Abstract
The AURORA pathway participates in mitosis and cell division, and alterations in mitosis and cell division can lead to carcinogenesis. Therefore, genetic variants in the AURORA pathway genes may be associated with susceptibility to pancreatic cancer. To test this hypothesis, we used three large publically available pancreatic cancer genome-wide association study (GWAS) datasets (PanScan I, II/III and PanC4) to assess the associations of 7168 single nucleotide polymorphisms (SNPs) in a set of 62 genes of this pathway with pancreatic cancer risk in 8477 cases and 6946 controls of European ancestry. We identify 15 significant pancreatic cancer risk-associated SNPs in three genes (SMC2, ARHGEF7 and TP53) after correction for multiple comparisons by a false discovery rate < 0.20. Through further linkage disequilibrium analysis, SNP functional prediction and stepwise logistic regression analysis, we focused on three SNPs: rs3818626 in SMC2, rs79447092 in ARHGEF7 and rs9895829 in TP53. We found that these three SNPs were associated with pancreatic cancer risk [odds ratio (OR) = 1.12, 95% confidence interval (CI) = 1.07–1.17 and P = 2.20E-06 for the rs3818626 C allele; OR = 0.76, CI = 0.66–0.88 and P = 1.46E-04 for the rs79447092 A allele and OR = 0.82, CI = 0.74–0.91 and P = 1.51E-04 for the rs9895829 G allele]. Their joint effect as the number of protective genotypes also showed a significant association with pancreatic cancer risk (trend test P ≤ 0.001). Finally, we performed an expression quantitative trait loci analysis and found that rs3818626 and rs9895829 were significantly associated with SMC2 and TP53 messenger RNA expression levels in 373 lymphoblastoid cell lines, respectively. In conclusion, these three representative SNPs may be potentially susceptibility loci for pancreatic cancer and warrant additional validation.
Introduction
Pancreatic cancer is a highly lethal malignancy and estimated to cause approximately 43 090 cancer-related deaths in the USA in 2017 (1). Some environmental factors, such as cigarette smoking, alcohol intake, diabetes, obesity and chronic pancreatitis have been identified as risk factors for pancreatic cancer (2,3). Genetic factors are also known to play an important role in pancreatic cancer etiology. For example, germline mutations in BRCA2, PALB2, CDKN2A, ATM, STK11, PRSS1, SPINK1 and DNA mismatch repair genes have been reported to be involved in pancreatic carcinogenesis (4–9).
Other genetic factors, such as common single nucleotide polymorphisms (SNPs), are also reported to be associated with pancreatic cancer risk, in several prior genome-wide association studies (GWASs) (10–13). Many pancreatic cancer susceptibility loci have been identified, such as 1q32.1(NR5A2), 2p13.3 (ETAA1), 3q29 (TP63), 5p15.33 (TERT, CLPTM1), 7p13 (SUGCT), 7q32.3(LINC-PINT), 8q24.21(MYC), 9q34.2(ABO), 13q12.2 (PDX1), 13q22.1(KLF5), 16q23.1(BCAR1), 17q25.1 (LINC00673) and 22q12.1 (ZNRF3), particularly in European populations (10–13). However, many of the SNPs identified by GWAS are not functionally related to possible mechanisms associated with the disease, and identification of the causal alleles that provide a clue to biologically plausible genes and pathways remains difficult. Therefore, we sought to perform a pathway-based analysis as a hypothesis-driven approach with fewer SNPs selected from available GWAS data sets to reduce multiple tests and also to identify possible functional SNPs associated with pancreatic cancer risk. We have applied this approach in lung cancer research, having identifed previously unreported susceptibility loci in genes involved in the pathways of centrosome (14), DNA repair (15), LncRNA (16) and RNA degradation (17). In the present study, we applied this pathway-based approach to investigate the associations between genetic variants of the gene set involved in the AURORA pathway and pancreatic cancer risk.
Genomic instability is one of the known cancer hallmarks that provide a driving power for cancer initiation and development. Aneuploidy and chromosome instability are two forms of genomic instability, regulated by a number of cell-cycle-dependent kinases (18–20), of which mitotic kinases play a key role in mitosis checkpoints and the maintenance of chromosome integrity and segregation. The Aurora kinases are a family of mitotic serine threonine/kinases including three members: Aurora A, B and C that participate in mitosis and cell division, including centrosome duplication, spindle formation, chromosome alignment, checkpoint activation and cytokinesis (21,22). Studies showed that overexpression of one mitotic kinase, Aurora A, can lead to centrosome amplification, inducing chromosomal instability (23,24). The Aurora kinases have been reported to be overexpressed in a wide range of human cancers, including pancreatic cancer (25,26), and thus targeted for the treatment of pancreatic cancer (27). Previous studies also revealed that a genetic variant in Aurora A was associated with risks of multiple cancers (28).
Some studies suggested that TP53 (29,30) and BIRC5 (31) play a role in the AURORA signaling pathway and thus are likely to be involved in pancreatic carcinogenesis. Other studies showed that MDM2 (32) and AKT1 (33) in this pathway were associated with tumor progression of pancreatic cancer. However, these studies did not include other related genes or SNPs of genes involved in the AURORA pathway. In the present study, we comprehensively investigated associations between common genetic variants of all possible genes likely to be involved in the AURORA pathway and pancreatic cancer risk.
