Summary
In this study we compare the genetic ancestry of individuals from two as yet genetically unstudied cultural traditions in Estonia in the context of available modern and ancient datasets: 15 from the Late Bronze Age stone-cist graves (1200–400 BC) (EstBA), and 6 from the Pre-Roman Iron Age tarand cemeteries (800/500 BC–50 AD) (EstIA). We also included 5 Pre-Roman to Roman Iron Age Ingrian (500 BC–450 AD) (IngIA) and 7 Middle Age Estonian (1200–1600 AD) (EstMA) individuals to build a dataset for studying the demographic history of the northern parts of the Eastern Baltic from the earliest layer of Mesolithic to modern times. Our findings are consistent with EstBA receiving gene flow from regions with strong Western hunter-gatherer (WHG) affinities, and EstIA from populations related to modern Siberians. The latter inference is in accordance with Y chromosome (chrY) distributions in present-day populations of the Eastern Baltic, as well as patterns of autosomal variation in the majority of the westernmost Uralic speakers [1–5]. This ancestry reached the coasts of the Baltic Sea no later than the mid-first millennium BC; i.e. in the same time window as the diversification of west Uralic/Finnic languages [6]. Furthermore, phenotypic traits often associated with modern Northern Europeans like light eyes, hair and skin as well as lactose tolerance can be traced back to the Bronze Age in the Eastern Baltic.
Keywords: ancient DNA, shotgun sequencing, population genetics, phenotype, kinship, Bronze Age, Iron Age, Middle Ages, Eastern Baltic, Estonia
Results and Discussion
The Eastern Baltic has witnessed several population shifts since people reached its southern part during the Final Paleolithic ~11,000–10,000 BC [7,8], and its northern part during the Mesolithic ~9000 BC [9]. No genetic information is available from Paleolithic populations but Mesolithic hunter-gatherers of the Kunda and Narva cultures were genetically most similar to WHGs widespread in Europe [10–12]. A genetic shift towards Eastern hunter-gatherer (EHG) genetic ancestry occurred with the arrival of the Neolithic Comb Ceramic culture (CCC) people ~3900 BC [10–13]. The Late Neolithic (LN) Corded Ware culture (CWC) people of Ponto-Caspian steppe origin [10–13] brought farming into the Eastern Baltic ~2800 BC, contrary to most of Europe where the Neolithic transition was mediated by Aegean early farmers [14–19]. Human remains radiocarbon dated to the Early Bronze Age (ca 1800–1200 BC) are rare from this region and no ancient DNA (aDNA) data is currently available. Genetic data from succeeding Bronze Age (BA) layers in Latvia and Lithuania indicate some genetic affinities with modern Eastern Baltic populations, but also notable differences [11].
In this study we present new genomic data from Estonian Late Bronze Age stone-cist graves (1200–400 BC) and Pre-Roman Iron Age tarand cemeteries (800/500 BC–50 AD). The cultural background of stone-cist graves indicates strong connections both to the west and the east [20,21]. The Iron Age (IA) tarands have been proposed to mirror ‘houses of the dead’ found among Uralic peoples of the Volga-Kama region [22]. As this time window matches the proposed diversification period of western Uralic languages [6] and the arrival of Proto-Finnic language in the Eastern Baltic from the east [23,24], our study considers linguistic, archaeological and genetic data to inform on this.
One of the most notable genetic features of Eastern Baltic populations is a high frequency of chrY haplogroup (hg) N3a (nomenclature of Karmin et al. [25]); a characteristic shared mostly with Finno-Ugric-speaking groups in Europe and several populations all over Siberia [1–5]. The rapid expansion of people carrying these lineages likely took place within the last 5,000 years [1] but their arrival time in the Eastern Baltic remains unresolved. The gene flow from Siberia to western-Uralic-speaking populations has also recently been inferred using autosomal data [5,26]. However, available aDNA data has not revealed chrY hg N lineages in Eastern Baltic individuals [10–13].
To characterize the genetic ancestry of people from the so far unstudied cultural layers, we extracted DNA from the tooth roots of 56 individuals (Figure 1A, Table S1, Methods). No individuals were included from later IA in Estonia because people were mostly cremated during that period. Individuals morphologically sexed as males were prioritized in sampling to make comparisons using autosomal and both sex chromosomes. We shotgun sequenced all samples and they formed 3 groups: 1) 15 with low endogenous DNA content and resulting coverage, which were excluded from further analyses; 2) 8 with sufficient mtDNA (and in some cases, chrY) coverage for determining hgs, but not for informative autosomal analyses; 3) 33 that yielded sufficient autosomal data for informative analyses. The 33 individuals included 15 from EstBA, 6 from EstIA, 5 from IngIA and 7 from EstMA, and yielded endogenous DNA ~4–88%, average genomic coverages ~0.017–0.734x and contamination estimates <4% (Table S1). We analysed the data in the context of modern and other ancient individuals, including from Neolithic Estonia [13].
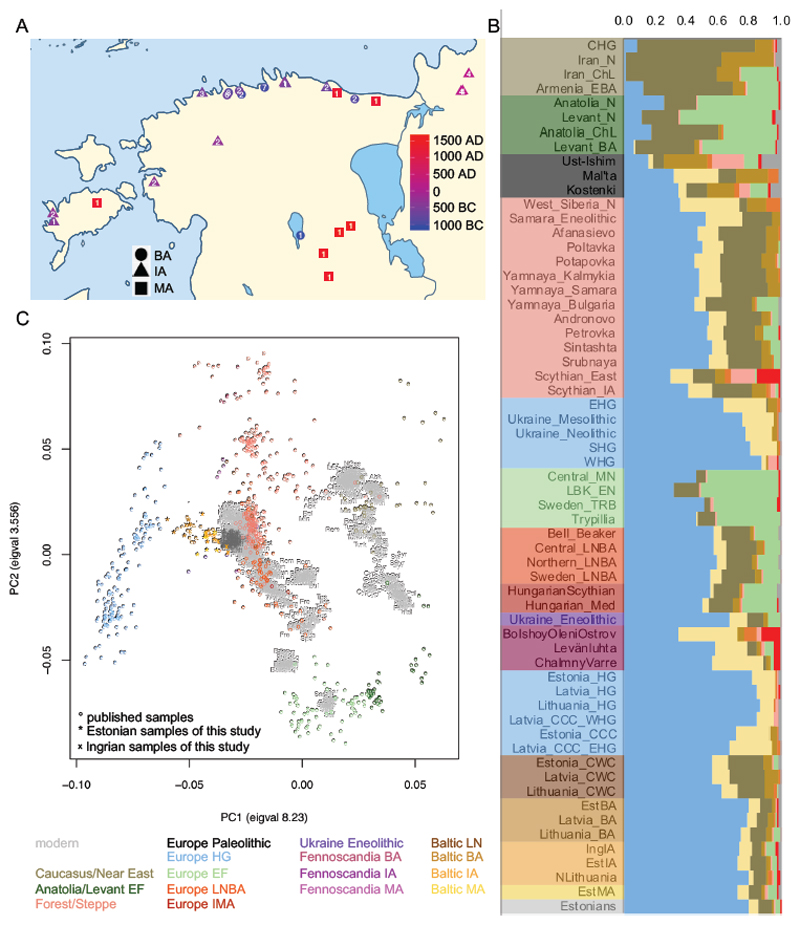
Figure 1. Geographical locations, ADMIXTURE and principal component analyses’ results.
EF – early farmers; HG – hunter-gatherers; LNBA – Late Neolithic/Bronze Age; IMA – Iron/Middle Ages; LN – Late Neolithic; BA – Bronze Age; IA – Iron Age; MA – Middle Ages. A. Map of the geographical locations of the individuals of this study. B. ADMIXTURE analysis results for a selection of ancient population averages at K9 with ancient individuals projected onto the modern genetic structure. The X axis shows the proportions of the ancestral components. C. Principal component analysis results of modern West Eurasians with ancient individuals projected onto the first two components (PC1 and PC2). See also Figure S1, Table S3.
Temporal Dynamics of Maternal and Paternal Lineages in Estonia
We identified mtDNA hgs for 41 individuals (Table 1). We then compared these with over 2,000 present-day Estonian whole mtDNA sequences (unpublished; cohort [29]) and found that all the hgs are also present in modern Estonia, and are not restricted to a particular region.
Table 1. Archaeological information, mtDNA and Y chromosome haplogroups and genetic sex of the individuals of this study.
* – typo-chronological date, ** – 14C date, *** – combined 14C date of multiple dates using OxCal v4.2.4 [27] R_combine, **, *** – calibrated using OxCal v4.2.4 [27] and IntCal13 atmospheric curve [28]; Morph. – morphological, M – male, F – female; Gen. – genetic; MT hg – mitochondrial DNA haplogroup; Y hg – Y chromosome haplogroup; Av. cov. – average genomic coverage, <0.017 – not included in autosomal analyses. See also Figure S3, Tables S1 and S2 and Data S2.
| Sex | ||||||||
|---|---|---|---|---|---|---|---|---|
| Individual | Location | Period | Date | Morph. | Gen. | MT hg | Y hg | Av. cov. |
| X02 | Iru, Harju, EST | BA | 1090–910 BC** | M | XY | T1a1b | R1a | 0.031 |
| 0LS11 | Jõelähtme, Harju, EST | BA | 1060–850 BC** | M | XY | H1c | R1a1 | 0.214 |
| V9 | Jõelähtme, Harju, EST | BA | 1220–1010 BC** | M | XY | K1c1h | R1a1'2 | 0.474 |
| V14 | Muuksi, Harju, EST | BA | 1280–1050 BC** | M | XY | U2e2a1 | R1a1'2 | 0.443 |
| X05 | Muuksi, Harju, EST | BA | 1210–1010 BC** | M | XY | T2a1b1a1 | R1a1'2 | 0.029 |
| X08 | Muuksi, Harju, EST | BA | 930–810 BC** | M | XY | T2a1b1a2 | R1a1c | 0.306 |
| X09 | Muuksi, Harju, EST | BA | 820–770 BC** | M | XY | J1b1a | R1a | <0.017 |
| X10 | Muuksi, Harju, EST | BA | 1220–1020 BC** | M | XY | U5a2a1 | R1a1'2 | 0.22 |
| X11 | Napa, Ida-Viru, EST | BA | 1030–890 BC** | M | XY | J1c2k | R1a | 0.224 |
| X12 | Napa, Ida-Viru, EST | BA | 900–790 BC** | M | XY | W6 | R1a1'6 | 0.023 |
| X13 | Rebala, Harju, EST | BA | 780–480 BC** | M | ? | K1b2a | - | <0.017 |
| X14 | Rebala, Harju, EST | BA | 780–430 BC** | M | XY | H1b2 | R1a1c | 0.307 |
| V16 | Väo, Harju, EST | BA | 730–390 BC** | M | XY | H1b2 | R1a1'2 | 0.22 |
| X16 | Väo, Harju, EST | BA | 1080–910 BC** | M? | XY | J1c4 | R1a | 0.018 |
| X17 | Väo, Harju, EST | BA | 930–810 BC** | M | XY | U4a2b | R1a1c | 0.387 |
| X18 | Väo, Harju, EST | BA | 1200 BC–1700 AD* | M | XY | U3b2a | ? | <0.017 |
| X19 | Väo, Harju, EST | BA | 1200–400 BC* | ? | XX | U | - | <0.017 |
| X20 | Väo, Harju, EST | BA | 900–800 BC** | ? | XY | U4a2b | R1a | 0.085 |
| X15 | Vehendi, Tartu, EST | BA | 1210–1000 BC** | M? | XY | U5b1b1 | R1a1c | 0.339 |
| 0LS09 | Ilmandu, Harju, EST | IA | 540–380 BC** | F | XX | H6a1a | - | <0.017 |
| V7 | Ilmandu, Harju, EST | IA | 790–430 BC** | M | XY | T2a1b1a1 | R1a | <0.017 |
| V8 | Ilmandu, Harju, EST | IA | 730–400 BC*** | M? | XX | HV0 | - | <0.017 |
| 0LS10 | Kunda, Lääne-Viru, EST | IA | 770–430 BC*** | M | XY | H13a1a1a | N3a3’5 | 0.319 |
| V10 | Kunda, Lääne-Viru, EST | IA | 790–430 BC** | M | XY | H1a | R1a1c | 0.403 |
| V11 | Kurevere, Saare, EST | IA | 390–200 BC** | M? | XX | W3a1d | - | 0.277 |
| V12 | Kurevere, Saare, EST | IA | 360–40 BC** | M? | XY | I1a1c | N3a3a | 0.245 |
| X04 | Loona, Saare, EST | IA | 480–360 BC** | M | XY | H1c | R1a1'2 | 0.256 |
| VII3 | Poanse, Pärnu, EST | IA | 380–180 BC** | M | XY | U5a1d | ? | <0.017 |
| VII4 | Võhma, Lääne-Viru, EST | IA | 760–400 BC** | M | XY | T1a1b | N3a3a | 0.342 |
| VII15 | Kerstovo, Ingria, RUS | IA | 45 BC–77 AD** | ? | XY | U5a2a1 | R1a | 0.244 |
| VIII7 | Kerstovo, Ingria, RUS | IA | 75–200 AD* | ? | XX | H2a1a | - | 0.062 |
| VIII8 | Kerstovo, Ingria, RUS | IA | 75–200 AD* | ? | XY | H3h | R1a1c | 0.0517 |
| VIII9 | Kerstovo, Ingria, RUS | IA | 75–200 AD* | ? | XX | U4a2 | - | 0.3 |
| VIII5 | Malli, Ingria, RUS | IA | 75–300 AD* | ? | XX | T1a1b | - | 0.398 |
| IIa | Karja, Saare, EST | MA | 1230–1300 AD* | M | XY | H3h1 | N3a3a | 0.734 |
| 0LS03 | Kukruse, Ida-Viru, EST | MA | 1180–1220/1240 AD* | M | XY | U4d1 | R1a1a'b | 0.0696 |
| IVLS09KT | Mäletjärve, Tartu, EST | MA | 1570–1600 AD* | M | XY | H2a1 | J2b2 | 0.332 |
| IIf | Otepää, Valga, EST | MA | 1360–1390 AD* | M | XY | T2b | N3a3a | 0.206 |
| IIg | Pada, Lääne-Viru, EST | MA | 1210–1230/1240 AD* | M | XY | U4a2b | N3a3a | 0.102 |
| IIIt | Vaabina, Võru, EST | MA | 1250–1450 AD* | F | XX | U5a2a1 | - | 0.0413 |
| ILS01 | Vana-Kuuste, Tartu, EST | MA | 1500–1625 AD* | M | XY | H11a1 | R1a | 0.0827 |
We identified chrY hgs for 30 male individuals (Table 1, Table S2, Methods). All 16 successfully haplogrouped EstBA males belonged to hg R1a, showing no change from the CWC period when this was also the only chrY lineage detected in the Eastern Baltic [11,13,30,31]. Three EstIA and two IngIA individuals also belonged to hg R1a but three EstIA males belonged to hg N3a; the earliest so far observed in the Eastern Baltic. Three EstMA individuals belonged to hg N3a, two to hg R1a and one to hg J2b. ChrY lineages found in the Baltic Sea region before the CWC belong to hgs I, R1b, R1a5 and Q [10–13,17,32]. Thus, it appears that these lineages were substantially replaced in the Eastern Baltic by hg R1a [10–13], most likely through Steppe migrations from the east [30,31]. Although we did not detect N3a chrYs in our BA sample, unlike in BA Fennoscandia [26], we cannot rule out its presence, due to small sample size. However, the frequency should not exceed 0.17 with 95% and 0.25 with 99% confidence [33]. The frequency of hg N3a was significantly higher in our IA than our BA group (Fisher’s exact test p-value 0.013). Our results enable us to conclude that although the expansion time for R1a1 and N3a3’5 in Eastern Europe is similar [25], hg N3a likely reached Estonia or at least became comparably frequent to modern Estonia [1] only during the BA-IA transition.
Autosomal Ancestries in Estonia from the Bronze Age Onward
To assess if the Eastern Eurasian influence indicated by chrY hg N3a is apparent elsewhere in the genome, we first applied principal component analysis (PCA). We projected ancient genomes from previous studies (Table S3) and this study on two axes inferred using Estonian Biocentre Illumina genotyping array data (EBC-chipDB) of modern Western Eurasian individuals (Table S3) (Figure 1C). A clear shift towards West Eurasian hunter-gatherers is visible between European LN/BA (including Baltic CWC) and EstBA individuals, the latter clustering together with Latvian and Lithuanian BA individuals [11]. EstIA, IngIA and EstMA individuals project between BA individuals and modern Estonians, partially overlapping with both.
We performed ADMIXTURE analysis by projecting aDNA data on world-wide EBC-chipDB modern data (Figure S1C–D, Table S3) and present results at K=9 (Figure 1B, Figure S1A–B, Methods). EstBA individuals are clearly distinguishable from Estonian CWC individuals as the former have more of the blue component most frequent in WHGs and less of the brown and yellow components maximized in Caucasus hunter-gatherers and modern Khanty, respectively. The individuals of EstBA, EstIA, IngIA, EstMA and modern Estonia are quite similar to each other on average, indicating that the relatively high proportion of WHG ancestry in modern Eastern Baltic populations compared to other present-day Europeans [15] traces back to the BA.
