Bruce Baker's laboratory made a huge impact on our understanding of Drosophila sex determination mechanisms. To celebrate these accomplishments, members of Bruce's laboratory describe the trailblazing science that led to mechanistic understanding of how sex differences...
Keywords: behavior, development, doublesex, Drosophila, fruitless, genital disc, mRNA splicing, sex, sex determination, transformer
Abstract
Bruce Baker, a preeminent Drosophila geneticist who made fundamental contributions to our understanding of the molecular genetic basis of sex differences, passed away July 1, 2018 at the age of 72. Members of Bruce’s laboratory remember him as an intensely dedicated, rigorous, creative, deep-thinking, and fearless scientist. His trainees also remember his strong commitment to teaching students at every level. Bruce’s career studying sex differences had three major epochs, where the laboratory was focused on: (1) sex determination and dosage compensation, (2) the development of sex-specific structures, and (3) the molecular genetic basis for sex differences in behavior. Several members of the Baker laboratory have come together to honor Bruce by highlighting some of the laboratory’s major scientific contributions in these areas.
THE discovery of sex chromosomes and their sex-determining roles, made over a century ago, opened the door to our current understanding of how sex differences arise at the molecular level [reviewed in Miko (2008)]. In the early to mid-20th century, the finding that single-gene mutations could cause sexual transformations lead to the idea that sex chromosomes and genic control are both important for sex determination [review for Drosophila genes: Baker and Belote (1983)]; however, the tools to go from genetic mutations to molecular mechanism had yet to be developed. It would not be until the late 1970s and early 1980s that the molecular genetic mechanisms of sex determination in animals would be revealed through the work of a small handful of laboratories studying Drosophila melanogaster and Caenorhabditis elegans. Bruce Baker’s laboratory entered the sex determination field at this time, switching focus from the genetic analysis of Drosophila chromosome mechanics to Drosophila sex determination. One major question in the field was how sex chromosome differences direct the formation of secondary sexual characteristics—both morphological and behavioral—at the molecular level. Another major question was how expression of X-linked genes are equalized between males and females, which differ in X chromosome number. In Drosophila, it was known that the number of X chromosomes was the primary determinant of sex—two in females and one in males—rather than the presence of a Y chromosome, which is the primary determinant of sex in humans. There were also known mutations that caused an animal that is chromosomally female (XX) to look and behave like a male, as well as mutations that caused intersexual phenotypes. Bruce, an expert geneticist, carried out epistasis analyses with the existing sex determination mutations to establish the order of gene activity. His laboratory subsequently used molecular biology to gain a mechanistic understanding of how each step of the pathway impacted the next. The molecular mechanisms underlying sex differences, which were often quite surprising, are now fully integrated into the scientific canon, appearing in almost all genetics textbooks. This perspective highlights the contributions of Bruce Baker’s laboratory to our nearly complete understanding of how Drosophila sex chromosome number is interpreted to activate downstream pathways controlling both the morphological and behavioral differences between the sexes, and X chromosome dosage compensation.
Sex Determination and Dosage Compensation in Drosophila
The title of Bruce Baker’s first publication on sex determination “Sex and the Single Cell” had been inspired by the book “Sex and the Single Girl” by Helen Gurley Brown, which sold two million copies in 3 weeks. Though not that kind of blockbuster, Bruce’s paper was the cornerstone of a research program that extended for almost 4 decades, establishing the critical role of genetics in codifying regulatory pathways (Baker and Ridge 1980). Key findings revealed in this study were: (1) that the four genes known at the time to cause dramatic sex-transformation phenotypes in Drosophila—transformer (tra), transformer-2 (tra-2), doublesex (dsx), and intersex (ix)—function in a cell-autonomous manner, unlike mammalian sex determination where diffusible hormones play critical roles; (2) that these genes function in a shared genetic pathway (Figure 1, A and B); (3) that the default product of the bifunctional dsx gene is the male-specifying form; (4) that tra and tra-2 act to convert products of the bifunctional dsx gene to the female-specifying form; and (5) that ix works in concert with the female product encoded by dsx to establish and maintain sexual dimorphism. The cloning and molecular characterization of each of these genes confirmed every aspect of this genetics-based pathway. The surprise would come in the discovery of the molecular mechanism by which the products of the bifunctional dsx gene are converted by its upstream regulators into a female modus operandi.
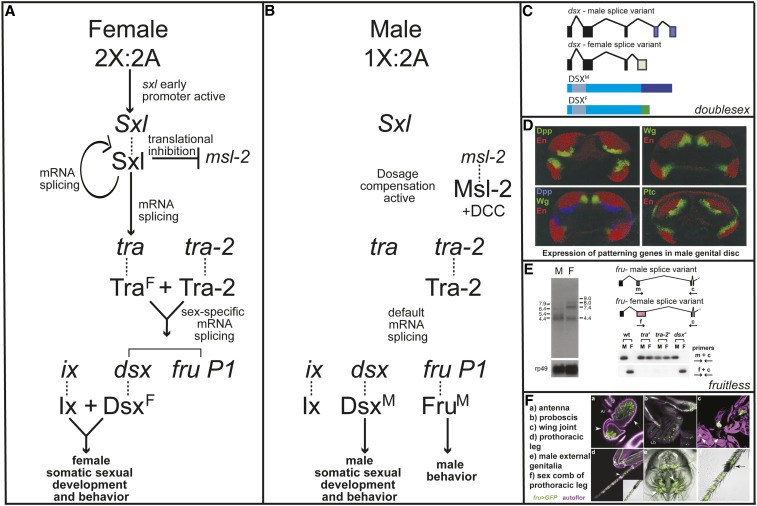
Figure 1.
Sex and the single fly: the somatic sex determination hierarchy. Bruce Baker and his laboratory, as well colleagues in the field, made many key discoveries about the genetic pathway that directs somatic sexual development. (A) Females with two X chromosomes produce Sxl, a pre-mRNA splicing factor. Sxl regulates the splicing of its own pre-mRNA and the tra pre-mRNA, resulting in the production of Sxl and TraF. TraF and Tra-2 regulate the pre-mRNA splicing of doublesex (dsx) and fruitless (fru; the product of the P1 promoter), resulting in the production of female-specific (DsxF) that binds Ix; the Dsx/Ix complex regulates transcription in females. Sxl also inhibits translation of msl-2, so dosage compensation is not active in females. (B) Males with one X chromosome do not produce Sxl. dsx and fru pre-mRNAs are spliced by the default pathway, resulting in the production of male-specific transcription factors DsxM and FruM. In males, DsxM does not require ix to direct male development. Dosage compensation is active in males, resulting in upregulation of genes that reside on the X chromosome in males, due to translation of msl-2. Msl-2 acts with a set of protein and noncoding RNAs (dosage compensation complex; DCC) to mediate upregulation of gene expression on the male X chromosome. In both males and females, Dsx directs somatic sex-specific development and aspects of behaviors. In males, FruM is required to specify the potential for male reproductive behaviors. (C) Alternative pre-mRNA splicing of dsx transcripts produces sex-specific transcription factors that share a common N-terminal region, containing the DNA-binding domain, but differ in the C-termini. This image is based on the original discovery (Burtis and Baker 1989). (D) The somatic sex determination hierarchy and conserved developmental patterning genes coordinate to bring about sex differences in genital morphology [reproduced from Chen and Baker (1997)]. Expression of patterning genes in the male genital disc is shown. Top is Engrailed (En) antibody staining (red), and decapentaplegic (dpp)-lacZ or wingless (wg)-LacZ reporter expression (green). Bottom left is En (red) and Wg (green) antibody staining, and dpp-LacZ reporter expression (blue). Bottom right is En antibody staining (red) and patched (ptc)-LacZ reporter expression (green). lacZ expression was detected with an anti-β-galactosidase antibody. (E) fruitless produces sex-specific transcripts due to sex hierarchy-regulated alternative pre-mRNA splicing. On the left, an RNA blot, with polyA(+) RNA from adult male and female heads; sex-specific transcripts are detected using a probe made from the ‘Broad-complex, Tramtrack, Bric-a-brac’ (BTB) domain coding region of a fru cDNA. The region of alternative pre-mRNA splicing of fru P1 transcripts is shown at the top right; splicing in females occurs downstream of where splicing occurs in males, due to sex hierarchy regulation. Primers used in RT-PCR are shown. Sex-specific splicing is downstream of tra and tra-2, but not dsx, as detected by Southern blot analysis of RT-PCR products [reproduced from Ryner et al. (1996)]. (F) fruitless is expressed in both the peripheral and central nervous system, in neurons important for reproductive behaviors in males and females [reproduced from Manoli et al. (2005)]. fruM-Gal4 drives expression of membrane-GFP (green) and autofluorescence (magenta). (a–f) Expression of fruM in olfactory neurons in the antenna (a), in gustatory neurons of the proboscis (b), in proprioceptive neurons in the wing joint (c), in gustatory and mechanosensory neurons of the prothoracic leg (d), in neurons of the male external genitalia (e), and in gustatory and mechanosensory neurons at base of male sex comb (f).
