Abstract
The mitochondrial proteome encompasses more than a thousand proteins, which are encoded by the mitochondrial and nuclear genomes. Mitochondrial biogenesis and network health relies on maintenance of protein import pathways and the protein-folding environment. Cell-extrinsic or -intrinsic stressors that challenge mitochondrial proteostasis negatively affect organellar function. During conditions of stress, cells use impaired protein import as a sensor for mitochondrial dysfunction to activate a stress response called the mitochondrial unfolded protein response (UPRmt). UPRmt activation leads to an adaptive transcriptional program that promotes mitochondrial recovery, metabolic adaptations, and innate immunity. In this review, we discuss the regulation of UPRmt activation as well as its role in maintaining mitochondrial homeostasis in physiological and pathological scenarios.
Mitochondria are double-membrane-bound organelles of endosymbiotic origin (Gray 2012). Often, they are described as the “powerhouse of the cell” for their bioenergetic function of cellular ATP generation by oxidative phosphorylation (OXPHOS). In addition, mitochondria are required for innate immunity, maintaining calcium homeostasis, synthesis of numerous cofactors, including heme and iron-sulfur clusters, as well as regulating apoptosis (Vandecasteele et al. 2001; Wang and Youle 2009; Stehling and Lill 2013; Weinberg et al. 2015). Mitochondria also act as platforms for metabolic pathways, including the TCA cycle, the urea cycle, β-oxidation, and lipid synthesis. The functional versatility and adaptability of the organelle depends on the concerted action by numerous proteins, protein complexes, and stress-response pathways.
Mitochondria are composed of more than 1000 proteins. Only 13 proteins are encoded by the mitochondrial genome (mitochondrial DNA [mtDNA]); all of which are components of the respiratory chain or ATP synthase (Anderson et al. 1981). All other mitochondrial proteins are encoded by nuclear genes, synthesized on cytosolic ribosomes, and subsequently imported into mitochondria via the TIM/TOM complexes (translocase of inner and outer membrane) (Chacinska et al. 2009). Given the high-protein concentration within mitochondria, coupled with complications deriving from the endosymbiotic origin of the organelle, maintenance of organellar protein homeostasis is challenging. Perturbations to proteostasis are caused by localized reactive oxygen species (ROS) production during normal and aberrant OXPHOS as well as uncoordinated expression of mitochondrial proteins encoded by either the mtDNA or nuclear genome (Curtis et al. 2012). Additionally, mutations in mtDNA and accumulation of damaged mtDNA adversely affect mitochondrial proteostasis (Moehle et al. 2018). Ultimately, such perturbations impair cellular health and are implicated in aging and several age-related diseases. For example, multiple studies have shown that mice harboring a mutation that annuls the proofreading activity of the mtDNA polymerase POLGγ have increased mtDNA mutations concomitant with an accelerated aging phenotype (Trifunovic et al. 2004; Kujoth et al. 2005). Thus, understanding pathways in place to maintain mitochondrial proteostasis and function not only provide insights into disease etiology but may also lead to new therapeutic approaches.
MITOCHONDRIAL BIOGENESIS, PROTEIN IMPORT, AND PROTEOSTASIS
Mitochondrial biogenesis is an elaborate process involving membrane synthesis, mtDNA replication, as well as import and trafficking of those proteins synthesized in the cytosol to the appropriate intramitochondrial compartments (outer membrane, intermembrane space, inner membrane, or matrix). Here, we focus on the trafficking of proteins to the matrix, which typically requires a mitochondrial targeting sequence (MTS) on the amino terminus of the protein to be imported. The MTS is a positively charged, amphipathic ∼25–35 amino acid sequence that first interacts with the TOM complex before import (Mayer et al. 1995; Meisinger et al. 2001; Saitoh et al. 2007). On passing through the TOM complex in an unfolded state, precursor proteins translocate through the TIM23 complex, which requires the inner membrane potential generated by a functional respiratory chain (Martin et al. 1991). Finally, preproteins translocate into the mitochondrial matrix, which also requires the matrix-localized molecular chaperone mtHsp70 (Mapa et al. 2010). mtHsp70 interacts directly with the TIM23 complex via cochaperones and facilitates import by either actively pulling proteins into the matrix or passively trapping preproteins to prevent retrograde diffusion out of the matrix (Gaume et al. 1998; Voisine et al. 1999; D'Silva et al. 2003). Once in the matrix, MTS is cleaved by the mitochondrial processing peptidase (MPP). Thus, mtHsp70 plays a dual role during protein import, both at the import channel and in the matrix to assist in proper protein handling. In yeast, it has been shown that the distinct functional roles of mtHsp70 in protein import and protein folding in the matrix can be attributed to its differential interactions with cochaperones forming two different complexes (Horst et al. 1997). At the import complex, mtHsp70 associates with Tim44 of the TIM23 complex, whereas in the matrix it interacts with Mdj1, the DnaJ homolog (Horst et al. 1997).
Chaperone-assisted protein folding following import is key to maintaining proteostasis in the matrix. In addition to mtHsp70, the chaperonin Hsp60 also interacts with imported proteins in the matrix to facilitate protein folding (Cheng et al. 1989). Hsp60s in cooperation with Hsp10 undergoes ATP-dependent conformational changes to form a protective socket that allows substrate proteins to fold within them by preventing contact with other surrounding proteins. Hsp60 also plays a protective role under conditions of heat stress by preventing aggregate formation (Bender et al. 2011).
Moreover, mitochondrial-localized quality-control proteases maintain proteostasis by degrading damaged or terminally misfolded proteins. The AAA proteases LON and ClpXP are largely responsible for removing damaged proteins from the mitochondrial matrix (Tatsuta and Langer 2009). Also, paraplegin and YMEL1 primarily prevent misfolding of respiratory chain complex proteins on or near the mitochondrial inner membrane (Leonhard et al. 1996).
In addition to OXPHOS activity and mitochondrial proteostasis, numerous additional factors in multiple cellular compartments contribute to the regulation and efficiency of mitochondrial protein import. For example, phosphorylation of an MTS can reduce import efficiency (Colombo et al. 2005; Boopathi et al. 2008; Rao et al. 2011). In the cytosol, protein import can be regulated by the binding of protein partners or metabolites to precursor proteins (Vongsamphanh et al. 2001; Munakata et al. 2004). For instance, import of the mitochondrial protein 5-aminolevulinate is impaired on heme binding in the cytosol (Dailey et al. 2005). Folding of the precursor proteins before interacting with the TOM channel can inhibit entry (Hill et al. 1998; Truscott et al. 2001; Strobel et al. 2002). As the majority of protein import requires the TOM complex, mechanisms that modulate the activity of the import machinery influence the entry of precursor proteins (Harbauer et al. 2014). Studies in yeast have shown that casein kinases (CKs) and protein kinase A (PKA) can phosphorylate TOM to modulate protein import. CKs phosphorylate Tom22 to promote import, whereas PKA phosphorylates Tom40, Tom22, as well as Tom70 to impair protein import (Schmidt et al. 2011; Rao et al. 2012; Gerbeth et al. 2013).
Mitochondrial function and proteostasis are integrally linked with mitochondrial protein import. For example, mitochondrial uncoupling or dissipation of the inner membrane potential because of OXPHOS defects directly inhibits protein translocation into the mitochondria. Furthermore, proteostasis perturbations require mtHsp70 to facilitate protein folding, which may impact the chaperones’ role in mitochondrial protein import. Because protein import capacity and proteostasis correlate with mitochondrial function, import efficiency can serve as a sensor for detecting perturbations in mitochondrial homeostasis. At least two mitochondrial stress responses are regulated by mitochondrial import capacity. Severe impairment of protein import leads to degradation of the entire compartment via mitochondrial autophagy (mitophagy) (Pickles et al. 2018). Impaired import also leads to activation of an adaptive transcriptional response known as the mitochondrial unfolded protein response (UPRmt), which promotes recovery of mitochondrial function (Shpilka and Haynes 2018). In this review, we focus on the mechanisms that regulate UPRmt activation and its role in diverse physiological scenarios.
UPRmt REGULATION
The UPRmt was initially discovered in cultured mammalian cells in which depletion of mtDNA or overexpression of a mitochondrial-targeted protein harboring a deletion rendering the protein terminally misfolded (ΔOTC) elicited a transcriptional response that included mitochondrial molecular chaperones and quality-control proteases (Martinus et al. 1996; Zhao et al. 2002). Over the ensuing years, findings in Caenorhabditis elegans have identified components in multiple subcellular compartments providing insight into how cells detect mitochondrial dysfunction and communicate with the nucleus to adapt transcription accordingly. Importantly, homologous UPRmt regulatory components have been identified in mammalian systems, demonstrating the evolutionary conservation of the adaptive mitochondrial stress response (Fiorese et al. 2016; Münch and Harper 2016; Quirós et al. 2017).
