Abstract
Bacterial cold shock proteins (CSPs) function as RNA chaperones. To assess CSP's roles in the intracellular human pathogen Salmonella Typhimurium, we analyzed their expression in varied stress conditions. We found that cold shock protein E (cspE or STM14_0732) is up-regulated during bile salt-induced stress and that an S. Typhimurium strain lacking cspE (ΔcspE) displays dose-dependent sensitivity to bile salts, specifically to deoxycholate. We also found that an uncharacterized gene, yciF (STM14_2092), is up-regulated in response to bile stress in WT but not in the ΔcspE strain. Complementation with WT CspE, but not with a F30V CspE variant, abrogated the bile sensitivity of ΔcspE as did multicopy overexpression of yciF. Northern blotting experiments with rifampicin disclosed that the regulation of yciF expression is, most likely, due to the RNA-stabilizing activity of CspE. Importantly, electrophoretic mobility shift assays indicated that purified CspE, but not the F30V variant, directly binds yciF mRNA. We also observed that the extra-cytoplasmic stress-response (ESR) pathway is augmented in the bile-treated ΔcspE strain, as judged by induction of RpoE regulon genes (rpoE, degP, and rybB) and downstream ESR genes (hfq, rne, and PNPase). Moreover, the transcript levels of the porin genes, ompD, ompF, and ompC, were higher in bile salts-stressed ΔcspE and correlated with higher intracellular accumulation of the fluorescent DNA stain bisBenzimide H 33258, indicating greater cell permeability. In conclusion, our study has identified YciF, a CspE target involved in the regulation of porins and in countering bile stress in S. Typhimurium.
Keywords: permeability, Salmonella enterica, stress response, RNA–protein interaction, bile acid, cell stress, Cold shock protein, porins, RNA chaperone, YciF
Introduction
Microbes face myriad stresses due to changes in the environment. Consequently, gene regulation plays important roles during their adaptation to different environmental milieu. For example, most bacteria react to a sudden decrease in temperature, i.e. a shift from 30 to 10 °C, by a cold-shock response (1). This temperature adaptation causes major physiological changes in the cell, including changes in composition and organization of lipids, leading to alterations in membrane fluidity, increase in superhelicity, compaction of DNA by the introduction of negative supercoils, and overall decreased metabolic rate (2). A family of genes involved in this response are the cold shock proteins (CSPs)3 consisting of 67–73 amino acids that are evolutionarily conserved across all three domains of life and are implicated to function as RNA chaperones (3).
The cold shock domain–containing proteins are characterized structurally and functionally in several organisms, e.g. Escherichia coli (4) and Bacillus subtilis (5). Various binding preferences have been reported for CSPs, namely for poly(dT) and poly(dC) ssDNAs (6), for the cold shock promoter sequence ATTGG (7, 8), or for AT-rich regions (9); however, in vivo evidence for substrate specificity is lacking. Numerous amino acid residues have been identified to be important for nucleotide binding, which largely center around two motifs termed the ribonucleoprotein (RNP) sites, RNP1 (KGFGF) and RNP2 (VFVH) (10). Not all CSPs are cold-inducible, and some are reported to fulfill noncold stress-related functions. E. coli harbors nine CspA paralogs (CspA–I), of which CspA, CspB, CspG, and CspI are cold-inducible (11). cspA is also constitutively expressed at “normal” growth temperatures (37 °C), as are cspC and cspE (12), whereas cspD is induced during nutrient starvation (13). The major CSP from E. coli, CspA, has been described as a multifunctional nucleic acid–binding protein and essential for mRNA stabilization after temperature downshift (14). In Listeria monocytogenes, CspA enables hemolysis by regulating the production of the pore-forming cytolysin listeriolysin (15). A cold shock–induced RNA-binding protein CspR plays a post-transcriptional function in the Gram-positive opportunistic human pathogen Enterococcus faecalis and has a role in the virulence, organ colonization, and its survival in macrophages (16).
The three-dimensional structure of CSPs is fairly similar, e.g. the E. coli–encoded CspE superimposes appropriately with the Salmonella Typhimurium CspE. It includes five anti-parallel β-strands forming a classic OB fold with the characteristic RNP1 and RNP2 motifs conserved on the nucleic acid–interacting surfaces (8). CspE is characterized to function in varied conditions. The nucleic acid melting ability and transcription anti-terminator activity of CspE is critical for growth at low temperature (17). It functions as a “housekeeping RNA chaperone” under general stress conditions (18), enhances translation of several mRNAs (9), and is important in imparting camphor resistance (19). It interferes with bacteriophage λ Q-mediated transcription anti-termination (20), regulation of the poly(A)-mediated 3′–5′-exonuclease activity of polynucleotide phosphorylase, and cleavage and poly(A) tail removal by RNase E (21).
The intracellular pathogen, S. Typhimurium, causes typhoid-like infection in mice and is a well-established model to study the roles of proteins during stress and infection. In high-income countries, nontyphoidal salmonellae mainly cause a self-limiting enterocolitis in immunocompetent individuals (22). Up to 5% of patients develop secondary bacteremia, but attributable mortality is low (1–5%) (23). In sub-Saharan Africa, bacteremia is commonly presented by invasive nontyphoidal salmonellae, in both children and adults, especially in regions of HIV and malaria prevalence. Despite anti-microbial treatments, fatality rates are 22–47% in African adults and children (24). Another troubling observation is the rise of multidrug-resistant strains in S. Typhimurium. In fact, DT104, is one of the leading causes of animal and human salmonellosis (25). DT104 is resistant to the commonly used antibiotics, e.g. ampicillin, chloramphenicol, streptomycin, sulfonamides, and tetracycline (26).These aspects reinforce the notion that it is important to study all aspects of investigations with respect to the biology of Salmonella.
In S. Typhimurium, six CSPs have been identified: CspA–E and CspH (8). Of these, cold inducibility of CspA, CspB, and CspH has been reported (3, 27–29), although their functions have not yet been elucidated. A recent study (30) in S. Typhimurium SL1344 identified a plethora of downstream targets and physiological functions that CspE and CspC may regulate. In this study, we focus on the role of cold shock protein E (STM14_0732) from S. Typhimurium 14028s in imparting bile resistance. Bile is synthesized by the liver, stored in the gallbladder, and plays a major role in dissolution and absorption of fats. In addition, bile demonstrates antimicrobial activity by solubilizing membrane lipids, disrupting membranes, damaging proteins, DNA, and RNA, and leading to the death of bacteria (31, 32). Commensal bacteria are also able to modify bile salts both in the small and large intestine (31). This balance between the bile salt abundance and modification is beneficial to both the host and microbiome, especially in the cases of invading microbes that can be competed out by the commensals. There is an inverse correlation between bile concentration and bacterial colonization; areas of high bile concentration (gallbladder) have low bacterial communities. A striking feature of the chronic residence of Salmonella is the host distal ileum, an area of low bile concentration (33), and biliary tract or gallbladder, areas of high bile concentration (34). To escape the high concentrations of bile, Salmonella either forms biofilms on the gallstone surface (35) or invades the gallbladder epithelium (36). Importantly, the colonization of the gallbladder by Salmonella is thought to be essential for the chronic asymptomatic carrier state, which is observed in 3–5% of infected individuals (34). These aspects underscore the importance of bile in the biology of Salmonella. Here, we show the mechanistic regulation of bile resistance controlled by S. Typhimurium–encoded CspE. Furthermore, the identification of YciF (STM14_2092) as a CspE-regulated protein and its role in regulating the outer membrane porins and cell permeability during bile stress are the highlights of this study.
Results
Transcript levels of S. Typhimurium–encoded cspE increases in the presence of bile salts
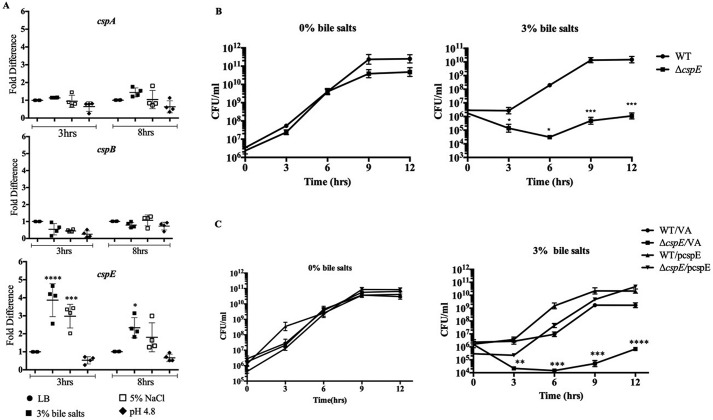
Salmonella faces a plethora of stresses within the host: acidic pH in the stomach, detergent-like stress from bile, and osmotic stress in the intestine. We attempted to replicate these stresses in vitro and study the involvement of CSPs in these stresses. The WT strain was grown in a rich media (LB) and subjected to several stress-inducing agents: bile stress (3% bile salts (w/v)), osmotic stress (5% NaCl (w/v)), and acidic pH stress (LB at pH 4.8). Transcript levels of four cold shock genes cspA, cspB, cspC, and cspE were analyzed at the 3rd and 8th h of growth. cspA and cspB did not show any quantifiable changes. cspC transcripts were not detected (Fig. S1A). As a control, we showed that the primers used were able to amplify the genomic copy of cspC (Fig. S1B). However, only cspE seemed consistently and significantly up-regulated at the 3rd and 8th h of growth, under the bile salt stress (Fig. 1A). The observations indicated that bile stress induced the expression of cspE.
Figure 1.
S. Typhimurium–encoded cspE is up-regulated during bile stress and is essential for resistance. A, transcript levels of cspA, cspB, and cspE were determined using qRT-PCR for the WT strain, under the indicated stress conditions, at the 3rd and 8th h of growth. The relative quantities of transcripts were calculated against the mean of the reference gene (rrlC). Transcript levels of target genes in the control set (WT cells grown in LB), at the 3rd and 8th h time point, were normalized to 1, and all other samples were calculated as fold-change to this reference value. B, kinetic growth analysis of WT and ΔcspE strains were analyzed in terms of CFU/ml over a period of 12 h of growth, in the absence (0%) or presence (3%) of bile salts. C, kinetic growth analysis, calculated in terms of CFU/ml WT/VA-, ΔcspE/VA-, and cspE-complemented (WT/pcspE and ΔcspE/pcspE) was studied for 12 h in the absence (0%) and presence (3%) of bile salts. Data are presented as mean ± S.E. and representative of three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
cspE deletion strain is highly susceptible to bile stress
To better understand its functional role, a single gene deletion of cspE was generated (ΔcspE). The WT and ΔcspE strains did not show any growth differences in LB or other stress conditions studied (Fig. S2). However, with increasing concentrations of bile salts, i.e. 3% (w/v) and above, ΔcspE showed significant growth reduction (Fig. S3A). The growth of WT and ΔcspE in the absence (0%) and presence (3%) of bile salts was monitored in a kinetic manner. The ΔcspE strain showed growth retardation in colony-forming units (CFU), compared with the WT, in the presence of 3% bile salts (Fig. 1B). The specificity was also attributed to the deoxycholate component of bile salts (Fig. S3B). The functional involvement of cspE in imparting bile resistance was determined by using an expression construct of cspE (pcspE–cspE was under the control of the native promoter in the pRS424 plasmid). As seen in Fig. 1C, the bile-sensitive phenotype of ΔcspE was rescued by the complementation of cspE but not by the vector-alone transformed cells. Also, this bile resistance regulation by CspE was not a general CSP-dependent phenotype observed, because multicopy overexpression of another CSP, namely CspA did not show any phenotype suppression effect in ΔcspE (Fig. S4). In S. Typhimurium 14028s, among the CSPs studied, cspE is essential for bile resistance.
