SUMMARY
Type 2 lymphocytes promote both physiologic tissue remodeling and allergic pathology, yet their physical tissue niches are poorly described. Here we used quantitative imaging to define tissue niches of group 2 innate lymphoid cells (ILC2s), critical instigators of type 2 immunity. We identified a dominant adventitial niche around lung bronchi and larger vessels in multiple tissues, where ILC2s localized with subsets of dendritic and regulatory T cells. However, ILC2s were most intimately associated with adventitial stromal cells (ASC), a mesenchymal fibroblast-like subset that expressed Interleukin-33 (IL-33) and thymic stromal lymphopoietin (TSLP). In vitro, ASCs produced TSLP that supported ILC2 accumulation and activation. ILC2s and IL-13 drove reciprocal ASC expansion and IL-33 expression. During helminth infection, ASC depletion impaired lung ILC2 and Th2 cell accumulation and function, in part dependent on ASC-derived IL-33. These data indicate that adventitial niches are conserved sites where ASCs regulate type 2 lymphocyte expansion and function.
eTOC Blurb
Tissue-resident type 2 lymphocytes are involved in both physiologic and pathologic responses, yet their physical tissue-niches are poorly described. Here, Dahlgren and colleagues identify a population of perivascular fibroblast-like stromal cells that express IL-33 and TSLP as local regulators of ILC2s and type 2 immunity.
INTRODUCTION
Type 2 immunity drives both beneficial responses that restrict helminth infections and pathologic responses that promote asthma, atopic dermatitis, and allergy. Group 2 innate lymphoid cells (ILC2s) are critical initiators of type 2 allergic immunity, defined by elevated tissue eosinophils, M2 alternatively activated macrophages, systemic IgE, and epithelial cell subsets (e.g. goblet cells, tuft cells) that ultimately mediate tissue remodeling (Klose and Artis, 2016; Schuijs and Halim, 2018). Recent work supports a broadening of the physiologic roles for ILC2s and type 2 immunity, including promoting tissue development, metabolic homeostasis, physiologic remodeling, and wound healing (Vivier et al., 2018).
Similar to many innate lymphoid cells and innate-like T cells, ILC2s are predominantly developmentally-allocated, tissue-resident lymphocytes that are long lived and integrate multiple signals to rapidly initiate local immune responses (Klose and Artis, 2016; Schuijs and Halim, 2018; Vivier et al., 2018). ILC2s are present at epithelial barriers, including the skin, gastrointestinal (GI), and respiratory tracts; however, ILC2s also reside in deep ‘non-barrier’ tissues such as adipose, liver, central nervous system meninges, pancreas, uterus, and kidney (Nussbaum et al., 2013). After allergic challenge or helminth infection, subsets of adaptive CD4+ T helper type 2 cells become tissue-resident memory cells (Th2 TRMs), can respond to tissue signals independent of antigen, and are amplifiers of allergic immunity (Endo et al., 2015; Guo et al., 2015; Van Dyken et al., 2016). Many of the upstream signals controlling ILC2s and Th2 TRM cells, including the cytokines IL-33 and thymic stromal lymphopoietin (TSLP), are locally released by tissue resident cells and are critical regulators of allergic physiology and pathology (Cayrol and Girard, 2018; Molofsky et al., 2015a; Ziegler, 2012). This raises the question of which cells produce these and other signals that locally regulate ILC2s and Th2 TRMs positioning and function.
Most adaptive lymphocytes interact in secondary lymphoid organs (SLOs) such as lymph nodes and spleen, where microanatomic niches regulate specialized immune functions (Chang and Turley, 2015; Rodda et al., 2018). However, the stromal niches of tissue-resident lymphocytes in non-SLO tissues have been difficult to define due to limited cell numbers and suboptimal reagents for tracking lymphocyte subsets. Stromal cells are diverse, including pericytes that support capillaries, epithelial-like mesothelial cells that form body-cavity serosal surfaces, and heterogenous fibroblast-like mesenchymal cells (Han et al., 2018). Adventitial stromal cells (ASCs) are the major constituents of perivascular adventitial ‘cuffs’, comprising the outermost layer of intermediate-to-large blood vessels and other tubular structures such as lung airways (Benias et al., 2018; Schraufnagel et al., 2003; Stenmark et al., 2013). Adventitial cuffs are diverse interstitial spaces rich in collagens, extracellular matrix components, small blood vessels, neurons, progenitor cells, and immune cells, providing both vascular support and conduits for interstitial fluid to accumulate and drain into lymphatics. In addition to roles in vascular support and remodeling, ASCs also participate in both vascular and tissue immune responses, engaging in bi-directional conversations with macrophages and dendritic cells in settings of inflammation, and ultimately contributing to the expansion of tertiary lymphoid organs (TLO) (Stenmark et al., 2013).
Here we used tissue clearing with 3D imaging (Oldham et al., 2008), image quantitation (i.e. histocytometry) (Gerner et al., 2012), transcriptomics, and functional assays to define ILC2 niches in the lung and multiple ‘non-barrier’ tissues. We describe a dominant ILC2 niche in adventitial cuffs, where ILC2s reside in proximity to subset(s) of regulatory T (Treg) cells, dendritic cells (DCs), and lymphatics. We identified IL-33 and TSLP producing ASCs that are intimately associated with ILC2s. Single-cell RNA sequencing confirmed ASCs as a fibroblast-like subset enriched for pathways involved in extracellular matrix remodeling, but also immune sensing and regulation. In vitro, ASCs selectively supported ILC2 and Th2 TRM proliferation and cytokine production, predominantly via production of TSLP. In vivo depletion of ASCs impaired lung ILC2s, Th2 TRMs, and the induction of helminth-driven type 2 immunity, in part via ASC-derived IL-33. Together, these data characterize ASCs as a stromal subset that reside in anatomically conserved perivascular niches and demonstrate the importance of ASCs for lung type 2 immune responses.
RESULTS
ILC2 localize to adventitial cuffs
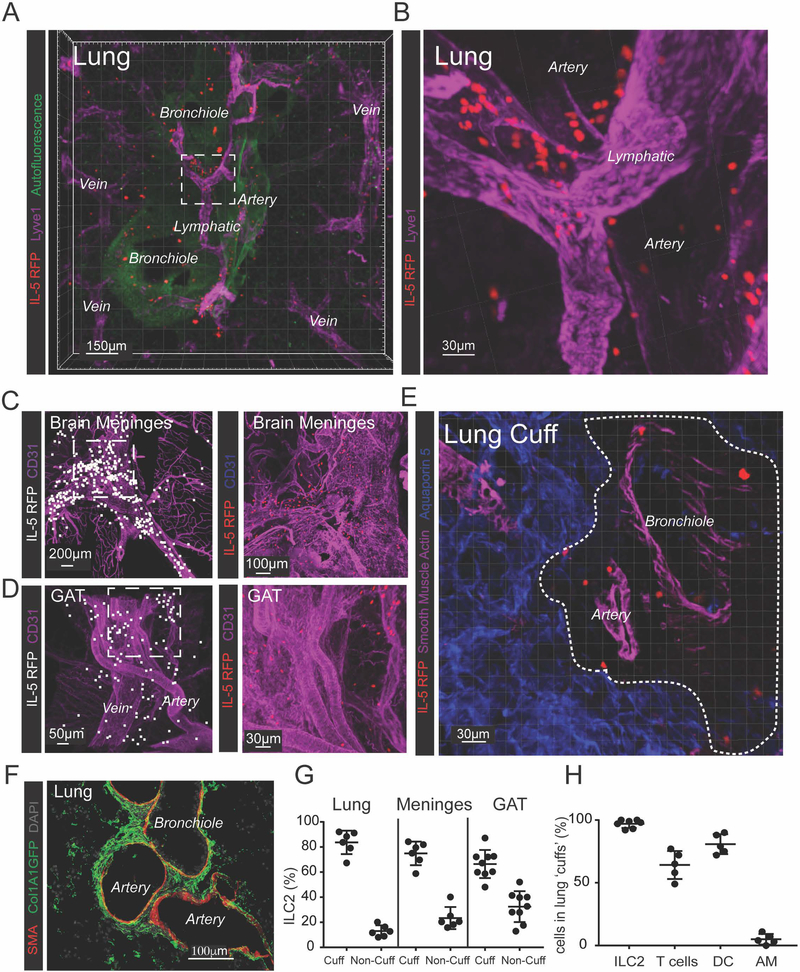
To determine the tissue localization of ILC2s, we imaged IL-5 reporter mice (ll5-cre; R26-CAG-RFP, Fig S1A) (Molofsky et al., 2013; Nussbaum et al., 2013). The IL-5 lineage-tracker was both sensitive and specific, with ~90% of tissue ILC2s lineage-marked by RFP, and ILC2s constituting ~90% of RFP+ cells; T cells accounted for the remaining RFP+ events (Fig S1B,C). Here we focused on lung ILC2 niches but confirmed results in multiple additional non-barrier tissues. Lung ILC2s localized to bronchovascular areas, with a particular enrichment around arteries (Fig 1A), and often in proximity to lymphatic vessels (Fig 1B). ILC2s also localized to intermediate-to-large vessels in brain meninges (Fig 1C, Movie S1) and perigonadal adipose tissue (GAT; Fig 1D). ILC2s were not within vessel or airway lumens (Fig 1E), instead residing in perivascular adventitial cuff spaces. Lung adventitial cuffs expressed structural Col1A1 but lacked basement-membrane associated ColIV and were internally bounded by smooth muscle actin (SMA)-positive sheaths (Fig 1F, S1D). Although cuffs accounted for a small fraction of the tissue volume, ~80% of the total ILC2s from lung and brain meninges resided in adventitial cuffs, with slightly lower frequencies in GAT (Fig 1G). When lung pleural surfaces were excluded, >95% of lung ILC2s were in adventitial cuffs, whereas T cells (CD3e+) and DCs were present in both cuff and parenchymal locations, and alveolar macrophages were exclusively present in the lung alveolar parenchyma (Fig 1H). ILC2s were an enriched population within resting lung cuffs, accounting for ~30–50% of non-B lymphocytes, compared with 1–2% in the total lung. ILC2s were similarly distributed in multiple non-barrier tissues including the pancreas (movie S2), mesenteric adipose tissue (MAT), liver, spleen, and uterus, as were rare IL-5+ Th2 TRM cells (Fig S1E). CCL19, a chemokine involved in CCR7-dependent immune cell trafficking to lymph nodes, also labeled lung cuff portions proximal to ILC2s (Fig S1F).
Figure 1: ILC2s localize to adventitial cuffs in multiple organs.
(A and B) 3D rendering with zoom of a 200μm thick lung slice with IL-5+ ILC2 (RFP), bronchioles (autofluor+ Lyve−), arteries (Lyve1dim+, Autofluor+, bronchiole−adjacent), veins (Lyve1dim+ Autofluor+, non-bronchiole adjacent) and lymphatics (Lyve1bright+, Autofluor−) visualized. (C and D) IL-5 RFP+ ILC2 and CD31+ vessels visualized in (C) brain meninges and (D) perigonadal adipose tissue (GAT). (E and F) IL-5 RFP+ ILC2 visualized in the lung adventitial cuff, highlighted by a lack of (E) Aquaporin 5 and SMA, and positively by (F) Col1A1 rich cuff regions surrounding bronchioles and arteries. (G) 3D image quantification of IL-5 RFP+ ILC2s localized within the cuff volume, using automatic surfacing (lungs), or found within 40μm of SMA+ vasculature (meninges, GAT). (H) Percent of cells in lung cuffs: ILC2 (IL-5 RFP+), T-cells (CD3ε+), DCs (CD11c+ MHCII+ co-positive) and alveolar macrophages (AM, CD11c+ MHCII−). Analysis in (G) included lung cuff, parenchyma, and pleural regions, whereas (H) excluded pleura. (A-E) Images are representative of 3 or more mice or (F) 2 mice. (G and H) Data points represent averages of individual mice with n ≥5 animals with ≥55 cells of each type analyzed per mouse. See also Figures S1–3 and Movies S1 and S2.
