Abstract
A rickettsial isolate was obtained from a partially engorged Ixodes pacificus female, which was collected from Humboldt County, California. The isolate was provisionally named Rickettsia endosymbiont Ixodes pacificus (REIP). The REIP isolate displayed the highest nucleotide sequence identity to Rickettsia species phylotype G021 in I. pacificus (99%, 99%, and 100% for ompA, 16S rRNA, and gltA, respectively), a bacterium that was previously identified in I. pacifiucs by PCR. Analysis of sequences from complete opening frames of five genes, 16S rRNA, gltA, ompA, ompB, and sca4, provided inference to the bacteria’s classification among other Rickettsia species. The REIP isolate displayed 99.8%, 99.4%, 99.2%, 99.5%, and 99.6% nucleotide sequence identity for 16S rRNA, gltA, ompA, ompB, and sca4 gene, respectively, with genes of ‘R. monacensis’ str. IrR/Munich, indicating the REIP isolate is closely related to ‘R. monacensis’. Our suggestion was further supported by phylogenetic analysis using concatenated sequences of 16S rRNA, gltA, ompA, ompB, and sca4 genes, concatenated sequences of dksA-xerC, mppA-purC, and rpmE-tRNAfMet intergenic spacer regions. Both phylogenetic trees implied that the REIP isolate is most closely related to ‘R. monacensis’ str. IrR/Munich. We propose the bacterium be considered as ‘Rickettsia monacensis’ str. Humboldt for its closest phylogenetic relative (=DSM 103975T =ATCC TSD-94T).
Keywords: ‘Rickettsia monacensis’ str. Humboldt, Ixodes pacificus, phylogenetic analysis, multilocus sequence typing, multi-spacer sequence typing
Introduction
Ixodes pacificus, the Western black-legged tick, is a predominant vector of Borrelia burgdorferi sensu stricto and Anaplasma phagocytophilum, the etiologic agents of Lyme borreliosis and anaplasmosis, respectively, in the Pacific West Coast region of the United States (Burgdorfer et al., 1985; Foley et al., 2008; Piesman et al., 1999). Ixodid ticks, as well as other arthropods, often harbor rickettsiae that are known as obligate intracellular bacteria and arthropod-borne pathogens of vertebrate hosts (Hardstone Yoshimizu and Billeter, 2018). Presently, 32 Rickettsia species within the spotted fever, typhus, Rickettsia bellii, and Rickettsia canadensis groups have been validated (Abdad et al., 2017; http://www.bacterio.net/rickettsia.html). Utilization of molecular techniques has facilitated the continual detection and isolation of rickettsiae from ticks around the world (Parola et al., 2013; Cazorla et al., 2008; Richards, 2012). Several serological studies have noted that I. pacificus harbors a spotted fever group Rickettsia that is antigenically related to pathogenic species (Hughes et al., 1976; Philip et al., 1978; Philip et al., 1981). We have previously identified Rickettsia species phylotype G021 and Rickettsia species phylotype G022 via PCR amplification using genomic DNA extracted from I. pacificus ticks; of which, the bacterial prevalence was found to be 100% and 2%, respectively (Phan et al., 2011; Cheng et al., 2013b). Transmission electron microscope and Fluorescence in situ hybridization assays detected the presence of Rickettsia species in midgut and ovaries of I. pacificus (Phan et al., 2011; Bagheri et al., 2017). It was later noted that Rickettsia species phylotype G021 was found to possess 100% transovarial and transstadial efficiency as well as the genetic capacity to synthesize tetrahydrofolate, indicating a possible nutritional relationship between the host and endosymbiont (Bodnar et al., 2018; Cheng et al., 2013a; Kurlovs et al., 2014; Hunter et al., 2015).
Material and methods
Source of ticks and tick cell culture
To isolate Rickettsia phylotype G021 from a mixed population of microorganisms in I. pacificus, we collected a partially engorged I. pacificus female that was donated from McKinleyville Animal Care Center, McKinleyville, CA, in March 2013. The standard Universal Transverse Mercator Grid System coordinate for the tick collection site is 10N 429550 4530570. The isolation of rickettsiae from the engorged tick followed methods reported previously (Kurtti et al., 1996; Kurtti et al. 2015). In brief, ovaries were extirpated from a surface disinfected, partially engorged, female. Ovarian fragments were cultured with tick cell line IRE11 and incubated in a candle jar at 25°C. Once established, the isolate was maintained in tick cell line ISE6 at 34°C (Simser et al., 2002; Munderloh et al., 1994). In the sixth serial transfer, genomic DNA was prepared and submitted to BioProject PRJNA232537 for genome sequencing (SRX476329; SRX476328 and SRX476327).
The isolate was initially named Rickettsia endosymbiont Ixodes pacificus (REIP). Infection of IRE11 and ISE6 cell lines was conducted by mixing 106 cells with 0.3–0.5 mL of REIP-infected IRE11 lysate, resulting in 20–100 rickettsiae per tick cell, in 25 cm2 flasks (Fisher Scientific, USA). Cells of rickettsiae and IRE11 were counted using a Petroff Hauser counting chamber under a Leica DM750 phase-contrast microscope with Leica highresolution camera module (Leica Microsystems, USA). Infected cells were cultured for three weeks at 25°C in L-15B300 complete medium supplemented with 10% fetal bovine serum, 5% tryptose phosphate broth, 0.1% bovine lipoprotein cholesterol concentrate, and 10 mM HEPES (Kurtti et al., 2005; Munderloh and Kurtti, 1989). Giemsa stain (KaryoMAX®, Thermo Fisher Scientific, USA) was performed on a weekly basis to monitor infection status. Giemsa stained REIP-infected ISE6 cells (or IRE11) were observed under the Leica DM750 microscope.
