Abstract
Iron supplements are widely consumed; however most of the iron is not absorbed and enters the colon where potentially pathogenic bacteria can utilise it for growth. This study investigated the effect of iron availability on human gut microbial composition and function using an in vitro colonic fermentation model inoculated with faecal microbiota from healthy adult donors, as well as examining the effect of iron on the growth of individual gut bacteria. Batch fermenters were seeded with fresh faecal material and supplemented with the iron chelator, bathophenanthroline disulphonic acid (BPDS). Samples were analysed at regular intervals to assess impact on the gut bacterial communities. The growth of Escherichia coli and Salmonella typhimurium was significantly impaired when cultured independently in iron-deficient media. In contrast, depletion of iron did not affect the growth of the beneficial species, Lactobacillus rhamnosus, when cultured independently. Analysis of the microbiome composition via 16S-based metataxonomics indicated that under conditions of iron chelation, the relative abundance decreased for several taxa, including a 10% decrease in Escherichia and a 15% decrease in Bifidobacterium. Metabolomics analysis using 1 H-NMR indicated that the production of SCFAs was reduced under iron-limited conditions. These results support previous studies demonstrating the essentiality of iron for microbial growth and metabolism, but, in addition, they indicate that iron chelation changes the gut microbiota profile and influences human gut microbial homeostasis through both compositional and functional changes.
Keywords: Chelation, Dysbiosis, Gut microbiota, Iron, Short chain fatty acids
Graphical Abstract
1. Introduction
Humans are host to a wide variety of microbes which develop ecosystems that are specific to any one individual but which are subjected to a life-long process of colonisation by foreign microbes [1]. Through evolution, an environment of mutualism has been created whereby many host-bacterial associations have become beneficial relationships; such as is found in the large intestine (colon), which is home to the highest number of microbes (> 100 trillion bacteria) in our body. Commensal bacteria of the mammalian gut have long been recognised for the advantages they confer to the host. The benefits include defence against opportunistic pathogens, metabolism of indigestible compounds, provision of essential nutrients and an involvement in the development of the intestinal structure [2], [3], whilst also producing metabolites, such as SCFAs that contribute towards providing energy to host cells [4], [5], [6]. The human gut microbiome also contributes towards the basic developmental features and functions of the immune system [7]. Conversely, perturbations (e.g. antibiotic treatment and/or overgrowth of pathogenic bacteria) in these symbiotic relationships, termed dysbiosis, can lead to a negative impact on the host's health [8]. Dysbiosis has been associated with a range of human disease states, such as autoimmune disorders [9], [10] increased vulnerability to cancers [11], irritable bowel syndrome [12], [13], [14], [15], and obesity [16], and research on these associations, including an understanding of the mechanisms involved, is a high priority. The number of studies examining the relationship between diet and the gut microbiome has increased over the past years and has shown that the gut microbiome composition and metabolism are significantly affected by diet [17].
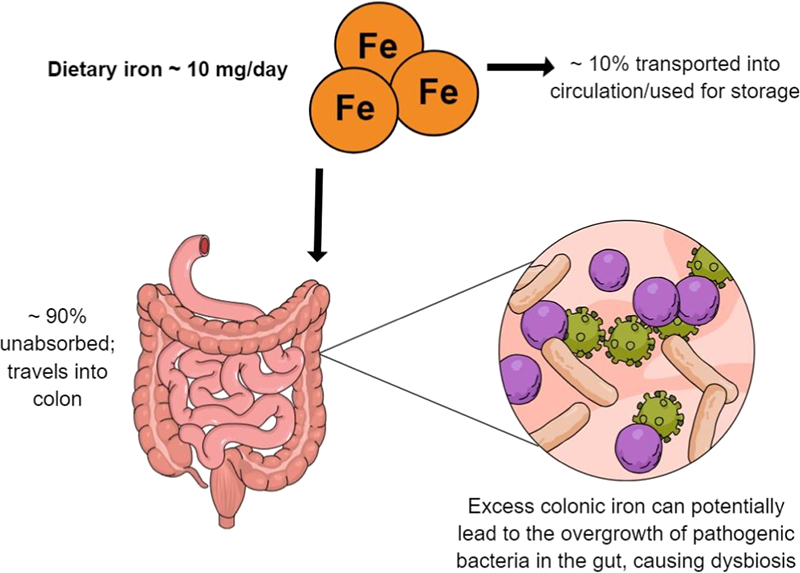
Iron, an essential nutrient for humans, is highly abundant in the environment and involved in numerous biological processes such as hydrogen production, respiration, and DNA biosynthesis. It also functions as a co-factor in numerous metabolic pathways within a host cell and is essential for nearly all prokaryotic and eukaryotic cells. In humans, the absorption of iron by the epithelial cells of the small intestine is a very tightly regulated process due to the absence of excretory pathways [18]. The efficiency of absorption depends on the potential availability of iron in the diet, and is regulated by physiological iron requirements, including body iron stores, with hepcidin having a central role in the control of absorption [19].
Dietary iron has been broadly classified into two types, non-haem and haem iron. Both forms of dietary iron have a separate pathway of uptake by enterocytes, where haem iron is better absorbed (25–30%) compared to the poorly absorbed non-haem iron, (0–15% absorbed) despite being the most common form of iron present in the diet [20]. The unabsorbed iron travels to the large intestine, and there is accumulating evidence to suggest that this unabsorbed colonic iron can facilitate the growth of intestinal pathogens [21].
The vast majority of bacteria in the gut require iron for growth and development, and they have formulated many strategies to acquire this nutrient, which in its more common state (Fe3+) has low or zero solubility. Potentially pathogenic bacteria make use of a continuous supply of micronutrients, such as iron, for metabolism and replication. Thus, there is constant competition for iron between various bacteria, many of which have developed mechanisms, such as siderophore production, to acquire iron, particularly when availability is limited. Unlike most bacteria, members of the Bifidobacteriaceae and Lactobacilliaceae families (two families that are believed to be beneficial to the host [22], [23], [24], [25]) have a very limited need for iron [26]. Lactobacilli do not produce siderophores to sequester iron, and their growth is similar in media with and without iron [27]. Bifidobacterium breve, an important member of the Bifidobacterial species in breast-fed infants, can acquire luminal iron with the help of a divalent metal permease, however, many of the Bifidobacterium species do not synthesise siderophores or other forms of iron-carriers. The human body has created many ways to promote the growth of these beneficial bacteria, such as the presence of lactoferrin in breast milk, which travels intact to the large intestine [28], [29], [30], which with its high affinity for iron, renders the iron unavailable to potentially pathogenic bacteria [31]. Along with high amounts of dietary iron in the gut lumen, the oral administration of iron supplements also results in freely available “unbound” iron in the colon [32], [33], [34]. This disturbs iron homeostasis consequently modifying the gut microbial composition [35], [36], [37].
