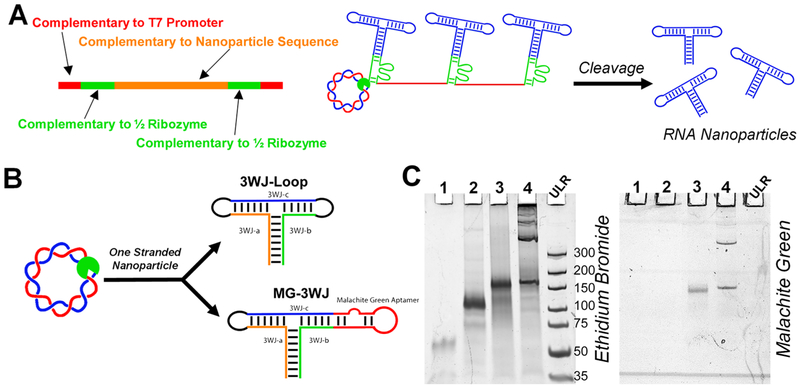
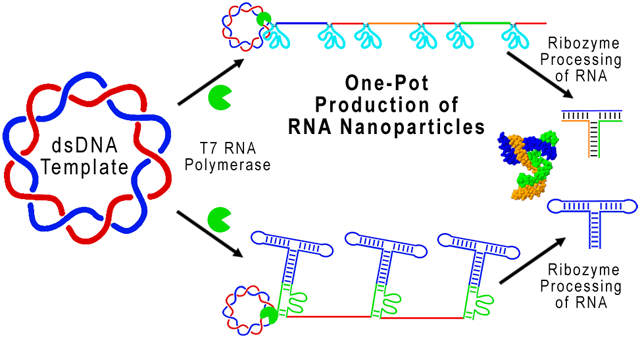
Abstract
From the original sequencing of the human genome, it was found that about 98.5% of the genome did not code for proteins. Subsequent studies have now revealed that a much larger portion of the genome is related to short or long noncoding RNAs that regulate cellular activities. In addition to the milestones of chemical and protein drugs, it has been proposed that RNA drugs or drugs targeting RNA will become the third milestone in drug development (Shu, Y.; et al. Adv. Drug Deliv. Rev. 2014, 66, 74.). Currently, the yield and cost for RNA nanoparticle or RNA drug production requires improvement in order to advance the RNA field in both research and clinical translation by reducing the multiple tedious manufacturing steps. For example, with 98.5% incorporation efficiency of chemical synthesis of a 100 nucleotide RNA strand, RNA oligos will result with 78% contamination of aborted byproducts. Thus, RNA nanotechnology is one of the remedies, because large RNA can be assembled from small RNA fragments via bottom-up self-assembly. Here we report the one-pot production of RNA nanoparticles via automated processing and self-assembly. The continuous production of RNA by rolling circle transcription (RCT) using a circular dsDNA template is coupled with self-cleaving ribozymes encoded in the concatemeric RNA transcripts. Production was monitored in real-time. Automatic production of RNA fragments enabled their assembly either in situ or via one-pot co-transcription to obtain RNA nanoparticles of desired motifs and functionalities from bottom-up assembly of multiple RNA fragments. In combination with the RNA nanoparticle construction process, a purification method using a large-scale electrophoresis column was also developed.
Keywords: rolling circle transcription, RNA nanoparticles, pRNA 3WJ motif, nanotechnology, nanobiotechnology, RNA therapeutics
Graphical Abstract
The field of RNA research has been expanded dramatically because of the finding that RNA plays a major role in the regulation of cellular activities. Once thought as a molecule that only transfers information between DNA and protein, RNA has now been found to partake in activities such as catalysis,1-3 recognition,4,5 regulation,6,7 translation,8-10 interference,11,12 processing,13-15 splicing, 16-18 silencing19,20 binding,21-23 switching,17,24, and so forth.25 RNA therapeutics, including RNA nanoparticles,26-28 RNA binding small molecules,29,30 and small RNAs such as siRNA,31 ribozymes,32 miRNA,33 riboswitches,24 or aptamers34 have been predicted to become the third milestone in drug development, following chemical drugs and protein drugs.27
RNA nanotechnology, with its concept proven in 1998 by showing the assembly of RNA dimers, trimers, tetramers, and hexamers from engineered RNA oligos,35 has been a field that continues to emerge rapidly. The versatility, complexity, and diversity of RNA makes it an attractive biomaterial to construct nanostructures with defined shape, structure, stoichiometry, and physical or biological properties.28,36-39 Many issues of RNA instability have been overcome to achieve a nanoparticle platform with potential for treatment of disease, especially cancers.26,40-42 Targeting aptamers and functional RNAs such as siRNA, miRNA, ribozymes, and riboswitches are available to construct diverse multifunctional nanoparticles.43-48 Development of RNA nanotechnology has been accelerated by the finding of an unusually stable three-way junction (3WJ) RNA motif from the packaging RNA (pRNA) of the phi29 DNA packaging motor.49 The 3WJ has been used for targeted delivery of therapeutic modalities to multiple cancer types and has been used as a scaffold for the construction of RNA nanoparticles with controllable size and shape.50-54
Although RNA nanoparticles show high clinical potential, the yield of RNA nanoparticle construction is an area that could be improved to aid in the advancement of the RNA nanotechnology field. For example, assembly of a small three-stranded RNA nanoparticle requires synthesis of three DNA templates, followed by purification of ssRNA monomers, assembly of the particles, and finally an additional particle purification.
RNA is commonly synthesized in vitro using runoff transcription of a linear dsDNA template. In vitro runoff transcription is limited by yield, time efficiency, homogeneity, and purity.55-57 Rolling circle transcription (RCT) is increasing in popularity as a result of its high production capabilities.39,58-66 Many functional RNA motifs, such as aptamers, miRNA, siRNA, and ribozymes can be continually synthesized in a normal RCT reaction.62,64 Short single-stranded (ss) circular DNAs encoding for ribozymes that selfprocess into unit length functional ribozymes have been synthesized. These ribozymes show biological functionality in trans, cleaving HIV-1 RNA targets.67,68 RCT has been used to synthesize siRNA loaded microsponges that show successful gene knockdown in vivo.39,62,63 Showing the versatility of RCT, nanowires,69 millimeter sized RNA membranes,65 mRNA nanoparticles,66 and tandem repeats of fluorogenic RNA aptamers have all been synthesized.70
RCT’s increased transcription efficiency over that of traditional runoff transcription could help to increase the production yield of RNA oligomers and RNA nanoparticles.71 Additionally, in vitro transcription is not limited by length, and one-stranded or multistranded nanoparticle assembly can occur co-transcriptionally, reducing the total number of steps required for RNA nanoparticle preparation.43,49,72 However, previous methods of circular DNA preparation for RCT are not amenable for ssDNA templates displaying stable secondary structure. Bacterial RNA polymerases are sensitive to secondary structure, falling off template DNA when encountering DNA hairpins and loops.73
Previous methods to create defined RNA oligomers using RCT include the use of ssDNA oligomers and RNase H during RCT, allowing site-specific cleavage.74 Although elegant, it could be possible to improve upon current technologies by avoiding the addition of enzymes or DNA oligomers to catalyze RNA cleavage. Encoding sequence-specific selfcleaving ribozymes75 in the DNA template, alongside RNA nanoparticle sequences, would allow simpler experimental processes and more widespread application of the RCT process to synthesize RNA oligomers with defined sequences.77-79 Additionally, in vivo expression of artificial RNA oligomers and RNA nanoparticles could be possible.80,81 Ribozymes have a stable secondary structure, and when combined in the same ssDNA template as an RNA nanoparticle sequence, the ΔG of the ssDNA template is quite low and thus not conducive to RCT using ssDNA templates.
Herein, we describe a method for the construction of circular dsDNA templates that code for self-cleaving ribozymes and RNA oligomers. Upon in vitro transcription, the ribozymes self-cleave with high efficiency, producing large amounts of product RNA and RNA nanoparticles. The phi29 pRNA-3WJ was assembled from its three component strands both co-transcriptionally and by self-assembly after RNA oligomer purification.49 Single-stranded RNA nanoparticles were synthesized via RCT by addition of loops to link adjacent 3WJ strands. The Malachite green fluorogenic RNA aptamer (MGA)82 was fused to one helix of the 3WJ, and Malachite green dye (MG)49,81 fluorescence was monitored in real-time80,81 during in vitro transcription. RCT produced a 3.2 times higher yield of fully assembled RNA nanoparticles compared to that of traditional in vitro runoff transcription.
