Abstract
How intracellular organelles acquire their characteristic sizes is a fundamental question in cell biology. Given stereotypical changes in nuclear size in cancer, it is important to understand the mechanisms that control nuclear size in human cells. Using a high‐throughput imaging RNAi screen, we identify and mechanistically characterize ELYS, a nucleoporin required for post‐mitotic nuclear pore complex (NPC) assembly, as a determinant of nuclear size in mammalian cells. ELYS knockdown results in small nuclei, reduced nuclear lamin B2 localization, lower NPC density, and decreased nuclear import. Increasing nuclear import by importin α overexpression rescues nuclear size and lamin B2 import, while inhibiting importin α/β‐mediated nuclear import decreases nuclear size. Conversely, ELYS overexpression increases nuclear size, enriches nuclear lamin B2 at the nuclear periphery, and elevates NPC density and nuclear import. Consistent with these observations, knockdown or inhibition of exportin 1 increases nuclear size. Thus, we identify ELYS as a novel positive effector of mammalian nuclear size and propose that nuclear size is sensitive to NPC density and nuclear import capacity.
Keywords: ELYS, nuclear pore complex, nuclear size, nucleocytoplasmic transport, nucleus
Subject Categories: Cell Cycle, Membrane & Intracellular Transport
Introduction
A fundamental question in cell biology is how intracellular organelles acquire their characteristic sizes. Organelle size almost certainly impacts function. For instance, expansion of the endoplasmic reticulum (ER) occurs in response to the need for increased protein synthesis or folding 1, 2, 3. Depending on cell size and cell type, Golgi and mitochondrial volumes must be sufficient to support cellular metabolic requirements 4, 5, 6, 7, 8. The functional significance of nuclear size is a particularly important problem. Changes in nuclear size are used to diagnose and stage many forms of cancer 9, 10, 11, 12, yet it is unknown if these nuclear size changes contribute to the disease pathology or result from it. Addressing this question requires a comprehensive understanding of the mechanisms that control nuclear size in normal and cancerous mammalian cells.
The nuclear envelope (NE) is composed of an enclosed double lipid bilayer that is continuous with the ER on the exterior and lined on the inside by multiple proteins that constitute the nuclear lamina, most notably nuclear lamins 13. Inserted into the NE are nuclear pore complexes (NPCs) that mediate nucleocytoplasmic transport of proteins and RNA 14, 15, 16, 17. The NPC is a large protein complex composed of multiple copies of ~30 different proteins termed nucleoporins (Nups), and distinct mechanisms are responsible for assembling NPCs into the NE after mitosis and for inserting NPCs into the NE during interphase 18, 19. Classical nuclear import is mediated by importin α/β karyopherins that transport cargos through the NPC and into the nucleoplasm where RanGTP induces cargo release through importin β binding. The three major nuclear lamins (lamin A/C, B1, and B2) are imported through this pathway. Conversely, exportins complexed with RanGTP mediate cargo export. While nuclear Ran is GTP bound due to the chromatin localization of its guanine nucleotide exchange factor (RCC1), cytoplasmically localized RanGAP converts RanGTP to RanGDP. Cytoplasmic Ran is recycled back into the nucleus by its dedicated import factor, NTF2 20, 21, 22, 23, 24.
Nuclear sizing mechanisms have been identified in a variety of model systems 11, 25, 26. In yeast, nuclear size scales with cell size and blocking nuclear export leads to an increase in nuclear size 27, 28, 29. In Xenopus egg extracts, differences in the levels of importin α and NTF2 account for nuclear size differences in two different Xenopus species 30. Over early Xenopus development, changes in cytoplasmic importin α levels and protein kinase C activity contribute to reductions in nuclear size 30, 31, 32. In C. elegans, nuclear size is sensitive to the levels of importin α, NTF2, and RCC1 33, 34. Altering lamin expression levels impacts nuclear size in Xenopus, C. elegans, and mammalian cells 34, 35, 36. Cytoskeletal elements also affect nuclear size. In confined Xenopus egg extracts, nuclear size scales with the size of microtubule asters 37. In mammalian cell culture, nuclear filamentous actin promotes nuclear growth 38, while connections between cytoplasmic actin and nesprins in the outer nuclear membrane tend to restrict nuclear growth 39, 40. Because nuclear and ER membranes are continuous, changes in ER morphology can also impact nuclear size 41, 42.
While yeast screens have been performed to identify nuclear size effectors 28, 43 and model systems such as Xenopus and C. elegans have begun to reveal some conserved mechanisms of nuclear size regulation 44, 45, 46, questions remain about how nuclear size is regulated in human cells. Beyond testing known mechanisms of nuclear size regulation in mammalian cells, imaging‐based RNAi screens offer an opportunity to identify novel nuclear size effectors 47. We have performed a high‐throughput imaging RNAi screen for nuclear size effectors in breast epithelial cells and here describe our mechanistic analysis of one candidate identified in the screen, ELYS (also known as MEL‐28 and AHCTF1), one of the first Nups recruited to chromatin for post‐mitotic NPC assembly 48, 49, 50, 51, 52. Previous work demonstrated that nuclei assembled in X. laevis egg extract failed to assemble NPCs when ELYS was immunodepleted or upon addition of a dominant negative fragment of ELYS and, as expected for import‐deficient nuclei, no nuclear growth was observed 53, 54. Here, we demonstrate that NPC densities are sensitive to ELYS protein levels in cultured mammalian cells. In turn, nuclear import capacity and nuclear size scale as a function of ELYS expression. In addition to identifying a novel modulator of nuclear size, our data suggest that NPC density and nuclear import capacity can impact nuclear size in mammalian cells.
Results
A high‐throughput imaging‐based siRNA screen identifies ELYS and SEC13 as nuclear size effectors
We carried out a high‐throughput imaging RNAi screen in a premalignant breast epithelial cell line (MCF‐10AT1k.cl2) to identify factors that affect nuclear size (Fig 1A), with an emphasis on factors whose loss results in smaller nuclei. Briefly, cells were transfected in 384‐well format with an siRNA oligo library targeting a total of 867 genes implicated in NE function, chromatin structure, and epigenetic mechanisms (for details see Materials and Methods). To minimize the frequency of both false negatives and false positives, we used the standard approach of employing three independent siRNA oligo sequences per target gene. The screen was performed in two biological replicates. As a positive control, lamin B1 (LMNB1) was knocked down to decrease nuclear size 36, and a non‐targeting siRNA was used as a negative control on each plate (Fig EV1A). 48 h after siRNA oligo transfection, cells were fixed, stained for DNA and nuclear lamins, and imaged using high‐throughput confocal microscopy (see Materials and Methods). Automated high content image analysis generated measurements of the nuclear cross‐sectional area, a reliable proxy for detecting changes in nuclear volume (see Materials and Methods) 30, 32, 42, 55. Statistical analysis of the image analysis output was performed, and genes for which silencing with at least 2 out of the 3 siRNA oligos led to a z‐score of < −1.5 (i.e., decreased nuclear size) were classified as putative nuclear size effectors (see Materials and Methods).
Figure 1. An imaging‐based siRNA screen for gene knockdowns that reduce nuclear size.
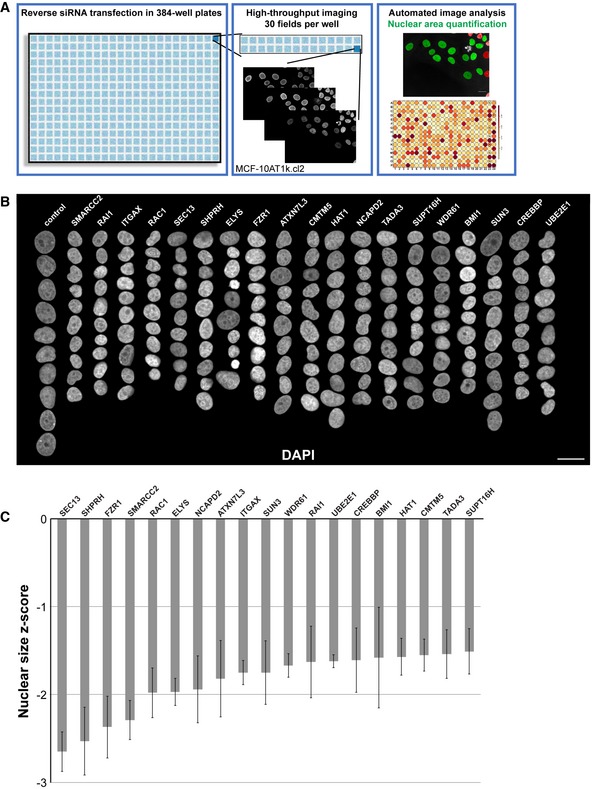
- The screening approach is depicted, and details are available in the Materials and Methods. Throughout the study, a premalignant breast cancer cell line was used (MCF‐10AT1k.cl2), unless otherwise indicated. Images and data shown in this figure were obtained directly from the screen. Scale bar, 20 μm.
- Montages of representative DAPI‐stained nuclei are shown for gene knockdowns that led to reduced nuclear size. Ten nuclei per column, organized by maximum nuclear size z‐score. Scale bar, 25 μm.
- Median nuclear size z‐scores are plotted for gene knockdowns that led to reduced nuclear size. Data are based on three different siRNA oligo sequences for each gene and two biological replicates. Error bars represent the SEM for biological replicates.
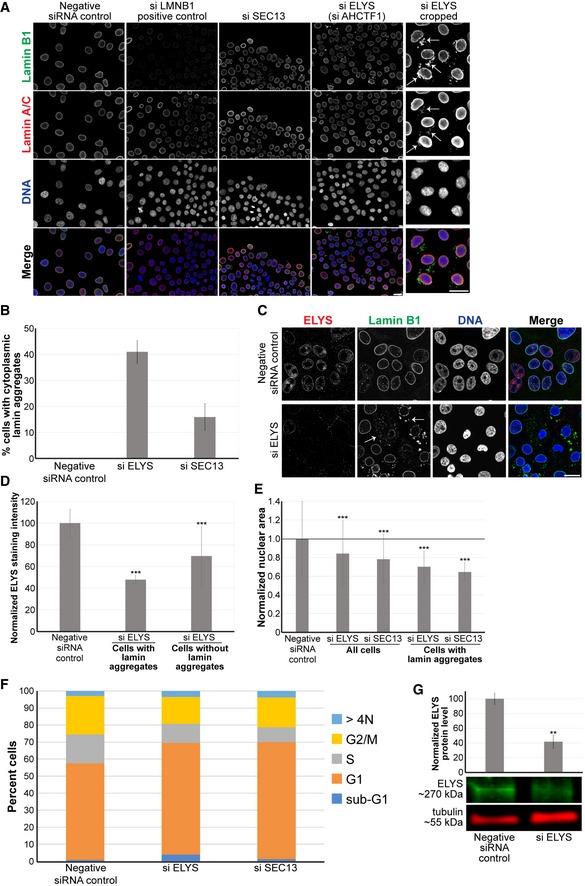
Figure EV1. ELYS and SEC13 knockdown lead to smaller nuclei and the formation of cytoplasmic lamin aggregates.
-
ARepresentative images from the screen are shown. To the far right, ELYS knockdown nuclei were cropped to highlight cytoplasmic lamin aggregates indicated with arrows.
-
BCells with and without cytoplasmic lamin B1 aggregates were counted, and the percentage of cells with aggregates was calculated. For each experiment, 698–1,213 cells were examined per condition. Data are shown for two siRNA sequences for each gene and two biological replicates.
-
C, DCells were transfected with control or ELYS siRNA and stained with ELYS and lamin B1 antibodies as indicated. Representative images are shown. Some examples of cytoplasmic lamin aggregates are indicated with arrows. For each experiment, nuclear ELYS staining intensity was quantified for 90–143 nuclei per condition and normalized to the negative control. Data from two biological replicates are shown. ELYS knockdown was greater in cells with lamin aggregates.
-
ENuclear cross‐sectional areas were quantified for 70–762 nuclei per condition, averaged, and normalized to the negative control. Six biological replicates, data from one representative experiment shown. While not all ELYS and SEC13 knockdown cells exhibited cytoplasmic lamins, those that did had smaller nuclei compared to knockdown cells without cytoplasmic lamin accumulations. This suggests that accumulation of lamins in the cytoplasm is associated with smaller nuclei.
-
FMCF‐10AT1k.cl2 cells were transfected with control siRNA or with siRNA against the indicated genes. Cells were stained with DAPI, and quantification of DNA staining intensity was used to estimate the fraction of cells in various stages of the cell cycle by high‐throughput imaging as previously described 123 (see Materials and Methods). The stacked bars represent the means of the fractions for each cell cycle phase calculated over 3 biological replicates. The control data are the same shown in Fig. 6D. Median cell number z‐scores were 0.35 (P‐value 0.62) and 0.41 (P‐value 0.34) for ELYS and SEC13 knockdown, respectively, indicating no significant effect on cell numbers.
-
GCell lysates from control and ELYS siRNA‐transfected cells were analyzed by Western blot and probed for ELYS and tubulin. One representative Western blot is shown. ELYS band intensity was normalized to tubulin. Quantification from two biological replicates is shown.
