Abstract
Antimicrobial peptides are promising alternative antimicrobial agents. However, little is known about whether resistance to small-molecule antibiotics leads to cross-resistance (decreased sensitivity) or collateral sensitivity (increased sensitivity) to antimicrobial peptides. We systematically addressed this question by studying the susceptibilities of a comprehensive set of 60 antibiotic-resistant Escherichia coli strains towards 24 antimicrobial peptides. Strikingly, antibiotic-resistant bacteria show a high frequency of collateral sensitivity to antimicrobial peptides, whereas cross-resistance is relatively rare. We identify clinically relevant multidrug-resistance mutations that increase bacterial sensitivity to antimicrobial peptides. Collateral sensitivity in multidrug-resistant bacteria arises partly through regulatory changes shaping the lipopolysaccharide (LPS) composition of the bacterial outer membrane. These advances allow the identification of antimicrobial peptide-antibiotic combinations that enhance antibiotic activity against multidrug-resistant bacteria and slow down de novo evolution of resistance. In particular, when co-administered as an adjuvant, the antimicrobial peptide PGLA caused up to 30-fold decrease in the antibiotic resistance level of resistant bacteria. Our work provides guidelines for the development of efficient peptide-based therapies of antibiotic-resistant infections.
Introduction
Evolution of resistance towards antibiotics, or any other drug, can simultaneously increase (cross-resistance) or decrease (collateral sensitivity) fitness to multiple other drugs1–5. The molecular mechanisms driving cross-resistance are well-described2,4,5. In contrast, it remains unclear how frequently genetic adaptation to a single drug increases bacterial sensitivity to other drugs and what the underlying molecular mechanisms of collateral sensitivity are. The issue is important as collateral sensitivity could direct future multidrug therapeutic strategies6. However, such strategies are limited by the scarcity of available drug-pairs showing collateral sensitivity that could also be used in clinical settings. Clearly, the concept of collateral sensitivity needs to be expanded by studying a broader scope of antimicrobial agents.
Here we systematically study the effect of antibiotic-resistance mechanisms on susceptibility to antimicrobial peptides, a promising class of new antibacterial compounds. Antimicrobial peptides are short peptides with a broad spectrum of antibacterial activities7. Such peptides are found among all classes of life, and are part of the defense mechanisms against microbial pathogens7. Because antimicrobial peptides have diverse chemical features and cellular targets, they are promising antibacterial agents8,9. However, the degree of similarity between the resistance mechanisms to peptides and to small-molecule antibiotics remains disputed10. This is relevant as some peptides have now reached advanced stages in clinical trials8.
We had previously initiated laboratory evolutionary experiments starting with a single clone of Escherichia coli K122,3. Parallel evolving populations were exposed to gradually increasing concentrations of one of 12 clinically relevant antibiotics (Supplementary Table 1), leading to up to 328-fold increases in their minimum inhibitory concentrations (MICs) relative to the wild-type2. The resistance levels were equal to or above the EUCAST clinical breakpoints, and 52% of the antibiotic-resistant strains showed resistance to multiple antibiotics2. Here, we focus on a representative set of 60 antibiotic-resistant strains (five strains per antibiotic) that have been subjected to whole-genome sequence analysis in order to identify the molecular mechanisms underlying antibiotic resistance2. Many of the observed mutations have also been detected in clinical drug-resistant isolates, and are known to target cellular systems involved in inner-and outer membrane transport and permeability (e.g. efflux pumps, porins), and cell envelope biogenesis2.
We hypothesized that such membrane-altering antibiotic-resistance mutations not only influence susceptibility to other antibiotics, but to antimicrobial peptides as well. Why should it be so? The action of most antimicrobial peptides relies on the interaction between the positively charged peptide and the negatively charged membrane components, for two reasons. First, the structural and physicochemical properties of antimicrobial peptides, (e.g. net positive charge, hydropathicity) and their capacity to adapt an amphipathic conformation upon membrane binding influence this interaction8,11,12. Second, the insertion of antimicrobial peptides into the hydrophobic core of the membrane depends on the general properties of the bacterial outer membrane11,12. For instance, lipopolysaccharide (LPS) composition has a major impact on the killing efficiency of cationic antimicrobial peptides13. For these reasons, it is plausible that membrane-affecting antibiotic-resistance mutations shape genetic susceptibility to antimicrobial peptides. However, no systematic study has been devoted to address this problem in detail.
Here we first measured the susceptibility of the 60 antibiotic-resistant strains2,3 against a set of 24 antimicrobial peptides of diverse origin and various modes of action. We found widespread bacterial collateral sensitivity towards antimicrobial peptides. Analysis of the mutational and transcriptome profiles of the antibiotic-resistant strains revealed that antibiotic-resistance mutations increase sensitivity to peptides via regulatory changes in lipopolysaccharide biosynthesis. The consequent alteration in the surface charge presumably strengthens the interaction of cationic antimicrobial peptides with the outer membrane, and thus enhances the killing efficiency of these peptides. Finally, we demonstrated that the antimicrobial peptide PGLA, when co-administered as an adjuvant, restores antibiotic activity against resistant bacteria and slows down antibiotic-resistance evolution.
Results
Widespread collateral sensitivity to antimicrobial peptides
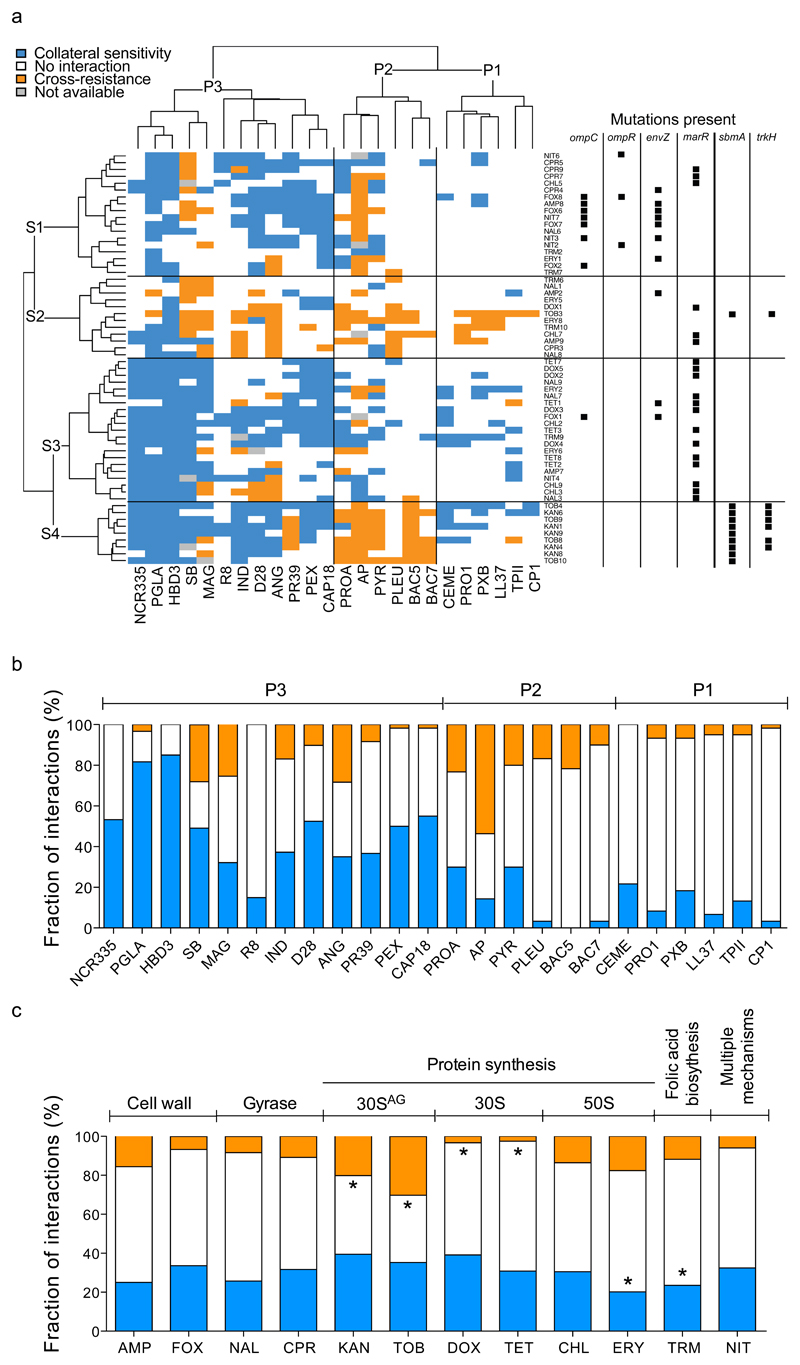
To study whether antibiotic resistance in E. coli leads to cross-resistance or collateral sensitivity towards antimicrobial peptides, we measured the changes in the susceptibilities of 60 antibiotic-resistant strains to a set of 24 peptides. Peptides were chosen based on the following criteria: diverse sources (synthetic/natural), different putative mechanisms of action, structural diversity, and clinical relevance (Supplementary Table 2). The obtained results allowed us to chart the map of cross-resistance/collateral sensitivity of the antibiotic-resistant strains towards the 24 antimicrobial peptides, and identify several general patterns (Figure 1A, Supplementary Table 3).
Figure 1. Susceptibility profiles of 60 laboratory-evolved antibiotic-resistant E. coli strains.
a, Hierarchical clustering of 60 antibiotic-resistant strains (rows) and a set of 24 antimicrobial peptides (columns) based on the cross-resistance and collateral sensitivity interactions between them (for abbreviations of antibiotics and antimicrobial peptides see Supplementary Tables 1 and 2, respectively). Hierarchical clustering was performed separately on rows and columns, using Ward’s method67. Black squares on the right side of each antibiotic-resistant strain denote previously identified mutations in antibiotic-resistance genes2,3 that were significantly enriched in one or more strain clusters (p<0.05, two-sided Fisher’s exact test). S1 strains were enriched in envZ, ompR and ompC mutations, whereas S3 strains were enriched in marR mutations (P<0.05 for all cases, two-sided Fisher’s exact test). While S3 strains show widespread collateral sensitivity to antimicrobial peptides, especially to P1 and P3 peptides, aminoglycoside-resistant strains (S4) show extensive cross-resistance to proline-rich peptides (P2) (p<0.0001, two-sided Fisher’s exact test). b, Efficiency of antimicrobial peptides against antibiotic-resistant bacteria expressed as the percentage of strains showing collateral sensitivity (blue), no interaction (white), or cross-resistance (orange) against each peptide. A total of 56 to 60 strains per antimicrobial peptide was employed for the analysis. c, Relative frequency of collateral sensitivity and cross-resistance interactions towards antimicrobial peptides upon adaptation to single antibiotics (n=5 strains per antibiotic). The frequency of interactions for each peptide was calculated by counting the number of cross-resistance (orange), no interaction (white) and collateral sensitivity (blue) interactions displayed by all strains adapted to a given antibiotic. Antibiotic modes-of-action are shown on the top of the figure. 30SAG refers to aminoglycosides. Asterisks (*) mark significant deviations from hypergeometric distribution models calculated from all the interactions of all peptide-strain combinations separately for cross-resistance and collateral sensitivity, respectively. Strains adapted to DOX and TET were depleted, whereas strains adapted to TOB and KAN were enriched in cross-resistance interactions towards peptides (p=0.005, p<0.008, p=0.003 and p<0.001, respectively, two-sided Fisher's combined probability test). Furthermore, strains adapted to ERY and TRM were significantly depleted in collateral sensitivity interactions (p=0.003 and p<0.001 respectively, two-sided Fisher's combined probability test).
