SUMMARY
The temporal order of DNA replication (replication timing, RT) is highly coupled with genome architecture, but cis-elements regulating either remain elusive. We created a series of CRISPR-mediated deletions and inversions of a pluripotency-associated topologically associating domain (TAD) in mouse embryonic stem cells. Domain boundaries were dispensable for RT. CTCF protein depletion weakened most TAD boundaries but had no effect on RT or A/B compartmentalization genome-wide. By contrast, deletion of three intra-TAD CTCF-independent 3D contact sites caused a domain-wide early to late RT shift, an A to B compartment switch, weakening of TAD architecture and loss of transcription. The dispensability of TAD boundaries and the necessity of these “early replication control elements” (ERCEs) was validated by deletions and inversions at additional domains. Our results demonstrate that discrete cis-regulatory elements mediate domain-wide RT, A/B compartmentalization, TAD architecture and transcription, representing a major advance in dissecting genome structure and function.
Keywords: Replication timing, sub-nuclear compartment, topologically-associating domain, ERCEs, CTCF, chromatin interactions
Editor In Brief Sentence:
Cis-regulatory elements control the three-dimensional compartmentalization and architecture of the genome to affect replication timing in a CTCF-independent manner.
INTRODUCTION
Eukaryotic DNA replication follows a defined temporal order (replication timing; RT). In mammals, developmentally regulated changes in the RT program occur in units of 400–800kb (replication domains; RDs) that correlate with changes in transcriptional potential (Rivera-Mulia and Gilbert, 2016b). Defects in RT occur in several diseases, correlated with mis-regulation of genes and associated with defects in chromosome condensation, sister chromatid cohesion, and genome instability (Platt et al., 2018; Rivera-Mulia et al., 2017a; Sasaki et al., 2017). RT is also closely aligned with genome architecture. Early cytological studies observed sites of briefly labeled DNA synthesis as punctate foci that persist as stable units of chromosome structure throughout multiple cell cycles (Solovei et al., 2016). These units are spatio-temporally segregated such that early replicating foci are dispersed throughout the interior of the nucleus, while late replicating foci are in close proximity to the nuclear lamina and nucleoli.
The structure and spatio-temporal segregation of RDs can now be mapped molecularly. High throughput chromosome conformation capture (Hi-C) reveals two major compartments of chromatin interaction (A and B) (Dixon et al., 2012; Lieberman-Aiden et al., 2009) that correlate with genome-wide molecular maps of RT, and proximity to the nuclear lamina (Ryba et al., 2010; Yaffe et al., 2010; Pope et al., 2014). Hi-C also detects units of self-interacting chromatin called “topologically associating domains” (TADs) (Dixon et al., 2012; Nora et al., 2012) that align with RDs defined by units of developmental change (Pope et al., 2014). A/B compartments often consist of several adjacent RDs that replicate at similar times, while individual RDs change their RT and compartment during differentiation (Dixon et al., 2015; Ryba et al., 2010; Takebayashi et al., 2012). Both TAD structure and A/B compartment organization are dismantled during mitosis and re-assembled during early G1 phase, coincident with the re-establishment of an RT program (Dileep et al., 2015a). The heterogeneity of TADs and the nomenclature reflecting their mechanisms of establishment are still being debated. Here, we define TADs as they were originally defined (Dixon et al., 2012; Nora et al., 2012): self-interacting domains that manifest as a significant change in the average directionality of interactions and visualized as a triangle on typical Hi-C contact maps. TADs are heterogenous and often nested, consisting of at least two categories distinguished by whether their architecture is primarily driven by CTCF/cohesin-dependent loops vs. CTCF/cohesin-independent compartmentalization of chromatin (Dixon et al., 2012; Nora et al., 2012; Rao et al., 2017; Rowley and Corces, 2018). Since some TAD boundaries align much more strongly to RD boundaries than others (Pope et al., 2014), it is possible that a specific category of TADs is more tightly aligned with RT.
The molecular mechanisms regulating the spatio-temporal regulation of replication are poorly understood. Perturbations of many chromatin regulatory proteins have only partial or localized effects on RT or A/B Hi-C compartments (Dileep et al., 2015b; Nora et al., 2017; Rao et al., 2017; Weintraub et al., 2017) and native cis-acting elements regulating mammalian RT or A/B compartments have not been identified. Thus, it is not clear whether such large-scale regulation is genetic or epigenetic and whether any role for DNA sequence would be via discrete elements or more complex features such as overall nucleotide composition or secondary structure. Early studies suggested that the β-globin Locus control region (LCR) is sufficient to advance RT at ectopic loci (Goren et al., 2008; Simon et al., 2001), but targeted deletion of the LCR at its native locus had no effect on RT (Cimbora et al., 2000). Other studies have identified DNA segments that regulate sub-nuclear localization and/or RT (Sima et al., 2017; van de Werken et al., 2017; Zullo et al., 2012), and a human chromosome has been shown to retain its RT regulation when carried in a mouse host (Pope et al., 2012), consistent with a role for the underlying DNA sequence in spatio-temporal regulation. To search for putative cis-acting elements, we performed CRISPR-mediated (clustered regularly interspaced short palindromic repeats) deletions and inversions at a pluripotency-associated RD in mouse embryonic stem cells (mESCs). Deletion of the boundaries of this RD or CTCF protein depletion, which weakens the boundaries of most domains, had no consequence on RT. However, we identified cis-regulatory elements within the domain that are responsible for early replication, compartmentalization and local genome architecture. These elements have properties of super-enhancers and their prediction genome-wide was validated at multiple loci. These results demonstrate the existence of cis-acting elements controlling the replication timing and 3D architecture of chromosome domains and suggest mechanisms by which they function.
RESULTS
Epigenetic landscape of the Dppa2/4 replication domain
The murine Dppa2/4 domain (Figure 1A and S1) contains 3 active genes: Dppa4, Dppa2 and Morc1/Morc (Figure 1A). Dppa4 and Dppa2 are important pluripotency markers (Hernandez et al., 2018) and essential for embryogenesis (Madan et al., 2009; Nakamura et al., 2011). Morc1 is also highly expressed in ESCs and contains an alternative transcription start site (TSS) 180kb downstream (Morc) of the Morc1 TSS (Figure S1B). The Dppa2/4 domain corresponds to a TAD whose boundaries contain CTCF and cohesin binding sites (Nora et al., 2017; Yue et al., 2014; Dowen et al., 2014; Mumbach et al., 2016) and also align with compartment boundaries (Bonev et al., 2017). It is flanked on both sides by lamina-associated domains (LADs) (Peric-Hupkes et al., 2010) (Figure S1B). Early to late RT regulation of the Dppa2/4 domain during loss of pluripotency coincides with repression of all three genes, spatial compaction of the domain, movement to the nuclear periphery, and a change in A/B compartment (Hiratani et al., 2010; Takebayashi et al., 2012) and it is one of the most difficult domains to reprogram when making induced pluripotent stem cells (iPSCs) (Hiratani et al., 2010).
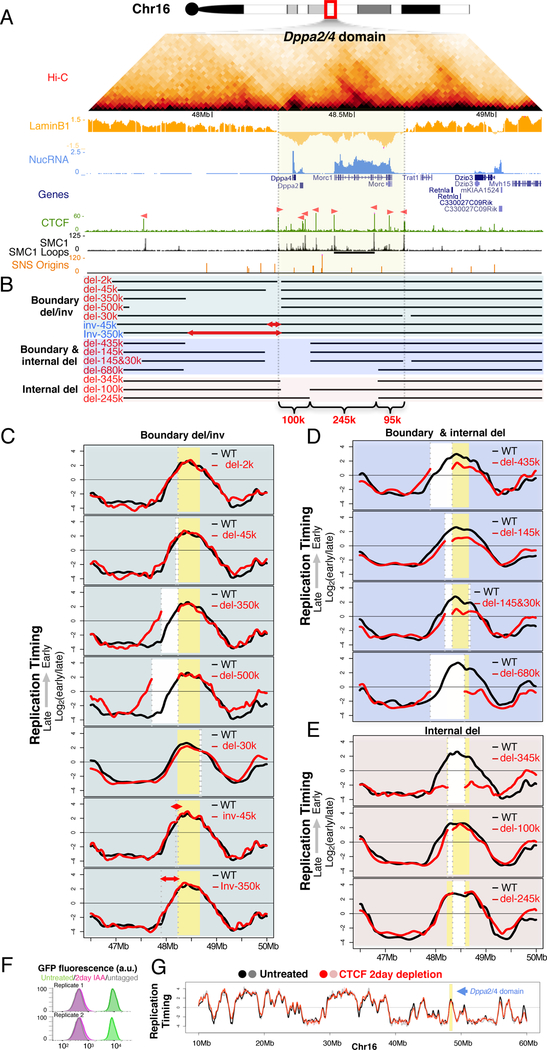
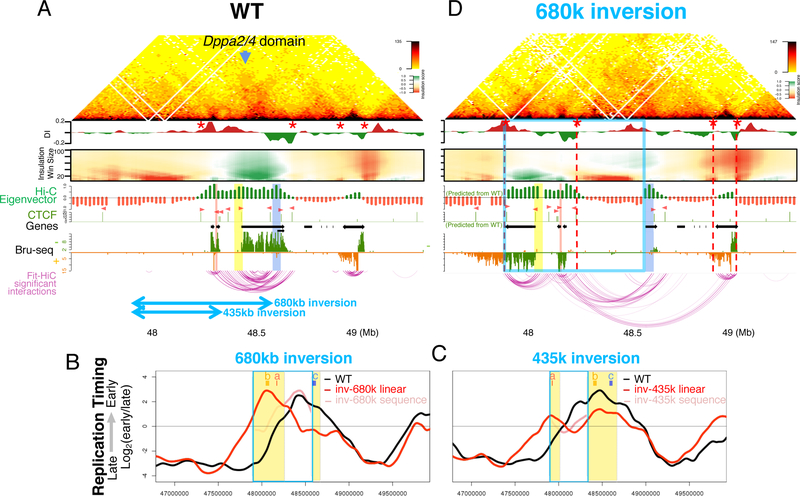
Figure 1. Internal segments contribute partially to early replication.
A. The Dppa2/4 domain in mESCs: Hi-C heatmap, LaminB1 DamID (LaminB1), nuclear RNA (NucRNA), reference genes (Genes), CCCTC-binding factor ChIP (CTCF; motif orientation in pink), SMC1 ChIP signal in ChIAPET datasets (SMC1), SMC1 ChIAPET identified loops (SMC1 loops, horizontal black bars), short nascent strand mapped replication origins (SNS origins), domain boundaries (grey vertical lines).
B. Positions of deletions and inversions.
C,D,E. RT profiles of corresponding deletions or inversions (red lines) vs. WT control (black lines), showing domain boundaries (yellow), deleted regions (white), inversions are indicated by (red arrows) and breakpoints (grey dashed lines).
F. FACS plots of GFP-tagged CTCF depletion.
G. RT profile in untreated (black and grey lines), and 2-day CTCF depleted samples (red and pink lines).
Domain boundaries and CTCF protein are dispensable for RT
We first investigated the role of CTCF by deleting or inverting boundary sequences harboring CTCF binding sites (Figure 1B & S1). The Dppa2/4 domain is flanked by CTCF peaks, with the right border peak co-localizing with cohesin (Figure 1A). Homozygous deletions of 2kb, 45kb, 335kb, or 500kb in 46C mESCs (Ying et al., 2003), encompassing the left boundary, or 30kb of the right boundary had no effect on RT (Figure 1C). Inversion of 45kb and 335kb harboring the orientation-dependent CTCF sites within the left boundary, which can create new loops in the reverse orientation (Lupiáñez et al., 2015), also had no effect on RT (Figure 1C). Circularized chromosome conformation capture (4C) analysis of the deletions using a bait within the domain also did not detect evidence of change in preferential chromatin interactions (Figure S2). These results suggest that the domain boundary is not necessary for maintaining RT or border insulation at the Dppa2/4 domain.
