The receptor-like cytoplasmic kinase BSK5 interacts with transmembrane receptors and is required for proper execution of plant immune responses.
Abstract
Plants utilize cell surface-localized pattern recognition receptors (PRRs) to detect pathogen- or damage-associated molecular patterns (PAMP/DAMPs) and initiate pattern-triggered immunity (PTI). Here, we investigated the role of Arabidopsis (Arabidopsis thaliana) BRASSINOSTEROID-SIGNALING KINASE5 (BSK5), a member of the receptor-like cytoplasmic kinase subfamily XII, in PRR-initiated immunity. BSK5 localized to the plant cell periphery, interacted in yeast and in planta with multiple receptor-like kinases, including the ELONGATION FACTOR-TU RECEPTOR (EFR) and PEP1 RECEPTOR1 (PEPR1) PRRs, and was phosphorylated in vitro by PEPR1 and EFR in the kinase activation loop. Consistent with a role in PTI, bsk5 mutant plants displayed enhanced susceptibility to the bacterial pathogen Pseudomonas syringae and to the fungus Botrytis cinerea. Furthermore, bsk5 mutant plants were impaired in several immune responses induced by the elf18, pep1, and flg22 PAMP/DAMPs, including resistance to P. syringae and B. cinerea, production of reactive oxygen species, callose deposition at the cell wall, and enhanced PATHOGENESIS-RELATED1 gene expression. However, bsk5 plants were not affected in PAMP/DAMP activation of mitogen-activated protein kinases and expression of the FLG22-INDUCED RECEPTOR-LIKE KINASE1 or the WRKY domain-containing gene WRKY29. BSK5 variants mutated in the BSK5 myristoylation site, ATP-binding site, and kinase activation loop failed to complement defective PTI phenotypes of bsk5 mutant plants, suggesting that localization to the cell periphery, kinase activity, and phosphorylation by PRRs are critical for the function of BSK5 in PTI. These findings demonstrate that BSK5 plays a role in PTI by interacting with multiple immune receptors.
Plants are sessile organisms and rely only on an innate immune system to ward off pathogens. Plant immune responses are triggered by the recognition of potential pathogenic invaders by membrane-localized pattern recognition receptors (PRRs; Boller and Felix, 2009). PRRs consist of a variety of receptor-like kinases (RLKs) and receptor-like proteins that recognize conserved components of pathogens, commonly referred to as pathogen-associated molecular patterns (PAMPs; Tang et al., 2017). RLKs typically contain an ectodomain, a transmembrane domain, and a cytoplasmic kinase domain, whereas receptor-like proteins contain similar domains to RLKs with the exception of the cytoplasmic kinase domain. Examples of extensively studied RLKs that recognize bacterial PAMPs are the Arabidopsis (Arabidopsis thaliana) FLAGELLIN SENSITIVE2 (FLS2) receptor, which binds the flg22 epitope of flagellin (Zipfel et al., 2004), and the ELONGATION FACTOR-TU (EF-Tu) RECEPTOR (EFR), which binds the elf18 epitope of EF-Tu (Zipfel et al., 2006). In addition to PAMPs, PRRs also perceive endogenous damage-associated molecular patterns (DAMPs) that are released by plant cells during pathogen attack. Two closely related Arabidopsis RLKs, PEP1 RECEPTOR1 (PEPR1) and PEPR2, recognize the DAMP pep1, a peptide that matures from the propeptide PROPEP1 and triggers immune responses (Krol et al., 2010). Recognition of PAMP/DAMPs by PRRs initiates pattern-triggered immunity (PTI), which is the first line of plant defense (Jones and Dangl, 2006). Early PTI responses include Ca2+ influx, generation of reactive oxygen species (ROS), activation of mitogen-activated protein kinases (MAPKs), and deposition of callose at the plant cell wall (Boller and Felix, 2009; Couto and Zipfel, 2016). Late responses include production of ethylene and salicylic acid and transcriptional induction of a large number of defense-related genes (van Loon et al., 2006; Vlot et al., 2009; Bigeard et al., 2015). Collectively, these PTI responses play a crucial role in defending plants against pathogen invasion.
Adapted pathogens have evolved effector proteins that suppress PTI by targeting plant proteins involved in PTI activation and signaling (Macho and Zipfel, 2015). Plants in turn have evolved a second mode of pathogen recognition, in which effectors or products of their activity are recognized by protein receptors that typically contain nucleotide-binding site and Leu-rich repeat domains. This recognition results in effector-triggered immunity, which is the second line of plant defense (Jones and Dangl, 2006).
Plant PRRs recruit receptor-like cytoplasmic kinases (RLCKs) for linking extracellular ligand perception and downstream signaling (Couto and Zipfel, 2016; Liang and Zhou, 2018). RLCKs are evolutionarily related to RLKs but lack an ectodomain and a transmembrane domain (Lehti-Shiu et al., 2009). The Arabidopsis and rice (Oryza sativa) genomes encode 149 and 379 RLCKs, respectively (Lehti-Shiu et al., 2009; Liang and Zhou, 2018). Several RLCKs have been reported to have a role in PTI (Couto and Zipfel, 2016; Liang and Zhou, 2018). For example, BOTRYTIS-INDUCED KINASE1 (BIK1), a member of the Arabidopsis RLCK subfamily VII, associates with the FLS2, EFR, PEPR1, and CHITIN ELICITOR KINASE1 (CERK1) PRRs (Lu et al., 2010; Zhang et al., 2010; Liu et al., 2013). Upon PAMP/DAMP elicitation, the regulatory coreceptor BRASSINOSTEROID INSENSITIVE1 (BRI1)-ASSOCIATED KINASE1 (BAK1) associates with FLS2, EFR, and PEPR1 and phosphorylates BIK1 (Sun et al., 2013; Couto and Zipfel, 2016). BIK1 in turn promotes the production of ROS by direct interaction and phosphorylation of the NADPH oxidase RESPIRATORY BURST OXIDASE HOMOLOG PROTEIN D (RBOHD; Kadota et al., 2014; Li et al., 2014). Like BIK1, the closely related members of the RLCK subfamily VII AVRPPHB SUSCEPTIBLE1 (PBS1), PBS1-LIKE1 (PBL1), PBL2, and PBL5 also contribute to PTI (Zhang et al., 2010; Liu et al., 2013). Two other RLCKs from subfamily VII, PTI COMPROMISED RLCK1 (PCRK1) and PCRK2, also function in PTI signaling downstream of FLS2, EFR, and PEPR1 (Sreekanta et al., 2015; Kong et al., 2016). In rice, OsRLCK176 and OsRLCK185, which are members of the rice RLCK subfamily VII, associate with the chitin receptor OsCERK1 and are required for chitin-induced immune responses (Yamaguchi et al., 2013; Ao et al., 2014). Similarly, PBL27, an ortholog of OsRLCK185, regulates chitin-induced defense responses in Arabidopsis (Yamada et al., 2016).
Brassinosteroid signaling kinases (BSKs) belong to the RLCK subfamily XII that includes 12 members in Arabidopsis (BSK1–BSK12; Tang et al., 2008; Sreeramulu et al., 2013). BSKs contain a putative kinase domain at the N terminus and tetratricopeptide repeats at the C terminus (Tang et al., 2008). Several of them have an established role in brassinosteroid signaling and growth (Tang et al., 2008; Sreeramulu et al., 2013; Zhang et al., 2016). Recent investigation indicates that certain BSKs are also involved in plant immunity. BSK1 associates with FLS2 and is required for flg22-induced ROS production and MAPK signaling (Shi et al., 2013a; Yan et al., 2018). OsBSK1-2, an ortholog of BSK1 in rice, regulates flg22- and chitin-induced defense responses and is involved in rice immunity (Wang et al., 2017). BSK3 interacts with multiple immunity-related RLKs in Arabidopsis (Xu et al., 2014). BSK8 physically associates with FLS2 (Qi et al., 2011) and is phosphorylated upon flg22 treatment (Benschop et al., 2007). Finally, SlBSK7 from tomato (Solanum lycopersicum) interacts with two Pseudomonas syringae effectors, and its silencing compromises PTI (Singh et al., 2014).
In this study, we provide evidence that Arabidopsis BSK5 plays a role in PTI initiated by multiple immune receptors, including FLS2, EFR, and PEPR1. By loss-of-function analysis, we show that BSK5 is important for PTI against Pseudomonas syringae pv tomato strain DC3000 (Pst) and Botrytis cinerea and is required for proper PAMP/DAMP-induced ROS production, callose deposition at the cell wall, and expression of the defense gene PR1 but not for MAPK activation. We also provide evidence suggesting that membrane association, kinase activity, and phosphorylation are required for the biological function of BSK5.
RESULTS
Arabidopsis BSK5 Interacts with Multiple RLKs
Members of the Arabidopsis BSK family of RLCKs were previously implicated in brassinosteroid signaling and plant immunity (Tang et al., 2008; Shi et al., 2013a; Sreeramulu et al., 2013; Yan et al., 2018). To start investigating their molecular properties and interacting partners, BSKs were individually used as baits to screen a yeast two-hybrid Arabidopsis complementary DNA (cDNA) library (Kim et al., 1997). In these screens, 94 BSK5-interacting proteins were identified, including partial cytoplasmic domains of a striking number of RLKs (44) and RLCKs (11; Supplemental Table S1). Among them were 24 proteins previously reported to play a role in pathogen perception and immune signaling (Table 1). A selected group of 11 partial cDNA clones encoding immunity-associated RLKs and a protein phosphatase 2C (PP2C) were isolated from yeast, retransformed, and confirmed as BSK5 interactors (Fig. 1A). Their interaction with BSK5 was specific, as none of them interacted with any of 10 other BSK family members (Supplemental Fig. S1). Expression of bait and prey proteins in yeast was confirmed by western-blot analysis (Supplemental Fig. S2A).
Table 1. Immunity-associated proteins interacting with BSK5 in yeast.
EGF, Epidermal growth factor; LRR, Leu-rich repeat; LysM, Lys motif.
| Name | Family | Accession No. | Amino Acids | No. of Hitsa | References |
|---|---|---|---|---|---|
| SRF7 | LRR-RLK | At3g14350 | 511–636 | 5 | Alcázar et al. (2010) |
| RIPK | RLCK | At2g05940 | 104–359 | 4 | Liu et al. (2011) |
| FERONIA | Malectin-RLK | At3g51550 | 741–843 | 3 | Stegmann et al. (2017) |
| WAKL8 | EGF-like-RLK | At1g16260 | 568–697 | 3 | Delteil et al. (2016) |
| BIR1 | LRR-RLK | At5g48380 | 334–566 | 2 | Gao et al. (2009) |
| BSK1 | RLCK | At4g35230 | 167–502 | 2 | Shi et al. (2013a) |
| IOS1 | LRR-RLK | At1g51800 | 601–879 | 2 | Yeh et al. (2016) |
| LYK5 | LysM-RLK | At2g33580 | 426–592 | 2 | Cao et al. (2014) |
| PBL17 | RLCK | At2g07180 | 167–249 | 2 | Rao et al. (2018) |
| PBL30 | RLCK | At4g35600 | 106–236 | 2 | Rao et al. (2018) |
| PEPR1 | LRR-RLK | At1g73080 | 881–1,034 | 2 | Liu et al. (2013) |
| SRF6 | LRR-RLK | At1g53730 | 461–597 | 2 | Alcázar et al. (2010) |
| WAKL14 | EGF-Like-RLK | At2g23450 | 378–501 | 2 | Delteil et al. (2016) |
| BIR2 | LRR-RLK | At3g28450 | 367–589 | 1 | Halter et al. (2014) |
| ERECTA | LRR-RLK | At2g26330 | 746–895 | 1 | Sánchez-Rodríguez et al. (2009) |
| LYK3 | LysM-RLK | At1g51940 | 299–598 | 1 | Paparella et al. (2014) |
| LecRK-S.5 | Lec-RLK | At5g06740 | 191–488 | 1 | Woo et al. (2016) |
| NIK1 | LRR-RLK | At5g16000 | 378–596 | 1 | Santos et al. (2010) |
| PBL2 | RLCK | At1g14370 | 154–376 | 1 | Zhang et al. (2010) |
| PBL19 | RLCK | At5g47070 | 279–393 | 1 | Rao et al. (2018) |
| PBL27 | RLCK | At5g18610 | 97–339 | 1 | Yamada et al. (2016) |
| PP2C | PP2C | At1g16220 | 228–411 | 1 | Couto et al. (2016) |
| SOBIR1 | LRR-RLK | At2g31880 | 422–558 | 1 | Albert et al. (2015) |
| WAKL18 | EGF-like-RLK | At4g31110 | 492–617 | 1 | Delteil et al. (2016) |
Number of independent cDNA clones encoding the same protein identified in the yeast two-hybrid screen as BSK5 interactors.
Figure 1.
