SUMMARY
How disseminated tumor cells (DTCs) engage specific stromal components in distant organs for survival and outgrowth is a critical but poorly understood step of the metastatic cascade. Previous studies have demonstrated the importance of the epithelial-mesenchymal transition (EMT) in promoting the cancer stem cell properties needed for metastasis initiation, while the reverse process of mesenchymal-epithelial transition (MET) is required for metastatic outgrowth. Here we report that this paradoxical requirement for simultaneous induction of both MET and cancer stem cell traits in DTCs is provided by bone vascular niche E-selectin, whose direct binding to cancer cells promotes bone metastasis by inducing MET and activating Wnt signaling. E-selectin binding activity mediated by α1–3 Fucosyltransferases Fut3/Fut6 and Glg1 are instrumental to the formation of bone metastasis. These findings provide unique insights into the functional role of E-selectin as a component of the vascular niche critical for metastatic colonization in bone.
Keywords: E-selectin, mesenchymal-epithelial transition, tumor-stromal interaction, vascular niche, Golgi glycoprotein 1, HCELL, α1-3 Fucosyltransferase, Wnt signaling, breast cancer, bone metastasis
INTRODUCTION
A critical gap in our understanding of cancer metastasis lies between the initial dissemination of circulating tumor cells (CTCs) to a secondary organ and their successful outgrowth to metastatic lesions1, 2. From recent studies, two molecular themes central to metastasis initiation have emerged: (1) Cellular plasticity mediated by epithelial-to-mesenchymal and mesenchymal-to-epithelial transitions (EMT and MET)3, 4 and (2) Tumor-stromal niche interactions, which activate tumor initiating cell (TIC) or cancer stem cell (CSC) properties through Wnt, Notch, and other pathways5, 6. Despite numerous studies, we do not yet understand how these programs interact to initiate metastasis.
The acquisition of mesenchymal traits through EMT has been shown to promote many pro-metastatic properties7,8,9. However, secondary metastatic tumors often match the epithelial state of the primary tumor10 and studies have suggested a requirement for MET during metastatic colonization3, 11–14. Thus, it has been proposed that sequential EMT-MET are needed for tumor cells to colonize a distant organ3. Yet this hypothesis does not explain the paradox of how MET induction and stem cell identity coexist, when essential stem- and metastasis-associated factors such as Sox2 are suppressed in an epithelial state15, 16. Furthermore, the causes of these sequential epithelial-mesenchymal-epithelial transitions remain only half described; inducers of EMT are well-studied4 yet few inducers of MET are known.
For metastatic cells to colonize an organ, inducers of MET must act in a context-dependent manner, suggesting that tumor-stromal interactions are instructive to MET. Clues from previous studies suggest the bone vascular niche that hosts hematopoietic stem/progenitor cells (HSCs/HPCs) may contain such stromal signals, as this niche is co-opted by cancer cells during bone metastasis17,18. One factor essential to the function of the HSC/HPC vascular niche is endothelial selectin (E-selectin, Sele, CD62e). As originally defined, E-selectin functions to capture circulating leukocytes or tumor cells onto vascular endothelium under hemodynamic shear flow19, yet leukocyte recruitment is not altered by ablation of E-selectin20. Further studies have shown that E-selectin plays a critical role in the creation of the bone marrow vascular niche that drives hematopoiesis, and engagement of E-selectin with its ligands on hematopoietic progenitor cells21 leads to their proliferation22.
Whereas it is known that certain tumor cells bind to E-selectin in vitro19, 23, 24, genetic ablation of E-selectin has not been shown to affect metastasis in vivo; furthermore, our knowledge of the role of E-selectin receptor/ligand interactions in metastasis is incomplete. Here we report that Golgi glycoprotein 1 (Glg1, “E-selectin Ligand-1”) and glycoprotein E-selectin ligands created by the α1–3 Fucosyltransferases 3 or 6 (Fut3/6) play key roles in mediating metastasis to bone by binding E-selectin which induces a mesenchymal-to-epithelial transition (MET) followed by stemness-enhancing Wnt signaling.
RESULTS
Enriched expression of E-selectin in bone specifically promotes bone metastasis
We first tested the correlation of in vitro E-selectin binding to metastatic propensity in vivo. To accurately quantify E-selectin binding across different cell lines, we designed an internally-controlled flow cytometry assay (Supplementary Fig. 1a). The fidelity of this assay to detect E-selectin binding was verified using isotype-control IgG, a glycomimetic inhibitor of E-selectin (GMI-1271, Uproleselan), and EDTA (Supplementary Fig. 1b). By applying this assay to a panel of isogenic MDA-MB-231 sublines with different organotropic metastatic abilities25, 26, we found that sublines with high bone or lung metastatic abilities generally have increased E-selectin binding ability (Supplementary Fig. 1c, d).
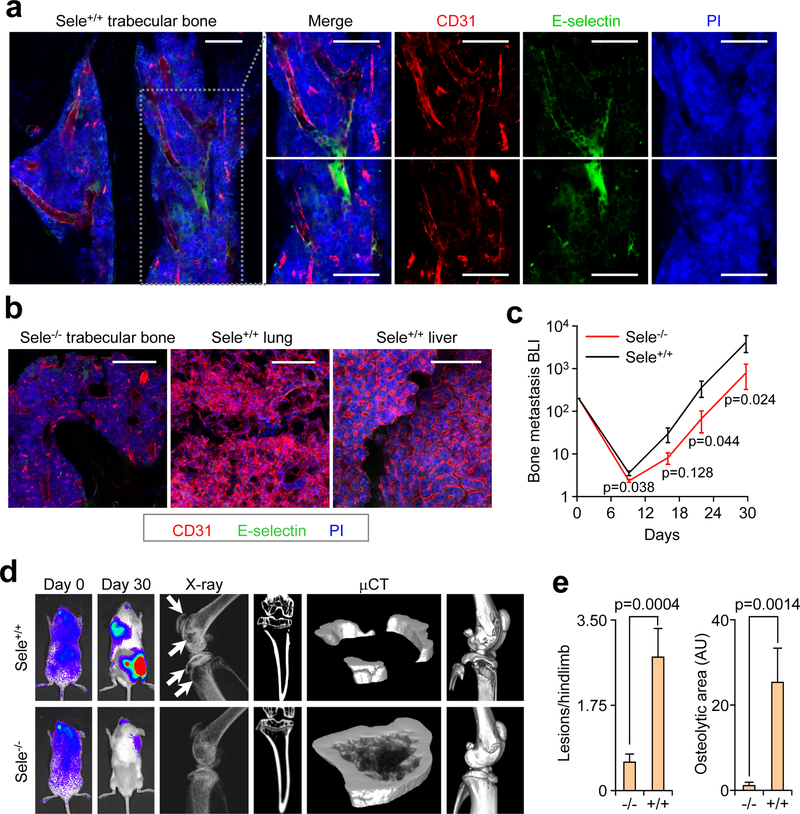
Whereas mouse and xenograft models of colon cancer27 and melanoma28 tested in E-selectin knockout (Sele−/−) mice20, 29 have demonstrated that E-selectin is not essential for lung metastasis or primary tumor formation, the effect of genetic ablation of E-selectin on bone metastasis has not been evaluated. Therefore, we crossed Sele−/− mice into the NOD/SCID background for xenograft studies of bone metastasis. Immunofluorescent staining of E-selectin and the endothelial marker CD31 revealed co-localization in the trabecular and cortical bone marrow space in wild type mice (Fig.1a), which is consistent with previous studies30, 31, and E-selectin staining was absent in the bone vasculature of Sele−/− NOD/SCID mice (Fig.1b). Immunostaining and qPCR demonstrated that E-selectin expression is significantly lower in the lung and liver vasculature compared to bone, in both normal (Fig. 1b, Supplementary Fig. 1e), and inflammatory conditions (Supplementary Fig.1e) known to induce E-selectin expression30. Finally, qPCR analysis of pooled RNA collected from human tissues showed a two-fold greater expression of CD31-normalized E-selectin mRNA in bone compared to lung (Supplementary Fig. 1f).
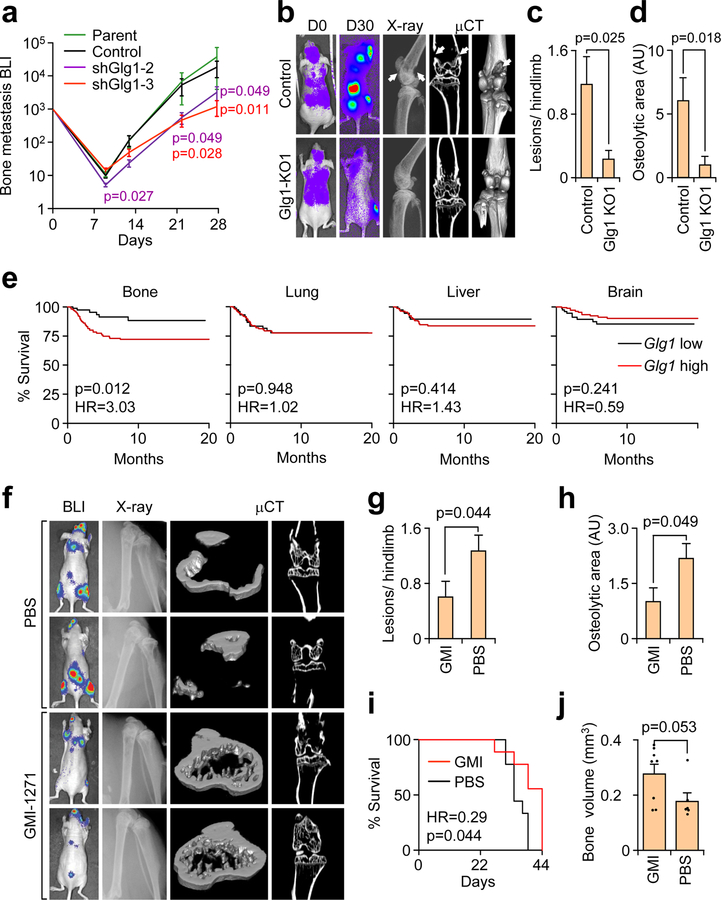
Figure 1. E-selectin is critical for bone but not lung metastasis.
(a,b) Bone, lung, and liver sections from WT or Sele−/− mice were assessed for E-selectin expression and CD31 co-localization by immunofluorescence. Scale bars: 100 μm. Data representative of three independent experiments. (c) BLI quantification of bone metastasis burden following intracardiac injection of the BM2 cell line into WT or Sele−/− SCID mice. n = 12 mice/group, Mann-Whitney U test, two-sided. (d) Representative BLI, X-ray, and μCT images of bone metastasis in WT and Sele−/− SCID mice. Images representative of median signal from (c). White arrows indicate osteolytic lesions. (e) Quantification of osteloytic area and the number of osteolytic lesions in the hind limbs of animals from (c). n = 23 Hindlimbs (WT), n = 24 hindlimbs (KO), Mann-Whitney U test, two-sided. Data representative of two independent experiments (c,d,e). Data represent mean ± SEM.
We next analyzed bone metastasis of the bone-tropic BM2 (1833) subline of MDA-MB-23125 in E-selectin knockout or wild type NOD/SCID mice. Bioluminescent imaging (BLI), X-ray and micro-CT (μCT) analyses revealed that genetic knockout of E-selectin significantly attenuated bone metastatic tumor burden (Fig. 1c–e). A similar result was observed when an in vivo-derived subline of the SUM159 breast cancer line that shows strong bone metastatic propensity, named SUM159-M1a, was injected in Sele−/− compared to WT mice (Supplementary Fig. 1g–j). SUM159-M1a primary tumor growth was largely unaffected by E-selectin knockout, although an increase in tumor volume in Sele−/− mice emerged on the final day of measurement (Supplementary Fig. 1k), consistent with a previous report28.
