Abstract
Approximately 60% of human cancers exhibit enhanced activity of ERK1 and ERK2, reflecting their multiple roles in tumor initiation and progression. Acquired drug resistance, especially mechanisms associated with the reactivation of the MAPK (RAF/MEK/ERK) pathway represent a major challenge to current treatments of melanoma and several other cancers. Recently, targeting ERK has evolved as a potentially attractive strategy to overcome this resistance. Herein, we report the design and synthesis of novel series of fused naphthofuro[3,2-c] quinoline-6,7,12-triones 3a-f and pyrano[3,2-c]quinoline-6,7,8,13-tetraones 5a,b and 6, as potential ERK inhibitors. New inhibitors were synthesized and identified by different spectroscopic techniques and X-ray crystallography. They were evaluated for their ability to inhibit ERK½ in an in vitro radioactive kinase assay. 3b and 6 inhibited ERK1 with IC50s of 0.5 and 0.19 μM, and inhibited ERK2 with IC50s of 0.6 and 0.16 μM respectively. Kinetic mechanism studies revealed that the inhibitors are ATP-competitive inhibitors where 6 inhibited ERK2 with a Ki of 0.09 μM. Six of the new inhibitors were tested for their in vitro anticancer activity against the NCI-60 panel of tumor cell lines. Compound 3b and 6 were the most potent against most of the human tumor cell lines tested. Moreover, 3b and 6 inhibited the proliferation of the BRAF mutant A375 melanoma cells with IC50s of 3.7 and 0.13 μM, respectively. In addition, they suppressed anchorage-dependent colony formation. Treatment of the A375 cell line with 3b and 6 inhibited the phosphorylation of ERK substrates p-90RSK and ELK-1 and induced apoptosis in a dose dependent manner. Finally, a molecular docking study showed the potential binding mode of 3b and 6 within the ATP catalytic binding site of ERK2.
Keywords: Naphthafuro-quinoline, Naphthafuro-pyranoquinoline, ERK inhibitors, NCI-60 panel, BRAF mutant, Melanoma
1. Introduction
The fused pyranoquinolines have a common structural motif, existing in many naturally occurring and biologically active alkaloids, such as flindersine, oricine, verprisine, and Araliopsis tabouensis alkaloids [1,2]. Several derivatives of these alkaloids are being used as pharmaceuticals and agrochemicals. They have been reported to exhibit a wide range of biological activities including anti-inflammatory, anti-allergic, psychotropic, and estrogenic effects [3]. Their broad biological activity has turned them into popular compounds for pharmacological studies [4–7]. Recent studies reported the successful construction of fused tricyclic pyranoquinolines, such as polycyclic using 4-hydro-xyquinolone together with different activated ylidenes [8]. Moreover, a multicomponent system has also been used for synthesis based on fused pyranoquinolines [9,10]. Derivatives of the 1,4-naphthoquinone pharmacophore are known to exhibit pronounced biological and pharmacological activities, including anti-tumor [11], anti-proliferative [12], anti-mycobacterial [13], anti-platelet, anti-inflammatory, anti-allergic[14], anti-malarial [15] and anti-leishmanial activities [16]. Substituted naphtho[2,3-b]furan-4,9-diones inhibited hyperproliferation of human keratinocytes and could therefore be potentially useful in the treatment of psoriasis, a common, immune-mediated inflammatory disease [17].
The high frequency of aberrant activity of the RAS/RAF/MEK/ERK cascade found in human cancers, makes this pathway a potential target for treatment [18,19]. Several potent inhibitors, targeting RAF and MEK kinases have been widely used for BRAF- and RAS- mutant tumors, including vemurafenib, dabrafenib, trametinib and selumetinib [18]. However, tumor regrowth after an initial response is common and is attributed to several mechanisms [20,21], commonly all involving ERK reactivation. ERK corresponds to a pair of downstream kinases (ERK1 and ERK2) and represents a terminal signal node for this pathway [22]. Several ERK inhibitors have been discovered and showed an ability to overcome resistance [23]. For example, the potent ERK inhibitor BVD-523 (Ulixertinib) showed encouraging antitumor activity in in vitro and in vivo models of acquired resistance to BRAF and/or MEK targeted inhibitors [24]. A recent dose escalation in-human study of BVD-523, exhibited early evidence for clinical activity in NRAS- and BRAF V600-solid tumor malignancies with acceptable safety and pharmacokinetic behavior [19]. The apparent promise of ERK modulators in human cancer, especially the ones with acquired resistance to BRAF and/or MEK inhibition, encouraged our efforts towards developing a potent and selective ERK inhibitor.
We have utilized the 2,3-dichloro-1,4-naphthoquinone (DCHNQ) moiety in several heterocyclic synthesis programs, including the synthesis of heterocycles having creatinine [25] and thiobarbituric acid[26]. In addition, we synthesized several naphtho-1,3-dithiole derivatives [27] and dibenzo[b,i]-thianthrene-5,7,12,14-tetraone derivatives[28]. In our previous studies, we synthesized a series of ethyl pyrano [3,2-c]quinoline-4-carboxylates [29], spiro(indoline-3,4′-pyrano[3,2-c]-quinoline)-3′-carbonitriles [30], 2,3-bis-(4-hydroxy-2-oxo-1,2-dihydroquinolin-3-yl)succinates and arylmethylene-bis-3,3′-quinoline-2-ones [31]. In addition to the synthesis of two series of N-2,3-bis(6-substituted-4-hydroxy-2-oxo-1,2-dihydroquinolin-3-yl)naphthalene-1,4-diones and substituted N-(methyl/ethyl)bisquinolinone triethyl-ammonium, that been reported to inhibit ERK2 activity in an vitro kinase assay [32].
In this current project we synthesized a novel series of fused naphthofuro[3,2-c]quinoline-6,7,12-triones 3a-f and pyrano[3,2-c]quinoline-6,7,8,13-tetraones 5a-b and 6 from the reaction of 1,6-disubstituted-4-hydroxy-quinoline-2-ones 1a-f and 4-hydroxy-pyrano[3,2-c]quinoline-4,5(6H)-diones 4a-c with 2,3-dichloro-1,4-naphthoquinone (DCHNQ). The ability of the new compounds to inhibit the ERK kinase activity was assessed in an in vitro kinase assay and in-cells using western blotting. The cytotoxic activity of the synthesized compounds towards the NCI-60 panel of cancer cell lines was determined and the cellular mechanism of the most potent inhibitors was further investigated in A375 melanoma cell line. Finally, the binding features of the synthesized compounds within the ATP-binding site of ERK2 was estimated using molecular docking studies.
2. Results and Discussion
2.1. Chemistry
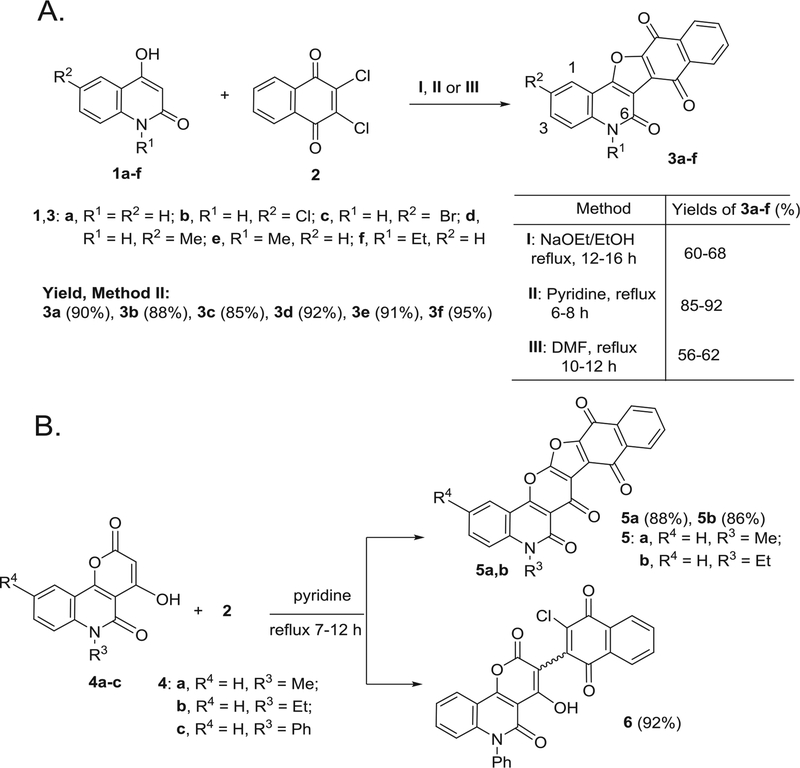
Upon mixing equivalent amounts of 2,3-dichloro-1,4-naphthoqui-none (DCHNQ, 2) and 1,6-disubstituted-4-hydroxy-quinoline-2-ones 1a-f, followed by refluxing in different solvents, such as: NaOEt/EtOH (Method I), pyridine (Method II) or DMF Method (III), naphtho [2′,3′:4,5]furo[3,2-c]quinoline-6,7,12(5H)-trione derivatives 3a-f were obtained as single products (Scheme 1A). Noticeably, the reaction in pyridine (Method II) afforded 3a-f in better yields (85–92%), if compared to the other two methods (Scheme 1A). Structure elucidation of compounds 3a-f was carried out by IR, 1H NMR, 13C NMR and mass spectrometry, as well as elemental analyses. The structures of 3c and 3f, were confirmed by X-ray structural analysis (Fig. 1A and B). The mass spectrmetry and elemental analysis proved the molecular formula of 3a to be C19H9NO4, representing a product from one molecule of 1a and one molecule of DCHNQ, 2 with elimination of two HCl molecules. The IR spectrum of 3a showed four characteristic bands at ν = 3210 (NH), 1700, 1660 (CO), 1596 (Ar–C=C). In addition, the 1H NMR spectrum of 3b; the H-3, H-1 and NH protons were appeared as doublet-doublet at δH 7.65 (J = 7.0, 0.8 Hz), doublet at δH 7.50 (J = 0.7 Hz) and singlet at δH 8.10 ppm. In the 13C NMR spectrum of 3f, the two carbonyl carbons appeared at δC = 176.0 and 173.0 ppm. Seven carbon signals were also distinguished in 13C NMR of 3f absorbed at δC = 160.4, 132.7, 128.6, 127.7, 126.8 and 115.5.5 ppm assigned to C-6, C-9 (C-10), C-3, C-1, C-8 (C-11) and C-4, respectively (Experimental section). The ethyl carbons appeared in the 13C NMR spectrum at δC = 13.7 (CH3) and 47.8 ppm (CH2) (Experimental section).
Scheme 1.
(A) Different methods for preparation of naphthofuro[3,2-c]quinoline-6,7,12-triones 3a-f. (B) Reaction of 4-hydroxy-pyrano[3,2-c]quinoline-4,5(6H)-diones 4a-c with 2.
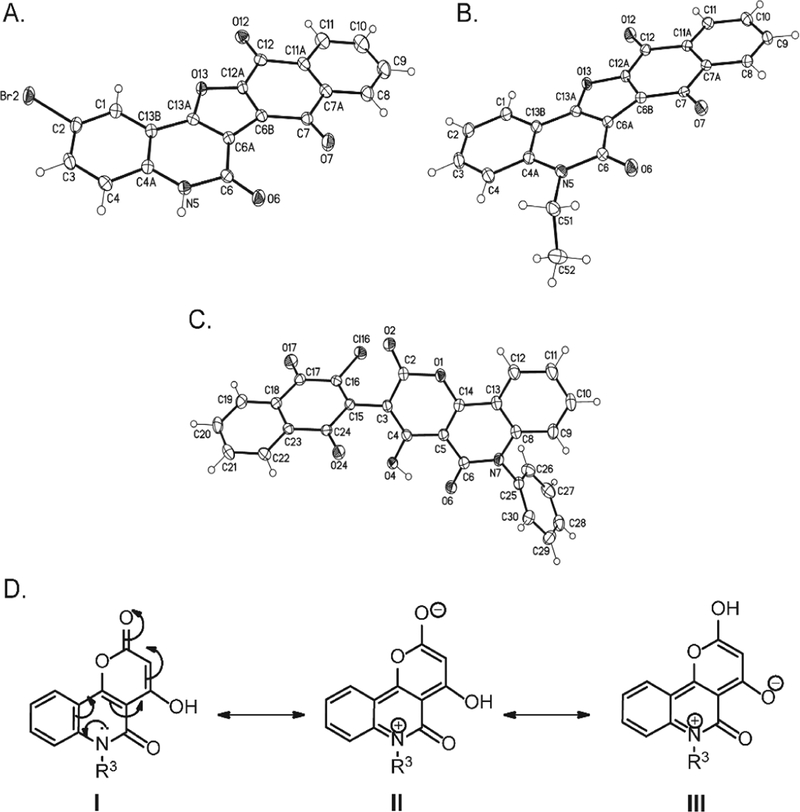
Fig. 1.
The X-ray structural analysis of (A) 3c and (B) 3f and (C) 6. The X-ray structure of one of the two crystallographic independent molecules of 6, solvent omitted for clarity and displacement parameters are drawn at 50% probability level. (D) Resonance structures of quinolones 4a,b.
Afterward, we investigated the reactions of DCHNQ (2) and 4-hydroxy-pyrano[3,2-c]quinoline-4,5(6H)-diones (4a-c) under the same conditions mentioned above (Scheme 1B). The reaction of 2 and 4a,b afforded two 5H-naphthalen-[2″,3″:4′,5′]-furo[3′,2′:4′,5,6]pyrano-[3,2-c]quinoline-6,7,8,13-tetraones derivatives 5a,b as single products. In the same time, the reaction of 2 and 4c afforded a different product of syn/anti-3-(1,4-dioxo-1,4-dihydronaphthalen-2-yl)-4-hydroxy-6-phenyl-2H-pyrano[3,2-c]-quinoline-2,5(6H)-dione (6). The chemical identities of 5a,b and 6 was confirmed by IR, 1H NMR, 13C NMR, mass spectrometry and elemental analyses. Compound 6 was proved by its X-ray structure analysis as shown in Fig. 1C. Compound 6 was obtained as characteristically orange crystals. Its molecular structure formula was prove to be C28H14ClNO6, represents a product from one molecule of DCHNQ, 2 and one molecule of 4c with the loss of one HCl molecule. In the 1H NMR spectrum, the low-field OH- attached to pyran ring is at δH = 13.90 ppm. In the 13C NMR spectrum, the distinct carbons assigned as CO (C-1′), C-OH, CO (C-2) and C-O, (C-5), resonated at δC = 176.9, 166.1, 163.2 and 162.3 ppm, respectively.