An emerging link between the gut microbiome and iron availability has been observed, however several studies that have investigated the effects of iron on the human gut microbiota are mainly focussed on the infant microbiome [6], [33], [34], [35], [38], [39], [40]. Other studies have looked at the role of iron in the gut microbiota in individuals with either metabolic [41] or iron disorders and therefore, this study was designed to use in vitro colon models that mimic the microbiological conditions of the human large intestine to assess the impact of iron limitation on structure and function of the complex gut bacterial communities of healthy adults.
2. Materials and methods
2.1. Bacterial strains and growth conditions
To evaluate the role of iron on bacterial growth, the following bacterial strains were chosen: Escherichia coli 1BO4 (isolated from human faeces), Bifidobacterium longum B78 F110564 (isolated from human faeces), Lactobacillus rhamnosus GG F111027, Salmonella Typhimurium (ATCC SL1344), Clostridium perfringens (NCTC 3110) and Bacteroides thetaiotaomicron VP1–5482 (ATCC 29148). Bacteria were cultured in the following media: Luria (L) Broth (E. coli and S. Typhimurium); Brain Heart Infusion (BHI; B. longum); BHI supplemented with vitamin K (0.5% v/v), hemin (0.5 mg/mL), resazurin (0.02% w/v) and L-cysteine (0.5 g/L) (C. perfringens) and BHI with hemin (B. thetaiotaomicron); De Man, Rogosa and Sharpe (MRS) supplemented with glucose (L. rhamnosus). All bacteria were grown under anaerobic conditions at 37 °C. E. coli and S. Typhimurium were agitated at medium speed throughout the growth period, whilst the remaining bacteria were grown under static conditions.
2.2. The effect of iron chelation on bacterial pure cultures
Bacterial cultures were grown overnight in rich media specified above. Cultures were seeded in 100-well honeycomb plates and cells were then provided with bathophenanthrolinedisulfonic acid disodium salt hydrate (BPDS; B1375, Sigma, United Kingdom) at final concentrations ranging from 0–50 μM. Samples were analysed using a Magellan Microplate Reader (Tecan Life Sciences, Switzerland), which monitors the growth of microorganisms by measuring the turbidity of the culture (optical density, OD600) of the liquid growth medium. The experiments were run for 24 h, with OD600, taken every 10 min. The experiments were performed at 37 °C under anaerobic conditions.
2.3. In vitro colonic fermentations
Ten grams of fresh faecal samples obtained from healthy volunteers were diluted 1:10 in deoxygenated phosphate buffered saline (pH 7.7), and homogenised using a Stomacher 400 (Seward, United Kingdom) at 230 rpm for 45 s. All samples were processed within 4 h of stool collection.
The working volume of each vessel was set at 150 mL, of which 15 mL was processed faecal inocula and 135 mL was nutritive media (peptone water 2 g/l, yeast extract 2 g/l, NaCl 0.1 g/l, K2HPO4 0.04 g/l, KH2PO4 0.04 g/l, MgSO4.7H2O 0.01 g/l, CaCl2.6H2O 0.01 g/l, NaHCO3 2 g/l, Tween 80 2 ml, glucose 10 g/l, vitamin K1 10 μl, cysteine HCl 0.5 g/l, bile salts 0.5 g/l, pH 7.0). The pH was controlled and maintained between 6.6–7.0 using pH control units Fermac 260 (Electrolab, United Kingdom), connected to 1 M NaOH and 1 M HCL solutions. Vessels were kept at 37 °C by a circulating water jacket and anaerobiosis was maintained by continuous bubbling of the system with oxygen-free nitrogen gas. Conditions tested were either nutritive media with faecal inocula only (Control), or with faecal inocula and BPDS (70 μM). The concentration of BPDS was chosen as a result of preliminary experiments investigating the baseline faecal iron concentrations in multiple healthy donors (data not shown). We used 70 μM BPDS as we calculated that this concentration would be sufficient to chelate most of the iron in the colon. Per donor, 1 vessel was used for each condition. Samples were taken at 0, 4, 8, and 24 h from each vessel, serially diluted in PBS and enumerated on selective agar plates [42]. Faecal samples used in the colon model experiments were obtained from participants recruited onto the QIB Colon Model study. Men and women aged 18 years or older who live or work within 10 miles of the Norwich Research Park were recruited onto the QIB Colon Model study if they satisfied the following criteria. Participants who were assessed to have a normal bowel habit, regular defecation between three times a day and three times a week, with an average stool type of 3–5 on the Bristol Stool Chart, and no diagnosed chronic gastrointestinal health problems, such as irritable bowel syndrome, inflammatory bowel disease, or coeliac disease were eligible to enrol onto the study. Demographic information was collected, and a brief health questionnaire was completed during the eligibility screening. Participants were asked additional questions immediately prior to donating a stool sample to confirm that they had not taken antibiotics or probiotics within the last four weeks, had not experienced a gastrointestinal complaint, such as vomiting or diarrhoea, within the last 72 h, were not currently pregnant or breast-feeding, had not recently had an operation requiring general anaesthetic, and were not taking iron or multivitamin supplements. The study was approved by the Quadram Institute Bioscience (formally Institute of Food Research) Human Research Governance committee (IFR01/2015), and London - Westminster Research Ethics Committee (15/LO/2169). The informed consent of all participating subjects was obtained, and the trial is registered at http://www.clinicaltrials.gov (NCT02653001).
2.4. DNA extraction for 16S rDNA based metagenomics
DNA was extracted from all samples collected from in vitro colonic fermentations using a commercially available kit (FastDNA spin kit for soil; MP Biomedicals, USA, Cat No. 6560200). Samples were thawed on ice, homogenised, and approximately 200 mg of each were used to extract DNA following manufacturer's instruction, with an additional bead beating step using FastPrep (MP Biomedicals, USA), as previously described by Kellingray et al. [43].
2.5. 16S rRNA gene amplification and sequencing
The impact of iron on the composition of the human gut microbiome was investigated using high throughput 16S rRNA gene (V4 region) sequencing using the Illumina Miseq platform, followed by data analysis using the Quantitative Insights into Microbial Ecology (QIIME, V1.9) pipeline. Each sample sequence was filtered to read lengths between minimum 200 and maximum 1000 bp. ChimeraSlayer was used to filter trimmed reads for chimeric sequences after which Shannon index were used to create alpha diversity plots. Weighted UniFrac data were used to produce beta diversity PCoA plots, using the XLSTAT add-on package on Microsoft Excel.
2.6. Short chain fatty acid quantification in cultured microbiomes
Faecal water was prepared to quantify short chain fatty acids in stool. Briefly, samples (13 mL) taken from colonic batch fermentations were centrifuged at 3220×g for 15 min at 4 °C. 100 μl NMR buffer (0.26 g NaH2PO4 and 1.41 g K2HPO4 made up in 100 ml D2O, containing 0.1% NaN3 (100 mg), and 1 mM sodium 3-(Trimethylsilyl)-propionate-d4, (TSP) (17 mg) as a chemical shift reference) was added to 900 μl supernatant and analysed using 1H NMR spectroscopy (this mixture is defined as ‘faecal water’). The 1H NMR spectra were recorded at 600 MHz on a Bruker Advance spectrometer (Bruker BioSpin GmbH, Germany) running Topspin 2.0 software and fitted with a cryoprobe and a 60-slot autosampler. Each 1H NMR spectrum was acquired with 256 scans, a spectral width of 12,300 Hz, and an acquisition time of 2.67 s. The “noesypr1d” pre-saturation sequence was used to suppress the residual water signal with a low-power selective irradiation at the water frequency during the recycle delay and a mixing time of 10 ms. Spectra were transformed with a 0.3 Hz line broadening, and were manually phased, baseline corrected, and referenced by setting the TSP methyl signal to 0 ppm. The metabolites were quantified using the software Chenomx® NMR Suite 7.0™.
