells by cytotoxic T cells and NK cells, and SIGLEC11, which mediates anti-inflammatory signaling.
Genes within cluster 7, which mostly consisted of ISGs (OAS1, IFIT2, TRIM9, RSAD2, ISG15, and HERC5), were highly expressed by both the S and VS animals on the day of challenge compared to the VNS animal (Fig. 2). The expression of genes in cluster 7 increased dramatically 3 DPC in the VS animal, while ISG expression did not increase in the VNS animal until 3 DPC. Interestingly, genes in cluster 6 were highly expressed in the VS animal 3 DPC and play a role in monocyte chemotaxis (CCL2) and lymphocyte migration (JAM2).
Genes that were highly expressed in viremic animals (both VS and VNS) DPC 0 (clusters 2 and 10) play a role in type I IFN-induced apoptosis (IFI27), antibacterial defense (DEFB108B), and TRIM40, an E3 ubiquitin ligase that promotes proteasomal degradation of RIG-I and MDA5 (Fig. 2). Genes that were only upregulated in the VNS animal (clusters 8 and 9) are involved in angiogenesis (POU5F1), coagulation (FGG), vasodilation (XPNPEP2), apoptosis (BAK1), and ion transport (SLC1A3 and SLC22A4). Interestingly IL-27, which has been shown to regulate T-cell-mediated immunity and promote production of anti-inflammatory cytokine interleukin-10 (IL-10), was also elevated only in the VNS animal on the day of challenge (18) (Fig. 2).
By 14 DPC, no viremia was detected in VS and S; therefore, these two animals were grouped for DEG analysis at subsequent time points with DPC 0 of the day −28 group serving as the baseline again (see Fig. S2 in the supplemental material). Most transcriptional changes 14 DPC were upregulated, and DEGs with the highest fold change (FC) were enriched to GO term “Immune system process” and play a role in innate immunity (Fig. S2A, B, and C). Although most DEGs 28 DPC encoded innate immunity-related transcripts, their expression was downregulated, suggesting resolution of host response (Fig. S2A, B, and D).
Surviving animals in the day −3 group exhibit transcriptional changes consistent with resolution of immune response. (A) Number of DEGs in the 2 animals from the day −3 group that survived challenge 14, 28, and 42 DPC. (B) Functional enrichment of DEGs detected 14 and 28 DPC in the surviving animals from the day −3 group. Color intensity represents statistical significance (shown as −log10 of the FDR-corrected P value). The range of colors is based on the lowest and highest –log10 (FDR) values for the entire set of GO processes, the number of DEGs enriching to each GO process each day is listed within each box, and blank boxes represent lack of significant enrichment to a given GO term (C) Heat map of DEGs detected 14 DPC in the surviving animals from the day −3 group that enriched to “Immune system process” with a fold change (FC) of >6. Each column represents 1 animal; BL represents transcriptional profile of the day −28 animals at 0 DPC. (D) Heat map of DEGs detected 28 DPC in the surviving animals from the day −3 group that enriched to “Immune system process” with an FC of >3.8. Each column represents 1 animal; BL represents the transcriptional profile of the day −28 animals at 0 DPC. Download FIG S2, TIF file, 1.5 MB (1.5MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
EBOV-Makona challenge 1 week after vaccination induces a robust adaptive immune response.
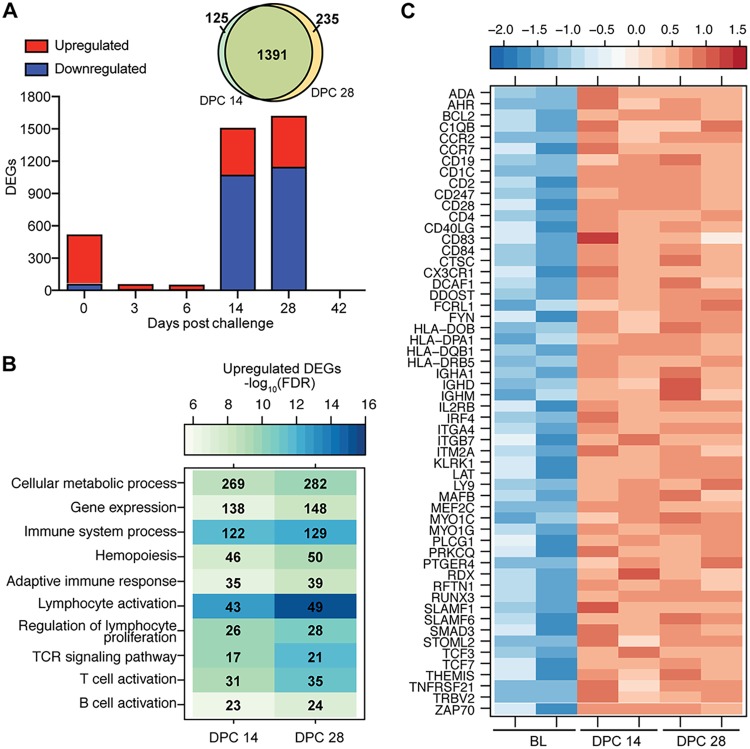
Samples collected from animals vaccinated 7 days prior to challenge were compared to BL. Following challenge, the number of DEGs detected 3 and 6 DPC initially decreased, followed by a large increase 14 and 28 DPC before a return to baseline by 42 DPC (Fig. 3A). DEGs detected 3 and 6 DPC enriched to “Innate immune response” and “Type I interferon signaling pathway” (see Fig. S3A in the supplemental material) and consisted mostly of upregulated ISGs (ISG15, MX2, OAS2, and IFIT2) (Fig. S3B).
FIG 3.
EBOV-Makona challenge 1 week after vaccination induces a recall response. (A) Number of DEGs detected in day −7-vaccinated animals following EBOV-Makona challenge. A Venn diagram displays overlap between DEGs detected 14 and 28 DPC in the day −7 group. (B) Functional enrichment of upregulated DEGs detected 14 and 28 DPC in the day −7 group. (C) Heat map representing upregulated DEGs detected 14 and 28 DPC in the day −7 group that enriched to “T-cell differentiation,” “B-cell activation,” and “Adaptive immune response.” Each column represents 1 animal; BL represents the transcriptional profile of the day −28 animals at 0 DPC.
