Abstract
Early detection of ovarian cancer has the potential to impact mortality. A multi-modal screening strategy where rising CA125 values over time, analyzed with the risk of ovarian cancer algorithm (ROCA), triggers transvaginal sonography and possible surgery has high sensitivity and specificity, but still fails to detect the 20% of early stage cases that do not express CA125. Use of multiple biomarkers could detect cases missed by CA125. We have studied the sensitivity and lead time of a multi-marker panel (CA125, HE4, MMP-7 and CA 72–4) compared to CA125 alone. We used PRoBE design principles to select pre-clinical longitudinal specimens from 75 women (50 screen-positive, 25 screen-negative) who developed invasive epithelial ovarian cancer (3–5 serial specimens each) and 547 corresponding healthy controls (1–10 serial specimens each) from the ovarian cancer screening trial, UKCTOCS, in a blinded fashion. We measured the multi-marker concentrations in ultra-low serum volumes (16 μL) utilizing multiplexed bead-based immunoassays with low detection limits, high inter- and intra-assay precision, negligible cross-reactivity and good correlation with standard immunoassays. While, at least one of the complementary biomarkers rose with CA125 in 44% (22/50) of screen-positive cases, there was no advantage in lead time over CA125. Therefore, we developed single-marker longitudinal algorithms (ROCA-like) to determine the presence of a change point to distinguish between the cases and controls. Using these algorithms, at 98% specificity, HE4 and CA72–4 identified 16% (4/25) of screen-negative cases, while MMP-7 identified none. Taken together, HE4 and CA72–4 show promise as complementary biomarkers to CA125 for longitudinal screening.
Introduction
Ovarian cancer remains the most lethal gynecologic cancer with a five year survival rate of approximately 47% across all stages (1). Localized disease that is confined to the ovary or the pelvic region has a high survival rate of 93% with current treatment strategies, whereas advanced stage disease that has spread beyond the pelvic region has a poor survival rate of 29% (1). Early detection remains the key challenge to realizing survival benefits since only 15–20% are diagnosed at an early stage (1). Ovarian cancer is neither a common nor a rare disease with a prevalence of 1 in 2500 in postmenopausal women at highest risk. As a result, a very high specificity (>99.6%) and at least moderate sensitivity (>75%) is required for a screening strategy in the general population to achieve a positive predictive value (PPV) of 10% i.e., 10 operations per case of ovarian cancer (2). Currently, while the multimodal screening strategy achieves these performance characteristics, it is not recommended by the United States Preventive Services Task Force (USPSTF) in asymptomatic women at normal risk for ovarian cancer as there is as yet no evidence of a definitive mortality benefit (3).
Cancer Antigen 125 (CA125) is a well-established ovarian cancer biomarker for disease progression and response to therapy (4). However, CA125 interpreted using a cut-off at the 97th percentile, is only elevated in 50–60% of early stage ovarian cancer patients. Use of longitudinal algorithms such as risk of ovarian cancer algorithm (ROCA), that incorporate change in levels over time, increase sensitivity to 87% but specificity remains below 90%, making it an suboptimal biomarker on its own for early detection (5,6). Transvaginal Ultrasound (TVS), while well-suited for detecting ovarian morphological and volume abnormalities does not generally visualize fallopian tubes and is not adequately specific or sensitive on its own as a screening methodology for ovarian cancer (7). However, a multi-modal approach utilizing annual screening with CA125 (interpreted using ROCA) as a first line screen, with TVS as a second-line screen in a large randomized controlled trial of 202,638 post-menopausal women in the United Kingdom Collaborative Trial of Ovarian Cancer Screening (UKCTOCS) achieved a PPV of 35.1% for the multi-modal screening arm, compared to 2.8% for the TVS only arm (8). Further analysis indicated a mortality reduction of up to 20% with the multimodal strategy when prevalent cases were excluded, indicating promise for this approach, although further follow-up is required to confirm this estimate (9). A single-arm prospective study utilizing the same approach, in a smaller cohort of 4051 women in the United States achieved a similar PPV of 40% (10).
Despite the promise of two-stage multi-modal strategies, 15–20% of ovarian cancers do not express CA125 and will be missed by CA125 alone (2). Several combinations of multiple marker panels have been proposed (most in combination with CA125) that improve sensitivity, while maintaining specificity compared to CA125 alone (11,12). Also, multiple biomarkers have shown lead times in prediagnostic specimens and improved lead times, in addition to complementarity. In a study of prediagnostic specimens from the Carotene and Retinol Efficacy Trial, serum concentrations of CA125, HE4 and mesothelin were found to rise in ovarian cancer patients approximately 3 years before diagnosis, highlighting the importance of biomarkers measured longitudinally (13). Also, algorithms that account for longitudinal trajectories of biomarker measurements have shown improved biomarker performance in multiple diseases (14,15).