2.7. Quantification of iron levels in fermentation samples
A 20 μl aliquot of the faecal water prepared in Section 2.6 was used to quantify iron concentrations of the fermentation samples at 0, 8 and 24 h using the ferrozine assay as per manual instructions (Iron Assay Kit ab83366, Abcam, UK). Briefly, iron in the sample is reduced using an Fe reducer, provided by the kit, after which iron reacts with Ferene S (an iron chromogen) to produce a stable coloured complex. Absorbance measurements were taken at 593 nm.
2.8. Statistical analysis
Data from pure culture experiments and in vitro colonic fermentations were expressed as means±S.E.M. Pure cultures were analysed using one-way ANOVA followed by Bonferroni post-tests with GraphPad Prism software (Version 5.04), whilst in vitro colonic batch fermentations were analysed using unpaired t tests assuming unequal variances on Microsoft Excel. P<.05 was considered statistically significant.
3. Results
3.1. Iron chelation significantly reduced the growth of selected bacterial species grown in pure cultures
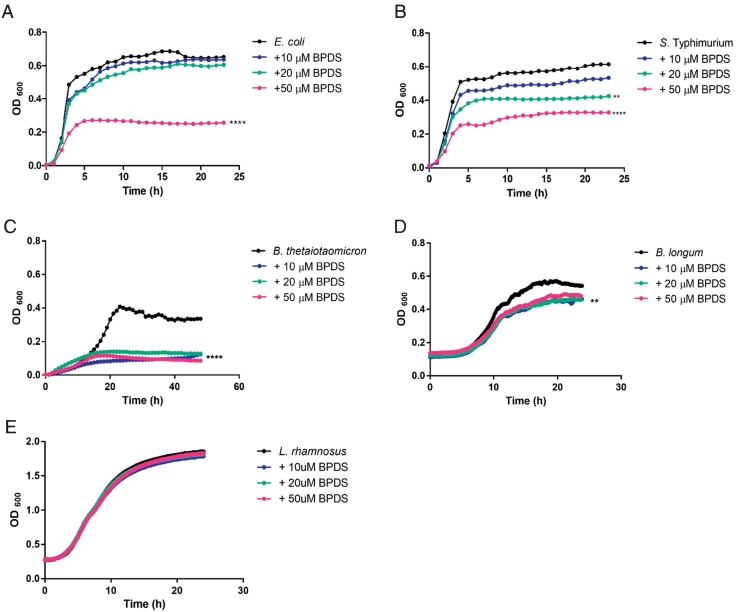
For pure culture experiments, we wanted to test a range of gut bacteria that are known to be affected or unaffected by iron, but the selection, to some extent, depended on what was available in the laboratory. Therefore, various bacterial species were treated with different concentrations (0, 10, 20, and 50 μM) of BPDS in triplicate for 24 h to determine the effect of iron chelation on their growth. A dose-dependent reduction in the growth of E. coli, S. Typhimurium and B. thetaiotaomicron (P<.0001), were observed in the presence of BPDS, at 24 h (Fig. 1A-C). As expected, L. rhamnosus, a bacterial species known to be able to grow in the absence of iron, exhibited comparable growth to the BPDS-free (0 μM) culture, confirming previous observations that iron is not an essential nutrient for the survival of this bacterial species (Fig. 1E). A decrease was also observed in the growth of B. longum (P<.01) upon iron chelation (Fig. 1D). These results support the idea that iron is an essential nutrient for bacteria, and in its absence the growth of many bacteria is restricted.
Fig. 1.
The effect of iron chelation with BPDS on pure bacterial cultures.
A strain of E. coli isolated from human gut (1A), S. Typhimurium (1B), B. thetaiotaomicron (1C) B. longum (1D) and L. rhamnosus (1E) were cultured anaerobically overnight in selective rich media. Cultures were supplemented with BPDS, at 0 (control), 10, 20 and 50 μM. Optical density measurements were taken every 10 min for a period of 24 h and growth curves were plotted for each strain. Data shown = mean and are representative of 3 independent experiments. The data was statistically analysed using, one-way ANOVA, comparing the different concentrations of BPDS to the control, over a period of 24 h; **P<.01, ****P<.0001 versus control (0 μM BPDS).
3.2. Reduced iron availability decreased the growth of potentially pathogenic bacteria under in vitro colonic fermentation conditions
Batch fermentations were performed for 24 h to elucidate the effects of iron depletion on the human gut microbiome by enumerating specific bacterial groups using selective agar plates. Counts from the control vessel of the three individual experiments were first normalised to 100% and thereafter, viable counts from the iron-chelated vessels were shown as percentages in relation to the Control. Finally, the data from the three individual experiments were combined and average percentages (±SEM) presented (Table 1). Results showed a decrease in the viable counts of a number of bacterial groups, including Enterobacteriaceae (P<.05 at 8 and 24 h), Bifidobacteria (P<.001 at 8 h) and Lactobacilli (P<.01 at 8 and 24 h), upon iron chelation at 8 h and 24 h (Table 1). It should be noted that the high viable counts of Bifidobacteria at 24 h is attributed to a single donor. The remaining two donors displayed a substantial reduction in the viable counts of Bifidobacteria under iron-chelated conditions.
Table 1.
The effect of iron chelation via BPDS on gut-derived bacterial communities
|
Bacterial Group |
Viable Counts % (“Control” normalised to 100%, ±SEM) |
|||
|---|---|---|---|---|
| T8 |
T24 |
|||
| Control | (+)BPDS | Control | (+)BPDS | |
| Total Anaerobes | 100 | 27.83±9.47 * | 100 | 29.30±7.46 * |
| Bacteroides | 100 | 68.71±31.61 | 100 | 29.70±18.47 |
| Bifidobacteria | 100 | 5.38±1.05 *** | 100 | 110±106.30 |
| Clostridia | 100 | 63.58±25.84 | 100 | 432.89±299.18 |
| Lactobacilli | 100 | 8.21±5.10 ** | 100 | 35.47±5.92 ** |
| Enterobacteriaceae | 100 | 19.01±11.19 * | 100 | 17.65±15.04 * |
Two vessels were set up containing nutritive media and faecal inocula; one vessel was supplemented with BPDS (70 μM BPDS) whilst the other was used as a control (0 μM BPDS). Samples were taken from each vessel at 0, 8, and 24 h, and then enumerated using selective agar plates. Data shown = mean of 3 individual experiments±S.E.M. The data was statistically analysed using unpaired t-test, assuming unequal variance; *P<.05, **P<.01, ***P<.001 and ****P<.0001 for Control versus viable counts at the relevant time-point.
