Zhang et al. identify human IL-2Rβ deficiency as a cause of severe immune dysregulation. The hypomorphic gene mutations reveal variable IL-2Rβ expression and function between different lymphocyte subsets as a means of selectively modulating immune responses.
Abstract
Interleukin-2, which conveys essential signals for immunity, operates through a heterotrimeric receptor. Here we identify human interleukin-2 receptor (IL-2R) β chain (IL2RB) gene defects as a cause of life-threatening immune dysregulation. We report three homozygous mutations in the IL2RB gene of eight individuals from four consanguineous families that cause disease by distinct mechanisms. Nearly all patients presented with autoantibodies, hypergammaglobulinemia, bowel inflammation, dermatological abnormalities, lymphadenopathy, and cytomegalovirus disease. Patient T lymphocytes lacked surface expression of IL-2Rβ and were unable to respond to IL-2 stimulation. By contrast, natural killer cells retained partial IL-2Rβ expression and function. IL-2Rβ loss of function was recapitulated in a recombinant system in which IL2RB mutations caused reduced surface expression and IL-2 binding. Stem cell transplant ameliorated clinical symptoms in one patient; forced expression of wild-type IL-2Rβ also increased the IL-2 responsiveness of patient T lymphocytes in vitro. Insights from these patients can inform the development of IL-2–based therapeutics for immunological diseases and cancer.
Introduction
The IL-2 receptor (IL-2R) complex plays a central role in control of the immune response by integrating signals from the key cytokines IL-2 and IL-15. Three distinct receptors for IL-2 are generated by combinations of three IL-2R subunits: IL-2Rα (CD25), IL-2Rβ (CD122), and IL-2Rγ (CD132). The last, known as the common γ chain, is also necessary for signaling by IL-4, IL-7, IL-9, IL-15, and IL-21. All three chains combine to form the high-affinity IL-2R, which is constitutively expressed on CD4+ regulatory T (T reg) cells, and induced upon activation of CD4 and CD8 T cells, B cells, and some myeloid-derived subsets (Busse et al., 2010; Liao et al., 2013). A second receptor, which binds IL-15 and IL-2 with intermediate affinity, is comprised of only the IL-2Rβ and IL-2Rγ subunits; it is constitutively expressed on resting CD8+ T cells and natural killer (NK) cells. The α subunit alone is a low-affinity receptor. Upon ligand binding, the IL-2Rβ and IL-2Rγ subunits undergo tyrosine phosphorylation, which, in turn, induces the phosphorylation of the associated Janus tyrosine kinases (JAK) 1 and 3, which phosphorylate the signal transducer and activator of transcription 5 (STAT5) transcription factor (Waldmann, 2006). STAT5, once dimerized and translocated to the nucleus, induces a pro-survival and proliferative transcription program. IL-2 is primarily produced by CD4+ T helper (Th) cells following TCR engagement with costimulation (Boyman and Sprent, 2012). It potently stimulates T cell proliferation, differentiation (promoting Th1, Th2, and Th9 cells, and suppressing Th17 cell polarization), and cytolytic effector activity. It also plays a key role in peripheral tolerance by promoting the generation and maintenance of T reg and antigen-specific peripheral T cell clonal deletion (Hatakeyama et al., 1989; Lenardo, 1991; Takeshita et al., 1992). CD25-deficient mice demonstrate grossly normal early B and T cell development, but lymphadenopathy and impaired T cell activation and clonal deletion. As they age, these mice develop autoimmune and inflammatory disease (e.g., hemolytic anemia and inflammatory enteropathy; Willerford et al., 1995). Humans with CD25 deficiency have a similar phenotype, developing prominent autoimmune disease with less consistent evidence of immunodeficiency, and resembling patients with immunodysregulation polyendocrinopathy enteropathy X-linked (IPEX), due to forkhead box P3 (FOXP3) deficiency, thus indicating the impact of loss of the high-affinity IL-2R can be ascribed to a loss of peripheral tolerance (Sharfe et al., 1997; Caudy et al., 2007).
The role in immunity of IL-2Rβ is less well understood, and no monogenic cause of human IL-2Rβ deficiency has yet been described. Although a case of NK-SCID was previously reported in which the patient did not express IL-2Rβ, no mutation within IL2RB was identified (Gilmour et al., 2001). IL-2Rβ–deficient mice had severe autoimmunity and diminished cytolytic effector function, with splenomegaly, lymphadenopathy, elevated IgG1 and IgE levels, antinuclear antibodies, and anti–double stranded DNA autoantibodies (Suzuki et al., 1995). They succumbed to autoimmunity around 12 wk unless rescued by the adoptive transfer of T reg cells (Malek et al., 2002). Despite showing evidence of activation, e.g., increased CD69, the T cells of IL-2Rβ–deficient mice failed to respond to stimuli including IL-2, PMA, and ionomycin, and had diminished CD8+ T cell cytolytic activity when rechallenged (Suzuki et al., 1995). This, plus the observation that they have reduced NK cell numbers (Suzuki et al., 1997), implies that IL-2Rβ deficiency in mice could produce susceptibility to infection in addition to T cell activation and autoimmunity. IL-2Rβ–mediated signaling is implicated in pathways known to be important in human autoimmune disease, and loci containing it have been associated with asthma and juvenile-onset arthritis in genome-wide association studies (Moffatt et al., 2010; Hinks et al., 2013). Moreover, high-dose IL-2 therapy is approved for use in renal cell carcinoma and malignant melanoma, and encouraging early-phase studies of low-dose IL-2 in type 1 diabetes, graft-versus-host disease, and systemic lupus erythematosus have led to >14 ongoing phase 2 and 3 trials (Ahmadzadeh and Rosenberg, 2006; Ye et al., 2018). It will thus be important to understand the biology of IL-2Rβ, and the impact of IL-2Rβ deficiency, in humans. To this end, we describe human homozygous recessive IL-2Rβ deficiency in four consanguineous families with eight affected individuals, which was associated with autoimmunity and immunodeficiency.
Results
Clinical phenotype and genotype of patients in a combined immunodeficiency/autoimmunity cohort
We investigated the medical records and clinical data of eight affected individuals from four consanguineous families with South Asian, Middle Eastern, and Eastern European origins, residing in countries on three different continents. All the patients have a history of severe immunodeficiency and autoimmunity (Fig. 1 and Table S1). Kindred A includes a 6-yr-old boy (A1) and his 3-yr-old sister (A2) born to first cousin Pakistani parents (Fig. 1 A). A1 was initially hospitalized at the age of 2 yr for thyrotoxicosis secondary to Graves’ disease, and A2 was hospitalized at the age of 6 mo for failure to thrive and persistent diarrhea (Table S1). Since their initial hospital course, A1 has developed severe gastroenteritis and dermatitis, and A2 has had pulmonary, gastrointestinal, and urinary infections as well as antineutrophil cytoplasmic antibody–positive vasculitis. Patient A1 has improved with rituximab treatment but continues to be intermittently ill. Patient A2 received an allogeneic hematopoietic stem cell transplant (HSCT) and has recovered with no sequelae. Kindred B consists of a girl (B1) born to related parents of South Asian origin. B1 initially presented in a collapsed state with severe diarrhea at the age of 4 wk and was found to have enteropathy, dermatitis, and later CMV viremia with hepatitis. She improved with immunosuppression and antiviral therapy but ultimately succumbed to probable CMV pneumonitis and respiratory failure after HSCT. Kindred C includes a boy (C1) and his first female cousin (C2) born to consanguineous Saudi Arabian parents. C1 presented with suppurative ear infections at the age of 6 mo. C2 presented with chest and ear infections and diarrhea at the age of 2 mo. Subsequently, both suffered recurrent otitis, severe dermatitis, CMV viremia, and food allergies. C1 and C2 died from probable CMV pneumonitis at the ages of 3 yr and 18 mo, respectively.
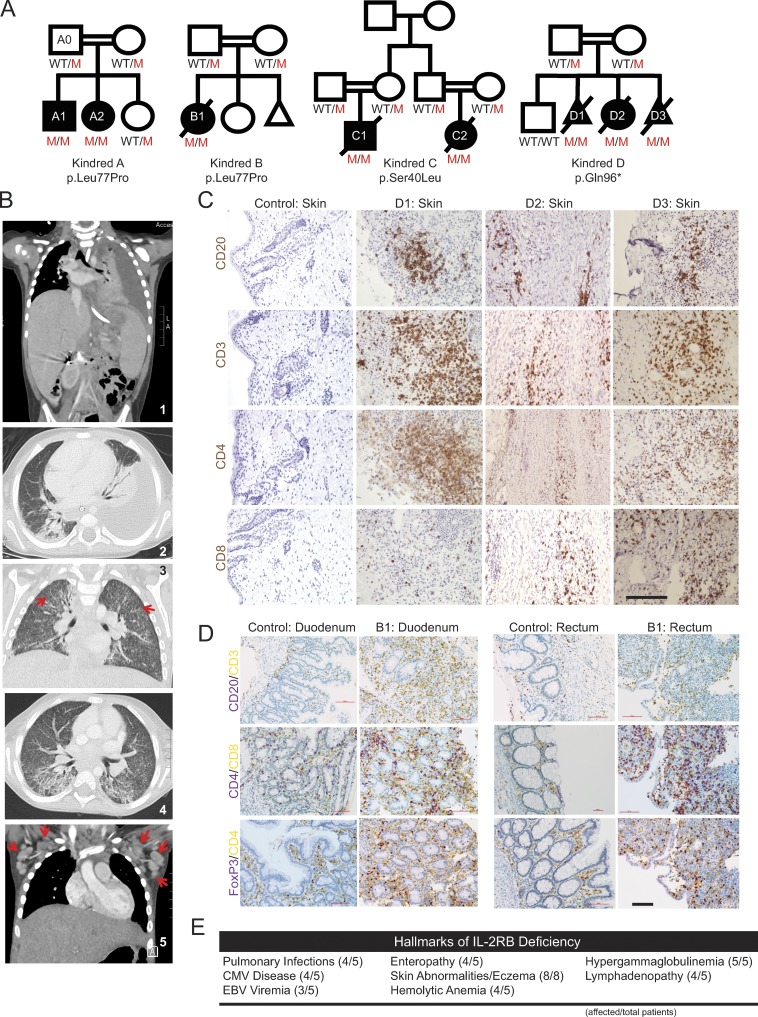
Figure 1.
Genetic and clinical features of the disease cohort. (A) Four consanguineous pedigrees of eight affected individuals (A1–D3) with three different homozygous recessive IL2RB mutations. (B) Radiographical evidence for pulmonary disease in kindred A. Panels 1 and 2 show a left pleural effusion. Hepatosplenomegaly can also be seen in panel 1. Panels 3 and 4 show numerous small pulmonary nodules and tree-in-bud changes suggestive of CMV pneumonia. Red arrows highlight two small lung nodules. Panel 5 shows enlarged axillary lymph nodes (red arrows). (C) Immunohistochemistry of fetal skin from kindred D; patients D1, D2, and D3 stained in brown for the lymphocyte markers as indicated. Bar, 50 µm. (D) Immunohistochemistry of duodenal (left) and rectal (right) biopsies of patient B1 and healthy control, stained with indicated markers. Purple: CD20, CD4, and FoxP3; yellow: CD3, CD8, and CD4 for the respective panels. Bar, 100 µm. (E) Summary of clinical hallmarks of IL-2Rβ deficiency in the five pediatric patients. Skin abnormalities were observed in the individuals in kindred D in addition to the pediatric patients (eight total).
