Abstract
Biologists marvel at the powers of adaptive convergence, when distantly related animals look alike. While mimetic wing patterns of butterflies have fooled predators for millennia, entomologists inferred that mimics were distant relatives despite similar appearance. However, the obverse question has not been frequently asked. Who are the close relatives of mimetic butterflies and what are their features? As opposed to close convergence, divergence from a non-mimetic relative would also be extreme. When closely related animals look unalike, it is challenging to pair them. Genomic analysis promises to elucidate evolutionary relationships and shed light on molecular mechanisms of divergence. We chose the firetip skipper butterfly as a model due to its phenotypic diversity and abundance of mimicry. We sequenced and analysed whole genomes of nearly 120 representative species. Genomes partitioned this subfamily Pyrrhopyginae into five tribes (1 new), 23 genera and, additionally, 22 subgenera (10 new). The largest tribe Pyrrhopygini is divided into four subtribes (three new). Surprisingly, we found five cases where a uniquely patterned butterfly was formerly placed in a genus of its own and separately from its close relatives. In several cases, extreme and rapid phenotypic divergence involved not only wing patterns but also the structure of the male genitalia. The visually striking wing pattern difference between close relatives frequently involves disappearance or suffusion of spots and colour exchange between orange and blue. These differences (in particular, a transition between unspotted black and striped wings) happen recurrently on a short evolutionary time scale, and are therefore probably achieved by a small number of mutations.
Keywords: biodiversity, museomics, higher classification, mimicry rings, skipper butterflies, sinimustvalge pattern
1. Introduction
Deciphering evolutionary relationships between animals is a non-trivial task. Careful comparative analysis of morphology was the only approach available a century ago. However, obstructed by adaptive convergence and rapid divergence, evolutionary relationships do not always follow morphological similarity. For several decades, sequencing of gene markers offered a successful orthogonal strategy to complement morphological analysis [1]. A set of standard gene markers was hugely important to probe phylogeny in essentially all branches of life. Yet, stymied by homoplasies, short DNA segments are not ideal for phylogenetic studies [2]. Next-generation sequencing technologies decreased the cost of DNA sequence by several orders of magnitude [3]. At the beginning of this century, the first human genome required nearly 3 billion dollars to complete (1 dollar per base pair) [4], whereas subsequent human genomes can be sequenced for around a thousand dollars today (3 million base pairs per dollar) [5].
Armed with these new methods, researchers can obtain nearly complete genomes of butterflies at a price they paid for only a dozen gene markers a decade ago [6–12]. Half a billion base pairs of genomic sequence dwarf several thousand base pairs of gene markers, and are more successful at distillation of phylogenetic signal from the noise of homoplasies [13]. After all, a genome contains the entire genetic information of an animal, and if its evolutionary history cannot be reconstructed with this comprehensive information, it may be truly lost with time. Thus, comparative analysis of complete genomes holds the best promise to reveal the most complete picture of evolutionary relationships. Moreover, comparison of genotypes and phenotypes suggests evolutionary mechanisms of adaptations [6–8,14]. Overlaying the differences and similarities of the phenotypes on our confident phylogeny reveals the cases of adaptive convergence and extreme divergence of close relatives.
Despite the great promise, genomic analysis is plagued with complexities. In addition to sheer volumes of data that require extensive and lengthy computation, universal phylogenetic difficulties such as long branch attraction are amplified and can lead to strong support for a wrong tree. Moreover, phylogenetic signal is heterogeneous across the genome due to incomplete lineage sorting and introgression [15]. The problem is particularly severe in the ‘anomaly zone' [16]. Finally, short genomic reads produced by Illumina are challenging to stitch together and obtain sequences long enough for orthology detection and alignment in the absence of a close reference genome.
Among butterflies, the skipper family (Hesperiidae) is one of the most speciose and the least studied [13,17]. While many skippers are rather small and dull-coloured, those from the Neotropical subfamily Pyrrhopyginae are large, robust and gaudy. Dubbed ‘firetips' for the frequent presence of a prominent tuft of red or orange scales at the end of the abdomen, many species are endowed with shiny metallic colours and bright spots and stripes not only on the wings but also on the body [18,19]. It is unusual for butterfly caterpillars to be hairy, but both caterpillars and pupae of firetips are characterized by long hairs covering their bodies [19]. Firetips are monophyletic, unified by the narrow first abdominal tergum, which appears compressed between the thorax and the second segment [20].