3.3. 16S rRNA gene sequencing revealed a decrease in the relative abundance of potentially pathogenic bacterial genera in response to iron limitation
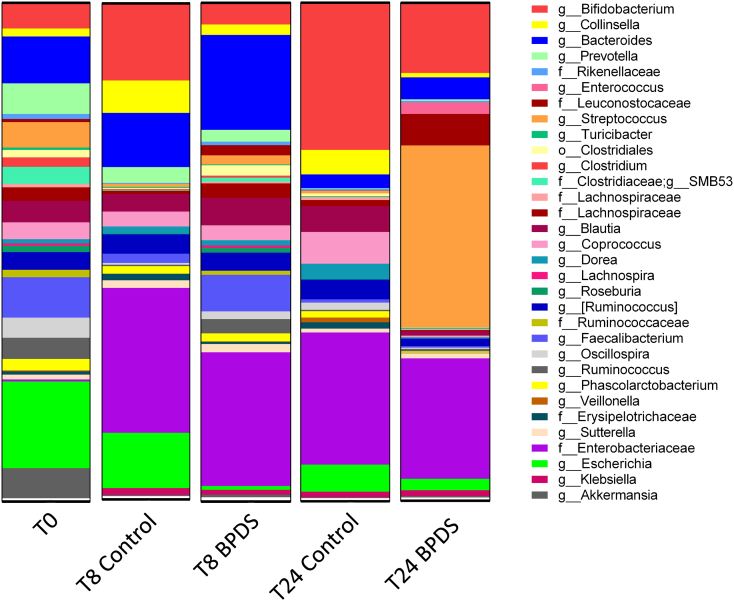
High-throughput paired-end sequencing of the 16S rRNA gene (V4 region) was performed on fermentation samples using the Illumina Miseq platform, and microbial communities present in relative abundances≥0.5% are illustrated in Fig. 2. After combining and averaging the abundance values from the three individual experiments, the data revealed that the most abundant genera at 0 h were Escherichia (16.1%), followed by Bacteroides (8.7%), (Fig. 2, SEM values can be found in Table 2). It is worth noting that the relative abundance of Escherichia is entirely attributed to one donor only, as the remaining two donors had no more than 0.1% Escherichia present at T0.
Fig. 2.
Microbial community profiles assessed by 16S rRNA gene analysis.
Microbial composition of both vessels (Control and BPDS). Proportions shown are of the most abundant genera (≥0.5%, identification to genus level where possible) identified by sequencing of the V4 hypervariable regions of the 16S rRNA gene. Data shown is the average relative abundance from the 3 individual experiments from 3 different donors and analysed using the QIIME pipeline. SEM values can be found in Table 2.
Table 2.
Proportions of different bacterial taxa (SEM) illustrated in 16S rRNA gene analysis
| Taxa | SEM |
||||
|---|---|---|---|---|---|
| T0 | T8 Control | T8 BPDS | T24 Control | T24 BPDS | |
| g__Bifidobacterium | 3.54 | 6.91 | 2.09 | 13.43 | 6.47 |
| g__Collinsella | 0.66 | 3.55 | 1.32 | 3.49 | 0.71 |
| g__Bacteroides | 3.17 | 3.33 | 10.06 | 0.80 | 2.90 |
| g__Prevotella | 5.45 | 3.07 | 2.33 | 0.13 | 0.30 |
| f__Rikenellaceae | 0.41 | 0.06 | 0.32 | 0.19 | 0.12 |
| g__Enterococcus | 0.03 | 0.03 | 0.03 | 0.02 | 1.25 |
| f__Leuconostocaceae | 0.60 | 0.03 | 1.97 | 0.03 | 3.92 |
| g__Streptococcus | 4.45 | 0.35 | 0.19 | 0.30 | 15.70 |
| g__Turicibacter | 0.32 | 0.19 | 0.15 | 0.00 | 0.03 |
| o__Clostridiales | 0.52 | 0.15 | 1.83 | 0.12 | 0.06 |
| g__Clostridium | 1.38 | 0.06 | 0.22 | 0.06 | 0.09 |
| f__Clostridiaceae;g__SMB53 | 1.56 | 0.10 | 0.47 | 0.17 | 0.07 |
| f__Lachnospiraceae | 0.30 | 0.04 | 0.18 | 0.29 | 0.08 |
| f__Lachnospiraceae | 0.54 | 0.44 | 1.96 | 0.55 | 0.17 |
| g__Blautia | 1.25 | 2.10 | 4.19 | 2.77 | 0.55 |
| g__Coprococcus | 1.39 | 2.78 | 2.17 | 3.42 | 0.17 |
| g__Dorea | 0.15 | 0.64 | 0.55 | 0.42 | 0.28 |
| g__Lachnospira | 0.20 | 0.00 | 0.45 | 0.00 | 0.00 |
| g__Roseburia | 0.82 | 0.17 | 0.85 | 0.13 | 0.00 |
| g__[Ruminococcus] | 1.10 | 1.84 | 2.66 | 1.91 | 1.39 |
| f__Ruminococcaceae | 0.78 | 0.00 | 0.70 | 0.03 | 0.00 |
| g__Faecalibacterium | 1.66 | 1.37 | 5.58 | 0.35 | 0.12 |
| g__Oscillospira | 1.69 | 0.19 | 0.67 | 0.44 | 0.22 |
| g__Ruminococcus | 2.32 | 0.25 | 1.75 | 0.15 | 0.45 |
| g__Phascolarctobacterium | 0.88 | 1.15 | 1.31 | 0.44 | 0.21 |
| g__Veillonella | 0.12 | 0.00 | 0.03 | 0.55 | 0.10 |
| f__Erysipelotrichaceae | 0.34 | 1.30 | 0.29 | 1.23 | 0.07 |
| g__Sutterella | 0.47 | 0.93 | 1.14 | 0.40 | 0.73 |
| f__Enterobacteriaceae | 0.37 | 28.13 | 25.50 | 25.70 | 23.90 |
| g__Escherichia | 16.08 | 5.86 | 0.23 | 2.61 | 1.72 |
| g__Klebsiella | 0.03 | 1.36 | 0.90 | 1.06 | 1.17 |
| g__Akkermansia | 7.95 | 0.22 | 0.39 | 0.15 | 0.21 |
Briefly, the relative abundance of Escherichia was reduced substantially in the iron chelated fermentation vessel in comparison to the control vessel at 8 h (0.8% vs. 10.7%) and 24 h (2.3% vs. 5.3%) (Fig. 2). This correlates well with the reduction in the viable counts for Enterobacteriaceae (Table 1). Again, it is worth noting that the high relative abundance of Enterobacteriaceae at T8 and T24 for both control and chelator conditions, is entirely attributed to the levels observed in one donor only, since in the other two donors, the levels were below the detectable limit. A similar trend was observed for Bifidobacterium, where the relative abundance was much lower at T8 and T24 under iron-chelated conditions (4.1% vs. 15% and 14% vs. 29%, respectively), which is also reflected in the viable counts results for Bifidobacteria at 8 h. Interestingly, 16S rDNA analysis indicated that at 24 h, the relative abundance of Streptococcus increased to 36% in the iron chelated condition compared to the starting proportion of 4.7%. Iron-depleted conditions have been shown to proportionally decrease many bacterial groups, and this may provide other bacteria, such as Streptococcus, with a competitive advantage resulting in their increased growth. This could also be true for Bacteroides, as the relative abundances of this genus increased at both 8 h (10.6% to 18.1%) and 24 h (2.7% to 4.4%) under iron chelated conditions. Finally, 16S rDNA analysis indicated that Clostridium abundance was largely unaffected by iron removal (Fig. 2), and this was reflected in the viable counts for Clostridia (Table 1).
