Abstract
Genetic alterations that might lead to colorectal cancer involve essential genes including those involved in DNA repair, inclusive of base excision repair (BER). Thymine DNA glycosylase (TDG) is one of the most well characterized BER genes that catalyzes the removal of thymine moieties from G/T mismatches and is also involved in many cellular functions, such as the regulation of gene expression, transcriptional coactivation, and the control of epigenetic DNA modification. Mutation of the TDG gene is implicated in carcinogenesis. In the present study, we aimed to investigate the association between TDG gene polymorphisms and their involvement in colon cancer susceptibility. One hundred blood samples were obtained from colorectal cancer patients and healthy controls for the genotyping of seven SNPs in the TDG gene. DNA was extracted from the blood, and the polymorphic sites (SNPs) rs4135113, rs4135050, rs4135066, rs3751209, rs1866074, and rs1882018 were investigated using TaqMan genotyping. One of the six TDG SNPs was associated with an increased risk of colon cancer. The AA genotype of the TDG SNP rs4135113 increased the risk of colon cancer development by more than 3.6-fold, whereas the minor allele A increased the risk by 1.6-fold. It also showed a 5-fold higher risk in patients over the age of 57. SNP rs1866074 showed a significant protective association in CRC patients. The GA genotype of TDG rs3751209 was associated with a decreased risk in males. There is a significant relationship between TDG gene function and colorectal cancer progression.
1. Introduction
The development of cancer is a multistep process involving aberrations in many cellular processes, including differentiation, cell cycle regulation, cell death, proliferation, and genomic conservation due to functional alterations in a variety of genes. Thymine DNA glycosylase (TDG) is a member of the mismatch uracil glycosylase subfamily. All of these uracil DNA glycosylase (UDG) enzymes have a monofunctional approach of action [1]. UDGs recruit a common base-flipping, DNA intercalation method for substrate identification and catalyze the removal of the N-glycosidic bond of the flipped base, thus creating an abasic site [2]. TDG has a crucial role in DNA repair, particularly BER, in which it specifically identifies G: U and G: T mismatches resulting from the impulsive deamination of 5-methylcytosine. In addition to its DNA repair function, TDG is also involved in other critical cellular processes, such as the regulation of gene expression, transcriptional coactivation, and the regulation of epigenetic DNA modification [3]. TDG has been shown to interact with some transcription factors and especially with nuclear receptors. TDG initiates the BER pathway, which utilizes the base-flipping method to delete the target bases from the DNA forming an AP site. This happens when TDG binds to the promoters of the BER proteins APE, DNA ligase, and Pol β [4].
The role of TDG in cancer progression is a hotly debated issue [5]. Its interaction with tumor suppressor P53 (TP53) proteins initially suggested that TDG merely acts as a tumor suppressor. Overexpression of TDG recruits TP53 proteins to the cyclin dependent kinase inhibitor 1A (p21Waf1) gene promoter and increases its transcriptional activity [6]. Moreover, TP53 binding to the TDG promoter will transcriptionally regulate its expression and control the nuclear translocation of TDG [7]. The relationship between TDG and cancer has been studied by a number of research groups who have suggested that genetic variants in TDG and other DNA repair genes confer susceptibility to colorectal cancer [8]. Xu and colleagues showed that TDG positively regulates the Wnt signaling pathway and is a key driver necessary for the progression of CRC [9]. They also reported that hypermethylation of TDG in multiple myeloma cell lines reduced its gene expression. As a result, DNA repair activity became less efficient [10] in pancreatic adenocarcinoma [11]. Finally, a lack of the DNA mismatch repair protein PMS2 (PMS2) and reduced TDG expression in rectal cancer has been found to produce a supermutator phenotype at CpG sites [12].
Recent studies reported that the SNP rs2888805 (Val367Met) in TDG might be implicated in nonmelanoma skin cancer [13]. The TDG SNPs rs167715 and rs4135087 might also be associated with the progression of ovarian cancer in most of the BRCA1/2 mutation carriers [14]. The coding region SNP rs369649741 (Arg66Gly) has been reported to be associated with a high risk in familial colorectal cancer patients [8]. Significant associations have been demonstrated between the risk of cancers, including esophageal squamous cell carcinoma and gastric cancer, and the rs4135054 SNP in TDG [15]. This study was conducted to determine the association of the DNA repair gene TDG SNPs and colon cancer risk in the Saudi population.
2. Materials and Methods
2.1. Study Population and Patient Selection
The study population was composed of 100 colorectal cancer patients and 192 control subjects from a Saudi population. Patients were recruited from King Saud Medical City. CRC was confirmed via histopathological examination. The age of the CRC cases varied from 21 to 90 years, with a mean age of 61.10 ± 12.17 years. The main exclusion conditions were autoimmune disorders, hereditary nonpolyposis colorectal cancer (HNPCC), or a previous history of any other disorders. CRC patients who had undergone prior chemoradiotherapy were also excluded. A total of 192 controls were recruited. The age of the controls varied from 21 to 87 years with a mean of 57.2 ± 8.34 years. The primary details of the volunteers were collected by a prestructured questionnaire. Each participant was informed in detail about the present study and signed standard consent. The Ethics Committee of King Saud Medical City approved the present study.
2.2. Single Nucleotide Polymorphisms (SNPs) Selection, DNA Extraction, and Genotyping
Genomic DNA was extracted from blood samples using a blood DNA kit (QIAGEN DNeasy Blood & Tissue Kit). According to previous reports, six SNPs located in the TDG gene were analyzed: rs4135113 (C__31582396_10), rs4135050 (C___1970689_10), rs4135066 (C___1970695_10), rs3751209 (C__11162283_20), rs1866074 (C___3152280_10), and rs1882018 (C__11490839_10). The preliminary data on the SNPs are shown in Table 1. These SNPs were also selected based on literature reviews of SNP associations with various diseases in diverse ethnic groups. The genotyping analysis was conducted using QuantStudio™ 7 Flex Real-Time PCR System (Applied Biosystems) with an endpoint reading of the genotypes [16].