Deletion of cspC does not affect bile resistance of S. Typhimurium 14028s
A previous study using S. Typhimurium SL1344 has shown that both CspC and CspE are responsible for resistance to multiple stress, e.g. bile, polymyxin B sulfate, and H2O2 (30). However, in the WT strain (S. Typhimurium 14028s) used in this study, we were unable to detect any cspC transcripts with either qRT or semi qRT-PCR (Fig. S1A). Deletion strains of cspE, cspC, and cspEcspC were generated in the background of both 14028s and SL1344 WT strains. We observed that the ΔcspE in SL1344 background was marginally more resistant to bile stress than that in the 14028s background. However, in our system single deletion of cspC (ΔcspC) did not show any significant effect on bile resistance in either strain (Fig. S5A). Accordingly, the double-deletion strain ΔcspEΔcspC showed a cspE-dependent effect.
To further confirm the role of cspC, a cloning and overexpression approach similar to that of cspE was utilized. The cspC gene, unlike cspE, occurs in an operon with an uncharacterized upstream gene, yobF. The full-length cspC operon (pcspC++) was cloned from SL1344 strain (30) in the pRS424 vector. Functional complementation assays were performed with the cspC construct, but no phenotypic rescue was seen with respect to the bile sensitivity of ΔcspE (Fig. S5B). Therefore, we conclude that in the S. Typhimurium strain used in this study, CspE is mainly responsible for combating bile stress.
CspE regulates an uncharacterized protein, YciF, during bile salt stress
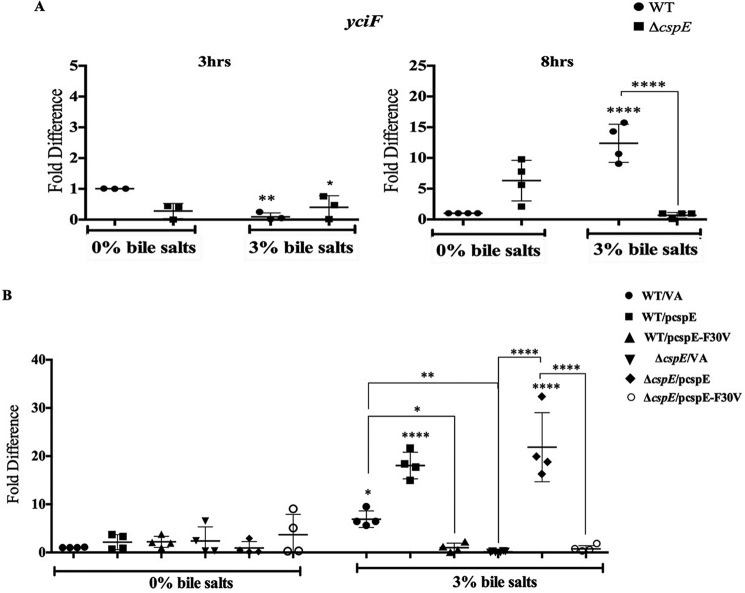
It was important to address the possible mediators involved in the CspE-mediated rescue of bile stress. An earlier study of a bile-modulated proteome of S. Typhimurium had identified several novel proteins. Of these, an uncharacterized protein YciF was up-regulated, whereas another protein PagC was down-regulated, by bile (37). In this study, the qRT-PCR data demonstrated a concomitant down-regulation of pagC in WT and ΔcspE with bile salts stress (Fig. S6). Notably, the bile-dependent up-regulation of yciF was observed in the WT strain with bile treatment but was absent in the ΔcspE strain (Fig. 2A). Consequently, we hypothesized that yciF might be a downstream target of CspE.
Figure 2.
S. Typhimurium–encoded cspE positively regulates yciF. A, transcript levels of yciF was determined using qRT-PCR for the WT and ΔcspE strains, in the absence (0%) and presence (3%) of bile salts, at the 3rd and 8th h of growth. In all panels, values are normalized by those obtained for the WT strain grown in 0% bile salts, at the indicated time point. B, transcript levels of endogenous yciF in the WT/VA-, ΔcspE/VA-, and cspE-complemented (WT/pcspE and ΔcspE/pcspE) and cspE-F30V-complemented (WT/pcspE-F30V and ΔcspE/pcspE-F30V) strains were determined using qRT-PCR, in the absence (0%) and presence (3%) of bile salts, at the 8th h of growth. In all panels, values are normalized by those obtained for the WT strain grown in 0% bile salts, at the indicated time point. Data are presented as mean ± S.E. and representative of three independent experiments. *, p < 0.05; **, p < 0.01; ****, p < 0.0001.
Overexpression of YciF suppresses the bile sensitivity of ΔcspE
To delineate the role of YciF, the ΔyciF and ΔcspEΔyciF strains were generated, and the growth kinetics of WT, ΔcspE, ΔyciF, and ΔcspEΔyciF in the absence (0%) or presence (3%) of bile salts was performed. In the absence of bile salts, the growth kinetics was indistinguishable for all four strains (Fig. S7). In the presence of 3% bile salts, the WT and ΔyciF strains showed similar growth while the ΔcspE and ΔcspEΔyciF strains showed severe growth attenuation with an approximate 6-h growth delay. To elaborate on the importance of yciF, we utilized the yciF overexpression system (WT/pyciF and ΔcspE/pyciF). Upon yciF overexpression, phenotype suppression was observed in the bile salts–treated ΔcspE (Table 1). These observations indicated two major aspects of YciF. First, the genetic data with the four strains demonstrated that YciF may not be the sole downstream player in the bile regulation pathway of CspE. Second, increases in YciF levels are capable of imparting bile resistance even in the absence of CspE. Most likely, CspE regulates multiple proteins, and this study has identified one target, i.e. YciF, which is important to counter bile stress.
Table 1.
Overexpression of yciF suppresses the bile sensitivity of ΔcspE
Quantification of kinetic growth determined in terms of log10 (CFU/ml) values of WT and ΔcspE with the following vectors: VA, pcspE, pcspE-F30V, and pyciF grown with and without 3% bile salts. Values are represented as mean ± S.E. Statistical analysis was performed using two-way ANOVA, and the different comparisons have been represented using the following symbols: #, p < 0.001. Comparison of each strain treated with 3% bile salts with respect to WT/VA grown in 3% bile salts is shown. a, p < 0.0001; b, p < 0.01. Comparison of each strain grown in 3% bile salts with respect to ΔcspE/VA grown in 3% bile salts is shown.
| Time | WT/VA | WT/pcspE | WT/pcspE-F30V | WT/pyciF | ΔcspE/VA | ΔcspE/pcspE | ΔcspE/pcspE-F30V | ΔcspE/pyciF |
|---|---|---|---|---|---|---|---|---|
| h | ||||||||
| 0% bile salts | ||||||||
| 0 | 5.72 ± 0.59 | 5.52 ± 0.48 | 6.25 ± 0.07 | 5.51 ± 0.53 | 5.83 ± 0.28 | 5.54 ± 0.11 | 6.35 ± 0.07 | 5.33 ± 0.29 |
| 6 | 9.35 ± 0.18 | 9.48 ± 0.17 | 8.68 ± 0.38 | 9.42 ± 0.13 | 9.65 ± 0.11 | 9.57 ± 0.08 | 8.63 ± 0.22 | 9.51 ± 0.06 |
| 12 | 11.14 ± 0.44 | 11.48 ± 0.15 | 10.55 ± 0.13 | 10.72 ± 0.15 | 10.58 ± 0.02 | 11.36 ± 0.42 | 10.47 ± 0.16 | 10.81 ± 0.42 |
| 3% bile salts | ||||||||
| 0 | 5.69 ± 0.64 | 4.59 ± 0.15 | 6.47 ± 0.12 | 5.1 ± 0.52 | 4.06 ± 0.82 | 5.15 ± 0.44 | 4.01 ± 0.33 | 5.73 ± 0.23 |
| 6 | 8.52 ± 0.23 | 8.80 ± 0.26 | 8.37 ± 0.18 | 7.79 ± 0.51 | 4.59 ± 0.73# | 8.50 ± 0.414a | 5.1 ± 0.34# | 7.61 ± 0.2b |
| 12 | 10.55 ± 0.24 | 11 ± 0.21 | 10.56 ± 0.31 | 10.04 ± 0.5 | 5.71 ± 0.34# | 10.71 ± 0.72a | 5.68 ± 0.47# | 11.31 ± 0.26a |
CspE increases the stability of yciF mRNA
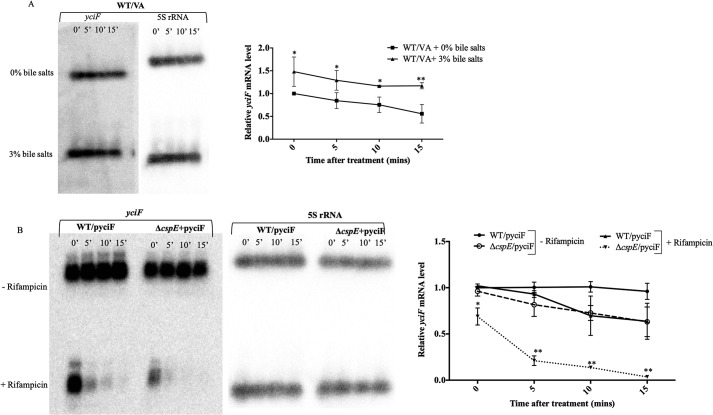
To understand the exact mechanism of YciF regulation by CspE and to distinguish between transcriptional regulation and mRNA stability, we utilized a rifampicin-based approach (38). Overexpression of yciF (under the trc promoter in the pTrc99A plasmid) was utilized. Initial experiments were performed with qRT-PCR, which revealed a reduced level of yciF mRNA in ΔcspE irrespective of rifampicin (Fig. S8). Further confirmation was obtained from Northern hybridization experiments, which demonstrated that in the absence of cspE the cells harbored lesser amounts of yciF mRNA despite using an overexpression system. Furthermore, in the presence of rifampicin, i.e. in the absence of any active transcription, there was hastened decay of the yciF mRNA. It is unlikely that CspE regulates the transcription of yciF. Despite the overexpression of yciF from a heterologous promoter, there is a significantly lesser amount of yciF transcript in the ΔcspE strain (ΔcspE/pyciF), compared with the WT, even in the absence of rifampicin (Fig. 3B). Most likely, CspE regulates YciF by mRNA stabilization and preventing its degradation.
Figure 3.