Methods and materials
Study subjects
We used the genotyping data of participants of European ancestry from two published GWASs, which were downloaded from the dbGaP (the database of Genotypes and Phenotypes) website: the PanScan study (dbGap#: phs000206.v5. p3) and the Pancreatic Cancer Case Control Association Study (dbGaP #: phs000648. v1. p1) (34,35). The ancestry information was imputed based on principal component analysis and self-reported in former and latter studies, respectively. The PanScan GWAS was previously performed in three phases: PanScan I, II and III (1921 cases and 2016 controls in PanScan I; 1754 cases and 1889 controls in PanScan II; 1538 cases and 0 controls in PanScan III) (10–12). We merged the PanScan II and PanScan III into one data set ‘PanScan II/III’ because the control data in PanScan III were not found in dbGaP. The other Pancreatic Cancer Case Control Association Study from the Pancreatic Cancer Case-Control consortium (PanC4) includes 4168 cases and 3814 controls (13,36,37). Therefore, these three data sets (PanScan I, PanScan II/III and PanC4) from dbGAP included a total of 15 423 individuals (8477 cases and 6946 controls) for the final analysis. All the cases were diagnosed with a primary adenocarcinoma of the exocrine pancreas. A written informed consent was obtained from all participants in the original GWASs. All the original studies were performed in accordance with the relevant guidelines and regulations for each of the participating institutions, and the present study followed the study protocols approved by the Duke University Health System Institutional Review Board. Supplementary Table S1, available at Carcinogenesis Online, showed the distributions of demographic characteristics of the three GWAS data sets.
Selection of SNPs in the gene set of the AURORA pathway
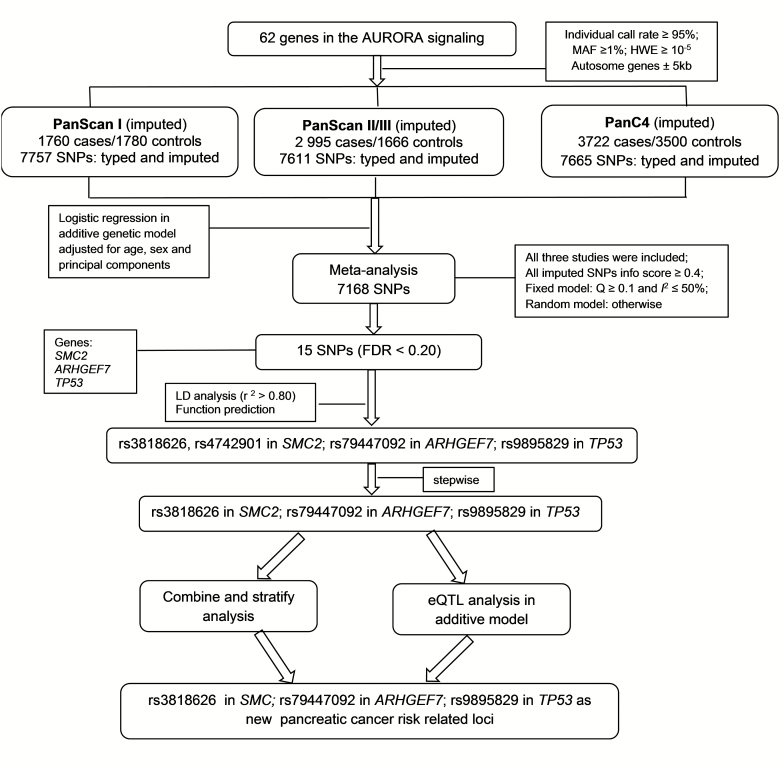
Genes in the AURORA pathway were selected from the Molecular Signatures Database (C2) (38). Overall, a set of 62 genes involved in the AURORA pathway from the PID data set were selected (details presented in Supplementary Table S2, available at Carcinogenesis Online). SNPs located in these genes and their ±5 kb flanking regions were extracted from the original GWAS data sets based on the following selection criteria (1): minor allele frequency ≥1% (2), genotyping rate ≥95% and (3) Hardy–Weinberg Equilibrium exact P-value ≥10–5. We used IMPUTE2 v2.1.1 software to impute untyped SNPs in our target regions, using a 500 kb buffer in our case-control data and the 1000 Genomes Project data (phase 3, released October 2015) as the imputation reference panel. After imputation, we extracted 7757, 7611 and 7665 SNPs within 5 kb up- and down-streams of genes in the AURORA pathway from populations of the PanScan I, PanScan II/III and PanC4, respectively. The final studies meta-analysis contained 7168 SNPs for each of the data sets with imputation quality (info) >0.4. The detailed workflow is shown in Figure 1.
Figure 1.
Flowchart of SNP selection among the AURORA pathway genes.
Functional prediction and validation
SNPinfo (39), RegulomeDB (40) and HaploReg (41) were used to predict SNP-associated potential functions. The expression quantitative trait loci (eQTL) analysis was performed by using the genotyping and expression data from the lymphoblastoid cell lines of 373 European individuals from Genetic European Variation in Health and Disease Consortium (GEUVADIS) and the 1000 Genomes Project (phase I integrated release 3, March 2012) (42). We also tested the correlations between the identified SNPs and the corresponding genes’ expression levels in normal pancreatic tissues using the online GTEx database (https://gtexportal.org)
Statistical analysis
We performed an unconditional logistic regression analysis with the PLINK (v1.90) software to estimate odds ratios (ORs) and their 95% confidence intervals (CIs) by using the genotyping data and the best-guess genotypes from imputation (43,44). Age, sex and top significant principal components were adjusted for in logistic regression models, including the top five and seven significant principal components in the analysis of PanScan I/II/III data and PanC4 data, respectively. A meta-analysis was performed for the selected 7168 SNPs with Stata software (v12; StataCorp, State College, TX). We tested for the heterogeneity among the data sets by using the Cochran’s Q statistic and investigated the proportion of the total variation by the I2 statistic. When there was no heterogeneity among the GWAS data sets (Q-test P > 0.100 and I2 < 50%), we used the fixed-effects model; otherwise, we used the random-effects model. We also performed the gene-based test by using the versatile gene-based association study (VEGAS) approach integrated in the VEGAS2 program (45,46). Briefly, for a given gene with n low linkage disequilibrium (LD) SNPs, the association P-values were first converted to one chi-squared statistics with one degree of freedom. The gene-based test statistic was then calculated by adding up all of the chi-squared statistics within that gene. A large number of simulations were performed by using the multivariate normal distribution, and the empirical gene-based P-value is the proportion of the simulated test statistics that exceeded the observed gene-based test statistic. We controlled for multiple testing with a threshold of a false discovery rate (FDR) of <0.20. LocusZoom (http://locuszoom.sph.umich.edu/locuszoom/) (reference version: 1000 Genomes, Nov 24, 2014; EUR) was applied to generate the regional association plots (47). Manhattan plot and LD plot were generated by Haploview v4.2 (48). Finally, the joint effect analysis, stratified analysis and stepwise analysis were conducted with SAS (Version 9.3; SAS Institute, Cary, NC).