Kindred D consists of two fetuses (D1 and D3) and a premature female neonate (D2) conceived by a Romany family living in France. D1, D2, and D3 were found to have intra-uterine growth retardation and reduced fetal movement; skin-like floating membranes were also present in the amniotic fluid in all three cases. While it was not possible to evaluate the fetuses for immunodeficiency, autoimmune skin desquamation in utero is consistent with echogenic debris findings in prenatal IPEX patients (Louie et al., 2017). D2 was delivered prematurely by emergency cesarean section at 31-wk gestation, but she died 2 h later of respiratory failure. D1 and D3 pregnancies were terminated due to fetal abnormalities at 25 wk and 30 wk, respectively. In summary, all the patients, who had survived the neonatal period, had recurrent infections, as well as autoimmune disease, leading to early death in most cases.
Immune dysregulation was a key shared feature across these four kindreds, manifest as enteropathy, dermatitis, autoimmune hemolytic anemia, and hypergammaglobulinemia (Fig. 1). All the children with IL-2Rβ deficiency also had recurrent infections, including defective handling of herpesviruses (CMV or EBV viremia in all; CMV disease in four of five; Table S1). Chest radiographs of patient A2 revealed a pleural effusion and numerous small pulmonary nodules and tree-in-bud opacities suggestive of CMV pneumonia in the context of CMV viremia (Fig. 1 B). Computed tomography imaging also revealed hepatosplenomegaly and marked lymphadenopathy in A2 (Fig. 1 B), a clinical feature that was noted in all five patients.
Skin abnormalities are a key hallmark of this disease. A1, A2, B1, C1, and C2 all had severe dermatitis, and D1, D2, and D3 had patches of hyperkeratosis and significant infiltration of B and T lymphocytes on skin immunohistochemistry (Fig. 1 C). Four out of the five children have also had severe diarrhea and infectious/autoimmune enteropathy. Endoscopy of patient B1 showed villous atrophy, and gastrointestinal biopsies revealed chronic inflammatory infiltration of the duodenum and rectum (Fig. 1 D). Additional hallmarks of disease include autoimmune hemolytic anemia (four of five patients) and hypergammaglobulinemia (five of five patients) comprising predominantly class-switched isotypes: IgA, IgG, and IgE (Fig. 1 E and Table S2). Overall, CD4 cell numbers were normal, but two of the patients had low CD8 T cell counts, while NK numbers were increased (Table S2). T cell proliferative responses measured in B1 were also globally impaired (Fig. S1). Detailed clinical histories can be found in the Materials and methods.
Identification of protein-coding mutations in the gene encoding IL-2Rβ (CD122)
Because of the early onset of disease in consanguineous families, we sought a genetic cause by whole exome DNA sequence analysis of the four kindreds. We identified three different IL2RB gene mutations (Fig. 2 A). For kindreds A and B, the IL2RB chr22: g.37538526 A>G (p.Leu77Pro) missense variant was prioritized. The mutation occurs in exon 4 (out of 10) and is not found in the Single Nucleotide Polymorphism, NHLBI Grand Opportunity Exome Sequencing Project, or Exome Aggregation Consortium databases, but has a minor allele frequency of 0.00001218 in the Genome Aggregation Database. The p.Leu77Pro mutation introduces a restrictive proline–proline motif in the extracellular D1 domain of IL-2Rβ (Fig. 2 B). For kindred C, the g.37539634 C>T (p.Ser40Leu) missense variant was prioritized and not found in any databases of genomic variation. The mutation appears to be located at the interface of IL-2Rβ and IL-2 (Fig. 2 B). For kindred D, a g.37537259 G>A (p.Gln96*) stop-gain mutation was identified and also not found in any databases. This mutation would lead to significant truncation of the 552–amino acid protein. Due to the predicted deleterious nature of these variants, their segregation with disease, and the similarity in phenotype with a mouse KO model, IL2RB represented an attractive candidate disease gene. Other prioritization criteria that were taken into consideration include Combined Annotation Dependent Depletion score, quality of reads, Genomic Evolutionary Rate Profiling conservation score, cosegregation of alleles, sorting intolerant from tolerant score, polymorphism phenotyping-2 score, tissue specific expression levels, structural modeling, and primary literature reviews leading to the conclusion that these variants were likely responsible for the disease. Other rare variants that were candidate disease alleles in individual families are listed in Table S3; the deleterious IL2RB alleles were the only variants that correlated with disease in all families.
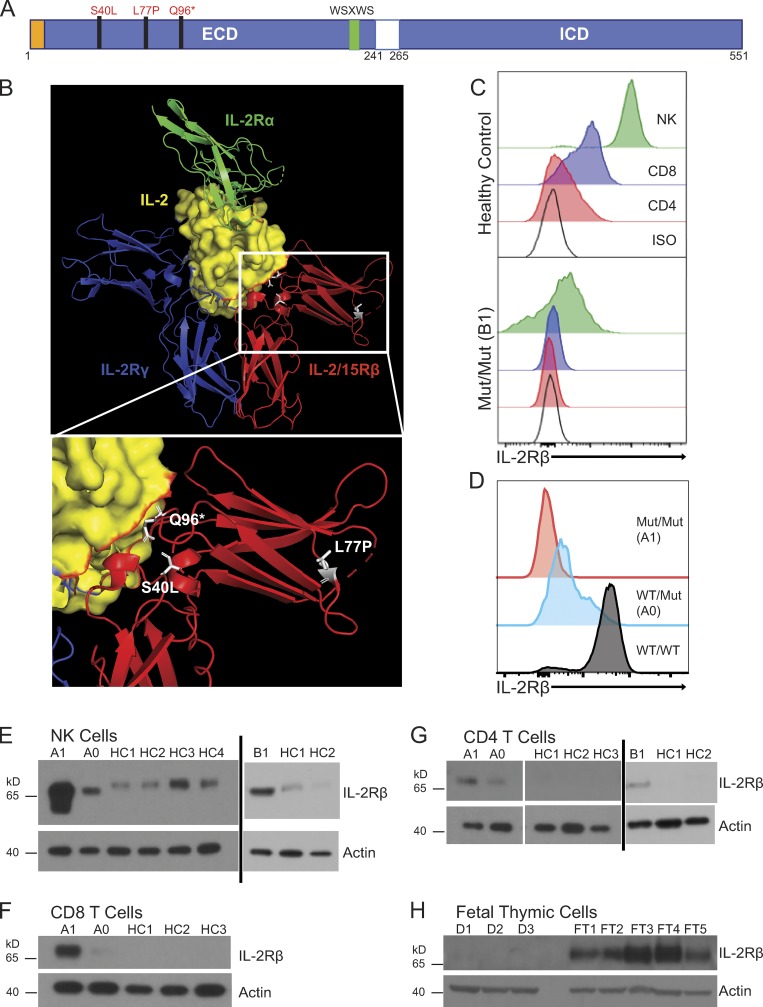
Figure 2.
IL2RB coding mutations cause IL-2Rβ surface receptor deficiency. (A) Schematic of intracellular (ICD) and extracellular domains (ECD) of the IL-2Rβ protein depicting the location of the three mutations in the ECD. The signal peptide is highlighted in orange, and the canonical WSXWS motif is highlighted in green. (B) Crystal structure of IL-2:IL-2R complex with the expanded view showing the position of the three mutations in white: L77P, S40L, and Q96*; (modified from PDB 2B5I, Wang et al., 2005). Red: IL-2/15Rβ; blue: IL-2Rγ; green: IL-2Rα; and yellow: IL-2 with IL-2Rβ interface colored in red. (C) Histogram of IL-2Rβ surface expression in CD3+ CD4+ (red), CD3+ CD8+ (blue), and CD3− CD56+ NK cells (green) and isotype control–stained cells (black) from healthy control (top panel) and patient B1 (bottom panel). (D) Histogram of IL-2Rβ surface expression in NK cells (CD3− CD56+; red, homozygous affected A1; blue, heterozygote healthy A0; black, healthy control). Data representative of four independent experiments. Mut, mutation. (E) Western blot of FACS-sorted CD3− CD56+ NK cells from A1, heterozygote parent (A0), B1, and four healthy controls (HC1–HC4). (F) Western blot of FACS-sorted CD3+ CD8+ T cells from A1, heterozygote parent (A0), and three healthy controls (HC1–HC3). (G) Western blot of FACS-sorted CD3+ CD4+ T cells from A1, heterozygote parent (A0), B1, and three healthy controls (HC1–HC3). (H) Western blot of fetal thymuses from kindred D (D1–D3) and five fetal thymic controls from 25-wk-old (FT3 and FT4) and 31-wk-old (FT1, FT2, and FT5). E–H, loading control: actin. Western blots (E–H) were repeated in triplicate.
The L77P IL2RB missense mutation causes loss of surface expression and function in T cells
At baseline, IL-2Rβ is normally highly expressed on the surface of NK cells and to a lesser extent on T cells; however, patients with the L77P mutation have markedly decreased surface expression of IL-2Rβ on NKs, CD4 T cells, and CD8 T cells as assessed by flow cytometry (Fig. 2, C and D). A healthy heterozygous parent showed intermediate surface expression of IL-2Rβ (Fig. 2 D). Despite diminished cell surface IL-2Rβ expression, immunoblotting of cytosolic lysates of patient NKs, CD4 T cells, and CD8 T cells revealed strikingly more IL-2Rβ than healthy controls (Fig. 2, E–G). This implied that the mutant L77P IL-2Rβ protein was sequestered intracellularly due to misfolding and an inability to properly traffic to the cell surface for subsequent turnover. In keeping with this hypothesis, the faster migration of L77P IL-2Rβ protein relative to WT IL-2Rβ is likely due to incomplete glycosylation branching modifications that are added post-translationally outside the ER (Fig. 2 E). In addition, the affected neonate (D1) and fetuses (D2 and D3) from kindred D with the more severe p.Q96* stop-gain mutation, resulting in a significant truncation, had no IL-2Rβ protein expression (Fig. 2 H).
Reduced signaling by mutant IL-2Rβ proteins encoded by patient alleles
We reconstituted the IL-2R complex in HEK293T cells via transfection of expression plasmids encoding IL-2Rβ, and IL-2Rγ, JAK-3, and STAT5; this system can transduce a signal from IL-2 to intracellular mediators such as the STAT signaling proteins (John et al., 1999; Majri et al., 2018). We used this system to compare the protein-coding sequences of the WT or L77P mutant IL-2Rβ and GFP (separated by a P2A sequence) under the control of a tetracycline (Tet)-inducible promoter. As expected, cells transfected with the WT plasmid showed increasing IL-2Rβ surface expression that correlated with increasing GFP expression after Tet induction (Fig. 3 A). However, cells transfected with the mutIL2RB plasmid showed very low surface IL-2Rβ expression, except at very high levels as judged by GFP coexpression (Fig. 3 A). Given similar levels of expression of the blue fluorescent protein (BFP) control for Tet-inducible system and GFP expression, WT IL-2Rβ is expressed at much greater abundance on the cell surface than the L77P mutant (Fig. 3 B). As observed in the L77P mutant patient lymphocytes, there is an increase in total cytoplasmic IL-2Rβ protein, despite decreased surface expression, in cells transfected with the mutant (Fig. 3 C). Confocal imaging of the live HEK293T cells transfected with an ER marker (KDEL-BFP) and wtIL-2RB-GFP or mutIL-2RB-GFP showed that mutIL-2RB-GFP colocalized with KDEL-BFP and therefore was being sequestered in the ER (Fig. 3 D), as we hypothesized from the patient data. Together these experiments demonstrate that even when the L77P IL-2Rβ is reconstituted in an exogenous HEK293T cell line, the allele encodes a mutant protein that is sequestered in the ER and inefficiently reaches the cell surface, thus recapitulating the patients’ cellular phenotype.
