Abstract
The t(4;11)(q21;q23) translocation is associated with high-risk infant pro-B-cell acute lymphoblastic leukemia and arises prenatally during embryonic/fetal hematopoiesis. The developmental/pathogenic contribution of the t(4;11)-resulting MLL-AF4 (MA4) and AF4-MLL (A4M) fusions remains unclear; MA4 is always expressed in patients with t(4;11)+ B-cell acute lymphoblastic leukemia, but the reciprocal fusion A4M is expressed in only half of the patients. Because prenatal leukemogenesis manifests as impaired early hematopoietic differentiation, we took advantage of well-established human embryonic stem cell-based hematopoietic differentiation models to study whether the A4M fusion cooperates with MA4 during early human hematopoietic development. Co-expression of A4M and MA4 strongly promoted the emergence of hemato-endothelial precursors, both endothelial- and hemogenic-primed. Double fusion-expressing hemato-endothelial precursors specified into significantly higher numbers of both hematopoietic and endothelial-committed cells, irrespective of the differentiation protocol used and without hijacking survival/proliferation. Functional analysis of differentially expressed genes and differentially enriched H3K79me3 genomic regions by RNA-sequencing and H3K79me3 chromatin immunoprecipitation-sequencing, respectively, confirmed a hematopoietic/endothelial cell differentiation signature in double fusion-expressing hemato-endothelial precursors. Importantly, chromatin immunoprecipitation-sequencing analysis revealed a significant enrichment of H3K79 methylated regions specifically associated with HOX-A cluster genes in double fusion-expressing differentiating hematopoietic cells. Overall, these results establish a functional and molecular cooperation between MA4 and A4M fusions during human hematopoietic development.
Introduction
The mixed-lineage leukemia (MLL) gene encodes for an H3K4 histone methyltransferase important in hematopoietic development.1 The human MLL gene is frequently rearranged in acute leukemia and typically confers a dismal outcome.2,3 Of particular interest is the t(4;11)(q21;q23) translocation, which encodes the fusion proteins MLL-AF4 (MA4) and AF4-MLL (A4M), and is associated with infant B-cell acute lymphoblastic leukemia (B-ALL). This t(4;11)+ infant leukemia is characterized by a very brief latency, raising the question of how it evolves so quickly.4 Moreover, the exceptionally high concordance rate of t(4;11)+ B-ALL in monozygotic twin infants5,6 suggests that all the necessary (epi)genetic events required for leukemogenesis are accomplished prenatally, during embryonic/fetal hematopoietic development.7 However, our understanding of t(4;11)-mediated developmental effects is limited due, at least in part, to the variety of phenotypes and long latency observed in currently available t(4;11) mouse models.2,8–17 These different phenotypes likely result from targeting a cell in the wrong developmental stage, or not addressing the impact of secondary hits, leaving open questions about the developmental impact of the t(4;11) translocation during early human development.
The functional and molecular contribution of the reciprocal fusion genes resulting from the derivative translocated chromosomes remains obscure in cancer. The MA4 fusion is always expressed in t(4;11)+ B-ALL patients, whereas the reciprocal fusion A4M is expressed in only half of the patients.18–20 Importantly, t(4;11)+ cell lines display addiction to MA4 but not to A4M,21,22 and although A4M was not sufficient to initiate leukemia in cord blood-derived CD34+ cells,23 it was nevertheless capable of initiating B-ALL in mice without the requirement of MA4, indicating that it contributes to t(4;11)-driven leukemogenesis.11,24,25 Strikingly, a very recent clinical study has unraveled an independent prognostic value for MA4 expression in t(4;11)+ infant B-ALL, thus adding a new piece to the puzzle.19 Thus, the developmental/pathogenic contribution of the t(4;11)− resulting reciprocal fusion A4M remains enigmatic.
Human embryonic stem cells (hESC) represent a powerful tool for modeling different developmental aspects of human disease that cannot otherwise be addressed by analyses of patients’ samples or mouse models.7,26,27 Given that prenatal leukemogenesis manifests as impaired early hematopoietic differentiation, modeling hematopoietic differentiation in hESC may represent a promising in vitro approach to study the onset of hematopoiesis and the mechanisms underlying early human hematopoietic development.7 During hESC differentiation, a primitive population of CD45− hemato-endothelial precursors (HEP) arises and further differentiates into CD45+ hematopoietic and mature endothelial cells.28–30 Beyond its pathogenic role in acute leukemias, the MLL gene has also been implicated in endothelial cell maturation,31 and endothelial dysfunction was recently linked to disease outcomes in childhood leukemias.32 We previously reported that MA4 favors the emergence of endothelial-primed HEP but not hemogenic HEP from hESC.10 Here, we took advantage of well-established hESC-based differentiation systems to study whether the A4M fusion cooperates with MA4 during early human hematopoietic and endothelial development. We report a functional and molecular cooperation between MA4 and A4M fusions, which results in enhanced hemato-endothelial output during human embryonic development.
Methods
Vector construction and lentiviral transduction
The cDNA for MA4 and A4M were subcloned into the pRRL-EF1α-PGK-NEO vector.11,16 Both fusions have been described previously (Online Supplementary Figure S1A).11,23 We used the following lentivectors containing either neomycin or dTo for cell selection: pRRL-EF1α-PGK-NEO (empty vector; EV), pRRL-EF1α-MA4-PGK-NEO (MA4) and pRRL-EF1α-A4M-PGK-dTo (A4M). VSV-G-pseudotyped lentiviral particles were generated in 293T cells using standard transfection protocols and concentrated by ultracentrifugation.33 hESC were infected overnight with concentrated EV or MA4 lentivirus plus 8 μg/mL polybrene. Viral super-natants were washed away the next day, and EV- and MA4-trans-duced hESC were then selected with G418 (50-100 μg/mL) for 3 weeks. For dual transduction of MA4 and A4M fusions, G418-resistant MA4-expressing hESC were infected with A4M-expressing viruses. EV/G418-selected hESC were also transduced with A4M alone. Transgene expression was confirmed for all the genotypes (Figure 1).
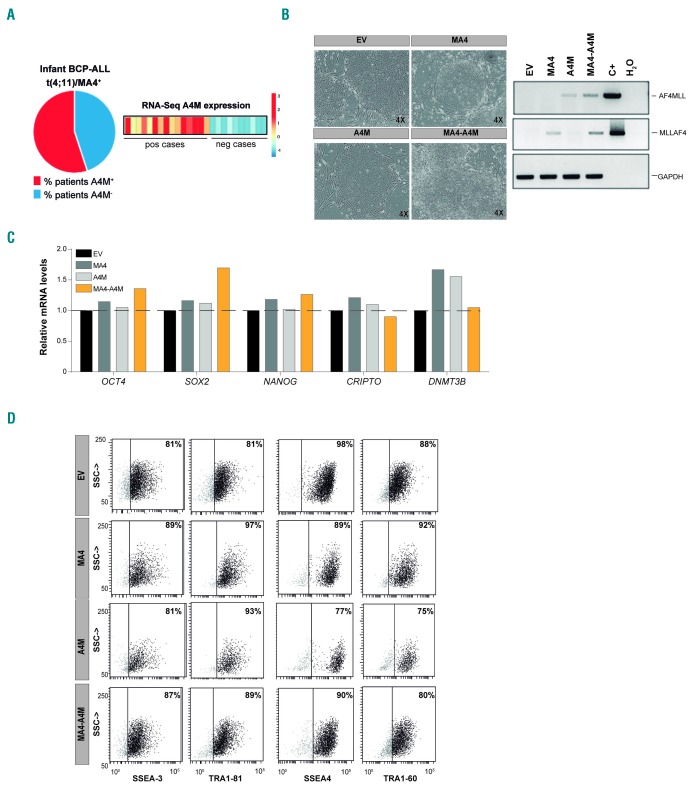
Figure 1.
Characterization of transgenic human embryonic stem cells expressing the reciprocal fusion A4M together with MA4. (A) RNA-sequencing and quantitative real-time polymerase chain reaction (qRT-PCR) validation revealed that ~45% (11/25) of the patients with t(4;11)+ B-cell precursor acute lymphoblastic leukemia do not express the reciprocal fusion A4M.18 (B) Left, Phase-contrast morphology of representative colonies from each transgenic human embryonic stem cell (hESC) line. Right, Reverse transcriptase polymerase chain reaction analysis (RT-PCR) confirming expression of both fusions in undifferentiated hESC. (C) qRT-PCR expression of the pluripotency genes OCT4, SOX2, NANOG, CRIPTO, and DNMT3B. (D) Representative FACS data confirming expression of the pluripotency surface markers SSEA-3, SSEA-4, TRA-1-60, and TRA-1-81. BCP-ALL: B-cell precursor acute lymphoblastic leukemia; RNA-seq: RNA-sequencing; pos: positive; neg: negative; EV: empty vector; C+: positive control.