DNA extraction and PCR amplification
To classify REIP into genus and species, genomic DNA was extracted from 5×105 REIP-infected IRE11 cells using DNeasy Blood & Tissue Kit for both MLST and MST (Qiagen, Germany). Amplification of 16S rRNA, gltA, ompA, dksA-xerC, mppA-purC, and rpmE-tRNAfMet was conducted using PCR using primers specified in Table 1 (Simser et al., 2002; Fournier and Raoult, 2007; Cheng et al., 2013b). Specifically, 2 μL genomic DNA (50 ng/μL), 1 μL 5 mM forward and reverse primers, 10 μL GoTaq® Green Master Mix (Promega, USA), and 6 μL PCR grade water were added in PCR reactions. Negative controls contained all components with the exception of REIP genomic DNA. The β-actin-coding gene was amplified simultaneously to serve as positive control using β-actin specific primers and field-collected I. pacificus (Cheng et al., 2013b). PCR reactions were performed in an Applied Biosystems 2720 Thermal Cycler (Applied Biosystems, USA) with an initial denaturation at 95°C for 5 minutes; this was followed by 30–45 cycles of denaturation at 95°C for 30 seconds, annealing at 50–60°C for 30 seconds, and extension at 72°C for 30 seconds. The final extension was completed at 72°C for ten minutes. Following amplification, PCR products were run on 2% agarose gel with 1× TAE buffer and stained in 10 μg/mL ethidium bromide. AlphaImager® HP was used to visualize and photograph gels (ProteinSimple, USA).
Table 1.
PCR reaction primers, annealing temperatures, and amplicon sizes.
| Gene name | Primer (5’−3’) | Annealing temperature | Amplicon size (bp) | Reference |
|---|---|---|---|---|
| 16S rRNA | F-AGAGTTTGATCCTGGCTCAG | 58°C | 436 | Simser et al., 2002 |
| R-AACGTCATTATCTTCCTTGC | ||||
| gltA | F-GGGGGCCTGCTCACGGCGG | 50°C | 382 | Simser et al., 2002 |
| R-ATTGCAAAAAGTACAGTGAACA | ||||
| ompA | F-ATGGCGAATATTTCTCCAAAAA | 60°C | 530 | Simser et al., 2002 |
| R-AGTGCAGCATTCGCTCCCCCT | ||||
| dksA-xerC | F-TTTTCATGACGCTCTTGAGC | 58°C | 165 | Fournier and Raoult, 2007 |
| R-GTAAAGAAAGAATAATTCCGTGGTT | ||||
| mppA-purC | F-GCATATGCRGTRGGTAGTTAT | 59°C | 1000 | Fournier and Raoult, 2007 |
| R-CACACGCCCAAATTCTAATT | ||||
| rpmE-tRNAfMet | F-TTCCGGAAATGTAGTAAATCAATC | 57°C | 368 | Fournier and Raoult, 2007 |
| R-TCAGGTTATGAG CCTGACGA | ||||
| β-actin | F-TTGTCCGCGACATCAAGGA | 57°C | 110 | Cheng et al., 2013b |
| R-CGGGAAGCTCGTAGGACTTCT |
Cloning and DNA sequencing
PCR amplicons were ligated into StrataClone’s pSC-A-amp/kan cloning vector (Agilent Technologies, USA). Transformants were plated on LB agar containing 100 μg/mL ampicillin and 40 μl of 2% X-gal. White colonies were then restreaked on LB/AMP/X-gal plates. Following restreaking, white colonies were grown in LB broth containing 100 mg/mL ampicillin overnight at 37°C. Plasmids were purified by E.Z.N.A.® Plasmid Mini Kit (Omega, USA) and constructs were confirmed by running EcoRI restriction enzyme digests (Promega, USA) on 1% agarose gel with 1× TAE buffer and stained in ethidium bromide. Sequencing of the PCR amplicons was conducted by Elim Biopharmaceuticals, Inc. using M13 reverse primer (5’-GGAAACAGCTATGACCATG-3’). The GenBank accession numbers for the 16S rRNA, gltA, and ompA genes, dksA-xerC, mppA-purC, and rpmE- tRNAfMet regions for determining the numbers of species and genotypes of the REIP isolate are KX505845, KX505846, KX505847, KX505844, KX505842, KX505843, respectively.
Phylogenetic analysis
We utilized the completed genome sequence of REIP, which is available in NCBI (accession number NZ_LAOP01000001). Homologous sequences of the whole open reading frame of each of the five genes of REIP, 16S rRNA (NZ_LAOP01000001), gltA (KJW02430), ompA (KJW02278), ompB (KJW03404), and sca4 (KJW03052), were obtained by BLAST analysis. For phylogenetic reconstructions, homologous sequences for each gene (Supplemental Table) were aligned using Clustal Omega at EMBL-EBI. The resulting aligned sequences of 16S rRNA (1,440 bp), gltA (1,233 bp), ompA (1,330 bp), ompB (2,847 bp) and sca4 (2,691 bp) genes of REIP and 21 validated Rickettsia species were concatenated to generate a sequence of 9,829 characters for each species in Mesquite (version 3.04). Sequences from other validated Rickettsia species were not included in this study since their ompA gene is either truncated or lacking. The concatenated nucleotide sequences were translated to amino acid sequences, with a purpose of aligning the nucleotide sequences to match the amino acid alignments in Mesquite. Phylogeny of the concatenated sequence was constructed by SeaView (version 4.4.3) (http://pbil.univlyon1.fr/software/seaview.html). Gouy and Guindon S. & Gascuels’ method was used to determine evolutionary distance values, which were then used to construct phylograms by neighbor-joining method and maximal parsimony method (Gouy et al., 2010). The pairwise nucleotide sequence identity of each gene between REIP and validated Rickettsia species was calculated in SIAS (http://imed.med.ucm.es/Tools/sias.html) using aligned sequences from Clustal Omega.