Perhaps the most interesting feature of firetips is a frequent and possibly mimetic convergence in wing patterns between distant relatives [18,19]. These relatives are not restricted to the subfamily and come from multiple subfamilies of skippers: Eudaminae and Hesperiinae. The resemblance is so noticeable that it is reflected by their names (e.g. Hesperiinae genera Pseudosarbia and Pyrrhopygopsis are named to contrast with the firetip genera Sarbia and Pyrrhopyge). Within firetips, the most prominent is the sinimustvalge (blue-black-white, as in the Estonian flag) pattern of Jemadia, Elbella, Parelbella, Protelbella, Croniades, Nosphistia, Zonia and Granila, similar to that of representatives in Eudaminae genera Phocides and Tarsoctenus.
Although potentially confusing to a novice, these similarities did not fool taxonomists who placed these distant relatives in different taxonomic groups. Mielke [21,22] revised the taxonomy of firetips, proposing 14 new genera to add to 21 used previously. Out of 35 recognized genera, 16 (45%) are monotypic (i.e. consist of a single species) [23]. It is unclear whether this abundance of monotypy reflects an unusual uniqueness of many firetips, poor fieldwork in discovering additional species, or an oversplit classification. To better understand the evolutionary relationships within Pyrrhopyginae and to shed light on the mechanism of convergence, we obtained and analysed genomic sequences of 119 species and subspecies, which covers about 60% of all known Pyrrhopyginae species (178 per Mielke [23]) representing all major species groups, and 100% of genera. On one hand, we confirm the convergent origins of wing patterns; on the other hand, genomic analysis reveals many surprising instances of close relationships that were not apparent in morphological analyses due to drastic divergence of phenotypes. We conclude that taxonomists are better at splitting similar-looking but distantly related taxa than at bringing together different-looking but closely related butterflies.
2. Material and methods
Methods and protocols used in the work follow the ones published previously [10,13,24]. Additional details are provided in the electronic supplementary material, appendix. Briefly, DNA was extracted using MN kit from a leg taken off a dry pinned specimen and placed in 0.5 µl plastic tube. The leg was soaked in DNA extraction buffer overnight and the buffer was retained for further procedures. This method is not destructive and the leg is kept intact for future morphological analysis. The quality of extracted DNA was evaluated using gel electrophoresis and, if needed, DNA was fragmented enzymatically using NEBNext Ultra II FS Kit. Genomic libraries were prepared using NEBNext Ultra DNA Library Prep Kit and sequenced on HiSeq x10 at GENEWIZ. Protein-coding regions were assembled from reads using TBLASTN with Cecropterus lyciades [25] exons as a queries followed by a filtering procedure that removes possible paralogs and contamination. Mitogenomes were assembled as we reported previously [26,27]. Phylogenetic trees were constructed from DNA sequences (not translated proteins) using RAxML, BEAST and TreeMix, and gene trees in partitioned analyses (actual gene sequences were taken) were combined using ASTRAL (see electronic supplementary material, appendix, Methods for references to these programs). Celaenorrhinus syllius (tribe Celaenorrhinini) and Tagiades gana meetana (tribe Tagiadini) were chosen as outgroups because the two tribes they belong to are the closest to Pyrrhopyginae according to published phylogenies [13,17,20]. In addition to traditional bootstrap, we also used a more stringent method that tends to give lower support values to less confident nodes. While still using concatenated alignment, the trees are built from 1% of the data (0.1 million positions). The concatenated alignment was cut into 100 consecutive non-overlapping segments and each segment was used to construct the tree. The number of trees (out of 100) having a certain node is used as a measure of node's reliability (analogous to bootstrap). Genomic data have been deposited to NCBI with Bioproject ID PRJNA532323 and the alignments used for phylogeny construction available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.q0sr5p5 [28]. This work has been registered with ZooBank as http://zoobank.org/214D0E4D-3FC5-4E93-9F5F-EA1294D38A4C and published on 22 April 2019.