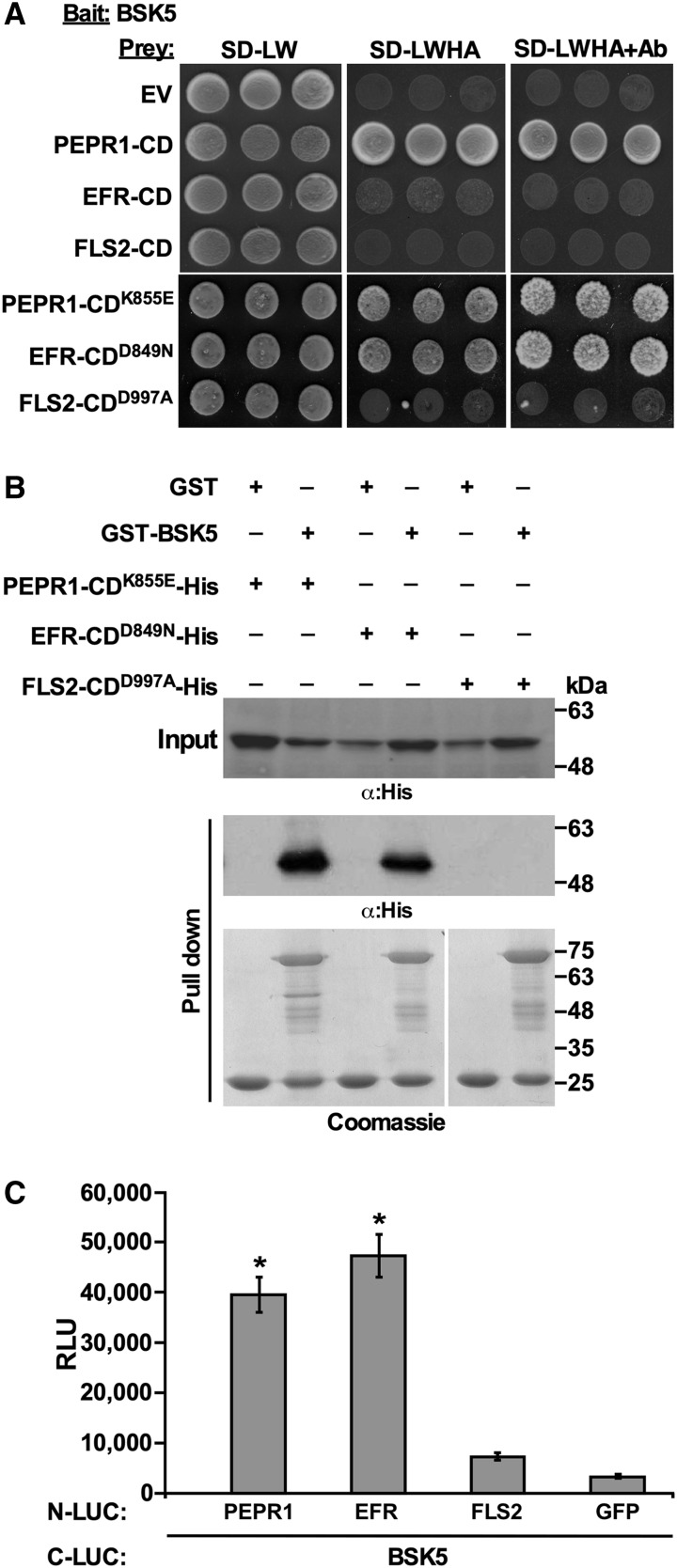
Interaction of BSK5 with multiple RLKs. A, Yeast expressing BSK5 fused to the GAL4 DNA-binding domain (Bait) and partial cDNA clones of the indicated proteins (Table 1) fused to the GAL4 DNA activation domain (Prey) were grown on synthetically defined (SD) medium lacking Leu and Trp (SD-LW), SD-LW lacking His and adenine (SD-LWHA), or SD-LW supplemented with aureobasidin A (SD-LW+Ab). Empty vectors (EV) were used as controls. B, The cytoplasmic domain (CD) of the indicated proteins fused to N-LUC or C-LUC were expressed in N. benthamiana leaves via A. tumefaciens. Luciferase activity was quantified as relative luminescence units (RLU) at 48 h post infiltration. Data are means ± se of three biological repeats. Asterisks indicate statistically significant differences (Student’s t test, P < 0.05) relative to N-LUC-GFP.
Next, we used split luciferase complementation assays to validate in planta protein-protein interactions that were observed in yeast. In these experiments, BSK5 or the closely related BSK6 (Sreeramulu et al., 2013) was fused to the C-terminal half of the luciferase protein (C-LUC) and coexpressed via Agrobacterium tumefaciens in Nicotiana benthamiana leaves with the cytoplasmic domain of selected immunity-associated RLKs (i.e. BAK1-INTERACTING RECEPTOR-LIKE KINASE1 [BIR1], SUPPRESSOR OF BIR1-1 [SOBIR1], LYSIN-MOTIF RECEPTOR KINASE5 [LYK5], ERECTA, STRUBBELIG-RECEPTOR FAMILY7 [SRF7], WALL-ASSOCIATED RECEPTOR KINASE-LIKE14 [WAKL14], or PP2C) fused to the N-terminal half of luciferase (N-LUC). As negative controls, C-LUC-BSK5 was coexpressed with either the N-LUC empty vector or N-LUC-GFP. Protein-protein interactions in planta were quantified by measurements of luminescence at 48 h after agroinfiltration. Coexpression of all the tested proteins with BSK5, but not with BSK6, resulted in the emission of significantly higher luminescence than the negative controls (Fig. 1B). Expression in planta of all the fusion proteins was validated by western-blot analysis (Supplemental Fig. S2B). Together, these results indicate that BSK5 physically interacts with multiple RLKs in yeast and in planta.
BSK5 Interacts with the PEPR1 and EFR PRRs
Because the cytoplasmic domains of PEPR1, receptor of the DAMP pep1 (Yamaguchi et al., 2006), and other PRRs were among the BSK5 interactors, we tested in yeast the interaction of BSK5 with the cytoplasmic domain of the EFR (EFR-CD) and FLS2 (FLS2-CD) RLKs, which are well-characterized receptors of the bacterial PAMPs EF-Tu and flagellin, respectively (Zipfel et al., 2004, 2006). The cytoplasmic domain of PEPR1 (PEPR1-CD) was included in these experiments as a positive control. BSK5 was used as bait and EFR-CD, FLS2-CD, or PEPR1-CD was used as prey. The catalytically inactive forms EFR-CDD849N, FLS2-CDD997A, and PEPR1-CDK855E were also used as prey to stabilize interactions that otherwise may be transient and not detectable. As shown in Figure 2A, BSK5 interacted with both kinase-active and -inactive forms of PEPR1-CD and with the kinase-inactive EFR-CDD849N. No interaction was detected between FLS2-CD variants and BSK5. Expression in yeast of prey proteins was confirmed by western-blot analysis (Supplemental Fig. S3A). To assess whether the interaction of BSK5 with PEPR1 and EFR is direct or if it involves additional proteins, the glutathione-S-transferase (GST)-BSK5 fusion protein was expressed in Escherichia coli and immobilized onto glutathione agarose beads. The His-tagged catalytically inactive forms EFR-CDD849N, FLS2-CDD997A, and PEPR1-CDK855E were used for pull-down assays. PEPR1-CDK855E and EFR-CDD849N, but not FLS2-CDD997A, were pulled down by GST-BSK5 (Fig. 2B), indicating a direct interaction between BSK5 and PEPR1 or EFR.
Figure 2.
Interaction of BSK5 with PEPR1, EFR, and FLS2. A, Yeast expressing BSK5 fused to the GAL4 DNA-binding domain (Bait) and the cytoplasmic domain (CD) of PEPR1, EFR, and FLS2 in wild-type or kinase-deficient forms fused to the GAL4 DNA activation domain (Prey) were grown on synthetically defined (SD) medium lacking Leu and Trp (SD-LW), SD-LW lacking His and adenine (SD-LWHA), or SD-LW supplemented with aureobasidin A (SD-LW+Ab). An empty prey vector (EV) was used as a control. B, Kinase-deficient PEPR1-CD-His, EFR-CD-His, and FLS2-CD-His were incubated with GST or GST-BSK5 bound to glutathione agarose beads. After washes, bound proteins were analyzed by immunoblot with anti-His antibodies (α:His) and stained with Coomassie Blue. Experiments were repeated three times with similar results. C, The indicated proteins (full length) fused to N-LUC or C-LUC were expressed in Arabidopsis protoplasts. Luciferase activity was quantified as relative luminescence units (RLU) at 8 h post transfection. Data are means ± se of three biological repeats. Asterisks indicate significant differences (Student’s t test, P < 0.05) relative to N-LUC-GFP.
The interaction between BSK5 and full-length PEPR1, EFR, and FLS2 was then examined in Arabidopsis protoplasts by the split luciferase complementation assay. BSK5 was fused to C-LUC and coexpressed in protoplasts along with PEPR1, EFR, or FLS2 fused to N-LUC. As the negative control, C-LUC-BSK5 was coexpressed with N-LUC-GFP. Expression of all the examined fusion proteins was validated by western-blot analysis (Supplemental Fig. S3B). The interaction of BSK5 with each PRR was monitored by measuring luminescence at 8 h after cotransfection of the tested protein pairs. C-LUC-BSK5 interacted with N-LUC-PEPR1 and N-LUC-EFR but not with N-LUC-FLS2 (Fig. 2C). Similar interactions were observed when the same protein pairs were expressed via A. tumefaciens in N. benthamiana leaves (Supplemental Figs. S3C and S4). Taken together, these results obtained in different experimental systems indicate that BSK5 physically interacted with the PEPR1 and EFR PRRs but not with FLS2.
PEPR1 and EFR Phosphorylate BSK5 in Vitro
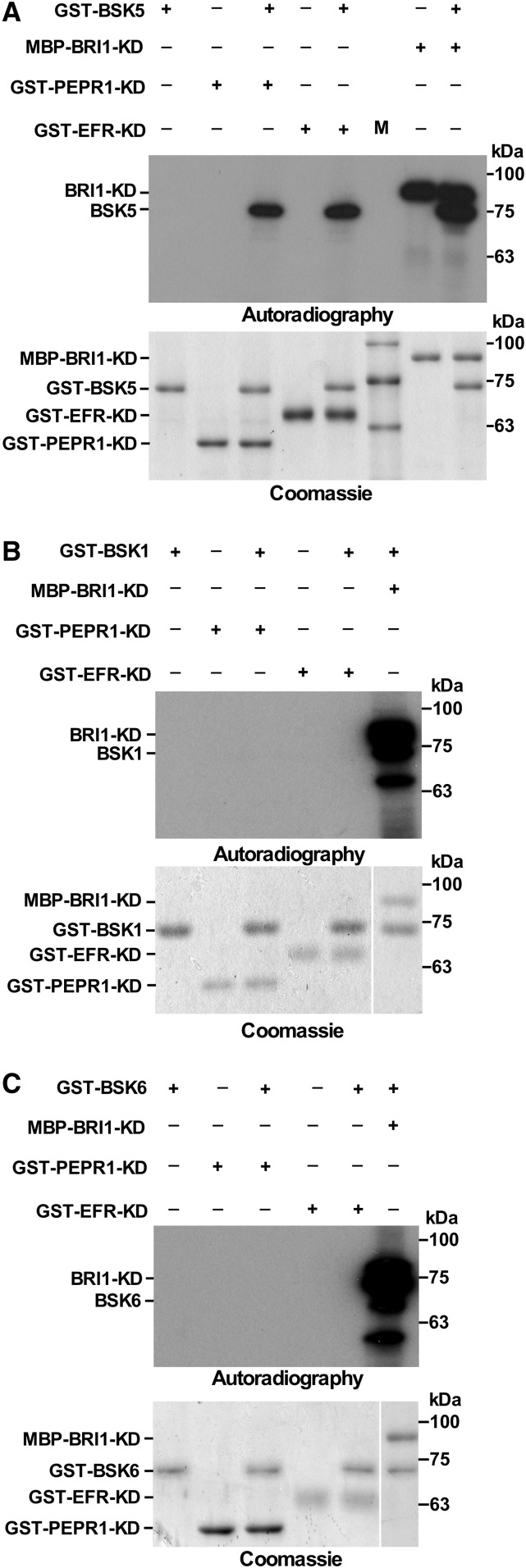
To address the hypothesis that BSK5 is a signaling component acting downstream of PRRs, we tested whether BSK5 is a substrate of PRR phosphorylation. We used a kinase assay to assess whether PEPR1 and EFR phosphorylate BSK5 in vitro. FLS2 was not included in these experiments because it was previously reported to display a weak kinase activity in vitro (Zhang et al., 2010; Schwessinger et al., 2011). BSK1 and BSK6, which did not interact with the tested PRRs, were used to asses phosphorylation specificity, whereas BRI1, which was previously shown to phosphorylate BSKs (Tang et al., 2008; Sreeramulu et al., 2013), was used as a positive control. BSK1, BSK5, and BSK6, and kinase domains of PEPR1 (PEPR1-KD) and EFR (EFR-KD), were fused to GST, while the BRI1 kinase domain was fused to maltose-binding protein (MBP). The fusion proteins were expressed in E. coli and purified. GST-PEPR1-KD, GST-EFR-KD, and MBP-BRI1-KD were incubated with each of the GST-BSK fusions in the presence of [γ-32P]ATP. Reactions were then fractionated by SDS-PAGE, transferred onto a polyvinylidene difluoride (PVDF) membrane, and exposed to autoradiography. As expected, MBP-BRI1-KD autophosphorylated and phosphorylated the GST-BSK fusions (Fig. 3). In agreement with the protein-protein interaction studies (Fig. 1; Supplemental Fig. S1), GST-BSK5, but not GST-BSK1 or GST-BSK6, was phosphorylated by both GST-PEPR1-KD and GST-EFR-KD (Fig. 3). To exclude the possibility that phosphorylation of GST-BSK5 was forced by the formation of GST dimers between the analyzed fusion proteins, an MBP-BSK5 fusion protein was generated and shown to be phosphorylated in vitro by both GST-PEPR1-KD and GST-EFR-KD (Supplemental Fig. S5).