Results and discussion
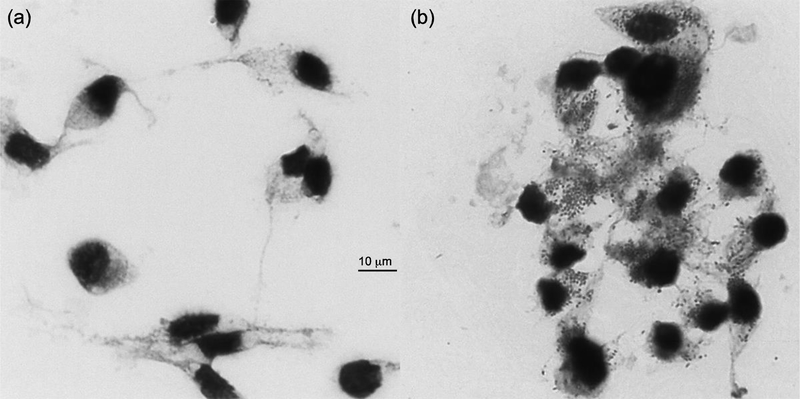
We successfully propagated the REIP isolate using embryonic tick cell lines IRE11 and ISE6 (Simser et al., 2002; Munderloh and Kurtti, 1989), in L15B300 media formulated for tick cell culture (Munderloh and Kurtti, 1989; Oliver et al., 2014). Giemsa stained REIP-infected ISE6 cells were observed under light microscope. Coccobacillus-shaped bacteria were viewed residing in the cytoplasm or outside lysed cells; uninfected controls had no visible signs of bacterial inhabitance (Fig. 1).
Fig. 1. Giemsa stain of Rickettsia endosymbiont Ixodes pacificus (REIP)-infected ISE6 cells propagated for three weeks at 26°C.
(a) Uninfected ISE6 control without bacteria residing in the cytoplasm. (b) Heavily infected ISE6 cells three weeks post infection with REIP. Original magnification: x1000. Bar, the scale bar in μm
To confirm that there is only one bacterial species and/or genotype in the REIP isolate, we utilized multilocus sequence typing (MLST) using three genes—16S rRNA, gltA, and ompA—allowing for the detection of single nucleotide polymorphisms that distinguish Rickettsia species (Fournier et al., 2003; Zhu et al., 2005a). However, because coding DNA may not provide enough nucleotide sequence variation to differentiate within some rickettsial species, we also used multi-spacer sequence typing (MST)—utilizing intergenic spacers such as dksA-xerC, mppA-purC, and rpmE-tRNAfMet—as this method is capable of differentiating at the intraspecies level (Fournier et al., 2004; Zhu et al., 2005b). The three genes and three intergenic spacers were amplified via PCR and amplicons were ligated into StrataClone’s pSC-A-amp/kan cloning vector. Sequencing data determined the insert size of clones for gltA, ompA, 16S rRNA genes of REIP to be 382, 530 and 436 base pair (bp)-long, respectively. PCR amplification and cloning of intergenic spacers determined the insert size of dksA-xerC, mppA-purC, and rpmE-tRNAfMet regions were 165, 121, 134 and 368 bp, respectively (data not shown). Additional, sequences from 30 clones of 16S rRNA, gltA, ompA, dksA-xerC, mppA-purC, and rpmE-tRNAfMet genes of REIP were aligned in ClustalX version 2.1. Nucleotide sequence alignments of thirty clones for each of the genes and intergenic spacers determined that the nucleotide sequences were identical to one another in the amplified regions. Additionally, no difference was noted when comparing amplified 16S rRNA, gltA, and ompA sequences to sequences obtained from the genome sequencing project of REIP (Supplemental Table). Considering REIP’s nucleotide homology among all thirty clones, we assumed the REIP isolate possessed one genotype.
We utilized complete open reading frames of 16S rRNA, gltA, ompA, ompB, and sca4 nucleotide sequences of REIP obtained from the genome sequencing project in NCBI (accession number NZ_LAOP01000001) to classify the REIP isolate. The pairwise nucleotide sequence identity comparison was conducted between 1,440 bp 16S rRNA, 1,233 bp gltA, 1,330 bp ompA, 2,847 bp ompB, and 2,691 bp sca4 genes of REIP and homologous sequences of 21 validated Rickettsia species in GenBank. The nucleotide sequence identities of 16S rRNA, gltA, ompA, ompB, and sca4 genes of REIP with homologous sequences of R. buchneri str. ISO7 (Kurtti et al., 2015) was 99.7%, 99.5%, 91.7%, 76.4%, and 47.3%, respectively (Table 2). Based on Fournier et al.’s classification scheme (Fournier et al., 2003), these values confirmed the isolate is a member of the Rickettsia genus. For all genes, REIP shared the highest nucleotide sequence identity with ‘R. monacensis’ str. IrR/Munich (99.8, 99.4, 99.2, 99.5, and 99.6% for 16S rRNA, gltA, ompA, ompB, and sca4, respectively). The identities between the two species for all genes indicate that the REIP isolate is closely related to ‘R. monacensis’. Moreover, the isolate also displayed similarity to Rickettsia species phylotype G021. For instance, the isolate shared 99%, 99%, and 100% nucleotide identity to Rickettsia species phylotype G021’s ompA, 16S rRNA, and gltA sequences, respectively (Phan et al., 2011). Considering the nucleotide sequences of 30 clones for each gene were identical, we ruled out the possibility that the discrepancies observed between nucleotide sequences for REIP and phylotype G021’s genes could have resulted from random errors during PCR. The incongruity is more likely due to bacterial genetic diversity within species or an error when sequencing phylotype G021. Therefore, we are in favor of the conclusion that REIP is the same species as phylotype G021, however possesses a different genotype.
Table 2.
