Abstract
Regulatory variants are often context-specific, modulating gene expression in a subset of possible cellular states. Although these genetic effects can play important roles in disease, the molecular mechanisms underlying context-specificity are poorly understood. Here, we identify shared quantitative trait loci (QTLs) for chromatin accessibility and gene expression in human macrophages exposed to IFNγ, Salmonella and IFNγ + Salmonella. We observe that ~60% of stimulus-specific eQTLs with a detectable effect on chromatin alter chromatin accessibility in naive cells, suggesting they perturb enhancer priming. We show that such variants probably influence the binding of cell type specific transcription factors (TFs), such as PU.1, which can then indirectly alter the binding of stimulus-specific TFs, such as NF-κB or STAT2. Thus, although chromatin accessibility assays are powerful for fine mapping causal regulatory variants, detecting their downstream impact on gene expression will be challenging, requiring profiling of large numbers of stimulated cellular states and timepoints.
Introduction
Genetic differences between individuals can profoundly alter how their immune cells respond to environmental stimuli1. At the molecular level, these differences manifest as expression quantitative trait loci (eQTLs) that alter the magnitude of gene expression change after stimulation (response eQTLs)2–7. Although response eQTLs have been implicated in modulating risk for complex immune-mediated disorders8,9, the molecular mechanisms that give rise to these context specific effects are poorly understood. The majority of eQTLs also alter chromatin accessibility, presumably reflecting disruption of transcription factor (TF) binding10. Because cellular response to external stimuli is regulated by stimulus-specific transcription factors (TFs), response eQTLs might directly disrupt their binding (Fig. 1a). In support of this model, a number of studies have observed that response eQTLs are enriched at the binding sites of stimulation-specific TFs such as NF-κB and STAT25–7. However, a single stimulus or a developmental cue can upregulate alternate sets of genes in different cell types, even when the activated signalling pathways and TFs remain the same11. To explain these observations, multiple studies have proposed a hierarchical enhancer activation model11–14, under which cell type specific TFs bind to a subset of enhancers without a direct effect on target gene expression. This enhancer ‘priming’ can facilitate their subsequent activation by signal specific TFs, producing a cell type specific response (Fig. 1b). Thus, genetic variants could modulate stimulus specific effects on gene expression indirectly, by altering the binding of a cell type specific TF, for example PU.1 in macrophages, that regulate chromatin accessibility (Fig. 1b). However, the genome-wide prevalence of enhancer priming is currently unclear because directed genome editing studies have been limited to handful of loci15,16. Previous studies have highlighted that profiling chromatin accessibility is a good proxy for measuring TF binding without necessarily identifying the underlying TFs involved10,17,18. Furthermore, TF binding can be predicted with high accuracy from chromatin accessibility data19,20. Thus, shared genetic associations at chromatin and gene expression level provide a powerful alternative to probe the relationships between enhancer accessibility, TF binding and gene transcription.
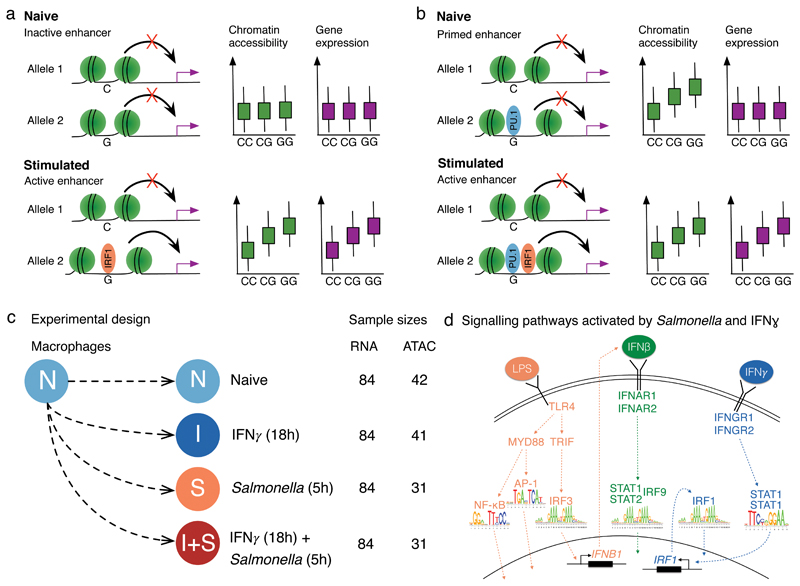
Figure 1. Regulation of gene expression in macrophage immune response.
(a) Genetic variant has a direct effect on the binding of a stimulation-specific TF (IRF1) and target gene activation. (b) Genetic variant in a primed enhancer disrupts the binding of a cell type specific TF (e.g. PU.1) that indirectly influences stimulation-specific TF (IRF1) binding via modulation of chromatin accessibility. (c) Overview of the experimental design. (d) TLR4 recognises lipopolysaccharide (LPS) on Salmonella cell wall and activates NF-κB, AP-1 and IRF3 transcription factors (TFs)53. IRF3 stimulates IFNβ production that culminates with the activation of STAT1-STAT2-IRF9 complex. IFNγ binds to IFNγ receptor and activates STAT1 and IRF1 TFs54.
Results
Genetics of gene expression and chromatin accessibility
We focussed on enhancer priming in the context of human macrophage immune response. To ensure sufficient numbers of cells, we differentiated macrophages from a panel of 123 human induced pluripotent cell lines (iPSCs) obtained from the HipSci project21,22. We profiled gene expression (RNA-seq) and chromatin accessibility (ATAC-seq23) in a subset of 86 successfully differentiated lines (Supplementary Fig. 1, Supplementary Table 1) in four experimental conditions: naive (N), 18 hours IFNγ stimulation (I), 5 hours Salmonella enterica serovar Typhimurium (Salmonella) infection (S), and IFNγ stimulation followed by Salmonella infection (I+S) (Fig. 1c). We chose these stimuli because they activate distinct, well characterised signalling pathways (Fig. 1d, Supplementary Fig. 2) and pre-stimulating macrophages with IFNγ prior to bacterial infection is known to lead to enhanced microbial killing and stronger activation of the inflammatory response24,25.
We identified common genetic variants that were associated with either gene expression (eQTLs) or chromatin accessibility (caQTLs). Using an allele-specific method implemented in RASQUAL26, we detected at least one QTL for up to 3,431 genes and 20,788 chromatin regions (caQTL regions) in each condition (10% FDR) (Supplementary Fig. 3, Supplementary Fig. 4), 50-75% of which were shared between conditions (Supplementary Fig. 3). Consistent with a previous report10, we found that caQTLs were associated with allele-specific TF binding (Supplementary Fig. 5). Furthermore, only 8% of the caQTL regions overlapped annotated promoters and 42% overlapped regions marked with H3K27ac histone modifications25 in macrophages (Supplementary Note). Next, using a statistical interaction test followed by filtering on effect size, we identified 387 response eQTLs and 2247 response caQTLs with a small or undetectable effect (fold change < 1.5) in the naive state that increased at least 1.5 fold after stimulation (see Methods). The use of an interaction test meant that our analysis should be robust to false positive response QTLs that could arise due to, for example, weak, undetected QTLs in the naive cell state. We verified this by down-sampling from a larger Fairfax et al3 monocyte response eQTL dataset (Supplementary Tables 2 and 3, Supplementary Fig. 6). These genetic effects displayed a variety of activity patterns (Fig. 2a, Supplementary Fig. 7a). Strikingly, 18% of the response eQTLs appeared only after the cells were exposed to both stimuli (cluster 1), exceeding the number that appeared after IFNγ stimulation alone (clusters 5 and 6). Response caQTL regions harboured closed chromatin in the naive cells (median transcripts per million (TPM) = 0.49) and became 3.8-fold more accessible only after the relevant stimulus (Supplementary Fig. 7b). Furthermore, response caQTLs were enriched for disrupting stimulus-specific TF motifs (Supplementary Fig. 7c), suggesting that they are largely driven by TFs that bind to DNA only after stimulation.
