Abstract
The arrangement of root hair and non-hair cells in the root epidermis provides a useful model for understanding the cell fate determination system in plants. A network of related transcription factors, including GLABRA3 (GL3), influences the patterning of cell types in Arabidopsis. GL3 is expressed primarily in root hair cells and encodes a bHLH transcription factor, which inhibits root hair differentiation in Arabidopsis root epidermis. By transforming the GL3 promoter::GFP into tomato, we demonstrated that the Arabidopsis GL3 promoter can function in tomato root epidermis. GFP fluorescence was observed in almost all root epidermal cells in the GL3::GFP transgenic tomato plants, indicating that all root epidermal cells of tomato possess root hair cell identity similar to that of Arabidopsis root hair cells. This is consistent with the phenotype of the tomato root, in which all epidermal cells produce root hairs. Moreover, we observed the localization of a GL3:GFP fusion protein in GL3::GL3:GFP transgenic tomato; although GL3 is known to exclusively localize in non-hair cell nuclei in Arabidopsis root epidermis, GL3:GFP fluorescence was detected not in the nuclei but in the cytoplasm of transgenic tomato epidermal cells. These results suggest that the nuclear localization mechanism differs between tomato and Arabidopsis.
Keywords: Arabidopsis, epidermis, GL3, root hair, tomato
Control of root hair and non-hair cell fate determination is a critical issue in plant developmental biology. The difference between root hair cell and non-hair cell identities is evident prior to root hair initiation (Brena-Medina et al. 2014). Three types of root hair patterns are defined (Dolan and Costa 2001). Tomato exhibits a type I root hair pattern, in which root hairs develop in a random pattern; all of the epidermal cells have potential to produce root hairs. Arabidopsis, in contrast, shows a type III pattern, in which root hairs are arranged in cell files that are interspersed with non-hair cell files (Pemberton et al. 2001). In Arabidopsis, root hair or non-hair cell identity is precisely controlled by several transcription factors, including GLABRA3 (GL3) (Payne et al. 2000). The GL3 gene encodes a bHLH transcription factor, which is involved in the inhibition of root hair differentiation in Arabidopsis root epidermis. Previously, we identified a GL3 homologous gene from the tomato (Solanum lycopersicum) genome and named it SlGL3 (Tominaga-Wada et al. 2013). To elucidate the functions of SlGL3, we introduced SlGL3 into Arabidopsis. However, we could not detect any remarkable effect of SlGL3 on the root hair phenotype in Arabidopsis (Tominaga-Wada et al. 2013). Tomato has root hairs in all epidermal cells. Thus, we wondered whether GL3 could repress root hair formation in the tomato. In this study, we adopted the reverse approach and examined the promoter activity and protein behavior of the Arabidopsis GL3 gene in tomato. We introduced GL3::GFP into tomato to clarify GL3’s promoter activity and examined GL3:GFP fluorescence in GL3::GL3:GFP transgenic tomato plants to elucidate the protein localization and movement ability of GL3 in tomato root epidermal cells.
Tomato (Solanum lycopersicum L. ‘Micro-Tom’) seeds were surface-sterilized and incubated as previously described (Tominaga-Wada et al. 2013). The generation of GL3::GL3:GFP transgenic tomato plants has been described previously (Wada et al. 2014). To create the GL3::GFP construct, a 3.2-kb promoter region of the GL3 gene from the Arabidopsis genome was PCR-amplified, fused to GFP, and then sub-cloned into a modified pPZP212 binary vector. The GL3::GFP construct was introduced into tomato according to a highly efficient transformation protocol for Micro-Tom using Agrobacterium (Sun et al. 2006). Homozygous transgenic lines were selected based on kanamycin resistance.
Root images were observed using a Leica MZ16FA stereomicroscope (Leica Microsystems GmbH, Wetzlar, Germany). GFP fluorescence was observed using a Zeiss LSM-510 Meta confocal laser scanning microscope (CLSM).
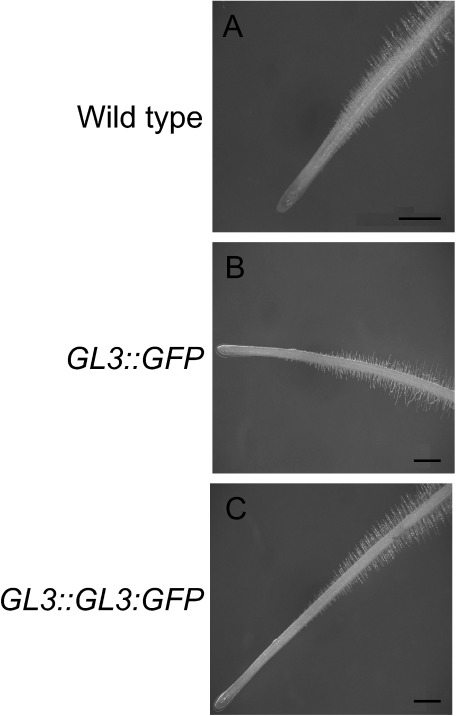
The GL3::GFP and GL3::GL3:GFP transgenic tomato plants were phenotypically similar to the wild-type plants. We did not detect any notable differences in root hair phenotype between GL3::GFP or GL3::GL3:GFP transgenic tomato plants and the wild-type tomato plant (Figure 1). GL3 is known to inhibit root hair formation in Arabidopsis (Bernhardt et al. 2003); however, as previously reported, the GL3::GL3:GFP transgenic tomato plants did not show the expected hairless phenotype (Supplemental Figure 1) (Wada et al. 2014). In addition, we did not detect any notable differences in trichome formation between GL3::GL3:GFP transgenic tomato and wild-type tomato plants (Wada et al. 2014)
Figure 1. Root epidermal phenotypes of transgenic tomato plants. (A) Five-day-old seedling roots of wild-type plant. (B) Five-day-old seedling roots of GL3::GFP transgenic plant. (C) Five-day-old seedling roots of GL3::GL3:GFP transgenic plant. Scale bars: 1 mm.