Results
Association analysis using three GWAS data sets
We first performed logistic regression analysis to estimate the associations between common SNPs (minor allele frequency > 0.01) and pancreatic cancer risk in the three available pancreatic cancer GWAS data sets. There were 7757, 7611 and 7665 SNPs in PanScan I, PanScan II/III and PanC4 data sets, respectively. As a result, 7168 SNPs from a set of 62 genes were included in a meta-analysis. All the associations between SNPs of these genes and pancreatic cancer risk as identified by the single-locus analysis are presented in a Manhattan plot (Supplementary Figure S1, available at Carcinogenesis Online). Overall, 15 SNPs in three genes (SMC2, ARHGEF7 and TP53) passed the multiple-testing correction by FDR <0.20. It should be mentioned that only the six SNPs in SMC2 had passed the Bonferroni correction, which is a more stringent test assuming that all the tested SNPs are independent. The SNPs’ locations and their associations with pancreatic cancer risk are presented in Table 1. The results of the meta-analysis with a random-effects model and the imputation qualities of these SNPs are shown in Supplementary Table S3, available at Carcinogenesis Online. The chromosome regions of the three genes are novel findings, because they were not previously reported by any of these three pancreatic cancer GWASs, and therefore we performed further functional analysis. By using the VEGAS method, we performed the gene-based test and found seven genes (SMC2, KIF20A, RHOA, TP53, EVI5, AURKC and NCAPD2) with an empirical P-value <0.05 (Supplementary Table S4, available at Carcinogenesis Online), four of which passed the multiple testing correction with an FDR <0.2. However, no significance was found for ARHGEF7.
Table 1.
Associations between SNPs in the AURORA pathway and pancreatic cancer risk with FDR <0.20
| SNP | Gene | Chr | Allelea | Position (hg19) |
I 2 | EAFb | OR (95% CI)c | P c | FDR |
|---|---|---|---|---|---|---|---|---|---|
| rs10820603 | SMC2 | 9 | A/G | 106877939 | 0 | 0.44 | 1.12 (1.07–1.18) | 8.39E-07 | 0.003 |
| rs7872034 | SMC2 | 9 | A/G | 106896809 | 0 | 0.44 | 1.12 (1.07–1.17) | 9.97E-07 | 0.003 |
| rs3818626 | SMC2 | 9 | T/C | 106856633 | 0 | 0.44 | 1.12 (1.07–1.17) | 2.20E-06 | 0.003 |
| rs4743687 | SMC2 | 9 | T/C | 106856910 | 0 | 0.44 | 1.12 (1.07–1.17) | 1.97E-06 | 0.003 |
| rs4742906 | SMC2 | 9 | G/A | 106857078 | 0 | 0.44 | 1.12 (1.07–1.17) | 1.33E-06 | 0.003 |
| rs7028408 | SMC2 | 9 | A/G | 106859811 | 0 | 0.44 | 1.12 (1.07–1.17) | 2.12E-06 | 0.003 |
| rs4742901 | SMC2 | 9 | T/C | 106856043 | 8.87 | 0.29 | 1.10 (1.04–1.15) | 2.92E-04 | 0.149 |
| rs79447092 | ARHGEF7 | 13 | T/A | 111809308 | 0 | 0.03 | 0.76 (0.66–0.88) | 1.46E-04 | 0.108 |
| rs17884306 | TP53 | 17 | C/T | 7572101 | 0 | 0.06 | 0.82 (0.74–0.91) | 1.45E-04 | 0.108 |
| rs9891744 | TP53 | 17 | C/T | 7574864 | 0 | 0.06 | 0.81 (0.73–0.90) | 1.26E-04 | 0.108 |
| rs9895829 | TP53 | 17 | A/G | 7578679 | 0 | 0.06 | 0.82 (0.74–0.91) | 1.51E-04 | 0.108 |
| rs17883323 | TP53 | 17 | G/T | 7579619 | 0 | 0.06 | 0.82 (0.74–0.91) | 1.77E-04 | 0.111 |
| rs8079544 | TP53 | 17 | C/T | 7580052 | 0 | 0.06 | 0.82 (0.74–0.91) | 1.86E-04 | 0.111 |
| rs75732100 | TP53 | 17 | C/T | 7576348 | 0 | 0.06 | 0.82 (0.74–0.91) | 2.29E-04 | 0.126 |
| rs17879377 | TP53 | 17 | C/T | 7574721 | 0 | 0.05 | 0.82 (0.73–0.91) | 3.28E-04 | 0.157 |
Chr, chromosome; EAF, effect allele frequency; FDR, false discovery rate.
aReference allele/effect allele.
bEAF in the controls of three studies (PanScan I, PanScan II/III and PanC4).
cMeta-analysis of the three studies: Fixed effect models were used when no heterogeneity was found between studies (Q test P > 0.10 and I2 < 50.0%); otherwise, random effect models were used.