Stressors that Perturb Mitochondria and Activate the UPRmt
A diverse number of stressors, which impair mitochondrial function, activate the UPRmt. Stressors that perturb mitochondrial proteostasis components, such as molecular chaperones and quality-control proteases, are among the strongest inducers of the UPRmt, possibly owing to the accumulation of misfolded proteins in the mitochondria, which exceeds the protein-handling capacity of chaperones and proteases, thereby hindering protein import (Yoneda et al. 2004). Consistent with this model, inhibiting import directly by depleting the import complex TIM (timm-23) strongly activates the UPRmt (Nargund et al. 2012). Furthermore, inhibition of mitochondrial protein synthesis via RNA interference (RNAi) or mitochondrial ribosome inhibitors, such as doxycycline or chloramphenicol, activates the UPRmt (Yoneda et al. 2004; Houtkooper et al. 2013).
In addition to perturbations in proteostasis, impairment of mitochondrial metabolic functions also elicits UPRmt activation. For example, mtDNA depletion by treatment with ethidium bromide activates the UPRmt (Yoneda et al. 2004; Houtkooper et al. 2013). Inhibition of OXPHOS by multiple compounds, including rotenone (complex I inhibitor), antimycin (complex III inhibitor), oligomycin (complex V inhibitor), or paraquat, also activates the UPRmt (Yoneda et al. 2004; Runkel et al. 2013). Lastly, inhibition of fumarate hydratase, the enzyme responsible for converting fumarate to malate in the TCA cycle, activates the UPRmt, indicating that the mitochondrial stress response is engaged by both mitochondrial proteostasis and metabolic perturbations (Wang et al. 2016b).
Transcriptional Program Mediated by UPRmt Activation
Mitochondrial Network Recovery and Function
The mitochondrial recovery program elicited by UPRmt activation includes induction of mitochondrial chaperones Hsp60 and mtHsp70, which promote protein folding and prevent aggregate formation. To eliminate the accumulation of damaged or misfolded proteins, the ATP-dependent m-AAA and i-AAA proteases, such as paraplegin and YMEL-1, are induced as are superoxide dismutases (SODs) to limit the accumulation of ROS from dysfunctional OXPHOS.
In addition to a transcriptional response induced for repair and recovery of damaged mitochondria, the UPRmt activation also induces genes that promote mitochondrial biogenesis and function, including iron–sulfur cluster and ubiquinone synthesis required for OXPHOS complex biogenesis. UPRmt activation also induces genes involved in mitochondrial fission, such as Dynamin-related protein (Drp1), to promote mitochondrial biogenesis as well as clearance of defective mitochondria.
Metabolic Adaptations
Most OXPHOS components are encoded by nuclear genes and assembled into complexes via chaperones and assembly factors. UPRmt activation limits the transcription of OXPHOS components possibly to reduce the load on the proteostasis machinery during stress (Nargund et al. 2015). Concomitantly, UPRmt activation induces all glycolysis genes, suggesting that UPRmt plays a role in balancing the metabolic landscape of the cell during mitochondrial stress likely by promoting an alternative means of ATP synthesis to facilitate mitochondrial recovery.
Mitochondria-to-Nuclear Communication
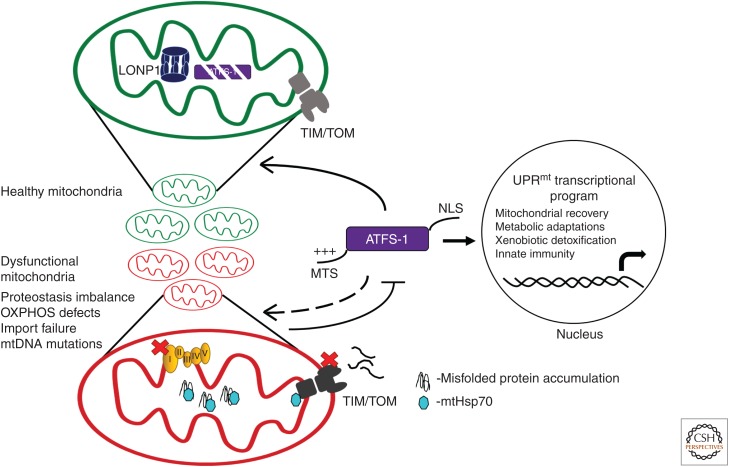
Genetic screens in C. elegans have revealed multiple components required for UPRmt activation, including the bZIP transcription factor associated with Stress-1 (ATFS-1) (Fig. 1). In addition to its bZIP DNA-binding domain, ATFS-1 also harbors both an MTS and a nuclear localization sequence (NLS) (Nargund et al. 2012). The net positive charge of the MTS facilitates its import into functional mitochondria, in which the MTS is cleaved and the remainder of the protein is degraded by the matrix-localized protease LONP1. However, during mitochondrial stress, a decline in mitochondrial function reduces import efficiency of ATFS-1 causing a fraction of the transcription factor to remain in the cytosol. Because ATFS-1 harbors an NLS, it then traffics to the nucleus to initiate a transcriptional program (Nargund et al. 2012). Thus, mitochondrial import efficiency of ATFS-1 allows cells to sense and monitor perturbations in mitochondrial network health and homeostasis (Haynes et al. 2010). In line with this model, mutations that reduce the strength of ATFS-1's MTS limit its mitochondrial import efficiency and constitutively activate the UPRmt (Rauthan et al. 2013).
Figure 1.
Regulation of the mitochondrial unfolded protein response (UPRmt) by ATFS-1. UPRmt activation is mediated by the transcription factor ATFS-1, which harbors a mitochondrial targeting sequence (MTS) as well as a nuclear localization sequence (NLS). When mitochondria are healthy, ATFS-1 is imported via the TIM/TOM (translocase of the inner and outer membrane) complex and subsequently degraded by the matrix-localized protease LONP1. Mitochondrial import of most precursor proteins and efficient folding within the matrix requires the matrix-localized chaperone mtHsp70 located at both the import channel as well as in the matrix. Mitochondrial dysfunction because of impaired proteostasis or oxidative phosphorylation (OXPHOS) reduces mitochondrial protein import efficiency of ATFS-1, allowing it to traffic to the nucleus to initiate a transcriptional program, which promotes the recovery of mitochondrial function.
ATFS-1 is activated by impaired mitochondrial protein import. But, how are the induced proteostasis components, such as chaperones and proteases, imported into mitochondria to facilitate recovery? One possibility is that ATFS-1 activation transcriptionally induces genes encoding the import machinery, thereby gradually increasing mitochondrial protein import as well as protein-handling capacity. Another possibility is that because mitochondrial chaperones, such as mtHsp70, play a dual role at the import complex and in the matrix for protein folding, reduced import capacity during stress may be concomitant with spatial redistribution of mtHsp70 and other chaperones from the sites of protein import to the sites of accumulated or misfolded proteins. Alternatively, ATFS-1 may simply have a relatively weak MTS, and the proteins induced by ATFS-1 may have strong MTSs, enabling them to be imported into dysfunctional mitochondria (Melber and Haynes 2018).
In addition to ATFS-1, several other proteins required for UPRmt activation have been identified. The homeobox domain transcription factor DVE-1 forms a complex with the ubiquitin-like protein UBL-5, and remodels chromatin by binding to UPRmt gene promoters such as hsp-60 (Benedetti et al. 2006; Haynes et al. 2007; Tian et al. 2016). Presumably, chromatin remodeling enables ATFS-1 to bind the UPRmt element in the promoters of UPRmt genes (Nargund et al. 2015).
The Role of Chromatin Remodeling in UPRmt Activation
Epigenetic regulation by chromatin remodeling plays an important role in UPRmt activation during mitochondrial dysfunction. A recent study has shown that mitochondrial stress caused by knockdown of prohibitin or a respiratory chain complex IV subunit leads to nuclear localization of the chromatin modifier LIN-65 (Tian et al. 2016). LIN-65 function requires the H3K9 methyltransferase MET-2. Combined, these factors reorganize global chromatin structure and alter compaction to enable transcription of UPRmt genes in response to stress (Tian et al. 2016).
The histone demethylases jmjd-1.2 and jmjd-3.1, members of the H3K27 family containing jmC-domain, are also necessary to activate the UPRmt when OXPHOS is impaired (Merkwirth et al. 2016). Furthermore, jmjd-1.2 and jmjd-3.1 overexpression is sufficient to induce the UPRmt and increase longevity. Both demethylases remove repressive chromatin marks and, therefore, positively regulate gene expression by promoting access of the transcriptional machinery to UPRmt gene promoters. The mechanisms that stimulate the activity of JMJD-1.2 and JMJD-3.1 during mitochondrial stress are unclear, but may relate to the regulation of these demethylases by metabolic cofactors, including α-ketoglutarate (a TCA-cycle intermediate) and iron (Tsukada et al. 2006). Production of such metabolic cofactors largely depends on healthy mitochondrial function and ATP generation (Teperino et al. 2010). During mitochondrial dysfunction, changes in metabolic pathways may alter the level of these metabolites, consequently affecting the activity of histone demethylases to promote UPRmt activation.
UPRmt Regulation and the Integrated Stress Response
Transcriptional regulation of the UPRmt in mammalian cells is comparable to that in C. elegans and is mediated by multiple bZIP transcription factors including ATF5 (Fiorese et al. 2016). Much like ATFS-1, ATF5 harbors an MTS and is regulated by organelle partitioning. Importantly, ATF5 rescues UPRmt activation in nematodes lacking ATFS-1 and cultured cells, ATF5 transcriptionally up-regulates mitochondrial chaperones and proteases during mitochondrial stress (Fiorese et al. 2016).