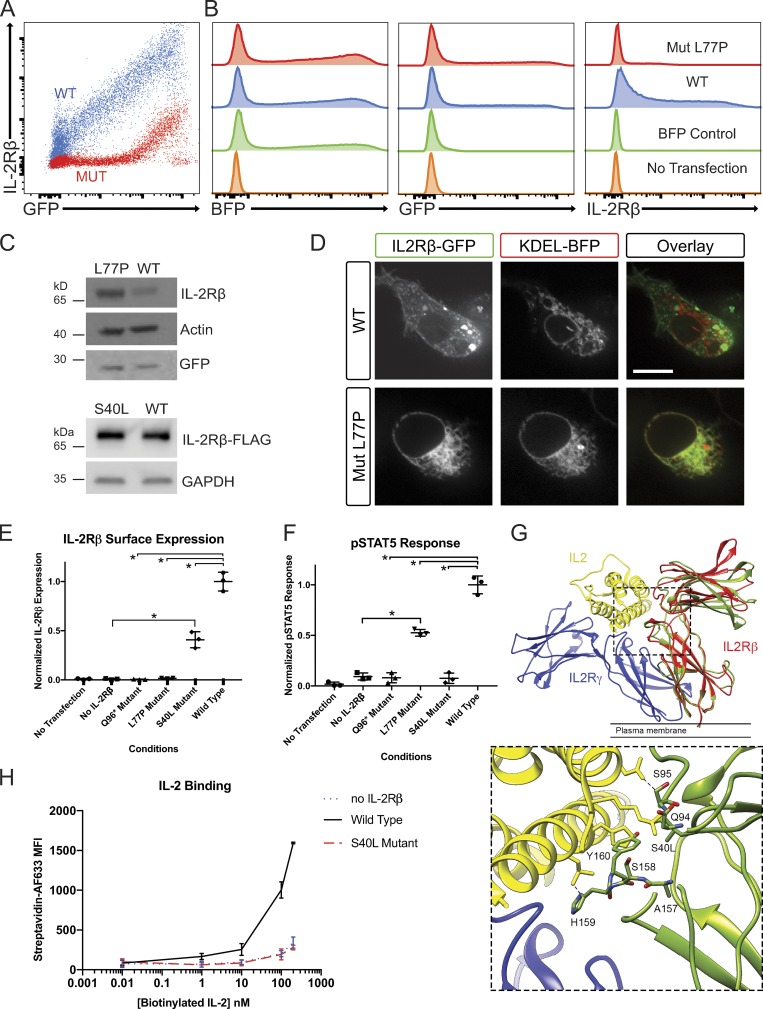
Figure 3.
Investigation of IL-2Rβ deficiency mechanisms in a HEK293T transfection model. (A) FACS plot of GFP and IL-2Rβ expression by HEK293T cells transfected with pHTC-wtIL2RB (red) and pHR-TetON-BFP or transfected with pHTC-mutIL2RB (blue) and pHR-TetON-BFP. (B) Histograms of BFP, GFP, or IL-2Rβ expression given the listed four transfection conditions: WT, mutant (Mut), TetON only, and no transfection. (C) Western blot of HEK293T cells transfected with pHTC-wtIL2RB-GFP or pHTC-mutIL2RB-GFP. Loading controls: actin and GFP. (D) Confocal images of live HEK293T cells cotransfected with KDEL-BFP (ER localization marker) and WT-IL2RB-GFP or Mut-IL2RB-GFP. Bar, 10 µm. (E) Graph of normalized surface IL-2Rβ expression in HEK293T cells with exogenous IL-2R system for the three disease-causing IL2RB mutations. *, P < 0.05, Mann-Whitney U test. (F) Graph of pSTAT5 response to high-dose IL-2 in HEK293T cells with exogenous IL-2R system. *, P < 0.05, Mann-Whitney U test. (G) MD simulation of the receptor cytokine binding interface in WT IL-2Rβ and the S40L variant. The IL-2R subunit is colored in blue, IL-2Rβ in red, and IL-2 in yellow (PDB: 5M5E). The structure of S40L mutant after 100 ns of MD simulation (green) is shown superimposed on the structure of the WT IL-2Rβ after 100 ns MD simulation (red). The leucine side chain clashes with main chain atoms in the BC2 loop (residues 157–165) of the D2 domain, which contributes directly to IL-2 binding. A zoomed in panel of the IL-2 and MD simulated S40L mutant IL-2Rβ interface is provided. (H) Graph of IL-2 binding by WT IL-2Rβ, S40L mutant, and no IL-2Rβ negative control in HEK293T cells measured by flow cytometry using a biotin–streptavidin system. MFI, mean fluorescence intensity. Experiments A–F and H were repeated in triplicate with graphs showing mean ± SEM.
Using our reconstituted receptor system, we also compared the Q96* and S40L alleles to the L77P allele for IL-2Rβ surface expression and phosphorylation of STAT5 (pSTAT5) after IL-2 stimulation (Fig. 3, E and F). As expected, the Q96* allele, which encodes an early stop codon and truncation of IL-2Rβ before the transmembrane domain, generated no IL-2Rβ surface expression and showed no pSTAT5 response to IL-2 stimulation (Fig. 3, E and F). The low level of L77P expressed at the cell surface was nonetheless capable of supporting some phosphorylation of STAT5 in response to IL-2, indicating hypomorphic behavior of this allele. By contrast, the S40L IL2RB allele promoted IL-2Rβ surface expression but conferred no response to IL-2 stimulation (Fig. 3, E and F). Molecular modeling of the S40L mutant showed that the substitution introduces steric clashes with main chain atoms in the BC2 loop (residues 157–165) in the D2 domain, which we predict would disrupt the IL-2 binding interface of IL-2Rβ (Fig. 3 G), consistent with this variant’s lack of responsiveness to IL-2 (Fig. 3 F). In addition, we performed molecular dynamics (MD) simulations on WT IL-2Rβ and the L77P and S40L mutants. After 100 ns of simulation, residues 76–78 in the L77P mutant adopt a different backbone conformation and do not contribute a β-strand to one of the β-sheets in the D1 domain as in WT (Fig. S2), which is consistent with our functional evidence that the L77P mutant is misfolded and sequestered in the ER. In an MD simulation of the S40L mutant, the BC2 loop and specifically key IL-2 binding residues His159 and Tyr160 adopted a conformation that would clash with an IL-2 molecule bound in the same position as in the WT complex. The D1 domains also rotated relative to the D2 domain in the S40L mutant by ∼15°, altering the shape of the IL-2 binding surface (Fig. 3 G). As predicted by the MD simulation, the S40L mutant had significantly decreased affinity to IL-2—comparable to levels in the negative control without IL-2Rβ based on a biotinylated cytokine-streptavidin fluorophore flow cytometry assay (Fig. 3 H). Thus, by using this reconstituted system, we define three distinct mechanisms in humans for IL-2Rβ deficiency by showing that it can occur due to an absence of IL-2Rβ (Q96*), impaired surface expression (L77P), and decreased binding of IL-2 (S40L).
Patient T cells show impaired IL-2Rβ-dependent signaling
We next explored the impact of IL2RB mutations on interleukin signaling in patient T lymphocytes by measuring STAT3 and STAT5 phosphorylation after IL stimulation (Fig. 4). High-dose IL-2 normally triggers tyrosine phosphorylation of the cytoplasmic tails of the IL-2Rβ and IL-2Rγ and downstream STAT1, STAT3, and STAT5 phosphorylation via JAK1 and JAK3. Consistent with a loss of function phenotype, we found that CD4+ and CD8+ T cells, which were available for laboratory analysis from patients A1 and B1, failed to phosphorylate STAT5 in response to IL-2 or IL-15 stimulation, whereas robust phosphorylation was observed in cells from healthy controls (Fig. 4, A–E). By contrast, patient T cells were fully responsive to IL-7 stimulation, indicating that the defect of IL-2Rβ–dependent signaling was selective (Fig. 4 E). Interestingly, CD4+ T cells from A0, the father with a heterozygous L77P genotype, have enough surface expression of IL2-Rβ to phosphorylate STAT3 and STAT5 at a comparable level to healthy controls (Fig. 4, A and B), although there was a trend toward reduced STAT3 and STAT5 phosphorylation in heterozygous CD8+ T cells (Fig. 4, C and D). Thus, surface IL-2Rβ deficiency impairs downstream STAT phosphorylation in response to IL-2 and IL-15 stimulation in a cell type– and receptor expression–dependent manner.
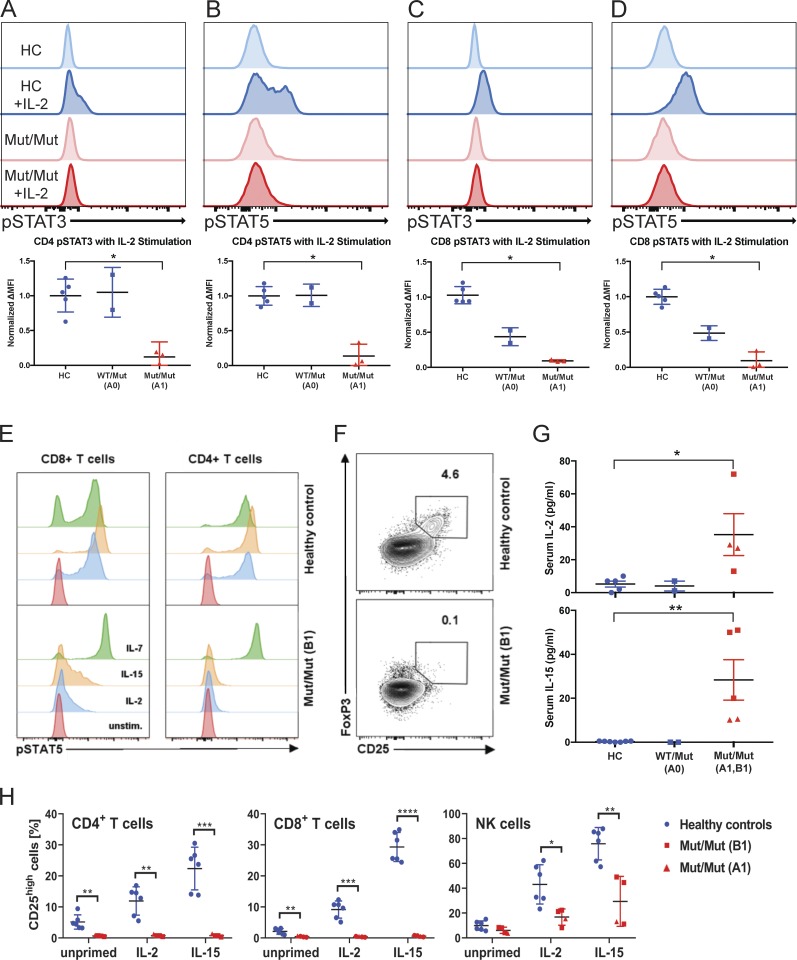
Figure 4.
IL-2Rβ deficiency abrogates IL-2 induced STAT3 and STAT5 phosphorylation in peripheral T cells. (A) Flow cytometry–based measure of STAT3 phosphorylation in CD3+ CD4+ T cells from healthy controls (HC), heterozygote parent A0 (WT/Mut), and homozygous affected A1 (Mut/Mut). (B) STAT5 phosphorylation in CD3+ CD4+ T cells. (C) STAT3 phosphorylation in CD8 T cells. (D) STAT5 phosphorylation in CD8+ T cells (red, representative healthy control; blue, representative affected; lighter shade, unstimulated; darker shade, stimulated with 1,000 U IL-2). (E) STAT5 phosphorylation in CD4+ and CD8+ T cells (B1) in response to IL-2, IL-7, and IL-15 stimulation. Unstim., unstimulated. (F) Flow cytometry plot of CD25 and FoxP3 expression in healthy control and homozygous affected (B1). Data representative of three independent experiments. (G) Graphs of IL-2 and IL-15 levels in healthy controls, A0 (healthy heterozygous father of A1), patient A1, and patient B1 serum. (H) Graphs of percent of CD25high cells in healthy control (blue), patient B1 (red square), and patient A1 (red triangle) CD4+ T cells, CD8+ T cells, and NK cells that have been primed with IL-2 or IL-15 for 12 h or left unprimed. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; Mann-Whitney U tests were performed in A–D, and G. Student t test was performed in H. Experiments A–H were performed in triplicate with graphs showing mean ± SD.
