ABSTRACT
A previous study reported that miR-155-5p knockout mice were more resistant to herpes simplex virus type I (HSV-1) infection. However, the exact underlying molecular mechanism remains to be elucidated. Here, we demonstrated that HSV-1 infection upregulates miR-155-5p expression. By binding to the promoter of serine/arginine-rich splicing factor 2 (SRSF2), which is an important transcriptional activator of HSV-1 genes that was previously reported by our group, and altering the histone modification located near the transcription start site (TSS) of the SRSF2 gene, miR-155-5p promotes the transcription of the SRSF2 gene, ultimately increasing viral replication and viral gene expression. Our results provide insight for an understanding of the roles and molecular mechanism of miR-155-5p in HSV-1 replication and the epigenetic control of SRSF2 gene expression.
KEYWORDS: Mir-155-5p, HSV-1, SRSF2, epigenetic regulation, gene transcription
Introduction
Herpes simplex virus type I (HSV-1) infection is widespread worldwide [1]. Epidemiological investigations have shown that HSV-1 infection has exceeded Herpes simplex virus type II (HSV-2) infection in becoming the main cause of genital herpes [2]. Additionally, HSV-1 infection has been reported to be closely related to Alzheimer’s disease [3,4] and can lead to encephalitis and even death [5]. HSV-1 is a human alpha herpesvirus that contains a double-stranded DNA that encodes more than 80 genes transcribed by RNA polymerase II (RNAP II) [6]. After infection, HSV-1 uses a series of host cell factors, such as Host cell factor 1 [7,8], DNA methyltransferase DNMT3A [9] and early growth response protein 1 [8], to facilitate its life cycle. Specifically, several cellular microRNAs (miRNAs) have been identified to be involved in HSV-1 infection [10–13].
MiRNAs are small noncoding RNAs with a length of approximately 22 bp that have been reported to be associated with a variety of fundamental biological and pathological processes, such as cell autophagy, proliferation, apoptosis, differentiation, metabolism and cancer [14–19]. By recruiting Argonaut protein complexes to bind the 3’UTR of mRNAs, miRNAs promote the degradation, inhibition or promotion of the translation of these mRNAs [20–22]. Of these miRNAs, miR-155-5p is among the best characterized miRNAs. Under hypoxic conditions, upregulated miR-155-5p was demonstrated to induce autophagy by regulating the expression of several autophagy-related genes [23]. In tumor cells, miR-155-5p functions as a promotor of proliferation in oral squamous cell carcinoma [24] and bladder cancer cells [25]. In addition, researchers have found that miR-155-5p mediates T lymphocyte differentiation [26] and inhibits cell apoptosis [27]. In pathogenesis, miR-155-5p is associated with many diseases, such as cancer [28,29], cardiovascular disease [30], autoimmune disease [31], metabolic disease [32], and neurological disorders [33]. During HSV-1 infection, several studies have shown that the expression level of miR-155-5p is markedly increased by the viral infection [34,35]. However, to date, the effect of miR-155-5p on HSV-1 replication and the underlying molecular mechanism are unclear [36,37].
In the current study, we found that HSV-1 infection increases the expression level of miR-155-5p and upregulated miR-155-5p, in turn, enhances HSV-1 replication. Furthermore, miR-155-5p was revealed to be involved in the HSV-1 infection-mediated induction of serine/arginine-rich splicing factor 2 (SRSF2), which is an important transcriptional activator of viral genes expression that we have previously reported [38]. By interacting with the SRSF2 promoter, miR-155-5p alters the histone modification located near the TSS of the SRSF2 promoter and enhances the transcription of the SRSF2 gene, ultimately increasing HSV-1 gene expression. These findings provide insight into the transcriptional activity of miRNAs and broaden our views concerning the mechanisms by which gene expression is regulated by miRNAs.
Results
miR-155-5p enhances HSV-1 replication
It has been reported that blocking the expression of miR-155-5p could reduce the severity of stromal keratitis (SK) lesion caused by HSV-1 infection [37]. To illustrate the roles of miR-155-5p in HSV-1 replication, we first examined the expression level of miR-155-5p in HeLa cells infected with or without HSV-1 and found that miR-155-5p expression in these cells was increased by approximately 20 times after HSV-1 infection for 4 hours (Figure 1(a)). To explore miR-155-5p expression patterns during HSV-1 infection, we determined the miR-155-5p levels in HeLa cells infected with HSV-1 at different time points. The results demonstrated that miR-155-5p expression increased 2 hours after HSV-1 infection and peaked at 4 hours before gradually declining thereafter (Figure 1(b)). Then, we overexpressed and knocked down miR-155-5p using miR-155-5p mimics (miR-155-5p) and inhibitors (miR-155-5pi) (Figure 1(c)) and examined HSV-1 glycoprotein expression using fluorescence microscopy. The HSV-1 glycoprotein density and intensity in the HeLa cells transfected with the miR-155-5p mimics were much higher than those in the cells transfected with the negative control siRNA (Figure 1(d–e)). In contrast, the downregulation of miR-155-5p expression by the transfection with the miR-155-5p inhibitors resulted in a much lower density and intensity of HSV-1 glycoproteins (Figure 1(d–e)), suggesting that miR-155-5p affected both HSV-1 replication and spread. Additionally, we determined the effects of miR-155-5p on HSV-1 plaque formation. HeLa cells transfected with miR-155-5p mimics or inhibitors were infected with HSV-1 at an MOI of 1 for 12, 24, or 36 hours. The viral supernatants were collected, and the viral titers were determined using plaque assays. The HSV-1 titers in the culture media from the miR-155-5p-overexpressed HeLa cells were markedly higher than the titers in the HeLa cells transfected with the negative control siRNA (Figure 1(f)). The opposite results were observed in the miR-155-5p-downregulated cells (Figure 1(f)). Taken together, these results demonstrated that HSV-1 infection increases miR-155-5p expression and upregulated miR-155-5p, in turn, enhances HSV-1 replication.
