Abstract
In this review, we describe a path for translation of gene editing into therapy for cystic fibrosis (CF). Cystic fibrosis results from mutations in the CFTR gene, with one allele predominant in patient populations. This simple, genetic etiology makes gene editing appealing for treatment of this disease. There already have been success in applying this approach to cystic fibrosis in cell and animal models, although these advances have been modest in comparison to advances for other disease.
Less than six years after its first demonstration in animals, CRISPR/Cas gene editing is in early clinical trials for several disorders. Most clinical trials, thus far, attempt to edit genes in cells of the blood lineages. The advantage of the blood is that the stem cells are known, can be isolated, edited, selected, expanded, and returned to the body. The likely next trials will be in the liver, which is accessible to many delivery methods. For cystic fibrosis, the biggest hurdle is to deliver editors to other, less accessible organs. We outline a path by which delivery can be improved.
The translation of new therapies doesn't occur in isolation, and the development of gene editors is occurring as advances in gene therapy and small molecule therapeutics are being made. The advances made in gene therapy may help develop delivery vehicles for gene editing, although major improvements are needed. Conversely, the approval of effective small molecule therapies for many patients with cystic fibrosis will raise the bar for translation of gene editing.
Keywords: CFTR gene, CRISPR/Cas9, Cystic fibrosis, Gene editing, Gene therapy
Cystic fibrosis
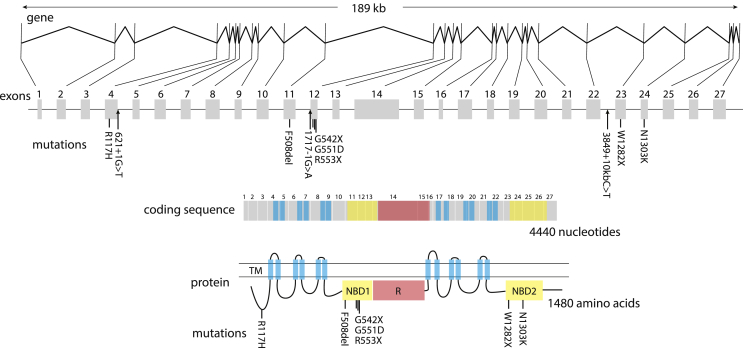
Cystic fibrosis (OMIM 219700) is an autosomal recessive disease caused by mutations in CFTR, a 1480 amino acid-long anion transporter1, 2 (Fig. 1). Cystic fibrosis causes severe impairment of lung function, serious pathology of the pancreas and gut, male infertility and reduced growth. The disease affects about 70,000 individuals worldwide, and is more prevalent in people of Northern European descent.3
Figure 1.
The structure of the human CFTR transcription unit, coding sequence and protein. The CFTR transcription unit is slightly less than 189,000 bases long with 27 exons. The most common disease-causing mutations are shown, as are three intronic mutations which lead to aberrant splicing (arrows). The CFTR protein forms a membrane channel with twelve transmembrane domains (TM), two nucleotide-binding domains (NBD1 and NBD2) and a regulatory domain (R).
The most common mutation (70% allele frequency with ∼50% of patients homozygous for this allele) is an in-frame deletion of a phenylalanine (F508del, also known as ΔF508) which impairs protein folding, maturation and transport to the surface of the cell. The remaining disease-causing mutations are diverse nonsense, missense, partial duplication and splice site mutations.1, 2 The severity of the disease varies both with the CFTR allele and with variants elsewhere in the genome.4, 5
Absence of the anion channel CFTR (cystic fibrosis transmembrane conductance regulator) causes electrolyte and transepithelial fluid imbalance resulting in viscous mucus along the epithelial lining of organs impairing both the pulmonary and gastrointestinal systems. The difficult-to-clear, thick sticky airway mucus in patients with cystic fibrosis contributes to lung infections and inflammation eventually leading to lung failure. Early pancreatic duct obstruction causes pancreatic insufficiency (PI), which is common is CF patients (∼85%), leading to impaired secretion of digestive enzymes, disrupting digestion, absorption and growth. PI, along with excessive mucus in the intestine, leads to intestinal obstructions in CF such as meconium ileus, distal intestinal obstruction syndrome and constipation.6
Survival and quality of life for cystic fibrosis patients have improved with the development of therapies, with some of the most significant resulting from pancreatic enzyme replacement and, more recently, CFTR modulators.7, 8, 9, 10, 11, 12, 13, 14 The FDA-approved CFTR modulator drugs have two different mechanisms of action: correctors increase the amount of protein which makes it to the cell surface (VX-809 or lumacaftor and VX-661 or tezacaftor), and potentiators which improve channel function (VX-770 or ivacaftor).
New drugs (VX-445 and VX-659) that increase CFTR at the cell surface by a mechanism complementary to the first generation of correctors have been developed. The two types of corrector and the potentiator have additive effects in cells and the triple combination therapies have been shown to be safe and efficacious in clinical trials.13, 14
The treatments for cystic fibrosis are expensive, and the associated therapies are time-consuming. Moreover, an estimated 10% of patients will not be responsive to these classes of drugs, most notably the nonsense mutations which eliminate protein expression. The hope is that somatic gene editing would provide a one-time cure, without the need for continuous expensive and time-consuming therapies. Although the costs of gene editing therapies are likely to be high (current gene therapies, for example, cost about $1 million), a successful one-time cure would be cheaper than continuous drug therapy at current prices (about $250,000 per year).
In the following few sections, the mechanisms of gene editing are described to illuminate why current editors have limitations for therapeutic application to cystic fibrosis, and how these limitations will be overcome.
The origins of gene editing
It was studies in yeast that laid the foundation for gene editing.15 Although this work was focused on the mechanisms of DNA repair, it showed that gene editing would be possible if one could direct DNA double-strand breaks to a single site in the genome.
Haploid baker's yeast (S. cerevisiae) can switch sex (mating type) at every division.16 The mRNA for HO endonuclease segregates into the daughter cell, where the HO protein cuts a single site in the daughter-cell genome. This DNA double-strand break leads to replacement of sequences around the break with DNA from donor sites, one donor for each sex. The sequences of both DNA donors have homology to the recipient site. Once incorporated into the mating type locus, the recombined DNA is expressed as a protein which determines the mating type.
James Haber and his laboratory used the HO-induced break to study the mechanisms of DNA repair.15 There were experimental advantages to studying DNA repair at one specific site rather than at multiple, arbitrary sites in the genome as induced by DNA damaging agents. The Haber lab determined the components and mechanism of homologous recombination (HR) that lead to local sequence replacement. Haber also showed that DNA repair by a second mechanism--non-homologous end-joining (NHEJ)--can generate small insertions and deletions (indels) at the DNA double-strand break. Subsequently, Maria Jasin engineered mammalian tissue culture cells to have a single site for a meganuclease called I-SceI.17 Jasin was able to show that the same two repair mechanisms (NHEJ and HR) are at work in mammalian cells. The investigations of Haber and Jasin led to the prediction that if one could program DNA endonucleases to cut at desired sites in the genome, one could use cells' DNA repair machinery to mutate any gene with NHEJ, and could direct desired sequence changes by HR at the DNA double-strand break.
This prediction was fulfilled shortly after when designer nucleases were developed. Four different families of designer nucleases have been developed: meganucleases, zinc finger nucleases (ZFNs), TALE nucleases (TALENs) and CRISPR/Cas nucleases.18, 19 All four types of nuclease can be programmed to create double-strand breaks at sequences sufficiently long and complex to be unique in the genome. All four families of nuclease have been used for gene editing. However, these designer nucleases differ in their ease of programming with CRISPR/Cas nucleases by far the easiest to program. It was their simplicity of design which led to widespread adoption in basic research, which in turn led to further improvements in specificity and activity, making them candidates for therapeutic use.20, 21
What is CRISPR/Cas?