3. Results and discussion
(a). DNA preparation and specimen age
All genomic sequences were obtained from dry specimens stored pinned in standard insect collections. None were preserved specifically for genomic work. Specimen age ranged from several years to more than a century. The oldest specimens were collected more than 140 years ago (electronic supplementary material, table S1). The genomic DNA from samples that are more than 30 years old is frequently fragmented to segments shorter than 150 bp before our library preparation, and therefore their 150 bp sequence reads contain a large fraction of Illumina adapters. Comparing the quality of sequences at different ages, we observe that the true read length after removal of adapters decreases with age and so does the completeness of protein-coding genes (electronic supplementary material, figure S1). To test the quality of sequences in old specimens, we obtained segments of the COI barcode for several primary type specimens by traditional PCR followed by Sanger sequencing. These amplified sequenced segments resulted in barcodes that were 100% identical to those obtained from next-generation sequencing, similar to the results reported previously [29]. Moreover, in phylogenetic trees, older samples of the same species grouped together with more recently collected specimens, suggesting that these older specimens still contain DNA suitable for phylogenetic analysis. We consider the ability to obtain usable genomic reads and partial assemblies from specimens of essentially any age very important for this study. Every butterfly specimen in a traditional museum collection is a source of unique genomic information that can be harvested.
(b). 120 genomes
We selected representatives of all 35 currently recognized genera, and of 4 genera that are considered subjective junior synonyms with the type species different from those for their senior synonyms [23]. The majority of these genera are represented by their type species or their close relatives as judged by the COI barcode and morphology of the type species. Additionally, we included species with type specimens available for DNA analysis and species with most distinctive morphology. In total, 119 Pyrrhopyginae specimens were sequenced. We used 12 618 protein-coding genes from the Hesperiidae genomes we have assembled previously [6,25] to detect genes from shotgun genomic reads of these 119 specimens plus two outgroups (electronic supplementary material, table S1). Each specimen was represented in the alignment by 8 853 912 ± 3 455 622 positions. We also assembled mitochondrial genomes that were more than 75% complete in 113 specimens and covered 9464 ± 910 positions.
(c). Dated genomic tree of pyrrhopyginae classification
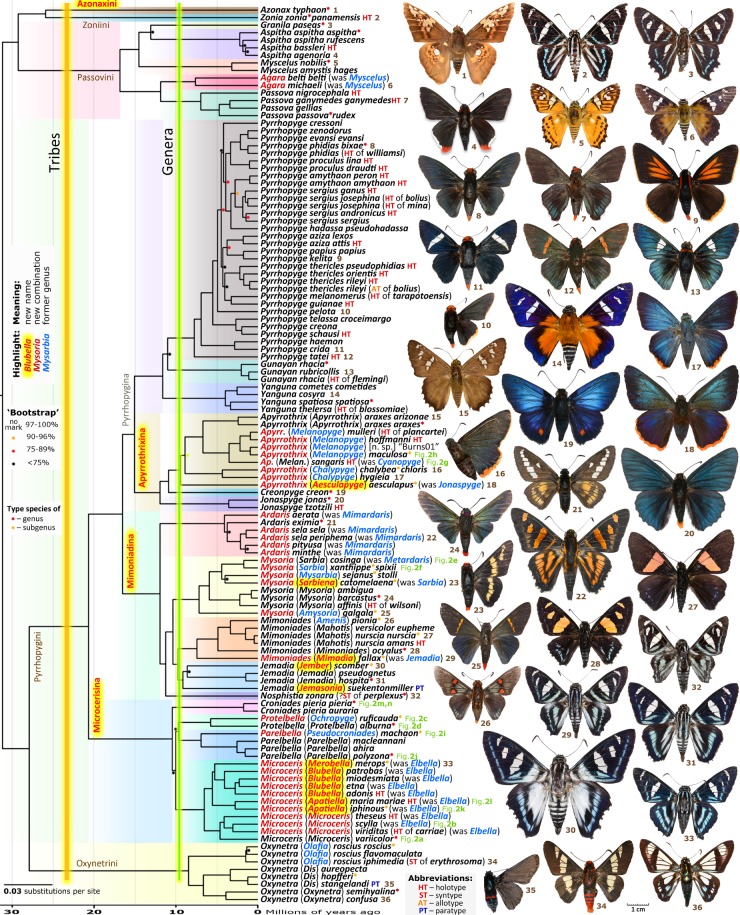
A genomic tree constructed on the concatenated alignment of protein-coding genes was dated and is shown in figure 1. Overall, we observe excellent agreement between our phylogenetic tree and the current classification of Pyrrhopyginae [23], thus largely confirming it. Our results are consistent in all different trees we have obtained (electronic supplementary material, figures S2–S7). Major branches near the base of the tree (figure 1) correspond to the tribes Passovini, Pyrrhopygini and Oxynetrini. Interestingly, Azonax is not placed in Passovini, but instead is confidently grouped with Zonia. Both of these genera diverged soon after their divergence from Passovini. Therefore, we agree that Zonia is best classified in a monotypic tribe Zoniini. Azonax is equidistant from other taxa, and a new monotypic tribe Azonaxini is proposed for it below. We found several polyphyletic (Myscelus, Jonaspyge, Sarbia, Jemadia) and paraphyletic (Mimoniades, Elbella) genera that we refine to ensure monophyly of all firetip genera. As a result, we outline 23 genera and, additionally, 22 subgenera, 10 of which are named here as new (see below and figure 1). The logic behind these changes is detailed in the electronic supplementary material, Taxonomic appendix.