Human embryonic stem cell culture and characterization of transgenic human embryonic stem cell lines
hESC (AND1 cell line) were maintained undifferentiated on a layer of irradiated human mesenchymal stem cells in complete knockout Dulbecco modified Eagle medium containing 20% knockout serum replacement and 8 ng/mL basic fibroblast growth factor.34,35 The medium was changed daily, and cells were passaged weekly by dissociation with 1:1 collagenase IV:dispase. Cultures were visualized daily by phase contrast microscopy. Approval for hESC work was obtained from the Spanish National Embryo Ethical Committee. The pluripotency of transgenic hESC was characterized by flow cytometry using antibodies against SSEA-3, SSEA-4 TRA-1-60 and TRA-1-81 (BD Biosciences).36 Expression of the pluripotency-associated transcription factors OCT4, NANOG, SOX2, CRIPTO, and DNMT3B as well as transgene expression (MA4 and A4M) were analyzed by quantitative real-time polymerase chain reaction (PCR) (Online Supplementary Table S1 shows the primers and PCR conditions used).23,37,38
Hematopoietic differentiation from human embryonic stem cells by embryoid body formation
Undifferentiated hESC were treated with collagenase IV:dispase for 1 h at 37°C. To examine embryoid body (EB) formation, cells were transferred to low-attachment plates and incubated overnight in differentiation medium (knockout Dulbecco modified Eagle medium supplemented with 20% fetal bovine serum, 1% non-essential amino acids, 1 mmol/L L-glutamine, and 0.1 mmol/L β-mercaptoethanol). The medium was changed the next day to the same differentiation medium supplemented with the following hematopoietic cytokines: 300 ng/mL stem cell factor, 300 ng/mL Flt3L, 10 ng/mL interleukin-3, 10 ng/mL interleukin-6, 50 ng/mL granulocyte - colony-stimulating factor and 25 ng/mL bone morphogenetic protein-4 (all from R&D).9,29,39–41 EB were dissociated at different time points during development using collagenase B and enzyme-free Cell Dissociation Buffer (Invitrogen). Dissociated cells were stained with anti-CD34-PE, anti-CD31-FITC, anti-CD45-APC or anti-CD34-PE-Cy7, CD31-BV510, anti-glycophorin A, anti-CD43-FITC, anti-CD45-APC antibodies (all from BD Biosciences) and 7-actinomycin D, and analyzed using a FACS Canto flow cytometer.9,29,39–41 Colony-forming unit assays were performed on days 10 and 15 of EB differentiation by plating 60×104 EB cells onto serum-free methylcellulose H4435 (Stem Cell Technologies). Colonies were scored after 12 days.9,29,42–44
Cell cycle and apoptosis analysis
For cell cycle analysis of hESC-derived HEP and CD45+ cells, day 15 EB were dissociated and harvested cells were fixed overnight in 70% ice-cold ethanol. Cells were then washed in phosphate-buffered saline and incubated with anti-CD31-FITC, anti-CD34-PE-Cy7 and anti-CD45-APC antibodies for 15 min. Cells were then suspended in propidium iodide-containing buffer and acquired and analyzed on a FACS Canto-II using Modfit LT4.0 software, discriminating between quiescent cells (G0/G1), cycling cells (S-phase) and G2/M cells.45,46 Apoptosis was assessed with an Annexin-V Apoptosis Detection kit (BD Biosciences).16
Human embryonic stem cell-OP9 co-cultures
hESC-OP9 co-cultures were performed as described elsewhere.47,48 OP9 stroma was prepared by plating OP9 cells in gelatin-coated dishes, and allowing them to overgrow as a monolayer. hESC were prepared as a suspension of small aggregates using collagenase IV:dispase. One-tenth of this suspension was plated on top of the 8-day overgrown OP9 stroma. Media were replaced on the next day and one-half volume media changes were performed every other day thereafter. Hematopoietic differentiation was assessed by flow cytometry at day 9 of co-culture. Accordingly, hESC-OP9 co-cultures were treated with collagenase IV/TrypLE and cells were dissociated and filtered through a 70-μm strainer. Cell suspensions were stained with anti-mouse CD29-FITC and anti-human CD34-PE and CD45-APC antibodies. The proportion of HEP (CD34+CD31+CD45−) and total blood cells (CD45+) were analyzed within the CD29− hESC-derived cell population. Hemogenic and endothelial HEP were distinguished based on CD34 and CD43 expression.40
Culture of FACS-isolated hemato-endothelial precursors in MS5 stroma or liquid culture
Day 9 human hESC-OP9 co-cultures were dissociated as above and both CD45+ cells and HEP were analyzed. FACS-purified HEP (CD29−CD34+CD31+CD45−) were plated onto MS5 stroma or in liquid culture for 30 or 16 days, respectively, in differentiation medium with hematopoietic cytokines (50 ng/mL stem cell factor, 50 ng/mL Flt3L, 10 ng/mL interleukin-3, 20 ng/mL interleukin-7). The medium was changed every 7 days, and the emergence of CD45+ hematopoietic cells was analyzed by FACS.
Endothelial differentiation of hemato-endothelial precursors
HEP (2×104) from day 9 human hESC-OP9 co-cultures were seeded onto 0.1% gelatin-coated plates in complete EGM-2 medium with microvasculature supplements (Lonza) for 7 days. Cells were then fixed, permeabilized and stained with rabbit anti-human VE-cadherin (Cayman), mouse anti-human endothelial nitric oxide synthase (BD Biosciences), and mouse anti-human von Willebrand factor (DAKO) followed by Alexa 488-conjugated anti-rabbit or Cy3-conjugated anti-mouse (Jackson Immunoresearch) antibodies. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole. Images were obtained using an inverted fluorescence microscope. Day 7 differentiating cells were trypsinized and cell suspensions were stained with anti-human CD31-FITC and CD144-PerCP-Cy5.5 antibodies.
Mouse transplantation and analysis of hematopoietic-endothelial engraftment
NOD/LtSz-scid IL-2Rγ−/− (NSG) mice were housed under sterile conditions. The Animal Care Committee approved all mouse protocols. Briefly, cord blood-derived CD34+ hematopoietic stem and progenitor cells (3×104 cells) or cells from day 15 EB (5×105 cells) were transplanted into the bone marrow as described previously.49 Animal health was monitored throughout the entire experiment. Mice were killed 10 weeks after transplantation and cell suspensions were analyzed by FACS for human chimerism using anti-HLA-ABC-FITC, anti-CD31-PE, CD144-PerCP-Cy5.5, and anti-CD45-APC antibodies.
RNA- and chromatin immunoprecipitation-sequencing
Details of the RNA- and chromatin immunoprecipitation-sequencing and analysis are provided in the Online Supplementary Methods.
Statistical analysis
All data are expressed as mean ± standard error of mean. Statistical comparisons were performed using GraphPad Prism software with the nonparametric Mann-Whitney test, two-tailed P-value (with 95% confidence interval). Statistical significance was defined as a P-value <0.05.
Results
Co-expression of A4M and MA4 does not hijack pluripotency
We showed very recently that only 45% of t(4;11)+ BALL patients express the reciprocal fusion A4M, whereas MA4 is consistently expressed in all t(4;11)+ B-ALL patients (Figure 1A).18–20 Here, we generated transgenic hESC lines expressing “MA4 alone”, “A4M alone” or MA4+A4M (double fusion), (Figure 1B and Online Supplementary Figure S1B). EV (control)- and MA4-hESC were established by G418 selection.9 G418-resistant EV- or MA4-expressing hESC were then transduced with A4M/dTo-expressing lentiviruses and greater than 90% transduction efficiency was achieved. Transgenic hESC lines were maintained for more than 50 passages and retained hESC-like morphology (Figure 1B, left), transgene expression (Figure 1B, right), and expression of pluripotency-associated transcription factors (Figure 1C) and surface markers (Figure 1D). All hESC genotypes formed teratomas in NSG mice (data not shown).9,50 Thus, (co-)expression of A4M and/or MA4 is compatible with hESC pluripotency.
