Abstract
Mammalian sensory systems detect sweet taste through the activation of a single heteromeric T1R2/T1R3 receptor belonging to class C G-protein-coupled receptors. Allosteric ligands are known to interact within the transmembrane domain, yet a complete view of receptor activation remains elusive. By combining site-directed mutagenesis with computational modeling, we investigate the structure and dynamics of the allosteric binding pocket of the T1R3 sweet-taste receptor in its apo form, and in the presence of an allosteric ligand, cyclamate. A novel positively charged residue at the extracellular loop 2 is shown to interact with the ligand. Molecular dynamics simulations capture significant differences in the behavior of a network of conserved residues with and without cyclamate, although they do not directly interact with the allosteric ligand. Structural models show that they adopt alternate conformations, associated with a conformational change in the transmembrane region. Site-directed mutagenesis confirms that these residues are unequivocally involved in the receptor function and the allosteric signaling mechanism of the sweet-taste receptor. Similar to a large portion of the transmembrane domain, they are highly conserved among mammals, suggesting an activation mechanism that is evolutionarily conserved. This work provides a structural basis for describing the dynamics of the receptor, and for the rational design of new sweet-taste modulators.
Keywords: allosteric binding site, class C GPCR, cyclamate, mammalian, sweet-taste receptor, taste modulator
Introduction
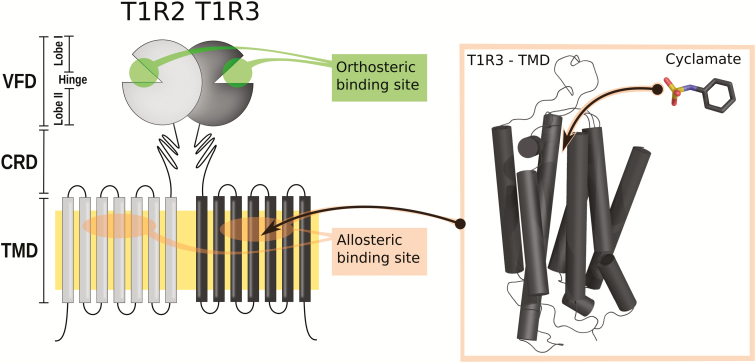
Our innate preference for sweet foods acts as a major determinant in the overconsumption of sugar, which is considered a significant public health problem in industrial countries. We perceive sweetness via the activation of a single receptor (Nelson et al. 2001, 2002) expressed at the surface of gustatory cells located in taste buds. Behind this apparent simplicity, the sweet-taste receptor is composed of 2 different subunits: T1R2 and T1R3, for Taste 1 Receptor member 2 and 3, respectively. These subunits contain extracellular and transmembrane domains (TMDs) that assume different functional roles (Li et al. 2002; Xu et al. 2004). Though only 1 receptor is devoted to sweet taste, multiple ligand-binding sites have been reported (DuBois 2016), which explains the wide chemical space of sweet compounds (Figure 1).
Figure 1.
Schematic of the sweet-taste receptor structure. VFD, CRD, TMD. This figure is reproduced in color in the online version of the issue.
The sweet-taste receptor belongs to the class C G protein-coupled receptor (GPCR) family that features a common structure comprised a large extracellular domain, called the venus flytrap domain (VFD), which is connected to a 7-helix TMD by a cysteine-rich domain (CRD) (Figure 1). The canonical activation mechanism of class C GPCRs follows a multiple-step process that requires communication between the VFDs (housing the orthosteric-binding site) and the TMDs via the CRDs (Rondard and Pin 2015; Xue et al. 2015; Kim et al. 2017). High sequence identity among mammalian sweet-taste receptors suggests a conserved activation mechanism across evolution despite some carnivores, such as cats, which have an inactivated T1R2 gene and have lost carbohydrate preferences (Jiang et al. 2012).
Natural sweeteners interact with the orthosteric binding pocket located in T1R2 (Xu et al. 2004). The closure of the T1R2 extracellular domain involves the rotation of both T1R2 and T1R3 VFDs. The signal is then transmitted to the TMDs via the CRDs (Rondard and Pin 2015). The presence of disulfide bonds rigidifies the CRD structure and amplifies the mechanical constraints like a lever (Chéron et al. 2017). It has also been shown that sweet proteins modulate the receptor by interacting with the CRD (Jiang et al. 2004). The coupling between the VFDs and the TMDs initiates a modification of the transmembrane protein–protein interface (Xue et al. 2015). A truncated class C GPCR (without its extracellular domain) still behaves like a GPCR in the presence of allosteric modulator (Goudet et al. 2004). This suggests that the TMDs contain molecular switches that control G protein coupling, but the final steps of the activation mechanism are still under debate.
To date, no experimental structure of the TMD of the sweet-taste receptor has been available. Nevertheless, recent X-ray crystal structures of other class C GPCRs (Doré et al. 2014; Wu et al. 2014), the metabotropic glutamate receptors mGluR1 and mGluR5, represent a significant breakthrough for the study of structure–function relationships of the sweet-taste receptor. We have previously shown the high conservation of residues involved in ligand recognition and receptor activation (Chéron et al. 2017). Allosteric modulators are known to interact within the TMD (Jiang et al. 2005a, 2005b; Winnig et al. 2007; Zhang et al. 2008), yet the allosteric modulation mechanism remains unclear. Therefore, it is only by unveiling the molecular basis of allosteric modulation that we can obtain a complete view of the receptor activation, which is fundamental for identifying new sweet-taste modulators.
