Abstract
The full gene sequence encoding for the Trypanosoma equiperdum ortholog of the cAMP-dependent protein kinase (PKA) regulatory (R) subunits was cloned. A poly-His tagged construct was generated [TeqR-like(His)8], and the protein was expressed in bacteria and purified to homogeneity. The size of the purified TeqR-like(His)8 was determined to be ~57,000 Da by molecular exclusion chromatography indicating that the parasite protein is a monomer. Limited proteolysis with various proteases showed that the T. equiperdum R-like protein possesses a hinge region very susceptible to proteolysis. The recombinant TeqR-like(His)8 did not bind either [3H] cAMP or [3H] cGMP up to concentrations of 0.40 and 0.65 µM, respectively, and neither the parasite protein nor its proteolytically generated carboxy-terminal large fragments were capable of binding to a cAMP-Sepharose affinity column. Bioinformatics analyses predicted that the carboxy-terminal region of the trypanosomal R-like protein appears to fold similarly to the analogous region of all known PKA R subunits. However, the protein amino-terminal portion seems to be unrelated and shows homology with proteins that contained Leu-rich repeats, a folding motif that is particularly appropriate for protein-protein interactions. In addition, the three-dimensional structure of the T. equiperdum protein was modeled using the crystal structure of the bovine PKA RIα subunit as template. Molecular docking experiments predicted critical changes in the environment of the two putative nucleotide binding clefts of the parasite protein, and the resulting binding energy differences support the lack of cyclic nucleotide binding in the trypanosomal R-like protein.
Keywords: Trypanosoma equiperdum, Regulatory subunits of the cAMP-dependent protein kinase, Gene cloning, Protein expression in bacteria, Protein purification, Biochemical characterization, Dourine, Trypanosomatid parasites
1. Introduction
Salivarian trypanosomes are intravascular, extracellular kinetoplastid haemoprotozoan parasites of a wide range of mammals including humans. Three species, Trypanosoma brucei, Trypanosoma evansi and Trypanosoma equiperdum, conform the subgenus Trypanozoon. T. equiperdum infects horses and other equids under natural conditions, and causes a venereal disease called dourine [1,2].
Cyclic AMP appears to be crucial in Kinetoplastida. For instance, T. brucei possess more than 80 different genes for adenylyl cyclases (ACs), the enzyme responsible for synthesizing cAMP from ATP [3–5]. In higher eukaryotes, the majority of the cAMP signals are mediated via the activation of the cAMP-dependent protein kinase (aka protein kinase A or PKA) and subsequent phosphorylation of various downstream factors, such as transcription factors, signal transduction components, cytoskeletal proteins or metabolic enzymes [6,7]. In most organisms, PKA is an inactive holoenzyme consisting of two catalytic (C) and two regulatory (R) subunits. PKA activation is achieved by the release of the C subunits from the holoenzyme complex caused by cAMP-induced conformational changes in the R subunits.
Complete or draft genomes of various trypanosomatids have been reported, providing both a huge achievement for trypanosome biology and an opportunity to consider a large amount of questions at the genome level [8–11]. Trypanosomatids possess a large set of protein kinases, comprising approximately 2% of each genome, and suggesting a key role for phosphorylation in parasite biology [12]. The AGC group of kinases is named after the PKA, the cGMP-dependent protein kinase or PKG, and protein kinase C (PKC) families, which are cytoplasmic serine/threonine ATP:-phosphotransferase enzymes that are regulated by cAMP (PKA), cGMP (PKG), or Ca2+ and diacylglycerol (PKC). The parasite genomes encode three AGC kinases that are related to the C subunits of PKA (PKAC1, PKAC2, and PKAC3), and a gene sharing high homology to the R subunits of PKA. We have identified a ~57-kDa polypeptide band corresponding to the PKA R subunit-like protein from the T. equiperdum TeAp-N/D1 isolate [13]. Antibody cross-reactivity clearly demonstrated the high level of conservation exhibited between the parasite protein and its mammalian counterparts [13]. Here, the T. equiperdum PKA R-like protein was cloned, expressed in bacteria, purified to homogeneity and biochemically characterized. Similar to higher eukaryotic PKA R subunits [14–16], this novel parasite protein possessed a hinge region very susceptible to proteolysis with a variety of proteases. However, the T. equiperdum PKA R-like protein differed from other PKA R subunits since it was not dimeric and was not capable of binding cyclic nucleotides.
2. Materials and methods
2.1. Materials
Reagents were purchased from the following sources: [2,8-3H] cAMP (33.0 Ci/mmol), [8-3H] cGMP (13.5 Ci/mmol), Perkin-Elmer NEN® Radiochemicals; anti-mouse IgG horseradish peroxidase conjugate, SuperSignal™ West Pico chemiluminescent substrate, Thermo Scientific; mouse monoclonal anti-His tag antibody, antimouse IgG alkaline phosphatase conjugate, benzamidine, iodoacetamide, phenyl methyl sulfonyl fluoride (PMSF), 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF), pepstatin, leupeptin, N-α-tosyl-L-lysinyl-chloromethyl ketone hydrochloride (TLCK), N-p-tosyl-L-phenylalaninyl chloromethyl ketone (TPCK), ampicillin, kanamycin, fibrous DEAE-cellulose, DEAE-Sepharose, isopropyl-1-thio-β-d-galactopyranoside (IPTG), imidazole, complete and incomplete Freund’s adjuvant, lipid A, acetylmuramyl-alanyl-isoglutamine (muramyl dipeptide), bovine serum albumin (BSA), cGMP, TPCK-treated trypsin, TLCK-treated chymotrypsin, thermolysin, Staphylococcus aureus V8 protease, papain, cysteine, Sigma; 8-(2-aminoethylamino) adenosine 3’,5’-cyclic monophosphate (8-AEA cAMP), Biolog; N-hydroxysuccinimide (NHS)-activated Sepharose 4 fast flow, Superdex S75 gel filtration column, Superdex 200 gel filtration column, Mono Q HR 16/10 anionic exchange column, GE Healthcare; nitrocellulose (0.45 mm pore size), Pierce; 5-bromo-4-chloro-3-indolyl phosphate (BCIP), nitro blue tetrazolium (NBT), Wizard® DNA extraction kit, Promega; OptiPhase Hisafe II (scintillation liquid), LKB; pQETrySystem vector, Qiagen; Yeast extract and tryptone (YT) medium, Luria-Bertani (LB) medium, MP Biomedicals; Probond™ nickel-chelating resin, Mark12™ unstained standard (molecular weight protein markers), BenchMark™ pre-stained protein ladder, precast NuPAGE® Novex 4e12% Bis-Tris sodium dodecyl sulfate (SDS) gels, Life Technologies; 96-well equilibrium dialyzer with a molecular weight cut off of 10,000 Da, Harvard Apparatus. All other chemicals were of the highest quality grade available.
2.2. Parasites
Cryopreserved samples of T. equiperdum from the Venezuelan TeAp-N/D1 isolate (aka TEVA1) were expanded in adult Sprague Dawley albino rats [17]. When the number of parasites reached ≥ 106 trypanosomes/ml, the blood was extracted from the rats by cardiac puncture using 0.5 M EDTA as anticoagulant. Parasites were purified by anion exchange chromatography using a fibrous DEAE-cellulose column [18].
2.3. Cloning of a T. equiperdum ortholog of the PKA R subunits
T. equiperdum genomic DNA was obtained using the Wizard® DNA extraction kit, according to the instructions of the manufacturer. Primers were designed to amplify the coding region of the gene for the PKA R-like protein from T. equiperdum genomic DNA by means of the polymerase chain reaction (PCR): a forward primer (5’-ACC ATG GCT GAA AAG GGA ACA TCG T −3’) with an annealing temperature of 68 °C and a Nco I restriction site (underlined), and a reverse primer (5’-CCT CGA GCG ACT TCC TCC CCT CTG CCC TTA −3’) with an annealing temperature of 74 °C and a Xho I restriction site (underlined) were utilized. The PCR reaction parameters were standard (Stage 1: 94 °C for 2 min; Stage 2: 35 cycles of 94 °C for 15 s, 68 °C for 20 s and 68 °C for 2 min; Stage 3: 1 cycle at 68 °C for 5 min). The PCR amplified PKA R-like protein coding region was ligated into the pQE-TrySystem vector, previously digested with the Nco I and Xho I restriction enzymes, upstream of an 8 × His tag region from the vector. The identity of the construct was confirmed by means of diagnostic restriction analysis and DNA sequencing. The amplified coding region sequence of the T. equiperdum PKA R-like protein has been lodged in the NCBI GenBank database (https://www.ncbi.nlm.nih.gov/genbank/) with the Accession No. KJ636459 (NCBI ID: AID53025.1).
2.4. Construction of a site-directed mutant of the T. equiperdum ortholog of the PKA R subunits tagged with 8 His residues at its COOH-terminal [TeqR-like(His)8]
A poly-His tagged construct of the T. equiperdum PKA complete R-like protein was built by modifying its stop codon to a tryptophan codon, and incorporating two additional amino acids followed by 8 His at its COOH-terminus. Thus, the following residues are present at the COOH-terminal end of the generated site-directed TeqR-like(His)8 mutant: Trp-Leu-Glu-His-His-His-His-His-His-His-His. The gene sequence for the poly-His tagged R-like protein was then inserted in the pQE-TriSystem vector for its expression in bacterial cells. Escherichia coli competent cells (strain M15[pREP4]) were transformed with the TeqR-like(His)8 DNA construct. Expression of the TeqR-like(His)8 protein in bacteria was evaluated by western blotting using commercial monoclonal anti-His tag antibodies.
2.5. eterologous overproduction of the TeqR-like(His)8 protein
YT broth cultures (50 ml) prepared in the presence of 100 mg/ml ampicillin and 50 mg/ml kanamycin were inoculated with a single colony of E. coli M15[pREP4] containing the gene sequence for TeqR-like(His)8 inserted in the pQE-TriSystem vector, and bacteria cells were grown overnight at 37 °C with shaking. The overnight cultures were diluted into 12 flasks of fresh YT broth (1 L each) containing 100 mg/ml ampicillin and 50 mg/ml kanamycin and grown at 37 °C with shaking until cultures reached mid-log phase (OD600 nm 0.6–0.7). The expression of TeqR-like(His)8 was initiated by the addition of IPTG to a final concentration of 0.5 mM and grown at 16e17 °C with shaking. The broth cultures were allowed to grow for 16–24 h before harvesting the cells by centrifugation (6000×g) for 30 min at 4 °C. Bacteria pellets were stored at − 20 °C until further use.
2.6. Purification of the TeqR-like(His)8 protein
The TeqR-like(His)8 protein was purified by combining the following chromatography steps: 1) a ProBond nickel-chelating affinity resin, 2) two sequential separations throughout a Super-dex 200 gel filtration column, and 3) a Mono Q anionic exchange column. During the purification steps, the purity and integrity of the TeqR-like(His)8 protein was analyzed using SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and western blots revealed with anti-polyHis antibodies.