A4M and MA4 co-operate to promote emergence of hemato-endothelial precursors and enhance blood production
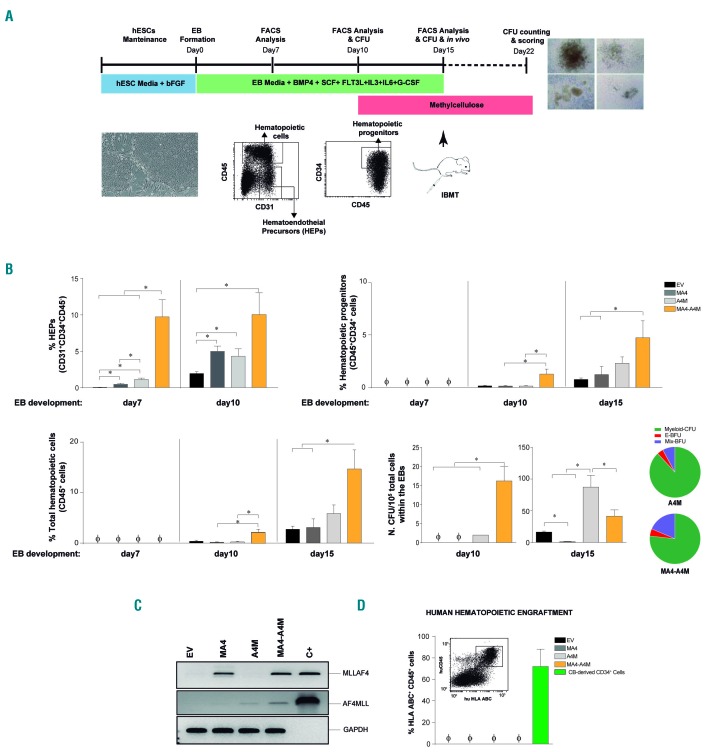
Hematopoietic differentiation was assessed using two distinct and well-established differentiation systems: EB formation43,47 (Figure 2) and OP9 co-culture47,48 (Figure 3). During differentiation, a population of primitive HEP arises, which is responsible for further hematopoietic and endothelial commitment10,30 (Figures 2A and 3A). We investigated whether co-expression of A4M and MA4 affects hESC-derived hematopoiesis by analyzing the emergence of HEP during EB development in hESC individually expressing the single fusions or the double fusion. We observed a pronounced (~5- to 10-fold; P<0.05) increase in HEP at days 7 and 10 of development in EB expressing the double A4M and MA4 fusion over those expressing single fusions (Figure 2B, upper-left panel). We next assessed whether co-expression of A4M and MA4 influences subsequent hematopoietic commitment of HEP. The kinetics of emergence and output of both total CD45+ hematopoietic cells and CD45+CD34+ hematopoietic progenitors was faster (EB day 10) from double fusion-expressing hESC than from equivalent single fusion-expressing cells, achieving a 2- to 3-fold higher hematopoietic output by day 15 of EB development (Figure 2B). Furthermore, double fusion-expressing HEP massively accelerated (EB day 10) the emergence of clonogenic hematopoietic progenitors as compared to single fusion-expressing HEP (Figure 2B, bottom-right panel). According to our previous work, if the kinetics of human EB differentiation is extended, allowing for a continuum of HEP-to-blood transition, MA4-expressing human EB display enhanced HEP production coupled to impaired blood output (EB day 20) (Online Supplementary Figure S2A) and clonogenic potential (EB day 15) (Figure 2B). We confirmed stable expression of ectopic MA4 and A4M upon EB differentiation, supporting the link between genotype and phenotype (Figure 2C).
Figure 2.
A4M cooperates with MA4 to accelerate human embryonic stem cell/erythroid body specification towards hemato-endothelial precursors and subsequent hematopoietic differentiation. (A) Schematic of erythroid body hematopoietic differentiation of human embryonic stem cells (hESC) and end-point analyses. (B) Upper left, specification into hemato-endothelial precursors (HEP; CD31+CD34+CD45−) is accelerated in double fusion-expressing hESC. Subsequent differentiation of HEP into hematopoietic progenitors (upper right) and mature CD45+ blood cells (bottom left) is enhanced in double fusion-expressing HEP. Bottom right, Colony-forming unit read-out and scoring (pie charts) confirming accelerated and enhanced hematopoietic progenitor potential from double fusion-expressing blood derivatives. (C) Reverse transcriptase polymerase chain reaction analysis confirming stable expression of MA4 and A4M upon erythroid body differentiation. (D) Neither MA4- nor double fusion-expressing blood derivatives display in vivo hematopoietic engraftment potential in irradiated NSG mice. Data are presented as mean ± standard error of mean from at least three independent experiments. *P<0.05. EB: erythroid body; bFGF: basic fibroblast growth factor; BMP4: bone morphogenetic protein-4; SCF: stem cell factor; IL: interleukin; G-CSF: granulocyte colony-stimulating factor; CFU: colony-forming unit; EV: empty vector; IBMT; intra-bone marrow transplantation; C+: positive control; CB: cord blood.
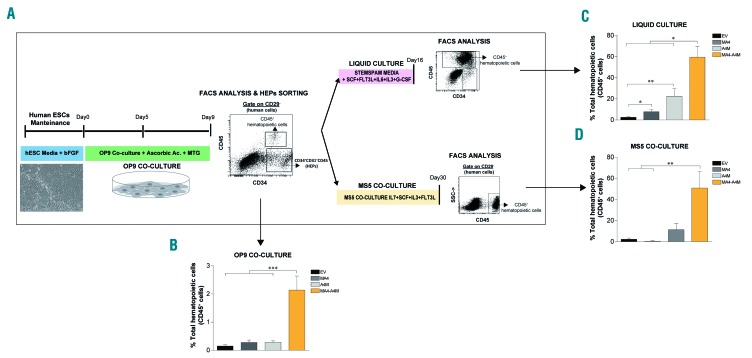
Figure 3.
Co-expression of MA4 and A4M enhances hematopoietic differentiation of human embryonic stem cells in OP9 co-culture. (A) Experimental design of OP9-based human embryonic stem cell (hESC) differentiation towards hemato-endothelial precursors (HEP) and further hematopoietic commitment of HEP maintained in either liquid culture for 16 days or in MS5 co-culture for 30 days. (B) Frequency of total CD45+ blood cells after 9 days in OP9 co-culture. (C,D) CD45-CD31+CD34+ HEP were FACS-purified at day 9 of OP9 co-culture and allowed to differentiate into CD45+ cells in liquid culture (C) or in MS5 co-culture (D). Data are represented as the mean ± standard error of mean from independent experiments. bFGF: basic fibroblast growth factor; MTG: monothioglycerol; SCF: stem cell factor; IL: interleukin; G-CSF: granulocyte colony-stimulating factor; EV: empty vector.
We also investigated hematopoietic differentiation using the OP9 differentiation system (Figure 3A,B), and by plating FACS-sorted HEP in either hematopoietic liquid culture (Figure 3C) or onto MS5 feeders (Figure 3D). After 10 days on OP9 stroma, double fusion-expressing hESC yielded a 10-fold higher number of CD45+ hematopoietic cells than did single fusion-expressing hESC (Figure 3B). Moreover, when HEP were FACS-sorted from day 9 OP9 co-cultures and allowed to differentiate into CD45+ blood cells, the yield of CD45+ cells was up to 60-fold higher in double fusion-expressing HEP than in single fusion-expressing HEP (Figure 3C,D). Encouraged by these results, we next investigated whether ectopic expression of both A4M and MA4 confers in vivo engraftment capacity to hESC-derived hematopoietic derivatives. To do this, we transplanted 5×105 hESC hematopoietic derivatives from each genotype into myeloablated NSG mice,4,43,47 finding that, despite regulating hematopoietic development in vitro, double fusion-expression did not confer in vivo engraftment to hESC hematopoietic derivatives (Figure 2D).