Animals vaccinated 1 week before challenge exhibit large transcriptional changes associated with adaptive immunity. (A) Functional enrichment of DEGs detected 3 and 6 DPC in day −7-vaccinated animals. (B) Heat map of common DEGs detected 3 and 6 DPC in the day −7 group that enrich to “Immune system process.” Each column represents 1 animal; BL represents day −28 animals at 0 DPC. (C) Immquant DCQ analysis using the IRIS database of gene expression was used to predict changes in immune cell subsets based on DEGs detected 14 DPC in the day −7 group. *, **, ***, and **** denote P values of ≤0.05, ≤0.01, ≤0.001, and ≤0.0001, respectively, at 14 DPC compared to baseline (BL) by unpaired t test. The mean and SEM are shown. (D) Functional enrichment of downregulated DEGs detected 14 and 28 DPC in the day −7 group. (E) Heat map of the most downregulated DEGs detected 14 and 28 DPC in the day −7 group that enriched to “Myeloid leukocyte activation” with an FC of >5. Each column represents 1 animal; BL represents the transcriptional profile of the day −28 animals at 0 DPC. Download FIG S3, TIF file, 1.4 MB (1.4MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
DEGs detected 14 and 28 DPC were largely shared between these two time points (>90%) (Fig. 3A). DEGs upregulated 14 and 28 DPC enriched to GO terms associated with metabolism and adaptive immunity (Fig. 3B). DEGs that mapped to either “Adaptive immune response,” “T-cell activation,” or “B-cell activation” play a role in antigen presentation (HLA-DQB1, CD83, and CD1C), lymphocyte proliferation and development (IL2RB, TCF7, and TCF3), T-cell signaling and activation (CD2, CD4, CD28, LAT, TRBV2, and ZAP70), and B-cell signaling and activation (CD19, SLAMF1, CD40LG, and IGHA1) (Fig. 3C). In silico analysis using Immquant indicates these transcriptional changes are predicted to be associated with a significant increase in monocytes, CD4 and CD8 T cells, and stimulated CD4 Th1 T cells, as well as memory B cells, 14 DPC (Fig. S3C). DEGs downregulated 14 and 28 DPC also enriched to metabolism, cell cycle, and immunity, in addition to signaling and cell death (Fig. S3D). The most downregulated DEGs that enriched to “Myeloid leukocyte activation” play a role in chemotaxis and inflammation (MMP9, TNFAIP6, and S100A9), neutrophil-mediated immunity and chemotaxis (FPR2 and GCA, CXCR1, and CXCL8), and pathogen recognition (TLR 2 and 4) (Fig. S3D and E).
Animals vaccinated at least 2 weeks prior to challenge show limited gene expression changes following challenge.
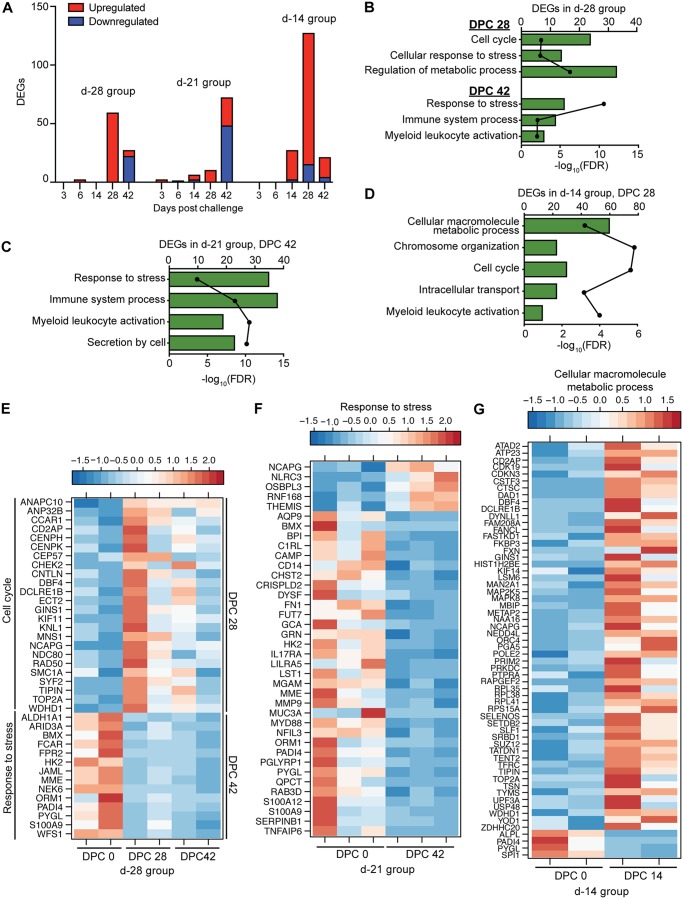
Consistent with the lack of clinical symptoms, we detected few DEGs in blood samples collected from the day −28 group following EBOV-Makona challenge (Fig. 4A). DEGs detected 28 DPC, which were mostly upregulated, play a role in cell cycle (e.g., CHEK2, ANAPC10, and KIF11) (Fig. 4B and E). At 42 DPC, DEGs were mostly downregulated and included genes involved in innate immunity (FPR1, S100A9, PADI4, and FCAR) (Fig. 4B and E). Similarly, a small number of DEGs was detected 42 DPC in the day −21 group (Fig. 4A). These DEGs, which were mostly downregulated, play a role in inflammation (e.g., S100A9, C1RL, CD14, and MYD88) (Fig. 4A, C, and F). A larger number of DEGs was detected in the day −14 group 28 DPC (Fig. 4A). These DEGs are associated with metabolic processes and include genes involved in ATP synthesis (ATP23), mitochondrial respiration (FXN), immunity (FKBP3 and CTSC), and DNA replication (ORC4, POLE2, and PRIM2) (Fig. 4D and G).