In this study, we provide an atomistic-level view of the T1R3-specific allosteric signaling pathway by combining computational modeling and new mutagenesis experiments. Molecular dynamics (MD) simulations capture the dynamics of the ligand–receptor association process and sample the differential behavior of residues N7375.47, Y7716.46, and W7756.50 in the presence of cyclamate. Comparing the receptor response to orthosteric and allosteric agonists, in vitro experiments confirm the role of these residues in the allosteric regulation of receptor signaling. These residues form the inner core of the TMD, which is highly conserved in mammals, suggesting a crucial role in sweet-taste transduction.
Results and discussion
A novel positively charged residue on extracellular loop 2 is involved in the recognition of allosteric modulators
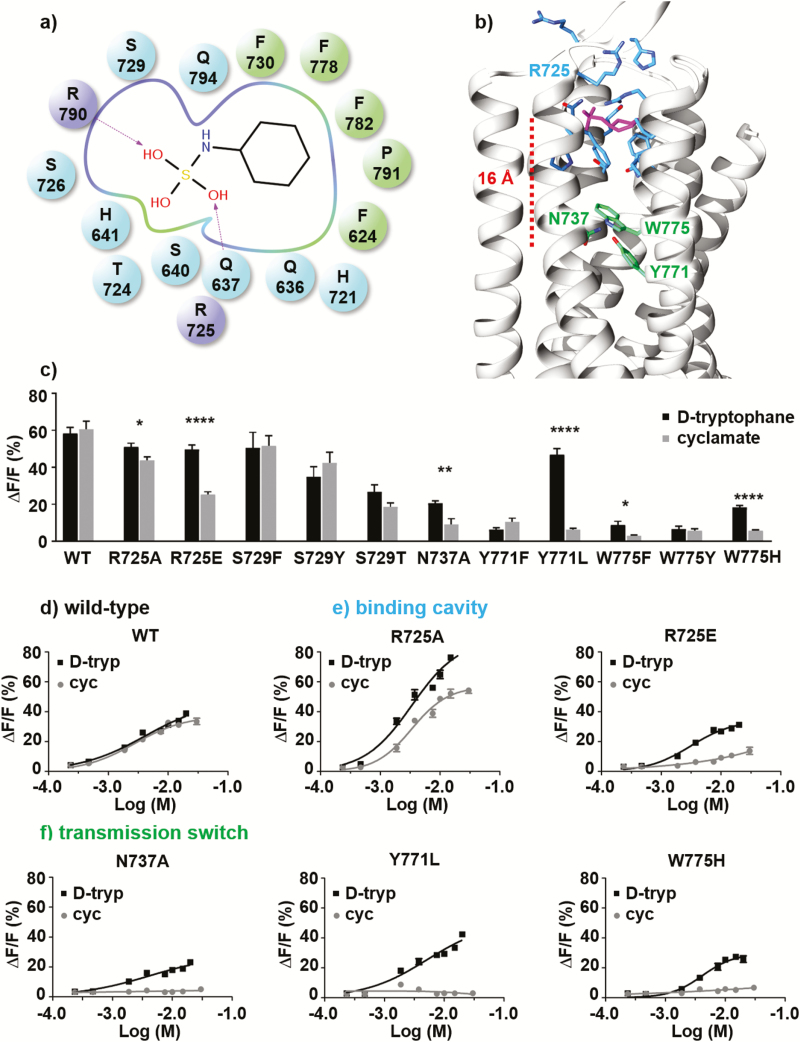
The binding mode of cyclamate within the allosteric binding cavity of the receptor, as captured by computational modeling, is fully consistent with previously published experimental data (Jiang et al. 2005b; Winnig et al. 2007) (Figure 2 and Supplementary Table 1). To sample the position of the agonist in the allosteric binding cavity without a priori assumptions of its exact location, MD simulations were performed starting with the apo form of the receptor and 5 ligands placed in the bulk phase. During the simulations 1 cyclamate molecule enters the binding pocket after 200 ns and reproduces a similar position to that found in the docking simulations (final root mean square deviation ~1.5 Å, Supplementary Figure 1). The sulfamate functional group of cyclamate interacts with the polar part of the binding pocket, whereas the apolar cyclic fragment interacts with the most hydrophobic part of the binding pocket (Figure 2a, b). To enhance the sampling of the ligand–receptor interactions, multiple independent MD simulations have been performed considering the ligand already docked in the binding pocket, as detailed in the Experimental procedures section and in Supplementary Table 2.
Figure 2.
(a) Schematic representation of the ligand–receptor interactions. Charged (R725ecl2 and R7907.28), polar (Q6363.32, Q6373.33, S6403.36, H6413.37, H721ecl2, S726ecl2, S7295.39, and Q7947.32), and hydrophobic residues (F6242.56, F7305.40, F7786.53, L7826.57, and P7917.29) are represented as purple, blue and green spheres (dark and light grey in the print version), respectively. (b) Representative structure of the cyclamate-bound T1R3 receptor obtained from molecular dynamics simulations. The binding mode is consistent with the best docking solution and recapitulates available experimental data. Residues of the allosteric binding pocket and those involved in the transmission switch are respectively shown as blue and green (light grey in the print version) sticks. (c) Activity of wild-type sweet-taste receptor and single-point mutants of functional residues upon application of 10-mM d-tryptophan (■) and cyclamate (●); each experiment was repeated 3 times. (Statistical significance: *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001.) (d-f) d-tryptophan (■) and cyclamate (●) dose–response curves obtained for wild-type sweet-taste receptor (d) and single-point mutants of residue R725ecl2 (e), N7375.47, Y7716.46, and W7756.50 (f); each experiment was repeated twice.