It is not clear what upstream events diverted Bruce’s research efforts toward the study of sex determination and dosage compensation. His graduate work had focused on meiosis, training under Larry Sandler at the University of Washington who, along with Bruce’s father William Baker, another prominent Drosophila biologist, had major influences on Bruce’s approach to science and to training the next generation of scientists. During his early years as an assistant professor, first at the University of North Carolina Chapel Hill and then at the University of California San Diego, Bruce had focused on mitosis, recombination, mutagenesis, and chromosome mechanics, working with Maurizio Gatti, Sergio Pimpinelli, and David Smith, among others. One theory about Bruce’s move into the sex determination field was shared with us by Adam Wilkins, who, like Bruce, attended Reed College as an undergraduate. “I was a year ahead of Bruce and took the course in genetics the year before he did. One of the lab exercises was a “Drosophila unknown” that pairs of students had to solve. Our teacher, Stephen Karashian, gave my partner and me the one that he said was the toughest. It was tra and, believe me, it was interesting and fun to solve; neither of us had heard of, or imagined, this kind of sexual transformation. The next year, Bruce and his partner, Ann Roman (the daughter of Herschel Roman, the founder and first chairman of the Department of Genetics at the University of Washington), were given the same unknown, with Dr. Karashian again telling them that it was the hardest. Of course, they solved it too. But might this have sown the seed of interest in matters of sex determination, gender-specific behavior, etc. for Bruce?”
Adelaide Carpenter, Bruce’s significant other through graduate school, and his postdoctoral and early years as an independent investigator, says Bruce demonstrated “no particular interest in sex determination or dosage compensation during [their] early years together.” She is unsure about what diverted Bruce into the sex determination/dosage compensation field, but her best hypothesis is that his interest could have emerged from their observation that in gynandromorphs (flies that are part female and part male because of loss of one of the two X chromosomes during development) only either very large or very small male clones are ever observed, when X chromosome loss should be equal across cell divisions. A third more speculative theory is that Bruce had devised a clever genetic screen for obtaining pure populations of sex-specific larvae and pupae at biochemical levels, and just needed to study a pathway where such a tool would be useful (Baker 1973), although this idea may be a bit too whimsical for how Bruce approached science. We will never know exactly why Bruce dove into the genetics of sex and dosage compensation, but his doing so opened up a new, exciting, and enduring area of investigation for him and his trainees, many of whom continue to address related questions in their own laboratories.
Following the publication of Baker and Ridge (1980), Bruce was on a mission to clone all four sex determination genes. This task that would take > 20 years to complete and would also involve the molecular characterization of another key regulator of sex determination, fruitless (fru), a gene that controls sex differences in behavior, and whose activity state is also regulated by tra and tra-2. Cloning of Drosophila genes was no small feat in the early days of molecular biology. The process often involved taking an already cloned gene coupled with nearby chromosomal aberrations to obtain DNA “close” to the gene of interest. A chromosomal walk would then ensue wherein one would screen for and map overlapping pieces of genomic DNA, phage-clone by phage-clone, to move into and across the molecularly defined interval containing the gene of interest. Chromosomal walking also often involved generating new mutant alleles to help position clones within the chromosome walk.
Thus, the expertise of both classical geneticists and molecular biologists was required; whereas Bruce had all the genetics experience necessary, he had no molecular expertise. So, in addition to John Belote, another expert geneticist who had trained in John Lucchesi’s laboratory, Bruce brought on Mariana Wolfner, a postdoctoral scholar who had trained in the laboratory of David Hogness. Together, they began to clone the dsx gene, with Bruce learning molecular biology at the bench from Mariana. Bruce then brought on another molecular biologist postdoctoral scholar, Michael McKeown, who trained in the laboratory of Rick Firtel, and who began the cloning of tra and tra-2. The inexperienced new graduate students who joined the laboratory around this time (Geoff Carson, Nelson Scott, and Deborah Andrew) were assigned more peripheral, albeit related, projects, a move that in retrospect served to not only allow these neophytes to develop a sense of scientific autonomy but that also did not impede progress.
The cloning of tra, the first of the sex determination genes to be reported, was done in both Bruce’s laboratory and that of Rolf Nöthiger, with the corresponding publications coming out within 2 months of one another (Butler et al. 1986; McKeown et al. 1987). The loss-of-function tra phenotype is the complete transformation of chromosomally XX flies into phenotypic males, with no effect on chromosomally XY males. The molecular characterization of tra revealed that transcripts of ∼1.2 kb existed in both sexes, but in addition, there was a ∼1-kb female-specific transcript. This finding suggested that either alternative transcriptional start sites or alternative mRNA processing might be at the root of the differences. The next year, Michael McKeown’s newly formed laboratory at the Salk Institute, along with Bruce and John Belote, reported on an alternatively spliced form of tra, the only transcript isoform that encoded a significant open reading frame (Boggs et al. 1987). They also demonstrated that driving expression of tra with a heat shock promoter was sufficient to rescue gene function, ruling out tra regulation through sex-specific transcriptional events.
Within a year of the reports on the cloning of tra, the cloning of dsx—a gene whose loss in both chromosomally XX and XY results in intersexual flies, with both male and female morphological features—was reported. The study uncovered developmentally regulated, sex-specific dsx transcripts, which in this study were detected during the pupal stages, when the remodeling that gives rise to the adult sexually dimorphic phenotypes occurs (Baker and Wolfner 1988).
As the cloning of both tra and dsx was being completed (1986), Bruce moved from the position and laboratory he had shared with Adelaide Carpenter at the University of California San Diego to Stanford University, where he was joined by a larger team of postdoctoral scholars and students. Many in this group would continue the charge of elucidating the molecular mechanisms of sex determination; others would focus on the cloning and characterization of genes required in males for dosage compensation.
Another major intellectual step was the cloning and characterization of Sex lethal (Sxl) by Thomas Cline’s laboratory [reviewed in Cline (2005)]. Sxl was shown to function upstream of both sex determination and dosage compensation. With molecular knowledge of tra, dsx, and Sxl, much of the regulatory interactions among the pathway members began to quickly unfold. Rod Nagoshi, and others from the Baker laboratory, showed that tra splicing was not affected by loss of tra-2 or dsx, and that female-specific splicing of dsx requires Sxl, tra, and tra-2, but not ix (Nagoshi et al. 1988). The Cline laboratory discovered that Sxl also undergoes sex-specific RNA splicing and that the Sxl protein has sequence similarity to RNA-binding proteins (Bell et al. 1988). Altogether, the findings suggested a model wherein Sxl regulates its own pre-mRNA splicing and that of downstream genes in the sex determination pathway, providing a simple molecular mechanism for both Sxl autoregulation that had been uncovered through genetic studies in Thomas Cline’s laboratory and Sxl regulation of downstream genes (Cline 1983). A series of key splicing events were at the core of Drosophila sex determination, quite a novel and exciting mechanism for a time when most of those in the field expected the differences to be at either the transcriptional, or the translational, level.
The cloning of tra-2 and the discovery that the Tra-2 protein is related to known RNA-binding proteins, by the Baker and Nöthiger laboratories (Amrein et al. 1988; Goralski et al. 1989), further supported this mode of gene regulation. The cloning and sequencing of dsx cDNAs revealed that the male and female transcript isoforms encode proteins with a common N-terminus, which contains the DNA-binding domain, but with distinct C-termini (Figure 1C), providing a molecular explanation for the bifunctionality of Dsx (Burtis and Baker 1989). Detailed mapping of the various lesions in the dsx gene revealed that sequences in the female-specific exons functioned with Tra and Tra-2 to divert the splicing machinery away from use of the default consensus splice acceptor sites. Dsx was subsequently revealed to function as a transcription factor through experiments showing that both the male and female Dsx protein isoforms can directly bind yolk protein gene enhancers, with female-specific Dsx activating transcription and male-specific Dsx repressing transcription (Burtis and Baker 1989; Burtis et al. 1991). Direct regulation of dsx splicing by Tra and Tra-2 in S2 cells confirmed the proposed mechanisms for dsx regulation (Ryner and Baker 1991). It would take another 11 years for the cloning of ix, which revealed its function as a transcriptional coactivator that physically interacts with regions present only in the Dsx female-specific isoform (Garrett-Engele et al. 2002).