Table 1.
Primary information for TDG polymorphisms.
| Genotyped SNP | rs4135113 | rs4135050 | rs4135066 | rs3751209 | rs1882018 | rs1866074 |
|
| ||||||
| Chromosome | 12 | 12 | 12 | 12 | 12 | 12 |
|
| ||||||
| Chromosome Position | 103982915 | 103968698 | 103972562 | 103979822 | 103969403 | 103980664 |
|
| ||||||
| Base change | G>A (Gly199Ser) | T>A | C>T | G>A | C>T | A>G |
|
| ||||||
| MAF in our controls | 0.10 | 0.21 | 0.77 | 0.31 | 0.23 | 0.66 |
|
| ||||||
| p-value for HWE | 0.11 | 0.11 | 0.09 | 0.4 | 0.09 | 0.52 |
MAF: minor allele frequency.
HWE: Hardy–Weinberg equilibrium.
3. Results
A total of 100 colorectal cancer patients and 192 normal controls from a Saudi Arabia population were included in the present study. The clinical and the demographic features of the study subjects are described in Supplementary Table1 (Suppl. Table1). Both CRC and normal samples were classified based on demographic parameters such as age and gender. Colorectal cancer samples were further classified based on tumor location, namely, colon or rectum. The average age of the CRC samples was 57.10 ± 12.17 years and of the controls was 58.2 ± 8.34 years.
All six SNPs in the normal control and CRC patient group obeyed Hardy-Weinberg equilibrium (HWE) (Table 1). Table 1 depicts the details of the SNPs used in the present study including the minor allele frequency and the HWE p-value. Out of the six SNPs, two SNPs, rs4135113 and rs1866074, showed a significant association with colorectal cancer. The genotypic distribution of rs4135113 was 75% GG, 18% GA, and 7% AA in colorectal cancer patients and 82% GG, 16% GA, and 2% AA in normal samples. SNP rs4135113 (Gly199Ser) showed a significant risk association with colorectal cancer in Saudi patients for its genotype AA (OR: 3.640, CI: 1.034–12.819, p = 0.03286) (Table 2). The frequency of the minor allele A in patient samples also showed a significant difference compared with that in the healthy controls (OR: 1.675, CI: 1.013–2.769, p = 0.04264) (Table 2).
Table 2.
Genotype frequencies of TDG gene polymorphism in colorectal cases and controls.
| SNP | Variant | Patients Cases | Controls | OR | CI | χ 2 Value | p-value |
|---|---|---|---|---|---|---|---|
| rs4135050 | TT | 58 (0.58) | 124 (0.65) | Ref | |||
| TA | 34 (0.34) | 55 (0.29) | 1.322 | 0.779–2.244 | 1.07 | 0.30107 | |
| AA | 8 (0.08) | 12 (0.06) | 1.425 | 0.553–3.676 | 0.54 | 0.46173 | |
| TA+AA | 42 (0.42) | 67 (0.35) | 1.340 | 0.816–2.201 | 1.34 | 0.24664 | |
| T | 150 (0.75) | 303 (0.79) | Ref | ||||
| A | 50 (0.25 | 79 (0.21) | 1.278 | 0.853–1.916 | 1.42 | 0.23347 | |
|
| |||||||
| rs1882018 | CC | 58 (0.58) | 118 (0.62) | Ref | |||
| CT | 32 (0.32) | 59 (0.31) | 1.103 | 0.648–1.880 | 0.13 | 0.71725 | |
| TT | 10 (0.10) | 14 (0.07) | 1.453 | 0.609–3.470 | 0.71 | 0.39800 | |
| CT+TT | 42 (0.42) | 73 (0.38) | 1.171 | 0.715–1.916 | 0.39 | 0.53104 | |
| C | 148 (0.74) | 295 (0.77) | Ref | ||||
| T | 52 (0.26) | 87 (0.23) | 1.191 | 0.802–1.771 | 0.75 | 0.38613 | |
|
| |||||||
| rs4135066 | CC | 4 (0.04) | 14 (0.08) | Ref | |||
| CT | 38 (0.38) | 58 (0.30) | 2.293 | 0.702–7.493 | 1.96 | 0.16114 | |
| TT | 58 (0.58) | 119 (0.62) | 1.706 | 0.538–5.413 | 0.84 | 0.35998 | |
| CT+TT | 96 (0.96) | 177 (0.92) | 1.898 | 0.608–5.927 | 1.25 | 0.26277 | |
| C | 46 (0.23) | 86 (0.23) | Ref | ||||
| T | 154 (0.77) | 296 (0.77) | 0.973 | 0.647–1.462 | 0.02 | 0.89402 | |
|
| |||||||
| rs3751209 | GG | 51 (0.51) | 87 (0.46) | Ref | |||
| GA | 38 (0.38) | 88 (0.46) | 0.737 | 0.441–1.232 | 1.36 | 0.24320 | |
| AA | 11 (0.11) | 16 (0.08) | 1.173 | 0.505–2.722 | 0.14 | 0.71041 | |
| GA+AA | 49 (0.49) | 104 (0.54) | 0.804 | 0.495–1.305 | 0.78 | 0.37654 | |
| G | 140 (0.70) | 262 (0.69) | Ref | ||||
| A | 60 (0.30) | 120 (0.31) | 0.936 | 0.645–1.357 | 0.12 | 0.72602 | |
|
| |||||||
| rs1866074 | AA | 22 (0.22) | 24 (0.12) | Ref | |||
| AG | 39 (0.39) | 82 (0.43) | 0.519 | 0.260–1.037 | 3.50 | 0.06152 | |
| GG | 39 (0.39) | 85 (0.45) | 0.501 | 0.251–1.000 | 3.91 | 0.04799 | |
| AG+GG | 78 (0.78) | 167 (0.88) | 0.510 | 0.269–0.964 | 4.39 | 0.03615 | |
| A | 83 (0.42) | 130 (0.34) | Ref | ||||
| G | 117 (0.58) | 252 (0.66) | 0.727 | 0.511–1.034 | 3.16 | 0.07567 | |
|
| |||||||
| rs4135113 | GG | 75 (0.75) | 156 (0.82) | Ref | |||
| GA | 18 (0.18) | 31 (0.16) | 1.208 | 0.635–2.297 | 0.33 | 0.56458 | |
| AA | 7 (0.07) | 4 (0.02) | 3.640 | 1.034–12.819 | 4.55 | 0.03286 | |
| GA+AA | 25 (0.25) | 35 (0.18) | 1.486 | 0.830–2.660 | 1.79 | 0.18130 | |
| G | 168 (0.84) | 343 (0.90) | Ref | ||||
| A | 32 (0.16) | 39 (0.10) | 1.675 | 1.013–2.769 | 4.11 | 0.04264 | |
The genotypic distribution of rs1866074 was 22% AA, 39% AG, and 39% GG in colorectal cancer patients and 12% AA, 43% AG, and 45% GG in the normal samples. The GG allele frequency was low in colorectal cancer patients compared with that in the controls. SNP rs1866074 showed a protective association of the GG allele (OR: 0.501, CI: 0.251–1, p = 0.047) and the additive (AG+GG) allele (OR: 0.51, CI: 0.269–0.964, p = 0.036) (Table 2). The remaining SNPs, rs4135050, rs4135066, rs3751209, and rs1882018, did not show any association with colorectal cancer in the overall analysis (Table 2).
