Abstract
Background
Dysregulation of immune checkpoint molecules leads to immune evasion in human tumours but has become a viable target for tumour therapy. Here, we examined expression of Herpes virus entry mediator (HVEM), an immune checkpoint molecule, in human glioblastoma (GBM) to assess its potential as a molecular target for treatment.
Methods
Molecular and clinical data from publicly available genomic databases containing WHO grade II-IV human glioma cases (n = 1866) were analyzed. Immunohistochemistry was applied to assess HVEM protein levels in primary tumour sections. Statistical analysis was performed using Matlab and R language.
Findings
HVEM was found to be elevated in aggressive gliomas, particularly in the mesenchymal and isocitrate dehydrogenase (IDH) wild-type molecular subtypes of GBM. HVEMhigh tumours tended to be associated with amplification of EGFR and loss of PTEN, while HVEMlow tumours harbored mutations in IDH1 (93%). HVEM exhibited potential as a prognostic marker based on Cox regression and nomogram models. HVEM displayed intra-tumour heterogeneity and was more highly expressed in peri-necrotic and microvascular regions. Gene ontology and pathway analysis revealed enrichment of HVEM in multiple immune regulatory processes, such as suppression of T cell mediated immunity in GBM. Finally, in cell lineage analysis, HVEM was found to be tightly associated with several infiltrating immune and stromal cell types which localized to the tumour microenvironment.
Interpretation
Our data highlights the importance of HVEM in the development of GBM and as a potential molecular target in combination with current immune checkpoint blockades for treatment of GBM.
Keywords: Glioma, HVEM, Immune response, Prognosis, Tumour microenvironment
Research in context.
Evidence before this study
The dysregulation of immune checkpoint molecules within tumors has been hotly investigated in recent years as a potential therapeutic target in human cancer. Herpes virus entry mediator (HVEM), also known as tumour necrosis factor receptor (TNFR) superfamily 14 (TNFRSF14), is a novel immune checkpoint molecule which plays essential roles in both innate and adaptive immunity. While its function has been illuminated in diverse cancers, the clinical relevance of HVEM in human gliomas remains largely unknown.
Added value of this study
We found that HVEM was elevated in aggressive gliomas, especially in GBM with wild-type isocitrate dehydrogenase (IDH). Patients with HVEMhigh expressing tumors were associated with poorer prognosis, and HVEM expression levels showed promise as a prognostic marker based on Cox regression and nomogram models. HVEM protein was localized to the peri-necrotic zone and areas of microvascular proliferation in sections from primary human tumors. Gene ontology and pathway analysis revealed that HVEM plays an important role in the regulation of immune response and inflammatory activation, especially in modulating suppression of antitumor T cell immunity. HVEM was also tightly associated with several infiltrating immune and stromal cell lineages in the microenvironment, as well as several classical immune checkpoint molecules.
Implications of all the available evidence
Our study highlights the clinical and immunological importance of HVEM in GBM. Targeting HVEM combined with current immune checkpoint blockades might therefore be a novel therapeutic strategy for GBM in the future.
Alt-text: Unlabelled Box
1. Introduction
Glioblastoma (GBM) is the most prevalent adult primary malignant tumour of the central nervous system (CNS). Despite advances in therapeutic methods, the life expectancy of GBM patients beyond primary diagnosis is about 15 months [1]. Poor survival is considered in large part to be due to the infiltrative nature [2] and microscopic spread [3] of the cancer cells which are the basis for recurrence.
The tremendous progress in understanding the molecular mechanisms underlying the development of GBM has been driving current directions in GBM therapy [4,5]. Unfortunately, although many molecular targeted therapies appeared promising in preclinical models, they failed to show a desirable effect in phase I, II and III clinical trials for patients [6]. The dilemma of molecular targeted therapies mainly arises from the clonal, evolutionary and cellular complexity of GBM [7,8]. Furthermore, the blockade of the blood-brain barrier (BBB) and rapidly developed drug resistance also attenuate the effect of drugs [9]. Given the poor outcome of patients even under treatment with surgical and molecular targeted therapies, immunotherapy is appealing for further exploration as a new treatment strategy.
Immunotherapy for cancers harnesses the immune system to destroy cancer cells. Emerging evidence demonstrates that, among the dynamic effects of immunity, immune checkpoints are a crucial mechanism in the interaction between the immune system and GBM. Defined as a cohort of co-stimulatory and co-inhibitory molecules modulating T-cell activity, immune checkpoints orchestrate as regulatory circuits to make the immune system self-tolerable under normal physiological circumstances [10,11]. The functions of many classical checkpoints in GBM immunity are now known. CTLA-4 (CD152), for example, is a co-inhibitory checkpoint molecule expressed on T cells, which interacts with CD80 and CD86 on antigen-presenting cells (APCs) to inhibit co-stimulatory pathways [12]. This mechanism affects GBM antigen presentation [13]. PD-1, expressed on T cells, mediates the interaction between activated T cells and tumour cells when activated T cells return to the CNS, and PD-1 ligand (PD-L1) expressed on GBM cells binds to PD-1 to suppress T cell immune response [14]. Tim 3 is another checkpoint molecule expressed on CD4+ and CD8+ T cells. High levels suppress immune response through T cell exhaustion, leading to immune escape in cancer [15]. Immune checkpoints are often exploited by GBM cells to escape antitumour immunity. Thus, inhibitory immune checkpoints have been investigated as targets for GBM treatment. Studies are currently examining the feasibility of targeting immune checkpoint inhibitors, such as humanized CTLA-4 antibody Ipilimumab and PD-1 suppressive antibodies Nivolumab and Pembrolizumab, and their efficacies are under different phase clinical trials (from I to III) for GBM [16].
Herpes Virus Entry Mediator (HVEM), also known as tumour necrosis factor receptor (TNFR) superfamily 14 (TNFRSF14), is another essential immune checkpoint molecule critical in immune surveillance for cancer [17]. HVEM is expressed on T cells, and it mainly interacts with B and T lymphocyte attenuator (BTLA), CD160, glycoprotein D (gD), lymphotoxin-like, inducible expression competes with Herpes Simplex Virus glycoprotein D for HVEM, a receptor expressed by T lymphocytes (LIGHT/TNFSF14) and lymphotoxin α (LTα3). HVEM appears to be a “molecular switch” as it exerts a co-stimulatory effect on T cells when it binds to LIGHT or LTα but a co-inhibitory effect when it binds to BTLA or CD160 [18,19].
HVEM has been implicated in widespread development of human cancers. HVEM is a candidate tumour suppressor as one of the most frequently mutated genes in germinal center lymphomas [20]. It is highly expressed and negatively correlated with PD-1 expression in non-small cell lung cancer [17] and in hepatocellular carcinoma where only limited expression has been observed in normal liver tissue. High HVEM expression has also been correlated with poorer recurrence-free survival (RFS) and overall survival (OS) in hepatocellular carcinoma [21].
Here, we investigated a potential role for HVEM in promoting the development of human gliomas using analysis of publicly available databases. Our results demonstrated that HVEM is highly expressed in GBM and predicted to contribute to suppressed T cell immunity, which might be an underlying tumour promoting activity. Our study supports HVEM as a potential prognostic marker or molecular target in the clinical management of GBM.
2. Materials and methods
2.1. Ethics statement
The research strategy was approved by the Research Ethics Committee of Shandong University and the Ethics Committee of Qilu Hospital (Shandong, China). All experiments were performed in accordance with the relevant guidelines and regulations, and written informed consent was obtained from all patients.
2.2. Clinical specimens
Archived paraffin embedded glioma tissues (WHO grades II-IV) were collected from patients (n = 34) who underwent surgery in the Department of Neurosurgery, Qilu Hospital of Shandong University. Normal brain tissue samples (n = 6) were taken from trauma patients who underwent partial resection of normal brain as decompression treatment for severe head injuries.