To assess the role of CTCF on RT genome wide, we profiled RT in a cell line in which CTCF protein is acutely depleted using the auxin-inducible degron (AID) system (Nora et al., 2017) (Figure 1F&G & S3). CTCF depletion did not change the spatio-temporal appearance of cytogenetic replication foci patterns (Figure S3A) and had almost no effect on RT genome-wide (0.44% of the genome with RT delay; 0.14% with RT advance) (Figure 1F&G & S3B–E). This result is consistent with the lack of effect of CTCF depletion on A/B compartments (Nora et al., 2017), which correlate strongly with RT (Ryba et al., 2010).
Multiple internal segments contribute to early replication
We next deleted elements internal to the domain. To avoid potential loss of pluripotency caused by homozygous deletion of Dppa2/4 genes and to directly compare the deleted and wild-type alleles within the same datasets, we generated heterozygous deletions in mESCs derived from a cross between M. casteneus (CAST/Ei) and M. musculus (129/sv) (Dupont et al., 2016), from which maternal and paternal chromosomes can be distinguished based on single nucleotide polymorphisms (SNPs) at an average density of one per 150 base pairs (Adams et al., 2015). Some deletions were also performed in mESC line V6.5, derived from a cross between the 129/sv and C57BL/6 strains of M. musculus (Rideout et al., 2000) and harboring SNPs at an average density of one per kilobase (kb). Table S1 lists which deletions were made in each of the cell lines studied in this report.
We first extended two prior boundary deletions (335 and 45kb) by an additional 100kb into the interior of the domain (435 and 145kb) resulting in a slight delay in RT (Figure 1D). Deletion of 30kb encompassing the right boundary in the context of the 145kb left boundary deletion did not cause any further effect. We next extended the 435kb deletion 245 kb further into the Dppa2/4 domain (680 kb) removing the Morc1 TSS, additional sites of active histone marks, an internal CTCF/cohesin loop anchor, most of the inter-LAD chromatin as well as a cluster of replication origins (Cayrou et al., 2015). This invasive deletion caused a substantial delay in RT of the remaining 95kb of the Dppa2/4 domain, which nonetheless remained the locally earliest replicating region. Deleting only the 345kb interior portion of the 680kb deletion had a similar effect on RT as the 680kb deletion (Figure 1E), demonstrating that domain boundaries are not sufficient for early RT. An extensive series of smaller deletions (Figure 1E and Figure S1) had little to no detectable effect on RT, suggesting the existence of functionally redundant segments within the interior of the Dppa2/4 domain.
Identification of Early Replication Control Elements (ERCEs)
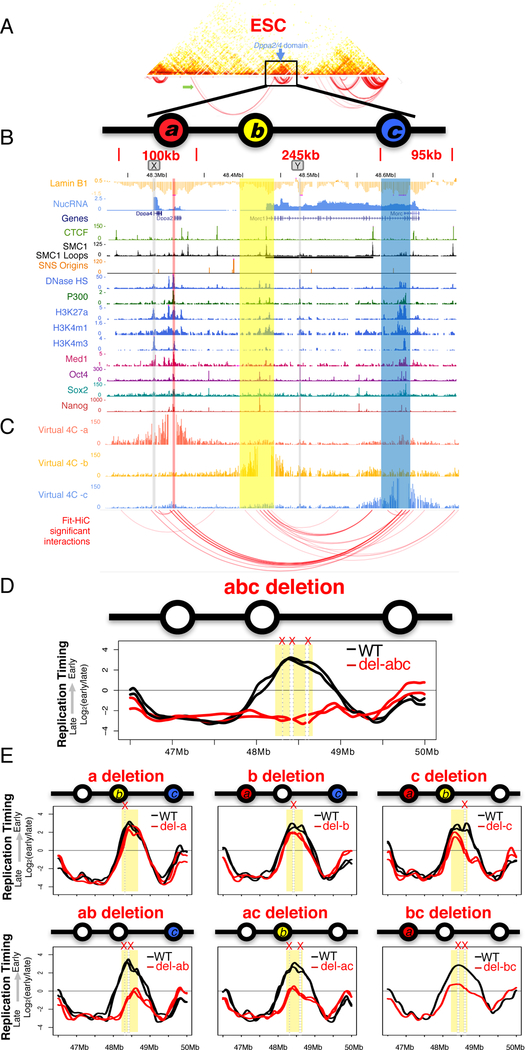
Since RT strongly correlates with chromatin 3D architecture (Dileep et al., 2015a; Pope et al., 2014; Ryba et al., 2010), we reasoned that a high-resolution 3D contact map of the Dppa2/4 domain could reveal features of domain structure consistent with the effects of cis-deletions and inversions described above. We designed a set of capture Hi-C (cHi-C) (Dryden et al., 2014) oligonucleotides hybridizing to ~8,000 SNP-containing MboI fragments covering ~5Mb region surrounding Dppa2/4 domain (Figure 2A). The resulting contact maps at 5kb resolution recapitulated the chromatin folding changes during ESC to NPC differentiation seen previously by FISH and 4C (Hiratani et al., 2008; Takebayashi et al., 2012), which eliminate compartment boundaries while maintaining TAD boundaries (Figure S4A&B). We applied Fit-Hi-C (Ay et al., 2014) to identify significant contacts (FDR > 0.1%) within the Dppa2/4 domain in ESCs. Several sites emerged, including a pair previously identified by cohesin ChIA-PET (Dowen et al., 2014) (Figure 2C and Figure S4A-C). Since CTCF is dispensable for RT (Figure 1G & S3), we focused on 3 contacts that were maintained after CTCF depletion (Nora et al., 2017) (Figure S4D). These sites were enriched in active epigenetic enhancer marks (DNase1 HS, P300, H3K27ac, H3K4m1, H3K4m3) (Yue et al., 2014) and transcription factor binding sites (OCT4, SOX2, NANOG) (King and Klose, 2017)(Figure 2B & S5). One site corresponds to an ESC-annotated super-enhancer (Dowen et al., 2014; Khan and Zhang, 2016), virtually all (229/231) of which we found to be early replicating. Targeted deletion of all 3 sites (designated a, b and c), caused a complete switch from early to late replication of the Dppa2/4 domain, similar in magnitude to that seen in NPCs (Figure 2D). This phenotype was reproduced in 2 independent CRISPR deletion clones, eliminating concerns of off target effects or other sources of clonal variation. We designate these sites Early Replication Control Elements (ERCEs). Notably, two sites outside of ERCEs (X and Y in Figure 2B) that shared many of the same chromatin marks but did not interact strongly with ERCEs were neither necessary nor sufficient for early replication (Figure S1E) suggesting the importance of both chromatin composition and CTCF-independent interactions for ERCE activity.
Figure 2. Identification of Early Replication Control Elements (ERCEs).
A. Capture Hi-C heatmap centered on the Dppa2/4 domain. Arc plot shows significant interaction pairs, including a long-range interaction (green arrow).
B. Chromatin features of the Dppa2/4 domain highlighting ERCEs (a, b, c). Site X and Y show similar marks but are not ERCEs.
C. Virtual 4C profiles from the viewpoints of sites a, b or c. Bait regions are removed and red arcs show significant interactions.
D. RT profile for abc triple deletion from two independent CRISPR clones, with mutant allele in red and WT allele in black.
E. Individual or pairwise deletions of the three ERCEs, as in D.
Consistent with the large deletions containing either a or b (Figure 2E and S1), deletion of either site a or b alone did not significantly alter RT. However, when site c alone was deleted, the right section of the domain acquired the gradually later replicating slope of a timing transition region (TTR) (Dileep et al., 2015b), while the left section containing a and b replicated early. The left and right sections are not totally independent, since all pairwise deletions had significant effects on RT. Also, all ERCEs exhibited activity on their own; the portion of the domain retaining one ERCE remained the locally earliest replicating segment. Targeted double deletion of a and b fully accounted for the effects of previous large deletions encompassing site a and b (Figure 1D&E). Thus, redundancy between ERCEs fully accounts for the complex results of the unbiased nested deletion approach (Figure 1 and S1).
ERCEs control compartmentalization and TAD strength
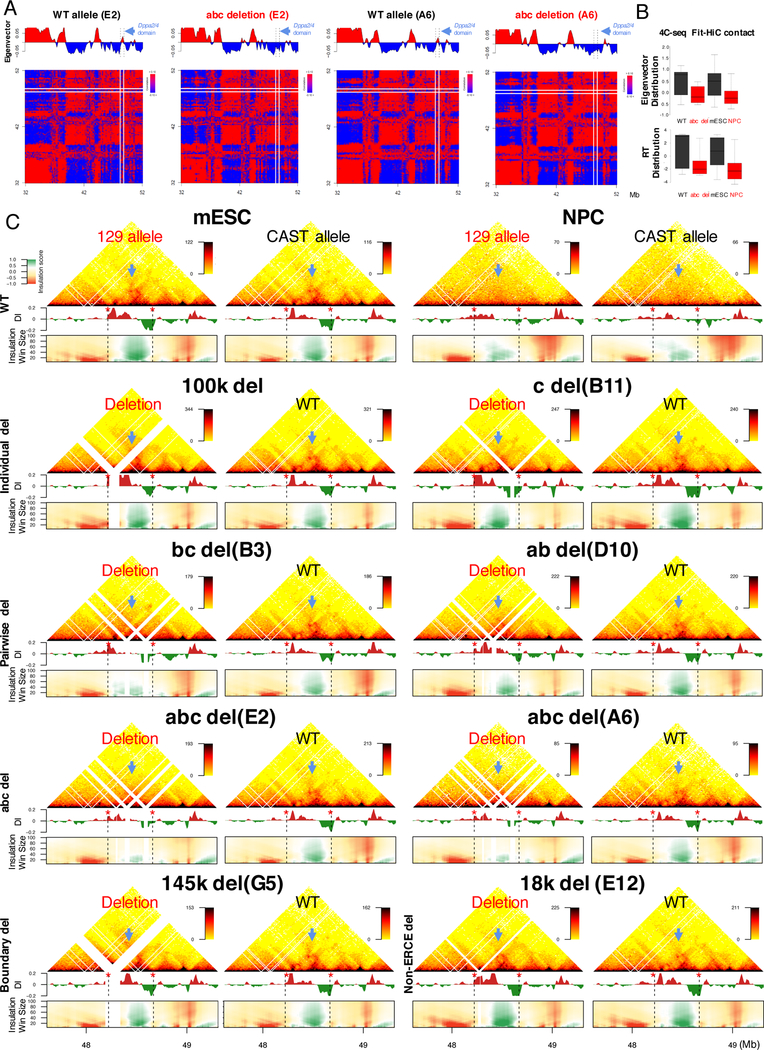
Allele-specific Hi-C eigenvector (Figure 3A), validated by 4C-seq (Figure 3B and S4E), revealed a domain-wide shift from the A to B compartment in the abc deleted allele, demonstrating that ERCEs are also necessary for interaction with the A compartment. In addition, high resolution cHi-C (Figure 3C and S4F) detected a weakening of TAD architecture for the pair-wise (ab and bc) deletions, which was more pronounced in the abc deletion clones (Figure 3C). Both directionality index (DI) and domainogram analyses of the cHi-C data revealed disruption of the local chromatin folding patterns (Figure 3C), which could also be visualized by 4C (Figure S4E). The effect of abc deletions on Dppa2/4 TAD architecture was considerably more pronounced than the effect of the left TAD boundary deletion (Figure 3C, 145k deletion) or an interior non-ERCE deletion (18k deletion), resembling that seen after differentiation of cells to NPCs (Figure 3C) or CTCF depletion (Figure S3D). Therefore, ERCEs are required for proper sub-nuclear compartmentalization and contribute along with CTCF to TAD architecture at the Dppa2/4 locus.