3.1. Stratification Analysis
After an overall analysis, we compared the TDG genotype frequencies based on gender. The genotype distributions of male (n = 58) and female (n = 42) patients were compared with those of matched healthy individuals (Tables 3 and 4). Only rs3751209 showed a protective association in female colon cancer patients with the GA genotype (OR, 0.407; CI: 0.196–0.847, p = 0.01495). The heterozygous GA genotype frequency was low in colorectal cancer patients compared with that in the controls (Table 3). No other SNPs showed any significant association with colorectal cancer based on gender (Tables 3 and 4). The frequency of the A allele in patient samples also showed a significant difference compared with that of the healthy individuals (OR: 2.238, CI: 1.059–4.729, p = 0.03159).
Table 3.
Genotype frequencies of TDG gene polymorphisms in male colorectal cases and controls.
| SNP | Variant | Patients Cases | Controls | OR | CI | χ 2 Value | p-value |
|---|---|---|---|---|---|---|---|
| rs4135050 | TT | 32 (0.55) | 60 (0.63) | Ref | |||
| TA | 21 (0.36) | 27 (0.28) | 1.458 | 0.714–2.977 | 1.08 | 0.29910 | |
| AA | 5 (0.09) | 8 (0.08) | 1.172 | 0.354–3.879 | 0.07 | 0.79493 | |
| TA+AA | 26 (0.45) | 35 (0.37) | 1.393 | 0.717–2.707 | 0.96 | 0.32771 | |
| T | 85 (0.73) | 147 (0.77) | Ref | ||||
| A | 31 (0.27) | 43 (0.23) | 1.247 | 0.731–2.126 | 0.66 | 0.41728 | |
|
| |||||||
| rs1882018 | CC | 35 (0.60) | 60 (0.63) | Ref | |||
| CT | 18 (0.31) | 27 (0.29) | 1.143 | 0.552–2.366 | 0.13 | 0.71901 | |
| TT | 5 (0.09) | 8 (0.08) | 1.071 | 0.325–3.531 | 0.01 | 0.90971 | |
| CT+TT | 23 (0.40) | 35 (0.37) | 1.127 | 0.576–2.204 | 0.12 | 0.72787 | |
| C | 88 (0.76) | 147 (0.77) | Ref | ||||
| T | 28 (0.24) | 43 (0.23) | 1.088 | 0.631–1.875 | 0.09 | 0.76200 | |
|
| |||||||
| rs4135066 | CC | 3 (0.05) | 7 (0.07) | Ref | |||
| CT | 21 (0.36) | 30 (0.32) | 1.633 | 0.378–7.054 | 0.44 | 0.50827 | |
| TT | 34 (0.59) | 58 (0.61) | 1.368 | 0.332–5.643 | 0.19 | 0.66390 | |
| CT+TT | 55 (0.95) | 88 (0.93) | 1.458 | 0.362–5.877 | 0.28 | 0.59390 | |
| C | 27 (0.23) | 44 (0.23) | Ref | ||||
| T | 89 (0.77) | 146 (0.77) | 0.993 | 0.575–1.716 | 0.001 | 0.98108 | |
|
| |||||||
| rs3751209 | GG | 36(0.62) | 44 (0.46) | Ref | |||
| GA | 15 (0.26) | 45 (0.47) | 0.407 | 0.196–0.847 | 5.92 | 0.01495 | |
| AA | 7 (0.12) | 6 (0.07) | 1.426 | 0.440–4.622 | 0.35 | 0.55296 | |
| GA+AA | 22 (0.38) | 51 (0.54) | 0.527 | 0.271–1.027 | 3.58 | 0.05840 | |
| G | 87 (0.75) | 133 (0.70) | Ref | ||||
| A | 29 (0.25) | 57 (0.30) | 0.778 | 0.461–1.311 | 0.89 | 0.34517 | |
|
| |||||||
| rs1866074 | AA | 12 (0.20) | 12 (0.13) | Ref | |||
| AG | 23 (0.40) | 40 (0.42) | 0.575 | 0.222–1.487 | 1.32 | 0.25137 | |
| GG | 23 (0.40) | 43 (0.45) | 0.535 | 0.208–1.379 | 1.70 | 0.19227 | |
| AG+GG | 46 (0.80) | 83 (0.87) | 0.554 | 0.230–1.333 | 1.77 | 0.18362 | |
| A | 47 (0.41) | 64 (0.34) | Ref | ||||
| G | 69 (0.59) | 126 (0.66) | 0.746 | 0.463–1.202 | 1.45 | 0.22776 | |
|
| |||||||
| rs4135113 | GG | 45 (0.78) | 75 (0.79) | Ref | |||
| GA | 9 (0.16) | 18 (0.19) | 0.833 | 0.345–2.012 | 0.16 | 0.68491 | |
| AA | 4 (0.07) | 2 (0.02) | 3.333 | 0.587–18.937 | 2.05 | 0.15266 | |
| GA+AA | 13 (0.22) | 20 (0.21) | 1.083 | 0.492–2.387 | 0.04 | 0.84257 | |
| G | 99 (0.85) | 168 (0.88) | Ref | ||||
| A | 17 (0.15) | 22 (0.12) | 1.311 | 0.664–2.588 | 0.61 | 0.43370 | |
Table 4.