RESULTS AND DISCUSSION
Sequence Design and Optimization for Ribozyme Cleavage Using Linear dsDNA.
Our goal was to produce short RNA oligomers that would self-assemble into RNA nanoparticles. To induce oligomer release, self-cleaving ribozymes were incorporated into the DNA templates. To enhance ribozyme cleavage efficiency, sequence optimization was carried out experimentally by gel analysis using linear dsDNA. The optimized sequences were then incorporated into circular constructs.
Linear dsDNA (dsDNA Lin) containing the T7 promoter, the 5′ and 3′ ribozymes, the RNA sites for ribozyme self-cleavage, and the product RNA sequences were constructed using PCR (Figure 1). The sequences of RNA products for ribozyme optimization were 3WJ-a, 3WJ-b, and 3WJ-c, which then assemble to form the pRNA-3WJ.49 Hammerhead ribozyme sequences were chosen, as they are well characterized and display high cleavage efficiency.76-78,83 Although the ribozyme core sequence must be conserved to maintain cleavage, non-core sequences can be modified, aiding in cleavage optimization.77,78
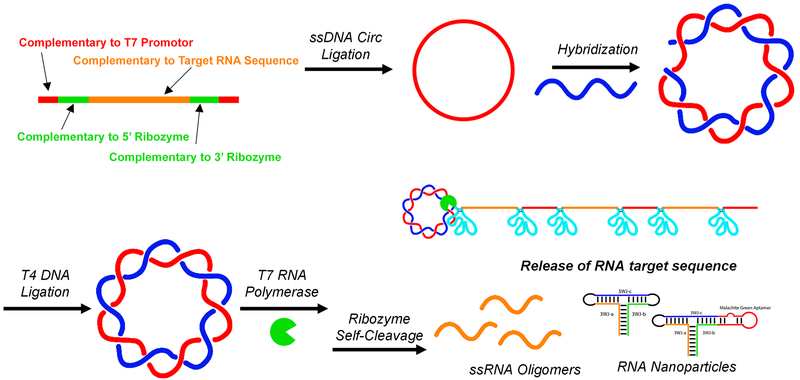
Figure 1.
Assembly scheme for the construction of double-stranded circular DNA encoding for T7 RNA promoter (red), self-cleaving ribozymes (green), and product RNA sequence (orange).
The 5′ and 3′ disabled ribozyme, the 5′ disabled ribozyme, the 3′ disabled ribozyme, and ribozyme only were used as RNA size controls (Supp Figure 1A). Ribozyme activity can be abolished by sequence mutation.77 The product RNA strands were chemically synthesized for size and assembly controls. Cleavage efficiency was calculated by comparing the product RNA band intensity (in green frame) to the total intensity (in green plus red frame) per lane over the 2 h after the initiation of transcription (Supp Figure 1B). ImageJ software was used to integrate the gel band intensity. Cleavage kinetics assays were done using truncated versions of the dsDNA. For example, 5′ ribozyme efficiency was assayed using the dsDNA template with no 3′ ribozyme. The cleavage efficiency of each ribozyme, 5′ and 3′ of the product RNA, was calculated independently (Supp Figure 1B).
A two base-pair (bp) “clamping” duplex led to a cleavage efficiency ranging from 36 to 65% cleavage. Upon lengthening the “clamping” duplex to five bp (Supp Figure 1C) to enhance the stability of the ribozyme sequence, cleavage efficiencies increased to 65 to 78%. When full length constructs with both active ribozymes were tested, cleavage efficiencies were more than 80% (Supp Table 1).
Circular dsDNA Construction.
RCT offers many advantages over traditional in vitro transcription methods including higher transcription rate,70,71 template DNA economy, and the potential for in vivo expression of artificial RNA sequences and RNA nanoparticles.80 Previously published RCT methods for circular DNA preparation called for assembly of a short splint ssDNA to longer phosphorylated ssDNA followed by DNA ligation, resulting in circular DNA with a double-stranded RNA promoter region and a single-stranded antisense region to the desired RNA sequence.39,58-66 Although this method is amenable for ssDNA templates with little secondary structure (ΔG of self-folding close to zero or a positive value, Supp Table 2)68,71,84,85 or stable dumbbell sequences,39,62,70,86 it was not amenable for ssDNA templates encoding for RNA with strong secondary structure such as the pRNA-3WJ.
The ΔG of self-folding for the ssDNA templates used in this study ranges from −11.6 to −27 kcal/mol (Supp Table 2), resulting in stable secondary structures hindering transcription by bacterial RNA polymerases. The published methods39,58-66 to prepare circular DNA template were attempted; yet, even in the presence of single-stranded binding proteins and elevated transcription temperatures at 42 °C, the stable secondary structure of the sequences hindered transcription (data not shown). Removing the issue of stable secondary structure in the DNA template was addressed by making the circular template entirely double-stranded. Therefore, circular dsDNA was made starting with two complementary and phosphorylated ssDNAs (Figure 1).
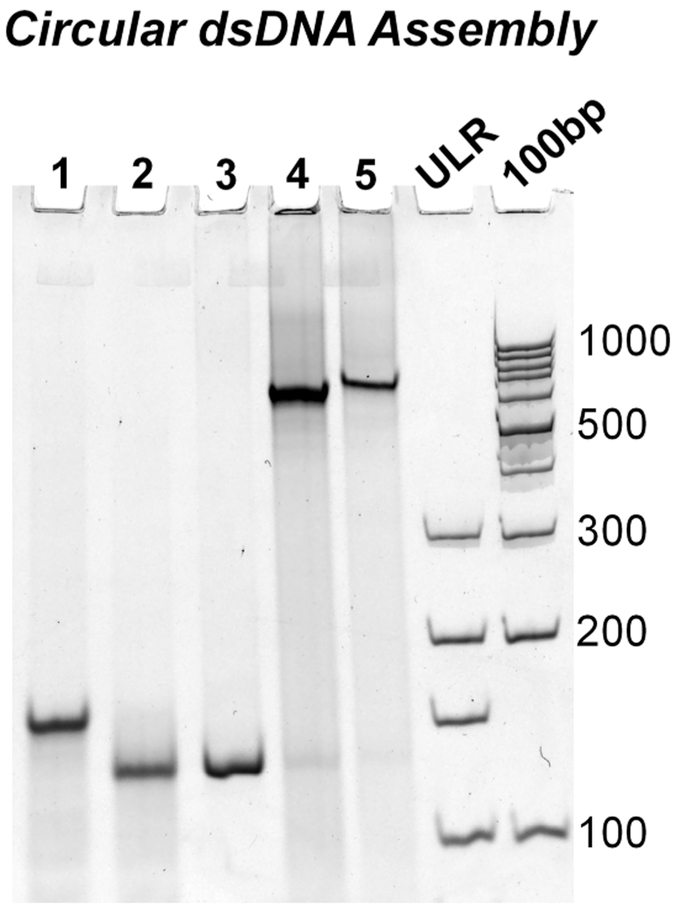
Phosphorylated ssDNA complementary to the T7 promoter, 5′ and 3′ ribozymes, and product RNA sequence was self-ligated using Epicenter ssDNA Circ Ligase to form circular ssDNA. To confirm cyclization, polyacrylamide gel electrophoresis (PAGE) was used to visualize DNA bands before and after single-stranded circular (ssDNA Circ) ligation (Figure 2, lanes 1 vs 2). Upon single-stranded cyclization, an increase in migration rate was seen presumably due to the compact structure of the now self-ligated ssDNA. Assembly of all dsDNA constructs can be found in Supp Figure 2.
Figure 2.