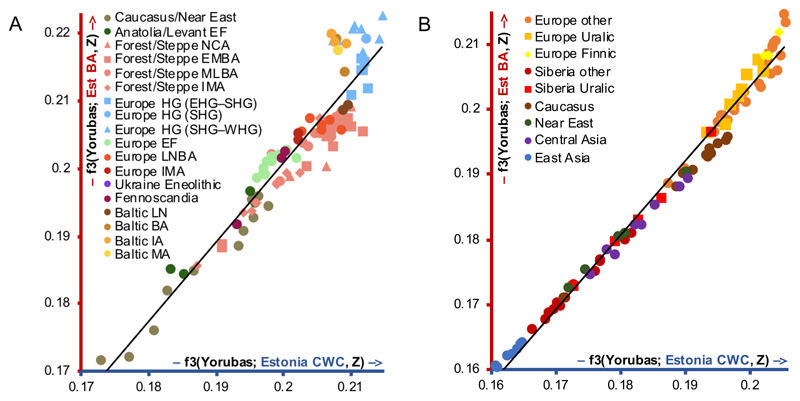
When comparing Estonian CWC and EstBA using autosomal outgroup f3 and Patterson’s D statistics (Table S3), the latter is more similar to other Baltic BA populations, to Baltic IA and Middle Age (MA) populations and also to populations similar to WHGs and Scandinavian hunter-gatherers (SHGs), but not to Estonian CCC (Figure 2A, Figure S2A, Data S1). The increase in WHG/SHG ancestry could be connected to western influences seen in material culture [20,21] and facilitated by a decline in local population after the CCC/CWC period [20]. A slight trend of bigger similarity of Estonian CWC to Forest/Steppe populations and of EstBA to European early farmer populations can also be seen. These differences remain when over 900,000 positions of the 1240k capture [16] are used instead of ~500,000 positions of the EBC-chipDB (Figure S2B, Data S1). When comparing to modern populations, Estonian CWC is slightly more similar to Caucasus individuals, but EstBA to Baltic populations and Finnic speakers (Figure 2B, Data S1). Outgroup f3 and D statistics do not reveal apparent differences when comparing EstBA/EstIA, EstIA/IngIA and EstIA/EstMA (Data S1). These results highlight how uniparental and autosomal data can lead to different demographic inferences – the genetic change between CWC and BA not seen in uniparental lineages is clear in autosomal data while the appearance of chrY hg N in the IA is not matched by a clear shift in autosomal profiles.
Figure 2. Outgroup f3 statistics' results.
Estonian Corded Ware culture (Estonia CWC; blue axis) and Estonian Bronze Age (Est BA; red axis) plotted against each other. A. Outgroup f3 statistics' values of form f3(Yorubas; Estonia CWC/Est BA, ancient). EF – early farmers; NCA – Neolithic/Copper Age; EMBA – Early/Middle Bronze Age; MLBA – Middle/Late Bronze Age; IMA – Iron/Middle Ages; HG – hunter-gatherers; LNBA – Late Neolithic/Bronze Age; LN – Late Neolithic; BA – Bronze Age; IA – Iron Age; MA – Middle Ages. B. Outgroup f3 statistics' values of form f3(Yorubas; Estonia CWC/Est BA, modern). See also Figure S2, Table S3, Data S1.
We also tested for sex biases by comparing outgroup f3 statistics calculated on autosomal (A) and X chromosomal (X) data. The high X to A ratio of European-early-farmer-related ancestry observed in Estonian CWC [13] decreases over time and disappears by the MA (Figure S2C–F, Data S1).
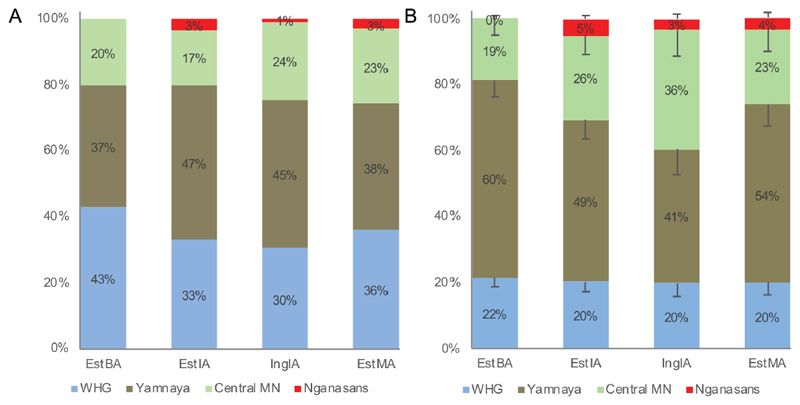
We used ChromoPainter/NNLS in the unlinked mode and qpAdm to infer mixing proportions of proxy source populations forming the genetic structure of the study populations. The best model for both analyses included WHG, Yamnaya, Central European Middle Neolithic (Central MN) and modern Nganasans as sources (Methods, Data S1). The study populations have on average 36%/20% WHG-, 42%/51% Yamnaya- and 21%/26% Central-MN-related ancestry as estimated by ChromoPainter/qpAdm (Figure 3, Data S1). The differences in WHG- and Yamnaya-related ancestry of the two methods could be due to the large amount of shared ancestry between those populations. Importantly, both analyses differentiate EstBA from other study populations – EstBA individuals have no Nganasan-related ancestry while EstIA, IngIA and EstMA individuals on average have 2%/4% (Figure 3, Data S1). The differentiation remains when using BA or IA Fennoscandian populations [26] instead of Nganasans (Data S1). Notably, the proportion of Nganasan-related ancestry varies between 0–12% among sampled EstIA/IngIA/EstMA individuals (Data S1), which may suggest its relatively recent admixture into the target population. Moreover, two individuals from Kunda (0LS10, V10) have the highest proportions of Nganasan ancestry among EstIA (6%, 8%), one of them has chrY hg N3a and isotopic analysis suggests neither individual being born in Kunda [34].
Figure 3. ChromoPainter/NNLS and qpAdm results.
EstBA – Estonian Bronze Age; EstIA – Estonian Iron Age; IngIA – Ingrian Iron Age; EstMA – Estonian Middle Ages; WHG – Western hunter-gatherers; Central MN – Central European Middle Neolithic. A. ChromoPainter/NNLS unlinked mode summarised results. B. qpAdm results. See also Table S3, Data S1.
To consolidate the previously described evidence of genetic input from Siberia, we applied f4 statistics (Data S1). A direct comparison between EstBA and EstIA suggests a closer affinity of EstIA to Siberian proxy Nganasan but the result is non-significant (|Z|=2.6). However, modern Estonians are significantly closer to Nganasan than EstBA (|Z|=5.6), while there is no significant difference between modern Estonians and EstIA in that regard (|Z|=1.2). Tests where Nganasans are replaced with Koryaks yield similar results, consistent with the signal relating to Siberian ancestry in general (Data S1). Additionally, the difference between EstBA and EstIA in their affinity to Nganasan can be seen through comparisons with preceding Central European LN/BA (|Z|=0.2/3.2 respectively). Furthermore, EstBA had a significantly higher affinity to WHGs than preceding CWC (|Z|=8.7) or modern Estonians (|Z|=5.1). We also tested the increase in affinity to Near Eastern populations between EstBA and modern Estonians seen on PCA and found that the latter share significantly more drift with modern Syrians than either EstBA or EstIA (|Z|=4.9/3.9). We then replaced Syrians with Yamnaya Kalmykia (|Z|=1.2/0.6) and Central MN (|Z|=3/2.6). This indicates a slight increase in early farmer ancestry from EstBA and EstIA to modern Estonians.
Finally, we performed formal tests of continuity between individual genomes of this study and modern Estonians [19]. We found that population continuity can be rejected for most scenarios (Data S1; p-value <0.05, colored grey, Figure S3E). Taking into account modern Estonian effective population size (Methods), continuity cannot be fully rejected only if the ancient sampling populations had an effective size of a few hundreds (Data S1; p-value >0.05, colored yellow to red, Figure S3E).
A Case of Close Genetic Relatedness Between Two Stone-cist Grave Groups
We screened the BA, IA and MA individuals for the presence of closely related pairs using READ and discovered that two BA individuals, X14 and V16, were 2nd degree relatives (Figure S3A–C). These individuals also shared mtDNA hg H1b2 and – like all EstBA males – chrY hg R1a. While chrY coverage is not sufficiently high to determine how closely related these individuals are patrilineally, their haplotypes matching across the entire mtDNA genome suggests that they were half-brothers sharing their mother or an uncle and his sister’s son. Notably, the two related individuals were not buried in the same site but 13 km apart. Given the small number of just sixteen stone-cist burials available for kinship analyses from a time span of ~500 years, the finding of cross-site relatedness supports the notion that these structures were built for a limited circle of people [35], possibly the elite.
The plateau in the calibration curve hinders precisely estimating the chronological separation between the radiocarbon dates of X14 (2481±30 BP) and V16 (2399±27 BP), with a 95% HPD -76–344 years (V16 dying 76 years earlier to 344 years later than X14). Given the estimated ages at death (35–45 for X14, 30–40 for V16 (Table S1)), female reproductive age 13–40, and assuming X14 to be the uncle of V16, the biologically plausible difference in time of the two individuals dying is -29–72 years (Methods). This interval is associated with a probability of 0.15 and is within the 95% HPD; hence the radiocarbon dates do not reject the relatedness inferred from aDNA. The plausible range of difference in time of deaths in case of V16 being the uncle is -82–19, whilst in case V16 and X14 were half-brothers this becomes -42–32. Both temporal intervals are less likely than the scenario described above (probability 0.08).
Frequency Changes of Phenotype Informative Alleles in the Eastern Baltic
We imputed the genotypes of 37 phenotype informative SNVs from the HIrisPlex-S system, two from TLR1 and one from MCM6 gene, and a 32-bp deletion (rs333) in the CCR5 gene for Mesolithic and Neolithic individuals from Latvia and Estonia [10,13] and the individuals of this study. We inferred a sharp increase to >50% in the frequency of the lactase persistence variant (MCM6/rs4988235) in the Baltic area after the LN (Data S2), in line with previous indications of this variant becoming common in Europe in the last 4,000–3,500 years [31,36] and of its fast increase in populations with Steppe ancestry due to local adaptation [37]. In contrast, the rs333, responsible for HIV resistance, which we first detect in a CWC individual, remains at low 5–17% frequency since then (Data S2), comparable to its present-day 14.8% frequency in Estonia [38]. Both TLR1 variants involved in the protection against leprosy were already present in Europe at medium-high frequencies since the Mesolithic ([16,39], Data S2). Notably, we infer a high proportion (>60%) of dark skin pigmentation in the hunter-gatherers and CWC farmers (Data S2). We infer dark skin and blue eyes for two individuals, similarly to another European Mesolithic individual [39]. However, from BA onward we infer pale/intermediate skin pigmentation for all individuals and an increase in the proportion of blue eyes and lighter shades of hair (Data S2). This is in line with previous suggestions that light skin pigmentation alleles reached high frequencies in Europe only recently [40].
Conclusions
We show that a component of possibly Siberian ancestry was added to the gene pool of the Eastern Baltic during the Bronze to Iron Age transition at the latest. This component is present in the autosomes and chrY of many northeastern European Uralic-speaking populations today [5,26], but arrived in the Eastern Baltic probably later than 3,500 ya when it reached Fennoscandia [26]. Considering the archaeological context of the individuals, this seems to have followed the so-called southwestern route from the Volga-Ural region [20,21]. Notably, the Bronze to Iron Age transition period also coincides with the hypothesized arrival of westernmost Uralic/Finnic languages [6] in the Eastern Baltic, supporting the idea that the spread of these languages was mediated by IA migrants from the east.
The EstBA individuals of this study, as other Baltic BA individuals [11], display more WHG ancestry compared to both earlier CWC and modern Estonians. Interestingly, we do not detect this change in their uniparental lineages. However, half of the admittedly small EstIA sample and over one third of modern Estonian men [1] share a hg N3a chrY – common in other Uralic-speaking populations living much further east [1–5] and not found in the Eastern Baltic earlier – while the autosomes of EstIA individuals only show 3–5% Siberian ancestry on average.
Furthermore, phenotypic characteristics often associated with modern Northern Europeans (light eyes, hair and skin pigmentation, lactose tolerance) can be traced back to the Bronze Age in the Eastern Baltic.
STAR Methods
Contact for reagent and resource sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Lehti Saag (lehti.saag@ut.ee).
Experimental model and subject details
The teeth used for DNA extraction were obtained with relevant institutional permissions from the University of Tartu, Institute of History and Archaeology; Tallinn University Archaeological Research Collection; the Museum of Anthropology and Ethnography (Kunstkamera) in St. Petersburg.
DNA was extracted from the teeth of 56 individuals – 23 from Late Bronze Age Estonia (EstBA; 1200–400 BC), 14 from Pre-Roman Iron Age Estonia (EstIA; 800/500 BC–50 AD), 12 from Pre-Roman to Roman Iron Age Ingria, Russia (IngIA; 500 BC–450 AD) and 7 from Middle Age Estonia (EstMA; 1200–1600 AD) (Figure 1, Table 1, Table S1). More detailed information about the archeological periods and the specific sites and burials of this study is given below.
Information about the archaeological time periods, sites and individuals of this study
In the archaeological record of Estonia, inhumation burials, which make the extraction of aDNA possible with current methods, date mainly from three major periods: 1) the Stone Age (9000–1800 BC), 2) the Bronze Age and Pre-Roman Iron Age (1800 BC–50 AD), 3) 2nd millennium AD. Thereby, inhumations from the Stone Age are presently known from the 7th millennium to the 3rd millennium BC, and those from the Bronze and Pre-Roman Iron Age from ca. 1200 BC until the beginning of CE, with a few exceptions from later Iron Age. From the third major period, individuals from ca. 1200–1600 AD, conventionally regarded as ’medieval’, have been involved in this study.
If the information in question was lacking beforehand, the preliminary estimation of the age at death of the individuals of this study was made at sample collection and the Bronze and Iron Age individuals who were included in autosomal analyses (having at least 10,000 overlapping SNPs with the EBC-chipDB) were radiocarbon dated.
Late Bronze Age stone-cist graves
Late Bronze Age (1100–500 BC) in Estonia is a period where first stone graves, extensive permanent field systems and cup-marked stones appear in the near-coastal alvar areas. Settlement sites of the period are small and poor in both finds and construction remains. The main settlement units were probably single farms, inhabited by small family or kin groups who subsisted on agriculture. Around 900–800 BC a few so-called fortified settlements were established on the island of Saaremaa and on the northern coast. These probably functioned as centres of (bronze) trade and were inhabited by larger groups. Late Bronze Age inland Estonia, on the other hand, is considerably poorer in archaeological sites, apart from settlement sites in open landscape and, in smaller numbers, on hilltops. Some hilltop settlements may have been similar to the coastal fortified settlements in function and nature.
Stone-cist graves are above-ground burial/mortuary structures, built of limestone or granite stones, or a combination thereof. They are round in shape, with a diameter from a few up to a few dozen metres, and their height rarely exceeds a metre. The graves feature one or more stone circles or drystone walls, which surround one or more human-length stone cists. A cist usually encloses several inhumations, both adults and children, with no clear patterning in age or sex categories. Some cists contain also or only cremated bones. Burials (or secondary bone deposits), both burnt and unburnt, are also common outside the cists. Grave goods are usually few and can rarely be associated with a particular skeleton. The most characteristic finds are bone pins; pottery is also commonplace (except for the earliest stone-cist graves); metal, amber and stone items are infrequent. While the grave type and one group of grave goods (imported bronzes) refer to dense contacts with Scandinavia, the other items (e.g. bone pins, temple ornaments, some of the pottery) witness the contacts with people in the East-European Forest Belt.
Stone-cist graves are distributed along the near-coastal zone of northern and western Estonia, including the largest islands. The graves usually come in groups. One such group is believed to have served a single family or kin group for several centuries. It is possible that only selected members of a household or kin group were accorded a burial in a stone-cist grave. Radiocarbon dating of bones have shown that the stone-cist graves appeared in what today is Estonia between 1200 and 1100 BC and were built until ca 400 BC at the latest. Many of them, however, contain occasional burials from the Iron and even the Middle Ages.
Besides stone-cist graves, cairn graves may have been occasionally built. The main difference of this grave type from stone-cist graves is the absence of cist(s) and, in some cases, ring wall(s). At the end of the period, early tarand graves appeared (see below). Most probably other types of burial sites existed, for instance burials in pits, but information on such sites is very limited.
In view of the hypothesis that a group of stone-cist graves represents a single kin group, aDNA samples were collected from as wide variety of such groups as possible. Males were targeted, since the aim was to focus on Y chromosome diversity. In this article, twenty-three burials from Late Bronze Age in Estonia are analysed.
Kangru at Väo
Location: Väo, Harjumaa, Estonia
Excavations: 1959 [41], 1976–1977 [42], 1980 [43]
Cemetery: A minimum of nine stone-cist graves (numeration of graves differs in publications; we follow the numeration as in Lõugas 1981 [43]). Beside inhumations within cists, occasional cremations and inhumations outside cists were also present. No proper osteological analysis has been performed (but see Lang & Ligi 1991 [44]). Bronze Age artefact finds were rare and included a bronze razor, dated to the IV or V period of the Nordic Bronze Age, and a few bone pins. Some Iron Age objects were also uncovered. The scarce evidence for dating suggests that the grave group was established around 900 BC at the latest.
DNA-analysed individuals:
X16: Male(?) from the cist of grave 1 (AI 4303), age unknown (the bones were too fragmented and intermingled for a preliminary age estimation). Sampled tooth right lower second molar (r M2), date 2834 ± 26 BP (SUERC-80019 (GU47830); 1080–910 cal BC). The cist contained remains of at least one other individual.
X17: Male from grave 8 (skeleton 1; AI 4939), age 25–35 years. In the absence of excavation records, location of the skeleton within the grave is indeterminate; position in one of the grave’s two cists is likely. Sampled tooth r M2, date 2732 ± 28 (SUERC-80020 (GU47831); 930–810 cal BC).
X18: Male from grave 8 (skeleton 3; AI 4939), age 17–22 years. Location of the skeleton within the grave cannot be established, but there are grounds to suggest that it was a burial outside the cists. The excavators dated the burial outside the cists to the Middle Ages [42], but an earlier date cannot be excluded.Sampled tooth left upper canine (l C1).
Jaani at Väo
Location: Väo, Harjumaa, Estonia
Excavations: 1982 [45]
Cemetery: Two stone-cist graves and a ship grave, attached to one another. A minimum of thirty-eight individuals, predominantly inhumations, had been interred in to the stone-cist graves; original cremation deposit of the ship grave has been lost since excavations [46]. The bones were heavily intermingled. Radiocarbon dates of the bones [46] show that the first of the stone-cist graves (B) was probably erected between 800 and 500 BC; grave A was attached in the 5th century BC at the latest. The graves were used for burial also in the Pre-Roman Iron Age and even later, until at least the 7th century AD. Artefact finds comprise pottery, a bone pin, and several poorly datable metal objects from the Iron Age and even later periods. The distance from the Pärna graves (see below) was ca 190 m, which means that the separation of the grave groups may be artificial.
DNA-analysed individual:
V16: Male from the cist of grave A (skeleton 1; AI 5220), age 30–40 years [46], date 2399 ± 27 BP (UBA-24124; 730–390 cal BC) [46]. Sampled tooth r M1. The cist contained an iron knife and, most probably, an indeterminate number of other skeletons.