The nucleotide sequence identity of the gltA, ompA, ompB, sca4, and 16S rRNA genes between ‘Rickettsia monacensis’ str. Humboldt and Rickettsia buchneri str. ISO7 and R. monacensis str. IrR/Munich.
| Strains | ‘R. monacensis’ str. Humboldt | ||||
|---|---|---|---|---|---|
| 16S rRNA | gltA | ompA | ompB | sca4 | |
| R. buchneri str. IS07 | 99.65 | 99.51 | 91.71 | 76.41 | 47.32 |
| R. monacensis str. IrR/Munich | 99.79 | 99.35 | 99.17 | 99.50 | 99.55 |
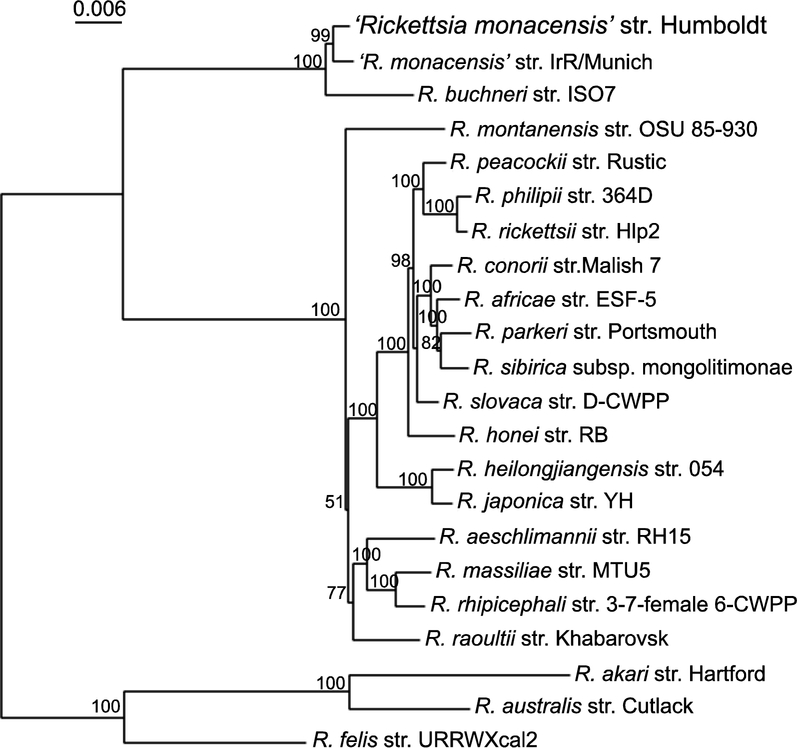
Sequence similarities present between REIP, R. buchneri, ‘R. monacensis’, and Rickettsia species phylotype G021 were also reflected via phylogenetic analysis. Phylogenetic analysis was conducted by comparing the REIP isolate’s concatenated gltA, ompA, ompB, sca4, and 16S rRNA nucleotide sequences to homologous sequences of rickettsiae available in GenBank. Trees generated from the concatenated sequences reflected those previously generated by our lab for Rickettsia species phylotype G021 (Phan et al., 2011). The tree topology indicated that REIP was clustered with ‘R. monacensis’ str. IrR/Munich and that the two bacteria were placed in the same clade as R. buchneri str. ISO7. This monophyletic group forms a sister group to the rest of a clade that contains 16 spotted fever group rickettsiae. The concatenated sequences of the five genes of the spotted fever group rickettsiae are distant from the sequences of R. akari, R. australis, and R. felis (Fig. 2). The neighbor joining tree indicated that the monophyletic groups, in which REIP resides, are distinct from the rest of the spotted fever group rickettsiae. We inferred that the REIP isolate is closely related to ‘R. monacensis’ and shares similar evolutionary history with R. buchneri and ‘R. monacensis’ str. IrR/Munich. As such, based on our nucleotide sequence identities and phylogenetic analyses, we propose that the REIP isolate constitutes a new strand of ‘R. monacensis’ different from ‘R. monacensis’ strain IrR/Munich. Therefore, we propose the name ‘Rickettsia monacensis’ str. Humboldt, for the bacterium.
Fig. 2. Phylogenetic tree of concatenated gltA, ompA, ompB, sca4, and 16S rRNA gene sequences of ‘Rickettsia monacensis’ str. Humboldt.
The tree was generated using neighbor-joining distance method and 1000 bootstrap replicates. Bootstrap values >50% are shown at the nodes. Bar, nucleotide distance. ‘R. monacensis’ str. Humboldt (or Rickettsia endosymbiont Ixodes pacificus) is represented at the top of the tree. Other selected sequences on the tree include: R. aeschlimannii, R. africae, R. akari, R. australis, R. buchneri, R. conorii, R. felis, R. heilongjiangensis, R. honei, R. japonica, R. massiliae, ‘R. monacensis’ str. IrR/Munich, R. montanensis, R. parkeri, R. peacockii, R. philipii, R. raoultii, R. rhipicephali, R. rickettsii, R. sibirica, and R. slovaca. The accession numbers of genes of all Rickettsia species are listed in Supplemental Table.