The same conclusion was confirmed by single crystal X-ray analysis (Fig. 1C). The basicity of the N-quinolone lone pair in compounds 4a,b, is responsible for the nucleophilic character of the OH group that enables the cyclization process. The former is related to the resonance structures I to III (Fig. 1D). Since the N-Ph derivative 4c is of lower basicity compared to 4a,b, the reaction stops at the step of replacement in the nucleophilic position-3 of the pyrano moiety (Fig. 1D).
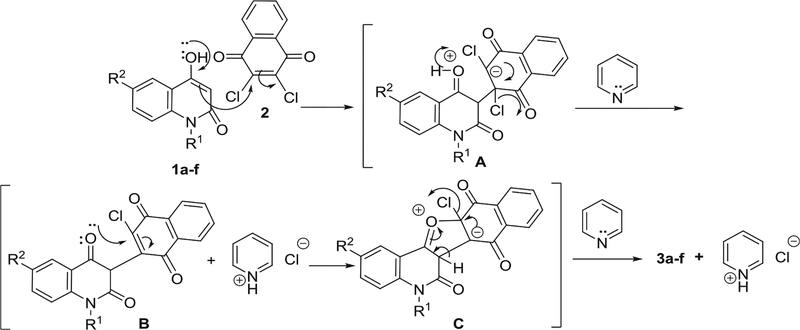
The reaction mechanism can be described as nucleophilic attack of CH-3 in 1a-f to C-2 in 2 to form the Zwitter ion A (Scheme 2). Pyridine molecule would then enhance elimination of HCl from A to give intermediate B and pyridinium salt. Further nucleophilic attack from the oxygen lone pair to vinylic carbon in B would give Zwitter ionic intermediate C (Scheme 2). Ultimately, a second molecule of pyridine would catalyze the elimination of HCl to give 3a-f and pyridinium salt (Scheme 2).
Scheme 2.
Mechanism described formation of compounds 3a-f.
2.2. Biological evaluation
2.2.1. Evaluation of the in vitro anticancer activity of the new compounds
Among the synthesized compounds, NCI selected compounds 3a, 3b and 3e from the fused naphthofuro[3,2-c]quinoline-6,7,12-trione derivatives and 5a, 5b and 6 from the fused naphthofuro-pyrano[3,2-c] quinoline-6,7,8,13-tetraone derivatives. Anticancer assays were performed according to the US NCI protocol (The procedures of the assay are described at (http://www.dtp.nci.nih.gov). The compounds were tested against a panel of 60 cancer cell lines, derived from different tumors. Including leukemia, melanoma, lung, colon, central nervous system (CNS), ovarian, renal, prostate and breast cancer. The compounds were incubated with the cells at a concentration of 10 μM for 48 h and the cell density was estimated using sulforhodamine B (SRB), a protein binding dye. The results were reported as the growth percent (G%) of the cells that were treated by each compound relative to the cells treated with vehicle, and relative to the number of cells present when the treatment with the compounds started (Table 1). On average, 3a, 3b and 3e exhibited inhibitory effects on most of the leukemia cell lines tested with varied responses. While, in the rest of the tested tumors, compound 3b showed much more pronounced growth suppressive activity, compared to 3a and 3e with an average G% of −4.7, 68 and 74% respectively.
Table 1.
Growth percent (G%) of the NCI-60 panel tumor cell lines at 10−5 M concentration of compounds 3a, 3b, 3e, 5a, 5b and 6.
| Tumor cell lines | Growth percent (G%) | |||||
|---|---|---|---|---|---|---|
| 3a | 3b | 3e | 5a | 5b | 6 | |
| Leukemia | ||||||
| CCRF−CEM | 20.72 | 29.04 | 19.07 | 92.93 | 2.47 | 17.76 |
| HL−60(TB) | 2.26 | 94.52 | ND | 104.29 | −18.8 | 7.05 |
| K−562 | 55.20 | 12.90 | 68.79 | 89.61 | 28.57 | 26.84 |
| MOLT−4 | 5.55 | 18.44 | 2.39 | 87.37 | −24.49 | 16.13 |
| RPMI−8226 | 62.72 | 4.83 | 51.06 | 100.23 | 33.49 | 19.65 |
| SR | 43.80 | 33.72 | ND | 92.01 | 80.76 | 11.44 |
| Non−small cell lung cancer | ||||||
| A549/ATCC | 97.18 | −0.05 | 81.67 | 85.17 | 82.36 | 37.73 |
| EKVX | 27.35 | 15.17 | 70.67 | ND | ND | ND |
| HOP−62 | 118.63 | −38.70 | 81.77 | 80.81 | 80.29 | 36.07 |
| HOP−92 | 71.42 | −37.24 | 100.92 | 99.17 | 50.81 | 80.01 |
| NCI−H226 | 78.47 | −28.80 | 73.22 | 88.55 | 99.44 | 86.30 |
| NCI−H23 | 65.95 | 21.21 | 78.33 | 86.15 | 66.79 | 38.22 |
| NCI−H322M | 89.28 | 13.98 | 96.16 | 91.04 | 97.72 | 63.53 |
| NCI−H460 | 68.27 | −69.73 | 78.50 | 108.09 | 83.68 | 33.57 |
| NCI−H522 | 54.81 | −16.76 | 50.55 | 78.37 | 65.5 | 19.42 |
| Colon cancer | ||||||
| HCC−2998 | 102.57 | 65.07 | 101.82 | 103.64 | 100.59 | 42.85 |
| HCT−116 | 55.46 | −12.24 | 71.69 | 99.77 | 34.6 | 41.72 |
| HCT−15 | 44.70 | −31.10 | 85.01 | 94.13 | 79.63 | 66.56 |
| HT29 | 102.52 | 66.37 | 73.69 | 96.96 | 63.02 | 84.18 |
| KM 12 | 66.22 | 0.12 | 91.54 | 95.63 | 86.61 | 18.86 |
| SW−620 | 89.23 | −53.04 | 86.24 | 103.1 | 73.47 | 18.61 |
| CNS cancer | ||||||
| SF−268 | 91.30 | −31.69 | 95.35 | 101.09 | 84.32 | 34.32 |
| SF−295 | 92.71 | −1.34 | ND | 101.32 | 95.07 | 18.38 |
| SF−539 | 90.30 | −27.99 | 97.17 | 93.83 | 89.92 | −5.45 |
| SNB−19 | 103.72 | 7.11 | 93.29 | 110.38 | 90.17 | 27.87 |
| SNB−75 | 112.98 | −9.01 | 64.39 | 101.36 | 119.73 | 31.77 |
| U251 | 79.04 | −77.55 | 90.22 | 104.35 | 57.67 | 21.15 |
| Melanoma | ||||||
| LOX IMVI | 56.12 | 11.13 | 76.59 | 95.89 | 32.39 | 42.99 |
| MALME−3M | 124.29 | −51.66 | 113.45 | 113.29 | 112.5 | 42.00 |
| M14 | 73.38 | 10.44 | 85.59 | 98.25 | 89.81 | 44.74 |
| MDA−MB−435 | 83.72 | 10.25 | 103.73 | 105.3 | 95.88 | 87.80 |
| SK−MEL−2 | 76.92 | 31.83 | 92.64 | 86.85 | 79.89 | 23.06 |
| SK−MEL−28 | 112.03 | −11.84 | 111.89 | 107.21 | 106.96 | 39.37 |
| SK−MEL−5 | 79.73 | 12.31 | 76.95 | 97.56 | 67.42 | 17.66 |
| UACC−62 | 88.73 | 75.08 | 77.03 | 93.45 | 63.95 | 13.17 |
| Ovarian cancer | ||||||
| IGROV1 | 55.11 | −3.63 | 68.65 | 65.92 | 68.31 | 71.85 |
| OVCAR−3 | −14.81 | −68.37 | 21.58 | 98.93 | −16.31 | 2.15 |
| OVCAR−4 | −5.49 | −42.04 | 4.38 | 97.38 | −99.52 | 18.80 |
| OVCAR−5 | 102.37 | 52.70 | 119.92 | 102.02 | 117.77 | 88.70 |
| OVCAR−8 | 35.61 | −27.45 | 85.32 | 100.1 | 10.25 | 48.24 |
| SK−OV−3 | 104.46 | 1.64 | 93.55 | 95.03 | 102.05 | 67.28 |
| Renal cancer | ||||||
| A498 | 103.58 | 80.44 | 93.29 | ND | ND | ND |
| ACHN | 65.70 | −14.18 | 75.49 | 85.7 | 79.61 | 28.55 |
| RXF 393 | 97.24 | −46.34 | 100.91 | 87.68 | 72.63 | 69.20 |
| SN12C | 70.40 | −26.26 | 81.84 | 125.68 | 106.07 | −14.11 |
| UO−31 | 60.44 | −54.91 | 54.76 | 137.02 | 112.5 | 64.18 |
| Prostate cancer | ||||||
| PC−3 | 65.49 | −33.26 | 64.38 | 79.23 | 81.82 | 49.16 |
| DU−145 | 82.02 | −45.39 | 90.97 | 81.38 | 42.4 | 34.21 |
| Breast cancer | ||||||
| MCF7 | 48.82 | 0.70 | 68.38 | 76.73 | 92.37 | 51.60 |
| BT−549 | 73.94 | 37.99 | 77.38 | 104.21 | 93.67 | 12.26 |
| T−47D | 22.52 | −58.11 | 9.70 | 83.56 | 85.06 | 4.47 |
| MDA−MB−468 | −8.43 | −32.41 | −9.91 | 64.82 | −9.96 | −27.29 |
| Average G% | 68.19 | −4.69 | 74.32 | 95.45 | 65.81 | 35.49 |
ND, Not determined.
Nevertheless, in a few cell lines strong growth inhibition and/or lethality were observed by 3a, specifically in the two leukemia cell lines HL-60(TB) and MOLT-4 and the ovarian cancer cell lines OVCAR-3 and OVCAR-4. Whereas 3e strongly suppressed the growth of the leukemia cell line MOLT-4 and the ovarian cancer cell line OVCAR-4. Interestingly, 3b was able to induce a broad tumor cell death for most of the non-small cell lung cancer, colon cancer, CNS cancer, melanoma, ovarian cancer, renal cancer and breast cancer cell lines (Table 1). Even in the cell lines where it was not toxic, 3b typically exhibited considerably more potent growth inhibition than 3a and 3e. Upon testing 5a, 5b and 6, compound 6 showed a major anti-proliferative effect on most of the tested cell lines, with average G% of 35%, whereas compound 5a and 5b showed little growth suppressive activity against most of the tested human cancer cell lines with average G1% of 65 and 95% respectively. For example, 5a induced cell death in few cell lines, including two ovarian cancer cell lines OVCAR-3 and OVCAR-4, two leukemia cell lines HL-60 and MOLT-4, and one breast cancer cell line MDA-MB-468. In addition, the growth of the leukemia cell line CCRF-CEM was almost completely inhibited by 5a.
As compound 3b exhibited the most pronounced inhibitory activity, it was further selected for advanced five dose testing, in which the cytotoxic and/or growth inhibitory effects of the compound were quantified in vitro against the full NCI-60 panel of human tumor cell lines using a series of five 10-fold dilutions, ranging from 10−4 M to 10−8 M. Three dose response parameters were calculated for each cell line, GI50 (molar concentration required for 50% growth inhibition), TGI (molar concentration leading to total growth inhibition), and LC50 (molar concentration resulting in 50% cell death).
As Table 2 demonstrates, the results indicated that the selected active compound 3b showed broad spectrum anti-tumor activity against the nine tumor sub-panels tested. It exhibited strong growth inhibitory activity against all human cancer cell lines tested, with GI50 values mostly in the submicromolar range (0.14–10.47 μM, with an average across all cell lines tested of 0.24 μM and only 7 cell lines showing GI50 values > 1 μM) and TGI values typically in the low micromolar range (0.35–27.54 μM, with an average of 1.02 μM). Importantly, compound 3b was also able to induce cell death in the vast majority of the cancer cell lines. The LC50 values ranged between 0.85 and 72.44 μM, with an average of 7.58 μM. The cytotoxic effects were strongest in the non-small cell lung cancer lines NCI-H226 (LC50 = 0.87 μM), HOP-92 (LC50 = 1.00 μM) and NCI-H460 (LC50 = 1.48 μM), the CNS cancer line SNB-75 (LC50 = 1.29 μM), the melanoma line MALME-3M (LC50 = 0.85 μM), the ovarian cancer lines OVCAR-4 (LC50 = 1.51 μM) and OVCAR-3 (LC50 = 1.82 μM), the renal cancer line SN12C (LC50 = 1.35 μM) and the prostate cancer line PC-3 (LC50 = 1.58 μM). Notably, compound 3b was also lethal to nearly all of the remaining cancer cell lines, with only five of the lines in the NCI-60 panel not reaching 50% lethality within the concentration range tested (Table 2). In line with the results from the single dose screen, compound 3b was least efficient against leukemia cells, four of which exhibited an LC50 > 100 μM. The promising anticancer activity observed by these new compounds may be attributed to their activity towards the ERK kinase pathway in these tested tumor cell lines. Accordingly, we decided to go further and test this hypothesis.
Table 2.