Pärna at Väo
Location: Väo, Harjumaa, Estonia
Excavations: 1895 [47], 1972–1973 [48]
Cemetery: The original number of the stone graves is unknown. At least four graves have been excavated, but the information on the results is poor. No osteological analysis has been carried out. Artefact finds include pottery and a bone pin; a few Roman-period metal objects were also uncovered. The artefacts and the radiocarbon date suggest that the graves were present before 800 BC. There is a possibility that the Pärna and Jaani graves (see above) were built and used by the same community.
DNA-analysed individuals:
X19: Individual from the cist of grave 1 (AI 4620: L44), sex and age unknown (the bones were too poorly preserved for a preliminary estimation during sample collection). Sampled tooth r M1.
X20: Individual from the cist of grave 1 (AI 4620: L46), sex and age unknown (the bones were too poorly preserved for a preliminary estimation during sample collection). Sampled tooth r M2, date 2677 ± 26 BP (SUERC-80018 (GU47829); 900–800 cal BC).
The cist also contained a clay vessel and remains of at least one sub-adult [44].
Iru
Location: Iru, Harjumaa, Estonia
Excavations: 1936 [49], 1974 [50]
Cemetery: Nine dispersed stone-cist graves, all excavated. The original number of graves was greater, and the graves possibly formed several (sub-)groups. Excavations yielded more than twenty inhumations [44], predominantly but not exclusively from the cists. A few deposits of cremated bone were also recorded. A proper osteological analysis is still to be done. The most characteristic grave inclusions were bone pins and pottery. The finds and a few radiocarbon dates (unpublished) suggest that the cemetery was established around 900 BC at the latest. Some burials or bone deposits outside cists may be later than Bronze Age in date.
DNA-analysed individuals:
X01: Male from the cist of grave 6 (AI 4808: L10), age 17–25 years. Sampled tooth l M2. The cist also contained remains of at least two sub-adults, a bone pin and pottery.
X02: Male from the cist of grave 14 (AI 4810: L5), age 17–25 years. Sampled tooth l M3, date 2834 ± 28 (SUERC-80017 (GU47828); 1090–910 cal BC). The cist also enclosed skeletons of at least two children and a bone pin.
X03: Male from the cist of grave 18 (AI 4811: L11), age 35–45 years, date 2595 ± 30 BP (Hela-2413; 830–590 cal BC [Laneman, unpublished]). Sampled tooth l M3. The cist contained a skeleton of another adult and two bone pins.
Lastekangrud at Rebala
Location: Rebala, Harjumaa, Estonia
Excavations: 1982 [51], 2000 [52]
Cemetery: Six stone-cist graves, one of them almost completely destroyed before excavations. The five remaining graves contained at least 40 inhumations, both inside and outside cists; cremated human bones were also present in almost each grave [52,53]. A quarter of the inhumations were infants, interred in grave 2 in the 15th century AD. The cist burials date from ca 800–400 BC, and a few individuals outside cists from the following centuries (unpublished radiocarbon data). Bronze Age artefact finds include clay vessels and bone pins, mostly in cists. Other areas of the graves contained occasional poorly datable metal items from various periods of the Iron Age and even beyond.
DNA-analysed individuals:
X13: Male from the cist of grave 2 (AI 5229), age 18–22 years [53], date 2485 ± 30 BP (Hela-2127; 780–480 cal BC) [Laneman, unpublished]. Sampled tooth l M1.
X14: Male from the cist of grave 2 (AI 5229), age 35–45 years [53], date 2481 ± 30 BP (Hela-2061; 780–430 cal BC) [Laneman, unpublished]. Sampled tooth l M2.
The cist also contained inhumed remains of an infant, cremated human bones, and a poorly preserved iron object.
Jõelähtme
Location: Jõelähtme, Harjumaa, Estonia
Excavations: 1982–1984 [54]
Cemetery: A dense cluster of thirty-six stone-cist graves with the remains of roughly a hundred inhumations (osteological analysis is incomplete, see Varul 2016 [55]). The cemetery was originally even bigger, as part of it has been destroyed by road construction and was in use between ca 1200/1100 and 800 BC (unpublished radiocarbon data). Grave goods include small bronze items, mostly of Scandinavian origin (razors, tweezers, buttons), bone pins and a few amber beads.
DNA-analysed individuals:
0LS11: Male from grave 34 (AI 5306), age 30–50 years [34], date 2815 ± 33 BP (Hela-2361; 1060–850 cal BC) [34]. The bones were commingled with the remains of at least one other adult and were located both inside and outside the cist. The DNA-analysed tooth was found outside the cist, but it is likely that the original location of the skeleton was in the cist. Fragments of two bone pins were found alongside. Isotope (Sr and O) analysis showed that the man had been born locally [34]. Sampled tooth left upper second premolar (l P2).
V9: Male from the cist of grave 7 (AI 5306), age 30+ years [34], date 2924 ± 32 BP (Hela-2365; 1220–1010 cal BC) [56]. Bronze tweezers and a bronze razor were found together with the skeleton. Isotope (Sr and O) analysis showed that the man had been born locally [34]. Sampled tooth r P1.
Toomani at Muuksi (Hundikangrud)
Location: Muuksi, Harjumaa, Estonia
Excavations: 1924–1926 [57], 1936 [58], 1976–1983 [59], 1995–1996 [60]
Cemetery: About forty closely spaced stone-cist graves, five of which have been excavated in their entirety and twelve partially [56]. Inhumations occur both inside and outside of cists; the same applies to the few cremation deposits. The number of excavated inhumations is well over thirty. Artefact finds comprise a few items of flint, quartz and bone. Radiocarbon data from the completely excavated grave 5 shows that burial began around 1100 BC at the latest, and ceased around 900 BC at the latest [56]. No such data is available for other graves, but in view of their uniform characteristics it is likely that the whole group dates from ca 1200–800 BC. Distance from the Lõokese graves (see below) is ca 1 kilometre.
DNA-analysed individuals:
V14: Male from cist 1 of grave 5 (AM 365: T4), age 50–60 years [61], date 2966 ± 29 BP (SUERC-44064 (GU29245); 1280–1050 cal BC) [56]. Isotope (Sr and O) analysis showed that the man had been born locally [34]. Sampled tooth l P1. The cist also housed remains of an adult female.
X05: Male from cist 2 of grave 5 (AI 6320: L135), age 20–25 years [62], date 2908 ± 26 BP (SUERC-44069 (GU29247); 1210–1010 cal BC) [56]. Sampled tooth l M3. A tooth of a dog was found nearby.
X06: Male from cist 4 of grave 5 (AI 6320: L176), age 25–35 years [62], date 2906 ± 25 BP (SUERC-44070 (GU29248); 1200–1010 cal BC) [56]. Sampled tooth l M3. The cist also housed remains of a child.
X07: Male from cist 2 of grave 12 (AM 365: T15), age 30–40 years [61]. Sampled tooth l M3.
Lõokese at Muuksi
Location: Muuksi, Harjumaa, Estonia
Excavations: 1921 [63]
Cemetery: Six stone graves, only one (partially) excavated. The grave had three parallel cists (A, B, C) built crosswise over the fourth (D). The upper cists housed a single skeleton each; cist D housed two inhumations. Remains of a child were uncovered outside the cists. Artefact finds comprise only three potsherds. Distance from the Toomani graves (see above) is ca 1 km.
DNA-analysed individuals:
X08: Male from cist A (AM ?: L1), age 50 years [61]. Sampled tooth l M3, date 2733 ± 26 BP (SUERC-80021 (GU47832); 930–810 cal BC).
X09: Male from cist B (AM ?: L2), age 18–20 years [61]. Sampled tooth l M2, date 2606 ± 28 BP (SUERC-80025 (GU47833); 820–770 cal BC).
X10: Male from cist C (AM ?: L3), age 60 years [61]. Sampled tooth l M1, date 2926 ± 28 BP (SUERC-80026 (GU47834); 1220–1020 cal BC). A potsherd and a tooth of a dog were reported nearby.
Napa
Location: Napa, Ida-Virumaa, Estonia
Excavations: 1927–1928 [64–66]
Cemetery: Around fifteen or twenty stone graves, of which partially excavated were at least five stone-cist graves and a probable tarand grave. Numeration of graves differs in publications; in this paper we generally follow Friedenthal 1932 [61]. The excavated cists housed a minimum of fourteen inhumations, and some cists had an assemblage of cremated bones beneath the cist floor. Both inhumations and cremations were observed outside the cists. Osteological analysis is available for only the cist inhumations [61]. Grave goods included a few bone pins and items of flint, bronze, and iron. The finds and radiocarbon dates show that the cemetery was present in the 9th century BC at the latest; it may also contain a few centuries older as well as a few centuries younger burials.
DNA-analysed individuals:
X11: Male from the cist of grave 3 (5 in other referred sources) (AM 331: N10), age 50 years [61]. Sampled tooth r M2, date 2805 ± 26 BP (SUERC-80010 (GU47824); 1030–890 cal BC). A single potsherd was found nearby.
X12: Male from the cist of grave 4 (6 in other referred sources) (AM 331: N11), age 40–50 years [61]. Sampled tooth r M2, date 2652 ± 26 BP (SUERC-80011 (GU47825); 900–790 cal BC). The cist also contained an infant. Burnt bones and a fragment of a bone pin were found under the cist floor.
Vehendi
Location: Vehendi, Tartumaa, Estonia
Excavations: 1894 [67], 1975–1976 [68]
Cemetery: Eleven stone mounds distributed within a one kilometre long stretch along the coast of Lake Võrtsjärv. Two mounds, nos 11 and probably 1, have been excavated, but information on the 19th-century digs is poor. The available evidence suggests that the graves are probably cairn and not stone-cist graves, i.e. their structure includes a stone circle but no cists. Grave 11 contained an inhumation in the centre (not available for analysis) and a few other bone deposits, both burnt and unburnt, in other parts of the cairn. The burials were poorly preserved, and no osteological analysis has been applied to the bones. No artefact finds were uncovered. The radiocarbon date obtained for the current project shows that the grave(s) must have been present around 1000 BC at the latest.
DNA-analysed individual:
X15: Male(?) from the eastern periphery of grave 11 (skeleton 3; AI 6950). The teeth indicate a relatively aged person. Sampled tooth l M1, date 2899 ± 28 BP (SUERC-80016 (GU47827); 1210–1000 cal BC).
Pre-Roman Iron Age early tarand and other cemeteries
In the Pre-Roman Iron Age (500 BC–50 AD), new developments took place in the culture and settlement pattern in what today is Estonia. The fortified sites were abandoned around 500 BC and an open settlement pattern (most likely in the form of single households) spread everywhere, both in coastal and interior regions. In the later Pre-Roman Iron Age, a new short-lived fortification wave can be observed all over the country. The building of new stone-cist graves was terminated around 400 BC at the latest. At some point of time within the period of ca 800–500 BC (due to difficulties in calibration of radiocarbon dates of that period it is not known when exactly), a new form of burying sites was introduced in coastal zone – the so-called early tarand cemeteries. Some of the earliest tarands were erected side by side with, or in close proximity to, stone-cist graves, the rest of them were built separately from other burial sites. In addition, burial sites of other forms are known, such as cairn graves, pit graves with either inhumations or cremations, and burial sites where cremated bones have been scattered over an open surface of the ground.
Early tarand cemeteries form a peculiar and diverse group of burial sites that were spread in Estonia, northern and western Latvia, south-western Finland, Ingria, and eastern central Sweden. Tarands are quadrangular stone enclosures for individual or collective burials built on the ground, with the straight flat sides of the walls facing outwards. The number of tarands in a cemetery can vary from one to a few dozens, and if there is more than a single tarand they are joined together. Inhumation was the original and most common burial custom in the earliest cemeteries during the early Pre-Roman Iron Age; cremation was introduced later, at the end of this period, but inhumation did not disappear. The number of burials in one tarand can vary greatly: in earlier cemeteries with smaller tarands this number rarely exceeds two or three; in later cemeteries, one tarand can house up to a dozen or even more individuals.
Grave goods were quite rare in the earliest graves that can be dated to the period of ca 800–500 BC by the radiocarbon method. The only grave goods of that time were clay pots of Ilmandu type, a new style in Estonian Final Bronze Age pottery, which was formed under the influences from the Oka and Moscow rivers’ region [20]. During the 5th–3rd centuries BC, many metal artefacts appeared among the grave goods, such as neck-rings and bracelets of bronze, massive bracelets of iron, temple ornaments with spoon-shaped ends, a variety of decorative pins (of bronze and iron, and bimetallic), etc. A distinguished group of grave goods originates in the East-European Forest Belt [69] but artefacts imported from central and northern Europe were not unique either. During the last centuries BC and the first century AD, the finds in tarand graves became more numerous: ornaments (shepherd’s crook pins, bracelets, finger-rings, etc.), small-sized tools (knives), and pottery (incl. cord- and comb-decorated vessels).
In this article, fourteen burials from Pre-Roman Iron Age in Estonia are analysed.
Loona
Location: Loona, Saaremaa, Estonia
Excavations: 1958–1959 [70]
Cemetery: Two stone graves, one of them excavated; four other stone graves at a distance of 300 metres [59]. The excavated grave was a stone-cist grave which contained at least seventeen inhumations outside the empty cist in generally lower layers and numerous deposits of cremated bone in upper layers. No osteological analysis has been performed. Artefact finds include various bone and amber objects (probably ornaments), iron and bronze bracelets, temple ornaments, and pottery. The finds, the majority of which have close parallels in early tarand graves, suggest that the grave was built in the Bronze Age, and was used for burial also in the Pre-Roman Iron Age. More precise dates are difficult to establish.
DNA-analysed individual:
X04: Male in the south-western part of grave 1 (skull 10; AI 4210). In preliminary examination, teeth and bones yielded contradictory evidence on age at death estimate (17–25 and 40+ years, respectively). Sampled tooth r M1, date 2331 ± 26 BP (SUERC-80015 (GU47826); 480–360 cal BC).
Tandemägi IV at Võhma
Location: Võhma, Lääne-Virumaa, Estonia
Excavations: 1969–1972 [71–73]
Cemetery: Tandemägi is a long ridge with seven stone settings. In the north-western part of this ridge there were three stone-cist graves (I–III) of the Late Bronze Age. The tarand cemetery (IV), dated from the Pre-Roman Iron Age, had been built on the south-eastern end of the ridge, 76 m apart from the stone-cist graves. It consisted of three quadrangular enclosures with altogether at least fifty inhumations and five cremations [74]. The cemetery was rather rich in grave goods, which mostly belonged to the late Pre-Roman Iron Age: ceramics, shepherd’s crook pins of iron, bracelets of bronze, knives and an axe of iron, etc. In contrast to generally very fragmentary and intermingled skeletons there was a well-preserved triple burial in tarand 2. It consisted of a 30–35 years old male, a 20–25 years old (fe)male, and a 6–7 years old child. The adults were richly furnished with grave goods: the older male had a neck-ring and a decorative pin of bronze, two bracelets of iron and one more of bronze; the other adult had a similar neck-ring and three bronze bracelets whereas the child had only a bronze temple ornament [72,73]. All these grave goods have an early Pre-Roman Iron Age date.
DNA-analysed individual:
VII4: Male from the triple burial (AI 5074: L64), age 30–35 years [74]. Sampled tooth r M3, date 2425 ± 35 BP (Poz-98210; 760–400 cal BC).
Hiiemägi at Kunda
Location: Kunda, Lääne-Virumaa, Estonia
Excavations: 2004–2006 [34]
Cemetery: The cemetery is located on a ridge called Hiiemägi in the outskirts of the town of Kunda. The cemetery has been ca 50 m long but was largely destroyed by quarrying. Only a small part of the cemetery was excavated but the results are not properly published as yet [34]. There were eleven small cist-like tarands distinguished in the excavated area, each of them contained one or more inhumation burials (altogether 32). Grave goods were very poor: a few potsherds, animal bones, a knife and three small decorative pins of iron from the early Pre-Roman Iron Age.
DNA-analysed individuals:
0LS10: Male from tarand III (burial 9; TÜ 1325: L777), age 17–25 years [34]. He had a fragment of a sheep/goat bone and ceramics as grave goods. This burial has two radiocarbon dates: 2430 ± 35 BP (Poz-10801; 760–400 cal BC) and 2530 ± 41 BP (UBA-26114; 800–530 cal BC) [34]. According to the isotopic analysis, the person was not born in the vicinity of Kunda; his place of birth is still unknown (but south-western Finland and Sweden are excluded) [34]. Sampled tooth r P1.
V10: Male from tarand XI (burial 24; TÜ 1325: L1925), age 25–35 years [34], date 2484 ± 40 BP (UBA-26115; 790–430 cal BC) [34]. He had a few potsherds near the skull. Likewise, this person was not locally born [34]. Sampled tooth l P1.
Kurevere
Location: Kurevere, Saaremaa, Estonia
Excavations: 1974–1975 [75,76]
Cemetery: It was one of the stone settings in a larger group and consisted of three structural parts: (1) a round-shaped grave surrounded with two concentric stone circles (but no cist in the centre), (2) a much larger stone circle around the former, and (3) ca 20 tarand-like enclosures by the northern, southern and western sides of the large stone circle. The majority of burials were inhumations, but the bones were rather fragmentary and intermingled. Cremated bones occurred sporadically and can be connected with the latest stage in the use of this burial site. Osteological material has not been analysed so far, however. Grave goods were quite numerous consisting mainly of pottery, various ornaments of bronze and iron (shepherd’s crook pins, a pin with a spiral-shaped head, bracelets, various temple ornaments, decorative mounts, etc.), tools (knives, awls, an axe, and a grinding stone), and a few weapons (fragments of a spearhead and a battle knife). The earliest part of the cemetery (the two concentric circles) was already built in the Late Bronze Age, while the rest of the cemetery belongs to the Pre-Roman Iron Age.
DNA-analysed individuals:
V11: Male(?) buried in the northern portion of the large stone circle (AI 4780: L17), age 25–35 years. Sampled tooth r M2, date 2220 ± 35 BP (Poz-98256; 390–200 cal BC).
V12: Male(?) buried in tarand VII (AI 4780: L118), age 25–35 years. Close to the bones there were also pieces of a clay pot with cord decoration found. Sampled tooth r M3, date 2125 ± 35 BP (Poz-98257; 360–40 cal BC).
