Abstract
As many as 30% to 40% of locally advanced rectal cancer (LARC) patients experience metastatic progression of the disease. Recognizing the potential of the genetic cargo in tumor-derived exosomes, we hypothesized that plasma exosomal microRNA (miRNA) may reflect biological aggressiveness in LARC and provide new markers for rectal cancer aggressiveness and risk stratification. In a prospective LARC cohort (NCT01816607), plasma samples were collected from 29 patients at the time of diagnosis, before neoadjuvant therapy and surgery. Exosomes, precipitated from plasma using a commercial kit, were verified by cryo-electron microscopy, nanoparticle tracking analysis, and western blotting. Expression of exosomal miRNAs was profiled using a miRCURY LNA miRNA microarray and validation of six miRNAs associated with pathological and clinical end-points was undertaken in plasma collected at the time of diagnosis from 64 patients in an independent prospective LARC cohort (NCT00278694). In both cohorts, exosomal miR-141-3p and miR-375 were higher in patients with synchronous liver metastasis than in those without (P = .010 and P = .017 respectively in the investigative cohort, and P < .001 for both in the validation cohort). Further, high exosomal miR-141-3p was associated with post-operative metastatic liver progression in the investigative cohort (P = .034). Because both miRNAs are associated with tumor angiogenesis and immune modulation, we propose that these miRNAs in circulating exosomes may reflect rectal cancer aggressiveness and accordingly be candidate biomarkers for further investigations.
Introduction
A considerable number of rectal cancer patients experience poor disease outcome due to metastatic progression [1], with the liver typically being the first distant organ affected, followed by the lungs [2]. Since total mesorectal excision became the standard surgical procedure in the mid-nineties and following the introduction of neoadjuvant therapy for the locally-advanced cases, systemic dissemination has remained the main challenge in rectal cancer management, with distant metastasis reported in approximately 30% to 40% of cases in recent trials [3]. For patients with locally advanced rectal cancer (LARC), defining tumors with threatened or involved margin to the planned resection planes or with extensive lymph node involvement in the pelvic cavity, neoadjuvant chemoradiotherapy (CRT) is given in an effort to enable complete surgical removal and reduce the probability of local recurrence [4]. Even though the selection of patients to undergo neoadjuvant treatment is stringent, the treatment responses are highly heterogeneous. Currently, the utility of markers to predict the effect of the neoadjuvant treatment is debated [5] and extensive efforts are invested in understanding the biological basis behind tumor aggressiveness and treatment response.
LARC patients presenting with distant metastasis at the time of diagnosis make up a sub-group of approximately 15% of the total population [6]. Although lung metastasis seem to be more frequent in rectal compared to colon cancer [7], liver metastasis is considered more aggressive than its thoracic counterpart, and associated with considerably shorter life expectancy [2]. A circulating, stable marker of tumor aggressiveness and disease progression could improve diagnostic precision. Additionally, it could give insight into the underlying mechanisms involved in resistance to the established LARC therapy and the early dissemination of tumor cells to distant organs.
Exosomes, defined as extracellular vesicles measuring 30 to 150 nm, are secreted from cells for intercellular communication in normal and pathologic conditions [8]. Their cargo containing nucleic acids and proteins that are specific for the cells of origin, together with their high biological stability, contributes to their emergence as a rich source of biomarkers in cancer patients [8]. MicroRNAs (miRNAs) constitute a portion of the nucleic acid cargo of exosomes. Since they regulate the expression of target genes at the post-transcriptional level [9], miRNAs are believed to have a profound impact on the pathogenesis of most, if not all, human malignancies [10]. Specific miRNAs have been found to regulate a variety of critical processes in tumor physiology including angiogenesis [11] and metastatic progression [12].
In this study, we investigated associations between circulating exosomal miRNA at the time of diagnosis and patient- and tumor-characteristics, as well as therapeutic outcome. We used an investigation cohort consisting of plasma samples collected at the time of diagnosis from 29 LARC patients who underwent neoadjuvant CRT or radiotherapy (RT) alone and corresponding specimens from an independent cohort of 64 LARC patients to validate the results.
Material and Methods
Patient Characteristics (Table 1) and Study Approvals
Table 1.
Patient characteristics with comparisons using Student's t test and chi-square test. Among the patients included in the analysis, one patient in the Investigation Cohort and 12 patients in the Validation Cohort did not undergo surgery on the primary tumor.
| Investigation Cohort |
Validation Cohort |
P | ||
|---|---|---|---|---|
| Median (Range) | Median (Range) | |||
| Age | 63 (41-80) | 60 (30-85) | 0.130 | |
| n (%) | n (%) | |||
| Gender | Female | 11 (38) | 28 (44) | |
| Male | 18 (62) | 36 (56) | 0.598 | |
| cTNM status | T2-3 | 18 (62) | 33 (52) | |
| T4 | 11 (38) | 31 (48) | 0.346 | |
| N0-1 | 8 (28) | 20 (31) | ||
| N2 | 20 (69) | 44 (69) | 0.265 | |
| ND | 1 (3) | 0 | ||
| M0 | 24 (83) | 43 (67) | ||
| M1 | 5 (17) | 21 (33) | 0.121 | |
| ypTN status | ypT0-2 | 8 (28) | 21 (33) | |
| ypT3-4 | 20 (69) | 31 (48) | ||
| ND | 1 (3) | 12 (19) | 0.295 | |
| ypN0 | 12 (41) | 32 (50) | ||
| ypN1-2 | 16 (56) | 20 (31) | ||
| ND | 1 (3) | 12 (19) | 0.109 | |
| TRG score | Good | 11 (38) | 28 (44) | |
| Poor | 17 (59) | 24 (37) | ||
| ND | 1 (3) | 12 (19) | 0.214 | |
cTNM = clinical Tumor-Node-Metastasis status assessed by magnetic resonance imaging (T and N) and computed tomography (M), yp = histological response to neoadjuvant therapy, TRG = tumor regression grade.
