The plasma membrane Na+/H+ exchanger SOS1 plays a major role in the salt tolerance of rice by controlling Na+ uptake and root-shoot partitioning.
Abstract
Rice (Oryza sativa) stands among the world's most important crop species. Rice is salt sensitive, and the undue accumulation of sodium ions (Na+) in shoots has the strongest negative correlation with rice productivity under long-term salinity. The plasma membrane Na+/H+ exchanger protein Salt Overly Sensitive 1 (SOS1) is the sole Na+ efflux transporter that has been genetically characterized to date. Here, the importance of SOS1-facilitated Na+ flux in the salt tolerance of rice was analyzed in a reverse-genetics approach. A sos1 loss-of-function mutant displayed exceptional salt sensitivity that was correlated with excessive Na+ intake and impaired Na+ loading into the xylem, thus indicating that SOS1 controls net root Na+ uptake and long-distance Na+ transport to shoots. The acute Na+ sensitivity of sos1 plants at low NaCl concentrations allowed analysis of the transcriptional response to sodicity stress without effects of the osmotic stress intrinsic to high-salinity treatments. In contrast with that in the wild type, sos1 mutant roots displayed preferential down-regulation of stress-related genes in response to salt treatment, despite the greater intensity of stress experienced by the mutant. These results suggest there is impaired stress detection or an inability to mount a comprehensive response to salinity in sos1. In summary, the plasma membrane Na+/H+ exchanger SOS1 plays a major role in the salt tolerance of rice by controlling Na+ homeostasis and possibly contributing to the sensing of sodicity stress.
Rice (Oryza sativa) is the most important salt-sensitive cereal crop (Hoang et al., 2016). Rice is grown in a diverse range of environments with varying soil-water conditions. Salinity is an intrinsic problem in coastal areas and river deltas where rice farming predominates, but is also a worsening problem in inland areas due to the buildup of secondary salinization of the soil surface as a consequence of excessive use of irrigation with poor quality water and insufficient drainage (Thomson et al., 2010). Excessive amounts of salts in the soil solution cause substantial declines in the productivity of many agricultural plants, and in rice as little as 3.5 dS/m (2.1 g/L of salts) is sufficient to curtail 10% of yield, and 7.2 dS/m (4.3 g/L) producing 50% yield loss (Munns and Tester, 2008; Hoang et al., 2016).
Salinity poses two main challenges to plants. One is osmotic stress due to excess solutes outside the roots that reduce the ability of plants to extract soil water, which affects cell turgor and expansion immediately. Second, the ionic stress that is most often caused by excessive influx of sodium ions (Na+) into the plant leads to interruption of metabolic processes (Hasegawa et al., 2000; Munns and Tester, 2008). Physiological and genetic studies indicate that a range of traits, such as low shoot Na+ content, preservation of the K+ status of the whole plant, vacuolar compartmentation of salts in older leaves, intrinsic cellular tolerance, and plant vigor, could incrementally improve the ability of rice plants to cope with salinity (Yeo et al., 1990; Thomson et al., 2010). Integration of all these physiological mechanisms for salinity tolerance by pyramiding superior alleles controlling different processes is the long-term goal to increase the level of salt tolerance in rice (Gregorio et al., 2002).
Sustained plant growth under salt stress requires tight control of Na+ uptake, Na+ redistribution among organs by long-distance transport, and Na+ accumulation into cell vacuoles for detoxification and osmotic balance (Munns and Tester, 2008). The ability to manage Na+ content has partly a cellular basis since rice protoplasts derived from cultivars with contrasting salt tolerance showed that halotolerance depended on reduced Na+ uptake and a fast compartmentation of Na+ into vacuoles (Kader and Lindberg, 2005), whereas enhancement of Na+ efflux yielded salt-tolerant calli (Zhu and Wu, 2008). At the whole-plant level, comparative studies have shown that salinity-tolerant rice cultivars accumulate less Na+ in leaves and shoots compared with that in salinity-sensitive rice cultivars (Golldack et al., 2003; Lee et al., 2003; Lin et al., 2004; Ren et al., 2005) and that shoot Na+ accumulation had the strongest negative correlation with the performance of contrasting rice genotypes under long-term salinity (Coskun et al., 2013). Hence, the quantitative measure of the Na+ and K+ concentrations in the shoot in salinized conditions has been often used as the trait screened in quantitative trait loci mapping and breeding programs (Hoang et al., 2016). Known proteins governing the partitioning of Na+ between roots and shoots are members of the so-called High-affinity Potassium Transporters (HKT), albeit they have repeatedly been shown to behave as Na+ transporters mediating either channel-like Na+ uniport (class I or HKT1 group) or Na+-K+ symport (class II or HKT2 group). Generally, dicot species have only a few HKT genes in their genomes preferably encoding class-I HKT proteins, whereas monocots have multiple HKT genes of both classes. The rice protein OsHKT1;5/SCK1 and the wheat proteins TmHKT1;4/ and TmHKT1;5, all of them class-I proteins, mediate the retrieval of Na+ from the xylem sap, thereby restricting the transfer of Na+ to shoots. Rice OsHKT1;4 also restricts leaf sheath-to-blade Na+ transfer under salinity stress (Cotsaftis et al., 2012; Suzuki et al., 2016).
Although several candidate proteins have been suggested, no major plasma membrane transporters responsible for the bulk influx of Na+ into root cells have as yet been conclusively identified at the genetic level, and it seems likely that multiple transport proteins and systems incrementally contribute to Na+ influx (Kronzucker and Britto, 2011; Nieves-Cordones et al., 2016). In rice, the HKT2;1 protein that is expressed in the root cortex and endodermis may provide an entry pathway for 'nutritional' Na+ uptake under K+ limitation to support cell expansion and plant growth (Garciadeblás et al., 2003; Horie et al., 2007). Sodium may also leak into the root stele of rice plants through anatomical discontinuities in the endodermis produced by the emergence of secondary roots (Yeo et al., 1987), or through the lateral roots themselves (Faiyue et al., 2010a, 2010b). For Na+ transport in the opposite (efflux) direction, only the SOS1 transporter (Salt Overly Sensitive 1) has been genetically characterized (Shi et al., 2002; Oh et al., 2009; Olías et al., 2009). In dicots, SOS1 controls net Na+ uptake by roots and long-distance transport to shoots (Oh et al., 2009; Olías et al., 2009). Thus, the opposing yet concerted activity of SOS1 in xylem loading and of HKT1-like proteins in xylem unloading might determine the amount of Na+ that is eventually exported from roots to shoots. Although HKT proteins of cereal crops are well established to play a major role in their salt tolerance, the contribution of Na+ loading into the xylem by specific transporters remains to be determined. Likewise, the regulatory cross talk of these complementary transport systems has not been explored (Ji et al., 2013).
Little is known about the regulation of HKT proteins. HKT gene expression is often modulated by K+ availability and soil salinity, but the posttranscriptional regulation of HKT transporters remains entirely unknown. In rice, the Na+ uptake gene HKT2;1 is rapidly repressed in roots by as little as 30 mm NaCl to curtail Na+ entry (Horie et al., 2007). Surprisingly, HKT1;4 expression is also down-regulated proportionally to the severity of salinity treatment during the vegetative growth stage, and RNA interference plants showed no differences in salt sensitivity or in Na+ content (Suzuki et al., 2016). A contribution of HKT1;4 toward the prevention of Na+ accumulation in flag leaf blades was found in the reproductive stage only (Suzuki et al., 2016). OsHKT1;1, involved in Na+ recirculation, is expressed mainly in the phloem of leaf blades and up-regulated by salt stress (Wang et al., 2015). The rice MYB-type transcription factor OsMYBc binds to the OsHKT1;1 promoter and loss of OsMYBc resulted in a reduction in NaCl-induced expression of OsHKT1;1 and salt sensitivity.
More progress has been made toward the understanding of SOS1 regulation. The SOS1 gene is up-regulated by salinity (Shi et al., 2003; Martínez-Atienza et al., 2007) and, in Arabidopsis (Arabidopsis thaliana), the mRNA is stabilized by an ill-defined process mediated by reactive oxygen species (ROS; Chung et al., 2008). The transport activity of SOS1 is enhanced by protein phosphorylation that counteracts autoinhibition (Quintero et al., 2002, 2011). The minimal SOS functional module in both dicots and monocots comprises proteins SOS1, SOS2/CBL-INTERACTING PROTEIN KINASE24 (CIPK24), and SOS3/CALCINEURIN-B LIKE4 (CBL4; Martínez-Atienza et al., 2007; Quintero et al., 2011). SOS2/CIPK24 is a protein kinase that belongs to the Suc non-fermenting-1-related protein kinase-3 (SnRK3) family. SOS3 is a myristoylated Ca2+-binding protein that likely perceives the increase in cytosolic Ca2+ elicited by excess Na+ and recruits SOS2 to the plasma membrane to achieve phosphorylation and activation of SOS1 (Quintero et al., 2002, 2011). In Arabidopsis, SOS3 functions primarily in roots, whereas the related protein SCaBP8/CBL10 operates by a similar mechanism in shoots (Quan et al., 2007). CBL10 may also regulate Na+ uptake into the vacuole (Kim et al., 2007).
The rice complement of SOS proteins has been characterized by heterologous expression in yeast and Arabidopsis (Martínez-Atienza et al., 2007), but a precise description of their role in the physiology and salinity stress management of rice plants has not been achieved. Here we show that a sos1 loss-of-function mutant displays exceptional salt sensitivity that correlates with excessive Na+ intake and impaired Na+ loading into the xylem. We took advantage of the acute sensitivity of sos1 plants to Na+ to inspect the sodicity-stress transcriptome with little interference by the osmotic challenge intrinsic to high salinity treatments. Surprisingly, sos1 mutant roots showed a marked down-regulation of genes despite the greater intensity of the stress they suffered, which suggested impaired stress detection or an inability to mount a comprehensive response to salinity stress in the absence of SOS1.
RESULTS
Selection of Mutant Lines
Public repositories of rice mutant lines were surveyed for entries with putative T-DNA or Tos17 insertions in genes SOS1 (Os12g44360), CIPK24/SOS2 (Os06g40370), and CBL4/SOS3 (Os05g45810). Mutants in the Nipponbare genetic background were preferred because this temperate japonica cultivar enabled the lines of interest to complete their life cycle in our experimental paddies. Mutant line codes, annotated insertion points of mutagenic DNA, and insertions experimentally confirmed by diagnostic PCR and sequencing of amplicons are listed in Table 1. Detailed description of the molecular characterization of all insertional lines tested herein is given in the Supplemental Figures S1 to S6. As contrasting materials for mutant phenotyping, congenic plants in which the mutagen was segregated out (null segregants) were kept as controls for subsequent experimentation.
Table 1. Description of rice lines containing Tos17 or T-DNA insertions in the SOS loci.
Insertion points (nt, nucleotide) are expressed relative to the ATG initiation codon in the genomic sequences, corresponding to nucleotides 27508321 in SOS1, 24043690 in CIPK24, and 26535878 in CBL4When confirmed by sequencing of the PCR amplicon containing the mutagenic insertion, the actual insertion point is also given; otherwise it is noted as not determined (n.d.). OTL, Oryza Tag Line database; NIAS, National Institute of Agrobiological Sciences, Japan.
| Gene | Source | Line Code | Mutagen | Insertion | |
|---|---|---|---|---|---|
| Annotated | Confirmed | ||||
| SOS1 | NIAS | NC2588 | Tos17 | Intron 5, nt 1228 | Intron 5, nt 1228 |
| SOS1 | OTL | AVIB03 | Tos17 | Intron 19, nt 9798 | Intron 19, nt 9802 |
| SOS1 | OTL | ARBF06 | Tos17 | Exon 20, nt 9951 | No insertion |
| SOS1 | OTL | AKFB09 | Tos17 | Exon 21, nt 11232 | Intron 20, nt 11096 |
| SOS1 | OTL | AKFG08 | Tos17 | Exon 21, nt 11312 | Intron 20, nt 11096 |
| SOS1 | OTL | AFNB04 | Tos17 | Exon 21, nt 11402 | No insertion |
| SOS1 | OTL | AUJE11 | Tos17 | Intron 21, nt 11410 | Intron 21, nt n.d. |
| CIPK24 | OTL | AMHC11 | |||
| AMHC12 | T-DNA | Exon 1, nt -84 | Exon 1, nt -84 | ||
| AMHE12 | |||||
| CBL4 | OTL | AJBC12 | T-DNA | 3′-UTR | n.d. |
Out of seven mutant lines tested for putative mutagenic insertions at SOS1 (Piffanelli et al., 2007), only AVIB03 carrying an insertion of transposon Tos17 at intron 19 was confirmed not to produce a wild-type SOS1 mRNA (Supplemental Figs. S1 to S4). Molecular analyses demonstrated that homozygous plants of this lineage produced a truncated mRNA whose elongation was interrupted by the transposon and that, in contrast with that in other lines (Supplemental Figs. S1 to S4), salinity treatment did not promote the correct processing of the mRNA and eviction of the Tos17 insertion (Fig. 1B). Of note is the significant induction of SOS1 mRNA by salinity in Nipponbare plants. Sequencing of the reverse transcription (RT)-PCR amplicon with the intervening Tos17 (Supplemental Fig. S4D) demonstrated that transposon insertion in AVIB03 had occurred after nucleotide 9802, relative to the ATG initiation codon in the genomic sequence of SOS1. The in silico read-through translation of the putative mRNA predicted the synthesis of a SOS1 protein with a frame-shift after residue K767 and the addition of a short C-terminal extension of sequence VCFCFKTTLSH. This truncated protein was predicted inactive based on the removal of essential amino acids and functional domains of the wild-type protein that are downstream the truncation point (Fig. 1A; Quintero et al., 2011; Núñez-Ramírez et al., 2012). To demonstrate this point, a synonymous mutation was created in the rice SOS1 complementary DNA (cDNA) by introducing a stop codon after residue K767. Expression of this truncated SOS1 protein (SOS1-∆767) in the yeast (Saccharomyces cerevisiae) strain AXT3K that is devoid of all endogenous Na+ transporters and is salt sensitive (Quintero et al., 2002) failed to restore salt tolerance upon the coexpression of SOS2 and SOS3 proteins of Arabidopsis (Fig. 1C). We have shown elsewhere the competence of the Arabidopsis SOS2/SOS3 complex to activate the rice SOS1 protein (Martínez-Atienza et al., 2007), and Figure 1C illustrates this fact. By contrast, the rice SOS1 protein with a C-terminal truncation after residue E970 yielded a hyperactive and SOS2/SOS3-independent exchanger (Fig. 1C). Truncation at residue E970 in the rice SOS1 protein mimics the Arabidopsis mutant protein SOS1-∆998, which lacks the autoinhibitory domain of SOS1 that is targeted by the SOS2/SOS3 kinase complex (Quintero et al., 2011).
