Abstract
Mycobacterium tuberculosis possesses an unusually large representation of type II toxin–antitoxin (TA) systems, whose functions and targets are mostly unknown. To better understand the basis of their unique expansion and to probe putative functional similarities among these systems, here we computationally and experimentally investigated their sequence relationships. Bioinformatic and phylogenetic investigations revealed that 51 sequences of the VapBC toxin family group into paralogous sub-clusters. On the basis of conserved sequence fingerprints within paralogues, we predicted functional residues and residues at the putative TA interface that are useful to evaluate TA interactions. Substitution of these likely functional residues abolished the toxin's growth-inhibitory activity. Furthermore, conducting similarity searches in 101 mycobacterial and ∼4500 other prokaryotic genomes, we assessed the relative conservation of the M. tuberculosis TA systems and found that most TA orthologues are well-conserved among the members of the M. tuberculosis complex, which cause tuberculosis in animal hosts. We found that soil-inhabiting, free-living Actinobacteria also harbor as many as 12 TA pairs. Finally, we identified five novel putative TA modules in M. tuberculosis. For one of them, we demonstrate that overexpression of the putative toxin, Rv2514c, induces bacteriostasis and that co-expression of the cognate antitoxin Rv2515c restores bacterial growth. Taken together, our findings reveal that toxin sequences are more closely related than antitoxin sequences in M. tuberculosis. Furthermore, the identification of additional TA systems reported here expands the known repertoire of TA systems in M. tuberculosis.
Keywords: Mycobacterium tuberculosis, structure–function, molecular modeling, protein evolution, bioinformatics, phylogenetics, protein sequence, genome analysis, homology, pathogenesis, toxin–antitoxin, VapBC
Introduction
Mycobacterium tuberculosis is a major human pathogen that causes an estimated million deaths every year (1). Toxin–antitoxin systems (TAs)8 are believed to be important for pathogenesis, stress survival, and bacterial persistence (2). They consist of a stable toxin that can interfere with vital cellular functions and a labile antitoxin that can inhibit the activity of the toxin. TA systems have been categorized into different types depending on the nature of the toxin and antitoxin (3, 4). Type I TA systems consist of an antisense RNA antitoxin complementary to toxin mRNA. Type II TA systems have a protein antitoxin that directly interacts with a protein toxin. Type III TA systems comprise an RNA antitoxin that directly binds the protein toxin. Type IV TA systems are composed of toxins and antitoxins that do not directly interact with each other but bind to the same cellular target. Type V antitoxins are endoribonucleases that specifically degrade the toxin mRNA (5). Type VI TA systems possess an antitoxin that neutralizes the toxin by promoting its proteolytic cleavage (6). An atypical tripartite type II toxin–antitoxin-chaperone with a chaperone that protects the antitoxin and controls its folding has also been reported (7). Functional roles of TAs have expanded since their initial association with post-segregational killing and plasmid maintenance (8–10). Type II toxin systems are associated with a broad array of targets and affect central dogma reactions. These include inhibition of DNA gyrase, ribosomes, and elongation factors or acting as ribonucleases (11–13). Their roles in persistence regulation, biofilm formation, antibiotic tolerance, stress adaptation, and virulence have been reported (9, 12, 14–16), although not demonstrated in M. tuberculosis.
Type II TA systems are widespread among Bacteria and Archaea (4) and are reported in the human microbiome as well (17–19). In M. tuberculosis alone, 81 TA systems have been reported (5). These include TAs of various types such as 51 VapBC, 10 MazEF, two RelBE, ParDE, and HigBA, and one YefM/YoeB, in addition to tripartite toxin–antitoxin–chaperone (7) and three type IV systems (5, 20). The large representation of type II TAs in M. tuberculosis is intriguing, with actual roles unknown for most. Also, little is known about the evolutionary and functional relationships among them. Of these, the TA systems with the PIN domain (originally identified in the N terminus of PilT), known as VapBC (virulence-associated proteins (12, 21–23)), constitute the largest family with 51 members. Targets of these toxins include mRNA, tRNA, and rRNA (24–27). In this study, we performed a comprehensive sequence analysis of the 51 VapBC TA. Using the results of this analysis, we explored the structural and functional similarities among these TAs. Strikingly, we observed that although the VapB and VapC homologues are divergent in sequence, they can be grouped into distinct sub-clusters, suggesting paralogous relationships. We noticed that VapC sequences in each sub-cluster share better overall sequence similarity than across the clusters. Careful analysis of the paralogous sequences revealed amino acid residues that are conserved within each sub-cluster with potential functional roles. Indeed, we show that mutation of these residues abrogated the activity associated with the toxins. Next, to identify orthologues of the M. tuberculosis type II TA systems, we probed 101 mycobacterial genomes and nearly 4500 prokaryotic genomes. Strikingly, we find that although only members of the MTB complex conserve the majority of homologues, some TAs are conserved in other mycobacteria as well. Furthermore, our searches uncovered several TAs of soil-dwelling, free-living bacteria from the class Actinobacteria that are homologous to the M. tuberculosis TA systems. Finally, we report five novel TAs and show that the total number of TA systems in M. tuberculosis may be even greater than previously thought. In this study, Rv2514c–Rv2515c was experimentally validated as a TA system. Recently, we have reported that Rv0366c–Rv0367c encodes a noncanonical PezAT-like toxin–antitoxin pair (28). Our findings may stimulate more detailed experimental characterization of these novel systems and expand the understanding of this complex machinery.
Results
Recognition of related TA systems
The large representation of TA systems in M. tuberculosis prompted us to examine their relationships. To understand their divergence at the sequence level, we probed for similarities within the known VapBC TA sequences (see under “Experimental procedures” for details). Here, we found that not all VapC toxins (Table S1) are closely related to one another (Fig. S1A and Tables S2 and S3). Indeed, VapC16, VapC45, and VapC50 do not find any homologues among the toxins, suggesting very-high-sequence divergence. We find that toxins such as VapC8, VapC9, VapC34, and VapC35 are closely related to only a few other VapCs. In contrast, 30 VapC toxins (e.g. VapC2 and VapC4) are homologous to over 40 other VapC toxins (Fig. S1A and Tables S2 and S3). Similarly, none of the VapB antitoxins were able to identify all the other antitoxins as homologues, exceptions being VapB31 and VapB29 that could identify 17 homologous antitoxins in individual searches (Table S3 and Fig. S1B). These results show that the VapC toxins are more closely related among themselves than their cognate antitoxins, which have diverged extensively.
Analysis of 10 MazEF TAs also showed that MazF toxins are more closely related (sequence identities in the range of 16–60%) (Fig. S2 and Table S2) than MazE antitoxins (Table S3). The high-divergence of antitoxin sequences in the MazEF TA systems implies specific recognition of their cognate toxins. However, earlier findings have reported noncognate interactions between MazF6–VapB27 and MazF9–VapB40 antitoxins, through in vivo toxicity, rescue experiments, and in vitro interaction experiments (29). Consistent with these findings, we report several close similarities between some VapB antitoxins (VapB19, -25, -29, -31, and -33 and others listed in Table S3) and MazE6 at a sequence identity of 23–29%. These newly identified relationships raise the possibility of previously unknown noncognate interactions as part of the complex TA network of M. tuberculosis.
Gene amplification is an important mechanism of adaptive evolution in response to environmental stresses in prokaryotes (30). The close relationships between some of the toxins and antitoxins identified here might correspond to TA systems that respond in unison to specific types of stress, as reported earlier for some TAs (31, 32), or likely expanded from a common ancestor, although this remains to be verified. This might suggest redundancy in the available complement of TA, which can prove advantageous for critical functions such as pathogenesis. Such systems may account for enhanced fidelity at times of variations in the external environment, necessitating the need for redundant virulent strategies (33).
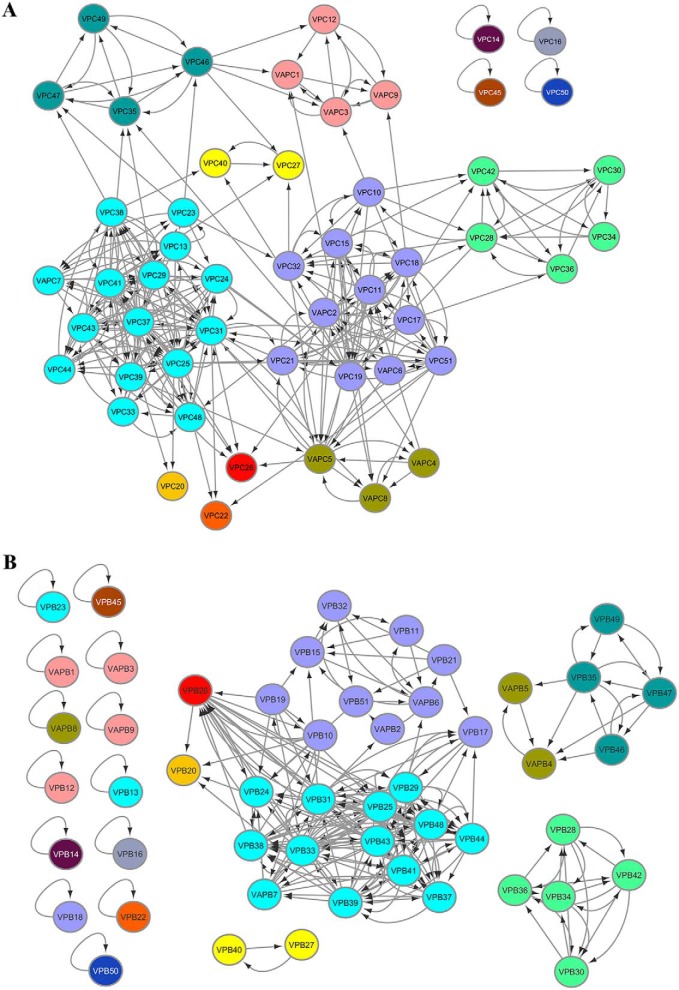
Paralogous VapBC TA group into distinct sub-clusters
We grouped closely-related toxins using a clustering approach (Fig. S2, B and C; see under “Experimental procedures” for details) (34). We obtained seven sub-clusters that group 44 of the 51 VapC toxins and two types of “orphan” VapC toxin clusters (Fig. 1A). These orphan toxins include VapC14, VapC16, VapC45, VapC50, VapC20, VapC22, and VapC26 that are only distantly related to the remaining toxins (Table S2).When antitoxins were clustered, they formed even more isolated sub-clusters (Fig. 1B) due to higher sequence divergence between their sequences (Fig. S2A). Indeed, 15 of the VapB antitoxins could not find any other hits in the searches (Table S3). In general, we observe that detection of paralogous relationships is more robust within the toxins than the antitoxins.
