CENTRORADIALIS interacts with FLOWERING LOCUS T-like genes to control spikelet initiation, floret development, and grain number in barley
Abstract
CENTRORADIALIS (CEN) is a key regulator of flowering time and inflorescence architecture in plants. Natural variation in the barley (Hordeum vulgare) homolog HvCEN is important for agricultural range expansion of barley cultivation, but its effects on shoot and spike architecture and consequently yield have not yet been characterized. Here, we evaluated 23 independent hvcen, also termed mat-c, mutants to determine the pleiotropic effects of HvCEN on developmental timing and shoot and spike morphologies of barley under outdoor and controlled conditions. All hvcen mutants flowered early and showed a reduction in spikelet number per spike, tiller number, and yield in the outdoor experiments. Mutations in hvcen accelerated spikelet initiation and reduced axillary bud number in a photoperiod-independent manner but promoted floret development only under long days (LDs). The analysis of a flowering locus t3 (hvft3) hvcen double mutant showed that HvCEN interacts with HvFT3 to control spikelet initiation. Furthermore, early flowering3 (hvelf3) hvcen double mutants with high HvFT1 expression levels under short days suggested that HvCEN interacts with HvFT1 to repress floral development. Global transcriptome profiling in developing shoot apices and inflorescences of mutant and wild-type plants revealed that HvCEN controlled transcripts involved in chromatin remodeling activities, cytokinin and cell cycle regulation and cellular respiration under LDs and short days, whereas HvCEN affected floral homeotic genes only under LDs. Understanding the stage and organ-specific functions of HvCEN and downstream molecular networks will allow the manipulation of different shoot and spike traits and thereby yield.
Shoot architecture is a major determinant of grain yield and therefore a primary target for crop improvement. Shoot architecture is largely defined by branching (tillering) patterns, plant height, leaf shape and arrangement, and inflorescence morphologies. These traits are controlled by the combined activities of the shoot apical meristem (SAM) and the axillary meristems (AXMs; Nakagawa et al., 2002; Teichmann and Muhr, 2015). A vegetative SAM gives rise to leaves and AXMs that form in the leaf axils (Turnbull, 2005). As plants transition from vegetative to reproductive growth, the SAM forms an inflorescence, flowers, and eventually seeds. Grasses exhibit a striking diversity in their inflorescence architectures that is determined by meristem initiation and determinacy decisions, the acquisition of spikelet meristem identity, and the determinacy of the spikelet meristem (Bommert and Whipple, 2018). The spikes of barley (Hordeum vulgare) display a raceme-like branchless shape and consist of triple spikelets produced on two opposite sides along the main axis (rachis). Although the barley inflorescence is indeterminate, the barley spikelet is determinate as a defined number of florets is produced, a maximum of a single floret and grain, per spikelet. The triple spikelet meristem of barley consists of one central spikelet meristem and two lateral spikelet meristems. Most studies on barley spike architecture have focused on the genetic basis of two-rowed versus six-rowed spikes, which is determined by differential development of the lateral spikelets (Komatsuda et al., 2007; Ramsay et al., 2011; Koppolu et al., 2013; Bull et al., 2017; van Esse et al., 2017; Youssef et al., 2017). However, recent studies have shown that spike architecture is also affected by genetic factors controlling the timing of preanthesis development in barley and wheat (Triticum aestivum). The flowering time regulators PHOTOPERIOD1 (PPD-H1) and its downstream target FLOWERING LOCUS T1 (FT1), homolog of Arabidopsis (Arabidopsis thaliana) FT, affect the number of spikelets produced on the main inflorescence in barley and wheat, likely by affecting the rate and duration of spikelet initiation (Boden et al., 2015; Digel et al., 2015). FT-like genes belong to phosphatidylethanolamine-binding proteins, whose homolog in humans (Homo sapiens) was characterized as a Raf kinase inhibitor protein, mediating the rapidly accelerated fibrosarcoma (RAF)/mitogen-activated protein kinase kinase (MEK)/extracellular signal-regulated kinases (ERK) signal transduction pathway (Bradley et al., 1996, 1997; Ohshima et al., 1997; Kardailsky et al., 1999; Kobayashi et al., 1999; Yeung et al., 1999). In plants, the phosphatidylethanolamine-binding proteins family comprises proteins with antagonistic effects on development as they either promote or inhibit floral transition. FT-like genes generally induce flowering while CENTRORADIALIS (CEN), first described in Garden Snapdragon (Antirrhinum mayus), represses the initiation of floral meristems (Bradley et al., 1996; Danilevskaya et al., 2008; Komiya et al., 2008; Turck et al., 2008; Digel et al., 2015; Kaneko-Suzuki et al., 2018). In Arabidopsis and rice (Oryza sativa), FT is expressed in the leaf vascular tissue and the FT protein is transported through the phloem to the SAM (Corbesier et al., 2007; Tamaki et al., 2007). In rice, the FT homolog HEADING DATE 3a (Hd3a) forms a florigen-activating complex with 14-3-3 proteins and FLOWERING LOCUS D (FD) homolog OsFD1 to activate the expression of meristem identity genes (Taoka et al., 2011). By contrast, TERMINAL FLOWER1 (TFL1), the Arabidopsis homolog of CEN, functions as a hypothetical competitor of FT in binding to FD and 14-3-3 proteins in the shoot apex, thereby preventing the induction of flowering (Abe et al., 2005; Wigge et al., 2005; Ahn et al., 2006; Hanano and Goto, 2011; McGarry and Ayre, 2012; Kaneko-Suzuki et al., 2018). An acceleration in flowering time and the formation of a terminal flower are observed in both tfl1 mutants and plants overexpressing FT in Arabidopsis (Bradley et al., 1997; Kardailsky et al., 1999; Kobayashi et al., 1999). The function of TFL1 homologs in controlling flowering time and inflorescence architecture are conserved to some extent between grasses and eudicots. For example, the Antirrhinum TFL1 homolog CENTRORADIALIS controls the determinacy of the inflorescence with no effects on flowering time (Bradley et al., 1996), whereas LpTFL1 in ryegrass (Lolium perenne) represses flowering and controls AXM identity (Jensen et al., 2001). RICE CENTRORADIALIS (RCN1 and RCN2), rice homologs of TFL1, delay flowering and alter the panicle architecture (Nakagawa et al., 2002). Likewise, ectopic expression of the RCN1-CORN CENTRORADIALIS/TFL1-like protein ZCN2, ZCN4, ZCN5 in maize (Zea mays) leads to late flowering and a bushy tassel with denser spikelets (Danilevskaya et al., 2010).
HvCEN, the barley homolog of Antirrhinum CEN and Arabidopsis TFL1, contributed to the expansion of barley cultivation into diverse habitats (Comadran et al., 2012). A natural Pro-135Ala substitution in HvCEN has been selected in spring barley cultivars and prolongs vegetative growth, while hvcen mutations lead to early flowering under natural long-day (LD) conditions (Comadran et al., 2012). The authors also show that induced hvcen mutants, originally designated as maturity-c (mat-c) mutants, flowered a few days earlier under natural LD conditions (Druka et al., 2011; Comadran et al., 2012; Lundqvist, 2014). However, the effects of HvCEN on shoot and inflorescence architecture and its interaction with FT-like genes have not been characterized so far.
Barley has six different FT-like homologs, of which only HvFT1 and HvFT3 have been functionally characterized (Schmitz et al., 2000; Yan et al., 2006; Hemming et al., 2008; Sasani et al., 2009; Casao et al., 2011; Chen and Dubcovsky, 2012; Halliwell et al., 2016). Elevated HvFT1 expression in leaves is correlated with a strong acceleration of floral development, whereas HvFT3 only induces spikelet initiation without effects on later floret development (Digel et al., 2015; Mulki et al., 2018). In this study, we analyzed a large collection of independent hvcen mutants to (1) identify pleiotropic effects of HvCEN on developmental timing and shoot and spike morphology, (2) determine transcriptional targets of HvCEN in the developing SAM under different photoperiods, and (3) investigate the genetic interactions between HvCEN and the FT-like genes HvFT1 and HvFT3.
We demonstrate that HvCEN has pleiotropic effects on several shoot traits, as it delays reproductive development and flowering, promotes axillary bud initiation/tillering, and increases the number of spikelet primordia and plant height. Mutations in HvCEN shortened the vegetative phase under both LDs and short days (SDs) but accelerated inflorescence development only under LDs. These photoperiod-specific effects of HvCEN were likely dependent on antagonistic interactions with different HvFT-like proteins during development. Global transcriptome analysis in developing SAMs suggested that spikelet initiation, as promoted by mutations in hvcen, coincided with a strong reprogramming of transcriptional networks, the induction of cell proliferation, and changes in the energy metabolism under SDs and LDs. The subsequent rapid floral development in the hvcen mutant was linked to the LD-specific up-regulation of floral homeotic genes.
RESULTS
HvCEN Controls Shoot and Spike Architecture in Barley
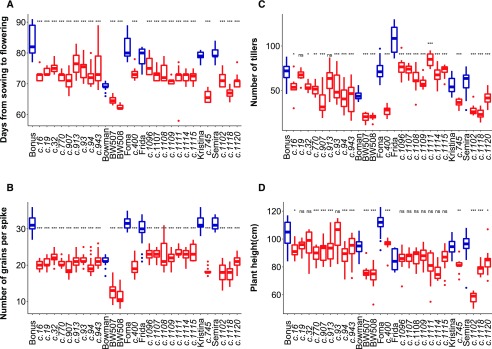
To characterize the effects of different HvCEN mutations on shoot development, we analyzed flowering time, grains per spike, plant height, tiller number, and different seed parameters in 23 allelic hvcen mutants in six different spring barley backgrounds under outdoor conditions (Fig. 1; Supplemental Table S1). All the hvcen mutants flowered significantly earlier and produced fewer grains per main spike than the respective wild-type genotypes (Fig. 1, A and B). In addition, all hvcen mutants, except for mat-c.19 and mat-c.913, produced fewer tillers at flowering time compared with the corresponding parental genotypes (Fig. 1C). Likewise, plant height of most hvcen mutants, excluding mat-c.19, mat-c.32, mat-c.93 in the Bonus background and all mutants in the Frida background, was reduced compared with the respective wild-type genotypes (Fig. 1D). Seed traits, including length, width, area, and thousand kernel weight (TKW), were not significantly different between all mutants versus wild-type plants (Supplemental Figure S1).
Figure 1.
Phenotypes of hvcen (mat-c) mutants trialed under outdoor conditions over 2 consecutive years. Mutants (red) in six different genetic backgrounds are shown next to their respective parental lines (blue; Supplemental Table S1). Various traits including flowering time (A), grain number per main spike (B), number of tillers at flowering time (C), and plant height (D) were measured over years. Flowering time was scored as time from sowing to heading when the awn of the main spike emerged from the flag leaf. The line within each box denotes the median. The sample size per genotype is given in Supplemental Table S1. Error bars: sd. Significant differences between the mutants and their wild-type parents were calculated by a one-way ANOVA using a Tukey honestly significant difference as a post hoc test, ***P < 0.001, **P < 0.01, *P < 0.05. ns, Not significant.
We calculated the degree of amino acid conservation across taxa for individual mutations. The analysis revealed that all single amino acid substitutions are located in conserved positions within the protein (Supplemental Table S2). The mutations were positioned in the potential ligand-binding pocket (mat-c.913, mat-c.93, mat-c.32, mat-c.907, mat-c.1115), the 14-3-3 protein interaction site (mat-c.943, mat-c.745), and the external loop (mat-c.400; Supplemental Figure S2; Ahn et al., 2006; Ho and Weigel, 2014). The external loop has been shown to represent the most substantial differences between FT and TFL1 and is critical for FT versus TFL1 activity in vivo (Ahn et al., 2006). The binding pocket has previously been suggested to play an important role in binding to phosphorylated interacting partners (Ahn et al., 2006; Ho and Weigel, 2014), whereas the 14-3-3 protein interaction site is important for the formation of the flowering activation complex (Taoka et al., 2011). All analyzed mutants showed a significant difference in most of the scored developmental traits, suggesting that these all altered the function of the protein.
