Abstract
Aberrant wound healing presents as inappropriate or insufficient tissue formation. Using a model of musculoskeletal injury, we demonstrate that loss of TGF-β activated kinase 1 (TAK1) signaling reduces inappropriate tissue formation (heterotopic ossification) through reduced cellular differentiation. Upon identifying increased proliferation with loss of TAK1 signaling, we considered a regenerative approach to address insufficient tissue production through coordinated inactivation of TAK1 to promote cellular proliferation, followed by re-activation to elicit differentiation and extracellular matrix (ECM) production. While the current regenerative medicine paradigm is centered on the effects of drug treatment (“drug on”), the impact of drug withdrawal (“drug off”) implicit in these regimens are unknown. Because current TAK1 inhibitors are unable to phenocopy genetic Tak1 loss, we introduce the dual-inducible COmbinational Sequential Inversion ENgineering (COSIEN) mouse model. The COSIEN mouse model, which allows us to study the response to targeted drug treatment (“drug on”) and subsequent withdrawal (“drug off”) through genetic modification, was used here to inactivate and re-activate Tak1 with the purpose of augmenting tissue regeneration in a calvarial defect model. Our study reveals the importance of both the “drug on” (Cre-mediated inactivation) and “drug off” (Flp-mediated re-activation) states during regenerative therapy using a mouse model with broad utility to study targeted therapies for disease.
INTRODUCTION
Normal tissue regeneration requires coordination between cellular proliferation and subsequent differentiation. Any disturbance of this coordinated balance leads to pathologic wound healing after injury, as observed in patients with heterotopic ossification (HO). Current state of knowledge on tissue engineering-based approaches for wound regeneration is based on cell transplantation and biomaterials.1–3 Additionally, approaches with small-molecules modify pathways responsible for cellular proliferation/apoptosis4–7 or differentiation8–13, but typically not both.
Heterotopic ossification is a condition in which extra-skeletal bone forms in response to local tissue injury; while HO has primarily been studied in the context of genetic mutations in type I bone morphogenetic protein receptors14–17, it also is known to form in patients after severe trauma without genetic mutations (e.g. trauma-induced HO).14,18–20 Recently, we have shown that trauma-induced HO is caused by pathologic cellular proliferation and subsequent differentiation through a cartilaginous intermediary14,20, prompting us to study transforming growth factor (TGF-β), a known mediator of cartilage formation21–27, in HO. Here we specifically focus on the TGF-β activated kinase 1 (TAK1) signaling pathway23,24,26,28 to coordinate cellular proliferation and differentiation to reduce heterotopic ossification or improve bony calvarial healing.
Transforming growth factor-beta activating kinase 1 (TAK1) is a key regulator of mitogen activated protein kinases (MAPK) kinase activation in TGF-β and BMP signaling pathways.27 In adults, TAK1 has diverse roles spanning inflammation and the immune response, wound healing, fibrosis, and oncogenesis.23,24,26,28–31 During development, TAK1 is critical for the proliferation and maturation of bone, cartilage, skin and vascular endothelium and is a major regulator of the condensation, proliferation, and differentiation of early mesenchymal population. Mechanistically, TAK1 transduces signals to several downstream signaling cascades, including Mitogen-Activated Protein Kinase Kinase 4/7 (MKK4/7)-c-Jun N-terminal kinases (JNK), MKK3/6-p38 MAPK, and Nuclear Factor-kappa β (NF-κB)-inducing kinase (NIK)-IκB kinase (IKK). TAK1 is necessary for propagation of both SMAD-dependent and independent (p38 MAPK) BMP signaling pathways. 23,24,26,28 Furthermore, TAK1 Is central to cytokine-induced activation of NF-κB via interleukin 1-beta (IL-1β).32
To coordinate the TAK1 signaling pathway, we generated a novel genetic mouse model (Cre/Flp mouse) which allows Tak1 to be knocked out using Cre/lox technology (fx) and subsequently re-activated using Flp/Frt technology (frt).32 The resulted COmbinational Sequential Inversion ENgineering (COSIEN) mouse allows us to elucidate a “drug on”/ “drug off” therapeutic paradigm to optimize bone regeneration. Our findings suggest that precise regulation of TAK1 allows for control of the proliferation-differentiation switch in stem cell/progenitor population at the wound site (Sup Fig. 1). Our system demonstrates the potential for therapeutic leverage of a central regulatory protein to affect wound healing in both physiologic and pathologic models.
RESULTS
TAK1 signaling mediates pathologic wound healing after musculoskeletal injury
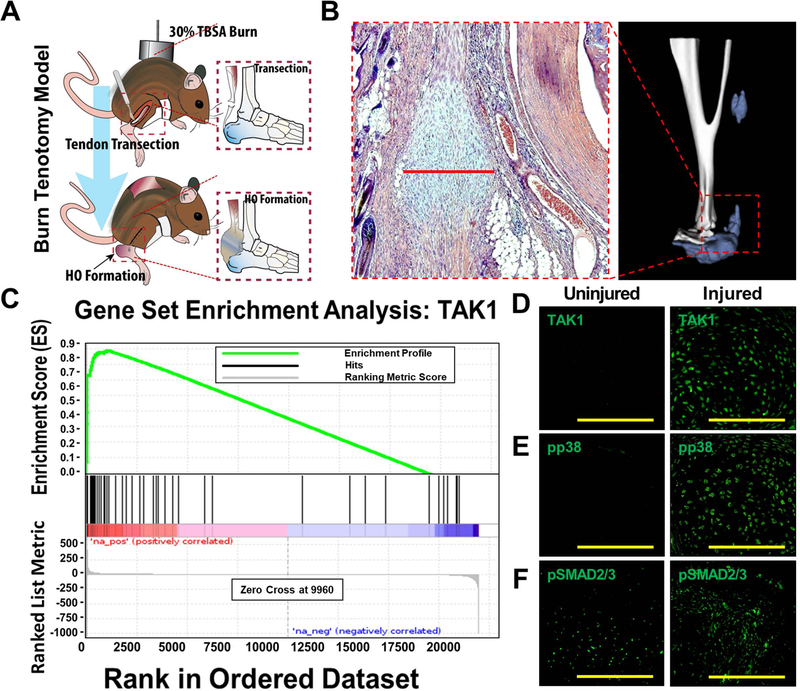
To study aberrant wound healing, we employed a model of trauma-induced heterotopic ossification (tHO) consisting of hindlimb Achilles’ tendon transection with dorsal burn injury (Fig. 1A,B) (16). Gene set enrichment analysis (GSEA) of RNA sequencing data from hindlimb tissue samples obtained from mice three weeks after injury identified up-regulation of the TAK1 pathway when compared with the uninjured hindlimb (Enrichment Score (ES) = 0.88, FDR <0.001) (Fig. 1C). Immunostaining also confirmed up-regulation of TAK1 protein expression at the injury site 3 weeks after injury (Fig. 1D), and importantly up-regulation of phosphorylated proteins downstream in the TAK1 pathway including pp38 and pSMAD 2/3 (Fig. 1E–F). Previous studies have shown that TAK1, an intracellular mediator of TGF-β and bone morphogenetic protein (BMP) signaling, plays a key role in normal postnatal endochondral ossification (24–26), making it a potential target to eliminate HO.
Figure 1. TAK1 signaling mediates pathologic wound healing after musculoskeletal injury.
(A) Mouse model of musculoskeletal injury with hindlimb tenotomy and dorsal 30% total body surface area (TBSA) burn injury – tenotomy at midpoint of Achilles’ tendon; burn ipsilateral to tenotomy. Histologic sections are collected from region between tibial mid-point and calcaneus at the site of highest chondrogenic and osteogenic differentiation; (B) Representative 3D reconstruction (9 weeks) and H&E (3 weeks) demonstrating localization of HO; (C) Gene set enrichment analysis (GSEA) demonstrates up-regulation of TAK1 signaling at the tendon transection site 3 weeks after injury (Enrichment Score (ES) = 0.88, FDR <0.001); (D) Expression of TAK1 in the uninjured and injured hindlimb – expression of TAK1 highest at areas undergoing early chondrogenic differentiation; (E) Expression of pp38 in the uninjured and injured hindlimb – expression of pp38 highest in area undergoing early chondrogenic differentiation; (F) Expression of pSMAD2/3 in the uninjured and injured hindlimb – expression of pSMAD2/3 highest in areas of mesenchymal condensation adjacent to areas of chondrogenic differentiation. Histology at 3 weeks post-injury; 40x magnification. Scale bars = 200 μm.
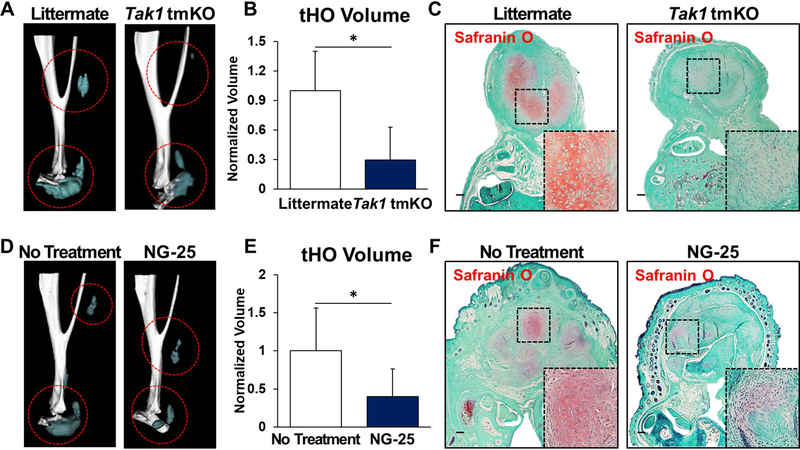
We first developed a mouse allowing targeted de-activation and re-activation of Tak1 (Takfx-frt/fx-frt) (Sup Fig. 2) and confirmed that genetic loss of Tak1 by tamoxifen inducible Ubiqutin-CreER (Ub.CreERT) significantly reduces tHO radiographically and histologically (Fig. 2A–C; Sup Fig. 3A). Systemic treatment of wild type mice with NG-25, a TAK1 inhibitor, which occupies the ATP binding pocket of TAK1 (33), significantly reduced tHO volume (Fig. 2D,E; Sup Fig. 3B) and decreased cartilage presence 3 weeks post-injury (Fig. 2F). Correspondingly, treatment with NG-25 reduced pSMAD 1/5, pSMAD 2/3, and SOX9 expression on immunostaining (Sup Fig. 4A–C).
Figure 2. TAK1 signaling is associated with heterotopic ossification after musculoskeletal injury.
(A) 3D microCT reconstruction of Tak1 tmKO (tamoxifen-inducible postnatal Tak1 knockout (Tak1 tmKO: Ub.CreERT/Tak1fx-frt/fx-frt)) and littermate control hindlimbs showing heterotopic bone 9 weeks after injury (red circles around HO), tamoxifen was injected 7 and 3 days before injury and 3 days after injury; (B) Quantification of heterotopic bone volume in Tak1 tmKO and littermate control hindlimbs showing heterotopic bone 9 weeks after injury (1.0 v. 0.29, p<0.05); (C) Safranin O staining (red stain) in Tak1 tmKO and littermate control hindlimbs showing cartilage 3 weeks after injury(4x magnification; dotted box indicates site of magnified image in right bottom corner); (D) 3-D microCT reconstruction of NG-25 and treatment control hindlimbs showing heterotopic bone 9 weeks after injury; (E) Quantification of heterotopic bone volume in NG-25 and treatment control hindlimbs showing heterotopic bone 9 weeks after injury (1.0 v. 0.35, p<0.05); (F) Safranin O staining (red) in NG-25 and treatment control hindlimbs showing cartilage 3 weeks after injury (4x magnification; dotted box indicates site of magnified image in right bottom corner). All scale bars =200 μm; n≥5 for all quantifications; * = p<0.05.