The increased hematopoietic output of double fusion-expressing hESC might be the consequence of transgene-mediated proliferation/survival of the emerging HEP or CD45+ cells. To address this, we analyzed cell cycle distribution (Online Supplementary Figure S2B) and apoptosis (Online Supplementary Figure S2C) within both HEP and the CD45+ cell population. No differences in the proportions of either cycling HEP or CD45+ cells were detected between genotypes (25–36% for HEP and 35–41% for CD45+ cells) (Online Supplementary Figure S2B). Apoptotic levels were similarly low in the different genotypes of HEP (6–8%) and CD45+ cells (5–7%) (Online Supplementary Figure S2C). Collectively, these results show that A4M cooperates with MA4 to induce HEP specification and blood commitment, without hijacking proliferation or survival of HEP.
A4M and MA4 cooperate to enhance endothelial cell fate from hemato-endothelial precursors
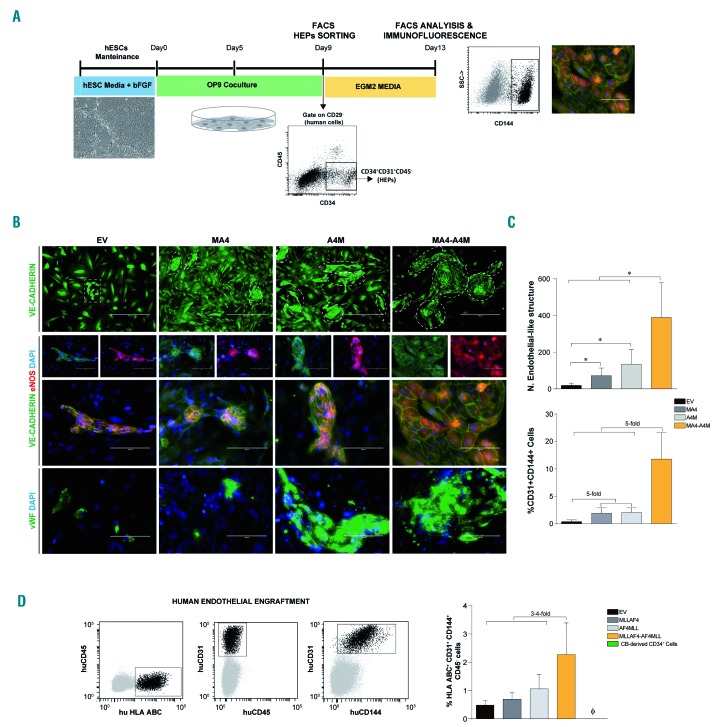
We next addressed the developmental impact of A4M in endothelial maturation from HEP.10,47 We hypothesized that co-expression of A4M and MA4 in HEP may concomitantly promote subsequent endothelial and hematopoietic commitment or skew the hemato-endothelial commitment in favor of hematopoiesis. To test this, we analyzed the ability of HEP to differentiate into mature endothelial cells. OP9-hESC co-cultures were dissociated on day 9 of development and HEP were FACS-sorted and cultured for 1 week in endothelial-promoting conditions (Figure 4A). The expression of the mature endothelial markers VE-cadherin (CD144), von Willebrand factor, endothelial nitric oxide synthase and CD31 was then analyzed. Irrespective of the genotype, HEP cultured in endothelial conditions attached, became spindle-shaped, and formed VE-cadherin+ endothelial-like structures co-expressing endothelial nitric oxide synthase, von Willebrand factor and CD31 (Figure 4B,C, top panel). However, double fusion-expressing HEP were more prone to differentiate into mature endothelial cells than were single fusion-expressing HEP. Accordingly, they yielded a 20-fold higher number of VE-cadherin+ endothelial-like structures (Figure 4C, top panel) and CD144+CD31+ endothelial cells (Figure 4C, bottom panel). Interestingly, endothelial cells (HLA.ABC+CD31+CD34+CD144+CD45-CD43−) were found in the bone marrow of mice transplanted with double fusion-expressing hESC blood derivatives at levels ~4-fold higher than those in mice transplanted with single fusion-expressing cells (Figure 4D).
Figure 4.
Enhanced endothelial cell fate from hemato-endothelial precursors co-expressing MA4 and A4M. (A) Scheme of hemato-endothelial precursor (HEP) endothelial differentiation and phenotypic characterization. (B) FACS-sorted HEP from day 9 human ESC-OP9 co-cultures were cultured in EGM2 medium for 5 days and analyzed by immunofluorescence for VE-cadherin, endothelial nitric oxide synthase and von Willebrand factor. (C) Top, Endothelial-like structures were identified and quantified based on VE-cadherin staining (white dotted-lined areas in B, top panel). Bottom, Frequency of CD45−CD31+CD144+ endothelial cells quantified by flow cytometry. (D) In vivo endothelial engraftment potential (HLA.ABC+CD31+CD144+CD45−) analyzed in bone marrow of NSG mice 8 weeks after transplantation of HEP. Data are presented as mean ± standard error of mean from five independent experiments. *P<0.05. bFGF: basic fibroblast growth factor; EV: empty vector; CB: cord blood.
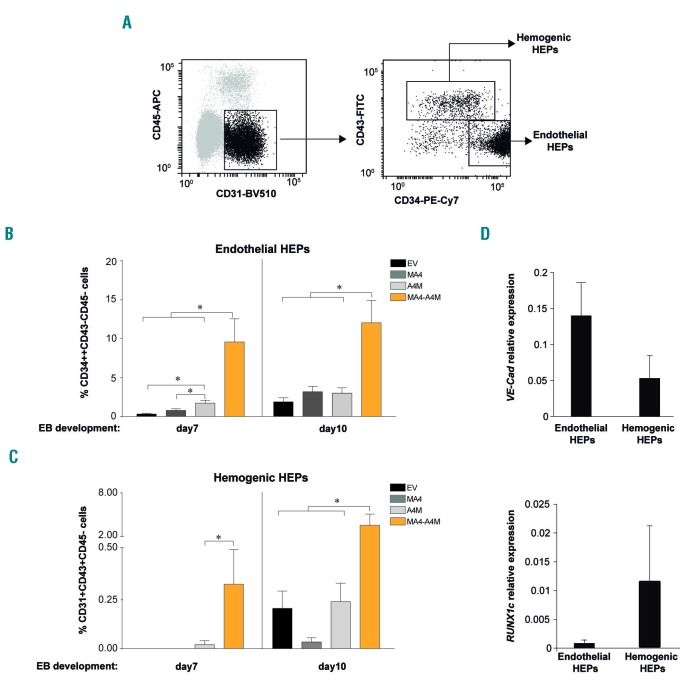
Within CD34+CD31+CD45− HEP, two subpopulations of phenotypically and functionally distinct HEP can be distinguished based on the expression of CD34 and CD43: hemogenic HEP (CD34low/+CD43+CD45−) and endothelial HEP (CD34++CD43−CD45−) (Figure 5A).40,48,51 We thus analyzed the contribution of both HEP populations to the superior hematopoietic and endothelial differentiation observed in double fusion-expressing HEP. Co-expression of A4M and MA4, but not single fusions, robustly enhanced the emergence of both endothelial and hemogenic HEP (Figure 5B,C). The identity of hemogenic and endothelial HEP was confirmed by the specific expression of early hematopoietic and endothelial master genes (Figure 5D). Thus, A4M cooperates with MA4 to promote hematopoietic and endothelial cell fate.
Figure 5.
Co-expression of MA4 and A4M significantly enhances the emergence of both endothelial and hemogenic hemato-endothelial precursors. (A) Representative flow cytometry analysis of hemato-endothelial precursors (HEP) with hemogenic (CD45-CD31+CD43+CD34dim/+) and endothelial (CD45−CD31+CD43−CD34++) potential. (B,C) A4M co-operates with MA4 to boost the emergence of both endothelial (B) and hemogenic (C) HEP. Data are presented as the mean ± standard error of mean from three independent experiments. (D) Expression of RUNX1c and Ve-Cad in hemogenic and endothelial HEP. *P<0.05. EB: erythroid body; EV: empty vector.