Out of 867 genes screened, knockdown of 19 resulted in decreased nuclear size with median z‐scores < −1.5 (Appendix Tables S1 and S2, Fig 1B and C). The hit rate of 2.2% indicates high specificity of the screen. Interestingly, two related Nups, SEC13 and ELYS, were the top and sixth hits with median z‐scores of −2.7 and −2.0, respectively. SEC13 and ELYS are components of the Nup107‐160 complex that has known roles in NPC assembly 56, 57, 58, 59, 60, 61, 62, 63. We were prompted to further investigate these proteins because their expression levels had not previously been implicated in nuclear size control. While nuclear transport factors are known to regulate nuclear size, less is known about how Nups might affect nuclear size. In addition, siRNA knockdown of these Nups not only induced smaller nuclei but also resulted in formation of cytoplasmic lamin puncta containing both A‐ and B‐type lamins (Fig EV1A–E). The observed effects of SEC13 and ELYS are specific and not a general property of Nups since, out of 33 Nups tested in the screen, siRNA oligos against only these two Nups decreased nuclear size. The reason for this may be because some Nups are particularly long‐lived and/or because of differences in post‐mitotic versus interphase NPC assembly (see Discussion).
While SEC13 and ELYS knockdown might be expected to have pleiotropic effects, there was no pronounced change in the cell cycle profiles and cell numbers were not affected (Fig EV1F), suggesting that observed nuclear size reductions were not indirectly due to altered cell proliferation or cell cycle progression. Because SEC13 plays dual roles in NPC assembly and protein trafficking 62, 64, we focused our subsequent analysis on ELYS. Near complete knockdown of ELYS in HeLa cells was previously reported to cause cytokinesis defects 50; however, we did not observe obvious cell cycle effects (Fig EV1F), possibly because overall ELYS protein levels were typically only reduced by ~60% (Fig EV1G) and/or because we are studying a breast epithelial cell line rather than a cancer cell line. It is likely that partial ELYS knockdown was important in allowing us to observe and characterize the nuclear size phenotype.
ELYS knockdown cells stained for various lamin types revealed that the cytoplasmic lamin puncta contain lamin B1, B2, and A/C (Fig 2A and B). Nuclear levels of lamins A/C and B1 appeared unaltered, whereas lamin B2 nuclear levels were reduced by about 30% (Fig 2C; P < 0.005). Dynamic cytoplasmic lamins were apparent by live cell imaging (Fig EV2A and Movies EV1, EV2 and EV3), and knockdown of the putative lamin protein phosphatase PPP1CA 65 largely dispersed cytoplasmic lamin puncta without affecting nuclear size (Fig EV2B–D). Thus, cytoplasmic lamin aggregates formed in ELYS knockdown cells are likely not the underlying cause of reduced nuclear size.
Figure 2. ELYS knockdown reduces NPC density and nuclear levels of lamin B2, Ran, and NTF2.
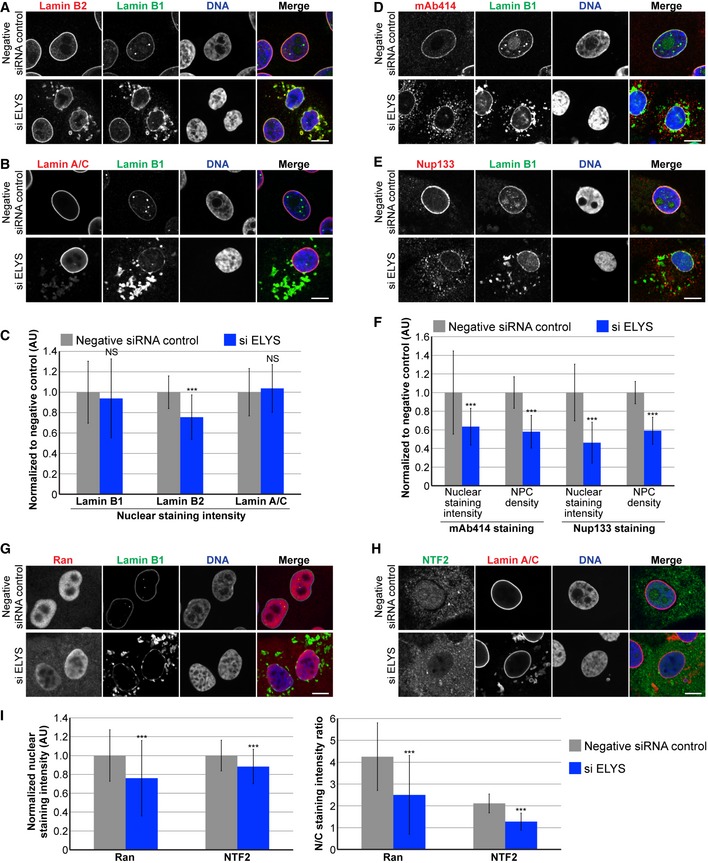
- Representative images of cells stained for lamin B2 and B1 are shown.
- Representative images of cells stained for lamin A/C and B1 are shown.
- For each experiment, nuclear lamin B1, B2, and A/C staining intensities were quantified for 44–159 nuclei per condition (96 nuclei on average) and normalized to the negative control. Three biological replicates, data from one representative experiment shown.
- Representative images of cells stained for FG‐Nups (mAb414) and lamin B1 are shown.
- Representative images of cells stained for Nup133 and lamin B1 are shown.
- For each experiment, nuclear mAb414 and Nup133 staining intensities were quantified for 31–83 nuclei per condition (60 nuclei on average) and normalized to the negative control. To measure NPC densities, confocal NE surface images were acquired for mAb414‐ and Nup133‐stained nuclei. NPC numbers were counted per unit area for 18–50 nuclei per condition (30 nuclei on average) and normalized to the negative control. Two biological replicates, data from one representative experiment shown.
- Representative images of cells stained for Ran and lamin B1 are shown.
- Representative images of cells stained for NTF2 and lamin A/C are shown.
- Nuclear Ran and NTF2 staining intensities were quantified and normalized to the negative control. To measure the N/C staining intensity ratio of a given cell, the average fluorescence intensity of a nuclear region was divided by the average fluorescence intensity of a cytoplasmic region. For each experiment, 54–216 nuclei were quantified per condition (112 nuclei on average). Two biological replicates, data from one representative experiment shown.
Figure EV2. Protein phosphatase PP1α knockdown rescues the lamin aggregation phenotype in ELYS knockdown cells.
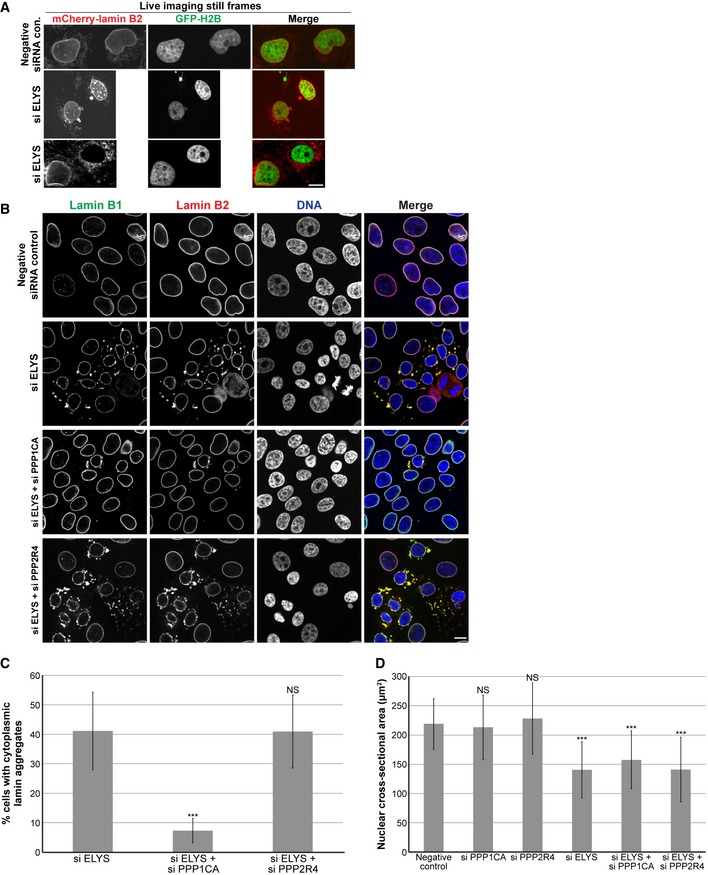
- To confirm that lamin puncta were not an artifact of fixation, we performed live cell imaging. MCF‐10AT1k.cl2 cells were co‐transfected with control or ELYS siRNA and with plasmids expressing mCherry‐lamin B2 and H2B‐GFP. Representative images of live cells are shown. Also see Movies EV1, EV2 and EV3. Cytoplasmic lamin puncta were apparent in ELYS knockdown cells but not in control cells. In addition, time‐lapse imaging showed that cytoplasmic lamin aggregates exhibited dynamic movements, often appeared immediately after mitosis regardless of whether or not the mother cell contained lamin puncta, and coalesce or dissolve during interphase. In some extreme cases, cells contained abundant cytoplasmic lamin foci but no NE‐localized mCherry‐lamin B2.
- Because lamins are known to be regulated by phosphorylation 32, 125, 126, 127, we asked whether the appearance of cytoplasmic lamin aggregates might depend on the lamin phosphorylation state. To test this hypothesis, we knocked down two putative lamin protein phosphatases, PPP1CA (protein phosphatase 1 catalytic subunit alpha) and PPP2R4 (protein phosphatase 2A activator regulatory subunit 4) 65. MCF‐10AT1k.cl2 cells were transfected with control siRNA, ELYS siRNA, ELYS siRNA plus PPP1CA siRNA, or ELYS siRNA plus PPP2R4 siRNA and stained with antibodies against lamin B1 and B2. Representative images are shown.
- Cells with and without cytoplasmic lamin B aggregates were counted and the percentage of cells with aggregates was calculated. For each experiment, 28–120 cells were examined per condition. Two biological replicates, data from one representative experiment shown. While over 40% of ELYS knockdown cells exhibited cytoplasmic lamins, less than 10% of cells co‐transfected with siRNA against ELYS and PPP1CA showed cytoplasmic lamins. Knockdown of PPP2R4 had no impact on the appearance of cytoplasmic lamin accumulations.
- For each experiment, nuclear cross‐sectional areas were quantified for 70–184 nuclei per condition (117 nuclei on average) and averaged. Two biological replicates, data from one representative experiment shown. Dissolving cytoplasmic lamin aggregates through PPP1CA knockdown did not result in a significant increase in nuclear size in ELYS knockdown cells. Thus, cytoplasmic lamin aggregates formed in ELYS knockdown cells are not sufficient to cause reduced nuclear size.
ELYS levels control NPC density and nuclear localization of Ran and NTF2
To begin to address the mechanism by which ELYS influences nuclear size, we stained cells for FG‐Nups and Nup133, another component of the Nup107‐160 complex, to assess nuclear pore number and integrity. Nuclear Nup staining intensity and NPC density were both reduced in ELYS knockdown cells (Fig 2D–F), as was nuclear localization of Ran and NTF2 (Fig 2G–I). Because NTF2 associates with the NPC and mediates Ran import 66, 67, 68, 69, 70, 71, 72, reduced levels of nuclear Ran and NTF2 likely reflect reduced NPC numbers, consistent with our observation of reduced nuclear lamin B2 (Fig 2C). We also noted cytoplasmic staining for FG‐Nups and Nup133 in ELYS knockdown cells that was not evident in control cells and did not appear to colocalize with lamin puncta (Fig 2D–E). Co‐transfections with ELYS siRNA and a plasmid expressing an siRNA‐resistant form of GFP‐ELYS rescued the effects of ELYS knockdown alone, resulting in increased nuclear size, nuclear lamin B2 staining, and NPC density as well as a dramatic reduction in the percentage of cells with cytoplasmic lamin aggregates (Fig 3). These data indicate that the phenotypes observed for ELYS knockdown cells are not due to off‐target effects.
Figure 3. ELYS overexpression increases nuclear size, NPC density, and nuclear levels of lamin B2, Ran, and NTF2.
