Children with mitochondrial diseases often present with slow growth and short stature, but the underlying mechanism remains unclear. In this study, Holzer et al. provide in vivo evidence that mitochondrial respiratory chain dysfunction induces cartilage degeneration coincident with altered metabolism, impaired extracellular matrix formation, and cell death at the cartilage–bone junction.
Abstract
In childhood, skeletal growth is driven by transient expansion of cartilage in the growth plate. The common belief is that energy production in this hypoxic tissue mainly relies on anaerobic glycolysis and not on mitochondrial respiratory chain (RC) activity. However, children with mitochondrial diseases causing RC dysfunction often present with short stature, which indicates that RC activity may be essential for cartilage-mediated skeletal growth. To elucidate the role of the mitochondrial RC in cartilage growth and pathology, we generated mice with impaired RC function in cartilage. These mice develop normally until birth, but their later growth is retarded. A detailed molecular analysis revealed that metabolic signaling and extracellular matrix formation is disturbed and induces cell death at the cartilage–bone junction to cause a chondrodysplasia-like phenotype. Hence, the results demonstrate the overall importance of the metabolic switch from fetal glycolysis to postnatal RC activation in growth plate cartilage and explain why RC dysfunction can cause short stature in children with mitochondrial diseases.
Introduction
Patients suffering from mitochondrial damage causing respiratory chain (RC) dysfunction, both due to mitochondrial DNA (mtDNA) mutations or defects in nuclear genes encoding mitochondrial proteins, are reported to often present with short stature, but the pathomechanism of the impaired skeletal growth remains unclear (Koenig, 2008; Wolny et al., 2009).
Skeletal growth is driven by the transformation of cartilage into bone tissue as a result of unidirectional cell proliferation within the growth plate cartilage. Chondrocytes are the only cells of the growth plate and it is current belief that these cells rely on anaerobic glycolysis to promote skeletal growth in the avascular, severely hypoxic growth plate (Martin et al., 2012). However, this hypothesis is in conflict with the observation that respiratory dysfunction in patients reduces skeletal growth. Recent ex vivo studies have also reported that mitochondrial dysfunction could act as a pathogenic factor in degenerative cartilage disease, but the in vivo evidence is missing (Blanco et al., 2011).
A major experimental limitation is the lack of models to study RC function in cartilage. Access to growth plate cartilage from patients with mitochondrial diseases is limited and genetic approaches to study RC function in vivo failed because of embryonal lethality when genes essential for mitochondrial homeostasis were manipulated. Only recently, genetic tools were developed to selectively inactivate the RC in mice (Dogan and Trifunovic, 2011).
The aim of this study was to use these novel genetic tools and determine if the RC dysfunction is a major cause for growth retardation and degenerative cartilage disease in the presence of mitochondrial damage. To achieve this goal, we first analyzed the RC activity during development. Interestingly, the RC is hardly active in growth plate cartilage in newborns, but growth plate RC activity markedly increases in juvenile mice, when secondary ossification centers are formed and vascular networks are established at the proximal and distal end of the growth plate. We then generated transgenic mice, which have an inactivated RC only in cartilage, using the cartilage-specific expression of an mtDNA helicase Twinkle mutant (Baris et al., 2015; Weiland et al., 2018). Here, we show that these mice, as a consequence of the lack of RC activation after birth, develop postnatal growth retardation and growth plate cartilage degeneration caused by energy deficiency, altered metabolic signaling, destabilization of the hypertrophic ECM, and increased chondrocyte death at the cartilage–bone junction.
These findings illustrate that glycolysis is sufficient to drive fetal cartilage growth and, in contrast to the current view, a metabolic switch from fetal glycolysis to respiration in growth plate cartilage after birth is essential to promote postnatal skeletal growth. Moreover, the results provide an explanation at the molecular level why loss of RC dysfunction in mitochondrial diseases can cause growth plate cartilage degeneration and impaired skeletal growth.
Results
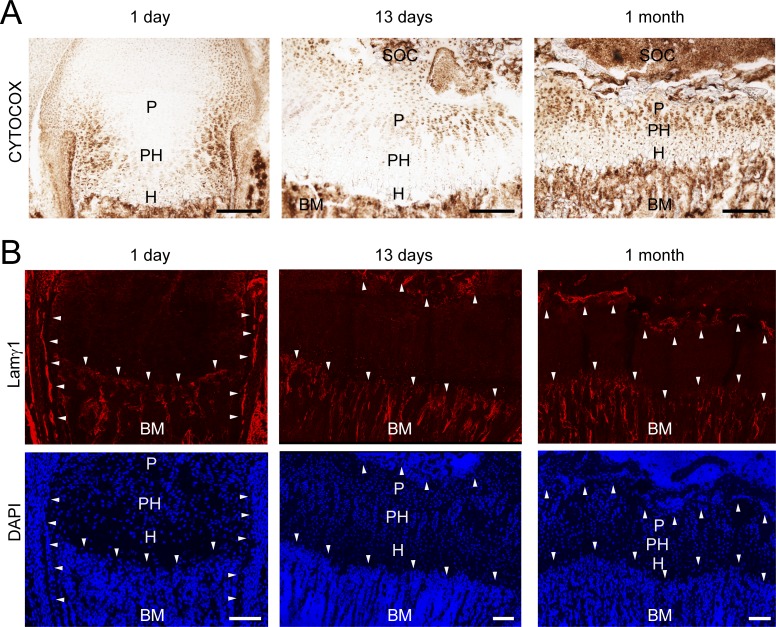
It was earlier proposed that the metabolism in cartilage is entirely anaerobic (Bywaters, 1936), but to our knowledge RC activity was never studied in situ during growth plate cartilage development. Hence, we applied cytochrome c oxidase (CYTOCOX; complex IV) activity staining to femoral sections of newborn, 13-d-old, and 1-mo-old mice to determine the complex IV activity in growth plate cartilage. In newborns, CYTOCOX staining was restricted to the lateral growth plate and the perichondrium (Fig. 1 A) close to laminin γ1–positive blood vessels (Fig. 1 B, upper panel, arrowheads), while it was hardly detectable in chondrocytes in the center (for cellularity, see DAPI staining; Fig. 1 B, lower panel). After formation of the secondary ossification center in 13-d-old mice, CYTOCOX staining was mainly detected in proliferating, but not in prehypertrophic or hypertrophic, chondrocytes. In 1-mo-old mice most cells of the growth plate were CYTOCOX positive but the strongest staining was still found in proliferating chondrocytes. At this stage, a vascular plexus has developed at the apical and distal growth plate (Fig. 1 B, arrowheads). These results unexpectedly indicate that the cartilage metabolism is partially aerobic and that respiration is activated at later stages of postnatal development when blood vessels surround the growth plate to supply sufficient oxygen and/or nutrient levels.
Figure 1.
Characterization of RC activity in the developing growth plate cartilage. (A) Femoral growth plate sections from 1-d-, 13-d-, and 1-mo-old mice were stained for CYTOCOX activity (brown; BM, bone marrow; H, hypertrophic chondrocytes; P, proliferative; PH, prehypertrophic; SOC, secondary ossification center). (B) Laminin γ1 chain immunostaining was used to visualize the vascular network in the proximity of the growth plate cartilage and DAPI to detect nuclei. Arrowheads show the blood vessels close to the growth plate cartilage. Bars: (A) 200 µm; (B) 100 µm. The brightness of the red signal for laminin γ1 staining and the blue signal for DAPI was adjusted for visualization.
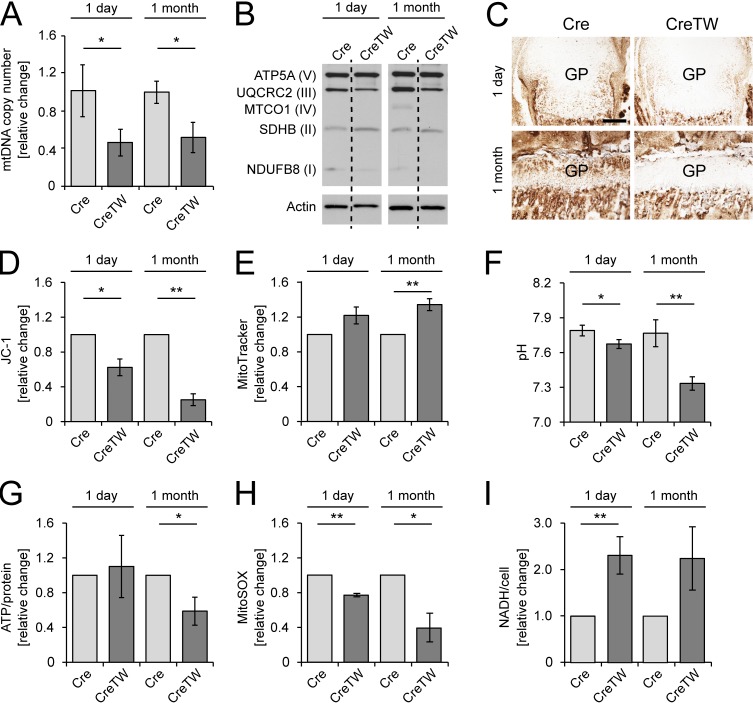
We next generated transgenic mice expressing a patient-derived dominant-negative mutant of the Twinkle helicase in chondrocytes to inhibit the RC and thus determine its function in growth plate–mediated skeletal growth. We crossed R26-K320E-TwinkleloxP/+ mice (Weiland et al., 2018) with mice expressing Cre recombinase driven by the Col2a1 promotor (Cre; Ovchinnikov et al., 2000; Bluhm et al., 2017) and induced the expression of the Twinkle mutant protein in cartilage during early embryonal development. In these mice (CreTW), a marked reduction in the multicopy mtDNA molecules was found and consequently a decrease in index subunits of those RC complexes, which contain essential mtDNA-encoded subunits (complexes I, III, and IV), was observed in cartilage from 1-mo-old mice (Fig. 2, A and B). CYTOCOX activity was absent in newborns and 1-mo-old CreTW mice (Fig. 2 C), which indicates that the RC was successfully inactivated in growth plate cartilage. The consequences for chondrocyte metabolism were then studied in detail using quantitative flow cytometry approaches. As expected, the mitochondrial membrane potential was decreased while mitochondrial mass was slightly, but significantly increased in isolated and cultured chondrocytes from newborn and 1-mo-old CreTW mice (Fig. 2, D and E). The pH in the cell culture medium was decreased at both time points, indicating increased lactate production upon RC dysfunction (Fig. 2 F). More pronounced changes (e.g., significantly reduced ATP levels and reduced reactive oxygen species [ROS] levels) were found in chondrocytes isolated from 1-mo-old mice compared with newborns (Fig. 2, G and H), whereas NADH levels were increased at both time points (Fig. 2 I). Hence, mutant Twinkle expression in chondrocytes leads to a depletion of mtDNA, followed by a decrease in RC complexes containing mtDNA-encoded subunits and reduced energy production in newborns and more pronounced in older CreTW mice.
Figure 2.
Characterization of mitochondrial function in chondrocytes of CreTW mice. (A) Relative mtDNA copy number was determined by qPCR analysis (n = 3 biological samples per group). (B) Immunoblot analysis of RC complexes I to V in femoral head cartilage of 1-d- and 1-mo-old Cre and CreTW mice. (C) Femoral growth plate (GP) sections were stained for CYTOCOX in 1-d- and 1-mo-old CreTW mice. Bar: 200 µm. (D and E) Flow cytometric assessment of mitochondrial membrane potential (D) and mitochondrial mass (E) in cultured primary chondrocytes from 1-d- and 1-mo-old Cre and CreTW mice using JC-1 and MitoTracker Deep Red staining, respectively. (F) The pH of the cell culture supernatant was analyzed. (G) A bioluminescence assay was used to determine the ATP content in isolated chondrocytes cultured for 3 d in DMEM-F12 medium. (H) Relative ROS (MitoSOX) changes were assessed by flow cytometry analysis. (I) Relative NADH levels were determined by MTS measurements. Quantitative data are mean ± SD (n = 3 mice per group). *P < 0.05, **P < 0.01. No SD is given for control when calculating relative changes of independent experiments.