By the time Bruce’s very “punny”-titled review in Nature came out “Sex in Flies: the Splice of Life” (Baker 1989), the Baker laboratory was already embarking on the cloning and molecular characterization of the male-specific lethal (msl) loci, with the expectation that the activity of these genes could be controlled by Sxl, possibly through splicing. This prediction would turn out to be true, just not exactly. The Baker laboratory cloned and characterized three of the original four known msl genes: male lethal (mle) (Kuroda et al. 1991), male-specific lethal-3 (msl-3) (Gorman et al. 1995), and male-specific lethal-2 (msl-2) (Bashaw and Baker 1995). The fourth msl—male-specific lethal-1—was cloned and characterized in the laboratory of Mitzi Kuroda, after she had established her own laboratory at Baylor College of Medicine (Palmer et al. 1993). mle was found to encode a protein with homology to RNA–DNA helicases, with transcripts and proteins of the same size, and abundance, in males and females. However, MLE accumulated on only the X chromosomes of males and not females in preparations of salivary gland polytene chromosomes, suggesting a direct role for MLE in male X chromosome hyper-transcription. The characterization of msl-1 and msl-3 also revealed no differences in transcript size or levels, and no differences in protein size between the two sexes; however, both proteins were much less abundant in females. All three proteins showed strong sex-specific localization to hundreds of sites along the male X chromosomes, dependent on the presence of the other MSLs and the absence of Sxl [Gorman et al. (1993); reviewed in Baker et al. (1994), Gorman and Baker (1994), and Gorman et al. (1995)]. These findings supported previous ideas that the MSLs were likely to function as a complex to control X chromosome transcription levels, but provided no molecular link to Sxl.
The cloning of male-specific lethal-2 (msl-2), the last of the four msl genes (known at the time) to be molecularly characterized, would provide the link between MSLs and Sxl (Bashaw and Baker 1995). The male msl-2 transcript is larger than the female transcript and splicing of the female-specific form requires Sxl. Interestingly, although Sxl-dependent splicing removes sequences in only the 5′-UTR, MSL-2 protein is produced only in males. Follow-up studies revealed that Sxl acts directly to block the translation of msl-2 through its binding to poly-U sequences present in both the 5′- and 3′-UTRs (Bashaw and Baker 1996). This showed that Sxl has two molecular functions: splicing and translational regulation, two sites of action: the nucleus and cytoplasm, and jobs in two different pathways: sex determination and dosage compensation.
Other discoveries on dosage compensation in flies that were published around this time include the Kuroda laboratory’s finding that an acetylated histone (H4Ac16) also associates with MSLs on the male X chromosome (Bone et al. 1994), and the identification and cloning of a new msl, males on first (mof), which encodes the histone acetylase that generates H4Ac16 (Hilfiker et al. 1994; Gu et al. 2000). Two long noncoding RNAs were discovered that associate with the male X chromosome, the roX1 and roX2 RNAs (Meller et al. 1997). The Baker laboratory would demonstrate that these RNAs colocalize with the MSLs and are redundantly required for the association of the MSL proteins to the male X chromosome (Franke and Baker 1999), and that the X chromosome can recruit the dosage complex machinery in the absence of a set of experimentally defined entry sites (Fagegaltier and Baker 2004). The Kuroda and Meller laboratories would uncover more details on the regulation, function, and molecular mechanisms by which these roX RNAs contribute to dosage compensation (Meller and Kuroda 2002; Park et al. 2002, 2003, 2005; Meller 2003; Stuckenholz et al. 2003; Rattner and Meller 2004; Deng and Meller 2006). Only a few additional papers from the Baker laboratory on dosage compensation appear after the turn of the century. Bruce’s attention was now focused on fru, and discovering how the transcriptional regulators at the end of the pathway control sex-specific differentiation and neuronal wiring.
The Development of Sexually Dimorphic Structures and the Evolution of Sex
By the 1990s, with most of the core genes of the sex determination pathway identified and characterized, Bruce focused on two additional major questions: how does the sex determination pathway direct the development of sexually dimorphic morphologies and how did sex differences evolve? Even in his first paper on sex determination (Baker and Ridge 1980), Bruce recognized that studying sex determination would lead to insights about developmental mechanisms. He wrote “Since sex determination affects the developmental fate of numerous organ primordia, information as to the nature of the genetic events involved in sex determination should contribute not only to our understanding of sex determination, but also to the elucidation of the mechanisms by which eukaryotes effect the expression of alternative developmental pathways.” To address these questions, Bruce’s laboratory began work in three general areas: (1) a molecular–genetic investigation of genital imaginal disc development; (2) genome-wide approaches to find downstream targets of the transcription factors found at the end of the sex determination pathway, Dsx and FruM; and (3) an examination of the evolutionary conservation of genes in the sex hierarchy.
Drosophila adult tissues arise from imaginal discs, sac-like structures that form during embryogenesis and develop inside the larva. Bruce chose to study development of the genital imaginal disc because it gives rise to the most sexually dimorphic structures in the adult fly, the internal and external genitalia. Bruce recruited three graduate students, Elizabeth Chen, Shaad Ahmad, and Eric Keisman, and later a postdoctoral scholar, Audrey Christiansen, to work on the genital disc project. These studies began during an exciting period for developmental biologists, when critical genes controlling both embryonic and imaginal disc patterning were being discovered and characterized. What was unclear was how the activities of the patterning genes intersected with those of the sex determination hierarchy to give rise to adult sexual dimorphism. The genital disc is different from the other imaginal discs in that it is a compound disc with three distinct primordia: cells that give rise to the female genitalia, male genitalia, and the analia. While colleagues including Nöthiger, Schüpbach, and Wieschaus had identified many of the fundamental principles regarding the organization of the genital disc (Nöthiger et al. 1977; Schüpbach et al. 1978; Dübendorfer and Nöthiger 1982), the molecular underpinnings that directed the distinct genital disc primordia to their fate were not known.
Elizabeth Chen showed that each of the three primordia is divided into an anterior (A) and a posterior (P) compartment, and she demonstrated the conserved functions of the pattern formation genes in setting up these compartments within the genital disc (Chen and Baker 1997). The next major challenge was to determine how the sex hierarchy interacts with the pattern formation regulatory networks. A study by Eric Keisman demonstrated that Dsx works together with the pattern formation genes wingless and decapentaplegic, which are expressed along the A/P compartment border, to establish the sex-specific expression of a critical downstream gene known as dachshund throughout each primordium (Keisman and Baker 2001). The sex determination pathway was shown to control sex-specific growth and identity of the genital disc by regulating the activity of a stripe of cells along the A/P border, known as the A/P organizer. Also, in contrast to earlier views that the sex determination hierarchy represses development of the inappropriate genital primordium in each sex, it was revealed that the pathway actually directs the growth and differentiation of both genital primordia in both males and females, but in a sex-specific manner (Keisman et al. 2001).
Up until this point, somatic sex determination had been shown to function entirely in a cell-autonomous manner, in keeping with the original findings reported in Baker and Ridge (1980). Shaad Ahmad’s graduate work revealed an exception to this. He showed that FGF receptor-expressing mesodermal cells are recruited into the male genital primordium by male-specific expression of the corresponding FGF ligand in the genital disc. The FGF receptor-expressing cells form a new compartment in the disc, distinct from the previously characterized A and P compartments (Ahmad and Baker 2002). Elizabeth Chen, working with Audrey Christiansen, also identified the pattern formation genes required for the initial establishment of the genital disc precursor cells in developing embryos (Chen et al. 2005). Through these studies, the sex determination pathway was intimately connected to the tissue patterning systems and some of the mysteries of the relatively obscure genital imaginal disc were solved [reviewed in Christiansen et al. (2002)].
During the mid-1990s, Bruce’s Stanford University colleagues were pioneering the use of genomic technologies to examine global gene expression. Bruce, with a postdoctoral scholar in his laboratory, Michelle Arbeitman, and a team of Stanford colleagues, played an instrumental role in the first study to use genome-scale approaches to examine gene expression across metazoan development and between the sexes (Arbeitman et al. 2002). This study defined the major transcriptional programs that direct progression through the Drosophila life cycle, and showed that 50% of the genome is deployed in a sex differential manner, mostly due to differences in germline gene expression. Further genomic analyses of somatic sex differences revealed genes regulated by Dsx and FruM (Arbeitman et al. 2004). Bruce had used large-scale molecular screening approaches to find sex differentially expressed genes even prior to the Stanford genome-wide studies. Work with Deborah Andrew and with Mariana Wolfner’s new laboratory, at Cornell, used genomic and cDNA hybridization screens to search for additional genes regulated by the sex hierarchy (DiBenedetto et al. 1987; Andrew and Baker 2008). Bruce’s laboratory, after his move to the Janelia Research Campus in 2008, and Michelle Arbeitman’s laboratory would continue to elucidate Dsx and FruM targets with novel genomic technologies (Goldman and Arbeitman 2007; Dalton et al. 2009, 2013; Luo et al. 2011; Arbeitman et al. 2016; Meissner et al. 2016; Newell et al. 2016).