The molecular model proposed here recovers all the ligand–receptor interactions suggested in the previous models by one of us (Jiang et al. 2005b) and by Winnig et al. (2007), except for R723ecl2, which is replaced here by R725ecl2 (Supplementary Table 1). The negatively charged sulfamate component of the molecule preferentially interacts with the positively charged side chains of residues R723ecl2 or R725ecl2 in addition to R7907.28. In the presence of cyclamate, interacting residues of the extracellular loop 2 (ecl2) lock the ligand within the allosteric pocket. MD simulations of the wild-type receptor and the R725Aecl2 mutant show that cyclamate rotates and samples different interacting modes within the binding pocket (Supplementary Figures 2 and 4). The loss of 1 interacting guanidinium functional group, acting as a strong anchor point for cyclamate, is compensated by the presence of charged or polar residues (Supplementary Figure 4).
Consistent with the in silico model, in vitro mutants show that R725ecl2 is involved in cyclamate recognition. In vitro, the charge reversal mutation R725Eecl2 abolishes the receptor response to cyclamate but not to D-tryptophan (d-trp), an orthosteric ligand that binds within the extracellular part of the receptor and not within the transmembrane region (Figure 2e and Table 1). The R725Eecl2 mutation likely leads to a repulsion of the negatively charged sulfamate functional group of cyclamate, thus confirming that the cyclamate interacts with ECL2 at R725ecl2. Dose–response curves of R725Aecl2 mutants (Figure 2e) show no significant effect on ligand potencies. The EC50 of d-trp and cyclamate are within the same range of values for wild type and the mutant (Table 1). However, the efficacy of cyclamate is slightly reduced compared to d-trp in the R725Aecl2 in vitro mutant. It refers to the ability of cyclamate to activate the receptor whereas its affinity is similar between wild-type and mutant receptor. The stabilizing role of R725ecl2 can be partly compensated by the proximity of other polar or positively charged side-chain residues (for instance Q6363.32, Q6373.33, H721ecl2, R723ecl2, or R7907.28).
Table 1.
EC50 and IC50 values of the sweet-taste receptor ligands
| EC50 (mM) | IC50 (mM) | ||
|---|---|---|---|
| D-tryptophan | Cyclamate | D-tryptophan (10 mM) + lactisole | |
| WT | 4.1 | 2.7 | 0.016 |
| R725A | 3.4 | 3.3 | 0.165 |
| R725E | 3.1 | NR | 0.065 |
| N737A | 4.2 | NR | NT |
| Y771L | 5.1 | NR | NT |
| W775F | NR | NR | NT |
| W775H | 4.1 | NR | NT |
WT, wild type. NR means that EC50 cannot be determined because the receptor is not responsive. NT means that the system has not been tested.
To confirm the crucial role of ECL2 in the allosteric signaling pathway, the effect of R725Aecl2 and R725Eecl2 single-point mutations have been tested in the presence of lactisole, a sweet inhibitor. Dose–response curves show that the receptor is less sensitive to lactisole for both mutants (Supplementary Figure 6). The IC50 values are increased 4- to 10-fold than that of wild-type receptor (Table 1). Although the cyclamate binding is much more affected by the R725Eecl2 mutation, the inhibitory effect of lactisole is less altered by this mutation than the R725Aecl2 one. Despite both ligands interact within the T1R3 TMD, their chemical structures and their binding modes are different. For instance, it has been shown that single mutation on residues A7335.43 and L7987.36 affects receptor response to lactisole (Jiang et al. 2005a), whereas these mutants have no significant effect on cyclamate (Jiang et al. 2005b). On the other hand, single mutation on residues Q6363.32, H721ecl2, R723ecl2, S7295.39, and F7305.40 modifies receptor response to cyclamate but have no effect on the inhibitory role of lactisole. Only the following residues interact with both allosteric modulators: S6403.36, H6413.37, R725ecl2, R7907.28, F7786.53, and L7826.57.
A network of interacting residues controls the mechanism of the allosteric modulation
To explore the dynamics of the ligand–receptor complex, MD simulations of the receptor in apo form and in complex with the allosteric modulator cyclamate have been compared. Each simulation was analyzed separately to check the convergence and the stability of the molecular system (Supplementary Table 2). The structural analyses show that the simulations rapidly converged and that the complexes form stable macromolecular edifices (Supplementary Figure 2). The bundle part of the receptor, as well as its allosteric binding pocket, keeps its structural integrity throughout the MD simulations. As mentioned earlier, the rotation of the cyclamate within the binding pocket leads to structural fluctuations, as was also observed throughout the MD simulations (Supplementary Figures 2 and 4).