Previously, we screened 96 potential biomarkers utilizing multiplex xMAP-based screening methodologies and identified several promising biomarker panels for early detection with 86% sensitivity at 98% specificity for early stage ovarian cancer (16). We further validated the most promising individual biomarkers from the study (CA125, HE4, MMP-7, CA 72–4, CA19–9, CA15–3, CEA and sVCAM) utilizing conventional immunoassays in pretreatment sera from 142 stage I ovarian cancer patients and longitudinal healthy specimens (5 annual samples) from 217 controls to validate a multi-marker panel suitable for longitudinal algorithm development (17). Our efforts resulted in the identification of a four-marker panel comprising CA125, HE4, MMP-7 and CA72–4 with 83.2% sensitivity at 98% specificity for the early detection of ovarian cancer. In addition, each individual biomarker had its own baseline in the healthy controls, indicating suitability as longitudinal biomarkers.
In this study, we combined the multi-marker strategy that has shown promising improvements in sensitivity with longitudinal biomarker algorithms,that have shown improvements in lead time and specificity, to develop a longitudinal biomarker panel for the early detection of ovarian cancer. Specifically, we evaluated lead times and CA125 complementarity for a multi-marker panel including CA125, HE4, MMP-7 and CA72–4 utilizing an in-house developed multiplexed bead-based immunoassay in retrospective preclinical longitudinal cases and controls obtained from the UKCTOCS study.
Materials and Methods
Study Design and Patient Population
Study samples were obtained as blinded retrospective specimens from the United Kingdom Collaborative Trial for Ovarian Cancer Screening (UKCTOCS). The study population included pre-clinical longitudinal specimens (3–5 each) from 75 post-menopausal women (cases) who went on to develop invasive epithelial ovarian cancer and 547 corresponding controls (1–10 longitudinal specimens each). Of the 75 cases, 50 were CA125 ROCA screen-positive and 25 were CA125 ROCA screen-negative (patients did not have an elevated CA125, but went on to develop ovarian cancer). The sample set was specifically constructed to include an overrepresentation of screen-negative cases (where improvement is most needed). The patient demographics are summarized in Table 1. These studies were conducted in accordance with the International Ethical Guidelines for Biomedical Research Involving Human Subjects (CIOMS). Approval was obtained from the relevant Institutional Review Boards. Informed written consent was obtained from participants in the UKCTOCS trial.
Table 1.
Characterisitics of the patient population
| Age (years) |
No. Longitudinal Samples |
||||
|---|---|---|---|---|---|
| Histology | No. Patients |
Mean | Median | Mean | Median |
| Healthy postmenopausal | 547 | 62.9 | 62.4 | 4.3 | 4 |
| Ovarian cancer stages IA-IC | 19 | 63.6 | 63.0 | 3.4 | 3 |
| Ovarian cancer stages IIA-IIC | 8 | 69.6 | 70.3 | 4.3 | 5 |
| Ovarian cancer stages IIIA-IIIC | 41 | 66.9 | 67.5 | 4.0 | 4 |
| Ovarian cancer stages IV | 7 | 69.4 | 68.8 | 4.3 | 3 |
| Serous cystadenocarcinoma | 39 | 67.5 | 67.6 | 4.1 | 5 |
| Papillary adenocarcinoma | 9 | 64.7 | 65.5 | 2.9 | 3 |
| Mucinous cystadenocarcinoma | 1 | 59.4 | 59.4 | 1 | 1 |
| Clear cell adenocarcinoma | 4 | 70 | 70 | 6.3 | 6 |
| Endometroid carcinoma | 7 | 61.4 | 59.6 | 3.3 | 3 |
| Carcinosarcoma | 6 | 63.4 | 63.5 | 4.8 | 5 |
| Adenocarcinoma | 6 | 66.5 | 66.9 | 3.2 | 2.5 |
| Unknown | 3 | 75 | 73.8 | 5.7 | 6 |
Sample Collection and Processing
Samples were collected in the UKCTOCS study following an established study protocol (8). Briefly, blood samples collected in 8 mL gel separation serum tubes at trial centers were transported overnight to a central laboratory, where they were centrifuged at 1500 g for 10 minutes to separate serum. Sera were measured for CA125 concentrations utilizing a electrochemiluminescence sandwich immunoassay on Roche Elecsys 2010 (Roche Diagnostics, Burgess Hill, UK) and the remaining sera were aliquoted and banked in 500 microlitre straws in liquid nitrogen for future studies. For our study, banked serum samples were retrieved, thawed, aliquoted and shipped from the University College London to MD Anderson Cancer Center on dry ice with temperature monitoring to ensure sample integrity during shipment. Samples were stored at - 80◦C until ready for analysis. Prior to multiplex immunoassay analysis, samples were thawed at 4◦C and centrifuged at 1000 × g for 10 minutes.