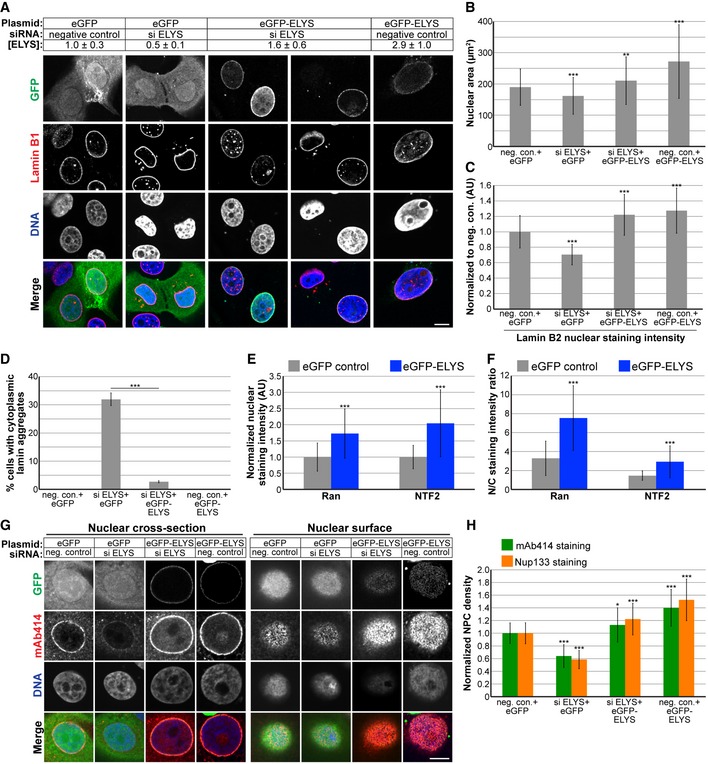
-
AMCF‐10AT1k.cl2 cells were co‐transfected with control or ELYS siRNA and plasmids expressing eGFP alone or siRNA‐resistant eGFP‐ELYS. Cell lysates were analyzed by Western blot and probed for ELYS and tubulin. ELYS band intensity was normalized to tubulin and then normalized to the eGFP/negative siRNA control. ELYS concentrations are indicated in the row labeled [ELYS] (average ± SD). Quantification was performed on two biological replicates. Cells were stained with an antibody against lamin B1. Representative images are shown.
-
BFor each experiment, nuclear cross‐sectional areas were quantified for 68–298 nuclei per condition (162 nuclei on average) and averaged. Three biological replicates, data from one representative experiment shown.
-
CFor each experiment, nuclear lamin B2 staining intensities were quantified for 29–81 nuclei per condition (60 nuclei on average) and normalized to the eGFP/negative siRNA control. Two biological replicates, data from one representative experiment shown.
-
DCells with and without cytoplasmic lamin B2 aggregates were counted, and the percentage of cells with aggregates was calculated. For each experiment, 58–299 cells were examined per condition. Three biological replicates, data from one representative experiment shown.
-
E, FCells were stained with antibodies against Ran and NTF2. For each experiment, 62–130 nuclei were quantified per condition (92 nuclei on average). Two biological replicates, data from one representative experiment shown. (E) Nuclear Ran and NTF2 staining intensities were quantified and normalized to the eGFP control. (F) To measure the N/C staining intensity ratio of a given cell, the average fluorescence intensity of a nuclear region was divided by the average fluorescence intensity of a cytoplasmic region.
-
GTransfections were performed as in (A) and cells were stained with an antibody against FG‐Nups (mAb414) and Nup133. Representative images for mAb414 staining are shown. Confocal imaging was performed through nuclear cross‐sections as well as on the surface of nuclei.
-
HTo measure NPC densities, confocal NE surface images were acquired for mAb414‐ and Nup133‐stained nuclei. NPC numbers were counted per unit area for 25–116 nuclei per condition (53 nuclei on average) and normalized to the negative control. Two biological replicates, data from one representative experiment shown.
We also examined the effect of ELYS overexpression by transfecting cells with GFP‐ELYS (Figs 3A and EV3A). An ~1.9‐fold ELYS overexpression led to a > 40% increase in nuclear cross‐sectional area with no apparent lamin aggregates (Fig 3B–D). Furthermore, NPC density was increased in ELYS overexpressing cells, along with a concomitant increase in nuclear localization of Ran, NTF2, and lamin B2 (Figs 3C–H and EV3B and C). Consistent with results from the screen, knockdown of Nup153 and the longer‐lived Nup107 did not affect nuclear size, likely because NPC numbers were not reduced to the same extent as upon knockdown of ELYS (Fig EV3D and E, see Discussion). Taken together, these data show that modulating ELYS levels affects NPC numbers and the localization of nuclear transport factors and lamins, suggesting that changes in nuclear import capacity underlie observed changes in nuclear size.
Figure EV3. ELYS overexpression, Nup153 and Nup107 knockdowns, and importin α overexpression.
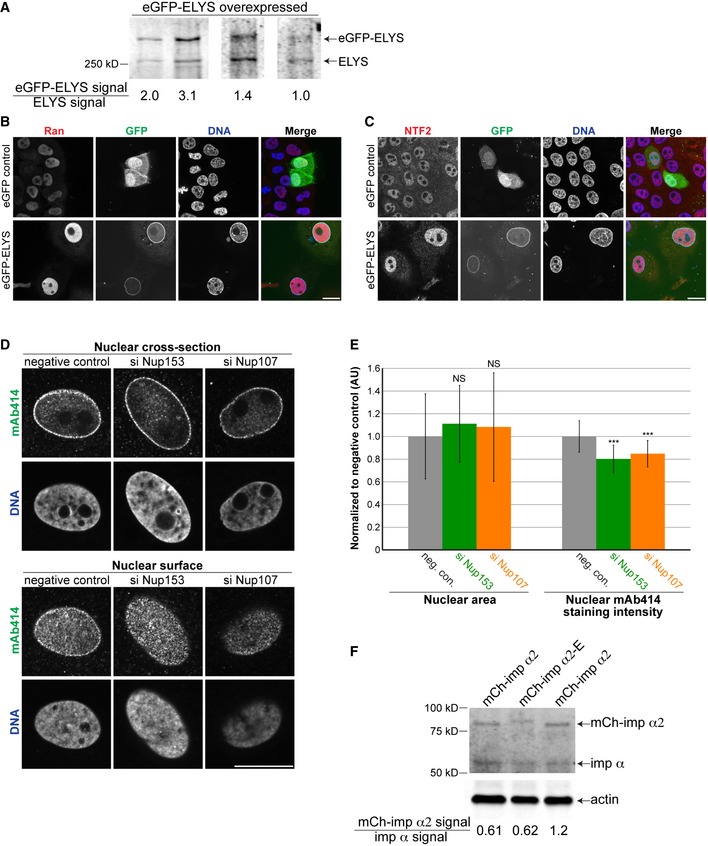
- MCF‐10AT1k.cl2 cells were transfected with a plasmid expressing eGFP‐ELYS. Cell lysates were analyzed by Western blot and probed for ELYS. The signal from the ectopically expressed eGFP‐ELYS band was divided by the signal from the endogenous ELYS band, indicated below the blots. Ectopically expressed eGFP‐ELYS was expressed 1.9 ± 0.9‐fold (average ± SD) above endogenous ELYS levels, based on four biological replicates.
- MCF‐10AT1k.cl2 cells were transfected with plasmids expressing eGFP alone or eGFP‐ELYS. Cells were stained with a Ran antibody. Representative images are shown.
- MCF‐10AT1k.cl2 cells were transfected with plasmids expressing eGFP alone or eGFP‐ELYS. Cells were stained with an NTF2 antibody. Representative images are shown.
- MCF‐10AT1k.cl2 cells were transfected with control, Nup153, or Nup107 siRNA and stained with an antibody against FG‐Nups (mAb414). Cells were co‐transfected with BLOCK‐iT Alexa Fluor red fluorescent control to identify transfected cells. Representative images are shown. Confocal imaging was performed through nuclear cross‐sections as well as on the surface of nuclei. Cytoplasmic staining for FG‐Nups apparent in ELYS knockdown cells (Fig 2D) was not evident upon knockdown of Nup153 or Nup107.
- Nuclear cross‐sectional areas were quantified for 36–49 nuclei per condition (41 nuclei on average), averaged, and normalized to the siRNA negative control. Nuclear mAb414 staining intensity was quantified for 36–49 nuclei per condition (41 nuclei on average), averaged, and normalized to the siRNA negative control. Data from three biological replicates are shown.
- MCF‐10AT1k.cl2 cells were transfected with plasmids expressing mCherry‐importin α2 or mCherry‐importin α2‐E 30. Cell lysates were analyzed by Western blot and probed for importin α and β‐actin. The signal from the ectopically expressed mCherry‐importin α2 band was divided by the signal from the endogenous importin α band, indicated below the blots. Ectopically expressed mCherry‐importin α2 was expressed at 81% ± 35% (average ± SD) of endogenous importin α levels, based on three biological replicates.
Nuclear import capacity tunes nuclear size
ELYS knockdown reduced NPC density, potentially affecting nucleocytoplasmic transport. While both nuclear import and export could be affected, reduced nuclear lamin B2 in ELYS knockdown cells suggested an import defect. Indeed, increasing bulk import in ELYS knockdown cells by importin α overexpression resulted in nuclear sizes comparable to control cells and a reduction in the percentage of cells with cytoplasmic lamins (Figs 4A–C and EV3F). These data indicate that small nuclear size and the formation of lamin aggregates in ELYS knockdown cells are due to limited nuclear import capacity. Consistent with this notion, importin α overexpression increased nuclear levels of both lamin B2 and GFP‐3x SV40 NLS, a reporter of importin α/β‐mediated nuclear import (Fig. 4D). We also observed that importin α overexpression alone resulted in a 40% increase in nuclear cross‐sectional area compared to control cells (Fig 4A and B).
Figure 4. Increasing nuclear import by overexpressing importin α rescues nuclear size and lamin aggregation phenotypes in ELYS knockdown cells.
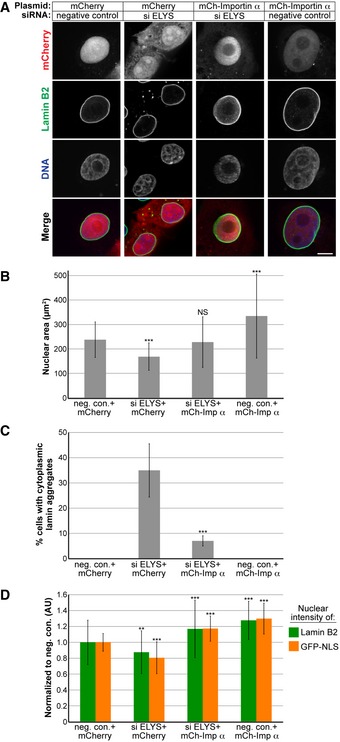
- MCF‐10AT1k.cl2 cells were co‐transfected with control or ELYS siRNA and plasmids expressing mCherry alone or mCherry‐importin α. Cells were stained with an antibody against lamin B2. Representative images are shown. Scale bar, 10 μm.
- For each experiment, nuclear cross‐sectional areas were quantified for 30–172 nuclei per condition (84 nuclei on average) and averaged. Two biological replicates, data from one representative experiment shown.
- Cells with and without cytoplasmic lamin B2 aggregates were counted, and the percentage of cells with aggregates was calculated. For each experiment, 74–218 cells were examined per condition. Two biological replicates, data from one representative experiment shown.
- For each experiment, nuclear lamin B2 staining intensities were quantified for 34–187 nuclei per condition (85 nuclei on average) and normalized to the mCherry/negative siRNA control. To measure bulk import capacity for importin α/β cargos, cells were co‐transfected as in (A) along with a plasmid expressing GFP‐3x SV40 NLS. For each experiment, nuclear GFP‐NLS intensities were quantified for 22–188 nuclei per condition (73 nuclei on average) and normalized to the mCherry/negative siRNA control. Two biological replicates, data from one representative experiment shown.
To test more generally the impact of nuclear import capacity on nuclear size, we inhibited nuclear import in two ways: (i) by expressing RanQ69L that is constitutively GTP bound and acts as a dominant negative inhibitor of importin β‐mediated nuclear import 73, 74, and (ii) by treating cells with importazole 75, a small molecule inhibitor of the interaction between importin β and RanGTP (Fig 5A and B). Both experimental approaches resulted in reduced bulk import capacity for importin α/β cargos, as expected, with concomitant reductions in nuclear size and the appearance of cytoplasmic lamin aggregates in a subset of cells (Fig 5C–E). Taken together, these data show that limiting import capacity, either by reducing NPC density or targeting nuclear transport factors, results in reduced nuclear size and the cytoplasmic accumulation of nuclear lamin proteins that have the propensity to aggregate.
Figure 5. Inhibiting nuclear import leads to smaller nuclei and cytoplasmic lamin aggregates.
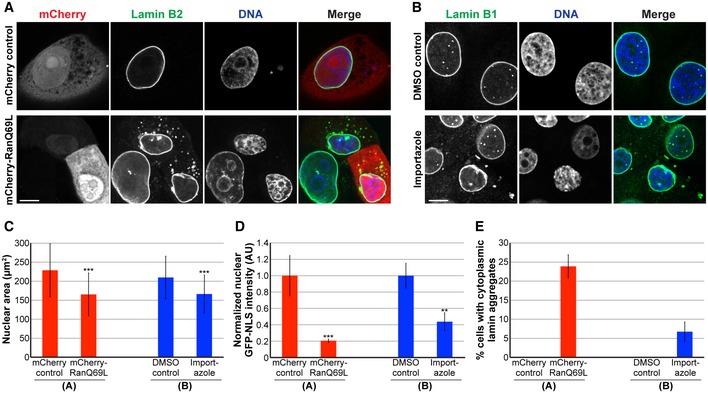
- MCF‐10AT1k.cl2 cells were transfected with plasmids expressing mCherry alone or mCherry‐RanQ69L and stained with an antibody against lamin B2. Representative images are shown.