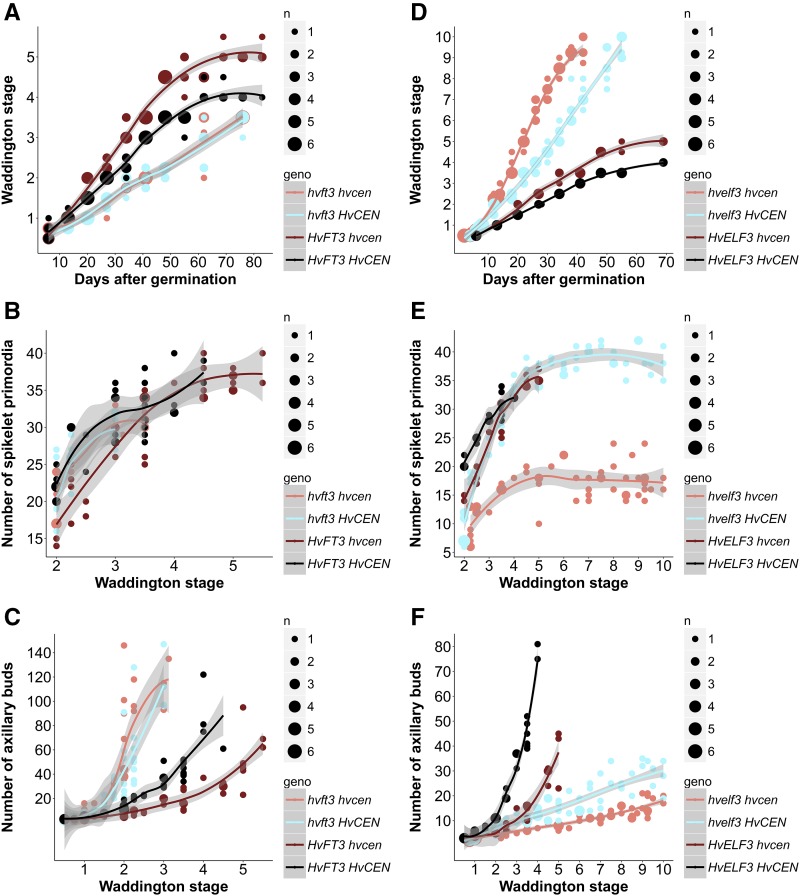
To further illustrate the effects of HvCEN on reproductive development, we focused on the development of primary shoots of three selected hvcen mutants (mat-c.907, mat-c.94, and mat-c.943) and cv Bonus under different photoperiods. We also compared the timing of spikelet initiation and inflorescence development between the hvcen mutants and the wild-type parent Bonus (Fig. 2). The hvcen mutants initiated spikelet primordia (W2.0) significantly earlier under both LDs and SDs compared with the wild-type cv Bonus (Fig. 3, A and B). However, hvcen mutants exhibited photoperiod-specific patterns of growth and floret development. Particularly under LDs, floret development was greatly accelerated in hvcen mutants starting from the stamen primordium stage (W3.5; Fig. 3A). Consequently, the hvcen mutant plants flowered (W9.0-10.0) about 18 d earlier than Bonus under LDs (Fig. 3A). In contrast, floret development under SDs did not proceed beyond the stages W4.05.5 in the mutants and the stagesW3.5-W4.5 in Bonus (Figs. 2B and 3B).
Figure 2.
Representative shoot apices of the hvcen mutant and the wild type. Apices were scored under LD (A) and SD (B) conditions. Under SDs, the MSA did not develop beyond the stages W3.5 and W4.5 in the mutant and Bonus, respectively. DAG, days after germination; W, Waddington stage. White bar = 1 mm.
Figure 3.
Phenotyping of three hvcen mutants (mat-c.907, mat-c.94, mat-c.943) and the wild-type Bonus under controlled LD and SD conditions. Development of the MSA and number of spikelet primordia and of axillary buds at different Waddington stages under LDs (A, C, E) and SDs (B, D, F) are shown. The number of spikelet primordia refers to the number of primordia per main spike. The axillary buds scored include all the axillary buds, primary, secondary, and higher order buds. Five or six plants per genotype were dissected at each time point under LDs (16 h light/8 h night) and SDs (8 h/16 h, light/dark). Statistical differences (P < 0.05) were calculated using a polynomial regression model at 95% confidence interval (Loess smooth line) shown in gray-shaded regions.
Because hvcen mutants displayed a reduction in grain number per main spike (Fig. 1B), we also evaluated the number of spikelet primordia initiated on the main shoot spike during development. Under LDs, the number of initiated spikelet primordia reached its maximal level at different developmental stages, in the mutant at W4.0 with 24.40 ± 1.58 spikelet primordia and in the wild type at W5.0 with 40.00 ± 0.76 primordia (Fig. 3C). Although the mutants developed fewer spikelet primordia, we did not observe the formation of a terminal spikelet as has been described for the Arabidopsis tfl1 mutant and is typical for the determinate wheat inflorescence (Supplemental Figure S3). Under SDs, the reduction in the number of spikelet primordia of hvcen mutants was only apparent before the lemma primordium stage (W3.0), and no differences were observed between the mutants and wild type after this stage (Fig. 3D). Furthermore, hvcen mutants produced fewer and shorter leaves on the main shoot (Supplemental Figure S4) and exhibited a reduced number of axillary buds under both LDs and SDs (Fig. 3, E and F). No significant differences in leaf width were observed in the mutants compared with wild type except for mat-c.943 with wider leaves under LDs (Supplemental Figure S4).
Taken together, mutations in the three hvcen (mat-c) mutants accelerated the initiation of spikelet primordia under both LDs and SDs, whereas floret development of the mutants was only accelerated under LDs. Moreover, hvcen mutants exhibited a reduced number of spikelet primordia on the main shoot apex (MSA). Finally, HvCEN affected the total number of leaves and leaf size on the main culm and tiller number.
HvCEN Interacts with HvFT1 and HvFT3 to Control Reproductive Development under LDs and SDs
The HvCEN homolog TFL1 acts antagonistically to FT in the meristem, and the relative abundance of FT and TFL1 proteins controls floral development and shoot architecture in Arabidopsis and other crop plants (McGarry and Ayre, 2012; Kaneko-Suzuki et al., 2018). Therefore, we tested if and how the effects of HvCEN on development are dependent on the function of FT-like genes in barley. Barley has several FT-like genes (Halliwell et al., 2016); however, only HvFT1 and HvFT3 have been functionally analyzed and integrated into flowering pathways. HvFT1 is only transcribed under LDs, and its protein promotes spikelet initiation and floret development (Yan et al., 2006; Digel et al., 2015). In contrast, HvFT3 is expressed under SDs and LDs and specifically accelerates the timing of spikelet initiation but has no effects on floret development (Mulki et al., 2018). To test whether the effects of HvCEN on spikelet initiation are dependent on HvFT3, we examined the development of single and double hvft3 hvcen mutants under SDs. The hvft3 single and the hvft3 hvcen double mutants did not differ in development and architecture, and both showed a delayed development and an increase in the number of tillers and spikelet primordia (Fig. 4, A–C). Consequently, the hvcen allele advanced spikelet initiation and decreased the number of spikelet primordia and axillary buds, but only in the presence of a functional HvFT3. We, therefore, inferred that the photoperiod-independent effects of HvCEN on spikelet initiation are dependent on HvFT3. However, there was no clear effect of HvCEN on the expression of HvFT3 (Supplemental Figure S5), suggesting that both interact on the protein level.
Figure 4.
Microscopic phenotypes of hvft3, hvelf3, and hvcen single mutants and hvft3 hvcen and hvelf3 hvcen double mutants in Bonus under SDs. Development of the MSA, number of spikelet primordia, and number of axillary buds in HvFT3 or hvft3 background (A, B, C) and HvELF3 or hvelf3 background (C, D, E) at different Waddington stages under SDs (8 h/16 h, light/dark) are shown. The hvcen line was mat-c.907. The number of spikelet primordia is referring to the number of primordia per main spike. The axillary buds scored include all the axillary buds, primary, secondary, and higher order buds. Three to six plants per genotype were dissected at each time point. Statistical differences (P < 0.05) were calculated using a polynomial regression model at 95% confidence interval (Loess smooth line) shown in gray-shaded regions.
Next, we examined if the LD-specific effects on floret development could be explained by the interaction of HvCEN with HvFT1 as HvFT1 is only expressed under LDs and not SDs. For this purpose, we crossed the hvcen mutant with a mutant line carrying a nonfunctional EARLY FLOWERING 3 (HvELF3) allele. Arabidopsis ELF3 is a circadian clock gene that modulates light signal transduction downstream of phytochromes and mediates the circadian gating of light perception and responses (Hicks et al., 1996; Zagotta et al., 1996; Liu et al., 2001). Barley hvelf3 mutants are characterized by photoperiod-independent expression of HvFT1 and early flowering (Faure et al., 2012). We confirmed that expression levels of HvFT1 were comparable between HvELF3 wild-type plants grown under LDs and hvelf3 mutant plants grown under SDs (Supplemental Figure S6). We then examined MSA development in single and double hvelf3 hvcen mutants under SDs. The plants carrying hvelf3 mutations developed significantly faster when compared with those carrying the wild-type HvELF3 allele irrespective of the HvCEN allele (Fig. 4D). More interestingly, variation at HvCEN strongly affected floral development under SDs in the background of the hvelf3 mutant but had only a minor effect on floral development in HvELF3 wild-type plants. This suggested that under conditions where HvFT1 is expressed, either under LDs or in the hvelf3 mutant background, HvCEN genetically interacted with HvFT1 to modulate floral development. The mutation in hvcen reduced the number of spikelets per MSA and axillary buds in the background of hvelf3 as it did in the HvELF3 background under LDs (Fig. 4, E and F). The genotype and photoperiod-specific expression patterns suggested that the photoperiod-dependent effects of hvcen on floret development and spikelet number were likely regulated via HvFT1. However, amutations in the clock gene HvELF3 modify the expression of a large number of genes (Faure et al., 2012), we therefore could not rule out that the hvelf3-specific effect of hvcen might be caused by genes other than HvFT1.
Molecular Characterization of HvCEN
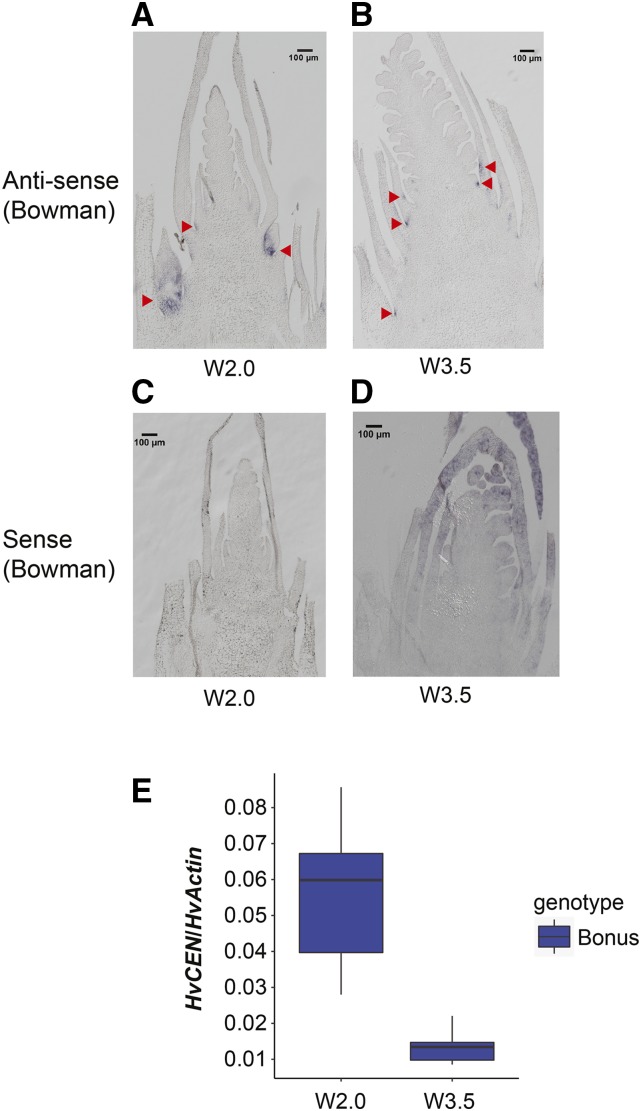
To further characterize the function of HvCEN, we examined the spatial expression patterns of HvCEN in the main shoot apex and crown tissue of cv Bonus and cv Bowman by RNA in situ hybridization at the spikelet initiation and stamen primordium stages. HvCEN RNA was localized in the AXMs and leaf axils, but no signals were detected in the inflorescence meristems (Fig. 5, A–D; Supplemental Figure S7). To test if HvCEN was also expressed in the inflorescence but at levels too low for detection by in situ hybridization, we dissected the inflorescence meristems only and tested for HvCEN expression by real-time quantitative PCR (RT-qPCR). HvCEN mRNA was expressed in the inflorescence at both spikelet initiation (W2.0) and stamen primordium stages (W3.5) with a higher level at spikelet initiation (Fig. 5E). These results indicate that HvCEN is expressed in both the AXMs and leaf axils as well as the main inflorescence.