Multiplex Assay Development and Validation
Multiplexed flow cytometric bead-based immunoassays (Luminex, Austin, TX) were developed and utilized for simultaneous measurements of soluble serum concentrations of CA125, HE4, MMP-7 and CA72–4 in the study population. Individual bead-based immunoassay kits were purchased from MilliporeSigma (Billerica, MA) for CA125 and MMP-7. Individual bead-based immunoassays were developed in-house for HE4 and CA72–4 and multiplexed to be compatible with the commercially acquired assays for CA125 and MMP-7. Briefly, for in-house assay development, antibody pairs (donated generously by Fujirebio Diagnostics Incorporated, Malvern, PA) were conjugated to internally dye-coded microbeads (Luminex, Austin, TX) using 2-step carbodiimide coupling at various concentrations following standard Luminex recommended protocols. Each analyte was assigned a different spectral bead region for multiplexing compatibility. The detecting antibody was biotinylated at various concentrations and screened against the capture antibody coated beads to establish optimal assay conditions to measure the dynamic range and detection limits needed for early stage cases and healthy normal samples. Cross-reactivity experiments were conducted to compare the individual ‘singleplex’ assay to the ‘multiplex assay’ to determine the influence of multiplexing on the dose-response curves. Incubation times, assay buffers, blocking buffers and bead diluent buffers for CA72–4 and HE4 were optimized for compatibility with commercially obtained kits. Samples containing low, medium and high concentrations of CA125, HE4, MMP-7 and CA72–4 were assessed in 5 replicates over 5 days to obtain inter- and intra-assay precision for the assays in the multiplex format (Supplemental Table 4). Limits-of-detection (LOD) were evaluated at three standard deviations (SDs) above the zero analyte blank. In addition, early stage cancer samples (n=40) previously measured with standard ELISA methods (17) were assessed with the multiplexed assay to evaluate correlation (Pearson) between methods.
Multiplex Analysis of Patient Samples
Retrospective blinded sera from longitudinal cases and corresponding controls obtained from UKCTOCS were measured on the Luminex Magpix instrument (Luminex, Austin, TX) utilizing the aforementioned, validated, multiplexed immunoassay to determine serum concentrations of CA125, HE4, MMP-7 and CA72–4. Samples were thawed at 4◦C and centrifuged prior to analysis. Blinded samples were coded at the source such that all the longitudinal cases and corresponding controls would be assessed on the same day to minimize the impact of inter-assay variations on biomarker concentrations. Briefly, diluted serum samples (1:4) and standards, prepared in serum matrix were incubated overnight on a plate shaker (4◦C) along with the multiplexed capturing antibody-coated bead cocktail. Following overnight incubation, the plates were brought to RT on the plate shaker for 1 hour and the sandwich immunoassay was completed with a detection cocktail incubation for 1 hour. Final visualization of the sandwich immunoassay complex was achieved with a half an hour incubation with streptavidin-phycoerythrin (SAPE). Following SAPE incubation, the plates were read on the Luminex Magpix reader (Luminex, Austin, TX). Sample read outs were obtained in terms of mean fluorescence intensity (MFI). Utilizing standard curves run on each plate for known standard concentrations, fitted to a 4-parameter logistic curve, MFIs were converted into concentrations for the unknown samples. QCs were run on each plate to ensure the validity of the run and assays that failed QC were repeated. The concentrations for the four biomarkers CA125 (U/mL), CA72–4 (U/mL), MMP-7 (pg/mL) and HE4 (pg/mL) were reported to the UKCTOCS study team for unblinding. The unblinded results were sent to the biostatistics team for analysis.
Data Analysis
The longitudinal trajectories for the log-transformed concentrations for individual biomarkers were plotted to visually assess lead-times of biomarkers over CA125. Individual single-marker algorithms were developed using Bayesian methods described in detail in the section below. Briefly, the data was divided into a training set and a test set. For the training set, the Bayesian model was applied to the longitudinal biomarker trajectory and the probability of the biomarker trajectory to have a change point was estimated. When no-change point was estimated, the marker trajectory was assessed the same property as control (within-person variation). For marker trajectories where change points were identified, the time point of linear change and rate of increase for each subject was utilized (within-person variation, change point, rate of rise). Utilizing the training model, unknown cases were fitted and prediction was completed to see if the model resembled a ‘case’ or a ‘control’. The percentages of cases identified by biomarkers that were missed by CA125 (screen-negatives) were assessed utilizing these individual algorithms.
Single Marker Algorithm Development – Bayesian method
To analyze the data generated from the multi-marker screening approach, the Bayesian method was applied in a two-step approach. In step 1, the training step, m1 cases and (m2-m1) controls with longitudinal marker data were used, in the training set ST. The Bayesian model was applied to fit the marker trajectory on a log scale and the probability that the marker trajectory (log-marker concentration over time) contains a change point was estimated. For control subjects, the marker value has constant mean over time. Each subject has her own mean and her own magnitude of variation. For patient i at the jth is the baseline mean.
The random jitters ϵij are assumed to have independent t-distributions with mean 0 and 5 degrees of freedom. The standard deviation (SD) of each subject is different and is drawn from a log-normal prior distribution. For case subjects, the marker trajectory has a nonzero probability of containing a change point (i.e., the tumor sheds the marker). The indicator grpi is used to represent whether the ith case has a change point; grpi = 1, in the presence of a change point, and grpi = 0, in the absence of a change point. If the marker trajectory has no change point, the marker trajectory is distributionally the same as that of a control. If the marker trajectory does have a change point, the marker trajectory is flat at the baseline. (The baseline mean αi shares a common normal prior distribution with those of the controls and those cases who have no change point.) The expected marker trajectory of such a case will start linearly increasing from a time point τi. Each subject has her own change point and her own rate of increase. For case i at the jth time point tij.