Assembly of circular dsDNA encoded for a single pRNA 3WJ strand surrounded by the ribozyme for cleavage. Representative gel image of the assembly process. (1) Single-stranded linear DNA (ssDNA Lin); (2) single-stranded circular DNA (ssDNA Circ), (3) double-stranded linear DNA (dsDNA Lin); (4) double-stranded circular DNA (dsDNA Circ) + Nick; (5) dsDNA Circ + T4 Ligation to close nick. Ultralow range DNA Ladder (ULR) and 100bp DNA ladder for size comparison.
To form circular dsDNA (dsDNA Circ), cyclized ssDNA and its phosphorylated complement were mixed at equimolar concentrations and annealed by thermal denaturation followed by slowly cooling to 4 °C over 1 h. Assembly of the complement strands resulted in a dramatic decrease in migration rate, indicating successful hybridization (Figure 2, lanes 2 vs 4/5). Compared to linear dsDNA (dsDNA Lin) controls, circular dsDNA migrates much slower, indicating circular conformation. After assembly, T4 DNA ligase was used to ligate the nicked circular dsDNA (Figure 2, lanes 4 vs 5). No apparent shift is observed by gel analysis after T4 ligation. Assembly of all dsDNA constructs can be found in Supp Figure 2.
RCT Reaction.
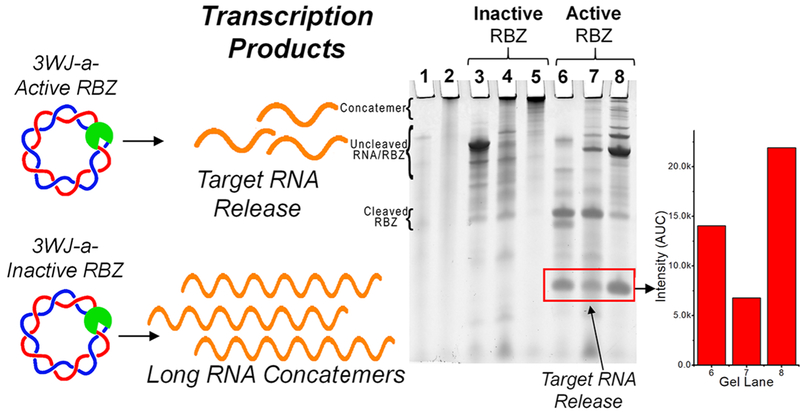
To show successful ribozyme processing, ribozyme activity was disabled by sequence mutation.77 Disabling of ribozyme resulted in full template length RNA product using linear dsDNA, and long concatemeric RNA using circular dsDNA, respectively. The encoded RNA sequence was 3WJ-a for both active and inactive ribozyme constructs. After in vitro transcription and termination by DNase, PAGE analysis was used to visualize RNA transcripts (Figure 3). Heavy accumulation of RNA transcripts in the well of the gel indicate long RNA concatemers and successful RCT (Figure 3, lane 5), compared to the transcription of both the linear dsDNA template (Figure 3, lane 3) and the nicked circular dsDNA template (Figure 3, lane 4). When ribozyme activity was restored, successful cleavage and release of product RNA strand was observed (Figure 3, lanes 6–8), evidenced by the appearance of short RNA transcripts not seen in inactivated ribozyme constructs.
Figure 3.
RCT assessment. Inactive ribozymes lead to concatemerized RNA, whereas cleavage results in short oligomers. RNA products of transcriptions using (1) single-stranded linear DNA (ssDNA Lin); (2) single-stranded circular DNA (ssDNA Circ); (3) double-stranded linear DNA (dsDNA Lin); (4) double-stranded circular DNA (dsDNA Circ) + Nick; (5) dsDNA Circ; (6) dsDNA Lin; (7) dsDNA Circ + Nick; (8) dsDNA Circ. Lanes 3–5 use inactivated ribozyme (RBZ) sequences; lanes 6–8 use active ribozyme sequences. Gel bands were quantified for desired products from lanes 6–8. Additional band details are provided in Supp Figure 1.
One-Pot Co-Transcriptional Assembly of 3WJ Nanoparticles.
Here, ribozymes were engineered, and their catalytic property leveraged to induce self-cleavage of specified RNA oligomers, which then co-transcriptionally assemble the pRNA-3WJ. This allows a hands-off method for transcriptional production of short and defined RNA oligomers in high yield without the use of additional enzymes and will aid in the future scale-up of this method for large-scale synthesis of RNA nanoparticles in vitro and in vivo.
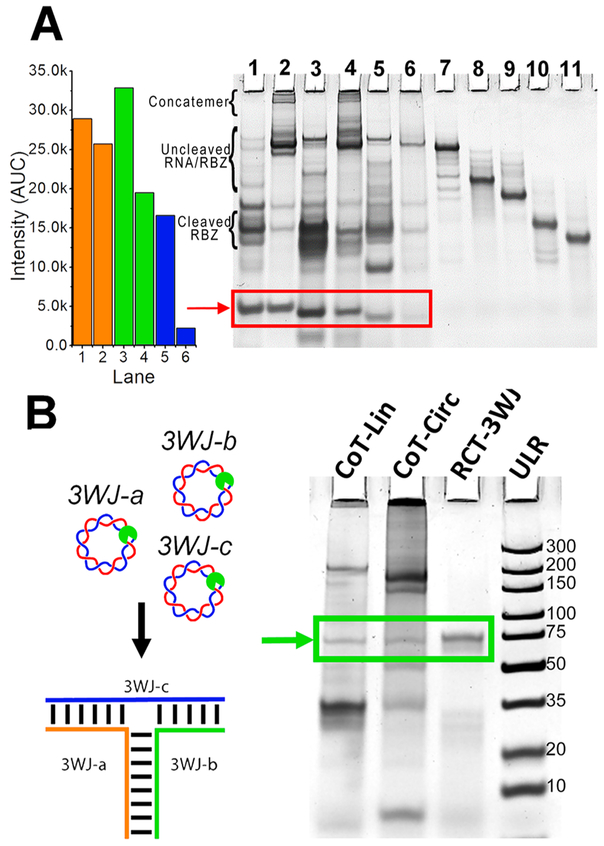
RNA 3WJ and other nanoparticles have the ability to self-assemble co-transcriptionally under isothermal conditions.43,49,72,87-89 To assemble the pRNA-3WJ co-transcriptionally, 3WJ-a, 3WJ-b, and 3WJ-c sequences were incorporated in separate circular dsDNA constructs. All constructs demonstrated cleavage and release of the product RNA (Figure 4A, red box). Product RNA strands were equal in size to that of their chemically synthesized size controls, indicating successful cleavage and release (Supp Figure 3A). To confirm that the sequences of product RNA were correct, each fragment was isolated, and their assembly was tested. Formation of the 3WJ from isolated RNA bands indicates sequence-specific cleavage and release of product RNAs (Supp Figure 3B). Additionally, it appears that there was low expression of the 3WJ-c strand from the transcription; however, this is due to the unstable secondary structure of the 3WJ-c monomer strand by itself, resulting in reduced EB intercalation.42,49
Figure 4.
In situ or one-pot co-transcriptional nanoparticles. (A) Circular (Circ) and linear (Lin) transcription for dsDNA constructs encoding for release of the 3WJ ssRNA oligomers.49 3WJ-a-Lin; (2) 3WJ-a-Circ; (3) 3WJ-b-Lin; (4) 3WJ-b-Circ; (5) 3WJ-c-Lin; (6) 3WJ-c-Circ; (7) 5′/3′Rbz-IN; (8) 3′Rbz-IN; (9) 5′Rbz-IN; (10) 5′Rbz Only; (11) 3′Rbz Only. Gel bands were quantified for desired products from lanes 1–5. Additional band details are provided in Supp Figure 1. (B) Assembly of the 3WJ occurs co-transcriptionally (CoT) by mixing equimolar amounts of dsDNA encoding for the release of 3WJ-a, -b, and -c strands. Rolling circle transcription (RCT)-3WJ is assembled from purified RNA fragments from RCT reaction. Ultralow range DNA Ladder (ULR) for comparison.
pRNA-3WJ nanoparticles were assembled co-transcriptionally by mixing 3WJ-a, -b, and -c dsDNA constructs in an equimolar ratio followed by in vitro transcription.49 PAGE analysis of both linear and circular co-transcription products indicates successful assembly of 3WJ nanoparticles when compared to the assembled 3WJ from the gel purified RCT product (Figure 4B).
