ABSTRACT
Lifestyle interventions, including exercise and dietary supplementation, can modify DNA methylation and exert health benefits; however, the underlying mechanisms are poorly understood. Here we investigated the impact of acute aerobic exercise and the supplementation of omega-3 polyunsaturated fatty acids (n-3 PUFA) and extra virgin olive oil (EVOO) on global and gene-specific (PPARGC1A, IL6 and TNF) DNA methylation, and DNMT mRNA expression in leukocytes of disease-free individuals. Eight trained male cyclists completed an exercise test before and after a four-week supplementation of n-3 PUFA and EVOO in a double-blind, randomised, repeated measures design. Exercise triggered global hypomethylation (Pre 79.2%; Post 78.7%; p = 0.008), alongside, hypomethylation (Pre 6.9%; Post 6.3%; p < 0.001) and increased mRNA expression of PPARGC1A (p < 0.001). Associations between PPARGC1A methylation and exercise performance were also detected. An interaction between supplement and trial was detected for a single CpG of IL6 indicating increased DNA methylation following n-3 PUFA and decreased methylation following EVOO (p = 0.038). Global and gene-specific DNA methylation associated with markers of inflammation and oxidative stress. The supplementation of EVOO reduced DNMT1 mRNA expression compared to n-3 PUFA supplementation (p = 0.048), whereas, DNMT3a (p = 0.018) and DNMT3b (p = 0.046) mRNA expression were decreased following exercise. In conclusion, we demonstrate that acute exercise and dietary supplementation of n-3 PUFAs and EVOO induce DNA methylation changes in leukocytes, potentially via the modulation of DNMT mRNA expression. Future studies are required to further elucidate the impact of lifestyle interventions on DNA methylation.
KEYWORDS: PPARGC1A, IL6, TNFa, DNMT, DNA methylation, exercise, inflammation, n-3 PUFA
Introduction
Environmental stimuli, including exercise and dietary interventions, can modify the DNA methylome at a global and gene-specific level [1]. Exercise training studies have demonstrated hypomethylation of the genome following exercise in both skeletal muscle [2–4] and blood leukocytes [5–7]. Within skeletal muscle, acute exercise has been demonstrated to induce hypomethylation [4,8–10]; however, the only investigation of DNA methylation in leukocytes following acute exercise failed to detect any changes in DNA methylation [11]. Despite the scarcity of literature surrounding the impact of acute exercise on DNA methylation in leukocytes, an epigenetic consequence is suggested by the remodelling of the leukocyte transcriptome [12–14].
Acute exercise is associated with adjustments in the expression of genes involved in a variety of cellular processes, including immune response mitochondrial biogenesis, metabolism and muscle remodelling [14–16]. The PPARGC1A gene, which encodes for peroxisome proliferator-activated receptor gamma, co-activator alpha (PGC1-a), is known as the master regulator of mitochondrial biogenesis and plays an important role in aerobic training adaptation [17]. In immune cells, PPARGC1A is associated with anti-inflammatory [18,19] and anti-oxidant defence [20]; however, the impact of exercise-induced inflammation and oxidative stress on PPARGC1A DNA methylation is unknown. Epigenetic studies have linked a CpG site −260 bases from the promoter of PPARCG1A with the regulation of mRNA expression. In skeletal muscle, exercise can demethylate the PPARGC1A −260 CpG site which has been shown to concurrently upregulate PPARGC1A mRNA expression [8,10,21]. Although well characterised in skeletal muscle, the regulation of PPARGC1A expression in other cells and tissues, including immune cells is poorly understood [22].
Exercise of sufficient intensity and duration can cause tissue injury and lead to a systemic inflammatory response [14,23]. Increased circulating levels of the inflammatory cytokines IL-6 and TNFa are strongly correlated with the progression of sarcopenia and measures of physical performance [24,25]. Acute exercise can also increase the production of reactive oxygen species, in both skeletal muscle and immune cells [26], potentially leading to the development of oxidative stress and damage to lipids, proteins and DNA [27]. Increases in markers of oxidative stress and circulating levels of inflammatory cytokines, such as IL-6 and TNFa, have been shown to alter the expression of DNA methyltransferases (DNMTs) [28–32] and influence DNA methylation patterns [11,33]. DNA methylation of inflammatory cytokines have been associated with various inflammatory diseases including IL6 with Rheumatoid Arthritis [34] and obesity [35]; TNF DNA methylation with type 2 diabetes [36] and Alzheimer’s disease [37]. Despite increased circulating levels of inflammatory cytokines post-exercise [14,23], the impact of exercise on the DNA methylation of genes encoding inflammatory cytokines such as IL6 and TNF remains unknown.
There is the potential for the dietary supplementation of fatty acids (FAs) to prevent the exercise-induced inflammation via the modulation of DNA methylation. Supplementation of FAs, including omega-3 polyunsaturated FAs (n-3 PUFAs) and extra virgin olive oil (EVOO), are consumed to reduce levels of inflammation [38,39], however, the impact of these supplements on exercise-induced inflammation is equivocal. Some studies have detected reductions in inflammation post-exercise with FA supplementation [40,41], whereas, others have reported no change in inflammation [42,43]. An emerging mechanism for the anti-inflammatory impact of FA supplementation is via epigenetic modifications [44–47]. The supplementation of the diet with krill oil, high in n-3 PUFAs, has been demonstrated to reduce PPARGC1A mRNA expression and the change in mRNA expression was negatively correlated to the change in plasma n-3 PUFAs [48]. Total n-3 PUFA content is negatively correlated to both IL6 DNA methylation and IL-6 protein concentration [47].
EVOO is a commonly used control in exercise studies to assess the impact of n-3 PUFA; however, the supplementation of EVOO has also been reported to modify the DNA methylation of genes associated with inflammation [49]. It remains to be identified whether the supplementation of FAs have an epigenetic impact on exercise-induced inflammation.
The present study investigated the impact of aerobic exercise on global and gene-specific (PPARGC1A, IL6 and TNF) DNA methylation and DNMT mRNA expression in leukocytes of disease-free individuals. We also investigated whether these relationships could be modified by the supplementation of FAs. The association between physiological markers related to exercise performance, inflammation and oxidative stress post exercise and DNA methylation were also investigated.
Results
Global cytosine methylation and DNMT mRNA expression
One-hour of cycling reduced global methylation, assessed by the Luminometric Methylation Assay (LUMA; Figure 1(a); Pre 79.2%; Post 78.7%, p = 0.008), and the mRNA expression of both DNMT3a (Figure 1(c)); p = 0.018) and DNMT3b (Figure 1(d)); p = 0.046). Supplementation of FAs did not alter global methylation or mRNA expression of DNMT3a or DNMT3b (Figure 2; p > 0.05). While DNMT1 mRNA expression was unaffected by exercise, a significant interaction was identified between supplement and trial (p = 0.048; Figure 2(b)) indicating differential effects on mRNA expression with the two supplements. No correlation was detected between global DNA methylation values and DNMT mRNA expression.
Figure 1.
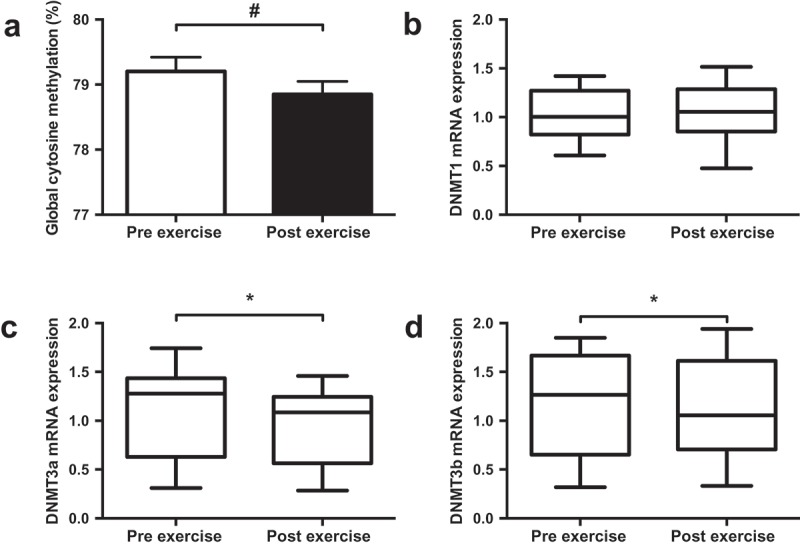
Effect of exercise on global DNA methylation (a) and mRNA expression of DNMT1 (b), DNMT3a (c) and DNMT3b (d). Data presented as the mean value of all trials for each time point. * p < 0.05, # p < 0.01.
Figure 2.