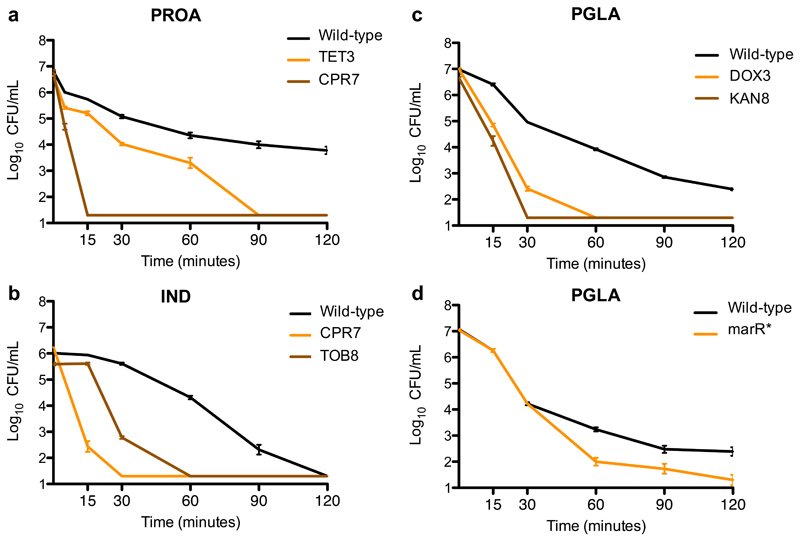
First, cross-resistance to antimicrobial peptides was relatively rare: only 12% of all possible antibiotic-resistant strain and peptide pairs showed cross-resistance, whereas 31% showed collateral sensitivity. The observed strength of cross-resistance and collateral sensitivity was usually a 2-fold change in MIC relative to the wild-type (Supplementary Figure 1, Supplementary Table 4). In typical clinical settings, drug concentrations well above the wild-type MIC are applied14. Therefore, we asked how collateral sensitivity shapes the killing kinetics when peptides were applied at a concentration 10-15 fold above the wild-type MIC. We focused on five resistant strains that showed collateral sensitivity to at least one of the peptides studied (Figure 2A-C). Upon antimicrobial peptide stress, collateral sensitive populations showed a far more rapid decline in size than the wild-type. For example, 15 minutes of protamine exposure nearly completely eradicated ciprofloxacin-resistant bacteria, whereas the size of the wild-type population declined only by 10-fold (Figure 2A).
Figure 2. Survival of collateral-sensitive antibiotic-resistant strains under lethal antimicrobial peptide stress.
a-c, The wild-type and antibiotic-resistant strains were exposed to high concentrations of antimicrobial peptides: (a) the tetracycline-resistant TET3 and the ciprofloxacin-resistant CPR7 strains were exposed to 15-fold (3000 μg/mL) MIC of protamine (PROA), (b) the ciprofloxacin-resistant CPR7 and the tobramycin-resistant TOB8 were exposed to 10-fold MIC (125 μg/mL) of indolicidin (IND), and (c) the doxycycline-resistant DOX3 and the kanamycin-resistant KAN8 were exposed to 15-fold MIC (1500 μg/mL) of PGLA. All antibiotic-resistant strains exhibited collateral sensitivity towards the applied peptide. d, A strain containing a single point mutation in marR (marR*) was also exposed to 15-fold MIC of PGLA. This strain exhibits resistance to multiple antibiotics and collateral sensitivity to many of the peptides tested, including PGLA (Table 1). Cells were incubated with the particular peptide for 120 minutes. Samples were taken at defined time points and plated in LB agar plates. Percentage of survival was calculated by counting the colony forming units (CFU). Each data point shows the mean ± standard error of the mean of 3 biological replicates.
Second, collateral sensitivity and cross-resistance patterns clustered the antimicrobial peptides into three main groups (Figure 1) with major differences in their physicochemical properties and modes of action. Antimicrobial peptides belonging to the P1 and P3 groups generally insert themselves into membrane bilayers to form pores and thus induce cell lysis (i.e. pore formers). However, these two groups differ both in their physicochemical properties and their interactions with antibiotic-resistant strains: P1 peptides have lower isoelectric point and hydropathicity index compared to P3 peptides (see Supplementary Figure 2) and are depleted in cross-resistance and collateral sensitivity interactions, whereas collateral sensitivity towards P3 peptides is prevalent (Figure 1A, Supplementary Table 5).
On the contrary, members of the P2 group typically penetrate into the cell and have intracellular targets (Supplementary Table 2). Unlike P1 and P3, the P2 group consists of proline-rich peptides which are characterized by unstable secondary peptide structure, high propensity for aggregation in aqueous solutions, and relatively low aliphatic and hydropathicity indices (Supplementary Figure 3, Supplementary Table 6). Notably, cross-resistance towards P2 peptides is frequent compared to the other peptide groups (Figure 1A, Supplementary Table 5).
Third, not all peptides were equally effective against antibiotic-resistant bacteria (Figure 1B). Most notably, 82% of the antibiotic-resistant strains showed collateral sensitivity to the peptide glycine-leucine-amide (PGLA), a member of the magainin family15 (Figure 1C). Consistent with their conserved evolutionary roles, human peptides, such as beta-defensin HBD3 and cathlecidin LL37, also showed few, if any, cross-resistance interactions (Figure 1B, Supplementary Table 3). In contrast, various antibiotic-resistant strains showed cross-resistance to the proline-rich peptide apidaecin IB (AP). This result could be of clinical relevance, as AP is currently being investigated for therapeutic usage16.
We next tested the evolutionary conservation of collateral sensitivity by measuring the susceptibility of clinically-derived E. coli strains towards 9 relevant antimicrobial peptides. Three clinical isolates were previously allowed to adapt to three different antibiotics (gentamicin, nalidixic acid and ampicillin) in the laboratory1. The peptide susceptibility profiles of these antibiotic-resistant strains showed good agreement with those presented in Figure 1, indicating that collateral sensitivity is partly conserved across multiple genetic backgrounds (Supplementary Table 7).
Aminoglycoside resistance induces cross-resistance to antimicrobial peptides
Whereas cross-resistance to antimicrobial peptides was generally much less common than collateral sensitivity, the distribution of cross-resistance interactions was far from random (Figure 1C). Specifically, strains resistant to aminoglycosides (tobramycin and kanamycin) showed cross-resistance to proline-rich peptides (P2 group, see Figure 1A). These strains are reminiscent of aminoglycoside-resistant small-colony variants observed in clinical settings, as they accumulated membrane potential altering mutations17. Moreover, they uniquely carried a loss-of function mutation in the gene encoding the inner-membrane transport protein sbmA2 (Figure 1A). sbmA is commonly mutated in response to aminoglycoside stress18, and is involved in the uptake of proline-rich peptides10,19.
Here we asked whether a loss-of-function mutation in sbmA contributes to the observed cross-resistance pattern of the aminoglycoside-resistant strains against P2 peptides. As expected, deletion of this gene in E. coli BW25113 and in the clinical isolate E. coli ATCC 25922 conferred mild, but significant resistance to both aminoglycosides and proline-rich peptides (Table 1, Supplementary Text 1, Supplementary Figure 4A-F). For a possible contribution of other mutations to cross-resistance between aminoglycosides and proline-rich peptides, see Supplementary Text 2.
Multidrug-resistance mutations confer collateral sensitivity to antimicrobial peptides
We next explored the mutations underlying collateral sensitivity. To this end, we first clustered the antibiotic-resistant strains based on their peptide susceptibility profiles and compiled the set of mutations shared among them (Figure 1A). This procedure revealed four main groups of resistant strains (S1-S4), each of them carrying distinct sets of mutations.
Strains belonging to group S1 were adapted to a range of different antibiotics, including cefoxitin (FOX), a cell-wall inhibitor, and nitrofurantoin (NIT) that targets several biochemical processes within the bacterial cell. However, they mostly exhibited collateral sensitivity to P3 peptides (Figure 1A), and accumulated mutations in a similar set of genes. Enrichment analysis revealed that they typically carry mutations in the EnvZ/OmpR two-component regulatory system, and in the outer membrane porin C (ompC)2; for details and statistics, see Figure 1A. The EnvZ/OmpR system, through regulating the outer membrane porin genes ompC and ompF, mediate bacterial defense against antimicrobials by reducing the uptake of hydrophilic antibiotics20,21. Consistent with a causal role in collateral sensitivity, inserting an ompC loss-of function mutation into wild-type E. coli conferred resistance to nitrofurantoin and cell-wall inhibitor antibiotics, and simultaneously increased sensitivity to multiple antimicrobial peptides (see Table 1 and Supplementary Text 3 for a proposed mechanism).
We further identified a group of strains (S3, see Figure 1A) that typically showed collateral sensitivity to pore-forming peptides (clusters P1 and P3). These strains carry mutations in marR, a transcriptional repressor of antibiotic stress response22 (Figure 1A). MarR represses the mar regulon that controls genes involved in membrane permeability and efflux of toxic chemicals23. Elevated expression of the mar regulon is frequently found in multidrug-resistant clinical isolates23. Insertion of a recurrently observed marR mutation2 either into wild-type E. coli BW25113 or into the antibiotic-sensitive clinical isolate E. coli ATCC 25922 conferred mild, but significant resistance to several antibiotics, and simultaneously enhanced sensitivity to P3 antimicrobial peptides (Table 1, Figure 2D, Supplementary Text 1, Supplementary Figure 4G-J).
Overall, these findings demonstrate that collateral sensitivity to antimicrobial peptides is induced by multidrug-resistant mutations that appeared repeatedly in response to various antibiotic stresses (for further examples, see Table 1).
Gene expression changes in lipopolysaccharide biosynthesis contribute to collateral sensitivity
We next hypothesized that changes in the outer membrane composition of antibiotic-resistant bacteria underlie the observed collateral sensitivity to antimicrobial peptides.
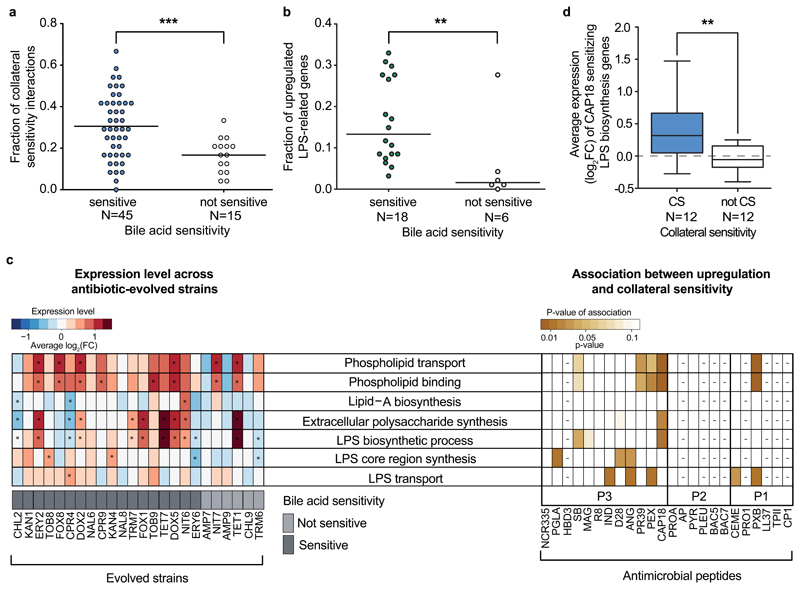
To test this possibility, we started by determining the sensitivity of the antibiotic-resistant strains to bile acid, a membrane-damaging agent. As expected, 75% of the antibiotic-resistant strains were more susceptible to bile acid than the wild-type, and the bile-acid sensitive strains showed collateral sensitivity to an especially high number of antimicrobial peptides (Figure 3A). To explore the common mechanistic basis of this pattern, we compared the transcriptome profiles of the wild-type and 24 antibiotic-resistant strains covering all investigated 12 antibiotics (see Materials and Methods). All strains were grown in antibiotic-free medium to ensure comparability between strains that display vastly different resistance levels.