In keeping with current understanding of the critical role of IL-2 signaling in the maintenance of T reg cells in the periphery (Fontenot et al., 2005), the CD25hiFoxP3+ CD4+ T cell compartment was almost empty (Fig. 4 F). Consistent with the inability of peripheral T cells to use and consume IL-2 and IL-15, the patients had elevated levels of serum IL-2 and IL-15, and their T cells did not up-regulate CD25 expression when stimulated with the same cytokines (Fig. 4, G and H). Taken together, the profound reduction of STAT5 signaling within the CD4+ T cell compartment, the inability to up-regulate CD25, and the absence of CD25hiFOXP3+ T reg cells closely mirror the situation in IL2RB-KO mice and other known disorders of T reg cells such as monogenic deficiencies of FOXP3 and CD25. Therefore, this could explain at least in part the various autoimmune manifestations we observed early in life.
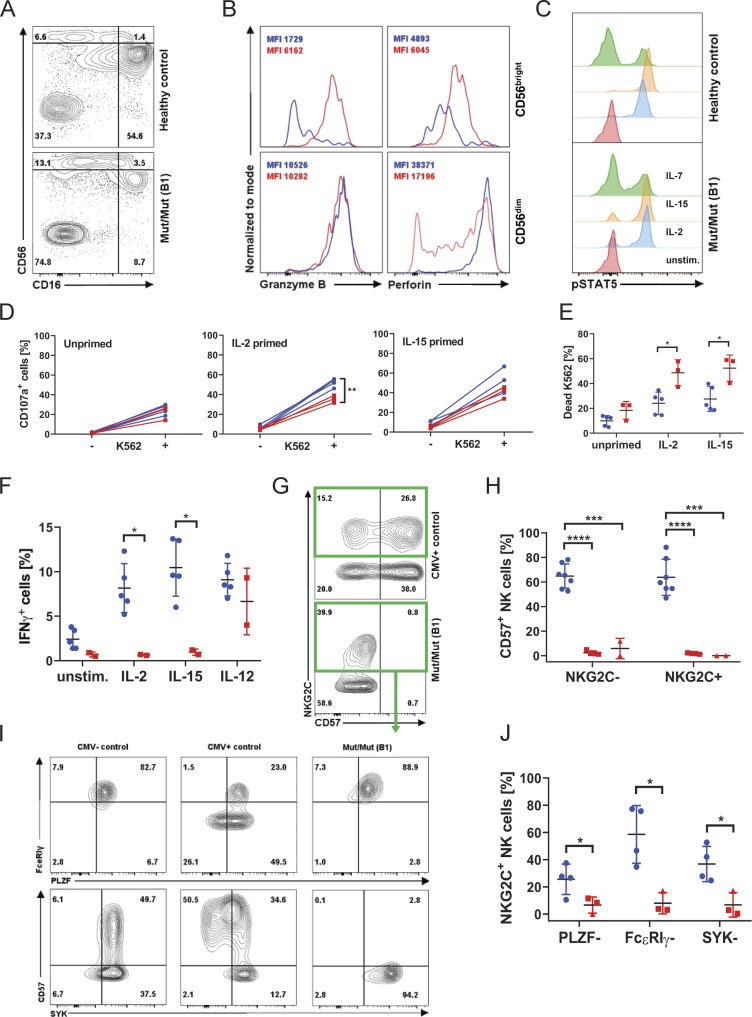
Hypomorphic nature of L77P IL2RB mutation in NK cells
The NK compartment of IL2RB-KO mice is almost completely depleted, but our patients bearing hypomorphic mutations instead showed an expansion of NK cells (Table S2) and an increase in CD56bright relative to CD56dim NKs (Fig. 5 A; Suzuki et al., 1995). These unusually abundant CD56bright NK cells expressed abnormally high levels of the cytotoxic effector proteins perforin and granzyme B (Fig. 5 B). A similar increase of the human CD56bright NK cell compartment has been observed in patients experiencing chronic cytokine exposure through low-dose IL-2 therapy (Ito et al., 2014), CD25 blockade (Bielekova et al., 2006), or low-dose IL-15 treatment (Dubois et al., 2017), associated with increased ex vivo cytotoxic function. Indeed, residual expression of IL-2RβL77P was clearly detectable (Fig. 2, C and D), just as it had been on the surface of transfected 293T cells. Moreover, this residual IL-2Rβ expression could sustain IL-2 and IL-15 signal transduction and downstream STAT5 phosphorylation (Fig. 5 C).
Figure 5.
NK cells retain IL-2/IL-15 responsiveness and effector function, but an adaptive “memory-like” subset is lacking. (A) Flow cytometry plot of CD16 and CD56 expression in CD3− CD19− lymphocytes (patient B1), representative of four independent experiments. (B) Histograms of granzyme B and perforin content in CD56bright versus CD56dim NK cells. Healthy control in blue, patient B1 in red. Experiment displayed representative of three independent experiments. (C) STAT5 phosphorylation in NK cells (B1) in response to IL-2, IL-7, and IL-15 stimulation. (D) CD107a expression (degranulation) in healthy control and patient B1 NK cells co-cultured with K562 cells after 12 h of priming with IL-2 or IL-15 or left unprimed. (E) Percentage of 7-AAD–positive, i.e., dead, K562 cells as a measure of cytotoxicity when co-cultured with healthy control (blue circles) PBMCs or patient B1 (red squares) PBMCs. (F) IFNγ production in response to the indicated stimuli in control NK cells (blue circles) or patient B1 NK cells (red squares). (G) Expression of NKG2C and CD57 on NK cells of CMV+ control and patient B1. (H) Summary graph displaying the percentage of CD57+ positivity within the NKG2C+ and NKG2C− NK cell subsets (mean ± SD) in patient A1 (red triangles) and B1 (red squares). Data representative of six independent experiments. (I) FACS plots of FcεRIγ, PLZF, and SYK expression in CMV− and CMV+ healthy controls and well as patient B1, gated on NKG2C-expressing NK cells. (J) Summary graphs showing the percentage of NKG2C+ NK cells from patient A1 (red triangles) and B1 (red squares) down-regulating the indicated proteins. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; Student t tests were performed. Experiments D–F were performed in duplicate and A–C and G–J were performed in triplicate with graphs showing mean ± SD.
We were able to examine NK cell effector function systematically in patient B1. When provided with the target cell line K562, patient NK cells degranulated to a similar extent to control NK cells, with evidence of priming by both IL-2 and IL-15 (Fig. 5 D and Fig. S3 B). Moreover, cytotoxicity toward K562 was increased in patient NK cells compared with control with IL-2 or IL-15 priming (Fig. 5 E). However, patient NK cells were poor producers of IFNγ in response to the same cytokines (Fig. 5 F and Fig. S3 C). This effect was IL-2Rβ dependent, because the same cells could produce normal levels of IFNγ when stimulated with IL-12. These data support the conclusion that L77P is a hypomorphic mutation of IL2RB that all but abolishes IL-2 signaling in T cells but still transduces residual signaling in high IL-2R–expressing cell subsets like NK cells. As a result, NK cells persist and can respond to the IL-2 and IL-15 that we speculate are normally produced but not consumed by IL-2Rβ–deficient T cells. Our data show that various outcomes of IL-2Rβ signaling are differentially affected by the L77P variant—from absent (IFNγ production; Fig. 5 F and Fig. S3 C) to reduced (CD25 up-regulation; Fig. 4 H and Fig. S3 A) to supra-normal (cytotoxicity; Fig. 5 E); however, these findings are based only on a single patient, and firm conclusions are difficult due to donor variability.
With this evidence of altered NK phenotype in mind, we hypothesized that NK differentiation might be perturbed in other ways. In particular, the NK compartment is known to adapt to the presence of CMV by expansion of memory-like cells that coexpress killer cell lectin like receptor C2, also known as KLRC2 (NKG2C), and CD57 on their surface (Lopez-Vergès et al., 2010; Foley et al., 2012). Such cells were strikingly absent from both A1 and B1 (Fig. 5, G and H), even though an NKG2C+ population was present, suggesting CMV recognition. To explore whether there was a general problem with NK maturation, we examined the expression of the transcription factor promyelocytic leukemia zinc finger (PLZF) and its downstream targets spleen tyrosine kinase (SYK) and FcεRIγ (Lee et al., 2015; Schlums et al., 2015). This analysis confirmed a failure of patient NKG2C+ NK cells to complete their differentiation by down-regulating PLZF, SYK, and FcεRIγ, as well as up-regulating CD57 (Fig. 5, I and J). We conclude that the NK compartment and its response to CMV are systematically altered in the context of the hypomorphic IL2RB mutation. We suggest that this is at least in part responsible for the observed clinical susceptibility to CMV disease.
Lentiviral rescue of IL2RB and STAT phosphorylation in patient T cells
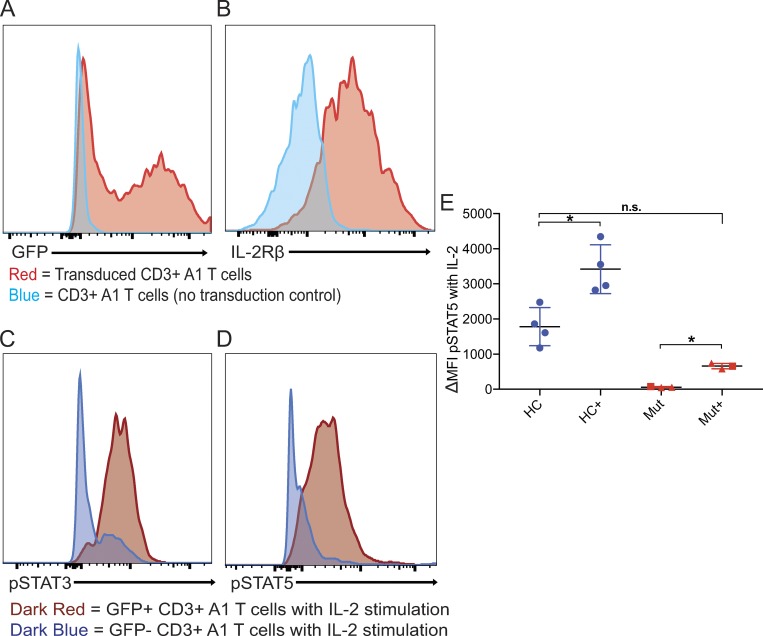
Finally, we performed complementation by lentiviral transduction of patient T cells with WT IL2RB (Fig. 6). Patient T cells were activated with anti-CD3 and anti-CD28 beads, transduced with lentiviral WT-IL2RB and GFP, and cultured in the presence of IL-7 rather than IL-2. Patient CD3+ T cells were transduced with >50% transfection efficiency (Fig. 6 A) and expressed high levels of IL-2Rβ on the cell surface (Fig. 6 B). While patient T cells that were not transduced with WT-IL2RB failed to respond to high-dose IL-2 stimulation as expected, transduced patient T cells, when stimulated with IL-2, were able to phosphorylate STAT3 and STAT5 (Fig. 6, C and D). Transduction of WT-IL2RB did not restore IL-2 response back to control levels (Fig. 6 E), but this may be due to heterodimerization of L77P IL-2Rβ and WT IL-2Rβ, leading to partial ER sequestration of the WT chain (Pillet et al., 2008). Nonetheless, lentiviral transduction of WT-IL2RB partially rescues STAT phosphorylation in response to IL-2 in patient T cells, indicating that the failure of IL-2 response in this patient is due to the genetic abnormality of IL2RB.
Figure 6.