However, in mammalian systems, UPRmt activation requires at least two additional bZIP proteins, ATF4 and CHOP, as rotenone exposure transcriptionally up-regulates both ATF4 and CHOP (Horibe and Hoogenraad 2007; Silva et al. 2009; Quirós et al. 2017). Furthermore, mtDNA depletion causes induction of ATF4 and ATF5. All three transcription factors are linked to the integrated stress response (ISR), which is a conserved adaptive response activated by a wide variety of stressors. The ISR was first characterized in yeast as the “general control pathway,” which is activated during amino acid deprivation. The yeast pathway was later found to be a highly conserved stress-response mechanism in mammalian systems (Harding et al. 2000). ISR initiation is mediated by kinases that respond to specific stressors and phosphorylate serine 51 of the translation initiation factor subunit eIF2α (Pathak et al. 1988). The mammalian ISR kinases are RNA-dependent protein kinase (PKR)-like ER kinase (PERK), heme-regulated eIF2α kinase (HRI), double-stranded PKR, and general control nonderepressible 2 (GCN2). PERK, HRI, and PKR are specifically activated in response to perturbations in ER homeostasis, heme availability, and the presence of cytosolic double-stranded RNA, respectively (Meurs et al. 1990; Chen and London 1995; Harding et al. 1999). GCN2 is activated by amino acid or glucose deprivation, ROS, ribosome stalling, mitochondrial dysfunction, and UV light (Dever et al. 1992; Deng et al. 2002; Baker et al. 2012). Ultimately, eIF2α phosphorylation causes attenuation of global protein synthesis while promoting the selective translation of messenger RNAs (mRNAs) harboring upstream open reading frames (uORFs) in the 5′ untranslated region (5′UTR). The bZIP transcription factors CHOP, ATF4, and ATF5 are among the mRNAs selectively translated on eIF2α phosphorylation.
In sum, ISR activation in mammals during stress causes eIF2α phosphorylation, which promotes activation of the transcription factors ATF4, CHOP, and ATF5. However, the nuances of the interrelationship between the transcription factors during mitochondrial stress or which kinases are specifically activated by which mitochondrial perturbations is not yet fully understood.
UPRmt and Translation Regulation within Mitochondria
mtDNA-encoded transfer RNAs (tRNAs) are essential for translation and RNA maturation within mitochondria. Following transcription, mitochondrial tRNAs are processed at the 5′ and 3′ ends by the RNase P and RNase Z complexes (Rossmanith 2012). In humans, RNase P is comprised of the proteins MRPP1, MRPP2, and MRPP3. During mitochondrial stress, UPRmt activation results in decreased MRPP3 caused by increased degradation by LON protease (Münch and Harper 2016). Therefore, UPRmt activation reduces protein synthesis within the cytosol by phosphorylating eIF2α and mitochondria by degradation of MRPP3 likely facilitating proteostasis and organelle recovery.
Cell-Nonautonomous UPRmt Regulation
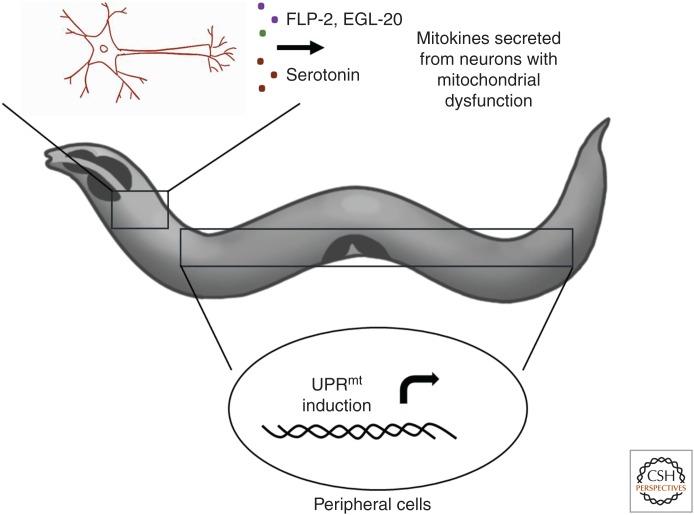
In addition to mitochondrial-to-nuclear communication within each cell, mitochondrial dysfunction can be communicated between tissues to induce the UPRmt. Recent studies in C. elegans and mammals indicate that intercellular communication involves secretion of signaling factors, called mitokines, from neurons with dysfunctional mitochondria. The mitokine(s) then act on peripheral cells, such as the intestinal cells, to activate the UPRmt (Fig. 2) (Durieux et al. 2011; Kim et al. 2013b; Berendzen et al. 2016; Shao et al. 2016). For example, FLP-2, a neuropeptide secreted from interneurons, acts as a signaling factor to activate the UPRmt in intestinal cells when mitochondrial stress is induced specifically in neurons (Shao et al. 2016). A separate study showed that during neuronal proteotoxic stress, secretion of the neurotransmitter serotonin is also required for communication with intestinal cells to activate the UPRmt (Berendzen et al. 2016). In addition to FLP-2 and serotonin, Wnt signaling pathway is required for cell-nonautonomous UPRmt activation. Mitochondrial stress induced in neurons leads to secretion of the Wnt ligand EGL-20, which acts as a mitokine to induce the UPRmt in peripheral cells (Zhang et al. 2018).
Figure 2.
Cell-nonautonomous mitochondrial unfolded protein response (UPRmt) signaling. Neuronal activation of the UPRmt is communicated cell nonautonomously to the intestine by the secretion of mitokines (FLP-2, serotonin, EGL-20) to activate the UPRmt in distal tissues such as the intestine.
In mammalian systems, the potential mitokine fibroblast growth factor 21 (FGF21), a metabolic hormone expressed in multiple mammalian tissues, is induced by the ISR regulator ATF4 during mitochondrial stress caused by autophagy deficiency (Kim et al. 2013a,b). Growth differentiation factor 15 (GDF15) is also a potential mitokine, which is induced during mitochondrial stress in skeletal muscles (Chung et al. 2017). In mice, GDF15 induction causes changes in lipid metabolism and insulin sensitivity, thereby conferring tolerance to onset of obesity (Chung et al. 2017).
Physiologies and Pathologies Impacted by UPRmt Signaling
UPRmt Signaling and Organismal Longevity
Decline in mitochondrial function is a hallmark of aging in most organisms and associated with accumulation of mtDNA mutations as well as reduced OXPHOS (Bratic and Larsson 2013). Surprisingly, considerable evidence indicates that modest mitochondrial perturbations cause increased longevity (Feng et al. 2001; Dillin et al. 2002; Liu et al. 2005; Copeland et al. 2009). In C. elegans, impairment of respiratory chain complex IV function (cco-1 RNAi) both activates the UPRmt and extends life span by ∼50% (Fig. 3) (Durieux et al. 2011). Interestingly, the life-span modulation is dependent on the developmental stage during which mitochondrial function is impaired, that is, increased longevity occurs only when cco-1 RNAi is administered before adulthood. This suggests that temporal regulation of mitochondrial function is associated with longevity, possibly owing to epigenetic level changes during different stages of development (Dillin et al. 2002; Durieux et al. 2011). Consistent with the role of mitochondria as central regulators of metabolic homeostasis, changes in metabolic inputs affecting mitochondrial function influence UPRmt activation and, in turn, impact life span. For example, modulating levels of the metabolic cofactor NAD+ activates the UPRmt and extends life span (Mouchiroud et al. 2013).
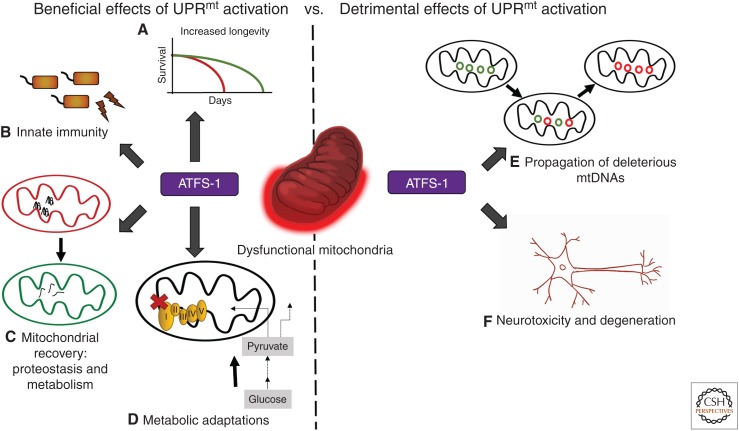
Figure 3.
Functions and consequences of mitochondrial unfolded protein response (UPRmt) activation. UPRmt activation has beneficial, as well as detrimental effects, depending on the physiological scenario as well as duration and strength of activation. Beneficial outcomes include (A) increased longevity, (B) increased resistance against pathogen infection, (C) efficient recovery of the mitochondrial network, and (D) metabolic adaptations, including the induction of glycolysis. However, prolonged UPRmt activation can result in (E) amplification of deleterious mitochondrial DNAs (mtDNAs), and (F) neurotoxicity, emphasizing the importance of proper UPRmt regulation. ATFS-1, Activated transcription factor associated with Stress-1.
