Summary
Investigators have long suspected that pathogenic microbes might contribute to the onset and progression of Alzheimer’s disease (AD) although definitive evidence has not been presented. Whether such findings represent a causal contribution, or reflect opportunistic passengers of neurodegeneration is also difficult to resolve. We constructed multiscale networks of the late onset AD-associated virome, integrating genomic, transcriptomic, proteomic, and histopathological data across four brain regions from human postmortem tissue. We observed increased human herpesvirus 6A (HHV-6A) and human herpesvirus 7 (HHV-7) from subjects with AD compared with controls. These results were replicated in two additional, independent and geographically dispersed cohorts. We observed regulatory relationships linking viral abundance and modulators of APP metabolism, including induction of APBB2, APPBP2, BIN1, BACE1, CLU, PICALM, and PSEN1 by HHV-6A. This study elucidates networks linking molecular, clinical, and neuropathological features with viral activity and is consistent with viral activity constituting a general feature of AD.
eTOC
Readhead et al. construct multiscale networks of the late onset Alzheimer’s disease (AD) associated virome, and observe pathogenic regulation of molecular, clinical and neuropathological networks by several common viruses, particularly human herpesvirus 6A and human herpesvirus 7.
Introduction
Important roles for microbes and antimicrobial defenses in the pathogenesis of Alzheimer’s disease (AD) have been postulated or evaluated for at least six decades, beginning with Sjögren in 1952(Sjogren et al., 1952). “Slow virus” was one of the early names used for the illness that eventually came to be known as prion disease, referring to the hypothesis that conventional viruses might be capable of acting to cause not only acute encephalitis, but also a progressive neuronal destruction process that might engender less inflammation because of its slowly progressive nature(Sigurðsson, 1954). Measles (MV) is a conventional virus that can act through acute inflammatory and slow neurodegenerative processes, occasionally re-emerging as a fatal brain disease known as subacute sclerosing panencephalitis (SSPE) up to a decade after a typical acute MV infection. (Murphy et al., 1976).
Beginning with Crapper McLachlan in 1980(Middleton et al., 1980), several investigators have proposed that AD is an SSPE-like illness, caused by a slow virus form of herpes simplex(Itzhaki, 2014). Hundreds of reports have associated AD with diverse bacterial and viral pathogens(Itzhaki et al., 2016, Mastroeni et al., 2018), most frequently implicating Herpesviridae (particularly HSV-1(Lovheim et al., 2015a, Lovheim et al., 2015b), EBV, HCMV and HHV-6(Westman et al., 2017, Carbone et al., 2014)). The results of these studies, taken in aggregate, are suggestive of a viral contribution to AD, though findings offer little insight into potential mechanisms, and a consistent association with specific viral species has not emerged.
Recent reports demonstrate that diverse classes of microbes can stimulate amyloid-beta (Aβ) aggregation and deposition as part of an intra-CNS anti-microbial innate immune response whereby the amyloidosis triggered by various microbes results in the coating of the infectious particles by the growing amyloid aggregate(Soscia et al., 2010, Kumar et al., 2016). These microbes coated with aggregated Aβ become unable to interact with cell surfaces, thereby arresting the infectious process.
We designed this study to map and compare biological networks underlying two distinct AD-associated phenotypes using multiple independent data sets collected from human subjects. We began with a computational network characterization of a specific endophenotype of AD: brains meeting neuropathological criteria for AD from individuals who were cognitively intact at the time of death(Liang et al., 2010), which we refer to here as ‘preclinical AD’(Sperling et al., 2011). We presumed that a network model of preclinical AD (and its comparison with networks built from cognitively intact persons without neuropathology) might provide novel insights into the molecular context of neuropathology in the absence of clinical symptoms. Alternatively, since these individuals had eluded cognitive decline despite significant AD pathology, we reasoned that this might illuminate protective or resilience mechanisms. Functional genomic analysis of preclinical AD network alterations revealed multiple lines of evidence consistent with viral activity. We then directly evaluated viral activity in a multiscale network analysis of four large, multi-omic data sets (comprising samples from individuals with ‘clinical AD’ as well as neuropathologically and cognitively normal controls) that included next generation sequencing data, enabling direct examination of viral DNA and RNA sequences.
This study presents novel evidence linking the activity of specific viruses with AD. This has been enabled by comprehensive molecular profiling of large patient cohorts, facilitating the integration of diverse biomedical data types into an expansive view spanning multiple disease stages, brain regions, and -omic domains. This has also allowed us to direct our analysis in an entirely data-driven manner, and benefit from a form of data capable of implicating specific viral species. Our results offer evidence of complex viral activity in the aging brain, including changes specific to AD clinical traits and genetic factors, particularly implicating Herpesviridae, HHV-6 and HHV-7. Taken together, these data provide compelling evidence that specific viral species contribute to the development of neuropathology and AD.
Results
Preclinical AD networks indicate early changes in G-quadruplex and C2H2 zinc finger activity
We constructed, mapped, and compared differences between distinct gene regulatory networks to investigate functional molecular changes underlying the etiology of AD (Figure 1). We used laser captured neuronal gene expression data to construct probabilistic causal networks (PCN) representing preclinical AD and also healthy control (individuals without cognitive impairment or neuropathology)(Liang et al., 2008, Liang et al., 2007) states. We focused on samples derived from brain regions associated with the most profound neuronal loss: the entorhinal cortex(Gomez-Isla et al., 1996) (EC) and the hippocampus(Hyman et al., 1984) (HIP). We built each PCN using a modified “inductive causation with latent variables” procedure(Pearl, 2009), constructing separate preclinical AD and control (CON) networks including paired samples from EC and HIP for each donor, and for each network, nominated genes that regulate the expression of unexpectedly large subnetworks as “network drivers” (See Methods and Table S1). Edge reproducibility in PCNs has been demonstrated to be strongly dependent on sample size, however detection of highly connected nodes is more robust across a range of sample sizes(Cohain et al., 2016). We therefore focused initially on characterizing the set of drivers present only in the CON network (“Lost in preclinical AD”) and those present only in the preclinical AD network (“Gained in preclinical AD”) as a means to prioritize differences between preclinical AD and CON states. We found that promoters of both sets of drivers are strongly enriched (compared with the rest of the network) for a shared set of C2H2 zinc finger transcription factor (C2H2-TF) binding motifs (Figure 1e), especially SP1, MAZ, NRF1 and EGR1. This prompted us to evaluate other co-regulatory features associated with C2H2-TF activity that might explain a general shift in their collective activity. We found that G-quadruplex (G4) sequences are strongly enriched among the promoters of both sets of drivers in the network, but that the “Lost in preclinical AD” drivers have significantly more G4 motifs within their introns, exons and 3’-UTR on both coding, and non-coding strands (Figure 1f–g). We concordantly found a strong negative correlation between gene G4 density (G4 motif count, normalized by gene length), and gene expression in the EC in preclinical AD and clinical AD samples (Figure 1h). Given the complex roles for G4 in dynamically regulating mRNA transcription, stability, translation and localization(Rhodes et al., 2015), we hypothesized that a global shift in G4 regulation or stability could explain the differences in C2H2-TF regulatory programs—for example, changes in expression and network influence of genes with especially high G4 density in locations like the 3’-UTR that are associated with alternative polyadenylation and miRNA-mediated regulation(Beaudoin et al., 2013).
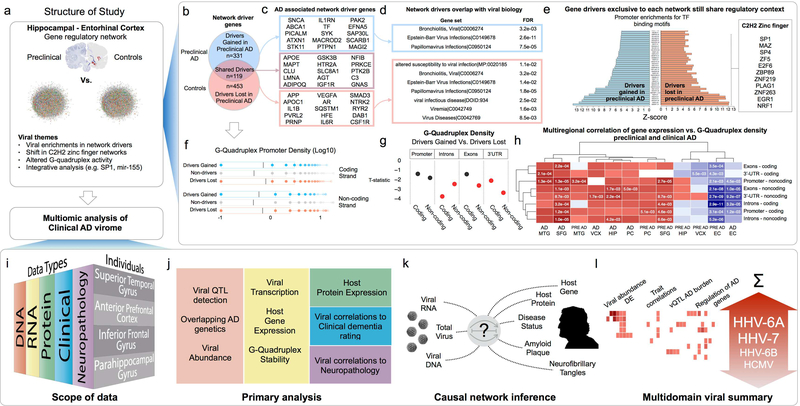
Figure 1: Multiomic evaluation of Alzheimer’s Disease associated virome.
(a) This study was performed in two phases, an initial exploration into the earliest network changes associated with preclinical AD, which identified multiple shifts in network biology consistent with viral perturbations and prompted a systematic, multiomic evaluation of viral biology on larger data sets focused on clinical AD. (b) Regulatory networks built across EC and HIP samples showed differences in gene drivers for preclinical AD vs. control networks (c) including many AD associated genes, (d) although drivers exclusive to each network shared functional characteristics, such as association with viral biology, and (e) promoter enrichments for the same C2H2 zinc finger transcription factors binding motifs. (f) G-quadruplex sequence motifs were strongly enriched in the promoters of the drivers “lost in preclinical AD”, and those “gained in preclinical AD”, however (g) drivers “lost in preclinical AD” had much higher G-Quadruplex density in introns, exons and 3’-UTR. (h) Genes with high G-Quadruplex density in these locations were uniformly down regulated in the EC of preclinical AD and AD samples, suggesting some significant alteration of G-Quadruplex regulation. (i) To investigate the possibility of an AD-associated virome, we incorporated clinical, neuropathological, RNA, DNA and proteomic data from individuals with clinical AD (and controls) across four brain regions to (j-k) identify and characterize viral activity associated with AD biology, (l) highlighting Roseoloviruses HHV-6A and HHV7. (Region abbreviations: EC: entorhinal cortex, HIP: hippocampus, MTG: medial temporal gyrus, PC: posterior cingulate cortex, SFG: superior frontal gyrus. VCX: visual cortex)
Functional analysis of network patterns suggests roles for viral mediators in AD
Identification of strong C2H2-TFs and G4 functional patterns in the differential network analysis suggested a potential role for virus-mediated network activities in AD (Figure 1d, see Table S1). Enrichments among the network drivers (gained and lost) in preclinical AD, implicated viral infection susceptibility risk genes, as well as host differential gene expression changes associated with viral infection. We also noted findings around C2H2-TF and G4 sequences that are implicated in a range of proviral and antiviral contexts, including SP1: (1) binding with Epstein-Barr Virus (EBV) protein Rta to regulate host and viral SP1 target genes(Chang et al., 2005); (2) regulating Human Immunodeficiency Virus 2 (HIV-2) LTR transcription(Harrich et al., 1989), and (3) mediating antiviral effects against Human Cytomegalovirus (HCMV) (Scholz et al., 2004). In addition, G4 sequences are recognized as important regulatory features for viral pathogens such as EBV and HSV-1(Artusi et al., 2016), dynamically impacting translation of viral proteins(Murat et al., 2014).
Patterns of miRNA target enrichments identified by the differential network analysis of preclinical AD vs. CON, as well as multiregional differential gene expression changes in clinical AD samples, offered an additional line of evidence for virally-mediated network activity. We looked for significant overlap between experimentally validated gene targets of human miRNAs(Hsu et al., 2011) and multiregional differential gene expression signatures from preclinical AD samples(Liang et al., 2010), clinical AD samples(Liang et al., 2008), as well as “Lost in preclinical AD” and “Gained in preclinical AD” drivers. This identified a number of miRNA with gene networks overlapping many of these AD contexts, particularly miR-155, a multifunctional miR with associations to malignancy, innate immunity, and DNA virus activity. Interactions between miR-155 and viral biology are well established, including perturbation of miR-155 by EBV to stabilize viral latency(Gatto et al., 2008), inhibition of miR-155 by HHV-6A(Caselli et al., 2017) and coding of a miR-155 functional ortholog by Kaposi’s sarcoma-associated herpesvirus (KSHV)(Gottwein et al., 2007) and Marek’s Disease Virus(Zhao et al., 2009). Considering these findings, we sought to directly evaluate viral DNA and RNA sequences in the context of clinical AD. We performed this investigation using four large, independent multiomic data sets from individuals with clinical AD as well as neuropathologically and cognitively normal controls.
Human Herpesvirus 6A and 7 are more abundant in Alzheimer’s disease in multiple brain regions across three independent clinical cohorts
To evaluate differential viral abundance in AD, we initially performed RNA-seq on samples from a cohort of AD and Control brains from the Mount Sinai Brain Bank (MSBB) and quantified differences in viral sequences between groups. We began by profiling MSBB sample transcriptomes across four brain regions, (STG: superior temporal gyrus, n=137, APFC: anterior prefrontal cortex, n=213, IFG: inferior frontal gyrus, n=186 and PHG: parahippocampal gyrus, n=107), which we used to quantify the presence and abundance of 515 viral species known or suspected to infect humans as a primary host(Brister et al., 2015). We applied a viral mapping approach (Figure S1), based on a modified ViromeScan workflow(Rampelli et al., 2016), optimized for detection specificity, rather than sensitivity to allow us to discriminate between viruses with highly homologous regions, and to ensure we were not falsely including human derived transcripts when summarizing viral abundance.
We estimated viral abundance at two levels. We summarized RNA reads to the level of the entire viral sequence, with the aim of estimating ‘total viral transcription’. We then summarized the RNA reads to the level of individual genomic features based on counting reads that overlap any of the genomic features included in the NCBI annotations for that viral sequence(Brister et al., 2015). Throughout this study, we have evaluated viral abundance according to these two levels separately.