In bacteria and archaea, CRISPR/Cas is a diverse set of adaptive immune systems which battles infections by degrading nucleic acids.22, 23 Cas9 is one of the nucleases of CRISPR/Cas, an RNA-guided DNA endonuclease.24 The infectious nucleic acid is degraded if a short segment of its sequence is represented in the cell's Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR). Transcripts of CRISPR are processed into short RNAs which have complementary segments to the infectious nucleic acids. The short RNAs bind to an endonuclease (Cas9 in some systems) and hybridize to the incoming nucleic acids, activating a nuclease to trigger degradation. New sequences can be added to the array in a nonlethal infection, for example those in which the host's Restriction-Modification system, a second non-adaptive nuclease-based immune system, inactivated the phage.25 Sequences in the host CRISPR are not cut because the genomic versions lack a short, shared motif adjacent to the unique sequence. This short motif, or proto-spacer adjacent motif (PAM), is present in the original infectious DNA, but is not copied into the genome. A PAM sequence can be very simple, for example NGG. The absence or presence of the PAM sequence is the determinant by which the cell distinguishes self from non-self.
Genes adjacent to CRISPR--the CRISPR associated (Cas) genes--encode the proteins necessary for acquisition of new sequences and execution of immunity.22, 23 The endonuclease most frequently used by researchers, Cas9, is an RNA-guided DNA endonuclease from Streptococcus pyogenes, a strep throat bacterium.24 Sp Cas9 has practical advantages over some other CRISPR nucleases in that it is a single protein, and it has high activity and specificity at mammalian body temperature. There are many different CRISPR systems to be explored which may be a source of nucleases with their own unique and advantageous properties that can be utilized for future gene editing strategies.
How Cas9 cuts DNA in a sequence-specific manner
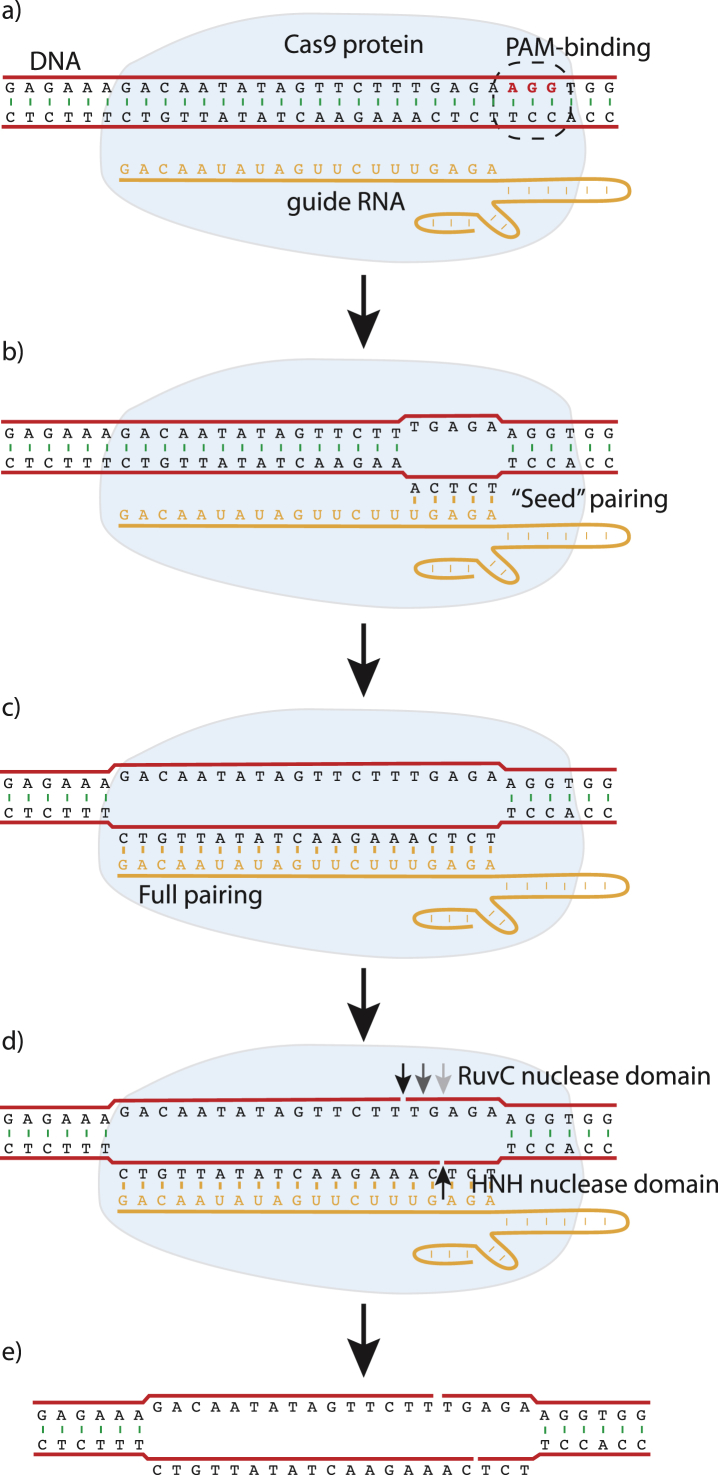
The mechanism of Cas9's endonuclease activity determines both its specificity and mutagenicity24 (Fig. 2). Cas9 is a large (1368 amino acids) protein with several functional domains. When Cas9 binds a guide RNA (gRNA), Cas9's C-terminal domain gains the ability bind to double-stranded DNA with a PAM (NGG in Cas9).24 When binding the PAM, Cas9 induces a kink in the DNA strand complementary to the NGG (Fig. 2a). This kink locally melts the DNA double helix and frees the bases of the kinked strand to rotate and attempt to pair with the gRNA (Fig. 2b). If hybridization occurs between the nucleotides in the free strand adjacent to the PAM, then the DNA strands separate and hybridization with the remainder of the guide is attempted (Fig. 2c). If hybridization is complete through the entire 20 base pairs of the gRNA, then Cas9's two nuclease domains (the HNH— and RuvC-like nuclease domains) are activated24 (Fig. 2d). Each DNA strand is cut close to the PAM, within the region bound by the gRNA. The DNA strand paired with the gRNA is cut 3 nucleotides away from the PAM, while the unhybridized strand is cut 3 to 6 nucleotides away.26, 27, 28, 29 The location of the cut on the free DNA strand varies with the target sequence. In some cases, Cas9 only cuts at one position, while in others Cas9 cuts at several. Thus, although the break made by Cas9 is commonly thought of as a double-strand break, in many cases it is not a clean, blunt-ended break, but has short 5′ overhangs (Fig. 2e). The ragged break contributes to Cas9's mutagenicity by preferentially engaging the error-prone alternative NHEJ repair pathway, instead of the more accurate, canonical NHEJ pathway.29
Figure 2.
Cas9 sequence recognition and cleavage. a) The complex of Cas9 protein and guide RNA bind DNA at the sequence NGG (the protospacer-adjacent motif or PAM). b–c) If the DNA strand opposite NGG and immediately downstream can base-pair with the guide RNA, then hybridization with the remainder of the guide RNA is attempted. c–d) If hybridization to the guide RNA is complete, then the two nuclease domains of Cas9 are activated. The HNH nuclease domain of Cas9 cuts the DNA strand paired with the guide RNA 3 nucleotides from the PAM. The Cas9 RuvC nuclease domain cuts the unpaired strand 3 to 6 nucleotides from the PAM. e) The most frequent product is a DNA double-strand break with short 5′ overhangs.
The sequence specificity of Cas9 is related to the sequence of molecular events: the earlier the event, the more essential it is for Cas9 activity.24 Cas9 will not bind a PAM without first binding a guide RNA, PAM binding is required before hybridization to the seed, and seed hybridization is required for hybridization at the 5′ half of the guide RNA. Thus, for nuclease activation, a gRNA is absolutely required, PAM binding is absolutely required, stable hybridization to seed sequences is very important, and stable hybridization to the remainder of the gRNA is important, but less so than the preceding step.
Although prediction of off-target activity has some subtleties, usually only one or two sequence mismatches are compatible with nuclease activation and only if the mismatches are outside the seed.30, 31, 32 Together, this results in zero nuclease activity at unrelated sites, and off-target cutting only at sites having a PAM and closely related sequences adjacent to the PAM. This means that off-target cutting and mutagenesis is predictable, and can either be avoided by selection of appropriate targets or at least monitored if unavoidable.