Since the lung metastatic LM2 subline of MDA-MB-23126 has substantial E-selectin binding ability (Supplementary Fig. 1c), we used this cell line to test the effect of E-selectin knockout on lung metastasis. E-selectin knockout did not attenuate lung metastasis; rather, we observed a trend toward increasing total lung metastasis burden in Sele−/− NOD/SCID mice (Supplementary Fig. 1l–n). We observed a similar result when the same lung metastasis experiment was performed using SUM159-M1a cells (Supplementary Fig. 1o–p). Taken together, these data establish that E-selectin is expressed at an elevated level in bone vasculature and is specifically important for the development of bone metastasis.
α1–3 Fucosyltransferase-3 and -6 promote E-selectin-mediated bone metastasis
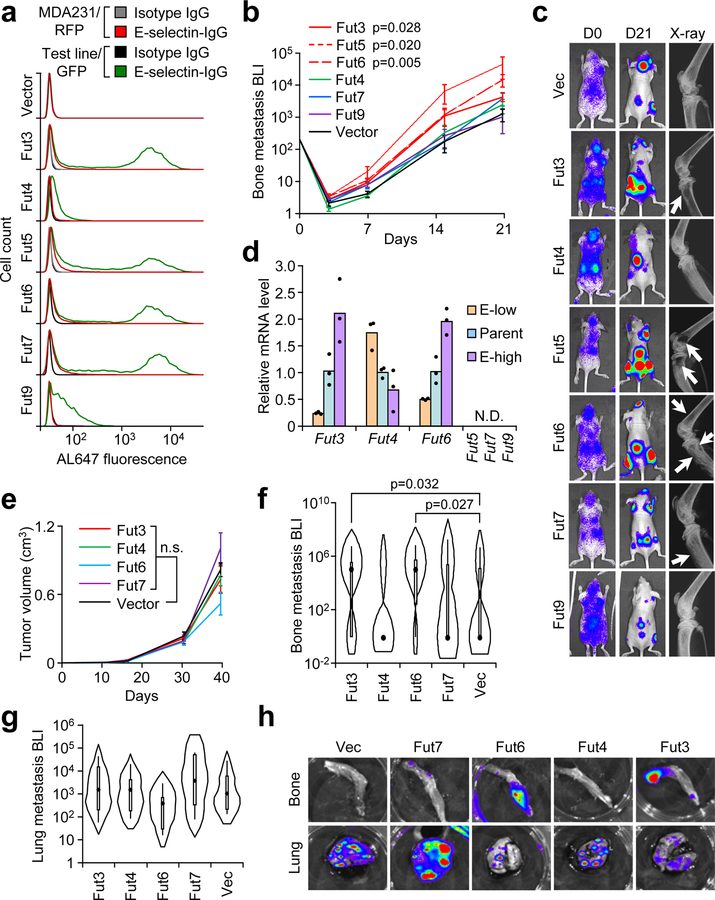
E-selectin binding to a cell requires the presence of sialyl Lewis X or A (sLeX/A) tetrasaccharides at the termini of cell surface glycolipids, or Glycoprotein O-glycans (Ser/Thr-linked) or N-glycans (Asn-linked)31. Six fucosyltransferases (Fut3/4/5/6/7/9) are competent to perform the final α1–3 or α1–4 fucosylation to generate sLeX/A 32,33, 34,35,36. Importantly, Fut4/7/9 are present in both the human and murine genomes, while Fut3/5/6 are present only in humans37. The contributions of these enzymes to tumorigenesis and organotropic metastasis have not been systematically analyzed. Ectopic expression of each Fut in the MDA-MB-231 cell line did not affect proliferation rates (Supplementary Fig. 2a–b) while flow cytometry showed that Fut3/5/6/7 increase E-selectin binding by several orders of magnitude while Fut4/9 increase binding by 2–5 fold (Fig. 2a). A similar binding pattern was observed in the SUM159-M1a cell line upon ectopic expression of Futs (Supplementary Fig. 2c–d).
Figure 2. Specific α1–3 Fucosyltransferases (Fut3 and Fut6) promote bone metastasis.
(a) Comparative flow cytometry analysis of E-selectin binding to MDA-MB-231 cells with stable ectopic expression of α1–3 Fut enzymes. MDA-MB-231-RFP was used as an internal control. Data representative of four independent experiments. (b) BLI quantification of bone metastasis burden following intracardiac injection of M1a cells stably expressing each Fut enzyme into Nu/Nu mice. n = 6 (Fut3, Fut4, Fut6), 5 (Fut5, Fut9), 3 (Fut7), and 10 (Vector) mice. Statistics by Mann-Whitney U test at Day 21, two-sided. (c) Representative BLI and X-ray images of bone lesions from (b). White arrows indicate osteolytic lesions. (d) qPCR analysis of endogenous α1–3 Fut mRNA levels in the parental and sorted MDA-MB-231 cells with differential E-selectin binding abilities. Fut5/7/9 were not detectable (N.D.) in all cell lines. n = 3 technical replicates. (e) Tumor volume measurements after orthotopic injection of M1a cells stably-expressing each relevant Fut enzyme into NSG mice. p > 0.05 for all between-group comparisons by two-sided Student’s t-test at Day 39. n = 6 mice/group. (f,g) Violin plot showing BLI quantification of spontaneous bone (f) and lung (g) metastasis burden. Plot elements include median, box for interquartile range, spikes to upper- and lower- adjacent values. Statistics by Mann Whitney U test, one-sided. n = 11 lung/22 hindlimb (Fut3 and Fut 4), 6 lung/12 hindlimb (Fut 6 and Fut 7), and 12 lung/24 hindlimb (Vector). (h) Representative BLI images of bone and lung tissues from each experimental group. Experiment performed once (e-h). Data represent mean ± SEM. No statistically significant difference (p>0.05) between Fut4, Fut7, and Fut9 groups vs Vector group in b, Fut 4 and Fut 7 groups vs Vector group in f, and any group vs Vector group in e and g.
Intracardiac injection of M1a-Fut cells demonstrated that only the human-specific Futs (Fut3/5/6) and not Fut7 promoted bone metastasis (Fig. 2b–c, Supplementary Fig. 2e–f), which is supported by previous findings that Fut6 mediates increased engraftment of human MSCs and human prostate cancer cells in bone38, 39. Clinical analysis of TCGA data next revealed that Fut5 is expressed at low levels in both normal and cancerous human breast tissues compared to more abundant expression of other Futs (Supplementary 2g).
To analyze which Fut enzymes endogenously contribute to increased E-selectin binding, the MDA-MB-231 cell line was FACS-sorted into the top and bottom decile of E-selectin binding (Supplementary Fig. 2h). After five passages, E-selectin binding levels were maintained at respectively higher or lower levels (Supplementary Fig. 2i) and qPCR revealed that Fut3 and Fut6 expression positively correlated with E-selectin binding while Fut4 was negatively correlated (Fig. 2d). A similar pattern was observed between different sublines of MDA-MB-231 cells, with elevated Fut6 and lower Fut4 expression in a strongly bone metastatic subline (SCP25) (Supplementary Fig. 2j).
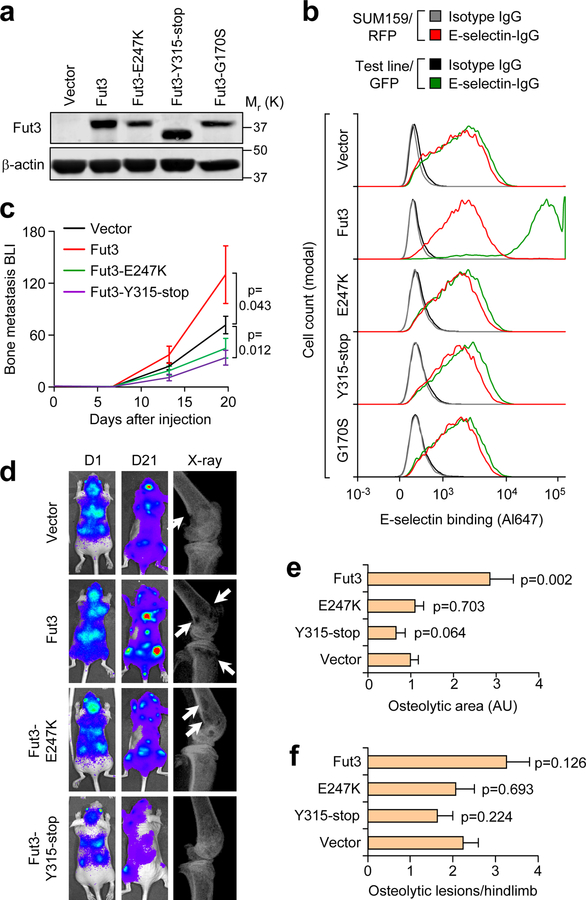
We next tested if the bone metastasis-promoting effects of Fut3/6 replicated in a spontaneous multi-organ metastasis model. SUM159-M1a cells expressing either of these bone metastasis-promoting Futs or Fut4/7 as negative controls were implanted in the mammary fat pad of NSG mice. Ectopic Fut3/4/6/7 expression did not affect primary tumor growth (Fig. 2e), while only Fut3 and Fut6 promoted spontaneous bone metastasis and none of the enzymes affected spontaneous lung metastasis (Fig. 2f–h, Supplementary Table 1.). To test whether these bone metastasis effects depend on Fut catalytic activity, we generated three catalytic mutants of Fut3 (Fig. 3a). E-selectin binding analysis confirmed that each mutant was catalytically inactive (Fig. 3b). In contrast to the increased bone metastasis caused by overexpression of Fut3, mutant forms of Fut3 did not promote bone metastasis formation (Fig. 3c–f). In particular, the Y315-stop mutant reduced bone metastasis based on BLI measurements, possibly due to dominant negative effects on wild type Fut3/6.
Figure 3. In vitro and in vivo characterization of Fut3 catalytic mutants.
(a) Western blot detection of three Fut3 catalytic mutants compared to wild-type Fut3 ectopically expressed in SUM159-M1a cells. (b) Comparative flow cytometry analysis of E-selectin binding to M1a cells expressing each Fut3 mutant using SUM159-RFP as an internal control. Data representative of 3 independent experiments (a,b). (c) BLI quantification of bone metastasis burden following intracardiac injection of M1a cells stably expressing each Fut3 mutant compared to vector and wild-type Fut3. n = 8 (Fut3 and Y315-stop) and 9 (E247K and Vector) mice/group. Mann-Whitney U test, two-sided. (d) Representative BLI and X-ray images from (c). (e,f) Quantification of osteloytic area (e) and the number of osteolytic lesions (f) in the hind limbs of Nu/Nu mice receiving an intracardiac injection of M1a cells expressing each Fut3 mutant. n = 16 (Fut3 and Y315-stop) and 18 (E247K and Vector) hindlimbs/group. Mann-Whitney U test, two-sided. Experiment performed once (c–f). Data represent mean ± SEM. Unprocessed original scans of the blots in a are shown Supplementary Fig. 9.
N-glycoprotein capture mass spectrometry identifies candidate substrates of human Fut enzymes
Given the data showing that increased expression of Fut3 or Fut6 promotes bone metastasis, non-fucosylated glycan substrates of these enzymes must exist in both cell lines (Supplementary Fig. 3a, top). We determined that these are displayed on N-glycans, as M1a and BM2 cells grown with Tunicamycin or 1-Deoxymannojirimycin (DMJ), which block N-linked glycosylation, have reduced E-selectin binding, while D,L-Threo PDMP, an inhibitor of glucosylceramide synthase, moderately reduced E-selectin binding to BM2 and did not affect E-selectin binding to M1a cells (Supplementary Fig. 3b–c). Furthermore, GMI-1271, a sLeX/A mimetic, was capable of blocking E-selectin binding to M1a cells expressing each Fut enzyme (Supplementary Fig. 3d), indicating these were conventional E-selectin ligands.