- MCF‐10AT1k.cl2 cells were treated with 20 μM importazole or an equivalent volume of DMSO as a control for 24 h and stained with an antibody against lamin B1. Representative images are shown.
- For each experiment, nuclear cross‐sectional areas were quantified for 32–271 nuclei per condition (123 nuclei on average) and averaged. Three biological replicates, data from one representative experiment shown.
- To measure bulk import capacity for importin α/β cargos, experiments were performed as in (A and B) except that cells were also transfected with a plasmid expressing GFP‐3x SV40 NLS. For each experiment, nuclear GFP‐NLS intensities were quantified for 11–75 nuclei per condition (35 nuclei on average) and normalized to the appropriate control. Two biological replicates, data from one representative experiment shown.
- Cells with and without cytoplasmic lamin B aggregates were counted, and the percentage of cells with aggregates was calculated. For each experiment, 31–130 cells were examined per condition. Three biological replicates, data from one representative experiment shown.
Given that reducing nuclear import decreased nuclear size, we predicted that inhibiting nuclear export might lead to an increase in nuclear size. We first knocked down the nuclear export factor XPO1 76, 77, 78, which increased nuclear levels of lamins B1 and A/C and increased nuclear size (Fig 6A–C), without grossly perturbing cell cycle progression (Fig 6D). Knockdown of lamin B1 and A in combination with XPO1 slightly reduced nuclear size, but not to the size of nuclei in control cells, indicating that increased lamin import in XPO1 knockdown cells cannot fully account for the increased nuclear size (Fig 6C). Indeed, staining for total protein showed increased accumulation of nuclear proteins in XPO1‐depleted cells (Fig 6E and F). Consistent with these data, inhibition of XPO1 with the small molecule leptomycin B also increased nuclear size and total nuclear protein levels (Fig 6E and F), as observed in fission yeast 27, 28. These data suggest that blocking nuclear export leads to increased accumulation of proteins in the nucleus, potentially driving increased nuclear size. Furthermore, in human skin fibroblasts (Fig EV4A), breast adenocarcinoma cells (Fig EV4B), breast epithelial cells (Fig EV4C), and breast epithelial cells with atypical hyperplasia (Fig EV4D), XPO1 knockdown increased nuclear size while SEC13 and ELYS knockdown led to smaller nuclei (Fig EV4E). Taken together, these results are consistent with a model in which modulating protein transport across the NPC represents a key mechanism of nuclear size control. We believe this is the first demonstration in mammalian cells that nuclear transport capacity can tune nuclear size.
Figure 6. Reducing nuclear export increases nuclear size.
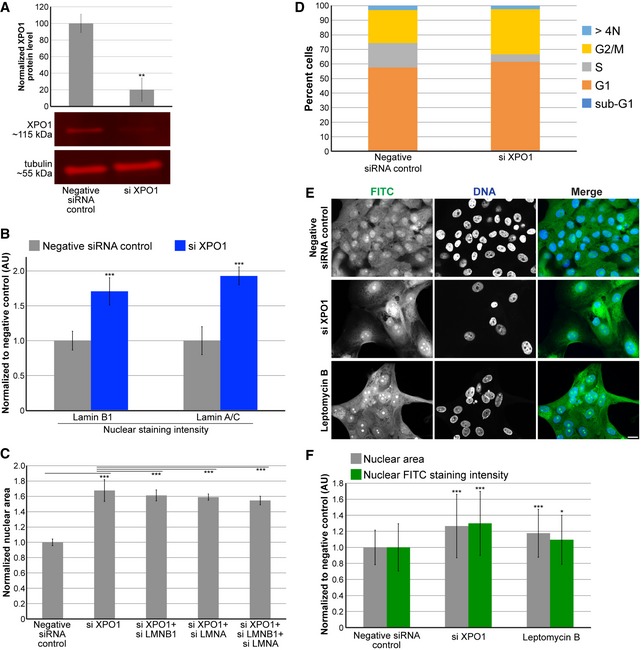
- Cell lysates from control and XPO1 siRNA‐transfected MCF‐10AT1k.cl2 cells were analyzed by Western blot and probed for XPO1 and tubulin. One representative Western blot is shown. XPO1 band intensity was normalized to tubulin. Quantification from two biological replicates is shown.
- MCF‐10AT1k.cl2 cells were transfected with control or XPO1 siRNA. Lamin B1 and lamin A/C nuclear staining intensities were quantified based on experiments using two different XPO1 siRNA sequences and two biological replicates. For each experiment, 108–865 nuclei were quantified per condition (416 nuclei on average). Average and SD values based on averages from these four experiments are plotted, normalized to the negative siRNA control.
- MCF‐10AT1k.cl2 cells were transfected with the indicated siRNAs, and nuclear cross‐sectional areas were quantified. For each experiment, 203–604 nuclei were quantified per condition (403 nuclei on average). Average and SD values based on averages from four biological replicates are plotted, normalized to the negative siRNA control.
- MCF‐10AT1k.cl2 cells were transfected with control siRNA or with siRNA against XPO1. Cells were stained with DAPI and quantification of DNA staining intensity was used to estimate the fraction of cells in various stages of the cell cycle by high‐throughput imaging as previously described 123 (see Materials and Methods). The stacked bars represent the means of the fractions for each cell cycle phase calculated over 3 biological replicates. The control data are the same shown in Fig EV1F.
- MCF‐10AT1k.cl2 cells were transfected with control or XPO1 siRNA. For the bottom row of images, cells transfected with control siRNA were treated with 20 ng/ml leptomycin B for 24 h. Cells were fixed and stained with 2 μg/ml FITC. Representative images are shown. Scale bar, 20 μm.
- For each experiment, nuclear cross‐sectional areas and nuclear FITC staining intensities were quantified for 105–620 nuclei per condition (267 nuclei on average) and normalized to the negative control. Two biological replicates, data from one representative experiment shown.
Figure EV4. ELYS, SEC13, and XPO1 knockdown in different cell lines.
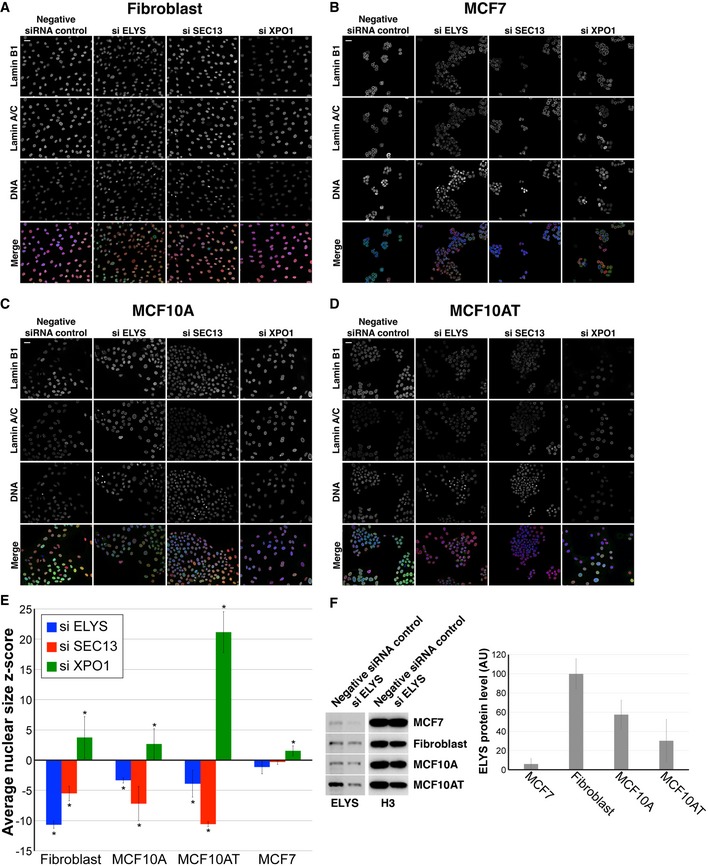
- Representative images for hTERT immortalized CRL‐1474 cells, roughly normal human skin fibroblasts.
- Representative images for MCF7 cells, breast adenocarcinoma cells.
- Representative images for MCF‐10A cells, roughly normal breast epithelial cells.
- Representative images for MCF‐10AT1k.cl2 cells, breast epithelial cells with atypical hyperplasia.
- Nuclear cross‐sectional areas were quantified, and z‐scores were calculated as described in Materials and Methods. For each cell line and condition, > 500 cells were quantified and the average z‐score and SD from three biological replicates are plotted.
- Cell lysates were analyzed by Western blot and probed for ELYS and histone H3. Band intensities were quantified, and ELYS signal was normalized to the histone H3 signal to obtain the relative ELYS protein levels in each cell line. Average data from three biological replicates are plotted. Notably, ELYS levels are lowest in MCF7 cells and ELYS was efficiently knocked down by siRNA treatment, eliminating high ELYS expression or poor ELYS knockdown as reasons for why ELYS knockdown minimally affected nuclear size in MCF7 cells.
Discussion
We have used an imaging‐based high‐throughput RNAi screen for nuclear size effectors to identify ELYS as a novel modulator of mammalian nuclear size. Out of 867 screened genes, knockdown of only 19 decreased nuclear size. Of those, two out of 33 Nups were identified (ELYS and SEC13), demonstrating specificity of the hits. Why did knockdown of only these two Nups decrease nuclear size in our screen? We found that ELYS knockdown decreased NPC numbers to a much greater extent than knockdown of other Nups, including Nup153 and Nup107. One possibility is that some long‐lived scaffold Nups, like Nup107, were not efficiently depleted over the 2‐day siRNA treatment typically used in our experiments 79, 80, 81. In line with this interpretation, twelve continuous days of Nup107 knockdown were required to reduce Nup107 protein levels to ~30% 80, while we found that ELYS levels were reduced to ~40% after only 2 days of depletion and SEC13 is known to turn over rapidly 79. Another possibility is that the effect of Nup depletion on nuclear size might depend on whether the Nup is involved in post‐mitotic versus interphase NPC assembly 80, 82. ELYS is critical for post‐mitotic NPC assembly, while nuclear import of Nup153 and subsequent recruitment of the Nup107‐160 complex is required for interphase, but not post‐mitotic, NPC assembly 82, 83. Post‐mitotic NPC assembly occurs within minutes of nuclear formation, while interphase NPC assembly is more sporadic and much slower, on the order of an hour 84, 85, 86, 87. Reducing post‐mitotic NPC assembly by ELYS depletion might influence nuclear size more strongly because those are the pores that will drive early nuclear growth. On the other hand, reducing interphase NPC assembly by Nup153 depletion might have less of an effect on NPC numbers and nuclear size, especially if post‐mitotic NPCs primarily drive the acquisition of a steady‐state size. In support of this hypothesis, blocking interphase NPC insertion had no effect on nuclear volume 88. Lastly, depletion of some Nups may have resulted in a modest reduction in nuclear size, but because we defined hits as having a z‐score < −1.5 for at least 2 out of 3 siRNA oligo sequences, we naturally focused on the stronger hits. In that sense, our screen was not saturating. It is worth noting that chromatin and epigenetic regulators were also identified as hits in our screen, and future work will focus on how changes in transcription and chromatin structure affect nuclear size.
Toward mechanism, we showed that decreasing ELYS expression levels resulted in a concomitant decrease in NPC density, importin α/β‐mediated nuclear import, and nuclear size, while ELYS overexpression increased NPC density, nuclear import, and nuclear size. Although we cannot formally exclude the possibility that ELYS levels affect the import capacity of individual NPCs, our data suggest that NPC number can limit nuclear import, thereby scaling nuclear size. Consistent with this model, we find that modulating nuclear import capacity alters nuclear size. One particularly striking observation was that importin α overexpression was sufficient to overcome the NPC density defect in ELYS knockdown cells, highlighting how import capacity might be dictated by a balance between NPC numbers and transport factor levels. Furthermore, inhibiting nuclear export in favor of import led to larger nuclei, suggesting that there is competition between import and export through limited numbers of NPCs. While the identities of the imported cargos required for nuclear growth are still unknown, nuclear lamins have previously been implicated in this process 30, 34, 35, 36, 89, 90. Our data also support a role for lamins, as their nuclear levels were decreased upon ELYS knockdown and increased upon XPO1 knockdown, and lamin knockdown mitigated some of the nuclear growth observed in XPO1‐depleted cells. In cells with reduced nuclear import, we observed cytoplasmic lamin accumulation, suggesting that relatively high lamin import kinetics are necessary to avoid cytoplasmic aggregation. These lamin aggregates could be relevant to disease states where nuclear import is compromised 91, 92, 93, 94, 95, laminopathies 96, 97, 98, 99, 100, 101, 102, or cellular stress 103.