Figure 1.
Functionality of truncated SOS1 protein. A, Diagram of functional domains of SOS1. The junction domain is the C-terminal portion of SOS1 that has sequence homology with NHX8 and no known functional role; further downstream is the regulatory domain that is specific and characteristic of SOS1-like proteins that has been dissected in three functional modules: the activation domain with sequence similarities to cyclic-nucleotide binding site (amino acids 740– 820), the autoinhibitory domain (977–1023), and the CIPK24 phosphorylation site (Ser-1135) that counteracts autoinhibition (Quintero et al., 2011). The arrowhead marks the truncation point after K767 in the mutant protein. B, RT-PCR of SOS1 mRNA of wild-type Nipponbare plants (WT, wild type) and homozygous plants of line AVIB03 (sos1), with primers S1-E19-F and S1-E20-R annealing at exons 19 and 20. Detached leaves were treated (salt) or not (unt) with 150 mm NaCl for 24 h. Insertion of Tos17 at the intervening intron 19 prevented the amplification in the sos1 mutant. As control, the cDNA of SOS1 was used as template in the PCR reaction (cDNA). C- indicates a mock reaction without template. C, Truncated SOS1 proteins of rice were expressed in the yeast strain AXT3K, with and without the coexpression of the Arabidopsis SOS2–SOS3 kinase complex, and compared with the wild-type SOS1. Decimal dilutions of saturated liquid cultures in selective media were plated in Arginine-Phosphate medium with 1 mm KCl (1K) supplemented with the indicated millimolar concentrations of NaCl.
In accordance with the yeast data demonstrating a defective SOS1 protein in line AVIB03, plants of that line showed a remarkable salt-sensitive phenotype in hydroponic culture that correlated with much greater Na+ contents in shoots and roots (Fig. 2). Representative pictures of sos1 plants at different time points after stress imposition (25, 50, and 75 mm NaCl) are shown in Figure 2A. The sos1 mutant plants showed growth retardation as early as 5 d after treatment with 50 and 75 mm NaCl. After 10 d, sos1 plants displayed severe symptoms of stress also at 25 mm NaCl, the lowest salt concentration tested. The experiment was terminated after 24 d when all sos1 plants had died irrespective of the salt concentration. Quantitative growth data at 7 d after transfer to 50 mm NaCl confirmed the salt sensitivity of homozygous sos1 plants compared with azygous and wild-type Nipponbare controls (Fig. 2B). At that point, sos1 roots were severely dehydrated and after 14 d sos1 shoots had bleached. Salt sensitivity correlated with greater Na+ contents in roots and shoots of the sos1 mutant.
Figure 2.
Growth parameters of sos1 mutant in salt. The 5-d-old seedlings of wild type (WT), sos1 mutant, and a congenic segregant without the sos1 mutation were transferred to hydroponic culture in Miyamoto's medium supplemented with the indicated amounts of NaCl. A, Depicted are three representative plants per genotype of wild type and sos1 mutant at 5 and 10 d after transfer to 25, 50, and 75 mm NaCl. B, Fresh weight (FW) and dry weight (DW) of shoot and roots, the chlorophyll content (SPAD units), and the shoot and root Na+ content of wild type (black columns), sos1 (white), and the congenic azygous control (gray) before and after 1 week in 50 mm NaCl. The chlorophyll content at 14 d is also given. Plants after 2-week growth in salt were not further processed because roots of the sos1 mutant were severely damaged. Values are the mean and SD of five plants per genotype. Different letters indicate significantly different values relative to that in the wild type determined by one-way ANOVA (a = P < 0.01).
The SOS1 protein activity accounts for ∼60% of the Na+/H+ exchange measured in plasma membrane vesicles of Arabidopsis (Qiu et al., 2003). Measurement of Na+/H+ exchange in purified plasma membrane fractions (Supplemental Fig. S5) demonstrated 39% reduction in exchange activity in vesicles derived from the AVIB03 line relative to that in the wild-type plants. Quenching of fluorescence units per mg of protein per minute was 34.73 ± 2.56 and 56.74 ± 11.34, respectively (P = 0.107 by one-way ANOVA test, n = 4 technical replicas in two different assays). Together, these results signify that line AVIB03 is deficient in SOS1 activity and hence this lineage was selected for further experimentation.
We next searched for mutants in the rice genes CIPK24 and CBL4, which we have previously described as functional homologs of Arabidopsis SOS2(CIPK24) and SOS3(CBL4; Martínez-Atienza et al., 2007). Mutant lines in the Nipponbare background AMHC11, AMHC12, and AMHE12 were annotated to carry T-DNA insertions 84 nucleotides upstream of the translation initiation codon of locus Os06g40370 (CIPK24; Sallaud et al., 2004). Molecular characterization confirmed identical T-DNA insertion points at the 5′-untranslated region (UTR) of CIPK24 in these lines (Supplemental Fig. S6). Homozygous plants produced a mature CIPK24 mRNA carrying the T-DNA insertion, and no wild-type mRNA was detected. Mutant seedlings were sensitive to NaCl in hydroponic culture. Supplemental Figure S6 illustrates the end-point growth of homozygous cipk24 plants (L24H), azygous CIPK24 controls (L24A), and wild-type Nipponbare plants after 7 d in hydroponic medium supplemented with 40 and 80 mm NaCl. These data indicate that the rice kinase CIPK24, which has been shown to activate SOS1 (Martínez-Atienza et al., 2007), also plays a role in the salt tolerance of rice.
At the time this research was conducted, there were no available mutants with mutagenic insertions in the coding region of gene CBL4 (Os05g45810) in the Nipponbare background. Line AJBC12 was annotated to bear a T-DNA insertion right downstream the 3′-UTR of CBL4 (Sallaud et al., 2004). RT-PCR analyses with T2 plants of this lineage showed that the CBL4 transcript was not affected by the adjacent T-DNA insertion, and no further experimentation was done with this line.
Salt Tolerance Assays in Rice Paddies
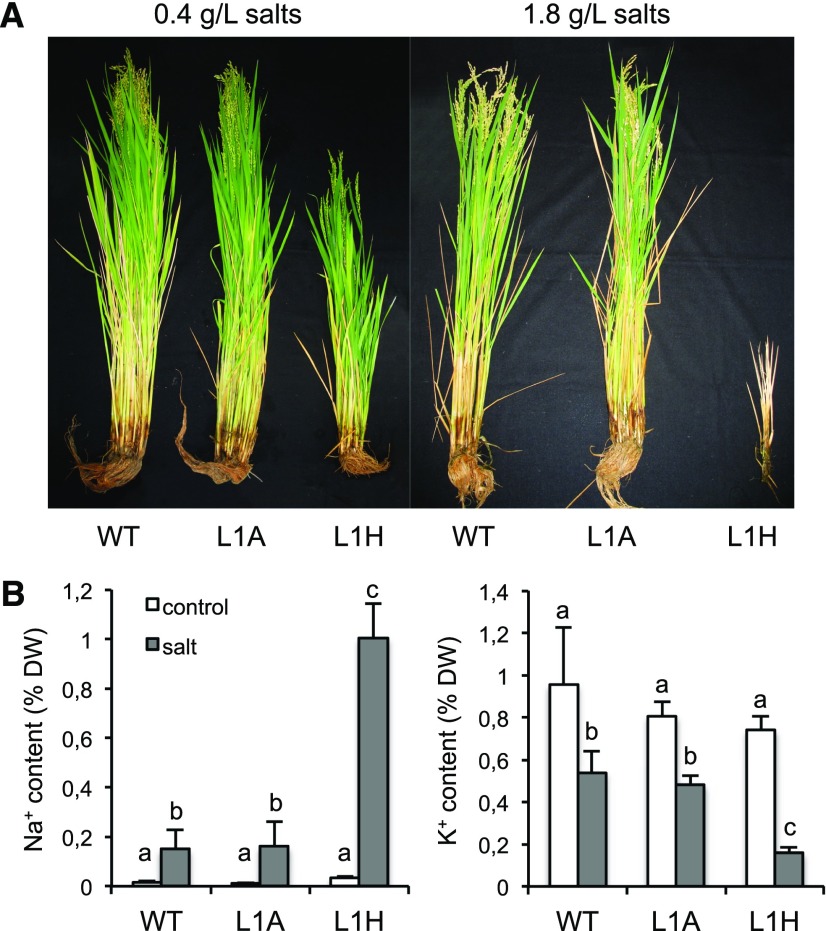
To examine in detail the relevance of the SOS system in the salt tolerance of cultivated rice, we next sought to define the performance of sos1 and cipk24 mutant plants in near-agronomical conditions in experimental rice paddies. Homozygous mutant lines of genotype sos1 and cipk24 (denoted henceforth as L1H and L24H, respectively) were compared with congenic segregant lines selected during the genotyping as lacking the mutagenic insertion (denoted here as azygous segregant lines L1A and L24A) and to control plants of the Nipponbare background. In lines L1H and L1A the T-DNA of the original transformation was also segregated out. Four-week-old plants were transferred to paddies irrigated with water from an underground well whose salinity content was on average 0.42 g/L, of which Na+ was at 5.09 mM, K+ at 0.14 mM, and Ca2+ at 4.0 mM. For the saline treatment, underground water was supplemented with seawater salts to a final concentration of 1.8 g/L salts during irrigation to mimic frequent salinity episodes in rice farms in Spain. The final concentrations of Na+, K+, and Ca2+ in the salinized irrigation water were 27.6, 0.24, and 4.1 mM, respectively. Chloride was 30.5 mM. As depicted in Figure 3 and Supplemental Figure S7, sos1 plants showed statistically significant growth retardation even at the low salt content of irrigation water, conditions in which the Na+ content in sos1 shoots was low but still triple that of the congenic SOS1 control and the wild-type Nipponbare (0.035% vs 0.01% of dry matter, respectively). Growth arrest of salinized sos1 plants at 1.8 g/L salt correlated with massive Na+ accumulation in shoots and notable K+ loss (Fig. 3). The cipk24 mutant plants showed a modest growth reduction in these conditions that was statistically significant only at P = 0.095 (n = 10) relative to that in the congenic azygous control (Supplemental Fig. S7), in agreement with the salt sensitivity that mutant cipk24 showed at early developmental stages in hydroponic culture at 80 mm NaCl but not at 40 mm (Supplemental Fig. S6).
Figure 3.
Plant growth in experimental paddies. A, Representative plants of sos1 (L1H), SOS1 congenic segregant (L1A), and wild type Nipponbare (WT, wild type) genotypes grown for 2 months in experimental paddies irrigated with underground water (left) or supplemented with salt (right). Salt contents in the irrigation water are indicated on top. B, Na+ and K+ content in shoots of control (white columns) and salinized plants (gray columns). Genotypes are labeled as in (A). Plotted are the mean and SD of five plants per genotype and condition after 30 d of saline treatment. Different letters indicate significantly different values at P < 0.001 determined by one-way ANOVA. DW, dry weight.
Complementation and Over-Expression Lines
Because only one putative sos1 mutant line was available, we sought to confirm that the exceptional salt sensitivity of AVIB03 plants was due to the lack of function of SOS1 and not to another uncharacterized mutation carried over through the mutant selection process. Genetically complemented lines were obtained by transformation of AVIB03 plants with the SOS1 cDNA under the control of the constitutive and moderately expressed UBIQUITIN1 (UBQ1) gene promoter of maize. Restoration of salt tolerance was confirmed by scoring the growth of geneticin-resistant T1 plants (n = 32), belonging to four independent transformation lines, in hydroponic culture with 50 and 75 mm NaCl for 3 weeks (Supplemental Fig. S8A). This test showed that 13 out 16 plants regained tolerance to 50 mm NaCl, and that 12 out of 16 withstood the 75 mm NaCl treatment. None of the untransformed sos1 plants survived the assay (n = 8). To further confirm the basis for the suppression of the salt-sensitive phenotype, the complemented line C11 that had a low expression level of the UBQ1:SOS1 transgene (Supplemental Fig. S8B) was chosen for additional analyses. Two-week-old seedlings of wild type, sos1 mutant, and transformed line C11 were transferred to 25 and 50 mm NaCl in Miyamoto's medium for 3 weeks, after which plant weight, and Na+ and K+ contents were determined. Results showed that shoot growth in saline media was restored to wild-type levels, whereas root growth lagged behind that of control plants (Fig. 4). Moreover, Na+ content in C11 plants largely matched that in wild-type plants.
Figure 4.
Complementation of sos1 mutant. The 10-d-old seedlings of wild type, sos1 mutant, and the complementation line C11 were transferred to hydroponic culture in Miyamoto's medium supplemented with the indicated amounts of NaCl. Plants were harvested 2 weeks later. A, Depicted are representative plants of genotypes Nipponbare (N), the sos1 mutant (S), and line C11 (C). B, Dry weight (DW) and Na+ contents in shoots and roots of plants harvested after 7 d of the salinity treatment. Represented are the mean values and SD of wild type (black columns), sos1 (white), and complementation line C11 (gray) of four plants per genotype and growth condition. Letters indicate significantly different values relative to that in the wild type determined by one-way ANOVA (a = P < 0.01, b = P < 0.05).