Figure 5.
Expression pattern of HvCEN under LDs. A and B, RNA in situ hybridization using the antisense probe. Red arrows indicate the HvCEN expression domains. C and D, Negative controls using the sense probe of HvCEN in the shoot apex of cv Bowman at Waddington stages 2.0 and 3.5. Scale bars = 100 µm. E, Expression of HvCEN in the pure inflorescence meristem of cv Bonus. Pure meristems, with six replicates per stage, were obtained by carefully removing leaf primordia and stem tissue beneath the inflorescence under the microscope. LDs are 16 h/8 h light/dark. W, Waddington stage. Error bars = sd.
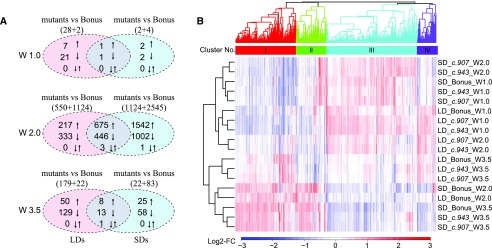
To further understand how HvCEN regulates the development of MSA in a photoperiod-dependent and independent manner, we performed genome-wide transcriptome profiling in developing MSA in two allelic hvcen mutants (mat-c.907 and mat-c.943) and the wild type (cv Bonus). We focused on three developmental stages during which genotypes exhibited phenotypic differences under LDs and SDs (Fig. 3; Supplemental Figure S4), including the vegetative stage (W1.0, MSA enriched tissue), the spikelet initiation stage (W2.0, MSA enriched tissue), and the stamen primordium stage (W3.5, MSA tissue). Transcriptome analysis revealed the expression of 24703 and 25037 transcripts, 62.2% and 63.0% of the total number of annotated transcripts in barley (Mascher et al., 2017), at levels greater than 5 counts in at least 2 libraries under LDs and SDs, respectively (Supplemental Table S3). Principle component (PC) analysis on all expressed genes clustered the samples according to the developmental stage under LDs and SDs and separated wild-type and mutant samples at all stages under SDs but not under LDs (Supplemental Figure S8). To determine differentially expressed transcripts (DETs), we performed individual pairwise comparisons between each mutant (two allelic mutants) with the wild-type background Bonus at each developmental stage (three stages) within each photoperiod treatment (LDs and SDs), yielding 12 sets of DETs. A large number of DETs were identified at the spikelet initiation stage (W2.0) with 3177 DETs in mat-c.907 and 2876 DETs in mat-c.943 under LDs and 5200 DETs in mat-c.907 and 5585 DETs in mat-c.943 under SDs. At the vegetative stage (W1.0), only 50 DETs in mat-c.907 and 419 DETs in mat-c.943 under LDs and under SDs 14 DETs in mat-c.907 and 17 DETs in mat-c.943 were found. At the stamen primordium stage (W3.5), we identified 503 DETs in mat-c.907 and 477 DETs in mat-c.943 under LDs and 281 DETs in mat-c.907 and 489 DETs in mat-c.943 under SDs (Supplemental Figure S9).
Differences in the number of genes that were affected in the two allelic hvcen mutants may be due to other background mutations in the mutants. Variant calling revealed that 12 and 78 transcripts contain mutations in the mutants mat-c.907 and mat-c.943, respectively, compared to Bonus. Among them, only one transcript (HvCEN) carried different mutations in both mutants (Supplemental Table S4). In order to minimize possible effects of other background mutations, we focused further analyses on transcripts that were differentially regulated in both hvcen mutants, with 33, 1926, and 229 DETs under LDs and 7, 4310, and 117 DETs under SDs at W1.0, W2.0, and W3.5, respectively (Supplemental Figure S9). Most DETs under both LDs and SDs were observed at W2.0, which corresponded to the developmental stage where the highest expression of HvCEN was observed in the MSA (Figs. 5E and 6A; Supplemental Figure S9). Hierarchical cluster analysis separated the developmental stages on the first PC and the photoperiods on the second PC. Only samples harvested at W2.0 were separated for mutants and wild type (Fig. 6B; Supplemental Figure S8C). The majority of genes showed photoperiod-specific regulation. At W2.0 ∼73% of the DETs were photoperiod specific, whereas at W3.5 92% of the DETs were photoperiod specific (Fig. 6A).
Figure 6.
DETs regulated between two hvcen mutants (mat-c.907 and mat-c.943) and wild type (Bonus) in at least one photoperiod condition (LD/SD) and coexpression patterns of the DETs. A, Number of DETs regulated in both mutants (mat-c.907 and mat-c.943) compared with wild type (Bonus) under LDs and/or SDs. The red circles display DETs detected under LDs, and blue circles show DETs detected under SDs. B, Heatmap of coexpression clusters for 4527 DETs. Colors represent log2-fold changes (log2-FC) in expression levels relative to the mean transcript abundance across the tested conditions, i.e. apex (enriched) samples of mat-c.907, mat-c.943, and Bonus grown under LD and SD conditions and harvested at different developmental stages (W1.0, W2.0, W3.5). LDs are 16 h/8 h, light/dark; SDs are 8 h/16 h, light/dark. W, Waddington stage. Transcripts with false discovery rate (FDR) < 0.01 were considered as DETs.
Taken together, HvCEN had the strongest effect on gene expression at spikelet initiation, specifically under SDs. In addition, the majority of transcripts showed a photoperiod- and stage-specific regulation in the mutant and the wild-type plants.
Transcripts Regulated at the Spikelet Initiation Stage in hvcen Mutants
Because spikelet initiation was accelerated in the mutants under both photoperiods, we first focused on all transcripts that were regulated at W2.0 independent of the photoperiod. Gene ontology enrichment analysis suggested that HvCEN primarily affected transcripts involved in chromatin modification, ribosome biogenesis, response to cytokinin, cell proliferation, and metabolic and biosynthetic processes (Supplemental Table S5).
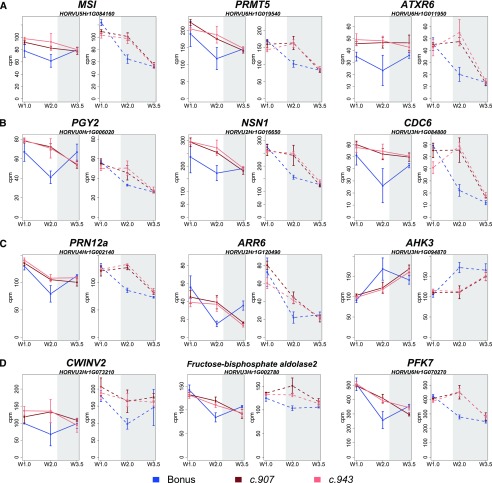
Among the genes with roles in chromatin modification (selected genes in Supplemental Table S6), we observed, for instance, the up-regulation of two homologs of Arabidopsis MULTICOPY SUPRESSOR OF IRA1 (AtMSI1), HORVU5Hr1G084160 (Fig. 7A) and HORVU5Hr1G093230. MSI1 is involved in de novo nucleosome assembly during DNA replication and SAM organization and promotes floral transition in Arabidopsis by inducing the expression of CONSTANS (CO) and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS (SOC1; Bouveret et al., 2006; Steinbach and Hennig, 2014). In addition, the expression of homologs of PROTEIN ARG METHYLTRANSFERASES (AtPRMT5, HORVU6Hr1G019540 [Fig. 7A], and AtPRMT10, HORVU7Hr1G020620) was increased in the mutants compared with wild type. In Arabidopsis, PRMT genes promote flowering by mediating the epigenetic silencing of FLOWERING LOCUS C (AtFLC) and by affecting pre-mRNA splicing (Schmitz et al., 2008; Deng et al., 2010). A putative target of epigenetic factors and repressor of flowering, a homolog of FLC, the grass-specific MADS-box gene HvODDSOC2 (HORVU3Hr1G095240), was downregulated in the mutants. In addition, we observed a higher expression of a homolog of ARABIDOPSIS TRITHORAX-RELATED PROTEIN 6 (ATXR6; HORVU6Hr1G011950; Fig. 7A), which encodes a [Su(var)3-9, Enhancer-of-zeste and Trithorax]-domain protein, a H3K27 monomethyltransferase required for chromatin structure and gene silencing (Jacob et al., 2008). Among the upregulated genes were also ribosomal proteins (selected genes in Supplemental Table S6) with functions in inflorescence development, vascular patterning, and adaxial cell fate, such as two homologs of PIGGYBACK 2 (PGY2, HORVU0Hr1G006020; Fig. 7B; HORVU3Hr1G001140), and a homolog of PGY3 (HORVU5Hr1G092630). A nucleolar GTPase NUCLEOSTEMIN-LIKE 1 (AtNSN1)-like transcript (HORVU2Hr1G016650; Fig. 7B), which plays a role in the maintenance of inflorescence meristem identity and floral organ development by modulating ribosome biogenesis in Arabidopsis (Wang et al., 2012; Jeon et al., 2015), was upregulated in the mutants. Furthermore, we found DETs with putative roles in cytokinin response and cell cycle regulation (Supplemental Table S6). For example, we observed the up-regulation of a barley 26S proteasome non-ATPase regulatory subunit 8 homolog A (RPN12a, HORVU4Hr1G002140; Fig. 7C), which regulates cytokinin responses by up-regulating type A Arabidopsis thaliana RESPONSE REGULATOR proteins (ARRs), which are in turn negative regulators of cytokinin signaling (Ryu et al., 2009). In addition, barley homologs of type A and B RESPONSE REGULATORS (ARR2, HORVU5Hr1G097560.5; ARR3, HORVU2Hr1G077000, HORVU3Hr1G108540; ARR6, HORVU2Hr1G120490, Fig. 7C) that act as regulators in the two-component cytokinin signaling pathway were up-regulated. In contrast, barley homologs of His kinases and putative cytokinin receptors, HIS KINASE 3 and HIS KINASE 4 (AHK3, HORVU3Hr1G094870, Fig. 7C; AHK4, HORVU6Hr1G077070), were down-regulated in the mutants compared with wild-type plants. Alterations in the expression of cytokinin response genes were accompanied by the up-regulation in the expression of genes involved in cell division. These included, for example, homologs of CELL DIVISION CONTROL 6 (CDC6, HORVU3Hr1G084800, Fig. 7B), PROLIFERATING CELL NUCLEAR ANTIGEN 2 (PCNA2, HORVU6Hr1G088120, HORVU0Hr1G031140), and REPLICON PROTEIN A2 (HORVU6Hr1G094080). A faster transition to a reproductive MSA and induction of cell cycle genes coincided with the differential expression of genes involved in cellular respiration, including glycolysis, pyruvate metabolism, and citrate cycle (Supplemental Table S6). For example, transcripts with roles in glycolysis and carbohydrate metabolism were up-regulated in the mutant compared with wild type, i.e. CELL WALL INVERTASE 2 (HORVU2Hr1G073210; Fig. 7D), a RAFFINOSE SYNTHASE family protein (HORVU7Hr1G027930), and a TREHALOSE-6-PHOSPHATASE (TPS1, HORVU1Hr1G013450). Further proteins with roles in glycolysis, such as a PYRUVATE KINASE family protein (HORVU3Hr1G039200), three homologs of FRU-BISPHOSPHATE ALDOLASE 2 (FBA2, HORVU3Hr1G002780; Fig. 7D; HORVU3Hr1G088540, HORVU3Hr1G088570), and three homologs of ATP-dependent 6-PHOSPHOFRUCTOKINASES (PFK2, HORVU2Hr1G081670; PFK3, HORVU3Hr1G070300; PFK7, HORVU6Hr1G070270; Fig. 7D) were up-regulated in the mutant versus wild-type plants. The up-regulation of genes involved in cellular respiration might be a consequence of changes in the source sink balance and a stronger energy demand required for the accelerated vegetative to reproductive stage transitions in the mutant plants compared with the wild type. Taken together, HvCEN had the strongest molecular effects on the MSA transcriptome at spikelet initiation, and these involved genes with functions in chromatin remodeling activities, cytokinin and cell cycle, and regulation and cellular respiration independent of the photoperiod.