The main cytotoxic parameters (GI50, TGI and LC50) of compound 3b in μM.
| Tumor cell lines | GI 50 | TGI | LC 50 |
|---|---|---|---|
| Leukemia | |||
| CCRF-CEM | 0.41 | 4.86 | > 100 |
| HL-60 (TB) | 1.70 | 5.25 | 34.76 |
| K-562 | 0.37 | 2.00 | 50.12 |
| MOLT-4 | 0.44 | 3.16 | > 100 |
| RPMI-8226 | 0.39 | 2.63 | > 100 |
| SR | 0.50 | 3.98 | > 100 |
| Non-small cell lung cancer | |||
| A549/ATCC | 1.02 | 2.88 | 8.32 |
| EKVX | 0.25 | 2.45 | 26.30 |
| HOP-62 | 0.20 | 0.65 | 3.24 |
| HOP-92 | 0.15 | 0.39 | 1.00 |
| NCI-H226 | 0.14 | 0.35 | 0.87 |
| NCI-H23 | 0.35 | 1.82 | 7.08 |
| NCI-H322M | 0.32 | 1.45 | 7.24 |
| NCI-H460 | 0.22 | 0.49 | 1.48 |
| NCI-H522 | 0.32 | 1.38 | 8.13 |
| Colon cancer | |||
| COLO 205 | 10.47 | 27.54 | 72.44 |
| HCC-2998 | 063 | 2.57 | 9.12 |
| HCT-116 | 0.31 | 1.10 | 4.57 |
| HCT-15 | 0.21 | 0.51 | 2.00 |
| HT29 | 3.72 | 12.88 | 39.81 |
| KM 12 | 0.45 | 3.02 | 27.54 |
| SW-620 | 0.33 | 1.00 | 7.59 |
| CNS cancer | |||
| SF-268 | 0.46 | 3.98 | 48.98 |
| SF-295 | 0.63 | 2.29 | 6.61 |
| SF-539 | 0.23 | 0.56 | 2.09 |
| SNB-19 | 0.31 | 1.10 | 6.17 |
| SNB-75 | 0.16 | 0.42 | 1.29 |
| U251 | 0.27 | 0.76 | 2.82 |
| Melanoma | |||
| LOX IMVI | 0.32 | 1.32 | 3.98 |
| MALME-3M | 0.23 | 044 | 0.85 |
| M14 | 0.40 | 1.51 | 4.57 |
| MDA-MB-435 | 0.30 | 1.20 | 4.07 |
| SK-MEL-2 | 1.74 | 5.13 | 19.95 |
| SK-MEL-28 | 0.34 | 1.23 | 4.68 |
| SK-MEL-5 | 1.30 | 1.20 | 3.72 |
| UACC-257 | 1.32 | 3.39 | 8.51 |
| UACC-62 | 0.28 | 1.15 | 3.98 |
| Ovarian cancer | |||
| IGROV1 | 0.22 | 0.74 | 3.47 |
| OVCAR-3 | 0.24 | 0.56 | 1.82 |
| OVCAR-4 | 0.17 | 0.45 | 1.51 |
| OVCAR-5 | 3.47 | 14.79 | 40.74 |
| OVCAR-8 | 0.39 | 2.29 | 30.20 |
| NCI/ADR-RES | 0.35 | 1.78 | 34.67 |
| SK-OV-3 | 0.35 | 1.78 | 5.62 |
| Renal cancer | |||
| 786–0 | 0.22 | 0.54 | 2.45 |
| A498 | 0.21 | 0.95 | 3.55 |
| ACHN | 0.20 | 0.58 | 2.63 |
| RXF 393 | 0.21 | 0.58 | 2.51 |
| SN12C | 0.20 | 0.47 | 1.35 |
| TK-10 | 0.38 | 1.10 | 6.03 |
| UO-31 | 0.19 | 0.60 | 2.51 |
| Prostate cancer | |||
| PC-3 | 0.18 | 0.46 | 1.58 |
| DU-145 | 0.28 | 0.78 | 12.30 |
| Breast cancer | |||
| MCF7 | 0.23 | 1.05 | 18.62 |
| MDA-MB-231/ATCC | 0.17 | 0.51 | 2.82 |
| HS 578 T | 0.26 | 1.38 | > 100 |
| BT-549 | 0.30 | 1.32 | 6.76 |
| T-47D | 0.26 | 0.98 | 5.13 |
| MDA-MB-468 | 0.25 | 0.87 | 4.57 |
2.2.2. The new compounds selectively inhibited ERK compared to other MAP kinases
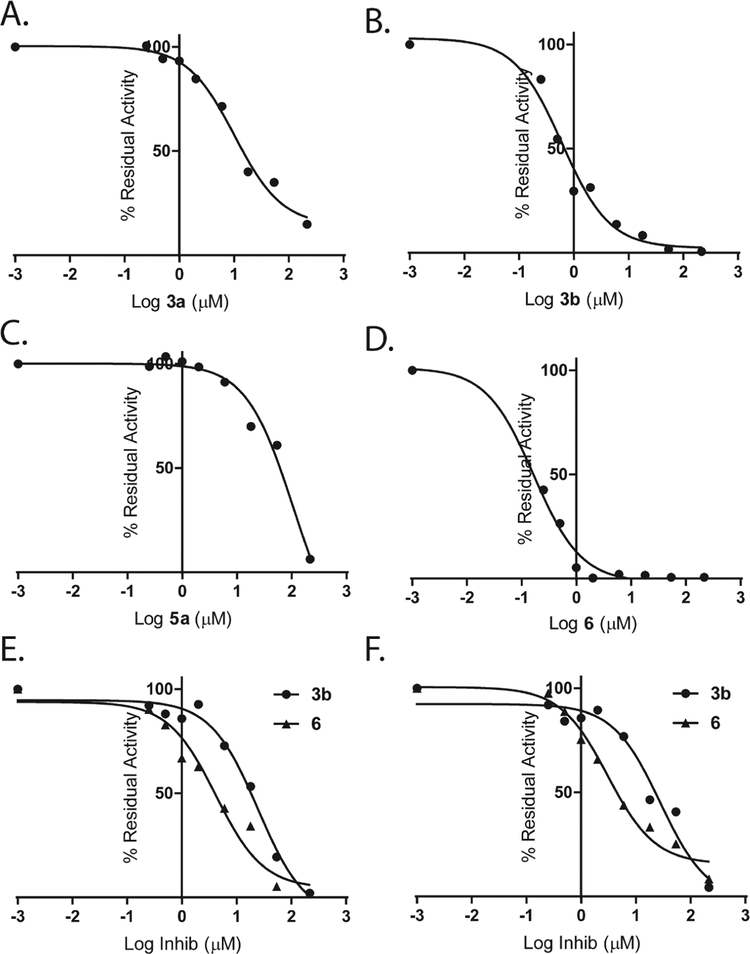
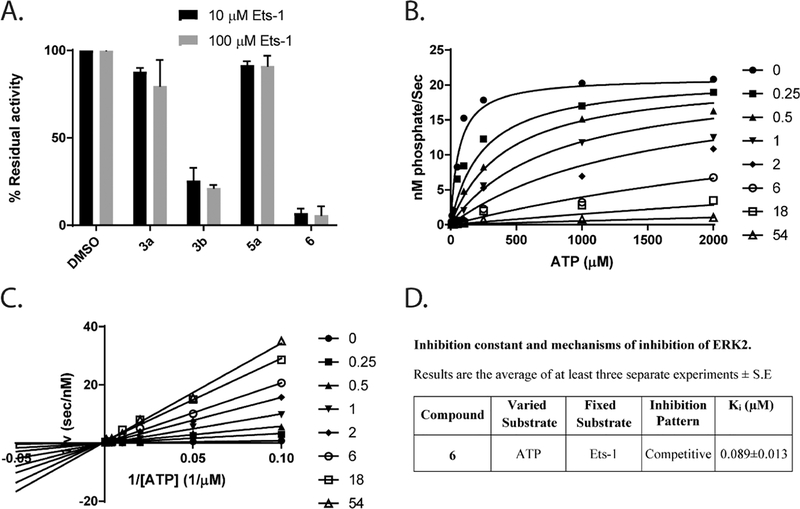
From the anticancer screening results (Table 1), we selected compounds 3a, 3b and 6 to study their molecular mechanism in vitro and in cells. Compound 5a was selected as a negative control. First, in an in vitro kinase assay, we tested the ability of these compounds to inhibit the activity of ERK1 and ERK2 towards a downstream substrate Ets-1 in the presence of 100 μM ATP. Fig. 2A–D show dose response curves for ERK2 inhibition.
Fig. 2.
3b and 6 are potent ERK inhibitors in in vitro cell-free kinase assays. 3a (A), 3b (B), 5a (C) and 6 (D) inhibited Ets-1 phosphorylation by ERK2 in the in vitro kinases assay. (E) The effect of 3b and 6 on the ability of JNK2 to phosphorylate GST-c-Jun (1–221). (F) The effect of 3b and 6 on the ability of p38MAPKα to phosphorylate GST-ATF2 (1–115). Data were fitted to Eq. (1) (Methods section).
As shown in Table 3, 3a, 3b and 6 inhibited ERK1 with IC50s of 9.12, 0.5 and 0.19 μM respectively while 3c inhibited ERK1 with much higher IC50 of 95 μM. Similar activities were observed for ERK2 with IC50s of 10.2, 0.6 and 0.16 μM respectively while 5a inhibited ERK2 with IC50 > 100 μM. To profile the selectivity of these compounds we examined their ability to inhibit other MAP kinases, including JNK2 or p38MAPKα. Compounds 3a and 5a did not show any ability to inhibit JNK2 or p38MAPKα (Table 3). Whereas compounds 3b and 6 showed little tendency to inhibit JNK2 and p38MAPKα (Fig. 2E and F). Compound 3b inhibited JNK2 and p38MAPKα with IC50s of 24 μM and 28 μM, respectively, revealing a selectivity of around 50-fold towards ERK. Compound 6 inhibited JNK2 and p38MAPKα with IC50s with IC50s of 4.2 and 3.1 μM respectively, corresponding to around 20-fold selectivity towards ERK.
Table 3.
Selectivity of the tested compounds.
| 3a IC50a (μM) |
3b IC50a (μM) |
5a IC50a (μM) |
6 IC50a (μM) |
|
|---|---|---|---|---|
| ERK1 (100 μiM ATP) | 9.12 ± 1.1 | 0.5 ± 0.09 | 95 ± 10.9 | 0.19 ± 0.01 |
| ERK2 (100 μM ATP) | 10.2 ± 1.2 | 0.6 ± 0.1 | 108 ± 12.2 | 0.16 ± 0.02 |
| ERK2 (10 μM ATP) | 0.2 ± 0.01 | 0.086 ± 0.005 | ||
| JNK2 | > 200 | 24 ± 1.24 | > 200 | 4.2 ± 0.9 |
| p38MAPKα | > 200 | 27.9 ± 1.5 | > 200 | 3.1 ± 0.5 |
| A375 Cell Viability | 3.7 ± 0.6 | 0.13 ± 0.03 |
IC50 Determined as described in Methods section, Data were fitted to Eq. (1).
Interestingly, the differential ability of the tested compounds to inhibit ERK activity in-vitro correlates to their cytotoxic activity that shown in Table 1. 3b and 6 showed the most potent ERK inhibition and the most pronounced anticancer activity against most of the tested cell lines (average G% of −4.5% and 35% respectively) while compound 3a showed a modest ability to inhibit ERK activity with IC50 of around 9 μM (Table 3) and weak ability to impact the cancer cells growth at 10 μM concentration with average %G of 68% (Table 1). The inability of 5a to exhibit any considerable cytotoxic or anti-proliferative activity (average %G of 95%) was mainly attributed to its limited potency towards inhibiting ERK with IC50s ≥ 100 μM (Tables 1 and 3).
2.2.3. In vitro studies of the new inhibitors identify a mechanism of ATP-competitive inhibition
To understand the mechanism of ERK inhibition by the new compounds, we initially tested the effect of varying the concentration of Ets-1 on the ability of the inhibitors to suppress ERK2 activity. Using excess of Ets-1 (100 μM) did not rescue the inhibition of ERK2 by 5 μM of 3a, 3b or 6 (Fig. 3A), as it showed a similar susceptibility to the inhibitors in presence of 10 μM Ets-1 (the Km of Ets-1 towards ERK2 ~ 10 μM[33]), suggesting that the binding of the inhibitor does not impact the binding of Ets-1 to ERK2, which binds at the D and F-recruitment sites of ERK2 [34,35]. In order to examine the possibility that the inhibitors may bind the active site of ERK where ATP is known to bind, we determined the IC50s of 3b and 6 towards ERK2 in the presence of 100 μM or 10 μM of ATP (the Km of ATP towards ERK2 ~ 100 μM[34]).
Fig. 3.
Mechanism of ERK2 inhibition by the newly synthesized compounds. (A) Higher concentrations of Ets-1 did not impact the ability of 3a, 3b, 5a and 6 to inhibit its phosphorylation by ERK2. 10 μM of each inhibitor were added to 2 nM of activated ERK2 in the presence of either 1 or 10-fold excess over the Km of Ets-1 and 100 μM ATP (B) Inhibition of ERK2 activity by 6 at varied fixed concentrations of Mg-ATP (10–2000 μM) and 50 μM Ets-1 and (C) the double reciprocal plot of 1/ V 0 vs 1/[Mg-ATP] at varied fixed concentrations of 6 (0–54 μM) and 50 μM Ets-1. Initial velocities were measured using various (10–2000 μM) concentrations of Mg-ATP. The data were fitted to a model of competitive inhibition according to Eq. (2), where , , (D) Inhibition constant and mechanism of inhibition of ERK2 by 6.
The results shown in Table 3 suggest that decreasing the concentration of ATP increased the ‘apparent’ potency of the inhibitor towards ERK2. Compound 3b showed an IC50 of 0.6 μM at 100 μM ATP and 0.2 μM in the presence of 10 μM ATP, while 6 showed an IC50 of 0.16 μM at 100 μM ATP and 0.09 μM in presence of 10 μM ATP. These observations represent the first evidence that these inhibitors bind ERK in the active site, where they are able to prevent ATP from reaching ERK. Furthermore, and to confirm this mechanism, we decided to determine the observed rate constant, kobs, over a range of ATP concentrations at different fixed concentrations of 6, in the presence of a saturating concentration of Ets-1 (50 μM) (Fig. 3B). The double reciprocal plot derived from this study (Fig. 3C) is consistent with a mechanism of competitive inhibition, where Ki ~ 0.09 ± 0.01 μM. According to this model (Fig. 3D). Compound 6 binds ERK and abrogates its affinity for ATP without affecting kcat. The binding mode of 6 to the ERK active site impedes the recognition of ATP by the enzyme.
2.2.4. Structure activity relationship
Based on the structural features of the newly synthesized compounds, their cytotoxic activities and their ability to inhibit ERK2 activity in vitro (Table 4), it is obvious that substituents on the quinolone moiety can alter the biological activity of these inhibitors. For example, substituent at N-quinoline moiety (R1 of 3a-f and R3 of 5a-b) seems to decrease their ability to inhibit ERK2 and to suppress their cytotoxic activity. Compound 3a (R1 = H) showed better ability to inhibit ERK2 and to suppress tumor cell growth, compared to 3e and 3f that (R1 = methyl and ethyl respectively). This demonstrates the important role played by the free NH in the ability of the compounds to inhibit ERK2 and to exhibit anti-tumor activity. Noticeably, the size of R1 is inversely proportion to the ability to inhibit ERK2 in vitro, especially if we compared the biological activities of 3a, 3e and 3f. Moreover, there is evidence that substituents at the position-2 of the quinolone ring can alter the biological activities of these new derivatives. Interestingly, 3b, that carries an electron withdrawing Cl at the R2 position showed very high potency towards ERK2 inhibition and tumor cell growth suppression (Table 4), while changing this group to Br (3c) or Me (3d) completely abrogated the ability of these inhibitors to affect ERK2 activity in vitro. Finally, compound 6 that formed of a mixture of syn/anti isomers of 3-(1,4-dioxo-1,4-dihydronaphthalen-2-yl)-4-hydroxy-6-phenyl-2H-pyrano[3,2-c]-quinoline-2,5(6H)-dione, represents a different scaffolds from 3a-f or 5a-b, its potent inhibition of ERK2 activity (IC50 ~ 0.16 μM) and tumor growth (average G% ~ 35% at 10 μM treatment) suggest that its binding mode to ERK2 and its selectivity profile can be different from compounds 3 and 5.