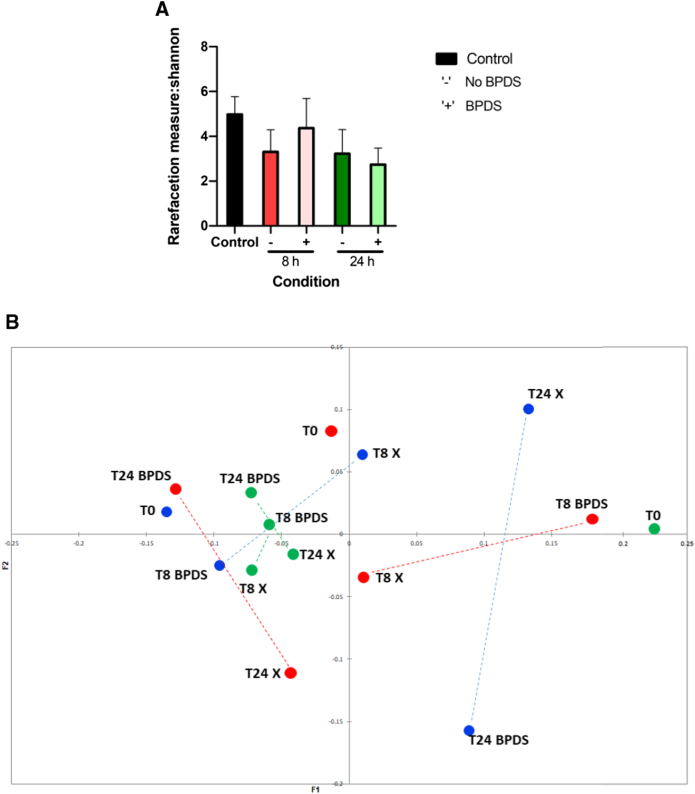
3.4. Iron chelation resulted in a shift in bacterial diversity profiles
Alpha (α) – and beta (β) – diversity was measured for all in vitro colonic fermentation samples tested. The Shannon index, which is a measure of α- diversity, showed a minimal difference in diversity within the population in the absence of the chelator (Fig. 3a). However, an increase in population diversity was observed at 8 h in the presence of BPDS. The principal coordinates analysis (PCoA) plot (Fig. 3b), using the weighted UniFrac metric, which depicts β- diversity, indicates that the microbiotas of the subjects before and after iron chelation did not have a large range of taxa in common. Interestingly, at both 8 h and 24 h, a shift in β-diversity was observed upon iron chelation when compared to the control at the respective time points, depicted by the dashed line.
Fig. 3.
Bacterial diversity profiles of colonic fermentation samples.
(a) α- diversity analysis of batch fermentation samples with (BPDS) and without iron chelator (No BPDS) using the Shannon index and (b) β- diversity analysis of batch fermentation samples portraying weighted analysis of samples with (BPDS) and without (control) iron chelator using the UniFrac metric and presented as a PCoA plot. Data shown is the average Eigenvalues from the 3 individual experiments from 3 different donors. Each colour represents a different donor. Analysis was performed using QIIME (V1.9) and visualised using the XLSTAT add-on package in Microsoft Excel.
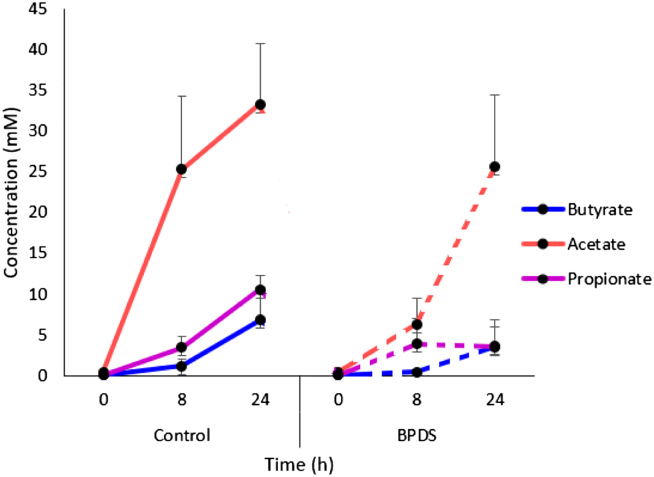
3.5. Levels of short chain fatty acids decrease upon iron chelation during in vitro colonic fermentation
1H-NMR spectroscopy was used to determine the levels of over 70 metabolites from samples taken from the fermenters at 0, 8, and 24 h. The three metabolites which showed the most substantial changes between treatments, were the short chain fatty acids (SCFAs), acetate, propionate and butyrate (Fig. 4).
Fig. 4.
Metabolite concentrations of batch fermentation samples.
Short chain fatty acid concentrations in both vessels were measured using 1H-NMR spectroscopy, with samples being screened for multiple metabolites against a spiked standard (TSP) and a validated reference library. Data shown is the average±S.E.M. metabolite concentrations from the 3 individual experiments from 3 different donors.
Acetate concentrations was~25.3 mM under control conditions (i.e. 0 μM BPDS) at 8 h with lower levels (~6.3 mM) found when iron was chelated (with 70 μM BPDS) at the same time-point. The same trend was observed at 24 h where acetate concentrations were~33.3 mM in the control vessel whereas under iron-limitation, its levels were reduced to~25.6 mM. Levels of propionate and butyrate exhibited the same pattern under control and iron-chelated conditions. In the control condition, ~1.1 mM (8 h) and~6.8 mM (24 h) of butyrate was measured whereas under iron-chelation, butyrate levels were reduced (~0.4 mM at 8 h and~3.6 mM 24 h) representing 47% reduction at 24 h. This is reflected in the reduction observed for the relative abundances of the members of the Ruminococcus (3.9% to 1.5%) genera, which are common butyrate producers.
Although similar propionate levels were observed between the different conditions at 8 h, propionate concentrations were lower in the iron-chelated vessel compared to the control at 24 h (~10.5 mM vs. ~3.5 mM). This represents a 67% decrease in production at 24 h under iron-limiting conditions. This is in line with the 70% decrease observed in the viable counts of Bacteroides (a genus containing propionate producers) under low iron conditions (Table 1).