2.3. Immunohistochemistry
Sections (4 μm) were obtained from formalin-fixed, paraffin-embedded tissues of different grades of human gliomas (WHO grades II-IV). Sections were boiled in sodium citrate buffer (pH 6.0) for antigen retrieval, and endogenous HRP activity was blocked with 3% H2O2. Slides were blocked with 10% normal goat serum and incubated with primary antibody (rabbit polyclonal anti-HVEM antibody, 1:50; Proteintech; Wuhan, China) at 4 °C overnight. Signal was visualized using standard protocols with horse radish peroxidase conjugated secondary antibody and 3, 3′-diaminobenzidine (DAB) as the substrate. For negative controls, sections were incubated with normal mouse serum rather than primary antibody. Slides were counterstained with hematoxylin, and representative images were obtained using an Olympus inverted microscope.
2.4. Biological function and gene set enrichment analysis
Correlation analysis of HVEM was performed in gene expression profiles available in the TCGA and CGGA datasets with Matlab software (https://cn.mathworks.com) and R language (https://www.r-project.org/). To identify biological processes and the KEGG signaling pathways associated with HVEM expression in gliomas, genes correlated with HVEM (P < .01) were analyzed using the DAVID web tool (http://david.abcc.ncifcrf.gov/home.jsp). Association between HVEM expression and hallmark gene sets from the Molecular Signatures Database (MSigDB) were analyzed using gene set enrichment analysis (GSEA) software (http://software.broadinstitute.org/). Somatic mutations and somatic copy number alternations (CNAs) were downloaded from the TCGA database. Copy number alternations associated with HVEM expression were analyzed using GISTIC 2.0 (https://gatkforums.broadinstitute.org).
2.5. Statistical analysis
Kaplan-Meier survival curves were generated and compared using the log-rank test. A two-tailed χ2 test was used to determine the association between HVEM expression and pathological characteristics. The Pearson correlation was applied to evaluate the linear relationship between gene expression levels. The Kolmogorov–Smirnov test was used to assess the normal distribution of data. The one-way ANOVA test or t-test was used for all other data comparisons using GraphPad Prism 7.0 (https://www.graphpad.com/; La Jolla, CA, USA). Data for each treatment group were represented as the mean ± SEM and compared with other groups for significance by one-way ANOVA followed by Bonferroni's post hoc test (multiple comparison tests). All tests were two-sided, and P-values <.05 were considered to be statistically significant.
3. Results
3.1. HVEM expression is elevated in aggressive gliomas
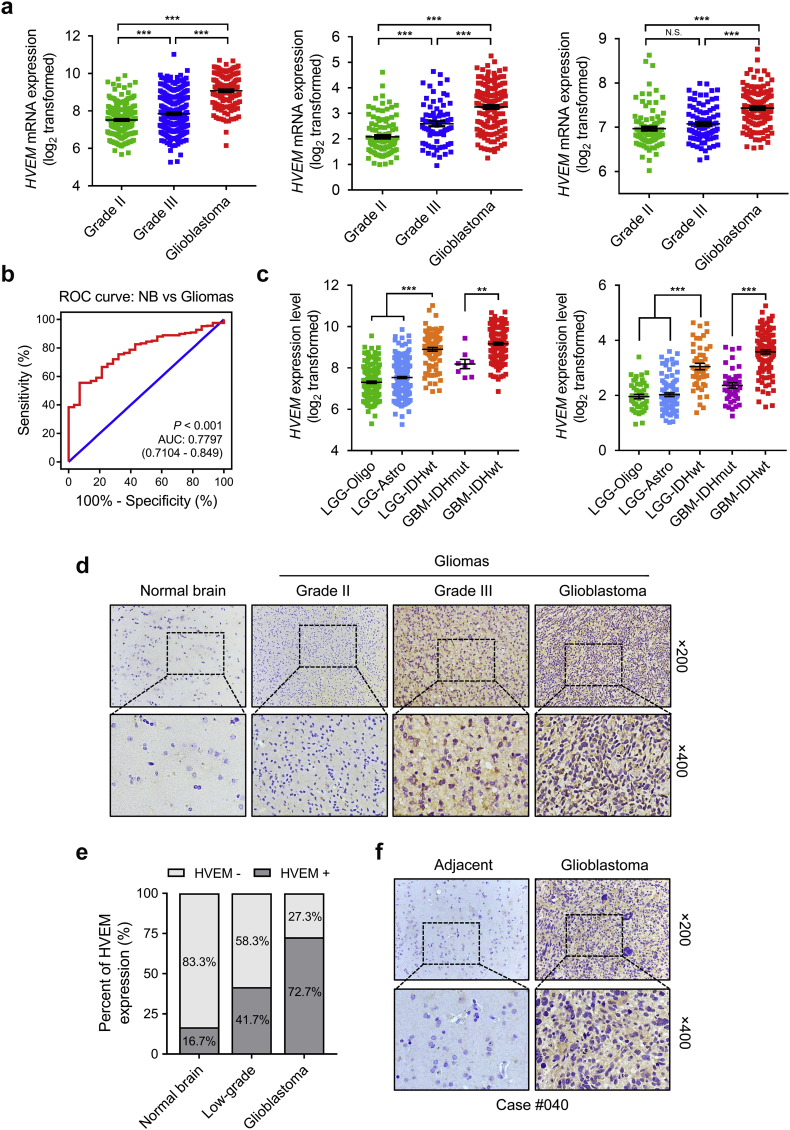
The mRNA expression levels of HVEM in different WHO grade gliomas was evaluated using expression data from publicly available databases containing genomic data from over 1500 gliomas samples: TCGA, n = 669; CGGA, n = 325; and Rembrandt, n = 510. HVEM was observed to be significantly up-regulated in GBM compared to low grade glioma (LGG) samples (P < .001, respectively; Fig. 1a). The expression of HVEM was also higher in WHO grade III than WHO grade II cases in the TCGA and CGGA cohorts (P < .001, respectively; Fig. 1a). Receiver operating characteristic (ROC) curve analysis further indicated that the expression of HVEM discriminated gliomas from normal brain tissue (the area under the curve (AUC) value = 0.7797; P < .001; Fig. 1b). Thus, HVEM might be a potential diagnostic marker in gliomas.
Fig. 1.
HVEM expression is elevated in aggressive gliomas.
a. Analysis of HVEM mRNA levels (log2) in WHO grade II-IV gliomas from TCGA (n = 669), CGGA (n = 325), and Rembrandt (n = 510) datasets.
b. Receiver operating characteristic (ROC) curve to assess sensitivity and specificity of HVEM expression as a diagnostic biomarker in gliomas.
c.HVEM mRNA levels (log2) in gliomas from TCGA and CGGA datasets based on the 2016 WHO classification.
d. Representative images of IHC staining for HVEM in normal brain and different pathological grades of gliomas (n = 40).
e. Quantification of HVEM IHC staining in normal brain (n = 6) and different pathological grades of gliomas (n = 34).
f. Representative images of HVEM IHC staining in GBM and adjacent brain tissues in a specific case from our cohort.
Data are shown as the mean ± the standard error of the mean (SEM) for each group. *P < .05, **P < .01, ***P < .001.
We also evaluated HVEM levels based on the 2016 WHO classification of CNS tumours. HVEM was downregulated in low grade oligodendroglioma (LGG-Oligo; IDHmut, 1p/19q codeletion), low grade astrocytoma (LGG-Astro; IDHmut, 1p/19q non-codeletion) but upregulated in the LGG-IDHwt group in both TCGA and CGGA datasets (Fig. 1c). Similarly, the GBM-IDHwt subtype, which is associated with poorer clinical outcome, was associated with higher HVEM levels (Fig. 1c).