Priors of the parameters are:
All training case and control data were fit in the training model, and the posterior distributions of the parameters (αi, βi, τi, grpi) and the hyper parameters (α0, σα, β0, σβτ0, τ0, στ, p0) were obtained. Convergence of select hyper parameters for three chains is included in Supplemental Figure 1.
In step 2, the detection step, the case/control status of a subject is unknown. Again, the marker trajectory is modeled in the same way as in step 1, except that an additional indicator Ii representing the probability of a subject trajectory being a case. The model tries to detect multiple subjects in the detection set SD simultaneously in one model. The trajectory of the marker is represented by the following formula
In addition to modeling the marker trajectory in the detection set, the posterior distributions of the parameters (αi, βi, τi, grpi), i ∈ ST (obtained in the training step) are loaded, with the assumption that they follow the same distribution. This approach ensures the parameters of controls and cases in SD behave similarly to those in the training set. The probability for a subject to be a case is estimated by the probability for Ii=1. When this probability exceeds a detection threshold, the patient is considered a case, where the detection threshold is set such that a given level of specificity is maintained among known controls.
This single marker algorithm was implemented using WinBUGS software (18). The posterior distributions of model parameters and the probability for being a case were based on ten thousand Monte Carlo Markov Chain (MCMC) draws with a burn-in of 500 draws. The detection threshold for the posterior probability of being a case was set such that 98% specificity was maintained among 547 known controls. The 50 screen-positive cases and the 547 controls were used as the training set and the 25 screen-negative cases were used as the test set.
Results
Multiplexed Assay Development and Validation
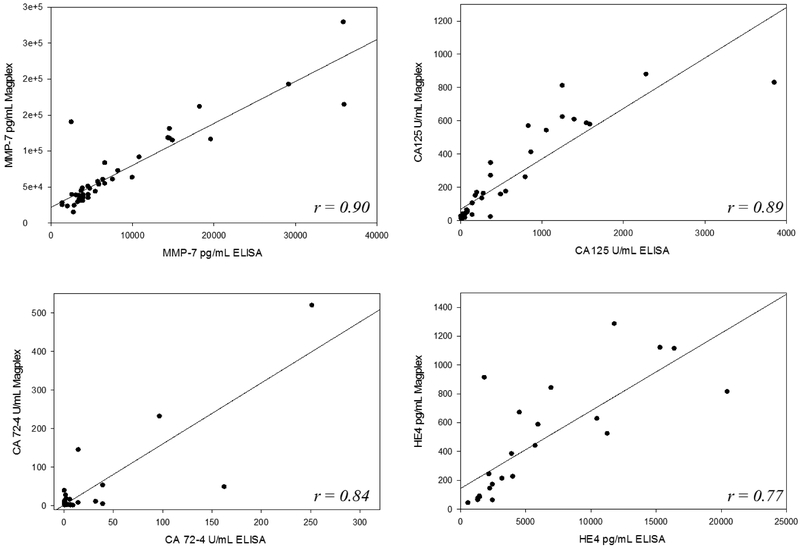
Multiplexed flow-cytometric bead-based immunoassays were established to simultaneously measure CA125, HE4, MMP-7 and CA72–4 on the Luminex Magpix system. The final optimized assay was able to utilize ultra-low volumes of serum (16 μL) to assay all four biomarkers simultaneously, critical to validation utilizing precious pre-clinical sera. The individual biomarker assays showed negligible cross-reactivity with each other, permitting implementation in a multiplexed format (Supplemental Figure 2). Effectively, the marker concentrations measured in the singleplex format correlated well with those measured in the multiplex format. Results of the inter- and intra-assay precision studies covering low, medium and high concentrations of the biomarkers in five replicates conducted over five days are reported in Table 2. The measured inter- and intra-assay precisions for the individual biomarker assays were below the mean within-person and between-person variation previously reported for these biomarkers on longitudinal samples (17) (Supplemental Table 1). This result is critical to the suitability of the assays for longitudinal patient analysis. The limits of detection (LOD) over 5 replicates at three standard deviations above the zero blank were 0.3 U/mL for CA125, 0.27 U/mL for CA72–4, 249.1 pg/mL for HE4 and 203.3 pg/mL for MMP-7. These detection limits were suitable for measuring early stage ovarian cancer samples and healthy controls. The multiplex assays, when validated against ‘gold standard’ ELISA methods utilizing early stage ovarian cancer samples (n=40) showed good correlation (Figure 1). Taken together, the validated multiplexed immunoassay methods for CA125, HE4, MMP-7 and CA72–4 was deemed suitable for simultaneous measurement of longitudinal biomarkers in retrospective ovarian cancer clinical specimens and healthy controls.
Table 2.