The investigation cohort consisted of 29 patients prospectively enrolled onto the OxyTarget biomarker study (NCT01816607) between October 28th 2013 and September 22nd 2015 (Table 1). Patients were referred to neoadjuvant treatment consisting of either 1 week of short-course RT with daily 5-Gy fractions to a total dose of 25 Gy (n = 5) or long-course CRT with daily 2-Gy fractions over 5 weeks to a total dose of 50 Gy with concomitant capecitabine on days of RT (n = 24). In the latter group, one patient had RT only because of cardiac comorbidity. In the former group, one patient was considered to be in a palliative setting and received the local treatment to obtain local tumor control (Supplementary Figure 1). Five OxyTarget patients in the investigation cohort presented with synchronous liver metastases.
The validation cohort consisted of 52 patients from the prospective non-randomized phase II study LARC-RRP (NCT00278694) and 12 additional patients from the OxyTarget study who were not previously analyzed. Nine LARC-RRP patients and all the OxyTarget patients in the validation cohort presented with synchronous liver metastasis. LARC-RRP patients received 4 weeks of neoadjuvant chemotherapy (the Nordic FLOX regimen, bolus 5-FU and oxaliplatin) followed by long-course CRT with concomitant oxaliplatin weekly and capecitabine on days of RT. [13] All of the 52 LARC-RRP patients underwent neoadjuvant treatment with curative intent, while only five OxyTarget patients in the validation cohort was found eligible for treatment with curative intent. These five patients were referred to neoadjuvant treatment consisting of either 1 week of short-course RT with daily 5-Gy fractions to a total dose of 25 Gy (n = 3) or long-course CRT with daily 2-Gy fractions over 5 weeks to a total dose of 50 Gy with concomitant capecitabine on days of RT (n = 1), and one patient was referred directly to surgery on both the primary tumor and the liver metastases. The remaining 7 OxyTarget cases were considered to be in a palliative setting and received local treatment to obtain local tumor control or systemic treatment when found unsuitable for surgery (Supplementary Figure 1).
The resected primary tumor specimens were histologically evaluated according to standard criteria (ypTN-status) as well as tumor regression grade (TRG) [14]. TRG scores spanned categories from the absence of residual tumor cells in the resected specimen (pathological complete response) to the lack of morphologic signs of tissue response to treatment [14], [15]. The scoring system for the two cohorts was either the College of American Pathologists (CAP) scoring system [15] with TRG ranging from 0 to 3 was used (for the OxyTarget cases) or the system proposed by Bouzourene [14] with TRG ranging from 1 to 5 (for the LARC-RRP cases). TRG1-2 (Bouzourene)/0-1 (CAP) and TRG3-5 (Bouzourene)/2-3 (CAP) were considered as good and poor tumor responses, respectively.
When last censored on July 31, 2018, patients in the investigation cohort had a median follow-up period of 35 months (range 2-57). Patients in the validation cohort were last censored on August 8, 2013 and had a median follow-up of 65 months (range 4-66). Both studies were approved by the Institutional Review Board and Regional Committee for Medical and Health Research Ethics of South-East Norway (reference numbers REK 2013/152 and S-05059, respectively). Written informed consent was required for participation.
Blood Samples
Routine clinical blood samples were collected at study enrollment (the time of diagnosis), and were analyzed by the hospital laboratory according to custom procedures. At the same time, additional citrate plasma samples were collected for research purposes and were prepared according to standardized protocols with centrifugation at 2000 g for 10 minutes, then aliquotation and storage at −80 ° C until analysis.
Exosome Isolation and Characterization
Exosome isolation was conducted at Exiqon Services (Vedbæk, Denmark). Exosomes were precipitated from 500 μl plasma (investigation cohort) or 1000 μl (validation cohort; to improve stability of the analysis) using the miRCURY Exosome Isolation Kit – Serum and Plasma (Exiqon), according to the protocol. Additionally, three random samples from the investigation cohort were selected for characterization of the isolated vesicles. The exosomes were re-suspended in 100 μl resuspension buffer and alliquoted for analysis with cryo-electron microscopy (cryo-EM), nanoparticle tracking analysis (NTA), and western immunoblotting. Procedures for exosome isolation and characterization have been described in detail previously [16].
Exosomal miRNA Analysis
All wet-laboratory procedures were conducted at Exiqon Services. Total RNA was extracted from exosomes using the miRCURY RNA isolation kit - bio fluids (Exiqon), and stored at −80 °C. For each sample, 10 μl (investigation cohort) or 2 μl (validation cohort) RNA were reverse transcribed in 50 μl (investigation cohort) or 10 μl (validation cohort) reactions using the miRCURY LNA Universal RT miRNA PCR, polyadenylation and cDNA synthesis kit (Exiqon). cDNA was diluted 50-fold and assayed in 10 μl PCR reactions; each miRNA was assayed once by qPCR on the miRNA Ready-to-Use PCR Human panel 1 (Exiqon; investigation cohort) or in triplicates in the miRCURY LNA Universal RT miRNA PCR Custom Pick and mix panel with assays for 14 human miRNAs (Exiqon; validation cohort), using ExiLENT SYBR Green master mix (Exiqon). UniSp2, UniSp4, and UniSp5 were used for RNA isolation controls, UniSp6 was used for cDNA synthesis control, and UniSp3 as a DNA spike-in for plate-to-plate variation control. Negative controls excluding the template from the reverse transcription reaction were included and treated identically to the patient samples. The amplification was performed in a LightCycler 480 Real-Time PCR System (Roche, Basel, Switzerland) in 384-well plates. The amplification curves were analyzed using the Roche LC software. All assays detected at 3 Cqs lower than the negative control were considered for further analysis; however, if an assay was found below the cut-off in at least 20% of the samples, all Cq values were included. For assays that did not yield any signal in the negative control, the upper limit of detection was set to Cq = 37. Normalization of sample readouts was performed with global mean [17], using 44 miRNAs detected in all of the samples in the investigation cohort. Seven miRNAs which displayed high stability in the initial investigation were included in the validation analysis as references for calculation of the global mean.