Parallel studies in multiple organisms have shown that modest mitochondrial perturbations, which cause UPRmt activation, also extend life span. A study in flies showed that inducing mitochondrial stress in muscle cells by perturbing respiratory chain function activates the UPRmt, specifically in muscles, and promotes longevity (Owusu-Ansah et al. 2013). In mice, impaired function of mitochondrial ribosomes by genetic or pharmacological means both activates the UPRmt and extends life span (Houtkooper et al. 2013). Although considerable data indicate a role for the UPRmt in extending life span, the cellular processes impacted by UPRmt activation, which influence life span, remain to be identified.
UPRmt Signaling in Propagation of Deleterious Mitochondrial Genome
Most cells harbor hundreds of copies of the mitochondrial genome distributed among the mitochondrial network, which are inherited via the maternal germline (Miller et al. 2003). As mtDNA harbors 13 protein-coding genes that are all essential for OXPHOS, mtDNA mutations can cause a wide range of pathologies, including neuropathies and myoclonic epilepsy (Tuppen et al. 2010). mtDNA acquires mutations at a higher rate than the nuclear genome, possibly because of the lack of efficient DNA repair pathways or the presence of a mutagenic environment. Given the high copy number of mtDNA, such mutations are generally well tolerated as they coexist with wild-type mtDNAs in a state known as heteroplasmy. However, as cells such as muscle and neurons age, the deleterious mtDNA (ΔmtDNA) often accumulates faster than the wild-type mtDNA, resulting in loss of OXPHOS. Although the mechanisms that underlie the preferential accumulation of ΔmtDNAs remain largely unknown, recent work in two heteroplasmic C. elegans strains indicates a requirement for ATFS-1 and the UPRmt in maintaining the ΔmtDNA (Gitschlag et al. 2016; Lin et al. 2016).
One such strain harbors ∼60% ΔmtDNA and 40% wild-type mtDNA. The ΔmtDNA harbors a 3.1-kb deletion lacking four essential OXPHOS genes and perpetually activates the UPRmt as a result of reduced OXPHOS. Impressively, deletion of the atfs-1 gene results in nearly a complete loss of the ΔmtDNA, while having no effect on the wild-type genomes. Alternatively, hyperactivation of the UPRmt causes a preferential accumulation of the ΔmtDNA, indicating that UPRmt activation is both necessary and sufficient for the preferential accumulation of ΔmtDNAs (Fig. 3; Gitschlag et al. 2016; Lin et al. 2016). Although the outputs of UPRmt activation that promote ΔmtDNA propagation remain unclear, evidence suggests that imbalanced mitochondrial biogenesis and mitophagy are involved in the process (Lin et al. 2016).
UPRmt Signaling in Mitochondrial Diseases
Dysfunctional mitochondria are manifested in several diseases that are currently incurable (Suomalainen 2015). An example of this is mitochondrial myopathy, a progressive disease associated with defective respiratory chain function and muscular abnormalities often resulting from accumulating mtDNA mutations (Pfeffer and Chinnery 2013). A recent study showed the link between mTORC1 (mechanistic target of rapamycin complex 1) and UPRmt regulation in a manner dependent on uORFs during mitochondrial myopathy. mTORC1 is a regulatory kinase that responds to nutrient availability, ATP production, and regulates protein synthesis through downstream effectors such as S6 kinase (Rohde et al. 2001; Magnuson et al. 2012). Using a mouse model of adult-onset mitochondrial myopathy caused by accumulating mtDNA deletions, the study showed that mitochondrial dysfunction stimulates mTORC1 (Khan et al. 2017). Interestingly, mTORC1 activation was required for induction of a transcriptional program, including cytokines such as FGF21 and GDF15, consistent with the UPRmt. Importantly, mTORC1 activation caused transcriptional up-regulation of ATF4 and ATF5, which is impaired by treatment with the known mTORC1 inhibitor rapamycin (Khan et al. 2017). However, the mechanistic details of mTORC1-mediated translational regulation of ATF4 and ATF5 remains elusive (Khan et al. 2017).
UPRmt Signaling in Neurodegenerative Diseases
Alzheimer's disease, a progressive neurodegenerative condition associated with aging, is characterized by mitochondrial dysfunction along with proteotoxic aggregation of amyloid–β (Aβ) peptides (Hashimoto et al. 2003). Recent work has suggested that the UPRmt is activated in neurons of Alzheimer's disease patients (Sorrentino 2017). Consistent with this data, expression of Aβ in C. elegans results in UPRmt activation (Sorrentino 2017). Impressively, enhanced UPRmt activation reduces the toxicity associated with Aβ accumulation and prolongs survival. In mouse models of Alzheimer's disease treatment with nicotinamide riboside (NR), which boosts NAD+ levels and activates the UPRmt, alleviates proteoxicity, as well as improves memory (Sorrentino 2017). In brief, these findings suggest that strategies to enhance UPRmt activation may be useful in the treatment of Alzheimer's disease.
In contrast to the findings relating to Alzheimer's disease and the UPRmt, prolonged or hyperactivation of the UPRmt causes dopaminergic neuron death in a C. elegans model of Parkinson's disease in which the protein α-synuclein is overexpressed (Fig. 3) (Martinez et al. 2017). Combined, these results indicate that UPRmt activation can have either beneficial or detrimental effects in the context of neurodegenerative conditions.
UPRmt Signaling in Cancer
Cancer is a complex and multifactorial disease that is often associated with mitochondrial dysfunction because of the hypoxic environment within tumors, accumulation of mtDNA mutations, and relatively high levels of ROS (Chatterjee et al. 2006; Liou and Storz 2010; Boland et al. 2013). Tumor cells rely more heavily on glycolysis rather than OXPHOS despite the presence of oxygen, a phenomenon known as the Warburg effect. Defects in mitochondrial function of cancer cells may contribute to altered flux in metabolic pathways giving rise to the Warburg effect. Accumulating evidence suggests that UPRmt activation, in response to mitochondrial dysfunction, contributes to cancer cell growth and survival. For example, expression of Hsp60 is induced in multiple cancer types presumably to maintain mitochondrial function (Cappello et al. 2002; Tsai et al. 2008). Furthermore, activation of ISR kinases PERK, GCN2, and PKR has been implicated in tumor cell survival and neoplastic growth although the mechanistic consequences of mitochondrial dysfunction in tumor cells remain unclear (Kim et al. 2000, 2002; Ye et al. 2010; Nagelkerke et al. 2013). Specifically, in gastric cancers, evidence indicates that ROS activates ATF4 via GCN2 causing enhanced resistance to the chemotherapeutic drug cisplatin (Wang et al. 2016a). Numerous studies have shown a role for ATF5 in the growth of a variety of cancer cell types, suggesting UPRmt inhibition may be a viable means to limit cancer progression (Chen et al. 2012; Wang et al. 2012; Nukuda et al. 2016; Angelastro 2017).
UPRmt Signaling during Pathogenic Infection
Bacterial pathogens secrete a wide array of virulence effectors that curb host functions to establish a persistent infection. Multiple pathogen-secreted virulence factors specifically target host mitochondrial function. For example, the opportunistic pathogen Pseudomonas aeruginosa perturbs mitochondria by secreting toxins, such as cyanide and siderophores, that impair OXPHOS (Kirienko et al. 2015; Managò et al. 2015). Recent studies in C. elegans show that pathogenic and nonpathogenic strains of Pseudomonas perturb mitochondrial function and activate the UPRmt (Fig. 3) (Liu et al. 2014; Pellegrino et al. 2014). Surprisingly, these studies revealed that in addition to mitochondrial-protective genes, ATFS-1 mediates the induction of antibacterial genes, including secreted lysozymes and antimicrobial peptides, as well as xenobiotic detoxification genes (Pellegrino et al. 2014). Importantly, worms lacking ATFS-1 were susceptible to infection, whereas hyperactivation of the UPRmt limited infection and prolonged host survival. These findings indicate that the UPRmt functions as an innate immune response and one mechanism by which eukaryotes can detect the presence of bacterial species that produce mitochondrial toxins, including OXPHOS and mitochondrial ribosome inhibitors (tetracyclines), is by monitoring mitochondrial function via the UPRmt (Pellegrino et al. 2014). In turn, the UPRmt activates a response to maintain mitochondrial function, and rid the host of the pathogen as well as the toxins.
Additional Mechanisms for Mitochondrial Quality Control
In addition to the UPRmt, several other mechanisms are activated during mitochondrial dysfunction to promote recovery of the mitochondrial network as well as alleviate pleiotropic toxic effects throughout the cell (Fig. 4). Intriguingly, all the mechanisms are stimulated by impaired mitochondrial protein import. Severely or irrecoverably damaged mitochondria are eliminated via mitophagy, which is a form of selective autophagy. Damaged mitochondria are detected by the protein kinase PINK1, which, like ATFS-1, is regulated by mitochondrial import efficiency. PINK1 is imported into healthy organelles in which it is processed and then degraded (Narendra et al. 2010). During mitochondrial stress, if PINK1 fails to be imported across the mitochondrial inner membrane, it accumulates on the outer membrane with the kinase domain facing the cytosol. Here, PINK1 phosphorylates ubiquitin and the ubiquitin ligase, Parkin, which in turn leads to polyubiquitination of multiple mitochondrial outer membrane proteins (Kondapalli et al. 2012; Kane et al. 2014; Kazlauskaite et al. 2014; Koyano et al. 2014; Ordureau et al. 2014). The accumulation of ubiquitinated mitochondrial proteins recruits adaptor proteins, which facilitate cargo recognition by the autophagy machinery (Heo et al. 2015). In the physiological context, mitophagy plays a role in dynamically modulating the levels of deleterious mitochondrial genome in addition to the UPRmt. Mutations in genes encoding both PINK1 and Parkin cause early-onset Parkinson's disease, suggesting that mitophagy plays a prominent role in maintaining the mitochondrial network within dopaminergic neurons (Lucking et al. 2000; Valente et al. 2004).