Figure 7.
Expression profiles of selected DETs between the hvcen mutants (mat-c.907, mat-c.943) and wild type (Bonus) at W2.0. Genes related to chromatin modification regulation of development, and organ initiation (A); ribosomal proteins, leaf development, leaf patterning (B); regulation of hormone level and hormone response (C); and cellular respiration, sink strength, carbohydrate metabolism, and glycolytic processes (D) under both LDs and SDs. LDs and SDs are shown by white and gray colors. LDs are 16 h/8 h, light/dark; SDs are 8 h/16 h, light/dark; white, light period (16 h); gray, dark period (8 h); W, Waddington stage. Error bars = sd. Transcripts with FDR < 0.01 were considered as DETs. Ten MSAs were pooled for each of the three biological replicates for each time point and genotype.
HvCEN Controls Floral Homeotic Genes under LDs
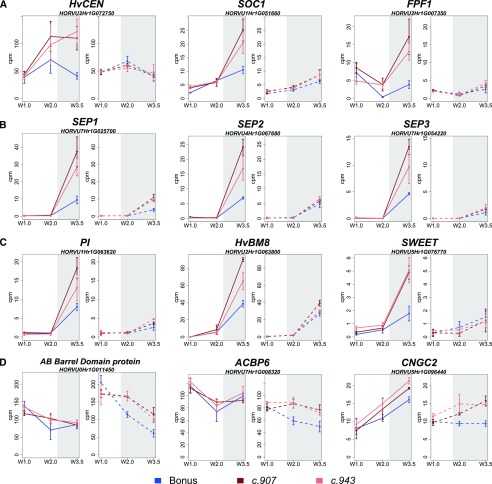
Spikelet initiation was advanced in the mutants under both LDs and SDs, but only florets developed and set seeds under LDs, whereas the MSAs were aborted under SDs. To gain a better understanding of the photoperiod-dependent effects of HvCEN on floret development, we analyzed the photoperiod-dependent transcripts at the stamen primordium stage (W3.5). The photoperiod-specific DETs, including 179 LD-specific and 83 SD-specific DETs, were enriched for transcripts with functions in reproductive processes, response to stimuli, and cell communication (Supplemental Table S7). Among the 179 LD-specific DETs at W3.5, 50 DETs were up-regulated and 129 DETs down-regulated in the mutants compared with wild type. Among the transcripts (selected genes in Supplemental Table S8) that were up-regulated to a higher extent in the mutants compared with wild type only under LD were HvCEN (HORVU2Hr1G072750; Fig. 8A) and flowering promoting genes such as barley homologs of SOC1 (HORVU1Hr1G051660; Fig. 8A), AGAMOUS-LIKE6 (AGL6, HORVU6Hr1G066140), and FLOWERING PROMOTING FACTOR 1 (HORVU2Hr1G007350; Fig. 8A). In addition, we observed the LD-dependent up-regulation of transcription factors that act in a combinatorial manner to achieve floral patterning in Arabidopsis (Coen and Meyerowitz, 1991). These are designated as class A, B, C, and E genes and, except for the class A gene AP2, encode members of the MADS intervening keratin-like and C-terminal type of MADS-box transcription factors. A class A protein, AP1-like (HvBM8, HORVU2Hr1G063800, FDR < 0.05; Fig. 8C; Supplemental Table S9), and a class B like gene, PISTILLATA (PI)-like (HORVU1Hr1G063620, FDR < 0.05; Fig. 8C; Supplemental Table S9), were up-regulated in the mutant lines under LDs but not SDs. In addition, the mutations in HvCEN caused an up-regulation of five E class genes, the SEP-like genes (SEP1, HORVU7Hr1G025700, HORVU5Hr1G095710; SEP2, HORVU4Hr1G067680; SEP3, HORVU7Hr1G054220, HORVU5Hr1G076400, FDR < 0.05) at the stamen primordium stage (Supplemental Table S9; Fig. 8B). An ABERRANT PANICLE ORGANIZATION (APO1)-like transcript (HORVU7Hr1G108970) was down-regulated in the mutants compared with wild type. Interestingly, the ABERRANT PANICLE ORGANIZATION protein positively controls spikelet number by suppressing the precocious conversion of inflorescence meristems to spikelet meristems in rice (Ikeda et al., 2007). In addition, barley homologs of a SWEET Suc transporter (HORVU5Hr1G076770; Fig. 8C), of a GLYCOGEN SYNTHASE (HORVU2Hr1G106410), and a TREHALOSE-6-PHOSPHATE PHOSPHATASE (HORVU6Hr1G074960) were up-regulated specifically at W3.5 in the mutants compared with the wild type. Likewise, a barley homolog of a SHAGGY-related kinase (HORVU3Hr1G034440) required for the establishment of tissue patterning and cell fate determination and a KNOTTED1-like homeobox gene (HORVU7Hr1G114650) involved in meristem differentiation were up-regulated in the mutants at W3.5 under LDs.
Figure 8.
Expression profiles of selected DETs between the hvcen mutants (mat-c.907, mat-c.943) and wild type (Bonus) at W3.5 under both LDs and SDs. A, Transcripts (selected genes in Supplemental Table S8) up-regulated to a higher extent in the mutants compared with wild type only under LD. B and C, Floral homeotic genes and SWEET Suc transporter upregulated in the mutants compared with wild type under LDs and SDs. D, transcripts down-regulated over development under SDs and higher expressed in the mutants vs wild type. LDs and SDs are shown by white and gray colors. LDs are 16 h/8 h, light/dark; SDs are 8 h/16 h, light/dark; white, light period (16 h); gray, dark period (8 h); W, Waddington stage. Error bars = sd. Ten MSAs were pooled for each of the three biological replicates for each time point and genotype. FPF1, FLOWERING PROMOTING FACTOR 1; AB, ALPHA/BETA BARREL domain protein; ACB, ACYL-COA-BINDING PROTEIN 6 (AtACBP6); CNG, CYCLIC NUCLEOTIDE-GATED ION CHANNEL2 (CNGC2).
Under SD, 83 DETs were detected at W3.5, and 58 among them were down-regulated and 25 up-regulated in the mutants compared with wild type. Among the upregulated genes (selected genes in Supplemental Table S8), we identified a number of stress-related genes involved in detoxification such as a stress responsive A/B Barrel Domain protein (DABB, HORVU0Hr1G011450; Fig. 8D) and a homolog of ACYL-COA-BINDING PROTEIN 6 (AtACBP6, HORVU7Hr1G008320; Fig. 8D). Furthermore, we recorded the mutant-specific up-regulation of transcripts with roles in cellular transport, such as a homolog of CYCLIC NUCLEOTIDE-GATED ION CHANNEL2 (AtCNGC2, HORVU5Hr1G096440; Fig. 8D), building nonselective cation channels and of PLASMA MEMBERANE INTRINSIC PROTEIN 3 (AtPIP3, HORVU5Hr1G125600) forming water channels (Fig. 8C). Finally, proteins with roles in starch and sugar metabolism, such as a TREHALOSE-6-PHOSPHATE PHOSPHATASE (TPPH, HORVU5Hr1G058300), a SUC SYNTHASE (HORVU7Hr1G033230), and a UDP-GLC 4-EPIMERASE (HORVU7Hr1G053260), showed a stronger up-regulation in the wild type than the mutants.
Taken together, the LD-specific expression patterns of floral homeotic genes with putative functions in inflorescence, spikelet, and flower development were linked to the differential floret development of mutants and wild type under LDs. Under SDs, the hvcen mutant lines were characterized by the differential regulation of genes implicated in abiotic stress responses, in cellular transport, and carbohydrate metabolism.
DISCUSSION
In the present work, we analyzed induced hvcen mutants to dissect the effects of HvCEN on spike development and plant architecture under different photoperiods and to identify potential molecular targets of HvCEN in the MSA. Mutations in HvCEN accelerated spikelet initiation under LDs and SDs, whereas subsequent floral development required LDs. Therefore, HvCEN showed stage and photoperiod-dependent effects on the development of the MSA. Previous studies have already indicated that spikelet initiation occurs under LDs and SDs, whereas floral development requires LDs in spring barley genotypes (Digel et al., 2015). Floral development, but not spikelet initiation, requires the expression of HvFT1 and TaFT in barley and wheat, respectively (Pearce et al., 2013; Digel et al., 2015). Spikelet initiation occurs in the absence of FT1 expression, likely promoted by FT3. However, the timing of spikelet initiation is affected by variation in FT1 expression (Digel et al., 2015; Dixon et al., 2018).
In Arabidopsis, TFL1 acts antagonistically to the FT protein to repress floral transition and development (Ruiz-García et al., 1997; Jaeger et al., 2013). In addition, rice TFL1-like proteins, such as RICE CENTRORADIALIS (RCN), compete with the rice FT ortholog Hd3a for the binding activities of the 14-3-3 protein, thereby antagonizing the activities of the florigen (Kaneko-Suzuki et al., 2018). Therefore, we also tested if HvCEN genetically interacts with HvFT1 and HvFT3, which affect different stages of preanthesis development (Yan et al., 2006; Casao et al., 2011; Halliwell et al., 2016; Mulki et al., 2018). Although HvFT1 primarily accelerates floral development under LDs (Hemming et al., 2008; Sasani et al., 2009), HvFT3 controls spikelet initiation under LDs and SDs (Mulki et al., 2018). The double mutant hvcen hvft3 did not differ in the timing of spikelet initiation under SDs from the HvCEN hvft3 line, suggesting that the repressive effect of HvCEN on the timing of spikelet initiation depends on a functional HvFT3 gene. In contrast to HvFT3, HvFT1 is only expressed under LDs, and HvFT1 expression is crucial for floral development and flowering (Digel et al., 2015). Consequently, we tested if the LD-specific effect of HvCEN on floral development was dependent on HvFT1 expression. For this purpose, we analyzed an HvCEN hvelf3 single and hvcen hvelf3 double mutant that both expressed HvFT1 under SDs to similar levels as seen under LDs. The strong delay in inflorescence development and higher number of the spikelets in the hvelf3 single mutant as compared with the hvcen hvelf3 double mutant under SDs indicated that the repressive effect of HvCEN on floral development depended on HvFT1 expression in the hvelf3 mutant. In addition, the interaction between HvCEN and FT1 had a strong effect on the determination of spikelet primordia number. However, as many light-dependent transcripts are misregulated in the hvelf3 mutant (Faure et al., 2012), we cannot rule out that other genes influenced the observed phenotype. Our results suggested that HvCEN genetically interacted with different FT-like genes in the shoot apical meristem to control different phases of inflorescence development: with HvFT3 to control spikelet initiation, and with HvFT1 to control floral development. The photoperiod-specific effects of HvCEN are therefore likely caused by the photoperiod-specific expression of its putative competitors HvFT3 and HvFT1.