Lentiviral rescue of IL-2Rβ and STAT phosphorylation in primary T cells. (A) Histogram showing GFP expression in patient A1 CD3+ T cells transduced (red) with lentiviral WT IL-2Rβ and GFP and no transduction control (blue). (B) Histogram showing IL-2Rβ expression in patient A1 CD3+ T cells transduced (red) with lentiviral WT IL2RB and GFP and no transduction control (blue). (C) Histograms showing STAT3 phosphorylation in response to IL-2 stimulation in transduced (dark red) and nontransduced (dark blue) CD3+ A1 T cells. (D) Histograms showing STAT5 phosphorylation in response to IL-2 stimulation in transduced (dark red) and nontransduced (dark blue) CD3+ A1 T cells. (E) Graph of delta mean fluorescence intensity (MFI) of pSTAT5 between IL-2 stimulated and unstimulated in healthy controls (HC) and L77P patients A1 and B1 transduced and no transduction control. *, P < 0.05, Mann-Whitney U test; n.s., not significant. Experiments A–E were performed in triplicate with the graph showing mean ± SEM.
Discussion
Here we provide the first report of autosomal recessive IL-2Rβ deficiency in four pedigrees harboring five affected liveborn children with immunodeficiency and autoimmune disease and three perinatally affected fatalities. Clinical hallmarks of the disease include prominent immune dysregulatory phenomena such as enteropathy, skin abnormalities, autoimmune hemolytic anemia, and hypergammaglobulinemia, together with susceptibility to respiratory and herpesvirus infections. We demonstrate that the three mutant alleles cause IL-2Rβ deficiency by different biochemical mechanisms. Kindreds A and B have the hypomorphic L77P IL-2Rβ mutant, which interferes with egress from the ER. We discovered that this abrogates surface expression and IL-2 signaling in T cells, but that NKs retain not only modest surface expression and responsiveness to IL-2 but also quite potent cytolytic activity. Kindred C possesses the S40L IL-2Rβ mutant, which has decreased responsiveness to IL-2 despite being expressed on the cell surface. Our analysis shows that this is due to an amino acid side group clash in the receptor:ligand interaction site. This mutation may also be hypomorphic in nature given IL-2Rβ expression, but studies to characterize this were limited by sample availability. Kindred D has the most severe Q96* IL-2Rβ stop-gain mutant. The severity of this mutation is reflected not only in the perinatal phenotype in the neonate and fetuses but also by the complete absence of IL-2Rβ expression and IL-2 signaling. However, it cannot be excluded that the severe phenotype in kindred D may be due to the added effects of other genetic variants. A NCF4 variant also cosegregated in the kindred, but the specific variant is reported in ClinVar to be likely benign and is not very rare, with a minor allele frequency > 0.001. Despite the differences in mechanism, all the mutations cause IL-2Rβ dysfunction in some manner and lead to a similar constellation of clinical features.
Specifically, the hypomorphic L77P IL2RB mutation highlights the significance of variable IL-2Rβ expression in different lymphocyte subsets as a means of modulating immune function. The L77P mutation causes ER sequestration and thus minimal IL-2Rβ surface expression in patient lymphocytes despite increased total IL-2Rβ protein. This decreased IL-2Rβ surface expression prevents downstream STAT3 and STAT5 phosphorylation following IL-2 stimulation in T cells. By contrast, NK cells are still capable of responding to IL-2 and maintain normal cytotoxic function, likely due to the cells’ intrinsic high expression of IL-2R. Nonetheless, abnormalities of NK adaptation to CMV and IFNγ production may contribute to the viral susceptibility observed in most patients.
While the human IL-2Rβ deficiency shares many similarities with the IL2RB KO mouse and FOXP3-deficient IPEX patients, there are interesting key differences. Like the KO mouse (Suzuki et al., 1995), the IL-2Rβ–deficient patients have autoimmune hemolytic anemia, elevated autoantibodies, hypergammaglobulinemia (IgG and IgE), lymphadenopathy, and splenomegaly. In vitro, both the IL2RB KO mouse and IL-2Rβ–deficient human T cells proliferate poorly in response to IL-2 and TCR stimulation. Human IL-2Rβ disease reveals that deficient IL-2Rβ also leads to skin abnormalities and enteropathy, which is not seen in the KO mouse. In addition, in the human patients, we observed an expansion of NK cells and skewing of the NK compartment, while a reduction of NK cells was recorded in the KO mouse (Suzuki et al., 1997). These observations might reflect the hypomorphic nature of the human disease alleles compared with complete loss of function in the mouse model and/or differences in the role of IL-2/15Rβ in NK maturation in the two species (Renoux et al., 2015). Mirroring the KO mouse, the IL-2Rβ–deficient patients also lack CD25+ FoxP3+ T reg cells, thus explaining the overlap in clinical features of immune dysregulation with IPEX syndrome. Enteropathy, dermatitis, and hemolytic anemia were seen in both disorders, although only one IL-2Rβ patient (A1) presented with any endocrinopathy—a hallmark of IPEX. A distinctive component of IL-2Rβ deficiency is the presence of β-herpesviral disease in addition to severe autoimmune/inflammatory disease. This combination recalls CD25 deficiency, where β-herpesviral susceptibility has also been noted (Sharfe et al., 1997; Caudy et al., 2007). The presence of both immunodeficiency and autoimmune/inflammatory disease as defining features of IL-2Rβ and CD25 deficiency states is consistent with the multi-faceted role of IL-2 signaling biology in the immune system.
The current definitive treatment for IL-2Rβ deficiency is HSCT. Patient A2 received an allogeneic HSCT and has had complete resolution of her symptoms. However, there are high risks associated with HSCT, as exemplified by patient B1, and the hope is that understanding the pathophysiological mechanism of IL-2Rβ deficiency can guide the development of novel therapeutics. One approach might be to repurpose IL-2 anti–IL-2 antibody complexes (Boyman et al., 2006), IL-2 superkine (Levin et al., 2012), orthoIL-2 analogues (Sockolosky et al., 2018), or IL-2 Fc fusion proteins (Vazquez-Lombardi et al., 2017) as a potential means of hyper-stimulating residual surface IL-2Rβ. Monoclonal anti-human IL-2 antibody MAB602 (mouse S4B6) in complex with IL-2 was found to selectively promote proliferation of effector T cells, while the antibody clone 5344 (mouse JES61) induced proliferation of T reg cells (Boyman et al., 2006). Similarly, the H9 IL-2 superkine was engineered to have enhanced binding to IL-2Rβ independent of CD25 (Levin et al., 2012). Another approach to hyper-stimulating the IL-2Rβ mutant would be to develop an orthoIL-2 with specific binding to the mutant (Sockolosky et al., 2018). The fact that we find hypomorphic mutations in the liveborn children suggests that an approach involving hyper-stimulating the IL-2 axis may ameliorate disease.
In summary, our identification of human IL-2Rβ deficiency as a monogenic cause of immunodeficiency and autoimmunity provides insight into one of the principal signaling pathways of the immune activation and peripheral tolerance and should prompt prenatal screening of IL2RB mutations and genetic counseling in families at risk.
Materials and methods
Human subjects
Written informed consent was provided by all human subjects or their legal guardians in accordance with the 1975 Helsinki principles for enrollment in research protocols that were approved by the Institutional Review Board of the National Institute of Allergy and Infectious Diseases, National Institutes of Health, and the Newcastle and North Tyneside Research Ethics Committee 1, UK. Patient and healthy control blood was obtained at Starship Children’s Hospital in Auckland, New Zealand, Addenbrooke’s Hospital in Cambridge, UK, and Great North Children’s Hospital in Newcastle, UK, under approved protocols.
Genetic analysis
DNA was obtained from probands and family members by isolation and purification from peripheral blood mononuclear cells (PBMCs) using Qiagen’s DNeasy Blood and Tissue Kit. The DNA was then submitted for whole exome sequencing by Illumina sequencers in the United States, UK, France, and Saudi Arabia. The reads were filtered for sequence quality and then mapped on to the h19 human genome reference by a Burrows-Wheeler Aligner with default parameters. Alignment, variant calling, and annotation were performed by the in-house bioinformatics core using the Genome Analysis Toolkit version 3.4 (Broad Institute) and GEMINI (GEnome MINIng). The IL2RB variant was confirmed by Sanger sequencing of PCR amplification products of cDNA, generated by reverse transcription of RNA using SuperScript IV VILO kit (Thermo Fisher Scientific) and the following PCR primers: forward, 5′-CCTGTGTCTGGAGCCAAGAT-3′ and reverse, 5′-GGGTGACGATGTCAACTGTG-3′ (Sigma-Aldrich) or forward, 5′-CCTCACAGTGGTTGGCACA-3′ and reverse, 5′-GCACTCTCTCCCTGGGTG-3′ (Sigma-Aldrich).
Cells and media
Primary patient or control PBMCs were obtained from whole blood subjected to Histopaque/Ficoll density gradient separation. The PBMCs were then washed with PBS and frozen in complete Roswell Park Memorial Institute (RPMI) media with 10% DMSO in liquid nitrogen for later use or −80°C for transport. HEK293T and K562 cells were obtained from the European Collection of Authenticated Cell Cultures and tested mycoplasma-free. Human cells were cultured in RPMI (Sigma-Aldrich) or DMEM (Sigma-Aldrich) supplemented with 10% heat-inactivated FBS (Sigma-Aldrich), 1% penicillin/streptomycin (Gibco), and 1% Glutamax (Gibco). Recombinant human IL-2, IL-7, and IL-15 (Peprotech) were used for stimulation. XVIVO 15 media (Lonza) supplemented with 1–10% human type AB serum (Sigma-Aldrich) was used for STAT phosphorylation assays.
Antibodies
The following monoclonal primary rabbit anti-human antibodies from Cell Signaling Technologies were used for Western blot analysis: anti-IL2RB, anti-GFP, anti-vinculin, and anti-IL2RA. Rabbit anti–β actin (Abcam) and goat anti-IL2RG (Thermo Fisher Scientific) were also used. Secondary HRP-linked anti-rabbit IgG and anti-goat IgG antibodies (Cell Signaling Technologies) were used to conjugate to the respective primary antibodies. The following flow cytometry antibodies are from BioLegend: CD3-AF700, CD3-PerCp-Cy5.5, CD3-BV705, CD4-Pacific Blue, CD56-PE-Cy7, CD122-PE-Dazzle, CD132-APC, CD25-APC-Cy7, CXCR5-FITC, CD45RA-PerCp-Cy5.5, CD127-APC, HLA-DR-Pacific Blue, Live/Dead-Zombie Aqua, pSTAT3-AF647, CD20 (2H7), PD-1 (EH12.2H7), CD45RA (HI100), CD127 (A019D5), TNFα (MAB11), CD56 (5.1H11), CD56 (HCD56), CD16 (3G8), CD19 (HIB19), CD122 (TU27), CD57 (QA17A04), Syk (4D10.2), and perforin (dG9); Thermo Fisher Scientific: CD4-APC-eF780, CD56-APC-eF780, TCRγδ (B1.1), and TCRVα24Jα18 (6B11); BD Biosciences: CD25-PE, pSTAT5-AF488, CD4 (SK3), CD3 (UCHT1), CD8 (RPA-T8), CD25 (2A3 or M-A251), CCR7 (3D12), CD45RO (UCHL1), granzyme B (GB11), IFNγ (B27), STAT5 (47/Stat5), S6 (N7-548), FoxP3 (259D/C7), CD127 (HIL-7R-M21), CD56 (NCAM16.2), CD28 (CD28.2), CD95 (DX2), CD16 (3G8), CD107a (H4A3), CD69 (FN50), IL-2 (5344.111), and PLZF (R17-809); Miltenyi: CD132 (REA313); Merck: FcεRIγ (polyclonal); and R&D Systems: NKG2C (134591). Cell trace violet (Thermo Fisher Scientific) was used to label K562 cells. Cell viability was assessed using Zombie Aqua or NIR, 7-AAD (all from BioLegend) or Live/Dead Fixable Green (Invitrogen, Thermo Fisher Scientific).
