Abstract
Reprogramming of fibroblasts into induced cardiomyocytes (iCMs) is a potentially promising strategy for regenerating a damaged heart. However, low fibroblast–cardiomyocyte conversion rates remain a major challenge in this reprogramming. To this end, here we conducted a chemical screen and identified four agents, insulin-like growth factor-1, Mll1 inhibitor MM589, transforming growth factor-β inhibitor A83–01, and Bmi1 inhibitor PTC-209, termed IMAP, which coordinately enhanced reprogramming efficiency. Using α–muscle heavy chain–GFP–tagged mouse embryo fibroblasts as a starting cell type, we observed that the IMAP treatment increases iCM formation 6-fold. IMAP stimulated higher cardiac troponin T and α-actinin expression and increased sarcomere formation, coinciding with up-regulated expression of many cardiac genes and down-regulated fibroblast gene expression. Furthermore, IMAP promoted higher spontaneous beating and calcium transient activities of iCMs derived from neonatal cardiac fibroblasts. Intriguingly, we also observed that the IMAP treatment repressed many genes involved in immune responses, particularly those in specific C-C chemokine signaling pathways. We therefore investigated the roles of C-C motif chemokine ligand 3 (CCL3), CCL6, and CCL17 in cardiac reprogramming and observed that they inhibited iCM formation, whereas inhibitors of C-C motif chemokine receptor 1 (CCR1), CCR4, and CCR5 had the opposite effect. These results indicated that the IMAP treatment directly suppresses specific C-C chemokine signaling pathways and thereby enhances cardiac reprogramming. In conclusion, a combination of four chemicals, named here IMAP, suppresses specific C-C chemokine signaling pathways and facilitates Mef2c/Gata4/Tbx5 (MGT)-induced cardiac reprogramming, providing a potential means for iCM formation in clinical applications.
Keywords: small molecule, cellular immune response, myocardial infarction, cardiomyocyte, regeneration, reprogramming, cardiovascular disease, fibrosis, cardiomyopathy, cell signaling, C-C chemokine, cardiac reprogramming, heart regeneration, immune response
Introduction
Cardiovascular disease (CVD) has remained the leading cause of death in the United States and the whole world in the last decades (1). Myocardial infarction (MI) is the most severe outcome caused by cardiovascular disease, which leads to impaired heart function because of massive cardiomyocyte (CM)3 loss and fibrosis (2). Existing treatments for myocardial infarction are primarily pharmacological and device-based, and do not address the underlying problem of CM loss (3). Indeed, the prevalence of chronic cardiomyopathy is steadily increasing worldwide, escalating the urgency of developing novel therapies for this morbid disease.
Direct reprogramming of fibroblasts into CM-like cells by introducing three cardiac transcription factors Gata4, Mef2C, and Tbx5 (G, M, and T) has emerged as an attractive strategy to generate induced CMs (iCMs) (4–6). One major advantage of this strategy is utilizing abundant cardiac fibroblasts (CFs) in the heart to generate functional CMs to replace the scar tissue. Other advantages include the immunologic compatibility of the reprogrammed cells and lower possibility of tumorigenicity. However, low conversion rate, poor purity, and lack of precise conversion of iCMs still present significant challenges.
Several strategies have been applied to address these challenges in both mouse and human cells (5, 7–27). It has been shown that by adding other core transcriptional factors or protein kinases such as Hand2, Akt1 could further improve cardiac reprogramming efficiency in several fibroblast lineage s (5, 12). Growth factors, such as fibroblast growth factor (FGF) 2, FGF10, and vascular endothelial growth factor (VEGF), can further promote cardiac reprogramming (17). Specific chemical compounds that influence different signaling pathways also enhance the efficiency and quality of cardiac reprogramming (16, 26). Moreover, recent studies have identified epigenetic barriers in the reprogramming process. For example, targeting the polycomb Ring finger oncogene Bmi1 and Mll1 H3K4 methyltransferase can enhance cardiac conversion (28). Single-cell study of cardiac reprogramming indicates that RNA splicing also plays an important role in the cardiac reprogramming (27, 29). Nevertheless, to bring this strategy closer to clinical studies, the iCM conversion and maturation need to be substantially improved.
Here we screened and identified four different chemical compounds, insulin-like growth factor-1 (IGF-1), Mll1 inhibitor MM589, transforming growth factor (TGF)-β inhibitor A83–01, and Bmi1 inhibitor PTC-209 (IMAP) that remarkably enhanced the conversion from fibroblasts into cardiomyocytes. We observed significant improvement in cardiac reprogramming including cardiac gene expression, sarcomere formation, calcium flux, and spontaneous beating. Importantly, RNA-Seq data showed that immune response–related genes, especially those involved in chemokine signaling pathways, were down-regulated. Therefore, we chose three chemokine axes, C-C motif chemokine ligand 6 (Ccl6)–C-C motif chemokine receptor 1 (Ccr1), Ccl17-Ccr4, and Ccl3-Ccr5, as the potential targets. We then examined chemokine signaling pathway ligands CCL3, CCL6, and CCL17 and observed decreased reprogramming efficiency. In contrast, corresponding chemokine receptor inhibitors significantly increased iCM formation, similar to IMAP treatment. Taken together, these findings identified specific immune responses as potential barriers for cardiac reprogramming and offered a promising strategy to resolve the low-efficiency challenge of iCM formation by providing a novel chemical combination that targets immune responses.
Results
IGF-1, MM589, A83–01, and PTC-209 enhanced cardiac reprogramming
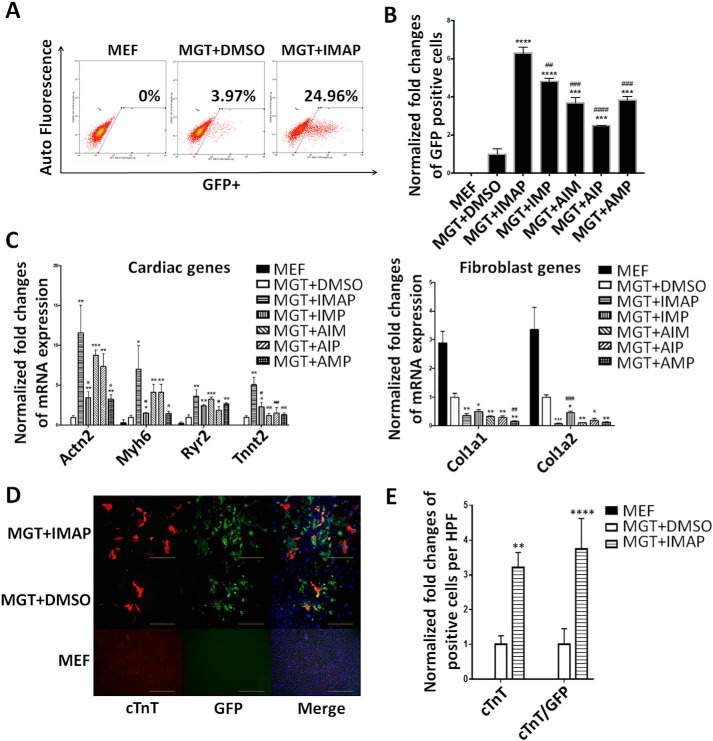
To identify small molecules that could enhance cardiac reprogramming, we used a screening platform reported previously (26, 28). In this screening strategy, we isolated mouse embryonic fibroblasts (MEF) from a transgenic α–muscle heavy chain (α-MHC)–green fluorescent protein (GFP) reporter mouse (4). MEFs were treated with chemicals every 3 days after the infection of retrovirus expressing polycistronic Mef2c/Gata4/Tbx5 (MGT) (18). 2 weeks later, the reprogramming efficiency was evaluated by percentage of GFP-positive cells and related gene expression (Fig. 1A). Four chemical compounds, namely, IGF-1, Mll1 inhibitor MM589, TGF-β inhibitor A83–01, and Bmi1 inhibitor PTC-209, were identified that could improve cardiac reprogramming efficiency individually 2- to 4-fold (Fig. 1, B and C). Quantitative PCR (qPCR) indicated that each chemical played an important role in reprogramming by up-regulation of cardiac-related genes and/or down-regulation of fibroblast-related genes (Fig. 1D).
