Abstract
Displaying the advantage of nanoparticles in cancer targeting and drug delivery, micelles have shown great potential in cancer therapy. The mechanism for micelle targeting to cancer without the need for ligands is due to the size advantage of micelles within the lower end of the nanometer scale that is the optimal size for favoring the enhanced permeability and retention (EPR) effect while escaping trapping by macrophages. MicroRNAs are ubiquitous and play critical roles in regulating gene expression, cell growth, and cancer development. However, their in vivo delivery in medical applications is still challenging. Here, we report the targeted delivery of anti-miRNA to cancers via RNA micelles. The phi29 packaging RNA three-way junction (pRNA-3WJ) was used as a scaffold to construct micelles. An oligo with 8nt locked nucleic acid (LNA) complementary to the seed region of microRNA21(miR21) was included in the micelles as an interference molecule for cancer inhibition. These RNA micelles carrying anti-miR21 exhibited strong binding and internalization to cancer cells, inhibited the function of oncogenic miR21, enhanced the expression of the pro-apoptotic factor, and induced cell apoptosis. Animal trials revealed effective tumor targeting and inhibition in xenograft models. The inclusion of folate as a targeting ligand in the micelles did not show significant improvement of the therapeutic efficacy in vivo, suggesting that micelles can carry therapeutics to a target tumor and inhibit its growth without ligands.
Keywords: RNA nanotechnology, RNA nanoparticle, RNA nanostructure, RNA micelles, anti-miRNA, cancer targeting and therapy
Graphical Abstract
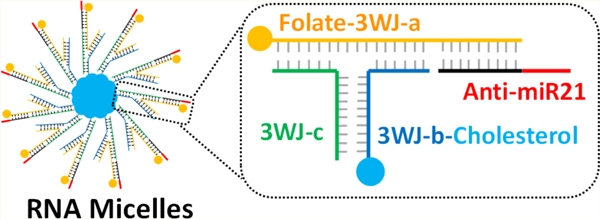
Micelles are spherical and supermolecular nano-constructs formed by amphiphilic block copolymers including polymer, peptide, and DNA.1–5 The micelles are a core–shell structure composed of both lipophilic and hydrophilic modules. They are able to either accommodate hydrophobic drugs interiorly or incorporate small interfering RNA as branches externally.6,7 The lipid core of micelles can solubilize hydrophobic drugs and enhance their bioavailability.8,9 In addition, the size of micelles is in nanometer scale, which promotes their extravasations at tumor sites while avoiding fast renal clearance in vivo.10 Longer circulation time also allows for greater accumulation of micelles in the tumor site by EPR effect, even without a targeting ligand.11,12 These micelles are widely used in biomedical fields due to their highly compact structure, easy therapeutics encapsulation, and enhanced cell permeability.4,13
RNA as a safe biomaterial can also be utilized for micelle construction. Because of the proof-of-concept of RNA nanotechnology in 1998,14 RNA molecules have been extensively investigated as building blocks for constructing nanoparticles via bottom up self-assembly.15–24 We have reported the 3WJ motif derived from the pRNA of the bacteriophage phi29 DNA packaging motor. The 3WJ core displays high thermodynamic stability with a melting temperature (Tm) of about 58 °C. It is chemically stable after 2′-fluoro (2’F) modification. Furthermore, the pRNA-3WJ complex is stable without dissociation in the presence of 8 M Urea or at extremely low concentration.18 Functional modules such as therapeutic molecules, targeting ligands, and imaging agents can be well-incorporated to the RNA scaffolds to formulate multifunctional nanoparticles.25–30 The physicochemical properties such as size, shape, and stoichiometry of RNA nanoparticles can be precisely controlled to achieve optimal therapeutic effects.31–33 Current studies have demonstrated that RNA nanoparticles are a safe and biocompatible delivery platform because they exhibit undetectable intrinsic immunogenicity and toxicity.25,31,32,34,35 Taking the advantage of favorable features of RNA, we have first designed and reported the construction of RNA micelles based on pRNA-3WJ.25 In aqueous solution, the amphiphilic oligonucleotide molecules, consisting of a hydrophobic cholesterol molecule and the hydrophilic pRNA-3WJ, spontaneously self-assemble into monodispersed micellar nanostructure with a lipid core and a pRNA-3WJ corona. We have indicated25 that the RNA-based micelles are capable of loading the chemotherapeutic drug paclitaxel, inhibiting cancer cell proliferation and targeting to tumor via the EPR effect. No obvious cytotoxicity and immunogenicity have been detected for our RNA micelle system.25 Here, we showed that the delivery capability of RNA micelles is not only limited to chemotherapeutic drugs but also can be expanded to the field of gene medicine such as siRNA and miRNA therapeutics.
MiRNAs are endogenously expressed small noncoding RNAs, regulating target mRNA by binding to imperfect complementarity in 3′-untraslated regions (3′-UTR).36 Early studies have demonstrated miRNAs are closely related to cell proliferation, differentiation, and apoptosis.37 Alterations in miRNA expression profile are correlated with tumor pathogenesis, cancer progression, and drug resistance.38–40 MiRNA21 is one of the most over-expressed miRNAs playing an important role in various cancer proliferation and apoptosis through targeting the PTEN (phosphatase and tensin homologue), PDCD4 (programmed cell death protein 4), and other signal transduction pathways.41–43 Previous studies have proven that the inhibition of miRNA21 by anti-miRNA21 can restore the suppressive functions of endogenous miRNA21, induce cell apoptosis, and enhance the anti-proliferation effects of anticancer drugs.44–46 Importantly, enhancing binding affinity of antisense oligo to oncogenic miRNA and developing a safe delivery platform are required for effective oncogenic miRNA knockdown. To favor the binding to miRNA21, chemical modifications, including 2′-O-methyl (2′-O-Me), 2′-O-methoxyethyl (2′-MOE), 2′-fluoro (2′F), and LNA, have been applied.47–50 Among these modifications, LNA modification stands out for enhanced miRNA21 binding by increasing thermodynamic stability significantly.27,49,51 Specifically, seed-targeting 8nt LNA modified oligonucleotides have been proven to silence miRNA without obvious off-target effects.27,49,68 As for delivery tools of gene medicine, the nanoparticle-based delivery system has been well studied and developed.52–54 Several nanoplatforms have been reported to deliver miRNA or anti-miRNA as therapeutic agents.27,55–58 However, the clinical translation remains challenging due to various barricades, including undefined size and shape, particle aggregation or dissociation, nonspecific targeting, poor tumor accumulation, and undesirable immunogenicity.54,59,60
Herein, we applied the RNA micelles platform for delivering anti-miR21. 8nt LNA modified anti-miR21 can be annealed to pRNA-3WJ scaffold via complemtation with an extended sequence. The cholesterol molecule attached to one 3WJ branch facilitates the micelles formation by hydrophobic force. In vitro studies demonstrated incorporation of folate was able to improve RNA micelles’ binding and internalization capability into cancer cells, delivery of anti-miR21 for miR21 inhibition and cell apoptosis induction. Compared with RNA nanoparticles without micelles formation, RNA micelles exhibited better tumor targeting ability and stronger tumor accumulation in mouse xenograft model. In vivo tumor regression study was performed to compare the therapeutic effects of RNA micelles carrying anti-miR21 with or without folate ligand.
RESULTS AND DISCUSSION
Design, Construction, And Characterization of RNA Micelles Incorporating Anti-miR21.
The RNA micelles were designed based on 3WJ scaffold from pRNA of bacteriophage phi29 DNA packaging motor (Figure 1A,B).18 The pRNA-3WJ is composed of three short RNA fragments (named as 3WJ-a, 3WJ-b, and 3WJ-c; Figure 1A). This multibranch property makes it feasible to incorporate different functional modules. We have solved the crystal structure of this pRNA-3WJ61 and determined the angle between the branches (60°, 120°, and 180°; Figure 1B). To facilitate the appropriate folding of the pRNA-3WJ and promote the formation of the hydrophobic core for producing the micelles while avoiding steric hindrance, a cholesterol molecule was attached to the 3′-end of the 3WJ-b strand. RNA micelles formation was visualized via the slower migration band in 2% agarose gel due to the increased size and mass, compared to a single 3WJ unit (Figure 1D). This hydrophobic force-driven formation was confirmed by the observed collapse of micelles band with the addition of sodium dodecyl sulfate (SDS) in Tris-acetate (TAE) running buffer because the SDS can interfere the core hydrophobic interaction (see Figure S2A). To further test whether RNA micelles were formed based on the hydrophobic force, an increasing volume percentage of dimethyl sulfoxide, (DMSO) was added into pRNA-3WJ micelles and the control group, which were subsequently loaded to 2% agarose gel. The existence of DMSO would dissolve the lipid core, disrupting the hydrophobic force and dissociating pRNA-3WJ micelles into single pRNA-3WJ (see Figure S2B). All of the evidence indicated that the RNA micelles were formed via hydrophobic force.