We also examined ILC2 localization in epithelial tissues and serosal body-cavity sites. As expected, gastrointestinal ILC2s localized to the lamina propria in adult mice (Fig S2A–B). Skin ILC2s localized to epithelial areas in proximity to hair follicles (Fig S2C). In SLOs (lymph nodes, peyer’s patches), rare ILC2s localized to capsular, interfollicular, and medullary regions (Fig S2D–E). In body cavities (lung pleura, peritoneal membrane, heart pericardium), ILC2s localized with the mesothelial lining (Fig S2F–I). These data indicate that ILC2s reside in perivascular cuff niches, although additional epithelial-, nodal-, and mesothelial- ILC2 tissue niches exist at unique micro-anatomic boundary sites, and consistent with recent evidence describing ILC2 heterogeneity both between and within tissues (Ricardo-Gonzalez et al., 2018). Here we focused on the ILC2 adventitial cuff niche that was present in virtually all tissues examined.
ILC2 localize with subsets of dendritic cells and regulatory T cells
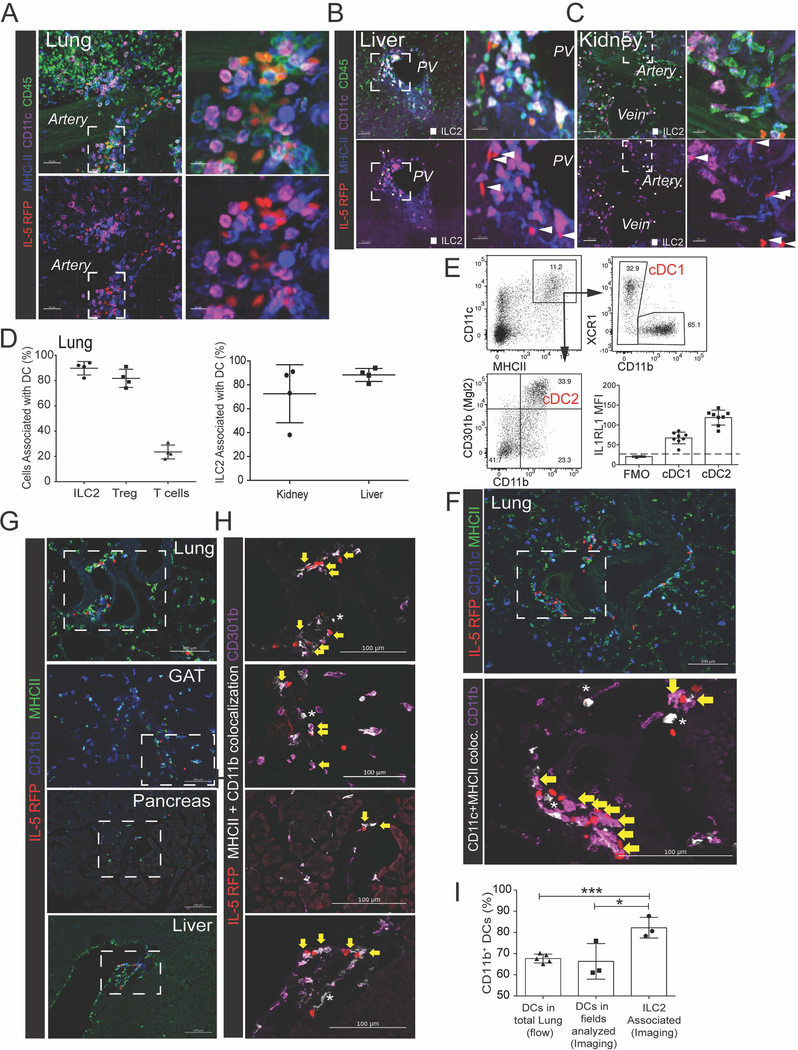
Next we identified the immune cells that were proximal to adventitial ILC2s. Lung ILC2s localized with CD11c+ MHCII+ myeloid cells, consistent with a dendritic cell identity (DCs, Fig 2A), as did ILC2s in the liver, kidney, and other tissues (Fig 2B–C, Fig S3A); 80–90% of ILC2s were in proximity to at least one DC (<1μm surface-to-surface), a degree of localization similar to lung Treg cell - DCs, and well above DC-association with T cells as a whole (~20%; Fig 2D). We found that ~35% of lung DCs were CD301b+ cDC2s expressing CD11c, MHCII, CD11b, CD301b (MGL2), and IL1RL1 (IL-33R, ST2), a subset associated with initiation of type 2 immunity (Durai and Murphy, 2016; Kumamoto et al., 2013) (Fig 2E). ILC2s in multiple tissues colocalized with CD11b+ CD301b+ DCs (Fig 2F–H), including in lung where we found a significant enrichment of CD11b+ DCs colocalized with ILC2 (Fig 2I). These findings suggest that previously described ILC2-cDC2 interactions (Halim et al., 2014; 2016) may also occur in adventitial perivascular niches.
Figure 2: ILC2s are associated with CD11b+ dendritic cells at cuff sites.
(A-C) 3D rendering with zoom of 200μm projections from (A) lung, (B) liver, or (C) kidney, highlighting IL-5 RFP+ ILC2 and CD11c+ MHCII+ myeloid cells, most consistent with dendritic cells (DCs). (D) Image analysis of 3D thick sections from lung, liver, and kidney for indicated populations; association defined as <900nm between the nearest ILC2 and DC surface vertices. (E) Flow cytometry analysis of lung DC subsets. (F-I) 2D-thin cut images of ILC2 localization (yellow arrows) with (F) CD11b+ DCs or (G and H) CD301b+ CD11b+ DCs from indicated tissues, with (I) quantification of co-localization. Occasional CD11b-neg DCs (F) or CD301b-neg DCs (G) are indicated with stars. (A-C, E-H) Representative of three or more mice. (D,I) Data points represent averages of individual mice n=3–4 with >n=25 cells analyzed for per animal. See also Figure S3.
IL-33 expressing adventitial stromal cells localize with ILC2
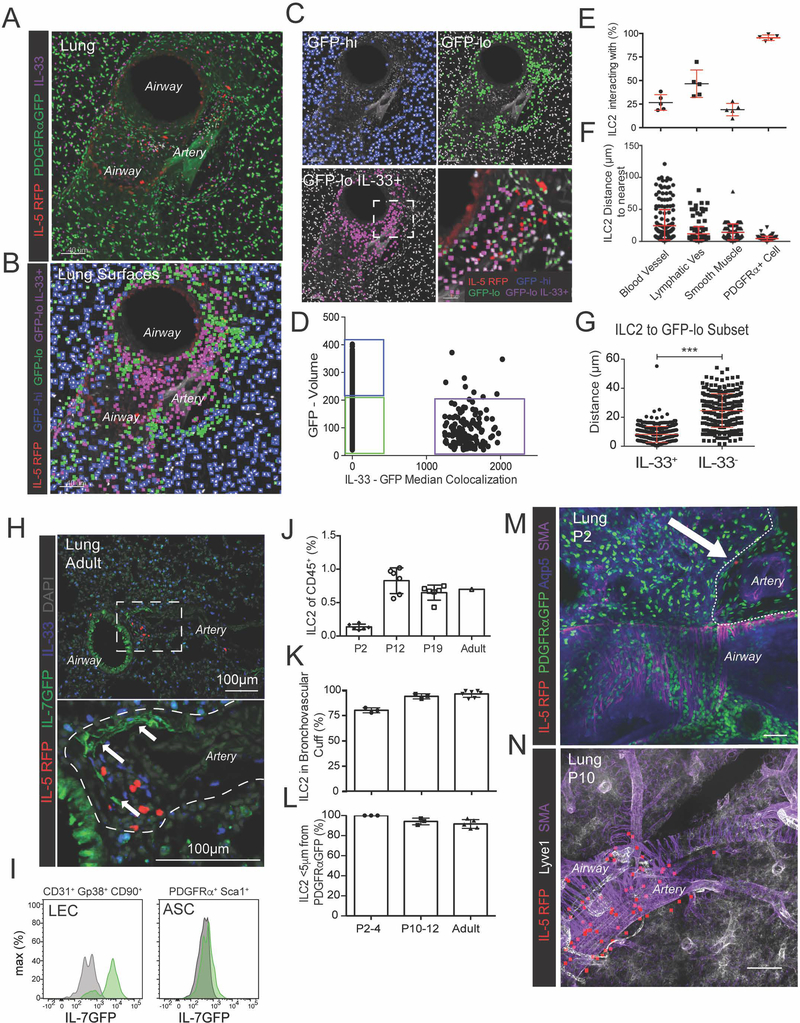
To determine which non-hematopoietic cells are positioned to locally regulate ILC2s, we first used a reporter that marks fibroblast-like stromal cells with a nuclear localized GFP (PDGFRαGFP). We identified a lung GFPlow/dim subset (~20% of GFP+ cells) that localized to adventitial cuffs and was distinct from the GFPhi/bright subset in the alveolar parenchyma (Fig 3A–C). Here we refer to these GFPlow cells as adventitial stromal cells (ASCs) although similar stromal subsets from diverse tissues have been called adventitial fibroblasts (AFs), mesenchymal stromal cells or mesenchymal stem cells (MSCs), fibro-adipogenic progenitors (FAPs), and adipocyte precursors (APs) (Corselli et al., 2012; Sitnik et al., 2016). Using 3D imaging with surfacing, we found that virtually all ILC2s resided in close proximity to ASCs relative to other cuff components such as smooth muscle cells and blood endothelial cells (Fig 3E–F). A subset of ILC2s were also in proximity to the lymphatic endothelium (Fig 3E–F).
Figure 3: ILC2s localize with adventitial stromal cells expressing IL-33.
(A) 3D rendering with zoom of a 200μm thick projection from a lung airway-artery region. (B-D) PDGFRαGFP+ stromal cells were surfaced and binned using GFP and IL-33 median colocalization into 3 groups: PDGFRαGFPhi, IL-33-neg parenchymal stroma (blue), GFPlow IL-33-neg cuff ASCs (green), and GFPlow IL-33+ ASCs (magenta). (E-F) Quantification of (E) percent ILC2s (IL-5 RFP+) less than 5μm apart from blood vessels (Lyve1+ SMA-proximal), lymphatic vessels (Lyve1+ SMA−), smooth muscle (SMA+), or PDGFRα+ stroma (GFP+), or (F) distance from ILC2 (IL-5 RFP+) to nearest cell type, or (G) distance from ILC2 to nearest PDGFRαGFPlow ASC subset (IL-33+ vs IL-33neg). (H) Lung 2D section highlighting ILC2 (IL-5 RFP+) and IL-7GFP+ lymphatics. (I) Flow cytometry analysis of IL-7GFP and wild-type controls for the indicated populations. (J) Flow cytometry analysis of ILC2s (Lin− Thy1+ Gata3hi) in developing wild-type (WT) lungs at indicated post-natal days (P2, P12, P19, >8wk old adults). (K and L) 3D imaging of 200μm thick section from pups at indicated ages, analyzing (K) percent ILC2s in the lung cuff and (L) localization between ILC2s and PDGFRαGFP+ stroma. (M and N) 3D rendering of (M) 25um or (N) 52um volume from lungs at indicated post-natal ages highlighting ILC2 localization. (E,F,J-L) Data points represent (E, J-L) average per mouse or (F) aggregation of five individual mice (N=237). (A-D, H-I, M-N) Representative images, surfaces, or flow cytometry analysis for three or more mice. See also Figure S3 and Movies S3 and S4.
Next we determined whether ASCs express signals known to support ILC2 function. First, we examined IL-33, a nuclear-localized IL-1 family cytokine that is released with cell death or stretch and activates ILC2s in cooperation with other tissue-derived signals (e.g. IL-7, TSLP) (Cayrol and Girard, 2018; Molofsky et al., 2015a). A subset of lung GFPlow ASCs (~60%) stained with nuclear IL-33 protein, whereas lung GFPhi parenchymal stroma did not (Fig 3A–D). Lung ILC2s preferentially localized to the IL-33+ ASC subset, frequently with two or more IL33+ ASCs in contact with a single ILC2 (Fig 3G, S3C, Movie S3), and with similar findings in the liver, pancreas, and adipose tissue (Fig S3D–E, Movie S4). Consistent with previous work (Halim et al., 2018; Molofsky et al., 2015b), Treg cell subset(s) from several tissues also localized to ILC2s and DC subsets, and to IL-33+ ASCs (Fig S3E–G). Lung ASCs did not express detectable IL-7 (IL7GFP), a signal required for ILC development and optimal lung Th2 TRM cell maintenance (Shinoda et al., 2016), although subset(s) of lung lymphatic cells were IL-7GFP+ (Fig 3H–I; (Miller et al., 2013; Shinoda et al., 2016). These data suggest that cuff ILC2s intimately associate with IL-33+ ASCs, raising the possibility that ASCs provide IL-33 or other signals that locally regulate ILC2s.
