Abstract
The immune response to melanoma improves survival in untreated patients and predicts response to immune checkpoint blockade. Here we report genetic and environmental predictors of the immune response in a large primary cutaneous melanoma cohort. Bioinformatic analysis of 703 tumor transcriptomes was used to infer immune cell infiltration and categorize tumors into immune subgroups, which were then investigated for association with biological pathways, clinicopathological factors, and copy number alterations. Three subgroups, with "Low”, “Intermediate", and "High" immune signals were identified in primary and replicated in metastatic tumors. Genes in the Low subgroup were enriched for cell cycle and metabolic pathways, whereas genes in the High subgroup were enriched for interferon and NF-κB signaling. We identified high MYC expression partially driven by amplification, HLA-B downregulation, and deletion of IFN-γ and NF-κB pathway genes as regulators of immune suppression. Furthermore, we showed that cigarette smoking, a globally detrimental environmental factor, modulates immunity, reducing survival primarily in patients with a strong immune response. Together these analyses identify a set of easily assessible factors that may serve as predictors of response to immunotherapy in melanoma patients.
Introduction
The presence of tumor infiltrating lymphocytes (TILs) predicts better outcomes from primary melanoma [1][2] and therapeutic benefit from checkpoint blockade is more likely if tumors are PD-L1 positive [3] in response to T cell infiltration. Data have been published suggesting that higher mutational load is predictive of response to immunotherapy, and some studies with small numbers of patients have reported gene expression signatures with some predictive value [4][5]. However, the crucial need remains to identify the biological processes underlying “cold” unresponsive tumors. Bioinformatic analysis of large-scale “omic” datasets such as The Cancer Genome Atlas (TCGA) increasingly contribute to our understanding of tumor immunology [6][7] but the tumors are highly selected/biased, at advanced stage and with limited clinical metadata. In this report, we have used transcriptomic data generated from 703 of the 2184 participants in a population-based primary melanoma cohort (the Leeds Melanoma Cohort, LMC) [8][9] to explore the drivers of immune responses/failure at diagnosis, with the aim, ultimately, of improving adjuvant therapeutic choices.
In a previous report, we applied an approach to inferring the tumour immune microenvironment described by Bindea et al. [10] and identified 6 immunologically different tumour subgroups [11]. The Immunome Compendium used in that study contained genes specific to 24 immune cells [11]. In the current study, we utilized a refined version of the immunome compendium derived from a more extensive literature screening and covering 31 immune cell subtypes as published by Angelova et al. [12]. We defined transcriptomic scores for these immune cells and used them to classify tumors with unsupervised methods to identify immunologically different subgroups. The classification was based on the immune cell scores generated from the expression of genes attributed to each cell subtype rather than on individual genes as we reported in our previous study [11]. We postulated that reducing the number of dimensions prior to classification analysis could identify tumor groupings with a clearer difference in survival, facilitating subsequent in depth biological and epidemiological characterization, directed towards the identification of candidates for therapeutic targets.
There is evidence that environmental factors may modify immune responses to tumors [13]. We have previously reported that smoking was associated with microscopic tumor ulceration and vitamin D was protective [9] and here we demonstrate the effect of smoking as a modifier of outcome within each immune subgroup.
Methods
The Leeds Melanoma Cohort transcriptomic data
The transcriptomic data from 703 tumors were generated and pre-processed as previously reported using the Illumina DASL whole genome array [14][15][11]. These data are accessible for the purposes of melanoma research from the European Genome-Phenome Archive with the accession number EGAS00001002922. All survival analyses used melanoma specific survival (MSS). The median follow-up for 703 samples at the time the data set was fixed, was 7.5 years. Detailed information about the cohort is provided in Supplementary Methods. The participants gave written informed consent and the study received ethical approval (MREC 1/03/57 and PIAG3-09(d)/2003) and was conducted in accordance with recognized ethical guidelines (Declaration of Helsinki).
Immune cell scoring
Angelova et al. generated a list of genes identified as specific to certain immune cells in the blood [12] (the compendium of immune genes). These 1,980 genes were identified from reports of 36 studies comprised of 813 microarrays generated from 30 purified immune cell subtypes (activated and memory B cells, activated CD4+ and CD8+ cells, central memory CD4+ and CD8+, cytotoxic cells, dendritic cells (DC), effector memory CD4+ and CD8+, eosinophils, immature B cells, macrophages, mast cells, monocytes, natural killer cells (NK), natural killer 56 bright, 56 dim and natural killer T, neutrophils, T cells, T follicular helper (TFH), T gamma delta (TGD), T helper 1 (Th), 2, 17, T regulatory cells (Treg), immature, plasmacytoid and memory dendritic cells (iDC, pDC, mDC)), as well as genes (searched from literature) for myeloid-derived suppressor cells (MDSCs), resulting altogether in 31 subtypes.
From the initial list of genes, we removed those also strongly expressed (in the top 25%) in a melanocyte cell line (GSE4570) and in melanoma cell lines, MEWO and SK-MEL28 (in-house data). In a second step, we removed cell subsets for which more than 90% of genes were eliminated (in the previous step) or if there was insufficient published evidence for the remaining genes to be considered representative of those cell types. We expected that expression of the majority of genes specific to a particular cell type would be positively correlated within the cell type, as this was the basis of gene selection in the Angelova et al. study. However, in our dataset this was not always the case, so in a third step of filtering, we removed genes negatively correlating with the majority within each cell subset to reduce the risk of noise due to technical factors. After applying the filters described above, we devised a score for each immune cell type, calculated as the mean of expression values of all genes attributed to that cell, after z-score normalization of the log2 transformed gene expression data as described before [11]. The scores were calculated in the LMC primary melanomas and in the TCGA metastatic melanomas. The reciprocal correlations of genes within each immune cell score were compared between these two datasets.
Clustering of LMC tumors
We applied consensus cluster analysis [16] within the R package ConsensusClusterPlus [17] to classify primary melanomas of LMC based on their immune cell scores. This approach generates a varying number of clusters (to a fixed maximum number, K) using resampling of the data. It is widely used to find stable sample subgroups in high-dimensional data as a better alternative to the standard one-off clustering, which might be affected by random variation. Additionally, consensus clustering offers useful metrics (see below) to indicate the optimal number of clusters, unavailable in standard clustering. K-means was chosen as the clustering algorithm with maximum K=12, Euclidean distance, 5000 repetitions, 80% genes and tumor resampling. Examination of the consensus cluster matrices, the cumulative density function (CDF) and delta CDF (the change in the area under the CDF curve) allowed definition of the optimal number of tumor clusters [16].
Clustering replication in TCGA
We downloaded RNAseq gene expression and survival data for The Cancer Genome Atlas (TCGA) metastatic melanoma data (xhttp://www.cbioportal.org/data_sets.jspx) (339 samples downloaded in 2016). We hypothesized that the immune subgroups observed in the primaries would be recapitulated in metastatic melanomas, and to test this hypothesis we calculated cluster centroids (vector of cell score means within clusters) in the LMC dataset and utilized them to classify TCGA metastatic melanomas using the nearest centroid method, as described elsewhere [15]. Briefly, immune cell scores were calculated in the TCGA data in a similar manner as in the LMC and each TCGA tumor was assigned to one of the new clusters, with which it had the strongest Spearman correlation.
Overrepresentation analysis (ORA) and networks
To test the whole transcriptome differences between the immune subgroups in the LMC, the Kruskal-Wallis test was used for 3 groups, the Mann-Whitney U test was used for 2 groups and Bonferroni correction was applied for multiple testing correction (0.05/29354=1.7x10-6, the number of probes tested was 29354). To visualize the expression of significantly differentially expressed genes (DEGs) (excluding the compendium of immune genes) among immune subgroups, these DEGs were hierarchically clustered and a heatmap plotted. Reactome FiViz [18] and Centiscape [19] in Cytoscape [20] were utilized to analyse the protein-protein interaction (PPI) network and infer pathways enriched in the DEGs characterizing each immune subgroup. The networks were created based on existing protein-protein interaction networks built in Reactome FiViz, which covers over 50% of human proteins. From the network, pathway enrichment was calculated at FDR <0.001. In order to identify the most influential (hub) genes in the networks, the “betweenness” metric (indicating a key role in communication between proteins) was used as a centrality measure in Centiscape [19]. Graphical adjustments for network visualization were made in Gephi software [21]. The Spearman rank correlation was used to evaluate the correlation between the expression of a hub gene and the whole transcriptome patient-derived primary melanoma cell line cultures.