Genetic loss of Tak1 within mesenchymal cells alters cellular proliferation and differentiation to prevent trauma-induced HO
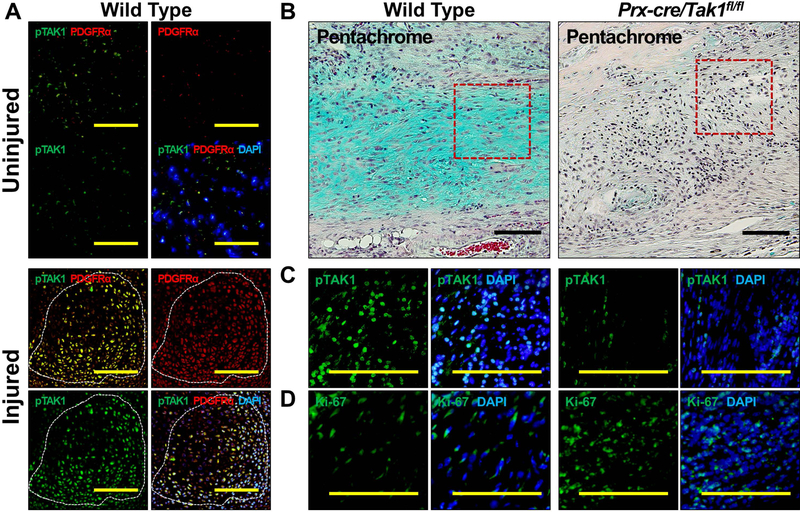
We have previously shown that Prx-Cre expressing cells, mesenchymal progenitors, contribute to tHO (14) induced by hindlimb Achilles’ tendon transection with dorsal burn injury. Because immunostaining of tissue from the injury site indicated that TAK1 signaling is present in MSCs (Fig. 3A) we generated a Tak1 conditional knockout mouse for MSCs (Tak1 cKO; Prxcre/Tak1fx-frt/fx-frt). Histologic analysis demonstrated little-to-no evidence of cartilage, compared to our WT control (Fig. 3B). Immunostaining confirmed reduction of pTAK1 at the injury site of these mice (Fig. 3C). Interestingly, when compared with littermate controls, Tak1 cKO mice had increased cellular proportion of K67+ cells at the injury site (Fig. 3D).
Figure 3. Genetic Loss of TAK1 signaling in mesenchymal cells increases cell proliferation and impairs chondrogenic differentiation to prevent trauma-induced HO.
(A) Co-expression of pTAK1 and PDGFRα+ in the injured and uninjured hindlimb 3 weeks after injury (20x magnification; Top Left Corner: pTAK1 – green overlay with PDGFRα – red; Bottom Left Corner: pTAK1 – green overlay with PDGFRα – red and DAPI - blue); (B) Pentachrome of injury site of Prx-cre/Tak1fx-frt/fx-frt and littermate control mice 3 weeks after injury (10x magnification; Alcian Blue represents cartilaginous tissue; red box shows areas of immunostaining); (C) Immunostaining for pTAK1 at the injury site of Prx-cre/Tak1fx-frt/fx-frt and littermate control mice 3 weeks after injury (10x magnification; Right Side: pTAK1 – green overlay with DAPI - blue); (D) Immunostaining for Ki67 at the injury site of Prx-cre/Tak1fx-frt/fx-frt and littermate control mice 3 weeks after injury (10x magnification; Right Side: Ki67 – green overlay with DAPI - blue). All scale bars = 200 μm.
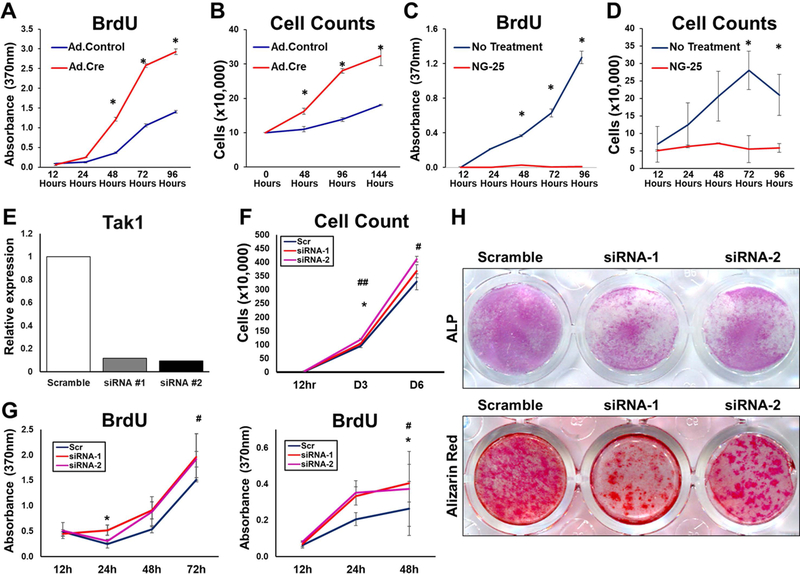
Consistent with these findings and previously published results, MSCs isolated from adipose tissues of Tak1fx-frt/fx-frt mice (Sup Fig. 5A–C) exhibited markedly reduced in vitro osteogenic and chondrogenic differentiation upon treatment with Adenovirus Cre (Ad.Cre) when compared with Adenovirus Control (Ad.LacZ) (Sup Fig. 6A–E). Additionally, Tak1fx-frt/fx-frt MSCs treated with Ad.Cre exhibited increased proliferation when compared with Tak1fx-frt/fx-frt MSCs treated with Ad.LacZ (Fig. 4A,B). NG-25 also reduced osteogenic differentiation and chondrogenic differentiation of MSCs in vitro (Sup Fig. 7A–F). These findings indicate that genetic or pharmacologic loss of TAK1 reduced pathologic wound healing, providing a novel target for therapeutic intervention. In contrast, NG-25-treated MSCs demonstrated reduced proliferation when compared with vehicle control-treated cells (Fig. 4C,D). Similarly, 5Z-7-Oxozeaenol (5Z-O) (33), a potent ATP competitive irreversible inhibitor of TAK1 significantly reduced proliferation when compared with vehicle control-treated cells (Sup Fig. 8A,B). The reduction in cell proliferation observed with currently developed TAK1 inhibitors may be due to their off target effects including inhibition of Abelson tyrosine kinase (ABL), an essential molecule that regulates cell proliferation (33). To test this, we developed a system to silence Tak1 with multiple siRNAs (Fig 4E; Sup Fig. 9A,B). Utilizing these specific siRNAs we found a similar increase in cell proliferation by BrdU assay and cell counting (Fig. 4F,G). Treatment with siRNA, similar to Tak1 knockout with our mouse model, decreased in vitro osteogenic differentiation by alkaline phosphatase and alizarin red (Fig 4H). Overall, these findings suggest that current pharmacologic inhibitors of TAK1 are unable to phenocopy genetic loss of Tak1 in the setting of tissue injury, indicating the need for more specific TAK1 inhibitors.
Figure 4. Comparison of Pharmacologic and Genetic Tak1 Inhibition.
(A) Cell proliferation (BrDU) of Ad.Cre and Ad.LacZ treated Tak1fx-frt/fx-frt adipose-derived stem cells (ASCs); (B) Cell proliferation (Cell counting) of Ad.Cre and Ad.LacZ treated Tak1fx-frt/fx-frt ASCs; (C) Cell proliferation (BrDU) of ASCs treated with NG25; (D) Cell proliferation (Cell counting) of ASCs treated with NG25. (E) ASCs were transfected with siRNAs for TAK1, and TAK1 expression level was analyzed by qPCR. (F) Cell counting showing that siRNAs for TAK1 transfection significantly promote cell proliferation in vitro. (G) BrdU proliferation assay showing that siRNAs for TAK1 transfection significantly promote cell proliferation in vitro (left: ASCs and right: tendon-derived cells (TdCs). (H) osteoblastic differentiation assay showing that siRNAs for TAK1 transfection significantly suppressed the differentiation in vitro (Upper: ALP stained TdCs at day 5 and lower: Alizarin red stained TdCs at day 12). * and # p<0.05; ##p<0.01. Student t-test (*: scramble vs. siRNA-1, #: scramble vs. siRNA-2).
In vitro validation of a novel dual-inducible model to knockout and rescue Tak1 signaling using Combinatorial Sequential Inversion Engineering (COSIEN)
The important finding that loss of Tak1 up-regulates proliferation led us to consider whether this property can be used to improve tissue regeneration. Typically, studies using pharmacologic agents to improve tissue regeneration have focused on the effects of these mediators during the “drug on” period. Interestingly, implicit in studies evaluating regenerative therapies is a period during which drug is not present within the organism – this “drug off” period may be in between doses or following completion of agent administration. The role of the “drug off” period, which proliferating cells may be able to undergo differentiation, has gone largely unrecognized in tissue regeneration. Therefore, because TAK1 is required for cell differentiation and extracellular matrix (ECM) production 23,24,26, we sought to determine whether TAK1 could be modulated to improve regeneration of tissues through a coordinated approach of TAK1 inhibition (proliferation) and subsequent re-activation (differentiation).
Based on our findings, we determined that current pharmacologic inhibitors of TAK1, like many pathway directed therapies, are not specific enough to provide insight into a “drug on”/”drug off” strategy to improve tissue healing. We decided to mimic this approach taking advantage of an important design aspect of the Tak1fx-frt/fx-frt mouse – in particular, this is a novel dual inducible Cre/Flp mouse model allowing for loss of gene function (Cre/lox) followed by rescue (Flp/Frt). We have named this the COmbinational Sequential Inversion ENgineering (COSIEN) mouse, as the targeted gene segment is initially inverted or “gene off” by Cre and then subsequently reinverted or “gene on” by Flp. Mutations were introduced into the Lox and Frt recognition sites to allow only one-time inversion (Cre) and reversion (Flp) of the targeted gene segment (Fig. 5A). This model was then validated genotypically in live animals (Sup Fig. 10A–D). While dual inducible models have been reported before to control temporal knockout of separate genes in neural development and tumor models (34–36), to our knowledge, use of a dual inducible approach to in-activate and subsequently re-activate gene activity has not been described. This ability to optimize gene disruption and reactivation in a timed fashion would allow therapeutic optimization for developmental and post-traumatic pathologies.