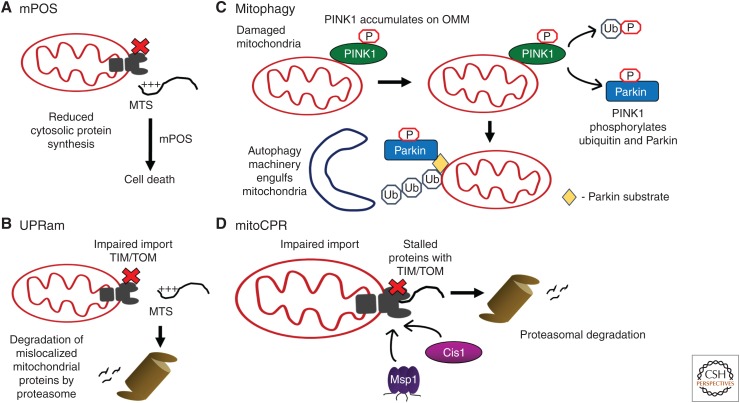
Figure 4.
Mitochondrial stress-response pathways that potentially interact with the mitochondrial unfolded protein response (UPRmt). (A) Impaired mitochondrial protein import caused by mitochondrial metabolic or proteostasis defects causes accumulation of mitochondrial precursor proteins in the cytosol and cell death; a pathway termed mPOS (mitochondrial precursor overaccumulation stress). (B) UPRam (unfolded protein response activated by mistargeted proteins) is activated during mitochondrial protein import failure, which causes accumulation of mitochondrial precursor proteins in the cytosol. To rid the cytosol of the toxic mislocalized proteins, proteasomal activity is increased. (C) Mitophagy is a process by which severely damaged mitochondria are degraded. The kinase PINK1 accumulates on the outer membrane of damaged mitochondria (OMM), where it phosphorylates ubiquitin and the E3 ubiquitin ligase Parkin, which leads to polyubiquitination of many OMM proteins. Subsequently, recognition by the autophagy machinery results in engulfment of the defective organelle, which is then delivered to lysosomes for degradation. (D) mitoCPR (mitochondrial compromised protein import response) is a stress-response activated by accumulation or stalling of mitochondrial precursor proteins within TOM (translocase of outer membrane) channels. The linker protein Cis1 is transcriptionally induced to recruit the ATP-dependent translocase Msp1 to the TOM channel to extract the stalled precursor proteins for proteasomal degradation. MTS, Mitochondrial targeting sequence; TIM, translocase of inner membrane.
Because nearly all mitochondrial-localized proteins are encoded on cytosolic ribosomes, impaired mitochondrial import results in the accumulation of mislocalized mitochondrial precursor proteins within the cytosol. If not reduced, the toxic mislocalized proteins cause cell death via a pathway dubbed mPOS (mitochondrial precursor overaccumulation stress) (Wang and Chen 2015). Although the mechanism by which mPOS causes cell death remains to be determined, at least two approaches are used to alleviate toxicity, including increased proteasomal activity to degrade the mislocalized proteins and reduced cytosolic translation to decrease the expression of mitochondrial precursors, which ultimately cannot be imported. These studies emphasize the damaging effects that mitochondrial dysfunction can have on the cytosol, and highlight cellular activities or pathways that serve to restore cytosolic proteostasis, collectively known as UPRam (unfolded protein response activated by mistargeting of proteins) (Wrobel et al. 2015).
Furthermore, a novel pathway that is activated specifically to resolve mitochondrial import stress known as mitoCPR (mitochondrial compromised protein import response) was discovered recently (Weidberg and Amon 2018). mitoCPR induces a transcriptional program that comprises more than 200 genes, including ABC transporters, and is regulated by a transcription factor PDR3 (pleiotropic drug resistance 3). Intriguingly, PDR3 induces the expression of Cis1, which recruits the ATP-dependent translocase Msp1 (mitochondrial sorting protein) to the TOM channel to facilitate the extraction and degradation of stalled proteins. Thus, mitoCPR functions to alleviate import stress and restore homeostasis by facilitating clearance of stalled precursor proteins.
The existence of multiple quality-control pathways, in addition to the UPRmt, illustrates the difficulty and importance of maintaining mitochondrial proteostasis. For the most part, each of these pathways has been studied in isolation. But, because all are regulated or impacted directly by impaired mitochondrial protein import, it will be exciting to understand how, or if, they interact both temporally and spatially.
PERSPECTIVES
Over the years, many studies from different organisms have shed light on the pathways that contribute to the maintenance of mitochondrial health and function. We have focused on the UPRmt, which monitors activity of the mitochondrial network and responds to a wide array of stressors, including proteostasis and metabolic perturbations. Reduced mitochondrial protein import, arising as a consequence of either mitochondrial proteostasis or metabolic stress, leads to nuclear translocation of the transcription factor ATFS-1, which mediates UPRmt activation, which in turn elicits an adaptive transcriptional program. ATFS-1 up-regulates expression of chaperones-encoding genes and promotes mitochondrial biogenesis. UPRmt activation also modulates protein import and restores mitochondrial proteostasis. This is mechanistically conserved in mammals as the transcription factor ATF5 is also regulated by impaired protein import and organelle partitioning. However, UPRmt regulation in mammals requires translation regulation, which necessitates one of four eIF2α kinases, as well as two additional transcription factors (ATF4 and CHOP), which are intimately associated with the ISR. The ISR has been best characterized as a response to ER stress, but how the pathway is specified to mitochondrial stress is yet to be discovered. It will also be of interest to further understand mechanisms that regulate ATF5 and ATFS-1 mitochondrial protein import efficiency, which may occur by direct protein modifications, such as phosphorylation, or by binding of metabolites and protein partners.
As discussed in this review, the diverse physiological consequences resulting from mitochondrial dysfunction repeatedly underscores the need for further research in understanding the UPRmt. UPRmt activation has been shown to have multiple beneficial effects such as promoting mitochondrial recovery, mounting an innate immune response against pathogenic infection, metabolic adaptations, and increasing longevity. However, prolonged UPRmt activation can result in neurodegeneration and propagation of deleterious mitochondrial genomes, suggesting that precise regulation of UPRmt activation is essential.
ACKNOWLEDGMENTS
This work was supported by the Howard Hughes Medical Institute (HHMI), the Mallinckrodt Foundation, and National Institutes of Health Grants (R01AG040061 and R01AG047182) to C.M.H.