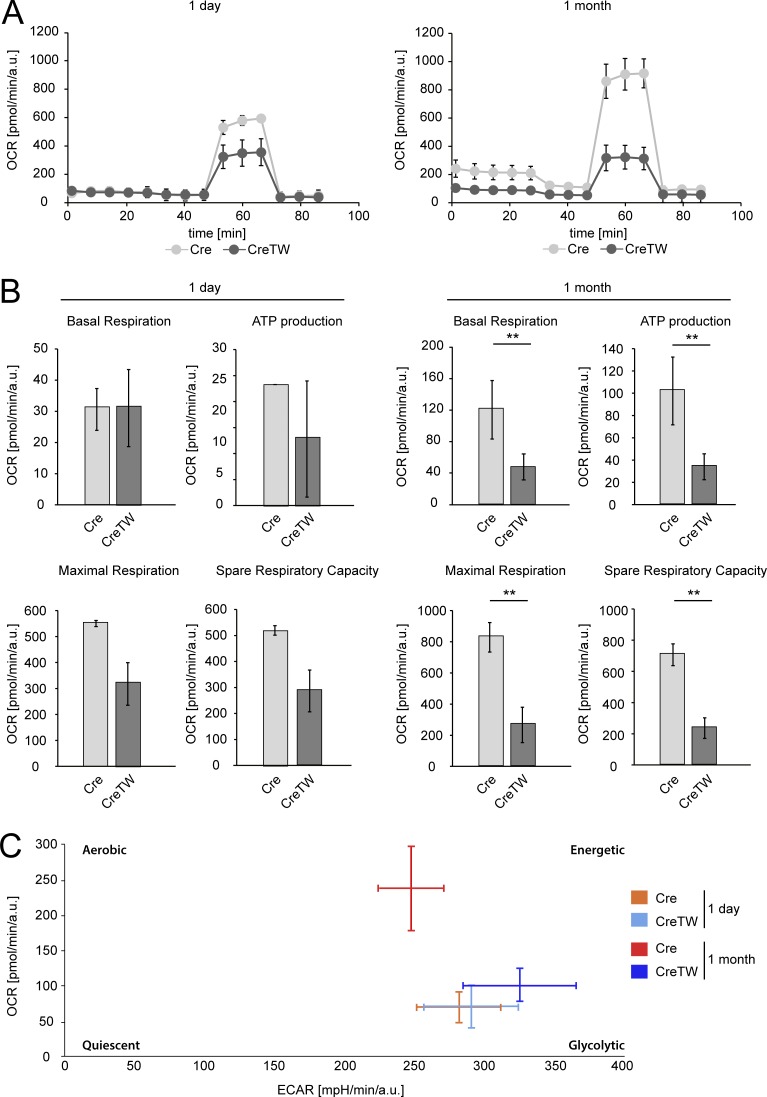
Respiratory capacity of chondrocytes was then characterized by the Seahorse XF96 analyzer. The oxygen consumption rate (OCR) and extracellular acidification rate were recorded over time, and basal respiration, ATP production–coupled respiration, and maximal and spare respiratory capacities were determined in chondrocytes isolated from newborn and 1-mo-old animals (Fig. 3). The data obtained showed that basal respiration and ATP production–coupled respiration were low in both Cre and CreTW chondrocytes isolated from newborns. The calculated maximal and spare respiratory capacities were, however, reduced in young CreTW chondrocytes compared with Cre chondrocytes, clearly indicating the lack of functional mitochondrial respiration in CreTW chondrocytes. Interestingly, basal respiration, ATP production, and maximal/spare respiration capacity were increased in older Cre chondrocytes compared with Cre chondrocytes isolated from newborns (Fig. 3, A and B). The aerobic energy production was activated in older control Cre chondrocytes, while mitochondrial respiratory dysfunction slightly stimulated glycolysis in old CreTW chondrocytes (Fig. 3 C). These results clearly showed that chondrocytes at early ages mainly use anaerobic glycolysis to cover cellular energetic demands and do not rely on mitochondrial respiration-coupled energy production. In contrast, at later stages, chondrocytes also use mitochondrial respiration and may be dependent on intact mitochondria.
Figure 3.
Analysis of the energy production in Cre and CreTW chondrocytes. The Seahorse XF Cell Mito Stress assay was used to determine the bioenergetics profile in isolated chondrocytes from 1-d-old (n = 2 Cre, n = 3 CreTW) and 1-mo-old Cre and CreTW mice (n = 3). (A) The OCR over time was studied. (B) Basal respiration, ATP production-coupled respiration, and maximal and spare respiratory capacities were determined by adding the ATP synthase inhibitor oligomycin A (2 µM), the uncoupler of the RC FCCP (1 µM), and the inhibitors of complex III and I antimycin A (2.5 µM) and rotenone (0.5 µM), respectively (Coutelle et al., 2014). (C) The OCR and extracellular acidification rate were plotted to obtain the energetic profile of the chondrocytes. *P < 0.05, **P < 0.01.
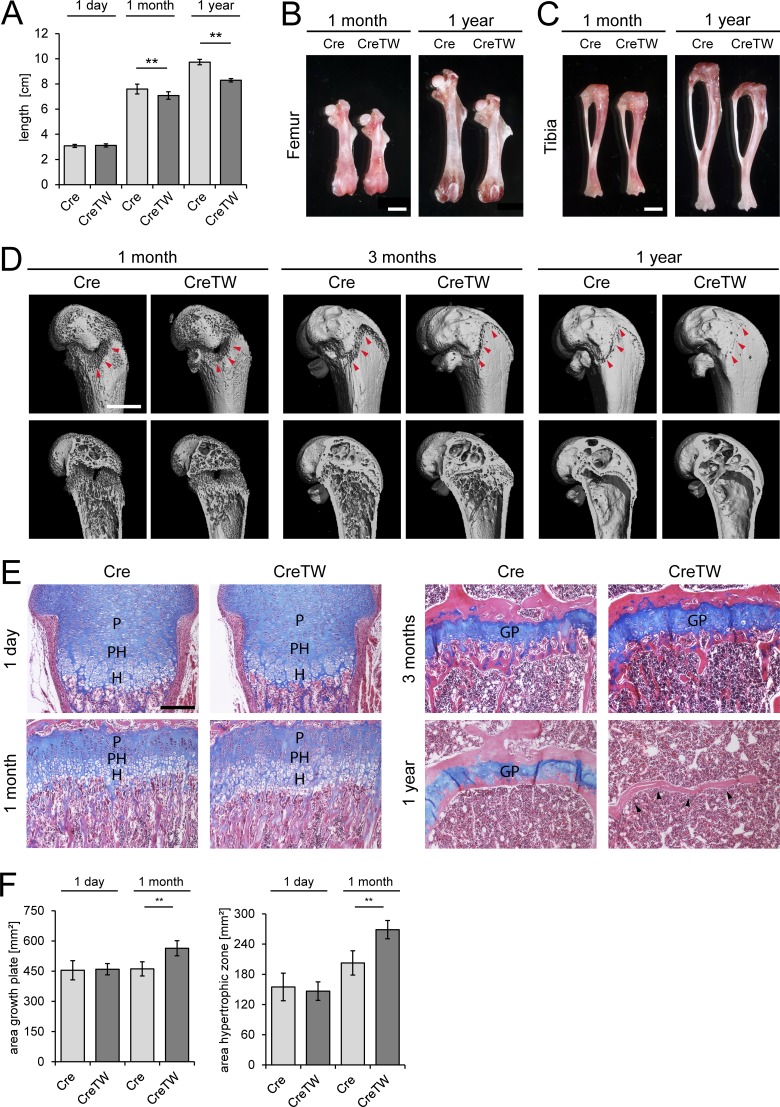
Next we studied the consequences of the lack of RC activation for skeletal growth in CreTW mice. No length alterations were observed in newborns, while in 1-mo-old and, more prominently, in 1-yr-old CreTW mice a significant reduction in body length was detected (Fig. 4 A). Accordingly, CreTW femora and tibiae were significantly shorter in 1-mo-old and 1-yr-old mice than in controls (Fig. 4, B and C). In addition, µCT measurements were performed on femora of 1-, 2-, 3-, 4-, and 6-mo-old mice and 1-yr-old mice. Quantification of the µCT measurements revealed that the trabecular bone mass at the distal femoral metaphysis is increased in 1-mo-old CreTW mice compared with the control mice, as indicated by a higher trabecular number (Tb.N) combined with a smaller trabecular separation (Tb.Sp). However, in older CreTW mice, the trabecular bone mass was decreased, and CreTW mice have fewer trabeculae with a greater distance between them (Fig. S1 B). In comparison to controls, the growth plate was smaller in 4-mo-old mice, partially lost in older animals, and completely closed in 1-yr-old CreTW animals (Fig. 4 D, arrowheads; Fig. S1). Hematoxylin/eosin and alcian blue–stained, in-plane matched sections showed that directly after birth there were no differences in the organization of the growth plate between Cre and CreTW mice (Fig. 4, E and F). At 1 mo, the growth plate was expanded and the prehypertrophic/hypertrophic area was even enlarged, whereas at 1 yr no cartilage remained in CreTW mice. Hence, lack of RC activation in chondrocytes severely disturbs growth plate organization, induces premature growth plate closure, and inhibits skeletal growth in older animals, mimicking the abnormal skeletal growth in patients with mitochondrial diseases.
Figure 4.
Analysis of the skeletal phenotype in mice with cartilage-specific RC inactivation. (A) Quantification of the body length (nose to tail) in 1-d-, 1-mo-, and 1-yr-old mice (n ≥ 10 mice per genotype and time point). (B and C) Representative femora (B) and tibiae (C) of 1-mo- and 1-yr-old mice. Bar: 2 mm. (D) µCT pictures of the femur of 1-mo-, 3-mo-, and 1-yr-old mice. The localization of the growth plate is indicated (red arrowheads). Bar: 1 mm. The pictures are also included in Fig. S1. (E) In-plane matched paraffin sections from Cre and CreTW femora of 1-d-, 1-mo-, 3-mo-, and 1-yr-old mice were stained with hematoxylin (nuclei, purple), eosin (cytoplasm, pink), and alcian blue (proteoglycans, blue). The proliferative (P), prehypertrophic (PH), and hypertrophic (H) zones and the growth plate (GP) are marked. Black arrowheads indicate loss of growth plate cartilage in 1-yr-old CreTW mice. Bar: 200 µm. (F) Quantification of the total area of growth plate cartilage and of the hypertrophic zone in femoral growth plate cartilage from 1-d- and 1-mo-old Cre and CreTW mice. Quantitative data are mean ± SD (n ≥ 5 animals per genotype and time point were analyzed). **P < 0.01.
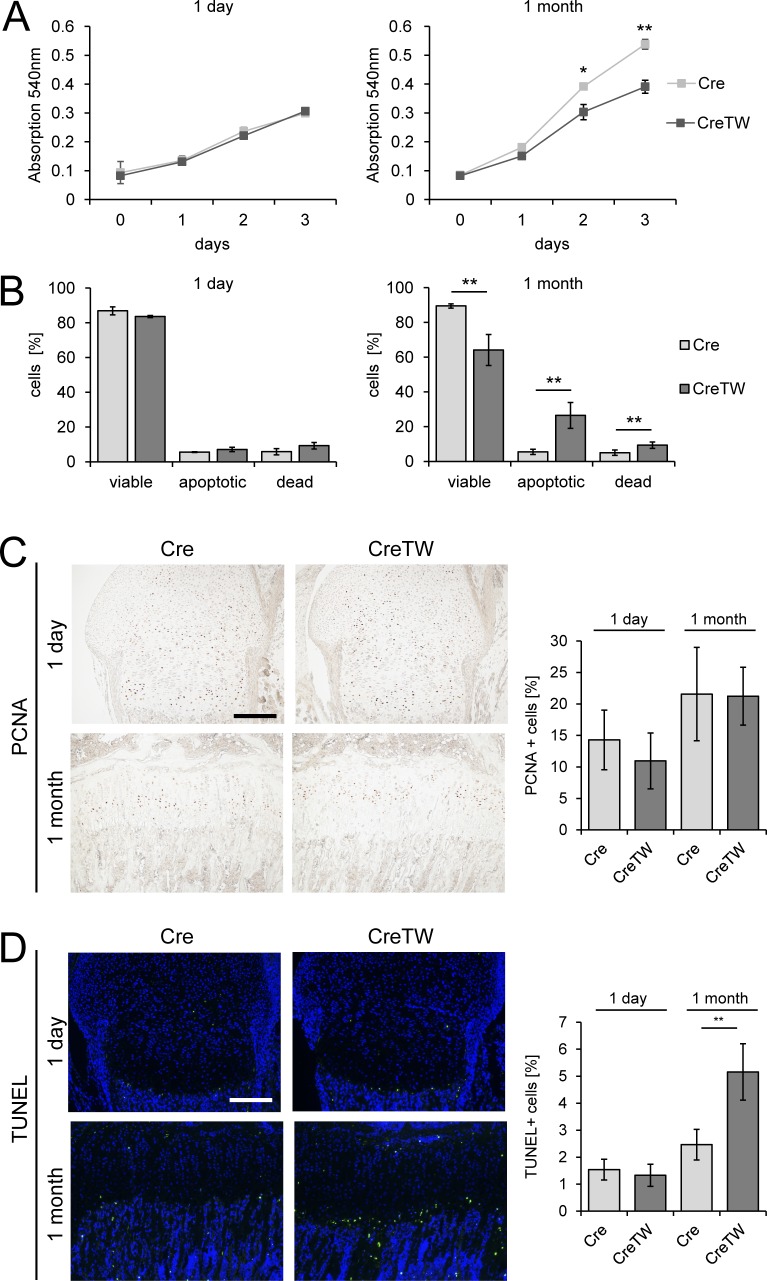
To understand the link between lack of RC activation and reduced skeletal growth, chondrocyte proliferation and cell death were analyzed in cartilage of CreTW mice. Chondrocytes isolated from newborns showed similar proliferation rates in culture, whereas cell numbers were significantly reduced in CreTW chondrocytes taken from 1-mo-old animals after 2 and 3 d in culture (Fig. 5 A). The proportion of apoptotic and necrotic cells was significantly increased in cultured chondrocytes from 1-mo-old CreTW mice, while no differences were detected in chondrocytes isolated from newborn mice (Fig. 5 B) as determined by AnxA5/propidium iodide (PI) staining and flow cytometry (Rosenbaum et al., 2011). In growth plate sections of newborn and 1-mo-old Cre and CreTW mice, a similar number of proliferating cell nuclear antigen (PCNA)–positive cells were detected (Fig. 5 C, graph). Only a few terminal TUNEL-positive dying cells were observed in newborn growth plates, whereas the numbers increased significantly in 1-mo-old CreTW mice compared with controls (Fig. 5 D, graph). Dying cells were only detected in the late hypertrophic zone, close to the cartilage–bone junction, but not in the proliferating zone. Therefore, lack of the RC activation does not impair proliferation of chondrocytes in vivo in older animals, but a higher number of CreTW chondrocytes are damaged along the differentiation process and undergo cell death in the late hypertrophic zone, where cartilage is replaced by bone.