Figure 3. ERCEs control A/B compartmentalization and TAD strength.
A. Hi-C matrix and eigenvector for 2 clones (E2 ands A6) of the abc deletions.
B. Eigenvector and RT distribution of 4C significant contacts from a bait within the Dppa2/4 domain in abc deletion or NPC differentiation.
C. Capture Hi-C results of the Dppa2/4 region in WT (mESC & NPC), individual (100k & c), pairwise (bc & ab), abc (clones E2&A6), boundary (145k) and interior non-ERCE (18k) deletions, with directionality index (DI), domainogram of insulation scores vs. window sizes (heatmap key to left of mESCs), Dppa2/4 TAD ( blue arrow) and boundaries (red stars and black dashed lines).
ERCEs regulate transcriptional activity
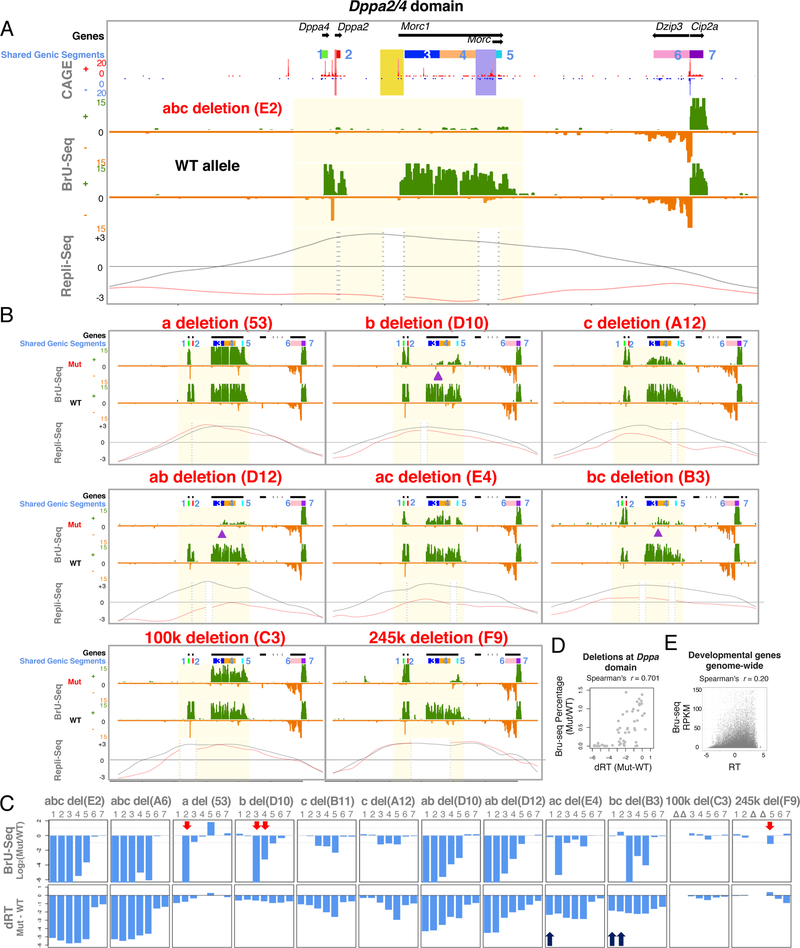
We evaluated the consequence of ERCE deletions on strand-specific and haplotype phased nascent RNA transcription using Bru-seq (Paulsen et al., 2014) (Figure 4 and Figure S6). Three genes and 6 TSSs are present within the Dppa2/4 domain. Deletion of all 3 ERCEs, which overlap 3 of the 6 TSSs, eliminated nearly all transcription throughout the domain (Figure 4A), suggesting that one or more of the ERCEs also play roles in transcription. Dppa4 transcription was maintained in the site a (containing a TSS for Dppa2) or site b (containing a TSS for Morc1) deletion (also 100k and 245k deletions), but lost in the ab deletion (Figure 4B), suggesting that a and b can both activate Dppa4. Site c contains a TSS for the short isoform of Morc1 (Morc) but may also stimulate transcription of the long isoform of Morc1, since its deletion led to ~2-fold reduction in Morc1 expression. Lastly, in the context of a b deletion (containing the Morc1 TSS), sites a and c were redundantly necessary to initiate transcription at an unannotated TSS within the Morc1 gene body (indicated with purple triangles in the b deletion, ab deletion and bc deletions). In sum, these results demonstrate the enhancer activities of ERCEs and their complex role in gene regulation.
Figure 4. Transcription after ERCE deletion.
A. Bru-seq profiles for abc deleted and WT alleles with CAGE (Cap Analysis of Gene Expression), positions of genic segments 1–7 shared among all a, b, c deletions (panel c), sites a, b, and c are highlighted (red, yellow and blue, respectively) negative (green) and positive (orange) strand expression, WT (black) and mutant (red) RT profiles.
B. Bru-seq profiles as in A. An alternative TSS (purple arrows) appears in Morc1 b deletion. (Remaining profiles found in Supplemental Figure 6)
C. Transcription and RT changes of the shared genic segments, indicating gene expression changes without RT changes (red arrows) and RT delay without gene expression changes (black arrows).
D. Transcription vs. RT changes of the shared genic segments in all mutants.
E. Transcription vs. RT for all genes within developmental domains.
To evaluate the relationship between transcription and RT, we quantified the changes in both activities for each mutant allele vs. its corresponding WT allele (Figure 4C), within the segments of the domain that are not removed by either a, b or c deletions, referred to as “shared genic segments” (bars at the top of Figure 4A and B). A scatter plot of transcription vs. RT changes in the Dppa2/4 domain showed a general positive correlation (Figure 4D). However, there are also examples of transcription inactivation with no change in RT and significantly delayed RT with no changes in transcription (red arrows in Figure 4C). These results suggest that transcription and RT are indirectly related. Consistently, a significant portion of developmentally regulated genes genome-wide were active while late replicating (Figure 4E), similar to prior reports in many cell types (Hiratani et al., 2010; Rivera-Mulia et al., 2015). In fact, the Dzip3 and CIP2A/C330027C09Rik genes adjacent to the Dppa2/4 domain (Figure 4A) are active but late replicating in mESCs.
Inversions demonstrate sufficiency of ERCE-containing segments
To determine whether ERCEs are sufficient to dictate early replication in ectopic chromosome environments, we generated a 680kb inversion consisting of 335kb of the left flanking late replicating gene desert and 345kb of the Dppa2/4 domain containing site a and site b leaving behind 95kb containing site c (Figure 5A). The inverted 345kb Dppa2/4 segment preserved early replication, while the remaining 95kb persisted as the locally earliest replicating segment (Figure 5B). Similarly, a 435kb inversion containing 100kb of the Dppa2/4 sequences maintained its early RT (Figure 5C). Bru-seq results with the 680k inversion showed that Dppa2 and Dppa4 gene transcription levels were maintained after inversion. This inversion also resulted in transcription of the Morc1 gene through the breakpoint into the flanking gene desert. cHi-C with 2 independent clones of the 680k inversion (Figure 5D) demonstrated two de novo or neo-TADs, one of which formed a new boundary within the extended and active Morc1 gene transcription unit. Thus, when a and b are re-located from their proximity to the right Dppa2/4 TAD boundary, they are sufficient to retain early replication independent of extensive architectural remodeling.
Figure 5. ERCE-containing inversions.
A. WT capture Hi-C heatmap, DI and domainograms as in Figure 3, showing positions of inversions (bottom blue arrows), sites a, b and c (red, yellow and blue, respectively), and TAD boundaries (red stars).
B,C. RT profile for the 680kb (B) and 435kb (D) inversions with WT (black) and inverted (red) alleles and inverted allele with WT coordinates (pink).
D. Capture Hi-C of the 680kb inversion with linear distance after inversion, indicating the inverted region (blue box) and newly formed boundaries (red stars and red dashed lines). Eigenvector and CTCF ChIP-seq peaks (pink arrowheads indicate orientation) are inverted from WT data.
Genome-wide properties and validation of ERCEs
To predict ERCEs genome-wide, we identified sites of significant CTCF-independent (Nora et al., 2017) interactions that overlap with P300, major pluripotency transcription factor binding sites for OCT4, SOX2, NANOG (OSN) and that reside within inter-LADs. 1835 ERCEs were predicted genome-wide, 33.9% of which overlap with annotated enhancers (Enhancer Atlas, mESC-Bruce4) and 20.2% with currently identified mESC super-enhancers (Figure 6A). Predicted ERCEs are also enriched in other active chromatin marks (Figure S5), and only a subset of P300 or OSN binding sites overlap with ERCEs (Figure 6A). Since YY1 regulates enhancer-promoter loops, we profiled RT in YY1 depleted cells, which leads to expression changes in 8234 genes (Nabet et al., 2018; Weintraub et al., 2017). We found no evidence of significant RT changes after YY1 depletion, further suggesting that ERCEs role in RT may not be directly linked to their role in transcription.
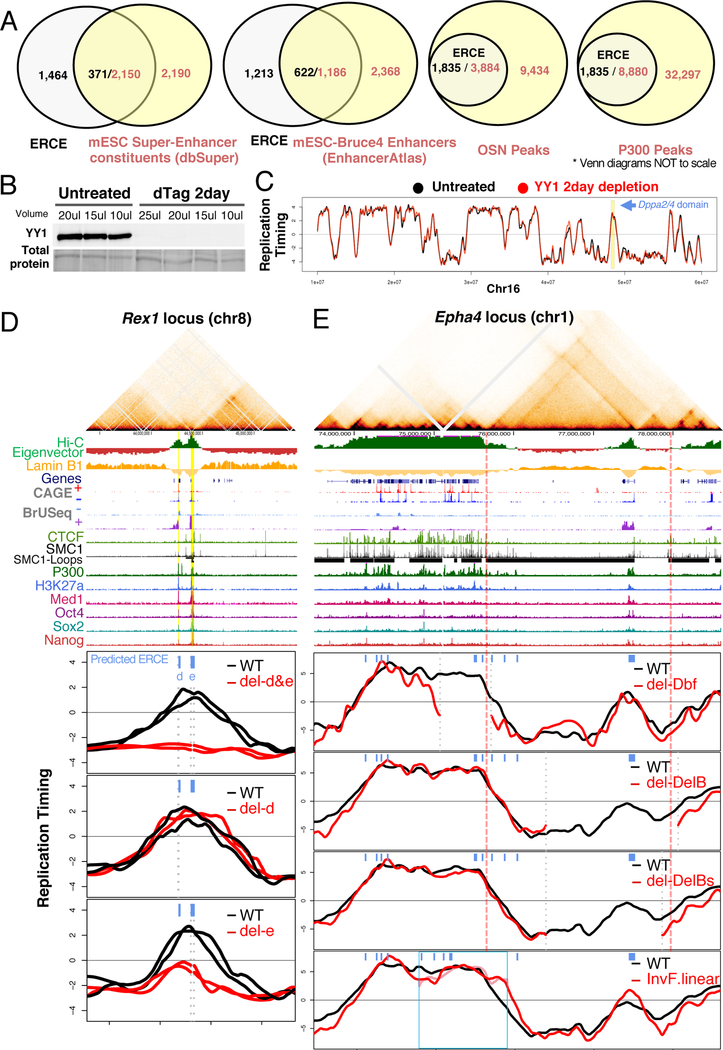
Figure 6. Genome-wide prediction and validation of ERCEs.