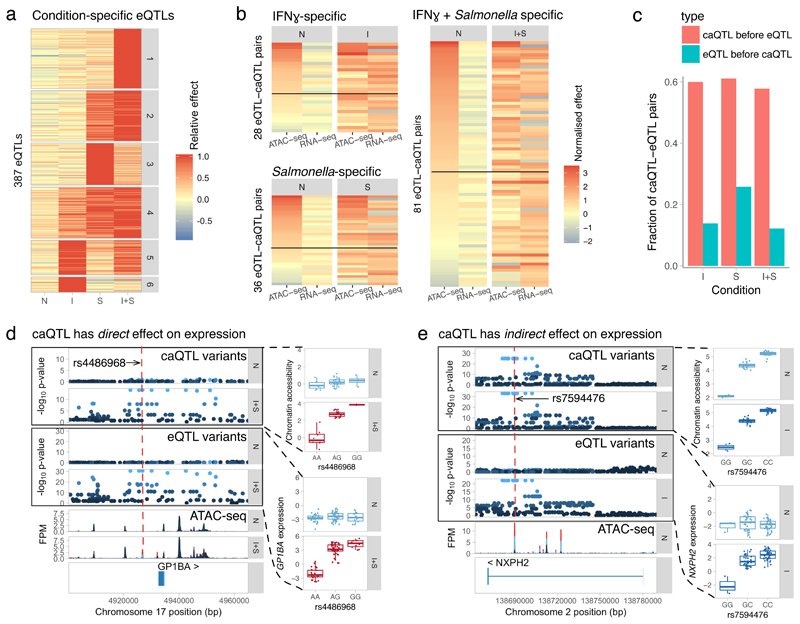
Figure 2. Quantifying the extent of enhancer priming in macrophage immune response.
(a) Clustering of response eQTLs by effect size. Response eQTLs appear after IFNγ stimulation (clusters 5 and 6), Salmonella infection (clusters 2-4) or only when both of the stimuli are present (cluster 1). Relative effect was calculated by dividing the log2 fold change values in each condition by the maximal fold change observed across conditions. (b) Quantile normalised log2 fold changes of eQTL-caQTL pairs in naive and stimulated conditions. The pairs are grouped by the condition in which the eQTL had the largest effect (I, S or I+S). The heat maps are sorted by caQTL effect size in the naive condition (first column). The solid lines represent the 1.5-fold threshold above which the caQTLs are considered to be present in the naive condition. (c) Comparison of our estimated rate of enhancer priming (caQTL precedes response eQTL) to a negative control (eQTL precedes a response caQTL). (d) Association between rs4486968 variant, chromatin accessibility and and gene expression (n = 84 independent donors) in the GP1BA locus. (e) Association between rs7594476 variant, chromatin accessibility and gene expression (n = 84 independent donors) in the NXPH2 locus. FPM, fragments per million. The - log10 p-values on panels d and e were calculated using RASQUAL. The caQTL analysis used n = 42 (N), n = 41 (I) and n = 31 (I+S) independent donors. Box plots show the median (center line) and the 25th and 75th percentiles (box edges), with whiskers extending to 1.5 times the interquartile range.
Enhancer priming in macrophage immune response
To quantify the extent of enhancer priming in macrophage immune response, we next focussed on how response eQTLs manifest on the chromatin level. We grouped response eQTLs (Fig. 2a) by the condition in which they had the largest effect size (I, S or I+S). We then used linkage disequilibrium (LD) (R2 > 0.8) between the lead variants to identify 145 caQTL-eQTL pairs that were likely to be driven by the same causal variant (Online Methods). For example, we identified a QTL upstream of GP1BA that had no effect in naive cells, but became simultaneously associated with chromatin accessibility and gene expression after IFNγ + Salmonella stimulation (Fig. 2d). The lead caQTL variant (rs4486968) was predicted to disrupt NF-κB binding motif (Supplementary Fig. 8), illustrating how a caQTL can directly affect stimulus-specific TF binding and gene expression. In contrast, a genetic variant in an intron of NXPH2 modulated the accessibility of a regulatory element both in naive and stimulated cells, but only became associated with gene expression after IFNγ stimulation (Fig. 2e). Genome-wide, we found that for approximately half of the response eQTLs with a linked caQTL, the caQTL was present in naive cells prior to stimulation (caQTL fold change > 1.5), suggesting that many response eQTLs regulate gene expression indirectly by first modulating the extent of enhancer priming in naive cells (Fig. 2b). One potential issue with our analysis is that using LD to identify eQTL-caQTL pairs will sometimes lead to false positives where two independent causal variants, one altering gene expression, the other chromatin accessibility, that are in strong LD with one another are mistaken for a single shared causal variant. We therefore performed a reverse analysis where we asked how often response caQTLs were linked to eQTLs that were present in the naive state, reasoning that these are likely to be false positives. Using the same fold change threshold as above, we estimated our false positive rate to be 15% (Fig. 2c). Consistent with this estimate, we found that 117/145 caQTLs-eQTL pairs (81%) showed concordant direction of effect in the stimulated cells (Fig. 2b). Furthermore, the difference in the number of eQTLs and caQTLs did not seem to bias our results (Supplementary Fig. 9). With a more stringent fold change threshold of two the false positive rate decreased further to 4% (Supplementary Fig. 9) while concordance in effect size direction increased to 90%. Finally, we performed the same analysis on a set of 26 caQTL-eQTL pairs that colocalised with each other and were able to confirm that most response eQTLs manifested as caQTLs in unstimulated cells (Supplementary Fig. 10).
Multiple TFs such as PU.1, AP-1 and CEBPα/β have been implicated in regulating enhancer priming in macrophages11–13. We speculated that response eQTLs that alter enhancer priming should be enriched for disrupting the motifs of those TFs. To test this, we focussed on the 145 eQTL-caQTL pairs (137 unique caQTLs) identified above (Fig. 2b). We found that 9/78 caQTLs present in the naive cells disrupted PU.1 motifs compared to none of the 59 caQTLs that appeared together with the response eQTL (Fisher’s exact test, p = 0.01). For example, the rs7594476 variant in the NXPH2 enhancer disrupted PU.1 binding in a direction consistent with the caQTL effect (Fig. 3a). Although the PU.1 enrichment is only nominally significant and does not survive multiple testing correction for other TFs we tested, our observation is consistent with the established role of PU.1 in defining the accessible chromatin landscape in macrophages that subsequently directs stimulation-specific TF binding11–13.