The hvcen mutants were altered in different shoot architecture traits, including the number of leaves on the main culm, leaf length, the number of tillers per plant, and the number of seeds per spike. The mutant plants developed 1 to 2 fewer leaves on the main culm under LDs and SDs, possibly as a consequence of the earlier transition from a vegetative to a reproductive meristem. Because AXMs initiate in the leaf axils, a reduction in the number of leaves may have caused the reduction in tiller number observed in the mutant lines. In addition, the leaves in the hvcen mutants were shorter, indicating that leaf size is controlled by HvCEN-dependent progression of plant development. It has already been demonstrated that flowering time genes may affect leaf size in barley by affecting the duration of leaf growth and consequent variation in leaf cell number (Digel et al., 2016). The earlier termination of leaf growth in the hvcen mutants was matched by an earlier termination of spikelet induction. However, the barley hvcen mutants did not form a terminal spikelet as has been described for the Arabidopsis tfl1 mutant (Shannon and Meeks-Wagner, 1991). Nevertheless, the mutation in hvcen reduced the period of spikelet primordia initiation and thereby decreased the number of spikelets on the MSA. The wild type reached the maximal number of spikelets at awn primordium stage as observed in previous studies (Riggs and Kirby, 1978; Waddington et al., 1983; Kirby and Appleyard, 1987; Kernich et al., 1997; Alqudah and Schnurbusch, 2014), whereas the hvcen (mat-c) mutants stopped to produce further spikelets at the pistil primordium stage under inductive LD conditions. It has already been shown before that variation in the developmental timing of the early reproductive phase has strong effects on the number of spikelet primordia on the MSA (Campoli and von Korff, 2014; Digel et al., 2015; Ejaz and von Korff, 2017). For instance, in the presence of a wild-type PPD-H1 allele, the development of the MSA is accelerated, and this is associated with a reduction in number of spikelet primordia and final grain number per main spike (Digel et al., 2015). In wheat, Alvarez et al. (2016) observed an acceleration of flowering time and an associated reduction in spikelet number in an elf3 mutant. Both PPD-H1 and HvELF3 likely affect developmental timing and spikelet number by inducing or repressing HvFT1, respectively. Taken together, the pleiotropic phenotypes of the hvcen mutants suggested that changes in the HvFT1/HvCEN ratios may control meristematic activity or its termination in different meristems of the barley shoot as has been proposed for tomato (Solanum lycopersicum; Krieger et al., 2010; Jiang et al., 2013; Lifschitz et al., 2014; Park et al., 2014). However, the molecular basis for these coordinated effects on meristems remain poorly understood.
In Arabidopsis, TFL1 mRNA is expressed in young AXMs but later becomes restricted to the central inflorescence meristem (Shannon and Meeks-Wagner, 1991; Bradley et al., 1997; Wigge et al., 2005; Conti and Bradley, 2007). By contrast, HvCEN mRNA signals were detected primarily in the AXMs, and expression levels were low in the shoot apex despite the strong effect of mutations in HvCEN on developmental timing and spike morphology, i.e. the number of spikelets and seeds per spike. Similarly, expression of the HvCEN homologs RCN1-4 in rice was not detected in the SAM, but in the vasculature of the subtending stem and young primordia and leaf blade (Kaneko-Suzuki et al., 2018). However, the authors demonstrated that the RCN protein is transported through the phloem to the inflorescence meristem where it controls phase transition and inflorescence determinacy. Consequently, HvCEN protein might also travel to the inflorescence meristem where it acts on spike development. Accordingly, the largest number of differentially transcribed genes was detected at the spikelet initiation stage when the SAM switches from vegetative to reproductive growth. This acceleration of spikelet initiation in the hvcen mutants under LDs and SDs coincided with the differential regulation of transcripts associated with chromatin modifications. Epigenetic genes putatively upstream of floral regulators included, for example, MSI1, which is part of the evolutionarily conserved Polycomb group chromatin-remodeling complex and controls spatial and temporal expression of several homeotic genes that regulate plant development and organ identity (Chanvivattana et al., 2004; Hennig et al., 2005; Derkacheva et al., 2013; Steinbach and Hennig, 2014). Similarly PRMT-like genes were upregulated in the mutants, and these control the epigenetic silencing of the floral repressor FLC and flowering time in Arabidopsis (Niu et al., 2007; Pei et al., 2007; Wang et al., 2007; Schmitz et al., 2008). The up-regulation of PRMT-like genes was linked to a down-regulation of a putative repressor of flowering and homolog of FLC, HvODDSOC2, suggesting that its expression might also be controlled epigenetically. Further, we recorded a strong up-regulation of Trithorax-like proteins that act as H3K27 methyltransferases required for transcriptional repression in Arabidopsis (Jacob et al., 2009). HvCEN-dependent regulation of epigenetic regulators in the MSA of barley, specifically at spikelet initiation, suggested that the transition from vegetative to reproductive meristem requires strong reprogramming of transcriptional networks by epigenetic modifiers. These modifiers and their role for developmental transitions in barley await further functional characterization.
Further, we observed the up-regulation of genes involved in cell cycle regulation, cytokinin signaling, and response and many ribosomal proteins that are thought to control cellular growth (Naora, 1999; Bhavsar et al., 2010; Schaller et al., 2014). The up-regulation of ribosomal proteins in the mutants was specific for the spikelet initiation stage, suggesting that the transition from a vegetative to a reproductive meristem requires a strong increase in translational efficiency and de novo protein synthesis. However, several of the ribosomal proteins up-regulated in the mutants may have more specific effects on organ development as these are also described as important regulators of leaf and inflorescence development, vascular patterning, and phase change, such as RPS13-like and PGY-like genes (Ito et al., 2000; Pinon et al., 2008). Spikelet initiation also coincided with the up-regulation of genes involved in carbon metabolism, glycolysis, cellular respiration, and tricarboxylic acid cycle. These transcripts may be important to prepare the plant for the subsequent fast inflorescence growth and increased energy demand. Ghiglione et al. (2008) have demonstrated that fast growing wheat inflorescences show strongly reduced soluble carbohydrate levels as compared with slowly developing spikes and suggested that these fast-growing tissues suffer from carbohydrate starvation. Consequently, the up-regulation of cellular respiration genes was possibly important to allow for a higher energy supply to the developing organs in the fast-growing mutants.
In contrast with spikelet initiation, floral development was photoperiod dependent, and the majority of transcripts in the MSA were regulated by HvCEN only under LDs. In particular, floral homeotic genes were primarily regulated under LD conditions by HvCEN, and these included homologs of floral patterning transcription factors designated as class A, B, and C genes (for review, see Theissen, 2001). The mutations in HvCEN caused an up-regulation of five E class, SEP-like, genes; a class B gene (PI, OsMADS4); and one AP1-like gene (HvBM8) at the stamen primordium stage. In rice, the SEP-like gene OsMADS34 (PANICLE PHYTOMER2) is important for controlling inflorescence and spikelet development (Gao et al., 2010; Kobayashi et al., 2010), whereas the other SEP-like genes OsMADS1, OsMADS5, OsMADS7, OsMADS8, and OsMADS34 control the development of different floral organs (Malcomber and Kellogg, 2005; Zahn et al., 2005; Arora et al., 2007). Interestingly, the barley homolog of OSMADS34 was regulated by HvCEN under both photoperiods, whereas the other SEP-like genes were controlled by HvCEN only under LDs. This suggested that genes important for floral development but not those involved in spikelet initiation were LD dependent. The repression of these genes by HvCEN is not only essential to delay floral development but also to prolong leaf initiation and leaf maturation, probably as a strategy to match vegetative and reproductive development of the plant.
MATERIAL AND METHODS
Germplasms of hvcen (praematurum-c, mat-c), hvelf3 hvcen, and hvft3 hvcen Double Mutants
All barley (Hordeum vulgare) mutants and parental lines were obtained from the Nordic Gene Bank (NordGen; http://www.nordgen.org/). The 21 allelic hvcen mutants were originally generated using different mutagens in various barley spring cultivars (wild-type parents): Bonus, Foma, Frida, Kristina, and Semira (Franckowiak and Lundqvist, 2011; Comadran et al., 2012; Matyszczak, 2014; Supplemental Table S1; Supplemental Figure S2). The different mutants were characterized by distinct changes in the gene body, including premature stop codons, changes in splice sites, amino acid replacement, frameshift, and whole gene deletions (Comadran et al., 2012). The mutation in mat-c.770 introduced a stop codon, resulting in a truncated protein. Mutations in mat-c.94, mat-c.1111, and mat-c.1114 caused splice site changes. Amino acid substitution at conserved sites were detected in mat-c.32, mat-c.770, mat-c.907, mat-c.943, mat-c.913, mat-c.93, mat-c.400, and mat-c.1115 (Supplemental Table S2). The mat-c.1109 genotype carried a 1-bp deletion that resulted in a frame shift. Sanger Sequencing of HvCEN in mat-c.1118 revealed a 12-bp deletion (GATGCAAACAAT) including 2 bp from the end of the 4th exon and 10 bp from the untranslated region (10 bp), leading to a larger protein consisting of 229 instead of 173 amino acids (Supplemental Figure S2). The mat-c.16, mat-c.19, mat-c.1096, mat-c.1107, mat-c.1102, mat-c.1108, and mat-c.1120 mutants were putative deletion mutants as HvCEN could not be amplified in these genotypes. In addition, two backcross-derived introgression lines BW508, with an introgression of mat-c.19, and BW507 with an introgression of mat-b.7 in the background of Bowman, were analyzed (Druka et al., 2011). BW507 likely contains a large deletion of HvCEN since no amplicons of this gene were found (Matyszczak, 2014). The effects of a single amino acid substitution in the point mutants were evaluated by PROVEAN (Protein Variation Effect Analyzer; http://provean.jcvi.org), which computationally predicts the influence of alternations in amino acids on the biological function of the protein (Supplemental Table S2). The program assesses the functional effects of protein variation using an alignment-based score (PROVEN score) that measures the change in sequence similarity between a query sequence and its variants to a homologous protein sequence (Choi et al., 2012).
To determine if HvCEN interacts with HvFT1 and HvFT3 genetically, hvelf3 hvcen (with HvFT1 expression under SDs) and hvft3 hvcen double mutants were produced. The hvelf3 hvcen double mutants were derived from the cross of the hvcen mutant in Bonus (mat-c.907) with the hvelf3 mutant in Bonus (mat-a.8, NGB110008). Three F2 progenies verified as homozygous hvelf3 hvcen double and two as hvelf3 HvCEN single mutants were propagated. F4 plants from these selected five lines were grown and dissected in a controlled climate chamber under SD conditions (8 h/16 h, light/dark, 20°C/18°C).
To obtain hvft3 hvcen double mutants, the hvcen mutant (mat-c.907) in the Bonus background was crossed to an introgressed line carrying a natural mutation in hvft3 in the background of Golden Promise. This introgression line was an F3 progeny derived from crosses between the winter barley cultivar Igri and the spring cultivar Golden Promise. Specifically, the introgression line carried a nonfunctional hvft3 allele from Igri and the natural mutation at Ppd-H1, a deletion in the first regulatory intron of VRN1, and a deletion of the VRN2 locus from Golden Promise. This introgression line shows a reduced photoperiod response and does not require vernalization. By genotyping, three F2 lines carrying homozygous hvft3 hvcen double mutations and two hvft3 HvCEN progenies were identified and propagated.