4. Discussion
Iron supplements are consumed widely, but the majority of the iron passes through to the colon where it can be utilised by iron-requiring bacteria. This study was designed to assess the effects of colonic iron chelation on the human gut microbiota. We found that the addition of BPDS, a chemical iron chelator, significantly reduced the growth of E. coli, S. Typhimurium, B. thetaiotaomicron and B. longum (Fig. 1). These results confirm well-established knowledge that iron promotes the growth of potentially pathogenic bacteria. However, Cronin et al., [44] reported that the growth of two B. longum strains were largely unaffected in the presence of an iron chelator, even at the highest chelator concentration (5 μM), suggesting that the effect of iron chelation on Bifidobacteria could be largely strain-dependent. In the same study [44], no effect on the growth of L. rhamnosus was observed and it is known that Lactobacillus does not require iron for growth due to an absence of haem-containing enzymes. However, they can substitute other metals for iron, such as cobalt and manganese [45], which may give these bacteria a competitive advantage in low-iron conditions.
The net effects of iron on the gut microbial composition are still inconclusive and need further research as the results reported are inconsistent [6], [39], [44], [45]. Our study showed a marked effect on the global microbiome composition when iron was chelated in the fermentation media with BPDS (Fig. 2). BPDS acts as a chelator of various metals and, in particular, it has been shown to bind iron with very high affinity and has therefore been used as an iron chelator in many studies [46], [47]. Comparison of the relative abundance of bacterial taxa between the two conditions (control and BPDS) illustrated that the most apparent differences were the decreased relative abundance of the potentially pathogenic Escherichia. The observed decrease of Bifidobacterium is interesting as other studies have reported different outcomes, which could be due to strain specificity, inter-individual variability of the host, as well as the effects of neighbouring taxa and metabolites. However, the relative abundance of Clostridium remained relatively stable, which has been observed in other studies. Notably, Streptococcus, a member of the lactic acid bacteria group to which Lactobacillus also belongs, was seen to increase upon iron removal, and this could potentially lead to restricted growth of other bacteria in the gut environment.
Studies investigating the effects of iron supplementation report comparable results to those found in this study in relation to the relative abundance of various bacterial taxa, including Escherichia. One study examined the effects of low and high doses of in-home iron supplementation on the gut microbiome of Kenyan children [39]. In this setting, provision of iron-fortified porridge led to an increase in pathogen abundance, with proportions of Enterobacteria, Clostridium and E. coli increasing, whilst Bifidobacteria decreased. In addition, children given iron-fortified porridge had elevated levels of faecal calprotectin, a marker of gut inflammation, which is likely to reflect an increased pathogenic profile.
Much of the published literature focuses on the effects of iron on the infant microbiome which may not be the same as adults. In addition, geographical location is probably important, as studies have shown that individuals from developing countries tend to have a greater pathogenic microbial profile due to the lack of clean food and water, and, in turn, a compromised gut function. This is reflected in two studies, where one investigated the effects of iron addition on the gut microbiota of children from Cote d'Ivoire [48], the second study focused on children from South Africa [6]. No detrimental effects of iron supplementation were observed in those that had access to clean water and food, yet the children that did not have access to clean water, had adverse side effects.
We also performed bacterial diversity analysis. α- diversity analysis using the Shannon index, which accounts for the distribution and richness of OTUs within a population, showed an increase in population diversity at 8 h in the presence of the iron chelator (Fig. 3a). We speculate that the chelation of iron may lead to certain taxa exploiting other metals as a means of replacing iron, and therefore temporarily facilitating growth in a micronutrient-restricted environment. However, by 24 h, a visible decrease in diversity was observed under iron-chelated conditions, suggesting an exhaustion of these metals and nutrients. β-diversity analysis demonstrated that there was not a large range of taxa in common when comparing the control samples to those that were cultured in iron-limiting conditions (Fig. 3b). Interestingly, at both 8 h and 24 h, there was a shift in β-diversity between the two groups. This is in line with a study performed by Dostal et al., [49], which investigated the effect of iron on butyrate production in the child gut microbiota, where altering the iron concentration in the medium affected microbial community structure as well as causing a shift in β-diversity.
Microbial metabolic activity contributes to human health. When iron was chelated, we observed a decrease in the three main short chain fatty acids (SCFAs) that are produced in the gut (Fig. 4), which presumably reflects poor growth of fermentative microbes. Based on the 16S rRNA gene sequencing data from this study and those published elsewhere, we can infer the potential mechanisms behind certain metabolic changes. Firstly, iron-dependent enzymes are critical operators of many metabolic pathways, and therefore these processes can be affected by differing iron concentrations. Moreover, any microbial fermentation that takes place requires the redox balance to be sustained. Due to the dual role of iron as an electron donor and acceptor, we speculate that changes in iron levels could have a large effect on redox balance. During the batch fermentation, the most prominent effect observed on metabolite production was that of acetate levels. Many gut bacteria produce acetate by either the reductive acetyl-CoA pathway, which uses H2 and CO2 [50], or via the regular glycolytic pathway through pyruvate metabolism [51]. The former pathway consists of numerous iron-dependent enzymes and can account for>25% of acetate produced in the gut [52]. It is therefore plausible to speculate a lack of conversion of H2 and CO2 to acetate under iron-limiting conditions, resulting in an overall decrease in acetate levels. This observation also correlated with the viable counts and 16S rRNA gene analysis, where a decrease in the members of Bifidobacterium, a prominent bacterial group which produces acetate, was observed under iron-limiting conditions. Although correlations of metabolite levels with the relative abundance of bacterial taxa do not provide a causal relationship, it may still provide some indications as to which taxa are responsible for any observed differences.
In conclusion, this in vitro study shows that the chelation of iron, achieved with BPDS, resulted in a decrease of the main intestinal SCFAs and a lower relative abundance of potentially pathogenic bacteria within the gut microbial community. This observation was also confirmed in pure culture studies with pathogenic bacteria. Taken together, these data suggest that iron could provide an advantageous niche for potentially pathogenic bacteria and highlights the importance of tight control of iron availability to avoid a pathogenic profile within the colon. The relationship between iron availability and the human gut microbiome is yet to be fully elucidated, and more extensive research on this topic will aid in expanding our knowledge and understanding of this relationship, and in extension, help define improved methods for maintaining gut microbial homeostasis.
Acknowledgments
Acknowledgements
We thank the participants who provided the faecal samples for the experiments performed. We also thank Dr. Melinda Mayer and Dr. Gary Barker for their contributions towards the experimental plan of this study.
Funding
This work was funded by the UK Biotechnology and Biological Sciences Research Council iCASE studentship (BB/M015122/1) with Intract Pharma and BBSRC Institute Strategic Programme Gut Microbes and Health BB/R012490/1.
Declarations
All authors read and approved the final manuscript. The authors declare that there are no conflicts of interest.
Footnotes
Declaration of interest: None.
Funding: This work was funded by the UK Biotechnology and Biological Sciences Research Council iCASE studentship (BB/M015122/1) with Intract Pharma and BBSRC Institute Strategic Programme Gut Microbes and Health BB/R012490/1.