Figure 1.
Dated genomic tree of Pyrrhopyginae. Specimens illustrated are the actual specimens sequenced. See electronic supplementary material, table S1 for specimen data. ‘Fig. 2’ refers to the illustration of this taxon (but not the specimen sequenced) in figure 2. ‘Bootstrap' is a bootstrap equivalent described in Material and methods.
(d). Convergence in wing patterns
Our phylogenetic analysis revealed an abundance of convergent wing patterns, confirming morphological studies. Most notably, the sinimustvalge pattern is present in representatives of 3 (out of 5) tribes and 9 genera: Zonia, Granila, Mimoniades (in M. fallax, formerly in Jemadia), Jemadia, Nosphistia, Croniades, Protelbella, Parelbella and Microceris [formerly Elbella]. Meanwhile, the other pattern, entirely black (sometimes shiny-metallic) wings with the firetip, and frequently orange head, is present in Protelbella, Microceris and other genera such as Passova, Pyrrhopyge, Jonaspyge, Mysoria and Oxynetra. At least one of these patterns (not clear which one) is convergent. Then there is a very close convergence between Microceris [formerly Elbella] iphinous and Mimoniades ocyalus (black wings with orange spots on the forewing and blue sprinkles on the hindwing), and Microceris [formerly Elbella] hegesippe and Mysoria [formerly Sarbia] xanthippe (black wings with yellow stripes crossed by black veins), among several others.
(e). Recurrent and pronounced phenotypic divergence
The most surprising result of this study is that several phenotypically distinct species are genetically close to others. Phenotypic distinctness hindered detection of their relationships with their close relatives and resulted in incorrect classification in the past. This rapid phenotypic divergence happened in several genera. The following are the five most prominent examples (figure 2).
Figure 2.

Rapidly diverging wing patterns in close relatives. Closely related or the same taxa are placed in the same row. Dorsal (left) and ventral (right) views are shown. A centimetre scale bar is shown on the left of each specimen.
Microceris variicolor (figure 2a) has an unusual and intricate pattern of wavy blue lines and yellow stripes. It is also characterized by rather unique genitalia with a compressed harpe. As indicated above, M. variicolor is closely related to the Elbella scylla group (figure 2b, black butterflies) and this relationship results in synonymizing the genus Elbella with Microceris. Compression of the harpe has been observed as individual variation in specimens of the same species [30], so apparently it is a modification that can be achieved on a short evolutionary time scale.
Protelbella [formerly Ochropyge] ruficauda (figure 2c) has black, shining, metallic-green wings with mostly white fringes, orange around the hindwing tornus. It does not resemble the sinimustvalge-patterned Protelbella alburna (figure 2d) and was previously placed in a monotypic genus Ochropyge thought to be more related to Crenopyge and Jonaspyge, metallic-black skippers. Genitalia of P. ruficauda and P. alburna are also rather distinct: highly elaborate with long prongs in P. ruficauda, but simpler and smoother in P. alburna.
In the next three examples, male genitalia are more similar. Mysoria cosinga (figure 2e) has almost all-yellow or orange wings with broad black margins, spots and veins. Placed in a monotypic genus Metardaris previously, it is a close kin of Mysoria [formerly Sarbia] xanthippe (figure 2f) that has mostly black wings with yellow stripes. Their genitalia are similar however: the harpe is distally bent. Apyrrothrix sangaris (figure 2g) has metallic greenish wings with a single red spot near the hindwing tornus. Its unique appearance earned it a monotypic genus Cyanopyge, because it is very different from other species in the subgenus Melanopyge with black white-spotted with orange wing bases below (figure 2h). Their genitalia are similar in the shape of valva and uncus. Parelbella [formerly Pseudocroniades] machaon (figure 2i) has yellow and white stripes and was placed in a monotypic genus before, but its relatives are sinimustvalge-patterned (figure 2j). Harpe of all these species is expanded in a T-shape, more crooked in P. machaon.