Figure 1.
VapBC toxins and antitoxins cluster distinctly. Clusters of VapC toxins (A) and VapB antitoxins (B) were derived using HIFIX, at 20% sequence identity and 60% query coverage. Cluster criteria were set after employing several identity and alignment thresholds on the VapC toxin alignments, to resolve the data into sub-clusters (Fig. S2). Each sub-cluster is indicated in a separate color. VapCs or VapBs within the same sub-cluster share at least 20% sequence identity. The antitoxin sub-clusters in B are color-coded based on the toxin clusters.
Sequence fingerprints in paralogous TA systems
To examine whether toxin sub-clusters that share distinct sequence similarity provide functional information, we analyzed the full-length sequence alignment of all the 51 VapC toxins (Fig. S3). A characteristic feature of all VapC toxins is the presence of a PIN domain (∼130 residues), which spans nearly the entire length of the toxin and is responsible for the RNase activity (21, 22). Inspection of the alignment shows that although the 51 VapC toxin sequences of M. tuberculosis show high-sequence divergence overall, the quartet of acidic amino acids typical of the PIN domain is well-conserved in all the toxins (Fig. S3). These residues, known to cluster at the active site and bind Mg2+ ions, are implicated in nucleotide cleavage. In addition, we find that the polar residue (Ser, Thr, or Asn), sequentially adjacent to the first conserved aspartate residue (21), is also well-conserved in the sequences (Fig. S3) (22). We find that in orphan toxins (VapC14, VapC16, VapC45, and VapC50), these residues are not conserved raising doubts on their inclusion as TAs. Notably, M. tuberculosis transcriptome analysis has shown that these loci are expressed differentially in response to drugs, implying a poor relationship between them (31). VapC14 and VapC45 in M. tuberculosis are also closely related to homologues in Mycobacterium marinum suggesting that they might have been acquired through lateral gene transfer (35). A recent unpublished report suggests that the putative VapC50 toxin sequence of M. tuberculosis (UniProt: L0TGF0) is altogether incorrect because it did not cleave any of the 45 tRNAs of M. tuberculosis,9 thus explaining its exclusion from the distinct sub-clusters.
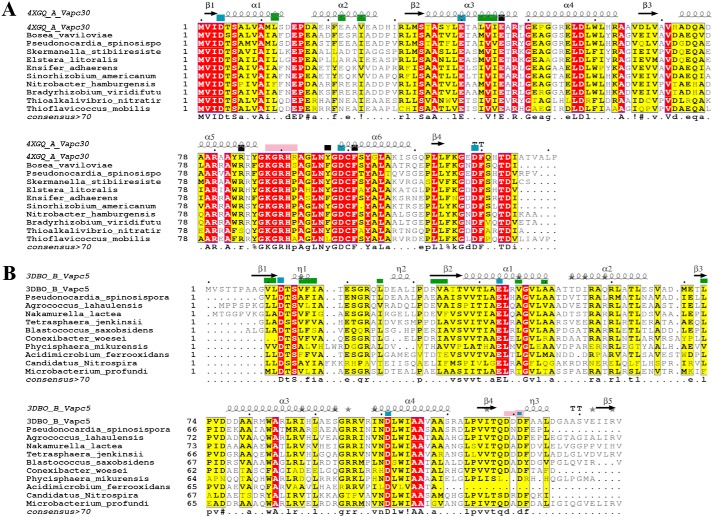
Next, we sought to identify sequence fingerprints of individual sub-clusters by analyzing each sub-cluster alignment (Fig. 2 and Figs. S3 and S4). Here, we observed better overall conservation of residues within each sub-cluster. Indeed, average sequence identities among the members of each sub-cluster vary from ∼24% (as in sub-clusters 1 and 2), ∼35% (sub-clusters 4 and 5) to ∼43% (sub-clusters 3 and 6). Careful analysis of the alignments shows that at some positions the residue preferences differ across the sub-clusters. We reckon that such positions are likely determinants of specificity that may be responsible for maintaining cognate toxin–antitoxin interactions or toxin–substrate specificity (Fig. S3). Therefore, we derived structure-guided alignments, for sub-clusters 2, 3, and 6, by employing the crystal structures of VapC2, VapC30, and VapC5 (36–39). Residue positions involved in antitoxin binding, maintenance of structural integrity, and substrate recognition in the available crystal structures were used to infer and predict residues that could assay similar roles in the paralogues within these sub-clusters (Fig. 2, A–C) (details under “Experimental procedures”) (36, 37, 39). Here, we observed that the putative antitoxin-binding residues, mapped to the alignments, show far less conservation across sub-clusters than within the sub-clusters (Fig. 2). This implies that toxins across sub-clusters may show different antitoxin-binding modes that could help TA systems achieve high specificity toward their cognate pairs. Strongly supporting this observation is the finding by Ramage et al. (20) that cross-talk between noncognate VapB and VapC TAs such as VapBC2, VapBC11, VapBC22, and VapBC47 is absent. Notably, these toxins occur in distinct sub-clusters in our analysis. This only goes to show that the proposed clusters of groups are meaningful.
Figure 2.
Multiple sequence alignments of VapC toxins from sub-clusters 2, 3, and 6 with representative structural templates are shown in A and C. The columns shaded dark red show conserved residues, and the columns shaded yellow show conservatively-substituted residues. Consensus sequence (at a threshold of >70%) for each sub-cluster alignment is shown at the bottom of the alignment. Green boxes at the top of the alignment point to antitoxin-binding residues. Blue boxes show the conserved acidic residues of the PIN domain. Black boxes point to residues that confer structural stability in the reference structures. Pink boxes at the top of the alignment are the substrate-binding residues in the reference structures. Secondary structure elements of the relevant reference structures are depicted at the top of the alignment.
In contrast, the conservation of residues in the interaction interface of the toxin and antitoxin within a sub-cluster could highlight residues that might enable potential cross-talk between the systems. Therefore, we examined each sub-cluster alignment for conserved residues at the predicted toxin–antitoxin interface (Fig. S4, D–F). Sub-cluster 6 alignment shows that antitoxin-binding residues of VapC5 are conserved in VapC4 and VapC8 (Fig. 2C) (39). This suggests that a mode of interaction similar to VapBC5 is possible in VapBC4 and VapBC8. Indeed, evidence for cross-talk between VapBC4 and VapBC5 has been reported in studies that identified the minimal domain of VapB4 and residues critical for antitoxin binding (40). These experiments showed that the C-terminal 31 residues of VapB4, including Asp-64 and Trp-48, are necessary for its interaction with VapC4 and antitoxin function. Other residues involved in this interaction include Thr-66, Leu-72, Gln-78, Thr-80, Ile-55, Leu-58, Leu-61, Gly-62, Leu-68, Glu-71, Glu-74, Thr-79, Asp-81, and Asp-82 (40). From the alignments, we find that in addition to Asp-64 and Trp-48, residues such as Leu-58, Leu-68, Glu-71, Leu-72, Thr-79, Asp-81, and Asp-82 are conserved in the alignments in all three members of the sub-cluster and support our predictions for potential cross-talk (Fig. S4F). We note similar trends in conservation of residues at the toxin/antitoxin interface in other sub-clusters as well (Fig. 2 and Fig. S4). In principle, to evaluate potential for cross-talk, such an analysis may also be extended to other paralogues in all sub-cluster alignments derived in this study.
Recognition of conserved residues with putative functional roles
Next, we sought to predict residues that are likely to play a functional role in the toxic activity of VapCs. Again, as for the antitoxin-binding sites, we employed the available structures of VapC2, VapC30, and VapC5 (in sub-clusters 2, 3, and 6) to map functional residues from the known crystal structures to the alignments (36, 37, 39). The VapC toxins require Mg2+ for their RNase activity. The coordination of the metal ion is mediated by the acidic aspartates that represent the canonical signature of the PIN domain. In each sub-cluster alignment, we found that these residues are well-conserved in the paralogues (Fig. 2 and Fig. S4). In addition, we specifically explored for the presence of positively charged and solvent-exposed residues in the reference structures, with roles in binding target RNAs (Fig. 2) (37). For the remaining sub-clusters (1, 4, and 5), where a reference template was not directly available, conserved and charged residues other than the quartet of Mg2+-interacting residues were also predicted as functionally important (Fig. S4). Finally, after all these considerations, conserved residues such as Arg-92 and Lys-128 for VapC13 (Rv1838c), Thr-121 and Lys-136 for VapC44 (Rv3320c) in sub-cluster 1, Lys-87 and Arg-91 for VapC21 (Rv2757c) in sub-cluster 2, and others listed in Table S4 were predicted as functional residues. Consistent with the toxin alignments in various sub-clusters, cognate antitoxin alignments also show conserved residues at both the N-terminal (DNA binding) and C-terminal (toxin binding) regions (Fig. S4). Therefore, careful analysis of the sequence alignments within each sub-cluster is a useful indicator to predict functionally conserved residues in toxins and antitoxins.