Figure 1.
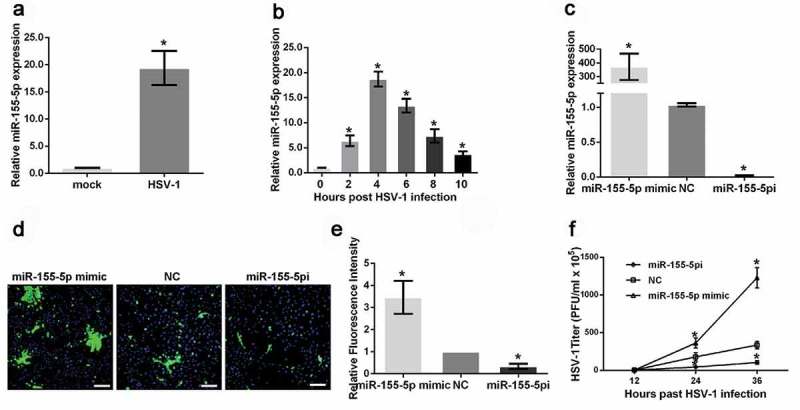
MiR-155-5p promotes HSV-1 replication. (a) HeLa cells were mock infected or infected with HSV-1 for 4 hours and harvested. The relative expression level of miR-155-5p was determined with real-time PCR. (b). HeLa cells were mock infected or infected with HSV-1 and harvested at the indicated time points. The relative expression level of miR-155-5p was determined with real-time PCR. (c) HeLa cells were transfected with miR-155-5p mimics, inhibitors or negative control for 36 hours. The relative expression level of miR-155-5p was determined with real-time PCR. (d and e). After 36 hours of transfection with the miR-155-5p mimics, inhibitors or negative control, the HeLa cells were infected with HSV-1 at an MOI of 1 for 12 hours, fixed, stained with an anti-HSV-1 glycoprotein antibody (green) and subjected to a confocal microscopy analysis. DAPI (blue) was used to stain the nuclei (d). The fluorescence signal value was measured using ImageJ software and normalized to a single cell. The data are normalized to the negative control level (e). (f) After 36 hours of transfection with the miR-155-5p mimics, inhibitors or negative control, the HeLa cells were infected with HSV-1 at an MOI of 1. The culture media were collected at the indicated time points after infection and subjected to a plaque assay. *p < 0.01.
miR-155-5p is involved in HSV-1 infection-mediated SRSF2 expression
Our previous study showed that SRSF2 mediates HSV-1 replication by regulating viral gene expression at the transcriptional and posttranscriptional levels [38]. In the current study, we examined the effects of HSV-1 infection on SRSF2 expression. Both the real-time PCR and immunoblot analyses revealed that HSV-1 infection caused an induction of SRSF2 gene expression (Figure 2(a–b)). Surprisingly, in the miR-155-5p-depletion cells, HSV-1 infection could not alter the expression of SRSF2 (Figure 2(a–b)). Additionally, the downregulation of miR-155-5p led to the inhibition of SRSF2 expression at the mRNA and protein levels (Figure 2(a–b)), and upregulation of miR-155-5p by miR-155-5p mimic increased the expression level of SRSF2 (Figure 2(c–d)). To further explore the role of miR-155-5p in HSV-1 infection-mediated SRSF2 expression, we generated luciferase reporter constructs by inserting the promoter fragments of SRSF2 into pGL3-enhancer vectors and measured the transcriptional activity of the SRSF2 promoter in HSV-1 infected HeLa cells transfected with miR-155-5p mimics or inhibitors using a luciferase assay. The results demonstrated that the overexpression of miR-155-5p increased the transcriptional activity of the SRSF2 promoter and that the downregulation of miR-155-5p inhibited the transcriptional activity of the SRSF2 promoter (Figure 2(e)). In addition, the results of another luciferase assay showed that HSV-1 infection promoted the transcription of SRSF2 (Figure 2(f)). However, the promotion effect of HSV-1 infection disappeared in these cells after the depletion of miR-155-5p (Figure 2(f)), indicating that miR-155-5p is involved in HSV-1 infection-mediated SRSF2 expression at the transcriptional level.
Figure 2.
