Abstract
Integrin-transmitted cellular forces are critical for platelet adhesion, activation, aggregation and contraction during hemostasis and thrombosis. Measuring and mapping single platelet forces are desired in both research and clinical applications. Conventional force-to-strain based cell traction force microscopies have low resolution which is not ideal for cellular force mapping in small platelets. To enable platelet force mapping with submicron resolution, we developed a force-activatable biosensor named integrative tension sensor (ITS) which directly converts molecular tensions to fluorescent signals, therefore enabling cellular force mapping directly by fluorescence imaging. With ITS, we mapped cellular forces in single platelets at 0.4 μm resolution. We found that platelet force distribution has strong polarization which is sensitive to treatment with the anti-platelet drug tirofiban, suggesting that the ITS force map can report anti-platelet drug efficacy. The ITS also calibrated integrin molecular tensions in platelets and revealed two distinct tension levels: 12~54 piconewton (nominal values) tensions generated during platelet adhesion and tensions above 54 piconewton generated during platelet contraction. Overall, the ITS is a powerful biosensor for the study of platelet mechanobiology, and holds great potential in antithrombotic drug development and assessing platelet activity in health and disease.
Keywords: Force-activatable biosensor, Integrin tension sensor, cellular force mapping, platelet adhesion, platelet contraction
1. Introduction
Platelets are small anucleate blood cells which aggregate and form the foundation of blood clots during hemostasis and thrombosis (Zucker and Nachmias 1985). Membrane receptors such as von Willebrand factor receptor (GPIb-V-IX), integrin α2β1 (glycoprotein Ia-IIa), integrin α5β1 (glycoprotein Ic-IIa) and integrin αIIbβ3 (glycoprotein IIb-IIIa) (Bennett et al. 2009; Moser et al. 2008; Rivera et al. 2009) play critical adhesive and regulative roles in platelet physiological functions (Gibbins 2004). Among the many adhesion receptors in platelets, integrin αIIbβ3 is the prominent one mediating platelet adhesion under low shear conditions, and is also the main integrin responsible for clot retraction (Moroi and Jung 1998). When platelets are activated by contact with ligands on the exposed subendothelium in injured blood vessels or by soluble factors including ADP (Adenosine diphosphate) and thrombin, platelet integrin αIIbβ3 in turn becomes activated allowing it to bind to a variety of ligands including von Willebrand factor, fibrinogen, fibronectin and vitronectin. Activated integrin αIIbβ3 ultimately mediates platelet adhesion, spreading, aggregation and blood clot retraction (Li et al. 2010). Being indispensable in hemostasis and thrombosis (Bennett 2005), Integrin αIIbβ3 is known to transmit cellular forces which are essential for the retraction and stabilization of the blood clot and thrombus (Liang et al. 2010; Rooney et al. 1998). Therefore, measuring, visualizing and mapping integrin αIIbβ3-transmitted platelet forces would provide valuable biomechanical information for the understanding of platelet function mechanisms and for the assessment of platelet activities. However, these forces have never been measured at the molecular level or mapped at submicron resolution.
Previously, multiple techniques including cell traction force microscopy (Henriques et al. 2012), atomic force microscopy (Lam et al. 2011; Lee and Marchant 2001), elastomers (Myers et al. 2017; Qiu et al. 2014) and micro-post assays (Feghhi et al. 2016; Liang et al. 2010) have been applied to the bulk force measurement in individual platelets. These pioneering methods have enabled the measurement of bulk platelet forces and advanced the study of platelet mechanobiology to a single cell level. These methods commonly adopted the force-to-strain approach for cellular force visualization and measurement: using elastic substrates with a pre-calibrated modulus to convert platelet forces to a visible and measurable substrate strain which is in turn used to compute cellular forces. While powerful at measuring the whole cellular forces, the force-to-strain approach typically yields a low resolution of about 2~5 micron in cellular force imaging (Polacheck and Chen 2016). This resolution is not ideal for force mapping in single platelets because platelets are small, averaging about 2~4 μm in diameter (Paulus 1975). As a result, platelet forces have not been mapped with an ideal resolution and very little is known about platelet force distribution at the subcellular level. Besides force mapping, molecular tensions transmitted by single integrins have never been calibrated in platelets. However, integrin tensions are the fundamental mechanical signals driving integrin signaling pathways which are essential for platelet functions (Chicurel et al. 1998). Thus to further the understanding of how platelets generate force and how the force in turn regulates platelet functions, it is imperative to study platelet forces at the molecular level.
To calibrate and map molecular tensions transmitted by integrin αIIbβ3 in platelets, we developed a force-activatable biosensor named integrative tension sensor (ITS). Instead of converting force to strain, the ITS directly converts molecular tensions to fluorescent signals (force-to-fluorescence conversion). As a molecular linker, the ITS is initially non-fluorescent and can be permanently activated to fluoresce by a tension. The tension threshold required for ITS activation is tunable in the range of 10–60 piconewton (pN), therefore enabling the selective visualization of molecular tensions at different force levels. On a surface where integrin ligands are tethered by the ITS, integrin tensions in platelets activate the ITS and can be directly mapped by fluorescence imaging without the need of post modeling and computation which is usually required by force-to-strain approaches. The force mapping resolution is determined by fluorescence microscopy and easily reaches the submicron level. Using the ITS, for the first time, we mapped integrin-transmitted platelet forces with submicron resolution (0.4 μm) and revealed the polarized cellular force distribution in single platelets. The integrin tension map was then tested to assess the change in platelet activation state in response to an anti-platelet drug tirofiban. Using the ITS, we also calibrated integrin tensions in platelets and revealed two distinct tension levels. We found that platelets produce low-level integrin tensions (12~54 pN. nominal values) during adhesion, while the tensions rise to a high level (>54 pN. nominal values) during contraction. This work paves the road for the study of mechanobiology of platelets at the molecular level by visualizing, calibrating and mapping integrin tensions, and demonstrates the potential of the ITS as a high-resolution force mapping tool for platelet function evaluation and anti-platelet drug assessment.
2. Materials and methods
2.1. ITS synthesis
ITS is a double stranded DNA (dsDNA) with 18 base pairs. The single-stranded DNAs for ITS synthesis were all customized and purchased from IDTDNA. Their sequences and modifications are shown below. ThioMC6-D/ denotes the thiol conjugation to the 5’ end of DNA. /BiosG/ and /Bio/ denote the DNA modifications with biotin conjugations at 5’ end and 3’ end, respectively. BHQ2 is the black hole quencher 2. The DNA sequences were selected and analyzed by DNA analysis tool available in IDTDNA company website to minimize the probability of self-dimer and hairpin formation in the ssDNA.