2.6.1. Affinity chromatography
Bacteria containing the overexpressed TeqR-like(His)8 protein were resuspended in 300 ml of native lysis buffer [50 mM sodium phosphate (pH 8.0), 500 mM NaCl, 5 mM dithiothreitol (DTT), 10 mM benzamidine, 0.4 mM AEBSF, 1 mM pepstatin, 1 mM leupeptin, 28 µM TPCK and 28 µM TLCK] and were disrupted by passing the solution twice through a Microfluidizer high shear fluid processor (model M-110P, Microfluidics, Westwood, USA). The cellular debris was harvested by means of centrifugation (12,000×g) for 30 min at 4°C and the supernatant was added to a Probond™ nickel-chelating resin (3 ml). Binding was allowed to occur overnight with gentle agitation at 4 °C. Following pelleting of the beads at 500×g for 5 min, the supernatant (flow through fraction) was removed and the sedimented beads were washed four times with three column volumes of native lysis buffer. The resin was poured into a column, and washed with native lysis buffer containing 20 mM imidazole. Finally, the TeqR-like(His)8 protein was eluted with native lysis buffer containing 250 mM imidazole.
2.6.2. Gel filtration chromatography
Fractions from the nickel-charged affinity resin containing the TeqR-like(His)8 protein were pooled, concentrated down to 2 ml, and loaded onto a Superdex 200 gel filtration column using a BioLogic DuoFlow™ chromatography system (Bio-Rad, Hercules, USA) with a 5 ml injection loop. The column was equilibrated and eluted with gel filtration buffer [50 mM 2-(N-morpholino)ethanesulfonic acid (MES, pH 5.8), 200 mM NaCl, 2 mM EDTA, 2 mM EGTA and 5 mM DTT]. The peak containing the separated TeqR-like(His)8 protein was pooled and re-chromatographed on the same Superdex 200 gel filtration column under identical conditions.
2.6.3. Ion exchange chromatography
Fractions from the Superdex 200 gel filtration column were pooled, concentrated down to 2 ml, and loaded on a Mono Q HR 16/10 anionic exchange column using a BioLogic DuoFlow™ low pressure liquid chromatography instrument with a 5 ml injection loop. The column was equilibrated with a low salt buffer [20 mM Tris-HCl (pH 8.0), 2 mM EDTA and 5 mM b-mercaptoethanol]. After injecting the sample containing the TeqR-like(His)8 protein, the column was initially thoroughly washed with the low salt buffer, and then the bound protein was eluted using a 200-ml salt gradient from the low salt buffer to a high salt buffer [20 mM Tris-HCl (pH 8.0), 500 mM NaCl, 2 mM EDTA and 5 mM b-mercaptoethanol].
2.7. Tandem mass spectrometry (MS/MS) analysis of TeqR-like(His)8 tryptic peptides
The purified TeqR-like(His)8 protein (5 µg) was digested with trypsin at a protein:enzyme ratio of 100:1. The resulting peptides were separated using Magic 2002 high-performance liquid chromatography (HPLC) system (Michrom BioResources, Inc.) and eluted into a LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific, San Jose, CA) using electrospray ionization.
2.8. Circular dichroism (CD) spectroscopy
CD measurements were performed with an Aviv 202 spectropolarimeter (Aviv Biomedical, Lakewood, USA). All the spectra were collected at a constant temperature of 25.1 °C.
2.9. Proteolysis of TeqR-like(His)8
Proteolysis was carried out at 4 °C or 37 °C for 30 min in 0.05 M ammonium bicarbonate (pH 8.3) using various ratios of protease to substrate (0, 1:500, 1:200, 1:100 and 1:50 w/w). The following proteases were used: trypsin, chymotrypsin, thermolysin, S. aureus V8 protease, and papain. Papain was preactivated by incubation for 30 min at 4 °C with 1.1 mM EDTA, 0.067 mM b-mercaptoethanol and 5.5 mM Cys. If the protein was run immediately on polyacrylamide gels, an appropriate aliquot of the sample buffer was added, and the tubes were placed immediately in a boiling water bath for 5 min. The samples were then loaded onto the polyacrylamide gels. For large scale proteolysis and subsequent isolation of the proteolytic fragments, the reaction was terminated by injecting the sample on a Superdex 200 or a Superdex 75 gel filtration column using a BioLogic low pressure liquid chromatography equipment and the same conditions described above.
2.10. Cyclic nucleotide binding assays
Cyclic nucleotide binding was measured by equilibrium dialysis. Equilibrium dialyses were carried out on a single plate Rotator (Harvard Apparatus, Holliston, USA) using a 96-well equilibrium dialyzer with a molecular weight cut off of 10,000 Da. Binding assays were performed in 50 mM Tris-HCl (pH 7.5), 2 mM EDTA, 5 mM b-mercaptoethanol, 1 mg/ml BSA, 30 nM of the purified TeqR-Like(His)8 protein and increasing concentrations of [3H] cAMP (0e400 nM) or [3H] cGMP (0–650 nM). The binding reaction mixture (200 µl) containing protein and cyclic nucleotide was placed in one side of the dialysis membrane, and 200 ml of just buffer [50 mM Tris-HCl (pH 7.5), 2 mM EDTA, 5 mM 2-mercaptoethanol and 1 mg/ml BSA] was placed on the opposite side. After an overnight incubation (16 h) at 4 °C, a 20-ml aliquot of each side was transferred into scintillation vials and counted in 6 ml of OptiPhase HiSafe II. Cyclic nucleotide binding to the protein was determined by subtraction of the free radioactive reagent. All reactions were performed in duplicates.
2.11. Preparation of polyclonal antibodies against the purified recombinant TeqR-like(His)8 protein
The purified recombinant TeqR-like(His)8 protein was used to produce polyclonal antibodies in ascitic fluid from 10 female BalbC mice. We pre-immunized the mice twice (at 7 and 9 weeks of age) with lipid A and muramyl dipeptide as adjuvants, under conditions that did not involve ascites fluid formation. Then, when the mice were 10 weeks old, we followed the standard protocol described by Tung et al. [19].
The animal protocol employed for the preparation of the antibodies was designed to minimize pain or discomfort to the mice, and was performed following the policies stated in the “National Institutes of Health guide for the care and use of laboratory animals (NIH Publications No. 8023, revised 1978)”.
2.12. Production and purification of recombinant bovine PKA RIα subunit, human PKA RIβ subunit, and mouse PKA Cα subunit
Escherichia coli BL21 (DE3) cells were transformed with the expression vectors pRSETB-RIa (containing the gene for the wildtype full-length Bos taurus PKA RIα subunit); pRSETB-RIβ (containing the gene for the wild-type full-length Homo sapiens PKA RIβ subunit); or pRSETB-Cat (containing the gene for the wild-type fulllength Mus musculus Ca subunit). RIα, RIβ and Cα subunits were expressed in bacteria using LB medium in the presence of ampicillin (100 mg/ml). Bacteria cells were grown at 37 °C with shaking until cultures reached mid-log phase (OD600 nm 0.6–0.7). The expression of the proteins was initiated by the addition of IPTG (0.5 mM) and cell growth was maintained with shaking. Broth cultures were allowed to grow overnight before harvesting the cells by centrifugation (6000×g) for 30 min at 4 °C.
R subunit isotypes were purified as previously described [20–22] using 8-AEA cAMP-Sepharose resin and cGMP elution. The Ca subunit was expressed and purified according to Gangal et al. [23]. Either peak I, which contains four phosphorylated residues (Ser10, Ser139, Thr197, and Ser338) or peak II, which contains three phosphorylation sites (Ser10, Thr197, and Ser338), was used to form the RIβ2C2 holoenzyme. Briefly, for holoenzyme regeneration, the human RIβ subunit was dialyzed with a 5% molar excess of mouse C subunit in 10 mM MES (pH 6.5), 50 mM NaCl, 5 mM MgCl2, 0.5 mM ATP, and 5 mM DTT. Following RIβ2C2 reconstitution, the remaining C subunit was removed by gel filtration chromatography on a Superdex 200 column.
2.13. Molecular docking
Modeling of cAMP and cGMP binding to both putative CNB domains of the TeqR-like protein was performed using the Auto-Dock Vina program [24]. Docking to the CNB B was chosen first given that this domain corresponds to the high affinity/low dissociation cAMP binding site in mammalian PKA R subunits. A homology model of the T. equiperdum protein was generated using the crystal structure of the PKA RIα subunit homodimer from B. taurus (PDB ID: 4MX3 [25]) as a framework template, and the SWISSMODEL server [26]. As a control, a model was also built for the PKA RIa subunit from B. taurus complexed with (Rp)-adenosine 3’, 5’-cyclic monophosphothioate (PDB ID: 1NE4, [27]. Since all cyclic nucleotide binding assays were carried out at pH 7.5, the protonation state of the ionizable residues was calculated using the web server H [28]. Ligand binding affinity was estimated by predicting empirical binding energy differences.
2.14. Other procedures
Protein concentration was determined using BSA as protein standard [29]. SDS-PAGE was carried out as reported by Laemmli [30] on 1.5-mm thick precast NuPAGE® Bis-Tris slab SDS gels containing a gradient of 4e12% polyacrylamide. Coomasie Blue R-250 was used for protein visualization on gels. For western blot analyses, proteins separated by SDS-PAGE were electrotransferred from the gels to nitrocellulose sheets as described by Towbin et al. [31]. To prepare 8-AEA cAMP-Sepharose, 100 mmol of 8-AEA cAMP was coupled to 25 ml of NHS-activated Sepharose fast flow using the protocol supplied by the vendor.