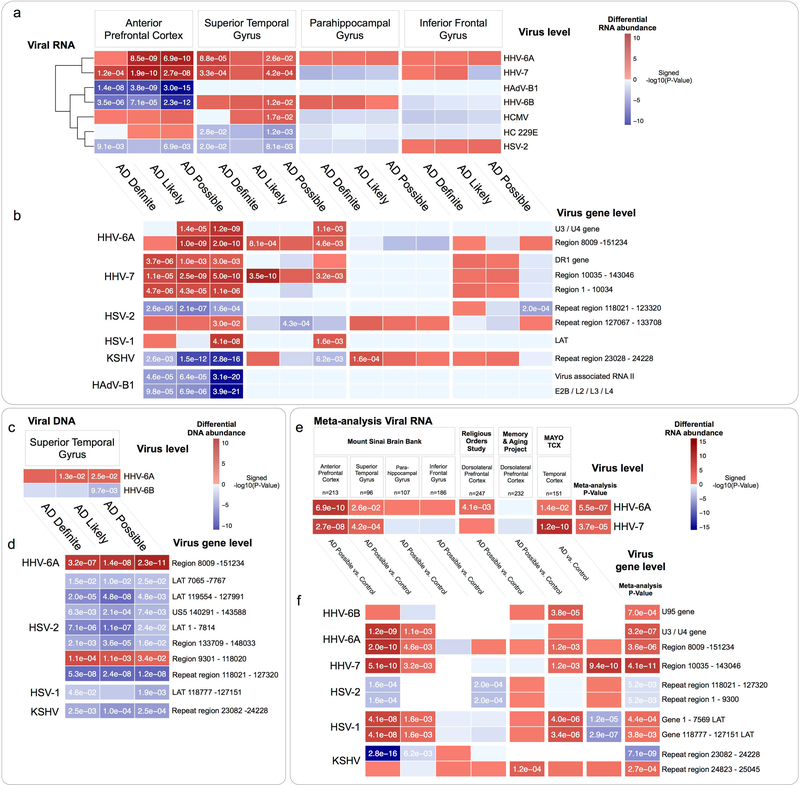
We identified differential abundance of multiple viral species in the APFC and STG (Figure 2a, Table S2). The most consistent difference we saw was an AD-associated increase in the abundance of two closely related Roseoloviruses, HHV-6A and HHV-7, across the APFC and STG. The third Roseolovirus, HHV-6B had a discordant profile with increased abundance in AD in the STG, and reduced in the APFC. Viral gene-level differential expression (Figure 2b, Table S2) identified increased abundance across the APFC and STG regions of the HHV-6A U3/U4 genes (positional homologs of human cytomegalovirus (HCMV) US22, with transactivating effects on other viral species(Mori et al., 1998)) and the HHV-7 direct repeat terminal gene, DR1.
Figure 2: Differential abundance of viral RNA and DNA in AD.
Multiregional comparison between AD vs. controls of viral RNA and DNA, summarized to the level of (a,c) full viral sequences, and (b,d) viral genomic features. Meta-analysis of differential abundance of viral RNA in AD (e-f), incorporating post-mortem brain RNA-seq data from the MSBB, ROS, MAP and MAYO TCX consortium studies also revealed increased HHV-6A and HHV-7 in AD.
(P-values shown in cells with FDR < 0.1, Features with a meta-analysis FDR < 0.1 shown in e-f)
To understand whether our observations of altered viral abundance in AD would also be preserved in additional cohorts we incorporated post-mortem brain RNA-seq data from three additional, independent consortium studies; (1) Religious Orders Study47 (ROS), a longitudinal clinico-pathological study comprising 300 samples from the dorsolateral prefrontal cortex (DLPFC) of individuals with AD and healthy controls, (2) Memory and Aging Project(Bennett et al., 2012a, Bennett et al., 2012b) (MAP) comprising 298 samples from the DLPFC of individuals with AD, as well as healthy controls, and (2) Mayo Temporal Cortex(Allen et al., 2016) (MAYO TCX) comprising 278 samples from the temporal cortex of individuals with AD, pathological aging (PA)(Dickson et al., 1992), progressive supranuclear palsy (PSP), and healthy controls. Our goal was to process these additional samples using the procedure described above, and integrate the results (Table S2) into a meta-analysis of viral abundance in AD (Figure 2e–f). Our main finding was a consistently increased abundance of HHV-6A and HHV-7, driven mainly by the “unique” region of each virus (the full viral sequence excluding the ~15kb flanking DRL and DRR terminal repeats). We also observed an increased abundance of the HSV-1 latency-associated transcript (LAT), supporting an increased rate of HSV-1 infection (latent or otherwise) in AD. These additional validations support the main findings originally observed in the MSBB cohort, that multiple viruses are at increased abundance in AD, across multiple brain tissues, with prominent roles for Roseoloviruses HHV-6A and HHV-7.
Increased HHV-6A and HHV-7 are not ubiquitous features of neurodegeneration
We utilized the PA and PSP samples available within the MAYO TCX cohort to perform comparisons against these additional neurodegenerative disorders, to understand whether our findings were specific to AD, or perhaps reflect a more general feature of neurodegenerative processes. In a comparison of AD vs. PA, we found an increased abundance of HHV-6A and HHV-7 (Figure S2). When we compared AD vs. PSP, we found increased HHV-7 in AD, but reduced HHV-6A. Taken together, these observations suggest that elevated HHV6-A and HHV-7 are not ubiquitous features of neurodegenerative disease, although HHV-6A may also be relevant to other diseases such as PSP.
Increased abundance of HHV-6A DNA in Alzheimer’s disease
We looked for evidence of viral DNA in whole exome sequencing (WES) data that was generated on STG samples (n=286) in the MSBB cohort, applying a similar procedure to that used in evaluating viral RNA abundance (Figure S1). We detected viral DNA for multiple viruses, and identified an increased abundance of HHV-6A (Figure 2c–d, Table S2). This was primarily due to reads mapping to HHV-6A Region 8009 – 151234, which comprises the “unique” region of HHV-6A, (and consistent with our findings in the RNA sequences). Chromosomal integration of HHV-6A into host subtelomeric regions is well described(Arbuckle et al., 2010), occurring via a mechanism involving homologous recombination between telomeric repeats and the DRR. Excision and reactivation of integrated HHV-6A is associated with preferential loss of the entire DRL, facilitating viral circularization and rolling circle replication(Prusty et al., 2013). This may indicate that the HHV-6A DNA that we find as more abundant in AD reflects HHV-6A that has undergone reactivation from a chromosomally integrated form, although we have not evaluated this directly.
Viral RNA abundance associates with clinical dementia and neuropathology traits
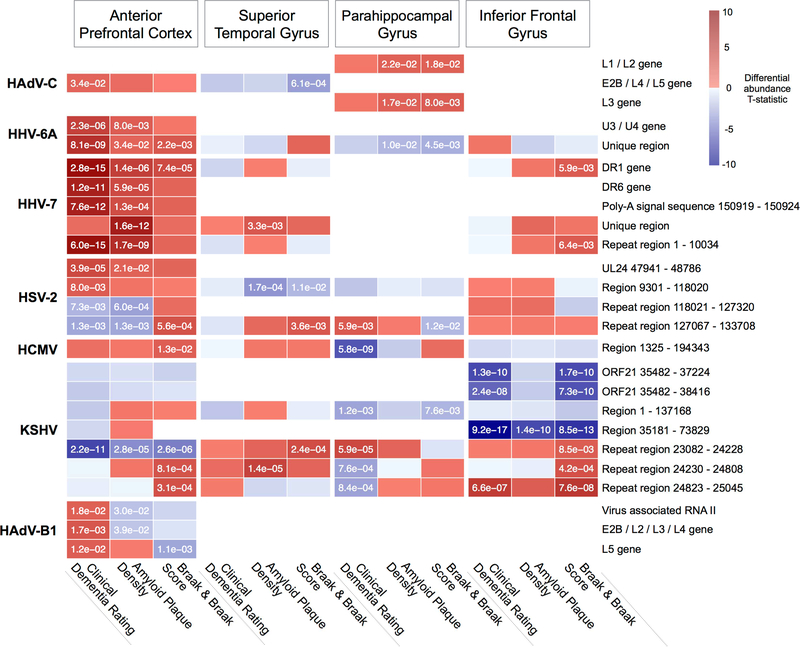
We extended our analysis to identify significant associations between virus level and viral gene level RNA abundance and AD-relevant clinical and neuropathological traits (“AD traits”) (Figure 3, Table S3), including a consensus based clinical dementia rating score(Morris, 1993) (CDR), multi-regional neuritic amyloid plaque density(Haroutunian et al., 1998) (APD), and Braak and Braak score(Braak et al., 2006) (Braak score). We identified several viral genes that significantly associate (FDR < 0.1) with multiple AD traits, including the HHV-7 DR1 gene, and HHV-6A unique region, both demonstrating a positive association with all three AD traits within the APFC.
Figure 3: Viral abundance associates with AD clinical and neuropathology traits.
Multiregional associations between abundance of viral genomic feature RNA and clinical and neuropathological traits. Viral features with significant (FDR < 0.1) associations across multiple traits or brain regions are shown. (P-values shown in cells with FDR < 0.1).
Human DNA variants that associate with viral abundance also associate with AD status, clinical dementia and neuropathological features of AD
Given the prolonged preclinical course of AD, a primary question for us was whether these viral species represent a truly informative, causal component of AD, or instead reflect an “opportunistic passenger” of a neurodegenerative process driven by other factors. To help address this, we integrated WES data with RNA-seq for each donor within the MSBB. Our goal was to identify host DNA variants that significantly associate with viral abundance, which we refer to as “viral quantitative trait loci” (vQTL). These vQTL might then be used in a causal inference paradigm(Millstein et al., 2009) to resolve directed regulatory interactions in virus-host networks and, for the fraction of vQTLs that are also AD risk-associated, evaluate whether viral abundance is an authentic risk factor for AD. Causal inference approaches like this have been used to elucidate molecular networks impacted by DNA loci across biological contexts as diverse as cardiometabolic disease(Franzen et al., 2016), COPD(Yoo et al., 2015) and meditation(Epel et al., 2016).
We identified host DNA markers(Shabalin, 2012) across all four brain regions with paired DNA and RNA samples (APFC: n=174, STG: n=86, PHG: n=80, IFG: n=147) that significantly associated with normalized viral abundance, for any viral species detected within that region (Figure 4, Table S4). DNA markers with a permutation based FDR < 0.25 were classified as vQTLs for that specific viral gene in the context of that region.
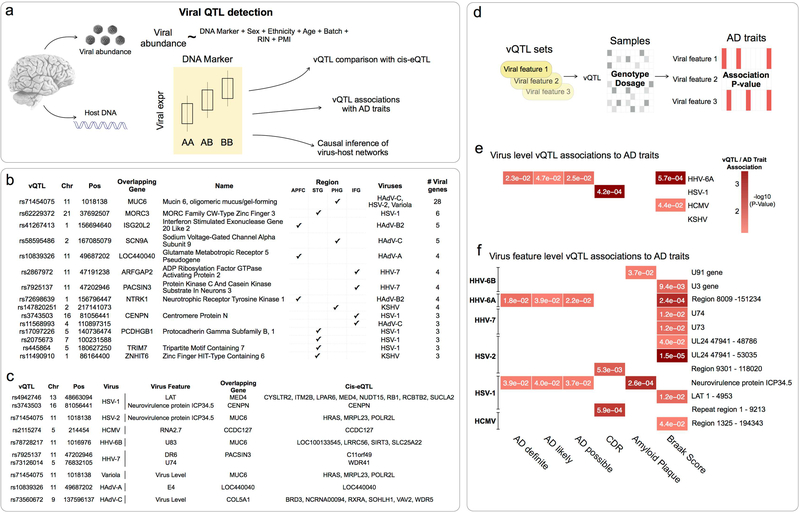
Figure 4: Viral QTL detection and association with AD genetics.
(a) Host DNA markers significantly associated (FDR < 0.25) with viral abundance were classified as vQTL for that feature-tissue combination. (b) Top multi-viral vQTL associations, with overlapping gene symbols and associated viruses. (c) Top vQTL associations where vQTL is also a cis-eQTL for at least one host gene. (d) Sequence kernel association test to evaluate whether vQTL markers are also associated with AD traits. (e) Virus level vQTL sets for HHV-6A were highlighted most strongly, (f) whereas viral feature level associations implicated features from HHV-6A and HSV-1.
(Panels e-f show associations for any viruses/viral features that were implicated in viral differential RNA abundance, P-values < 0.05 shown in cells)
We identified 1,672 vQTL associations across the four regions assayed (APFC: 883, STG: 479, PHG: 175, IFG: 135). This represented 747 non-independent vQTL markers that collectively associate with 16 separate viruses. The viruses with the largest number of separate vQTL markers were human adenovirus-A (HAdV-A), HHV-6A, HSV-2 and HSV-1 (222, 103, 91 and 87 markers respectively). The vQTL associated with the most viruses and regions (Figure 4b) (rs71454075) falls within the glycoprotein Mucin 6, Oligomeric Mucus/Gel-Forming gene (MUC6), expressed particularly in gastric epithelium, and with cytoprotective roles against pathogens, acids and proteases(Toribara et al., 1993). The genes that collectively overlapped vQTLs across the most regions and viruses (Figure 4c) indicated biological themes that plausibly relate to individual variability in virome composition, including mucosal immunity (MUC6, NTRK1), innate immunity and antiviral sensing (ISG20L2, MORC3, NTRK1, TRIM7).
We hypothesized that cis-eQTL associations could represent a potential mechanism linking vQTL with viral abundance, whereby a vQTL might alter the expression of a nearby host gene that has the potential to impact, or be impacted by viral abundance. We performed a cis-eQTL analysis across this same cohort, detecting significant associations (FDR < 0.1) between host gene expression, and markers within 1 MB of gene boundaries. Thirty five percent of the unique vQTL markers (263 of 747) were associated with at least one cisgene, inlcuding several with associations to AD and other dementias, for instance, rs4942746 is a vQTL for HSV-1 LAT, and is also a cis-eQTL for Integral Membrane Protein 2B (ITM2B), an endogenous inhibitor of Aβ aggregation and associated with familial British(Vidal et al., 1999) and Danish(Vidal et al., 2000) dementia.