From site-specific cut to site-specific mutagenesis
The Cas9-generated DNA break can be repaired accurately, and likely is repaired accurately most of the time. But, if Cas9 and guide RNA are present after the repair, precise repairs can be cut again. Thus, there may be cycles of cutting and repair while the cell is arrested by cell cycle checkpoint. The cycles of cutting and repair and cell cycle checkpoint are escaped when an error arises which prevents further cutting, releasing the cell to propagate a mutation.
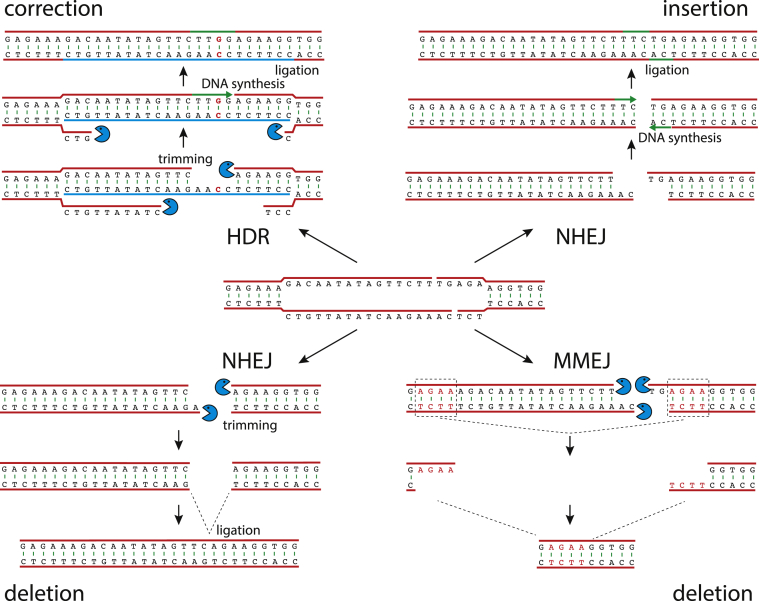
Most frequently, the mutation which arises from a Cas9-generated double-strand break is a small deletion of less than 10 base pairs33, 34, 35 (Fig. 3). The nature of some deletions is consistent with cellular enzymes removing the 5′ overhangs and ligating the two ends.26, 27, 28, 29 In other cases, double-strand breaks are repaired by microhomology-mediated end joining, where short sequences at the break are recombined with local similar sequence to generate deletions.
Figure 3.
The repair of DNA double-strand breaks generated by Cas9 occurs by multiple pathways that can lead to correction, insertion and deletion. The break can be bridged by hybridization with a transduced single-stranded oligonucleotide (HDR, top left), the overhangs of the double strand break are removed by exonucleolytic digestion followed by DNA synthesis and ligation leading to correction if nucleotide substitutions (in red) are included in the oligonucleotide. Small insertions (NHEJ, top right) can occur if the 5′ overhangs are filled in by cellular DNA synthesis, followed by ligation. Small deletions (NHEJ, bottom left) can occur if the overhangs are digested and the DNA ends ligated. Large deletions (MMEJ, bottom right) can occur if microhomologous sequences on either side of the break are recognized, leading to pairing and ligation.
Small insertions occur less frequently, and typically are duplications of a few nucleotides at the break.26, 27, 28, 29 In this case, cellular polymerases fill in both 5′ overhangs at the ends before ligation, leading to duplication of the nucleotides which were present in the overhangs.
When surveyed across many guide RNAs, indels appear somewhat random. However, some guide RNAs lead to highly reproducible mutations, while others have a more varied pattern. The nonrandom spectrum of indels generated by a given gRNA can mean that gRNAs cannot be assumed to generate null alleles by frameshift mutations, and indel patterns need to be empirically determined for each guide. Larger deletions, constituting up to 10% of all Cas9-induced mutations also occur.36 The mechanism that leads to the larger deletions may be microhomology-mediated end-joining (MMEJ), which generates deletions between very short homologous sequences on either side of the break.
When specific sequence changes are required, a single-stranded DNA oligonucleotide which can bridge the break is introduced at the same time as Cas9 and the gRNA.37 The oligonucleotide is designed to match sequences on both sides, but incorporates desired sequence changes. The likelihood that a new nucleotide will be incorporated into the final sequence is greatest if the change is within a handful of nucleotides of the break.38 This strong distance-dependence may be due to mismatch repair and processive cellular exonucleases. If DNA adjacent to the break is not removed, hybridization of the oligonucleotide with the endogenous strand will result in a mismatched base pair. The mismatched base will be preferentially removed from the oligonucleotide by mismatch repair, which is biased to preserve the “older” strand.
Recombination with the single-stranded oligonucleotide around the double-strand break is mechanistically distinct from homologous recombination (HR). HR is normally active only during G1 and S phases, requires cellular proteins dedicated to it, and requires double-stranded DNA.39 In contrast, recombination with the oligonucleotide can occur at any time in the cell cycle and in non-dividing cells.40 Because the mechanism of recombination with the bridging oligonucleotide is distinct, it is called homology-directed repair (HDR). Both HDR and HR (double-stranded targeting vector) are stimulated by double-strand breaks made by Cas9 and sometimes the designation is used interchangeably in the literature, causing lack of clarity.
The limitations of editing with double-strand breaks
Even under optimal conditions, HDR at a double-strand break occurs only one-tenth as often as indels.41 A number of strategies have been used to inhibit indel formation to favor HDR, but none yet has been able to make large changes in the ratio. While indel mutations in an already mutant allele might not seem to matter, these on-target, undesired mutations can convert druggable alleles into non-druggable alleles. With a disease like cystic fibrosis, where small molecule drugs are already effective against specific mutations (Table 1), converting a majority of a patient's druggable alleles to non-druggable nonsense alleles would be a step backward.
Table 1.
The ten most common CF disease alleles.
| Allele Frequencya | Mutation | DNA Change |
Class | CFTR Modulator |
|---|---|---|---|---|
| 70% | F508del | delCTT | deletion | VX-661/VX-770 VX-809/VX-770 |
| 2.5% | G542X | G > T | transversion | |
| 2.1% | G551D | G > A | transition | VX-770 |
| 1.5% | N1303K | C > G | transversion | |
| 1.3% | R117H | G > A | transition | VX-770 |
| 1.2% | W1282X | G > A | transition | |
| 0.93% | R553X | C > T | transition | |
| 0.93% | 621+1G > T | G > T | transversion | |
| 0.86% | 1717-1G > A | G > A | transition | |
| 0.82% | 3849 + 10kbC > T | C > T | transition | VX-661/VX-770 |
Data from the CFTR2 project.62
In some cell types, checkpoint is sensitive enough that one Cas9-generated double-strand break leads to activation of p53 and cell cycle arrest or apoptosis.42, 43, 44 The worry is that stem cells may have sensitive triggers for checkpoint, and thus the very cells that one is most interested in editing might be eliminated by apoptosis.
The precise sequence access afforded by Cas9 can be separated from double-strand breaks by mutating the nuclease domains of Cas9. In what follows, base editors are highlighted as an example of an innovative and imaginative approach which overcomes the limitations posed by indels and checkpoint by accomplishing editing without the mutagenic double-strand breaks.