3. Results
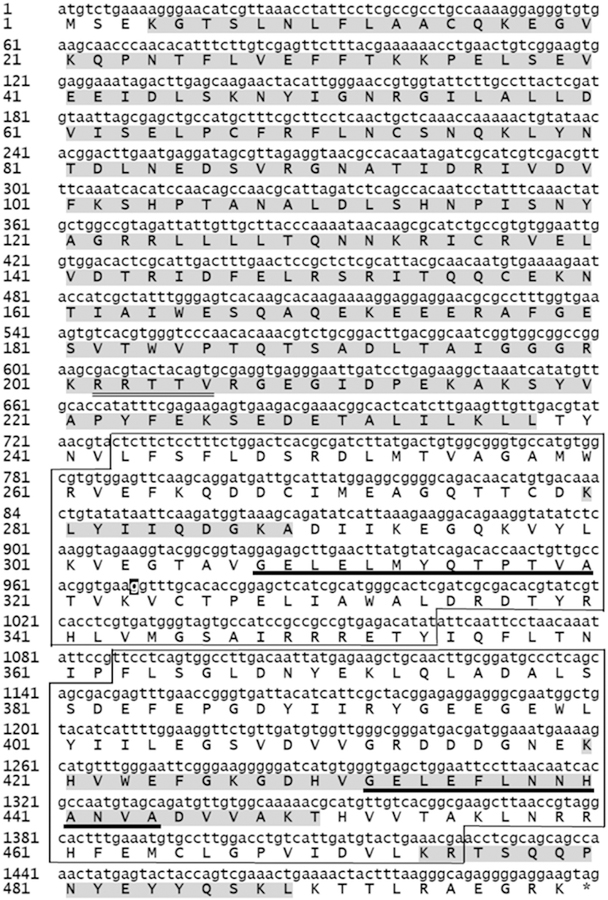
The DNA database of the T. b. brucei genome project was used to obtain the complete gene sequence encoding for the T. b. brucei ortholog of the PKA R subunits. On the basis of predicted homologies between T. brucei and T. equiperdum genes, appropriate oligonucleotides were designed to amplify the corresponding gene from the genomic DNA of T. equiperdum. The resulting PCR fragment was cloned and verified by sequencing. Fig. 1 illustrates that the gene for the T. equiperdum PKA R-like protein only possesses one nucleotide difference with respect to the corresponding sequence of the T. brucei Tb11.02.2210 PKA R-like protein [Tb927.11.4610 from the TriTrypDB database (http://tritrypdb.org); and accession No. AF326975.1, NCBI ID: AAG49383.1 from the NCBI GenBank database (https://www.ncbi.nlm.nih.gov/genbank/)]. Recently, a first draft of the T. equiperdum genome has been reported [11] [Accesion No. CZPT02000000 (https://www.ncbi.nlm.nih.gov/nuccore/)]; however, the last version lodged in the NCBI Nucleotide database does not include the gene encoding for the TeqR-like protein cloned here. The translated amino acid sequence of the parasite R-like protein is also shown in Fig. 1. At the protein level, the T. equiperdum protein was 100% identical to the T. brucei protein. The open reading frame of the T. equiperdum protein predicts an acidic protein (pI 5.24) of 499 amino acids, with a calculated molecular weight of 56,734.3. As also shown in Fig. 1, the trypanosomal R-like protein contains the inhibitor or pseudosubstrate site that in other eukaryotes is involved in binding and inactivation of the PKA C subunits, and two carboxy-terminally located putative cyclic nucleotide binding (CNB) domains containing their presumptive phosphate binding cassette (PBC) consensus motifs.
Fig. 1. Gene and amino acid sequence of the T. equiperdum R-like protein.
The single nucleotide difference between the T. equiperdum and the T. brucei PKA R-like protein genes is highlighted in black (nucleotide 969). Highlighted in grey is the sequence information gathered by MS/MS analysis of the tryptic peptides from the recombinant Teq R-like protein (Table SI). A potential PKA consensus phosphorylation sequence that resembles the inhibitor or pseudosubstrate site is double underlined. The putative CNB domains and PBC sites are boxed and underlined, respectively.
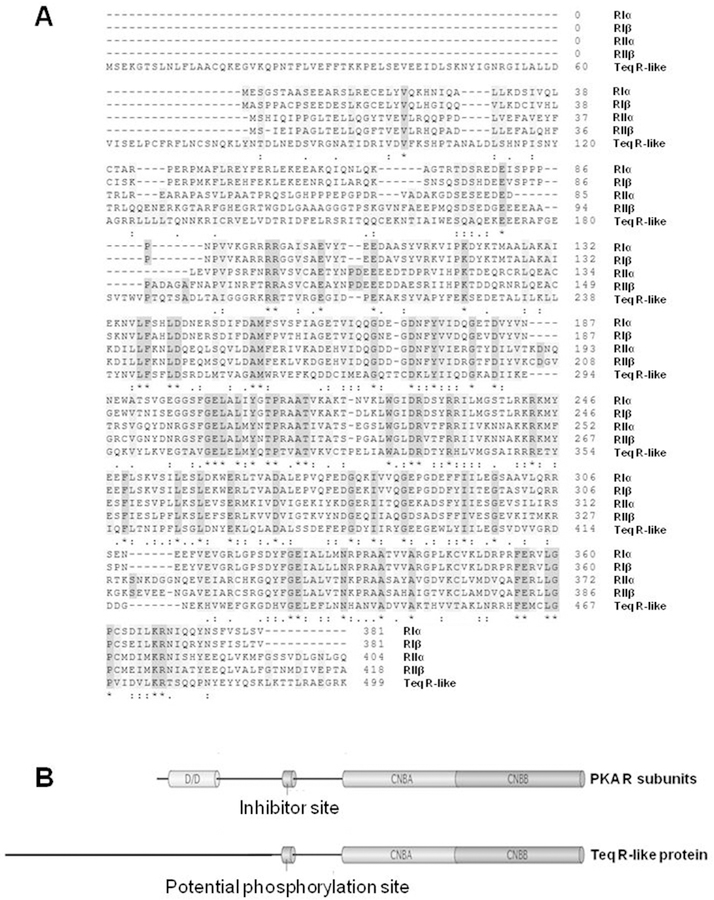
When the amino acid sequence of the T. equiperdum R-like protein was compared with the four human PKA R subunit isoforms, RIα, RIβ, RIIα, and RIIβ, it was clear that the parasite protein shares sequence homology with mammalian R subunits (Fig. 2A), especially at the carboxy-terminal region (residues 243–499) holding the hypothetical CNB domains. Although the four human R subunits possess high level of conservation at the amino-terminal region containing the dimerization/docking (D/D) domain, the corresponding region of the T. equiperdum R-like protein (residues 1e201) shares very little homology with the mammalian R subunits (Fig. 2A). Additionally, the N-terminal region is unusually long in the parasite protein. The results of the comparison of the domain organization between the T equiperdum R-like protein and the PKA R subunits are shown diagrammatically in Fig. 2B. The figure illustrates that in contrast to its mammalian orthologs, the parasite protein lacks the D/D domain at its amino-terminal region.
Fig. 2. Comparison of the primary structure and domain organization of the T. equiperdum R-like protein and the four human PKA R subunit isoforms.
A, Alignment of amino acid sequences of the R-like protein from T. equiperdum and the RIα, RIβ, RIIα, and RIIβ subunits from human [Accesion No. P10644.1, P31321.4, P13861.2 and P31323.3, respectively using the NCBI Protein database (https://www.ncbi.nlm.nih.gov/protein/)]. Identical residues are shown with an asterisk (*), conservative substitutions are labeled with a colon punctuation mark (:), and semi-conservative substitutions are labeled with a period (.). B, The standard domain structure of mammalian PKA R subunits is contrasted to the domain organization of the Teq R-like protein. CNBA and CNBB = cyclic nucleotide binding domains A and B, respectively; D/D = dimerization and docking domain.
A recombinant T. equiperdum R-like protein containing a poly-tag was generated by site-directed mutagenesis [TeqR-like(-His)8]. Once the IPTG-dependence on the induction of the expression of the recombinant protein was checked, cells were grown in 12 L of YT culture media. Although some protein remained in the particulate fraction, most of the recombinant protein was expressed in a soluble manner (Fig. S1A) and was purified to homogeneity by consecutively using the following chromatography steps: 1) a Probond ™ nickel-chelating affinity resin, 2) two sequential separations throughout a Superdex 200 gel filtration column, and 3) a Mono Q anionic exchange column (Figs. S1 and S2). A pure polypeptide band that migrated with an apparent molecular mass of ~56 kDa was obtained by SDS-PAGE, which corresponded to the expected size for the T. equiperdum ortholog protein.We obtained a yield of 5–6mg of the recombinant purified protein per liter of media, which was soluble up to concentrations higher than 40 mg/ml.
To verify the identity of the parasite PKA R-like protein, the recombinant polyHis-tagged protein was digested with trypsin and the resulting tryptic peptides were separated by HPLC and analyzed by mass spectrometry. The sequence information that was obtained is summarized in Fig. 1 and Table SI, showing coverage of ~64% of the parasite protein by MS/MS (317/499 residues). The N-terminal region of the protein was almost completely confirmed (residues 4–237) (Fig. 1). In contrast, the C-terminal region of the parasite R-like protein, containing the putative CNB domains, appeared to be more protected to proteolysis by trypsin and, consequently, fewer sequences related to this region were acquired by mass spectrometry (Fig. 1, Table SI).
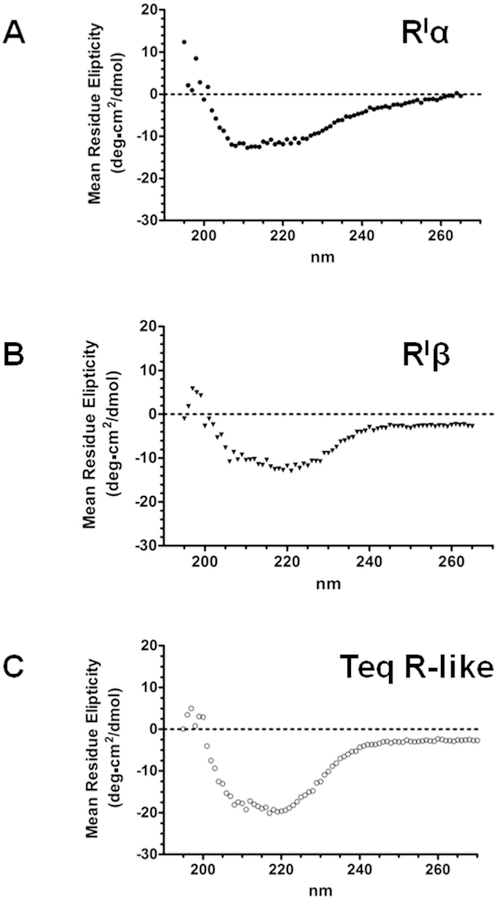
The conformation of the recombinant TeqR-like(His)8 protein was analyzed by CD, and the protein appeared to be correctly folded (Fig. 3). A comparison with the CD measurements obtained for the PKA bovine RIα and human RIβ subunits is also shown (Fig. 3A and B). Using the K2D3 program to estimate secondary structure from CD spectra [32], percentages of 36.78% and 4.36% of α-helix and β-strand, respectively, were predicted for the T. equiperdum protein.
Fig. 3. Analysis by circular dichroism spectroscopy of the recombinant TeqR-like(His)8 protein.
Comparison of CD measurements obtained for the bovine PKA RIα subunit (A), human PKA RIβ subunit (B), and T. equiperdum R-like protein (C).