Primary melanoma cell lines validation experiment
As previously described, primary melanoma cells were isolated from surplus surgical specimens of consenting patients (approved by the local IRB [EK.647/800]) at the University of Zurich, and maintained by the University Research Priority Program in Translational Cancer Research at the University of Zurich Hospital [22]. The cell lines were authenticated by Sanger sequencing for the oncogenes seen from the tumor and tested for mycoplasma infection using the PlasmoTest Kit (InvivoGen). Cells were passaged about 5 times before RNA extraction. Details are provided in Supplementary Methods.
Immunohistochemistry (IHC) validation
The most influential genes identified in our analyses were further examined by IHC staining of sections of available primary tumors from the LMC, to assess the protein-level and gene expression correlations. Primary antibodies were supplied by Abcam (UK): anti-MYC (ab32072), anti-NF-κB p105 (ab32360), anti-HLA-B (ab193415). The details of scoring are described in the supplementary methods. The Mann-Whitney U test was used to compare the mRNA level and the IHC scores. The nuclear staining scores in tumor cells and in TILs were compared using the Fisher’s exact test. The correlation of continuous scoring was tested using Spearman’s rank correlation. Images presented are of digitally scanned slides generated by the Digital Pathology Group, Leeds Institute of Medical Research at St. James’s, using Aperio technology (Leica Biosystems, UK).
Analysis of CNA among the immune subgroups
We extracted copy number profiles in the genomic regions spanning the hub genes from the network analysis and compared them between immune subgroups and with gene expression and patient survival. Next-Generation Sequencing (NGS)-derived copy number alteration (CNA) profiles were available from 276 tumor samples among the 703 transcriptomic-profiled. To evaluate the full extent of the role of structural variation, we expanded the gene list to other genes of the same family or the same pathway as the hub genes (plus NF-κB and its regulators, and IFN-γ signaling genes). The association between CNAs and gene expression was tested using Fisher’s exact test. Since we have previously reported β-catenin signaling pathway to be upregulated in 42% of primary melanomas overall and in 73% of those with the worst outcome [11], we tested the overlap between the CTNNB1 expression signature with the immunosuppressive mechanisms identified in this study and their joint effect on survival (Cox-proportional hazard regression). For the CNA visualization, the ComplexHeatmap package in R was used [23]. The CNA data analysis in detail is included in supplementary methods.
Statistical analyses
The univariable Cox proportional hazards model was used to test the association between tumor immune subgroups and melanoma specific survival (MSS) in the LMC and overall survival (OS) in TCGA datasets. To test independence between the tumor immune subgroups and clinico-pathological factors, Chi-square and Kruskal-Wallis tests were used. A univariable Cox proportional hazard model was used to test the prognostic value of the immune cell scores and the clinical and environmental factors (AJCC staging version 7, age at diagnosis (median: 58.34 years), sex, site of melanoma (limbs vs rest), smoking never/ever (median duration of smoking in the smoking group was 23 years), vitamin D levels at recruitment (median level in winter: 39.5 nmol/L)) and a social status/deprivation index measured by Townsend score [24] in the whole LMC dataset. Subsequently, the significant clinical and environmental predictors were included in a multivariate model (adjusting for the immune clusters). The predictors with the strongest degree of independence in the whole dataset were jointly tested within each immune subgroup.
Results
Devising a list of genes indicative of specific immune cells infiltrating melanoma
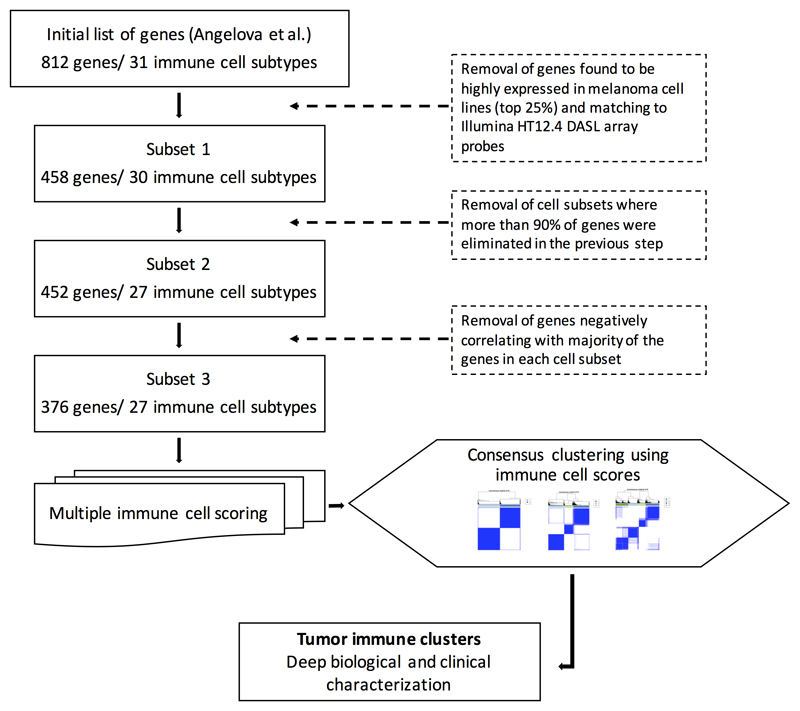
The first filtration step resulted in 458 genes representing 30 distinct immune cell subsets (Subset 1, Figure 1, Supplementary Data 1). The second step resulted in the elimination of scores for effector memory CD4+ T cells, activated CD8+ T cells and activated CD4+ T cells (Subset 2, Figure 1, Supplementary Data 1). The plasmacytoid dendritic cell score (pDCs) was retained despite having only 1 attributed gene (IL3RA), as in the previous version of the Immunome compendium [10] because it is known to be highly expressed in pDCs [25][26]. The final filtration step left 376 genes representing 27 immune cell subsets (Subset 3, Figure 1, Supplementary Data 1). We noted that when applied to TCGA transcriptomes, the correlation matrices between genes within each cell type also demonstrated a number of negatively correlated genes although fewer than in our primary melanoma cohort (data available upon request).
Figure 1. Devising a list of genes indicative of specific immune cells infiltrating melanoma.
From the initial gene list, genes found to be highly expressed in melanoma cell lines (in-house data) were removed, resulting in 458 genes retained representing 30 distinct immune cell types (Subset 1). In a second step, cell subsets where more than 90% of genes were lost in the previous step were removed, resulting in Subset 2. In the third step of filtering, all genes negatively correlating with the majority of the genes within each cell subset were removed, and thus the final number of genes was 376 representing 27 immune cell scores (Subset 3). Next, those genes were used to calculate a score for each immune cell, and these were used for consensus clustering of the tumors in immunologically different groups.
The association of 27 immune cell scores with survival
We tested the association of the immune cell scores with melanoma specific survival (MSS) (univariable analysis) and the results revealed that the majority of immune cell scores (17 out of 27) in the LMC and (23 out of 27) in TCGA were associated with improved survival after Bonferroni correction (27 tests, P<0.002). For 8 of the remaining 10 cell scores in the LMC, a similar protective effect was found but the effects did not withstand adjustment for multiple testing (Supplementary Table S1). The survival analysis was repeated after removal of the 16 participants known to have received checkpoint therapies and the results for all the immune cell scores were essentially unchanged.