Figure 5.
Characterization of proliferation and cell death in Cre and CreTW chondrocytes. (A) Cell numbers were determined as the incorporation of neutral red into the cytoplasm of viable cells over time in culture. (B) Cell viability was characterized by studying the proportion of AnxA5-Alexa647−/PI− viable, AnxA5-Alexa647+/PI− apoptotic, and AnxA5-Alexa647+/PI+ necrotic cells by flow cytometry after 3 d in culture. Quantitative data are mean ± SD (n = 3 mice per group). *P < 0.05, **P < 0.01. (C and D) The number of proliferating PCNA-positive (C) and apoptotic TUNEL-positive (D) cells was determined on paraffin sections from growth plate cartilage of 1-d- and 1-mo-old Cre and CreTW mice. DAPI staining was used to visualize the nuclei. Brightness was increased to visualize the green TUNEL signal. Bars: 200 µm. The numbers of proliferating PCNA-positive (C) and dying TUNEL-positive (D) cells in femoral growth plate cartilage from 1-d- and 1-mo-old Cre and CreTW mice were determined. The percentage of PCNA-positive or TUNEL-positive cells within the total cell population is shown. Quantitative data are mean ± SD (n ≥ 5 animals per genotype and time point). *P < 0.05, **P < 0.01.
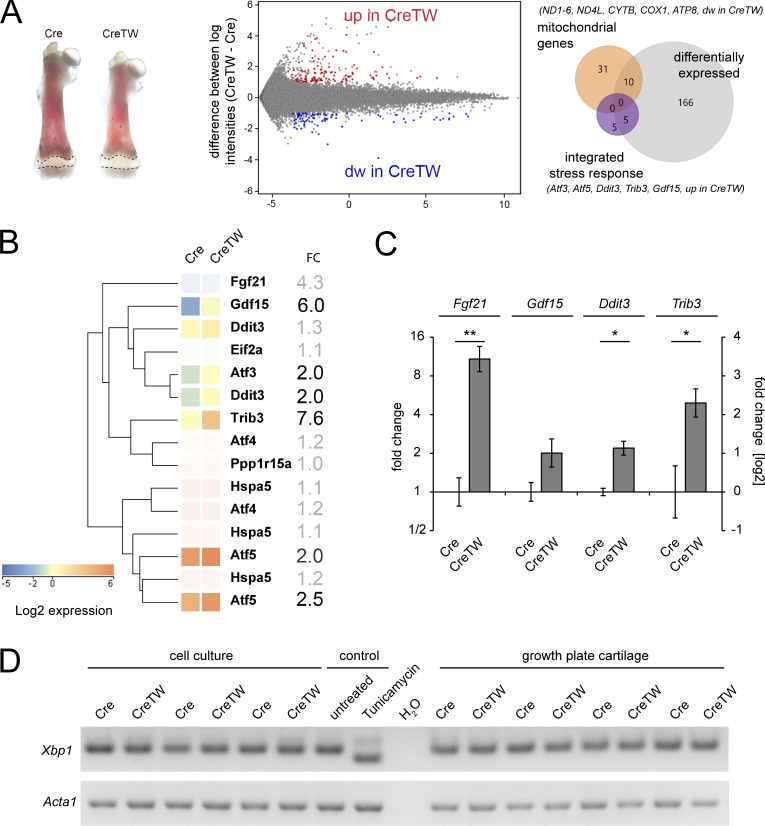
To define the molecular pathways leading to impaired chondrocyte survival, isolated femoral growth plates from 12-d-old Cre and CreTW mice were subjected to transcriptome analysis using established protocols (Belluoccio et al., 2010a; Bluhm et al., 2017; Bergmeier et al., 2018). At this time point, length differences between Cre and CreTW femora were already detected and still sufficient amounts of growth plate cartilage could be isolated. Bioinformatic analysis revealed that within the 181 entities, which were found differentially expressed in CreTW cartilage compared with control, many were described as mitochondrial genes. mRNA levels for several mtDNA-encoded subunits of the RC complexes (i.e., the NADH dehydrogenase complex [Ndx], cytochrome B [Cytb], and ATP-synthase 8 [Atp8]) were significantly lowered in CreTW mice (Fig. 6 A). Lower RC mRNA levels will result in a decrease in RC protein complexes and therefore cause RC inactivation in growth plate cartilage of CreTW mice. On the other hand, genes encoding main players of the integrated stress response (ISR; Quirós et al., 2016) were up-regulated in CreTW mice (Fig. 6 B). The activating transcription factor 3 (Atf3), Atf5, and DNA damage–inducible transcript 3 (Ddit3-Chop) were twofold enhanced, whereas the growth differentiation factor 15 (Gdf15) and tribbles homologue 3 (Trib3) were strongly increased in CreTW mice (Fig. 4 B). Fibroblast growth factor 21 (Fgf21) mRNA was increased fourfold, but expressed at only very low absolute levels. In contrast, Atf4 was significantly expressed but not differentially regulated. ISR genes were further validated by quantitative PCR (qPCR) analysis (Fig. 6 C). Here, the expression of Fgf21, Gdf15, Ddit3 (Chop), and Trib3 was clearly increased in CreTW chondrocytes.
Figure 6.
Determination of transcriptome changes in the growth plate cartilage and in cultured chondrocytes of CreTW mice. The transcriptome of femoral growth plate cartilage (dashed line) isolated from 12-d-old Cre and CreTW mice was compared using whole-genome mRNA arrays. (A) The growth plates of four individual animals per genotype were analyzed, and the corresponding expression intensity plots and Venn diagrams of differentially expressed genes are shown (fold-change ≥2, P < 0.05, Cre versus CreTW); mitochondrial genes as annotated in the array (ochre) and genes of the ISR are given (purple). (B) Cluster analysis: ISR genes. Entities not differentially expressed or below expression threshold are displayed in desaturated colors. For some genes, results of multiple probes are shown. FC, fold change. (C) The expression of selected genes was validated by qPCR using cDNA from growth plate cartilage of 12-d-old mice. Quantitative data are mean ± SD (n = 3 animals per genotype). *P < 0.05, **P < 0.01. (D) The induction of a UPR was studied by semiquantitative PCR using cDNA from isolated primary rib cage chondrocytes of 1-mo-old Cre and CreTW mice or femoral growth plate cartilage isolated from 12-d-old mice. The expression of Xbp1 and spliced sXbp1 mRNA was determined. Tunicamycin-treated chondrocytes were used as control.
The ISR can be induced by an unfolded protein response (UPR) in the ER. We therefore determined the processing of X-box binding protein 1 (Xbp1) mRNA, which is spliced to a shorter mRNA (sXbp1) during UPR. The RNA of growth plate cartilage from 12-d-old mice and from cultured chondrocytes of 1-mo-old mice was isolated and the relative abundance of the Xbp1 transcripts was analyzed by semiquantitative PCR. The unprocessed Xbp1 transcript was detected in growth plate cartilage and in isolated chondrocytes of Cre and CreTW mice, but not the processed spliced form (Fig. 6 D). The results show that an Xbp1-dependent UPR does not occur in CreTW mice.
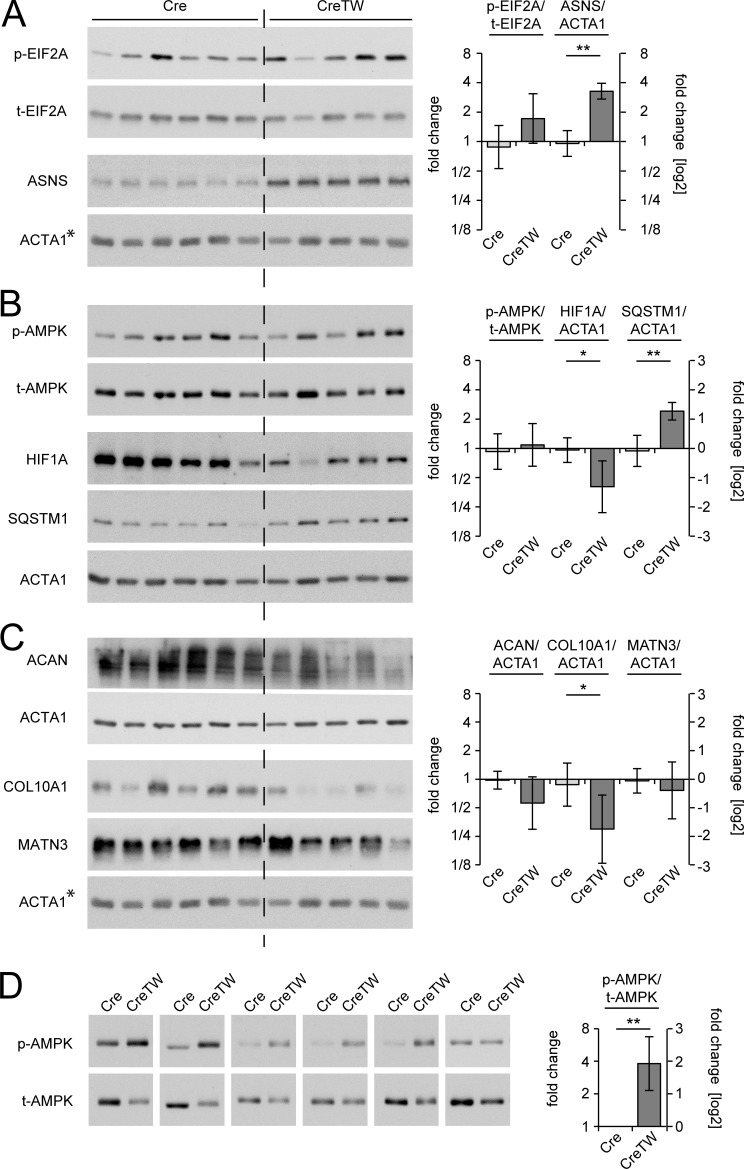
If the ISR causes the phenotype, its central mediators of the ISR should be activated. We therefore analyzed the phosphorylation of EIF2a and the expression levels of the stress response target gene asparagine synthetase (ASNS;Quirós et al., 2016; Lomelino et al., 2017) in growth plate cartilage from 12-d-old mice. The phosphorylation of EIF2a may have been increased in CreTW mice, but variations in the ratio of phosphorylated EIF2a to total EIF2a were detected (Fig. 7 A), and ASNS levels were significantly increased in lysates of CreTW mice compared with controls. The results point to the induction of an ISR in growth plate cartilage of CreTW mice. Energetic stress in old CreTW chondrocytes could also be sensed by AMP-activated protein kinase (AMPK). The phosphorylation of AMPK was determined in growth plate cartilage from 12-d-old mice (Fig. 7 B). Here, variations in the phosphorylation of AMPK were detected irrespective of the genotype. We next studied AMPK activation in chondrocytes isolated from older mice. Chondrocytes of 1-mo-old mice were cultured for 3 d in growth medium and total and phosphorylated AMPK levels were determined in extracts by immunoblot analysis (Fig. 7 D). A strong phosphorylation of AMPK was observed in cultured 1-mo-old CreTW chondrocytes compared with the Cre control. Hence, AMPK senses energy depletion when ATP levels were reduced in older CreTW chondrocytes (see Fig. 2 G).
Figure 7.
Regulation of metabolic sensitive pathways and ECM proteins. (A–C) Extracts of Cre (n = 6) and CreTW (n = 5) growth plates were analyzed for the activation of the integrated stress response (A; p-EIF2A, t-EIF2A, ASNS), the metabolic response (B; p-AMPK, t-AMPK, HIF1A, p62/SQSTM1), and for ECM proteins (C; ACAN, COL10A1, MATN3) by immunoblotting. Actin was used as loading control. *As all proteins shown in A and C were tested on a single blot, the same actin control is included in both panels. (D) The activation of AMPK was determined in extracts of cultured chondrocytes from individual isolations of 1-mo-old growth plate cartilage of Cre and CreTW mice (n = 6). ImageJ analysis was used for quantification, and fold changes in protein activation or expression between Cre and CreTW are shown (graphs). Quantitative data are mean ± SD. *P < 0.05, **P < 0.01. p-, phosphorylated protein; t-, total protein.