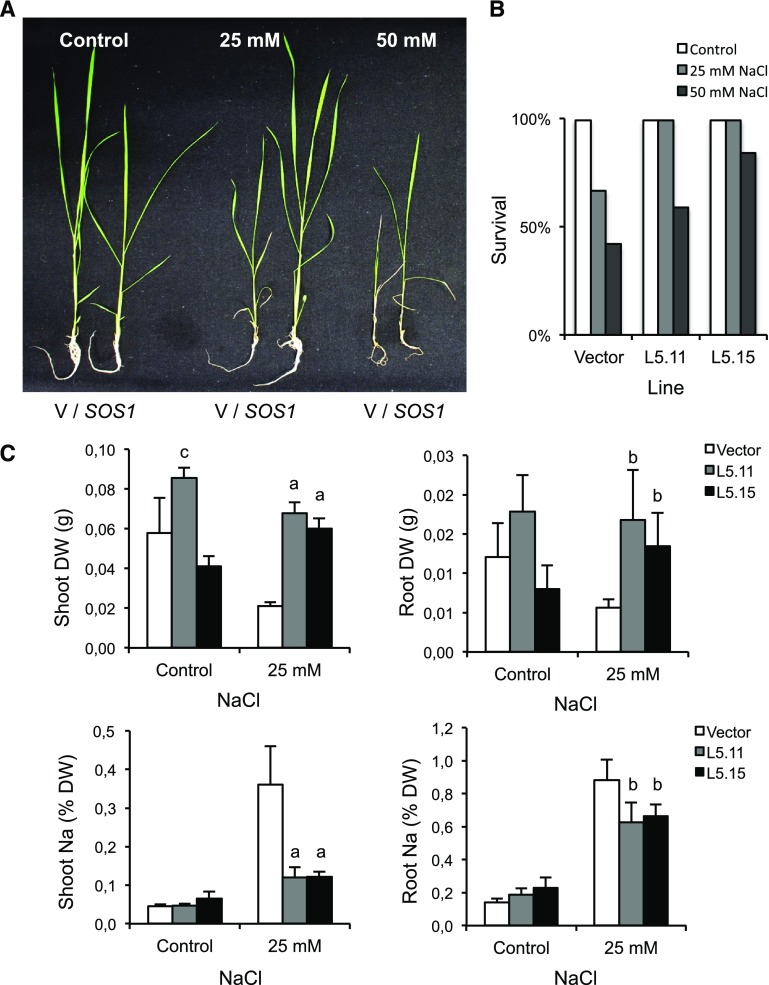
To test the potential of the SOS1 exchanger to increase the salt tolerance of rice, we also generated Nipponbare transgenic plants with expression of the SOS1 cDNA driven by the strong Cauliflower mosaic virus (CaMV) 35S gene promoter to create SOS1 overexpression. Transgenic plants showing high levels of transgene expression as determined by RT-PCR were selected and propagated to produce homozygous lines. Salt tolerance was evaluated in hydroponic cultures with Miyamoto's medium, which contains low levels of K+ (0.20 mM) and Na+ (0.14 mM) and thus could maximize the impact of the ionic component of moderate salt stress (25 to 50 mm NaCl). Seven-d-old seedlings of two independent 35S:SOS1 lines were cultivated in the presence of 25 and 50 mm NaCl for 4 weeks (n = 5 per genotype and treatment). Transgenic plants overexpressing SOS1 showed greater survival under salt treatments than that of control plants transformed with a mock empty vector (Fig. 5). Plants that survived the treatment at 25 mm were assessed for dry weight and Na+ content (control plants surviving 50 mm NaCl had damaged roots and were discarded for further analyses; see Fig. 5A). Plants overexpressing SOS1 sustained a robust growth at 25 mm NaCl that correlated with lower Na+ contents in both shoots and roots compared with that in control plants (Fig. 5C), as should be expected from the near ubiquitous expression of SOS1 driven by the 35S promoter and the consequently enhanced Na+ efflux in many cell types.
Figure 5.
Salt tolerance imparted by SOS1. The 7-d-old seedlings of two independent Nipponbare transgenic lines expressing 35S:SOS1 were cultivated alongside control seedlings transformed with an empty vector (V) in the absence or presence of 25 and 50 mm NaCl for 4 weeks. A, Representative control (V) and transgenic plants (SOS1) are depicted. B, Survival rate of plants after the completion of the experiment. C, Dry weight (DW) and Na+ content (as percentage of DW) of plants surviving the 25-mM NaCl treatment (n = 6). Letters indicate significantly different values relative to that in the wild type determined by one-way ANOVA (a = P < 0.01, b = P < 0.05, c = P < 0.1).
Expression Pattern of SOS1
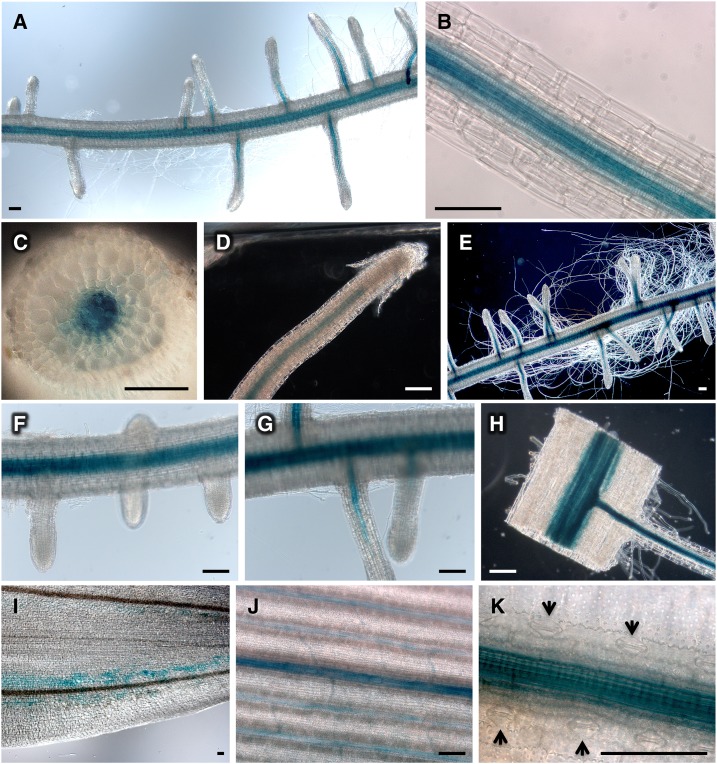
In dicots, the SOS1 protein has been implicated in Na+ fluxes relevant to net ion uptake by roots and to xylem loading for root/shoot ion partition (Shi et al., 2002; Oh et al., 2009; Olías et al., 2009). To understand the critical role of SOS1 in the salt tolerance of rice plants, the expression pattern of the SOS1 gene was investigated in transgenic rice transformed with the SOS1 gene promoter (defined as the 3.9 kb genomic fragment upstream the initiation codon) fused to the GUS reporter gene. Four independent transgenic lines were assayed for GUS expression, and they all produced consistent staining patterns, thereby ruling out artifacts by positional effects of the transgene. GUS expression was detected at all developmental stages tested, and no significant differences in the expression pattern were found between control and saline conditions (50 mm NaCl, 24 h). Maximal GUS staining occurred in the vasculature, both in shoots and roots (Fig. 6). In roots, longitudinal and transverse sections indicated a preferential expression in xylem parenchyma (Fig. 6, A–C). No expression was detected at the root apex or root hairs (Fig. 6, D and E). In secondary roots the staining was apparent only after the vascular bundle started to differentiate (Fig. 6, F–H). Likewise, the expression of SOS1:GUS was located in vascular bundles in aerial tissues. In the coleoptile, staining was restricted to undifferentiated cells around the two vascular bundles (Fig. 6I). In the flag leaf, all vascular bundles were stained, although the signal was stronger around older xylem vessels (Fig. 6, J and K). There was no detectable expression in stomata (Fig. 6K). Thus, the preferential expression pattern of rice SOS1 recapitulated the pattern described in Arabidopsis, with the exception that the rice reporter gene SOS1:GUS was not strongly expressed in root tips as reported for Arabidopsis plants using an equivalent approach (Shi et al., 2002). In conclusion, the expression pattern in rice suggested a role for SOS1 in Na+ xylem loading in addition to the cellular-based protection imparted by Na+ efflux.
Figure 6.
Tissue expression pattern of SOS1. A to K, Representative images of staining patterns resulting from GUS expression under the control of the SOS1 gene promoter. A and B, Preferential expression around the root vascular bundle. C, Root cross section. D and E, Lack of staining in undifferentiated tissues near the root cap and in root hairs. F to H, Staining indicates that expression of SOS1 correlates with the differentiation of vascular bundles in secondary roots. I, Scattered expression around the vasculature of the coleoptile. J, Vascular bundles of the flag leaf. K, Strong expression in leaf xylem parenchyma. Arrowheads point to nonstained stomata. Scale bars =100 µm.
Function of SOS1 in Na+ Efflux and Xylem Loading
The preferential expression pattern of the SOS1 gene promoter in parenchyma cells associated to the vasculature suggested a function for SOS1 in xylem loading since, at physiological Na+ and H+ concentrations, a plasma membrane Na+/H+ exchanger likely mediates the energetically uphill Na+ efflux out of parenchyma cells. Scanning electron microscopy coupled to energy dispersive x-ray microanalysis (SEM-EDX) was used to determine the mineral composition of various root cell types in cross sections of salt-treated rice plants (Fig. 7). In parallel, net tissue contents of Na+ and K+ were also determined. Roots of the sos1 plants treated with 50 mm for 2 d in hydroponic culture had a Na+ content that doubled the amount in the wild type (112.95 mm vs. 51.4 mM) and showed a 37% reduction in the K+ content (10.0 mm vs 15.8 mM). In agreement with these values, the amount of Na+ measured by SEM-EDX in the sos1 mutant was greater than that in the corresponding wild-type samples of all cell types tested (Fig. 7). In wild-type roots, the maximal Na+ content was found in the cortex, with a progressive decline in endodermis and xylem parenchyma cells. This profile is in accordance with the selectivity filter imposed by the root endodermis (and buildup of Na+ content in the cortex relative to that in the endodermis), and with the active loading of Na+ ions into the xylem vessels after ions entered the stele (minimal Na+ content in xylem parenchyma). By contrast, the Na+ distribution in the sos1 root followed an inward-decreasing gradient signifying the unrestricted transit of Na+ through all cell layers and the passive leak of Na+ into the xylem. Together, these results highlight the critical role that the Na+/H+ exchanger SOS1 plays in Na+ efflux and xylem loading in rice plants.
Figure 7.
Mineral content analysis by SEM-EDX. A, Representative SEM cross section of root with labeled arrows indicating the cell types whose elemental composition was analyzed by EDX. Scale bar = 0.1 mm. B, Percentage of total counts of Na+ and K+, measured in 4 to 5 cells of indicated cell types from 2 plants of each genotype (Exod, exodermis; Endo, endodermis; ParX, xylem parenchyma). Plants were treated with 50 mm NaCl for 2 d. Shown are the mean and SD values. Letters indicate significantly different values relative to that in the wild type (WT) for each cellular type determined by one-way ANOVA (a = P < 0.01; b = P < 0.05). C, Na/K ratios of net content values given in (B). The experiment was repeated twice with similar results.
On the other hand, the minimal K+ content in cortical cells in wild-type roots and the increasing K+ values toward the xylem reflected the active uptake of K+ by endodermal cells and its translocation toward the vasculature. A key parameter of the physiological status of individual cells and whole plants under salt stress is the Na+/K+ ratio. Figure 7B shows that the ion-selective barrier imposed by the endodermis to the centripetal movement of Na+ in the root caused a disproportionate Na+/K+ ratio of 11.43 in cortex cells of wild-type roots, in stark contrast with the low 0.49 ratio measured in xylem parenchyma cells presumably effected by the active Na+ loading in xylem vessels. In accordance with the critical role of SOS1 in Na+ export to the xylem, the Na+/K+ ratio in the xylem parenchyma of the sos1 mutant escalated to 9.40, a 19-fold increase relative to wild-type values. This change in the Na+/K+ ratio between wild type and the sos1 mutant signified the greatest difference among cell types. These results are also evidence of the profound effects that a deficient Na+-specific transport system may produce on the K+ status of individual cells and tissues.
Transcriptome of the sos1 Mutant
High salinity imposes both a water stress because the osmotic effect of the saline solution outside and an ionic imbalance resulting primarily from the accumulation of Na+ and the loss of K+ (Munns and Tester, 2008). Contrary to most analyses of the transcriptional response of plants to high salinity, in which it is not possible to separate the genetic response to the osmotic and the ionic components of high salinity (Rabbani et al., 2003; Kanwar et al., 2014; Wang et al., 2016), the extreme Na+-sensitivity of the sos1 mutant plant offered the unique advantage of inspecting the specific transcriptional response to sodicity stress at low external salt concentrations. Roots are the main defensive barrier against salinity and the organ that first senses this soil-derived stress (Munns and Tester, 2008). Hence, to identify genes responding primarily to the Na+ toxicity affecting sos1 plants, roots from mutant and control wild-type seedlings were harvested at the same time point (4 d) after a 75 mm NaCl treatment, and compared with roots harvested alongside from untreated seedlings. Total RNA was extracted and used for microarray experiments (see “Materials and Methods” for details).