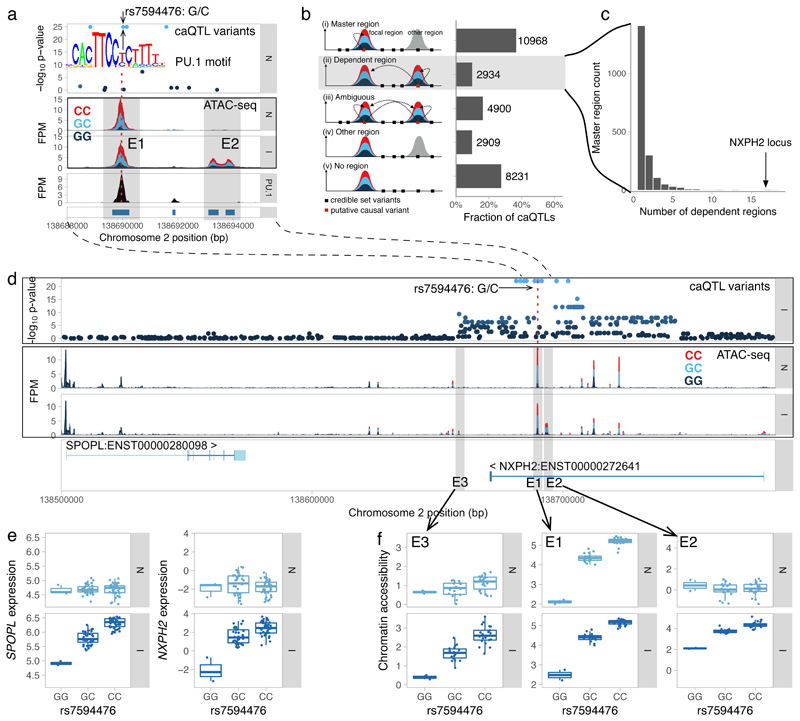
Figure 3. Identifying caQTLs that regulate chromatin accessibility at multiple independent regions.
(a) Fine mapping the putative causal variant at the NXPH2 locus. The master caQTL region (E1) overlaps a PU.1 ChIP-seq peak. Only one of the two variants (rs7594476) within the E1 region is predicted to disrupt the PU.1 motif (M6119_1.02). No associated variants overlap the IFNγ-specific dependent region E2. (b) Classifying caQTLs into master regions (i), dependent regions (ii) and ambiguous cases where the credible set overlaps either multiple regulated regions (iii) or does not overlap any regulated regions (iv-v). (c) Histogram of the number of associated dependent regions for each master region. (d) Multiple open chromatin regions regulated by a single caQTL at the NXPH2 locus. Master caQTL region (E1) and two IFNγ-specific dependent regions (E2 and E3) are highlighted by grey shadows. FPM, fragments per million. (e) Association between rs7594476 variant and expression of NXPH2 and SPOPL genes before and after IFNγ stimulation (n = 84 independent donors). (f) Association between rs7594476 variant, master caQTL region (E1) and two dependent regions (E2 and E3) before and after IFNγ stimulation (n = 42 and n = 41 independent donors, respectively). The -log10 p-values on panels a and d were calculated using RASQUAL. Box plots show the median (center line) and the 25th and 75th percentiles (box edges), with whiskers extending to 1.5 times the interquartile range.
Genetic effects on multiple chromatin regions
Recent evidence suggests that single genetic variants can modulate the activity of multiple regulatory elements within topologically associated domains (TADs)26–29. One plausible mechanism for these broad associations is that a single causal variant may directly regulate the accessibility of a “master” region, which subsequently influences neighbouring “dependent” regions26. We used caQTL summary statistics to heuristically identify likely master and dependent regions, assuming that the causal variant should reside within the master region itself, and this affects accessibility in dependent regions (Fig. 3b) (see Methods). We found a striking example of such a relationship at the NXPH2 locus, where a putative causal variant in the master region was also associated with the accessibility of neighbouring dependent region after IFNγ stimulation (Fig. 3b). Using this approach, we identified 2,934 dependent regions that belonged to 1,921 unique master regions (Fig. 3b). Master-dependent region pairs were enriched in TADs (odds ratio 1.5, Fisher’s exact test p = 1.26x10-6) (Supplementary Note) and in 95% of the cases, the caQTL had the same direction of effect on master and dependent regions. While 77% of the master regions had a single dependent region only a few kb away (Supplementary Fig. 11), we found many loci where master peaks were associated with multiple regions of open chromatin (Fig. 3c). One of the largest effect was observed in the NXPH2 locus introduced above, where we detected 18 dependent regions spanning 100 kilobases of DNA (Fig. 3c), six of which appeared only after IFNγ stimulation (Fig. 3d,f). Notably, the appearance of condition-specific dependent regions correlated with the caQTL becoming a response eQTL for both NXPH2 and SPOPL (Fig. 3e), suggesting that some of them might be required for gene activation. Using a linear model followed by strict filtering (see Online Methods), we found a total of 64 condition-specific dependent regions genome-wide, two of which are highlighted in Supplementary Fig. 12.
Colocalisation with disease associations
Because they can be engineered with high efficiency, iPSC-derived cells are promising cellular models of disease. Similarly to previous studies7, we found that macrophage eQTLs and caQTLs were enriched for GWAS hits of multiple immune-mediated disorders (Supplementary Fig. 13, Supplementary Table 4). However, observing a genome-wide enrichment has only limited utility and detailed follow up of a locus is only justified when there is evidence for a shared causal mechanism between GWAS and eQTL associations. Thus, we used coloc30 to identify cases where the gene expression and trait association signals were consistent with a model of a single, shared causal variant. We identified 22 eQTLs (Supplementary Table 5) that showed evidence of colocalisation (PP3 + PP4 > 0.8, PP4/PP3 > 9) with at least one disease (Online Methods). Consistent with our enrichment analysis, we found the largest number of overlaps with inflammatory bowel disease31 (IBD) and rheumatoid arthritis32 (RA) (Fig. 4a). Interestingly, only 10/22 of the colocalised eQTLs were detected in the naive cells and each additional stimulated state increased the number of overlaps by approximately 30% (Fig. 4b). However, coloc does not directly test condition-specificity of colocalisations and is thus subject to false positives due to limited power. To estimate the severity of this issue, we repeated the analysis on the Fairfax dataset3 and found that 2/3 of the additional overlaps were not detected in unstimulated cells even if the sample size was increased 5-fold (Supplementary Fig. 14). For example, we found an IFNγ + Salmonella specific response eQTL for TRAF1 that colocalised with an RA GWAS hit (Supplementary Fig. 15). Although the same colocalisation was previously reported in whole blood33, our data highlights the environmental condition in which the association is active. Furthermore, the same associations is specific to 2 hours LPS stimulation in the Fairfax dataset and not detected in unstimulated monocytes even with 414 samples (Supplementary Fig. 16).
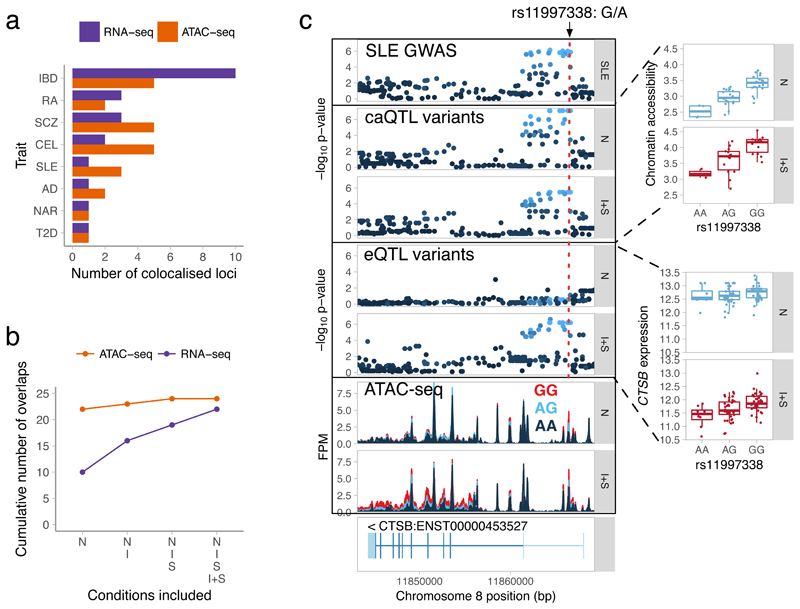
Figure 4. Identifying eQTLs and caQTLs that colocalise with complex disease risk loci.