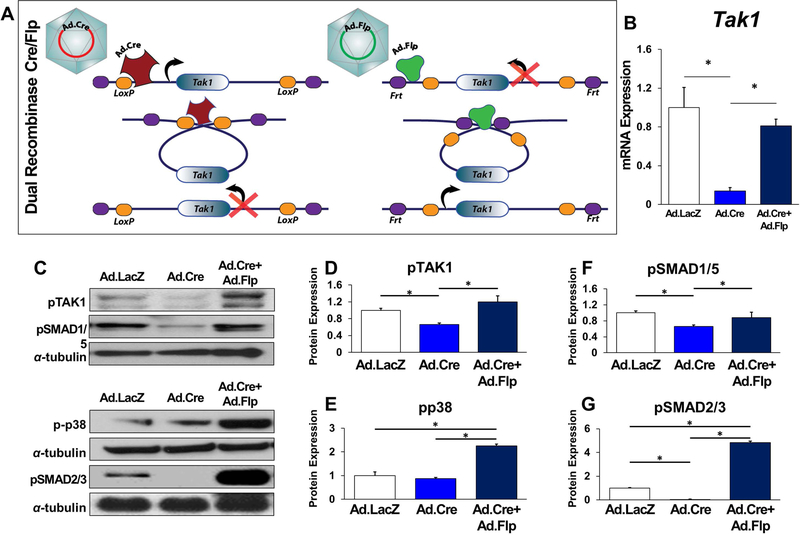
Figure 5. In vitro validation of a dual-inducible model to knockout and rescue Tak1 signaling using COSIEN.
(A) Schematic for Dual-Inducible Model within the Tak1fx-frt/fx-frt mouse: Mutations are introduced into the Lox and Frt sites to allow only one-time inversion and reversion of the targeted gene segment. Initial inversion (loss of gene function) is driven by a Cre/Lox system following addition of Ad.Cre. Rescue reversion (return of gene function) is driven by a Flp/Frt system following addition of Ad.Flp; (B) Normalized quantification of Tak1 gene expression from Ad.LacZ, Ad.Cre, and Ad.Cre+Ad.Flp treated mesenchymal cells (Ad.LacZ: 1.0; Ad.Cre: 0.41; Ad.Cre+Ad.Flp: 0.55). (C) Representative immunoblot of Ad.LacZ, Ad.Cre, and Ad.Cre+Ad.Flp treated mesenchymal cells for pTAK1, pSMAD 1/5, pp38, pSMAD 2/3 and α-tubulin; (D) Normalized quantification of pTAK1 protein expression from Ad.LacZ, Ad.Cre, and Ad.Cre+Ad.Flp treated mesenchymal cells (Ad.LacZ: 1.0; Ad.Cre: 0.66; Ad.Cre+Ad.Flp: 1.2); (E) Normalized quantification of pp38 protein expression from Ad.LacZ, Ad.Cre, and Ad.Cre+Ad.Flp treated mesenchymal cells (Ad.LacZ: 1.0; Ad.Cre: 0.65; Ad.Cre+Ad.Flp: 0.88); (F) Normalized quantification of pSMAD1/5 protein expression from Ad.LacZ, Ad.Cre, and Ad.Cre+Ad.Flp treated mesenchymal cells (Ad.LacZ: 1.0; Ad.Cre: 0.87; Ad.Cre+Ad.Flp: 2.25); (G) Normalized quantification of pSMAD 2/3 protein expression from Ad.LacZ, Ad.Cre, and Ad.Cre+Ad.Flp treated mesenchymal cells (Ad.LacZ: 1.0; Ad.Cre: 0.02; Ad.Cre+Ad.Flp: 4.81); All cells were treated with Ad.Cre (or Ad.LacZ) for 24 hours under serum deprivation conditions followed by 48 hours in serum replete and subsequently treated with Ad.LacZ (Ad.LacZ group), Ad.Cre (Ad.Cre group), or Ad.Flp (Ad.Cre+Ad.Flp) for 24 hours in serum deprived conditions followed by culture for an additional two days in serum replete conditions. Mesenchymal cells described are adipose-derived stem cells (ASCs). * = p<0.05.
MSCs isolated from Tak1fx-frt/fx-frt mice showed reduced Tak1 mRNA expression with Ad.Cre, which was subsequently rescued after Ad.Flp treatment (Fig. 5B). Protein level expression of pTAK1, pp38, pSMAD 1/5, and pSMAD 2/3 were reduced with Ad.Cre but increased with subsequent Ad.Flp treatment (Ad.Cre/Ad.Flp) (Fig. 5C–G). As expected, genetic loss of Tak1 (Ad.Cre) reduced osteogenic differentiation while subsequent rescue of Tak1 (Ad.Cre/Ad.Flp) improved osteogenic differentiation relative to Ad.LacZ treatment (Sup Fig. 11A,B). Taken together, these findings confirm that the COSIEN mouse provides an approach to study “drug on” (Ad.Cre) and “drug off” (Ad.Flp) with specificity for Tak1.
Improved bone regeneration with dual-inducible model to evaluate “drug on” and “drug off” therapeutic strategy targeting TAK1
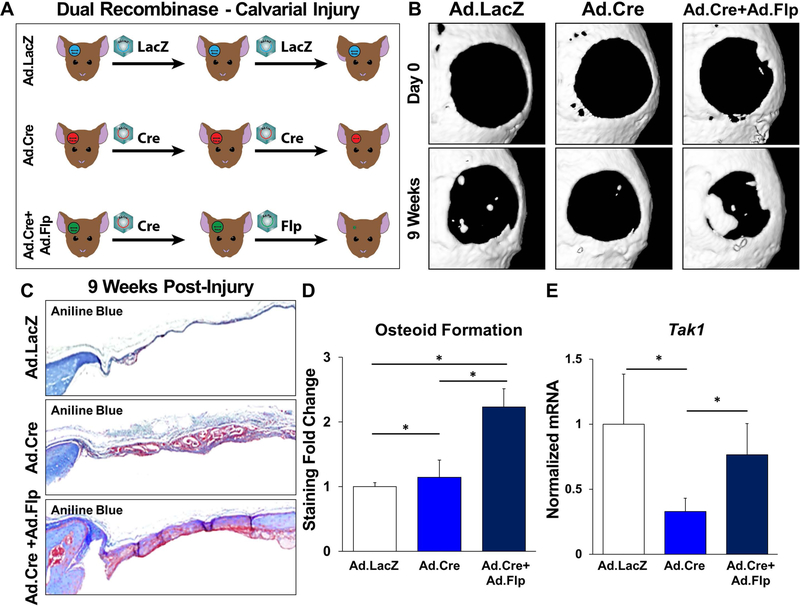
After validating the COSIEN model in vitro, we next studied its effect in vivo using a critical-size calvarial defect model. Calvarial defects are often used for the study of bone healing; while most studies describing critical-size calvarial defects use scaffolds impregnated with MSCs or growth factors, these approaches may be difficult to translate clinically. To validate the COSIEN model in vivo and to demonstrate that a regenerative medicine strategy emphasizing both the “drug on” and “drug off” periods of therapy could improve wound healing, we injected mice with either Ad.LacZ only, Ad.Cre only, or Ad.Cre followed by Ad.Flp (Ad.Cre/Ad.Flp), directly into the calvarial defect area for 9 weeks (Fig. 6A). For the third regimen, Ad.Flp was started to inject from the second week. As expected, mice treated with Ad.Cre/Ad.Flp had significantly more osteoid deposition on the basis of microCT (Fig. 6B) and histologically with aniline blue quantification (Fig. 6C,D). The Ad.Cre group developed similar amount of tissues with the Ad/Cre/Ad.Flp group, but osteoid formation remained similar to the control group (Fig. 6C,D). Calvarial samples obtained from Tak1fx-frt/fx-frt mice showed reduced Tak1 mRNA expression with Ad.Cre, which was subsequently restored after Ad.Flp treatment (Fig. 6E).
Figure 6. Improved regeneration with Cre/Flp dual inducible mouse model to simulate “drug on” and “drug off” therapeutic strategy targeting TAK1 signaling.
(A) Calvarial defect schematic with Ad.LacZ, Ad.Cre, or Ad.Cre/Ad.Flp; (B) Representative microCT scans showing healing of Ad.LacZ, Ad.Cre, and Ad.Cre/Ad.Flp treated calvarial defects at 9 weeks with corresponding baseline scans at day 1; (C) Representative aniline blue staining of the calvarial defect site 9 weeks post-injury (dashed black box marks defect site); (D) Normalized quantification of osteoid in Ad.LacZ, Ad.Cre, and Ad.Cre/Ad.Flp treated calvarial defects 9 weeks after injury (Ad.LacZ: 1.0; Ad.Cre: 1.14; Ad.Cre/Ad.Flp: 2.23); (E) Normalized quantification of Tak1 gene expression from Ad.LacZ, Ad.Cre, and Ad.Cre/Ad.Flp treated calvarial defects (Ad.LacZ: 1.0; Ad.Cre: 0.33; Ad.Cre/Ad.Flp: 0.76). In experimental groups, defects were treated with Ad.Cre every three days starting from the day of surgery until day 12; at day 12, defects were treated with either Ad.Cre (Ad.Cre group) or Ad.Flp (Ad.Cre/Ad.Flp group); in control group, defects were treated with Ad.LacZ every 3 days. Cells for protein extraction collected by harvest of the calvarial defect after removal of dura. * = p<0.05.
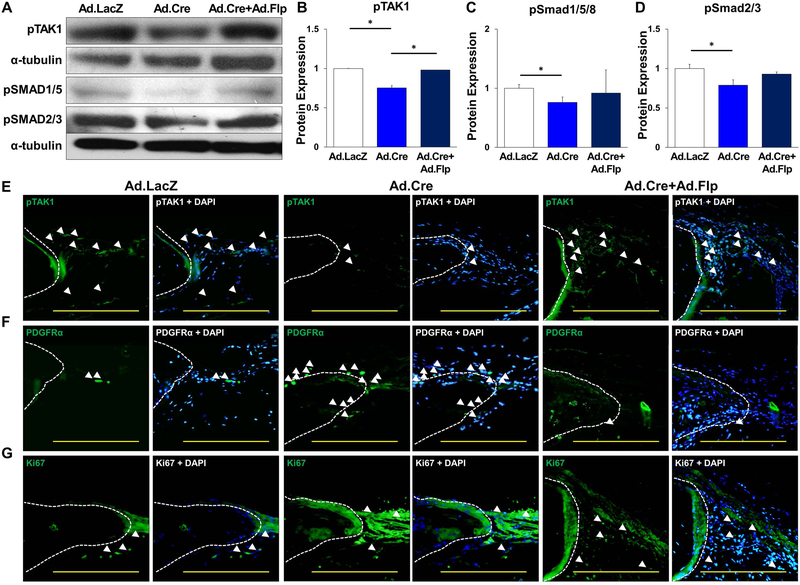
Levels of pTAK1, pSMAD 1/5, and pSMAD 2/3 from these samples were found reduced in the Ad.Cre group but increased with subsequent Ad.Flp treatment (Ad.Cre/Ad.Flp group) (Fig. 7A–D). Immunostaining confirmed a decrease of TAK1 (Ad.Cre) with subsequent rescue of TAK1 (Ad.Cre/Ad.Flp) (Fig. 7E) and also increased presence of MSCs (PDGFRα+) in both the Ad.Cre and Ad.Cre/Ad.Flp groups (Fig. 7F). Immunostaining for pSMAD2/3 was consistent with immunoblot results (Sup Fig. 12). Proliferation observed by Ki67 was increased in the Ad.Cre vs. Ad.LacZ group, with restoration of normal pattern on Ad.Flp administration (Fig. 7G). This was confirmed with immunoblotting for PCNA in protein obtained from Ad.LacZ, Ad.Cre, and Ad.Cre/Ad.Flp treated calvaria (Sup Fig. 13A,B).