5’- /ThioMC6-D/GGG CGG CGA CCT CAG CAT/BHQ2/ -3’
5’- /BiosG/T/iCy3/ATG CTG AGG TCG CCG CCC/ -3’
5’- /Cy3/ATG CTG AGG TCG CCG CCC/Bio/ -3’
5’- /Alexa647/ATG CTG AGG TCG CCG CCC/Bio/ -3’
Cyclic peptide RGD with an amine group (PCI-3696-PI, Peptides International) was conjugated with the DNA strand with thiol modification. The Cyclic RGD has a PEG linker which enhance the accessibility of the RGD to integrins on cell membrane. Conjugation was conducted according to a previously published protocol (Wang and Ha 2013). Briefly, the thiol modification on 5’ end of DNA was deprotected by TCEP (Tris(2-carboxyethyl)phosphine hydrochloride) and reacted with the maleimide group of a heterolinker SMCC-sulfo (22622, thermos scientific). The other end of SMCC-sulfo is a NHS ester (N-hydroxysuccinimide esters) which reacts with the amine of RGD. The RGD conjugated DNA was purified by electrophoresis. The 12 pN and 54 pN ITSs used in this paper were assembled by hybridizing the two ssDNAs with a concentration ratio of 1.1:1 (The strand with quencher: the DNA strand with fluorophore).
ITS with 54 pN threshold (with Cy3-BHQ2 pair)
5’- /RGD/GGG CGG CGA CCT CAG CAT/BHQ2/ -3’
5’- /BiosG/T/iCy3/ATG CTG AGG TCG CCG CCC/ -3’
ITS with 12 pN threshold (with Cy3-BHQ2 pair)
5’- /RGD/GGG CGG CGA CCT CAG CAT/BHQ2/ -3’
5’- /Cy3/ATG CTG AGG TCG CCG CCC/Bio/ -3’
ITS with 12 pN threshold (with Alexa647-BHQ2 pair)
5’- /RGD/GGG CGG CGA CCT CAG CAT/BHQ2/ -3’
5’- / Alexa647/ATG CTG AGG TCG CCG CCC/Bio/ -3’
2.2. Preparation of the ITS coated surface
The ITS surface yields a strong fluorescent signal due to the signal integration process, leading to a high tolerance for surface defects or fluorescence background. Moreover, the ITS immobilization starts with physical adsorption of bovine serum albumin-biotin on a surface. These two features enable the ITS to be coated directly on regular cell culture surfaces. In ITS surface preparation, glass-bottom dish (D35–10-1.5-N, In Vitro Scientific) or glass-bottom 96-well plate (P96G-0–5-F, MaTek Corp) was first immersed in 100 μg/ml BSA-biotin (biotinylated bovine serum albumin, A8549, Sigma-Aldrich) mixed with 5 μg/ml fibronectin for 30 min at room temperature, followed by three PBS washes. The fibronectin facilitates platelet adhesion and minimizes the influence of the ITS rupture on platelet activities. The dish was then incubated with 100 μg/ml neutravidin (31000, ThermoFisher Scientific) for 30 min at room temperature. Neutravidin is now immobilized on the surface through avidin-biotin interactions. After being washed with PBS three times, the dish was coated with 0.1 μM ITS-biotin and incubated for 30 mins at 4 °C. At this stage, the ITS is immobilized on the neutravidin surface (each neutravidin molecule has four biotin binding sites) with the binding scheme: glass:BSA-biotin:neutravidin:biotin-ITS. After being rinsed with PBS, the ITS surface is ready for platelet seeding and force mapping.
2.3. Preparation of Platelet-rich plasma (PRP)
The ITS methodology was developed using canine platelets because we had ready access to canine blood. Blood was drawn from healthy research dogs with permission from Iowa State University’s Institutional Animal Care and Use Committee. During a typical ITS assay, 1.8 mL blood was drawn from a dog’s cephalic vein into a 3-mL syringe containing 0.2 ml Acid Citrate Dextrose (ACD) buffer (ACD buffer: 85.3 mM sodium citrate, 41.6 mM citric acid, 136 mM glucose), and then transferred in to a 15-ml falcon tube containing 2 mL buffered saline glucose citrate (BSGC: 129 mM NaCl, 14 mM Na3citrate, 11 mM glucose, 10 mM NaH2PO4, pH 7.3). The mixture was centrifuged at 180 g for 8 min at room temperature. The supernatant was harvested as platelet-rich plasma (PRP) and kept at room temperature until future use.
2.4. Platelet isolation for the ITS
Platelet activity in PRP decreases by time. The freshly prepared PRP should be used for ITS assay within 5 hours. Because PRP contains high levels of fibrinogen and anti-coagulant reagents which may interfere with the ITS assay, platelets need be purified from PRP before experiments. During purification, 0.5 mL PRP was transferred to a new eppendorf tube and mixed with 0.5 mL Buffer 1 (recipes: 113 mM NaCl, 4.3 mM K2HPO4, 4.2 mM Na2HPO4, 24.3 mM NaH2PO4, 5.5 mM Dextrose, pH 6.3) spiked with 0.02 U/ml apyrase and 1 μM prostaglandin E1. After 8 min centrifugation at 800 g, the supernatant was removed and the platelet pellet was re-suspended in serum-free F-12 medium (HFL05, Caisson Laboratories). The platelets are quiescent at this stage and ready for the ITS assay.
2.5. ITS assay with platelets
Platelets purified from PRP were re-suspended in F-12 medium (HFL05, Caisson Laboratories) at a concentration of 100,000/μL. ADP (Adenosine diphosphate, 10 μM final concentration) was added to platelet suspension to activate the platelets. Activated platelets were plated on the ITS surfaces and incubated at 37°C for 30 min. The petri dishes were imaged by a Nikon Ti-E inverted microscope with controlled 37°C temperature and 5% CO2. ITS fluorescence images reporting platelet force maps were acquired using a 60× or 100× lens. Exposure time during imaging was 2 seconds. The force map movies were taken in 30 sec intervals for a period of 15 min. In some experiments, tirofiban (1 or 5 μg/mL), myosin inhibitor blebbistatin (25 μM), or Rho Kinase inhibitor Y27632 (25 μM) were added to the platelet suspensions.
2.6. ITS assay with human blood samples acquired by finger prick method
Human blood was sourced from a healthy volunteer that did not receive any pharmacological treatment in the last 10 days before sampling. The donor gave informed written consent in accordance with the Declaration of Helsinki and the Institutional Review Board (Iowa State University). The tip of the index finger was lanced with a blood lancet (Medipoint). The fingertip was gently squeezed to form a blood droplet on the finger. 10 μL blood was drawn from the blood droplet using a pipette. The blood was added to 100 μL ACD-BGSC solution and spun at 250 g for 3 min. The supernatant with platelets was carefully transferred to an Eppendorf tube containing 100 μL Buffer 1. After centrifuging at 800 g for 3 min, the platelet pellet was re-suspended in 80 μL serum-free F-12 medium with 10 μM ADP. 40 μL platelet suspension was seeded on one well of 12 pN ITS coated 96-well glass bottom plate (P96G-0–5-F, MaTek Corp), the other 40 μL platelet solution was treated with 100 μg/ml 7E3 antibody against human integrin αIIbβ3 and plated on another well coated with 12 pN ITS. The 96-well plate was incubated at 37°C for 30 min, and then imaged with a Nikon fluorescence microscope.