While Bruce and colleagues were identifying the critical sex determination genes in D. melanogaster, genes specifying sex in C. elegans were also being discovered using a similar strategy of cloning genes that when mutated caused sex transformations [for genetic screens see Hodgkin and Brenner (1977) and Hodgkin (1980)]. The molecular-level comparative studies revealed that the genes and molecular mechanisms specifying sex differences were not conserved [reviewed in Cline and Meyer (1996)]. The scientific community at the time recognized that sex determination mechanisms evolve rapidly, as compared to other processes (Bull 1983), but how such different sex determining pathways could evolve was not clear, especially for pathways with multiple steps in the genetic regulatory hierarchy. An important hypothesis was that these pathways could evolve in a step-wise fashion from the bottom of a sex hierarchy to the top, with the rationale that the most downstream step of a genetic pathway is the critical output step (Wilkins 1995). Furthermore, it was proposed that there might be preferential functional conservation of genes at the bottom of the sex determining regulatory hierarchy, given that it is the important output step, with novel mechanisms to achieve the regulation of the output step arising during evolution (Wilkins 1995; Pomiankowski et al. 2004). This hypothesis was proven true with the cloning of C. elegans male-determining mab-3, a gene that is related to Drosophila dsx (Raymond et al. 1998). More recently, dsx-related genes have been shown to have a role in sex determination in all animals that have been studied [reviewed in Kopp (2012)]. It was in this context that Ignacio Marín, a postdoctoral scholar in Bruce’s laboratory, reviewed the field (Marín and Baker 1998). Then, along with other Baker laboratory members, investigated the evolution of dosage compensation to understand sex chromosome evolution [Marín et al. (1996) and reviewed in Marín et al. (2000)]. Mark Siegal, a postdoctoral scholar in the Baker laboratory, examined the conservation of the interaction of Dsx with Ix to gain insight into the evolution of sex determination across different species (Siegal and Baker 2005). Bruce remained interested in evolutionary questions through the rest of his career. His laboratory would later turn their attention to the evolution of reproductive behaviors and make interesting discoveries about species discrimination during courtship behaviors (Fan et al. 2013; Vaughan et al. 2014).
Sex Behavior Meets the Sex Determination Regulatory Hierarchy: The Genetic Control of Sexual Behavior
Bruce’s entry into the genetics of sexual behavior came from the unexpected discovery that the fruitless gene (fru), a gene known to be involved in controlling aspects of male sexual behavior, defined a new branch in the sex determination regulatory hierarchy. By the early 1990s, it was clear that tra and tra-2 have regulatory control over the doublesex (dsx) gene, but Bruce was interested in knowing if there were yet undiscovered genes in the hierarchy. These efforts, combined with those of collaborators Barbara Taylor and Jeffrey Hall, led to the discovery of fru’s position in the sex determination hierarchy. This discovery was the critical finding that led Bruce’s laboratory into the study of behavioral genetics (Ryner et al. 1996).
Barbara Taylor had published findings showing that development of a male-specific abdominal muscle, the Muscle of Lawrence (MOL), depended on the function of tra and tra-2, but not dsx. This finding suggested the existence of a previously unrecognized branch in the sex-determination hierarchy (Taylor 1992). Lisa Ryner had found that a 13-nt sequence, repeated six times in dsx in a noncoding region, was necessary and sufficient to direct sex-specific splicing (Ryner and Baker 1991). Lisa Ryner, with input from William Mattox, devised a genomic screening method that used a tandem repeat of the dsx 13-nt sequence to “fish out” the gene or genes that might function at the top of this predicted new branch in the hierarchy. This approach led to the identification of a genomic region containing the 13-nt repeat sequences that was not the dsx region. When this genomic fragment was used to probe a northern blot, sex-specific transcripts were revealed. In situ hybridization to the giant salivary gland polytene chromosomes mapped the sequences to the genomic interval known to contain the fru gene.
The fru gene was being extensively studied in the laboratory of Jeffrey Hall at Brandeis University. Jeffrey Hall and Bruce were graduate students contemporaneously in the Sandler laboratory. fru was known to be involved in male sexual behavior based on analysis of mutant flies exhibiting aberrant courtship. In addition, it had been shown that fru was involved in sex-specific development of the MOL (Gailey et al. 1991). Interestingly, development of the MOL was dependent on the sex of the innervating neurons (Lawrence and Johnston 1986). Furthermore, earlier work by Belote and Baker showed that tra-2 is required in adult females to block male-specific courtship behaviors, linking control of sexual behavior to this part of the hierarchy (Belote and Baker 1987). Hence, fru was a good candidate for a gene residing at the top of the new branch in the sex determination regulatory hierarchy and this branch appeared to control sex-specific neuronally controlled functions, such as courtship behavior and the development of the MOL.
While work in Bruce’s laboratory was ongoing to identify and understand fru’s transcripts and genomic structure, Bruce met Steven Wasserman and Diego Castrillon at a Drosophila conference, where he learned about their work to identify genes involved in spermatogenesis (Castrillon et al. 1993). Several P element “hits” landed in the fru locus. Even though fru probably was not directly involved in spermatogenesis, it was nonetheless an interesting gene, so they had set out to clone it. By the time they met with Bruce, Wasserman and Castrillon had cloned 180 kb of genomic DNA that spanned the chromosomal lesions that defined the fru locus, but they had not yet succeeded at identifying transcripts in the region. Through a subsequent collaboration, it was shown that the genomic fragment that Lisa Ryner identified with the 13-nt repeat sequence was within their genomic walk.
Lisa Ryner recalls that Bruce was brilliant in bringing all four laboratories with an interest in fru into a collaboration referred to as the “Fruitless Consortium.” The Consortium included Jeffrey Hall’s group, with Stephen Goodwin and Adriana Villella dissecting fru’s role in male courtship behavior; Barbara Taylor studying the role of fru in MOL development; Steven Wasserman and Diego Castrillon providing additional fru alleles, and the clones spanning the fru genomic interval; and Bruce’s own laboratory, including Anuranjan Anand and Lisa Ryner, identifying the sex-specific transcripts (Anand et al. 2001). The Fruitless Consortium was very fruitful (pun intended); the consortium showed that fru encodes transcription factors whose sex-specific transcripts are generated by alternative splicing controlled by the sex hierarchy proteins Tra and Tra-2 (Figure 1E). This splicing was shown to be necessary for male-specific courtship behavior. Strikingly, fru’s sex-specific transcripts were only expressed in the adult nervous system and in a very distinct pattern (Ryner et al. 1996). This was the first sex hierarchy gene to function exclusively in the nervous system and fit well with the notion that fru, like dsx, was a master regulatory gene, but in the case of fru, controlling sex-specific functions of the nervous system. Around the same time, Daisuke Yamamoto’s laboratory had also independently determined that fruitless (called satori in their paper) is important for male courtship behaviors through a P element mutant screen and cloning project (Ito et al. 1996).
The combined work of the consortium had built a very strong case that a single gene, fru, had highly specific control of Drosophila sexual orientation and sex behavior. However, at that time, the idea that genes could control sexual orientation and behavior in humans was quite controversial, so much so that a national press conference was held at Stanford on the eve before publication of the initial work on fru in December of 1996. The press conference resulted in a very wide audience hearing about fru with many additional public airings and interpretations of the findings in the popular press. The story ran on local TV, radio, in newspapers, and there was even a reference to the study in a lifestyle magazine (Self). Even among behavioral scientists, the fru story was paradigm changing.
With the cloning and molecular characterization of the fru locus (Ryner et al. 1996), the stage was set for Bruce and the Fruitless Consortium to tackle head on the historically Manichean debate, as to whether or not behavioral repertoires could in fact be genetically specified, or were predominantly shaped by environmental factors. In a landmark review, “Are Complex Behaviors Specified by Dedicated Regulatory Genes? Reasoning from Drosophila,” Bruce, Barbara Taylor, and Jeffrey Hall elegantly laid out the theoretical framework for the concept of the genetic control of behavior (Baker et al. 2001). They argued that despite the complexity of social interactions and displays, innate behaviors that underlie reproduction and survival are likely to be under the strongest genetic control, because they are so important for the survival of the species. They further observed that previous approaches focused on determining the genetic or environmental influences that cause variation in behavior had overlooked the very basis of the innate behaviors being examined. The essay articulated an experimental roadmap that would seek to demonstrate how the example of fru function could directly support this idea.