To confirm that HVEM expression was also up-regulated at the protein level, we performed IHC staining for HVEM on an independent cohort of primary glioma (n = 34) and normal brain tissue (n = 6) samples from our institution. HVEM was located in the cytoplasm and membrane in tumours, with lower expression in normal brain tissue (1/6; 16.7%) compared to LGG (5/12; 41.7%) and GBM (16/22; 72.7%). Expression also increased with increasing tumour grade (Fig. 1d and e). IHC staining in GBM case #040 showed that HVEM was positively expressed in GBM but lower in adjacent normal brain tissue (Fig. 1f). To determine whether HVEM was expressed on immune infiltrates, immunofluorescence staining was performed with antibodies against HVEM and CD45, a marker for microglia/macrophages. We found no obvious co-localization of HVEM and CD45-positive cells in tumour sections (Fig. S1), indicating that HVEM was mainly expressed on tumour cells in GBM. Taken together, the expression of HVEM was up-regulated and positively correlated with increasing tumour grade in gliomas based on large-scale analysis in silico analysis and in an independent set of primary glioma specimens at the protein level.
3.2. Inter-tumour and intra-tumour heterogeneous characteristics of HVEM in gliomas
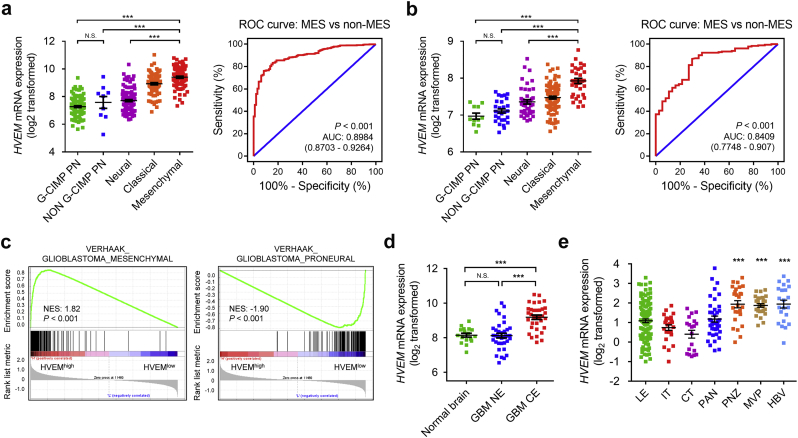
Human gliomas have been molecularly categorized into distinct subclasses: classical (CL), mesenchymal (MES), proneural (PN), and neural (NE). CL and MES subtypes are associated with more aggressive behavior relative to PN or NE subtypes [22]. We subsequently investigated the inter-tumour heterogeneity of HVEM among different molecular subtypes based on the VERHAAK_2010 classification scheme [23]. In the TCGA dataset, increased HVEM expression was associated with the MES molecular subtype compared to the NE, PN CpG island methylator phenotype (G-CIMP) or non-G-CIMP molecular subtypes (P < .001; Fig. 2a). The ROC curve further indicated that HVEM might be effective as an MES subtype predictor among gliomas (AUC value = 0.8984; P < .001; Fig. 2a). Similar results were observed in the Rembrandt cohort (AUC value = 0.8409; P < .001; Fig. 2b). Gene Set Enrichment Analysis (GSEA) in TCGA GBM patients further confirmed this observation (Fig. 2c).
Fig. 2.
Inter-tumour and intra-tumour heterogeneous expression characteristics of HVEM in gliomas.
HVEM mRNA expression (log2) using VERHAAK_2010 molecular classification scheme, including CpG island methylator phenotype (G-CIMP) proneural, non–G-CIMP proneural, neural, classical, and mesenchymal subtypes, from the a. TCGA and and b. Rembrandt datasets. ROC curve indicating sensitivity and specificity of HVEM expression as a diagnostic biomarker for the MES molecular subtype.
c. GSEA enrichment analysis of MES and PN signatures in HVEM high vs low samples in the TCGA GBM dataset. Normalized enrichment score (NES) and FDR are shown for each plot.
d.HVEM mRNA expression (log2) in different radiographical regions of GBM and normal brain from the Gill dataset.
e. Intra-tumour analysis of HVEM expression using IVY GBM RNA-seq data. Anatomic structures analysed are the following: LE (Leading Edge), IT (Infiltrating Tumour), CT (Cellular Tumour), PAN (Pseudopalisading Cells Around Necrosis), PNZ (Perinecrotic Zone), MVP (Microvascular Proliferation), and HBV (Hyperplastic Blood Vessels).
The intra-tumour distribution of HVEM in GBM tissues was evaluated. Radiographically, GBM tissues contained within the T1 contrast-enhancing (CE) regions have different compositions relative to the non-enhancing (NE; abnormal T2/FLAIR signal) GBM margins, which represent oedematous tissues with infiltrating tumour cells. Analysis of RNA sequencing data from 93 samples [24] revealed that HVEM was highly expressed in GBM-CE regions compared to NE or normal brain areas (P < .001, respectively; Fig. 2d). Furthermore, based on the Ivy Glioblastoma Atlas Project data, HVEM was found to be abundant in peri-necrotic zones, microvascular proliferation and hyperplastic blood vessels compared to other pathological areas (P < .001, respectively; Fig. 2e).
3.3. HVEM expression is associated with poor survival in glioma patients
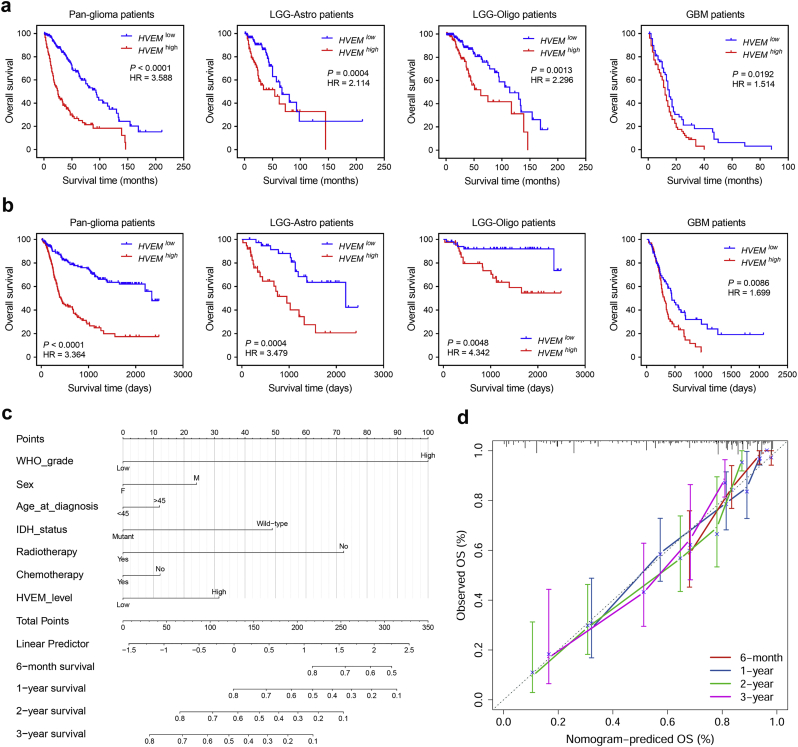
We next assessed the prognostic value of HVEM expression in human gliomas using Kaplan-Meier analysis. Kaplan-Meier survival curves were generated based on median values of HVEM expression in gliomas. In pan-glioma analysis of both TCGA and CGGA datasets, HVEMhigh patients exhibited significantly shorter overall survival (OS) than HVEMlow patients (P < .001, respectively; Fig. 3a). Unfavorable OS was also associated with higher HVEM expression in analysis performed on patient data on the basis of different histological subtypes alone (Fig. 3a and b). In addition, higher HVEM expression was associated with poorer progression-free survival (PFS) among glioma patients overall as well as LGG-Astro and GBM patients (Fig. S2a). HVEM status remained informative among IDHwt GBM samples (P = .0382), but was not prognostic for IDHmut GBM samples (Fig. S2b).