Table 4.
Structure Activity Relationship.
 |
 |
ERK2 IC50a(μM) at 100 μM ATP /10mM Ets-1 |
NCI Cytotoxicity Screening Average G% |
|||
|---|---|---|---|---|---|---|
| R1 | R2 | R3 | R4 | |||
| 3a | −H | −H | 10.2 ± 1.2 | 68 | ||
| 3b | −H | −Cl | 0.6 ± 0.1 | −4.5 | ||
| 3c | −H | −Br | > 100 | ND | ||
| 3d | −H | −Me | 59 ± 8.2 | ND | ||
| 3e | −Me | −H | 21 ± 5.6 | 74 | ||
| 3f | −Et | −H | > 100 | ND | ||
| 5a | −Me | −H | > 100 | 95 | ||
| 5b | −Et | −H | 8.3 ± 1.5 | 65 | ||
| 6 | 0.16 ± 0.02 | 35 | ||||
2.2.5. Cellular studies in the BRAFV600E mutant human melanoma cell line A375
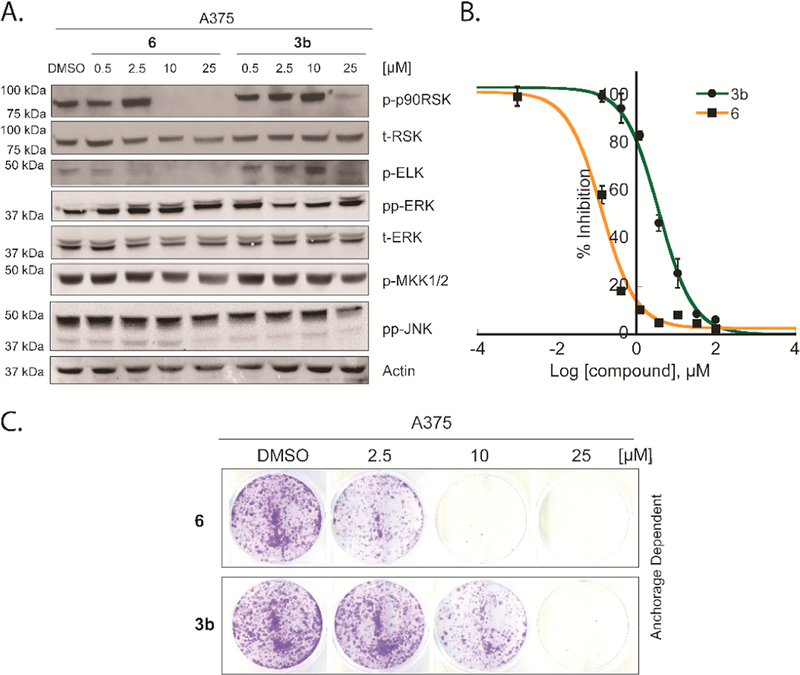
Inhibition of ERK activity towards the downstream substrates – Melanomas that carry activating BRAF and NRAS mutations exhibit constitutive ERK activity that is known to drive tumorigenesis. Inhibition of ERK activity has been reported to alter the proliferation of melanoma cell lines and inhibit the growth of melanoma tumors [36].
Treatment of A375 cells (melanoma cell line expressing BRAFV600E) with 3b or 6, followed by EGF stimulation resulted in a dose-dependent inhibition of the ERK substrates (p90RSK and ELK) phosphorylation. With no effect on ERK or MKK½ phosphorylation or the JNK pathway (Fig. 4A). 10 μM of compound 6 was enough to inhibit p-90RSK phosphorylation while 2.5 μM was enough to inhibit ELK phosphorylation. Compound 3b inhibited p90RSK at a concentration of 25 μM, while inhibited p-ELK at 10 μM. Notably, the cellular potencies of the tested compounds matched their relative activity in the in vitro kinase assay.
Fig. 4.
Effect of compounds on BRAF-mutant A375 melanoma cell line A. 6 and 3b inhibited Elk and p90RSK phosphorylation in the BRAF-mutant A375 melanoma cell line. A375 melanoma cells were treated with a DMSO control, different concentrations of 6 or 3b (0.5–25 μM) for 12 h in serum free media. The MAP kinase pathway was then induced by the addition of EGF (100 nM) for 15 min before lysing the cells. Lysates were fractionated by SDS PAGE (10% gel) and subjected to western blot analysis in order to detect the phosphorylated forms of p-90RSK, ELK, ERK, MEK½ and JNK. B. 6 is a more potent inhibitor of A375 melanoma cell viability than 3b. Cells were treated with different doses of each inhibitor and incubated for 72 h before analysis by the MTS assay. C. Compounds 6 and 3b inhibited BRAF-mutant A375 cells anchorage-dependent growth in a dose dependent manner (Cells were treated as described in the methods section).
Effect of the inhibitors on A375 cell viability and colonies formation – It has been reported that the clinical efficacy that can be produced by any drug targeting the ERK pathway becomes significant after at least 80% ERK signaling is suppressed [37] and the degree of ERK inhibition is directly correlated to the inhibition of melanoma cell proliferation [36]. The western blotting experiments emphasized the ability of the new inhibitors to inhibit ERK activity in the A375 cell line. To assess the selectivity of 3b and 6 in this cell line, we evaluated their antiproliferative activity. 3b and 6 inhibited A375 proliferation (MTS assay) with IC50s of 3.7 and 0.13 μM, respectively (Fig. 4B and Table 3). Next, we tested the biological effect of these inhibitors on anchorage dependent colony formation assay (Fig. 4C) that is known to be regulated by ERK kinase activity in melanoma cell lines [38]. Compound 6 started to inhibit melanoma cell colony forming ability at a concentration of 2.5 μM while 3b showed inhibition at 10 μM.
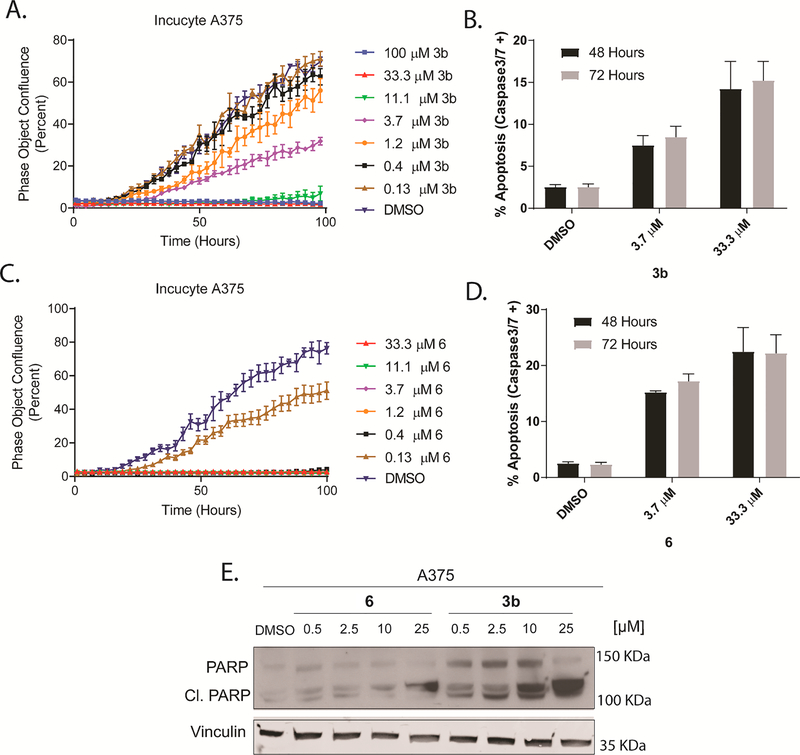
Effect of the compounds on the Proliferation rate of A375 Melanoma cell line – ERK has been shown to directly regulate over 100 known cellular targets (e.g. as RSK, MYC and BIM) that together control tumor cell proliferation, survival, and apoptosis [22]. To test the new inhibitors in a real time proliferation study A375 cells were treated with different doses of each inhibitor and the real time growth confluence was recorded every 1 h for a total of 90 h using an IncuCyte Zoom microscope. 3b and 6 showed dose-dependent inhibition of A375 proliferation (Fig. 5A and C) with compound 6 exhibiting higher potency, if compared to 3b.
Fig. 5.
The tested compounds inhibited BRAF-mutant A375 melanoma cell proliferation and induced apoptosis. (A and C) IncuCyte analysis of real time growth confluence (imaged every 1 h) showed dose-response inhibition of A375 growth rates by 3b and 6, cells were treated with different doses of each inhibitor before starting the IncuCyte imaging. Error bars: SEM (n = 3). (B and D) Treatment of 3b and 6 increases the rate of apoptosis of BRAF-mutant A375 melanoma cells in a dose-dependent manner. Cells were treated with different doses of each inhibitor, apoptosis was assessed using the lncuCyte Caspase 3/7 reagent and lncuCyte® ZOOM equipment with a × 10 objective at indicated time points. (E) 6 and 3b induced PARP cleavage in A375 cells. Cells were exposed to different doses of each inhibitor for 12 h before cell lysis. Protein expression of PARP and cleaved PARP were analyzed by western blot using vinculin as a loading control. (Experiment was repeated two times).
Apoptosis induction – Melanomas that carry BRAF mutations tend to be more resistant to apoptosis. ERK inhibition has been shown to induce apoptosis in such resistant melanoma [39].
To determine the ability of the new compounds to induce apoptosis, we performed a single cell assay using the IncuCyte microscope. A375 cells were treated with different doses of each inhibitor, plus a fixed concentration of the IncuCyte Caspase3/7 reagent that quantifies the activity of cellular caspase 3/7. Stained cells were imaged and counted using the IncuCyte microscope. Interestingly, compounds 3b and 6 exhibited activation of caspase 3/7 in A375 cell line, at doses of 3.7 and 33.3 μM after 48 and 72 h of incubation (Fig. 5B and D). Moreover, treatment of A375 cells with compound 3b or 6 resulted in an elevated level of the cleaved PARP (Fig. 5E), another marker of apoptosis induction [39]. These data support the notion that the induction of apoptosis by these inhibitors is a major mechanism of death that results from ERK pathway inhibition.
2.3. Molecular docking calculations
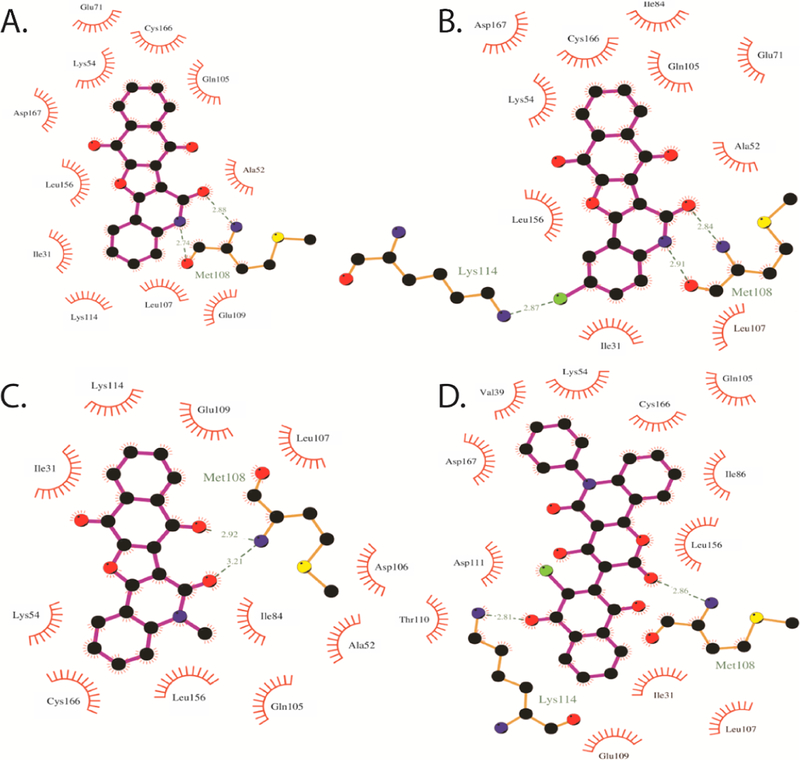
To reveal the binding features of the synthesized compounds with ERK2, molecular docking calculations were performed on the most potent compounds towards the ATP-active site of ERK2 using Autodock4.2 software.[40]. Autodock performance was first validated on an ERK2 crystal structure in complex with a tetra-hydropyridopyrimidine amine derivative (N-[(1S)-1-(3-chloro-4-fluorophenyl)-2-hydroxyethyl]-2-(tetrahydro-2H-pyran-4-ylamino)-5,8-dihydropyrido[3,4-d]pyrimidine-7(6H)-carboxamide) or 2SH that been reported previously as an ATP competitive inhibitor of ERK [41] (PDB code: 4O6E [41]). Self-docking of 2SH was performed and the corresponding binding mode was investigated. Compared to the crystal structure of 2SH-ERK2 complex, Autodock accurately predicted the binding mode of 2SH inside the ERK2 active site, forming five essential hydrogen bonds with Lys42, Met108, Lys114 and Asp167. Molecular docking of the synthesized compounds 3a, 3b, 3e and 6 towards the active site of ERK2 was then performed and the corresponding binding energies and features were investigated. According to the docked 3a-ERK2 structure (Fig. 6), 3a forms two essential hydrogen bonds with Met108 with bond lengths of 2.74 and 2.88 Å. Further interactions including van der Waals and hydrophobic interactions were observed, giving a total docking score of −8.34 kcal/mol. Introduction of a chlorine atom in the 2-position of the quinolone ring increased the binding energy of compound 3b, relative to the unsubstituted compound 3a, giving a binding score of −8.82 kcal/mol. The higher binding energy of compound 3b was attributed to the formation of a further hydrogen bond between chlorine atom and the Lys114.
Fig. 6.
Molecular docking of the synthesized compounds 3a, 3b, 3e and 6 towards ATP-active site of ERK2.
Additionally, introduction of a methyl group on the N-quinoline moiety resulted in lower binding energy of compound 3e (calc. −7.88 kcal/mol). This demonstrates the important role played by the free NH for the formation of the essential hydrogen bond with Met108. Compared to compound 3 derivatives, compound 6 showed better affinity towards ERK2 with docking score of −9.22 kcal/mol. The investigation of the binding mode of compound 6 with ERK2, suggested a different binding mode from compound 3 derivatives. The molecular docking model revealed that compound 6 formed two hydrogen bonds between the pyrano ring and Met108 and the naphthalene ring and Lys114, with bond lengths of 2.86 and 2.81 Å, respectively (Fig. 6).