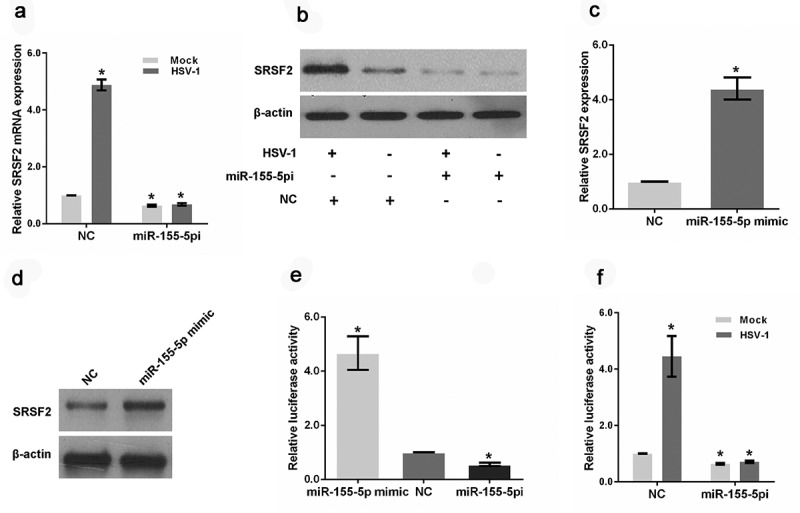
MiR-155-5p is involved in HSV-1 infection-mediated SRSF2 expression. (a). HeLa cells transfected with miR-155-5p inhibitors or negative control were mock infected or infected with HSV-1 for 4 hours. Relative SRSF2 level was analyzed with real-time PCR. The data were normalized to the negative control level in the mock-infected cells. (b). HeLa cells transfected with miR-155-5p inhibitors or negative control were mock infected or infected with HSV-1 for 4 hours. The cells were harvested and subjected to a western blot analysis to analyze the expression of SRSF2 and β-actin. (c). HeLa cells were transfected with a miR-155-5p mimic or negative control. The relative SRSF2 level was analysed with real-time PCR. The data were normalized to the negative control level. (d). HeLa cells were transfected with a miR-155-5p mimic or negative control. The cells were harvested and subjected to western blotting analysis to analyse SRSF2 and β-Actin expression. (e). After co-transfection with the miR-155-5p mimics, inhibitors or negative control and the pGL3 enhancer plasmid containing the SRSF2 promoter for 36 hours, the HeLa cells were infected with HSV-1 at an MOI of 1 for 4 hours. The relative transcriptional activities of the SRSF2 promoter were determined with a luciferase assay. (f). After co-transfection with the miR-155-5p inhibitors or negative control and the pGL3 enhancer plasmid containing the SRSF2 promoter for 36 hours, the HeLa cells were infected with HSV-1 or mock infected. The relative transcriptional activities of the SRSF2 promoter were determined with a luciferase assay. The data were normalized to the negative control level in the mock-infected cells. *p < 0.01.
miR-155-5p associates with the SRSF2 promoter
To explore the mechanism by which miR-155-5p regulates SRSF2 transcription, we analyzed the SRSF2 promoter sequence to search for potential miR-155-5p binding motifs and identified a motif located between 222 and 232 bp upstream of the human SRSF2 transcription start site (TSS) (Figure 3(a)). To determine whether miR-155-5p binds this site, first, we synthesized Cy3-labeled miR-155-5p at the 5ʹ end of the miRNA and FAM-labeled fragments of the SRSF2 promoter with or without potential miR-155-5p binding motifs in vitro. Then, HeLa cells were co-transfected with Cy3-labeled miR-155-5p, and these fragments were subjected to an immunofluorescence assay. As shown in Figure 3(b), miR-155-5p largely co-localized with the fragments containing the miR-155-5p-binding motif. The intensity plot shows yellow fluorescence, indicating the co-localization of miR-155-5p and fragments containing the SRSF2-binding motif (Figure 3(b), upper panels). Conversely, miR-155-5p did not co-localize with the fragments with a deletion in the miR-155-5p-binding motif. The intensity plot showed red and green fluorescence, indicating no co-localization between miR-155-5p and the fragments due to the deletion of the miR-155-5p-binding motif (Figure 3(b), lower panels). To confirm the binding sites, we synthetized a short fragment (TTCATGCCAAT) complementary to the miR-155-5p binding sequence (Blocker) and then performed qRT-PCR, western blotting and immunofluorescence assays to determine whether miR-155-5p could enhance SRSF2 expression and HSV-1 replication when the miR-155-5p binding sites were blocked. The results demonstrated that the synthetized short fragment blocked activation of SRSF2 expression (Figure 3(c–d)) and HSV-1 replication (Figure 3(e–f)) by miR-155-5p.
Figure 3.