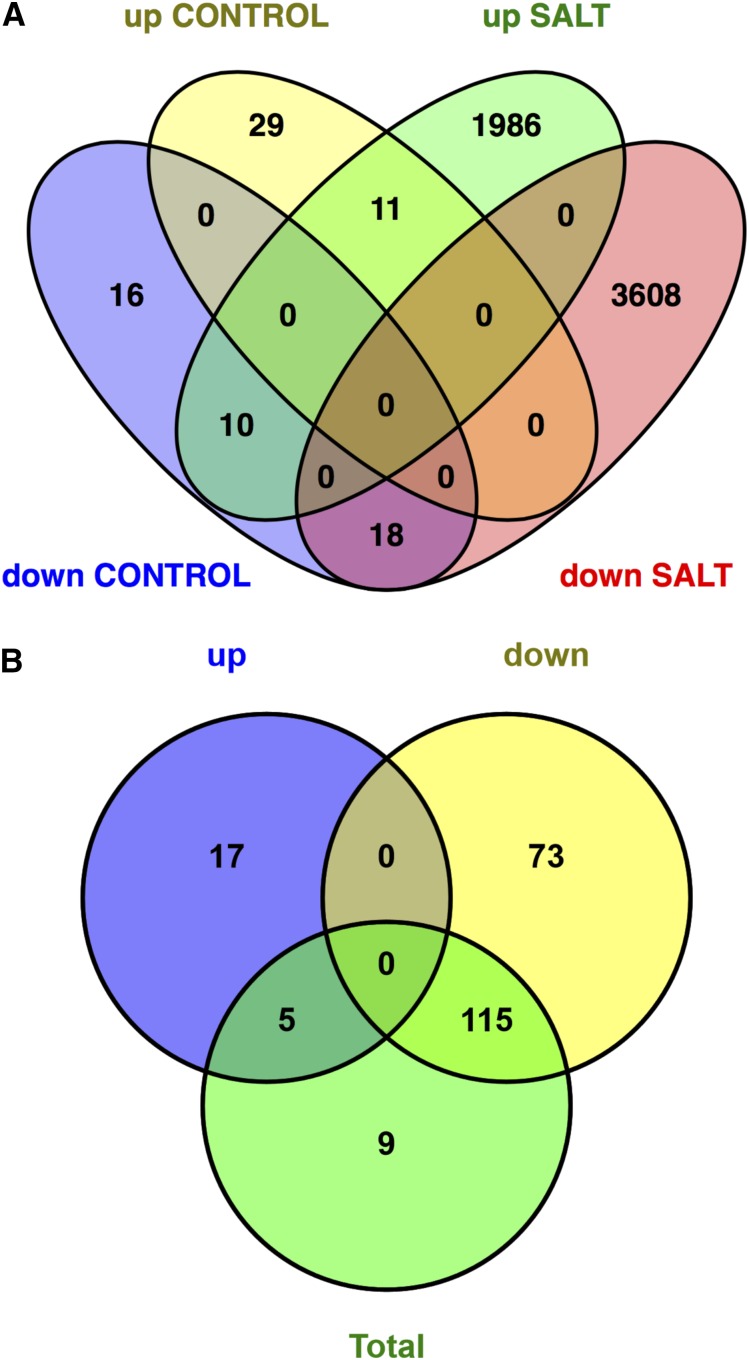
Based on the standard criteria of greater than 2-fold changes with statistical significance at P < 0.05 in t tests, we identified differentially expressed genes (DEGs) in sos1 mutant roots under salt and control conditions. Supplemental Table S1 online summarizes the genes up- and down-regulated in the mutant roots when compared with expression levels in the control line under the two treatments. Only 84 DEGs, representing 0.3% of total genes annotated, were detected between the two contrasting lines cultivated in control conditions. This result points to a minor role of SOS1 during the root development in the absence of sodicity stress. Of note is that HKT2;1 (Os06g0701700), encoding a transporter protein that mediates Na+ influx into K+-starved roots and thereby improves growth (Horie et al., 2007), was the second most up-regulated DEG in the sos1 mutant without salinity treatment (Supplemental Table S1). By contrast, many more genes were differentially expressed in the mutant relative to that in the wild type when salt stress was imposed, in accordance with the substantial impact of salinity on sos1 roots. Of the 5678 total DEGs identified in the roots of the sos1 mutant under salinity, only 11 genes were commonly up-regulated and 18 were down-regulated in the sos1 mutant relative to expression levels in the wild type irrespective of the salt treatment (Fig. 8), indicating that the transcriptional impact of the sos1 mutation was largely specific to the salt-induced response. The large number of DEGs (5633 genes, 19.48% of total) that presented an altered response in sos1 roots relative to that in wild-type roots under salt stress demonstrates the drastic effect of the sos1 mutation in the transcriptional response to sodicity. Notably, the majority (64%, 3608 genes) of these salt-responsive DEGs were down-regulated in the mutant root, which may represent a biased transcriptional response toward either repressed or less-induced transcription in the sos1 mutant in response to salt.
Figure 8.
Summary of differentially expressed genes. A, Four-way Venn diagram indicating the number of up-regulated and down-regulated genes found in the comparison between the sos1 rice mutant and the wild-type Nipponbare in the nonstress-treated (left; Control: 0 mM) and salt-stress (right; Salt: 75 mM)–treated roots. B, Three-way Venn diagram indicating the number of significant GO terms from the gene-ontology analysis of the up-regulated, down-regulated, and total DEGs found in the sos1 rice mutant and the wild-type Nipponbare in salt-stressed roots.
To evaluate the potential biological function of the differentially expressed genes in the sos1 mutant, Gene Ontology (GO) analysis was performed using the AgriGO service (http://bioinfo.cau.edu.cn/agriGO) for the up-regulated, down-regulated, and total DEGs identified in the salt-stressed or control-cultivated roots (Supplemental Table S2). The 5633 salt-responding DEGs fell into 129 significantly enriched GO categories corresponding to 66 biological processes, 28 functional categories, and 35 cellular components. The top over-represented GO categories included “cellular nitrogen compound metabolic process” (137 genes; False Discovery Rate [FDR] of 7.2 × 10−16), and those related to the biological process “transport” (FDR = 2.2 × 10−4; Supplemental Table S2). Further analysis showed again that these GO categories were constituted by far more down-regulated than up-regulated genes. This bias explained that most (n = 115) of the 129 significant GO terms were also enriched specifically in the group of the down-regulated DEGs (Fig. 8), including the 25 top enriched GO categories (Supplemental Table S3). Moreover, 73 of the significant GO terms identified were only present among the enriched categories of the down-regulated DEGs. For example, among the 97 total DEGs classified under the term “ion transmembrane transporter activity” (GO:0015075), 79 of them were down-regulated DEGs. The GO cellular component “membrane” and “membrane part” ranked top among GO terms enriched in down-regulated DEGs.
The MapMan software (Thimm et al., 2004) was also used to assign genes to functional categories and to further analyze the metabolic pathways and cellular responses altered in sos1 roots. Over-representation analysis of the up- and down-regulated categories (Supplemental Table S4; Supplemental Fig. S9) indicated a general repression of the glycolysis, the TCA cycle, nitrogen metabolism, amino acid synthesis, redox metabolism, protein targeting (secretory pathway) and protein degradation (ubiquitin proteasome). Among the genes related to “abiotic stress” responses, only the “drought/salt” and “unspecified” subcategories were significantly different between the two genotypes, with an over-representation of unspecified stress genes among the up-regulated salt-responding DEGs. Together, these results support the notion that the transcriptional response of the sos1 mutant to salt involves a global alteration of the cell physiology due to preferential down-regulation of genes that otherwise would be part of the normal response of wild-type roots to salinity.
The majority of the DEGs showing the largest variations in transcript abundance corresponded to proteins of unknown function or with sequence similarity to other proteins but whose specific function remains to be determined. Moreover, the differential hybridization method used in microarrays only captures genes whose transcript abundance at sampling are substantially different between samples; genes whose transcriptional dynamics are of similar magnitude between samples are not detected even if they responded to the stimulus. This was best exemplified by CIPK24, which presumably was not identified as a DEG by the microarray data because this gene was similarly induced in wild-type and sos1 roots, as demonstrated by RT-quantitative PCR (RT-qPCR; Fig. 9). Thus, we analyzed in greater detail groups of genes encoding proteins with a known role in processes that are likely to be relevant to the ionic component of salinity stress, mainly Na+ and K+ transporters and their regulators CIPKs and CBLs. Among the 100 genes selected for this analysis (Supplemental Table S5), the rice SalT locus was the most transcriptionally induced gene in sos1 roots relative to that in control salt-treated roots (log2 fold change [FC] = 4.79). Gene SalT, encoding a 15-kD jacalin-related Man-binding lectin, is located on a quantitative trait loci responsible for the bulk of genetic variation in ion uptake under saline conditions at the seedling stage (Thomson et al., 2010). The strong induction of this gene was confirmed by RT-qPCR (Fig. 9). Likewise, SIMILAR TO RCD-ONE1c (SRO1c; Os03g0230300) was highly induced in sos1 roots in the transcriptomic data set (log2 FC = 4.56) and verified by RT-qPCR (168-fold induction at 7 d, compared with 2.6-fold in the wild type). SRO1c encodes a putative poly-[ADP-ribose] polymerase and is the most stress-responsive gene within the rice SRO family (You et al., 2013). Notably, SRO1c is closely related to the Arabidopsis protein RADICAL-INDUCED CELL DEATH1 (RCD1), an inactive putative poly-[ADP-ribose] polymerase that shuttles between nucleus and cytoplasm to interact with SOS1 (Katiyar-Agarwal et al., 2006).
Figure 9.
Quantitation of expression levels of genes relevant to salinity tolerance. RT-qPCR analysis was performed using the 2−∆∆CT method and the gene expression data normalized for the quantity of UBQ5 gene transcript before (0) and 4 and 7 d after salt treatment with 75 mm NaCl. All y axes are mean values of normalized expression levels ± sd. All x axes are time of treatment and genotype. WT, wild type.
Among the CIPK-encoding genes, CIPK21 showed the largest transcriptional up-regulation by salt in sos1 vs. wild-type roots according to the microarray data (log2 FC = 2.22), which was confirmed by RT-qPCR. A modest 1.93-fold transcriptional induction was measured in wild-type roots after 7 d in salt, whereas in sos1 the 6.05-fold induction after 4-d treatment was followed by a striking 21.47-fold induction after 7 d (Fig. 9). CIPK15, which has been implicated in the salt stress response and enhances the salinity tolerance of transgenic rice plants (Xiang et al., 2007), was also up-regulated in the sos1 mutant (log2 FC = 1.75; Supplemental Table S5). CIPK1, 31, 10, and 17 also showed differential up-regulation in sos1 roots relative to that in the wild type, whereas CIPK23, 29, 25, and 4 were down-regulated by at least 2-fold. CBL genes showed little differential responsiveness in microarray data, with CBL9 being the most highly induced (log2 FC = 2.04) and CBL7 the most down-regulated (log2 FC = −2.46). RT-qPCR data confirmed the up-regulation of CBL9 and showed that the down-regulation of CBL7 in sos1 was only transient at 4 d, with a return to prestress level after 7 d (Fig. 9). Although not identified as statistically significant DEGs, the rice homologs of CBL4/SOS3 and CBL10, the two CBLs regulating CIPK24/SOS2 and SOS1 in Arabidopsis, showed complementary kinetics in sos1 roots so that the transient induction of OsCBL10 was followed by up-regulation of OsCBL4 (Fig. 9).
Successful adaptation to salinity requires the accumulation of inorganic and organic osmolytes inside vacuoles for osmoregulation and ion detoxification (Munns and Tester, 2008). Among these osmolytes, Na+, K+, Cl−, and organic acids such as malate are most abundant. In Arabidopsis, NHX1 and NHX2 are vacuolar Na,K/H exchangers that function in planta to drive the uphill accumulation of K+ into the vacuolar lumen for osmotic regulation, generation of cellular turgor, and stomatal movements (Leidi et al., 2010; Barragán et al., 2012; Andrés et al., 2014). The rice genes encoding exchangers OsNHX1 and OsNHX2 showed a 3- to 4-fold transcriptional up-regulation in sos1 roots. Gene Os09g0484900, encoding a putative protein 75% similar to the tonoplast dicarboxylate transporter (sTDT) of Arabidopsis involved in the uptake of malate into the vacuole (Emmerlich et al., 2003), was also highly induced in the salinized sos1 roots according to both the microarray and RT-qPCR data (8-fold induction at 7 d in the wild type vs. 85-fold in sos1). Malate storage within the vacuole allows the plant to accumulate this metabolite to very high concentrations (up to > 300 mM), and this large gene induction likely reflects the need to accumulate anions in the vacuolar lumen for osmotic adjustment and charge compensation of compartmentalized Na+. These results suggest that in response to salinity, the sos1 mutant differentially modulates multiple K+ and malate transporters to achieve cytosolic ion homeostasis and osmoregulation in intracellular compartments.
Other genes encoding K+-transport proteins showed minor transcriptional responses. No HIGH-AFFINITY K+ UPTAKE (HAK) genes showed differential up-regulation in sos1 roots compared with that in the wild type according to microarray data. On the other hand, HAK1, HAK5, and HAK16 were down-regulated 3- to 4-fold in sos1 relative to that in the wild type. OsHAK5 mediates high-affinity K+ uptake by roots under nutrient limitation and in saline conditions (Yang et al., 2014), whereas OsHAK1 plays a role in K+ uptake and translocation to shoots over low- and high-K+ concentration ranges (Bañuelos et al., 2002; Chen et al., 2015). Closer inspection by RT-qPCR showed that both OsHAK1 and OsHAK5 transcript abundance displayed complex kinetics (Fig. 9). OsHAK1 transcripts showed opposite kinetics after stress imposition in control and sos1 plants. A transient induction of OsHAK5 was also observed in control plants, but the up-regulation of OsHAK5 was sustained throughout the treatment in the sos1 mutant. The RT-qPCR data are in agreement with the expected role of OsHAK5 in sustaining K+ uptake in a saline environment (Horie et al., 2011; Yang et al., 2014) and probably reflect the need of sos1 roots to re-establish K+ homeostasis under salinity.
The HKT proteins of cereals have been involved in the uptake of Na+ by roots and in the root-to-shoot partition of Na+ and K+ (Garciadeblás et al., 2003; Yao et al., 2010; Munns et al., 2012; Hamamoto et al., 2015). Out of eight HKT genes in the rice genome, only HKT8/HKT1;5/SHOOT K+ CONTENT1 (SKC1; Os01t0307500) showed a modest yet statistically significant down-regulation in sos1 roots under salt (log2 FC = −0.81), whereas HKT1/HKT2;1 (Os06t0701700) was up-regulated in sos1 roots in the absence of stress (log2 FC = 2.10). The rice transporter HKT8/SKC1 regulates Na+/K+ levels in shoots by recirculating Na+ from shoots to roots (Ren et al., 2005). The significance of the HKT8/HKT1;5/SKC1 down-regulation might be related to the excessive accumulation of Na+ in the sos1 roots and represents a root-protecting mechanism limiting the back-flow of Na+ to roots.