Genome-wide transcriptomic and H3K79 methylation profiles support the developmental cooperation between A4M and MA4
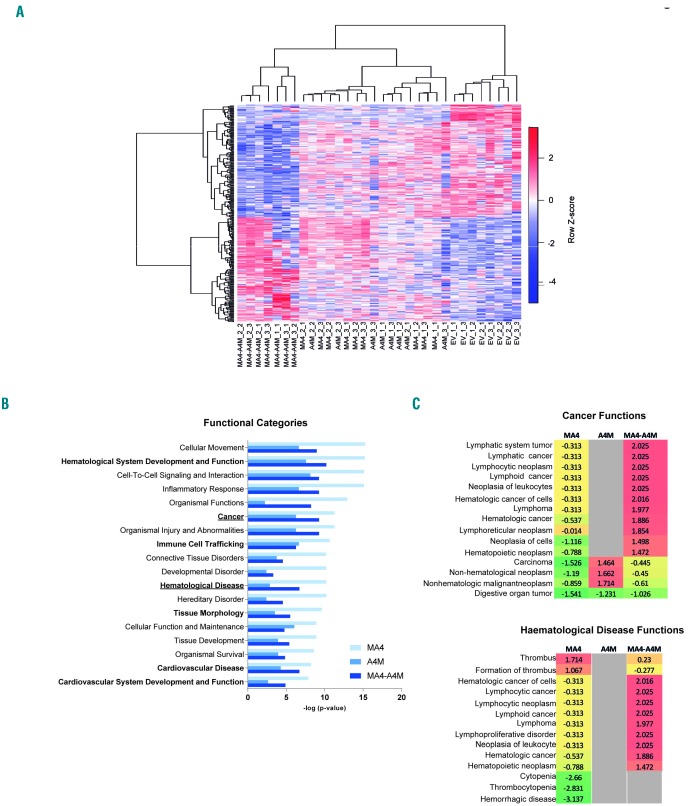
To identify patterns of gene expression that might provide a molecular explanation of the functional cooperation between A4M and MA4 in hematopoietic specification, we performed RNA-sequencing analysis on FACS-purified EV-, MA4-, A4M- and double fusion-expressing HEP from day 15 EB. Figure 6A shows a heatmap representation of the hierarchical clustering of the 335 genes differentially expressed between the four genotypes (Online Supplementary Table S2). There is a clear transcriptomic transition towards a hematopoietic/endothelial gene signature from EV-HEP to double fusion-expressing HEP. Single fusion-expressing HEP clustered interspersed between EV and double-fusion HEP. The biological functions affected by genes differentially expressed in MA4-, A4M- and double fusion-expressing HEP relative to EV were classified by Ingenuity Pathway Analysis software;47,52 the top significantly enriched functional categories included “hematological system development and function”, “cancer” and “hematological disease” (Figure 6B). Statistical (−logp-value) power shows distant effects of MA4 and A4M; however, co-expression of both fusions seems to establish a molecular balance/developmental cooperation in promoting blood-endothelial specification from hPSC. Strikingly, biofunctions specifically associated with hematologic lymphoid malignancies (not with non-hematologic cancer) were predicted to be activated (positive z-score) exclusively in double fusion-expressing HEP, further suggesting a molecular cooperation between MA4 and A4M in development and infant leukemia. (Figure 6C).
Figure 6.
Transcriptional transition towards a hematopoietic/endothelial gene signature in double fusion-expressing hemato-endothelial precursors. (A) Heatmap representation of hierarchical clustering of genes differentially expressed between empty vector (EV)-, single fusions- and double fusion-expressing hemato-endothelial precursors (HEP). Each column represents a technical replicate from three independent experiments. (B,C) Statistically significant functional categories (B) and cancer/leukemia-associated biofunctions (C) identified using Ingenuity Pathway Analysis on genes differentially expressed in single fusions-, and double fusion-expressing HEP relative to EV. They are ranked by z-score. Functional categories associated with “hematological system development and function” and “cardiovascular system development” are shown in bold. All significant biofunctions are associated with blood cell differentiation, homeostasis and migration/movement.
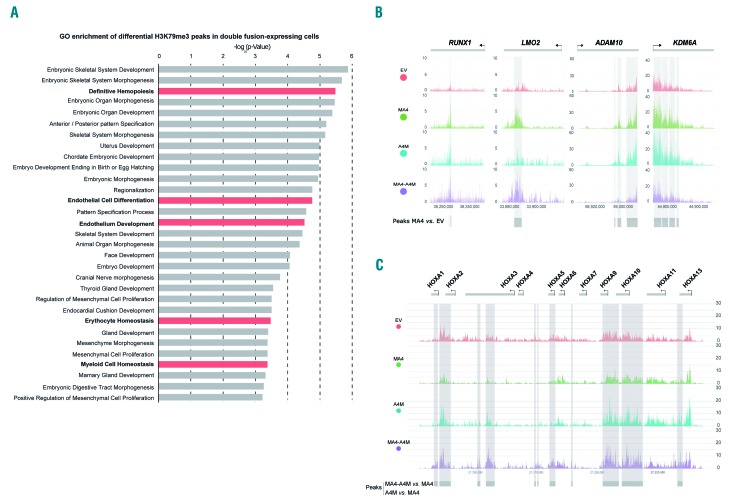
The C-terminal partners of MLL fusions normally interact with the histone methyltransferase DOT1L, which is the sole histone methyltransferase catalyzing histone 3 lysine 79 (H3K79) methylation, a chromatin modification widely associated with the dysregulated expression of HOX-A cluster genes in MLL leukemias.13,53 We thus performed genome-wide chromatin immunoprecipitation-sequencing analysis of the H3K79 trimethylation (H3K79me3) profiles in control, MA4-, A4M- and double fusion-expressing hESC-derived blood derivatives (Figure 7, Online Supplementary Figure S3A, Online Supplementary Table S3). In line with the RNA-sequencing data, functional analysis of the differential H3K79me3 peaks specific for double fusion-expressing cells revealed significant gene ontology functional categories associated with ”definitive hematopoiesis”, “myeloid and erythroid differentiation/homeostasis” and “endothelial cell development” (Figure 7A, Online Supplementary Figure S3B). This further supports the developmental co-operation between A4M and MA4 in promoting hemato-endothelial specification.
Figure 7.
H3K79 methylation profiles at genomic loci of MLL targets in MA4-, A4M- and double fusion-expressing human embryonic stem cell-derived blood derivatives. (A) Gene ontology enrichment of differential H3K79me3 peaks specific for double fusion-expressing cells. (B,C) Representative profiles for chromatin immuno-precipitation-sequencing using anti-H3K79me3 antibody at genomic regions of typical non-HOXA (B) and HOXA MLL targets (C).