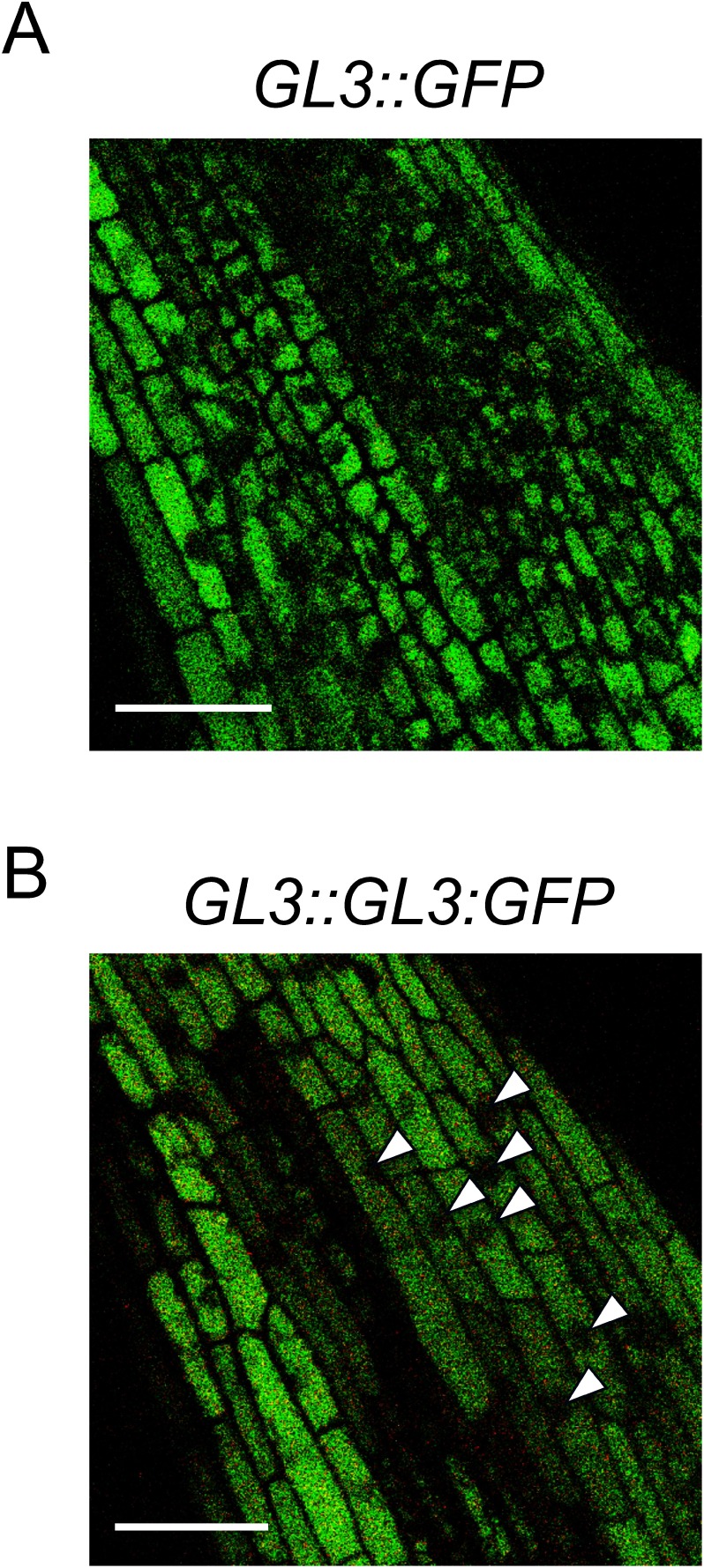
The GL3 gene is preferentially expressed in root hair cells in the Arabidopsis root epidermis (Bernhardt et al. 2005). Transgenic tomato plants carrying the GL3::GFP construct, in which the expression of GFP was driven by the GL3 promoter, showed GFP fluorescence in almost all epidermal cells (Figure 2A). This result indicates that the GL3 promoter sequence from Arabidopsis is evidently also functional in tomato (Figure 2A). Because all root epidermal cells produce root hairs in tomato (Figure 1), they must possess the identity of root hair cells. Thus, the GL3 promoter, which is specific to root hair cells in Arabidopsis, should be active in all root epidermal cells in tomato (Figure 2A). We confirmed that GFP was not detectable without the epidermis in the root on CSLM observation (Supplemental Figure 2).