S. Typhimurium–encoded CspE increases the stability of yciF mRNA. The yciF mRNA stability was determined in terms of the relative amounts of its transcript levels. A, relative mRNA levels of yciF were determined upon bile treatment in the WT/VA strain, by Northern blotting and quantitation. B, mRNA levels of yciF were determined with or without transcription inhibition by rifampicin. The mRNA levels were quantitated relative to the amounts at 0-min post-addition of rifampicin, in the yciF overexpressing strains (WT/pyciF and ΔcspE/pyciF), using 500 μg/ml rifampicin, added at the end of the 8th of growth. Data are presented as mean ± S.E. and are representative of three independent experiments. *, p < 0.05; **, p < 0.01.
YciF regulation is imparted through the function of Phe-30 residue in CspE during bile salts stress
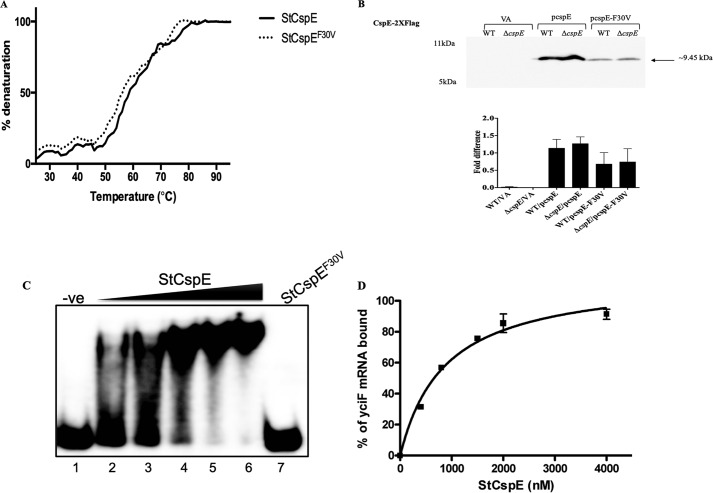
To better understand the regulation of YciF by CspE, we utilized the cspE complementation system (pcspE) along with previously known data about CSP functional mutants. Mutations in residues in the RNP motifs have been reported to abrogate nucleic acid melting functions (39). Mutation studies of the B. subtilis CspB had identified the F30A mutation in the RNP2 motif to prevent ssDNA binding (10). This residue appeared to be conserved in CspE across several species in eubacteria and archaebacteria (Fig. S9A). To identify whether the same would play a role in the S. Typhimurium–encoded CspE function, we generated an F30V mutation (Fig. S9B). We verified the effect of this mutation on the protein stability both in vitro and in vivo. In vitro stability of the protein was determined by quantitating thermal stability of the proteins from 25 to 95 °C and following the denaturation kinetics at 219 nm wavelength (40). There appeared to be negligible change in the melting temperature (Tm ∼53 °C) of CspE upon F30V mutation (Fig. 4A). For in vivo stability analysis, Western blotting and quantitation were performed on the steady-state levels of FLAG-tagged CspE and its F30V mutant. The CspEF30V was expressed but in lesser amounts, compared with the WT CspE, in both the WT and ΔcspE strains (Fig. 4B). Although there were sufficient amounts of CspEF30V present after 8 h of growth, it was unable to rescue the bile sensitivity of ΔcspE. This experiment further validates the essential role of the interplay of CspE and YciF in bile resistance (Table 1).
Figure 4.
S. Typhimurium–encoded CspE binds to yciF mRNA, and the Phe-30 residue is essential for this role. A, thermal stability curve of purified StCspE and StCspEF30V proteins obtained using CD spectroscopy at 219-nm wavelength. B, qualitative and quantitative analysis of in vivo stability of 2× FLAG-tagged StCspE and StCspEF30V obtained using Western blotting with antibody against the FLAG tag. C, binding of StCSpE and StCspEF30V to full-length yciF mRNA. Lane 1, mRNA alone; lanes 2–6, increasing concentration of StCspE; lane 7, StCspEF30V. The filled triangle represents increasing concentrations of purified protein. D, graphical representation of complex formation by StCspE, as a function of protein concentration. Data are presented as mean ± S.E. and representative of three independent experiments.
To obtain validation of the physical binding of purified CspE to a nucleic acid substrate, we performed electrophoretic mobility shift assay (EMSA) using a 60-mer nonspecific ssDNA substrate. The StCspE binds with a high affinity (kd 836 ± 0.0902 nm) to the oligonucleotide, although the StCspEF30V showed no binding even at 10 μm protein concentration (Fig. S9C). qRT-PCR revealed the induction of yciF transcripts upon complementation of WT cspE, but not the F30V mutant of CspE (pcspE-F30V), in the bile salts–treated ΔcspE (Fig. 2B).
We further validated the direct binding of CspE to yciF mRNA using the in vitro-transcribed full-length mRNA as a substrate in EMSA. StCspE showed robust binding to the yciF mRNA with a kd of 626 ± 0.021 nm. The StCspEF30V, however, showed no binding even at a concentration of 10 μm (Fig. 4, C and D). These data demonstrated that CspE directly binds to the yciF mRNA through the Phe-30 residue in RNP2 that plays an important role, effectuating its nucleic acid binding property.
Bile salts stress triggers the envelope stress response (ESR) in S. Typhimurium
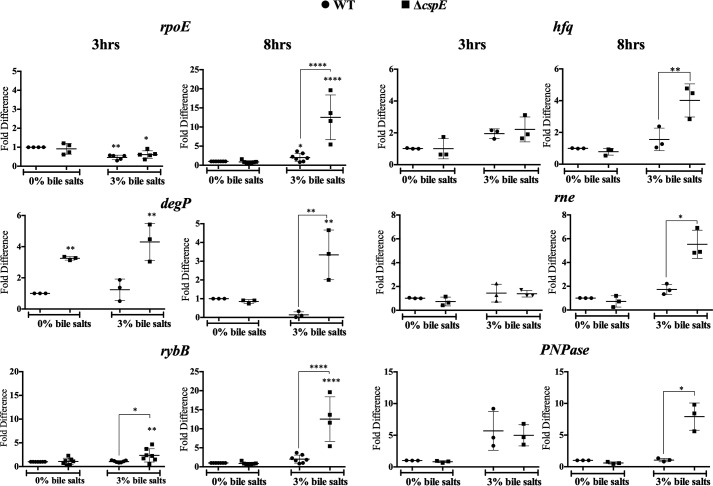
To gain a better understanding of the mechanisms involved in the bile sensitivity of ΔcspE, the transcript levels of several master regulators, such as dps, uspA, or rpoS, were assayed (data not shown). Only rpoE was significantly and kinetically induced in a bile salts stress scenario, in both the WT and ΔcspE strains (Fig. 5); however, the levels were much greater in ΔcspE. Upon membrane damage, the bacterial ESR is triggered, mediated by the alternative σ factor RpoE (41). Subsequently, the transcript levels of the downstream players in the ESR pathway, namely degP, rybB, etc., were significantly induced in the bile salts–treated ΔcspE strain, at 8 h of growth (Fig. 5). Downstream factors that are not classically defined as part of the ESR regulon, namely hfq, rne, and PNPase, but are essential in the appropriate outcome of the ESR pathway were also assayed. mRNA levels of these genes also appeared up-regulated much more in the bile salts–treated ΔcspE strain. These observations indicated that not only the ESR but downstream players of the pathway were also up-regulated to combat possible membrane damage occurring in the ΔcspE strain upon exposure to bile salts. Overall, this indicated a greater insult to membrane integrity of the ΔcspE strain.
Figure 5.
ESR pathway is greatly induced in the ΔcspE strain, upon bile stress. Transcript levels of the ESR components, degP, rpoE, rybB, hfq, rne, and PNPase were determined using qRT-PCR for the WT and ΔcspE strains in the absence (0%) or presence (3%) of bile salts at the 3rd and 8th h of growth. In all panels, values are normalized by those obtained for the WT strain grown in 0% bile salts, at the indicated time point. Data are presented as mean ± S.E. and representative of three independent experiments. *, p < 0.05; **, p < 0.01; ****, p < 0.0001.
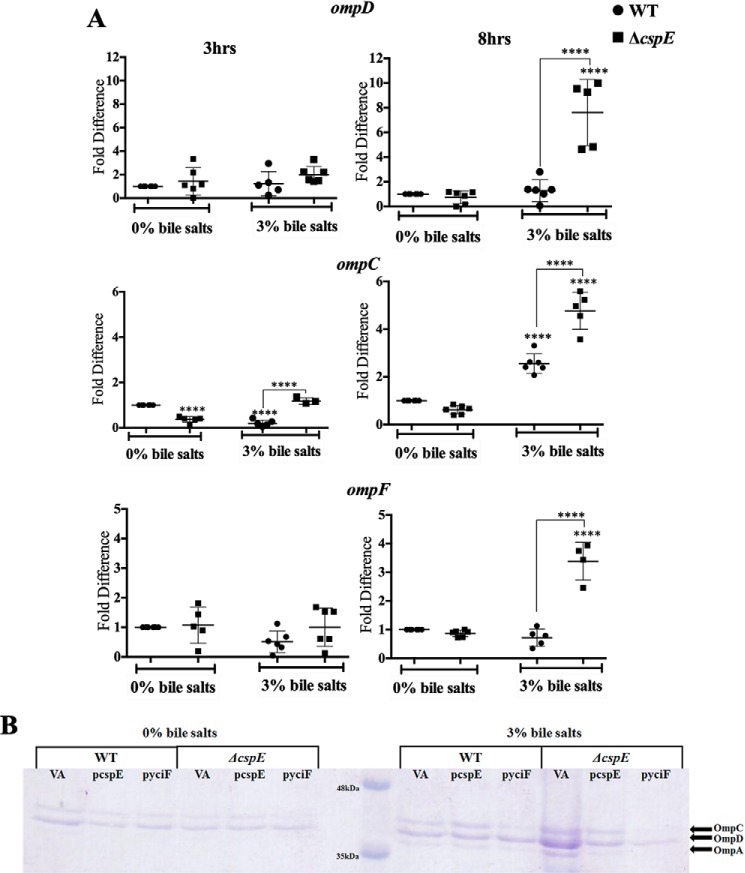
ΔcspE harbors elevated outer membrane protein (OMP)/porin amounts
Next, the expression of genes involved in influx and efflux that are likely to be affected during ESR were studied. Reduced expression of efflux pumps has been reported to lead to bile sensitivity. Consequently, a strain lacking acrAB has been shown to be highly sensitive to bile salts (33). The major efflux pump genes, acrA, acrB, and tolC, transcript showed a similar up-regulation upon bile salts stress in WT and ΔcspE strains (Fig. S11). This indicates that the efflux mechanism was functional and likely to be similar in both the strains. The ESR generally culminates in the degradation of the OMP transcripts, leading to reduction of OMP protein amounts and lowering cell permeability (42). However, the bile salts–treated ΔcspE displayed significantly higher levels of the porin transcripts, namely ompD, ompC, and ompF (Fig. 6A). Concurrently, the qualitative estimation of porins revealed a similar result (Fig. 6B). The major OMPs were identified using trypsin digestion followed by MS. Upon cspE complementation and yciF-mediated phenotype suppression, there was a lesser induction of the ESR pathway components, rpoE and rybB (Fig. S12), and the porin transcripts were significantly lower during bile stress (Fig. 6B). These experiments demonstrated a clear link between CspE and porin mRNA amounts during bile stress.