Plant Growth Conditions and Phenotyping in Outdoor Conditions and Climate Chambers
All mutants and their parental lines (Supplemental Table S1) were evaluated under outdoor conditions over 2 consecutive years for the number of tillers and spikelets per main spike, plant height, and flowering time. Plants were sown in 96-well trays in mid-February in 2014 and early March in 2015, germinated in the greenhouse and then transferred outside (Cologne, Germany). After 5 weeks in 2014 and 3 weeks in 2015, plants were transferred to 12 L pots with one plant per pot during late March, each filled with a custom-made peat and clay soil mixture (EinheitsErde ED73 Osmocote, Einheitserdewerke Werkverband e.V., Sinntal-Altengronau, Germany) containing a long-term fertilizer. The pots were arranged in 22 rows with a distance of 1 m between rows where each row contained 54 pots with a distance of 10 cm. To avoid edge effects, the plot was surrounded by border pots containing cv Morex barley plants. The plot was irrigated by a sprinkling robot and treated with additional fertilizer or pesticides when necessary. Flowering time was scored as the time from sowing to heading, when the awns emerge from the flag leaf of the main culm. Tiller number was recorded at heading, whereas the number of grains per main spike and plant height (soil to base of topmost spike) were measured at full maturity, 2 weeks before harvest. The numbers of mat-c mutants and wild-type plants grown in each year are indicated in Supplemental Table S1.
In parallel, phenotyping was conducted in environment-controlled growth chambers using selected hvcen mutants. Specifically, three hvcen mutants in the Bonus background (mat-c.907, mat-c.94, and mat-c.943) and Bonus were grown in 96-well trays using “Mini Tray” (Einheitserde) soil. To synchronize germination, the trays were stratified in dark at 4°C for 3 d followed by growth under LDs (16 h, 22°C day; 8 h, 18°C night) or SDs (8 h, 22°C day; 16 h, 18°C night). The developmental stage of the MSA was determined using the Waddington quantitative scale of shoot apex development that is based on the progression of spike initiation and then the most advanced floret primordium and pistil of the inflorescence (Waddington et al., 1983). Five to six individual plants for each focal accession were scored for their MSA development every 3 d, and the number of spikelet primordia per main spike, the number of axillary buds, and leaf primordia number were scored during the dissection. The axillary buds scored include all the axillary buds, primary, secondary, and higher order buds. Each leaf was dissected to see the axillary bud under the leaf sheath. Leaf size and visible leaf number on the main culm were scored in 20-well trays as described by Digel et al. (2016).
The hvelf3 hvcen and hvft3 hvcen double mutants and their control or parental lines were scored for differences in preanthesis development, number of spikelet primordia per main spike, and number of axillary buds, including all the axillary buds, primary, secondary, and higher order buds by dissecting the lines every 2/4 d (hvelf3 hvcen) or weekly (hvft3 hvcen) in 96-well trays under SDs.
Statistical Analysis
One-way ANOVA was conducted followed by a post hoc Tukey honestly significant difference test to test for differences in flowering time, plant height, tiller number, and number of seeds per main spike of the mutants compared with their respective parental lines. In addition, a one-way ANOVA was conducted to compare HvFT3 expression in HvCEN wild-type and hvcen mutant backgrounds. In the dissection experiments, differences between the parental lines, the mutant, and double mutant lines were revealed by calculating a polynomial regression model at a 95% confidence interval (Loess smooth line).
RNA Isolation and Sample Preparation for RNA Sequencing
Total mRNA was isolated from plants grown under LDs and SDs. MSA-enriched tissues were harvested and pooled for three distinct developmental stages: the vegetative stage (W1.0), the spikelet initiation stage (W2.0), and the anther primordium stage (W3.5; Waddington et al., 1983). The samples were harvested 2 h before dark under both LD and SD conditions. To enrich shoot apex-specific mRNA, leaves surrounding the MSA were removed manually using a microsurgical stab knife (5-mm blade at 15° [SSC#72-1551]). The enriched MSA tissue was cut from the base of the MSA and still included leaf primordia. At least 10 MSAs were pooled for each of the three biological replicates per time point. All samples harvested for RNA extraction were frozen immediately in liquid nitrogen and stored at −80°C. Total mRNA was extracted using TRIzol reagent (Invitrogen) and further purified using an RNA easy Micro Kit (Qiagen). The residual DNA was removed using a DNA-free kit (Ambion), and the quality of the RNA was assessed using a bioanalyzer (Agilent 2100 Bioanalyzer). The Illumina complementary DNA (cDNA) libraries were prepared according to the TruSeq RNA sample preparation (version 2; Illumina). A cBot (Illumina) was used for clonal sequence amplification and generation of sequence clusters. Single-end sequencing was performed using a HiSeq 3000 (Illumina) platform by multiplexing 8 libraries resulting in ∼ 18 million reads per library. The requested single end read length was 100 bp for LD samples and 150 bp for SD samples. The initial quality control of the raw reads was performed using the FastQC software (version 0.10.1; http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/).
Transcriptome Profiling and Variant Calling
The obtained RNA sequencing reads were mapped to a barley High Confidence (HC) transcripts reference (Mascher et al., 2017) using Salmon in quasi-mapping-based mode. When the quasi-mapping-based index was being built, an auxiliary k-mer hash over k-mers of length 31 was used. U (unstranded single end read) was chosen as library type to quantify the reads of each library. The expected number of reads (NumReads) that have originated from each transcript given the structure of the uniquely mapped and multimapped reads and the relative abundance estimates for each transcript and transcripts per million values were extracted using Salmon (Patro et al., 2017). Transcripts with expression levels greater than 5 NumReads in at least two libraries under LDs or SDs were retained. Tables with expected NumReads (raw counts) and expression levels (normalized cpm) are provided in a supplementary table (Supplemental Table S10).
To identify DETs, pairwise comparisons, including mat-c.907 vs Bonus and mat-c.943 vs Bonus at each stage and photoperiod condition were done using the R bioconductor package Limma-vroom with a Benjamin & Hochberg adjustment for multiple testing (FDR; Ritchie et al., 2015). The comparisons between the stages and between the photoperiods were not conducted to avoid false positive DETs due to differences in the sample types and potential effects of diurnal gene expression differences between photoperiods. The FDR value of 0.01 was used as initial cutoff value for the selection of DETs. In the end, the DETs were extracted per mutant per developmental stage per photoperiod. To minimize the background mutation effects in the two hvcen mutants, the DETs regulated in both mutants were focused on compared with wild type. To visualize the number of genotype- or stage- or photoperiod-dependent DETs, Venn diagrams were drawn using the R package VennDiagram (Chen and Boutros, 2011). The DETs that were observed in both mutants were considered as candidate DETs regulated by HvCEN under each condition. Total, 4527 DETs regulated in the two hvcen mutant at least at one stage compared with wild type under LDs or SDs were obtained. The expression patterns were then examined using hierarchical cluster analysis (using Pearson correlation coefficients) and principle component analysis of the DETs with R.
To determine which gene categories were enriched under each treatment, de novo Gene Ontology (GO) annotations were first produced for High Confidence (HC) transcripts using Blast2Go local blast (e value cutoff 1 X 10−5; Conesa et al., 2005). To assess the effect of HvCEN on biological process, the overrepresentation analysis of particular GO terms was performed based on the Fisher's Exact Test (significant cutoff 0.05) using TopGo package in R (Alexa et al., 2006). The redundant GO categories were removed by first filtering annotated transcripts and allowing similarity as 0.5 (Supek et al., 2011). Then the representative GO categories were retained by keeping the enriched transcripts number ≥ 10 and ≥ 5 at W2.0 and W3.5, respectively.
Variant calling to verify the HvCEN mutation in mat-c.907 and mat-c.943 and evaluate the number of mutated transcripts in the RNA sequencing reads was done as described in van Esse et al. (2017). Briefly, RNA sequencing reads were mapped to a barley High Confidence (HC) coding sequence reference (Mascher et al., 2017) using Burrows-Wheeler-Aligner using maximal exact matches (version 0.7.15; Li, 2013), allowing a mismatch penalty of 3. Mapping was evaluated using PicardTools (version 1.1.00; http://picard.sourceforge.net) CollectAlignmentSummaryMetrics, and SAMtools (version 1.1.3; Li et al., 2009) was used to determine the number of reads mapped with good mapping quality scores (MAPQ > 1). Read duplicate removal and indel realignment were done using PicardTools MarkDuplicates and GATK IndelRealigner (version 3.1-1; McKenna et al., 2010), respectively. Variant calling was done with GATK UnifiedGenotyper using 30.0 as a minimum confidence threshold and 10.0 for emitting of called single nucleotide polymorphisms and 1 for ploidy. Filtered variants with a depth of coverage ≥ 100, a quality of the assigned genotype ≥ 98, and a value of Phred-scaled likelihood ≥ 2000 were taken into consideration. Mutation types included single nucleotide polymorphism and insertion/deletion. The number of mutations and number of mutated transcripts were summarized in Supplemental Table S3.
Gene Expression using RT-qPCR and RNA in Situ Hybridization
RNA was isolated from leaf tissues harvested from plants grown under LD and SD conditions. Under LD conditions, the second youngest leaf on the main shoot of Bonus (HvCEN HvELF3) and mat-c.907 (hvcen HvELF3) at W3.5 was harvested at 2 h before dark. Under SD conditions, the second youngest leaf of three hvcen hvelf3, two HvCEN hvelf3, mat-a.8 (HvCEN hvelf3 -3), mat-c.907, and Bonus were harvested 7 h after the beginning of the dark period at the four- to five-leaf stage. Total RNA extraction, first-strand cDNA synthesis, and RT-qPCR were performed as described in Campoli et al. (2012). The primer for HvFT1 expression is listed in Supplemental Table S10. Two technical replicates were used for each cDNA sample, and starting amounts for each data point were calculated based on the titration curve for each target gene and the reference (HvACTIN) gene using the LightCycler 480 Software (Roche; version 1.5).
The expression of HvCEN (HORVU2Hr1G072750.4) was detected using RNA in situ hybridization on SAMs of Waddington stages W2 and W3.5 as described in Kirschner et al. (2018). The probes were prepared using the whole gene sequences followed by carbonate hydrolysis to ∼200 bp fragments. The antisense probe was used for detecting HvCEN expression, and the sense probe was used for the negative control.
Images were taken using a plan-neofluar 10× objective with a numerical aperture of 0.30 using the Zeiss Axioskop light microscope, and image processing, i.e. stitching, was performed with the Stitching Plugin in Fiji (Preibisch et al., 2009; Schindelin et al., 2012).
Data availability was Illumina data in the European Short Read Archive: E-MTAB-7807.
Accession Numbers
Accession Numbers of major flowering time genes are listed in Supplemental Table S10.
Supplemental Data
Supplemental Figure S1. Seed parameters of hvcen (mat-c) mutants trialled under outdoor condition.
Supplemental Figure S2. The mutation sites of the 14 mat-c mutants on the schematic structure of the HvCEN gene.
Supplemental Figure S3. Determinate spike with a terminal spikelet in wheat (Chinese Spring) and indeterminate spike without terminal spikelet in barley (mat-c.907 and Bonus). W: Waddington stage; white bar: 1 mm.
Supplemental Figure S4. Leaf length and number of leaves on the main culm of hvcen mutants (mat-c.907, mat-c.94, mat-c.943) and Bonus under LDs and SDs.
Supplemental Figure S5. HvFT3 expression in leaves of Bonus, mat-c.907 and mat-c.943 at W2.0 and W3.5 under SDs.
Supplemental Figure S6. HvFT1 expression in leaves.
Supplemental Figure S7. Expression pattern of HvCEN in cv Bonus under LDs.
Supplemental Figure S8. Principal component analysis (PCA) of normalized expression profiles for all expressed genes under LDs and SDs.
Supplemental Figure S9. DETs in each mutant (mat-c.907 or mat-c.943) compared with the wild type (Bonus) under each photoperiod (LDs, SDs) at W1.0, W2.0 and W3.5.