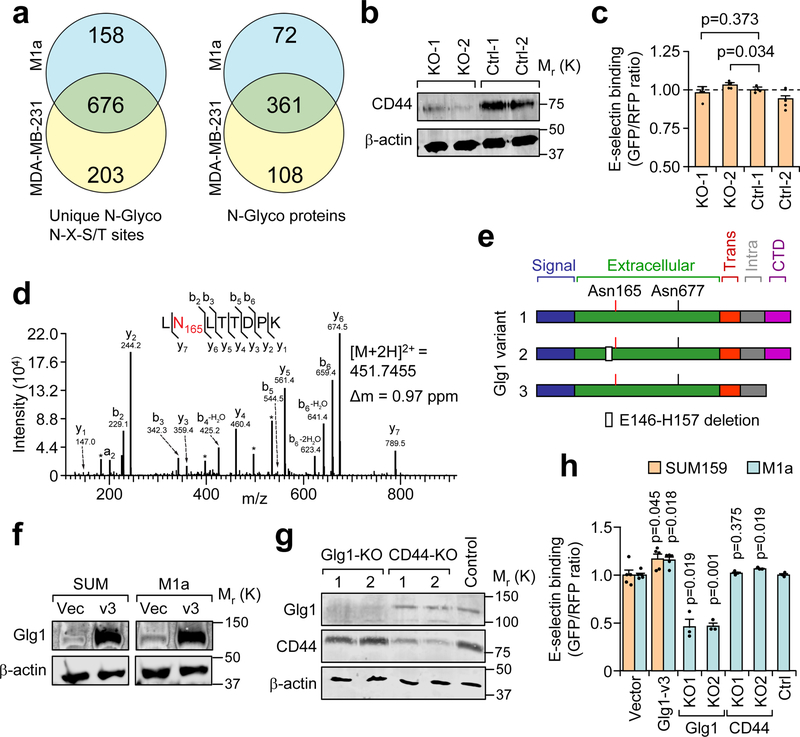
We therefore adapted a solid-phase N-glycoprotein enrichment protocol to comprehensively profile cell surface glycoproteins and identify candidate Fut enzyme substrates (Supplementary Fig. 3a, bottom)40, 41. Imaging and flow cytometry controls demonstrated effective ligation of extracellular oxidized glycoproteins (Supplementary Fig. 4a–c). N-glycoproteins were then isolated from MDA-MB-231 and SUM159-M1a cells via 1) trypsin digestion to remove non-glycosylated peptides, 2) N-glycopeptide release with PNGase F, 3) LC-MS/MS analysis, 4) Computational filtering if peptides contained one asparagine that was deamidated from PNGase F digestion, matched the known N-X-S/T glycosylation motif, and were detected in either all four samples or detected in both samples from one cell line. Filtering by these criteria resulted in the identification of 1037 unique N-glycosites that mapped to 541 proteins (Fig. 4a, Supplementary Table 2).
Figure 4. Cell surface N-glycan analysis reveals candidate E-selectin ligands in metastatic breast cancer cells.
(a) Summary of mass spectrometry results of N-glycosites and N-glyco proteins using two biological replicates of each cell line collected over independent isolations. (b) Western blot confirming CD44 knockout by CRISPR-Cas9 in the BM2 cell line. Data representative of 3 independent experiments. (c) Comparative flow cytometry analysis of E-selectin binding to CD44-KO and control BM2 cell lines. MDA-MB-231-RFP used as an internal control and binding levels normalized to Ctrl-1. n = 4 independent biological replicates, Student’s t-test, two-sided. (d) Representative fragmentation spectra of the doubly-charged Glg1 tryptic peptide LNLTTDPK containing deamidated Asn(N)165 (red). N- and C-terminal (b and y) ions matching the predicted peptide fragment are indicated with their respective mass-to-charge (m/z) values. A complete y ion series was observed, including the site-determining y7 ion, localizing the deamidation to the Asn residue. The measured doubly-charged monoisotopic precursor ion m/z and its mass deviation (Δm) relative to the theoretical m/z are indicated. * interfering m/z from lower abundance co-isolated ion(s). Similar fragmentation spectra were obtained across independent isolations. (e) Schematic diagram of the three major Glg1 splice variants in humans. (f) Western blot analysis of Glg1 expression in SUM159 and M1a cells after stable transduction of retroviral vector expressing Glg1 variant 3. (g) Western blot analysis of Glg1 and CD44 after population-level CRISPR/Cas9 knockout of Glg1 and CD44 in M1a cells. Data representative of three independent experiments (f, g). (h) Comparative flow cytometry analysis of E-selectin binding to genetically manipulated SUM159 and M1a cells using SUM159-RFP as an internal control. Binding levels were normalized to respective controls. n = 5 (Glg1 or Vector), n = 3 (KO cell lines) independent biological replicates. Student’s t-test, two-sided, compares Glg1v3 cells to Vector cells and CRISPR-KO cells to CRISPR-Control cells. Data represent mean ± SEM. Unprocessed original scans of the blots in b, f, g are shown Supplementary Fig. 9.
The 361 N-glycosylated proteins shared between the MDA-MB-231 and M1a cells were cross-referenced with reported E-selectin ligands and ranked according to individual N-glycopeptide detection across all four runs by their respective normalized intensities (Supplementary Table 3). The top candidate identified was CD44, which, when decorated with sLeX is known as Hematopoietic Cell E-/L-selectin Ligand (HCELL). HCELL has been attributed as the major E-selectin binding ligand on metastatic cells23, 24, 42, 43 and HPCs21. To test if HCELL was responsible for E-selectin binding, we utilized lentiviral CRISPR/Cas9 system to knock out CD44 in the BM2 cell line (Fig. 4b). Loss of CD44 expression did not decrease E-selectin binding in vitro (Fig. 4c), or inhibit bone metastasis in vivo (Supplementary Fig. 4d–e).
Glg1 is functionally important to E-selectin binding of bone-metastatic breast cancer cells
N-glycosylation of the second candidate, Golgi glycoprotein 1 (Glg1, also called E-selectin Ligand 1, Esl1) (Fig. 4d–e), were detected at two different residues, with Asn165 detected across all samples (Supplementary Table 3). The site and peptide was confirmed by MS/MS fragmentation and similar elution profiles/ extracted ion chromatograms from both cell lines (Fig. 4d, Supplementary Fig. 4f). Previous research has identified a role for Glg1 by HPCs in maintaining homeostasis or proliferation, possibly in an E-selectin-dependent manner22, 44. While Glg1 has been detected in metastatic prostate cells23, 45, the functional importance of Glg1 in cancer has not been investigated.
To test if Glg1 expression affected with E-selectin binding, isoform-specific primers for the three main Glg1 variants (Fig. 4e) were used for qPCR. We found that variants 1 and 3 were enriched in E-selectin high-binding cells (E-high and SCP25), while variant 2, which harbors an internal deletion located 8 amino acids upstream from the putative Asn165 modification site, is inversely correlated to E-selectin binding ability (Supplementary Fig. 4g–h). We ectopically expressed Glg1 variants 1 and 3 in SUM159, M1a, MDA-MB-231, and BM2 cells, and knocked out Glg1 in M1a and BM2 cells using lentiviral CRISPR/Cas9 (Fig. 4f–g, Supplementary Fig. 4i–k). The comparative flow cytometry assay revealed that ectopic expression of Glg1 variant 1 and variant 3 increased E-selectin binding to varying degrees in different cell lines, while population-level CRISPR-mediated knockout of Glg1 reduced E-selectin binding by ~50% in the M1a cells and ~20% in the BM2 cell line (Fig. 4h, Supplementary Fig. 4l). Consistent with the result using BM2 cells (Fig. 4c), we observed that CD44 knockout also did not affect E-selectin binding in the M1a cell line (Fig. 4h).
In order to serve as a bona fide E-selectin ligand, Glg1 must be expressed on the cell membrane and be capable of binding E-selectin. TIRF imaging was performed at the critical angle on M1a cells probed with anti-Glg1 antibody and E-selectin-IgG demonstrated that Glg1 and E-selectin ligands are displayed on the cell surface, co-localized to the same puncta (Fig. 5a). Furthermore, western blotting of Glg1 revealed its presence in membrane fractions of M1a cells (Supplementary Fig. 5a). Confocal microscopy of permeabilized M1a cells also demonstrated colocalization of Glg1 and E-selectin binding throughout the cell, both at the cell membrane and within internal structures (including the Golgi/ER) (Fig. 5b). To assess whether Glg1 directly binds E-selectin, we performed western blot under denaturing conditions on cell membrane fractions from M1a Glg1 knockout, Glg1 overexpression, and Fut3–7,9 overexpression cells using E-selectin-IgG as a probe with Fut6-transfected prostate cancer cells (PCR1) serving as a positive control46. While E-selectin-IgG-reactive bands were detectable in the cell membrane fractions of Fut3–5-6–7 expressing cells, E-selectin-IgG-reactive bands were not detected in control, Glg1 knockout, Glg1 overexpression, or Fut4/9 membrane fractions (Supplementary Fig. 5b) despite E-selectin binding evident by flow cytometry and immunofluorescence microscopy. Immunoprecipitation of M1a lysates by E-selectin-IgG, followed by probing with Glg1 antibody, further confirmed that an E-selectin interaction with Glg1 was not detectable using traditional biochemical methods (Supplementary Fig. 5c). Thus, although the data indicate that (1) N-glycan decoration of proteins, but not glycosphingolipids, are critical to E-selectin binding, (2) Glg1 is expressed on the cell surface, is N-glycosylated, and co-localizes with E-selectin binding, and (3) Genetic manipulation of Glg1 leads to significant changes in E-selectin binding levels, we did not obtain evidence that Glg1 directly binds E-selectin.
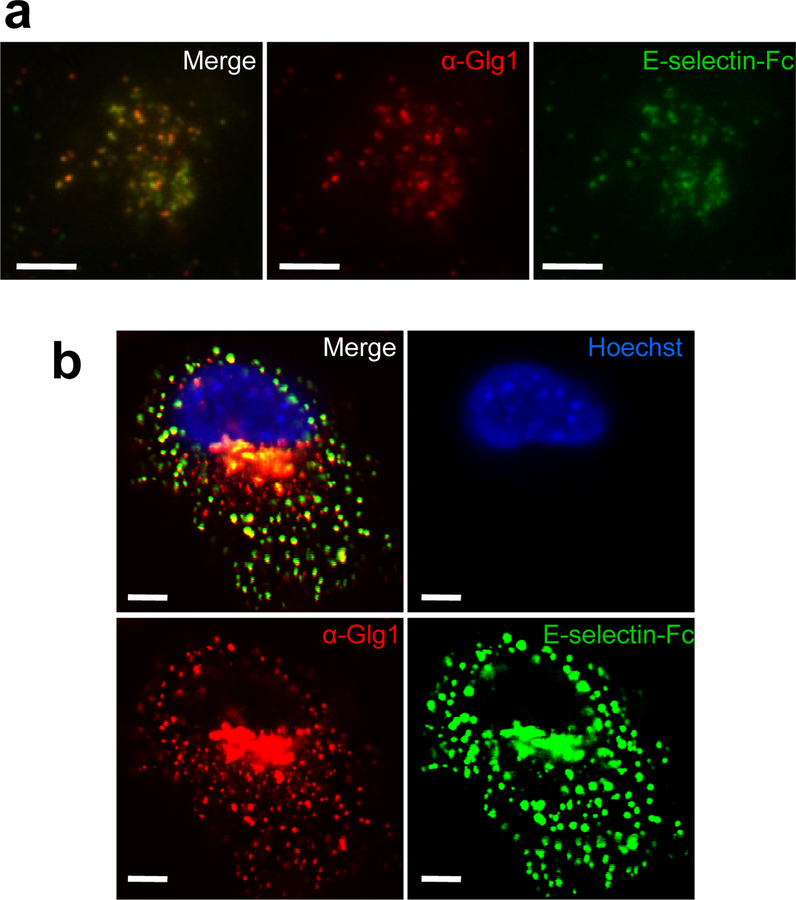
Figure 5. E-selectin ligands co-localize with Glg1 at the cell surface.