Figure 3.
In vitro phosphorylation of BSK5 by PEPR1 and EFR. BSK1, BSK5, BSK6, and the kinase domain (KD) of PEPR1 and EFR fused to GST and BRI1 fused to MBP were expressed in E. coli and purified. Phosphorylation of GST-BSK5 (A), GST-BSK1 (B), and GST-BSK6 (C) by GST-PEPR1-KD, GST-EFR-KD, and MBP-BRI1-KD was assayed in vitro in the presence of [γ-32P]ATP. Proteins were fractionated and either transferred to a PVDF membrane and exposed to autoradiography or stained with Coomassie Blue. Experiments were repeated three times with similar results.
To determine the sites of BSK5 phosphorylated in vitro by PEPR1 and EFR, GST-BSK5 and GST-PEPR1-KD or GST-EFR-KD were incubated in the presence or absence of ATP, digested by trypsin, and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). In this analysis, Ser-209 and Thr-210 of BSK5 were found to be phosphorylated by both GST-PEPR1-KD and GST-EFR-KD in the presence of ATP (Supplemental Table S2). To validate this finding, GST-BSK5 Ser-209 and Thr-210 were substituted with Ala individually or together and phosphorylation of the mutant proteins by GST-PEPR1-KD and GST-EFR-KD was tested in vitro. As shown in Figure 4, phosphorylation of GST-BSK5 by GST-PEPR1-KD and GST-EFR-KD was significantly decreased or abolished by individual and combined mutations of Ser-209 and Thr-210, respectively, suggesting that these residues represent the main BSK5 sites phosphorylated by the two PRRs in vitro.
Figure 4.
Ser-209 and Thr-210 are major BSK5 sites phosphorylated in vitro by PEPR1 and EFR. BSK5, BSK5S209A, BSK5T210A, BSK5S209A/T210A, and the kinase domain (KD) of PEPR1 and EFR fused to GST were expressed in E. coli and purified. Phosphorylation of GST-BSK5, GST-BSK5S209A, GST-BSK5T210A, and GST-BSK5S209A/T210A by GST-PEPR1-KD (A) and GST-EFR-KD (B) was assayed in vitro in the presence of [γ-32P]ATP. Proteins were fractionated and either transferred to a PVDF membrane and exposed to autoradiography or stained with Coomassie Blue. Experiments were repeated three times with similar results.
BSK5 Contributes to Immunity against Pst and B. cinerea
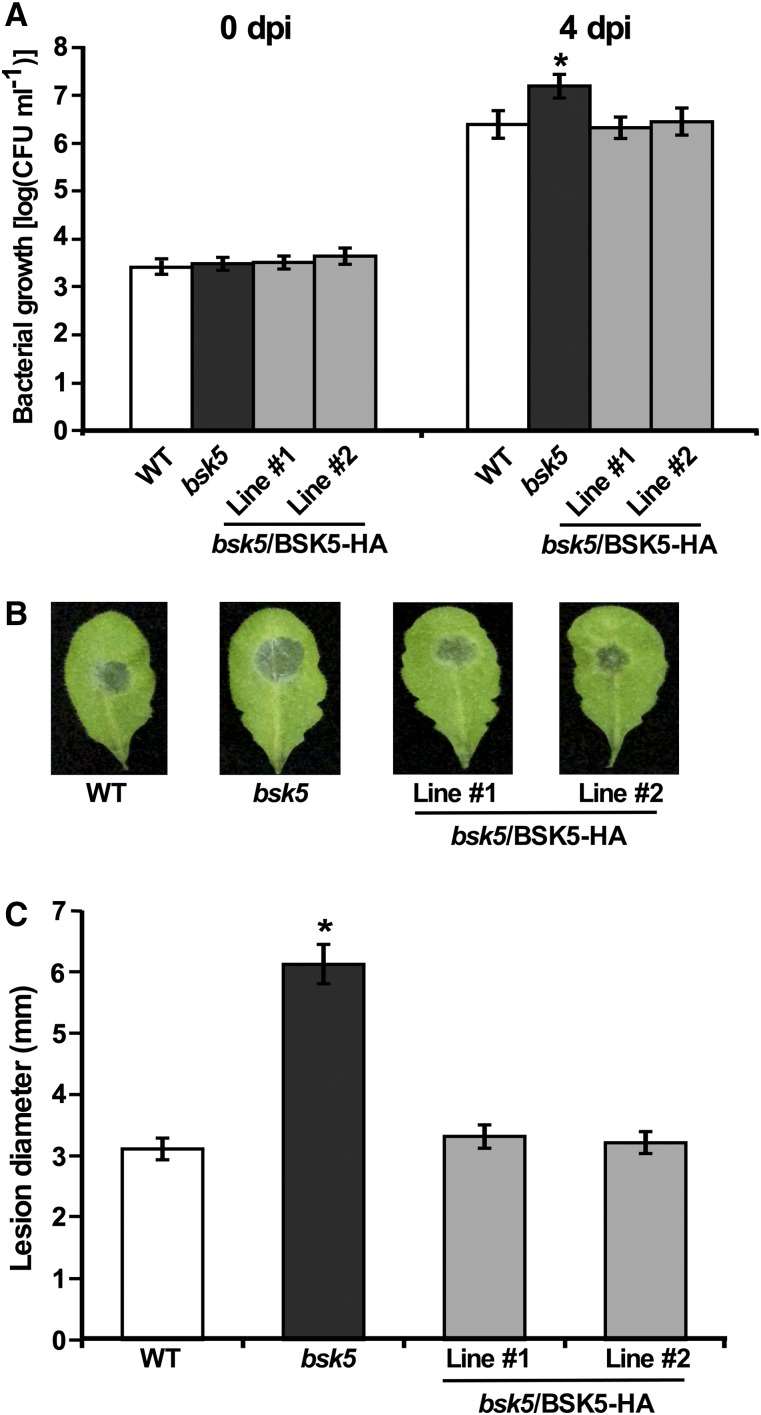
To investigate the role of BSK5 in plant immunity, we tested the susceptibility of an Arabidopsis bsk5 transfer DNA (T-DNA) insertion mutant to the biotrophic bacterial pathogen Pst. Leaves of 5-week-old Arabidopsis Columbia-0 (Col-0) wild-type and bsk5 homozygous mutant plants were inoculated by syringe infiltration with a Pst bacterial suspension (1 × 105 colony-forming units [CFU] mL−1). Bacterial populations were determined in leaf tissues sampled at 0 and 4 d post inoculation (dpi). Pst bacteria displayed a significantly higher growth in bsk5 mutant plants as compared with wild-type plants (Fig. 5A). To confirm that the observed phenotype was caused by BSK5 loss of function, transgenic plants expressing the BSK5 gene driven by its native promoter were generated in the bsk5 mutant background (bsk5/BSK5-HA). Bacterial growth in bsk5/BSK5-HA transgenic lines challenged with Pst was comparable to that observed in the wild-type plants (Fig. 5A), indicating that the enhanced susceptibility phenotype of the bsk5 mutant was complemented by the BSK5 transgene.
Figure 5.
Enhanced susceptibility of bsk5 mutant plants to Pst and B. cinerea. A, Leaves of wild-type (WT), bsk5, and bsk5/BSK5-HA (two lines) plants were inoculated by infiltration with a suspension of Pst (1 × 105 CFU mL−1). Bacterial growth was measured at 0 and 4 dpi. Data are means ± se of three biological replicates each including five plants. The asterisk indicates a significant difference (Student’s t test, P < 0.05) compared with wild-type plants. B and C, Leaves of wild-type, bsk5, and bsk5/BSK5-HA plants were droplet inoculated with a suspension of B. cinerea spores (5 × 105 conidia mL−1). Representative leaves were photographed (B) and the size of disease lesions was measured at 3 dpi (C). Data are means ± se of four biological replicates each including five plants. The asterisk indicates a significant difference (Student’s t test, P < 0.05) compared with wild-type plants.
Next, we tested the susceptibility of bsk5 mutant plants to the necrotrophic fungal pathogen B. cinerea. Wild-type and bsk5 mutant plants were inoculated by placing a droplet of B. cinerea spore suspension (5 × 105 conidia mL−1) on their leaves. Infected leaves were monitored for the development of lesions whose diameter was measured at 3 dpi. Symptoms developed more rapidly and lesions were significantly larger in leaves of bsk5 mutant plants than in wild-type plants (Fig. 5, B and C). Transgenic plants expressing BSK5 in the bsk5 mutant background (bsk5/BSK5-HA) showed similar disease symptoms to wild-type plants (Fig. 5, B and C). These results indicate that BSK5 is involved in Arabidopsis immunity to both Pst bacteria and the fungus B. cinerea.
BSK5 Is Required for PAMP/DAMP-Induced Immunity against Pst and B. cinerea
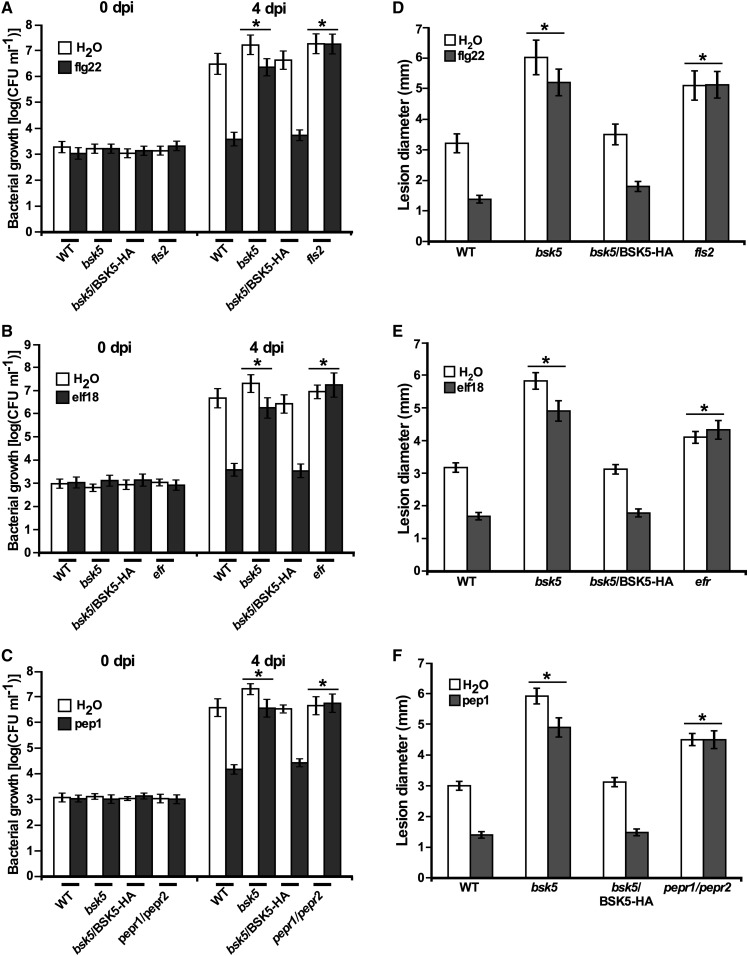
We then tested whether bsk5 mutant plants were affected in PTI. Plants pretreated with a PAMP/DAMP are more resistant to a subsequent pathogen infection because of PTI induction, while immunocompromised plants are equally susceptible if treated with a PAMP/DAMP or untreated (Zipfel et al., 2004). To test the requirement of BSK5 for PTI triggered by PAMP/DAMPs, wild-type (Col-0), bsk5 mutant, and complemented bsk5/BSK5-HA plants were pretreated with flg22, elf18, pep1, or water and 24 h later were infected either by infiltrating leaves with a Pst bacterial suspension (1 × 105 CFU mL−1) or by placing a droplet of B. cinerea spore suspension (5 × 105 conidia mL−1) on the leaves. The fls2, efr, and pepr1/pepr2 mutant plants, which are mutated in the FLS2, EFR, and PEPR1/PEPR2 receptors and insensitive to flg22, elf18, and pep1, respectively, were used as controls. Pst bacterial populations were determined in leaf tissues sampled at 0 and 4 dpi. In wild-type plants pretreated with PAMP/DAMPs, bacterial growth at 4 dpi was lower compared with plants pretreated with water (Fig. 6, A–C). However, in bsk5 mutant plants, the effect of pretreatment with flg22, elf18, and pep1 on bacterial growth compared with water was significantly reduced compared with wild-type plants (Fig. 6, A–C). In bsk5/BSK5-HA complemented plants, the growth pattern of Pst was very similar to that observed in wild-type plants (Fig. 6, A–C). In plants inoculated with B. cinerea, lesion size was measured at 3 dpi. In wild-type and bsk5/BSK5-HA complemented plants pretreated with flg22, elf18, or pep1, lesion size was significantly smaller as compared with plants pretreated with water (Fig. 6, D–F). However, in bsk5 mutant plants, the effect of flg22, elf18, and pep1 pretreatment on lesion size was significantly reduced compared with that in wild-type plants (Fig. 6, D–F). flg22-, elf18-, and pep1-induced immunity to Pst and B. cinerea was completely abolished in fls2, efr, and pepr1/pepr2 mutant plants, respectively, which showed similar bacterial growth and lesion size when treated with PAMP/DAMPs or water (Fig. 6). Together, these results demonstrated that BSK5 plays an important role in PTI induced by multiple PAMP/DAMPs against different pathogens.