Finally, we analyzed the H3K79me3 profiles at genomic loci of the classical MLL target genes reported by Guenther et al.54 Non-HOX-A classical MLL targets such as RUNX1, LMO2, ADMA10, and KDM6A showed a slight although non-significant, enrichment of H3K79me3 in MA4-expressing hESC, validating our chromatin immunoprecipitation-sequencing approach (Figure 7B). However, enrichment of H3K79me3 in HOX-A cluster genes was observed exclusively in A4M-expressing cells although it was statistically significant only in double fusion-expressing differentiating hESC (false discovery rate <0.1) (Figure 7C). As such, HOX-A genes were upregulated in double fusion-expressing hematopoietic clonogenic progenitors (Online Supplementary Figure S3C). No differential enrichment of the repressive H3K4me2/3 mark was observed in either HOX-A or non-HOX genes in double fusion-expressing cells (Online Supplementary Figure S4). Collectively, these data suggest that the deregulated expression of HOX-A genes in MLL leukemias may be imposed by the reciprocal A4M fusion through H3K79 methyltransferase activity. In support of this, a recent RNA-sequencing study performed in 42 infants with t(4;11)+ B-ALL enrolled in the Interfant treatment protocol, revealed that 45% of t(4;11)+ patients express the A4M fusion, and that HOX-A cluster genes are exclusively expressed in this AF4-MLL-expressing subgroup of t(4;11)+ patients, who do in fact have a significantly more favorable clinical outcome.19
Discussion
From etiological and pathogenic standpoints, infant cancer is distinct from adult cancer and should be studied as a developmental disease.4,7,16 A biologically and clinically intriguing infant cancer is the t(4;11)+ B-ALL, which is associated with a dismal outcome.4,21,23 Evidence in support of its prenatal origin comes from studies in monozygotic twins and archived blood spots, providing compelling evidence of a single prenatal cell as the origin for t(4;11),5 and also from recent genome-wide studies demonstrating that this infant leukemia has one of the lowest frequencies of somatic mutations of any sequenced cancer.55 The stable genome of these patients suggests that in infant developmental cancer, one “big-hit” might be sufficient to cause overt disease, supporting a key contribution of the prenatal cell-of-origin during a critical developmental window of stem cell vulnerability in leukemogenesis. However, despite its aggressiveness and short latency, our current understanding about its etiology, pathogenesis and cellular origin is still limited.2,4,14,16,52 Importantly, a recently developed xenograft model which represents the most bona fide model for t(4;11)+ B-ALL so far, has revealed the instructive role of MLL-Af4 in cord blood-derived CD34+ cells.14
Studies using primary cells from t(4;11)+ B-ALL patients are incapable of addressing the developmental genesis of the hematopoietic system. Recent data suggest that fetal liver lymphoid-primed multipotent progenitors may provide the developmental prerequisites for the initiation of t(4;11)+/MLL-AF4 infant leukemia.56 Because leukemogenesis manifests as a blockage or altered cell differentiation, the hematopoietic differentiation of hESC may represent a promising in vitro model for studying the onset of hematopoiesis and the earliest events leading to the specification of the hematopoietic cells.36 Previous studies have addressed the oncogenic role of leukemic fusion genes in hESC-derived hematopoiesis.57–59 We previously explored the developmental impact of the prenatal fusion MA4 in hESC hemato-endothelial development,10 and found that MA4 expression promotes the emergence of endothelial-primed HEP and further endothelial commitment, but hijacks the specification of hemogenic-primed HEP, impairing hematopoietic output.10
The functional and molecular contribution of the reciprocal fusion genes resulting from the derivative translocated chromosomes remains obscure in cancer.18,60 The MA4 fusion is always expressed in t(4;11)+ B-ALL patients, whereas the reciprocal fusion A4M is expressed in only 45-50% of the patients.18–20,60 Here, we took advantage of well-established hESC-based differentiation systems to study whether the A4M fusion cooperates with MA4 during early human hematopoietic and endothelial development. Co-expression of A4M and MA4 strongly promoted the emergence of both endothelial-primed and hemogenic-primed HEP. Moreover, the double fusion-expressing HEP specified into significantly higher numbers of both hematopoietic and endothelial cells, irrespective of the in vitro differentiation protocol used and without affecting survival or proliferation, indicating a functional (developmental) cooperation between MA4 and A4M fusions during human hematopoietic development. This notion was confirmed by genome-wide transcriptomic analysis of differentiating HEP. These developmental biology studies support previous evidence suggesting that A4M contributes to the pathobiology of t(4;11)+ BALL. Accordingly, Bursen et al.11 reported that A4M-trans-duced murine hematopoietic stem cells developed pro-BALL, whereas co-transduction with MA4 and A4M resulted in mixed lineage leukemia. Moreover, studies from Milne’s laboratory demonstrated that RUNX1 is directly activated by MA4 and the RUNX1 protein interacts with the A4M protein, suggesting a mechanism of cooperation between the two fusion genes at the molecular level.25
In the embryo, definitive hematopoiesis cannot occur in the absence of endothelial cell development. Definitive hematopoietic stem cells in both mice and humans emerge from the hemogenic endothelium by a process known as endothelial-to-hematopoietic transition.61 Hematopoietic differentiation of hESC occurs through the generation of HEP, from which both endothelial and hematopoietic cells then originate. Here, co-expression of A4M and MA4 in HEP concomitantly promoted endothelial and hematopoietic commitment rather than skewing the hemato-endothelial commitment in favor of one lineage over the other. This finding is important because beyond its pathogenic role in acute leukemias, the MLL gene is implicated in endothelial cell maturation, and endothelial dysfunction was recently linked to disease outcome in childhood leukemias.31,32 Furthermore, other leukemia fusion oncogenes as well as lymphoma-specific genetic aberrations have been found in endothelial cells from patients with chronic myeloid leukemia and B-cell lymphoma,10,43,44 suggesting that endothelial cells may be part of the neoplastic clone. In addition, bone marrow-derived mesenchymal stem cells from infant t(4;11)+ B-cell ALL were recently found to harbor and express the t(4;11) translocation.33 The existence of a common embryonic precursor for mesenchymal stem cells and endothelial cells has been recently demonstrated by hESC-directed differentiation.10,45 The finding of such a common embryonic precursor, and the presence of t(4;11) in both leukemic blasts and bone marrow mesenchymal stem cells of infant patients, suggests that the t(4;11) translocation arises and has a developmental impact on early pre-hematopoietic precursors. As a technical caveat, it is important to emphasize that MA4 and A4M were sequentially transduced to allow for antibiotic selection and homogeneously-transduced hESC cultures. However, in double fusion-expressing differentiating blood cells, MA4 was never individually expressed in the absence of A4M. When hematopoietic differentiation of hESC was induced by EB formation both fusions were readily co-expressed, ruling out a biased functional phenotype driven by the sequential expression of each transgene in distinct developmental windows.
Mechanistically, a putative function of A4M is to activate chromatin, rendering a chromatin landscape similar to that present during stem cell development. It is currently unknown how A4M is able to reprogram chromatin, but it does contain the SET domain disrupted from its “specification domain”, the N-terminal portion of MLL, which binds to MEN1 and LEDGF, shaping the gene targeting module of the MLL gene. When A4M is expressed, the N-terminal portion is substituted by the AF4 N-terminus, which is the crucial domain (AF4N) that binds to and strongly activates RNA polymerase II (RNAP II) for transcriptional elongation. Overexpression of either AF4, AF4N or the fusion protein A4M induces robust RNAP II-dependent gene transcription by overwriting the elongation control process in a dominant fashion.62–64 Since gene transcription per se and in particular “sterile” transcription is a powerful mechanism for chromatin activation, A4M could potentiate MA4 to skew normal and leukemic hematopoietic cell fate decisions. This also explains why MA4 has a more prominent role in the disease than the reciprocal A4M. If A4M functioned to initiate this process only by itself, then it would become obsolete after fulfilling the “chromatin opening job”. However, transcription factors such as MA4, RUNX1 or others then establish the transcriptional program leading to leukemogenesis. This is reflected in the enhanced hematopoietic potential of double fusion-expressing hESC and the enriched H3K79me3 activation mark in HOX-A cluster genes exclusively when MA4 and A4M are co-expressed. Thus, A4M prepares other transcription factors to become oncoproteins.
Molecularly, C-terminal-partners of MLL fusions (AF4, AF9, ENL) interact with DOT1L, which is the sole histone methyltransferase catalyzing H3K79 methylation, a chromatin modification widely associated with the dysregulated expression of the HOX-A gene cluster in MLL-rearranged leukemias.13,53 Here, chromatin immunoprecipitation-sequencing analysis of differentially enriched H3K79me3 genomic regions confirmed a hematopoietic/endothelial cell differentiation signature in double fusion-expressing HEP, and revealed a significant enrichment of H3K79 methylated regions specifically associated with HOX-A cluster MLL target genes (but not with non-HOX-A MLL targets) in double fusion-expressing differentiating hematopoietic cells. This is in line with the recently found significant positive correlation between the upregulation of the HOX-A gene cluster and the expression of A4M in primary t(4;11)+ infant B-cell ALL samples, and with previous studies identifying that approximately one-half of t(4;11)+ patients do not have an activated HOX-A signature.20,65,66 This may explain why MA4 failed recently to bind to HOX-A genes to regulate HOXA gene expression.14 Collectively, MA4 and A4M might cooperate through a complex molecular interaction to control HOX-A gene regulation.25 In conclusion, we describe a functional and molecular cooperation between MA4 and A4M fusions during human hematopoietic development, and demonstrate how hESC-based hematopoietic differentiation represents a promising system to explore the developmental impact of the chimeric proteins resulting from chromosomal translocations, which remains obscure in human leukemia.
Supplementary Material
Acknowledgments
Financial support for this work was obtained from the European Research Council (CoG-2014-646903 and PoC-2018-811220) and the Generalitat de Catalunya (SGR330 and PERIS 2017-2019) to PM, the Spanish Ministry of Economy and Competitiveness (SAF2016-80481-R and SAF2016-76758-R) to PM and IV, the Spanish Association against Cancer (AECC-CI-2015) and Fero Foudation to CB, the Health Institute Carlos III (ISCIII/FEDER, PI17/01028 and PI17/01028) to CB and PJR, the NIHR GOSH BRC and Great Ormond Street Hospital Children’s Charity to J.dB, and Bloodwise and Cancer Research UK to BG. RM and PM were also supported by the Deutsche José Carreras Leukämie Stiftung. PM also acknowledges financial support from the Obra Social La Caixa-Fundaciò Josep Carreras. R-T-R is supported by a fellowship from the Spanish Association of Cancer Research (AECC). RV-M is supported by a Torres Quevedo fellowship from the Spanish Ministry of Science and Innovation (PTQ-16-08623). P.M is an investigator of the Spanish Cell Therapy cooperative network (TERCEL).