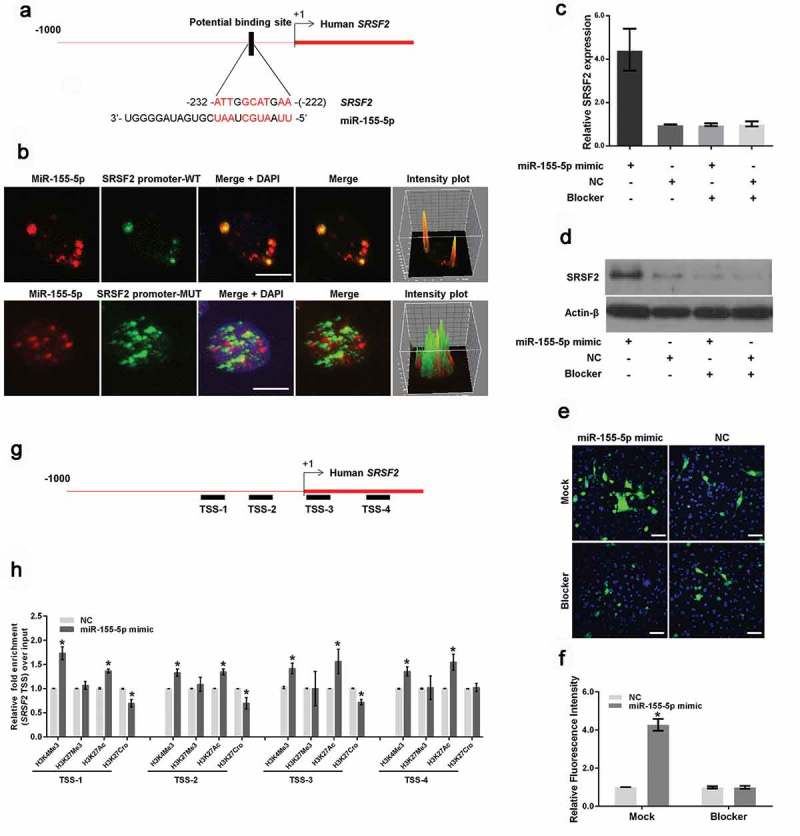
MiR-155-5p interacts with the SRSF2 promoter. (a). Schematic representation of the miR-155-5p-binding site in the human SRSF2 gene. The black box shows the potential binding site, and the red characters indicate matching sequences. (b). HeLa cells were cotransfected with Cy3-labeled miR-155-5p (red) and FAM-labeled fragments of the SRSF2 promoter containing potential miR-155-5p binding motifs (green) or a fragment with a deletion in the miR-155-5p-binding motif (green). The cells were infected with HSV-1, fixed and subjected to a confocal microscopy analysis. The intensity plots of the red and green channels were analyzed with ImageJ software. DAPI (blue) was used to stain the nuclei. Scale bars, 10 μm. (c and d). 12 hours after transfection with the Blocker or Mock control, the HeLa cells were transfected with a miR-155-5p mimic or negative control and incubated for 24 hours. The SRSF2 expression levels were measured with real-time PCR (c) and western blotting (d). (e and f). 12 hours after transfection with the Blocker or Mock control, the HeLa cells were transfected with a miR-155-5p mimic or negative control for 24 hours, infected with HSV-1 for 12 hours, fixed, stained with an anti-HSV-1 glycoprotein antibody (green) and subjected to confocal microscopy analysis. DAPI (blue) was used to stain the nuclei (e). The fluorescence signal value was measured using the ImageJ software and normalized to a single cell. The data are normalized to the negative control level (f). (g). Schematic diagram showing the gene structure of SRSF2 in which the black boxes represent the primer-amplified regions. (h). HeLa cells transfected with the miR-155-5p mimics or negative control were collected for ChIP assays to analyze the relative fold enrichment of the SRSF2 promoter by an anti-H3K4Me3, H3K27Me3, H3K27Ac or H3K27Cro antibody. *p < 0.01.
We further determined whether miR-155-5p altered the histone modifications near the TSS of the SRSF2 gene. We designed sets of primer pairs that recognized the corresponding TSS regions of the SRSF2 gene (Figure 3(c)) and performed chromatin immunoprecipitation (ChIP) experiments with antibodies against tri-methylated histone H3 at lysine 4 (H3K4Me3), tri-methylated histone H3 at lysine 27 (H3K27Me3), acetylated histone H3 at lysine 27 (H3K27Ac) or crotonylated histone H3 at lysine 27 (H3K27Cro) in HeLa cells transfected with miR-155-5p or negative control. In these histone modifications, H3K4Me3 and H3K27Ac at transcription start sites (TSS) are markers of actively transcribed genes, and H3K27Me3 at TSS is associated with gene repression [39]; H3K27Cro is a newly identified modification with unknown functions [40]. The results showed that the overexpression of miR-155-5p increased the enrichment of H3K4Me3 and H3K27Ac at the TSS of the promoter and reduced the enrichment of H3K27Cro at TSS1, TSS2 and TSS3 of the promoter (Figure 3(d)). These data are the first to show that miR-155-5p interacts with the SRSF2 promoter and alters histone modifications near the TSS of the SRSF2 promoter.
MiR-155-5p increases HSV-1 gene expression via the regulation of SRSF2
Furthermore, we measured the expression levels of the HSV-1 infected cell polypeptide 0 (ICP0), infected cell polypeptide 27 (ICP27) and thymidine kinase (TK) genes required for the appropriate expression of viral early and late gene products in HeLa cells transfected with a miR-155-5p mimic or negative control. The results showed that the upregulation of miR-155-5p enhanced ICP0, ICP27 and TK expression at the mRNA (Figure 4(a-c)) and protein levels (Figure 4(d)), respectively. To better understand whether SRSF2 is a key factor in miR-155-5p mediated-viral gene expression, we depleted SRSF2 expression with SRSF2 specific siRNAs in HeLa cells transfected with a miR-155-5p mimic or negative control and examined the expression level of ICP0, ICP27 and TK. As shown in Figure 4, in the cells with the depletion of SRSF2, the upregulation of miR-155-5p by the transfection with the miR-155-5p mimic resulted in a loss of the promotion effects on viral gene expression, indicating that SRSF2 functions as an important mediator of miR-155-5p regulated viral gene expression.
Figure 4.