To reinforce the conclusion of rapid phenotypic divergence, we found that Microceris [formerly Elbella] iphinous (figure 2k, yellow-spotted forewings, blue-sprinkled hindwing) and Microceris [formerly Elbella] mariae (figure 2l, solid black wings) are very closely related (figure 1). Moreover, the difference between two morphs in females—orange, black-striped (figure 2m) and sinimustvalge (figure 2n)—of Croniades is equally striking, as are the two morphs of Microceris [formerly Elbella] luteizona—with yellow stripes and solid black—reported previously [18,21].
(f) Description of new taxa
See electronic supplementary material for additional information, sequences with diagnostic characters and species included in subgenera. All names of subgenera are treated as nouns in the nominative singular. Genera included in the tribes and subtribes are shown in figure 1 in the main text and are listed in electronic supplementary material, Taxonomic appendix section T2. ZooBank registration URL should be preceded by http://zoobank.org/ and is given for each taxon. ‘Description’ gives a taxon definition that states in words diagnostic characters to differentiate the taxon and gives a bibliographic reference to such a published statement. Words for DNA sequence characters from different genes are separated by semicolon, different sites in the same gene are separated by comma. Characters without a dot are for the COI barcode region and only their combination is diagnostic to distinguish from relatives. The word A79T means position 79 is T, changed from A in the ancestor; 59C means position 59 is C, but the ancestral state is unclear. Sequence characters with a dot are from nuclear genes, word 272.1.2:A192G means position 192 in exon 2 of the gene 1 on the scaffold 272 (Cecropterus lyciades reference) is G, changed from A in the ancestor.
Azonaxini Grishin, trib. n.
ZooBank: 6E3B9F8E-91C0-45BD-AF1F-78C5F2769392
Type genus: Azonax Godman & Salvin, 1893
Description: C83T, A139T, C206T, T208A, T376C, A400C, T484C, G512T, A526T, A583T; keys to A.15 in Evans (1951:5); uncus divided, gnathos I-shaped
Apyrrothrixina Grishin, subtr. n.
ZooBank: 8EEE17EE-CCD5-4A4A-9105-0F81E498C6FD
Type genus: Apyrrothrix Lindsey, 1921
Description: 300.8.1:G95T; 1838.7.1:T90C; 60.16.9: C66T; 2612.6.2:T640C; 2548.11.2:A71G; 822.26.1:A174G; A238Y, 286Y; keys to A.1.1 or 1.42a (except 44, 48 & 49b) in Evans (1951)
Mimoniadina Grishin, subtr. n.
ZooBank: D71E4FF9-F89D-4BE1-ABB1-F86F08B98817
Type genus: Mimoniades Hübner, 1823
Description: 7758.8.1:C31A; 318.14.16:T4044C; 990.1.14:A84G; 851.9.1:A186G; 502A; keys to A.4a, 6a (except 9 & 13) or A.1.44 in Evans (1951)
Microcerisina Grishin, subtr. n.
ZooBank: 2D1DB769-9A47-4BD1-A66D-9CD937825113
Type genus: Microceris E. Watson, 1893
Description: 276558.16.1:T219C, T222C; 536.39.1: G60A, 115.1:A576G; 207.4.1:T58C; 2954.5.2:C185G; 301A, 415T; keys to A.2, 9, 13, 14 or A.1.48 in Evans (1951); lateral lobe at distal end of aedeagus, apparently to support vesica
Aesculapyge Grishin, subgen. n.
ZooBank: D6952953-3744-402D-9A01-9D88246DAB47
Type species: Pyrrhopyge aesculapus Staudinger, 1876
Description: 31A, T59C, 85C, C206T, T208A, T250C, A280G, T379A, T490C, T581C; keys to A.1.46 in Evans (1951:32)
Derivation: Feminine, a blend of Aescula[pus] and [Pyrrho]pyge
Sarbiena Grishin, subgen. n.
ZooBank: D76F2A12-DB82-46E3-A06E-3EA038A0B0E0
Type species: Sarbia catomelaena Mabille & Boullet, 1908
Description: T25C, T97C, 130T, 214C, 352C, 364T, T367C, 412A, T428C, T616C, T622C; keys to A.10.2 in Evans (1951:63)
Derivation: Feminine, a blend of Sarbi[a] and [catomela]ena
Santea Grishin, subgen. n.