Base editors: editing without mutagenic double-strand breaks
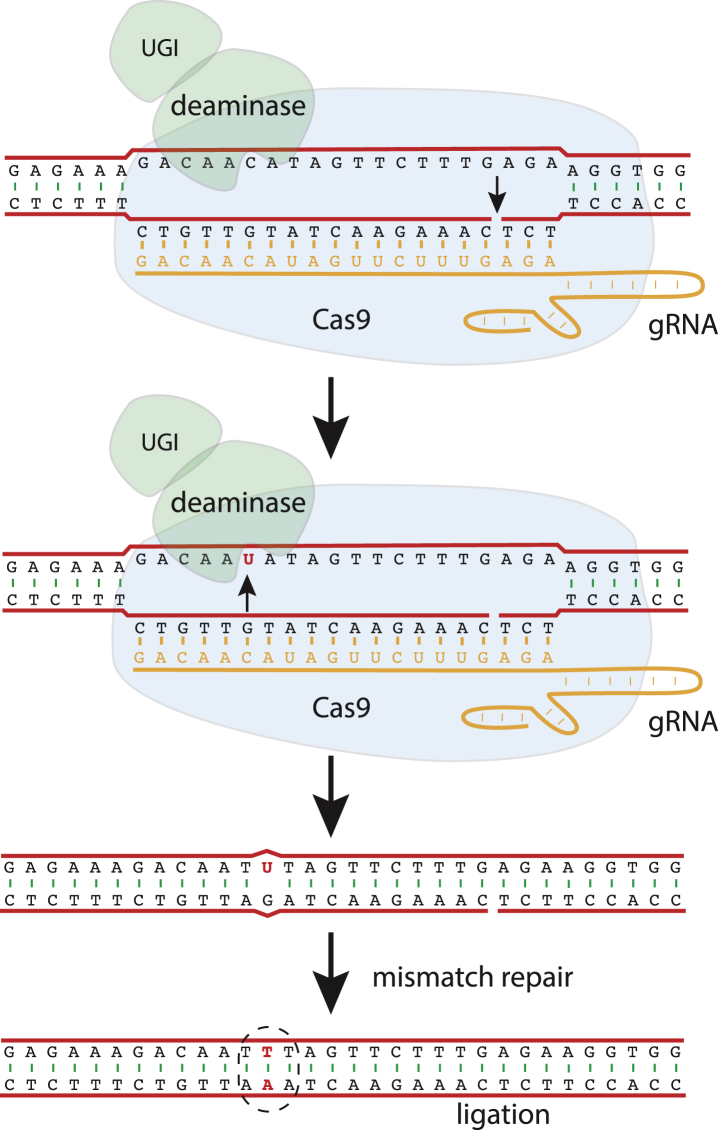
Researchers have used gRNA/Cas9 complexes to deliver other proteins to precise targets in the genome. Cargoes have included transcriptional activators and repressors, epigenetic modifiers and base deaminases. When Cas9 is fused to a base deaminase, these have been termed base editors (Fig. 4). Base editors have been constructed which can convert C to T, or A to G (through U and I, respectively).45, 46, 47, 48, 49, 50, 51, 52, 53 When targeted to the complementary strand, they convert G to A, or T to C.
Figure 4.
The mechanism of a C to T base editor (BE3 of 48). The base editor, consisting of a protein fusion of Cas9, cytidine deaminase and uracil glycosylase inhibitor (UGI), binds to a specific DNA target as shown in Fig. 2. The RuvC-like nuclease domain of Cas9 is inactivated and thus only the DNA strand paired with the gRNA is cut. The cytidine deaminase is only able to reach the sixth nucleotide on the unpaired strand, and if this base is a cytidine it is deaminated to uracil. The uracil glycosylase inhibitor prevents cellular enzymes from removing the uracil, and the break on the opposite strand stimulates removal of the mis-paired G and thus the templated change to T.
A base deaminase can act only on single-stranded DNA, since DNA bases are not accessible in double-stranded DNA. In the gRNA/Cas9/DNA complex, one DNA strand is unpaired and the bases are accessible to the enzyme. If the linker between Cas9 and the deaminase is of an appropriate length, the deaminase can reach only one or a few bases in the unpaired strand. The restriction of deaminase activity to a few unpaired bases in the target sequence combined with the high specificity of the gRNA/Cas9 complex yields off-target mutation frequencies that are below the limit of detection.
In addition to being highly specific, base editors can be remarkably efficient. Base conversion rates can be as high as 80% of alleles, substantially greater than the 30% achieved thus far with double-strand break-mediated HDR.48, 52 To reach the highest rates of base editing, one DNA strand, the one opposite the edited base, is cut by Cas9 (Fig. 4). The break on the unedited strand biases mismatch repair such that the base on the cut strand is removed, leaving the edited base to be used as a template.
The DNA single-strand break used to bias mismatch repair incurs a cost in the form of indel formation. However, because single-strand breaks are much less mutagenic than double strand breaks, the indel mutation rate is much lower. This phenomenon is reflected in the ratios of correction to indels. With a Cas9 double-strand break and a bridging oligonucleotide to drive HDR, the ratio of correction to indels is about 1–10. With base editors and a single-strand break, the ratio is the inverse or greater, about 10 to 1 for current C to T base editors and about 200 to 1 for A to G52
While base editors are powerful, base editors cannot correct all disease-causing mutations. Base editors only can exchange one purine base for another, or one pyrimidine base for another. Disease-causing mutations are much more diverse, including deletions, insertions and exchanges of purines for pyrimidines (Table 1). In addition, the extremely high specificity of base editors is currently a limitation. Because the C or A which can be edited must be a precise distance from, and on the same strand as the NGG PAM, only a small subset of C to T and A to G edits are currently feasible. As there has been rapid progress in this area, it is likely that at least some of the limitations of base editors will be overcome.
Other editing technologies
In addition to CRISPR/Cas-based systems, there are other approaches to editing. The other nuclease families--particularly the ZFNs and TALENs--still are applicable to therapeutic gene editing. In the development of a therapeutic, the time required to program new specificities is not rate-limiting, thus the advantage provided by Cas9 in a research setting is lost.
There is a non-nuclease form of editing, wherein a triple-helix forming oligonucleotide-based clamp binds close to the site of the desired edit, and the edit is directed by a second, single-stranded oligonucleotide.54, 55, 56 Although the efficiencies of editing achieved by this approach may not yet be as high as needed in a therapeutic setting, it is a promising technology. The clamp approach has significant advantages in that it appears to have greater flexibility than base editors with respect to the classes of mutations (small indels, transition and transversions) it can correct, and it does not convert druggable alleles to non-druggable alleles at a high rate.
CF allele frequencies
The distribution of CFTR mutant alleles in the patient population is unusual for a recessive genetic disease in that one allele is much more prevalent than others (Table 1). There are two competing hypotheses for the high prevalence of the F508del mutation. It has been proposed that one copy of F508del provided a survival advantage in the face of an infectious disease, similar to HbS and malaria.57, 58 As appealing as this idea is, both the disease and mechanism of resistance are hypothetical at this point. Alternatively, it has been suggested that F508del was driven to high frequency by population bottlenecks in Europe.59 Whatever its origins, the three nucleotides removed by the F508del mutation constrain strategies to correct it--as pointed out, base editors cannot correct this mutation. On the other hand, an editing approach which does work for this allele can be applied to most CF patients.
The six next most common CFTR mutations are single nucleotide missense or nonsense mutations followed by three single nucleotide splicing mutations (Table 1). Three of the ten most common mutations are base transversions--not the transitions which are correctable by base editors. Several hundred other disease-causing mutations are dispersed along the length of the gene and are a diverse set of substitution, insertion and deletion mutations.2, 60, 61, 62
Genetic strategies
For many in this field, the greatest promise of gene editing lies in precise correction of disease variants. However, ongoing and proposed clinical trials have focused not on precise corrections, but on insertions and deletions, many carried out with ZFNs and TALENs.63, 64, 65, 66, 67, 68 With large insertions and deletions of coding sequences, the small indels created by double-strand breaks are less problematic. Small indels are inconsequential, because either the intention is to delete a gene, or to insert DNA into an intron where precise conservation of the target sequence does not matter. Another significant advantage is that one insertion or deletion strategy can be applied to multiple alleles of the same gene.
A single, targeted insertion strategy could be applied to many different CF disease alleles.69 The goal is to insert coding sequences into an intron in the CFTR gene.70 The inserted DNA would have a splice acceptor and a continuous open reading frame for the downstream exons. The earlier the intron in the gene, the greater the number of different alleles which could be corrected by this strategy.