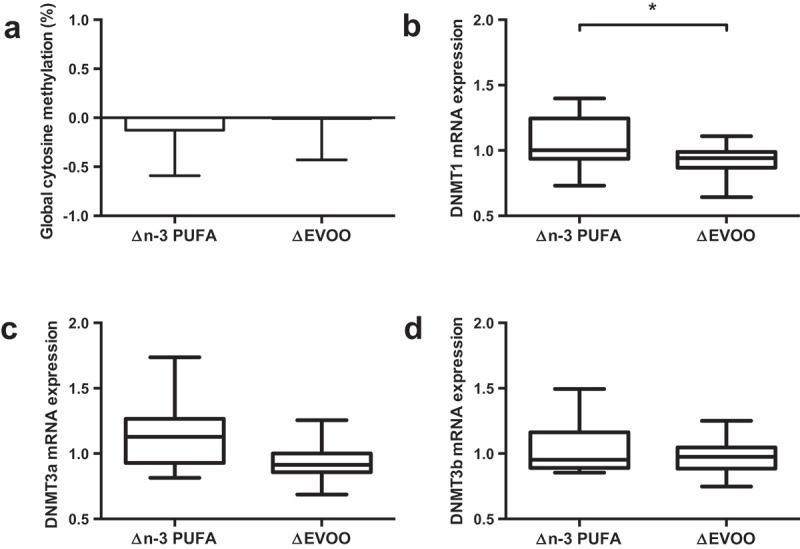
The impact of supplementation of n-3 PUFA and EVOO on global DNA methylation (a) and mRNA expression of DNMT1 (b), DNMT3a (c) and DNMT3b (d). Data presented as the relative change (Δ) between pre and post supplementation trials (post supplementation – pre supplementation) for each supplement. n-3 PUFA, n-3 polyunsaturated fatty acid; EVOO, extra virgin olive oil. * p < 0.05.
Gene-specific DNA methylation and mRNA expression
PPARGC1A
A reduction in PPARGC1A DNA methylation (Pre 6.9%; Post 6.3%, Figure 3(a); p < 0.001) and an increase in mRNA expression (Figure 3(b)); p < 0.001) were detected following exercise. The supplementation of FAs had no impact on PPARGC1A DNA methylation or mRNA expression (p > 0.05). Moderate but non-significant negative correlations were detected between PPARGC1A DNA methylation and DNMT3a and DNMT3b mRNA expression (Figure 5).
Figure 3.
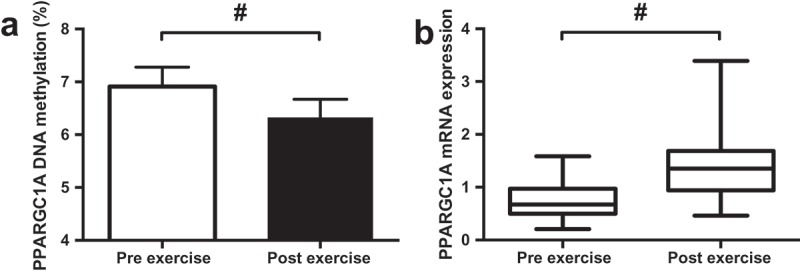
Effect of exercise on DNA methylation of CpG-260 (a) and mRNA expression (b) of PPARGC1A. Data presented as the mean value of all trials for each time point. # p < 0.01.
Figure 5.
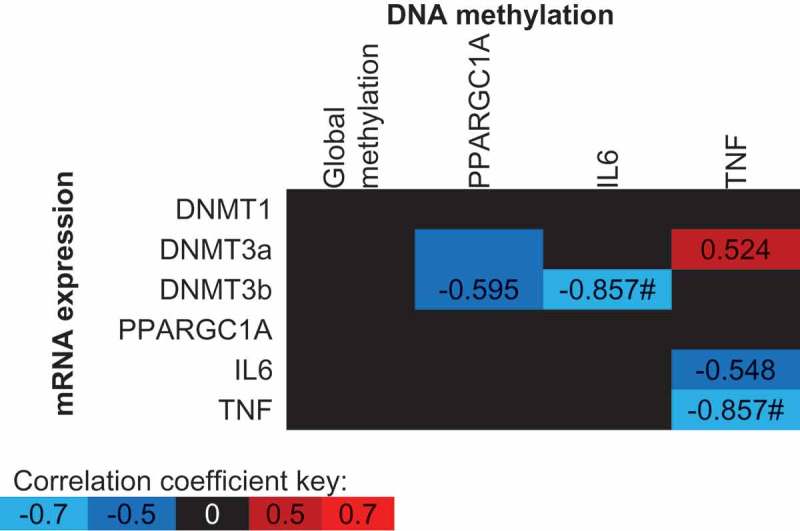
Spearman’s Rho correlation coefficients between mean DNA methylation values and gene expression values across all conditions (supplement, time and trial). The mean of all CpG sites assessed for each gene has been used to provide an overall view of the region of interest. Blue indicates a negative correlation, red indicates a positive correlation and black indicates correlation coefficients between −0.5 and 0.5. * p < 0.05, # p < 0.01.
IL6
Despite an increase in IL-6 protein concentrations following exercise (Pre: 0.63 ± 0.24 pg/mL, Post: 3.78 ± 0.55 pg/mL; p < 0.001), there was no change in IL6 DNA methylation (p > 0.05) or mRNA expression (p > 0.05) following exercise. A significant interaction was detected between supplement and trial for CpG3 (−1094) indicating increased DNA methylation following n-3 PUFA and decreased methylation following EVOO (Figure 4(a)); p = 0.038). A similar, non-significant (p = 0.080) trend was detected for IL6 mRNA expression following supplementation (Figure 4(b)). A significant correlation was detected between the mean IL6 methylation across all CpG sites and DNMT3b mRNA expression (Figure 5, p = 0.007).
Figure 4.
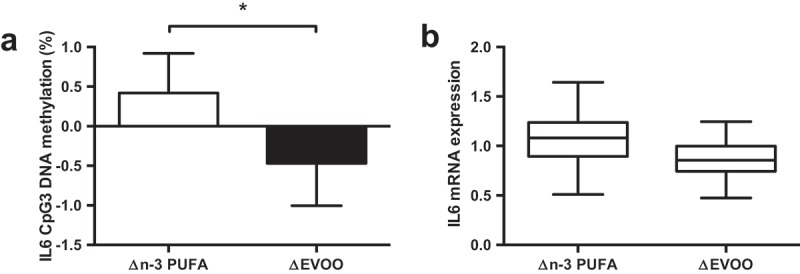
The impact of n-3 PUFA and EVOO supplementation on IL6 CpG3 DNA methylation (a) and IL6 mRNA expression (b). Data presented as the change (Δ) between pre and post supplementation trials (post supplementation – pre supplementation). n-3 PUFA, n-3 polyunsaturated fatty acid; EVOO, extra virgin olive oil. * p < 0.05.
TNF
Neither exercise or the supplementation of FAs altered TNF DNA methylation or mRNA expression. Trends were identified between 3 TNF CpG sites and differential methylation following supplementation (CpG2 p = 0.069; CpG3 p = 0.098; CpG4 p = 0.067; CpGmean p = 0.077). TNF DNA methylation was negatively correlated with TNF mRNA expression (Figure 5; p = 0.007). Moderate, however, non-significant correlations were detected between both IL6 and DNMT3a mRNA expression, and TNF DNA methylation (Figure 5).
Associations between DNA methylation and post-exercise physiological markers
Figure 6 demonstrates the association between post-exercise DNA methylation and physiological markers related to exercise, oxidative stress and inflammation. Prior to FA supplementation, both PPARGC1A and TNF methylation post-exercise are significantly correlated with Time Trial (TT) performance (Figure 6, p < 0.05). Following the supplementation of n-3 PUFA and EVOO, correlations between TT performance and both PPARGC1A and TNF DNA methylation are weakened and no longer significant (Figure 6). A negative correlation was detected between peripheral blood mononuclear cell (PBMC) protein carbonyl (PC) concentration, an intracellular measure of oxidative stress, and both global and PPARGC1A methylation prior to supplementation of FAs, however, no association was detected following n-3 PUFA supplementation (Figure 6). The concentration of PC in serum, a systemic measure of oxidative stress, was uncorrelated with DNA methylation at baseline, however, following EVOO supplementation significant correlations existed between serum PCs and both PPARGC1A and TNF DNA methylation (Figure 6). The only significant correlation between DNA methylation and serum IL-6 concentration was a negative correlation with global DNA methylation following n-3 PUFA supplementation (Figure 6).
Figure 6.