ELYS is a multifunctional protein which has been shown to interact with chromatin, enhancers, and promoters 53, 104, 105. While we cannot eliminate the possibility that ELYS depletion affects transcription, the fact that reduced nuclear import and size resulting from ELYS depletion were rescued by importin α overexpression strongly argues that the nuclear size effects are import‐mediated. ELYS knockdown decreased nuclear lamin B2 import and nuclear size, while ELYS overexpression gave the opposite result. It seems unlikely that these reciprocal effects reflect ELYS‐mediated changes in chromatin or gene regulation. At least some of the nuclear size effects we observe must result from ELYS‐mediated effects on NPC number and import.
Our data suggest that the number of NPCs can modulate nuclear size and that nuclear transport can tune nuclear size in mammalian cells. Experiments using Xenopus egg extract and early embryos indicated that NPC numbers are not limiting for nuclear import or nuclear size 30, 86, 106, likely because nuclear import capacity is extremely high in these systems. In HeLa cells, cyclin‐dependent kinase inhibition blocked interphase NPC assembly leading to a reduction in NPC density, yet nuclear growth was unaffected 88, 107, and ELYS depletion did not significantly affect nuclear lamin localization or import capacity 108. These results might be explained by an upregulation of nuclear import that is frequently observed in cancer cells 109, 110, 111, 112. These studies highlight the importance of cell type and disease state when considering mechanisms of nuclear size control. Notably, NPC densities may dominate nuclear size control in normal somatic cells, while transport factor levels and activities may play a more important role in early development and cancer. Consistent with this idea, ELYS, SEC13, and XPO1 knockdown had variable effects on nuclear size in different cell lines (Fig EV4E). In particular, ELYS and SEC13 knockdown significantly reduced nuclear size in three roughly normal cell lines but minimally affected nuclear size in MCF7 breast cancer cells in which ELYS expression was the lowest (Fig EV4E and F).
In conclusion, using an siRNA screening approach, we have uncovered a novel mammalian nuclear size effector. Changes in nuclear size are commonly used in cancer diagnosis and prognosis, yet it is unknown whether altered nuclear size contributes to, or is simply a consequence of, the pathology. Levels of nuclear transport factors are frequently altered in cancer and have begun to be targeted for cancer treatments 10, 109, 110, 112, 113, 114, 115. Our results suggest that it may also be important to examine changes in NPC densities in cancer.
Materials and Methods
Cell culture and small molecule treatments
The MCF‐10AT1k.cl2 and MCF‐10A cell lines were obtained from the Barbara Ann Karmanos Cancer Institute 116, 117, 118. MCF‐10AT1k.cl2 cells were passaged every ~3 days and were cultured at 37°C and 5% CO2 in DMEM/F12 (1:1), 1.05 mM CaCl2, 4.9% horse serum, 10 mM HEPES, 10 μg/ml insulin, 20 ng/ml EGF, and 0.5 μg/ml hydrocortisone. MCF‐10A cells were passaged every ~5 days and were cultured at 37°C and 5% CO2 in DMEM/F12 (1:1), 1.05 mM CaCl2, 4.9% horse serum, 10 mM HEPES, 10 μg/ml insulin, 20 ng/ml EGF, 0.5 μg/ml hydrocortisone, and 0.1 μg/ml cholera toxin. We used previously described hTERT immortalized dermal fibroblast cells 119 maintained in MEM supplemented with 15% fetal bovine serum, 2 mM l‐glutamine, 1 mM sodium pyruvate, 100 U/ml penicillin, and 100 μg/ml streptomycin. The MCF7 cell line was obtained from ATCC and cultured in MEM supplemented with 100 μg/ml insulin, 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin. Cells were cultured at 37°C and 5% CO2. Importazole (MedChemExpress # HY‐101091) was dissolved in DMSO, and cells were treated with a final concentration of 20 μM for 24 h. Leptomycin B (Santa Cruz Biotechnology # sc‐358688) was dissolved in ethanol, and cells were treated with a final concentration of 20 ng/ml for 24 h.
Plasmids and siRNA sequences
The following plasmids were obtained commercially: pEGFP‐C2 (Clontech #6083‐1), pcDNA‐EGFP‐ELYS‐polyA (Addgene #59746) was a gift from Yi Zhang 120, pCMV/myc/nuc/GFP which consists of GFP fused to 3x SV40 nuclear localization signals whose nuclear import is mediated by importin α/β (Invitrogen V821‐20), pmCherry‐C1‐RanQ69L (Addgene #30309) was a gift from Jay Brenman 121, and H2B‐GFP (Addgene #11680) was a gift from Geoff Wahl 122. Plasmid pmCherry‐C2 is a derivative of pEGFP‐C2 and was a gift from Anne Schlaitz. Plasmid pmCherry‐C2‐lamin B2 (pDL24) was described previously 36. Plasmid pmCherry‐C2‐importin alpha was constructed by cloning hSRP1a into pmCherry‐C2 at EcoRI and KpnI (pDL21). The following siRNAs were ordered from Life Technologies:
ELYS (siRNA ID # s24727): sense 5′ GCGAUUGUCUGCUUACAGAtt 3′, antisense 5′ UCUGUAAGCAGACAAUCGCtc 3′ (note that this siRNA does not target ELYS in pcDNA‐EGFP‐ELYS‐polyA)
SEC13 (siRNA ID # s12662): sense 5′ CAAUUACAUCAAGAGGUUUtt 3′, antisense 5′ AAACCUCUUGAUGUAAUUGgg 3′
PPP1CA (siRNA ID # s10930): sense 5′ CAUCUAUGGUUUCUACGAUtt 3′, antisense 5′ AUCGUAGAAACCAUAGAUGcg 3′
PPP2R4 (siRNA ID # s10978): sense 5′ GGAUUCAUCCUUACCCUCAtt 3′, antisense 5′ UGAGGGUAAGGAUGAAUCCga 3′
XPO1 (siRNA ID # s14939): sense 5′ CCAAUAUUCGACUUGCGUAtt 3′, antisense 5′ UACGCAAGUCGAAUAUUGGta 3′
LMNA (siRNA ID # s8222): sense 5′ GAAGGAGGGUGACCUGAUAtt 3′, antisense 5′ UAUCAGGUCACCCUCCUUCtt 3′
LMNB1 (siRNA ID # s8225): sense 5′ GAAUCGUUGUCAGAGCCUUUU 3′, antisense 5′ AAGGCUCUGACAACGAUUCUC 3′
NUP107 was knocked down with siRNA ID #s32727. NUP153 was knocked down with siRNA ID #s19376.
Negative control No. 1 siRNA (catalog number 4390843).
Transfections
For siRNA transfection, reverse transfection was performed in 6‐ or 24‐well plates using Lipofectamine RNAiMAX (Thermo Fisher Scientific) with a final siRNA concentration of 20 nM according to the manufacturer's protocol. For plasmid transfection, reverse transfection was performed in 24‐well plates using Lipofectamine 3000 (Thermo Fisher Scientific) with 500 ng of plasmid per well according to the manufacturer's protocol. For co‐transfection of siRNA and plasmid, reverse transfection was performed in 24‐well plates using Lipofectamine 3000 (Thermo Fisher Scientific) with 500 ng of plasmid per well and a final siRNA concentration of 30 nM according to the manufacturer's protocol. In some cases, BLOCK‐iT Alexa Fluor red fluorescent control (Thermo Fisher Scientific) was used to identify transfected cells.
High‐throughput siRNA oligo transfections
MCF‐10AT1k.cl2 cells were reverse transfected with the oligo siRNA libraries (siRNA Silencer Select, Thermo Fisher), positive control (LMNB1, Thermo Fisher, s8225), and negative non‐targeting control (Thermo Fisher cat#4390847) in 384‐well imaging plates (CellCarrier‐384 PerkinElmer, 6057300). The RNAi libraries used were a custom library targeting gene products predicted to localize to the nuclear envelope (346 genes, Thermo Fisher, lot# AMO20JUZ), and an off the shelf library targeting proteins involved in epigenetic and chromatin regulation (521 genes, Thermo Fisher, cat# A30085, lot# AMO20K2X). Each gene was targeted with 3 individual siRNAs, for a total of 2601 siRNAs in the screen. Assay ready plates were prepared by first spotting 2 μl of a 400 nM solution of siRNA oligos at the bottom of a dry 384‐well plate using the MDT module of a Janus automated liquid handler (PerkinElmer) and then air‐drying under a laminar sterile air‐flow for 2 h. For cell cycle experiments (Figs 6D and EV1F) and experiments with different cell lines (Fig EV4), 150 nl of an siRNA oligo solution at 5 μM concentration was spotted at the bottom of imaging plates using an Echo525 instrument (Labcyte). The negative control No. 1 siRNA (Life Technologies, catalog number 4390843) non‐targeting siRNA control was used. The assay ready plates were then sealed and stored at −80°C until the day of the transfection, when they were thawed, equilibrated at room temperature, and centrifuged at 1,000 rpm for 20 min. 20 μl of a solution containing 0.035 μl of RNAiMax transfection reagent (Thermo Fisher, cat#13778) was dispensed in each well of the assay ready plates using a Multidrop Combi Reagent Dispenser (Thermo Fisher). The siRNA oligo/RNAiMax solution was then incubated at room temperature for 20 min. 20 μl of a cell suspension containing 1,200 cells in DMEM/F12 (1:1), 1.05 mM CaCl2, 9.8% horse serum, 10 mM HEPES, 10 μg/ml insulin, 20 ng/ml EGF, and 0.5 μg/ml hydrocortisone was then dispensed in each well of the plate using a Multidrop Combi Reagent Dispenser. The final concentration of the siRNA oligos in each well was 20 nM. Cells were then incubated for 48 h at 37°C. The screen was performed in two biological replicates on different days. Cell cycle experiments (Figs 6D and EV1F) and experiments with different cell lines (Fig EV4) were performed in three technical replicates over 3 independent biological replicates, for a total of 9 wells per experimental condition.
High‐throughput immunofluorescence
siRNA oligo‐transfected cells were fixed in 4% PFA in PBS for 20 min at room temperature, washed 3 × 5 min in PBS, permeabilized with 0.5% Triton X‐100 in PBS for 15 min, washed 3 × 5 min in PBS, and blocked in PBS with 0.05% Tween‐20 (PBST) and 5% BSA. To visualize the periphery of the cell nucleus, cells were immunostained with primary antibodies against lamin A/C (Santa Cruz, sc‐376248, mouse, 1:1,000) and lamin B1 (Santa Cruz, sc‐6217, goat, 1:500) in PBST with 1% BSA for 4 h at room temperature or overnight at 4°C. Cells were washed 3 × 5 min with PBST and incubated for one hour at room temperature with secondary antibodies diluted in 1% BSA in PBST containing DAPI (5 ng/μl). Secondary antibodies were 1:500 dilutions of Alexa Fluor 488 anti‐goat IgG (Molecular Probes, A‐11055) and Alexa Fluor 568 anti‐mouse IgG (Molecular Probes, A‐11031). Plates were then washed 3 × 5 min in PBST, sealed, and stored at 4°C until imaging. All the automated liquid handling steps necessary for high‐throughput immunofluorescence staining of the 384‐well plates were performed using a Biotek EL406 plate washer.
High‐throughput image acquisition
Image acquisition was performed using an Opera QEHS (PerkinElmer) high‐throughput dual spinning‐disk confocal microscope. Images were acquired using the 40× water immersion lens (N.A. 0.9) and two CCD cameras (1.3 MPixels) with pixel binning of 2 (Pixel size: 323 nm). For the DAPI channel, the 405‐nm laser line was used for excitation, and a 450/50‐nm bandpass filter was used for acquisition. For the lamin B1 channel, the 488‐nm laser line was used for excitation, and a 520/35‐nm bandpass filter was used for acquisition. For the lamin A channel, the 561‐nm laser line was used for excitation, and a 600/40‐nm bandpass filter was used for acquisition. All the channels included a primary excitation dichroic (405/488/561/640 nm), a primary emission dichroic longpass mirror (650/HT 660–780 nm, HR 400–640 nm), and a secondary emission dichroic shortpass mirror (568/HT 400–550 nm, HR 620–790 nm). The three channels were acquired at a single focal plane in three sequential acquisitions in 30 randomly selected fields of view per well. > 250 cells were acquired per well. For cell cycle experiments (Figs 6D and EV1F) and experiments with different cell lines (Fig EV4), a Yokogawa CV7000 high‐throughput dual spinning‐disk microscope was used. In particular, the DAPI channel was imaged in a single plane by using the CV7000 in epifluorescence mode, with a 20× air objective (NA 0.75), a 405‐nm laser as the excitation light source, a 405/488/561/640‐nm excitation dichroic mirror, a 561‐nm emission dichroic mirror, a 445/45‐nm bandpass emission filter, and a 2,550 × 2,160 pixel (5.5 MPixel) sCMOS camera with binning set to 2 (Pixel size: 650 nm). Nine randomly selected fields of view were imaged per well.