Supplemental Table S1. mat-c (hvcen) mutants carrying different types of mutations in HvCEN.
Supplemental Table S2. Effects of amino acid substitutions in the mat-c (hvcen) mutants.
Supplemental Table S3. Normalized expression value (cpm), log2FC, FDR and annotation of all expressed transcripts.
Supplemental Table S4. Number of transcripts mutated in the two mutants compared to wild type.
Supplemental Table S5. GO enrichment analysis of photoperiod-independent DETs at W2.0.
Supplemental Table S6. Selected transcripts differentially regulated in the mutant MSA at W2.0 under LDs and SDs (FDR < 0.01).
Supplemental Table S7. GO enrichment analysis of photoperiodic-specific DETs (FDR < 0.01) at W3.5.
Supplemental Table S8. Selected photoperiod-independent/dependent DETs at W3.5 (FDR < 0.01).
Supplemental Table S9. Selected DETs involved in floral development and identity at W3.5 (FDR < 0.05).
Supplemental Table S10. Primers used in RT-qPCR and genotyping.
Acknowledgments
We cordially thank Kerstin Luxa, Caren Dawidson, Thea Rütjes, and Andrea Lossow for excellent technical assistance and Artem Pankin for uploading the raw sequencing data to the European Nucleotide Archive.
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) (German Research Foundation) under Germany´s Excellence Strategy – EXC 2048/1 – 390686111; the Priority Programme (SPP1530 Flowering time control-from natural variation to crop improvement) and the Max-Planck-Gesellschaft (Max Planck Society); and a Chinese Scholarship Council fellowship (to X.B.).
Articles can be viewed without a subscription.
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056 [DOI] [PubMed] [Google Scholar]
- Ahn JH, Miller D, Winter VJ, Banfield MJ, Lee JH, Yoo SY, Henz SR, Brady RL, Weigel D (2006) A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO J 25: 605–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexa A, Rahnenführer J, Lengauer T (2006) Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22: 1600–1607 [DOI] [PubMed] [Google Scholar]
- Alqudah AM, Schnurbusch T (2014) Awn primordium to tipping is the most decisive developmental phase for spikelet survival in barley. Funct Plant Biol 41: 424–436 [DOI] [PubMed] [Google Scholar]
- Alvarez MA, Tranquilli G, Lewis S, Kippes N, Dubcovsky J (2016) Genetic and physical mapping of the earliness per se locus Eps-A m 1 in Triticum monococcum identifies EARLY FLOWERING 3 (ELF3) as a candidate gene. Funct Integr Genomics 16: 365–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora R, Agarwal P, Ray S, Singh AK, Singh VP, Tyagi AK, Kapoor S (2007) MADS-box gene family in rice: Genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 8: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhavsar RB, Makley LN, Tsonis PA (2010) The other lives of ribosomal proteins. Hum Genomics 4: 327–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boden SA, Cavanagh C, Cullis BR, Ramm K, Greenwood J, Jean Finnegan E, Trevaskis B, Swain SM (2015) Ppd-1 is a key regulator of inflorescence architecture and paired spikelet development in wheat. Nat Plants 1: 14016. [DOI] [PubMed] [Google Scholar]
- Bommert P, Whipple C (2018) Grass inflorescence architecture and meristem determinacy. Semin Cell Dev Biol 79: 37–47 [DOI] [PubMed] [Google Scholar]
- Bouveret R, Schönrock N, Gruissem W, Hennig L (2006) Regulation of flowering time by Arabidopsis MSI1. Development 133: 1693–1702 [DOI] [PubMed] [Google Scholar]
- Bradley D, Carpenter R, Copsey L, Vincent C, Rothstein S, Coen E (1996) Control of inflorescence architecture in Antirrhinum. Nature 379: 791–797 [DOI] [PubMed] [Google Scholar]
- Bradley D, Ratcliffe O, Vincent C, Carpenter R, Coen E (1997) Inflorescence commitment and architecture in Arabidopsis. Science 275: 80–83 [DOI] [PubMed] [Google Scholar]
- Bull H, Casao MC, Zwirek M, Flavell AJ, Thomas WTB, Guo W, Zhang R, Rapazote-Flores P, Kyriakidis S, Russell J, et al. (2017) Barley SIX-ROWED SPIKE3 encodes a putative Jumonji C-type H3K9me2/me3 demethylase that represses lateral spikelet fertility. Nat Commun 8: 936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campoli C, von Korff M (2014) Genetic control of reproductive development in temperate cereals. In Fornara F, ed. Advances in botanical research. Elsevier, pp 131–158 [Google Scholar]
- Campoli C, Drosse B, Searle I, Coupland G, von Korff M (2012) Functional characterisation of HvCO1, the barley (Hordeum vulgare) flowering time ortholog of CONSTANS. Plant J 69: 868–880 [DOI] [PubMed] [Google Scholar]
- Casao MC, Karsai I, Igartua E, Gracia MP, Veisz O, Casas AM (2011) Adaptation of barley to mild winters: A role for PPDH2. BMC Plant Biol 11: 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanvivattana Y, Bishopp A, Schubert D, Stock C, Moon YH, Sung ZR, Goodrich J (2004) Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development 131: 5263–5276 [DOI] [PubMed] [Google Scholar]
- Chen A, Dubcovsky J (2012) Wheat TILLING mutants show that the vernalization gene VRN1 down-regulates the flowering repressor VRN2 in leaves but is not essential for flowering. PLoS Genet 8: e1003134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Boutros PC (2011) VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics 12: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Sims GE, Murphy S, Miller JR, Chan AP (2012) Predicting the functional effect of amino acid substitutions and indels. PLoS One 7: e46688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen ES, Meyerowitz EM (1991) The war of the whorls: Genetic interactions controlling flower development. Nature 353: 31–37 [DOI] [PubMed] [Google Scholar]
- Comadran J, Kilian B, Russell J, Ramsay L, Stein N, Ganal M, Shaw P, Bayer M, Thomas W, Marshall D, et al. (2012) Natural variation in a homolog of Antirrhinum CENTRORADIALIS contributed to spring growth habit and environmental adaptation in cultivated barley. Nat Genet 44: 1388–1392 [DOI] [PubMed] [Google Scholar]
- Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676 [DOI] [PubMed] [Google Scholar]
- Conti L, Bradley D (2007) TERMINAL FLOWER1 is a mobile signal controlling Arabidopsis architecture. Plant Cell 19: 767–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, Coupland G (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Danilevskaya ON, Meng X, Hou Z, Ananiev EV, Simmons CR (2008) A genomic and expression compendium of the expanded PEBP gene family from maize. Plant Physiol 146: 250–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilevskaya ON, Meng X, Ananiev EV (2010) Concerted modification of flowering time and inflorescence architecture by ectopic expression of TFL1-like genes in maize. Plant Physiol 153: 238–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Gu L, Liu C, Lu T, Lu F, Lu Z, Cui P, Pei Y, Wang B, Hu S, et al. (2010) Arginine methylation mediated by the Arabidopsis homolog of PRMT5 is essential for proper pre-mRNA splicing. Proc Natl Acad Sci USA 107: 19114–19119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkacheva M, Steinbach Y, Wildhaber T, Mozgová I, Mahrez W, Nanni P, Bischof S, Gruissem W, Hennig L (2013) Arabidopsis MSI1 connects LHP1 to PRC2 complexes. EMBO J 32: 2073–2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digel B, Pankin A, von Korff M (2015) Global transcriptome profiling of developing leaf and shoot apices reveals distinct genetic and environmental control of floral transition and inflorescence development in barley. Plant Cell 27: 2318–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digel B, Tavakol E, Verderio G, Tondelli A, Xu X, Cattivelli L, Rossini L, von Korff M (2016) Photoperiod1 (Ppd-H1) controls leaf size. Plant Physiol 172: 405–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon LE, Farré A, Finnegan EJ, Orford S, Griffiths S, Boden SA (2018) Developmental responses of bread wheat to changes in ambient temperature following deletion of a locus that includes FLOWERING LOCUS T1. Plant Cell Environ 41: 1715–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druka A, Franckowiak J, Lundqvist U, Bonar N, Alexander J, Houston K, Radovic S, Shahinnia F, Vendramin V, Morgante M, et al. (2011) Genetic dissection of barley morphology and development. Plant Physiol 155: 617–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejaz M, von Korff M (2017) The genetic control of reproductive development under high ambient temperature. Plant Physiol 173: 294–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure S, Turner AS, Gruszka D, Christodoulou V, Davis SJ, von Korff M, Laurie DA (2012) Mutation at the circadian clock gene EARLY MATURITY 8 adapts domesticated barley (Hordeum vulgare) to short growing seasons. Proc Natl Acad Sci USA 109: 8328–8333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franckowiak JD, Lundqvist U (2011) Descriptions of barley genetic stocks for 2011. Barley Genet Newsl 41: 11–53 [Google Scholar]
- Gao X, Liang W, Yin C, Ji S, Wang H, Su X, Guo C, Kong H, Xue H, Zhang D (2010) The SEPALLATA-like gene OsMADS34 is required for rice inflorescence and spikelet development. Plant Physiol 153: 728–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiglione HO, Gonzalez FG, Serrago R, Maldonado SB, Chilcott C, Curá JA, Miralles DJ, Zhu T, Casal JJ (2008) Autophagy regulated by day length determines the number of fertile florets in wheat. Plant J 55: 1010–1024 [DOI] [PubMed] [Google Scholar]
- Halliwell J, Borrill P, Gordon A, Kowalczyk R, Pagano ML, Saccomanno B, Bentley AR, Uauy C, Cockram J (2016) Systematic Investigation of FLOWERING LOCUS T-like poaceae gene families identifies the short-day expressed flowering pathway gene, TaFT3 in wheat (Triticum aestivum L.). Front Plant Sci 7: 857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanano S, Goto K (2011) Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. Plant Cell 23: 3172–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemming MN, Peacock WJ, Dennis ES, Trevaskis B (2008) Low-temperature and daylength cues are integrated to regulate FLOWERING LOCUS T in barley. Plant Physiol 147: 355–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig L, Bouveret R, Gruissem W (2005) MSI1-like proteins: An escort service for chromatin assembly and remodeling complexes. Trends Cell Biol 15: 295–302 [DOI] [PubMed] [Google Scholar]
- Hicks KA, Millar AJ, Carré IA, Somers DE, Straume M, Meeks-Wagner DR, Kay SA (1996) Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science 274: 790–792 [DOI] [PubMed] [Google Scholar]
- Ho WW, Weigel D (2014) Structural features determining flower-promoting activity of Arabidopsis FLOWERING LOCUS T. Plant Cell 26: 552–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Ito M, Nagasawa N, Kyozuka J, Nagato Y (2007) Rice ABERRANT PANICLE ORGANIZATION 1, encoding an F-box protein, regulates meristem fate. Plant J 51: 1030–1040 [DOI] [PubMed] [Google Scholar]
- Ito T, Kim GT, Shinozaki K (2000) Disruption of an Arabidopsis cytoplasmic ribosomal protein S13-homologous gene by transposon-mediated mutagenesis causes aberrant growth and development. Plant J 22: 257–264 [DOI] [PubMed] [Google Scholar]