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/104/6/1189
References
- 1.Milne TA. Mouse models of MLL leukemia: recapitulating the human disease. Blood. 2017;129(16):2217–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Montes R, Ayllon V, Prieto C, et al. Ligand-independent FLT3 activation does not cooperate with MLL-AF4 to immortalize/transform cord blood CD34+ cells. Leukemia. 2014;28(3):666–674. [DOI] [PubMed] [Google Scholar]
- 3.Stam RW, den Boer ML, Schneider P, et al. Targeting FLT3 in primary MLL-gene-rearranged infant acute lymphoblastic leukemia. Blood. 2005;106(7):2484–2490. [DOI] [PubMed] [Google Scholar]
- 4.Sanjuan-Pla A, Bueno C, Prieto C, et al. Revisiting the biology of infant t(4;11)/MLL-AF4+ B-cell acute lymphoblastic leukemia. Blood. 2015;126(25):2676–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ford AM, Ridge SA, Cabrera ME, et al. In utero rearrangements in the trithorax-related oncogene in infant leukaemias. Nature. 1993;363(6427):358–360. [DOI] [PubMed] [Google Scholar]
- 6.Greaves M. Infection, immune responses and the aetiology of childhood leukaemia. Nat Rev Cancer. 2006;6(3):193–203. [DOI] [PubMed] [Google Scholar]
- 7.Bueno C, Montes R, Catalina P, Rodriguez R, Menendez P. Insights into the cellular origin and etiology of the infant pro-B acute lymphoblastic leukemia with MLL-AF4 rearrangement. Leukemia. 2011;25(3):400–410. [DOI] [PubMed] [Google Scholar]
- 8.Barrett NA, Malouf C, Kapeni C, et al. Mll-AF4 confers enhanced self-renewal and lymphoid potential during a restricted window in development. Cell Rep. 2016;16(4):1039–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bueno C, Ayllon V, Montes R, et al. FLT3 activation cooperates with MLL-AF4 fusion protein to abrogate the hematopoietic specification of human ESCs. Blood. 2013;121(19):3867–3878, S3861–3863. [DOI] [PubMed] [Google Scholar]
- 10.Bueno C, Montes R, Melen GJ, et al. A human ESC model for MLL-AF4 leukemic fusion gene reveals an impaired early hematopoietic-endothelial specification. Cell Res. 2012;22(6):986–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bursen A, Schwabe K, Ruster B, et al. The AF4.MLL fusion protein is capable of inducing ALL in mice without requirement of MLL.AF4. Blood. 2010;115(17):3570–3579. [DOI] [PubMed] [Google Scholar]
- 12.Chen W, Li Q, Hudson WA, et al. A murine Mll-AF4 knock-in model results in lymphoid and myeloid deregulation and hematologic malignancy. Blood. 2006;108(2):669–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krivtsov AV, Feng Z, Lemieux ME, et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell. 2008;14(5):355–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin S, Luo RT, Ptasinska A, et al. Instructive role of MLL-fusion proteins revealed by a model of t(4;11) pro-B acute lymphoblastic leukemia. Cancer Cell. 2016;30(5):737–749. [DOI] [PubMed] [Google Scholar]
- 15.Metzler M, Forster A, Pannell R, et al. A conditional model of MLL-AF4 B-cell tumourigenesis using invertor technology. Oncogene. 2006;25(22):3093–3103. [DOI] [PubMed] [Google Scholar]
- 16.Montes R, Ayllon V, Gutierrez-Aranda I, et al. Enforced expression of MLL-AF4 fusion in cord blood CD34+ cells enhances the hematopoietic repopulating cell function and clonogenic potential but is not sufficient to initiate leukemia. Blood. 2011;117(18): 4746–4758. [DOI] [PubMed] [Google Scholar]
- 17.Tamai H, Miyake K, Takatori M, et al. Activated K-Ras protein accelerates human MLL/AF4-induced leukemo-lymphomogenicity in a transgenic mouse model. Leukemia. 2011;25(5):888–891. [DOI] [PubMed] [Google Scholar]
- 18.Kowarz E, Burmeister T, Lo Nigro L, et al. Complex MLL rearrangements in t(4;11) leukemia patients with absent AF4.MLL fusion allele. Leukemia. 2007;21(6):1232–1238. [DOI] [PubMed] [Google Scholar]
- 19.Agraz-Doblas A, Bueno C, Bashford-Rogers R, et al. Unraveling the cellular origin and clinical prognostic markers of infant B-cell acute lymphoblastic leukemia using genome-wide analysis. Haematologica. 2019;104(6):1176–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trentin L, Giordan M, Dingermann T, et al. Two independent gene signatures in pediatric t(4;11) acute lymphoblastic leukemia patients. Eur J Haematol. 2009;83(5):406–419. [DOI] [PubMed] [Google Scholar]
- 21.Kumar AR, Yao Q, Li Q, Sam TA, Kersey JH. t(4;11) leukemias display addiction to MLL-AF4 but not to AF4-MLL. Leuk Res. 2011;35(3):305–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanders DS, Muntean AG, Hess JL. Significance of AF4-MLL reciprocal fusion in t(4;11) leukemias? Leuk Res. 2011;35(3):299–300. [DOI] [PubMed] [Google Scholar]
- 23.Prieto C, Marschalek R, Kuhn A, et al. The AF4-MLL fusion transiently augments multilineage hematopoietic engraftment but is not sufficient to initiate leukemia in cord blood CD34(+) cells. Oncotarget. 2017;8(47):81936–81941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rego EM, Pandolfi PP. Reciprocal products of chromosomal translocations in human cancer pathogenesis: key players or innocent bystanders? Trends Mol Med. 2002;8(8):396–405. [DOI] [PubMed] [Google Scholar]
- 25.Wilkinson AC, Ballabio E, Geng H, et al. RUNX1 is a key target in t(4;11) leukemias that contributes to gene activation through an AF4-MLL complex interaction. Cell Rep. 2013;3(1):116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Menendez P, Bueno C, Wang L. Human embryonic stem cells: a journey beyond cell replacement therapies. Cytotherapy. 2006;8(6):530–541. [DOI] [PubMed] [Google Scholar]
- 27.Romero-Moya D, Santos-Ocana C, Castano J, et al. Genetic rescue of mitochondrial and skeletal muscle impairment in an induced pluripotent stem cells model of coenzyme Q10 deficiency. Stem Cells. 2017;35(7): 1687–1703. [DOI] [PubMed] [Google Scholar]
- 28.Menendez P, Vargas A, Bueno C, et al. Quantitative analysis of bcl-2 expression in normal and leukemic human B-cell differentiation. Leukemia. 2004;18(3):491–498. [DOI] [PubMed] [Google Scholar]
- 29.Ramos-Mejia V, Melen GJ, Sanchez L, et al. Nodal/activin signaling predicts human pluripotent stem cell lines prone to differentiate toward the hematopoietic lineage. Mol Ther. 2010;18(12):2173–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L, Li L, Shojaei F, et al. Endothelial and hematopoietic cell fate of human embryonic stem cells originates from primitive endothelium with hemangioblastic properties. Immunity. 2004;21(1):31–41. [DOI] [PubMed] [Google Scholar]
- 31.Diehl F, Rossig L, Zeiher AM, Dimmeler S, Urbich C. The histone methyltransferase MLL is an upstream regulator of endothelial-cell sprout formation. Blood. 2007;109(4): 1472–1478. [DOI] [PubMed] [Google Scholar]
- 32.Hatzipantelis ES, Athanassiou-Metaxa M, Gombakis N, et al. Thrombomodulin and von Willebrand factor: relation to endothelial dysfunction and disease outcome in children with acute lymphoblastic leukemia. Acta Haematol. 2011;125(3):130–135. [DOI] [PubMed] [Google Scholar]
- 33.Menendez P, Catalina P, Rodriguez R, et al. Bone marrow mesenchymal stem cells from infants with MLL-AF4+ acute leukemia harbor and express the MLL-AF4 fusion gene. J Exp Med. 2009;206(13):3131–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramos-Mejia V, Fernandez AF, Ayllon V, et al. Maintenance of human embryonic stem cells in mesenchymal stem cell-conditioned media augments hematopoietic specification. Stem Cells Dev. 2012;21(9):1549–1558. [DOI] [PubMed] [Google Scholar]