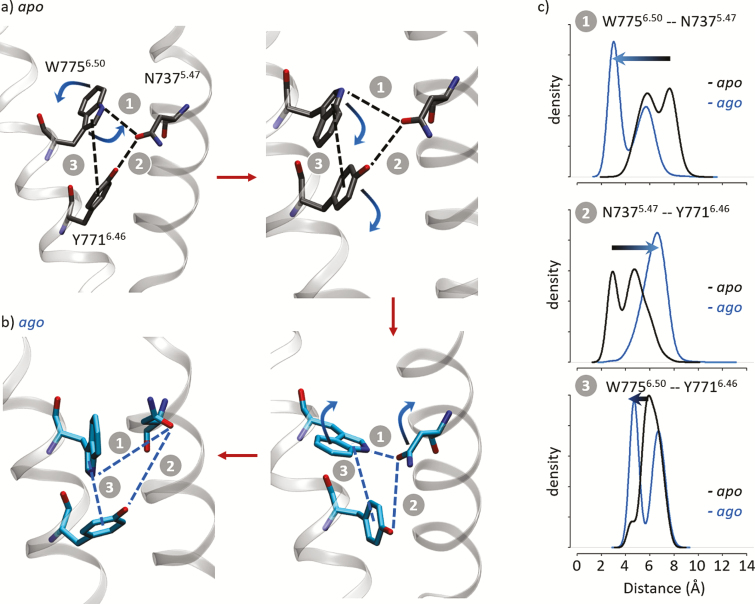
Interestingly, a network of interacting residues that do not directly interacts with cyclamate, including N7375.47, Y7716.46, and W7756.50 (Figure 2b), display different behaviors in the presence and absence of the ligand (Figure 3 and Supplementary Figure 5). Residue Y7716.46 is part of the triad (with L6383.34 and C8017.39) that forms the transmission switch of class C GPCRs (Doré et al. 2014; Chéron et al. 2017), that is, a molecular switch that acts as a ligand sensor and communicates the chemical signal to the cytosolic side of the receptor through a conformational change of the transmembrane helix 6 (TM6). In the apo state simulations, a network of 3 residues show strong interactions. W7756.50 side chain rotates to form a π–π interaction with Y7716.46 (Figure 3a). The latter interacts with the side chain of N7375.47 through a hydrogen bond. This network is modified when agonists are present within the cavity, 16 Å removed from this site. Figure 3b shows that the N7375.47-Y7716.46 hydrogen bond is disrupted in the presence of cyclamate. In the meantime, the W7756.50-Y7716.46 π-stacking is partially lost (Figure 3c). This leads to structural modifications of the receptor involving TM3, 5, and 6 that may correspond to conformational changes from inactive to active-like states (Supplementary Figure 3). Consequently, W7756.50 is more flexible and can adopt an alternate conformation to preferentially form a hydrogen bond with N7375.47 (Figure 3c).
Figure 3.
Structural analysis of the apo (a) and the agonist-bound (b) forms of the T1R3 receptor. Representative structures display the conformations of residues N7375.47, Y7716.46, and W7756.50. Red arrows suggest a putative activation mechanism. (c) Distribution of the inter-residue N7375.47-Y7716.46 and N7375.47-W7756.50 distances (1 and 2). Distribution of the Y7716.46-W7756.50 side-chain center-of-mass distances (3). Distances have been calculated between the Oδ1, Oη, and Nε1 atoms of N7375.47, Y7716.46, and W7756.50, respectively. Y7716.46 center of mass has been calculated considering the aromatic ring including Cε1, Cε2, Cδ1, Cδ2, Cγ, and Cζ atoms. W7756.50 center of mass has been calculated considering the pyrrole ring including Nε1, Cε2, Cδ1, Cδ2, Cγ atoms. See Supplementary Figure 4 for time series plots.
The effect of the Y771A6.46 and W775A6.50 substitutions in vitro has already been tested in previous studies (Jiang et al. 2005b; Winnig et al. 2007). The amino acid mutation series have been extended to decipher the role of these residues. Consistently, the Y771A6.46, Y771F6.46, W775A6.50, W775F6.50, and W775Y6.50 substitutions abolish the in vitro response of the receptor to both orthosteric and allosteric agonists (Figure 2c, f, Supplementary Figure 7 and Supplementary Table 3). This indicates that the hydrogen bond network involving N7375.47, Y7716.46, and W7756.50 is crucial for maintaining the functionality of the sweet-taste receptor regardless of the ligand. Dose–response curves of in vitro mutations N737A5.47, Y771L6.46, and W775H6.50 show a response to d-trp, but not to cyclamate (Figure 2c, f). Although the substitutions abolish the receptor activity toward cyclamate, they reduce the response to d-trp by up to 50%. In agreement with the in silico model, site-directed mutagenesis data of N7375.47, Y7716.46, and W7756.50 residues show differential effects on the receptor’s response to allosteric and orthosteric ligands, which confirms the role of these residues in the allosteric modulation pathway.
Allosteric modulation of the mammalian sweet-taste receptor involves conserved residues
We have shown how a network of residues within the T1R3 TMD affects the human sweet-taste receptor, upon allosteric ligand binding. These residues are highly conserved in mammalian sweet-taste receptor gene sequences, suggesting a conserved mechanism across species.