We employed the sequence kernel association testing(Lee et al., 2012, Wu et al., 2016) approach to understand whether vQTLs are genetic risk markers for AD status, clinical dementia rating or neuropathology (AD traits) (Figure 4d, Table S5). Iterating over each viral gene feature, we combined vQTL from all regions into a single set, and calculated a P-value associating each vQTL set with each AD trait. We detected multiple viral features with significant associations to AD traits (Figure 4e–f), supporting the hypothesis that DNA variants that predispose to AD also predispose to abundance of key viral species - most notably HHV-6A. We also found multiple viral genes with vQTL sets that associate with multiple AD traits, including the HHV-6A unique region, and the HSV-1 Neurovirulence protein ICP34.5.
These results indicate a significant overlap between the genetic basis for AD traits, and the abundance of specific viral species and viral genomic features. This supports our broader hypothesis that viral activity plays a role in the development and progression of AD, and is consistent with a role for HHV-6A in particular.
Viral regulation of AD associated host networks
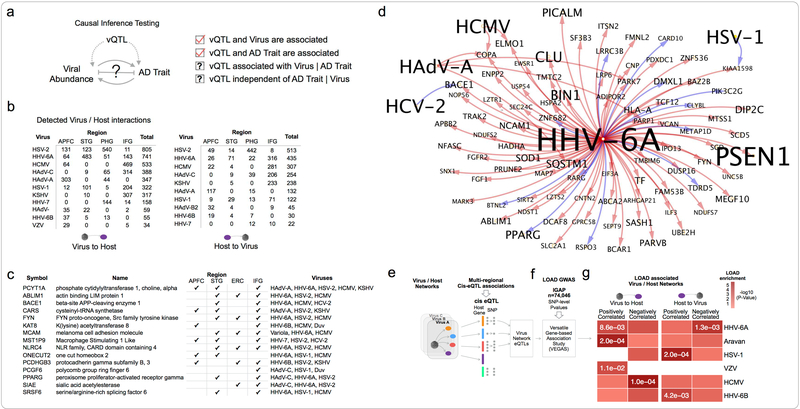
We generated virus-host gene regulatory networks to understand the biological context of viral activity across samples. We constructed informative models by detecting the set of host genes that are correlated with vQTL/virus pairs within each tissue, and iterating over each candidate trio (each trio comprising a vQTL marker, virus feature abundance, and host gene expression), and then testing the mathematical conditions required to demonstrate causal mediation of an association between a DNA marker and a trait(Millstein et al., 2009) (Figure 5a). We built tissue-specific virus-host gene networks for all detected viral features (Table S6) identifying interactions in all four brain tissues (Figure 5b), comprising a total of 4,110 “virus to host” interactions, and 2,255 “host to virus” interactions, collectively associating 16 different viral species with 4,929 host genes. Only three viruses had detected interactions in all four tissues: HSV-1, HSV-2 and HHV-6A. Several viruses only had interactions detected in a single tissue: Aravan Virus, HCV-2, Variola Virus, and Wesselbron Virus, which may reflect higher regional tropism for the species, or the potential for contamination to be driving the detection of these viruses in a subset of samples.
Figure 5: Viral regulation of AD associated host networks.
(a) Integration of vQTL with viral abundance, and host gene expression was used to infer directed virus / host subnetworks. (b) Virus / host gene network sizes for viruses with interactions detected in multiple tissues (c) Host genes that are most frequently perturbed by viruses (d) Host genes upregulated by HHV-6A are enriched for a heterogeneous set of AD risk and biomarker associated genes. Shown here is the HHV-6A / AD-associated gene subnetwork, and detected interactions with additional viruses. (e-g) Brain cis-eQTLs associated with expression of specific virus/host networks are enriched for AD GWAS risk loci.
(P-values shown in cells with FDR < 0.1)
Host genes that are most commonly perturbed by viruses are shown (Figure 5c), ranked according to the number of unique viruses that we detected perturbing that gene across all four regions surveyed. This includes several genes implicated in regulation of APP processing and AD, including β-site amyloid precursor protein cleaving enzyme 1 (BACE1), FYN Proto-Oncogene, Src Family Tyrosine Kinase (FYN), and Peroxisome Proliferator Activated Receptor Gamma (PPARG). Several of these genes are also associated with pro and antiviral signalling, including positive regulation of interferon-λ1 genes by FYN during viral infection(Nousiainen et al., 2013), negative regulation of viral replication by PPARG(Bernier et al., 2013), and promotion of viral translation by SRSF6(Swanson et al., 2010).
We evaluated the set of genes causally regulated by each virus, against a set of known AD-associated genes, including risk genes for early and late onset AD, as well as AD associated traits (such as β-amyloid plaque density, rate of disease progression, neurofibrillary tangle density) from multiple human genetics disease resources(Rouillard et al., 2016). We found that multiple viruses interact with AD risk genes. HHV-6A stood out as notable with significant overlap (FDR < 3e-3) between the set of host genes it collectively induces across all tissues and AD-associated genes (Figure 5d, Table S7). This includes several regulators of APP processing and AD risk-associated genes, including gamma-secretase subunit presenilin-1 (PSEN1), BACE1, amyloid beta precursor protein binding family B member 2 (APBB2), Clusterin (CLU), Bridging Integrator 1 (BIN1) and Phosphatidylinositol Binding Clathrin Assembly Protein (PICALM). We also found that several other viruses regulate, or are regulated by AD risk genes, including: (1) HAdV-C induced expression of Complement Receptor 1 (CR1), and inhibition of Solute Carrier Family 24 Member 4 (SLC24A4), (2) inhibition of KSHV by Fermitin Family Member 2 (FERMT2), and (3) inhibition of HSV-2 by Translocase Of Outer Mitochondrial Membrane 40 (TOMM40). These findings indicate multiple points of overlap between virus-host interactions and AD risk genes.
Expression QTL of virus-host networks are enriched for AD GWAS risk loci
To investigate the broader context of how virus-host interactions might overlap with AD genetic risk, we integrated cis-eQTL data with AD GWAS summary statistics (Figure 5e–f). Our hypothesis was that identifying virus-host networks that are enriched for AD GWAS loci could provide a natural means to prioritize the relevance of individual viruses to AD, as well as provide useful functional context for specific viruses. Our approach was to use cis-eQTL identified in any of the four brain tissues within the MSBB data, to define the set of markers that are associated with the host genes in each virus/host network (“virus network eQTLs”), and then determine whether virus network eQTLs are enriched for AD risk-associated loci(Lambert et al., 2013) using the versatile gene-based association study (VEGAS) approach(Liu et al., 2010b, Mishra et al., 2015). We observed that multiple viruses have host networks that are enriched for AD risk-associated eQTLs (Figure 5g), most consistently for HHV-6A, but also HCMV, HSV-1, Aravan, HHV-6B, and VZV. This suggests that loci that alter the expression of genes that regulate, or are regulated by specific viruses are in aggregate associated with AD risk.
Viral mediation of neuronal loss in AD
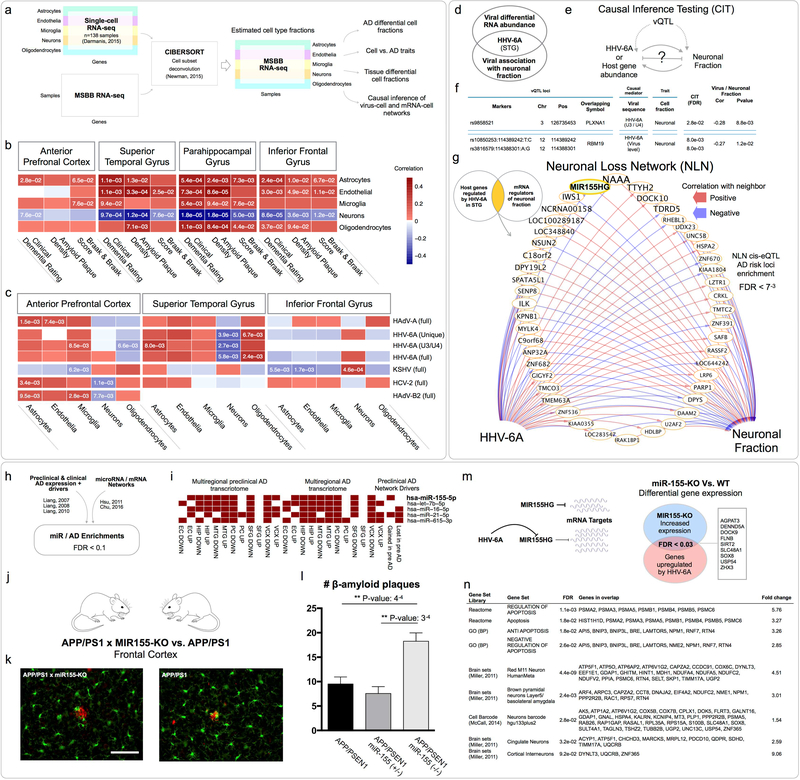
Progressive neuronal dysfunction and eventual death is a hallmark feature of AD and is the primary driver of the striking cortical atrophy associated with the disease. Diverse findings implicate aberrant regulation of apoptosis(Su et al., 1994), necroptosis(Caccamo et al., 2017), and autophagy(Nixon et al., 2005) although findings are conflicting and a unified understanding of the mechanisms underlying neuronal loss is lacking. Given the potential for viral infection to modulate cellular death pathways through these mechanisms and others(Upton et al., 2014), we were interested in understanding whether viral abundance might be associated with specific brain cell type fractions within our data set, particularly neurons. We used CIBERSORT(Newman et al., 2015) to deconvolute each MSBB RNA-seq sample into estimated fractions for major brain cell types (neurons, astrocytes, microglia, endothelial cells, and oligodendrocytes), based on comparison with a reference panel of single cell RNA-seq in cortical samples from neurologically normal, middle-aged adults(Darmanis et al., 2015) (Figure 6a). Our goal was to identify significant associations between estimated cell fractions, AD traits and viral abundance.
Figure 6: Viral mediation of neuronal loss in AD.
(a) Cell fraction estimates in the MSBB RNA-seq data, linking cell fractions with (b) AD traits, and (c) viral RNA abundance. (d) HHV-6A was negatively correlated with neuronal fraction, while also at increased abundance in the STG. (e-f) Causal inference testing identified HHV-6A vQTL associated with neuronal fraction, which also have their effect mediated by HHV-6A sequences. (g) Neuronal loss network of host genes regulated by HHV-6A, which also exert an effect on neuronal fraction, includes MIR155HG. Gene label size is inversely proportional to the smallest AD risk P-value for its associated cis-eQTL. (h-i) We initially prioritized miR-155 during our analysis of the preclinical AD networks, due to strong associations between known miR155 gene targets, and a variety of multiregional AD and preclinical AD transcriptomic changes, as well as differential drivers of the preclinical AD network. (j-l) Four month old miR-155-KOxAPP/PS1 produce increased cortical β-amyloid plaques and oligomers compared with APP/PS1 mice. (m) We hypothesized that HHV-6A inhibition of miR-155 might cause a disinhibition of miR-155 targets. Cortical RNA-seq of miR-155KO vs. WT mice demonstrated significant overlap between genes upregulated (FDR < 0.1) in the absence of miR-155, and host genes detected as upregulated by HHV-6A in the virus/host networks. (n) Gene set enrichments for miR-155-KO vs. WT differentially expressed genes identified multiple enrichments (FDR < 0.1) consistent with a role for miR-155 in neuronal loss.
The most consistent AD-associated change (Table S8) was a significant (FDR < 0.1) decrease in neuronal fraction, and an increased endothelial cell fraction in the PHG, IFG, and STG, which we confirmed using an alternative cell signature enrichment method, xCell(Aran et al., 2017) (Table S8). We observed strong negative correlation between neuronal fractions and CDR in all four brain tissues, and with Braak score and neuritic plaque density in three (PHG, IFG, STG) (Figure 6b, Table S8).
We found multiple correlations between cell fractions and viral RNA abundance (Figure 6c, Table S9), with associations detected in the APFC, STG and IFG, collectively implicating all cell types with five viruses. Only a single virus (HHV-6A) was associated with cell fraction changes in multiple tissues (APFC and STG; the same two tissues in which HHV-6A was observed at increased abundance in AD), although the specific cell profiles were different between tissues. We were most interested in focusing on viruses or viral genes that are negatively correlated with neuronal fraction, particularly if they were also detected as differentially abundant in that same brain tissue (Figure 6d). Only HHV-6A met these criteria demonstrating strong negative correlation with neuronal fraction in the STG, as well as being highlighted in the viral RNA analyses of the MSBB, MAP, MAYO TCX cohorts and meta-analysis.
We applied causal inference testing to evaluate the hypothesis that abundance of HHV-6A exerts a significant effect on neuronal fraction in the STG, and detected multiple instances consistent with the HHV-6A virus, and the HHV-6A U3/U4 gene mediating such an effect (Figure 6e–f, Table S9). We reasoned that part of this effect might be through the direct impact of viral products on biological mechanisms underlying the neuronal loss, however there might also be informative, indirect effects mediated through a host subnetwork (i.e. host genes regulated by HHV-6A, and which also causally regulate neuronal fraction). We iterated over the set of host genes we detected as regulated by each HHV-6A sequence in the STG, and for each gene, tested whether it also correlated with neuronal fraction, and whether the vQTL marker for that host gene was associated with neuronal fraction in the STG. We performed causal inference testing, and detected genes that are regulated by HHV-6A virus (121 genes), HHV-6A unique region (86 genes) and HHV-6A U3/U4 (6 genes), and which exert an effect on neuronal fraction (Figure 6g, Table S9). To determine whether these “neuronal loss networks” (NLN) might overlap with AD GWAS risk loci, we used the VEGAS approach described above, and found a significant enrichment within the cis-eQTL of the HHV-6A virus level NLN (FDR < 7e-3).