Figure 3. Altered membrane composition in antibiotic-resistant bacteria contributes to increased sensitivity to antimicrobial peptides.
a, Antibiotic-resistant strains sensitive towards a membrane-damaging agent (bile acid) show especially large numbers of collateral sensitivity interactions to antimicrobial peptides. For antibiotic abbreviations see Supplementary Table 1. Strains sensitive to bile acid show significantly more collateral sensitivity interactions to peptides than those showing wild-type bile acid sensitivity (not sensitive group) (p<10-3, two-sided generalized linear mixed model with binomial response distributions, see Materials and Methods section). Relative frequency was calculated by dividing the number of the collateral sensitivity interactions by the number of all tested peptides (N=24). b, Resistant strains with sensitivity to bile acid have significantly more LPS-related synthesis genes being transcriptionally upregulated than those showing wild-type bile acid sensitivity (p<10-3, two-sided GLMM with binomial response distributions). Relative frequency was calculated by dividing the number of the upregulated genes by the number of all LPS-related genes (N=100). c, Left heatmap shows the average log2(fold change) of genes related to selected membrane-associated GO processes. Many antibiotic-resistant strains are enriched in significantly up- or downregulated genes (fold change>2 or <0.5, FDR-corrected p-value<0.05, two-sided Fisher’s exact test), associated with membrane-related functions. Significant enrichments (p<0.05) are marked with an asterisk (*). Strains sensitive to a given peptide show significant upregulations in specific GO groups compared to non-sensitive strains (right heatmap, two-sided Student’s t–test; for further details, see Supplementary Figure 5). Peptides with either too few or too many collateral sensitivity interactions (n<4 or n>21, respectively) were excluded from the statistical analysis based on sample size calculation with alpha=0.1, power=0.8, delta=2, SD=1, and are indicated with a minus sign (-). Sample size used in this analysis is provided in Supplementary Table 3 d, Upregulation of LPS-related genes sensitize to CAP18. CAP18-sensitive antibiotic-resistant strains (CS, n=12) have significantly higher expression levels of CAP18 sensitizing genes within the ‘LPS biosynthetic process’ GO category than non-sensitive strains (not CS, n=12) (p=0.008, two-sided Wilcoxon rank-sum test). Boxplots show the median, first and third quartiles, with whiskers showing the 5th and 95th percentile. Significant difference (p<0.01) is marked with asterisks (**).
Several lines of evidence support the hypothesis that antibiotic resistance leads to alterations in outer membrane composition. In 75% of the analyzed antibiotic-resistant strains, transcriptional changes are significantly enriched in LPS- and outer membrane-related genes (Figure 3C, left panel), including genes involved in LPS biosynthesis, and phospholipid binding and transfer. Importantly, strains with an especially high number of upregulated LPS-related genes (Supplementary Table 8 and Supplementary Table 9) display an enhanced susceptibility to bile acid (Figure 3B). Finally, strains with upregulated genes in membrane-related functions were more likely to show collateral sensitivity to specific antimicrobial peptides (Figure 3C and Supplementary Figure 5).
Chemogenomic analysis of collateral sensitivity
To directly investigate the causality between upregulation of membrane-related genes and collateral sensitivity, we carried out a chemogenomic screen to identify genes that, when overexpressed, sensitize E. coli to the membrane-interacting 18-kDa cationic antimicrobial peptide (CAP18). CAP18 is an ideal choice for two reasons. First, it displaces divalent cations in the lipopolysaccharide layer and thereby permeabilizes the outer membrane24. Second, strains that are sensitive to CAP18 displayed upregulation of genes involved in LPS biosynthesis, and phospholipid binding and transport (Figure 3C, Supplementary Tables 8 and 9).
Chemogenomic screen of CAP18 was carried out by applying an established plasmid collection overexpressing all E. coli ORFs25 in a pooled fitness assay with a deep sequencing readout (for details see Supplementary Figure 6). Following growth of the pooled collection both in the presence and absence of CAP18, we identified genes that produce a growth defect when overexpressed in CAP18-treatment only (see Supplementary Figure 6C). Such growth defects indicate increased CAP18 sensitivity upon overexpression of single genes. As a control, we performed the same assay on CP1, a peptide to which antibiotic-resistant strains rarely show collateral sensitivity (Figure 1B). Our chemogenomic screen revealed 624 and 258 sensitizing genes for CAP18 and CP1, respectively (see Supplementary Table 10). As expected, LPS-related genes were highly enriched among genes that sensitize to CAP18, but not to CP1 when overexpressed (Supplementary Table 11). For example, LPS biosynthesis genes were especially likely to sensitize the bacteria to CAP18 when overexpressed (odds ratio=4.4, p=0.004, two-sided Fisher’s exact test). Furthermore, CAP18–sensitizing LPS biosynthesis genes, as inferred by chemogenomics, showed higher average expression levels in those antibiotic-resistant strains that are sensitive to CAP18 compared to the rest (p=0.008, two-sided Wilcoxon rank-sum test, Figure 3D). We note that, although also frequently upregulated in CAP18-sensitive strains (Figure 3C), genes with phospholipid-related functions were not enriched among sensitizing genes in the chemogenomic screen (Supplementary Table 11). Taken together, these analyses indicate that altered LPS biosynthesis plays a causal role in the widespread collateral sensitivity of antibiotic-resistant strains to antimicrobial peptides.
A marR mutation induces collateral sensitivity
We next deciphered a mechanistic link between a specific regulatory mutation and its impact on antibiotic resistance and collateral sensitivity. We focused on marR, as mutation in this gene affects bacterial response to antimicrobials in two opposite ways: it increases resistance to multiple antibiotics, but simultaneously sensitizes to membrane-interacting peptides (Table 1). Why is it so?
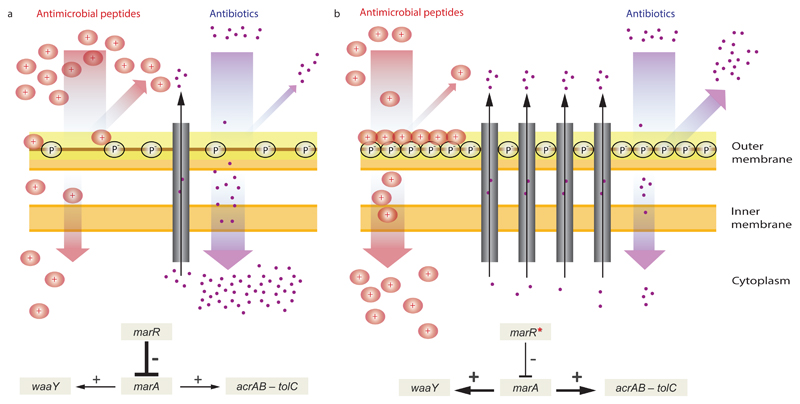
Lipopolysaccharide (LPS) is a major component of the bacterial outer membrane that stabilizes the membrane structure, regulates its permeability and contributes to its negative charge26. The mar regulon is known to mediate LPS modification by positively regulating WaaY, a kinase responsible for phosphorylation of the inner core of LPS, leading to an increased negative surface charge of the bacterial outer membrane27,28 (Figure 4). As an increased negative surface charge of the membrane generally promotes antimicrobial peptide killing efficiency9,27, upregulation of waaY is expected to enhance bacterial susceptibility to peptides.
Figure 4. A putative mechanism underlying collateral sensitivity of antibiotic-resistant bacteria to cationic antimicrobial peptides.
a, The wild-type MarR represses marA, which leads to the reduced expression of the AcrAB-TolC efflux pump and hence the cytosolic accumulation of antibiotics. b, Upon a canonical resistance mutation in the marR gene (marR*), repression of marA is substantially decreased leading to the upregulation of the AcrAB-TolC efflux pump, and hence an increased resistance to multiple antibiotics. On the other hand, MarA simultaneously promote the upregulation of WaaY, a kinase responsible for phosphorylation of the inner core of lipopolysaccharides (LPS), which increases the net negative surface charge of the bacterial outer membrane. This in turn enhances susceptibility to membrane-interacting cationic antimicrobial peptides (CAP+). In addition, phosphorylation of the LPS core may also enhance the permeability barrier of the outer membrane by cross-linking of neighboring LPS molecules. Such changes promote both 1) a decreased uptake and increased efflux of antibiotics, thereby contributing to antibiotic resistance, and 2) a higher affinity of the positively charged antimicrobial peptides to the negative phosphoryl groups of the LPS core, leading to enhanced sensitivity to antimicrobial peptides.
Based on these facts, we hypothesized that mutations in marR increase antimicrobial peptide susceptibility through upregulation of waaY. Such upregulation should lead to an increase in phosphorylation of LPS and, as a consequence, elevated negative surface charge of the bacterial outer membrane (Figure 4). Several lines of evidence support this hypothesis. First, the antimicrobial peptide susceptibility profiles of the marR single-mutant and the waaY-overexpressing strain showed substantial overlap (Table 1). Second, qRT-PCR analysis confirmed that waaY is nearly 4-fold upregulated in the marR single-mutant strain when compared to the wild-type (fold-change: 3.88 ± 0.026 SEM, see Materials and Methods). Third, we carried out zeta potential measurements and demonstrated that both the marR mutant and the waaY-overexpressing strain display relatively high negative surface charge compared to the wild-type (Supplementary Figure 7). Finally, inactivation of waaY abolished collateral sensitivity to P3 peptides in marR mutants (Supplementary Figure 8). To summarize, these results indicate that collateral sensitivity of the marR mutant to peptides occurs via modulation of the LPS phosphorylation pathway. We propose that the consequent altered outer membrane composition facilitates the interaction of antimicrobial peptides with the cell membrane and thereby enhances their killing efficiency (Figure 4).
Peptide PGLA restores antibiotic activity in antibiotic-resistant bacteria
The above results have important implications for future development of drug combinations. Recently, two complementary mechanisms were proposed to contribute to the selective eradication of drug-resistant bacteria29. First, resistance to one drug can induce hypersensitivity to the other (collateral sensitivity, see also Supplementary Text 4)1,2; and second, interactions can become more synergistic with the evolution of resistance, making the mutant more sensitive to the combination (resistance mutation-induced synergy)30.
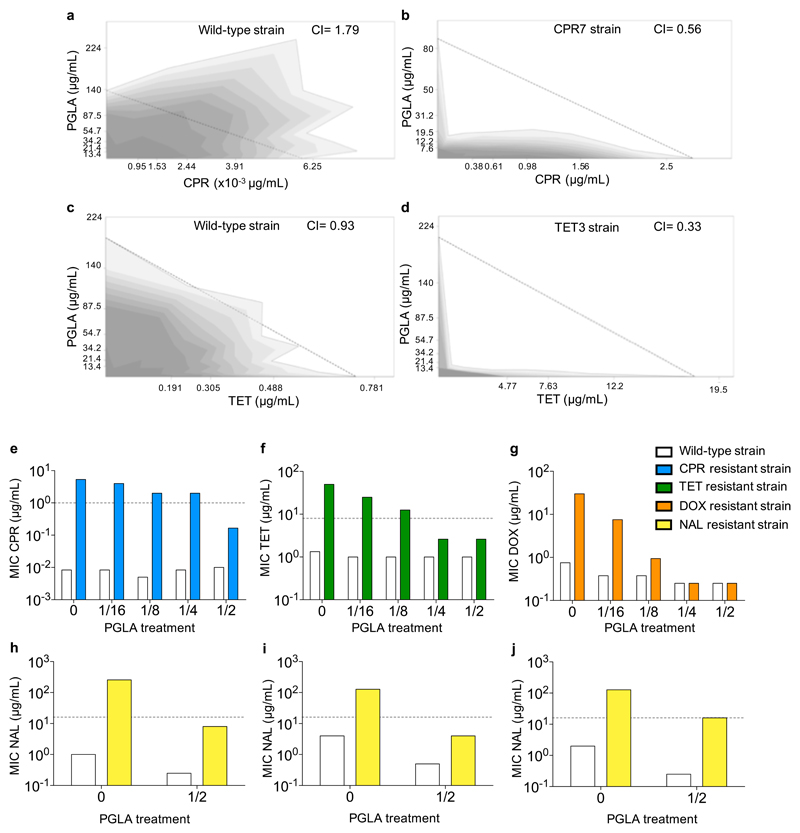
We focused on PGLA for two reasons. First, it shows an exceptionally high number of collateral sensitivity interactions (Figure 1). Second, when used in combination, antibiotic-PGLA pairs show strong synergism in antibiotic-resistant, but not in the corresponding wild-type bacteria (Figure 5A-D, Supplementary Table 12). Remarkably, strong synergism is prevalent in antibiotic-resistant strains carrying mutations in marR, envZ or ompF. As these genes influence membrane permeability, we speculate that PGLA may interfere with the activity of multidrug efflux systems and/or porins responsible for the observed antibiotic-resistance in these strains.
Figure 5. Interaction of PGLA and antibiotics, when applied in combination.