One goal was to elucidate the function of subsets of neurons that express the male-specific products encoded by fru (fruM), which turned out to be more challenging than anticipated, given that fru is a large complex gene. This complexity made it challenging to develop tools to modify subsets of fruM-expressing neurons, which relied on identifying discrete enhancer DNA sequences. Instead, Devanand Manoli developed an RNA interference (RNAi)-based approach that allowed the manipulation of fruM expression levels using existing Gal4 expression tools (Manoli and Baker 2004). This approach helped elucidate the function of fru in neurons; even when the Gal4 line was broadly expressed, the RNAi would impact only the neurons that expressed fruM. This RNAi-based approached illuminated a role for the median bundle cluster of neurons in the appropriate initiation and gating of courtship, coordination of courtship behaviors to members of the right species, and species-specific discrimination of courtship targets (Manoli and Baker 2004; Fan et al. 2013; Tran et al. 2014). Geoffrey Meissner and Devanand Manoli next carried out a large genetic screen using the fruM RNAi transgene, and a large collection of Gal4 lines in collaboration with the laboratory of Ulrike Heberlein, Bruce’s long-time friend and colleague. The goal was to identify neurons that had behavioral phenotypes due to loss of fruM, thus further defining the role of subsets of fruM neurons (Meissner et al. 2011).
Devanand Manoli also employed a recently developed gene replacement approach (Gong and Golic 2003), using homologous recombination to insert the Gal4 coding region into fru, as a means to visualize and manipulate fruM-expressing neurons (Manoli et al. 2005). This study revealed the full repertoire of fruM expression and, unexpectedly, that fruM is expressed in regions of the peripheral nervous system in every sensory system implicated in courtship behavior (Figure 1F). Around the same time, Barry Dickson’s laboratory reported on a Gal4 insertion into fru that also revealed the function and anatomy of fruM-expressing neurons (Stockinger et al. 2005). Up until that point, fruM was thought to be expressed almost exclusively in the central nervous system. The observation of fruM in such “labeled lines,” that is primary sensory neurons as well as downstream sensory pathways, suggested that fruM is important for the detection of ethologically relevant sensory information and for transmission of this information to the central nervous system. Before this study, it was thought that the primary role of fruM was to specify the fate of neurons that integrated sensory information common to both sexes to initiate and execute sex-specific behavioral programs. By controlling the sensory perception, as well as the behavioral response to cues that elicit species-specific behaviors like courtship, fruM provided developmental and evolutionary flexibility in this circuitry, independent of anatomic sex.
This study also demonstrated that males and females share much, but not all, of the neuroanatomy that expresses fruM. Up until this point, it was not clear if fruM was expressed in neurons that are only present in males. Additionally, the study showed that fruM is expressed in the regions of the fly brain that underlie learning and memory, providing a mechanistic explanation for how behavior can be both genetically specified and modified by experience. Using fruM as a model, Bruce and his colleagues proposed that such genetic specification of neural circuits likely occurred across many species, controlling both basic patterns of innate behavior, but also higher-order social behaviors and relationships that were modifiable across an animal’s life [reviewed in Manoli et al. (2006)]. The critical insight that one could find anatomical targets of peripheral fruM neurons, and also that one could screen for critical neurons using behavioral phenotypes, led to work by Alex Vaughan and Chuan Zhou in the auditory system. They identified neurons that process and respond to cues relevant to conspecific song, and neurons leading to the display of song-induced behaviors (Vaughan et al. 2014; Zhou et al. 2014, 2015).
Carmen Robinett employed a similar gene-targeting approach for dsx that revealed restricted expression throughout the body and brain (Robinett et al. 2010), as did Stephen Goodwin’s laboratory (Rideout et al. 2010). These studies fit well with previous studies examining Dsx expression with antibodies (Lee et al. 2002; Sanders and Arbeitman 2008). Further studies of the role of dsx in behavior revealed a role for dsx-expressing neurons in female receptivity behaviors in response to auditory and olfactory cues (Zhou et al. 2014). Studies by David Mellert, Carmen Robinett, and additional Baker laboratory members uncovered the interwoven role of both the dsx and fru branches of the sex hierarchy in controlling anatomical sex differences in neural circuitry (Mellert et al. 2010, 2012).
More recent work from the Baker laboratory included a deeper exploration of the neuronal mechanisms underlying sexual behavior, as well as revisiting earlier observations of behavior through the lens of molecular genetics. Consistent with the observation that the fruM circuitry was dedicated to sexual behaviors, activation of these neurons drove courtship behaviors in multiple contexts (Pan et al. 2011). Even more striking, these studies also revealed that activation of dsx-expressing neurons—a much smaller set of neurons than fruM neurons, with limited overlap— alone could also drive courtship and preserve species discrimination, and that, quite surprisingly, courtship driven by dsx+ neurons could occur even in the absence of FruM. Using the activation of distinct subsets of fruM neurons to “sensitize” this circuitry to additional cues that facilitate sexual behaviors, Yufeng Pan and other members of the laboratory demonstrated a role for specific patterns of motion in the elicitation of courtship (Pan et al. 2012). Pan further showed that fruM-independent, learned courtship displays required male-specific dsx function, lending credence to Bruce’s original notion that Dsx and its orthologs are central to the sexual differentiation of most animals, with adjacent genetic programs modifying their effects for specific aspects of development (Pan and Baker 2014). The Baker laboratory reported on the coordinated role of fruM and dsx in courtship, including an enhancer trap screen that identified a direct gene target of FruM regulation that is repressed in males and is controlled by DsxF in neurons that control female receptivity (Meissner et al. 2016).
Returning full circle to the original genetic insights through which Bruce began to dissect sex determination in the fly, the Baker laboratory demonstrated that although fruM is clearly necessary to specify the circuitry controlling the elicitation and coordinated display of courtship and other sexual behaviors, it appears that the basic patterning of the nervous system for the behaviors that underlie this ritual is coordinated with and regulated by the same developmental program responsible for all other aspects of somatic sexual differentiation, i.e., Dsx. Devanand Manoli recalls that “Bruce often wondered aloud whether such a branched control of core behaviors and the circuitry controlling their coordinated display permitted significant flexibility in the adaptation of the behavioral circuits, and separately allow behavioral variation and distinct social experiences to exert pressure on the evolution of these neural pathways. The observation lended new insight into the evolution of the nervous system and the bodies and parts it serves.”
Concluding Thoughts
Bruce had an exceptionally rich academic career. He made major, revelatory contributions to scientific knowledge, with the publication of > 100 academic papers from his laboratory. While he and his trainees examined several research topics using different approaches, he spent 4 decades deeply focused on elaborating upon the theme of his first major sex determination paper “Sex and the single cell. I. On the action of major loci affecting sex determination in Drosophila melanogaster” (Baker and Ridge 1980). This theme took the laboratory on a journey through a range of intellectual topics, from reproductive behavior, to alternative pre-mRNA splicing, to sex chromosome evolution, sex differences in developmental biology, and many more. Each topic was approached with a high level of scientific rigor, curiosity, and ingenuity, through Bruce’s mentoring and training the next generation of scientists. Bruce told members of his laboratory that he had planned for a series of “Sex and the single cell” papers. However, time passed, and the second one arrived a full 30 years after the first, titled “Sex and the single cell. II. There is a time and place for sex” (Robinett et al. 2010). It is unfortunate for the scientific community that we will not know the next chapters, as they would have been written by Bruce. We are all saddened by the unexpected loss of our mentor, colleague, and friend Bruce Baker. We have lost a deep thinker and an intensely dedicated scientist. We are all grateful and happy to have been part of the fantastic adventure of scientific discovery as trainees in the Baker laboratory, and hope to continue his legacy of imagination and rigor.
Acknowledgments
The authors thank Barbara Taylor, Adam Wilkins, Mariana Wolfner, and anonymous reviewer two for their helpful input and comments. This perspective is not meant to be a comprehensive review, but highlights contributions of Bruce Baker’s laboratory, and findings from other laboratories that were similar and published contemporaneously. The authors are funded by NIH R01 DE013899 (D.J.A); NIH R01 GM073039 and NIH R01 GM116998 (M.N.A); NIH R01 AR053173 and NIH R01 GM098816, American Heart Association Established Investigator Award, and HHMI Faculty Scholar Award (E.H.C).