Figure 1.
Drug screening identified four chemicals enhancing cardiac reprogramming. A, schematic diagram of the chemical screening using α-MHC–GFP MEFs. B, bar graph showing % α-MHC–GFP+ cells 2 weeks after GMT infection with or without indicated chemical treatment. C, representative FACS plots showing iCMs with α-MHC–GFP+ cells after 2 weeks of reprogramming and quantification. D, bar graph representing major cardiac and fibroblast genes as determined by qPCR. Error bars indicate mean ± S.E.; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001 compared with MGT+DMSO group.
The combination of four chemicals IMAP achieved high reprogramming efficiency of MEFs
To explore the significance of each chemical in combination, we applied minus-one strategy to eliminate possible overlapping effects of these compounds. Using the same screening platform, we found that the combination of all four chemicals showed the highest efficiency and increased iCM formation 6-fold compared with MGT+DMSO treatment (Fig. 2A). Eliminating every single compound in the IMAP caused a significant reduction of the reprogramming efficiency (Fig. 2B). qPCR results indicated that IMAP maximally enhanced the expression of cardiac genes and repressed fibroblast genes (Fig. 2C). After 2 weeks of reprogramming with IMAP treatment, immunofluorescence staining showed 3- to 4-fold increases in the expression of cardiac troponin T (cTnT) and α-actinin as well as significantly increased cTnT/GFP–double positive cells (Fig. 2D and Fig. S1A). Cell counting in several high power fields (HPFs) showed similar increase in cTnT, α-actinin, and cTnT/GFP–double positive cells (Fig. 2E and Fig. S1B).
Figure 2.
IMAP enhanced cardiac reprogramming of MEFs. A, representative FACS plots showing MEFs (control) and iCMs with α-MHC–GFP+ cells after 2 weeks of reprogramming with IMAP or DMSO treatment. B, bar graph showing normalized -fold changes of α-MHC–GFP+ cells 2 weeks after GMT infection with indicated chemical treatment. C, bar graph representing major cardiac and fibroblast genes as determined by qPCR in cells after 2 weeks of reprogramming and indicated chemical treatment. D and E, representative images of immunofluorescence staining (D) and quantification of (E) cardiac markers α-MHC–GFP (green), cTnT (red), and double positive cells in fibroblasts (control) or cells treated with either MGT+DMSO or MGT+IMAP (scale bar, 500 μm). Error bars indicate mean ± S.E.; #, p < 0.05; ##, p < 0.01; ###, p < 0.001; ####, p < 0.0001 compared with MGT+IMAP group; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001 compared with MGT+DMSO group.
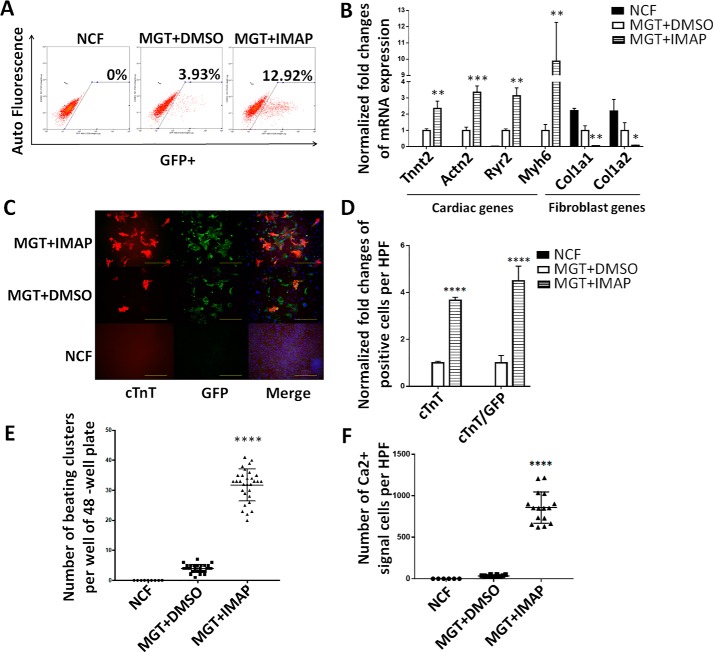
IMAP also enhanced cardiac reprogramming of neonatal cardiac fibroblasts (NCFs)
Considering cardiac fibroblasts are the major in vivo target cell type for iCM reprogramming, we next determined the effect of IMAP on conversion of NCFs to iCMs. NCFs were isolated from the same α-MHC–GFP mouse line from which we obtained MEFs (30). Flow cytometry results indicated that the conversion efficiency was elevated 4- to 5-fold by IMAP compared with MGT+DMSO group (Fig. 3A). qPCR showed significant increase of cardiac genes and decrease of fibroblast lineage–related genes (Fig. 3B). Immunofluorescence staining revealed a more significant increase of cTnT, α-actinin expression, and cTnT/GFP–double positive cells in NCFs compared with MEFs (Fig. 3, C and D and Fig. S1, C–E) and clearly visualized sarcomere formation in iCMs derived from NCFs (Fig. S1C).
Figure 3.
IMAP improved cardiac reprogramming efficiency of NCFs. A, representative FACS plots showing NCFs (control) and iCMs with α-MHC–GFP+ cells after 2 weeks of reprogramming with MGT plus IMAP or DMSO treatment. B, bar graph representing major cardiac and fibroblast genes as determined by qPCR in cells after 2 weeks of reprogramming and indicated chemical treatment. C and D, representative images of immunofluorescence staining (C) and quantification of (D) cardiac markers α-MHC–GFP (green), cTnT (red), and double positive cells in NCFs (control) or cells treated with either MGT+DMSO or MGT+IMAP (scale bar, 500 μm). E, quantification of spontaneous beating iCMs per well of 48-well plate after 2 weeks of reprogramming and another 2 weeks with mature medium (n = 10). F, quantification of NCFs (control) or iCMs with calcium transient activities stained by Rhod-3 dye after 2 weeks of reprogramming followed by another 2 weeks of mature medium treatment. Error bars indicate mean ± S.E.; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001 compared with MGT+DMSO group.
Next, we tested spontaneous beating and calcium transient of iCMs because these two features represent characteristic cell functions of CMs. After treating NCFs with IMAP for 2 weeks and maturation medium for another 2 weeks, 8- to 9-fold increase in spontaneous beating of iCMs was observed (Fig. 3E and Movies S1 and S2). Additionally, spontaneous calcium transient was measured by Ca2+ imaging with Rhod-3 staining (4). With Rhod-3 staining in HPFs, hundreds of cells with spontaneous calcium transients were observed in MGT+IMAP group whereas only dozens were observed in MGT+DMSO group (Fig. 3F and Movies S3 and S4).
It is possible that potential CM contamination could contribute to the iCM population. To address this potential issue, MEFs or NCFs without GMT infection and IMAP treatment were examined for GFP-positive signals from initial fibroblast isolation to 4 weeks after iCM induction. With microscopic examination, flow cytometry or immunofluorescence staining (Figs. 2, A, B, D, and E and 3, A, C, and D and Fig. S1, A–D), no GFP signals were detected in those MEF or NCF control groups, indicating that CM contamination is highly unlikely in our experiments. Likewise, no cells in MEF or NCF control groups were observed to exhibit any spontaneous calcium transient or other CM properties (Fig. 3, E and F).