Fig. 3.
HVEM expression is associated with poor survival in glioma patients.
Kaplan-Meier analysis of overall survival (OS) based on high vs low expression of HVEM in pan-glioma analysis, and LGG and GBM patients in a. TCGA and and b. CGGA datasets. The median value of HVEM expression was used as the cut-off value. P-values were obtained from the log-rank test. c. Nomogram for predicting the proportion of glioma patients with OS. d. Plots depict the calibration of each model in terms of agreement between predicted and observed OS. Model performance is shown by the plot, relative to the 45-degree line, which represents perfect prediction.
Furthermore, we investigated whether HVEM could serve as a marker for the prediction of patient response to radiation and chemotherapy. HVEMlow GBM patients receiving radiotherapy exhibited longer OS than HVEMhigh patients (P = .0135; Fig. S2c). However, HVEMlow GBM patients displayed only a trend towards better prognosis compared to the HVEMhigh group (Fig. S2c). HVEM expression was subsequently validated as an independent prognostic marker after adjusting for several risk factors including age, IDH status and radiotherapy in univariate and multivariate Cox regression analysis in both LGG (HR = 2.621, 95% CI = 1.329 to 5.169, P = .005; Table S1) and GBM (HR = 1.795, 95% CI = 1.086 to 2.967, P = .023; Table S1) in the CGGA dataset. The prognostic value was only significant in univariate Cox regression analysis in the TCGA dataset (Table S1).
To better predict patient prognosis in the clinic, HVEM expression along with clinicopathological risk factors were integrated to develop a prognostic nomogram model (Fig. 3c). Nomograms are a graphical representation of predictive statistical models, which generate the probability of a clinical event, such as response to therapy, and have been widely used for cancer patients. Calibration plots showed that the nomograms did well in predicting patient survival according to an ideal model (Fig. 3d).
3.4. HVEM expression levels are associated with distinct genomic alterations
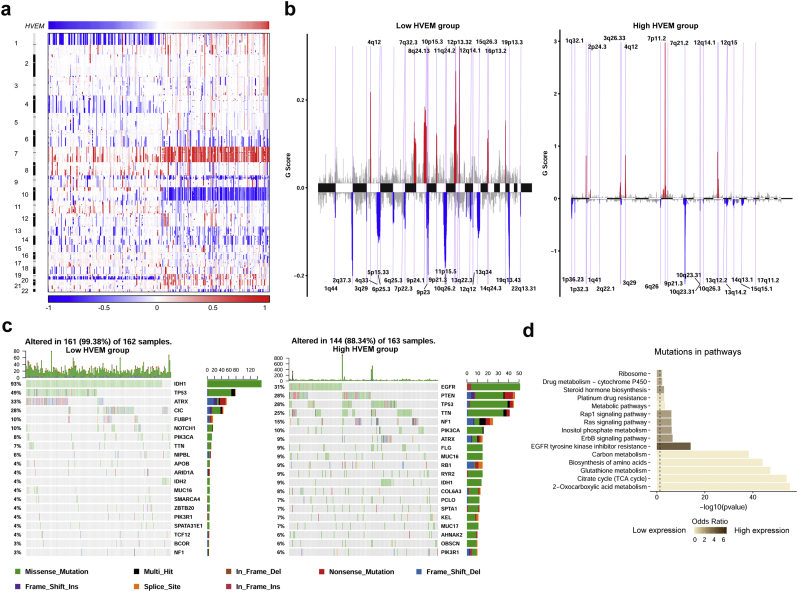
To determine whether HVEM expression levels were associated with specific genomic characteristics in gliomas, we performed copy number variation (CNV) and somatic mutation analysis using the TCGA dataset. A distinct overall CNV profile emerged from the comparison of the HVEMlow (n = 162) vs the HVEMhigh (n = 163) cluster (Fig. 4a and b). Amplification of chr7 and deletion of chr10, which are both common genomic events in GBM, frequently occurred in the HVEMhigh cluster (Fig. 4a). Deletion of 1p and 19q, a genomic hallmark of oligodendroglioma, however, more frequently appeared associated with the HVEMlow cluster (Fig. 4b). Using GSITIC analysis, we identified 33 and 38 genomic events to be enriched in either the HVEMhigh or HVEMlow group, respectively (Fig. 4b and Table S2). In HVEMhigh samples, frequently amplified genomic regions included oncogenic driver genes such as EGFR (7p11.2), PDGFRA (4q12) and CDK4 (12q14.1), while deleted regions contained tumour suppressor genes including CDKN2A/CDKN2B (9p21.3) and PTEN (10q23.3). Analysis of somatic mutation profiles based on HVEM expression levels revealed a high frequency of mutations in EGFR (31%), PTEN (28%), TTN (25%) and NF1 (15%) in the HVEMhigh group (n = 163), while IDH1 (93%), ATRX (33%), CIC (28%) were more frequently mutated in the HVEMlow group (n = 162; Fig. 4c). Finally, genomic mutations associated with HVEM expression levels were linked to functionally distinct biological pathways. HVEMlow expressing tumours, for example, harbored mutations associated with 2-oxocarboxylic acid metabolism, the TCA cycle, and glutathione metabolism. EGFR tyrosine kinase inhibitor resistance, the ErbB signaling pathway, and the Ras signaling pathway were the significant differential pathways which had higher mutation rates among HVEMHigh expressing tumours (Fig. 4d and Table S3).
Fig. 4.
HVEMhigh or low expression is associated with distinct genomic alterations.
a. Overall copy number variation (CNV) profile according to high vs low HVEM expression. Blue (deletion); red (amplification).
b. Frequency of specific changes based on HVEMlow and HVEMhigh groups. The Y-axis represents the frequency of chromosomal deletion (blue) or amplification (red).
c. Spectrum of somatic mutations in gliomas from HVEMlow and HVEMhigh groups.
d. KEGG pathway analysis for mutations based on HVEM expression levels. Deeper brown represents functions highly associated with HVEMhigh expressing tumours.
Since the HVEM gene is located on 1p36, a chromosomal band deleted in 20–40% of all types of gliomas [[25], [26], [27]], we also examined the relationship between HVEM copy number (CN) and HVEM expression levels. In both LGGs and GBMs, tumours with HVEM CN loss expressed significantly lower levels of HVEM mRNA (P < .001, respectively; Fig. S3a and S3b). Moreover, in an independent dataset, glioma samples with 1p deletion expressed significantly lower levels of HVEM than 1p intact tumours (P < .01; Fig. S3c). Thus, these data demonstrated that HVEM expression could be regulated by gross chromosomal changes in human gliomas.