Figure 1.
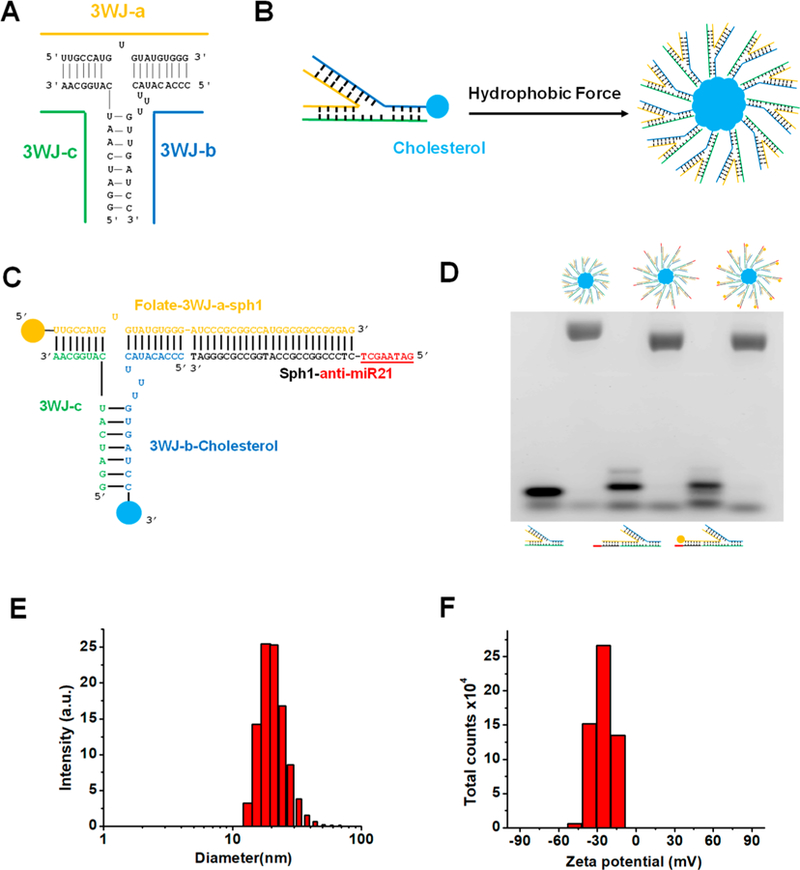
Design, construction and characterization of RNA micelles. (A) 3WJ motif of pRNA from bacteriophage phi29 DNA packaging motor. (B) Illustration of pRNA-3WJ micelles formation by hydrophobic force. (C) 2D structure of 3WJ/FA/anti-miR21 micelles (underlined sequence is 8nt LNA modified anti-miR21 seed region). (D) Assembly of RNA micelles assayed by 2% agarose gel. (Lane from left to right: 3WJ, 3WJ micelles, 3WJ/anti-miR21, 3WJ/anti-miR21 micelles, 3WJ/FA/anti-miR21, and 3WJ/FA/anti-miR21 micelles). (E) Size distribution of 3WJ/FA/anti-miR21 micelles. (F) ζ potential of 3WJ/FA/anti-miR21 micelles.
Due to the multivalent nature of the 3WJ scaffold, RNA micelles can be rationally designed to be multifunctional. As such, anti-miR21 as a therapeutic agent was incorporated by a complementary extension sequence (Figure 1C). Besides, folate as a specific targeting ligand was introduced to single-stranded 3WJ-a-sph1 by CuAAC “click reaction” prior to assembly of RNA micelles (see Figure S1A). The reaction was carried out in mild reaction conditions and resulted in high yield. Successful conjugation of folate onto the RNA strand was evidenced by the upshifting band in 20% 8 M UREA PAGE, compared to an unconjugated strand (see Figure S1B).
Noticeably, the multifunctional 3WJ/FA/anti-miR21 micelles showed slightly faster migration band in 2% agarose gel compared with pRNA-3WJ micelles without any modules (Figure 1D). One possible explanation is that the extension on the RNA oligo will increase structural hindrance and result in a smaller number of pRNA-3WJ copies in micellar structure. DLS determined that the average hydrodynamic diameter of 3WJ/FA/anti-miR21 micelles was about 21.51 ± 6.136 nm (Figure 1E). This nanoparticle size has been demonstrated to be within the favorable size range for in vivo biodistribution. Additionally, the ζ potential was determined to be −25 ± 8.35 V, which is consistent with the polyanionic nature of RNA.
Determination of Critical Micelle Formation Concentration.
Critical micelle formation conentration (CMC) is one of the key factors for micelle construction. To calculate the CMC of RNA micelles, Nile red assay was used as previously reported.62 Nile red is a commonly used fluorescent probe that is very sensitive to hydrophobicity change.63 Specifically, Nile red displays very weak fluorescence in hydrophilic environment but strong fluorescence in hydrophobic environment. Therefore, the CMC can be determined by the measurement of fluorescence intensity of Nile red when co-incubated with serial diluted RNA micelle solution (Figure 2A). To estimate the value of CMC, the fluorescence intensity of Nile Red was plotted as a function of the sample concentration. At the concentrations below CMC, the hydrophobic interaction of cholesterol molecules was insufficient to drive the formation of micelles, and the fluorescence intensity of Nile Red is almost a constant. However, the fluorescence increases dramatically when Nile Red is entrapped in the micelle core once amphiphilic RNA concentration exceeds CMC. Thus, the CMC value could be estimated as 100 nM, which is the intersection of tangents to the horizontal line of intensity ratio with relatively constant value (Figure 2A). To ensure the successful formation, all 3WJ/FA/anti-miR21 micelles were assembled at the concentration higher than 100 nM in this study.
Figure 2.
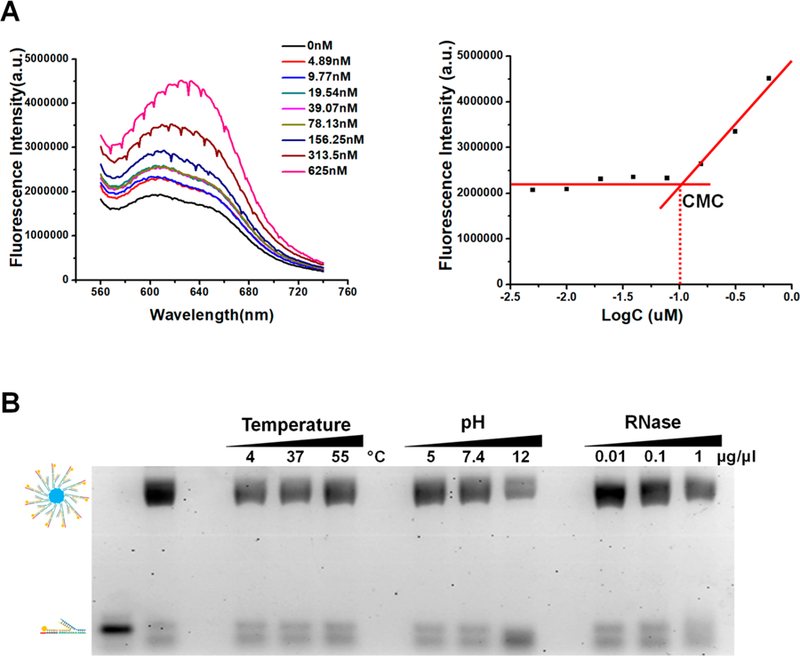
CMC determination and stability study of RNA micelles. (A) CMC determined by Nile red encapsulation assay. (B) Stability study of RNA micelles in different temperature, pH, and RNase condition.
RNA Micelle Stability Study.
To evaluate the stability, 3WJ/FA/anti-miR21 micelles were incubated in solution with different pH values, temperatures, and RNase concentrations, respectively. After incubation, the samples were loaded to 2% agarose gel electrophoresis for comparison (Figure 2B). It can be found that the 3WJ/FA/anti-miR21 micelles remained stable in a wide range of temperatures (4, 37, and 55 °C). In addition, when 3WJ/FA/anti-miR21 micelles were incubated at acidic (pH = 5) and neutral buffer (pH = 7.4), no obvious micellar structure changes were observed. Interestingly, at pH12, there’s little dissociation detected from the lower band. Lastly, to validate the chemical stability, RNA micelles incubated with RNase (0.01 μg/μL, 0.1 μg/μL and 1 μg/μL) did not show much degradation. These results demonstrate the advantages for RNA micelles construction, and more importantly, suggested that 3WJ/FA/anti-miR21 micelles is a competitive nanoparticle for in vivo applications.