FIG 4.
Animals that were vaccinated at least 2 weeks prior to EBOV challenge show limited gene expression changes. (A) Number of DEGs detected following EBOV-Makona challenge in animals vaccinated 28, 21, and 14 days before challenge. (B to D) Functional enrichment of DEGs observed in animals vaccinated 28 (B), 21 (C), and 14 (D) days before challenge. (E) Heat map of DEGs detected 28 and 42 DPC in the day −28 group that enriched to “Cell cycle” or “Response to stress,” respectively. Each column represents 1 animal. (F) Heat map of DEGs detected 42 DPC in the day −21 group that enriched to “Response to stress.” Each column represents 1 animal. (G) Heat map representing DEGs detected 28 DPC in the day −14 group that enriched to the GO term “Cellular macromolecule metabolic process.” Each column represents 1 animal.
EBOV-Makona challenge results in gene expression changes consistent with excessive inflammation and lymphopenia in unprotected animals.
A substantial number of DEGs (380) were detected 3 days postchallenge (DPC) with EBOV-Makona in negative-control animals, with the largest changes observed 5 or 6 DPC (1,569 DEGs), consistent with a large increase in viral transcript counts (see Fig. S4A in the supplemental material). Overall transcriptional changes detected in negative-control animals are indicative of excessive inflammation and lymphopenia consistent with EVD development. Expression of the majority of genes upregulated 3 DPC remained increased 5 or 6 DPC and enriched to host defense, inflammation, cardiovascular disease, and cell death (Fig. S4B). Notable upregulated genes play a role in inflammation, such as transcription factor genes RELB, STAT1, CEBPB, and IRF7, signaling molecule genes MYD88 and IRAK 2 and 3, and chemokine/cytokine genes CCL2, CXCL10, and TNF (Fig. S4C). Additionally, ISGs (such as ISG15, IFIT2, OAS1 and -2, and MX1), as well as genes that play a role in cell death (e.g., FAS, BAK1, and TNFSF10A), were upregulated (Fig. S4C). Downregulated DEGs were only detected 5 or 6 DPC and mapped to cell cycle, cellular metabolism, and T-cell activation (see Fig. S5A in the supplemental material). Decreased transcripts that enriched to “TCR signaling pathway” included HLA-DMB, TCF7, THEMIS, and ZAP70 (Fig. S5B). The second major group of downregulated genes enriched to cell cycle, such as ANAPC10, CCND2, and CHEK2 (Fig. S5B).
Negative-control animals exhibit gene expression changes consistent with EVD. (A) Number of DEGs following EBOV-Makona challenge in animals that received VSV-MARV. The line graph represents the number of normalized viral transcripts (RPKM). *, P ≤ 0.05 compared to DPC 0. A Venn diagram displays overlap between DEGs detected 3 and 5 or 6 DPC in negative-control animals. (B) Functional enrichment of upregulated DEGs detected 3 and 5 or 6 DPC. (C) Network image of DEGs upregulated 5 or 6 DPC in negative-control animals that enriched to the GO term “Immune response,” with an FC of ≥5, and which directly interact with each other. Download FIG S4, TIF file, 2.0 MB (2MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Negative-control animals display gene expression changes consistent with lymphopenia and exhibit distinct transcriptional changes compared to naïve EBOV-Makona-challenged animals. (A) Functional enrichment of downregulated DEGs 5 or 6 DPC. Horizontal bar graphs represent the number of genes that mapped to each GO term listed, while the line graph represents the FDR-corrected P value. (B) Heat map of genes downregulated 5 or 6 DPC in negative-control animals that map to “TCR signaling” and “Cell cycle” (fold change of ≥4). Each column represents 1 animal. (C) Functional enrichment of DEGs detected 6 DPC in animals challenged with EBOV-Makona (19) or animals given VSV-MARV prior to EBOV-Makona challenge (present study). Color intensity represents the statistical significance; bubble size is based on the number of DEGs enriched. Download FIG S5, TIF file, 1.1 MB (1.1MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
We compared DEGs detected 6 DPC in negative controls to those reported in our recent study in which cynomolgus macaques were intramuscularly challenged with the same isolate of EBOV-Makona (19). Approximately ∼60% of DEGs detected 6 DPC (872 DEGs) were common, while the remaining 40% of DEGs represented unique transcriptional changes (660 DEGs in our previous study, 573 DEGs in this study). DEGs detected in both studies were associated with defense response to virus, response to type I IFN, cell death, and lymphopenia (Fig. S5C). DEGs detected only in our previous study (challenged with EBOV-Makona only) enriched to similar GO terms as the shared DEGs, while those detected exclusively in this study (vaccinated with VSV-MARV before EBOV-Makona challenge) also enriched to GO terms involved in cell cycle, DNA repair, and reactive oxygen species metabolism (Fig. S5C).
DISCUSSION
In this study, we performed a longitudinal transcriptome analysis in peripheral blood samples obtained postvaccination and post-EBOV-Makona challenge from cynomolgus macaques that were vaccinated 28, 21, 14, 7, and 3 days before challenge in order to identify mechanisms of rapid protection conferred by VSV-EBOV. We previously reported gene expression changes in NHP PBMCs following vaccination with VSV-EBOV (13). The present study provided us with an opportunity to gain a broader understanding of the immune response to VSV-EBOV by identifying transcriptional changes in whole-blood samples following vaccination with the human vaccine. Our analysis showed DEGs detected in WB post-VSV-EBOV vaccination are associated with antiviral innate immunity, notably ISGs, which had the highest degree of expression 7 DPV. This is in accordance with the elevated levels of IFN-α detected 3 and 7 DPV in these animals (12). Consistent with these transcriptional changes, ImmGen analysis indicates that gene expression changes detected 7 DPV originate mostly from monocytes and dendritic cells, which were also predicted to increase in frequency based on Immquant analysis. The production of IFN-α and expression of ISGs prior to infection correlate with survival in both day −7 and day −3 animals by potentially creating an antiviral state that restricts viral loads, thus providing enough time for development of an adaptive immune response.