2.7. Vinculin immunostaining
Canine platelets were incubated on an ITS surface for 30 min and then fixed with 4% paraformaldehyde plus 0.5% Triton X in PBS for 30 min. After washing with PBS three times, the platelets were immersed in 2% BSA (bovine serum albumin) for 1 hour at room temperature. 2.5 μg/ml vinculin antibody (FAK100, Millipore, mouse anti-Vinculin) was added to platelet surface and incubated for 1 hour. After washing, 2.5 μg/ml secondary antibody (AP192SA6, Millipore) was added to the platelet surface and incubated for 1 hour at room temperature. After washing, the ITS force map and vinculin were co-imaged with the fluorescence microscope.
3. Results and discussion
3.1. Integrative tension sensor (ITS): converting force signal to fluorescence signal
The ITS is a synthetic force-activatable molecule: The initially non-fluorescent ITS molecule becomes fluorescent after sustaining a tension above a certain level (selectable from 10~60 pN). A surface coated with the ITS can be locally activated to fluoresce by the cellular forces of platelets, and therefore report the force distribution through fluorescence imaging. This force-to-fluorescence conversion enables cellular force imaging with the advantages of fluorescence microscopy: high sensitivity, high resolution (submicron), fast imaging speed and high data throughput.
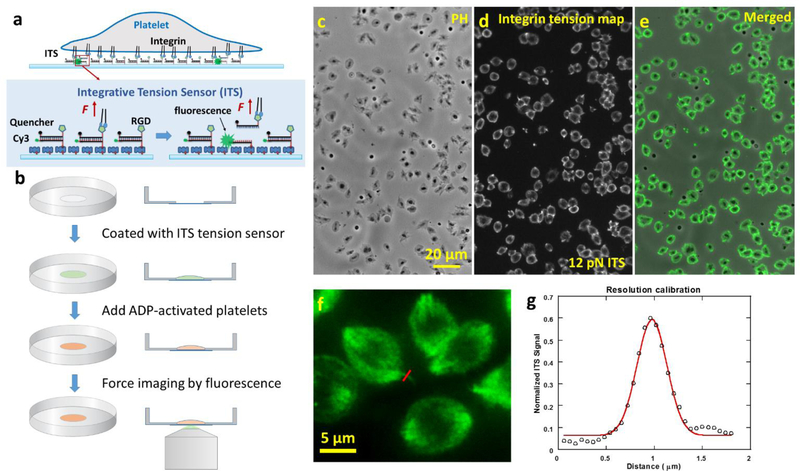
As shown in Fig. 1a, the ITS is an 18 base-paired (bp) dsDNA decorated with a biotin tag, a Cy3 dye, a fluorophore quencher (black hole quencher 2) and an arginine-glycine-aspartic acid (RGD) peptide. dsDNA can be mechanically dissociated and the force required for rupturing dsDNA is well-defined, making dsDNA an ideal biomaterial for the construction of the molecular tension sensor(Albrecht et al. 2003). The biotin tag in the ITS enables the immobilization of the ITS on a surface through biotin-neutravidin interaction. The Cy3 in the ITS is initially quenched by the fluorophore quencher with 95% quenching efficiency (Marras et al. 2002), causing initial near-zero fluorescence level. The RGD peptide in the ITS serves as an integrin ligand. RGD is the critical peptide motif in fibrinogen, vitronectin, von Will brand factor, and fibronectin for integrin binding (Bennett 2001; Pankov and Yamada 2002). Many types of integrins including integrin αIIbβ3 bind to RGD (Sanchez-Cortes and Mrksich 2009). On an ITS-coated surface, integrins on the platelet membrane bind to RGD and transmit tensions to the ITS during platelet adhesion and contraction. These tensions may mechanically separate the dsDNA and free Cy3 from quenching, thus causing the ITS to fluoresce (ITS activation by a tension). The tension level required for dsDNA separation is tunable and dependent on the dsDNA geometry (Wang and Ha 2013). In this paper, we developed the ITS with 12 pN and 54 pN thresholds, based on dsDNAs in unzipping and shear geometries, respectively (Cocco et al. 2001; Hatch et al. 2008). Note that the tension thresholds for dsDNA mechanical separation is also dependent on force loading time (Strunz et al. 1999) which is unknown in platelets. Therefore the 12 pN and 54 pN thresholds are subject to change and should be considered as nominal values for integrin tension calibration in platelets. However, the monotonicity between the low and high thresholds remains unchanged regardless of force loading time. Because the ITS remains fluorescent once activated, all integrin tensions higher than the threshold will be recorded on the ITS surface. This signal integration process greatly improves the sensitivity of integrin tension mapping, and preserves the force footprint even after the cells are fixed or removed from the surface. Because of the signal integration process, this force sensor was named the integrative tension sensor.
Fig. 1. The integrative tension sensor (ITS) maps integrin-transmitted platelet forces with 0.4 micron resolution and high data throughput.
a, molecular structure of ITS (12 pN tension threshold). The ITS converts molecular tension above a selected threshold (here 12 pN) to a fluorescent signal. The ITS is an 18 base pair DNA decorated with a fluorophore, a quencher, a biotin tag and an integrin ligand RGD peptide. The initial non-fluorescent DNA can be pulled apart by a tension transmitted by integrin-RGD bonds and become fluorescent if the tension is higher than the DNA unzipping force (12 pN), thereby reporting the integrin tension level and location. b, The ITS assay procedure. The ITS was coated on a glass-bottom petri dish through biotin-neutravidin interaction. Platelets activated by ADP were plated in a 100 μL well in the petri dish. After 30 min, the force map reported by the ITS was imaged by a fluorescence microscope. c-e, platelet morphology and integrin tension map (above 12 pN) of hundreds of canine platelets were simultaneously imaged in phase contrast (PH, c) and fluorescence channels (d, with 60× lens). e is the merged image of images c and d. f, platelet force map with a fine feature for resolution calibration. g, resolution of force imaging is calibrated to be 0.4 μm based on one section of the force map marked by the red line in image f. The linear profile of fluorescence image marked by the red line is plotted and fitted with Gaussian function. The full width of half maximum of the Gaussian function is determined to be 0.4 μm which is treated as the spatial resolution of platelet force map.
Compared to the contemporary integrin molecular tension sensors which are based on polyethylene glycol polymer (Stabley et al. 2012), elastic peptide (Morimatsu et al. 2013) and hairpin DNA (Blakely et al. 2014; Zhang and Salaita 2014), and detect molecular tensions in the range of 0~20 pN in real time, ITS has a broader dynamic range (nominal values: 10–60 pN) which is advantageous to study integrin tensions in platelets which can exceed 54 pN as shown in the following study. Moreover, the tension signal integration due to the permanent activation of ITS gives rise to strong fluorescence intensity and high tolerance for surface defects or fluorescence background, and therefore enables platelet force mapping with a common fluorescence microscope and minimal surface preparation. These features make the ITS an appealing method for the potential application as a technically simple platelet lab test.