In Vitro Cell Binding and Internalization Assay of RNA Micelles.
Micellar nanoparticles as drug carriers have been reported to enhance cell permeability. In this study, we evaluated the in vitro cell binding and internalization efficiency between RNA micelles and a single 3WJ unit as well as RNA micelles with and without folate ligand. Alexa 647 labeled 3WJ/FA/anti-miR21 micelles and control nanoparticles (3WJ/anti-miR21 micelles, 3WJ/FA/anti-miR21, and 3WJ/anti-miR21) were incubated with KB cells, respectively, before being assayed by flow cytometry. KB cells, identified as folate receptor over-expressed cancer cells, were used for all in vitro and in vivo studies. Strong cell binding was observed for 3WJ/FA/anti-miR21 micelles (94.2% positive cells) compared with 3WJ/anti-miR21 micelles (49.2% positive cells), 3WJ/FA/anti-miR21(65.8% positive cells), and 3WJ/anti-miR21 (0.38% positive cells) (Figure 3A). Similarly, images taken by confocal microscope after treatments also revealed consistent result (Figure 3B). These results demonstrated that micelles formation contributed to strong and favorable cell binding and internalization to KB cells in vitro, while the folate ligand further enhanced the events when they were decorated on the RNA micelles. Folate receptors have been identified as targets highly expressed on the surface of various cancer cells including ovarian, breast, colon, and malignant nasopharyngeal carcinomas.64,65 The folate-decorated RNA micelles would recognize the folate receptor on the KB cell surface and bring the nanoparticles to cells by endocytosis. We also tested the binding and internalization profile on human colon cancer HT29 cells, which have high folate receptor expression.66,67 The results (see Figure S3) were consistent with the results from the in vitro assay of KB cells. Flow cytometry data (see Figure S3A) revealed that RNA micelles bound to HT29 cells efficiently. 3WJ/FA/anti-miR21 with folate but without the cholesterol did not show such strong binding affinity to HT29 cells as KB cells. It may be due to less folate receptor expression on HT29 cell surface than KB cells. Confocal microscopy images (see Figure S3B) also provided evidence of the efficient internalization of RNA micelles into HT29 cells as KB cells.
Figure 3.
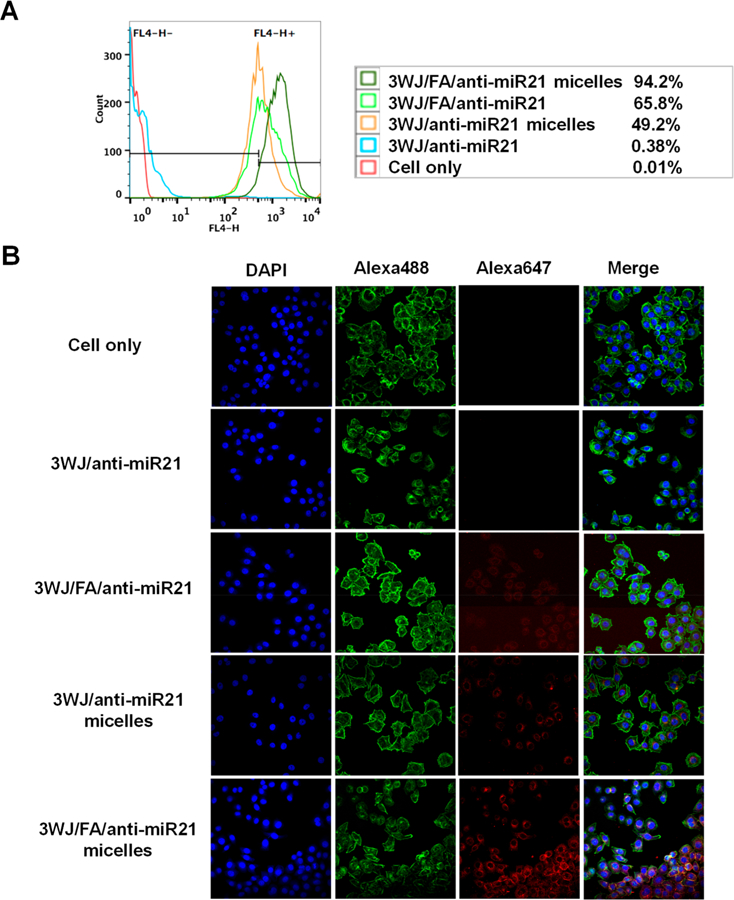
In vitro binding and internalization of RNA micelles to cancer cells. (A) Flow cytometry comparing binding affinity to KB cells after treatment for 1 h. (B) Confocal microscopy showing internalization profile. Blue: nuclei; green: cytoskeleton; red: RNA nanoparticles.
Delivery of Anti-miR21 by RNA Micelles into Cancer Cells.
We hypothesized that the RNA micelles as a delivery system can improve the potency of anti-miR21 in cancer cells. To study this assumption, the delivery efficiency of anti-miR21 by RNA micelles was first assayed by dual-luciferase-based miR21 reporter system, as we previously reported.27,49,68 When anti-miR21 are delivered into the cancer cells, it will competitively bind to miR21 that used to bind to the 3′-UTR region of the Renilla luciferase gene and block its translation, resulting in an increased expression of Renilla luciferase. The co-transfected firefly luciferase gene would express as an internal control. Therefore, higher Renilla-to-firefly luciferase ratio (R/F luc) is correlated with stronger miR21 inhibition. Expectedly, 3WJ/FA/anti-miR21-treated cells showed higher a R/F luc than 3WJ/anti-miR21, proving folate as a ligand can improve anti-miR21 delivery efficiency and inhibit miR21 (Figure 4A). In addition, 3WJ/anti-miR21 micelles also performed better miR21 inhibition effects than 3WJ/anti-miR21 as a result of higher delivery efficacy of anti-miR21 by RNA micelles. The active targeting by folate combined with the passive targeting by micelles formation contribute to the delivery of anti-miR21 in vitro synergistically.
Figure 4.
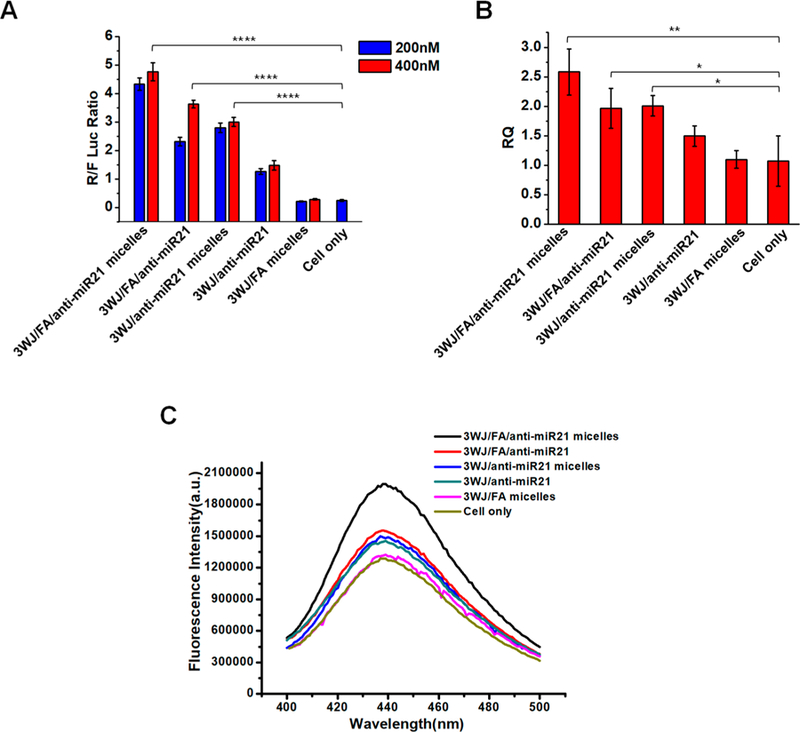
In vitro studies of RNA micelles carrying anti-miR21. (A) Dual-luciferase assay demonstrating delivery of anti-miR21 to KB cells. (B) qRT-PCR showing effect of miR21 knock-down on the target gene PTEN expression. (C) Caspase-3 assay exhibiting cell apoptosis induction after treatment.