References
- 1.Dethlefsen L., McFall-Ngai M., Relman D.A. An ecological and evolutionary perspective on human–microbe mutualism and disease. Nature. 2007;449(7164):811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hooper L.V., Gordon J.I. Commensal Host-Bacterial Relationships in the Gut. Science. 2001;292(5519):1115–1118. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 3.Round J.L., Mazmanian S.K. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9(5):313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sekirov I., Russell S.L., Antunes L.C., Finlay B.B. Gut microbiota in health and disease. Physiol Rev. 2010;90(3):859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 5.Flint H.J., Scott K.P., Louis P., Duncan S.H. The role of the gut microbiota in nutrition and health. Nat Rev Gastroenterol Hepatol. 2012;9(10):577–589. doi: 10.1038/nrgastro.2012.156. [DOI] [PubMed] [Google Scholar]
- 6.Dostal A., Baumgartner J., Riesen N., Chassard C., Smuts C.M., Zimmermann M.B. Effects of iron supplementation on dominant bacterial groups in the gut, faecal SCFA and gut inflammation: a randomised, placebo-controlled intervention trial in South African children. Br J Nutr. 2014;112(4):547–556. doi: 10.1017/S0007114514001160. [DOI] [PubMed] [Google Scholar]
- 7.Kau A.L., Ahern P.P., Griffin N.W., Goodman A.L., Gordon J.I. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474(7351):327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tamboli C.P., Neut C., Desreumaux P., Colombel J.F. Dysbiosis in inflammatory bowel disease. Gut. 2004;53(1):1–4. doi: 10.1136/gut.53.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collado M.C., Rautava S., Isolauri E., Salminen S. Gut microbiota: a source of novel tools to reduce the risk of human disease? Pediatr Res. 2015;77(1-2):182–188. doi: 10.1038/pr.2014.173. [DOI] [PubMed] [Google Scholar]
- 10.McLean M.H., Dieguez D., Jr., Miller L.M., Young H.A. Does the microbiota play a role in the pathogenesis of autoimmune diseases? Gut. 2015;64(2):332–341. doi: 10.1136/gutjnl-2014-308514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Viaud S., Daillere R., Boneca I.G., Lepage P., Pittet M.J., Ghiringhelli F. Harnessing the intestinal microbiome for optimal therapeutic immunomodulation. Cancer Res. 2014;74(16):4217–4221. doi: 10.1158/0008-5472.CAN-14-0987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kostic A.D., Xavier R.J., Gevers D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology. 2014;146(6):1489–1499. doi: 10.1053/j.gastro.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cammarota G., Ianiro G., Cianci R., Bibbò S., Gasbarrini A., Currò D. The involvement of gut microbiota in inflammatory bowel disease pathogenesis: Potential for therapy. Pharmacol Ther. 2015;149:191–212. doi: 10.1016/j.pharmthera.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 14.Han S.W., McColl E., Steen N., Barton J.R., Welfare M.R. The inflammatory bowel disease questionnaire: a valid and reliable measure in ulcerative colitis patients in the North East of England. Scand J Gastroenterol. 1998;33 doi: 10.1080/003655298750026994. [DOI] [PubMed] [Google Scholar]
- 15.DuPont H.L. Review article: evidence for the role of gut microbiota in irritable bowel syndrome and its potential influence on therapeutic targets. Aliment Pharmacol Ther. 2014;39(10):1033–1042. doi: 10.1111/apt.12728. [DOI] [PubMed] [Google Scholar]
- 16.Moran C.P., Shanahan F. Gut microbiota and obesity: role in aetiology and potential therapeutic target. Best Pract Res Clin Gastroenterol. 2014;28(4):585–597. doi: 10.1016/j.bpg.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 17.Power S.E., O'Toole P.W., Stanton C., Ross R.P., Fitzgerald G.F. Intestinal microbiota, diet and health. Br J Nutr. 2014;111(3):387–402. doi: 10.1017/S0007114513002560. [DOI] [PubMed] [Google Scholar]
- 18.Miret S., Simpson R.J., McKie A.T. Physiology and molecular biology of dietary iron absorption. Annu Rev Nutr. 2003;23:283–301. doi: 10.1146/annurev.nutr.23.011702.073139. [DOI] [PubMed] [Google Scholar]
- 19.Wallace D.F. The Regulation of Iron Absorption and Homeostasis. Clin Biochem Rev. 2016;37(2):51–62. [PMC free article] [PubMed] [Google Scholar]
- 20.Hurrell R., Egli I. Iron bioavailability and dietary reference values. Am J Clin Nutr. 2010;91(5):1461S–1467S. doi: 10.3945/ajcn.2010.28674F. [DOI] [PubMed] [Google Scholar]
- 21.Kortman G.A.M., Raffatellu M., Swinkels D.W., Tjalsma H. Nutritional iron turned inside out: intestinal stress from a gut microbial perspective. FEMS Microbiol Rev. 2014;38(6):1202–1234. doi: 10.1111/1574-6976.12086. [DOI] [PubMed] [Google Scholar]
- 22.O'Callaghan A., van Sinderen D. Bifidobacteria and Their Role as Members of the Human Gut Microbiota. Front Microbiol. 2016;7:925. doi: 10.3389/fmicb.2016.00925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vlasova A.N., Kandasamy S., Chattha K.S., Rajashekara G., Saif L.J. Comparison of probiotic lactobacilli and bifidobacteria effects, immune responses and rotavirus vaccines and infection in different host species. Vet Immunol Immunopathol. 2016;172:72–84. doi: 10.1016/j.vetimm.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.LeBlanc J.G., Chain F., Martín R., Bermúdez-Humarán L.G., Courau S., Langella P. Beneficial effects on host energy metabolism of short-chain fatty acids and vitamins produced by commensal and probiotic bacteria. Microb Cell Fact. 2017;16:79. doi: 10.1186/s12934-017-0691-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martín R., Miquel S., Ulmer J., Kechaou N., Langella P., Bermúdez-Humarán L.G. Role of commensal and probiotic bacteria in human health: a focus on inflammatory bowel disease. Microb Cell Fact. 2013;12:71. doi: 10.1186/1475-2859-12-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larsen P.E., Dai Y. Metabolome of human gut microbiome is predictive of host dysbiosis. GigaScience. 2015;4(1) doi: 10.1186/s13742-015-0084-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imbert M., Blondeau R. On the iron requirement of lactobacilli grown in chemically defined medium. Curr Microbiol. 1998;37(1):64–66. doi: 10.1007/s002849900339. [DOI] [PubMed] [Google Scholar]