A targeted insertion is similar to gene therapy, but has several advantages. The promoter, enhancer and most other cis-acting regulatory sequences derive from the insertion site and thus neither need to be characterized nor incorporated into the incoming DNA. Reduction of the size of the incoming DNA will increase the frequency of insertion. Targeted integration would eliminate the widespread stochastic insertional mutagenesis that occurs with gene therapy. Lastly, insertion into a permissive locus prevents the epigenetic silencing that is seen at some sites with arbitrary insertions.71, 72, 73
DNA sequences can be inserted at a double-strand break by three different mechanisms: HDR, HR and NHEJ. The large size of the CFTR coding sequence likely makes the single-stranded DNA vectors necessary for HDR impractical. HR only occurs in G1 or S phase of dividing cells, so HR may be too inefficient for the slowly dividing airway epithelium. Although NHEJ is thought of as the mechanism of closing a double-strand break, it also can be used to insert DNA. In the case of NHEJ-mediated insertion, half of insertions would be inverted relative to the gene. A strategy called homology-independent targeted integration (HITI) has been developed to rescue inverted insertions.74 In HITI, the Cas9 cleavage target in the genome is added to the incoming DNA in an inverted orientation. If the incoming DNA is inserted in reverse orientation, the two cleavage sites will flank the insertion. If Cas9 is still present, the reversed insertion can be cut out and may re-integrate in the correct orientation. With HITI, insertion in the correct orientation can be as high as nearly 60% of all mutational events.
With a strategy of insertion into an intron, small indels should be benign as long as cis-acting sequences are avoided when choosing the target site. Large deletions, if they do occur, can lead to gene inactivation, but large deletions constitute less than 10% of induced mutations at least in the small number of loci examined thus far in mice.36
Since the majority of disease-causing variants are loss-of-function mutations, the utility of deletions for correction is usually either indirect or allele-specific. Indirect correction by deletion of another gene is the current strategy for the globinopathies, which result from mutations to the beta-globin gene. In these diseases, drugs which increase fetal beta-globin lessen disease symptoms. Accordingly, globinopathy deletion strategies inactivate a repressor of fetal globin expression.75 An allele-specific deletion strategy has been proposed for Duchenne Muscular Dystrophy (DMD).67 A number of DMD disease-causing mutations cause premature termination. Reading-frame-conserving deletion of the mutated exons allows expression of an internally deleted dystrophin that partially restores function.
For cystic fibrosis, there are both possible allele-specific and indirect approaches which could be taken. There are specific CFTR alleles with intronic mutations that lead to disruption of splicing. Deletion of these mutations has led to restoration of CFTR splicing and function in vitro.76 However, of the common mutations which affect CFTR splicing (Table 1), two are 1 base pair from an intron-exon junction (Fig. 1) and thus are not candidates for correction by deletion. For indirect approaches, it has been proposed that reduced epithelial sodium channel function might restore ion balance in the airway epithelium and ameliorate disease pathology. However, small molecule inhibitors of the sodium channels have not had great success in clinical trials, so it is not clear if genetic deletion will do better.77
Correction of mutations can be carried out by HDR or base editors. Of the seven most prevalent CF mutations, one is a three base pair deletion (F508del), two are transversions, and four are transitions (Table 1). The three base pair deletion and transversions currently can be precisely repaired with Cas9 by generating a double-strand break and utilizing HDR. There are current approved drugs for one of these alleles, and the predominant outcome would be re-mutated alleles which may be non-druggable. The three most common transition mutations are correctable by base editors, in principle. However, the base editors published thus far cannot correct these mutations because the combined requirement for an NGG PAM and an editable base positioned a precise distance from the PAM is too constraining. This constraint will be relaxed if base editors can be combined with Cas9 variants that recognize a greater variety of PAM sequences or lack a PAM requirement. Thus, the therapeutic potential of direct correction of mutated alleles is limited currently due to indels converting druggable alleles to non-druggable alleles, and the limited sequence addressability of base editors. However, improvements in the specificity and activity editors have occurred at a steady rate, and by the time that delivery vehicles have been developed, perhaps these problems will have been solved.
Achieving persistent correction
Part of the appeal of gene editing lies in the possibility of a single treatment which ameliorates patient health without need for subsequent treatments. Fulfilling this goal for cystic fibrosis likely will require correcting sufficient numbers of stem cells to sustain long term functional restoration of the airway epithelium.
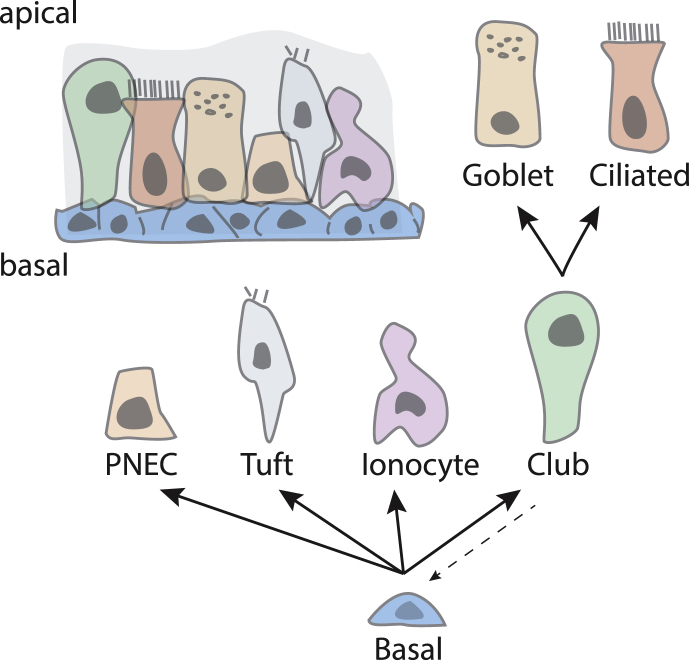
The airway is covered by a stratified epithelium consisting of multiple different cell types (Fig. 5). Most apical airway epithelial cells turn over slowly, with some localized populations turning over more rapidly.78, 79, 80 Both long- and short-lived apical cell types derive from stem cells located more basally in the stratified epithelium (Fig. 5).79, 80, 81 After injury, apical airway epithelial cells regenerate from basal stem cells.78, 79 If injury results in severe depopulation, apical club cells can dedifferentiate to become stem cells.78 In the lifetime of a treated patient, it is reasonable to assume that, between normal turnover and injuries due to infection or physical insult, replacement and regeneration of apical epithelial cells from basal stem cells will occur. Thus, for long term functional restoration from a single treatment, gene editing needs to be targeted to the basal stem cells.
Figure 5.
The airway epithelium and stem cell hierarchy (modified from Montoro et al80). The airway epithelium is a stratified epithelium with apical club, ciliated, goblet, tuft, ionocyte, pulmonary neuroendocrine cells (PNEC) as well as basal cells. The basal cells are stem cells which produce all apical cell types. Club cells are progenitor cells for goblet and ciliated cells. Under conditions of severe depopulation, club cells can dedifferentiate to basal stem cells (dotted arrow).
The apical cells are relatively accessible to airborne delivery vehicles, but basal cells normally are not. The more abundant, thicker mucus, infection and inflammation of CF pathology present additional challenges to delivery from the airway.
Apical airway cells consist of several different types including ionocytes, which have much higher CFTR expression than other epithelial cells.80, 82 Like other apical cells, ionocytes descend from basal stem cells. Thus, if correction in basal stem cells can be accomplished, then all apical cell types, including ionocytes, should have function restored. However, it is not clear how many ionocytes or other cell types need to have CFTR corrected to restore function.
Delivering to stem cells
Gene therapy for treatment of the cystic fibrosis lung has been an active area of research. Advances made with delivery vehicles for gene therapy can inform development of delivery vehicles for gene editing. It is important to note that the cargoes for gene therapy and gene editing likely will differ. Gene therapy delivers DNA with the goal of the introduced DNA persisting. In gene editing the gene editor need only be present long enough to accomplish the correction. Of course, in targeted insertion of large open reading frames, DNA would also be part of the cargo. Likewise, in strategies utilizing HDR an oligo would be included. However, with some strategies like deletion, base editing or oligo clamps, the cargo would not necessarily include DNA. Perhaps the ideal cargo would be RNPs composed of the Cas9 and guide RNA. The different cargoes make new demands on the delivery vehicles but may also lessen some of the constraints, particularly removing the need to transduce large DNA molecules.