Identification of three prognostic immune subgroups
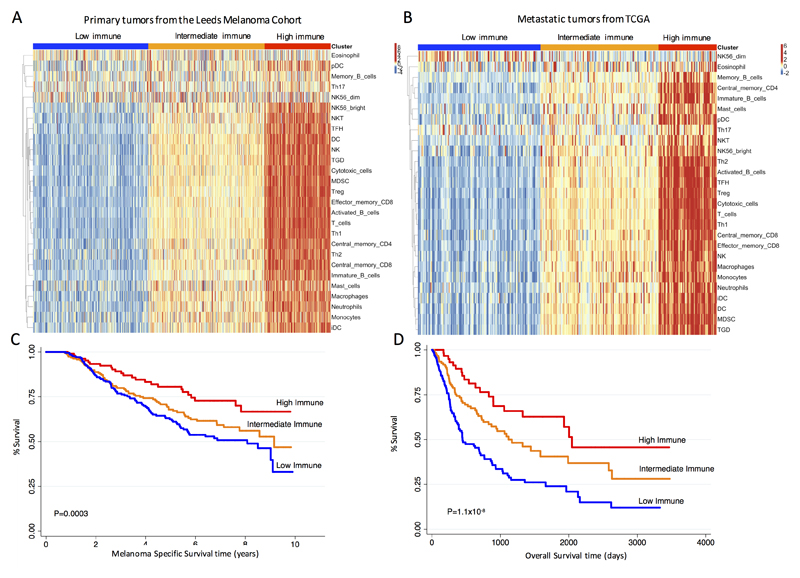
Consensus clustering analysis of tumor samples using the 27 immune cell scores identified 3 clusters with distinct immune phenotypes (Supplementary Fig. S1), which we termed Low, Intermediate and High Immune Subgroups (Figure 2A). Importantly, by classifying the TCGA metastatic melanomas in these 3 immune subgroups (supervised classification), we were able to replicate the results obtained in LMC with strong similarities in overall immunological profiles (Figure 2B). Furthermore, the three immune subgroups were associated with survival in both datasets: in the LMC, a significantly lower hazard of melanoma death was observed for patients assigned to the High compared to Low and Intermediate Immune Subgroups (Hazard Ratio (HR)=0.5, P=0.001 (95% CI 0.3-0.7); HR=0.6, P=0.05 (95% CI 0.4-1.0), respectively) (Figure 2C).
Figure 2. Identification of three prognostic immune subgroups in LMC primaries and TCGA metastatic melanoma datasets.
(A) Heatmap showing the three identified immune subgroups from the consensus clustering of immune cell scores in LMC (N=703). (B) Similar heatmap in TCGA (N=339). (C) Kaplan Meier survival curves for melanoma-specific survival (MSS) in LMC by the three immune subgroups. (D) Similar curves for overall survival (OS) in TCGA. P values from likelihood ratio test.
For TCGA, tumors classified as High Immune also exhibited a lower overall death hazard with HR=0.3, P=1.1x10-7 (95% CI 0.2-0.5) compared to those classified in the Low Immune Subgroup. Tumors in the Intermediate Immune Subgroup had a HR=0.5, P=4.6x10-5 (95% CI 0.4-0.7) when compared to those of the Low Immune Subgroup (Figure 2D).
The 3 class signature reported here (High, Intermediate and Low Immune) was concordant with the 6 consensus immunome clusters (CICs) we published previously [11], (Cramer’s V=0.72) (see Supplementary Fig. S2A).
However, there was only moderate concordance with another 3-class signature (immune, keratin and MITF low) published by TCGA, (Cramer’s V=0.47). In essence, our High Immune group overlapped well with the TCGA immune class but the Intermediate and Low Immune subgroups had much less overlap with TCGA groups (Supplementary Fig. S2B). Kaplan Meier curves for our three immune subgroups were more clearly/significantly separated than the three TCGA classes. Generally, we observed the expected prognostic trend of the immune signature but there was no difference between the immune and keratin groups of the TCGA signature (Supplementary Fig. S2C). There were 70 genes identified that were shared between our and TCGA signature (listed in Supplementary Data 2X).
The immune subgroups were associated with tumor thickness, TILs and mitotic number
In the LMC data, the High Immune, in comparison to the Low and Intermediate Subgroups, featured consistently thinner tumors (Kruskall Wallis P=0.004) and, crucially, more TILs reported by both clinical dermato-pathologists (Chi2 P=4.0x10-7) and a single observer from our research group who was blinded to the transcriptomic data (Chi2 P=3.6x10-8) (see Supplementary Table S2). The mitotic number was significantly lower in the High Immune Subgroup (Kruskal Wallis P=2x10-4). The Low Immune Subgroup had the lowest proportion of tumors harboring a BRAF mutation (40%) and the highest proportion with an NRAS mutation (30%), although these observations were only marginally significant (Supplementary Table S2). The recorded site of melanoma was significantly different across the immune subgroups, with primary tumors located at “rare” sites (not exposed to the sun) most frequently classified in the Low (19%) compared to the Intermediate (9.5%) and the High Immune Subgroups (8%) (Chi2 P=0.02). AJCC stage, patient sex, age at diagnosis, smoking status and levels of season-adjusted serum vitamin D were not significantly different between the subgroups (Supplementary Table S2).
Identification of MYC as the regulator of immune response in network analyses
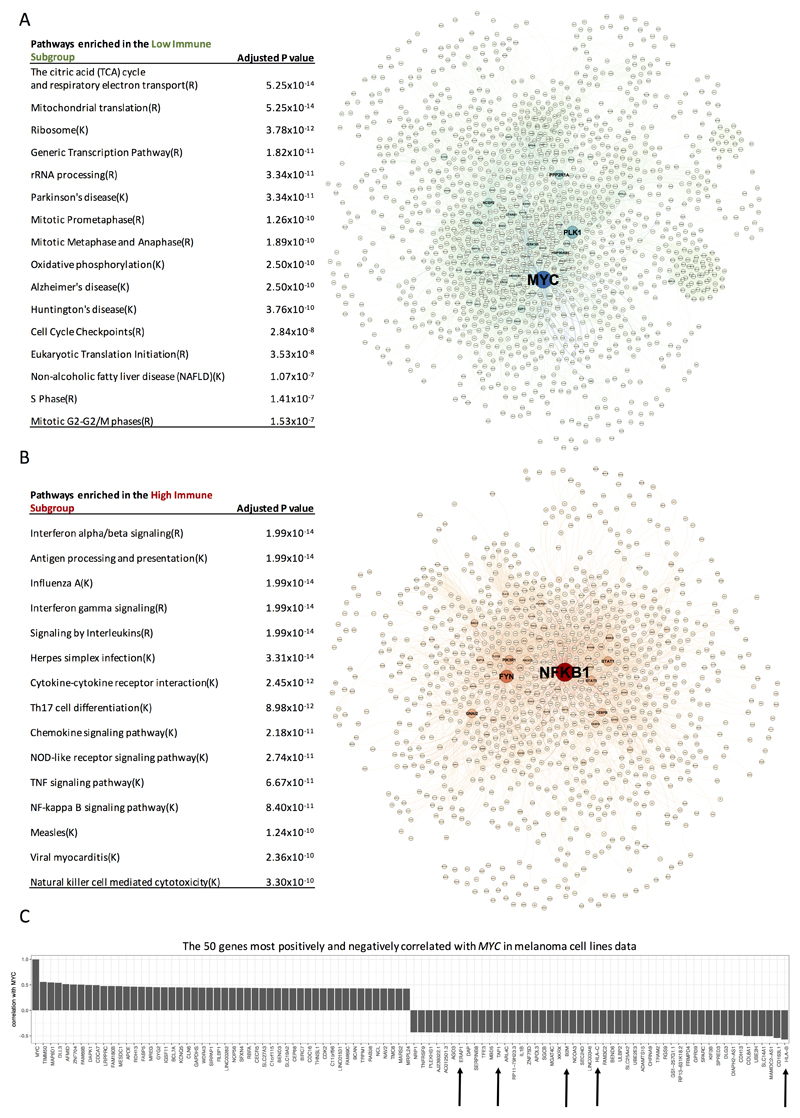
We compared gene expression among the 3 immune subgroups (Supplementary Fig. S3). 5607 differentially expressed genes across the genome were identified between the High vs Low Immune Subgroups. The genes upregulated in the Low Immune Subgroup (n=3324) were predominantly associated with high proliferation and metabolic activity (Hypergeometric test adjusted P value 10-14 to 10-7) with lower levels of expression of the genes coding immune checkpoint molecules (expressed by tumor) such as CD274 coding for PD-L1. The most enriched pathways were the citric acid (TCA) cycle and respiratory electron transport, mitochondrial translation and mitosis pathways (Figure 3A and Supplementary Data 2A). Network analysis of genes enriched in the Low Immune Subgroup revealed that the proto-oncogene MYC had the highest centrality (Figure 3A). Unsurprisingly, network analysis indicated that the genes upregulated in the High Immune Subgroup (n=2283) were mostly involved in immune pathways (Hypergeometric test adjusted P value 10-14 to 10-10). The top enriched pathways were: Interferon alpha/beta signaling, antigen processing and presentation, interferon gamma and NF-κB signaling (Figure 3B and Supplementary Data 2B), with the nodal gene in this network being NFKB1 encoding the p105/p50 subunit of NF-κB (Figure 3B).