(a) Total number of colocalised GWAS hits identified for each trait across the four conditions. (b) Cumulative number of colocalised GWAS hits identified by starting with overlaps in the naive condition and sequentially adding IFNγ, Salmonella and IFNγ + Salmonella conditions. (c) Colocalisation between an SLE GWAS hit (rs11997338), chromatin accessibility and CTSB gene expression (n = 84 independent donors) before and after IFNγ + Salmonella stimulation. The caQTL results are based on n = 42 (naive) and n = 31 (IFNγ + Salmonella) independent donors. FPM, fragments per million. Disease acronyms: SLE, systemic lupus erythematosus; IBD, inflammatory bowel disease; RA, rheumatoid arthritis; SLE, systemic lupus erythematosus; AD, Alzheimer’s disease; SCZ, schizophrenia; T2D, type 2 diabetes; NAR, narcolepsy; CEL, celiac disease. The -log10 p-values on panels c were calculated using FastQTL55. Box plots show the median (center line) and the 25th and 75th percentiles (box edges), with whiskers extending to 1.5 times the interquartile range.
Our analysis of enhancer priming suggested that many disease associations might manifest at the level of chromatin without an apparent effect on expression. To explore this further, we focussed on colocalisation between caQTLs and GWAS hits. We detected 24 caQTLs that colocalised with a GWAS hit (Supplementary Table 6), but only two of these also colocalised with an eQTL (PTK2B eQTL with Alzheimer’s disease34 (Supplementary Fig. S17) and WFS1 eQTL with type 2 diabetes35). Since genes often have multiple independent eQTLs36, we reasoned that some caQTLs might be secondary eQTLs for their target genes. To capture these secondary effects, we first identified four additional genes that were associated with a caQTL lead variant at FDR < 10%, even though the caQTL and eQTL lead variants were not in strong LD (i.e. R2 < 0.8). We repeated the colocalisation analysis on these loci and identified two additional overlaps (Supplementary Table 5), including a secondary eQTL for CTSB that colocalised with a GWAS hit for systemic lupus erythematosus37 (SLE) (Fig. 4C). Interestingly, although the CTSB eQTL appeared after IFNγ + Salmonella stimulation, the caQTL was already present in naive cells. Although some caQTL colocalisation with eQTLs might remain undetected due to lack of power, the CTSB example suggests that a fraction of disease-associated caQTLs might correspond to primed enhancers that regulate gene expression in some other yet unknown conditions. Although the majority (22/24) of caQTL overlaps with disease were detected in the naive cells (Fig. 4C), this is confounded with a smaller ATAC-seq sample size in Salmonella and IFNγ + Salmonella conditions that limited our power to detect colocalisations (Supplementary Fig. 14a).
Discussion
Multiple reports have highlighted that, although disease loci from association studies are strongly enriched in gene regulatory elements38,39, a relatively small fraction are explained by known eQTLs, even those identified in trait-relevant tissues33,40,41. Even recent systematic analysis by the GTEx Consortium over 44 tissues from 449 individuals found that only 52% of the trait-associated variants colocalised with an eQTL in one or more tissues42. Our results suggest that one reason for this apparent contradiction could be that many disease risk variants affect chromatin structure in a broad range of cellular states, but their impact on expression is highly context-specific. This interpretation is further supported by studies of 3D chromatin structure linking GWAS loci to putative target genes but with no observable effect on gene expression43, in particular because enhancer-promoter interactions are known to precede transcription44,45. We believe our result has important implications for future studies of human disease. First, it is likely that a large range of cellular states will need to be profiled in order to capture the effects of disease-associated variants on expression. Second, overlap of disease variants with open chromatin, while likely to be informative regarding the identity of the causal variant, may be less useful predictors of the disease relevant cell state.
One limitation of our study is that we were underpowered to detect caQTLs. Although previous studies have estimated that more than 55% of eQTLs are also associated with changes in chromatin accessibility10, we were only able to detect a linked caQTL for 145/387 (37%) of our response eQTLs, limiting the the number of enhancer priming events that we could detect. Another possibility is that a subset of the response eQTLs are mediated by chromatin-independent mechanisms such as stimulation-specific regulation of mRNA stability, which is estimated to be responsible for ~10% of the eQTLs46. Finally, we found that current colocalisation methods are not well suited to assess the condition-specificity of eQTL-disease overlaps and can lead to a large number (~30%) of false positives.
Although our study suggests that many human disease associated variants impact enhancer priming, the functional relevance of this is currently not well understood. First, enhancer priming may facilitate cell type specific response to ubiquitous signals11,47,48. Although specificity can also be achieved by cooperative binding to newly established enhancers49, TFs differ in their intrinsic ability to bind to closed chromatin50. Thus, enhancer priming might be a preferred mechanism of cooperation between ‘pioneer’ TFs that can independently open up chromatin (e.g. PU.1 in macrophages) and ‘settlers’ (e.g. NF-κB) that predominantly bind to accessible regions20. Alternatively, enhancer priming might facilitate rapid response to external stimuli. In support of this model, promoters of immediate early response genes are already accessible in naive cells51 and TF binding to primed enhancers peaks minutes after stimulation while the activation of de novo enhancers can take several hours49. Thus, response eQTLs that appear rapidly after stimulation might be enriched for primed enhancers relative to those that appear later. Finally, enhancer priming might not be limited to single regulatory elements. Our results (Fig. 3d) together with previous reports16,52 suggest that some regulatory elements can act as ‘seed’ enhancers that allow other neighbouring enhancers to become active after stimulation and lead to upregulation of gene expression. Although we have identified a small number of such examples, future caQTL mapping studies in multiple cell types and conditions have a potential to systematically identify and characterise these hierarchical relationships between enhancers.
In summary, our results illustrate how pre-existing genetic effects on chromatin propagate to gene expression during immune activation, and highlights the relevance of these hidden genetic effects for deciphering the molecular architecture of disease-associated variants. Our study is also the first that we are aware of to utilise iPSC-derived cells to study genetic effects in immune response. We believe a major future use of this system will be the systematic exploration of gene-environment interactions across large numbers of cell states. Furthermore, because iPSCs are readily engineered, the identity of causal variants and their downstream consequences can be directly tested in exactly the same cell types and conditions where they were discovered.
Online Methods
Donors and cell lines
Human induced pluripotent stem cells (iPSCs) from 123 healthy donors (72 female and 51 male) (Supplementary Table 1) were obtained from the HipSci project22. Of these lines, 57 were initially grown in feeder-dependent medium and 66 were grown in feeder-free E8 medium. The cell lines were screened for mycoplasma by the HipSci project22. All samples for the HipSci resource were collected from consented research volunteers recruited from the NIHR Cambridge BioResource (http://www.cambridgebioresource.org.uk). Samples were collected initially under ethics for iPSC derivation (REC Ref: 09/H0304/77, V2 04/01/2013), with later samples collected under a revised consent (REC Ref: 09/H0304/77, V3 15/03/2013).