The delivery vehicles for gene therapy have included diverse viral and non-viral vectors. Viral vectors have included adeno-associated virus, adenovirus, lentivirus, pseudotyped Sendai virus and others.83, 84, 85, 86, 87, 88 These viral vectors have transduced apical cells for the most part and have had limited efficiency at transducing basal stem cells. Because of the difficulty of transducing basal cells, gene therapy for CF has largely been predicated on repeat administration. Repeat administration leads to the possibility of immune responses developing to the therapeutics. There has been some progress recently transducing basal cells with adenoviruses when co-administered with a surfactant.89, 90
Clinical trials of nebulized aerosol delivery of liposomes were carried out but demonstrated little efficacy.90, 91 Pre-clinical research has made some advances in non-viral penetration of mucus.92 However, thus far, non-viral delivery has not been much more successful than viral means in transducing basal stem cells.
Lifelong administration of gene editors to treat CF likely will be a non-starter by the time gene editing is more mature, and so the goal will need to be a single administration that targets enough stem cells to populate the airway and restore function throughout the lifespan.
Tools and knowledge to advance single treatment gene editing
Specific tools and knowledge are needed to advance gene editing into therapeutic use. First and foremost, improved editors are needed. An ideal editor would not generate mutagenic double strand breaks, nor would it require the transduction of DNA. The base editors and oligo-clamp editors fulfill these requirements but are limited in the mutations they can correct. However, rapid evolution of editors, particularly the base editor class, likely will correct these deficits.
There is a need for delivery vehicles which can efficiently transduce airway basal stem cells. Given that only a minority of short-lived apical cells--ionocytes--may be the locus of airway disease, the delivery systems need to transduce their lineal precursors. There are some vehicles, like adenovirus co-administered with surfactant, which appear to have had some success, but there is much room for improvement.
A greater understanding of the lineal relationships and localization of the different airway cell populations is needed with a definite understanding of which cells need to be corrected to restore function. The understanding of lineage hierarchies and function need to include all developmental stages, the effect of cystic fibrosis on hierarchies and function, and the effect of other challenges to the airway.
There is a great need for animal models which report on gene editing. With such animals, the vehicles, editors, conditions and stages that yield persistent corrected cells can be identified. Ideally, editing reporter animals would report on editing that occurred in stem cells, since these are of the greatest interest. With editing reporter animals, delivery vehicles and editors can be rapidly improved.
Experiments to address when in the life-cycle the most benefit can accrue from gene editing are also necessary. Can treatment earlier, perhaps at fetal stages, yield the highest rates of restoration of health? There is reason to think that transduction at fetal stages may transduce more organs, may be more efficient and may give greater persistence.93, 94, 95
Lastly, animals are needed which can demonstrate functional restoration by editing, in the presence of CF pathology. With the continued development of effective small molecule therapeutics for CF, the bar for translation will be higher, and will likely require the restoration of health in an animal model which has representative pathology and anatomy.
Conclusions and future directions
We have presented here what we believe is a reasonable path toward somatic gene editing for the treatment of cystic fibrosis. Animal models are likely to play a much larger role in the translation of gene editing to the clinic than they did for small molecule drugs. For the development and refinement of delivery vehicles, animal models that accurately and sensitively report on gene editing events will be needed. And for translation from pre-clinical research to the patient, animal models that model the challenges of pathology, and can be used to demonstrate functional restoration and amelioration of pathology will be needed. Although there is much to be done there is reason to be optimistic, since the hurdles appear to be well-defined and addressable.
Funding sources
Cystic Fibrosis Foundation (HODGES15XX0, CONLON15XX0, CONLON18G0).
Conflict of interest
The authors declare no conflicts of interest.
Acknowledgements
We thank our colleagues for Mitch Drumm and Ann Harris for discussions leading us to write this review. We thank also an anonymous reviewer for their extensive constructive comments, and the Cystic Fibrosis Foundation, United States for funding (HODGES15XX0, CONLON15XX0, CONLON18G0).
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Cutting G.R. Cystic fibrosis genetics: from molecular understanding to clinical application. Nat Rev Genet. 2015;16(1):45–56. doi: 10.1038/nrg3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sosnay P.R., Siklosi K.R., Van Goor F. Defining the disease liability of variants in the cystic fibrosis transmembrane conductance regulator gene. Nat Genet. 2013;45(10):1160–1167. doi: 10.1038/ng.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bobadilla J.L., Macek M., Jr., Fine J.P., Farrell P.M. Cystic fibrosis: a worldwide analysis of CFTR mutations--correlation with incidence data and application to screening. Hum Mutat. 2002;19(6):575–606. doi: 10.1002/humu.10041. [DOI] [PubMed] [Google Scholar]
- 4.Drumm M.L., Ziady A.G., Davis P.B. Genetic variation and clinical heterogeneity in cystic fibrosis. Annu Rev Pathol. 2012;7:267–282. doi: 10.1146/annurev-pathol-011811-120900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Egan M.E. Genetics of cystic fibrosis: clinical implications. Clin Chest Med. 2016;37(1):9–16. doi: 10.1016/j.ccm.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 6.De Lisle R.C., Borowitz D. The cystic fibrosis intestine. Cold Spring Harb Perspect Med. 2013;3(9):a009753. doi: 10.1101/cshperspect.a009753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis P.B. Cystic fibrosis since 1938. Am J Respir Crit Care Med. 2006;173(5):475–482. doi: 10.1164/rccm.200505-840OE. [DOI] [PubMed] [Google Scholar]
- 8.Ramsey B.W., Davies J., McElvaney N.G. A CFTR potentiator in patients with cystic fibrosis and the G551D mutation. N Engl J Med. 2011;365(18):1663–1672. doi: 10.1056/NEJMoa1105185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rowe S.M., Daines C., Ringshausen F.C. Tezacaftor-ivacaftor in residual-function heterozygotes with cystic fibrosis. N Engl J Med. 2017;377(21):2024–2035. doi: 10.1056/NEJMoa1709847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sawicki G.S., McKone E.F., Pasta D.J. Sustained Benefit from ivacaftor demonstrated by combining clinical trial and cystic fibrosis patient registry data. Am J Respir Crit Care Med. 2015;192(7):836–842. doi: 10.1164/rccm.201503-0578OC. [DOI] [PubMed] [Google Scholar]
- 11.Taylor-Cousar J.L., Munck A., McKone E.F. Tezacaftor-Ivacaftor in Patients with Cystic Fibrosis Homozygous for Phe508del. N Engl J Med. 2017;377(21):2013–2023. doi: 10.1056/NEJMoa1709846. [DOI] [PubMed] [Google Scholar]
- 12.Wainwright C.E., Elborn J.S., Ramsey B.W. Lumacaftor-Ivacaftor in Patients with Cystic Fibrosis Homozygous for Phe508del CFTR. N Engl J Med. 2015;373(3):220–231. doi: 10.1056/NEJMoa1409547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davies J.C., Moskowitz S.M., Brown C. VX-659-Tezacaftor-Ivacaftor in Patients with Cystic Fibrosis and One or Two Phe508del Alleles. N Engl J Med. 2018;379(17):1599–1611. doi: 10.1056/NEJMoa1807119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keating D., Marigowda G., Burr L. VX-445-Tezacaftor-Ivacaftor in Patients with Cystic Fibrosis and One or Two Phe508del Alleles. N Engl J Med. 2018;379(17):1612–1620. doi: 10.1056/NEJMoa1807120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallagher D.N., Haber J.E. Repair of a site-specific DNA cleavage: old-school lessons for cas9-mediated gene editing. ACS Chem Biol. 2018;13(2):397–405. doi: 10.1021/acschembio.7b00760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee C.S., Haber J.E. Mating-type gene switching in Saccharomyces cerevisiae. Microbiol Spectr. 2015;3(2) doi: 10.1128/microbiolspec.MDNA3-0013-2014. MDNA3-0013-2014. [DOI] [PubMed] [Google Scholar]