In mice, lung ILC2s expand in the first two weeks of life, a postnatal period associated with alveolar and vascular growth and activated lung ILC2s and type 2 immunity (Fig 3J) (de Kleer et al., 2016; Saluzzo et al., 2017). Recent data also indicate that ASC-like stromal cells can promote terminal differentiation of mesenteric ILC precursors to mature ILC2s (Koga et al., 2018). As such, we examined ILC2 localization during early postnatal development. We found that by postnatal day (P)2, rare IL-5+ ILC2 were already in close proximity to ASCs in developing lung cuffs, a relationship that was maintained during the next two weeks of ILC2 expansion (Fig 3K–N, Fig S3H). In the liver and small intestine, we identified ILC2s that also associated with IL-33+ stromal cells (Fig S3I–K). These data indicate that postnatally expanding ILC2s localize with IL-33 expressing stromal cells, often in adventitial cuff structures, and suggest that the distribution of ILC2s begins early in life.
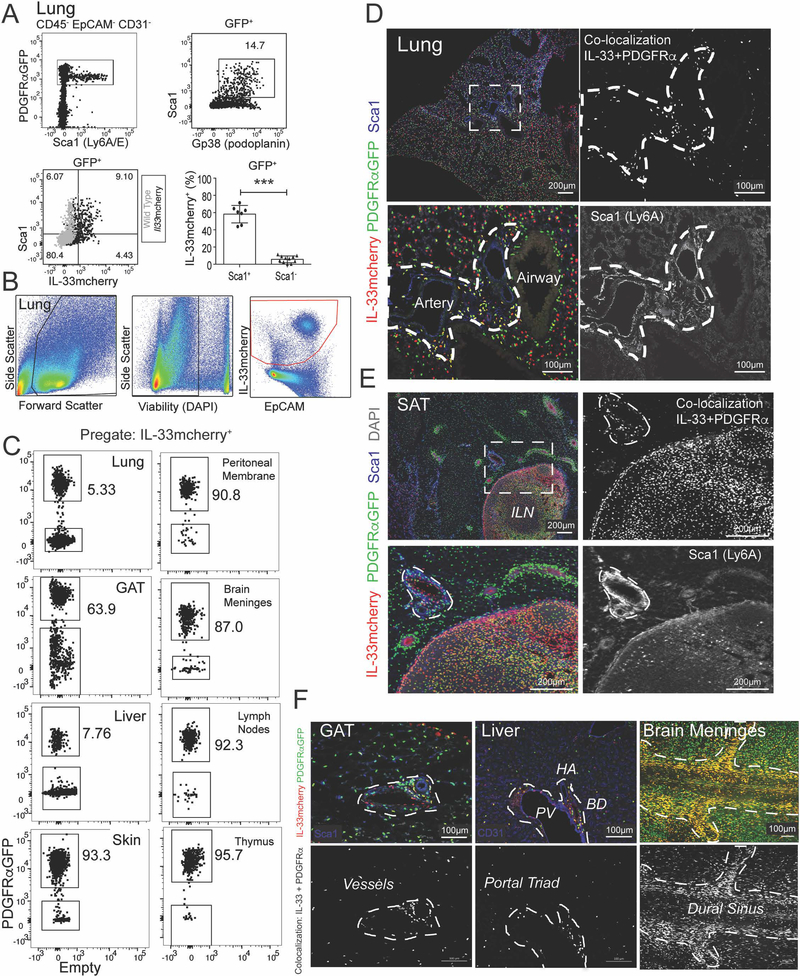
IL-33mcherry identifies ASCs in multiple tissues
To better characterize the cellular sources of IL-33 within the ILC2 niche, we used IL-33-H2B-mcherry reporter mice (Fig S4A) (Vainchtein et al., 2018). PDGFRαGFPlow ASCs preferentially expressed Sca1 (Ly6A) and Gp38 (podoplanin) and ~60% expressed IL-33mcherry (Fig 4A), consistent with IL-33 protein expression (Fig 3A–D). ASCs were the most abundant IL-33+ cell type in many tissues (Fig 4B–C) and IL-33+ ASCs were always a subset of total PDGFRαGFP+ stroma (Fig S4D). However, other lung cells expressed both IL-33mcherry+ and Il33 transcript, including epithelial (EpCAM+), endothelial (CD31+), and other stromal (EpCAM−CD31− Sca1−) subsets (Fig S4B–C). Small numbers of alveolar macrophages were dimly-positive for IL-33mcherry, whereas other hematopoietic cells examined were negative in naïve lung, peritoneal fluid, and skin (not shown). Imaging confirmed that IL-33mcherry+ ASCs expressed Sca1 and were the primary IL-33+ cell type in adventitial cuffs of multiple tissues (Fig 4D–F, Fig S4E–H). Other IL-33 sources were also confirmed, including lung epithelial type 2 pneumocytes (AT2), lymph node stromal cells, and endothelial cell subset(s) in lung, liver, and adipose tissues (Fig 4C–F, not shown). These data demonstrate that ASCs are a conserved IL-33 expressing stromal subset in adventitial niches in proximity to ILC2s, although additional IL-33 expressing cell types are present in a tissue- and niche-dependent manner.
Figure 4: ASCs express IL-33mcherry, Sca1 and Gp38 and are widely distributed in tissues.
(A-C) Flow cytometry plots and quantification from (A) lung of PDGFRαGFP+ IL-33mcherry mice (black) or wild-type gating controls (grey) or (B and C) indicated tissues, including pre-gating strategy. (D and E) 2D thin-cut images with zooms from (D) lung or (E) subcutaneous adipose tissue (SAT) with inguinal lymph node (ILN) or (F) gonadal adipose tissue (GAT), liver, and brain meninges, with antibody stains indicated and PDGFRαGFP – IL33mcherry colocalization highlighted (white). Flow cytometry plots and images are representative of three or more mice. See also Figure S4.
ASCs are a molecularly distinct stromal subset associated with matrix remodeling and inflammatory response
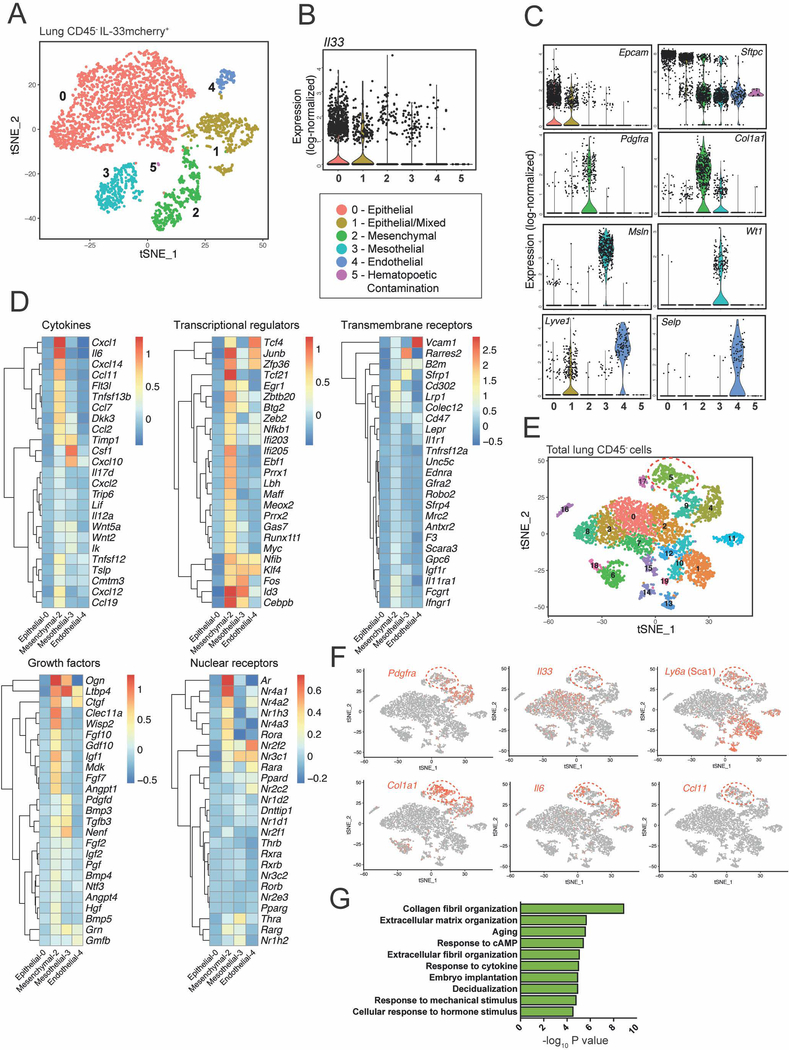
To gain an unbiased insight into the molecular identity of ASCs, we performed droplet-based single cell RNA-sequencing (scRNAseq). In sorted lung IL33mcherry+ cells from pooled mice we identified four Il33 expressing subsets, consistent with our flow cytometry and imaging data. These were epithelial AT2s (cluster 0), mesenchymal ASCs (cluster 2), mesothelial cells (serosal lining, cluster 3), and endothelial cell subsets (cluster 4) (Fig 5A–C). ASCs (cluster 2) expressed high amounts of Pdgfra and Col1a1 and were enriched for expression of several cytokines (Tslp, Il6, Csf1), chemokines (Ccl11, Ccl2, Ccl7, Cxcl12, Ccl19), growth factors (Igf1, Igf2), and cytokine receptors (Ifngr1, Il11ra1, Il1r1) (Fig 5D). Several transcription factors (Prxx1, Prxx2, Klf4) and nuclear receptors (Ar, Nr4a1–3, Rora) were also enriched (Fig 5D, Fig S5A, Table S1). IL-33mcherry+ ASCs were also marked by expression of multiple early-response genes and markers of NF-κB pathway activation, including Nr4a1–3, Fos, Junb, Egr1, Nfkbia, Nfkbiz and Zfp36 (Fig 5D, Table S1). These findings suggested that lung ASCs have potential roles in immune responses, expressing genes implicated in the regulation of type 2 immunity including Il33, Tslp, and Ccl11 (Eotaxin-1).
Figure 5: ASCs are a distinct IL-33 expressing fibroblast-like subset expressing genes associated with matrix remodeling and inflammation.
(A) Unsupervised clustering analysis of scRNAseq on IL-33mcherry+ CD45− cells from naïve lung visualized with t-SNE. Each dot indicates an individual cell (total cell number: 3584). A mixed subset comprising apparent epithelial with endothelial cell doublets (cluster 1) and a small hematopoietic contaminating subset (cluster 5) were identified and not analyzed further. (B and C) Violin plots of (B) Il33 or (C) selected genes used to assign cluster identity. (D) Heatmaps of mean expression values of top 25 genes in ASCs (cluster 2) in indicated gene category across clusters identified in (A). Data is log-transformed. Genes were assigned to categories using Ingenuity Pathway Analysis (IPA). (E) Unsupervised clustering of aggregated total CD45-negative and IL-33+ (mCherry+) cells from naïve lung visualized with t-SNE. Each dot indicates an individual cell (total cell number: 5392). (F) Gene expression of ASC-associated genes projected onto tSNE plots. (G) Gene ontology analysis in DAVID of cluster 5 differentially expressed genes. See also Figure S5 and Table S1.
To identify unique molecular markers of IL-33+ ASCs versus other IL-33− stromal subsets, we compared ASCs with all non-hematopoietic cells (CD45−) in the lung (Fig 5E). Cluster 5 cells were enriched for genes encoding Sca1 (Ly6A), Gp38 (Pdpn), and IL-33, consistent with an ASC identity. In addition, this subset expressed relatively elevated amounts of collagens, chemokines, and was enriched for gene ontology (GO) terms associated with extracellular matrix remodeling and immune response (Fig 5F–G, Table S1, Fig S5B). Cluster 5 ASCs did not express detectable Il7 (Table S1), although Il7 was detectable in a lymphatic endothelial sub-cluster, consistent with the IL-7GFP reporter results (Fig 3H–I). ASCs were distinct from putative smooth muscle cells (cluster 17) or pericytes (cluster 14), which lacked Il33 and Col1a1 (Table S1).
We identified a similar stromal subset in gonadal adipose tissue (GAT), where IL-33mcherry+ cells clustered into four groups (Fig S5C–D), including mesothelial cells (cluster 1; expressing Wt1, Msln) and three stromal subsets (clusters 0,2,3; all expressing Pdgfra and Col1a1). The largest stromal subset (cluster 0) was enriched for Ly6a, Cd34, Pi16, Ccl11, and Cxcl1, and was similar to lung ASCs (Fig S5D–F). These data indicate that lung and adipose tissue IL-33mcherry+ ASCs are transcriptionally related subsets. Similar ASC-like populations expressing transcripts for CD34, Ly6A, TSLP, and IL-33 were identified in scRNAseq datasets from multiple tissues (Han et al., 2018), suggesting ASCs have conserved functions that include the regulation of local immune responses.