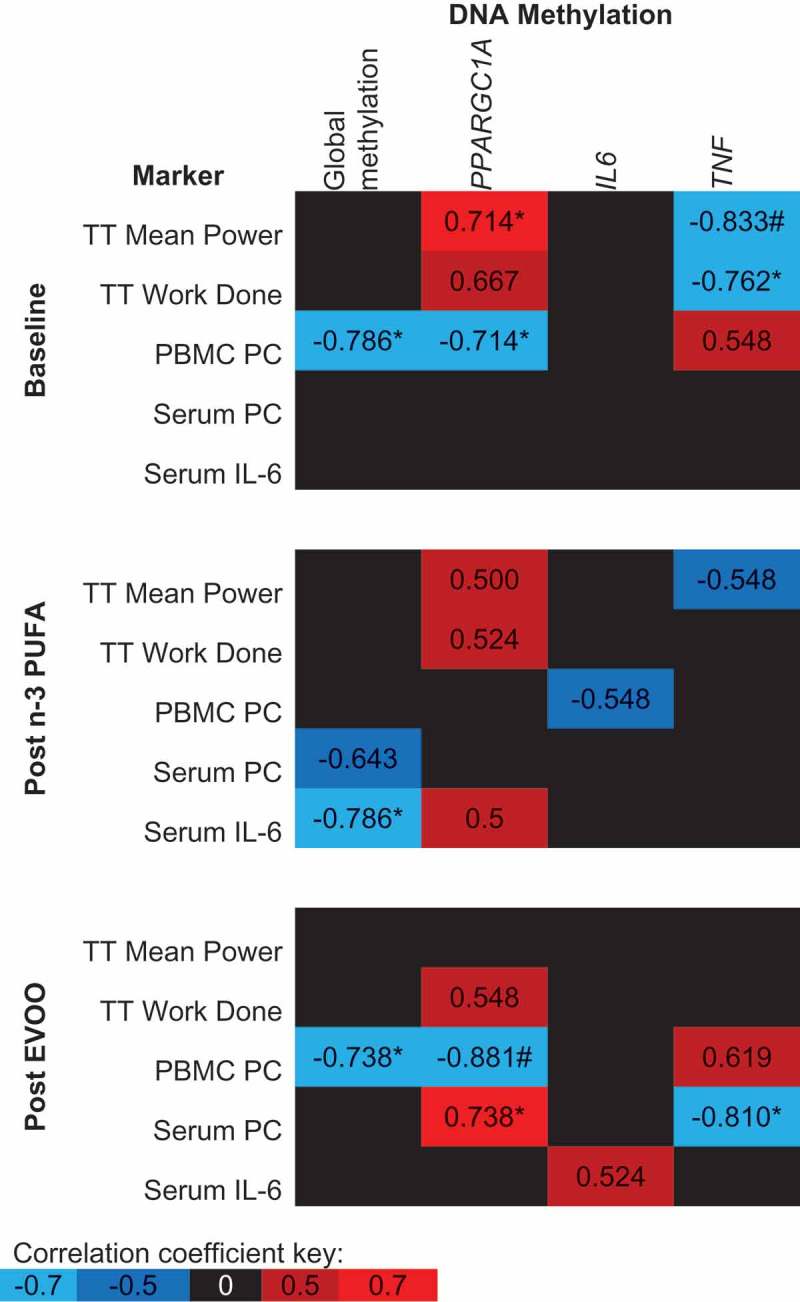
Spearman’s Rho between post-exercise DNA methylation and physiological markers related to exercise performance, oxidative stress and inflammation. The mean of all CpG sites assessed for each gene has been used to provide an overall view of the region of interest. Blue indicates a negative correlation, red indicates a positive correlation and black indicates correlation coefficients between −0.5 and 0.5. n-3 PUFA, omega-3 polyunsaturated fatty acid; EVOO, extra virgin olive oil; TT, Time trial; PC, protein carbonyl. *p < 0.05, # p < 0.01.
Discussion
A single bout of aerobic exercise and supplementation of FAs can modulate leukocyte DNA methylation and mRNA expression patterns. A one-hour cycling bout decreased global and PPARGC1A DNA methylation and mRNA expression of DNMT3a, DNMT3b and PPARGC1A. The supplementation of FAs induced differential effects on the DNA methylation of a CpG site in the promoter region of IL6; n-3 PUFA increased methylation, whereas, EVOO supplementation decreased methylation. The same result was identified for mRNA expression of DNMT1 and trends existed for 3 CpG sites in the promoter region TNF. Significant correlations were identified between global DNA methylation; PPARGC1A, IL6 and TNF DNA methylation post-exercise; and physiological markers related to exercise performance, inflammation and oxidative stress indicating that the epigenetic modifications have functional effects.
For the first time we report, global hypomethylation in leukocytes following an acute bout of exercise. The only previous study to investigate the impact of acute exercise in blood cells failed to detect any change in DNA methylation following correction for multiple testing [11]. The results of the present study are in accordance with previous reports of a net hypomethylation following chronic exercise training [2–7] and acute bouts of exercise in plasma [50] and skeletal muscle [4,8]. Other studies have failed to detect any change in global DNA methylation [51,52]; however, this can be explained by a similar number of CpG sites increasing and decreasing in DNA methylation [51]. It has also been demonstrated that exercise-induced hypomethylation is retained during periods of detraining, allowing it to become further hypomethylated following further training [4]. These data suggest that both acute and chronic exercise is sufficient to alter DNA methylation patterns typically resulting in hypomethylation.
In the present study, a 4-week supplementation of FAs did not influence global DNA methylation. In contrast, a 6-month supplementation of n-3 PUFA decreased LINE-1 DNA methylation, a surrogate for global DNA methylation, in Alzheimer’s patients [53]. However, LINE-1 methylation is increased in Alzheimer’s patients compared to healthy controls [54], therefore, the supplementation of n-3 PUFA in these individuals may act to restore global DNA methylation to the normal level detected in healthy individuals. The use of different surrogate measures of global methylation (LUMA vs LINE-1) prevents the direct comparison between studies because of the different region which these assays investigate. Two separate studies have indicated that the methylation estimates provided by LINE-1 and LUMA are poorly correlated [55,56].
For the first time, decreased methylation and concurrent increased mRNA expression of PPARGC1A following a bout of aerobic exercise have been detected in leukocytes. The results from the present study match previous reports of aerobic exercise-induced hypomethylation in skeletal muscle [2,8,10] potentially indicating a systemic impact of exercise on PPARGC1A DNA methylation. The mRNA expression profile of skeletal muscle and PBMCs have been shown to be highly associated following an 8-week supplementation of n-3 PUFAs [57]. Although we do not find any association with PPARGC1A methylation/mRNA expression and n-3 PUFA supplementation in the present study, the hypomethylation detected in the present study is consistent with the impact of exercise in skeletal muscle providing further evidence for blood-derived expression profiles to be used as a surrogate for skeletal muscle.
The only previous report of PPARGC1A methylation from leukocytes failed to detect an association with physical activity [58]. The lack of previous association could be the result of the investigation of different CpG sites in the promoter region of PPARGC1A. Alternatively, the discordance in these results could reflect the heterogeneity in methylation pattern of immune cells [59]. Exercise increases the number of circulating leukocytes, therefore, changes in methylation may be the result of different proportions of leukocytes rather than a change in DNA methylation patterns [60]. The present study has adjusted DNA methylation values to account for the number of leukocytes (lymphocytes, neutrophils, monocytes, basophils and eosinophils) [59], whereas, previous reports have failed to account for this critical variable.
The positive correlation between leukocyte PPARGC1A methylation and exercise performance indicates that increased DNA methylation may provide a performance advantage. PPARGC1A is thought to upregulate mitochondrial biogenesis in monocytes to induce a shift towards an anti-inflammatory phenotype [18,19] and antioxidant defence in lymphocytes [20]. Although we did not find an association with IL-6 protein concentration, a negative association was detected between PPARGC1A DNA methylation and PC concentration indicating epigenetic control of the antioxidant role of PPARGC1A. There is limited literature comparing mitochondrial function in leukocytes and skeletal muscle following exercise; however, the association between gait speed and mitochondrial function in both skeletal muscle tissue and PBMCs provides a conserved mechanism between mitochondrial function in skeletal muscle and blood-derived mitochondria [61]. Further evidence of a conserved mechanism is suggested with genes related to mitochondrial structure and function found to be co-expressed in skeletal muscle and neutrophils following aerobic exercise [62]. Future studies are required to detect if the same phenotypic associations exist in skeletal muscle as detected in leukocytes in the present study.
Aerobic exercise did not alter the DNA methylation or mRNA expression of either IL6 or TNF. The epigenetic impact of exercise on inflammatory cytokines is relatively unknown, however, several studies have indicated a role for cytokine DNA methylation in inflammatory disease [34–37]. Although no association between TNF DNA methylation and mRNA expression was detected in the present study, n-3 PUFAs have previously been demonstrated to reverse the epigenetic changes observed with inflammation in skeletal muscle cells. The administration of TNF induced hypermethylation and decreased mRNA expression of MyoD [63], whereas the supplementation of EPA dampens the impact of TNF in muscle and restores MyoD mRNA expression [44]. Despite an increase in the circulating protein concentration of IL-6 in the present study, the exercise bout may have not increased TNFa protein concentration and induced an inflammatory response sufficient to modify DNA methylation patterns of inflammatory cytokines. TNF hypermethylation is reported in elderly individuals who maintained or increased their energy expenditure by 500 kcal/wk over an 8-year period compared to those who decreased energy expenditure over the same period [64]. The same TNF CpG sites as the present study have previously been shown to negatively associate with mRNA expression, plasma concentrations and measures of adiposity [65,66]. In the present study, a significant negative correlation was detected between TNF DNA methylation post-exercise and BMI, exercise performance and TNF mRNA expression. These data suggest an acute bout of exercise may not regulate TNF DNA methylation, however, the long-term benefits of regular exercise, such as reduced adiposity, may subsequently increase TNF DNA methylation levels and as a result, reduce TNF mRNA expression and the chronic low-grade inflammation levels associated with increased adiposity.