3.2. ITS maps integrin tensions in platelets with submicron resolution and high data throughput
To map integrin tensions in platelets, we coated glass-bottom petri dishes with 12 pN ITS which can be activated by a tension above 12 pN. Fibronectin was coated along with the ITS on the surface to ensure stable platelet adhesion and minimize the influence of the rupture of the ITS on platelet adhesion and contraction. Here we mostly used canine platelets to test the performance of the ITS because dog blood is more accessible than human blood while dog platelets bear similar biological traits to human platelets (LeVine et al. 2014). Dogs have sizes close to humans, resulting in similar rheology and metabolic rates in blood (Son et al. 2010). Dogs and humans have similar platelet counts, signaling pathways, and platelet sizes (Boudreaux et al. 2007). For these reasons, canine platelets are good cell models for testing the ITS performance in the platelet force study. In our experiments, canine platelets were isolated from whole dog blood, activated by ADP and plated on the ITS surface. The sample was incubated at 37°C for 30 min. Fluorescence imaging was then performed to map how ITS was activated by integrin tensions. Fig. 1b shows the standard ITS assay procedure. With direct force-to-fluorescence conversion, the ITS maps cellular forces without the need of computation or modeling, and therefore promises rapid data acquisition with high data throughput (Fig. 1c–e). Using a 60× objective lens, we collect thousands of platelet force maps in a few seconds (the exposure time of fluorescence imaging). Because ITS coverts force to fluorescence, in principle, the resolution of platelet force map is determined by the optical resolution of fluorescence microscopy which can easily reach 0.2–0.3 μm. Nonetheless, we calibrated the force map resolution on our platform by examining the force map feature of one platelet. The linear profile of a fine feature in Fig. 1f was fitted by Gaussian function. The full width half maximum (FWHM) of the Gaussian function was determined to be 0.4 micron that can be treated as the resolution of platelet force map (Fig. 1f–g). To confirm that ITS was activated by integrins, we tested and found that 0.1 mM soluble RGD blocked platelets from adhering on fibrinogen or fibronectin surfaces (sFig. 1). In another control experiment, platelets were plated on a surface coated with the ITS without the RGD conjugation. The adherent platelets caused zero fluorescence signal on the ITS surface (sFig. 2), suggesting that RGD is required to activate the ITS and the tensions activating the ITS are indeed transmitted by integrins. Collectively, we mapped integrin-transmitted cellular forces in platelets for the first time with 0.4 micron resolution, and show that the integrin tensions are above 12 pN in platelets during adhesion.
3.3. Mapping integrin αIIbβ3-transmitted platelet forces with 10 μL human blood
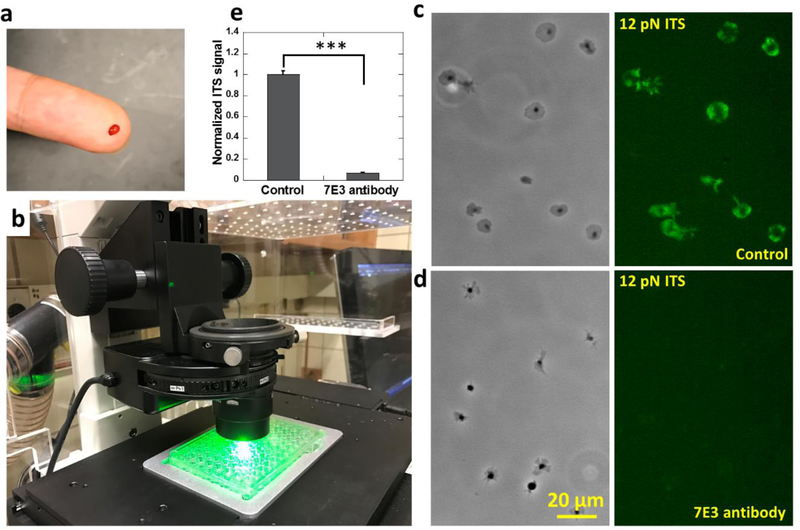
The ITS has the potential to be applied as a biomechanical lab test for the examination of human platelets. Here we demonstrated the feasibility of the ITS assay in human platelet force mapping, and showed that the ITS is compatible with commercially available cell culture apparatuses and requires minimal blood sample volume. The ITS can be conveniently coated on regular surfaces intended for cell culture and therefore is highly compatible with commercially available petri dishes or 96-well plates which enables a large number of assays under different biochemical conditions. Another advantage of the ITS assay is the high sample utilization efficiency. During ITS force mapping, nearly 100% of platelets in the medium adhere on the surface and produce ITS force maps. Therefore, the ITS assay only requires a minuscule amount of blood which can be sufficiently provided by a simple finger prick method. In principle, 1 nanoliter (10−9 L) blood containing about 150~400 platelets(Daly 2011) should suffice for an examination of platelet forces. In practice, we tested a 10 μL (10−5 L) blood sample acquired by a finger prick on ITS coated 96-well glass bottom plate (P96G-0–5-F, MaTek Corp). The blood on a fingertip following pricking with a blood lancet (Medipoint) was collected with a 10 μL pipette tip and added to anti-coagulant medium (Fig. 2a). The platelets were then purified by centrifuge and re-suspended in 80 μL F-12 cell culture medium and activated by 10 μM ADP. Platelet suspension (40 μL) was added to one well in a 96-well plate that was coated with 12 pN ITS to map integrin tensions higher than 12 pN (Fig. 2b). Platelet force maps were acquired successfully after 30 minutes of incubation. Human platelets exhibit integrin-transmitted force patterns similar to canine platelets (Fig. 2c). To confirm that the platelet forces mapped on the ITS surface are transmitted by integrin αIIbβ3, we tested platelets treated with 100 μg/ml monoclonal 7E3 antibody against human integrin αIIbβ3 on a 12 pN ITS surface. With integrin αIIbβ3 blocked by the antibody, platelets exhibited poorer adhesion and produced near zero ITS signals (Fig. 2d–e), confirming that the platelet forces reported by ITS are mainly transmitted by integrin αIIbβ3.
Fig. 2. Platelet force mapping with human blood obtained by finger prick method.
a, 10 μL blood collected by finger prick method. b, Platelet force mapping conducted in a 12 pN ITS coated 96-well plate. Integrin tensions higher than 12 pN were fluorescently recorded on the surface. c-d, Platelet morphology and force map of platelets in a control group (c) and platelets treated with 100 μg/ml 7E3 antibody which blocks integrin αIIbβ3 (d). e, ITS signal intensity of platelets in the control group and the 7E3 antibody-treated group. Error bar represents standard error. (48 platelets for each data points) The two-tailed P value is less than 0.001 between ITS signals of platelets in control group and in 7E3 antibody treated group.