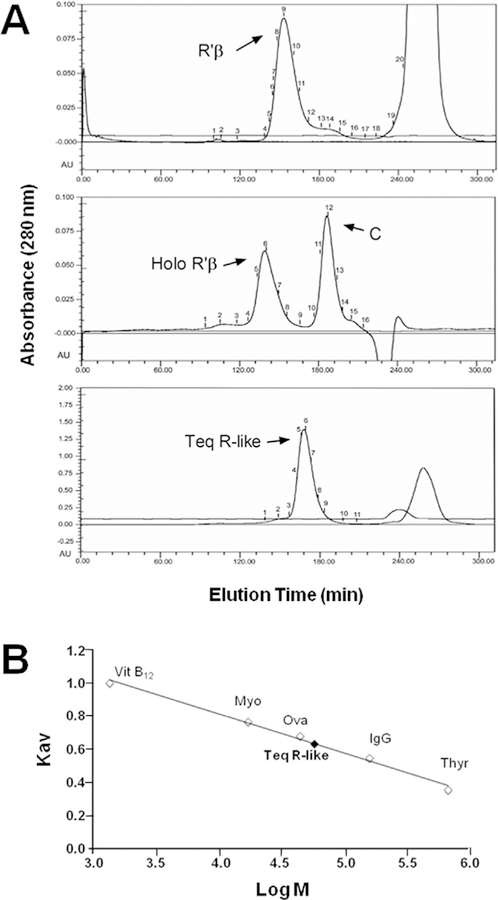
A comparative analysis by gel filtration chromatography was carried out using the tetrameric RIβ2C2 holoenzyme, the dimeric RIβ2 subunit, and the monomeric C subunit with native molecular weights of ~172,000, 96,000 and 38,000, respectively. As illustrated in Fig. 4A, these experiments showed that the recombinant parasite R-like protein possesses a size of approximately 60,000 Da. An HPLC separation by gel filtration chromatography of TeqR-like(His)8 was also performed in the presence of various molecular weight markers (Fig. 4B). The molecular mass of the parasite protein was empirically estimated to be 57,323 Da by plotting the Kav value of each standard versus the logarithm of its molecular weight (Fig. 4B). All these results demonstrated that the T. equiperdum R-like protein is a monomer, differing from R subunits of higher eukaryotic cells, which are dimeric.
Fig. 4. Size of the TeqR-like protein.
A, Separation by molecular exclusion column chromatography of the TeqR-like(His)8 protein (Teq R-like). Parallel experiments were performed with the RIβ2C2 holoenzyme (Holo RIβ), the RIβ2 subunit (RIβ), and the C subunit (C). B, Separation by gel filtration chromatography of TeqR-like(His)8 in the presence of various molecular weight markers: thyroglobulin (Thyr), immunoglobulin G (IgG), ovalbumin (Ova), myoglobin (Myo) and vitamin B12 (VitB12). The size of the unknown protein was estimated by plotting the Kav value of each standard versus the logarithm of its molecular weight (Mr).
A calibration curve of (-log Kav)1/2 versus the Stokes radii of each standard was also obtained. The elution volume yielded the (-log Kav)1/2 for TeqR-like(His)8, and a Stokes radius of 3.45 nm was determined by interpolation (data not shown). The frictional coefficient f/fo can also be evaluated from the molecular mass and the Stokes radius [33]. Deviations of the ratio of f/f0 from unity reflect non-spherical molecular shape and/or hydration effects. The calculated frictional ratio for TeqR-like(His)8 was 1.36. This f/fo value suggested some asymmetry in the molecule and indicated that the native parasite R-like monomeric protein folds into a conformation that lies in the boundary between globular and moderately elongated, when compared with compact spheres [34]. Since the frictional coefficient contains information not only about the molecular shape of the protein, but also about the hydration effects, an upper limit on the contributions of hydration can be calculated by assuming that all deviations of f/f0 from unity are due to hydration. Then, the maximal value of hydration (wmax) in grams of H2O bound per gram of TeqR-like(His)8 can be calculated using the following equation [35]:
where is the partial specific volume of TeqR-like(His)8 which is assumed to be 0.703 cm3 g−1, is the partial specific volume of bound water (1 cm3 g−1), Rs is the experimental Stokes radius, and R0 corresponds to the minimal radius for a perfect sphere possessing the calculated molecular weight. The wmax for TeqR-like(-His)8 is 1.07 g of H2O per gram of protein, which deviates from the value of 0.3–0.5 g of H2O per gram of a typical medium sized compact protein.
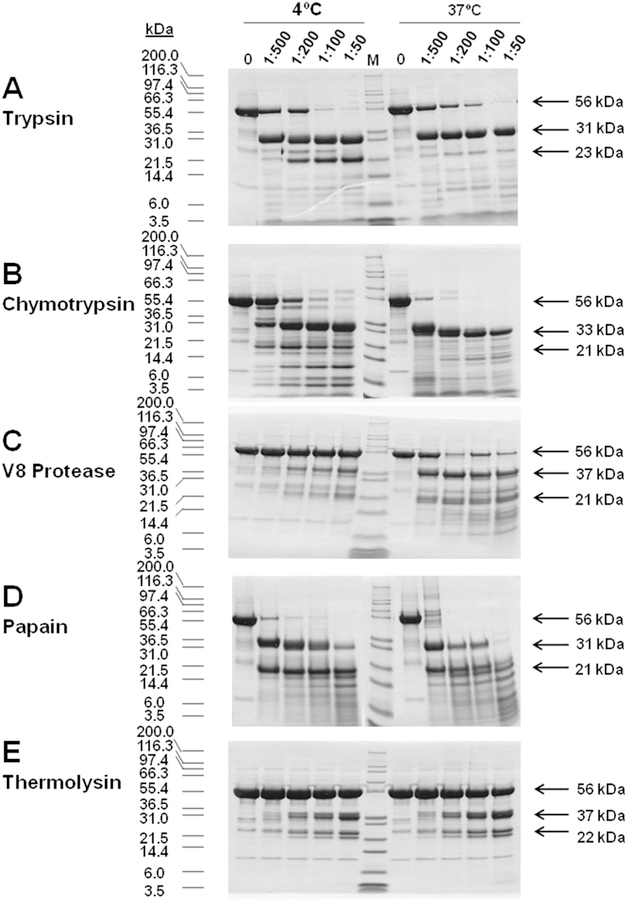
Higher eukaryotes RI and RII subunits contain a hinge region that is very susceptible to proteolysis by a variety of enzymes [14–16]. In order to assess whether the parasite R-like protein also contained this hinge region, and to define which fragments were more stable, limited proteolysis experiments were carried out with trypsin, chymotrypsin, the V8 protease from S. aureus, papaín, and thermolysin at two temperatures, 4 °C and 37 °C. As shown in Fig. 5, the T. equiperdum R-like protein appears to possess the characteristic hinge region that has been reported for mammalian R subunits. At 4 °C, enzymatic digestions showed that the protein that originally had a molecular mass of ~56 kDa was predominantly cleaved into two fragments; a large fragment of 31e37 kDa and a small fragment of 21–23 kDa. At 37 °C, a similar proteolytic pattern was observed, especially at low concentrations of the proteases (Fig. 5). However, at higher levels of proteases, the 21–23 kDa fragments were further cleaved into smaller fragments at both temperatures (Fig. 5). Therefore, the resulting 31–37 kDa fragments seemed to be more resistant to proteolysis than the 21–23 kDa fragments as the amount of proteases increased in the reaction mixtures. Given that the T. equiperdum protein contained a polyHis tag at its COOH-terminus, the large proteolytic fragment was identified as the corresponding carboxy-terminal region of the protein by affinity chromatography throughout a Probond ™ nickel-chelating resin (data not shown). From these results, we concluded that the C-terminal fragment appears to be more stable than the small N-terminal fragment. These findings coincided with the results previously attained by sequencing using MS/MS (Fig. 1, Table SI). Western blotting showed that both fragments were recognized by polyclonal antibodies raised in mice ascites against the recombinant T. equiperdum R-like protein (data not shown).
Fig. 5. Limited proteolysis analyses of the T. equiperdum R-like protein.
Assays were carried out at 4 °C and 37 °C, using various amounts of trypsin (A), chymotrypsin (B), the V8 protease from S. aureus (C), papaín (D), and thermolysin (E). Arrows indicate the molecular masses of the undigested TeqR-like(His)8 protein and of the major proteolytic fragments obtained following the various enzymatic digestions.
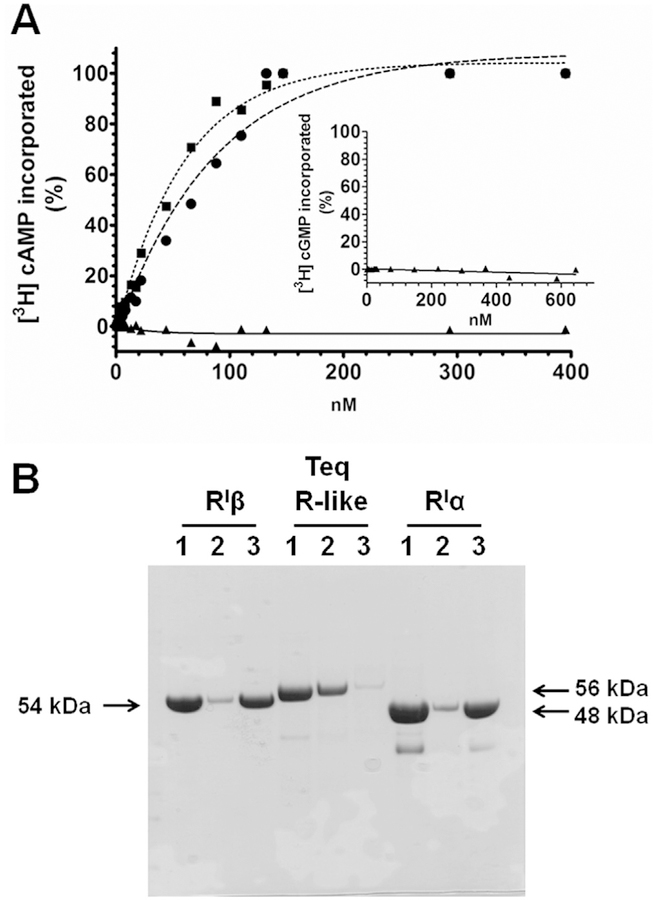
Since amino acids 243e354 and 363e475 of the T. equiperdum R-like protein appeared to form the putative CNB domains A and B, respectively (Fig. 1), the cyclic nucleotide binding properties of the parasite protein were also measured here. In contrast to the PKA bovine RIα and human RIβ subunits, which showed dissociation constant (Kd) values for cAMP of 40.1 nM and 56.9 nM, respectively, the T. equiperdum R-like protein did not bind [3H] cAMP up to concentrations of 0.4 mM (Fig. 6A). The Kd’s (cAMP) obtained for the mammalian RI subunits were comparable to published values (Kd 22 nM for the PKA bovine RIα [36]). The generation of such reproducible digestion patterns using various proteases (Fig. 5), prompted us to purify the resulting proteolytic fragments from the T. equiperdum R-like protein using gel filtration liquid chromatography (data not shown). Similar to the non-digested R-like protein, none of the COOH-terminal proteolytic large fragments containing the putative CNB sites were capable of binding [3H] cAMP (data not shown). The inability to bind cAMP was also analyzed employing chromatography throughout a cAMP-Sepharose affinity column. Neither the entire parasite R-like protein (Fig. 6B) nor the C-terminal large fragments resulting from enzymatic limited proteolysis (data not shown) were able to bind to a cAMP-Sepharose column. These experiments confirmed that the T. equiperdum R-like protein is not capable of binding cAMP, which agrees with the results reported by Shalaby et al. [37] for the T. brucei R-like protein that is 100% identical to the T. equiperdum protein. Given that Shalaby et al. [37] reported that the T. brucei R-like protein was capable of binding cGMP rather than cAMP, we also evaluated the binding of [3H] cGMP to the T. equiperdum R-like protein. As shown in Fig. 6A (inset), the T. equiperdum protein was not capable of binding [3H] cGMP up to concentrations of 0.65 mM, which curiously argued against the results reported by Shalaby et al. [37]. In addition, none of the generated C-terminal proteolytic fragments that contain the putative CNB sites were capable of binding [3H] cGMP (data not shown). Our experiments clearly showed that neither the T. equiperdum R-like protein nor its large proteolytic fragments were capable of binding either cAMP or cGMP.