Figure 6.
BSK5 is important for PTI induced by multiple PAMP/DAMPs against Pst and B. cinerea. Plants of the indicated genotype were treated with 1 μm flg22 (A and D), elf18 (B and E), pep1 (C and F), or water. A to C, At 24 h, plants were inoculated by infiltration with a suspension of Pst (1 × 105 CFU mL−1). Bacterial growth was measured at 0 and 4 dpi. D to F, At 24 h after pretreatment, plants were droplet inoculated with a suspension of B. cinerea spores (5 × 105 conidia mL−1). The diameter of disease lesions was measured at 3 dpi. Data are means ± se of three biological replicates each consisting of five plants. PTI was measured by subtracting bacterial growth or lesion diameter in PAMP/DAMP-treated plants from that in respective water-pretreated plants. Asterisks indicate significant differences (Student’s t test, P < 0.05) compared with PTI in wild-type (WT) plants.
bsk5 Mutant Plants Display Reductions in ROS Production, Callose Deposition, and Expression of PR1
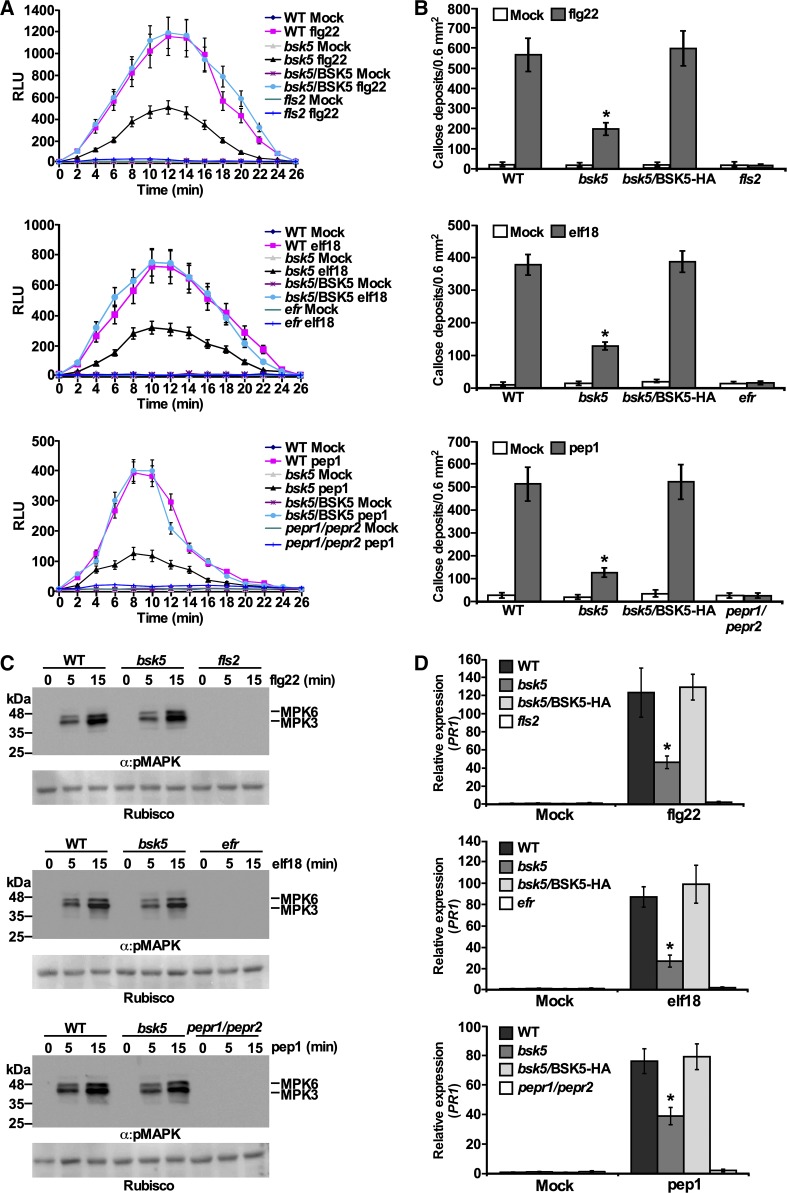
Recognition of PAMP/DAMPs by their receptors triggers PTI resulting in typical immune responses (Gómez-Gómez et al., 1999; Boller and Felix, 2009). To test the involvement of BSK5 in the activation of PTI-associated responses, wild-type, bsk5 mutant, and bsk5/BSK5-HA complemented plants were treated with flg22, elf18, or pep1 and monitored for the accumulation of ROS, callose deposition at the cell wall, and MAPK phosphorylation. The fls2, efr, and pepr1/pepr2 mutants were used as controls in these experiments. Upon treatment with flg22, elf18, or pep1, bsk5 mutant plants produced less ROS and accumulated less callose compared with wild-type and bsk5/BSK5-HA complemented plants (Fig. 7, A and B). However, in bsk5 mutant plants, the MAPKs MPK3 and MPK6 were rapidly phosphorylated in response to PAMP/DAMPs as in wild-type plants (Fig. 7C). As expected, the fls2, efr, and pepr1/pepr2 mutants were unable to respond to flg22, elf18, and pep1, respectively, and did not accumulate ROS or callose or phosphorylate the MAPKs (Fig. 7, A–C).
Figure 7.
PAMP/DAMP-induced PTI responses in bsk5 mutant plants. A, ROS production. Leaf discs from plants of the indicated genotypes were treated with flg22 (100 nm), elf18 (100 nm), pep1 (1 μm), or water and incubated with luminol and horseradish peroxidase. Luminescence was measured as relative luminescence units (RLU) for 26 min after treatment every 2 min. Data are means ± se of three biological repeats each including 10 samples. B, Callose deposition. Leaves were treated with 1 μm flg22, elf18, pep1, or water, and samples were collected 16 h later. Callose deposits were visualized by fluorescence microscopy and counted. Data are means ± se of four biological replicates each with five leaves. C, MAPK phosphorylation. Leaf discs were floated overnight in water and treated with 1 μm flg22, elf18, pep1, or water. Samples were harvested at 0, 5, and 15 min after treatment and analyzed by immunoblots with anti-pMAPK antibody (α:pMAPK). Ponceau S staining of Rubisco is shown as a loading control. Data are representative of three biological repeats. D, PR1 mRNA expression. Leaves were sprayed with 100 nm flg22, elf18, pep1, or water. After 12 h, PR1 mRNA levels were measured by RT-qPCR analysis relative to wild-type (WT) mock-inoculated plants. ACTIN2 was used as a normalizer. Data are means ± se of three biological repeats. In B and D, asterisks indicate significant differences (Student’s t test, P < 0.05) compared with wild-type plants.
We then tested the expression pattern of defense-related genes in bsk5 mutant plants treated with PAMP/DAMPs. The genes included in this analysis were FRK1 and WRKY29, which act downstream of MPK3 and MPK6 and are induced at early time points after PAMP/DAMP treatment (Asai et al., 2002), and PR1, which is induced by salicylic acid and pathogen infection at later time points (Lebel et al., 1998). Reverse transcription quantitative PCR (RT-qPCR) analysis revealed that FRK1 and WRKY29 mRNA accumulated similarly in bsk5 mutant and wild-type plants treated with flg22, elf18, and pep1 but not in fls2, efr, and pepr1/pepr2 mutants treated with the corresponding PAMP/DAMP (Supplemental Fig. S6, A and B). However, upon flg22, elf18, and pep1 treatment, the PR1 mRNA level was significantly lower in bsk5 mutant plants than in wild-type and bsk5/BSK5-HA complemented plants (Fig. 7D). Taken together, these results suggest that BSK5 plays a role in PTI-associated signaling pathways that control ROS production, callose deposition, and expression of PR1 but not MAPK phosphorylation and FRK1 or WRKY29 expression.
Localization to the Cell Periphery Is Required for the Function of BSK5 in Immunity
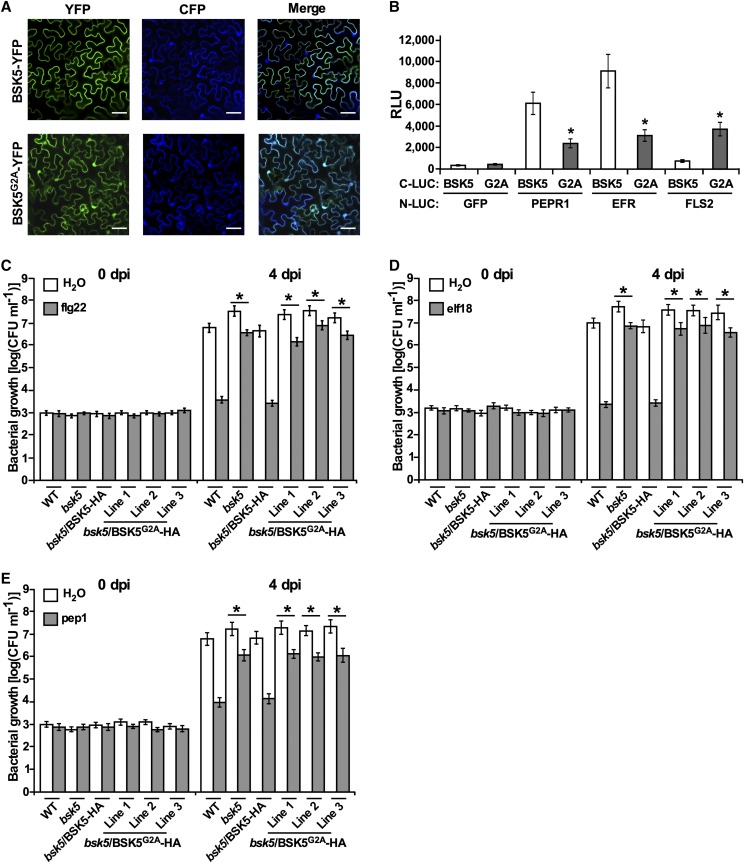
Several Arabidopsis BSK family members (i.e. BSK1, BSK3, BSK6, and BSK8) were shown to localize to the cell plasma membrane (Tang et al., 2008; Shi et al., 2013b; Sreeramulu et al., 2013; Xu et al., 2014). Among them, BSK1 was shown to anchor to the plasma membrane by N-terminal myristoylation (Shi et al., 2013b). Similarly, BSK5 was predicted to contain a myristoylation site at the N terminus. To investigate BSK5 subcellular localization, the BSK5 coding region, either in the wild-type form or carrying a mutation (G2A) in the putative myristoylation site, was fused upstream to the yellow fluorescent protein (YFP). The BSK5-YFP and BSK5G2A-YFP fusions were transiently expressed in leaves of N. benthamiana plants via A. tumefaciens, and their localization was monitored by fluorescence microscopy. A cyan fluorescent protein (CFP), which localizes to the cytoplasm and nucleus (Kruse et al., 2010), was used as a control. As shown in Figure 8A, the BSK5-YFP fusion protein localized exclusively to the cell periphery, while BSK5G2A-YFP was distributed in the cytoplasm and nucleus, similar to CFP. These results suggest that BSK5 anchors to the plasma membrane through N-terminal myristoylation.
Figure 8.
BSK5 is anchored to the plasma membrane through myristoylation. A, The BSK5-YFP and BSK5G2A-YFP fusion proteins were coexpressed with CFP in N. benthamiana leaves via A. tumefaciens. After 36 h, fluorescence was monitored in epidermal cells by confocal microscopy. YFP, CFP, and merged fluorescence images are shown. Bars = 50 μm. B, The indicated proteins fused to N-LUC or C-LUC were expressed in N. benthamiana leaves via A. tumefaciens. After 48 h, luciferase activity was quantified as relative luminescence units (RLU). Data are means ± se of three biological repeats. Asterisks indicate a significant difference (Student’s t test, P < 0.05) relative to C-LUC-BSK5. C to E, Wild-type (WT), bsk5, bsk5/BSK5-HA, and bsk5/BSK5G2A-HA (three lines) plants were treated with 1 μm flg22 (C), elf18 (D), pep1 (E), or water and 24 h later were inoculated by infiltration with a suspension of Pst (1 × 105 CFU mL−1). Bacterial growth was measured at 0 and 4 dpi. Data are means ± se of three biological replicates each consisting of five plants. PTI was measured by subtracting bacterial growth at 4 dpi in PAMP/DAMP-treated plants from that in water-treated plants. Asterisks indicate significant differences compared with the PTI in wild-type plants based on Student’s t test (P < 0.05).