A. Overlap between predicted ERCEs and enhancers, super enhancers, P300 and OSN (OCT4, SOX2, NANOG) binding sites. Please note that the Venn diagrams are not drawn to scale.
B. Western blot of YY1 protein depletion.
C. RT profile in untreated (black and grey lines), and YY1 depleted samples (red and pink lines). Dppa2/4 domain is highlighted in yellow.
D. Validation of predicted ERCEs at the Zfp42/Rex1 domain with Hi-C heatmap as in Fig, 1 with predicted ERCEs (d&e) highlighted in yellow and RT profiles of two independent CRISPR clones.
E. RT of boundary containing deletions/inversions at the Epha4/Wnt6 locus in mESCs. Hi-C heatmap and chromatin features as in D.
To validate predicted ERCEs, we generated CRISPR deletions of predicted ERCEs at an independent locus and analyzed RT of these and other mESC deletions and inversions in the literature that harbored predicted ERCEs. The Zfp42/Rex1 domain on chromosome 8 contains 3 active genes (Triml1, Triml2 and Zfp42/Rex1), its TAD is an A compartment inter-LAD flanked by LADs and it contains two predicted ERCEs, designated d&e (Figure 6D). Deletion of both predicted ERCEs caused a complete shift of the domain to late replication. Deletion of d alone had no effect on RT, and deletion of e caused the right side of the domain to shift to late, as for the c deletion in Dppa2/4. These results validate our ability to predict ERCEs and reproduce the partial redundancy and interdependency among ERCEs within a domain. We also profiled a Klf4 super-enhancer deletion (Xie et al., 2017) that contained a predicted ERCE (Figure S7). This deletion led to a shift of the TTR deeper within the domain, while the rest of the domain remained early replicating, possibly maintained by two other predicted ERCEs (Figure 6E). Analysis of deletions and inversions at the constitutively early replicating Wnt6/Ihh domain on chr1 (Lupiáñez et al., 2015) confirmed that RT was retained after TAD boundary deletions (Dbf and DelB deletions) (Figure 6E) and after a 1.07Mb inversion (InvF/InvF) that detached the Ihh domain from its TAD boundary. Altogether, these results validate the principles regulating RT and the existence of ERCEs in other domains throughout the genome.
DISCUSSION
Cis-elements regulating RT in mammalian cells have been sought for decades. While studies involving integrated or extra-chromosomal vectors provided preliminary evidence for a genetic influence on RT control and sub-nuclear organization (Pope et al., 2012; Sinclair et al., 2010; van de Werken et al., 2017;Sima et al., 2017;Goren et al., 2008; Hassan-Zadeh et al., 2012), it has been challenging to identify functional cis-elements in situ. ERCEs do not coincide with the most efficient replication origins / initiation zones, measured by ensemble small nascent strand methods (Figure 2B) (Cayrou et al., 2015), consistent with prior studies demonstrating that RT is regulated independently of initiation sites (Dileep et al., 2015b). ERCEs are also necessary for sub-nuclear compartmentalization and 3D architecture of replication domains, and they play roles in transcription. Our results pave the way for a systematic approach to dissect mechanisms regulating temporal and spatial control of DNA replication and its relationship to gene expression.
ERCEs form prominent CTCF-independent interactions and are sites of p300 and pluripotency transcription factor binding (Oct4, Sox2, Nanog; OSN), which are known to mediate long-range interactions (Fang et al., 2014). Some ERCEs also contain DHSs, H3K27ac, H3K4m1, H3K4m3, or Med1, all hallmarks of enhancers and/or promoters (Bogu et al., 2016; Whyte et al., 2013; Yue et al., 2014). Enhancers are critical for polyoma and papilloma virus replication (Chandrasekharappa and Subramanian, 1987; Spalholz et al., 1985), but their role in cellular DNA replication is unknown. We show that not all active promoters or enhancers are ERCEs and their characteristic chromatin features alone are not sufficient to constitute an ERCE: several deletions containing these features but lacking strong 3D interactions had no effect on RT, even in combination with other ERCE deletions (Figure 2B and S1E). It will be important to determine whether looping to an ERCE is sufficient to convey ERCE function to sites containing enhancer/promoter chromatin features.
Consistent with the discovery of many genes that are active in late replicating regions (Rivera-Mulia and Gilbert, 2016b; Rivera-Mulia et al., 2015), delayed RT can occur without loss of transcription and loss of transcription does not necessarily lead to a delay in RT. Moreover, studies in fly, fish, frog embryos have shown that an RT program precedes activation of transcription (Kaaij et al., 2018; Pourkarimi et al., 2016; Seller and O’Farrell, 2018; Siefert et al., 2017). Together, these studies imply a close but indirect liaison between transcription and replication timing.
ERCE redundancy offers a plausible explanation for the previous failure to identify such elements at the human β-globin locus, which is controlled by a super-enhancer-like element (LCR) that is able to partially advance RT at ectopic locations but is dispensable for early RT at the native locus (Cimbora et al., 2000;Simon et al., 2001). ERCEs are sufficient to partially advance RT (Figure 2 & 6), and hence are likely to advance late replication at ectopic sites, but are often dispensable for early replication at the native locus due to presence of redundant ERCEs. We therefore predict that additional ERCEs are present at the β-globin locus in erythroid cells.
Our results also provide two important new insights into genome architecture and function. First, we demonstrate that TAD boundaries play little to no role in RT, while ERCEs play a role in chromatin domain architecture, including TAD boundaries. TADs whose structure is primarily driven by CTCF-cohesin (Nora et al., 2017; Rao et al., 2017; Schwarzer et al., 2017) have a convergent pair of CTCF binding sites at their boundaries forming corner peaks in HI-C heat maps and loss of CTCF or cohesin disrupts these “loop domains” (Nora et al., 2017; Rao et al., 2017). The Dppa2/4 domain is demarcated by a pair of convergent CTCF binding sites, and CTCF depletion has a measurable effect on Dppa2/4 local chromatin interactions (Figure S3D), even though it does not exhibit strong corner peaks in the contact map, and is thus unlikely to be a loop domain. In fact, ERCE deletion affected local chromatin interactions at least as strongly as CTCF depletion or boundary deletions, and inversion of ERCEs was sufficient to create a new boundary, demonstrating that domain structure can be dictated by both CTCF and ERCEs. However, TAD boundaries were also dispensable for RT at the Epha4 locus, which does exhibit corner peaks, and inversions detaching the interior of the Epha4 or Dppa2/4 domains from their boundaries retain their RT. Finally, weakening of TAD boundaries genome-wide following CTCF depletion does not affect RT (Figure 1F&G and S3D&E). Second, our results implicate ERCEs as drivers of large-scale chromatin compartmentalization. While CTCF and cohesin are necessary for loop domain formation, their depletion does not affect compartments (Nora et al., 2017; Rao et al., 2017) or RT (Figure 1F&G). We show that YY1, involved in enhancer-promoter loops, is also dispensable for RT. Thus, while there are several drivers of loop formation, compartment drivers have not been identified. ERCEs form robust long-range CTCF-independent interactions with predicted ERCEs within other domains (Figure 7A&B) and ERCEs are required for compartmentalization (Figure 3A). This suggests the existence of an ERCE 3D network that could underpin A/B compartmentalization.
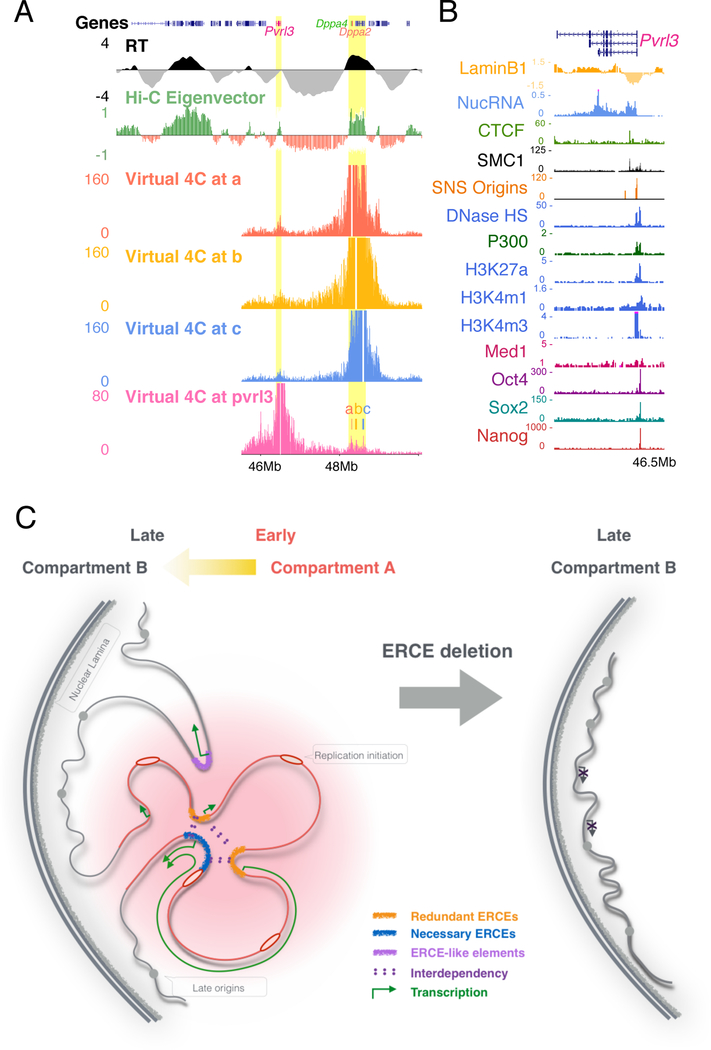
Figure 7. Model for ERCE function.
A. ERCEs interact strongly with predicted ERCEs outside the domain. Virtual 4C profiles (bait regions removed) from Capture Hi-C data viewpoints of site a, b, c and a predicted ERCE.
B. Chromatin features of the predicted ERCE from A.
C. ERCEs regulate RT, A/B compartmentalization, TAD architecture and gene transcription.
We have shown that ERCEs are responsible for A/B compartmentalization, TAD architecture, transcription and RT of a chromosome domain, all chromatin properties that are highly correlated but not necessarily causally linked. In budding yeast, Fkh1,2 transcription factors interact to cluster a set of early replication origins but, mutants of Fkh1,2 that disrupt their ability to dimerize and cluster origins fail to direct early RT but they retain their ability to activate transcription (Ostrow et al., 2017). The ability of Fkh1,2 TFs to promote early replication has been traced to their ability to recruit Dbf4, the activating partner of the essential replication initiation kinase Cdc7 (Fang et al., 2017). ERCEs may serve a similar function in mammalian cells. The essential initiation protein Treslin, which cooperates with Cdc7 to activate the replicative helicase, interacts with acetylated histone binding proteins Brd2 and Brd4 (Sansam et al., 2018). ERCEs are co-occupied by core transcriptional regulatory network (TRN) factors (such as OSN) that promote local histone acetylation. On their own, they may be able to create a large enough histone acetylation landing pad for Brd2/4-Treslin to partially advance RT, as with the pair-wise deletions in Dppa2/4 or single deletions in ß. However, there is also evidence that OSN factors can self-interact to form loops (Jerabek et al., 2017; Lyu et al., 2018), which would not only contribute to the manifestation of a TAD but would form a chromatin hub enriched in histone acetylation and thus Brd2/4 and Treslin. Concentrated Brd4 is known to drive phase separation (Sabari et al., 2018), which promotes transcription and, we suggest, by way of Treslin recruitment, replication as well (Gilbert, 2001). Such sub-nuclear domains may also tend to self-interact, driving the formation of larger chromatin compartments. In metazoans, cell-type specific TFs and the regions of acetylated histones that they create may promote both transcription and replication initiation activity in a cell type specific manner, enhanced by their 3D interaction (Figure 7C). Now that ERCEs can be predicted genome-wide, testing these predictions and further probing how ERCEs regulate RT and link RT to transcription and genome architecture should finally be within our grasp.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to the Lead Contact, David M. Gilbert (gilbert@bio.fsu.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse ESCs 46C (XY, 129P2/Ola), Cas/129 or F121–9 (XX, CAST/Ei X 129/sv) and V6.5 (XY, C57BL/6 X 129/sv) were maintained on 1% gelatin coated dishes. 46C was grown in GMEM supplemented with 10% FBS and LIF; hybrid cells (Cas/129 and V6.5) in DMEM supplemented with 2i (1-Azakenpaullone, Sigma; PD0325901, APExBIO) plus LIF. Mouse ESCs occasionally display chromosome losses and gains that can be seen in our NGS data; confirming reproducible results with two independent clones ensures that these karyotypic alterations do not affect our data. ESC to NPC differentiation was carried out as previously described (Abranches et al., 2009). Briefly, mESCs were plated on 0.1% gelatin-coated dishes in serum-free ESGRO Complete Clonal Grade medium (Millipore Inc.) at high density. After 24 hours, cells were passaged to RHB-A media (Stem Cell Science Inc., Y40001) at the density of 1×104cells/cm2 and maintained for 12 days with media changed every other day. CTCF and YY1 depleted cells, Epha4 and Klf4 mutants were handled in the same conditions as previously published (Lupiáñez et al., 2015; Nora et al., 2017; Weintraub et al., 2017).