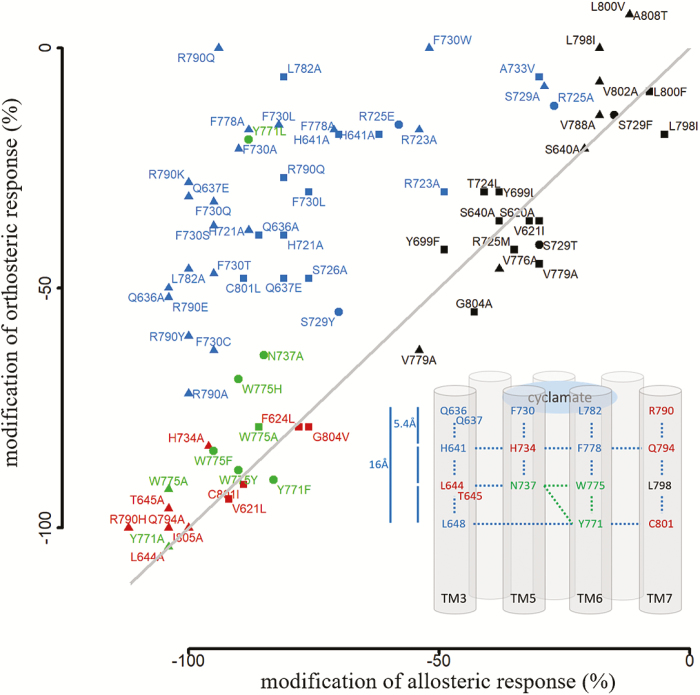
A meta-analysis of site-directed mutagenesis experiments shows a correlation between the position of a given residue in the structure and its effect on receptor response. We have reported the variation in receptor responses for each single-point mutation that was tested with both orthosteric (i.e., 1 that binds the T1R2 extracellular domain) and allosteric agonists. In the case of orthosteric ligands, d-trp and aspartame were considered (Jiang et al. 2005b; Winnig et al. 2007). The in vitro effect of each single-point mutation is summarized in Figure 4 and Supplementary Table 3. As expected, residues that form the allosteric binding pocket have a differential effect on allosteric and orthosteric ligands (in blue in Figure 4). Interestingly, residues that control the signaling pathway completely abolish the receptor response (in red and green in Figure 4). As a control, residues that are part of T1R3 bundle and do not participate in ligand recognition nor receptor activation, have no effect, or only a weak effect on receptor response (in black in Figure 4).
Figure 4.
Meta-analysis of T1R3 TMD site-directed mutagenesis data. Experimental data have been summarized from Jiang et al. (2005b) and Winnig et al. (2007), represented by triangle and square symbols. New site-directed mutagenesis experiments reported in the present study are indicated by filled circles. Receptor responses indicate the effect of mutations on the receptor activity, expressed as a percentage compared to the wild-type receptor, to the allosteric (cyclamate in x axis) and orthosteric (d-trp and aspartame in y axis) ligands. Cyclamate-specific interacting residues are in blue. Residues that control the signaling pathway or abolish the receptor response are colored in green and red, respectively. Residues that have no or weak-specific effect on receptor response are in black. All data are summarized in Supplementary Table 3.
An alignment of 41 mammalian T1R3 amino acid sequences indicates a correlation between the position of a given residue and its conservation among species (Supplementary Figure 8). The closer the residue is to the cradle of the allosteric binding cavity, the higher the conservation is (Supplementary Table 1). Deep inside the binding pocket, residues that form the core of the TM bundle, N7375.47, Y7716.46, W7756.50 (in green in Supplementary Figure 4; Y7716.46 corresponds to the transmission switch), and L6483.44 and C8017.39, are 97–100% conserved. This emphasizes that these residues are crucial for receptor functionality and suggests that a common allosteric activation mechanism has been conserved throughout evolution. In contrast, to activate the transmission switch located 3 helix turns (at roughly 16 Å) above the allosteric ligands, the chemical signal must propagate along the binding cavity through interacting side-chain residues (Figure 4). Residues that delimit the allosteric binding pocket of the sweet-taste receptor belong to TM3, 5, 6, and 7. Comparing the mammalian T1R3 sequences, these residues are highly conserved (>80% identity), except for 7 residues that are in close contact with cyclamate (Supplementary Figure 8 and Supplementary Table 1). They are located in the extracellular loop 2 (R723ecl2, T724ecl2, R725ecl2) and at the top of TM3, 5, 6, and 7 (S6403.36, F7305.40, L7826.57, and R7907.28). Interestingly, species-specific sweeteners such as cyclamate or neohesperidin dihydrochalcone (NHDC) are not recognized by rodents. Rodents possess a functional sweet-taste receptor and interspecies sequence variability within the T1R3 transmembrane region provides a rationale for the phenotypic differences in sweetener recognition (Li et al. 2011).
A model of sweet-taste receptor activation for the design of new taste modulators
To date, although a few sweet-taste modulators are known, such as cyclamate, lactisole, or NHDC, mechanistic clues at the molecular level of the sweet-taste modulation remain elusive. In this study, new experimental data, combined with novel structural details, reveal new crucial residues in the allosteric signaling pathway of the sweet-taste modulator, cyclamate.
The crystal structures of class C GPCRs mGluR1 (Wu et al. 2014) and 5 (Doré et al. 2014) clearly show that the 6.46 residue is part of the residue triad that defines the transmission switch (Doré et al. 2014; Chéron et al. 2017). It is very likely that residue W7756.50, 1 helical turn above the Y7716.46 toggle switch, participates in the ligand sensing, as already explored for equivalent residues in other class C members ((Chéron et al. 2017) and references therein), and in the TM6 deformation that leads to the activation of the receptor (Dalton et al. 2017; Pérez-Benito et al. 2017). These results are also compatible with the crucial role of TM6 at the dimer interface in the activated form of class C GPCR (Xue et al. 2015). This suggests regulation of the communication between the T1R2 and T1R3 subunits of the receptor through TM6.
Although cyclamate acts as an agonist for the sweet-taste receptor, it also enhances the response of the umami receptor (T1R1-T1R3) (Zhang et al. 2008). It has been suggested that cyclamate binding induces conformational changes in the T1R3 TMD, which in turn leads to intersubunit rearrangement between the 2 TMDs. This is fully compatible with our new experimental data and with our model of sweet-taste receptor activation.