Ilmandu III
Location: Ilmandu, Harjumaa, Estonia
Excavations: 1994 [77]
Cemetery: The cemetery belongs to a larger group of burial sites (stone-cist graves and early tarand cemeteries), which are dispersed over the lands of Ilmandu and Rannamõisa villages close to northern Estonian limestone cliff. Cemetery III of Ilmandu was partially destroyed by building a house. Altogether six tarands and two cist-like constructions were distinguished in the preserved part of the cemetery. All burials were inhumations, except a few cremated bones that were of later date. Osteological material is properly not analysed but during excavations at least seventeen adult individuals were distinguished. Grave goods were very poor consisting of pottery of Ilmandu type and a temple ornament.
DNA-analysed individuals:
0LS09: Female from cist I (AI 6009: L180), age 19–25 years [34], date 2361 ± 29 BP (SUERC-44060 (GU29241); 540–380 cal BC) [56], most likely locally born [34]. Sampled tooth r P1.
V7: Male from tarand IV (burial 1; AI 6009: L166), age 35–45 years [34], date 2484 ± 41 BP (UBA-26113; 790–430 cal BC) [34]. According to isotopic analyse, this person was most likely locally born [34]. Sampled tooth l M3.
V8: Male(?) from tarand IV (burial 9; AI 6009: L184), age 17–25 years, date of right femur 2413 ± 29 BP (SUERC-44062 (GU29243); 750–400 cal BC) [56]. Furnished with a clay pot of Ilmandu type and a bronze temple ornament (fragment). Sampled tooth l M1, date 2405 ± 35 BP (Poz-98215; 750–390 cal BC).
Tõugu II
Location: Tõugu, Lääne-Virumaa, Estonia
Excavations: 1993–1995 [72,73]
Cemetery: There is a group of at least eleven stone settings at Tõugu but only one of them is excavated. Cemetery II consisted of three separate parts: a stone-cist grave from the Bronze Age (IIA), topped with a large single tarand (IIB) of the Pre-Roman Iron Age, and a chain of five tarands (IIC) that was erected next to the latter structures also in the Pre-Roman Iron Age. According to Jonathan Kalman [78], there were altogether at least twenty-five inhumations excavated from the Tõugu II cemetery, sixteen of them from the series of five interconnected tarands IIC. Grave goods were rather poor, including pottery, iron knives, some bracelets of bronze, pieces of quartz and a few grinding stones.
DNA-analysed individual:
V15: Male from tarand 1 of the cemetery IIC (AI 6003: L637), age 25–35 years [78]. Sampled tooth l M2.
Poanse I
Location: Poanse, Läänemaa, Estonia
Excavations: 1975–1976 [79,80]
Cemetery: There were two Pre-Roman tarand cemeteries close to each other. Cemetery I consisted of seven enclosures. Kalman [81] identified forty-four burials in this burial site, whereas most remains were commingled and fragmentary. In some cases, the skeletons were preserved well enough to make the identification of individual burials possible. The majority of burials were without grave goods, but some were furnished quite remarkably with bracelets of iron and bronze, shepherd’s crook pins, temple ornaments with spoon-shaped ends, and cord-decorated pottery; as an extraordinary find for tarands also a sickle should be mentioned. Cemetery II was smaller than cemetery I, it consisted of two tarands and housed altogether thirty-four burials. Judging from grave goods – a spearhead, bracelets, shepherd’s crook pins, knives, and pottery – cemetery II was at least partly contemporary with cemetery I in the mid- and late Pre-Roman Iron Age.
DNA-analysed individuals (cemetery I):
VII2: Male(?) buried in tarand 1 (AM A483: L18), age 17–25 years. Sampled tooth r M3, date 2275 ± 35 BP (Poz-98208; 410–200 cal BC). Tarand 1 was built as the first enclosure in this cemetery. Together with this male person there were also two juveniles (14–18 and 16–18 years old) and a 50+ years old male, and a few subadults buried.
VII3: Male from tarand 4 (AM A483: L30), age 30–40 years [81]. Sampled tooth r M3, date 2205 ± 35 BP (Poz-98209; 380–180 cal BC). Buried together with four adults and two children. Tarand 4 was built some time (perhaps a few generations) later than tarand 1.
Alu
Location: Alu/Kalevi, Raplamaa, Estonia
Excavations: 2015 [82]
Cemetery: The site, a low moraine hump covered in field clearance stones, contained only two inhumations, a few metres apart from each other. The clearance cairn, which was of a later date, made it difficult to determine the original appearance and type of the burial site. One individual had been interred in a shallow earth-cut grave which, possibly, may have been surrounded and/or covered with stones, including sizeable boulders. The other burial structure possibly also included a shallow pit grave, and most certainly boulders and smaller rocks had been used in its construction. The stone structure had been disturbed and the bones were scattered. Both of the skeletons belonged to adults, perhaps mature adults, but a more precise age-at-death estimation was impossible due to poor preservation of bones; sex determination by osteological methods was not possible. No grave goods were found, though some of the pottery, scattered over the site, may have been contemporary with the burials.
DNA-analysed individuals:
0LS07: Individual in the earth-cut grave (TÜ 2525: L264), adult [82], dates 2209 ± 33 BP (SUERC-63659 (GU38997); 380–190 cal BC), 2213 ± 33 BP (SUERC-63660 (GU38998); 380–190 cal BC) [82]. Sampled tooth l P2?.
0LS08: Individual in the stone structure (TÜ 2525: L291), adult [82], dates 2162 ± 31 BP (SUERC-63661 (GU38999); 360–110 cal BC), 2166 ± 33 BP (SUERC-63665 (GU39000); 360–110 cal BC), 2145 ± 31 BP (SUERC-63666 (GU39002); 360–50 cal BC) [82]. Sampled tooth r M2.
Pre-Roman and Roman Iron Age cemeteries in Ingria, Russia
Archaeological material from Pre-Roman (500 BC–50 AD) and Roman Iron Age (50–450 AD) in Ingria, south-western part of Leningrad district in Russia, are quite limited and studied only a little more than 30 years [83]. Most common type of archaeological sites is so-called tarand cemeteries. The tarand cemeteries have been excavated more widely at the burial sites of Kerstovo 1 and Malli, but similar structures are found also at the cemetery of Valgovitsy and Velikino. Isolated finds, possibly originating from disturbed burials, were found in the villages of Ratchino, Georgiyevsky, Voynosolovo and Ropsha. The walls of tarands were built of granite stones and limestone, while the inner space was filled with smaller stones and limestone gravel.
The overwhelming number of finds from Ingrian tarands is dated to the Early Roman Period, that is, to the time span from ca 75 to 200 AD. The grave goods included different types of fibulas, bracelets, rings, temple rings, weapons and iron tools for everyday life (spearheads and javelin heads, socketed axes, razors, awls, needles, scythes, knives). In Kerstovo 1 and Malli plaques imported from more eastern regions of the East-European Forest Belt (basins of the Upper Volga, Mologa, Middle Volga and the Kama region) were found. The tarand cemeteries in Ingria represent a local variant that finds its closest parallels at sites in north-eastern Estonia. The easternmost site in Estonia – the cemetery of Utria – is located some 40 km to the west of the sites on the Izhora Heights. The tarands in Ingria have a distinctive difference compared to those in Estonia by the presence of numerous weapons (spearheads, javelin heads, axes) and objects imported from the more eastern areas.
Among other findings there are three hoards of Roman coins that were discovered near the village of Koporye worth mentioning [83].
In this study, twelve burials from Pre-Roman and Roman Iron Age in Ingria are analysed.
Kerstovo I
Location: Kerstovo, district of Kingisepp, north-western Russia
Excavations: 2008–2009 [84], 2016 [83]
Cemetery: The burial ground is situated on an arable field and its upper level was partly disturbed. A funerary installation, elongated along the west-east line, consisting of a chain of four tarands was investigated. Numerous skeletal remains were discovered – altogether ca. 19 kg of bones, mostly calcined. The bones were found within the structures both as isolated pieces and in associations. The rite of an outside cremation prevailed and the skeletal remains are predominantly represented by small calcined fragments. Also fragments of unburned bones were found; these were lying in no anatomical order. At least 38 persons were interred at the site. The grave goods from the excavations – 155 items altogether – included parts of garments and bronze ornaments, among others different types of fibula. Other ornaments include bronze bracelets, rings, temple rings, large beads, with a lug, and an iron clasp. In addition, weapons and iron tools for everyday life were found (spearheads and javelin heads, half-moon-shaped razors, a scythe, a needle, awls, and knives), as well as a gold-glass bead, a bronze needle, and fragments of ceramic vessels. The surface finds (150 metal objects) included bronze ornaments – eye brooches, profile fibulae, rings, and a plaque in the form of a rosette, as well as iron javelin heads and spearheads, socketed axes, and knives. The materials from the excavations and the surface finds suggest that also other tarand cemeteries can be found here.
DNA-analysed individuals:
VII15: Adult from tarand 3 (horizon 3). Sampled tooth l C1, date 1980 ± 30 BP (Poz-103328; 45 cal BC–77 cal AD).
VIII7: Adult from tarand 3(horizon 2), No. 219. Sampled tooth l P1.
VIII8: Adult from tarand 2 (horizon above bedrock), No. 2979. Sampled tooth r C1.
VIII9: Adult from tarand 2 (horizon 3). Sampled tooth r P2.
Malli
Location: Malli, district of Kingisepp, north-western Russia
Excavations: 2010–2011, 2013 [83,85–88]
Cemetery: The burial structure was consisting of two tarands and stone pavements. The westernmost tarand (NNE-SSW) was evidently built first. After destroying its eastern wall, a new tarand was constructed there in a slightly different orientation. The walls were joined by a lateral mound constructed of limestone gravel and granite pavement. The lateral mound was well preserved along the western wall of the western tarand. To the south and east of the tarands, a stone pavement was discovered.
The calcined and unburned bones (ca 116 kg) were deposited within the structures both dispersed and in accumulations but with no anatomical order.
The grave goods – 850 artefacts in total – are distinctly subdivided into two chronologically different groups. The first group is dated to the time of construction of the tarand cemeteries, i.e. the Roman Iron Age; the second group derives from the third quarter of the 1st millennium AD. The finds of the Roman Iron Age are represented by bronze and iron ornaments (fibulae and their parts), as well as ceramics. The discovered bronze ornaments also included closed and spiral finger rings, bracelets, spiral beads, spiral temple rings, possible fragments of neck rings and some other rare specimens. These objects are typical of the Pyanobor archaeological culture and were evidently imported from the Kama River region. The weaponry and tools included iron spearheads, scythes and knives with a curved back, awls, a miniature pick-axe, and a miniature knife. Fragments of ceramics with striated and smoothed surfaces belong to the same period.
DNA-analysed individuals:
VII14: Adult from pit No. 8, No. 2479. Sampled tooth r P1.
VIII4: Adult from the stone pavement, 2011, No. 2348 (horizon 4). Sampled tooth ? P?.
VIII5: Adult from the eastern tarand, 2011, No. 1622 (horizon 2). Sampled tooth r C1.
VIII6: Adult from the eastern tarand, 2013 (horizon 2). Sampled tooth l I2.
Udosolovo
Location: Udosolovo, district of Kingisepp, north-western Russia
Excavations: 2013 [89,90], 2016-2017 [Stasyuk, unpublished]
Cemetery: The cemetery was originally a low flat stone mound of approximately rectangular shape (oriented NW-SE), badly damaged. The lower layer of burials in this mound reveals some inhumations in single stone cists, six of which were investigated. The skeletons were lying stretched on the back, head to the north. The cists were fragmentarily preserved, the bones were crushed into pieces by the weight of the stones and soil, some of the bones were displaced. Only a few items were found in graves: a narrow bronze bracelet, a javelin head and a fragment of an iron plate. Numerous small fragments of pottery (including those with striated surfaces) were found in this layer. The lower layer of burials in Udosolovo cemetery should be dated by the late Pre-Roman Iron Age (1st century BC – the first half of the 1st century AD).
The upper layer of burials in the cemetery contained some scatterings of cremated bones mixed with gravel and soil, lying directly under the present turf. Between the two stages of the use of this cemetery there was a chronological gap, during which the stone cists were destroyed. No stone structures were found in the upper layer of the mound. There were almost no ceramics in the upper layer, but there were numerous metal items, often melted: an iron razor, iron knives, spirals of bronze wire, pieces of narrow bronze bracelets, a fragment of a silver neck-ring, etc. Finds from the upper layer with cremations are similar to those of tarand cemeteries in Northern Estonia and allow to date the assemblage to the 3rd century or even later, to the 5th–7th centuries.
DNA-analysed individuals:
VII16: Male from burial 1, age 25–35 years [89]. Sampled tooth l M3.
VIII10: Male from burial 5, age 20–40 years [89]. Sampled tooth l M1.
VIII11: Adult [89] from square 4 (upper horizon). Sampled tooth l M1.
VIII12: Adult [89] from square 4 (horizon on stone layer). Sampled tooth r M2.
Medieval rural cemeteries in Estonia
During the entire first millennium AD cremation burials were practised in Estonia. Inhumations with potential for aDNA analysis re-appear in the late 10th/11th century. The 11th and 12th centuries belong to the High Middle Ages in the historical chronology of western and central Europe, but Iron Age societies and culture still continued in the eastern Baltic area in that time.
The territory of Estonia was gradually conquered by German and Danish crusaders in the wars of 1208–1227. This conquest and forced Christianization mark the end of the Iron Age and the birth of medieval Livonia – a confederation of small states: the bishoprics of Tartu and Ösel-Wiek in Estonia, those of Riga and Couronia in Latvia, and the Livonian branch of the Teutonic Order in a part of both countries. Northern Estonia belonged to Denmark until 1346, then it was sold to the Order. Although the end of the Middle Ages is usually dated around 1500 AD in Western Europe, for the area of medieval Livonia it is defined by the war with Russia (1558–1561).
In the rural archaeology of Estonia, the borders of the medieval period are, however, flowing and conventional. Burials from pre-conquest decades cannot clearly be distinguished from post-conquest ones. Until the transition of the country to Lutheran Sweden (since 1583 in Northern Estonia, since 1625 in Southern Estonia), the archaeological record of native Estonian population preserves features characteristic for medieval times. Thus, in the context of present research, the Middle Ages are regarded in a long-term perspective and individuals from ca 1200–1600 AD are conventionally regarded as ‘medieval’.
As the Christianization of Estonia took place in a forced and violent way, the acceptance of Christian practices remained limited and a lot of pre-Christian traditions survived in medieval times. While in medieval Christian Europe people were normally buried in consecrated churchyards, in Livonia the dead were often buried at the home place, near villages and hamlets until the early 18th century [91,92]. Although cremation as a pagan practice was banned and greatly abandoned together with Christianization, the non-churchyard village cemeteries existed parallel to churchyards. As the Livonian nobility of German origin was buried in churches and churchyards, individuals from Estonian village cemeteries represent the native Estonian population.
The village cemeteries lie usually 200/300–600/700 m from medieval village centres. If the landscape allows, they are located on low hummocks with the diameter usually from 15–20 to 40–60 metres, sometimes more. In Estonia, there were usually ca 20–30 village cemeteries per parish. The number of people buried there depends on the local situation and duration of use, but it usually comprises several hundreds. Most of rural people were probably buried in village cemeteries in medieval time. The hinterlands of a local cemetery may have comprised from one to 2–4 villages/hamlets, the number probably increasing in time, in parallel to population growth and settlement expansion. In Northern Estonia, the size of a village was mostly between 5–15 ploughlands in the mid-13th century, whereby each unit might roughly correspond to the number of farms, probably inhabited on the average by 5–8 people (incl. children) [93]. Villages of Southern Estonia were often of similar size in the 16th century (earlier data are missing) but in areas with dispersed settlement there were small hamlets based on a few farms only.
Culturally, Estonia can be divided into coastal (sea-oriented) and inland (southern and eastern) areas. This distinction is clearly expressed in Estonian dialects [94], ethnography [95], folklore and traditional popular culture [96], as well as in present-day population genetic data [97]. The difference between the two macro-regions distinctly appears in the archaeological record also in the medieval period.
In the present study, Estonia’s coastal areas are represented by the cemeteries of Karja, Pada and Kukruse, the inland areas by those of Otepää, Vana-Kuuste, Mäletjärve and Vaabina. In coastal Northern and Western Estonia, inhumations appear on some of the village cemeteries (e.g. Pada and Kukruse) some decades before the crusades, as a sign of transition to Christian religion and burial traditions. Some of these sites may have been deserted already soon after the conquest in the 13th century. In that region, grave goods almost disappeared on rural cemeteries since the 2nd half of the 13th century but re-appeared again in the 16th century. In inland Estonia, the pre-Christian practice of burying the dead dressed, together with jewellery items (brooches, rings, necklaces) and furnished with some minor grave goods – coins, knives, needles and other small utensil, survived continuously until the early 18th century. The dead were buried mostly with the head towards west or south-west, according to medieval Christian practices, but in south-eastern Estonia the opposed orientation of men and women, a tradition of pre-Christian origin, lasted until the 17th century.
Considering the presence of well-datable grave goods and coins, as well as relative chronology – in case of cemeteries of long-term use, earlier graves are often cut by later ones – the dates of 2nd millennium AD inhumation burials are not based on radiocarbon samples which often provide a vague and wide date range, but on artefact chronology.
Karja
Location: Karja, Saaremaa, Estonia
Excavations: 1955 [98]
Cemetery: Village cemetery on flat land, studied with rescue excavations (ca 150 m2, 32 burials). The cemetery (full number of graves estimated as ca 70) with graves mainly from the 13th century was probably founded soon after the Christianization (1227) and seems to have been deserted in the early 14th century or by its middle. Burials of both sexes were oriented with the head towards W or SW. Some graves were furnished with jewellery (brooches, bracelets, rings), knives and belt accessories, some were unfurnished.
DNA-analysed individual:
IIa: Male (burial 16; AI 4115), 45+ years old, orientation WSW, furnished with a knife sheath. Sampled tooth r M1, date 1230–1300 AD.
Pada
Location: Pada, Lääne-Virumaa, Estonia
Excavations: 1987–1989 [99,100]
Cemetery: Cemetery on flat land beside large 12th and 13th cc. Pada hill fort, a Final Iron Age district centre, separated from it by a deep valley. The cemetery (investigated 171 burials and 253 m2) which dates from ca 1180–1250 probably belonged to the inhabitants of the hill fort and was deserted when the churchyard of Viru-Nigula was founded. Burials of both sexes were irregularly oriented with the head towards W, SW, E and NE. Graves were rich furnished with jewellery (brooches, bracelets, neck rings, breast chains with pins, rings, necklaces), tools (axes, senses, knives), weapons (spears) and belt accessories. In four graves Gotlandic coins from 1140–1210/1220 were found.