(a) Total internal reflectance fluorescence (TIRF) microscopy at the critical angle using a spinning disc was performed on SUM159-M1a cells probed with recombinant E-selectin-Fc (green) and anti-Glg1 (red). Scale bar represents 5 μm. (b) Confocal Z-slice of M1a cells probed with E-selectin-Fc (green), anti-Glg1 (red) and Hoechst (blue). Scale bar represents 5 μm. Data representative of 3 independent experiments (a, b).
We next tested whether Glg1 ablation affects metastasis in vivo. Two lentiviral shRNA constructs targeting all splice variants of Glg1 reduced Glg1 expression levels in BM2 cells (Supplementary Fig. 6a). Bioluminescent imaging following intracardiac injection confirmed that Glg1 knockdown reduced bone metastasis burden (Fig. 6a). CRISPR/Cas9-mediated knockout of Glg1 in BM2 and M1a cells similarly resulted in a significant reduction of bone metastasis based on BLI, X-ray and μCT analysis of bone lesions (Fig. 6b–d, Supplementary Fig. 6b–f). In contrast, Glg1 knockdown in LM2 cells (Supplementary Fig. 6g) did not affect lung metastasis progression or survival of animals (Supplementary Fig. 6h–i). The results were in line with the same observations made with either E-selectin knockout (Fig. 1) or Fut overexpression (Fig. 2).
Figure 6. Glg1 is required to support bone metastasis progression.
(a) BLI quantification of bone metastasis burden after intracardiac injection of BM2 cells stably expressing Glg1-targeting or control shRNA or untransduced (parental) BM2 cells into Nu/Nu mice. n = 8 mice/group. Mann-Whitney U test, two-sided, compares shGlg1 BLI signal to control BLI signal. Data representative of two independent experiments. (b) Representative BLI, X-ray, and μCT images from mice injected with BM2 cells modified by CRISPR-mediated Glg1 knockout. White arrows indicate osteolytic bone lesions. (c,d) The number of lesions/ hindlimb (c) and average osteolytic area (d) were quantified from X-ray images in b. n = 17 hindlimbs/group. Mann-Whitney U test, two-sided. Data representative of two independent experiments (a-d). (e) Kaplan-Meier organ-specific metastasis-free survival curves of ER− breast cancer patients in the EMC-MSK data set stratified by median expression level of Glg1 mRNA. n = 244, Cox’s proportional hazards model, two-sided. (f) Representative BLI, X-ray, and μCT images of mice treated with either GMI-1271 or PBS control after intracardiac injection of BM2 cells to generate bone metastasis in Nu/Nu mice. (g, h) Total number of lesions per hindlimb (g) and total osteolytic area (h) were quantified between treatments. n = 12 hindlimbs/group. Mann Whitney U test, two-sided. (i) Mice from f were censored upon becoming moribund and followed for Kaplan-Meier survival curve analysis. n = 9 mice/group, Cox’s proportional hazards model, two-sided. (j) Bones from moribund mice collected at day 39 (PBS, n = 3) or day 44 (GMI-1271, n = 4) were analyzed by μCT and trabecular bone volume from 4 mm above and below the knee joint was quantified. Mann-Whitney U test, two-sided. Experiment performed once (f–j). Data represent mean ± SEM.
Pharmaceutical targeting of E-selectin reduces bone metastasis
Given the identification of E-selectin as a pro-metastatic receptor of the bone vascular niche whose binding is predominantly governed by Fut3/6 and Glg1, we sought to assess if this relationship could predict bone metastasis in human breast cancer. Patients in the NKI-29547 or EMC-MSK26, 48 datasets were stratified by median levels of Glg1 and Fut3 or Fut6 and grouped into four cohorts. Those with high Glg1 and high Fut3 or Fut6 suffered from worse prognosis for distant metastasis-free relapse in the NKI295 datasets (Supplementary Fig. 6j). Next, organ-specific metastasis data from the EMC-MSK dataset indicated that Glg1 is a significant prognostic marker of bone metastasis but not of metastasis to brain, liver or lung (Supplementary Fig. 6k). Here we analyzed data from ER− breast cancer as metastases in these patients often manifest at multiple organ sites. We further assessed the prognostic potential of each candidate E-selectin ligand identified by mass spectrometry (Supplementary Table 3) in organ-specific metastasis, and found that the only significant relationship was Glg1 as a poor prognosis indicator in bone metastasis (Fig. 6e; Supplementary Table 4).
These results suggest that therapeutic inhibition of E-selectin in patients with tumors expressing a high level of Fut3/6 and Glg1 may slow down or prevent bone metastasis progression. To test this notion, mice were treated with GMI-1271, a glycomimetic inhibitor of E-selectin (Supplementary Fig. 1b), twice daily at 20 mg/kg for 14 days immediately following intracardiac injection with BM2 cells. Bone metastasis-associated bone degradation was attenuated in mice treated with GMI-1271 (Fig.6f–h). A distinct survival advantage for those mice receiving GMI-1271 was observed (Fig. 6i) while post-mortem μCT bone volume scans revealed greater preservation of bone tissue in those mice receiving treatment (Fig. 6j).
E-selectin binding promotes a specific mesenchymal-to-epithelial transition program
While the majority of in vitro studies have speculated that E-selectin may promote metastasis by arresting tumor cells in the vasculature of target organs through adhesive interactions19, 27, 46, this mechanism is unlikely to be important to bone metastasis as the blood flow in sinusoidal endothelium of bone marrow has been noted to be particularly slow49, 50, or even stationary51. Instead, we considered the possibility that E-selectin engagement of metastatic cells to the endosteal endothelium during the initial phase of bone metastatic colonization (as visualized in Supplementary Fig. 7a) may induce molecular changes in cancer cells that promote bone colonization. This hypothesis was supported by the observation that the growing edge of BM2 bone lesions showed strong Glg1 expression and were associated with a high density of E-selectin+ vasculature (Supplementary Fig. 7b).
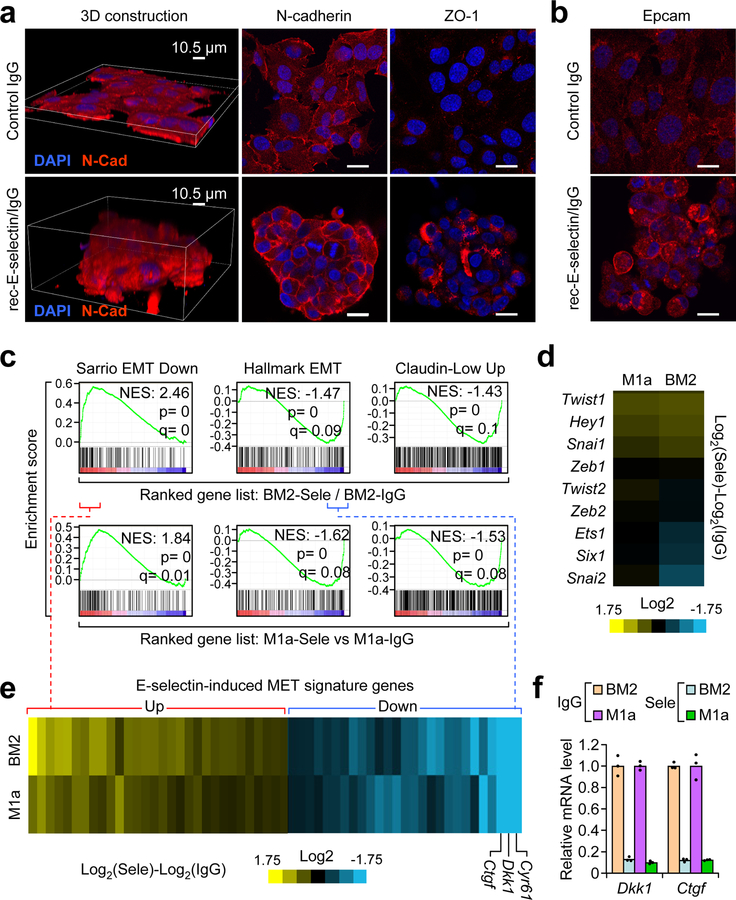
To mimic the engagement of tumor cells to E-selectin, BM2 or M1a cells were seeded on E-selectin- or IgG-coated plates, and were subjected to spinning-disc microscopy over a period of 40 hours. Dramatic differences in cell behavior and morphology emerged within 3 hours of seeding (Supplementary Movie 1). Both BM2 and M1a cells migrated along the E-selectin-coated dishes, forming 3-dimensional clusters ranging from tens to hundreds of cells thick, while cells seeded on IgG plates formed simple monolayers as in normal tissue culture conditions (Fig. 7a). Immunofluorescence analysis of M1a spheres revealed enriched N-cadherin localization at cell-cell boundaries compared to diffuse staining in cells seeded over IgG. Furthermore, Tight Junction Protein-1 (TJP-1, ZO-1) was only detectable in cells seeded on E-selectin (Fig. 7a). Staining for Epcam and Keratin-14 revealed an enhanced shift toward an epithelial state in BM2 spheres (Fig. 7b, Supplementary Fig. 7c).
Figure 7. E-selectin binding to tumor cells induces a non-canonical mesenchymal-to-epithelial transition.
(a) Confocal imaging of E-selectin-induced morphological changes in M1a cells after culturing on E-selectin- compared to IgG-coated plates (10 μg/mL) for 24 h. Z-slice of confocal immunofluorescent images demonstrating N-cadherin and Tight Junction Protein-1 (ZO-1) localization/expression changes in M1a cells. Scale bars represent 20 μm. (b) Confocal imaging of EpCam immunofluorescence in BM2 cells seeded over E-selectin or IgG coated plates. Scale bars represent 20 μm. Images representative of 3 independent experiments (a–b). (c) GSEA of the Sarrio EMT Down (GSE8430), Hallmark EMT Up (M5930), and Claudin-low Up61 gene sets in the ranked gene list of the BM2 and M1a cells cultured in E-selectin- vs. IgG-coated plates. Single mRNA isolation used for each condition in each cell line. p and q statistics by GSEA software, n = 108 gene sets queried. (d) Heat map of the expression of known EMT transcriptional regulators in BM2 and M1a cells cultured in E-selectin vs. IgG coated plates. (e) Overlapping enrichment core genes from the Sarrio EMT down gene set (positive) and Hallmark EMT Up gene set (negative) between the BM2 and M1a cells were compiled to form the E-selectin-induced MET gene signature shown in the heatmap. The gene identities are listed in Supplementary Table 5. (f) qPCR analysis of Dkk1 and Ctgf mRNA levels in BM2 and M1a cells seeded over IgG or E-selectin for 24h. n = 3 technical replicates. Data represent mean ± SEM.