To assemble nanoparticles from one ssRNA oligomer, circular dsDNAs encoding for the full sequence of the 3WJ, with the helix ends closed with loops or malachite green aptamer (MGA) sequences, were constructed (Figure 5). Including an aptamer in the sequence serves two purposes: (1) it monitors transcription kinetics by fluorescence, (2) and it shows accurate cleavage and correct folding of the RNA nanoparticle, as malachite green (MG) will not bind to MGA unless the sequences are correct. Following transcription, the RCT reaction mixture was analyzed by PAGE (Figure 5C). RCT-3WJ assembled from purified 3WJ monomers and one-piece 3WJ with loops were used as size controls. The one-stranded nanoparticles migrate slower due to increased size from the incorporation of loop sequences used to connect helix ends. Gel staining with MG shows binding of the MG-3WJ nanoparticle to its fluorophore, indicating correct sequence and folding of the MG-3WJ. No MG signal from the 3WJ, which lacks the MGA, indicates specific binding of MG to MGA. Higher-order concatemers are present in the RCT reactions, indicating that ribozyme cleavage is not 100%.
Figure 5.
Assembly of one-stranded RNA nanoparticle with functionalities. (A) Schematic for modified design of self-cleaving ribozyme. (B) Circular (Circ) dsDNA was assembled to code for the 3WJ and Malachite green aptamer (MG)-3WJ nanoparticles assembled from one long piece of RNA. (C) PAGE analysis of transcription of one-piece RNA nanoparticles. (1) RCT-3WJ; (2) 3WJ-Loop; (3) MG-3WJ-Lin; (4) MG-3WJ-Circ. Ultralow range DNA Ladder (ULR) for comparison.
Real-Time Monitoring of Transcription by Fluorescence.
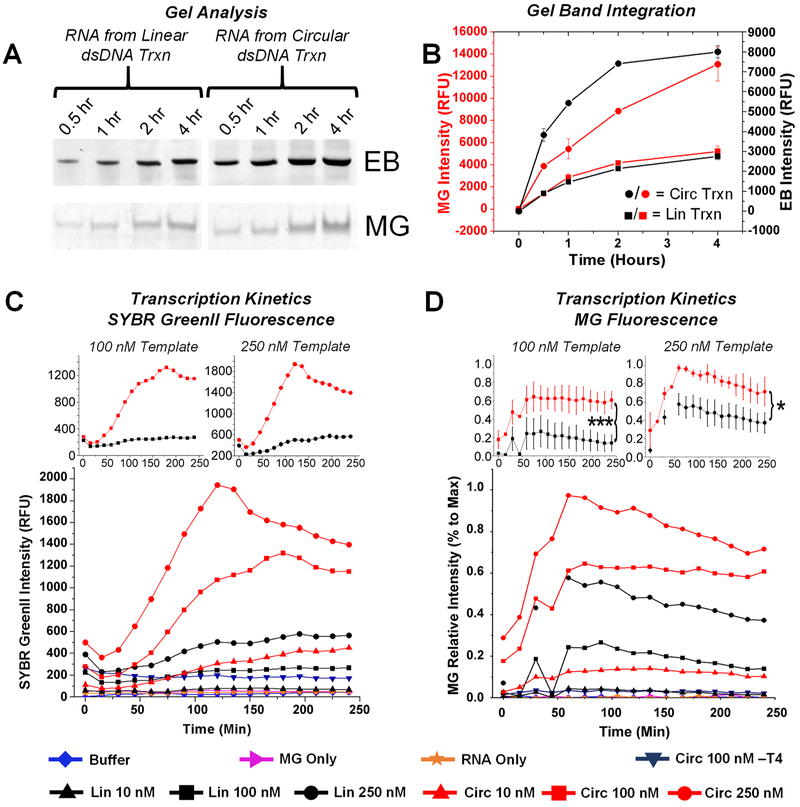
The goal of preparing circular dsDNA for transcription is the hypothesis that RCT will result in higher amounts of RNA nanoparticles from the same starting DNA concentrations, therefore being more efficient and faster in nanoparticle production. Thus, gel analysis was first used to compare the transcription rate of linear and circular dsDNA by analyzing transcription time points after termination by DNaseI. Gels were stained for total RNA using ethidium bromide (EB) and for MG signal using MG dye (Figure 6A). Comparison of gel band intensity revealed both faster production and a higher yield of MG-3WJ by RCT compared to that of linear transcription (Figure 6B). As the ribozyme cleavage efficiency was not 100%, gel staining represents an accurate comparison of RNA nanoparticle production, because the quantified gel band was the produced RNA nanoparticle.
Figure 6.
Linear vs circular DNA transcription kinetics. (A,B) Gel analysis of transcription on Malachite green (MG) fluorescence (specific to nanoparticle folding) and ethidium bromide (EB) (RNA-specific) channels. Bands were integrated and plotted. (C,D) Monitoring of transcription using MG fluorescence (specific to nanoparticle folding) and SYBR Green II (RNA-specific). MG experiments were repeated a minimum of three times; error is shown as standard deviation. *p < 0.05, ***p < 0.005.
Fluorescence measurements were used to monitor transcription rates in real-time.80,81 MGA fluorescence was monitored by adding MG dye to a final concentration of 5 μM in the transcription mixture, whereas total RNA production was monitored by adding RNA-specific SYBR GreenII to a final concentration of 1×. Both MG and MGA are not fluorescent by themselves, but fluorescence will appear when the MG binds to the MGA.80-82 Fluorescence measurements were taken every 15 min, and intensity was plotted versus time using OriginPro (Figure 6C,D). Among three different DNA template concentrations of 10 (▲), 100 (■), and 250 (●) nM, RCT reactions (red) consistently outperformed linear transcription (black) in both nanoparticle production rate and overall transcription yield. It is important to note that identical sequences were used in all constructs in comparison experiments, the only difference being circular or linear dsDNA. RCT produced on average 10 times more RNA at the termination of the transcription reaction, consistent with previous findings on the rate of RCT.70,71 The nicked circular dsDNA template without T4 ligation was similar in transcription rate to linear dsDNA at the same concentration, suggesting that T7 polymerase does not proceed to RCT with nicked circular dsDNA.
Substrate-concentration-dependent kinetics of total RNA transcription were analyzed by comparing SYBR GreenII fluorescence values at different time points (values from Figure 6C). Time points from 60–165 min were chosen, as these were during and after the highest increase of observed fluorescence. Fluorescence values were plotted and fit linearly (y = mx + b) (Supp Figure 4A,B). A strong linear correlation is seen between DNA template concentrations and their fluorescence output (Supp Figure 4C). The slope (x) values for the circular DNA template display an average value of 5.27, higher than those observed for the linear DNA template, with an average equal to 1.27. These values can be correlated to fluorescence output per nM (RFU/nM) of the DNA template. Thus, these results support that the idea that the RCT reaction results in an increase in transcription efficiency. At these DNA template concentrations, the relationship appears to be linear.
Large-Scale Purification Post-RCT.