3.5. Genes positively associated with HVEM are enriched in immune related processes
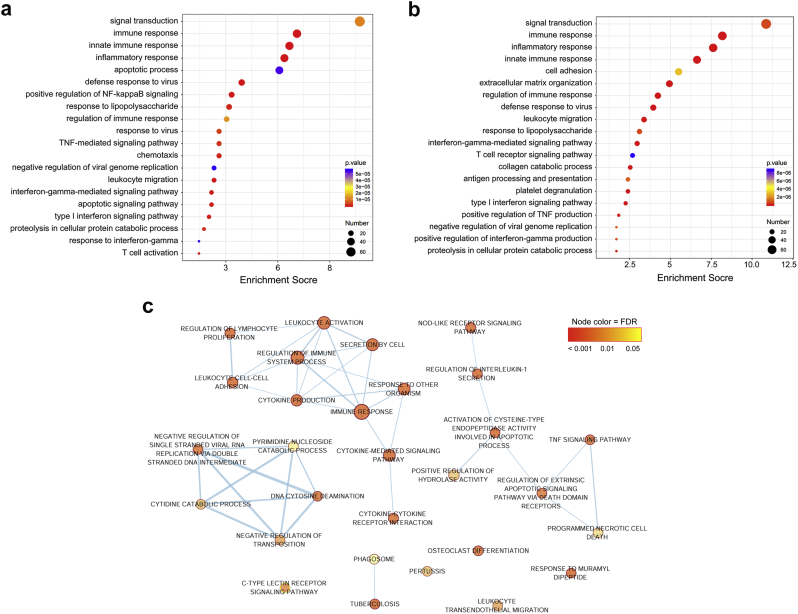
We investigated the potential function of HVEM in the development of human glioma by using GO analysis on genes positively associated with HVEM expression. GO results with the genes from either dataset (TCGA, n = 756 genes; CGGA, n = 733 genes; Table S4) revealed that they were enriched in a wide variety of immune-related functions, including innate immune response, inflammatory response, leukocyte migration, NF-κB signaling regulation, TNF mediated signaling, and type I interferon (Fig. 5a and b). The signaling network that resulted from KEGG pathway analysis further established the association between HVEM and immune-related pathways, such as TNF signaling and apoptosis (Fig. 5c). These data suggested that HVEM might play essential roles in immune related processes in gliomas.
Fig. 5.
HVEM-related biological functions in gliomas.
Biological processes assessed using the set of HVEM-associated genes in a. TCGA and b. CGGA datasets. Results are based on the GO databases. c. Network representation of pathway terms enriched in HVEM-positively associated gene signatures. Categorical enrichment was calculated using DAVID, and enrichment results were plotted using Cytoscape.
3.6. HVEM is associated with T cell immunity in gliomas
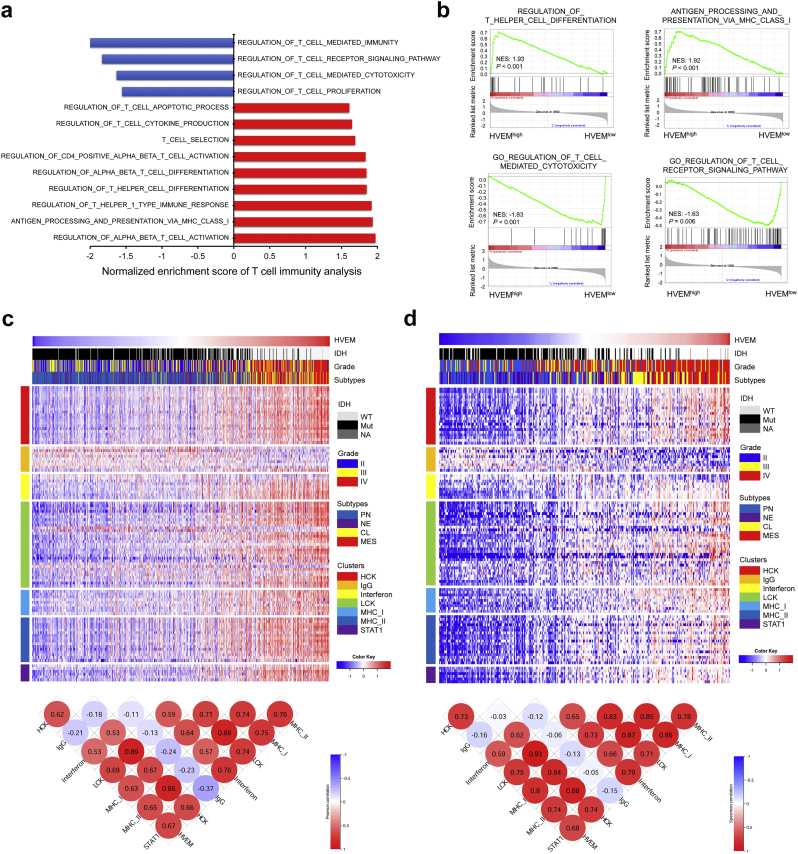
HVEM on tumour cells has been reported to suppress cytokine production and proliferation of CD8+ T cells through BTLA [28]. In hematopoietic cells, HVEM is found to be involved in both T-cell activation and inhibition processes depending on the ligands [29,30]. We thus investigated whether HVEM might have a role in T cell immunity in gliomas using GSEA analysis. We found that HVEM was negatively associated with regulation of T cell mediated immunity, regulation of T cell receptor signaling, regulation of T cell mediated cytotoxicity, and T cell proliferation (Fig. 6a and b). In contrast, HVEM was positively associated with regulation of T helper cell differentiation, regulation of T helper 1 type immune response, antigen processing and presentation through MHC class I molecules, and regulation of alpha beta T cell activation. Thus, HVEM may contribute to the suppression of T cell associated anti-tumour immunity in the glioma microenvironment.
Fig. 6.
HVEM is associated with T cell immunity and inflammatory activities in gliomas.
a. GSEA analysis links HVEM high vs low gene signatures to T cell immunity. The X-axis indicates the normalized enrichment score (NES). Blue, positive association; red, negative association.
b. GSEA plots for enrichment of T cell immunity in HVEMhighvs HVEMlow samples in the TCGA GBM dataset. NES and FDR are shown for each plot.
Heatmaps illustrating HVEM related inflammatory activities in gliomas. Corrgrams illustrate Pearson r values for analysis between HVEM and inflammatory metagenes in c. TCGA and d. CGGA datasets. Expression values are z-transformed and are colored red for high expression and blue for low expression, as indicated in the scale bar. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.7. HVEM is involved in inflammatory activities in gliomas
Based on our in silico analysis which implicated HVEM in the inflammatory response in gliomas, we examined the association of the molecule with various inflammatory activity signatures [31]. HVEM expression was found to be positively associated with HCK, interferon, LCK, MHC-I, MHC-II, and STAT1 metagenes, but negatively associated with the IgG metagene in pan-glioma analysis (Fig. 6c and d) and GBM alone (Fig. S4a and S4b). These findings indicated that HVEM was enriched in macrophage activation, T cell related signaling transduction, and antigen presenting cells, but not interaction with B lymphocytes during immunosuppression and glioma progression.
3.8. HVEM is associated with increased immune and stromal cell populations in the tumour microenvironment
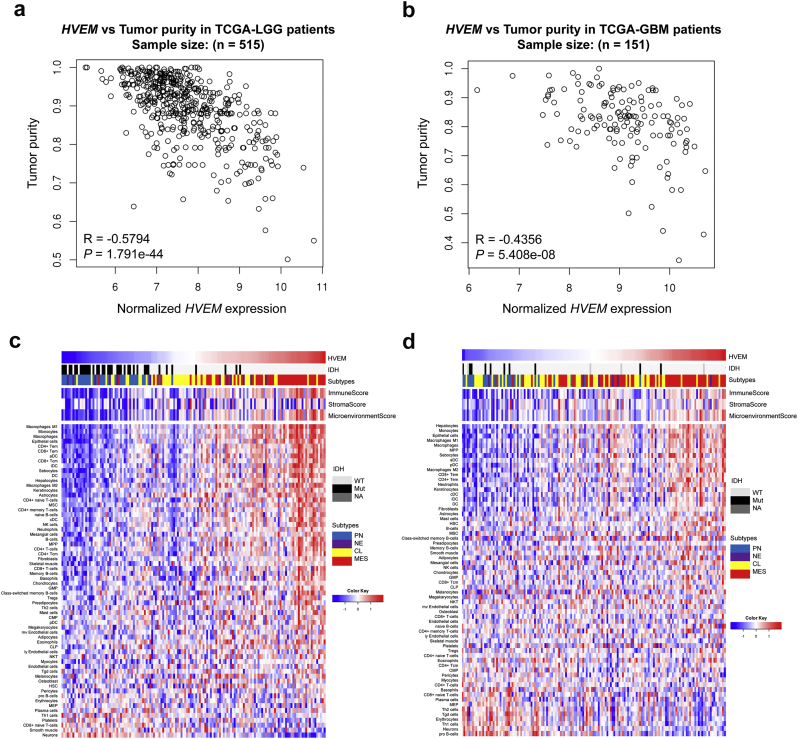
To further examine the significance of increased HVEM in gliomas, we performed analysis to identify the cell types or activities in the tumour microenvironment that might be influenced by the molecule. In this analysis, HVEM was found to be negatively correlated with glioma cell purity for both LGG (Fig. 7a) and GBM (Fig. 7b) samples in independent analyses. Moreover, a tight association was found between HVEM levels and the immune or microenvironment score in GBM samples from both TCGA and CGGA datasets (Fig. 7c and d).