DNA-analysed individual:
IIg: Male (burial 151; AI 5366), 25–35 years old, WSW-oriented, richly furnished – horse harness, 4 silver coins (1140/60–1210/20), knife, belt accessories. Sampled tooth l M3, date 1210–1230/1240 AD.
Kukruse
Location: Kukruse, Ida-Virumaa, Estonia
Excavations: 2009–2010 [101]
Cemetery: Cemetery on flat land, ca 300 m SE of Kukruse manor centre. Rescue excavations (ca 600 m2) revealed 44 inhumations mainly from the late 12th and 13th century and traces of earlier cremations. Burials of both sexes were of diverse orientation W, NW, SW, SSW, S, SE, E, N. Until Christianization (in 1220), and maybe also somewhat later, burials were rich in grave goods. A group of W-oriented graves (inc. grave 9) was most richly furnished with jewellery (brooches, bracelets, neck rings, breast chains with pins, rings, necklaces with silver sheet pendants), tools (axes, senses, knives), weapons (spears, a sword), and metal accessories of the costume. Special publications relate to burial rites [102,103] and artefacts [104].
DNA-analysed individual:
0LS03: Male (burial 9; TÜ 1977), 25–30 years old, oriented towards W, richly furnished (clay vessel, sense, spearhead, knife, fire steel, neck rings, bracelets, brooch etc). Sampled tooth l M3, date 1180–1220/1240 AD.
Otepää
Location: Otepää, Tartumaa, Estonia
Excavations: 1928 [105], 1929 [106], 1938 [107], 1996 [108]
Cemetery: Located on flat land, studied with rescue excavations (ca 330 m2; 136 burials). Otepää was a main castle of Tartu bishopric, with a big urban settlement at its foot in the 13th and 14th cc., the cemetery belongs to its inhabitants. Graves in parallel irregular rows were oriented with the head between W and SW. Judging by the almost total lack of disturbed graves, the site was of short-time use, dated by coin finds to the last third of the 14th century. As most graves contained 2–4 skeletons, the site seems to relate to some epidemic, maybe the plague of 1378 in which 5/6 [109] or even about 9/10 [110] of the people of the bishopric died. Judging by finds typical for the village cemeteries of the region – jewellery (brooches, rings, necklaces of cowry shells, glass beads, bells), knives, and belt accessories, the cemetery belongs to Estonian population.
DNA-analysed individual:
IIf: Male (burial 1; AI 3680), 25–35 years old, oriented towards SW, finds: belt buckle, belt ring, knife. Sampled tooth r M3, date 1360–1390 AD.
Vana-Kuuste
Location: Vana-Kuuste, Tartumaa, Estonia
Excavations: 1982 [111]
Cemetery: Village cemetery on a low hummock in a forest, excavated (ca 75 m2, 99 burials) to identify the character of the site. Investigated burials from the late 13th or 14th to the late 17th century were oriented with the head towards W and SW, furnished with jewellery (brooches, rings, necklaces), knives, coins and belt accessories.
DNA-analysed individual:
ILS01: Male (burial 73; TM A 153), 25–35 years old, oriented towards WSW, finds: knife, penannular brooch. Sampled tooth l M1, date 1500–1625 AD.
Mäletjärve
Location: Mäletjärve, Tartumaa, Estonia
Excavations: 1984 [112]
Cemetery: Village cemetery on flat land, founded beside a Roman Iron Age tarand cemetery. Trial excavations (50 m2, 50 burials) in 1984 to establish the preservation/destruction state of the cemetery. Investigated graves from the late 14th to the early 17th century were oriented towards W and SW, furnished with jewellery (brooches, rings, necklaces), knives, coins and belt accessories.
DNA-analysed individual:
IVLS09KT: Male (burial 18; TM A 155), 30–40 years old, oriented towards SSW, finds: coin from 157?, penannular brooch, knife. Sampled tooth l M3, date 1570–1600 AD.
Vaabina
Location: Vaabina, Võrumaa, Estonia
Excavations: 1985 [113]
Cemetery: Village cemetery on top of a high hummock, studied with rescue excavations (ca 350 m2, remains of 64 skeletons), dates from the mid-13th–late 17th century. Male graves were oriented with the head towards W, female, according to local regional tradition, towards E. Burials were furnished with jewellery (brooches, rings, necklaces), knives, coins and belt accessories.
DNA-analysed individual:
IIIt: Female (burial 43; AI 5354), 40+ years old, oriented towards E, finds: knife, 13th–14th cc. brooch. Sampled tooth r M1, date 1250–1450 AD.
Method details
All of the laboratory work was performed in dedicated ancient DNA laboratories of the Institute of Ecology and Earth Sciences, University of Tartu or the Department of Archaeology and Anthropology, University of Cambridge. The library quantification and sequencing were performed at the Estonian Biocentre Core Laboratory. The main steps of the laboratory work are detailed below.
DNA extraction
The teeth of 56 individuals were used to extract DNA.
Tooth roots were broken off and used for extraction since root cementum has been shown to contain more endogenous DNA than crown dentine [114]. The roots were used whole to avoid heat damage during powdering with a drill and to reduce the risk of cross-contamination between samples. Contaminants were removed from the surface of tooth roots by soaking in 6% bleach for 15 minutes, then rinsing twice with water and lastly soaking in 70% ethanol for 2 minutes, shaking the tubes during each round to dislodge particles. Finally, the samples were left to dry under a UV light for 30 minutes on both sides.
Next, the samples were weighed, [20 * sample mass (mg)] μl of EDTA and [sample mass (mg) / 2] μl of proteinase K was added and the samples were left to digest for 72 hours on a slow shaker at 20 °C to compensate for the smaller surface area of the whole root compared to powder. Undigested material was stored for a second DNA extraction if need be.
The DNA solution was concentrated to 250 μl (Amicon Ultra-15 30 kDa, Merck Millipore) and purified in large volume columns (High Pure Viral Nucleic Acid Large Volume Kit, Roche) using 2.5 ml of PB buffer, 1 ml of PE buffer and 50 μl of EB buffer (MinElute PCR Purification Kit, QIAGEN).
Library preparation
Sequencing libraries were built using NEBNext DNA Library Prep Master Mix Set for 454 (E6070, New England Biolabs) and Illumina-specific adaptors [115] following established protocols [115–117]. The end repair module was implemented using 18.75 μl of water, 7.5 μl of buffer and 3.75 μl of enzyme mix, incubating at 20 °C for 30 minutes. The samples were purified using 500 μl PB and 650 μl of PE buffer and eluted in 30 μl EB buffer (MinElute PCR Purification Kit, QIAGEN). The adaptor ligation module was implemented using 10 μl of buffer, 5 μl of T4 ligase and 5 μl of adaptor mix [115], incubating at 20 °C for 15 minutes. The samples were purified as in the previous step and eluted in 30 μl of EB buffer (MinElute PCR Purification Kit, QIAGEN). The adaptor fill-in module was implemented using 13 μl of water, 5 μl of buffer and 2 μl of Bst DNA polymerase, incubating at 37 °C for 30 and at 80 °C for 20 minutes. The libraries were amplified and both the indexed and universal primer (NEBNext® Multiplex Oligos for Illumina, New England Biolabs) were added by PCR using HGS Diamond Taq DNA polymerase (Eurogentec). The samples were purified and eluted in 35 μl of EB buffer (MinElute PCR Purification Kit, QIAGEN). Three verification steps were implemented to make sure library preparation was successful and to measure the concentration of dsDNA/sequencing libraries – fluorometric quantitation (Qubit, Thermo Fisher Scientific), parallel capillary electrophoresis (Fragment Analyser, Agilent Technologies) and qPCR.
DNA sequencing
DNA was sequenced using the Illumina NextSeq 500 platform with the 75 bp single- or paired-end method. As a norm, 12 samples were sequenced together on one flow cell; additional data was generated for 6 samples on one flow cell to increase coverage.
Quantification and statistical analysis
Mapping
Before mapping, the sequences of adaptors and indexes and poly-G tales occurring due to the specifics of the NextSeq 500 technology were cut from the ends of DNA sequences using cutadapt 1.11 [118]. Sequences shorter than 30 bp were also removed with the same program to avoid random mapping of sequences from other species.
The sequences were mapped to reference sequence GRCh37 (hs37d5) using Burrows-Wheeler Aligner (BWA 0.7.12) [119] and command mem with re-seeding disabled.
After mapping, the sequences were converted to BAM format and only sequences that mapped to the human genome were kept with samtools 1.3 [120]. Next, data from different flow cell lanes was merged and duplicates were removed with picard 2.12 (http://broadinstitute.github.io/picard/index.html). Indels were realigned with GATK 3.5 [121] and lastly, reads with mapping quality under 10 were filtered out with samtools 1.3 [120].
The average endogenous DNA content (proportion of reads mapping to the human genome) for EstBA samples was 21%, for EstIA samples 23%, for IngIA samples 15% and for EstMA samples 36% (Table S1). The variation in the endogenous DNA content was variable as is common in aDNA studies, ranging from under 1% in most sample groups to at least over 60% in all four groups (Table S1).
aDNA authentication
As a result of degrading over time, aDNA can be distinguished from modern DNA by certain characteristics: short fragments and a high frequency of C=>T substitutions at the 5' ends of sequences due to cytosine deamination. The program mapDamage2.0 [122] was used to estimate the frequency of 5' C=>T transitions.
mtDNA contamination was estimated using the method from Fu et al. 2013 [123].This included calling an mtDNA consensus sequence based on reads with mapping quality at least 30 and positions with at least 5x coverage, aligning the consensus with 311 other human mtDNA sequences from Fu et al. 2013 [123], mapping the original mtDNA reads to the consensus sequence and running contamMix 1.0-10 with the reads mapping to the consensus and the 312 aligned mtDNA sequences while trimming 7 bases from the ends of reads with the option trimBases.
For the male individuals, contamination was also estimated based on X chromosome using the two contamination estimation methods first described in Rasmussen et al. 2011 [124] and incorporated in the ANGSD software [125] in the script contamination.R.
The Bronze and Iron Age samples on average showed 14% C=>T substitutions at the 5' ends while for the considerably more recent Middle Age samples this result was 7% (Tabel S1). The mtDNA contamination point estimate for samples with >6x mtDNA coverage ranged from 0.05% to 3.65% with an average of 0.6% (Table S1). The average of the two X chromosome contamination methods of male individuals with average X chromosome coverage >0.1x was between 0.07% and 3.02% with an average of 1.07% (Table S1).
Calculating general statistics and determining genetic sex
Samtools 1.3 [120] option stats was used to determine the number of final reads, average read length, average coverage etc.
Genetic sex was calculated using the script sexing.py from Skoglund et al. 2013 [126], estimating the fraction of reads mapping to Y chromosome out of all reads mapping to either X or Y chromosome.
The average coverage of the whole genome for the samples was between 0.0001x and 0.7x (Table S1). Genetic sexing confirmed morphological sex estimates or provided additional information about the sex of the individuals involved in the study. The sex of 12 of the samples could not be reliably estimated due to low coverage. Apart from those samples, the study involves 12 females and 32 males (Table 1) since a focal point of the study is chromosome Y.
Variant calling
Variants were called with the ANGSD software [125] command doHaploCall, sampling a random base for the positions that are present in the EBC-chipDB [5,127–135] (Table S3).
Determining mtDNA haplogroups
mtDNA haplogroups were determined by submitting mtDNA BAM files to mtDNA-Server [136] which uses HaploGrep2 [137,138] for assigning haplogroups. Subsequently, the results were checked visually by aligning mapped sequences to reference sequence rCRS [139] with samtools 0.1.19 [120] command tview and confirming the haplogroup assignments in PhyloTree [138].
41 of the 56 individuals were successfully haplogrouped (Table 1).
Y chromosome variant calling and haplogrouping
Y chromosome variants were called from the BAM files of the samples using ANGSD [125] doHaploCall. The resulting VCF files were filtered for regions of a total length of 8.8 Mbp of sequence that uniquely maps to human Y chromosome when using short read sequencing technology [25]. Variants called within this 8.8 Mbp region were further filtered for 113,217 haplogroup informative positions [25,29,140–142] using BEDTools 2.19.0 [143] intersect option. Haplogroup assignments of each individual sample were made by determining the haplogroup with the highest proportion of informative positions called in derived state in the given sample. Y chromosome haplogrouping was performed on all samples to check if any of the samples estimated to be female also give a result.
None of the female samples were successfully haplogrouped as expected. 30 out of the 32 males were successfully haplogrouped (Table 1).
Preparing the datasets for autosomal analyses
The EBC-chipDB [5,127–135] was used as the modern DNA background. Individuals from Lazaridis et al. 2016 [14], Jones et al. 2017 [10], Unterländer et al. 2017 [144], Saag et al. 2017 [13], Mittnik et al. 2018 [11], Mathieson et al. 2018 [12], two Damgaard et al. 2018 [145,146] papers, Narasimhan et al. 2018 [147] and Lamnidis et al. 2018 [26] were used as the ancient DNA background. The full genome sequencing data of the aDNA background dataset [10,13,145,146] in the form of FASTQ files was called as described in the Variant calling section. The 1240k capture data of the aDNA background dataset [11,12,14,26,144,147] was downloaded in EIGENSTRAT format. The data of the two comparison datasets and of the individuals of this study was converted to BED format using PLINK 1.90 (http://pngu.mgh.harvard.edu/purcell/plink/) [148], the datasets were merged and the 503,714 SNPs of the modern comparison dataset were kept. Due to low coverage (<0.017x) resulting in a low number of SNPs (<10,000 of the 503,714), 23 of the individuals of this study were removed from further autosomal analyses, leaving 15 individuals from Bronze Age Estonia, 6 from Iron Age Estonia, 5 from Iron Age Ingria and 7 from Middle Age Estonia to be used in autosomal analyses (Table S1).
Principal component analysis
To prepare for principal component analysis (PCA), a reduced comparison sample-set composed of 817 modern individuals from 46 populations of Europe, Caucasus and Near East and 645 ancient individuals from 97 populations was assembled (Table S3). The data was converted to EIGENSTRAT format using the program convertf from the EIGENSOFT 7.2.0 package [149]. PCA was performed with the program smartpca from the same package, projecting ancient individuals onto the components constructed based on the modern genotypes using the option lsqproject and trying to account for the shrinkage problem introduced by projecting by using the option autoshrink.
Outgroup f3 statistics
For calculating autosomal outgroup f3 statistics, the same ancient sample-set as for PCA was used and the modern sample-set was increased to 1490 individuals from 92 populations from Europe, Caucasus, Near East, Siberia, Central Asia and East Asia and Yorubas as outgroup (Table S3). Heterozygous positions were converted to homozygous by randomly choosing one of the alleles at each position to enable comparison between pseudo-haploid ancient samples and diploid modern samples. The data was converted to EIGENSTRAT format using the program convertf from the EIGENSOFT 5.0.2 package [149]. Outgroup f3 statistics of the form f3(Yorubas; EstBA/EstIA/IngIA/EstMA, modern/ancient) were computed using the ADMIXTOOLS 1.1 [150] program qp3Pop.
To allow for X chromosome versus autosomes comparison, outgroup f3 statistics using X chromosome SNPs were computed. However, the overlap between the X chromosome positions of the EBC-chipDB [5,127–135] and the 1240k capture data of the ancient comparison sample-set was only 17,852 SNPs. To be able to use the whole ancient comparison dataset for this analysis, the full genome sequencing data of that dataset and the individuals of this study were called as described in the Variant calling section but using the positions of the Lazaridis et al. [14] ancient dataset. To allow for the use of the bigger number of positions in the ancient over the modern dataset from Lazaridis et al. [14], Mbuti from Panel C of the Simons Genome Diversity Project [151] was used as the outgroup. The outgroup f3 analyses of the form f3(Mbuti; EstBA/EstIA/IngIA/EstMA, ancient) were run both using 991,166 autosomal SNPs and also 40,185 X chromosome positions available in the Lazaridis et al. [14] ancient dataset. Since all children inherit half of their autosomal material from their father but only female children inherit their X chromosome from their father then in this comparison X chromosome data gives more information about the female and autosomal data about the male ancestors of a population.
The autosomal outgroup f3 results of the two different SNP sets were compared to see whether the SNPs used affect the trends seen.
D statistics
D statistics of the form D(Yorubas, EstBA/EstIA/IngIA/EstMA; Estonians, modern/ancient) were calculated on the same EBC-chipDB [5,127–135] as outgroup f3 statistics (Table S3). The ADMIXTOOLS 1.1 [150] package program qpDstat was used.
Admixture analysis
Three Paleolithic individuals were added to the ancient sample-set used for previous analyses and the modern sample-set was increased to 1799 individuals from 115 populations from all over the world for Admixture analysis [152] (Table S3). The analysis was carried out using ADMIXTURE 1.3 [152] with the P option, projecting ancient individuals into the genetic structure calculated on the modern dataset due to missing data in the ancient samples. The dataset of modern individuals was pruned to decrease linkage disequilibrium using the option indep-pairwise with parameters 1000 250 0.4 in PLINK 1.90 (http://pngu.mgh.harvard.edu/purcell/plink/) [148]. This resulted in a set of 216,398 SNPs. Admixture was run on this set using K=3 to K=18 in 100 replicates. This enabled us to assess convergence of the different models. K=10 and K=9 were the models with the largest number of inferred genetic clusters for which >10% of the runs that reached the highest Log Likelihood values yielded very similar results. This was used as a proxy to assume that the global Likelihood maximum for this particular model was indeed reached. Then the inferred genetic cluster proportions and allele frequencies of the best run at K=9 were used to run Admixture to project the aDNA individuals on the inferred clusters. The same projecting approach was taken for all models for which there is good indication that the global Likelihood maximum was reached (K3–18). We present all individuals on Figure S1 but only population averages of those aDNA samples on Figure 1 for which the intersection with the LD pruned modern dataset yielded data for more than 10,000 SNPs. The resulting membership proportions to K genetic clusters are sometimes called “ancestry components” which can lead to over-interpretation of the results. The clustering itself is, however, an objective description of genetic structure and as such a valuable tool in population comparisons.