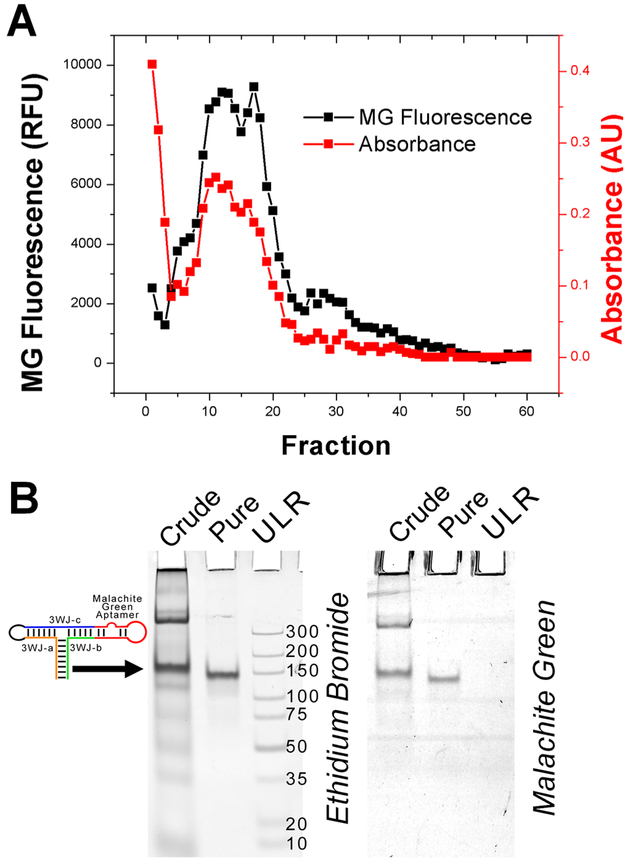
Purification of the MG-3WJ nanoparticles was carried out using a preparative version of typical gel electrophoresis. A BioRad Model 491 Gel Electrophoresis Prep Cell was used to purify large-scale transcription products of RNA nanoparticles transcribed from circular dsDNA encoding for the MG-3WJ. Fractions were analyzed for both MG fluorescent signal and absorbance at 260 nm (Figure 7A). Three distinct peaks were seen by MG fluorescence: fractions 5–7 (peak 1), 7–23 (peak 2), and 25–32 (peak 3). PAGE analysis was used to determine the identity of each peak compared to the crude transcription mixture. Peak 1 was smaller than the product RNA, peak 2 contained the product RNA, and peak 3 contained both the product RNA and larger RNA bands. Peak 2 fractions were then combined and analyzed by PAGE (Figure 7B). Of note is the large absorbance value of the first fractions, which has been attributed to remaining nucleotides from the transcription mixture, as no band was seen by gel analysis.
Figure 7.
Large-scale purification of RNA by gel-electrophoresis column. (A) Malachite green (MG) fluorescence and absorbance at 260 nm were used to analyze fractions after purification. (B) PAGE analysis demonstrating purity before and after gel purification. Ultralow range DNA Ladder (ULR) for comparison.
Here we were able to construct self-folding RNA nanoparticles by combining several technologies: rolling circle transcription, ribozyme self-cleavage, and gel column electrophoresis. By combining these techniques, RNA nanoparticle construction was simplified, in that the complete nanoparticles were produced through three simple steps of template construction, RNA transcription, and nanoparticle purification. This methodology of RNA nanoparticle production removes several steps, producing and purifying each RNA strand within the nanoparticle, nanoparticle assembly, and nanoparticle purification. Furthermore, through the development of this method set, RNA production was proven to be produced at a higher yield than traditional linear in vitro transcription, and RNA nanoparticles were shown to self-assemble with original and authentic folding. Although this system is not fully optimized, it may lead to breakthroughs in current industrial techniques for RNA production, leading to a much needed reduced cost in RNA nanoparticle production.
CONCLUSIONS
This study establishes a solution for the simplification of RNA nanoparticle production as well as an approach for higher-yield assembly of RNA nanoparticles in vitro. The method for circular dsDNA preparation is broadly applicable to the field of RNA biology and RNA nanotechnology for the production of functional self-folding nanoparticles. To release RNA nanoparticles transcribed during RCT, self-cleaving ribozymes were coded in the template, allowing bottom-up assembly either in situ or one-pot co-transcriptional release of the product RNA fragments. Production was monitored in real-time. RNA nanotechnology is emerging as a new drug delivery platform and shows great promise to help advance the current state of nanomedicine. The methods introduced here are a step toward the large-scale production of RNA nanoparticles and could be helpful for future clinical applications of RNA nanotechnology. As well as large-scale batch synthesis, in vivo expression and production of RNA nanoparticles could be possible using the methods introduced here.
MATERIALS AND METHODS
Sequence Design.
Sequences for product oligomers were derived from the pRNA-3WJ nanoparticle as reported previously.49 Additional nucleotides were added to the 5′ and 3′ ends of the native 3WJ sequences, 5′-GAC-3′ and 5′-GUC-3′, respectively, aiding in ribozyme stability and cleavage efficiency. For the synthesis of single-stranded RNA nanoparticles, loops (5′-GAGA-3′) were used to join adjacent 5′ and 3′ ends of the multistranded RNA nanoparticles. Ribozyme sequences were adapted from previously solved structures of the hammerhead ribozyme.77 For ribozyme efficiency assays, ribozyme sequences were mutated to abolish activity as previously described.77 The Malachite green aptamer sequence was adapted from previously published sequences.82 All sequences are summarized in Supporting Table 3.
DNA and RNA Preparation.
RNA oligomers were prepared in vitro using T7 RNA polymerase from linear and circular dsDNA containing the T7 promoter. Linear dsDNA was prepared by PCR using ssDNA primers purchased from Integrated DNA Technologies (IDT). Circular dsDNA was prepared from Ultramer oligomers purchased from IDT. The assembly method of circular dsDNA is detailed in Figure 1. ssDNA antisense to the RNA strand (including T7 promoter, ribozymes, and product RNA) was self-cyclized intramolecularly using Epicenter CircLigase following the manufacturer’s protocol. Remaining linear ssDNA was removed by addition of DNA Exonuclease I. ssDNA complementary to the cyclized ssDNA was annealed by thermal denaturation at 85 °C for 5 min followed by slow cooling to 4 °C at a rate of —2 °C/min at 4 μM. Following annealing, T4 DNA ligase closed the nick in the circular dsDNA. Ligations were performed at a DNA concentration of 2.5 μM in 1× DNA ligase buffer, 5% (w/v) PEG4000, and a ligase concentration of 0.25 U/μL. The ligation mixture was incubated at 25 °C for 4 h followed by heating at 65 °C to heat denature the enzyme. Control constructs without T4 ligation were also assembled.
RNA transcriptions were completed using T7 RNA polymerase isolated from E. coli containing plasmid encoding for the polymerase through the use of a His tag on the protein. All transcriptions, linear and circular, were completed at 37 °C with the following final concentrations of reagents: 40 mM HEPES-KOH (pH 7.5), 12 mM MgCl2, 30 mM DTT, 1 mM spermidine, 5 mM rNTPs (rATP, rCTP, rGTP, and rUTP) along with DNA template and T7 RNA polymerase. Reactions were incubated for varying times depending on the desired experiment, but complete reactions were carried out for 4 h.
Gel Analysis and Quantification.
Assembly of linear and circular dsDNA constructs and RNA products was confirmed using 10% (29:1) PAGE in a buffer containing 50 mM TRIS, 100 mM NaCl, and 5 mM EDTA, and DNA or RNA was stained using ethidium bromide (EB) solution or MG dye solution (final concentration of 5 μM MG Dye, HEPES pH = 7.4, 100 mM KCl, 5 mM MgCl2. Gel images were then quantified using ImageJ and plotted using OriginPro 8.5.
Ribozyme Cleavage Efficiency.
Time course experiments were run to analyze ribozyme cleavage efficiency. A typical transcription reaction was quenched at specific time points by addition of DNase. Equal aliquots of each time point were then analyzed on PAGE, and gel band intensity was integrated using ImageJ software.90 Ribozyme cleavage occurred during the transcription process and thus is at 37 °C and in T7 RNA polymerase transcription buffer. Cleavage efficiency was calculated by dividing the band intensity of the cleaved fractions by total band intensity per lane. Cleavage percentage versus time was then plotted using OriginPro 8.5.
Gel Analysis of Transcription Kinetics.
dsDNA constructs, both linear and circular, were transcribed following typical T7 in vitro transcription protocols with a 250 nM final DNA concentration. At 0.5, 1, 2, and 4 h, transcription reactions were quenched using DNase. Equal aliquots of transcription from each time point were analyzed on PAGE, and product RNA bands were integrated using ImageJ software. Gels were stained separately for total RNA (EB) and MG fluorescence. Gel band intensity versus time was then plotted using OriginPro 8.5. Gel assays were carried out in triplicate.
Fluorescence Monitoring of RNA Transcription.