LD and functional prediction
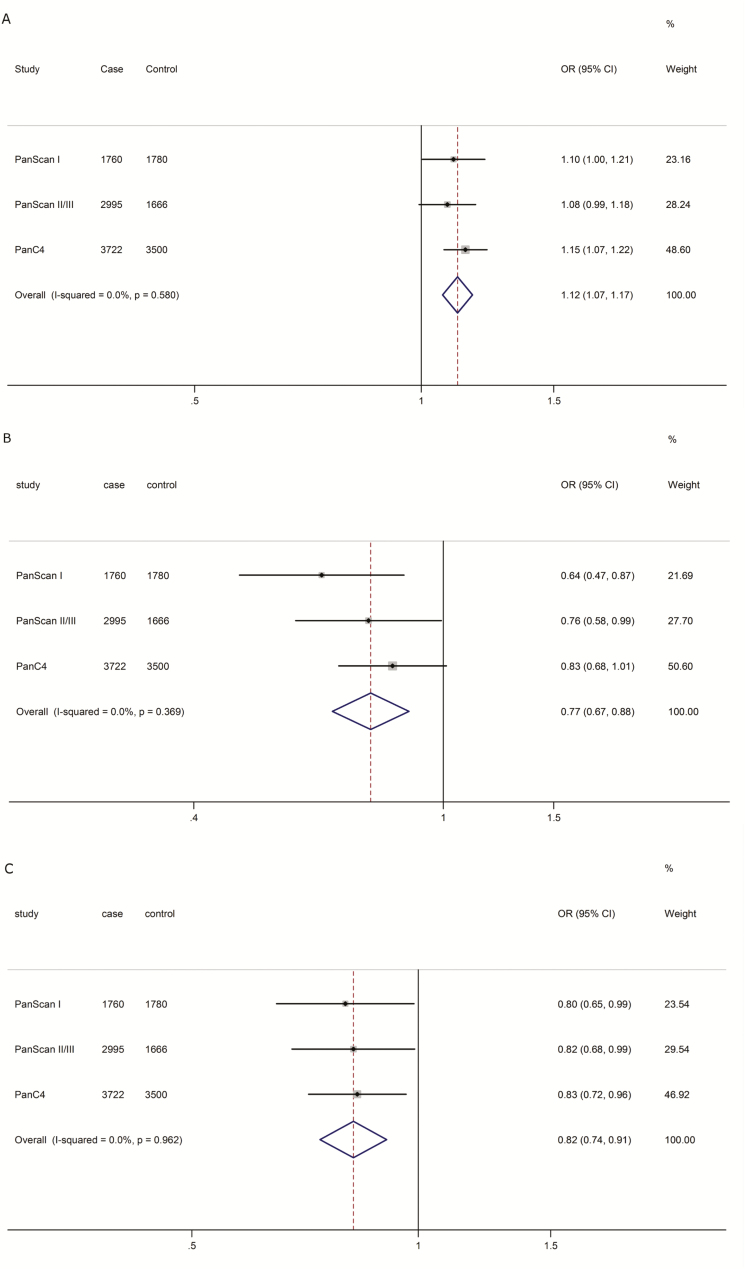
Based on the LD analysis (r2 > 0.80) (Supplementary Figure S2 d and e, available at Carcinogenesis Online) and in silico SNP functional prediction (SNPinfo, RegulomeDB and HaploReg) (Supplementary Table S5, available at Carcinogenesis Online), we chose four representative SNPs (i.e. rs3818626 and rs4742901 in SMC2, rs79447092 in ARHGEF7 and rs9895829 in TP53) for further analyses. Then, we employed the multivariate stepwise logistic regression analysis with adjustment for age, sex and data source (components). As a result, three representative SNPs (i.e. rs3818626 in SMC2, rs79447092 in ARHGEF7 and rs9895829 in TP53) remained statistically significantly associated with pancreatic cancer risk (Supplementary Table S6, available at Carcinogenesis Online). Regional association plots of the three SNPs in the 200 kb up- and down-stream regions are shown in Supplementary Figure S2 a––c , available at Carcinogenesis Online. The final meta-analysis results of three representative SNPs are summarized in Figure 2. We also presented individual association results for each of the three identified SNPs in the three GWAS data sets, which shows that our results are consistent across the three data sets (Supplementary Table S7, available at Carcinogenesis Online) and thus reliable.
Figure 2.
Forest plots of the effect sizes and directions for the three representative SNPs [(a) rs3818626 in SMC2; (b) rs79447092 in ARHGEF7; (c) rs9895829 in TP53] in the AURORA pathway.
Potentially functional SNPs and pancreatic cancer risk
We performed risk analysis with different genetic models for each of representative SNPs by using logistic regression analysis (Table 2). We found that rs3818626 in SMC2 was associated with an increased pancreatic cancer risk, whereas rs79447092 in ARHGEF7 and TP53 rs9895829 were associated with a decreased pancreatic cancer risk in both additive and dominant models.
Table 2.