Expression of several genes important for cell cycle progression was increased 3 and 7 DPV, consistent with the proliferative burst of lymphocytes observed 7 DPV in our previous studies (11). Additionally, our analysis revealed increased expression of genes important for antigen presentation, T-cell activation, and B-cell activation 7 DPV, in line with robust antibody responses detected 14 DPV. Indeed, we detected increased expression of LYN, BAFF, and BLIMP1, which are important for B-cell activation, proliferation, and differentiation into plasma cells (20–22). Moreover, Immquant predicted that transcriptional changes 7 DPV would be associated with an increase of CD4 Th2 T cells, a subclass of helper T cells that stimulate B cells to produce antibodies. A previous study showed that vaccination with a single dose of VSV-EBOV can confer partial protection when administered 20 to 30 min postexposure (5). However, another study showed that 6 cynomolgus macaques treated with 2 doses of either VSV-EBOV or VSV-MARV 1 h and 24 h after EBOV challenge had similar survival rates (6). Whether VSV-mediated innate immune responses solely contribute to rapid protection or if there is a role for EBOV-GP-specific immune responses should be further investigated.
In comparison to our recent transcriptional analysis in PBMCs following vaccination with VSV-EBOV (13), we detected a much larger number of DEGs in WB (467) compared to PBMCs (60) 7 DPV. Interestingly, only 12 DEGs were shared between the 2 studies and consisted of ISGs (IFIT2, IFIT3, IFI44, IFI44L, OAS2, OASL, and GBP6). This discrepancy could be mediated by differences in cellular composition of WB versus PBMCs, use of a higher vaccine dose (5 × 107 versus 1 × 107), and the immunogen (EBOV-Kikwit GP in the present study versus EBOV-Mayinga GP in our previous study).
Immunization 3 days prior to EBOV challenge was not uniformly protective against viremia or fatality and resulted in distinct disease outcomes (S, VS, and VNS) for each animal. Although transcriptional changes that enriched to type I interferon signaling and innate immunity were detected on the day of challenge in the day −3 group, the number of genes and magnitude of fold changes were much smaller than those observed in day −7-vaccinated animals. Gene signatures associated with survival include high expression of ISGs, as well as genes important for innate immunity on the day of challenge. In contrast, viremia and/or fatality was associated with high expression of genes that play a role in regulating immunity, coagulation, and apoptosis. Specifically, the animal that did not experience clinical disease (S) exhibited the highest number of transcripts involved in host defense as well as the highest plasma levels of IFN-α on the day of challenge (12). By 3 DPC, expression of these genes was reduced in this animal, while expression of genes involved in humoral immunity and effector responses increased, suggesting a regulated innate immune response and development of an adaptive immune response. This animal also developed EBOV GP-specific antibodies 7 DPC. As observed in the S animal, ISG transcripts were increased 0 DPC and peaked 3 DPC in the VS animal, contributing to the lower level of viremia. On the other hand, expression of genes associated with humoral immunity was not as high as that observed in the S animal. In line with this observation, development of EBOV GP-specific antibodies was delayed and reduced in magnitude in the VS compared to the S animal.
Unlike the two animals from the day −3 group that survived EBOV challenge, expression of ISGs and innate immune-related transcripts in the VNS animal was significantly delayed, appearing 3 DPC. This pattern of expression is reminiscent of that seen in unvaccinated cynomolgus macaques and patients who succumbed to EVD (19, 23, 24). Only 6 DEGs were common between the VNS animal and the day −7 animals on the day of challenge, including IFI27, TRIM22, BAK1, CHD7, PLA2G4C, and SLFN13, suggesting these genes do not correlate with survival or fatality. Interestingly, transcripts associated with negative regulation of innate immune responses (TRIM40 and IL-27) or promoting vasodilation and coagulation (XPNPEP2 and FGG) were only highly expressed by the VNS. Thus, it is reasonable that the absence of an early innate immune response in addition to indications of vascular disease may have resulted in development of EVD in this animal. Although there was limited overlap in the number of DEGs between VNS at 0 to 3 DPC and the negative-control animals at 6 DPC (11 DEGs), increased expression of FGG, the gamma component of fibrinogen, was detected in both the VNS animal and negative-control animals, which may be a potential marker that correlates with fatal disease outcome. In contrast to what we observed in animals vaccinated 7 days before challenge, we did not detect gene expression changes consistent with the generation of a recall response in the animals that were vaccinated 3 days before challenge and survived. These data are reminiscent of decreased immune responses observed when booster injections are administered too soon following priming vaccination (25).
Although animals in the day −7 group had weak IgM responses and no detectable EBOV GP-specific IgG antibodies at the time of challenge, they were protected against lethal infection (12). Our analysis showed an increased number of antiviral defense- and B-cell activation-related transcripts in these animals on the day of challenge, suggesting innate defenses engendered by VSV-EBOV coupled with the initiation of a humoral response contribute to initial protection. Following challenge, transcriptional changes in these animals are highly suggestive of the development of a recall adaptive immune response 14 to 28 DPC. Highly expressed genes play a role in antigen presentation and T- and B-cell activation, as well as T- and B-cell differentiation. Moreover, Immquant predicted an increase in CD4 and CD8 T cells and memory B cells 14 DPC. Although the GP-specific antibody titer did not increase after the detection of these transcriptional changes 14 to 28 DPC, it is possible the antibody repertoire and the avidity of the antibody response could have improved. Additionally, the quantity and quality of the T-cell response to EBOV could have also changed. Unfortunately, we are unable to address these questions due to a lack of cryopreserved cells. The reason for the delayed appearance of these transcriptional changes (14 DPC) is also not understood. These questions will be the focus of future studies.