In technical-validation experiments conducted at Akershus University Hospital, an equal amount of the RNA was reversely transcribed using miScript II RT Kit (Qiagen, Hilden, Germany) and analyzed using the miScript PCR Assay (Qiagen). The cDNA was diluted 20-fold and analyzed in triplicates in 10 μl PCR reactions. Primers used were miR-375 (MS00003507) and miR-23a-5p (MS00007336) as reference. The PCR reactions were run in 96-well plates using 7900 HT Fast Real-Time PCR system (Applied Biosystem, Foster City, CA, USA). Data normalization was performed using (normalizer assay Ct – assay Ct = dCt).
Statistical Methods
A panel of exosomal miRNA selected on the basis of known associations to cancer was chosen for analysis. Exosomal miRNA levels were correlated to the pathological and clinical characteristics of the patients using the Significance Analysis of Microarrays (SAM) software version 5.0, employing unpaired analysis and a false discovery rate cut-off of 10% [18]. Herein, differences in expression levels within the study population which were significant when separating according to Tumor (T) Node (N) Metastatis (M) status, ypTN status, and TRG score were highlighted on the basis of the individual miRNA's expression relative to the standard deviation (SD) of repeat measurements. The software handles any missing data by imputation using the K-nearest neighbor method [18]. The findings from SAM were verified individually using Student's t test to eliminate miRNA candidates with many missing values, using IBM SPSS Statistics for Mac version 25. Based on significantly different expression of exosomal miRNA according to M status and TRG score, a set of miRNAs from the investigation cohort was selected for further validation in an independent cohort (Supplementary Table 1). Correlations between continuous data were determined by Pearson product correlation analysis after transformation to natural logarithms for symmetric distribution. Continuous data were described with mean and SD, and groups were compared using Student's t test or Mann–Whitney U test when groups with few cases were examined. Differences between categorical data were determined using Chi squared test. Sample size determination in the validation cohort was done by power analysis based on miRNA association with M-status as the most interesting finding (Supplementary Table 1). Associations between selected variables and progression-free survival (PFS) or overall survival (OS) were modeled with univariate Cox regression analysis, and results were expressed as hazard ratio with 95% confidence interval. P-values were adjusted for multiple testing using the Benjamini-Hochberg method. All tests were two-sided. P-values less than 0.05 were considered statistically significant.
Results
Exosomal miRNA versus Patient and Tumor Characteristics at Baseline
Of the initial 372 exosomal miRNAs tested in the investigation cohort, 44 were detected in all samples, with an average of 135 miRNAs per sample (Supplementary Table 2 and Supplementary Table 3A). Analysis with SAM uncovered 37 miRNAs with significantly altered expression according to N- or M-status, or TRG (Supplementary Table 1). Of these, six miRNAs with a strong probability of relation to M-status and TRG were verified using single parameter Student's t test (Supplementary Table 1).
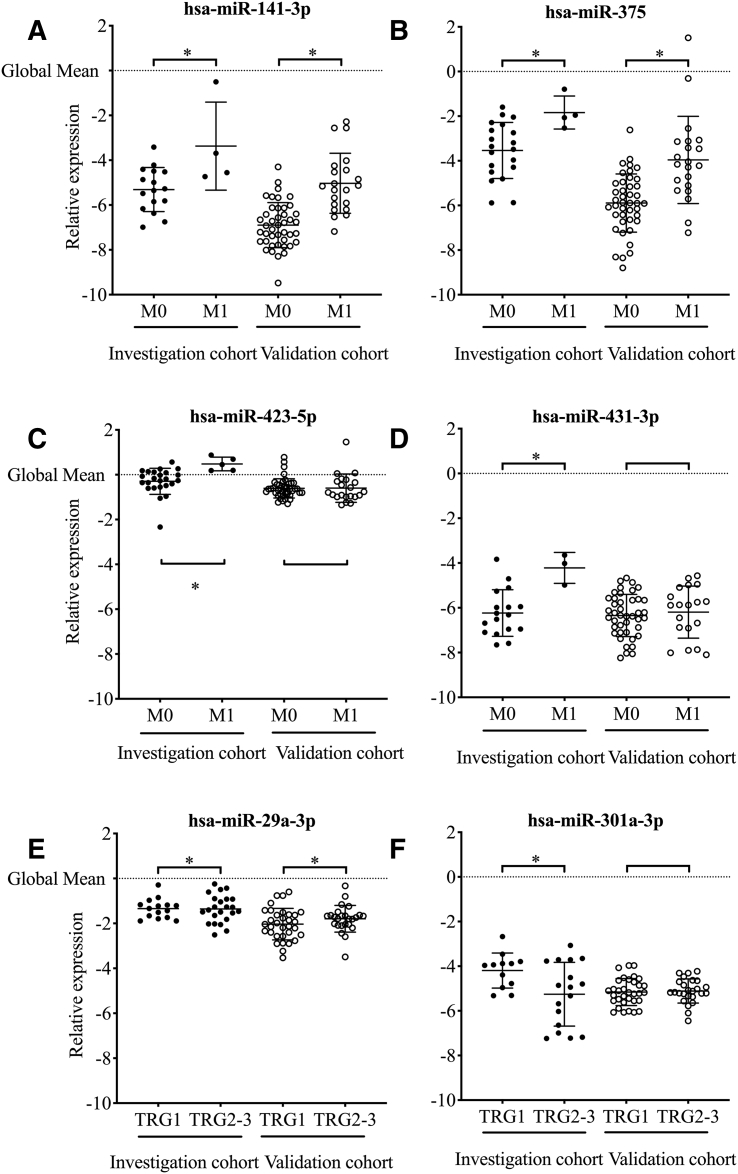
The observed significant associations presented in Figure 1 show that high expression of miR-29a-3p and low levels of miR-301a-3p were found in patients with poor tumor response (TRG 2-3) to neoadjuvant therapy in the investigation cohort. Further, miR-141-3p, miR-375, miR-423-5p, and miR-431-3p were all found to be higher expressed in patients with synchronous liver metastasis. All six miRNAs were selected for validation along with seven miRNAs as reference for normalization.