METHOD DETAILS
CRISPR-mediated genome editing
CRISPR guide RNAs (gRNAs) (listed in Supplementary Table 2) were designed using http://crispr.mit.edu/, and cloned into pX330 (for Dppa2/4 domain) or px458 (for Rex1 domain). CRISPR-mediated genetic engineering pipeline was carried out as previously described (Byrne et al., 2015) with modifications. Two CRISPR plasmids with gRNAs targeting each breakpoint were transiently transfected into mESCs by nucleofection (Lonza, P3 primary kit) along with a pCAG-mCherry plasmid. Two days post-transfection, mCherry positive cells were then sorted as single cells into 96-well plates. After colonies appeared, cells in each well were dissociated and the plate was duplicated; genomic DNA from one of the 96-well plates was prepared as previously described (Ramírez-Solis et al., 1992), and mutations were screened by polymerase chain reaction (PCR) using primers across the breakpoint junctions. PCR fragments were also Sanger sequenced to identify allele-specific mutation based on SNPs on both sides of the breakpoints. Positive clones were then further expanded for downstream experiments.
Repli-seq
Repli-seq was performed as described in detail (Marchal et al., 2017). Briefly, growing cells were pulse labeled with BrdU (Sigma Aldrich, B5002) for 2 hours, fixed in 75% ice-cold ethanol with gentle rotation, and sorted into early and late S-phase fractions by flow cytometry based on PI staining of DNA content. BrdU-substituted DNA from 40k early or late S-phase cells was immuno-precipitated using anti-BrdU antibody (Becton Dickinson 347580), and purified after proteinase K digestion overnight. Repli-seq libraries were constructed using NEBNext Ultra DNA Library Prep Kit for Illumina (E7370) and sequenced on Illumina HiSeq 2500.
Hi-C and Capture Hi-C
Hi-C and capture Hi-C enriching 45488946–50879849 on chromosome 16 was performed as previously described (Dryden et al., 2014). 25 million cells were fixed within 2% formaldehyde at RT for 10 minutes, and quenched with 0.125M glycine. Cells were then lysed in 50ml ice-cold lysis buffer (10 mM Tris-HCl pH 8, 10 mM NaCl, 0.2% Igepal CA-630, one tablet protease inhibitor cocktail (Roche complete, EDTA-free, 11873580001) on ice for 30 minutes. Collected cell pellet were then incubate in 2.8% SDS with NEBuffer 2 at 37°C for 60 min to remove pr oteins that were not directly cross-linked to the DNA. SDS was quenched by Triton X-100 at 37°C for 60 min. Chromatin for each sample were then divided into 4 tubes digested with 1500 units of MboI (NEB R0147M) for each tube at 37°C overnight while rotat ing at 950 rpm. To fill in the restriction fragment overhangs and mark the DNA ends with biotin, 6µl 10x NEB2, 2µl H2O, 1.5µl 10 mM dCTP, 1.5µl 10 mM dGTP, 1.5µl 10 mM dTTP, 37.5µl 0.4 mM biotin-14-dATP (Life Technologies 19524–016), and 10μl 5U/μl Klenow (DNA polymerase I large fragment, NEB M0210L) were added to each tube, and incubated for 60 minutes at 37°C. Nuclei were then centrifuged and resuspend in 1ml ligation mix (100μl 10x ligation buffer (NEB B0202S), 10 μl 20mg/ml BSA (NEB B9001S), and 885 μl H2O). For ligation, 5μl 400U/μl T4 DNA ligase (NEB M0202S) was added to each tube for 4 hours at 16°C, followed by 30 min RT. Crosslinks were reversed and protein was degraded by adding 60μl 10 mg/ml proteinase K (Roche 03115879001) per tube and incubating the tubes overnight at 65°C. DNA was then purified by 2 rounds of phenol:chloroform (Sigma P3803) purification, and Hi-C libraries from all 4 tubes for each sample were pooled. Hi-C ligation efficiency was controlled by PCR using cell-type specific primers. Biotin-14-dATP at non-ligated DNA ends was removed from 40μg of Hi-C libraries for each sample with the exonuclease activity of T4 DNA polymerase. Hi-C libraries were divided into 8 tubes, and 5μg of Hi-C library with 0.5μl 10 mg/ml BSA, 5μl 10× NEBuffer 2, 2μl 2.5mM dATP, and 5μl T4 DNA polymerase (NEB M0203L) were added and incubated at 20°C for 4 hours. The reaction was sto pped by adding 2ul 0.5 M EDTA pH 8.0 to each tube. DNA was then purified by phenol:chloroform extraction, sheared using Covaris E220, end repaired and size selected between 200 and 650 bp using double-sided SPRI beads. Generate PE adapters were ligated to the fragments and Biotin-streptavidin pulldown was performed using Dynabeads MyOne Streptavidin C1 beads (Life Technologies 650.01). Hi-C libraries were then amplified using 5–9 PCR cycles using Phusion polymerase (NEB F531). In the case of capture Hi-C, Target enrichment was then performed using Agilent SureSelect RNA oligo system with custom designed capture probes (50bp RNA targeting Mbo1 fragments containing at least 1 SNP to increase the percentage of SNP overlapping reads), following the manufacture’s protocol. A final round of PCR amplification (~4 cycles) for capture Hi-C libraries was performed before the libraries were purified using SPRI beads, quantified using KAPA and sequenced with one 150bp pair end mid-output lane on Illumina NextSeq 550.
4C-seq
4C-seq was performed as previously described (Splinter et al., 2012) with minor modifications. 10 million cells were fixed in 1% formaldehyde for 10 minutes at room temperature and quenched with glycine (0.125 M final conc.). Nuclei were isolated in lysis buffer (50mM Tris-HCl ph7.5, 150mM NaCl, 5mM EDTA, 0.5% NP-40, 1% TX-100 containing 1X Roche cOmplete Mini protease inhibitors) and dounced 15 times on ice. Nuclei resuspended in 260 μl warm 1X NEB 2.1 buffer were permeabilized using 7.5 μl 20% SDS at 65°C for 10 minutes, and then 20 Triton X100 for 60min at 37°C. Chromatin was digested using 500U HindIII (NEB) for 8 hours at 37°C, and the digestion was repeated twice. Enzyme was deactivated at 65°C for 20 mins and digestion was determined by gel electrophoresis. Chromatin samples were diluted and ligated using 8000 CEU NEB T4 DNA Ligase (M0202M) at 16°C overnight. Ligation efficiency was checked by gel electrophoresis. Chromatin was de-crosslinked with proteinase K overnight at 65°C, and treated with RN ase A at 37°C for 1 hour. DNA was extracted by Phenol:Choroform and precipitated with Ethanol. Purified DNA was treated with 500U NEB DpnII at 37°C for 8 hours, twice. DNA circularization was performed using 8000 CEU NEB T4 DNA Ligase overnight at 16°C. 4C amplification used 3.2 ug 3C template with inverse PCR primers containing Illumina forward and reverse sequencing adapters. PCR was performed using NEB Q5 Hot Start High Fidelity DNA polymerase with the following thermocycler program: 94°C for 2 min; 94°C for 30 seconds; 61–65°C for 1 min; 72°C for 3 min; repeat for 29 cycles; 72°C for 5 min; hold at 4°C. 4C-seq libraries were size-selected then quant ified using RT-PCR (KAPA Biosystems) and sequenced using 50bp single-end on Illumina HiSeq 2500.
Bru-seq
Bru-seq was performed as previously described (Paulsen et al., 2014). Cells were labeled with 2 mM Bromouridine (Sigma Aldrich, 850187) for 30 min, and lysed in 3ml of Trizol. Total RNA was extracted using chloroform and dissolved in DEPC-treated water. To pull down the Bromouridine incorporated RNA, total RNA was denatured at 80˚C for 10 minutes, incubated with anti-BrU antibodies conjugated magnetic beads (anti-BrU: Becton Dickinson, 555627) for 1 hour at room temperature with gentle mixing, and washed 3 times with 0.1% BSA in PBS. The beads were resuspended in 35μl DEPC-treated water, mixed and incubated at 96˚C for 10 min to elute the Bru-RNA. Strand-specific cDNA libraries were then prepared with the exclusion of polyA RNA isolation. Briefly, 300ng of Bru-RNA was fragmented by mixing with first strand buffer and random primers and incubated at 85˚C for 10 min. The first strand cDNA was synthesized in the presence of Actinomycin D to result in strand specific libraries. This cDNA was purified using AMPure RNAclean beads (Beckman Coulter). The second strand cDNA was synthesized and then purified using AMPure XP beads (Beckman Coulter). Next, end repair, adenylation, and adaptor ligation was performed, followed by a size selection in which the cDNA was run on a 3% NuSieve 3:1 agarose gel (Lonza) and gel slices excised in the 300bp region. The gel slices were purified using the QIAEX II Gel Extraction Kit (Qiagen) and then PCR amplified. Resulting libraries were purified with AmPure XP beads, quantified and sequenced using 100bp single-end on Illumina HiSeq 2500.
BrdU Immunofluorescence Staining
Asynchronous cell populations were grown on glass coverslips in a 4 well dish in 0.5ml media. 1.5ul of BrdU was added to the cells either untreated or treated with Auxin for 2 days and incubated at 37˚C for 30 minutes. Cell were washed with PBS and fixed with ice cold 70% EtOH. Cells were incubated with 1.5N HCl for 30 minutes at room temperature. Cells were permeabilized with PBS+0.5% Tween20 for 5 minutes. Anti-BrdU antibody was added at 1:20 in PBS+10% goat serum for 1 hour at room temperature in the dark. 647-anti-mouse secondary antibody (Lifetechnology A21239) was added at 1:100 in PBS+10% goat serum and incubated for 1 hour at room temperature in the dark. Cell were counter stained with DAPI for nuclei.