Respiration inactivation was described to repress HIF1A protein synthesis through AMPK-dependent signaling pathways (Hsu et al., 2013; Martínez-Reyes et al., 2016) and to inhibit the autophagic flux (López de Figueroa et al., 2015). As p62/SQSTM1 accumulates when autophagy is inhibited (Bjørkøy et al., 2009) we next determined HIF1A and p62/SQSTM1 protein levels in extracts of growth plate cartilage isolated from 12-d-old Cre and CreTW mice (Fig. 7 B). Interestingly, full-length HIF1A protein levels were decreased and p62/SQSTM1 levels were increased in lysates of the growth plate derived from 12-d-old CreTW mice compared with control. A decrease in HIF1A and inhibition of autophagy can suppress ECM production (Pfander et al., 2003; Kang et al., 2017). We therefore studied aggrecan, collagen X, and matrilin 3 levels in growth plate cartilage of 12-d-old mice (Fig. 7 C). Here a significant reduction in the protein amounts of collagen X was detected in CreTW mice compared with control.
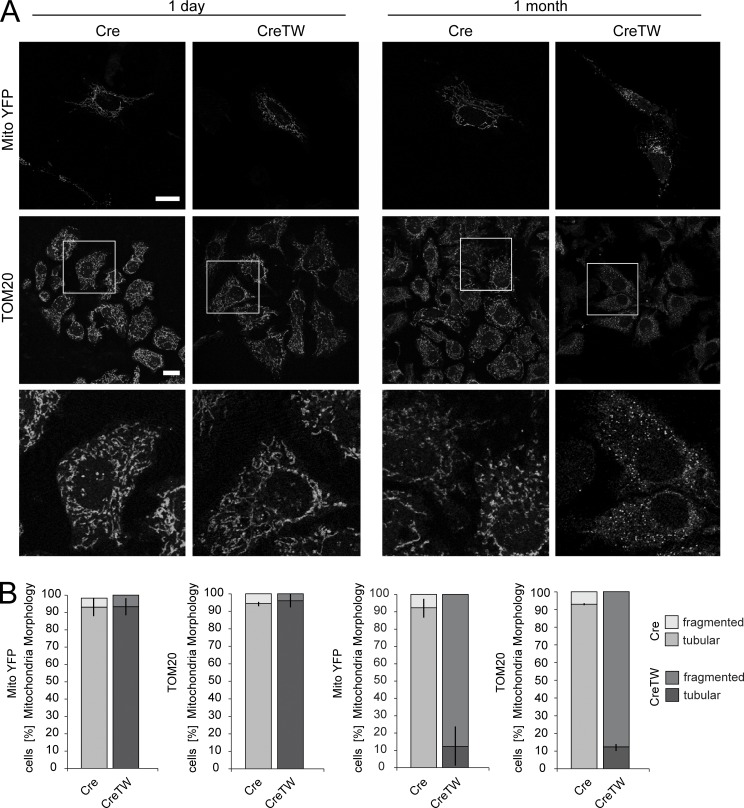
Accumulation of p62/SQSTM1 protein points to a reduction in the autophagic flux and to an accumulation of damaged mitochondria in older CreTW chondrocytes. Therefore, mitochondrial fragmentation in cultured chondrocytes from newborn or 1-mo-old Cre and CreTW mice was characterized. Mitochondrial membrane organization was visualized by transfection of a reporter construct encoding the YFP linked to a mitochondrial targeting sequence (pOCT-YFP; Neuspiel et al., 2008) or by immunofluorescence detection of the endogenous mitochondrial outer membrane protein TOM20 (Fig. 8 A). Mitochondria were elongated and formed an extended membrane network in young chondrocytes and in older Cre chondrocytes. In contrast, massive mitochondrial fragmentation was observed in older CreTW chondrocytes accompanied by a pronounced increase in vesicle-like structures, irrespective of the detection assay. Quantification of mitochondrial fragmentation per cell (Fig. 8 B) confirmed the strong increase in older CreTW chondrocytes. The results demonstrate that loss of membrane potential inhibits the removal of damaged mitochondria by autophagy and induces severe mitochondrial fragmentation in older chondrocytes.
Figure 8.
Mitochondrial membrane organization in Cre and CreTW chondrocytes. (A) Cultured chondrocytes from 1-d- or 1-mo-old Cre and CreTW mice were transfected with a MitoYFP reporter gene construct or immunostained with a TOM20-specific antibody to visualize mitochondrial membrane organization. (B) Quantification of the tubular and fragmented mitochondria (n ≥ 3 per genotype) is given as mean ± SD. Significant changes (P < 0.01) between genotypes were detected for isolated chondrocytes of 1-mo-old mice. The brightness and contrast of the fluorescence signal was adjusted to enable visualization. Bars: 10 µm.
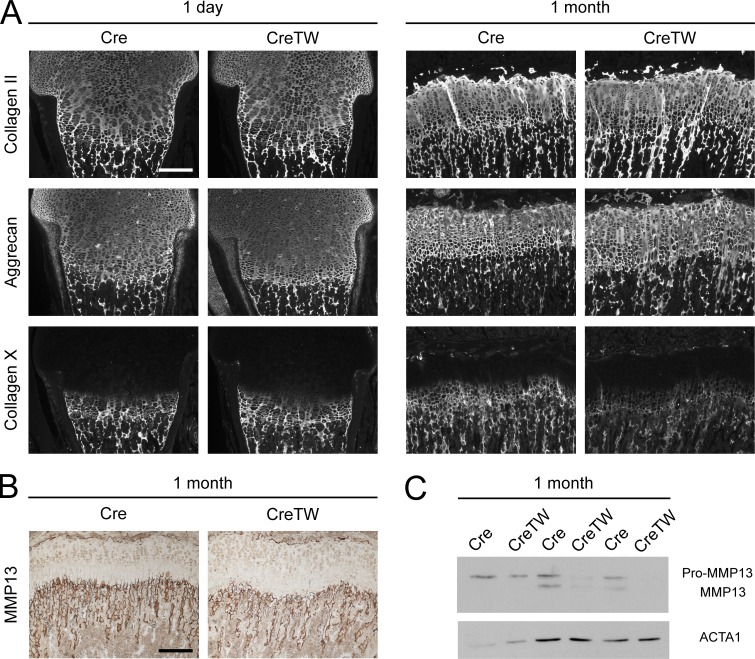
Mitochondrial damage and energy loss can be translated into cellular damage responses and cell death (Quirós et al., 2016). More important, damage responses in chondrocytes have been shown to destabilize the organization of the ECM, which then results in the expansion of the hypertrophic growth plate cartilage and reduced skeletal growth (Rajpar et al., 2009; Tsang et al., 2010). We could detect a reduction in collagen X in cultured older CreTW chondrocytes and hypothesized that mitochondrial damage and energy loss in chondrocytes also disturbs hypertrophic ECM organization in situ. Hence, the composition of the ECM in the growth plate of CreTW mice was studied by immunofluorescence analysis. In newborns, the main components of the cartilage ECM, the fibrillar collagen II, the extrafibrillar proteoglycan aggrecan, and the network-forming collagen X were normally distributed within the growth plate of CreTW mice (Fig. 9 A and Fig. S2, A and B). In 1-mo-old CreTW mice, the collagen II+/aggrecan+ growth plate area was enlarged, which shows that RC inactivation results in a transiently expanded growth plate (see also Fig. 4, E and F) accompanied by an enlargement of the cartilage ECM. Collagen X levels were strongly reduced (Fig. 9 A and Fig. S2B) and metallopeptidase 13 (MMP13; Fig. 9 B and Fig. S2 B) levels were decreased in the hypertrophic growth plate of 1-mo-old CreTW mice compared with Cre control. The decrease in MMP13 was confirmed by immunoblot analysis of rib cage chondrocytes from 1-mo-old CreTW mice (Fig. 9 C). Collagen X and MMP13 are specifically expressed in hypertrophic growth plate cartilage to stabilize and remodel the transition zone between cartilage and bone. Loss of collagen X and MMP13 is linked to expanded growth plates (Inada et al., 2004; Grskovic et al., 2012). Therefore, RC dysfunction in CreTW mice induces severe damage in the hypertrophic chondrocytes with a transient increase in hypertrophy, destabilization of the ECM at the cartilage–bone junction, and growth plate degeneration in older CreTW mice.
Figure 9.
Consequences of RC inactivation for ECM organization in CreTW mice. (A) Distribution of collagen II, aggrecan, and collagen X protein was analyzed by immunofluorescence staining. (B) Immunohistochemical staining was used to localize MMP13 protein in 1-mo-old growth plate. Pictures are representative of n ≥ 3 per genotype and time point and are also included in Fig. S2. Bars: 200 µm. The pictures are also included in Fig. S1. (C) Immunoblot analysis of MMP13 and actin (ACTA1) in cultured chondrocytes of 1-mo-old Cre and CreTW mice.
Collectively, these results provide a molecular scenario that explains why a RC defect impairs skeletal growth in CreTW mice and may cause short stature in many patients with mitochondrial diseases. Moreover, the results illustrate the importance of the metabolic switch from fetal glycolysis to RC activation in growth plate cartilage after birth to promote postnatal growth, which may apply to various other cellular systems that are challenged with the transition from fetal to postnatal life.
Discussion
Anaerobic glycolysis was considered the main energy source for skeletal growth due to the avascular nature of growth plate cartilage with consequently low oxygen supply, but surprisingly, mitochondrial diseases and RC dysfunction often present with slow growth and short stature. We proposed that RC activity is important for cartilage-mediated skeletal growth, but experimental systems to define the role of the RC in vivo were missing and, to our knowledge, the activity of the RC in growth plate cartilage was never determined.
Blood vessel infiltration supports the activation of respiration in the postnatal growth plate cartilage
We now provide the first evidence that the RC is active in growth plate cartilage at late stages of postnatal development. In newborns, complex IV activity was restricted to cartilage of the lateral growth plate and the chondro-osseous junction, both in close vicinity to the vasculature, while no complex IV activity was detectable in the zone of proliferating chondrocytes. In contrast, complex IV is active in proliferating chondrocytes of 13-d-old mice and, even more markedly, of 1-mo-old mice. At this stage of development, the growth plate cartilage normally decreases in size (Sivaraj and Adams, 2016) and blood vessels invade the distal end of the femur cartilage to form the secondary ossification center, providing oxygen and nutrients to the distal side of the growth plate. Therefore, the locally increased oxygen tension or nutrient availability in older mice may induce a metabolic switch from glycolytic energy production to respiration in these chondrocytes. This is in line with the lack of changes in skeletal growth, cartilage organization, or ECM deposition in CreTW newborn mice, despite a similar degree of mtDNA depletion and reduction in mtDNA-encoded RC subunits as in 1-mo-old CreTW mice. Hence, glycolysis is sufficient to promote cartilage-mediated skeletal growth in the hypoxic fetal and neonatal environment, but not in later postnatal stages with increasing energy demand, when the growth plate cartilage becomes less hypoxic. Here the activation of the RC is crucial for energy production and for the inhibition of premature growth plate cartilage differentiation and degradation as demonstrated by the later onset and progression of the cartilage phenotype in older CreTW mice. Therefore, respiration is essential to provide energy for postnatal cartilage growth but is also needed to control cartilage differentiation and prevent premature cartilage degeneration and its transformation into bone.
Respiration promotes growth plate differentiation and hypertrophic ECM formation in aging mice
In 3–4-mo-old CreTW mice, growth plate cartilage is reduced in size and completely resorbed and transformed into bone in older CreTW mice, while still present in control animals. In mice, the growth plates are not fully degraded (Hirai et al., 2011), in contrast to humans. The phenotype of CreTW mice shows that the activation of the RC is important to prevent the complete resorption of the growth plate. Presumably, RC activity provides sufficient energy to maintain the growth plate cartilage, but the detailed analysis of the organization of the growth plate also showed that the RC is needed to regulate hypertrophic chondrocyte differentiation, survival, and ECM stabilization.
RC function may be important for progenitor cells (Mizuhashi et al., 2018) that are normally responsible for sustaining growth plate chondrocytes and growth plate–derived osteoblasts. These progenitors could also be targeted by Col2-Cre expression and may increasingly suffer from mtDNA depletion during aging in CreTW mice and give rise to chondrocytes (and osteoblasts) with a higher load of mtDNA damage compared with younger chondrocytes. Progenitor-derived older CreTW chondrocytes may accumulate further mitochondrial damage during replication cycles within the stacks of the growth plate and then reach a critical threshold in hypertrophic chondrocytes. A previous study indicated that terminally differentiated chondrocytes may undergo a maturation-dependent loss of mitochondrial function and become sensitive to signals for programmed cell death (Rajpurohit et al., 1999). This sensitivity may be increased in the presence of mtDNA damage to promote specifically the death of older hypertrophic CreTW chondrocytes and to destabilize the hypertrophic ECM.