Antibiotic-PGLA interactions were determined in E. coli K12 BW25133 wild-type and corresponding antibiotic-resistant strains. For antibiotic abbreviations see Supplementary Table 1. Figures a-d show the combination effect of PGLA and ciprofloxacin (CPR) or tetracycline (TET) on the wild-type strain (a and c), ciprofloxacin-resistant strain (CPR7) (b) and tetracycline-resistant strain (TET3) (d). While the combination shows strong antagonism (a) or no interaction (c) in the wild-type strain, the interaction shifted to strong synergism in the resistant strain (b and d). Dashed line represents no interaction calculated based on the Loewe additivity model (see Materials and Methods). Growth rate is represented in the combination space by the shade of the grey color with darker shades denoting higher growth rates Figures e-j show the effect of subinhibitory concentrations of PGLA on antibiotic activity. Ciprofloxacin-resistant CPR7 (e), tetracycline-resistant TET3 (f) and doxycycline-resistant DOX3 (g) strains, derived from E. coli K12 BW25133 were treated with subinhibitory concentrations of PGLA, while measuring the MIC for the given antibiotic to which they were adapted. The concentrations of PGLA used were 1/16, 1/8, 1/4 and 1/2 of its MIC against the wild-type strain. The minimal inhibitory concentration of nalidixic acid (NAL) was measured in E. coli clinical isolates 0370 (h), 3539 (i) and CFT073 (j), and their corresponding nalidixic acid-resistant strains in the presence of 1/2 of the MIC for PGLA. None of the PGLA concentrations, when applied alone, affected the growth of the wild-type or the resistant strains (the only exception being the 40% growth rate reduction of the tetracycline (TET) resistant strain in response to ½ MIC PGLA). Dashed lines represent the clinical breakpoints for the antibiotics in E. coli (not available for doxycycline (DOX)). Data in this figure is representative of at least 2 biological replicates.
We then tested whether PGLA could be used as an adjuvant and selectively potentiate antibiotic activity against antibiotic-resistant bacteria. We tested ciprofloxacin, nalidixic acid, tetracycline and doxycycline resistant E. coli, including laboratory-evolved strains (Figure 5E-G) and clinical isolates (Figure 5H-J, Supplementary Figure 9). The dataset was also augmented with nalidixic acid resistant Klebsiella pneumoniae and Shigella flexneri isolates (Supplementary Figure 10). PGLA was administered at subinhibitory concentrations, i.e. it allowed growth of the wild-type and the antibiotic-resistant strains alike. Strikingly, when used in combination, PGLA caused up to 30-fold decrease in the antibiotic resistance level of the resistant strains (Figure 5E-G, Supplementary Figures 9 and 10, and Supplementary Table 13). We conclude that PGLA can restore antibiotic susceptibility against resistant bacteria when administered as an adjuvant.
PGLA inhibits de novo evolution of resistance to antibiotics
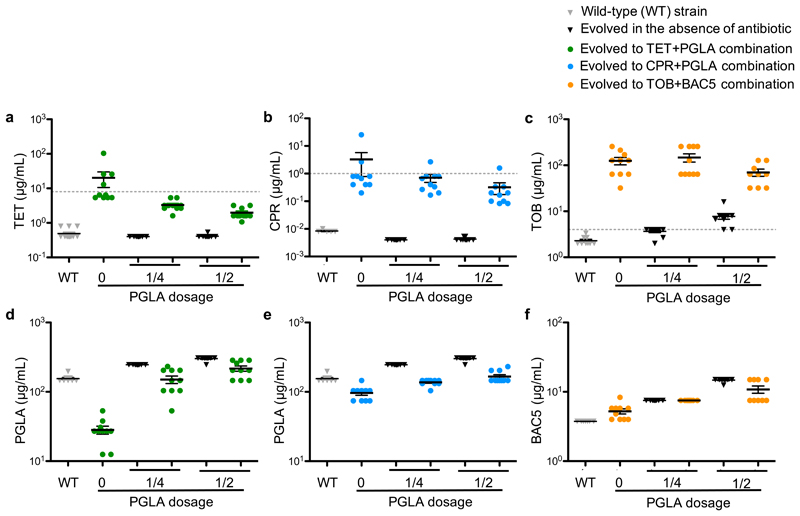
Finally, we asked whether concurrent administration of antibiotics with subinhibitory dosages of PGLA hinders de novo resistance evolution in the laboratory (Figure 6). To this end, we focused on two antibiotic-peptide combinations, ciprofloxacin-PGLA and tetracycline-PGLA. We evolved the wild-type E. coli strain in the presence of these antibiotic-PGLA combinations and their corresponding single drug components. The protocol aimed to maximize the level of antibiotic resistance in the evolving populations during a fixed time period. For each of the 8 drug conditions, 10 parallel bacterial lineages were evolved (Materials and Methods).
Figure 6. Minimum inhibitory concentrations (MICs) of laboratory-evolved lines adapted to antibiotics in the absence and the presence of subinhibitory dosage of antimicrobial peptides.
MIC was measured following a laboratory evolution of the wild-type E. coli strain to tetracycline (TET, green), ciprofloxacin (CPR, blue) and tobramycin (TOB, orange) in the absence or in the presence of ¼ or ½ of the MIC of the antimicrobial peptides PGLA (a, b, d, e) or BAC5 (c, f) against the wild-type strain. MICs of the wild-type and both PGLA and BAC5 evolved lines (in the absence of antibiotic) are represented by grey and white colored bars, respectively. Each data point represents the MIC value of one of each ten parallel-evolved lines. Error bars represent the mean ± standard error of the mean for each experimental condition. Dashed lines represent clinical breakpoints for TET, CPR or TOB in E. coli. Both the CPR-PGLA and the TET-PGLA combinations, which are representatives of collateral sensitive interactions (Figure 1), significantly slowed down the evolution of resistance towards the given antibiotic when administered together. Reassuringly, the control combination (BAC5-TOB), representing a cross-resistance interaction, did not reduce the rate of TOB resistance evolution (P=0.0834, 1-way ANOVA).
After only 160 generations, bacterial populations evolving in the presence of a single antibiotic reached 40- and 390-fold increase in tetracycline and ciprofloxacin MIC levels relative to their ancestor, respectively (Figure 6 A-B). Reassuringly, collateral sensitivity to PGLA arose in these lineages (Figure 6 D-E). In contrast, antibiotic-PGLA co-treatment significantly slowed down the evolution of antibiotic resistance. In the presence of subinhibitory dosages of PGLA, the level of antibiotic resistance reached during the course of laboratory evolution was 10-fold lower than in the absence of PGLA, and it remained consistently below the suggested EUCAST clinical breakpoints (Figure 6 A-B). In addition, evolution of resistance to PGLA was marginal (Figure 6 D-E). Reassuringly, no substantial cross-resistance was observed between PGLA and the antibiotics ciprofloxacin or tetracycline (Figure 6). These patterns are not due to variations in population size across treatments (Supplementary Figure 11).
We suspect that the efficiency of the antibiotic-PGLA co-treatment reflects an elevated fitness cost of antibiotic resistance mutations under such conditions. In other words, collateral sensitivity to PGLA may reduce the number of available resistance-conferring mutations under ciprofloxacin-PGLA or tetracycline-PGLA co-treatment (see also Supplementary Figure 12). A full answer to this question will require detailed molecular and phenotypic characterization of laboratory-evolved bacteria. As a preliminary test, we used the drug pair tobramycin-BAC5, as tobramycin-resistant strains showed cross-resistance rather than collateral sensitivity to the peptide BAC5 (Figure 1). As expected, the tobramycin-BAC5 combination did not reduce the rate of antibiotic resistance evolution (Figure 6C and Figure 6F).
Discussion
How do mutations conferring antibiotic resistance change the susceptibility to antimicrobial peptides? This question is all the more relevant as antimicrobial peptides are promising antibacterial alternatives to antibiotics currently used in the clinics8,9. In this work, we applied an integrated approach to study the susceptibilities of antibiotic-resistant Escherichia coli strains towards antimicrobial peptides.
First, we found that antibiotic-resistant bacteria generally exhibited collateral sensitivity (increased susceptibility) to antimicrobial peptides, while cross-resistance was relatively rare. Several prior works have investigated cross-resistance and collateral sensitivity interactions between conventional antibiotics1–3,31,32. Despite substantial differences in the protocols, these systematic studies agreed that cross-resistance is generally 2-3 times more frequent than collateral sensitivity. By contrast, we see nearly three times more collateral sensitivity than cross-resistance towards antimicrobial peptides (Figure 1).
Second, although we studied bacteria adapted to a diverse set of antibiotics with major differences in their mechanisms of action (Supplementary Table 1), bacterial susceptibilities to antimicrobial peptides revealed several general trends. This is partly due to mutations in canonical resistance genes that emerged repeatedly in response to various antibiotic stresses. By introducing these mutations into a wild-type genetic background, we showed that mutations conferring resistance to one or more antibiotics simultaneously increase sensitivity to several antimicrobial peptides (Table 1). The most noteworthy example is the transcriptional repressor of the mar regulon (marR). Mutations in this gene are frequently observed both in the laboratory and in clinical settings, and increase resistance to multiple unrelated antibiotics2,22. Here we showed that a mutation in this gene increases the negative surface charge of the bacterial outer membrane (Supplementary Figure 7), and eventually leads to elevated susceptibility to several antimicrobial peptides.
More generally, our findings indicate that susceptibility to antimicrobial peptides arises as a by-product of genomic expression changes in antibiotic-resistant bacteria (Figure 3), presumably because these alterations modify the chemical structure of the bacterial outer membrane. Importantly, these conclusions do not hold for all combinations of antimicrobial peptides and resistant bacteria studied. Most notably, aminoglycoside-resistant bacteria accumulated a distinct set of mutations showing practically no overlap with other laboratory-evolved antibiotic-resistant bacteria2. They uniquely carried deleterious mutations in the gene encoding the inner membrane transport protein sbmA2,3. This mutation delivered resistance to proline-rich peptides and aminoglycosides as well (Figure 1 and Table 1).
These considerations could be important for the development of combination therapies. The fact that antibiotic-resistant strains showed extensive collateral sensitivity to certain antimicrobial peptides led us to study one of them, PGLA, in more detail. We identified two important properties of PGLA. First, antibiotic resistance generally resulted in collateral sensitivity to this peptide (Figure 1) and second, antibiotic-PGLA combinations induced synergism in certain antibiotic-resistant bacteria (resistance mutation-induced synergy, Figure 5A-D). Based on these properties, we hypothesized that PGLA could be used for the eradication of drug-resistant bacteria as well as the inhibition of de novo resistance evolution. When used in combination, a subinhibitory dose of PGLA caused up to 30-fold increase in susceptibility in antibiotic-resistant bacteria (Figure 5E-J). Furthermore, the co-administration of the same subinhibitory dose of PGLA efficiently slowed down the evolution of antibiotic resistance and kept resistance levels under the EUCAST breakpoints (Figure 6). Future works should examine whether such a combination strategy can be maintained over longer time scales without the appearance of resistance mutations that diminish the antibiotic-PGLA synergy33. Needless to say, we only consider PGLA as a first step towards the development of an adjuvant therapy. By studying the structural and functional properties of promising antimicrobial peptides, peptidomimetic molecules could be developed that are less prone to resistance, stable against human proteases8,34 and efficient against a broad range of drug-resistant pathogens.
An important unresolved issue is whether collateral sensitive peptide - antibiotic combinations could also be employed sequentially to select against resistance. This strategy, termed collateral sensitivity cycling, has remained controversial1,35. Our preliminary results indicate that the two key ingredients of this strategy are present. First, evolving resistance to an antimicrobial peptide (CAP18) results in collateral sensitivity to several conventional antibiotics, and this collateral sensitivity is reciprocal (Supplementary Figure 13). Second, both antibiotic- and CAP18-resistant bacteria can be selectively eradicated by deploying the collateral sensitive drug partner (Supplementary Figure 14).