Western Blot
YY1 2-day dTAG treated and untreated cells were lysed with Laemelli buffer and boiled for 10 minutes. Lysate was run on an 8% polyacrylamide gel and transferred to nitrocellulose membrane overnight at 4C. Total protein loading control was assayed with Ponceau S. The membrane was blocked with PBS+0.1% Tween20+10% nonfat milk for 1 hour at room temperature. Anti-YY1 antibody (Santa Cruz Biotech sc-7341, lot A2318) was added at 1:200 in PBS+0.1% Tween20+5% nonfat milk for 1 hour at room temperature. HRP-anti mouse secondary antibody (Abcam, Ab6789) was added at 1:5000 in PBS+0.1% Tween20+5% nonfat milk for 1 hour at room temperature.
List of published datasets used in this study
Replication domain boundary calls for Dppa2/4 domain were retrieved from (Pope et al., 2014). Hi-C data from ESC and NPC (Bonev et al., 2017) were visualized as heatmaps in HiGlass (Kerpedjiev et al., 2017). Hi-C eigenvector was retrieved from (Nora et al., 2017). Mouse ESC and NPC laminB1 DamID (Peric-Hupkes et al., 2010); nucRNA (Rivera-Mulia et al., 2017b), DNaseI HS, H3K27ac, H3K4m1,H3K4m3,H3K36m3, P300 ChIP (Yue et al., 2014), CTCF ChIP (Nora et al., 2017; Yue et al., 2014), SMC1-ChIAPET (Dowen et al., 2014), SMC1- HiChIP (Mumbach et al., 2016), Med1 (Whyte et al., 2013), Oct4, Sox2, Nanog (King and Klose, 2017), CAGE coverage (Arner et al., 2015), SNS mapped origins (Cayrou et al., 2015) used in this study were visualized in UCSC genome browser (Kent et al., 2002).
QUANTIFICATION AND STATISTICAL ANALYSIS
Determination of CTCF binding site orientation
CTCF MACSs peaks was called with qvalue < 0.05; CTCF MACSs peaks that were present in both datasets (Nora et al., 2017; Yue et al., 2014) were used. CTCF orientation was determined using PWMEnrich package based on its maximum forward or reverse enrichment score. Mouse CTCF binding sequence motif information was retrieved from http://hocomoco11.autosome.ru/motif/CTCF_MOUSE.H11MO.0.A.
Repli-seq data analysis
For inbred cells, reads were quality trimmed (q score =20) and mapped to mm10 reference genome using bowtie2 (Langmead and Salzberg, 2012). For cas/129 hybrid cells, reads were initially mapped to svim genome with quality score above 10, and SNPs parsed to the corresponding allele using HARP (Rivera-Mulia, 2018). Reads overlapping SNPs between the two genomes were filtered and sorted to each allele according to the exact nucleotide at the SNP positions. Reads that do not overlap with SNPs or do not match the exact nucleotide for either genome at the SNP position were discarded. For V6.5 mESCs, reads were mapped to mm10 reference genome and sorted to either allele using same method with SNPs between reference genome and svim. Log2 ratios of early vs late read counts were calculated for 5kb non-overlapping windows, Loess smoothed at 400–500kb windows. Coordinates were corrected for deletions by removing the deleted region in Loess smoothing process. Inversions were smoothed according to actual linear distance in the mutants. Chromosome 16 RT profiles for inbred cell lines were quantile normalized. Chromosome 16 RT profiles for hybrid cells were rescaled to the same interquartile range, and mutant allele was compared to WT allele.
Genotype confirmation with repli-seq coverage
Reads from both early and late fractions for a given profile (inbred) or each allele (hybrid cell) are binned at 5kb non-overlapping windows, and normalized to reads per million (RPM).
in silico RT compute in heterozygous mutants
Log2 ratios of heterozygous deletions in inbred mESCs (46C, and the Dbf/+ in Epha4 deletion series) was calculated as follows: 2 × RTheterozygous − RTWT.
RT Fingerprinting in CTCF depletion
RT signatures are identified as previously described at 150kb windows (Ryba et al., 2011). The fingerprints were filtered with qvalue cutoff at 0.01, and RT value difference >1. Constitutive and developmentally regulated domains are defined as in (Dileep et al., 2015a).
Hi-C and Capture Hi-C data processing
Reads were trimmed by Trim Galore! (https://github.com/FelixKrueger/TrimGalore); mapped using HiCUP v0.5.9 (Wingett et al., 2015), and allele parsed using SNPsplit v0.3.0 (Krueger and Andrews, 2016) under standard settings. Hi-C eigenvector was calculated using Juicer (Durand et al., 2016) at 200Kb resolution. Capture Hi-C interaction matrix was converted in R, and heatmaps were plotted in R using interaction matrix for 5kb windows. Significant interactions were called using Fit-Hi-C (Ay et al., 2014), and visualized as Arcs in WashU browser (https://epigenomegateway.wustl.edu/). Directionality Index (DI) for all the capture-Hi-C and other Hi-C samples was calculated as previously described (Dixon et al., 2012) at 10kb resolution and with a window size ranging from 300kb to 450kb that matches the actual size of the Dppa2/4 domain in the deletions. Insulation scores at multiple window sizes for capture-Hi-C and Hi-C data at 10kb resolution were calculated as previously described (Crane et al., 2015) and the domainograms were plotted using the GENOVA package (https://github.com/robinweide/GENOVA).
Genome-wide association of ERCE chromatin features
Raw sequencing reads for all the individual ChIP-seq datasets from GSE31039 (H3K4me1, H3K4me3, H3K36me3, H3K27me3, and H3K27ac), GSE36027 (P300 and CTCF), and GSE44288 (Oct4, Sox2, Nanog, and Med1) (Whyte et al., 2013) were aligned using bowtie2 program (Langmead and Salzberg, 2012). We allowed two mismatches relative to the reference and only retained the alignments with Phred quality score greater than 30. The datasets were mapped against the mm10 version of the mouse genome. MACS2 (Feng et al., 2012) peak calling was performed using the following settings ‘-q 0.05 –extsize 200 –m 5 50’ gainst the respective ChIP-seq input files as control. DNaseI peaks were retrieved directly from GSE37074 study and converted to mm10 coordinates from the mm9 assembly using liftOver tool (Tyner et al., 2017). We then mapped the WT lamin B1 (GSE51334) (Pope et al., 2014) and RT signal (this study) surrounding +/−200Kb of each peak from every sample. In case of Oct4, Sox2 and Nanog data; we identified the common triple peak regions among themselves using bedtools (Quinlan and Hall, 2010) and termed them as “OSN” peaks to plot lamin B1 and RT signal.
4C-seq
For inbred cells, reads were quality trimmed (q score =20) and mapped to mm10 reference genome using bowtie2 (Langmead and Salzberg, 2012). For cas/129 hybrid cells, reads were initially mapped to svim genome with quality score above 10, and SNPs parsed to the corresponding allele using the same method as described for repli-seq. Reads overlapping SNPs between the two genomes were filtered and sorted to each allele according to the exact nucleotide at the SNP positions. Reads that did not overlap with SNPs or did not match the exact nucleotide for either genome at the SNP position were discarded. 4C reads mapped at HindIII restriction site resolution were further normalized for library sizes across the WT and ABC Del samples for chromosome 16. Library normalization was performed by ‘calcNormFactors’ function from edgeR package that minimizes the log-fold changes between the samples (Robinson and Oshlack, 2010). The scale-factors were calculated using a trimmed mean of M-values (TMM) between each pair of samples. After library normalization, the raw signal from each 4C sample was converted to counts per million (CPM) values for further downstream analysis. The relative enrichment of 4C interactions in a sample was calculated by comparing a sliding window of 100 (w=100) restriction sites compared to a background window of 3000 restriction fragments (W=3000). To calculate a false discovery rate (FDR) for each enrichment, random permutation on the dataset was performed to determine the threshold enrichment at which the FDR was 0.01 (Splinter et al., 2012). 4C interactions above the FDR threshold of 0.01 were considered as significant and successively used to measure their RT and eigenvalue feature distribution (excluding the surrounding 48.20–48.65Mb region). For mESC and NPC virtual 4C profile, we first mapped the Hi-C data from (Bonev et al., 2017) at 5Kb resolution on mm10 genome using HiC-Pro pipeline (Servant et al., 2015). Once mapped, we extracted the interaction corresponding to the chr16: 48455,000-48555000 region (equivalent to the +/−50Kb 4C-bait position of 48507782) from both mESC and NPC samples to create the virtual 4C profile. mESC profile was down-sampled to the read count of NPC profile for further analysis. We then repeated the same procedure as mentioned for the real 4C data analysis to calculate CPM values and determined the enriched interactions for virtual 4C profiles above the FDR threshold of 0.01 to measure their RT and eigenvalue feature distribution (excluding the surrounding 48.20–48.65Mb region). 4C profile plots were created using custom R scripts. For visualization purpose, we smoothed both the real and virtual 4C profile with a moving average of 300 HindIII site CPM values.
BrU-seq
Reads were mapped using bowtie2, and sorted into either allele using the same script as for repli-seq with SNPs for 129/Sv and CAST/Ei from the Mouse Genomes Project, version 5 (Adams et al., 2015). RPM-normalized read densities were calculated with bedtools2 (Quinlan and Hall, 2010) genomecov, using a scaling factor of 1000000/(number of parsed reads in library). The profiles are shown at 1kb window bins.
Genome-wide prediction of ERCEs
For genome-wide prediction of ERCEs, CTCF independent Hi-C interactions (20kb resolution, FDR 5%) were retrieved using the auxin-induced CTCF depletion HiC (Nora et al., 2017) with Fit-Hi-C program (Ay et al., 2014). The significant interactions were then filtered based on their overlap with P300 binding sites, non-lamin associated domains and Oct4, Sox2 and Nanog (OSN) triple peaks on both sides of the interaction. The final set of interactions were then flattened into 1D regions, which were further processed to merge multiple sites as the predicted ERCEs.
3D modeling
3D modeling of cHi-C data was performed using Chrom3D modeling package (Paulsen et al. 2017). We used FitHiC (Ay et al., 2014) derived significant interactions (FDR < 0.01) from cHi-C data and their lamin-associated domain (LAD) information (Pope et al. 2014) as positional constraints to build the model at 5Kb resolution. UCSF chimera was used for model visualization and image generation (Pettersen et al. 2004).
Supplementary Material
Highlights.
Early replicating control elements (ERCEs) regulate replication timing
ERCEs regulate A/B compartmentalization and TAD architecture
ERCEs form CTCF-independent loops and have features of enhancer/promoters
ERCEs enable genetic dissection of large-scale chromosome structure and function.
ACKNOWLEDGMENTS
We thank G. Church lab for advice on CRISPRology; S. Schoenfelder for cHi-C; S. Wingett for HiCUP; S. Andrews for cHi-C probe design; F. Krueger for SNPsplit; D. Vera for bioinformatics; R. Didier for FACS; C. Pye & K. Poduch for molecular cloning; C. Huffstetler for figure graphics; R. Tjian for Klf4 enhancer deletion; R. Young & N. Behnam for YY1 samples; B. Chadwick for px458; M. Mechali for SNS origin data; A. Belmont for helpful discussions. F.A. and A.C. were partially funded by Institute Leadership Funds from La Jolla Institute for Allergy and Immunology and NIH grant R35GM128938 (F.A.). This work was supported by NIH grant R01GM083337 and U54DK107965 (D.M.G.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
DATA AND SOFTWARE AVAILABILITY
The Repli-seq, capture Hi-C, Hi-C, 4C-seq, Bru-seq data, and Sanger sequencing confirmation for allele specific mutants from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) with accession number GSE114139. Sanger sequencing confirmation for allele specific mutants was deposited at www.replicationtiming.com under CRISPR INDELs sequencing.