Figure 1.
Differential expression of exosomal miRNAs selected with significance analysis of microarrays according to synchronous metastasis (M-status) or tumor regression grade (TRG). The relative expression of each miRNA was normalized according to the global mean, and significant differences between the groups are marked with *.
hsa = Homo Sapiens.
In the validation cohort, of the initial 13 miRNAs on the array, 9 miRNAs were detected in all samples, with an average of 13 miRNAs per sample (Supplementary Table 2 and Supplementary Table 3B). When correlating to pathological and clinical baseline parameters, two of the validated exosomal miRNAs, miR-141-3p and miR-375, showed similar and significant results in both of the two cohorts. A technical validation of one of the six miRNAs was conducted in ten patients in the investigation cohort. miR-375 analyzed with the miScript PCR system correlated with the miRCURY PCR system (R = 0.8623, P = .0028; Supplementary Figure 2).
Exosomal miR-141-3p was detected in 20 of the 29 cases in the investigation cohort and in all of the 64 validation cohort cases. For miR-375, the corresponding numbers were 24 of 29 and 63 of 64. In the investigation cohort, 18 patients had readable results for both miRNAs; two had results for miR-141-3p only and 6 for miR-375 only, adding up to a total of 26 patients eligible for inclusion in further analysis (Supplementary Table 2). No significant difference was found according to TN-status for either miRNA (Supplementary Table 4). Exosomal miR-141-3p and miR-375 expressions were higher in four patients presenting with synchronous liver metastasis.
Of the 64 patients in the validation cohort, 32 cases developed liver metastasis, where 21 cases were synchronous with diagnosis of the primary tumor. The association between synchronous liver metastasis and higher plasma exosomal miR-141-3p and miR-375 was confirmed in the validation cohort (Figure 1 and Supplementary Table 4). Receiver-operating curve analysis enabled the separation of patients with and without liver metastasis in the validation cohort with an area under the curve of 0.88 (SD 0.04) for miR-141-3p and 0.82 (SD 0.061) for miR-375.
Table 2 shows that in the validation cohort, which was almost twice the size of the investigation cohort, both miR-141-3p and miR-375 were negatively correlated with blood lymphocyte count and bilirubin levels and positively correlated with lactate dehydrogenase levels. miR-375 was also negatively correlated with monocyte count. In addition, miR-141-3p correlated positively with aspartate-aminotransferase and γ-glutamyl transferase, typically associated with liver metastasis, and carcinoembryonic antigen, indicative of unfavorable tumor biology.
Table 2.
Associations between miRNA expression and clinical parameters and standard blood measures. P-values were calculated by Pearson's correlation coefficient (significant findings in italic). P-values were adjusted for multiple testing calculated by the Benjamini-Hochberg method.
| miR-141-3p |
||||||||
|---|---|---|---|---|---|---|---|---|
| Investigation cohort |
Validation cohort |
|||||||
| n | r | P | P adjusted | n | r | P | P adjusted | |
| miR-375 | 18 | 0.520 | 0.027 | 0.342 | 63 | 0.689 | 0.000 | 0.000 |
| Age | 20 | −0.156 | 0.511 | 0.826 | 64 | −0.013 | 0.916 | 0.996 |
| Hemoglobin | 20 | −0.054 | 0.822 | 0.868 | 64 | −0.051 | 0.689 | 0.854 |
| Thrombocytes | 18 | 0.188 | 0.455 | 0.826 | 64 | −0.046 | 0.719 | 0.854 |
| Leukocytes | 20 | −0.110 | 0.645 | 0.826 | 64 | −0.056 | 0.661 | 0.854 |
| Neutrophils | 17 | −0.197 | 0.448 | 0.826 | 60 | 0.007 | 0.955 | 0.996 |
| Lymphocytes | 17 | 0.112 | 0.667 | 0.826 | 59 | −0.328 | 0.011 | 0.042 |
| Monocytes | 17 | 0.116 | 0.658 | 0.826 | 58 | −0.241 | 0.068 | 0.162 |
| AST | 15 | −0.044 | 0.875 | 0.875 | 12 | 0.740 | 0.006 | 0.033 |
| ALT | 18 | −0.171 | 0.491 | 0.826 | 64 | 0.158 | 0.211 | 0.364 |
| GT | 18 | −0.099 | 0.696 | 0.826 | 47 | 0.300 | 0.041 | 0.111 |
| ALP | 18 | −0.382 | 0.118 | 0.561 | 64 | 0.199 | 0.115 | 0.243 |
| LDH | 13 | −0.081 | 0.792 | 0.868 | 58 | 0.348 | 0.007 | 0.033 |
| Bilirubin | 18 | −0.158 | 0.532 | 0.826 | 64 | −0.459 | 0.000 | 0.000 |
| Albumin | 18 | −0.172 | 0.495 | 0.826 | 62 | 0.058 | 0.657 | 0.854 |
| CRP | 19 | 0.449 | 0.054 | 0.342 | 54 | 0.164 | 0.237 | 0.375 |
| CEA | 20 | 0.451 | 0.046 | 0.342 | 64 | 0.303 | 0.015 | 0.047 |
| miR-375 | ||||||||
| Investigation Cohort | Validation Cohort | |||||||
| n | r | P | P adjusted | n | r | P | P adjusted | |
| miR-141-3p | 18 | 0.520 | 0.027 | 0.394 | 63 | 0.689 | 0.000 | 0.000 |
| Age | 24 | 0.137 | 0.524 | 0.974 | 63 | 0.027 | 0.863 | 0.964 |
| Hemoglobin | 24 | −0.030 | 0.889 | 0.974 | 63 | 0.031 | 0.808 | 0.960 |
| Thrombocytes | 20 | −0.072 | 0.763 | 0.974 | 63 | −0.183 | 0.152 | 0.413 |
| Leukocytes | 23 | 0.018 | 0.934 | 0.974 | 63 | 0.014 | 0.916 | 0.967 |
| Neutrophils | 19 | 0.121 | 0.620 | 0.974 | 59 | 0.168 | 0.203 | 0.429 |
| Lymphocytes | 19 | 0.091 | 0.711 | 0.974 | 58 | −0.417 | 0.001 | 0.006 |
| Monocytes | 19 | 0.460 | 0.048 | 0.394 | 57 | −0.297 | 0.025 | 0.095 |
| AST | 17 | −0.188 | 0.469 | 0.974 | 12 | 0.183 | 0.568 | 0.771 |
| ALT | 22 | −0.222 | 0.322 | 0.874 | 63 | 0.139 | 0.278 | 0.480 |
| GT | 20 | 0.149 | 0.530 | 0.974 | 46 | 0.227 | 0.130 | 0.412 |
| ALP | 21 | 0.009 | 0.969 | 0.974 | 63 | 0.053 | 0.679 | 0.860 |
| LDH | 13 | 0.083 | 0.787 | 0.974 | 57 | 0.337 | 0.010 | 0.047 |