Macrophage differentiation outcomes
We performed 138 macrophage differentiation attempts from 123 distinct HipSci iPSC lines (Supplementary Note, Supplementary Table 1). We were able to differentiate macrophages from 101/123 (82%) of the iPSC lines. For 97/101 lines, we further confirmed the cell surface expression of CD14, CD16 and CD206 macrophage markers using flow cytometry (Supplementary Fig. 1). However, some of the differentiated lines did not produce enough macrophages to perform all of the experimental assays or the differentiated cells were not pure enough to be used in stimulation experiments. In total, we obtained high quality RNA-seq data from 89 differentiations corresponding to 85 unique donors and ATAC-seq data from up to 42 unique donors in up to four experimental conditions (Supplementary Table 1). The final sample size was decided based on similar gene expression and chromatin QTL mapping studies performed previously2,7,26–28.
RNA-seq preprocessing and quality control
RNA-seq reads were aligned to the GRCh38 reference genome and Ensembl 79 transcript annotations using STAR v2.4.0j56. Subsequently, VerifyBamID v1.1.257 was used to detect and correct any potential sample swaps and cross-contamination between donors. We did not detect any cross-contamination, but we did identify one sample swap between two donors. We used featureCounts v1.5.058 to count the number of uniquely mapping fragments overlapping GENCODE59 basic annotation from Ensembl 79. We excluded short RNAs and pseudogenes from the analysis leaving 35,033 unique genes of which 19,796 were protein coding. Furthermore, we only used 15,797 genes with mean expression in at least one of the conditions greater than 0.5 transcripts per million (TPM)60 in all downstream analyses. We quantile-normalised the data and corrected for sample-specific GC content bias using the conditional quantile normalisation (cqn)61 R package. To detect hidden confounders in gene expression, we applied PEER62 to each condition separately allowing for at most 10 hidden factors. We found that the first 3-5 factors explained the most variation in the data and the others remained close to zero. Although we performed replicate macrophage differentiations and RNA-seq from four iPSC lines, for simplicity we decided to use only one of the replicates in downstream analyses. We further excluded samples from one donor (qaqx_1) from downstream analysis because they appeared as outliers in principal component analysis (PCA). The final dataset consisted of 336 RNA-seq samples from 84 donors.
ATAC-seq data analysis
Read alignment
Illumina Nextera sequencing adapters were trimmed using skewer v0.1.12763 in paired end mode. Trimmed reads were aligned to GRCh38 human reference genome using bwa mem v0.7.1264. Reads mapping to the mitochondrial genome and alternative contigs were excluded from all downstream analysis. Picard 1.134 MarkDuplicates was used to remove duplicate fragments. We used verifyBamID57 1.1.2 to detect and correct potential sample swaps between individuals. Fragment coverage BigWig files were constructed using bedtools v2.17.065.
Peak calling
We used MACS266 v2.1.0 with ‘--nomodel --shift -25 --extsize 50 -q 0.01’ to identify open chromatin regions (peaks) that were enriched for transposase integration sites compared to the background at 1% FDR level. With these parameters we detected between 31,658 and 208,330 peaks per sample. We constructed consensus peak sets in each condition separately by pooling all of the peak calls from all of the samples. For each peak, we first counted the number of samples in which that peak was identified. We then calculated the union of all peaks that were detected in at least three samples. Finally, we pooled the consensus peaks from all four conditions to obtain the final set of 296,220 unique peaks that were used for all downstream analyses. We used featureCounts58 v.1.5.0 to count the number of fragments overlapping consensus peak annotations and ASEReadCounter67 from Genome Analysis Toolkit (GATK) to quantify allele-specific chromatin accessibility.
Sample quality control
We used the following criteria summarised in Supplementary Table 8 to assess the quality of ATAC-seq samples:
Assigned fragment count - the total number of paired end fragments assigned to peaks by featureCounts.
Mitochondrial fraction - fraction of total fragments aligned to the mitochondrial genome.
Assigned fraction - fraction of non-mitochondrial reads assigned to consensus peaks. A measure of signal-to-noise ratio.
Duplicated fraction - fraction of fragments that were marked as duplicates by Picard MarkDuplicates.
Peak count - number of peaks called by MACS2.
Length ratio - # of short fragments (< 150 nt) / # long fragments (>= 150 nt). This measures if the read length distribution has characteristic ATAC-seq profile with clearly visible mono-nucleosomal and di-nucleosomal peaks.
We used these criteria to exclude 5 samples from downstream analysis (Supplementary Table 8). One sample was excluded because of very low assigned fraction (~10%) and peak count, two more were excluded because of extremely large length ratio (>7) and a fragment length distribution uncharacteristic for ATAC-seq library. The final two samples were excluded because they appeared to be outliers in the principal component analysis (PCA).
QTL mapping
Preparing genotype data
We obtained imputed genotypes for all of the samples from the HipSci22 project. We used CrossMap v0.1.868 to convert variant coordinates from GRCh37 reference genome to GRCh38. Subsequently, we filtered the VCF file with bcftools v.1.2 to retain only bi-allelic variants (both SNPs and indels) with IMP2 score > 0.4 and minor allele frequency (MAF) > 0.05 in our 86 samples. The same VCF file was used for all subsequent analyses. The VCF file was imported into R using the SNPRelate69 package.
Quantifying allele-specific expression and chromatin accessibility
We used ASEReadCounter67 from the Genome Analysis ToolKit (GATK) to count the number of allele-specific fragments overlapping each variant in the RNA-seq and ATAC-seq datasets. We used the following flags with ASEReadCounter: ‘-U ALLOW_N_CIGAR_READS -dt NONE -- minMappingQuality 10 -rf MateSameStrand’. We removed indels from the VCF file prior to quantifying allele-specific expression because they are not supported by the RASQUAL model.
Detecting QTLs using RASQUAL
We wrote a collection of python scripts and a rasqualTools R package to simplify running RASQUAL on large number of samples and work with large RASQUAL output files (see URLs). We used the vcfAddASE.py script to add allele-specific counts calculated in the previous step into the VCF file. We ran RASQUAL26 independently for each experimental condition using sex and first two PEER factors as covariates (sex and first 3 PCs for caQTLs). In contrast to standard linear model, covariates seemed to have only a minor effect on the number of QTLs detected by RASQUAL. We only included variants that were either in the gene body or within +/- 500 kb from the gene (+/- 50kb from the accessible region). We specified ‘--imputation-quality > 0.7’. As a result, variants with imputation quality of < 0.7 were used as feature SNPs in allele-specific analysis but were not considered as possible causal variants. We also used RASQUAL’s GC correction option to correct for sample-specific GC bias in the feature-level read count data. To correct for multiple testing, we picked one minimal p-value per feature, used eigenMT70 to estimate the number of independent tests performed in the cis-region of each feature and then performed Bonferroni correction to obtain the corrected p-value. We also ran RASQUAL once with the ‘--random-permutation’ option to obtain empirical null p-values from data with permuted sample labels. We performed the same eigenMT multiple testing procedure on the permuted p-values and compared the true association p-values to the empirical null distribution to identify QTLs with FDR < 10%.