Fig. 6. The T. equiperdum R-like protein is not capable of binding cyclic nucleotides.
A, Cyclic nucleotide binding was determined by equilibrium dialysis. Shown is the [3H] cAMP binding of the PKA bovine RIα subunit (■), the human RIβ subunit (●), and the T. equiperdum R-like protein (▲). Also shown is the [3H] cGMP binding of the parasite R-like protein (Inset). B, cAMP binding was also analyzed by chromatography throughout a cAMP-Sepharose affinity column. Gel lanes contained the following samples: 1, original protein; 2, flow through fraction; 3, bound protein. The bovine RIα subunit and human RIβ subunit were included as controls. Arrows indicate the molecular masses of the three proteins used in the experiment.
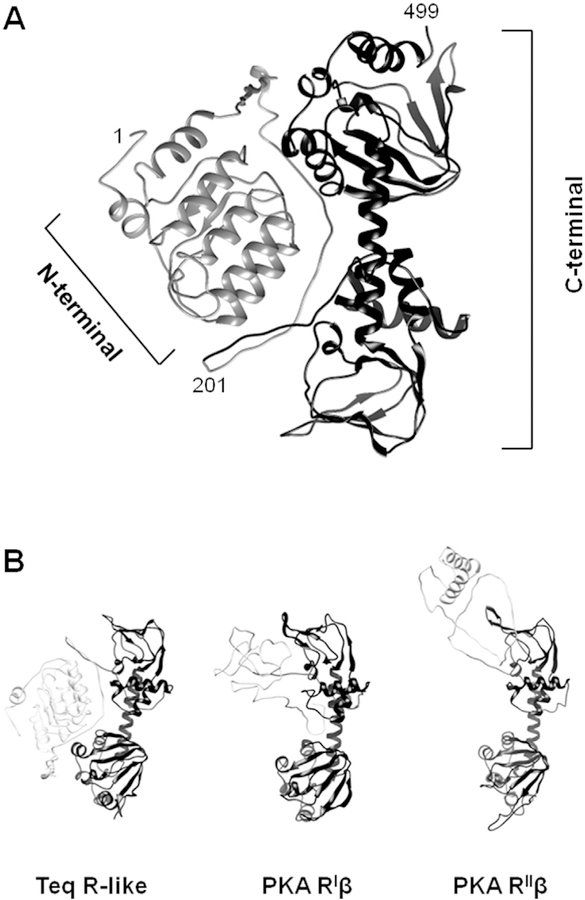
The primary structures of the T. equiperdum R-like protein and PKA R subunits are homologous, especially at the COOH-terminal region bearing the CNB domains (Fig. 2). On the basis of this homology, the Phyre2 server that is available on the web [38] was employed to predict and analyze the structure of the trypanosome protein. The model retrieved is shown in Fig. 7A. The NH2-terminal portion of the trypanosomal R-like protein appears to be unrelated to PKA R subunits (Fig. 7B). Yet, bioinformatics analyses predicted that the carboxy-terminal region of the parasite protein folds in the same way that the corresponding region of all known PKA R subunits from higher eukaryotes (Fig. 7B). Similarly, we carried out a secondary structure prediction bioinformatics analysis of the first 201 residues of the protein using the Phyre2 web portal [38] in order to obtain structural information on the amino-terminal region of the parasite protein. As shown in Fig. S3, the analysis predicted that the N-terminal portion of the parasite R-like protein is enriched in α-helices, and appears to possess a structural unit consisting of a small β-strand followed by an α-helix. When this result was compared with proteins stored in the data bank, the N-terminal sequence of the parasite R subunit had homology with proteins that contained Leu-rich (LRR) repeats or LRR proteins. The LRR motif is a structural fold vastly spread in proteins, which consists of 20–30 residues with a characteristic repetitive sequence pattern that is enriched in Leu [39]. The consensus sequence defining the LRR motif is LxxLxLxxNxL (x any aminoacid), although other hydrophobic residues may substitute Leu and Asn at the consensus positions. In LRR proteins, two or more repetitions in tandem form a horseshoe-shaped or curved solenoid type of structure that is particularly appropriate for protein-protein interactions.
Fig. 7. Structural model of the T. equiperdum R-like protein.
A, The Phyre2 bioinformatics tool [38] was employed to predict the structure of the T. equiperdum R-like protein. Ribbon diagram structures of the modeled N-terminal (residues 1–201) and C-terminal (residues 202e499) regions are shown in light grey and dark grey, respectively. B, The model retrieved for the trypanosome protein is compared to those retrieved for PKA human RIβ (PDB ID: 4DIN [22]) and rat RIIβ (PDB ID: 3TNQ [61]). N-terminal and C-terminal regions are shown in light grey and dark grey, respectively.
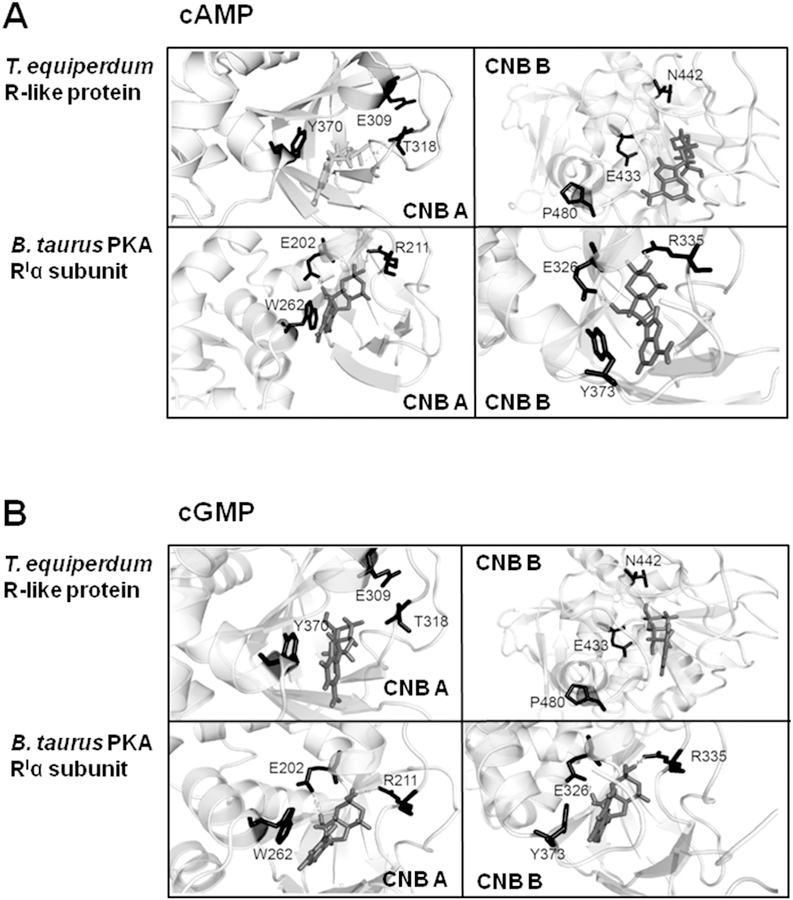
The AutoDock Vina docking program [24] was employed to model how the CNB domains are arrayed in the TeqR-like protein, and to predict differences in the environment that outline the putative binding pockets in the parasite protein. Docking was carried out using the crystal structure of the B. taurus PKA RIα subunit as a template. In order to calculate the deviation between the template and the structure modeled by the SWISS-MODEL server [26], a model for the B. taurus PKA RIα subunit complexed with the (Rp)-adenosine 3’, 5’-cyclic monophosphothioate analog of cAMP was also built. A root mean-square deviation of 1.863 Å was found between the two resulting structures of the mammalian proteins. In the case of the RIα subunit, both cAMP (Fig. 8A) and cGMP (Fig. 8B) fitted comfortably and tightly inside the cyclic nucleotide binding clefts. The conserved residues, Glu202 and Arg211 in CNB domain A and Glu326 and Arg335 in CNB domain B, which have been reported to be involved in cyclic nucleotide binding by crystallography studies [40], consistently appeared to participate directly in the binding of both cAMP and cGMP. Crystal structures have shown that Glu residues interact with the 2’eOH of the ribose ring while the Arg residues interact with the negative charge of the phosphate group. In both domains, an aromatic side chain, Trp262 in CNB domain A and Tyr373 in CNB domain B seemed to stack with the ring of the nitrogenous base moieties, as also has been shown by X-ray diffraction [40]. Interestingly, when photoaffinity labeling with 8-N3cAMP was used to identify residues that are located close to the cAMP binding sites, both Trp262 in CNB site A and Tyr373 in CNB site B were labeled in RIα [41]. These two aromatic side chains in RIα are optimally aligned [40], which presumably accounts for the exceptionally high efficiency in photolabeling. Molecular predictions illustrated that cAMP (Fig. 8A) and cGMP (Fig. 8B) can also be accommodated into the two CNB sites of the TeqR-like protein. However, chemical ligation appears to be unfavorable in the para-site protein since a loose and weak fit was obtained between the cyclic nucleotide ligands and all the relevant residues that participate in binding, showing much empty space in both binding clefts. Specifically, in the modeled structure for the bovine PKA RIα subunit bound to either cAMP or cGMP, all distances were in the range of 1.9–3.4 Å between the side chains of the functional residues and the various moieties of the cyclic nucleotide. In contrast, the same distances were 4.6 Å in the modeled structure of the TeqR-like protein. Moreover, even though the parasite protein possesses residues Glu309 and Glu433 that in mammalian PKA R subunits are involved in hydrogen bonding with the ribose group, surprisingly, the strongly conserved Arg residue, which makes a salt bridge interaction with the nucleotide cyclic phosphate, is replaced by Thr at position 318 and Asn at position 442 in the CNB domains A and B, respectively. Therefore, the electrostatic/ionic balance within the nucleotide binding pockets is clearly destabilized. In the TeqR-like protein, Trp262 of RIa is replaced with Tyr370 in CNB site A, maintaining the aromatic features of this side chain; however, Tyr373 of RIα is swapped by Pro480 in CNB site B of the TeqR-like protein, which introduced a kink that affects the structural conformation and chemical nature of this region in the nucleotide binding cleft (Fig. 8). However, Tyr482 of the parasite protein appears to be located close to where Tyr373 of RIα is located in the predicted three-dimensional structures (Fig. S4), which is interesting since these residues are not at the same position in the amino acid sequences of the two proteins. Therefore, the aromatic characteristics of this particular section of the CNB domain B also seem to be retained. In addition, the COOH-terminal regions of the mammalian PKA RIα subunit and the trypanosomal R-like protein were super-imposed to reveal the similarities and differences between both modeled structures within the two putative cyclic nucleotide binding sites (Fig. S5).