2.4. Drug likeliness and oral bioavailability
Application of in silico computational technology during drug discovery and development offers considerable potential for reducing the number of experimental studies required for compound selection and development. Lipinski’s rule of five is commonly used in drug design and development to predict oral bioavailability of potential lead or drug molecules.
Molinspiration cheminformatics, (http://www.molinspiration.com) online software calculation toolkit, was used to determine the physicochemical parameters e.g. lipinski’s parameters, topological polar surface area (TPSA) and percentage of absorption (% ABS) of the synthesized compounds. The absorption percentage was calculated by using the following formula: % ABS = 109 − [0.345× TPSA] [42]. According to Lipinski’s “rule of five”, a candidate molecule will likely to be orally active, if the molecule satisfies the following rule [43]: (i) the calculated octanol/water partition coefficient (Log P) < 5, (ii) the molecular weight is under 500, (iii) number of hydrogen bond acceptors (notably N and O) ≤10, (iv) number of hydrogen bond donors (OH and NH groups) ≤5. Molecules violating more than one of these rules may have problems with bioavailability (Table 5).[43] Gratifyingly, on close inspection of Table 5, all the synthesized compounds were found to be in compliance with Lipinski rule of five. The milog Pa values of all compounds were found below five, suggesting that the molecules have good permeability across the cell membrane which in turn is needed for generation of bioactivity. Molecular weight was found to be less than 500 and thus these molecules are predicted to be easily transported, diffused and absorbed. In addition, the number of hydrogen bond donors and acceptors were less than 5 and 10, respectively, thus these values are in accordance with Lipinski’s rules. TRSA of all titled compounds are observed in the range of (80.14–106.59 Å) which is below the limit 160 Å, which is a very good indicator of the bioavailability of the synthesized compounds. Meanwhile, the calculated percentage of absorption of all derivatives ranged between (72.23–85.09%), demonstrating that they may have good cell membrane permeability and oral bioavailability.
Table 5.
Pharmacokinetic prediction of the synthesized compounds by Molinspiration v2016.10.
| Compound | miLog Pa | TPSAb | nONc | nOHNHd | nviolation | Nrotbe | Mol Volf | MWtg | %ABSh |
|---|---|---|---|---|---|---|---|---|---|
| 3a | 3.08 | 80.14 | 5 | 1 | 0 | 0 | 256.10 | 315.28 | 81.35% |
| 3b | 3.73 | 80.14 | 5 | 1 | 0 | 0 | 269.63 | 349.73 | 81.35% |
| 3c | 3.86 | 80.14 | 5 | 1 | 0 | 0 | 273.98 | 394.18 | 81.35% |
| 3d | 3.50 | 80.14 | 5 | 1 | 0 | 0 | 272.66 | 329.31 | 81.35% |
| 3e | 3.59 | 69.29 | 5 | 0 | 0 | 0 | 273.04 | 329.31 | 85.09% |
| 3f | 3.97 | 69.29 | 5 | 0 | 0 | 1 | 289.84 | 343.34 | 85.09% |
| 5a | 4.39 | 99.50 | 7 | 0 | 0 | 1 | 334.38 | 411.37 | 74.67% |
| 5b | 4.02 | 99.50 | 7 | 0 | 0 | 0 | 317.58 | 397.34 | 74.67% |
| 6 | 4.93 | 106.59 | 7 | 1 | 0 | 2 | 395.84 | 495.87 | 72.23% |
Logarithm of partition coefficient between n-octanol and water (miLogP).
Topological polar surface area (TPSA).
Number of hydrogen bond acceptors (n-ON).
Number of hydrogen bond donors (n-OHNH).
Number of rotatable bonds (n-rotb).
Molecular Volume.
Molecular weight (MWt).
Percentage of absorption (%ABS).
3. Conclusions
A group of novel fused naphthofuro[3,2-c]quinoline-6,7,12-triones and naphthofuropyrano [3,2-c]quinoline-6,7,8,13-tetraones were synthesized and characterized based on different spectral data, as well as X-ray crystallography for some of the compounds. Six compounds were selected for in vitro anticancer screening against the NCI-60 panel of cancer cell lines. Most of the tested compounds showed growth inhibitory activity against at least a subset of the human tumor cell lines at a concentration of 10 μM. Compounds, 3b and 6 had the strongest cytotoxic activity, against the majority of the tested cell lines. To confirm the targeting of the ERK pathway by these inhibitors, we tested all the derivatives in an in vitro kinase assay for ERK2. 3b and 6 showed the most potent activity towards ERK2 with IC50s of 0.6 and 0.16 μM respectively. 3b exhibited 50-fold selectivity towards ERK2, when compared to other MAP kinases while 6 showed 20-fold selectivity. The in vitro kinetic mechanism studies revealed an ATP competitive mechanism of inhibition by the new compounds towards ERK. The structure-activity study emphasized that substitution within the quinolone can alter the activity of the compounds towards ERK. The promising potency of 3b and 6 towards ERK, makes it important to investigate the possible utilization of these inhibitors in RAF mutant melanoma. As a model, we used the A375 cell line that carries the BRAFV600E activating mutant that is known to hyperactivate the ERK pathway. 3b and 6 inhibited ERK substrate phosphorylation in this cell line in a dose-dependent manner. Treating the cells with these inhibitors resulted in potent inhibition of cell proliferation and real time cell growth. 3b and 6 were potent enough to suppress A375 colony formation (a marker for tumorigenesis), that is known to be regulated by the ERK pathway and induced apoptosis. Finally, the molecular docking results indicated that compounds 3b and 6 have better affinity towards the ATP-active site of ERK2 with docking scores of −8.82 and −9.22, respectively. Furthermore, this work demonstrates that all the synthesized compounds adhere to the Lipinski rule of five, and the synthesized compounds are expected to have good oral bioavailability and a good pharmacokinetic profile. The inhibitor’s ability to manipulate ERK signaling in this type of melanoma cell line that harbors constitutive ERK activity, suggests that they represent a valuable group of ERK inhibitors that can be improved by further medicinal chemistry efforts.
4. Material and methods
4.1. Chemistry
Melting points, using open glass capillaries on a Gallenkamp melting point apparatus (Weiss-Gallenkamp, Loughborough, UK), were measured without applying any corrections. The IR spectra were recorded from potassium bromide disks with a FT device. 1H and 13C NMR spectra were measured with a Bruker Avance (400 MHz for 1H, and 100 MHz for 13C), Institute of Organic Chemistry, Karlsruhe Institute of Technology (KIT), Karlsruhe, Germany. The 1H and 13C chemical shifts are given relative to internal standard TMS. Coupling constants are stated in Hz, 1H-coupled 13C spectra were measured using gated decoupling. Correlations were established using 1H–1H COSY, HMBC and HSQC experiments. Mass spectrometry was performed by electron impact at 70 eV, (FAB-MS): Finnigan MAT 95, Institute of Organic Chemistry, Karlsruhe Institute of Technology, Karlsruhe, Germany. Mass spectra were recorded on Finnigan Fab, 70 eV. TLC was performed on analytical Merck 9385 silica aluminum sheets (Kieselgel60) with Pf254 indicator; TLC’s were viewed λmax = 254 nm. Elemental analyses were carried out at the Microanalytical Center, Cairo University, Egypt.
4.1.1. Starting materials
1,6-Disubstituted-4-hydroxy-quinoline-2-ones 1a-f were prepared as previously described. [44,45] 4-Hydroxy-pyrano[3,2-c]quinoline-4,5(6H)-diones 4a-c were prepared according to published procedures[46]. 2,3-Dichloro-1,4-naphthoquinone (2) (Aldrich) was purchased from Aldrich.
4.1.2. General procedure
A mixture of 1a-f (0.1 mol) or 4a-c (0.1 mol) and DCHNQ (2, 0.1 mol) in 20 mL (10% NaOEt as 1 g Na in 10 mL EtOH), pyridine (10 mL) or DMF (30 mL), was refluxed with stirring. The time peroid until the reactants had disappeared, as mentioned in Scheme 1A and B, was monitored by TLC. The resulting precipitate was filtered off, washed with ethanol, and recrystallized to give pure crystals of 3a-f, 5a,b and 6.
4.1.2.1. Naphtho[2′,3′:4,5]furo[3,2-c]quinoline-6,7,12(5H)-trione (3a).
Yellow crystals (DMF/EtOH), yield: 0.284 g (90%), mp > 360 °C; IR (KBr): 3210 (NH), 1700, 1660 (CO), 1596 (Ar–C=C) cm−1; 1H NMR (400 MHz, DMSO-d6): δH 7.50–7.56 (m, 2H, Ar–H), 7.65–7.70 (m, 2H, Ar–H), 7.75–7.80 (m, 2H, Ar–H), 8.00 (s, 1H, NH), 8.12–8.20 (m, 2H, H-9,10); 13C NMR (100, DMSO-d6): δC 176.2 (C-7), 173.0 (C-12), 169.1 (C-13a), 160.8 (C-6), 156.3 (C-12a), 136.4 (C-4a), 133.1 (C-6b), 132.7 (C-9, C-10), 132.0 (C-11a), 130.7 (C-6a), 128.9 (C-3), 128.4 (C-7a, C-13b), 127.7 (C-1), 126.6 (C-8, C-11), 124.8 (C-2), 116.0 (C-4); MS (Fab, 70 eV,%): m/z 315 (M+, 100). Anal. Calcd. for C19H9NO4: C, 72.38; H, 2.88; N, 4.44. Found: C, 72.51; H,3.01; N, 4.39.
4.1.2.2. 2-Chloronaphtho[2′,3′:4,5]furo[3,2-c]quinoline-6,7,12(5H)-trione (3b).
Yellow crystals (DMF/H2O), yield: 0.307 g (88%), mp = 330–2 °C; IR (KBr): 3210 (NH), 3065 (Ar–CH), 1700, 1667 (CO), 1593 (Ar–C=C) cm−1; 1H NMR (400 MHz, DMSO-d6): δH 7.50 (d, 1H, J = 0.7 Hz, H-1), 7.65 (dd, 1H, J = 7.0, 0.8 Hz, H-3), 7.86–7.90 (m, 3H, Ar–H), 8.10 (s, 1H, NH), 8.15–8.20 (m, 2H, Ar–CH); 13C NMR (100 MHz, DMSO-d6): δC 176.4 (C-7), 173.0 (C-12), 169.2 (C-13a), 160.2 (C-6), 156.3 (C-12a), 134.5 (C-4a), 133.1 (C-6b), 132.7 (C-9), 132.0 (C-10), 132.2 (C-11a), 130.3 (C-6a), 130.4 (C-2), 129.8 (C-13b), 129.0 (C-3), 128.4 (C-7a), 127.6 (C-1), 126.6 (C-8, C-11), 123.5 (C-4); MS (Fab, 70 eV,%): m/z 351 (M+2, 38), 349 (M+, 100). Anal. Calcd. for C19H8ClNO4: C, 65.25; H, 2.31; N, 4.01. Found: C, 65.36; H, 2.19; N, 3.98.
4.1.2.3. 2-Bromonaphtho[2′,3′:4,5]furo[3,2-c]quinoline-6,7,12(5H)-trione (3c).
Yellow crystals (DMF/DMSO), yield: 0.334 g (85%), mp = 330–2 °C; IR (KBr): 3195 (NH), 3065 (Ar–CH), 1696, 1667 (CO), 1593 (Ar–CH) cm−1; 1H NMR (400 MHz, DMSO-d6): δH 7.37 (d, 1H, J = 0.8 Hz, H-1), 7.75 (dd, 1H, J = 7.0, 0.8 Hz, H-3), 7.86–7.92 (m, 3H, Ar–H), 8.10–8.15 (m, 2H, Ar–H), 8.25 (bs, 1H, NH); 13C NMR (100 MHz, DMSO-d6): δC 176.5 (C-7), 173.0 (C-12), 169.2 (C-13a), 160.1 (C-6), 156.0 (C-12a), 135.2 (C-4a), 133.1 (C-1), 132.8 (C-6b), 132.4 (C-9, C-10), 131.6 (C-11a), 131.2 (C-3), 130.5 (C-6a), 130.0 (C-13b), 128.2 (C-7a), 126.6 (C-8, C-11), 124.2 (C-4), 117.4 (C-2); MS (Fab, 70 eV,%): m/z 392 (M+, 100), 394 (M+, 36), 351 (65), 352 (55). Anal. Calcd. for C19H8BrNO4: C, 57.89; H, 2.05; N, 3.55. Found: C, 58.02; H, 2.11; N, 3.47.
4.1.2.4. 2-Methylnaphtho[2′,3′:4,5]furo[3,2-c]quinoline-6,7,12(5H)-trione (3d).
Yellow crystals (CHCl3/hexane), yield: 0.302 g (95%), mp > 360 °C; IR (KBr): 3194 (NH), 3060 (Ar–CH), 2921−2810 (Aliph.–CH), 1687, 1665 (CO), 1590 (Ar–C=C) cm−1; 1H NMR (400 MHz, DMSO-d6): δH 2.30 (s, 3H, CH3), 7.20 (dd, 1H, J = 7.0,1.0 Hz, H-3), 7.72 (d, 1H, J = 0.8 Hz, H-1), 7.80–7.90 (m, 3H, Ar–CH),8.20–8.25 (m, 3H, Ar–H, NH); 13C NMR (100 MHz, DMSO-d6): δC 176.0 (C-7), 173.0 (C-12), 169. 0 (C-13a), 160.8 (C-6), 156.3 (C-12a), 134.1 (C-5), 133.2 (C-4a), 133.0 (C-6b), 132.6 (C-9, C-10), 132.0 (C-11a), 130.4 (C-6a), 130.2 (C-1), 129.0 (C-3), 128.2 (C-7a), 128.0 (C-13b), 126.5 (C-8, C-11), 122.0 (C-4), 21.6 (CH3); MS (Fab, 70 eV,%): m/z 329 (M+, 100), 90 (10). Anal. Calcd. for C20H11NO4: C, 72.95; H, 3.37; N, 4.25. Found: C, 72.88; H, 3.55; N, 4.13.
4.1.2.5. 5-Methylnaphtho[2′,3′:4,5]furo[3,2-c]quinoline-6,7,12(5H)-trione (3e).