Detecting QTLs using a linear model
We used linear regression implemented in the FastQTL55 software to map cis-QTLs in each experimental condition. We used the ‘--permute 100 10000’ option to obtain permutation p-values for each association. The size of the cis windows was set to +/- 500 kb around each gene and +/- 50kb around each ATAC-seq peak. Prior to QTL mapping, the read count data was quantile normalised using the cqn package with GC-content of the feature (gene or peak) included as a covariate. For eQTL analysis, we used sex and the first six PEER factors as covariates in the model. For caQTL analysis we used sex and the first three principal components (PCs) as covariates in the model. Although FastQTL reported feature-level permutation p-values, obtaining those was computationally not feasible for RASQUAL. Therefore, to be able to faithfully compare the number of QTLs detected by FastQTL and RASQUAL, we decided to apply exactly the same multiple testing correction procedure (eigenMT + single permutation of sample labels) to both methods. We further restricted the comparison to features that were tested by both methods. This affected a small number of genes that were tested by FastQTL but filtered out by RASQUAL, because the raw read count was exactly zero in all samples.
Detecting response eQTLs
In each condition, we first identified all genes and corresponding lead variants that displayed significant association at 10% FDR level from RASQUAL. For each gene, we only kept independent lead variants (R2 < 0.8). Finally, we used all independent pairs of genes and corresponding lead variants to test if the eQTL effect size was significantly different between conditions. This was equivalent to testing the significance of the interaction term between condition and lead eQTL variant for each gene. Furthermore, to take advantage of the fact that gene expression was profiled in the same 84 lines in the four conditions, we also included the cell line as a random effect and fitted a linear mixed model using the lme471 package. Specifically, for each gene and lead variant pair we compared the following two models:
H0: expression ~ genotype + condition + covariates + (1|cell_line)
H1: expression ~ genotype + condition + genotype:condition + covariates + (1|cell_line)
where (1|cell_line) denotes the cell line specific random effect. We then calculated empirical p-values for the interaction test by permuting the conditions within each individual line 1,000 times. Subsequently, we used Benjamini-Hochberg FDR correction on the permutation p-values to identify 1,950 significant interactions at 10% FDR level. We used the same normalised data and covariates for interaction testing that were previously used for eQTL mapping in each condition separately.
Detecting response caQTLs
The procedure to identify response caQTLs was almost identical to the one used to detect response eQTLs above. However, instead of a linear mixed model we decided to use standard linear model without the random effect for cell line because not all lines were measured in all conditions. Furthermore, we found that our strategy to permute conditions within individual lines was not reliable when the number of measured conditions was not the same for each individual. Therefore, we decided to apply Benjamini-Hochberg FDR correction to nominal p-values from the linear model and identify significant interactions at 10% FDR level. With this approach we identified significant interactions for 6,591 caQTL regions.
Filtering and clustering QTLs based on effect size
Next, we focussed on all significant response eQTLs and response caQTLs that were detected with the interaction test above. We extracted the RASQUAL QTL effect size estimates π for each feature-variant pair in each condition and converted them to log2 fold changes between the two homozygotes using the formula log2FC = -log2(π/(1-π)). Multiplication with -1 was necessary because RASQUAL uses alternative allele dosage to represent genotypes while the SNPRelate package that the we used to import genotypes into R uses reference allele dosage. For a QTL to be considered condition specific we required the absolute log2FC in the naive condition to be less than 0.59 (1.5-fold) and the absolute difference in log2FC between naive and any one of the stimulated conditions to be greater than 0.59 (~1.5 fold). We further required the absolute log2FC to be greater than 0.59 in at least one condition. To demonstrate that our result were not sensitive to the exact fold change threshold used, we also repeated the same analysis using log2FC threshold of 1 (= 2-fold) for all three filters. To obtain relative log2FC, we divided the log2FC values in each condition by the maximal log2FC value observed across conditions. This scaling was necessary to make QTLs with different absolute effect size comparable to each other. Finally, we used k-means clustering to identify six groups of QTLs that had similar activity patterns across conditions.
Identifying master and dependent regions
For each caQTL region, we defined its credible set of causal variants as those with R2 > 0.8 to the lead variant. We then classified the focal caQTL region as a master region (i - Fig. 3b), if the credible set overlapped the region itself, suggesting that the caQTL is directly caused by a variant within the region disrupting transcription factor binding. Alternatively, if the credible set overlapped some other regulated region but not the focal region, then we classified it as a dependent region (ii - Fig. 3b). We also excluded ambiguous cases where the credible set overlapped either multiple regulated regions (iii - Fig. 3b) or it did not overlap any regulated regions (iv-v - Fig. 3b). To estimate the fraction of master-dependent region pairs that had the same direction of effect, we limited our analysis to region pairs with nominal p-value of the lead master caQTL variant for both master and dependent regions < 10-4. This filtering was necessary to ensure that master and dependent caQTLs were both active in the same condition.
Motif disruption analysis
We limited motif disruption analysis to caQTL regions that did not contain associated indels and had <= 3 overlapping single nucleotide polymorphisms (SNPs) in them. For each SNP and peak pair we focussed on the sequence +/- 25 bp from the SNP. We constructed both reference and alternative versions of the sequence and used TFBSTools v1.10.472 to calculate the relative binding scores for both alleles (expressed as percentage from 0-100%). The TF motifs were downloaded from CIS-BP73 database. We considered the variant to be motif disrupting if the difference in relative binding score between the two alleles was > 3 percentage points. We also required the relative binding score for at least one of the alleles to be >= 85% of the theoretical maximum. This filter was necessary to exclude potential motif disruption events in very weak motif matches that were not likely to correspond to binding in vivo and is similar to the default thresholds recommended by TFBSTools. We used Fisher’s exact test to identify motifs that were significantly more often disrupted in one of the six condition-specific caQTL clusters compared to all caQTLs. For condition-specific caQTLs we further limited the analysis to putative master caQTL regions, because they were more likely to harbour the causal caQTL variant. We did not use that filter for caQTLs regulating putative primed enhancers, because the number of primed enhancers was much smaller.
Identifying condition-specific dependent regions
To identify condition-specific dependent regions, we tested if the effect size of the caQTL changed differently for master and dependent regions (2,023 unique pairs) between two conditions. This was equivalent to testing the significance of a three-way interactions between genotype, region (master or dependent) and condition. We implemented this as the comparison of two standard linear models in R:
H0: accessibility ~ genotype + region + condition + region*condition + genotype*region + genotype*condition + covariates
H1: accessibility ~ genotype + region + condition + region*condition + genotype*region + genotype*condition + genotype*condition*region + covariates
Similarly to condition-specific caQTL analysis, we used the first three principal components calculated separately for each condition as covariates in the model. For each master and dependent region pair we picked the minimal p-value from three tests (naive vs each simulated condition) and used Bonferroni correction to correct for multiple testing. We then applied the Benjamini-Hochberg FDR correction to the Bonferroni-corrected p-values to identify all master-dependent region pairs that showed significant interaction at 10% FDR. We used the log2FC from RASQUAL as the measure of caQTL effect size. To identify true condition-specific dependent regions, we further filtered the results by requiring the absolute log2FC of the master region to be > 0.59 (1.5-fold) in the naive condition and the change in the log2FC for the dependent region between the naive and stimulated condition to be > 0.59. We also required the change in the log2FC for the master peak to be < 1.