Footnotes
Communicating editor: A. S. Wilkins
Literature Cited
- Ahmad S. M., Baker B. S., 2002. Sex-specific deployment of FGF signaling in Drosophila recruits mesodermal cells into the male genital imaginal disc. Cell 109: 651–661. 10.1016/S0092-8674(02)00744-4 [DOI] [PubMed] [Google Scholar]
- Amrein H., Gorman M., Nothiger R., 1988. The sex-determining gene tra-2 of Drosophila encodes a putative RNA binding protein. Cell 55: 1025–1035. 10.1016/0092-8674(88)90247-4 [DOI] [PubMed] [Google Scholar]
- Anand A., Villella A., Ryner L. C., Carlo T., Goodwin S. F., et al. , 2001. Molecular genetic dissection of the sex-specific and vital functions of the Drosophila melanogaster sex determination gene fruitless. Genetics 158: 1569–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew D. J., Baker B. S., 2008. Expression of the Drosophila secreted cuticle protein 73 (dsc73) requires Shavenbaby. Dev. Dyn. 237: 1198–1206. 10.1002/dvdy.21512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbeitman M. N., Furlong E. E., Imam F., Johnson E., Null B. H., et al. , 2002. Gene expression during the life cycle of Drosophila melanogaster. Science 297: 2270–2275. 10.1126/science.1072152 [DOI] [PubMed] [Google Scholar]
- Arbeitman M. N., Fleming A. A., Siegal M. L., Null B. H., Baker B. S., 2004. A genomic analysis of Drosophila somatic sexual differentiation and its regulation. Development 131: 2007–2021. 10.1242/dev.01077 [DOI] [PubMed] [Google Scholar]
- Arbeitman M. N., New F. N., Fear J. M., Howard T. S., Dalton J. E., et al. , 2016. Sex differences in Drosophila somatic gene expression: variation and regulation by doublesex. G3 (Bethesda) 6: 1799–1808. 10.1534/g3.116.027961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker B. S., 1973. The maternal and zygotic control of development by cinnamon, a new mutant in Drosophila melanogaster. Dev. Biol. 33: 429–440. 10.1016/0012-1606(73)90148-6 [DOI] [PubMed] [Google Scholar]
- Baker B. S., 1989. Sex in flies: the splice of life. Nature 340: 521–524. 10.1038/340521a0 [DOI] [PubMed] [Google Scholar]
- Baker B. S., Belote J. M., 1983. Sex determination and dosage compensation in Drosophila melanogaster. Annu. Rev. Genet. 17: 345–393. 10.1146/annurev.ge.17.120183.002021 [DOI] [PubMed] [Google Scholar]
- Baker B. S., Ridge K. A., 1980. Sex and the single cell. I. On the action of major loci affecting sex determination in Drosophila melanogaster. Genetics 94: 383–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker B. S., Wolfner M. F., 1988. A molecular analysis of doublesex, a bifunctional gene that controls both male and female sexual-differentiation in drosophila-Melanogaster. Genes Dev. 2: 477–489. 10.1101/gad.2.4.477 [DOI] [PubMed] [Google Scholar]
- Baker B. S., Gorman M., Marin I., 1994. Dosage compensation in Drosophila. Annu. Rev. Genet. 28: 491–521. 10.1146/annurev.ge.28.120194.002423 [DOI] [PubMed] [Google Scholar]
- Baker B. S., Taylor B. J., Hall J. C., 2001. Are complex behaviors specified by dedicated regulatory genes? Reasoning from Drosophila. Cell 105: 13–24. 10.1016/S0092-8674(01)00293-8 [DOI] [PubMed] [Google Scholar]
- Bashaw G. J., Baker B. S., 1995. The msl-2 dosage compensation gene of Drosophila encodes a putative DNA-binding protein whose expression is sex specifically regulated by sex-lethal. Development 121: 3245–3258. [DOI] [PubMed] [Google Scholar]
- Bashaw G. J., Baker B. S., 1996. Dosage compensation and chromatin structure in Drosophila. Curr. Opin. Genet. Dev. 6: 496–501. 10.1016/S0959-437X(96)80073-6 [DOI] [PubMed] [Google Scholar]
- Bell L. R., Maine E. M., Schedl P., Cline T. W., 1988. Sex-lethal, a Drosophila sex determination switch gene, exhibits sex-specific RNA splicing and sequence similarity to RNA binding proteins. Cell 55: 1037–1046. 10.1016/0092-8674(88)90248-6 [DOI] [PubMed] [Google Scholar]
- Belote J. M., Baker B. S., 1987. Sexual behavior: its genetic control during development and adulthood in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 84: 8026–8030. 10.1073/pnas.84.22.8026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggs R. T., Gregor P., Idriss S., Belote J. M., McKeown M., 1987. Regulation of sexual differentiation in D. melanogaster via alternative splicing of RNA from the transformer gene. Cell 50: 739–747. 10.1016/0092-8674(87)90332-1 [DOI] [PubMed] [Google Scholar]
- Bone J. R., Lavender J., Richman R., Palmer M. J., Turner B. M., et al. , 1994. Acetylated histone H4 on the male X chromosome is associated with dosage compensation in Drosophila. Genes Dev. 8: 96–104. 10.1101/gad.8.1.96 [DOI] [PubMed] [Google Scholar]
- Bull J. J., 1983. Evolution of Sex Determining Mechanisms. Benjamin-Cummings, Menlo Park, CA. [Google Scholar]
- Burtis K. C., Baker B. S., 1989. Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell 56: 997–1010. 10.1016/0092-8674(89)90633-8 [DOI] [PubMed] [Google Scholar]
- Burtis K. C., Coschigano K. T., Baker B. S., Wensink P. C., 1991. The doublesex proteins of Drosophila melanogaster bind directly to a sex-specific yolk protein gene enhancer. EMBO J. 10: 2577–2582. 10.1002/j.1460-2075.1991.tb07798.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler B., Pirrotta V., Irminger-Finger I., Nothiger R., 1986. The sex-determining gene tra of Drosophila: molecular cloning and transformation studies. EMBO J. 5: 3607–3613. 10.1002/j.1460-2075.1986.tb04689.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrillon D. H., Gonczy P., Alexander S., Rawson R., Eberhart C. G., et al. , 1993. Toward a molecular-genetic analysis of spermatogenesis in Drosophila-Melanogaster - characterization of male-sterile mutants generated by single P element mutagenesis. Genetics 135: 489–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. H., Baker B. S., 1997. Compartmental organization of the Drosophila genital imaginal discs. Development 124: 205–218. [DOI] [PubMed] [Google Scholar]
- Chen E. H., Christiansen A. E., Baker B. S., 2005. Allocation and specification of the genital disc precursor cells in Drosophila. Dev. Biol. 281: 270–285. 10.1016/j.ydbio.2005.02.032 [DOI] [PubMed] [Google Scholar]
- Christiansen A. E., Keisman E. L., Ahmad S. M., Baker B. S., 2002. Sex comes in from the cold: the integration of sex and pattern. Trends Genet. 18: 510–516. 10.1016/S0168-9525(02)02769-5 [DOI] [PubMed] [Google Scholar]
- Cline T. W., 1983. The interaction between daughterless and sex-lethal in triploids: a lethal sex-transforming maternal effect linking sex determination and dosage compensation in Drosophila melanogaster. Dev. Biol. 95: 260–274. 10.1016/0012-1606(83)90027-1 [DOI] [PubMed] [Google Scholar]
- Cline T. W., 2005. Reflections on a path to sexual commitment. Genetics 169: 1179–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline T. W., Meyer B. J., 1996. Vive la difference: males vs. females in flies vs. worms. Annu. Rev. Genet. 30: 637–702. 10.1146/annurev.genet.30.1.637 [DOI] [PubMed] [Google Scholar]
- Dalton J. E., Lebo M. S., Sanders L. E., Sun F., Arbeitman M. N., 2009. Ecdysone receptor acts in fruitless- expressing neurons to mediate drosophila courtship behaviors. Curr. Biol. 19: 1447–1452. 10.1016/j.cub.2009.06.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton J. E., Fear J. M., Knott S., Baker B. S., McIntyre L. M., et al. , 2013. Male-specific Fruitless isoforms have different regulatory roles conferred by distinct zinc finger DNA binding domains. BMC Genomics 14: 659 10.1186/1471-2164-14-659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Meller V. H., 2006. roX RNAs are required for increased expression of X-linked genes in Drosophila melanogaster males. Genetics 174: 1859–1866. 10.1534/genetics.106.064568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiBenedetto A. J., Lakich D. M., Kruger W. D., Belote J. M., Baker B. S., et al. , 1987. Sequences expressed sex-specifically in Drosophila melanogaster adults. Dev. Biol. 119: 242–251. 10.1016/0012-1606(87)90225-9 [DOI] [PubMed] [Google Scholar]