ASCs produce TSLP to support ILC2s
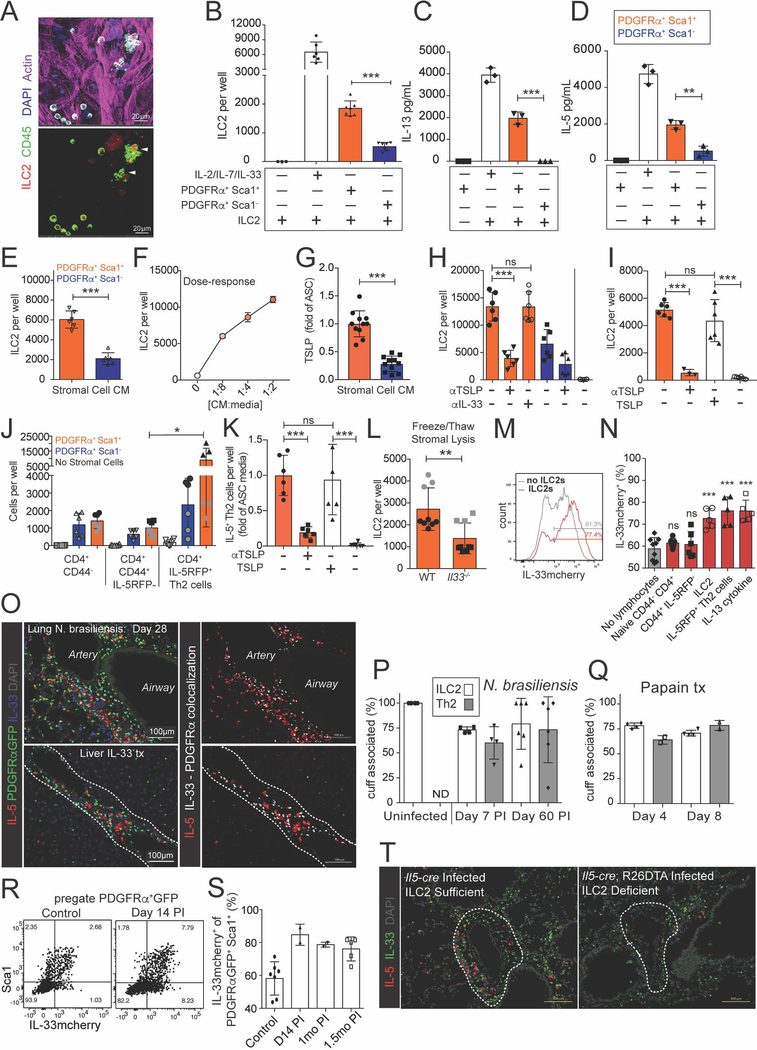
To test if lung ASCs were sufficient to support ILC2s in vitro, we sorted lung ILC2s and co-cultured them with subsets of lung stromal cells. In vitro, ILC2s require STAT5-activating cytokine supplement (i.e. IL-2, IL-7, TSLP) for survival and proliferation, and IL-33 synergizes with these signals to provide increased ILC2 activation and proliferation. Surprisingly, bulk lung stroma was sufficient to maintain ILC2s in the absence of exogenous cytokines (Fig 6A). We found that sorted ASCs (PDGFRα+ Sca1+) and parenchymal-enriched stromal cells (PDGFRα+ Sca1−) formed similar monolayers and stabilized at similar cell densities (Fig S6A–B), yet ASC co-cultures yielded ~4-fold more ILC2s (Fig 6B). In vivo, tissue ILC2s produce constitutive IL-5 and variable IL-13 (Molofsky et al., 2013; Nussbaum et al., 2013; Van Dyken et al., 2014). Similarly, in vitro ASCs supported low amounts of ILC2 IL-5 and IL-13 production (Fig 6C–D).
Figure 6: ASCs produce TSLP to support ILC2s, whereas ILC2s and IL-13 in turn promote ASC accumulation and IL-33 expression.
(A) Bulk lung stroma was co-cultured with ILC2s for 3 days and imaged with IL-5 RFP+ ILC2 clusters indicated. (B-D) PDGFRα Sca1+ (ASCs, orange) and PDGFRα+ Sca1− (parenchymal-enriched, blue) stromal cells were cultured with lung ILC2s for 7 days and ILC2s were (B) counted and (C and D) IL-5 and IL-13 amounts measured from culture supernatants. (E-I) Culture media (CM) from ASCs (Sca1+, orange), control parenchymal-enriched SCs (Sca1−, blue), or fresh culture media (white) was plated with freshly sorted ILC2s for (F) 3 days or (E,H,I) 5 days +/− the indicated blocking antibodies or TSLP supplementation (10ng/mL), or (G) assayed for TSLP. (J and K) Indicated lung T cell subsets sorted from IL-33 treated mice were (J) co-cultured with the stromal cell subsets for 5 days without TCR stimulation or (K) cultured with ASC culture media (orange) or fresh culture media (white) +/− the indicated blocking antibodies or TSLP (10ng/mL). (L) ASCs from WT or IL-33 deficient mice were subjected to 3 rounds of freeze-thaw lysis, after which ILC2s were co-cultured for 5 days. (M and N) IL-33mcherry+ ASCs were cultured for 5 days post confluency with the indicated cells or cytokines (10ng/mL) and assessed for IL-33mcherry, showing (M) representative histogram or (N) percent of IL-33mcherry expressing cells. (O) Mice were infected with 500 N. brasiliensis or treated x3 with 500ng of IL-33 and examined for the indicated markers. (P) 200um thick tissue slices were analyzed at the indicated points post N. brasiliensis infection or (Q) post papain treatment (x3 daily doses), ILC2s (IL-5 RFP+ CD3e−), Th2 cells (IL-5 RFP+ CD3e+), and cuff regions (Aqp5−, bronchovascular-associated) were defined and the association enumerated. (R-S) Flow cytometry analysis of lungs on N. brasiliensis post-infection (PI) days indicated with (R) representative ASC flow cytometry plots and (S) quantification of ASC IL-33mcherry expression. (T) 2D thin-cut lung images at day 28 PI from ILC2-deleter (Il5-cre; R26-DTA) and control Il5-cre) mice with IL-5+ cells and IL-33+ cells highlighted. Data are (A,C,D,I,K,M,R) representative or (B,E-H,J,L,N) aggregated from 2–3 independent cohorts. (O,T) Representative images of two independent experiments. (P,Q,S) Aggregated data from two independent experiments, each data point represents one mouse. Independent cohorts indicated in (J and L) with black and grey symbols. See also Figure S6.
ASC-conditioned media (CM) was also sufficient to support ILC2s in a dose-dependent manner (Fig 6E–F), suggesting the presence of ASC-secreted factor(s). ASCs expressed Tslp transcript (Fig S6C) and ASC-CM contained TSLP protein, but not IL-7 or IL-33 (Fig 6G, Fig S6E, not shown). Blockade of TSLP impaired the ability of ASC-CM and ASCs to support ILC2s and their proliferation (Fig 6H, Fig S6D–F), whereas TSLP alone was sufficient to support ILC2s (Fig 6I). ASCs and ASC-CM also supported IL-5+ Th2 cells (CD4+ CD44+ IL-5RFP+; Fig 6J,K) in a TSLP-dependent manner. In contrast, other lung CD4+ T cell naïve or memory subsets were not supported by ASCs. Although ASCs did not release detectable IL-33 protein under the in vitro conditions tested, ASC-derived IL-33 was bioactive and conferred additional support to ILC2s after ASC lysis (Fig 6L). We conclude that ASCs can support both ILC2 and Th2 TRM cells via TSLP production, consistent with the ability of purified TSLP and related STAT5-activating cytokines (i.e. IL-2, IL-7) to support these type 2 lymphocytes (Halim et al., 2012; Mohapatra et al., 2016). However, additional ASC-derived signal(s) may also contribute to ILC2 and Th2 TRM regulation.
ILC2s and IL-13 promote ASC IL-33 expression
Next we tested if ILC2-derived signals might reciprocally regulate ASCs. In vitro, both ILC2s and IL-5+ Th2s, but not other lung CD4+ T-cells, promoted increased ASC IL-33mcherry expression (Fig 6M,N, Fig S6I). These type 2 lymphocytes expressed IL-13 after activation, and ASCs express both components of the IL-13 receptor (Fig S6G–H). IL-13 treatment significantly increased ASC IL-33mcherry expression, in contrast to several other cytokines and signals for which ASCs expressed receptors and which have been reported to increase IL-33 production (IL-1β, PDGF-BB, or IL-17A + TNFα; Fig S6J). PDGF-BB increased ASC numbers (~2.3x, not shown) and IL-1β and IL17A + TNFα promoted ASC TSLP production (Fig S6K), indicating that these compounds were bioactive and likely regulate additional aspects of ASC biology. In vivo, repetitive intranasal administration of IL-13 or IL-4 increased both ASC and epithelial cell IL-33mcherry expression and promoted lung eosinophil accumulation (Fig S6L; not shown).
Type 2 immune challenges drive ILC2 and Th2 cell expansion in cuffs
To examine the relationship of ILC2s and Th2 cells with the adventitial cuff during type 2 inflammation, we determined the localization of these type 2 lymphocytes after helminth infection. N. brasiliensis is a model helminth that traffics from skin to lung and is cleared through the intestine by day 7 post-infection (PI) (Molofsky et al., 2013; Nussbaum et al., 2013). IL-5+ Th2 TRM cells are rare in naïve lungs, but expand along with ILC2s and persist for extended periods post-helminth infection (Nussbaum et al., 2013; Van Dyken et al., 2016). At 4 weeks PI, lung IL-5+ cells were a mix of ILC2s and Th2 TRM cells that localized predominantly to adventitial cuffs in proximity to IL-33+ ASC (Fig 6O, Fig S6M,N). The percentage of ILC2s in adventitial cuffs modestly changed from >95% in naïve mice to ~75% post N. brasiliensis infection, similar to Th2 TRM cells (Fig 6P). ILC2s and Th2 cells showed similar cuff localization in a papain model of type 2 lung inflammation (Fig 6Q), and after systemic IL-33 injections, which expanded ILC2s in cuff structures in multiple tissues, including liver, spleen, adipose tissue, and lung (Fig 6O, S6O, not shown). In the tissues tested, we conclude that during type 2 inflammatory challenges ILC2s and Th2 TRM cells are primarily localized to adventitial cuff niches and in proximity to ASCs, although minor subet(s) of type 2 lymphocytes expand into parenchymal niches.
Type 2 inflammation promotes IL-33+ ASC expansion dependent on ILC2s
Concurrent with ILC2 and Th2 cell expansion, we found increased IL-33mcherry expression in ASCs (PDGFRαGFPlo Sca1+) and increased frequency and IL-33 expression in GFPlow Gp38+ Sca1− SCs, both localized predominantly to adventitial cuffs post helminth infection (Fig 6R–S, Fig S6P,Q). scRNAseq indicated that the four IL-33mcherry+ populations identified in naïve lungs were also present at day 30 PI, although both ASCs and lymphatic endothelial cells relatively increased (Fig S5G). Modest transcriptional changes in IL-33mcherry+ ASCs were noted post-infection, including decreased expression of several collagen and matrix-related genes (Col1a1, Col1a2, Dpt1, Table S1). Loss of ILC2s (Il5-cre; R26-DTA; ~80–90% deletion) impaired the helminth-driven expansion of IL-33+ PDGFRαGFPlow ASCs (Fig 6T, Fig S6R–U). These results were supported by decreased cuff-localized IL-33 protein in ILC2 deleter mice, analyzed by quantitative imaging (controls 3.6×105/mm3 ± 6.8×104; ILC2-deficient 1.3×105/mm3 ± 3.7×104, n=2/group, D14 PI). These findings suggest that ASCs dynamically expand and increase IL-33 expression after type 2 inflammation, and ILC2s promote this niche remodeling.