To explore whether membrane association is important for the interaction of BSK5 with PRRs, we used split luciferase complementation assays to examine in planta the physical interaction between the myristoylation mutant BSK5G2A and FLS2, EFR, or PEPR1. In these experiments, C-LUC-BSK5 or C-LUC-BSK5G2A was coexpressed with N-LUC-FLS2, N-LUC-EFR, and N-LUC-PEPR1 in N. benthamiana leaves. The interaction of BSK5G2A with PEPR1 and EFR was significantly weaker than that of the wild-type BSK5 (Fig. 8B). Conversely, BSK5G2A interacted with FLS2 more strongly than wild-type BSK5 (Fig. 8B). These results indicate that membrane localization favors the interaction of BSK5 with PEPR1 and EFR but not with FLS2.
To further study the role of myristoylation in the BSK5 biological function, we generated Arabidopsis transgenic plants expressing BSK5G2A-HA under the control of the BSK5 native promoter in the bsk5 mutant background (bsk5/BSK5G2A-HA). Independent transgenic lines expressed the BSK5G2A-HA fusion protein of the correct size, as detected by anti-HA immunoblot analysis (Supplemental Fig. S7A). We then tested whether BSK5G2A-HA was able to complement the defective PTI phenotype of the bsk5 mutant. To this aim, wild-type, bsk5 mutant, and bsk5/BSK5-HA plants, as well as three independent bsk5/BSK5G2A-HA lines, were pretreated with flg22, elf18, pep1, or water, and after 24 h they were infected with Pst. As observed above, bacterial populations at 4 dpi were lower in wild-type plants pretreated with PAMP/DAMPs as compared with water, while in bsk5 mutant plants, the effect of PAMP/DAMP pretreatment on bacterial growth was significantly reduced (Fig. 8, C–E). As opposed to BSK5-HA, BSK5G2A-HA failed to complement the phenotype observed in bsk5 mutant plants (Fig. 8, C–E), suggesting that membrane localization is required for the BSK5 immune function.
The Putative BSK5 ATP-Binding Site and Kinase Activation Loop Play Roles in the BSK5 Immune Function
Next, we investigated the importance of BSK5 kinase activity and phosphorylation for the BSK5 immune function. To this aim, we used BSK5 variants carrying mutations either at a Lys residue conserved in the ATP-binding site of protein kinases and critical for their activity (Lys-83; Hanks et al., 1988) or at the major BSK5 sites phosphorylated in vitro by PEPR1 and EFR in the kinase activation loop (Ser-209 and Thr-210). BSK5K83E and BSK5S209A/T210A were then tested for their ability to complement the defective PTI response of bsk5 mutant plants to PAMP/DAMPs. For these experiments, we generated transgenic plants expressing BSK5K83E-HA or BSK5S209A/T210A-HA driven by the BSK5 native promoter in the bsk5 mutant background (bsk5/BSK5K83E-HA or bsk5/ BSK5S209A/T210A-HA). Independent transgenic lines expressed the mutant proteins of the correct size, as assessed by anti-HA immunoblot analysis (Supplemental Fig. S7, B and C). Wild-type, bsk5, and bsk5/BSK5-HA plants and three independent bsk5/BSK5K83E-HA or bsk5/ BSK5S209A/T210A-HA lines were pretreated with flg22, elf18, or pep1. Twenty-four hours later, plants were infected with Pst and bacterial populations were determined at 4 dpi. Bacterial growth was lower in wild-type plants pretreated PAMP/DAMPs as compared with water, while in bsk5 mutant plants, the effect of PAMP/DAMPs on bacterial growth was significantly reduced (Fig. 9). The phenotype observed in bsk5 mutant plants was completely complemented by BSK5-HA, but it was not at all or only partially complemented by BSK5K83E-HA and BSK5S209A/T210A-HA, respectively (Fig. 9). These results suggest that BSK5 kinase activity and phosphorylation at Ser-209 and Thr-210 play important roles in the BSK5 immune function.
Figure 9.
Kinase activity and phosphorylation at Ser-209 and Thr-210 play roles in the BSK5 immune function. Wild-type (WT), bsk5, bsk5/BSK5-HA, bsk5/BSK5K83E-HA (three lines), and bsk5/BSK5S209A/T210A-HA (three lines) plants were treated with flg22 (A), elf18 (B), pep1 (C), or water and 24 h later were inoculated by infiltration with a suspension of Pst (1 × 105 CFU mL−1). Bacterial growth was measured at 0 and 4 dpi. Data are means ± se of three biological replicates each consisting of five plants. PTI was measured by subtracting bacterial growth at 4 dpi in PAMP/DAMP-treated plants from that in water-treated plants. Asterisks indicate significant differences compared with the PTI of wild-type plants based on Student’s t test (P < 0.05).
DISCUSSION
A function for Arabidopsis BSK5 in plant immunity was first revealed by the analysis of bsk5 T-DNA insertion mutant plants that showed enhanced susceptibility to Pst bacteria and to the fungus B. cinerea. Subsequent analysis demonstrated that BSK5 is involved in PTI, as resistance to Pst and B. cinerea induced by the flg22 and elf18 PAMPs, as well as the pep1 DAMP, was compromised in bsk5 mutant plants. Its requirement for PTI induced by multiple PAMP/DAMPs and physical interaction in yeast and in planta with a large group of RLKs and RLCKs support the notion that BSK5 is an important molecular hub of plant immunity acting in association with multiple PRRs. Among the RLKs that interacted with BSK5 were the PRRs PEPR1, EFR, and LYK5 as well as other RLKs that were previously shown to form complexes with PRRs or the PRR coreceptor BAK1, including IMPAIRED OOMYCETE SUSCEPTIBILITY1, BIR1, BIR2, SOBIR1, and FERONIA (Tang et al., 2017). BSK3 was also reported to associate with multiple immunity-related RLKs, but the functional relevance of these interactions was not investigated any further (Xu et al., 2014). Similar to BSK5, the RLCK BIK1 was found to interact with multiple PRRs (i.e. FLS2, EFR, PEPR1, and CERK1) and to regulate flg22-, elf18-, pep1-, and chitin-mediated responses (Lu et al., 2010; Zhang et al., 2010; Liu et al., 2013). In addition, the RLCKs PBL1, PBS1, and PCRK1 were shown to be important for PTI responses induced by multiple PAMP/DAMPs (Zhang et al., 2010; Sreekanta et al., 2015). However, other PTI-associated RLCKs appear to play a role only in association with specific PRRs. For example, BSK1 interacts with FLS2 in planta but not with any of the BSK5-interacting RLKs in yeast (Supplemental Fig. S1), and bsk1 mutant plants are insensitive to flg22 treatment but still responsive to elf18 (Shi et al., 2013a). Similarly, PBL27 physically interacts with the chitin receptor CERK1 and contributes to the regulation of chitin-induced immunity but not to flg22 signaling (Shinya et al., 2014). It is interesting that the interaction of PRRs with multiple RLCKs may provide a mechanism for signal amplification and activation of a variety of different outputs. Based on the interaction of BSK5 with multiple PRRs and its requirement for proper PTI responses and in vitro phosphorylation by EFR and PEPR1, it is possible that BSK5 is a signaling component acting downstream of PRRs. Alternatively, it might act upstream of the PRRs and play a role in processes important for the setup of the PTI system, possibly as a scaffold protein coordinating the assembly of immune complexes.
BSK5 also interacted with immunity-associated RLCKs, including BSK1, PBL2, PBL17, PBL19, PBL27, PBL30, and RPM1-INDUCED PROTEIN KINASE (RIPK), suggesting the formation of higher order complexes that contain multiple RLKs and RLCKs (Liang and Zhou, 2018). It is possible that BSK5 and interacting RLCKs cooperate and act at the same level or participate in a sequential cascade that includes an upstream RLCK that activates one or multiple other RLCKs, causing branching of the pathway. The interaction of BSK5 with the sibling BSK1 protein, together with previous findings that BSK1, BSK3, BSK6, BSK8, and BSK11 interact with other BSK family members (Sreeramulu et al., 2013; Xu et al., 2014), suggest that BSKs may function as homodimers or heterodimers. Finally, the interaction of BSK5 with RLKs and additional proteins not involved in plant immunity indicates that BSK5 may also participate in other physiological and developmental processes. In support of this possibility, a mutation in the BSK5 gene was previously reported to affect the response of Arabidopsis seedlings to salinity and abscisic acid (Li et al., 2012).
Despite the fact that a bsk5 mutation has a similar effect on PEPR1, EFR, and FLS2 signaling, protein-protein interaction studies revealed an association of BSK5 with EFR and PEPR1 but not with FLS2. This discrepancy may be the result of a weak or unstable interaction between BSK5 and FLS2 that escapes detection in our assays. It is also possible that the dynamics of the interaction of BSK5 with FLS2 are different than with EFR and PEPR1 and may require activation of the receptor by the corresponding ligand. A ligand-induced RLK-RLCK interaction was reported for the RLCK RIPK that is recruited to a complex with the FERONIA RLK in response to the plant-derived small regulatory peptide rapid alkalinization factor1 (Du et al., 2016).
Consistent with its interaction with transmembrane receptors, subcellular localization studies revealed that BSK5 is associated with the plasma membrane through N-terminal myristoylation. Moreover, membrane association was required for the function of BSK5 in PTI and its interaction with EFR and PEPR1. These observations are in line with previous reports on the requirement of membrane association for the biological function of BSK family members in growth and immunity (Shi et al., 2013b; Zhang et al., 2016). For example, a mutation in the myristoylation site impaired BSK1 interaction with FLS2 and its function in disease resistance displayed by Arabidopsis edr2 mutant plants (Shi et al., 2013a). In addition, a similar mutation in rice OsBSK3 impaired its function in brassinosteroid signal transduction (Zhang et al., 2016). Surprisingly, the myristoylation mutant BSK5G2A, although not functional in flg22-induced PTI, significantly interacted with FLS2. It is possible that localization of BSK5G2A to the cytoplasm facilitates its interaction with FLS2 when internalized from the plasma membrane into endosomes (Robatzek et al., 2006), but the dynamics of this interaction remain to be elucidated further.
BSK5 was found to be phosphorylated in vitro by PEPR1 and EFR in the kinase activation loop at Ser-209 and Thr-210, which were required for the function of BSK5 in PTI. This evidence supports the notion that BSK5 is activated by PRRs through phosphorylation, as previously proposed for other RLCKs acting downstream of PRRs. For example, BIK1 phosphorylation is rapidly induced by flg22 treatment in Arabidopsis protoplasts and dependent on the kinase activity of FLS2 and its coreceptor BAK1 (Lu et al., 2010). BIK1 was also shown to be phosphorylated in vitro by PEPR1 and EFR in the kinase activation loop at residues that are required for BIK1 function in PTI signaling (Liu et al., 2013; Lal et al., 2018). Similarly, Arabidopsis PBL27 and its rice ortholog OsRLCK185 were found to be phosphorylated and thereby possibly activated by the chitin receptor CERK1 of the respective plant species (Yamaguchi et al., 2013; Shinya et al., 2014). It is interesting that BSK5 Ser-209, which was phosphorylated in vitro by PEPR1 and EFR, corresponds to BSK1 Ser-230, the major BSK1 site phosphorylated by the BR receptor BRI1 (Tang et al., 2008). Remarkably, Ser-230 was shown to be important for the role of BSK1 in brassinosteroid signaling (Kim et al., 2009) but not for the disease resistance phenotype displayed by edr2 Arabidopsis mutant plants (Shi et al., 2013a).
It was previously reported that BSK1 and OsBSK3 display kinase activity in vitro requiring Mn2+, rather than Mg2+, as a divalent cation cofactor (Shi et al., 2013a; Zhang et al., 2016). However, we did not detect BSK5 autophosphorylation activity in standard in vitro kinase assays with either Mg2+ or Mn2+, in agreement with previous biochemical analysis of five other Arabidopsis BSKs (Grütter et al., 2013; Sreeramulu et al., 2013). In this study, the requirement of kinase activity for the BSK5 immune function was assessed by mutagenizing Lys-83 in the BSK5 ATP-binding pocket that is predicted to be critical for kinase activity. Amino acid substitution of the corresponding residue Lys-104 in BSK1 and Lys-89 in OsBSK3 abolished kinase activity of the respective proteins (Shi et al., 2013a; Zhang et al., 2016). Expression of BSK5K83E as a transgene in the bsk5 mutant background failed to complement the bsk5-defective PTI phenotype, suggesting that BSK5 kinase activity or ATP binding to BSK5 is required for its function. In support of this notion, BSK1 was recently reported to interact with the MAP kinase kinase kinase MAPKKK5 and to phosphorylate a Ser residue in the N-terminal domain of the protein to regulate immunity in Arabidopsis (Yan et al., 2018). Further investigation will be needed to determine what proteins act downstream of BSK5 and whether they represent substrates for BSK5 phosphorylation.