Figure 6.
Bile salts treated ΔcspE exhibits higher OMP amounts, which is lowered upon cspE complementation or yciF overexpression. A, transcript levels of outer membrane proteins (ompD, ompF, and ompC). B, 12.5% SDS-PAGE analysis of isolated OMPs. Data are presented as mean ± S.E. and representative of three independent experiments. **** p < 0.0001.
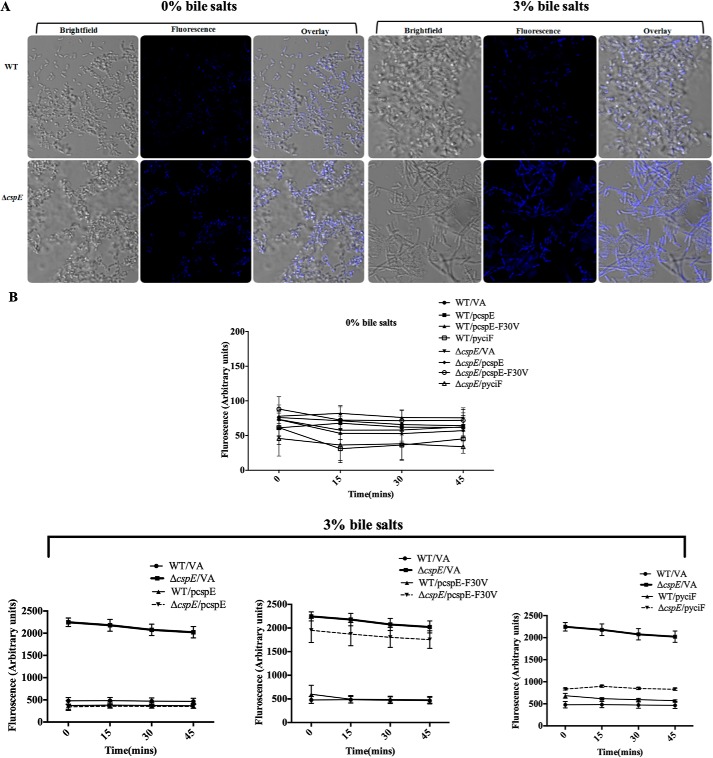
Bile salts–treated ΔcspE manifests increased permeability, which is regulated by CspE and YciF
Higher levels of porins would make the cell more porous, thereby allowing increased entry of damaging agents. This aspect was functionally tested by studying the intracellular accumulation of the DNA-staining dye bisBenzimide H 33258 (43). The accumulation of the dye was greater in the bile salts–treated ΔcspE compared with WT and untreated controls (Fig. 7A). Correlating with earlier data, cspE complementation and yciF-mediated phenotype suppression was able to reduce the permeability significantly, whereas the CspEF30V mutant was not effective (Fig. 7B). These data suggest that CspE and YciF are likely to be involved in the OMP mRNA degradation steps during the ESR pathway.
Figure 7.
ΔcspE exhibits increased permeability upon bile salts stress, which is rescued upon cspE complementation, but not by the F30V mutant of CspE. A, representative fluorescence image 45 min after addition of bisBenzimide H 33258 in WT and ΔcspE, without (0%) or with (3%) pretreatment with bile salts for 5 h. B, quantification of bisBenzimide H 33258 accumulation in WT/VA-, ΔcspE/VA-, cspE-complemented strains (WT/pcspE and ΔcspE/pcspE) and cspE-F30V-complemented strains (WT/pcspE-F30V and ΔcspE/pcspE-F30V), and yciF overexpressing strains (WT/pyciF and ΔcspE/pyciF) without (0%) or with (3%) pretreatment with bile salts for 5 h. Data are presented as mean ± S.E. and representative of three independent experiments.
Discussion
Genetic and biochemical studies have identified several factors that determine bile resistance in enteric bacteria: lipopolysaccharide (44); enterobacterial common antigen (45); efflux pumps (46); regulatory genes such as phoPQ (47), toxR--toxT (48), and marAB (49); and porins (50). In this study, we demonstrate the importance of a cold shock protein, CspE, in imparting bile resistance in S. Typhimurium 14028s. Compared with physiologically relevant stress conditions such as acidic pH and high-salt concentrations, cspE was moderately up-regulated in a bile salts environment (Fig. 1A) and was functionally important for resistance to bile salts (Fig. 1, B and C, and Fig. S3A), specifically to the bile component deoxycholate (Fig. S3B). Moreover, this regulation is specific to CspE, because an overexpression of another cold shock protein (CspA) did not show a functional rescue (Fig. S4). A previous study had shown that both cspC and cspE were required to counter several stresses in the SL1344 strain (30). Several investigations have reported strain-specific bile tolerance, which cannot be extended as a generalized behavior of the species (32). Most likely strain differences exist, and this study demonstrates that CspE plays a major role in countering bile stress in S. Typhimurium. In addition, we identified the F30V variant in S. Typhimurium CspE (11, 41) to be important for substrate binding and bile resistance (Fig. 4, Fig. S9, B and C, and Table 1).
It was important to identify CspE-regulated proteins that play important roles during bile stress. Increased transcript levels of yciF in a bile salts-supplemented milieu were detected (Fig. 3A) in a CspE-dependent manner (Fig. 2A).The functional characterization of YciF in this study was conducted in two ways: genetic deletion of yciF and multicopy overexpression. The single gene deletion of yciF did not exemplify any effect in bile salts stress, and neither was there an added effect of the yciF deletion in the cspE deletion background (Fig. S7). If yciF was the only downstream effector of CspE, its gene deletion should have presented a phenotype. On the contrary, multicopy overexpression of yciF suppressed the bile sensitivity of ΔcspE (Table 1). Also, this phenotypic suppression by yciF was not due to alteration of plasmid copy number of the yciF harboring pTrc99A vector in WT and ΔcspE (plasmid copy numbers were same in both strains, ∼900 copies). This indicated that there are multiple pathways that CspE regulates, one of which might be through the regulation of YciF. Consequently, shifting the equilibrium by overexpressing YciF facilitates suppression of bile sensitivity in the ΔcspE strain.
YciF is a hypothetical protein (51), belonging to the yciGFE–katN (52) operon, and was first identified as a member of the RpoS regulon in Salmonella (53). The functions of yciG (53) and yciF (52) are unascertained, although yciE encodes an acid-shock protein and katN codes for a nonheme catalase (52). The structure of E. coli YciF reveals a dimeric organization in solution and is structurally similar to the di-iron–binding proteins ruberythrin and sulerythrin (54). Notably, CspE is widely distributed and present in diverse bacteria, including Enterobacteriaceae, Archaea, and Firmicutes (Fig. S9A). In contrast, a global alignment revealed a restricted presence of YciF in only six prokaryotic genera. Of these, four belong to family Enterobacteriaceae (Salmonella, Escherichia, Citrobacter, and Klebsiella) and two belong to the nonenterobacteriales (Achromobacter and Vibrio). Hence, this regulatory pathway could be very specific to those genera that encounter high amounts of bile salts. The CspEF30V did not show any effect on the regulation of yciF mRNA levels (Fig. 2B). Finally, the F30V-complemented ΔcspE did not show any phenotypic rescue in bile sensitivity unlike the WT-complemented ΔcspE (Table 1). Overall, it appears that in S. Typhimurium CspE is a nodal regulator of multiple pathways of bile resistance, and YciF is a downstream player in one such pathway. Further studies are likely to identify other targets of CspE that play important roles during various conditions.
CspE is important for mRNA binding and stabilizing a plethora of genes (30). The transcription inhibitor, rifampicin, was used in Northern hybridization experiments to understand the mechanisms by which CspE regulates YciF. In the absence of rifampicin, the ΔcspE strain harbored lesser yciF transcripts compared with the WT. This reduction in transcript levels was exaggerated in the presence of rifampicin (upon transcription inhibition), and the kinetics of this degradation was faster in ΔcspE as compared with the WT (Fig. 3B). It is possible that deletion of CspE leads to increased nonspecific activity of RNases, leading to major down-regulation of several transcripts. To address this issue, qRT-PCR and Northern blotting experiments showed that acrB was up-regulated with bile in the WT strain. However, no difference was observed in the mRNA stability in the presence or absence of CspE (Fig. S10). This was unlike the results obtained with yciF (Fig. 3) and demonstrates that CspE does not regulate global mRNA stability.
Additionally, structured RNA has been reported to be degraded by PNPase and RNase E, the universal degraders of structured RNA in vivo (41, 55). In the present context, it is possible that CspE increases the half-life of the yciF mRNA by binding to it, unwinding its secondary structure, and preventing subsequent degradation. Furthermore, it is unlikely that CspE expends the YciF regulatory role through its anti-transcription terminator property: the yciF mRNA does not harbor any Rho-independent transcription terminators. The software ARNold (http://rna.igmors.u-psud.fr/toolbox/arnold/)4 is specific for identification of Rho-independent transcription terminators in mRNA (56, 57). The software was unable to identify any possible Rho-independent transcription terminators either in the 5′ end of the yciF mRNA or even in the upstream region to the transcription start site or anywhere else in the entire length of the mRNA. Using EMSA with the full-length yciF mRNA, we were able to show a direct, cooperative, and robust binding of CspE to the mRNA mediated by the Phe-30 residue (Fig. 4C). Overall, the data indicate that CspE directly binds to and stabilizes the mRNA of yciF thereby preventing its degradation in a bile salts stress condition.
Bile exhibits multifaceted deleterious effects on bacteria (32). LPS is known to be important for resistance to bile (44). We isolated LPS as described by Hitchcock and Brown (58) and checked for the components on a 15% SDS-PAGE. However, there was no qualitative difference in LPS in the strains lacking cspE (data not shown). We assayed for several other membrane-associated parameters that would physically regulate the bile tolerance, and we detected transcript level alterations of the extra-cytoplasmic σ factor RpoE (Fig. 5). Any insult to the membrane integrity activates the ESR cascade involving the extra cytoplasmic σ factor RpoE, which plays fundamental roles in bacterial virulence and survival. RpoE remains in an inactive membrane-bound state in cells. The induction of the ESR leads to controlled proteolysis and release of the active RpoE into the cytosol (59). The cytosolic RpoE governs expression of >80 transcription units in E. coli (60) and Salmonella (61). Most genes of the RpoE core regulon act to synthesize and assemble lipopolysaccharides and OMPs to maintain envelope homeostasis. Activation of the ESR pathway results in the rapid down-regulation of major OMP mRNAs involving multiple mechanisms. First, the major periplasmic protease DegP gets activated upon a stress to the membrane (accumulation of misfolded OMPs), and this triggers other periplasmic proteases such as RseP and DegS to cleave and release active RpoE from the membrane-sequestered ensemble of RseA–RpoE (62). Active RpoE then enables transcription of rpoE-controlled major small noncoding RNA (sRNA), rybB, which binds to the 5′ end of its target mRNA. Second, Hfq binds to the sRNAs and stabilizes their interaction with the target mRNA. Third, RNase E and PNPase act to degrade the target mRNAs, thereby preventing further translation and accumulation of unfolded OMPs (41). Our results show that in ΔcspE upon bile salts treatment all the conventional RpoE-mediated ESR pathway components (rpoE, degP, and rybB) and the downstream nonconventional players (hfq, rne, and PNPase) are up-regulated (Fig. 5).