Figure 3. Identification of “hub” genes and enriched pathways in the network analyses.
(A) The most enriched pathways in the Low Immune Subgroup, in the network. Protein-protein interaction network of genes upregulated in the Low Immune Subgroup. (B) The most enriched pathways in the High Immune Subgroup, in the network. (K=KEGG, R=Reactome). Protein-protein interaction network of genes upregulated in the High Immune Subgroup. The size of nodes (protein from the interaction network) indicates the importance in the network (betweenness). (C) The 50 genes most positively and negatively correlated with MYC in melanoma cell lines data (Spearman’s rank correlation). The arrows point to the genes coding for proteins involved in antigen processing and presentation via HLA.
The identification of MYC as the gene with the highest centrality in the Low Immune network suggested that it might fulfil a key role in immune evasion. We took an agnostic approach to testing correlations between MYC expression and the rest of the genome in transcriptomes from patient-derived primary melanoma cell lines (lacking immune cells) [22]. Genes were ranked according to their correlation with MYC and of 50 genes most significantly negatively correlated with MYC one tenth were involved in antigen processing and presentation (HLA-B, HLA-C, B2M, TAP1 and ERAP1), with HLA-B representing the strongest results (R=-0.57, P=1.6x10-10) (Figure 3C and Supplementary Data 2C). The correlation of MYC with HLA-B gene expression in the LMC was: R=-0.3 (P=5.5x10-14).
An immunosuppressive effect of oncogenic MYC has previously been demonstrated, although by different mechanisms than suggested in this study: MYC was reported to increase expression of genes encoding CD47 and PD-L1 on lymphoblastic leukemia cells [27]. We tested this observation using Spearman’s rank correlation, but MYC expression did not significantly correlate or correlated negatively with CD47 or PD-L1 expression in either the primary melanoma cell lines (R=-0.17, P=0.09; R=-0.16, P=0.1, respectively) or in the LMC tumors (R=0.04, P= 0.3; R=-0.17, P=2.3x10-6, respectively).
mRNA gene expression correlation with protein scores – Immunohistochemistry (IHC)
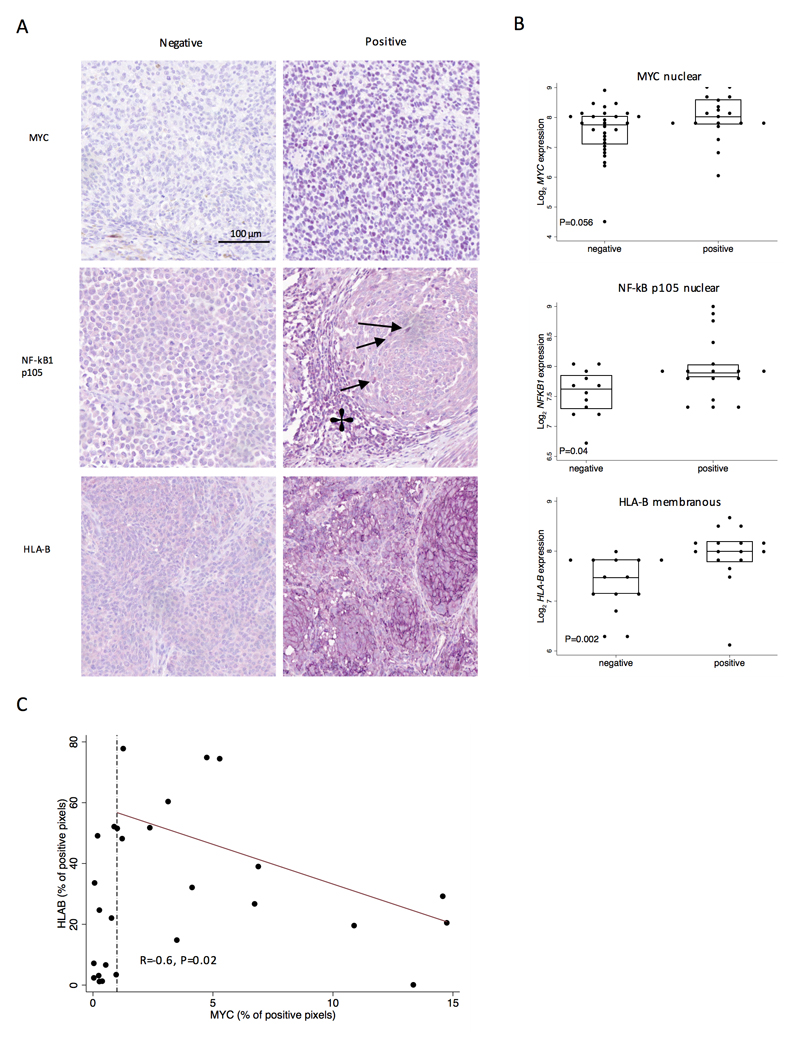
A subset of tumors was immunohistochemically stained using antibodies for key proteins. We found that the protein expression of MYC localized to the tumor cell nuclei while HLA-B localized to the tumor cell membrane and the expression of both proteins was positively associated with their mRNA transcripts (P=0.056 and P=0.002 respectively) (Figure 4). MYC staining was only detected in tumor, not immune cells. Using the Nuance software for calculation of number pixels of positive staining per analyzed image of MYC and HLAB we observed a negative correlation (R=-0.6, P=0.02) only for samples where MYC was detected (N=15) (Figure 4C). For samples where MYC was barely detected (<1%) the correlation was not seen, which indicated that there are other factor regulating HLAB expression in the absence of MYC. NF-κB p105 was detectable in the nuclei of both tumor cells and TILs and the levels of expression from these were positively correlated (P=3x10-5). Importantly, mRNA expression of NFKB1 was positively correlated with tumor NF-κB p105 nuclear staining (P=0.02) (Figure 4).
Figure 4. Gene expression levels correlate with protein scores - Immunohistochemistry (IHC).
(A) Representative images of positive and negative staining for MYC (nuclear), HLA-B (membranous), NF-κB p105 (Tumor and TILs nuclei), 20x magnification. (B) Dot and box plots show comparisons of mRNA level (y axis) and staining level (x axis), using Mann-Whitney U test (MYC: N=48, HLA-B: N=30, NF-κB p105: N=29). NF-κB p105 nuclear staining is indicated by arrows, NF-κB p105 in tumor infiltrating lymphocytes by a star. Purple chromogen was used for staining, therefore the color representing the positive staining is lavender-purple. (C) The scatter plot represents the scoring values for HLA-B (percentage of positive pixels for chromagen in outputs from Nuance software) on the y-axis and on the x-axis MYC (percentage of positive pixels for both haematoxylin and chromagen). The dashed line indicates MYC detection at less than 1%, which we considered as very low/absent expression. The red line is fitted for the MYC values higher that 1% of positive pixels.