Fig. 8. View of the predicted structures obtained by molecular docking for the cyclic nucleotide binding pockets of the T. equiperdum R-like protein.
Shown is a comparison of the models attained for both cyclic nucleotide binding sites of the T. equiperdum R-like protein and the bovine PKA RIα subunit. CNB A and CNB B= cyclic nucleotide binding domains A and B, respectively. Highlighted are the functionally important residues involved in cAMP binding in RIα, and the corresponding residues in the parasite protein based on amino acid sequence alignment. Docking was performed using both cAMP (A) and cGMP (B).
Docking results were also used to determine the binding affinities for cAMP and cGMP of the TeqR-like protein and PKA RIα subunit CNB domains. While the mammalian protein showed binding energies for cAMP of −9.4 and −9.6 kcal/mol in CNB domains A and B, respectively, the parasite protein showed binding energies for cAMP of −5.9 and −5.5 kcal/mol in CNB domains A and B, respectively. Similarly, the mammalian protein showed binding energies for cGMP of −10.2 and −9.5 kcal/mol in CNB domains A and B, respectively, whilst the parasite protein showed binding energies for cGMP of −6.1 and −5.5 kcal/mol in CNB domains A and B, respectively. On the basis of the predicted binding affinities, no preference of the TeqR-like protein to bind cGMP over cAMP can be inferred. On top, a loss in binding energy of approximately 4 kcal/ mol was acquired in the two CNB domains of the parasite protein for both cAMP and cGMP, which supports and might account for the lack of cyclic nucleotide binding that was obtained experimentally (Fig. 6).
4. Discussion
In this work, a PKA R-like protein from the Venezuelan TeAp-N/ D1 isolate of T. equiperdum was cloned, expressed in bacteria, purified to homogeneity and biochemically characterized. At the protein level, a similarity and identity of 100% was achieved when the R-like proteins from T. equiperdum and the TREU 927 strain of T. b. brucei were compared. The similarity and identity between the R-like protein from T. equiperdum and its counterpart from the T. evansi STIB 805 strain stored in TriTrypDB (http://tritrypdb.org) (TevSTIB805.11_01.4760) were 100% and 99.6%, respectively, and only two conservative substitutions were detected at positions 215 and 409 from a total of 499 residues [13].
Similar to mammalian PKA R subunits, this novel parasite protein possessed a hinge region very susceptible to proteolysis with a variety of proteases. Yet, the T. equiperdum R-like protein differed from PKA R subunits since it was not dimeric. This result is in agreement with the fact that the trypanosomal protein bears no identifiable functional D/D domain in its amino acid sequence, justifying the monomeric state of the protein. Another major difference with mammalian PKA R subunits is that the parasite TeqR-like protein did not bind cAMP. In agreement with these results, Calabokis et al. [13] have also shown that a partially purified fraction of the T. equiperdum native PKA R-like protein was not capable of binding cAMP. These findings clearly suggest that the TeqR-like protein is not a direct downstream cAMP effector in this parasite. Absence of cAMP binding is probably caused by replacements on specific amino acids that are functionally essential for the ligation of the nucleotide phosphate in both PBC motifs [13]. It has been reported that the variety observed within PBC sequences in several organisms suggests that the CNB domain has evolved as a scaffold for binding not only cAMP, but also an ample diversity of other ligands [42,43]. Since both CNB sites of the T. equiperdum R-like protein hold non-canonical PBC sequences, these structural motifs might be used to bind a different kind of ligand that has not been identified to date.
Shalaby et al. [37] reported that the T. brucei R-like protein was capable of binding cGMP rather than cAMP, yet the T. equiperdum R-like protein, which is 100% identical to the T. brucei protein, was not able of binding either nucleotide. Thus, a rigorous comparison of our findings with those previously published by Shalaby et al. [37] is essential. Most likely, technical variations in the methodological conditions employed in the two studies might account for the discrepancies that were found. Firstly, Shalaby et al. [37] expressed the T. brucei recombinant protein in bacteria as a glutathione S-transferase (GST)-fusion molecule, but it was all produced as an insoluble protein. Attempts to solubilize the recombinant fusion protein with urea were not successful, and the protein remained insoluble and denatured. Therefore, none of the cyclic nucleotide binding experiments that they performed was actually carried out using the full-length recombinant T. brucei protein. Instead, deletion mutants restricted to only one of the two CNB domains (either CNB A or CNB B) and that carried a V5 immunological tag and a His6-tag at their C-termini, were expressed individually in the Drosophila cell line Schneider 2 (S2) and purified by affinity chromatography using a nickel-chelating resin. These truncated CNB sites A and B constructs were capable of binding cGMP but not cAMP, and exhibited Kd values for cGMP of about 8 µM and 11 µM, respectively [37]. On the basis of these findings, the authors concluded about the cGMP binding ability of the parasite protein. In contrast, our experiments were performed using the full-length T. equiperdum R-like protein containing all its functional domains.
Another dissimilar issue is the concentration of cAMP and cGMP used by Shalaby et al. [37], which was much higher than the concentration of cyclic nucleotides employed in the present study. It is critical to state which is the estimated amount of cyclic nucleotide that is physiologically relevant. Strickler and Patton [44] measured the intracellular concentrations of cAMP in Trypanosoma lewisi, a blood protozoan of rats that undergoes differentiation from a rapidly reproducing form to a nonreproducing form in response to the host antibody ablastin. They found that the concentration of cAMP was different in the two developmental stages of T. lewisi (0.167 µM in the reproducing form versus 0.328 µM in the nonreproducing form). Similar low micromolar levels of cAMP have been found in other cellular systems [45], and multiple studies have established that the basal concentration of cAMP in several cell types is about 1 µM [46–49], which is well above the reported concentration of cAMP required to half-maximally activate PKA that is in the 100–300 nM range [50,51]. Moreover, intracellular cGMP levels have been reported to be even lower than cAMP levels in certain cells or tissues, for example in cultures of fibroblast cells [52], and the reported concentration of cGMP required to half-maximally activate PKG is also in the nanomolar range (~90–200 nM) [53]. Therefore, we strongly believe that 20 µM cGMP, which is the concentration used by Shalaby et al. [37], is extremely high and non-physiological, and under these conditions nonspecific binding may occur. Conversely, the cAMP and cGMP concentration range that was assayed in the present manuscript is much lower (up to 0.4 µM for cAMP and up to 0.65 µM for cGMP), which lie in the expected physiological range. It is known that ligand binding proteins bind their ligand molecules in a concentration dependent manner, which allows the description of two parameters: i) the maximum number of ligand molecules bound per mole of protein and ii) the Kd of the reversible binding process. So, for a putative cyclic nucleotide binding protein as the T. equiperdum PKA R-like protein, and under the pretense that the protein is capable of ligating cyclic nucleotides, it would be expected to measure increasing number of ligand molecules bound to the protein as the concentration of the cyclic nucleotide increases. Even if we assume a threshold concentration of ~1 µM cyclic nucleotide to elicit a response [54], the cAMP and cGMP concentrations used here should be more than enough to evaluate cyclic nucleotide binding on a purified protein that contains two putative CNB domains, and is homologous to the mammalian PKA R sub-units and PKG enzymes. Therefore, the lack of cAMP and cGMP binding to the parasite protein clearly demonstrates that the CNB domains of the TeqR-like protein are defective for cyclic nucleotide binding.
It is commonly known that cAMP and cGMP behaved as cyclic nucleotide analogs in structure-activity studies. PKAs and PKGs are defined as cAMP-dependent protein kinases and cGMP-dependent protein kinases, respectively, on the basis of their affinity for each nucleotide; explicitly, PKA exhibits a >50-fold higher affinity for cAMP than for cGMP and preferentially binds cAMP, similarly, PKG has a >50-fold higher affinity for cGMP than for cAMP and prefer-entially binds cGMP [55–57]. Accordingly, PKAs and PKGs are activated relatively specifically by cAMP and cGMP, respectively. However, either cyclic nucleotide can cross-activate the other enzyme in vitro and due to the lack of absolute specificity of either enzyme, PKAs and PKGs are capable of binding both nucleotides. It has even been postulated that it is possible that either cyclic nucleotide could cross-activate the other protein kinase in certain tissues [58]. Hence, it seems unusual to find a cyclic nucleotide binding protein that is capable of binding only cGMP but not cAMP as reported by Shalaby et al. [37], especially at such a high concentration as 20 µM. It appears more convincing to find that the trypanosomal protein is not capable of binding neither cAMP nor cGMP, as we are reporting here. Our results from docking experiments are consistent and strengthen these findings since they revealed binding energy differences that support the lack of both cAMP and cGMP binding to the TeqR-like protein. Moreover, no preference of the parasite protein to bind cGMP over cAMP can be presumed based on the predicted binding affinities disclosed by molecular modeling.
Besides the previous report of Shalaby et al. [37] showing that the T. brucei PKA is stimulated by cGMP, not any other role of cGMP has been proposed in these parasites. Likewise, the cGMP dependence of the T. brucei PKA R-like protein has not been confirmed by any other group. It is commonly known that guanylyl cyclases (GC) are the enzymes responsible for synthesizing cGMP from GTP, and cGMP-dependent phosphodiesterases (PDE) are the enzymes responsible for degrading cGMP to GMP. Strikingly, no GC activities have been identified in trypanosomes, nor is there any measurable cGMP-dependent PDE activity in trypanosomes [59]. These results also argue against a physiological role of cGMP in these parasites.
A plausible explanation for the inability of the recombinant trypanosomal R-like protein to bind cyclic nucleotides might be that the protein is not correctly folded. Although analysis by CD demonstrated the presence of secondary structures, it is known that the occurrence of secondary structures does not necessary mean that the protein possesses its functional tertiary conformation given that some secondary structures could be resistant to denaturation. However, the purified protein seemed to be properly folded because it remained soluble up to concentrations higher than 40 mg/ml. It would not be thermodynamically viable for a partially unfolded protein to stay soluble at such a high concentration. In addition, formation of stable fragments by limited proteolysis with trypsin, chymotrypsin, V8 protease, papaín, and thermolysin, argues in favor of the recombinant protein possessing its native conformation. In all cases, the protein that originally had a molecular mass of ~56 kDa was primarily digested into two fragments; a large piece of 31–37 kDa and a small piece of 21–23 kDa. A partially unfolded protein would yield randomly generated proteolytic peptides following incubation with such different proteases.