Footnotes
Editors: Richard I. Morimoto, F. Ulrich Hartl, and Jeffery W. Kelly
Additional Perspectives on Protein Homeostasis available at www.cshperspectives.org
REFERENCES
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. 1981. Sequence and organization of the human mitochondrial genome. Nature 290: 457–465. 10.1038/290457a0 [DOI] [PubMed] [Google Scholar]
- Angelastro JM. 2017. Targeting ATF5 in cancer. Trends Cancer 3: 471–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BM, Nargund AM, Sun T, Haynes CM. 2012. Protective coupling of mitochondrial function and protein synthesis via the eIF2α kinase GCN-2. PLoS Genet 8: e1002760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender T, Lewrenz I, Franken S, Baitzel C, Voos W. 2011. Mitochondrial enzymes are protected from stress-induced aggregation by mitochondrial chaperones and the Pim1/LON protease. Mol Biol Cell 22: 541–554. 10.1091/mbc.e10-08-0718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti C, Haynes CM, Yang Y, Harding HP, Ron D. 2006. Ubiquitin-like protein 5 positively regulates chaperone gene expression in the mitochondrial unfolded protein response. Genetics 174: 229–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendzen KM, Durieux J, Shao LW, Tian Y, Kim HE, Wolff S, Liu Y, Dillin A. 2016. Neuroendocrine coordination of mitochondrial stress signaling and proteostasis. Cell 166: 1553–1563.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland ML, Chourasia AH, Macleod KF. 2013. Mitochondrial dysfunction in cancer. Front Oncol 3: 292 10.3389/fonc.2013.00292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boopathi E, Srinivasan S, Fang JK, Avadhani NG. 2008. Bimodal protein targeting through activation of cryptic mitochondrial targeting signals by an inducible cytosolic endoprotease. Mol Cell 32: 32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratic A, Larsson N-G. 2013. The role of mitochondria in aging. J Clin Invest 123: 951–957. 10.1172/JCI64125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappello F, Bellafiore M, Palma A, Marciano V, Martorana G, Belfiore P, Martorana A, Farina F, Zummo G, Bucchieri F. 2002. Expression of 60-kD heat shock protein increases during carcinogenesis in the uterine exocervix. Pathobiology 70: 83–88. 10.1159/000067304 [DOI] [PubMed] [Google Scholar]
- Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. 2009. Importing mitochondrial proteins: Machineries and mechanisms. Cell 138: 628–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Mambo E, Sidransky D. 2006. Mitochondrial DNA mutations in human cancer. Oncogene 25: 4663–4674. 10.1038/sj.onc.1209604 [DOI] [PubMed] [Google Scholar]
- Chen JJ, London IM. 1995. Regulation of protein synthesis by heme-regulated eIF-2α kinase. Trends Biochem Sci 20: 105–108. 10.1016/S0968-0004(00)88975-6 [DOI] [PubMed] [Google Scholar]
- Chen A, Qian D, Wang B, Hu M, Lu J, Qi Y, Liu DX. 2012. ATF5 is overexpressed in epithelial ovarian carcinomas and interference with its function increases apoptosis through the downregulation of Bcl-2 in SKOV-3 cells. Int J Gynecol Pathol 31: 532–537. [DOI] [PubMed] [Google Scholar]
- Cheng MY, Hartl FU, Martin J, Pollock RA, Kalousek F, Neuper W, Hallberg EM, Hallberg RL, Horwich AL. 1989. Mitochondrial heat-shock protein hsp60 is essential for assembly of proteins imported into yeast mitochondria. Nature 337: 620–625. 10.1038/337620a0 [DOI] [PubMed] [Google Scholar]
- Chung HK, Ryu D, Kim KS, Chang JY, Kim YK, Yi HS, Kang SG, Choi MJ, Lee SE, Jung SB, et al. 2017. Growth differentiation factor 15 is a myomitokine governing systemic energy homeostasis. J Cell Biol 216: 149–165. 10.1083/jcb.201607110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo S, Longhi R, Alcaro S, Ortuso F, Sprocati T, Flora A, Borgese N. 2005. N-myristoylation determines dual targeting of mammalian NADH-cytochrome b5 reductase to ER and mitochondrial outer membranes by a mechanism of kinetic partitioning. J Cell Biol 168: 735–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland JM, Cho J, Lo T Jr, Hur JH, Bahadorani S, Arabyan T, Rabie J, Soh J, Walker DW. 2009. Extension of Drosophila life span by RNAi of the mitochondrial respiratory chain. Curr Biol 19: 1591–1598. [DOI] [PubMed] [Google Scholar]
- Curtis JM, Hahn WS, Stone MD, Inda JJ, Droullard DJ, Kuzmicic JP, Donoghue MA, Long EK, Armien AG, Lavandero S, et al. 2012. Protein carbonylation and adipocyte mitochondrial function. J Biol Chem 287: 32967–32980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey TA, Woodruff JH, Dailey HA. 2005. Examination of mitochondrial protein targeting of haem synthetic enzymes: In vivo identification of three functional haem-responsive motifs in 5-aminolaevulinate synthase. Biochem J 386: 381–386. 10.1042/BJ20040570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Harding HP, Raught B, Gingras AC, Berlanga JJ, Scheuner D, Kaufman RJ, Ron D, Sonenberg N. 2002. Activation of GCN2 in UV-irradiated cells inhibits translation. Curr Biol 12: 1279–1286. 10.1016/S0960-9822(02)01037-0 [DOI] [PubMed] [Google Scholar]
- Dever TE, Feng L, Wek RC, Cigan AM, Donahue TF, Hinnebusch AG. 1992. Phosphorylation of initiation factor 2α by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell 68: 585–596. 10.1016/0092-8674(92)90193-G [DOI] [PubMed] [Google Scholar]
- Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C. 2002. Rates of behavior and aging specified by mitochondrial function during development. Science 298: 2398–2401. 10.1126/science.1077780 [DOI] [PubMed] [Google Scholar]
- D'Silva PD, Schilke B, Walter W, Andrew A, Craig EA. 2003. J protein cochaperone of the mitochondrial inner membrane required for protein import into the mitochondrial matrix. Proc Natl Acad Sci 100: 13839–13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durieux J, Wolff S, Dillin A. 2011. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell 144: 79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Bussière F, Hekimi S. 2001. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev Cell 1: 633–644. 10.1016/S1534-5807(01)00071-5 [DOI] [PubMed] [Google Scholar]
- Fiorese CJ, Schulz AM, Lin YF, Rosin N, Pellegrino MW, Haynes CM. 2016. The transcription factor ATF5 mediates a mammalian mitochondrial UPR. Curr Biol 26: 2037–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaume B, Klaus C, Ungermann C, Guiard B, Neupert W, Brunner M. 1998. Unfolding of preproteins upon import into mitochondria. EMBO J 17: 6497–6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbeth C, Schmidt O, Rao S, Harbauer AB, Mikropoulou D, Opalińska M, Guiard B, Pfanner N, Meisinger C. 2013. Glucose-induced regulation of protein import receptor Tom22 by cytosolic and mitochondria-bound kinases. Cell Metab 18: 578–587. [DOI] [PubMed] [Google Scholar]
- Gitschlag BL, Kirby CS, Samuels DC, Gangula RD, Mallal SA, Patel MR. 2016. Homeostatic responses regulate selfish mitochondrial genome dynamics in C. elegans. Cell Metab 24: 91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW. 2012. Mitochondrial evolution. Cold Spring Harb Perspect Biol. 4: a011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbauer AB, Zahedi RP, Sickmann A, Pfanner N, Meisinger C. 2014. The protein import machinery of mitochondria-a regulatory hub in metabolism, stress, and disease. Cell Metab 19: 357–372. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Ron D. 1999. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397: 271–274. 10.1038/16729 [DOI] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell 6: 1099–1108. 10.1016/S1097-2765(00)00108-8 [DOI] [PubMed] [Google Scholar]
- Hashimoto M, Rockenstein E, Crews L, Masliah E. 2003. Role of protein aggregation in mitochondrial dysfunction and neurodegeneration in Alzheimer's and Parkinson's diseases. Neuromol Med 4: 21–36. 10.1385/NMM:4:1-2:21 [DOI] [PubMed] [Google Scholar]
- Haynes CM, Petrova K, Benedetti C, Yang Y, Ron D. 2007. ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Dev Cell 13: 467–480. [DOI] [PubMed] [Google Scholar]
- Haynes CM, Yang Y, Blais SP, Neubert TA, Ron D. 2010. The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Mol Cell 37: 529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo JM, Ordureau A, Paulo JA, Rinehart J, Harper JW. 2015. The PINK1-PARKIN mitochondrial ubiquitylation pathway drives a program of OPTN/NDP52 recruitment and TBK1 activation to promote mitophagy. Mol Cell 60: 7–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K, Model K, Ryan MT, Dietmeier K, Martin F, Wagner R, Pfanner N. 1998. Tom40 forms the hydrophilic channel of the mitochondrial import pore for preproteins [see comment]. Nature 395: 516–521. 10.1038/26780 [DOI] [PubMed] [Google Scholar]
- Horibe T, Hoogenraad NJ. 2007. The chop gene contains an element for the positive regulation of the mitochondrial unfolded protein response. PLoS ONE 2: e835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst M, Oppliger W, Rospert S, Schönfeld HJ, Schatz G, Azem A. 1997. Sequential action of two hsp70 complexes during protein import into mitochondria. EMBO J 16: 1842–1849. 10.1093/emboj/16.8.1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, Williams RW, Auwerx J. 2013. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature 497: 451–457. 10.1038/nature12188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane LA, Lazarou M, Fogel AI, Li Y, Yamano K, Sarraf SA, Banerjee S, Youle RJ. 2014. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol 205: 143–153. 10.1083/jcb.201402104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazlauskaite A, Kondapalli C, Gourlay R, Campbell DG, Ritorto MS, Hofmann K, Alessi DR, Knebel A, Trost M, Muqit MM. 2014. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem J 460: 127–141. 10.1042/BJ20140334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan NA, Nikkanen J, Yatsuga S, Jackson C, Wang L, Pradhan S, Kivelä R, Pessia A, Velagapudi V, Suomalainen A. 2017. mTORC1 regulates mitochondrial integrated stress response and mitochondrial myopathy progression. Cell Metab 26: 419–428.e5. [DOI] [PubMed] [Google Scholar]