In bsk5 mutants, increased susceptibility to bacterial and fungal pathogen was associated with defective PTI responses, including reduced ROS production, callose deposition at the cell wall, and expression of PR1 upon treatment with flg22, elf18, and pep1. However, the bsk5 mutation did not affect PAMP/DAMP-induced activation of MPK3 and MPK6 or expression of the FRK1 and WRKY29 genes, which act downstream of MPK3 and MPK6 (Asai et al., 2002). A direct link between PAMP perception and ROS production in Arabidopsis was recently established, as the BIK1 and PBL1 RLCKs were shown to associate with and directly phosphorylate the NADPH oxidase RBOHD upon flg22 perception by the PRR FLS2 (Kadota et al., 2014; Li et al., 2014). However, other signaling components, in addition to BIK1 and PBL1, appear to be involved in RBOHD phosphorylation and thereby in the activation of ROS production. For example, a number of calcium-dependent protein kinases (CPKs), including CPK4, CPK5, CPK6, and CPK11, have been shown to phosphorylate RBOHD and to be required for PAMP-induced ROS production (Dubiella et al., 2013; Gao et al., 2013). It is possible that BSK5 and other RLCKs, which are required for the PAMP/DAMP-triggered ROS burst, affect RBOHD activation either by direct phosphorylation or by regulating the activity of BIK1, PBL1, or CPKs.
Most of the Arabidopsis RLCKs currently known to be involved in PTI have not demonstrated a role in MAPK activation, possibly because of functional redundancy among family members (Zhang et al., 2010; Liu et al., 2013; Shi et al., 2013a; Sreekanta et al., 2015). In support of this hypothesis, a line carrying a combination of mutations in six members of the RLCK family VII (i.e. PBL19/PBL20/PBL37/PBL38/PBL39/PBL40) was recently shown to be impaired in MAPKKK5 phosphorylation and MAPK activation induced specifically by chitin treatment but not by other PAMPs (Bi et al., 2018; Rao et al., 2018). It should also be noted that Arabidopsis PBL27 and its rice homolog RLCK185 were previously reported to be specifically impaired in MAPK activation in response to chitin but not flg22 (Yamaguchi et al., 2013; Shinya et al., 2014; Wang et al., 2017). However, subsequent analysis of pbl27 mutant plants failed to reproduce this phenotype (Rao et al., 2018). Additional members of the RLCK family VII involved in MAPK activation are BIK1, PBL1, and PBL11, as a triple mutant carrying mutations in the corresponding genes displayed a reduction in pep2-triggered MAPK activation (Rao et al., 2018). Our results related to the PTI responses affected in bsk5 mutant plants indicate that BSK5 is required for a subset of PTI responses, including ROS production and callose deposition but not MAPK activation. However, it is still possible that BSK5 also plays a role in MAPK activation, but this is masked by functional redundancy with other BSK family members. In support of this hypothesis, BSK1, which forms a complex with FLS2, was shown to interact with and phosphorylate MAPKKK5, which is required for disease resistance to bacterial and fungal pathogens (Yan et al., 2018).
Together, our data shed new light on the RLCK XII family member BSK5, a novel component of PTI initiated by multiple immune receptors that plays a critical role in the induction of PTI-induced responses. Future identification of BSK5 substrates will lead to the identification of downstream PTI signaling components and to the understanding of their activation mechanisms.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Plant cultivars used were Arabidopsis (Arabidopsis thaliana) ecotype Col-0 and Nicotiana benthamiana (Goodin et al., 2008). The Arabidopsis T-DNA insertion mutants used were bsk5 (Salk_074467; Sreeramulu et al., 2013), fls2 (Salk_141277; Xiang et al., 2008), efr (Salk_044334; Zipfel et al., 2006), and pepr1/pepr2 (kindly provided by Jian-Min Zhou; Liu et al., 2013). Mutants were obtained from the Arabidopsis Biological Resource Center. Arabidopsis transgenic lines containing the constructs ProBSK5:BSK5-HA, ProBSK5:BSK5G2A-HA, ProBSK5:BSK5K83E-HA, and ProBSK5:BSK5S209A/T210A-HA in the bsk5 background were generated by Agrobacterium tumefaciens-mediated transformation of homozygous bsk5 mutant lines. Arabidopsis plants were grown in phytochambers at 20°C to 22°C with 40% to 60% humidity and approximately 120 μE m–2 s–1 light intensity in short-day conditions (8 h of light/16 h of dark) for PTI assays or in long-day conditions (16 h of light/8 h of dark) for seed set. N. benthamiana plants were grown in a growth room in long-day conditions at 25°C.
Bacterial, Yeast, and Fungal Strains and Growth Conditions
The strains used were Escherichia coli DH5α (Invitrogen) and Rosetta (Merck), Pseudomonas syringae pv tomato DC3000 (Guo et al., 2009), A. tumefaciens GV2260 (Deblaere et al., 1985), yeast (Saccharomyces cerevisiae) Y190 and Y2HGold (Clontech Laboratories), and Botrytis cinerea B05.10 (Ma et al., 2017). Bacterial, yeast, and fungal strains were grown with the appropriate antibiotics as follows: E. coli in Luria-Bertani (LB) medium at 37°C; Pst and A. tumefaciens in LB medium at 28°C; yeast at 30°C in selective synthetic complete medium supplemented with 2 % (w/v) Glc; and B. cinerea in potato dextrose broth at 20°C. Antibiotics were used at the following concentrations (μg mL−1): ampicillin, 100; kanamycin, 50; rifampicin, 100; spectinomycin, 50; streptomycin, 100; and aureobasidin A, 20.
Peptide Elicitors
Peptides of flg22 (QRLSTGSRINSAKDDAAGLQIA; Krol et al., 2010), elf18 (Ac-SKEKFERTKPHVNVGTIG; Kunze et al., 2004), and pep1 (ATKVKAKQRGKEKVSSGRPGQHN; Krol et al., 2010) were obtained from EZBiolab, dissolved in water to stock solutions of 1 mm, and diluted to the working concentration.
Construction of Vectors
For yeast two-hybrid assays, genes encoding full-length BSK1 to BSK11 and cytoplasmic domains of PEPR1 (amino acids 781–1,123; PEPR1-CD), EFR (amino acids 680–1,031; EFR-CD), and FLS2 (amino acids 828–1,173; FLS2-CD) were amplified from Arabidopsis Col-0 cDNA using Phusion DNA Polymerases (Thermo Fisher Scientific) and cloned into the pAS1 (bait), pGBT9 (bait), or pGADT7 (prey) vector (Clontech Laboratories) in frame with the GAL4 DNA-binding domain or GAL4 activation domain. The kinase-deficient forms PEPR1-CDK855E, EFR-CDD849N, and FLS2-CDD997A were generated by site-directed mutagenesis using the QuikChange II kit (Agilent Technologies).
For split luciferase complementation assays in N. benthamiana leaves, the BSK5, BSK5G2A, BSK6, PEPR1, EFR, FLS2, and GFP genes were cloned into pCAMBIA1300:C-LUC fused to the C-terminal amino acids 398 to 550 of firefly luciferase or into pCAMBIA1300:N-LUC fused to the N-terminal amino acids 2 to 416 of firefly luciferase and driven by the Cauliflower mosaic virus (CaMV) 35S promoter (Chen et al., 2008). For split luciferase complementation assays in Arabidopsis protoplasts, the BSK5 gene fused to C-LUC and PEPR1, EFR, FLS2, and GFP genes fused to N-LUC were excised using restriction enzymes or PCR amplified from the pCAMBIA vectors and inserted into the pTEX vector under the control of the CaMV 35S promoter (Frederick et al., 1998).
For expression of recombinant proteins in E. coli, BSK1, BSK5, BSK6, and kinase domains of PEPR1 (amino acids 832–1,105; PEPR1-KD) and EFR (amino acids 712–1,031; EFR-KD) were fused to the C terminus of GST in the pGEX-4T-1 vector (GE Healthcare); BSK5 and the kinase domain of BRI1 (amino acids 814–1,196; BRI1-KD) was fused to the C terminus of MBP in the pMAL-c2x vector (New England Biolabs); PEPR1-CD, EFR-CD, and FLS2-CD were cloned into the pET-16b vector with a 10×His tag (Novagen).
For subcellular localization, BSK5 or BSK5G2A coding sequence was fused upstream to the gene encoding the YFP in the pBTEX binary vector (Frederick et al., 1998) under the control of the CaMV 35S promoter.
For transgene complementation, a BSK5 genomic fragment containing its promoter and terminator regions (ProBSK5:BSK5-HA) was amplified from Col-0 genomic DNA and cloned into the binary vector pCAMBIA3300 (Cambia).
The mutant BSK5 genomic fragments ProBSK5:BSK5G2A-HA, ProBSK5:BSK5K83E-HA, and ProBSK5:BSK5S209A/T210A-HA were generated by site-directed mutagenesis and cloned into the binary vector pCAMBIA3300 for complementation assays. All constructs were verified by DNA sequencing to ensure introduction of the desired mutations and exclude undesired mutations. Sequences of oligonucleotides used in this study are listed in Supplemental Table S3.
Protoplast Preparation and Transfection
Protoplasts were prepared from leaves of 5-week-old Arabidopsis plants and transfected with plasmid DNA as described by Popov et al. (2016).
A. tumefaciens-Mediated Transient Expression
For transient expression, A. tumefaciens overnight cultures were pelleted, washed three times with 10 mm MgCl2, resuspended in induction medium (10 mm MgCl2, 10 mm MES [pH 5.6], and 200 mm acetosyringone), and incubated at 28°C with shaking for 3 to 4 h. A. tumefaciens cultures were diluted to OD600 = 0.2 and infiltrated into leaves of 6-week-old N. benthamiana plants using a needleless syringe.
Arabidopsis Transformation
Arabidopsis plants were transformed using the floral dip method with A. tumefaciens strain GV2260 (Zhang et al., 2006). Transformant seeds were germinated on plates (one-half-strength Murashige and Skoog salts with vitamins supplemented with MES, Suc [pH 5.7], and 0.8% [w/v] agar) supplemented with Basta (10 μg mL−1) and timentin (150 μg mL−1). After 2 weeks, resistant plants were transferred to soil.
Expression and Purification of GST and MBP Fusion Proteins in E. coli
BSK1, BSK5, BSK6, PEPR1-KD, and EFR-KD were cloned into the pGEX-4T-1 vector, while BSK5 and BRI1-KD were cloned into the pMAL-c2x vector. Plasmids were transformed into E. coli Rosetta strain. Bacterial cultures were grown at 37°C to OD600 = 0.4 to 0.6, supplemented with 0.1 mm isopropyl β-d-1-thiogalactopyranoside, and incubated overnight at 16°C with shaking. Bacteria were pelleted, resuspended in column binding buffer (25 mm Tris-HCl [pH 7.5], 150 mm NaCl, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride [PMSF], 5 μg mL−1 leupeptin, and 5 μg mL−1 aprotinin), lysed using a French press, and centrifuged. Supernatants were incubated with glutathione agarose beads (Sigma-Aldrich) or amylose resin beads (New England Biolabs), and proteins were purified according to the manufacturer’s instructions.
Yeast Two-Hybrid Assay
Yeast two-hybrid interactions and library screening were conducted as described (Sreeramulu et al., 2013).
Split Luciferase Complementation Assay
Gene fragments encoding the cytoplasmic domain of BIR1 (amino acids 309–620), SOBIR1 (351–566), LYK5 (382–616), ERECTA (652–876), SRF7 (415–641), WAKL14 (476–708), and PP2C (227–478) and full-length genes of PEPR1, EFR, FLS2, BSK5, BSK5G2A, BSK6, and GFP were cloned in frame to firefly luciferase fragments in the binary vector pCAMBIA1300:N-LUC or pCAMBIA1300:C-LUC. The obtained vectors were transformed into A. tumefaciens and coexpressed in N. benthamiana leaves. Split luciferase complementation assays were performed as described by Chen et al. (2008) with minor modifications. Three-millimeter-diameter leaf discs were harvested at 48 h after agroinfiltration and floated in 100 µL of water on a white 96-well plate. Samples were supplemented with 1 mm d-luciferin (Sigma-Aldrich) and incubated in the dark for 10 min to quench fluorescence. Luminescence was measured using a Veritas Microplate Luminometer (Promega).
In Vitro Pull-Down Assay
GST and GST-BSK5 were expressed in E. coli, and bacteria were lysed in column binding buffer (25 mm Tris-HCl [pH 7.5], 150 mm NaCl, 1 mm EDTA, 1 mm PMSF, 5 μg mL−1 leupeptin, and 5 μg mL−1 aprotinin). The soluble protein was incubated with glutathione agarose beads (Sigma-Aldrich) at 4°C for 2 h. After three washes with binding buffer, beads were incubated with equal amounts of bacterial lysate containing PEPR1-CD-His, EFR-CD-His, or FLS2-CD-His recombinant protein with constant rotation at 4°C for 3 h. The beads were washed five times with wash buffer (25 mm Tris-HCl [pH 7.5], 150 mm NaCl, and 1 mm EDTA), and the bound protein was eluted with elution buffer (25 mm Tris-HCl [pH 7.5], 150 mm NaCl, and 1 mm EDTA containing 10 mm reduced glutathione). Input and pulled-down proteins were fractionated by 10% (v/v) SDS-PAGE and detected by western-blot analysis with anti-His antibodies.
Protein Extraction
For protein extraction from yeast, overnight-grown cultures were pelleted, resuspended in ice-cold lysis buffer (4% [v/v] 5 n NaOH and 0.5% [v/v] β-mercaptoethanol), and incubated with SDS sample buffer (30% [v/v] glycerol, 15% [v/v] β-mercaptoethanol, 37.5% [v/v] 500 mm Tris-HCl [pH 6.8], 0.15% [w/v] SDS, and a few grains of Bromophenol Blue) for 10 min at 95°C.