Multiplex Assay Analytical Validation
| Intra-assay Precision (%CV) | Inter-assay Precision (%CV) | LOD | Units | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Low | Medium | High | Avg | Low | Med | High | Avg | |||
| CA125 | 3.6% | 5.0% | 10.6% | 7.4% | 9.2% | 4.7% | 3.9% | 5.9% | 0.27 | U/mL |
| HE4 | 5.9% | 21.0% | 5.4% | 9.3% | 9.2% | 9.6% | 6.4% | 8.4% | 0.3 | U/mL |
| MMP-7 | 2.5% | 8.2% | 9.4% | 7.7% | 4.2% | 6.8% | 4.2% | 7.8% | 249 | pg/mL |
| CA72-4 | 3.3% | 14.0% | 7.6% | 8.3% | 53.1% | 15.4% | 13.7% | 27.4% | 203 | pg/mL |
Figure 1:
Comparison of multiplex immunoassays for CA125, HE4, MMP-7 and CA72-4 against ELISA methods utilizing early stage ovarian cancer patient samples show good correlation between methods.
Evaluation of the biomarker panel
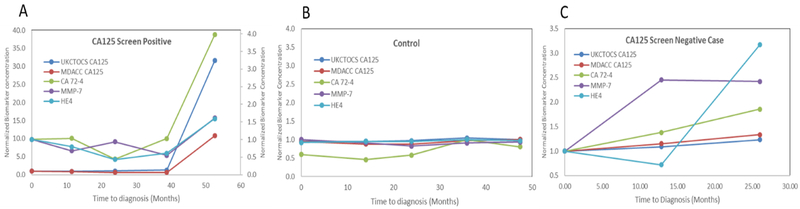
We used a nested case-control study design that involves prospective collection of specimens before outcome ascertainment from a cohort that is relevant to the clinical application and retrospective-blinded-evaluation (PRoBE design) (19). The validated multiplex immunoassays were then utilized to measure the blinded longitudinal sera from 75 women (3–5 serial samples) who later developed ovarian cancer and 547 healthy controls (1–10 serial samples) from the UKCTOCS trial. Of the 75 cases, 50 women were CA125 screen-positive and 25 women were CA125 screen-negative. The screen-positives were chosen to determine if individual biomarkers from the biomarker panel offered improved lead times over CA125. The screen-negatives were chosen to determine if cases that were missed by CA125 are identified by the other biomarkers. The screen-positives included 3–5 time points per case depending on the time of diagnosis. The time points were chosen to encompass the time of diagnosis, time of CA125 inflection, time point between CA125 inflection and diagnosis, time point 1 year prior to CA125 inflection and one time point multiple years prior to CA125 inflection. The controls were women who were enrolled in the study, but did not develop ovarian cancer and the time points were chosen to match the number of annual samples per case and age at recruitment into the trial, instead of matching at collection time points. Serial specimens for cases and controls were blocked (blinded) for analysis on the same day to mitigate the impact of inter-assay variation on longitudinal biological variation of biomarker concentrations. Unblinding of case/control status and UKCTOCS CA125 concentrations was completed by the UKCTOCS team upon completion of biomarker evaluation of the sample set. The results were reported as concentrations of individual biomarkers per case per time point. The log-biomarker concentrations for CA125, HE4, MMP-7 and CA72–4 were plotted against time to diagnosis to generate longitudinal biomarker plots. Representative longitudinal biomarker plots are shown in Figure 2 for a screen-positive case (A), screen-negative case (B) and control (C). The CA125 concentration profiles measured utilizing the bead-based multiplexed immunoassays at MD Anderson paralleled longitudinal profiles for CA125 measured in the UKCTOCS trial.
Figure 2:
Representative multi-marker longitudinal profiles of a CA125 screen-positive case (A), Control (B) and CA125 screen-negative case (C).
Longitudinal Profiles and Lead Times
The longitudinal profiles of the controls for all four biomarkers remained flat over time, indicating that each woman had her own baseline that did not vary over time, consistent with our previous observations (17).
Among the 50 CA125 screen-positive cases, where inflection in CA125 concentration interpreted with ROCA triggered TVS, one or more complementary biomarkers rose with CA125 in 22 cases, indicating co-amplification with CA125 (Supplemental Figure 3). These longitudinal profiles in preclinical sera of women destined to develop ovarian cancer suggest that multiple biomarkers may be elevated along with CA125 prior to diagnosis. The inflection point in the longitudinal profiles of HE4, CA72–4 and MMP-7 coincided with that of CA125, indicating co-amplification and reinforcement of CA125 inflection. Nevertheless, the biomarker levels were not elevated visually prior to CA125. Hence, no lead time benefits were observed for HE4, CA72–4 or MMP-7.
However, among the 25 screen-negative cases, where the women developed ovarian cancer, but no CA125 inflection was observed in the biomarker trajectory, one or more markers (CA72–4, HE4, MMP-7) rose when CA125 was not elevated, indicating possible complementarity.
Single marker algorithms for longitudinal profiling and complementarity
Since lead times were not observed, but complementarity advantages were feasible, the longitudinal profiles of the logarithmic biomarker concentration trajectories were modeled using a Bayesian approach to develop single-marker algorithms, akin to the CA125 ROCA algorithm, employed in the UKCTOCS study. Development of these algorithms have been detailed in the methods section. Where the Bayesian model predicted the longitudinal biomarker trajectory to have no change point (the inflection point), the patient’s marker trajectory was designated as a control and when the model predicted the presence of a change point, the time point of change and rate of increase were utilized to determine if the patient’s marker trajectory resembled a case.