Previously decreased methylation in a region ~600 bp upstream of the IL6 promoter has been associated with increased erythrocyte n-3 PUFA concentrations and mRNA expression [47]. In the present study, the supplementation of EVOO and n-3 PUFA had contrasting effects on a single CpG (−1094) of IL6 (increased methylation following n-3 PUFA and decreased methylation with EVOO). The region ~1,000 bp from upstream of was investigated in the present study because of previous associations between DNA methylation and both inflammatory diseases [34,35] and mRNA expression [34]. Conflicting results between studies may indicate that distinct regions of the promoter regulate IL6 expression differently. Supplementation of n-3 PUFA and OO have been shown to induce differential methylation of elongase and desaturase enzymes which are responsible for the metabolism of FAs [67]. The differential DNA methylation of these enzymes indicates the potential for n-3 PUFAs to switch towards the production of less inflammatory eicosanoids. Although the DNA methylation of desaturase and elongase enzymes have not been measured in the present study, a switch towards n-3 PUFA derived eicosanoid production, such as 3-series rather than 2-series prostaglandins, has been shown to reduce cytokine expression [38] which is potentially indicated by the increased DNA methylation of IL6 following n-3 PUFA, but not EVOO, supplementation.
The impact of exercise and FA supplementation on DNMT mRNA expression was investigated to identify whether changes in DNMT mRNA expression could be a potential mechanism underlying modulated DNA methylation. DNMT1 mRNA expression was modulated by FA supplementation, whereas, exercise reduced the expression of both DNMT3a and DNMT3b. This is the first demonstration of reduced expression of DNMT3a following acute exercise, whereas, the reduction in DNMT3b expression has previously been reported [31,68]. The inclusion of DNA methylation assessment in the present study allows the confirmation that following a single bout of aerobic exercise DNMT expression is decreased alongside decreases in global and gene-specific DNA methylation. The only previous report of concurrent assessment of exercise-induced DNMT expression and DNA methylation was following an 8-week resistance training program [6]. The genome-wide method of methylation does not identify a net increase or decrease in global methylation; therefore, further studies are required to identify whether the modulation of DNMT3b causes hypomethylation or if it is important in both hyper- and hypomethylation.
The present study detects contrasting effects of n-3 PUFA and EVOO supplementation on DNMT1 mRNA expression. There is a paucity of literature surrounding the impact of FA supplementation on DNMT expression in humans, whereas, animal models have associated supplementation of alpha-linolenic acid supplementation, a n-3 PUFA, with changes in DNMT mRNA expression [69,70]. Interestingly, similar to the present study, no change in global DNA methylation was detected alongside modulated DNMT1 expression [69]. A change in global DNA methylation potentially would not be expected with increased in DNMT1 mRNA expression because DNMT1 functions to maintain DNA methylation. The impact of EVOO on DNMT expression is unknown, however, EVOO contains phenolic compounds, including decarboxymethyl oleuropein aglycone (DOA) [71], which reduce DNMT activity via competitive inhibition [72]. The absence of a measure of DNMT activity is a limitation of the present study, however, parallel changes in DNMT mRNA expression and activity have previously been reported [73]. A measure of activity could potentially explain the lack of association between altered DNMT mRNA expression and modulated DNA methylation following supplementation which should be considered in future studies.
While exercise and FA supplementation may directly influence DNMT expression, these interventions may modulate DNMT expression by intermediary mechanisms. The expression of several miRNAs, including miRNA-29 −130 and −148, are associated with: DNMT expression [74–77], exercise [78] and FA supplementation [79–81]. IL-6 protein levels have been reported to regulate DNMT mRNA expression [30–32] via the modulation of miRNA [29]. The small increase in IL-6 protein expression following exercise in the present study may be insufficient to modulate DNMT expression explaining the lack of agreement with previous reports. Future studies should use a bout of exercise with a greater inflammatory response, such as eccentric exercise, to examine the effect of exercise-induced inflammation on DNMT expression. The capability of exercise and n-3 PUFA supplementation to modify the expression of the same miRNAs which control the expression of DNMTs suggests miRNA expression could be one of the underlying mechanisms controlling DNA methylation.
The use of a homogenous population of trained cyclists in the present study potentially limits the generalisability of the results to other populations. Trained male cyclists were selected as the population for the present study because they are the most familiar with the exercise stimuli and we would expect this to reflect in the smallest epigenetic response. Previously a single bout of exercise was sufficient to reduce global DNA methylation in plasma of COPD patients; however, following a training intervention the exercise bout was no longer sufficient to reduce global DNA methylation [50]. Exercise training has previously been demonstrated to alter DNA methylation patterns differently depending on family history of diabetes [2]. Future studies should compare the impact of exercise in trained athletes and sedentary individuals or a disease cohort to determine whether exercise-induced alterations to the DNA methylome are contributors to health and disease in diverse populations.
In conclusion, the present study highlights the impact of an acute bout of aerobic exercise and the supplementation of FAs on DNA methylation and mRNA expression in leukocytes of trained male cyclists. Alterations in the epigenetic control of these genes are associated with physiological markers related to exercise performance and inflammation/oxidative stress, however, a more extensive study is required to confirm these associations. The observational nature of the present study prevents the identification of the underlying mechanisms controlling altered DNA methylation following exercise and FA supplementation, therefore, future mechanistic studies are required to identify such mechanisms. Here we suggest that modulation of DNMT mRNA expression may be one such mechanism for future research. Future studies should compare multiple tissue types to examine whether exercise and supplementation of FAs have systemic effects on DNA methylation.
Methods
Participants
Complete sets of data were available for eight participants whose characteristics are described in Table 1. Prior to participation, informed written consent was provided by each participant. Participants were healthy, non-smokers with no history of metabolic or cardiovascular disease. In the six-months prior to the study, participants had no history of n-3 PUFA, anti-oxidant or anti-inflammatory supplementation. Participants recorded their physical activity and maintained habitual diet throughout the study. The experimental protocol was approved by the Loughborough University Ethics Human Participants sub-committee and performed in accordance with the Declaration of Helsinki 1975.
Table 1.
Participant characteristics. Wmax, maximal aerobic work rate.
| Variable | All participants (n = 8) |
|---|---|
| Age (yrs) | 39.50 ± 5.90 |
| Body Mass (kg) | 73.04 ± 8.31 |
| Height (cm) | 174.26 ± 8.41 |
| Wmax (W) | 321.63 ± 28.15 |
| V̇O2max (mL∙kg∙min−1) | 53.88 ± 5.24 |
Study overview
The study consisted of a pre-test and four experimental trials. Experimental trials were completed before and after a four-week supplementation of n-3 PUFA and EVOO in a double-blind, randomised, repeated measures design. A four-week washout was included between each supplementation period (Figure 7).
Figure 7.
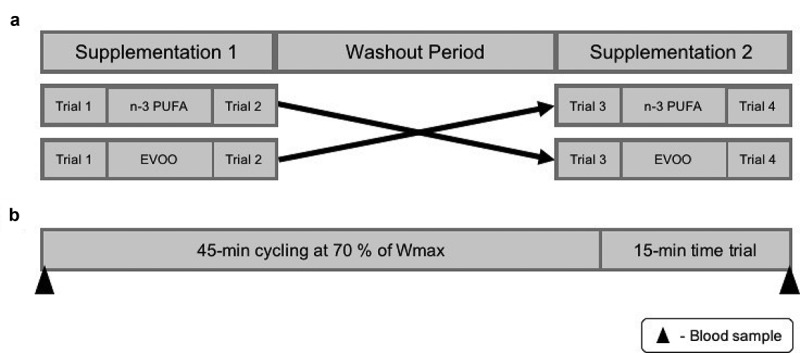
Schematic representation of study outline (a) and trial day (b). n-3 PUFA, omega-3 polyunsaturated fatty acid; EVOO, extra virgin olive oil; Wmax, maximal aerobic work rate.
Pre-test
Participants underwent anthropometric assessment for height, body mass and eight-skinfold measurements prior to the start of the study. Maximal aerobic work rate (Wmax) and maximal oxygen uptake (V̇O2max) were determined using a graded exercise test on a Lode Excalibur Sport ergometer (Lode B.V, Netherlands). The exercise test began with a warm-up period of 5-min cycling at 100 W. Workload then increased by 50 W every 3-min until volitional fatigue (decrease in self-selected cadence of 20 revs∙min−1). Expired air was collected in the final minute of each stage to allow V̇O2max determination using primary and secondary criteria [82]. Wmax was calculated using the formula:
Where t is the time in seconds completed in the final stage. Following the completion of the incremental cycling test, participants received a 10-minute rest before completing a 15-minute TT familiarization.
Experimental trials
Trials were conducted in the morning (7–9 am) following a 10-hour overnight fast. Participants were asked to complete a 3-day food diary, refrain from strenuous exercise and the consumption of alcohol or caffeine for the 24-hours prior to the trial. The performance test consisted of 45-minutes cycling at 70% Wmax, followed by a 15-minute TT [83].
Supplementation
Both n-3 PUFA (Holland and Barrett, Warwickshire, UK) and EVOO (Puritan’s Pride, New York, USA) supplements were provided in capsule form. Participants were instructed to take 6 capsules per day providing 5.7g of n-3 PUFA and 0.01g per day of α-Tocopherol or 6 g per day of EVOO. The n-3 PUFA dose was chosen based on previous findings showing the dose was sufficient to induce changes in the lipid profile of human blood over four weeks [84,85]. Compliance of supplementation was monitored by capsule counts.