For protein extraction from leaves, three to six leaf discs (1 cm diameter) were frozen in liquid nitrogen, homogenized in extraction buffer (100 mm Tris [pH 7.4], 1% [v/v] Triton X-100, 1 mm PMSF, 5 μg mL−1 leupeptin, 5 μg mL−1 aprotinin, 50 mm NaF, and 1 mm Na3VO4), and centrifuged at 17,000g for 30 min at 4°C. The protein concentration of the supernatants was determined using Bradford protein assay solution (Bio-Rad), and equal amounts of protein were fractionated by 10% (v/v) SDS-PAGE and then subjected to western-blot analysis with specific antibodies.
In Vitro Kinase Assay
GST and MBP fusion proteins (0.1–0.5 µg) were incubated in a kinase assay solution (50 mm Tris-HCl [pH 7], 1 mm DTT, 10 mm MgCl2, 20 µm ATP, and 10 µCi of [γ-32P]ATP [3,000 Ci mmol−1; Perkin-Elmer]) at 25°C for 1 h. Reactions were stopped by the addition of SDS sample buffer. Half of the reaction volume was fractionated by SDS-PAGE and stained with Coomassie Blue. The second half was fractionated by SDS-PAGE and transferred onto a PVDF membrane, and the membrane was exposed to autoradiography.
LC-MS/MS
Mass spectrometry analysis was performed at The Nancy & Stephen Grand Israel National Center for Personalized Medicine, Weizmann Institute of Science. LC-MS/MS analysis was carried out as described (Gillette and Carr, 2013).
Subcellular Localization
To visualize BSK5 subcellular localization, the BSK5-YFP and BSK5G2A-YFP fusion proteins were expressed via A. tumefaciens in leaves of 6-week-old N. benthamiana plants. Protein localization was visualized by a confocal laser scanning microscope (LSM 510 META; Zeiss). Images were processed with AxioVision software (Zeiss). CFP was used as a control for colocalization (Kruse et al., 2010). YFP and chlorophyll were excited with an argon laser at 488 nm, while CFP was excited with a diode laser at 405 nm. Emission was detected with a spectral detector set between 420 and 490 nm for CFP and between 505 and 550 nm for YFP.
Bacterial Growth Assay
Four-week-old Arabidopsis plants were inoculated by infiltration with a suspension (1 × 105 CFU mL−1) of Pst in a 10 mm MgCl2 solution using a needleless syringe. Three 1-cm-diameter leaf discs were sampled at 0 and 4 dpi from five inoculated plants and ground in 1 mL of 10 mm MgCl2. Samples were then 10-fold serially diluted and plated on LB plates supplemented with 25 mm rifampicin. The colony counts were recorded 2 d after incubation at 28°C.
B. cinerea inoculation
B. cinerea spores were diluted to 5 × 105 spores mL−1 in 0.5× potato dextrose broth. Droplets (10 µL) of 0.5× potato dextrose broth with B. cinerea spores were deposited on leaf surfaces of 4-week-old Arabidopsis plants (three leaves per plant). After incubation of the inoculated plants at high humidity for 3 d, the size of the disease lesion was measured. At least 15 lesion diameters were evaluated for each independent treatment (five plants).
ROS Assay
Leaf discs from 4-week-old Arabidopsis plants were placed on 96-well white plates floating on 100 μL of deionized water overnight. PAMP/DAMPs (100 nm flg22, 100 nm elf18, or 1 μm pep1) or water was then added with 20 μm luminol (Sigma-Aldrich) and 2 μg mL−1 horseradish peroxidase (Sigma-Aldrich). Luminescence was measured with a Veritas Microplate Luminometer (Promega) over a period of 30 min at 2-min intervals. At least three independent biological repeats were carried out for each PAMP/DAMP.
Callose Deposition Assay
Four-week-old Arabidopsis plants were syringe infiltrated with 1 µm flg22, elf18, pep1, or water, and samples were collected 16 h later. Callose deposition assays were performed as described (Kim and Mackey, 2008). Leaves were cleared of chlorophyll, and callose deposits were stained with an Aniline Blue solution (0.01% [w/v]) and were visualized by a fluorescence microscope with AxioVision software (Zeiss), with excitation at 365 nm and emission at 445 nm. A leaf from each of five plants was photographed in four fields (0.6 mm2). The number of callose deposits in the photographed fields was automatically counted using the Icy software (http://icy.bioimageanalysis.org). The number of callose deposits counted in mock-inoculated leaves was subtracted from the mean of each treatment. At least four independent biological repeats were carried out for each PAMP/DAMP.
MAPK Phosphorylation Assay
Leaf discs from 4-week-old Arabidopsis plants were floated overnight in 5 mL of deionized water and then treated for 0, 5, or 15 min with 1 µm flg22, elf18, pep1, or water. Discs were ground in extraction buffer (50 mm Tris-HCl [pH 7.5], 200 mm NaCl, 1 mm EDTA, 10 mm NaF, 1 mm Na2MoO4, 2 mm Na3VO4, 10% [v/v] glycerol, 2 mm DTT, and 1 mm PMSF) and centrifuged, and the supernatants were recovered. Proteins were fractionated by 10% (v/v) SDS-PAGE and subjected to western-blot analysis with rabbit anti-p44/42 MAPK antibodies (Cell Signaling Technology).
RNA Isolation and RT-qPCR
Total RNA was isolated from leaves (60 mg) using the SV Total RNA Isolation System (Promega). RNA samples (1 µg) were reverse transcribed with oligo(dT) primers using a RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific) and subjected to quantitative PCR using gene-specific primers (Supplemental Table S3). cDNAs were amplified using the SYBR Premix Ex Taq II (Clontech Laboratories) and the Mx3000P System (Agilent Technologies). The ACTIN2 gene was used for normalization, and gene expression was calculated by the comparative Ct method (Pfaffl, 2001).
Accession Numbers
Sequence data from this article can be found in Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: Arabidopsis ACTIN2 (At3g18780), BIR1 (At5g48380), BRI1 (At4g39400), BSK1 (At4g35230), BSK2 (At5g46570), BSK3 (At4g00710), BSK4 (At1g01740), BSK5 (At5g59010), BSK6 (At3g54030), BSK7 (At1g63500), BSK8 (At5g41260), BSK9 (At3g09240), BSK10 (At5g01060), BSK11 (At1g50990), EFR (At5g20480), ERECTA (At2g26330), FERONIA (At3g51550), FLS2 (At5g46330), FRK1 (At2g19190), LYK5 (At2g33580), PEPR1 (At1g73080), PEPR2 (At1g17750), PP2C (At1g16220), PR1 (At2g14610), SRF6 (At1g53730), SRF7 (At3g14350), SOBIR1 (At2g31880), WAKL8 (At1g16260), WAKL14 (At2g23450), WAKL18 (At4g31110), and WRKY29 (At4g23550).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. BSK1 to BSK11 do not interact with BSK5-interacting proteins in yeast.
Supplemental Figure S2. Expression of BSKs and BSK-interacting proteins in yeast and in planta.
Supplemental Figure S3. Expression of BSK5, FLS2, EFR, and PEPR1 in yeast and/or in planta.
Supplemental Figure S4. Interaction of BSK5 with PEPR1, EFR, and FLS2 in N. benthamiana.
Supplemental Figure S5. In vitro phosphorylation of BSK5 by PEPR1 and EFR.
Supplemental Figure S6. FRK1 and WRKY29 mRNA expression in leaves of bsk5 mutant plants treated with PAMP/DAMPs.
Supplemental Figure S7. Protein expression in transgenic plants.
Supplemental Table S1. RLKs and RLCKs encoded by partial cDNA clones interacting with BSK5 in yeast.
Supplemental Table S2. BSK5 residues phosphorylated in vitro by the PEPR1 and EFR kinase domains.
Supplemental Table S3. Oligonucleotides used in this study.
Acknowledgments
We thank Georgy Popov and Dor Salomon for fruitful suggestions and discussions.
Footnotes
This work was supported by grants of the Israel Science Foundation (309/15), the United States-Israel Binational Agricultural Research and Development Fund (IS-4931-16C), and the European Cooperation in Science and Technology (EuroXanth CA16107) from the European Union.
References
- Albert I, Böhm H, Albert M, Feiler CE, Imkampe J, Wallmeroth N, Brancato C, Raaymakers TM, Oome S, Zhang H, et al. (2015) An RLP23-SOBIR1-BAK1 complex mediates NLP-triggered immunity. Nat Plants 1: 15140. [DOI] [PubMed] [Google Scholar]
- Alcázar R, García AV, Kronholm I, de Meaux J, Koornneef M, Parker JE, Reymond M (2010) Natural variation at Strubbelig Receptor Kinase 3 drives immune-triggered incompatibilities between Arabidopsis thaliana accessions. Nat Genet 42: 1135–1139 [DOI] [PubMed] [Google Scholar]
- Ao Y, Li Z, Feng D, Xiong F, Liu J, Li JF, Wang M, Wang J, Liu B, Wang HB (2014) OsCERK1 and OsRLCK176 play important roles in peptidoglycan and chitin signaling in rice innate immunity. Plant J 80: 1072–1084 [DOI] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Benschop JJ, Mohammed S, O’Flaherty M, Heck AJR, Slijper M, Menke FLH (2007) Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol Cell Proteomics 6: 1198–1214 [DOI] [PubMed] [Google Scholar]
- Bi G, Zhou Z, Wang W, Li L, Rao S, Wu Y, Zhang X, Menke FLH, Chen S, Zhou JM (2018) Receptor-like cytoplasmic kinases directly link diverse pattern recognition receptors to the activation of mitogen-activated protein kinase cascades in Arabidopsis. Plant Cell 30: 1543–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigeard J, Colcombet J, Hirt H (2015) Signaling mechanisms in pattern-triggered immunity (PTI). Mol Plant 8: 521–539 [DOI] [PubMed] [Google Scholar]
- Boller T, Felix G (2009) A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Cao Y, Liang Y, Tanaka K, Nguyen CT, Jedrzejczak RP, Joachimiak A, Stacey G (2014) The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. eLife 3: e03766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM (2008) Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 146: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couto D, Zipfel C (2016) Regulation of pattern recognition receptor signalling in plants. Nat Rev Immunol 16: 537–552 [DOI] [PubMed] [Google Scholar]
- Couto D, Niebergall R, Liang X, Bücherl CA, Sklenar J, Macho AP, Ntoukakis V, Derbyshire P, Altenbach D, Maclean D, et al. (2016) The Arabidopsis protein phosphatase PP2C38 negatively regulates the central immune kinase BIK1. PLoS Pathog 12: e1005811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deblaere R, Bytebier B, De Greve H, Deboeck F, Schell J, Van Montagu M, Leemans J (1985) Efficient octopine Ti plasmid-derived vectors for Agrobacterium-mediated gene transfer to plants. Nucleic Acids Res 13: 4777–4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delteil A, Gobbato E, Cayrol B, Estevan J, Michel-Romiti C, Dievart A, Kroj T, Morel JB (2016) Several wall-associated kinases participate positively and negatively in basal defense against rice blast fungus. BMC Plant Biol 16: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C, Li X, Chen J, Chen W, Li B, Li C, Wang L, Li J, Zhao X, Lin J, et al. (2016) Receptor kinase complex transmits RALF peptide signal to inhibit root growth in Arabidopsis. Proc Natl Acad Sci USA 113: E8326–E8334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubiella U, Seybold H, Durian G, Komander E, Lassig R, Witte CP, Schulze WX, Romeis T (2013) Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc Natl Acad Sci USA 110: 8744–8749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick RD, Thilmony RL, Sessa G, Martin GB (1998) Recognition specificity for the bacterial avirulence protein AvrPto is determined by Thr-204 in the activation loop of the tomato Pto kinase. Mol Cell 2: 241–245 [DOI] [PubMed] [Google Scholar]
- Gao M, Wang X, Wang D, Xu F, Ding X, Zhang Z, Bi D, Cheng YT, Chen S, Li X, et al. (2009) Regulation of cell death and innate immunity by two receptor-like kinases in Arabidopsis. Cell Host Microbe 6: 34–44 [DOI] [PubMed] [Google Scholar]
- Gao X, Chen X, Lin W, Chen S, Lu D, Niu Y, Li L, Cheng C, McCormack M, Sheen J, et al. (2013) Bifurcation of Arabidopsis NLR immune signaling via Ca2+-dependent protein kinases. PLoS Pathog 9: e1003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillette MA, Carr SA (2013) Quantitative analysis of peptides and proteins in biomedicine by targeted mass spectrometry. Nat Methods 10: 28–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gómez L, Felix G, Boller T (1999) A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J 18: 277–284 [DOI] [PubMed] [Google Scholar]
- Goodin MM, Zaitlin D, Naidu RA, Lommel SA (2008) Nicotiana benthamiana: Its history and future as a model for plant-pathogen interactions. Mol Plant Microbe Interact 21: 1015–1026 [DOI] [PubMed] [Google Scholar]
- Grütter C, Sreeramulu S, Sessa G, Rauh D (2013) Structural characterization of the RLCK family member BSK8: A pseudokinase with an unprecedented architecture. J Mol Biol 425: 4455–4467 [DOI] [PubMed] [Google Scholar]