High‐throughput image analysis
The images generated by the Opera QEHS were analyzed using Columbus 2.6 (PerkinElmer). Briefly, nuclei regions of interest (ROI) were segmented using the DAPI channel, and nucleus ROIs adjacent to the image edges were excluded from subsequent image analysis steps. The area of the nucleus ROI was measured in square microns. The mean fluorescence intensity in the lamin A and lamin B1 channels for the nucleus ROI was also measured. All single cell values were then aggregated on a mean per well basis. Columbus per well results were exported as tab‐separated text files. Nuclear cross‐sectional area serves as a reliable proxy for detecting differences in nuclear volume (Fig EV5A), consistent with previous reports 30, 32, 42, 55. Furthermore, measurements of nuclear cross‐sectional areas for the same DAPI‐ and lamin B1‐stained nuclei indicate that DAPI staining provides a generally reliable method for detecting nuclear size differences (Fig EV5B). In the case of ELYS and SEC13 knockdown, DAPI staining was preferred to lamin staining because the presence of cytoplasmic lamins complicated measurements of nuclear size. For the analysis of cell cycle experiments (Figs 6D and EV1F) and experiments with different cell lines (Fig EV4), images generated by the CV7000 were imported and analyzed in Columbus 2.8.1. Nuclei were segmented as described above, and the integrated fluorescence intensity in the DAPI channel was calculated over the nucleus ROI on a per single cell basis. Analysis of cell cycle distributions obtained from single cell measurements of normalized DAPI integrated intensity was performed using R (v. 3.4.4) as previously described 123. > 650 cells per well were analyzed.
Figure EV5. Validation of nuclear size quantification methods.
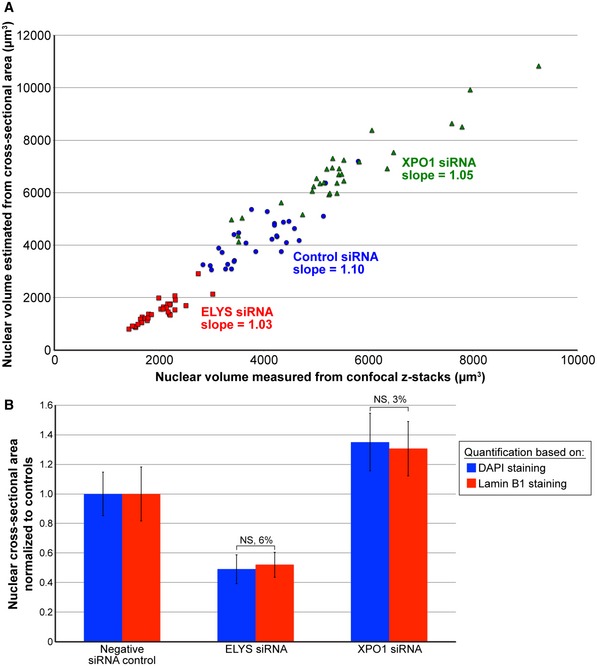
- MCF‐10AT1k.cl2 cells were transfected with control siRNA, ELYS siRNA, or XPO1 siRNA and stained with DAPI and an antibody against lamin B1. Confocal z‐stacks were acquired with a z‐slice thickness of 1 μm and a total of 21 z‐slices. Nuclear volume was quantified from the DAPI confocal z‐stacks and plotted on the x‐axis. For the same DAPI‐stained nuclei, nuclear volume was estimated from the maximum cross‐sectional nuclear area assuming a spherical nucleus and plotted on the y‐axis. Data from three biological replicates are shown. For each experimental condition, 30 nuclei were quantified, linear regression analysis was performed, and the slope of the fitted line is indicated. A t‐test calculated by comparing all measured nuclear volumes to estimated nuclear volumes returned a non‐significant P‐value of 0.25.
- Maximum nuclear cross‐sectional areas were quantified from the confocal z‐stacks described in (A) for both the DAPI and lamin B1 channels. For each experimental condition, three biological replicates were performed and 30 nuclei were quantified. For a given staining method, nuclear areas were normalized to the negative siRNA control. The percent difference between the DAPI and lamin B1 staining methods is indicated above each pair of bars.
RNAi screen statistical analysis
The statistical analysis was performed using R (v 3.3.2) and the cellHTS2 R package (v 2.36.0) 124. Per well results were normalized on a per plate basis using the B‐score method (calculation based on the siRNA oligo library samples) in the cellHTS2 package. Normalized values for each biological replicate were then scored across all the different screen plates by taking the z‐score of the B‐scores distribution for the siRNA oligo library samples. The biological replicates z‐score values were then aggregated by calculating their mean, which is the value reported for each siRNA oligo. Putative positive hits in the RNAi screen were defined as genes that showed a z‐score value of < −1.5 for at least 2 out of the 3 targeting siRNA oligos. Results for ELYS and SEC13 were validated by ordering 2 independent siRNA oligo sequences that were different from the ones used in the screen against these genes. For the validation of siRNA oligo knockdowns in different cell lines (Fig EV4E), the z‐score was calculated using the mean and the standard deviation of the non‐targeting negative control siRNA wells on the same plate. Data obtained using the high‐throughput screening approach are reported as z‐scores, a convenient way to normalize these data. For follow‐up experiments, nuclear area measurements are reported.
Immunofluorescence and antibodies
In general, cells were reverse transfected on poly‐d‐lysine‐coated glass coverslips (Electron Microscopy Sciences #72294‐04). 48 h post‐transfection, cells were washed briefly twice with PBS and fixed with 4% PFA in PBS for 20 min at room temperature. All subsequent steps were performed at room temperature. After three 5‐min PBS washes, fixed cells were permeabilized with 0.5% Triton X‐100 in PBS for 15 min at room temperature. After three 5‐min PBS washes, cells were blocked with PBS containing 0.05% Tween‐20 and 5% BSA (5% PBStBSA) for 40 min. Cells were then incubated with primary antibodies diluted in 1% PBStBSA for four hours at room temperature or 4°C overnight. After three 5‐min washes in PBSt, cells were incubated with secondary antibodies diluted in 1% PBStBSA along with 10 μg/ml Hoechst for one hour at room temperature. After three 5‐min washes in PBSt, coverslips were mounted in Vectashield (Vector Laboratories) onto glass slides and sealed with nail polish. The following primary antibodies were used: ELYS (Santa Cruz Biotechnology, # sc‐81265, mouse, 1:100), ELYS (Bethyl Laboratories, # A300‐166A, rabbit, 1:50), lamin B1 (Abcam, # ab16048, rabbit, 1:1,000), lamin B2 (GeneTex, # GTX628803, mouse, 1:500), lamin A/C (Santa Cruz Biotechnology, # sc‐376248, mouse, 1:1,000), mAb414 (BioLegend, # 902901, mouse, 1:2,000), Nup133 (Santa Cruz Biotechnology, # sc‐376763, mouse, 1:50), Ran (BD Transduction Laboratories, # 610341, mouse, 1:500), NTF2 (ABclonal, # A7057, rabbit, 1:200). Secondary antibodies were 1:500 dilutions of Alexa Fluor 488 and 568 anti‐mouse IgG (Molecular Probes, A‐11001 and A‐11004) and Alexa Fluor 488 and 568 anti‐rabbit IgG (Molecular Probes, A‐11008 and A‐11011). For FITC staining of total protein, a 10 mg/ml stock solution of FITC (Life Technologies, F1906) dissolved in DMSO was diluted to 2 μg/ml in PBS and used to stain fixed cells for one hour at room temperature.
Microscopy and image quantification
Confocal imaging was performed on a spinning‐disk confocal microscope based on an Olympus IX71 microscope stand equipped with a five‐line LMM5 laser launch (Spectral Applied Research) and switchable two‐fiber output to facilitate imaging through either a Yokogawa CSU‐X1 spinning‐disk head or TIRF illuminator. Confocal images were acquired with an EM‐CCD camera (ImagEM, Hamamatsu). Z‐axis focus was controlled using a piezo Pi‐Foc (Physik Instrumentes), and multiposition imaging was achieved using a motorized Ludl stage. Olympus objectives included UPlanFLN 40x (NA 1.30, oil) and UPlanSApo 60x (NA 1.35, oil). Image acquisition and all system components were controlled using Metamorph software. In most cases, nuclei were imaged through the largest cross‐section; however, in some cases the nuclear surface was imaged, for instance for measuring NPC densities. Images for measuring fluorescence intensity were acquired using the same exposure times. Total nuclear fluorescence intensities and cross‐sectional nuclear areas were measured from original thresholded images using Metamorph software. Nuclear sizes were generally quantified based on images of Hoechst‐stained nuclei. For NPC density measurements, images acquired on the NE surface were used to count the number of NPCs per unit area (within 5‐μm2 regions). N/C staining intensity ratios were quantified by measuring the average staining intensity within 5‐μm2 regions in the nucleus and cytoplasm and dividing the nuclear value by the cytoplasmic value. For publication, images were cropped and merged using ImageJ, but were otherwise unaltered. For live cell imaging, transfected cells were seeded onto chambered coverslips (Ibidi # 80826, μ‐Slide 8 Well ibiTreat: #1.5 polymer coverslip). Cells were imaged 36 h post‐transfection by confocal time‐lapse microscopy using objective UPlanFLN 60x (NA 0.90, air) or LCPlanFL 40x (NA 0.60, air) and a Tokai Hit stage incubator to maintain 37°C and 5% CO2. Images were acquired every 10–15 min. Time‐lapse movies were analyzed and assembled using ImageJ.
Western blots
Whole‐cell lysates from tissue culture cells were prepared using SDS–PAGE sample buffer supplemented with benzonase nuclease (Sigma, E1014) and boiled for 5 min. Proteins were separated on SDS–PAGE gels (4–20% gradient or 7%) and transferred onto PVDF membrane. Membranes were blocked in Odyssey PBS Blocking Buffer (Li‐Cor, 927‐40000). The primary antibodies used were rabbit anti‐ELYS at 1:100 (Bethyl Laboratories, # A300‐166A), mouse anti‐CRM1/XPO1 at 1:100 (Santa Cruz Biotechnology, # sc‐74454), mouse anti‐importin α at 1:500 (Sigma I1784), rabbit anti‐β‐actin at 1:1000 (RevMAb Biosciences 31‐1013‐00), and DM1A mouse anti‐α‐tubulin at 1:2000 (Santa Cruz Biotechnology, # sc‐32293). The secondary antibodies were IRDye‐680RD‐conjugated anti‐mouse‐IgG (Li‐Cor 925‐68070) and IRDye‐800CW‐conjugated anti‐rabbit‐IgG (Li‐Cor 926‐32211) used at 1:20,000. Blots were scanned on a Li‐Cor Odyssey CLx instrument, and band quantification was performed with ImageStudio, normalizing to the tubulin signal. For analysis of ELYS expression in different cell lines, cells were lysed in buffer containing 8 M urea, 10 mM Tris–HCl pH 6.8, 10% glycerol, 1% SDS, and boiled in SDS–PAGE sample buffer for 5 min. Proteins were separated on SDS–PAGE gels (4–20% gradient), transferred onto PVDF membrane, and blocked in 5% non‐fat milk in TBST. The primary antibodies used were rabbit anti‐ELYS at 1:100 (Bethyl Laboratories, # A300‐166A) and rabbit anti‐histone H3 at 1:10,000 (Abcam, #ab1791). Blots were scanned on a Bio‐Rad ChemiDoc, and band quantification was performed in ImageStudio.
Statistics
Where indicated, nuclear area and intensity measurements were normalized to controls. Averaging and statistical analysis were performed for independently repeated experiments. Two‐tailed Student's t‐tests assuming equal variances were performed in Excel (Microsoft) to evaluate statistical significance. The P‐values, number of independent experiments, number of nuclei quantified, and error bars are denoted in the figure legends.
Author contributions
Conceptualization, PJ, ACS, GP, TM, DLL; Methodology, ACS, GP, TM; Formal Analysis, GP; Investigation, PJ, ACS, GP, CCW; Writing—Original Draft, PJ, DLL; Writing—Review & Editing, PJ, ACS, CCW, GP, TM, DLL; Funding Acquisition, TM, DLL; Supervision, TM, DLL.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Movie EV1
Movie EV2
Movie EV3
Review Process File
Acknowledgements
Research in the Levy laboratory is supported by funding from the National Institutes of Health/National Institute of General Medical Sciences (R01GM113028) and the American Cancer Society (RSG‐15‐035‐01‐DDC). Research in the Misteli lab and HiTIF is supported by funding from the Intramural Research Program of the National Institutes of Health (NIH), National Cancer Institute, and Center for Cancer Research.
EMBO Reports (2019) 20: e47283
Contributor Information
Tom Misteli, Email: mistelit@mail.nih.gov.
Daniel L Levy, Email: dlevy1@uwyo.edu.