3.4. Heterogeneity and polarization of platelet forces
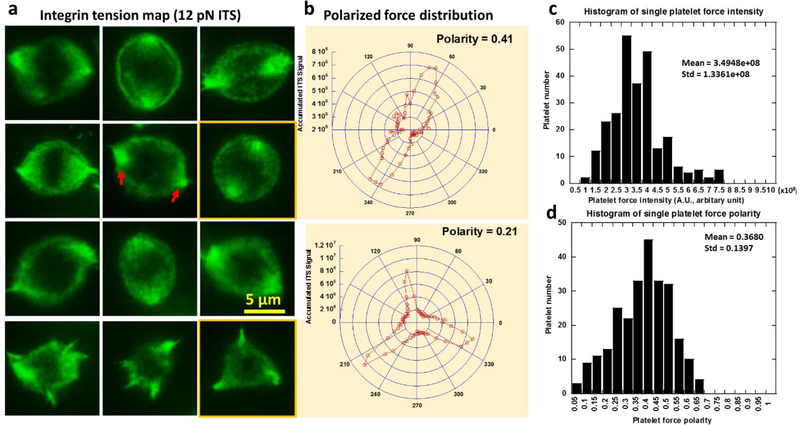
Canine platelets were used in all the following tests. We analyzed the cellular force maps of 548 platelets on the 12 pN ITS surface as shown in sFig. 3. Because fluorescence intensity is proportional to the number of ITS molecules activated by integrin tensions on a unit area, the ITS fluorescence intensity reflects the activity of integrin-transmitted cellular forces and can be used to evaluate local cellular force intensity. By analyzing the force map of hundreds of platelets, we found that the platelet force is mainly distributed on the peripheral region in platelet-substrate contact area with little force at the center, appearing in a ring pattern (Fig. 3a). However, platelet force is not isotropically distributed as reported by a previous platelet force study using cell traction force microscopy (Henriques et al. 2012). With high resolution, the ITS revealed that the distribution of platelet forces is polarized. There are usually two or three dot regions with strong ITS signal under each adherent platelet (Fig. 3a), showing the inhomogeneous and polarized force distribution in platelets. The nature of polarized force distribution may be related to the function of platelet contraction during which platelets link to the fibrin network in a blood clot and contract to strengthen the clot. Therefore, the platelet force polarization may be potentially used as a biomarker to evaluate platelet contraction ability. We quantified the polarity of platelet force distribution by analyzing the ITS image of single platelets. The polar distribution of fluorescence intensity was calculated and plotted in polar coordinates as shown in Fig. 3b. The polarity of force distribution was further calculated based on the intensity curve in the polar coordinates. The data analysis algorithm is detailed in sFig. 4. The polarity of platelet force distribution is typically 0.4 (0 means isotropic distribution and 1.0 means force distribution in one straight line). We analyzed the histogram of force intensity and polarity over 256 platelets and found wide distributions of both factors (Fig. 3c–d), indicating a strong heterogeneity among platelet forces. The coefficient of variation (CV, ratio between standard deviation and mean) is 0.382 and 0.379 for intensity and polarity, respectively. Because of the heterogeneity, platelet analysis must be statistically performed over many platelets in order to acquire an accurate evaluation of platelet functions for each host. This requirement is met by the high throughput offered by the ITS method.
Fig. 3. Polarization and intensity of platelet forces contributed by integrin tensions higher than 12 pN.
a, force distribution as measured with the ITS in platelets is inhomogeneous and polarized. Cellular forces transmitted by integrins in platelets are distributed in a ring pattern, with two or three force foci in which the force signal is more concentrated (shown by red arrows). b, Platelet force distribution curve translated from (a) into polar coordinates. Polarity of the force distribution is quantified based on the curves. c-d, histograms of single platelet force intensity shows a wide distribution of platelet force intensity and polarity, suggesting strong heterogeneity in individual platelet forces. Force intensity is calculated by summing grayscale values of all pixels under single platelets in the ITS images.
3.5. Anti-platelet drug tirofiban reduces platelet force polarity and intensity
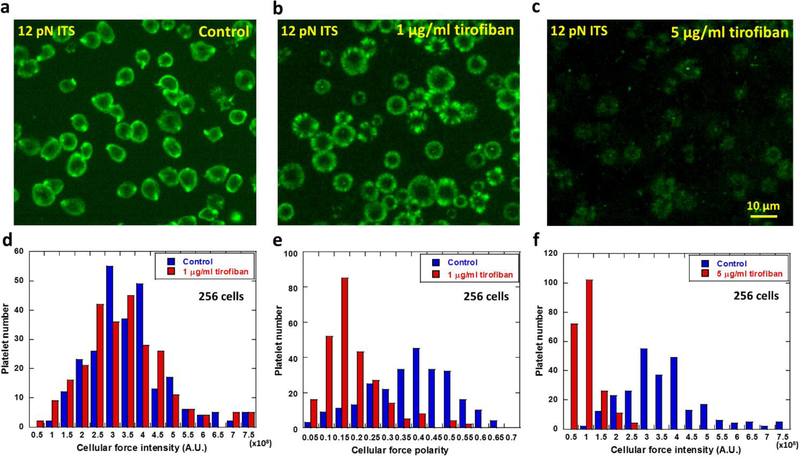
Next we tested that how the anti-platelet drug tirofiban changes platelet forces. Tirofiban is an antiplatelet agent that inhibits integrin αIIbβ3 (Moser et al. 2003; Topol et al. 2001) and is commonly used to reduce the rate of thrombotic cardiovascular events in patients. To study if and how tirofiban changes integrin αIIbβ3-transmitted platelet force distribution and intensity, we plated ADP-activated platelets on 12 pN ITS surface in media spiked with 0, 1 and 5 μg/ml tirofiban respectively (Fig 4a–c). After 30 min incubation at 37°C, platelets in the control group yielded the polarized force map (Fig. 4a), consistent with previous data. Platelets treated with 1 μg/ml tirofiban displayed force maps with similar fluorescence intensity as control samples (Fig. 4b, 4d and sFig. 5), suggesting that total platelet adhesion force was not significantly influenced by tirofiban at this concentration. However, the force distribution pattern was significantly changed as the force map shows nearly isotropic force distribution in a ring pattern. The polarity of the platelet force map was significantly reduced from a mean value of 0.4 in control platelets to 0.15 in tirofiban-treated platelets (Fig. 4e). It remains unknown to us how tirofiban reduces the platelet force polarity. However, this experiment clearly demonstrates that platelet force distribution can be altered by an antiplatelet drug while, at this drug concentration, the change of the overall force level remains insignificant. Therefore, mapping platelet forces at high resolution can yield new information and report an antiplatelet drug’s efficacy on platelets which may be undetectable by bulk force measurement of platelets.
Fig. 4. Polarization and intensity of platelet forces (integrin tension higher than 12 pN) altered by the anti-platelet drug tirofiban.
a, Platelet force map in the control group. b, force map of platelets treated with 1 μg/ml tirofiban. c, force map of platelets treated with 5 μg/ml tirofiban. d, Force intensity histograms of platelets treated with 1 μg/ml tirofiban and the control group. p value = 0.125 between the two groups of platelet force intensities. e, Force polarity histograms of platelets treated with 1 μg/ml tirofiban and the control group. p value < 0.001 between the two groups of platelet force polarities. f, Force intensity histograms of platelets treated with 5 μg/ml tirofiban and the control group. p value <0.001 between the two groups of platelet force intensities.