Figure 7. Increased cellular proliferation during Tak1 in-activation followed by differentiation during Tak1 reactivation.
(A) Representative immunoblot of Ad.LacZ, Ad.Cre, and Ad.Cre/Ad.Flp treated calvarial defects for pTAK1, pSMAD 1/5, pSMAD 2/3 and α-tubulin; (B) Normalized quantification of pTAK1 protein expression from Ad.LacZ, Ad.Cre, and Ad.Cre/Ad.Flp treated calvarial defects (Ad.LacZ: 1.0; Ad.Cre: 0.75; Ad.Cre/Ad.Flp:0.98); (C) Normalized quantification of pSMAD 1/5 protein expression from Ad.LacZ, Ad.Cre, and Ad.Cre/Ad.Flp treated calvarial defects (Ad.LacZ: 1.0; Ad.Cre: 0.82; Ad.Cre/Ad.Flp: 1.43); (D) Normalized quantification of pSMAD 2/3 protein expression from Ad.LacZ, Ad.Cre, and Ad.Cre/Ad.Flp treated calvarial defects (Ad.LacZ: 1.0; Ad.Cre: 0.82; Ad.Cre/Ad.Flp: 1.43); (E) Representative immunostaining of Ad.LacZ, Ad.Cre, and Ad.Cre/Ad.Flp treated calvarial defects for pTAK1. (F) Representative immunostaining for PDGFRα in Ad.LacZ, Ad.Cre, Ad.Cre/Ad.Flp treated calvarial defects 9 weeks after injury. (G) Representative immunostaining for Ki67 in Ad.LacZ, Ad.Cre, Ad.Cre/Ad.Flp treated calvarial defects 9 weeks after injury. White dotted line marks edge of native calvaria. Cells for protein extraction collected by harvest of the calvarial defect after removal of dura. All scale bars = 200 μm; * = p < 0.05.
DISCUSSION
Trauma-induced HO (tHO) is a clinical phenomenon which occurs in patients following severe trauma, and is characterized by the development of extra-skeletal bone in soft tissues. Our findings indicate that TAK1 can be targeted pharmacologically to prevent tHO. We then use this model of pathologic wound healing to elucidate a critical aspect of therapeutic regimens – the contribution of the “drug on” and the “drug off” state to therapeutic impact. Taken together, our findings using the COSIEN mouse (Tak1fx-frt/fx-frt) suggest that a coordinated “drug on” and “drug off” strategy may be critical for pharmacologic improvement of tissue regeneration, thereby transforming the paradigm of regenerative medicine. Although current studies focus primarily on the “drug on” state, implicit in these treatment strategies is a “drug off” state, which also contributes to healing. The dual inducible approach may allow for validation of gene targets considering both the “drug on” and “drug off” effects prior to pharmacologic development in the context of wound healing.
Patients with tHO are faced with debilitating pain, non-healing wounds, and joint contractures limiting function.17,18,37 We have previously shown that tHO forms via endochondral ossification, and that inhibition of hypoxic signaling reduces tHO through its effects on cartilage production.14 We focused on TAK1 signaling due to the prominent role of TGF-β signaling during normal cartilage development.20–26 Mice with genetic loss of Tak1 driven by the Col2 or Prx promoters (Col2-cre or Prx-cre) exhibit impaired cartilage formation and loss of secondary ossification centers.25 In this study, we found that mice with tamoxifen-inducible systemic loss of Tak1 exhibit significantly reduced levels of tHO, consistent with the role of TAK1 in the development of ossification centers. Because tHO forms through an inflammatory stimulus (e.g. trauma), we subsequently focused on genetic loss of Tak1 only in the mesenchymal cells which contribute directly to the developing ectopic anlagen of tHO using the Prx promoter (Prx-cre)14; indeed, we found that in vivo genetic loss of Tak1 in mesenchymal cells is sufficient to eliminate tHO. Use of the pharmacologic agent NG-25 to inhibit TAK1 confirmed an approach targeting TAK1 to prevent tHO. As expected, in vitro experiments confirmed that genetic loss of Tak1 or treatment with NG-25 reduced chondrogenic differentiation. However, while NG-25 and 5Z-O, another pharmacologic TAK1 inhibitor, reduced mesenchymal cell proliferation in vitro, genetic loss of Tak1 increased cellular proliferation. These findings may be attributed to off-target effects of TAK1 inhibitors such as NG-25 which also inhibit other kinases including epidermal growth factor receptor (EGFR) and ABL.33 These findings also indicate that improved TAK1 inhibitors are required to achieve the therapeutic potential, which is highly encouraged by our genetic data.
Initially, we designed the dual-recombinase mouse model to examine whether a single gene could be targeted to knock out and subsequently reactivate the gene of interest. Several previous studies have combined Cre/lox and Flp/Frt technologies to generate dual-inducible mouse models, these have been used to target different genes 33,34. Therefore, these technologies have been used to temporally control different genes. The LoxP-FRT Trap (LOFT) method has been reported to disrupt a floxed copy of a gene and subsequently re-activate another copy of the same gene that has been inactivated via gene-trap; however, in that model, gene knockout is caused by Cre-induced removal of the floxed copy with “re-activation” of a second copy of the gene which is silent until treated with Flp. Since gene-trap is used to initially inactivate the second copy, genes applicable for this method need to be expressed in embryonic stem cells, and it may be difficult to predict efficiency of inactivation of the gene trapped allele that is known to vary gene to gene. There have been no other reports using the LOFT approach, and no reports of a gene knockout/re-activation strategy for tissue regeneration.35
In this study we invoke a similar principle of gene in-activation and re-activation using Cre/lox and Flp/Frt technologies but with a simpler and more straight-forward targeting strategy (COSIEN technology). Serendipitously, we found that genetic disruption of Tak1 increases proliferation and reduces differentiation, leading us to wonder whether reactivation of Tak1 could rescue cellular differentiation after a period of knockout-induced proliferation. The dual recombinase mouse model allowed us to study this, as pharmacologic inhibition of TAK1 with available agents does not increase proliferation. Though an inducible siRNA strategy can be used to silence and reactivate a gene, there are important distinctions between siRNA and our COSIEN technology: 1) The COSIEN technology completely knocks out gene function in a given cell whereas siRNA has a more limited knockdown. 2) Even though COSIEN recombination efficiency is not perfect, there is a mixture of knockout cells and wild type cells that is advantageous when cell fate specification is in question and proliferative ability of each cell is in question.
Finally, our findings shed light on the importance of considering the effects of target inhibition and subsequent re-activation in the context of wound healing. Current efforts at tissue regeneration are focused on reducing apoptosis and/or increasing proliferation (2–5). However, this strategy does not account for the contribution of non-cellular components to tissue regeneration, primarily extracellular matrix (ECM) which is laid down by cells and forms an integral part of tissue morphology. Other strategies which include cell transplantation or scaffold use face hurdles to long-term incorporation and regulatory clearance. Here we show that an approach including target inhibition and re-activation may allow for a period of proliferation followed by subsequent return of cell differentiation, thereby reconstituting the functional tissue of interest.
MATERIALS AND METHODS
Ethics statement
All animal experiments described were approved by the University Committee on Use and Care of Animals at the University of Michigan-Ann Arbor (Protocols: #05909, 05182, 05716 and 07715). This study was carried out in strict accordance with the recommendations in the Guide for the Use and Care of Laboratory Animals from the Institute for Laboratory Animal Research (ILAR, 2011). All animals were housed in IACUC-supervised facilities, not to exceed five mice housed per cage at 18–22ºC, 12-hour light-dark cycle with ad libitum access to food and water.
Animals
All mice used in this study were derived from a C57BL/6 background. Adult (6–8) week-old wild-type C57BL/6 (Charles River Laboratory) mice were included as controls. Mutant mice used in this study included tamoxifen-inducible postnatal Tak1 knockout (Tak1 tmKO: Ub.CreERT/Tak1fl-frt/fl-frt), conditional Tak1 knockout (Tak1 cKO: Prx.Cre/Tak1fl-frt/fl-frt), dual inducible Tak1 knockout (Tak1fl-frt/fl-frt), and their respective littermate controls. All breeding was performed at the University of Michigan in facilities managed by the Unit for Laboratory Animal Medicine (ULAM). Tail genomic DNA was used for genotyping.
Generation of dual inducible COmbinational Sequential Inversion ENgineering (COSIEN) mouse for inactivation and re-activation of Tak1
Exon 2 of Tak1 was targeted because removal of exon 2 has been shown to be sufficient to disrupt gene function(30). A 4.5-kb fragment containing intron 1 of Tak1 locus was PCR amplified from 129SvEv genomic DNA with Phusion polymerase (New England Biolabs, Inc. MA). A 2.8-kb fragment containing exon 2 and 3.6-kb fragment containing intron 2 were PCR amplified. After amplification, these fragments were ligated with mutant loxP sites, mutant FRT sites, a PGK-Neo cassette, and DTA cassette to generate a targeting vector (Sup Fig. 1,2). The positions of the probes used for Southern analysis and positions of PCR genotyping primers are shown. The sizes of the restriction fragments detected by these probes in WT and targeted DNA are shown above or below the locus. A 5’ and 3’ loxP–FRT sites are marked with an EcoRV and a HindIII sites, respectively.
Since both loxP sites and FRT sites are placed in the locus in an opposite direction, recombinase-mediated DNA recombination flips the sequence between instead of its deletion. To avoid continuous flipping and allow for one-time recombination, we introduced mutations in each recognition site. Lox66 has mutations in 5 bases at the most 3’ region whereas Lox72 has mutations in the most 5’ region (38). FRT GS1–1 has a single base change in 5’ arm whereas FRT GS2–1 has a single base change in 3’ arm (39). Cre-mediated DNA recombination flips the sequence between Lox66 and Lox72, but after recombination, one of the resulted LoxP sequences bears mutations in both 5’ and 3’ arms, which no longer can be a substrate for Cre recombinase. Thus, exon 2 will be flipped by Cre recombinase only one time and gene function will be lost (Sup Fig. 2). Flippase-mediated DNA recombination flips the sequence between FRT GS1–1 and FRT GS1–2. After recombination, one of the resulted FRT sequences bears mutations in both 5’ and 3’ arms to stop further recombination. Thus, exon 2 will be flip back and gene function will be restored (Sup Fig. 2). We named this system as COmbinational Sequential Inversion ENgineering (COSIEN).
Linearized targeting vector was electroporated into 1.6 × 107 clones A3 of UG347 ES cells, which we established from 129SvEv blastocysts. Three hundred G418-resistant ES cell clones were initially screened by Southern blot and targeted ES cell clones were identified (Sup Fig. 10A). The targeted ES clones were injected into blastocysts from C57BL/6 albino mice. The resulting chimeras were bred to C57BL/6 females and F1 agouti offspring were genotyped by Southern analyses. Three targeted clones were used for injection and one of them underwent germline transmission. Subsequently, mice heterozygous for Tak1 floxed-FRTed allele (Tak1fl-frt/wt) (Sup Fig. 10B) were intercrossed to obtain homozygous mice for Tak1 floxed-FRTed allele (Tak1fl-frt/fl-frt). The homozygous mice were obtained as an expected ratio (25%, n>100), suggesting that presence of the loxP, FRT and the neo cassette does not influence gene activity.