Figure 2. Distribution of GFP fluorescence in transgenic tomato plants. (A) CLSM images showing GFP fluorescence in the root epidermis of a GL3::GFP transgenic tomato plant. (B) CLSM images showing GFP fluorescence in the root epidermis of a GL3::GL3:GFP transgenic tomato plant. Arrowheads indicate the nucleus. Scale bars: 50 µm.
The GL3 gene encodes a bHLH transcription factor, and therefore it is expected that the GL3 protein is transported into and functions in the nucleus. In fact, GL3:YFP protein has been observed in the nuclei of Arabidopsis root epidermal cells (Bernhardt et al. 2005). However, in the present study, we did not detect GL3:GFP fluorescence in the nuclei of tomato epidermal cells, although fluorescence was observed in most parts of the cytoplasm (Figure 2B).
In this study, we demonstrated that the Arabidopsis GL3 promoter is also functional in tomato. This result was shown to be useful in a plant engineering experiment, in which specific gene expression in tomato root epidermis was achieved by using the GL3 promoter. We also confirmed that no GFP fluorescence was present in the root epidermis of wild-type tomato plants, indicating that the GFP fluorescence detected in this study was not due to the influence of auto-fluorescence of tomato tissue (Supplemental Figure 3).
The SlGL3 gene was identified to be homologous to GL3 in tomato (Tominaga-Wada et al. 2013). Although the precise functions of the tomato SlGL3 gene have not been determined, it is hypothesized that the SlGL3 has a function similar to that of GL3 in the tomato root epidermis owing to their amino acid sequence similarity. Therefore, GL3 was expected to function in tomato root epidermis as an inhibitor of root hair formation, as observed in Arabidopsis root epidermis. However, the introduction of the GL3 gene into tomato had no effect on the root hair phenotype of tomato (Figure 1). This might be due to the inability of the GL3 protein to translocate into the nucleus (Figure 2B), perhaps because it is missing a specific signal sequence or binding of another component that guides the protein to the nucleus. This inability of GL3 to move to and function in the nucleus indicates that it probably does not function as a transcription factor. The GL3 promoter functioned in the epidermal cells of both in Arabidopsis and tomato. However, the GL3 protein was non-functional in tomato because GL3 did not move from the cytoplasm to the nucleus. It is therefore hypothesized that the function of GL3 in tomato is entirely different from that in Arabidopsis. Our study suggests that the cell differentiation system of tomato is different from that of Arabidopsis at present and that the tomato evolved to lose its bHLH transcription factor-mediated root-hair inhibitory mechanism. SlGL3 has the same origin as GL3 but might have evolved with a different function in tomato.
Acknowledgments
We wish to thank Y. Nukumizu for providing technical support. This work was partly funded by a Japan Society for the Promotion of Science KAKENHI (grant numbers 15K14656 and JP16K07644).
Supplementary Data
References
- Bernhardt C, Lee MM, Gonzalez A, Zhang F, Lloyd A, Schiefelbein J (2003) The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 130: 6431–6439 [DOI] [PubMed] [Google Scholar]
- Bernhardt C, Zhao M, Gonzalez A, Lloyd A, Schiefelbein J (2005) The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development 132: 291–298 [DOI] [PubMed] [Google Scholar]
- Brena-Medina V, Champneys AR, Grierson C, Ward MJ (2014) Mathematical modeling of plant root hair initiation: Dynamics of localized patches. SIAM J Appl Dyn Syst 13: 210–248 [Google Scholar]
- Dolan L, Costa S (2001) Evolution and genetics of root hair stripes in the root epidermis. J Exp Bot 52(suppl 1): 413–417 [DOI] [PubMed] [Google Scholar]
- Payne CT, Zhang F, Lloyd AM (2000) GL3 encodes a bHLH protein that regulates trichome development in arabidopsis through interaction with GL1 and TTG1. Genetics 156: 1349–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton LMS, Tsai SL, Lovell PH, Harris PJ (2001) Epidermal patterning in seedling roots of eudicotyledons. Ann Bot (Lond) 87: 649–654 [Google Scholar]
- Sun HJ, Uchii S, Watanabe S, Ezura H (2006) A highly efficient transformation protocol for Micro-Tom, a model cultivar for tomato functional genomics. Plant Cell Physiol 47: 426–431 [DOI] [PubMed] [Google Scholar]
- Tominaga-Wada R, Nukumizu Y, Sato S, Wada T (2013) Control of plant trichome and root-hair development by a tomato (Solanum lycopersicum) R3 MYB transcription factor. PLoS One 8: e54019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Kunihiro A, Tominaga-Wada R (2014) Arabidopsis CAPRICE (MYB) and GLABRA3 (bHLH) control tomato (Solanum lycopersicum) anthocyanin biosynthesis. PLoS One 9: e109093. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.