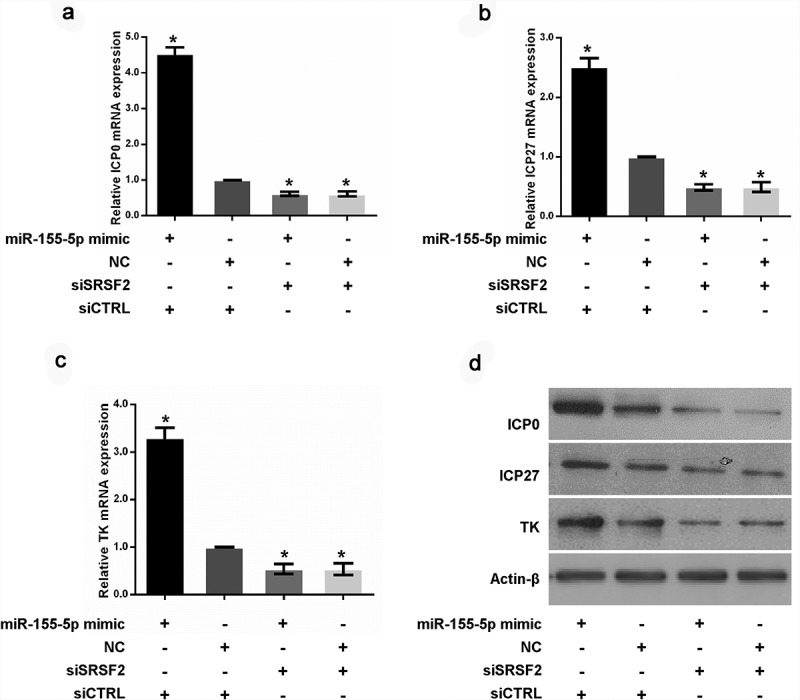
MiR-155-5p increases HSV-1 gene expression via the regulation of SRSF2. After co-transfecting HeLa cells with miR-155-5p mimic and SRSF2 siRNAs or negative control, the cells were infected with HSV-1, and the ICP0, ICP27 and TK expression levels were measured with real-time PCR (a-c) and western blotting (d). *p < 0.01.
Discussion
Upon entry into host cells, HSV-1 uses a series of host cell factors to enable efficient viral replication. MiRNAs are regulatory molecules that control the virus life cycle. Many miRNAs have been shown to promote viral replication by targeting factors involved in host antiviral activity and survival [41–43]. However, few reports have investigated the direct regulation of miRNAs in viral gene products that are important for viral infection. MiR-155-5p is one of the well characterized miRNAs and associated with multiple biological processes and diseases, including HSV-1 infection. Study showed that miR-155-5p KO mice presented diminished stromal keratitis lesions caused by HSV-1 infection [37]. However, the exact mechanism remained unclear. In the current study, we demonstrated that HSV-1 infection increases the expression of miR-155-5p and upregulated miR-155-5p, in turn, enhanced HSV-1 replication. Furthermore, miR-155-5p was revealed to attribute to induction of SRSF2 by HSV-1 infection. Through binding to SRSF2 promoter and altering the histone modification located nearby the TSS of SRSF2 promoter, miR-155-5p enhanced the transcription of the SRSF2 gene and finally increased HSV-1 genes expression.
SRSF2, one of serine/arginine-rich (SR) protein family member, is an important component of cell structure speckle. SRSF2 consists of an RNA recognition motif and a motif rich in serine and arginine residues [44]. The RNA recognition motif binds to exon of target pre-mRNA and the motif rich in serine and arginine residues bind to other SR) protein family member [44], finally completing the RNA splicing. Our previous study found that SRSF2 not only participated in the splicing of the HSV-1 ICP0 pre-mRNA, but also mediated the transcription of several genes, such as ICP 0, ICP27 and TK, through the interaction with these genes promoters [38]. In the current study, we further study the effects of HSV-1 infection on expression of SRSF2 and found that HSV-1 infection induced SRSF2 expression and miR-155-5p involved in this promotion. In SRSF2 promoter, a miR-155-5p potential binding motif was identified. Through binding to the promoter, miR-155-5p altered levels of histone modification located nearby the TSS of SRSF2 promoter, including H3K4Me3, H3K27Ac and H3K27Cro. In which, H3K4Me3 and H3K27Ac are markers for actively transcribed genes, and H3K27Cro is a newly identified modification with unknown functions for genes regulation. These results suggested the epigenetic regulation of miR-155-5p in SRSF2 expression.
Traditionally, miRNAs regulated the degradation or translation of target mRNAs through recruiting argonaut protein complexes to bind to the 3’UTR of these mRNAs and there has been no report that miRNAs can regulate gene expression at the transcriptional level. In recent years, a class of small activating RNAs (saRNAs) that can activate gene transcription has been discovered, such as some double-stranded RNAs (dsRNAs) and piwi-related RNAs (piRNAs) [45–48]. These saRNAs specifically recognize and bind to the promoter motifs, thereby altering the local conformation of chromatin and initiating the transcription of the corresponding genes. There are two similarities between miRNAs and saRNAs: (1) their length is about 22 bp and (2) Argonaut2 protein involved in their function in genes regulation. Therefore, we speculated that some miRNAs (such as miR-155-5p) might belong to saRNAs or at least had the potential to function as transcription regulators like saRNAs.
In this study, we investigated the role of miR-155-5p in HSV-1 replication. We found that HSV-1 infection upregulates miR-155-5p expression and miR-155-5p, in turn, enhances HSV-1 replication and viral genes expression. Also, we found that miR-155-5p involved in the induction of SRSF2 by HSV-1 infection. Through binding with motif in SRSF2 promoter, miR-155-5p altered significantly levels of multiple histone modification near the TSS of SRSF2 gene and finally increased the transcription of SRSF2. Overall, our study demonstrated that miR-155-5p regulated HSV-1 replication via the epigenetic regulation of SRSF2 expression.