Genotype frequencies of TDG gene polymorphisms in female colorectal cases and controls.
| SNP | Variant | Patients Cases | Controls | OR | CI | χ 2 Value | p-value |
|---|---|---|---|---|---|---|---|
| rs4135050 | TT | 26 (0.62) | 64 (0.67) | Ref | |||
| TA | 13 (0.31) | 28 (0.29) | 1.143 | 0.513–2.544 | 0.11 | 0.74356 | |
| AA | 3 (0.07) | 4 (0.04) | 1.846 | 0.386–8.828 | 0.60 | 0.43682 | |
| TA+AA | 16 (0.38) | 32 (0.33) | 1.231 | 0.579–2.615 | 0.29 | 0.58890 | |
| T | 65 (0.77) | 156 (0.81) | Ref | ||||
| A | 19 (0.23) | 36 (0.19) | 1.267 | 0.677–2.370 | 0.55 | 0.45905 | |
|
| |||||||
| rs1882018 | CC | 23 (0.55) | 58 (0.60) | Ref | |||
| CT | 14 (0.33) | 32 (0.34) | 1.103 | 0.500–2.436 | 0.06 | 0.80789 | |
| TT | 5 (0.12) | 6 (0.06) | 2.101 | 0.584–7.568 | 1.33 | 0.24858 | |
| CT+TT | 19 (0.45) | 38 (0.40) | 1.261 | 0.606–2.623 | 0.39 | 0.53475 | |
| C | 60 (0.71) | 148 (0.77) | Ref | ||||
| T | 24 (0.29) | 44 (0.23) | 1.345 | 0.753–2.405 | 1.01 | 0.31578 | |
|
| |||||||
| rs4135066 | CC | 1 (0.02) | 8 (0.08) | Ref | |||
| CT | 17 (0.40) | 28 (0.29) | 4.857 | 0.558–42.3 | 2.40 | 0.12134 | |
| TT | 24 (0.57) | 60 (0.63) | 3.200 | 0.379–26.983 | 1.26 | 0.26149 | |
| CT+TT | 41 (0.98) | 88 (0.92) | 3.727 | 0.451–30.794 | 1.70 | 0.19254 | |
| C | 19 (0.23) | 44 (0.23) | Ref | ||||
| T | 65 (0.77) | 148 (0.77) | 1.017 | 0.552–1.876 | 0.0012 | 0.95677 | |
|
| |||||||
| rs3751209 | GG | 15 (0.36) | 43 (0.45) | Ref | |||
| GA | 23 (0.54) | 43 (0.45) | 1.533 | 0.706–3.331 | 1.17 | 0.27879 | |
| AA | 4 (0.10) | 10 (0.10) | 1.147 | 0.313–4.207 | 0.04 | 0.83645 | |
| GA+AA | 27 (0.64) | 53 (0.55) | 1.460 | 0.691–3.087 | 0.99 | 0.32021 | |
| G | 53 (0.63) | 129 (0.67) | Ref | ||||
| A | 31 (0.37) | 63 (0.33) | 1.198 | 0.701–2.047 | 0.44 | 0.50919 | |
|
| |||||||
| rs1866074 | AA | 10 (0.24) | 12 (0.14) | Ref | |||
| AG | 16 (0.38) | 42 (0.44) | 0.457 | 0.165–1.265 | 2.32 | 0.12761 | |
| GG | 16 (0.38) | 42 (0.44) | 0.457 | 0.165–1.265 | 2.32 | 0.12761 | |
| AG+GG | 32 (0.76) | 84 (0.88) | 0.457 | 0.180–1.162 | 2.79 | 0.09493 | |
| A | 36 (0.43) | 66 (0.34) | Ref | ||||
| G | 48 (0.57) | 126 (0.66) | 0.698 | 0.413–1.180 | 1.80 | 0.17917 | |
|
| |||||||
| rs4135113 | GG | 30 (0.71) | 81 (0.84) | Ref | |||
| GA | 9 (0.21) | 13 (0.14) | 1.869 | 0.725–4.821 | 1.71 | 0.19133 | |
| AA | 3 (0.07) | 2 (0.02) | 4.050 | 0.645–25.439 | 2.56 | 0.10991 | |
| GA+AA | 12 (0.29) | 15 (0.16) | 2.160 | 0.908–5.140 | 3.11 | 0.07773 | |
| G | 69 (0.82) | 175 (0.91) | Ref | ||||
| A | 15 (0.18) | 17 (0.09) | 2.238 | 1.059–4.729 | 4.62 | 0.03159 | |
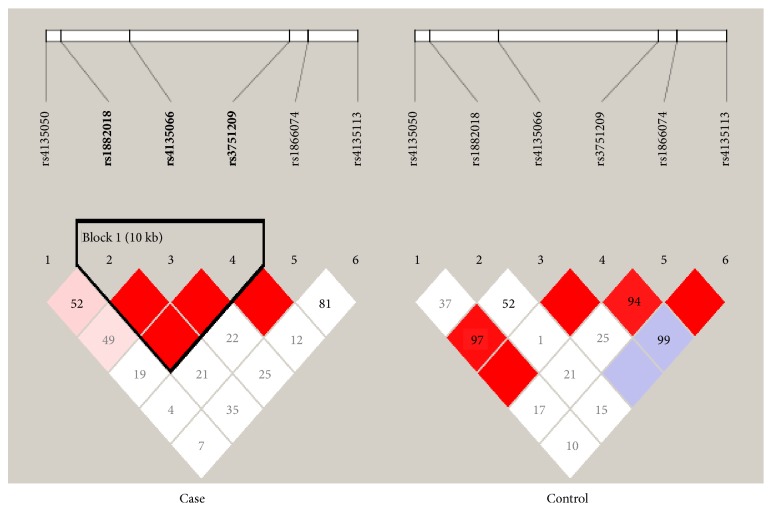
The TDG genotype distribution was further correlated with the age at colon cancer diagnosis and tumor location. To assess the association of the analyzed SNPs with age at colon cancer diagnosis, we divided the patients into two groups based on the median age of the samples: ≤57 (n = 53) or >57 (n = 47) years of age. The distributions of genotype and allele frequencies for each SNP are shown in Tables 5 and 6. SNP rs4135113, which showed a significant association with CRC in the overall analysis, showed a significant risk association in CRC patients in the group of