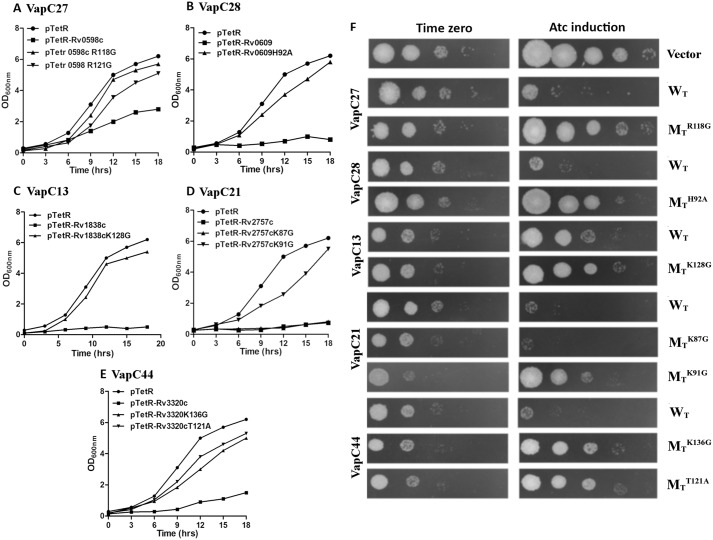
Site-directed mutagenesis of predicted functional residues rescues growth defect
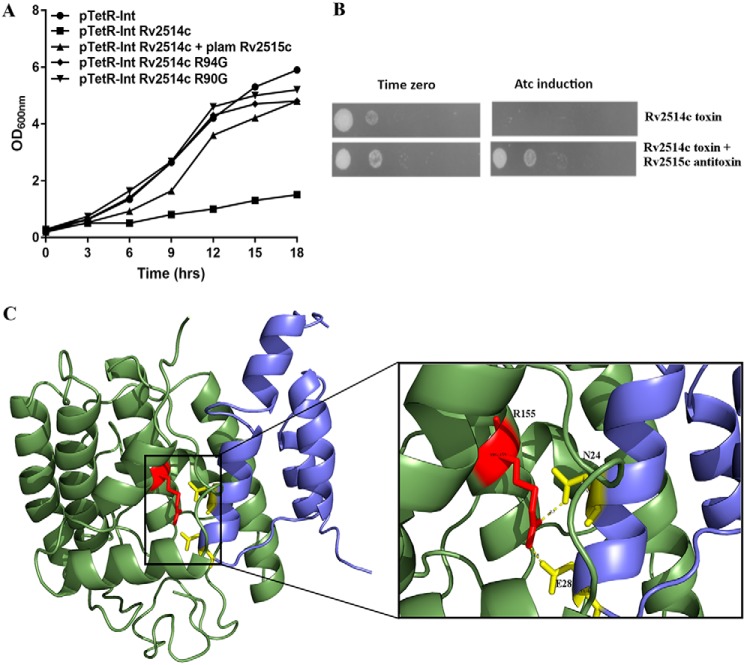
To understand the significance of the putative functional residues recognized in the various sub-clusters derived from the alignments, we performed growth inhibition assays in Mycobacterium smegmatis. For this, both WT and mutant toxins were cloned and overexpressed in M. smegmatis using an anhydrotetracyline-based expression system, pTetR (32). As shown in Fig. 3, we observed severe growth inhibition in M. smegmatis, with expression of the WT toxins. In concordance with our predictions, no growth inhibition was seen in M. smegmatis overexpressing most of the mutant toxins (Fig. 3). We observed that mutation of Arg-118 and Arg-121 in Rv0598c, His-92 in Rv0609, Lys-128 in Rv1838c, Arg-91 in Rv2757c, and Lys-136 and Thr-121 in Rv3320c abrogated the toxicity associated with their WT counterparts in liquid medium (Fig. 3, A–E). In concordance, we observed in spotting assays that overexpression of WT toxin resulted in bacteriostasis, whereas no growth inhibition was seen in strains overexpressing mutant proteins (Fig. 3F). The mutation of Lys-87 residue in Rv2757c alone retained its toxic effect indicating that it may not have a functional role in Rv2757c RNase activity (Fig. 3). In each case, we checked to ensure that the predicted mutations did not destabilize the toxin structure and truly signified a functional role through mutational free-energy calculations (41) (details under “Experimental procedures”). For this purpose, structural models were generated for Rv0598c, Rv0609, Rv1838c, and Rv3320c (details under “Experimental procedures”) (Fig. S5) (38, 42). None of the above mutations caused a ΔΔG >1 kcal/mol, and hence we conclude that they are unlikely to destabilize the protein. Supporting this finding, inter-residue interaction analysis, based on the number of main-chain and side-chain contacts of the residues in the protein, showed that the mutated residues are not involved in any extensive contacts with other residues in the structure (data not shown). Furthermore, except for Arg-91 of Rv2757c, none of the other mutated residues were found to lie at the toxin dimer interface, thus supporting our predictions of the potential involvement of these residues in the toxin function. Overall, the predictions show a high-success rate in the recognition of residues that are essential for either substrate-binding or RNase activity, although further experiments will be required to determine their intracellular targets.
Figure 3.
Effect of overexpression of WT and mutant proteins on the growth of M. smegmatis. A–E, recombinant strains were grown to an OD600 nm of 0.2, and the expression of toxins was induced by the addition of 50 ng/ml Atc. The growth of various strains was monitored by measuring absorbance at 3-h intervals. The graphs also show the growth curves when predicted functional residues were mutated (Table S4). F, for spotting assays 10.0-fold serial dilutions of induced and uninduced cultures were prepared and spotted on MB7H11 plates. The plates were incubated at 37 °C for 2 days, and the images were recorded. The data shown in these panels are representative of three independent experiments. Images in F correspond to the growth curves in A–E. These experiments were also supported by computational alanine-scanning predictions of the modeled proteins harboring the individual mutations, using FoldX (Fig. S5).
M. tuberculosis TA systems are better conserved in M. tuberculosis complexes (MTBC) than in mycobacteria and other prokaryotic genomes
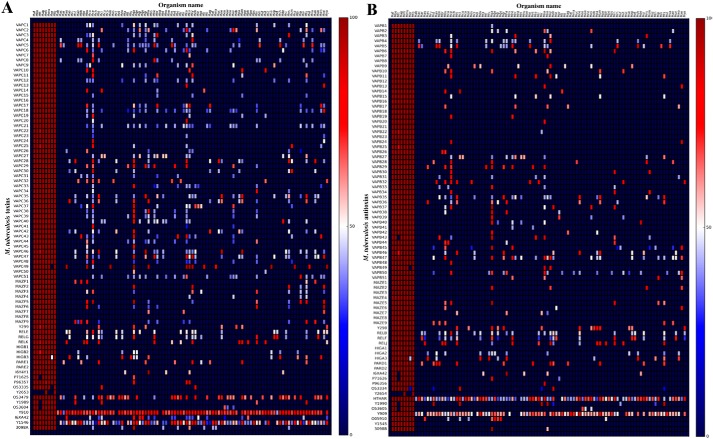
The MTBC constitute a group of mycobacteria that cause tuberculosis in humans/other animal hosts. Previously, homologues of the M. tuberculosis TA systems were probed in 18 genomes of mycobacteria (that included five from MTBC) and revealed orthologues for 65 toxins (20). We have extended this analysis to the now-available 101 genome sequences, with three additional MTBC, namely Mycobacterium orygis, Mycobacterium caprae, and Mycobacterium mungi. We find well-conserved orthologous VapBC and MazEF TA pairs in all MTBC, with Mycobacterium microti conserving all 81 TAs (Figs. 4 and Figs. S6 and S7, Table S5, and details in supporting data S1). Our finding on the expanded set of mycobacteria is consistent with earlier reports that most of the TA systems were present in the MTBC progenitor (20). Comparison with the search results in the larger dataset of 101 genomes shows that four MazEF TAs (MazEF1, MazEF3, MazEF4, and MazEF8) and eight VapBCs (VapBC41, VapBC25, VapBC23, VapBC21, VapBC16, VapBC18, VapBC12, and VapBC8) uniquely occur only in MTBC with no homologues in other mycobacterial genomes (Fig. 5 and Figs. S6 and S8 and Tables S5 and S6). A few TA such as MazEF10, VapBC47, VapBC5, and VapBC4 have orthologues in MTBC and many mycobacteria. TAs such as VapBC49, VapBC48 and a few others show a more limited distribution (Fig. 5 and Fig. S6 and Tables S5 and S6), with homologues only in one or two mycobacteria other than MTBC. Clearly, these findings support the view that the expansion of TA is a conserved feature of all members of MTBC. Although we probed for the distribution of TA homologues that occur as pairs, our searches also identified that two toxin sequences find many orthologues in mycobacteria (Rv2035 (O53479) and Rv1546 (P9WLU7)) (Fig. 4 and Fig. S8, Tables S5 and S6, and supporting data S1).
Figure 4.
Distribution of toxins and antitoxins within the genus Mycobacterium. Heat map shows the distribution of VapC toxins (A) and VapB antitoxins (B) within the 101 mycobacterial species in the study (Table S5). A dot represents the occurrence of a sequence homologue of the M. tuberculosis query (shown on the y axis) for each genome (listed on the x axis). For the identification of a hit, cutoff thresholds of e-value <0.0001 and alignment length of >60% query length coverage were applied. The color convention shows the sequence identity of each hit in the range of 5% (blue) to 100% (red). The first eight points on the x axis are mycobacteria that constitute the MTBC. All other organisms listed on the x axis have been designated a three-letter code (mapping between three-letter code and organism name is provided in Table S7) and are arranged in alphabetical order. Comparisons between the left and right panels in A and B emphasize that a majority of the TA homologues of M. tuberculosis are found in the MTBC. They also show that several toxin and antitoxin homologues, in the genomes studied here, are lone toxins or antitoxins.
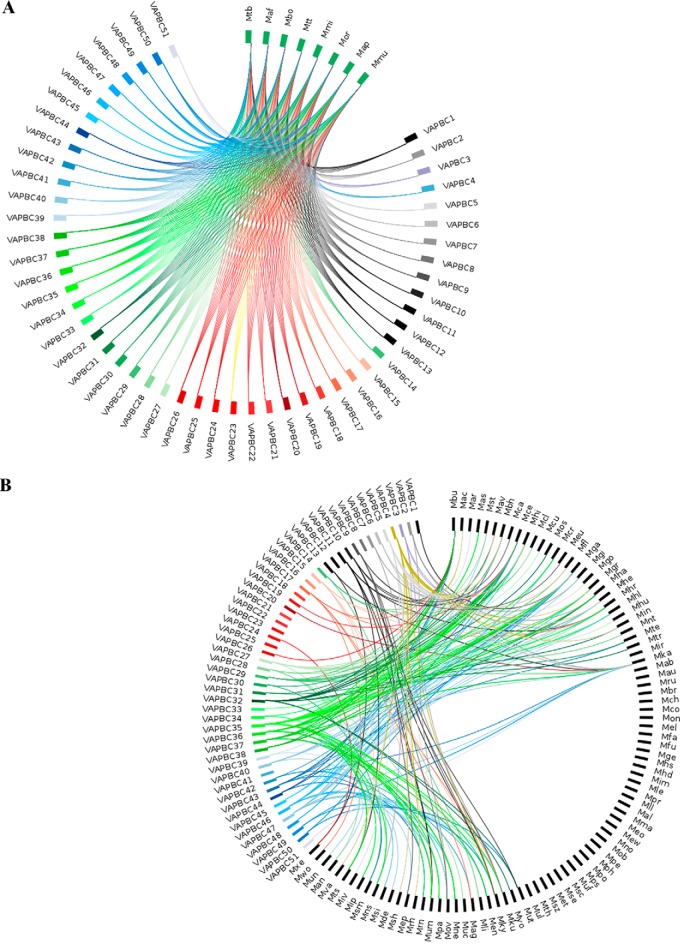
Figure 5.