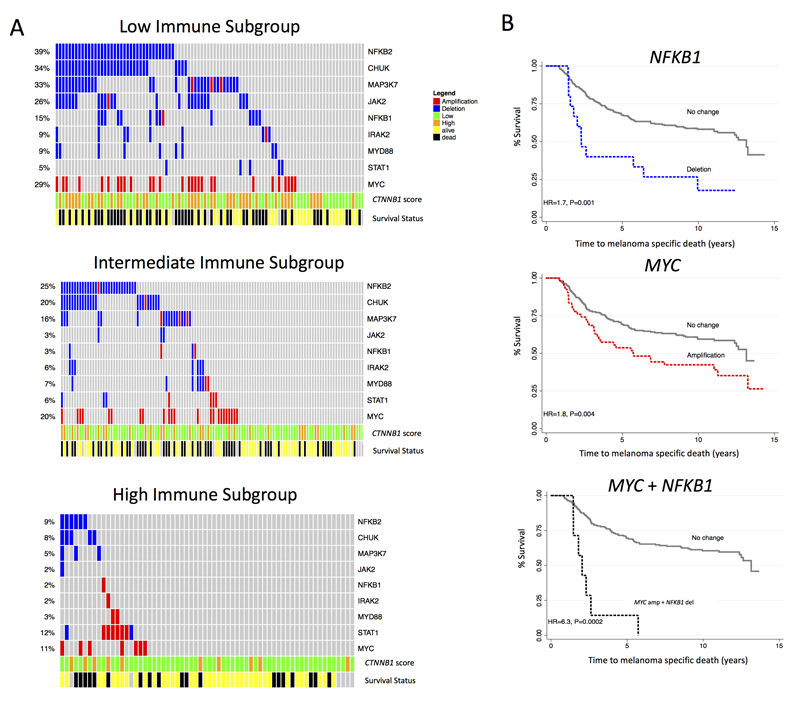
MYC was more frequently amplified, while NF-κB and IFN-γ signaling genes was more frequently deleted, in the Low Immune Subgroup
Given that we observed upregulation of MYC and downregulation of NFKB1 expression (the nodal genes) in the Low Immune Subgroup, we hypothesized that MYC amplifications and NFKB1 deletions would be more common in this immune subgroup in the LMC. Using next-generation sequencing derived copy number data from a subset of the LMC tumors, we observed that 29% had amplifications of MYC and 14% deletions of NFKB1 in the Low Immune Subgroup, more than in the Intermediate or the High Immune Subgroup (P= 0.02 for MYC, P=0.0003 for NFKB1) (Figure 5A, Supplementary Data 2D). Interestingly both of these copy number changes were strongly predictive of poor prognosis overall (adjusted for AJCC stage) separately (MYC amplifications: HR=1.8 (95% CI 1.8-2.6), P=0.006; NFKB1 deletions: HR=1.5 (95% CI 1.1-2.1), P=0.007) and when combined (HR=3.7 (95% CI 1.6-8.5), P=0.002, adjusted for AJCC) (Figure 5B, Supplementary Data 2E and F).
Figure 5. MYC is amplified while NF-κB and IFN-γ signaling genes are deleted in the Low Immune Subgroup.
(A) Oncoprint figure for Low, Intermediate and High Immune group representing CNA of MYC, NF-κB and genes in the IFN-γ pathway with annotation of CTNNB1 expression and survival status. (B) Kaplan Meier plots for participants whose tumors showed MYC amplifications, NFKB1 deletions, and for a combination of MYC and NFKB1 CNAs in the whole dataset. Hazard ratios were calculated using the univariable Cox proportional hazard model.
Because the NF-κB and IFN-γ pathways were amongst the most enriched pathways in the High Immune Subgroup, we then asked if other genes within these pathways were deleted in the Low Immune Subgroup. Indeed, we found evidence of deletion of NFKB2 (26% of whole dataset), CHUK (22%), MYD88 (5%), IRAK2 (5%), MAP3K7 (17%), JAK2 (10%), and STAT1 (4%). These copy number changes were not mutually exclusive (Figure 5A) but were much more frequent in the Low Immune than in other subgroups (Figure 5B, Supplementary Data 2D). Deletion of CHUK, MYD88, IRAK2 or JAK2 each were predictive of death from melanoma after adjusting for AJCC stage (Figure 5B, Supplementary Data 2E and F). As expected, these copy number changes were highly correlated with mRNA expression of corresponding genes (Supplementary Data 2F).
In our previous study [11] we demonstrated that CTNNB1 expression alone was overexpressed in 30% of all primaries and 59% of in the CIC4 (Immune Low/β-catenin high). Here, we observed a similar pattern across the three immune subgroups: CTNNB1 was more commonly overexpressed in the Low Immune Subgroup compared to other groups (Figure 5A). Comparing the copy number alteration of genes in the NF-κB pathway and CTNNB1 expression, we found some overlap but also heterogeneity in the Low Immune Subgroup. Specifically, 15% of tumors had evidence of increased CTNNB1 expression alone, 32% had a deletion in at least one gene of the NF-κB pathway in the absence of CTNNB1 overexpression, whilst 31% had both (i.e. increased β-catenin and a deletion in at least one gene). In prognostic terms, in the whole dataset the HR for melanoma death in the presence of CTNNB1 upregulation was HR=2.2, P=5x10-5, 1.5-3.1; for NF-κB pathway deletions was HR=2.03, P=2x10-4, 1.4-3.0; and for the combination of these two pathways was HR=3.4, P=5x10-5, 95% CI 2.2-5.5.
These data demonstrate the involvement of genetic factors in modulating immunity and shaping the tumor immuno-phenotype. However, it is commonly postulated that environmental factors also play a role in this process, and we therefore tested this hypothesis.