ASCs support helminth-driven induction of ILC2s and type 2 immunity
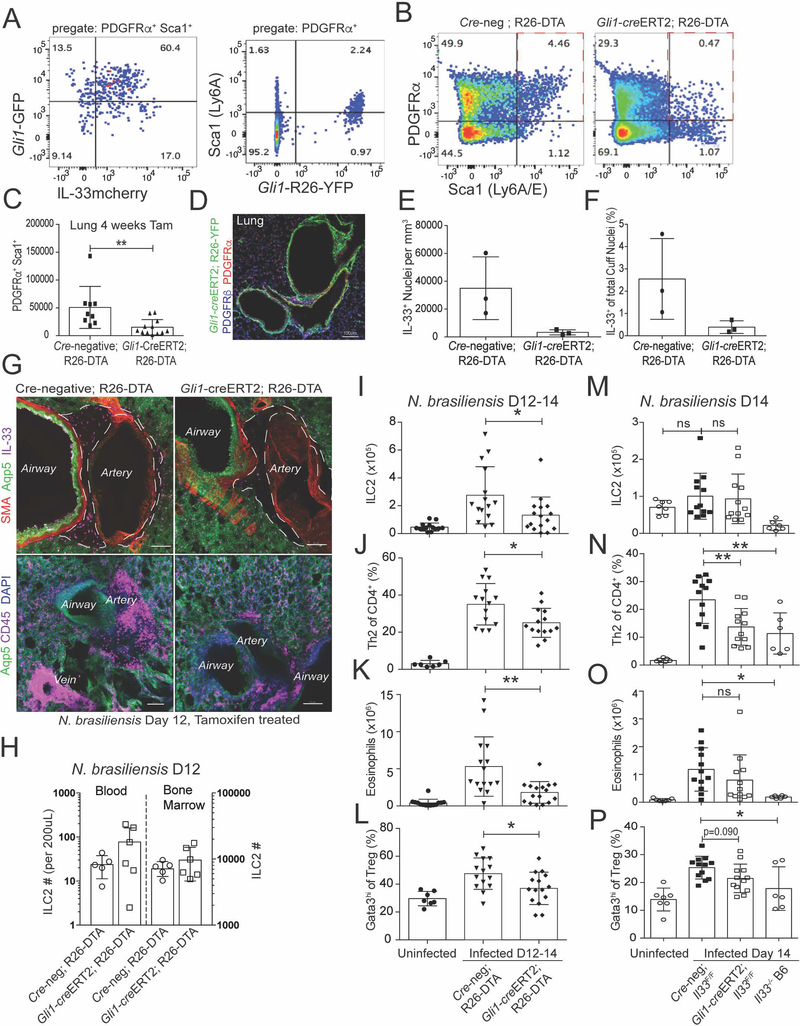
Next we tested if ASCs support type 2 inflammation in vivo. scRNAseq identified a number of potential markers expressed in ASCs (e.g. Col1a1, Pdgfra, Pdgfrb), but many of these were broadly expressed during development or lacked specificity for ASCs. The hedgehog family effector Gli1 was enriched in our dataset of IL-33mcherry+ ASCs, and previous work has defined Gli1 expression in similar lung adventitial stroma (Kramann et al., 2015). We confirmed that ASCs co-expressed Gli1, Sca1, and Gp38 in bronchovascular cuffs and several other perivascular regions (Kramann et al., 2015), accounting for over 80% of total lung Gli1+ cells at rest and after type 2 stimuli (Fig 7A,D, Fig S7A–D).
Figure 7: ASCs support ILC2s and the induction of lung type 2 inflammation.
(A) Flow cytometry analysis of lungs from Gli1-GFP mice (left) or Gli1-creERT2; R26-YFP tamoxifen treated lineage trackers (right). (B and C) Gli1-creERT2; R26-DTA mice (tamoxifen treated) with (C) enumeration of ASC depletion. (D) Lung 2D thin-cut image from Gli1-creERT2; R26-YFP tamoxifen treated mice. (E-G) 3D imaging with quantitation of ASC-replete (Cre-neg; R26DTA) or ASC-deplete (Gli1-creERT2+; R26-DTA) IL-33+ nuclei in adventitial cuff spaces at day 12 PI with N. brasiliensis (H-P) Flow cytometry quantification of (H) blood and bone marrow or (I-P) lungs from tamoxifen treated N. brasiliensis infected mice at PI D12–14 from the indicated strains (ASC-depleted, ASC-IL-33 Flox), quantifying ILC2s, eosinophils, Th2 cells, and Gata3hi Treg cells. (A,B,D,G,H) Representative plots, images, or experiments from 2 or more mice. (C, I-P) Pooled data from greater than 3 experiments, with individual mice shown. (E,F) Three individual mice analyzed for each genotype. See also Figure S7.
To deplete lung ASCs, we used tamoxifen inducible expression of Diphtheria toxin-a in Gli1-expressing cells (Gli1-creERT2; R26-DTA), with an overall deletion efficiency ranging from 50–75% (Fig 7B–C). We found no differential impact on mouse viability in the time frame analyzed, as previously reported (Kramann et al., 2015). In naïve mice, ILC2s were not reduced after Gli1+ cell deletion (Fig S7E–F), potentially consistent with the slow turnover of adult tissue ILC2s (Nussbaum et al., 2013). However, N. brasiliensis infection with concurrent depletion of Gli1+ cells reduced lung IL-33+ ASCs, as expected, and the accumulation of adventitial hematopoietic cells (Fig 7E–G). Flow cytometry analysis showed preserved numbers of blood and bone marrow ILC2s (Fig 7H), but reduced accumulation of lung ILC2s, Th2 cells, eosinophils, and Gata3hi type-2 skewed Treg cells (Fig 7I–L, Fig S7G–H, Day 12–14PI), with preserved lung CD8+ T cell numbers (Fig S7I) and lymph node CD4+ and CD8+ T cells (not shown). These data indicated that ASCs are required for optimal accumulation of lung ILC2s, Th2 TRM cells, and helminth-driven type 2 immune responses.
IL-33 and TSLP are not required for ILC2 development or tissue residency, although they synergize to promote ILC2 activation at rest and after type 2 immune challenge (Ricardo-Gonzalez et al., 2018; Van Dyken et al., 2014). Therefore, we tested if ASC-derived IL-33 contributed to allergic inflammation-driven type 2 lymphocyte accumulation and function. First, we treated ASC-deficient or littermate controls with exogenous IL-33 (Molofsky et al., 2015b). In mice deficient in Gli1+ ASCs, we observed a modest reduction in IL-33 driven eosinophil and Gata3hi Treg cell accumulation, and a trend towards fewer ILC2s (Fig S7J–L), suggesting exogenous IL-33 was unable to fully bypass the loss of ASCs, and consistent with a possible role for additional ASC-derived signals (e.g. TSLP). Next, we tested the converse, if ASC-derived IL-33 contributes to type 2 immune function in vivo. We conditionally deleted Il33 from ASCs and validated loss of IL-33 protein and message (Gli1-creERT2; Il33F/F, Fig S7M–N). Mice with IL-33 deficient ASCs showed impaired helminth-driven induction of Th2 cells and trends towards reduced eosinophils and Gata3-hi Treg cells (Fig 7M–P), despite limited expansion of ILC2s in this mixed genetic background (Chen et al., 2015), and potentially consistent with mouse strain-dependent variations in ILC2 activation to type 2 challenges (Molofsky et al., 2013; Oboki et al., 2010). However, additional cellular sources of lung IL-33 (Fig 5A–B) may also cooperate with ASCs to coordinately promote ILC2 expansion. Together, our data indicate that ASCs are an important potentiator of lung type 2 immune responses, promoting type 2 lymphocyte expansion and the orchestration of allergic immune responses via both IL-33 dependent and independent pathways.
DISCUSSION
Here we have defined a conserved microanatomical niche of ILC2s and demonstrated its role in supporting type 2 immunity after helminth infection. We found that ILC2s predominantly resided in the adventitial cuffs of lung vessels and airways, with similar cuff localization in the vascular tree of adipose tissue, liver, kidney, spleen, meninges, and pancreas. Within this niche, ILC2s localized with a population of mesenchymal, fibroblast-like adventitial stromal cells (ASCs). ASCs are transcriptionally defined by matrix deposition and remodeling functions, but we also identified an immune-related program that included expression of the cytokines IL-33 and TSLP. Lung ASCs were responsive to type 2 lymphocyte derived signals, including IL-13, and expanded with ILC2s and Th2 TRM cells after helminth infection. After helminth infection, ASCs were partially necessary, and entirely sufficient, to support ILC2 survival, proliferation, and cytokine production. ASCs also supported tissue-resident Th2 cells, establishing ASCs as a niche-cell supporting type 2 lymphocytes and the induction of allergic immune responses. ILC2s are long-lived and slowly proliferating cells, and although we found minimal impact of short-term ASC depletion on adult ILC2 numbers, differences in homeostatic maintenance may not be revealed within this time-frame. It is also possible that other niche cells provide redundant signals that support ILC2s at rest.
Lung perivascular cuffs were originally defined in models of pulmonary edema as conduits for excess fluid drainage from the alveolar parenchyma to lymphatics. Similar spaces have been identified in humans, defined by intermittent compression (e.g. arterial pressure waves), pre-lymphatic fluid flow, the presence of CD34+ fibroblast-like stromal cells, and well-organized collagen fibrils (Benias et al., 2018). Niches for hematopoietic cells often include stromal cells that cooperate with specialized endothelial cells, with examples including bone marrow hematopoietic stem cells, lymphocytes in SLOs, and inflammation-induced TLOs. Our work demonstrates that tissue-resident type 2 lymphocytes localize to adventitial niche sites that also include specialized stromal (ASCs) and both lymphatic and blood endothelial components, and further emphasize that the concept of tissue-niches is relevant to understanding the regulation of tissue resident lymphocytes and the initiation of tissue immune responses.
The potential for immune cell interactions within the cuff raises interesting questions about the regulation of type 2 immunity. A subset of tissue Treg cells are in close proximity to cuff ILC2s and expand in response to IL-33 via both direct and ILC2-dependent effects (Halim et al., 2018; Molofsky et al., 2015b; Panduro et al., 2016), suggesting that ASCs and their IL-33 production may also regulate Treg cell subset(s). We also demonstrated here that cDC2s, previously implicated in the induction of type 2 immune responses, were enriched at cuff sites and localized with ILC2s. Both cDC2 and Treg cell subsets can also respond directly to IL-33 and TSLP to promote their type 2 and/or tissue reparative functions (Kashiwagi et al., 2017; Molofsky et al., 2015a; Panduro et al., 2016; Ziegler, 2012). Further work is required to understand these intriguing cellular interactions occurring in adventitial spaces.
Key open questions remain regarding the identity of the signals that establish the ILC2-niche during development, the crosstalk between niche immune and stromal cells, and importantly, to what extent this is a conserved physical location for tissue type 2 immunity in mouse and human. For example, what is the contribution of other components to the ILC2 cuff-niche? Microanatomic localization of ILC2 in both cuff and serosal boundary spaces closely mirrors that of tissue lymphatics and peripheral nerves. What is the mechanism by which ILC2s and IL-13 feedback to impact ASCs and help shape tissue (re)modeling? ASCs themselves are likely heterogenous, including cells with mesenchymal progenitor capacity (Cano et al., 2017; Kramann et al., 2015; Sitnik et al., 2016) that directly respond to type 2 cytokines (Heredia et al., 2013) and could contribute to niche and tissue remodeling after challenge. What other signals regulate ASC function, including IL-33 and TSLP expression and release? Although ASCs contain bioactive IL-33 protein and upregulate Il33 message in response to ILC2-derived signals such as IL-13, no IL-33 release was detected in vitro, suggesting additional niche components (e.g. cytokines, mechanical stretch) are required. In contrast, TSLP was constitutively expressed by ASCs, but could be further driven by IL-1β and TNFα + IL-17A, findings that are similar to cancer associated fibroblasts (De Monte et al., 2011). What signals localize ILC2s and Th2s to adventitial niches? Lung ILC2s expand postnatally during a period of development that correlates with the first three years of human life, a critical window for the development of allergic asthma in children (Lambrecht and Hammad, 2017). Although ASCs express both TSLP and IL-33, neither of these signals are required for ILC2 or Th2 TRM cell tissue colonization or identity (Ricardo-Gonzalez et al., 2018), suggesting other niche-derived signals may recruit and expand ILC2s (or ILC precursors) and Th2s at these sites. The relationship between ASCs and the stromal cells recently reported to promote fetal-liver derived ILC2 development remains to be determined (Koga et al., 2018), but along with our data raise the possibility that ASCs are involved in ILC2-development and/or postnatal deposition in tissues.