Plots showing the distribution of M. tuberculosis VapBC TA. Circos plots of homology searches for VapBC TA pairs (A) in eight MTBC (B) in 93 other mycobacteria. In the plots, the nodes represent organism names and TA types that are grouped distinctly, to facilitate interpretation of the figures. Each TA system is designated a separate color. A line is drawn between a TA node and that of an organism if both the toxin and antitoxin query find a homologue in PSI-BLAST and TBLASTN searches. The plots emphasize which of the TA systems of M. tuberculosis find homologues in other mycobacteria. Query types with large number of links (e.g. VapBC47 and VapBC5) are well-conserved with many homologues in various mycobacteria, whereas query types with few/nil links are MTBC-specific (VapBC41 and VapBC16, etc.), as also shown in Figs. S6 and S7.
Such similarity searches when extended to other prokaryotes showed that although homologues of toxins or antitoxins occur in nearly 2200 prokaryotes, their occurrence as a pair is largely in members of the order Corynebacteriales that includes Actinobacteria of which M. tuberculosis is a member (Figs. S9 and S10 and for details see supporting data S2 and Tables S8–S10). The majority of TA pairs was detected in the class Acidimicrobiia (five pairs) and Actinobacteria (11 pairs). Interestingly, of the TA pairs that we found to be unique to MTBC with no homologues in other mycobacteria, the TA systems MazEF5, MazEF6, MazEF9, MazEF10, VapBC12, and VapBC41 do report similarities in other prokaryotes. This suggests that they might have been shared through horizontal gene transfer. In contrast, we report that VapBC16, an orphan toxin that did not detect paralogues within the M. tuberculosis genome, is unique and MTBC-specific with no orthologues in any of the other prokaryotic genomes (Figs. 1 and 5 and Fig. S11). Also, our data show that Rv2654–Rv2653 is present only in M. tuberculosis and M. microti with no homologues in other MTBC or prokaryotes (Figs. S8 and S11). An important finding is that Rv0909–Rv0910, which is considered as a universal TA system for mycobacteria, is found to be unique to it and is absent in other prokaryotic genomes (Figs. S8 and S11) (20).
Overall, we have provided a comprehensive inventory of several putative TA pairs, orphan toxin, or antitoxin homologues of the M. tuberculosis type II TA in all mycobacteria (Tables S5 and S8). In general, we observed that toxin sequences find more homologues than antitoxin sequences, which show high-sequence divergence among antitoxins. Importantly, we show that TA systems are not confined to the virulent mycobacteria of the MTBC, in contrast to the report by Ramage et al. (20).
Conservation of predicted functional residues among M. tuberculosis orthologues
We identified putative M. tuberculosis TA-like homologues encoded in several prokaryotic genomes through sequence similarity searches. Here, we evaluated whether the similarity was by virtue of characteristic hallmarks of the toxins, as in the case of the VapC toxins. Specifically, we assessed whether it extended to functional residues recognized from crystal structures of various toxins such as VapC30 and VapC5 (37, 39). Because many of the orthologues are as yet uncharacterized proteins, we examined sequence alignments of representative homologues of VapC30 (highest sequence identity between members was 64%). Here, we found that the orthologues conserved the PIN domain, residues involved in antitoxin binding, substrate binding, and those known to maintain the structural integrity of VapC30 toxins (Fig. 6) (37, 39). In VapC5 alignments, antitoxin-binding residues were observed to be only partially conserved in the homologues (Fig. 6) (39). Furthermore, examination of surrounding genomic regions in each orthologue of the toxin revealed a putative antitoxin-like sequence upstream of the putative toxin. Alignments of the putative antitoxins showed that the positively charged DNA-binding residues in the N terminus are better conserved than the toxin-binding sequences, which are implicated in specific recognition of their cognate toxins (Fig. S12). Based on these studies, a potential TA function may be predicted in the orthologues. Importantly, these results suggest that, in these cases, signatures of the toxin and antitoxin sequences (both DNA binding and toxin binding) have been stably inherited in various species during evolution.
Figure 6.
Multiple sequence alignment of M. tuberculosis VapC30 (PDB code 4xgq) with selected orthologues from the prokaryotic genomes (A) and VapC5 (PDB code 3dbo) with selected orthologues from the prokaryotic genomes (B). The columns shaded dark red show conserved residues, and the columns shaded yellow show conservatively substituted residues. Secondary structure elements of the reference structures are depicted at the top of each alignment. Green boxes at the top of the alignment point to antitoxin-binding residues. Blue boxes show the conserved acidic residues of the PIN domain. Black boxes point to residues that confer structural stability in the reference structures. Pink boxes at the top of the alignment are the substrate-binding residues in the reference structures. Consensus sequence (at a threshold of >70%) for the alignment is shown at the bottom of the alignment.
Expanding the TA repertoire in M. tuberculosis
Although 81 TA are already reported in M. tuberculosis, searches for homologues of TA, previously reported in other organisms, uncovered nearly 199 hits that were further screened, as described under “Experimental procedures.” Ultimately, after employing several criteria, five novel TA pairs (Table 1) were recognized that were studied further using a combination of bioinformatics and laboratory investigations. We have already characterized one of these predicted systems as a novel, noncanonical PezAT TA system through experiments (28). Here, we provide detailed analysis for two other predicted TA systems.
Table 1.
List of novel toxins identified in M. tuberculosis H37Rv
9000 queries (that included experimentally characterized and predicted TA) from TADB were queried in the M. tuberculosis genome using TBLASTN. 2000 hits obtained from these searches were subjected to various filters. Each of the 199 hits from this set was manually evaluated to identify five novel cases for further detailed study.
| S. no | Putative toxin | Genomic location of toxin | Putative antitoxin | Genomic location of antitoxin | Query ID | Query source | Fold (HHPred and Phyre2 ) | Potential function |
|---|---|---|---|---|---|---|---|---|
| 1 | Rv2514c | 2,830,877–2,831,338 | Rv2515c | 2,831,344–2,832,591 | TADB_3732229 | C. chlorochromatii also identified as a hit when queried with VAPC1_HAEI6, VAPC1_RICFE, and Y4JK_SINFN | Toxin: VapC templates (4chg and 1w8i); Antitoxin: HTH regulator (3cec) | Putative VapBC |
| 2 | Rv0366c | 444,222–444,815 | Rv0367c | 444,844–445,233 | Pnap_4620 | Polaromonas naphthalenivorans CJ2 plasmid pPNAP04 | Toxin: nucleotide kinase fold; Antitoxin: ParD (2an7, E. coli) | PezAT-like |
| 3 | Rv3641c | 4,079,925–4,080,560 | Rv3642c | 4,080,571–4,080,765 | gi|157148945 | C. koseri | Toxin: VbhT (3shg_A); Antitoxin: VbhA (3shg_B) | Putative FicTA |
| 4 | Rv1128c | 1,251,617–1,252,972 | Rv1129c | 1,253,074–1,254,534 | gi|166364680 | Microcystis aeruginosa | Toxin: endonuclease (4h9d); Antitoxin: HigA (4mct) | Putative VapBC or HigBA |
| 5 | Rv0024 | 28,362–29,707 | Rv0023 | 27,595–28,365 | gi|194289065 | Cupriavidus taiwanensis | Toxin: hydrolase; Antitoxin: HigA (4mct) | Putative toxin is an endopeptidase |
Rv2514c–Rv2515c may function as a novel VapBC in M. tuberculosis
Rv2514c was identified as a homologue of VapC toxins from Chlorobium chlorochromatii and Haemophilus influenzae at a sequence identity of ∼17%. Its gene neighbor, Rv2515c, was predicted to harbor a putative DNA-binding motif and is situated on the same operon. Domain assignments for both putative toxin and antitoxin recognized the PIN domain in Rv2514c, a helix-turn-helix (residues 27–83) and peptidase domain (residue 152–332) in Rv2515c. We first studied the effect of overexpression of putative toxin on the growth of M. smegmatis, using the anhydrotetracycline-based expression system. As shown in Fig. 7, A and B, we observed that overexpression of Rv2514c inhibited the growth of M. smegmatis in a bacteriostatic manner. As expected, no growth inhibition was observed in M. smegmatis harboring pTetR vector control. We also observed that this growth defect was restored by co-expressing Rv2514c along with its putative cognate antitoxin, Rv2515c (Fig. 7). These experiments affirm that Rv2514c–Rv2515c functions as a novel type II TA in M. tuberculosis. Furthermore, based on alignments with VapC toxins of known structure (36, 37, 43), we predicted that Arg-90 and Arg-94 are functionally important (Fig. S13). Indeed, mutation of these residues abrogated the bacteriostatic effect of Rv2514c (Fig. 7), suggesting a role for these residues in Rv2514c-mediated growth inhibition. These investigations point to the detection of a novel TA system, although further experiments are required to (i) understand the regulation of this novel TA system and (ii) investigate its contribution to M. tuberculosis physiology and pathogenesis.
Figure 7.
Rv2514c–Rv2515c and Rv3642c–Rv3641c are novel TA in M. tuberculosis. A shows the growth curves for predicted toxin Rv2514c when expressed alone and in the presence of its potential, cognate antitoxin partner Rv2515c. Graph also shows the growth curves of the bacteria when predicted functional residues in the toxin were mutated. B shows spotting images corresponding to the growth curves. C, model of the predicted TA complex of Rv3641c–Rv3642c showing the putative TA interaction interface. Inset shows the predicted interactions between Arg-155–Asn-24 and Arg-155–Glu-28.