Flow cytometry
Cells were pelleted by centrifugation and stained with antibodies in FACS Buffer (1–2% FBS, 0.05% sodium azide, and 2–5 mM EDTA in PBS) at 4°C for 30–60 min. The stained cells were then washed with PBS or FACS buffer, pelleted, and resuspended at ∼106 cells/ml in FACS Fix Buffer (FACS Buffer with or without 1% paraformaldehyde) for flow cytometry analysis (Fortessa, Symphony A5, or FACS Aria Fusion system). The flow data were analyzed using FlowJo or Treestar.
Western blot
Cells were lysed with NuPage lithium dodecyl sulfate sample buffer (Thermo Fisher Scientific) at the concentration of 105 cells per 15 μl lithium dodecyl sulfate supplemented with 10% β-mercaptoethanol and benzonase nuclease (Sigma-Aldrich). The samples were then denatured at 70°C. Protein lysates were separated by SDS-PAGE on 4–12% Bis-Tris precase gels (Invitrogen) and transferred to a polyvinylidene difluoride membrane (Invitrogen) by iBlot (Thermo Fisher Scientific) or wet transfer. Membranes were then blocked in milk with 5% Tris-buffered saline with 0.01% Tween-20; TBST) for 1 h at room temperature and then incubated with primary antibody in milk or 5% BSA overnight at 4°C. The membrane was washed for 3 × 10 min with TBST at room temperature and then stained with HRP-linked secondary antibody in milk for 1 h at room temperature. After 3 × 10–min washes with TBST and 1 × 10–min wash with PBS, the membrane was exposed to enhanced chemiluminescent substrates (Thermo Fisher Scientific) and developed by film.
Flow cytometry–based STAT phosphorylation assay
At the National Institutes of Health, PBMCs were thawed in XVIVO media (Lonza) with 10% human AB serum (Sigma-Aldrich), pelleted, washed with XVIVO, and resuspended in XVIVO media with 1% human AB serum at the concentration of 106 cells/ml. Then the cells were stimulated with 1,000 U IL-2 (Peprotech) for 10 min at 37°C, fixed with BD Fix/Lyse buffer (BD Biosciences) for 10 min at 37°C, and then washed with cold PBS with 0.2% BSA. Next, the fixed cells were permeabilized with −20°C methanol for 20 min on ice, washed five times with cold PBS with 0.2% BSA, and then stained with surface and intracellular flow cytometry antibodies for 30 min at 4°C. The fixed, permeabilized, and stained cells were washed with PBS and resuspended in PBS with 0.2% BSA for flow cytometry analysis. In Newcastle, thawed PMBCs were rested for 4 h in serum-free RPMI media. After the addition of surface markers and a fixable viability dye, 2 × 105 cells were stimulated for 10 min at 37°C with 100 ng/ml of IL-2, IL-7, or IL-15, or left unstimulated. The Transcription Factor Phospho Buffer set (BD Biosciences) was used to fix and permeabilize cells according to the manufacturer’s instructions. Cells were stained with the remaining surface as well as intracellular markers for 45 min at 4°C before cells were washed in TFP Perm/Wash buffer and finally resuspended in FACS buffer for acquisition.
Site-directed mutagenesis
The WT pME18S-IL2RB template plasmid (∼5,000 bp) was obtained from the National Institutes of Health. Site-directed mutagenesis of T230C (p.L77P) was performed using the In-Fusion HD Cloning Kit (Takara Clontech) and following PCR primers (Sigma-Aldrich): forward, 5′-AGCTGCCCCCCGTGAGTCAA-3′ and reverse, 5′-TCACGGGGGGCAGCTCACAGGTTT-3′.
The linearized vector was generated by PCR using the CloneAmp HiFi PCR master mix (Takara Clontech), plasmid template, and primers with the following thermocycling conditions: 35 cycles of 10 s at 98°C, 5 s at 55°C, and 25 s at 72°C. The PCR products were separated on a 1% agarose gel by gel electrophoresis, and the desired mutagenized product band was cut out. The PCR product was purified using the NucleoSpin Gel and PCR Clean Up (Takara Clontech) from the InFusion Cloning Kit. The linearized, mutagenized product was ligated using the InFusion Enzyme (Takara Clontech) to generate the L77P mutant pME18S-IL2RB plasmid. Stellar cells (Takara Clontech) were transformed with the new plasmid by heat shock; the transformed cells were plated on ampicillin plates and incubated overnight at 37°C. Plasmid was extracted from individual colonies using the QIAprep Spin MiniPrep Kit (Qiagen). The mutation was confirmed by Sanger sequencing.
Cloning
Using WT and mutant pME18S-IL2RB plasmids as the template, wtIL2RB and mutIL2RB PCR products with AsiSI and SpeI restriction sites were generated using the following primers: forward, 5′-TAGTAGGCGATCGCGCCACCATGGCGGCCCCTGCTCTGTC-3′ and reverse, 5′-CTACTAACTAGTCACCAAGTGAGTTGGGTCCTGAC-3′. The PCR products were purified by gel electrophoresis. Next the gel-purified PCR products and pHTC-P2A plasmid (provided by John R. James) were digested with AsiSI and SpeI restriction enzymes in CutSmart Buffer (NEB) for 2 h at 37°C and then purified by gel electrophoresis. IL2RB WT and mutant were ligated into separate custom-made pHTC/pBR322-P2A vectors using T4 DNA ligase (NEB). DH5α competent bacteria (NEB) were transformed with pHTC-wtIL2RB and pHTC-mutIL2RB and plated on Amp plates overnight. Individual colonies were Sanger-sequenced to confirm successful cloning. pHTC-wtIL2RB, pHTC-mutIL2RB, and pGFP (provided by John R. James) were digested with mLuI and BamHI in NEB3.1 buffer and then purified by gel electrophoresis. Similar to above, GFP was ligated in to the pHTC vectors to generate pHTC-wtIL2RB-P2A-GFP and pHTC-mutIL2RB-P2A-GFP. The final plasmids were transformed into DH5α bacteria, and individual colonies were Sanger-sequenced again.
HEK293T transfection and confocal imaging
HEK293T cells were cultured in complete DMEM or RPMI at 37°C in T75 flasks. 4 × 105 cells in 2 ml media were seeded into 6-well plates and grown overnight at 37°C. At 40–50% confluence, the cells were transfected using 97 μl OPTI-MEM (Gibco) and 3 μl GeneJuice Transfection Reagent (VWR) per 1 μg DNA. Cells were transfected with 1:pHTC-wtIL2RB-P2A-GFP and pHR-TetON-P2A-BFP, 2:pHTC-mutIL2RB-P2A-GFP and pHR-TetON-P2A-BFP, 3:pHTC-wtIL2RB-P2A-GFP, 4:pHTC-mutIL2RB-P2A-GFP, and 5:pHR-TetON-P2A-BFP. 6 h after transfection with pHTC-IL2RB-P2A-GFP and pHR-TetON-P2A-BFP, cells were dosed with doxycycline (1 μg/ml). The transfected cells were cultured overnight at 37°C, pelleted, washed with PBS, and stained with CD122-PE-Dazzle antibody for flow cytometry analysis. Similarly, HEK293T cells were transfected with pHR-wtIL2RB-GFP or pHR-mutIL2RB-GFP and pBFP-KDEL in the same conditions in fibronectin-coated dishes for confocal imaging. An Andor spinning disc confocal microscope system was used to image the live cells at 37°C. Under the same conditions, HEK293T cells were also transfected with pME-IL2RG, pME-JAK3, pME-STAT5-HA, pBFP, and different IL2RB plasmids to reconstitute IL-2R. After successful transfection, the cells were stimulated with high-dose IL-2, and STAT phosphorylation was measured by flow cytometry as described above.
IL-2 binding assay
HEK293T cells were transfected with WT and mutant IL-2RB and IL-2RG using the same transfection conditions as described above. 105 transfected cells in 0.2 ml 0.2% BSA-PBS were incubated with 200, 100, 10, 1, or 0.01 nM biotinylated IL-2 (R&D Systems) for 30 min at 4°C. Then streptavidin–Alexa Fluor 633 (1 μg/ml; Invitrogen) was added and incubated for 15 min at 4°C. Each sample was then washed three times with 1 ml PBS. The cells were resuspended in 300 μl FACS buffer for subsequent FACS analysis. This protocol was adopted from Pillet et al. (2008).
NK degranulation and cytotoxicity assays
PBMCs were seeded at 105 per well in a 96-well plate and primed with either IL-2 or IL-15 (100 ng/ml each) for 12 h or left unprimed. After the priming period, cells were coincubated with K562 target cells (E:T ratio of 10:1) for 3 h. Alongside K562 exposure, the CD107a-antibody was added to the wells. Cells were harvested, and surface staining was performed for 60 min on ice. Degranulation was measured by means of CD107a surface expression. Cytotoxicity was assessed by calculating the percentage of 7-AAD–positive K562 after coincubation minus the spontaneous K562 death rate when cultured alone. The addition of IL-2 or IL-15 was found to not affect the viability of K562 cells.
IFNγ production
PBMCs (105 in 100 μl) were stimulated with either IL-2, IL-15, or IL-12 (100 ng/ml each, all from Peprotech) for 6 h with brefeldin A (BD GolgiStop) added for the final 5 h. Cells were fixed and permeabilized using Cytofix/Cytoperm buffers (BD Biosciences) according to the manufacturer’s instructions, and surface as well as intracellular staining were performed for 30 min at 4°C.
ELISA
Serum was analyzed for IL-2 using an IL-2 ELISA kit (Invitrogen) and for IL-15 using IL-15 High Performance Luminex kit (R&D Systems).
T cell proliferation
2 × 105 PBMCs were incubated in triplicate per experimental condition with the indicated stimuli for 3 d before being pulsed with tritiated thymidine for 8 h. A scintillation β-counter was used to measure incorporation into dividing cells.
Molecular modeling
Starting models were derived from a crystal structure of IL-2RB in complex with IL2–IL-2RB and IL-2 determined at 2.3 Å resolution (PDB: 5M5E; Klein et al., 2017). For the S40L variant, the Leu40 side chain was modeled with COOT (Emsley and Cowtan, 2004) without MD simulation. For the L77P variant, the Pro77 side chain was placed in the experimental electron density of Leu77 with COOT while minimizing clashes with surrounding atoms to achieve a favorable initial geometry. The GROMACS software package (Abraham et al., 2015) was used to set up and run MD simulations. The AMBER99SB-ILDN force field (Lindorff-Larsen et al., 2010) and TIP3P water model were used and the structures placed in dodecahedral boxes with 10 Å padding and surrounded with solvent including water and 150 mM NaCl. After steepest-gradient energy minimization, a modified Berendsen thermostat (two groups, time constant 0.1 ps, temperature 310 K) followed by a Berendsen barostat (isotropic, coupling constant 0.5 ps, reference pressure 1 bar) were coupled to the system over 100 ps. 100-ns runs of unrestrained MD trajectories were produced. After removal of periodic boundary condition artifacts, MD runs were visualized and analyzed in UCSF Chimera (Pettersen et al., 2004) and bulk statistics extracted using GROMACS analysis routines.
Lentiviral transduction
WT IL2RB was cloned into a pHR-GFP transfer plasmid. Lenti-X 293T cells (Clontech) were transfected with the pHR-IL2RB-GFP transfer plasmid, pSPAX2, and pMD2.g plasmid in OPTI-MEM and polyethylenimine. Lentivirus was harvested at 24, 48, and 72 h after transfection. The viral supernatants were pooled and concentrated using Lenti-X concentrator (Clontech). An aliquot of the concentrated lentivirus was quantified by quantitative PCR using the Lenti-X qRT-PCR Titration Kit (Clontech). PBMCs were thawed and cultured in complete RPMI with IL-7. The cells were activated with CD3/CD28 Dynabeads (Gibco) for at least 24 h before lentiviral transduction. After activation, PBMCs were transduced with a ratio of 100:1 WT-IL2RB lentivirus:PBMCs on retronectin-coated plated by spinfection. The cells were transduced for 72 h in complete RPMI with IL-7. Transduction efficiency was measured by FACS, and transduced cells were subsequently assessed for STAT phosphorylation after IL-2 stimulation.