Associations between the top three representative SNPs and pancreatic cancer risk in the combined data set of PanScan and PanC4 studies
| Genotype | PanScan + PanC4 | OR (95% CI)a | P a | ||
|---|---|---|---|---|---|
| Case (%) | Control (%) | ||||
| rs3818626 T>C | |||||
| TT | 2402 (28.4) | 2182 (31.5) | 1.00 | ||
| TC | 4247 (50.2) | 3427 (49.4) | 1.13 (1.05–1.21) | 0.001 | |
| CC | 1808 (21.4) | 1323 (19.1) | 1.24 (1.13–1.36) | <0.001 | |
| Trend test | <0.001 | ||||
| Dominant model | |||||
| TT | 2402 (28.4) | 2182 (31.5) | 1.00 | ||
| TC+CC | 6055 (71.6) | 4750 (68.5) | 1.16 (1.08–1.24) | <0.001 | |
| rs79447092 T>A | |||||
| TT | 7944 (95.2) | 6421 (94.0) | 1.00 | ||
| TA | 394 (4.7) | 395 (5.8) | 0.80 (0.69–0.93) | 0.003 | |
| AA | 4 (0.1) | 12 (0.2) | 0.29 (0.09–0.90) | 0.032 | |
| Trend test | <0.001 | ||||
| Dominant model | |||||
| TT | 7944 (95.2) | 6421 (94.0) | 1.00 | ||
| TA+AA | 398 (4.8) | 407 (6.0) | 0.79 (0.68–0.91) | 0.001 | |
| rs9895829 A>G | |||||
| AA | 7675 (90.6) | 6153 (88.6) | 1.00 | ||
| AG | 775 (9.2) | 769 (11.1) | 0.81 (0.73–0.90) | <0.001 | |
| GG | 17 (0.2) | 20 (0.3) | 0.62 (0.32–1.19) | 0.149 | |
| Trend test | <0.001 | ||||
| Dominant model | |||||
| AA | 7675 (90.6) | 6153 (88.6) | 1.00 | ||
| AG+GG | 792 (9.4) | 789 (11.4) | 0.81 (0.73–0.90) | <0.001 |
aAdjusted for age, sex and data source.
To evaluate the joint effect of these three representative SNPs on pancreatic cancer risk, we combined the number of protective genotypes (NPGs) of rs3818626 TT, rs79447092 TA+AA and rs9895829 AG+GG into a genetic score and divided all the patients into four groups: 0–3 risk genotypes, and we found that there was a significant association between increased NPGs and pancreatic cancer risk in a dominant model (Table 3). Then, all participants were divided into a low-protection group (0 NPGs) and a high-protection group (1–3 NPGs). We found that the high-protection group had a significantly decreased cancer risk (Table 3), compared with the low-protection group.
Table 3.
Associations between number of protective genotypes (NPGs) and risk of pancreatic cancera
| NPGb | PanScan + PanC4 | OR (95% CI)c | P c | |
|---|---|---|---|---|
| Case (%) | Control (%) | |||
| 0 | 5137 (61.8) | 3900 (57.3) | 1.00 | |
| 1 | 2827 (34.0) | 2505 (36.8) | 0.86 (0.80–0.92) | <0.001 |
| 2 | 342 (4.1) | 397 (5.8) | 0.66 (0.56–0.76) | <0.001 |
| 3 | 7 (0.1) | 8 (0.1) | 0.78 (0.28–2.16) | 0.634 |
| Trend test | <0.001 | |||
| 0 | 5137 (61.8) | 3900 (57.3) | 1.00 | |
| 1–3 | 3176 (38.2) | 2910 (42.7) | 0.83 (0.78–0.88) | <0.001 |
NPG, number of protective genotypes.
aThe logistic regression analysis was performed in the combined data set of PanScan and PanC4 studies.
bProtective genotypes were rs3818626 TT, rs79447092 TA+AA and rs9895829 AG+GG.
cLogistic regression analyses with adjustment for age, sex and data source.
Stratified analysis of combined protective genotypes and pancreatic cancer
To further analyze the interactive effect in associations between genotypes and pancreatic cancer risk, we performed stratified analysis by age and sex. In subgroup analysis by age (Supplementary Table S8, available at Carcinogenesis Online), we found that the high-protection group had a significantly decreased cancer risk in all age subgroups (<60, 60–70 and >70) and both sex groups (male: OR = 0.86, 95% CI = 0.78–0.94 and P < 0.001; female: OR = 0.80, 95% CI = 0.72–0.88 and P < 0.001; Supplementary Table S8, available at Carcinogenesis Online) compared with the low-protection group. There were no differences in the risk among these subgroups.
Functional validation by the eQTL analysis
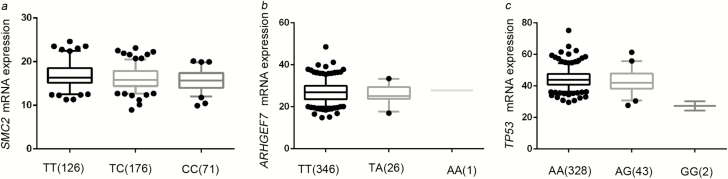
We further performed the eQTL analysis to assess the associations between the representative SNPs and their messenger RNA (mRNA) expression levels, and we found that SMC2 mRNA expression levels significantly decreased as the number of the rs3818626 risk alleles (C) increased in an additive model (P = 0.0007) (Figure 3a). The eQTL analysis result of rs9895829 in TP53 was also significant in an additive model, demonstrating that the protective (A) allele was associated with higher TP53 expression levels (P = 0.005) (Figure 3c). However, we did not find such a correlation for rs79447092 in an additive model (Figure 3b). We have also tested the correlations between genotypes of the three identified SNPs and the corresponding genes’ expression levels in the GTEx database (https://gtexportal.org), but we did not find significant results (Supplementary Table S9, available at Carcinogenesis Online).
Figure 3.
The correlations between the identified SNPs (a. rs3818626; b. rs79447092; c. rs9895829) and mRNA expression levels of the associated genes.
Discussion
In the present study, we investigated the associations between genetic variants in the AURORA pathway and pancreatic cancer risk using two published GWASs (PanScan study and PanC4 study). We found that three novel SNPs associations (i.e. rs3818626 in SMC2, rs79447092 in ARHGEF7 and rs9895829 in TP53) were independently and jointly associated with pancreatic cancer risk. Further functional analyses showed that rs3818626 and rs9895829 were significantly associated with decreased mRNA expression levels of SMC2 and TP53, respectively.