Microarray and gene set enrichment analysis (GSEA) performed on cells seeded for 24 h on either E-selectin- or IgG-coated plates revealed inverse enrichment of gene signatures related to epithelial-to-mesenchymal transition (EMT) (Fig. 7c, Supplementary Fig. 7d–e). Unlike traditional models of EMT, E-selectin-induced MET did not affect the RNA expression of the master transcriptional regulators of EMT, such as Snail1/2, Twist1/2 and Zeb1/2 (Fig. 7d). Furthermore, staining of the EMT maker N-cadherin did not show reduced expression, but rather altered localization and apparent molecular weight (Fig.7a, Supplementary Fig. 7f), while protein levels of the Slug transcription factor were much lower after binding E-selectin (Supplementary Fig. 7f). Taken together, these observations pointed toward a non-canonical MET program that is not the binary opposite of traditional EMT programs. Supporting this, extraction of the enrichment core genes in the Sarrio EMT and Hallmark EMT gene sets (Fig. 7c) showed that the majority of upregulated EMT-associated genes were involved in immune-related processes while the down-regulated genes were largely secreted or extracellular proteins (Fig. 7e and Supplementary Table 5). Taken together, these results indicate that E-selectin binding induces a non-canonical MET-like shift in cancer cells. Finally, staining for epithelial markers revealed ubiquitous E-cadherin and intermittent EpCam staining in BM2 bone lesions (Supplementary Fig. 7g–h), supporting the occurrence of MET during bone metastasis in vivo.
E-selectin-induced MET activates Wnt signaling
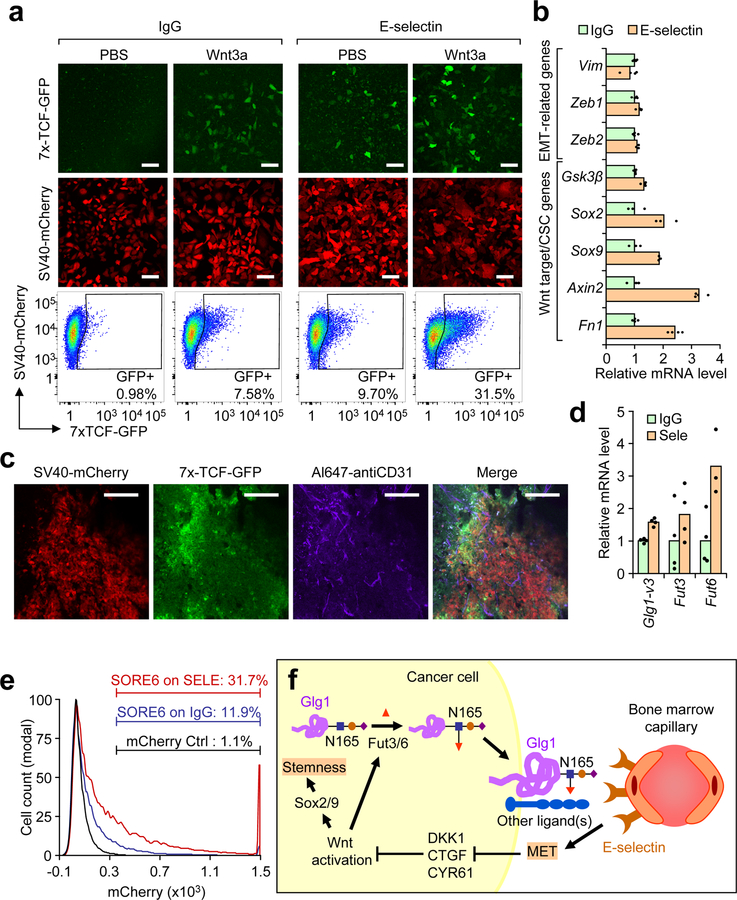
The three most down-regulated mesenchymal-associated genes (Dkk1, Ctgf, and Cyr61) after E-selectin-induced MET encode secreted Wnt repressors52 (Fig. 7e–f). We next asked if repression of these Wnt inhibitors by E-selectin-induced MET could indeed activate Wnt signaling, as Wnt signaling has been reported to promote cancer stem cell activities during metastasis55. To this end, we introduced a 7x-TCF-GFP Wnt reporter plasmid53 into BM2 cells (BM2-TGC). Cells seeded on E-selectin for 48 h activated this Wnt reporter to similar levels as those seeded on IgG and treated with recombinant Wnt3a (Fig. 8a). Live imaging revealed that MET occurred in the first several hours while Wnt activation occurred 30 hours after binding (Supplementary Movie 1). To confirm that E-selectin activated canonical Wnt signaling, qPCR revealed that multiple Wnt target genes, including those related to CSCs (e.g. Sox2 and Sox9) were induced by plating tumor cells on E-selectin, while canonical EMT markers Vim, Zeb1, and Zeb2 remained mostly unchanged (Fig. 8b), consistent with results from gene expression profiling analysis (Fig. 7d). Ex vivo microscopy of BM2-TGC bone lesions demonstrated that Wnt signaling was active in cells that were in a high density of CD31+ vasculature (Fig. 8c, Supplementary Fig. 8a).
Figure 8. E-selectin-induced MET activates Wnt signaling.
(a) BM2 cells stably expressing the 7x TCF-GFP reporter and SV40-mCherry as internal control (BM2-TGC) were plated on either E-selectin or IgG coated (10 μg/mL) plates with or without recombinant Wnt3a (100 ng/mL). Fluorescence was assessed by confocal microscopy after 48 h and quantified by flow cytometry. Scale bars represent 100 μm. Data representative of >5 independent experiments. (b) qPCR analysis of EMT- or Wnt-associated genes from the BM2 cells plated on E-selectin or IgG (10 μg/mL) for 48 h. n= 3 technical replicates. (c) Ex vivo confocal images of bone metastasis in Nu/Nu mice injected with BM2-TGC. Live animal labeling with anti-CD31 was conducted to visualize vasculature. Scale bar represents 100 μm. Data representative of 4 biological replicates. (d) qPCR of Glg1-variant 3, Fut3 and Fut6 levels in BM2 cells seeded on either IgG or E-selectin (10 μg/mL) for 48 h. n = 4 technical replicates. (e) BM2 cells stably-expressing the SORE6-mCherry stemness reporter or control mCherry reporter were plated on E-selectin or control IgG (10 μg/mL) for 48 h. mCherry fluorescence was assessed by flow cytometry after 48 h. Data representative of 3 independent biological replicates. (f) Proposed model of E-selectin function during bone metastasis. Data represent mean ± SEM.
Ectopic expression of Dkk1-Flag in BM2-TGC cells (Supplementary Fig. 8b) blocked Wnt activation following exposure to Wnt3a-conditioned media but exerted a lesser effect on E-selectin-mediated Wnt signaling. In contrast, inhibitors of β-catenin-TCF binding (ICG-001 and LF3) disrupted E-selectin mediated Wnt signaling while exerting less of an effect on paracrine-mediated Wnt activation (Supplementary Fig. 8c). Treatment of BM2 bone metastases with LF3 did not exhibit observable toxicity but reduced the number of osteolytic lesions (Supplementary Fig. 8d–f).
Recent glycomic analysis has found a strong correlation between extracellular fucosylation and epithelial features54. qPCR analysis of Glg1-variant 3, Fut3, and Fut6 levels 48 h after seeding on E-selectin confirmed that E-selectin-induced MET led to increased expression of the genes responsible for the binding of tumor cells to E-selectin (Fig. 8d), indicating the existence of a positive feedback loop. This finding is consistent with the stronger Glg1 staining in metastatic cancer cells in close contact with bone vasculature (Supplementary Fig. 7b). To investigate whether this Wnt activation contributed to cancer stem cell identity, we transduced BM2 cells transduced with a Sox2/Oct4-mCherry (SORE6-mCherry) cancer stem cell reporter55. These cells were 11% positive for Sox2/Oct4 when plated on IgG, but increased to 31.7% positive upon binding to E-selectin-coated plates, indicating an increase in cancer stemness activity upon binding to E-selectin (Fig. 8e).
Finally, we wanted to understand whether this mechanism extended to other models of bone metastasis and whether the discovered mechanism is relevant to clinical bone metastasis patients. Therefore, we used in vivo selection to derive a bone-metastatic subline (named ob1) from the DU145 prostate cancer cell line56. Flow cytometry revealed this subline binds E-selectin (Supplementary Fig. 8g). Similar to breast cancer studies, E-selectin knockout mice exhibited lower bone metastasis colonization and survived longer than wild-type hosts following intracardiac injection of DU145-ob1 (Supplementary Fig. 8h–i). Plating these cells on E-selectin-coated plates revealed a similar phenomenon of clustering and a shift toward epithelial features (Supplementary Fig. 8j). Furthermore, immunostaining of E-selectin, Glg1 and Ki67 in bone metastasis biopsies from prostate cancer patients demonstrated an association between E-selectin expression and Glg1+ tumor cells that are proliferative (Ki67+) (Supplementary Fig. 8k).
DISCUSSION
While numerous studies have established that EMT is often necessary for the escape from a primary tumor7, and others have shown that these cells must revert to an epithelial state to successfully colonize an organ11–13, the lack of evidence for how MET is induced, especially in the context-dependent manner required for metastatic colonization in a distant organ, has resulted in considerable controversy within the field. Here we provide evidence that a unique stromal cue – binding to E-selectin via Fut3/6 and Glg1 expressed by bone metastatic cells – induces MET to facilitate bone metastasis. The data further indicate that this non-canonical MET activates Wnt signaling to promote stemness via Sox2/9 expression and further increases Glg1 and Fut3/6 expression (Fig. 8f).
E-selectin-induced MET is a non-canonical program compared to MET-inducers such as miR-200, which target the master transcription factors of EMT57. Rather, a key function of E-selectin-induced MET is the activation of Wnt signaling, a pathway linked to self-renewal, CSC traits, and EMT induction58. This reported link between Wnt-induced stemness and EMT is incompatible with the requirement for MET during metastatic colonization, which therefore raises an interesting paradox: how can MET and cancer stemness coexist during metastatic colonization? By linking E-selectin induced MET with Wnt signaling and Sox2/9 induction, we show how E-selectin engagement could resolve this paradox in bone metastasis, which is akin to recent findings showing Prrx1 uncouples EMT from stemness traits during lung metastasis11.
Our discovery of the crucial role of E-selectin in bone metastasis raises the question of why no prior studies have shown a functional effect of E-selectin. Three discoveries made here explain a possible cause: 1) Fut3/6 are responsible for generating functional E-selectin ligands, implying that murine models of bone metastasis may not be dependent on E-selectin. 2) E-selectin-induced Wnt activation occurred 30 hours after binding, whereas prior in vivo studies of E-selectin were performed within 6 hours of injection 42, 59. 3) The organ-specific expression pattern of E-selectin indicates it is most relevant to bone metastasis as compared to lung or liver metastasis27. Interestingly, in a transgenic mouse model of E-selectin under the control of the β-actin promoter, the murine B16F10 cell line could be redirected to the liver only if both Fut3 and E-selectin were ectopically expressed in the cell line and liver endothelium60.
Our discovery that Glg1 is instrumental to MET during bone metastasis and co-localizes with E-selectin ligands opens numerous avenues for further research to elucidate whether it chaperones ligand maturation or expression, and how it may function as an intermediate between E-selectin binding and MET induction. The common dependence of MDA-MB-231, SUM159 breast cancer cells and DU145 prostate cancer cells on E-selectin binding to achieve bone metastasis, together with the prognostic significance of Glg1/Fut3/Fut6 in clinical bone metastasis, indicate that the discovered pathway is likely a conserved mechanism that facilitates tumor-endothelial interaction during metastatic seeding in bone. Therapeutic agents targeting the E-selectin ligand-mediated pathway, such as LF3 or GMI-1271, may be developed for the treatment of bone metastasis.