Treated with 5 μg/ml tirofiban, platelets still adhere. At this concentration, the ITS map shows that integrin αIIbβ3 transmitted force intensity was significantly reduced (Fig. 4c and 4f).
3.6. Two levels of integrin molecular tensions in platelets
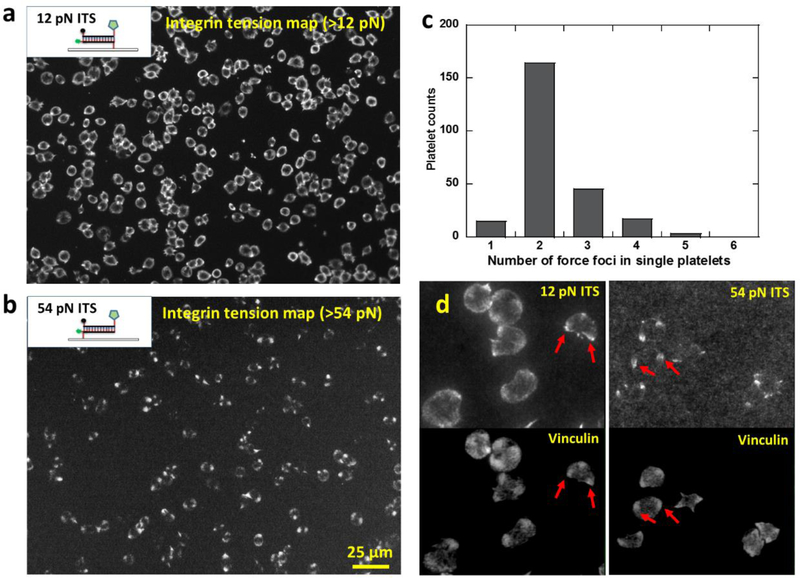
By tuning the tension threshold for ITS activation, we can selectively image integrin tensions at different force levels in platelets. We have acquired platelet force maps of integrin tensions above 12 pN using 12 pN ITS, and shown that integrin tensions in platelets are able to mechanically separate 18 bp DNA in an unzipping geometry. To study the existence and distribution of higher level integrin tensions, we prepared the ITS with a 54 pN threshold (the critical force to rupture 18bp dsDNA in a shear geometry) to report the distribution of integrin tensions higher than 54 pN. We chose 54 pN as the threshold because it was previously reported that integrin tensions in focal adhesions (integrin clusters) in regular cells can go beyond 54 pN (Wang et al. 2015). The 12 pN and 54 pN ITS structures based on dsDNA in unzipping and shear geometries respectively are shown in Fig. 5a and 5b. Platelets activated by ADP were added on both 12 pN and 54 pN ITS surfaces and incubated for 30 min. 12 pN ITS shows the typical ring-shaped force distribution with two or three force foci, consistent with previous tests. In contrast, 54 pN ITS force map does not show this ring pattern but instead showed a dot pattern, indicating that the high-level integrin tensions are more concentrated than the low-level tensions (12~54 pN) (Fig. 5a–b). We verified the co-localization of force foci on 12 pN and 54 pN ITS force maps by plating platelets on a multiplex ITS surface where 12 pN ITS and 54 pN ITS were coated with 1:1 molar ratio, and labeled with Cy3 and Alexa647 respectively (sFig. 6). Collectively, these experiments show that integrin tensions at a low level (12~54 pN) are distributed in a ring pattern while the high-level tensions (>54 pN) are located in 2–3 dot regions in platelets (Fig. 5c).
Fig. 5. Platelet force maps of low-level (>12 pN) and high-level (>54 pN) integrin tensions.
a, platelet force map of low-level integrin tensions (above 12 pN) reported by the ITS with a 12 pN tension threshold. The dsDNA of the 12 pN ITS is in an unzipping geometry. b, platelet force map of high-level integrin tensions (above 54 pN) reported by the ITS with a 54 pN tension threshold. The dsDNA of the 54 pN ITS is in a shear geometry. c, histogram of force foci number in the high-level tension map (>54 pN). d, Force foci on both 12 pN and 54 pN ITS surfaces generally co-localize with vinculin clusters which mark the sites of focal adhesions. Examples of co-localizations are shown by red arrows.
The ITS surface preserves the platelet force map even if the platelets are fixed, immunostained or removed. This feature allows us to co-image cellular force and cell structure by immunostaining. By staining vinculin with an anti-vinculin antibody, we found that the force foci in the ITS force map is co-localized with vinculin which are the common markers of focal adhesions (FAs) where integrins cluster (Fig. 5d), suggesting that FAs are the force transduction sites of high level integrin tensions in platelets. This is consistent with regular adherent cells in which FAs transmit high level integrin tensions (Wang et al. 2015), suggesting that platelets may use the same mechanism to generate high-level integrin tensions.
3.7. Temporal dynamics of integrin tensions at different force levels
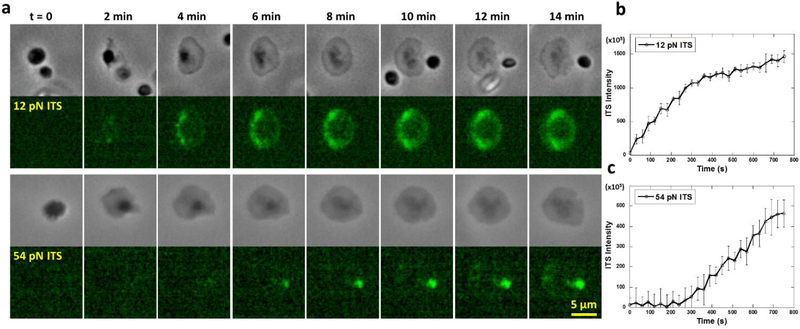
Next, we studied the temporal dynamics of the two levels of integrin tensions. We performed time-series imaging of platelet forces on 12 pN and 54 pN ITS surfaces (Supplementary movies 1 and 2). Phase contrast (PH) imaging was performed simultaneously with ITS fluorescence imaging to determine the platelet adhesion status (Fig. 6a). We found that 12 pN ITS signal started to appear once platelets adhered. In the first 5 min, 12 pN ITS signal continued to increase with a steady rate which reflects integrin tension activity. After 5 min, the ITS signal rate of increase was reduced to 30% of initial rate, indicating that the low-level integrin tensions are more active in the first five minutes after platelet adhesion (Fig. 6b). In contrast, the 54 pN ITS signal reporting high-level integrin tensions was undetectable in the initial 4 min after cell adhesion and spreading (Fig. 6c). The 54 pN ITS signal started to appear after 4 min of platelet adhesion and increased at a stable rate, suggesting that high-level integrin tensions are not produced during platelet initial adhesion and spreading. Considering that platelet contraction takes place after platelet adhesion, we speculate that 4 min might be the time for platelets to transit from adhesion state to contraction state and platelets may produce higher integrin tensions during contraction.