When bred with a germ line deleter Cre strain Meox2-Cre, all mice positive for the Cre showed a flipped band by genomic PCR (Sup Fig. 10C). After segregation of Cre from the flipped Tak1 allele (designated as Tak1fc allele, Tak1fc/+ mice were bred with Meox2-Cre mice again. None of the Tak1fc/+ mice carrying Cre showed the floxed band (200bp) with Tak1 G1/G2 primers indicating that the Cre-mediate DNA inversion occurs only one time (n>20, data not shown). Homozygous mice for Tak1 Cre-flipped allele (Tak1fc/fc) were generated by intercross of Tak1fc/+ mice and resulted homozygous mice showed embryonic lethality around E9.5 similar to the Tak1 homozygous null embryos reported earlier (data not shown). These suggesting that the Tak1 floxed-FRTed allele can flip one time with Cre recombinase to disrupt gene function.
When bred with Flipper mice (carrying FLPe gene) (32), mice carrying both the Tak1fx-frt alleles and Flpe showed a flipped band by genomic PCR using TAK1 F6/G2 primers (Sup Fig. 10D). Unlike the case of Meox2-Cre, we found some of them showed both floxed and flipped bands suggesting that DNA inversion mediated by FLPe may be less efficient than that by Cre (Sup Fig. 10D, sample #4).
Injury models
All mice received pre-surgical analgesia consisting of 0.1 mg/kg buprenorphine, followed by anesthesia with inhaled isoflurane, and close post-operative monitoring with analgesic administration. Experimental trauma model 1: Burn/tenotomy (B/T) mice received a 30% total body surface area (TBSA) partial-thickness burn on the shaved dorsum followed by left hindlimb Achilles’ tendon transection(16). The dorsum was burned using a metal block heated to 60°C and applied to the dorsum for 18 seconds continuously. The tenotomy site was closed with a single 5–0 vicryl suture placed through the skin only. Ub.Cre/Tak1fl-frt/fl-frt and littermate control mice received 175 mg/kg tamoxifen 7 and 3 days prior the surgery and 7, 14, 21 days after the B/T surgery. Experimental trauma model 2: Critical-sized (4-mm) calvarial defects were created in Tak1fl-frt/fl-frt mice to assess TAK1 in bone healing with local injection of either 1) Control adenovirus (Ad.control; 9 × 1010 PFU/injection site for 9 weeks); 2) Cre adenovirus (Ad.cre; 1.6 × 1011 PFU/injection site for 9 weeks), or 3) Cre/Flp (1 × 1010 PFU/injection site ) virus (Ad.cre for 1 week followed by re-constitution of TAK1 expression with Ad.Flp for 8 weeks). Respective adenoviruses were injected into the calvarial defects.
In vivo drug treatment: NG25, TAK1 inhibitor
C57BL/6 mice underwent burn/tenotomy as described above. Following injury mice received either PBS vehicle control or TAK1 inhibitor (2 mg/kg) in 500 μL via intraperitoneal injection. Mice in both groups were euthanized at 3-, and 9-weeks after injury for further analysis. Each group had n≥3 animals.
Cell harvest
Mesenchymal cells, local tissue, and osteoblasts from Tak1fl-frt/fl-frt, and corresponding littermate controls were harvested from: a) the inguinal fat pad (adipose-derived stem cells (ASCs), b) from the Achilles’ tendon (tendon-derived cells; TdCs), and c) from femur, tibia, and fibula (osteoblasts). All tissue was mechanically minced, digested with collagenase A and dispase. Cells were separated via 100 μm cell strainer and digestive enzyme were quenched in standard growth medium (DMEM supplemented with 10% FBS and 1% penicillin/ streptomycin). Cells were spun down at 1000 rpm for 5 minutes. The supernatant was discarded, and the cell pellet was resuspended in standard growth media and subsequently plated. Isolation of adipose and bone-marrow derived mesenchymal cells in this manner has previously been validated as demonstrating tri-lineage differentiation consistent with mesenchymal stem cells.36–39
Cell culture and transfection
Cells were grown in standard growth medium (DMEM supplemented with 10% FBS and 1% penicillin/ streptomycin). Cells used were all passage 2 through 6. Tak1fl-frt/fl-frt cells were treated with either Ad.LacZ (MOI 500), Ad. Cre (MOI 500) in DMEM free of FBS (serum deprived) for 24 hours. Cells were then cultured in standard growth media (serum-replete) for 48 hours. Subsequent tranfection was then performed with either Ad.LacZ, Ad.Cre or Ad. FLP (MOI 500). As with the initial transfection, 2nd transfection was performed in DMEM free of FBS for 24 hours before being transitioned to standard growth media for 48 hours. At this point cells were ready for RNA/protein harvest or for use in proliferation and differentiation assays.
Flow Cytometric Preparation and Confirmation of Purity
Adipose-derivced stem cells were harvested as above and suspended in Hanks Balanced Salt Solution (HBSS) prior to filtration through a 70-micron sterile strainer and centrifuged at 800 rpm for 5 minutes before removing the supernatant and washing in HBSS. This process was repeated three times before incubation with fluorescently labeled antibodies. AmCyan viability dye used as a marker to gate for live vs. dead cells. Lineage defined by the following myeloid markers CD45-PE; MHCII-PE; B220-PE; CD11b-FITC; CD34-FITC (eBioscience, Thermo Fisher). Following 1 hour of incubation at 4°C, sample were washed and filtered through a 45-micron mesh filter before being run on a FACSAria II (BD Biosciences) Cell Sorter at the University of Michigan Flow Cytometry Core in the Biomedical Science Research Center. Samples were gated to separate debris and autofluorescent signals from the cell population. Data were then analyzed using the FlowJo software (TreeStar). Flow cytometric data was normalized to account for differences in aggregate number between cell types.
siRNA treatment
To generate TAK1 knockdown cells, ASCs and tendon-derived cells (TdCs) were transfected with siRNA (s77092, s77094 and negative control No.1, Ambion) using Lipofectamine RNAiMAX transfection reagent (Thermo). For cell proliferation assays, siRNAs were transfected when cells were plated after 3–4 hr and the medium was changed. For cell differentiation assays, siRNAs were transfected when the medium was changed to ODM and every two days.
Proliferation assays
Cells were seeded in 12-well plates at a density of 5×103 cells per well (n = 3). Cells were grown in standard growth medium (DMEM supplemented with 10% FBS and 1% penicillin/ streptomycin). Treatment groups had their media supplemented as follows: NG25) 4μM NG25/DMSO in DMEM; 5Z-O) 1μM 5Z-O/DMSO in DMEM. Media changed every 3 days. At 12, 24, 48, 72, and 96 or 144 hours cells were lifted following trypsin-EDTA treatment and were manually enumerated using Trypan blue stain and a hemocytometer. Additionally, cell proliferation was assessed by bromodeoxyuridine (BrdU) incorporation.
Differentiation assays
Cells were seeded in 12-well plates at a density of 3×103 cells per well (n = 3). Prior to differentiation cells were maintained in standard growth media (DMEM supplemented with 10% FBS and 1% penicillin/streptomycin). Differentiation was performed in Osteogenic differentiation media (ODM: DMEM supplemented with 10% FBS, 1% penicillin/streptomycin, 10 mM β-glycerophosphate, 100 μg/ml ascorbic acid). ODM media in isolation was used for control groups. The NG25 test group was treated with 4μM NG25/DMSO in ODM. Differentiation media were changed every 3 days. Cells for early RNA or protein quantification were collected after 3 days of differentiation. Early functional osteogenic differentiation was assessed by alkaline phosphatase (ALP) stain and quantification of ALP enzymatic activity after 7 days. Alizarin red staining for bone mineral deposition and colorimetric quantification was completed at 14 days.
MicroCT analysis
MicroCT scans (Siemens Inveon using 80 kVp, 80 mA, and 1,100 ms exposure) were used to quantify:1) heterotopic ossification volume in mice with burn/tenotomy. Images were reconstructed and HO volume quantified using a calibrated imaging protocol as previously described with the MicroView μCT viewer (Parallax Innovations, Ilderton, Canada)(14). The calculation of the threshold for regenerating calvarial bone was performed in MicroView and determined equivalent to 800 Houndsfield Units. Percentage healing on the parietal bone containing the defect was determined by dividing the rest-defect area by the mean of the defect size at day 1 post surgery. TAK1 mice were scanned at 24 hour and 9 weeks post surgery.
Preparation of tissue for histology
Histologic evaluation was performed at indicated time points in hind limbs of burn/tenotomy mice (Wild type, tmKO and respective littermate controls) and calvarial defects of Tak1fl-frt/fl-frt mice. Hind limbs and calvaria were fixed in formalin overnight at 4°C and subsequently decalcified in 19% EDTA solution for 3–5 weeks at 4°C until X-ray verification of decalcification. Hindlimbs were embedded in paraffin, and 5–7 μm sections were cut and mounted on Superfrost Plus slides (Fisherbrand, Hampton, NH) and stored at room temperature.
Histology and immunostaining
Hematoxylin/eosin and Movat’s pentachrome staining were performed of the ankle region and calvaria, respectively. Immunostaining of extra-skeletal ectopic bone was performed on rehydrated wax sections with the following primary antibodies: rabbit anti-PDGFR (antibody sc-338, Santa Cruz Biotechnology), mouse anti-PDGFR (antibody sc-398206, Santa Cruz Biotechnology) goat anti-Sox9 (antibody sc-17341, Santa Cruz Biotechnology), rabbit anti-Ki67 (antibody AB9260, Millipore), goat anti-mouse anti-pSmad1/5 (antibody sc-12353, Santa Cruz Biotechnology), polyclonal rabbit anti-TAK1 (NB100–56363, Novus Biologicals, Littleton, CO, USA), rabbit anti-pTAK1 (antibody sc-4508, Cell Signaling), goat anti-pSmad1/5/8 (antibody sc-12353, Santa Cruz Biotechnology), rabbit anti-pSmad 2/3 (antibody sc-11769 Santa Cruz Biotechnology), rabbit anti-pTAK1 (antibody NBP1–9609, Novus Biologicals, Littleton, CO, USA), rabbit anti-pp38 (antibody sc-9211, Cell Signaling). Appropriate dilutions were determined before achieving final images. The appropriate fluorescent secondary antibody was applied and visualized using fluorescent microscopy. Secondary antibodies consisted of anti-rabbit or anti-goat Alexafluor-488 (green) or Alexafluor-594 (red)(A21206, A11055, A21207, A11058, Life technologies).
Quantification of calvarial defect healing
Histomorphometric measurements to quantify the area of regenerate bone were performed using Image J on every 10th Aniline Blue stained slide of the defect (Ad.LacZ: 6 defects, 118 total images; Ad.Cre: 6 defects, 120 total images; Ad.Cre/Ad.Flp: 8 defects, 120 total images). Regenerative bone was manually selected and isolated from each section and the area calculated using the measure function on ImageJ. For each defect, measured areas were summed to estimate total new bone formation
Microscopy
All fluorescently stained images were taken using an Olympus BX-51 upright light microscope equipped with standard DAPI, 488 nm, and TRITC cubes attached to an Olympus DP-70 high resolution digital camera. Each site was imaged in all channels and overlaid in DPViewer before examination in Adobe Photoshop. H&E, safranin O, pentachrome, and aniline blue sections were imaged at 10x and 20x magnification. Immunofluorescent images were taken at either 20x or 40x magnification. Immunocytochemical images were taken at 60x and 100x magnification under oil. Scale bars were placed for all images with a standard 200 μm diameter.