Smoking as a strong independent risk factor for melanoma death in the High Immune Subgroup
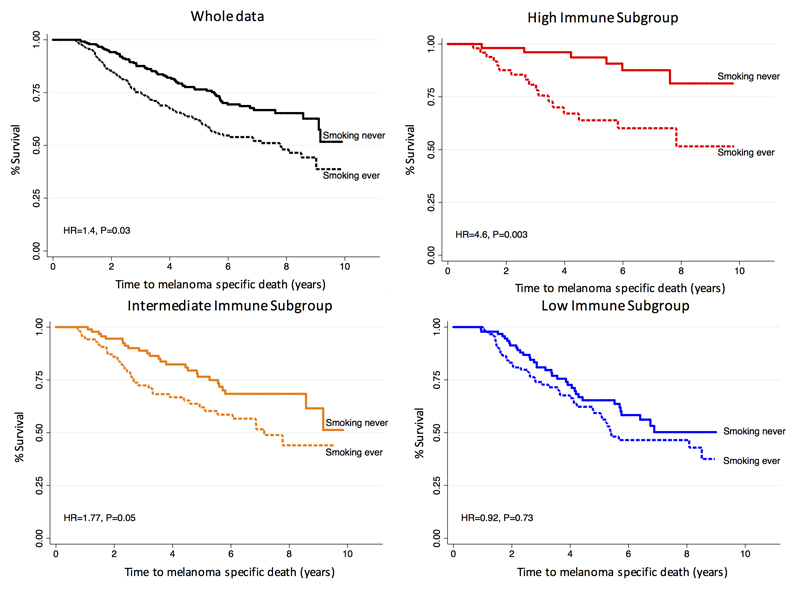
In addition to clinico-pathological tumor characteristics, the LMC has a record of patient smoking behaviors, a vitamin D level from a blood sample at diagnosis and a deprivation index measured by the Townsend score [24]). In a univariable Cox proportional hazard model, AJCC staging, mitotic number, site of primary melanoma, age at diagnosis, sex and smoking (never/ever), were significantly predictive of MSS in the whole dataset while season-adjusted vitamin D was not. Among these variables, AJCC stage, mitotic number, site of melanoma, age at diagnosis, and smoking remained significant in multivariable analysis of the whole dataset but different sets of variables were significant in each of the three immune subgroups (Table 1). Body site of the primary melanoma was a strong predictor of MSS in the Low Immune Subgroup along with AJCC stage, driven by tumors arising in sun-protected body sites which were predominantly classified within this group and are known to have a particularly bad outcome [28] (Table 1). The prognostic effect of smoking was heterogeneous across the three immune subgroups (P<0.03 for equal HRs across the subgroups). In the High Immune Subgroup (HR=4.6 for “ever smoked”, P=0.003, N=156), compared to within the Intermediate Immune Subgroup (HR=1.8, P=0.05, N=275) and the Low Immune Subgroup (HR=0.9, P=0.7, N=272) (Figure 6). The deleterious effect of smoking in the High Immune Subgroup was reproduced when the analysis was repeated using two alternative definitions of smoking habits: duration of smoking (number of years) and the cumulative number of smoked cigarettes (packs per year) (Supplementary Data 2G and H). The negative prognostic effect of cigarette smoking in the High Immune Subgroup remained significant after adjusting for the deprivation index (HR=1.6, P=0.001). To gain a deeper insight into the interplay between smoking and immune responses, we assessed the expression of GPR15 which has previously been described as a biomarker of exposure to tobacco smoke, with increased expression previously demonstrated in a number of immune cell types measured in the peripheral blood [29][30]. In the tumors, we found no significant association between GPR15 expression with smoking (never/ever) across the whole LMC dataset (fold change=1.07, P=0.12). However, the association was stronger in the High Immune Subgroup (fold change=1.32, P=0.02) (Supplementary Fig. S4A). Furthermore, GPR15 expression was the highest in the High Immune Subgroup when testing a subset of data of ever smokers: P=5x10-5, while the result was not significant for the second subset - never smokers: P=0.3 (Supplementary Fig. S4B). GPR15 expression in the blood is reported to decrease after cessation of smoking [30] and therefore we assessed its expression in ‘still smokers’ compared to ‘non-smokers’. We observed a markedly stronger differential expression in the High Immune Subgroup for still smoking (fold change=1.9, P=0.002) than in the whole dataset (fold change=1.32, P=0.01) (Supplementary Fig. S4B). We tested the differences in immune cell scores between never/ever smokers (in the High Immune Subgroup), but we did not find any statistically significant results (Supplementary Table S3). We also examined the association between smoking and tumor histological features (Supplementary Table S4) as well as the whole genome expression, including cytokine genes, but no significant associations were identified after multiple testing.
Table 1.
Multivariable Cox proportional hazard model for MSS in different immune subgroups, showing variables statistically significant in the whole data analysis. Smoking categories: never/ever; AJCC stage was categorized as stage 1, 2 or stage 3. Site of melanoma was sun exposed vs non-sun exposed. Mitotic number was the count of mitoses per mm2. Significant associations are shown in bold.
| Characteristics (risk category) | HR | P-value | 95% CI |
|---|---|---|---|
| Whole dataset (N=703) | |||
| AJCC stage | 2.05 | 2.4x10-8 | 1.59-2.64 |
| Smoking (ever) | 1.4 | 0.032 | 1.03-2.04 |
| Site of melanoma (non-sun exposed) | 1.64 | 0.012 | 1.11-2.41 |
| Age at diagnosis (per year) | 1.03 | 2.0x10-5 | 1.01-1.05 |
| Mitotic number (per mitosis) | 1.02 | 0.008 | 1.00-1.03 |
| High Immune (N=156) | |||
| AJCC stage | 3.99 | 0.0002 | 1.93-8.22 |
| Smoking (ever) | 4.58 | 0.003 | 1.68-12.53 |
| Site of melanoma (non-sun exposed) | 2.52 | 0.075 | 0.91-6.7 |
| Age at diagnosis (per year) | 1.05 | 0.025 | 1.00-1.10 |
| Mitotic number (per mitosis) | 1.02 | 0.71 | 0.93-1.11 |
| Intermediate Immune (N=275) | |||
| AJCC stage | 1.75 | 0.012 | 1.13-2.71 |
| Smoking (ever) | 1.77 | 0.05 | 1.01-3.12 |
| Site of melanoma (non-sun exposed) | 1.36 | 0.31 | 0.75-2.46 |
| Age at diagnosis (per year) | 1.03 | 0.021 | 1.00-1.06 |
| Mitotic number (per mitosis) | 1.04 | 0.0004 | 1.02-1.06 |
| Low Immune (N=272) | |||
| AJCC stage | 2.01 | 1.3x10-4 | 1.40-2.87 |
| Smoking (ever) | 0.92 | 0.73 | 0.56-1.50 |
| Site of melanoma (non-sun exposed) | 1.97 | 0.016 | 1.13-3.43 |
| Age at diagnosis (per year) | 1.03 | 0.002 | 1.01-1.06 |
| Mitotic number (per mitosis) | 1.01 | 0.26 | 0.99-1.03 |
Figure 6. Kaplan Meier plots of smoking (never vs ever).
In the whole data set (N=703) - black, High Immune Subgroup (N=156) - red, Intermediate Immune Subgroup (N=275) - yellow, and Low Immune Subgroups (N=272) - blue. Hazard ratios were calculated using a Cox proportional hazard model for MSS. Figure shows that the detrimental effect of smoking on survival increases with the strength of the patient’s tumor immune signal.
Discussion
The dramatic survival benefit of checkpoint blockade in melanoma, in around half of patients with advanced disease [31], has highlighted the need to understand the drivers of Low Immune (“cold” tumors) which are less likely to respond to treatment. In silico immune cell analysis in cancer has been adopted in recent years in order to better understand these drivers [32][12][33] although these approaches do not allow distinction to be made between weak signals coming from numerous infiltrating immune cells and strong signals from fewer cells.
Previously we reported the survival analysis of 24 immune cell scores [11], derived from an earlier version of the immunome [10]. In the current analysis, we used an updated version of the immunome [12] allowing inference of 31 immune cell scores, which we reduced to 27 after the gene filtration. We noted that in the earlier report, 10/24 cells were significantly prognostic and replicated in TCGA (41% of cells) but in this study the number increased 17/27 (63%).
In evaluating the updated Immunome, we showed that almost half of the genes proposed to be specific to particular immune cell subsets were also expressed at significant levels in melanoma cell lines, disqualifying them as immune specific. The fact that not all of the genes postulated to characterize a particular immune cell type were positively correlated may represent in part a technical feature of our dataset, as we observed that the correlations between these genes were slightly higher in the TCGA dataset sourced from fresh-frozen tumors rather than archived Formalin-Fixed Paraffin-Embedded (FFPE) specimens. These observations suggest that use of “off the shelf” algorithms to infer immune activity may have limited application.
We have identified three immune subgroups with distinct survival profiles indicating better survival in the presence of stronger immune responses. We also showed that these subgroups were stronger in terms of prognosis prediction than the 3-class identified in metastatic melanoma from TCGA, which we applied to our primary tumors from LMC. We recapitulated three immune subgroups in the TCGA metastatic melanomas, suggesting that similar immune infiltration and exclusion mechanisms span the whole spectrum of disease progression.
All the immune scores were highly correlated with each other (including those known to play an immunosuppressive role), the majority being upregulated in the High Immune Subgroup. We did not observe increasing representation of immunosuppressive cells e.g. Tregs, nor a relative increase in expression of checkpoint molecules in the Low Immune Subgroup. Rather, our data suggest a coordination of the immune cell populations as a whole. This is entirely in keeping with previously published observations of increased Treg numbers and accompanying expression of checkpoint molecules as a results of homeostatic mechanisms driven by melanoma infiltrating CD8+ T cells [34]. We cannot however exclude the possibility that the inference of immune cell subgroup infiltration from transcriptomic datasets may be insensitive to subtle variations that nevertheless might have an impact on immune function.