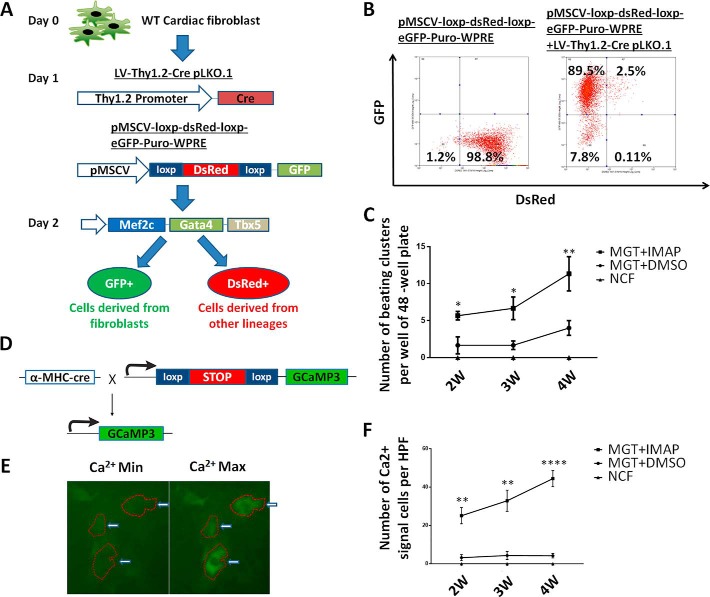
Lineage-tracing further validated that IMAP significantly enhanced iCMs formation from NCFs
To determine unequivocally that iCMs were indeed reprogrammed from NCFs, we performed iCM experiments with two complementary lineage-tracing experiments. First, isolated NCFs were infected at day 1 with lentiviruses produced with two lentiviral plasmids LV-Thy1.2-Cre pLKO.1 and pMSCV-loxp-dsRed-loxp-eGFP-Puro-WPRE (Fig. 4A). Because Cre is driven by fibroblast-specific Thy1.2 promoter, the Cre expression will convert the original dsRed cells with pMSCV-loxp-dsRed-loxp-eGFP-Puro-WPRE into GFP green cells. Indeed, flow cytometry data showed that ∼99% of the NCFs were Red without LV-Thy1.2-Cre pLKO.1 co-infection and ∼90% of NCFs showed strong GFP expression with LV-Thy1.2-Cre pLKO.1 co-infection (Fig. 4B). At day 2, NCFs were infected with retrovirus encoding Mef2c, Gata4, and Tbx5 to induce the cardiac reprogramming. Green cells with spontaneous beating were counted 2, 3, and 4 weeks after infection and iCMs derived from MGT+IMAP groups showed much better spontaneous beating, one of the most representative CM characteristics, than those obtained from MGT+DMSO groups (Fig. 4C and Movies S5 and S6).
Figure 4.
Lineage-tracing system showed that reprogrammed iCMs were derived from fibroblasts rather than other lineage s and could be enhanced by IMAP. A, schematic diagram of lineage-tracing system using another two lentiviral plasmids, LV-Thy1.2-Cre pLKO.1 and pMSCV-loxp-dsRed-loxp-eGFP-Puro-WPRE, in which iCMs derived from fibroblasts showed green and iCMs derived from other lineages showed red under fluorescence microscope. B, representative FACS plots showing the validation and viral infection efficiency of lineage-tracing system. C, quantification of spontaneous beating iCMs (green) per well of 48-well plate after 2, 3, and 4 weeks of reprogramming in lineage-tracing system under fluorescence microscope. D, schematic representation of the strategy for generating a calcium transient detection mouse line by crossing α-MHC–Cre mouse line with Rosa26A-Flox-Stop-Flox-GCaMP3 mouse line. E, representative images of iCMs exhibiting calcium transient. F, quantification of NCFs (control) or iCMs with calcium transient activities after 2 weeks of reprogramming followed by another 2 weeks of mature medium treatment. (n = 30 from 10 wells). Error bars indicate mean ± S.E.; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001compared with MGT+DMSO group.
To characterize further the iCM lineage conversion and CM functionality, αMHC-Cre/Rosa26A-Flox-Stop-Flox-GCaMP3 NCFs were isolated from corresponding transgenic mice (7) and calcium flux was measured during reprogramming(Fig. 4, D and E). This system is able to trace lineage conversion from non-CM lineages to CM lineage. No cells exhibiting spontaneous calcium transient under fluorescence microscope were observed in control groups, indicating no CM contamination in our experiments (Fig. 4F). Importantly, IMAP promoted calcium flux as early as 2 weeks after treatment (Fig. 4F). Four-week treatment of IMAP resulted in 10-fold increase of calcium flux (Fig. 4F and Movies S7 and S8), indicating that iCM matured further under extended IMAP treatment. Altogether, these data indicated that iCMs were reprogrammed from originally isolated fibroblasts and showed much better CM functionality such as spontaneous beating and calcium transient in MGT+IMAP groups than those obtained from MGT+DMSO groups.
RNA-Seq data showed significant changes in gene expression profiles defining cell identity and immune responses
To identify the downstream targets of IMAP in iCM formation, we first extracted the total RNAs and performed RNA-Seq analyses using cells reprogrammed from α-MHC–GFP NCFs after 2 weeks of GMT and IMAP treatment. Cells reprogrammed with IMAP showed a much more robust cardiomyocyte gene expression profile compared with MGT+DMSO group (Fig. 5A). iCMs derived from MEFs showed similar results with 1442 differentially expressed genes when comparing MGT+IMAP group with MGT+DMSO group (Fig. 5B and Fig. S2A). 528 overlapping genes were identified in both datasets, and gene ontology (GO) enrichment analysis revealed that IMAP stimulated gene expression related to muscle structure formation and muscle contraction as well as decreased gene expression in extracellular matrix formation (Fig. 5B and Fig. S2, B and C). Intriguingly, GO analysis identified significant gene repression in immune system process and inflammation response by IMAP (Fig. 5B and Fig. S2C). We next examined in more detail the effect of IMAP on immune response gene expression. Several immune response representative genes were tested in reprogramming. Il6, Tnf, Ccl2, and Ptgs2 expression decreased as early as day 3 after IMAP treatment (Fig. 5C). RNA-Seq analysis further identified many immune-related genes that were significantly down-regulated after IMAP treatment in both NCFs (Fig. 5D) and MEFs (Fig. S2D). GO molecular function analysis revealed that these down-regulated genes shared similar biological function in C-C chemokine signaling pathways (Fig. 5E and Fig. S2E). These data suggested that C-C chemokine signaling pathways could be potential novel targets in improving iCM formation and may provide mechanistic insights into IMAP-mediated cardiac reprogramming.
Figure 5.
RNA-Seq data showed significant changes of gene expression profiles defining cell identity and immune responses. A, heat map showing differential expression of representative cardiac and fibroblast-related genes among NCFs (control), MGT+DMSO, and MGT+IMAP group. B, Venn diagram showing the number of differentially expressed genes between MGT+DMSO and MGT+IMAP group in MEFs and NCFs. Bar graph showing the top gene ontology (GO) terms of the up-regulated (red) or down-regulated (blue) genes expressed between MGT+DMSO group and MGT+IMAP group. C, bar graph showing representative immune response–related gene expression between MGT+DMSO group and MGT+IMAP group at day 3 after MGT infection. D, heat map showing differential expression of immune-related genes among NCFs (control), MGT+DMSO, and MGT+IMAP group. E, bar graph for GO molecular function analysis terms of IMAP down-regulated immune-related genes compared with MGT+DMSO group. Error bars indicate mean ± S.E.; ***, p < 0.001; ****, p < 0.0001 compared with MGT+DMSO group.