The delivery efficacy of anti-miR21 was further assessed by measuring miR21 downstream tumor suppressor PTEN gene expression by qualitative real-time polymerase chain reaction (qRT-PCR). The up-regulation of PTEN can reduce cancer cell proliferation and increase cell death. After incubation with KB cells for 72 h, RNA nanoparticles carrying anti-miR21 groups showed the up-regulation of PTEN. Among them, folate-decorated micelles group exhibited highest PTEN expression resulting from better anti-miR21 delivery capability (Figure 4B). The effective miR21 silence was achieved by both enhancing miR21 binding via LNA-modified anti-miR21 and improving delivery efficacy by multifunctional RNA micelles. Combining the results in Figure 3, it can be concluded that RNA micelles showed the high delivery efficiency of anti-miR21 to cells and subsequently contributed to potent gene regulation.
The enhanced induction of cell apoptosis by the knockdown of miR21 is considered as an important feature of cancer cells inhibition. Particularly, caspase-3 is an early cellular apoptotic marker that is closely related to cell apoptosis. The elevation of caspase-3 activity can be reflected by increased fluorescence intensity from released fluorescent substrate. We found that 3WJ/FA/anti-miR21 micelle treated cells showed highest fluorescence emission of caspase-3 substrate in contrast with control groups (3WJ/anti-miR21 micelles, 3WJ/FA/anti-miR21, 3WJ/anti-miR21, and 3WJ/FA micelles; Figure 4C). The activation of the caspase-3 pathway will trigger cancer cell apoptosis, suggesting a stronger inhibitory effect on cancer cells.
Targeting of RNA Micelles to Cancer Xenograft in Vivo.
To study whether pRNA-3WJ micelles will further enhance tumor targeting compared with pRNA-3WJ nanoparticles without micelles formation, Alexa 647 labeled 3WJ/FA/anti-miR21 micelles, 3WJ/FA/anti-miR21, and 3WJ/anti-miR21 were systemically administered via the tail vein into KB tumor bearing mice. It can be clearly observed that the RNA nanoparticles distributed to the whole body at 1 h post-injection. At 4 and 8 h post-injection, the fluorescent RNA nanoparticles showed specific accumulation in tumor while it is almost completely cleared in other organs (Figure 5A). Ex vivo images of the main organs (heart, liver, spleen, lung, and kidney) and tumors were taken after 8 h post-injection. Compared to 3WJ/anti-miR21 treatment, 3WJ/FA/anti-miR21-treated mice exhibited stronger fluorescence signal in tumor due to the additional active tumor targeting mediated by folate. More importantly, RNA micelles group showed stronger accumulation compared with RNA nanoparticles without micelle formation (Figure 5B). The micelle formation prolongs the nanoparticle retention in tumor site. This might benefit from the advantageous nanometer size and spherical shape of RNA micelles that facilitate them to penetrate the tumor site by EPR effects.
Figure 5.
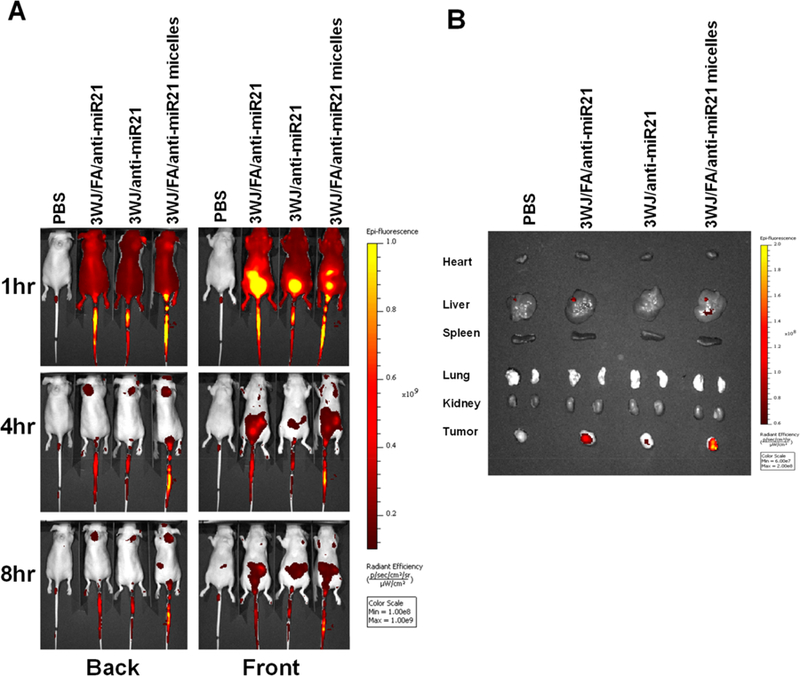
In vivo biodistribution study in mice with xenograft. (A) Whole-body image. (B) Ex vivo organ image 8 h post-injection.
In Vivo Tumor Inhibition Study by RNA Micelles Carrying Anti-miR21.
Delivery of anti-miR21 by RNA micelles was studied in mice model bearing KB tumor xenograft. The RNA micelles with or without folate ligand incorporating anti-miR21 were intravenously injected into mice model for a total of five doses every other day. Compared to the phosphate-buffered saline (PBS) group, both treatment groups showed tumor inhibition effects (Figure 6). The difference of tumor regression level between the two treatment groups appeared after the 4th injection on day 7 (Figure 6A). Folate decorated micelles showed slightly higher efficacy in tumor regression but not a significant one, thus demonstrating that the passive tumor targeting played the key role in the RNA micelles system. The weight of the tumor harvested at day 10 also showed a slight difference between RNA micelles carrying anti-miR21 with or without folate ligand (Supplementary Figure 3).
Figure 6.
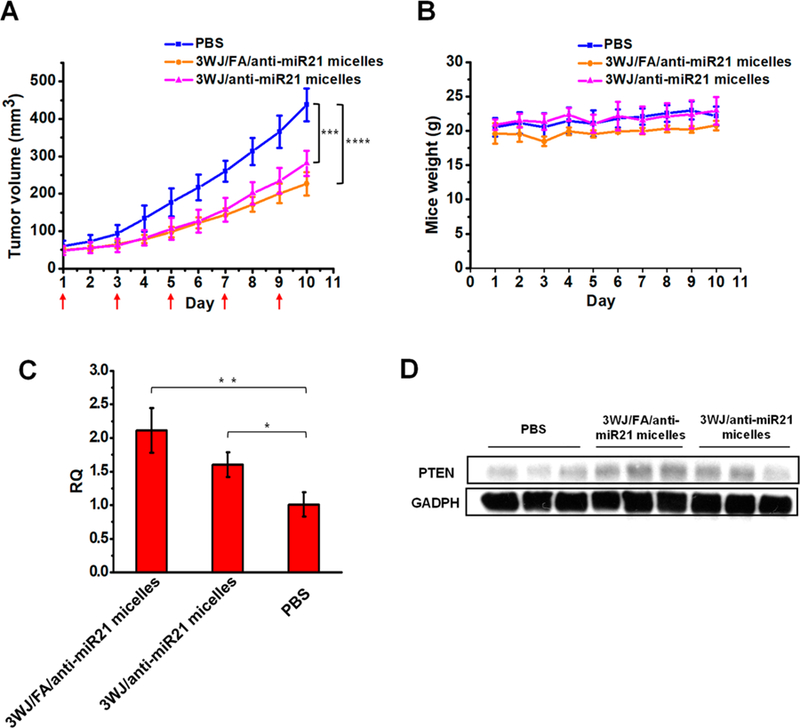
In vivo therapeutic effect of RNA micelles in mice with xenograft. (A) Tumor regression curve over the course of five injections (red arrow shows the day of injection). (B) Mice weight curve during treatment period. (C) qRT-PCR and (D) Western blot analysis showing the up-regulation of PTEN after the in vivo delivery of anti-miR21.
To examine the regulatory effect on PTEN level in tumor tissues, qRT-PCR was also used to measure the expression in all harvested tumor samples. The result showed that both treatment groups increased PTEN level of tumor tissues compared to the PBS control. 3WJ/FA/anti-miR21 micelles treated tumors have up-regulated PTEN in the mRNA and protein level, which was slightly higher than those treated by 3WJ/anti-miR21 micelles (Figure 6C,D). Furthermore, the RNA micelles are biocompatible and well-tolerated in vivo because there are no obvious changes in mouse weight during the experiment (Figure 6B).
Our studies revealed that RNA micelles were able to deliver the anti-miR21 and inhibited cancer growth efficiently even without ligands. The micelles with folate decoration can improve the therapeutic effects but not significantly. Both size-dependent EPR effects and ligand-mediated specific targeting affect therapeutic delivery, although the detailed correlation between these two factors was not investigated in the current study. Based on our results, RNA micelles were capable of delivering therapeutics for cancer therapy mainly by EPR effects.