The protein-protein network analysis in Reactome FIViz of genes upregulated in the Low Immune Subgroup revealed enrichment for genes in cell proliferative pathways with MYC as the major node (the gene with the highest centrality). MYC is a pro-proliferative oncogene which has in recent years been reported to have various immunosuppressive functions [27][35][36][37], and to have specific involvement in melanoma metastasis and invasiveness [38]. However, the relation of MYC and immune response within melanoma is unclear.
In our study we were able to show evidence for a negative relationship between MYC and antigen processing and presentation machinery especially with HLA-B in tumors and patient derived melanoma cell lines. An inverse relationship between HLA class I genes and MYC expression has previously been reported [39] in melanoma cell lines. Moreover, it was described that MYC down-regulates the expression of HLA-B by directly binding to its proximal promoter [40]. Our data therefore provide strong evidence that MYC contributes significantly to immune evasion in primary melanoma making it a therapeutic target, notwithstanding the difficulties of achieving this [41]. The requirement for MYC in T cells suggests that a more targeted approach may be required, or that regulators/effectors of MYC activity might prove more appropriate targets [41].
NFKB1 was the network hub gene in the High Immune Subgroup. A number of important NF-κB family genes (NFKB1, NFKB2, c-REL, RELB) were also upregulated in this group suggesting activation of the pathway. RELA was stable across the immune subgroups, reflecting its constitutive expression in different tissues. IHC staining showed that tumor and TILs nuclear localization of NF-κB significantly correlated with lymphocytic infiltration, suggesting a reciprocal NF-κB-driven phenotype generated between the tumor and its immediate microenvironment, as described in other cancers [42][43]. Conversely in the Low Immune Subgroup we found loss of genes important in NF-κB and IFN-γ signaling, resulting in decreased gene expression. JAK2 mutations have recently been reported to be involved in acquired and primary resistance to anti PD-1 therapy [44][45]. Our hypothesis therefore is that a significant proportion of melanoma tumors in the Low Immune Subgroup may have primary resistance to this therapy even in adjuvant usage.
In our study, we report for the first time the association of smoking with immune responses to primary melanoma. Our results implied that smoking had an adverse effect on outcome by reducing the protective value of immune infiltration. That there were no obvious transcriptomic differences between melanomas in smokers and non-smokers may however suggest that the immune infiltrate in smokers may simply represent non-specific systemic inflammation or even that we see similar transcriptomic signals from pro-tumorigenic (cigarette driven) and anti-tumour immune responses.
We did observe a positive correlation between smoking and the expression of the GPR15 gene which codes for a chemo-attractant receptor which is regarded as a biomarker of smoking known to be hypo-methylated and hence overexpressed in circulating immune cells in smokers [46][29]. The GPR15 protein is reported to play a role in the trafficking of T cells [47][48], but its full biological function and significance with respect to smoking is still unknown. The overall pattern of association between reported smoking and death from melanoma however reinforces the view that discontinuation of smoking should be strongly recommended in melanoma patients. As it is not known whether the adverse effects of smoking in melanoma are mediated by nicotine or other components of cigarettes, the recommendation should probably be to avoid vaping [49], despite the acknowledged difficulty of smoking cessation for many.
In conclusion we report the use of bioinformatics to define broad prognostic immunophenotypes of primary melanoma, with evidence of a prominent role of NF-κB and IFN-γ signaling downregulation (including by deletion) and MYC overexpression (including amplification) in driving immunosuppression. We report evidence that a key mechanism in this process is perturbation of the antigen presentation machinery and that smoking predicted significantly worse melanoma specific survival in patients with evidence for a stronger immune responses.
Supplementary Material
Acknowledgements
We are thankful to Professor Ulf Klein for providing knowledge and insight into NF-κB signalling. We are grateful to all the participants whose samples have been used in this study. This work was funded by Cancer Research UK C588/A19167, C8216/A6129, and C588/A10721 and NIH CA83115. J.P., J.M.D, and S.M. were funded by Horizon 2020 Research and Innovation Programme no. 641458 (MELGEN).
Footnotes
Declaration of interests
The authors declare no potential conflict of interest.
References
- [1].Clark WH, Elder DE, Guerry D, Braitman LE, Trock BJ, Schultz D, et al. Model Predicting Survival in Stage I Melanoma Based on Tumor Progression. JNCI J Natl Cancer Inst. 1989;81:1893–904. doi: 10.1093/jnci/81.24.1893. [DOI] [PubMed] [Google Scholar]
- [2].Day CL, Sober AJ, Kopf AW, Lew RA, Mihm MC, Golomb FM, et al. A prognostic model for clinical stage I melanoma of the trunk. Am J Surg. 1981;142:247–51. doi: 10.1016/0002-9610(81)90286-5. [DOI] [PubMed] [Google Scholar]
- [3].Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Cowey CL, Lao CD, et al. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N Engl J Med. 2015;373:23–34. doi: 10.1056/NEJMoa1504030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, et al. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell. 2017;168:542. doi: 10.1016/j.cell.2017.01.010. [DOI] [PubMed] [Google Scholar]
- [5].Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. Genetic Basis for Clinical Response to CTLA-4 Blockade in Melanoma. N Engl J Med. 2014;371:2189–99. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, et al. The Immune Landscape of Cancer. Immunity. 2018;48:812–830.e14. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Charoentong P, Finotello F, Angelova M, Mayer C. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. BioRxiv. 2016 doi: 10.1101/056101. 056101. [DOI] [PubMed] [Google Scholar]
- [8].Newton-Bishop JA, Beswick S, Randerson-Moor J, Chang YM, Affleck P, Elliott F, et al. Serum 25-hydroxyvitamin D3 levels are associated with Breslow thickness at presentation and survival from melanoma. J Clin Oncol. 2009;27:5439–44. doi: 10.1200/JCO.2009.22.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Newton-Bishop JA, Davies JR, Latheef F, Randerson-Moor J, Chan M, Gascoyne J, et al. 25-Hydroxyvitamin D2/D3 levels and factors associated with systemic inflammation and melanoma survival in the Leeds Melanoma Cohort. Int J Cancer. 2015;136:2890–9. doi: 10.1002/ijc.29334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–95. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- [11].Nsengimana J, Laye J, Filia A, O’Shea S, Muralidhar S, Poźniak J, et al. β–Catenin-mediated immune evasion pathway frequently operates in primary cutaneous melanomas. J Clin Invest. 2018;128:2048–63. doi: 10.1172/JCI95351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Angelova M, Charoentong P, Hackl H, Fischer ML, Snajder R, Krogsdam AM, et al. Characterization of the immunophenotypes and antigenomes of colorectal cancers reveals distinct tumor escape mechanisms and novel targets for immunotherapy. Genome Biol. 2015;16:64. doi: 10.1186/s13059-015-0620-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541:321–30. doi: 10.1038/nature21349. [DOI] [PubMed] [Google Scholar]