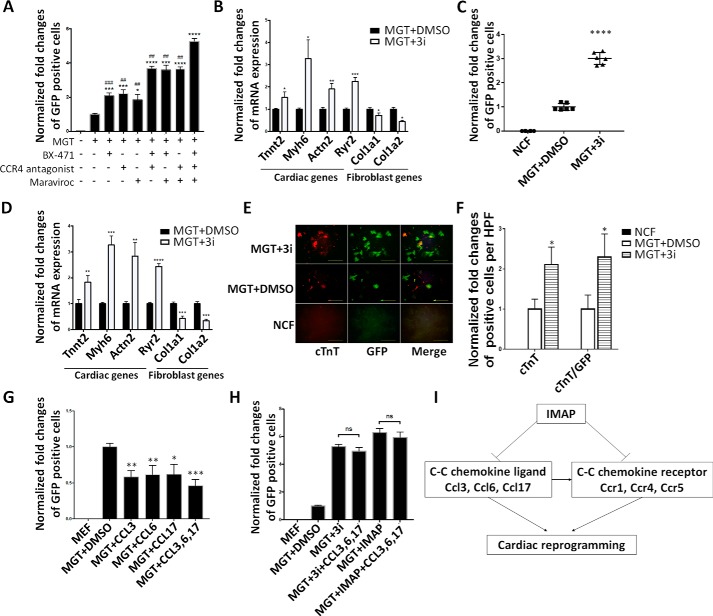
Specific C-C chemokine signaling pathways were likely important barriers in cardiac reprogramming that were overcome by IMAP treatment
Based on the above results, we examined C-C chemokine signaling pathways Ccl6-Ccr1, Ccl17-Ccr4 and Ccl3-Ccr5 in cardiac reprogramming, as they were significantly down-regulated in MGT+IMAP–treated group. We first performed loss-of-function studies by testing the inhibitors of corresponding C-C chemokine receptors in cardiac reprogramming. We applied CCR1 inhibitor BX471, a CCR4 antagonist, and CCR5 inhibitor Maraviroc in α-MHC–GFP MEFs. In MGT-induced cardiac reprogramming, all three inhibitors enhanced reprogramming individually whereas the three inhibitors together, called 3i, achieved a combinatorial improvement (Fig. 6A). These chemokine receptor inhibitors promoted the expression of cardiac-related genes and repressed the expression of fibroblast-related genes, similar to the effect of IMAP (Fig. 6B). Similar results were observed in NCF reprogramming (Fig. 6, C and D). Immunofluorescence staining showed much higher number of cTnT, α-actinin, and cTnT/GFP–double positive cells in MGT+3i–treated group compared with MGT+DMSO group (Fig. 6, E and F and Fig. S3, A and B).
Figure 6.
Specific C-C chemokine signaling pathways were likely important barriers in cardiac reprogramming that were overcome by IMAP treatment. A and C, bar graph or scatter plots showing normalized -fold changes of α-MHC–GFP+ cells 2 weeks after GMT infection with indicated chemical treatment in (A) MEFs and (C) NCFs. B and D, bar graph representing major cardiac and fibroblast genes as determined by qPCR in cells after 2 weeks of reprogramming and indicated chemical treatment in (B) MEFs and (D) NCFs. E and F, representative images of immunofluorescence staining (E) and quantification of (F) cardiac markers α-MHC–GFP (green), cTnT (red), and double positive cells in NCFs (control) or cells treated with either MGT+DMSO or MGT+3i. G, bar graph showing normalized -fold changes of α-MHC–GFP+ cells 2 weeks after GMT infection with indicated C-C chemokine ligands treatment in MEFs (scale bar, 500 μm). H, bar graph showing normalized -fold changes of α-MHC–GFP+ cells 2 weeks after GMT infection with indicated treatment in MEFs. I, schematic diagram showing IMAP-enhanced cardiac reprogramming by suppressing the expression of several specific C-C chemokine ligands and receptors. Error bars indicate mean ± S.E.; #, p < 0.05; ##, p < 0.01; ###, p < 0.001; ####, p < 0.0001 compared with MGT+3i group; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001 compared with MGT+DMSO group.
To further validate our findings that blocking the activities of specific C-C chemokine signaling pathways increased iCM reprogramming, we also performed gain-of-function experiments to examine the effect of chemokine ligands in reprogramming. Addition of chemokine ligands Ccl3, 6, and 17 in MGT-mediated reprogramming significantly reduced reprogramming efficiency (Fig. 6G and supporting Fig. S3C). In contrast, the inhibitory effect of these ligands was abolished when their receptor expressions or activities were inhibited in the presence of IMAP and 3i (Fig. 6H and Fig. S3D), indicating that these ligands' inhibitory effect on reprogramming was specifically through C-C chemokine signaling pathways. Taken together, our research showed that IMAP enhanced cardiac reprogramming by eliminating iCM reprogramming barriers imposed by specific C-C chemokine pathways (Fig. 6I).
Discussion
We report here a chemical mixture that enhances cardiac reprogramming by suppressing specific C-C chemokine signaling pathways. Administration of chemicals IGF-1, MM589, A83–01, and PTC-209 (IMAP), significantly enhances the reprogramming efficiency of MEFs or NCFs to iCMs. IMAP promotes cardiac gene expression, sarcomere formation, spontaneous beating, and calcium transient in reprogramming, indicating a significant role of IMAP in promoting maturation and functionality of iCMs. Importantly, repression of Ccr1, Ccr4, and Ccr5 signaling pathways is likely to be responsible for cardiac reprogramming augmentation mediated by IMAP. Direct inhibition of these chemokine receptors has achieved similar positive effect on cardiac reprogramming whereas their corresponding ligands repress reprogramming. Therefore, our study provides a novel platform of identifying chemicals that repress specific C-C chemokine signaling pathways for cardiac reprogramming.
In this study, we have developed a new platform enhancing cardiac reprogramming by combining four chemicals named IMAP. Two chemicals IGF-1 (12) and A83–01 (13), targeting IGF growth factor and TGF-β signaling, enhance reprogramming, as reported previously. PTC-209 is an inhibitor of polycomb Ring finger oncogene Bmi1, which functions through chromatin remodeling as an essential epigenetic regulator (31) and has been identified as a barrier in cardiac reprogramming (28). Importantly, our previous work reveals that targeting WD repeat domain 5 (WDR5) and blocking the WDR5 mixed-lineage leukemia (MLL) protein-protein interaction enhance cardiac reprogramming (28). In this study, we have identified that MM589, a newly invented version of WDR5-MLL1 inhibitor, promotes reprogramming. IMAP combination shows significant enhancement of MGT-induced cardiac reprogramming, and elimination of each of them decreases reprogramming efficiency. Our data thus indicate that these four chemicals work together to convert fibroblasts into iCMs.