ChromoPainter/NNLS
In order to infer the admixture proportions of ancient individuals, the ChromoPainter/NNLS pipeline [19,153,154] was applied. Due to the low coverage of the ancient data, it is not possible to infer haplotypes and the analysis was performed in unlinked mode (option -u). Only samples with more than 20,000 SNPs were used in the analyses. Since ChromoPainter [155] does not tolerate missing data, every ancient target individual was iteratively painted together with one representative individual from potential source populations as recipients. All the remaining modern individuals from the sample-set used for Admixture analysis were used as donors (Table S3). Subsequently, we reconstructed the profile of each target individual as a combination of three or more ancient individuals, using the non-negative least square approach. Let Xg and Yp be vectors summarising the proportion of DNA that source and target individuals copy from each of the modern donor groups as inferred by ChromoPainter. Yp = β1X1 + β2X2 + β3X3 + … + βzXz was reconstructed using a slight modification of the nnls function in R [156] and implemented in GlobeTrotter [157] under the conditions βg ≥ 0 and ∑βg = 1. In order to evaluate the fitness of the NNLS estimation, we inferred the sum of the squared residual for every tested model and reported the one with the lowest value [158]. The model with the smallest residual values included WHG (Loschbour [15]), Yamnaya (Yamnaya [146]), Central MN (I0172 [16]) and modern Nganasans (Nganassan11 [132]) as sources (see other models in Data S1). The resulting painting profiles, which summarise the fraction of the individual’s DNA inherited by each donor individual, were summed over individuals from the same population.
qpAdm
The ADMIXTOOLS 1.1 [150] package programs qpWave and qpAdm were used to estimate which populations and in which proportions are suitable proxies of admixture to form the populations of this study. Only samples with more than 100,000 SNPs were used in the analyses. The best model tested (taking into account p-values, standard errors and the presence of negative values for proportions) included EstBA/EstIA/IngIA/EstMA, WHG, Yamnaya Kalmykia, Central MN and Nganasans as left populations and Yorubas, Ust-Ishim, Mal’ta, Kostenki, SHG and Han as right populations (see other models in Data S1).
f4 statistics
f4 statistics of the form f4(Yorubas, Nganasans; period in Estonia/Central LNBA, period in Baltics), f4(Yorubas, Koryaks; period in Estonia, period in Baltics), f4(Yorubas, WHG; period in Estonia, period in Baltics) and f4(Yorubas, Syrians/Yamnaya Kalmykia/Central MN; period in Estonia, period in Baltics) were calculated on the same EBC-chipDB [5,127–135] as outgroup f3 statistics (Table S3). The ADMIXTOOLS 1.1 [150] package program qpDstat and the option f4mode: YES was used.
Population continuity tests
We applied the forward simulation method described in Hofmanová et al. 2016 [19] to test whether individual genomes from the ancient Estonian populations can be considered as sampled from a population directly ancestral to modern Estonians under a model of full continuity. We used the overlapping positions between the pseudo-haploid calls of our ancient genomes and the biallelic calls of ten modern Estonian genomes extracted from the Human Origins dataset [14] to estimate their population allele frequencies and infer the site frequency spectrum (SFS). In order to preserve the SFS shape, we only tested ancient genomes for which more than 100,000 SNPs overlapped with the modern dataset (Data S1). Alleles were then polarized into ancestral and derived by comparing them with the alleles in the chimpanzee to obtain the derived folded SFS.
For each combination of an ancient genome and the ten modern genomes we performed the steps described in Hofmanová et al. 2016 [19]. Briefly, we first incorporated uncertainty on the allele frequencies by sampling 100 frequency vectors using a beta distribution and the Jeffreys’ prior [159] from the distribution of allele frequencies of the SFS of the modern Estonian individuals. We then use binomial sampling in forward simulations to emulate a genetic drift process and generate possible allele trajectories given the age of the ancient sample in generations. We explored two parameters, ancient (Nea) and modern (Nem) effective population sizes, assuming a model of exponential growth between them. For each simulation we sampled a haploid genome from the initial frequency vector and another one from each simulated final frequency vector. We compared the observed calls with the simulated ones using an allelic sharing classification consisting of six possible classes formed by all possible combinations of haploid calls of an ancient genome (t0) and the biallelic calls of each modern genome (tn) for the same position: 1) match A: t0 ancestral (A) and tn AA, 2) match D: t0 derived (D) and tn DD, 3) mismatch AD: t0 A and tn DD, 4) mismatch DA: t0 D and tn AA, 5) half match A: t0 A and tn AD, and 6) half match D: t0 D and tn AD (see Hofmanova et al. 2016 [19]). Allele sharing fraction values are calculated for both the observed and simulated data as the proportion of all analyzed positions that fall into each one of these six classes. Finally, we calculated an overall p-value for the null-hypothesis of rejection of population continuity for each combination of parameters by combining the individual p-values for each allelic sharing fraction using Fisher’s and Voight’s methods [160] as described in Hofmanová et al. 2016 [19].
We explored a wide parameter space of Nea and Nem for each ancient genome and the modern individuals by performing the test in a 50x50 grid composed for values of these effective population sizes ranging from 10 to 10 million individuals on a log scale. For each combination of parameters, we performed 1,000 simulations, thus the total number of simulations per test (each ancient genome vs modern Estonians) was 2.5 million. The ranges of realistic effective population sizes in which continuity could not be rejected were examined by slicing the parameter grid by the mean, upper and lower CI of the effective population size of modern Estonians, estimated on over 2,000 modern Estonian full genomes (unpublished; cohort [29]) using the program IBDNe [161]. The two-tail p-values of the test for each ancient effective size are reported.
Kinship analysis
A total of 4,375,438 biallelic single nucleotide variant sites, with MAF>0.1 in a set of over 2,000 high coverage genomes of EGC (unpublished; cohort [29]) were identified and called with ANGSD-0.916 [125] command doHaploCall from the BAM files of 12 Bronze Age, 11 Iron Age and 6 medieval individuals with coverage >0.03x. The ANGSD output files were converted to .tped format as an input for the analyses with READ script to infer pairs with 1st and 2nd degree relatedness [162].
Radiocarbon date difference probability estimation
Probabilistic estimates of the temporal distance between the radiocarbon dates associated with X14 (2481±30 BP) and V16 (2399±27 BP) have been obtained by: 1) calibrating both dates using the IntCal13 calibration curve [28] (using the rcarbon R package [163]); and 2) sampling one million pairs of random dates from each distribution and calculating their differences. The resulting distribution of differences had a 95% HPD between between -76 (i.e. V16 earlier) and 344 years (i.e. V16 later). We then calculated the expected difference in time between the date of death of X14 and V16, assuming that: 1) the former was the uncle, the latter the nephew; 2) an age of death between 35 and 45 for X14 [53] and between 30 and 40 for V16 [46]; 3) a reproductive age span between 13 and 40 years old; and 4) a maximum age difference between X14 and his sister of 27 years (i.e. 40-13). The difference in the date of death can then be calculated using the following formula (a+b)-(c+d), where a is the age at death of V16, b is the age at which X14’s sister gave birth to V16, c is the age at death of X14 and d is the difference in age between X14’s sister and X14 (i.e. negative if X14 is assumed to be older, positive if his sister was born first). It follows that the difference in time between the date of death of X14 and V16 could range between -29 (i.e. V16 dying before X14) and 72 years (V16 dying later). We then computed that the probability that difference in the age of the radiocarbon dates is within this interval computing the proportion of dates within -29 to 72, which was equivalent to 0.15. We also calculated the ranges and probabilities if V16 was X14’s uncle and if the two were half brothers sharing a mother.
Phenotyping
The phenotype prediction was performed only on the samples with an average genomic coverage greater than 0.1x, for a total of 23 subjects (Data S2).
In order to predict eye, hair and skin colour in the ancient individuals (Data S2), we selected all the 41 variants from 19 genes in 9 autosomes in the HIrisPlex-S system [164] and, for each autosome, we selected the region to be analysed adding 5 Mb at each side of the chromosomal segment delimited by the first and the last SNP. We analysed the three genes on chromosome 15 in two different regions (OCA2-HERC2 region and SLC24A5 region), because the distance between the two nearest SNPs of the two chromosomal segments was greater than 20 Mb. We obtained a total of 10 regions ranging from about 10 Mb to about 15 Mb. We chose as reference panel a set of 606 modern individuals, from all the European (EUR) populations and one Asian (CHB) population of the 1000 Genomes Phase 3 [165]. The Chinese outgroup was added to include also the variants that are very rare in Europe. The variant sites in the 10 chromosomal regions were extracted from the phased VCF files of the modern individuals with VCFtools [166], discarding the indels. The resulting VCF files were filtered using bcftools [167] to keep only the biallelic SNPs with a minor allele frequency (MAF) above a chosen threshold. We set the MAF threshold to 1% for all the genes, with the exception of the region on chromosome 16, for which the MAF threshold was set to 0.1% to retain as many rare SNPs as possible from the MC1R gene. These settings allowed us to exclude only 3 SNPs and one indel out of the 41 HIrisPlex-S informative markers. The final VCFs were manipulated with PLINK 1.9 (http://pngu.mgh.harvard.edu/purcell/plink/) [148] to obtain a list of variant sites and a map file for each region.
We calculated the genotype likelihoods for the variant sites in the ancient individuals using the ANGSD [125] -GL command, with the -dopost 1 option and a reference sequence in the FASTA format. We then performed the imputation step using the Beagle 4.1 and Beagle 5.0 software [168]. First, we loaded on Beagle the ANGSD VCF output and we used the -gl command to infer genotype probabilities (GP). We obtained a VCF output including a GT value in the FORMAT field and filtered it to discard all variants with a GP ≤0.99. The filtered VCF was loaded again on Beagle 5.0 for a second run with the -gt command to impute at ungenotyped sites. The resulting VCF files were filtered again to keep only variants with a GP ≥0.85, with the exception of the HERC2 rs12913832 variant: since the lack of this SNP will not produce an eye colour prediction result, we set the GP threshold to 0.6. The resulting VCF files were subset with VCFtools [166] to extract the SNPs relevant for the phenotype prediction. The SNPs were recoded and organised with PLINK 1.9 (http://pngu.mgh.harvard.edu/purcell/plink/) [148] and R [156] and the missing variants were coded as “NA” to produce a csv input file for the HIrisPlex-S webtool (https://hirisplex.erasmusmc.nl/), that was used to perform the phenotype prediction [164,169,170].
We used the same approach to extract the allele information for the lactase persistence variant (rs4988235 in the MCM6 gene) and two variants involved in the protection against leprosy (rs5743618 and rs4833095 in the TLR1 gene). For the TLR1 variants, a more relaxed GP threshold of 0.60 was used. We also used the same approach to impute rs333 (CCR5-32bp deletion) but used a larger local Estonian reference panel of over 2,000 EGC high coverage genomes (unpublished; cohort [29]).
We tested the accuracy of our imputation pipeline by downsampling a high-coverage sample and comparing the variants imputed in the downsampled samples to the variants in the original one. To this aim, we selected the high-coverage (20x) NE1 individual from Gamba et al. 2014 [164] and randomly downsampled it to a coverage of 0.05x and 0.1x using SAMtools [120]. We applied the same ANGSD commands described above to calculate the genotype likelihoods for the variants in the chromosome 20 region in both the high-coverage and low-coverage NE1 bams. We then followed the same pipeline described above, obtaining an overall concordance rate of about 95%.
Supplementary Material
Acknowledgements
This work was funded by research projects of the Estonian Research Council IUT24-1 (R.V., T.K., En.M., M.Me., Al.K., La.S, K.T.), IUT20-7 (V.L., Ai.K., H.V., M.L., M.Ma.), IUT34-11 (M.R.), PUT1217 (K.T., La.S., Le.S.), PRG243 (M.Me., La.S., C.L.S., R.M., Le.S., A.S.), PUT1339 (Al.K.); of the EU European Regional Development Fund 2014-2020.4.01.16-0030, 2014-2020.4.01.16-0125, 2014-2020.4.01.15-0012; European Research Council Starting Investigator Grant FP7-261213 (T.K.); Wellcome Trust Senior Research Fellowship Grant 100719/Z/12/Z (M.G.T.); Sapienza University of Rome fellowship “borsa di studio per attività di perfezionamento all’estero 2017” (E.D’A.); Arheograator Ltd. (L.V., Ai.K.). The authors would like to thank the University of Tartu Development Fund for support to the Collegium for Transdisciplinary Studies in Archaeology, Genetics and Linguistics. We thank Ajai Kumar Pathak and Tarmo Puurand for help with analyses. Analyses were carried out using the facilities of the High Performance Computing Center of the University of Tartu.
The DNA sequences are available through the data depository of the EBC (http://evolbio.ut.ee) and through European Nucleotide Archive (accession code PRJEB31893).
Footnotes
Data and software availability
The DNA sequences are available through the data depository of the EBC (http://evolbio.ut.ee) and through European Nucleotide Archive (accession code PRJEB31893).
Author contributions
Le.S., Ai.K., R.V., V.L., M.Me. and K.T. conceived the study. M.L., L.V., M.Ma., H.V., I.G.S., V.I.K., E.R.M., Ai.K. and V.L. assembled skeletal samples and performed osteological analyses. Le.S., Al.K., C.L.S., A.S., T.R., J.P. and K.T. performed aDNA extraction and sequencing. Le.S., La.S., E.M., S.R., F.M., M.R., R.M., E.D’A., E.R.C., D.D-del-M., M.G.T., T.K. and K.T. analysed data. Le.S., M.L., H.V., M.A.R., Ai.K., T.K., V.L. and K.T. wrote the manuscript with input from remaining authors.
Declaration of Interests
The authors declare no competing financial interests.