Clinical histories
Patient A1
A1 was born to consanguineous parents. He was initially referred at the age of 2 yr due to feeding difficulties. On investigation, he was found to have Graves’ thyrotoxicosis, hypergammaglobulinemia, lymphadenopathy, splenomegaly, large tonsils with history of obstruction during sleep, and eczema. An adenotonsillectomy and skin biopsy were performed. 6 mo later, he was doing well with normal growth and improved appetite. Skin was improving with occasional 1% hydrocortisone cream, but he continued to have significant lymphadenopathy.
When he was 3 yr old, after a trip to Pakistan, he had significant weight loss, diarrhea, otitis media, and otorrhea, as well as continuing lymphadenopathy and splenomegaly. At the age of 4 yr, he presented to the emergency department with abdominal pain, fever, and elevated respiratory rate. He was treated with oral amoxicillin. 1 mo after this emergency department visit, he was referred to dermatology due to worsening periorbital eczema. His dermatological exam revealed generalized dermatitis over most of his torso and limbs. He was found to have post-inflammatory hyperpigmentation changes in numerous regions of his body. 6 mo later, he had loss of one third of his outer eyebrows as well as nonscarring alopecia.
At age 5 yr, he presented with a cough, abdominal pain, and fever. Chest x ray showed peribronchial changes, and he was treated with oral amoxicillin. Although he has not been positive for CMV, he has had persistent Epstein-Barr viremia. He was also found to be positive for mycoplasma on bronchoalveolar lavage. Last year he was started on rituximab with some improvement in his skin and lymphadenopathy. He continues to have a widespread rash.
Patient A2
A2 was born to consanguineous parents and referred for poor weight gain likely due to poor oral intake at the age of 5 mo. At 9 mo, following a trip to Pakistan, she was admitted due to diarrhea and failure to thrive and found to have mucocutaneous candidiasis, urinary tract infection, ear infection with otorrhea, and lymphadenopathy. She also had CMV viremia, positive celiac serology, and villous blunting on biopsy. She was treated with ganciclovir, and the resulting neutropenia was treated with G-CSF. Her diarrhea resolved and she was able to tolerate nasogastric feeding. In the following months she had mild eczema that developed into a more generalized papular rash and was referred to dermatology.
At 15 mo, she developed CMV pneumonia with large pleural effusion, panuveitis, and proctocolitis. She was restarted on ganciclovir, which was later changed to valganciclovir. At 19 mo, she developed a complicated extended spectrum β lactamases urinary tract infection, which necessitated prolonged i.v. antibiotics treatment.
At 26 mo, she was admitted with fever and respiratory distress. She initially improved on intravenous antibiotics but then deteriorated and needed supplemental oxygen. Bronchoscopy and bronchoalveolar lavage showed a petechial appearance of the airway mucosa. She was positive for influenza A and also found to have antineutrophil cytoplasmic antibody vasculitis. At 30 mo, she was admitted for an HSCT. After the first dose of alemtuzumab, she had hematuria and declining renal function. Renal biopsy revealed crescentic glomerulonephritis. She was treated with plasmapheresis, rituximab, and steroids. After her glomerular filtration rate normalized, she received an HSCT. After transplant, she had CMV reactivation, which was successfully treated. 6 mo after transplant, she is doing well.
Patient B1
B1 was born to consanguineous parents at 37 wk by emergency cesarean section following failed induction of labor for intrauterine growth retardation. Dry skin and loose stools were noted from early days but progressed to severe secretory diarrhea, culminating in her emergency readmission to hospital in hypovolemic shock at 4 wk of age. At this time, the patient weighed 1.85 kg with a severe metabolic acidosis (pH 7.0), and imaging subsequently confirmed thrombus within the great veins and right atrium. She was resuscitated and treated with bowel rest and parenteral nutrition along with low-molecular-weight heparin.
Endoscopic examination and biopsies of the upper gut showed severe architectural distortion and effacement of duodenal villi, associated with chronic inflammatory infiltration and marked apoptosis of epithelial cells. Biopsy of an associated skin rash revealed inflammatory changes suggestive of acrodermatitis enteropathica, although serum zinc was normal. In keeping with a possible autoimmune diathesis, the patient also developed a positive direct antiglobulin test (2+) and moderate thrombocytopenia along with hypergammaglobulinemia (maximum IgG 26.9, IgM 6.6 g/liter) and positive autoantibodies (antinuclear antibody 1/160, smooth muscle antibody 1/160). Inflammatory markers were raised, and the patient required respiratory support, but criteria for hemophagocytic lymphohistiocytosis were never satisfied. She developed a hepatitis associated with CMV viremia that was successfully treated with ganciclovir. At this time her peripheral blood immunophenotype revealed normal numbers of T cells, with a reduced CD4:CD8 ratio and relatively low naive T cell percentage yet a high proportion of class-switched memory B cells. T reg cells were present but reduced in number. In whole blood assays, poor production of IL-10 was noted as well as overall poor T cell cytokine responses that were not boasted by IL-2. T cell proliferative responses were globally reduced, but again, insensitivity to IL-2 was noted.
The patient was immunosuppressed using steroids, infliximab, and sirolimus with significant improvement in her diarrhea, dermatitis, and general status (including nutrition and growth). However, she remained total parental nutrition dependent and experienced periods of presumed inflammatory pneumonitis and Staphylococcus aureus sepsis. Owing to the severity of her immune dysregulation and the suspicion of a monogenic etiology, she underwent peripheral blood HSCT. She engrafted but, despite anti-infective prophylaxis, had reactivated CMV and developed severe pneumonitis with respiratory failure. She died, and postmortem examination was declined.
Patient C1
C1 was born to consanguineous parents. He presented at the age of 3 yr with severe autoimmune hemolytic anemia with high reticulocyte count and positive Coombs test.
At 3.5 yr old, he presented with severe eczematous scaly dermatitis and recurrent suppurative ear infection since the age of 6 mo. He has a history of recurrent CMV and EBV infections associated with lymphadenopathy and hepatosplenomegaly. In addition, he has a history of food allergies to milk, soybean, wheat, and peanut with a positive radioallergosorbent test. He died at a local hospital from pneumonitis and sepsis.
Patient C2
C2 was born to consanguineous parents. She presented at the age of 18 mo with a history of recurrent chest and ear infections and chronic diarrhea, requiring frequent intensive care unit admissions since the age of 2 mo. At the age of 4 mo, she started to have generalized scaly eruption and multiple eczematous plaques associated with redundant skin over body folds and neck, generalized lymphadenopathy, and hepatosplenomegaly. She had evidence of autoimmune hemolytic anemia with high reticulocyte count and positive Coombs test. Bone marrow biopsy was normal. She had a history suggestive of IgE-mediated food allergy to milk, soybean, wheat, and peanut with a positive radioallergosorbent test. During her admissions to the hospital, she had positive blood cultures to Klebsiella pneumoniae and CMV viremia. Skin biopsy showed superficial neutrophil rich dermal inflammation with few eosinophils suggestive of Sweet syndrome, and loss of elastic fibers in papillary dermis consistent with cutis laxa. She died from progressive pneumonitis and respiratory failure.
Online supplemental material
Fig. S1 shows healthy control and patient T cell proliferative responses to PHA, anti-CD3, anti-CD3 and IL-2, and PMA and ionomycin. Fig. S2 shows the molecular dynamic simulation results for WT, S40L, and L77P IL-2Rβ. Fig. S3 shows the FACS plots for CD25 up-regulation, CD107a degranulation, and IFNγ expression in healthy control and patient NK cells. Table S1 shows patient mutations and clinical manifestations. Table S2 shows lab values and absolute cell counts. Table S3 shows rare variants that were identified and cosegregated in each kindred.
Supplementary Material
Acknowledgments
We thank John Sowerby, Iosifina Foskolou, Lixin Zheng, Francesco Colucci, Morgan Similuk, Warren Leonard, and Helen Su for their advice and insight. We thank Daniil Prigozhin for advice on MD simulations and Danny Lim for blood processing at Hematology LabPlus at Auckland City Hospital. We thank Claudia Gonzaga-Jauregui, Alan Shuldiner, John Overton, Aris Baris and the Regeneron Genetics Center, Genomics Core Facility, Newcastle University and Patricia Fergelot at the Genome Transcriptome Facility of Bordeaux Biodiversité, Genes, & Communautés for genomic support and DNA sequencing. We acknowledge the Cambridge Institute of Medical Research and Newcastle University Flow Cytometry Core Facility for assistance with the generation of flow cytometry data. Finally, we thank all the patients described in this manuscript and their families for facilitating this work.
This work was supported by the Wellcome Trust (Investigator Award 083650/Z/07/Z to K.G.C. Smith, 207556/Z/17/Z to S. Hambleton, 101908/Z/13/Z to Y. Modis, and 099966/Z/12/Z to J.R. James), the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, and the UK National Institute of Health Research Cambridge Biomedical Research Centre and the Sir Jules Thorn Charitable Trust (12/JTA to S. Hambleton). Z. Zhang was supported by the National Institutes of Health Oxford-Cambridge Scholarship in Biomedical Research program and the National Institutes of Health M.D./Ph.D partnership program with Harvard Medical School. F. Gothe was supported by the Deutsche Forschungsgemeinschaft (GO2955/1-1).
The authors declare no competing financial interests.
Author contributions: Z. Zhang and F. Gothe performed the majority of the experiments and analyses. Z. Zhang, F. Gothe, S. Hambleton, C. Rooryck, K.G.C. Smith, and M.J. Lenardo conceived and planned the experiments. J.E. Thaventhiran, K.R. Engelhardt., H. Al-Mousa, S. Hambleton, C. Rooryck, K.G.C. Smith, and M.J. Lenardo supervised the project. Z. Zhang, F. Gothe, S. Hambleton, K.G.C. Smith, and M.J. Lenardo wrote the manuscript. All authors discussed and revised the manuscript. P. Pennamen and Z. Zhang performed in vitro experiments on kindred D samples. J.R. James assisted with imaging and reconstitution studies. D. McDonald assisted with flow cytometry studies. C.P. Mata and Y. Modis performed molecular dynamic simulations. A.M. Alazami, H. Alajlan, and Z. Zhang performed reconstitution experiments involving the S40L mutation. M. Acres, R. Döffinger, S. Naudion, F. Pelluard, and Y. Yamazaki assisted with experiments. Y. Zhang performed the initial genomic analyses. W. Haller, C. Bowen, J. Sinclair, S. Brothers, H.F. Matthews, and L.D. Notarangelo referred patients, provided clinical data, and coordinated sample collection.