Our IMAP studies identify an unexpected connection between C-C chemokine signaling pathways and cardiac reprogramming. In particular, we have observed that immune response–related genes, especially those involved in chemokine signaling pathways, were down-regulated in IMAP-mediated cardiac reprogramming. Subsequently, we have identified three chemokine axes, Ccl6-Ccr1, Ccl17-Ccr4, and Ccl3-Ccr5, as important reprogramming barriers. Administration of these three chemokine signaling pathway ligands, CCL3, CCL6, and CCL17, significantly decreases reprogramming efficiency. In contrast, addition of corresponding chemokine receptor inhibitors in our reprogramming assay significantly increased iCM formation, similar to IMAP treatment. Thus, our study reveals that immune responses may act as barriers for cardiac reprogramming. Mechanistically, the effect of chemokine signaling pathways in reprogramming could be linked to profibrotic signaling, which has been identified to inhibit cardiac reprogramming as shown by others (13, 15) and our studies. The three C-C chemokine pathways we have identified are all reported to promote fibrosis process both in rodents and humans (32–34). In addition, the effect of C-C chemokine pathways could be through Wnt pathway regulation. Wnt pathway–related genes also show significant decrease in our MGT+IMAP treatment group compared with MGT+DMSO group, and Wnt pathway is required for chemokine signaling–mediated functions in cardiac reprogramming (26, 35). Future studies will be required to further reveal the underlying mechanisms.
Even though the identification of chemokine signaling pathways in inhibiting cardiac reprogramming is surprising, studies of immune response and chemokine signaling pathways in cell fate change and development provide strong rationale that chemokine signaling pathways are indeed very likely a key barrier in cardiac reprogramming. Traditionally, C-C chemokine signaling pathways were initially considered merely as low-molecular-weight proteins that stimulate recruitment of leukocytes (36). However, recent studies show that specific C-C chemokine signaling pathways, such as stromal cell–derived factor-1–cxcr4 axis, can exert negative feedback on the Fgf pathway at the beginning of fin regeneration (37) and regulate myoD and myf5 expression and fast fiber formation in muscle differentiation of zebrafish (38). Importantly, C-C chemokine signaling pathways can significantly impact cardiac development. In particular, Cxcr4 is considered an important marker for cardiomyocyte progenitors and may play a functional role in their differentiation (39). Further studies also show that repressing Cxcl10 and Ccl5 by WT1 is also required during cardiac development whereas Cxcr4 is required for spontaneous beating of hiPSC-CMs (40, 41). In addition, targeting C-C chemokine signaling pathways has been proposed in cardiac repair after injury (42). Finally, anti-inflammatory drugs also facilitate induced pluripotent stem cell reprogramming (43, 44), and anti-inflammation treatment may benefit cardiac reprogramming (25). Therefore, further investigation of chemokine pathways in cardiac reprogramming may provide key insights into cell fate change.
Direct cardiac reprogramming provides a promising strategy for repairing injured heart after massive cardiomyocyte loss. Application of chemical approaches for reprogramming represents a highly desirable approach to improve the efficiency of iCM formation, as reported previously by us (28) and others (13, 15, 16, 21, 26, 45, 46). Importantly, our studies have identified specific C-C chemokine signaling pathways regulated by a combination of four chemicals IMAP in cardiac reprogramming. We expect that future studies toward combined chemical approaches in targeting chemokine signaling pathways will further improve the iCM formation and bring cardiac reprogramming closer to clinical applications.
Experimental procedures
Mouse lines
The α-MHC–GFP transgenic mice were used to derive MEFs and NCFs as described previously (30, 47). All animal-related procedures were approved by the Institutional Animal Care and Use Committee of the University of Michigan and are consistent with the National Institutes of Health Guide for Use and Care of Animals.
Plasmids
The pMXs-based retroviral polycistronic vector encoding Mef2c, Gata4, Tbx5 were provided from Dr. Li Qian's lab (18). This polycistronic constructs vector was constructed by DNA fragment containing Mef2c, Gata4, and Tbx5 sequentially, which were separated by oligonucleotides encoding P2A and T2A peptides. This polycistronic vector also contains puromycin selection marker for cell purification.
The LV-Thy1.2-Cre pLKO.1 plasmid was constructed by cloning Cre fragment from Thy1.2-Roxed-Cre vector (Addgene, 51276) into LV-Cre pLKO.1 (Addgene, 25997) to replace the cytomegalovirus enhance and promoter. pMSCV-loxp-dsRed-loxp-eGFP-Puro-WPRE was also acquired from Addgene (32702), as well as lentiviral packaging plasmid psPAX2(12260) and VSV-G envelope expressing plasmid pMD2.G (12259).
Primary cell isolation
Preparation of MEFs (isolated at E13.5) was reported previously (47). Briefly, embryos were harvested from α-MHC–GFP transgenic mice at 13.5 days post coitum followed by decapitation and removal of internal organs. The tissue was minced and digested with 0.05% trypsin/EDTA (Gibco, Thermo Fisher Scientific). Cells were resuspended in MEF medium (DMEM containing 10% FBS, 1% penicillin/streptomycin, 10 μl/ml GlutaMAX, and 1 mm sodium pyruvate) and plated onto one 10-cm dish per embryo. Cells were passaged at the ratio of 1:3 (passage 1). Passage 3 MEFs were used for reprogramming.
NCFs were isolated from P2-P3 α-MHC–GFP transgenic or αMHC-Cre/Rosa26A-Flox-Stop-Flox-GCaMP3 mice as described previously (30). Briefly, heart tissue was isolated, minced, and digested with 0.05% trypsin-EDTA. Then NCFs were collected with type II collagenase (0.5 mg/ml) in Hanks' Balanced salt solution. After washing and resuspending in Dulbecco's PBS with 2.5 g BSA and 0.5 m EDTA, cells were incubated with CD90.2 microbeads (Miltenyi Biotec, MidiMACS Starting Kit) at 4 °C for 30min. Positive cells were isolated by magnetic-activated cell sorting and plated onto a 10-cm dish with FB medium (Iscove's modified Dulbecco's medium with 20% FBS and 1% penicillin/streptomycin) for future use. Isolated fibroblasts were routinely examined under fluorescence microscope or with GFP immunostaining to determine potential CM contamination.
Chemicals information
IGF-1was from Peprotech (100–11), MM589 was from Dr. Shaomeng Wang's lab, A83–01 was from Stemgent (04–0014), PTC-209 was from Sigma (SML1143–5MG). C-C chemokine receptor inhibitors including CCR1 inhibitor BX471, CCR4 antagonist, and CCR5 inhibitor Maraviroc were from Cayman (18503, 21885, 14641). Recombinant mouse CCL3, CCL6, and CCL17 were from BioLegend (593802, 758502, 581402).
Retrovirus preparation
80% confluent 10-cm plates of Plat-E cells were transfected by using Lipofectamine 2000 (Thermo Fisher Scientific) with 10 μg retrovirus vectors in additional 1.5 ml Opti-MEM (Thermo Fisher Scientific). 24 h later, medium was changed into 10 ml fresh MEFs medium. 48 and 72 h after transfection, viral medium was collected twice and filtered through a 0.45-mm cellulose filter. The virus containing medium was added 1/5 volume of 40% PEG8000 solution to make a final concentration of 8% PEG8000. The mixture was kept at 4 °C overnight and spun at 3000 × g, 4 °C, 30 min to get concentrated. Virus was resuspended by fresh MEFs medium with 8 μg/ml Polybrene (Sigma).