individuals above 57 years of age. The AA genotype frequency was higher in patients than in healthy individuals. This genotype showed a 5-fold increased risk of colon cancer in the Saudi Arabian population (OR: 5.588; CI: 1.032–30.254; p = 0.02745). In addition to this, the rs4135113 minor allele A also showed a 2-fold increased risk for colorectal cancer in the Saudi population (OR: 2.184, CI: 1.077–4.431; p = 0.02778) (Table 6). A linkage disequilibrium analysis revealed that there was a difference in strength among the SNP associations in cases and controls (Figure 1).
Table 5.
Genotype frequencies of TDG gene polymorphisms in colorectal cases and controls in the below-57-year-old group.
| SNP | Variant | Patients Cases | Controls | OR | CI | χ 2 Value | p-value |
|---|---|---|---|---|---|---|---|
| rs4135050 | TT | 31 (0.58) | 67 (0.68) | Ref | |||
| TA | 19 (0.36) | 25 (0.25) | 1.643 | 0.789–3.418 | 1.78 | 0.18271 | |
| AA | 3 (0.06) | 7 (0.07) | 0.926 | 0.224–3.824 | 0.01 | 0.91567 | |
| TA+AA | 22 (0.42) | 32 (0.32) | 1.486 | 0.745–2.962 | 1.27 | 0.25944 | |
| T | 81 (0.76) | 159 (0.80) | Ref | ||||
| A | 25 (0.24) | 39 (0.20) | 1.258 | 0.712–2.223 | 0.63 | 0.42813 | |
|
| |||||||
| rs1882018 | CC | 30 (0.57) | 61 (0.62) | Ref | |||
| CT | 17 (0.32) | 31 (0.31) | 1.115 | 0.534–2.327 | 0.08 | 0.77161 | |
| TT | 6 (0.11) | 7 (0.07) | 1.743 | 0.538–5.642 | 0.87 | 0.34986 | |
| CT+TT | 23 (0.43) | 38 (0.38) | 1.231 | 0.625–2.423 | 0.36 | 0.54797 | |
| C | 77 (0.73) | 153 (0.77) | Ref | ||||
| T | 29 (0.27) | 45 (0.23) | 1.281 | 0.745–2.200 | 0.80 | 0.36989 | |
|
| |||||||
| rs4135066 | CC | 2 (0.04) | 9 (0.09) | Ref | |||
| CT | 21 (0.40) | 31 (0031) | 3.048 | 0.598–15.547 | 1.93 | 0.16466 | |
| TT | 30 (0.56) | 59 (0.60) | 2.288 | 0.465–11.265 | 1.08 | 0.29768 | |
| CT+TT | 51 (0.96) | 90 (0.91) | 2.550 | 0.530–12.260 | 1.45 | 0.22791 | |
| C | 25 (0.24) | 49 (0.25) | Ref | ||||
| T | 81 (0.76) | 149 (0.75) | 1.066 | 0.613–1.851 | 0.05 | 0.82191 | |
|
| |||||||
| rs3751209 | GG | 26 (0.49) | 46 (0.46) | Ref | |||
| GA | 20 (0.38) | 44 (0.44) | 0.804 | 0.394–1.643 | 0.36 | 0.54978 | |
| AA | 7 (0.13) | 9 (0.10) | 1.376 | 0.459–4.128 | 0.33 | 0.56807 | |
| GA+AA | 27 (0.51) | 53 (0.54) | 0.901 | 0.462–1.758 | 0.09 | 0.76037 | |
| G | 72 (0.68) | 136 (0.69) | Ref | ||||
| A | 34 (0.32) | 62 (0.31) | 1.036 | 0.624–1.719 | 0.02 | 0.89161 | |
|
| |||||||
| rs1866074 | AA | 13 (0.25) | 13 (0.13) | Ref | |||
| AG | 22 (0.42) | 42 (0.42) | 0.524 | 0.208–1.322 | 1.90 | 0.16815 | |
| GG | 18 (0.33) | 44 (0.43) | 0.409 | 0.159–1.052 | 3.53 | 0.06029 | |
| AG+GG | 40 (0.75) | 86 (0.87) | 0.465 | 0.198–1.094 | 3.16 | 0.07536 | |
| A | 48 (0.45) | 68 (0.34) | Ref | ||||
| G | 58 (0.55) | 130 (0.66) | 0.632 | 0.390–1.023 | 3.50 | 0.06132 | |
|
| |||||||
| rs4135113 | GG | 41 (0.77) | 80 (0.81) | Ref | |||
| GA | 10 (0.19) | 17 (0.17) | 1.148 | 0.482–2.732 | 0.10 | 0.75528 | |
| AA | 2 (0.04) | 2 (0.02) | 1.951 | 0.265–14.35 | 0.45 | 0.50442 | |
| GA+AA | 12 (0.23) | 19 (0.19) | 1.232 | 0.546–2.784 | 0.25 | 0.61496 | |
| G | 92 (0.87) | 177 (0.89) | Ref | ||||
| A | 14 (0.13) | 21 (0.11) | 1.283 | 0.623–2.639 | 0.46 | 0.49826 | |
Table 6.