The chemical structure of the histidine and tryptophan side chains and the dynamics of these residues are the key to explaining why the W775H6.50 mutant retains partial receptor activity. The side chain of W7756.50 can adopt different conformations: the conformation observed in the experimental structures pointing toward TM5 and an alternative conformation reoriented inside the binding cavity (Figure 3). The differences observed in the W775H6.50 and W775F/Y6.50 mutants reveal that the hydrogen bond site of the nitrogen-containing aromatic ring is crucial in maintaining the function of the receptor. However, the missing 6-membered aromatic ring appears to be crucial for allosteric signaling. One can speculate that the hydrophobic interactions are lost between W7756.50 and the surrounding residues, which then breaks the communication between the transmission switch Y7716.46 and cyclamate, which is located roughly 16 Å above.
The homology model of the human T1R3 subunit based on the mGluR1 template (PDB 4OR2) shows a ~16-Å-long allosteric cavity, which is able to bind ligands that are larger than cyclamate (Supplementary Figure 9). The mGluR1 allosteric site has a volume of 760 Å3, which is comparable to the volume of the site identified in the initial model of the sweet-taste receptor (730 Å3). Although the FITM ligand is curved and extended such that it fits perfectly in the mGluR1 binding cavity, cyclamate does not fill the T1R3 extended cavity. However, the MD simulations of the sweet-taste receptor capture alternate conformational states of the residues forming the T1R3 bundle. In particular, the side chain of F7305.40 and H7345.44 TM5 residues and W7756.50, F7786.53, and L7826.57 TM6 residues fill in the empty space of the initial homology model, restoring the communication between the cyclamate-binding site and the cradle of the TMD cavity.
Allosteric ligands that are able to bind deep inside the cavity and interact with TM6 residues above the toggle switch should be more potent sweet-taste modulators. The size and orientation of NHDC docked in the allosteric-binding site (Supplementary Figure 9) indicates that its sweet-taste potency can be compared to cyclamate. Indeed, cyclamate is a less potent sweetener than NHDC, as shown by their EC50 values (0.2 and 2.2 μM for NHDC and cyclamate, respectively) (Winnig et al. 2007). This work provides a structural basis for the rational design of novel sweet-taste modulators that are able to explore the elongated shape of the TMD cavity.
Experimental procedures
Initial coordinates and system setup
The homology model of the T1R3 TMD was generated using Modeller9.15 (Sali and Blundell 1993) based on a previously published class C GPCR sequence alignment (Chéron et al. 2017) and the mGluR1 structure (PDB identifier 4OR2) (Wu et al. 2014). The canonical disulfide bridge between residues C633 and C722 of the transmembrane helix 3 (TM3) and the extracellular loop 2 (ecl2) was constrained during the modeling. The best model according to the discrete optimized protein energy was validated with PROCHECK prior to molecular docking. The structure of the cyclamate and NHDC ligand as well as the receptor structure were prepared using AutoDockTools (Morris et al. 2009). The docking search space was defined to encompass the binding cavity in the TMD of T1R3. Docking simulations were performed using AutoDock4 (Morris et al. 2009). Top-ranking solutions were selected according to site-directed mutagenesis data (Jiang et al. 2005b). Residue numbering written in superscript corresponds to the Ballesteros–Weinstein definition proposed by Pin et al. (2003) for class C GPCRs. It consists of 2 numbers, for instance X.Y, where X denotes the transmembrane segment (1–7), and Y is the residue position relative to the most conserved residue in the helix, defined as number 50.
MD simulation protocol
The GAFF parameters for the cyclamate were obtained with the Antechamber module of AMBER14 (Case et al. 2014) and the AM1-bcc charges. The AMBER force fields ff03.r1 and lipid11 were used for the protein and the membrane, respectively. The receptor orientation in the membrane was predicted by the OPM server (Lomize et al. 2012). The POPC membrane bilayer and the TIP3P water molecules were generated using VMD (Humphrey et al. 1996). The protonation states of titratable residues were computed at pH 6.5 through the H++ server (Gordon et al. 2005). Then, the system was neutralized with 3 Cl− ions using the leap module of AMBER14. The TIP3P solvent phase was extended to a distance of 10 Å from any solute atom in z direction, resulting in a 77 × 75 × 94 Å3 box. First, 3 steps of minimization (5000 steps of steepest descent and 5000 steps of conjugate gradient) were applied to the protein side chains, the membrane, and the solvent, respectively, with positional restraints of 50 kcal/mol/Å2 on the rest of the system. MD calculations in periodic boundary conditions were performed using the PMEMD.cuda module of AMBER14 in the NPT ensemble at 310 K and 1 bar (0.987 atm)—anisotropic pressure—controlled with the Langevin thermostat and the Berendsen barostat. The SHAKE bond length constraint was applied on the bonds involving hydrogen atoms and a time step of 2 fs was chosen. A cutoff of 8 Å was used to compute the nonbonded interactions with an update of the neighbor list every 25 steps. Long-range electrostatic interactions were computed using the Particle Mesh Ewald summation method. After the minimization, a heating step from 100 to 310 K of 20 ps and an equilibration of 1 ns were performed on the membrane and the solvent with a restraint of 50 kcal/mol/Å2 on the protein. Then, 4 cycles of minimization and 20 ps of equilibration at 100 K were applied with a diminished restraint from 15 to 0 kcal/mol/Å2 on the protein atoms. Finally, the whole system was heated to 310 K more than 20 ps and equilibrated for 40 ns prior to the production runs. We performed 2 independent MD simulations (labeled s1 and s2 in the Supplementary Table 2) in of the wild-type receptor in apo form for a total of 6.0 μs, 2 of the wild-type receptor in complex with cyclamate for a total of 7.9 μs, and 2 of the R725Aecl2 and W775H6.50 mutants for a total of 1.5 and 1.0 μs, respectively. The MD simulations are summarized in Supplementary Table 2. To study the dynamics of the ligand entrance into the receptor, we performed another 0.65 μs MD simulation of the apo receptor in the presence of 5 cyclamate molecules located initially in the solvent phase.