The RNA micelles take advantage of the high thermodynamic stability and multivalence of pRNA-3WJ for therapeutics delivery. This established RNA micelles system enables the delivery of miRNA, anti-miRNA, siRNA, or chemical drugs for cancer therapy, especially for the cancer types whose targeting markers or ligands are not available. The addition of chemical drugs, either being encapsulated inside the lipid core or conjugated to the corona outside, provides feasibility for combining chemotherapy with gene therapy.
CONCLUSIONS
Multifunctional RNA micellar nanoparticles were constructed using the pRNA-3WJ scaffold with the aid of a hydrophobic cholesterol molecule. RNA micelles harboring anti-miR21 were stable over a wide range of pH, temperature, and RNase environment. The CMC was estimated to be 100 nM. The resulting micelles can deliver anti-miR21 specifically to cancer cells, regulate gene expression, and induce cell apoptosis. Furthermore, RNA micelles exhibited stronger tumor accumulation compared with RNA nanoparticles without micelles formation via in vivo biodistribution study. The anti-miR21-carrying micelles by themselves without a ligand were able to deliver the anti-miR21 and inhibit cancer growth, while the folate as a targeting ligand did not improve the therapeutic efficacy significantly.
METHODS AND EXPERIMENTAL DETAILS
Design and Construction of Folate-Decorated RNA Micelles Incorporating Anti-miR21.
The 3WJ/FA/anti-miR21 micelles are composed of four strands: folate-3WJ-a-sph1, Sph1-anti-miR21, and 3WJ-b-cholesterol, 3WJ-c, which were prepared by RNA synthesizer using solid-phase synthesis or followed by chemical reaction (home-made or by ExonanoRNA, LLC). The sequences are described as below:
folate-3WJ-a-sph1:5′-(Folate) uuG ccA uGu GuA uGu GGG Auc ccG cGG ccA uGG cGG ccG GGA G-3′;
Sph1-anti-miR21:5′-+G+A+T+A+A+G+C+T CTC CCG GCC GCC ATG GCC GCG GGA T-3′ (underlined sequence is 8nt LNA modified anti-miR21 seed region);
3WJ-b-cholesterol: 5′-ccc AcA uAc uuu Guu GAu cc (TEG-cholesterol)-3′; and
3WJ-c: 5′-GGA ucA Auc AuG GcA A-3′.
Cytosine(C) and uracil(U) in lowercase were 2′fluoro (2′F)-modified to make RNA more stable.69,70 Cholesterol was synthesized on the RNA strand (home-made or by ExonanoRNA, LLC) using 3′-cholesteryl-TEG controlled-pore glass (CPG) column (Glen Research Corp.). Folate-TEG-azide (Berry and Associates) was conjugated to alkyne-3WJ-a-sph1 by copper(I)-catalyzed azide–alkyne cycloaddition (CuAAC), called “click reaction” (home-made or by ExonanoRNA, LLC). Briefly, 50 μL of 2 mM alkyne-3WJ-a-sph1 solution in water and 20 μL of 50 mM folate-TEG-azide [dissolved in 3:1 (v/v) DMSO, extra dried, Acros Organics]/tBuOH (tert-butanol, anhydrous, Sigma-Aldrich)] at a molar ratio of 1:10 were mixed with 30 μL of freshly prepared “click solution” containing 0.1 M CuBr [copper(I) bromide, Sigma-Aldrich] and 0.1 M Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (TBTA, Sigma-Aldrich) in a 1:2 molar ratio. The solution was mixed thoroughly and reacted in room temperature for 4 h. After the reaction, the conjugate and control strand were evaluated by 20% UREA PAGE. Reverse-phase high-performance liquid chromatography (Agilent 1260 Infinity II) was utilized to purify the conjugated products. 3WJ/FA/anti-miR21 micelles were first prepared by mixing four strands at the same molar concentration in PBS buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4, pH 7.4) heated to 90 °C for 5 min and slowly cooled to 37 °C with a subsequent 1 h incubation at 37 °C.
To study the RNA micelles formation relying on hydrophobic interaction by cholesterol molecule, the pRNA-3WJ micelles as well as pRNA-3WJ and pRNA-3WJ-c were loaded on 2% (w/v) agarose gel and run in 40 mM TAE, 1 mM EDTA buffer with or without 0.5% (w/v) SDS. Besides, pRNA-3WJ micelles and pRNA-3WJ were prepared in increasing percentage (0%, 20%, 40%, 60%, 80%, 90%, and 100%) of DMSO solution before loading to 2% (w/v) agarose gel. The gel was stained by ethidium bromide and visualized by Typhoon FLA 7000 (GE Healthcare).
Characterization of RNA Micelles Containing 3WJ/FA/Anti-miR21.
pRNA-3WJ micelles and pRNA-3WJ nanoparticles without micelle formation were loaded onto 2% (w/v) agarose gel in TAE buffer, stained by ethidium bromide, and visualized by Typhoon FLA 7000 (GE healthcare). The gel was run at 100 V for about 50 min.
The apparent hydrodynamic diameter and ζ potential of assembled 3WJ/FA/anti-miR21 micelles (10 μM in PBS buffer) were measured by Zetasizer nano-ZS (Malvern Instrument, LTD) at 25 °C.
Determination of Critical Micelle Formation Concentration.
The CMC of RNA micelles was determined by fluorescent Nile red encapsulation assay.62 The 2-fold serial diluted RNA micelle samples (Range from 625 to 4.89 nM) were incubated with 100 μM of Nile red (Thermo Fisher Scientific) in 50 μL of PBS buffer. The samples were heated to 90 °C for 5 min and slowly cooled to 37 °C over 40 min followed by another 1 h of incubation at 37 °C. The fluorescence intensity of Nile red versus RNA micelles concentration was measured by Fluorolog spectrofluorometer (Horiba Jobin Yvon) with an excitation wavelength of 535 nm and emission spectra taken from 560 to 760 nm. The CMC could be calculated by tracking the fluorescence intensity of Nile Red as a function of the sample concentration.
RNA Micelles Stability Study.
For stability assay in different condition, 3WJ/FA/anti-miR21 micelles were incubated in acidic (pH = 5), neutral (pH = 7.4), and basic (pH = 12) PBS buffers at 37 °C for 1.5 h. These micelles were also incubated at different temperatures (4, 37, and 55 °C) for 1.5 h. Besides, they were incubated with different concentration of RNase (0.01, 0.1, and 1 μg/μL) for 1.5 h. The samples after incubation were loaded onto a 2% (w/v) agarose gel for electrophoresis under 100 V in TAE buffer. The gel was stained by EB and visualized by Typhoon FLA 7000 (GE Healthcare).
Cell Culture.
Human KB cells and human colon cancer HT29 cells (American Type Culture Collection) were grown and cultured in RPMI-1640-folate-deficient medium (Life Technologies) containing 10% fetal bovine serum (FBS) in a 37 °C incubator under 5% CO2 and a humidified atmosphere.
In Vitro Cell Binding and Internalization Assay of RNA Micelles.
To evaluate micelles binding capability, Alexa647 was used to label the 3′ end of pRNA-3WJ-c strand by solid-phase synthesis. Alexa 647 labeled 3WJ/FA/anti-miR21 micelles (100 nM) and the control groups (3WJ/anti-miR21 micelles, 3WJ/FA/anti-miR21, and 3WJ/anti-miR21) were incubated with KB cells at 37 °C for 1 h. After washing with PBS twice, the cells were resuspended in PBS for analysis. Flow cytometry was performed by FACSCalibur (Becton Dickinson) of the OSUCCC Analytical Cytometry Shared Resource.
For cell internalization study, KB cells were grown on glass slides overnight. Alexa 647 labeled 3WJ/FA/anti-miR21 micelles (100 nM) and the control groups as described above were incubated with the KB cells at 37 °C for 1 h. After washing with PBS, the cells were fixed by 4% paraformaldehyde and washed by PBS. The cytoskeleton of the fixed cells was stained by Alexa Fluor 488 Phalloidin (Invitrogen) for 25 min after pretreated with 0.1% Triton-X100 (Sigma) in PBS for 5 min. The glass slides with cells were then mounted with Prolong Gold antifade reagent with DAPI (Life Technologies) for nucleus staining. The images of internalization to cells were attained by FluoView FV1000-Filter Confocal Microscope System (Olympus Corp.).
For binding and internalization studies on HT29 cells, 200 nM Alexa647 labeled nanoparticles described above were incubated with HT29 cells at 37 °C for 1 h. Flow cytometry and Confocal microscope assays were conducted following the same procedures described above.
Dual Luciferase Assay to Study the Delivery Efficacy of Anti-miR21 by RNA Micelles.