Rv3641c–Rv3642c may function as a FicTA in M. tuberculosis
The presence and biochemical characterization of Fic domain proteins in MtbH37Ra (O06366) with roles in transfer of NMPs from NTPs (ATP, GTP, CTP, and UTP) to specific target proteins have been demonstrated elsewhere (44). These experiments, tailored with structural modeling, highlighted the importance of key residues that are conserved in all FIC domain proteins. In the study here, using the cell filamentation Fic protein from Citrobacter koseri as query (gi|157148945|ref|YP_001456264.1), we identified Rv3641c (4,079,925–4,080,560) as the M. tuberculosis H37Rv homologue of the FicT toxin, at a sequence identity of 34%. Domain assignments of Rv3641c and its gene neighbor Rv3642c (64 residues: 4,080,571–4,080,765), using the Pfam (45) and conserved domain assignments (46), predict the Fic/Doc-like domain for the putative toxin and VbhA-like domain for the putative antitoxin. 3D-fold of VbhTA (a type of FicTA) from Bartonella schoenbuchensis (3shg: E6Z0R3) was predicted with over 95% confidence for Rv3641c. Alignments of Rv3641c–Rv3642c with VbhTA sequences (Fig. S14) show that the signature motif HXFX(D/E)GNGRXXR of FIC domain proteins is observed in Rv3641c. The inhibitory motif (S/T)XXE(G/N) (47), present in FicA, is also observed in Rv3642c suggesting that Rv3641c–Rv3642c might function as a FicTA in M. tuberculosis (details in supporting data S3 and Fig. S14, A and B).
A secretory signal present in the C terminus of VbhT of B. schoenbuchensis is absent in Rv3641c, and similarities are limited to the N-terminal Fic domain. Therefore, we predict that although it is likely to share the 3D-fold with VbhTA, Rv3641c is unlikely to function as VbhTA and is closer to other FicTA proteins (47). Models of the toxin, antitoxin, and their complex (Fig. 7 and Fig. S15, “Experimental procedures,” and supporting data S3) show that the amino acid residues critical for the formation of the complex in the template (Arg-147, VbhT; Ser-20 and Glu-24, VbhA) are conserved (Arg-155, Rv3641c; Asn-24 and Glu 28, Rv3642c) (Fig. 7 and Figs. S14 and S15) in all closely related homologues (Fig. S14). Based on this, we predict conserved hydrogen bond and salt-bridge interactions (Arg-155–Asn-24 and Arg-155–Glu-28) in the predicted toxin/antitoxin interface of Rv3641c and Rv3642c (47). Furthermore, as in the template, Glu-28 in Rv3642c is found to lie on a potential inhibitory motif (αinh), with a role in interfering with ATP binding (47) (details in supporting data S3). As shown in Fig. 7, this residue can form a salt bridge with Arg-155, implying that the mode of toxin neutralization by Rv3642c could be as in the template. Future work on this TA system will be important to characterize its function and targets and confirm its biological role in M. tuberculosis.
Discussion
Paralogues result from expansion of a protein through extensive gene duplication; although some duplicates become pseudogenes due to gene mutations, others are known to acquire new functional roles by undergoing sequence alterations. The large complement of TAs in M. tuberculosis has prompted many studies that have probed the functional and evolutionary basis of the expansion of this gene family (48). Whether these genes were independently acquired through horizontal gene transfer or result from extensive gene amplification of select TAs remains an unanswered question. To address this, we used a novel approach of clustering and identifying closely related paralogues in the currently known complement of M. tuberculosis TA. We demonstrate that the 51 VapBC family of TAs could be grouped into seven distinct sub-clusters with members of each cluster sharing >20% sequence identity with each other. Careful analysis of the alignments both within and across clusters shows that the members of each cluster share similarities at multiple levels. Although the characteristic residues of the PIN domain are well-conserved across the cluster members, sequences within each cluster conserve putative antitoxin and target-binding residues suggesting functional redundancy within each cluster. To demonstrate this, we used the available crystal structures of known VapBC TA sequences to predict the location of potential functional residues in each cluster. Using bacterial overexpression systems, we confirmed that by mutating these conserved residues in VapC13, VapC21, VapC27, VapC28, and VapC44, the growth defect associated with these toxins could be restored. Specifically, these results demonstrate the merit and predictive power of computational clustering of TA sequences in the recognition of functionally important residues. Our findings also support the results of Winther et al. (49), who identified the cellular targets of 12 VapCs through UV cross-linking and deep sequencing. Phylogenetic clustering-based prediction of substrates of select VapCs in their study showed that target RNA-cleavage specificities were conserved in some instances. In concordance with their results, we also observed that some of our paralogues occur in the same phylogenetic subgroups identified by them. Our analysis suggests that similar targets may be recognized by paralogous members, although further experiments are required to validate these observations.
Views on the specificity of interactions in bacterial TA systems have been ambivalent with arguments both in favor and against their specificity (29, 50, 51). Although few experiments have been successful in demonstrating noncognate interactions between toxin and antitoxin (52), many more experiments have shown through co-expression studies on specific VapBC families that such interactions are unlikely to be significant (20). Indeed, the TA systems that did not show cross-talk in other experiments such as VapBC2, VapBC11, VapBC22, and VapBC47 were observed in different paralogous clusters in our analysis as well. In contrast, in sub-cluster 6, which groups VapC4, VapC5, and VapC8, many similarities at the toxin/antitoxin interaction interface have been observed in VapC4 and VapC5 (40). Close examination of alignments within this cluster shows conservation of residues that play key roles in the interaction interface. We anticipate that careful analysis of alignments between paralogous toxins and antitoxins may be useful in predicting potential interaction interfaces between them and in evaluating the possibility of cross-talk between select systems in the future. Studies on co-evolution trends will also be useful to gain insight into the evolution of interaction specificity between cognate partners. Conserved features in various TA pairs can also be employed to identify potential targets for the design of polyspecific inhibitors or activators. Of course, although our approach may help in focusing experiments to assess cross-talk between specific systems, we must bear in mind that the failure to observe it may be due to several reasons, as also reported elsewhere (53, 54). It is interesting to note that most of the members within a given sub-cluster occur at different genomic locations. Such a distribution of paralogous TAs throughout the M. tuberculosis genome may prove advantageous for the organism to efficiently encounter and adapt to stress conditions. A few such as VapBC4, -5, and -8, VapBC17–19, VapBC46, and VapBC 47 are present in proximity within the genome.
Ramage et al. (20) have earlier reported that homologues of the M. tuberculosis are well-conserved in members of the MTBC. Our extension of these searches in the 101 mycobacterial genomes serves to identify two TA modules, Rv0901–Rv0910 and Rv2034–Rv2035, which are universally conserved in many of the mycobacterial species, suggesting that these TA modules were present in mycobacteria before the speciation of the MTBC. Our comprehensive survey also finds that this TA has no homologues in other bacterial and archaeal taxa. On the contrary, VapBC12 and VapBC41 find more hits in other prokaryotic genomes than in the MTBC, suggesting closer evolutionary proximity or recent acquisition from other genomes. Furthermore, this study also finds that Rv2654c–Rv2653c has only one other homologue in M. microti and no hits in other MTBC or other prokaryotic genomes surveyed here. Many of the hits that we identified through the searches reported here are unannotated and remain unverified in the prokaryotic genomes, and this study offers clues to their functional roles. An interesting finding is that most orthologues of the M. tuberculosis TA are found in soil-dwelling, slow-growing pathogens from the same class. Mycobacteria are known to survive for a long time in the environment and can be found in great numbers in rivers, sediments, and in soil. It has been shown that M. tuberculosis remains virulent while in the soil, outside its hosts, for extended periods of time (55). This raises the possibility that many of these TA genes may be transferred from and to other microorganisms residing in similar environments.
One of our objectives was to probe for novel TA in M. tuberculosis. The discovery of a novel TA in M. tuberculosis, Rv3641c–Rv3642c, which can likely adenylate enzymes that control DNA topology leading to reversible growth arrest, is the first report of this TA system in this organism. For another novel TA, Rv2514c–Rv2515c, we have shown signatures of a VapBC-like TA system for which we were able to confirm expression experimentally. For a third case, we have described, for the first time, the presence of a noncanonical PezAT-like TA system in M. tuberculosis (28). In all, these systems add to the list of TA modules that may contribute to its survival in the human host. It must be mentioned here that although nearly 81 TA systems have been predicted in M. tuberculosis, efforts to functionally express and demonstrate their existence has not always been successful due to factors such as high toxicity or the lack of appropriate expression systems. Although our computational analysis has helped us recognize these five systems, future experimental studies targeting these novel TAs will help validate their roles and the predicted interactions between the toxin–antitoxins, as well as explore cross-talk between different structurally related TA systems.
Experimental procedures
Relationships among known M. tuberculosis TA systems: identification of paralogous clusters and generation of sub-cluster alignments
162 sequences of either experimentally characterized or computationally predicted type II TA systems of M. tuberculosis were obtained from the UniProt (Table S1) (56). All sequences were manually verified to remove any potentially incorrect entries (fragmented/truncated sequences). Each sequence was queried against a protein sequence database (comprising all bacterial and archaeal protein sequences) using PSI-BLAST, until convergence, if applicable, or for a maximum of 20 iterations (57). Hits from the M. tuberculosis proteome were extracted at an E-value cutoff of 1e−06. These searches were performed in a large database embedded with sequences of M. tuberculosis proteins to improve confidence in the search results of M. tuberculosis homologues and to minimize the risk of false-positives. Some of the hits were obtained in common for more than one query, suggesting that certain toxins were closer in sequence than others (Table S2). To group them, clusters of homologous toxins were generated using HiFiX (34), at 20% sequence identity and 60% alignment length criteria. HiFiX takes the PSI-BLAST output of an all-versus-all sequence comparison as input and clusters sequences based on single-linkage with alignment coverage constraints. In this study, a threshold of 20% sequence identity was employed because it resolved the VapC toxins into sub-clusters with most connected members (Fig. S2). Because pairwise alignments were used to group the toxins, the extent of sequence divergence within each cluster was determined by averaging all pairwise identity scores of members within each sub-cluster. Average identity scores for each sub-cluster were as follows: sub-cluster 1 (23.8%), sub-cluster 2 (24.4%), sub-cluster 3 (42.3%), sub-cluster 4 (32.5%), sub-cluster 5 (35.2%), and sub-cluster 6 (42.6%). Similarly, VapB antitoxins were also clustered at 20% identity and 60% alignment length criteria. Resulting sub-clusters, in both toxins and antitoxins, were numbered from 1 to 7. Sub-cluster alignments were derived using MAFFT (58). For the alignments that included a toxin/antitoxin of known structure, structure-based multiple sequence alignments were derived using Promals 3D (59). These included sub-clusters 2, 3, and 6 with reference structures available for VapC2, VapC30, and VapC5, respectively (36, 37, 39). When sub-clusters did not possess a member with known structure, secondary structure predictions for every sequence in each cluster were performed using PSI-PRED (60). Also, domain boundaries in each sequence was defined using Superfamily (61). All alignments were visually represented using ESPript (62).