Materials and methods
Cell culture, HSV-1 infection, and plaque assay
HeLa cells (American Type Culture Collection, ATCC) were grown in Dulbecco’s modified Eagle’s medium (Gibco/Invitrogen Ltd, 12,800–017) containing 10% fetal bovine serum (PAA, A15-101) and 10 U/ml penicillin-streptomycin (Gibco/Invitrogen Ltd, 15,140–122) in a humidified 5% CO2 incubator at 37°C. The HeLa cells were infected with HSV-1 strain SM44 at an MOI of 1. To conduct the plaque-forming assay, Vero cells (Chinese Academy of Sciences) were inoculated with 100 μl of serially diluted viral fluid for 1 hour. After viral adsorption, the HSV-1-infected cells were overlaid with medium containing 1% human serum for 48 hours and stained with 1% crystal violet in 10% formaldehyde for 15 min to visualize the plaques.
Cells transfection, RNA isolation, reverse transcription and qPCR
All synthetic miRNAs, siRNAs and the negative control were purchased from Shanghai GenePharma Co. Ltd. The siRNA sequences were as follows: SRSF2 siRNA, 5ʹ- GUGAGAAGUUGCUUAGAAA-3ʹ (sense) and 5ʹ- UUUCUAAGCAACUUCUCAC-3ʹ (antisense); Negative control siRNA, 5ʹ- UUCUCCGAACGUGUCACGU-3ʹ (sense) and 5ʹ- ACGUGACACGUUCGGAGAA-3ʹ (antisense). All miRNAs and siRNAs were transfected with lipofectamine™ 2000 (Invitrogen, 11,668–019) according to the manufacturer’s protocol. Total RNA was isolated using RNAiso Plus (Takara, D9108B) according to the manufacturer’s protocol. Real-time qRT-PCR was performed using ReverTra Ace® qPCR RT Master Mix with gDNA remover (TOYOBO, FSQ-301) and SYBR Green PCR Master Mix (TOYOBO, QPK-201). All mRNA levels were measured and normalized to beta-actin. To detect the expressional level of miR-155-5p, miRNA qPCR were performed using the Hairpin-it™ miRNA qPCR kit according to manufacturer’s instructions. MiR-155-5p expression level were normalized to U6 snoRNA. The primers for RT-PCR analysis were as follows: SRSF2, 5ʹ- GAGAACCAAAGGGAGGGGTG-3ʹ (sense) and 5ʹ- TGCTGCGTATGCAAGTCTGA-3ʹ (antisense); ICP0, 5ʹ-CCCACTATCAGGTACACCAGCTT-3ʹ (sense) and 5ʹ-CTGCGCTGCGACACCTT-3ʹ (antisense); ICP27, 5ʹ-CGATGACTTACTGGCGGGTGT −3ʹ (sense) and 5ʹ-GCGTCGGTCACGGCATAA-3ʹ (antisense); TK, 5ʹ-TGTGTGACACCATTCATTGATGC-3ʹ (sense) and 5ʹ-TCCTCACATGGGGGAGGTAG-3ʹ (antisense); Actin-beta, 5ʹ-GTACCCAGGCATTGCTGACA-3ʹ (sense) and 5ʹ-CGCAGCTCAGTAACAGTCCG-3ʹ (antisense).
Western blotting
Cells were lysed in ice-cold whole cell extract buffer B (50 mM TRIS-HCl, pH 8.0, 4 M urea and 1% Triton X-100), supplemented with complete protease inhibitor mixture. Cell extracts were resolved by SDS-PAGE, and analyzed by Western blot. Protein bands were visualized using ECL Blotting Detection Reagents. Antibodies used for western blotting include anti-SRSF2 antibody (Santa Cruz Biotechnology, sc-10,252), anti-HSV-1 ICP0 antibody (Abcam, ab6513), anti-HSV-1 ICP27 antibody (Abcam, ab31631), an anti-HSV-1 TK antibody (Santa Cruz Biotechnology, sc-28,037), and anti-beta-actin antibody (Proteintech, 60,008–1-Ig).
Immunofluorescence microscopy
To evaluate the influence of miR-155-5p on HSV-1 replication, HeLa cells were transfected with miR-155-5p mimics, inhibitors or negative control for 36 h. After infection with HSV-1 for 12 hours, the cells were fixed with 4% paraformaldehyde for 10 min. After blocking, the cells incubated with an anti-HSV-1 antibody (Abcam, ab9533). The cells were washed, counterstained with DAPI and observed with an Olympus FV1000 confocal laser microscope. The fluorescence signal value was measured with Image J software and normalized to a single cell. To validate the interaction between miR-155-5p and the SRSF2 promoter fragments, HSV-1-infected HeLa cells were co-transfected with Cy3-labeled miR-155-5p and FAM-labeled fragments of SRSF2 promoter containing miR-155-5p potentially binding motifs or not for 24 hours. For the co-localization studies, the cells were fixed, washed, counterstained with DAPI and observed with an Olympus FV1000 confocal laser microscope. The intensity plots for the red and green channels were analyzed with ImageJ software.
Luciferase assay
For generation of luciferase reporters for promoter assay, a luciferase reporter constructs that inserted the sequences from −500 bp to +500 bp relative to the TSS of SRSF2 were purchased from Shanghai GenePharma Co. Ltd. Luciferase activities were assayed using a Dual-Luciferase Reporter System (Promega, E1960) according to the manufacturer’s protocol.