Direct reprogramming of fibroblasts to iCMs
The protocol of direct cardiac reprogramming was similar to previous studies with minor optimization (30). Fresh fibroblasts were seeded on tissue culture dishes at a density of 10,000 cells/cm2 before virus infection. Fibroblasts were infected with freshly made viral mixture containing 8 μg/ml Polybrene (Sigma) 24 h post seeding. 24 h later, the viral medium was then replaced with induction medium composed of DMEM/199 (4:1) (Gibco, Thermo Fisher Scientific) containing 10% FBS, 1% penicillin/streptomycin, and 10 μl/ml GlutaMAX (Gibco, Thermo Fisher Scientific). Medium was changed every 2–3 days with or without indicated chemicals for 2 weeks before cells were examined. 1 μg/ml puromycin (SIGP8833–25MG, Sigma) was added into the medium 3 days after infection to eliminate noninfected cells. For spontaneous beating and calcium transient assessment experiments, induction medium was replaced by mature medium containing StemPro-34 SF medium (Gibco, Thermo Fisher Scientific), GlutaMAX (10 μl/ml, Gibco, Thermo Fisher Scientific), ascorbic acid (50 μg/ml, Sigma), recombinant human VEGF165 (5 ng/ml, R&D Systems), recombinant human FGF basic146 aa (10 ng/ml, R&D Systems), and recombinant human FGF10 (25 ng/ml, R&D Systems). Then mature medium was changed every 2–3 days for another 2 weeks as described previously (17).
Immunocytochemistry
Cells were fixed with 4% formaldehyde for 15 min and permeabilized with 0.1% Triton X-100 in PBS for 15 min at room temperature. Cells were blocked with 4% horse serum in PBS for 1 h and then incubated with primary antibodies against cTnT (Thermo Fisher Scientific), α-actinin (Abcam), and GFP (Thermo Fisher Scientific) overnight at 4 °C followed by incubation with appropriate Alexa fluorogenic secondary antibodies (Thermo Fisher Scientific) at room temperature for 1 h. cTnT, α-actinin, and cTnT/GFP–double positive cells were manually quantified by single-blind method from 10 randomly HPFs within each well.
Lineage-tracing experiments
Two lentiviral plasmids LV-Thy1.2-Cre pLKO.1 and pMSCV-loxp-dsRed-loxp-eGFP-Puro-WPRE were used in our cardiac fibroblast lineage-tracing experiments. The plasmids were transfected with psPAX2 and pMD2.G into HEK-293 cells. Lentivirus was harvested 2 days after transfection and purified using the method described above for retrovirus preparation. Isolated NCFs were infected with these two lentiviruses at day 1 of cardiac reprogramming. At day 2, NCFs were infected with retrovirus encoding Mef2c, Gata4, and Tbx5 to induce cardiac reprogramming and medium was changed every 2–3 days. Green cells with spontaneous beating were counted under fluorescence microscope after 2, 3, and 4 weeks.
Flow cytometry
Fluorescence flow cytometry data were from 10,000 single-cell events which were collected using a standard MoFloAstrios flow cytometer (Immunocytometry Systems; BD Biosciences). Data were analyzed using Summit (BD Biosciences).
Quantitative real time PCR (qPCR)
Total RNAs from all induced cells with 2-week GMT and IMAP treatment were extracted using TRIzol Reagent (Thermo Fisher Scientific) following the manufacturer's instructions. RNA integrity was determined using formaldehyde denaturalization agarose gel electrophoresis. RNA concentrations were measured with the NanoDrop spectrophotometer (Thermo Fisher Scientific). RNA was reverse transcribed using iScript cDNA Synthesis Kit (Bio-Rad). qPCR was performed using StepOne Real-Time PCR System (Thermo Fisher Scientific). Primer oligonucleotides were synthesized by Sigma and are listed in Table S1.
Spontaneous beating and calcium transient assessment
Spontaneous beating assessment was performed by light microscopy at room temperature after indicated treatment of NCFs at 4-week time point. Beating cell number was manually quantified by single-blind method after isoproterenol treatment in each well of 48-well plate.
Calcium transient was measured with Rhod-3 Calcium Imaging Kit (Thermo Fisher Scientific) according to the manufacturer's instructions. αMHC-Cre/Rosa26A-Flox-Stop-Flox-Gcamp3 NCFs were also used for calcium transient measurements as reported previously (12) which were performed by fluorescence microscopy at room temperature after the indicated treatment of retrovirus and chemicals. Three HPFs of view were randomly selected within each well, video recorded for 3 min, and manually quantified.
RNA-Seq
Total RNAs from all induced cells with 2-week GMT and IMAP treatment were isolated using TRIzol following the provider's instructions. RNA (RIN >8.5) was used for RNA-Seq library preparation using NEBNext® UltraTM II Directional RNA Library Prep Kit for Illumina (E7760S). The libraries were sequenced using HiSeq 4000 by the University of Michigan Sequencing Core. The quantification of RNA expression was estimated by Kallisto (48). Differential gene expression analysis was done using the R package DESeq2. The abundance of genes was used to calculate -fold change and p values. Cutoff values of -fold change greater than 2 and p values less than 0.05 were then used to select for differentially expressed genes between sample group comparisons. Significant pathway enrichment analysis was performed using PANTHER Overrepresentation Test (release 20150430, http://geneontology.org)4 (49, 50).
Statistic analysis
Results were presented as mean ± S.E. Statistically significant difference between groups was analyzed by one-way analysis of variance (ANOVA) followed by the Student-Newman-Keuls multiple comparisons tests. A p value <0.05 was regarded as significant. Each experiment was performed at least twice.
Author contributions
Y. G., L. L., and Z. W. conceptualization; Y. G., S. T., W. G., and L. L. data curation; Y. G. and I. L. formal analysis; Y. G. and L. L. validation; Y. G. visualization; Y. G. writing-original draft; I. L. software; I. L., S. T., W. G., K. H., Y. L., and S. W. methodology; K. H., Y. L., S. W., and Z. W. resources; L. L. and Z. W. investigation; L. L. and Z. W. project administration; L. L. and Z. W. writing-review and editing; Z. W. supervision; Z. W. funding acquisition.
Supplementary Material
This work was supported by the National Institutes of Health Grant HL109054 and a Pilot Award from the Joint Institute of University of Michigan Health System and Peking University Health Science Center (to Z. W.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S3, Table S1, and Movies S1–S8.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- CM
- cardiomyocyte
- iCM
- induced CM
- FGF
- fibroblast growth factor
- IGF
- insulin-like growth factor
- TGF
- transforming growth factor
- IMAP
- insulin-like growth factor-1, Mll1 inhibitor MM589, transforming growth factor-β inhibitor A83–01, and Bmi1 inhibitor PTC-209
- MEF
- mouse embryonic fibroblasts
- α-MHC
- α-muscle heavy chain
- MGT
- Mef2c/Gata4/Tbx5
- qPCR
- quantitative PCR
- cTnT
- cardiac troponin T
- HPF
- high power fields
- NCFs
- neonatal cardiac fibroblasts
- GO
- gene ontology.