The individual algorithms were then applied to the biomarker trajectories among the 50 screen-positives. For these 50 screen-positive cases, along with CA125, HE4 alone was elevated in 28% (14/50) of the cases, MMP-7 alone was elevated in 2% (1/50) of cases and CA 72–4 alone was elevated in 2% (1/50) of cases (Supplemental Table 2). Along with CA125, HE4 and CA72–4 together were increased in 10% (5/50) and HE4 and MMP-7 together in 2% (1/50) of cases. Among the CA125 screen-positives studied, there were no cases where all four markers were elevated together. Altogether, HE4 was elevated in 40% of screen-positives, CA 72–4 in 12% of screen-positives and MMP-7 in 4% of screen-positives.
The individual algorithms were then applied to the screen-negative cases to identify percentages of cases identified by biomarkers that were missed by CA125. At 98% specificity, 16% (4/25; 95%CI 4.5–36.1) of cases missed by CA125 were independently detected by HE4 and CA72–4, suggesting complementarity in the absence of lead time (Supplemental Table 3). MMP-7 did not identify a single screen-negative case, indicating lack of utility for identifying CA125 negative cases. There was no significant difference in the histologies represented or detected among the screen-positives and screen-negatives (Supplemental Table 4).
Discussion
Our goal in this study was to combine the increased sensitivity achieved using a multi-marker panel, with the specificity afforded by longitudinal monitoring of biomarkers using longitudinal algorithms to develop an early detection strategy for ovarian cancer. We evaluated this longitudinal multi-marker algorithm-based strategy in preclinical sera from women destined to develop ovarian cancer and in corresponding control sera from women who did not develop cancer. To determine whether a longitudinal multiple biomarker panel improved upon lead times, we evaluated screen-positives, women who were detected on annual screening in UKCTOCS as a result of an inflection in their CA125 biomarker trajectories. To find whether a multi-marker panel identified cases that are missed by CA125, we evaluated screen-negatives, women who were missed on screening trial as their CA125 was not elevated prior to diagnosis of ovarian cancer. Also, for development of such algorithms, there needs to be little variability of marker levels in healthy individuals and cases in the years prior to development of the cancer. Such a multi-marker longitudinal strategy will need to offer improved diagnostic performance at 98% specificity so that it may be utilized as a first-line screen followed by confirmatory transvaginal ultrasound.
For validation of such multi-marker panels in longitudinal sera of women destined to develop ovarian cancer and corresponding healthy controls, it was crucial to develop multiplexed assay methodologies that could allow for such validation to be completed in small volumes of valuable preclinical sera. The multiplexed xMAP bead-based methodology utilized in this study allowed us to utilize ultra-low volumes (16 μL) of preclinical serum specimens, while maintaining negligible cross-reactivity, important to measuring multiple biomarkers simultaneously. Also, extensive optimization allowed for high inter- and intra-assay precision, critical while quantifying serial biomarker measures. The biological variations can only be meaningful when the assay-related bioanalytical variations are lower in comparison. We had previously identified the normal biological variations of these biomarkers (17) and had developed multiplex immunoassays that exhibited analytical precision (%CV) below these within-individual biomarker variations, permitting careful delineation of biological perturbations in biomarker concentrations compared to assay variations. Nevertheless, between-day precision may confound serial sample measures assessed over multiple weeks or months in the course of the study. We mitigated such effects by assembling, together, the blinded serial samples from the cases and the corresponding controls at shipment from the University College London, to permit assays at MD Anderson Cancer Center on the same day. We undertook these steps to improve the reliability of retrospective longitudinal biomarker measurements from prospective specimens to derive a longitudinal algorithm reflective of healthy and cancer induced biological biomarker variations.
Development of single-marker Bayesian algorithms to model trajectories of log-concentrations of biomarkers allowed us to determine biomarker performance in screen-positive and screen-negative cases. It should be noted that due to the limited sample size of this study, a multi-marker algorithm approach was not feasible. Biomarker trajectory inflections were present only in cases, while baselines remained flat in controls permitting the development of such an algorithm. Among the screen-positives, while reinforcement of the CA125 inflection was observed with the other biomarkers, indicating co-amplification, lead times were not observed. Lead times of up to 3 years observed in preclinical specimens in a previous report, was not observed in our study (13). In the future, when such a multi-marker longitudinal algorithm is developed, a coordinate increase in two or more biomarkers may increase confidence in obtaining TVS based on a rising CA125 concentration. HE4 was the most co-amplified biomarker followed by CA72–4. MMP-7 offered the least utility among the markers chosen in screen-positives.
Among the screen-negatives, 16% of cases missed by CA125 were identified by HE4 and CA72–4 while MMP-7 offered minimal utility as a complementary biomarker and should be dropped from future studies. Considering that 20% of ovarian cancers do not express CA125, HE4 and CA72–4 clearly show promise in preclinical specimens, warranting further investigation utilizing commercial immunoanalyzers. Indeed, prospective evaluation of preclinical specimens of multiple biomarkers in the European EPIC cohort also offered improvements with multiple biomarkers combined with CA125, consistent with our results (20).