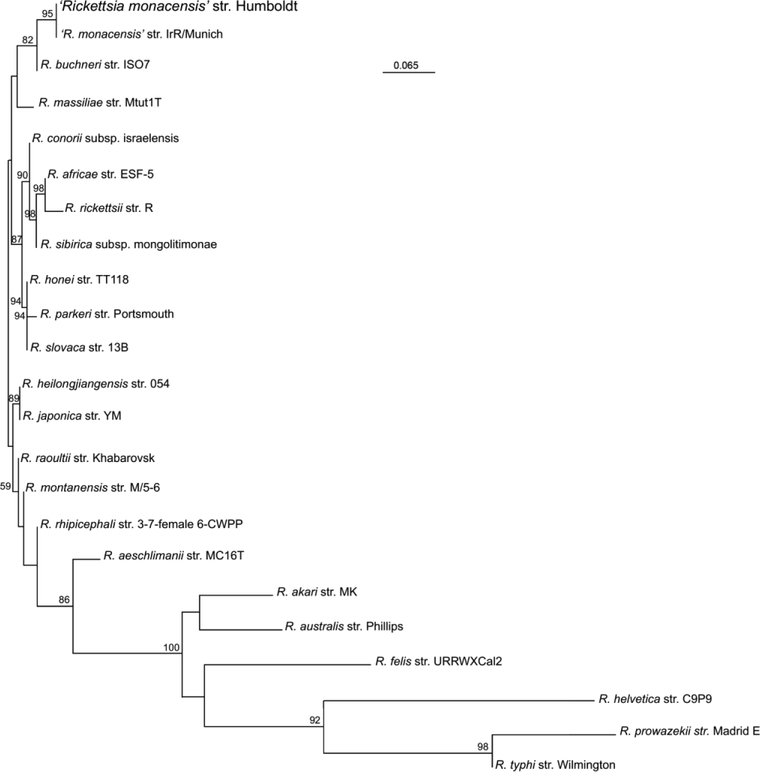
Previous studies have demonstrated the ability of MST to differentiate between genotypes of some Rickettsia species by at least one nucleotide mutation (Fournier and Raoult, 2007; Zhu et al., 2005b; Znazen et al., 2013). As such, MST was used to further understand the evolutionary relationship between ‘R. monacensis’ str. Humboldt and other Rickettsia species. A neighbor joining tree was generated based on the concatenated nucleotide sequence of dksA-xerC, mppA-purC, and rpmE-tRNAfMet regions and their homologous sequences from 22 validated Rickettsia species in GenBank (Supplemental Table). There was a limit to what rickettsial strains could be used to generate the tree as some strains don’t have all three spacer sequences and these regions for other strains have yet to be sequenced and made available in GenBank. As shown in Figure 3, the phylogram indicated that ‘R. monacensis’ str. Humboldt and ‘R. monacensis’ str. IrR/Munich form a unique clade. The phylogram also confirmed that ‘R. monacensis’ str. Humboldt is closely related to ‘R. monacensis’ str. IrR/Munich and that ‘R. monacensis’ str. Humboldt, ‘R. monacensis’ str. IrR/Munich, R. buchneri str. ISO7, and R. massiliae str. Mtut1T form a monophyletic group that is distinct from other spotted fever group rickettsiae (Fig. 3).
Fig. 3. Phylogenetic tree of concatenated nucleotide sequences of dksA-xerC, mppA-purC, and rpmE-tRNAfMet intergenic spacers of ‘Rickettsia monacensis’ str. Humboldt.
The tree was generated using neighbor-joining distance method and 1000 bootstrap replicates. Bootstrap values >50% are shown at the nodes. Bar, nucleotide distance. ‘R. monacensis’ str. Humboldt (or Rickettsia endosymbiont Ixodes pacificus) is represented at the top of the tree. Other selected sequences on the tree include: R. aeschlimannii, R. africae, R. akari, R. australis, R. buchneri, R. conorii, R. felis, R. heilongjiangensis, R. helvetica, R. japonica, R. massiliae, ‘R. monacensis’ str. IrR/Munich, R. montanensis, R. parkeri, R. prowazekii, R. raoultii, R. rhipicephali, R. rickettsii, R. sibirica, R. slovaca, and R. typhi. The accession numbers of genes of all Rickettsia species are listed in Supplemental Table.
Based on MLST, MST, and phylogenetic tree constructions of this study, we have drawn a conclusion that the REIP isolate from I. pacificus is ‘R. monacensis’. Our conclusion is supported by a recent genome-wide comparison among genomes of Rickettsia species using core genome alignment sequence identity (CGASI), a method using both length and sequence identity of genome for species classification. Using a CGASI cutoff of equal or greater than 96.8%, ‘R. monacensis’ str. Humboldt from I. pacificus and ‘R. monacensis’ str. IrR/Munich are classified as the same species. There are only two Rickettsia species in the spotted fever group based on CGASI reclassification. In addition to ‘R. monacensis’ species, all other Rickettsia species in the spotted fever group are classified as the second Rickettsia species (Chung et al., 2018). Our conclusion that ‘R. monacensis’ str. Humboldt and ‘R. monacensis’ str. IrR/Munich are two different strains within the same species is also supported by CGASI since the core genome alignment sequence identity between ‘R. monacensis’ str. Humboldt and ‘R. monacensis’ str. IrR/Munich is 99.6% (Chung et al., 2018). In addition, the different genome content between the two strains is reflected by unique plasmids that exist in their genomes. Pulse-field gel electrophoresis and Southern blot identified pRM plasmid of ‘R. monacensis’ str. IrR/Munich (Baldridge et al., 2007) (GenBank Accession Number EF564599), whereas genome sequencing of ‘R. monacensis’ str. Humboldt revealed the presence of pREIP plasmid (KR611317). Although the size of pREIP (102 kb) differs dramatically with that of pRM (23.5 kb), parts of the pREIP share 99% nucleotide sequence identity with pRM (T. Kurtti, personal communication). However, the pREIP and pRM plasmids have low nucleotide homology with other rickettsial plasmids from other spotted fever group rickettsia, including R. amblyommii, R. buchneri, R. helvetica, R. hoogstraalii, R. massiliae, and R. peacockii (Baldridge et al., 2008; Gillespie et al., 2012; T. Kurtti, personal communication).