- Guo M, Tian F, Wamboldt Y, Alfano JR (2009) The majority of the type III effector inventory of Pseudomonas syringae pv. tomato DC3000 can suppress plant immunity. Mol Plant Microbe Interact 22: 1069–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halter T, Imkampe J, Mazzotta S, Wierzba M, Postel S, Bücherl C, Kiefer C, Stahl M, Chinchilla D, Wang X, et al. (2014) The leucine-rich repeat receptor kinase BIR2 is a negative regulator of BAK1 in plant immunity. Curr Biol 24: 134–143 [DOI] [PubMed] [Google Scholar]
- Hanks SK, Quinn AM, Hunter T (1988) The protein kinase family: Conserved features and deduced phylogeny of the catalytic domains. Science 241: 42–52 [DOI] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kadota Y, Sklenar J, Derbyshire P, Stransfeld L, Asai S, Ntoukakis V, Jones JD, Shirasu K, Menke F, Jones A, et al. (2014) Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol Cell 54: 43–55 [DOI] [PubMed] [Google Scholar]
- Kim MG, Mackey D (2008) Measuring cell-wall-based defenses and their effect on bacterial growth in Arabidopsis. Methods Mol Biol 415: 443–452 [DOI] [PubMed] [Google Scholar]
- Kim J, Harter K, Theologis A (1997) Protein-protein interactions among the Aux/IAA proteins. Proc Natl Acad Sci USA 94: 11786–11791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Guan S, Sun Y, Deng Z, Tang W, Shang JX, Sun Y, Burlingame AL, Wang ZY (2009) Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat Cell Biol 11: 1254–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Q, Sun T, Qu N, Ma J, Li M, Cheng YT, Zhang Q, Wu D, Zhang Z, Zhang Y (2016) Two redundant receptor-like cytoplasmic kinases function downstream of pattern recognition receptors to regulate activation of SA biosynthesis. Plant Physiol 171: 1344–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol E, Mentzel T, Chinchilla D, Boller T, Felix G, Kemmerling B, Postel S, Arents M, Jeworutzki E, Al-Rasheid KAS, et al. (2010) Perception of the Arabidopsis danger signal peptide 1 involves the pattern recognition receptor AtPEPR1 and its close homologue AtPEPR2. J Biol Chem 285: 13471–13479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse T, Gehl C, Geisler M, Lehrke M, Ringel P, Hallier S, Hänsch R, Mendel RR (2010) Identification and biochemical characterization of molybdenum cofactor-binding proteins from Arabidopsis thaliana. J Biol Chem 285: 6623–6635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze G, Zipfel C, Robatzek S, Niehaus K, Boller T, Felix G (2004) The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16: 3496–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal NK, Nagalakshmi U, Hurlburt NK, Flores R, Bak A, Sone P, Ma X, Song G, Walley J, Shan L, et al. (2018) The receptor-like cytoplasmic kinase BIK1 localizes to the nucleus and regulates defense hormone expression during plant innate immunity. Cell Host Microbe 23: 485–497.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel E, Heifetz P, Thorne L, Uknes S, Ryals J, Ward E (1998) Functional analysis of regulatory sequences controlling PR-1 gene expression in Arabidopsis. Plant J 16: 223–233 [DOI] [PubMed] [Google Scholar]
- Lehti-Shiu MD, Zou C, Hanada K, Shiu SH (2009) Evolutionary history and stress regulation of plant receptor-like kinase/pelle genes. Plant Physiol 150: 12–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Li M, Yu L, Zhou Z, Liang X, Liu Z, Cai G, Gao L, Zhang X, Wang Y, et al. (2014) The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15: 329–338 [DOI] [PubMed] [Google Scholar]
- Li ZY, Xu ZS, He GY, Yang GX, Chen M, Li LC, Ma YZ (2012) A mutation in Arabidopsis BSK5 encoding a brassinosteroid-signaling kinase protein affects responses to salinity and abscisic acid. Biochem Biophys Res Commun 426: 522–527 [DOI] [PubMed] [Google Scholar]
- Liang X, Zhou JM (2018) Receptor-like cytoplasmic kinases: Central players in plant receptor kinase-mediated signaling. Annu Rev Plant Biol 69: 267–299 [DOI] [PubMed] [Google Scholar]
- Liu J, Elmore JM, Lin ZJD, Coaker G (2011) A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe 9: 137–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Wu Y, Yang F, Zhang Y, Chen S, Xie Q, Tian X, Zhou JM (2013) BIK1 interacts with PEPRs to mediate ethylene-induced immunity. Proc Natl Acad Sci USA 110: 6205–6210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Wu S, Gao X, Zhang Y, Shan L, He P (2010) A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc Natl Acad Sci USA 107: 496–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Salas O, Bowler K, Oren-Young L, Bar-Peled M, Sharon A (2017) Genetic alteration of UDP-rhamnose metabolism in Botrytis cinerea leads to the accumulation of UDP-KDG that adversely affects development and pathogenicity. Mol Plant Pathol 18: 263–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho AP, Zipfel C (2015) Targeting of plant pattern recognition receptor-triggered immunity by bacterial type-III secretion system effectors. Curr Opin Microbiol 23: 14–22 [DOI] [PubMed] [Google Scholar]
- Paparella C, Savatin DV, Marti L, De Lorenzo G, Ferrari S (2014) The Arabidopsis LYSIN MOTIF-CONTAINING RECEPTOR-LIKE KINASE3 regulates the cross talk between immunity and abscisic acid responses. Plant Physiol 165: 262–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov G, Fraiture M, Brunner F, Sessa G (2016) Multiple Xanthomonas euvesicatoria type III effectors inhibit flg22-triggered immunity. Mol Plant Microbe Interact 29: 651–660 [DOI] [PubMed] [Google Scholar]
- Qi Y, Tsuda K, Glazebrook J, Katagiri F (2011) Physical association of pattern-triggered immunity (PTI) and effector-triggered immunity (ETI) immune receptors in Arabidopsis. Mol Plant Pathol 12: 702–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S, Zhou Z, Miao P, Bi G, Hu M, Wu Y, Feng F, Zhang X, Zhou JM (2018) Roles of receptor-like cytoplasmic kinase VII members in pattern-triggered immune signaling. Plant Physiol 177: 1679–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek S, Chinchilla D, Boller T (2006) Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev 20: 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Rodríguez C, Estévez JM, Llorente F, Hernández-Blanco C, Jordá L, Pagán I, Berrocal M, Marco Y, Somerville S, Molina A (2009) The ERECTA receptor-like kinase regulates cell wall-mediated resistance to pathogens in Arabidopsis thaliana. Mol Plant Microbe Interact 22: 953–963 [DOI] [PubMed] [Google Scholar]
- Santos AA, Lopes KVG, Apfata JAC, Fontes EPB (2010) NSP-interacting kinase, NIK: A transducer of plant defence signalling. J Exp Bot 61: 3839–3845 [DOI] [PubMed] [Google Scholar]
- Schwessinger B, Roux M, Kadota Y, Ntoukakis V, Sklenar J, Jones A, Zipfel C (2011) Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet 7: e1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Shen Q, Qi Y, Yan H, Nie H, Chen Y, Zhao T, Katagiri F, Tang D (2013a) BR-SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in Arabidopsis. Plant Cell 25: 1143–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Yan H, Li J, Tang D (2013b) BSK1, a receptor-like cytoplasmic kinase, involved in both BR signaling and innate immunity in Arabidopsis. Plant Signal Behav 8: e24996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinya T, Yamaguchi K, Desaki Y, Yamada K, Narisawa T, Kobayashi Y, Maeda K, Suzuki M, Tanimoto T, Takeda J, et al. (2014) Selective regulation of the chitin-induced defense response by the Arabidopsis receptor-like cytoplasmic kinase PBL27. Plant J 79: 56–66 [DOI] [PubMed] [Google Scholar]
- Singh DK, Calviño M, Brauer EK, Fernandez-Pozo N, Strickler S, Yalamanchili R, Suzuki H, Aoki K, Shibata D, Stratmann JW, et al. (2014) The tomato kinome and the tomato kinase library ORFeome: Novel resources for the study of kinases and signal transduction in tomato and Solanaceae species. Mol Plant Microbe Interact 27: 7–17 [DOI] [PubMed] [Google Scholar]
- Sreekanta S, Bethke G, Hatsugai N, Tsuda K, Thao A, Wang L, Katagiri F, Glazebrook J (2015) The receptor-like cytoplasmic kinase PCRK1 contributes to pattern-triggered immunity against Pseudomonas syringae in Arabidopsis thaliana. New Phytol 207: 78–90 [DOI] [PubMed] [Google Scholar]
- Sreeramulu S, Mostizky Y, Sunitha S, Shani E, Nahum H, Salomon D, Hayun LB, Gruetter C, Rauh D, Ori N, et al. (2013) BSKs are partially redundant positive regulators of brassinosteroid signaling in Arabidopsis. Plant J 74: 905–919 [DOI] [PubMed] [Google Scholar]
- Stegmann M, Monaghan J, Smakowska-Luzan E, Rovenich H, Lehner A, Holton N, Belkhadir Y, Zipfel C (2017) The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 355: 287–289 [DOI] [PubMed] [Google Scholar]
- Sun Y, Li L, Macho AP, Han Z, Hu Z, Zipfel C, Zhou JM, Chai J (2013) Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 342: 624–628 [DOI] [PubMed] [Google Scholar]
- Tang D, Wang G, Zhou JM (2017) Receptor kinases in plant-pathogen interactions: More than pattern recognition. Plant Cell 29: 618–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Kim TW, Oses-Prieto JA, Sun Y, Deng Z, Zhu S, Wang R, Burlingame AL, Wang ZY (2008) BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321: 557–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon LC, Geraats BPJ, Linthorst HJM (2006) Ethylene as a modulator of disease resistance in plants. Trends Plant Sci 11: 184–191 [DOI] [PubMed] [Google Scholar]
- Vlot AC, Dempsey DA, Klessig DF (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol 47: 177–206 [DOI] [PubMed] [Google Scholar]
- Wang J, Shi H, Zhou L, Peng C, Liu D, Zhou X, Wu W, Yin J, Qin H, Ma W, et al. (2017) OsBSK1-2, an orthologous of AtBSK1, is involved in rice immunity. Front Plant Sci 8: 908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo JY, Jeong KJ, Kim YJ, Paek KH (2016) CaLecRK-S.5, a pepper L-type lectin receptor kinase gene, confers broad-spectrum resistance by activating priming. J Exp Bot 67: 5725–5741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang T, Zong N, Zou Y, Wu Y, Zhang J, Xing W, Li Y, Tang X, Zhu L, Chai J, et al. (2008) Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr Biol 18: 74–80 [DOI] [PubMed] [Google Scholar]
- Xu P, Xu SL, Li ZJ, Tang W, Burlingame AL, Wang ZY (2014) A brassinosteroid-signaling kinase interacts with multiple receptor-like kinases in Arabidopsis. Mol Plant 7: 441–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Yamaguchi K, Shirakawa T, Nakagami H, Mine A, Ishikawa K, Fujiwara M, Narusaka M, Narusaka Y, Ichimura K, et al. (2016) The Arabidopsis CERK1-associated kinase PBL27 connects chitin perception to MAPK activation. EMBO J 35: 2468–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K, Yamada K, Ishikawa K, Yoshimura S, Hayashi N, Uchihashi K, Ishihama N, Kishi-Kaboshi M, Takahashi A, Tsuge S, et al. (2013) A receptor-like cytoplasmic kinase targeted by a plant pathogen effector is directly phosphorylated by the chitin receptor and mediates rice immunity. Cell Host Microbe 13: 347–357 [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Pearce G, Ryan CA (2006) The cell surface leucine-rich repeat receptor for AtPep1, an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells. Proc Natl Acad Sci USA 103: 10104–10109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Zhao Y, Shi H, Li J, Wang Y, Tang D (2018) BRASSINOSTEROID-SIGNALING KINASE1 phosphorylates MAPKKK5 to regulate immunity in Arabidopsis. Plant Physiol 176: 2991–3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh YH, Panzeri D, Kadota Y, Huang YC, Huang PY, Tao CN, Roux M, Chien HC, Chin TC, Chu PW, et al. (2016) The Arabidopsis malectin-like/LRR-RLK IOS1 is critical for BAK1-dependent and BAK1-independent pattern-triggered immunity. Plant Cell 28: 1701–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Wang X, Zhao Z, Wang R, Huang X, Zhu Y, Yuan L, Wang Y, Xu X, Burlingame AL, et al. (2016) OsBRI1 activates BR signaling by preventing binding between the TPR and kinase domains of OsBSK3 via phosphorylation. Plant Physiol 170: 1149–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li W, Xiang T, Liu Z, Laluk K, Ding X, Zou Y, Gao M, Zhang X, Chen S, et al. (2010) Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7: 290–301 [DOI] [PubMed] [Google Scholar]
- Zhang X, Henriques R, Lin SS, Niu QW, Chua NH (2006) Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc 1: 641–646 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JDG, Felix G, Boller T (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JDG, Boller T, Felix G (2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760 [DOI] [PubMed] [Google Scholar]