Materials
Cyclamate, d-trp, and lactisole were obtained from Sigma-Aldrich. Unless otherwise specified, 10 mM cyclamate and d-trp were used for cell-based assays.
Mutants
Human T1R2 and T1R3 constructs were generated as described previously (Jiang et al. 2004). T1R3 mutants were designed based on the molecular modeling simulations and generated by site-directed mutagenesis. Briefly, polymerase chain reaction reactions were done using Pfu DNA polymerase, pcDNA3.1(+)-T1R3 plasmid, and mutagenic primers containing desired mutation. After DpnI digestion of parental DNA template, mutated molecules were transformed into competent bacteria cells for nick repair. Single colonies were picked and grew for preparation of mutant constructs. As many T1R3 mutants have already been published (Jiang et al. 2005b; Winnig et al. 2007), we have chosen to complete the series of amino acid substitutions accordingly R725Aecl2, R725Eecl2, S729Fecl2, S729Tecl2, S729Yecl2, N737A5.47, Y771F6.46, Y771L6.46, W775F6.50, W775Y6.50, W775H6.50. All mutant constructs were verified by direct sequencing (Penn Genomic Analysis Core).
Cell culture and calcium imaging
Human embryonic kidney 293 cells (PEAKrapid, ATCC CRL-2828) were cultured and maintained as described previously (Lei et al. 2015). Cells were seeded in 96-well plates (Corning 3904) at a density of 25 000 cells per well. After 16–24 h, cells were transiently transfected with T1R2, T1R3 (or mutants), and a coupling chimeric G protein Gα16-gust44 (Jiang et al. 2004; Lei et al. 2015). DNA (0.06 µg) of each construct was transfected using Lipofectamine 2000 (Thermo Fisher 1168019) (0.5 µL/well) into each well by following the manufacture’s protocol. After 40–48 h, cells were washed with Hanks’ balanced salt solution (HBSS) supplemented with 10 mM HEPES (assay buffer), and loaded with 2.5µg/mL Fluo-4 (Invitrogen, F14201) and 2.5µg/mL Pluoronic (Invitrogen, P3000MP) for 1 h. After 3 washes and a 30-min incubation in 50 µL/well in assay buffer, cells were assayed for their responses to cyclamate and d-trp using a Flexstation III (Molecular Probes). Relative fluorescence units (excitation at 494, emission at 516, and cutoff at 515) were recorded every 2 s for 200 s. Cyclamate and d-trp were dissolved in HBSS at 2× the final concentration. 50 µL of each solution was added via Flexstation 30 s after the initial start. Responses were quantified as previously described (Lei et al. 2015). In short, changes in fluorescence (ΔF) were quantified as peak fluorescence minus the baseline level (F) and are expressed as percent ΔF relative to F. Receptor responses have not been corrected by cell surface expression since preceding experiments have shown that all mutants exhibit similar levels of cell surface expression (Jiang et al. 2005a, 2005b; Winnig et al. 2007). In addition, the modification of the allosteric response to a mutation is systematically compared to an orthosteric ligand control and such comparison is less sensible to expression levels. Bar graphs were generated using GraphPad Prism 5. Statistical analysis was performed by using 2-tailed Student’s t-tests.
Supplementary Material
Acknowledgments
We thank Dr Xiaojing Cong for fruitful discussions and Abby Cuttriss for careful proofreading.
Funding
This work was supported by the French government, through the UCAJEDI investments in the future project managed by the National Research Agency (ANR) with the reference number [ANR-15-IDEX-01]; by the French Ministry of Higher Education and Research [PhD Fellowship of J.-B.C.], by GIRACT (Geneva, Switzerland) [Promoting Flavor Research first year bursary award, J.-B.C]; by the Gen Foundation [Registered UK Charity No. 1071026, J.-B.C.] (a charitable trust that principally provides grants to students/researchers in natural sciences, in particular food sciences/technology); and by National Institutes of Health Grant [DC010842 to P.J.].