‘R. monacensis’ was originally isolated in an I. ricinus tick in Munich, Germany (Simser et al., 2002). Later, ‘R. monacensis’ infection in I. ricinus was identified all along Europe, including Hungary (Sreter-Lancz et al., 2006), Portugal (de Carvalho et al., 2008), Switzerland (Boretti et al., 2009), Sweden (Elfving et al., 2010), Italy (Corrain et al., 2012), the Netherlands (Koetsveld et al., 2016), Turkey (Gargili et al., 2012), Spain (Palomar et al., 2012), Poland (Rymaszewska and Piotrowski, 2013), Romania (Ionita et al., 2013), Croatia (Tijsse-Klasen et al., 2013), Slovakia (Spitalska et al., 2014), and Estonia (Katargina et al., 2015). In addition to Europe, ‘R. monacensis’ has been reported in Africa, Asia, and South and Central America. In Africa, ‘R. monacensis’ was isolated from I. ricinus in Tunisia (Sfar et al., 2008) and Algeria (Dib et al., 2009). Also, the bacterium infection was documented in I. nipponensis ticks in Korea (Lee et al., 2013) and in I. sinensis ticks in China (Ye et al., 2014). In South and Central America, I. boliviensis and Rhipicephalus sanguineus sensu lato were tested positive for ‘R. monacensis’ in Nicaragua and Costa Rica (Springer et al., 2018; Troyo et al., 2014). The global distribution of ‘R. monacensis’ is, at least partially, attributed to migrating birds, which serve as possible dispersers of the bacterium (Elfving et al., 2010; Biernat et al., 2016). Many not yet isolated rickettsia in Ixodes ticks have been reported. Some of them are genetically close to ‘R. monacensis’ str. Humboldt and ‘R. monacensis’. For example, the Rickettsia species detected in Amblyomma ovale (Springer et al., 2018). Future isolation and characterization of the bacteria, especially genomic sequencing and CGASI analysis, are required to confirm that they belong to ‘R. monacensis’.
Supplementary Material
Funding and acknowledgements
This research was supported by National Institutes of Health grant 1R15AI099902-01. We would like to express our great appreciation to the veterinarian and staff at McKinleyville Animal Care Center, McKinleyville, CA, for donating partially engorged Ixodes pacificus females for our research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interest: None
References
- Abdad MY, Abdallah RA, Karkouri KE, Beye M, Stenos J, Owen H, Unsworth N, Robertson I, Blacksell SD, Nguyen TT, Nappez C, Raoult D, Fenwick S, Fournier PE, 2017. Rickettsia gravesii sp. nov.: a novel spotted fever group rickettsia in Western Australian Amblyomma triguttatum triguttatum ticks. Int. J. Syst. Evol. Microbiol 67, 3156–3161. [DOI] [PubMed] [Google Scholar]
- Bagheri G, Lehner JD, Zhong J, 2017. Enhanced detection of Rickettsia species in Ixodes pacificus using highly sensitive fluorescence in situ hybridization coupled with Tyramide Signal Amplification. Ticks Tick Borne Dis. 8, 915–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biernat B, Stanczak J, Michalik J, Sikora B, Cieniuch S, 2016. Rickettsia helvetica and R. monacensis infections in immature Ixodes ricinus ticks derived from sylvatic passerine birds in west-central Poland. Parasitol. Res 115, 3469–3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldridge GD, Burkhardt NY, Felsheim RF, Kurtti TJ, Munderloh UG, 2007. Transposon insertion reveals pRM, a plasmid of Rickettsia monacensis. Appl. Environ. Microbiol 73, 4984–4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldridge GD, Burkhardt NY, Felsheim RF, Kurtti TJ, Munderloh UG, 2008. Plasmids of the pRM/pRF family occur in diverse Rickettsia species. Appl. Environ. Microbiol 74, 645–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodnar JL, Fitch S, Rosati A, Zhong J, 2018. The folA gene from the Rickettsia endosymbiont of Ixodes pacificus encodes a functional dihydrofolate reductase enzyme. Ticks Tick Borne Dis. 9, 443–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boretti FS, Perreten A, Meli ML, Cattori V, Willi B, Wengi N, Hornok S, Honegger H, Hegglin D, Woelfel R, Reusch CE, Lutz H, Hofmann-Lehmann R, 2009. Molecular investigations of Rickettsia helvetica infection in dogs, foxes, humans, and Ixodes ticks. Appl. Environ. Microbiol 75, 3230–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgdorfer W, Lane RS, Barbour AG, Gresbrink RA, Anderson JR, 1985. The western black-legged tick, Ixodes pacificus: a vector of Borrelia burgdorferi. Am. J. Trop. Med. Hyg 34, 925–930. [DOI] [PubMed] [Google Scholar]
- Cazorla C, Socolovschi C, Jensenius M, Parola P, 2008. Tick-borne diseases: tick-borne spotted fever rickettsioses in Africa. Infect. Dis. Clin. North. Am 22, 531–544, ix–x. [DOI] [PubMed] [Google Scholar]
- Cheng D, Lane RS, Moore BD, Zhong J, 2013a. Host blood meal-dependent growth ensures transovarial transmission and transstadial passage of Rickettsia sp. phylotype G021 in the western black-legged tick (Ixodes pacificus). Ticks Tick Borne Dis. 4, 421–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng D, Vigil K, Schanes P, Brown RN, Zhong J, 2013b. Prevalence and burden of two rickettsial phylotypes (G021 and G022) in Ixodes pacificus from California by real-time quantitative PCR. Ticks Tick Borne Dis. 4, 280–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung M, Munro JB, Dunning Hotopp JC, 2018. Using core genome alignments to assign bacterial species. bioRxiv doi: 10.1101/328021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrain R, Drigo M, Fenati M, Menandro ML, Mondin A, Pasotto D, Martini M, 2012. Study on ticks and tick-borne zoonoses in public parks in Italy. Zoonoses Public Health 59, 468–476. [DOI] [PubMed] [Google Scholar]