Fig. 6. Temporal dynamics of low-level (>12 pN) and high-level integrin tensions (>54 pN) in platelets.
a, Time series of a single platelet force map on 12 pN ITS and 54 pN ITS surfaces, respectively. b, 12 pN ITS signal intensity per platelet increases by time. The platelet adhered at t=0. c, 54 pN ITS signal intensity per platelet increases by time, but has a 4 min delay after platelet adhesion. The platelet adhered at t=0. The time trace curves are each averaged over 16 platelets (the error bar represents standard error).
3.8. 54 pN integrin tensions are abolished by platelet contraction inhibition
To further determine if platelet contraction produces the higher-level integrin tensions (>54 pN), we inhibited platelet contraction with blebbistatin or ROCK inhibitor Y-27632, which inhibit myosin II and Rho kinases, respectively. Myosin II is the main force source for platelet contraction and Rho kinases are required for the regulation of platelet contraction (Calaminus et al. 2007; Myers et al. 2017; Ono et al. 2008). Under the treatment of blebbistatin and Y-27632, platelets adhered and spread with normal morphology, and low-level integrin tensions in platelets mapped by 12 pN ITS displayed the regular ring-shaped force distribution with the similar force intensity as the control group. However, the intensity of high-level integrin tensions reported by 54 pN ITS was significantly reduced to 26% (blebbistatin group) and 15 % (Y-27632 group) of the control group, suggesting that high-level integrin tension activity was impaired by platelet contraction inhibition (sFig. 7). This experiment further suggests that integrin tensions above 54 pN are generated during platelet contraction stage. Overall, we determined two distinct integrin tension levels in platelets. The 12~54 pN integrin tensions are generated during platelet adhesion and >54 pN integrin tensions are generated during platelet contraction.
4. Conclusion
Hemostasis and thrombosis are mediated by both biochemical and biomechanical cues. Integrin-transmitted forces in platelets regulate a series of platelet functions and mediate the formation and solidification of blood clots. A variety of methods have been developed and applied to force measurement in single platelets, pioneering the mechanobiology study of platelets at single cell level. To advance platelet force mapping and calibration to submicron resolution and molecular sensitivity, we adopted a force-to-fluorescence strategy and developed the biosensor ITS which directly converts molecular tensions to fluorescent signal. Using the ITS, we unprecedentedly mapped cellular forces in platelets with 0.4 micron resolution. The high-resolution force mapping revealed the polarized force distribution in platelets and demonstrated that the platelet force polarity is sensitive to the treatment with the antiplatelet drug tirofiban. The ITS also revealed two levels of integrin tensions in platelets, with low-level tensions generated in a ring pattern during platelet adhesion, and high-level tensions generated in a dot pattern during platelet contraction. Because the ITS faithfully preserves the footprint of integrin tensions, cellular forces now can be co-visualized with cellular structures with comparable resolution and sensitivity even if the cells are fixed and immunostained. By co-visualization of cellular force and focal adhesions, we determined that the high-level tensions are transmitted by integrins in focal adhesions in platelets. These experiments demonstrated the capability and versatility of the ITS in platelet force study and platelet activity assessment.
Besides the high resolution and molecular tension sensitivity, the ITS is also a highly accessible technique that could be performed in most biomedical laboratories. ITS can be directly coated on commercially available cell culture apparatuses such as a petri dish or 96-well plate. This feature is valuable for the ITS to be adopted as a standard lab test for platelet function examination or for anti-platelet drug screening. Because of the signal integration process, the ITS force map has high fluorescence intensity and therefore can be imaged by a routine fluorescence microscope. The ITS also requires minimal blood sample volume because of its high efficiency of sample utilization. Collectively, the ITS maps integrin-transmitted platelet forces with submicron resolution, rapid data acquisition, high data throughput, low sample volume and excellent compatibility with commercial cell culture apparatuses. With these features, the ITS will facilitate both basic research and drug development of platelet inhibitors. The ITS also holds great potential as a diagnostic tool for rapid examination of platelet function or dysfunction in patients, helping to better diagnose and treat bleeding and thrombotic disorders.
Supplementary Material
Acknowledgements
This work was supported by startup fund provided by Iowa State University. We thank Dr. Barry Coller for the generous gift of 7E3 antibody. We thank Dr. Matthew Ellinwood and Jackie Jens for the generous use of their research dogs for blood samples.
References
- Albrecht C, Blank K, Lalic-Multhaler M, Hirler S, Mai T, Gilbert I, Schiffmann S, Bayer T, Clausen-Schaumann H, Gaub HE, 2003. DNA: a programmable force sensor. Science 301(5631), 367–370. [DOI] [PubMed] [Google Scholar]
- Bennett JS, 2001. Platelet-fibrinogen interactions. Ann N Y Acad Sci 936, 340–354. [DOI] [PubMed] [Google Scholar]
- Bennett JS, 2005. Structure and function of the platelet integrin alphaIIbbeta3. Journal of Clinical Investigation 115(12), 3363–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett JS, Berger BW, Billings PC, 2009. The structure and function of platelet integrins. J Thromb Haemost 7 Suppl 1, 200–205. [DOI] [PubMed] [Google Scholar]
- Blakely BL, Dumelin CE, Trappmann B, McGregor LM, Choi CK, Anthony PC, Duesterberg V, Baker BM, Block SM, Liu DR, Chen CS, 2014. A DNA-based molecular probe for optically reporting cellular traction forces. Nature Methods 11(12), 1229–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreaux MK, Catalfamo JL, Klok M, 2007. Calcium-Diacylglycerol Guanine Nucleotide Exchange Factor I gene mutations associated with loss of function in canine platelets. Translational Research 150(2), 81–92. [DOI] [PubMed] [Google Scholar]
- Calaminus SD, Auger JM, McCarty OJ, Wakelam MJ, Machesky LM, Watson SP, 2007. MyosinIIa contractility is required for maintenance of platelet structure during spreading on collagen and contributes to thrombus stability. J Thromb Haemost 5(10), 2136–2145. [DOI] [PubMed] [Google Scholar]
- Chicurel ME, Singer RH, Meyer CJ, Ingber DE, 1998. Integrin binding and mechanical tension induce movement of mRNA and ribosomes to focal adhesions. Nature 392(6677), 730–733. [DOI] [PubMed] [Google Scholar]