Analytic procedures
Blood sampling
Venous blood was sampled via an intravenous catheter inserted into an antecubital vein of the non-dominant arm for the collection of whole blood pre and immediately post-exercise (Figure 7) for DNA methylation analysis, mRNA expression and a whole blood cell count using the COULTER® Ac·T™ 5diff (Beckman Coulter, UK). PBMCs were isolated from whole blood by density gradient centrifugation using Ficoll-Paque Premium (GE healthcare, USA) according to manufacturer’s instructions. The resulting PBMC cell pellet was suspended in 200μl RIPA buffer for analysis of protein carbonyls. Whole blood collected in vacutainers (Becton, Dickson & Company, UK) that contained no anticoagulant was allowed to clot at room temperature and centrifuged at 2800 rpm for 15 minutes for analysis of serum protein carbonyls and IL-6.
Nucleic acid isolation
Genomic DNA (gDNA) was isolated from 2mL of whole blood using the QIAamp DNA Blood Midi kit (Qiagen, Germany) according to the manufacturer’s instructions. RNA was isolated from whole blood collected in Tempus Blood RNA tubes using the Tempus Spin RNA Isolation Kit (Applied Biosystems, USA) according to the manufacturer’s instructions. The concentration (mean ± SD) and purity (absorbance ratio A260/A280 ± SD) of isolated DNA and RNA were determined using a Nanodrop 2000 (ThermoScientific, USA). The mean concentration of isolated gDNA was 183.50 ± 54.48 ng/μL with a A260/A280 ratio of 1.90 ± 0.02, whereas, RNA concentration was 120.32 ± 41.02 ng/μL with an A260/A280 ratio of 2.09 ± 0.02. Following extraction, DNA and RNA were stored at −20°C and −80°C respectively.
Luminometric methylation assay
LUMA was used as a marker of global DNA methylation as previously described [86], with minor adjustments. Briefly, two reactions containing 200 ng of gDNA were set up per sample, one with the methylation-sensitive enzyme FastDigest HpaII and one FastDigest MspI (Thermo Scientific, USA) and incubated for 20 min at 37 °C. Following incubation, 13 µL of each reaction were mixed with annealing buffer and added to a separate well of a Pyromark Q24 plate and analyzed using a PyroMark Q24 MDx system (Qiagen, Germany) with the following dispensation order: ACTCGA. Peak heights were exported, and methylation percentage was calculated using the following formula:
Methylation = (1 – (HpaII peak 2/HpaII peak 1)/(MspI peak 2/MspI peak 1)) x 100.
Bisulfite pyrosequencing
gDNA samples were bisulfite converted using the EpiTect Fast Bisulfite Conversion Kit (Qiagen, Germany) according to the manufacturer’s instructions. PCR of bisulfite converted DNA samples was performed using the PyroMark PCR Kit (Qiagen, Germany) according to the manufacturer’s instructions. For all assays, an initial activation period of 15 min at 95°C was followed by a 3-stage cycling process of denaturation (95°C for 30s), annealing (56°C for 30 s) and extension (72°C for 30 s) for 45 cycles. The PCR process was finished with a final extension period of 72°C for 10 min. Pyromark custom assay (Qiagen, Germany) genomic location, primer sequences and the sequence to analyze are presented in Table 2. To confirm a single PCR product, amplicons were analyzed by gel electrophoresis and visualised by ultraviolet trans-illuminator (BioRad, USA). The absence of PCR amplification of non-bisulfite converted DNA confirmed the specificity of each assay for bisulfite converted DNA. DNA methylation was assessed using a PyroMark Q48 Autoprep system (Qiagen, Germany) using PyroMark Q48 Advanced CpG Reagents (Qiagen, Germany). The nucleotide dispensation order was generated by entering the sequence to analyze into the PyroMark Q48 Autoprep software version 2.4.2 (Qiagen, Germany). A non-CpG cytosine was included in the nucleotide dispensation order to detect incomplete bisulfite conversion. The methylation at each CpG site was determined using the PyroMark Q48 Autoprep software set in CpG mode. The mean methylation of all CpG sites within the target region was determined using the methylation at the individual CpG sites. Standards of known methylation percentages (0%, 12.5%, 25%, 50%, 75%, 87.5%, 100%) were created using the EpiTect PCR control DNA set (Qiagen, Germany) and underwent pyrosequencing analysis to generate standard curves between the expected and observed methylation percentage to check the assays for PCR bias. A high coefficient of determination (R2 > 0.99) was determined for each assay indicating the absence of PCR bias.
Table 2.
Details of pyrosequencing assays used to determine DNA methylation. Genomic location identified using genome reference consortium human build 38 patch release 12. CpG sites are indicated in the sequence to analyze by Y. For, forward primer; Rev, reverse primer, Seq, sequencing primer; TSS, transcription start site; bp, base pair.
| Assay ID [Genomic location] |
Primer | Sequence | No. of CpG sites (distance from TSS; bp) |
|---|---|---|---|
| PPARGC1A [chr4:23,890,308–23,890,372] | For: | 5ʹ-TGTAGGGGATTTTGGTTATTATATGGT-3’ | 1 (−260) |
| Rev: | 5ʹ-biotin-ACCAACTTTAAATACCACAAACTCTA-3’ | ||
| Seq: | 5ʹ-GGTTATTATATGGTTAGGGT-3’ | ||
| Sequence to analyze: | TTYGTTTAGAGTTTGTGGTATTTAAAGTT | ||
|
IL6 [chr7:22,726,051–22,726,198] |
For: | 5ʹ-GGGAAGAGGGTTTTTGAATTAG-3’ | 6 (−1099, −1096, −1094, -1069, −1061 & −1057) |
| Rev: | 5ʹ-biotin-CTCCCTCTCCCTATAAATCTTAATTTAA-3’ | ||
| Seq: | 5ʹ-TTGAATTAGTTTGATTTAATAAGAA-3’ | ||
| Sequence to analyze: | ATTTTTGGGTGTYGAYGYGGAAGTAGATTTAGAGTTTAGAG TYGTGTTTGYGTTYGTAGTTTTTTTTTAGTTTTTTTTGATTT |
||
|
TNF [chr6:31,575,730–31,575,816] |
For: | 5ʹ-GGAAAGGATATTATGAGTATTGAAAGTATG-3’ | 4 (+197, +202, +214 & +222) |
| Rev: | 5ʹ-biotin-CTAAAACCCCCCTATCTTCTTAAA-3’ | ||
| Seq: | 5ʹ-ATTATGAGTATTGAAAGTATGAT-3’ | ||
| Sequence to analyze: | TYGGGAYGTGGAGTTGGTYGAGGAGGYGTTTTTTAAGAA GATAGGGGGGTTT |
mRNA expression
A minimum of 1 µg of RNA was reverse transcribed into complementary DNA (cDNA) using the High-Capacity RNA-to-cDNA™ Kit (Applied Biosystems, USA) according to the manufacturer’s instructions and diluted to a concentration of 5 ng/µL in deionised water. Relative mRNA expression was performed by quantitative PCR (qPCR) for each gene of interest and normalised to the expression of GAPDH using a Viia7 Real-Time PCR system (Applied Biosystems, USA). Each reaction contained 5 µL of SybrGreen PrecisionPlus qPCR Master Mix (PrimerDesign, UK), 0.5 µL of forward and reverse primer (Table 3) and 4 µL of 5 ng/µL cDNA. All samples were run in duplicate using the following cycling conditions: initial denaturation at 95°C for 2 min, followed by 40 cycles of 95°C for 15 s and 60°C for 60 s. Melt curves were visually inspected for a single peak indicating the generation of a single product. The relative mRNA expression of the genes of interest were calculated using the 2−(ΔΔCt) formula; the pooled group mean pre-exercise Ct from the initial trial was used as the control. The mean Ct value of GAPDH across all participants and experimental conditions was 17.13 ± 0.41 with low variation of 2.40%. The efficiency of each mRNA expression assay was determined (Table 3) using standard curves generated from a serial dilution of a cDNA sample. The efficiency was calculated using the formula:
Table 3.
` Details of assays used to determine mRNA expression. For, forward primer; Rev, reverse primer; bp, base pairs.
| Assay ID | Accession No. | Sequence | Product length (bp) | PCR efficiency (%) | |
|---|---|---|---|---|---|
| GAPDH | NM_001289745.2 | For: | 5ʹ- GCCTCAAGATCATCAGCAATGCCT-3’ | 104 | 98.1 |
| Rev: | 5ʹ- TGTGGTCATGAGTCCTTCCACGAT-3’ | ||||
| PPARGC1A | NM_001330751.1 | For: | 5ʹ-CAGCCTCTTTGCCCAGATCTT-3’ | 101 | 104.0 |
| Rev: | 5ʹ-TCACTGCACCACTTGAGTCCAC-3’ | ||||
| IL6 | NM_000600.4 | For: | 5ʹ-GCAGAAAAAGGCAAAGAATC-3’ | 178 | 100.9 |
| Rev: | 5ʹ-CTACATTTGCCGAAGAGC-3’ | ||||
| TNF | NM_000594.3 | For: | 5ʹ-AGGCAGTCAGATCATCTTC-3’ | 142 | 99.5 |
| Rev: | 5ʹ- TTATCTCTCAGCTCCACG-3’ | ||||
| DNMT1 | NM_001130823.2 | For: | 5ʹ-TACCTGGACGACCCTGACCTC-3’ | 103 | 94.5 |
| Rev: | 5ʹ-CGTTGGCATCAAAGATGGACA-3’ | ||||
| DNMT3a | NM_175629.2 | For: | 5ʹ-TATTGATGAGCGCACAAGAGAGC-3’ | 111 | 95.9 |
| Rev: | 5ʹ-GGGTGTTCCAGGGTAACATTGAG-3’ | ||||
| DNMT3b | NM_006892.3 | For: | 5ʹ-GGCAAGTTCTCCGAGGTCTCTG-3’ | 113 | 96.2 |
| Rev: | 5ʹ-TGGTACATGGCTTTTCGATAGGA-3’ | ||||
, where the slope is the gradient of the linear regression fitted to the standard curve. The efficiency of each assay was between 90 and 105% with a R2 > 0.99.