KB cells were seeded on a 24-well plate at 70% confluence in RPMI-1640-folate-deficient medium with 10% FBS 1 day prior to the treatment. The following day, cells were transfected with 300 ng of psi-Check 2 plasmid (Promega), which contains an oncogenic miR-21 binding sequences at the 3′-UTR region of the Renilla luciferase gene using Lipofectamine 2000 (Life Technologies). A total of 4 h after transfection, the medium was replaced with complete medium, and the cells were incubated for another 2 h. Final concentrations of 200 nM and 400 nM 3WJ/FA/anti-miR21 micelles and the control groups (3WJ/anti-miR21, 3WJ/FA/anti-miR21, 3WJ/anti-miR21, and 3WJ/FA micelles) were then incubated with cells in compete medium at 37 °C, respectively. After incubating overnight, dual-luciferase assay (Promega) was used to evaluate the anti-miR21 effects following manufacturer’s instruction.
qRT-PCR Assay to Study Downstream Gene Expression.
To assay the downstream target gene PTEN expression of miR-21, TaqMan Gene Expression Assays was performed according to the manufacturer’s instructions (Life Technologies). The nanoparticles described above were incubated with KB cells at 400 nM final concentration for 72 h. After incubation, the total RNA was extracted by TRIzol reagent (Life Technologies). Then, the first cDNA strand was synthesized from total RNA (1 μg) using SuperScript III First-Strand Synthesis System (Life Technologies). RT-PCR was performed using Taqman assay. All reactions were carried out in a final volume of 20 μL using Taqman Universal PCR Master mix, primers and probe, and synthesized cDNA. The primer and probe set for human PTEN and GADPH (housekeeping gene) were purchased from Life Technologies. PCR was performed on StepOne and StepOnePlus systems (Applied Biosystem). The data were analyzed by the ΔΔCT method.
Induction of Cell Apoptosis by 3WJ/FA/Anti-miR21 Micelles.
KB cells were seeded on 24-well plate in 1640-folate-deficient plus 10% FBS medium overnight at 37 °C in a humidified 5% CO2 atmosphere. Cells were then treated with 400 nM 3WJ/FA/anti-miR21 micelles and the control groups (3WJ/anti-miR21 micelles, 3WJ/FA/anti-miR21, 3WJ/anti-miR21, and 3WJ/FA micelles). To measure the early cell apoptosis marker caspase-3 level, a caspase-3 assay kit (BD Pharmingen) was used for the assay. Cells, after 24 h of treatment, were lysed using the cold cell lysis buffer in the kit. For each group, 25 μL of cell lysate was added with 2 μL of reconstituted Ac-DEVD-AMC in 80 μL of HEPES buffer and incubated at 37 °C for 1 h. The amount of caspase-3 substrate AMC released from Ac-DEVD-AMC was measured by Fluorolog fluorospectrometer (Horiba Jobin Yvon) using an excitation wavelength of 380 nm and an emission wavelength range of 400–500 nm.
Animal Models.
All protocols involving animals are performed under the supervision of the Ohio State University Institutional Animal Care and Use Committee. To generate a xenograft model, female athymic nu/nu mice, 4–6 weeks old, were purchased from Taconic. Subcutaneous tumor xenografts were established by injecting 2.5 × 106 KB cells per site into the shoulder of nude mice.
Targeting of RNA Micelles to Cancer Xenograft in Vivo.
To compare the biodistribution profile between RNA micelles and RNA nanoparticles without micelle formation, fluorescence imaging study was performed. A total of 100 μL of 20 μM Alexa 647 labeled 3WJ/FA/anti-miR21 micelles, 3WJ/FA/anti-miR21 without micelle formation, and 3WJ-anti-miR-21 without either folate ligand or micelle formation were injected into mice bearing KB tumor by tail vein injection. PBS-injected mice were used as fluorescence negative controls. The whole body imaging of mice was conducted at 1, 4, and 8 h using an IVIS Spectrum station (Caliper Life Sciences) with excitation at 640 nm and emission at 680 nm. The mice were sacrificed at 8 h post-injection by the inhalation of CO2 followed by cervical dislocation, and major organs including heart, lungs, liver, spleen, and kidneys together with tumors were collected and subjected to fluorescence imaging for the assessment of biodistribution profiles.
In Vivo Tumor Inhibition Study by the RNA Micelles Carrying Anti-miR21.
When the tumor size reached about 50 mm3, the mice were randomly divided into three groups (n = 5 each group). The mice were injected with 3WJ/FA/anti-miR21 micelles as well as 3WJ/anti-miR21 micelles without folate ligand and PBS as control groups. There were a total of 5 injections (RNA dose: 5 mg/kg and LNA dose: 0.26 mg/kg) conducted every other day. The tumor volume and mouse weight were measured and recorded every day. The tumor volume was calculated as (length × width2) /2. At day 10, the mice were sacrificed, followed by tumor extraction and weight measurement.
To quantify the miR21 target gene PTEN expression, the tumor tissues harvested were homogenized completely. For qRT-PCR assay, TRIzol (Life Technologies) was added to extract total RNA from tumor tissue, and the PTEN level was further quantified using Taqman Assay (Life Technologies) as described above. For Western blot assay, tumor tissues were lysed in RIPA buffer with protease inhibitor cocktail and protein concentration was quantified by BCA Protein Assay Kit (Pierce). A total of 15 μg of protein was loaded to 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis, transferred to polyvinylidene fluoride membrane (Biored), and followed by blocking in 5% fat-free milk in room temperature for 2 h. The membrane was then stained with primary antibody (rabbit-PTEN: 1:1000; mouse-GADPH: 1:10 000) at 4 °C overnight and washed 3 times for 15 min using TBST. The membrane was then stained with secondary antibody (goat pAb to rabbit IgG and goat pAb to mouse IgG: 1:20 000) in room temperature for 40 min and washed 3 times for 15 min using TBST. Membranes were then incubated with ECL substrate (Biored), exposed to Amersham HyperfilmTM (GE Healthcare) together, and processed with a Series 2000A Processor film developer (TiBA).
Statistical Analysis.
Each experiment was repeated 3 times with triplication for each sample tested. The results are presented as mean plus or minus the standard deviation unless otherwise indicated. Statistical differences were evaluated using a Student’s t test, and p < 0.05 was considered significant. Single asterisks indicate p < 0.05, double asterisks indicate p < 0.01, triple asterisks indicate p < 0.001, and quadruple asterisks indicate p < 0.0001
Supplementary Material
ACKNOWLEDGMENTS
The research was supported by NIH grants (nos. R01EB019036, U01CA151648, and U01CA207946) to P.G. as well as the DOD Award (no. W81XWH-15-1-0052) to D.S. We thank Yi Shu for project discussion as well as Sijin Guo, Lora E. McBride, and Dana Driver for manuscript preparation. P.G.’s Sylvan G. Frank Endowed Chair position in Pharmaceutics and Drug Delivery is supported by the C.M. Chen Foundation.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsnano.8b07948.
Figures showing the conjugation of folate to RNA, an evaluation of micelle formation by hydrophobic force, in vitro binding and internalization of RNA micelles to colon cancer HT29 cells, and the weight of tumors harvested from mice at day 10 (PDF)
The authors declare the following competing financial interest(s): P.G. is the consultant of Oxford Nanopore Technologies and Nanobio Delivery Pharmaceutical Co. Ltd. He is the co-founder of Shenzhen P&Z Biomedical Co. Ltd and its subsidiary US P&Z Biological Technology LLC as well as ExonanoRNA, LLC and its subsidiary ExonanoRNA (Foshan Wei-Na) Biomedicine Co., Ltd.