Genotype frequencies of TDG gene polymorphisms in colorectal cases and controls in the above-57-year-old group.
| SNP | Variant | Patients Cases | Controls | OR | CI | χ 2 Value | p-value |
|---|---|---|---|---|---|---|---|
| rs4135050 | TT | 27 (0.57) | 57 (0.62) | Ref | |||
| TA | 15 (0.32) | 30 (0.33) | 1.056 | 0.488–2.281 | 0.02 | 0.89062 | |
| AA | 5 (0.11) | 5 (0.05) | 2.111 | 0.563–7.914 | 1.27 | 0.25994 | |
| TA+AA | 20 (0.43) | 35 (0.38) | 1.206 | 0.590–2.466 | 0.26 | 0.60699 | |
| T | 69 (0.73) | 144 (0.78) | Ref | ||||
| A | 25 (0.27) | 40 (0.22) | 1.304 | 0.733–2.321 | 0.82 | 0.36543 | |
|
| |||||||
| rs1882018 | CC | 28 (0.60) | 57 (0.62) | Ref | |||
| CT | 15 (0.32) | 28 (0.30) | 1.091 | 0.503–2.363 | 0.05 | 0.82605 | |
| TT | 4 (0.08) | 7 (0.08) | 1.163 | 0.314–4.307 | 0.05 | 0.82075 | |
| CT+TT | 19 (0.40) | 35 (0.38) | 1.105 | 0.539–2.267 | 0.07 | 0.78518 | |
| C | 71 (0.76) | 142 (0.77) | Ref | ||||
| T | 23 (0.24) | 42 (0.23) | 1.095 | 0.612–1.962 | 0.09 | 0.75960 | |
|
| |||||||
| rs4135066 | CC | 2 (0.04) | 5 (0.05) | Ref | |||
| CT | 17 (0.36) | 27 (0.29) | 1.574 | 0.274–9.045 | 0.26 | 0.60894 | |
| TT | 28 (0.60) | 60 (0.65) | 1.167 | 0.213–6.387 | 0.03 | 0.85883 | |
| CT+TT | 45 (0.94) | 87 (0.95) | 1.293 | 0.241–6.930 | 0.09 | 0.76356 | |
| C | 21 (0.22) | 37 (0.20) | Ref | ||||
| T | 73 (0.78) | 147 (0.80) | 0.875 | 0.478–1.602 | 0.19 | 0.66485 | |
|
| |||||||
| rs3751209 | GG | 25 (0.53) | 41 (0.45) | Ref | |||
| GA | 18 (0.38) | 44(0.47) | 0.671 | 0.320–1.407 | 1.12 | 0.28959 | |
| AA | 4 (0.09) | 7 (0.08) | 0.937 | 0.249–3.527 | 0.01 | 0.92351 | |
| GA+AA | 22 (0.47) | 51 (0.55) | 0.707 | 0.349–1.432 | 0.93 | 0.33531 | |
| G | 68 (0.72) | 126 (0.68) | Ref | ||||
| A | 26 (0.28) | 58 (0.32) | 0.831 | 0.480–1.438 | 0.44 | 0.50706 | |
|
| |||||||
| rs1866074 | AA | 9 (0.19) | 11 (0.12) | Ref | |||
| AG | 17 (0.36) | 4 (0.43) | 0.519 | 0.182–1.481 | 1.52 | 0.21694 | |
| GG | 21 (0.45) | 41 (0.45) | 0.626 | 0.224–1.747 | 0.81 | 0.36892 | |
| AG+GG | 38 (0.81) | 81 (0.88) | 0.573 | 0.219–1.500 | 1.31 | 0.25305 | |
| A | 35 (0.37) | 62 (0.34) | Ref | ||||
| G | 59 (0.63) | 122 (0.66) | 0.857 | 0.510–1.438 | 0.34 | 0.55817 | |
|
| |||||||
| rs4135113 | GG | 34 (0.72) | 76 (0.83) | Ref | |||
| GA | 8 (0.17) | 14 (0.15) | 1.277 | 0.490–3.33 | 0.25 | 0.61607 | |
| AA | 5 (0.11) | 2 (0.02) | 5.588 | 1.032–30.254 | 4.86 | 0.02745 | |
| GA+AA | 13 (0.28) | 16 (0.17) | 1.816 | 0.787–4.191 | 1.99 | 0.15870 | |
| G | 76 (0.81) | 166 (0.90) | Ref | ||||
| A | 18 (0.19) | 18 (0.10) | 2.184 | 1.077–4.431 | 4.84 | 0.02778 | |
Figure 1.
Pairwise LD among the six SNPs in colon cancer and controls. The bright red color indicates a high D′.