Western blot analysis
Tissue/cells were lysed with RIPA lysis buffer (Santa Cruz Biotechnology, Dallas, TX) containing protease inhibitors, 1 nM sodium orthovanadate, 1 mM PMSF. The protein concentration was determined using the BCA Plus protein assay kit (Pierce, Rockford, IL, USA). SDS-PAGE was used to separate the protein extract (40 μg). After transfer to a polyvinylidene fluoride (PVDF) membrane (EMD, Millipore, Darmstadt, Germany), and blocking with 5% milk in TBS with 0.1% Tween-20 (TBST) for 1 hour, then incubated overnight with the following antibodies at 4°C: rabbit anti-pTak1 (antibody sc-9339, Cell Signaling), rabbit anti-TAK1 (NB100–56363, Novus Biologicals, Littleton, CO, USA), rabbit anti-pSmad1/5/8 (antibody 9516, Cell signaling), rabbit anti-Smad2/3 (antibody sc-3102, Cell Signaling), rabbit anti-pp38 (antibody 9211, Cell Signaling), rabbit anti-PCNA (antibody 2586, Cell Signaling), and rabbit anti-α-tubulin (antibody 2144, Cell Signaling). After washing with TBST 5 times, the membrane was incubated with appropriate Horseradish peroxidase (HRP)-conjugated secondary antibody (antibody 7074, Cell Signaling) for 30 minutes at room temperature and detected using chemiluminescence PICO substrate (Pierce, Rockford, IL, USA).
Gene analysis
To assess the recombination efficiency of the floxed Tak1 locus (Exon 2) and gene expression in the total RNAs were isolated from AdMSCs transfected with Ad.LacZ, Ad.Cre, Ad.FLP, and Ad.Cre/Ad.FLP (RNeasy mini kit; Qiagen, Germantown, MD) per manufacturer’s specifications, and 1 μg RNA using High capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA) according to manufacturer’s protocols. Quantitative real-time PCR was carried out using the Applied Biosystems Prism 7900HT Sequence Detection System and SybrGreen PCR Master Mix (Applied Biosystems, Foster City, CA). Specific primers for these genes were:
| Tak1 | Forward: | GGTTGTCGGAAGAGGAGCTTTT |
| Tak1 | Reverse: | AACTGCCGGAGCTCCACAAT |
| Gapdh | Forward: | TCTCCTGCGACTTCAACAGCAA |
| Gapdh | Reverse: | CCCACATACCAGGAAATGAGCTTG |
| Alp | Forward: | TCTGCCTTGCCTGTATCTGGAATC |
| Alp | Reverse: | GTGCTTTGGGAATCTGTGCAGTCT |
| Runx2 | Forward: | CACCGAGACCAACCGAGTCATTTA |
| Runx2 | Reverse: | AAGAGGCTGTTTGACGCCATAG |
| Sox9 | Forward: | GGAGGAAGTCGGTGAAGAAC |
| Sox9 | Reverse: | AGCGCCTTGAAGATAGCATT |
The PCR protocol included a 95°C denaturation (20s), annealing (20s), and 72°C extension (30s). Detection of the fluorescent product was carried out at the end of the 72°C extension period. Each sample was tested at least in triplicate and repeated for three independent cell/tissue preparations.
Statistical analysis
Means and SDs were calculated from numerical data, as presented in the text, figures, and figure legends. In figures, bar graphs represent mean, whereas error bars represent one SD. Statistical analysis was performed using a Student’s t test to directly compare two groups. p-values are included in figure legends.
Supplementary Material
Supp Fig 1. Tak1 expression during wound regeneration
(A) A schematic expression profile of proposed Tak1 expression during wound regeneration. Tak1 expression is lower at the beginning when undifferentiated cells proliferate and subsequently becomes higher when cells differentiate with reduced proliferation.
Supp Fig 2. Design of the COmbinatorial Sequential Inversion ENgineering (COSIEN) mouse targeting Tak1 Exon 2.
Design of the floxed-FRTed allele for Tak1. Exon 2 of Tak1 is flanked by mutant loxP and FRT sites, respectively, with reverse orientation to invert one time with Cre or Flippase. Approximate positions of Southern probes and PCR primers for genotyping are shown. H, HindIII, RV, EcoRV.
Supp Fig 3. microCT cross sections of HO after trauma with TAK1 inhibition.
(A) Representative microCT cross-sections from the mid-tibia and calcaneus of Tak1 tmKO (tamoxifen-inducible postnatal Tak1 knockout (Tak1 tmKO: Ub.CreERT/Tak1fx-frt/fx-frt)) and littermate control hindlimbs showing heterotopic bone 9 weeks after injury, tamoxifen was injected 7 and 3 days before injury and 3 days after injury; (B) Representative microCT cross-sections from the mid-tibia and calcaneus of NG-25 and treatment control hindlimbs showing heterotopic bone 9 weeks after injury. Red lines and shading indicate areas of HO.
Supp Fig 4. Diminished osteogenic and chondrogenic signaling at the injury site with pharmacologic inhibition of TAK1 with NG-25.
(A) Representative pSMAD 1/5 immunostaining of untreated and NG-25 treated mice 3 weeks after injury; (B) Representative pSMAD 2/3 immunostaining of untreated and NG-25 treated mice 3 weeks after injury; (C) Representative Sox9 immunostaining of untreated and NG-25 treated mice 3 weeks after injury. Mice were treated with daily NG25 (2mg/kg) in PBS solution via IP injection for 3 weeks. Scale bars = 100 μm
Supp Fig 5. Confirmation of Purity of Adipose-Derived Mesenchmyal Stem Cells (ASCs)
(A) Flow gate schematic demonstrating isolation of single cell populations; (B) Flow gate schematic demonstrating isolation of viable cells (AmCyan Negative) and purity analysis. Lineage Negative (PE – CD45, MHCII, B220; FITC – CD11b, CD34); (C) Frequency of lineage negative ASCs vs. Isotype controls. Mesenchymal cells described are adipose-derived stem cells (ASCs). AmCyan viability dye used as a marker to gate for live vs. dead cells. Lineage defined by the following myeloid markers CD45-PE; MHCII-PE; B220-PE; CD11b-FITC; CD34-FITC.
Supp Fig 6. Diminished osteogenic and chondrogenic differentiation with loss of Tak1.
(A) Representative ALP stain of Ad.LacZ and Ad.Cre treated Tak1fl/fl mesenchymal cells; (B) Normalized quantification of Alp gene expression from Ad.LacZ and Ad.Cre treated Tak1fl/fl mesenchymal cells (Ad.LacZ: 1.0; Ad.Cre: 0.05); (C) Representative Alizarin Red stain of Ad.LacZ and Ad.Cre treated Tak1fl/fl mesenchymal cells; (D) Normalized quantification of Runx2 gene expression from Ad.LacZ and Ad.Cre treated Tak1fl/fl mesenchymal cells (Ad.LacZ: 1.0; Ad.Cre: 0.32); (E) Normalized quantification of Sox9 gene expression from Ad.LacZ and Ad.Cre treated Tak1fl/fl mesenchymal cells (Ad.LacZ: 1.0; Ad.Cre: 0.46). AR = Alizarin red; n≥3 for all quantification. Mesenchymal cells described are adipose-derived stem cells (ASCs). For differentiation assay, all ASCs were treated with 4uM NG25/DMSO in ODM, changed every 3 days prior to differentiation (7 days for ALP, 14 days for AR, 3 days for RNA collection). *p<0.05.
Supp Fig 7. Pharmacologic inhibition of TAK1 with NG-25 reduces osteogenic and chondrogenic differentiation.
(A) Representative ALP stain of Vehicle Control and NG-25 treated mesenchymal cells; (B) Normalized quantification of Alp gene expression from Vehicle Control and NG-25 treated mesenchymal cells (Vehicle Control: 1.0; NG-25: 0.26); (C) Representative Alizarin Red stain of Vehicle Control and NG-25 treated mesenchymal cells; (D) Normalized quantification of Runx2 gene expression from Vehicle Control and NG-25 treated mesenchymal cells (Vehicle Control: 1.0; NG-25: 0.12); (E) Representative Alcian Blue stain of Vehicle Control and NG-25 treated mesenchymal cells (F) Normalized quantification of Sox9 gene expression from Vehicle Control and NG-25 treated mesenchymal cells (Vehicle Control: 1.0; NG-25: 0.16). ALP = alkaline phosphatase; AR = Alizarin red; n≥3 for all quantification; AB = Alcian blue; All normalization performed to Vehicle Control group. Mesenchymal cells described are adipose-derived stem cells (ASCs). For differentiation assay, all ASCs were treated with 4uM NG25/DMSO in ODM, changed every 3 days prior to differentiation (7 days for ALP, 14 days for AR, 3 days for RNA collection). *p<0.05.
Supp Fig 8. In vitro proliferation with pharmacologic inhibition of TAK1 using 5Z-7-Oxozeaenol (5Z-O). (A) Cell proliferation (BrDU) of 5Z-O and vehicle treated mesenchymal cells; (B) Cell proliferation (Cell counting) of 5Z-O and vehicle treated mesenchymal cells. Mesenchymal cells described are adipose-derived stem cells (ASCs). For differentiation assay, all ASCs were treated with 1μM 5Z-O/DMSO in DMEM, changed every 3 days prior to differentiation (7 days for ALP, 14 days for AR, 3 days for RNA collection). *p<0.05.
Supp Fig 9. siRNA targeted for Tak1 at separate exons effectively decreases the expression of Tak1 in multiple cell lines. (A) Schematic demonstrating the targeting of siRNA against specific sites on the Tak1 gene. (B) Decrease in the relative expression of Tak1 between a control scramble siRNA and two siRNAs targeting the Tak1 gene in 3 different cell lines. β-actin used as internal control. ASCs – Adipose-derived stem cells; TdCs – Tendon-derived cells; Obs – Osteoblasts.
Supp Fig 10. Genetic validation of COSIEN mouse model for Tak1.
(A) Identification of targeted clones for the Tak1fl-frt/wt allele by genomic Southern blot using designated restriction endonucleases; (B) Intercrossing Tak1fl-frt/wt mice to generate Tak1fl-frt/fl-frt mice (W, Takfwt/wt, H, Tak1fl-frt/wt, m, Tak1fl-frt/fl-frt); (C) Genotyping of mice from Tak1fl-frt/wt x Meox2-Cre breeding strategy showing efficient flipping of the Tak1fl-frt allele (samples 1,2,5, positive for Meox2-Cre), persistence of the unrecombined floxed allele in mice negative for Meox2-Cre (samples 3,4,6,7,) Wild type littermates for Tak1 are also shown (samples 8,9); (D) Genotyping of mice from Tak1fl-frt/wt x FLPe breeding strategy showing efficient flipping of the Tak1fl-frt allele (samples 4,5,7,8, white asterisks, positive for Flpe), persistence of the floxed allele in mice negative for FLPe (sample 6). Wild type littermates for Tak1 are also shown (samples 1,2,3,9). Sample #4 shows mosaicism of the floxed and flipped alleles.
Supp Fig 11. In vitro differentiation studies using a dual-inducible model to knockout and rescue Tak1 signaling using COSIEN.