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
References
- Case DA, Babin V, Berryman JT, Betz RM, Cai Q, Cerutti DS, Cheatham TE, Darden TA, Duke RE, Gohlke H, et al. 2014. Amber 14. San Francisco (CA): University of California. [Google Scholar]
- Chéron JB, Golebiowski J, Antonczak S, Fiorucci S. 2017. The anatomy of mammalian sweet taste receptors. Proteins. 85:332–341. [DOI] [PubMed] [Google Scholar]
- Dalton JAR, Pin JP, Giraldo J. 2017. Analysis of positive and negative allosteric modulation in metabotropic glutamate receptors 4 and 5 with a dual ligand. Sci Rep. 7:4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doré AS, Okrasa K, Patel JC, Serrano-Vega M, Bennett K, Cooke RM, Errey JC, Jazayeri A, Khan S, Tehan B, et al. 2014. Structure of class C GPCR metabotropic glutamate receptor 5 transmembrane domain. Nature. 511:557–562. [DOI] [PubMed] [Google Scholar]
- DuBois GE. 2016. Molecular mechanism of sweetness sensation. Physiol Behav. 164:453–463. [DOI] [PubMed] [Google Scholar]
- Gordon JC, Myers JB, Folta T, Shoja V, Heath LS, Onufriev A. 2005. H++: a server for estimating pKas and adding missing hydrogens to macromolecules. Nucleic Acids Res. 33:W368–W371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet C, Gaven F, Kniazeff J, Vol C, Liu J, Cohen-Gonsaud M, Acher F, Prézeau L, Pin JP. 2004. Heptahelical domain of metabotropic glutamate receptor 5 behaves like rhodopsin-like receptors. Proc Natl Acad Sci USA. 101:378–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. 1996. VMD: visual molecular dynamics. J Mol Graph. 14:33–8, 27. [DOI] [PubMed] [Google Scholar]
- Jiang P, Cui M, Zhao B, Liu Z, Snyder LA, Benard LM, Osman R, Margolskee RF, Max M. 2005a. Lactisole interacts with the transmembrane domains of human T1R3 to inhibit sweet taste. J Biol Chem. 280:15238–15246. [DOI] [PubMed] [Google Scholar]
- Jiang P, Cui M, Zhao B, Snyder LA, Benard LM, Osman R, Max M, Margolskee RF. 2005b. Identification of the cyclamate interaction site within the transmembrane domain of the human sweet taste receptor subunit T1R3. J Biol Chem. 280:34296–34305. [DOI] [PubMed] [Google Scholar]
- Jiang P, Ji Q, Liu Z, Snyder LA, Benard LM, Margolskee RF, Max M. 2004. The cysteine-rich region of T1R3 determines responses to intensely sweet proteins. J Biol Chem. 279:45068–45075. [DOI] [PubMed] [Google Scholar]
- Jiang P, Josue J, Li X, Glaser D, Li W, Brand JG, Margolskee RF, Reed DR, Beauchamp GK. 2012. Major taste loss in carnivorous mammals. Proc Natl Acad Sci USA. 109:4956–4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Chen Y, Abrol R, Goddard WA III, Guthrie B. 2017. Activation mechanism of the G protein-coupled sweet receptor heterodimer with sweeteners and allosteric agonists. Proc Natl Acad Sci USA. 114:2568–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei W, Ravoninjohary A, Li X, Margolskee RF, Reed DR, Beauchamp GK, Jiang P. 2015. Functional analyses of bitter taste receptors in domestic cats (Felis catus). PLoS One. 10:e0139670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Bachmanov AA, Maehashi K, Li W, Lim R, Brand JG, Beauchamp GK, Reed DR, Thai C, Floriano WB. 2011. Sweet taste receptor gene variation and aspartame taste in primates and other species. Chem Senses. 36:453–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Staszewski L, Xu H, Durick K, Zoller M, Adler E. 2002. Human receptors for sweet and umami taste. Proc Natl Acad Sci USA. 99:4692–4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomize MA, Pogozheva ID, Joo H, Mosberg HI, Lomize AL. 2012. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 40:D370–D376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. 2009. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 30:2785–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson G, Chandrashekar J, Hoon MA, Feng L, Zhao G, Ryba NJ, Zuker CS. 2002. An amino-acid taste receptor. Nature. 416:199–202. [DOI] [PubMed] [Google Scholar]
- Nelson G, Hoon MA, Chandrashekar J, Zhang Y, Ryba NJ, Zuker CS. 2001. Mammalian sweet taste receptors. Cell. 106:381–390. [DOI] [PubMed] [Google Scholar]
- Pérez-Benito L, Doornbos MLJ, Cordomí A, Peeters L, Lavreysen H, Pardo L, Tresadern G. 2017. Molecular switches of allosteric modulation of the metabotropic glutamate 2 receptor. Structure. 25:1153–1162.e4. [DOI] [PubMed] [Google Scholar]
- Pin JP, Galvez T, Prézeau L. 2003. Evolution, structure, and activation mechanism of family 3/C G-protein-coupled receptors. Pharmacol Ther. 98:325–354. [DOI] [PubMed] [Google Scholar]
- Rondard P, Pin JP. 2015. Dynamics and modulation of metabotropic glutamate receptors. Curr Opin Pharmacol. 20:95–101. [DOI] [PubMed] [Google Scholar]
- Sali A, Blundell TL. 1993. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 234:779–815. [DOI] [PubMed] [Google Scholar]
- Winnig M, Bufe B, Kratochwil NA, Slack JP, Meyerhof W. 2007. The binding site for neohesperidin dihydrochalcone at the human sweet taste receptor. BMC Struct Biol. 7:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Wang C, Gregory KJ, Han GW, Cho HP, Xia Y, Niswender CM, Katritch V, Meiler J, Cherezov V, et al. 2014. Structure of a class C GPCR metabotropic glutamate receptor 1 bound to an allosteric modulator. Science. 344:58–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Staszewski L, Tang H, Adler E, Zoller M, Li X. 2004. Different functional roles of T1R subunits in the heteromeric taste receptors. Proc Natl Acad Sci USA. 101:14258–14263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L, Rovira X, Scholler P, Zhao H, Liu J, Pin JP, Rondard P. 2015. Major ligand-induced rearrangement of the heptahelical domain interface in a GPCR dimer. Nat Chem Biol. 11:134–140. [DOI] [PubMed] [Google Scholar]
- Zhang F, Klebansky B, Fine RM, Xu H, Pronin A, Liu H, Tachdjian C, Li X. 2008. Molecular mechanism for the umami taste synergism. Proc Natl Acad Sci USA. 105:20930–20934. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.