The HHV-6A virus level NLN includes many genes with roles in cellular viral response, as well as associations to biological mechanisms that could help account for neuronal death, including upregulation of Poly(ADP-Ribose) Polymerase 1 (PARP1) by HHV-6A (and negative regulation of neuronal fraction by PARP1), consistent with its reported activation by EBV(Mattiussi et al., 2007), HIV(Ha et al., 2001) and HSV-1(Grady et al., 2012), and induction of caspase-independent apoptosis in the latter(Grady et al., 2012). AD risk-associated haplotypes for PARP1 have also been reported(Liu et al., 2010a), and suggested as a mechanism mediating cell death following cytotoxic response(Liu et al., 2010a). The NLN gene with the lowest eQTL AD risk P-values (Figure 6g) is N-acylethanolamine acid amidase (NAAA), a close homolog of Acid Ceramidase (AC), suggested to regulate neuronal apoptosis in AD(Huang et al., 2004).
We were also interested to find that MIR155 Host Gene (MIR155HG) was within the HHV-6A NLN and associated with multiple low AD risk P-value eQTLs. We had prioritized miR-155 during our analysis of the preclinical AD networks (Figure 6h–i, also Table S1) due to strong associations between its mRNA targets and a variety of multiregional AD and preclinical AD transcriptomic changes as well as “gained in preclinical AD” drivers. In addition to diverse associations with viral biology including EBV(Gatto et al., 2008), HHV6A(Caselli et al., 2017), KSHV(Gottwein et al., 2007) and MDV(Zhao et al., 2009), miR-155 has also been reported as a regulator of T-cell response in AD(Song et al., 2015), a mediator of inflammation induced neurogenic dysfunction and apoptosis(Woodbury et al., 2015), and an effector of TREM2-APOE regulation of a microglial neurodegenerative phenotype(Krasemann et al., 2017). Reports of viral tropism have also demonstrated that HHV-6 and HHV-7 can infect microglia(Albright et al., 1998), and macrophages(Zhang et al., 2001). In the NLN, we found that HHV-6A suppressed miR-155, as described in recent reports of miR-155 inhibition by HHV-6A in infected T-cells(Caselli et al., 2017).
These findings show that computational deconvolution of RNA-seq in AD recapitulates the expected neuronal loss. The negative correlations between STG neuronal fraction and HHV-6A sequences (that are also at increased abundance in the STG), in combination with the findings of causal testing, and the association of the NLN cis-eQTLs with AD risk are consistent with HHV-6A exerting an effect on neuronal fraction in AD.
miR-155 is suppressed by HHV-6A, and alters Aβ oligomer levels and amyloid plaque density in a genetically manipulated mouse model
Given the convergence of multiple analyses upon miR-155 (Figure 6g,i), we crossed miR-155-KO mice(Thai et al., 2007) with a standard APP/PS1 amyloidosis strain(Jankowsky et al., 2004) to evaluate the effect of miR-155 depletion on molecular and tissue pathology. At age four months, we observed that the brains of miR-155-KO/APP/PS1 mice displayed larger, more frequent cortical amyloid plaques and higher levels of certain Aβ conformers as compared with APP/PS1 mice (Figure 6k–l). We hypothesized that if HHV-6A was inhibiting the expression of miR-155, then this might cause the de-repression of certain miR-155 mRNA targets (Figure 6m). We generated RNA-seq on cortical samples comparing miR-155-KO vs. wildtype mice to identify the differentially expressed genes (DEG, FDR < 0.1, Table S10), and found a significant overlap between upregulated DEGs, and the set of host genes we had identified as upregulated by HHV-6A (FDR < 0.03), suggesting that some component of our detected HHV-6A network is mediated through an effect on miR-155, or a viral ortholog of miR-155 (which has not been described for HHV-6A).
Given the context of HHV-6A induced inhibition of miR-155 within the NLN, we wondered whether we would observe transcriptomic evidence for mechanisms associated with neuronal loss. Molecular and functional enrichments of the miR-155-KO DEGs (Table S10), highlighted multiple enrichments for apoptosis regulatory pathways and neuronal signature gene sets (Figure 6n).
Collectively, these findings support the role of miR-155 as a key node in host response to AD-relevant viral perturbation, and as a potential mediator of neuronal loss. This is also consistent with a contribution of viral perturbation in driving the preclinical AD transcriptional phenotype given that our prioritization of miR155 was informed by findings in the preclinical AD networks. The finding that miR-155-KO causes increased Aβ plaque deposition in the presence of APP/PS1 mutations also suggests a pathway linking viral perturbation with AD-associated neuropathology. This line of evidence is also consistent with findings linking viral infection with antimicrobial innate immune response, proamyloidogenesis, and microbe entombment(Kumar et al., 2016, Eimer et al., 2017).
Viral perturbation of transcription factor regulatory networks
Viruses demonstrate remarkable strategies to effectively co-opt endogenous host factors necessary to their survival. Through context-dependent transcription of viral gene products, viruses hijack host transcription, signaling networks and cellular machinery to orchestrate all aspects of their life cycle.
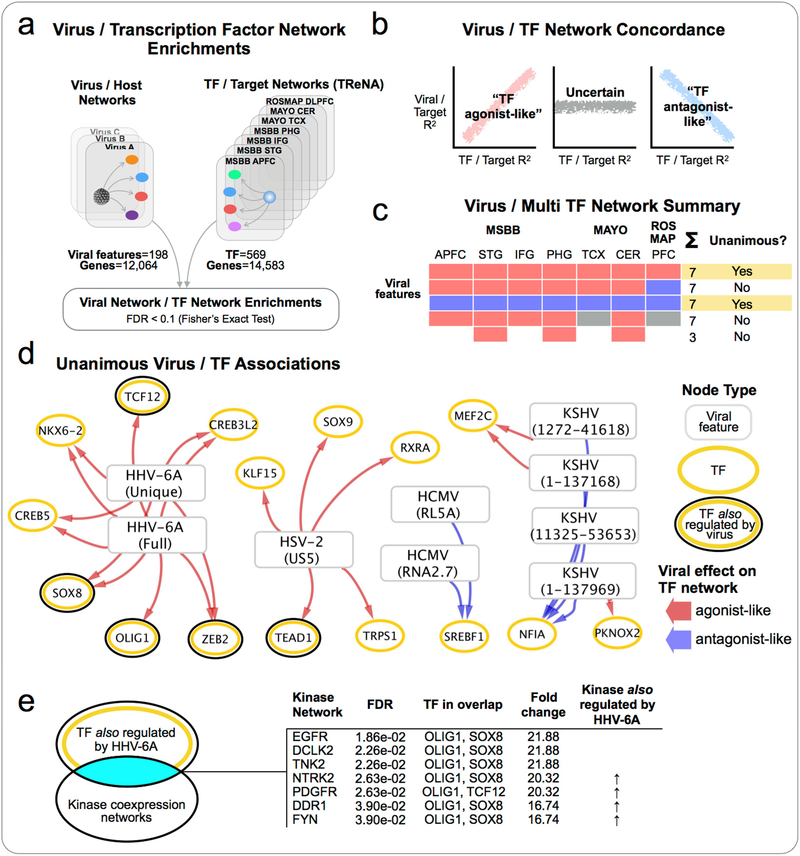
Given our earlier findings of widespread changes in C2H2-TF in the preclinical networks we were interested in determining whether the virus-host networks might indicate viral targeting (directly or indirectly) of specific TFs (Figure 7). We constructed a diverse collection of TF-target networks, generated from the MSBB, MAYO and ROSMAP consortium data, using the Transcriptional Regulatory Network Analysis (TReNA) approach comprising seven TF-target networks in total (reflecting tissue-specific networks within each cohort), which collectively model transcriptional relationships between 569 unique TFs, and 14,583 target genes.
Figure 7: Viral perturbation of transcription factor regulatory networks.
(a) Comparison of virus/host networks with diverse TF-Target networks built from multiple independent AD data sets, to identify virus / TF network enrichments. (b) Examination of the concordance of effect exerted by associated virus and TF upon target genes, and (c) summarization of results across all TF-target networks (d) to identify unanimous virus/TF associations. (e) Kinase enrichment analysis of the most strongly implicated TF identified several kinases that we also detected as regulated by HHV-6A, with known associations to AD and HHV-6A, indicating a potential mechanism for viral co-option of TF networks in AD.
Within each virus-host network, we compared the “virus to host” genes with the target genes for each TF, across all seven TF-target networks to identify significantly (FDR < 0.1) associated virus/TF pairs (Figure 7a). For each enriched pair, we examined the concordance of effect that the virus and TF demonstrate on the genes driving the enrichment (Figure 7b). We calculated the Pearson’s correlation between the individual virus-host gene correlations and the TF/target gene correlations to identify instances where the virus and TF both exerted a similar effect (“TF agonist-like”, Pearson Corr > 0, FDR < 0.1), and instances where they exerted an opposing effect (“TF antagonist-like”, Pearson Corr < 0, FDR < 0.1). We then identified the set of virus/TF pairs that were associated in all seven TF-networks, and which demonstrated a consistent status as TF agonistlike/antagonist-like in each case (Figure 7c), reasoning that these “unanimous virus/TF associations” would represent the most robust candidates for considering viral perturbation of a TF network.
This unanimous virus/TF network (Figure 7d) includes 26 associations between four different viruses (HSV-2, HHV-6A, HCMV and KSHV) and 14 TFs. The majority of connections (20 of 26) indicate associations where a viral feature exerts an agonist-like effect on TF targets. We found multiple virus-TF interactions where the TF was also directly detected as being regulated by the virus, including agonist-like interactions between HHV-6A and OLIG1, TCF12, SOX8 and ZEB2. In each of these cases, the TF was upregulated by the virus, consistent with the agonist-like associations inferred from the TF network enrichments. We hypothesized that a potential mechanism to link viral perturbation of multiple TF networks might be mediated through viral mimicry or modulation of host kinase activity(Shugar, 1999). We compared the set of four HHV-6A regulated TF (that were also directly detected as regulated by HHV-6A) with a library of kinase coexpression networks(Lachmann et al., 2017), and identified significant (FDR < 0.1) enrichments with seven different human kinases (Figure 7e). This includes four kinases that were also detected as directly upregulated by HHV-6A. For example, Neurotrophic Receptor Tyrosine Kinase 2 (NTRK2) expression is linked with neuronal survival in AD(Wong et al., 2012) and associated with AD risk(Chen et al., 2008). FYN tyrosine kinase, linked with synaptic dysfunction in AD via a range of Aβ- and/or tau-related mechanisms(Nygaard, 2017), is also bound directly by the HHV-6A U24 protein potentially disrupting interactions with endogenous ligands(Sang et al., 2014).
These observations indicate the potential for viral perturbation of host TFs and TF-Target networks in the context of AD, and offer an explanatory mechanism that could account for the large number of virus-host interaction detected for species such as HHV-6A and HSV-2. The described kinase-TF enrichments represent a potential upstream mechanism whereby HHV-6A modulation of kinase activity could alter the activity of specific endogenous TFs, thus perturbing host regulatory programs in the manner reflected in the HHV-6A host networks.
Virus-host protein networks indicate perturbation of cellular nucleotide pools, tRNA synthesis and protein translation
We constructed virus-host protein networks to evaluate proteomic consequences of viral activity in AD. We generated protein expression profiles for a subset of APFC samples (n=152), performing liquid-chromatography-mass spectrometry (LC-MS) and using MaxQuant(Cox et al., 2008) for quantifying label free protein. Using a similar procedure as outlined for generation of virus-host RNA networks, we identified proteins that are associated with vQTLs, and which are regulated by (or which regulate) each virus. Of the viruses that we found as differentially abundant in the APFC RNA data, we detected interactions with host proteins for HSV-1 (14 proteins), HSV-2 (34 proteins) and HHV-6A (28 proteins) (Figure S3, Table S11). Protein regulators of cellular nucleotide pools, especially purine biosynthesis (NUDT16, GMPR2), guanine nucleotide binding proteins (GNAS, GNAO1, GNG3, GNG5), aminoacyl-tRNA synthetases (SARS, VARS; AARS), mitochondrial function (MT-ND5, MFF), nuclear organization (NCL, LMNA) and cytoskeletal disruption (CAMK2D, LMNA, MYLK, PRKCB, TF) are among the most prominent biological themes of the networks. For instance, we found that several viruses alter expression of nucleotide regulating proteins, including HSV-2 induction of reductase enzyme GMPR2, which catalyzes conversion of G to A nucleotides, and HHV-6A induction of inosine diphosphatase NUDT16 which depletes the cellular pool of non-canonical purines IDP and ITP. Collectively, this suggested a picture of virally induced dysregulation of nucleotide pool metabolism, especially purine bases, consistent with several metabolomics studies in AD(Kaddurah-Daouk et al., 2013, Kaddurah-Daouk et al., 2011). This is notable, given our observations of G4 activity as key regulatory features among preclinical AD driver genes, which are primarily features of guanine enriched sequences. Nucleotide pool depletion or imbalance is known to induce replicative stress, and mediated through inadequate unwinding of stabilized G4 sequences, with associated genomic and epigenetic instability(Papadopoulou et al., 2015).
We also found that HHV-6A induced expression of multiple AARS enzymes (VARS and SARS), responsible for charging their cognate tRNAs with valine and serine respectively. Increased d-serine has been reported in the CSF of patients with AD, as well cellular and mouse AD models(Madeira et al., 2015). Some viruses possess endogenous tRNA and AARS sequences, which appear critical to viral protein synthesis throughout the infectious cycle(Nishida et al., 1999). Multiple viruses recruit host tRNAs into virions for use as primers in reverse transcription(Mak et al., 1997) and some such as HIV also selectively incorporate host AARSs as well(Cen et al., 2001). Recent works have also demonstrated the role of tRNA in shifting the G4 conformational equilibrium towards a hairpin conformer(Rode et al., 2016).
These results indicate novel molecular mediators and associated pathways that might help shed light on mechanisms of viral pathogenicity, such as a potential role for AARSs in HHV-6A co-option of host protein synthesis machinery. These findings may also suggest novel molecular mediators and mechanisms for more widespread changes we have observed, such as dysregulation of G4 activity.