Fig. 7.
HVEM is associated with tumour purity and immune and stromal cell populations in the tumour microenvironment.
Correlation analysis of HVEMhigh or HVEMlow expression levels and tumour purity in TCGA a. LGG and b. GBM samples. Heatmaps illustrating the relationship between HVEM and 64 immune and stromal cell populations based on c. TCGA and d. CGGA GBM data. Expression values are z-transformed and are colored red for high expression and blue for low expression, as indicated in the scale bar. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To comprehensively evaluate its putative role in tumour-immune cell interactions, we investigated the relationship between HVEM and 64 immune and stromal cell populations using cell type enrichment analysis [32]. HVEM was tightly associated with multiple infiltrating immune cell types, including monocytes, macrophages, CD8+ T effector memory cells (TEM), CD4+ TEM, neutrophils, DCs and NK cells; however, HVEM was negatively associated with pro B-cells, plasma cells and CD8+ naive T cells (heatmaps in Fig. 7c and d). In addition, specific stromal cell types, such as epithelial cells, astrocytes and fibroblasts, were also enriched in HVEMhigh GBMs. These findings were further validated based on a 9-immune cell lineage analysis, confirming the enrichment of multiple immune cell lineages [33] in HVEMhigh glioma samples (Fig. S5a-S5d). All together, our data suggested that HVEMhigh glioma samples tend to recruit infiltrating immune and stromal cells into the tumour microenvironment.
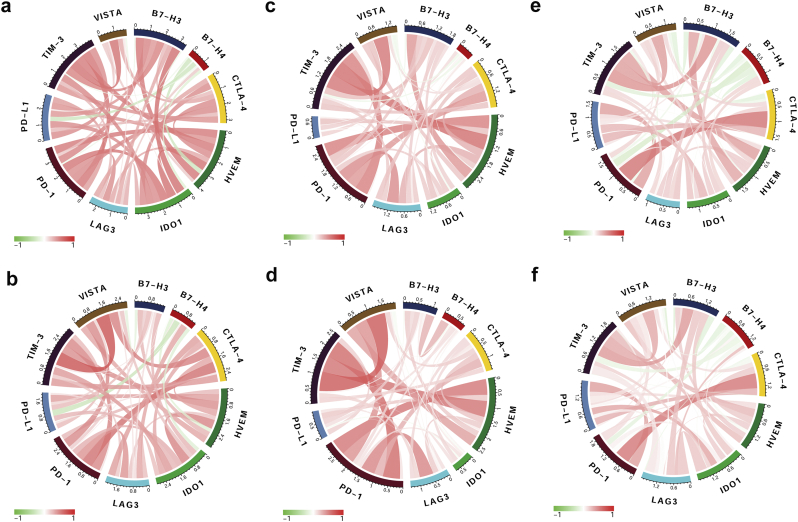
3.9. HVEM is associated with other immune checkpoint molecules in gliomas
Given the vital functions of immune checkpoint molecules in the regulation of immune processes, we performed correlation analysis to assess the relationship between HVEM and several well-known immune checkpoint genes in glioma samples. HVEM was highly correlated with TIM-3, PD-1, PD-L1, CTLA-4, LAG3 and VISTA in pan-glioma analysis and GBM samples alone in all 3 datasets (Fig. 8a - 8f). We also observed a synergistic association between HVEM and its receptors (BTLA and CD160) or ligands (LIGHT and LTA). A strong positive association emerged between HVEM and LIGHT or BTLA in pan-glioma analysis and GBM samples alone. No significant relationship was observed between HVEM and CD160 or LTA (Fig. S6a–S6f). These results highlight HVEM/LIGHT and HVEM/BTLA potentially as major signaling pathways involved in immunosuppression in the glioma microenvironment.
Fig. 8.
HVEM is associated with other immune checkpoint molecules in gliomas.
Correlation of HVEM and immune checkpoint molecules in pan-glioma analysis (upper row) and GBM samples (lower row) in (a, b) TCGA, (c, d) CGGA, and (e, f) Rembrandt datasets.
Based on these findings, we used univariate Cox regression analysis to explore the prognostic value of HVEM, LIGHT or BTLA in GBM patients. A risk score formula for survival prediction was constructed based on a linear combination of the mRNA expression levels of the three genes and weighted by the regression coeffcient from the univariate Cox regression analyses (β) [34]: risk value = (0.304 × HVEM expression) + (0.331 × LIGHT expression) + (0.049 × BTLA expression). Using the median risk score as the cutoff value, the patients were successfully divided into high- and low-risk groups for each grade. In Kaplan-Meier survival analysis of the LGG and GBM cohorts from CGGA and TCGA datasets, patients with high-risk scores exhibited shorter OS than those with low-risk scores (Fig. S7a and S7b). Taken together, the HVEM risk score may serve as a useful gene-signature marker for selecting high-risk patients to receive more personalized treatments in the future.
4. Discussion
Based on a comprehensive, large-scale bioinformatic analysis, we characterized the landscape of HVEM among gliomas. HVEM mRNA expression levels were found to be highly upregulated in GBM, especially in the GBM-IDHwt subtype based on the 2016 WHO classification. HVEMhigh or low expression served as a sensitive diagnostic marker with higher expression in the MES molecular subtype of gliomas. HVEM was localized to peri-necrotic regions, microvascular proliferation and hyperplastic blood vessels. Moreover, a nomogram predictive model was established based on HVEM and several risk factors, which appears promising as a tool for clinical management of patients.
GBM attracts a variety of immune cell types from both the innate and adaptive immune systems, thus defying assumptions about immune privilege with regard to the human brain. The discovery of a unique system of lymphatic vessels in the CNS in 2015 provides a direct channel for T cells to interact with antigens in the CNS [35]. In addition, it has been demonstrated that APCs can present antigens in the CNS to T cells, and the antigen-specific T cells can respond to CNS antigens [13]. Furthermore, tumour development interferes with the integrity of the BBB, which can enable a direct immune response to GBM [13]. A mechanism regulating immune response to GBM is immune checkpoints. Immune checkpoint molecules are primarily utilized to regulate the activity of T lymphocytes, and they can stimulate or inhibit function. Inhibitory checkpoint molecules are often exploited by GBM, as expression levels are increased in the disease as a mechanism of escape from immune surveillance [13]. Checkpoint molecules PD-1 and CTLA-4, for example, play a critical role in debilitating the immune response against GBM. PD-1 expressed on T cells receives an inhibitory signal from PD-L1 on APCs or GBM cells which leads to reduced T cell activity, cytotoxic effects of T cells and cytokine production [36,37]. CTLA-4 mainly functions between APCs and T cells to inhibit other stimulatory pathways of T cells [12].