REFERENCES
- (1).Shuai X; Ai H; Nasongkla N; Kim S; Gao J Micellar Carriers Based on Block Copolymers of Poly(Epsilon-Caprolactone) and Poly(Ethylene Glycol) for Doxorubicin Delivery. J. Controlled Release 2004, 98, 415–426. [DOI] [PubMed] [Google Scholar]
- (2).Qin B; Chen Z; Jin W; Cheng K Development of Cholesteryl Peptide Micelles for SiRNA Delivery. J. Controlled Release 2013, 172, 159–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Liu H; Zhu Z; Kang H; Wu Y; Sefan K; Tan W DNA-Based Micelles: Synthesis, Micellar Properties and Size-Dependent Cell Permeability. Chem. - Eur. J 2010, 16, 3791–3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Wu Y; Sefah K; Liu H; Wang R; Tan W DNA Aptamer-Micelle As an Efficient Detection/Delivery Vehicle Toward Cancer Cells. Proc. Natl. Acad. Sci. U. S. A 2010, 107, 5–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Carrillo-Carrion C; Atabakhshi-Kashi M; Carril M; Khajeh K; Parak WJ Taking Advantage of Hydrophobic Fluorine Interactions for Self-Assembled Quantum Dots As a Delivery Platform for Enzymes. Angew. Chem., Int. Ed 2018, 57, 5033–5036. [DOI] [PubMed] [Google Scholar]
- (6).Alemdaroglu FE; Alemdaroglu NC; Langguth P; Herrmann A DNA Block Copolymer Micelles-A Combinatorial Tool for Cancer Nanotechnology. Adv. Mater 2008, 20, 899–902. [Google Scholar]
- (7).Roh YH; Lee JB; Kiatwuthinon P; Hartman MR; Cha JJ; Um SH; Muller DA; Luo D DNAsomes: Multifunctional DNA-Based Nanocarriers. Small 2011, 7, 74–78. [DOI] [PubMed] [Google Scholar]
- (8).Cho H; Lai TC; Tomoda K; Kwon GS Polymeric Micelles for Multi-Drug Delivery in Cancer. AAPS PharmSciTech 2015, 16, 10–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Lu Y; Park K Polymeric Micelles and Alternative Nanonized Delivery Vehicles for Poorly Soluble Drugs. Int. J. Pharm 2013, 453, 198–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Torchilin VP Structure and Design of Polymeric Surfactant-Based Drug Delivery Systems. J. Controlled Release 2001, 73, 137–172. [DOI] [PubMed] [Google Scholar]
- (11).Matsumura Y; Maeda H A New Concept for Macromolecular Therapeutics in Cancer Chemotherapy: Mechanism of Tumoritropic Accumulation of Proteins and the Antitumor Agent Smancs. Cancer Res 1986, 46, 6387–6392. [PubMed] [Google Scholar]
- (12).Salzano G; Navarro G; Trivedi MS; De Rosa G; Torchilin VP Multifunctional Polymeric Micelles Co-Loaded With Anti-Survivin SiRNA and Paclitaxel Overcome Drug Resistance in an Animal Model of Ovarian Cancer. Mol. Cancer Ther 2015, 14, 1075–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Jin JO; Park H; Zhang W; de Vries JW; Gruszka A; Lee MW; Ahn DR; Herrmann A; Kwak M Modular Delivery of CpG-Incorporated Lipid-DNA Nanoparticles for Spleen DC Activation. Biomaterials 2017, 115, 81–89. [DOI] [PubMed] [Google Scholar]
- (14).Guo P; Zhang C; Chen C; Trottier M; Garver K Inter-RNA Interaction of Phage Phi29 PRNA to Form a Hexameric Complex for Viral DNA Transportation. Mol. Cell 1998, 2, 149–155. [DOI] [PubMed] [Google Scholar]
- (15).Guo P The Emerging Field of RNA Nanotechnology. Nat. Nanotechnol 2010, 5, 833–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Khisamutdinov EF; Jasinski DL; Li H; Zhang K; Chiu W; Guo P Fabrication of RNA 3D Nanoprism for Loading and Protection of Small RNAs and Model Drugs. Adv. Mater 2016, 28, 10079–10087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Jasinski D; Khisamutdinov EF; Lyubchenko YL; Guo P Physicochemically Tunable Poly-Functionalized RNA Square Architecture With Fluorogenic and Ribozymatic Properties. ACS Nano 2014, 8, 7620–7629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Shu D; Shu Y; Haque F; Abdelmawla S; Guo P Thermodynamically Stable RNA Three-Way Junctions for Constructing Multifuntional Nanoparticles for Delivery of Therapeutics. Nat. Nanotechnol 2011, 6, 658–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Grabow WW; Zakrevsky P; Afonin KA; Chworos A; Shapiro BA; Jaeger L Self-Assembling RNA Nanorings Based on RNAI/II Inverse Kissing Complexes. Nano Lett 2011, 11, 878–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Afonin KA; Bindewald E; Yaghoubian AJ; Voss N; Jacovetty E; Shapiro BA; Jaeger L Vitro Assembly of Cubic RNA-Based Scaffolds Designed in Silico. Nat. Nanotechnol 2010, 5, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Severcan I; Geary C; Chworos A; Voss N; Jacovetty E; Jaeger L A Polyhedron Made of TRNAs. Nat. Chem 2010, 2, 772–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Geary C; Chworos A; Verzemnieks E; Voss NR; Jaeger L Composing RNA Nanostructures From a Syntax of RNA Structural Modules. Nano Lett 2017, 17, 7095–7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Jaeger L; Westhof E; Leontis NB TectoRNA: Modular Assembly Units for the Construction of RNA Nano-Objects. Nucleic Acids Res 2001, 29, 455–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Boerneke MA; Dibrov SM; Hermann T Crystal-Structure-Guided Design of Self-Assembling RNA Nanotriangles. Angew. Chem., Int. Ed 2016, 55, 4097–4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Shu Y; Yin H; Rajabi M; Li H; Vieweger M; Guo S; Shu D; Guo P RNA-Based Micelles: A Novel Platform for Paclitaxel Loading and Delivery. J. Controlled Release 2018, 276, 17–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Afonin KA; Viard M; Koyfman AY; Martins AN; Kasprzak WK; Panigaj M; Desai R; Santhanam A; Grabow WW; Jaeger L; Heldman E; Reiser J; Chiu W; Freed EO; Shapiro BA Multifunctional RNA Nanoparticles. Nano Lett 2014, 14, 5662–5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Shu D; Li H; Shu Y; Xiong G; Carson WE; Haque F; Xu R; Guo P Systemic Delivery of Anti-MiRNA for Suppression of Triple Negative Breast Cancer Utilizing RNA Nanotechnology. ACS Nano 2015, 9, 9731–9740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Cui D; Zhang C; Liu B; Shu Y; Du T; Shu D; Wang K; Dai F; Liu Y; Li C; Pan F; Yang Y; Ni J; Li H; Brand-Saberi B; Guo P Regression of Gastric Cancer by Systemic Injection of RNA Nanoparticles Carrying Both Ligand and SiRNA. Sci. Rep 2015, 5, 10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Jasinski DL; Yin H; Li Z; Guo P Hydrophobic Effect From Conjugated Chemicals or Drugs on In Vivo Biodistribution of RNA Nanoparticles. Hum. Gene Ther 2018, 29, 77–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Haque F; Li J; Wu H-C; Liang X-J; Guo P Solid-State and Biological Nanopore for Real-Time Sensing of Single Chemical and Sequencing of DNA. Nano Today 2013, 8, 56–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Guo S; Li H; Ma M; Fu J; Dong Y; Guo P Size, Shape and Sequence-dependent Immunogenicity of RNA Nanoparticles. Mol. Ther.-Nucleic Acids 2017, 9, 399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Khisamutdinov E; Li H; Jasinski D; Chen J; Fu J; Guo P Enhancing Immunomodulation on Innate Immunity by Shape Transition Among RNA Triangle, Square, and Pentagon Nano-vehicles. Nucleic Acids Res 2014, 42, 9996–10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Li H; Zhang K; Pi F; Guo S; Shlyakhtenko L; Chiu W; Shu D; Guo P Controllable Self-Assembly of RNA Tetrahedrons With Precise Shape and Size for Cancer Targeting. Adv. Mater 2016, 28, 7501–7507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Abdelmawla S; Guo S; Zhang L; Pulukuri SM; Patankar P; Conley P; Trebley J; Guo P; Li QX Pharmacological Characterization of Chemically Synthesized Monomeric Phi29 PRNA Nanoparticles for Systemic Delivery. Mol. Ther 2011, 19, 1312–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Hong E; Halman JR; Shah AB; Khisamutdinov EF; Dobrovolskaia MA; Afonin KA Structure and Composition Define Immunorecognition of Nucleic Acid Nanoparticles. Nano Lett 2018, 18, 4309–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Lee RC; Feinbaum RL; Ambros V The C. Elegans Heterochronic Gene Lin-4 Encodes Small RNAs With Antisense Complementarity to Lin-14. Cell 1993, 75, 843–854. [DOI] [PubMed] [Google Scholar]