DISCUSSION
We showed previously that the rice SOS system comprises the OsSOS1 transporter, the OsCIPK24 protein kinase that is the closest homolog to the Arabidopsis SOS2/AtCIPK24, and OsCBL4 that is most similar to SOS3/AtCBL4 (Martínez-Atienza et al., 2007). The fact that rice OsSOS1 is encoded by the single copy gene Os12g44360 allowed a reverse-genetics approach to determine the importance of this protein in the salt tolerance of rice. Here, we have shown that the sos1 mutant plant displays an exceptional salt sensitivity that is akin to the Arabidopsis sos1 mutant phenotype. By contrast to the uniqueness of OsSOS1, the modest salt sensitivity of the rice cipk24 mutant relative to sos1 plants is likely due to functional redundancy. The genetic complement of CIPK-CBL modules in the rice genome comprises 10 CBLs and 33 CIPKs, which is similar to the 10 CBLs and 25 CIPKs known in Arabidopsis (Kolukisaoglu et al., 2004; Gu et al., 2008; Kanwar et al., 2014). Most of the CIPK genes of rice showed transcriptional responses to various abiotic stresses including salinity (Xiang et al., 2007; Kanwar et al., 2014). Because transcription of CIPK8, the closest homolog to CIPK24, was induced by salt (Xiang et al., 2007; Kanwar et al., 2014), a functional relatedness of CIPK8 and CIPK24 appears likely regarding the salt-stress response of rice. Our transcriptomic analysis also flagged CIPK15, CIPK21, and CIPK31 as potential players in the sodicity response of roots based on the greater up-regulation in sos1 roots compared with that in the wild type (Supplemental Table S5). Indeed, CIPK15 has been reported to increase the salinity tolerance of transgenic rice (Xiang et al., 2007), whereas a loss-of-function cipk31 mutant was salt sensitive (Piao et al., 2010). There is no reverse genetics data for CIPK21, but RT-qPCR confirmed a 21.47-fold up-regulation of CIKP21 transcripts after 7 d in sos1 roots (Fig. 8). Likely, this large transcriptional induction of CIPK21 in the sos1 mutant reflects its major role in Na+ homeostasis. Thus, transcriptomic and genetic data, together with the enormous difference between the salt-sensitive phenotype of the cipk24 and the sos1 mutants, suggest that several CIPKs may target SOS1 for posttranscriptional activation in rice.
Recently, a genetic screen for reduced cesium (Cs+) accumulation in rice identified a putative loss-of-function allele of SOS2/CIPK24 as the underlying genetic lesion (Ishikawa et al., 2017). The lower Cs+ content in the lcs1/sos2 mutant correlated with reduced expression of K+ transporters HAK1, HAK5, and ARABIDOPSIS K+ TRANSPORT1 (AKT1) under low external K+/Na+ ratios, suggesting that Cs+ uptake occurred through root K+ transporters. Similar to our findings, the lcs1/sos2 mutant accumulated Na+ in roots and was moderately salt sensitive.
Entry and Redistribution of Na+
The salt tolerance imparted by SOS1 is so intimately related to cellular tolerance that overexpression of SOS1 and resistance to NaCl has been used as selection system for rice transformation (Zhu and Wu, 2008). Here, we show that nearly ubiquitous overexpression of SOS1 driven by the 35S promoter yielded salt tolerance that correlated with reduced Na+ contents in shoots and roots, as should be expected from enhanced efflux of cellular Na+ (Fig. 4). In addition, we have shown that SOS1 also plays a systemic role by redistributing Na+ ions between roots and shoots at the xylem-parenchyma boundary (Fig. 6). Rice roots develop Casparian bands and suberin lamellae on the walls of the exodermal and endodermal cell layers that act as physical barriers against the apoplastic flow of water and dissolved minerals (Krishnamurthy et al., 2009). However, lateral roots emerge from the pericycle and disrupt the continuity of the endodermal barrier as they grow toward the cortex and exodermis. This rupture of the apoplastic barrier generates sites through which salts could leak into the main root. The contribution of this ‘bypass-flow’ to Na+ uptake in rice is thought to constitute a major component of the salt sensitivity of this crop, particularly at high transpiration rates (Yeo et al., 1987). Opposing this view, later studies found no evidence indicative of Na+ entry at the sites of secondary root emergence in rice plants, although the tissue still remained open to apoplastic flow tracers (Faiyue et al., 2010a, 2010b). Rather, it was concluded that bypass-flow occurred through the lateral roots themselves (Faiyue et al., 2010a). Regardless of whether bypass flow occurs through lateral roots or the endodermal rupture points caused by them, our data support the notion that net Na+ fluxes into and across the root are far from being an uncontrolled process. The combination of the tissue expression pattern of the SOS1 gene promoter and the profile of Na+ content along a cross section of rice roots from wild-type and sos1 mutant plants submitted to a moderate salinity stress (2 d in 50 mm NaCl) indicated that SOS1 prevented, in the first place, the excessive accumulation of Na+ in all cell types of the root and that this protein specifically directed the flow of Na+ past the endodermal filter toward the xylem vessels for the acropetal evacuation of Na+ out of the root (Figs. 5 and 6). Nevertheless, it should be emphasized that SOS1 is primarily a cellular-based Na+ efflux system (Shi et al., 2002; Oh et al., 2009) and Na+ will accumulate over time in root tissues to greater levels in the sos1 mutant than that in wild-type plants, which would eventually lead to Na+ leaking into shoots by mass flow. Moreover, discontinuities in the root endodermis may also allow Na+ export out of roots without a biochemical control by dedicated transporters. In other words, Na+ will accumulate in the shoot of the sos1 mutant as a consequence of uncontrolled uptake by roots even if SOS1 contributes to transporter-mediated Na+ loading into the xylem.
The sensitivity of rice plants to salinity is strongly associated with the accumulation of Na+ in the leaves (Coskun et al., 2013). Like SOS1, the gene encoding the Na+-selective transporter SKC1/HKT8/HKT1;5 is preferentially expressed in the parenchyma cells surrounding the xylem vessels, where the HKT protein is thought to mediate the recirculation of Na+ by unloading Na+ from the root xylem (Ren et al., 2005). Our data show that SOS1 counteracts HKT function by actively exporting Na+ from parenchyma cells into the xylem vessels. Thus, the amount of Na+ that is eventually delivered to shoots will be determined by the interplay between HKT and SOS1 proteins acting antagonistically. The optimal set-points for Na+ root content and the net transfer of Na+ from shoots to roots must be commensurate with the ability of shoot tissues to compartmentalize the incoming Na+ to avert ion toxicity while achieving full osmotic adjustment (Pardo, 2010). The down-regulation of SKC1/HKT8/HKT1;5 in salinized sos1 roots could represent a root-protecting mechanism limiting the back-flow of Na+ to roots that are already exposed to high Na+ contents. The reciprocally balanced activity of HKT and SOS1 proteins has been evidenced also in wheat. Two loci, termed Na+ EXCLUSION1 (Nax1) and Nax2 encoding proteins HKT1;4 and HKT1;5, respectively, are genetic determinants of enhanced Na+ retrieval from the xylem and reduced rates of Na+ delivery to the shoot (Huang et al., 2006; James et al., 2006; Byrt et al., 2007). Notably, the Nax loci also reduced the activity and expression level of the SOS1 exchanger in both root cortical and stele tissues, which contributed further toward reducing the xylem Na+ load (Zhu et al., 2016). In Arabidopsis, a functional hierarchy of HKT- and SOS1-dependent transport processes was established by comparing the salt sensitivity of hkt1 and sos1-sos3 mutants (Rus et al., 2004). The mutant lacking HKT1 function was sensitive to long-term salinity due to the excessive accumulation of Na+ in shoots, and the hkt1 mutation suppressed the NaCl hypersensitivity of the sos3 mutant by achieving a balanced distribution of Na+ that was closer to that of wild-type plants (Rus et al., 2001, 2004). Together, these data demonstrate that a finely regulated interplay between HKT and SOS systems at the xylem parenchyma is a key component of the salt tolerance of vascular plants.
An unresolved question is the mechanistic basis for the intimate relationship of Na+-specific transporters acting at the xylem parenchyma with the K+ status of the plant. In rice, both SOS1 and SKC1/HKT8/HKT1;5 selectively transport Na+ and yet they affect the accumulation of K+ in shoots. Wheat plants carrying the Nax1 and Nax2 loci encoding the Na+ transporters HKT1;4 and HKT1;5 also had higher rates of K+ transport from roots to shoots (James et al., 2006). Root xylem loading is a key step for the delivery of nutrients to the shoot (Drew et al., 1990; Engels and Marschner, 1992; Gaymard et al., 1998). In Arabidopsis, the voltage-dependent K+-channel STELAR K+ OUTWARD RECTIFIER (SKOR) mediates xylem loading and root-to-shoot translocation of K+ (Gaymard et al., 1998). The SKOR gene is primarily expressed in pericycle and root xylem parenchyma cells. The SKOR channel directs outward K+ currents upon membrane depolarization facilitating the release of the K+ from the parenchyma cells into the xylem vessels. The Arabidopsis sos1 mutant was initially described as impaired in K+ uptake (Wu et al., 1996), but subsequent research has shown that SOS1 is a highly specific Na+/H+ exchanger with no ability to transport K+ (Qiu et al., 2002; Shi et al., 2002; Quintero et al., 2011). Here we show that removal of SOS1 from its preferential expression tissue at the xylem parenchyma cells leads to the accumulation of Na+ ions while in transit to the xylem vessels, with the concomitant reduction in K+ content in these same cells. The Na+/K+ ratio in xylem parenchyma cells climbed from 0.49 in the wild type to 9.40 in the sos1 mutant, i.e. a 19-fold change (Fig. 6). The coupling of Na+ unloading by the xylem-localized HKT transporters with K+ loading into xylem vessels via depolarization-activated K+ channels has been hypothesized (Horie et al., 2009). HKT-mediated Na+ uptake from xylem vessels could induce membrane depolarization of xylem parenchyma cells, which in turn would activate SKOR, the depolarization-activated outward-rectifying K+ channel mediating K+ release from xylem parenchyma cells. On the other hand, cytosolic Na+ has been shown to inhibit the K+ channel AKT1 involved in K+ uptake by Arabidopsis roots (Qi and Spalding, 2004). Greater cytosolic Na+ concentration in sos1 plants resulted in impaired K+ uptake by roots and poor growth under salt stress. Our data suggest a combined scenario in which the high Na+/K+ ratio in cortex cells may interfere with root K+ uptake and the symplastic movement toward the stele (note that AKT1 an AKT2 genes are differentially repressed in sos1 roots; Supplemental Table S5), whereas the exorbitant Na+/K+ ratio in xylem parenchyma cells of the sos1 may interfere with K+ loading into the xylem for nutrient partitioning between shoots and roots. This model may also explain the initial observation that the growth of the Arabidopsis sos1 mutant was impaired under low K+ availability. This hypothesis about the mechanistic links between long-distance transport of Na+ and K+ nutrition deserves further research.
Transcriptome of the sos1 Mutant
Nitrate (NO3−) is the quantitatively most important anion counter-balancing xylem loading of K+ (Engels and Marschner, 1993). In Arabidopsis, the low-affinity, bidirectional NO3− transporter NITRATE TRANSPORTER1.5 (NRT1.5) mediates NO3− efflux from pericycle cells to the xylem vessels of Arabidopsis, and SKOR and NRT1.5 cooperate to ensure K+ delivery to the shoot via xylem (Drechsler et al., 2015). Alternatively, NRT1.5 could load K+ into the xylem directly (Li et al., 2017; Ragel et al., 2019). A similarly complex relationship is also apparent between Na+ and K+ loading into the xylem (Álvarez-Aragón and Rodríguez-Navarro, 2017). Our transcriptomic data of the sos1 mutant suggests a broad molecular connection between Na+/K+ homeostasis and nitrogen metabolism under salinity stress. The biological process, as categorized by GO terms, “Cellular Nitrogen Compound Metabolic Process” was the most severely down-regulated category in the sos1 mutant under salt relative to that in control roots (FDR = 1.6 × 10−18). Key proteins involved in nitrate uptake by roots and nitrate assimilation were repressed in the sos1 mutant. Genes encoding four NRT2-like transporters and two Nitrate Transporter-Activating protein (NAR)2-like regulators have been identified in the rice genome. NAR2.1 interacts with the nitrate transporters OsNRT2.1, OsNRT2.2, and OsNRT2.3a to activate the uptake and distribution of nitrate within the plant (Feng et al., 2011). The strong expression of OsNAR2.1, OsNRT2.1, and OsNRT2.2 in root epidermal cells and that OsNAR2.1 knockdown greatly impaired both high- and low-affinity nitrate transport systems demonstrated the key role of these proteins in nitrate uptake (Yan et al., 2011). Notably, these three key genes were substantially repressed in sos1 roots by salinity. The log2 fold changes in transcript abundance for NAR2.1 (Os02g0595900), NRT2.1 (Os02g0112100), and NRT2.2 (Os02g0112600) were −4.24, −3.33, and −2.91, respectively. Similarly, the majority of enzymatic genes involved in the nitrate assimilation by its incorporation to Gln (nitrate and nitrite reductases, and Gln synthetases) were down-regulated in salt-treated sos1 roots. OsCIPK23 (Os07t0150700), the closest rice homolog to the protein kinase CIKP23 that regulates positively nitrate uptake in Arabidopsis (Ho et al., 2009), was also down-regulated in salinized sos1 roots relative to that in the control (log2 FC = −1.66), in agreement with a general repression of nitrogen uptake and metabolism. Consequently, the transcriptional repression of these key genes in the sos1 mutant root should have a profound effect on nitrate uptake from the mineral medium and subsequent nitrogen assimilation, as already indicated by the GO term and MapMan analyses. The physiological implications of this response are presently unclear, but might be related to the mutual stimulation of nitrate and Na+ uptakes that has been observed in various species including rice (Kaburagi et al., 2015; Nie et al., 2015; Gao et al., 2016). In Arabidopsis, nitrate-dependent transport systems mediate the uptake and load of Na+ into the xylem, and they may constitute a major pathway for the accumulation of Na+ in Arabidopsis shoots (Álvarez-Aragón and Rodríguez-Navarro, 2017). Thus, repression of nitrate uptake by sos1 roots could have helped to curtail Na+ entry and/or distribution.
An intriguing observation is that the GO term 'Response to Stress' was down-regulated in the sos1 mutant in the two conditions tested, with and without salt, which in principle meant that the sos1 mutant was less responsive to stress. A number of genes whose function could be reasonably predicted as being beneficial to overcome salinity stress (CBL4, NHX1, NHX2, and the putative vacuolar malate transporter encoded by Os09g0484900) or genes that are known molecular markers of stress intensity (SalT) were more highly expressed in the sos1 mutant than that in wild-type roots. These molecular markers and the severely disturbed Na+/K+ profiles of sos1 roots demonstrate that the mutant roots were indeed suffering sodicity stress more intensely than the wild type. Thus, the large proportion of genes down-regulated in the sos1 mutant might be the consequence of impaired stress detection or an inability to mount a full-fledged response, despite the greater intensity of the stress imposed upon sos1 roots. Previous studies (Zhu, 2002; Shabala et al., 2005; Ji et al., 2013) have suggested that SOS1 may perform regulatory or sensory functions that may or may not be strictly linked to its function as a Na+ transporter. The exceedingly high salt sensitivity of sos1 mutants of various species (Zhu, 2002; Oh et al., 2009; Olías et al., 2009) and the interaction of the very large C-terminal domain of SOS1 with other stress signaling intermediaries (Katiyar-Agarwal et al., 2006) lend indirect support to this hypothesis. Precedents of this dual function as enzymes and key regulators are the nitrate transporter NRT1.1 and the hexokinase HXK1 that serve as sensor proteins of their substrates (Moore et al., 2003; Ho et al., 2009; Bouguyon et al., 2015).