HVEM, also known as CD270, is an immune checkpoint receptor belonging to the tumour necrosis factor receptor (TNFR) superfamily [38]. The HVEM gene has been found to be mutated or over-expressed in diverse cancers. Higher expression of HVEM is also associated with poorer prognosis in cancers including germinal center lymphomas [20], non-small cell lung cancer [17], hepatocellular carcinoma [21], breast cancer [39], colorectal cancer [40], ovarian cancer [41] and gastric cancer [42]. These results indicate that HVEM widely participates in oncogenic processes.
HVEM is unique among TNFRSF proteins, in part due to that fact it transduces both pro-inflammatory and inhibitory signals [43]. As a canonical TNF receptor, HVEM signals through TNF receptor-associated factors (TRAFs) to activate nuclear factor-κB (NF-κB) transcription factors, which control transcription of genes essential for cell survival and inflammation [[44], [45], [46]]. Through GO and KEGG analysis, we found that HVEM function is closely associated with inflammation and immune signaling pathways, such as NF-κB signaling regulation and TNF mediated signaling. Our results are thus consistent with the reported function of HVEM.
Previous studies have also identified a tight association between HVEM and T cell activity; HVEM was found to be involved in both T cell activation and inhibition depending on the ligands [18,19]. In our study, we found HVEM to be negatively associated with T cell immunity as well as T helper cell activity. Furthermore, the correlation analysis between HVEM and microenvironment components suggested that HVEMhigh GBM cells tend to recruit more infiltrating immune and stromal cells into the tumour microenvironment, including monocytes, macrophages, CD8+ and CD4+ TEM, neutrophils, DCs and NK cells. These data support a function for HVEM as a contributor to the suppressive anti-tumour immunity in the glioma microenvironment. However, further studies are needed to elucidate the interaction network between HVEM and infiltrating immune cells.
Several immune checkpoint inhibitors have exhibited preclinical benefits, such as Ipilimumab and Tremelimumab for CTLA-4, and Nivolumab and Pembrolizumab for PD-1. We also analyzed the interaction between HVEM and immune checkpoint molecules. HVEM was highly correlated with TIM-3, PD-1, PD-L1, CTLA-4, LAG3 and VISTA in both pan-glioma analysis and GBM samples alone. The correlation between HVEM and so many classic immune checkpoint molecules establishes a solid basis for combined immune checkpoint blockade, targeting HVEM or other molecules, as a novel approach for clinical management of glioma.
Based on protein structures, immune checkpoint proteins are divided into two families, the immunoglobulin (Ig) or tumour necrosis factor receptor superfamily (TNFRSF). As a canonical TNFR checkpoint, HVEM mainly interacts with molecules including BTLA, CD160, gD, LIGHT and LTα3 [18]. We found a strong correlation between HVEM and LIGHT or BTLA in gliomas. However, no significant relationship was observed between HVEM and CD160 or LTα3. Thus, LIGHT and BTLA may dominate HVEM-mediated immunosuppression activities in glioma. Consistent with our findings, previous studies have reported that HVEM transduces a pro-inflammatory signal through activation of the NF-κB pathway after interacting with/binding to BTLA. The activation of HVEM/BTLA signaling has been observed in cancer, including gastric cancer and hepatocellular carcinoma [21,42], which can lead to inhibition of T cell activity in the disease. Thus, a blockade of this pathway could possibly enhance/reactivate the anticancer effect of T cells [47]. However, further study is needed to elucidate the function of HVEM signaling in the development of human glioma.
Taken together, our work illuminates a possible role for HVEM in the development and treatment of human gliomas. Future studies are warranted for further investigation of HVEM as a new immune-therapeutic target or prognostic marker for GBM.
The following are the supplementary data related to this article.
Univariate and multivariate Cox regression of HVEM expression for overall survival in TCGA and CGGA glioma patients
GSITIC analysis identified 33 and 38 genomic events to be enriched in either the HVEM high or HVEM low group
Genomic mutations associated with HVEM expression levels were linked to functionally distinct pathways
Genes positively associated with HVEM expression in CGGA and TCGA
Datasets summary
Supplementary fig S1-S7 and Supplementary methods and materials
Funding sources
This work was supported by the National Natural Science Foundation of China (81702474 and 81702475), the Department of Science & Technology of Shandong Province (2017CXGC1502, 2017CXGC1504 and 2018GSF118082), the Special Foundation for Taishan Scholars (ts20110814 and tshw201502056), the Shandong Provincial Natural Science Foundation (ZR2017MH116), the China Postdoctoral Science Foundation (2018M642666), the Jinan Science and Technology Bureau of Shandong Province (201704096 and 201704124), the Norwegian Cancer Society, the Norwegian Research Council, Haukeland University Hospital, Helse-Vest and the University of Bergen.
Declaration of interests
The authors declare that there are no potential conflicts of interest.
Author contributions
D.W., X.L. and J.W. conceived the project, designed the experiments, and wrote the paper; M.H., N.Y. and S.W. performed bioinformatic analysis; W.Z. and Y.K. collected patient samples; and S.N., A.C. and B.H. provided the statistical support.
References
- 1.Carlsson S.K., Brothers S.P., Wahlestedt C. Emerging treatment strategies for glioblastoma multiforme. EMBO Mol Med. 2014;6(11):1359–1370. doi: 10.15252/emmm.201302627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Claes A., Idema A.J., Wesseling P. Diffuse glioma growth: a guerilla war. Acta Neuropathol. 2007;114(5):443–458. doi: 10.1007/s00401-007-0293-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang R. High precision imaging of microscopic spread of glioblastoma with a targeted ultrasensitive SERRS molecular imaging probe. Theranostics. 2016;6(8):1075–1084. doi: 10.7150/thno.13842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen R., Cohen A.L., Colman H. Targeted therapeutics in patients with high-grade gliomas: past, present, and future. Curr Treat Options in Oncol. 2016;17(8):42. doi: 10.1007/s11864-016-0418-0. [DOI] [PubMed] [Google Scholar]
- 5.Puduvalli V.K. Beyond alkylating agents for gliomas: quo vadimus? Am Soc Clin Oncol Educ Book. 2017;(37):175–186. doi: 10.14694/EDBK_175003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Touat M. Glioblastoma targeted therapy: updated approaches from recent biological insights. Ann Oncol. 2017;28(7):1457–1472. doi: 10.1093/annonc/mdx106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramirez Y.P. Glioblastoma multiforme therapy and mechanisms of resistance. Pharmaceuticals (Basel) 2013;6(12):1475–1506. doi: 10.3390/ph6121475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellis H.P. Current challenges in glioblastoma: intratumour heterogeneity, residual disease, and models to predict disease recurrence. Front Oncol. 2015;5:251. doi: 10.3389/fonc.2015.00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stavrovskaya A.A., Shushanov S.S., Rybalkina E.Y. Problems of glioblastoma multiforme drug resistance. Biochemistry (Mosc) 2016;81(2):91–100. doi: 10.1134/S0006297916020036. [DOI] [PubMed] [Google Scholar]
- 10.Driessens G., Kline J., Gajewski T.F. Costimulatory and coinhibitory receptors in anti-tumor immunity. Immunol Rev. 2009;229(1):126–144. doi: 10.1111/j.1600-065X.2009.00771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Syn N.L. De-novo and acquired resistance to immune checkpoint targeting. Lancet Oncol. 2017;18(12):e731–e741. doi: 10.1016/S1470-2045(17)30607-1. [DOI] [PubMed] [Google Scholar]