In 1-mo-old CreTW mice we detected a transient enlargement of the area of the hypertrophic zone. This enlargement was accompanied by decreased amounts of important ECM-related proteins in this zone, e.g., collagen X and MMP13, while the amounts of main ECM components like collagen II or aggrecan were not affected by RC inactivation. Mitochondrial dysfunction can modulate MMP expression and inhibit MMP13 production in human chondrocytes cultures (Cillero-Pastor et al., 2013), and the decrease in collagen X and MMP13 is indicative of a general reduction of the hypertrophic ECM and ECM-remodeling proteins in cartilage of CreTW mice. Collagen II and aggrecan molecules have an exceptionally long lifetime and slow turnover (Verzijl et al., 2000; Sivan et al., 2006), while collagen X and MMP13 are located in the chondro-osseous junction where the ECM is remodeled and cartilage is continuously replaced by bone. Therefore, collagen X and MMP13 production in the hypertrophic growth plate cartilage could be more susceptible to RC dysfunction in 1-mo-old CreTW mice.
Collagen X forms hexagonal networks (Kwan et al., 1991) and interacts with collagen II fibrils (Poole and Pidoux, 1989; Schmid et al., 1990) and proteoglycans (Chen et al., 1992) to stabilize the hypertrophic growth plate and the cartilage–bone junction, whereas MMP13 degrades various substrates to remodel the cartilage ECM. The lack of MMP13 or collagen X in mice results in expanded growth plates (Inada et al., 2004; Grskovic et al., 2012), like in our mice, and mutations in collagen X are often associated with short stature and waddling gait in humans (Chan and Jacenko, 1998; Bateman et al., 2005). Interestingly, increased hypertrophic differentiation and decreased collagen X or MMP13 expression are also believed to be induced by stress responses in cartilage (Tsang et al., 2010; Cameron et al., 2011; Yan et al., 2016). Hence, collagen X and MMP13 are required to maintain the integrity of the ECM and the growth plate cartilage at the chondro-osseous junction. We now show that RC inactivation induces a similar damage response in chondrocytes to destabilize the growth plate cartilage and consequently impair skeletal growth.
RC inactivation provokes a cellular stress response
Lack of RC activation has to be translated into cellular responses in CreTW mice. The transcriptome analysis of growth plate cartilage indicates that a form of mitonuclear ISR is induced in these chondrocytes. A mitochondrial UPR (UPRmt) has been mainly described in Caenorhabditis elegans, which is commonly induced as a response to mitochondrial stress signals in order to activate a compensatory gene expression program (Quirós et al., 2016; Shpilka and Haynes, 2018). The mammalian counterpart is driven by ATFs (Khan et al., 2017; Shpilka and Haynes, 2018). Interestingly, we could detect an increased expression of Atf3, which facilitates cell cycle exit and terminal differentiation of chondrocytes (James et al., 2006), modulates Mmp13 expression in chondrocytes (Chan et al., 2017), and can act as an adaptive response gene with proapoptotic functions (Thompson et al., 2009). The expression of Ddit3/Chop and Atf5 is also increased in CreTW cartilage, and DDIT3/CHOP protein has been reported before to mediate stress-induced apoptosis in cartilage (Cameron et al., 2011) and directly induce Atf5 expression to potentiate DDIT3/CHOP-dependent apoptosis (Teske et al., 2013). ATF5 is capable of activating the ISR in nematodes and presumably in mammals (Melber and Haynes, 2018). It harbors a mitochondrial targeting sequence potentially allowing it to immediately translate mitochondrial damage into cellular responses (Fiorese et al., 2016). These three transcriptional regulators may act in concert to induce the expression of damage-response genes, e.g., Asns, Gdf15, and Fgf21, but the molecular interactions remain to be determined. The secreted proteins GDF15 and FGF21 are described as mitokines produced in response to mitochondrial damage (Suomalainen et al., 2011; Kim et al., 2013), and interestingly, both are up-regulated in chondrocytes of CreTW mice. FGF21 directly acts on chondrocytes of the growth plate to inhibit collagen X expression and to mediate growth hormone insensitivity in hypertrophic chondrocytes (Wu et al., 2012; Wang et al., 2017), which could contribute to reduced collagen X protein levels in 1-mo-old mice. At later stages, FGF21 as well as GDF15 may act cooperatively to inhibit cartilage-mediated skeletal growth as both are known to exert effects on systemic energy homeostasis and growth hormone–mediated body growth (Chung et al., 2017). Patients with mitochondrial diseases often present with increased GFD15 and FGF21 serum levels (Yatsuga et al., 2015; Davis et al., 2016; Lehtonen et al., 2016; Montero et al., 2016; Ji et al., 2017), although clinical studies correlating growth retardation with these factors are needed. These mitokines could be produced locally in growth plate cartilage in response to RC dysfunction to inhibit cartilage-mediated skeletal growth, but they also to contribute to systemic metabolic effects on body growth in these patients.
Multiple factors recognize mitochondrial dysfunction, and it is well described that mtDNA damage and respiration failure lead to increased intracellular partial presure of oxygen (pO2), destabilized HIF1A levels, and impaired cell survival (Doege et al., 2005; Martínez-Reyes et al., 2016). Interestingly, we indeed observed reduced HIF1A in extracts of growth plate from 12-d-old CreTW mice. HIF1A was originally described to activate Sox9 in prechondrogenic mesenchymal cells and maintain differentiation and ECM production in response to hypoxia (Amarilio et al., 2007) during early embryonic development. However, the same group demonstrated that, in differentiated chondrocytes, HIF1A directly controls collagen hydroxylation and secretion during later embryonal development independently of Sox9 (Bentovim et al., 2012). HIF1A directly drives prolyl-4-hydroxylase expression, a central stabilizer of collagen triple-helix formation (Wagner et al., 2000), and lack of HIF1A leads to reduced collagen hydroxylation, arrested ECM secretion, and ER stress in differentiated growth plate chondrocytes in vivo (Bentovim et al., 2012). This is in line with the decrease in collagen X levels at the chondro-osseous junction and the occurrences of cellular stress in the growth plate of older CreTW mice. Moreover, it was shown that chondrocyte metabolism and HIF1A signaling control collagen modification and matrix resistance to protease digestion (Stegen et al., 2019). Hence, HIF1A can exert important effects on ECM homeostasis and the chondrocyte phenotype in the differentiated growth plate cartilage. Considering that terminal differentiated chondrocytes have a unique sensitivity to mitochondrial function (Rajpurohit et al., 1999), an increase in intracellular pO2 levels in older CreTW mice could decrease HIF1A levels to directly impair collagen hydroxylation and secretion, and chondrocyte survival at the chondro-osseous junction.
Diminished respiration can also inhibit ROS production, which is necessary for the activation of HIF1A (Martínez-Reyes et al., 2016) and the induction of autophagy (Marambio et al., 2010; Wang et al., 2011). We could also observe reduced ROS levels in extracts of the growth plate from 12-d-old CreTW mice and detect the accumulation of fragmented mitochondria and the reduction in the autophagic flux in older CreTW chondrocytes. Autophagy was previously described to promote chondrocyte differentiation, survival (Vuppalapati et al., 2015), and ECM production (Pfander et al., 2003) and is likely to contribute to cell death and ECM dysregulation in CreTW mice.
The consequences of RC inactivation in mitochondria of growth plate cartilage may not necessarily be explained by the simplistic view of a single key mechanism or pathway, but in our view, a unique damage response is evoked due to energy depletion in older CreTW mice. Energy depletion is first sensed in hypertrophic chondrocytes based on their unique sensitivity to respiration dysfunction. High pO2 and reduced ROS levels can decrease HIF1A levels to impair hydroxylation of collagens and ECM secretion. In addition, inhibition of autophagy can induce a stress response to impair ECM homeostasis and chondrocyte differentiation, and the combination of these factors destabilizes the ECM and promotes chondrocyte death and growth plate degradation and closure in older CreTW animals.
In conclusion, this study demonstrates, to our knowledge for the first time, that, in contrast to the current view, postnatal cartilage development requires RC activity in chondrocytes and indicates that RC activation protects from premature growth plate closure and growth retardation. Glycolysis is sufficient to drive fetal cartilage development but cannot compensate for the lack of RC activation during postnatal skeletal growth in CreTW mice and most likely in humans with mitochondrial diseases and RC dysfunction. Mechanistically, the phenotype is caused by a defect in energy production, which disturbs metabolic signaling and chondrocyte hypertrophy, destabilizes the ECM, and finally induces cell death. Hence, the metabolic switch in cartilage from anaerobic glycolysis to RC activation is crucial to drive postnatal skeletal development, while RC dysfunction accelerates premature growth plate closure and induces mitochondria-related skeletal pathologies. In general, this study illustrates the importance of metabolic adaptation for the coordination of postnatal cartilage development and homeostasis.
Materials and methods
Animals
Previously established R26-K320E-TwinkleloxP/+ C57BL/6J mice were crossed with Col2a1-Cre C57BL/6N mice (Ovchinnikov et al., 2000; Bluhm et al., 2017) to induce cartilage-specific TwK320E expression in heterozygous Col2a1-Cre-R26-K320E-TwinkleloxP/+ mice (TW). Col2a1-Cre C57BL/6 mice of the same litter (Cre) were used as controls. For neonatal assessment of skeletal development, newborns of both sexes were analyzed. All experiments were performed in accordance with the guidelines of the German animal protection law (institutional review board Landesamt für Natur, Umwelt und Verbraucherschutz Nordrhein-Westfalen).
Histology and immunohistochemistry
Microtome sections (5 µm) of decalcified right femora from mice were used for morphological and immunohistological analyses. The organization of the growth plate was assessed by hematoxylin/eosin/alcian blue staining and the distribution of ECM proteins was determined by immunofluorescence microscopy. To detect collagen II and X, sections were pretreated with 0.025% pepsin for 15 min at 37°C, 5 mg/ml hyaluronidase (Sigma-Aldrich) for 30 min at 37°C, and 10 µg/ml proteinase K (Sigma-Aldrich) for 10 min at 50°C. To study aggrecan distribution, sections were preincubated with 10 µg/ml proteinase K for 10 min at 50°C, and to localize the laminin γ1 chain, sections were incubated with 0.005% Trypsin for 10 min at 37°C. The following primary antibodies were used: mouse anti-collagen II (1:500; Calbiochem), mouse anti-collagen X (1:100; hybridoma clone X53; Girkontaite et al., 1996), rabbit anti-aggrecan (1:100; Millipore/Chemicon), and rabbit anti–laminin γ1 chain (1:2,000; Tunggal et al., 2003). Corresponding secondary antibodies coupled to Cy3 (1:800; Jackson ImmunoResearch) were applied. To determine the distribution of MMP13 and PCNA, immunohistochemical stainings were performed. Sections were pretreated with citrate buffer (pH 6.0) for 1 h at 60°C. Primary antibodies against MMP13 (1:500; Abcam) or PCNA (1:200; Abcam) were applied, and appropriate secondary antibodies labeled with horseradish peroxidase (Dako) were used for detection (Zytomed Systems). Sections were analyzed using a Nikon Eclipse TE2000-U microscope.
mtDNA copy number
DNA of femoral head cartilage was extracted using phenol-chloroform extraction, and the relative mtDNA content was analyzed by qPCR (Baris et al., 2011). The CT values of the mtDNA-encoded Nd2 and the nuclear DNA–encoded Pecam were determined using the QuantiTect SYBR Green PCR assay (Qiagen). Pecam amplification was used for normalization and the fold change was calculated with the ΔΔCT method (Pfaffl, 2001).
In situ detection assays
The DeadEnd Fluorometric TUNEL assay (Promega) was applied to paraffin sections (5 µm) as previously described (Bluhm et al., 2017). Briefly, sections were digested with 20 µg/ml proteinase K for 8 min at room temperature and then incubated with 4% PFA for 10 min. The rTdT labeling reaction was performed for 1 h at 37°C. Nuclei were detected with DAPI and then sections were embedded in Mowiol. The number of TUNEL-positive cells was quantified by ImageJ software (National Institutes of Health). Deparaffinized sections (5 µm) were permeabilized with 0.2% Triton X-100 in TBS for 10 min, washed with TBS for 10 min, and incubated with the 5-bromo-4-chloro-3′-indolyphosphate (BCIP)/nitro-blue tetrazolium (NBT) staining solution (86 µg/ml BCIP, 162 µg/ml NBT, and 5 mM MgCl2 in 10% polyvinyl alcohol/0.1 M Tris [pH 9.0]/0.1 M NaCl at 30°C) until staining was visible. Sections were extensively washed with H2O, stained with 0.001% Fast Green FCFfor 5 min, and embedded in gelatin. To visualize complex IV activity in situ, cryotome sections (10 µm) of right mouse femora were prepared using Kawamoto's film method (Kawamoto, 2003) and stained for complex IV (CYTOCOX) activity as previously described (Sciacco and Bonilla, 1996).