IL-33 and TSLP are both implicated in human studies and multiple mouse models of allergic asthma (Molofsky et al., 2015a). Our data demonstrates that lung ILC2s reside in ASC-rich cuff niches early in development and are distant from epithelial, IL-33 producing type 2 pneumocytes. Although epithelial cells likely contribute to ILC2 activation and the pathogenesis of allergic lung disease, the contribution of IL-33 and TSLP from mesenchymal sources such as ASCs may also be important. Further, given the close proximity of ILC2s to ASCs in multiple tissues, additional ASC-derived contact-dependent or secreted signals are likely involved in ILC2 regulation. Recent studies have identified subsets of stromal cells and their IL-33 production in the regulation of tumor-associated macrophages, pancreatic insulin production, and neuronal synapse remodeling (Dalmas et al., 2017; Yang et al., 2016; Vainchtein et al., 2018), suggesting that stromal-immune crosstalk regulates both physiologic and pathologic tissue immunity. Ultimately, a precise anatomic understanding of the regulation of tissue-resident immune cell niche size, composition, and expansion could have profound implications for a variety of diseases, including allergic asthma, fibrosis, diabetes, cancer, neurodevelopmental disorders, and cardiovascular disease.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be delivered to and will be fulfilled by the Lead Contact, Ari B. Molofsky (ari.molofsky@ucsf.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Red5 (Il5-tdtomato-cre) cytokine reporter mice were used for tracking IL-5-producing cells (Jackson 030926) (Nussbaum et al., 2013). For imaging Red5 mice were crossed to R26-CAG-RFP mice (Ai14) containing a flox-stop-flox sequence upstream of an CAG-RFP-WPRE- cassette in the constitutively expressed ROSA26 (R26) locus (Jackson 007914)). This strain marks all cells that have ever produced Il5-cre with tdtomato expression, serving as an IL-5 lineage-reporter. Additional mice utilized include genetically targeted IL-33mcherry (Vainchtein et al., 2018), PDGFRα-H2B-eGFP nuclear-localized GFP (Jackson 007669), FoxP3eGFP (tm2Tch) bi-cistronic mice (Jackson 006772), IL-7eGFP mice (Miller et al., 2013), IL-33 LacZ mice (Pichery et al., 2012), Gli1-GFP (Gong et al., 2003), Ccl19-Cre (Chai et al., 2013); R26-YFP, and Gli1-cre-ERT2 mice (Jackson 007913) crossed with either R26-DTA (Diptheria toxin A, tamoxifen inducible cell deletion, Jackson 009669), R26-YFP mice (tamoxifen inducible lineage tracking, Jackson 006148), or IL-33 flox mice (Chen et al., 2015). To delete ILC2s, Red5 (Il5-tdtomato-cre) mice were intercrossed with R26-DTA, an approach that specifically deletes ~80% of ILC2s, as described previously (Nussbaum et al., 2013). All genetically targeted reporter strains were used in heterozygous state. Mice were mixed gender animals backcrossed on C57BL/6 for at least 10 generations, or on a mixed genetic background (IL-33 flox, Gli1-creERT2). Mice were maintained in the UCSF specific pathogen–free animal facility in accordance with guidelines established by the Institutional Animal Care and Use Committee and Laboratory Animal Resource Center.
METHOD DETAILS
Flow cytometry
Whole lung was prepared by harvesting lung lobes into 5 ml HBSS with 0.2 mg/ml Liberase TM and 25 μg/ml DNase I, followed by automated tissue dissociation (GentleMacs; Miltenyi Biotec) using the “lung1” program, followed by tissue digestion for 30 min at 37°C with gentle agitation. Samples were subsequently processed on the GentleMacs using the “lung2” program, passed through 70μm filters, washed, and subjected to red blood cell lysis (PharmLyse; BD Biosciences) before final suspension in FACS buffer (PBS, 3% FCS, 0.05% NaN3). ILC2s were identified as lineage negative (CD11b−, CD3ε−, CD4−, CD8α−, CD19−, NK1.1−), FSClow, SSClow, CD45+, Thy1.2 (CD90.2)+, and Gata3hi, IL1RL1 (ST2)+, CD25 (IL-2Rα)+, or KLRG1+, as indicated. In some cases, IL-5Cretdtomato; R26RFP mice were used to identify ILC2 as Lineage-, CD90+ RFP+. CD4+ T cells were identified as FSClow SSClow, CD45+, CD3ε+, CD4+. CD8+ T cells were identified as FSClow SSClow, CD45+, CD3ε+, CD8+. Eosinophils were identified as CD45+, side-scatter high, DAPI-lo, CD11c−, CD11b+, and Siglec F+. Tregs were identified as CD45+ CD3ε+ CD4+ FoxP3+. In some cases, FoxP3eGFP mice were used to identify Treg cell as GFP+ CD3e+ CD4+ cells. DCs were identified as CD45+, CD19−, NK1.1−,SiglecF−, Ly6G−, Ly6C−,CD11c+ and MHCIIhigh. Epithelial cells were identified as CD45−, EpCAM+, CD31−. Endothelial cells were identified as CD45−, CD31+, EpCAM−. Lymphatic endothelial cells were identified as CD45−, CD31+, EpCAM−, Gp38+. Mesenchymal stromal cells were identified as CD45− CD31− EpCAM−, PDGFRα+. In some cases, PDGFRαGFP mice were used to identify mesenchymal cells as CD45− CD31− EpCAM− GFP+. Putative mesothelial cells were identified as CD45− EpCAM− CD31− PDGFRα− Sca1− Gp38hi. Populations were back-gated to verify purity and gating. Samples were analyzed on an LSR II or, for cell sorting, a FACSAria II (both BD Biosciences). Live lymphocytes were gated by DAPI (4′,6-diamidine-2′-phenylindole dihydrochloride; Roche), Draq7 (Biolegend) (extracellular) or Zombie NIR fixable (Biolegend) (intracellular) exclusion, size and granularity based on forward- and side-scatter. Data were analyzed using FlowJo software (TreeStar, USA) and compiled using Prism (Graphpad Software). Cell counts were performed using flow cytometry counting beads (CountBright Absolute; Life Technologies) per manufacturer’s instructions.
Flow cytometry Antibodies
Monoclonal antibodies used for flow cytometry include anti-CD4 (RM4–5, eBioscience and Biolegend), anti-Siglec-F (E50–2440, BD Pharmingen), anti-CD11b (M1/70; Biolegend), anti-F4/80 (BM8; eBioscience), anti-pan-NK (CD49b)(DX5; eBioscience), anti-NK1.1 (PK136, Biolegend or eBioscience or BD Bioscience), anti-FceR1a (MAR-1; eBioscience or Biolegend), anti-CD3ε (17A2, BioLegend or eBioscience), anti-CD8α (53–6.7, Biolegend), anti-CD19 (1D3; BD-Pharmingen or Biolegend), anti-CD25 (IL2Ra, PC61, Biolegend), anti-CD127 (IL7Rα)(A7R34, eBioscience), anti-T1/ST2 (DJ8, MD Biosciences), anti-CD45 (30-F11, Biolegend), anti-KLRG1 (MAFA, Biolegend and eBioscience), and anti-CD90.2 (thy1.2)(30-H12, Biolegend), anti-301b (URA-1, Biolegend), anti-XCR1 (ZET, Biolegend), anti-CD11c (N418, Biolegend), anti-I-A/I-E (MHCII)(M5/114.15.2, Biolegend), anti-Ly6G (1A8, Biolegend), anti-Ly6C (HK1.4, Biolegend), anti-TSLPR (polyclonal, R&D systems). For non-hematopoietic cells, antibodies used include anti-CD31 (390, Biolegend, eBioscience), anti-EpCAM (CD326, G8.8, Biolegend), anti-Ly6A/E (Sca1, D7, eBioscience), anti-Gp38 (podoplanin)(8.1.1, Biolegend), anti-Lyve1 (ALY7, eBioscience), anti-PDGFRα (CD140a, APA5, Biolegend), anti-CD124 (IL4Rα)(REA235, Miltenyi Biotech). Anti-FoxP3 (FJK-16S, eBiosciences), anti-Ki-67 (eBioscience) and anti-Gata3 (TWAJ, eBiosciences) were utilized after first using a fixable live/dead stain (Invitrogen), then fixing and permeabilizing cells per manufacturer’s instructions.
Thin-Section Immunofluorescence microscopy
Animals were anesthetized and injected via intra-cardiac injection with 4% paraformaldehyde (PFA). Tissues were harvested and fixed for 2–4 h in 2% PFA, washed overnight with PBS, cryoprotected with 30% sucrose for 12–36 h, and embedded in OCT (Sakura Finetek) prior to freezing in blocks. For whole-mounts, tissues were fixed as above and imaged after permeabilization with 0.4% triton X and DAPI nuclear counterstaining. Frozen sections were processed on a Leica CM 3050S cryomicrotome (45 μm in VAT, 8 μm all others), dried on slides for 30 min, and kept at −80° C until staining. Tissues were blocked with 5% goat or horse serum (or other serum of secondary antibody) and maintained in PBS + 5% serum + 0.4% triton X throughout antibody treatments. Primary and secondary antibodies were incubated for 1 h at room temperature. Primary antibodies used include goat-anti IL-33 (R&D Systems, 1:100), anti-CD4 (RM4–5, Biolegend, 1:100), chicken anti-GFP (Aves labs, 1:500), rabbit anti-dsRed (Clontech, 1:500), rabbit anti-SMA (Abcam, 1:200), anti-Aqp5 (GR3200850–1, 1:200; Abcam, pAb), anti-Lyve1 (ALY7, 1:500 eBioscience), anti-Ly6A/E (Sca-1, MSCA21, Life Technologies), anti-CD3e (17A2, Biolegend, 1:100), anti-ColIV (diluted 1:200; Abcam), anti-MHCII I-A/I-E (M5/114.15.2, 1:100 eBiosiences), anti-CD11c (N418, Biolegend, 1:100), anti-CD45 (30-F11, Biolegend, 1:100), anti-human/mouse PDGFRα (BD Biosciences, 1:100), anti-PDGFRb (APB5, Biolegend, 1:100). As necessary secondary antibodies were used at 1:1000 dilution at RT for 1 hour, conjugated to A488, A555, and A647 (Life Technologies, Thermo-Fisher). Slides were mounted with Vectashield hardset mounting media. Whole-mount tissue or slides were examined with a Zeiss AxioVision M2 fluorescent microscope.
3D Tissue Preparation and Imaging
Animals were sacrificed by lethal dose injection of avertin followed by cutting of the femoral artery. The lungs were subsequently inflated with 4% paraformaldehyde (PFA) via the trachea followed by clamping the trachea for 5min. The lungs were then removed and further fixed in agitated PFA for 10min. During lung fixation other organs were removed and fixed in 4% PFA overnight at 4C. Fixed tissues were cut into slices of approximately 300um using an in house built vibratome (Compresstome). Tissue slices were blocked, and permeabilized with 0.05% triton X, 10% FBS, and 1% BSA for 1–12hrs at 4C. Tissue was stained in fresh 700μL of 0.05 triton X, +10% FBS, 5% rat serum for primary antibodies, or appropriate species serum for secondary antibodies supplemented with antibodies and agitated at 4° C for 24hrs. For localization of ILC2s to anatomical structures, lung tissues were stained with anti-Lyve1 (ALY7, 1:500 eBioscience) and anti-SMA (clone IA4 1:500; Invitrogen) and in some cases, anti-CD31 (MEC13.3, 1:100, Biolegend). For localization of ILC2 cells to cuff regions the same stain was used with anti-Aquaporin 5 (1:200; Abcam, pAb, GR3200850–1). For localization of ILC2 cells with DCs, tissue slices were stained with anti-MHCII I-A/I-E (M5/114.15.2, 1:400 eBiosiences), anti-CD45 (30-F11, Biolegend, 1:250), anti-CD11c (N418, Biolegend, 1:200). For localization of ILC2s with stromal cell types, anti-SMA, Lyve1, and anti-IL-33 were used as described above. As necessary, species specific secondary antibodies were used (1:1000, Life Technologies) as described above. After overnight staining lung tissue was washed in PBS for 2hrs followed by clearing and mounting in refractive index matching solution (RIMS). For adipose tissue, pancreas, meninges, kidney, and liver sections; slices were dehydrated in an ascending ethanol series (20–100%, 10 min each) and then cleared by soaking in methyl salicylate. All preparations were scanned using a Nikon A1R laser scanning confocal including 405, 488, 561, and 650 laser lines for excitation and imaging with 16X/0.8 or 25X1.1, NA Plan Apo long working distance water immersion objectives. Z steps were acquired every 2.5μm with an average of 200μm from the center of each slice imaged. Large surveys were acquired in resonant mode with pixel sizes of 900nm per pixel. High-resolution images were acquired in galvo mode with pixel sizes of 300nm and 2X frame averaging.