- 12.Lee K.M. Molecular basis of T cell inactivation by CTLA-4. Science. 1998;282(5397):2263–2266. doi: 10.1126/science.282.5397.2263. [DOI] [PubMed] [Google Scholar]
- 13.Huang J. Immune checkpoint in glioblastoma: promising and challenging. Front Pharmacol. 2017;8:242. doi: 10.3389/fphar.2017.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Razavi S.M. Immune evasion strategies of glioblastoma. Front Surg. 2016;3:11. doi: 10.3389/fsurg.2016.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Das M., Zhu C., Kuchroo V.K. Tim-3 and its role in regulating anti-tumor immunity. Immunol Rev. 2017;276(1):97–111. doi: 10.1111/imr.12520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Romani M. Immune checkpoints and innovative therapies in glioblastoma. Front Oncol. 2018;8:464. doi: 10.3389/fonc.2018.00464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ren S. The immune checkpoint, HVEM may contribute to immune escape in non-small cell lung cancer lacking PD-L1 expression. Lung Cancer. 2018;125:115–120. doi: 10.1016/j.lungcan.2018.09.004. [DOI] [PubMed] [Google Scholar]
- 18.Murphy T.L., Murphy K.M. Slow down and survive: enigmatic immunoregulation by BTLA and HVEM. Annu Rev Immunol. 2010;28:389–411. doi: 10.1146/annurev-immunol-030409-101202. [DOI] [PubMed] [Google Scholar]
- 19.Cheung T.C. Evolutionarily divergent herpesviruses modulate T cell activation by targeting the herpesvirus entry mediator cosignaling pathway. Proc Natl Acad Sci U S A. 2005;102(37):13218–13223. doi: 10.1073/pnas.0506172102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boice M. Loss of the HVEM tumor suppressor in lymphoma and restoration by modified CAR-T cells. Cell. 2016;167(2):405–418.e13. doi: 10.1016/j.cell.2016.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hokuto D. Clinical impact of herpesvirus entry mediator expression in human hepatocellular carcinoma. Eur J Cancer. 2015;51(2):157–165. doi: 10.1016/j.ejca.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 22.Phillips H.S. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 23.Verhaak R.G. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gill B.J. MRI-localized biopsies reveal subtype-specific differences in molecular and cellular composition at the margins of glioblastoma. Proc Natl Acad Sci U S A. 2014;111(34):12550–12555. doi: 10.1073/pnas.1405839111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burton E.C. Genetic aberrations defined by comparative genomic hybridization distinguish long-term from typical survivors of glioblastoma. Cancer Res. 2002;62(21):6205–6210. [PubMed] [Google Scholar]
- 26.Smith J.S. Localization of common deletion regions on 1p and 19q in human gliomas and their association with histological subtype. Oncogene. 1999;18(28):4144–4152. doi: 10.1038/sj.onc.1202759. [DOI] [PubMed] [Google Scholar]
- 27.Weller M. Molecular predictors of progression-free and overall survival in patients with newly diagnosed glioblastoma: a prospective translational study of the German Glioma Network. J Clin Oncol. 2009;27(34):5743–5750. doi: 10.1200/JCO.2009.23.0805. [DOI] [PubMed] [Google Scholar]
- 28.Derre L. BTLA mediates inhibition of human tumor-specific CD8+ T cells that can be partially reversed by vaccination. J Clin Invest. 2010;120(1):157–167. doi: 10.1172/JCI40070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sedy J.R. B and T lymphocyte attenuator regulates T cell activation through interaction with herpesvirus entry mediator. Nat Immunol. 2005;6(1):90–98. doi: 10.1038/ni1144. [DOI] [PubMed] [Google Scholar]
- 30.Cai G. CD160 inhibits activation of human CD4+ T cells through interaction with herpesvirus entry mediator. Nat Immunol. 2008;9(2):176–185. doi: 10.1038/ni1554. [DOI] [PubMed] [Google Scholar]
- 31.Rody A. T-cell metagene predicts a favorable prognosis in estrogen receptor-negative and HER2-positive breast cancers. Breast Cancer Res. 2009;11(2):R15. doi: 10.1186/bcr2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aran D., Hu Z., Butte A.J. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18(1):220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Donson A.M. Increased immune gene expression and immune cell infiltration in high-grade astrocytoma distinguish long-term from short-term survivors. J Immunol. 2012;189(4):1920–1927. doi: 10.4049/jimmunol.1103373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lossos I.S. Prediction of survival in diffuse large-B-cell lymphoma based on the expression of six genes. N Engl J Med. 2004;350(18):1828–1837. doi: 10.1056/NEJMoa032520. [DOI] [PubMed] [Google Scholar]
- 35.Louveau A. Structural and functional features of central nervous system lymphatic vessels. Nature. 2015;523(7560):337–341. doi: 10.1038/nature14432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng X. Structure and interactions of the human programmed cell death 1 receptor. J Biol Chem. 2013;288(17):11771–11785. doi: 10.1074/jbc.M112.448126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Francisco L.M. PD-L1 regulates the development, maintenance, and function of induced regulatory T cells. J Exp Med. 2009;206(13):3015–3029. doi: 10.1084/jem.20090847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shui J.W., Kronenberg M. HVEM is a TNF receptor with multiple regulatory roles in the mucosal immune system. Immune Netw. 2014;14(2):67–72. doi: 10.4110/in.2014.14.2.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsang J.Y.S. Expression and clinical significance of herpes virus entry mediator (HVEM) in breast cancer. Ann Surg Oncol. 2017;24(13):4042–4050. doi: 10.1245/s10434-017-5924-1. [DOI] [PubMed] [Google Scholar]
- 40.Inoue T. HVEM expression contributes to tumor progression and prognosis in human colorectal cancer. Anticancer Res. 2015;35(3):1361–1367. [PubMed] [Google Scholar]
- 41.Zhang T. Knockdown of HVEM, a lymphocyte regulator gene, in ovarian cancer cells increases sensitivity to activated T cells. Oncol Res. 2016;24(3):189–196. doi: 10.3727/096504016X14641336229602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lan X. Increased BTLA and HVEM in gastric cancer are associated with progression and poor prognosis. Onco Targets Ther. 2017;10:919–926. doi: 10.2147/OTT.S128825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Steinberg M.W., Cheung T.C., Ware C.F. The signaling networks of the herpesvirus entry mediator (TNFRSF14) in immune regulation. Immunol Rev. 2011;244(1):169–187. doi: 10.1111/j.1600-065X.2011.01064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheung T.C. Unconventional ligand activation of herpesvirus entry mediator signals cell survival. Proc Natl Acad Sci U S A. 2009;106(15):6244–6249. doi: 10.1073/pnas.0902115106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marsters S.A. Herpesvirus entry mediator, a member of the tumor necrosis factor receptor (TNFR) family, interacts with members of the TNFR-associated factor family and activates the transcription factors NF-kappaB and AP-1. J Biol Chem. 1997;272(22):14029–14032. doi: 10.1074/jbc.272.22.14029. [DOI] [PubMed] [Google Scholar]
- 46.Shui J.W., Kronenberg M. HVEM: An unusual TNF receptor family member important for mucosal innate immune responses to microbes. Gut Microbes. 2013;4(2):146–151. doi: 10.4161/gmic.23443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spodzieja M. Design of short peptides to block BTLA/HVEM interactions for promoting anticancer T-cell responses. PLoS One. 2017;12(6) doi: 10.1371/journal.pone.0179201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Univariate and multivariate Cox regression of HVEM expression for overall survival in TCGA and CGGA glioma patients
GSITIC analysis identified 33 and 38 genomic events to be enriched in either the HVEM high or HVEM low group
Genomic mutations associated with HVEM expression levels were linked to functionally distinct pathways
Genes positively associated with HVEM expression in CGGA and TCGA
Datasets summary
Supplementary fig S1-S7 and Supplementary methods and materials