- Dübendorfer K., Nöthiger R., 1982. A clonal analysis of cell lineage and growth in the male and female genital disc of Drosophila melanogaster. Wilehm Roux Arch. Dev. Biol. 191: 42–55. 10.1007/BF00848545 [DOI] [PubMed] [Google Scholar]
- Fagegaltier D., Baker B. S., 2004. X chromosome sites autonomously recruit the dosage compensation complex in Drosophila males. PLoS Biol. 2: e341 10.1371/journal.pbio.0020341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan P., Manoli D. S., Ahmed O. M., Chen Y., Agarwal N., et al. , 2013. Genetic and neural mechanisms that inhibit Drosophila from mating with other species. Cell 154: 89–102. 10.1016/j.cell.2013.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke A., Baker B. S., 1999. The rox1 and rox2 RNAs are essential components of the compensasome, which mediates dosage compensation in Drosophila. Mol. Cell 4: 117–122. 10.1016/S1097-2765(00)80193-8 [DOI] [PubMed] [Google Scholar]
- Gailey D. A., Taylor B. J., Hall J. C., 1991. Elements of the fruitless locus regulate development of the muscle of Lawrence, a male-specific structure in the abdomen of drosophila-melanogaster adults. Development 113: 879–890. [DOI] [PubMed] [Google Scholar]
- Garrett-Engele C. M., Siegal M. L., Manoli D. S., Williams B. C., Li H., et al. , 2002. Intersex, a gene required for female sexual development in Drosophila, is expressed in both sexes and functions together with doublesex to regulate terminal differentiation. Development 129: 4661–4675. [DOI] [PubMed] [Google Scholar]
- Goldman T. D., Arbeitman M. N., 2007. Genomic and functional studies of Drosophila sex hierarchy regulated gene expression in adult head and nervous system tissues. PLoS Genet. 3: e216 10.1371/journal.pgen.0030216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong W. J., Golic K. G., 2003. Ends-out, or replacement, gene targeting in Drosophila. Proc. Natl. Acad. Sci. USA 100: 2556–2561. 10.1073/pnas.0535280100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goralski T. J., Edstrom J. E., Baker B. S., 1989. The sex determination locus transformer-2 of Drosophila encodes a polypeptide with similarity to RNA binding proteins. Cell 56: 1011–1018. 10.1016/0092-8674(89)90634-X [DOI] [PubMed] [Google Scholar]
- Gorman M., Baker B. S., 1994. How flies make one equal two: dosage compensation in Drosophila. Trends Genet. 10: 376–380. 10.1016/0168-9525(94)90135-X [DOI] [PubMed] [Google Scholar]
- Gorman M., Kuroda M. I., Baker B. S., 1993. Regulation of the sex-specific binding of the maleless dosage compensation protein to the male X chromosome in Drosophila. Cell 72: 39–49. 10.1016/0092-8674(93)90048-U [DOI] [PubMed] [Google Scholar]
- Gorman M., Franke A., Baker B. S., 1995. Molecular characterization of the male-specific lethal-3 gene and investigations of the regulation of dosage compensation in Drosophila. Development 121: 463–475. [DOI] [PubMed] [Google Scholar]
- Gu W., Wei X., Pannuti A., Lucchesi J. C., 2000. Targeting the chromatin-remodeling MSL complex of Drosophila to its sites of action on the X chromosome requires both acetyl transferase and ATPase activities. EMBO J. 19: 5202–5211. 10.1093/emboj/19.19.5202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilfiker A., Yang Y., Hayes D. H., Beard C. A., Manning J. E., et al. , 1994. Dosage compensation in Drosophila: the X-chromosomal binding of MSL-1 and MLE is dependent on Sxl activity. EMBO J. 13: 3542–3550. 10.1002/j.1460-2075.1994.tb06661.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J., 1980. More sex-determination mutants of Caenorhabditis elegans. Genetics 96: 649–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J. A., Brenner S., 1977. Mutations causing transformation of sexual phenotype in the nematode Caenorhabditis elegans. Genetics 86: 275–287. [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fujitani K., Usui K., Shimizu-Nishikawa K., Tanaka S., et al. , 1996. Sexual orientation in Drosophila is altered by the satori mutation in the sex-determination gene fruitless that encodes a zinc finger protein with a BTB domain. Proc. Natl. Acad. Sci. USA 93: 9687–9692. 10.1073/pnas.93.18.9687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keisman E. L., Baker B. S., 2001. The Drosophila sex determination hierarchy modulates wingless and decapentaplegic signaling to deploy dachshund sex-specifically in the genital imaginal disc. Development 128: 1643–1656. [DOI] [PubMed] [Google Scholar]
- Keisman E. L., Christiansen A. E., Baker B. S., 2001. The sex determination gene doublesex regulates the A/P organizer to direct sex-specific patterns of growth in the Drosophila genital imaginal disc. Dev. Cell 1: 215–225. 10.1016/S1534-5807(01)00027-2 [DOI] [PubMed] [Google Scholar]
- Kopp A., 2012. Dmrt genes in the development and evolution of sexual dimorphism. Trends Genet. 28: 175–184. 10.1016/j.tig.2012.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda M. I., Kernan M. J., Kreber R., Ganetzky B., Baker B. S., 1991. The maleless protein associates with the X chromosome to regulate dosage compensation in Drosophila. Cell 66: 935–947. 10.1016/0092-8674(91)90439-6 [DOI] [PubMed] [Google Scholar]
- Lawrence P. A., Johnston P., 1986. The muscle pattern of a segment of Drosophila may be determined by neurons and not by contributing myoblasts. Cell 45: 505–513. 10.1016/0092-8674(86)90282-5 [DOI] [PubMed] [Google Scholar]
- Lee G., Hall J. C., Park J. H., 2002. Doublesex gene expression in the central nervous system of Drosophila melanogaster. J. Neurogenet. 16: 229–248. 10.1080/01677060216292 [DOI] [PubMed] [Google Scholar]
- Luo S. Z. D., Shi G. W., Baker B. S., 2011. Direct targets of the D. melanogaster DSXF protein and the evolution of sexual development. Development 138: 2761–2771. 10.1242/dev.065227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoli D. S., Baker B. S., 2004. Median bundle neurons coordinate behaviours during Drosophila male courtship. Nature 430: 564–569. 10.1038/nature02713 [DOI] [PubMed] [Google Scholar]
- Manoli D. S., Foss M., Villella A., Taylor B. J., Hall J. C., et al. , 2005. Male-specific fruitless specifies the neural substrates of Drosophila courtship behaviour. Nature 436: 395–400. 10.1038/nature03859 [DOI] [PubMed] [Google Scholar]
- Manoli D. S., Meissner G. W., Baker B. S., 2006. Blueprints for behavior: genetic specification of neural circuitry for innate behaviors. Trends Neurosci. 29: 444–451. 10.1016/j.tins.2006.06.006 [DOI] [PubMed] [Google Scholar]
- Marín I., Baker B. S., 1998. The evolutionary dynamics of sex determination. Science 281: 1990–1994. 10.1126/science.281.5385.1990 [DOI] [PubMed] [Google Scholar]
- Marín I., Franke A., Bashaw G. J., Baker B. S., 1996. The dosage compensation system of Drosophila is co-opted by newly evolved X chromosomes. Nature 383: 160–163. 10.1038/383160a0 [DOI] [PubMed] [Google Scholar]
- Marín I., Siegal M. L., Baker B. S., 2000. The evolution of dosage-compensation mechanisms. Bioessays 22: 1106–1114. [DOI] [PubMed] [Google Scholar]
- McKeown M., Belote J. M., Baker B. S., 1987. A molecular analysis of transformer, a gene in Drosophila melanogaster that controls female sexual differentiation. Cell 48: 489–499. 10.1016/0092-8674(87)90199-1 [DOI] [PubMed] [Google Scholar]
- Meissner G. W., Manoli D. S., Chavez J. F., Knapp J. M., Lin T. L., et al. , 2011. Functional dissection of the neural substrates for sexual behaviors in Drosophila melanogaster. Genetics 189: 195–211. 10.1534/genetics.111.129940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner G. W., Luo S. D., Dias B. G., Texada M. J., Baker B. S., 2016. Sex-specific regulation of Lgr3 in Drosophila neurons. Proc. Natl. Acad. Sci. USA 113: E1256–E1265. 10.1073/pnas.1600241113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller V. H., 2003. Initiation of dosage compensation in Drosophila embryos depends on expression of the roX RNAs. Mech. Dev. 120: 759–767. 10.1016/S0925-4773(03)00157-6 [DOI] [PubMed] [Google Scholar]