To monitor transcription in real-time, MG dye or SYBR GreenII was added at a final concentration of 5 μM to transcription reactions. Solutions were incubated at 37 °C in 96-well microplates, and fluorescent signal was monitored every 15 min using a BioTek Synergy 4 Microplate Reader. MG signal was read from excitation and emission wavelengths of 590 and 630 nm, respectively. SYBR GreenII fluorescence was read from excitation and emission wavelengths of 496 and 520 nm, respectively. OriginPro was used to plot fluorescent signal versus time. Transcription analysis was performed at DNA concentrations of 10, 100, and 250 nM. MG fluorescent studies were completed a minimum of three times and in most cases a minimum of four repeats. Error was calculated by standard deviation, and statistics were calculated by a t-test using GraphPad comparing fluorescent values of the same DNA template concentrations.
Large-Scale Purification Using Gel-Electrophoresis Column.
Transcription reactions were purified on a BioRad model 491 Prep Cell using continuous-elution gel electrophoresis following the manufacturer’s standard protocol. A 20 mL aliquot of 8% polyacrylamide gel was prepared in a buffer containing 50 mM TRIS, 100 mM NaCl, and 5 mM EDTA. The gel was polymerized to a column height of 5.5 cm and prerun at 300 V for 1 h. Large-scale, 1 mL transcriptions had a final DNA concentration of 250 nM, and transcriptions were diluted to 2 mL with 2× gel loading dye and loaded onto the column. After 2 h of electrophoresis at 300 V, fractions were collected at a rate of 0.25 mL/min for 4 min for a total fraction volume of 1 mL. A total of 60 fractions were collected for 4 h. Fractions were analyzed by adding 20 μL of 10× MG binding buffer (final concentration of 5 μM MG Dye, HEPES pH = 7.4, 100 mM KCl, 5 mM MgCl2) to 180 μL of each fraction. Fluorescence was analyzed as described previously. RNA concentrations was measured by reading absorbance at 260 nm.
Supplementary Material
ACKNOWLEDGMENTS
The research in P.G.’s lab was supported by NIH grants R01EB019036, U01CA151648, and U01CA207946. P.G.’s Sylvan G. Frank Endowed Chair position in Pharmaceutics and Drug Delivery is funded by the CM Chen Foundation.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsnano.9b00649.
The supplemental figures provide detailed validation of bands seen in gel images, quantification of ribozyme cleavage and validation of target strands, and validation of circular dsDNA templates and comparison of stability to published literature as well as DNA sequences used for each of our designs and target sequences incorporated into the circular DNA template. These figures and tables provide more robust details to the figures presented here in the main text (PDF)
The authors declare the following competing financial interest(s): P.G. is the consultant of Oxford Nanopore Technologies, cofounder of Shenzhen P&Z Biomedical Co. Ltd and its subsidiary US P&Z Biological Technology LLC as well as cofounder of ExoNano RNA LLC and its subsidiary Weina Biomedical (GD) Ltd.
REFERENCES
- (1).Westhof E Ribozymes, Catalytically Active RNA Molecules. Introduction. Methods Mol. Biol. 2012, 848, 1–4. [DOI] [PubMed] [Google Scholar]
- (2).Cech TR; Zaug AJ; Grabowski PJ In Vitro Splicing of the Ribosomal RNA Precursor of Tetrahymena: Involvement of a Guanosine Nucleotide in the Excision of the Intervening Sequence. Cell 1981, 27, 487–496. [DOI] [PubMed] [Google Scholar]
- (3).Guerrier-Takada C; Gardiner K; Marsh T; Pace N; Altman S The RNA Moiety of Ribonuclease P Is the Catalytic Subunit of the Enzyme. Cell 1983, 35, 849–857. [DOI] [PubMed] [Google Scholar]
- (4).Holley RW Structure of an Alanine Transfer Ribonucleic Acid. JAMA 1965, 194, 868–871. [PubMed] [Google Scholar]
- (5).Suddath FL; Quigley GJ; McPherson A; Sneden D; Kim JJ; Kim SH; Rich A Three-Dimensional Structure of Yeast Phenylalanine Transfer RNA at 3.0 Angstroms Resolution. Nature 1974, 248, 20–24. [DOI] [PubMed] [Google Scholar]
- (6).Lee RC; Feinbaum RL; Ambros V The C. Elegans Heterochronic Gene Lin-4 Encodes Small RNAs With Antisense Complementarity to Lin-14. Cell 1993, 75, 843–854. [DOI] [PubMed] [Google Scholar]
- (7).Croce CM; Calin GA miRNAs, Cancer, and Stem Cell Division. Cell 2005, 122, 6–7. [DOI] [PubMed] [Google Scholar]
- (8).Correll CC; Freeborn B; Moore PB; Steitz TA Metals, Motifs, and Recognition in the Crystal Structure of a 5S RRNA Domain. Cell 1997, 91, 705–712. [DOI] [PubMed] [Google Scholar]
- (9).Shoji S; Walker SE; Fredrick K Ribosomal Translocation: One Step Closer to the Molecular Mechanism. ACS Chem. Biol. 2009, 4, 93–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Yusupov MM; Yusupova GZ; Baucom A; Lieberman K; Earnest TN; Cate JH; Noller HF Crystal Structure of the Ribosome at 5.5 A Resolution. Science 2001, 292, 883–896. [DOI] [PubMed] [Google Scholar]
- (11).Fire A; Xu S; Montgomery MK; Kostas SA; Driver SE; Mello CC Potent and Specific Genetic Interference by Double-Stranded RNA in Caenorhabditis Elegans. Nature 1998, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- (12).Parrish S; Fleenor J; Xu S; Mello C; Fire A Functional Anatomy of a dsRNA Trigger: Differential Requirement for the Two Trigger Strands in RNA Interference. Mol. Cell 2000, 6, 1077–1087. [DOI] [PubMed] [Google Scholar]
- (13).Jady BE; Kiss TA Small Nucleolar Guide RNA Functions Both in 2 ’-O-Ribose Methylation and Pseudouridylation of the U5 Spliceosomal RNA. EMBO J. 2001, 20, 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Bachellerie JP; Cavaille J; Huttenhofer A The Expanding SnoRNA World. Biochimie 2002, 84, 775–790. [DOI] [PubMed] [Google Scholar]
- (15).Mercer TR; Dinger ME; Mattick JS Long Non-Coding RNAs: Insights into Functions. Nat. Rev. Genet. 2009, 10, 155–159. [DOI] [PubMed] [Google Scholar]
- (16).Scott WG Ribozymes. Curr. Opin. Struct. Biol. 2007, 17, 280–286. [DOI] [PubMed] [Google Scholar]
- (17).Strobel SA; Cochrane JC RNA Catalysis: Ribozymes, Ribosomes, and Riboswitches. Curr. Opin. Chem. Biol. 2007, 11, 636–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Mulhbacher J; St-Pierre P; Lafontaine DA Therapeutic Applications of Ribozymes and Riboswitches. Curr. Opin. Pharmacol. 2010, 10, 551–556. [DOI] [PubMed] [Google Scholar]
- (19).Hamilton AJ; Baulcombe DC A Species of Small Antisense RNA in Posttranscriptional Gene Silencing in Plants. Science 1999, 286, 950–952. [DOI] [PubMed] [Google Scholar]
- (20).Elbashir SM; Harborth J; Lendeckel W; Yalcin A; Weber K ; Tuschl T Duplexes of 21-Nucleotide RNAs Mediate RNA Interference in Cultured Mammalian Cells. Nature 2001, 411, 494–498. [DOI] [PubMed] [Google Scholar]
- (21).Bunka DH; Stockley PG Aptamers Come of Age - at Last. Nat. Rev. Microbiol. 2006, 4, 588–596. [DOI] [PubMed] [Google Scholar]
- (22).Zhu H; Li J; Zhang XB; Ye M; Tan W Nucleic Acid Aptamer-Mediated Drug Delivery for Targeted Cancer Therapy. ChemMedChem 2015, 10, 39–45. [DOI] [PubMed] [Google Scholar]
- (23).Keefe AD; Pai S; Ellington A Aptamers as Therapeutics. Nat. Rev. Drug Discovery 2010, 9, 537–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Tucker BJ; Breaker RR Riboswitches as Versatile Gene Control Elements. Curr. Opin. Struct. Biol. 2005, 15, 342–348. [DOI] [PubMed] [Google Scholar]