ZooBank: 86F43126-2F5D-491C-A6A5-C0CCC584E278
Type species: Pyrrhopyga antias C. & R. Felder, 1859
Description: 130A, 286G, T301C, 352C, 364T, T367C, 412G, A470G, T616C, T622A; keys to A.10.1 in Evans (1951:63)
Derivation: Feminine, a mix of Sarbia and antias, avoiding a homonym
Mimadia Grishin, subgen. n.
ZooBank: D9680514-89C4-42B8-BA11-F36A628652C8
Type species: Pyrrhopyga fallax Mabille, 1878
Description: T106C, T133C, A190T, T232C, A290C, T379A, T428C, C497T; keys to A.5.7 in Evans (1951:54)
Derivation: Feminine, a blend of Mim[oniades] and [Jem]adia
Jematus Grishin, subgen. n.
ZooBank: 8CE439F0-2FC8-4D67-BA9A-CB90CB47B447
Type species: Papilio gnetus Fabricius, 1781
Description: A38G, A55T, 82G, T121A, T358C, 391A, T394C, A415T, G512T, T553A; keys to A.5.6 in Evans (1951:54)
Derivation: Masculine, a blend of Jema[dia] and [gne]tus
Jember Grishin, subgen. n.
ZooBank: BD5E8AE8-1F65-4580-8288-2507928612D4
Type species: Jemadia scomber Druce, 1908
Description: T4A, A296G, 374A, A520T, T568C; keys to A.5.3a in Evans (1951:52); ampulla toothed, harpe bent dorsad
Derivation: Masculine, a blend of Jem[adia] and [scom]ber
Jemasonia Grishin, subgen. n.
ZooBank: 06E23C76-CCCF-4DBF-9129-6207C8315FCD
Type species: Pyrrhopyga [sic] hewitsonii Mabille, 1878
Description: T50C, A52T, G87A, T334A, T361C, A412T, G474A, T478C; keys to A.5.5 in Evans (1951:52)
Derivation: Feminine, a blend of Jema[dia] and [hewit]soni[i] + a
Merobella Grishin, subgen. n.
ZooBank: 227201CF-9B7E-4413-9624-A976A5045620
Type species: Jemadia merops E. Bell, 1934
Description: T46A, 190T, 223A, T319A, 506A, G512T; keys to A.2.10 in Evans (1951:43); harpe broadly bulbous
Derivation: Feminine, a blend of Mero[ps] and [El]bella
Blubella Grishin, subgen. n.
ZooBank: 373B8338-3A00-418E-A196-FF6312C88C2C
Type species: Pyrrhopyga [sic] patroclus Plötz, 1879
Description: 46T, 190A, 223A, 290A, 319T, 506G, 512G, 565A, 578T; keys to A.2.7, 12 or 13a in Evans (1951); harpe tapered, with small projection(s) at the base
Derivation: Feminine, a blend of Blu[e] and [El]bella
Apatiella Grishin, subgen. n.
ZooBank: 62010BD5-793C-429F-90DB-AA702B3C91EB
Type species: Hesperia iphinous Latreille, [1924]
Description: T91A, T205A, 220C, 223C, C284T, C343G, 346G, A520T, 544C, 565T; keys to A.2.8 or 9 in Evans (1951:42); harpe forked, tegumen processes short and rounded
Derivation: Feminine, a blend of Apat[e] (for deceipt) through i with [Elb]ella
Supplementary Material
Acknowledgements
We are grateful to Robert K. Robbins, John M. Burns and Brian Harris (National Museum of Natural History, Smithsonian Institution), David A. Grimaldi and Courtney Richenbacher (American Museum of Natural History), Weiping Xie (Los Angeles County Museum of Natural History), John Rawlins (Carnegie Museum of Natural History), John R. MacDonald and Richard L. Brown (Mississippi Entomological Museum), Wolfram Mey and Viola Richter (Berlin Museum für Naturkunde) for facilitating access to collections in their care and stimulating discussions, and to the late Edward C. Knudson for sampled specimens (now at the McGuire Center for Lepidoptera and Biodiversity). Special thanks to Olaf H. H. Mielke and Carlos Mielke for discussions, comments and sampling specimens for DNA analysis. We acknowledge the Texas Advanced Computing Center (TACC) at the University of Texas at Austin (http://www.tacc.utexas.edu) for providing invaluable HPC resources that were essential to carry out this study, which has been supported by the grants from the National Institutes of Health GM094575 and GM127390 and the Welch Foundation I-1505.