References
- 1. Federovitch CM, Ron D, Hampton RY (2005) The dynamic ER: experimental approaches and current questions. Curr Opin Cell Biol 17: 409–414 [DOI] [PubMed] [Google Scholar]
- 2. Walter P, Ron D (2011) The unfolded protein response: from stress pathway to homeostatic regulation. Science 334: 1081–1086 [DOI] [PubMed] [Google Scholar]
- 3. Schuck S (2016) On keeping the right ER size. Nat Cell Biol 18: 1118–1119 [DOI] [PubMed] [Google Scholar]
- 4. Sengupta D, Linstedt AD (2011) Control of organelle size: the Golgi complex. Annu Rev Cell Dev Biol 27: 57–77 [DOI] [PubMed] [Google Scholar]
- 5. Ferraro F, Kriston‐Vizi J, Metcalf DJ, Martin‐Martin B, Freeman J, Burden JJ, Westmoreland D, Dyer CE, Knight AE, Ketteler R et al (2014) A two‐tier Golgi‐based control of organelle size underpins the functional plasticity of endothelial cells. Dev Cell 29: 292–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rafelski SM, Viana MP, Zhang Y, Chan YH, Thorn KS, Yam P, Fung JC, Li H, Costa Lda F, Marshall WF (2012) Mitochondrial network size scaling in budding yeast. Science 338: 822–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Miettinen TP, Bjorklund M (2017) Mitochondrial function and cell size: an allometric relationship. Trends Cell Biol 27: 393–402 [DOI] [PubMed] [Google Scholar]
- 8. DuBoff B, Feany M, Gotz J (2013) Why size matters – balancing mitochondrial dynamics in Alzheimer's disease. Trends Neurosci 36: 325–335 [DOI] [PubMed] [Google Scholar]
- 9. Zink D, Fischer AH, Nickerson JA (2004) Nuclear structure in cancer cells. Nat Rev Cancer 4: 677–687 [DOI] [PubMed] [Google Scholar]
- 10. Chow KH, Factor RE, Ullman KS (2012) The nuclear envelope environment and its cancer connections. Nat Rev Cancer 12: 196–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jevtic P, Levy DL (2014) Mechanisms of nuclear size regulation in model systems and cancer. Adv Exp Med Biol 773: 537–569 [DOI] [PubMed] [Google Scholar]
- 12. Dey P (2010) Cancer nucleus: morphology and beyond. Diagn Cytopathol 38: 382–390 [DOI] [PubMed] [Google Scholar]
- 13. Dittmer TA, Misteli T (2011) The lamin protein family. Genome Biol 12: 222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rothballer A, Kutay U (2012) SnapShot: the nuclear envelope II. Cell 150: 1084.e1081 [DOI] [PubMed] [Google Scholar]
- 15. Rothballer A, Kutay U (2012) SnapShot: the nuclear envelope I. Cell 150: 868.e861 [DOI] [PubMed] [Google Scholar]
- 16. Wilson KL, Berk JM (2010) The nuclear envelope at a glance. J Cell Sci 123: 1973–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Misteli T, Spector DL (2011) The nucleus. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; [Google Scholar]
- 18. Rothballer A, Kutay U (2013) Poring over pores: nuclear pore complex insertion into the nuclear envelope. Trends Biochem Sci 38: 292–301 [DOI] [PubMed] [Google Scholar]
- 19. Hetzer MW, Wente SR (2009) Border control at the nucleus: biogenesis and organization of the nuclear membrane and pore complexes. Dev Cell 17: 606–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Madrid AS, Weis K (2006) Nuclear transport is becoming crystal clear. Chromosoma 115: 98–109 [DOI] [PubMed] [Google Scholar]
- 21. Stewart M (2007) Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol 8: 195–208 [DOI] [PubMed] [Google Scholar]
- 22. Fried H, Kutay U (2003) Nucleocytoplasmic transport: taking an inventory. Cell Mol Life Sci 60: 1659–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dickmanns A, Kehlenbach RH, Fahrenkrog B (2015) Nuclear pore complexes and nucleocytoplasmic transport: from structure to function to disease. Int Rev Cell Mol Biol 320: 171–233 [DOI] [PubMed] [Google Scholar]
- 24. Hutten S, Kehlenbach RH (2007) CRM1‐mediated nuclear export: to the pore and beyond. Trends Cell Biol 17: 193–201 [DOI] [PubMed] [Google Scholar]
- 25. Heald R, Gibeaux R (2018) Subcellular scaling: does size matter for cell division? Curr Opin Cell Biol 52: 88–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Levy DL, Heald R (2012) Mechanisms of intracellular scaling. Annu Rev Cell Dev Biol 28: 113–135 [DOI] [PubMed] [Google Scholar]
- 27. Neumann FR, Nurse P (2007) Nuclear size control in fission yeast. J Cell Biol 179: 593–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kume K, Cantwell H, Neumann FR, Jones AW, Snijders AP, Nurse P (2017) A systematic genomic screen implicates nucleocytoplasmic transport and membrane growth in nuclear size control. PLoS Genet 13: e1006767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jorgensen P, Edgington NP, Schneider BL, Rupes I, Tyers M, Futcher B (2007) The size of the nucleus increases as yeast cells grow. Mol Biol Cell 18: 3523–3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Levy DL, Heald R (2010) Nuclear size is regulated by importin alpha and Ntf2 in Xenopus. Cell 143: 288–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wilbur JD, Heald R (2013) Mitotic spindle scaling during Xenopus development by kif2a and importin alpha. eLife 2: e00290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Edens LJ, Levy DL (2014) cPKC regulates interphase nuclear size during Xenopus development. J Cell Biol 206: 473–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ladouceur AM, Dorn JF, Maddox PS (2015) Mitotic chromosome length scales in response to both cell and nuclear size. J Cell Biol 209: 645–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Meyerzon M, Gao Z, Liu J, Wu JC, Malone CJ, Starr DA (2009) Centrosome attachment to the C. elegans male pronucleus is dependent on the surface area of the nuclear envelope. Dev Biol 327: 433–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu J, Rolef Ben‐Shahar T, Riemer D, Treinin M, Spann P, Weber K, Fire A, Gruenbaum Y (2000) Essential roles for Caenorhabditis elegans lamin gene in nuclear organization, cell cycle progression, and spatial organization of nuclear pore complexes. Mol Biol Cell 11: 3937–3947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jevtic P, Edens LJ, Li X, Nguyen T, Chen P, Levy DL (2015) Concentration‐dependent effects of nuclear lamins on nuclear size in xenopus and mammalian cells. J Biol Chem 290: 27557–27571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hara Y, Merten CA (2015) Dynein‐based accumulation of membranes regulates nuclear expansion in xenopus laevis egg extracts. Dev Cell 33: 562–575 [DOI] [PubMed] [Google Scholar]
- 38. Baarlink C, Plessner M, Sherrard A, Morita K, Misu S, Virant D, Kleinschnitz EM, Harniman R, Alibhai D, Baumeister S et al (2017) A transient pool of nuclear F‐actin at mitotic exit controls chromatin organization. Nat Cell Biol 19: 1389–1399 [DOI] [PubMed] [Google Scholar]
- 39. Lu W, Schneider M, Neumann S, Jaeger VM, Taranum S, Munck M, Cartwright S, Richardson C, Carthew J, Noh K et al (2012) Nesprin interchain associations control nuclear size. Cell Mol Life Sci 69: 3493–3509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Luke Y, Zaim H, Karakesisoglou I, Jaeger VM, Sellin L, Lu W, Schneider M, Neumann S, Beijer A, Munck M et al (2008) Nesprin‐2 Giant (NUANCE) maintains nuclear envelope architecture and composition in skin. J Cell Sci 121: 1887–1898 [DOI] [PubMed] [Google Scholar]
- 41. Anderson DJ, Hetzer MW (2008) Reshaping of the endoplasmic reticulum limits the rate for nuclear envelope formation. J Cell Biol 182: 911–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jevtic P, Levy DL (2015) Nuclear size scaling during Xenopus early development contributes to midblastula transition timing. Curr Biol 25: 45–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cantwell H, Nurse P (2019) A systematic genetic screen identifies essential factors involved in nuclear size control. PLoS Genet 15: e1007929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vukovic LD, Jevtic P, Edens LJ, Levy DL (2016) New insights into mechanisms and functions of nuclear size regulation. Int Rev Cell Mol Biol 322: 1–59 [DOI] [PubMed] [Google Scholar]
- 45. Mukherjee RN, Chen P, Levy DL (2016) Recent advances in understanding nuclear size and shape. Nucleus 7: 167–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jevtic P, Levy DL (2018) Elucidating nuclear size control in the Xenopus model system. Vet Glas 72: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pegoraro G, Misteli T (2016) High‐throughput imaging. Methods 96: 1–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gillespie PJ, Khoudoli GA, Stewart G, Swedlow JR, Blow JJ (2007) ELYS/MEL‐28 chromatin association coordinates nuclear pore complex assembly and replication licensing. Curr Biol 17: 1657–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Galy V, Askjaer P, Franz C, Lopez‐Iglesias C, Mattaj IW (2006) MEL‐28, a novel nuclear‐envelope and kinetochore protein essential for zygotic nuclear‐envelope assembly in C. elegans . Curr Biol 16: 1748–1756 [DOI] [PubMed] [Google Scholar]
- 50. Rasala BA, Orjalo AV, Shen Z, Briggs S, Forbes DJ (2006) ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proc Natl Acad Sci USA 103: 17801–17806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bilokapic S, Schwartz TU (2013) Structural and functional studies of the 252 kDa nucleoporin ELYS reveal distinct roles for its three tethered domains. Structure 21: 572–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gomez‐Saldivar G, Fernandez A, Hirano Y, Mauro M, Lai A, Ayuso C, Haraguchi T, Hiraoka Y, Piano F, Askjaer P (2016) Identification of conserved MEL‐28/ELYS domains with essential roles in nuclear assembly and chromosome segregation. PLoS Genet 12: e1006131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rasala BA, Ramos C, Harel A, Forbes DJ (2008) Capture of AT‐rich chromatin by ELYS recruits POM121 and NDC1 to initiate nuclear pore assembly. Mol Biol Cell 19: 3982–3996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Franz C, Walczak R, Yavuz S, Santarella R, Gentzel M, Askjaer P, Galy V, Hetzer M, Mattaj IW, Antonin W (2007) MEL‐28/ELYS is required for the recruitment of nucleoporins to chromatin and postmitotic nuclear pore complex assembly. EMBO Rep 8: 165–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vukovic LD, Jevtic P, Zhang Z, Stohr BA, Levy DL (2016) Nuclear size is sensitive to NTF2 protein levels in a manner dependent on Ran binding. J Cell Sci 129: 1115–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Walther TC, Alves A, Pickersgill H, Loiodice I, Hetzer M, Galy V, Hulsmann BB, Kocher T, Wilm M, Allen T et al (2003) The conserved Nup107‐160 complex is critical for nuclear pore complex assembly. Cell 113: 195–206 [DOI] [PubMed] [Google Scholar]
- 57. Belgareh N, Rabut G, Bai SW, van Overbeek M, Beaudouin J, Daigle N, Zatsepina OV, Pasteau F, Labas V, Fromont‐Racine M et al (2001) An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J Cell Biol 154: 1147–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vasu S, Shah S, Orjalo A, Park M, Fischer WH, Forbes DJ (2001) Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export. J Cell Biol 155: 339–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Harel A, Orjalo AV, Vincent T, Lachish‐Zalait A, Vasu S, Shah S, Zimmerman E, Elbaum M, Forbes DJ (2003) Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol Cell 11: 853–864 [DOI] [PubMed] [Google Scholar]
- 60. Loiodice I, Alves A, Rabut G, Van Overbeek M, Ellenberg J, Sibarita JB, Doye V (2004) The entire Nup107‐160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol Biol Cell 15: 3333–3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Krull S, Thyberg J, Bjorkroth B, Rackwitz HR, Cordes VC (2004) Nucleoporins as components of the nuclear pore complex core structure and Tpr as the architectural element of the nuclear basket. Mol Biol Cell 15: 4261–4277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Enninga J, Levay A, Fontoura BM (2003) Sec13 shuttles between the nucleus and the cytoplasm and stably interacts with Nup96 at the nuclear pore complex. Mol Cell Biol 23: 7271–7284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Siniossoglou S, Wimmer C, Rieger M, Doye V, Tekotte H, Weise C, Emig S, Segref A, Hurt EC (1996) A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell 84: 265–275 [DOI] [PubMed] [Google Scholar]
- 64. Stagg SM, Gurkan C, Fowler DM, LaPointe P, Foss TR, Potter CS, Carragher B, Balch WE (2006) Structure of the Sec13/31 COPII coat cage. Nature 439: 234–238 [DOI] [PubMed] [Google Scholar]
- 65. Thompson LJ, Bollen M, Fields AP (1997) Identification of protein phosphatase 1 as a mitotic lamin phosphatase. J Biol Chem 272: 29693–29697 [DOI] [PubMed] [Google Scholar]
- 66. Paschal BM, Gerace L (1995) Identification of NTF2, a cytosolic factor for nuclear import that interacts with nuclear pore complex protein p62. J Cell Biol 129: 925–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Clarkson WD, Corbett AH, Paschal BM, Kent HM, McCoy AJ, Gerace L, Silver PA, Stewart M (1997) Nuclear protein import is decreased by engineered mutants of nuclear transport factor 2 (NTF2) that do not bind GDP‐Ran. J Mol Biol 272: 716–730 [DOI] [PubMed] [Google Scholar]