- (25).Lieberman J; Slack F; Pandolfi PP; Chinnaiyan A; Agami R; Mendell JT Noncoding RNAs and Cancer. Cell 2013, 153, 9–10. [DOI] [PubMed] [Google Scholar]
- (26).Guo P The Emerging Field of RNA Nanotechnology. Nat. Nanotechnol. 2010, 5, 833–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Jasinski D; Haque F; Binzel DW; Guo P Advancement of the Emerging Field of RNA Nanotechnology. ACS Nano 2017, 11, 1142–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Afonin KA; Viard M; Koyfman AY; Martins AN; Kasprzak WK; Panigaj M; Desai R; Santhanam A; Grabow WW; Jaeger L; et al. Multifunctional RNA Nanoparticles. Nano Lett. 2014, 14, 5662–5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Thomas JR; Hergenrother PJ Targeting RNA with Small Molecules. Chem. Rev. 2008, 108, 1171–1224. [DOI] [PubMed] [Google Scholar]
- (30).Rizvi NF; Howe JA; Nahvi A; Klein DJ; Fischmann TO ; Kim HY; McCoy MA; Walker SS; Hruza A; Richards MP; et al. Discovery of Selective RNA-Binding Small Molecules by Affinity-Selection Mass Spectrometry. ACS Chem. Biol. 2018, 13, 820–831. [DOI] [PubMed] [Google Scholar]
- (31).Yang D; Buchholz F; Huang Z; Goga A; Chen CY; Brodsky FM; Bishop JM Short RNA Duplexes Produced by Hydrolysis With Escherichia Coli RNase III Mediate Effective RNA Interference in Mammalian Cells. Proc. Natl. Acad. Sci. U. S. A. 2002, 99, 9942–9947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Hampel A The Hairpin Ribozyme: Discovery, Two-Dimensional Model, and Development for Gene Therapy. Prog. Nucleic Acid Res. Mol. Biol. 1997, 58, 1–39. [DOI] [PubMed] [Google Scholar]
- (33).Dennis C Small RNAs: the Genome’s Guiding Hand? Nature 2002, 420, 732. [DOI] [PubMed] [Google Scholar]
- (34).Zhou J; Bobbin ML; Burnett JC; Rossi JJ Current Progress of RNA Aptamer-Based Therapeutics. Front. Genet. 2012, 3, 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Guo P; Zhang C; Chen C; Trottier M; Garver K Inter-RNA Interaction of Phage Phi29 pRNA to Form a Hexameric Complex for Viral DNA Transportation. Mol. Cell 1998, 2, 149–155. [DOI] [PubMed] [Google Scholar]
- (36).Afonin KA; Viard M; Tedbury P; Bindewald E; Parlea L; Howington M; Valdman M; Johns-Boehme A; Brainerd C; Freed EO; et al. The Use of Minimal RNA Toeholds to Trigger the Activation of Multiple Functionalities. Nano Lett. 2016, 16, 1746–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Khisamutdinov EF; Jasinski DL; Guo P RNA as a Boiling-Resistant Anionic Polymer Material to Build Robust Structures with Defined Shape and Stoichiometry. ACS Nano 2014, 8, 4771–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Grabow WW; Jaeger L RNA Self-Assembly and RNA Nanotechnology. Acc. Chem. Res. 2014, 47, 1871–1880. [DOI] [PubMed] [Google Scholar]
- (39).Lee JB; Hong J; Bonner DK; Poon Z; Hammond PT Self-Assembled RNA Interference Microsponges for Efficient siRNA Delivery. Nat. Mater. 2012, 11, 316–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Liu J; Guo S; Cinier M; Shlyakhtenko LS; Shu Y; Chen C; Shen G; Guo P Fabrication of Stable and RNase-Resistant RNA Nanoparticles Active in Gearing the Nanomotors for Viral DNA Packaging. ACS Nano 2011, 5, 237–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Binzel DW; Khisamutdinov EF; Guo P Entropy-Driven One-Step Formation of Phi29 pRNA 3WJ From Three RNA Fragments. Biochemistry 2014, 53, 2221–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Binzel DW; Khisamutdinov E; Vieweger M; Ortega J; Li J; Guo P Mechanism of Three-Component Collision to Produce Ultrastable pRNA Three-Way Junction of Phi29 DNA-Packaging Motor by Kinetic Assessment. RNA 2016, 22, 1710–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Afonin KA; Kireeva M; Grabow WW; Kashlev M; Jaeger L; Shapiro BA Co-Transcriptional Assembly of Chemically Modified RNA Nanoparticles Functionalized With siRNAs. Nano Lett. 2012, 12, 5192–5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Boerneke MA; Dibrov SM; Hermann T Crystal-Structure-Guided Design of Self-Assembling RNA Nanotriangles. Angew. Chem., Int. Ed. 2016, 55, 4097–4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Lagoja IM; Herdewijn P Use of RNA in drug design. Expert Opin. Drug Discovery 2007, 2, 889–903. [DOI] [PubMed] [Google Scholar]
- (46).Afonin KA; Kasprzak WK; Bindewald E; Kireeva M; Viard M; Kashlev M; Shapiro BA In Silico Design and Enzymatic Synthesis of Functional RNA Nanoparticles. Acc. Chem. Res. 2014, 47, 1731–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Liu Y; Kuan CT; Mi J; Zhang X; Clary BM; Bigner DD; Sullenger BA Aptamers Selected Against the Unglycosylated EGFRvIII Ectodomain and Delivered Intracellularly Reduce Membrane-Bound EGFRvIII and Induce Apoptosis. Biol. Chem. 2009, 390, 137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Esposito CL; Passaro D; Longobardo I; Condorelli G; Marotta P; Affuso A; de Franciscis V; Cerchia L A Neutralizing RNA Aptamer against EGFR Causes Selective Apoptotic Cell Death. PLoS One 2011, 6, No. e24071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Shu D; Shu Y; Haque F; Abdelmawla S; Guo P Thermodynamically Stable RNA Three-Way Junctions for Constructing Multifuntional Nanoparticles for Delivery of Therapeutics. Nat. Nanotechnol. 2011, 6, 658–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Lee TJ; Haque F; Shu D; Yoo JY; Li H; Yokel RA; Horbinski C; Kim TH; Kim S-H; Kwon C-H; et al. RNA Nanoparticles as a Vector for Targeted siRNA Delivery into Glioblastoma Mouse Model. Oncotarget. 2015, 6, 14766–14776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Cui D; Zhang C; Liu B; Shu Y; Du T; Shu D; Wang K; Dai F; Liu Y; Li C; et al. Regression of Gastric Cancer by Systemic Injection of RNA Nanoparticles Carrying Both Ligand and siRNA. Sci. Rep. 2015, 5, 10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Khisamutdinov EF; Jasinski DL; Li H; Zhang K; Chiu W; Guo P Fabrication of RNA 3D Nanoprism for Loading and Protection of Small RNAs and Model Drugs. Adv. Mater. 2016, 28, 10079–10087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Khisamutdinov E; Li H; Jasinski D; Chen J; Fu J; Guo P Enhancing Immunomodulation on Innate Immunity by Shape Transition Among RNA Triangle, Square, and Pentagon Nanovehicles. Nucleic Acids Res. 2014, 42, 9996–10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Jasinski D; Khisamutdinov EF; Lyubchenko YL; Guo P Physicochemically Tunable Poly-Functionalized RNA Square Architecture with Fluorogenic and Ribozymatic Properties. ACS Nano 2014, 8, 7620–7629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Maslak M; Martin CT Kinetic Analysis of T7 RNA Polymerase Transcription Initiation from Promoters Containing Single-Stranded Regions. Biochemistry 1993, 32, 4281–4285. [DOI] [PubMed] [Google Scholar]
- (56).Chamberlin M; Ring J Characterization of T7-Specific Ribonucleic Acid Polymerase. 1. General Properties of the Enzymatic Reaction and the Template Specificity of the Enzyme. J. Biol. Chem. 1973, 248, 2235–2244. [PubMed] [Google Scholar]