- [14].Jewell R, Conway C, Mitra A, Randerson-Moor J, Lobo S, Nsengimana J, et al. Patterns of expression of DNA repair genes and relapse from melanoma. Clin Cancer Res. 2010;16:5211–21. doi: 10.1158/1078-0432.CCR-10-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Nsengimana J, Laye J, Filia A, Walker C, Jewell R, Van den Oord JJ, et al. Independent replication of a melanoma subtype gene signature and evaluation of its prognostic value and biological correlates in a population cohort. Oncotarget. 2015;6:11683–93. doi: 10.18632/oncotarget.3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Monti S, Tamayo P, Mesirov J, Golub T. Consensus clustering: A resampling based method for class discovery and visualization of gene expression microarray data. Mach Learn. 2003;52:91–118. [Google Scholar]
- [17].Wilkerson MD, Hayes DN. ConsensusClusterPlus: A class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–3. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wu G, Feng X, Stein L, Hanahan D, Weinberg R, Vogelstein B, et al. A human functional protein interaction network and its application to cancer data analysis. Genome Biol. 2010;11:R53. doi: 10.1186/gb-2010-11-5-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Scardoni G, Petterlini M, Laudanna C. Analyzing biological network parameters with CentiScaPe. Bioinformatics. 2009;25:2857–9. doi: 10.1093/bioinformatics/btp517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Shannon P. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bastian M, Heymann S, Jacomy M. Gephi: An Open Source Software for Exploring and Manipulating Networks. Third Int AAAI Conf Weblogs Soc Media; 2009. pp. 361–2. [DOI] [Google Scholar]
- [22].Raaijmakers MIG, Widmer DS, Maudrich M, Koch T, Langer A, Flace A, et al. A new live-cell biobank workflow efficiently recovers heterogeneous melanoma cells from native biopsies. Exp Dermatol. 2015;24:377–80. doi: 10.1111/exd.12683. [DOI] [PubMed] [Google Scholar]
- [23].Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–9. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- [24].Townsend P. Deprivation *. 2018:125–46. [Google Scholar]
- [25].Masten BJ, Olson GK, Tarleton CA, Rund C, Schuyler M, Mehran R, et al. Characterization of Myeloid and Plasmacytoid Dendritic Cells in Human Lung. J Immunol. 2006;177:7784–93. doi: 10.4049/jimmunol.177.11.7784. [DOI] [PubMed] [Google Scholar]
- [26].Fening K, Parekh V, McKay K. CD123 immunohistochemistry for plasmacytoid dendritic cells is useful in the diagnosis of scarring alopecia. J Cutan Pathol. 2016;43:643–8. doi: 10.1111/cup.12725. [DOI] [PubMed] [Google Scholar]
- [27].Casey SC, Tong L, Li Y, Do R, Walz S, Fitzgerald KN, et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science (80-) 2016;352:227–31. doi: 10.1126/science.aac9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Asgari MM, Shen L, Sokil MM, Yeh I, Jorgenson E. Prognostic factors and survival in acral lentiginous melanoma. Br J Dermatol. 2017;177:428–35. doi: 10.1111/bjd.15600. [DOI] [PubMed] [Google Scholar]
- [29].Bauer M, Fink B, Thürmann L, Eszlinger M, Herberth G, Lehmann I. Tobacco smoking differently influences cell types of the innate and adaptive immune system—indications from CpG site methylation. Clin Epigenetics. 2016;8:83. doi: 10.1186/s13148-016-0249-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kõks S, Kõks G. Activation of GPR15 and its involvement in the biological effects of smoking. Exp Biol Med. 2017;242:1207–12. doi: 10.1177/1535370217703977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wolchok JD, Chiarion-Sileni V, Gonzalez R, Rutkowski P, Grob J-J, Cowey CL, et al. Overall Survival with Combined Nivolumab and Ipilimumab in Advanced Melanoma. N Engl J Med. 2017;377:1345–56. doi: 10.1056/NEJMoa1709684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–7. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Becht E, Giraldo NA, Lacroix L, Buttard B, Elarouci N, Petitprez F, et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17:218. doi: 10.1186/s13059-016-1070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Spranger S, Spaapen RM, Zha Y, Williams J, Meng Y, Ha TT, et al. Up-Regulation of PD-L1, IDO, and Tregs in the Melanoma Tumor Microenvironment Is Driven by CD8+ T Cells. Sci Transl Med. 2013;5:200ra116–200ra116. doi: 10.1126/scitranslmed.3006504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kortlever RM, Sodir NM, Wilson CH, Burkhart DL, Pellegrinet L, Brown Swigart L, et al. Myc Cooperates with Ras by Programming Inflammation and Immune Suppression. Cell. 2017;171:1301–1315.e14. doi: 10.1016/j.cell.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Topper MJ, Vaz M, Chiappinelli KB, DeStefano Shields CE, Niknafs N, Yen R-WC, et al. Epigenetic Therapy Ties MYC Depletion to Reversing Immune Evasion and Treating Lung Cancer. Cell. 2017;171:1284–1300.e21. doi: 10.1016/j.cell.2017.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Schaub FX, Dhankani V, Berger AC, Trivedi M, Richardson AB, Shaw R, et al. Pan-cancer Alterations of the MYC Oncogene and Its Proximal Network across the Cancer Genome Atlas. Cell Syst. 2018:282–300. doi: 10.1016/j.cels.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Schlagbauer-Wadl H, Griffioen M, van Elsas A, Schrier PI, Pustelnik T, Eichler HG, et al. Influence of increased c-Myc expression on the growth characteristics of human melanoma. J Invest Dermatol. 1999;112:332–6. doi: 10.1046/j.1523-1747.1999.00506.x. [DOI] [PubMed] [Google Scholar]
- [39].Versteeg R, Noordermeer IA, Krüse-Wolters M, Ruiter DJ, Schrier PI. c-myc down-regulates class I HLA expression in human melanomas. EMBO J. 1988;7:1023–9. doi: 10.1002/j.1460-2075.1988.tb02909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Peltenburg LTC, Schrier PI. Transcriptional suppression of HLA-B expression by c-Myc is mediated through the core promoter elements. Immunogenetics. 1994;40:54–61. doi: 10.1007/BF00163964. [DOI] [PubMed] [Google Scholar]
- [41].Chen H, Liu H, Qing G. Targeting oncogenic Myc as a strategy for cancer treatment. Signal Transduct Target Ther. 2018;3:5. doi: 10.1038/s41392-018-0008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Hopewell EL, Zhao W, Fulp WJ, Bronk CC, Lopez AS, Massengill M, et al. Lung tumor NF-kB signaling promotes T cell–mediated immune surveillance. J Clin Invest. 2013;123:2509–22. doi: 10.1172/JCI67250DS1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Muthuswamy R, Berk E, Junecko BF, Zeh HJ, Zureikat AH, Normolle D, et al. NF-B Hyperactivation in Tumor Tissues Allows Tumor-Selective Reprogramming of the Chemokine Microenvironment to Enhance the Recruitment of Cytolytic T Effector Cells. Cancer Res. 2012;72:3735–43. doi: 10.1158/0008-5472.CAN-11-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N Engl J Med. 2016;375:819–29. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Shin DS, Zaretsky JM, Escuin-Ordinas H, Garcia-Diaz A, Hu-Lieskovan S, Kalbasi A, et al. Primary Resistance to PD-1 Blockade Mediated by JAK1/2 Mutations. Cancer Discov. 2017;7:188–201. doi: 10.1158/2159-8290.CD-16-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Bauer M, Linsel G, Fink B, Offenberg K, Hahn AM, Sack U, et al. A varying T cell subtype explains apparent tobacco smoking induced single CpG hypomethylation in whole blood. Clin Epigenetics. 2015;7:1–11. doi: 10.1186/s13148-015-0113-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Bilsborough J, Viney JL. GPR15: a tale of two species. Nat Immunol. 2015;16:137–9. doi: 10.1038/ni.3084. [DOI] [PubMed] [Google Scholar]
- [48].Lahl K, Sweere J, Pan J, Butcher E. Orphan chemoattractant receptor GPR15 mediates dendritic epidermal T-cell recruitment to the skin. Eur J Immunol. 2014;44:2577–81. doi: 10.1002/eji.201444628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Martin EM, Clapp PW, Rebuli ME, Pawlak EA, Glista-Baker E, Benowitz NL, et al. E-cigarette use results in suppression of immune and inflammatory-response genes in nasal epithelial cells similar to cigarette smoke. Am J Physiol Cell Mol Physiol. 2016;311:L135–44. doi: 10.1152/ajplung.00170.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.