Impact of viral activity on genetic, transcriptomic, clinical and neuropathology networks in AD
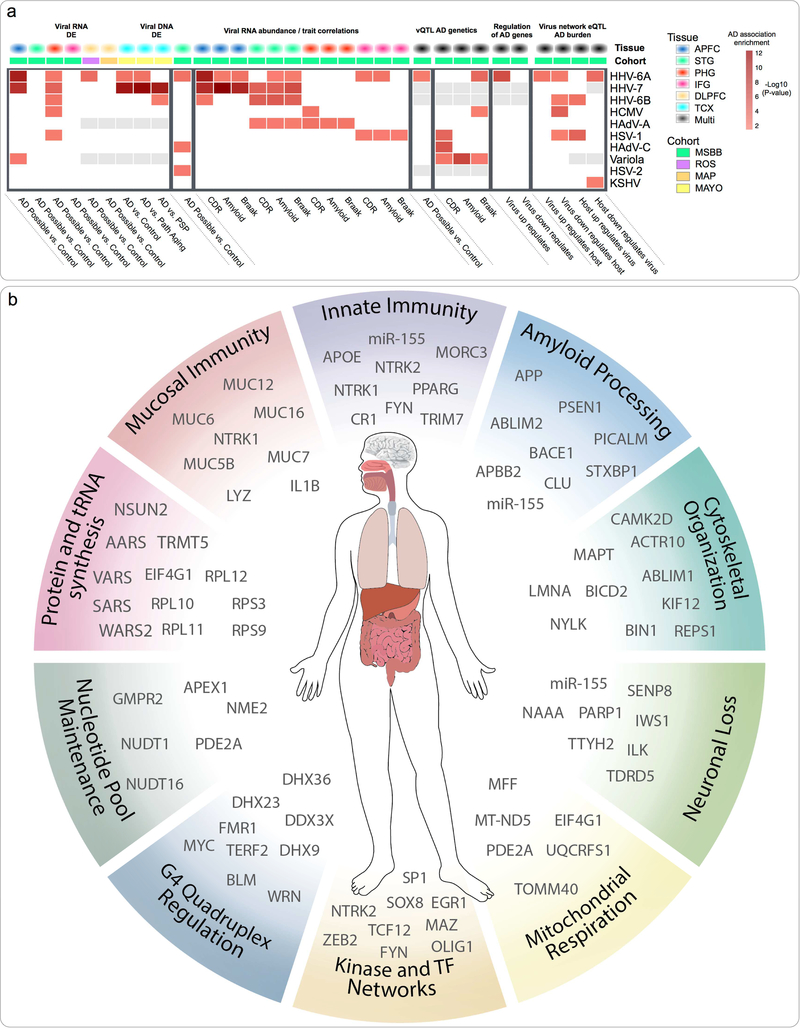
We aggregated the results across several analyses to summarize the systems-level impact of individual viral species on AD biology (Figure 8a). This included multiregional viral RNA differential abundance, multiregional correlations of RNA abundance with AD traits, associations between vQTL sets and AD genetics, viral DNA differential abundance and AD risk gene enrichment scores for virus-host subnetworks. iral species implicated in any of these analyses were assigned a combined score based on the summed -log10(P-value) of each individual association. This view indicated that multiple viruses impact on AD associated biology, across multiple -omic domains. The most strongly implicated viruses, were Roseoloviruses HHV-6A, HHV-7 and HHV-6B. HHV-6A in particular was robustly prioritized on the basis of: (1) RNA and (2) DNA differential abundance, (3) RNA abundance association with AD traits (4) vQTL markers association with AD traits, (5) HHV-6A/host network eQTL enrichments for AD risk loci, and (6) inducing the expression of a significant fraction of AD risk genes within the HHV-6A/host network.
Figure 8: Multiomic evaluation of AD associated virome implicates Roseoloviruses HHV-6 and HHV-7.
(a) Summarized associations for each virus to diverse aspects of AD biology. Multiple viruses appear to have significant impacts on AD associated biology, particularly HHV-6A, HHV-7 and HHV-6B. (b) Findings from this study indicate complex relationships between viral and host factors that are likely to be relevant across a range of time scales and organ systems. Key biological processes that have been highlighted are shown, along with top candidate molecular mediators.
Discussion
Developing a sophisticated understanding of the causal basis for AD is complicated by its protracted preclinical course, and the inability to routinely sample brain tissue. Distinguishing the earliest drivers of disease from the “opportunistic passengers” of a multi-decade neurodegenerative process is especially formidable given the profound changes in transcriptomic, proteomic, and histopathological profiles(Zhang et al., 2013).
We report a multi-stage study that aims to reconcile nascent changes in preclinical AD with findings made in the context of AD. Our strategy began by examining transcriptomes from brain regions that undergo the earliest changes in AD with the goal of identifying novel biology that could offer a frame for understanding the more dramatic changes seen in later stages of AD. Exploration of the preclinical AD networks sensitized us to pervasive dysregulation of C2H2-TF, G4 sequences, and a possible role for viral perturbations in driving network changes. This informed a focused evaluation of viral perturbations in clinical AD. We examined four, large multi-omic data sets that included next-generation sequence data that enabled direct examination of viral sequences. We observed the presence of many viral species in the ageing brain, and linked multiple viral species with AD biology, including regulation of AD genetic risk networks, AD gene expression changes, and association with clinical dementia rating and neuropathology burden. We found prominent roles for Roseoloviruses HHV-6A and HHV-7, both implicated across multiple domains, and in 3 independent cohorts. Importantly, the inclusion of the MAYO TCX data set allowed us to perform comparisons between AD and other neuropathological controls. These additional comparisons suggest that HHV-6A and HHV-7 are not ubiquitous features of neuropathology, and appear at least partly specific to AD. Comparison against additional neuropathological diseases and brain regions would help clarify this specificity, for instance although PSP is associated with accumulation of cortical neurofibrillary tangles, the most severe manifestations are typically seen throughout the basal ganglia, brainstem and cerebellar structures, regions that were not profiled in this study, yet which might harbor different viral species and abundances than those observed.
Additional focused sequencing efforts are required to gain further resolution on the role of HHV-6 and HHV-7 in AD. Previous reports suggest an increased prevalence of HHV-6 in AD based on PCR of postmortem brain tissue(Lin et al., 2002) and seroprevalence(Carbone et al., 2014), though subsequent serological studies did not find increased HHV-6 seropositivity(Agostini et al., 2016). Importantly, not all of the methods employed in these studies distinguished between HHV-6A and HHV-6B, which may account for some discrepancy. We found considerable inter-regional variability in AD associated viral abundance, finding differences in four of the six regions assayed. Additionally, we found a discordant profile for HHV-6B (increased in STG, reduced in APFC), indicating the importance of distinguishing HHV-6B from HHV-6A (increased in the STG, APFC, DLPFC and TCX). Given the near universal seropositivity for HHV-6 in the general population, seropositivity is likely too non-specific to reliably distinguish AD-relevant states of viral activity within the brain.
The miR-155 network offers a targeted area for further investigation of viral activity and neuronal loss in AD. We first identified miR-155 during investigation of the preclinical AD networks, and later identified that HHV-6A negatively regulates neuronal fraction in the STG, and that the cis-eQTLs of the host genes that mediate this are enriched for AD risk loci, leading to the re-emergence of miR-155 (via MIR155HG) in our analysis. Our finding that four month miR-155-KO x APP/PS1 mice develop increased cortical amyloid plaque density and increased levels of Aβ oligomers provides further evidence linking miR-155 to AD pathology. These findings support the view of miR-155 as a regulator of complex anti- and pro-viral actions, and offer a mechanism linking viral activity with AD neuropathology and supports the hypothesis that viral activity contributes to AD.
The integrated findings of this study suggest that AD biology is impacted by a complex constellation of viral and host factors acting across different time scales and physiological systems (Figure 8b). This includes host mucosal defense and modulation of innate immune response by virus and host. It also includes disturbance of core biological processes, including some that are well described in AD (e.g. APP processing, cytoskeletal organization, mitochondrial respiration, protein synthesis and cell cycle control) and some that are less well characterized (e.g. widespread shifts in G4 activity and C2H2-TF regulatory programs). We note potential mechanisms (and candidate molecular mediators) that we find perturbed by viral species, and which have known impacts on these altered processes, for instance virally driven changes in protein synthesis machinery, tRNA synthetase activity, and nucleotide pool maintenance, which collectively exert complex effects on G4 regulation and C2H2-TF activity.
Our interpretation of the changes seen in the preclinical AD networks rests partly on the true disease relevance of neuropathology in the absence of cognitive impairment. We cannot readily discriminate molecules involved in disease progression from molecules that are responsible for resilience and for maintaining brain function in the face of advanced AD pathology. Despite the uncertainty around the eventual health trajectory of these donors, our reasoning was that by that conditioning our analysis on changes in AD vulnerable brain regions we might still find instructive biological themes, even if uncertainty remained around whether those changes were disease associated, opportunistic or somehow adaptive. Despite this reasoning, and supportive circumstantial evidence (e.g. miR-155) we have not yet confirmed in an equivalent data set whether the viral findings associated with clinical AD have predecessors in the preclinical AD context.
Investigating the subcellular distribution of viral DNA, especially for the key species of interest (HHV6A and HHV-7) would add valuable context to these findings. Unlike most viruses discussed in this study, HHV-6(Tanaka-Taya et al., 2004) and HHV-7(Prusty et al., 2016) can integrate into subtelomeric regions of host chromosomes during latent infection, and are excised into an episomal form during lytic infection and replication. Characterizing the extent and distribution of integrated vs. episomal Roseolovirus in AD, would be an important step in further understanding the mechanisms that connect viral abundance with molecular aspects of AD biology, and would have implications for therapeutic targeting of latent viral reservoirs relevant to AD.
It is important to note that the findings reported in this study are not sufficient to definitively demonstrate that viral activity causally contributes to the onset or progression of AD which would be most naturally established in a prospective, intervention-based study. We do report on multiple streams of indirect evidence, however, which enabled us to partially address this with the available data, including: (a) causal inference testing that supports a role for HHV-6A in contributing to neuronal loss in AD, (b) AD GWAS risk loci enrichments in virus-host network eQTLs, (c) emergence of molecules such as miR-155 from preclinical AD networks and virus-host networks, and (d) relative specificity of HHV-6A and HHV-7 for AD, compared with other neurodegenerative diseases. Follow-up studies that evaluate the onset and progression of AD phenotypes in virally infected AD model systems would be one approach to better delineate the causal and mechanistic relationships that link pathogen activity with the evolution of AD associated behavioral, molecular and neuropathological changes.
In summary, we find evidence that links the activity of specific viral species with molecular, genetic, clinical, and neuropathological aspects of AD. Interpretation of these findings in light of the disturbances in G4 and C2H2-TF regulation in the preclinical AD samples that prompted our evaluation of viral activity is supportive of an important role for viral activity, especially Roseoloviruses HHV-6A and HHV-7, in the development and progression of AD.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Joel T. Dudley (joel.dudley@mssm.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
APPKM670/671NL/PSEN1Δexon9 (APP/PS1)(Jankowsky et al., 2004), and miR-155-KO(Thai et al., 2007) mice were obtained from Jackson Laboratories. APPKM670/671NL/PSEN1Δexon9 were crossed with miR-155-KO mice to obtain APPKM670/671NL/PSEN1Δexon9 heterozygous or KO for miR-155. Mouse lines were maintained on a C57Bl6/J background. 4-month-old male and female mice were sacrificed by decapitation. Brains were dissected into right and left hemispheres. One hemisphere was collected and fixed in 4% paraformaldehyde for immunohistochemistry analysis. The second hemisphere was dissected and prefrontal cortex (PC) was collected for transcriptomic analysis. PC and hemibrains were then snap-frozen and stored at −80°C prior to RNA isolation or biochemistry analysis. Male and female mice were used in the experiments. The experimental procedures were conducted in accordance with NIH guidelines for animal research and were approved by the Institutional Animal Care and Use Committee (IACUC) at Icahn School of Medicine at Mount Sinai.
METHOD DETAILS
RNA sequencing human gene expression
RNA sequencing data was obtained from the Accelerating Medicines Partnership - Alzheimer’s Disease (AMP-AD) Knowledge Portal (MSSB synapse ID: syn3157743, MAYO TCX: syn8612203, ROS/MAP: syn8612097). For the MSBB cohort, post-mortem samples were collected from the STG (Brodmann Area 22), APFC (Brodmann Area 10), PHG (Brodmann Area 36) and IFG (Brodmann Area 44) by the Mount Sinai NIH Brain and Tissue Repository. MAYO TCX cohort samples were collected from the temporal cortex, as previously described(Allen et al., 2016). ROS and MAP cohort samples were collected from the DLPFC as previously described(Bennett et al., 2012a, Bennett et al., 2012b).
For all cohorts, RNA-seq samples with RIN less than 6, were removed from the analysis.
For host gene expression (MSSB cohort), single end reads were aligned to human genome reference (GRCh37 ensembl version 70(Kersey et al., 2016)), using STAR-RNAseq (2.4.0g1) read aligner(Dobin et al., 2013), and accepted mapped reads were summarized to gene level counts using the featureCounts function of the subread software package(Liao et al., 2013, Liao et al., 2014). Genes with at least 1 count per million mapped reads(Robinson et al., 2010) in at least half of the sample libraries were retained, and normalized using the voom function in the Limma package(Law et al., 2014, Ritchie et al., 2015).
Whole exome sequencing
Whole exome sequencing data (MSBB) used in this study can be obtained from the Accelerating Medicines Partnership - Alzheimer’s Disease (AMP-AD) Knowledge Portal (synapse ID: syn4645334). Reads were aligned to human genome hg19 using BWA aligner(Li et al., 2009a). DNA sequence variants were called using the DNAseq Variant Analysis workflow of GATK Best Practices version 3(Van der Auwera et al., 2013). Variants with a minor allele frequency < 0.05, or with missing calls in > 10 samples were removed from further analysis. Common variants were imputed using IMPUTE2(Howie et al., 2009, Howie et al., 2011) using 1000 Genomes Phase 3 reference genotypes(Genomes Project et al., 2015).