REFERENCES
- Abranches E, Silva M, Pradier L, Schulz H, Hummel O, Henrique D, and Bekman E (2009). Neural Differentiation of Embryonic Stem Cells In Vitro: A Road Map to Neurogenesis in the Embryo. PLoS One 4, e6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams DJ, Doran AG, Lilue J, and Keane TM (2015). The Mouse Genomes Project: a repository of inbred laboratory mouse strain genomes. Mamm. Genome 26, 403–412. [DOI] [PubMed] [Google Scholar]
- Arner E, Daub CO, Vitting-Seerup K, Andersson R, Lilje B, Drablos F, Lennartsson A, Ronnerblad M, Hrydziuszko O, Vitezic M, et al. (2015). Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science (80-. ) 347, 1010–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ay F, Bailey TL, and Noble WS (2014). Statistical confidence estimation for Hi-C data reveals regulatory chromatin contacts. Genome Res 24, 999–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogu GK, Vizán P, Stanton LW, Beato M, Di Croce L, and Marti-Renom MA (2016). Chromatin and RNA Maps Reveal Regulatory Long Noncoding RNAs in Mouse. Mol. Cell. Biol 36, 809–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev B, Mendelson Cohen N, Szabo Q, Fritsch L, Papadopoulos GL, Lubling Y, Xu X, Lv X, Hugnot J-P, Tanay A, et al. (2017). Multiscale 3D Genome Rewiring during Mouse Neural Development. Cell 171, 557–572.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne SM, Ortiz L, Mali P, Aach J, and Church GM (2015). Multi-kilobase homozygous targeted gene replacement in human induced pluripotent stem cells. Nucleic Acids Res 43, e21–e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayrou C, Ballester B, Peiffer I, Fenouil R, Coulombe P, Andrau J-C, van Helden J, and Méchali M (2015). The chromatin environment shapes DNA replication origin organization and defines origin classes. Genome Res 25, 1873–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekharappa SC, and Subramanian KN (1987). Effects of position and orientation of the 72-base-pair-repeat transcriptional enhancer on replication from the simian virus 40 core origin. J. Virol 61, 2973–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimbora DM, Schübeler D, Reik A, Hamilton J ., Francastel C, Epner EM, and Groudine M (2000). Long-distance control of origin choice and replication timing in the human beta-globin locus are independent of the locus control region. Mol. Cell. Biol 20, 5581–5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane E, Bian Q, McCord RP, Lajoie BR, Wheeler BS, Ralston EJ, Uzawa S, Dekker J, and Meyer BJ (2015). Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature 523, 240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dileep V, Ay F, Sima J, Vera DL, Noble WS, and Gilbert DM (2015a). Topologically associating domains and their long-range contacts are established during early G1 coincident with the establishment of the replication-timing program. Genome Res 25, 1104–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dileep V, Rivera-Mulia JC, Sima J, and Gilbert DM (2015b). Large-Scale Chromatin Structure-Function Relationships during the Cell Cycle and Development: Insights from Replication Timing. Cold Spring Harb. Symp. Quant. Biol 80, 53–63. [DOI] [PubMed] [Google Scholar]
- Dimitrova DS, and Gilbert DM (1999). The spatial position and replication timing of chromosomal domains are both established in early G1 phase. Mol. Cell 4, 983–993. [DOI] [PubMed] [Google Scholar]
- Dimitrova DS, Prokhorova TA, Blow JJ, Todorov IT, and Gilbert DM (2002). Mammalian nuclei become licensed for DNA replication during late telophase. J. Cell Sci 115, 51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, and Ren B (2012). Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY, Ye Z, Kim A, Rajagopal N, Xie W, et al. (2015). Chromatin architecture reorganization during stem cell differentiation. Nature 518, 331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen JM, Fan ZP, Hnisz D, Ren G, Abraham BJ, Zhang LN, Weintraub AS, Schuijers J, Lee TI, Zhao K, et al. (2014). Control of Cell Identity Genes Occurs in Insulated Neighborhoods in Mammalian Chromosomes. Cell 159, 374–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryden NH, Broome LR, Dudbridge F, Johnson N, Orr N, Schoenfelder S, Nagano T, Andrews S, Wingett S, Kozarewa I, et al. (2014). Unbiased analysis of potential targets of breast cancer susceptibility loci by Capture Hi-C. Genome Res 24, 1854–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Chen T, Zou X, Xiong B, and Lu G (2010). Dppa2 knockdown-induced differentiation and repressed proliferation of mouse embryonic stem cells. J. Biochem 147, 265–271. [DOI] [PubMed] [Google Scholar]
- Dupont C, Maduro C, Den Braanker H, Boers R, Kurek D, and Gribnau J (2016). Characterization of Histone Modifications Associated with Inactive X-Chromosome in Trophoblast Stem Cells, eXtra-Embryonic Endoderm Cells and in In Vitro Derived Undifferentiated and Differentiated Epiblast Like Stem Cells. PLoS One 11, e0167154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand NC, Shamim MS, Machol I, Rao SSP, Huntley MH, Lander ES, and Aiden EL (2016). Juicer Provides a One-Click System for Analyzing Loop-Resolution Hi-C Experiments. Cell Syst 3, 95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang D, Lengronne A, Shi D, Forey R, Skrzypczak M, Ginalski K, Yan C, Wang X, Cao Q, Pasero P, et al. (2017). Dbf4 recruitment by forkhead transcription factors defines an upstream rate-limiting step in determining origin firing timing. Genes Dev 31, 2405–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang F, Xu Y, Chew K-K, Chen X, Ng H-H, and Matsudaira P (2014). Coactivators p300 and CBP Maintain the Identity of Mouse Embryonic Stem Cells by Mediating Long-Range Chromatin Structure. Stem Cells 32, 1805–1816. [DOI] [PubMed] [Google Scholar]
- Feng J, Liu T, Qin B, Zhang Y, and Liu XS (2012). Identifying ChIP-seq enrichment using MACS. Nat. Protoc 7, 1728–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foti R, Gnan S, Cornacchia D, Dileep V, Bulut-Karslioglu A, Diehl S, Buness A, Klein FA, Huber W, Johnstone E, et al. (2016). Nuclear Architecture Organized by Rif1 Underpins the Replication-Timing Program. Mol. Cell 61, 260–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DM (2001). Nuclear position leaves its mark on replication timing. J. Cell Biol 152, F11–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DM (2010). Evaluating genome-scale approaches to eukaryotic DNA replication. Nat. Rev. Genet 11, 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goren A, Tabib A, Hecht M, and Cedar H (2008). DNA replication timing of the human beta-globin domain is controlled by histone modification at the origin. Genes Dev 22, 1319–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan-Zadeh V, Chilaka S, Cadoret J-C, Ma MK-W, Boggetto N, West AG, and Prioleau M-N (2012). USF binding sequences from the HS4 insulator element impose early replication timing on a vertebrate replicator. PLoS Biol 10, e1001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez C, Wang Z, Ramazanov B, Tang Y, Mehta S, Dambrot C, Lee Y-W, Tessema K, Kumar I, Astudillo M, et al. (2018). Dppa2/4 Facilitate Epigenetic Remodeling during Reprogramming to Pluripotency. Cell Stem Cell 23, 396–411.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Yokochi T, Schwaiger M, Chang C-WW, Lyou Y, Townes TM, Schübeler D, Gilbert DM, et al. (2008). Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol 6, e245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Takebayashi S, Lu J, and Gilbert DM (2009). Replication timing and transcriptional control: beyond cause and effect—part II. Curr. Opin. Genet. Dev 19, 142–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Rathjen J, Kulik M, Papp B, Fussner E, Bazett-Jones DP, Plath K, Dalton S, et al. (2010). Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome Res 20, 155–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, Schafer X, Lun Y, and Lemischka IR (2006). Dissecting self-renewal in stem cells with RNA interference. Nature 442, 533–538. [DOI] [PubMed] [Google Scholar]
- Jerabek S, Ng CK, Wu G, Arauzo-Bravo MJ, Kim K-P, Esch D, Malik V, Chen Y, Velychko S, MacCarthy CM, et al. (2017). Changing POU dimerization preferences converts Oct6 into a pluripotency inducer. EMBO Rep 18, 319–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaaij LJT, van der Weide RH, Ketting RF, and de Wit E (2018). Systemic Loss and Gain of Chromatin Architecture throughout Zebrafish Development. Cell Rep 24, 1–10.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Y, Xu Y, Chen X, Feng S, Liu Z, Sun Y, Yao X, Li F, Zhu W, Gao L, et al. (2017). 3D Chromatin Structures of Mature Gametes and Structural Reprogramming during Mammalian Embryogenesis. Cell 170, 367–381.e20. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, and Haussler D (2002). The human genome browser at UCSC. Genome Res 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerpedjiev P, Abdennur N, Lekschas F, McCallum C, Dinkla K, Strobelt H, Luber JM, Ouellette SB, Azhir A, Kumar N, et al. (2017). HiGlass: Web-based Visual Exploration and Analysis of Genome Interaction Maps. bioRxiv 121889. [DOI] [PMC free article] [PubMed]
- Khan A, and Zhang X (2016). dbSUPER: a database of super-enhancers in mouse and human genome. Nucleic Acids Res 44, D164–D171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind J, Pagie L, Ortabozkoyun H, Boyle S, de Vries SS, Janssen H, Amendola M, Nolen LD, Bickmore WA, and van Steensel B (2013). Single-cell dynamics of genome-nuclear lamina interactions. Cell 153, 178–192. [DOI] [PubMed] [Google Scholar]
- King HW, and Klose RJ (2017). The pioneer factor OCT4 requires the chromatin remodeller BRG1 to support gene regulatory element function in mouse embryonic stem cells. Elife 6, e22631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren A, Handsaker RE, Kamitaki N, Karlić R, Ghosh S, Polak P, Eggan K, and McCarroll SA (2014). Genetic Variation in Human DNA Replication Timing. Cell 159, 1015–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, and Andrews SR (2016). SNPsplit: Allele-specific splitting of alignments between genomes with known SNP genotypes. F1000Research 5, 1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande-Diner L, Zhang J, and Cedar H (2009). Shifts in Replication Timing Actively Affect Histone Acetylation during Nucleosome Reassembly. Mol. Cell 34, 767–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Chen J, Solessio E, and Gilbert DM (2003). Spatial distribution and specification of mammalian replication origins during G1 phase. J. Cell Biol 161, 257–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. (2009). Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Li F, Murphy CS, Davidson MW, and Gilbert DM (2010). G2 phase chromatin lacks determinants of replication timing. J. Cell Biol 189, 967–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, et al. (2015). Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161, 1012–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu X, Rowley MJ, and Corces VG (2018). Architectural Proteins and Pluripotency Factors Cooperate to Orchestrate the Transcriptional Response of hESCs to Temperature Stress. Mol. Cell [DOI] [PMC free article] [PubMed]
- Madan B, Madan V, Weber O, Tropel P, Blum C, Kieffer E, Viville S, and Fehling HJ (2009). The pluripotency-associated gene Dppa4 is dispensable for embryonic stem cell identity and germ cell development but essential for embryogenesis. Mol. Cell. Biol 29, 3186–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchal C, Sasaki T, Vera D, Wilson K, Sima J, Rivera-Mulia J-C, Trevilla Garcia C, Nogues C, Nafie E, and Gilbert DM (2017). Repli-seq: genome-wide analysis of replication timing by next-generation sequencing. bioRxiv [DOI] [PMC free article] [PubMed]
- Masaki H, Nishida T, Kitajima S, Asahina K, and Teraoka H (2007). Developmental pluripotency-associated 4 (DPPA4) localized in active chromatin inhibits mouse embryonic stem cell differentiation into a primitive ectoderm lineage. J. Biol. Chem 282, 33034–33042. [DOI] [PubMed] [Google Scholar]