(A) Representative ALP stain of Ad.LacZ, Ad.Cre, and Ad.Cre+Ad.Flp treated mesenchymal cells undergoing osteogenic differentiation with quantification (Ad.LacZ: 1.0; Ad.Cre: 0.34; Ad.Cre+Ad.Flp: 0.60); (B) Representative Alizarin red of Ad.LacZ, Ad.Cre, and Ad.Cre+Ad.Flp treated mesenchymal cells undergoing differentiation with quantification (Ad.LacZ: 1.0; Ad.Cre: 0.31; Ad.Cre+Ad.Flp: 0.75). All cells were treated with Ad.Cre (or Ad.LacZ) for 24 hours under serum deprivation conditions followed by 48 hours in serum replete and subsequently treated with Ad.LacZ (Ad.LacZ group), Ad.Cre (Ad.Cre group), or Ad.Flp (Ad.Cre+Ad.Flp) for 24 hours in serum deprived conditions followed by culture for an additional two days in serum replete conditions. Mesenchymal cells described are adipose-derived stem cells (ASCs). * = p<0.05.
Supp Fig 12. pSMAD 2/3 expression in calvarial defects during Tak1 in-activation followed by differentiation during Tak1 reactivation
Representative immunostaining of Ad.LacZ, Ad.Cre, and Ad.Cre/Ad.Flp treated calvarial defects for pSMAD 2/3. White dotted line marks edge of native calvaria. All scale bars = 200 μm.
Supp Fig 13. PCNA in calvarial defects during Tak1 in-activation followed by differentiation during Tak1 reactivation
(A) Representative immunoblot of Ad.LacZ, Ad.Cre, and Ad.Cre/Ad.Flp treated calvarial defects for PCNA and α-tubulin; (B) Normalized quantification of PCNA protein expression from Ad.LacZ, Ad.Cre, and Ad.Cre/Ad.Flp treated calvarial defects (Ad.LacZ: 1.0; Ad.Cre: 2.34; Ad.Cre/Ad.Flp:1.26). Cells for protein extraction collected by harvest of the calvarial defect after removal of dura. * = p < 0.05.
SIGNIFICANCE STATEMENT.
We target the TAK1 pathway to reduce heterotopic ossification, a pathologic condition in which bone develops within muscle or soft tissues. We show that Tak1 knockout leads to cellular proliferation; this can be harnessed to increase the number of cells present at the injury site. Using a mouse model, we inactivate and reactivate the Tak1 gene. We show that inactivation and reactivation of Tak1 can improve bony healing through the coordination of increased proliferation (inactivation) followed by differentiation (reactivation). This approach elucidates a new paradigm in regenerative medicine in which coordination between treatment and withdrawal of treatment can augment healing.
ACKNOWLEDGEMENTS/ FUNDING
We would like to thank the University of Michigan Center for Molecular Imaging and Amanda Welton for her assistance. The core is supported, in part, by the National Institutes of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health (NIH) (P30 AR069620 to Karl Jepsen).
SA funded by NIH F32 AR066499, NIH Loan Repayment Program; SJL and JD funded by Howard Hughes Medical Institute (HHMI) Medical Fellows Program; KR funded by NIH F32 AR068902; YM funded by NIH R01DE020843, DoD W81XWH-11–2-0073; MTL funded by: California Institute for Regenerative Medicine (CIRM) Clinical Fellow training grant TG2–01159, American Society of Maxillofacial Surgeons (ASMS)/Maxillofacial Surgeons Foundation (MSF) Research Grant Award, the Hagey Laboratory for Pediatric Regenerative Medicine and e Oak Foundation, NIH grant U01 HL099776 and the Gunn/Olivier fund.; MS funded by the Plastic Surgery Foundation National Endowment Award; BL funded by NIH, NIGMS K08GM109105, NIH R01GM123069, American Association of Plastic Surgery Research Fellowship, Plastic Surgery Foundation/AAPS Pilot Research Award, ACS Clowes Award, International Fibrodysplasia Ossificans Progressiva Association Research Award.
DISCLOSURES/CONFLICT OF INTEREST
B.L. began a collaboration with Boehringer Ingleheim after data collection and final submission of this manuscript was complete. N.G. declare employment/patient holder with Dana Farber Cancer Institute; advisory role with Syros, Gatekeeper, Soltego, B2S, Petra, C4 therapeutics; research funding with Taiho, Astellas, Takeda, Vivid Biosciences, Kinogen, Aduro; ownership interest with Syros, Gatekeeper, Petra, C4, Soltego, Aduro. All of the other declared no potential conflict of interest.
Footnotes
Data Availability Statement: The datasets generated during and/or analysed during the current study are available in the GEO repository, GEO Submission (GSE126118) [NCBI tracking system #19738436].
REFERENCES
- 1.Levi B et al. In vivo directed differentiation of pluripotent stem cells for skeletal regeneration. Proceedings of the National Academy of Sciences of the United States of America 109, 20379–20384, doi: 10.1073/pnas.1218052109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levi B et al. Nonintegrating knockdown and customized scaffold design enhances human adipose-derived stem cells in skeletal repair. Stem cells 29, 2018–2029, doi: 10.1002/stem.757 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koria P Delivery of growth factors for tissue regeneration and wound healing. BioDrugs : clinical immunotherapeutics, biopharmaceuticals and gene therapy 26, 163–175, doi: 10.2165/11631850-000000000-00000 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Fan F et al. Pharmacological targeting of kinases MST1 and MST2 augments tissue repair and regeneration. Science translational medicine 8, 352ra108, doi: 10.1126/scitranslmed.aaf2304 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Peng T et al. Hedgehog actively maintains adult lung quiescence and regulates repair and regeneration. Nature 526, 578–582, doi: 10.1038/nature14984 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y et al. TISSUE REGENERATION. Inhibition of the prostaglandin-degrading enzyme 15-PGDH potentiates tissue regeneration. Science 348, aaa2340, doi: 10.1126/science.aaa2340 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y et al. Drug-induced regeneration in adult mice. Science translational medicine 7, 290ra292, doi: 10.1126/scitranslmed.3010228 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng F et al. Vimentin coordinates fibroblast proliferation and keratinocyte differentiation in wound healing via TGF-beta-Slug signaling. Proceedings of the National Academy of Sciences of the United States of America 113, E4320–4327, doi: 10.1073/pnas.1519197113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dutta P et al. Myocardial Infarction Activates CCR2(+) Hematopoietic Stem and Progenitor Cells. Cell stem cell 16, 477–487, doi: 10.1016/j.stem.2015.04.008 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leung Y et al. Bifunctional ectodermal stem cells around the nail display dual fate homeostasis and adaptive wounding response toward nail regeneration. Proceedings of the National Academy of Sciences of the United States of America 111, 15114–15119, doi: 10.1073/pnas.1318848111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Velasquez LS et al. Activation of MRTF-A-dependent gene expression with a small molecule promotes myofibroblast differentiation and wound healing. Proceedings of the National Academy of Sciences of the United States of America 110, 16850–16855, doi: 10.1073/pnas.1316764110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeitouni S et al. Human mesenchymal stem cell-derived matrices for enhanced osteoregeneration. Science translational medicine 4, 132ra155, doi: 10.1126/scitranslmed.3003396 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou X et al. Blockage of neddylation modification stimulates tumor sphere formation in vitro and stem cell differentiation and wound healing in vivo. Proceedings of the National Academy of Sciences of the United States of America 113, E2935–2944, doi: 10.1073/pnas.1522367113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agarwal S et al. Inhibition of Hif1alpha prevents both trauma-induced and genetic heterotopic ossification. Proceedings of the National Academy of Sciences of the United States of America 113, E338–347, doi: 10.1073/pnas.1515397113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dey D et al. Two tissue-resident progenitor lineages drive distinct phenotypes of heterotopic ossification. Science translational medicine 8, 366ra163, doi: 10.1126/scitranslmed.aaf1090 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatsell SJ et al. ACVR1R206H receptor mutation causes fibrodysplasia ossificans progressiva by imparting responsiveness to activin A. Science translational medicine 7, 303ra137, doi: 10.1126/scitranslmed.aac4358 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu PB et al. BMP type I receptor inhibition reduces heterotopic [corrected] ossification. Nature medicine 14, 1363–1369, doi:nm.1888 [pii] 10.1038/nm.1888 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agarwal S, Sorkin M & Levi B Heterotopic Ossification and Hypertrophic Scars. Clinics in plastic surgery 44, 749–755, doi: 10.1016/j.cps.2017.05.006 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peterson JR et al. Treatment of heterotopic ossification through remote ATP hydrolysis. Science translational medicine 6, 255ra132, doi: 10.1126/scitranslmed.3008810 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ranganathan K et al. Heterotopic Ossification: Basic-Science Principles and Clinical Correlates. The Journal of bone and joint surgery. American volume 97, 1101–1111, doi: 10.2106/JBJS.N.01056 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bush JR & Beier F TGF-beta and osteoarthritis--the good and the bad. Nature medicine 19, 667–669, doi: 10.1038/nm.3228 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Fortier LA, Barker JU, Strauss EJ, McCarrel TM & Cole BJ The role of growth factors in cartilage repair. Clinical orthopaedics and related research 469, 2706–2715, doi: 10.1007/s11999-011-1857-3 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greenblatt MB, Shim JH & Glimcher LH TAK1 mediates BMP signaling in cartilage. Annals of the New York Academy of Sciences 1192, 385–390, doi: 10.1111/j.1749-6632.2009.05222.x (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gunnell LM et al. TAK1 regulates cartilage and joint development via the MAPK and BMP signaling pathways. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research 25, 1784–1797, doi: 10.1002/jbmr.79 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leah E Osteoarthritis: TGF-beta overload at bones of cartilage degeneration. Nature reviews. Rheumatology 9, 382, doi: 10.1038/nrrheum.2013.81 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Shim JH et al. TAK1 is an essential regulator of BMP signalling in cartilage. The EMBO journal 28, 2028–2041, doi: 10.1038/emboj.2009.162 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhen G et al. Inhibition of TGF-beta signaling in mesenchymal stem cells of subchondral bone attenuates osteoarthritis. Nature medicine 19, 704–712, doi: 10.1038/nm.3143 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao T et al. Inhibition of HIF-1alpha by PX-478 enhances the anti-tumor effect of gemcitabine by inducing immunogenic cell death in pancreatic ductal adenocarcinoma. Oncotarget 6, 2250–2262 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mihaly SR, Ninomiya-Tsuji J & Morioka S TAK1 control of cell death. Cell death and differentiation 21, 1667–1676, doi: 10.1038/cdd.2014.123 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato S et al. Essential function for the kinase TAK1 in innate and adaptive immune responses. Nature immunology 6, 1087–1095, doi: 10.1038/ni1255 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Ninomiya-Tsuji J et al. A resorcylic acid lactone, 5Z-7-oxozeaenol, prevents inflammation by inhibiting the catalytic activity of TAK1 MAPK kinase kinase. The Journal of biological chemistry 278, 18485–18490, doi: 10.1074/jbc.M207453200 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Minoda Y, Sakurai H, Kobayashi T, Yoshimura A & Takaesu G An F-box protein, FBXW5, negatively regulates TAK1 MAP3K in the IL-1beta signaling pathway. Biochemical and biophysical research communications 381, 412–417, doi: 10.1016/j.bbrc.2009.02.052 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Moding EJ et al. Atm deletion with dual recombinase technology preferentially radiosensitizes tumor endothelium. The Journal of clinical investigation 124, 3325–3338, doi: 10.1172/JCI73932 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schonhuber N et al. A next-generation dual-recombinase system for time- and host-specific targeting of pancreatic cancer. Nature medicine 20, 1340–1347, doi: 10.1038/nm.3646 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaiyachati BH et al. LoxP-FRT Trap (LOFT): a simple and flexible system for conventional and reversible gene targeting. BMC biology 10, 96, doi: 10.1186/1741-7007-10-96 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levi B et al. Divergent modulation of adipose-derived stromal cell differentiation by TGF-beta1 based on species of derivation. Plast Reconstr Surg. 2010. August;126(2):412–25. doi: 10.1097/PRS.0b013e3181df64dc. [DOI] [PubMed] [Google Scholar]
- 37.James AW, et al. Paracrine interaction between adipose-derived stromal cells and cranial suture-derived mesenchymal cells. Plast Reconstr Surg. 2010. September;126(3):806–21. doi: 10.1097/PRS.0b013e3181e5f81a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levi B, et al. Differences in osteogenic differentiation of adipose-derived stromal cells from murine, canine, and human sources in vitro and in vivo. Plast Reconstr Surg. 2011. August;128(2):373–86. doi: 10.1097/PRS.0b013e31821e6e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levi B, Longaker MT. (2011). Osteogenic differentiation of adipose-derived stromal cells in mouse and human: in vitro and in vivo methods. J Craniofac Surg. 2011 March;22(2):388–91. doi: 10.1097/SCS.0b013e318207b72b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supp Fig 1. Tak1 expression during wound regeneration
(A) A schematic expression profile of proposed Tak1 expression during wound regeneration. Tak1 expression is lower at the beginning when undifferentiated cells proliferate and subsequently becomes higher when cells differentiate with reduced proliferation.