Data accessibility
Genomic data have been deposited to NCBI with Bioproject ID PRJNA532323 and the alignments used for phylogeny construction available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.q0sr5p5 [28]. This work has been registered with ZooBank as http://zoobank.org/214D0E4D-3FC5-4E93-9F5FEA1294D38A4C.
Authors' contributions
J.Z. developed the methods and performed the computations. Q.C. developed the methods. J.S. conducted DNA extraction and library preparation. E.B. and N.G. sampled specimens for DNA extraction. N.G. conceived the idea and supervised this work. All authors discussed the results and wrote the manuscript.
Competing interests
We declare we have no competing interests.
References
- 1.Patwardhan A, Ray S, Roy A. 2014. Molecular markers in phylogenetic studies: a review. J. Phylogen Evol. Biol. 2, 131 ( 10.4172/2329-9002.1000131) [DOI] [Google Scholar]
- 2.Chenuil A. 2006. Choosing the right molecular genetic markers for studying biodiversity: from molecular evolution to practical aspects. Genetica 127, 101–120. ( 10.1007/s10709-005-2485-1) [DOI] [PubMed] [Google Scholar]
- 3.Muir P, et al. 2016. The real cost of sequencing: scaling computation to keep pace with data generation. Genome Biol. 17, 53 ( 10.1186/s13059-016-0917-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.National Human Genome Research Institute. 2016. The cost of sequencing a human genome. Bethesda, MD: NHGRI.
- 5.Phillips KA, Pletcher MJ, Ladabaum U. 2015. Is the “$1000 Genome” really $1000? Understanding the full benefits and costs of genomic sequencing. Technol. Health Care 23, 373–379. ( 10.3233/THC-150900) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cong Q, Borek D, Otwinowski Z, Grishin NV. 2015. Skipper genome sheds light on unique phenotypic traits and phylogeny. BMC Genomics 16, 639 ( 10.1186/s12864-015-1846-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cong Q, Borek D, Otwinowski Z, Grishin NV. 2015. Tiger swallowtail genome reveals mechanisms for speciation and caterpillar chemical defense. Cell Rep. 10, 910–919. ( 10.1016/j.celrep.2015.01.026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cong Q, Shen J, Borek D, Robbins RK, Otwinowski Z, Grishin NV. 2016. Complete genomes of Hairstreak butterflies, their speciation, and nucleo-mitochondrial incongruence. Sci. Rep. 6, 24863 ( 10.1038/srep24863) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cong Q, Shen J, Li W, Borek D, Otwinowski Z, Grishin NV. 2017. The first complete genomes of Metalmarks and the classification of butterfly families. Genomics 109, 485–493. ( 10.1016/j.ygeno.2017.07.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cong Q, Shen J, Borek D, Robbins RK, Opler PA, Otwinowski Z, Grishin NV. 2017. When COI barcodes deceive: complete genomes reveal introgression in hairstreaks. Proc. R. Soc. B 284, 20161735 ( 10.1098/rspb.2016.1735) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Triant DA, Cinel SD, Kawahara AY. 2018. Lepidoptera genomes: current knowledge, gaps and future directions. Curr. Opin. Insect Sci. 25, 99–105. ( 10.1016/j.cois.2017.12.004) [DOI] [PubMed] [Google Scholar]
- 12.Cong Q, Shen J, Warren AD, Borek D, Otwinowski Z, Grishin NV. 2016. Speciation in cloudless sulphurs gleaned from complete genomes. Genome Biol. Evol. 8, 915–931. ( 10.1093/gbe/evw045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li W, Cong Q, Shen J, Zhang J, Hallwachs W, Janzen DH, Grishin NV. 2019. Genomes of skipper butterflies reveal extensive convergence of wing patterns. Proc. Natl Acad. Sci. USA 116, 6232–6237. ( 10.1073/pnas.1821304116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Radwan J, Babik W. 2012. The genomics of adaptation. Proc. R. Soc. B 279, 5024–5028. ( 10.1098/rspb.2012.2322) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wen D, Yu Y, Hahn MW, Nakhleh L. 2016. Reticulate evolutionary history and extensive introgression in mosquito species revealed by phylogenetic network analysis. Mol. Ecol. 25, 2361–2372. ( 10.1111/mec.13544) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu L, Edwards SV. 2009. Phylogenetic analysis in the anomaly zone. Syst. Biol. 58, 452–460. ( 10.1093/sysbio/syp034) [DOI] [PubMed] [Google Scholar]