Cell culture
To analyze sufficient cell numbers in downstream experiments, rib cage growth plates from newborn and 1-mo-old mice were incubated in collagenase II solution (450 U/ml; Biochrom) for 6 h at 37°C. Femoral growth plates could not be used due to the insufficient yield in cell numbers, especially from 1-mo-old mice. To obtain adequate numbers of cells, rib cage–derived chondrocytes from two to three animals of both sexes were pooled unless otherwise stated (Fig. 7 B) and cultured in DMEM-F12 supplemented with 10% FCS (Biochrom), penicillin (100 U/ml; Biochrom), streptomycin (100 µg/ml; Biochrom), ascorbate (44 µg/ml; Sigma-Aldrich), and L-ascorbate-2-phosphate (130 µg/ml; Sigma-Aldrich). After 24 h, chondrocytes were detached with collagenase II in DMEM-F12 (2 mg/ml; Biochrom), centrifuged, and resuspended in medium. Cells were cultured for another 3 d and then used for experiments. The pH of the cell culture supernatant was determined using a Seven-Easy-pH-Meter (Mettler).
Mitochondrial mass, membrane potential, ATP production, MitoSOX, and NADH
To assess mitochondrial mass, chondrocytes were detached with collagenase II, washed with cell culture medium, and stained with 50 nM MitoTracker Deep Red dye in cell culture medium for 30 min at 37°C. Cells were washed and resuspended in 5% FCS/PBS containing PI. The mean fluorescence intensity of the MitoTracker Deep Red signal (APC-A; 660 ± 20 nm) was determined using a FACS Canto II flow cytometer (Becton Dickinson). To evaluate the mitochondrial membrane potential, chondrocytes were resuspended in cell culture medium containing 5 µM JC-1 and then incubated for 15 min at 37°C. Cells were washed with 5% FCS/PBS and resuspended in 1 µM SYTOX blue (Thermo Fischer Scientific)/5% FCS/PBS; after 15 min of incubation on ice, they were analyzed by flow cytometry. The ratio of the mean PE-A (mitochondrial JC-1+) and the mean FITC signal (cytoplasmic JC-1+) from SYTOX blue− viable chondrocytes was calculated. The ATP content of chondrocytes was studied using the ATP Bioluminescence Assay Kit CLS (Sigma-Aldrich). Briefly, 9 × 104 chondrocytes were cultured in a 12-well plate for 3 d, detached with collagenase II, washed with PBS, and resuspended in 100 µl PBS. The protein content of 4.5 × 104 cells was determined with the bicinchoninic acid assay (Interchim). The remaining 4.5 × 104 cells were dissolved in 450 µl of hot 100 mM Tris/4 mM EDTA (pH 7.8) and incubated for 2 min at 100°C. The luciferase reagent was added, and the ATP-dependent production of a bioluminescence signal was determined (Infinite M1000; Tecan) and normalized to the protein amount. Reactive oxygen levels of chondrocytes were analyzed by the MitoSOX red staining assay. Chondrocytes were incubated for 15 min at 37°C in 250 µl of 5 µM MitoSOX red (Thermo Fisher Scientific) in DMEM without phenol red, and the mean fluorescence intensity was determined by flow cytometry. Reducing equivalents were analyzed using the CellTiter 96 AQueous One Solution (Promega) according to the manufacturer’s specification. Cells were cultured for 2, 24, 48, and 72 h; substrate was added; and production of colored formazan product was detected at 492 nm using a 96-well plate reader (Sunrise; Tecan).
Bioenergetics profiling
For bioenergetics profiling of chondrocytes, 10,000 cells/well were seeded on Seahorse 96-well plates and analyzed according to Agilent protocols (MitoStress Kit) with a Seahorse XF96 analyzer. Parameters of metabolic function depicted as bar charts in the figures were calculated according to Agilent protocols (Coutelle et al., 2014). Basal respiration: (last rate measurement before oligomycin injection) − (minimum rate measurement after rotenone/antimycin A injection) = nonmitochondrial oxygen consumption); proton leak: (minimum rate measurement after oligomycin injection) − (nonmitochondrial respiration); ATP production: (last rate measurement before oligomycin injection) − (minimum rate measurement after oligomycin injection); spare respiratory capacity: [(maximum rate measurement after FCCP injection) − (nonmitochondrial respiration) = maximal respiration)] − (basal respiration). Values were normalized to protein concentrations/well determined by DC Protein Assay according to the manufacturer’s protocol (BioRad).
µCT analysis
A high-resolution µCT scanner (µCT 35; Scanco Medical AG) was used to determine the microarchitecture of the left femora. Isolated bones were scanned with an isotropic voxel size of 7 × 7 × 7 µm using 70-kVp tube voltage, 114-µA tube current, and 400-ms integration time. Image noise was removed by preprocessing of the grayscale data of the raw CT images using a 3D Gaussian filter algorithm and separation of mineralized tissue from the soft tissue by a global thresholding procedure (Stauber and Müller, 2008). The segmentation steps were applied with support = 1, sigma = 0.8. For assessment of trabecular bone, 142 (Cre mice, 1 mm) or 107 (CreTW mice, 0.75 mm) slices were evaluated at the femoral metaphysis 1 mm (Cre mice) or 0.75 mm (CreTW mice) below the growth plate, which included only secondary spongiosa (dependent on the different femur length of the Cre and CreTW mice). The image data were segmented using a threshold (dependent on the age of the mice) of 19% (1 and 2 mo), 21% (3 mo), and 22% (4 mo, 6 mo, and 1 yr) of the maximum grayscale value. Trabecular parameters included bone volume fraction (BV/TV; 1), connectivity density (Conn-Dens; 1/mm3), Tb.N (1/mm), trabecular thickness (Tb.Th; mm), and Tb.Sp (mm). Statistical analysis (Mann–Whitney U test) was performed using IBM SPSS Statistics (version 21; IBM Deutschland GmbH). For whole-bone overview pictures, the image data were segmented using a threshold of 21% (1 mo), 24% (2 mo), 26% (4 mo), and 27% (3 mo, 6 mo, and 1 yr) of the maximum grayscale value.
Cell viability
Proliferation of chondrocytes was analyzed using the neutral red assay (Repetto et al., 2008). 7.5 × 103 chondrocytes were cultured in 96-well plates for 1, 2, or 3 d. Neutral red dye solution (0.5 mg/ml in 0.9% NaCl) was added and incubated for 2 h at 37°C. Cells were washed twice with PBS, and then the dye was extracted with 0.05 M Na2HPO4 in 50% ethanol overnight at −20°C. Absorbance was measured at 540 nm (Infinite M1000; Tecan). Apoptosis was studied by Annexin A5 (ANXA5) staining as described (Rosenbaum et al., 2011). Briefly, chondrocytes were detached with collagenase II, washed, and stained with ANXA5-Alexa647 and PI. The number of ANXA5-Alexa647−/PI− viable, ANXA5-Alexa647+/PI− apoptotic, and ANXA5-Alexa647+/PI+ dead cells was determined by flow cytometry (FACS Canto II; Becton Dickinson).
Gene expression analysis
The RNA from femoral growth plates of 12-d-old Cre and CreTW littermate mice or cultured chondrocytes was isolated by phenol-chloroform extraction, and RNA integrity was confirmed by microcapillary electrophoresis (2100 Bioanalyzer; Agilent). For microarray analysis, 100 ng RNA of growth plate cartilage from four Cre and CreTW mice, respectively, was amplified, labeled, and hybridized to a 8 × 60-K whole-genome mRNA microarray using the Agilent protocol. Microarrays were scanned (G2595C scanner; Agilent) and data were extracted and analyzed by GeneSpring V14.9 software (Agilent). mRNAs were selected according to their differential expression, their similar expression levels in all four microarrays of a given genotype, and their statistical significance. The arrays of the expression analysis have been deposited in GEO under accession number GSE130191.
qPCR
RNA was reverse transcribed into cDNA with the Omniscript RT assay (Qiagen) and used for semiquantitative PCR or mRNA SYBR Green– (Gdf15; Qiagen) or probe-based qPCR assays (Fgf21, Ddit3, Trib3; Roche, Invitrogen; Belluoccio et al., 2010a). The expression was normalized to Mapk7, constantly expressed in the proliferative, prehypertrophic, and hypertrophic zone of growth plate cartilage (Belluoccio et al., 2010b; Etich et al., 2015). The fold change was calculated with the ΔΔCT method (Pfaffl, 2001). The following primers were used: Actin (forward: 5′-GACGAGGCCCAGAGCAAGAG-3′; reverse: 5′-CTAGAGCAACATAGCACAGC-3′), Ddit3 (forward: 5′-ATCTCATCCCCAGGAAACG-3′; reverse: 5′-ATGTGCGTGTGACCTCTGTT-3′), Fgf21 (forward: 5′-AAGACACTGAAGCCCACCTG-3′; reverse: 5′-GAATGACCCCTGGCTTCAA-3′), Gdf15 (forward: 5′-GAGAGGACTCGAACTCAGAAC-3′; reverse: 5′-GACCCCAATCTCACCTCTG-3′; Wang et al., 2017), Mapk7 (forward: 5′-GGTTCATCTCAGACCCACCTT-3′; reverse: 5′-CTGTTATGGCTCGGTGGTG-3′), Trib3 (forward: 5′-GCTATCGAGCCCTGCACT-3′; reverse: 5′-ACATGCTGGTGGGTAGGC-3′), and Xbp1 (forward: 5′-GATCCTGACGAGGTTCCAGA-3′; reverse: 5′-ACAGGGTCCAACTTGTCCAG-3′).
Immunoblotting
Femoral growth plate, femur head cartilage, or cultured cells were taken up in radioimmunoprecipitation assay buffer containing 5% NP-40, 0.25% Triton X-100, 2.5% 750 mM NaCl, 100 mM Tris-HCl (pH 7.4), and cOmplete Protease Inhibitor (for metabolic signaling) and disrupted by sonication (Branson). Protein concentration was determined using the BC Protein Assay (Interchim) according to the manufacturer's recommendations, and equal amounts were resolved on SDS-polyacrylamide gels. After transfer onto a nitrocellulose membrane (Whatman), blots were incubated for 30–60 min at RT with 5% milk powder or 5% BSA in TBS–0.1% Tween20, and then a primary antibody cocktail was added. Corresponding secondary antibodies coupled to horseradish peroxidase (Dako) were applied, and antibody binding was visualized by enhanced chemiluminescence. Membranes were incubated in 0.2 M NaOH for 2 min, rinsed with water and TBS–0.1% Tween20, and after blocking reused for additional immunoblot experiments. The following antibodies were used: actin (1:2,000; Millipore), aggrecan (1:1,000; Millipore), p-/t-AMPK (1:1,000; CST), ASNS (1:500; Biorbyt), collagen X (1:500; (Girkontaite et al., 1996), p-/t-EIF2A (1:1,000; CST), HIF1A (1:500; Genetex), SQSTM1 (1:1,000; MBL), Matrilin3 (1:2,000; Klatt et al., 2000), MMP13 (1:1,000; Abcam), and RC complexes (1:500; Abcam). For quantification, band intensities were analyzed using ImageJ software.
Mitochondrial fragmentation
For immunofluorescence detection of the outer mitochondrial membrane protein, TOM20 chondrocytes were cultured for 24 h on glass coverslips in a 24-well plate, fixed with 4% PFA/PBS for 10 min at RT, permeabilized with 0.5% NP-40/PBS for 10 min, and incubated with 1% FCS/PBS for 30 min at RT. The TOM20 antibody (Santa Cruz) was added, and after 1 h at RT cells were washed three times with blocking buffer and incubated with appropriate Alexa-Fluor–conjugated secondary antibodies and DAPI for an additional 45 min at RT. In addition, chondrocytes were transfected with 1 µg plasmid DNA (pOCT-YFP; Neuspiel et al., 2008) per well using XtremeGENE HP (Roche) according to the manufacturer’s protocol in a 1:2 ratio (DNA:XtremeGENE HP) to fluorescently label mitochondria (MitoYFP). 24 h after transfection cells were fixed in 4% PFA/PBS for 10 min at RT. After mounting (Dako), images were acquired using a TCS SP5 microscope controlled with LAS AF Software (Leica).
Statistical analysis
Statistical significance was determined for n ≥ 3 using the unpaired two-tailed method of the Student's t test or Mann–Whitney U test (Fig. 4 D). P values <0.05 (*) were considered to be significant and P values <0.01 (**) as highly significant. Data are shown as mean ± SD. For relative changes, a one-sample Student’s t test was applied.
Online supplemental material
Fig. S1 demonstrates changes in femur organization and bone formation in 1-, 2-, 3-, 4-, and 6-mo-old and 1-yr-old Cre and CreTW mice using µCT analysis. In Fig. S2, the distribution of collagen II, aggrecan, collagen X, and MMP13 protein was determined in sections from three individual newborn and/or from 1-mo-old Cre and CreTW mice using immunostainings. Table S1 reports trabecular parameters of BV/TV and Conn-Dens (1/mm3).
Supplementary Material
Acknowledgments
This work was funded by grants from the Deutsche Forschungsgemeinschaft (BR2304/9-1, BR2304/12-1, and FOR2722 to B. Brachvogel; Wi889/6-3, CRC1218, TP B07, CRC829, and TP A14 to R.J. Wiesner; and CRC1218 and TP B06 to H. Kashkar).