METHODS
Cell lines, cell culture and human prostate cancer specimens
MDA-MB-231 cells and its derivatives25,26 and DU145 cells56 and its derivatives were cultured in DMEM and RPMI media, respectively, supplemented with 10% FBS and pen/strep. The SUM159 cell line62, its highly metastatic M1a subline, and all of the derivative sublines were grown in F12 media supplemented with 10% FBS, 10 μg/mL Insulin and 20 ng/mL EGF. The M1a cell line was derived from sequential in vivo selection, first from a SUM159 primary tumor which was dissociated and cultured, and then from a lung metastatic node following tail-vein injection of this culture. Both selections were performed in athymic Nu/Nu female mice. The ob1 cell line was derived from an osteoblastic lesion in Nu/Nu mice injected with DU145-GFP, expanded in culture, and validated to have increased bone metastatic ability in vivo. Cell lines were labeled with retroviral vectors with bi-cistronic expression of GFP/firefly luciferase or RFP/renilla luciferase to facilitate imaging and flow cytometry experiments. The PCR1 cell line that was used to prepare cell lysates was a gift from Steven Barthel46. All cell lines were verified negative for mycoplasma contamination by monthly PCR analysis. No cells lines used here appear in the database of commonly misidentified cell lines (ICLAC). All cell lines were validated with STR analysis and compared to NCBI repository data. Major characteristics of key cell lines used in the study are listed in Supplementary Table 6.
Human prostate cancer bone metastasis specimens were archived, anonymized specimens obtained from MDACC Prostate Cancer Tissue Bank through an IRB protocol approved by the MD Anderson Cancer Center Institutional Review Board. The study is compliant with all relevant ethical regulations regarding research involving human participants, and informed consent was obtained by all participants.
Mouse models and xenografts
All procedures involving mice and experimental protocols were approved by the University Institutional Animal Care and Use Committee (IACUC). The study is compliant with all relevant ethical regulations regarding animal research. E-selectin knockout mice were ordered from the Jackson Laboratory (B6.129S4-Seletm1/DmiL) and were backcrossed for 5 generations into the NOD/SCID strain. Genotyping for E-selectin knockout was performed as recommended by the Jackson Laboratory. Briefly, ear punches were digested in 75 μL 25 mM NaOH, 0.2 mM EDTA at 95°C for 20 min, followed by 10 min at 4°C and neutralization with 75 μL 40 mM Tris-HCL. The PCR reaction mixture contained 2 μL ear punch digest, 0.2 mM dNTP, 1 μM each primer (5’- AATGTCCTATGACTCACAGGAAGCC-3’, 5’- TTCTGTTCAGTGGAATTGTTGTTCTGGCGTT-3’, 5’- GCAGCCTCTGTTCCACATACACTT C-3’), 1x PCR reaction buffer, and 0.5 μL Taq. Annealing temperature of 65°C was used for 37 cycles followed by 1.5% agarose electrophoresis. PCR product of wild type SELE resolves at 182 bp while the mutant resolves at 220 bp. One founder pair of heterozygotes was used to generate homozygous knockout and wild-type breeder pairs. Age-matched offspring of these breeder pairs were used for xenograft experiments.
All xenograft experiments were conducted on 8-week-old female mice (athymic Nu/Nu, NOD/SCID, NOD/SCID Gamma) except in prostate cancer xenografts, which were performed on male mice of the same age. All mice were originally ordered from the Jackson Laboratory and breeding was conducted in an SPF barrier facility. No statistical method was used to predetermine the number of animals needed. Rather, studies in E-selectin WT or KO SCID mice were performed if the number of age-matched mice per genotype was ≥8. Xenograft experiments were conducted using 50k cells suspended in 10 μL PBS for mammary gland injection or 100k cells in 100 μL PBS for tail vein or intracardiac injection. Bioluminescent imaging (BLI) was conducted as previously described26, however only bioluminescent signal from the lower third of each mouse was quantified to exclude signal from metastasis in non-bone tissues. LPS was administered at 0.25 mg/30 g body mass via IP injection 6 h prior to tissue isolation from 8 week-old wild-type NOD/SCID mice. Bone marrow was isolated via femur flush, while lung tissue was extracted from a single lung lobe. Total RNA quantification via qPCR and statistical comparison was performed using tissues from the same mouse within the control or LPS condition. GMI-1271 (Glycomimetics Inc.) was dissolved in PBS at 4 mg/mL and dosed at 20 mg/kg body mass twice daily for 14 days immediately following intracardiac injection of BM2 cells. Wnt inhibitors LF3 (Selleck) were dissolved at 80 mg/mL in DMSO, diluted to 40 mg/mL in Cremaphor EL, and then diluted 1:5 in PBS. Mice were treated at 40 mg/kg via daily IP injection. Bioluminescence of spontaneous metastasis to the lungs or bones was conducted by retro-orbital injection of luciferin followed by euthanasia, dissection and imaging. Bones or lung isolated from mice were fixed in 10% buffered formalin or Bouin’s solution for 12 h at 22°C. X-ray images were collected at an exposure of 15 sec, 35 keV. Bone lesion count and area were calculated manually using ImageJ and single-blinded review of images. μCT was performed as previously described63 using core facility-specified threshold values for bone versus non-bone tissues.
Cloning, viral production, and transduction
The coding sequence of Glg1 variants 1 and 3, Fut3–7,9 were cloned from either the cDNA of the BM2 cell line or the cDNA of a pooled human reference RNA sample. Cloned sequences flanked by EcoR1 or Hpa1 restriction sites were inserted into the pMSCV retroviral plasmid. Clones were sequenced and compared against NCBI expressed sequence tags (ESTs) for accuracy. Catalytic mutants of Fut3 were generated as Glu247Lys (GAG>AAG), Tyr315stop (TAC>TAA), and Gly170Ser (GGC>AGC) according to mutations observed in patients64, 33. Viral production of Fut enzyme or Glg1 variant pMSCV viruses was performed by transfection into the H29 packaging cell line using PEI. Viruses were collected and filtered at 0.45 μm, then cells were transduced using polybrene (8 μg/mL) for 12 hours, followed by culture with 1 μg/mL puromycin for the duration of experiments. Dkk1-Flag was obtained (Addgene 16690) and subcloned into the pLex vector for lentiviral production. Knockdowns were performed using lentiviral shRNA vectors purchased from Sigma (Supplementary Table 7), and knockouts performed using the lentiviral CRISPR-Cas9 vector system pLentiCrispr-v2 (Supplementary Table 7)65. Virus was created via PEI transfection of vectors together with the lentiviral packaging plasmids psPax2 and VSVG in 293-T cells. Transduction was conducted as described for retroviral plasmids. The SORE6 reporter system55 used to detect Sox2 transcription by flow cytometry was a gift from Lalage Wakefield (NCI) and the Wnt reporter system (TGC)66 used to detect Wnt signaling activation was a gift from Roel Nusse (Stanford). Both reporters were inserted into BM2 cells using the described techniques for lentivirus production and delivery. All viral transduction and selection was performed on a cell population-wide basis.
Flow cytometry
For E-selectin binding experiments, a RFP-labeled internal control line and GFP-labeled cells were co-cultured at equal ratios for 48 h, then cells were harvested at 80% confluence by non-enzymatic dissociation buffer (Life Sciences) at 37°C, washed once with PBS, and suspended in flow buffer (10% FBS in PBS supplemented with 1 mM Ca2+/Mg2+) at 1 million cells/mL. Recombinant mouse E-selectin/IgG Fc or Isotype IgG (RD systems) was added at 10 μg/mL for 1.5 h with vortexing every 15 min. Cells were washed with PBS and incubated with anti-human IgG-AL647-conjugated antibody (Biolegend) at 0.1μg/mL for 45 min. Cells were washed with PBS, resuspended in flow buffer with DAPI (1 μg/mL), and analyzed with the BD LSRII flow cytometer. Internal control cell lines (RFP-labeled) corresponded to the parental population of each cell line – either MDA-MB-231-RFP or SUM159-RFP. E-selectin binding ratios were quantified using the formula (SELEGFP/IgGGFP)/(SELERFP/IgGRFP). Gating for DAPI, RFP, and GFP were performed using negative controls of the corresponding cell line using FlowJo version X. For pharmacological treatments, cells were grown in Tunicamycin (Cayman), 1-Deoxymannojirimycin (Cayman) or D,L-threoPDMP for 24h. Following differential treatment, internal control RFP-labeled cells were added to the cell mixture. For FACS sorting, MDA-MB-231 cells were labeled using this protocol and were sorted into the top and bottom 10% of binding intensities. E-selectin-sorted MDA-MB-231 cells were passaged 5 times prior to assessing E-selectin binding levels to generate a large enough pool both for flow cytometry and sub-culturing. Negative control treatments (EDTA or GMI-1271) were tested in every cell line to ensure that E-selectin binding was specific. For Wnt signaling activation analysis, BM2 cells stably transduced with the 7x-TCF-GFP-SV40-mCherry (TGC) reporter were trypsinized, washed once with PBS, and suspended in PBS + 1 μg/mL DAPI. Cells were analyzed with the LSRII instrument, and gating analysis was performed using unlabeled BM2 cells using the FlowJo version X software. All flow cytometry experiments were repeated a minimum of three times.
qPCR, western blot, and proliferation assay
Quantitative real-time PCR (qRT-PCR) was performed using the SYBR green mastermix protocol on cDNA generated using the SuperScript III cDNA assembly kit (Invitrogen) and the listed primers (Supplementary Table 7). RNA and DNA were purified using the respective Qiagen kits. Human bone marrow and lung RNA pooled from recently deceased patients was purchased from Takara Bio (636591, 636531). Western blotting was performed using standard reducing conditions followed by transfer to PVDF membranes. Membranes were blocked in 5% milk in TBS-T, probed with antibodies listed in Supplementary Table 8, and imaged on the Licor Odyssey CLX system using Licor-supplied IRDye680 and IRDye800 secondary antibodies. Anti-Glg1 was detected with the Chemifluorescent substrate kit (Licor). E-selectin immunoprecipitation western blots were performed in the presence of 1 mM CaCl2 for E-selectin IgG and, along with anti-Slug, were detected with traditional HRP detection. Cell proliferation rates were quantified using the EZQuant reagent (Alstem Bio).
Cell surface protein isolation
Cell surface proteins were biotinylated using Pierce Cell Surface Protein Isolation Kit (Thermo Scientific, Waltham, MA). Briefly, WT and variant M1a cell lines were harvested from two 175 cm2 flasks by trypsinization, rinsed with PBS 3 times and suspended in 0.25 mg/ml solution of Sulfo-NHS-SS-Biotin at 1x106 cells/ml at 4°C. After 30 min of incubation, 500μl quenching solution was added to each tube followed by centrifugation at 1000 rpm for 5 min. Cell pellet was further washed twice in 10 ml Tris-buffered saline. Cell pellets were then lysed with buffer containing 1% NP40. Lysates were equilibrated with Neutravidin agarose beads for 2 hours at 4°C. Cell membrane proteins were eluted by boiling the beads with 100μl western blot sample buffer with 50mM DTT for 5 min at 95°C.
Immunoprecipitation with E-selectin-IgG
5x106 M1a cell variants were harvested from tissue culture flasks and were washed three times with PBS. Cell lysates were prepared in Selectin wash/lysis buffer (2%NP40, 150mM NaCl, 50mM Tris-HCl (pH7.4), 2mM CaCl2, 20μg/mL PMSF, and 1x protease inhibitor cocktail (Roche)). Cell lysates were precleared by incubating overnight with Protein G agarose beads (Invitrogen) pre-blocked with Bovine Serum Albumin (BSA). The precleared lysates were then mixed with 10μg/ml E-selectin-Ig for 4 hours at 4°C. E-selectin-Ig binding proteins were pulled down via incubation with Protein G Agarose beads pre-blocked with BSA, and eluted via boiling in 1.5x reducing sample buffer with SDS.