- Monkhorst K, Jonkers I, Rentmeester E, Grosveld F, and Gribnau J (2008). X Inactivation Counting and Choice Is a Stochastic Process: Evidence for Involvement of an X-Linked Activator. Cell 132, 410–421. [DOI] [PubMed] [Google Scholar]
- Mumbach MR, Rubin AJ, Flynn RA, Dai C, Khavari PA, Greenleaf WJ, and Chang HY (2016). HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat. Methods 13, 919–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabet B, Roberts JM, Buckley DL, Paulk J, Dastjerdi S, Yang A, Leggett AL, Erb MA, Lawlor MA, Souza A, et al. (2018). The dTAG system for immediate and target-specific protein degradation. Nat. Chem. Biol 14, 431–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Nakagawa M, Ichisaka T, Shiota A, and Yamanaka S (2011). Essential Roles of ECAT15–2/Dppa2 in Functional Lung Development. Mol. Cell. Biol 31, 4366–4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. (2012). Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Goloborodko A, Valton A-L, Gibcus JH, Uebersohn A, Abdennur N, Dekker J, Mirny LA, and Bruneau BG (2017). Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell 169, 930–944.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrow AZ, Kalhor R, Gan Y, Villwock SK, Linke C, Barberis M, Chen L, and Aparicio OM (2017). Conserved forkhead dimerization motif controls DNA replication timing and spatial organization of chromosomes in S. cerevisiae. Proc. Natl. Acad. Sci. U. S. A 114, E2411–E2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen MT, Veloso A, Prasad J, Bedi K, Ljungman EA, Magnuson B, Wilson TE, and Ljungman M (2014). Use of Bru-Seq and BruChase-Seq for genome-wide assessment of the synthesis and stability of RNA. Methods 67, 45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SWM, Solovei I, Brugman W, Gräf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al. (2010). Molecular Maps of the Reorganization of Genome-Nuclear Lamina Interactions during Differentiation. Mol. Cell 38, 603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt EJ, Smith L, and Thayer MJ (2018). L1 retrotransposon antisense RNA within ASAR lncRNAs controls chromosome-wide replication timing. J. Cell Biol 217, 541–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope BD, Chandra T, Buckley Q, Hoare M, Ryba T, Wiseman FK, Kuta A, Wilson MD, Odom DT, and Gilbert DM (2012). Replication-timing boundaries facilitate cell-type and species-specific regulation of a rearranged human chromosome in mouse. Hum. Mol. Genet 21, 4162–4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope BD, Ryba T, Dileep V, Yue F, Wu W, Denas O, Vera DL, Wang Y, Hansen RS, Canfield TK, et al. (2014). Topologically associating domains are stable units of replication-timing regulation. Nature 515, 402–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourkarimi E, Bellush JM, and Whitehouse I (2016). Spatiotemporal coupling and decoupling of gene transcription with DNA replication origins during embryogenesis in C. elegans. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prioleau M-N, Gendron M-C, and Hyrien O (2003). Replication of the chicken beta-globin locus: early-firing origins at the 5’ HS4 insulator and the rho- and betaA-globin genes show opposite epigenetic modifications. Mol. Cell. Biol 23, 3536–3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-Solis R, Rivera-Pérez J, Wallace JD, Wims M, Zheng H, and Bradley A (1992). Genomic DNA microextraction: a method to screen numerous samples. Anal. Biochem 201, 331–335. [DOI] [PubMed] [Google Scholar]
- Rao SSP, Huang S-C, Glenn St Hilaire B, Engreitz JM, Perez EM, Kieffer-Kwon K-R, Sanborn AL, Johnstone SE, Bascom GD, Bochkov ID, et al. (2017). Cohesin Loss Eliminates All Loop Domains. Cell 171, 305–320.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik A, Telling A, Zitnik G, Cimbora D, Epner E, and Groudine M (1998). The locus control region is necessary for gene expression in the human beta-globin locus but not the maintenance of an open chromatin structure in erythroid cells. Mol. Cell. Biol 18, 5992–6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rideout WM, Wakayama T, Wutz A, Eggan K, Jackson-Grusby L, Dausman J, Yanagimachi R, and Jaenisch R (2000). Generation of mice from wild-type and targeted ES cells by nuclear cloning. Nat. Genet 24, 109–110. [DOI] [PubMed] [Google Scholar]
- Rivera-Mulia JC, and Gilbert DM (2016). Replication timing and transcriptional control: beyond cause and effect—part III. Curr. Op in. Cell Biol 40, 168–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Mulia JC, Buckley Q, Sasaki T, Zimmerman J, Didier RA, Nazor K, Loring JF, Lian Z, Weissman S, Robins AJ, et al. (2015). Dynamic changes in replication timing and gene expression during lineage specification of human pluripotent stem cells. Genome Res 25, 1091–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Mulia JC, Desprat R, Trevilla-Garcia C, Cornacchia D, Schwerer H, Sasaki T, Sima J, Fells T, Studer L, Lemaitre J-M, et al. (2017a). DNA replication timing alterations identify common markers between distinct progeroid diseases. Proc. Natl. Acad. Sci. U. S. A 114, E10972–E10980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Mulia JC, Dimond A, Vera D, Trevilla-Garcia C, Sasaki T, Zimmerman J, Dupont C, Gribnau J, Fraser P, and Gilbert DM (2017b). Allele-specific control of replication timing and genome organization during development. bioRxiv 221762. [DOI] [PMC free article] [PubMed]
- Robinson MD, and Oshlack A (2010). A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley MJ, and Corces VG (2018). Organizational principles of 3D genome architecture. Nat. Rev. Genet 19, 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J, Schulz TC, Robins AJ, Dalton S, and Gilbert DM (2010). Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res 20, 761–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T, Hiratani I, Sasaki T, Battaglia D, Kulik M, Zhang J, Dalton S, and Gilbert DM (2011). Replication timing: a fingerprint for cell identity and pluripotency. PLoS Comput. Biol 7, e1002225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabari BR, Dall’Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, Abraham BJ, Hannett NM, Zamudio AV, Manteiga JC, et al. (2018). Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansam CG, Pietrzak K, Majchrzycka B, Kerlin MA, Chen J, Rankin S, and Sansam CL (2018). A mechanism for epigenetic control of DNA replication. Genes Dev 32, 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Rivera-Mulia JC, Vera D, Zimmerman J, Das S, Padget M, Nakamichi N, Chang BH, Tyner J, Druker BJ, et al. (2017). Stability of patient-specific features of altered DNA replication timing in xenografts of primary human acute lymphoblastic leukemia. Exp. Hematol 51, 71–82.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schübeler D, Francastel C, Cimbora DM, Reik A, Martin DI, and Groudine M (2000). Nuclear localization and histone acetylation: a pathway for chromatin opening and transcriptional activation of the human beta-globin locus. Genes Dev 14, 940–950. [PMC free article] [PubMed] [Google Scholar]
- Schwarzer W, Abdennur N, Goloborodko A, Pekowska A, Fudenberg G, Loe-Mie Y, Fonseca NA, Huber WH Haering C, Mirny L, et al. (2017). Two independent modes of chromatin organization revealed by cohesin removal. Nature 551, 51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seller CA, and O’Farrell PH (2018). Rif1 prolongs the embryonic S phase at the Drosophila mid-blastula transition. PLOS Biol 16, e2005687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servant N, Varoquaux N, Lajoie BR, Viara E, Chen C-J, Vert J-P, Heard E, Dekker J, and Barillot E (2015). HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol 16, 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siefert JC, Georgescu C, Wren JD, Koren A, and Sansam CL (2017). DNA replication timing during development anticipates transcriptional programs and parallels enhancer activation. Genome Res 27, 1406–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sima J, Bartlett DA, Gordon MR, and Gilbert DM (2017). Bacterial artificial chromosomes establish replication timing and sub-nuclear compartment de novo as extra-chromosomal vectors. Nucleic Acids Res [DOI] [PMC free article] [PubMed]
- Simon I, Tenzen T, Mostoslavsky R, Fibach E, Lande L, Milot E, Gribnau J, Grosveld F, Fraser P, and Cedar H (2001). Developmental regulation of DNA replication timing at the human beta globin locus. EMBO J 20, 6150–6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair P, Bian Q, Plutz M, Heard E, and Belmont AS (2010). Dynamic plasticity of large-scale chromatin structure revealed by self-assembly of engineered chromosome regions. J. Cell Biol 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovei I, Thanisch K, and Feodorova Y (2016). How to rule the nucleus: divide et impera. Curr. Opin. Cell Biol 40, 47–59. [DOI] [PubMed] [Google Scholar]
- Spalholz BA, Yang YC, and Howley PM (1985). Transactivation of a bovine papilloma virus transcriptional regulatory element by the E2 gene product. Cell 42, 183–191. [DOI] [PubMed] [Google Scholar]
- Splinter E, de Wit E, van de Werken HJG, Klous P, and de Laat W (2012). Determining long-range chromatin interactions for selected genomic sites using 4C-seq technology: From fixation to computation. Methods 58, 221–230. [DOI] [PubMed] [Google Scholar]
- Takebayashi S, Dileep V, Ryba T, Dennis JH, and Gilbert DM (2012). Chromatin-interaction compartment switch at developmentally regulated chromosomal domains reveals an unusual principle of chromatin folding. Proc. Natl. Acad. Sci. U. S. A 109, 12574–12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyner C, Barber GP, Casper J, Clawson H, Diekhans M, Eisenhart C, Fischer CM, Gibson D, Gonzalez JN, Guruvadoo L, et al. (2017). The UCSC Genome Browser database: 2017 update. Nucleic Acids Res 45, D626–D634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub AS, Li CH, Zamudio AV, Sigova AA, Hannett NM, Day DS, Abraham BJ, Cohen MA, Nabet B, Buckley DL, et al. (2017). YY1 Is a Structural Regulator of Enhancer-Promoter Loops. Cell 171, 1573–1588.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Werken HJG, Haan JC, Feodorova Y, Bijos D, Weuts A, Theunis K, Holwerda SJB, Meuleman W, Pagie L, Thanisch K, et al. (2017). Small chromosomal regions position themselves autonomously according to their chromatin class. Genome Res 27, 922–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, and Young RA (2013). Master Transcription Factors and Mediator Establish Super-Enhancers at Key Cell Identity Genes . Cell 153, 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingett S, Ewels P, Furlan-Magaril M, Nagano T, Schoenfelder S, Fraser P, and Andrews S (2015). HiCUP: pipeline for mapping and processing Hi-C data. F1000Research 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Torigoe SE, Xiao J, Mai DH, Li L, Davis FP, Dong P, Marie-Nelly H, Grimm J, Lavis L, et al. (2017). A dynamic interplay of enhancer elements regulates Klf4 expression in naïve pluripotency. Genes Dev 31, 1795–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe E, Farkash-Amar S, Polten A, Yakhini Z, Tanay A, and Simon I (2010). Comparative analysis of DNA replication timing reveals conserved large-scale chromosomal architecture. PLoS Genet 6, e1001011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Q-L, Stavridis M, Griffiths D, Li M, and Smith A (2003). Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat. Biotechnol 21, 183–186. [DOI] [PubMed] [Google Scholar]
- Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, Sandstrom R, Ma Z, Davis C, Pope BD, et al. (2014). A comparative encyclopedia of DNA elements in the mouse genome. Nature 515, 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zullo JM, Demarco I. a, Piqué-Regi R, Gaffney DJ, Epstein CB, Spooner CJ, Luperchio TR, Bernstein BE, Pritchard JK, Reddy KL, et al. (2012). DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell 149, 1474–1487. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.