- 28.Jakaitis B.M., Denning P.W. Human Breast Milk and the Gastrointestinal Innate Immune System. Clin Perinatol. 2014;41(2):423–435. doi: 10.1016/j.clp.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turin C.G., Zea-Vera A., Pezo A., Cruz K., Zegarra J., Bellomo S. Lactoferrin for prevention of neonatal sepsis. Biometals. 2014;27(5):1007–1016. doi: 10.1007/s10534-014-9754-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ochoa T.J., Cleary T.G. Effect of lactoferrin on enteric pathogens. Biochimie. 2009;91(1):30–34. doi: 10.1016/j.biochi.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mevissen-Verhage E.A., Marcelis J.H., Harmsen-Van Amerongen W.C., de Vos N.M., Verhoef J. Effect of iron on neonatal gut flora during the first three months of life. Eur J Clin Microbiol. 1985;4(3):273–278. doi: 10.1007/BF02013651. [DOI] [PubMed] [Google Scholar]
- 32.Paganini D., Zimmermann M.B. Effects of iron fortification and supplementation on the gut microbiome and diarrhea in infants and children: a review. Am J Clin Nutr. 2017;106(6):1688–1693. doi: 10.3945/ajcn.117.156067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dostal A., Lacroix C., Pham V.T., Zimmermann M.B., Del'homme C., Bernalier-Donadille A. Iron supplementation promotes gut microbiota metabolic activity but not colitis markers in human gut microbiota-associated rats. Br J Nutr. 2014;111(12):2135–2145. doi: 10.1017/S000711451400021X. [DOI] [PubMed] [Google Scholar]
- 34.Dostal A., Chassard C., Hilty F.M., Zimmermann M.B., Jaeggi T., Rossi S. Iron depletion and repletion with ferrous sulfate or electrolytic iron modifies the composition and metabolic activity of the gut microbiota in rats. J Nutr. 2012;142(2):271–277. doi: 10.3945/jn.111.148643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee T., Clavel T., Smirnov K., Schmidt A., Lagkouvardos I., Walker A. Oral versus intravenous iron replacement therapy distinctly alters the gut microbiota and metabolome in patients with IBD. Gut. 2016;66(5):863–887. doi: 10.1136/gutjnl-2015-309940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kortman G.A.M., Reijnders D., Swinkels D.W. Oral iron supplementation: Potential implications for the gut microbiome and metabolome in patients with CKD. Hemodial Int. 2017;21:S28–S36. doi: 10.1111/hdi.12553. [DOI] [PubMed] [Google Scholar]
- 37.Alexeev E.E., He X., Slupsky C.M., Lönnerdal B. Effects of iron supplementation on growth, gut microbiota, metabolomics and cognitive development of rat pups. PLoS One. 2017;12(6) doi: 10.1371/journal.pone.0179713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dostal A., Fehlbaum S., Chassard C., Zimmermann M.B., Lacroix C. Low iron availability in continuous in vitro colonic fermentations induces strong dysbiosis of the child gut microbial consortium and a decrease in main metabolites. FEMS Microbiol Ecol. 2013;83(1):161–175. doi: 10.1111/j.1574-6941.2012.01461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jaeggi T., Kortman G.A., Moretti D., Chassard C., Holding P., Dostal A. Iron fortification adversely affects the gut microbiome, increases pathogen abundance and induces intestinal inflammation in Kenyan infants. Gut. 2015;64(5):731–742. doi: 10.1136/gutjnl-2014-307720. [DOI] [PubMed] [Google Scholar]
- 40.Kortman G.A.M., Dutilh B.E., Maathuis A.J.H., Engelke U.F., Boekhorst J., Keegan K.P. Microbial metabolism shifts towards an adverse profile with supplementary iron in the TIM-2 in vitro model of the human colon. Front Microbiol. 2016;6 doi: 10.3389/fmicb.2015.01481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dekker Nitert M., Gomez-Arango L.F., Barrett H.L., McIntyre H.D., Anderson G.J., Frazer D.M. Iron supplementation has minor effects on gut microbiota composition in overweight and obese women in early pregnancy. Br J Nutr. 2018:1–7. doi: 10.1017/S0007114518001149. [DOI] [PubMed] [Google Scholar]
- 42.Mandalari G., Nueno Palop C., Tuohy K., Gibson G.R., Bennett R.N., Waldron K.W. In vitro evaluation of the prebiotic activity of a pectic oligosaccharide-rich extract enzymatically derived from bergamot peel. Appl Microbiol Biotechnol. 2007;73(5):1173–1179. doi: 10.1007/s00253-006-0561-9. [DOI] [PubMed] [Google Scholar]
- 43.Kellingray L., Tapp H.S., Saha S., Doleman J.F., Narbad A., Mithen R.F. Consumption of a diet rich in Brassica vegetables is associated with a reduced abundance of sulphate-reducing bacteria: A randomised crossover study. Mol Nutr Food Res. 2017;61(9) doi: 10.1002/mnfr.201600992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cronin M., Zomer A., Fitzgerald G., van Sinderen D. Identification of iron-regulated genes of Bifidobacterium breve UCC2003 as a basis for controlled gene expression. Bioengineered Bugs. 2012;3(3):157–167. doi: 10.4161/bbug.18985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pandey A., Bringel F., Meyer J.-M. Iron requirement and search for siderophores in lactic acid bacteria. Appl Microbiol Biotechnol. 1994;40(5):735–739. [Google Scholar]
- 46.Koh E.-I., Robinson A.E., Bandara N., Rogers B.E., Henderson J.P. Copper import in Escherichia coli by the yersiniabactin metallophore system. Nat Chem Biol. 2017;13:1016. doi: 10.1038/nchembio.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Askwith C., Kaplan J. An oxidase-permease-based iron transport system in Schizosaccharomyces pombe and its expression in Saccharomyces cerevisiae. J Biol Chem. 1997;272(1):401–405. doi: 10.1074/jbc.272.1.401. [DOI] [PubMed] [Google Scholar]
- 48.Zimmermann M.B., Chassard C., Rohner F., N’Goran E.K., Nindjin C., Dostal A. The effects of iron fortification on the gut microbiota in African children: a randomized controlled trial in Cote d’Ivoire. Am J Clin Nutr. 2010;92 doi: 10.3945/ajcn.110.004564. [DOI] [PubMed] [Google Scholar]
- 49.Dostal A., Lacroix C., Bircher L., Pham V.T., Follador R., Zimmermann M.B. Iron Modulates Butyrate Production by a Child Gut Microbiota In Vitro. MBio. 2015;6(6) doi: 10.1128/mBio.01453-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leclerc M., Bernalier A., Donadille G., Lelait M. H2/CO2 metabolism in acetogenic bacteria isolated from the human colon. Anaerobe. 1997;3(5):307–315. doi: 10.1006/anae.1997.0117. [DOI] [PubMed] [Google Scholar]
- 51.Macfarlane G.T., Macfarlane S. Fermentation in the human large intestine: its physiologic consequences and the potential contribution of prebiotics. J Clin Gastroenterol. 2011;45(Suppl):S120–S127. doi: 10.1097/MCG.0b013e31822fecfe. [DOI] [PubMed] [Google Scholar]
- 52.Rey F.E., Faith J.J., Bain J., Muehlbauer M.J., Stevens R.D., Newgard C.B. Dissecting the in vivo metabolic potential of two human gut acetogens. J Biol Chem. 2010;285(29):22082–22090. doi: 10.1074/jbc.M110.117713. [DOI] [PMC free article] [PubMed] [Google Scholar]