Chip assay
ChIP assay was conducted according to Dahl’s protocol [49]. In brief, cells were fixed with 1% formaldehyde and sonicated to shear DNA. After centrifugation, the supernatants were incubated with H3K4Me3 antibody (Abcam, ab8580), H3K27Me3 antibody (Abcam, ab6002), H3K27Ac antibody (Abcam, ab4729) or H3K27Cro antibody (Jingjie PTM Biolab, PTM-501) Chromatin DNA was purified by Dynabeads protein G (Invitrogen, 10004D) and subjected to real-time PCR. The region-specific primers were as follows: TSS-1, 5ʹ- TCATTTGCCTCTCCCTGTGAC-3ʹ (sense) and 5ʹ-CTCTGGCGAAAGGGGGTTG-3ʹ (antisense); TSS-2, 5ʹ- CGGAATTAGCGGGCAGTTG-3ʹ (sense) and 5ʹ- CCCGATGTGGAGGGTATGAC-3ʹ (antisense); TSS-3, 5ʹ- GCTAGCGCACCTGAGTAACA-3ʹ (sense) and 5ʹ- AGGTTTCATTTCCGGGTGGC-3ʹ (antisense); TSS-4, 5ʹ- GAAAGCGAACGAAGGCGAAG-3ʹ (sense) and 5ʹ- CCCGATGTGGAGGGTATGAC-3ʹ (antisense).
Statistical analysis
Each experiment was repeated three times. The results are presented as the mean ±SD. *p < 0.01. Comparisons between two groups were evaluated with a two-sample t test. For three or more groups, standard one-way analysis of variance (ANOVA) followed by Bonferroni’s test for multiple comparisons was completed. A 2-tailed probability value <0.05 was considered statistically significant.
Funding Statement
This work was supported by the China Postdoctoral Research Foundation of China [2018M633216].
Acknowledgments
This work was supported by the China Postdoctoral Research Foundation of China [2018M633216].
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Looker KJ, Garnett GP.. A systematic review of the epidemiology and interaction of herpes simplex virus types 1 and 2. Sex Transm Infect. 2005;81:103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Hofstetter AM, Rosenthal SL, Stanberry LR. Current thinking on genital herpes. Curr Opin Infect Dis. 2014;27:75–83. [DOI] [PubMed] [Google Scholar]
- [3].Wozniak MA, Mee AP, Itzhaki RF. Herpes simplex virus type 1 DNA is located within Alzheimer‘s disease amyloid plaques. J Pathol. 2009;217:131–138. [DOI] [PubMed] [Google Scholar]
- [4].Harris SA, Harris EA. Herpes simplex virus type 1 and other pathogens are key causative factors in sporadic Alzheimer’s disease. J Alzheimers Dis. 2015;48:319–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bustos DE, Atherton SS. Detection of herpes simplex virus type 1 in human ciliary ganglia. Invest Ophthalmol Vis Sci. 2002;43:2244–2249. [PubMed] [Google Scholar]
- [6].Rice SA, Long MC, Lam V. Schaffer PA and Spencer CA. Herpes simplex virus immediate-early protein ICP22 is required for viral modification of host RNA polymerase II and establishment of the normal viral transcription program. J Virol. 1995;69:5550–5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].O‘Reilly D, Hanscombe O, O‘Hare P. A single serine residue at position 375 of VP16 is critical for complex assembly with Oct- 1and HCF and is a target of phosphorylation by casein kinase II. Embo J. 1997;16:2420–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hsia SC, Graham LP, Bedadala GR, et al. Induction of transcription factor early growth response protein 1 during HSV-1 infection promotes viral replication in corneal cells. Br Microbiol Res J. 2013;3:706–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Rowles DL, Tsai YC, Greco TM, et al. DNA methyltransferase DNMT3A associates with viral proteins and impacts HSV-1 infection. Proteomics. 2015;15:1968–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zheng SQ, Li YX, Zhang Y, et al. MiR-101 regulates HSV-1 replication by targeting ATP5B. Antiviral Res. 2011;89:219–226. [DOI] [PubMed] [Google Scholar]
- [11].Ru J, Sun H, Fan H, et al. MiR-23a facilitates the replication of HSV-1 through the suppression of interferon regulatory factor 1. PLoS One. 2014;9:e114021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Jm H, Zhao Y, Clement C, et al. HSV-1 infection of human brain cells induces miRNA-146a and Alzheimer-type inflammatory signaling. Neuroreport. 2009;20:1500–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Piedade D, Azevedo-Pereira JM. The Role of microRNAs in the pathogenesis of herpesvirus infection. Viruses. 8(6):E156 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Mendell JT. MicroRNAs: critical regulators of development, cellular physiology and malignancy. Cell Cycle. 2005;4:1179–1184. [DOI] [PubMed] [Google Scholar]
- [15].Ventura Aand Jacks T.. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136:586–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Dumortier O, Hinault C, Van OE. MicroRNAs and metabolism crosstalk in energy homeostasis. Cell Metab. 2013;18:312–324. [DOI] [PubMed] [Google Scholar]
- [17].Krol J, Loedige I and Filipowicz W.. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. [DOI] [PubMed] [Google Scholar]
- [18].Frankel LB, Lund AH. MicroRNA regulation of autophagy. Carcinogenesis. 2012;33:2018–2025. [DOI] [PubMed] [Google Scholar]
- [19].Pourrajab F, Babaei Zarch M, BaghiYazdi M, et al. MicroRNA-based system in stem cell reprogramming; differentiation/dedifferentiation. Int J Biochem Cell Biol. 2014;55:318–328. [DOI] [PubMed] [Google Scholar]
- [20].Ma V-S, Liu J, Gj H, et al. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. [DOI] [PubMed] [Google Scholar]