Last, we need to emphasize the limitations of our work. The molecular mechanisms underlying collateral sensitivity need to be addressed in more detail. It also remains to be established whether antimicrobial peptide–antibiotic combinations generally outperform conventional antibiotic combinations in their ability to hinder resistance evolution. At best, we made the first step in these directions. An important objection to clinical usage of antimicrobial peptides is that acquisition of resistance against synthetic antimicrobial peptides could drive cross-resistance towards antimicrobial peptides of the host innate immune system36,37. While this is certainly a realistic danger - at least in laboratory settings - two notes must be made. First, the diverse susceptibility profiles of antibiotic-resistant strains towards antimicrobial peptides (Figure 1) confirm major differences both in the mechanisms of action of antimicrobial peptides and the potential routes to antimicrobial peptide resistance. Second, our data indicates that certain peptide-antibiotic combinations could select against the de novo evolution of resistance against both agents (Figure 6).
In summary, our work establishes that antibiotic-resistant bacteria frequently show collateral sensitivity to antimicrobial peptides, a finding that can be utilized to identify peptide-antibiotic combinations that effectively eradicate resistant bacteria and slow down the de novo evolution of resistance to antibiotics.
Materials and Methods
Medium, antimicrobial agents and strains used in the study
Antimicrobial peptides
24 cationic antimicrobial peptides were used in this study: SB006 (SB), human beta defensin 3 (HBD3), protamine (PROA), PR-39, Peptide Glycine-Leucine Amide (PGLA), pexiganan (PEX), indolicidin (IND), bactenecin 5 (BAC5) and bactenecin 7 (BAC7), NCR335, magainin 2 (MAG), R8, D28, anginex (ANG), apidaecin IB (AP), rabbit 18-kDa cationic antimicrobial protein (CAP18), pyrrhocoricin (PYR), pleurocidin (PLEU), synthetic cecropin-melittin hybrid (CEME), protegrin-1 (PRO1), polymyxin B (PXB), LL-37 cathelicidin (LL37), tachyplesin II (TPII), cecropin P1 (CP1) (see also Supplementary Table 2). Antimicrobial peptides were custom synthesized by ProteoGenix, except for PROA and PXB, which were purchased from Sigma-Aldrich. The antimicrobial peptide solutions were prepared in sterile water and kept in -80ºC until usage.
Antibiotics
The following antibiotics (ABs) were used in this study (see also Supplementary Table 1): chloramphenicol (CHL), tetracycline (TET), ampicillin (AMP), cefoxitin (FOX), ciprofloxacin (CPR), erythromycin (ERY), doxycycline (DOX), trimethoprim (TRM), tobramycin (TOB), kanamycin (KAN), nitrofurantoin (NIT) and nalidixic acid (NAL). Most of the antibiotics were purchased from Sigma-Aldrich, except for ERY (AMRESCO) and DOX (AppliChem). Fresh antibiotic solutions were prepared from powder stocks on a weekly basis, kept at -20ºC and were filter sterilized before use.
Strains
Escherichia coli K12 BW25113 was used as the wild-type strain. The 60 laboratory-evolved resistant strains, adapted to twelve different clinically relevant antibiotics (5 evolved lines per antibiotic with the exception of AMP and NAL, for which 4 and 6 replicates were used, respectively) were established in our previous work3. Briefly, the parallel-evolved populations reached up to 328-fold increases in the minimum inhibitory concentrations relative to the ancestor. In all cases, the resistance levels were equal to or above the current clinical breakpoints according to The European Committee on Antimicrobial Susceptibility Testing (EUCAST) or the European Reference Laboratory for Antimicrobial Resistance (EURL-AR). The evolution of multidrug resistance (i.e. resistance to two or more drugs) was frequent under a single antibiotic pressure: on average, 52% of all investigated antibiotic pairs showed cross-resistance in at least one direction. Parallel-evolved lines were subjected to whole-genome sequencing to characterize the mutations responsible for multidrug resistance. On average, we detected 4.2 point mutations, 1.2 deletions, 0.26 insertions and 0.07 duplications per clone. Details of the laboratory evolution experiments as well as the results of the whole-genome sequencing of the evolved strains are available in our previous works2,3.
The four E. coli single-mutant strains (envZ[Ala396Thr], ompC[Met1Ile], marR[Val84Glu], trkH[Thr350Lys]) were established using a highly precise allele replacement protocol described in our previous studies2,3,38. The sbmA single-gene deletion mutant was derived from the KEIO collection39. Both the wild-type and ompC[Met1Ile] strains were transformed with the waaY overexpression and the empty plasmids isolated from the ASKA collection25.
The three E. coli uropathogen clinical isolates (0370, 3538, CFT073) before and after adaptation to three different antibiotics (gentamicin (GEN), nalidixic acid (NAL) and ampicillin (AMP)), the Klebsiella pneumoniae r1 (A) and the Shigella flexneri 668 clinical isolates and their NAL evolved strains were kindly provided by Morten Sommer, Technical University of Denmark, Hørsholm. Four ciprofloxacin-resistant Escherichia coli isolates 1 to 4 were obtained from the local hospital and kindly provided by Edit Urbán, Department of Clinical Microbiology, University of Szeged, Hungary. Strains 1 to 3 were isolated from urine samples, while Strain 4 was isolated from intraperitoneal punction. E. coli ATCC 25922, BAA 2469 and BAA 2340 strains were obtained from Microbiologics.
Medium
In line with previous, methodologically relevant laboratory evolution studies31,32,40, the physical conditions (including the medium) were the same during the course of laboratory evolution and all forthcoming assays on the resulting resistant lines. The logic is to minimize any potential confounding effects unrelated to adaptation to the antibiotic studied (i.e. the only difference being the presence/absence of a given drug). Therefore, as a general rule, the same chemically defined medium and growth conditions were employed throughout the experiments in which the investigated antibiotic-resistant strains were evolved. This medium was optimized for a large set of antimicrobial peptides by decreasing the sodium citrate and the magnesium sulfate concentrations. This optimized medium was minimal salts (MS) medium (1g/L (NH4)2SO4, 3g/L KH2PO4 and 7g/L K2HPO4) supplemented with 0.1mM MgSO4, 0.54 μg/mL FeCl3, 1 μg/mL thiamine hydrochloride, 0.2% Cas amino-acids and 0.2% glucose). Phosphate was available in the medium. This is important as scarcity of phosphate can modify the membrane composition and therefore the activity of antimicrobial peptides41,42. Unlike Muller-Hinton broth, the minimal salts medium used in our work is a chemically defined, highly controlled and reproducible environment, a characteristic that is particularly important in laboratory evolution and high-throughput screenings. As an exception, note that Muller-Hinton II medium was used to study drug susceptibility of Shigella flexneri 668 strain, as this strain cannot grow in MS medium. Reassuringly, the applied medium did not have an effect on the activity of PGLA when used as an adjuvant (see Supplementary Figure 10 where experiments with K. pneumoniae were done in MS medium while the ones using S. flexneri were performed in Muller-Hinton II medium). All components were obtained from Sigma-Aldrich.
Systematic measurement of antimicrobial peptide susceptibilities
We aimed at detecting changes in the sensitivities of a large number of antibiotic-resistant strains compared to the wild-type strain towards a wide variety of antimicrobial peptides as well as bile acid. To this end, we developed a high-throughput screening and a robust statistical analysis methodology for the systematic detection of cross-resistance and collateral sensitivity interactions in E. coli. We compared the susceptibility of 60 antibiotic-resistant strains (5 evolved lines per antibiotic, established in our previous work3) to the wild-type strain across the entire set of antimicrobial peptides (n=24) and bile acid (Figure 1 and Supplementary Table 3). The three main steps of our methodology were i) high-throughput measurement of growth inhibition of strains in liquid cultures in 96-well microtiter plates with the aim of detecting collateral sensitive and cross-resistant phenotypes without precise MIC measurements (see below); ii) validation of the high-throughput results by standard MIC measurements on a subset of strain-peptide condition pairs and using receiver operating characteristic (ROC) curve analysis; iii) further validation of collateral sensitivity interactions at high antimicrobial peptide dosages using kill curve assays.
High-throughput estimation of collateral sensitivity and cross-resistance
To infer collateral sensitivity and cross-resistance interactions, we compared the growth sensitivity of antibiotic-resistant and wild-type strains by measuring their propensity to be arrested in growth by two specific dosages. In this screen, approximately 103 bacteria/ml were inoculated into each well of the 96-well microtiter plate with a 96-pin replicator, and were propagated at 30°C shaken at 300 r.p.m. Bacterial growth was monitored by measuring optical density (OD600) of the liquid cultures at a single time point after 24h of incubation in the presence of a given antimicrobial peptide. Growth arrest was defined as the failure to obtain growth at a given peptide concentration (i.e. OD was below the mean + 2 x standard deviations of OD values of bacteria-free wells containing only growth medium). In order to be able to discern condition-specific growth arrest from slow growth caused by a general cost of resistance, we chose an incubation time of 24 hours where all populations reached detectable growth in medium devoid of antimicrobial peptide. In order to maximize reproducibility and accuracy, a robotic liquid handling system and an automatic incubator were used.
Antibiotic-resistant strains and wild-type strain were inoculated in four and twelve replicate populations on the same 96-well plate, respectively. Growth arrest was determined for each of them and an inhibition score (IS) was calculated based on the replicate measurements as follows: IS = (NGAcontrol/12) - (NGAABstrain/4). NGA is the number of replicates showing growth arrest out of the 12 control populations (NGAcontrol) or out of the 4 antibiotic-resistant populations (NGAABstrain). Two different peptide concentrations were chosen as one above (~1.2x) and one below (~0.8x) the minimum inhibitory concentration (MIC) of the wild-type strain. The lower and the higher concentrations were applied to calculate IS for collateral sensitivity and cross-resistance interactions (CR), respectively. Specifically, to detect collateral sensitivity against a given peptide, we tested whether a peptide dosage below the wild-type MIC (0.8x) fully inhibits growth of a particular antibiotic-resistant strain, but not the wild-type. To detect cross-resistance, we applied a peptide dosage somewhat above the wild-type MIC (1.2x), and asked whether antibiotic-resistant strains can grow. We conducted at least 4 independent experimental runs for each combination of strains and peptide conditions. Next, to filter out unreliable measurements for the detection of cross-resistance (CR) / collateral sensitivity interactions (CS), we excluded cases where (i) cross-contamination might have occurred on the plate during susceptibility measurements (based on growth in non-inoculated wells), (ii) the control wells devoid of peptide showed large variations (coefficient of variation was above 20%), iii) in the case of CS interactions where the applied dosages were too high and arrested the growth of more than 50% of the control populations, iv) in the case of CR interactions where the applied dosages were too low and only arrested the growth of less than 50% of the control populations. This quality control procedure resulted in 2-4 replicates for each combination of strains and peptide conditions. Then, we calculated the average of the inhibition score (IS) derived from the multiple independent experimental runs, and defined cut-off values of the IS to detect cross-resistance and collateral sensitivity interactions, respectively (see below).
To define a robust cut-off value on the IS, we first performed a detailed analysis of minimum inhibitory concentrations (MIC) on a subset of strain-peptide condition pairs (n=110) (Supplementary Table 4). Each antibiotic-resistant strain was characterized as to show cross-resistance (CR) or collateral sensitivity (CS) to a certain antimicrobial peptide if it showed at least 20% difference in its MIC value compared to the wild-type strain. Next, we chose an inhibition score threshold that maximized the fit between the results of the high-throughput susceptibility screen and those of the traditional MIC measurements. The fit between the two datasets was defined as the area under the receiver operating characteristic (ROC) curve calculated from the inhibition score (IS) of the high-throughput susceptibility screen and the CS / CR classification of the detailed MIC measurements. The area under the ROC curve (AUC) is a measure of how well the IS can distinguish between CR/CS interactions and the absence of interactions. The calculated AUC was 0.89 and 0.93 for the collateral sensitivity and cross-resistance interactions, respectively. A random classifier has an AUC of 0.5, while AUC for a perfect classifier is equal to 1. The cut-off point of the IS was chosen as the value that minimizes the Euclidean distance between the ROC curve and the optimum point of the graph (true positive rate=1, false positive rate=0).