| Bilirubin | 21 | −0.008 | 0.974 | 0.974 | 63 | −0.455 | 0.000 | 0.000 |
| Albumin | 19 | −0.415 | 0.077 | 0.394 | 61 | 0.103 | 0.429 | 0.627 |
| CRP | 22 | 0.267 | 0.230 | 0.728 | 53 | −0.122 | 0.386 | 0.611 |
| CEA | 24 | 0.298 | 0.157 | 0.597 | 63 | 0.152 | 0.234 | 0.445 |
AST = aspartate-aminotransferase, ALT = alanine-aminotransferase, GT = γ-glutamyl transferase, ALP = alcalic phosphatase, LDH = lactate dehydrogenase, CRP = C-reactive protein, CEA = carcinoembryonic antigen, miRNA/miR = micro Ribonucleic Acid.
Exosomal miR-141-3p and miR-375 versus Treatment Outcome
No significant association was found between histopathological treatment outcome and plasma levels of the two miRNAs in the two cohorts (Figure 1 and Supplementary Table 4). However, for long-term outcome, low exosomal miR-141-3p in the investigation cohort was associated with longer time to metastatic progression. This was not found in the validation cohort (Supplementary Table 5).
Discussion
Through miRNA profiling of exosomes isolated from LARC patients' plasma, we identified significant associations between several miRNAs and clinical traits associated with tumor aggressiveness. We selected a limited number of miRNAs for validation in an independent cohort, and verified higher plasma exosomal miR-141-3p and miR-375 in patients presenting with synchronous liver metastasis.
It has been shown that cancer cells secrete much higher quantities of exosomes than normal cells, which enables malignant cells to transfer oncogenic signals and promote tumorigenesis through interactions with other cell types locally and in distant organs [19]. Furthermore, it has been indicated that exosomes may carry genetic cargo, paving the way for circulating tumor cells to metastasize to distant organs [20]. Exosomes have been found to be highly stable and a rich source of biomarkers in biofluids [8]. Although many studies have presented findings from circulating miRNA, the ones detected in exosomes may be of special interest. Measurements of miRNA in exosomes provide higher stability and possibly more specificity towards the tumor environment than measurements directly in serum or plasma, probably due to the higher resemblance of the exosomal content with that of the cancer cells [21].
High tissue miR-141-3p expression has been reported to target the phosphatase and tensin homolog (PTEN) [22], which in turn results in activation of the phosphoinositide 3-kinase (PI3K)-Akt pathway [23]. Mutations in the PI3K-Akt pathway are frequent in human cancers [24]. This pathway is unique in the sense that every step of the pathway has been found to be either mutated or amplified in a broad range of cancers [24]. Elevated circulating levels of miR-141-3p have been shown to be associated with metastatic colorectal cancer development and adverse prognosis [25], though also with opposite roles in tumors originating at other sites [26]. miR-375 tissue upregulation has been associated with inhibition of the PI3K-Akt pathway [27] and often exhibits a protective effect against metastasis [28], [29]. However, in spite of these conflicting findings, a simultaneous upregulation of these two miRNAs in circulation and tissues have been associated with the presence, aggressiveness, and treatment responsiveness in several malignancies [30], [31], [32]. As we have demonstrated here, when comparing LARC patients with or without liver metastasis, higher levels of circulating exosomal miR-375 and miR-141-3p were associated with metastatic disease.
Elaborating on the role of miR-375 in PI3K-Akt regulation, Biton et al. showed that PTEN inhibition and subsequent PI3K-Akt activation resulted in an increase in miR-375 expression [33]. Chen et al. also demonstrated this mechanism, where miR-375 inhibited the yes-associated protein 1 (YAP1), a modulator of PTEN, and where miR-375 increased when YAP1 was blocked [34]. Furthermore, miR-375 upregulation has been found to inhibit autophagy [35] and reduce activity of tumor antigen-presenting dendritic cells [36] to promote an immunosuppressive environment in the tumor [37]. Studies also indicate that miR-375 is involved in inflammatory and neoangiogenic responses [38]. Taken together, it is tempting to postulate that the rapidly evolving environment of aggressive LARC increases the secretion of miR-141-3p [39], which in turn up-regulates the PI3K-Akt pathway [22]. This further leads to an increase of miR-375 [33], which subsequently enhances the immunosuppressive environment [37] and lets the tumor escape immunosurveillance, as often seen in advanced cancers. This would be in line with the theory proposed by Kuttke et al. [40], where a constitutional activation of the PI3K-Akt pathway leads to a shift of the immune cell populations towards a tolerogenic environment for the tumor. Consistent with the above, the immunosuppressive effect of the PI3K-activation is also supported by two recent studies by Kaneda and De Henau et al. [41], [42]. Interestingly, we have previously demonstrated an association between high activity of PI3K-Akt signaling in tumor tissue from several of the patients included in the current validation cohort and treatment resistance and long-term outcome [43]. In the same cohort, the release into the circulation of factors associated with proliferation and maturation of antigen-presenting dendritic cells during the induction neoadjuvant chemotherapy was associated with longer PFS [44], [45].