Prediction of putative functional residues
The availability of crystal structures of several VapC toxins from M. tuberculosis is useful to identify residues with importance in substrate recognition, antitoxin binding, and structural roles. Because these VapC toxins are also members of some of the sub-clusters, such residue positions may be obtained from the known crystal structures and mapped to the alignment. If uniformly conserved, they may be predicted to assay similar roles in the uncharacterized members within each cluster. Functional residues of known VapBC TA were obtained from the literature, in sub-clusters 2, 3, and 6, and mapped to the relevant alignments (36, 37, 39). Where this was not immediately available, due to the absence of a reference structural template, charged residues that were uniformly conserved and in the vicinity of conserved acidic residues were identified from the alignments. Also, residues predicted to lie in loops in each of the paralogues and therefore surface-exposed were determined based on PSI-PRED predictions. It must be noted that the alignments involved the full length of the toxin sequences, and each residue position in the alignment was examined systematically. Based on these criteria, few residues were predicted with functional roles, as listed in Table S4, and were mutated to validate their functional relevance.
Bacterial strains, plasmids, and growth conditions
All bacterial strains and plasmids used in the study are listed in Table S11. M. smegmatis cells were grown in Middlebrook MB7H9 medium with shaking at 200 rpm, 37 °C. Spotting or measuring optical densities at 600 nm was used to monitor bacterial growth.
DNA manipulations
The culturing of Escherichia coli and M. smegmatis strains was performed as per standard protocols, as described previously (32). The WT VapC toxins were overexpressed in M. smegmatis using anhydrotetracycline-based expression systems as described previously (32). Site-directed mutagenesis, for creating desired mutations in selected VapC toxins, was performed by overlap PCR as per standard protocols. The ORF for VapC proteins harboring a desired mutation were cloned in the anhydrotetracyline-based inducible vector pTetR (32). Various pTetR constructs were electroporated in mc2155, and transformants were selected on MB7H11 plates supplemented with hygromycin. The ORF encoding Rv2514c was PCR-amplified and cloned into integrative vector pTetR-Int, and the ORF encoding antitoxin Rv2515c was PCR-amplified and cloned into acetamide-inducible vector plam (63). The final construct plam-Rv2515c was transformed in mc2155 harboring an integrative copy of Rv2514c, and transformants were selected on MB7H11 plates supplemented with kanamycin and hygromycin. The sequences of all recombinant constructs were verified by DNA sequencing. The effect of overexpression of toxins either alone or in the presence of antitoxin on the growth of M. smegmatis was determined by either measuring OD600 nm or by spotting assays, as described previously (32).
Comparative modeling of select TA systems for mutational free-energy calculations
Models were derived for selected members of the clusters for which mutation of predicted functional residues was carried out. This included VapC28 (sub-cluster 3), VapC44 and VapC13 (both in sub-cluster 1), and VapC27 (sub-cluster 7). The toxin sequences show between 15 and 42% sequence identity with their templates, identified by fold recognition using Phyre2 (42), and the models were generated by multitemplate modeling, using Modeler version 9.14 (64). In each case, a maximum of three templates was used to model the protein. Template details, prediction scores, and sequence identities with the toxin sequences are listed in Table S12. The templates in all the cases are VapBC systems from either M. tuberculosis or other organisms. All toxin sequences were modeled as dimers. 100 models were generated for every query toxin, and the model with minimum energy (DOPE score) was chosen as the final structure. Because the sequence identity of the templates and toxin targets was not very high, the side-chain positions were further optimized using the side-chain rotamer library from SCWRL4.0 (65). The final models were subjected to quality check using ProQ (66). To determine whether mutation of functional residues could have an impact on the structural stability of the protein, each model was subjected to the AlaScan module of the FoldX package (41). The FoldX force field is empirical in nature with terms for desolvation energies, coulombic interactions, van der Waal's forces, hydrogen bonding, entropic changes, and others. AlaScan module of FoldX mutates every residue to alanine one-by-one and estimates the difference in energy between the “mutant” and the “WT” structure. A value of ΔΔG >1 kcal mol−1 is considered significant to classify a mutation as a destabilizing mutation. Prior to running AlaScan, all the structures were repaired using repairPDB module of FoldX. Protein interactions calculator server, which recognizes various kinds of interactions within a protein or between proteins in a complex, was employed to determine whether the functional residues that we predicted and that were mutated were involved in any dense intra- or inter-chain interactions (67).
Searches for homologues in mycobacteria and other prokaryotic genomes
PSI-BLAST searches were performed for 162 TA sequences against a database of 1672 archaeal and bacterial protein sequences, downloaded from the UniProt proteomes resource (56). In parallel, TBLASTN was used to search 4500 prokaryotic genomes, including 101 mycobacterial genomes, obtained from the NCBI genome database (68), using the same set of queries. These databases include the gene and protein sequences of the MTBC, namely M. tuberculosis, Mycobacterium canetti, M. orygis, M. caprae, M. mungi, M. microti, Mycobacterium africanum, and Mycobacterium bovis. In either search, hits were identified for the queries at E-values lower than 10−6 and a query coverage of >60%. Potential homologues for each of the queries, from the individual mycobacterial species, were extracted using scripts developed for the purpose. The proximity of the identified toxin and antitoxin homologues was carefully analyzed and reported as a TA pair, if they were less than 200 bp apart. This distance cutoff was employed because we observed that for 98% of the hits, the toxin and cognate antitoxin were less than 150 bp and for the remaining 2% either 150–170 or 250 bp as in the VapBC50 and VapBC6 TA systems. Also, search methods such as BLAST perform local alignments and therefore may split an alignment and report it as two separate alignments, both satisfying the search thresholds. Setting a larger cutoff of 200 bp to identify the putative toxin and antitoxin sequences was one approach to overcome the inherent limitation of the search. To avoid false-positives, for the cases where the detected TA pair was observed at a distance >150 bp, we manually examined the region between the putative TA pairs to verify that they are immediate gene neighbors. A histogram of the distances between the detected toxin and antitoxin homologues in mycobacteria and other prokaryotes is available in Fig. S2, D and E.
Visualization of phylogenetic trees and circos plots
A phylogenetic tree for the 2220 genomes, in which homologues of M. tuberculosis TA were found, was derived using phyloT (https://phylot.biobyte.de/).10 Visual inspection was performed using the Interactive Tree of Life online tool (iTOL) (69), to determine the distribution of M. tuberculosis type II TA in the various organisms. Plots showing the number of individual toxins, antitoxins, and TA pairs in the various genomes were included, to annotate the tree using the iTOL tool. The unrooted tree, available in Figs. S9 and S10 has also been made searchable online (https://itol.embl.de/tree/1413912822138041518174018 and https://itol.embl.de/tree/1413912822265121518597108)10 (69).
Leaves are labeled by organism name, colored by class, and contain popup windows that list the data pertaining to bar plots. In Fig. S10, class-specific pie charts on the nodes provide the number of TA pairs in each constituent family (Table S10).
The circos plots were generated using Circos software (70). These plots represent data in a circular layout and are used to represent relationships between organisms and various TA systems. For each circos plot in this study, organisms are grouped distinctly from the listed TA pairs to facilitate interpretation of the figures. Three-letter codes, used to list the organisms, are provided in Table S7. Links are drawn between the TA and an organism only if a homologue is detected for a query TA pair in the organism, through the sequence searches performed here. For example, a line is drawn between VapBC24 and Mxe (Mycobacterium xenopi) to show that similarity searches revealed the presence of a homologue for this TA in this organism (Fig. 5B). Likewise, lines between VapBC35 and various mycobacteria show that more homologues are detected for this TA query pair in a number of organisms (Fig. 5). The absence of a line linking a TA and any of the listed organisms depicts that no homologues were found during the similarity searches for the query TA pair. Because of the sheer scale of the data, these plots may look crowded at first sight, but they are extremely useful in locating TA pairs that find homologues in a maximum number of organisms or no hits at all. Other trends are also relatively easily identified.
Identification of novel TA pairs in M. tuberculosis
The TADB serves as a repository of predicted and experimentally verified TA loci from 2786 prokaryotic species (71). Nearly 10,000 sequences of toxins and antitoxins from the TADB (version 1.1) (9584 representative sequences derived by clustering 11,370 sequences from TADB at 90%) and from UniProt (613; based on keyword searches and cases reported in literature) were queried against the M. tuberculosis genome using TBLASTN. This search attempts to identify homologues for toxins and antitoxins that are previously reported only in other organisms. The searches were complemented with PSI-BLAST searches in the UniProt database, and hits from M. tuberculosis were identified. In both searches, an E-value cutoff of 10−6 and alignment length coverage of 60% of the query length were imposed. Depending on the query, the hit was examined for the presence of a putative toxin or antitoxin either upstream or downstream of the detected hit. To allow for genomic rearrangements in the neighborhood of the putative toxin/antitoxin, our search criteria were relaxed in terms of distance between the two identified genes. However, we confirmed to check that they were immediate gene neighbors and consulted predictions to lie on an operon from OperonDB (72).