Minimum inhibitory concentration (MIC) measurements
Minimum inhibitory concentrations (MIC) were determined using a standard serial broth dilution technique43. In order to maximize reproducibility and accuracy, we used a robotic liquid handling system to prepare 12-step serial dilutions automatically in 96-well microtiter plates. In case of determination of relative MIC involving the measurement of small changes, 1.15 to 1.5 times dilutions were used, and approximately 104 bacteria/ml were inoculated into each well using a 96-pin replicator, and were propagated at 30°C shaken at 300 r.p.m. (3 replicates per strain per antimicrobial peptide concentration). This protocol was previously established with the aim of identifying small changes in relative MICs in a high-throughput and reproducible manner2,3. Otherwise, experiments on the impact of PGLA co-treatment of antibiotic resistant levels or determination of antibiotic and peptide MIC levels upon evolution of resistance used 2 times dilutions and an inoculum of 5x105 bacteria/ml as suggested by the CLSI guidelines. To avoid possible edge effects, rows A and H contained only media devoid of cells. The environment during incubation was also set to minimize evaporation and hence edge effects. After 24h of incubation at 30ºC, raw OD values were measured in a Biotek Synergy microplate reader. MIC was defined by a cutoff OD value (i.e., mean+2 standard deviations of OD values of bacteria-free wells containing only growth medium). Relative MIC was calculated as follows: MICrelative = MICABresistant / MICcontrol.
Kill curve assay
Tolerance of antibiotic-resistant strains to antimicrobial peptides was measured by determining changes in population size upon exposure to lethal concentrations of PGLA (15xMIC), PROA (15xMIC), IND (10xMIC) in the wild-type, various antibiotic-resistant and single-mutant strains. The experiments were conducted in 96 deep-well plates (1000 μl medium supplemented with the peptide). Each well initially contained approximately 106 cells. During antibiotic treatment, 3 samples were taken at multiple time points (0, 15, 30, 60, 90 and 120 minutes post-exposure) from each 3 parallel populations per strain. The total number of viable cells (CFU) was estimated from colony counts after plating the diluted cells on agar plates and incubating them for 24 hours.
Membrane integrity measurements
Membrane integrity of the antibiotic-resistant strains was investigated by measuring their sensitivity to the amphipathic LPS and outer membrane related genes-damaging agent bile acid (see Materials and Methods: High-throughput estimation of collateral sensitivity and cross-resistance). Antibiotic-resistant strains were then grouped by their sensitivity to bile acid: sensitive strains could not grow on medium supplemented with 6% bile acid while non-sensitive strains were able to grow on this concentration. We compared the number of antibiotic-resistant strains that showed collateral sensitivity to antimicrobial peptides between the above two groups (Figure 3A). Because the number of antibiotic-resistant strains qualifies as a count data, we assessed statistical significance in this comparison using a generalized linear mixed model (GLMM) with binomial response distributions and logit link functions as implemented in the R package “lme4”44. We note that the variable representing the number of antibiotic-resistant strains was set as an observational-level random effect to account for overdispersion45.
Total RNA isolation
Total RNA was extracted, as described previously46, from samples grown in minimal medium devoid of any antibiotic, to ensure comparability of strains that show vastly different resistance levels across antibiotics. Samples were collected in log-phase growth (optical density ≈ 0.5 for RT-PCR experiments and optical density ≈ 1 for RNA sequencing) applying the QIAGEN RNA Protect bacteria reagent and stored at -80°C until RNA isolation. RNA isolation was performed using NucleoSpin RNA extraction kit (Macherey-Nagel) according to the manufacturer’s protocol. Purity of total RNA was determined as 260nm/280nm absorbance ratio with expected value of 2.0 by the NanoDrop 1000 spectrophotometer (Thermo Scientific). RNA integrity was confirmed by gel electrophoresis using 1% agarose with GelRed staining. Total RNA was DNase I treated by Ambion DNase I to eliminate residual genomic DNA.
Whole transcriptome sequencing and data analysis
Whole transcriptome sequencing was performed on 24 antibiotic-resistant strains (2 strains per antibiotic, 3 biological replicates per strain) as described previously47. Reference genome sequence (U00096.3) as well as genome annotation data was downloaded from the EcoGene 3.0 database (http://www.ecogene.org/)48 for E. coli strain K-12 MG1655. Raw sequence data was size-selected by discarding reads shorter than 50 bp. This filtering was applied to ensure maximum read quality and consistent read mapping parameters. CLC Genomic Workbench tool version 7.5.1 (CLC Bio now part of Qiagen) was used to obtain gene expression estimates (mapped read counts) for each annotated gene in all samples using the following CLC RNA-Seq analysis parameters: the maximum number of mismatches was set to 2 with minimum length and similarity fractions both set to 0.8 and unspecific match hit set to 10. It is important to note, that the applied alignment length parameter is expressed as a fraction of the raw read length. This could result in a more permissive mapping for shorter reads. In order to ensure the same mapping stringency for all reads, we decided to exclude reads shorter than 50 bp. Please also note, that the excluded short raw read fraction is generated by the SOLiD analysis pipeline due to the removal of low quality read parts. SOLiD low quality readouts, as well as the resulting short reads are expected to appear randomly within the sequencing run. That way, the removal of reads shorter than 50 bp is not expected to cause any bias in the subsequent differential gene expression analysis. Read count data was then imported into R version 3.0.249 excluding rDNA genes. Genes were filtered based on their expression levels, keeping only those features that were detected by at least 5 mapped reads in at least 25% of the samples included in the study. Subsequently, “calcNormFactors” from package “edgeR” version 3.4.250 was used to perform data normalization based on the “trimmed mean of M-values” (TMM) method51. Log transformation and quantile normalization was carried out by the “voom” function of the “limma” package version 3.18.152. “ComBat” tool from the “sva” package version 3.8.053 was applied to correct for systematic batch effects that was caused by growing subsets of the samples in separate batches. Linear modeling, empirical Bayes moderation as well as the calculation of differentially expressed genes were carried out using “limma”.
The selection of differentially expressed genes was based on the comparison of the normalized counts per reads of the control and the antibiotic-resistant strains. Genes with significant differences (false-discovery rate corrected p-values < 0.05) and with fold change (FC) greater than a chosen threshold (|log2(FC)|>1) were regarded as differentially expressed. The log2 fold change values of the normalized expression data and the false-discovery rate corrected p-values are provided in Supplementary Table 8. Finally, we note that although obtaining RNA samples at optical density ≈ 1 may be close to stationary phase, the inferred differential expressions of LPS and phospholipid-related genes are unlikely to be byproducts of the applied RNA sample extraction condition. Specifically, our conclusions remain after excluding genes with potential stationary phase dependent expression from our analyses (see Supplementary Figure 15). The set of stationary phase regulated genes (N=449) was compiled from literature54–56 and by identifying those genes in our dataset whose expression level was significantly correlated (p<0.05) with OD at the time of sample collection in our experiments.
The transcriptome data can be accessed from the Gene Expression Omnibus (GEO) repository (https://www.ncbi.nlm.nih.gov/geo/) with the following access number: GSE96706.
Functional analyses of differentially expressed genes
To obtain a detailed functional annotation of the genes associated with outer membrane LPS and phospholipid synthesis, we selected the following Gene Ontology categories: lipid-A biosynthetic process (GO:0009245), extracellular polysaccharide biosynthetic process (GO:0045226), lipopolysaccharide biosynthetic process (GO:0009103), lipopolysaccharide core region biosynthetic process (GO:0009244), lipopolysaccharide transport (GO:0015920), phospholipid transport (GO:0015914), phospholipid binding (GO:0005543). The selection was based on EcoGene 3.0 database48.
The category ‘All LPS-related genes’ includes all the genes that are associated with at least one of the following GO categories: lipid-A biosynthetic process, extracellular polysaccharide biosynthetic process, lipopolysaccharide biosynthetic process, lipopolysaccharide core region biosynthetic process and lipopolysaccharide transport. The category ‘All Phospholipid-related genes’ includes all the genes that are associated with phospholipid transport or phospholipid binding GO categories. Further information about the full gene sets is provided in Supplementary Table 9.
To investigate the upregulated LPS-related genes in more detail, we performed Fisher’s exact tests on all resistant strains and all subcategories of LPS-related genes (Figure 3C, left heatmap). In addition, student’s t-tests were used to identify differences in the average gene expression in a given GO category among antibiotic-resistant strains grouped according to their antimicrobial peptide sensitivity (Figure 3C, right heatmap, Supplementary Figure 5).
Chemogenomic screen
Competition experiment
A high-throughput gene overexpression screen was performed to study the impact of upregulating each E. coli ORF on the susceptibility to a particular antimicrobial peptide (CAP18 and CP1). For a schematic workflow see Supplementary Figure 6A. The screen was carried out using a well-established plasmid library (A complete set of E. coli K-12 Open Reading Frame Archive library (ASKA)), where each E. coli ORF is cloned into a high copy number expression plasmid (pCA24N-ORF-GFP(-))25. A previously described, pooled version of the plasmid collection57 was transformed into E. coli K12 BW25113 strain by electroporation, as well as the empty plasmid pCA24N (without an ORF). The empty plasmid serves as a control to measure read counts that come from genomic DNA contamination during plasmid preparation (background). The resulting strain collection and the strain carrying the empty plasmid were grown in parallel in MS medium and expression of the ORFs was induced mildly with a relatively low inducer concentration (100μM isopropyl-ß-D-thiogalactopyranoside (IPTG)). Following 1h of incubation at 30°C, 500,000 cells from the library were used to inoculate each well of a 96-well microtiter plate prefilled with a concentration gradient of the peptide in MS media supplemented with 10μg/mL chloramphenicol and 100μM IPTG, the same way as for a standard MIC measurement (described above). As a further control, both the library and the empty plasmid were also grown in the absence of peptide. Growth was monitored in a microplate reader (Biotek Synergy 2). Wells that showed 50% growth inhibition in late log phase were transferred into 20 mL MS media supplemented with the corresponding peptide. Peptide concentration was set again to result in 50% growth inhibition. Growth was stopped at late log phase (OD600>1). Following plasmid isolation, the samples were digested overnight with a mixture of lambda exonuclease and exonuclease I (Fermentas) at 37°C to remove genomic DNA contamination. The cleaned samples were subjected to next-generation sequencing with the SOLiD System (Life Technologies) to determine the diversity of the pooled library as described previously57. Each treatment was carried out in two replicates, except the untreated sample (in the absence of peptide), which had 5 replicates. The experiment with the empty plasmid was also carried out twice.
Data analysis
Raw sequence data processing and mapping onto E. coli ORFs were carried out the same way as in the whole transcriptome analysis. Raw sequence data was also mapped to the plasmid backbone. In order to make the mapped read counts comparable between the different samples, we carried out the following data processing workflow based on established protocols58,59, using a custom made R script: The extra read counts coming from the genomic DNA contamination (background) was estimated by assuming that the reads mapping to the unit length of the plasmid and the ORFs should have a ratio of 1:1. The total extra read count estimated thereof was partitioned among the ORFs based on their background frequency (i.e. their relative frequency obtained from the experiment involving the empty plasmid). Next, these ORF-specific backgrounds were subtracted from the read counts. Then, a loglinear transformation was carried out on the background-corrected relative read counts. This transformation has the advantage over canonical logarithmic transformation of avoiding the inflation of data variance for ORFs with very low read counts60. The transformed relative read counts showed bimodal distributions (Supplementary Figure 6B-C). The lower mode (peak) of the distribution corresponds to ORFs that were not present in the sample. The upper mode represents those ORFs that grew unaffected by the overexpression. To make the different samples comparable, the two modes of the distribution of each sample were set to two predefined values. These values were chosen such as the original scale of the data was retained. In order to align the modes between samples, we introduced two normalization steps: one before and one after the loglinear transformation. The first normalization step identified the lower mode corresponding to the absent strains and shifted it to zero. Next, we performed the loglinear transformation step described above. The second normalization step was a linear transformation moving the upper mode to a higher predefined value. Following this normalization step, genes that were close to the lower mode in the untreated samples were discarded from the analysis as these represent strains that displayed poor growth even in the absence of drug treatment (i.e. antimicrobial peptide sensitivity could not be reliably detected). Differential growth was calculated as the ratio of the normalized relative read counts in the treated and the non-treated samples at the end of the competition. Genes showing at least a 2-fold decrease in relative abundance in both replicates upon peptide treatment were considered as sensitizing genes.