Yellow crystals DMF/MeOH, yield: 0.282 g (91%), mp 350–2 °C; IR (KBr): 3060 (Ar–CH), 2940–2870 (Aliph.-CH). 1699, 1671 (CO), 1590 (Ar–C=C) cm−1; 1H NMR (400 MHz, DMSO-d6): δH 3.75 (s, 3H, CH3), 7.40–7.50 (m, 2H, Ar–CH), 7.70–7.73 (m, 2H, Ar–CH), 8.00–8.34 (m, 4H, Ar–H); 13C NMR (100 MHz, DMSO-d6): δC 176.5 (C-7), 173.0 (C-12), 169.2 (C-13a), 161.1 (C-6), 156.3 (C-12a), 134.3 (C-4a), 133.1 (C-6b), 132.7 (C-9, C-10), 132.0 (C-11a), 130.7 (C-6a), 128.9 (C-3), 128.4 (C-7a, C-13b), 127.7 (C-1), 126.6 (C-8, C-11), 124.8 (C-2), 115.5 (C-4), 37.9 (CH3); MS (Fab, 70 eV,%): m/z 329 (M+, 100), 90 (15). Anal. Calcd. for C20H11NO4: C, 72.95; H, 3.37; N, 4.25. Found: C, 73.08; H, 3.33; N, 4.43.
4.1.2.6. 2-Ethylnaphtho[2′,3′:4,5]furo[3,2-c]quinoline-6,7,12(5H)-trione (3f).
Yellow crystals, yield: 0.326 g (95%), mp 310 °C; IR (KBr): 3036 (Ar–CH), 2290 (Aliph.-CH), 1688, 1670 (CO), 1582 (C=C) cm−1; 1H NMR (400 MHz, DMSO-d6): δH 1.30 (t, 3H, J = 7.0 Hz, CH3), 4.47 (q, 2H, CH2, J = 7.0 Hz), 7.45–7.50 (m, 2H, Ar–CH), 7.65–7.70 (m, 2H, Ar–H), 7.88–7.94 (m, 4H, Ar–H); 13C NMR (100 MHz, DMSO-d6): δC 176.0 (C-7), 173.0 (C-12), 169.0 (C-13a), 160.4 (C-6), 156.2 (C-12a), 134.2 (C-4a), 133.0 (C-6b), 132.7 (C-9, C-10), 132.0 (C-11a), 130.8 (C-6a), 128.6 (C-3), 128.4 (C-7a, C-13b), 127.7 (C-1), 126.6 (C-8, C-11), 124.8 (C-2), 115.5 (C-4), 47.8 (CH2), 13.7 (CH3); MS (Fab, 70 eV,%): m/z 343 (M+, 100). Anal. Calcd. for C21H13NO4: C, 73.46; H, 3.82; N, 4.08. Found: C, 73.29; H, 3.75; N, 4.25.
4.1.2.7. 5-Methyl-5H-naphthalen[2″,3″:4′,5′]furo[3′,2′:4′,5,6]pyrano [3,2-c]quinoline-6,7,8,13-tetraone (5a).
Yellow crystals, yield: 0.342 g (86%), mp 315 °C; IR (KBr): 3044 (Ar–CH), 2985 (Aliph.-CH), 1730, 1715, 1679 (CO), 1600, 1594 (Ar–C=C) cm−1; 1H NMR (400 MHz, DMSO-d6): δH 3.4 (s, 3H, N-CH3), 7.45–8.21 (m, 8H, Ar–H); 13C NMR (100 MHz, DMSO-d6): δC 176.0, 173.0, 170.2, 162.0 (CO), 160.0, 156.0, 135.1, 134.2, 133.0 (Ar–C), 132.4 (Ar–2CH), 128.0 (Ar–C), 127.7, 126.8, 126.6 (Ar–CH), 126.0, 125.0 (Ar–C), 124.0, 122.8, 116.2 (Ar–CH), 115.8, 115.4 (Ar–C), 29.0 (CH3-N-methyl); MS (Fab, 70 eV, %): m/z 397 (M+, 100). Anal. Calcd. for C23H11NO6: C, 69.53; H, 2.79; N, 3.53. Found: C, 69.66; H, 2.81; N, 3.45.
4.1.2.8. 5-Ethyl-5H-naphthalen[2″,3″:4′,5′]furo[3′,2′:4′,5,6]pyrano[3,2-c]quinoline-6,7,8,13-tetraone (5b).
Yellow crystals, yield: 0.361 g (88%), mp 308 °C; IR (KBr): 3190 (Ar–CH), 2980 (Aliph-CH), 1730, 1720, 1669 (CO), 1605, 1585 (Ar–C=C) cm−1; 1H NMR (400 MHz, DMSO-d6): δH = 8.19−8.16 (m, 3H), 7.94−7.92 (m, 2H), 7.80−7.78 (m, 2H), 7.47 (dd, 1H, J = 6.8, 6.4), 4.42 (q, 2H, J = 6.9 Hz), 1.29 (t, 3H, J = 6.9 Hz); 13C NMR (100 MHz, DMSO-d6): δC = 177.9 (C=O, C-8), 174.7 (C=O, C-13), 173.6 (C=O, C-7), 162.3 (C=O, C-6), 158.1 (C-15A), 155.6 (C-14A), 153.1 (C-13A), 138.9 (C-4A), 134.5 (C-7B, HSQC δH 7.94), 134.0 (C-12A), 133.3 (C-11), 132.7 (C-10, HSQC δH 7.80), 126.9 (C-3), 126.8 (C-9, HSQC: δH = 8.19), 126.0 (C-12), 125.9 (C-2), 124.2 (C-2), 122.8 (C-8A), 122.3 (C-7A), 115.8 (C-6A, HSQC: δH = 7.47), 111.5 (C-6A), 110.7 (C-4), 37.1 (CH2, HSQC: δH = 4.42), 12.8 (CH3, HSQC: δH = 1.29). MS (Fab, 70 eV, %): m/z 411 (M+, 25), 89 (22). Anal. Calcd. for C24H13NO6: C, 70.07; H, 3.19; N, 3.40. Found: C, 70.22; H, 3.33; N, 3.55.
4.1.2.9. 3-(3-Chloro-1,4-dioxo-1,4-dihydronaphthalen-2-yl)-4-hydroxy-6-phenyl-2H-pyrano[3,2-c]quinoline-2,5(6H)-dione (6).
Orange crystals, yield: 0.457 g (92%), mp 280 °C; IR (KBr): 3441 (OH), 3044 (Ar–CH), 1727, 1676 (CO), 1594 (Ar–C=C), cm−1; 1H NMR (400 MHz, DMSO-d6): δH 13.90 (br, s, 1H, OH), 8.50−7.50 (m, 13H, Ar–H); 13C NMR (100 MHz, DMSO-d6): δC 179.9, 177.0 (CO), 166.1 (C-OH), 163.2 (CO, C-2), 162.3 (CO, C-5), 157.8 (C-10a), 145.9 (C-2′), 140.3 (Ph-C), 138.4 (C-6a), 135.3 (C-4′a, 8′a), 132.0, 131.2, 131.0, 130.3 (Ar–CH), 130.0, 128.2, 127.1, 124.6 (Are2CH), 122.6 (Ar–CH-p), 117.0 (C-9), 113.0 (C-4b), 99.5 (C-4a), 95.8 (C-3); MS (Fab, 70 eV,%): m/z 497 (M+2, 34), 495 (M+2, 100), 89 (22). Anal. Calcd. for C28H14ClNO6: C, 67.84; H, 2.85; N, 2.82. Found: C, 67.77; H, 2.99; N, 2.76.
4.2. Biological activity assessment
4.2.1. Anticancer activity
The methodology of the NCI anticancer screening has been described in detail at (http://www.dtp.nci.nih.gov). Briefly, the primary anticancer assay was performed against the NCI-60 panel of approximately 60 human tumor cell lines, derived from nine neoplastic diseases, in accordance with the protocol of the Developmental Therapeutics Program, National Cancer Institute, Bethesda. In the one-dose screen, cells were seeded into 96-well plates at densities between 5000 and 40 000 cells (depending on the doubling time of the cell line) and 24 h later the test compounds were added to the culture at a single concentration (10−5 M) for 48 h. End point determinations were made spectrophotometrically, based on the protein binding dye, sulforhodamine B (SRB). Results for each tested compound were reported as the percent of growth of the treated cells when compared to the untreated control cells and to the number of cells at time zero. A growth percent value between 0 and 100 indicates growth inhibition, e.g. a growth percent of 30 corresponds to 70% growth inhibition; negative values indicate lethality (lower cell number than at time zero), with −30% meaning 30% lethality.
The cytotoxic and/or growth inhibitory effects of the most active compounds in the one-dose screen were tested in vitro at five different concentrations ranging from 10−4 to 10−8 M (five-dose screen). Seven absorbance measurements for each cell line were made: time zero (Tz), control growth in the absence of drug (C), and test growth in the presence of the drug at the five concentration levels (Ti)]. The percentage growth was calculated at each of the drug concentrations levels. Percentage growth inhibition was calculated as: [(Ti − Tz)/(C − Tz)] × 100 for concentrations for which Ti ≥ Tz, and as [(Ti − Tz)/Tz] × 100 for concentrations for which Ti < Tz. Three-dose response parameters were calculated for each compound. Growth inhibition of 50% (GI50) was calculated from [(Ti − Tz)/(C − Tz)] × 100 = 50, which is the drug concentration resulting in a 50% lower net protein increase in the treated cells (measured by SRB staining) as compared to the net protein increase seen in the control cells. The drug concentration resulting in total growth inhibition (TGI) was calculated from Ti = Tz. The LC50 (concentration of drug resulting in a 50% reduction in the measured protein at the end of the drug treatment as compared to that at the beginning), indicating a net loss of cells following treatment, was calculated from [(Ti − Tz)/Tz] × 100 = −50. Values were calculated for each of these three parameters if the level of activity was reached; however, if the effect was not reached, the value for that parameter was expressed as more than the maximum concentration tested. The lowest values are obtained with the most sensitive cell lines.
4.2.2. Biochemistry
Expression, Purification and Activation of MAP Kinases –
Activated ERK2 (Rattus norvegicus mitogen activated protein kinase 1, GenBank accession number NM_053842) was expressed, purified and activated as previously reported in Kaoud et al. [33]. Activated ERK1 were expressed, purified and activated as mentioned in Callaway et al.[47]. Full length human JNK2α2 (GenBank accession number NM_002752) was expressed and purified following previously published protocol [48,49]. p38MAPKα was expressed, purified and activated as described previously [50]. The activated kinases were all stored in buffer [25 mM HEPES (pH 7.5), 50 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 2 mM DTT and 10–20% (v/v) glycerol] at −80 °C.
Expression, Purification of protein substrates His-Ets-1 (1–138), GST-C-Jun (1–221) and GST-ATF2 (1–115) –
His-Ets-1 (1–138) was expressed and purified according to Kaoud et. al previously published protocol [33]. GST-c-Jun (1–221) was expressed and purified according to Yan et al. method [49]. GST-ATF2 (1–115) was expressed and purified according to Szafranska et al. protocol [50].
Kinase activity assay –
MAP kinase assays were performed following Kaoud et al. previously reported method [51]. To summarize, all assays were conducted at 28–30 °C in kinase assay buffer (25 mM HEPES buffer-pH 7.5, 50 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 2 mM DTT, 11 mM MgCl2 and 10 μg mL−1 BSA) containing different concentrations of each inhibitor with a final DMSO concentration of 5%. To estimate the IC50 of each tested inhibitor towards the activity of different MAP kinases, 2 nM active ERK1 or ERK2 was assayed with 10 μM Ets1 (1–138) protein substrate. 20 nM active JNK2α2 was assayed with 2 μM GST-c-Jun (1–221) as protein substrate. 10 nM active p38MAPKα, was assayed with 10 μM GST-ATF2 (Δ 1–115) protein substrate. In all the assays, the reaction was started by the addition of radio labeled 10 or 100 μM [γ−32P] ATP (100–1000 c.p.m. pmol−1). In order to estimate the IC50 of each tested inhibitor, dose-response curves for data confirming to inhibition were fitted to Eq. (1).
| (1) |
The parameters used in deriving Eq. (1) are defined as follows; i, concentration of inhibitor I; V0, observed rate; V00, is the observed rate in the absence of inhibitor, V′ is the observed rate constant at saturating inhibitor, I, K50 is the concentration that leads to half the maximal change in V0.
Steady-state kinetic experiments –
All assays were performed as described in the kinase activity assay section and as previously reported by Kaoud et al. [51], the concentrations of Mg-ATP and compound 6 were varied at fixed Ets-1 concentration level of Km. Plots of the product against time were employed to calculate the initial rate for each tested condition. Reciprocal plots of 1/v against 1/s were checked for linearity. Kinetic constants values were estimated using GraphPad Prism software where the kinetic data were globally fitted to Eq. (2);
| (2) |
The parameters used in deriving Eq. (2) are defined as follows; V0, observed rate; , apparent maximum rate; s, concentration of substrate S; , apparent Michaelis constant for substrate S; i, concentration of inhibitor I; , apparent competitive inhibition constant for inhibitor I.
4.2.3. Anticancer activity assays:
Cell culture –
BRAF-mutant A375 melanoma cell line was maintained in RPMI media (Invitrogen) containing 10% (v/v) heat inactivated FBS-US grade (Invitrogen), 1X Glutamax (Invitrogen), 100 U mL−1 penicillin and 100 g mL−1 streptomycin (Invitrogen). Cells were incubated in a humidified 5% CO2 incubator at 37 °C.
Viability assays –
To estimate the effect of each tested inhibitor on A375 cell viability, 2000 cells were seeded in each well of 96 well plate, cells were treated with different doses of each compound and incubated for 72 h before performing the MTS assay (CellTiter 96® AQueous One Solution Cell Proliferation Assay from Promega) following the manufacturer assay. All experiments were done in triplicate.
Colony formation assay –
For every tested inhibitor, 1000 A375 cells were seeded in each 60 mm TC plate. Cells were incubated for 24 h to adhere then treated with different doses of each compound and incubated for additional 10–14 days. Plates were washed with 1X PBS, fixed by 4% paraformaldehyde for 15 min at room temperature, washed with PBS then stained with 0.2% crystal violet in 20% methanol/80% water for 10 more minutes. Plates were washed excessively by distilled water then imaged under microscope.