To date, very few clinical studies have explored the utility of multiple biomarkers concomitantly in longitudinal preclinical sera of women destined to develop ovarian cancer. Often, sera from a single time point are evaluated allowing lead time evaluation based on an individual measurement (21). The biomarkers chosen in this study and most other studies for early detection ovarian cancer markers are often obtained from screening multiple biomarkers in a large cohort of early stage cancers or even late stage cancers and corresponding benign and healthy controls and are then eventually validated in preclinical specimens prior to diagnosis. In order to achieve lead times and address the flaws with such a strategy, discovery paradigms will need to include samples prior to diagnosis to identify markers that will offer lead times. Based on our results, we have remedied our discovery strategies to be implemented in preclinical sera. Beyond improvements in discovery strategy, it may be possible that tumors simply do not shed antigen biomarkers in sufficient concentrations to be measured utilizing current methodologies, multiple years prior to diagnosis. A mathematical model developed for ovarian tumor growth and CA125 shedding data revealed that tumors may grow for up to a decade unnoticed before being detected by current clinical biomarker assays (22). Other biomarkers such as autoantibodies may have greater sensitivity than shed protein antigens, where small amounts of tumor associated antigen could evoke an immune response that is detectable prior to diagnosis. Indeed, autoantibodies to TP53 have been elevated, on average, 8 months prior to an increase in CA125 and 22 months prior to clinical diagnosis in patients who did not have an elevated CA125 (23).
CA125 continues to do most of the ‘heavy-lifting’ in terms of diagnostic performance, leaving a small (15–20%) window of opportunity for other biomarkers to improve upon a first line screening strategy. Whether the increased sensitivity of 16% provided by HE4 and CA72–4 can be achieved without compromising specificity will require a clinical trial. Such a trial will be sponsored by the Early Detection Research Network. The present study suggests that HE4 and CA72–4 are promising complementary biomarkers that deserve further evaluation for their ability to improve upon detection of ovarian cancer over serial measurement of CA125 alone.
Supplementary Material
Acknowledgments:
We are grateful to the UKCTOCS participants who donated their samples for use in secondary studies. The authors are solely responsible for the design of the study, the analysis and interpretation of the data, the writing of the article, and the decision to submit the article for publication. The authors acknowledge Weiqun Mao and Maojie Yang for their generous help with sample aliquoting in this study.
Grant and Philanthropic Support: This work was supported by funds from the Early Detection Research Network (5 U01 CA200462–02) and the MD Anderson Ovarian SPORE (P50 CA83639 and P50 CA217685), National Cancer Institute, Department of Health and Human Services; the Cancer Prevention Research Institute of Texas (RP101382 and RP160145); Golfer’s Against Cancer, the Mossy Foundation, the Roberson Endowment, National Foundation for Cancer Research; The K Yao Foundation; UT MD Anderson Women’s Moon Shot; and generous donations from Stuart and Gaye Lynn Zarrow. S.J. Skates received additional support from the NCI Early Detection Research Network (U01 CA152990). UKCTOCS was core funded by the Medical Research Council, Cancer Research UK, and the Department of Health with additional support from the Eve Appeal, Special Trustees of Bart’s and the London, and Special Trustees of UCLH and supported by researchers at the National Institute for Health Research University College London Hospitals Biomedical Research Centre. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Disclosure of Potential Conflicts of interest: Dr. Bast receives royalties from Fujirebio Diagnostics Inc., for the discovery of CA125. Prof Menon has shares awarded to her by UCL in Abcodia Pvt Ltd.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68(1):7–30 doi 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Patriotis C, Simmons A, Lu KH, Bast RC and Skates SJ Ovarian Cancer In: Srivastava S, editor. Biomarkers in Cancer Screening and Early Detection Chichester, UK: : John Wiley & Sons, Ltd; 2017. [Google Scholar]
- 3.Force USPST, Grossman DC, Curry SJ, Owens DK, Barry MJ, Davidson KW, et al. Screening for Ovarian Cancer: US Preventive Services Task Force Recommendation Statement. JAMA 2018;319(6):588–94 doi 10.1001/jama.2017.21926. [DOI] [PubMed] [Google Scholar]
- 4.Pignata S, Cannella L, Leopardo D, Bruni GS, Facchini G, Pisano C. Follow-up with CA125 after primary therapy of advanced ovarian cancer: in favor of continuing to prescribe CA125 during follow-up. Ann Oncol 2011;22 Suppl 8:viii40–viii4 doi 10.1093/annonc/mdr470. [DOI] [PubMed] [Google Scholar]
- 5.Rauh-Hain JA, Krivak TC, Del Carmen MG, Olawaiye AB. Ovarian cancer screening and early detection in the general population. Rev Obstet Gynecol 2011;4(1):15–21. [PMC free article] [PubMed] [Google Scholar]