In the next step, we examined the enrichment of CAP18- and CP1-sensitizing genes (Supplementary Table 10) in various GO categories using Fisher’s exact test. Obtained p-values were corrected for multiple testing using the Benjamini & Hochberg method61 (Supplementary Table 11).
Real-time PCR analysis
The waaY gene expression of the wild-type and marR mutant strains was determined by quantitative reverse-transcription Real-Time PCR (qRT-PCR) as described previously38. Briefly, quantitation of cysG expression level was applied as a reference-standard across both samples62. qRT-PCR was performed in a Bio-Rad CFX96 Real Time System (Bio-Rad Laboratories). The PCR mixtures consisted of ~300 ng of total RNA sample, 0.1 μM of the waaY_FW-waaY_REV primer pair (5’-ATCGATCTCTCCGGAAAGC, 5’-CCTTTCAAACGCCGCATA) or 0.1 μM of the cysG_FW-cysG_REV reference primer pair (5'-TTGTCGGCGGTGGTGATGTC, 5'- ATGCGGTGAACTGTGGAATAAACG)62 and Verso one-step RT-qPCR Master Mix with low ROX reference dye (Thermo Scientific) in a final volume of 25 μL. The assay included “nontemplate” and “non-RT” controls to detect reagent contamination and presence of gDNA. RT reaction was performed at 50ºC for 15 min and the thermal profile of the PCR procedure repeated for 40 cycles was: 95ºC for 10 min; 20 sec denaturation at 95ºC, 20 sec annealing at 60ºC and 60 sec at 72ºC coupled to data collection at the end of each amplification step. Dissociation curve consisted of 10 sec incubation at 95ºC, 5 sec incubation at 65ºC, and ramp up to 95ºC. Melting curves were used to validate product specificity. All samples were amplified in triplicates from the same total RNA preparation and the mean value was used for further analysis. Cycle threshold (Ct) values were determined using Bio-Rad CFX96 software.
The waaY expression level was calculated by relative mRNA quantitation, based on the mean of technical replicates, normalized to the expression level of the cysG control, taking into consideration the efficiency level of each primer pair. Each primer efficiency (E) was determined as E=10−1/slope−1 based on the slope of four point standard curves with 10 times concentration intervals, in duplicate, using wild-type strain RNA. Efficiency of waaY and cysG primer pairs was determined as 1.6 and 1.8 respectively. waaY expression level in the marR mutant strain was calculated relative to that of the wild-type strain as follows: Expression level=(1.6(CtwaaY_WT-CtwaaY_marR*)/(1.8(CtcysG_WT)-(CtcysG_marR*)); and is presented as the mean of triplicates ± standard error of the mean.
Measurement of antibiotic and PGLA combinations
We selected the peptide glycine-leucine-amide (PGLA) antimicrobial peptide, which interacts with the bacterial lipopolysaccharide layer, and 10 different antibiotics with a wide range of mechanisms of action (Supplementary Table 1). All experiments were conducted in the antibiotic sensitive wild-type Escherichia coli K-12 (BW25113) strains and in 21 different antibiotic-resistant strains adapted to one of 10 antibiotics (established in our previous work3) (Supplementary Table 12). Combination screens were performed in 96-well plates, using a liquid-handling robotic system (Hamilton Star workstation) to improve reproducibility.
Interactions between the two drugs were classified as synergistic, independent (additive) or antagonistic based on growth measurements at multiple concentration combinations (see below). Drug interaction was defined as deviation from non-interaction under the Loewe additivity model63, which assumes that a drug does not interact with itself. We followed a previously published protocol64 with two important modifications. First, instead of examining all pairwise combinations of a pre-defined number of linearly increasing concentration points, we focused on a set of different antibiotic-PGLA relative concentration ratios and their dilution series (see Supplementary Figure 16). This setup enabled us to efficiently sample the most informative regions of the two-dimensional concentration space. Second, we inferred drug interactions based on concentration combinations that led to 90% growth inhibitions (Supplementary Figure 16). This enabled an especially robust detection of growth inhibition for antimicrobial peptides, which often exhibit steep dose-response curves that hinders precise measurement of, say, 50% inhibition concentrations.
As a first step, for each single agent (antibiotic and peptide alike), a 1.6-fold 8-step dilution series was prepared with dose points determined based on the minimal inhibitory concentration (MIC) of the agents. The concentration range for each agent was between 10.5 times lower and 2.6 times higher than the MIC of the strain. Then for each antibiotic-peptide pair, we setup a 96-well plate as follows: we defined 7 different antibiotic:peptide relative concentration ratios (7:1, 3:1, 5:3, 1:1, 3:5, 1:3, 1:7) and generated dilution series thereof across the plate. As a result, each plate contained dilution series of 7 antibiotic:peptide ratio, dilution series from the given antibiotic or peptide alone, 4 bacteria-free wells (no growth control) and 4 wells containing only medium without any drugs (growth control) (see Supplementary Figure 16A).
Combination screen plates were inoculated with 5×104 cells/well from overnight culture (grown at 30°C, with shaking at 300 rpm). The culture volume was 100 μl. Assay plates were incubated at 30°C, with shaking at 300 rpm and bacterial growth was monitored by measuring the optical density (OD600) of the liquid cultures after 24 h. We chose an incubation time of 24 hours in order to be able to discern condition-specific fitness defects from general costs of resistance of the antibiotic-resistant strains.
Identifying interactions between antibiotics and PGLA
To assess antagonism and synergy between pairs of antibiotics and PGLA in the sensitive wild-type and in the antibiotic-resistant strains, we used the Loewe additivity model63 which assumes that a drug does not interact with itself. To identify interactions for each pair of antibiotics and PGLA we first calculated relative inhibition values based on the initial OD (maximum inhibition) and the average OD of antibiotic-free control wells (maximum growth). Then we identified those two concentration points for each antibiotic:peptide ratio where the inhibition of the growth was just above and below 90%, respectively. By fitting a linear model between these two concentration points we could interpolate the dosages for each antibiotic:peptide ratio that is responsible for the 90% growth inhibition (90% effective dosage: EC90%). Based on the Loewe model from the EC90% values of the single agents, we then calculated the theoretical EC90% dosages for each of the 7 antibiotic:peptide ratio. Geometrically, the theoretical EC90% based on the Loewe model can be represented as a straight line between the EC90% of the single agents in the two-dimensional linear concentration space. Deviation of the shape of the lines connecting the experimentally measured EC90% from linearity indicates either synergy (concave isoboles) or antagonism (convex isoboles) (Supplemetary Figure 12). For each of the 7 antibiotic:peptide ratio we defined the expected and the experimentally measured EC90% values. The combination index was calculated as: [theoretical EC90%] / [experimental EC90%] for each antibiotic:peptide ratio (CIr). The combination index (CI) for a given antibiotic and peptide pair was defined as the average of the combination index of the 7 antibiotic:peptide ratio (mean[CIr1,CIr2…CIr7]). Where multiple independent experimental runs were available, we calculated the average value of the measured CIs.
Measurement errors of interaction screens were estimated from two independent experimental runs of 24 combinations by calculating the pooled variance (SD) of CI of the replicate experiments. The cut-off values were defined as 1.95xSD value of the CI. The cut-off values were as follows: CI≥1.14 for antagonism; CI≤0.86 for synergism; and 0.86<CI<1.14 for no interaction.
Experimental evolution of resistance
Experimental evolution was preformed based on a previously established automated evolution experiment65,66 during approximately 160 generations (24 transfers). 10 parallel independent cultures were propagated in 8 experimental conditions such as tetracycline (TET) or ciprofloxacin (CPR) in the presence or absence of ½ and ¼ of the MIC of the peptide glycine-leucine-amide (PGLA); tobramycin (TOB) in the presence or absence of ½ and ¼ of the MIC of the bactenecin 5 (BAC5); as well as ½ and ¼ of the MIC of PGLA and ½ and ¼ of the MIC of BAC5, alone. Chess-board layout was used on the plate to monitor potential cross-contamination events. Starting with subinhibitory drug concentration, each culture was allowed to grow for 24 hours. 20 μl of culture was transferred to four independent wells containing fresh medium and increasing dosages of antibiotic (0.5x, 1x, 1.5x and 2.5x the concentration of the previous step). At each transfer, cell growth was monitored by measuring the optical density at 600 nm (OD600 value, Biotek Synergy 2 microplate reader was used for this purpose). Only populations of the highest drug concentration that reached OD600 > 0.2 were selected for further evolution. Accordingly, only one of the four populations was retained for each independently evolving lineage. This protocol was designed to avoid population extinction and to ensure that populations with the highest level of resistance were propagated further during evolution.
Supplementary Material
Acknowledgements
The authors thank to Éva Kondorosi for providing the cationic antimicrobial peptide NCR335, and to Morten O.A. Sommer for the clinical isolates. The authors also acknowledge the following financial support: Hungarian Academy of Sciences Postdoctoral Fellowship Programme (VL), Hungarian Scientific Research Fund NKFI PD 116222 (AM), NKFI 120220 (BK), OTKA PD 109572 (BC) and NKFI FK 124254 (OM), 'Lendület' Programme of the Hungarian Academy of Sciences and The Wellcome Trust (BP and CP), H2020-ERC-2014-CoG (CP), GINOP-2.3.2-15-2016-00014 (EVOMER), GINOP-2.3.2-15-2016-00020 (MolMedEx TUMORDNS) and GINOP-2.3.3-15-2016-00001. IN and BK were supported by the János Bolyai Research Scholarship of the Hungarian Academy of Sciences and ÁN by the Ph.D. fellowship of the Boehringer Ingelheim Fonds.
Footnotes
Data availability
The transcriptome data can be accessed from the Gene Expression Omnibus (GEO) repository (https://www.ncbi.nlm.nih.gov/geo/) with access number GSE96706.
Authors Contributions
CP and BP conceived and supervised the project; VL and AM designed the experiments and developed data analysis procedures; CP, BP, VL, AM and RS wrote the paper; RS, AK, AD, FW and MD performed the zeta potential measurements; GO, ZH and TAM synthetize the peptides magainin 2 and anginex; RS and OM purified RNA for transcriptomic analysis; BB and IN performed RNA-Seq experiments; PKJ, GF, MS and BK generated and analyzed the chemogenomic data; EU isolated and identified E. coli clinical isolates; BC, AN prepared the mutant strains; VL, LD, AM, RS, BC contributed to all other experiments. VL, AM, GG, GF and AG analyzed and interpreted the data.
Competing interests
The authors declare no competing interests. BB and IN had consulting positions at SeqOmics Biotechnology Ltd. at the time the study was conceived. SeqOmics Biotechnology Ltd. was not directly involved in the design and execution of the experiments or in the writing of the manuscript. This does not alter the author’s adherence to all the Nature policies on sharing data and materials.