Linking response eQTLs to caQTLs
First, we grouped all response eQTLs into three groups according to the condition in which they had the maximal effect size (IFNγ, Salmonella or IFNγ + Salmonella). For each response eQTL, we then identified all caQTLs that were in high linkage disequilibrium (LD) with it in any of the four conditions (R2 > 0.8 between the lead variants) (Supplementary Fig. 18). If there was more than one caQTL in high LD, we picked the one with the smallest association p-value to obtain at most one caQTL corresponding to each response eQTL. Next, to estimate the prevalence of enhancer priming, we asked how often was the corresponding caQTL present already in the naive condition. Since response eQTLs were required to have RASQUAL log2FC < 0.59 in the naive condition (see above), we used the same threshold to decide if the caQTL was present (log2FC > 0.59) or absent (log2FC < 0.59) in the naive condition. We also repeated this analysis using a more stringent threshold of log2FC > 1. Since there are various reasons why this analysis might lead to false positives, we decided to quantify our false positive rate by performing a reverse analysis where we started with response caQTLs, identified corresponding eQTLs (R2 > 0.8) and asked how often were the eQTLs present already in the naive condition (log2FC > 0.59).
Colocalisation with GWAS hits
We used coloc v2.3-130 to test for colocalisation between molecular QTLs and GWAS hits. In the colocalisation analysis we used summary statistics from the linear model (rather than RASQUAL), because RASQUAL summary statistics could not be easily converted to approximate Bayes factors required by coloc. We ran coloc on a 400kb region centered on each lead eQTL and caQTL variant (200kb for the secondary eQTLs) that was less than 100kb away from at least one GWAS variant with nominal p-value < 10-5. We then applied a set of filtering steps to identify a stringent set of eQTLs and caQTL that colocalised with GWAS hits. Similarly to a published analysis40, we first removed all cases where PP3 + PP4 < 0.8, to exclude loci where we were underpowered to detect colocalisation. We then required PP4/(PP3+PP4) > 0.9 to only keep loci where coloc strongly prefered the model of a single shared causal variant driving both association signals over a model of two distinct causal variants. We excluded all colocalisation results from the MHC region (GRCh38: 6:28,510,120-33,480,577) because they were likely to be false positives due to complicated LD patterns in this region. We only kept results where the minimal GWAS p-value was < 10-6. Finally, we manually excluded 11 potential eQTL overlaps and 6 potential caQTL overlaps where on visual inspection the LD block exceeded the 400kb window that we used for colocalisation testing.
Supplementary Material
Acknowledgements
We thank Leopold Parts, Jeremy Schwartzentruber, Chris Wallace, Lili Milani, Kaido Lepik and Hedi Peterson for helpful comments on the manuscript. We thank Rachel Nelson for assistance and early access to HipSci iPSC lines. We thank Roman Kreuzhuber for providing access to the imputed genotype data from the Fairfax study. We also thank WTSI DNA Pipelines and Cytometry Core Facility for their sequencing and flow cytometry services. This work was supported by the Wellcome Trust grant WT098051. K.A. was supported by a PhD fellowship from the Wellcome Trust (WT099754) and a postdoctoral fellowship from the Estonian Research Council (MOBJD67). The iPSC lines were generated at the Wellcome Trust Sanger Institute, under the Human Induced Pluripotent Stem Cell Initiative funded by a strategic award (WT098503) from the Wellcome Trust and Medical Research Council. We also acknowledge Life Science Technologies Corporation as the provider of cytotune.
Footnotes
Code availability
Most figures were made in R using ggplot274 and wiggleplotr75 was used to produce RNA-seq and ATAC-seq read coverage plots. Custom data analysis scripts are available in the accompanying GitHub repository (see URLs). Full list of published software used in this study is presented in Supplementary Table 9.
Data availability
Imputed genotype data from the HipSci cell lines is available from ENA (ERP013161) and EGA (EGAD00010000773). Raw RNA-seq and ATAC-seq data has been deposited to ENA (ERP020977) and EGA (EGAS00001002236) and processed read counts are available from Zenodo (doi: 10.5281/zenodo.259661). Raw flow cytometry data is also available from Zenodo (doi: 10.5281/zenodo.234214).
URLs
Data analysis scripts: https://github.com/kauralasoo/macrophage-gxe-study
Processed data: https://zenodo.org/communities/macrophage-gene-expression-genetics/
wiggleplotr: https://bioconductor.org/packages/wiggleplotr/
rasqualTools: https://github.com/kauralasoo/rasqual
Author Contributions
KA, DG: Wrote the paper with input from all authors. KA, JR: Performed the macrophage differentiation experiments. JR, AK: Performed the chromatin accessibility assays. AM, KK: Assisted with disease colocalisation and enrichment analysis. KA, SM, CH: Optimised the stimulation experiments. KA: Analysed the data. KA, SM, GD, DG: Designed the experiments. GD, DG: Supervised research.
Competing interests
Authors declare no competing financial interests.
References
- 1.Li Y, et al. Inter-individual variability and genetic influences on cytokine responses to bacteria and fungi. Nat Med. 2016;22:952–960. doi: 10.1038/nm.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barreiro LB, et al. Deciphering the genetic architecture of variation in the immune response to Mycobacterium tuberculosis infection. Proc Natl Acad Sci U S A. 2012;109:1204–1209. doi: 10.1073/pnas.1115761109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fairfax BP, et al. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science. 2014;343 doi: 10.1126/science.1246949. 1246949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim S, et al. Characterizing the genetic basis of innate immune response in TLR4-activated human monocytes. Nat Commun. 2014;5:5236. doi: 10.1038/ncomms6236. [DOI] [PubMed] [Google Scholar]
- 5.Lee MN, et al. Common genetic variants modulate pathogen-sensing responses in human dendritic cells. Science. 2014;343 doi: 10.1126/science.1246980. 1246980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Çalışkan M, Baker SW, Gilad Y, Ober C. Host genetic variation influences gene expression response to rhinovirus infection. PLoS Genet. 2015;11:e1005111. doi: 10.1371/journal.pgen.1005111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nédélec Y, et al. Genetic Ancestry and Natural Selection Drive Population Differences in Immune Responses to Pathogens. Cell. 2016;167:657–669.e21. doi: 10.1016/j.cell.2016.09.025. [DOI] [PubMed] [Google Scholar]
- 8.de Lange KM, et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet. 2017 doi: 10.1038/ng.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim-Hellmuth S, et al. Genetic regulatory effects modified by immune activation contribute to autoimmune disease associations. Nat Commun. 2017;8:266. doi: 10.1038/s41467-017-00366-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Degner JF, et al. DNase I sensitivity QTLs are a major determinant of human expression variation. Nature. 2012;482:390–394. doi: 10.1038/nature10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin F, Li Y, Ren B, Natarajan R. PU.1 and C/EBP(alpha) synergistically program distinct response to NF-kappaB activation through establishing monocyte specific enhancers. Proc Natl Acad Sci U S A. 2011;108:5290–5295. doi: 10.1073/pnas.1017214108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heinz S, et al. Effect of natural genetic variation on enhancer selection and function. Nature. 2013;503:487–492. doi: 10.1038/nature12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang A, et al. Epigenetic priming of enhancers predicts developmental competence of hESC-derived endodermal lineage intermediates. Cell Stem Cell. 2015;16:386–399. doi: 10.1016/j.stem.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chow NA, Jasenosky LD, Goldfeld AE. A Distal Locus Element Mediates IFN-γ Priming of Lipopolysaccharide-Stimulated TNF Gene Expression. Cell Rep. 2014 doi: 10.1016/j.celrep.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shin HY, et al. Hierarchy within the mammary STAT5-driven Wap super-enhancer. Nat Genet. 2016;48:904–911. doi: 10.1038/ng.3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Banovich NE, et al. Impact of regulatory variation across human iPSCs and differentiated cells. Genome Res. 2017 doi: 10.1101/gr.224436.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pique-Regi R, et al. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res. 2011;21:447–455. doi: 10.1101/gr.112623.