The structural maintenance of chromosome 2 (SMC2) protein product belongs to the condensin complex and plays an important role in packaging of chromatin before cell division and DNA damage response, which is required for proper chromosome segregation and maintenance of chromosomal stability (49). SMC2 plays a dual role in development and progression of cancer. For example, emerging evidence has showed that SMC2 may have a pro-oncogenic function and that SMC2 is involved in the mitotic cell division and also a direct transcriptional target of the oncogenic WNT signaling (50). Experimental studies suggested that SMC2 knockdown would suppress tumor growth in colorectal cancer (50) and increase apoptosis in neuroblastoma cells (49). Many studies also showed significantly higher SMC2 mRNA expression levels in human pancreatic cancer tissues than in adjacent non-neoplastic pancreas tissues (25,51). On the other hand, tumor suppressor p53-binding protein 1 known as p53-binding protein 1 or 53BP1, is a tumor suppressor, and 53BP1 nuclear bodies are partially suppressed by knocking down SMC2 (52). The PanC4 GWAS previously reported that rs10991043 near SMC2 reached 7.00 × 10–8 in association with pancreatic cancer risk, but this association was not observed in other pancreatic GWASs (13). In the present study, we found that the representative SMC2 SNP rs3818626 was associated with pancreatic cancer risk in both PanC4 and PanScan GWAS data sets, but in moderate LD with the previously reported rs10991043 (r2 = 0.53). More importantly, the SNP rs3818626 was predicted to be involved in TFBS/Splicing with a Regulome DB Score 2b and also associated with SMC2 mRNA expression levels in 373 lymphoblastoid cell lines by the eQTL analysis. A similar trend was found between rs3818626 and the mRNA expression levels of SMC2 in normal pancreatic tissues in the GTEx data, but the correlation was not significant, which might be due to small sample size or transcription specificity between tissues. Therefore, the finding of the association between SMC2 rs3818626 and pancreatic cancer risk is biologically plausible.
P53 protein (encoded by TP53 gene) is responsive to DNA damage, hypoxia, metabolic stress and oncogenic activation. The P53 protein suppresses cancer formation through its role in regulating cell cycle and apoptosis. This tumor suppressor gene is frequently mutated in various solid tumors, including pancreatic cancer (53,54). Although most of the mutations lead to loss of p53 function in inducing apoptosis and senescence, recent evidence shows that p53 inactivation/dysfunction would also directly or indirectly lead to promote tumorigenesis (55–57). In the present study, seven SNPs were found to be significantly associated with pancreatic cancer risk after multiple-testing correction by an FDR <0.20. The representative SNP rs9895829 (with a Regulome DB Score 1f) was associated with TP53 mRNA expression levels in 373 lymphoblastoid cell lines by the eQTL analysis, potentially affecting p53 activation and function. Similarly, a non-significant trend was found in normal pancreatic tissues in the GTEx data. These observations suggest that the association between rs9895829 and pancreatic cancer risk is also biologically plausible.
While a large proportion of cancer genomics research has been focusing on somatic mutations in TP53, a well-studied tumor suppresser, this gene does have a number of germline variants. Significantly, the majority of somatic mutations in TP53 occur in the codons for amino acid positions 175, 245, 248, 273 and 282 of the exons (58). In the present study, however, the SNP rs9895829, although located in an intron, was found to be associated with a decreased pancreatic cancer risk, as a result of an effect of the G allele that was associated with higher TP53 mRNA expression levels.
ARHGEF7 has many aliases, such as Rho Guanine Nucleotide Exchange Factor (GEF) 7, PAK-Interacting Exchange Factor Beta, BETA-PIX, COOL-1 and P85SPR (http://www.genecards.org/cgi-bin/carddisp.pl?gene=ARHGEF7). ARHGEF7 participates in the Hippo pathway to promote the tumorigenesis (59). Although we also found that ARHGEF7 SNP rs79447092 was associated with pancreatic cancer risk, we did not find an association between the representative SNP rs79447092 and ARHGEF7 mRNA expression levels.
The present study has some limitations. First of all, although we found two AURORA pathways from the Molecular Signatures Database, there may be other relevant genes that we failed to include. Second, we cannot get detailed clinical data for the study populations, such as family history, smoking status, alcohol intake, diabetes, obesity and chronic pancreatitis in the publically available GWAS data sets, to perform either further adjustment or stratified analysis. Finally, although we chose the representative SNPs by in silico SNP functional prediction tools and assessment by the eQTL analysis, more direct functional validations are needed to support our findings.
In conclusion, the present study revealed three potentially susceptibility loci in SMC2, ARHGEF7 and TP53, which were associated with pancreatic cancer risk in 8463 cases and 6970 controls of European descent. The joint effect analysis demonstrated a significant association between increased NPGs and pancreatic cancer risk. Further validations and functional evaluations of these genetic variants are warranted to support these findings.
Funding
PanScan
The PanScan project was funded in whole or in part with federal funds from the National Cancer Institute (NCI), US National Institutes of Health (NIH) under contract number HHSN261200800001E. Additional support was received from the National Institutes of Health/National Cancer Institute under K07 CA140790, the American Society of Clinical Oncology Conquer Cancer Foundation, the Howard Hughes Medical Institute, the Lustgarten Foundation, the Robert T. and Judith B. Hale Fund for Pancreatic Cancer Research and Promises for Purple. A full list of acknowledgments for each participating study is provided in the Supplementary Note of the manuscript with PubMed ID: 25086665. The dbGaP accession number for this study used in this manuscript is phs000206.v5. p3.