References
- 1. Benjamin E. J., Blaha M. J., Chiuve S. E., Cushman M., Das S. R., Deo R., de Ferranti S. D., Floyd J., Fornage M., Gillespie C., Isasi C. R., Jiménez M. C., Jordan L. C., Judd S. E., Lackland D., et al. (2017) Heart disease and stroke statistics—2017 update: A report from the American Heart Association. Circulation 135, e146–e603 10.1161/CIR.0000000000000485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cahill T. J., Choudhury R. P., and Riley P. R. (2017) Heart regeneration and repair after myocardial infarction: Translational opportunities for novel therapeutics. Nat. Rev. Drug Discov. 16, 699–717 10.1038/nrd.2017.106 [DOI] [PubMed] [Google Scholar]
- 3. Kim B. S., Yang J. H., Jang W. J., Song Y. B., Hahn J. Y., Choi J. H., Kim W. S., Lee Y. T., Gwon H. C., Lee S. H., and Choi S. H. (2015) Clinical outcomes of multiple chronic total occlusions in coronary arteries according to three therapeutic strategies: Bypass surgery, percutaneous intervention and medication. Int. J. Cardiol. 197, 2–7 10.1016/j.ijcard.2015.06.017 [DOI] [PubMed] [Google Scholar]
- 4. Ieda M., Fu J. D., Delgado-Olguin P., Vedantham V., Hayashi Y., Bruneau B. G., and Srivastava D. (2010) Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142, 375–386 10.1016/j.cell.2010.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Song K., Nam Y. J., Luo X., Qi X., Tan W., Huang G. N., Acharya A., Smith C. L., Tallquist M. D., Neilson E. G., Hill J. A., Bassel-Duby R., and Olson E. N. (2012) Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature 485, 599–604 10.1038/nature11139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Qian L., Huang Y., Spencer C. I., Foley A., Vedantham V., Liu L., Conway S. J., Fu J. D., and Srivastava D. (2012) In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature 485, 593–598 10.1038/nature11044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Addis R. C., Ifkovits J. L., Pinto F., Kellam L. D., Esteso P., Rentschler S., Christoforou N., Epstein J. A., and Gearhart J. D. (2013) Optimization of direct fibroblast reprogramming to cardiomyocytes using calcium activity as a functional measure of success. J. Mol. Cell Cardiol. 60, 97–106 10.1016/j.yjmcc.2013.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Christoforou N., Chakraborty S., Kirkton R. D., Adler A. F., Addis R. C., and Leong K. W. (2017) Core transcription factors, microRNAs, and small molecules drive transdifferentiation of human fibroblasts towards the cardiac cell lineage. Sci. Rep. 7, 40285 10.1038/srep40285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fu J. D., Stone N. R., Liu L., Spencer C. I., Qian L., Hayashi Y., Delgado-Olguin P., Ding S., Bruneau B. G., and Srivastava D. (2013) Direct reprogramming of human fibroblasts toward a cardiomyocyte-like state. Stem Cell Reports 1, 235–247 10.1016/j.stemcr.2013.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wada R., Muraoka N., Inagawa K., Yamakawa H., Miyamoto K., Sadahiro T., Umei T., Kaneda R., Suzuki T., Kamiya K., Tohyama S., Yuasa S., Kokaji K., Aeba R., Yozu R., Yamagishi H., Kitamura T., Fukuda K., and Ieda M. (2013) Induction of human cardiomyocyte-like cells from fibroblasts by defined factors. Proc. Natl. Acad. Sci. U.S.A. 110, 12667–12672 10.1073/pnas.1304053110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Christoforou N., Chellappan M., Adler A. F., Kirkton R. D., Wu T., Addis R. C., Bursac N., and Leong K. W. (2013) Transcription factors MYOCD, SRF, Mesp1 and SMARCD3 enhance the cardio-inducing effect of GATA4, TBX5, and MEF2C during direct cellular reprogramming. PLoS One 8, e63577 10.1371/journal.pone.0063577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhou H., Dickson M. E., Kim M. S., Bassel-Duby R., and Olson E. N. (2015) Akt1/protein kinase B enhances transcriptional reprogramming of fibroblasts to functional cardiomyocytes. Proc. Natl. Acad. Sci. U.S.A. 112, 11864–11869 10.1073/pnas.1516237112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhao Y., Londono P., Cao Y., Sharpe E. J., Proenza C., O'Rourke R., Jones K. L., Jeong M. Y., Walker L. A., Buttrick P. M., McKinsey T. A., and Song K. (2015) High-efficiency reprogramming of fibroblasts into cardiomyocytes requires suppression of pro-fibrotic signalling. Nat. Commun. 6, 8243 10.1038/ncomms9243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Muraoka N., Yamakawa H., Miyamoto K., Sadahiro T., Umei T., Isomi M., Nakashima H., Akiyama M., Wada R., Inagawa K., Nishiyama T., Kaneda R., Fukuda T., Takeda S., Tohyama S., et al. (2014) MiR-133 promotes cardiac reprogramming by directly repressing Snai1 and silencing fibroblast signatures. EMBO J. 33, 1565–1581 10.15252/embj.201387605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ifkovits J. L., Addis R. C., Epstein J. A., and Gearhart J. D. (2014) Inhibition of TGFβ signaling increases direct conversion of fibroblasts to induced cardiomyocytes. PLoS One 9, e89678 10.1371/journal.pone.0089678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Abad M., Hashimoto H., Zhou H., Morales M. G., Chen B., Bassel-Duby R., and Olson E. N. (2017) Notch inhibition enhances cardiac reprogramming by increasing MEF2C transcriptional activity. Stem Cell Reports 8, 548–560 10.1016/j.stemcr.2017.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yamakawa H., Muraoka N., Miyamoto K., Sadahiro T., Isomi M., Haginiwa S., Kojima H., Umei T., Akiyama M., Kuishi Y., Kurokawa J., Furukawa T., Fukuda K., and Ieda M. (2015) Fibroblast growth factors and vascular endothelial growth factor promote cardiac reprogramming under defined conditions. Stem Cell Reports 5, 1128–1142 10.1016/j.stemcr.2015.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang L., Liu Z., Yin C., Asfour H., Chen O., Li Y., Bursac N., Liu J., and Qian L. (2015) Stoichiometry of Gata4, Mef2c, and Tbx5 influences the efficiency and quality of induced cardiac myocyte reprogramming. Circ. Res. 116, 237–244 10.1161/CIRCRESAHA.116.305547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jayawardena T. M., Finch E. A., Zhang L., Zhang H., Hodgkinson C. P., Pratt R. E., Rosenberg P. B., Mirotsou M., and Dzau V. J. (2015) MicroRNA induced cardiac reprogramming in vivo: Evidence for mature cardiac myocytes and improved cardiac function. Circ. Res. 116, 418–424 10.1161/CIRCRESAHA.116.304510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jayawardena T. M., Egemnazarov B., Finch E. A., Zhang L., Payne J. A., Pandya K., Zhang Z., Rosenberg P., Mirotsou M., and Dzau V. J. (2012) MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ. Res. 110, 1465–1473 10.1161/CIRCRESAHA.112.269035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cao N., Huang Y., Zheng J., Spencer C. I., Zhang Y., Fu J. D., Nie B., Xie M., Zhang M., Wang H., Ma T., Xu T., Shi G., Srivastava D., and Ding S. (2016) Conversion of human fibroblasts into functional cardiomyocytes by small molecules. Science 352, 1216–1220 10.1126/science.aaf1502 [DOI] [PubMed] [Google Scholar]