- Jacob Y, Feng S, LeBlanc CA, Bernatavichute YV, Stroud H, Cokus S, Johnson LM, Pellegrini M, Jacobsen SE, Michaels SD (2009) ATXR5 and ATXR6 are H3K27 monomethyltransferases required for chromatin structure and gene silencing. Nat Struct Mol Biol 16: 763–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger KE, Pullen N, Lamzin S, Morris RJ, Wigge PA (2013) Interlocking feedback loops govern the dynamic behavior of the floral transition in Arabidopsis. Plant Cell 25: 820–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen CS, Salchert K, Nielsen KK (2001) A TERMINAL FLOWER1-like gene from perennial ryegrass involved in floral transition and axillary meristem identity. Plant Physiol 125: 1517–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon Y, Park YJ, Cho HK, Jung HJ, Ahn TK, Kang H, Pai HS (2015) The nucleolar GTPase nucleostemin-like 1 plays a role in plant growth and senescence by modulating ribosome biogenesis. J Exp Bot 66: 6297–6310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K, Liberatore KL, Park SJ, Alvarez JP, Lippman ZB (2013) Tomato yield heterosis is triggered by a dosage sensitivity of the florigen pathway that fine-tunes shoot architecture. PLoS Genet 9: e1004043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko-Suzuki M, Kurihara-Ishikawa R, Okushita-Terakawa C, Kojima C, Nagano-Fujiwara M, Ohki I, Tsuji H, Shimamoto K, Taoka KI (2018) TFL1-like proteins in rice antagonize rice FT-like protein in inflorescence development by competition for complex formation with 14-3-3 and FD. Plant Cell Physiol 59: 458–468 [DOI] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D (1999) Activation tagging of the floral inducer FT. Science 286: 1962–1965 [DOI] [PubMed] [Google Scholar]
- Kernich GC, Halloran GM, Flood RG (1997) Variation in duration of pre-anthesis phases of development in barley (Hordeum vulgare). Aust J Agric Res 48: 59–66 [Google Scholar]
- Kirby EJM, Appleyard M (1987) Development and structure of the wheat plant. In Lupton FGH, ed. Wheat breeding. Springer, pp 287–311 [Google Scholar]
- Kobayashi K, Maekawa M, Miyao A, Hirochika H, Kyozuka J (2010) PANICLE PHYTOMER2 (PAP2), encoding a SEPALLATA subfamily MADS-box protein, positively controls spikelet meristem identity in rice. Plant Cell Physiol 51: 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962 [DOI] [PubMed] [Google Scholar]
- Komatsuda T, Pourkheirandish M, He C, Azhaguvel P, Kanamori H, Perovic D, Stein N, Graner A, Wicker T, Tagiri A, et al. (2007) Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc Natl Acad Sci USA 104: 1424–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya R, Ikegami A, Tamaki S, Yokoi S, Shimamoto K (2008) Hd3a and RFT1 are essential for flowering in rice. Development 135: 767–774 [DOI] [PubMed] [Google Scholar]
- Koppolu R, Anwar N, Sakuma S, Tagiri A, Lundqvist U, Pourkheirandish M, Rutten T, Seiler C, Himmelbach A, Ariyadasa R, et al. (2013) Six-rowed spike4 (Vrs4) controls spikelet determinacy and row-type in barley. Proc Natl Acad Sci USA 110: 13198–13203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger U, Lippman ZB, Zamir D (2010) The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nat Genet 42: 459–463 [DOI] [PubMed] [Google Scholar]
- Li H. (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv: 1303.3997
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup (2009) The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifschitz E, Ayre BG, Eshed Y (2014) Florigen and anti-florigen---a systemic mechanism for coordinating growth and termination in flowering plants. Front Plant Sci 5: 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XL, Covington MF, Fankhauser C, Chory J, Wagner DR (2001) ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell 13: 1293–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundqvist U. (2014) Scandinavian mutation research in barley - a historical review. Hereditas 151: 123–131 [DOI] [PubMed] [Google Scholar]
- Malcomber ST, Kellogg EA (2005) SEPALLATA gene diversification: brave new whorls. Trends Plant Sci 10: 427–435 [DOI] [PubMed] [Google Scholar]
- Mascher M, Gundlach H, Himmelbach A, Beier S, Twardziok SO, Wicker T, Radchuk V, Dockter C, Hedley PE, Russell J, et al. (2017) A chromosome conformation capture ordered sequence of the barley genome. Nature 544: 427–433 [DOI] [PubMed] [Google Scholar]
- Matyszczak I. (2014). Characterization of early maturity barley mutants praematurum-a, -b and –c. PhD. thesis. Aarhus University, Department of Molecular Biology and Genetics, Faculty of Science and Technology, Aarhus, Denmark. [Google Scholar]
- McGarry RC, Ayre BG (2012) Manipulating plant architecture with members of the CETS gene family. Plant Sci 188-189: 71–81 [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. (2010) The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulki MA, Bi X, von Korff M (2018) FLOWERING LOCUS T3 controls spikelet initiation but not floral development. Plant Physiol 178: 1170–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Shimamoto K, Kyozuka J (2002) Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J 29: 743–750 [DOI] [PubMed] [Google Scholar]
- Naora H. (1999) Involvement of ribosomal proteins in regulating cell growth and apoptosis: Translational modulation or recruitment for extraribosomal activity? Immunol Cell Biol 77: 197–205 [DOI] [PubMed] [Google Scholar]
- Niu L, Lu F, Pei Y, Liu C, Cao X (2007) Regulation of flowering time by the protein arginine methyltransferase AtPRMT10. EMBO Rep 8: 1190–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima S, Murata M, Sakamoto W, Ogura Y, Motoyoshi F (1997) Cloning and molecular analysis of the Arabidopsis gene Terminal Flower 1. Mol Gen Genet 254: 186–194 [DOI] [PubMed] [Google Scholar]
- Park SJ, Jiang K, Tal L, Yichie Y, Gar O, Zamir D, Eshed Y, Lippman ZB (2014) Optimization of crop productivity in tomato using induced mutations in the florigen pathway. Nat Genet 46: 1337–1342 [DOI] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C (2017) Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14: 417–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce S, Vanzetti LS, Dubcovsky J (2013) Exogenous gibberellins induce wheat spike development under short days only in the presence of VERNALIZATION1. Plant Physiol 163: 1433–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Y, Niu L, Lu F, Liu C, Zhai J, Kong X, Cao X (2007) Mutations in the Type II protein arginine methyltransferase AtPRMT5 result in pleiotropic developmental defects in Arabidopsis. Plant Physiol 144: 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinon V, Etchells JP, Rossignol P, Collier SA, Arroyo JM, Martienssen RA, Byrne ME (2008) Three PIGGYBACK genes that specifically influence leaf patterning encode ribosomal proteins. Development 135: 1315–1324 [DOI] [PubMed] [Google Scholar]
- Preibisch S, Saalfeld S, Tomancak P (2009) Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics 25: 1463–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay L, Comadran J, Druka A, Marshall DF, Thomas WT, Macaulay M, MacKenzie K, Simpson C, Fuller J, Bonar N, et al. (2011) INTERMEDIUM-C, a modifier of lateral spikelet fertility in barley, is an ortholog of the maize domestication gene TEOSINTE BRANCHED 1. Nat Genet 43: 169–172 [DOI] [PubMed] [Google Scholar]
- Riggs TJ, Kirby EJM (1978) Developmental consequences of two-row and six-row ear type in spring barley: 1. Genetical analysis and comparison of mature plant characters. J Agric Sci 91: 199–205 [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-García L, Madueño F, Wilkinson M, Haughn G, Salinas J, Martínez-Zapater JM (1997) Different roles of flowering-time genes in the activation of floral initiation genes in Arabidopsis. Plant Cell 9: 1921–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu MY, Cho SK, Kim WT (2009) RNAi suppression of RPN12a decreases the expression of type-A ARRs, negative regulators of cytokinin signaling pathway, in Arabidopsis. Mol Cells 28: 375–382 [DOI] [PubMed] [Google Scholar]
- Sasani S, Hemming MN, Oliver SN, Greenup A, Tavakkol-Afshari R, Mahfoozi S, Poustini K, Sharifi HR, Dennis ES, Peacock WJ, Trevaskis B (2009) The influence of vernalization and daylength on expression of flowering-time genes in the shoot apex and leaves of barley (Hordeum vulgare). J Exp Bot 60: 2169–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller GE, Street IH, Kieber JJ (2014) Cytokinin and the cell cycle. Curr Opin Plant Biol 21: 7–15 [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012) Fiji: An open-source platform for biological-image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz J, Franzen R, Ngyuen TH, Garcia-Maroto F, Pozzi C, Salamini F, Rohde W (2000) Cloning, mapping and expression analysis of barley MADS-box genes. Plant Mol Biol 42: 899–913 [DOI] [PubMed] [Google Scholar]
- Schmitz RJ, Sung S, Amasino RM (2008) Histone arginine methylation is required for vernalization-induced epigenetic silencing of FLC in winter-annual Arabidopsis thaliana. Proc Natl Acad Sci USA 105: 411–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon S, Meeks-Wagner DR (1991) A mutation in the Arabidopsis TFL1 gene affects inflorescence meristem development. Plant Cell 3: 877–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbach Y, Hennig L (2014) Arabidopsis MSI1 functions in photoperiodic flowering time control. Front Plant Sci 5: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Bošnjak M, Škunca N, Šmuc T (2011) REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6: e21800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K (2007) Hd3a protein is a mobile flowering signal in rice. Science 316: 1033–1036 [DOI] [PubMed] [Google Scholar]
- Taoka K, Ohki I, Tsuji H, Furuita K, Hayashi K, Yanase T, Yamaguchi M, Nakashima C, Purwestri YA, Tamaki S, et al. (2011) 14-3-3 Proteins act as intracellular receptors for rice Hd3a florigen. Nature 476: 332–335 [DOI] [PubMed] [Google Scholar]
- Teichmann T, Muhr M (2015) Shaping plant architecture. Front Plant Sci 6: 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen G. (2001) Development of floral organ identity: Stories from the MADS house. Curr Opin Plant Biol 4: 75–85 [DOI] [PubMed] [Google Scholar]
- Turck F, Fornara F, Coupland G (2008) Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol 59: 573–594 [DOI] [PubMed] [Google Scholar]
- Turnbull CGN. (2005) Plant Architecture and Its Manipulation. John Wiley & Sons, Hoboken, NJ [Google Scholar]
- van Esse GW, Walla A, Finke A, Koornneef M, Pecinka A, von Korff M (2017) Six-rowed Spike3 (VRS3) is a histone demethylase that controls lateral spikelet development in barley. Plant Physiol 174: 2397–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington SR, Cartwright PM, Wall PC (1983) A quantitative scale of spike initial and pistil development in barley and wheat. Ann Bot 51: 119–130 [Google Scholar]
- Wang X, Zhang Y, Ma Q, Zhang Z, Xue Y, Bao S, Chong K (2007) SKB1-mediated symmetric dimethylation of histone H4R3 controls flowering time in Arabidopsis. EMBO J 26: 1934–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Gingrich DK, Deng Y, Hong Z (2012) A nucleostemin-like GTPase required for normal apical and floral meristem development in Arabidopsis. Mol Biol Cell 23: 1446–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059 [DOI] [PubMed] [Google Scholar]
- Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Yasuda S, Dubcovsky J (2006) The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc Natl Acad Sci USA 103: 19581–19586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung K, Seitz T, Li S, Janosch P, McFerran B, Kaiser C, Fee F, Katsanakis KD, Rose DW, Mischak H, Sedivy JM, Kolch W (1999) Suppression of Raf-1 kinase activity and MAP kinase signalling by RKIP. Nature 401: 173–177 [DOI] [PubMed] [Google Scholar]
- Youssef HM, Eggert K, Koppolu R, Alqudah AM, Poursarebani N, Fazeli A, Sakuma S, Tagiri A, Rutten T, Govind G, et al. (2017) VRS2 regulates hormone-mediated inflorescence patterning in barley. Nat Genet 49: 157–161 [DOI] [PubMed] [Google Scholar]
- Zagotta MT, Hicks KA, Jacobs CI, Young JC, Hangarter RP, Meeks-Wagner DR (1996) The Arabidopsis ELF3 gene regulates vegetative photomorphogenesis and the photoperiodic induction of flowering. Plant J 10: 691–702 [DOI] [PubMed] [Google Scholar]
- Zahn LM, Kong H, Leebens-Mack JH, Kim S, Soltis PS, Landherr LL, Soltis DE, Depamphilis CW, Ma H (2005) The evolution of the SEPALLATA subfamily of MADS-box genes: A preangiosperm origin with multiple duplications throughout angiosperm history. Genetics 169: 2209–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]