Liquid Chromatography tandem mass spectrometry
Proteomics data (MSBB) used in this study can be obtained from the Accelerating Medicines Partnership - Alzheimer’s Disease (AMP-AD) Knowledge Portal (synapse ID : syn5759470). All samples were from a single brain region (APFC), and underwent Liquid Chromatography tandem Mass Spectrometry (LC-MS/MS) and MaxQuant(Cox et al., 2008) was used to quantify label free protein counts. Protein counts were normalized by the total counts detected for that sample, and log transformed (log2) using an offset of 0.25 for null values.
Immunohistochemistry
Paraformaldehyde-fixed mouse brains were cut (30μm thick) with a vibratome VT1000S (Leica Microsystems, Germany). Sagittal free-floating sections were pre-treated with 70% formic acid for 15min at room temperature. Sections were then blocked for 1h at room temperature (PBS with 0.1% v/v Tween-20 and 10% goat serum) and incubated overnight with anti-Iba1 (1:500;Wako, Richmond, VA) and 6E10 (1:1000;Covance, Princeton, NJ) antibodies (PBS with 0.1% v/v Tween-20 and 1% goat serum). Sections were then incubated for 1h with fluorescent conjugated secondary antibodies (PBS with 0.1% v/v Tween-20 and 1% goat serum) (anti-rabbit Alexa 488 (1:400) for Iba1 and anti-mouse Alexa 568 (1:400) for 6E10) and mounted in Superfrost Plus slides. Images were acquired on an Olympus BX61 microscope with an attached Olympus DP71 camera.
Mouse RNA isolation and library preparation
RNA isolation and library preparation were performed as previously described(Readhead et al., 2016). Briefly, snap frozen samples from male and female mice were homogenized in QIAzol Lysis Reagent (Qiagen). Total RNA purification was performed with the miRNeasy Mini kit (Qiagen), according to the manufacturer’s instructions. RNA quantification and quality were evaluated by Agilent BioAnalyzer and processed for RNA library preparation. RNA integrity was checked by either the Fragment Analyzer (Advanced Analytical, IA, USA) or the 2100 Bioanalyzer using the RNA 6000 Nano assay (Agilent, CA, USA). All processed total RNA samples had RQN/RIN value of 8.8 or greater.
QUANTIFICATION AND STATISTICAL ANALYSIS
Construction of entorhinal cortex and hippocampal preclinical AD and control regulatory networks
Preclinical AD and Control networks were generated from post mortem microarray gene expression data downloaded from Gene Expression Omnibus (Preclinical AD: accession GSE9770(Liang et al., 2010), Control accession: GSE5281(Liang et al., 2007)). In both data sets, samples were derived from layer 2 entorhinal cortex neurons, and hippocampal CA1 pyramidal neurons. For each group, we built single-tissue networks (HIP and EC), as well as a separate cross-tissue HIP-EC network, with the goal of combining these three individual networks afterwards into a single union network that is able to capture intra-tissue, as well as inter-tissue connectivity.
Networks were constructed using a modification of the “inductive causation with latent variables” procedure(Pearl, 2009). This approach is a constraint-based method for building causal networks, using conditional dependence tests between nodes to first identify a network skeleton (the set of undirected edges), learn v-structures (a triplet of nodes where two non-adjacent nodes share a target node), and output a partially directed graph (PDG). The final PDG contains four types of edges: undirected (indicating uncertain causation), bi-directed (spurious or latent causation), directed (that directed edge is present in at least one Markov equivalent graph, and none contain the opposite arrow for that edge), and marked-directed edges (that directed edge exists in every Markov-equivalent graph). For each network, we retained all edges marked as directed, or marked-directed in the PDG.
Each of the networks we generated was constructed across the top 15,000 most differentially expressed probes within that brain region (Preclinical AD vs. Controls). For the HIP-EC network, we combined HIP and EC samples from each donor, into a single expression set, comprising the top 7,500 most differentially expressed probes from each tissue. We constructed the final preclinical AD and control networks by combining discovered edges from the EC, HIP and HIP-EC networks into a final structure comprising 30,000 tissue-annotated probes. We only included HIP-EC cross-tissue edges if the edge was not present as an intra-tissue edge in either of the HIP or EC networks.
We then identified network probes with unusually high downstream influence, iterating over Gene Ontology (GO) biological process terms, and inducing connected subgraphs from our network, according to their annotation with that specific term. Within that subgraph (containing a minimum of two connected probes) for each probe, we identified the number of probes within the network diameter of the induced subgraph that can be reached via incoming connections, and outgoing connections respectively. A GO term score was calculated for each probe, based on the difference between Z-scores of total network connectivity. A final network driver score was calculated based on the scaled sum of individual scores associated for all GO biological process terms. More formally, the driver score for probe (P) is the sum of individual differences between upstream neighborhood size and downstream neighborhood size, for each subgraph (G) induced according to each GO biological process term:
We converted these driver scores to standard scores, and nominated probes with a Z-score ≥ 2 as network drivers. Drivers that were seen in the context of the preclinical AD network (but not the control network) were classified as “Gained in preclinical AD”, and those seen only in the control network, were classified as “Lost in preclinical AD”.
G-quadruplex sequence prediction
G4 sequence prediction within human genes was based on pattern matching against the human reference genome (hg19). Genomic coordinates for each gene in the UCSC hg19 “Known genes” were used to download nucleotide sequences for gene promoter regions, 5’-UTR, introns, exons and 3’-UTR. Promoters were defined as 2000 base pairs upstream and downstream from the transcription start site. We used regular expressions to identify the genomic coordinates corresponding to occurrences of four runs of at least three guanine bases, interspersed with between one to seven other bases (including guanine) based on similar approaches(Garant et al., 2015). We used separate patterns to retrieve matches located on the coding strand (G(3,).(1,7)?G(3,).(1,7)?G(3,).(1,7)?G(3,)) and non-coding strand (C(3,).(1,7)?C(3,).(1,7)?C(3,).(1,7)?C(3,)) respectively. Normalized G4 motif density within a particular feature was calculated by dividing the number of predicted motifs by the length (in base pairs) of that feature.
Generation of viral 31mer database
We downloaded nucleotide sequences for 515 unique viruses with humans as known or suspected hosts(Brister et al., 2015). We segmented each viral sequence into its set of unique 31mers using Jellyfish count and dump commands(Marcais et al., 2011), followed by removal of any 31mers present in multiple viral species. We then generated bowtie2 indices(Langmead et al., 2012) for the viral sequences, and the corresponding 31mer database for use in subsequent viral mapping steps.
Detection of viral transcription in RNA and whole exome sequencing
We quantified viral transcription through a modified workflow based on ViromeScan(Rampelli et al., 2016), which proceeds as follows: preliminary alignment of fastq files to a viral reference database using bowtie2(Langmead et al., 2012), identifying candidate viral sequences. Mapped reads were filtered through BMtagger(Rotmistrovsky et al., 2011) to remove likely human reads. Any putatively non-human reads with low quality scores were trimmed, and reads with a trimmed read length < 60 bases were discarded. Reads were again filtered using BMtagger(Rotmistrovsky et al., 2011) to remove likely bacterial reads. Filtered, trimmed, non-human reads are then mapped to the viral 31mer database with bowtie2(Langmead et al., 2012), using a very sensitive, local alignment, outputting all valid alignments. Bam files were then sorted and indexed, using Samtools(Li et al., 2009b). BAM files were processed manually to output a single best alignment for each read (randomly outputting a single alignment in cases where multiple best alignments were found). To minimize misclassification of human reads as viral, we then performed an additional BLAST(Altschul et al., 1990) search for any 31mers with homology to the combined hg19 and cDNA sequences(Lander et al., 2001) (blastn e-value < 1e-3), and removed those 31mers from further analysis.
We generated a 31mer count matrix for all samples, and then summarized these separately to the level of viral species, and also for individual viral genomic features. Genomic features and coordinates were derived from NCBI gene transfer format files for each sequence. Although each viral 31mer is unique to a single viral sequence, some 31mers might occur multiple times within that sequence. In these instances, counts for 31mers were assigned to all genomic features containing that 31mer. We merged any overlapping genomic features that also shared the same total counts in the genomic feature count matrix.
Differential abundance of viral transcription in RNA sequencing
We performed differential viral abundance analysis at the level of viral species, and separately, at the level of individual genomic features. Due to differences in the study design and availability of covariates in each of the three AD cohorts, there were slight differences in the procedure for estimating differential viral abundance, reflecting differences in approaches for the (a) classification of AD vs. Control status, and (b) availability of technical covariates to incorporate into linear modelling. In the MSBB, ROS and MAP cohorts, for each of the brain regions assayed, we compared normalized viral abundance between AD cases and controls, using three different definitions of AD within each comparison. Definitions of AD were based on the multiple levels of CERAD neuropathology classification(Mirra et al., 1991), specifically we performed comparisons between “Definite AD Vs. Controls” (NP score: 2 Vs. NP score: 1), “Likely AD Vs. Controls” (NP score: 2 / 3 Vs. NP score: 1) and “Possible AD Vs. Controls” (NP score: 2 / 3 / 4 Vs. NP score: 1).
Diagnosis within the MAYO TCX cohort was based on neuropathological evaluation and classification as follows:
AD was based on “Definite AD” diagnosis according to the NINCDS-ADRDA criteria and a Braak NFT stage of IV or greater.
Control subjects had Braak NFT stage of <= III, CERAD neuritic and cortical plaque densities of 0 (none) or 1 (sparse) and the absence of any of the following pathologic diagnoses: AD, Parkinson’s disease, dementia with Lewy Bodies, vascular dementia, PSP, motor neuron disease, corticobasal degeneration, Pick’s disease, Huntington’s disease, frontotemporal lobar degeneration, hippocampal sclerosis, or dementia lacking distinctive histology.
PA subjects had a Braak NFT stage of <= III, CERAD neuritic and cortical plaque densities of >= 2, and the absence of the above diagnoses, as well as the absence of any dementia or mild cognitive impairment (MCI).
PSP subjects were identified via semiquantitative distribution of NFT(Hauw et al., 1994) at autopsy.
Within each comparison, we retained viral features with multiple mapped reads in at least 10 samples. Viral feature counts adjusted for the total number of reads in each fastq file, and quantile normalized using the Voom function in the Limma package(Law et al., 2014, Ritchie et al., 2015). Linear models were fit for each of the viral features, for analysis of the MSBB cohort, we included covariates for: AD status, age of death, sex, ethnicity, RIN, post-mortem interval (PMI), and batch. For the ROS and MAP cohorts, we included covariates for AD status, age of death, sex, ethnicity, RIN, PMI, batch and years of education. For the MAYO TCX cohort, we included covariates for diagnosis, age of death, sex, RIN, batch and study center. Differential abundance between AD and control groups were estimated using the eBayes function(Smyth, 2004), setting robust=TRUE to minimize the effect of outliers in variance.
Viral QTL detection
Host DNA markers that associate with viral abundance (vQTL) were identified separately for each brain region. Within each region, data was included from donors with paired whole exome sequencing and RNA-seq data. Viral genomic feature counts were normalized using the same procedure used in analyzing differential viral abundance, and we used the Matrix eQTL(Shabalin, 2012) software package to identify DNA variants associated with normalized viral abundance (controlling for age, sex, ethnicity, RNA-seq batch, RIN and PMI as covariates), assuming an additive linear model for associating genotype dosage with viral abundance. We generated a distribution of null association P-values for each viral feature by shuffling sample labels for viral abundance (1000 permutations), retaining the minimum association P-value for that feature, across all markers. We estimated the empirical FDR by comparison of each observed association P-value with a distribution of null association P-values generated separately for each viral feature (1000 permutations of viral abundance labels). DNA markers with an empirical association FDR < 0.25 were classified as vQTLs for that specific viral feature in the context of that brain region.
AD GWAS enrichments for virus network eQTLs
We identified cis-acting ( < 1 MB) DNA markers that associate with gene expression (cis-eQTL) separately for each brain region. Within each region, we included data from donors with paired whole exome sequencing and RNA-seq data. We used the Matrix eQTL(Shabalin, 2012) software package to identify DNA variants associated with normalized gene expression (controlling for age, sex, ethnicity, RNA-seq batch, RIN and PMI as covariates), assuming an additive linear model for associating genotype dosage with gene expression. We classified DNA markers with an association FDR < 0.1 as cis-eQTL, and used these to define the set of markers that are collectively associated with each virus/host subnetwork. For each virus, we pooled host interactions from all four tissues, classified each according to direction (i.e. “virus to host” or “host to virus”), and sign of correlation (i.e. positively or negatively correlated with viral abundance), and then calculated enrichments for AD risk-associated loci(Lambert et al., 2013) using the versatile gene-based association study (VEGAS) approach(Liu et al., 2010b, Mishra et al., 2015).
Estimating cell type fractions from RNA-seq
We used CIBERSORT(Newman et al., 2015) to deconvolute RNA-seq samples (MSBB) into estimated fractions for major brain cell types (neurons, astrocytes, microglia, endothelial cells, oligodendrocytes and oligodendrocyte precursor cells). This requires an independent reference panel of presumed relevant cell-types, usually derived from transcriptomic studies of individual cell types. We utilized a single cell RNA-seq dataset generated by Darmanis et al(Darmanis et al., 2015), using 138 individual samples derived from the cortex of middle-aged adults (age range: 47 – 63 years) representing a variety of brain cell types (astrocytes n=52, neurons n=43, oligodendrocytes n=19, endothelial n=15, microglia n=7, oligodendrocyte precursor cells n=2). Genes with at least 1 count per million mapped reads(Robinson et al., 2010) in at least n sample libraries were retained, where n=the size of the smallest group of cell types. We then subset this single cell type expression, and the MSBB expression to the set of unique gene identifiers present in both, and supplied these as inputs to CIBERSORT, identifying signature genes according to default parameters: kappa: 999, qvalue: 0.3, min: 50, max: 150, quantile normalization=disabled, and specifying 1000 permutations.