Animals vaccinated 2 or more weeks before challenge exhibited few DEGs—most likely due to the presence of EBOV GP-specific IgG and neutralizing antibodies at the time of challenge (12). Animals vaccinated 28 days before challenge displayed limited transcriptional changes post-EBOV challenge, suggestive of resolution of an immune response. This is in contrast to our previous study (13), where we reported upregulation of genes involved in antiviral defense in cynomolgus macaques vaccinated with VSV-EBOV and challenged with EBOV-Kikwit 28 days later. The difference in the transcriptional changes and the lack of viral transcripts in this study could potentially be attributed to the use of different EBOV GP, increased vaccine dose, or different challenge strains (Makona versus Kikwit) (26). Similarly, expression of genes involved in response to stress and host defense was reduced in the day −21-vaccinated animals, indicative of immune regulation. Although we observed a larger number of DEGs in day −14-vaccinated animals compared to day −28- and −21-vaccinated animals, functional enrichment analysis revealed these DEGs are not consistent with EVD.
In contrast to VSV-EBOV-vaccinated animals, we detected robust gene expression changes in negative-control animals following EBOV-Makona challenge that correlated with viremia and clinical scores. The transcriptional profile of these animals was dominated by increased transcripts of genes that positively regulate production of inflammatory cytokines and chemokines. These transcriptional changes contribute to a cytokine storm, vascular leakage, and cell death. Moreover, we detected lymphopenia consistent with severe EVD and similar to that reported in recent NHP and clinical studies (19, 23, 24, 27, 28). We compared DEGs detected in the control animals with those we recently reported in a separate study investigating the transcriptional response to EBOV-Makona (19). Transcriptional changes reported in both studies were consistent with development of EVD. Interestingly, DEGs detected only in the negative-control animals in this study that were vaccinated with VSV-MARV prior to EBOV-Makona challenge enriched to cell cycle and DNA repair. In contrast, transcriptional changes detected exclusively in the earlier study enriched to immune terms. Since these animals received the same EBOV-Makona isolate (Guinea C07) and had similar disease progression, it is possible that these differences are due to previous exposure to the VSV-MARV vector prior to challenge, which resulted in less immune dysregulation and more pronounced changes in cell cycle that contributed to disease by promoting cell death.
In summary, the data presented here demonstrate that the robust innate antiviral immune response elicited by VSV-EBOV is followed by the development of a humoral response. This analysis provides novel insight into the possible role of innate immune responses and ISG expression in dictating disease outcome after EBOV challenge. Future studies should be repeated with a larger cohort and focus on (i) delineating the relative contribution of innate versus early adaptive immunity in mediating rapid VSV-EBOV protection, (ii) evaluating the durability of this immune response, (iii) determining effector functions elicited by antibodies generated using B-cell repertoire sequencing analysis, and (iv) investigating the role of T-cell responses in protection against subsequent infections.
MATERIALS AND METHODS
Study design.
We leveraged samples collected during our previous study (12), in which 15 cynomolgus macaques were divided into groups of 2 or 3 animals that were immunized with a single intramuscular injection of 5 × 107 PFU of VSV-EBOV at 28, 21, 14, 7, or 3 days before lethal EBOV-Makona challenge (Fig. S1A). Additionally, a negative-control group was immunized with the VSV-Marburg virus GP vaccine (VSV-MARV), which does not provide cross protection against EBOV, 28 days before EBOV challenge.
Animal ethics statement.
All animals from the previous study from which these samples were derived (12) were handled in strict accordance with the recommendations described in the Guide for the Care and Use of Laboratory Animals (29) of the National Institutes of Health, the Office of Animal Welfare, and the United States Department of Agriculture. Animal procedures were carried out under ketamine anesthesia by trained personnel under the supervision of veterinary staff, and all efforts were made to ameliorate the welfare and to minimize animal suffering in accordance with the recommendations of the Weatherall Report on the Use of Non-human Primates in Research (30). All animal work was approved by the Institutional Animal Care and Use Committee at the Rocky Mountain Laboratories (RML). RML is accredited by the American Association for Accreditation of Laboratory Animal Care.
Library generation and sequencing.
RNA was isolated from whole blood using the QIAmp viral RNA kit (Qiagen, Valencia, CA). RNA concentration and integrity were determined using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). rRNA was depleted and libraries were constructed using the TruSeq stranded total RNA LT-LS kit (Illumina, San Diego, CA). First, rRNA-depleted RNA was fragmented and converted to cDNA. Adapters were ligated, and the ∼300-bp-long fragments were then amplified by PCR and selected by size exclusion. In order to ensure proper sizing, quantitation, and quality prior to sequencing, libraries were analyzed on the Agilent 2100 Bioanalyzer. Multiplexed libraries were subjected to single-end 100-bp sequencing using the Illumina HiSeq2500.
Bioinformatic analysis.
Data analysis was performed with the RNA-Sequencing (RNA-Seq) workflow of systemPipeR (31). RNA-Seq reads were demultiplexed, quality filtered, and trimmed using Trim Galore (average phred score of 30 and minimum length of 50 bp). In order to quantify longitudinal changes in viral reads, the EBOV-Makona genome (H.sapiens-wt/GIN/2014/Makona-Gueckedou-C07) from Virus Pathogen Resource was adjoined to the Macaca fascicularis reference genome (Macaca_fascicularis.Macaca_fascicularis_5.0.dna.toplevel.fa) as previously described (19, 32). Trimmed RNA-Seq reads were aligned using HISAT2 against the reference genome containing both Macaca fascicularis and EBOV-Makona genomes. Raw expression values in the form of gene-level read counts were generated with the summarizeOverlaps function, counting only the reads overlapping exonic regions of genes using the annotation file from Ensembl (Macaca_fascicularis.Macaca_fascicularis_5.0.94.gtf).
Our previous studies using VSV-EBOV demonstrated host transcriptional responses to vaccination are resolved 28 days postvaccination (13). Thus, day 0 (day of challenge) of the day −28 group (n = 2) served as a baseline (BL). Gene expression changes induced by VSV-EBOV were determined by comparing samples collected on the day of challenge from animals vaccinated 21 (n = 3), 14 (n = 2), 7 (n = 2), and 3 (n = 3) days to those from BL. Following challenge, each time point was compared to the respective day 0 for groups at days −28, −21, and −14 or BL for groups at days −7 and −3.