- Kim SH, Forman AP, Mathews MB, Gunnery S. 2000. Human breast cancer cells contain elevated levels and activity of the protein kinase, PKR. Oncogene 19: 3086–3094. 10.1038/sj.onc.1203632 [DOI] [PubMed] [Google Scholar]
- Kim SH, Gunnery S, Choe JK, Mathews MB. 2002. Neoplastic progression in melanoma and colon cancer is associated with increased expression and activity of the interferon-inducible protein kinase, PKR. Oncogene 21: 8741–8748. 10.1038/sj.onc.1205987 [DOI] [PubMed] [Google Scholar]
- Kim KH, Jeong YT, Kim SH, Jung HS, Park KS, Lee HY, Lee MS. 2013a. Metformin-induced inhibition of the mitochondrial respiratory chain increases FGF21 expression via ATF4 activation. Biochem Biophys Res Commun 440: 76–81. [DOI] [PubMed] [Google Scholar]
- Kim KH, Jeong YT, Oh H, Kim SH, Cho JM, Kim YN, Kim SS, Kim DH, Hur KY, Kim HK, et al. 2013b. Autophagy deficiency leads to protection from obesity and insulin resistance by inducing Fgf21 as a mitokine. Nat Med 19: 83–92. 10.1038/nm.3014 [DOI] [PubMed] [Google Scholar]
- Kirienko NV, Ausubel FM, Ruvkun G. 2015. Mitophagy confers resistance to siderophore-mediated killing by Pseudomonas aeruginosa. Proc Natl Acad Sci 112: 1821– 1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondapalli C, Kazlauskaite A, Zhang N, Woodroof HI, Campbell DG, Gourlay R, Burchell L, Walden H, Macartney TJ, Deak M, et al. 2012. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol 2: 120080 10.1098/rsob.120080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, Kimura Y, Tsuchiya H, Yoshihara H, Hirokawa T, et al. 2014. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature 510: 162–166. 10.1038/nature13392 [DOI] [PubMed] [Google Scholar]
- Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, Jobling WA, et al. 2005. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science 309: 481–484. 10.1126/science.1112125 [DOI] [PubMed] [Google Scholar]
- Leonhard K, Herrmann JM, Stuart RA, Mannhaupt G, Neupert W, Langer T. 1996. AAA proteases with catalytic sites on opposite membrane surfaces comprise a proteolytic system for the ATP-dependent degradation of inner membrane proteins in mitochondria. EMBO J 15: 4218–4229. 10.1002/j.1460-2075.1996.tb00796.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YF, Schulz AM, Pellegrino MW, Lu Y, Shaham S, Haynes CM. 2016. Maintenance and propagation of a deleterious mitochondrial genome by the mitochondrial unfolded protein response. Nature 533: 416–419. 10.1038/nature17989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou GY, Storz P. 2010. Reactive oxygen species in cancer. Free Radic Res 44: 479–496.Liu X, Jiang N, Hughes B, Bigras E, Shoubridge E, Hekimi S. 2005. Evolutionary conservation of the clk-1-dependent mechanism of longevity: Loss of mclk1 increases cellular fitness and lifespan in mice. Genes Dev 19: 2424–2434. 10.1101/gad.1352905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Samuel BS, Breen PC, Ruvkun G. 2014. Caenorhabditis elegans pathways that surveil and defend mitochondria. Nature 508: 406–410. 10.1038/nature13204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucking CB, Durr A, Bonifati V, Vaughan J, De Michele G, Gasser T, Harhangi BS, Meco G, Denefle P, Wood NW, et al. 2000. Association between early-onset Parkinson's disease and mutations in the parkin gene. New Engl J Med 342: 1560–1567. [DOI] [PubMed] [Google Scholar]
- Magnuson B, Ekim B, Fingar DC. 2012. Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem J 441: 1–21. 10.1042/BJ20110892 [DOI] [PubMed] [Google Scholar]
- Managò A, Becker KA, Carpinteiro A, Wilker B, Soddemann M, Seitz AP, Edwards MJ, Grassmé H, Szabò I, Gulbins E. 2015. Pseudomonas aeruginosa pyocyanin induces neutrophil death via mitochondrial reactive oxygen species and mitochondrial acid sphingomyelinase. Antioxid Redox Signal 22: 1097–1110. 10.1089/ars.2014.5979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapa K, Sikor M, Kudryavtsev V, Waegemann K, Kalinin S, Seidel CA, Neupert W, Lamb DC, Mokranjac D. 2010. The conformational dynamics of the mitochondrial Hsp70 chaperone. Mol Cell 38: 89–100. [DOI] [PubMed] [Google Scholar]
- Martin J, Mahlke K, Pfanner N. 1991. Role of an energized inner membrane in mitochondrial protein import. ΔΨ drives the movement of presequences. J Biol Chem 266: 18051–18057. [PubMed] [Google Scholar]
- Martinez BA, Petersen DA, Gaeta AL, Stanley SP, Caldwell GA, Caldwell KA. 2017. Dysregulation of the mitochondrial unfolded protein response induces non-apoptotic dopaminergic neurodegeneration in C. elegans models of Parkinson's disease. J Neurosci 37: 11085–11100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinus RD, Garth GP, Webster TL, Cartwright P, Naylor DJ, Høj PB, Hoogenraad NJ. 1996. Selective induction of mitochondrial chaperones in response to loss of the mitochondrial genome. Eur J Biochem 240: 98–103. 10.1111/j.1432-1033.1996.0098h.x [DOI] [PubMed] [Google Scholar]
- Mayer A, Nargang FE, Neupert W, Lill R. 1995. MOM22 is a receptor for mitochondrial targeting sequences and cooperates with MOM19. EMBO J 14: 4204–4211. 10.1002/j.1460-2075.1995.tb00094.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisinger C, Ryan MT, Hill K, Model K, Lim JH, Sickmann A, Muller H, Meyer HE, Wagner R, Pfanner N. 2001. Protein import channel of the outer mitochondrial membrane: A highly stable Tom40-Tom22 core structure differentially interacts with preproteins, small tom proteins, and import receptors. Mol Cell Biol 21: 2337–2348. 10.1128/MCB.21.7.2337-2348.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melber A, Haynes CM. 2018. UPRmt regulation and output: A stress response mediated by mitochondrial-nuclear communication. Cell Res 28: 281–295. 10.1038/cr.2018.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkwirth C, Jovaisaite V, Durieux J, Matilainen O, Jordan SD, Quiros PM, Steffen KK, Williams EG, Mouchiroud L, Tronnes SU, et al. 2016. Two conserved histone demethylases regulate mitochondrial stress-induced longevity. Cell 165: 1209–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurs E, Chong K, Galabru J, Thomas NS, Kerr IM, Williams BR, Hovanessian AG. 1990. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell 62: 379–390. 10.1016/0092-8674(90)90374-N [DOI] [PubMed] [Google Scholar]
- Miller FJ, Rosenfeldt FL, Zhang C, Linnane AW, Nagley P. 2003. Precise determination of mitochondrial DNA copy number in human skeletal and cardiac muscle by a PCR-based assay: Lack of change of copy number with age. Nucleic Acids Res 31: e61 10.1093/nar/gng060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehle EA, Shen K, Dillin A. 2018. Mitochondrial proteostasis in the context of cellular and organismal health and aging. J Biol Chem 10.1074/jbc.TM117.000893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchiroud L, Houtkooper RH, Moullan N, Katsyuba E, Ryu D, Canto C, Mottis A, Jo YS, Viswanathan M, Schoonjans K, et al. 2013. The NAD+/sirtuin pathway modulates longevity through activation of mitochondrial UPR and FOXO signaling. Cell 154: 430–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munakata H, Sun JY, Yoshida K, Nakatani T, Honda E, Hayakawa S, Furuyama K, Hayashi N. 2004. Role of the heme regulatory motif in the heme-mediated inhibition of mitochondrial import of 5-aminolevulinate synthase. J Biochem 136: 233–238. 10.1093/jb/mvh112 [DOI] [PubMed] [Google Scholar]
- Münch C, Harper JW. 2016. Mitochondrial unfolded protein response controls matrix pre-RNA processing and translation. Nature 534: 710–713. 10.1038/nature18302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagelkerke A, Bussink J, Mujcic H, Wouters BG, Lehmann S, Sweep FC, Span PN. 2013. Hypoxia stimulates migration of breast cancer cells via the PERK/ATF4/LAMP3-arm of the unfolded protein response. Breast Cancer Res 15: R2 10.1186/bcr3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. 2010. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol 8: e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM. 2012. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science 337: 587–590. 10.1126/science.1223560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargund AM, Fiorese CJ, Pellegrino MW, Deng P, Haynes CM. 2015. Mitochondrial and nuclear accumulation of the transcription factor ATFS-1 promotes OXPHOS recovery during the UPRmt. Mol Cell 58: 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nukuda A, Endoh H, Yasuda M, Mizutani T, Kawabata K, Haga H. 2016. Role of ATF5 in the invasive potential of diverse human cancer cell lines. Biochem Biophys Res Commun 474: 509–514. [DOI] [PubMed] [Google Scholar]