4. Discussion
To the best of our knowledge, very few studies have been reported which correlate variation in the TDG gene with cancer [16–18]. With the aim of studying the role played by the polymorphisms in the TDG gene in CRC risk, we investigated six SNPs (rs4135113, rs4135050, rs4135066, rs3751209, rs1866074, and rs1882018) distributed in different regions of the TDG gene. The SNPs were selected based on their location in the TDG gene: rs4135113 is located in exon 5; rs4135050, rs4135066, and rs1882018 are in intron 1; and rs3751209 and rs1866074 are in intron 2 and intron 3, respectively. We chose these SNPs to study the effect of mutations in exons and introns. Mutations in an exon might affect the synthesized protein, whereas intron mutations might affect the RNA processing machinery and RNA splicing and stability, which could impact the level of expression and/or protein output [17]. Five of the SNPs were located in intronic region and four of them are in regulatory regions. SNPs rs4135066, rs4135050, and rs1882018 are located in aligned intronic regions flanking alternative conserved exon region (ACE). SNPs rs4135050 and rs1882018 are in exonic splicing silencer (ESS) region, and rs1866074 is in exonic splicing enhancer region.
All six SNPs in the normal control and CRC patient group obeyed the Hardy-Weinberg equilibrium (HWE). Out of the six SNPs, two showed a significant association with CRC. SNP rs4135113 showed a significant risk association of its genotype AA (OR: 3.640, CI: 1.0341–2.819, p = 0.03286) and of the minor allele A (OR: 1.675, CI: 1.013–2.769, p = 0.04264) with colorectal cancer in Saudi patients. The SNP rs1866074 showed a protective association of the GG allele (OR: 0.501, CI: 0.251–1, p = 0.047) and the additive (AG+GG) allele (OR: 0.51, CI: 0.269–0.964, p = 0.036). Our genotyping results showed that there was no association of the other four SNPs (rs4135050, rs4135066, rs3751209, and rs1882018) with CRC patients in the Saudi population in the overall analysis.
The SNP located in the coding region of the TDG gene, rs4135113, a G/A transition (missense mutation, Gly199Ser), was studied to detect if there was any association with CRC. There is recent evidence supporting an association between this polymorphism and the development of cancer. Sjolund et al. [15] reported that the Gly199Ser polymorphism occurs in approximately 10% of the global population and the expression of TDG with the G199S variant in human breast epithelial cells might lead to an increased number of DNA double-strand breaks. Thus, it initiates and activates DNA damage that induces cellular transformation and chromosomal aberrations [18]. Our results showed that the A/A genotype variation increases the risk of CRC by approximately fourfold in Saudi patients and is statistically significant (OR= 3.64, p-value = 0.03) (Table 2). Further investigation was conducted to explore the correlation of this polymorphic site with the clinicopathological factors and we observed that rs4135113 showed a fivefold increased risk in old aged patients. A study carried out by Wen-Bin and colleagues (2009) on a Chinese population showed a significant association of rs4135113 with an increased micronucleus in the Chinese population. A few other studies have reported that this SNP has no association with an increased risk of lung cancer, rectal cancer, or gastric adenocarcinoma in a Polish population and a Chinese population [19–21].
We also investigated the effect of rs4135050 on the risk of CRC when the T was substituted by A. The genotype AA in our study showed an elevated CRC risk, although the difference was not statistically significant (Table 2). In an urban Puerto Rican population, the one-carbon nutrient status was not associated with the DNA uracil concentration in this SNP [22].
The SNP rs4135066 has the C substituted by a T. In this investigation, the homozygous TT showed an increased risk of CRC; however, this was not statistically significant (Table 2). A recent study by Barry et al. in an American population showed that the SNP rs4135066 was not statistically associated with prostate cancer [23]. In the rs3751209 polymorphism, the A/G variation in our study showed a reduction in the CRC risk, but the difference did not reach statistical significance (Table 2). A recent study by Osorio et al. showed that this SNP was not associated with breast cancer risk in BRCA1/2 mutation carriers [14]. Another SNP studied was rs1866074, which is located in the intronic region and results from a transition mutation where A is substituted by G. A recent case control study showed that the increase in the frequency of micronuclei in bladder cancer among the AG and GG carriers improved patient prognosis [24]. In this investigation, we observed that the GG genotype and the AG+GG additive genotype decreased the risk of CRC (Table 2). Finally, rs1882018 was studied during this investigation, which is also located in the intronic region and is produced as a result of a transition mutation where A is substituted by G. Our results showed that the GG genotype increased the risk of BC, but the finding did not reach statistical significance (Table 2). A previous study carried out by Wei et al. showed that this SNP had a protective effect against the development of bladder cancer [25].
5. Conclusions
In conclusion, the present study showed a significant association between the TDG gene and colorectal cancer progression in a Saudi population. One of the six TDG SNPs showed an increased risk of colon cancer. TDG rs4135113 increased the risk of colon cancer development by more than 3.6- and 1.6-fold in CRC patients in general, and 5-fold in patients aged more than 57 years. SNP rs1866074 showed a significant protective association in CRC patients. The GA genotype of TDG rs3751209 showed a decreased risk of CRC in males. Thus, there is a significant relationship between TDG gene function and colorectal cancer progression. However, further studies are required to determine the exact effect of amino acid (Gly199Ser) replacement using in vitro methods.
Acknowledgments
This project is financially supported by King Saud University through the vice deanship of scientific research.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
All authors declare no conflicts of interest.
Supplementary Materials
Suppl. Table 1: General clinical parameters of the study participants.