Supp Fig 2. Design of the COmbinatorial Sequential Inversion ENgineering (COSIEN) mouse targeting Tak1 Exon 2.
Design of the floxed-FRTed allele for Tak1. Exon 2 of Tak1 is flanked by mutant loxP and FRT sites, respectively, with reverse orientation to invert one time with Cre or Flippase. Approximate positions of Southern probes and PCR primers for genotyping are shown. H, HindIII, RV, EcoRV.
Supp Fig 3. microCT cross sections of HO after trauma with TAK1 inhibition.
(A) Representative microCT cross-sections from the mid-tibia and calcaneus of Tak1 tmKO (tamoxifen-inducible postnatal Tak1 knockout (Tak1 tmKO: Ub.CreERT/Tak1fx-frt/fx-frt)) and littermate control hindlimbs showing heterotopic bone 9 weeks after injury, tamoxifen was injected 7 and 3 days before injury and 3 days after injury; (B) Representative microCT cross-sections from the mid-tibia and calcaneus of NG-25 and treatment control hindlimbs showing heterotopic bone 9 weeks after injury. Red lines and shading indicate areas of HO.
Supp Fig 4. Diminished osteogenic and chondrogenic signaling at the injury site with pharmacologic inhibition of TAK1 with NG-25.
(A) Representative pSMAD 1/5 immunostaining of untreated and NG-25 treated mice 3 weeks after injury; (B) Representative pSMAD 2/3 immunostaining of untreated and NG-25 treated mice 3 weeks after injury; (C) Representative Sox9 immunostaining of untreated and NG-25 treated mice 3 weeks after injury. Mice were treated with daily NG25 (2mg/kg) in PBS solution via IP injection for 3 weeks. Scale bars = 100 μm
Supp Fig 5. Confirmation of Purity of Adipose-Derived Mesenchmyal Stem Cells (ASCs)
(A) Flow gate schematic demonstrating isolation of single cell populations; (B) Flow gate schematic demonstrating isolation of viable cells (AmCyan Negative) and purity analysis. Lineage Negative (PE – CD45, MHCII, B220; FITC – CD11b, CD34); (C) Frequency of lineage negative ASCs vs. Isotype controls. Mesenchymal cells described are adipose-derived stem cells (ASCs). AmCyan viability dye used as a marker to gate for live vs. dead cells. Lineage defined by the following myeloid markers CD45-PE; MHCII-PE; B220-PE; CD11b-FITC; CD34-FITC.
Supp Fig 6. Diminished osteogenic and chondrogenic differentiation with loss of Tak1.
(A) Representative ALP stain of Ad.LacZ and Ad.Cre treated Tak1fl/fl mesenchymal cells; (B) Normalized quantification of Alp gene expression from Ad.LacZ and Ad.Cre treated Tak1fl/fl mesenchymal cells (Ad.LacZ: 1.0; Ad.Cre: 0.05); (C) Representative Alizarin Red stain of Ad.LacZ and Ad.Cre treated Tak1fl/fl mesenchymal cells; (D) Normalized quantification of Runx2 gene expression from Ad.LacZ and Ad.Cre treated Tak1fl/fl mesenchymal cells (Ad.LacZ: 1.0; Ad.Cre: 0.32); (E) Normalized quantification of Sox9 gene expression from Ad.LacZ and Ad.Cre treated Tak1fl/fl mesenchymal cells (Ad.LacZ: 1.0; Ad.Cre: 0.46). AR = Alizarin red; n≥3 for all quantification. Mesenchymal cells described are adipose-derived stem cells (ASCs). For differentiation assay, all ASCs were treated with 4uM NG25/DMSO in ODM, changed every 3 days prior to differentiation (7 days for ALP, 14 days for AR, 3 days for RNA collection). *p<0.05.
Supp Fig 7. Pharmacologic inhibition of TAK1 with NG-25 reduces osteogenic and chondrogenic differentiation.
(A) Representative ALP stain of Vehicle Control and NG-25 treated mesenchymal cells; (B) Normalized quantification of Alp gene expression from Vehicle Control and NG-25 treated mesenchymal cells (Vehicle Control: 1.0; NG-25: 0.26); (C) Representative Alizarin Red stain of Vehicle Control and NG-25 treated mesenchymal cells; (D) Normalized quantification of Runx2 gene expression from Vehicle Control and NG-25 treated mesenchymal cells (Vehicle Control: 1.0; NG-25: 0.12); (E) Representative Alcian Blue stain of Vehicle Control and NG-25 treated mesenchymal cells (F) Normalized quantification of Sox9 gene expression from Vehicle Control and NG-25 treated mesenchymal cells (Vehicle Control: 1.0; NG-25: 0.16). ALP = alkaline phosphatase; AR = Alizarin red; n≥3 for all quantification; AB = Alcian blue; All normalization performed to Vehicle Control group. Mesenchymal cells described are adipose-derived stem cells (ASCs). For differentiation assay, all ASCs were treated with 4uM NG25/DMSO in ODM, changed every 3 days prior to differentiation (7 days for ALP, 14 days for AR, 3 days for RNA collection). *p<0.05.
Supp Fig 8. In vitro proliferation with pharmacologic inhibition of TAK1 using 5Z-7-Oxozeaenol (5Z-O). (A) Cell proliferation (BrDU) of 5Z-O and vehicle treated mesenchymal cells; (B) Cell proliferation (Cell counting) of 5Z-O and vehicle treated mesenchymal cells. Mesenchymal cells described are adipose-derived stem cells (ASCs). For differentiation assay, all ASCs were treated with 1μM 5Z-O/DMSO in DMEM, changed every 3 days prior to differentiation (7 days for ALP, 14 days for AR, 3 days for RNA collection). *p<0.05.
Supp Fig 9. siRNA targeted for Tak1 at separate exons effectively decreases the expression of Tak1 in multiple cell lines. (A) Schematic demonstrating the targeting of siRNA against specific sites on the Tak1 gene. (B) Decrease in the relative expression of Tak1 between a control scramble siRNA and two siRNAs targeting the Tak1 gene in 3 different cell lines. β-actin used as internal control. ASCs – Adipose-derived stem cells; TdCs – Tendon-derived cells; Obs – Osteoblasts.
Supp Fig 10. Genetic validation of COSIEN mouse model for Tak1.
(A) Identification of targeted clones for the Tak1fl-frt/wt allele by genomic Southern blot using designated restriction endonucleases; (B) Intercrossing Tak1fl-frt/wt mice to generate Tak1fl-frt/fl-frt mice (W, Takfwt/wt, H, Tak1fl-frt/wt, m, Tak1fl-frt/fl-frt); (C) Genotyping of mice from Tak1fl-frt/wt x Meox2-Cre breeding strategy showing efficient flipping of the Tak1fl-frt allele (samples 1,2,5, positive for Meox2-Cre), persistence of the unrecombined floxed allele in mice negative for Meox2-Cre (samples 3,4,6,7,) Wild type littermates for Tak1 are also shown (samples 8,9); (D) Genotyping of mice from Tak1fl-frt/wt x FLPe breeding strategy showing efficient flipping of the Tak1fl-frt allele (samples 4,5,7,8, white asterisks, positive for Flpe), persistence of the floxed allele in mice negative for FLPe (sample 6). Wild type littermates for Tak1 are also shown (samples 1,2,3,9). Sample #4 shows mosaicism of the floxed and flipped alleles.
Supp Fig 11. In vitro differentiation studies using a dual-inducible model to knockout and rescue Tak1 signaling using COSIEN.
(A) Representative ALP stain of Ad.LacZ, Ad.Cre, and Ad.Cre+Ad.Flp treated mesenchymal cells undergoing osteogenic differentiation with quantification (Ad.LacZ: 1.0; Ad.Cre: 0.34; Ad.Cre+Ad.Flp: 0.60); (B) Representative Alizarin red of Ad.LacZ, Ad.Cre, and Ad.Cre+Ad.Flp treated mesenchymal cells undergoing differentiation with quantification (Ad.LacZ: 1.0; Ad.Cre: 0.31; Ad.Cre+Ad.Flp: 0.75). All cells were treated with Ad.Cre (or Ad.LacZ) for 24 hours under serum deprivation conditions followed by 48 hours in serum replete and subsequently treated with Ad.LacZ (Ad.LacZ group), Ad.Cre (Ad.Cre group), or Ad.Flp (Ad.Cre+Ad.Flp) for 24 hours in serum deprived conditions followed by culture for an additional two days in serum replete conditions. Mesenchymal cells described are adipose-derived stem cells (ASCs). * = p<0.05.
Supp Fig 12. pSMAD 2/3 expression in calvarial defects during Tak1 in-activation followed by differentiation during Tak1 reactivation
Representative immunostaining of Ad.LacZ, Ad.Cre, and Ad.Cre/Ad.Flp treated calvarial defects for pSMAD 2/3. White dotted line marks edge of native calvaria. All scale bars = 200 μm.
Supp Fig 13. PCNA in calvarial defects during Tak1 in-activation followed by differentiation during Tak1 reactivation
(A) Representative immunoblot of Ad.LacZ, Ad.Cre, and Ad.Cre/Ad.Flp treated calvarial defects for PCNA and α-tubulin; (B) Normalized quantification of PCNA protein expression from Ad.LacZ, Ad.Cre, and Ad.Cre/Ad.Flp treated calvarial defects (Ad.LacZ: 1.0; Ad.Cre: 2.34; Ad.Cre/Ad.Flp:1.26). Cells for protein extraction collected by harvest of the calvarial defect after removal of dura. * = p < 0.05.