Alternatively, SOS1 may not be a bonafide sensor but serve as a molecular scaffold to facilitate the cross talk of various signaling pathways needed to mount salt tolerance. The SRO proteins are a group of plant-specific proteins that have important functions in stress adaptation and development. In Arabidopsis, SOS1 interacts with RCD1, a nucleo-cytoplasmic protein that together with AtSRO1 functions to regulate oxidative stress, hormonal, and developmental responses (Katiyar-Agarwal et al., 2006; Jaspers et al., 2010). Moreover, RCD1 interacts with DEHYDRATION RESPONSIVE ELEMENT BINDING2 (DREB2) protein and a number of transcription factors belonging to several protein families. The rice genome contains five SRO genes (SRO1a to 1e), and transcription of SRO1c is highly responsive to various stresses including salinity. As in Arabidopsis, rice SRO1c has a negative role in resistance to oxidative stress, which seems to be associated to the repression of ROS-scavenging enzyme genes (You et al., 2013). Surprisingly, transcriptomic and RT-qPCR data showed a staggering 55- and 168-fold induction of SRO1c in sos1 roots after 4 and 7 d of salt treatment, which runs counterintuitive to the expected output since ROS scavenging is vital to overcome salinity-induced oxidative stress in rice (Hoang et al., 2016) and OsSRO1c-overexpressing rice suffers from greater sensitivity to oxidative stress (You et al., 2013). The observed under-representation of DEGs belonging to the GO Biological Process 'Oxidation Reduction' (FDR = 2.3 × 10−6; Supplemental Table S2) and the GO Molecular Function Oxidoreductase Activity (FDR = 2.6 10−5) in sos1 roots under salinity are likely related to over-induction of the negative transcription regulator SRO1c. Our MapMan analysis of the sos1 mutant transcriptional response also pointed to a significant enrichment of genes involved in the redox cellular metabolism among the down-regulated DEGs, with 61 down-regulated genes grouped in this category. Detailed analysis of the repressed genes indicates that detoxification enzymes thioredoxins, ascorbate-peroxidases, catalases, and superoxide dismutases were mainly repressed in the mutant relative to that in the wild type. Moreover, further analysis of the mutant transcriptome indicated that the expression of genes grouped in the “drought/salt abiotic stress” category is significantly altered in response to salt (corrected p-value = 0.037). The differential expression was surprisingly not due to a higher level of induction of the genes of this category in response to salt, but again to a general repression in the sos1 mutant relative to the wild-type expression level. Thus, it appears that the lack of SOS1 triggers a run-away process leading to unsuitable repression of ROS-controlling genes by OsSRO1c and exacerbated salt damage. Taken globally, the transcriptomic profile of the sos1 mutant root is evidence of an as yet undefined but superior regulatory role of SOS1 in the salinity stress response.
MATERIALS AND METHODS
Plant Culture, Salinity Treatments, and Chemical Analyses
Rice seeds were germinated onto wet filter paper for 5 d and then transferred to decapped and perforated Eppendorf tubes prefilled with rockwool. Culture was continued in a hydroponics setting with aeration in modified Miyamoto's medium (Miyamoto et al., 2001), with weekly transfers to fresh medium. Nutrient medium consisted of 0.09 mm (NH4)2SO4, 0.05 mm KH2PO4, 0.05 mm KNO3, 0.03 mm K2SO4, 0.06 mm Ca(NO3)2, 0.07 mm MgSO4, 0.11 mm Fe-EDTA, 4.6 µM H3BO3, 1.8 µM MnSO4, 0.3 µM ZnSO4, 0.3 µM CuSO4 (pH 5.7), plus 0.3 µM Na2MoO4. The actual concentrations of K+ and Na+ in the medium as measured by Atomic Absorption Spectrophotometry were 0.20 mm K+ and 0.14 mm Na+. The hydroponic containers were placed in a growth chamber set to a daily light/dark cycle of 16/8 h, photosynthetically active radiation 300 µmol m−2 s−1, 30°C/22°C day/night temperature, and 40–60% relative humidity. Salt treatments were given by transferring the plants to fresh medium supplemented with the indicated amounts of NaCl.
Contrary to Nipponbare plants, seedlings of mutant (L1H and L24H) and congenic azygous lines (L1A and L24A) showed asynchronous germination and heterogeneous growth rates, which presumably arose from somaclonal variation. Therefore, after 1 week in hydroponic culture, outlier plants were removed and only those with sizes similar to Nipponbare controls were transferred to fresh media with and without supplemental NaCl.
For halotolerance tests in experimental paddies in the field, seeds were first germinated in pots with peat. Four-week-old plants of mutant genotypes sos1 and cipk24, their respective congenic null segregants, and of wild-type Nipponbare were then transferred to paddies with clay-loam soil. Plants were arranged in paddies with 5 rows (one per genotype) of 10 plants each, to have 4 repetitions (individual paddies) of each growth condition (control and salinized) and 40 plants per genotype and treatment. Genotypes were arranged differently in each individual paddy. Control paddies were irrigated with water from an underground well with a salt content of 0.4 g/L. Salinity treatment was imposed by automatic mixing of underground water with a salty brine to give a final concentration of 1.8 g/L of seawater salts in the irrigation water.
Chlorophyll contents were determined with a Minolta SPAD-502 Chlorophyll Meter and values are given as SPAD units. To measure Na+ and K+ contents in tissues, plants exposed to salt stress were separated in shoots and roots and weighed. Roots were washed thoroughly 2 to 3 times over 2 min with cold distilled water to remove surface-contaminating salts, blotted dry, and deep frozen until use. Dried ground material was autoclaved to extract minerals and the supernatant submitted to Atomic Absortion Spectrophotometry.
Rice Mutant Lines
Nomenclature and genes codes are according to the Rice Genome Annotation Project, version 6.1 (http://rice.plantbiology.msu.edu/). Putative mutant lines annotated to carry T-DNA or Tos17 insertions in genes SOS1 (Os12g44360), SOS2/CIPK24 (Os06g40370), and SOS3/CBL4 (Os05g45810) in the Nipponbare genetic background were screened for in the public repositories OryGenesDB of Génoplante (http://orygenesdb.cirad.fr/), the Rice Functional Genomic Express Database (http://signal.salk.edu/cgi-bin/RiceGE), the Rice Tos17 Insertion Mutant Database (https://tos.nias.affrc.go.jp/), and the Rice Mutants Database (http://rmd.ncpgr.cn/).
Seeds received from repositories were considered as T2 generation, comprising both heterozygous and homozygous individuals. Plants emerging from these primary T2 seed pools were subjected to diagnostic PCR to identify individuals homozygous for the mutagenic insertion based on the presence of the amplicon predicted from the line annotation and by the absence of the wild-type amplicon when oligonucleotides flanking the insertion point were used as primers in the diagnostic PCR. Primers used for diagnostic PCR and RT-PCR are listed in Supplemental Table S6. Congenic null-segregants without the mutagenic insertion (denoted here as azygous plants) were also selected as controls for further experiments to account for possible epigenetic or phenotypic alterations arising during plant transformation and regeneration procedures. Plants selected after diagnostic PCR were used to produce T3 seeds, and T3 plants were subsequently used for RT-PCR and phenotypic analyses.
Nucleic Acids, Plasmids, and Transformation
For RT-PCR analyses, total RNA was isolated from approximately 100 mg of frozen plant leaves with TRIZOL (Invitrogen). Synthesis of cDNA with integrated genomic DNA removal was done from 1 μg of total RNA using the QuantiTect Reverse Transcription Kit (Qiagen). PCR was done with DNA purified as described in Murray and Thompson (1980). For sequencing of amplicons and precise identification of insertion points, RT-PCR was performed with Accuzyme (Bioline Reagents Ltd) and amplicons were ligated to vector pCR-Blunt and transformed into Escherichia coli. Recombinant plasmids were purified using the Perfectprep Gel Cleanup kit (Eppendorf), diluted at 100 ng/µL in water, and sequenced with M13 reverse and forward primers.
Constructs made to test the function of the rice SOS1 protein in yeast were done in vector pDR195 carrying the PLASMA MEMBRANE ATPase1 (PMA1) gene promoter. Coexpression of Arabidopsis SOS2 and SOS3 was achieved with plasmid pFL32T (Quintero et al., 2002). The open reading frame encoding the truncated protein OsSOS1∆970 was obtained by PCR using as template the wild-type cDNA (Martínez-Atienza et al., 2007) and with oligonucleotides that included BamHI cutting sites and a stop codon after amino acid L969 of SOS1. The resulting amplicon was digested with BamHI and inserted in pDR195. Yeast transformation followed the Lithium-Acetate method. The Saccharomyces cerevisiae strain AXT3K (Δena1::HIS3::ena4, Δnha1::LEU2, Δnhx1::KanMX4) has been described elsewhere (Guo et al., 2004). Sodium tolerance tests were performed in the alkali cation-free Arginine-Phosphate medium (Rodríguez-Navarro and Ramos, 1984) supplemented with 1 mm KCl and with NaCl as indicated in each experiment.
To determine the expression pattern of SOS1, a 3.9-kb fragment in the intergenic region between SOS1 (Os12g44360) and the adjacent gene (Os12g44370) and proximal to the coding region of SOS1 was amplified by PCR and inserted as a SalI and XbaI fragment into vector pBIG containing a promoter-less GUS reporter gene (Becker, 1990). GUS chemical staining with X-gal in whole tissues was as described in Jefferson et al. (1987). To test complementation of the sos1 mutant, the full-length cDNA of rice SOS1 was cloned in a pCAMBIA1300-derived vector containing the maize UBIQUITIN1 gene promoter and the nopaline synthase terminator (Campo et al., 2014). For SOS1 overexpression, the cDNA was cloned into the vector pCAMBIA2300 under the control of the CaMV 35S promoter and terminator and transformed in Nipponbare plants. Rice transformation by Agrobacterium tumefaciens EHA105 has been described (Campo et al., 2014).
Measurement of Na+/H+ Exchange in Membrane Vesicles
Plasma membrane vesicles were isolated from 2-month-old rice roots from 24-h salinized (50 mm NaCl) plants using two-phase partitioning as described by Qiu et al. (2003). All steps were carried out at 4°C or on ice. Roots were homogenized in buffer containing 0.33 m Suc, 10% (w/v) glycerol, 0.2% (w/v) bovine serum albumin, 5 mm EDTA, 5 mm dithiothreitol (DTT), 5 mm ascorbate, 0.2% (w/v) casein, 0.6% (w/v) polyvinylpyrrolidone, 1 mm phenylmethylsulfonyl fluoride, 1 µg/mL pepstatin A, and 50 mm HEPES-KOH (pH 7.5). Eight milliliters of homogenization buffer were used per gram of tissue. The homogenate was filtered through two layers of Miracloth (Merck) and centrifuged at 10,000 g for 10 min. The supernatant then was centrifuged for 50 min at 100,000 g to obtain a microsomal pellet that was resuspended in a buffer containing 0.33 m Suc, 3 mm KCl, 0.1 mm EDTA, 1 mm DTT, 1 mm phenylmethylsulfonyl fluoride, 1 µg/mL pepstatin A, and 5 mm potassium phosphate (pH 7.8). The suspension was added to a phase mixture to obtain a phase system consisting of 6.2% (w/w) Dextran T-500 and 6.2% (w/w) polyethylene glycol 3350 in 5 mm potassium phosphate (pH 7.8), 0.33 m Suc, and 3 mm KCl. Phase separation was facilitated by centrifugation at 1,000 g. The upper phase was then repartitioned twice in this system. The final upper phase was collected, diluted with suspension buffer [0.33 m Suc, 10% (w/v) glycerol, 0.1 mm EDTA, 2 mM DTT, 1 µg/mL pepstatin A, 20 mm HEPES-KOH (pH 7.5)], and centrifuged for 45 min at 100,000 g. The resulting pellet was collected and resuspended with the above-described suspension buffer containing 1 mm EDTA, frozen in liquid nitrogen, and stored at −80°C until use. Before membrane isolation, plants were treated for 24 h with 50 mm NaCl in Miyamoto’s medium to induce SOS1 activity. Vesicles purified from nontreated plants produced very low rates of Na+/H+ antiport activity, in agreement with previous reports (Qiu et al., 2003).
The measurement of Na+/H+ proton exchange was as described by Qiu et al. (2003), except that fluorescence quenching of 9-amino-6-chloro-2-methoxy-acridine was used to monitor the formation and dissipation of pH gradients. Fluorescence was recorded with a Hitachi fluorescence spectrophotometer (FL-2500) in a thermostated cell (25°C) at excitation and emission wavelengths of 415 and 485 nm, respectively (slit-width, 10 nm). Purified plasma membranes (50 µg of protein) were added to a buffer containing 250 mm mannitol, 10 mm bistris-propane-MES (pH 6.8), 100 mm KCl, 3 mm MgSO4, and 1 µM 9-amino-6-chloro-2-methoxy-acridine (1 mL final volume). The ∆pH was generated with the activity of the plasma membrane H+-ATPase and recorded as fluorescence quenching after ATP addition (2.5 mm ATP, pH 6.8). The Na+-induced dissipation of ∆pH was initiated after adding 100 mm NaCl into the cuvette, and recorded as the recovery of fluorescence. Exchange rates are calculated as fluorescence recovery relative to that before ATP addition (∆F/Fmax) per minute and milligram of protein.