- 35.Sanchez L, Gutierrez-Aranda I, Ligero G, et al. Maintenance of human embryonic stem cells in media conditioned by human mesenchymal stem cells obviates the requirement of exogenous basic fibroblast growth factor supplementation. Tissue Eng Part C Methods. 2012;18(5):387–396. [DOI] [PubMed] [Google Scholar]
- 36.Bueno C, Montes R, Martin L, et al. NG2 antigen is expressed in CD34+ HPCs and plasmacytoid dendritic cell precursors: is NG2 expression in leukemia dependent on the target cell where leukemogenesis is triggered? Leukemia. 2008;22(8):1475–1478. [DOI] [PubMed] [Google Scholar]
- 37.Castano J, Menendez P, Bruzos-Cidon C, et al. Fast and efficient neural conversion of human hematopoietic cells. Stem Cell Reports. 2014;3(6):1118–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Munoz-Lopez A, Romero-Moya D, Prieto C, et al. Development refractoriness of MLL-rearranged human B cell acute leukemias to reprogramming into pluripotency. Stem Cell Reports. 2016;7(4): 602–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bueno C, Sardina JL, Di Stefano B, et al. Reprogramming human B cells into induced pluripotent stem cells and its enhancement by C/EBPalpha. Leukemia. 2016;30(3):674–682. [DOI] [PubMed] [Google Scholar]
- 40.Giorgetti A, Castano J, Bueno C, et al. Proinflammatory signals are insufficient to drive definitive hematopoietic specification of human HSCs in vitro. Exp Hematol. 2017;45:85–93 e82. [DOI] [PubMed] [Google Scholar]
- 41.Menendez P, Wang L, Chadwick K, Li L, Bhatia M. Retroviral transduction of hematopoietic cells differentiated from human embryonic stem cell-derived CD45(neg)PFV hemogenic precursors. Mol Ther. 2004;10(6):1109–1120. [DOI] [PubMed] [Google Scholar]
- 42.Bueno C, Roldan M, Anguita E, et al. Bone marrow mesenchymal stem cells from patients with aplastic anemia maintain functional and immune properties and do not contribute to the pathogenesis of the disease. Haematologica. 2014;99(7):1168–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramos-Mejia V, Navarro-Montero O, Ayllon V, et al. HOXA9 promotes hematopoietic commitment of human embryonic stem cells. Blood. 2014;124(20):3065–3075. [DOI] [PubMed] [Google Scholar]
- 44.Toscano MG, Navarro-Montero O, Ayllon V, et al. SCL/TAL1-mediated transcriptional network enhances megakaryocytic specification of human embryonic stem cells. Mol Ther. 2015;23(1):158–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bueno C, Montes R, Menendez P. The ROCK inhibitor Y-27632 negatively affects the expansion/survival of both fresh and cryopreserved cord blood-derived CD34+ hematopoietic progenitor cells: Y-27632 negatively affects the expansion/survival of CD34+HSPCs. Stem Cell Rev. 2010;6(2): 215–223. [DOI] [PubMed] [Google Scholar]
- 46.Rubio R, Garcia-Castro J, Gutierrez-Aranda I, et al. Deficiency in p53 but not retinoblastoma induces the transformation of mesenchymal stem cells in vitro and initiates leiomyosarcoma in vivo. Cancer Res. 2010;70(10):4185–4194. [DOI] [PubMed] [Google Scholar]
- 47.Ayllon V, Bueno C, Ramos-Mejia V, et al. The Notch ligand DLL4 specifically marks human hematoendothelial progenitors and regulates their hematopoietic fate. Leukemia. 2015;29(8):1741–1753. [DOI] [PubMed] [Google Scholar]
- 48.Vodyanik MA, Slukvin II. Hematoendothelial differentiation of human embryonic stem cells. Curr Protoc Cell Biol. 2007;Chapter 23:Unit 23 26. [DOI] [PubMed] [Google Scholar]
- 49.Bueno C, Montes R, de la Cueva T, Gutierrez-Aranda I, Menendez P. Intra-bone marrow transplantation of human CD34(+) cells into NOD/LtSz-scid IL-2rgamma(null) mice permits multilineage engraftment without previous irradiation. Cytotherapy. 2010;12(1):45–49. [DOI] [PubMed] [Google Scholar]
- 50.Gutierrez-Aranda I, Ramos-Mejia V, Bueno C, et al. Human induced pluripotent stem cells develop teratoma more efficiently and faster than human embryonic stem cells regardless the site of injection. Stem Cells. 2010;28(9):1568–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vodyanik MA, Bork JA, Thomson JA, Slukvin II. Human embryonic stem cell-derived CD34+ cells: efficient production in the coculture with OP9 stromal cells and analysis of lymphohematopoietic potential. Blood. 2005;105(2):617–626. [DOI] [PubMed] [Google Scholar]
- 52.Prieto C, Stam RW, Agraz-Doblas A, et al. Activated KRAS cooperates with MLL-AF4 to promoteextramedullary engraftment and migration of cord blood CD34+ HSPC but is insufficient to initiate leukemia. Cancer Res. 2016;76(8):2478–2489. [DOI] [PubMed] [Google Scholar]
- 53.Deshpande AJ, Deshpande A, Sinha AU, et al. AF10 regulates progressive H3K79 methylation and HOX gene expression in diverse AML subtypes. Cancer Cell. 2014;26(6):896–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guenther MG, Lawton LN, Rozovskaia T, et al. Aberrant chromatin at genes encoding stem cell regulators in human mixed-lineage leukemia. Genes Dev. 2008;22(24):3403–3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Andersson AK, Ma J, Wang J, et al. The landscape of somatic mutations in infant MLL-rearranged acute lymphoblastic leukemias. Nat Genet. 2015;47(4):330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Malouf C, Ottersbach K. The fetal liver lymphoid-primed multipotent progenitor provides the prerequisites for the initiation of t(4;11) MLL-AF4 infant leukemia. Haematologica. 2018. June 14 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peters DG, Klucher KM, Perlingeiro RC, et al. Autocrine and paracrine effects of an ES-cell derived, BCR/ABL-transformed hematopoietic cell line that induces leukemia in mice. Oncogene. 2001;20(21): 2636–2646. [DOI] [PubMed] [Google Scholar]
- 58.Ji J, Risueno RM, Hong S, et al. Brief report: ectopic expression of NUP98-HOXA10 augments erythroid differentiation of human embryonic stem cells. Stem Cells. 2011; 29(4):736–741. [DOI] [PubMed] [Google Scholar]
- 59.Tan YT, Ye L, Xie F, et al. Respecifying human iPSC-derived blood cells into highly engraftable hematopoietic stem and progenitor cells with a single factor. Proc Natl Acad Sci U S A. 2018;115(9):2180–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marschalek R. Mechanisms of leukemogenesis by MLL fusion proteins. Br J Haematol. 2011;152(2):141–154. [DOI] [PubMed] [Google Scholar]
- 61.Boisset JC, van Cappellen W, Andrieu-Soler C, et al. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature. 2010;464(7285):116–120. [DOI] [PubMed] [Google Scholar]
- 62.Ahmad K, Scholz B, Capelo R, et al. AF4 and AF4-MLL mediate transcriptional elongation of 5-lipoxygenase mRNA by 1, 25-dihydrox-yvitamin D3. Oncotarget. 2015;6(28):25784–25800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Benedikt A, Baltruschat S, Scholz B, et al. The leukemogenic AF4-MLL fusion protein causes P-TEFb kinase activation and altered epigenetic signatures. Leukemia. 2011;25(1):135–144. [DOI] [PubMed] [Google Scholar]
- 64.Muck F, Bracharz S, Marschalek R. DDX6 transfers P-TEFb kinase to the AF4/AF4N (AFF1) super elongation complex. Am J Blood Res. 2016;6(3):28–45. [PMC free article] [PubMed] [Google Scholar]
- 65.Driessen EM, van Roon EH, Spijkers-Hagelstein JA, et al. Frequencies and prognostic impact of RAS mutations in MLL-rearranged acute lymphoblastic leukemia in infants. Haematologica. 2013;98(6):937–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kuhn A, Loscher D, Marschalek R. The IRX1/HOXA connection: insights into a novel t(4;11)- specific cancer mechanism. Oncotarget. 2016;7(23):35341–35352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.