Image Analysis
Imaris Bitplane 8.1 was used for all 3D image analysis with the Imaris Matlab plugin Sortomato. For localization of ILC2s to anatomical structures in the lung colocalization channels were generated to represent different structures; airways (SMA alone), lymphatics (Lyve1 alone), blood vessels (SMA + Lyve1dim). Each new channel was than surfaced and arteries and veins were further split based on anatomical location with arteries running parallel to the bronchioles and veins separated from bronchioles. ILC2s were then surfaced, and nearest neighbor analysis and distance analysis was conducted.
For ILC2 cuff localization in adults, SMA surfaces, Lyve1 surfaces (vessels) and Aquaporin 5 surfaces (parenchyma) were combined and pixels outside of the combined surfaces were masked to create a new channel representing the original empty spaces including, cuffs, lumens and alveolar spaces. Surfaces were analyzed on Sortomato, and non-cuff surfaces could be removed by a combination of volume, sphericity, surface area and distance to SMA.
For localizations of ILC2s with DCs, surfaces were generated on CD45 followed by analysis with Sortomato where DCs were selected as CD11c+ and MHCII+, AMs were selected as CD11c+ and MHCII− with a sphericity greater than 0.8. ILC2s were identified by RFP expression. Localizations were measured using the surfaces interaction function of Sortomato with a 2X optical resolution cut off.
To localize ILC2s with different stromal cell types, colocalization channels were generated between DAPI+ and green+ pixels to represent PDGFRα+ nuclei and DAPI- green+ for SMA. Surfaces could then be generated as described above. To parse PDGFRα subsets, a colocalization channel was made between PDGFRα and anti-IL-33. Surfaces were generated on PDGFRα+ pixels. These surfaces were then sorted based on size and median colocalization of IL-33 and PDGFRα, which effectively excluded parenchymal stromal cells that appeared IL-33+ due to a bright adjacent cell. This reveals the three PDGFRα+ subsets of GFP-large IL-33−, GFP-small IL-33−, and GFP-small IL-33+.
For developmental images, due to small numbers of ILC2s and compact airspaces, ILC2s were hand scored for localization within cuffs by examining their localization relative to the cuff surfaces in serial thin sections from confocal stacks. For distance quantitation for thin-cut sections, analysis was performed with a combination of ImageJ to define individual cells and python3 to measure the shortest distance between cells.
scRNAseq
Cells were purified from lung or adipose tissue and sorted for 1) all CD45− or 2) CD45− IL-33mcherry+ cells and were run concurrently on the 10x Chromium (10X Genomics). The cells were partitioned into Gel Beads in Emulsion in the instrument, where cell lysis and barcoded reverse transcription of RNA occurred, followed by amplification, shearing and 5’ adaptor and sample index attachment. Libraries were sequenced on an Illumina HiSeq 4000.
Data from 10x CellRanger were analyzed using R and the R package Seurat for single cell analysis (Satija et al., 2015). Cells were processed via the Seurat workflow to remove doublets and unwanted sources of variation by removing cells with more than 7000 genes per cell and regressing on number of UMIs. All genes expressed in fewer than three cells were filtered out, as were all cells that expressed fewer than 200 genes. The matrices of data were log normalized in a sparse data matrix and principal component analysis was performed to reduce dimensionality. The first 10 PCA components were used to cluster the cells by Louvain clustering implemented in Seurat while tSNE plots were independently generated to aid in 2D representation of multidimensional data independent of the clustering. This clustering method uses an optimization based on a KNN graph on Euclidean distance in the PCA space and processes the edge weights of pairs of cells based on their local neighbors. The ‘bimod’, likelihood-ratio test for single cell gene expression was used for differential gene analysis between the clusters to determine significant genes (McDavid et al., 2013). In cases where independent data sets were combined, we applied linear regression to the data sets to correct for batch effects prior to running the pipeline described above. Log-normalized gene expression data was used for visualizations with violin plots (VlnPlot), tSNE plots (TSNEPlot), expression comparison plots (FeaturePlot), and generation of heatmaps (pheatmap (1.0.8)).
Induction of type 2 immune responses
For infections, 500 third-stage larvae of N. brasiliensis were injected subcutaneously as described (Molofsky et al., 2015b). Mice were killed at the indicated timepoints and tissues were harvested and analyzed. For cytokine injections, Interleukin-33 (R&D Systems) was given as 500 ng in 0.2 ml PBS i.p. every other day for three doses. Intranasal (i.n.) cytokine treatments was given for seven consecutive days, using 4μg of IL-13 and 2μg of IL-4 (both Peprotech). IL-4 was given in complex with anti-IL-4 mAb (BioXcell, clone 11B11, 10μg). 10ug of papain was given i.n. in a total volume of 40μL for three consecutive days and lungs were analyzed four or eight days from the last treatment.
RNA preparation and qRT-PCR
Indicated populations were sorted into RLT Plus lysis buffer (Qiagen) and stored at −80°C, then processed using Allprep DNA/RNA micro kit (Qiagen) per manufacturer’s protocol. For qPCR analyses, RNA was reverse transcribed using SuperScript III cDNA synthesis kit (ThermoFisher) and amplified using Power SYBR Green PCR master mix (ThermoFisher).
Mesenchymal – ILC2 co-cultures and cytokine analysis
Lungs from wild-type C57Bl/6 mice were harvested, manually dissociated with a razorblade, and subsequently digested with 7.5U/ml Dispase II, 114U/ml Collagenase I and 25μg/ml DNase I in PBS for 30 min at 37°C with gentle agitation. Samples were subsequently 1) washed with DMEM (supplemented with 10% FBS, 50U/mL penicillin and 50 μg /mL streptomycin) to obtain crude fibroblasts using a “walk-out” approach to culture adherent lung stromal cells or 2) passed through 70μm filters, washed, and subjected to red blood cell lysis (PharmLyse; BD Biosciences) before final suspension in FACS buffer (PBS, 3% FCS, 50U/mL penicillin and 50μg/mL streptomycin) to sort of PDGFRα+ Sca1+ and PDGFRα+ Sca1− stroma (>98% purity). Stromal cells were seeded in flatbottomed 96-well plates in 200μl DMEM (supplemented with 10% FBS, 50U/mL penicillin and 50 μg/mL streptomycin) at a density of 12,000 cells per well and allowed to form monolayers over 5–8 days. Lungs from IL-33-injected, IL-5Cretdtomato; R26RFP animals were pooled for ILC2 and T cell purification. Sorted ILC2s (lin−, CD4−, CD3−, CD45+ RFP+), IL-5+ Th2 cells (CD4+, CD3+, CD45+ RFP+), non-Th2 ‘antigen-exposed’ CD4+ T cells (CD4+, CD3+, CD44+, CD45+ RFP−) or non-Th2 ‘naïve’ CD4+ T cells (lin−, CD4+, CD3+, CD44−, CD45+ RFP−, confirmed CD62L+), were seeded onto the stromal monolayers or cultured with conditioned media from the stromal cell cultures in a total volume of 200μl, at a density of 3,000–10,000 ILC2s per well. In indicated experiments, ILC2s were labelled with 2.5uM CellTrace Violet (Invitrogen) to track proliferation. IL-2 (200U/mL), IL-7 (20ng/mL), IL-33 (10ng/mL) were added to culture media as a positive control and culture media only was used as negative control. After 3–8 days of culture, supernatants were collected, and cells were liberated, stained for CD45 and viability dye for dead cell exclusion, and analyzed and enumerated using CountBright Absolute counting beads (Life Technologies) by flow cytometry. In some experiments, wells were imaged by confocal microscopy or stained for AnnexinV to identify apoptotic cells. Indicated cytokines (TSLP (R&D Systems), IL-1β, IL-13, PDGF-BB, TNFα and IL-17A (Peprotech)) were added at a final concentration of 10ng/mL. TSLP and IL-33 were neutralized by addition of 1ug/mL anti-TSLP (R&Dsystems, clone #152614) or 1ug/mL anti-IL-33 (R&D systems, Polyclonal goat IgG), respectively. Amounts of IL-5, IL-7, IL-13, TSLP and IL-33 in cell culture supernatants were analyzed using LEGENDplex (BioLegend) according to the manufacturer’s protocol.
QUANTIFICATION AND STATISTICAL ANALYSIS
All data were analyzed by comparison of means using unpaired two-tailed Student’s t-tests using Prism, or for multiple comparisons ANOVA with HolmSidak post hoc test (GraphPad Software, La Jolla, CA), with * = p<0.05, ** = p<0.01, *** = p<0.001. Figures display means ± standard deviation unless otherwise noted. When possible, results from independent experiments were pooled. All data points reflect individual biological replicates.
Supplementary Material
Highlights.
3D imaging defines ILC2 niches in perivascular regions of multiple tissues.
ILC2s localize with fibroblast-like adventitial stromal cells (ASCs).
Lung ASCs produce IL-33 and TSLP to support ILC2 and Th2s.
ILC2s promote ASC expansion and IL-33 production after helminth infection.