Interleukin-6 (IL-6)
Serum IL-6 concentrations prior to and immediately post-exercise were determined using high sensitivity enzyme immunoassay kits (R & D Systems, USA). Haematocrit and haemoglobin were used to ascertain plasma volume changes that were used to adjust serum IL-6 values [87].
Protein carbonyls (PC)
PC was assessed by an in-house ELISA [88,89]. Serum samples, PBMC lysates and standards were diluted in coating buffer (50mM sodium carbonate, pH = 9.2) to a concentration of 0.05mg/mL using the bicinchoninic assay method. Protein carbonyls groups were derivatised with 2, 4-dinitrophenylhydrazine (1mM, in 2M HCl) and incubated with monoclonal mouse anti-DNP antibody (Sigma Aldrich, UK) and rat anti-mouse IgE, conjugated to HRP (AbD Serotec, UK). Well absorbance was measured at 490nm and the PC concentration determined by using absorbance values of known PC standards made in our laboratory (1.28–5.20 nmol/mg protein). PC concentration in PBMCs was adjusted for changes in protein concentration and cell number (Beckman Coulter, UK) induced by acute exercise.
Statistical analysis
All statistical analysis was performed using IBM SPSS Statistics software (SPSS version 23). The data were assessed for normality by Shapiro-Wilk’s test. The composition of white blood cells from which the DNA is extracted is an important consideration in DNA methylation research; therefore, all DNA methylation analysis was conducted on cell heterogeneity adjusted values [59]. Analysis of mRNA expression was performed on log fold change data. DNA methylation and mRNA expression values were analyzed using a 2 (supplement) x 2 (trial) x 2 (time) repeated measures ANOVA. The impact of exercise is presented using the absolute values (mean of all trials for each time point), whereas, the impact of supplementation of FAs is presented as the relative change (Δ) between pre and post supplementation trials (post supplementation – pre supplementation). Values represented as mean ± 95% CI.
Spearman’s Rho correlation analysis was used to assess the relationship between DNA methylation values, mRNA expression values and physiological markers related to exercise performance, inflammation and oxidative stress. A p-value < 0.05 was considered as statistically significant. Moderate (>0.5) correlation coefficients were considered to be of interest; however, only large (> 0.7) correlation coefficients were deemed statistically significant.
Funding Statement
This research was supported by Loughborough University, School of Sport Exercise and Health Sciences and the NIHR Leicester Biomedical Research Centre.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Alegría-Torres JA, Baccarelli A, Bollati V.. Epigenetics and lifestyle. Epigenomics [Internet]. 2011;3:267–277. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22122337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Nitert MD, Dayeh T, Volkov P, et al. Impact of an exercise intervention on DNA methylation in skeletal muscle from first-degree relatives of patients with type 2 diabetes. Diabetes [Internet]. 2012;61:3322–3332. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3501844&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Rowlands DS, Page RA, Sukala WR, et al. Multi-omic integrated networks connect DNA methylation and miRNA with skeletal muscle plasticity to chronic exercise in Type 2 diabetic obesity. Physiol Genomics [Internet]. 2014;46:747–765. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25138607%5Cnhttp://physiolgenomics.physiology.org/content/physiolgenomics/46/20/747.full.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Seaborne RA, Strauss J, Cocks M, et al. Human skeletal muscle possesses an epigenetic memory of hypertrophy. Sci Rep. 2018;8:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Dimauro I, Scalabrin M, Fantini C, et al. Resistance training and redox homeostasis: correlation with age-associated genomic changes. Redox Biol [Internet]. 2016;10:34–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Denham J, Marques FZ, Bruns EL, et al. Epigenetic changes in leukocytes after 8 weeks of resistance exercise training. Eur J Appl Physiol [Internet]. 2016;116:1245–1253. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27155847 [DOI] [PubMed] [Google Scholar]
- [7].Denham J, O’Brien BJ, Marques FZ, et al. Changes in the leukocyte methylome and its effect on cardiovascular-related genes after exercise. J Appl Physiol [Internet]. 2015;118:475–488. DOI: 10.1152/japplphysiol.00878.2014 [DOI] [PubMed] [Google Scholar]
- [8].Barrès R, Yan J, Egan B, et al. Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metab [Internet]. 2012;15:405–411. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22405075 [DOI] [PubMed] [Google Scholar]
- [9].Lane SC, Camera DM, Lassiter DG, et al. Effects of sleeping with reduced carbohydrate availability on acute training responses. J Appl Physiol [Internet]. 2015;119:643–655. DOI: 10.1152/japplphysiol.00857.2014 [DOI] [PubMed] [Google Scholar]
- [10].Bajpeyi S, Covington JD, Taylor EM, et al. Skeletal muscle PGC1α −1 nucleosome position and −260 nt DNA methylation determine exercise response and prevent ectopic lipid accumulation in men. Endocrinology [Internet]. 2017;158:2190–2199. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28398573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Robson-Ansley PJ, Saini A, Toms C, et al. Dynamic changes in dna methylation status in peripheral blood Mononuclear cells following an acute bout of exercise: potential impact of exercise-induced elevations in interleukin-6 concentration. J Biol Regul Homeost Agents [Internet]. 2014;28:407–417. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25316129 [PubMed] [Google Scholar]
- [12].Connolly PH, Caiozzo VJ, Zaldivar F, et al. Effects of exercise on gene expression in human peripheral blood mononuclear cells. J Appl Physiol [Internet]. 2004;97:1461–1469. [DOI] [PubMed] [Google Scholar]
- [13].Büttner P, Mosig S, Lechtermann A, et al. Exercise affects the gene expression profiles of human white blood cells. J Appl Physiol [Internet]. 2007;102:26–36. Available from: http://jap.physiology.org/content/102/1/26 [DOI] [PubMed] [Google Scholar]
- [14].Gjevestad GO, Holven KB, Ulven SM.. Effects of exercise on gene expression of inflammatory markers in human peripheral blood cells: a systematic review. Curr Cardiovasc Risk Rep [Internet]. 2015;9:34 Available from: http://www.ncbi.nlm.nih.gov/pubmed/26005511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Booth FW, Chakravarthy MV, Spangenburg EE. Exercise and gene expression: physiological regulation of the human genome through physical activity. J Physiol [Internet]. 2002;543:399–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Egan B, Zierath JR. Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab [Internet]. 2013;17:162–184. [DOI] [PubMed] [Google Scholar]
- [17].Ventura-Clapier R, Garnier A, Veksler V. Transcriptional control of mitochondrial biogenesis: the central role of PGC-1alpha. Cardiovasc Res [Internet]. 2008;79:208–217. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18430751 [DOI] [PubMed] [Google Scholar]
- [18].Thomas AW, Davies NA, Moir H, et al. Exercise-associated generation of PPAR ligands activates PPAR signaling events and upregulates genes related to lipid metabolism. J Appl Physiol [Internet]. 2012;112:806–815. [DOI] [PubMed] [Google Scholar]
- [19].Yakeu G, Butcher L, Isa S, et al. Low-intensity exercise enhances expression of markers of alternative activation in circulating leukocytes: roles of PPAR?? and Th2 cytokines. Atherosclerosis [Internet]. 2010;212:668–673. [DOI] [PubMed] [Google Scholar]
- [20].Ferrer MD, Tauler P, Sureda A, et al. Antioxidant regulatory mechanisms in neutrophils and lymphocytes after intense exercise. J Sports Sci [Internet]. 2009;27:49–58. Available from: http://www.ncbi.nlm.nih.gov/pubmed/19031335 [DOI] [PubMed] [Google Scholar]
- [21].Alibegovic AC, Sonne MP, Højbjerre L, et al. Insulin resistance induced by physical inactivity is associated with multiple transcriptional changes in skeletal muscle in young men. Am J Physiol Endocrinol Metab [Internet]. 2010;299:E752–63. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20739510 [DOI] [PubMed] [Google Scholar]
- [22].Busquets-Cortés C, Capó X, Martorell M, et al. Training and acute exercise modulates mitochondrial dynamics in football players’ blood mononuclear cells. Eur J Appl Physiol [Internet]. 2017;117:1977–1987. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28748372 [DOI] [PubMed] [Google Scholar]
- [23].Gleeson M, Bishop NC, Stensel DJ, et al. The anti-inflammatory effects of exercise: mechanisms and implications for the prevention and treatment of disease. Nat Rev Immunol [Internet]. 2011;11:607–610. [DOI] [PubMed] [Google Scholar]
- [24].Cesari M, Penninx BWJH, Pahor M, et al. Inflammatory markers and physical performance in older persons: the InCHIANTI study. J Gerontol Ser A Biol Sci Med Sci. [Internet]. 2004;59:M242–M248. [DOI] [PubMed] [Google Scholar]