Bioinformatics analysis of the N-terminal region of the TeqR-like protein established its homology with LRR proteins, a series of molecules that contain structures involved in protein-protein interactions. In all mammalian PKA R subunit isoforms, the D/D domain allows binding to the so-called A-kinase anchoring proteins (AKAPs), which function as key-regulator scaffolds of cAMP intracellular specificity synchronizing spatial and temporal aspects of cell signaling. Since no conventional AKAPs have been reported in trypanosomes, and the parasite R-like protein lacks the putative D/ D domain, it is appealing to postulate that under certain conditions this R-like protein might be capable of working in an analogous manner as an AKAP, tethering and directing trypanosome kinases that may be related to the PKA C subunits to their downstream substrate targets. As illustrated above (Figs. 1 and 2), the R-like protein possesses a site that contains a substrate recognition sequence (inhibitor site), and as some AKAPs from higher eukaryotes [60], it could be a preferred substrate for its associated protein kinase. In any case, its presence in every trypanosomatid genome elucidated to date clearly suggests that PKA R-like proteins might possess a significant function in trypanosome physiology.
5. Conclusions
The T. equiperdum ortholog of the PKA R subunit is a monomer, which is incapable of ligating cyclic nucleotides and contains an exposed hinge region that is vulnerable to hydrolysis by several proteolytic enzymes. Analyses using bioinformatics tools predicted that the COOH-terminal regions of the parasite R-like protein and mammalian PKA R subunits are related and possess similar 3D structures; however, the NH2-terminal region of the trypanosomal protein is unrelated to that of its mammalian counterparts and shows homology with Leu-rich repeats-containing proteins. Predictions by molecular docking support the lack of cyclic nucleotide binding in the T. equiperdum R-like protein.
Supplementary Material
Acknowledgments
This research was supported by grant number S1-IC-CB-007–14 (Decanato de Investigación y Desarrollo, Universidad Simón Bolívar, Caracas, Venezuela).
Abbreviations:
- PKA
cAMP-dependent protein kinase
- R
PKA regulatory subunit
- C
PKA catalytic subunit
- TeqR-like(His)8
poly-His tagged construct of the Trypanosoma equiperdum ortholog of the PKA R subunit
- AC
adenylyl cyclase
- PMSF
phenyl methyl sulfonyl fluoride
- AEBSF
4-(2-Aminoethyl) benzenesulfonyl fluoride hydrochloride
- TLCK
N-α-tosyl-L-lysinyl-chloromethyl ketone hydrochloride
- TPCK
N-p-tosyl-L-phenylalaninyl chloromethyl ketone
- IPTG
isopropyl-1-thio-β-d-galactopyranoside
- muramyl dipeptide
acetylmuramyl-alanyl-isoglutamine
- BSA
bovine serum albumin
- 8-AEA
cAMP 8-(2-aminoethylamino) adenosine 3’,5’ -cyclic monophosphate
- NHS
N-hydroxysuccinimide
- BCIP
5-bromo-4-chloro-3-indolyl phosphate
- NBT
nitro blue tetrazolium
- YT
Yeast extract and tryptone
- LB
Luria-Bertani
- SDS
sodium dodecyl sulfate
- SDS-PAGE
SDS-polyacrylamide gel electrophoresis
- MS/MS
tandem mass spectrometry
- PCR
polymerase chain reaction
- MES
2-(N-morpholino)ethanesulfonic acid
- DTT
dithiothreitol
- HPLC
high-performance liquid chromatography
- Mr
molecular weight
- CD
circular dichroism
- CNB
cyclic nucleotide binding
- PBC
phosphate binding cassette
- D/D
dimerization/docking
- f/f0
frictional coefficient
partial specific volume
partial specific volume of bound water
- wmax
maximal hydration
- Rs
Stokes radius
- Ro
minimal radius
- LRR
Leu-rich repeats
- GST
glutathione S-transferase
- GC
guanylyl cyclases
- PDE
phosphodiesterase
- AKAP
A-kinase anchoring protein
Footnotes
Conflicts of interest
The authors declare that there is no conflict of interests regarding the publication of this article.
Appendix A. Supplementary data
Supplementary data related to this article can be found at https://doi.org/10.1016/j.biochi.2017.12.010.
References
- [1].Brun R, Hecker H, Lun Z-R, Trypanosoma evansi and T. equiperdum: distribution, biology, treatment and phylogenetic relationship, Vet. Parasitol 79 (1998) 95–107, 10.1016/S0304-4017(98)00146-0. [DOI] [PubMed] [Google Scholar]
- [2].Claes F, Büscher P, Touratier L, Goddeeris BM, Trypanosoma equiperdum: master of disguise or historical mistake? Trends Parasitol 21 (2005) 316–321, 10.1016/j.pt.2005.05.010. [DOI] [PubMed] [Google Scholar]
- [3].Alexandre S, Paindavoine P, Hanocq-Quertier J, Paturiaux-Hanocq F, Tebabi P, Pays E, Families of adenylate cyclase genes in Trypanosoma brucei, Mol. Biochem. Parasitol 77 (1996) 173–182, 10.1016/0166-6851(96)02591-1. [DOI] [PubMed] [Google Scholar]
- [4].Bridges DJ, Pitt AR, Hanrahan O, Brennan K, Voorheis HP, Herzyk P, de Koning HP, Burchmore RJ, Characterisation of the plasma membrane sub-proteome of bloodstream form Trypanosoma brucei, Proteomics 8 (2008) 83–99, 10.1002/pmic.200700607. [DOI] [PubMed] [Google Scholar]
- [5].Salmon D, Bachmaier S, Krumbholz C, Kador M, Gossmann JA, Uzureau P, Pays E, Boshart M, Cytokinesis of Trypanosoma brucei bloodstream forms depends on expression of adenylyl cyclases of the ESAG4 or ESAG4-like subfamily, Mol. Microbiol 84 (2012) 225–242, 10.1111/j.1365-2958.2012.08013.x. [DOI] [PubMed] [Google Scholar]
- [6].Krebs EG, Beavo JA, Phosphorylation-dephosphorylation of enzymes, Annu. Rev. Biochem 48 (1979) 923–959, 10.1146/annurev.bi.48.070179.004423. [DOI] [PubMed] [Google Scholar]
- [7].Taylor SS, Kim C, Cheng CY, Brown SH, Wu J, Kannan N, Signaling through cAMP and cAMP-dependent protein kinase: diverse strategies for drug design, Biochim. Biophys. Acta 1784 (2008) 16–26, 10.1016/j.bbapap.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Choi J, El-Sayed NM, Functional genomics of trypanosomatids, Parasite Immunol 34 (2012) 72–79, 10.1111/j.1365-3024.2011.01347.x. [DOI] [PubMed] [Google Scholar]
- [9].Jackson AP, Genome evolution in trypanosomatid parasites, Parasitology 142 (Suppl 1) (2015) S40–S56, 10.1017/S0031182014000894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Carnes J, Anupama A, Balmer O, Jackson A, Lewis M, Brown R, Cestari I, Desquesnes M, Gendrin C, Hertz-Fowler C, Imamura H, Ivens A, Kořený L, Lai DH, MacLeod A, McDermott SM, Merritt C, Monnerat S, Moon W, Myler P, Phan I, Ramasamy G, Sivam D, Lun ZR, Lukěs J, Stuart K, Schnaufer A, Genome and phylogenetic analyses of Trypanosoma evansi reveal extensive similarity to T. brucei and multiple independent origins for dyskinetoplasty, PLoS Neglected Trop. Dis 9 (2015) e3404, 10.1371/journal.pntd.0003404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hébert L, Moumen B, Madeline A, Steinbiss S, Lakhdar L, Van Reet N, Büscher P, Laugier C, Cauchard J, Petry S, First draft genome sequence of the dourine causative agent: Trypanosoma equiperdum strain OVI, J. Genom 5 (2017) 1–3, 10.7150/jgen.17904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Parsons M, Worthey EA, Ward PN, Mottram JC, Comparative analysis of the kinomes of three pathogenic trypanosomatids: Leishmania major, Trypanosoma brucei and Trypanosoma cruzi, BMC Genom 6 (2005) 127, 10.1186/1471-2164-6-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Calabokis M, González Y, Merchán A, Escalona JL, Araujo NA, Sanz-Rodríguez CE, Cywiak C, Spencer L, Martínez JC, Bubis J, Immunological identification of a cAMP-dependent protein kinase regulatory subunit-like protein from the Trypanosoma equiperdum TeAp-N/D1 isolate, J. Immunoassay Immunochem 37 (2016) 485–514, 10.1080/15321819.2016.1162799. [DOI] [PubMed] [Google Scholar]
- [14].Potter RL, Taylor SS, Relationships between structural domains and function in the regulatory subunit of cAMP-dependent protein kinases I and II from porcine skeletal muscle, J. Biol. Chem 254 (1979) 2413–2418. [PubMed] [Google Scholar]
- [15].Potter RL, Taylor SS, Correlation of the cAMP binding domain with a site of autophosphorylation on the regulatory subunit of cAMP-dependent protein kinase II from porcine skeletal muscle, J. Biol. Chem 254 (1979) 9000–9005. [PubMed] [Google Scholar]
- [16].Potter RL, Taylor SS, The structural domains of cAMP-dependent protein kinase I. Characterization of two sites of proteolytic cleavage and homologies to cAMP-dependent protein kinase II, J. Biol. Chem 255 (1980) 9706–9712. [PubMed] [Google Scholar]
- [17].Uzcanga G, Mendoza M, Aso PM, Bubis J, Purification of a 64 kDa antigen from Trypanosoma evansi that exhibits cross-reactivity with Trypanosoma vivax, Parasitology 124 (2002) 287–299, 10.1017/S0031182001001214. [DOI] [PubMed] [Google Scholar]
- [18].Lanham SM, Godfrey DG, Isolation of salivarian trypanosomes from man and other mammals using DEAE-cellulose, Exp. Parasitol 28 (1970) 521–534, 10.1016/0014-4894(70)90120-7. [DOI] [PubMed] [Google Scholar]
- [19].Tung AS, Ju ST, Sato S, Nisonoff A, Production of large amounts of antibodies in individual mice, J. Immunol 116 (1976) 676–681. [PubMed] [Google Scholar]
- [20].Kim C, Cheng CY, Saldanha SA, Taylor SS, PKA-I holoenzyme structure reveals a mechanism for cAMP-dependent activation, Cell 130 (2007) 1032–1043, 10.1016/j.cell.2007.07.018. [DOI] [PubMed] [Google Scholar]
- [21].Brown SH, Wu J, Kim C, Alberto K, Taylor SS, Novel isoform-specific interfaces revealed by PKA RIIb holoenzyme structures, J. Mol. Biol 393 (2009) 1070–1082, 10.1016/j.jmb.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ilouz R, Bubis J, Wu J, Yim YY, Deal MS, Kornev AP, Ma Y, Blumenthal DK, Taylor SS, Localization and quaternary structure of the PKA RIβ holoenzyme, Proceed. Natl. Acad. Sci. U. S. A 109 (2012) 12443–12448, 10.1073/pnas.1209538109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gangal M, Cox S, Lew J, Clifford T, Garrod SM, Aschbaher M, Taylor SS, Johnson DA, Backbone flexibility of five sites on the catalytic subunit of cAMP-dependent protein kinase in the open and closed conformations, Biochemistry 37 (1998) 13728–13735, 10.1021/bi980560z. [DOI] [PubMed] [Google Scholar]