- de Carvalho IL, Milhano N, Santos AS, Almeida V, Barros SC, De Sousa R, Nuncio MS, 2008. Detection of Borrelia lusitaniae, Rickettsia sp. IRS3, Rickettsia monacensis, and Anaplasma phagocytophilum in Ixodes ricinus collected in Madeira Island, Portugal. Vector Borne Zoonotic Dis. 8, 575–579. [DOI] [PubMed] [Google Scholar]
- Dib L, Bitam I, Bensouilah M, Parola P, Raoult D, 2009. First description of Rickettsia monacensis in Ixodes ricinus in Algeria. Clin. Microbiol. Infect 15 Suppl 2, 261–262. [DOI] [PubMed] [Google Scholar]
- Elfving K, Olsen B, Bergstrom S, Waldenstrom J, Lundkvist A, Sjostedt A, Mejlon H, Nilsson K, 2010. Dissemination of spotted fever rickettsia agents in Europe by migrating birds. PLoS One 5, e8572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley JE, Nieto NC, Adjemian J, Dabritz H, Brown RN, 2008. Anaplasma phagocytophilum infection in small mammal hosts of Ixodes ticks, western United States. Emerg. Infect. Dis 14, 1147–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier PE, Dumler JS, Greub G, Zhang J, Wu Y, Raoult D, 2003. Gene sequencebased criteria for identification of new rickettsia isolates and description of Rickettsia heilongjiangensis sp. nov. J. Clin. Microbiol 41, 5456–5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier PE, Raoult D, 2007. Identification of rickettsial isolates at the species level using multi-spacer typing. BMC Microbiol. 7, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier PE, Zhu Y, Ogata H, Raoult D, 2004. Use of highly variable intergenic spacer sequences for multispacer typing of Rickettsia conorii strains. J. Clin. Microbiol 42, 5757–5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargili A, Palomar AM, Midilli K, Portillo A, Kar S, Oteo JA, 2012. Rickettsia species in ticks removed from humans in Istanbul, Turkey. Vector Borne Zoonotic Dis. 12, 938–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie JJ, Joardar V, Williams KP, Driscoll T, Hostetler JB, Nordberg E, Shukla M, Walenz B, Hill CA, Nene VM, Azad AF, Sobral BW, Caler E, 2012. A Rickettsia genome overrun by mobile genetic elements provides insight into the acquisition of genes characteristic of an obligate intracellular lifestyle. J. Bacteriol 194, 376–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O, 2010. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol 27, 221–224. [DOI] [PubMed] [Google Scholar]
- Hardstone Yoshimizu M and Billeter SA, 2018. Suspected and Confirmed Vector-Borne Rickettsioses of North America Associated with Human Diseases. Trop. Med. Infect. Dis 3(1): 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes LE, Clifford CM, Gresbrink R, Thomas LA, Keirans JE, 1976. Isolation of a spotted fever group rickettsia from the Pacific Coast tick, Ixodes pacificus, in Oregon. Am. J. Trop. Med. Hyg 25, 513–516. [DOI] [PubMed] [Google Scholar]
- Hunter DJ, Torkelson JL, Bodnar J, Mortazavi B, Laurent T, Deason J, Thephavongsa K, Zhong J, 2015. The Rickettsia endosymbiont of Ixodes pacificus contains all the genes of de novo folate biosynthesis. PLoS One. 10, e0144552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionita M, Mitrea IL, Pfister K, Hamel D, Silaghi C, 2013. Molecular evidence for bacterial and protozoan pathogens in hard ticks from Romania. Vet. Parasitol 196, 71–76. [DOI] [PubMed] [Google Scholar]
- Katargina O, Geller J, Ivanova A, Varv K, Tefanova V, Vene S, Lundkvist Å,Golovljova I, 2015. Detection and identification of Rickettsia species in Ixodes tick populations from Estonia. Ticks Tick Borne Dis. 6, 689–694. [DOI] [PubMed] [Google Scholar]
- Koetsveld J, Tijsse-Klasen E, Herremans T, Hovius JW, Sprong H, 2016. Serological and molecular evidence for spotted fever group Rickettsia and Borrelia burgdorferi sensu lato co-infections in The Netherlands. Ticks Tick Borne Dis. 7, 371–377. [DOI] [PubMed] [Google Scholar]
- Kurlovs AH, Li J, Cheng D, Zhong J, 2014. Ixodes pacificus ticks maintain embryogenesis and egg hatching after antibiotic treatment of Rickettsia endosymbiont. PLoS One 9, e104815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtti TJ, Felsheim RF, Burkhardt NY, Oliver JD, Heu CC, Munderloh UG, 2015. Rickettsia buchneri sp. nov., a rickettsial endosymbiont of the blacklegged tick Ixodes scapularis. Int. J. Syst. Evol. Microbiol 65, 965–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtti TJ, Munderloh UG, Andreadis TG, Magnarelli LA, Mather TN, 1996. Tick cell culture isolation of an intracellular prokaryote from the tick Ixodes scapularis. J. Invertebr. Pathol 67, 318–321. [DOI] [PubMed] [Google Scholar]
- Kurtti TJ, Simser JA, Baldridge GD, Palmer AT, Munderloh UG, 2005. Factors influencing in vitro infectivity and growth of Rickettsia peacockii (Rickettsiales: Rickettsiaceae), an endosymbiont of the Rocky Mountain wood tick, Dermacentor andersoni (Acari, Ixodidae). J. Invertebr. Pathol 90, 177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KM, Choi YJ, Shin SH, Choi MK, Song HJ, Kim HC, Klein TA, Richards AL, Park KH, Jang WJ, 2013. Spotted fever group rickettsia closely related to Rickettsia monacensis isolated from ticks in South Jeolla province, Korea. Microbiol. Immunol 57, 487–495. [DOI] [PubMed] [Google Scholar]