- Cocco S, Monasson R, Marko JF, 2001. Force and kinetic barriers to unzipping of the DNA double helix. Proceedings of the National Academy of Sciences of the United States of America 98(15), 8608–8613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly ME, 2011. Determinants of platelet count in humans. Haematologica-the Hematology Journal 96(1), 10–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feghhi S, Munday AD, Tooley WW, Rajsekar S, Fura AM, Kulman JD, Lopez JA, Sniadecki NJ, 2016. Glycoprotein Ib-IX-V Complex Transmits Cytoskeletal Forces That Enhance Platelet Adhesion. Biophysical Journal 111(3), 601–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbins JM, 2004. Platelet adhesion signalling and the regulation of thrombus formation. Journal of Cell Science 117(16), 3415–3425. [DOI] [PubMed] [Google Scholar]
- Hatch K, Danilowicz C, Coljee V, Prentiss M, 2008. Demonstration that the shear force required to separate short double-stranded DNA does not increase significantly with sequence length for sequences longer than 25 base pairs. Physical Review E 78(1). [DOI] [PubMed] [Google Scholar]
- Henriques SS, Sandmann R, Strate A, Koster S, 2012. Force field evolution during human blood platelet activation. Journal of Cell Science 125(16), 3914–3920. [DOI] [PubMed] [Google Scholar]
- Lam WA, Chaudhuri O, Crow A, Webster KD, Li TD, Kita A, Huang J, Fletcher DA, 2011. Mechanics and contraction dynamics of single platelets and implications for clot stiffening. Nature Materials 10(1), 61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Marchant RE, 2001. Force measurements on the molecular interactions between ligand (RGD) and human platelet alpha(IIb) beta(3) receptor system. Surface Science 491(3), 433–443. [Google Scholar]
- LeVine DN, Birkenheuer AJ, Brooks MB, Nordone SK, Bellinger DA, Jones SL, Fischer TH, Oglesbee SE, Frey K, Brinson NS, Peters AP, Marr HS, Motsinger-Reif A, Gudbrandsdottir S, Bussel JB, Key NS, 2014. A novel canine model of immune thrombocytopenia: has immune thrombocytopenia (ITP) gone to the dogs? Br J Haematol 167(1), 110–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Delaney MK, O’Brien KA, Du X, 2010. Signaling during platelet adhesion and activation. Arterioscler Thromb Vasc Biol 30(12), 2341–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XM, Han SJ, Reems JA, Gao DY, Sniadecki NJ, 2010. Platelet retraction force measurements using flexible post force sensors. Lab on a Chip 10(8), 991–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marras SAE, Kramer FR, Tyagi S, 2002. Efficiencies of fluorescence resonance energy transfer and contact-mediated quenching in oligonucleotide probes. Nucleic Acids Research 30(21). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimatsu M, Mekhdjian AH, Adhikari AS, Dunn AR, 2013. Molecular Tension Sensors Report Forces Generated by Single Integrin Molecules in Living Cells. Nano Letters 13(9), 3985–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroi M, Jung SM, 1998. Integrin-mediated platelet adhesion. Front Biosci 3, d719–728. [DOI] [PubMed] [Google Scholar]
- Moser M, Bertram U, Peter K, Bode C, Ruef J, 2003. Abciximab, eptifibatide, and tirofiban exhibit dose-dependent potencies to dissolve platelet aggregates. J Cardiovasc Pharmacol 41(4), 586–592. [DOI] [PubMed] [Google Scholar]
- Moser M, Nieswandt B, Ussar S, Pozgajova M, Fassler R, 2008. Kindlin-3 is essential for integrin activation and platelet aggregation. Nat Med 14(3), 325–330. [DOI] [PubMed] [Google Scholar]
- Myers DR, Qiu Y, Fay ME, Tennenbaum M, Chester D, Cuadrado J, Sakurai Y, Baek J, Tran R, Ciciliano JC, Ahn B, Mannino RG, Bunting ST, Bennett C, Briones M, Fernandez-Nieves A, Smith ML, Brown AC, Sulchek T, Lam WA, 2017. Single-platelet nanomechanics measured by high-throughput cytometry. Nat Mater 16(2), 230–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A, Westein E, Hsiao S, Nesbitt WS, Hamilton JR, Schoenwaelder SM, Jackson SP, 2008. Identification of a fibrin-independent platelet contractile mechanism regulating primary hemostasis and thrombus growth. Blood 112(1), 90–99. [DOI] [PubMed] [Google Scholar]
- Pankov R, Yamada KM, 2002. Fibronectin at a glance. Journal of Cell Science 115(20), 3861–3863. [DOI] [PubMed] [Google Scholar]
- Paulus JM, 1975. Platelet size in man. Blood 46(3), 321–336. [PubMed] [Google Scholar]
- Polacheck WJ, Chen CS, 2016. Measuring cell-generated forces: a guide to the available tools. Nature Methods 13(5), 415–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu YZ, Brown AC, Myers DR, Sakurai Y, Mannino RG, Tran R, Ahn B, Hardy ET, Kee MF, Kumar S, Bao G, Barker TH, Lam WA, 2014. Platelet mechanosensing of substrate stiffness during clot formation mediates adhesion, spreading, and activation. Proceedings of the National Academy of Sciences of the United States of America 111(40), 14430–14435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera J, Lozano ML, Navarro-Nunez L, Vicente V, 2009. Platelet receptors and signaling in the dynamics of thrombus formation. Haematologica-the Hematology Journal 94(5), 700–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney MM, Farrell DH, van Hemel BM, de Groot PG, Lord ST, 1998. The contribution of the three hypothesized integrin-binding sites in fibrinogen to platelet-mediated clot retraction. Blood 92(7), 2374–2381. [PubMed] [Google Scholar]
- Sanchez-Cortes J, Mrksich M, 2009. The platelet integrin alphaIIbbeta3 binds to the RGD and AGD motifs in fibrinogen. Chem Biol 16(9), 990–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son KH, Lim CH, Song EJ, Sun K, Son HS, Lee SH, 2010. Inter-species hemorheologic differences in arterial and venous blood. Clinical Hemorheology and Microcirculation 44(1), 27–33. [DOI] [PubMed] [Google Scholar]
- Stabley DR, Jurchenko C, Marshall SS, Salaita KS, 2012. Visualizing mechanical tension across membrane receptors with a fluorescent sensor. Nature Methods 9(1), 64–U172. [DOI] [PubMed] [Google Scholar]
- Strunz T, Oroszlan K, Schafer R, Guntherodt HJ, 1999. Dynamic force spectroscopy of single DNA molecules. Proceedings of the National Academy of Sciences of the United States of America 96(20), 11277–11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topol EJ, Moliterno DJ, Herrmann HC, Powers ER, Grines CL, Cohen DJ, Cohen EA, Bertrand M, Neumann FJ, Stone GW, DiBattiste PM, Demopoulos L, 2001. Comparison of two platelet glycoprotein IIb/IIIa inhibitors, tirofiban and abciximab, for the prevention of ischemic events with percutaneous coronary revascularization. N Engl J Med 344(25), 1888–1894. [DOI] [PubMed] [Google Scholar]
- Wang XF, Ha T, 2013. Defining Single Molecular Forces Required to Activate Integrin and Notch Signaling. Science 340(6135), 991–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XF, Sun J, Xu Q, Chowdhury F, Roein-Peikar M, Wang YX, Ha T, 2015. Integrin Molecular Tension within Motile Focal Adhesions. Biophysical Journal 109(11), 2259–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Salaita K, 2014. DNA-Based “Digital” Tension Probes with Pn Sensitivity Reveal Early Cell Adhesion Mechanics at the Single Molecule Level. Biophysical Journal 106(2), 786a–786a. [Google Scholar]
- Zucker MB, Nachmias VT, 1985. Platelet Activation. Arteriosclerosis 5(1), 2–18. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.