- [24].Trott O, Olson AJ, AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading, J. Comput. Chem 31 (2010) 455–461, 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Bruystens JG, Wu J, Fortezzo A, Kornev AP, Blumenthal DK, Taylor SS, PKA RIa homodimer structure reveals an intermolecular Interface with implications for cooperative cAMP binding and Carney complex disease, Structure 22 (2014) 59–69, 10.1016/j.str.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M, Bordoli L, Schwede T, SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information, Nucleic Acids Res 42 (Web Server issue) (2014) W252–W258, 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wu J, Jones JM, Nguyen-Huu X, Ten Eyck LF, Taylor SS, Crystal structures of RIa subunit of cyclic adenosine 5’-monophosphate (cAMP)-dependent protein kinase complexed with (Rp)-adenosine 3’,5’-cyclic monophosphothioate and (Sp)-adenosine 3’,5’-cyclic monophosphothioate, the phosphorothioate analogues of cAMP, Biochemistry 43 (2004) 6620–6629, 10.1021/bi0302503. [DOI] [PubMed] [Google Scholar]
- [28].Gordon JC, Myers JB, Folta T, Shoja V, Heath LS, Onufriev A, H++ : a server for estimating pKas and adding missing hydrogens to macromolecules, Nucleic Acids Res 33 (Web Server issue) (2005) W368–W371, 10.1093/nar/gki464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Bradford MM, A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding, Anal. Biochem 72 (1976) 248–254, 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- [30].Laemmli UK, Cleavage of structurals proteins during the assembly of the head of bacteriophage T4, Nature 227 (1970) 680–685, 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- [31].Towbin H, Staehelin T, Gordon J, Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications, Proceed. Natl. Acad. Sci. U. S. A 76 (1979) 4350–4354, 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Louis-Jeune C, Andrade-Navarro MA, Perez-Iratxeta C, Prediction of protein secondary structure from circular dichroism using theoretically derived spectra, Prot. Struct. Funct. Bioinform 80 (2012) 374–381, 10.1002/prot.23188. [DOI] [PubMed] [Google Scholar]
- [33].Bloomfield V, Dalton WO, van Holde KE, Frictional coefficients of multi-subunit structures. I. Theory, Biopolymers 5 (1967) 135–148, 10.1002/bip.1967.360050202. [DOI] [PubMed] [Google Scholar]
- [34].Erickson HP, Size and shape of protein molecules at the nanometer level determined by sedimentation, gel filtration, and electron microscopy, Biol. Proced. Online 11 (2009) 32–51, 10.1007/s12575-009-9008-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Darling PJ, Holt JM, Ackers GK, Coupled energetics of λ cro repressor self-assembly and site-specific DNA operator binding I: analysis of cro dimerization from nanomolar to micromolar concentrations, Biochemistry 39 (2000) 11500–11507, 10.1021/bi000935s. [DOI] [PubMed] [Google Scholar]
- [36].Herberg FW, Taylor SS, Dostmann WR, Active site mutations define the pathway for the cooperative activation of cAMP-dependent protein kinase, Biochemistry 35 (1996) 2934–2942, 10.1021/bi951647c. [DOI] [PubMed] [Google Scholar]
- [37].Shalaby T, Liniger M, Seebeck T, The regulatory subunit of a cGMP-regulated protein kinase A of Trypanosoma brucei, Eur. J. Biochem 268 (2001) 6197–6206, 10.1046/j.0014-2956.2001.02564.x. [DOI] [PubMed] [Google Scholar]
- [38].Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ, The Phyre2 web portal for protein modeling, prediction and analysis, Nat. Protoc 10 (2015) 845–858, 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Bella J, Hindle KL, McEwan PA, Lovell SC, The leucine-rich repeat structure, Cell. Mol. Life Sci 65 (2008) 2307–2333, 10.1007/s00018-008-8019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Su Y, Dostmann WR, Herberg FW, Durick K, Xuong NH, Ten Eyck L, Taylor SS, Varughese KI, Regulatory subunit of protein kinase A: structure of deletion mutant with cAMP binding domains, Science 269 (1995) 807–813, 10.1126/science.7638597. [DOI] [PubMed] [Google Scholar]
- [41].Bubis J, Taylor SS, Correlation of photolabeling with occupancy of cAMP binding sites in the regulatory subunit of cAMP-dependent protein kinase I, Biochemistry 26 (1987) 3478–3486, 10.1021/bi00386a035. [DOI] [PubMed] [Google Scholar]
- [42].Kannan N, Wu J, Anand GS, Yooseph S, Neuwald AF, Venter JC, Taylor SS, Evolution of allostery in the cyclic nucleotide binding module, Genome Biol 8 (2007) R264, 10.1186/gb-2007-8-12-r264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Mohanty S, Kennedy EJ, Herberg FW, Hui R, Taylor SS, Langsley G, Kannan N, Structural and evolutionary divergence of cyclic nucleotide binding domains in eukaryotic pathogens: implications for drug design, Biochim. Biophys. Acta 1854 (2015) 1575–1585, 10.1016/j.bbapap.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Strickler JE, Patton CL, Adenosine 3’,5’-monophosphate in reproducing and differentiated trypanosomes, Science 190 (1975) 1110–1112. [DOI] [PubMed] [Google Scholar]
- [45].Otten J, Johnson GS, Pastan I, Cyclic AMP levels in fibroblasts: relationship to growth rate and contact inhibition of growth, Biochem. Biophys. Res. Commun 44 (1971) 1192–1198, 10.1016/S0006-291X(71)80212-7. [DOI] [PubMed] [Google Scholar]
- [46].Beavo JA, Bechtel PJ, Krebs EG, Activation of protein kinase by physiological concentrations of cyclic AMP, Proceed. Natl. Acad. Sci. U. S. A 71 (1974) 3580–3583, 10.1073/pnas.71.9.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Terasaki WL, Brooker G, Cardiac adenosine 3’:5’-monophosphate. Free and bound forms in the isolated rat atrium, J. Biol. Chem 252 (1977) 1041–1050. [PubMed] [Google Scholar]
- [48].Kameyama M, Hofmann F, Trautwein W, On the mechanism of b-adrenergic regulation of the Ca channel in the Guinea-pig heart, Pflügers Archiv Eur. J. Physiol 405 (1985) 285–293, 10.1007/BF00582573. [DOI] [PubMed] [Google Scholar]
- [49].Börner S, Schwede F, Schlipp A, Berisha F, Calebiro D, Lohse MJ, Nikolaev VO, FRET measurements of intracellular cAMP concentrations and cAMP analog permeability in intact cells, Nat. Protoc 6 (2011) 427–438, 10.1038/nprot.2010.198. [DOI] [PubMed] [Google Scholar]
- [50].Adams SR, Harootunian AT, Buechler YJ, Taylor SS, Tsien RY, Fluorescence ratio imaging of cyclic AMP in single cells, Nature 349 (1991) 694–697, 10.1038/349694a0. [DOI] [PubMed] [Google Scholar]
- [51].Mongillo M, McSorley T, Evellin S, Sood A, Lissandron V, Terrin A, Huston E, Hannawacker A, Lohse MJ, Pozzan T, Houslay MD, Zaccolo M, Fluorescence resonance energy transfer-based analysis of cAMP dynamics in live neonatal rat cardiac myocytes reveals distinct functions of compart-mentalized phosphodiesterases, Circ. Res 95 (2004) 67–75, 10.1161/01.RES.0000134629.84732.11. [DOI] [PubMed] [Google Scholar]
- [52].Moens W, Vokaer A, Kram R, Cyclic AMP and cyclic GMP concentrations in serum-and density-restricted fibroblast cultures, Proceed. Natl. Acad. Sci. U. S. A 72 (1975) 1063–1067, 10.1073/pnas.72.3.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Campbell JC, Henning P, Franz E, Sankaran B, Herberg FW, Kim C, Structural basis of analog specificity in PKG I and II, ACS Chem. Biol 12 (2017) 2388–2398, 10.1021/acschembio.7b00369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Vassella E, Reuner B, Yutzy B, Boshart M, Differentiation of African trypanosomes is controlled by a density sensing mechanism which signals cell cycle arrest via the cAMP pathway, J. Cell Sci 110 (1997), 2661–2571. [DOI] [PubMed] [Google Scholar]
- [55].Døskeland SO, Øgreid D, Ekanger R, Sturm PA, Miller JP, Suva RH, Mapping of the two intrachain cyclic nucleotide binding sites of adenosine cyclic 3’,5’-phosphate dependent protein kinase I, Biochemistry 22 (1983) 1094–1101, 10.1021/bi00274a016. [DOI] [PubMed] [Google Scholar]
- [56].Corbin JD, Øgreid D, Miller JP, Suva RH, Jastorff B, Døskeland SO, Studies of cGMP analog specificity and function of the two intrasubunit binding sites of cGMP-dependent protein kinase, J. Biol. Chem 261 (1986) 1208–1214. [PubMed] [Google Scholar]
- [57].Landgraf W, Hullin R, Göbel C, Hofmann F, Phosphorylation of cGMP-dependent protein kinase increases the affinity for cyclic AMP, Eur. J. Biochem 154 (1986) 113–117, 10.1111/j.1432-1033.1986.tb09365.x. [DOI] [PubMed] [Google Scholar]
- [58].Jiang H, Shabb JB, Corbin JD, Cross-activation: overriding cAMP/cGMP selectivities of protein kinases in tissues, Biochem. Cell. Biol 70 (1992) 1283–1289, 10.1139/o92-175. [DOI] [PubMed] [Google Scholar]
- [59].Laxman S, Beavo JA, Cyclic nucleotide signaling mechanisms in trypanosomes: possible targets for therapeutic agents, Mol. Interv 7 (2007) 203–215, 10.1124/mi.7.4.7. [DOI] [PubMed] [Google Scholar]
- [60].Smith FD, Langeberg LK, Scott JD, The where’s and when’s of kinase anchoring, Trends Biochem. Sci 31 (2006) 316–323, 10.1016/j.tibs.2006.04.009. [DOI] [PubMed] [Google Scholar]
- [61].Zhang P, Smith-Nguyen EV, Keshwani MM, Deal MS, Kornev AP, Taylor SS, Structure and allostery of the PKA RIIb tetrameric holoenzyme, Science 335 (2012) 712–716, 10.1126/science.1213979. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.