- Munderloh UG, Kurtti TJ, 1989. Formulation of medium for tick cell culture. Exp. Appl. Acarol 7, 219–229. [DOI] [PubMed] [Google Scholar]
- Munderloh UG, Liu Y, Wang M, Chen C, Kurtti TJ, 1994. Establishment, maintenance and description of cell lines from the tick Ixodes scapularis. J. Parasitol 80, 533–543. [PubMed] [Google Scholar]
- Oliver JD, Burkhardt NY, Felsheim RF, Kurtti TJ, Munderloh UG, 2014. Motility characteristics are altered for Rickettsia bellii transformed to overexpress a heterologous rickA gene. Appl. Environ. Microbiol 80, 1170–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomar AM, Santibanez P, Mazuelas D, Roncero L, Santibanez S, Portillo A, Oteo JA, 2012. Role of birds in dispersal of etiologic agents of tick-borne zoonoses, Spain, 2009. Emerg. Infect. Dis 18, 1188–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parola P, Paddock CD, Socolovschi C, Labruna MB, Mediannikov O, Kernif T, Abdad MY, Stenos J, Bitam I, Fournier PE, Raoult D, 2013. Update on tickborne rickettsioses around the world: a geographic approach. Clin. Microbiol. Rev 26, 657–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan JN, Lu CR, Bender WG, Smoak RM 3rd, Zhong J, 2011. Molecular detection and identification of Rickettsia species in Ixodes pacificus in California. Vector Borne Zoonotic Dis. 11, 957–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip RN, Casper EA, Burgdorfer W, Gerloff RK, Hughes LE, Bell EJ, 1978. Serologic typing of rickettsiae of the spotted fever group by microimmunofluorescence. J. Immunol 121, 1961–1968. [PubMed] [Google Scholar]
- Philip RN, Lane RS, Casper EA, 1981. Serotypes of tick-borne spotted fever group rickettsiae from western California. Am. J. Trop. Med. Hyg 30, 722–727. [DOI] [PubMed] [Google Scholar]
- Piesman J, Clark KL, Dolan MC, Happ CM, Burkot TR, 1999. Geographic survey of vector ticks (Ixodes scapularis and Ixodes pacificus) for infection with the Lyme disease spirochete, Borrelia burgdorferi. J. Vector Ecol 24, 91–98. [PubMed] [Google Scholar]
- Richards AL, 2012. Worldwide detection and identification of new and old rickettsiae and rickettsial diseases. FEMS Immunol. Med. Microbiol 64, 107–110. [DOI] [PubMed] [Google Scholar]
- Rymaszewska A, Piotrowski M, 2013. Use of DNA sequences for Rickettsia identification in Ixodes ricinus ticks: the first detection of Rickettsia monacensis in Poland. Microbes Infect. 15, 140–146. [DOI] [PubMed] [Google Scholar]
- Sfar N, M’Ghirbi Y, Letaief A, Parola P, Bouattour A, Raoult D, 2008. First report of Rickettsia monacensis and Rickettsia helvetica from Tunisia. Ann. Trop. Med. Parasitol 102, 561–564. [DOI] [PubMed] [Google Scholar]
- Simser JA, Palmer AT, Fingerle V, Wilske B, Kurtti TJ, Munderloh UG, 2002Rickettsia monacensis sp. nov., a spotted fever group Rickettsia, from ticks (Ixodes ricinus) collected in a European city park. Appl. Environ. Microbiol 68, 4559–4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitalska E, Boldis V, Derdakova M, Selyemova D, Rusnakova Taragelova V, 2014. Rickettsial infection in Ixodes ricinus ticks in urban and natural habitats of Slovakia. Ticks Tick Borne Dis. 5, 161–165. [DOI] [PubMed] [Google Scholar]
- Springer A, Montenegro VM, Schicht S, Wölfel S, Schaper SR, Chitimia-Dobler L, Siebert S, Strube C, 2018. Detection of Rickettsia monacensis and Rickettsia amblyommatis in ticks collected from dogs in Costa Rica and Nicaragua. Ticks Tick Borne Dis. 9, 1565–1572. [DOI] [PubMed] [Google Scholar]
- Sreter-Lancz Z, Szell Z, Kovacs G, Egyed L, Marialigeti K, Sreter T, 2006Rickettsiae of the spotted-fever group in ixodid ticks from Hungary: identification of a new genotype (‘Candidatus Rickettsia kotlanii’). Ann. Trop. Med. Parasitol 100, 229–236. [DOI] [PubMed] [Google Scholar]
- Tijsse-Klasen E, Sprong H, Pandak N, 2013. Co-infection of Borrelia burgdorferi sensu lato and Rickettsia species in ticks and in an erythema migrans patient. Parasit. Vectors 6, 347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyo A, Moreira-Soto A, Carranza M, Calderón-Arguedas O, Hun L, Taylor L, 2014. Detection of an undescribed Rickettsia sp. in Ixodes boliviensis from Costa Rica. Ticks Tick Borne Dis. 5, 883–886. [DOI] [PubMed] [Google Scholar]
- Ye X, Sun Y, Ju W, Wang X, Cao W, Wu M, 2014. Vector competence of the tick Ixodes sinensis (Acari: Ixodidae) for Rickettsia monacensis. Parasit. Vectors 7, 512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Fournier PE, Eremeeva M, Raoult D, 2005a. Proposal to create subspecies of Rickettsia conorii based on multi-locus sequence typing and an emended description of Rickettsia conorii. BMC Microbiol. 5, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Fournier PE, Ogata H, Raoult D, 2005b. Multispacer typing of Rickettsia prowazekii enabling epidemiological studies of epidemic typhus. J. Clin. Microbiol 43, 4708–4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Znazen A, Khrouf F, Elleuch N, Lahiani D, Marrekchi C, M’Ghirbi Y, Ben Jemaa M, Bouattour A, Hammami A, 2013. Multispacer typing of Rickettsia isolates from humans and ticks in Tunisia revealing new genotypes. Parasit. Vectors 6, 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.