- Ordureau A, Sarraf SA, Duda DM, Heo JM, Jedrychowski MP, Sviderskiy VO, Olszewski JL, Koerber JT, Xie T, Beausoleil SA, et al. 2014. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol Cell 56: 360–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu-Ansah E, Song W, Perrimon N. 2013. Muscle mitohormesis promotes longevity via systemic repression of insulin signaling. Cell 155: 699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak VK, Schindler D, Hershey JW. 1988. Generation of a mutant form of protein synthesis initiation factor eIF-2 lacking the site of phosphorylation by eIF-2 kinases. Mol Cell Biol 8: 993–995. 10.1128/MCB.8.2.993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino MW, Nargund AM, Kirienko NV, Gillis R, Fiorese CJ, Haynes CM. 2014. Mitochondrial UPR-regulated innate immunity provides resistance to pathogen infection. Nature 516: 414–417. 10.1038/nature13818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer G, Chinnery PF. 2013. Diagnosis and treatment of mitochondrial myopathies. Ann Med 45: 4–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickles S, Vigié P, Youle RJ. 2018. Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr Biol 28: R170–R185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirós PM, Prado MA, Zamboni N, D'Amico D, Williams RW, Finley D, Gygi SP, Auwerx J. 2017. Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J Cell Biol 216: 2027–2045. 10.1083/jcb.201702058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S, Gerbeth C, Harbauer A, Mikropoulou D, Meisinger C, Schmidt O. 2011. Signaling at the gate: Phosphorylation of the mitochondrial protein import machinery. Cell Cycle 10: 2083–2090. 10.4161/cc.10.13.16054 [DOI] [PubMed] [Google Scholar]
- Rao S, Schmidt O, Harbauer AB, Schonfisch B, Guiard B, Pfanner N, Meisinger C. 2012. Biogenesis of the preprotein translocase of the outer mitochondrial membrane: protein kinase A phosphorylates the precursor of Tom40 and impairs its import. Mol Biol Cell 23: 1618–1627. 10.1091/mbc.e11-11-0933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauthan M, Ranji P, Aguilera Pradenas N, Pitot C, Pilon M. 2013. The mitochondrial unfolded protein response activator ATFS-1 protects cells from inhibition of the mevalonate pathway. Proc Natl Acad Sci 110: 5981–5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde J, Heitman J, Cardenas ME. 2001. The TOR kinases link nutrient sensing to cell growth. J Biol Chem 276: 9583–9586. 10.1074/jbc.R000034200 [DOI] [PubMed] [Google Scholar]
- Rossmanith W. 2012. Of P and Z: Mitochondrial tRNA processing enzymes. Biochim Biophys Acta 1819: 1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runkel ED, Liu S, Baumeister R, Schulze E. 2013. Surveillance-activated defenses block the ROS-induced mitochondrial unfolded protein response. PLoS Genet 9: e1003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Igura M, Obita T, Ose T, Kojima R, Maenaka K, Endo T, Kohda D. 2007. Tom20 recognizes mitochondrial presequences through dynamic equilibrium among multiple bound states. EMBO J 26: 4777–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt O, Harbauer AB, Rao S, Eyrich B, Zahedi RP, Stojanovski D, Schonfisch B, Guiard B, Sickmann A, Pfanner N, et al. 2011. Regulation of mitochondrial protein import by cytosolic kinases. Cell 144: 227–239. [DOI] [PubMed] [Google Scholar]
- Shao LW, Niu R, Liu Y. 2016. Neuropeptide signals cell non-autonomous mitochondrial unfolded protein response. Cell Res 26: 1182–1196. 10.1038/cr.2016.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpilka T, Haynes CM. 2018. The mitochondrial UPR: Mechanisms, physiological functions and implications in ageing. Nat Rev Mol Cell Biol 19: 109–120. 10.1038/nrm.2017.110 [DOI] [PubMed] [Google Scholar]
- Silva JM, Wong A, Carelli V, Cortopassi GA. 2009. Inhibition of mitochondrial function induces an integrated stress response in oligodendroglia. Neurobiol Dis 34: 357–365. [DOI] [PubMed] [Google Scholar]
- Sorrentino V. 2017. Enhancing mitochondrial proteostasis reduces amyloid-β proteotoxicity. Nature 552: 187–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehling O, Lill R. 2013. The role of mitochondria in cellular iron–sulfur protein biogenesis: Mechanisms, connected processes, and diseases. Cold Spring Harb Perspect Biol 5: a011312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel G, Zollner A, Angermayr M, Bandlow W. 2002. Competition of spontaneous protein folding and mitochondrial import causes dual subcellular location of major adenylate kinase. Mol Biol Cell 13: 1439–1448. 10.1091/mbc.01-08-0396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomalainen A. 2015. Mitochondrial roles in disease: A box full of surprises. EMBO Mol Med 7: 1245–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuta T, Langer T. 2009. AAA proteases in mitochondria: Diverse functions of membrane-bound proteolytic machines. Res Microbiol 160: 711–717. [DOI] [PubMed] [Google Scholar]
- Teperino R, Schoonjans K, Auwerx J. 2010. Histone methyl transferases and demethylases; Can they link metabolism and transcription? Cell Metab 12: 321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Garcia G, Bian Q, Steffen KK, Joe L, Wolff S, Meyer BJ, Dillin A. 2016. Mitochondrial stress induces chromatin reorganization to promote longevity and UPRmt. Cell 165: 1197–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly YM, Gidlof S, Oldfors A, Wibom R, et al. 2004. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature 429: 417–423. 10.1038/nature02517 [DOI] [PubMed] [Google Scholar]
- Truscott KN, Kovermann P, Geissler A, Merlin A, Meijer M, Driessen AJ, Rassow J, Pfanner N, Wagner R. 2001. A presequence- and voltage-sensitive channel of the mitochondrial preprotein translocase formed by Tim23. Nat Struct Biol 8: 1074–1082. 10.1038/nsb726 [DOI] [PubMed] [Google Scholar]
- Tsai YP, Teng SC, Wu KJ. 2008. Direct regulation of HSP60 expression by c-MYC induces transformation. FEBS Lett 582: 4083–4088. [DOI] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. 2006. Histone demethylation by a family of JmjC domain-containing proteins. Nature 439: 811–816. 10.1038/nature04433 [DOI] [PubMed] [Google Scholar]
- Tuppen HA, Blakely EL, Turnbull DM, Taylor RW. 2010. Mitochondrial DNA mutations and human disease. Biochim Biophys Acta 1797: 113–128. [DOI] [PubMed] [Google Scholar]
- Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG, et al. 2004. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science 304: 1158–1160. 10.1126/science.1096284 [DOI] [PubMed] [Google Scholar]
- Vandecasteele G, Szabadkai G, Rizzuto R. 2001. Mitochondrial calcium homeostasis: Mechanisms and molecules. IUBMB Life 52: 213–219. [DOI] [PubMed] [Google Scholar]
- Voisine C, Craig EA, Zufall N, von Ahsen O, Pfanner N, Voos W. 1999. The protein import motor of mitochondria: Unfolding and trapping of preproteins are distinct and separable functions of matrix Hsp70. Cell 97: 565–574. 10.1016/S0092-8674(00)80768-0 [DOI] [PubMed] [Google Scholar]
- Vongsamphanh R, Fortier PK, Ramotar D. 2001. Pir1p mediates translocation of the yeast Apn1p endonuclease into the mitochondria to maintain genomic stability. Mol Cell Biol 21: 1647–1655. 10.1128/MCB.21.5.1647-1655.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chen XJ. 2015. A cytosolic network suppressing mitochondria-mediated proteostatic stress and cell death. Nature 524: 481–484. 10.1038/nature14859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Youle RJ. 2009. The role of mitochondria in apoptosis. Annu Rev Genet 43: 95–118. 10.1146/annurev-genet-102108-134850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SZ, Ou J, Zhu LJ, Green MR. 2012. Transcription factor ATF5 is required for terminal differentiation and survival of olfactory sensory neurons. Proc Natl Acad Sci 109: 18589–18594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SF, Chen MS, Chou YC, Ueng YF, Yin PH, Yeh TS, Lee HC. 2016a. Mitochondrial dysfunction enhances cisplatin resistance in human gastric cancer cells via the ROS-activated GCN2-eIF2α-ATF4-xCT pathway. Oncotarget 7: 74132–74151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Pandey AK, Mulligan MK, Williams EG, Mozhui K, Li Z, Jovaisaite V, Quarles LD, Xiao Z, Huang J, et al. 2016b. Joint mouse-human phenome-wide association to test gene function and disease risk. Nat Commun 7: 10464 10.1038/ncomms10464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H, Amon A. 2018. MitoCPR—A surveillance pathway that protects mitochondria in response to protein import stress. Science 360: eaan4146 10.1126/science.aan4146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg SE, Sena LA, Chandel NS. 2015. Mitochondria in the regulation of innate and adaptive immunity. Immunity 42: 406–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel L, Topf U, Bragoszewski P, Wiese S, Sztolsztener ME, Oeljeklaus S, Varabyova A, Lirski M, Chroscicki P, Mroczek S, et al. 2015. Mistargeted mitochondrial proteins activate a proteostatic response in the cytosol. Nature 524: 485–488. 10.1038/nature14951 [DOI] [PubMed] [Google Scholar]
- Ye J, Kumanova M, Hart LS, Sloane K, Zhang H, De Panis DN, Bobrovnikova-Marjon E, Diehl JA, Ron D, Koumenis C. 2010. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J 29: 2082–2096. 10.1038/emboj.2010.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda T, Benedetti C, Urano F, Clark SG, Harding HP, Ron D. 2004. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. J Cell Sci 117: 4055–4066. 10.1242/jcs.01275 [DOI] [PubMed] [Google Scholar]
- Zhang Q, Wu X, Chen P, Liu L, Xin N, Tian Y, Dillin A. 2018. The mitochondrial unfolded protein response is mediated cell-non-autonomously by retromer-dependent wnt signaling. Cell 174: 870–883.e17. 10.1016/j.cell.2018.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, Hoogenraad NJ. 2002. A mitochondrial specific stress response in mammalian cells. EMBO J 21: 4411–4419. 10.1093/emboj/cdf445 [DOI] [PMC free article] [PubMed] [Google Scholar]