The authors declare no competing financial interests.
Author contributions: T. Holzer designed the research studies, conducted experiments, acquired data, analyzed data, and wrote the manuscript. K. Probst conducted experiments, acquired data, and analyzed data. J. Etich conducted experiments, acquired data, and analyzed data and wrote the manuscript. M. Auler conducted experiments, acquired data, and analyzed data. V.S. Georgieva conducted experiments and acquired data. B. Bluhm, C. Frie, and J. Heilig conducted experiments and acquired data. A. Niehoff analyzed data and wrote the manuscript. J. Nüchel conducted experiments and acquired data. M. Plomann analyzed the data and wrote the manuscript. J.M. Seeger conducted experiments and acquired data. H. Kashkar analyzed the data and wrote the manuscript. O.R. Baris conducted experiments and acquired data. R.J. Wiesner designed research studies, analyzed data, provided reagents, and wrote the manuscript. B. Brachvogel designed research studies, conducted experiments, acquired data, analyzed data, and wrote the manuscript.
References
- Amarilio R., Viukov S.V., Sharir A., Eshkar-Oren I., Johnson R.S., and Zelzer E.. 2007. HIF1α regulation of Sox9 is necessary to maintain differentiation of hypoxic prechondrogenic cells during early skeletogenesis. Development. 134:3917–3928. 10.1242/dev.008441 [DOI] [PubMed] [Google Scholar]
- Baris O.R., Klose A., Kloepper J.E., Weiland D., Neuhaus J.F., Schauen M., Wille A., Müller A., Merkwirth C., Langer T., et al. 2011. The mitochondrial electron transport chain is dispensable for proliferation and differentiation of epidermal progenitor cells. Stem Cells. 29:1459–1468. [DOI] [PubMed] [Google Scholar]
- Baris O.R., Ederer S., Neuhaus J.F., von Kleist-Retzow J.C., Wunderlich C.M., Pal M., Wunderlich F.T., Peeva V., Zsurka G., Kunz W.S., et al. 2015. Mosaic deficiency in mitochondrial oxidative metabolism promotes cardiac arrhythmia during aging. Cell Metab. 21:667–677. 10.1016/j.cmet.2015.04.005 [DOI] [PubMed] [Google Scholar]
- Bateman J.F., Wilson R., Freddi S., Lamandé S.R., and Savarirayan R.. 2005. Mutations of COL10A1 in Schmid metaphyseal chondrodysplasia. Hum. Mutat. 25:525–534. 10.1002/humu.20183 [DOI] [PubMed] [Google Scholar]
- Belluoccio D., Etich J., Rosenbaum S., Frie C., Grskovic I., Stermann J., Ehlen H., Vogel S., Zaucke F., von der Mark K., et al. 2010a Sorting of growth plate chondrocytes allows the isolation and characterization of cells of a defined differentiation status. J. Bone Miner. Res. 25:1267–1281. 10.1002/jbmr.30 [DOI] [PubMed] [Google Scholar]
- Belluoccio D., Grskovic I., Niehoff A., Schlötzer-Schrehardt U., Rosenbaum S., Etich J., Frie C., Pausch F., Moss S.E., Pöschl E., et al. 2010b Deficiency of annexins A5 and A6 induces complex changes in the transcriptome of growth plate cartilage but does not inhibit the induction of mineralization. J. Bone Miner. Res. 25:141–153. 10.1359/jbmr.090710 [DOI] [PubMed] [Google Scholar]
- Bentovim L., Amarilio R., and Zelzer E.. 2012. HIF1α is a central regulator of collagen hydroxylation and secretion under hypoxia during bone development. Development. 139:4473–4483. 10.1242/dev.083881 [DOI] [PubMed] [Google Scholar]
- Bergmeier V., Etich J., Pitzler L., Frie C., Koch M., Fischer M., Rappl G., Abken H., Tomasek J.J., and Brachvogel B.. 2018. Identification of a myofibroblast-specific expression signature in skin wounds. Matrix Biol. 65:59–74. [DOI] [PubMed] [Google Scholar]
- Bjørkøy G., Lamark T., Pankiv S., Øvervatn A., Brech A., and Johansen T.. 2009. Monitoring autophagic degradation of p62/SQSTM1. Methods Enzymol. 452:181–197. 10.1016/S0076-6879(08)03612-4 [DOI] [PubMed] [Google Scholar]
- Blanco F.J., Rego I., and Ruiz-Romero C.. 2011. The role of mitochondria in osteoarthritis. Nat. Rev. Rheumatol. 7:161–169. 10.1038/nrrheum.2010.213 [DOI] [PubMed] [Google Scholar]
- Bluhm B., Ehlen H.W.A., Holzer T., Georgieva V.S., Heilig J., Pitzler L., Etich J., Bortecen T., Frie C., Probst K., et al. 2017. miR-322 stabilizes MEK1 expression to inhibit RAF/MEK/ERK pathway activation in cartilage. Development. 144:3562–3577. 10.1242/dev.148429 [DOI] [PubMed] [Google Scholar]
- Bywaters E.G.L. 1936. Metabolism of cartilage. Nature. 138:30–31. 10.1038/138030b0 [DOI] [Google Scholar]
- Cameron T.L., Bell K.M., Tatarczuch L., Mackie E.J., Rajpar M.H., McDermott B.T., Boot-Handford R.P., and Bateman J.F.. 2011. Transcriptional profiling of chondrodysplasia growth plate cartilage reveals adaptive ER-stress networks that allow survival but disrupt hypertrophy. PLoS One. 6:e24600 10.1371/journal.pone.0024600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan D., and Jacenko O.. 1998. Phenotypic and biochemical consequences of collagen X mutations in mice and humans. Matrix Biol. 17:169–184. 10.1016/S0945-053X(98)90056-7 [DOI] [PubMed] [Google Scholar]
- Chan C.M., Macdonald C.D., Litherland G.J., Wilkinson D.J., Skelton A., Europe-Finner G.N., and Rowan A.D.. 2017. Cytokine-induced MMP13 Expression in human chondrocytes is dependent on activating transcription factor 3 (ATF3) regulation. J. Biol. Chem. 292:1625–1636. 10.1074/jbc.M116.756601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Linsenmayer C., Gu H., Schmid T.M., and Linsenmayer T.F.. 1992. Domains of type X collagen: alteration of cartilage matrix by fibril association and proteoglycan accumulation. J. Cell Biol. 117:687–694. 10.1083/jcb.117.3.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H.K., Ryu D., Kim K.S., Chang J.Y., Kim Y.K., Yi H.S., Kang S.G., Choi M.J., Lee S.E., Jung S.B., et al. 2017. Growth differentiation factor 15 is a myomitokine governing systemic energy homeostasis. J. Cell Biol. 216:149–165. 10.1083/jcb.201607110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cillero-Pastor B., Rego-Pérez I., Oreiro N., Fernandez-Lopez C., and Blanco F.J.. 2013. Mitochondrial respiratory chain dysfunction modulates metalloproteases-1, -3 and -13 in human normal chondrocytes in culture. BMC Musculoskelet. Disord. 14:235 10.1186/1471-2474-14-235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutelle O., Hornig-Do H.T., Witt A., Andree M., Schiffmann L.M., Piekarek M., Brinkmann K., Seeger J.M., Liwschitz M., Miwa S., et al. 2014. Embelin inhibits endothelial mitochondrial respiration and impairs neoangiogenesis during tumor growth and wound healing. EMBO Mol. Med. 6:624–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R.L., Liang C., and Sue C.M.. 2016. A comparison of current serum biomarkers as diagnostic indicators of mitochondrial diseases. Neurology. 86:2010–2015. 10.1212/WNL.0000000000002705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doege K., Heine S., Jensen I., Jelkmann W., and Metzen E.. 2005. Inhibition of mitochondrial respiration elevates oxygen concentration but leaves regulation of hypoxia-inducible factor (HIF) intact. Blood. 106:2311–2317. 10.1182/blood-2005-03-1138 [DOI] [PubMed] [Google Scholar]
- Dogan S.A., and Trifunovic A.. 2011. Modelling mitochondrial dysfunction in mice. Physiol. Res. 60:S61–S70. [DOI] [PubMed] [Google Scholar]
- Etich J., Holzer T., Pitzler L., Bluhm B., and Brachvogel B.. 2015. MiR-26a modulates extracellular matrix homeostasis in cartilage. Matrix Biol. 43:27–34. 10.1016/j.matbio.2015.02.014 [DOI] [PubMed] [Google Scholar]
- Fiorese C.J., Schulz A.M., Lin Y.F., Rosin N., Pellegrino M.W., and Haynes C.M.. 2016. The transcription factor ATF5 mediates a mammalian mitochondrial UPR. Curr. Biol. 26:2037–2043. 10.1016/j.cub.2016.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girkontaite I., Frischholz S., Lammi P., Wagner K., Swoboda B., Aigner T., and Von der Mark K.. 1996. Immunolocalization of type X collagen in normal fetal and adult osteoarthritic cartilage with monoclonal antibodies. Matrix Biol. 15:231–238. 10.1016/S0945-053X(96)90114-6 [DOI] [PubMed] [Google Scholar]
- Grskovic I., Kutsch A., Frie C., Groma G., Stermann J., Schlötzer-Schrehardt U., Niehoff A., Moss S.E., Rosenbaum S., Pöschl E., et al. 2012. Depletion of annexin A5, annexin A6, and collagen X causes no gross changes in matrix vesicle-mediated mineralization, but lack of collagen X affects hematopoiesis and the Th1/Th2 response. J. Bone Miner. Res. 27:2399–2412. 10.1002/jbmr.1682 [DOI] [PubMed] [Google Scholar]
- Hirai T., Chagin A.S., Kobayashi T., Mackem S., and Kronenberg H.M.. 2011. Parathyroid hormone/parathyroid hormone-related protein receptor signaling is required for maintenance of the growth plate in postnatal life. Proc. Natl. Acad. Sci. USA. 108:191–196. 10.1073/pnas.1005011108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C.C., Wang C.H., Wu L.C., Hsia C.Y., Chi C.W., Yin P.H., Chang C.J., Sung M.T., Wei Y.H., Lu S.H., and Lee H.C.. 2013. Mitochondrial dysfunction represses HIF-1α protein synthesis through AMPK activation in human hepatoma HepG2 cells. Biochim. Biophys. Acta. 1830:4743–4751. 10.1016/j.bbagen.2013.06.004 [DOI] [PubMed] [Google Scholar]
- Inada M., Wang Y., Byrne M.H., Rahman M.U., Miyaura C., López-Otín C., and Krane S.M.. 2004. Critical roles for collagenase-3 (Mmp13) in development of growth plate cartilage and in endochondral ossification. Proc. Natl. Acad. Sci. USA. 101:17192–17197. 10.1073/pnas.0407788101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James C.G., Woods A., Underhill T.M., and Beier F.. 2006. The transcription factor ATF3 is upregulated during chondrocyte differentiation and represses cyclin D1 and A gene transcription. BMC Mol. Biol. 7:30 10.1186/1471-2199-7-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X., Zhao L., Ji K., Zhao Y., Li W., Zhang R., Hou Y., Lu J., and Yan C.. 2017. Growth differentiation factor 15 is a novel diagnostic biomarker of mitochondrial diseases. Mol. Neurobiol. 54:8110–8116. 10.1007/s12035-016-0283-7 [DOI] [PubMed] [Google Scholar]
- Kang X., Yang W., Feng D., Jin X., Ma Z., Qian Z., Xie T., Li H., Liu J., Wang R., et al. 2017. Cartilage-specific autophagy deficiency promotes ER stress and impairs chondrogenesis in PERK-ATF4-CHOP-dependent manner. J. Bone Miner. Res. 32:2128–2141. 10.1002/jbmr.3134 [DOI] [PubMed] [Google Scholar]
- Kawamoto T. 2003. Use of a new adhesive film for the preparation of multi-purpose fresh-frozen sections from hard tissues, whole-animals, insects and plants. Arch. Histol. Cytol. 66:123–143. 10.1679/aohc.66.123 [DOI] [PubMed] [Google Scholar]
- Khan N.A., Nikkanen J., Yatsuga S., Jackson C., Wang L., Pradhan S., Kivela R., Pessia A., Velagapudi V., and Suomalainen A.. 2017. mTORC1 regulates mitochondrial integrated stress response and mitochondrial myopathy progression. Cell Metab. 26:419–428. [DOI] [PubMed] [Google Scholar]
- Kim K.H., Jeong Y.T., Oh H., Kim S.H., Cho J.M., Kim Y.N., Kim S.S., Kim D.H., Hur K.Y., Kim H.K., et al. 2013. Autophagy deficiency leads to protection from obesity and insulin resistance by inducing Fgf21 as a mitokine. Nat. Med. 19:83–92. 10.1038/nm.3014 [DOI] [PubMed] [Google Scholar]
- Klatt A.R., Nitsche D.P., Kobbe B., Mörgelin M., Paulsson M., and Wagener R.. 2000. Molecular structure and tissue distribution of matrilin-3, a filament-forming extracellular matrix protein expressed during skeletal development. J. Biol. Chem. 275:3999–4006. 10.1074/jbc.275.6.3999 [DOI] [PubMed] [Google Scholar]