- [25].Visser M, Pahor M, Taaffe DR, et al. Relationship of interleukin-6 and tumor necrosis factor-alpha with muscle mass and muscle strength in elderly men and women: the Health. J Gerontol Biol Sci Med Sci. 2002;57:M326–32. [DOI] [PubMed] [Google Scholar]
- [26].Powers SK, Nelson WB, Hudson MB. Exercise-induced oxidative stress in humans: cause and consequences. Free Radic Biol Med [Internet]. 2011;51:942–950. [DOI] [PubMed] [Google Scholar]
- [27].He F, Li J, Liu Z, et al. Redox mechanism of reactive oxygen species in exercise. Front Physiol [Internet]. 2016;7:486 Available from: http://www.ncbi.nlm.nih.gov/pubmed/27872595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Angelini F, Pagano F, Bordin A, et al. The impact of environmental factors in influencing epigenetics related to oxidative states in the cardiovascular system. Oxid Med Cell Longev [Internet]. 2017;2017:2712751 Available from: http://www.ncbi.nlm.nih.gov/pubmed/28607629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Braconi C, Huang N, Patel T. MicroRNA-dependent regulation of DNA methyltransferase-1 and tumor suppressor gene expression by interleukin-6 in human malignant cholangiocytes. Hepatology [Internet]. 2010;9:NA–NA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Foran E, Garrity-Park MM, Mureau C, et al. Upregulation of DNA methyltransferase-mediated gene silencing, anchorage-independent growth, and migration of colon cancer cells by interleukin-6. Mol Cancer Res [Internet]. 2010;8:471–481. DOI: 10.1158/1541-7786.MCR-09-0496 [DOI] [PubMed] [Google Scholar]
- [31].Horsburgh S, Todryk S, Toms C, et al. Exercise-conditioned plasma attenuates nuclear concentrations of DNA methyltransferase 3B in human peripheral blood mononuclear cells. Physiol Rep [Internet]. 2015;3:1–10. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4760429&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hodge DR, Xiao W, Clausen PA, et al. Interleukin-6 regulation of the human DNA methyltransferase (HDNMT) gene in human erythroleukemia cells. J Biol Chem [Internet]. 2001;276:39508–39511. Available from: http://www.ncbi.nlm.nih.gov/pubmed/11551897 [DOI] [PubMed] [Google Scholar]
- [33].Sharples AP, Polydorou I, Hughes DC, et al. Skeletal muscle cells possess a ‘memory’ of acute early life TNF-α exposure: role of epigenetic adaptation. Biogerontology. 2016;17:603–617. [DOI] [PubMed] [Google Scholar]
- [34].Nile CJ, Read RC, Akil M, et al. Methylation status of a single CpG site in the IL6 promoter is related to IL6 messenger RNA levels and rheumatoid arthritis. Arthritis Rheum [Internet]. 2008;58:2686–2693. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18759290 [DOI] [PubMed] [Google Scholar]
- [35].Na YK, Hong HS, Lee WK, et al. Increased methylation of interleukin 6 gene is associated with obesity in korean women. Mol Cells [Internet]. 2015;38:452–456. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC4443287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zhang J, Wang C, Ha X, et al. DNA methylation of tumor necrosis factor-α, monocyte chemoattractant protein-1, and adiponectin genes in visceral adipose tissue is related to type 2 diabetes in the Xinjiang Uygur population. J Diabetes [Internet]. 2017;9:699–706. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27573980 [DOI] [PubMed] [Google Scholar]
- [37].Kaut O, Ramirez A, Pieper H, et al. DNA methylation of the TNF-α promoter region in peripheral blood monocytes and the cortex of human Alzheimer’s disease patients. Dement Geriatr Cogn Disord [Internet]. 2014;38:10–15. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24556805 [DOI] [PubMed] [Google Scholar]
- [38].Calder PC. Marine omega-3 fatty acids and inflammatory processes: effects, mechanisms and clinical relevance. Biochim Biophys Acta - Mol Cell Biol Lipids [Internet]. 2015;1851:469–484. Available from: http://linkinghub.elsevier.com/retrieve/pii/S1388198114001656 [DOI] [PubMed] [Google Scholar]
- [39].Rosignoli P, Fuccelli R, Fabiani R, et al. Effect of olive oil phenols on the production of inflammatory mediators in freshly isolated human monocytes. J Nutr Biochem [Internet]. 2013;24:1513–1519. [DOI] [PubMed] [Google Scholar]
- [40].Mickleborough TD, Sinex JA, Platt D, et al. The effects PCSO-524®, a patented marine oil lipid and omega-3 PUFA blend derived from the New Zealand green lipped mussel (Perna canaliculus), on indirect markers of muscle damage and inflammation after muscle damaging exercise in untrained men: a random. J Int Soc Sports Nutr [Internet]. 2015;12:1–17. Available from: http://www.jissn.com/content/12/1/10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Marques CG, Santos VC, Levada-Pires AC, et al. Effects of DHA-rich fish oil supplementation on the lipid profile, markers of muscle damage, and neutrophil function in wheelchair basketball athletes before and after acute exercise. Appl Physiol Nutr Metab [Internet. 2015;40:596–604. [DOI] [PubMed] [Google Scholar]
- [42].Nieman DC, Henson DA, McAnulty SR, et al. n-3 polyunsaturated fatty acids do not alter immune and inflammation measures in endurance athletes. Int J Sport Nutr Exerc Metab [Internet]. 2009;19:536–546. Available from: http://www.ncbi.nlm.nih.gov/pubmed/19910654 [DOI] [PubMed] [Google Scholar]
- [43].Martorell M, Capó X, Sureda A, et al. Effect of DHA on plasma fatty acid availability and oxidative stress during training season and football exercise. Food Funct [Internet]. 2014;5:1920–1931. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24955731 [DOI] [PubMed] [Google Scholar]
- [44].Saini A, Sharples AP, Al-Shanti N, et al. Omega-3 fatty acid EPA improves regenerative capacity of mouse skeletal muscle cells exposed to saturated fat and inflammation. Biogerontology. 2017;18:109–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Tremblay BL, Guénard F, Rudkowska I, et al. Epigenetic changes in blood leukocytes following an omega-3 fatty acid supplementation. Clin Clin Epigenet. 2017;9:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Tartibian B, Maleki BH, Abbasi A. 11.11: omega-3 fatty acids supplementation attenuates inflammatory markers after eccentric exercise in untrained men. Clin J Sport Med. 2011;21:131–137. [DOI] [PubMed] [Google Scholar]
- [47].Ma Y, Smith CE, Lai C, et al. The effects of omega-3 polyunsaturated fatty acids and genetic variants on methylation levels of the interleukin-6 gene promoter. Mol Nutr Food Res [Internet]. 2016;60:410–419. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26518637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Rundblad A, Holven KB, Bruheim I, et al. Effects of fish and krill oil on gene expression in peripheral blood mononuclear cells and circulating markers of inflammation: a randomised controlled trial. J Nutr Sci. 2018;7:e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Arpón A, Milagro FI, Razquin C, et al. Impact of consuming extra-virgin olive oil or nuts within a mediterranean diet on DNA methylation in peripheral white blood cells within the PREDIMED-navarra randomized controlled trial: a role for dietary lipids. Nutrients [Internet]. 2017;10 Available from: http://www.ncbi.nlm.nih.gov/pubmed/29295516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Da Silva IRV, de Araujo CLP, Dorneles GP, et al. Exercise-modulated epigenetic markers and inflammatory response in COPD individuals: A pilot study. Respir Physiol Neurobiol [Internet]. 2017;242:89–95. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28435027 [DOI] [PubMed] [Google Scholar]
- [51].Lindholm ME, Marabita F, Gomez-Cabrero D, et al. An integrative analysis reveals coordinated reprogramming of the epigenome and the transcriptome in human skeletal muscle after training. Epigenetics [Internet. 2015;9:1557–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].King-Himmelreich TS, Schramm S, Wolters MC, et al. The impact of endurance exercise on global and AMPK gene-specific DNA methylation. Biochem Biophys Res Commun [Internet]. 2016;474:284–290. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27103439 [DOI] [PubMed] [Google Scholar]
- [53].Karimi M, Vedin I, Freund Levi Y, et al. DHA-rich n–3 fatty acid supplementation decreases DNA methylation in blood leukocytes: the OmegAD study. Am J Clin Nutr [Internet]. 2017;106:1157–1165. Available from: https://academic.oup.com/ajcn/article/106/4/1157-1165/4652030 [DOI] [PubMed] [Google Scholar]
- [54].Di Francesco A, Arosio B, Falconi A, et al. Global changes in DNA methylation in Alzheimer’s disease peripheral blood mononuclear cells. Brain Behav Immun [Internet]. 2015;45:139–144. [DOI] [PubMed] [Google Scholar]
- [55].Wu H-C, Delgado-Cruzata L, Flom JD, et al. Global methylation profiles in DNA from different blood cell types. Epigenetics [Internet]. 2011;6:76–85. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20890131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Lisanti S, Omar WAW, Tomaszewski B, et al. Comparison of methods for quantification of global DNA methylation in human cells and tissues. PLoS One [Internet]. 2013;8:e79044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Rudkowska I, Raymond C, Ponton A, et al. Validation of the use of peripheral blood mononuclear cells as surrogate model for skeletal muscle tissue in nutrigenomic studies. Omi A J Integr Biol [Internet]. 2011;15:1–7. [DOI] [PubMed] [Google Scholar]