- [21].Carmell MA, Xuan Z, Mq Z, et al. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 2002;16:2733–2742. [DOI] [PubMed] [Google Scholar]
- [22].Vasudevan S, Tong Y and Steitz JA.. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. [DOI] [PubMed] [Google Scholar]
- [23].Wan G, Xie W, Liu Z, et al. Hypoxia-induced MIR155 is a potent autophagy inducer by targeting multiple players in the MTOR pathway. Autophagy. 2014;10:70–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zeng Q, Tao X, Huang F, et al. Overexpression of miR-155 promotes the proliferation and invasion of oral squamous carcinoma cells by regulating BCL6/cyclin D2. Int J Mol Med. 2016;37:1274–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Peng Y, Dong W, Lin TX, et al. MicroRNA-155 promotes bladder cancer growth by repressing the tumor suppressor DMTF1. Oncotarget. 2015;6:16043–16058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Yang J, Zhang P, Krishna S, et al. Unexpected positive control of NFκB and miR-155 by DGKα and ζ ensures effector and memory CD8+ T cell differentiation. Oncotarget. 2016;7:33744–33764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Xu L, Leng H, Shi X, et al. MiR-155 promotes cell proliferation and inhibits apoptosis by PTEN signaling pathway in the psoriasis. Biomed Pharmacother. 2017;90:524–530. [DOI] [PubMed] [Google Scholar]
- [28].Zhang XF, Tu R, Li K, et al. Tumor suppressor PTPRJ is a target of miR-155 in colorectal cancer. J Cell Biochem. 2017;118:3391–3400. [DOI] [PubMed] [Google Scholar]
- [29].Wallace JA, Kagele DA, Eiring AM, et al. miR-155 promotes FLT3-ITD–induced myeloproliferative disease through inhibition of the interferon response. Blood. 2017;129:3074–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hao L, Wang XG, Cheng JD, et al. The up-regulation of endothelin-1 and down-regulation of miRNA-125a-5p, −155, and −199a/b-3p in human atherosclerotic coronary artery. Cardiovasc Pathol. 2014;23:217–223. [DOI] [PubMed] [Google Scholar]
- [31].Ge J, Huang Z, Liu H, et al. Lower expression of MicroRNA-155 contributes to dysfunction of natural killer cells in patients with chronic hepatitis B. Front Immunol. 2017;8:1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lin X, Qin Y, Jia J, et al. MiR-155 enhances insulin sensitivity by coordinated regulation of multiple genes in Mice. PLoS Genet. 2016;12:e1006308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Huang LG, Zou J, Lu QC. Silencing rno-miR-155-5p in rat temporal lobe epilepsy model reduces pathophysiological features and cell apoptosis by activating Sestrin-3. Brain Res. 2018;1689:109–122. [DOI] [PubMed] [Google Scholar]
- [34].Majer A, Caligiuri KA, Gale KK, et al. Induction of multiple miR-200/182 members in the brains of mice are associated with acute herpes simplex virus 1 encephalitis. PLoS One. 2017;12:e0169081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Rajasagi NK, Bhela S, Sk V, et al. Frontline science: aspirin-triggered resolvin D1 controls herpes simplex virus-induced corneal immunopathology. J Leukoc Biol. 2017;102:1159–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Bhela S, Mulik S, Reddy PB, et al. Critical role of microRNA-155 in herpes simplex encephalitis. J Immunol. 2014;192:2734–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Bhela S, Mulik S, Gimenez F, et al. Role of miR-155 in the pathogenesis of herpetic stromal keratitis. Am J Pathol. 2015;185:1073–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Wang Z, Liu Q, Lu J, et al. Serine/Arginine-rich splicing factor 2 modulates herpes simplex virus type 1 replication via regulating viral gene transcriptional activity and pre-mRNA splicing. J Biol Chem. 2016;291:26377–26387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kimura H. Histone modifications for human epigenome analysis. J Hum Genet. 2013;58:439–445. [DOI] [PubMed] [Google Scholar]
- [40].Tan M, Luo H, Lee S, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zhang Y, Dai J, Tang J, et al. MicroRNA-649 promotes HSV-1 replication by directly targeting MALT1. J Med Virol. 2017;89:1069–1079. [DOI] [PubMed] [Google Scholar]
- [42].Lagos D, Pollara G, Henderson S, et al. miR-132 regulates antiviral innate immunity through suppression of the p300 transcriptional co-activator. Nat Cell Biol. 2010;12:513–519. [DOI] [PubMed] [Google Scholar]
- [43].Xie Y, He S, Wang J. MicroRNA-373 facilitates HSV-1 replication through suppression of type I IFN response by targeting IRF1. Biomed Pharmacother. 2018;97:1409–1416. [DOI] [PubMed] [Google Scholar]
- [44].Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. 2002;3(4):285–298. [DOI] [PubMed] [Google Scholar]
- [45].Portnoy V, Huang V, Rf P, et al. Small RNA and transcriptional upregulation. Wiley Interdiscip Rev RNA. 2011;2:748–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Li LC, Okino ST, Zhao H, et al. Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci USA. 2006;103:17337–17342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wang X, Arai S, Song X, et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Janowski BA, Younger ST, Hardy DB, et al. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat Chem Biol. 2007;3:166–173. [DOI] [PubMed] [Google Scholar]
- [49].Dahl JA, Collas P. A rapid micro chromatin immunoprecipitation assay (microChIP). Nat Protoc. 2008;3:1032–1045. [DOI] [PubMed] [Google Scholar]