To better understand the reasons for the perturbation of membrane homeostasis and induction of ESR, the involvement of other membrane-related factors, the status of the major efflux pump components (acrB, acrA, and tolC), and porins were studied. There was an induction in the mRNA levels of all the above genes (46), but a significant difference was observed only in porin genes between the bile-treated WT and ΔcspE (Fig. 6 and Fig. S11). We observed greater amounts of ompC mRNA but no major changes in the levels of ompF upon bile treatment in the WT strain. This is in congruence with reports that state the smaller pore OmpC is favored more than the larger pore OmpF in the presence of bile salts (50, 63). The mRNA levels of ompD, ompC, and ompF were higher in the ΔcspE strain than the WT upon bile treatment (Fig. 6A). In addition, OmpD was majorly detected in ΔcspE with bile stress (Fig. 6B). This observation is especially important because OmpD is the major porin of S. Typhimurium and amounts to almost 1% of the total cell proteome and half of the porins (64). Also, ompD and the surrounding genomic region is absent in Salmonella typhi and marks one of the major differences between typhi and the other Salmonella serovars (65), making it a potential serovar-specific drug target. Importantly, rybB is reported to target ompD, ompC, and ompF for degradation (41). In fact, the binding of these sRNAs to the 5′ end of the target mRNAs leads to their degradation by multiple mechanisms, e.g. recruitment of RNase E degradosome machinery (41, 61). In fact, the interactome of CspE, as determined by STRING (66), revealed that the foremost and confirmed interacting partners of CspE are RNA helicases, some of which are involved in the RNA degradation pathway, e.g. DeaD, RhlB, and DbpA (Fig. S13). Qualitative and quantitative evaluation revealed that the bile salts–treated Salmonella had more intracellular levels of bisBenzimide H 33258 compared with the untreated controls (Fig. 7). Most likely, the higher levels of porins in bile-treated ΔcspE rendered it susceptible to a greater influx of bile components. This compromised ESR status was rescued in terms of increased rpoE and rybB transcripts and consecutively reduced levels of the porins upon complementation with cspE or multicopy overexpression of yciF (Fig. 6B and Fig. S12). This observation was further confirmed by the rescue in permeability by multicopy yciF overexpression and WT cspE complementation but not by the F30V mutant of cspE (Fig. 7B). Most likely, CspE and its mediators, e.g. YciF, are involved in the pathway of porin mRNA decay (42, 61), resulting in lower OMPs and reducing permeability upon bile stress (Fig. 8). Therefore, the vast excess of bile accumulation occurs due to dysregulated amounts of OMPs, leading to lower survival of cells in the absence of CspE. Further studies need to be performed to address the precise mechanisms that are involved YciF-mediated bile resistance.
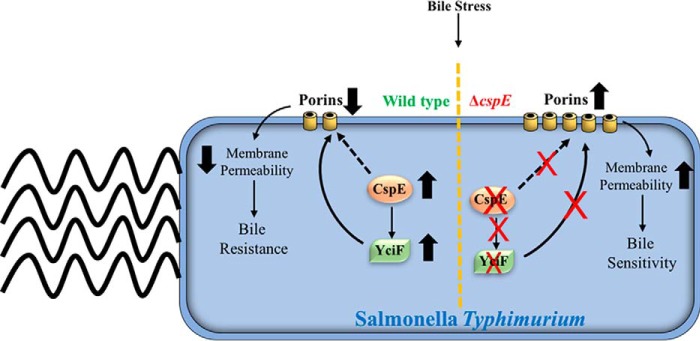
Figure 8.
Graphical representation of the CspE- and YciF-mediated down-regulation of porins to impart bile resistance in S. Typhimurium. Bile stress induces the up-regulation of cspE and the ESR pathway. CspE through multiple pathways regulates bile resistance, one of which involves YciF. CspE increases yciF mRNA stability, thereby enabling translation and further functions of YciF. Complementation of cspE or overexpression of yciF suppresses the bile sensitivity of the ΔcspE strain by establishing the appropriate culmination of the ESR pathway, with respect to porin degradation.
There is a need for identification of novel targets and mechanisms for combating bile resistance and genes involved in virulence/carrier status in Salmonella. Two points need to be highlighted. First, in a global expression analysis study (SalCom–Salmonella Compendium V1.0), among Csps, only CspE is majorly induced during bile stress (67). Second, several bile mutants, e.g. phoPQ and acrB etc., are highly sensitive to bile at low amounts (e.g. 1%) (33, 47). However, higher amounts of bile (3% and higher) were required for the increased sensitivity of the ΔcspE strain. The major thrust of the study was to identify the interplay of CspE and YciF and their effective role in regulating bile resistance. This study has identified and shed light on the involvement of CspE, which acts as the master regulator in bile resistance in S. Typhimurium. The strength of the study is that it combines genetic and biochemical approaches to establish the downstream players and the physiological mechanisms regulated by CspE. In addition, this is the first report of functional and mechanistic detailing of yciF. As part of the study, we uncovered the possible physiological mechanism that these two proteins may be regulating OMP porin degradation. Further studies are required to identify additional novel targets of CspE and the detailed mechanisms of porin regulation by CspE and YciF in regulating bile resistance.
Experimental procedures
Bacterial strains and growth conditions
The bacterial strains and plasmids used in this study are listed in Table S1. All cultures were grown in Luria-Bertani (LB) medium consisting of 10 g/liter tryptone (HiMedia Laboratories, Mumbai, India), 10 g/liter NaCl (Merck, Darmstadt, Germany), and 5 g/liter yeast extract (HiMedia Laboratories) at 37 °C, except for strains containing pKD46, which were grown at 30 °C with constant shaking at 160 rpm. Single-colony cultures grown for 8 h served as pre-inoculum cultures for all experiments. Antibiotics were used at the following concentrations: ampicillin, 100 μg/ml (HiMedia Laboratories); chloramphenicol, 30 μg/ml (HiMedia Laboratories); and kanamycin, 50 μg/ml (Sigma). Arabinose (HiMedia Laboratories) was used at 40 mm for induction of Red recombinase by pKD46.
Chemicals
Bile salts (Sigma) was used at concentrations of 1–5% (w/v). Sodium deoxycholate and cholate were procured from Sigma.
Generation of single- and double-gene deletion strains
To construct the single gene deletion strains, a one-step gene disruption strategy was employed (68). S. Typhimurium 14028s (WT) was used as the parent strain for all experiments, unless otherwise mentioned. Briefly, for construction of ΔcspE, primers listed in Table S2 (Sigma, Bangalore, India) were designed for amplification of the kan from pKD4 having 40-nucleotide flanking regions of cspE. The resulting PCR product was purified and electroporated into WT cells harboring pKD46, which expresses λ Red recombinase. A similar strategy was followed for the other gene deletions. The double gene deletion strain was generated by amplifying the region flanking the gene from its single-knockout strain and electroporating the amplicon into ΔcspE harboring pKD46 (Table S1). All gene deletions were confirmed by PCR amplification using primers designed to anneal ∼100 bp upstream and downstream of the gene (Table S2). Notably, the antibiotic resistance cassettes were removed by pCP20 transformation in the mutant strains used.
Cloning of genes for trans-complementation
The WT 14028s genomic DNA was used as the template for the PCR amplification of cspE, yciF, cspA, and cspC, using specific primers (Table S3) and Phusion DNA polymerase (New England Biolabs). cspE and its promoter were targeted for cloning between the SpeI and XhoI sites in the pRS424 plasmid, whereas yciF and cspA were cloned between the EcoRI and BamHI sites in the pTrc99A plasmid. For cspC, the entire operon comprising its native promoter (pcspC++) were cloned from the SL1344 strain. Positive clones were confirmed by sequencing (Aggrigenome, India). The positive clones (pcspE, pyciF, pcspA, and pcspC++–SL1344) and control vector pRS424 (VA) were then transformed into S. Typhimurium WT and ΔcspE by electroporation, to generate the following strains: WT/VA, ΔcspE/VA, WT/pcspE, ΔcspE/pcspE, WT/pyciF, ΔcspE/pyciF, WT/pcspA, ΔcspE/pcspA, WT/pcspC++–SL1344, and ΔcspE/pcspC++–SL1344.
Construction of F30V mutant of CspE
For mutagenesis, plasmid pcspE, which contains the structural gene of cspE under the control of its original promoter, was used. Single amino acid substitution was carried out by QuikChange site-directed mutagenesis, using high-fidelity Pfu Turbo DNA polymerase (Stratagene). The following pair of oligonucleotides was used for mutagenesis: FP- 5′-GGCAGCAAAGACGTGGCTGTACACTTCTCTGC; RP-5′-GCAGAGAAGTGTACAGCCACGTCTTTGCTGCC. The oligonucleotides were obtained from Sigma, Bangalore, India, and confirmed by sequencing.
Stress assays
Experiments were performed using overnight-grown cultures, using ∼5 × 106 CFU/ml for indicated stress conditions. For the dose assays, normalized cultures along with stress-inducing compounds were grown in a 96-well cell culture plate (SPL Life Sciences, Korea) for 8–9 h. A at 600 nm was measured using a Microplate Reader (Tecan, Männedorf, Switzerland). For kinetic assays, the Bioscreen-C automated growth curve analysis system (Bioscreen, Helsinki, Finland) was used over a period of 16 h, and A at 600 nm was measured every 1 h. For CFU experiments, cultures were collected every 3 h, and appropriate dilutions were plated on LB agar plates and incubated overnight at 37 °C.
RNA extraction
For total RNA preparation, bacterial cultures grown to the mentioned time points, and ∼2 A (600 nm) cells were mixed with 0.2 volumes of STOP solution (95% ethanol, 5% phenol), and RNA was extracted using the TRIzol reagent (Invitrogen) as per the manufacturer's instructions.
Northern hybridization
Rifampicin (final concentration 500 μg/ml) was added to bacterial liquid cultures (growth along-with 3% bile salts), to stop transcription, at 8 h of growth. Cells were collected at the indicated time points, and RNA was extracted as mentioned. 1% formaldehyde/TBE–agarose gel was run at 40 V to resolve the total extracted RNA. The RNA was then transferred onto a NytranTM membrane at 5 V for 3 h (Bio-Rad) and fixed by UV cross-linking at 120 mJ/cm2 (CL1000-UV products). The membranes were blocked using pre-hybridization buffer containing yeast total RNA and Denhardt's solution (1% BSA, 1% Ficoll, 1% polyvinylpyrrolidone 40). The Northern blot analysis was performed using 5′-32P-end–labeled DNA oligomers (69). For yciF, three independent oligomers from different regions of the gene were used (Y1, CAGTTTAATGCCAGATTCAG; Y2, ATTTCGATTTGCGTTGAGCAC; and Y3, CATTGGACGTGGCTCTGGCAAG). acrB was used as the negative control and two oligomers were used as probes (A1, AAGTCCCGCCTGGTCAATCAACTCGAAGTC; A2, CTTCACGAACGGCGTGGTGTC). 5S rRNA was used as the loading control, and a single oligomer (5S-CTACGGCGTTTCACTTCTGAGTTC) was used as probe. The blots were initially probed for yciF, exposed to phosphor-imager screen (GE Healthcare), and analyzed on BioImage Analyzer (FLA5100, Fuji Film). The yciF probes were then stripped and re-blotted for acrB and then 5S rRNA. The successive procedure was the same as mentioned. Bands were measured using ImageJ (version 1.51j8) and plotted using GraphPad Prism (version 5.0).