Molecular and functional enrichment analysis
Gene set enrichments for discrete groups of genes (i.e., virus-host subnetworks networks) were calculated using Fisher’s exact text, and one-sided P-values (to identify over-representation of genesets) were adjusted using the Benjamini-Hochberg method(Benjamini et al., 1995). Gene sets used throughout the enrichment analysis were derived from a combination of publicly available sources, such as the molecular signatures database(Subramanian et al., 2005), brain specific gene sets curated from publicly available data(Miller et al., 2011), protein-protein hubs interactor sets(Chen et al., 2012), Mirtarbase(Hsu et al., 2011) and ChipSeq based transcription factor target sets(Lachmann et al., 2010).
Transcription factor motif enrichments in the promoters of preclinical AD network drivers were calculated based on position weight matrix matching within the gene promoter for each driver (defined as the region within 2000 basepairs of the transcription start site) using MatInspector(Cartharius et al., 2005).
Inferring causal relationships between viral abundance and biomolecular, clinical and neuropathological traits
We applied a causal inference paradigm to multiple aspects of the analysis performed in this study, to determine directed relationships between viral abundance, with a variety of host traits, including molecular (e.g. gene or protein expression), clinical (clinical dementia rating) and neuropathological (Neuritic plaque density, Braak and Braak score) traits. We used a statistical framework introduced by Millstein et al(Millstein et al., 2009), which offers a causal inference test (CIT) that tests the hypothesis that a molecule (such as the normalized abundance of a viral species, or of a specific viral genomic feature) is mediating a causal association between a DNA locus (such as a vQTL for that specific virus), and some other quantitative trait (such as the expression of host genes that are correlated with the vQTL and the viral abundance). Causal relationships can be inferred from a chain of mathematical conditions, requiring that for a given trio of loci (L), a potential causal mediator (G) and a quantitative trait (T), the following conditions must be satisfied to establish that G is a causal mediator of the association between L and T:
L and G are associated
L and T are associated
L is associated with G, given T
L is independent of T, given G
Although CIT includes tests for linkage (conditions a and b), to control the number of candidate L / G / T trios that are submitted to the CIT function, we perform multiple pre-filtering steps, which are aimed at establishing association between L and G, and L and T, before we submit a particular trio for CIT. Association between L and G is established in the course of the viral QTL analysis (described above), where we classify variants with an association FDR < 0.1 as a vQTL for that specific viral feature. If T is a molecular species (gene expression, protein expression), nominal association between L and T is established using matrix eQTL(Shabalin, 2012) retaining candidate T molecules (for that specific vQTL) with an association P-value < 0.05.
Although CIT outputs what is ostensibly a P-value, it is actually the highest P-value of the four constituent hypothesis tests, reflecting each of the conditions required to establish causal mediation. This results in a non uniform CIT P-value distribution under null conditions, which can make appropriate multiple test correction unreliable. To overcome this, we employed a permutation based approach to assess the significance of candidate causal relationships, where candidate traits (T) are randomly shuffled, separately within each genotype dosage group (0, 1 or 2) for each permutation. The false discovery rate was estimated by counting the proportion of permutations (1000 per trio) with a CIT P-value lower than the test CIT P-value.
To minimize the number of false positive inferences, we performed two separate tests for each candidate trio. We tested models that include the viral feature (G) as causal for the host trait (T) (“causal model”) and separately, the G being regulated by the T (“reactive model”). We required that for G to be classified as regulating T, its permutation based FDR for the causal model be < 0.05, and reactive model be > 0.05. Conversely, for a G to be classified as being regulated by a T, we required that the FDR for the reactive model be < 0.05, and > 0.05 for the causal model.
Transcription Factor Regulatory Network Analysis
The conceptual framework for the transcriptional regulatory network using TReNA was described previously(Pearl et al., 2017). Briefly, DNase Hypersensitivity (DHS) fastq files from ENCODE for all available brain samples were downloaded and aligned using the SNAP method(Zaharia et al., 2011). Two alignments were performed using seed size 16 and 20 as the sequence data was typically > 50 bp in length. The peak calling algorithm F-seq was used to identify regions of open chromatin(Boyle et al., 2008). Footprinting algorithms for Wellington(Piper et al., 2013) and HINT(Gusmao et al., 2016) were generated using default parameters. For each individual gene model, footprints within the proximal promoter (+/−5 kb of the transcription start site) were considered as priors in assessing the relationship between the expression of the transcription factor and target gene. Using the R package trena(Ament et al., 2017), which utilizes several LASSO regression techniques, Pearson and Spearman correlation, and random forest to prioritize a list of putative transcription factor regulators for each gene. Scores from all these approaches were scaled and projected into PCA space and their principle components added together to produce a single composite score (pcaMax). This approach was applied to each of the RNA-seq datasets generated within the AMP-AD consortium to generate networks used for the virus / host analyses herein.
Kinase enrichment analysis of candidate virus-TF associations were performed as described in “Molecular and functional enrichment analysis”, while subsetting the gene background to the set of 569 TF within the scope of the TF networks.
DATA AND SOFTWARE AVAILABILITY
Viral abundance estimates, and scripts to reproduce results can be accessed at: https://www.synapse.org/#!Synapse:syn12177270.
Supplementary Material
KEY RESOURCES TABLE.
Highlights.
Common viral species frequently detected in normal, ageing brain
Increased HHV-6A and HHV-7 in brains of subjects with Alzheimer’s disease (AD)
Findings were replicated in two additional, independent cohorts
Multiscale networks reveal viral regulation of AD risk, and APP processing genes
Acknowledgments
MSBB RNA-seq, WES and proteomics data used for the evaluation of viral sequences in AD was generated from postmortem brain tissue collected through the Mount Sinai VA Medical Center Brain Bank and were provided by Dr. Eric Schadt from Mount Sinai School of Medicine. Proteomics data were also provided by Dr. Levey from Emory University.
Construction of TReNA networks was supported by U01AG046139 and U54EB020406.
The MAYO TCX RNAseq study data was led by Dr. Nilüfer Ertekin-Taner, Mayo Clinic, Jacksonville, FL as part of the multi-PI U01 AG046139 using samples from the following sources:
(1) The Mayo Clinic Brain Bank. Data collection was supported through funding by NIA grants P50 AG016574, R01 AG032990, U01 AG046139, R01 AG018023, U01 AG006576, U01 AG006786, R01 AG025711, R01 AG017216, R01 AG003949, NINDS grant R01 NS080820, CurePSP Foundation, and support from Mayo Foundation.
(2) Sun Health Research Institute Brain and Body Donation Program of Sun City, Arizona. The Brain and Body Donation Program is supported by the National Institute of Neurological Disorders and Stroke (U24 NS072026 National Brain and Tissue Resource for Parkinson’s Disease and Related Disorders), the National Institute on Aging (P30 AG19610 Arizona Alzheimer’s Disease Core Center), the Arizona Department of Health Services (contract 211002, Arizona Alzheimer’s Research Center), the Arizona Biomedical Research Commission (contracts 4001, 0011, 05–901 and 1001 to the Arizona Parkinson’s Disease Consortium) and the Michael J. Fox Foundation for Parkinson’s Research.
The ROS/MAP RNAseq study data was provided by the Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago. Data collection was supported through funding by NIA grants P30AG10161, R01AG15819, R01AG17917, R01AG30146, R01AG36836, U01AG32984, U01AG46152, the Illinois Department of Public Health, and the Translational Genomics Research Institute.
SG and MEE acknowledge the support of U01 AG046170.
BR, SG, MEE and JTD acknowledge the support of 1R56AG058469.
Philanthropic financial support was provided by Katherine Gehl.
The computational resources and staff expertise provided by the Department of Scientific Computing at the Icahn School of Medicine at Mount Sinai also contributed to the performance of this research.
Footnotes
Declarations of Interests
The authors declare that they have no competing financial interests in relation to the work described.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agostini S,et al. (2016). Lack of evidence for a role of HHV-6 in the pathogenesis of Alzheimer’s disease. J Alzheimers Dis, 49, 229–35. [DOI] [PubMed] [Google Scholar]
- Albright AV,et al. (1998). The effect of human herpesvirus-6 (HHV-6) on cultured human neural cells: oligodendrocytes and microglia. J Neurovirol, 4, 486–94. [DOI] [PubMed] [Google Scholar]
- Allen M,et al. (2016). Human whole genome genotype and transcriptome data for Alzheimer’s and other neurodegenerative diseases. Sci Data, 3, 160089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF,et al. (1990). Basic local alignment search tool. J Mol Biol, 215, 403–10. [DOI] [PubMed] [Google Scholar]
- Ament S,et al. (2017). TReNA: Fit transcriptional regulatory networks using gene expression, priors, machine learning. R package version 0.99.10 ed.
- Aran D,et al. (2017). xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol, 18, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbuckle JH,et al. (2010). The latent human herpesvirus-6A genome specifically integrates in telomeres of human chromosomes in vivo and in vitro. Proc Natl Acad Sci U S A, 107, 5563–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artusi S,et al. (2016). Visualization of DNA G-quadruplexes in herpes simplex virus 1-infected cells. Nucleic Acids Res, 44, 10343–10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin JD,et al. (2013). Exploring mRNA 3’-UTR G-quadruplexes: evidence of roles in both alternative polyadenylation and mRNA shortening. Nucleic Acids Res, 41, 5898–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y,et al. (1995). Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological, 57, 289–300. [Google Scholar]
- Bennett DA,et al. (2012a). Overview and findings from the religious orders study. Curr Alzheimer Res, 9, 628–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett DA,et al. (2012b). Overview and findings from the rush Memory and Aging Project. Curr Alzheimer Res, 9, 646–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier A,et al. (2013). Transcriptional profiling reveals molecular signatures associated with HIV permissiveness in Th1Th17 cells and identifies peroxisome proliferator-activated receptor gamma as an intrinsic negative regulator of viral replication. Retrovirology, 10, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP,et al. (2008). F-Seq: a feature density estimator for high-throughput sequence tags. Bioinformatics, 24, 2537–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak H,et al. (2006). Staging of Alzheimer disease-associated neurofibrillary pathology using paraffin sections and immunocytochemistry. Acta Neuropathol, 112, 389–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brister JR,et al. (2015). NCBI viral genomes resource. Nucleic Acids Res, 43, D571–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caccamo A,et al. (2017). Necroptosis activation in Alzheimer’s disease. Nat Neurosci, 20, 1236–1246. [DOI] [PubMed] [Google Scholar]
- Carbone I,et al. (2014). Herpes virus in Alzheimer’s disease: relation to progression of the disease. Neurobiol Aging, 35, 122–9. [DOI] [PubMed] [Google Scholar]
- Cartharius K,et al. (2005). MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics, 21, 2933–42. [DOI] [PubMed] [Google Scholar]
- Caselli E,et al. (2017). HHV-6A in vitro infection of thyrocytes and T cells alters the expression of miRNA associated to autoimmune thyroiditis. Virol J, 14, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cen S,et al. (2001). Incorporation of lysyl-tRNA synthetase into human immunodeficiency virus type 1. J Virol, 75, 5043–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang LK,et al. (2005). Activation of Sp1-mediated transcription by Rta of Epstein-Barr virus via an interaction with MCAF1. Nucleic Acids Res, 33, 6528–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY,et al. (2012). Expression2Kinases: mRNA profiling linked to multiple upstream regulatory layers. Bioinformatics, 28, 105–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z,et al. (2008). Genetic association of neurotrophic tyrosine kinase receptor type 2 (NTRK2) With Alzheimer’s disease. Am J Med Genet B Neuropsychiatr Genet, 147, 363–9. [DOI] [PubMed] [Google Scholar]
- Cohain A,et al. (2016). Exploring the Reproducibility of Probabilistic Causal Molecular Network Models. Pac Symp Biocomput, 22, 120–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J,et al. (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol, 26, 1367–72. [DOI] [PubMed] [Google Scholar]
- Darmanis S,et al. (2015). A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci U S A, 112, 7285–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson DW,et al. (1992). Identification of normal and pathological aging in prospectively studied nondemented elderly humans. Neurobiol Aging, 13, 179–89. [DOI] [PubMed] [Google Scholar]
- Dobin A,et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics, 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eimer WA,et al. (2017). Aβ protects against herpesviridae infections in brain.