- 22. Dal-Pra S., Hodgkinson C. P., Mirotsou M., Kirste I., and Dzau V. J. (2017) Demethylation of H3K27 is essential for the induction of direct cardiac reprogramming by miR combo. Circ. Res. 120, 1403–1413 10.1161/CIRCRESAHA.116.308741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bektik E., Dennis A., Prasanna P., Madabhushi A., and Fu J. D. (2017) Single cell qPCR reveals that additional HAND2 and microRNA-1 facilitate the early reprogramming progress of seven-factor-induced human myocytes. PLoS One 12, e0183000 10.1371/journal.pone.0183000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Umei T. C., Yamakawa H., Muraoka N., Sadahiro T., Isomi M., Haginiwa S., Kojima H., Kurotsu S., Tamura F., Osakabe R., Tani H., Nara K., Miyoshi H., Fukuda K., and Ieda M. (2017) Single-construct polycistronic doxycycline-inducible vectors improve direct cardiac reprogramming and can be used to identify the critical timing of transgene expression. Int. J. Mol. Sci. 18, E1805 10.3390/ijms18081805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhou H., Morales M. G., Hashimoto H., Dickson M. E., Song K., Ye W., Kim M. S., Niederstrasser H., Wang Z., Chen B., Posner B. A., Bassel-Duby R., and Olson E. N. (2017) ZNF281 enhances cardiac reprogramming by modulating cardiac and inflammatory gene expression. Genes Dev. 31, 1770–1783 10.1101/gad.305482.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mohamed T. M., Stone N. R., Berry E. C., Radzinsky E., Huang Y., Pratt K., Ang Y. S., Yu P., Wang H., Tang S., Magnitsky S., Ding S., Ivey K. N., and Srivastava D. (2017) Chemical enhancement of in vitro and in vivo direct cardiac reprogramming. Circulation 135, 978–995 10.1161/CIRCULATIONAHA.116.024692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu Z., Wang L., Welch J. D., Ma H., Zhou Y., Vaseghi H. R., Yu S., Wall J. B., Alimohamadi S., Zheng M., Yin C., Shen W., Prins J. F., Liu J., and Qian L. (2017) Single-cell transcriptomics reconstructs fate conversion from fibroblast to cardiomyocyte. Nature 551, 100–104 10.1038/nature24454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhou Y., Wang L., Vaseghi H. R., Liu Z., Lu R., Alimohamadi S., Yin C., Fu J. D., Wang G. G., Liu J., and Qian L. (2016) Bmi1 is a key epigenetic barrier to direct cardiac reprogramming. Cell Stem Cell 18, 382–395 10.1016/j.stem.2016.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhou Y., Alimohamadi S., Wang L., Liu Z., Wall J. B., Yin C., Liu J., and Qian L. (2018) A loss of function screen of epigenetic modifiers and splicing factors during early stage of cardiac reprogramming. Stem Cells Int. 2018, 3814747 10.1155/2018/3814747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang L., Liu Z., Yin C., Zhou Y., Liu J., and Qian L. (2015) Improved generation of induced cardiomyocytes using a polycistronic construct expressing optimal ratio of Gata4, Mef2c and Tbx5. J. Vis. Exp. 105, e53426 10.3791/53426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kreso A., van Galen P., Pedley N. M., Lima-Fernandes E., Frelin C., Davis T., Cao L., Baiazitov R., Du W., Sydorenko N., Moon Y. C., Gibson L., Wang Y., Leung C., Iscove N. N., et al. (2014) Self-renewal as a therapeutic target in human colorectal cancer. Nat. Med. 20, 29–36 10.1038/nm.3418 [DOI] [PubMed] [Google Scholar]
- 32. Belperio J. A., Dy M., Murray L., Burdick M. D., Xue Y. Y., Strieter R. M., and Keane M. P. (2004) The role of the Th2 CC chemokine ligand CCL17 in pulmonary fibrosis. J. Immunol. 173, 4692–4698 10.4049/jimmunol.173.7.4692 [DOI] [PubMed] [Google Scholar]
- 33. Seki E., De Minicis S., Gwak G. Y., Kluwe J., Inokuchi S., Bursill C. A., Llovet J. M., Brenner D. A., and Schwabe R. F. (2009) CCR1 and CCR5 promote hepatic fibrosis in mice. J. Clin. Invest. 119, 1858–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pignatti P., Brunetti G., Moretto D., Yacoub M. R., Fiori M., Balbi B., Balestrino A., Cervio G., Nava S., and Moscato G. (2006) Role of the chemokine receptors CXCR3 and CCR4 in human pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 173, 310–317 10.1164/rccm.200502-244OC [DOI] [PubMed] [Google Scholar]
- 35. Ghosh M. C., Collins G. D., Vandanmagsar B., Patel K., Brill M., Carter A., Lustig A., Becker K. G., Wood W. W. 3rd, Emeche C. D., French A. D., O'Connell M. P., Xu M., Weeraratna A. T., and Taub D. D. (2009) Activation of Wnt5A signaling is required for CXC chemokine ligand 12-mediated T-cell migration. Blood 114, 1366–1373 10.1182/blood-2008-08-175869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Graves D. T., and Jiang Y. (1995) Chemokines, a family of chemotactic cytokines. Crit. Rev. Oral Biol. Med. 6, 109–118 10.1177/10454411950060020101 [DOI] [PubMed] [Google Scholar]
- 37. Bouzaffour M., Dufourcq P., Lecaudey V., Haas P., and Vriz S. (2009) Fgf and Sdf-1 pathways interact during zebrafish fin regeneration. PLoS One 4, e5824 10.1371/journal.pone.0005824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chong S. W., Nguyet L. M., Jiang Y. J., and Korzh V. (2007) The chemokine Sdf-1 and its receptor Cxcr4 are required for formation of muscle in zebrafish. BMC Dev. Biol. 7, 54 10.1186/1471-213X-7-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Velecela V., Lettice L. A., Chau Y. Y., Slight J., Berry R. L., Thornburn A., Gunst Q. D., van den Hoff M., Reina M., Martínez F. O., Hastie N. D., and Martínez-Estrada O. M. (2013) WT1 regulates the expression of inhibitory chemokines during heart development. Hum. Mol. Genet. 22, 5083–5095 10.1093/hmg/ddt358 [DOI] [PubMed] [Google Scholar]
- 40. Liu W., and Foley A. C. (2011) Signaling pathways in early cardiac development. Wiley Interdiscip. Rev. Syst. Biol. Med. 3, 191–205 10.1002/wsbm.112 [DOI] [PubMed] [Google Scholar]
- 41. Ceholski D. K., Turnbull I. C., Pothula V., Lecce L., Jarrah A. A., Kho C., Lee A., Hadri L., Costa K. D., Hajjar R. J., and Tarzami S. T. (2017) CXCR4 and CXCR7 play distinct roles in cardiac lineage specification and pharmacologic β-adrenergic response. Stem Cell Res. 23, 77–86 10.1016/j.scr.2017.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cavalera M., and Frangogiannis N. G. (2014) Targeting the chemokines in cardiac repair. Curr. Pharm. Des. 20, 1971–1979 10.2174/13816128113199990449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yang C. S., Lopez C. G., and Rana T. M. (2011) Discovery of nonsteroidal anti-inflammatory drug and anticancer drug enhancing reprogramming and induced pluripotent stem cell generation. Stem Cells 29, 1528–1536 10.1002/stem.717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mah N., Wang Y., Liao M. C., Prigione A., Jozefczuk J., Lichtner B., Wolfrum K., Haltmeier M., Flöttmann M., Schaefer M., Hahn A., Mrowka R., Klipp E., Andrade-Navarro M. A., and Adjaye J. (2011) Molecular insights into reprogramming-initiation events mediated by the OSKM gene regulatory network. PLoS One 6, e24351 10.1371/journal.pone.0024351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang H., Cao N., Spencer C. I., Nie B., Ma T., Xu T., Zhang Y., Wang X., Srivastava D., and Ding S. (2014) Small molecules enable cardiac reprogramming of mouse fibroblasts with a single factor, Oct4. Cell Rep. 6, 951–960 10.1016/j.celrep.2014.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fu Y., Huang C., Xu X., Gu H., Ye Y., Jiang C., Qiu Z., and Xie X. (2015) Direct reprogramming of mouse fibroblasts into cardiomyocytes with chemical cocktails. Cell Res. 25, 1013–1024 10.1038/cr.2015.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jozefczuk J., Drews K., and Adjaye J. (2012) Preparation of mouse embryonic fibroblast cells suitable for culturing human embryonic and induced pluripotent stem cells. J. Vis. Exp. 64, e3854 10.3791/3854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bray N. L., Pimentel H., Melsted P., and Pachter L. (2016) Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- 49. Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., et al. (2000) Gene Ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. The Gene Ontology Consortium (2017) Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 45, D331–D338 10.1093/nar/gkw1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.