References
- 1.Imamovic L, Sommer MO. Use of collateral sensitivity networks to design drug cycling protocols that avoid resistance development. Sci Transl Med. 2013;5:204ra132. doi: 10.1126/scitranslmed.3006609. [DOI] [PubMed] [Google Scholar]
- 2.Lázár V, et al. Genome-wide analysis captures the determinants of the antibiotic cross-resistance interaction network. Nat Commun. 2014;5:4352. doi: 10.1038/ncomms5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lázár V, et al. Bacterial evolution of antibiotic hypersensitivity. Molecular Systems Biology. 2013;9:700–700. doi: 10.1038/msb.2013.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toprak E, et al. Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nat Genet. 2012;44:101–105. doi: 10.1038/ng.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palmer AC, Kishony R. Understanding, predicting and manipulating the genotypic evolution of antibiotic resistance. Nat Rev Genet. 2013;14:243–248. doi: 10.1038/nrg3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pál C, Papp B, Lázár V. Collateral sensitivity of antibiotic-resistant microbes. Trends in Microbiology. 2015;23:401–407. doi: 10.1016/j.tim.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jenssen H, Hamill P, Hancock REW. Peptide Antimicrobial Agents. Clinical Microbiology Reviews. 2006;19:491–511. doi: 10.1128/CMR.00056-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fjell CD, Hiss JA, Hancock RE, Schneider G. Designing antimicrobial peptides: form follows function. Nat Rev Drug Discov. 2012;11:37–51. doi: 10.1038/nrd3591. [DOI] [PubMed] [Google Scholar]
- 9.Mahlapuu M, Håkansson J, Ringstad L, Björn C. Antimicrobial Peptides: An Emerging Category of Therapeutic Agents. Frontiers in Cellular and Infection Microbiology. 2016;6:194. doi: 10.3389/fcimb.2016.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andersson DI, Hughes D, Kubicek-Sutherland JZ. Mechanisms and consequences of bacterial resistance to antimicrobial peptides. Drug Resist Updat. 2016;26:43–57. doi: 10.1016/j.drup.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 11.Melo MN, Ferre R, Castanho MARB. Antimicrobial peptides: linking partition, activity and high membrane-bound concentrations. Nature Reviews Microbiology. 2009;7:245. doi: 10.1038/nrmicro2095. [DOI] [PubMed] [Google Scholar]
- 12.Alves CS, et al. Escherichia coli cell surface perturbation and disruption induced by antimicrobial peptides BP100 and pepR. J Biol Chem. 2010;285:27536–27544. doi: 10.1074/jbc.M110.130955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Avitabile C, D'Andrea LD, Romanelli A. Circular Dichroism studies on the interactions of antimicrobial peptides with bacterial cells. Scientific Reports. 2014;4:4293. doi: 10.1038/srep04293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soriano F, Ponte C, Santamaria M, Jimenez-Arriero M. Relevance of the inoculum effect of antibiotics in the outcome of experimental infections caused by Escherichia coli. Journal of Antimicrobial Chemotherapy. 1990;25:621–627. doi: 10.1093/jac/25.4.621. [DOI] [PubMed] [Google Scholar]
- 15.Bechinger B, Zasloff M, Opella SJ. Structure and dynamics of the antibiotic peptide PGLa in membranes by solution and solid-state nuclear magnetic resonance spectroscopy. Biophys J. 1998;74:981–987. doi: 10.1016/S0006-3495(98)74021-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ostorhazi E, Nemes-Nikodem E, Knappe D, Hoffmann R. In Vivo Activity of Optimized Apidaecin and Oncocin Peptides Against a Multiresistant, KPC-Producing Klebsiella pneumoniae Strain. Protein & Peptide Letters. 2014;21:368–373. doi: 10.2174/09298665113206660107. [DOI] [PubMed] [Google Scholar]
- 17.Proctor RA, et al. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat Rev Micro. 2006;4:295–305. doi: 10.1038/nrmicro1384. [DOI] [PubMed] [Google Scholar]
- 18.Munck C, Gumpert HK, Wallin AI, Wang HH, Sommer MO. Prediction of resistance development against drug combinations by collateral responses to component drugs. Sci Transl Med. 2014;6:262ra156. doi: 10.1126/scitranslmed.3009940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mattiuzzo M, et al. Role of the Escherichia coli SbmA in the antimicrobial activity of proline-rich peptides. Molecular Microbiology. 2007;66:151–163. doi: 10.1111/j.1365-2958.2007.05903.x. [DOI] [PubMed] [Google Scholar]
- 20.Pages J-M, James CE, Winterhalter M. The porin and the permeating antibiotic: a selective diffusion barrier in Gram-negative bacteria. Nat Rev Micro. 2008;6:893–903. doi: 10.1038/nrmicro1994. [DOI] [PubMed] [Google Scholar]
- 21.Fernández L, Hancock REW. Adaptive and Mutational Resistance: Role of Porins and Efflux Pumps in Drug Resistance. Clinical Microbiology Reviews. 2012;25:661–681. doi: 10.1128/CMR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seoane AS, Levy SB. Characterization of MarR, the repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli. Journal of Bacteriology. 1995;177:3414–3419. doi: 10.1128/jb.177.12.3414-3419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davin-Regli A, et al. Membrane Permeability and Regulation of Drug "Influx and Efflux" in Enterobacterial Pathogens. Current Drug Targets. 2008;9:750–759. doi: 10.2174/138945008785747824. [DOI] [PubMed] [Google Scholar]
- 24.Gutsmann T, Fix M, Larrick JW, Wiese A. Mechanisms of Action of Rabbit CAP18 on Monolayers and Liposomes Made from Endotoxins of Phospholipids. The Journal of Membrane Biology. 2000;178 doi: 10.1007/s00232001092. a71-a71. [DOI] [PubMed] [Google Scholar]
- 25.Kitagawa M, et al. Complete set of ORF clones of Escherichia coli ASKA library (A Complete Set of E. coli K-12 ORF Archive): Unique Resources for Biological Research. DNA Research. 2006;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- 26.Delcour AH. Outer membrane permeability and antibiotic resistance. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 2009;1794:808–816. doi: 10.1016/j.bbapap.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bociek K, et al. Lipopolysaccharide Phosphorylation by the WaaY Kinase Affects the Susceptibility of Escherichia coli to the Human Antimicrobial Peptide LL-37. J Biol Chem. 2015;290:19933–19941. doi: 10.1074/jbc.M114.634758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee J-H, Lee K-L, Yeo W-S, Park S-J, Roe J-H. SoxRS-Mediated Lipopolysaccharide Modification Enhances Resistance against Multiple Drugs in Escherichia coli. Journal of Bacteriology. 2009;191:4441–4450. doi: 10.1128/JB.01474-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gonzales PR, et al. Synergistic, collaterally sensitive β-lactam combinations suppress resistance in MRSA. Nature Chemical Biology. 2015;11:855. doi: 10.1038/nchembio.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baym M, Stone LK, Kishony R. Multidrug evolutionary strategies to reverse antibiotic resistance. Science. 2016;351:aad3292. doi: 10.1126/science.aad3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oz T, et al. Strength of Selection Pressure Is an Important Parameter Contributing to the Complexity of Antibiotic Resistance Evolution. Molecular Biology and Evolution. 2014;31:2387–2401. doi: 10.1093/molbev/msu191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horinouchi T, et al. Prediction of Cross-resistance and Collateral Sensitivity by Gene Expression profiles and Genomic Mutations. Scientific Reports. 2017;7:14009. doi: 10.1038/s41598-017-14335-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pena-Miller R, et al. When the Most Potent Combination of Antibiotics Selects for the Greatest Bacterial Load: The Smile-Frown Transition. PLOS Biology. 2013;11:e1001540. doi: 10.1371/journal.pbio.1001540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Avan I, Hall CD, Katritzky AR. Peptidomimetics via modifications of amino acids and peptide bonds. Chemical Society Reviews. 2014;43:3575–3594. doi: 10.1039/c3cs60384a. [DOI] [PubMed] [Google Scholar]
- 35.Jiao YJ, Baym M, Veres A, Kishony R. Population diversity jeopardizes the efficacy of antibiotic cycling. bioRxiv. 2016 [Google Scholar]
- 36.Fleitas O, Franco OL. Induced Bacterial Cross-Resistance toward Host Antimicrobial Peptides: A Worrying Phenomenon. Frontiers in Microbiology. 2016;7:381. doi: 10.3389/fmicb.2016.00381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kubicek-Sutherland JZ, et al. Antimicrobial peptide exposure selects for Staphylococcus aureus resistance to human defence peptides. Journal of Antimicrobial Chemotherapy. 2016;72:115–127. doi: 10.1093/jac/dkw381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nyerges Á, et al. A highly precise and portable genome engineering method allows comparison of mutational effects across bacterial species. Proceedings of the National Academy of Sciences. 2016;113:2502–2507. doi: 10.1073/pnas.1520040113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100050. 2006 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tenaillon O, et al. Tempo and mode of genome evolution in a 50,000-generation experiment. Nature. 2016;536:165. doi: 10.1038/nature18959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malouin F, Chamberland S, Brochu N, Parr TR. Influence of growth media on Escherichia coli cell composition and ceftazidime susceptibility. Antimicrobial Agents and Chemotherapy. 1991;35:477–483. doi: 10.1128/aac.35.3.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yeaman MR, Yount NY. Mechanisms of Antimicrobial Peptide Action and Resistance. Pharmacological Reviews. 2003;55:27–55. doi: 10.1124/pr.55.1.2. [DOI] [PubMed] [Google Scholar]
- 43.Wiegand I, Hilpert K, Hancock REW. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protocols. 2008;3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 44.Bates D, Machler M, Bolker BM, Walker SC. Fitting Linear Mixed-Effects Models Using lme4. Journal of Statistical Software. 2015;67:1–48. [Google Scholar]
- 45.Bolker BM, et al. Generalized linear mixed models: a practical guide for ecology and evolution. Trends in Ecology & Evolution. 2009;24:127–135. doi: 10.1016/j.tree.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 46.Méhi O, et al. Perturbation of Iron Homeostasis Promotes the Evolution of Antibiotic Resistance. Molecular Biology and Evolution. 2014;31:2793–2804. doi: 10.1093/molbev/msu223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karcagi I, et al. Indispensability of Horizontally Transferred Genes and Its Impact on Bacterial Genome Streamlining. Molecular Biology and Evolution. 2016;33:1257–1269. doi: 10.1093/molbev/msw009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou J, Rudd KE. EcoGene 3.0. Nucleic Acids Research. 2013;41:D613–D624. doi: 10.1093/nar/gks1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.R: A language and environment for statistical computing. the R Foundation for Statistical Computing; Viena, Austria: 2014. [Google Scholar]
- 50.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biology. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research. 2015;43 doi: 10.1093/nar/gkv007. e47-e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang DE, Smalley DJ, Conway T. Gene expression profiling of Escherichia coli growth transitions: an expanded stringent response model. Molecular Microbiology. 2002;45:289–306. doi: 10.1046/j.1365-2958.2002.03001.x. [DOI] [PubMed] [Google Scholar]
- 55.Tjaden B, et al. Transcriptome analysis of Escherichia coli using high‐density oligonucleotide probe arrays. Nucleic Acids Research. 2002;30:3732–3738. doi: 10.1093/nar/gkf505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dong T, Schellhorn HE. Control of RpoS in global gene expression of Escherichia coli in minimal media. Molecular Genetics and Genomics. 2009;281:19–33. doi: 10.1007/s00438-008-0389-3. [DOI] [PubMed] [Google Scholar]
- 57.Notebaart RA, et al. Network-level architecture and the evolutionary potential of underground metabolism. Proceedings of the National Academy of Sciences. 2014;111:11762–11767. doi: 10.1073/pnas.1406102111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pierce SE, Davis RW, Nislow C, Giaever G. Genome-wide analysis of barcoded Saccharomyces cerevisiae gene-deletion mutants in pooled cultures. Nat Protocols. 2007;2:2958–2974. doi: 10.1038/nprot.2007.427. [DOI] [PubMed] [Google Scholar]
- 59.Robinson DG, Chen W, Storey JD, Gresham D. Design and Analysis of Bar-seq Experiments. G3: Genes|Genomes|Genetics. 2014;4:11–18. doi: 10.1534/g3.113.008565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rocke DM, Durbin B. Approximate variance-stabilizing transformations for gene-expression microarray data. Bioinformatics. 2003;19:966–972. doi: 10.1093/bioinformatics/btg107. [DOI] [PubMed] [Google Scholar]
- 61.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 62.Zhou K, et al. Novel reference genes for quantifying transcriptional responses of Escherichia coli to protein overexpression by quantitative PCR. BMC Molecular Biology. 2011;12:18. doi: 10.1186/1471-2199-12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Loewe S. The problem of synergism and antagonism of combined drugs. Arzneimittelforschung. 1953;3:285–290. [PubMed] [Google Scholar]
- 64.Cokol M, et al. Systematic exploration of synergistic drug pairs. Molecular Systems Biology. 2011;7:544–544. doi: 10.1038/msb.2011.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Suzuki S, Horinouchi T, Furusawa C. Prediction of antibiotic resistance by gene expression profiles. Nature Communications. 2014;5:5792. doi: 10.1038/ncomms6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bódi Z, et al. Phenotypic heterogeneity promotes adaptive evolution. PLOS Biology. 2017;15:e2000644. doi: 10.1371/journal.pbio.2000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Murtagh F, Legendre P. Ward’s Hierarchical Agglomerative Clustering Method: Which Algorithms Implement Ward’s Criterion? Journal of Classification. 2014;31:274–295. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.