- 17.Jasin M., Haber J.E. The democratization of gene editing: insights from site-specific cleavage and double-strand break repair. DNA Repair (Amst) 2016;44:6–16. doi: 10.1016/j.dnarep.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernandez A., Josa S., Montoliu L. A history of genome editing in mammals. Mamm Genome. 2017;28(7-8):237–246. doi: 10.1007/s00335-017-9699-2. [DOI] [PubMed] [Google Scholar]
- 19.Chandrasegaran S., Carroll D. Origins of programmable nucleases for genome engineering. J Mol Biol. 2016;428(5 Pt B):963–989. doi: 10.1016/j.jmb.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157(6):1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knott G.J., Doudna J.A. CRISPR-Cas guides the future of genetic engineering. Science. 2018;361(6405):866–869. doi: 10.1126/science.aat5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson S.A., McKenzie R.E., Fagerlund R.D., Kieper S.N., Fineran P.C., Brouns S.J. CRISPR-Cas: adapting to change. Science. 2017;356(6333) doi: 10.1126/science.aal5056. [DOI] [PubMed] [Google Scholar]
- 23.Leon L.M., Mendoza S.D., Bondy-Denomy J. How bacteria control the CRISPR-Cas arsenal. Curr Opin Microbiol. 2018;42:87–95. doi: 10.1016/j.mib.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang F., Doudna J.A. CRISPR-Cas9 structures and mechanisms. Annu Rev Biophys. 2017;46:505–529. doi: 10.1146/annurev-biophys-062215-010822. [DOI] [PubMed] [Google Scholar]
- 25.Hynes A.P., Villion M., Moineau S. Adaptation in bacterial CRISPR-Cas immunity can be driven by defective phages. Nat Commun. 2014;5:4399. doi: 10.1038/ncomms5399. [DOI] [PubMed] [Google Scholar]
- 26.Shou J., Li J., Liu Y., Wu Q. Precise and predictable CRISPR chromosomal rearrangements reveal principles of cas9-mediated nucleotide insertion. Mol Cell. 2018;71(4) doi: 10.1016/j.molcel.2018.06.021. 498-509 e494. [DOI] [PubMed] [Google Scholar]
- 27.Lemos B.R., Kaplan A.C., Bae J.E. CRISPR/Cas9 cleavages in budding yeast reveal templated insertions and strand-specific insertion/deletion profiles. Proc Natl Acad Sci U S A. 2018;115(9):E2040–E2047. doi: 10.1073/pnas.1716855115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taheri-Ghahfarokhi A., Taylor B.J.M., Nitsch R. Decoding non-random mutational signatures at Cas9 targeted sites. Nucleic Acids Res. 2018;46(16):8417–8434. doi: 10.1093/nar/gky653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richardson C.D., Kazane K.R., Feng S.J. CRISPR-Cas9 genome editing in human cells occurs via the Fanconi anemia pathway. Nat Genet. 2018;50(8):1132–1139. doi: 10.1038/s41588-018-0174-0. [DOI] [PubMed] [Google Scholar]
- 30.Chen J.S., Dagdas Y.S., Kleinstiver B.P. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature. 2017;550(7676):407–410. doi: 10.1038/nature24268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fu Y., Foden J.A., Khayter C. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31(9):822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu P.D., Scott D.A., Weinstein J.A. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31(9):827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cong L., Ran F.A., Cox D. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho S.W., Kim S., Kim J.M., Kim J.S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31(3):230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 35.Mali P., Yang L., Esvelt K.M. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kosicki M., Tomberg K., Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol. 2018;36(8):765–771. doi: 10.1038/nbt.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang H., Yang H., Shivalila C.S. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153(4):910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paquet D., Kwart D., Chen A. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature. 2016;533(7601):125–129. doi: 10.1038/nature17664. [DOI] [PubMed] [Google Scholar]
- 39.Capecchi M.R. Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat Rev Genet. 2005;6(6):507–512. doi: 10.1038/nrg1619. [DOI] [PubMed] [Google Scholar]
- 40.Nishiyama J., Mikuni T., Yasuda R. Virus-mediated genome editing via homology-directed repair in mitotic and postmitotic cells in mammalian brain. Neuron. 2017;96(4):755–768 e755. doi: 10.1016/j.neuron.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sander J.D., Joung J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32(4):347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haapaniemi E., Botla S., Persson J., Schmierer B., Taipale J. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat Med. 2018;24(7):927–930. doi: 10.1038/s41591-018-0049-z. [DOI] [PubMed] [Google Scholar]
- 43.Ihry R.J., Worringer K.A., Salick M.R. p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat Med. 2018;24(7):939–946. doi: 10.1038/s41591-018-0050-6. [DOI] [PubMed] [Google Scholar]
- 44.van den Berg J., Manjon A.G., Kielbassa K., Feringa F.M., Freire R., Medema R.H. A limited number of double-strand DNA breaks is sufficient to delay cell cycle progression. Nucleic Acids Res. 2018;46(19):10132–10144. doi: 10.1093/nar/gky786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gaudelli N.M., Komor A.C., Rees H.A. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature. 2017;551(7681):464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim K., Ryu S.M., Kim S.T. Highly efficient RNA-guided base editing in mouse embryos. Nat Biotechnol. 2017;35(5):435–437. doi: 10.1038/nbt.3816. [DOI] [PubMed] [Google Scholar]
- 47.Kim Y.B., Komor A.C., Levy J.M., Packer M.S., Zhao K.T., Liu D.R. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat Biotechnol. 2017;35(4):371–376. doi: 10.1038/nbt.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533(7603):420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gehrke J.M., Cervantes O., Clement M.K. An APOBEC3A-Cas9 base editor with minimized bystander and off-target activities. Nat Biotechnol. 2018;36(10):977–982. doi: 10.1038/nbt.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lei L., Chen H., Xue W. APOBEC3 induces mutations during repair of CRISPR-Cas9-generated DNA breaks. Nat Struct Mol Biol. 2018;25(1):45–52. doi: 10.1038/s41594-017-0004-6. [DOI] [PubMed] [Google Scholar]
- 51.Zafra M.P., Schatoff E.M., Katti A. Optimized base editors enable efficient editing in cells, organoids and mice. Nat Biotechnol. 2018;36(9):888–893. doi: 10.1038/nbt.4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Komor A.C., Badran A.H., Liu D.R. Editing the genome without double-stranded DNA breaks. ACS Chem Biol. 2018;13(2):383–388. doi: 10.1021/acschembio.7b00710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rees H.A., Liu D.R. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat Rev Genet. 2018;19(12):770–788. doi: 10.1038/s41576-018-0059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McNeer N.A., Anandalingam K., Fields R.J. Nanoparticles that deliver triplex-forming peptide nucleic acid molecules correct F508del CFTR in airway epithelium. Nat Commun. 2015;6:6952. doi: 10.1038/ncomms7952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ricciardi A.S., Bahal R., Farrelly J.S. In utero nanoparticle delivery for site-specific genome editing. Nat Commun. 2018;9(1):2481. doi: 10.1038/s41467-018-04894-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ricciardi A.S., Quijano E., Putman R., Saltzman W.M., Glazer P.M. Peptide nucleic acids as a tool for site-specific gene editing. Molecules. 2018;23(3) doi: 10.3390/molecules23030632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Piel F.B., Patil A.P., Howes R.E. Global distribution of the sickle cell gene and geographical confirmation of the malaria hypothesis. Nat Commun. 2010;1:104. doi: 10.1038/ncomms1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Romeo G., Devoto M., Galietta L.J. Why is the cystic fibrosis gene so frequent? Hum Genet. 1989;84(1):1–5. doi: 10.1007/BF00210660. [DOI] [PubMed] [Google Scholar]
- 59.Farrell P., Ferec C., Macek M. Estimating the age of p.(Phe508del) with family studies of geographically distinct European populations and the early spread of cystic fibrosis. Eur J Hum Genet. 2018;26(12):1832–1839. doi: 10.1038/s41431-018-0234-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsui L.C., Dorfman R. The cystic fibrosis gene: a molecular genetic perspective. Cold Spring Harb Perspect Med. 2013;3(2):a009472. doi: 10.1101/cshperspect.a009472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cystic Fibrosis Mutation Database (CFTR1). http://www.genet.sickkids.on.ca/cftr/Home.html.