Normalization and statistical analysis of differentially expressed genes (DEGs) were performed using edgeR. Host DEGs were defined as those with a fold change of ≥2 and a false-discovery rate (FDR)-corrected P value of ≤0.05. Only protein-coding genes with human homologs and an average of 5 reads per kilobase of transcript per million mapped reads (RPKM) were included for further analysis. Reads mapping to the EBOV-Makona genome were also normalized as RPKM. Statistical analysis of changes in normalized reads mapping to the EBOV-Makona genome was performed using edgeR. Heat maps and Venn diagrams were generated using R packages gplot and VennDiagram.
MaSigPro was used to obtain a list of genes that correlated significantly with disease outcomes in animals vaccinated 3 days before challenge and at early time points of infection. R2 of the regression model was set to 0.7, and vars was set to “all” in the “get.siggenes” function (17).
Functional enrichment of these genes was done to identify clusters of genes mapping to specific biological pathways, specifically Gene Ontology (GO) terms, using MetaCore (Thomson Reuters, New York, NY).
Data availability.
The RNA sequencing data presented in this article have been submitted to the National Center for Biotechnology Information Sequence Read Archive (accession no. PRJNA539939).
ACKNOWLEDGMENTS
This work was funded by the Division of Intramural Research (DIR), National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), and NIH grant 5U19A109945.
A.M., H.F., and I.M. conceived and designed the study. A.M. and H.F. performed the animal study and collected the samples. A.R.M. and A.J. conducted the experiments and analyzed the data. A.M., H.F., A.R.M., and I.M. wrote the manuscript. All authors approved the manuscript.
H.F. claims intellectual property regarding vesicular stomatitis virus-based vaccines for Ebola virus infections. All other authors declare no conflicts of interest.
Footnotes
Citation Menicucci AR, Jankeel A, Feldmann H, Marzi A, Messaoudi I. 2019. Antiviral innate responses induced by VSV-EBOV vaccination contribute to rapid protection. mBio 10:e00597-19. https://doi.org/10.1128/mBio.00597-19.
Contributor Information
Gary P. Kobinger, CHU de Quebec and Laval University.
Christine A. Biron, Brown University.
REFERENCES
- 1.CDC. 2017. 2014–2016 Ebola outbreak in West Africa. https://www.cdc.gov/vhf/ebola/history/2014-2016-outbreak/index.html. Accessed 5 March 2019.
- 2.Wang Y, Li J, Hu Y, Liang Q, Wei M, Zhu F. 2017. Ebola vaccines in clinical trial: the promising candidates. Hum Vaccin Immunother 13:153–168. doi: 10.1080/21645515.2016.1225637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garbutt M, Liebscher R, Wahl-Jensen V, Jones S, Möller P, Wagner R, Volchkov V, Klenk H-D, Feldmann H, Ströher U. 2004. Properties of replication-competent vesicular stomatitis virus vectors expressing glycoproteins of filoviruses and arenaviruses. J Virol 78:5458–5465. doi: 10.1128/JVI.78.10.5458-5465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones SM, Feldmann H, Stroher U, Geisbert JB, Fernando L, Grolla A, Klenk HD, Sullivan NJ, Volchkov VE, Fritz EA, Daddario KM, Hensley LE, Jahrling PB, Geisbert TW. 2005. Live attenuated recombinant vaccine protects nonhuman primates against Ebola and Marburg viruses. Nat Med 11:786–790. doi: 10.1038/nm1258. [DOI] [PubMed] [Google Scholar]
- 5.Feldmann H, Jones SM, Daddario-DiCaprio KM, Geisbert JB, Stroher U, Grolla A, Bray M, Fritz EA, Fernando L, Feldmann F, Hensley LE, Geisbert TW. 2007. Effective post-exposure treatment of Ebola infection. PLoS Pathog 3:e2. doi: 10.1371/journal.ppat.0030002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marzi A, Hanley PW, Haddock E, Martellaro C, Kobinger G, Feldmann H. 2016. Efficacy of vesicular stomatitis virus-Ebola virus postexposure treatment in rhesus macaques infected with Ebola virus Makona. J Infect Dis 214(Suppl 3):S360–S366. doi: 10.1093/infdis/jiw218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gunther S, Feldmann H, Geisbert TW, Hensley LE, Rollin PE, Nichol ST, Stroher U, Artsob H, Peters CJ, Ksiazek TG, Becker S, ter Meulen J, Olschlager S, Schmidt-Chanasit J, Sudeck H, Burchard GD, Schmiedel S. 2011. Management of accidental exposure to Ebola virus in the biosafety level 4 laboratory, Hamburg, Germany. J Infect Dis 204(Suppl 3):S785–S790. doi: 10.1093/infdis/jir298. [DOI] [PubMed] [Google Scholar]
- 8.Lai L, Davey R, Beck A, Xu Y, Suffredini AF, Palmore T, Kabbani S, Rogers S, Kobinger G, Alimonti J, Link CJ Jr, Rubinson L, Stroher U, Wolcott M, Dorman W, Uyeki TM, Feldmann H, Lane HC, Mulligan MJ. 2015. Emergency postexposure vaccination with vesicular stomatitis virus-vectored Ebola vaccine after needlestick. JAMA 313:1249–1255. doi: 10.1001/jama.2015.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong G, Mendoza EJ, Plummer FA, Gao GF, Kobinger GP, Qiu X. 2018. From bench to almost bedside: the long road to a licensed Ebola virus vaccine. Expert Opin Biol Ther 18:159–173. doi: 10.1080/14712598.2018.1404572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henao-Restrepo AM, Longini IM, Egger M, Dean NE, Edmunds WJ, Camacho A, Carroll MW, Doumbia M, Draguez B, Duraffour S, Enwere G, Grais R, Gunther S, Hossmann S, Kondé MK, Kone S, Kuisma E, Levine MM, Mandal S, Norheim G, Riveros X, Soumah A, Trelle S, Vicari AS, Watson CH, Kéïta S, Kieny MP, Røttingen J-A. 2015. Efficacy and effectiveness of an rVSV-vectored vaccine expressing Ebola surface glycoprotein: interim results from the Guinea ring vaccination cluster-randomised trial. Lancet 386:857–866. doi: 10.1016/S0140-6736(15)61117-5. [DOI] [PubMed] [Google Scholar]