References
- 1.Ilumäe A-M, Reidla M, Chukhryaeva M, Järve M, Post H, Karmin M, Saag L, Agdzhoyan A, Kushniarevich A, Litvinov S, et al. Human Y chromosome haplogroup N: a non-trivial time-resolved phylogeography that cuts across language families. Am J Hum Genet. 2016;99:163–173. doi: 10.1016/j.ajhg.2016.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tambets K, Rootsi S, Kivisild T, Help H, Serk P, Loogväli E-L, Tolk H-V, Reidla M, Metspalu E, Pliss L, et al. The western and eastern roots of the Saami – the story of genetic “outliers” told by mitochondrial DNA and Y chromosomes. Am J Hum Genet. 2004;74:661–682. doi: 10.1086/383203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pliss L, Tambets K, Loogväli E-L, Pronina N, Lazdins M, Krumina A, Baumanis V, Villems R. Mitochondrial DNA portrait of Latvians: towards the understanding of the genetic structure of Baltic-speaking populations. Ann Hum Genet. 2006;70:439–458. doi: 10.1111/j.1469-1809.2005.00238.x. [DOI] [PubMed] [Google Scholar]
- 4.Rootsi S, Zhivotovsky LA, Baldovic M, Kayser M, Kutuev IA, Khusainova R, Bermisheva MA, Gubina M, Fedorova SA, Ilumäe A-M, et al. A counter-clockwise northern route of the Y-chromosome haplogroup N from Southeast Asia towards Europe. Eur J Hum Genet EJHG. 2007;15:204–211. doi: 10.1038/sj.ejhg.5201748. [DOI] [PubMed] [Google Scholar]
- 5.Tambets K, Yunusbayev B, Hudjashov G, Ilumäe A-M, Rootsi S, Honkola T, Vesakoski O, Atkinson Q, Skoglund P, Kushniarevich A, et al. Genes reveal traces of common recent demographic history for most of the Uralic-speaking populations. Genome Biol. 2018;19:139. doi: 10.1186/s13059-018-1522-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Honkola T, Vesakoski O, Korhonen K, Lehtinen J, Syrjänen K, Wahlberg N. Cultural and climatic changes shape the evolutionary history of the Uralic languages. J Evol Biol. 2013;26:1244–1253. doi: 10.1111/jeb.12107. [DOI] [PubMed] [Google Scholar]
- 7.Zagorska I. Environmental and Cultural History of the Eastern Baltic Region. PACT 57. PACT. Strassbourg: European Council; 1999. The Earliest Settlement of Latvia; pp. 131–156. [Google Scholar]
- 8.Šatavičius E. A Hundred Years of Archaeological Discoveries in Lithuania. Vilnius: 2016. The First Palaeolithic Inhabitants and the Mesolithic in Lithuanian Territory; pp. 8–39. [Google Scholar]
- 9.Kriiska A, Lõugas L. Stone Age settlement sites on an environmentally sensitive coastal area along the lower reaches of the River Pärnu (south-western Estonia), as indicators of changing settlement patterns, technologies and economies. In: McCartan S, Schulting R, Warren G, Woodman P, editors. Mesolithic Horizons. Oxford-Oakville: Oxbow Books; 2009. pp. 167–175. [Google Scholar]
- 10.Jones ER, Zarina G, Moiseyev V, Lightfoot E, Nigst PR, Manica A, Pinhasi R, Bradley DG. The Neolithic transition in the Baltic was not driven by admixture with early European farmers. Curr Biol CB. 2017;27:576–582. doi: 10.1016/j.cub.2016.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mittnik A, Wang C-C, Pfrengle S, Daubaras M, Zariņa G, Hallgren F, Allmäe R, Khartanovich V, Moiseyev V, Tõrv M, et al. The genetic prehistory of the Baltic Sea region. Nat Commun. 2018;9 doi: 10.1038/s41467-018-02825-9. 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mathieson I, Alpaslan-Roodenberg S, Posth C, Szécsényi-Nagy A, Rohland N, Mallick S, Olalde I, Broomandkhoshbacht N, Candilio F, Cheronet O, et al. The genomic history of southeastern Europe. Nature. 2018;555:197–203. doi: 10.1038/nature25778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saag L, Varul L, Scheib CL, Stenderup J, Allentoft ME, Saag L, Pagani L, Reidla M, Tambets K, Metspalu E, et al. Extensive farming in Estonia started through a sex-biased migration from the Steppe. Curr Biol CB. 2017;27:2185–2193.e6. doi: 10.1016/j.cub.2017.06.022. [DOI] [PubMed] [Google Scholar]
- 14.Lazaridis I, Nadel D, Rollefson G, Merrett DC, Rohland N, Mallick S, Fernandes D, Novak M, Gamarra B, Sirak K, et al. Genomic insights into the origin of farming in the ancient Near East. Nature. 2016;536:419–424. doi: 10.1038/nature19310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lazaridis I, Patterson N, Mittnik A, Renaud G, Mallick S, Kirsanow K, Sudmant PH, Schraiber JG, Castellano S, Lipson M, et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature. 2014;513:409–413. doi: 10.1038/nature13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mathieson I, Lazaridis I, Rohland N, Mallick S, Patterson N, Roodenberg SA, Harney E, Stewardson K, Fernandes D, Novak M, et al. Genome-wide patterns of selection in 230 ancient Eurasians. Nature. 2015;528:499–503. doi: 10.1038/nature16152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skoglund P, Malmström H, Omrak A, Raghavan M, Valdiosera C, Günther T, Hall P, Tambets K, Parik J, Sjögren K-G, et al. Genomic diversity and admixture differs for Stone-Age Scandinavian foragers and farmers. Science. 2014;344:747–750. doi: 10.1126/science.1253448. [DOI] [PubMed] [Google Scholar]
- 18.Olalde I, Schroeder H, Sandoval-Velasco M, Vinner L, Lobón I, Ramirez O, Civit S, García Borja P, Salazar-García DC, Talamo S, et al. A common genetic origin for Early Farmers from Mediterranean Cardial and Central European LBK cultures. Mol Biol Evol. 2015;32:3132–3142. doi: 10.1093/molbev/msv181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hofmanová Z, Kreutzer S, Hellenthal G, Sell C, Diekmann Y, Díez-Del-Molino D, van Dorp L, López S, Kousathanas A, Link V, et al. Early farmers from across Europe directly descended from Neolithic Aegeans. Proc Natl Acad Sci U S A. 2016;113:6886–6891. doi: 10.1073/pnas.1523951113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lang V. Läänemeresoome tulemised. Tartu: University of Tartu Press; 2018. [Google Scholar]
- 21.Lang V. Formation of Proto-Finnic – an archaeological scenario from the Bronze Age/Early Iron Age. In: Mantila H, Leinonen K, Brunni S, Palviainen S, Sivonen J, editors. Congressus Duodecimus Internationalis Fenno-Ugristarum, Oulu 2015. Plenary papers. Oulu: University of Oulu; 2015. pp. 63–84. [Google Scholar]
- 22.Patrushev V. The Early History of the Finno-Ugric Peoples of European Russia. Oulu: 2000. [Google Scholar]
- 23.Kallio P. Suomen kantakielten absoluuttista kronologiaa. Virittäjä. 2006;1:2–25. [Google Scholar]
- 24.Häkkinen J. Kantauralin ajoitus ja paikannus: perustelut puntarissa. Vol. 92. Suom-Ugr Seuran Aikakauskirja; 2009. pp. 9–56. [Google Scholar]
- 25.Karmin M, Saag L, Vicente M, Wilson Sayres MA, Järve M, Talas UG, Rootsi S, Ilumäe A-M, Mägi R, Mitt M, et al. A recent bottleneck of Y chromosome diversity coincides with a global change in culture. Genome Res. 2015;25:459–466. doi: 10.1101/gr.186684.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamnidis TC, Majander K, Jeong C, Salmela E, Wessman A, Moiseyev V, Khartanovich V, Balanovsky O, Ongyerth M, Weihmann A, et al. Ancient Fennoscandian genomes reveal origin and spread of Siberian ancestry in Europe. Nat Commun. 2018;9 doi: 10.1038/s41467-018-07483-5. 5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bronk Ramsey C. Bayesian analysis of radiocarbon dates. Radiocarbon. 2009;51:337–360. [Google Scholar]
- 28.Reimer PJ, Bard E, Bayliss A, Beck JW, Blackwell PG, Bronk Ramsey C, Buck CE, Cheng H, Edwards RL, Friedrich M, et al. IntCal13 and Marine13 radiocarbon age calibration curves 0-50,000 years cal BP. Radiocarbon. 2013;55:1869–1887. [Google Scholar]
- 29.Mitt M, Kals M, Pärn K, Gabriel SB, Lander ES, Palotie A, Ripatti S, Morris AP, Metspalu A, Esko T, et al. Improved imputation accuracy of rare and low-frequency variants using population-specific high-coverage WGS-based imputation reference panel. Eur J Hum Genet EJHG. 2017;25:869–876. doi: 10.1038/ejhg.2017.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haak W, Lazaridis I, Patterson N, Rohland N, Mallick S, Llamas B, Brandt G, Nordenfelt S, Harney E, Stewardson K, et al. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature. 2015;522:207–211. doi: 10.1038/nature14317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allentoft ME, Sikora M, Sjögren K-G, Rasmussen S, Rasmussen M, Stenderup J, Damgaard PB, Schroeder H, Ahlström T, Vinner L, et al. Population genomics of Bronze Age Eurasia. Nature. 2015;522:167–172. doi: 10.1038/nature14507. [DOI] [PubMed] [Google Scholar]
- 32.Günther T, Malmström H, Svensson EM, Omrak A, Sánchez-Quinto F, Kılınç GM, Krzewińska M, Eriksson G, Fraser M, Edlund H, et al. Population genomics of Mesolithic Scandinavia: Investigating early postglacial migration routes and high-latitude adaptation. PLoS Biol. 2018;16:e2003703. doi: 10.1371/journal.pbio.2003703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burger J, Kirchner M, Bramanti B, Haak W, Thomas MG. Absence of the lactase-persistence-associated allele in early Neolithic Europeans. Proc Natl Acad Sci U S A. 2007;104:3736–3741. doi: 10.1073/pnas.0607187104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oras E, Lang V, Rannamäe E, Varul L, Konsa M, Limbo-Simovart J, Vedru G, Laneman M, Malve M, Price TD. Tracing prehistoric migration: isotope analysis of Bronze and Pre-Roman Iron Age coastal burials in Estonia. Est J Archaeol. 2016;20(1):3–32. [Google Scholar]
- 35.Lang V. Traceless death. Missing burials in Bronze and Iron Age Estonia. Est J Archaeol. 2011;15(2):109–129. [Google Scholar]
- 36.Olalde I, Brace S, Allentoft ME, Armit I, Kristiansen K, Booth T, Rohland N, Mallick S, Szécsényi-Nagy A, Mittnik A, et al. The Beaker phenomenon and the genomic transformation of northwest Europe. Nature. 2018;555:190–196. doi: 10.1038/nature25738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mathieson S, Mathieson I. FADS1 and the Timing of Human Adaptation to Agriculture. Mol Biol Evol. 2018;35:2957–2970. doi: 10.1093/molbev/msy180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adojaan M, Mölder T, Männik A, Kivisild T, Villems R, Krispin T, Ustav M. High prevalence of the CCR5Delta32 HIV-resistance mutation among Estonian HIV type 1-infected individuals. AIDS Res Hum Retroviruses. 2007;23:193–197. doi: 10.1089/aid.2006.0113. [DOI] [PubMed] [Google Scholar]
- 39.Olalde I, Allentoft ME, Sánchez-Quinto F, Santpere G, Chiang CWK, DeGiorgio M, Prado-Martinez J, Rodríguez JA, Rasmussen S, Quilez J, et al. Derived immune and ancestral pigmentation alleles in a 7,000-year-old Mesolithic European. Nature. 2014;507:225–228. doi: 10.1038/nature12960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skoglund P, Mathieson I. Ancient genomics of modern humans: the first decade. Annu Rev Genomics Hum Genet. 2018;19:381–404. doi: 10.1146/annurev-genom-083117-021749. [DOI] [PubMed] [Google Scholar]
- 41.Tõnisson E, Selirand J. Nõukogude Eesti arheoloogide välitööd aastail 1958–1962. Proc Acad Sci Est SSR Soc Sci. 1964;13(3):225–246. [Google Scholar]
- 42.Jaanits K, Lavi A. Über die Ausgrabungen eines Steinkistengrabes in Väo. Proc Acad Sci Est SSR Soc Sci. 1978;27(4):330–333. [Google Scholar]
- 43.Lõugas V. Archäologische Rettungsgrabungen im neuen Wohngebiet Lasnamäe in Tallinn. Proc Acad Sci Est SSR Soc Sci. 1981;30(4):390–393. [Google Scholar]
- 44.Lang V, Ligi P. Muistsed kalmed ajaloolise demograafia allikana. In: Jaanits L, Lang V, editors. Arheoloogiline kogumik. Muinasaja teadus, 1. Vol. 1. Tallinn: Eesti Arheoloogiaselts; 1991. pp. 216–238. [Google Scholar]
- 45.Lang V. Ein neues Steinschiffsgrab in Nordestland. Proc Acad Sci Est SSR Soc Sci. 1983;32(4):293–295. [Google Scholar]
- 46.Laneman M, Lang V, Malve M, Rannamäe E. New data on Jaani stone graves at Väo, Northern Estonia. Est J Archaeol. 2015;19(2):110–137. [Google Scholar]
- 47.Howen A. Ausgrabungen in Estland. Beitr Zur Kunde Est- Liv- Kurlands. 1900:92–96. [Google Scholar]
- 48.Lõugas V. Über die Entstehnung ortsgebundene Bodenbaukultur in Westestland. Proc Acad Sci Est SSR Soc Sci. 1975;24(1):85–88. [Google Scholar]
- 49.Vassar A. Kivikalme Nehatus. Eesti Rahvuslaste Klubid. 1936;7(8):190–195. [Google Scholar]
- 50.Lõugas V. Ausgrabungen der Steingräber und Flurrelikte in Iru. Proc Acad Sci Est SSR Soc Sci. 1976;25(1):48–52. [Google Scholar]
- 51.Lõugas V. Über die Steingräbergruppe Lastekangrud in Rebala. Proc Acad Sci Est SSR Soc Sci. 1983;32(4):295–297. [Google Scholar]
- 52.Lang V, Laneman M, Ilves K, Kalman J. Fossil fields and stone-cist graves of Rebala revisited. Archaeol Fieldwork Est. 2001;2000:34–47. [Google Scholar]
- 53.Kalman J. Human remains from the stone-cist graves of Rebala Lastekangrud, North Estonia. Est J Archaeol. 1999;3(1):19–34. [Google Scholar]
- 54.Kraut A. Die Steinkistengräber von Jõelähtme. Proc Acad Sci Est SSR Soc Sci. 1985;34(4):348–350. [Google Scholar]
- 55.Varul L. Jõelähtme kivikirstkalmete 1–9, 12–24, 34–36 inimluude analüüs. Manuscript in the archive of the archaeology department of the University of Tartu; Tartu: 2016. [Google Scholar]
- 56.Laneman M, Lang V. New radiocarbon dates for two stone-cist graves at Muuksi, northern Estonia. Est J Archaeol. 2013;17(2):89–122. [Google Scholar]
- 57.Friedenthal A. Ein Gräberfeld der Bronzezeit in Estland. Beitr Zur Kunde Est. 1927;XIII(1–2):47–52. [Google Scholar]
- 58.Vassar A. Drei Steinkistengräber aus Nordestland. Sitzungsberichte Gelehrt. Estnischen Ges. 1938;1937:304–364. [Google Scholar]
- 59.Lõugas V, Selirand J. Arheoloogiga Eestimaa teedel. Second. Tallinn: Valgus; 1989. [Google Scholar]
- 60.Vedru G. New settlement sites in the surroundings of Lake Kahala and revision excavations of stone-cist grave. Archaeol Fieldwork Est. 1997;1996:62–67. [Google Scholar]
- 61.Friedenthal A. Ein Beitrag zur vorgeschichtlichen Anthropologie Estlands. Z Für Ethnol. 1932;63:1–42. [Google Scholar]
- 62.Kalman J. Skeletal report. In: Vedru G, editor. Aruanne kivikirstkalmete kaevamistest Muuksi Hundikangrutes 1996–1997. Manuscript in the archive of the archaeology department of the University of Tartu; Tartu: 1998. [Google Scholar]
- 63.Spreckelsen A. Ausgrabungen in Neuenhof, Kirchsp. Kusal, Dorf Muuksi, Lõokese-Gesinde. Beitr Zur Kunde Est. 1926;XI:38–42. [Google Scholar]
- 64.Friedenthal A. Bericht über die im Auftrage der Estländischen Literärischen Gesellschaft im Sommer 1927 vorgenommenen archäologischen Untersuchungen. Manuscript in the archive of the archaeology department of the University of Tartu; 1927. [Google Scholar]
- 65.Friedenthal A. Bericht über die im Auftrage der Estländischen Literärischen Gesellschaft von Dr. A. Friedenthal im Juli 1928 vorgenommenen archäologischen Ausgrabungen. Manuscript in the archive of the archaeology department of the University of Tartu; 1928. [Google Scholar]
- 66.Schmiedehelm MH. Археологические памятники периода разложения родового строя на северо-востоке Эстонии. (Archaeological sites of the period of the disintegration of the tribal system in north-eastern Estonia. Tallinn: Academy of Sciences of Estonian SSR, Institute of History; Estonian State Publishing House; 1955. (In Russian) [Google Scholar]
- 67.von Schroeder L. Die Steinhügel-Gräber von Randen. Sitzungsberichte Gelehrt. Estnischen Ges. 1895;1894:75–81. [Google Scholar]
- 68.Laul S. Die Steinkistengräber von Vehendi. Proc Acad Sci Est SSR Soc Sci. 1978;27(1):76–77. [Google Scholar]
- 69.Lang V. Estonian Archaeology. Vol. 3 Tartu: University of Tartu Press; 2007. The Bronze and Early Iron Ages in Estonia. [Google Scholar]
- 70.Jaanits L, Laul S, Lõugas V, Tõnisson E. Eesti esiajalugu. Tallinn: ENSV Teaduste Akadeemia Ajaloo Instituut; Eesti Raamat; 1982. [Google Scholar]
- 71.Moora T. Раскопки каменного могильника у с. Выхма в Северной Эстонии (Excavations of a stone burial site in Võhma in Northern Estonia) Proc Acad Sci Est SSR Soc Sci. 1974;23(1):84–87. (In Russian) [Google Scholar]
- 72.Lang V. Viljelusmajandusliku asustuse kujunemine ja areng Vihasoo-Palmse piirkonnas Virumaal. Vol. 7 Muinasaja teadus; Tallinn: 2000. Keskusest ääremaaks. [Google Scholar]
- 73.Lang V. From centre to periphery. Establishment and history of the agricultural settlement in the Vihasoo–Palmse area (Virumaa, north Estonia) Acta Archaeol. 2003;74:123–188. [Google Scholar]
- 74.Kalman J. Tandemägi stone grave – osteological report. In: Lang V, editor. Keskusest ääremaaks. Viljelusmajandusliku asustuse kujunemine ja areng Vihasoo–Palmse piirkonnas Virumaal. Vol. 7. Muinasaja teadus; Tallinn: 2000. pp. 423–436. [Google Scholar]
- 75.Lõugas V. Ausgrabungen eines Steingräberfeldes von Kurevere. Proc Acad Sci Est SSR Soc Sci. 1977;26(1):48–52. [Google Scholar]
- 76.Vaab H. Peaseminaritöö. Manuscript in the archive of the archaeology department of the University of Tartu. Tartu: University of Tartu; 2003. Kurevere kivikalme Saaremaal. [Google Scholar]
- 77.Lang V. A Pre-Roman tarand-grave and Late Medieval fossil fields of Ilmandu, NW Estonia. Proc Est Acad Sci Humanit Soc Sci. 1995;44(4):429. [Google Scholar]
- 78.Kalman J. Stone grave II of Tõugu – skeletal report. In: Lang V, editor. Keskusest ääremaaks. Viljelusmajandusliku asustuse kujunemine ja areng Vihasoo–Palmse piirkonnas Virumaal. Vol. 7. Muinasaja teadus; Tallinn: 2000. pp. 387–407. [Google Scholar]
- 79.Mandel M. Über die Ausgrabungen der Tarandgräber von Poanse. Proc Acad Sci Est SSR Soc Sci. 1978;27(1):78–81. [Google Scholar]
- 80.Mandel M. Töid arheoloogia alalt II. Tallinn: Eesti Ajaloomuuseum; 2000. Poanse tarandkalmed; pp. 89–111. [Google Scholar]
- 81.Kalman J. Töid ajaloo alalt II. Tallinn: Eesti Ajaloomuuseum; 2000. Skeletal analysis of the graves of Kaseküla, Poanse I and Poanse II; pp. 17–40. [Google Scholar]
- 82.Laneman M, Lang V, Saage R. Burial site hidden in a clearance cairn at Alu. Raplamaa Archaeol Fieldwork Est. 2016;2015:35–46. [Google Scholar]
- 83.Yushkova MA. New group of sites of the 1st to 7th centuries AD in the south-west of Leningrad Oblast. New Sites, New Methods. The 14th Finnish-Russian Archaeological Symposium. ISKOS 21; Helsinki: 2016. pp. 143–159. [Google Scholar]
- 84.Yushkova MA, Kulešov VS. Kyorstovo 1: a new burial ground of the period of Roman influences in North-Western Russia. Archaeol Litu. 2011;12:99–121. [Google Scholar]
- 85.Yushkova MA. Археологические открытия 2010-2013 гг. (Archaeological discoveries of 2010-2013) Moskow: 2015. Раскопки могильника Малли. (Excavations of the burial ground of Malli) pp. 100–102. (In Russian) [Google Scholar]