- Epel ES,et al. (2016). Meditation and vacation effects have an impact on disease-associated molecular phenotypes. Transl Psychiatry, 6, e880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzen O,et al. (2016). Cardiometabolic risk loci share downstream cis- and trans-gene regulation across tissues and diseases. Science, 353, 827–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garant JM,et al. (2015). G4RNA: an RNA G-quadruplex database. Database (Oxford), 2015, bav059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatto G,et al. (2008). Epstein-Barr virus latent membrane protein 1 trans-activates miR-155 transcription through the NF-kappaB pathway. Nucleic Acids Res, 36, 6608–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomes Project, C.,et al. (2015). A global reference for human genetic variation. Nature, 526, 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Isla T,et al. (1996). Profound loss of layer II entorhinal cortex neurons occurs in very mild Alzheimer’s disease. J Neurosci, 16, 4491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwein E,et al. (2007). A viral microRNA functions as an orthologue of cellular miR-155. Nature, 450, 10969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady SL,et al. (2012). Herpes simplex virus 1 infection activates poly(ADP-ribose) polymerase and triggers the degradation of poly(ADP-ribose) glycohydrolase. J Virol, 86, 8259–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusmao EG,et al. (2016). Analysis of computational footprinting methods for DNase sequencing experiments. Nat Methods, 13, 303–9. [DOI] [PubMed] [Google Scholar]
- Ha HC,et al. (2001). Poly(ADP-ribose) polymerase-1 is required for efficient HIV-1 integration. Proc Natl Acad Sci U S A, 98, 3364–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haroutunian V,et al. (1998). Regional distribution of neuritic plaques in the nondemented elderly and subjects with very mild Alzheimer disease. Arch Neurol, 55, 1185–91. [DOI] [PubMed] [Google Scholar]
- Harrich D,et al. (1989). Role of SP1-binding domains in in vivo transcriptional regulation of the human immunodeficiency virus type 1 long terminal repeat. J Virol, 63, 2585–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauw JJ,et al. (1994). Preliminary NINDS neuropathologic criteria for Steele-Richardson-Olszewski syndrome (progressive supranuclear palsy). Neurology, 44, 2015–9. [DOI] [PubMed] [Google Scholar]
- Howie B,et al. (2011). Genotype imputation with thousands of genomes. G3 (Bethesda), 1, 457–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie BN,et al. (2009). A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet, 5, e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu SD,et al. (2011). miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res, 39, D163–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y,et al. (2004). Elevation of the level and activity of acid ceramidase in Alzheimer’s disease brain. Eur J Neurosci, 20, 3489–97. [DOI] [PubMed] [Google Scholar]
- Hyman BT,et al. (1984). Alzheimer’s disease: cell-specific pathology isolates the hippocampal formation. Science, 225, 1168–70. [DOI] [PubMed] [Google Scholar]
- Itzhaki RF (2014). Herpes simplex virus type 1 and Alzheimer’s disease: increasing evidence for a major role of the virus. Front Aging Neurosci, 6, 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzhaki RF,et al. (2016). Microbes and Alzheimer’s Disease. J Alzheimers Dis, 51, 979–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky JL,et al. (2004). Mutant presenilins specifically elevate the levels of the 42 residue beta-amyloid peptide in vivo: evidence for augmentation of a 42-specific gamma secretase. Hum Mol Genet, 13, 15970. [DOI] [PubMed] [Google Scholar]
- Kaddurah-Daouk R,et al. (2011). Metabolomic changes in autopsy-confirmed Alzheimer’s disease. Alzheimers Dement, 7, 309–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaddurah-Daouk R,et al. (2013). Alterations in metabolic pathways and networks in Alzheimer’s disease. Transl Psychiatry, 3, e244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersey PJ,et al. (2016). Ensembl Genomes 2016: more genomes, more complexity. Nucleic Acids Res, 44, D574–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasemann S,et al. (2017). The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases. Immunity, 47, 566–581 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar DK,et al. (2016). Amyloid-beta peptide protects against microbial infection in mouse and worm models of Alzheimer’s disease. Sci Transl Med, 8, 340ra72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachmann A,et al. (2017). Massive Mining of Publicly Available RNA-seq Data from Human and Mouse. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachmann A,et al. (2010). ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics, 26, 2438–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JC,et al. (2013). Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet, 45, 1452–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES,et al. (2001). Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- Langmead B,et al. (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods, 9, 357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CW,et al. (2014). voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol, 15, R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S,et al. (2012). Optimal tests for rare variant effects in sequencing association studies. Biostatistics, 13, 762–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H,et al. (2009a). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 25, 1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H,et al. (2009b). The Sequence Alignment/Map format and SAMtools. Bioinformatics, 25, 2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang WS,et al. (2008). Altered neuronal gene expression in brain regions differentially affected by Alzheimer’s disease: a reference data set. Physiol Genomics, 33, 240–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang WS,et al. (2010). Neuronal gene expression in non-demented individuals with intermediate Alzheimer’s Disease neuropathology. Neurobiol Aging, 31, 549–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang WS,et al. (2007). Gene expression profiles in anatomically and functionally distinct regions of the normal aged human brain. Physiol Genomics, 28, 311–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y,et al. (2013). The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res, 41, e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y,et al. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics, 30, 923–30. [DOI] [PubMed] [Google Scholar]
- Lin WR,et al. (2002). Herpesviruses in brain and Alzheimer’s disease. J Pathol, 197, 395–402. [DOI] [PubMed] [Google Scholar]
- Liu HP,et al. (2010a). Evaluation of the poly(ADP-ribose) polymerase-1 gene variants in Alzheimer’s disease. J Clin Lab Anal, 24, 182–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ,et al. (2010b). A versatile gene-based test for genome-wide association studies. Am J Hum Genet, 87, 139–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovheim H,et al. (2015a). Reactivated herpes simplex infection increases the risk of Alzheimer’s disease. Alzheimers Dement, 11, 593–9. [DOI] [PubMed] [Google Scholar]
- Lovheim H,et al. (2015b). Herpes simplex infection and the risk of Alzheimer’s disease: A nested case-control study. Alzheimers Dement, 11, 587–92. [DOI] [PubMed] [Google Scholar]
- Madeira C,et al. (2015). d-serine levels in Alzheimer’s disease: implications for novel biomarker development. Transl Psychiatry, 5, e561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak J,et al. (1997). Primer tRNAs for reverse transcription. J Virol, 71, 8087–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcais G,et al. (2011). A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics, 27, 764–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastroeni D,et al. (2018). Laser-captured microglia in the Alzheimer’s and Parkinson’s brain reveal unique regional expression profiles and suggest a potential role for hepatitis B in the Alzheimer’s brain. Neurobiol Aging, 63, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattiussi S,et al. (2007). Inhibition of Poly(ADP-ribose)polymerase impairs Epstein Barr Virus lytic cycle progression. Infect Agent Cancer, 2, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton PJ,et al. (1980). Herpes-simplex viral genome and senile and presenile dementias of Alzheimer and Pick. Lancet, 1, 1038. [DOI] [PubMed] [Google Scholar]
- Miller JA,et al. (2011). Strategies for aggregating gene expression data: the collapseRows R function. BMC Bioinformatics, 12, 322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millstein J,et al. (2009). Disentangling molecular relationships with a causal inference test. BMC Genet, 10, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirra SS,et al. (1991). The Consortium to Establish a Registry for Alzheimer’s Disease (CERAD). Part II. Standardization of the neuropathologic assessment of Alzheimer’s disease. Neurology, 41, 479–86. [DOI] [PubMed] [Google Scholar]
- Mishra A,et al. (2015). VEGAS2: Software for More Flexible Gene-Based Testing. Twin Res Hum Genet, 18, 86–91. [DOI] [PubMed] [Google Scholar]
- Mori Y,et al. (1998). Analysis of human herpesvirus 6 U3 gene, which is a positional homolog of human cytomegalovirus UL 24 gene. Virology, 249, 129–39. [DOI] [PubMed] [Google Scholar]
- Morris JC (1993). The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology, 43, 2412–4. [DOI] [PubMed] [Google Scholar]
- Murat P,et al. (2014). G-quadruplexes regulate Epstein-Barr virus-encoded nuclear antigen 1 mRNA translation. Nat Chem Biol, 10, 358–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy JV,et al. (1976). Encephalopathy following measles infection in children with chronic illness. J Pediatr, 88, 937–42. [DOI] [PubMed] [Google Scholar]
- Newman AM,et al. (2015). Robust enumeration of cell subsets from tissue expression profiles. Nat Methods, 12, 453–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida K,et al. (1999). Aminoacylation of tRNAs encoded by Chlorella virus CVK2. Virology, 263, 220–9. [DOI] [PubMed] [Google Scholar]
- Nixon RA,et al. (2005). Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J Neuropathol Exp Neurol, 64, 113–22. [DOI] [PubMed] [Google Scholar]
- Nousiainen L,et al. (2013). Human kinome analysis reveals novel kinases contributing to virus infection and retinoic-acid inducible gene I-induced type I and type III IFN gene expression. Innate Immun, 19, 51630. [DOI] [PubMed] [Google Scholar]
- Nygaard HB (2017). Targeting Fyn Kinase in Alzheimer’s Disease. Biol Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulou C,et al. (2015). Nucleotide Pool Depletion Induces G-Quadruplex-Dependent Perturbation of Gene Expression. Cell Rep, 13, 2491–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl J (2009). Causality, Cambridge university press. [Google Scholar]
- Pearl JR,et al. (2017). Genome-scale transcriptional regulatory network models of psychiatric and neurodegenerative disorders. bioRxiv. [DOI] [PubMed] [Google Scholar]
- Piper J,et al. (2013). Wellington: a novel method for the accurate identification of digital genomic footprints from DNase-seq data. Nucleic Acids Res, 41, e201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusty BK,et al. (2016). Possible Chromosomal and Germline Integration of Human Herpesvirus 7 (HHV-7). J Gen Virol. [DOI] [PubMed] [Google Scholar]
- Prusty BK,et al. (2013). Reactivation of chromosomally integrated human herpesvirus-6 by telomeric circle formation. PLoS Genet, 9, e1004033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampelli S,et al. (2016). ViromeScan: a new tool for metagenomic viral community profiling. BMC Genomics, 17, 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Readhead B,et al. (2016). Molecular systems evaluation of oligomerogenic APP(E693Q) and fibrillogenic APP(KM670/671NL)/PSEN1(Deltaexon9) mouse models identifies shared features with human Alzheimer’s brain molecular pathology. Mol Psychiatry, 21, 1099–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D,et al. (2015). G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res, 43, 8627–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME,et al. (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res, 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD,et al. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26, 139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rode AB,et al. (2016). tRNA Shifts the G-quadruplex-Hairpin Conformational Equilibrium in RNA towards the Hairpin Conformer. Angew Chem Int Ed Engl, 55, 14315–14319. [DOI] [PubMed] [Google Scholar]
- Rotmistrovsky K,et al. (2011). BMTagger: Best Match Tagger for removing human reads from metagenomics datasets.
- Rouillard AD,et al. (2016). The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database (Oxford), 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang Y,et al. (2014). Probing the interaction between U24 and the SH3 domain of Fyn tyrosine kinase. Biochemistry, 53, 6092–102. [DOI] [PubMed] [Google Scholar]
- Scholz M,et al. (2004). Thrombin induces Sp1-mediated antiviral effects in cytomegalovirus-infected human retinal pigment epithelial cells. Med Microbiol Immunol, 193, 195–203. [DOI] [PubMed] [Google Scholar]
- Shabalin AA (2012). Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics, 28, 1353–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shugar D (1999). Viral and host-cell protein kinases: enticing antiviral targets and relevance of nucleoside, and viral thymidine, kinases. Pharmacol Ther, 82, 315–35. [DOI] [PubMed] [Google Scholar]
- Sigurðsson B (1954). Rida, a chronic encephalitis of sheep: with general remarks on infections which develop slowly and some of their special characteristics.
- Sjogren T,et al. (1952). Morbus Alzheimer and morbus Pick; a genetic, clinical and patho-anatomical study. Acta Psychiatr Neurol Scand Suppl, 82, 1–152. [PubMed] [Google Scholar]
- Smyth GK (2004). Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol, 3, Article3. [DOI] [PubMed] [Google Scholar]
- Song J,et al. (2015). miR-155 is involved in Alzheimer’s disease by regulating T lymphocyte function. Front Aging Neurosci, 7, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soscia SJ,et al. (2010). The Alzheimer’s disease-associated amyloid beta-protein is an antimicrobial peptide. PLoS One, 5, e9505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling RA,et al. (2011). Toward defining the preclinical stages of Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement, 7, 280–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su JH,et al. (1994). Immunohistochemical evidence for apoptosis in Alzheimer’s disease. Neuroreport, 5, 2529–33. [DOI] [PubMed] [Google Scholar]
- Subramanian A,et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A, 102, 15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson CM,et al. (2010). SRp40 and SRp55 promote the translation of unspliced human immunodeficiency virus type 1 RNA. J Virol, 84, 6748–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka-Taya K,et al. (2004). Human herpesvirus 6 (HHV-6) is transmitted from parent to child in an integrated form and characterization of cases with chromosomally integrated HHV-6 DNA. J Med Virol, 73, 465–73. [DOI] [PubMed] [Google Scholar]
- Thai TH,et al. (2007). Regulation of the germinal center response by microRNA-155. Science, 316, 604–8. [DOI] [PubMed] [Google Scholar]
- Toribara NW,et al. (1993). Human gastric mucin. Identification of a unique species by expression cloning. J Biol Chem, 268, 5879–85. [PubMed] [Google Scholar]
- Upton JW,et al. (2014). Staying alive: cell death in antiviral immunity. Mol Cell, 54, 273–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Auwera GA,et al. (2013). From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics, 43, 11 10 1–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal R,et al. (1999). A stop-codon mutation in the BRI gene associated with familial British dementia. Nature, 399, 776–81. [DOI] [PubMed] [Google Scholar]
- Vidal R,et al. (2000). A decamer duplication in the 3’ region of the BRI gene originates an amyloid peptide that is associated with dementia in a Danish kindred. Proc Natl Acad Sci U S A, 97, 4920–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westman G,et al. (2017). Decreased HHV-6 IgG in Alzheimer’s Disease. Frontiers in Neurology, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J,et al. (2012). Amyloid beta selectively modulates neuronal TrkB alternative transcript expression with implications for Alzheimer’s disease. Neuroscience, 210, 363–74. [DOI] [PubMed] [Google Scholar]
- Woodbury ME,et al. (2015). miR-155 Is Essential for Inflammation-Induced Hippocampal Neurogenic Dysfunction. J Neurosci, 35, 9764–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B,et al. (2016). Sequence Kernel Association Test of Multiple Continuous Phenotypes. Genet Epidemiol, 40, 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S,et al. (2015). Integrative analysis of DNA methylation and gene expression data identifies EPAS1 as a key regulator of COPD. PLoS Genet, 11, e1004898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaharia M,et al. (2011). Faster and more accurate sequence alignment with SNAP. arXiv preprint arXiv:1111.5572. [Google Scholar]
- Zhang B,et al. (2013). Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell, 153, 707–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y,et al. (2001). Productive infection of primary macrophages with human herpesvirus 7. J Virol, 75, 10511–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y,et al. (2009). A functional MicroRNA-155 ortholog encoded by the oncogenic Marek’s disease virus. J Virol, 83, 489–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Viral abundance estimates, and scripts to reproduce results can be accessed at: https://www.synapse.org/#!Synapse:syn12177270.