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sherwood RI, et al. Discovery of directional and nondirectional pioneer transcription factors by modeling DNase profile magnitude and shape. Nat Biotechnol. 2014;32:171–178. doi: 10.1038/nbt.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alasoo K, et al. Transcriptional profiling of macrophages derived from monocytes and iPS cells identifies a conserved response to LPS and novel alternative transcription. Sci Rep. 2015;5 doi: 10.1038/srep12524. 12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kilpinen H, et al. Common genetic variation drives molecular heterogeneity in human iPSCs. Nature. 2017;546:370–375. doi: 10.1038/nature22403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu X, Ivashkiv LB. Cross-regulation of signaling pathways by interferon-gamma: implications for immune responses and autoimmune diseases. Immunity. 2009;31:539–550. doi: 10.1016/j.immuni.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiao Y, et al. Synergistic activation of inflammatory cytokine genes by interferon-γ-induced chromatin remodeling and toll-like receptor signaling. Immunity. 2013;39:454–469. doi: 10.1016/j.immuni.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumasaka N, Knights AJ, Gaffney DJ. Fine-mapping cellular QTLs with RASQUAL and ATAC-seq. Nat Genet. 2016;48:206–213. doi: 10.1038/ng.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grubert F, et al. Genetic Control of Chromatin States in Humans Involves Local and Distal Chromosomal Interactions. Cell. 2015;162:1051–1065. doi: 10.1016/j.cell.2015.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waszak SM, et al. Population Variation and Genetic Control of Modular Chromatin Architecture in Humans. Cell. 2015;162:1039–1050. doi: 10.1016/j.cell.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 29.Cheng CS, et al. Genetic determinants of chromatin accessibility and gene regulation in T cell activation across human individuals. bioRxiv. 2016 090241. [Google Scholar]
- 30.Giambartolomei C, et al. Bayesian Test for Colocalisation between Pairs of Genetic Association Studies Using Summary Statistics. PLoS Genet. 2014;10:e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu JZ, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okada Y, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu Z, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016;48:481–487. doi: 10.1038/ng.3538. [DOI] [PubMed] [Google Scholar]
- 34.Lambert JC, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morris AP, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jansen R, et al. Conditional eQTL analysis reveals allelic heterogeneity of gene expression. Hum Mol Genet. 2017;26:1444–1451. doi: 10.1093/hmg/ddx043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bentham J, et al. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat Genet. 2015;47:1457–1464. doi: 10.1038/ng.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Farh KK-H, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518:337–343. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo H, et al. Integration of disease association and eQTL data using a Bayesian colocalisation approach highlights six candidate causal genes in immune-mediated diseases. Hum Mol Genet. 2015;24:3305–3313. doi: 10.1093/hmg/ddv077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chun S, et al. Limited statistical evidence for shared genetic effects of eQTLs and autoimmune-disease-associated loci in three major immune-cell types. Nat Genet. 2017;49:600–605. doi: 10.1038/ng.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.GTEx Consortium et al. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Javierre BM, et al. Lineage-Specific Genome Architecture Links Enhancers and Non-coding Disease Variants to Target Gene Promoters. Cell. 2016;167:1369–1384.e19. doi: 10.1016/j.cell.2016.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jin F, et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghavi-Helm Y, et al. Enhancer loops appear stable during development and are associated with paused polymerase. Nature. 2014;512:96–100. doi: 10.1038/nature13417. [DOI] [PubMed] [Google Scholar]
- 46.Pai AA, et al. The contribution of RNA decay quantitative trait loci to inter-individual variation in steady-state gene expression levels. PLoS Genet. 2012;8:e1003000. doi: 10.1371/journal.pgen.1003000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mullen AC, et al. Master transcription factors determine cell-type-specific responses to TGF-β signaling. Cell. 2011;147:565–576. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trompouki E, et al. Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell. 2011;147:577–589. doi: 10.1016/j.cell.2011.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ostuni R, et al. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152:157–171. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 50.Magnani L, Eeckhoute J, Lupien M. Pioneer factors: directing transcriptional regulators within the chromatin environment. Trends Genet. 2011;27:465–474. doi: 10.1016/j.tig.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 51.Ramirez-Carrozzi VR, et al. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bojcsuk D, Nagy G, Balint BL. Inducible super-enhancers are organized based on canonical signal-specific transcription factor binding elements. Nucleic Acids Res. 2017;45:3693–3706. doi: 10.1093/nar/gkw1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 54.Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-gamma: an overview of signals, mechanisms and functions. J Leukoc Biol. 2004;75:163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- 55.Ongen H, Buil A, Brown AA, Dermitzakis ET, Delaneau O. Fast and efficient QTL mapper for thousands of molecular phenotypes. Bioinformatics. 2016;32:1479–1485. doi: 10.1093/bioinformatics/btv722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jun G, et al. Detecting and estimating contamination of human DNA samples in sequencing and array-based genotype data. Am J Hum Genet. 2012;91:839–848. doi: 10.1016/j.ajhg.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 59.Harrow J, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wagner GP, Kin K, Lynch VJ. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012;131:281–285. doi: 10.1007/s12064-012-0162-3. [DOI] [PubMed] [Google Scholar]
- 61.Hansen KD, Irizarry RA, Wu Z. Removing technical variability in RNA-seq data using conditional quantile normalization. Biostatistics. 2012;13:204–216. doi: 10.1093/biostatistics/kxr054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stegle O, Parts L, Piipari M, Winn J, Durbin R. Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat Protoc. 2012;7:500–507. doi: 10.1038/nprot.2011.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jiang H, Lei R, Ding S-W, Zhu S. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics. 2014;15:182. doi: 10.1186/1471-2105-15-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv [q-bio.GN] 2013 [Google Scholar]
- 65.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Castel S, Levy-Moonshine A, Mohammadi P, Banks E, Lappalainen T. Tools and best practices for data processing in allelic expression analysis. Genome Biol. 2015;16:195. doi: 10.1186/s13059-015-0762-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao H, et al. CrossMap: a versatile tool for coordinate conversion between genome assemblies. Bioinformatics. 2014;30:1006–1007. doi: 10.1093/bioinformatics/btt730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zheng X, et al. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics. 2012;28:3326–3328. doi: 10.1093/bioinformatics/bts606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Davis JR, et al. An Efficient Multiple-Testing Adjustment for eQTL Studies that Accounts for Linkage Disequilibrium between Variants. Am J Hum Genet. 2016;98:216–224. doi: 10.1016/j.ajhg.2015.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bates D, Mächler M, Bolker B, Walker S. Fitting Linear Mixed-Effects Models Using lme4. J Stat Softw. 2015;67:1–48. [Google Scholar]
- 72.Tan G, Lenhard B. TFBSTools: an R/bioconductor package for transcription factor binding site analysis. Bioinformatics. 2016;32:1555–1556. doi: 10.1093/bioinformatics/btw024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weirauch MT, et al. Determination and Inference of Eukaryotic Transcription Factor Sequence Specificity. Cell. 2014;158:1431–1443. doi: 10.1016/j.cell.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer; 2009. [Google Scholar]
- 75.Alasoo K. wiggleplotr: Make read coverage plots from BigWig files. Bioconductor; 2017. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.