PanC4
The patients and controls for this study were derived from the following PANC4 studies: Johns Hopkins National Familial Pancreas Tumor Registry, Mayo Clinic Biospecimen Resource for Pancreas Research, Ontario Pancreas Cancer Study (OPCS), Yale University, MD Anderson Case Control Study, Queensland Pancreatic Cancer Study, University of California San Francisco Molecular Epidemiology of Pancreatic Cancer Study, International Agency of Cancer Research and Memorial Sloan Kettering Cancer Center. This work is supported by the National Cancer Institute under R01CA154823. Genotyping services were provided by the Center for Inherited Disease Research. The Center for Inherited Disease Research is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number HHSN2682011000111. The dbGaP accession number for this study used in this manuscript is phs000648.v1. p1.
Supplementary Material
Acknowledgements
As Duke Cancer Institute member, Q.W. acknowledges support from the Duke Cancer Institute as part of the P30 Cancer Center Support Grant (Grant ID: NIH CA014236). Q.W. was also supported by a start-up fund from Duke Cancer Institute, Duke University Medical Center. Y.F. was supported by Shanghai Key Discipline for Respiratory Diseases (2017ZZ02014), Shanghai Jiao Tong University Medical Cross Project (YG2017MS64) and Shanghai Natural Science Foundation (18ZR1424000). The authors thank all the participants of the PanScan GWAS Study and Pancreatic Cancer Case Control Association Study. The authors would also like to acknowledge dbGaP repository for providing the cancer genotyping data set.
Glossary
Abbreviations
- CI
confidence interval
- eQTL
expression quantitative trait loci
- FDR
false discovery rate
- GWAS
genome-wide association studies
- LD
linkage disequilibrium
- mRNA
messenger RNA
- NPG
number of protective genotypes
- OR
odds ratio
- SNP
single nucleotide polymorphism
Conflict of Interest Statement: None declared.
References
- 1. Siegel R.L. et al. (2017) Cancer statistics, 2017. CA Cancer J. Clin., 67, 7–30. [DOI] [PubMed] [Google Scholar]
- 2. Wolfgang C.L. et al. (2013) Recent progress in pancreatic cancer. CA Cancer J. Clin., 63, 318–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stolzenberg-Solomon R.Z. et al. (2015) Epidemiology and inherited predisposition for sporadic pancreatic adenocarcinoma. Hematol. Oncol. Clin. North Am., 29, 619–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goggins M. et al. (1996) Germline BRCA2 gene mutations in patients with apparently sporadic pancreatic carcinomas. Cancer Res., 56, 5360–5364. [PubMed] [Google Scholar]
- 5. Jones S. et al. (2009) Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science, 324, 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kastrinos F. et al. (2009) Risk of pancreatic cancer in families with Lynch syndrome. JAMA, 302, 1790–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Murphy K.M. et al. (2002) Evaluation of candidate genes MAP2K4, MADH4, ACVR1B, and BRCA2 in familial pancreatic cancer: deleterious BRCA2 mutations in 17%. Cancer Res., 62, 3789–3793. [PubMed] [Google Scholar]
- 8. Vasen H.F. et al. (2000) Risk of developing pancreatic cancer in families with familial atypical multiple mole melanoma associated with a specific 19 deletion of p16 (p16-Leiden). Int. J. Cancer, 87, 809–811. [PubMed] [Google Scholar]
- 9. Roberts N.J. et al. (2012) ATM mutations in patients with hereditary pancreatic cancer. Cancer Discov., 2, 41–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Amundadottir L. et al. (2009) Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat. Genet., 41, 986–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Petersen G.M. et al. (2010) A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat. Genet., 42, 224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wolpin B.M. et al. (2014) Genome-wide association study identifies multiple susceptibility loci for pancreatic cancer. Nat. Genet., 46, 994–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Childs E.J. et al. (2015) Common variation at 2p13.3, 3q29, 7p13 and 17q25.1 associated with susceptibility to pancreatic cancer. Nat. Genet., 47, 911–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kang X., et al. ; Transdisciplinary Research in Cancer of the Lung (TRICL) Research Team (2016) Polymorphisms of the centrosomal gene (FGFR1OP) and lung cancer risk: a meta-analysis of 14,463 cases and 44,188 controls. Carcinogenesis, 37, 280–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang M., et al. ; TRICL Research Team (2016) Genetic variant in DNA repair gene GTF2H4 is associated with lung cancer risk: a large-scale analysis of six published GWAS datasets in the TRICL consortium. Carcinogenesis, 37, 888–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yuan H. et al. (2016) A novel genetic variant in long Non-coding RNA Gene NEXN-AS1 is associated with risk of lung cancer. Sci. Rep., 6, 34234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhou F. et al. (2016) Susceptibility loci of CNOT6 in the general mRNA degradation pathway and lung cancer risk-A re-analysis of eight GWASs. Mol. Carcinog, 56, 1227–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schvartzman J.M. et al. (2010) Mitotic chromosomal instability and cancer: mouse modelling of the human disease. Nat. Rev. Cancer, 10, 102–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Perez de Castro I. et al. (2012) Mitotic stress and chromosomal instability in cancer: the case for TPX2. Genes Cancer, 3, 721–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thompson S.L. et al. (2010) Mechanisms of chromosomal instability. Curr. Biol., 20, R285–R295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nigg E.A. (2001) Mitotic kinases as regulators of cell division and its checkpoints. Nat. Rev. Mol. Cell Biol., 2, 21–32. [DOI] [PubMed] [Google Scholar]
- 22. Salaun P. et al. (2008) Cdk1, Plks, Auroras, and Neks: the mitotic bodyguards. Adv. Exp. Med. Biol., 617, 41–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Maia A.R. et al. (2014) A growing role for Aurora A in chromosome instability. Nat. Cell Biol., 16, 739–741. [DOI] [PubMed] [Google Scholar]
- 24. Fu J. et al. (2007) Roles of Aurora kinases in mitosis and tumorigenesis. Mol. Cancer Res., 5, 1–10. [DOI] [PubMed] [Google Scholar]