Expression and purification of StCspE and StCspEF30V
StCspE and StCspEF30V were amplified using the following primers: FP, 5′-CGCGGATCCATGTCTAAGATTAAAGGTAACG; RP, 5′-CGCGTCGACTCAGTGGTGGTGGTGGTGGTGCAGAGCAGTTACGTTTGCAGCGG. The PCR products were cloned between the BamHI and SalI sites in the pRSFDuet-1 plasmid. The His-tagged proteins were overexpressed in the E. coli BL21(DE3) strain harboring plasmid constructs. E. coli cells were grown at 180 rpm in LB broth, supplemented with 50 μg/ml kanamycin, at 37 °C to an A600 of 0.5. Subsequently, the culture was incubated for 1 h at 4 °C. StCspE and StCspEF30V expression was initiated by the addition of isopropyl 1-thio-β-d-galactopyranoside to a final concentration of 0.1 mm, followed by incubation for an additional 12 h at 18 °C. Cells were collected by centrifugation for 10 min at 6000 rpm and 4 °C and resuspended in buffer A (50 mm Tris-HCl (pH 8), 100 mm NaCl, 1 mm EDTA, 5 mm β-mercaptoethanol, and 10% glycerol). Bacteria were then disrupted by sonication, and the cell debris was removed by centrifugation in a Beckman Ti-45 rotor at 30,000 rpm for 1 h at 4 °C. The supernatant was applied to 5 ml of a HiTrap column (GE Healthcare) pre-equilibrated with buffer A. The column was washed with 10 bed volumes of buffer B (50 mm Tris-HCl (pH 8.0), 100 mm NaCl, 1 mm EDTA, 10–20 mm imidazole, 5 mm β-mercaptoethanol, and 10% glycerol). The bound proteins were eluted with a linear gradient of imidazole (30–200 mm) in buffer A. All the fractions were analyzed on an SDS-polyacrylamide gel. The fractions containing the StCspE and StCspEF30V were pooled and dialyzed against buffer C (50 mm Tris-HCl (pH 8.0), 100 mm NaCl, 1 mm EDTA, 5 mm β-mercaptoethanol, and 10% glycerol). The proteins were further dialyzed against buffer D (50 mm Tris-HCl (pH 8.0), 100 mm NaCl, 1 mm EDTA, 5 mm β-mercaptoethanol, and 50% glycerol). The final purified protein was run on a SDS-PAGE and checked for purity, and the identity was confirmed by mass spectrometry. The purified proteins were then stored at −80 °C.
Thermal stability determination by circular dichroism
Purified enzymes (∼500 μm protein) in 400 μl of 20 mm phosphate buffer (pH 8.0) were used, and ellipticity was monitored from 200 to 250 nm in a 0.2-cm path length cuvette with a bandwidth of 1 nm and response time of 2 s at 20 °C. The thermal denaturation temperature (Tm) was calculated as reported previously (70).
In vitro transcription of full-length yciF mRNA and radiolabeling of Northern blotting and EMSA substrates
yciF was cloned between the BamHI and SalI sites in pRSFDuet-1 plasmid using primers listed in Table S3. T7 in vitro transcriptions were carried out using the MEGAscript T7 kit (Ambion). In vitro transcribed mRNA was quality checked by loading the sample on a 5% polyacrylamide, 7 m urea gel, and 5′-end–labeled with γ-32P. The probes for Northern blotting were radiolabeled at the 5′ end by using [γ-32P]ATP and T4 polynucleotide kinase (Thermo Fisher Scientific) as per the manufacturer's protocol. The DNA substrates were purified using the QIAquick nucleotide removal kit (Qiagen). Oligonucleotide probes for Northern hybridization were labeled by the same method.
EMSA
EMSAs were conducted as described previously (71). yciF mRNA binding reaction mixtures contained 1.25 mm Tris-HCl (pH 8.0), 0.05 mm DTT, 0.05 mm KCl, 0.2 mg/ml BSA, 2 mm vanadyl ribonucleoside complex, 2 nm 32P-labeled mRNA substrate in a volume of 20 μl made up with protein storage buffer. 0.2, 1.6, 3, and 10 μm StCspE was used and 10 μm StCspEF30V. Reaction mixtures were incubated at 37 °C for 5 min, and reactions were terminated by the addition of a loading dye (0.1% (w/v) bromphenol blue and xylene cyanol in 20% glycerol). Samples were resolved on a 6% native PAGE in a 0.25× TAE buffer at 80 V and 4 °C. For ssDNA binding, reaction mixtures contained 40 mm Tris-HCl (pH 8.6), 100 mm NaCl, 12% glycerol, 4 mm EDTA, 1 nm 32P-labeled substrate ssDNA and 0.2, 1.6, 3, 10 μm StCspE and StCspEF30V. Reaction mixtures were incubated at 37 °C for 20 min, and reactions were terminated by the addition of a loading dye (0.1% (w/v) bromphenol blue and xylene cyanol in 20% glycerol). Samples were resolved on a 10% native PAGE in a 0.5× TAE buffer at 80 V and 4 °C. Gels were dried, and the bands were visualized using a Fuji FLA-9000 phosphorimager. The band intensities were quantified in a UVItech gel documentation system using UVI-Band Map software (version 97.04) and plotted using GraphPad Prism (version 5.0).
Gene expression analysis by qRT-PCR
Total extracted RNA was treated with the Turbo DNA-free kit (Life Technologies, Inc.).The RNA integrity was analyzed by electrophoresis on a 1.5% agarose gel, and concentration was estimated using Nano-Drop (Thermo Fisher Scientific). 1 μg of DNase-treated RNA was reverse-transcribed using the iScript cDNA synthesis kit (Bio-Rad). The primers (Sigma, Bangalore, India) used for qRT-PCR are listed in Table S4. The expression level of each gene of interest was calculated as the average of three independent cDNA samples. Each cDNA sample and each gene were performed in triplicate. The cycling conditions were as follows: 95 °C for 5 min, followed by 40 cycles of 95 °C for 20 s, 57 °C for 10 s, and 72 °C for 20 s. At the end of the final cycle, the amplification specificity and absence of primer dimers were calculated by the melt curve acquired by 81 cycles of heating the PCR products from 55 to 95 °C for 20 s, with a 0.5 °C increase per cycle (CFX Connect, Bio-Rad). The relative quantities of transcripts were determined using the standard curve method and normalized against the mean of the reference gene (rrlC-23S rRNA). The WT cells grown in LB without any treatment at the early time point (3 h) was normalized to 1, and all other samples were calculated as fold-change to this reference value.
Western blotting
Cells were grown for 8 h without bile. Approximately 2 A (600 nm) cells were taken for lysis and subsequently run on a 12% SDS-PAGE. Western blots were prepared by electroblotting SDS-polyacrylamide gels onto polyvinylidene difluoride membranes, probed with 1:5000 mouse anti-FLAG primary antibody (E-bioscience), and incubated at 4 °C overnight. Prior to antibody addition, the membranes were blocked for 2 h at 25 °C with 5% skim milk in TBS (50 mm Tris-HCl (pH 8.0), 150 mm NaCl, and 0.1% Tween 20). The membranes were washed with TBS for 1 min and then incubated with the primary antibody for 12 h at 4 °C (Sigma). After washing with TBS five times for 15 min each, the membranes were incubated with the 1:10,000 horseradish peroxidase–conjugated anti-mouse secondary antibody (Sigma) for 3 h at 4 °C (70). Finally, the blots were developed using chemiluminescent substrates for horseradish peroxidase (Millipore) and imaged with ChemiDoc ImageQuant (GE LAS 4000). Bands were quantified using ImageJ (version 1.51j8) and plotted using GraphPad Prism (version 5.0).
bisBenzimide H 33258 accumulation assay
Briefly, cultures grown overnight were used to inoculate fresh medium with or without 3% bile salts for 5 h at 37 °C. Bacterial cells were collected by centrifugation at 4000 × g, washed twice with PBS, and resuspended in 1 ml of PBS. The A at 600 nm was normalized to 0.1 and 180 μl was transferred to wells (n = 8; 1 strain per column) of a 96-well plate (flat-bottomed, black, Greiner Bio-one, Kremsmünster, Austria). Every plate contained eight technical replicates (i.e. one column) of PBS alone and heat-inactivated WT (10 min, 90 °C). Replicates (i.e. one column) were analyzed for every strain and growth condition. The plate was transferred to the Microplate Reader and incubated at 37 °C, and 20 μl of 25 μm bisBenzimide H 33258 (Sigma) was added to each well using a multichannel pipette to attain a final concentration of 2.5 μm. Fluorescence was read from the top of the wells using excitation and emission filters of 355 and 460 nm, respectively, with 25 flashes/well; readings were taken for 45 cycles with a 75-s delay between cycles and a 75% gain (43). Raw fluorescence values were analyzed using Excel (Microsoft) that included subtraction of appropriate control blanks from each value of the well of the column, and each experiment was repeated three times.
Imaging of bisBenzimide H 33258-stained Salmonella
Samples were taken as replicates from the bisBenzimide H 33258 accumulation assay and processed for imaging. Briefly, ∼20 μl of the stained samples were collected at the 15-, 30-, and 45-min time points, after addition of H 33258. The samples were fixed with 4% paraformaldehyde (Sigma) for 30 min at room temperature, washed twice with PBS, and then added onto clean coverslips. The samples were allowed to dry at room temperature for 30 min. The coverslips were then mounted on a glass slide with 3–5 μl of mounting medium containing 1% 1,4-diazabicyclo(2.2.2)octane (DABCO) (Sigma) in 1× PBS. Images were acquired with the Zeiss LSM880 with Airy Scan (Carl Zeiss, Oberkochen, Germany) at ×63 magnification (IISc Confocal Imaging Facility).
Outer membrane protein isolation, purification, and quantitation
OMPs were isolated from S. Typhimurium strains grown in LB with or without 3% bile salts (72). Briefly, cells were harvested in the late-log phase (12 h) of growth and washed twice with 1× PBS. Approximately 4 A (600 nm) cells were used for the extraction. OMP concentrations were determined by the Bradford's assay, using BSA as standard. Equal volume of resuspended solution was loaded analyzed by a 12.5% SDS-PAGE and visualized by staining with Coomassie Brilliant Blue (Sigma).