An inverse correlation was found in the validation cohort between miR-141-3p and miR-375 and the blood lymphocyte and monocyte counts, further implying an association with patients' immune cell activity. Additionally, both miRNAs also correlated positively with lactate dehydrogenase, whereas miR-141-3p correlated with aspartate aminotransferase, γ-glutamyl transferase, and carcinoembryonic antigen, markers commonly associated with tumor evolution. Of note, there was an inverse correlation between miR-141-3p and bilirubin. An elevated bilirubin is often associated with the presence of and poor prognosis of liver metastases [46], perhaps due to cholestasis and hyperbilirubinemia caused by metastatic masses obstructing intrahepatic biliary ducts [47]. However, studies have shown a protective effect of bilirubin against colorectal cancer [48], and bilirubin seems to act as a crucial antioxidant in the cellular redox homeostasis [49], which may explain this finding.
Conclusions
To summarize, we have presented and validated an association between elevated expression of miR-375 and miR-141-3p in plasma exosomes and the presence of synchronous liver metastasis in rectal cancer. However, our cohorts were small, and since there was no comparison to a disease-free cohort, it is difficult to conclude on the absolute level of expression of these miRNAs and the onset of metastasis. Both miR-141-3p and miR-375 have been shown to be involved in the regulation of the PTEN/PI3K-Akt pathway. Further supporting our findings, it has been demonstrated that synchronous elevation of these two miRNAs is a sign of aggressiveness in several cancer types, including prostate and breast cancer [30], [31] and osteosarcoma [32]. We propose that high expression of miR-141-3p and miR-375 in plasma exosomes from LARC patients may be markers of a specific tumor trait related to disease progression to the liver, and that these two miRNAs should be further investigated as candidate biomarkers of rectal cancer aggressiveness and systemic dissemination.
The Role of the Funding Sources
The funding sources had no role in the study design; collection, analysis, and interpretation of data; writing of the report; or the decision to submit the article for publication.
Acknowledgements
This work was supported by the South-Eastern Norway Regional Health Authority (grant numbers 2013002, 2014010, 2016050, 2017109) and Akershus University Hospital (grant numbers 2016-266942, 2017-26794, 2017-267913, 2018-268925, 2018-268936).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tranon.2019.04.014.
Appendix A. Supplementary Data
Supplementary material
References
- 1.Adam R, de Gramont A, Figueras J, Kokudo N, Kunstlinger F, Loyer E, Poston G, Rougier P, Rubbia-Brandt L, Sobrero A. Managing synchronous liver metastases from colorectal cancer: a multidisciplinary international consensus. Cancer Treat Rev. 2015;41(9):729–741. doi: 10.1016/j.ctrv.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 2.Riihimaki M, Hemminki A, Sundquist J, Hemminki K. Patterns of metastasis in colon and rectal cancer. Sci Rep. 2016;6 doi: 10.1038/srep29765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valentini V, van Stiphout RG, Lammering G, Gambacorta MA, Barba MC, Bebenek M, Bonnetain F, Bosset JF, Bujko K, Cionini L. Nomograms for predicting local recurrence, distant metastases, and overall survival for patients with locally advanced rectal cancer on the basis of European randomized clinical trials. J Clin Oncol. 2011;29(23):3163–3172. doi: 10.1200/JCO.2010.33.1595. [DOI] [PubMed] [Google Scholar]
- 4.Lutz MP, Zalcberg JR, Glynne-Jones R, Ruers T, Ducreux M, Arnold D, Aust D, Brown G, Bujko K, Cunningham C. Second St. Gallen European Organisation for Research and Treatment of Cancer Gastrointestinal Cancer Conference: consensus recommendations on controversial issues in the primary treatment of rectal cancer. Eur J Cancer. 2016;63:11–24. doi: 10.1016/j.ejca.2016.04.010. [DOI] [PubMed] [Google Scholar]
- 5.Lopez NE, Peterson CY. Advances in Biomarkers: Going Beyond the Carcinoembryonic Antigen. Clin Colon Rectal Surg. 2016;29(3):196–204. doi: 10.1055/s-0036-1584289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manfredi S, Lepage C, Hatem C, Coatmeur O, Faivre J, Bouvier AM. Epidemiology and management of liver metastases from colorectal cancer. Ann Surg. 2006;244(2):254–259. doi: 10.1097/01.sla.0000217629.94941.cf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robinson JR, Newcomb PA, Hardikar S, Cohen SA, Phipps AI. Stage IV colorectal cancer primary site and patterns of distant metastasis. Cancer Epidemiol. 2017;48:92–95. doi: 10.1016/j.canep.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma A, Khatun Z, Shiras A. Tumor exosomes: cellular postmen of cancer diagnosis and personalized therapy. Nanomedicine (London, England) 2016;11(4):421–437. doi: 10.2217/nnm.15.210. [DOI] [PubMed] [Google Scholar]
- 9.Lai EC. Micro RNAs are complementary to 3' UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30(4):363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 10.Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiol Rev. 2011;91(3):827–887. doi: 10.1152/physrev.00006.2010. [DOI] [PubMed] [Google Scholar]