References
- Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., and Lindahl E.. 2015. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 1-2:19–25. 10.1016/j.softx.2015.06.001 [DOI] [Google Scholar]
- Ahmadzadeh M., and Rosenberg S.A.. 2006. IL-2 administration increases CD4+ CD25(hi) Foxp3+ regulatory T cells in cancer patients. Blood. 107:2409–2414. 10.1182/blood-2005-06-2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielekova B., Catalfamo M., Reichert-Scrivner S., Packer A., Cerna M., Waldmann T.A., McFarland H., Henkart P.A., and Martin R.. 2006. Regulatory CD56(bright) natural killer cells mediate immunomodulatory effects of IL-2Ralpha-targeted therapy (daclizumab) in multiple sclerosis. Proc. Natl. Acad. Sci. USA. 103:5941–5946. 10.1073/pnas.0601335103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyman O., and Sprent J.. 2012. The role of interleukin-2 during homeostasis and activation of the immune system. Nat. Rev. Immunol. 12:180–190. 10.1038/nri3156 [DOI] [PubMed] [Google Scholar]
- Boyman O., Kovar M., Rubinstein M.P., Surh C.D., and Sprent J.. 2006. Selective stimulation of T cell subsets with antibody-cytokine immune complexes. Science. 311:1924–1927. 10.1126/science.1122927 [DOI] [PubMed] [Google Scholar]
- Busse D., de la Rosa M., Hobiger K., Thurley K., Flossdorf M., Scheffold A., and Höfer T.. 2010. Competing feedback loops shape IL-2 signaling between helper and regulatory T lymphocytes in cellular microenvironments. Proc. Natl. Acad. Sci. USA. 107:3058–3063. 10.1073/pnas.0812851107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudy A.A., Reddy S.T., Chatila T., Atkinson J.P., and Verbsky J.W.. 2007. CD25 deficiency causes an immune dysregulation, polyendocrinopathy, enteropathy, X-linked-like syndrome, and defective IL-10 expression from CD4 lymphocytes. J. Allergy Clin. Immunol. 119:482–487. 10.1016/j.jaci.2006.10.007 [DOI] [PubMed] [Google Scholar]
- Dubois S., Conlon K.C., Müller J.R., Hsu-Albert J., Beltran N., Bryant B.R., and Waldmann T.A.. 2017. IL15 infusion of cancer patients expands the subpopulation of cytotoxic CD56bright NK cells and increases NK cell cytokine release capabilities. Cancer Immunol. Res. 5:929–938. 10.1158/2326-6066.CIR-17-0279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., and Cowtan K.. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132. 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- Foley B., Cooley S., Verneris M.R., Pitt M., Curtsinger J., Luo X., Lopez-Vergès S., Lanier L.L., Weisdorf D., and Miller J.S.. 2012. Cytomegalovirus reactivation after allogeneic transplantation promotes a lasting increase in educated NKG2C+ natural killer cells with potent function. Blood. 119:2665–2674. 10.1182/blood-2011-10-386995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenot J.D., Rasmussen J.P., Gavin M.A., and Rudensky A.Y.. 2005. A function for interleukin 2 in Foxp3-expressing regulatory T cells. Nat. Immunol. 6:1142–1151. 10.1038/ni1263 [DOI] [PubMed] [Google Scholar]
- Gilmour K.C., Fujii H., Cranston T., Davies E.G., Kinnon C., and Gaspar H.B.. 2001. Defective expression of the interleukin-2/interleukin-15 receptor beta subunit leads to a natural killer cell-deficient form of severe combined immunodeficiency. Blood. 98:877–879. 10.1182/blood.V98.3.877 [DOI] [PubMed] [Google Scholar]
- Hatakeyama M., Tsudo M., Minamoto S., Kono T., Doi T., Miyata T., Miyasaka M., and Taniguchi T.. 1989. Interleukin-2 receptor beta chain gene: generation of three receptor forms by cloned human alpha and beta chain cDNA’s. Science. 244:551–556. 10.1126/science.2785715 [DOI] [PubMed] [Google Scholar]
- Hinks A., Cobb J., Marion M.C., Prahalad S., Sudman M., Bowes J., Martin P., Comeau M.E., Sajuthi S., Andrews R., et al. 2013. Dense genotyping of immune-related disease regions identifies 14 new susceptibility loci for juvenile idiopathic arthritis. Nat. Genet. 45:664–669. 10.1038/ng.2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S., Bollard C.M., Carlsten M., Melenhorst J.J., Biancotto A., Wang E., Chen J., Kotliarov Y., Cheung F., Xie Z., et al. 2014. Ultra-low dose interleukin-2 promotes immune-modulating function of regulatory T cells and natural killer cells in healthy volunteers. Mol. Ther. 22:1388–1395. 10.1038/mt.2014.50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S., Vinkemeier U., Soldaini E., Darnell J.E. Jr., and Leonard W.J.. 1999. The significance of tetramerization in promoter recruitment by Stat5. Mol. Cell. Biol. 19:1910–1918. 10.1128/MCB.19.3.1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein C., Waldhauer I., Nicolini V.G., Freimoser-Grundschober A., Nayak T., Vugts D.J., Dunn C., Bolijn M., Benz J., Stihle M., et al. 2017. Cergutuzumab amunaleukin (CEA-IL2v), a CEA-targeted IL-2 variant-based immunocytokine for combination cancer immunotherapy: Overcoming limitations of aldesleukin and conventional IL-2-based immunocytokines. OncoImmunology. 6:e1277306 10.1080/2162402X.2016.1277306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Zhang T., Hwang I., Kim A., Nitschke L., Kim M., Scott J.M., Kamimura Y., Lanier L.L., and Kim S.. 2015. Epigenetic modification and antibody-dependent expansion of memory-like NK cells in human cytomegalovirus-infected individuals. Immunity. 42:431–442. 10.1016/j.immuni.2015.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenardo M.J. 1991. Interleukin-2 programs mouse alpha beta T lymphocytes for apoptosis. Nature. 353:858–861. 10.1038/353858a0 [DOI] [PubMed] [Google Scholar]
- Levin A.M., Bates D.L., Ring A.M., Krieg C., Lin J.T., Su L., Moraga I., Raeber M.E., Bowman G.R., Novick P., et al. 2012. Exploiting a natural conformational switch to engineer an interleukin-2 ‘superkine.’. Nature. 24:352–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W., Lin J.X., and Leonard W.J.. 2013. Interleukin-2 at the crossroads of effector responses, tolerance, and immunotherapy. Immunity. 38:13–25. 10.1016/j.immuni.2013.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindorff-Larsen K., Piana S., Palmo K., Maragakis P., Klepeis J.L., Dror R.O., and Shaw D.E.. 2010. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 78:1950–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Vergès S., Milush J.M., Pandey S., York V.A., Arakawa-Hoyt J., Pircher H., Norris P.J., Nixon D.F., and Lanier L.L.. 2010. CD57 defines a functionally distinct population of mature NK cells in the human CD56dimCD16+ NK-cell subset. Blood. 116:3865–3874. 10.1182/blood-2010-04-282301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie R.J., Tan Q.K., Gilner J.B., Rogers R.C., Younge N., Wechsler S.B., McDonald M.T., Gordon B., Saski C.A., Jones J.R., et al. 2017. Novel pathogenic variants in FOXP3 in fetuses with echogenic bowel and skin desquamation identified by ultrasound. Am. J. Med. Genet. A. 173:1219–1225. 10.1002/ajmg.a.38144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majri S.S., Fritz J.M., Villarino A.V., Zheng L., Kanellopoulou C., Chaigne-Delalande B., Grönholm J., Niemela J.E., Afzali B., Biancalana M., et al. 2018. STAT5B: A Differential Regulator of the Life and Death of CD4+ Effector Memory T Cells. J. Immunol. 200:110–118. 10.4049/jimmunol.1701133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malek T.R., Yu A., Vincek V., Scibelli P., and Kong L.. 2002. CD4 regulatory T cells prevent lethal autoimmunity in IL-2Rbeta-deficient mice. Implications for the nonredundant function of IL-2. Immunity. 17:167–178. 10.1016/S1074-7613(02)00367-9 [DOI] [PubMed] [Google Scholar]
- Moffatt M.F., Gut I.G., Demenais F., Strachan D.P., Bouzigon E., Heath S., von Mutius E., Farrall M., Lathrop M., and Cookson W.O.C.M.. GABRIEL Consortium . 2010. A large-scale, consortium-based genomewide association study of asthma. N. Engl. J. Med. 363:1211–1221. 10.1056/NEJMoa0906312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., and Ferrin T.E.. 2004. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Pillet A.H., Juffroy O., Mazard-Pasquier V., Moreau J.L., Gesbert F., Chastagner P., Colle J.H., Thèze J., and Rose T.. 2008. Human IL-Rbeta chains form IL-2 binding homodimers. Eur. Cytokine Netw. 19:49–59. [DOI] [PubMed] [Google Scholar]
- Renoux V.M., Zriwil A., Peitzsch C., Michaëlsson J., Friberg D., Soneji S., and Sitnicka E.. 2015. Identification of a Human Natural Killer Cell Lineage-Restricted Progenitor in Fetal and Adult Tissues. Immunity. 43:394–407. 10.1016/j.immuni.2015.07.011 [DOI] [PubMed] [Google Scholar]
- Schlums H., Cichocki F., Tesi B., Theorell J., Beziat V., Holmes T.D., Han H., Chiang S.C., Foley B., Mattsson K., et al. 2015. Cytomegalovirus infection drives adaptive epigenetic diversification of NK cells with altered signaling and effector function. Immunity. 42:443–456. 10.1016/j.immuni.2015.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharfe N., Dadi H.K., Shahar M., and Roifman C.M.. 1997. Human immune disorder arising from mutation of the alpha chain of the interleukin-2 receptor. Proc. Natl. Acad. Sci. USA. 94:3168–3171. 10.1073/pnas.94.7.3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sockolosky J.T., Trotta E., Parisi G., Picton L., Su L.L., Le A.C., Chhabra A., Silveria S.L., George B.M., King I.C., et al. 2018. Selective targeting of engineered T cells using orthogonal IL-2 cytokine-receptor complexes. Science. 359:1037–1042. 10.1126/science.aar3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Kündig T.M., Furlonger C., Wakeham A., Timms E., Matsuyama T., Schmits R., Simard J.J., Ohashi P.S., Griesser H., et al. 1995. Deregulated T cell activation and autoimmunity in mice lacking interleukin-2 receptor beta. Science. 268:1472–1476. 10.1126/science.7770771 [DOI] [PubMed] [Google Scholar]
- Suzuki H., Duncan G.S., Takimoto H., and Mak T.W.. 1997. Abnormal development of intestinal intraepithelial lymphocytes and peripheral natural killer cells in mice lacking the IL-2 receptor beta chain. J. Exp. Med. 185:499–505. 10.1084/jem.185.3.499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita T., Asao H., Ohtani K., Ishii N., Kumaki S., Tanaka N., Munakata H., Nakamura M., and Sugamura K.. 1992. Cloning of the gamma chain of the human IL-2 receptor. Science. 257:379–382. 10.1126/science.1631559 [DOI] [PubMed] [Google Scholar]
- Vazquez-Lombardi R., Loetsch C., Zinkl D., Jackson J., Schofield P., Deenick E.K., King C., Phan T.G., Webster K.E., Sprent J., and Christ D.. 2017. Potent antitumour activity of interleukin-2-Fc fusion proteins requires Fc-mediated depletion of regulatory T-cells. Nat. Commun. 8:15373 10.1038/ncomms15373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldmann T.A. 2006. The biology of interleukin-2 and interleukin-15: implications for cancer therapy and vaccine design. Nat. Rev. Immunol. 6:595–601. 10.1038/nri1901 [DOI] [PubMed] [Google Scholar]
- Wang X., Rickert M., and Garcia K.C.. 2005. Structure of the quaternary complex of interleukin-2 with its alpha, beta, and gammac receptors. Science. 310:1159–1163. 10.1126/science.1117893 [DOI] [PubMed] [Google Scholar]
- Willerford D.M., Chen J., Ferry J.A., Davidson L., Ma A., and Alt F.W.. 1995. Interleukin-2 receptor alpha chain regulates the size and content of the peripheral lymphoid compartment. Immunity. 3:521–530. 10.1016/1074-7613(95)90180-9 [DOI] [PubMed] [Google Scholar]
- Ye C., Brand D., and Zheng S.G.. 2018. Targeting IL-2: an unexpected effect in treating immunological diseases. Signal Transduct. Target. Ther. 3:2 10.1038/s41392-017-0002-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.