Immunofluorescence
Ex vivo immunofluorescence on bone, lung, and liver was conducted with or without retro-orbital injection of 10 μg anti-CD31 (Biolegend 102416) in 100 μL PBS to label vasculature followed by whole body perfusion with 10% buffered formalin. Tissues were removed and fixed for 12 hours with 10% buffered formalin at 4°C. These were equilibrated in 15% and 30% sucrose for 3 h each prior to embedding in SCEM (Section Lab Co) and cryo-sectioning using specialized adhesive film (Section Lab Co). Following evaporation at 22°C, sections were fixed 10 min with buffered formalin, permeabilized with 20% goat serum, 0.1% triton X-100 in PBS for one hour. E-selectin was stained with BV421-conjugated anti-E-selectin (BD Pharmigen clone 10E9.6, 1:100) for 12 h in 20% goat serum, 0.1% triton-X100 and nuclei were counterstained with Propidium iodide. Other markers, including EpCam, E-cadherin, and Glg1 (Supplementary Table 8) were used similarly except with detection by secondary antibodies and Hoescht counterstaining. Bones of mice injected with cells that expressed endogenous labels (e.g. BM2-TGC) were fixed, stained with Hoechst and mounted. Sections were imaged using a standard Nikon A1R confocal point-scanning system. For immunostaining of 2 clinical samples from patients with prostate cancer bone metastasis, sections from FFPE human prostate cancer bone metastasis specimens were dewaxed, rehydrated, treated with Target Retrieval solution (Dako) according to manufacturer’s instructions, blocked with 5% normal donkey serum, and incubated for 12h with anti-E-selectin (R & D Systems BBA18, 1:50) at 4°C followed by Alexafluor-conjugated secondary antibody (1:300). Sections were then sequentially incubated with anti-Glg1 (Sigma-Aldrich HPA 010815 1:50) and anti-Ki67 (Dako M7240 1:40), using the same procedures. Nuclei were counterstained by DAPI. Images were taken on an Olympus Confocal FV 1000 microscope and representative images from >10 images per specimen were selected.
Mass spectrometry
Protocol was adapted from previous published work41 with minor modifications. Substituted reagents include EZLink Alkoxyamine-PEG4-Biotin (Thermo), TCEP and chloroacetamide. Metaperiodate concentration was increased to 10 mM. Streptavidin ultralink resin (Pierce) and PNGase F (NEB) were used for isolation. Cells were grown in normal culture conditions and detached with non-enzymatic dissociation buffer. Cells were washed with PBS, suspended in 10 mM sodium metaperiodate at 2x106 cells/mL, and rotated at 4°C for 1 h. Reaction was quenched at 1 mM glycerol, followed by a PBS wash. Cells were resuspended in 100 μM EZLink-alkoxyamine-PEG4-biotin supplemented with 10 mM Aniline in PBS + 5% FBS followed by rotation at 4°C for 1 h. Cells were then washed with PBS and either conjugated with Streptavidin-AL647 (Biolegend, 10 μg/mL) for verification of efficient oxidation and conjugation via flow cytometry/microscopy or lysed (1% Triton X-100, 150 mM NaCl, 1x protease inhibitor, 5 mM chloroacetimide, 0.1 mg/mL PMSF, 10 mM Tris-HCl, ph 7.6) at 106 cells/mL end-over-end at 4°C for 30 min. Supernatants were then saved after sequential centrifugation at 2800 g and 16000g. Supernatants were incubated with 6 μL washed resin per 106 cells at 4°C for 12 h. Beads were then washed 10 times each with lysis buffer and 0.5% SDS in PBS. Beads were then incubated in 0.5% SDS in PBS, 50 mM TCEP for 20 min at 22°C, washed 10 times with 6 M urea in 100 mM Tris-HCl ph 8.5, then incubated with 6 M urea in 100 mM Tris-HCl (pH 8.5) and 50 mM chloroacetamide for 20 min. Beads were then washed 10x with each of the following: 6 M urea in 100 mM Tris-HCl ph 8.5, 5 M NaCl, 100 mM sodium carbonate, PBS, and water. Beads were subsequently digested with 5 μg sequencing-grade trypsin in 0.5 mL ammonium bicarbonate at 37°C for 24 h. Tryptic peptides were isolated by centrifugation and one wash with ammonium bicarbonate. Beads were then washed 10 times each with PBS and water, and digested with 2500 U PNGase F in glycobuffer 2 (NEB) for 10 h at 37°C. Supernatant and the first wash were pooled for mass spectrometry. The peptides released by PNGase F digestion were identified by nanoLC-tandem mass spectrometry using a previously described method67. The Skyline software was used for peptide relative quantification by calculating peptide peaks areas from MS1 extracted ion chromatograms, as previously described68. Total protein characterization is listed in Supplementary Table 2.
E-selectin plate coating and immunofluorescence
Ibidi culture-treated chamber slides were coated with E-selectin/Fc chimera at 10 μg/mL in PBS for 12 hours at 4°C. Indirect immunofluorescence for markers of MET was conducted by fixing cells that were grown for 24 h in coated Ibidi chamber slides with 10% buffered formalin solution for 20 min at 4°C followed by permeabilization with 0.1% Triton X-100 in PBS and incubation with each antibody for 12 h at 4°C. N-cadherin (BD Pharmigen 610920), ZO-1 (CST 5406), EpCAM (CST VU1D9) and K14 (Biolegend 905301) were used at 1:100 for immunofluorescence. Slides were washed with PBS, Hoechst, and probed with appropriate secondary antibodies (Invitrogen) followed by confocal imaging of chambers filled with PBS. Wnt activation by live cell imaging of BM2-TGC seeded over E-selectin in Ibidi culture slides was conducted from 0–48 h via TIRF microscopy and was first observable approximately 30 h post-seeding. Analysis of Wnt activation via flow cytometry was conducted at 48 h. Cells analyzed by flow cytometry were trypsinized, centrifuged, and immediately analyzed using DAPI as a cell death indicator. Various inducers and inhibitors of Wnt signaling were added to culture, these include ICG-001 (Selleck, 25 μM), LF3 (Selleck, 50 μM), Wnt3a- or control-conditioned media, or recombinant Wnt3a (RD systems, 100 ng/mL). Wnt3a and control-conditioned media were generated by collecting 72 h conditioned media from L cells with or without Wnt3a expression (ATCC CRL-2647 and CRL-2648) followed by filtration at 0.45 μm.
Microarray analysis
BM2 and M1a cells were seeded over 6 cm tissue culture-treated plates coated with E-selectin-IgG or control IgG (10 μg/mL) for 12 h at 37°C. RNA was isolated from cells using the RNAeasy Minikit (Qiagen) according to manufacturer’s instructions. Gene expression profiles influenced by E-selectin binding were analyzed using the Agilent human GE 8x60k two-color microarray system (Agilent G4858A-039494). Briefly, the RNA samples and a universal human reference RNA (Agilent) were labeled with CTP-cy5 and CTP-cy3 using the Agilent Quick Amp Labeling Kit. Labeled samples were mixed equally and hybridized to the array. The array was then scanned with the G2505C scanner (Agilent). Data was deconvoluted and analyzed with the Genespring v13 software (Agilent). Briefly, array controls, flagged values, and expression values falling below the median value were removed. Multiple values for any given gene were collapsed into the single highest expression value. Data was extracted as a Log2-transformed ratio of Cy5/Cy3.
Gene set enrichment analysis (GSEA)
Log2-transformed data was subtracted (Log2(E-selectin)-Log2(IgG)) and rank ordered for each cell line. Data was analyzed using GSEAv2.0. Interrogated signatures from the MySigDB v5.1 database included the Hallmark EMT data set (M5930), Sarrio-EMT (GSE8430), Claudin-low (GSE18229), Luminal (GSE22446), Bruno-hematopoiesis (M1492), and Blick-EMT69 as part of a broader, manually compiled set of EMT- and Stem cell-related signatures. Only data sets with a nominal p-value < 0.05 were considered in the analysis.
Statistics and Reproducibility
All statistical comparisons were conducted with Stata v13 (Mann-Whitney U and Cox’s Proportional Hazards), Microsoft Excel 2010 (t-test), or Broad Institute GSEA software (Pre-ranked GSEA analysis). Results are reported as mean + standard error of the mean (S.E.M.) for bar graphs. For all animal experiments, animals were only excluded if they died or had to be sacrificed according to the pre-defined criteria listed in the IACUC protocol. A two-sided Mann-Whitney U test was used for bone metastasis as this data does not exhibit a normal distribution. Osteolytic area and lesion analysis assumed each hindlimb as an independent event. Bioluminescent data from imaging of spontaneous bone metastasis in the Fut primary tumor injection or Ob1 bone metastasis experiment were set to zero for values below a threshold 100,000 ph/s or via visual inspection in order to avoid false ranking attributed only to background signal. For spontaneous bone and lung metastasis from M1a-Fut cells, one-sided Mann Whitney U and one-sided Fisher’s exact tests were used to test whether Fut overexpression promoted spontaneous metastasis to bone while one-sided Student’s t-test was used to assess whether Fut expression promoted lung metastasis. Two-sided, unpaired Student’s t-test was used for all other lung metastasis, primary tumor growth, qPCR, flow cytometry data, and other normal data. All statistical comparisons are non-significant at an alpha level of 0.05 if not marked by a specific p value. Patient expression values in the NKI295 and EMC-MSK datasets were stratified by median value for single gene analysis or grouped by median-stratified values according to (Glg1 and (Fut3 or Fut6)). Log2RPKM values of Exon1 from each Fut gene were extracted from the TCGA dataset and plotted. Survival data for mouse experiments and patient datasets were analyzed by Cox’s proportional Hazards model to generate hazard ratios and their associated p-values. All in vivo bone metastasis experiments were performed at least 2 times to ensure reproducibility and significance with the exception of the BM2-CD44 KO injection, catalytic Fut3 mutants and the GMI-1271 treatment, which was performed once. All lung metastasis and primary tumor growth experiments were performed once. All other experiments were repeated according to their corresponding figure legend with similar results.
Data availability
Microarray data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession codes GSE96754, and the mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD012942; mass spectrometry data is further available in Supplementary Table 2.
The human clinical breast cancer data are not available in GEO; these were derived from the TCGA Research Network and the data-set derived from this resource that supports the findings of this study is available in https://xenabrowser.net/datapages/?cohort=GDC%20TCGA%20Breast%20Cancer%20(BRCA) The human breast cancer data were also derived from the NKI-295 dataset47 which is available at https://xenabrowser.net/datapages/?cohort=Breast%20Cancer%20(Vijver%202002)
Finally, human breast cancer data from the EMC-MSK dataset is available in Bos et. al, 200970, Source data relevant for the clinical data analyses performed in this study are available in Supplementary Table 9.
Unprocessed western blot images are provided as Supplementary Fig. 9. Source data supporting the findings of this study are provided in Supplementary Table 9. All protocols, cell lines and reagents are available from the corresponding author upon request.
Supplementary Material
Acknowledgments
We thank L.M. Wakefield for providing the SORE6-mCherry stemness reporter, R. Nusse for providing the 7x-TCF-GFP Wnt reporter, G. Laevsky for assistance with microscopy, and C. DeCoste for assistance with flow cytometry. This work was supported by fellowships from the NIH (F31CA192461) and NJCCR to ME, the National Institutes of Health NHLBI Grant PO1 HL107146, the Program of Excellence in Glycosciences, and the Team Jobie Fund to RS, and grants from the Susan G. Komen Foundation (SAC160067), Glycomimetics Inc., Brewster Foundation, Department of Defense (BC123187), and the National Institutes of Health (R01CA141062) to YK. This research was also supported by the Preclinical Imaging, Genomic Editing and Flow Cytometry Shared Resources of the Cancer Institute of New Jersey (P30CA072720).
JLM is the Vice President and Chief Scientific Officer of Glycomimetics, Inc., which owns the patent to GMI 1271. YK received research support from Glycomimetics Inc. for experiments using GMI-1271.
Footnotes
Competing Financial Interests
No other authors declare potential conflicts of interest.