- 11.Pecot CV, Rupaimoole R, Yang D, Akbani R, Ivan C, Lu C, Wu S, Han HD, Shah MY, Rodriguez-Aguayo C. Tumour angiogenesis regulation by the miR-200 family. Nat Commun. 2013;4:2427. doi: 10.1038/ncomms3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olson P, Lu J, Zhang H, Shai A, Chun MG, Wang Y, Libutti SK, Nakakura EK, Golub TR, Hanahan D. MicroRNA dynamics in the stages of tumorigenesis correlate with hallmark capabilities of cancer. Genes Dev. 2009;23(18):2152–2165. doi: 10.1101/gad.1820109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dueland S, Ree AH, Groholt KK, Saelen MG, Folkvord S, Hole KH, Seierstad T, Larsen SG, Giercksky KE, Wiig JN. Oxaliplatin-containing Preoperative Therapy in Locally Advanced Rectal Cancer: Local Response, Toxicity and Long-term Outcome. Clin Oncol (R Coll Radiol) 2016;28(8):532–539. doi: 10.1016/j.clon.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 14.Bouzourene H, Bosman FT, Matter M, Coucke P. Predictive factors in locally advanced rectal cancer treated with preoperative hyperfractionated and accelerated radiotherapy. Hum Pathol. 2003;34(6):541–548. doi: 10.1016/s0046-8177(03)00176-x. [DOI] [PubMed] [Google Scholar]
- 15.Compton CC, Fielding LP, Burgart LJ, Conley B, Cooper HS, Hamilton SR, Hammond ME, Henson DE, Hutter RV, Nagle RB. Prognostic factors in colorectal cancer. College of American Pathologists Consensus Statement 1999. Arch Pathol Lab Med. 2000;124(7):979–994. doi: 10.5858/2000-124-0979-PFICC. [DOI] [PubMed] [Google Scholar]
- 16.Bjornetro T, Redalen KR, Meltzer S, Thusyanthan NS, Samiappan R, Jegerschold C, Handeland KR, Ree AH. An experimental strategy unveiling exosomal microRNAs 486-5p, 181a-5p and 30d-5p from hypoxic tumour cells as circulating indicators of high-risk rectal cancer. J Extracell Vesicles. 2019;8(1) doi: 10.1080/20013078.2019.1567219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mestdagh P, Van Vlierberghe P, De Weer A, Muth D, Westermann F, Speleman F, Vandesompele J. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009;10(6):R64. doi: 10.1186/gb-2009-10-6-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thind A, Wilson C. Exosomal miRNAs as cancer biomarkers and therapeutic targets. J Extracell Vesicles. 2016;5 doi: 10.3402/jev.v5.31292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H, Deng T, Liu R, Bai M, Zhou L, Wang X, Li S, Wang X, Yang H, Li J. Exosome-delivered EGFR regulates liver microenvironment to promote gastric cancer liver metastasis. Nat Commun. 2017;8 doi: 10.1038/ncomms15016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nedaeinia R, Manian M, Jazayeri MH, Ranjbar M, Salehi R, Sharifi M, Mohaghegh F, Goli M, Jahednia SH, Avan A. Circulating exosomes and exosomal microRNAs as biomarkers in gastrointestinal cancer. Cancer Gene Ther. 2017;24(2):48–56. doi: 10.1038/cgt.2016.77. [DOI] [PubMed] [Google Scholar]
- 22.Jin YY, Chen QJ, Xu K, Ren HT, Bao X, Ma YN, Wei Y, Ma HB. Involvement of microRNA-141-3p in 5-fluorouracil and oxaliplatin chemo-resistance in esophageal cancer cells via regulation of PTEN. Mol Cell Biochem. 2016;422(1-2):161–170. doi: 10.1007/s11010-016-2816-9. [DOI] [PubMed] [Google Scholar]
- 23.Jiang BH, Liu LZ. PI3K/PTEN signaling in tumorigenesis and angiogenesis. Biochim Biophys Acta. 2008;1784(1):150–158. doi: 10.1016/j.bbapap.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 24.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27(41):5497–5510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng H, Zhang L, Cogdell DE, Zheng H, Schetter AJ, Nykter M, Harris CC, Chen K, Hamilton SR, Zhang W. Circulating plasma MiR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PLoS One. 2011;6(3) doi: 10.1371/journal.pone.0017745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao Y, Feng B, Han S, Zhang K, Chen J, Li C, Wang R, Chen L. The Roles of MicroRNA-141 in Human Cancers: From Diagnosis to Treatment. Cell Physiol Biochem. 2016;38(2):427–448. doi: 10.1159/000438641. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, Tang Q, Li M, Jiang S, Wang X. MicroRNA-375 inhibits colorectal cancer growth by targeting PIK3CA. Biochem Biophys Res Commun. 2014;444(2):199–204. doi: 10.1016/j.bbrc.2014.01.028. [DOI] [PubMed] [Google Scholar]
- 28.Xu L, Wen T, Liu Z, Xu F, Yang L, Liu J, Feng G, An G. MicroRNA-375 suppresses human colorectal cancer metastasis by targeting Frizzled 8. Oncotarget. 2016;7(26):40644–40656. doi: 10.18632/oncotarget.9811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei R, Yang Q, Han B, Li Y, Yao K, Yang X, Chen Z, Yang S, Zhou J, Li M. microRNA-375 inhibits colorectal cancer cells proliferation by downregulating JAK2/STAT3 and MAP3K8/ERK signaling pathways. Oncotarget. 2017;8(10):16633–16641. doi: 10.18632/oncotarget.15114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madhavan D, Zucknick M, Wallwiener M, Cuk K, Modugno C, Scharpff M, Schott S, Heil J, Turchinovich A, Yang R. Circulating miRNAs as surrogate markers for circulating tumor cells and prognostic markers in metastatic breast cancer. Clin Cancer Res. 2012;18(21):5972–5982. doi: 10.1158/1078-0432.CCR-12-1407. [DOI] [PubMed] [Google Scholar]