- 11.Marzi A, Engelmann F, Feldmann F, Haberthur K, Shupert WL, Brining D, Scott DP, Geisbert TW, Kawaoka Y, Katze MG, Feldmann H, Messaoudi I. 2013. Antibodies are necessary for rVSV/ZEBOV-GP-mediated protection against lethal Ebola virus challenge in nonhuman primates. Proc Natl Acad Sci U S A 110:1893–1898. doi: 10.1073/pnas.1209591110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marzi A, Robertson SJ, Haddock E, Feldmann F, Hanley PW, Scott DP, Strong JE, Kobinger G, Best SM, Feldmann H. 2015. Ebola vaccine. VSV-EBOV rapidly protects macaques against infection with the 2014/15 Ebola virus outbreak strain. Science 349:739–742. doi: 10.1126/science.aab3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menicucci AR, Sureshchandra S, Marzi A, Feldmann H, Messaoudi I. 2017. Transcriptomic analysis reveals a previously unknown role for CD8(+) T-cells in rVSV-EBOV mediated protection. Sci Rep 7:919. doi: 10.1038/s41598-017-01032-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heng TS, Painter MW, Immunological Genome Project Consortium. 2008. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol 9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 15.Frishberg A, Brodt A, Steuerman Y, Gat-Viks I. 2016. ImmQuant: a user-friendly tool for inferring immune cell-type composition from gene-expression data. Bioinformatics 32:3842–3843. doi: 10.1093/bioinformatics/btw535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abbas AR, Baldwin D, Ma Y, Ouyang W, Gurney A, Martin F, Fong S, van Lookeren Campagne M, Godowski P, Williams PM, Chan AC, Clark HF. 2005. Immune Response In Silico (IRIS): immune-specific genes identified from a compendium of microarray expression data. Genes Immun 6:319–331. doi: 10.1038/sj.gene.6364173. [DOI] [PubMed] [Google Scholar]
- 17.Nueda MJ, Tarazona S, Conesa A. 2014. Next maSigPro: updating maSigPro bioconductor package for RNA-seq time series. Bioinformatics 30:2598–2602. doi: 10.1093/bioinformatics/btu333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freitas do Rosario AP, Lamb T, Spence P, Stephens R, Lang A, Roers A, Muller W, O'Garra A, Langhorne J. 2012. IL-27 promotes IL-10 production by effector Th1 CD4+ T cells: a critical mechanism for protection from severe immunopathology during malaria infection. J Immunol 188:1178–1190. doi: 10.4049/jimmunol.1102755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Versteeg K, Menicucci AR, Woolsey C, Mire CE, Geisbert JB, Cross RW, Agans KN, Jeske D, Messaoudi I, Geisbert TW. 2017. Infection with the Makona variant results in a delayed and distinct host immune response compared to previous Ebola virus variants. Sci Rep 7:9730. doi: 10.1038/s41598-017-09963-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gauld SB, Cambier JC. 2004. Src-family kinases in B-cell development and signaling. Oncogene 23:8001–8006. doi: 10.1038/sj.onc.1208075. [DOI] [PubMed] [Google Scholar]
- 21.Mackay F, Browning JL. 2002. BAFF: a fundamental survival factor for B cells. Nat Rev Immunol 2:465–475. doi: 10.1038/nri844. [DOI] [PubMed] [Google Scholar]
- 22.Cattoretti G, Angelin-Duclos C, Shaknovich R, Zhou H, Wang D, Alobeid B. 2005. PRDM1/Blimp-1 is expressed in human B-lymphocytes committed to the plasma cell lineage. J Pathol 206:76–86. doi: 10.1002/path.1752. [DOI] [PubMed] [Google Scholar]
- 23.Caballero IS, Honko AN, Gire SK, Winnicki SM, Mele M, Gerhardinger C, Lin AE, Rinn JL, Sabeti PC, Hensley LE, Connor JH. 2016. In vivo Ebola virus infection leads to a strong innate response in circulating immune cells. BMC Genomics 17:707. doi: 10.1186/s12864-016-3060-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X, Speranza E, Muñoz-Fontela C, Haldenby S, Rickett NY, Garcia-Dorival I, Fang Y, Hall Y, Zekeng E-G, Lüdtke A, Xia D, Kerber R, Krumkamp R, Duraffour S, Sissoko D, Kenny J, Rockliffe N, Williamson ED, Laws TR, N’Faly M, Matthews DA, Günther S, Cossins AR, Sprecher A, Connor JH, Carroll MW, Hiscox JA. 2017. Transcriptomic signatures differentiate survival from fatal outcomes in humans infected with Ebola virus. Genome Biol 18:4. doi: 10.1186/s13059-016-1137-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siegrist C-A. 2008. Vaccine immunology. Elsevier, Inc, Philadelphia, PA. [Google Scholar]
- 26.Reisler RB, Yu C, Donofrio MJ, Warren TK, Wells JB, Stuthman KS, Garza NL, Vantongeren SA, Donnelly GC, Kane CD, Kortepeter MG, Bavari S, Cardile AP. 2017. Clinical laboratory values as early indicators of Ebola virus infection in nonhuman primates. Emerg Infect Dis 23:1316–1324. doi: 10.3201/eid2308.170029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rubins KH, Hensley LE, Wahl-Jensen V, Daddario DiCaprio KM, Young HA, Reed DS, Jahrling PB, Brown PO, Relman DA, Geisbert TW. 2007. The temporal program of peripheral blood gene expression in the response of nonhuman primates to Ebola hemorrhagic fever. Genome Biol 8:R174. doi: 10.1186/gb-2007-8-8-r174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garamszegi S, Yen JY, Honko AN, Geisbert JB, Rubins KH, Geisbert TW, Xia Y, Hensley LE, Connor JH. 2014. Transcriptional correlates of disease outcome in anticoagulant-treated non-human primates infected with Ebolavirus. PLoS Negl Trop Dis 8:e3061. doi: 10.1371/journal.pntd.0003061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 30.Weatherall D. (ed). 2006. The Weatherall report on the use of non-human primates in research. https://royalsociety.org/-/media/Royal_Society_Content/policy/publications/2006/Weatherall-Report.pdf.