- (37).Bartel DP MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [DOI] [PubMed] [Google Scholar]
- (38).Lovat F; Valeri N; Croce CM MicroRNAs in the Pathogenesis of Cancer. Semin. Oncol 2011, 38, 724–733. [DOI] [PubMed] [Google Scholar]
- (39).Jansson MD; Lund AH MicroRNA and Cancer. Mol. Oncol 2012, 6, 590–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Ma J; Dong C; Ji C MicroRNA and Drug Resistance. Cancer Gene Ther 2010, 17, 523–531. [DOI] [PubMed] [Google Scholar]
- (41).Selcuklu SD; Donoghue MT; Spillane C MiR-21 As a Key Regulator of Oncogenic Processes. Biochem. Soc. Trans 2009, 37, 918–925. [DOI] [PubMed] [Google Scholar]
- (42).Chan JA; Krichevsky AM; Kosik KS MicroRNA-21 Is an Antiapoptotic Factor in Human Glioblastoma Cells. Cancer Res 2005, 65, 6029–6033. [DOI] [PubMed] [Google Scholar]
- (43).Lu Z; Liu M; Stribinskis V; Klinge CM; Ramos KS; Colburn NH; Li Y MicroRNA-21 Promotes Cell Transformation by Targeting the Programmed Cell Death 4 Gene. Oncogene 2008, 27, 4373–4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Dong G; Liang X; Wang D; Gao H; Wang L; Wang L; Liu J; Du Z High Expression of MiR-21 in Triple-Negative Breast Cancers Was Correlated With a Poor Prognosis and Promoted Tumor Cell in Vitro Proliferation. Med. Oncol 2014, 31, 57. [DOI] [PubMed] [Google Scholar]
- (45).Ma X; Conklin DJ; Li F; Dai Z; Hua X; Li Y; Xu-Monette ZY; Young KH; Xiong W; Wysoczynski M; Sithu SD; Srivastava S; Bhatnagar A; Li Y The Oncogenic MicroRNA MiR-21 Promotes Regulated Necrosis in Mice. Nat. Commun 2015, 6, 7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Wagenaar TR; Zabludoff S; Ahn SM; Allerson C; Arlt H; Baffa R; Cao H; Davis S; Garcia-Echeverria C; Gaur R; Huang SM; Jiang L; Kim D; Metz-Weidmann C; Pavlicek A; Pollard J; Reeves J; Rocnik JL; Scheidler S; Shi C; Sun F; Tolstykh T; Weber W; Winter C; Yu E; Yu Q; Zheng G; Wiederschain D Anti-MiR-21 Suppresses Hepatocellular Carcinoma Growth via Broad Transcriptional Network Deregulation. Mol. Cancer Res 2015, 13, 1009–1021. [DOI] [PubMed] [Google Scholar]
- (47).Lennox KA; Owczarzy R; Thomas DM; Walder JA; Behlke MA Improved Performance of Anti-MiRNA Oligonucleotides Using a Novel Non-Nucleotide Modifier. Mol. Ther.–Nucleic Acids 2013, 2, e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Davis S; Lollo B; Freier S; Esau C Improved Targeting of MiRNA With Antisense Oligonucleotides. Nucleic Acids Res 2006, 34, 2294–2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Obad S; dos Santos CO; Petri A; Heidenblad M; Broom O; Ruse C; Fu C; Lindow M; Stenvang J; Straarup EM; Hansen HF; Koch T; Pappin D; Hannon GJ; Kauppinen S Silencing of MicroRNA Families by Seed-Targeting Tiny LNAs. Nat. Genet 2011, 43, 371–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Stenvang J; Petri A; Lindow M; Obad S; Kauppinen S Inhibition of MicroRNA Function by AntimiR Oligonucleotides. Silence 2012, 3, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Di Martino MT; Gulla A; Gallo Cantafio ME; Altomare E; Amodio N; Leone E; Morelli E; Lio SG; Caracciolo D; Rossi M; Frandsen NM; Tagliaferri P; Tassone P In Vitro and in Vivo Activity of a Novel Locked Nucleic Acid (LNA)-Inhibitor-MiR-221 Against Multiple Myeloma Cells. PLoS One 2014, 9, e89659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Wu C; Li J; Wang W; Hammond PT Rationally Designed Polycationic Carriers for Potent Polymeric SiRNA-Mediated Gene Silencing. ACS Nano 2018, 12, 6504–6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Tatiparti K; Sau S; Kashaw SK; Iyer AK SiRNA Delivery Strategies: A Comprehensive Review of Recent Developments. Nanomaterials 2017, 7, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Chen Y; Gao DY; Huang L In Vivo Delivery of MiRNAs for Cancer Therapy: Challenges and Strategies. Adv. Drug Delivery Rev 2015, 81, 128–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Rui M; Qu Y; Gao T; Ge Y; Feng C; Xu X Simultaneous Delivery of Anti-MiR21 With Doxorubicin Prodrug by Mimetic Lipoprotein Nanoparticles for Synergistic Effect Against Drug Resistance in Cancer Cells. Int. J. Nanomed 2017, 12, 217–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Devulapally R; Sekar NM; Sekar TV; Foygel K; Massoud TF; Willmann JK; Paulmurugan R Polymer Nanoparticles Mediated Codelivery of AntimiR-10b and AntimiR-21 for Achieving Triple Negative Breast Cancer Therapy. ACS Nano 2015, 9, 2290–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Wu FL; Zhang J; Li W; Bian BX; Hong YD; Song ZY; Wang HY; Cui FB; Li RT; Liu Q; Jiang XD; Li XM; Zheng JN Enhanced Antiproliferative Activity of Antibody-Functionalized Polymeric Nanoparticles for Targeted Delivery of Anti-MiR-21 to HER2 Positive Gastric Cancer. Oncotarget 2017, 8, 67189–67202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Costa PM; Cardoso AL; Custodia C; Cunha P; Pereira de Almeida L; Pedroso de Lima MC MiRNA-21 Silencing Mediated by Tumor-Targeted Nanoparticles Combined With Sunitinib: A New Multimodal Gene Therapy Approach for Glioblastoma. J. Controlled Release 2015, 207, 31–39. [DOI] [PubMed] [Google Scholar]
- (59).Pereira DM; Rodrigues PM; Borralho PM; Rodrigues CM Delivering the Promise of MiRNA Cancer Therapeutics. Drug Discovery Today 2013, 18, 282–289. [DOI] [PubMed] [Google Scholar]
- (60).Pecot CV; Calin GA; Coleman RL; Lopez-Berestein G; Sood AK RNA Interference in the Clinic: Challenges and Future Directions. Nat. Rev. Cancer 2011, 11, 59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Zhang H; Endrizzi JA; Shu Y; Haque F; Sauter C; Shlyakhtenko LS; Lyubchenko Y; Guo P; Chi YI Crystal Structure of 3WJ Core Revealing Divalent Ion-Promoted Thermo-stability and Assembly of the Phi29 Hexameric Motor PRNA. RNA 2013, 19, 1226–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Zhang A; Zhang Z; Shi F; Ding J; Xiao C; Zhuang X; He C; Chen L; Chen X Disulfide Crosslinked PEGylated Starch Micelles As Efficient Intracellular Drug Delivery Platforms. Soft Matter 2013, 9, 2224–2233. [Google Scholar]
- (63).Kurniasih IN; Liang H; Mohr PC; Khot G; Rabe JP; Mohr A Nile Red Dye in Aqueous Surfactant and Micellar Solution. Langmuir 2015, 31, 2639–2648. [DOI] [PubMed] [Google Scholar]
- (64).Theti DS; Jackman AL The Role of Alpha-Folate Receptor-Mediated Transport in the Antitumor Activity of Antifolate Drugs. Clin. Cancer Res 2004, 10, 1080–1089. [DOI] [PubMed] [Google Scholar]
- (65).Toffoli G; Cernigoi C; Russo A; Gallo A; Bagnoli M; Boiocchi M Overexpression of Folate Binding Protein in Ovarian Cancers. Int. J. Cancer 1997, 74, 193–198. [DOI] [PubMed] [Google Scholar]
- (66).Rychahou P; Haque F; Shu Y; Zaytseva Y; Weiss HL; Lee EY; Mustain W; Valentino J; Guo P; Evers BM Delivery of RNA Nanoparticles into Colorectal Cancer Metastases Following Systemic Administration. ACS Nano 2015, 9, 1108–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Wang Y; Li P; Chen L; Gao W; Zeng F; Kong LX Targeted Delivery of 5-Fluorouracil to HT-29 Cells Using High Efficient Folic Acid-Conjugated Nanoparticles. Drug Delivery 2015, 22, 191–198. [DOI] [PubMed] [Google Scholar]
- (68).Binzel D; Shu Y; Li H; Sun M; Zhang Q; Shu D; Guo B; Guo P Specific Delivery of MiRNA for High Efficient Inhibition of Prostate Cancer by RNA Nanotechnology. Mol. Ther 2016, 24, 1267–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Behlke MA Chemical Modification of SiRNAs for in Vivo Use. Oligonucleotides 2008, 18, 305–319. [DOI] [PubMed] [Google Scholar]
- (70).Piao X; Wang H; Binzel DW; Guo P Assessment and Comparison of Thermal Stability of Phosphorothioate-DNA, DNA, RNA, 2’-F RNA, and LNA in the Context of Phi29 PRNA 3WJ. RNA 2018, 24, 67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