Comparative modeling of Rv3641c–Rv3642c
The templates for both Rv3641c (toxin) and antitoxin (Rv3642c) were identified using HHPred (73) and Phyre2 (42). VbhTA (a type of FicTA) of B. schoenbuchensis (3shg: E6Z0R3) was identified as the best template with over 95% confidence. The sequence identity of Rv3641c with the template toxin sequence (3shgA) is 30% and of Rv3642c with the antitoxin sequence (3shgB) is 17%. Because the structures for both toxin and antitoxin in the template are available as a complex, we performed a template-based modeling of the Rv3642c–Rv3641c complex, using Modeler version 9.14 with B. schoenbuchensis VbhTA (PDB code 3shg) as template. Of the 100 complexes generated, the model with the best DOPE score was assessed for quality using ProQ (66). Residues in the signature motifs of the toxin and antitoxin of the template complex structure were mapped to the derived model. Furthermore, residues in the interaction interface of the template were mapped to equivalent residues in the modeled complex.
Author contributions
H. T. data curation; H. T., A. S., S. W., and S. S. formal analysis; H. T. and S. S. investigation; H. T. and S. S. methodology; H. T., R. V., R. S., N. S., and S. S. writing-review and editing; R. V., R. S., and N. S. supervision; R. V. and N. S. funding acquisition; R. S., N. S., and S. S. conceptualization; N. S. project administration; S. S. writing-original draft.
Supplementary Material
Acknowledgments
We thank members of R. V., R. S., and N. S. labs for their inputs. We acknowledge funding for infrastructural support from the following agencies or programs of the Government of India, Ministry of Human Resource Development (MHRD), Department of Science and Technology–Fund for improvement of S&T infrastructure (DST-FIST), University Grants Commission (UGC) Center for Advanced Study, Mathematical Biology Program of the Department of Science and Technology, and the Department of Biotechnology–Indian Institute of Science (DBT-IISc) Partnership Program.
The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S15, Tables S1–S12, and supporting data S1–S3.
J. N. Cortese, unpublished data.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- TA
- toxin–antitoxin
- PIN
- N terminus of PilT
- MTBC
- M. tuberculosis complex
- PDB
- Protein Data Bank
- BLAST
- basic local alignment search tool
- PSI-BLAST
- position-specific iterative basic local alignment search tool
- TADB
- toxin–antitoxin database.
References
- 1. World Health Organization. (2018) WHO Global Tuberculosis Report 2018, World Health Organization, Geneva, Switzerland: 10.1016/j.pharep.2017.02.021 [DOI] [Google Scholar]
- 2. Gerdes K., Christensen S. K., and Løbner-Olesen A. (2005) Prokaryotic toxin–antitoxin stress response loci. Nat. Rev. Microbiol. 3, 371–382 10.1038/nrmicro1147 [DOI] [PubMed] [Google Scholar]
- 3. Magnuson R. D. (2007) Hypothetical functions of toxin–antitoxin systems. J. Bacteriol. 189, 6089–6092 10.1128/JB.00958-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yamaguchi Y., and Inouye M. (2016) Stress and Environmental Regulation of Gene Expression and Adaptation in Bacteria (de Bruijn F. J., ed) pp. 97–107, John Wiley & Sons, Inc., New York [Google Scholar]
- 5. Sala A., Bordes P., and Genevaux P. (2014) Multiple toxin–antitoxin systems in Mycobacterium tuberculosis. Toxins 6, 1002–1020 10.3390/toxins6031002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aakre C. D., Phung T. N., Huang D., and Laub M. T. (2013) A bacterial toxin inhibits DNA replication elongation through a direct interaction with the β sliding clamp. Mol. Cell 52, 617–628 10.1016/j.molcel.2013.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bordes P., Sala A. J., Ayala S., Texier P., Slama N., Cirinesi A. M., Guillet V., Mourey L., and Genevaux P. (2016) Chaperone addiction of toxin–antitoxin systems. Nat. Commun. 7, 13339 10.1038/ncomms13339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wen Y., Behiels E., and Devreese B. (2014) Toxin–antitoxin systems: their role in persistence, biofilm formation, and pathogenicity. Pathog. Dis. 70, 240–249 10.1111/2049-632X.12145 [DOI] [PubMed] [Google Scholar]
- 9. Hayes F., and Van Melderen L. (2011) Toxins-antitoxins: diversity, evolution and function. Crit. Rev. Biochem. Mol. Biol. 46, 386–408 10.3109/10409238.2011.600437 [DOI] [PubMed] [Google Scholar]
- 10. Kȩdzierska B., and Hayes F. (2016) Emerging roles of toxin–antitoxin modules in bacterial pathogenesis. Molecules 21, E790 10.3390/molecules21060790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Prozorov A. A., and Danilenko V. N. (2010) Toxin-antitoxin systems in bacteria: Apoptotic tools or metabolic regulators? Microbiology 79, 129–140 10.1134/S002626171002001320411670 [DOI] [Google Scholar]
- 12. Van Melderen L., and Saavedra De Bast M. (2009) Bacterial toxin–antitoxin systems: more than selfish entities? PLoS Genet. 5, e1000437 10.1371/journal.pgen.1000437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Van Melderen L. (2010) Toxin–antitoxin systems: why so many, what for? Curr. Opin. Microbiol. 13, 781–785 10.1016/j.mib.2010.10.006 [DOI] [PubMed] [Google Scholar]
- 14. Wang X., and Wood T. K. (2011) Toxin–antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl. Environ. Microbiol. 77, 5577–5583 10.1128/AEM.05068-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Coray D. S., Wheeler N. E., Heinemann J. A., and Gardner P. P. (2017) Why so narrow: distribution of anti-sense regulated, type I toxin–antitoxin systems compared with type II and type III systems. RNA Biol. 14, 275–280 10.1080/15476286.2016.1272747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Leplae R., Geeraerts D., Hallez R., Guglielmini J., Drèze P., and Van Melderen L. (2011) Diversity of bacterial type II toxin–antitoxin systems: a comprehensive search and functional analysis of novel families. Nucleic Acids Res. 39, 5513–5525 10.1093/nar/gkr131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Klimina K. M., Kjasova D. K., Poluektova E. U., Krügel H., Leuschner Y., Saluz H.-P., and Danilenko V. N. (2013) Identification and characterization of toxin–antitoxin systems in strains of Lactobacillus rhamnosus isolated from humans. Anaerobe 22, 82–89 10.1016/j.anaerobe.2013.05.007 [DOI] [PubMed] [Google Scholar]
- 18. Averina O., Alekseeva M., Shkoporov A., and Danilenko V. (2015) Functional analysis of the type II toxin–antitoxin systems of the MazEF and RelBE families in Bifidobacterium longum subsp. infantis ATCC 15697. Anaerobe 35, 59–67 10.1016/j.anaerobe.2015.07.007 [DOI] [PubMed] [Google Scholar]
- 19. Georgiades K., and Raoult D. (2011) Genomes of the most dangerous epidemic bacteria have a virulence repertoire characterized by fewer genes but more toxin–antitoxin modules. PLoS ONE 6, e17962 10.1371/journal.pone.0017962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ramage H. R., Connolly L. E., and Cox J. S. (2009) Comprehensive functional analysis of Mycobacterium tuberculosis toxin–antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet. 5, e1000767 10.1371/journal.pgen.1000767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Arcus V. L., Rainey P. B., and Turner S. J. (2005) The PIN-domain toxin–antitoxin array in mycobacteria. Trends Microbiol. 13, 360–365 10.1016/j.tim.2005.06.008 [DOI] [PubMed] [Google Scholar]
- 22. Arcus V. L., McKenzie J. L., Robson J., and Cook G. M. (2011) The PIN-domain ribonucleases and the prokaryotic VapBC toxin–antitoxin array. Protein Eng. Des. Sel. 24, 33–40 10.1093/protein/gzq081 [DOI] [PubMed] [Google Scholar]
- 23. Sevin E. W., and Barloy-Hubler F. (2007) RASTA-Bacteria: a web-based tool for identifying toxin–antitoxin loci in prokaryotes. Genome Biol. 8, R155 10.1186/gb-2007-8-8-r155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McKenzie J. L., Robson J., Berney M., Smith T. C., Ruthe A., Gardner P. P., Arcus V. L., and Cook G. M. (2012) A vapbc toxin–antitoxin module is a posttranscriptional regulator of metabolic flux in mycobacteria. J. Bacteriol. 194, 2189–2204 10.1128/JB.06790-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Winther K. S., and Gerdes K. (2011) Enteric virulence associated protein VapC inhibits translation by cleavage of initiator tRNA. Proc. Natl. Acad. Sci. U.S.A. 108, 7403–7407 10.1073/pnas.1019587108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Winther K. S., Brodersen D. E., Brown A. K., and Gerdes K. (2013) VapC20 of mycobacterium tuberculosis cleaves the sarcin-ricin loop of 23S rRNA. Nat. Commun. 4, 2796 10.1038/ncomms3796 [DOI] [PubMed] [Google Scholar]
- 27. Cruz J. W., Sharp J. D., Hoffer E. D., Maehigashi T., Vvedenskaya I. O., Konkimalla A., Husson R. N., Nickels B. E., Dunham C. M., and Woychik N. A. (2015) Growth-regulating Mycobacterium tuberculosis VapC-mt4 toxin is an isoacceptor-specific tRNase. Nat. Commun. 6, 7480 10.1038/ncomms8480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tandon H., Sharma A., Sandhya S., Srinivasan N., and Singh R. (2019) Mycobacterium tuberculosis Rv0366c–Rv0367c encodes a noncanonical PezAT-like toxin–antitoxin pair. Sci. Rep. 9, 1163 10.1038/s41598-018-37473-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhu L., Sharp J. D., Kobayashi H., Woychik N. A., and Inouye M. (2010) Noncognate Mycobacterium tuberculosis toxin–antitoxins can physically and functionally interact. J. Biol. Chem. 285, 39732–39738 10.1074/jbc.M110.163105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Romero D., and Palacios R. (1997) Gene amplification and genomic plasticity in prokaryotes. Annu. Rev. Genet. 31, 91–111 10.1146/annurev.genet.31.1.91 [DOI] [PubMed] [Google Scholar]