Statistical analysis
All data were analyzed using the GraphPad Prism (Version 6.0c). For analyzing bacterial growth, one-way ANOVA was performed; for comparison of steady-state RT-PCR quantification, one-way ANOVA was performed. The statistical significance of differences in the accumulation of H33258 was determined using two-way ANOVA, with each strain compared against appropriate controls such as the parent WT grown in LB medium alone.
Author contributions
S. R. validation; S. R. and M. D. investigation; S. R. and R. D. C. methodology; S. R. writing-original draft; S. R. and D. N. writing-review and editing; D. N. conceptualization; D. N. resources; D. N. formal analysis; D. N. supervision; D. N. funding acquisition; D. N. project administration.
Supplementary Material
Acknowledgments
We thank Umesh Varshney, Riyaz Shah, D. N. Rao, Utpal Nath, and Raghavan Varadarajan for facilitation of some studies. We appreciate the valuable comments and interest on this study by Sandeepa Eswarappa and Amit Singh. We thank Ayub Qadri (NII, New Delhi) for the generous gift of the SL1344 strain. The support of the Divisional Imaging facility, IISc, and all members of the Nandi laboratory is gratefully acknowledged. In addition, the infrastructural support from DBT-IISc program, DST-FIST and UGC CAS/SAP, is appreciated.
This work was supported in part by Grant 37(1670)16/EMR-II from the Council of Scientific and Industrial Research (CSIR) (to D. N.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S13, Tables S1–S4, supporting Methods, and supporting Refs. 1–7.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- CSP
- cold shock protein
- qRT
- quantitative RT
- ESR
- extracytoplasmic stress response
- OMP
- outer membrane protein
- ssDNA
- single-stranded DNA
- ANOVA
- analysis of variance
- CFU
- colony-forming unit
- RNP
- ribonucleoprotein
- PNPase
- polynucleotide phosphorylase.
References
- 1. Graumann P., and Marahiel M. A. (1996) Some like it cold: response of microorganisms to cold shock. Arch. Microbiol. 166, 293–300 10.1007/s002030050386 [DOI] [PubMed] [Google Scholar]
- 2. Phadtare S., Alsina J., and Inouye M. (1999) Cold-shock response and cold-shock proteins. Curr. Opin. Microbiol. 2, 175–180 10.1016/S1369-5274(99)80031-9 [DOI] [PubMed] [Google Scholar]
- 3. Horton A. J., Hak K. M., Steffan R. J., Foster J. W., and Bej A. K. (2000) Adaptive response to cold temperatures and characterization of cspA in Salmonella typhimurium LT2. Antonie Van Leeuwenhoek 77, 13–20 10.1023/A:1002055719798 [DOI] [PubMed] [Google Scholar]
- 4. Ermolenko D. N., and Makhatadze G. I. (2002) Bacterial cold-shock proteins. Cell. Mol. Life Sci. 59, 1902–1913 10.1007/PL00012513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Graumann P. L., and Marahiel M. A. (1999) Cold-shock response in Bacillus subtilis. J. Mol. Microbiol. Biotechnol. 1, 203–209 [PubMed] [Google Scholar]
- 6. Lopez M. M., and Makhatadze G. I. (2000) Major cold shock proteins, CspA from Escherichia coli and CspB from Bacillus subtilis, interact differently with single-stranded DNA templates. Biochim. Biophys. Acta 1479, 196–202 10.1016/S0167-4838(00)00048-0 [DOI] [PubMed] [Google Scholar]
- 7. Graumann P., and Marahiel M. A. (1997) Effects of heterologous expression of CspB, the major cold shock protein of Bacillus subtilis, on protein synthesis in Escherichia coli. Mol. Gen. Genet. 253, 745–752 10.1007/s004380050379 [DOI] [PubMed] [Google Scholar]
- 8. Morgan H. P., Wear M. A., McNae I., Gallagher M. P., and Walkinshaw M. D. (2009) Crystallization and X-ray structure of cold-shock protein E from Salmonella typhimurium. Acta Crystallogr Sect. F. Struct. Biol. Cryst. Commun. 65, 1240–1245 10.1107/S1744309109033788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Phadtare S., and Inouye M. (2001) Role of CspC and CspE in regulation of expression of RpoS and UspA, the stress response proteins in Escherichia coli. J. Bacteriol. 183, 1205–1214 10.1128/JB.183.4.1205-1214.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schröder K., Graumann P., Schnuchel A., Holak T. A., and Marahiel M. A. (1995) Mutational analysis of the putative nucleic acid-binding surface of the cold-shock domain, CspB, revealed an essential role of aromatic and basic residues in binding of single-stranded DNA containing the Y-box motif. Mol. Microbiol. 16, 699–708 10.1111/j.1365-2958.1995.tb02431.x [DOI] [PubMed] [Google Scholar]
- 11. Blattner F. R., Plunkett G. 3rd., Bloch C. A., Perna N. T., Burland V., Riley M., Collado-Vides J., Glasner J. D., Rode C. K., Mayhew G. F., Gregor J., Davis N. W., Kirkpatrick H. A., Goeden M. A., Rose D. J., et al. (1997) The complete genome sequence of Escherichia coli K-12. Science 277, 1453–1462 10.1126/science.277.5331.1453 [DOI] [PubMed] [Google Scholar]
- 12. Yamanaka K., Mitani T., Ogura T., Niki H., and Hiraga S. (1994) Cloning, sequencing, and characterization of multicopy suppressors of a mukB mutation in Escherichia coli. Mol. Microbiol. 13, 301–312 10.1111/j.1365-2958.1994.tb00424.x [DOI] [PubMed] [Google Scholar]
- 13. Yamanaka K., and Inouye M. (1997) Growth-phase–dependent expression of cspD, encoding a member of the CspA family in Escherichia coli. J. Bacteriol. 179, 5126–5130 10.1128/jb.179.16.5126-5130.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jiang W., Hou Y., and Inouye M. (1997) CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem. 272, 196–202 10.1074/jbc.272.1.196 [DOI] [PubMed] [Google Scholar]
- 15. Schärer K., Stephan R., and Tasara T. (2013) Cold shock proteins contribute to the regulation of listeriolysin O production in Listeria monocytogenes. Foodborne Pathog. Dis. 10, 1023–1029 10.1089/fpd.2013.1562 [DOI] [PubMed] [Google Scholar]
- 16. Michaux C., Martini C., Shioya K., Ahmed Lecheheb S., Budin-Verneuil A., Cosette P., Sanguinetti M., Hartke A., Verneuil N., and Giard J. C. (2012) CspR, a cold shock RNA-binding protein involved in the long-term survival and the virulence of Enterococcus faecalis. J. Bacteriol. 194, 6900–6908 10.1128/JB.01673-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Uppal S., Maurya S. R., Hire R. S., and Jawali N. (2011) Cyclic AMP receptor protein regulates cspE, an early cold-inducible gene, in Escherichia coli. J. Bacteriol. 193, 6142–6151 10.1128/JB.05728-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shenhar Y., Biran D., and Ron E. Z. (2012) Resistance to environmental stress requires the RNA chaperones CspC and CspE. Environ. Microbiol. Rep. 4, 532–539 10.1111/j.1758-2229.2012.00358.x [DOI] [PubMed] [Google Scholar]
- 19. Hu K. H., Liu E., Dean K., Gingras M., DeGraff W., and Trun N. J. (1996) Overproduction of three genes leads to camphor resistance and chromosome condensation in Escherichia coli. Genetics 143, 1521–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hanna M. M., and Liu K. (1998) Nascent RNA in transcription complexes interacts with CspE, a small protein in E. coli implicated in chromatin condensation. J. Mol. Biol. 282, 227–239 10.1006/jmbi.1998.2005 [DOI] [PubMed] [Google Scholar]
- 21. Feng Y., Huang H., Liao J., and Cohen S. N. (2001) Escherichia coli poly(A)-binding proteins that interact with components of degradosomes or impede RNA decay mediated by polynucleotide phosphorylase and RNase E. J. Biol. Chem. 276, 31651–31656 10.1074/jbc.M102855200 [DOI] [PubMed] [Google Scholar]
- 22. Hohmann E. L. (2001) Nontyphoidal salmonellosis. Clin. Infect. Dis. 32, 263–269 10.1086/318457 [DOI] [PubMed] [Google Scholar]
- 23. Feasey N. A., Dougan G., Kingsley R. A., Heyderman R. S., and Gordon M. A. (2012) Invasive non-typhoidal Salmonella disease: an emerging and neglected tropical disease in Africa. Lancet 379, 2489–2499 10.1016/S0140-6736(11)61752-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gordon M. A., Graham S. M., Walsh A. L., Wilson L., Phiri A., Molyneux E., Zijlstra E. E., Heyderman R. S., Hart C. A., and Molyneux M. E. (2008) Epidemics of invasive Salmonella enterica serovar Enteritidis and S. enterica serovar Typhimurium infection associated with multidrug resistance among adults and children in Malawi. Clin. Infect. Dis. 46, 963–969 10.1086/529146 [DOI] [PubMed] [Google Scholar]
- 25. Tadesse D. A., Singh A., Zhao S., Bartholomew M., Womack N., Ayers S., Fields P. I., and McDermott P. F. (2016) Antimicrobial resistance in Salmonella in the United States from 1948 to 1995. Antimicrob. Agents Chemother. 60, 2567–2571 10.1128/AAC.02536-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Poppe C., Smart N., Khakhria R., Johnson W., Spika J., and Prescott J. (1998) Salmonella typhimurium DT104: a virulent and drug-resistant pathogen. Can. Vet. J. 39, 559–565 [PMC free article] [PubMed] [Google Scholar]
- 27. Craig J. E., Boyle D., Francis K. P., and Gallagher M. P. (1998) Expression of the cold-shock gene cspB in Salmonella typhimurium occurs below a threshold temperature. Microbiology 144, 697–704 10.1099/00221287-144-3-697 [DOI] [PubMed] [Google Scholar]
- 28. Jeffreys A. G., Hak K. M., Steffan R. J., Foster J. W., and Bej A. K. (1998) Growth, survival and characterization of cspA in Salmonella enteritidis following cold shock. Curr. Microbiol. 36, 29–35 10.1007/s002849900275 [DOI] [PubMed] [Google Scholar]
- 29. Kim B. H., Bang I. S., Lee S. Y., Hong S. K., Bang S. H., Lee I. S., and Park Y. K. (2001) Expression of cspH, encoding the cold shock protein in Salmonella enterica serovar Typhimurium UK-1. J. Bacteriol. 183, 5580–5588 10.1128/JB.183.19.5580-5588.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Michaux C., Holmqvist E., Vasicek E., Sharan M., Barquist L., Westermann A. J., Gunn J. S., and Vogel J. (2017) RNA target profiles direct the discovery of virulence functions for the cold-shock proteins CspC and CspE. Proc. Natl. Acad. Sci. U.S.A. 114, 6824–6829 10.1073/pnas.1620772114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hay A. J., and Zhu J. (2016) In sickness and in health: the relationships between bacteria and bile in the human gut. Adv. Appl. Microbiol. 96, 43–64 10.1016/bs.aambs.2016.07.019 [DOI] [PubMed] [Google Scholar]
- 32. Begley M., Gahan C. G., and Hill C. (2005) The interaction between bacteria and bile. FEMS Microbiol. Rev. 29, 625–651 10.1016/j.femsre.2004.09.003 [DOI] [PubMed] [Google Scholar]