- 86.Shirobokov IG, Yushkova MA. Антропологические материалы из коллективных захоронений по обряду кремации и ингумации каменного могильника с оградками Малли (по результатам раскопок 2010 г.). (Anthropological materials from collective burials according to the rite of cremation and inhumation of a stone fence burial ground in Malli (according to the results of excavations in 2010)) Bull Archeol Anthropol Ethnogr. 2014;2:71–79. (In Russian) [Google Scholar]
- 87.Shirobokov IG, Yushkova MA. Результаты планиграфического и макроскопического анализа антропологических материалов из могильника с каменными оградками Малли. (Results of a planigraphic and macroscopic analysis of anthropological materials from a stone fence burial ground in Malli) Bull Anthropol New Ser. 2015;3:93–109. (In Russian) [Google Scholar]
- 88.Yushkova MA, Grigorieva OV, Grigorieva NV. Археология и история Пскова и Псковской земли. (Archeology and History of Pskov and Pskov region) Vol. 60. Moskow: 2015. Раскопки могильника Малли в 2013 г. (Excavations of the burial ground of Malli in 2013) pp. 245–258. (In Russian) [Google Scholar]
- 89.Mikhaylova ER. The population of the south-eastern coast of the Gulf of Finland and its contacts with the regions of the Baltic Sea in the 1st millennium AD. Archaeol Balt. 2016;23:181–198. [Google Scholar]
- 90.Mikhaylova ER. Археологические вести (Archaeological news) 21. Sankt-Peterburg: 2015. Древности Западной Ингрии I тыс. н.э.: Новые материалы. (Antiquities of Western Ingria I millennium BC: New materials) pp. 176–186. (In Russian) [Google Scholar]
- 91.Valk H. CCC Papers, 3. Second. Gotland University College, Centre for Baltic Studies, University of Tartu Archaeology Centre; Visby–Tartu: 2001. Rural Cemeteries of Southern Estonia 1225–1800 AD. [Google Scholar]
- 92.Muižnieks V. Latvijas Nacionāla Vēstures Muzeja Raksti. Vol. 21 Riga: 2015. Bēru tradīcijas Latvijā pēc arheologiski pētīto 14.–18. gadsimta apbedīšanas vietu materiāla. [Google Scholar]
- 93.Palli H. Eesti rahvastiku ajalugu aastani 1712. Tallinn: Eesti Teaduste Akadeemia Kirjastus; 1996. Third. [Google Scholar]
- 94.Pajusalu K, Hennoste T, Niit E, Päll P, Viikberg J. Eesti murded ja kohanimed. Tallinn: Eesti Keele Sihtasutus; 2018. Third. [Google Scholar]
- 95.Viires A. Tagasivaade. In: Viires A, Vunder E, editors. Eesti rahvakultuur. Tallinn: Eesti Entsüklopeediakirjastus; 2008. pp. 449–455. [Google Scholar]
- 96.Remmel M-A, Valk H. Muistis, koht ja pärimus 2. Pärimus ja paigad. Muinasaja teadus. 2. Vol. 26. Tartu: Tartu Ülikooli ajaloo ja arheoloogia instituut; 2014. Muistised, pärimuspaigad ja kohapärimus: ajalised ning ruumilised aspektid; pp. 305–398. [Google Scholar]
- 97.Nelis M, Esko T, Mägi R, Zimprich F, Zimprich A, Toncheva D, Karachanak S, Piskácková T, Balascák I, Peltonen L, et al. Genetic structure of Europeans: a view from the North-East. PloS One. 2009;4:e5472. doi: 10.1371/journal.pone.0005472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kustin A. Kalmistu XIII–XIV sajandist Karjas, Saaremaal. Proc Acad Sci Est SSR Soc Sci. 1958;7(1):47–57. [Google Scholar]
- 99.Tamla T. Zum Grabraub in vor- und frühgeschichtlichen Gräbern Estlands. In: Wesse A, editor. Studien zur Archäologie des Ostseeraumes. Von der Eisenzeit zum Mittelalter. Festschrift für Michael Müller-Wille. Mainz-Stuttgart: Akademie der Wissenschaften und der Litteratur, Franz Steiner Verlag; 1998. pp. 291–297. [Google Scholar]
- 100.Tamla T. Aruanne Pada maa-aluse kalmistu kaevamistest Rakvere rajoonis (Viru-Nigula kihelkond, Pada mõis; tänapäeval Viru-Nigula vald, Pada küla) 1989. Manuscript in the archive of the archaeology department of the University of Tartu. Tartu: University of Tartu; 1989. aastal. [Google Scholar]
- 101.Lõhmus M, Jonuks T, Malve M. Archaeological salvage excavations at Kukruse: a Modern Age road, cremation field and 12th–13th century inhumation cemetery. Preliminary results. Archaeol Fieldwork Est. 2011;2010:103–114. [Google Scholar]
- 102.Jonuks T, Oras E, Best J, Demarchi B, Mänd R, Presslee S, Vahur S. Multi-method analysis of avian eggs as grave goods: revealing symbolism in conversion period burials at Kukruse, NE Estonia. Environ Archaeol. 2018;23:109–122. [Google Scholar]
- 103.Oras E, Tõrv M, Jonuks T, Malve M, Radini A, Isaksson S, Gledhill A, Kekišev O, Vahur S, Leito I. Social food here and hereafter: Multiproxy analysis of gender-specific food consumption in conversion period inhumation cemetery at Kukruse, NE-Estonia. J Archaeol Sci. 2018;97:90–101. [Google Scholar]
- 104.Kurisoo T. Ajast ja ruumist. Uurimusi Mare Auna auks. Muinasaja teadus. Vol. 25. Tallinn-Tartu: Ajaloo Instituut; 2014. Pada kalmistu rinnakeed; pp. 79–92. [Google Scholar]
- 105.Schmiedehelm MH. Manuscript in the archive of Archaeological Research Collection of the University of Tallinn. Tallinn: Tallinn University; 1928. Aruanne kaevamisest külakalmel Otepääl 1. VI–5.VI 1928. [Google Scholar]
- 106.Schmiedehelm MH. Aruanne kaevamisest Otepääl 17.VI–21.VI 1929. a. (Manuscript in the archive of Archaeological Research Collection of the University of Tallinn) Tallinn: Tallinn University; 1929. [Google Scholar]
- 107.Saadre O. Aruanne kaevamistest Otepää külakalmel 13.–15. oktoobrini 1938. (Manuscript in the archive of Archaeological Research Collection of the University of Tallinn) Tallinn: Tallinn University; 1938. [Google Scholar]
- 108.Valk H. Archaeological investigations in Otepää and its Surroundings in 1996. In: Tamla U, editor. Stilus. Eesti Arheoloogiaseltsi väljaanne,Ü. Tallinn: Eesti Arheoloogiaselts; 1997. pp. 124–129. [Google Scholar]
- 109.Raudkivi P. Maa meie ema, ilm meie isa. Märkmeid looduse rollist Liivimaa 14. sajandi ajaloos. Acta Hist Tallinn. 2010;15:3–23. [Google Scholar]
- 110.Napiersky CE. Beitrag zur Geschichte des ehemaligen Bisthums Dorpat. Riga: Müllersche Buchdruckerei; 1846. [Google Scholar]
- 111.Valk H. Aruanne Vana-Kuuste 15.–17. sajandi külakalme arheoloogilistest kaevamistest 1982. a. (Manuscript in the archive of the archaeology department of the University of Tartu) Tartu: University of Tartu; 1983. [Google Scholar]
- 112.Valk H. Der Dorffriedhof von Mäletjärve. Proc Acad Sci Est SSR Soc Sci. 1985;34(4):376–379. [Google Scholar]
- 113.Valk H. Der Dorffriedhof von Vaabina. Proc Acad Sci Est SSR Soc Sci. 1986;35(4):389–393. [Google Scholar]
- 114.Damgaard PB, Margaryan A, Schroeder H, Orlando L, Willerslev E, Allentoft ME. Improving access to endogenous DNA in ancient bones and teeth. Sci Rep. 2015;5 doi: 10.1038/srep11184. 11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Meyer M, Kircher M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc. 2010;2010 doi: 10.1101/pdb.prot5448. pdb.prot5448. [DOI] [PubMed] [Google Scholar]
- 116.Orlando L, Ginolhac A, Zhang G, Froese D, Albrechtsen A, Stiller M, Schubert M, Cappellini E, Petersen B, Moltke I, et al. Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature. 2013;499:74–78. doi: 10.1038/nature12323. [DOI] [PubMed] [Google Scholar]
- 117.Malaspinas A-S, Lao O, Schroeder H, Rasmussen M, Raghavan M, Moltke I, Campos PF, Sagredo FS, Rasmussen S, Gonçalves VF, et al. Two ancient human genomes reveal Polynesian ancestry among the indigenous Botocudos of Brazil. Curr Biol CB. 2014;24:R1035–1037. doi: 10.1016/j.cub.2014.09.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet journal. 2011;17:10–12. [Google Scholar]
- 119.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinforma Oxf Engl. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinforma Oxf Engl. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jónsson H, Ginolhac A, Schubert M, Johnson PLF, Orlando L. mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinforma Oxf Engl. 2013;29:1682–1684. doi: 10.1093/bioinformatics/btt193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fu Q, Mittnik A, Johnson PLF, Bos K, Lari M, Bollongino R, Sun C, Giemsch L, Schmitz R, Burger J, et al. A revised timescale for human evolution based on ancient mitochondrial genomes. Curr Biol CB. 2013;23:553–559. doi: 10.1016/j.cub.2013.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rasmussen M, Guo X, Wang Y, Lohmueller KE, Rasmussen S, Albrechtsen A, Skotte L, Lindgreen S, Metspalu M, Jombart T, et al. An Aboriginal Australian genome reveals separate human dispersals into Asia. Science. 2011;334:94–98. doi: 10.1126/science.1211177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Korneliussen TS, Albrechtsen A, Nielsen R. ANGSD: analysis of next generation sequencing data. BMC Bioinformatics. 2014;15:356. doi: 10.1186/s12859-014-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Skoglund P, Storå J, Götherström A, Jakobsson M. Accurate sex identification of ancient human remains using DNA shotgun sequencing. J Archaeol Sci. 2013;40:4477–4482. [Google Scholar]
- 127.Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 128.Behar DM, Yunusbayev B, Metspalu M, Metspalu E, Rosset S, Parik J, Rootsi S, Chaubey G, Kutuev I, Yudkovsky G, et al. The genome-wide structure of the Jewish people. Nature. 2010;466:238–242. doi: 10.1038/nature09103. [DOI] [PubMed] [Google Scholar]
- 129.Yunusbayev B, Metspalu M, Järve M, Kutuev I, Rootsi S, Metspalu E, Behar DM, Varendi K, Sahakyan H, Khusainova R, et al. The Caucasus as an asymmetric semipermeable barrier to ancient human migrations. Mol Biol Evol. 2012;29:359–365. doi: 10.1093/molbev/msr221. [DOI] [PubMed] [Google Scholar]
- 130.Yunusbayev B, Metspalu M, Metspalu E, Valeev A, Litvinov S, Valiev R, Akhmetova V, Balanovska E, Balanovsky O, Turdikulova S, et al. The genetic legacy of the expansion of Turkic-speaking nomads across Eurasia. PLoS Genet. 2015;11:e1005068. doi: 10.1371/journal.pgen.1005068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Raghavan M, Skoglund P, Graf KE, Metspalu M, Albrechtsen A, Moltke I, Rasmussen S, Stafford TW, Orlando L, Metspalu E, et al. Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans. Nature. 2014;505:87–91. doi: 10.1038/nature12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Rasmussen M, Li Y, Lindgreen S, Pedersen JS, Albrechtsen A, Moltke I, Metspalu M, Metspalu E, Kivisild T, Gupta R, et al. Ancient human genome sequence of an extinct Palaeo-Eskimo. Nature. 2010;463:757–762. doi: 10.1038/nature08835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kushniarevich A, Utevska O, Chuhryaeva M, Agdzhoyan A, Dibirova K, Uktveryte I, Möls M, Mulahasanovic L, Pshenichnov A, Frolova S, et al. Genetic heritage of the Balto-Slavic speaking populations: a synthesis of autosomal, mitochondrial and Y-chromosomal data. PloS One. 2015;10:e0135820. doi: 10.1371/journal.pone.0135820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Behar DM, Metspalu M, Baran Y, Kopelman NM, Yunusbayev B, Gladstein A, Tzur S, Sahakyan H, Bahmanimehr A, Yepiskoposyan L, et al. No evidence from genome-wide data of a Khazar origin for the Ashkenazi Jews. Hum Biol. 2013;85:859–900. doi: 10.3378/027.085.0604. [DOI] [PubMed] [Google Scholar]
- 135.Fedorova SA, Reidla M, Metspalu E, Metspalu M, Rootsi S, Tambets K, Trofimova N, Zhadanov SI, Hooshiar Kashani B, Olivieri A, et al. Autosomal and uniparental portraits of the native populations of Sakha (Yakutia): implications for the peopling of Northeast Eurasia. BMC Evol Biol. 2013;13:127. doi: 10.1186/1471-2148-13-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Weissensteiner H, Forer L, Fuchsberger C, Schöpf B, Kloss-Brandstätter A, Specht G, Kronenberg F, Schönherr S. mtDNA-Server: next-generation sequencing data analysis of human mitochondrial DNA in the cloud. Nucleic Acids Res. 2016;44:W64–69. doi: 10.1093/nar/gkw247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Weissensteiner H, Pacher D, Kloss-Brandstätter A, Forer L, Specht G, Bandelt H-J, Kronenberg F, Salas A, Schönherr S. HaploGrep 2: mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic Acids Res. 2016;44:W58–63. doi: 10.1093/nar/gkw233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat. 2009;30:E386–394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- 139.Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 140.Poznik GD, Xue Y, Mendez FL, Willems TF, Massaia A, Wilson Sayres MA, Ayub Q, McCarthy SA, Narechania A, Kashin S, et al. Punctuated bursts in human male demography inferred from 1,244 worldwide Y-chromosome sequences. Nat Genet. 2016;48:593–599. doi: 10.1038/ng.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.ISOGG. Y-DNA Haplogroup Tree 2019. [Accessed January 3, 2019]; Available at: https://isogg.org/tree/
- 142.YFull. Analysis and comparing your NextGen Y-Chr sequencing data. [Accessed January 3, 2019]; Available at: https://www.yfull.com/
- 143.Quinlan AR. BEDTools: The Swiss-army tool for genome feature analysis. Curr Protoc Bioinforma. 2014;47:11.12.1–34. doi: 10.1002/0471250953.bi1112s47. Ed. Board Andreas Baxevanis Al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Unterländer M, Palstra F, Lazaridis I, Pilipenko A, Hofmanová Z, Groß M, Sell C, Blöcher J, Kirsanow K, Rohland N, et al. Ancestry and demography and descendants of Iron Age nomads of the Eurasian Steppe. Nat Commun. 2017;8 doi: 10.1038/ncomms14615. 14615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.de B Damgaard P, Marchi N, Rasmussen S, Peyrot M, Renaud G, Korneliussen T, Moreno-Mayar JV, Pedersen MW, Goldberg A, Usmanova E, et al. 137 ancient human genomes from across the Eurasian steppes. Nature. 2018;557:369–374. doi: 10.1038/s41586-018-0094-2. [DOI] [PubMed] [Google Scholar]
- 146.de Barros Damgaard P, Martiniano R, Kamm J, Moreno-Mayar JV, Kroonen G, Peyrot M, Barjamovic G, Rasmussen S, Zacho C, Baimukhanov N, et al. The first horse herders and the impact of early Bronze Age steppe expansions into Asia. Science. 2018;360 doi: 10.1126/science.aar7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Narasimhan VM, Patterson NJ, Moorjani P, Lazaridis I, Mark L, Mallick S, Rohland N, Bernardos R, Kim AM, Nakatsuka N, et al. The genomic formation of South and Central Asia. bioRxiv. 2018 292581. [Google Scholar]
- 148.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan Y, Genschoreck T, Webster T, Reich D. Ancient admixture in human history. Genetics. 2012;192:1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sudmant PH, Mallick S, Nelson BJ, Hormozdiari F, Krumm N, Huddleston J, Coe BP, Baker C, Nordenfelt S, Bamshad M, et al. Global diversity, population stratification, and selection of human copy-number variation. Science. 2015;349 doi: 10.1126/science.aab3761. aab3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Leslie S, Winney B, Hellenthal G, Davison D, Boumertit A, Day T, Hutnik K, Royrvik EC, Cunliffe B, Wellcome Trust Case Control Consortium 2 et al. The fine-scale genetic structure of the British population. Nature. 2015;519:309–314. doi: 10.1038/nature14230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Montinaro F, Busby GBJ, Pascali VL, Myers S, Hellenthal G, Capelli C. Unravelling the hidden ancestry of American admixed populations. Nat Commun. 2015;6 doi: 10.1038/ncomms7596. 6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lawson DJ, Hellenthal G, Myers S, Falush D. Inference of population structure using dense haplotype data. PLoS Genet. 2012;8:e1002453. doi: 10.1371/journal.pgen.1002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.R Core Team. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2017. Available at: https://www.R-project.org/ [Google Scholar]
- 157.Hellenthal G, Busby GBJ, Band G, Wilson JF, Capelli C, Falush D, Myers S. A genetic atlas of human admixture history. Science. 2014;343:747–751. doi: 10.1126/science.1243518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Raveane A, Aneli S, Montinaro F, Athanasiadis G, Barlera S, Birolo G, Boncoraglio G, Blasio AMD, Gaetano CD, Pagani L, et al. Population structure of modern-day Italians reveals patterns of ancient and archaic ancestries in Southern Europe. bioRxiv. 2018 doi: 10.1126/sciadv.aaw3492. 494898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Jeffreys Harold. An invariant form for the prior probability in estimation problems. Proc R Soc Lond Ser Math Phys Sci. 1946;186:453–461. doi: 10.1098/rspa.1946.0056. [DOI] [PubMed] [Google Scholar]
- 160.Voight BF, Adams AM, Frisse LA, Qian Y, Hudson RR, Di Rienzo A. Interrogating multiple aspects of variation in a full resequencing data set to infer human population size changes. Proc Natl Acad Sci U S A. 2005;102:18508–18513. doi: 10.1073/pnas.0507325102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Browning SR, Browning BL. Accurate non-parametric estimation of recent effective population size from segments of identity by descent. Am J Hum Genet. 2015;97:404–418. doi: 10.1016/j.ajhg.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Monroy Kuhn JM, Jakobsson M, Günther T. Estimating genetic kin relationships in prehistoric populations. PloS One. 2018;13:e0195491. doi: 10.1371/journal.pone.0195491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Bevan A, Crema ER. rcarbon v1.2.0: Methods for calibrating and analysing radiocarbon dates. 2018 Available at: https://CRAN.R-project.org/package=rcarbon.
- 164.Gamba C, Jones ER, Teasdale MD, McLaughlin RL, Gonzalez-Fortes G, Mattiangeli V, Domboróczki L, Kovári I, Pap I, Anders A, et al. Genome flux and stasis in a five millennium transect of European prehistory. Nat Commun. 2014;5 doi: 10.1038/ncomms6257. 5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. The variant call format and VCFtools. Bioinforma Oxf Engl. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Browning BL, Browning SR. Genotype imputation with millions of reference samples. Am J Hum Genet. 2016;98:116–126. doi: 10.1016/j.ajhg.2015.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Walsh S, Chaitanya L, Clarisse L, Wirken L, Draus-Barini J, Kovatsi L, Maeda H, Ishikawa T, Sijen T, de Knijff P, et al. Developmental validation of the HIrisPlex system: DNA-based eye and hair colour prediction for forensic and anthropological usage. Forensic Sci Int Genet. 2014;9:150–161. doi: 10.1016/j.fsigen.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 170.Walsh S, Chaitanya L, Breslin K, Muralidharan C, Bronikowska A, Pospiech E, Koller J, Kovatsi L, Wollstein A, Branicki W, et al. Global skin colour prediction from DNA. Hum Genet. 2017;136:847–863. doi: 10.1007/s00439-017-1808-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.