Microarray Analysis
Control and sos1 seedlings were grown hydroponically in Miyamoto's medium for 3 weeks in environmentally controlled conditions (12 h of light at 25°C and ∼450 µmol quanta m−2 s−1, and 12 h darkness at 21°C; relative humidity approximately 65%). Salt stress was imposed by transferring to fresh nutrient solution containing 75 mm NaCl for 4 d. Control plants received fresh nutrient solution without NaCl. No signs of plant deterioration were observed at the end of the salinity treatment. Total RNA was extracted from whole roots pooled from five plants per genotype and treatment. The quality and concentration of the RNA were determined using the Agilent 2100 Bioanalyzer, and samples with an RNA Integrity Number less than 6 were discarded. Three on-chip technical replicates per biological sample were analyzed. Gene expression profiling was performed by microarray hybridization using a custom rice transcriptomic array (Oryzon Genomics) containing 3 × 29450 probes (3 technical replicates per probe) representing 20750 genes. Microarray construction, probe labeling, and hybridization conditions were as in Campo et al. (2014). Results were normalized using self-to-self comparison Nipponbare vs Nipponbare, and fold change was calculated as log2 of the ratio of relative expression of sos1 (sample, Smp) versus wild-type (control, Ctr) plants grown in identical conditions. Only genes for which replicate spots showed differential expression of −0.5 > log2(Smp/Ctr) > 0.5 and P value < 0.05 were considered as differentially expressed. Bootstrap analysis with Significance Analysis of Microarrays was used to identify differentially expressed genes using a cutoff of 2 (Tusher et al., 2001). Significance Analysis of Microarrays calculates the fold change and significance of differences in expression. Data sets for microarray analyses have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus with accession number GSE129844.
Differential expression of a select number of DEGs was confirmed by reverse-transcription-quantitative PCR (RT-qPCR). Three-week-old control and sos1 seedlings were treated with 75 mm NaCl in Miyamoto's medium for 4 and 7 d. Total RNA was extracted from roots, and 500 ng were used to synthesize the cDNA using the Quantitec Reverse Transcription kit (Qiagen). Quantitative real-time PCR assays were performed using the CFX Connect Real-Time PCR Detection System (Bio-Rad). The PCR mixture (total volume of 20 μl) was composed of 5 μl of diluted cDNA, 10 μl iTaq Universal SYBR Green supermix (2×), 0.3 μm of each primer, and water. The specific oligonucleotides used are listed in the Supplemental Table S6. Thermal cycling consisted of 95°C for 3 min, followed by 40 cycles of 10 s at 95°C, 15 s at 58°C, and 15 s at 72°C. A melting curve was generated to check the specificity of the amplified fragment. The efficiency of all primers under these conditions was 85–100% for all samples tested. Rice UBIQUITIN5 was used as a house-keeping gene in expression analysis. Relative gene expression was determined using the 2−∆∆CT method (Livak and Schmittgen, 2001). Threshold cycle values were determined using ICYCLER Q software (Bio-Rad).
Statistical Analyses
Numerical data were analyzed with IBM SPSS Statistics v.21 (Armonk) using a completely randomized design model. Means and variances were compared by one-way ANOVA to find significant differences using the Fisher's Least Significant Differences test at P < 0.01 level, unless stated otherwise.
Accession Numbers
Sequence data from this article can be found in the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/) and UniProt (https://www.uniprot.org) databases under accession numbers SOS1 (LOC_Os12g44360, Q5ICN3), CIPK24/SOS2 (LOC_Os06g40370, Q69Q47), and CBL4/SOS3 (LOC_Os05g45810, Q75KU4).
Supplemental Data
The following supplemental information is available:
Supplemental Figure S1. Molecular characterization of rice lines AKFG08 and AKFB09.
Supplemental Figure S2. Molecular characterization of rice line NC2588.
Supplemental Figure S3. Molecular characterization of rice line AUJE11.
Supplemental Figure S4. Molecular characterization of the rice line AVIB03.
Supplemental Figure S5. Na+/H+ exchange activity in plasma membrane vesicles.
Supplemental Figure S6. Molecular characterization of the rice lines AMHC11 and AMHC12.
Supplemental Figure S7. Salinity tolerance test in experimental paddies.
Supplemental Figure S8. Complementation of the sos1 mutant.
Supplemental Figure S9. MapMan analysis of the metabolic and cellular responses of sos1 roots to salt.
Supplemental Table S1. Differentially expressed genes (DEGs) in the sos1 mutant roots when compared to the control line (Excel file).
Supplemental Table S2. Gene Ontology (GO) analysis of total DEGs identified in the salt-stressed or control-cultivated roots (Excel file).
Supplemental Table S3. Top 25 enriched GO categories (Excel file).
Supplemental Table S4. MapMap analysis of DEGs identified in the salt-stressed sos1 roots (Excel file).
Supplemental Table S5. Differential expression levels of select genes involved in Na+ and K+ transport processes (Excel file).
Footnotes
This work was supported by MINECO (grant nos. BIO2015-70946-R to F.J.Q., and BFU2015-64671-R and BIO2016-81957-REDT to J.M.P.); cofinanced by the European Regional Development Fund and by the Rural Development Administration (RDA), Republic of Korea (SSAC grant nos PJ01108101 to D.-J.Y. and PJ01318205 to J.M.P.). Part of this work was conducted at the Rice Functional Genomics Platform (REFUGE), Montpellier France, supported by an Agropolis Fondation grant to D.M. and E.G. F.G.-A. was supported by a 'Juan de la Cierva' aid from MINECO.
Articles can be viewed without a subscription.
References
- Álvarez-Aragón R, Rodríguez-Navarro A (2017) Nitrate-dependent shoot sodium accumulation and osmotic functions of sodium in Arabidopsis under saline conditions. Plant J 91: 208–219 [DOI] [PubMed] [Google Scholar]
- Andrés Z, Pérez-Hormaeche J, Leidi EO, Schlücking K, Steinhorst L, McLachlan DH, Schumacher K, Hetherington AM, Kudla J, Cubero B, et al. (2014) Control of vacuolar dynamics and regulation of stomatal aperture by tonoplast potassium uptake. Proc Natl Acad Sci USA 111: E1806–E1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bañuelos MA, Garciadeblas B, Cubero B, Rodríguez-Navarro A (2002) Inventory and functional characterization of the HAK potassium transporters of rice. Plant Physiol 130: 784–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barragán V, Leidi EO, Andrés Z, Rubio L, De Luca A, Fernández JA, Cubero B, Pardo JM (2012) Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. Plant Cell 24: 1127–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker D. (1990) Binary vectors which allow the exchange of plant selectable markers and reporter genes. Nucleic Acids Res 18: 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouguyon E, Brun F, Meynard D, Kubeš M, Pervent M, Leran S, Lacombe B, Krouk G, Guiderdoni E, Zažímalová E, et al. (2015) Multiple mechanisms of nitrate sensing by Arabidopsis nitrate transceptor NRT1.1. Nat Plants 1: 15015. [DOI] [PubMed] [Google Scholar]
- Byrt CS, Platten JD, Spielmeyer W, James RA, Lagudah ES, Dennis ES, Tester M, Munns R (2007) HKT1;5-like cation transporters linked to Na+ exclusion loci in wheat, Nax2 and Kna1. Plant Physiol 143: 1918–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campo S, Baldrich P, Messeguer J, Lalanne E, Coca M, San Segundo B (2014) Overexpression of a calcium-dependent protein kinase confers salt and drought tolerance in rice by preventing membrane lipid peroxidation. Plant Physiol 165: 688–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Hu Q, Luo L, Yang T, Zhang S, Hu Y, Yu L, Xu G (2015) Rice potassium transporter OsHAK1 is essential for maintaining potassium-mediated growth and functions in salt tolerance over low and high potassium concentration ranges. Plant Cell Environ 38: 2747–2765 [DOI] [PubMed] [Google Scholar]
- Chung JS, Zhu JK, Bressan RA, Hasegawa PM, Shi H (2008) Reactive oxygen species mediate Na+-induced SOS1 mRNA stability in Arabidopsis. Plant J 53: 554–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coskun D, Britto DT, Jean YK, Kabir I, Tolay I, Torun AA, Kronzucker HJ (2013) K+ efflux and retention in response to NaCl stress do not predict salt tolerance in contrasting genotypes of rice (Oryza sativa L.). PLoS One 8: e57767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotsaftis O, Plett D, Shirley N, Tester M, Hrmova M (2012) A two-staged model of Na+ exclusion in rice explained by 3D modeling of HKT transporters and alternative splicing. PLoS One 7: e39865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsler N, Zheng Y, Bohner A, Nobmann B, von Wirén N, Kunze R, Rausch C (2015) Nitrate-dependent control of shoot k homeostasis by the nitrate transporter1/peptide transporter family member NPF7.3/NRT1.5 and the stelar K+ outward rectifier SKOR in Arabidopsis. Plant Physiol 169: 2832–2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew M, Webb J, Saker L (1990) Regulation of K+ uptake and transport to the xylem in barley roots; K+ distribution determined by electron probe X-ray microanalysis of frozen-hydrated cells. J Exp Bot 41: 815–825 [Google Scholar]
- Emmerlich V, Linka N, Reinhold T, Hurth MA, Traub M, Martinoia E, Neuhaus HE (2003) The plant homolog to the human sodium/dicarboxylic cotransporter is the vacuolar malate carrier. Proc Natl Acad Sci USA 100: 11122–11126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels C, Marschner H (1992) Adaptation of potassium translocation into the shoot of maize (Zea mays) to shoot demand: Evidence for xylem loading as a regulating step. Physiol Plant 86: 263–268 [Google Scholar]
- Engels C, Marschner H (1993) Influence of the form of nitrogen supply on root uptake and translocation of cations in the xylem exudate of maize (Zea mays L). J Exp Bot 44: 1695–1701 [Google Scholar]
- Faiyue B, Al-Azzawi MJ, Flowers TJ (2010a) The role of lateral roots in bypass flow in rice (Oryza sativa L.). Plant Cell Environ 33: 702–716 [DOI] [PubMed] [Google Scholar]
- Faiyue B, Vijayalakshmi C, Nawaz S, Nagato Y, Taketa S, Ichii M, Al-Azzawi MJ, Flowers TJ (2010b) Studies on sodium bypass flow in lateral rootless mutants lrt1 and lrt2, and crown rootless mutant crl1 of rice (Oryza sativa L.). Plant Cell Environ 33: 687–701 [DOI] [PubMed] [Google Scholar]
- Feng H, Yan M, Fan X, Li B, Shen Q, Miller AJ, Xu G (2011) Spatial expression and regulation of rice high-affinity nitrate transporters by nitrogen and carbon status. J Exp Bot 62: 2319–2332 [DOI] [PubMed] [Google Scholar]
- Gao L, Liu M, Wang M, Shen Q, Guo S (2016) Enhanced salt tolerance under nitrate nutrition is associated with apoplast Na+ content in canola (Brassica. napus L.) and rice (Oryza sativa L.) plants. Plant Cell Physiol 57: 2323–2333 [DOI] [PubMed] [Google Scholar]
- Garciadeblás B, Senn ME, Bañuelos MA, Rodríguez-Navarro A (2003) Sodium transport and HKT transporters: The rice model. Plant J 34: 788–801 [DOI] [PubMed] [Google Scholar]
- Gaymard F, Pilot G, Lacombe B, Bouchez D, Bruneau D, Boucherez J, Michaux-Ferrière N, Thibaud JB, Sentenac H (1998) Identification and disruption of a plant shaker-like outward channel involved in K+ release into the xylem sap. Cell 94: 647–655 [DOI] [PubMed] [Google Scholar]
- Golldack D, Quigley F, Michalowski CB, Kamasani UR, Bohnert HJ (2003) Salinity stress-tolerant and -sensitive rice (Oryza sativa L.) regulate AKT1-type potassium channel transcripts differently. Plant Mol Biol 51: 71–81 [DOI] [PubMed] [Google Scholar]
- Gregorio G, Senadhira D, Mendoza R, Manigbas N, Roxas J, Guerta C (2002) Progress in breeding for salinity tolerance and associated abiotic stresses in rice. Field Crops Res 76: 91–101 [Google Scholar]
- Gu Z, Ma B, Jiang Y, Chen Z, Su X, Zhang H (2008) Expression analysis of the calcineurin B-like gene family in rice (Oryza sativa L.) under environmental stresses. Gene 415: 1–12 [DOI] [PubMed] [Google Scholar]
- Guo Y, Qiu QS, Quintero FJ, Pardo JM, Ohta M, Zhang C, Schumaker KS, Zhu JK (2004) Transgenic evaluation of activated mutant alleles of SOS2 reveals a critical requirement for its kinase activity and C-terminal regulatory domain for salt tolerance in Arabidopsis thaliana. Plant Cell 16: 435–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamoto S, Horie T, Hauser F, Deinlein U, Schroeder JI, Uozumi N (2015) HKT transporters mediate salt stress resistance in plants: From structure and function to the field. Curr Opin Biotechnol 32: 113–120 [DOI] [PubMed] [Google Scholar]
- Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ (2000) Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol 51: 463–499 [DOI] [PubMed] [Google Scholar]
- Ho CH, Lin SH, Hu HC, Tsay YF (2009) CHL1 functions as a nitrate sensor in plants. Cell 138: 1184–1194 [DOI] [PubMed] [Google Scholar]
- Hoang TML, Tran TN, Nguyen TKT, Williams B, Wurm P, Bellairs S, Mundree S (2016) Improvement of salinity stress tolerance in rice: Challenges and opportunities. Agronomy (Basel) 6: 54 [Google Scholar]
- Horie T, Costa A, Kim TH, Han MJ, Horie R, Leung HY, Miyao A, Hirochika H, An G, Schroeder JI (2007) Rice OsHKT2;1 transporter mediates large Na+ influx component into K+-starved roots for growth. EMBO J 26: 3003–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T, Hauser F, Schroeder JI (2009) HKT transporter-mediated salinity resistance mechanisms in Arabidopsis and monocot crop plants. Trends Plant Sci 14: 660–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T, Sugawara M, Okada T, Taira K, Kaothien-Nakayama P, Katsuhara M, Shinmyo A, Nakayama H (2011) Rice sodium-insensitive potassium transporter, OsHAK5, confers increased salt tolerance in tobacco BY2 cells. J Biosci Bioeng 111: 346–356 [DOI] [PubMed] [Google Scholar]