- 68. Clarkson WD, Kent HM, Stewart M (1996) Separate binding sites on nuclear transport factor 2 (NTF2) for GDP‐Ran and the phenylalanine‐rich repeat regions of nucleoporins p62 and Nsp1p. J Mol Biol 263: 517–524 [DOI] [PubMed] [Google Scholar]
- 69. Smith A, Brownawell A, Macara IG (1998) Nuclear import of Ran is mediated by the transport factor NTF2. Curr Biol 8: 1403–1406 [DOI] [PubMed] [Google Scholar]
- 70. Bayliss R, Leung SW, Baker RP, Quimby BB, Corbett AH, Stewart M (2002) Structural basis for the interaction between NTF2 and nucleoporin FxFG repeats. EMBO J 21: 2843–2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bayliss R, Ribbeck K, Akin D, Kent HM, Feldherr CM, Gorlich D, Stewart M (1999) Interaction between NTF2 and xFxFG‐containing nucleoporins is required to mediate nuclear import of RanGDP. J Mol Biol 293: 579–593 [DOI] [PubMed] [Google Scholar]
- 72. Morrison J, Yang JC, Stewart M, Neuhaus D (2003) Solution NMR study of the interaction between NTF2 and nucleoporin FxFG repeats. J Mol Biol 333: 587–603 [DOI] [PubMed] [Google Scholar]
- 73. Ren M, Drivas G, D'Eustachio P, Rush MG (1993) Ran/TC4: a small nuclear GTP‐binding protein that regulates DNA synthesis. J Cell Biol 120: 313–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Bischoff FR, Klebe C, Kretschmer J, Wittinghofer A, Ponstingl H (1994) RanGAP1 induces GTPase activity of nuclear Ras‐related Ran. Proc Natl Acad Sci USA 91: 2587–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Soderholm JF, Bird SL, Kalab P, Sampathkumar Y, Hasegawa K, Uehara‐Bingen M, Weis K, Heald R (2011) Importazole, a small molecule inhibitor of the transport receptor importin‐beta. ACS Chem Biol 6: 700–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Stade K, Ford CS, Guthrie C, Weis K (1997) Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90: 1041–1050 [DOI] [PubMed] [Google Scholar]
- 77. Fornerod M, Ohno M, Yoshida M, Mattaj IW (1997) CRM1 is an export receptor for leucine‐rich nuclear export signals. Cell 90: 1051–1060 [DOI] [PubMed] [Google Scholar]
- 78. Ossareh‐Nazari B, Bachelerie F, Dargemont C (1997) Evidence for a role of CRM1 in signal‐mediated nuclear protein export. Science 278: 141–144 [DOI] [PubMed] [Google Scholar]
- 79. D'Angelo MA, Raices M, Panowski SH, Hetzer MW (2009) Age‐dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell 136: 284–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Doucet CM, Talamas JA, Hetzer MW (2010) Cell cycle‐dependent differences in nuclear pore complex assembly in metazoa. Cell 141: 1030–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Rabut G, Doye V, Ellenberg J (2004) Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat Cell Biol 6: 1114–1121 [DOI] [PubMed] [Google Scholar]
- 82. Otsuka S, Ellenberg J (2018) Mechanisms of nuclear pore complex assembly – two different ways of building one molecular machine. FEBS Lett 592: 475–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Vollmer B, Lorenz M, Moreno‐Andres D, Bodenhofer M, De Magistris P, Astrinidis SA, Schooley A, Flotenmeyer M, Leptihn S, Antonin W (2015) Nup153 recruits the Nup107‐160 complex to the inner nuclear membrane for interphasic nuclear pore complex assembly. Dev Cell 33: 717–728 [DOI] [PubMed] [Google Scholar]
- 84. Haraguchi T, Koujin T, Hayakawa T, Kaneda T, Tsutsumi C, Imamoto N, Akazawa C, Sukegawa J, Yoneda Y, Hiraoka Y (2000) Live fluorescence imaging reveals early recruitment of emerin, LBR, RanBP2, and Nup153 to reforming functional nuclear envelopes. J Cell Sci 113(Pt 5): 779–794 [DOI] [PubMed] [Google Scholar]
- 85. Dultz E, Zanin E, Wurzenberger C, Braun M, Rabut G, Sironi L, Ellenberg J (2008) Systematic kinetic analysis of mitotic dis‐ and reassembly of the nuclear pore in living cells. J Cell Biol 180: 857–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. D'Angelo MA, Anderson DJ, Richard E, Hetzer MW (2006) Nuclear pores form de novo from both sides of the nuclear envelope. Science 312: 440–443 [DOI] [PubMed] [Google Scholar]
- 87. Otsuka S, Bui KH, Schorb M, Hossain MJ, Politi AZ, Koch B, Eltsov M, Beck M, Ellenberg J (2016) Nuclear pore assembly proceeds by an inside‐out extrusion of the nuclear envelope. Elife 5: e19071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Maeshima K, Iino H, Hihara S, Funakoshi T, Watanabe A, Nishimura M, Nakatomi R, Yahata K, Imamoto F, Hashikawa T et al (2010) Nuclear pore formation but not nuclear growth is governed by cyclin‐dependent kinases (Cdks) during interphase. Nat Struct Mol Biol 17: 1065–1071 [DOI] [PubMed] [Google Scholar]
- 89. Newport JW, Wilson KL, Dunphy WG (1990) A lamin‐independent pathway for nuclear envelope assembly. J Cell Biol 111: 2247–2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Jenkins H, Holman T, Lyon C, Lane B, Stick R, Hutchison C (1993) Nuclei that lack a lamina accumulate karyophilic proteins and assemble a nuclear matrix. J Cell Sci 106(Pt 1): 275–285 [DOI] [PubMed] [Google Scholar]
- 91. Freibaum BD, Lu Y, Lopez‐Gonzalez R, Kim NC, Almeida S, Lee KH, Badders N, Valentine M, Miller BL, Wong PC et al (2015) GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature 525: 129–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Jovicic A, Mertens J, Boeynaems S, Bogaert E, Chai N, Yamada SB, Paul JW III, Sun S, Herdy JR, Bieri G et al (2015) Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci 18: 1226–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, Daley EL, Miller SJ, Cunningham KM, Vidensky S et al (2015) The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 525: 56–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Gasset‐Rosa F, Chillon‐Marinas C, Goginashvili A, Atwal RS, Artates JW, Tabet R, Wheeler VC, Bang AG, Cleveland DW, Lagier‐Tourenne C (2017) Polyglutamine‐expanded huntingtin exacerbates age‐related disruption of nuclear integrity and nucleocytoplasmic transport. Neuron 94: 48–57.e44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Grima JC, Daigle JG, Arbez N, Cunningham KC, Zhang K, Ochaba J, Geater C, Morozko E, Stocksdale J, Glatzer JC et al (2017) Mutant huntingtin disrupts the nuclear pore complex. Neuron 94: 93–107.e106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Fairley EA, Riddell A, Ellis JA, Kendrick‐Jones J (2002) The cell cycle dependent mislocalisation of emerin may contribute to the Emery‐Dreifuss muscular dystrophy phenotype. J Cell Sci 115: 341–354 [DOI] [PubMed] [Google Scholar]
- 97. Janin A, Bauer D, Ratti F, Millat G, Mejat A (2017) Nuclear envelopathies: a complex LINC between nuclear envelope and pathology. Orphanet J Rare Dis 12: 147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Prokocimer M, Davidovich M, Nissim‐Rafinia M, Wiesel‐Motiuk N, Bar DZ, Barkan R, Meshorer E, Gruenbaum Y (2009) Nuclear lamins: key regulators of nuclear structure and activities. J Cell Mol Med 13: 1059–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Davidson PM, Lammerding J (2014) Broken nuclei–lamins, nuclear mechanics, and disease. Trends Cell Biol 24: 247–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Burke B, Stewart CL (2014) Functional architecture of the cell's nucleus in development, aging, and disease. Curr Top Dev Biol 109: 1–52 [DOI] [PubMed] [Google Scholar]
- 101. Worman HJ, Ostlund C, Wang Y (2010) Diseases of the nuclear envelope. Cold Spring Harb Perspect Biol 2: a000760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Wu D, Flannery AR, Cai H, Ko E, Cao K (2014) Nuclear localization signal deletion mutants of lamin A and progerin reveal insights into lamin A processing and emerin targeting. Nucleus 5: 66–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Zhang K, Daigle JG, Cunningham KM, Coyne AN, Ruan K, Grima JC, Bowen KE, Wadhwa H, Yang P, Rigo F et al (2018) Stress granule assembly disrupts nucleocytoplasmic transport. Cell 173: 958–971.e917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Zierhut C, Jenness C, Kimura H, Funabiki H (2014) Nucleosomal regulation of chromatin composition and nuclear assembly revealed by histone depletion. Nat Struct Mol Biol 21: 617–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Pascual‐Garcia P, Debo B, Aleman JR, Talamas JA, Lan Y, Nguyen NH, Won KJ, Capelson M (2017) Metazoan nuclear pores provide a scaffold for poised genes and mediate induced enhancer‐promoter contacts. Mol Cell 66: 63–76.e66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Theerthagiri G, Eisenhardt N, Schwarz H, Antonin W (2010) The nucleoporin Nup188 controls passage of membrane proteins across the nuclear pore complex. J Cell Biol 189: 1129–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Maeshima K, Iino H, Hihara S, Imamoto N (2011) Nuclear size, nuclear pore number and cell cycle. Nucleus 2: 113–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Mimura Y, Takagi M, Clever M, Imamoto N (2016) ELYS regulates the localization of LBR by modulating its phosphorylation state. J Cell Sci 129: 4200–4212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Mahipal A, Malafa M (2016) Importins and exportins as therapeutic targets in cancer. Pharmacol Ther 164: 135–143 [DOI] [PubMed] [Google Scholar]
- 110. Kau TR, Way JC, Silver PA (2004) Nuclear transport and cancer: from mechanism to intervention. Nat Rev Cancer 4: 106–117 [DOI] [PubMed] [Google Scholar]
- 111. Poon IK, Jans DA (2005) Regulation of nuclear transport: central role in development and transformation? Traffic 6: 173–186 [DOI] [PubMed] [Google Scholar]
- 112. Cagatay T, Chook YM (2018) Karyopherins in cancer. Curr Opin Cell Biol 52: 30–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Kau TR, Silver PA (2003) Nuclear transport as a target for cell growth. Drug Discov Today 8: 78–85 [DOI] [PubMed] [Google Scholar]
- 114. Kim J, McMillan E, Kim HS, Venkateswaran N, Makkar G, Rodriguez‐Canales J, Villalobos P, Neggers JE, Mendiratta S, Wei S et al (2016) XPO1‐dependent nuclear export is a druggable vulnerability in KRAS‐mutant lung cancer. Nature 538: 114–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Dickmanns A, Monecke T, Ficner R (2015) Structural basis of targeting the exportin CRM1 in cancer. Cells 4: 538–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Heppner GH, Wolman SR (1999) MCF‐10AT: a model for human breast cancer development. Breast J 5: 122–129 [DOI] [PubMed] [Google Scholar]
- 117. Dawson PJ, Wolman SR, Tait L, Heppner GH, Miller FR (1996) MCF10AT: a model for the evolution of cancer from proliferative breast disease. Am J Pathol 148: 313–319 [PMC free article] [PubMed] [Google Scholar]
- 118. McElwee JL, Mohanan S, Griffith OL, Breuer HC, Anguish LJ, Cherrington BD, Palmer AM, Howe LR, Subramanian V, Causey CP et al (2012) Identification of PADI2 as a potential breast cancer biomarker and therapeutic target. BMC Cancer 12: 500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Scaffidi P, Misteli T (2011) In vitro generation of human cells with cancer stem cell properties. Nat Cell Biol 13: 1051–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Inoue A, Zhang Y (2014) Nucleosome assembly is required for nuclear pore complex assembly in mouse zygotes. Nat Struct Mol Biol 21: 609–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Kazgan N, Williams T, Forsberg LJ, Brenman JE (2010) Identification of a nuclear export signal in the catalytic subunit of AMP‐activated protein kinase. Mol Biol Cell 21: 3433–3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Kanda T, Sullivan KF, Wahl GM (1998) Histone‐GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr Biol 8: 377–385 [DOI] [PubMed] [Google Scholar]
- 123. Zane L, Chapus F, Pegoraro G, Misteli T (2017) HiHiMap: single‐cell quantitation of histones and histone posttranslational modifications across the cell cycle by high‐throughput imaging. Mol Biol Cell 28: 2290–2302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Boutros M, Bras LP, Huber W (2006) Analysis of cell‐based RNAi screens. Genome Biol 7: R66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Heald R, McKeon F (1990) Mutations of phosphorylation sites in lamin A that prevent nuclear lamina disassembly in mitosis. Cell 61: 579–589 [DOI] [PubMed] [Google Scholar]
- 126. Mall M, Walter T, Gorjanacz M, Davidson IF, Nga Ly‐Hartig TB, Ellenberg J, Mattaj IW (2012) Mitotic lamin disassembly is triggeredby lipid‐mediated signaling. J Cell Biol 198: 981–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Hatch E, Hetzer M (2014) Breaching the nuclear envelope in development and disease. J Cell Biol 205: 133–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Movie EV1
Movie EV2
Movie EV3
Review Process File