- 31.Backman TWH, Girke T. 2016. systemPipeR: NGS workflow and report generation environment. BMC Bioinformatics 17:388. doi: 10.1186/s12859-016-1241-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menicucci AR, Versteeg K, Woolsey C, Mire CE, Geisbert JB, Cross RW, Agans KN, Jankeel A, Geisbert TW, Messaoudi I. 2017. Transcriptome analysis of circulating immune cell subsets highlight the role of monocytes in Zaire Ebola virus Makona pathogenesis. Front Immunol 8:1372. doi: 10.3389/fimmu.2017.01372. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Study timeline and ImmGen and DCQ analysis of transcriptional changes detected 7 days post-VSV-EBOV vaccination. (A) Study time line. (B) Heat map showing expression profile of DEGs detected 7 DPV in the day −7 group across various immune cell populations as predicted by ImmGen’s MyGeneSet application. Red indicates high while blue indicates low likelihood of expression within indicated immune cell population. B, B cells; T, T cells; NK, natural killer cells; DC, dendritic cells; MF, macrophages; MO, monocytes. (C) Changes in immune cell subsets based on DEGs detected in the day −7 group on the day of challenge were predicted using Immquant software, which implements the DCQ deconvolution algorithm using the IRIS database of gene expression. * and ** denote P values of ≤0.05 and ≤0.01, respectively, compared to baseline (BL) by unpaired t test. Mean and SEM are shown. Download FIG S1, TIF file, 1.4 MB (1.5MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Surviving animals in the day −3 group exhibit transcriptional changes consistent with resolution of immune response. (A) Number of DEGs in the 2 animals from the day −3 group that survived challenge 14, 28, and 42 DPC. (B) Functional enrichment of DEGs detected 14 and 28 DPC in the surviving animals from the day −3 group. Color intensity represents statistical significance (shown as −log10 of the FDR-corrected P value). The range of colors is based on the lowest and highest –log10 (FDR) values for the entire set of GO processes, the number of DEGs enriching to each GO process each day is listed within each box, and blank boxes represent lack of significant enrichment to a given GO term (C) Heat map of DEGs detected 14 DPC in the surviving animals from the day −3 group that enriched to “Immune system process” with a fold change (FC) of >6. Each column represents 1 animal; BL represents transcriptional profile of the day −28 animals at 0 DPC. (D) Heat map of DEGs detected 28 DPC in the surviving animals from the day −3 group that enriched to “Immune system process” with an FC of >3.8. Each column represents 1 animal; BL represents the transcriptional profile of the day −28 animals at 0 DPC. Download FIG S2, TIF file, 1.5 MB (1.5MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Animals vaccinated 1 week before challenge exhibit large transcriptional changes associated with adaptive immunity. (A) Functional enrichment of DEGs detected 3 and 6 DPC in day −7-vaccinated animals. (B) Heat map of common DEGs detected 3 and 6 DPC in the day −7 group that enrich to “Immune system process.” Each column represents 1 animal; BL represents day −28 animals at 0 DPC. (C) Immquant DCQ analysis using the IRIS database of gene expression was used to predict changes in immune cell subsets based on DEGs detected 14 DPC in the day −7 group. *, **, ***, and **** denote P values of ≤0.05, ≤0.01, ≤0.001, and ≤0.0001, respectively, at 14 DPC compared to baseline (BL) by unpaired t test. The mean and SEM are shown. (D) Functional enrichment of downregulated DEGs detected 14 and 28 DPC in the day −7 group. (E) Heat map of the most downregulated DEGs detected 14 and 28 DPC in the day −7 group that enriched to “Myeloid leukocyte activation” with an FC of >5. Each column represents 1 animal; BL represents the transcriptional profile of the day −28 animals at 0 DPC. Download FIG S3, TIF file, 1.4 MB (1.4MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Negative-control animals exhibit gene expression changes consistent with EVD. (A) Number of DEGs following EBOV-Makona challenge in animals that received VSV-MARV. The line graph represents the number of normalized viral transcripts (RPKM). *, P ≤ 0.05 compared to DPC 0. A Venn diagram displays overlap between DEGs detected 3 and 5 or 6 DPC in negative-control animals. (B) Functional enrichment of upregulated DEGs detected 3 and 5 or 6 DPC. (C) Network image of DEGs upregulated 5 or 6 DPC in negative-control animals that enriched to the GO term “Immune response,” with an FC of ≥5, and which directly interact with each other. Download FIG S4, TIF file, 2.0 MB (2MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Negative-control animals display gene expression changes consistent with lymphopenia and exhibit distinct transcriptional changes compared to naïve EBOV-Makona-challenged animals. (A) Functional enrichment of downregulated DEGs 5 or 6 DPC. Horizontal bar graphs represent the number of genes that mapped to each GO term listed, while the line graph represents the FDR-corrected P value. (B) Heat map of genes downregulated 5 or 6 DPC in negative-control animals that map to “TCR signaling” and “Cell cycle” (fold change of ≥4). Each column represents 1 animal. (C) Functional enrichment of DEGs detected 6 DPC in animals challenged with EBOV-Makona (19) or animals given VSV-MARV prior to EBOV-Makona challenge (present study). Color intensity represents the statistical significance; bubble size is based on the number of DEGs enriched. Download FIG S5, TIF file, 1.1 MB (1.1MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data Availability Statement
The RNA sequencing data presented in this article have been submitted to the National Center for Biotechnology Information Sequence Read Archive (accession no. PRJNA539939).