- 33. Prouty A. M., Brodsky I. E., Falkow S., and Gunn J. S. (2004) Bile-salt–mediated induction of antimicrobial and bile resistance in Salmonella typhimurium. Microbiology 150, 775–783 10.1099/mic.0.26769-0 [DOI] [PubMed] [Google Scholar]
- 34. Gonzalez-Escobedo G., and Gunn J. S. (2013) Gallbladder epithelium as a niche for chronic Salmonella carriage. Infect. Immun. 81, 2920–2930 10.1128/IAI.00258-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Prouty A. M., Schwesinger W. H., and Gunn J. S. (2002) Biofilm formation and interaction with the surfaces of gallstones by Salmonella spp. Infect. Immun. 70, 2640–2649 10.1128/IAI.70.5.2640-2649.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Menendez A., Arena E. T., Guttman J. A., Thorson L., Vallance B. A., Vogl W., and Finlay B. B. (2009) Salmonella infection of gallbladder epithelial cells drives local inflammation and injury in a model of acute typhoid fever. J. Infect. Dis. 200, 1703–1713 10.1086/646608 [DOI] [PubMed] [Google Scholar]
- 37. Prouty A. M., Brodsky I. E., Manos J., Belas R., Falkow S., and Gunn J. S. (2004) Transcriptional regulation of Salmonella enterica serovar Typhimurium genes by bile. FEMS Immunol. Med. Microbiol. 41, 177–185 10.1016/j.femsim.2004.03.002 [DOI] [PubMed] [Google Scholar]
- 38. Smirnov A., Wang C., Drewry L. L., and Vogel J. (2017) Molecular mechanism of mRNA repression in trans by a ProQ-dependent small RNA. EMBO J. 36, 1029–1045 10.15252/embj.201696127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Phadtare S., and Severinov K. (2005) Nucleic acid melting by Escherichia coli CspE. Nucleic Acids Res. 33, 5583–5590 10.1093/nar/gki859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kelly S. M., Jess T. J., and Price N. C. (2005) How to study proteins by circular dichroism. Biochim. Biophys. Acta 1751, 119–139 10.1016/j.bbapap.2005.06.005 [DOI] [PubMed] [Google Scholar]
- 41. Papenfort K., Pfeiffer V., Mika F., Lucchini S., Hinton J. C., and Vogel J. (2006) σE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol. Microbiol. 62, 1674–1688 10.1111/j.1365-2958.2006.05524.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vogel J., and Papenfort K. (2006) Small non-coding RNAs and the bacterial outer membrane. Curr. Opin. Microbiol. 9, 605–611 10.1016/j.mib.2006.10.006 [DOI] [PubMed] [Google Scholar]
- 43. Coldham N. G., Webber M., Woodward M. J., and Piddock L. J. (2010) A 96-well plate fluorescence assay for assessment of cellular permeability and active efflux in Salmonella enterica serovar Typhimurium and Escherichia coli. J. Antimicrob. Chemother. 65, 1655–1663 10.1093/jac/dkq169 [DOI] [PubMed] [Google Scholar]
- 44. Picken R. N., and Beacham I. R. (1977) Bacteriophage-resistant mutants of Escherichia coli K12 with altered lipopolysaccharide. Studies with concanavalin A. J. Gen. Microbiol. 102, 319–326 10.1099/00221287-102-2-319 [DOI] [PubMed] [Google Scholar]
- 45. Ramos-Morales F., Prieto A. I., Beuzón C. R., Holden D. W., and Casadesús J. (2003) Role for Salmonella enterica enterobacterial common antigen in bile resistance and virulence. J. Bacteriol. 185, 5328–5332 10.1128/JB.185.17.5328-5332.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rosenberg E. Y., Bertenthal D., Nilles M. L., Bertrand K. P., and Nikaido H. (2003) Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol. Microbiol. 48, 1609–1619 10.1046/j.1365-2958.2003.03531.x [DOI] [PubMed] [Google Scholar]
- 47. van Velkinburgh J. C., and Gunn J. S. (1999) PhoP–PhoQ-regulated loci are required for enhanced bile resistance in Salmonella spp. Infect. Immun. 67, 1614–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Provenzano D., and Klose K. E. (2000) Altered expression of the ToxR-regulated porins OmpU and OmpT diminishes Vibrio cholerae bile resistance, virulence factor expression, and intestinal colonization. Proc. Natl. Acad. Sci. U.S.A. 97, 10220–10224 10.1073/pnas.170219997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sulavik M. C., Dazer M., and Miller P. F. (1997) The Salmonella typhimurium mar locus: molecular and genetic analyses and assessment of its role in virulence. J. Bacteriol. 179, 1857–1866 10.1128/jb.179.6.1857-1866.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Thanassi D. G., Cheng L. W., and Nikaido H. (1997) Active efflux of bile salts by Escherichia coli. J. Bacteriol. 179, 2512–2518 10.1128/jb.179.8.2512-2518.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. McClelland M., Sanderson K. E., Spieth J., Clifton S. W., Latreille P., Courtney L., Porwollik S., Ali J., Dante M., Du F., Hou S., Layman D., Leonard S., Nguyen C., Scott K., et al. (2001) Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413, 852–856 10.1038/35101614 [DOI] [PubMed] [Google Scholar]
- 52. Robbe-Saule V., Coynault C., Ibanez-Ruiz M., Hermant D., and Norel F. (2001) Identification of a non-haem catalase in Salmonella and its regulation by RpoS (σS). Mol. Microbiol. 39, 1533–1545 10.1046/j.1365-2958.2001.02340.x [DOI] [PubMed] [Google Scholar]
- 53. Ibanez-Ruiz M., Robbe-Saule V., Hermant D., Labrude S., and Norel F. (2000) Identification of RpoS (σS))-regulated genes in Salmonella enterica serovar Typhimurium. J. Bacteriol. 182, 5749–5756 10.1128/JB.182.20.5749-5756.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hindupur A., Liu D., Zhao Y., Bellamy H. D., White M. A., and Fox R. O. (2006) The crystal structure of the E. coli stress protein YciF. Protein Sci. 15, 2605–2611 10.1110/ps.062307706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Oussenko I. A., Abe T., Ujiie H., Muto A., and Bechhofer D. H. (2005) Participation of 3′-to-5′ exoribonucleases in the turnover of Bacillus subtilis mRNA. J. Bacteriol. 187, 2758–2767 10.1128/JB.187.8.2758-2767.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gautheret D., and Lambert A. (2001) Direct RNA motif definition and identification from multiple sequence alignments using secondary structure profiles. J. Mol. Biol. 313, 1003–1011 10.1006/jmbi.2001.5102 [DOI] [PubMed] [Google Scholar]
- 57. Macke T. J., Ecker D. J., Gutell R. R., Gautheret D., Case D. A., and Sampath R. (2001) RNAMotif, an RNA secondary structure definition and search algorithm. Nucleic Acids Res. 29, 4724–4735 10.1093/nar/29.22.4724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hitchcock P. J., and Brown T. M. (1983) Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154, 269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ruiz N., Kahne D., and Silhavy T. J. (2006) Advances in understanding bacterial outer-membrane biogenesis. Nat. Rev. Microbiol. 4, 57–66 10.1038/nrmicro1322 [DOI] [PubMed] [Google Scholar]
- 60. Rhodius V. A., Suh W. C., Nonaka G., West J., and Gross C. A. (2006) Conserved and variable functions of the σE stress response in related genomes. PLoS Biol. 4, e2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Skovierova H., Rowley G., Rezuchova B., Homerova D., Lewis C., Roberts M., and Kormanec J. (2006) Identification of the σE regulon of Salmonella enterica serovar Typhimurium. Microbiology 152, 1347–1359 10.1099/mic.0.28744-0 [DOI] [PubMed] [Google Scholar]
- 62. Leiser O. P., Charlson E. S., Gerken H., and Misra R. (2012) Reversal of the ΔdegP phenotypes by a novel rpoE allele of Escherichia coli. PLoS One 7, e33979 10.1371/journal.pone.0033979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Nikaido H. (2003) Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67, 593–656 10.1128/MMBR.67.4.593-656.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Santiviago C. A., Toro C. S., Hidalgo A. A., Youderian P., and Mora G. C. (2003) Global regulation of the Salmonella enterica serovar Typhimurium major porin, OmpD. J. Bacteriol. 185, 5901–5905 10.1128/JB.185.19.5901-5905.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Santiviago C. A., Fuentes J. A., Bueno S. M., Trombert A. N., Hildago A. A., Socias L. T., Youderian P., and Mora G. C. (2002) The Salmonella enterica sv. Typhimurium smvA, yddG and ompD (porin) genes are required for the efficient efflux of methyl viologen. Mol. Microbiol. 46, 687–698 10.1046/j.1365-2958.2002.03204.x [DOI] [PubMed] [Google Scholar]
- 66. Szklarczyk D., Morris J. H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N. T., Roth A., Bork P., Jensen L. J., and von Mering C. (2017) The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 45, D362–D368 10.1093/nar/gkw937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kröger C., Colgan A., Srikumar S., Händler K., Sivasankaran S. K., Hammarlöf D. L., Canals R., Grissom J. E., Conway T., Hokamp K., and Hinton J. C. (2013) An infection-relevant transcriptomic compendium for Salmonella enterica serovar Typhimurium. Cell Host Microbe 14, 683–695 10.1016/j.chom.2013.11.010 [DOI] [PubMed] [Google Scholar]
- 68. Datsenko K. A., and Wanner B. L. (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Shetty S., and Varshney U. (2016) An evolutionarily conserved element in initiator tRNAs prompts ultimate steps in ribosome maturation. Proc. Natl. Acad. Sci. U.S.A. 113, E6126–E6134 10.1073/pnas.1609550113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bhosale M., Pande S., Kumar A., Kairamkonda S., and Nandi D. (2010) Characterization of two M17 family members in Escherichia coli, peptidase A and peptidase B. Biochem. Biophys. Res. Commun. 395, 76–81 10.1016/j.bbrc.2010.03.142 [DOI] [PubMed] [Google Scholar]
- 71. Thakur M., Kumar M. B., and Muniyappa K. (2016) Mycobacterium tuberculosis UvrB is a robust DNA-stimulated ATPase that also possesses structure-specific ATP-dependent DNA helicase activity. Biochemistry 55, 5865–5883 10.1021/acs.biochem.6b00558 [DOI] [PubMed] [Google Scholar]
- 72. Villarreal J. M., Becerra-Lobato N., Rebollar-Flores J. E., Medina-Aparicio L., Carbajal-Gómez E., Zavala-García M. L., Vázquez A., Gutierrez-Ríos R. M., Olvera L., Encarnación S., Martínez-Batallar A. G., Calva E., and Hernández-Lucas I. (2014) The Salmonella enterica serovar Typhi ltrR–ompR–ompC–ompF genes are involved in resistance to the bile salt sodium deoxycholate and in bacterial transformation. Mol. Microbiol. 92, 1005–1024 10.1111/mmi.12610 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.