REFERENCES
- 1.Valastyan S & Weinberg RA Tumor metastasis: molecular insights and evolving paradigms. Cell 147, 275–292 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Massague J & Obenauf AC Metastatic colonization by circulating tumour cells. Nature 529, 298–306 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang J & Weinberg RA Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell 14, 818–829 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Nieto MA, Huang RY, Jackson RA & Thiery JP Emt: 2016. Cell 166, 21–45 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Esposito M, Guise T & Kang Y The Biology of Bone Metastasis. Cold Spring Harbor perspectives in medicine 8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oskarsson T, Batlle E & Massague J Metastatic stem cells: sources, niches, and vital pathways. Cell stem cell 14, 306–321 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang J et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 117, 927–939 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Mani SA et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133, 704–715 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Labelle M, Begum S & Hynes RO Direct signaling between platelets and cancer cells induces an epithelial-mesenchymal-like transition and promotes metastasis. Cancer Cell 20, 576–590 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brabletz T et al. Variable beta-catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment. Proc Natl Acad Sci U S A 98, 10356–10361 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ocana OH et al. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell 22, 709–724 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Tsai JH, Donaher JL, Murphy DA, Chau S & Yang J Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell 22, 725–736 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korpal M et al. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat Med 17, 1101–1108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Celia-Terrassa T et al. Epithelial-mesenchymal transition can suppress major attributes of human epithelial tumor-initiating cells. J Clin Invest 122, 1849–1868 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y et al. Mouse fibroblasts lacking RB1 function form spheres and undergo reprogramming to a cancer stem cell phenotype. Cell stem cell 4, 336–347 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiou SH et al. Coexpression of Oct4 and Nanog enhances malignancy in lung adenocarcinoma by inducing cancer stem cell-like properties and epithelial-mesenchymal transdifferentiation. Cancer Res 70, 10433–10444 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Shiozawa Y et al. Human prostate cancer metastases target the hematopoietic stem cell niche to establish footholds in mouse bone marrow. J Clin Invest 121, 1298–1312 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghajar CM et al. The perivascular niche regulates breast tumour dormancy. Nature Cell Biology 15, 807–817 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walz G, Aruffo A, Kolanus W, Bevilacqua M & Seed B Recognition by ELAM-1 of the sialyl-Lex determinant on myeloid and tumor cells. Science 250, 1132–1135 (1990). [DOI] [PubMed] [Google Scholar]
- 20.Frenette PS, Mayadas TN, Rayburn H, Hynes RO & Wagner DD Double knockout highlights value of endothelial selectins. Immunol Today 17, 205 (1996). [DOI] [PubMed] [Google Scholar]
- 21.Dimitroff CJ, Lee JY, Rafii S, Fuhlbrigge RC & Sackstein R CD44 is a major E-selectin ligand on human hematopoietic progenitor cells. J Cell Biol 153, 1277–1286 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winkler IG et al. Vascular niche E-selectin regulates hematopoietic stem cell dormancy, self renewal and chemoresistance. Nat Med 18, 1651–1657 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Dimitroff CJ et al. Identification of leukocyte E-selectin ligands, P-selectin glycoprotein ligand-1 and E-selectin ligand-1, on human metastatic prostate tumor cells. Cancer Res 65, 5750–5760 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanley WD, Burdick MM, Konstantopoulos K & Sackstein R CD44 on LS174T colon carcinoma cells possesses E-selectin ligand activity. Cancer Res 65, 5812–5817 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Kang Y et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 3, 537–549 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Minn AJ et al. Genes that mediate breast cancer metastasis to lung. Nature 436, 518–524 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laubli H & Borsig L Selectins as mediators of lung metastasis. Cancer Microenviron 3, 97–105 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taverna D et al. Increased primary tumor growth in mice null for beta3- or beta3/beta5-integrins or selectins. Proc Natl Acad Sci U S A 101, 763–768 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Labow MA et al. Characterization of E-selectin-deficient mice: demonstration of overlapping function of the endothelial selectins. Immunity 1, 709–720 (1994). [DOI] [PubMed] [Google Scholar]
- 30.Khatib AM et al. Rapid induction of cytokine and E-selectin expression in the liver in response to metastatic tumor cells. Cancer Res 59, 1356–1361 (1999). [PubMed] [Google Scholar]
- 31.Lowe JB Glycan-dependent leukocyte adhesion and recruitment in inflammation. Curr Opin Cell Biol 15, 531–538 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Mondal N et al. Distinct human α(1,3)-fucosyltransferases drive Lewis-X/sialyl Lewis-X assembly in human cells. Journal of Biological Chemistry 293, 7300–7314 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mollicone R et al. Molecular basis for Lewis alpha(1,3/1,4)-fucosyltransferase gene deficiency (FUT3) found in Lewis-negative Indonesian pedigrees. Journal of Biological Chemistry 269, 20987–20994 (1994). [PubMed] [Google Scholar]
- 34.Oulmouden A et al. Molecular Cloning and Expression of a Bovine α(1,3)-Fucosyltransferase Gene Homologous to a Putative Ancestor Gene of the Human FUT3-FUT5-FUT6 Cluster. Journal of Biological Chemistry 272, 8764–8773 (1997). [DOI] [PubMed] [Google Scholar]
- 35.Goelz SE et al. ELFT: A gene that directs the expression of an ELAM-1 ligand. Cell 63, 1349–1356 (1990). [DOI] [PubMed] [Google Scholar]
- 36.Buffone A et al. Silencing α1,3-Fucosyltransferases in Human Leukocytes Reveals a Role for FUT9 Enzyme during E-selectin-mediated Cell Adhesion. Journal of Biological Chemistry 288, 1620–1633 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Costache M et al. Evolution of fucosyltransferase genes in vertebrates. J Biol Chem 272, 29721–29728 (1997). [DOI] [PubMed] [Google Scholar]
- 38.Li J et al. Human fucosyltransferase 6 enables prostate cancer metastasis to bone. British journal of cancer 109, 3014–3022 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sackstein R et al. Ex vivo glycan engineering of CD44 programs human multipotent mesenchymal stromal cell trafficking to bone. Nat Med 14, 181–187 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Ossola R et al. Biomarker validation in blood specimens by selected reaction monitoring mass spectrometry of N-glycosites. Methods Mol Biol 728, 179–194 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Weekes MP et al. Proteomic plasma membrane profiling reveals an essential role for gp96 in the cell surface expression of LDLR family members, including the LDL receptor and LRP6. J Proteome Res 11, 1475–1484 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Price TT et al. Dormant breast cancer micrometastases reside in specific bone marrow niches that regulate their transit to and from bone. Sci Transl Med 8, 340ra373 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morita Y et al. E-selectin Targeting PEGylated-thioaptamer Prevents Breast Cancer Metastases. Molecular therapy. Nucleic acids 5, e399 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Sreeramkumar V et al. Coordinated and unique functions of the E-selectin ligand ESL-1 during inflammatory and hematopoietic recruitment in mice. Blood 122, 3993–4001 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yasmin-Karim S, King MR, Messing EM & Lee YF E-selectin ligand-1 controls circulating prostate cancer cell rolling/adhesion and metastasis. Oncotarget 5, 12097–12110 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barthel SR et al. Definition of molecular determinants of prostate cancer cell bone extravasation. Cancer Res 73, 942–952 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van de Vijver MJ et al. A gene-expression signature as a predictor of survival in breast cancer. The New England journal of medicine 347, 1999–2009 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Wang Y et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet 365, 671–679 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Winkler IG et al. Positioning of bone marrow hematopoietic and stromal cells relative to blood flow in vivo: serially reconstituting hematopoietic stem cells reside in distinct nonperfused niches. Blood 116, 375–385 (2010). [DOI] [PubMed] [Google Scholar]
- 50.Pries AR & Kuebler WM Normal endothelium. Handbook of experimental pharmacology, 1–40 (2006). [DOI] [PubMed]
- 51.Bixel MG et al. Flow Dynamics and HSPC Homing in Bone Marrow Microvessels. Cell Rep 18, 1804–1816 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Macdonald BT, Semenov MV & He X SnapShot: Wnt/beta-catenin signaling. Cell 131, 1204 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Fuerer C & Nusse R Lentiviral vectors to probe and manipulate the Wnt signaling pathway. PLoS One 5, e9370 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Holst S et al. N-glycosylation Profiling of Colorectal Cancer Cell Lines Reveals Association of Fucosylation with Differentiation and Caudal Type Homebox 1 (CDX1)/Villin mRNA Expression. Mol Cell Proteomics 15, 124–140 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tang B et al. A flexible reporter system for direct observation and isolation of cancer stem cells. Stem Cell Reports 4, 155–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stone KR, Mickey DD, Wunderli H, Mickey GH & Paulson DF Isolation of a human prostate carcinoma cell line (DU 145). International journal of cancer 21, 274–281 (1978). [DOI] [PubMed] [Google Scholar]
- 57.Korpal M, Lee ES, Hu G & Kang Y The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem 283, 14910–14914 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anastas JN & Moon RT WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer 13, 11–26 (2013). [DOI] [PubMed] [Google Scholar]
- 59.Sipkins DA et al. In vivo imaging of specialized bone marrow endothelial microdomains for tumour engraftment. Nature 435, 969–973 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Biancone L, Araki M, Araki K, Vassalli P & Stamenkovic I Redirection of tumor metastasis by expression of E-selectin in vivo. The Journal of experimental medicine 183, 581–587 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Herschkowitz JI et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome biology 8, R76 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Forozan F et al. Molecular cytogenetic analysis of 11 new breast cancer cell lines. Br J Cancer 81, 1328–1334 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ell B et al. Tumor-induced osteoclast miRNA changes as regulators and biomarkers of osteolytic bone metastasis. Cancer Cell 24, 542–556 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Koda Y, Kimura H & Mekada E Analysis of Lewis fucosyltransferase genes from the human gastric mucosa of Lewis-positive and -negative individuals. Blood 82, 2915–2919 (1993). [PubMed] [Google Scholar]
- 65.Sanjana NE, Shalem O & Zhang F Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fuerer C & Nusse R Lentiviral vectors to probe and manipulate the Wnt signaling pathway. PLoS One 5, e9370 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Diner BA et al. The functional interactome of PYHIN immune regulators reveals IFIX is a sensor of viral DNA. Mol Syst Biol 11, 787 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Giguere SS et al. The Proteomic Profile of Deleted in Breast Cancer 1 (DBC1) Interactions Points to a Multifaceted Regulation of Gene Expression. Mol Cell Proteomics 15, 791–809 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Blick T et al. Epithelial mesenchymal transition traits in human breast cancer cell lines parallel the CD44(hi/)CD24 (lo/-) stem cell phenotype in human breast cancer. Journal of mammary gland biology and neoplasia 15, 235–252 (2010). [DOI] [PubMed] [Google Scholar]
- 70.Bos PD et al. Genes that mediate breast cancer metastasis to the brain. Nature 459, 1005–1009 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microarray data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession codes GSE96754, and the mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD012942; mass spectrometry data is further available in Supplementary Table 2.
The human clinical breast cancer data are not available in GEO; these were derived from the TCGA Research Network and the data-set derived from this resource that supports the findings of this study is available in https://xenabrowser.net/datapages/?cohort=GDC%20TCGA%20Breast%20Cancer%20(BRCA) The human breast cancer data were also derived from the NKI-295 dataset47 which is available at https://xenabrowser.net/datapages/?cohort=Breast%20Cancer%20(Vijver%202002)
Finally, human breast cancer data from the EMC-MSK dataset is available in Bos et. al, 200970, Source data relevant for the clinical data analyses performed in this study are available in Supplementary Table 9.
Unprocessed western blot images are provided as Supplementary Fig. 9. Source data supporting the findings of this study are provided in Supplementary Table 9. All protocols, cell lines and reagents are available from the corresponding author upon request.