- 25. Pei H. et al. (2009) FKBP51 affects cancer cell response to chemotherapy by negatively regulating Akt. Cancer Cell, 16, 259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Grützmann R. et al. (2004) Gene expression profiling of microdissected pancreatic ductal carcinomas using high-density DNA microarrays. Neoplasia, 6, 611–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Boss D.S. et al. (2009) Clinical experience with aurora kinase inhibitors: a review. Oncologist, 14, 780–793. [DOI] [PubMed] [Google Scholar]
- 28. Xu L. et al. (2014) STK15 rs2273535 polymorphism and cancer risk: a meta-analysis of 74,896 subjects. Cancer Epidemiol., 38, 111–117. [DOI] [PubMed] [Google Scholar]
- 29. Sonoyama T. et al. (2011) TP53 codon 72 polymorphism is associated with pancreatic cancer risk in males, smokers and drinkers. Mol. Med. Rep., 4, 489–495. [DOI] [PubMed] [Google Scholar]
- 30. Naccarati A. et al. (2010) Genotype and haplotype analysis of TP53 gene and the risk of pancreatic cancer: an association study in the Czech Republic. Carcinogenesis, 31, 666–670. [DOI] [PubMed] [Google Scholar]
- 31. Qin L. et al. (2015) Association between rs9904341 G<C gene polymorphism and susceptibility to pancreatic cancer in a Chinese population. Genet. Mol. Res., 14, 5197–5202. [DOI] [PubMed] [Google Scholar]
- 32. Staff P.O. (2015) Correction: impact of TP53 codon 72 and MDM2 SNP 309 polymorphisms in pancreatic ductal adenocarcinoma. PLoS One, 10, e0126295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Avan A. et al. (2014) AKT1 and SELP polymorphisms predict the risk of developing cachexia in pancreatic cancer patients. PLoS One, 9, e108057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mailman M.D. et al. (2007) The NCBI dbGaP database of genotypes and phenotypes. Nat. Genet., 39, 1181–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tryka K.A. et al. (2014) NCBI’s database of genotypes and phenotypes: dbGaP. Nucleic Acids Res., 42(Database issue), D975–D979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Borgida A.E. et al. (2011) Management of pancreatic adenocarcinoma in Ontario, Canada: a population-based study using novel case ascertainment. Can. J. Surg., 54, 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McWilliams R.R. et al. (2009) Nucleotide excision repair pathway polymorphisms and pancreatic cancer risk: evidence for role of MMS19L. Cancer Epidemiol. Biomarkers Prev., 18, 1295–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liberzon A. et al. (2015) The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst., 1, 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xu Z. et al. (2009) SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res., 37(Web Server issue), W600–W605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Boyle A.P. et al. (2012) Annotation of functional variation in personal genomes using RegulomeDB. Genome Res., 22, 1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ward L.D. et al. (2012) HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res., 40(Database issue), D930–D934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lappalainen T., et al. ; Geuvadis Consortium (2013) Transcriptome and genome sequencing uncovers functional variation in humans. Nature, 501, 506–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Purcell S. et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chang C.C. et al. (2015) Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience, 4, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu J.Z., et al. ; AMFS Investigators (2010) A versatile gene-based test for genome-wide association studies. Am. J. Hum. Genet., 87, 139–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mishra A. et al. (2015) VEGAS2: software for more flexible gene-based testing. Twin Res. Hum. Genet., 18, 86–91. [DOI] [PubMed] [Google Scholar]
- 47. Pruim R.J. et al. (2010) LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics, 26, 2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Barrett J.C. et al. (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics, 21, 263–265. [DOI] [PubMed] [Google Scholar]
- 49. Murakami-Tonami Y. et al. (2014) Inactivation of SMC2 shows a synergistic lethal response in MYCN-amplified neuroblastoma cells. Cell Cycle, 13, 1115–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dávalos V. et al. (2012) Human SMC2 protein, a core subunit of human condensin complex, is a novel transcriptional target of the WNT signaling pathway and a new therapeutic target. J. Biol. Chem., 287, 43472–43481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Badea L. et al. (2008) Combined gene expression analysis of whole-tissue and microdissected pancreatic ductal adenocarcinoma identifies genes specifically overexpressed in tumor epithelia. Hepatogastroenterology, 55, 2016–2027. [PubMed] [Google Scholar]
- 52. Lukas C. et al. (2011) 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat. Cell Biol., 13, 243–253. [DOI] [PubMed] [Google Scholar]
- 53. Barton C.M. et al. (1991) Abnormalities of the p53 tumour suppressor gene in human pancreatic cancer. Br. J. Cancer, 64, 1076–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mohamadkhani A., et al. (2013) Detection of TP53 R249 mutation in Iranian patients with pancreatic cancer. J. Oncol., 2013, 738915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Guo G. et al. (2013) Trp53 inactivation in the tumor microenvironment promotes tumor progression by expanding the immunosuppressive lymphoid-like stromal network. Cancer Res., 73, 1668–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Menendez D. et al. (2013) Interactions between the tumor suppressor p53 and immune responses. Curr. Opin. Oncol., 25, 85–92. [DOI] [PubMed] [Google Scholar]
- 57. Cui Y. et al. (2016) Immunomodulatory function of the tumor suppressor p53 in host immune response and the tumor microenvironment. Int J Mol Sci., 19, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Olivier M. et al. (2010) TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb. Perspect. Biol., 2, a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Heidary Arash E. et al. (2014) Arhgef7 promotes activation of the Hippo pathway core kinase Lats. EMBO J., 33, 2997–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.