- 31. Gupta A., Venkataraman B., Vasudevan M., and Gopinath Bankar K. (2017) Co-expression network analysis of toxin–antitoxin loci in Mycobacterium tuberculosis reveals key modulators of cellular stress. Sci. Rep. 7, 5868 10.1038/s41598-017-06003-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Agarwal S., Tiwari P., Deep A., Kidwai S., Gupta S., Thakur K. G., and Singh R. (2018) System wide analysis unravels differential regulation and in vivo essentiality of VapBC TA systems from Mycobacterium tuberculosis. J. Infect. Dis. 217, 1809–1820 10.1093/infdis/jiy109 [DOI] [PubMed] [Google Scholar]
- 33. Thomas J. H. (1993) Thinking about genetic redundancy. Trends Genet. 9, 395–399 10.1016/0168-9525(93)90140-D [DOI] [PubMed] [Google Scholar]
- 34. Miele V., Penel S., Daubin V., Picard F., Kahn D., and Duret L. (2012) High-quality sequence clustering guided by network topology and multiple alignment likelihood. Bioinformatics 28, 1078–1085 10.1093/bioinformatics/bts098 [DOI] [PubMed] [Google Scholar]
- 35. Stinear T. P., Seemann T., Harrison P. F., Jenkin G. A., Davies J. K., Johnson P. D., Abdellah Z., Arrowsmith C., Chillingworth T., Churcher C., Clarke K., Cronin A., Davis P., Goodhead I., Holroyd N., et al. (2008) Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res. 18, 729–741 10.1101/gr.075069.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Min A. B., Miallau L., Sawaya M. R., Habel J., Cascio D., and Eisenberg D. (2012) The crystal structure of the Rv03010–Rv0300 VapBC-3 toxin–antitoxin complex from M. tuberculosis reveals a Mg2+ ion in the active site and a putative RNA-binding site. Protein Sci. 21, 1754–1767 10.1002/pro.2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee I.-G., Lee S. J., Chae S., Lee K.-Y., Kim J.-H., and Lee B.-J. (2015) Structural and functional studies of the Mycobacterium tuberculosis VapBC30 toxin–antitoxin system: implications for the design of novel antimicrobial peptides. Nucleic Acids Res. 43, 7624–7637 10.1093/nar/gkv689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jardim P., Santos I. C., Barbosa J. A., de Freitas S. M., and Valadares N. F. (2016) Crystal structure of VapC21 from Mycobacterium tuberculosis at 1.31 Å resolution. Biochem. Biophys. Res. Commun. 478, 1370–1375 10.1016/j.bbrc.2016.08.130 [DOI] [PubMed] [Google Scholar]
- 39. Miallau L., Faller M., Chiang J., Arbing M., Guo F., Cascio D., and Eisenberg D. (2009) Structure and proposed activity of a member of the VapBC family of toxin–antitoxin systems VapBC-5 from Mycobacterium tuberculosis. J. Biol. Chem. 284, 276–283 10.1074/jbc.M805061200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jin G., Pavelka M. S. Jr., and Butler J. S. (2015) Structure–function analysis of VapB4 antitoxin identifies critical features of a minimal VapC4 toxin-binding module. J. Bacteriol. 197, 1197–1207 10.1128/JB.02508-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schymkowitz J., Borg J., Stricher F., Nys R., Rousseau F., and Serrano L. (2005) The FoldX web server: an online force field. Nucleic Acids Res. 33, W382–W388 10.1093/nar/gki387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kelley L. A., Mezulis S., Yates C. M., Wass M. N., and Sternberg M. J. (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Das U., Pogenberg V., Subhramanyam U. K., Wilmanns M., Gourinath S., and Srinivasan A. (2014) Crystal structure of the VapBc-15 complex from Mycobacterium tuberculosis reveals a two-metal ion dependent pin-domain ribonuclease and a variable mode of toxin–antitoxin assembly. J. Struct. Biol. 188, 249–258 10.1016/j.jsb.2014.10.002 [DOI] [PubMed] [Google Scholar]
- 44. Mishra S., Bhagavat R., Chandra N., Vijayarangan N., Rajeswari H., and Ajitkumar P. (2012) Cloning, expression, purification, and biochemical characterisation of the FIC motif containing protein of Mycobacterium tuberculosis. Protein Expr. Purif. 86, 58–67 10.1016/j.pep.2012.08.020 [DOI] [PubMed] [Google Scholar]
- 45. Finn R. D., Coggill P., Eberhardt R. Y., Eddy S. R., Mistry J., Mitchell A. L., Potter S. C., Punta M., Qureshi M., Sangrador-Vegas A., Salazar G. A., Tate J., and Bateman A. (2016) The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285 10.1093/nar/gkv1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Marchler-Bauer A., Derbyshire M. K., Gonzales N. R., Lu S., Chitsaz F., Geer L. Y., Geer R. C., He J., Gwadz M., Hurwitz D. I., Lanczycki C. J., Lu F., Marchler G. H., Song J. S., Thanki N., et al. (2015) CDD: NCBI's conserved domain database. Nucleic Acids Res. 43, D222–D226 10.1093/nar/gku1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Engel P., Goepfert A., Stanger F. V., Harms A., Schmidt A., Schirmer T., and Dehio C. (2012) Adenylylation control by intra-or intermolecular active-site obstruction in Fic proteins. Nature 482, 107–110 10.1038/nature10729 [DOI] [PubMed] [Google Scholar]
- 48. Pandey D. P., and Gerdes K. (2005) Toxin–antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 33, 966–976 10.1093/nar/gki201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Winther K., Tree J. J., Tollervey D., and Gerdes K. (2016) VapCs of Mycobacterium tuberculosis cleave RNAs essential for translation. Nucleic Acids Res. 44, 9860–9871 10.1093/nar/gkw781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fiebig A., Castro Rojas C. M., Siegal-Gaskins D., and Crosson S. (2010) Interaction specificity, toxicity and regulation of a paralogous set of ParE/RelE-family toxin–antitoxin systems. Mol. Microbiol. 77, 236–251 10.1111/j.1365-2958.2010.07207.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yang M., Gao C., Wang Y., Zhang H., and He Z. G. (2010) Characterization of the interaction and cross-regulation of three Mycobacterium tuberculosis RelBE modules. PLoS ONE 5, e10672 10.1371/journal.pone.0010672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wei Y., Li Y., Yang F., Wu Q., Liu D., Li X., Hua H., Liu X., Wang Y., Zheng K., and Tang R. (2017) Physical and functional interplay between MazF1Bif and its noncognate antitoxins from Bifidobacterium longum. Appl. Environ. Microbiol. 83, e03232 10.1128/AEM.03232-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Soo V. W., Cheng H. Y., Kwan B. W., and Wood T. K. (2014) De novo synthesis of a bacterial toxin/antitoxin system. Sci. Rep. 4, 4807 10.1038/srep04807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Aakre C. D., Herrou J., Phung T. N., Perchuk B. S., Crosson S., and Laub M. T. (2015) Evolving new protein–protein interaction specificity through promiscuous intermediates. Cell 163, 594–606 10.1016/j.cell.2015.09.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ghodbane R., Mba Medie F., Lepidi H., Nappez C., and Drancourt M. (2014) Long-term survival of tuberculosis complex mycobacteria in soil. Microbiology 160, 496–501 10.1099/mic.0.073379-0 [DOI] [PubMed] [Google Scholar]
- 56. The UniProt Consortium. (2017) UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45, D158–D169 10.1093/nar/gkw1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., and Lipman D. J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Katoh K., and Standley D. M. (2014) MAFFT: iterative refinement and additional methods. Methods Mol. Biol. 1079, 131–146 10.1007/978-1-62703-646-7_8 [DOI] [PubMed] [Google Scholar]
- 59. Pei J., Kim B. H., and Grishin N. V (2008) PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 36, 2295–2300 10.1093/nar/gkn072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. McGuffin L. J., Bryson K., and Jones D. T. (2000) The PSI-PRED protein structure prediction server. Bioinformatics 16, 404–405 [DOI] [PubMed] [Google Scholar]
- 61. Pandurangan A. P., Stahlhacke J., Oates M. E., Smithers B., and Gough J. (2019) The SUPERFAMILY 2.0 database: a significant proteome update and a new webserver. Nucleic Acids Res. 47, D490–D494 10.1093/nar/gky1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Robert X., and Gouet P. (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 10.1093/nar/gku316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. van Kessel J. C., and Hatfull G. F. (2007) Recombineering in Mycobacterium tuberculosis. Nat. Methods 4, 147–152 10.1038/nmeth996 [DOI] [PubMed] [Google Scholar]
- 64. Sali A., and Blundell T.L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 234, 779–815 10.1006/jmbi.1993.1626 [DOI] [PubMed] [Google Scholar]
- 65. Krivov G. G., Shapovalov M. V., and Dunbrack R. L. (2009) Improved prediction of protein side-chain conformations with SCWRL4. Proteins 77, 778–795 10.1002/prot.22488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wallner B., and Elofsson A. (2003) Can correct protein models be identified? Protein Sci. 12, 1073–1086 10.1110/ps.0236803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tina K. G., Bhadra R., and Srinivasan N. (2007) PIC: protein interactions calculator. Nucleic Acids Res. 35, W473–W476 10.1093/nar/gkm423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. NCBI Resource Coordinators. (2017) Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 45, D12–D17 10.1093/nar/gkw1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Letunic I., and Bork P. (2016) Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245 10.1093/nar/gkw290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S. J., and Marra M. A. (2009) Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645 10.1101/gr.092759.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Shao Y., Harrison E. M., Bi D., Tai C., He X., Ou H. Y., Rajakumar K., and Deng Z. (2011) TADB: a web-based resource for Type 2 toxin–antitoxin loci in bacteria and archaea. Nucleic Acids Res. 39, D606–D611 10.1093/nar/gkq908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Pertea M., Ayanbule K., Smedinghoff M., and Salzberg S. L. (2009) OperonDB: a comprehensive database of predicted operons in microbial genomes. Nucleic Acids Res. 37, D479–D482 10.1093/nar/gkn784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hildebrand A., Remmert M., Biegert A., and Söding J. (2009) Fast and accurate automatic structure prediction with HHpred. Proteins 77, 128–132 10.1002/prot.22499 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.