Western Blotting –
For Western blotting, cells were seeded at 1000,000 cells per well in a 6-well plate and incubated for 24 h before treatment with DMSO or different doses of each compound for 4 or 12 h. Cells were induced with 100 nM EGF for 10–15 min before lysis. Cells were washed by 1X PBS (Invitrogen) then lysates were prepared in M-PER™ Mammalian Protein Extraction Reagent (Thermo-Fisher) containing IX protease and phosphatase inhibitors cocktail (Pierce). After centrifuging the lysates, the protein concentration of each supernatant was estimated using Bradford analysis (Bio-Rad). 60 μg of each lysate were fractionated on a 10% SDS polyacrylamide gel (Bio-Rad) and transferred to Hybond-P PVDF Membrane (GE Healthcare). Primary antibodies were incubated overnight at 4 °C using 1:1000 anti-phospho-p90RSK (Thr573) rabbit polyclonal Abs (Cell Signaling Technology); 1:1000 anti- RSK1/RSK2/RSK3 (32D7) Rabbit mAb (Cell Signaling Technology); 1:200 anti-phospho-Elk-1 (Ser-383), rabbit polyclonal IgG (Santa Cruz Biotechnology, Inc); 1:2000 anti-phospho-p44/42 MAPK (ERK½) (Thr202/Tyr204) (E10) mouse mAb (Cell Signaling Technology); 1:1000 anti p44/42 MAPK (ERK½) (137F5) rabbit mAb (Cell Signaling Technology); 1:2000 anti-phospho-SAPK/JNK (Thr183/Tyr185) (G9) mouse mAb (Cell Signaling Technology); 1:1000 anti-PARP (46D11) rabbit mAB (Cell Signaling Technology); 1:1000 anti-cleaved-PARP (D214) rabbit polyclonal Abs (Cell Signaling Technology); ½000 anti-Vinculin (E1E9V) XP rabbit mAb (Cell Signaling Technology) and 1:5000 anti-actin, clone 4 mouse mAb (Millipore). Either anti-rabbit (Bio-Rad) or anti-mouse (Cell Signaling Technology) horseradish peroxidase-conjugated secondary antibodies and Western Bright ECL Western Blotting Reagents (Advansta) were used to develop the blots. All experiments were reproduced in independent experiments. All experiments were performed two times.
Incucyte proliferation assay –
2000 A375 cells were seeded in each well of 96 well plate. Cells were incubated for 24 h to adhere then treated with different doses of each inhibitor and incubated and analyzed using Incucyte ZOOM microscope (image recorder every 1 h with a 10× objective). The real time growth confluence was estimated and plotted using GraphPad Prism software.
IncuCyte Apoptosis assay –
2000 A375 cells were seeded in each well of 96 well plate. Cells were incubated for 24 h to adhere then treated with different doses of each inhibitor plus constant concentration of the lncuCyte Caspase-3/7 reagent) following the manufacturer protocol. Cells were incubated and imaged in the lncuCyte® ZOOM equipment with a × 10 objective at indicated time points. Number of apoptotic cells was normalized to the percentage area coverage (confluency) at the final time point to account for cell proliferation.
4.3. Computational methodology
The crystal structure of ERK2 receptor complexed with N-[(1S)-1-(3-chloro-4–fluorophenyl)–2–hydroxyethyl]–2–(tetrahydro–2H–pyran–4–ylamino)–5,8–dihydropyrido[3,4–d]pyrimidine-7(6H)-carboxamide (2SH) (PDB code: 4OE6 [41]) was taken as the template for all docking calculations. Modeller software [52] was used to construct and refine the missing residues (177–187, 202–204 and 331–333). The protonation state of the protein was predicted using H++ server [53]. All molecular docking calculations were carried out using Autodock4.2 software [40]. The pdbqt file for ERK2 was prepared according to the AutoDock protocol [54]. All docking parameters were kept to their default values. However, the number of GA runs was set to 250 and the maximum number of energy evaluation was set to 25,000,000. The docking grid was selected to encompass the ATP-active site of ERK2 receptor with a grid size of 70 Å × 70 Å × 70 Å and a spacing value of0.375 Å. The atomic charges of inhibitors were calculated by gasteiger method [55]. The grid center was placed at 16.20, 7.50, 12.40 (XYZ coordinates). Prior to molecular docking, the 3D structures of the studied compounds were energetically minimized using MMFF94S force field with the help of SZYBKI software [56].
4.4. Crystal structure determinations
The single-crystal X-ray diffraction study were carried out on a Bruker D8 Venture diffractometer with Photon100 detector at 123(2) K using Cu-Kα radiation (λ = 1.54178 Å for 3c, 3f) and Mo-Kα radiation (λ = 0.71073 Å for 6). Direct Methods for 3c, 3f (SHELXS-97) [57] and dual space methods for 6 (SHELXT) [58] were used for structure solution and refinement was carried out using SHELXL-2014 (full-matrix least-squares on F2) [57]. Hydrogen atoms were localized by difference electron density determination and refined using a riding model (H(N,O) free). Semi-empirical absorption corrections were applied.
3c: Yellow crystals, C19H8BrNO4 ∙ C2H6OS, Mr = 472.30, crystal size 0.18 × 0.12 × 0.03 mm, triclinic, space group P-1 (No. 2), a = 7.3950(2) Å, b = 10.0211(3) Å, c = 13.8015(4) Å, α = 70.268(1)°, β = 78.915(1)°, γ = 79.360(1)°, V = 936.79(5) Å3, Z = 2, ρ = 1.674 Mg/m−3, μ(Cu-Kα) = 4.347 mm−1, F(0 0 0) = 476, 2θmax = 144.2°, 14,049 reflections, of which 3661 were independent (Rint = 0.032), 267 parameters, 1 restraint, R1 = 0.026 (for 3325 I > 2σ(I)), wR2 = 0.065 (all data), S = 1.05, largest diff. peak/hole = 0.352/−0.359 e Å−3.
3f: Yellow crystals, C21H13NO4, Mr = 343.32, crystal size 0.26 × 0.06 × 0.04 mm, monoclinic, space group P21/c (No. 14), a = 8.0265(3) Å, b = 12.3658(5) Å, c = 15.3573(6) Å, β = 93.529(2)°, V = 1521.38(10) Å3, Z = 4, ρ = 1.499 Mg/m−3, μ(Cu-Kα) = 0.864 mm−1, F(0 0 0) = 712, 2θmax = 144.2°, 15,270 reflections, of which 2991 were independent (Rint = 0.024), 235 parameters, R1 = 0.034 (for 2800 I > 2σ(I)), wR2 = 0.091 (all data), S = 1.06, largest diff. peak/hole = 0.229/−0.249 e Å−3.
6: Orange crystals, C28H14ClNO6·½ (C3H7NO), Mr = 532.40, crystal size 0.40 × 0.30 × 0.20 mm, triclinic, space group P-1 (No. 2), a = 10.8841(4) Å, b = 12.3025(5) Å, c = 19.4831(8) Å, α = 108.007(1)°, β = 104.210(1)°, γ = 92.144(1)°, V = 2386.89(16) Å3, Z = 4, ρ = 1.482 Mg/m−3, μ(Mo-Kα) = 0.213 mm−1, F(0 0 0) = 1096, 2θmax = 55.0°, 48,590 reflections, of which 10,949 were independent (Rint = 0.027), 702 parameters, 2 restraints, R1 = 0.043 (for 9320 I > 2σ(I)), wR2 = 0.117 (all data), S = 1.04, largest diff. peak/hole = 0.875/−0.459 e Å−3 (in the solvent dmf).
CCDC 1542037 (3c), CCDC 1539309 (3f), and 1539310 (6) contain the supplementary crystallographic data for this paper. These data can be obtained free of charge from The Cambridge Crystallographic Data Centre via www.ccdc.cam.ac.uk/data_request/cif.
Acknowledgements
The authors also thank 3-MET Society for providing Prof Ashraf A. Aly, one-month fellowship enabling him to carry out the compounds analysis in Karlsruhe Institute of Technology, Karlsruhe, Germany. We also acknowledge the NCI for inclusion of our compounds in their screening program. Funding by The Cancer Prevention and Research Institute of Texas grant (RP160657), the National Institutes of Health (R01 GM123252) and The Welch Foundation (F-1390) to K.N. Dalby is also acknowledged. The computational work was completed in part with resources supported by the Science and Technology Development Fund, STDF, Egypt.
Footnotes
Appendix A. Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioorg.2018.10.044.
References
- [1].Ramesh M, Mohan PS, Shanmugam P, A convenient synthesis of flindersine, atanine and their analogs, Tetrahedron 40 (20) (1984) 4041–4049, 10.1016/0040-4020(84)85084-X. [DOI] [Google Scholar]
- [2].Wabo HK, Tane P, Connolly JD, Okunji CC, Schuster BM, Iwu MM, Tabouensinium chloride, a novel quaternary pyranoquinoline alkaloid from Araliopsis tabouensis, Nat. Prod. Res. 19 (6) (2005) 591–595, 10.1080/14786410412331280078. [DOI] [PubMed] [Google Scholar]
- [3].Johnson JV, Rauchman BS, Baccanari DP, Roth B, 2,4-Diamino-5-benzylpyrimidines and analogues as antibacterial agents. 12. 1,2-Dihydroquinolylmethyl analogues with high activity and specificity for bacterial dihydrofolate reductase, J. Med. Chem. 32 (8) (1989) 1942–1949, 10.1021/jm00128a042. [DOI] [PubMed] [Google Scholar]
- [4].Hanawa F, Fokialakis N, Skaltsounis A-L, Photo-activated DNA binding and antimicrobial activities of furoquinoline and pyranoquinolone alkaloids from rutaceae, Planta Med. 70 (6) (2004) 531–535, 10.1055/s-2004-827153. [DOI] [PubMed] [Google Scholar]
- [5].Martinez-Grau A, Marco JL, Friedlander reaction on 2-amino-3-cyano-4H-pyrans: synthesis of derivatives of 4H-pyran[2,3-b]quinoline, new tacrine analogs, Bioorg. Med. Chem. Lett. 7 (24) (1997) 3165–3170, 10.1016/S0960-894X(97)10165-2. [DOI] [Google Scholar]
- [6].Yamada N, Kadowaki S, Takahashi K, Umezu K, MY-1250, a major metabolite of the anti-allergic drug repirinast, induces phosphorylation of a 78-kDa protein in rat mast cells, Biochem. Pharmacol. 44 (6) (1992) 1211–1213. [DOI] [PubMed] [Google Scholar]
- [7].Chen J-J, Chen P-H, Liao C-H, Huang S-Y, Chen I-S, New Phenylpropenoids, Bis(1-phenylethyl)phenols, Bisquinolinone Alkaloid, and Anti-inflammatory Constituents from Zanthoxylum integrifoliolum, J. Nat. Prod. 70 (9) (2007) 1444–1448, 10.1021/np070186g. [DOI] [PubMed] [Google Scholar]
- [8].Gunasekaran P, Prasanna P, Perumal S, Almansour AI, ZnCl2-catalyzed three-component domino reactions for the synthesis of pyrano[3,2-c]quinolin-5(6H)-ones, Tetrahedron Lett. 54 (25) (2013) 3248–3252, 10.1016/j.tetlet.2013.04.022. [DOI] [Google Scholar]
- [9].Magedov IV, Manpadi M, Ogasawara MA, Dhawan AS, Rogelj S, Van Slambrouck S, Steelant WF, Evdokimov NM, Uglinskii PY, Elias EM, Knee EJ, Tongwa P, Antipin MY, Kornienko A, Structural simplification of bioactive natural products with multicomponent synthesis. 2. antiproliferative and antitubulin activities of pyrano[3,2-c]pyridones and pyrano[3,2-c]quinolones, J. Med. Chem. 51 (8) (2008) 2561–2570, 10.1021/jm701499n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lei M, Ma L, Hu L, A green, efficient, and rapid procedure for the synthesis of 2-amino-3-cyano-1,4,5,6-tetrahydropyrano[3,2-c]quinolin-5-one derivatives catalyzed by ammonium acetate, Tetrahedron Lett. 52 (20) (2011) 2597–2600, 10.1016/j.tetlet.2011.03.061. [DOI] [Google Scholar]
- [11].Song GY, Kim Y, Zheng XG, You YJ, Cho H, Chung JH, Sok DE, Ahn BZ, Naphthazarin derivatives (IV): synthesis, inhibition of DNA topoisomerase I and cytotoxicity of 2- or 6-acyl-5,8-dimethoxy-1, 4-naphthoquinones, Eur. J. Med. Chem. 35 (3) (2000) 291–298, 10.1016/S0223-5234(00)00129-X. [DOI] [PubMed] [Google Scholar]
- [12].Shchekotikhin AE, Buyanov VN, Preobrazhenskaya MN, Synthesis of 1-(omega-aminoalkyl)naphthoindolediones with antiproliferative properties, Bioorg. Med. Chem. 12 (14) (2004) 3923–3930, 10.1016/j.bmc.2004.04.042. [DOI] [PubMed] [Google Scholar]
- [13].Tran T, Saheba E, Arcerio AV, Chavez V, Li Q-Y, Martinez LE, Primm TP, Quinones as antimycobacterial agents, Bioorg. Med. Chem. 12 (18) (2004) 4809–4813, 10.1016/j.bmc.2004.07.015. [DOI] [PubMed] [Google Scholar]
- [14].Lien JC, Huang LJ, Wang JP, Teng CM, Lee KH, Kuo SC, Synthesis and antiplatelet, antiinflammatory, and antiallergic activities of 2-substituted 3-chloro-1,4-naphthoquinone derivatives, Bioorg. Med. Chem. 5 (12) (1997) 2111–2120, 10.1016/S0968-0896(97)00133-8. [DOI] [PubMed] [Google Scholar]
- [15].dos Santos EV, Carneiro JW, Ferreira VF, Quantitative structure-activity relationship in aziridinyl-1,4-naphthoquinone antimalarials: study of theoretical correlations by the PM3 method, Bioorg. Med. Chem. 12 (1) (2004) 87–93, 10.1016/j.bmc.2003.10.022. [DOI] [PubMed] [Google Scholar]
- [16].Valderrama JA, Zamorano C, Gonzalez MF, Prina E, Fournet A, Studies on quinones. Part 39: synthesis and leishmanicidal activity of acylchloroquinones and hydroquinones, Bioorg. Med. Chem. 13 (13) (2005) 4153–4159, 10.1016/j.bmc.2005.04.041. [DOI] [PubMed] [Google Scholar]
- [17].Bannwitz S, Krane D, Vortherms S, Kalin T, Lindenschmidt C, Zahedi Golpayegani N, Tentrop J, Prinz H, Muller K, Synthesis and structure-activity relationships of lapacho analogues. 2. Modification of the basic naphtho[2,3-b]furan-4,9-dione, redox activation, and suppression of human keratinocyte hyperproliferation by 8-hydroxynaphtho[2,3-b]thiophene-4,9-diones, J. Med. Chem. 57 (14) (2014) 6226–6239, 10.1021/jm500754d. [DOI] [PubMed] [Google Scholar]