- 31.Cheng HH, Mitchell PS, Kroh EM, Dowell AE, Chery L, Siddiqui J, Nelson PS, Vessella RL, Knudsen BS, Chinnaiyan AM. Circulating microRNA profiling identifies a subset of metastatic prostate cancer patients with evidence of cancer-associated hypoxia. PLoS One. 2013;8(7) doi: 10.1371/journal.pone.0069239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berlanga P, Munoz L, Piqueras M, Sirerol JA, Sanchez-Izquierdo MD, Hervas D, Hernandez M, Llavador M, Machado I, Llombart-Bosch A. miR-200c and phospho-AKT as prognostic factors and mediators of osteosarcoma progression and lung metastasis. Mol Oncol. 2016;10(7):1043–1053. doi: 10.1016/j.molonc.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Biton M, Levin A, Slyper M, Alkalay I, Horwitz E, Mor H, Kredo-Russo S, Avnit-Sagi T, Cojocaru G, Zreik F. Epithelial microRNAs regulate gut mucosal immunity via epithelium-T cell crosstalk. Nat Immunol. 2011;12(3):239–246. doi: 10.1038/ni.1994. [DOI] [PubMed] [Google Scholar]
- 34.Chen S, Zheng Y, Zhang S, Jia L, Zhou Y. Promotion Effects of miR-375 on the Osteogenic Differentiation of Human Adipose-Derived Mesenchymal Stem Cells. Stem Cell Reports. 2017;8(3):773–786. doi: 10.1016/j.stemcr.2017.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Y Chang, W Yan, X He, L Zhang, C Li, H Huang, G Nace, DA Geller, J Lin, A Tsung. miR-375 inhibits autophagy and reduces viability of hepatocellular carcinoma cells under hypoxic conditions. Gastroenterology. 2012;143(1):177-187 e178. [DOI] [PubMed]
- 36.Lin J, Xia J, Tu CZ, Zhang KY, Zeng Y, Yang Q. H9N2 Avian Influenza Virus Protein PB1 Enhances the Immune Responses of Bone Marrow-Derived Dendritic Cells by Down-Regulating miR375. Front Microbiol. 2017;8:287. doi: 10.3389/fmicb.2017.00287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Galluzzi L, Bravo-San Pedro JM, Demaria S, Formenti SC, Kroemer G. Activating autophagy to potentiate immunogenic chemotherapy and radiation therapy. Nat Rev Clin Oncol. 2017;14(4):247–258. doi: 10.1038/nrclinonc.2016.183. [DOI] [PubMed] [Google Scholar]
- 38.Garikipati VNS, Verma SK, Jolardarashi D, Cheng Z, Ibetti J, Cimini M, Tang Y, Khan M, Yue Y, Benedict C. Therapeutic inhibition of miR-375 attenuates post-myocardial infarction inflammatory response and left ventricular dysfunction via PDK-1-AKT signalling axis. Cardiovasc Res. 2017;113(8):938–949. doi: 10.1093/cvr/cvx052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Della Vittoria Scarpati G, Calura E, Di Marino M, Romualdi C, Beltrame L, Malapelle U, Troncone G, De Stefano A, Pepe S, De Placido S. Analysis of differential miRNA expression in primary tumor and stroma of colorectal cancer patients. Biomed Res Int. 2014;2014 doi: 10.1155/2014/840921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuttke M, Sahin E, Pisoni J, Percig S, Vogel A, Kraemmer D, Hanzl L, Brunner JS, Paar H, Soukup K. Myeloid PTEN deficiency impairs tumor-immune surveillance via immune-checkpoint inhibition. Oncoimmunology. 2016;5(7) doi: 10.1080/2162402X.2016.1164918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaneda MM, Messer KS, Ralainirina N, Li H, Leem CJ, Gorjestani S, Woo G, Nguyen AV, Figueiredo CC, Foubert P. PI3Kgamma is a molecular switch that controls immune suppression. Nature. 2016;539(7629):437–442. doi: 10.1038/nature19834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Henau O, Rausch M, Winkler D, Campesato LF, Liu C, Cymerman DH, Budhu S, Ghosh A, Pink M, Tchaicha J. Overcoming resistance to checkpoint blockade therapy by targeting PI3Kgamma in myeloid cells. Nature. 2016;539(7629):443–447. doi: 10.1038/nature20554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ree AH, Flatmark K, Saelen MG, Folkvord S, Dueland S, Geisler J, Redalen KR. Tumor phosphatidylinositol 3-kinase signaling in therapy resistance and metastatic dissemination of rectal cancer: Opportunities for signaling-adapted therapies. Crit Rev Oncol Hematol. 2015;95(1):114–124. doi: 10.1016/j.critrevonc.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 44.Meltzer S, Kalanxhi E, Hektoen HH, Dueland S, Flatmark K, Redalen KR, Ree AH. Systemic release of osteoprotegerin during oxaliplatincontaining induction chemotherapy and favorable systemic outcome of sequential radiotherapy in rectal cancer. Oncotarget. 2016;7(23):34907–34917. doi: 10.18632/oncotarget.8995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kalanxhi E, Meltzer S, Schou JV, Larsen FO, Dueland S, Flatmark K, Jensen BV, Hole KH, Seierstad T, Redalen KR. Systemic immune response induced by oxaliplatin-based neoadjuvant therapy favours survival without metastatic progression in high-risk rectal cancer. Br J Cancer. 2018;118(10):1322–1328. doi: 10.1038/s41416-018-0085-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lahr CJ, Soong SJ, Cloud G, Smith JW, Urist MM, Balch CM. A multifactorial analysis of prognostic factors in patients with liver metastases from colorectal carcinoma. J Clin Oncol. 1983;1(11):720–726. doi: 10.1200/JCO.1983.1.11.720. [DOI] [PubMed] [Google Scholar]
- 47.O'Brien MJ, Bronstein B, Zamcheck N, Saravis C, Burke B, Gottlieb LS. Cholestasis and hepatic metastases: a factor contributing to extreme elevations of carcinoembryonic antigen. J Natl Cancer Inst. 1980;64(6):1291–1294. doi: 10.1093/jnci/64.6.1291. [DOI] [PubMed] [Google Scholar]
- 48.Zucker SD, Horn PS, Sherman KE. Serum bilirubin levels in the U.S. population: gender effect and inverse correlation with colorectal cancer. Hepatology. 2004;40(4):827–835. doi: 10.1002/hep.20407. [DOI] [PubMed] [Google Scholar]
- 49.Ziberna L, Martelanc M, Franko M, Passamonti S. Bilirubin is an Endogenous Antioxidant in Human Vascular Endothelial Cells. Sci Rep. 2016;6 doi: 10.1038/srep29240. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material