References
- 1.Schormann N., Ricciardi R., Chattopadhyay D. Uracil-DNA glycosylases—structural and functional perspectives on an essential family of DNA repair enzymes. Protein Science. 2014;23(12):1667–1685. doi: 10.1002/pro.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aravind L., Koonin E. V. The alpha/beta fold uracil DNA glycosylases: a common origin with diverse fates. Genome Biology. 2000;1(4) doi: 10.1186/gb-2000-1-4-research0007.RESEARCH0007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cortázar D., Kunz C., Saito Y., Steinacher R., Schär P. The enigmatic thymine DNA glycosylase. DNA Repair. 2007;6(4):489–504. doi: 10.1016/j.dnarep.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 4.Métivier R., Gallais R., Tiffoche C. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- 5.Dalton S. R., Bellacosa A. DNA demethylation by TDG. Epigenomics. 2012;4(4):459–467. doi: 10.2217/epi.12.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim E.-J., Um S.-J. Thymine-DNA glycosylase interacts with and functions as a coactivator of p53 family proteins. Biochemical and Biophysical Research Communications. 2008;377(3):838–842. doi: 10.1016/j.bbrc.2008.10.058. [DOI] [PubMed] [Google Scholar]
- 7.Da Costa N. M., Hautefeuille A., Cros M.-P., et al. Transcriptional regulation of thymine DNA glycosylase (TDG) by the tumor suppressor protein p53. Cell Cycle. 2012;11(24):4570–4578. doi: 10.4161/cc.22843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broderick P., Bagratuni T., Vijayakrishnan J., Lubbe S., Chandler I., Houlston R. S. Evaluation of NTHL1, NEIL1, NEIL2, MPG, TDG, UNG, and SMUG1 genes in familial colorectal cancer predisposition. BMC Cancer. 2006;6, article 243(1) doi: 10.1186/1471-2407-6-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu X., Yu T., Shi J., et al. Thymine DNA glycosylase is a positive regulator of Wnt signaling in colorectal cancer. The Journal of Biological Chemistry. 2014;289(13):8881–8890. doi: 10.1074/jbc.M113.538835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng B., Hurt E. M., Hodge D. R., Thomas S. B., Farrar W. L. DNA hypermethylation and partial gene silencing of human thymine-DNA glycosylase in multiple myeloma cell lines. Epigenetics. 2006;1(3):138–145. doi: 10.4161/epi.1.3.2938. [DOI] [PubMed] [Google Scholar]
- 11.Yatsuoka T., Furukawa T., Abe T., et al. Genomic analysis of the thymine-DNA glycosylase (TDG) gene on 12q22–q24. 1 in human pancreatic ductal adenocarcinoma. International Journal of Pancreatology. 1999;25(2):97–102. doi: 10.1385/IJGC:25:2:97. [DOI] [PubMed] [Google Scholar]
- 12.Vasovcak P., Krepelova A., Menigatti M., et al. Unique mutational profile associated with a loss of TDG expression in the rectal cancer of a patient with a constitutional PMS2 deficiency. DNA Repair. 2012;11(7):616–623. doi: 10.1016/j.dnarep.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruczinski I., Jorgensen T. J., Shugart Y. Y., et al. A population-based study of dna repair gene variants in relation to non-melanoma skin cancer as a marker of a cancer-prone phenotype. Carcinogenesis. 2012;33(9):1692–1698. doi: 10.1093/carcin/bgs170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fogarty M. P., Cannon M. E., Vadlamudi S., Gaulton K. J. Identification of a regulatory variant that binds FOXA1 and FOXA2 at the CDC123/CAMK1D type 2 diabetes GWAS locus. PLoS Genetics. 2014;10(9) doi: 10.1371/journal.pgen.1004633.e1004633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li W.-Q., Hu N., Hyland P. L., et al. Genetic variants in DNA repair pathway genes and risk of esophageal squamous cell carcinoma and gastric adenocarcinoma in a Chinese population. Carcinogenesis. 2013;34(7):1536–1542. doi: 10.1093/carcin/bgt094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alanazi M., Pathan A. A. K., Shaik J. P., et al. The hOGG1 Ser326Cys gene polymorphism and breast cancer risk in saudi population. Pathology & Oncology Research. 2017;23(3):525–535. doi: 10.1007/s12253-016-0146-6. [DOI] [PubMed] [Google Scholar]
- 17.Penny D., Hoeppner M. P., Poole A. M., Jeffares D. C. An overview of the introns-first theory. Journal of Molecular Evolution. 2009;69(5):527–540. doi: 10.1007/s00239-009-9279-5. [DOI] [PubMed] [Google Scholar]
- 18.Sjolund A., Nemec A. A., Paquet N., et al. A Germline polymorphism of thymine DNA glycosylase induces genomic instability and cellular transformation. PLoS Genetics. 2014;10(11) doi: 10.1371/journal.pgen.1004753.e1004753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krześniak M., Butkiewicz D., Samojedny A., Chorazy M., Rusin M. Polymorphisms in TDG and MGMT genes - epidemiological and functional study in lung cancer patients from Poland. Annals of Human Genetics. 2004;68(4):300–312. doi: 10.1046/j.1529-8817.2004.00079.x. [DOI] [PubMed] [Google Scholar]
- 20.Curtin K., Ulrich C. M., Samowitz W. S., et al. Candidate pathway polymorphisms in one-carbon metabolism and risk of rectal tumor mutations. International Journal of Molecular Epidemiology and Genetics. 2011;2(1):1–8. [PMC free article] [PubMed] [Google Scholar]
- 21.Kohli R. M., Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502(7472):472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chanson A., Parnell L. D., Ciappio E. D., et al. Polymorphisms in uracil-processing genes, but not one-carbon nutrients, are associated with altered dna uracil concentrations in an urban puerto rican population. The American Journal of Clinical Nutrition. 2009;89(6):1927–1936. doi: 10.3945/ajcn.2009.27429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barry K. H., Koutros S., Berndt S. I., et al. Genetic variation in base excision repair pathway genes, pesticide exposure, and prostate cancer risk. Environmental Health Perspectives. 2011;119(12):1726–1732. doi: 10.1289/ehp.1103454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pardini B., Viberti C., Naccarati A., et al. Increased micronucleus frequency in peripheral blood lymphocytes predicts the risk of bladder cancer. British Journal of Cancer. 2017;116(2):202–210. doi: 10.1038/bjc.2016.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei H., Kamat A., Chen M., et al. Association of polymorphisms in oxidative stress genes with clinical outcomes for bladder cancer treated with bacillus calmette-guérin. PLoS ONE. 2012;7(6) doi: 10.1371/journal.pone.0038533.e38533 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Suppl. Table 1: General clinical parameters of the study participants.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.