- 6.Menon U, Ryan A, Kalsi J, Gentry-Maharaj A, Dawnay A, Habib M, et al. Risk Algorithm Using Serial Biomarker Measurements Doubles the Number of Screen-Detected Cancers Compared With a Single-Threshold Rule in the United Kingdom Collaborative Trial of Ovarian Cancer Screening. J Clin Oncol 2015;33(18):2062–71 doi 10.1200/JCO.2014.59.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Nagell JR Jr., DePriest PD, Reedy MB, Gallion HH, Ueland FR, Pavlik EJ, et al. The efficacy of transvaginal sonographic screening in asymptomatic women at risk for ovarian cancer. Gynecol Oncol 2000;77(3):350–6 doi 10.1006/gyno.2000.5816. [DOI] [PubMed] [Google Scholar]
- 8.Menon U, Gentry-Maharaj A, Hallett R, Ryan A, Burnell M, Sharma A, et al. Sensitivity and specificity of multimodal and ultrasound screening for ovarian cancer, and stage distribution of detected cancers: results of the prevalence screen of the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS). Lancet Oncol 2009;10(4):327–40 doi 10.1016/S1470-2045(09)70026-9. [DOI] [PubMed] [Google Scholar]
- 9.Jacobs IJ, Menon U, Ryan A, Gentry-Maharaj A, Burnell M, Kalsi JK, et al. Ovarian cancer screening and mortality in the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS): a randomised controlled trial. Lancet 2016;387(10022):945–56 doi 10.1016/S0140-6736(15)01224-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu KH, Skates S, Hernandez MA, Bedi D, Bevers T, Leeds L, et al. A 2-stage ovarian cancer screening strategy using the Risk of Ovarian Cancer Algorithm (ROCA) identifies early-stage incident cancers and demonstrates high positive predictive value. Cancer 2013;119(19):3454–61 doi 10.1002/cncr.28183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yurkovetsky ZR, Linkov FY, D EM, Lokshin AE. Multiple biomarker panels for early detection of ovarian cancer. Future Oncol 2006;2(6):733–41 doi 10.2217/14796694.2.6.733. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Z, Bast RC Jr., Yu Y, Li J, Sokoll LJ, Rai AJ, et al. Three biomarkers identified from serum proteomic analysis for the detection of early stage ovarian cancer. Cancer Res 2004;64(16):5882–90 doi 10.1158/0008-5472.CAN-04-0746. [DOI] [PubMed] [Google Scholar]
- 13.Anderson GL, McIntosh M, Wu L, Barnett M, Goodman G, Thorpe JD, et al. Assessing lead time of selected ovarian cancer biomarkers: a nested case-control study. J Natl Cancer Inst 2010;102(1):26–38 doi 10.1093/jnci/djp438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim Y, Kong L. Classification using longitudinal trajectory of biomarker in the presence of detection limits. Stat Methods Med Res 2016;25(1):458–71 doi 10.1177/0962280212460438. [DOI] [PubMed] [Google Scholar]
- 15.Fagan AM, Xiong C, Jasielec MS, Bateman RJ, Goate AM, Benzinger TL, et al. Longitudinal change in CSF biomarkers in autosomal-dominant Alzheimer’s disease. Sci Transl Med 2014;6(226):226ra30 doi 10.1126/scitranslmed.3007901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yurkovetsky Z, Skates S, Lomakin A, Nolen B, Pulsipher T, Modugno F, et al. Development of a multimarker assay for early detection of ovarian cancer. J Clin Oncol 2010;28(13):2159–66 doi 10.1200/JCO.2008.19.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simmons AR, Clarke CH, Badgwell DB, Lu Z, Sokoll LJ, Lu KH, et al. Validation of a Biomarker Panel and Longitudinal Biomarker Performance for Early Detection of Ovarian Cancer. Int J Gynecol Cancer 2016;26(6):1070–7 doi 10.1097/IGC.0000000000000737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fryback DG, Stout NK, Rosenberg MA. An elementary introduction to Bayesian computing using WinBUGS. Int J Technol Assess Health Care 2001;17(1):98–113. [DOI] [PubMed] [Google Scholar]
- 19.Pepe MS, Feng Z, Janes H, Bossuyt PM, Potter JD. Pivotal evaluation of the accuracy of a biomarker used for classification or prediction: standards for study design. J Natl Cancer Inst 2008;100(20):1432–8 doi 10.1093/jnci/djn326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Terry KL, Schock H, Fortner RT, Husing A, Fichorova RN, Yamamoto HS, et al. A Prospective Evaluation of Early Detection Biomarkers for Ovarian Cancer in the European EPIC Cohort. Clin Cancer Res 2016;22(18):4664–75 doi 10.1158/1078-0432.CCR-16-0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cramer DW, Bast RC Jr., Berg CD, Diamandis EP, Godwin AK, Hartge P, et al. Ovarian cancer biomarker performance in prostate, lung, colorectal, and ovarian cancer screening trial specimens. Cancer Prev Res (Phila) 2011;4(3):365–74 doi 10.1158/1940-6207.CAPR-10-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hori SS, Gambhir SS. Mathematical model identifies blood biomarker-based early cancer detection strategies and limitations. Sci Transl Med 2011;3(109):109ra16 doi 10.1126/scitranslmed.3003110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang WL, Gentry-Maharaj A, Simmons A, Ryan A, Fourkala EO, Lu Z, et al. Elevation of TP53 Autoantibody Before CA125 in Preclinical Invasive Epithelial Ovarian Cancer. Clin Cancer Res 2017;23(19):5912–22 doi 10.1158/1078-0432.CCR-17-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.