- Meller V. H., Kuroda M. I., 2002. Sex and the single chromosome. Adv. Genet. 46: 1–24. 10.1016/S0065-2660(02)46002-6 [DOI] [PubMed] [Google Scholar]
- Meller V. H., Wu K. H., Roman G., Kuroda M. I., Davis R. L., 1997. roX1 RNA paints the X chromosome of male Drosophila and is regulated by the dosage compensation system. Cell 88: 445–457. 10.1016/S0092-8674(00)81885-1 [DOI] [PubMed] [Google Scholar]
- Mellert D. J., Knapp J. M., Manoli D. S., Meissner G. W., Baker B. S., 2010. Midline crossing by gustatory receptor neuron axons is regulated by fruitless, doublesex and the Roundabout receptors. Development 137: 323–332. 10.1242/dev.045047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellert D. J., Robinett C. C., Baker B. S., 2012. Doublesex functions early and late in gustatory sense organ development. PLoS One 7: e51489 10.1371/journal.pone.0051489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miko I., 2008. Sex chromosomes and sex determination. Nature Educ. 1: 108. [Google Scholar]
- Nagoshi R. N., McKeown M., Burtis K. C., Belote J. M., Baker B. S., 1988. The control of alternative splicing at genes regulating sexual differentiation in D. melanogaster. Cell 53: 229–236. 10.1016/0092-8674(88)90384-4 [DOI] [PubMed] [Google Scholar]
- Newell N. R., New F. N., Dalton J. E., McIntyre L. M., Arbeitman M. N., 2016. Neurons that underlie Drosophila melanogaster reproductive behaviors: detection of a large male-bias in gene expression in fruitless-expressing neurons. G3 (Bethesda) 6: 2455–2465. 10.1534/g3.115.019265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nöthiger R., Dübendorfer A., Epper F., 1977. Gynandromorphs reveal two separate primordia for male and female genitalia inDrosophila melanogaster. Wilehm Roux Arch. Dev. Biol. 181: 367–373. 10.1007/BF00848062 [DOI] [PubMed] [Google Scholar]
- Palmer M. J., Mergner V. A., Richman R., Manning J. E., Kuroda M. I., et al. , 1993. The male-specific lethal-one (msl-1) gene of Drosophila melanogaster encodes a novel protein that associates with the X chromosome in males. Genetics 134: 545–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Baker B. S., 2014. Genetic identification and separation of innate and experience-dependent courtship behaviors in Drosophila. Cell 156: 236–248. 10.1016/j.cell.2013.11.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Robinett C. C., Baker B. S., 2011. Turning males on: activation of male courtship behavior in Drosophila melanogaster. PLoS One 6: e21144 10.1371/journal.pone.0021144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Meissner G. W., Baker B. S., 2012. Joint control of Drosophila male courtship behavior by motion cues and activation of male-specific P1 neurons. Proc. Natl. Acad. Sci. USA 109: 10065–10070. 10.1073/pnas.1207107109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y., Kelley R. L., Oh H., Kuroda M. I., Meller V. H., 2002. Extent of chromatin spreading determined by roX RNA recruitment of MSL proteins. Science 298: 1620–1623. 10.1126/science.1076686 [DOI] [PubMed] [Google Scholar]
- Park Y., Mengus G., Bai X., Kageyama Y., Meller V. H., et al. , 2003. Sequence-specific targeting of Drosophila roX genes by the MSL dosage compensation complex. Mol. Cell 11: 977–986. 10.1016/S1097-2765(03)00147-3 [DOI] [PubMed] [Google Scholar]
- Park Y., Oh H., Meller V. H., Kuroda M. I., 2005. Variable splicing of non-coding roX2 RNAs influences targeting of MSL dosage compensation complexes in Drosophila. RNA Biol. 2: 157–164. 10.4161/rna.2.4.2473 [DOI] [PubMed] [Google Scholar]
- Pomiankowski A., Nothiger R., Wilkins A., 2004. The evolution of the Drosophila sex-determination pathway. Genetics 166: 1761–1773. 10.1534/genetics.166.4.1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattner B. P., Meller V. H., 2004. Drosophila male-specific lethal 2 protein controls sex-specific expression of the roX genes. Genetics 166: 1825–1832. 10.1534/genetics.166.4.1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond C. S., Shamu C. E., Shen M. M., Seifert K. J., Hirsch B., et al. , 1998. Evidence for evolutionary conservation of sex-determining genes. Nature 391: 691–695. 10.1038/35618 [DOI] [PubMed] [Google Scholar]
- Rideout E. J., Dornan A. J., Neville M. C., Eadie S., Goodwin S. F., 2010. Control of sexual differentiation and behavior by the doublesex gene in Drosophila melanogaster. Nat. Neurosci. 13: 458–466. 10.1038/nn.2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinett C. C., Vaughan A. G., Knapp J. M., Baker B. S., 2010. Sex and the single cell. II. There is a time and place for sex. PLoS Biol. 8: e1000365 10.1371/journal.pbio.1000365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryner L. C., Baker B. S., 1991. Regulation of doublesex pre-mRNA processing occurs by 3′-splice site activation. Genes Dev. 5: 2071–2085. 10.1101/gad.5.11.2071 [DOI] [PubMed] [Google Scholar]
- Ryner L. C., Goodwin S. F., Castrillon D. H., Anand A., Villella A., et al. , 1996. Control of male sexual behavior and sexual orientation in Drosophila by the fruitless gene. Cell 87: 1079–1089. 10.1016/S0092-8674(00)81802-4 [DOI] [PubMed] [Google Scholar]
- Sanders L. E., Arbeitman M. N., 2008. Doublesex establishes sexual dimorphism in the Drosophila central nervous system in an isoform-dependent manner by directing cell number. Dev. Biol. 320: 378–390. 10.1016/j.ydbio.2008.05.543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüpbach T., Wieschaus E., Nöthiger R., 1978. The embryonic organization of the genital disc studied in genetic mosaics ofDrosophila melanogaster. Wilehm Roux Arch. Dev. Biol. 185: 249–270. 10.1007/BF00848355 [DOI] [PubMed] [Google Scholar]
- Siegal M. L., Baker B. S., 2005. Functional conservation and divergence of intersex, a gene required for female differentiation in Drosophila melanogaster. Dev. Genes Evol. 215: 1–12. 10.1007/s00427-004-0445-x [DOI] [PubMed] [Google Scholar]
- Stockinger P., Kvitsiani D., Rotkopf S., Tirian L., Dickson B. J., 2005. Neural circuitry that governs Drosophila male courtship behavior. Cell 121: 795–807. 10.1016/j.cell.2005.04.026 [DOI] [PubMed] [Google Scholar]
- Stuckenholz C., Meller V. H., Kuroda M. I., 2003. Functional redundancy within roX1, a noncoding RNA involved in dosage compensation in Drosophila melanogaster. Genetics 164: 1003–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor B. J., 1992. Differentiation of a male-specific muscle in Drosophila-Melanogaster does not require the sex-determining genes doublesex or intersex. Genetics 132: 179–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran D. H., Meissner G. W., French R. L., Baker B. S., 2014. A small subset of fruitless subesophageal neurons modulate early courtship in Drosophila. PLoS One 9: e95472 10.1371/journal.pone.0095472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan A. G., Zhou C., Manoli D. S., Baker B. S., 2014. Neural pathways for the detection and discrimination of conspecific song in D. melanogaster. Curr. Biol. 24: 1039–1049. 10.1016/j.cub.2014.03.048 [DOI] [PubMed] [Google Scholar]
- Wilkins A. S., 1995. Moving up the hierarchy - a hypothesis on the evolution of a genetic sex determination pathway. Bioessays 17: 71–77. 10.1002/bies.950170113 [DOI] [PubMed] [Google Scholar]
- Zhou C., Pan Y., Robinett C. C., Meissner G. W., Baker B. S., 2014. Central brain neurons expressing doublesex regulate female receptivity in Drosophila. Neuron 83: 149–163. 10.1016/j.neuron.2014.05.038 [DOI] [PubMed] [Google Scholar]
- Zhou C., Franconville R., Vaughan A. G., Robinett C. C., Jayaraman V., et al. , 2015. Central neural circuitry mediating courtship song perception in male Drosophila. Elife 4: e08477. 10.7554/eLife.08477 [DOI] [PMC free article] [PubMed] [Google Scholar]