- [58].Clarke-Harris R, Wilkin TJ, Hosking J, et al. PGC1α promoter methylation in blood at 5-7 years predicts adiposity from 9 to 14 years (EarlyBird 50). Diabetes [Internet]. 2014;63:2528–2537. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24622795 [DOI] [PubMed] [Google Scholar]
- [59].Jones MJ, Islam SA, Edgar RD, et al. Adjusting for cell type composition in DNA methylation data using a regression-based approach. Methods Mol Biol [Internet]. 2017;1589:99–106. [DOI] [PubMed] [Google Scholar]
- [60].Jaffe AE, Irizarry RA. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol [Internet]. 2014;15:R31 Available from. ;:. http://www.ncbi.nlm.nih.gov/pubmed/24495553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Tyrrell DJ, Bharadwaj MS, Van Horn CG, et al. Respirometric profiling of muscle mitochondria and blood cells are associated with differences in gait speed among community-dwelling older adults. J Gerontol Ser A Biol Sci Med Sci [Internet]. 2015;70:1394–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Broadbent J, Sampson D, Sabapathy S, et al. Gene networks in skeletal muscle following endurance exercise are coexpressed in blood neutrophils and linked with blood inflammation markers. J Appl Physiol [Internet]. 2017;122:752–766. [DOI] [PubMed] [Google Scholar]
- [63].Sharples AP, Stewart CE, Seaborne RA. Does skeletal muscle have an ’epi’-memory? The role of epigenetics in nutritional programming, metabolic disease, aging and exercise. Aging Cell [Internet]. 2016:1–14 DOI: 10.1111/acel.12486\nhttp://www.ncbi.nlm.nih.gov/pubmed/27102569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Shaw B, Leung WC, Tapp HS, et al. A change in physical activity level affects leukocyte DNA methylation of genes implicated in cardiovascular disease in the elderly. Proc Physiol Soc [Internet]. 2014;31:C46 Available from: http://www.physoc.org/proceedings/abstract/Proc Physiol Soc 31C46 [Google Scholar]
- [65].Marques-Rocha JL, Milagro FI, Mansego ML, et al. LINE-1 methylation is positively associated with healthier lifestyle but inversely related to body fat mass in healthy young individuals. Epigenetics [Internet]. 2016;11:49–60. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26786189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Hermsdorff HH, Mansego ML, Campión J, et al. TNF-alpha promoter methylation in peripheral white blood cells: relationship with circulating TNFα, truncal fat and n-6 PUFA intake in young women. Cytokine [Internet]. 2013;64:265–271. [DOI] [PubMed] [Google Scholar]
- [67].Hoile SP, Clarke-Harris R, Huang R-C, et al. Supplementation with N-3 long-chain polyunsaturated fatty acids or olive oil in men and women with renal disease induces differential changes in the DNA methylation of FADS2 and ELOVL5 in Peripheral blood mononuclear cells. PLoS One [Internet]. 2014;9:e109896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Laye MJ, Pedersen BK. Acute exercise and Ca2+ stimulation regulate enzymes involved in DNA methylation in human skeletal muscle. Med Sci Sport Exercise [Internet]. 2010;42:23Available from: https://insights.ovid.com/crossref?an=00005768-201010002-00053 [Google Scholar]
- [69].Niculescu MD, Lupu DS, Craciunescu CN. Alpha-linolenic acid alters cell cycle, apoptosis, and DNA methyltransferase expression in mouse neural stem cells, but not global DNA methylation. J Hum Nutr Food Sci [Internet]. 2014;2:1026 Available from: https://www.jscimedcentral.com/Nutrition/nutrition-2-1026.pdf [Google Scholar]
- [70].Niculescu MD, Lupu DS, Craciunescu CN. Perinatal manipulation of α-linolenic acid intake induces epigenetic changes in maternal and offspring livers. FASEB J [Internet]. 2013;27:350–358. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22997227 [DOI] [PubMed] [Google Scholar]
- [71].Montaño A, Hernández M, Garrido I, et al. Fatty acid and phenolic compound concentrations in eight different monovarietal virgin olive oils from extremadura and the relationship with oxidative stability. Int J Mol Sci. [Internet] 2016;17:1–20. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27886101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Corominas-Faja B, Cuyàs E, Lozano-Sánchez J, et al. Extra-virgin olive oil contains a metabolo-epigenetic inhibitor of cancer stem cells. Carcinogenesis [Internet]. 2018;39:601–613. Available from: http://www.ncbi.nlm.nih.gov/pubmed/29452350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Casillas MA, Lopatina N, Andrews LG, et al. Transcriptional control of the DNA methyltransferases is altered in aging and neoplastically-transformed human fibroblasts. Mol Cell Biochem. 2003;252:33–43. [DOI] [PubMed] [Google Scholar]
- [74].Duursma AM, Kedde M, Schrier M, et al. miR-148 targets human DNMT3b protein coding region. Rna[Internet]. 2008;14:872–877. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18367714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Fabbri M, Garzon R, Cimmino A, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A [Internet]. 2007;104:15805–15810. Available from: http://www.pnas.org/cgi/doi/10.1073/pnas.0707628104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Garzon R, Liu S, Fabbri M, et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood [Internet]. 2009;113:6411–6418. Available from: http://www.bloodjournal.org/content/113/25/6411.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Xu Y, Chao L, Wang J, et al. miRNA-148a regulates the expression of the estrogen receptor through DNMT1-mediated DNA methylation in breast cancer cells. Oncol Lett [Internet]. 2017;14:4736–4740. Available from: http://www.ncbi.nlm.nih.gov/pubmed/29085474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Silva GJJ, Bye A, El Azzouzi H, et al. MicroRNAs as important regulators of exercise adaptation. Prog Cardiovasc Dis [Internet]. 2017;60:130–151. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28666746 [DOI] [PubMed] [Google Scholar]
- [79].Roessler C, Kuhlmann K, Hellwing C, et al. Impact of polyunsaturated fatty acids on miRNA profiles of monocytes/macrophages and endothelial cells-a pilot study. Int J Mol Sci [Internet]. 2017;18:1–24. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28134837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Chakraborty N, Muhie S, Kumar R, et al. Contributions of polyunsaturated fatty acids (PUFA) on cerebral neurobiology: an integrated omics approach with epigenomic focus. J Nutr Biochem [Internet]. 2017;42:84–94. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28152499 [DOI] [PubMed] [Google Scholar]
- [81].D’Amore S, Vacca M, Cariello M, et al. Genes and miRNA expression signatures in peripheral blood mononuclear cells in healthy subjects and patients with metabolic syndrome after acute intake of extra virgin olive oil. Biochim Biophys Acta - Mol Cell Biol Lipids [Internet. 2016;1861:1671–1680. [DOI] [PubMed] [Google Scholar]
- [82].Howley ET, Bassett DR, Welch HG. Criteria for maximal oxygen uptake: review and commentary. Med Sci Sports Exercise. Internet]. 1995;27:1292–1301. Available from: http://www.ncbi.nlm.nih.gov/pubmed/8531628 [PubMed] [Google Scholar]
- [83].Jeukendrup A, Saris WH, Brouns F, et al. A new validated endurance performance test. Med Sci Sports Exercise [Internet]. 1996;28:266–270. Available from: https://insights.ovid.com/crossref?an=00005768-199602000-00017 [DOI] [PubMed] [Google Scholar]
- [84].Metherel AH, Armstrong JM, Patterson AC, et al. Assessment of blood measures of n-3 polyunsaturated fatty acids with acute fish oil supplementation and washout in men and women. Prostaglandins Leukot. Essent Fat Acids [Internet. 2009;81:23–29. [DOI] [PubMed] [Google Scholar]
- [85].McGlory C, Galloway SDR, Hamilton DL, et al. Temporal changes in human skeletal muscle and blood lipid composition with fish oil supplementation. Prostaglandins Leukot. Essent Fat Acids [Internet. 2014;90:199–206. [DOI] [PubMed] [Google Scholar]
- [86].Karimi M, Johansson S, Stach D, et al. LUMA (LUminometric Methylation Assay)–a high throughput method to the analysis of genomic DNA methylation. Exp Cell Res [Internet]. 2006. [Cited 2015 September 6];312:1989–1995. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16624287 [DOI] [PubMed] [Google Scholar]
- [87].Dill DB, Costill DL. Calculation of percentage changes in volumes of blood, plasma, and red cells in dehydration. J Appl Physiol [Internet] 1974;37:247–248. DOI: 10.1152/jappl.1974.37.2.247 [DOI] [PubMed] [Google Scholar]
- [88].Buss H, Chan TP, Sluis KB, et al. Protein carbonyl measurement by a sensitive ELISA method. Free Radic Biol Med [Internet]. 1997;23:361–366. Available from: http://www.ncbi.nlm.nih.gov/pubmed/9214571 [DOI] [PubMed] [Google Scholar]
- [89].Carty JL, Bevan R, Waller H, et al. The effects of vitamin C supplementation on protein oxidation in healthy volunteers. Biochem Biophys Res Commun. 2000;273:729–735. [Cited. 2000 June 30]. Available from http://www.ncbi.nlm.nih.gov/pubmed/8531628 [DOI] [PubMed] [Google Scholar]