- (57).Chamberlin M; Ring J Characterization of T7-Specific Ribonucleic Acid Polymerase. II. Inhibitors of the Enzyme and Their Application to the Study of the Enzymatic Reaction. J. Biol. Chem. 1973, 248, 2245–2250. [PubMed] [Google Scholar]
- (58).Guo P Rolling Circle Transcription of Tandem siRNA to Generate Spherulitic RNA Nanoparticles for Cell Entry. Mol. Ther.–Nucleic Acids 2012, 1, No. e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Mohsen MG; Kool ET The Discovery of Rolling Circle Amplification and Rolling Circle Transcription. Acc. Chem. Res. 2016, 49, 2540–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Mezger A; Ohrmalm C; Herthnek D; Blomberg J; Nilsson M Detection of Rotavirus Using Padlock Probes and Rolling Circle Amplification. PLoS One 2014, 9, No. e111874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Li X; Zheng F; Ren R Detecting miRNA by Producing RNA: a Sensitive Assay That Combines Rolling-Circle DNA Polymerization and Rolling Circle Transcription. Chem. Commun. (Cambridge, U. K.) 2015, 51, 11976–11979. [DOI] [PubMed] [Google Scholar]
- (62).Roh YH; Deng JZ; Dreaden EC; Park JH; Yun DS; Shopsowitz KE; Hammond PT A Multi-RNAi Microsponge Platform for Simultaneous Controlled Delivery of Multiple Small Interfering RNAs. Angew. Chem., Int. Ed. 2016, 55, 3347–3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Shopsowitz KE; Roh YH; Deng ZJ; Morton SW; Hammond PT RNAi-Microsponges Form Through Self-Assembly of the Organic and Inorganic Products of Transcription. Small 2014, 10, 1623–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Hsu BB; Hagerman SR; Jamieson K; Veselinovic J; O’Neill N; Holler E; Ljubimova JY; Hammond PT Multilayer Films Assembled From Naturally-Derived Materials for Controlled Protein Release. Biomacromolecules 2014, 15, 2049–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Han D; Park Y; Kim H; Lee JB Self-Assembly of Free-Standing RNA Membranes. Nat. Commun. 2014, 5, 4367–4373. [DOI] [PubMed] [Google Scholar]
- (66).Kim H; Park Y; Lee JB Self-Assembled Messenger RNA Nanoparticles (mRNA-NPs) for Efficient Gene Expression. Sci. Rep. 2015, 5, 12737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Daubendiek SL; Kool ET Generation of Catalytic RNAs by Rolling Transcription of Synthetic DNA Nanocircles. Nat. Biotechnol. 1997, 15, 273–277. [DOI] [PubMed] [Google Scholar]
- (68).Diegelman AM; Kool ET Generation of Circular RNAs and Trans-Cleaving Catalytic RNAs by Rolling Transcription of Circular DNA Oligonucleotides Encoding Hairpin Ribozymes. Nucleic Acids Res. 1998, 26, 3235–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Zheng HN; Ma YZ; Xiao SJ Periodical Assembly of Repetitive RNA Sequences Synthesized by Rolling Circle Transcription With Short DNA Staple Strands to RNA-DNA Hybrid Nanowires. Chem. Commun. (Cambridge, U. K.) 2014, 50, 2100–2103. [DOI] [PubMed] [Google Scholar]
- (70).Furukawa K; Abe H; Abe N; Harada M; Tsuneda S; Ito Y Fluorescence Generation From Tandem Repeats of a Malachite Green RNA Aptamer Using Rolling Circle Transcription. Bioorg. Med. Chem. Lett. 2008, 18, 4562–4565. [DOI] [PubMed] [Google Scholar]
- (71).Daubendiek S; Ryan K; Kool E Rolling-Circle RNA Synthesis: Circular Oligonucleotides as Efficient Substrates for T7 RNA Polymerase. J. Am. Chem. Soc. 1995, 117 (29), 7818–7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Afonin KA; Desai R; Viard M; Kireeva ML; Bindewald E; Case CL; Maciag AE; Kasprzak WK; Kim T; Sappe A; et al. Co-Transcriptional Production of RNA-DNA Hybrids for Simultaneous Release of Multiple Split Functionalities. Nucleic Acids Res. 2014, 42, 2085–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Ducani C; Bernardinelli G; Hogberg B Rolling Circle Replication Requires Single-Stranded DNA Binding Protein to Avoid Termination and Production of Double-Stranded DNA. Nucleic Acids Res. 2014, 42, 10596–10604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Wang X; Li C; Gao X; Wang J; Liang X Preparation of Small RNAs Using Rolling Circle Transcription and Site-Specific RNA Disconnection. Mol. Ther.–Nucleic Acids 2015, 4, No. e215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Cech TR RNA Chemistry. Ribozyme Self-Replication? Nature 1989, 339, 507–508. [DOI] [PubMed] [Google Scholar]
- (76).Murray JB; Terwey DP; Maloney L; Karpeisky A; Usman N; Beigelman L; Scott WG The Structural Basis of Hammerhead Ribozyme Self-Cleavage. Cell 1998, 92, 665–673. [DOI] [PubMed] [Google Scholar]
- (77).Ruffner DE; Stormo GD; Uhlenbeck OC Sequence Requirements of the Hammerhead RNA Self-Cleavage Reaction. Biochemistry 1990, 29, 10695–10702. [DOI] [PubMed] [Google Scholar]
- (78).McCall MJ; Hendry P; Jennings PA Minimal Sequence Requirements for Ribozyme Activity. Proc. Natl. Acad. Sci. U. S. A. 1992, 89, 5710–5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Ruffner DE; Dahm SC; Uhlenbeck OC Studies on the Hammerhead RNA Self-Cleaving Domain. Gene 1989, 82, 31–41. [DOI] [PubMed] [Google Scholar]
- (80).Shu D; Khisamutdinov E; Zhang L; Guo P Programmable Folding of Fusion RNA Complex Driven by the 3WJ Motif of Phi29 Motor pRNA. Nucleic Acids Res. 2014, 42, No. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Reif R; Haque F; Guo P Fluorogenic RNA Nanoparticles for Monitoring RNA Folding and Degradation in Real Time in Living Cells. Nucleic Acid Ther. 2012, 22 (6), 428–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Baugh C; Grate D; Wilson C 2.8 A Crystal Structure of the Malachite Green Aptamer. J. Mol. Biol. 2000, 301, 117–128. [DOI] [PubMed] [Google Scholar]
- (83).Blount KF; Uhlenbeck OC The Hammerhead Ribozyme. Biochem. Soc. Trans. 2002, 30, 1119–1122. [DOI] [PubMed] [Google Scholar]
- (84).Lindstrom UM; Chandrasekaran RA; Orbai L; Helquist SA; Miller GP; Oroudjev E; Hansma HG; Kool ET Artificial Human Telomeres From DNA Nanocircle Templates. Proc. Natl. Acad. Sci. U. S. A. 2002, 99, 15953–15958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Hartig JS; Fernandez-Lopez S; Kool ET Guanine-Rich DNA Nanocircles for the Synthesis and Characterization of Long Cytosine-Rich Telomeric DNAs. ChemBioChem 2005, 6, 1458–1462. [DOI] [PubMed] [Google Scholar]
- (86).Jang M; Kim JH; Nam HY; Kwon IC; Ahn HJ Design of a Platform Technology for Systemic Delivery of siRNA to Tumours Using Rolling Circle Transcription. Nat. Commun. 2015, 6, 7930–7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Halman JR; Satterwhite E; Roark B; Chandler M; Viard M ; Ivanina A; Bindewald E; Kasprzak WK; Panigaj M; Bui MN ; et al. Functionally-Interdependent Shape-Switching Nanoparticles With Controllable Properties. Nucleic Acids Res. 2017, 45, 2210–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Afonin KA; Bindewald E; Yaghoubian AJ; Voss N; Jacovetty E; Shapiro BA; Jaeger L In Vitro Assembly of Cubic RNA-Based Scaffolds Designed in Silico. Nat. Nanotechnol 2010, 5, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Geary C; Rothemund PW; Andersen ES RNA Nanostructures. A Single-Stranded Architecture for Cotranscriptional Folding of RNA Nanostructures. Science 2014, 345, 799–804. [DOI] [PubMed] [Google Scholar]
- (90).Collins TJ ImageJ for Microscopy. BioTechniques 2007, 43, S25–S30. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.