- 62.Clinical and Functional Translation of CFTR (CFTR2). https://www.cftr2.org.
- 63.Laoharawee K., DeKelver R.C., Podetz-Pedersen K.M. Dose-Dependent prevention of metabolic and neurologic disease in murine MPS II by ZFN-mediated in vivo genome editing. Mol Ther. 2018;26(4):1127–1136. doi: 10.1016/j.ymthe.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qasim W., Zhan H., Samarasinghe S. Molecular remission of infant B-ALL after infusion of universal TALEN gene-edited CAR T cells. Sci Transl Med. 2017;9(374) doi: 10.1126/scitranslmed.aaj2013. [DOI] [PubMed] [Google Scholar]
- 65.Li H., Haurigot V., Doyon Y. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature. 2011;475(7355):217–221. doi: 10.1038/nature10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sharma R., Anguela X.M., Doyon Y. In vivo genome editing of the albumin locus as a platform for protein replacement therapy. Blood. 2015;126(15):1777–1784. doi: 10.1182/blood-2014-12-615492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Amoasii L., Hildyard J.C.W., Li H. Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science. 2018;362(6410):86–91. doi: 10.1126/science.aau1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yin H., Xue W., Chen S. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol. 2014;32(6):551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bednarski C., Tomczak K., Vom Hovel B., Weber W.M., Cathomen T. Targeted Integration of a Super-Exon into the CFTR Locus Leads to Functional Correction of a Cystic Fibrosis Cell Line Model. PloS One. 2016;11(8) doi: 10.1371/journal.pone.0161072. e0161072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Harrison P.T., Hoppe N., Martin U. Gene editing & stem cells. J Cyst Fibros. 2018;17(1):10–16. doi: 10.1016/j.jcf.2017.11.018. [DOI] [PubMed] [Google Scholar]
- 71.Ichise H., Ichise T., Sasanuma H., Yoshida N. The Cd6 gene as a permissive locus for targeted transgenesis in the mouse. Genesis. 2014;52(5):440–450. doi: 10.1002/dvg.22779. [DOI] [PubMed] [Google Scholar]
- 72.Montoliu L., Chavez S., Vidal M. Variegation associated with lacZ in transgenic animals: a warning note. Transgenic Res. 2000;9(3):237–239. doi: 10.1023/a:1008995730285. [DOI] [PubMed] [Google Scholar]
- 73.Robertson G., Garrick D., Wu W., Kearns M., Martin D., Whitelaw E. Position-dependent variegation of globin transgene expression in mice. Proc Natl Acad Sci U S A. 1995;92(12):5371–5375. doi: 10.1073/pnas.92.12.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Suzuki K., Tsunekawa Y., Hernandez-Benitez R. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature. 2016;540(7631):144–149. doi: 10.1038/nature20565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bak R.O., Gomez-Ospina N., Porteus M.H. Gene editing on center stage. Trends Genet. 2018;34(8):600–611. doi: 10.1016/j.tig.2018.05.004. [DOI] [PubMed] [Google Scholar]
- 76.Sanz D.J., Hollywood J.A., Scallan M.F., Harrison P.T. Cas9/gRNA targeted excision of cystic fibrosis-causing deep-intronic splicing mutations restores normal splicing of CFTR mRNA. PloS One. 2017;12(9) doi: 10.1371/journal.pone.0184009. e0184009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Strug L.J., Stephenson A.L., Panjwani N., Harris A. Recent advances in developing therapeutics for cystic fibrosis. Hum Mol Genet. 2018;27(R2):R173–R186. doi: 10.1093/hmg/ddy188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hogan B.L., Barkauskas C.E., Chapman H.A. Repair and regeneration of the respiratory system: complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell. 2014;15(2):123–138. doi: 10.1016/j.stem.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Randell S.H. Airway epithelial stem cells and the pathophysiology of chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2006;3(8):718–725. doi: 10.1513/pats.200605-117SF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Montoro D.T., Haber A.L., Biton M. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature. 2018;560(7718):319–324. doi: 10.1038/s41586-018-0393-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rock J.R., Onaitis M.W., Rawlins E.L. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci U S A. 2009;106(31):12771–12775. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Plasschaert L.W., Zilionis R., Choo-Wing R. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature. 2018;560(7718):377–381. doi: 10.1038/s41586-018-0394-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hart S.L., Harrison P.T. Genetic therapies for cystic fibrosis lung disease. Curr Opin Pharmacol. 2017;34:119–124. doi: 10.1016/j.coph.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 84.Flotte T.R., Solow R., Owens R.A., Afione S., Zeitlin P.L., Carter B.J. Gene expression from adeno-associated virus vectors in airway epithelial cells. Am J Respir Cell Mol Biol. 1992;7(3):349–356. doi: 10.1165/ajrcmb/7.3.349. [DOI] [PubMed] [Google Scholar]
- 85.Rosenfeld M.A., Yoshimura K., Trapnell B.C. In vivo transfer of the human cystic fibrosis transmembrane conductance regulator gene to the airway epithelium. Cell. 1992;68(1):143–155. doi: 10.1016/0092-8674(92)90213-v. [DOI] [PubMed] [Google Scholar]
- 86.Olsen J.C., Johnson L.G., Stutts M.J. Correction of the apical membrane chloride permeability defect in polarized cystic fibrosis airway epithelia following retroviral-mediated gene transfer. Hum Gene Ther. 1992;3(3):253–266. doi: 10.1089/hum.1992.3.3-253. [DOI] [PubMed] [Google Scholar]
- 87.Stocker A.G., Kremer K.L., Koldej R., Miller D.S., Anson D.S., Parsons D.W. Single-dose lentiviral gene transfer for lifetime airway gene expression. J Gene Med. 2009;11(10):861–867. doi: 10.1002/jgm.1368. [DOI] [PubMed] [Google Scholar]
- 88.Mitomo K., Griesenbach U., Inoue M. Toward gene therapy for cystic fibrosis using a lentivirus pseudotyped with Sendai virus envelopes. Mol Ther. 2010;18(6):1173–1182. doi: 10.1038/mt.2010.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cooney A.L., Singh B.K., Loza L.M. Widespread airway distribution and short-term phenotypic correction of cystic fibrosis pigs following aerosol delivery of piggyBac/adenovirus. Nucleic Acids Res. 2018;46(18):9591–9600. doi: 10.1093/nar/gky773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cooney A.L., McCray P.B., Jr., Sinn P.L. Cystic fibrosis gene therapy: looking back, looking forward. Genes (Basel) 2018;9(11) doi: 10.3390/genes9110538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Alton E., Armstrong D.K., Ashby D. Repeated nebulisation of non-viral CFTR gene therapy in patients with cystic fibrosis: a randomised, double-blind, placebo-controlled, phase 2b trial. Lancet Respir Med. 2015;3(9):684–691. doi: 10.1016/S2213-2600(15)00245-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mastorakos P., da Silva A.L., Chisholm J. Highly compacted biodegradable DNA nanoparticles capable of overcoming the mucus barrier for inhaled lung gene therapy. Proc Natl Acad Sci U S A. 2015;112(28):8720–8725. doi: 10.1073/pnas.1502281112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Carlon M., Toelen J., Van der Perren A. Efficient gene transfer into the mouse lung by fetal intratracheal injection of rAAV2/6.2. Mol Ther. 2010;18(12):2130–2138. doi: 10.1038/mt.2010.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Keswani S.G., Balaji S., Le L. Pseudotyped AAV vector-mediated gene transfer in a human fetal trachea xenograft model: implications for in utero gene therapy for cystic fibrosis. PloS One. 2012;7(8) doi: 10.1371/journal.pone.0043633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rossidis A.C., Stratigis J.D., Chadwick A.C. In Utero CRISPR-mediated therapeutic editing of metabolic genes. Nat Med. 2018;24(10):1513–1518. doi: 10.1038/s41591-018-0184-6. [DOI] [PMC free article] [PubMed] [Google Scholar]