- Huang S, Spielmeyer W, Lagudah ES, James RA, Platten JD, Dennis ES, Munns R (2006) A sodium transporter (HKT7) is a candidate for Nax1, a gene for salt tolerance in durum wheat. Plant Physiol 142: 1718–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa S, Hayashi S, Abe T, Igura M, Kuramata M, Tanikawa H, Iino M, Saito T, Ono Y, Ishikawa T, et al. (2017) Low-cesium rice: Mutation in OsSOS2 reduces radiocesium in rice grains. Sci Rep 7: 2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James RA, Davenport RJ, Munns R (2006) Physiological characterization of two genes for Na+ exclusion in durum wheat, Nax1 and Nax2. Plant Physiol 142: 1537–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspers P, Brosché M, Overmyer K, Kangasjärvi J (2010) The transcription factor interacting protein RCD1 contains a novel conserved domain. Plant Signal Behav 5: 78–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Pardo JM, Batelli G, Van Oosten MJ, Bressan RA, Li X (2013) The salt overly sensitive (SOS) pathway: Established and emerging roles. Mol Plant 6: 275–286 [DOI] [PubMed] [Google Scholar]
- Kaburagi E, Yamada M, Fujiyama H (2015) Sodium, but not potassium, enhances root to leaf nitrate translocation in Swiss chard (Beta vulgaris var. cicla L.). Environ Exp Bot 112: 27–32 [Google Scholar]
- Kader MA, Lindberg S (2005) Uptake of sodium in protoplasts of salt-sensitive and salt-tolerant cultivars of rice, Oryza sativa L. determined by the fluorescent dye SBFI. J Exp Bot 56: 3149–3158 [DOI] [PubMed] [Google Scholar]
- Kanwar P, Sanyal SK, Tokas I, Yadav AK, Pandey A, Kapoor S, Pandey GK (2014) Comprehensive structural, interaction and expression analysis of CBL and CIPK complement during abiotic stresses and development in rice. Cell Calcium 56: 81–95 [DOI] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Zhu J, Kim K, Agarwal M, Fu X, Huang A, Zhu JK (2006) The plasma membrane Na+/H+ antiporter SOS1 interacts with RCD1 and functions in oxidative stress tolerance in Arabidopsis. Proc Natl Acad Sci USA 103: 18816–18821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B-G, Waadt R, Cheong YH, Pandey GK, Dominguez-Solis JR Jr., Schültke S, Lee SC, Kudla J, Luan S (2007) The calcium sensor CBL10 mediates salt tolerance by regulating ion homeostasis in Arabidopsis. Plant J 52: 473–484 [DOI] [PubMed] [Google Scholar]
- Kolukisaoglu U, Weinl S, Blazevic D, Batistic O, Kudla J (2004) Calcium sensors and their interacting protein kinases: Genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol 134: 43–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy P, Ranathunge K, Franke R, Prakash HS, Schreiber L, Mathew MK (2009) The role of root apoplastic transport barriers in salt tolerance of rice (Oryza sativa L.). Planta 230: 119–134 [DOI] [PubMed] [Google Scholar]
- Kronzucker HJ, Britto DT (2011) Sodium transport in plants: A critical review. New Phytol 189: 54–81 [DOI] [PubMed] [Google Scholar]
- Lee KS, Choi WY, Ko JC, Kim TS, Gregorio GB (2003) Salinity tolerance of japonica and indica rice (Oryza sativa L.) at the seedling stage. Planta 216: 1043–1046 [DOI] [PubMed] [Google Scholar]
- Leidi EO, Barragán V, Rubio L, El-Hamdaoui A, Ruiz MT, Cubero B, Fernández JA, Bressan RA, Hasegawa PM, Quintero FJ, et al. (2010) The AtNHX1 exchanger mediates potassium compartmentation in vacuoles of transgenic tomato. Plant J 61: 495–506 [DOI] [PubMed] [Google Scholar]
- Li H, Yu M, Du XQ, Wang ZF, Wu WH, Quintero FJ, Jin XH, Li HD, Wang Y (2017) NRT1.5/NPF7.3 functions as a proton-coupled H+/K+ antiporter for K+ loading into the xylem in Arabidopsis. Plant Cell 29: 2016–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HX, Zhu MZ, Yano M, Gao JP, Liang ZW, Su WA, Hu XH, Ren ZH, Chao DY (2004) QTLs for Na+ and K+ uptake of the shoots and roots controlling rice salt tolerance. Theor Appl Genet 108: 253–260 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Martínez-Atienza J, Jiang X, Garciadeblas B, Mendoza I, Zhu JK, Pardo JM, Quintero FJ (2007) Conservation of the salt overly sensitive pathway in rice. Plant Physiol 143: 1001–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto N, Steudle E, Hirasawa T, Lafitte R (2001) Hydraulic conductivity of rice roots. J Exp Bot 52: 1835–1846 [DOI] [PubMed] [Google Scholar]
- Moore B, Zhou L, Rolland F, Hall Q, Cheng WH, Liu YX, Hwang I, Jones T, Sheen J (2003) Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300: 332–336 [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59: 651–681 [DOI] [PubMed] [Google Scholar]
- Munns R, James RA, Xu B, Athman A, Conn SJ, Jordans C, Byrt CS, Hare RA, Tyerman SD, Tester M, et al. (2012) Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nat Biotechnol 30: 360–364 [DOI] [PubMed] [Google Scholar]
- Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8: 4321–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie L, Feng J, Fan P, Chen X, Guo J, Lv S, Bao H, Jia W, Tai F, Jiang P, et al. (2015) Comparative proteomics of root plasma membrane proteins reveals the involvement of calcium signalling in NaCl-facilitated nitrate uptake in Salicornia europaea. J Exp Bot 66: 4497–4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieves-Cordones M, Martínez V, Benito B, Rubio F (2016) Comparison between Arabidopsis and rice for main pathways of K+ and Na+ uptake by roots. Front Plant Sci 7: 992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Núñez-Ramírez R, Sánchez-Barrena MJ, Villalta I, Vega JF, Pardo JM, Quintero FJ, Martinez-Salazar J, Albert A (2012) Structural insights on the plant salt-overly-sensitive 1 (SOS1) Na+/H+ antiporter. J Mol Biol 424: 283–294 [DOI] [PubMed] [Google Scholar]
- Oh D-H, Leidi E, Zhang Q, Hwang S-M, Li Y, Quintero FJ, Jiang X, D’Urzo MP, Lee SY, Zhao Y, et al. (2009) Loss of halophytism by interference with SOS1 expression. Plant Physiol 151: 210–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olías R, Eljakaoui Z, Li J, De Morales PA, Marín-Manzano MC, Pardo JM, Belver A (2009) The plasma membrane Na+/H+ antiporter SOS1 is essential for salt tolerance in tomato and affects the partitioning of Na+ between plant organs. Plant Cell Environ 32: 904–916 [DOI] [PubMed] [Google Scholar]
- Pardo JM. (2010) Biotechnology of water and salinity stress tolerance. Curr Opin Biotechnol 21: 185–196 [DOI] [PubMed] [Google Scholar]
- Piao HL, Xuan YH, Park SH, Je BI, Park SJ, Park SH, Kim CM, Huang J, Wang GK, Kim MJ, et al. (2010) OsCIPK31, a CBL-interacting protein kinase is involved in germination and seedling growth under abiotic stress conditions in rice plants. Mol Cells 30: 19–27 [DOI] [PubMed] [Google Scholar]
- Piffanelli P, Droc G, Mieulet D, Lanau N, Bès M, Bourgeois E, Rouvière C, Gavory F, Cruaud C, Ghesquière A, Guiderdoni E (2007) Large-scale characterization of Tos17 insertion sites in a rice T-DNA mutant library. Plant Mol Biol 65: 587–601 [DOI] [PubMed] [Google Scholar]
- Qi Z, Spalding EP (2004) Protection of plasma membrane K+ transport by the salt overly sensitive1 Na+-H+ antiporter during salinity stress. Plant Physiol 136: 2548–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu QS, Guo Y, Dietrich MA, Schumaker KS, Zhu JK (2002) Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci USA 99: 8436–8441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu QS, Barkla BJ, Vera-Estrella R, Zhu JK, Schumaker KS (2003) Na+/H+ exchange activity in the plasma membrane of Arabidopsis. Plant Physiol 132: 1041–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan R, Lin H, Mendoza I, Zhang Y, Cao W, Yang Y, Shang M, Chen S, Pardo JM, Guo Y (2007) SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell 19: 1415–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero FJ, Ohta M, Shi H, Zhu JK, Pardo JM (2002) Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc Natl Acad Sci USA 99: 9061–9066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero FJ, Martinez-Atienza J, Villalta I, Jiang X, Kim WY, Ali Z, Fujii H, Mendoza I, Yun DJ, Zhu JK, et al. (2011) Activation of the plasma membrane Na/H antiporter Salt-Overly-Sensitive 1 (SOS1) by phosphorylation of an auto-inhibitory C-terminal domain. Proc Natl Acad Sci USA 108: 2611–2616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbani MA, Maruyama K, Abe H, Khan MA, Katsura K, Ito Y, Yoshiwara K, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol 133: 1755–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragel P, Raddatz N, Leidi EO, Quintero FJ, Pardo JM (2019) Regulation of K+ nutrition in plants. Front Plant Sci 10: 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Z-H, Gao J-P, Li L-G, Cai X-L, Huang W, Chao D-Y, Zhu M-Z, Wang Z-Y, Luan S, Lin H-X (2005) A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet 37: 1141–1146 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Navarro A, Ramos J (1984) Dual system for potassium transport in Saccharomyces cerevisiae. J Bacteriol 159: 940–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rus A, Yokoi S, Sharkhuu A, Reddy M, Lee BH, Matsumoto TK, Koiwa H, Zhu JK, Bressan RA, Hasegawa PM (2001) AtHKT1 is a salt tolerance determinant that controls Na+ entry into plant roots. Proc Natl Acad Sci USA 98: 14150–14155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rus A, Lee BH, Muñoz-Mayor A, Sharkhuu A, Miura K, Zhu JK, Bressan RA, Hasegawa PM (2004) AtHKT1 facilitates Na+ homeostasis and K+ nutrition in planta. Plant Physiol 136: 2500–2511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallaud C, Gay C, Larmande P, Bès M, Piffanelli P, Piégu B, Droc G, Regad F, Bourgeois E, Meynard D, et al. (2004) High throughput T-DNA insertion mutagenesis in rice: A first step towards in silico reverse genetics. Plant J 39: 450–464 [DOI] [PubMed] [Google Scholar]
- Shabala L, Cuin TA, Newman IA, Shabala S (2005) Salinity-induced ion flux patterns from the excised roots of Arabidopsis sos mutants. Planta 222: 1041–1050 [DOI] [PubMed] [Google Scholar]
- Shi H, Quintero FJ, Pardo JM, Zhu JK (2002) The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 14: 465–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Lee BH, Wu SJ, Zhu JK (2003) Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat Biotechnol 21: 81–85 [DOI] [PubMed] [Google Scholar]
- Suzuki K, Yamaji N, Costa A, Okuma E, Kobayashi NI, Kashiwagi T, Katsuhara M, Wang C, Tanoi K, Murata Y, et al. (2016) OsHKT1;4-mediated Na+ transport in stems contributes to Na+ exclusion from leaf blades of rice at the reproductive growth stage upon salt stress. BMC Plant Biol 16: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Thomson M, de Ocampo M, Egdane J, Rahman MA, Sajise A, Adorada D, Tumimbang-Raiz E, Blumwald E, Seraj Z, Singh R, Gregorio G, et al. (2010) Characterizing the saltol quantitative trait locus for salinity tolerance in rice. Rice (N Y) 3: 148–160 [Google Scholar]
- Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Jing W, Xiao L, Jin Y, Shen L, Zhang W (2015) The rice high-affinity potassium transporter1;1 is involved in salt tolerance and regulated by an MYB-type transcription factor. Plant Physiol 168: 1076–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WS, Zhao XQ, Li M, Huang LY, Xu JL, Zhang F, Cui YR, Fu BY, Li ZK (2016) Complex molecular mechanisms underlying seedling salt tolerance in rice revealed by comparative transcriptome and metabolomic profiling. J Exp Bot 67: 405–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SJ, Ding L, Zhu JK (1996) SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell 8: 617–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Huang Y, Xiong L (2007) Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant Physiol 144: 1416–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M, Fan X, Feng H, Miller AJ, Shen Q, Xu G (2011) Rice OsNAR2.1 interacts with OsNRT2.1, OsNRT2.2 and OsNRT2.3a nitrate transporters to provide uptake over high and low concentration ranges. Plant Cell Environ 34: 1360–1372 [DOI] [PubMed] [Google Scholar]
- Yang T, Zhang S, Hu Y, Wu F, Hu Q, Chen G, Cai J, Wu T, Moran N, Yu L, et al. (2014) The role of a potassium transporter OsHAK5 in potassium acquisition and transport from roots to shoots in rice at low potassium supply levels. Plant Physiol 166: 945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, Horie T, Xue S, Leung H-Y, Katsuhara M, Brodsky DE, Wu Y, Schroeder JI (2010) Differential sodium and potassium transport selectivities of the rice OsHKT2;1 and OsHKT2;2 transporters in plant cells. Plant Physiol 152: 341–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo A, Yeo M, Flowers T (1987) The contribution of an apoplastic pathway to sodium uptake by rice roots in saline conditions. J Exp Bot 38: 1141–1153 [Google Scholar]
- Yeo AR, Yeo ME, Flowers SA, Flowers TJ (1990) Screening of rice (Oryza sativa L.) genotypes for physiological characters contributing to salinity resistance, and their relationship to overall performance. Theor Appl Genet 79: 377–384 [DOI] [PubMed] [Google Scholar]
- You J, Zong W, Li X, Ning J, Hu H, Li X, Xiao J, Xiong L (2013) The SNAC1-targeted gene OsSRO1c modulates stomatal closure and oxidative stress tolerance by regulating hydrogen peroxide in rice. J Exp Bot 64: 569–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Wu R (2008) Regeneration of transgenic rice plants using high salt for selection without the need for antibiotics or herbicides. Plant Sci 174: 519–523 [Google Scholar]
- Zhu M, Shabala L, Cuin TA, Huang X, Zhou M, Munns R, Shabala S (2016) Nax loci affect SOS1-like Na+/H+ exchanger expression and activity in wheat. J Exp Bot 67:835–44 [DOI] [PMC free article] [PubMed] [Google Scholar]