ACKNOWLEDGEMENTS
We thank the UCSF Biological Imaging and Development Core and members Adam Fries and Kyle Marchuk (confocal microscopy and analysis); UCSF Institute for Human Genetics and members Eunice Wan and Dedeepya Vaka (scRNAseq); Satoru Joshita and Marco Colonna (IL-33mcherry mice); Richard M. Locksley (comments and discussion); Z.E. Wang (cell sorting); H.E. Liang (assistance with N. brasiliensis stocks). ABM is supported by the NIDDK (K08DK101604), NLHBI (R56HL142701), the Larry L. Hillblom Foundation, UCSF PBBR grant; SWJ is supported by the American Cancer Society post-doctoral fellowship (PF-15-157-01-CSM); MWD is supported by the Swedish Society for Medical Research and a UCSF Bold & Basic fellowship.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Benias PC, Wells RG, Sackey-Aboagye B, Klavan H, Reidy J, Buonocore D, Miranda M, Kornacki S, Wayne M, Carr-Locke DL, Theise ND, 2018. Structure and Distribution of an Unrecognized Interstitium in Human Tissues. Sci Rep 8, 4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano E, Gebala V, Gerhardt H, 2017. Pericytes or Mesenchymal Stem Cells: Is That the Question? Stem Cell 20, 296–297. [DOI] [PubMed] [Google Scholar]
- Cayrol C, Girard J-P, 2018. Interleukin-33 (IL-33): A nuclear cytokine from the IL-1 family. Immunol. Rev 281, 154–168. [DOI] [PubMed] [Google Scholar]
- Chai Q, Onder L, Scandella E, Gil-Cruz C, Perez-Shibayama C, Cupovic J, Danuser R, Sparwasser T, Luther SA, Thiel V, Rülicke T, Stein JV, Hehlgans T, Ludewig B, 2013. Maturation of Lymph Node Fibroblastic Reticular Cells from Myofibroblastic Precursors Is Critical for Antiviral Immunity. Immunity 38, 1013–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JE, Turley SJ, 2015. Stromal infrastructure of the lymph node and coordination of immunity. Trends Immunol 36, 30–39. [DOI] [PubMed] [Google Scholar]
- Chen W-Y, Hong J, Gannon J, Kakkar R, Lee RT, 2015. Myocardial pressure overload induces systemic inflammation through endothelial cell IL-33. Proc Natl Acad Sci USA 112, 201424236–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corselli M, Chen C-W, Sun B, Yap S, Rubin JP, Péault B, 2012. The tunica adventitia of human arteries and veins as a source of mesenchymal stem cells. Stem Cells Dev. 21, 1299–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmas E, Lehmann FM, Dror E, Wueest S, Thienel C, Borsigova M, Stawiski M, Traunecker E, Lucchini FC, Dapito DH, Kallert SM, Guigas B, Pattou F, Kerr-Conte J, Maechler P, Girard J-P, Konrad D, Wolfrum C, Böni-Schnetzler M, Finke D, Donath MY, 2017. Interleukin-33-Activated Islet-Resident Innate Lymphoid Cells Promote Insulin Secretion through Myeloid Cell Retinoic Acid Production. Immunity 47, 928–942.e7. [DOI] [PubMed] [Google Scholar]
- de Kleer IM, Kool M, de Bruijn MJW, Willart M, van Moorleghem J, Schuijs MJ, Plantinga M, Beyaert R, Hams E, Fallon PG, Hammad H, Hendriks RW, Lambrecht BN, 2016. Perinatal Activation of the Interleukin-33 Pathway Promotes Type 2 Immunity in the Developing Lung. Immunity 45, 1285–1298. [DOI] [PubMed] [Google Scholar]
- De Monte L, Reni M, Tassi E, Clavenna D, Papa I, Recalde H, Braga M, Di Carlo V, Doglioni C, Protti MP, 2011. Intratumor T helper type 2 cell infiltrate correlates with cancer-associated fibroblast thymic stromal lymphopoietin production and reduced survival in pancreatic cancer. J. Exp. Med 208, 469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durai V, Murphy KM, 2016. Functions of Murine Dendritic Cells. Immunity 45, 719–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo Y, Hirahara K, Iinuma T, Shinoda K, Tumes DJ, Asou HK, Matsugae N, Obata-Ninomiya K, Yamamoto H, Motohashi S, Oboki K, Nakae S, Saito H, Okamoto Y, Nakayama T, 2015. The interleukin-33-p38 kinase axis confers memory T helper 2 cell pathogenicity in the airway. Immunity 42, 294–308. [DOI] [PubMed] [Google Scholar]
- Gerner MY, Kastenmuller W, Ifrim I, Kabat J, Germain RN, 2012. Histocytometry: a method for highly multiplex quantitative tissue imaging analysis applied to dendritic cell subset microanatomy in lymph nodes. Immunity 37, 364–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N, 2003. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 425, 917–925. [DOI] [PubMed] [Google Scholar]
- Guo L, Huang Y, Chen X, Hu-Li J, Urban JF, Paul WE, 2015. Innate immunological function of TH2 cells in vivo. Nat Immunol 16, 1051–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halim TYF, MacLaren A, Romanish MT, Gold MJ, McNagny KM, Takei F, 2012. Retinoic-acid-receptor-related orphan nuclear receptor alpha is required for natural helper cell development and allergic inflammation. Immunity 37, 463–474. [DOI] [PubMed] [Google Scholar]
- Halim TYF, Rana BMJ, Walker JA, Kerscher B, Knolle MD, Jolin HE, Serrao EM, Haim-Vilmovsky L, Teichmann SA, Rodewald H-R, Botto M, Vyse TJ, Fallon PG, Li Z, Withers DR, McKenzie ANJ, 2018. Tissue-Restricted Adaptive Type 2 Immunity Is Orchestrated by Expression of the Costimulatory Molecule OX40L on Group 2 Innate Lymphoid Cells. Immunity 48, 1195–1207.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Wang R, Zhou Y, Fei L, Sun H, Lai S, Saadatpour A, Zhou Z, Chen H, Ye F, Huang D, Xu Y, Huang W, Jiang M, Jiang X, Mao J, Chen Y, Lu C, Xie J, Fang Q, Wang Y, Yue R, Li T, Huang H, Orkin SH, Yuan G-C, Chen M, Guo G, 2018. Mapping the Mouse Cell Atlas by Microwell-Seq. Cell 172, 1091–1107.e17. [DOI] [PubMed] [Google Scholar]
- Heredia JE, Mukundan L, Chen FM, Mueller AA, Deo RC, Locksley RM, Rando TA, Chawla A, 2013. Type 2 innate signals stimulate fibro/adipogenic progenitors to facilitate muscle regeneration. Cell 153, 376–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwagi M, Hosoi J, Lai J-F, Brissette J, Ziegler SF, Morgan BA, Georgopoulos K, 2017. Direct control of regulatory T cells by keratinocytes. Nat Immunol 18, 334–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose CSN, Artis D, 2016. Innate lymphoid cells as regulators of immunity, inflammation and tissue homeostasis. Nat Immunol 17, 765–774. [DOI] [PubMed] [Google Scholar]
- Koga S, Hozumi K, Hirano K-I, Yazawa M, Terooatea T, Minoda A, Nagasawa T, Koyasu S, Moro K, 2018. Peripheral PDGFRα +gp38 +mesenchymal cells support the differentiation of fetal liver–derived ILC2. J Exp Med 278, jem.20172310–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramann R, Schneider RK, DiRocco DP, Machado F, Fleig S, Bondzie PA, Henderson JM, Ebert BL, Humphreys BD, 2015. Perivascular Gli1+ progenitors are key contributors to injury-induced organ fibrosis. Cell Stem Cell 16, 51–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto Y, Linehan M, Weinstein JS, Laidlaw BJ, Craft JE, Iwasaki A, 2013. CD301b⁺ dermal dendritic cells drive T helper 2 cell-mediated immunity. Immunity 39, 733–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrecht BN, Hammad H, 2017. The immunology of the allergy epidemic and the hygiene hypothesis. Nat Immunol 18, 1076–1083. [DOI] [PubMed] [Google Scholar]
- McDavid A, Finak G, Chattopadyay PK, Dominguez M, Lamoreaux L, Ma SS, Roederer M, Gottardo R, 2013. Data exploration, quality control and testing in single-cell qPCR-based gene expression experiments. Bioinformatics 29, 461–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CN, Hartigan-O’Connor DJ, Lee MS, Laidlaw G, Cornelissen IP, Matloubian M, Coughlin SR, McDonald DM, McCune JM, 2013. IL-7 production in murine lymphatic endothelial cells and induction in the setting of peripheral lymphopenia. Int. Immunol. 25, 471–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra A, Van Dyken SJ, Schneider C, Nussbaum JC, Liang H-E, Locksley RM, 2016. Group 2 innate lymphoid cells utilize the IRF4-IL-9 module to coordinate epithelial cell maintenance of lung homeostasis. Mucosal Immunol 9, 275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AB, Nussbaum JC, Liang H-E, Van Dyken SJ, Cheng LE, Mohapatra A, Chawla A, Locksley RM, 2013. Innate lymphoid type 2 cells sustain visceral adipose tissue eosinophils and alternatively activated macrophages. J. Exp. Med 210, 535–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AB, Savage AK, Locksley RM, 2015a. Interleukin-33 in Tissue Homeostasis, Injury, and Inflammation. Immunity 42, 1005–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AB, Van Gool F, Liang H-E, Van Dyken SJ, Nussbaum JC, Lee J, Bluestone JA, Locksley RM, 2015b. Interleukin-33 and Interferonγ Counter-Regulate Group 2 Innate Lymphoid Cell Activation during Immune Perturbation. Immunity 43, 161–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussbaum JC, Van Dyken SJ, Moltke, von J, Cheng LE, Mohapatra A, Molofsky AB, Thornton EE, Krummel MF, Chawla A, Liang H-E, Locksley RM, 2013. Type 2 innate lymphoid cells control eosinophil homeostasis. Nature 502, 245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oboki K, Ohno T, Kajiwara N, Arae K, Morita H, Ishii A, Nambu A, Abe T, Kiyonari H, Matsumoto K, Sudo K, Okumura K, Saito H, Nakae S, 2010. IL-33 is a crucial amplifier of innate rather than acquired immunity. Proc Natl Acad Sci USA 107, 18581–18586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham M, Sakhalkar H, Oliver T, Allan Johnson G, Dewhirst M, 2008. Optical clearing of unsectioned specimens for three-dimensional imaging via optical transmission and emission tomography. J. Biomed. Opt 13, 021113–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panduro M, Benoist C, Mathis D, 2016. Tissue Tregs. Annu. Rev. Immunol 34, 609–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichery M, Mirey E, Mercier P, Lefrancais E, Dujardin A, Ortega N, Girard J-P, 2012. Endogenous IL-33 is highly expressed in mouse epithelial barrier tissues, lymphoid organs, brain, embryos, and inflamed tissues: in situ analysis using a novel Il-33-LacZ gene trap reporter strain. J. Immunol. 188, 3488–3495. [DOI] [PubMed] [Google Scholar]
- Ricardo-Gonzalez RR, Van Dyken SJ, Schneider C, Lee J, Nussbaum JC, Liang H-E, Vaka D, Eckalbar WL, Molofsky AB, Erle DJ, Locksley RM, 2018. Tissue signals imprint ILC2 identity with anticipatory function. Nat Immunol 19, 1093–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodda LB, Lu E, Bennett ML, Sokol CL, Wang X, Luther SA, Barres BA, Luster AD, Ye CJ, Cyster JG, 2018. Single-Cell RNA Sequencing of Lymph Node Stromal Cells Reveals Niche-Associated Heterogeneity. Immunity 48, 1014–1028.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saluzzo S, Gorki A-D, Rana BMJ, Martins R, Scanlon S, Starkl P, Lakovits K, Hladik A, Korosec A, Sharif O, Warszawska JM, Jolin H, Mesteri I, McKenzie ANJ, Knapp S, 2017. First-Breath-Induced Type 2 Pathways Shape the Lung Immune Environment. Cell Reports 18, 1893–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, Regev A, 2015. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 33, 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schraufnagel DE, Agaram NP, Faruqui A, Jain S, Jain L, Ridge KM, Sznajder JI, 2003. Pulmonary lymphatics and edema accumulation after brief lung injury. Am J Physiol Lung Cell Mol Physiol 284, L891–L897. [DOI] [PubMed] [Google Scholar]
- Schuijs MJ, Halim TYF, 2018. Group 2 innate lymphocytes at the interface between innate and adaptive immunity. Ann N Y Acad Sci. [DOI] [PubMed] [Google Scholar]
- Shinoda K, Hirahara K, Iinuma T, Ichikawa T, Suzuki AS, Sugaya K, Tumes DJ, Yamamoto H, Hara T, Tani-ichi S, Ikuta K, Okamoto Y, Nakayama T, 2016. Thy1+IL-7+ lymphatic endothelial cells in iBALT provide a survival niche for memory T-helper cells in allergic airway inflammation. Proc Natl Acad Sci USA 113, E2842–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitnik KM, Wendland K, Weishaupt H, Uronen-Hansson H, White AJ, Anderson G, Kotarsky K, Agace WW, 2016. Context-Dependent Development of Lymphoid Stroma from Adult CD34(+) Adventitial Progenitors. Cell Reports 14, 2375–2388. [DOI] [PubMed] [Google Scholar]
- Stenmark KR, Yeager ME, Kasmi El, K.C., Nozik-Grayck E, Gerasimovskaya EV, Li M, Riddle SR, Frid MG, 2013. The Adventitia: Essential Regulator of Vascular Wall Structure and Function. Annu Rev Physiol 75, 23–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainchtein ID, Chin G, Cho FS, Kelley KW, Miller JG, Chien EC, Liddelow SA, Nguyen PT, Nakao-Inoue H, Dorman LC, Akil O, Joshita S, Barres BA, Paz JT, Molofsky AB, Molofsky AV, 2018. Astrocyte-derived interleukin-33 promotes microglial synapse engulfment and neural circuit development. Science 359, 1269–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyken SJ, Mohapatra A, Nussbaum JC, Molofsky AB, Thornton EE, Ziegler SF, McKenzie ANJ, Krummel MF, Liang H-E, Locksley RM, 2014. Chitin activates parallel immune modules that direct distinct inflammatory responses via innate lymphoid type 2 and γδ T cells. Immunity 40, 414–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyken SJ, Nussbaum JC, Lee J, Molofsky AB, Liang H-E, Pollack JL, Gate RE, Haliburton GE, Ye CJ, Marson A, Erle DJ, Locksley RM, 2016. A tissue checkpoint regulates type 2 immunity. Nat Immunol 17, 1381–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivier E, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie ANJ, Mebius RE, Powrie F, Spits H, 2018. Innate Lymphoid Cells: 10 Years On. Cell 174, 1054–1066. [DOI] [PubMed] [Google Scholar]
- Yang Y, Andersson P, Hosaka K, Zhang Y, Cao R, Iwamoto H, Yang X, Nakamura M, Wang J, Zhuang R, Morikawa H, Xue Y, Braun H, Beyaert R, Samani N, Nakae S, Hams E, Dissing S, Fallon PG, Langer R, Cao Y, 2016. The PDGF-BB-SOX7 axis-modulated IL-33 in pericytes and stromal cells promotes metastasis through tumour-associated macrophages. Nature Communications 7, 11385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler SF, 2012. Thymic stromal lymphopoietin and allergic disease. J Allergy Clin Immunol 130, 845–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.