- [18].Blake JF, Burkard M, Chan J, Chen H, Chou KJ, Diaz D, Dudley DA, Gaudino JJ, Gould SE, Grina J, Hunsaker T, Liu L, Martinson M, Moreno D, Mueller L, Orr C, Pacheco P, Qin A, Rasor K, Ren L, Robarge K, Shahidi-Latham S, Stults J, Sullivan F, Wang W, Yin J, Zhou A, Belvin M, Merchant M, Moffat J, Schwarz JB, Discovery of (S)-1-(1-(4-Chloro-3-fluorophenyl)-2-hydroxyethyl)-4-(2-((1-methyl-1H-pyrazol-5-yl)amino)pyrimidin-4-yl)pyridin-2(1H)-one (GDC-0994), an extracellular signal-regulated kinase ½ (ERK½) inhibitor in early clinical development, J. Med. Chem. 59 (12) (2016) 5650–5660, 10.1021/acs.jmedchem.6b00389. [DOI] [PubMed] [Google Scholar]
- [19].Sullivan RJ, Infante JR, Janku F, Wong DJL, Sosman JA, Keedy V, Patel MR, Shapiro GI, Mier JW, Tolcher AW, Wang-Gillam A, Sznol M, Flaherty K, Buchbinder E, Carvajal RD, Varghese AM, Lacouture ME, Ribas A, Patel SP, DeCrescenzo GA, Emery CM, Groover AL, Saha S, Varterasian M, Welsch DJ, Hyman DM, Li BT, First-in-class ERK½ inhibitor Ulixertinib (BVD-523) in patients with MAPK mutant advanced solid tumors: results of a phase i dose-escalation and expansion study, Cancer Discov. 8 (2) (2018) 184–195, 10.1158/2159-8290.CD-17-1119. [DOI] [PubMed] [Google Scholar]
- [20].Kong X, Kuilman T, Shahrabi A, Boshuizen J, Kemper K, Song JY, Niessen HWM, Rozeman EA, Geukes Foppen MH, Blank CU, Peeper DS, Cancer drug addiction is relayed by an ERK2-dependent phenotype switch, Nature 550 (7675) (2017) 270–274, 10.1038/nature24037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Moriceau G, Hugo W, Hong A, Shi H, Kong X, Yu CC, Koya RC, Samatar AA, Khanlou N, Braun J, Ruchalski K, Seifert H, Larkin J, Dahlman KB, Johnson DB, Algazi A, Sosman JA, Ribas A, Lo RS, Tunable-combinatorial mechanisms of acquired resistance limit the efficacy of BRAF/MEK cotargeting but result in melanoma drug addiction, Cancer Cell 27 (2) (2015) 240–256, 10.1016/j.ccell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Morris EJ, Jha S, Restaino CR, Dayananth P, Zhu H, Cooper A, Carr D, Deng Y, Jin W, Black S, Long B, Liu J, Dinunzio E, Windsor W, Zhang R, Zhao S, Angagaw MH, Pinheiro EM, Desai J, Xiao L, Shipps G, Hruza A, Wang J, Kelly J, Paliwal S, Gao X, Babu BS, Zhu L, Daublain P, Zhang L, Lutterbach BA, Pelletier MR, Philippar U, Siliphaivanh P, Witter D, Kirschmeier P, Bishop WR, Hicklin D, Gilliland DG, Jayaraman L, Zawel L, Fawell S, Samatar AA, Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors, Cancer Discov. 3 (7) (2013) 742–750, 10.1158/2159-8290.CD-13-0070. [DOI] [PubMed] [Google Scholar]
- [23].Maik-Rachline G, Seger R, The ERK cascade inhibitors: towards overcoming resistance, Drug Resist. Updat. 25 (2016) 1–12, 10.1016/j.drup.2015.12.001. [DOI] [PubMed] [Google Scholar]
- [24].Germann UA, Furey BF, Markland W, Hoover RR, Aronov AM, Roix JJ, Hale M, Boucher DM, Sorrell DA, Martinez-Botella G, Fitzgibbon M, Shapiro P, Wick MJ, Samadani R, Meshaw K, Groover A, DeCrescenzo G, Namchuk M, Emery CM, Saha S, Welsch DJ, Targeting the MAPK signaling pathway in cancer: promising preclinical activity with the novel selective ERK½ inhibitor BVD-523 (Ulixertinib), Mol. Cancer Ther. 16 (11) (2017) 2351–2363, 10.1158/1535-7163.MCT-17-0456. [DOI] [PubMed] [Google Scholar]
- [25].Mourad A-FE, Hassan AA, Mohamed NK, Aly AA, Ali BA, Novel heterocycles from creatinine, Heterocycl. Commun. 7 (6) (2001) 541–548, 10.1515/HC.2001.7.6.541. [DOI] [Google Scholar]
- [26].Aly AA, Mourad AF, Hassan AA, Mohamed NK, Ali BA, El-Sayed MM, Novel reaction products from thiobarbituric Acid of biological interest, Arch. Pharm. (Weinheim) 337 (3) (2004) 133–139, 10.1002/ardp.200300802. [DOI] [PubMed] [Google Scholar]
- [27].Aly AA, Brown AB, El-Shaieb KM, Hassan AA, Bedair TMI, NMR study of the naphtho-1,3-dithioles formed from carbamodithioates and 2,3-dichloro-1,4-naphthoquinone, J. Chem. Res. 11 (2009) 689–691, 10.3184/030823409X12561978806846. [DOI] [Google Scholar]
- [28].Aly AA, Brȁse S, Oxidation-reduction and heterocyclization of the reactions of alkanedithiols with π-deficient compounds, J. Sulf. Chem. 38 (3) (2017) 291–302, 10.1080/17415993.2017.1278762. [DOI] [Google Scholar]
- [29].El-Sheref EM, Aly AA, Mourad A-FE, Brown AB, Brȁse S, Bakheet MEM, Synthesis of pyrano[3,2-c]quinoline-4-carboxylates and 2-(4-oxo-1,4-dihydroquinolin-3-yl)fumarates, Chem. Pap. 72 (1) (2018) 181–190, 10.1007/s11696-017-0269-6. [DOI] [Google Scholar]
- [30].Aly AA, El-Sheref EM, Mourad A-FE, Brown AB, Brȁse S, Bakheet MEM, Nieger M, Synthesis of spiro[indoline-3,4’-pyrano[3,2-c]quinolone]-3’-carbonitriles, Monatsh. Chem. 149 (3) (2018) 635–644, 10.1007/s00706-017-2078-6. [DOI] [Google Scholar]
- [31].Aly AA, El-Sheref EM, Mourad A-FE, Bakheet MEM, Brȁse S, Nieger M, One-pot synthesis of 2,3-bis-(4-hydroxy-2-oxo-1,2-dihydroquinolin-3-yl)succinates and arylmethylene-bis-3,3’-quinoline-2-ones, Chem. Pap (2018), 10.1007/s11696-018-0561-0. [DOI] [Google Scholar]
- [32].Aly AA, El-Sheref EM, Bakheet MEM, Mourad MAE, Brown AB, Bräse S, Nieger M, Ibrahim MAA, Synthesis of novel 1,2-bis-quinolinyl-1,4-naphthoquinones: ERK2 inhibition, cytotoxicity and molecular docking studies, Bioorg. Chem. 81 (2018) 700–712, 10.1016/j.bioorg.2018.09.017. [DOI] [PubMed] [Google Scholar]
- [33].Kaoud TS, Devkota AK, Harris R, Rana MS, Abramczyk O, Warthaka M, Lee S, Girvin ME, Riggs AF, Dalby KN, Activated ERK2 is a monomer in vitro with or without divalent cations and when complexed to the cytoplasmic scaffold PEA-15, Biochemistry 50 (21) (2011) 4568–4578, 10.1021/bi200202y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lee S, Warthaka M, Yan C, Kaoud TS, Ren P, Dalby KN, Examining docking interactions on ERK2 with modular peptide substrates, Biochemistry 50 (44) (2011) 9500–9510, 10.1021/bi201103b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Piserchio A, Warthaka M, Kaoud TS, Callaway K, Dalby KN, Ghose R, Local destabilization, rigid body, and fuzzy docking facilitate the phosphorylation of the transcription factor Ets-1 by the mitogen-activated protein kinase ERK2, Proc. Natl. Acad. Sci. USA 114 (31) (2017) E6287–E6296, 10.1073/pnas.1702973114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Warthaka M, Adelmann CH, Kaoud TS, Edupuganti R, Yan C, Johnson WH Jr., S. Ferguson, C.D. Tavares, L.J. Pence, E.V. Anslyn, P. Ren, K.Y. Tsai, K.N. Dalby, Quantification of a pharmacodynamic ERK end point in melanoma cell lysates: toward personalized precision medicine, ACS Med. Chem. Lett. 6 (1) (2015) 47–52, 10.1021/ml500198b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, Spevak W, Zhang C, Zhang Y, Habets G, Burton EA, Wong B, Tsang G, West BL, Powell B, Shellooe R, Marimuthu A, Nguyen H, Zhang KY, Artis DR, Schlessinger J, Su F, Higgins B, Iyer R, D’Andrea K, Koehler A, Stumm M, Lin PS, Lee RJ, Grippo J, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, Chapman PB, Flaherty KT, Xu X, Nathanson KL, Nolop K, Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma, Nature 467 (7315) (2010) 596–599, 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Qin J, Xin H, Nickoloff BJ, Specifically targeting ERK1 or ERK2 kills melanoma cells, J. Transl. Med. 10 (2012) 15, 10.1186/1479-5876-10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wong DJ, Robert L, Atefi MS, Lassen A, Avarappatt G, Cerniglia M, Avramis E, Tsoi J, Foulad D, Graeber TG, Comin-Anduix B, Samatar A, Lo RS, Ribas A, Antitumor activity of the ERK inhibitor SCH772984 [corrected] against BRAF mutant, NRAS mutant and wild-type melanoma, Mol. Cancer 13 (2014) 194, 10.1186/1476-4598-13-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ, AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility, J. Comput. Chem. 30 (16) (2009) 2785–2791, 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Blake JF, Gaudino JJ, De Meese J, Mohr P, Chicarelli M, Tian H, Garrey R, Thomas A, Siedem CS, Welch MB, Kolakowski G, Kaus R, Burkard M, Martinson M, Chen H, Dean B, Dudley DA, Gould SE, Pacheco P, Shahidi-Latham S, Wang W, West K, Yin J, Moffat J, Schwarz JB, Discovery of 5,6,7,8-tetrahydropyrido[3,4-d]pyrimidine inhibitors of Erk2, Bioorg. Med. Chem. Lett. 24(12) (2014) 2635–2639, 10.1016/j.bmcl.2014.04.068. [DOI] [PubMed] [Google Scholar]
- [42].Zhao YH, Abraham MH, Le J, Hersey A, Luscombe CN, Beck G, Sherborne B, Cooper I, Rate-limited steps of human oral absorption and QSAR studies, Pharm. Res. 19 (10) (2002) 1446–1457. [DOI] [PubMed] [Google Scholar]
- [43].Lipinski CA, Lombardo F, Dominy BW, Feeney PJ, Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings, Adv. Drug Deliv. Rev. 46 (1–3) (2001) 3–26. [DOI] [PubMed] [Google Scholar]
- [44].Buckle DR, Cantello BC, Smith H, Spicer BA, 4-hydroxy-3-nitro-2-quinolones and related compounds as inhibitors of allergic reactions, J. Med. Chem. 18 (7) (1975) 726–732. [DOI] [PubMed] [Google Scholar]
- [45].Rao VS, Darbarwar M, One-pot synthesis of 7-[1,2-dihydro-4-hydroxy-1-methyl/phenyl-2-oxo-3-quinolinyl]-5,7-dihydro-5-methyl/phenyl-6H-[1]benzopyrano[3,2-c]quinolin-6-ones, Synth. Commun. 18 (18) (1988) 2267–2272, 10.1080/00397918808082369. [DOI] [Google Scholar]
- [46].Kappe T, The “pyrono route” to 4-hydroxy-2-quinolones and 4-hydroxy-2-pyridones, Farmaco 54 (5) (1999) 309–315, 10.1016/S0014-827X(99)00030-0. [DOI] [Google Scholar]
- [47].Callaway K, Abramczyk O, Martin L, Dalby KN, The anti-apoptotic protein PEA-15 is a tight binding inhibitor of ERK1 and ERK2, which blocks docking interactions at the D-recruitment site, Biochemistry 46 (32) (2007) 9187–9198, 10.1021/bi700206u. [DOI] [PubMed] [Google Scholar]
- [48].Madsen JA, Kaoud TS, Dalby KN, Brodbelt JS, 193-nm photodissociation of singly and multiply charged peptide anions for acidic proteome characterization, Proteomics 11 (7) (2011) 1329–1334, 10.1002/pmic.201000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Yan C, Kaoud T, Lee S, Dalby KN, Ren P, Understanding the specificity of a docking interaction between JNK1 and the scaffolding protein JIP1, J. Phys. Chem. B 115 (6) (2011) 1491–1502, 10.1021/jp1073522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Szafranska AE, Luo X, Dalby KN, Following in vitro activation of mitogen-activated protein kinases by mass spectrometry and tryptic peptide analysis: purifying fully activated p38 mitogen-activated protein kinase alpha, Anal. Biochem. 336 (1) (2005) 1–10, 10.1016/j.ab.2004.09.039. [DOI] [PubMed] [Google Scholar]
- [51].Kaoud TS, Park H, Mitra S, Yan C, Tseng CC, Shi Y, Jose J, Taliaferro JM, Lee K, Ren P, Hong J, Dalby KN, Manipulating JNK signaling with (–)-zuonin A, ACS Chem. Biol. 7 (11) (2012) 1873–1883, 10.1021/cb300261e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Martí-Renom MA, Stuart AC, Fiser A, Sánchez R, Melo F, Šali A, Comparative protein structure modeling of genes and genomes, Annu. Rev. Biophys. Biomol. Struct. 29 (1) (2000) 291–325, 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- [53].Gordon JC, Myers JB, Folta T, Shoja V, Heath LS, Onufriev A, H++: a server for estimating pK(a)s and adding missing hydrogens to macromolecules, Nucleic Acids Res. 33 (Web Server issue) (2005) W368–W371, 10.1093/nar/gki464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Forli S, Huey R, Pique ME, Sanner MF, Goodsell DS, Olson AJ, Computational protein-ligand docking and virtual drug screening with the AutoDock suite, Nat. Protocols 11 (5) (2016) 905–919, 10.1038/nprot.2016.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Gasteiger J, Marsili M, Iterative partial equalization of orbital electronegativity - a rapid access to atomic charges, Tetrahedron 36 (22) (1980) 3219–3228, 10.1016/0040-4020(80)80168-2. [DOI] [Google Scholar]
- [56].SZYBKI, OpenEye Scientific Software, Santa Fe, NM, USA, 2016. [Google Scholar]
- [57].Sheldrick GM, A short history of SHELX, Acta Crystallogr A 64 (Pt 1) (2008) 112–122, 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- [58].Sheldrick GM, SHELXT - Integrated space-group and crystal-structure determination, Acta Crystallogr., Sect. A: Found. Adv. 71 (1) (2015) 3–8, 10.1107/S2053273314026370. [DOI] [PMC free article] [PubMed] [Google Scholar]