- Koenig M.K. 2008. Presentation and diagnosis of mitochondrial disorders in children. Pediatr. Neurol. 38:305–313. 10.1016/j.pediatrneurol.2007.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan A.P., Cummings C.E., Chapman J.A., and Grant M.E.. 1991. Macromolecular organization of chicken type X collagen in vitro. J. Cell Biol. 114:597–604. 10.1083/jcb.114.3.597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtonen J.M., Forsström S., Bottani E., Viscomi C., Baris O.R., Isoniemi H., Höckerstedt K., Österlund P., Hurme M., Jylhävä J., et al. 2016. FGF21 is a biomarker for mitochondrial translation and mtDNA maintenance disorders. Neurology. 87:2290–2299. 10.1212/WNL.0000000000003374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomelino C.L., Andring J.T., McKenna R., and Kilberg M.S.. 2017. Asparagine synthetase: Function, structure, and role in disease. J. Biol. Chem. 292:19952–19958. 10.1074/jbc.R117.819060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López de Figueroa P., Lotz M.K., Blanco F.J., and Caramés B.. 2015. Autophagy activation and protection from mitochondrial dysfunction in human chondrocytes. Arthritis Rheumatol. 67:966–976. 10.1002/art.39025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marambio P., Toro B., Sanhueza C., Troncoso R., Parra V., Verdejo H., García L., Quiroga C., Munafo D., Díaz-Elizondo J., et al. 2010. Glucose deprivation causes oxidative stress and stimulates aggresome formation and autophagy in cultured cardiac myocytes. Biochim. Biophys. Acta. 1802:509–518. 10.1016/j.bbadis.2010.02.002 [DOI] [PubMed] [Google Scholar]
- Martin J.A., Martini A., Molinari A., Morgan W., Ramalingam W., Buckwalter J.A., and McKinley T.O.. 2012. Mitochondrial electron transport and glycolysis are coupled in articular cartilage. Osteoarthritis Cartilage. 20:323–329. 10.1016/j.joca.2012.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Reyes I., Diebold L.P., Kong H., Schieber M., Huang H., Hensley C.T., Mehta M.M., Wang T., Santos J.H., Woychik R., et al. 2016. TCA cycle and mitochondrial membrane potential are necessary for diverse biological functions. Mol. Cell. 61:199–209. 10.1016/j.molcel.2015.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melber A., and Haynes C.M.. 2018. UPRmt regulation and output: a stress response mediated by mitochondrial-nuclear communication. Cell Res. 28:281–295. 10.1038/cr.2018.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuhashi K., Ono W., Matsushita Y., Sakagami N., Takahashi A., Saunders T.L., Nagasawa T., Kronenberg H.M., and Ono N.. 2018. Resting zone of the growth plate houses a unique class of skeletal stem cells. Nature. 563:254–258. 10.1038/s41586-018-0662-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero R., Yubero D., Villarroya J., Henares D., Jou C., Rodríguez M.A., Ramos F., Nascimento A., Ortez C.I., Campistol J., et al. 2016. GDF-15 is elevated in children with mitochondrial diseases and is induced by mitochondrial dysfunction. PLoS One. 11:e0148709 10.1371/journal.pone.0148709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuspiel M., Schauss A.C., Braschi E., Zunino R., Rippstein P., Rachubinski R.A., Andrade-Navarro M.A., and McBride H.M.. 2008. Cargo-selected transport from the mitochondria to peroxisomes is mediated by vesicular carriers. Curr. Biol. 18:102–108. 10.1016/j.cub.2007.12.038 [DOI] [PubMed] [Google Scholar]
- Ovchinnikov D.A., Deng J.M., Ogunrinu G., and Behringer R.R.. 2000. Col2a1-directed expression of Cre recombinase in differentiating chondrocytes in transgenic mice. Genesis. 26:145–146. [DOI] [PubMed] [Google Scholar]
- Pfaffl M.W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:45e 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfander D., Cramer T., Schipani E., and Johnson R.S.. 2003. HIF-1alpha controls extracellular matrix synthesis by epiphyseal chondrocytes. J. Cell Sci. 116:1819–1826. 10.1242/jcs.00385 [DOI] [PubMed] [Google Scholar]
- Poole A.R., and Pidoux I.. 1989. Immunoelectron microscopic studies of type X collagen in endochondral ossification. J. Cell Biol. 109:2547–2554. 10.1083/jcb.109.5.2547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirós P.M., Mottis A., and Auwerx J.. 2016. Mitonuclear communication in homeostasis and stress. Nat. Rev. Mol. Cell Biol. 17:213–226. 10.1038/nrm.2016.23 [DOI] [PubMed] [Google Scholar]
- Rajpar M.H., McDermott B., Kung L., Eardley R., Knowles L., Heeran M., Thornton D.J., Wilson R., Bateman J.F., Poulsom R., et al. 2009. Targeted induction of endoplasmic reticulum stress induces cartilage pathology. PLoS Genet. 5:e1000691 10.1371/journal.pgen.1000691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajpurohit R., Mansfield K., Ohyama K., Ewert D., and Shapiro I.M.. 1999. Chondrocyte death is linked to development of a mitochondrial membrane permeability transition in the growth plate. J. Cell. Physiol. 179:287–296. [DOI] [PubMed] [Google Scholar]
- Repetto G., del Peso A., and Zurita J.L.. 2008. Neutral red uptake assay for the estimation of cell viability/cytotoxicity. Nat. Protoc. 3:1125–1131. 10.1038/nprot.2008.75 [DOI] [PubMed] [Google Scholar]
- Rosenbaum S., Kreft S., Etich J., Frie C., Stermann J., Grskovic I., Frey B., Mielenz D., Pöschl E., Gaipl U., et al. 2011. Identification of novel binding partners (annexins) for the cell death signal phosphatidylserine and definition of their recognition motif. J. Biol. Chem. 286:5708–5716. 10.1074/jbc.M110.193086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid T.M., Popp R.G., and Linsenmayer T.F.. 1990. Hypertrophic cartilage matrix. Type X collagen, supramolecular assembly, and calcification. Ann. N. Y. Acad. Sci. 580:64–73. 10.1111/j.1749-6632.1990.tb17918.x [DOI] [PubMed] [Google Scholar]
- Sciacco M., and Bonilla E.. 1996. Cytochemistry and immunocytochemistry of mitochondria in tissue sections. Methods Enzymol. 264:509–521. 10.1016/S0076-6879(96)64045-2 [DOI] [PubMed] [Google Scholar]
- Shpilka T., and Haynes C.M.. 2018. The mitochondrial UPR: Mechanisms, physiological functions and implications in ageing. Nat. Rev. Mol. Cell Biol. 19:109–120. 10.1038/nrm.2017.110 [DOI] [PubMed] [Google Scholar]
- Sivan S.S., Tsitron E., Wachtel E., Roughley P.J., Sakkee N., van der Ham F., DeGroot J., Roberts S., and Maroudas A.. 2006. Aggrecan turnover in human intervertebral disc as determined by the racemization of aspartic acid. J. Biol. Chem. 281:13009–13014. 10.1074/jbc.M600296200 [DOI] [PubMed] [Google Scholar]
- Sivaraj K.K., and Adams R.H.. 2016. Blood vessel formation and function in bone. Development. 143:2706–2715. 10.1242/dev.136861 [DOI] [PubMed] [Google Scholar]
- Stauber M., and Müller R.. 2008. Micro-computed tomography: a method for the non-destructive evaluation of the three-dimensional structure of biological specimens. Methods Mol. Biol. 455:273–292. 10.1007/978-1-59745-104-8_19 [DOI] [PubMed] [Google Scholar]
- Stegen S., Laperre K., Eelen G., Rinaldi G., Fraisl P., Torrekens S., Van Looveren R., Loopmans S., Bultynck G., Vinckier S., et al. 2019. HIF-1α metabolically controls collagen synthesis and modification in chondrocytes. Nature. 565:511–515. 10.1038/s41586-019-0874-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomalainen A., Elo J.M., Pietiläinen K.H., Hakonen A.H., Sevastianova K., Korpela M., Isohanni P., Marjavaara S.K., Tyni T., Kiuru-Enari S., et al. 2011. FGF-21 as a biomarker for muscle-manifesting mitochondrial respiratory chain deficiencies: a diagnostic study. Lancet Neurol. 10:806–818. 10.1016/S1474-4422(11)70155-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teske B.F., Fusakio M.E., Zhou D., Shan J., McClintick J.N., Kilberg M.S., and Wek R.C.. 2013. CHOP induces activating transcription factor 5 (ATF5) to trigger apoptosis in response to perturbations in protein homeostasis. Mol. Biol. Cell. 24:2477–2490. 10.1091/mbc.e13-01-0067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M.R., Xu D., and Williams B.R.. 2009. ATF3 transcription factor and its emerging roles in immunity and cancer. J. Mol. Med. (Berl.). 87:1053–1060. 10.1007/s00109-009-0520-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang K.Y., Chan D., Bateman J.F., and Cheah K.S.. 2010. In vivo cellular adaptation to ER stress: survival strategies with double-edged consequences. J. Cell Sci. 123:2145–2154. 10.1242/jcs.068833 [DOI] [PubMed] [Google Scholar]
- Tunggal J., Wartenberg M., Paulsson M., and Smyth N.. 2003. Expression of the nidogen-binding site of the laminin gamma1 chain disturbs basement membrane formation and maintenance in F9 embryoid bodies. J. Cell Sci. 116:803–812. 10.1242/jcs.00293 [DOI] [PubMed] [Google Scholar]
- Verzijl N., DeGroot J., Thorpe S.R., Bank R.A., Shaw J.N., Lyons T.J., Bijlsma J.W., Lafeber F.P., Baynes J.W., and TeKoppele J.M.. 2000. Effect of collagen turnover on the accumulation of advanced glycation end products. J. Biol. Chem. 275:39027–39031. 10.1074/jbc.M006700200 [DOI] [PubMed] [Google Scholar]
- Vuppalapati K.K., Bouderlique T., Newton P.T., Kaminskyy V.O., Wehtje H., Ohlsson C., Zhivotovsky B., and Chagin A.S.. 2015. Targeted deletion of autophagy genes Atg5 or Atg7 in the chondrocytes promotes caspase-dependent cell death and leads to mild growth retardation. J. Bone Miner. Res. 30:2249–2261. 10.1002/jbmr.2575 [DOI] [PubMed] [Google Scholar]
- Wagner K., Pöschl E., Turnay J., Baik J., Pihlajaniemi T., Frischholz S., and von der Mark K.. 2000. Coexpression of alpha and beta subunits of prolyl 4-hydroxylase stabilizes the triple helix of recombinant human type X collagen. Biochem. J. 352:907–911. 10.1042/bj3520907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Liang B., Shirwany N.A., and Zou M.H.. 2011. 2-Deoxy-D-glucose treatment of endothelial cells induces autophagy by reactive oxygen species-mediated activation of the AMP-activated protein kinase. PLoS One. 6:e17234 10.1371/journal.pone.0017234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Liu J., McDonald C., Lupino K., Zhai X., Wilkins B.J., Hakonarson H., and Pei L.. 2017. GDF15 is a heart-derived hormone that regulates body growth. EMBO Mol. Med. 9:1150–1164. 10.15252/emmm.201707604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiland D., Brachvogel B., Hornig-Do H.T., Neuhaus J.F.G., Holzer T., Tobin D.J., Niessen C.M., Wiesner R.J., and Baris O.R.. 2018. Imbalance of Mitochondrial respiratory chain complexes in the epidermis induces severe skin inflammation. J. Invest. Dermatol. 138:132–140. 10.1016/j.jid.2017.08.019 [DOI] [PubMed] [Google Scholar]
- Wolny S., McFarland R., Chinnery P., and Cheetham T.. 2009. Abnormal growth in mitochondrial disease. Acta Paediatr. 98:553–554. 10.1111/j.1651-2227.2008.01148.x [DOI] [PubMed] [Google Scholar]
- Wu S., Levenson A., Kharitonenkov A., and De Luca F.. 2012. Fibroblast growth factor 21 (FGF21) inhibits chondrocyte function and growth hormone action directly at the growth plate. J. Biol. Chem. 287:26060–26067. 10.1074/jbc.M112.343707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan B., Zhang Z., Jin D., Cai C., Jia C., Liu W., Wang T., Li S., Zhang H., Huang B., et al. 2016. mTORC1 regulates PTHrP to coordinate chondrocyte growth, proliferation and differentiation. Nat. Commun. 7:11151 10.1038/ncomms11151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsuga S., Fujita Y., Ishii A., Fukumoto Y., Arahata H., Kakuma T., Kojima T., Ito M., Tanaka M., Saiki R., and Koga Y.. 2015. Growth differentiation factor 15 as a useful biomarker for mitochondrial disorders. Ann. Neurol. 78:814–823. 10.1002/ana.24506 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.