- 17.Toussaint EFA, et al. 2018. Anchored phylogenomics illuminates the skipper butterfly tree of life. BMC Evol. Biol. 18, 101 ( 10.1186/s12862-018-1216-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evans WH. 1951. A catalogue of the American Hesperiidae indicating the classification and nomenclature adopted in the British Museum (Natural History). Part I. Introduction and group A Pyrrhopyginae. London, UK: British Museum (Natural History; ). [Google Scholar]
- 19.Burns JM, Janzen DH. 2001. Biodiversity of pyrrhopygine skipper butterflies (Hesperiidae) in the Area de Conservación Guanacaste, Costa Rica. J. Lepidopterists' Society 55, 15–43. [Google Scholar]
- 20.Warren AD, Ogawa JR, Brower AVZ. 2009. Revised classification of the family Hesperiidae (Lepidoptera: Hesperioidea) based on combined molecular and morphological data. Syst. Entomol. 34, 467–523. ( 10.1111/j.1365-3113.2008.00463.x) [DOI] [Google Scholar]
- 21.Mielke OHH. 1995. Revisão de Elbella Evans e gêneros afins (Lepidoptera, Hesperiidae, Pyrrhopyginae). Revista brasileira de Zoologia 11, 395–586. ( 10.1590/S0101-81751994000300001) [DOI] [Google Scholar]
- 22.Mielke OHH. 2002. Pyrrhopyginae: gêneros novos e revalidados (Lepidoptera: Hesperiidae). Revista brasileira de Zoologia 19, 217–228. ( 10.1590/S0101-81752002000100020) [DOI] [Google Scholar]
- 23.Mielke OHH. 2005. Catalogue of the American hesperioidea: hesperiidae (lepidoptera). Curitiba, Paraná, Brazil: Sociedade Brasileira de Zoologia. [Google Scholar]
- 24.Janzen DH, Burns JM, Cong Q, Hallwachs W, Dapkey T, Manjunath R, Hajibabaei M, Hebert PDN, Grishin NV. 2017. Nuclear genomes distinguish cryptic species suggested by their DNA barcodes and ecology. Proc. Natl Acad. Sci. USA 114, 8313–8318. ( 10.1073/pnas.1621504114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen J, Cong Q, Borek D, Otwinowski Z, Grishin NV. 2017. Complete Genome of Achalarus lyciades, The First Representative of the Eudaminae Subfamily of Skippers. Curr Genomics 18, 366–374. ( 10.2174/1389202918666170426113315) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen J, Cong Q, Grishin NV. 2015. The complete mitochondrial genome of Papilio glaucus and its phylogenetic implications. Meta Gene 5, 68–83. ( 10.1016/j.mgene.2015.05.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen J, Cong Q, Grishin NV. 2016. The complete mitogenome of Achalarus lyciades (Lepidoptera: Hesperiidae). Mitochondrial DNA B Resour 1, 581–583. ( 10.1080/23802359.2016.1197070) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J, Cong Q, Shen J, Brockmann E, Grishin N. 2019. Data from: Genomes reveal drastic and recurrent phenotypic divergence in Firetip skipper butterflies (Hesperiidae: Pyrrhopyginae). Dryad Digital Repository ( 10.5061/dryad.q0sr5p5) [DOI] [PMC free article] [PubMed]
- 29.Prosser SW, deWaard JR, Miller SE, Hebert PD. 2016. DNA barcodes from century-old type specimens using next-generation sequencing. Mol. Ecol. Resour. 16, 487–497. ( 10.1111/1755-0998.12474) [DOI] [PubMed] [Google Scholar]
- 30.Grishin NV, Janzen DH, Hallwachs W. 2013. Hiding behind gaudy looks, a new Central American species of Phareas (Hesperiidae: Eudaminae). J. Lepidopterists’ Soc. 67, 161–174. ( 10.18473/lepi.v67i3.a3) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Zhang J, Cong Q, Shen J, Brockmann E, Grishin N. 2019. Data from: Genomes reveal drastic and recurrent phenotypic divergence in Firetip skipper butterflies (Hesperiidae: Pyrrhopyginae). Dryad Digital Repository ( 10.5061/dryad.q0sr5p5) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Genomic data have been deposited to NCBI with Bioproject ID PRJNA532323 and the alignments used for phylogeny construction available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.q0sr5p5 [28]. This work has been registered with ZooBank as http://zoobank.org/214D0E4D-3FC5-4E93-9F5FEA1294D38A4C.